- 1Department of Integrative Biotechnology, College of Biotechnology and Bioengineering, Sungkyunkwan University, Suwon, South Korea
- 2Department of Molecular Cell Biology, Sungkyunkwan University School of Medicine, Suwon, South Korea
- 3Samsung Medical Center, School of Medicine, Sungkyunkwan University, Suwon, South Korea
Upon intracellular cues, bacterial mRNA leaders often form secondary structures that determine expression of a downstream protein-coding region(s), thereby providing bacteria with a mechanism to control the amounts of necessary proteins in the right locales. Here we describe a polycistronic mRNA leader that secures bacterial growth by preventing dysregulated expression of the protein-coding regions. In Salmonella, the mgtCBR mRNA encodes the virulence protein MgtC and the Mg2+ transporter MgtB. A mutant designed to produce leaderless mgtCBR mRNA induced MgtC and MgtB in conditions that promote mgtC transcription. The dysregulated expression of MgtC and MgtB impaired bacterial growth under all such non-host environments. While MgtC, but not MgtB, normally reduces ATP levels in a process requiring the F1F0 ATP synthase, dysregulated MgtC and MgtB reduced ATP levels independently of the F1F0 ATP synthase, which correlated with the mutant’s growth defect. The mutant showed dysregulated MgtC expression and attenuated survival inside macrophages. While MgtB normally does not affect the phenotype, MgtB impaired intramacrophage survival of the mutant in the presence of MgtC. We provide an example showing that a polycistronic mRNA leader prevents the dysregulated function of protein-coding regions to allow bacteria to proliferate across complex niches.
Introduction
The bacterium Salmonella enterica serovar Typhimurium (hereafter referred to as Salmonella) is a facultative intracellular pathogen that can proliferate both inside and outside host cells. To survive under both conditions, Salmonella must tightly control the amounts of necessary proteins in each locale.
The PhoP/PhoQ two-component system in Salmonella consists of the response regulator PhoP and the sensor kinase PhoQ (Groisman, 2001). The PhoQ protein senses multiple signals, such as low (i.e., micromolar concentrations) Mg2+, acidic pH, and certain antimicrobial peptides, and phosphorylates the PhoP protein, rendering it functional as a transcriptional regulator (Garcia Vescovi et al., 1996; Bader et al., 2005; Prost et al., 2007). The gene expression programs regulated by PhoP/PhoQ enable Salmonella to adapt to low Mg2+ (Garcia Vescovi et al., 1996), survive in acidic pH (Foster and Hall, 1990), and acquire resistance against antimicrobial peptides (Fields et al., 1989). Moreover, activation of PhoP/PhoQ is a key event enabling Salmonella to survive inside the macrophage phagosome (Fields et al., 1989), which contains antimicrobial factors, including acidic pH and antimicrobial peptides.
The virulence protein MgtC contributes to the growth of Salmonella inside macrophages as well as in low Mg2+ conditions (Blanc-Potard and Groisman, 1997; Rang et al., 2007). These dual roles of MgtC are associated with its ability to reduce cytoplasmic ATP levels by directly acting on the bacterium’s own F1F0 ATP synthase (Lee et al., 2013; Pontes et al., 2015). MgtC also prevents hyperpolarization of Salmonella membrane (Lee et al., 2013). Given that MgtC plays this role independently of the F1F0 ATP synthase, this result suggests that MgtC could affect ATP levels by acting on a protein(s) other than the F1F0 ATP synthase (Lee et al., 2013).
The MgtC protein, together with the Mg2+ transporter MgtB (Snavely et al., 1991b) and the regulatory peptide MgtR (Alix and Blanc-Potard, 2008), are produced from the mgtCBR operon, which is regulated at multiple levels (Lee and Lee, 2015). When Salmonella is placed in low Mg2+ or in acidic pH environments, activated PhoP directly promotes mgtC transcription (Shin and Groisman, 2005; Choi and Groisman, 2016). However, this event alone does not ensure production of full length mgtCBR mRNA. The mgtCBR mRNA contains a 296 nucleotides long leader that harbors two short open reading frames (ORFs), mgtM and mgtP (Figure 1A and Supplementary Figure S1). In response to intracellular levels of ATP or an amino acid, these ORFs can promote formation of alternative structures of the mgtCBR mRNA leader that determine the degree of mgtC expression (Lee and Groisman, 2012a, b; Lee et al., 2014). For instance, an increase in ATP levels and a decrease in proline levels in the cytoplasm affects the coupling/uncoupling of transcription of the mgtCBR mRNA leader with translation of mgtM and mgtP, respectively, to induce structures of the leader that allow transcription elongation into the mgtC-coding region (Lee and Groisman, 2012a, b; Lee et al., 2014). Such mgtM- and mgtP regulation enables Salmonella to induce mgtC transcription at levels that ensure its survival inside macrophages (Lee and Groisman, 2012a, b; Lee et al., 2014).
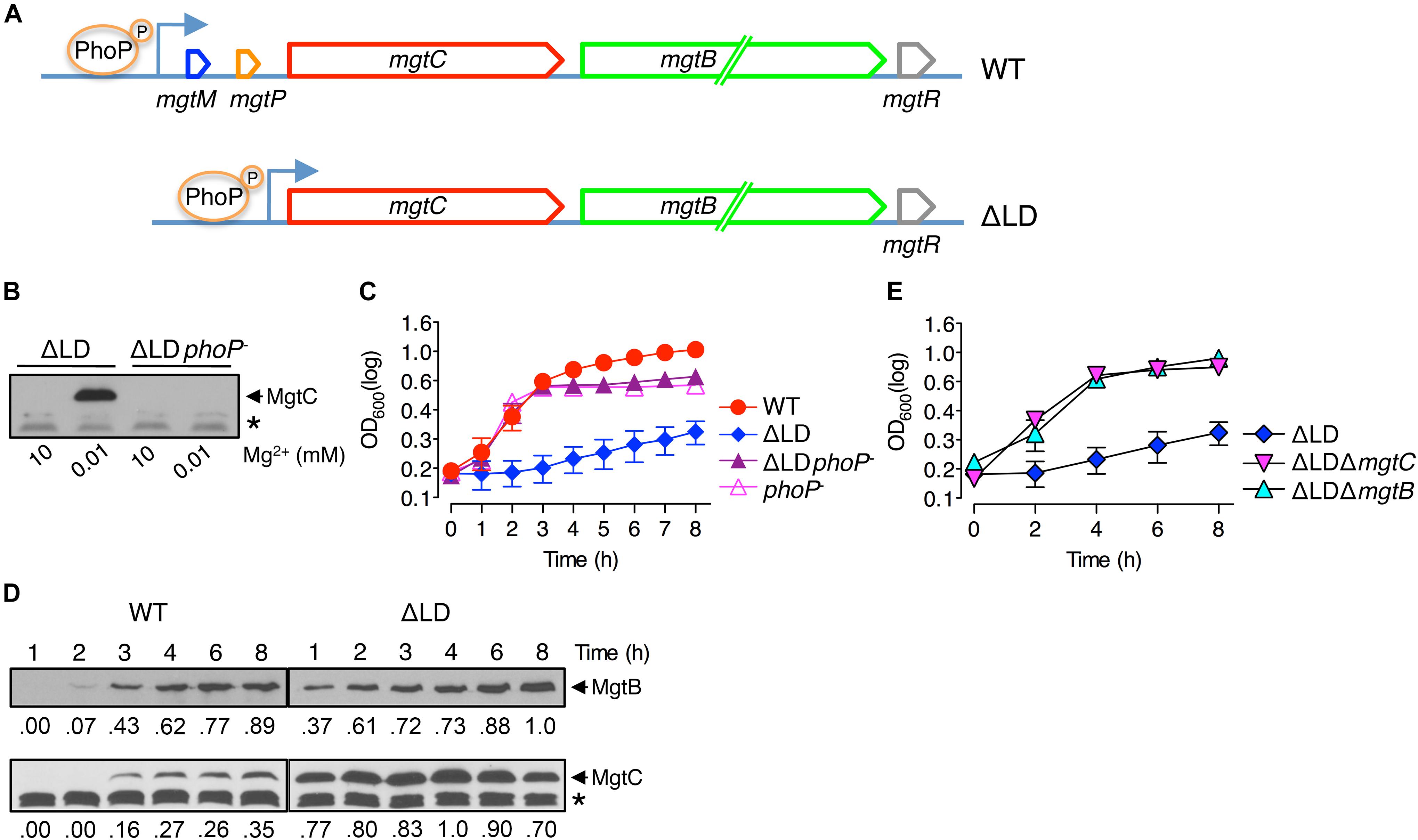
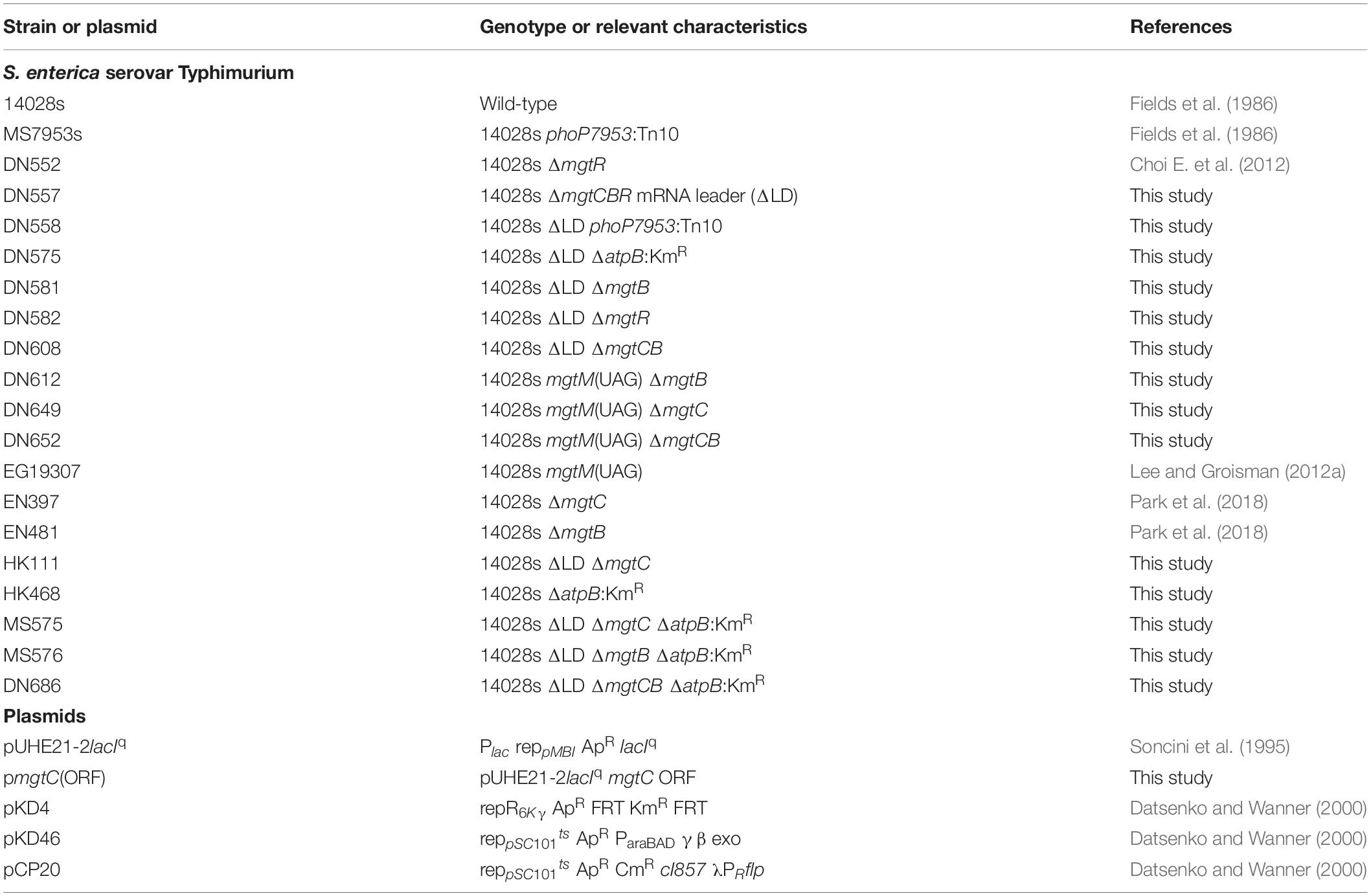
Figure 1. Dysregulated induction of MgtC and MgtB impairs growth of Salmonella in low Mg2+. (A) Schematic of the mgtCBR operon in the wild-type Salmonella (WT, 14028s) and ΔLD mutant (DN557). (B) Immunoblot analysis using anti-MgtC antibodies of crude extracts prepared from ΔLD and ΔLD phoP– (DN558) strains. Bacteria were grown in N-minimal medium with 10 mM or 10 μM Mg2+ at pH 7.5 for 4 h. (C,E) Growth curves of wild-type (WT), ΔLD, ΔLD phoP–, phoP– (MS7953s), ΔLD ΔmgtC (HK111), and ΔLD ΔmgtB (DN581) strains. Bacteria were grown in N-minimal medium with 10 μM Mg2+ at pH 7.5, and OD600 values were determined at the indicated time points. Means and standard deviations from three independent experiments are shown. (D) Immunoblot analysis using anti-MgtC and anti-MgtB antibodies of crude extracts prepared from wild-type (WT) and ΔLD strains grown in N-minimal medium with 10 μM Mg2+ at pH 7.5 and harvested at the indicated time points. The band indicated with an asterisk (∗) corresponds to a protein displaying cross-reactivity against the anti-MgtC antibody and serves as an internal loading control. Numbers below the blots correspond to relative levels of MgtC and MgtB at a given time point.
MgtC expression is also negatively controlled at the posttranscriptional level. In low Mg2+ conditions, PhoP binds to and activates the amgR promoter located in the mgtC–mgtB intergenic region (Lee and Groisman, 2010). This event produces the AmgR antisense RNA that promotes degradation of the mgtC mRNA in a process requiring the RNase E (Lee and Groisman, 2010). Inactivation of amgR transcription increases MgtC production and renders Salmonella more virulent in mice (Lee and Groisman, 2010). The regulatory peptide MgtR encoded by the mgtCBR operon binds to the MgtC protein and promotes MgtC degradation in a process dependent on the FtsH protease (Alix and Blanc-Potard, 2008). Although the mgtR mutant produces MgtC at much higher levels than the wild-type (WT) strain, the mutant is slightly attenuated for survival inside macrophages (Alix and Blanc-Potard, 2008).
In the present study, we designed a Salmonella mutant that produced leaderless mgtCBR mRNA. We found that in both host and non-host environments where the PhoP regulator is activated, removal of the mgtCBR mRNA leader caused Salmonella to induce MgtC and MgtB in a dysregulated manner, which impaired bacterial growth in such conditions. The dysregulated expression of MgtC and MgtB reduces ATP to abnormal levels in a process that does not require the F1F0 ATP synthase, and this reduction in ATP levels seems to be a cause for the Salmonella growth defect seen. Our study reveals that the role of the mgtCBR mRNA leader to prevent dysregulated expression of MgtC and MgtB is important for the growth of Salmonella both inside and outside host cells.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. All S. enterica serovar Typhimurium strains were derived from strain 14028s. Phage P22-mediated transductions were performed as described (Davis et al., 1980). Bacteria were grown at 37°C in N-minimal medium (Snavely et al., 1991a), pH 7.5 or pH 5.5, supplemented with 0.1% Casamino Acids, 38 mM glycerol, and the indicated concentrations of MgCl2. Ampicillin, kanamycin, and C18G peptide (AnaSpec) were used at 50, 30, and 5 μg/ml, respectively. For induction of genes from plasmids, isopropyl 1-thio-β-D-galatoside (IPTG) was used at the indicated concentrations.
Construction of Chromosomal Mutants and Plasmid
Salmonella enterica strains carrying a chromosomal gene deletion were constructed using the one-step gene inactivation method (Datsenko and Wanner, 2000) with necessary modifications. DN557 is a strain, in which the sequences encoding the mgtCBR mRNA leader regions were deleted from the chromosome. To construct DN557, the tetRA fragment was amplified from strain MS7953s using the primer pair Del701/Del702 and integrated into the chromosome of WT strain 14028s harboring the pKD46 plasmid (Datsenko and Wanner, 2000). The resulting strain HK514 keeps the tetRA genes in the mgtCBR mRNA leader-encoding region and pKD46 at 30°C. To obtain the engineered DNA fragment that is deleted for mgtCBR mRNA leader-encoding regions, the left and right arms of the DNA fragment were amplified from strain 14028s using primer pairs Del703/Del704 and Del705/Del706, respectively. Then, the second step of PCR was conducted on the mixture of the first step of PCR products using the primer pair Del703/Del706. The resulting PCR product was used to electroporate the HK514 strain, and the bacterial suspension was plated on medium containing fusaric acid and incubated at 42°C to select against the tetRA genes (Maloy and Nunn, 1981).
Strain HK111, in which the sequences corresponding to mgtCBR mRNA leader and mgtC-coding regions were removed from the chromosome, was similarly constructed. In the first step of PCR, two DNA fragments were amplified from strain 14028s using primer pairs Del703/Del704 and Del705/Del708. The second step of PCR was conducted on the mixture of the first step of PCR products using the primer pair Del703/Del708. The resulting PCR product was introduced into the strain HK514, and tetracycline-sensitive colonies were selected. Deletion of the sequences in the strains DN557 and HK111 was verified by nucleotide sequencing.
To delete the mgtB gene in the DN557 and HK111 strains, the kanamycin resistance gene (KmR) cassette from the pKD4 plasmid (Datsenko and Wanner, 2000) was amplified using the primer pair Del711/Del712. The resulting PCR product was used to electroporate the DN557 and HK111 strains carrying pKD46. To delete the mgtC, mgtB, and mgtCB genes in strain EG19307, the KmR cassette from plasmid pKD4 was amplified using primer pairs Del713/Del714, Del711/Del712, and Del713/Del712, respectively. The respective PCR products were used to electroporate the EG19307 strain carrying pKD46. To delete the mgtR gene in strain DN557, the KmR cassette from plasmid pKD4 was amplified using the primer pairs Del715/Del716. The respective PCR products were used to electroporate the DN557 strain carrying pKD46. The KmR cassette was removed using plasmid pCP20 (Datsenko and Wanner, 2000). To delete the atpB gene in the 14028s strain, the KmR cassette from plasmid pKD4 was amplified using the primer pair Del717/Del718, and the PCR product was used to electroporate the 14028s strain carrying pKD46. Deletion of the corresponding genes was verified by colony PCR. The sequences of all primers used for strain construction are listed in Supplementary Table S1.
Plasmid pmgtC(ORF) expresses the mgtC ORF from the lac promoter. The mgtC ORF was amplified using the primer pair Ex301/Ex302 and chromosomal DNA from the 14028s strain, and the mgtC ORF was then introduced between the EcoRI and PstI restriction sites of pUHE21-2lacIq (Soncini et al., 1995). Recombinant plasmid sequences were confirmed by nucleotide sequencing. The sequences of primers used for plasmid construction are listed in Supplementary Table S1.
Immunoblot Analysis
Bacteria were grown in N-minimal medium for the indicated amounts of time. Equivalent amounts of bacterial cells normalized by OD600 values were collected, washed with phosphate-buffered saline (PBS), suspended in 0.15 ml SDS sample buffer (Laemmli sample buffer), and boiled. Whole-cell lysates were resolved on 12% SDS polyacrylamide gels, transferred to nitrocellulose membranes, and analyzed by immunoblot using anti-MgtC (Park et al., 2018), anti-MgtB (Choi et al., 2017), or anti-RpoA (NeoClone) antibodies. Membranes were incubated with anti-rabbit IgG horseradish peroxidase-linked antibodies (GE Healthcare), and bands were visualized using the ECL detection system (GE Healthcare). Protein levels were quantified using the ImageJ program (NIH).
RNA Isolation and Quantitative Real-Time RT-PCR (qRT-PCR) Analysis
Bacteria were grown in N-minimal medium for the indicated amounts of time. The culture (0.5 ml) was removed and mixed with 1 ml RNAprotect Bacteria Reagent (Qiagen). RNA was isolated using the RNeasy Mini Kit (Qiagen) and treated with RNase-free DNase (Ambion). cDNA was synthesized from 0.1 μg of template RNA using the PrimeScript RT reagent Kit (Takara) and random primers (Life Technologies). Amounts of cDNA were quantified by real-time PCR using the SYBR Green Realtime PCR Master Mix (Toyobo) with an ABI7300 Sequence Detection System (Applied Biosystems). The following primer pairs were used for the detection of cDNA corresponding to mgtC, mgtB, and gyrB mRNAs: Q-mgtC-F/Q-mgtC-R, Q-mgtB-F/Q-mgtB-R, and Q-gyrB-F/Q-gyrB-R, respectively. Transcription levels of each gene were calculated from a standard curve obtained by PCR with the same primers and serially diluted genomic DNA. mRNA levels of target genes were normalized to gyrB mRNA levels. The sequences of primers are listed in Supplementary Table S1.
Measurement of Intracellular ATP Levels
ATP levels were determined as previously described (Lee et al., 2013). Bacteria were grown in N-minimal medium for 4 h. Equivalent amounts of bacterial cells (0.5 × OD600) were removed, washed with PBS, and suspended in 0.5 ml of PBS. Nucleic acids were extracted by adding 100 ml of ice-cold 3 M perchloric acid. After incubation for 5 min, extracts were neutralized with 225 ml of neutralization buffer (1 M KOH, 0.5 M Tris, 0.5 M KCl) and centrifuged at 13,000 rpm and 25°C for 10 min. Fifty microliters of the supernatant were diluted with 50 ml of L buffer (25 mM KCl, 50 mM MgSO4, 100 mM HEPES pH 7.4), and ATP levels were measured using an ATP Determination Kit (Life Technologies) according to the manufacturer’s instruction. Statistical analysis of the data was conducted using the GraphPad Prism program (version 5.0).
Macrophage Survival Assay
The experiment was conducted as previously described (Choi Y. et al., 2012). J774A.1 macrophage cells were grown in Dulbecco modified Eagle medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum (FBS) and 1% Antibiotic-Antimycotic (Life Technologies) at 37°C under 5% CO2. Prior to bacterial infection, a monolayer of 1 × 105 J774A.1 cells was prepared in a 24-well tissue culture plate and incubated in DMEM with 10% FBS for 1 h. Bacteria were cultured in Luria-Bertani (LB) medium for 18 h with aeration and opsonized in 20% normal mouse serum for 25 min at 37°C. Opsonized bacteria were diluted in DMEM with 10% FBS and added to the cell monolayer at a multiplicity of infection (MOI) of 10. After 1 h of incubation, the wells were washed three times with pre-warmed PBS to remove extracellular bacteria and then incubated for another 1 h with the pre-warmed medium with 100 μg/ml gentamicin to kill extracellular bacteria. The wells were washed three times with PBS and incubated in pre-warmed medium with 10 μg/ml of gentamicin. At the desired time points, the wells were washed three times with PBS and treated with 1% Triton X-100 for 10 min. The suspension was diluted in PBS and plated on LB agar plates to enumerate colony-forming units.
Determination of MgtC Levels Inside Macrophages
J774A.1 macrophage and bacterial cells were cultured as described above. A monolayer of 1 × 106 J774A.1 cells was prepared in a 6-well tissue culture plate. Opsonized bacteria were added to the cell monolayer at a MOI of 10. The wells were then treated as described above to remove extracellular bacteria. At the desired time points, the wells were washed three times with PBS and treated with 1% Triton X-100 for 30 min. The cell lysis mixture was centrifuged at 13,000 rpm for 10 min, and the bacterial cell pellet was suspended in 0.15 ml SDS sample buffer and boiled. MgtC levels in bacterial cell lysates were analyzed by immunoblot as described above.
Measurement of Bacterial Growth Using a Plate Reader
Bacteria were grown in N-minimal medium with 10 mM MgCl2 at pH 7.5 to saturation. One milliliter of bacterial cells were collected, washed twice with medium not supplemented with MgCl2, and diluted 1:100 into wells of a 24-well plate containing 1 ml of medium with 10 μM or 1 mM MgCl2 at pH 7.5 or pH 5.5. The C18G peptide was added to medium at 5 μg/ml. A plate was covered with a Breathe-Easy sealing membrane (Sigma) to prevent evaporation. By using an xMarkTM Microplate Spectrophotometer (Bio-Rad), bacteria were cultivated at 37°C with shaking, and the OD600 values were measured every 5 min up to 8 h.
Results
A Salmonella Strain Designed to Produce Leaderless mgtCBR mRNA Is Severely Impaired for Growth in Low Mg2+
We investigated the behaviors of Salmonella that produces leaderless mgtCBR mRNA. We deleted the sequences specifying the mgtCBR mRNA leader regions that participate in forming regulatory structures from the chromosome without leaving any heterologous sequences behind (Figure 1A and Supplementary Figure S1). The resulting strain (ΔLD) still possessed PhoP-dependent Mg2+ regulation of mgtC expression. The MgtC protein was detected in the ΔLD strain grown in defined medium supplemented with 10 μM Mg2+ but not in medium with 10 mM Mg2+ for 4 h, and the low Mg2+-induced MgtC production did not occur in the strain carrying a phoP– allele (Figure 1B). (Note that the medium pH was adjusted to 7.5, unless otherwise stated.)
While preparing bacterial cultures, we repeatedly observed that the ΔLD strain did not grow well in low Mg2+. During experiments, Salmonella strains were initially grown to saturation in medium with 10 mM Mg2+ and then diluted to OD600 values of ∼0.15 in medium with 10 μM Mg2+. After a growth lag for the first 1 h, the WT strain grew logarithmically up to 3 h and then displayed slow linear growth for the subsequent 5 h (Figure 1C), generating a typical growth curve in 10 μM Mg2+ (Soncini et al., 1996; Blanc-Potard and Groisman, 1997; Park et al., 2018). However, the ΔLD strain did not achieve logarithmic growth and only grew in a slow linear manner during the entire 8 h (Figure 1C).
The ΔLD Strain Expresses MgtC and MgtB in Low Mg2+ in a Dysregulated Manner
To understand the molecular basis underlying the growth defect seen in the ΔLD strain, we compared expression of mgtC and mgtB between the WT and ΔLD strains. When Salmonella is placed in low Mg2+, PhoP directly promotes transcription initiation of the mgtC gene (Shin and Groisman, 2005). However, Salmonella can produce MgtC protein at detectable levels after cytoplasmic Mg2+ levels drop to a certain threshold (Yeom et al., 2017; Park et al., 2018). Given that a secondary structure of the mgtCBR mRNA leader prevents transcription elongation from reaching the mgtC-coding region (Lee and Groisman, 2012a, b; Lee et al., 2014), we reasoned that the expression kinetics of mgtC and mgtB must be different between the WT and ΔLD strains.
When placed in medium with 10 μM Mg2+, the WT strain produced mgtC mRNA at very low levels for the first 2 h (Supplementary Figure S2A). The mgtC mRNA levels then increased at 3 h by ∼10-fold, and this high-level production continued for the following 5 h (Supplementary Figure S2A). By contrast, under the same growth condition, the ΔLD strain produced mgtC mRNA at levels similar to the maximum levels of the WT as early as at 1 h and maintained these levels for the following 7 h (Supplementary Figure S2A). Moreover, consistent with the notion that the mgtC and mgtB mRNAs constitute a polycistronic mRNA (Alix and Blanc-Potard, 2008; Lee and Groisman, 2010), the WT and ΔLD strains highly induced mgtB mRNA at the delayed (i.e., 3 h) and the early (i.e., 1 h) time points, respectively (Supplementary Figure S2B).
We also determined levels of the MgtC and MgtB proteins over time during bacterial growth in 10 μM Mg2+. Consistent with previous findings (Yeom et al., 2017; Park et al., 2018) and the mgtC mRNA levels (Supplementary Figure S2A), MgtC was detected in the WT strain only after 3 h (Figure 1D). In contrast, and as observed with mgtC mRNA production (Supplementary Figure S2A), the ΔLD strain produced detectable levels of MgtC as early as 1 h of growth (Figure 1D). Notably, despite the finding that the maximum levels of mgtC mRNA were comparable between the two strains (Supplementary Figure S2A), MgtC levels were ∼4-fold higher in the ΔLD strain than in the WT strain at 4 h (Figure 1D). As observed with mgtB mRNA production (Supplementary Figure S2B), the WT and ΔLD strains produced detectable levels of MgtB in parallel with MgtC (Figure 1D). However, in contrast to MgtC levels, MgtB levels detected after 4 h were similar between the two strains (Figure 1D).
In medium with 10 μM Mg2+, WT Salmonella grew logarithmically for 3 h by consuming Mg2+ in the medium (Figure 1C). During this time period, the ΔLD strain produced the Mg2+ transporter MgtB at much higher levels (Figure 1D) with even lower growth yields than the WT strain (Figure 1C), suggesting that cytoplasmic Mg2+ levels in the mutant are at least not lower than the WT levels. Taken together, these results suggest that removal of the leader from the mgtCBR mRNA causes Salmonella to induce MgtC and MgtB in a dysregulated manner even prior to its experiencing low cytoplasmic Mg2+ stress.
MgtC and MgtB Impair Growth of the ΔLD Strain in Low Mg2+
As previously observed (Soncini et al., 1996; Blanc-Potard and Groisman, 1997), in medium with 10 μM Mg2+, the phoP– strain grew logarithmically in a manner similar to the WT strain but showed defective growth in the slow-growth phase (Figure 1C). The ΔLD phoP– strain, which failed to induce MgtC expression (Figure 1B), showed logarithmic growth, with a growth curve similar to that of the phoP– strain (Figure 1C). This result suggests that the growth defect of the ΔLD strain may be associated with dysregulated MgtC induction. Consistent with this prediction, the ΔLD ΔmgtC strain recovered the ability of logarithmic growth (Figure 1E).
Because the ΔLD strain produced MgtB in a dysregulated manner as well as MgtC (Figure 1D), we next explored whether the mgtB gene was also responsible for the growth defect. Indeed, in medium with 10 μM Mg2+, mgtB deletion enabled the ΔLD strain to grow logarithmically (Figure 1E). In the ΔLD background, mgtC deletion did not affect MgtB production, and vice versa (Supplementary Figure S3), suggesting that the growth phenotypes of the ΔLD ΔmgtC and ΔLD ΔmgtB strains are due to lack of MgtC- and MgtB function, respectively.
The mgtR gene in the mgtCBR operon (Figure 1A) encodes the regulatory peptide MgtR that controls MgtC levels (Alix and Blanc-Potard, 2008). Because the ΔLD strain is also predicted to produce MgtR in a dysregulated manner, we further examined whether mgtR deletion affects growth of the ΔLD strain. However, the ΔLD ΔmgtR strain grew in a manner similar to the ΔLD strain (Supplementary Figure S4). Together these results indicate that when produced in a dysregulated manner, MgtC and MgtB impair the growth of Salmonella under low Mg2+ conditions.
In the ΔLD Strain, MgtC and MgtB Contribute to ATP Reduction to Abnormal Levels in a Process That Does Not Require F1F0 ATP Synthase
Given that MgtC functions to reduce cytoplasmic ATP levels by acting on the F1F0 ATP synthase (Lee et al., 2013) and that the ΔLD strain produced higher levels of MgtC (Figure 1D), we reasoned that ATP levels may be different between the ΔLD and WT strains. We thus determined ATP levels in Salmonella at 4 h after growth in medium with 10 μM Mg2+, in which both WT and ΔLD strains produced MgtC at detectable levels (Figure 1D). Similar to previous finding (Lee et al., 2013), ATP levels were higher in the ΔmgtC strain than in the WT strain (Figure 2A). In contrast, the ΔLD strain exhibited ∼2-fold lower ATP levels than the WT (Figure 2A). The ATP reduction in the ΔLD strain was dependent on MgtC, as deleting mgtC increased the ATP levels by ∼3-fold (Figure 2A).
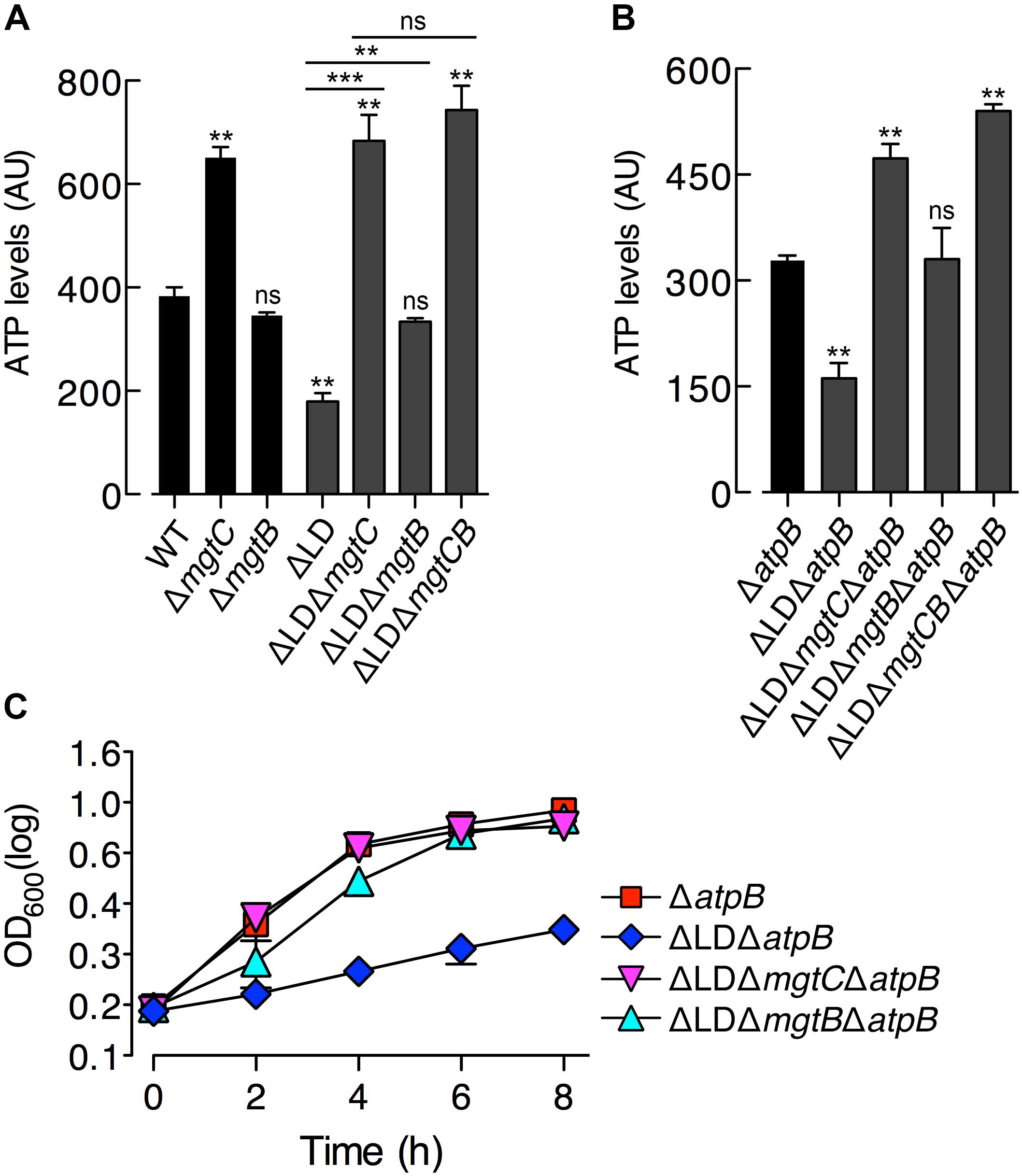
Figure 2. Dysregulated expression of MgtC and MgtB reduces ATP to abnormal levels in a process that does not require F1F0 ATP synthase. ATP levels were determined in wild-type (WT, 14028s), ΔmgtC (EN397), ΔmgtB (EN481), ΔLD (DN557), ΔLD ΔmgtC (HK111), ΔLD ΔmgtB (DN581), and ΔLD ΔmgtCB (DN608) strains (A) as well as in ΔatpB (HK468), ΔLD ΔatpB (DN575), ΔLD ΔmgtC ΔatpB (MS575), ΔLD ΔmgtB ΔatpB (MS576), and ΔLD ΔmgtCB ΔatpB (DN686) strains (B). Bacteria were grown in N-minimal medium with 10 μM Mg2+ at pH 7.5 for 4 h. Data depicted in arbitrary units (AU) are means and standard deviations from three independent experiments. ∗∗P < 0.01, ∗∗∗P < 0.001, two-tailed t-test with each sample vs. WT (A) and with each sample vs. ΔatpB (B), ns, not significant. (C) Growth curves of Salmonella strains, ΔatpB (HK468), ΔLD ΔatpB (DN575), ΔLD ΔmgtC ΔatpB (MS575), and ΔLD ΔmgtB ΔatpB (MS576). Bacteria were grown in N-minimal medium with 10 μM Mg2+ at pH 7.5, and OD600 values were determined at the indicated time points. Data are representative of three independent experiments, and means and standard deviations from three independent experiments.
Despite the findings that MgtB does not impact ATP levels in the WT strain (Lee et al., 2013; Figure 2A) and that MgtB levels were comparable between the WT and ΔLD strains (Figure 1D), we examined if MgtB affected ATP levels in the ΔLD strain. Interestingly, deletion of the mgtB gene elevated ATP levels in the ΔLD strain by ∼2-fold (Figure 2A). Moreover, additional deletion of mgtC further increased ATP levels of the ΔLD ΔmgtB strain to levels of the ΔLD ΔmgtC strain (Figure 2A). Together these results suggest that in the ΔLD strain, MgtB might contribute to ATP reduction in a manner dependent on MgtC.
The MgtC protein inhibits ATP synthesis by directly binding to the F0 a subunit of the ATP synthase (Lee et al., 2013). Consistent with this observation, MgtC-dependent ATP reduction is no longer observed in the absence of the atpB gene, which encodes the F0 a subunit (Lee et al., 2013). To explore whether ATP reduction in the ΔLD strain requires the F1F0 ATP synthase, we determined ATP levels in a set of ΔatpB strains grown in 10 μM Mg2+ for 4 h. We found that ATP levels were still ∼2-fold lower in the ΔLD ΔatpB strain than in the ΔatpB strain (Figure 2B). Moreover, deleting mgtC and mgtB increased ATP levels in the ΔLD ΔatpB strain by ∼3- and ∼2-fold, respectively (Figure 2B). Additional deletion of mgtC further increased ATP levels of the ΔLD ΔmgtB ΔatpB strain to levels of the ΔLD ΔmgtC ΔatpB strain (Figure 2B). Together these results suggest that dysregulated expression of MgtC and MgtB reduces ATP to abnormally low levels in a process that does not require F1F0 ATP synthase.
ATP Reduction Impairs Growth of the ΔLD Strain in Low Mg2+
In medium with 10 μM Mg2+, the ΔLD strain, which displayed lower ATP levels than the WT (Figure 2A), showed impaired growth (Figure 1C). In contrast, the ΔLD ΔmgtC strain, which displayed ATP levels as high as the ΔmgtC strain (Figure 2A), recovered growth to levels of the ΔmgtC strain (Supplementary Figure S5A). A similar relationship between ATP level and growth behavior was also observed between the ΔLD ΔmgtB and ΔmgtB strains (Figure 2A and Supplementary Figure S5B).
Growth behaviors of the strains carrying an atpB deletion also correlated with their ATP levels. In medium with 10 μM Mg2+, the ΔatpB strain grew in a manner similar to the WT strain (Figure 2C). However, the ΔLD ΔatpB strain, which exhibited lower ATP levels than the ΔatpB strain (Figure 2B), showed severely impaired growth (Figure 2C). Additionally, deleting mgtC and mgtB, which each increased ATP levels in the ΔLD ΔatpB strain (Figure 2B), recovered growth of the ΔLD ΔatpB strain (Figure 2C). Together these results suggest that the ATP reduction led by MgtC and MgtB independent of F1F0 ATP synthase caused impaired growth of the ΔLD strain in low Mg2+.
Dysregulated Induction of MgtC and MgtB Impairs Salmonella in High Mg2+ Environments With Acidic pH or an Antimicrobial Peptide
The PhoP regulator can be activated even in high (i.e., millimolar concentrations) Mg2+ by mildly acid pH (Prost et al., 2007; Choi and Groisman, 2016) or certain antimicrobial peptides (Bader et al., 2005). Given that PhoP promotes mgtC transcription at acidic pH (Choi and Groisman, 2016), we speculated about the behavior of the ΔLD strain under such conditions. We first determined MgtC levels in Salmonella grown in medium supplemented with 1 mM Mg2+ adjusted to pH 5.5. The ΔLD strain produced detectable levels of MgtC at 1 and 6 h after growth in a PhoP-dependent manner (Figure 3A). However, in the WT strain, MgtC was not detected at either time point (Figure 3A).
Figure 3. Dysregulated induction of MgtC and MgtB impairs growth of Salmonella in high Mg2+ with acidic pH or an antimicrobial peptide. Bacteria were grown in N-minimal medium with 1 mM Mg2+ at pH 5.5 (A–C) or with 1 mM Mg2+ and 5 μg/ml C18G peptide at pH 7.5 (D–F). (A,D) Immunoblot analysis of MgtC levels in wild-type (WT), ΔLD (DN557), and ΔLD phoP– (DN558) strains at 1 and 6 h after growth. The band indicated with an asterisk (∗) corresponds to a protein displaying cross-reactivity against the anti-MgtC antibody and serves as an internal loading control. (B,E) Growth curves of wild-type, ΔLD, ΔLD ΔmgtC (HK111), and ΔLD ΔmgtB (DN581) strains. OD600 values were determined at the indicated time points. Means and standard deviations from three independent experiments are shown. (C,F) ATP levels were determined in wild-type (WT), ΔLD, ΔLD ΔmgtC, and ΔLD ΔmgtB strains at 4 h after growth. Data depicted in arbitrary units (AU) are means and standard deviations from three independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, two-tailed t-test with each sample vs. WT; ns, not significant.
In the acidified medium, the ΔLD strain grew in a linear, slow fashion during the entire 8 h, which was in contrast to the WT strain that grew logarithmically for the first 4 h and continued to grow slowly for the remaining 4 h (Figure 3B). The growth defect seen in the ΔLD strain was also dependent on MgtC and MgtB, as evidenced by the finding that deleting mgtC and mgtB restored growth to the ΔLD strain (Figure 3B). Growth behaviors of the strains correlated with their ATP levels. ATP levels were ∼4-fold lower in the ΔLD strain than in the WT strain (Figure 3C). Deleting mgtC and mgtB increased ATP levels in the ΔLD strain by ∼6- and ∼2.5-fold, respectively (Figure 3C).
We next examined growth in medium containing 1 mM Mg2+ and sublethal concentrations of C18G, an antimicrobial peptide that activates the PhoP regulator (Bader et al., 2005). Again, only the ΔLD strain induced detectable MgtC (Figure 3D) and displayed a growth defect dependent on MgtC and MgtB (Figure 3E). Moreover, the ΔLD strain displayed reduced ATP levels in a manner associated with MgtC and MgtB (Figure 3F). Together, these results indicate that even if mgtC transcription is activated in high Mg2+ by acidic pH or an antimicrobial peptide, the mgtCBR mRNA leader prevents dysregulated induction of MgtC and MgtB, which is necessary for Salmonella to normally grow in these environments.
MgtC and MgtB Impair Growth of the ΔLD Strain Inside Macrophages in a Manner Not Associated With the F1F0 ATP Synthase
The notion that MgtC induction is necessary for Salmonella to survive inside macrophages (Blanc-Potard and Groisman, 1997) led us to explore behaviors of the ΔLD strain inside macrophages. We first determined MgtC levels in cell extracts prepared from Salmonella engulfed by macrophages. After entry into macrophages, the WT strain induced detectable MgtC at 6 h but not at 1 h, whereas the ΔLD strain induced MgtC at both time points (Figure 4A). This result indicates that the mgtCBR mRNA leader prevents earlier induction of MgtC during Salmonella infection of macrophages.
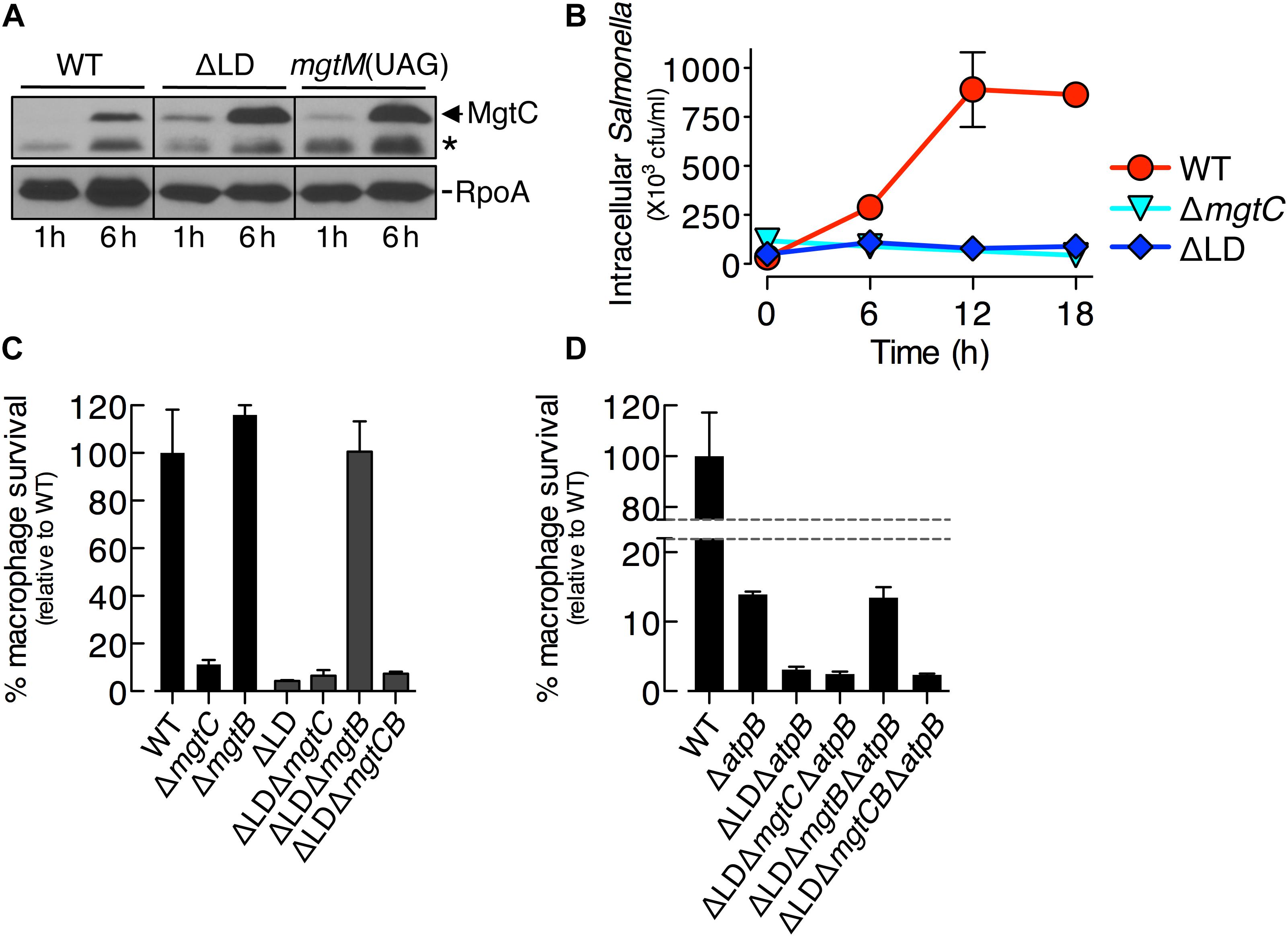
Figure 4. Dysregulated induction of MgtC and MgtB impairs Salmonella inside macrophages. (A) Immunoblot analysis of MgtC and RpoA determined in the wild-type (WT, 14028s), ΔLD (DN557), and mgtM(UAG) (EG19307) strains at 1 and 6 h after engulfment by J774A.1 macrophages. The band indicated with an asterisk (∗) corresponds to a protein displaying cross-reactivity against the anti-MgtC antibody. (B) Growth behaviors of wild-type, ΔmgtC (EN397), and ΔLD strains inside macrophages. After infection of macrophages, intracellular numbers of bacteria were determined at the indicated time points. (C,D) Intramacrophage survival of wild-type, ΔmgtC, ΔmgtB (EN481), ΔLD, ΔLD ΔmgtC (HK111), ΔLD ΔmgtB (DN581), ΔLD ΔmgtCB (DN608), ΔatpB (HK468), ΔLD ΔatpB (DN575), ΔLD ΔmgtC ΔatpB (MS575), ΔLD ΔmgtB ΔatpB (MS576), and ΔLD ΔmgtCB ΔatpB (DN686) strains. After infection of macrophages, the intracellular numbers of bacteria at 18 h were divided by those at 1 h. The percentage of survival of each mutant relative to the wild-type strain is presented. Means and standard deviations from three independent experiments are shown.
We then examined the growth of Salmonella inside macrophages. Intracellular numbers of WT Salmonella increased at 6 h after engulfment by ∼8-fold and peaked at 12 h by ∼25-fold, and these numbers were maintained at 18 h (Figure 4B). By contrast, during the entire experiment, the ΔLD strain was unable to grow inside macrophages, resembling the phenotype of the ΔmgtC strain (Figure 4B).
We also determined intramacrophage survival of Salmonella mutants relative to the WT strain at 18 h post infection. The ΔLD strain displayed only ∼5% survival compared with the WT strain (Figure 4C). The ΔLD ΔmgtC strain showed a low survival similar to the ΔLD strain, whereas the ΔLD ΔmgtB strain exhibited WT levels of survival (Figure 4C). These findings suggest that the phenotype of the ΔLD strain is dependent on MgtB and that MgtC alone supports survival of the ΔLD strain unless MgtB is present. Furthermore, the similarly low survival between the ΔLD ΔmgtC and the ΔLD ΔmgtCB strains (Figure 4C) excludes the possibility that MgtB alone might impair survival of the ΔLD strain in the absence of MgtC. Together these results suggest that, when induced inside macrophages in a dysregulated manner, MgtC and MgtB impair intramacrophage growth of Salmonella.
The role of MgtC in supporting survival of Salmonella inside macrophages is due to its ability to inhibit the F1F0 ATP synthase (Lee et al., 2013). Consistent with this notion, mgtC deletion did not further impair the ΔatpB strain, which showed much lower survival than the WT strain (Figure 4D; Lee et al., 2013). By contrast, the ΔLD ΔatpB strain displayed more attenuated survival than the ΔatpB strain (Figure 4D). The ΔLD ΔmgtC ΔatpB strain showed a low survival similar to the ΔLD ΔatpB strain, whereas survival of the ΔLD ΔmgtB ΔatpB strain increased to levels of the ΔatpB strain (Figure 4D). Intramacrophage survival was similarly low between the ΔLD ΔmgtC ΔatpB and the ΔLD ΔmgtCB ΔatpB strains (Figure 4D). These results suggest that MgtC and MgtB impair intramacrophage growth of the ΔLD strain in a manner not associated with F1F0 ATP synthase. Moreover, given that MgtC and MgtB reduced ATP to abnormal levels independently of the F1F0 ATP synthase in the ΔLD strain (Figure 2), this result suggests that such ATP reduction could also inhibit growth of Salmonella in host environments.
A Salmonella Mutant Lacking mgtM Translation Displays Behaviors Resembling Those of the ΔLD Strain
The mgtCBR mRNA leader harbors the two short ORFs mgtM and mgtP (Figure 1A). Translation of mgtM and mgtP affects mgtC expression by affecting conformational changes in the leader (Lee and Groisman, 2012a, b; Lee et al., 2014). Thus, we hypothesized mutations in the leader that cause dysregulated induction of MgtC and MgtB might confer ΔLD strain-like phenotypes to Salmonella. We focused on the role of mgtM, as mutations preventing normal mgtM translation greatly increased β-galactosidase activity produced by Salmonella carrying a chromosomal mgtC–lacZ fusion (Lee and Groisman, 2012a).
We determined levels of MgtC and MgtB in the mgtM(UAG) strain, in which the start codon UUG of mgtM was replaced with the amber stop codon UAG (Lee and Groisman, 2012a). When the mgtM(UAG) strain was placed in medium with 10 μM Mg2+, both MgtC and MgtB were induced to detectable levels as early as at 1 h, and these levels were maintained for the following 7 h (Figure 5A), resembling the temporal production of MgtC and MgtB in the ΔLD strain (Figure 1D). Four hours after growth, the mgtM(UAG) strain produced MgtC at levels ∼2-fold higher than the WT strain and ∼2-fold lower than the ΔLD strain (Figure 5B). However, MgtB levels were similar among the three strains (Figure 5B).
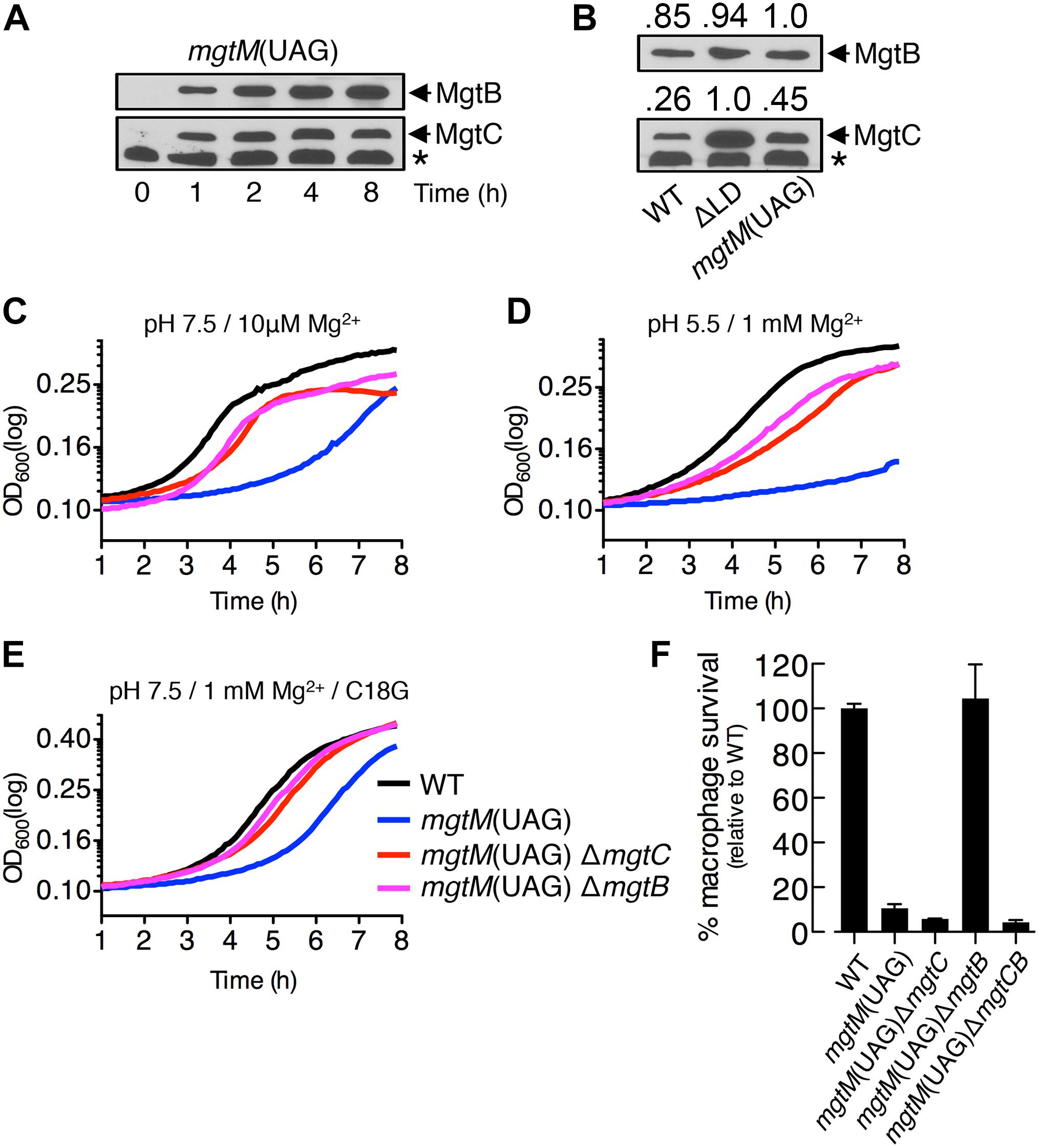
Figure 5. A conformation of the mgtCBR mRNA leader that causes dysregulated induction of MgtC and MgtB impairs Salmonella in host and non-host environments. (A,B) Immunoblot analysis of MgtC and MgtB in wild-type (WT, 14028s), ΔLD (DN557), and mgtM(UAG) (EG19307) strains. Bacteria were grown in N-minimal medium with 10 μM Mg2+ at pH 7.5 and harvested at the indicated time points (A) or at 4 h (B). The band indicated with an asterisk (∗) corresponds to a protein displaying cross-reactivity against the anti-MgtC antibody. In panel (B), numbers above the blots correspond to relative levels of MgtC and MgtB in each lane. (C–E) Growth curves of wild-type, mgtM(UAG) (EG19307), mgtM(UAG) ΔmgtC (DN649), and mgtM(UAG) ΔmgtB (DN612) strains. Bacteria were grown in N-minimal medium with 10 μM Mg2+ at pH 7.5 (C), with 1 mM Mg2+ at pH 5.5 (D), or with 1 mM Mg2+ and 5 μg/ml C18G peptide at pH 7.5 (E), and OD600 values were determined every 5 min using a plate reader. Data are representative of three independent experiments, which gave similar results. (F) Intramacrophage survival of wild-type, mgtM(UAG), mgtM(UAG) ΔmgtC, mgtM(UAG) ΔmgtB, and mgtM(UAG) ΔmgtCB (DN652) strains was determined as described in the legend of Figure 4. Means and standard deviations from three independent experiments are shown.
We also found that the mgtM(UAG) strain exhibited growth defects in all host and non-host environments that promote mgtC transcription. When placed in medium with 10 μM Mg2+, the mgtM(UAG) strain entered logarithmic growth later and displayed lower growth yields during logarithmic growth than the WT strain (Figure 5C). However, deleting mgtC and mgtB corrected the logarithmic growth of the mgtM(UAG) strain (Figure 5C). The growth defect was also observed in medium with 1 mM Mg2+ and pH 5.5 as well as in medium with 1 mM Mg2+ and the C18G antimicrobial peptide (Figures 5D,E). Again, deletions of mgtC and mgtB restored growth of the mgtM(UAG) strain under these conditions (Figures 5D,E).
Survival of the mgtM(UAG) strain inside macrophages was only ∼10% relative to the WT strain (Figure 5F). While mgtB deletion increased survival of the mgtM(UAG) strain to WT levels, deletion of mgtC or both mgtC and mgtB slightly decreased survival of the strain to similar levels (Figure 5F). These results suggest that MgtC and MgtB impair growth of the mgtM(UAG) strain inside macrophages. Taken together, our results emphasize that the function of the mgtCBR mRNA leader in preventing dysregulated induction of MgtC and MgtB is necessary for Salmonella to proliferate in both host and non-host environments.
Discussion
The mgtCBR mRNA leader, which contains two short ORFs for mgtM and mgtP, possesses tandem attenuators that sense two distinct signals (Lee and Groisman, 2012a, b; Lee et al., 2014). In response to acidic pH, an increase in ATP levels in the cytoplasm affects coupling/uncoupling of transcription of the leader with mgtM translation, inducing a conformation of the leader that promotes expression of the mgtC-coding region (Lee and Groisman, 2012a). Likewise, a decrease of intracellular proline promotes mgtC expression by affecting the events between leader transcription and mgtP translation (Lee and Groisman, 2012b; Lee et al., 2014).
In this study, we sought to further understand the physiological significance of the mgtCBR mRNA leader. We investigated two Salmonella mutants: the ΔLD strain, in which the sequences specifying the mgtCBR mRNA leader were removed (Figure 1A), and the mgtM(UAG) strain, in which the start codon of mgtM was replaced with a stop codon, resulting in the locking of the leader in a conformation allowing expression of the mgtC-coding region (Lee and Groisman, 2012a). Of note, MgtC expression was not constitutive but still inducible in these two strains; the ΔLD strain produced detectable levels of MgtC in 10 μM Mg2+ but not in 10 mM Mg2+ (Figure 1B), and MgtC was not detected in the mgtM(UAG) strain immediately at transfer from 10 mM to 10 μM Mg2+ (Figure 5A).
When Salmonella is placed in 10 μM Mg2+, the PhoP regulator promotes transcription initiation of mgtC (Shin and Groisman, 2005). However, this event alone does not ensure MgtC production. Salmonella logarithmically grows for the first few hours in 10 μM Mg2+ by consuming Mg2+ in the medium and then displays slow linear growth (Figure 1C; Soncini et al., 1996; Blanc-Potard and Groisman, 1997; Park et al., 2018). MgtC production is detectable after Salmonella enter into the slow-growth phase (Figures 1C,D; Yeom et al., 2017; Park et al., 2018). The decrease of cytoplasmic Mg2+ levels causes the onset of slow-growth phase (Pontes et al., 2016). The observation that MgtC induction is further delayed when Salmonella is grown in 50 μM Mg2+ (Park et al., 2018) further supports the relation between decreased cytoplasmic Mg2+ levels and MgtC induction. In contrast, both the ΔLD and mgtM(UAG) strains produced MgtC at detectable levels even during the time period corresponding to the logarithmic growth phase of the WT strain (Figures 1C,D, 5A). During this time period, growth yields of the two mutants, which both highly produced the Mg2+ transporter MgtB (Snavely et al., 1991b; Figures 1D, 5A), were lower than that of the WT (Figures 1C, 5C), suggesting that cytoplasmic Mg2+ levels in the mutants could be higher than the WT levels. Together, these results suggest that the ΔLD and mgtM(UAG) strains induce MgtC and MgtB in a dysregulated manner under conditions in which low cytoplasmic Mg2+ stress does not exist.
The F1F0 ATP synthase inhibitory protein MgtC (Lee et al., 2013) contributes to growth of WT Salmonella in low Mg2+ (Blanc-Potard and Groisman, 1997; Rang et al., 2007). MgtC directly acts on the F1F0 ATP synthase and reduces ATP levels, while MgtB has no effect (Figure 2A; Lee et al., 2013). Abnormally high ATP levels cause a growth defect of the mgtC mutant in low Mg2+ (Pontes et al., 2015). By contrast, in the ΔLD strain, MgtC and MgtB reduced ATP at levels lower than the WT levels in a process that did not require F1F0 ATP synthase (Figure 2), which appeared to impair bacterial growth (Figures 1E, 2C and Supplementary Figure S5).
The ΔLD and mgtM(UAG) strains produced ∼4- and ∼2-fold higher levels of MgtC than the WT, respectively, whereas MgtB levels were similar among the strains (Figure 5B). However, growth of the ΔLD strain was more impaired than the mgtM(UAG) strain (Figures 2A, 5). These findings suggest that overproduction of MgtC might impair growth of Salmonella in whatever environment it finds itself. However, this scenario is unlikely because of the following observations. The regulatory peptide MgtR specified by the mgtCBR operon binds to and promotes MgtC degradation (Alix and Blanc-Potard, 2008). Although the ΔmgtR strain produced ∼4-fold higher levels of MgtC than the WT (Supplementary Figure S6A), the mutant grew in a manner similar to the WT strain in 10 μM Mg2+ (Supplementary Figure S6B). The ΔmgtR strain also overproduced MgtC only in the slow-growth phase (Supplementary Figure S6A), resembling the temporal production of MgtC in the WT strain (Figure 1D). Moreover, even if MgtC was produced in a dysregulated manner, its inhibitory effect was not observed unless MgtB is produced in a dysregulated manner. We engineered the ΔmgtC and ΔLD ΔmgtC strains to express the mgtC ORF from the plasmid-linked lac promoter. When placed in medium with 10 μM Mg2+ and the same concentrations of IPTG, the two strains produced comparable levels of MgtC after 1 h (Supplementary Figure S7A). However, the ΔmgtC and ΔLD ΔmgtC strains produced MgtB in manners similar to the WT and ΔLD strains, respectively (Supplementary Figure S7A). Furthermore, ATP reduction and growth defect resembling the phenotypes of ΔLD strain were observed in the ΔLD ΔmgtC strain but not in the ΔmgtC strain (Supplementary Figures S7B,C). Together, all of these lines of evidence reinforce that dysregulated expression of MgtC and MgtB leads to ATP reduction, which impairs Salmonella growth.
The molecular basis of how the dysregulated induction of MgtC and MgtB reduces ATP levels independently of the F1F0 ATP synthase is currently unclear. MgtC affects membrane potential in a manner not associated with the ability to inhibit the F1F0 ATP synthase, suggesting that MgtC also affects ATP levels by acting on a protein(s) other than the F1F0 ATP synthase (Lee et al., 2013). The ΔmgtC strain harbors a hyperpolarized membrane, whereas mgtC overexpression causes membrane depolarization in WT Salmonella (Lee et al., 2013). Interestingly, mgtB overexpression in WT Salmonella also depolarizes a membrane (Lee et al., 2013). Moreover, the MgtB transporter is one of proteins that are crosslinked to the MgtC protein (Lee et al., 2013), suggesting direct interaction between these two proteins. Based on these lines of evidence, we hypothesize that when produced in a dysregulated manner, MgtC and MgtB together act on the protein(s) that affects membrane potential to cause membrane depolarization, which in turn reduces ATP to abnormal levels.
To survive inside the macrophage phagosome, Salmonella induces mgtC expression (Blanc-Potard and Groisman, 1997). MgtC acts on the F1F0 ATP synthase to reduce intracellular ATP, otherwise, ATP levels increase upon acidic pH inside the phagosome (Lee et al., 2013). The coupling/uncoupling of transcription of the mgtCBR mRNA leader with the translation of mgtM and mgtP responds to distinct intracellular cues (i.e., an increase in ATP and a decrease in proline, respectively), inducing a conformation of the leader that allows transcription elongation into the mgtC-coding region (Lee and Groisman, 2012a, b; Lee et al., 2014). These events occur independently but additively to each other and enable Salmonella to achieve mgtC expression at levels that ensure its survival inside macrophages (Lee et al., 2014). In addition to these findings, our data suggest that preventing dysregulated induction of MgtC by the mgtCBR mRNA leader is another important determinant for intramacrophage survival of Salmonella. During infection of macrophages, WT Salmonella induced detectable MgtC at 6 h but not at 1 h (Figure 4A), which is in good agreement with the previous detection of MgtC at 20 h but no detection at 5 h post-infection (Yeom et al., 2018). By contrast, the ΔLD and mgtM(UAG) strains induced MgtC at the early time point and showed greatly attenuated survival inside macrophages (Figures 4, 5).
The role of cis-acting regulatory RNAs in preventing dysregulated gene expression also impacts the pathogenesis of another bacterial species. In Yersinia pseudotuberculosis, the yscW–lcrF operon specifies the transcriptional regulator LcrF (Skurnik and Toivanen, 1992). The intergenic region of the yscW–lcrF mRNA functions as a thermosensor, forming a secondary structure that permits translation of lcrF at host body temperature (Bohme et al., 2012). Mutations that destabilize this structure enable LcrF production at lower temperatures, resulting in LcrF-dependent expression of virulence genes and attenuated bacterial virulence in mice (Bohme et al., 2012). However, the molecular basis underlying the virulence attenuation remains unknown. We determined that the Mg2+ transporter MgtB, which is not normally associated with the virulence phenotype (Figure 4C; Blanc-Potard and Groisman, 1997), impairs intramacrophage survival of the ΔLD and mgtM(UAG) strains in the presence of MgtC (Figures 4C, 5F). Moreover, given that the ΔLD strain exhibited the defective phenotype in the absence of normal F1F0 ATP synthase function (Figure 4D), these results suggest that the unusual ATP reduction by dysregulated expression of MgtC and MgtB could impair growth of Salmonella inside host cells.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
MP, HK, DN, and DS conceived the research. MP, HK, and DN performed the research. MP, HK, DN, D-HK, and DS analyzed the data. MP, HK, D-HK, and DS wrote the manuscript.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2015R1D1A1A01057723 and NRF-2018R1D1A1B07042525).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Eun-Jin Lee for kindly providing us the S. enterica strain EG19307 and anti-MgtB antibodies.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02831/full#supplementary-material
References
Alix, E., and Blanc-Potard, A. B. (2008). Peptide-assisted degradation of the Salmonella MgtC virulence factor. EMBO J. 27, 546–557. doi: 10.1038/sj.emboj.7601983
Bader, M. W., Sanowar, S., Daley, M. E., Schneider, A. R., Cho, U., Xu, W., et al. (2005). Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122, 461–472. doi: 10.1016/j.cell.2005.05.030
Blanc-Potard, A. B., and Groisman, E. A. (1997). The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16, 5376–5385. doi: 10.1093/emboj/16.17.5376
Bohme, K., Steinmann, R., Kortmann, J., Seekircher, S., Heroven, A. K., Berger, E., et al. (2012). Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 8:e1002518. doi: 10.1371/journal.ppat.1002518
Choi, E., Choi, S., Nam, D., Park, S., Han, Y., Lee, J. S., et al. (2017). Elongation factor P restricts Salmonella’s growth by controlling translation of a Mg2+ transporter gene during infection. Sci. Rep. 7:42098. doi: 10.1038/srep42098
Choi, E., Lee, K. Y., and Shin, D. (2012). The MgtR regulatory peptide negatively controls expression of the MgtA Mg2+ transporter in Salmonella enterica serovar Typhimurium. Biochem. Biophys. Res. Commun. 417, 318–323. doi: 10.1016/j.bbrc.2011.11.107
Choi, J., and Groisman, E. A. (2016). Acidic pH sensing in the bacterial cytoplasm is required for Salmonella virulence. Mol. Microbiol. 101, 1024–1038. doi: 10.1111/mmi.13439
Choi, Y., Choi, J., Groisman, E. A., Kang, D. H., Shin, D., and Ryu, S. (2012). Expression of STM4467-encoded arginine deiminase controlled by the STM4463 regulator contributes to Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 80, 4291–4297. doi: 10.1128/IAI.00880-12
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A 97, 6640–6645. doi: 10.1073/pnas.120163297
Davis, R. W., Bolstein, D., and Roth, J. R. (1980). Advanced Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Fields, P. I., Groisman, E. A., and Heffron, F. (1989). A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243(4894 Pt 1), 1059–1062. doi: 10.1126/science.2646710
Fields, P. I., Swanson, R. V., Haidaris, C. G., and Heffron, F. (1986). Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U.S.A. 83, 5189–5193. doi: 10.1073/pnas.83.14.5189
Foster, J. W., and Hall, H. K. (1990). Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172, 771–778. doi: 10.1128/jb.172.2.771-778.1990
Garcia Vescovi, E., Soncini, F. C., and Groisman, E. A. (1996). Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84, 165–174. doi: 10.1016/s0092-8674(00)81003-x
Groisman, E. A. (2001). The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183, 1835–1842. doi: 10.1128/jb.183.6.1835-1842.2001
Lee, E. J., Choi, J., and Groisman, E. A. (2014). Control of a Salmonella virulence operon by proline-charged tRNAPro. Proc. Natl. Acad. Sci. U.S.A. 111, 3140–3145. doi: 10.1073/pnas.1316209111
Lee, E. J., and Groisman, E. A. (2010). An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol. Microbiol. 76, 1020–1033. doi: 10.1111/j.1365-2958.2010.07161.x
Lee, E. J., and Groisman, E. A. (2012a). Control of a Salmonella virulence locus by an ATP-sensing leader messenger RNA. Nature 486, 271–275. doi: 10.1038/nature11090
Lee, E. J., and Groisman, E. A. (2012b). Tandem attenuators control expression of the Salmonella mgtCBR virulence operon. Mol. Microbiol. 86, 212–224. doi: 10.1111/j.1365-2958.2012.08188.x
Lee, E. J., Pontes, M. H., and Groisman, E. A. (2013). A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium’s own F1Fo ATP synthase. Cell 154, 146–156. doi: 10.1016/j.cell.2013.06.004
Lee, J. W., and Lee, E. J. (2015). Regulation and function of the Salmonella MgtC virulence protein. J. Microbiol. 53, 667–672. doi: 10.1007/s12275-015-5283-1
Maloy, S. R., and Nunn, W. D. (1981). Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol. 145, 1110–1111.
Park, M., Nam, D., Kweon, D. H., and Shin, D. (2018). ATP reduction by MgtC and Mg2+ homeostasis by MgtA and MgtB enables Salmonella to accumulate RpoS upon low cytoplasmic Mg2+ stress. Mol. Microbiol. 110, 283–295. doi: 10.1111/mmi.14105
Pontes, M. H., Lee, E. J., Choi, J., and Groisman, E. A. (2015). Salmonella promotes virulence by repressing cellulose production. Proc. Natl. Acad. Sci. U.S.A. 112, 5183–5188. doi: 10.1073/pnas.1500989112
Pontes, M. H., Yeom, J., and Groisman, E. A. (2016). Reducing ribosome biosynthesis promotes translation during low Mg2+ stress. Mol. Cell. 64, 480–492. doi: 10.1016/j.molcel.2016.05.008
Prost, L. R., Daley, M. E., Le Sage, V., Bader, M. W., Le Moual, H., Klevit, R. E., et al. (2007). Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol. Cell. 26, 165–174. doi: 10.1016/j.molcel.2007.03.008
Rang, C., Alix, E., Felix, C., Heitz, A., Tasse, L., and Blanc-Potard, A. B. (2007). Dual role of the MgtC virulence factor in host and non-host environments. Mol. Microbiol. 63, 605–622. doi: 10.1111/j.1365-2958.2006.05542.x
Shin, D., and Groisman, E. A. (2005). Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J. Biol. Chem. 280, 4089–4094. doi: 10.1074/jbc.m412741200
Skurnik, M., and Toivanen, P. (1992). LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Bacteriol. 174, 2047–2051. doi: 10.1128/jb.174.6.2047-2051.1992
Snavely, M. D., Gravina, S. A., Cheung, T. T., Miller, C. G., and Maguire, M. E. (1991a). Magnesium transport in Salmonella typhimurium. Regulation of mgtA and mgtB expression. J. Biol. Chem. 266, 824–829.
Snavely, M. D., Miller, C. G., and Maguire, M. E. (1991b). The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J. Biol. Chem. 266, 815–823.
Soncini, F. C., Garcia Vescovi, E., Solomon, F., and Groisman, E. A. (1996). Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178, 5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996
Soncini, F. C., Vescovi, E. G., and Groisman, E. A. (1995). Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177, 4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995
Yeom, J., Pontes, M. H., Choi, J., and Groisman, E. A. (2018). A protein that controls the onset of a Salmonella virulence program. EMBO J. 37:e96977. doi: 10.15252/embj.201796977
Keywords: mRNA leader, MgtC, MgtB, ATP, Salmonella
Citation: Park M, Kim H, Nam D, Kweon D-H and Shin D (2019) The mgtCBR mRNA Leader Secures Growth of Salmonella in Both Host and Non-host Environments. Front. Microbiol. 10:2831. doi: 10.3389/fmicb.2019.02831
Received: 04 September 2019; Accepted: 21 November 2019;
Published: 06 December 2019.
Edited by:
Benjamin Makepeace, University of Liverpool, United KingdomReviewed by:
Calvin A. Henard, University of North Texas, United StatesAudrey Chong, National Institute of Allergy and Infectious Diseases (NIAID), United States
Copyright © 2019 Park, Kim, Nam, Kweon and Shin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dae-Hyuk Kweon, ZGhrd2VvbkBza2t1LmVkdQ==; Dongwoo Shin, c2hpbmRAc2trdS5lZHU=
†These authors have contributed equally to this work
 Myungseo Park
Myungseo Park Hyunkeun Kim
Hyunkeun Kim Daesil Nam
Daesil Nam Dae-Hyuk Kweon
Dae-Hyuk Kweon Dongwoo Shin
Dongwoo Shin