- 1School of Life Sciences, Central South University, Changsha, China
- 2School of Chemistry and Biological Engineering, Changsha University of Science and Technology, Changsha, China
- 3State Key Laboratory of Coal Resources and Safe Mining, China University of Mining and Technology, Xuzhou, China
The special ecological environment of the Arctic has brought about a large number of salt-tolerant and psychrotolerant microorganisms. We isolated two culturable bacterial strains of the genus Mesonia; one from the Arctic ocean, Mesonia algae K4-1, and one from the tropical sea, Mesonia sp. HuA40. Our genome analysis and phenotypic experiments indicated that Mesonia algae K4-1 is a moderately halophilic and psychrophilic bacterium. Mesonia algae K4-1 can tolerate 3–14% NaCl and grow at a wide range of temperatures from 4 to 50°C. Mesonia sp. HuA40 is a mesophilic bacterium that can only grow with 3–9% NaCl. In addition, the salt adaptation strategy of Mesonia algae K4-1 accumulates organic osmolytes in the cell. RNA helicases, glutathione and organic compatible solutes may play important roles in maintaining the metabolism and physiological function of Mesonia algae K4-1 under cold stress. Moreover, the ability of Mesonia algae K4-1 to adapt to an oligotrophic marine environment is likely due to the synthesis of a large number of extracellular polysaccharides and the secretion of various families of extracellular proteases. This study systematically analyzed the relationship between genomic differentiation and environmental factors of the Mesonia genus and revealed the possible adaptation mechanism of Mesonia algae K4-1 in the extreme Arctic marine environment at the genomic level.
Introduction
The polar regions constitute up to 14% of the cold habitats on Earth. The Arctic is very cold and harsh for most of the year with little solar radiation. One of the unique characteristics of Arctic is the periodical melting of sea ice. Bacteria in the Arctic may face a sudden drop in ambient salt concentration when the ice melts (Thomas and Dieckmann, 2002). The capacity for polar microbes to exist and proliferate in this extreme environment means that they have overcome key intrinsic obstacles to survive in cold environments (D’Amico et al., 2006). Many halophilic and psychrophilic bacteria have been isolated from the Arctic ocean (Van Trappen et al., 2004; Yukimura et al., 2010). However, how these microbes are adapted to survive environmental changes in extreme conditions is largely unknown.
Different species of the same genus from different ecosystems may evolve different biochemical and physiological properties (Konstantinidis et al., 2009; Chen et al., 2010; Qin et al., 2010, 2014). Comparative genomics can offer detailed descriptions of bacterial growth, evolution, and phylogeny (Daubin et al., 2003; Descorps-Declere et al., 2008; Caputo et al., 2019). It has become a powerful method to exploit the relationship between environmental adaption and species evolution. Pittera et al. (2018) revealed a specific adaptability to the content of lipid desaturase by conducting comparative genomic studies on membrane lipid biosynthesis pathways of 53 marine Synechococcus strains from different ecotypes. Omae et al. (2017) used comparative genomic analysis to show the adaptability of C. maritimus KKC1 to submerged marine caldera; C. maritimus KKC1 lacks specific electron-bifurcating enzymes and possesses six CO dehydrogenases.
The genus Mesonia, belonging to the family Flavobacteriaceae within the phylum Bacteroidetes, was first discovered by Nedashkovskaya et al. (2003). Currently, a total of 8 species of Mesonia have been validly described (Nedashkovskaya et al., 2003, 2006; Kang and Lee, 2010; Lee et al., 2012; Choi et al., 2015; Kolberg et al., 2015; Wang et al., 2015; Sung et al., 2017). All species isolated from the ocean and some additional species can hydrolyze gelatin (Nedashkovskaya et al., 2003; Sung et al., 2017), casein (Nedashkovskaya et al., 2003; Choi et al., 2015; Sung et al., 2017), fucoidan (Urvantseva et al., 2006), Tween 80 (Nedashkovskaya et al., 2006; Kang and Lee, 2010; Choi et al., 2015; Wang et al., 2015). Earlier studies found that some strains of the genus can form biofilms (Jeong et al., 2016) and are multiresistant (Miranda et al., 2015). Biofilms afford bacterial stability in the growing environment, allow for the capture of nutrients, and provide a range of environmental challenges and pressure protection (Ricciardelli et al., 2019). However, the current research on this genus is still very limited, and most of the existing reports are about their uncultivable research. No genome of the genus Mesonia isolated from the Arctic has been sequenced. Therefore, a genome and physiological characteristic study of the genus Mesonia is beneficial for exploring its potential and development as a type of biological resource.
In this work, we isolated two strains of the genus Mesonia from different niches and found differences in their physiological and biochemical characteristics. Mesonia algae K4-1 is a typical psychrophilic and moderate halophilic bacterium isolated from the Arctic (12°07.553E, 78°55.464 N). Mesonia sp. HuA40 was isolated from hull attachments at the bottom of a ship located in the tropical marine Zhanjiang, Guangdong Province, China (110°96.191E, 20°88.073N). Strain K4-1 is an ideal material for studying how the Arctic bacteria of the genus Mesonia are adapted to survive in extreme conditions. Considering the species diversity and metabolic plasticity of polar bacteria, more representative sequences must be analyzed to fully describe the special characteristics. Therefore, we analyzed the genome sequence and phenotypic characterization focusing on the study of genes related to several metabolic processes that are crucial to environmental adaptability, such as salt tolerance, cold adaptation, exopolysaccharide production and degradation of proteinaceous organic nitrogen. This research provides insights into the survival mechanism of the Mesonia group from a genomic perspective.
Materials and Methods
Bacterial Strains and Genomic DNA Extraction
Mesonia algae K4-1 was collected at 117 m depth in the Arctic (12°07.553E, 78°55.464N). The strain Mesonia sp. HuA40 was isolated from hull attachments in Zhanjiang, Guangdong Province, China (110°96.191E, 20°88.073N). The two strains were cultured in 2216E medium containing 5 g of peptone, 1 g of yeast extract, and 0.01 g of Fe2(PO4)3 in 1 L of artificial seawater, pH 7.5 (Wu et al., 2016). The 16S rRNA genes were sequenced to validate the obtained strains. Genomic DNA was prepared using a genomic DNA extraction kit (Biospin, China) according to the manufacturer’s instructions. The two strains were deposited in the China Center for Type Culture Collection (CCTCC), the collection number for Mesonia algae K4-1 is CCTCC M 2018482, and the collection number for the Mesonia sp. HuA40 is CCTCC NO:M 2018481.
Draft Genome Sequence
Whole genome sequencing of the two strains was performed using the Illumina HiSeq 3000 (Illumina, Inc., United States) at Genergy Biotechnology (Shanghai, China). Skewer software was used to dynamically remove joint sequence fragments and tail segments with a mass value lower than Q30 from the 3′ end of the sequence, erase segments with a mean value lower than Q30, and remove segments with lengths less than 50 bp. Reads were assembled into contigs using SPAdes (version: 3.5.0), and contig less than 500 bp in length were removed. Gene prediction software Prokka1 was used to predict the assembled sequences.
Genome Annotation and Analysis
Putative coding sequences (CDSs) were identified by Glimmer 3.0 (Delcher et al., 2007). Subsequently, protein sequences were further analyzed on the basis of Swiss-Prot (Magrane and UniProt, 2011), KEGG (Kanehisa et al., 2008), and eggNOG databases (evalue < 0.00001) (Huerta-Cepas et al., 2016).
The average nucleotide identity (ANI) values between genomes were estimated by the online web server EzBioCloud (Yoon et al., 2017). tRNA genes were predicted by tRNAscan-SE (Lowe and Chan, 2016), and RNA genes were predicted by RNAmmer1.2 (Lagesen et al., 2007). Secondary metabolite-related genes were predicted by the online web server antiSMASH3.0.5 (Weber et al., 2015). Amino acid composition and protein isoelectric points were counted via the EMBOSS Pepstats website. Signal peptide prediction was performed with SignalP 3.0 (Bendtsen et al., 2004). The MEROPS peptidase database (Release 12.0)2 was used to classify the enzymes (Rawlings et al., 2014). A genome circle was constructed using Circos (Krzywinski et al., 2009).
Comparative Genome Analysis
Clusters of orthologous groups (COGs) of proteins were used for functional classifications performed with the eggNOG (version 4.5) database. Orthologous clusters (OCs) were assigned by grouping all protein sequences from the two genomes using OrthoMCL v2.0.9 software based on their sequence similarity (BLASTP E-value less than 1e–5, MCL inflation = 1.5) (Fischer et al., 2011). All sequences were first filtered, allowing a minimum protein length of 10 and a maximum stop codon of 20%.
Morphological, Phenotypic Characteristics and Comparison
Morphology
Colonies on marine agar were investigated under a light microscope (SMZ445, Nikon) using cells grown on 2216E agar for 96 h. The individual cells were observed by scanning electron microscopy. The strains were cultured at optimum growth temperature, and when the optical density (OD600 nm) reached 0.6, the culture medium was removed. The bacteria were fixed with 2.5% glutaraldehyde for 4 h, washed three times with 0.1M phosphate buffer solution (PBS, pH 7.4), and dehydrated in increasing concentrations of ethanol (30, 50, 70, 90, and 100%) for 20 min each time. Then, the samples were dried in a vacuum freeze dryer. The dried samples were sputter coated with gold and viewed under scanning electron microscopy (Mira 3, Tescan, Czechia).
Physiological and Biochemical Characteristics
Hydrolysis of Tween 80, casein, starch, gelatin, and agar was determined on 2216E medium supplemented with 1.0% (v/v) Tween 80, 1% (w/v) casein, 0.5% (w/v) starch, 1% (w/v) gelatin, or 2.0% (v/v) agar, respectively (Nedashkovskaya et al., 2003). Catalase activity was determined by pouring 3% H2O2 solution on bacterial colonies and observing bubble production. Oxidase activities were tested using 1% (w/v) N, N, N9, N9-tetramethyl-p-phenylenediamine solutions (Gerhardt, 1994).
Optimum Growth Temperature, pH and Salinity Tolerance
Mesonia algae K4-1 and Mesonia sp. HuA40 cells were cultivated in 2216E medium, and the optimum growth temperature was examined by growing cells at different temperatures (4, 10, 20, 30, 40, and 50°C). The growth effects of pH were measured at pH 4, 5, 6, 7, 8, 9,10, 11, and 12. Culture growth was followed at OD600 nm by Perkin Elmer (United States). The salinity tolerance tests were examined in 2216E liquid medium, Mesonia algae K4-1 were cultivated at 18°C and Mesonia sp. HuA40 were cultivated at 37°C overnight. When the optical density at 600 nm (OD600 nm) reached 0.6, 100 μl of each culture was transferred into 100 ml of 2216E medium containing different concentrations of NaCl (3, 7, 8, 9, 12, 13, 14, and 15%). The OD600 nm value was measured every 8 h on average.
Extracellular Protease Activity
The strains were cultured in 500 ml flasks at the optimum growth temperature with shaking at 200 rpm (Wu et al., 2016). The supernatant of the fermentation broth was collected by centrifugation (11,000 × g, 4°C, 30 min) after 120 h incubation. The protease activity of the culture supernatant on casein was detected by the Folin phenol method (He et al., 2004). One unit of enzyme activity was determined as the amount of enzyme that catalyzed the formation of 1 μg tyrosine per min.
Extracellular Polysaccharide Yield
The fermentation supernatant was added with five volumes of absolute ethanol, mixed, allowed to stand at 4°C for 30 min and centrifuged at 11,000 × g for 5 min. Anhydrous ethanol was added to the precipitate, shaken vigorously, and centrifuged at 4°C; the monosaccharide was washed away and repeated three times. The precipitate was dried at 60°C for 30 min and redissolved in distilled water. After the appropriate dilution, the yield of extracellular polysaccharide in the bacterial fermentation broth was determined by the phenol sulfuric acid method (Masuko et al., 2005).
Results and Discussion
Classification, Morphology and Characteristics of Strains
The strain K4-1 was collected in the Arctic ocean (12°07.553E, 78°55.464N), and HuA40 was isolated from hull attachments in Zhanjiang, Guangdong Province, China. The 16S rRNA gene sequences from the genomes of strains K4-1 and HuA40 were subjected to BLAST analysis to identify the closest reference sequences available in GenBank, which showed that the two strains are most closely related to the members of the genus Mesonia (Figure 1). The 16S rRNA gene homology between Mesonia algae K4-1 and its closest cultured member Mesonia algae strain NBRC 100447 is 99.86%. The OrthoANIu value between Mesonia algae K4-1 and Mesonia algae strain DSM 15361(Accession NZ_QKYV00000000) is 95.28% (>95%). Thus, the two strains are thought to be the same species (Goris et al., 2007). The 16S rRNA gene homology between Mesonia sp. HuA40 and its closest cultured member Mesonia sediminis MF32f is 99.04%. Because the genomic sequence of Mesonia sediminis has not been reported thus far, we defined strain HuA40 from the genus level, Mesonia sp. HuA40. In addition, the 16S rRNA gene homology between the two strains was 92%.
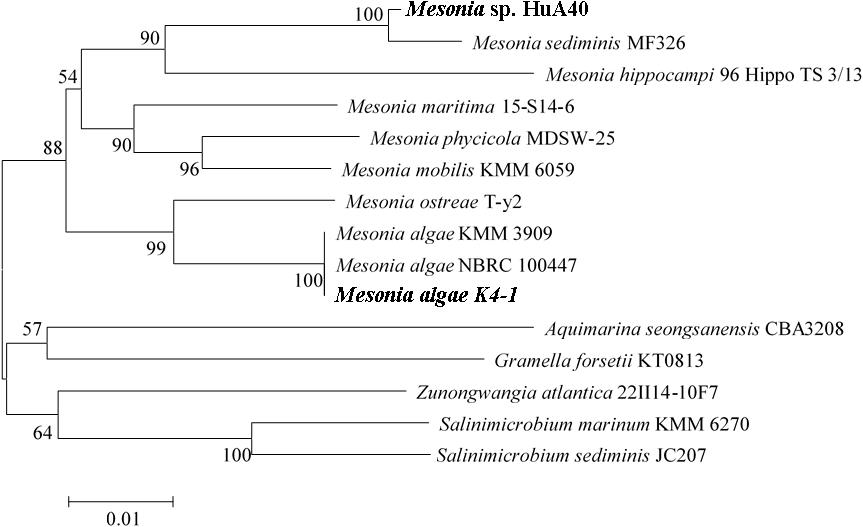
Figure 1. Phylogenetic tree based on the 16S rRNA gene sequences of Mesonia sp. HuA40, Mesonia algae K4-1 and related strains. The strains Mesonia sp. HuA40 and Mesonia algae K4-1 are indicated in bold font.
As shown in Figure 2, the morphological characteristics of the two strains were similar. Both strains were rod-shaped and ranged from 0.3 to 0.4 μm in width and from 0.7 to 1.2 μm in length (Figures 2A,B). Mesonia algae K4-1 cells were slightly larger than Mesonia sp. HuA40 cells. The colonies of these two types were 0.5–1.0 mm in diameter, and circular with entire edges when cultivated on 2216E agar at 16°C for 72 h (Figures 2C,D). Compared with Mesonia algae K4-1, the smooth surface of Mesonia sp. HuA40 appeared moister and the color was close to orange. On the other hand, the growth curve shows that Mesonia sp. HuA40 grew in 3-9% (w/v) NaCl and pH 6.0–8.5, whereas Mesonia algae K4-1 grew well even at 3-15% (w/v) NaCl (Table 1). Mesonia algae K4-1 could grow at 4°C, and the halo formed by hydrolysis of Tween 80 and the transparent ring formed by the hydrolysis of casein were obviously larger than Mesonia sp. HuA40 (Table 1). In addition, Mesonia algae K4-1 had a catalase that was missing in Mesonia sp. HuA40 (Table 1), which may be related to the tolerance to high concentrations of H2O2 in the Arctic ocean (Yu et al., 2015). The physiological and biochemical analysis of the two strains showed that the differences were mainly reflected in the tolerance of acid, alkali and salt, growth temperature range, hydrolysis activity of protein and lipids, and hydrogen peroxide decomposition.
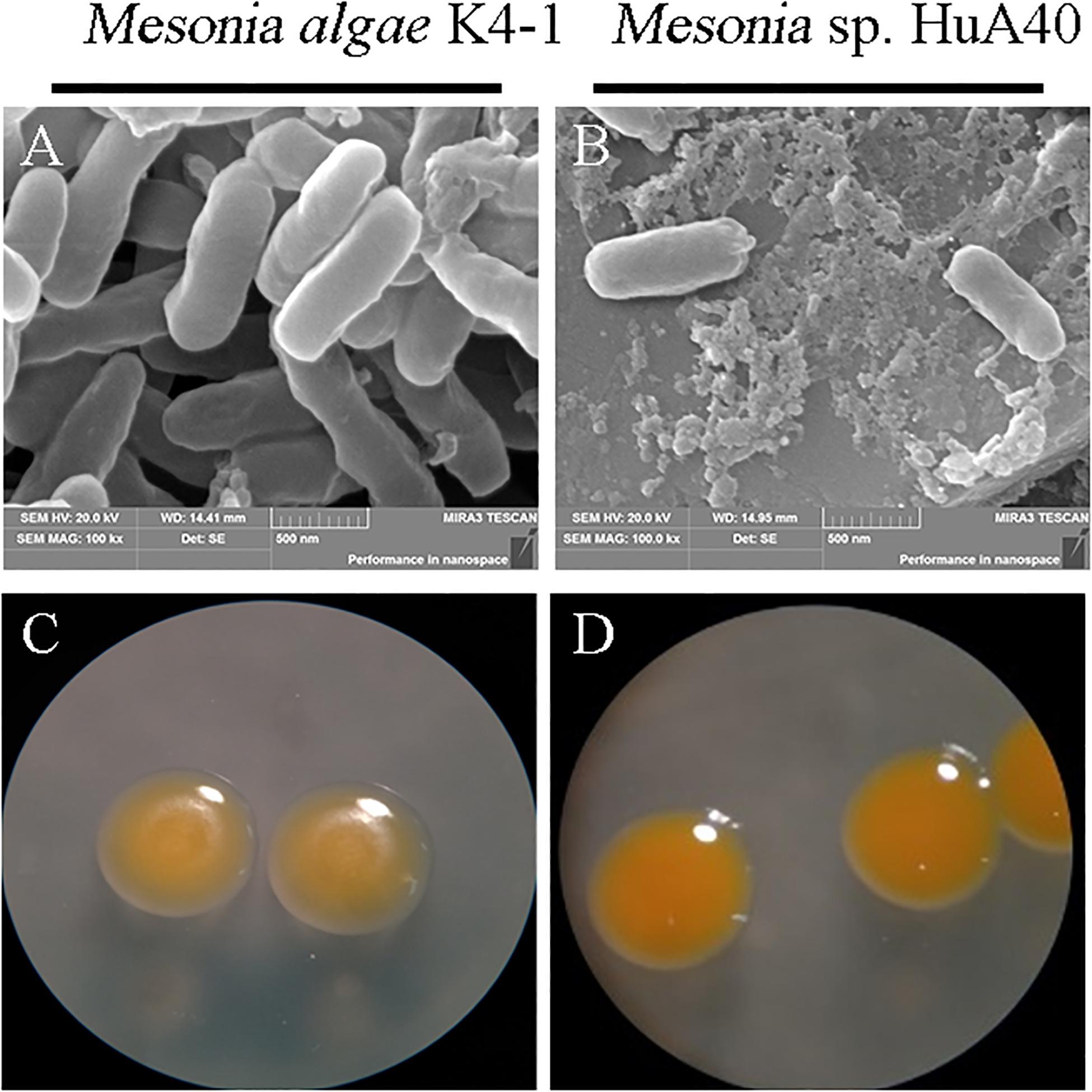
Figure 2. Images of cell morphology. [(A) Mesonia algae K4-1 and (B) Mesonia sp. HuA40] Scanning electron microscope (magnification, ×100,000); [(C) Mesonia algae K4-1 and (D) Mesonia sp. HuA40] light microscopy pictures in 2216E plate colony (magnification, ×4).

Table 1. Differential phenotypic characteristics of strain Mesonia algae K4-1 and Mesonia sp. HuA40.
General Genomic Features
Considering the phenotypic differences between the two strains, we further analyzed the genomic characteristics. General features of the Mesonia algae K4-1 and Mesonia sp. HuA40 genomes are summarized in Table 2 and Supplementary Figure S1. Overall, draft assemblies of the Mesonia algae K4-1 genome yielded 111 contigs with an average GC content of 33.9%. The draft genome is approximately 3.08 Mbp, and a total of 3,190 coding sequences (CDSs) were identified. The Mesonia sp. HuA40 genome was automatically assembled into 116 contigs, with an estimated genome size of 2.70 Mbp and a total of 2585 CDSs. The Arctic bacterium Mesonia algae K4-1 has a larger genome and a higher proportion of CDSs than the Mesonia sp. HuA40 genome. Bentkowski et al. (2015) established a correlation model to study the genome size evolution of populations at different environmental disturbance levels. It is proposed that different environmental variations lead to more genes in the genome. Konstantinidis and Tiedje (2004) proposed that strains with relatively large genomes could buffer environmental disturbances more effectively. The genome analysis results showed that Mesonia algae K4-1 may be more robust to variable conditions and equipped with an ecological advantage. Horizontal gene transfer (HGT) is a general way for bacteria to respond to different selective pressures and obtain an important competitive advantage (Chen et al., 2016). In HGT events, the acquisition or loss of MGE plays a key role in carrying functional genes that enhance fitness in the environment (Hall et al., 2017; Wang L. et al., 2017; Husnik and McCutcheon, 2018). In general, the number of transposable elements is relatively high in large genomes, indicating that gene turnover is relatively frequent and has metabolic diversity (Gweon et al., 2017). As shown in Table 3, 11 transposase- and 13 integrase-encoding genes were found in the genome of strain Mesonia algae K4-1, while Mesonia sp. HuA40 encodes 8 transposases and 3 integrases. The increased number of copies of the enzyme support that Mesonia algae K4-1 has a larger genome than Mesonia sp. HuA40. In addition to potential HGT activities, these adaptive genes move between different species, promoting the diversity of bacteria in different environments. Mesonia algae K4-1 and Mesonia sp. HuA40 encode 78 and 51 transcriptional regulators, respectively, suggesting that Mesonia algae K4-1 may be more adaptable in a wider range of external and gradient-affected environments.
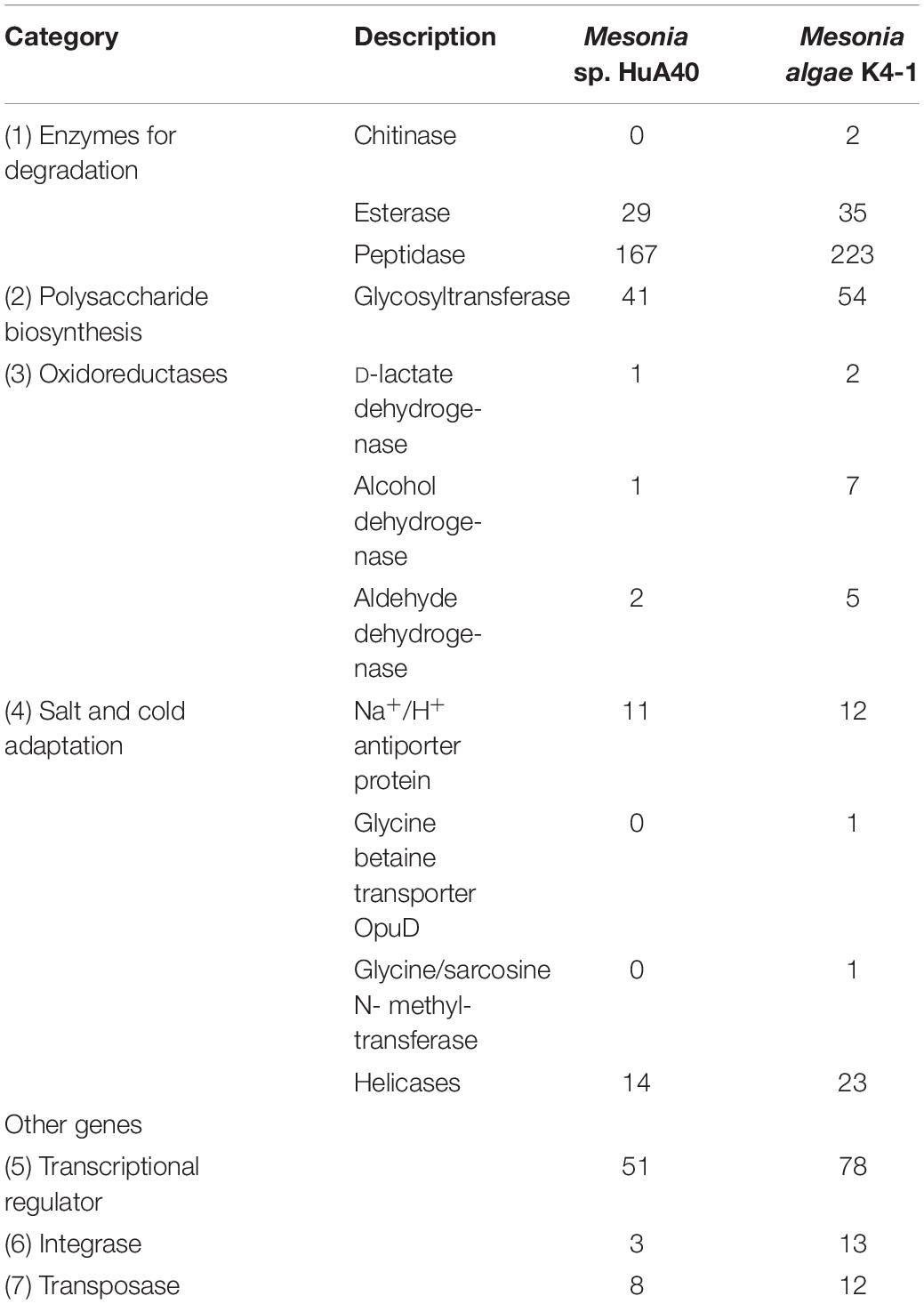
Table 3. Comparison of the numbers of selected proteins between Mesonia algae K4-1 and Mesonia sp. HuA40.
Differential Gene Content
All genes of Mesonia algae K4-1 and Mesonia sp. HuA40 were functionally classified according to the COG category. In total, 88.04% of Mesonia sp. HuA40 genes and 88.53% of Mesonia algae K4-1 genes were annotated with the COG database (Figure 3A). To evaluate the degree of genetic differences encoded by the two strains, the unique genes of each strain were classified by COG categories, and the frequencies of the two genomes were compared (Figure 3B and Supplementary Table S1). The results showed that Mesonia algae K4-1 has a higher number of unique genes in COG K (transcription), COG T (signal transduction mechanism), COG P (inorganic metal ion transport and metabolism), COG L (replication, recombination and repair), and COG M (cell wall, membrane and envelope biogenesis). The increase in COG K, COG T revealed that Mesonia algae K4-1 may possess a more complex transcriptional system and strong regulatory systems to respond to various environmental stimuli and change the transcription rate by altering the level of gene expression. Moreover, the grouping of COG M shows that Mesonia algae K4-1 encodes specific genes related to polysaccharide synthesis, outer membrane proteins, and lipid synthesis. The unique genes of COG P encoded by Mesonia algae K4-1 may suggest mechanisms that maintain the specificity of osmotic adjustment. Interestingly, genomic analysis showed that the strain Mesonia algae K4-1 encodes approximately 42.17% of the proteins belonging to the unknown functional COG, which contains 1,191 proteins. In contrast, Mesonia sp. HuA40 encodes 39.8% proteins of unknown function, which contains 879 proteins. This result indicated that the Mesonia algae K4-1 currently has large number of proteins of unknown functions.
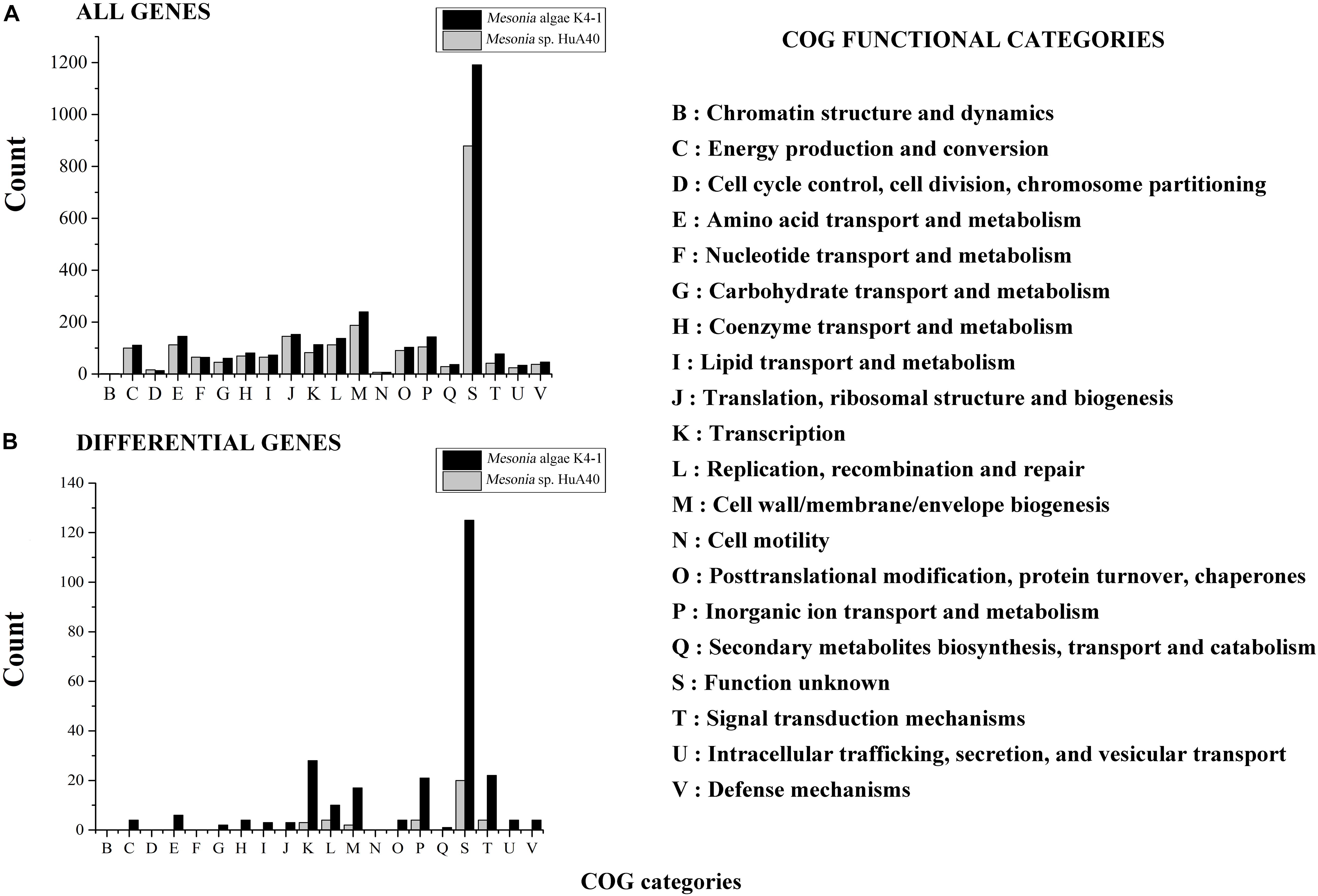
Figure 3. Distribution of COG functional classes. Count of COGs predicted in Mesonia algae K4-1 and Mesonia sp. HuA40 genomes. All genes (A) and genes found only in one of the genomes (B) are indicated. COG, clusters of orthologous groups.
Salinity Adaptation Strategy and pH Regulation
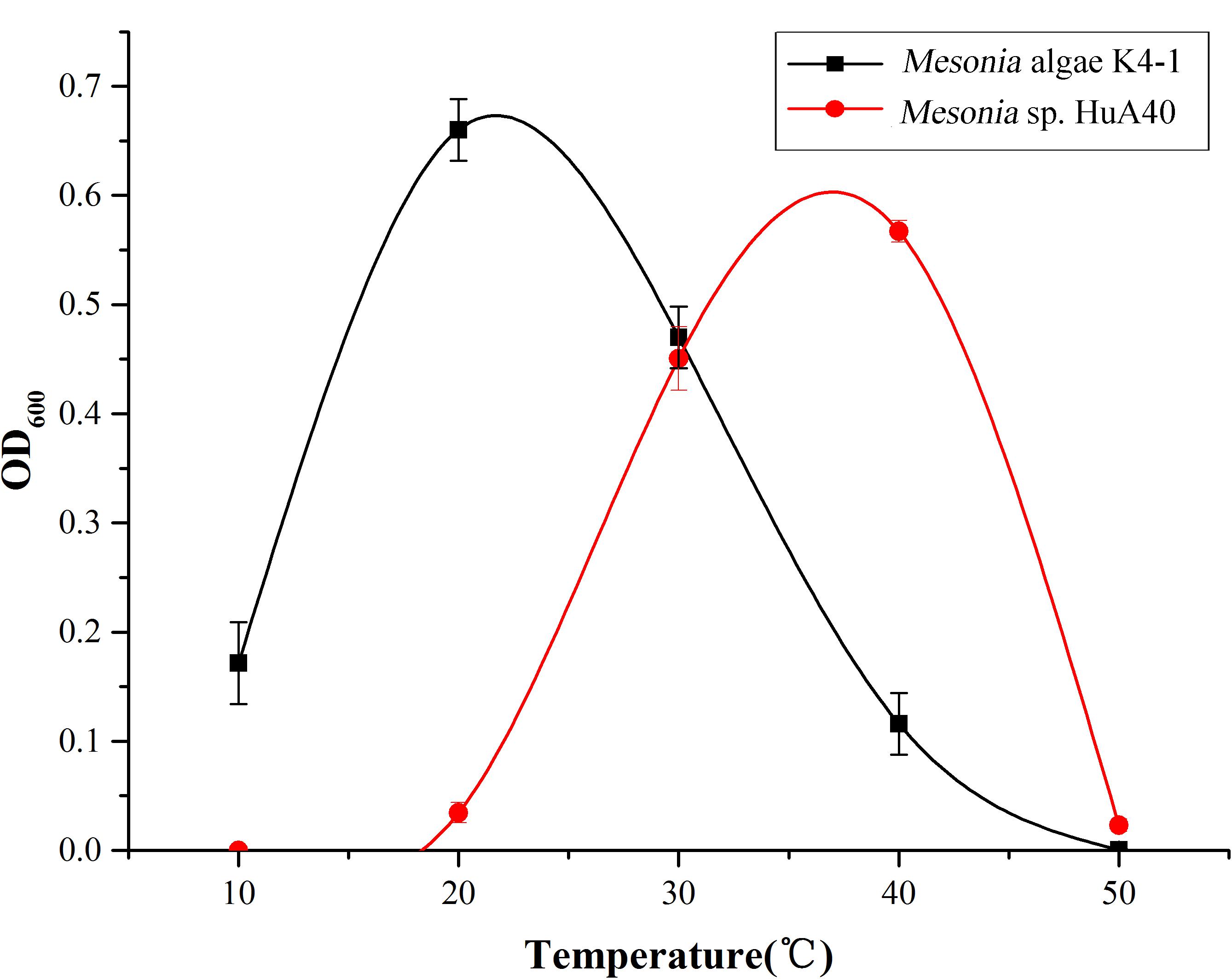
As shown in Figure 4, Mesonia algae K4-1 tolerated 3–14% NaCl, while strain Mesonia sp. HuA40 only tolerated 9% NaCl. At 3% and 7% NaCl, the lag phase of Mesonia algae K4-1 was shorter than that of Mesonia sp. HuA40. Moreover, Mesonia algae K4-1 had a much higher cell density at stationary phase when the NaCl concentration was increased to 14%. The Mesonia sp. HuA40 biomass gradually decreased in stationary phase when the NaCl concentration was increased to 9%. These results indicated that the salt tolerance of Mesonia sp. HuA40 was significantly lower than that of Mesonia algae K4-1. Due to the high salinity of seawater accompanied by high pH (Collins et al., 2010), we also tested the pH tolerance of the two strains. Mesonia algae K4-1 can tolerate pH 6–10, while Mesonia sp. HuA40 can only tolerate pH 6-8 (Supplementary Figure S2). This result is consistent with the expectation that salt-tolerant bacteria have strong alkali tolerance (Oren, 1999).
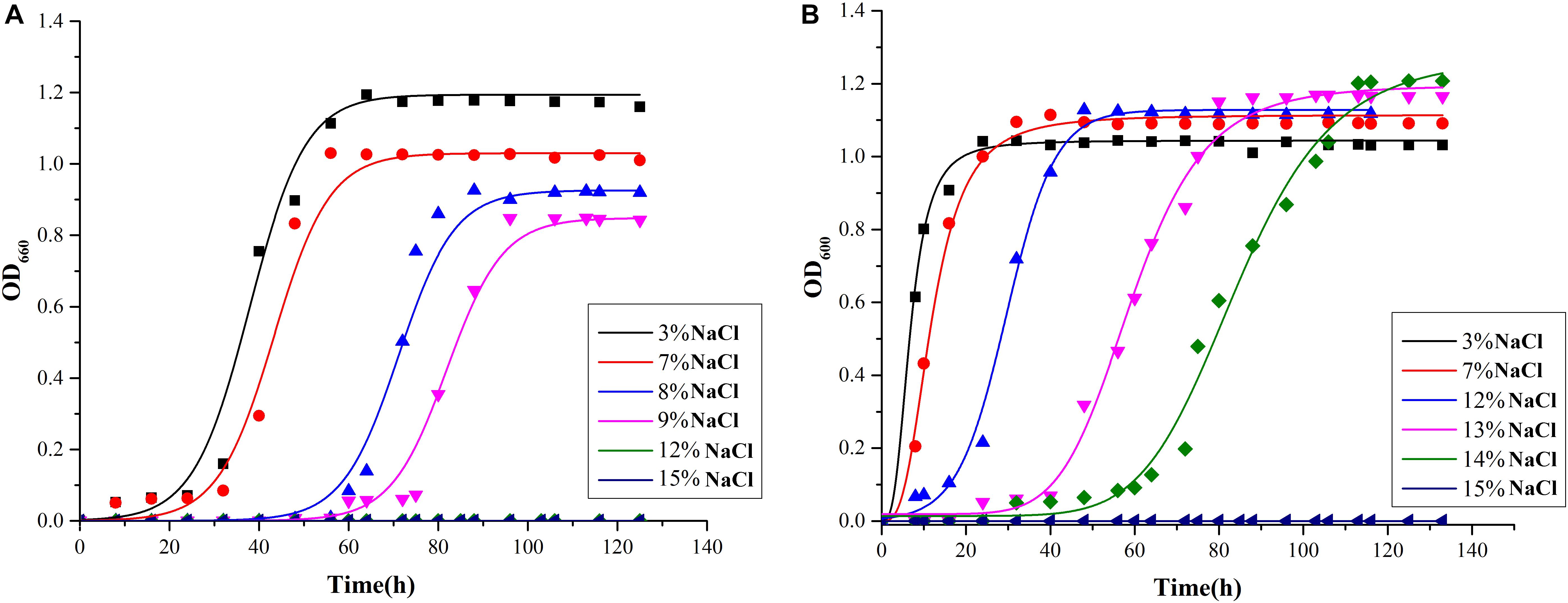
Figure 4. Growth curves of Mesonia sp. HuA40 (A) and Mesonia algae K4-1 (B) in different NaCl concentrations.
The moderate halophilic bacterium Mesonia algae K4-1 must have a highly efficient, salt-tolerance mechanism unique to Mesonia sp. HuA40. To further clarify the salt tolerance mechanism of Mesonia algae K4-1, we further analyzed and compared the adaptation mechanism of Mesonia algae K4-1 and Mesonia sp. HuA40 to the marine salt environment. Usually, halophilic microorganisms have two strategies for maintaining suitable osmotic pressure in the cytoplasm (Oren, 2008; Joghee and Jayaraman, 2016). The first strategy involves accumulation of inorganic ions (K+/Na+) of equal molar concentration in cells (Oren, 2013a). The intracellular proteins of bacteria relying on this strategy to maintain structural stability and physiological activity at high salt concentrations and exhibit remarkable instability at low salt concentrations (Lentzen and Schwarz, 2006). The proteomes of such bacteria exhibit a high degree of acidity, and microorganisms that normally rely on this strategy are obligate halophiles, such as Archaea (Siglioccolo et al., 2011). The second strategy is to synthesize or accumulate organic solutes such as secretin, glycine betaine, trehalose, amino acids and derivatives, which are osmotic substances that maintain the intracellular and extracellular osmotic balance and reduce the accumulation of intracellular salt (Welsh, 2000; Saum et al., 2013). Mesonia algae K4-1 has the glycine betaine transporter OpuD and the choline/glycine/valine betaine transporter BetT (driven by proton or sodium motive force, can also transport betaine or choline and, in some instances, proline), both of which are not annotated in Mesonia sp. HuA40. Glycine betaine is small in size and relatively low in energy (Gunasekera et al., 2008). Although glycine betaine is a relatively favorable organic solute, only a few archaea that live in a highly permeable environment can synthesize it. Additionally, one of the two methyltransferases that catalyze the glycine to betaine three-step reaction, encoded by the gsmT gene, was only found in the Mesonia algae K4-1 genome. Moreover, glycine/sarcosine N-methyltransferase (GSMT), a key protein for the biosynthesis of glycine betaine from glycine, allows the accumulation of sarcosine and is considered an osmotic protector (Nyyssola et al., 2000; Raiger Iustman et al., 2015). Secondary transporters of the betaine/choline/carnitine transporter family (BCCT) can also transport betaine or choline and, in some instances, proline (Ziegler et al., 2010). This indicated that Mesonia algae K4-1 can use an organic compatible solutes strategy to maintain its cell osmotic balance in the face of high osmotic pressure.
Bacteria that generally use this strategy require only a small amount of protein to adapt and have a wide range of salt ion tolerance (Roberts, 2005; Oren, 2013b). Therefore, we further compared the characteristics of intracellular and extracellular proteins between Mesonia algae K4-1 and Mesonia sp. HuA40. In Mesonia algae K4-1, the extracellular proteins with signal peptides had a higher ratio of acidic residues to basic residues than the intracellular proteins (Table 4). The predicted isoelectric point (pI) of the extracellular proteins with signal peptides was lower (p < 0.05) than that of the intracellular proteins without signal peptides (p < 0.05). Salinibacter ruber, which has been studied as an extreme halophilic bacterium, maintains cell osmotic balance by increasing the intracellular ion concentration in high salt environments (Oren, 2013a; Sanjukta et al., 2013). Both intracellular and extracellular proteins of S. ruber are tolerant to high salt environments, and the intracellular and extracellular pI was significantly lower than that of non-halophilic Escherichia coli (Qin et al., 2010). The salt-tolerance of the extracellular proteins of strain K4-1 and the genome annotation results indicated that Mesonia algae K4-1 can maintain its cell osmotic balance by using glycine betaine and proline, glutamate and other uptake systems. This strategy was also applied by the moderate halophilic strain Zunongwangia profunda SM-A87, which also contains a glycine betaine transporter (Qin et al., 2010). In addition, the pI of both intracellular and extracellular proteins of Mesonia algae K4-1 was lower than that of Mesonia sp. HuA40; the protein of the halophilic bacterium Mesonia algae K4-1 may have a certain preadaptation ability to a high salt environment. As previously demonstrated, the basic mechanism of salinity adaptation of Mesonia algae K4-1 is to prevent a large amount of inorganic salts from entering the cells and to use organic permeate to balance the high salinity of the environment. Further research at the transcriptional and translational levels is needed to elucidate the exact mechanism of salinity adaptation.
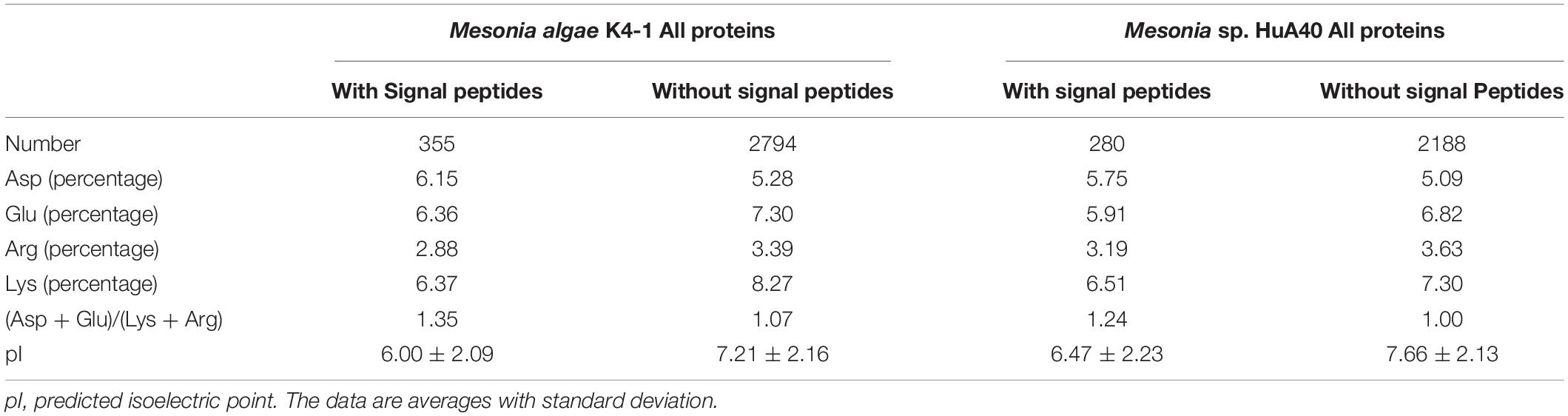
Table 4. Properties of intracellular and extracellular proteins of Mesonia algae K4-1 and Mesonia sp. HuA40.
Since the high salt environment is generally accompanied by a high alkali environment (Padan et al., 2005), to study the tolerance of the two strains to pH, we also measured the growth of the two strains under different pH conditions. The urease, arginine deaminase and arginine dehydrogenase systems (Ads), which help bacteria survive in acidic conditions by producing alkaline substances, were missing in Mesonia algae K4-1 and Mesonia sp. HuA40. This result may be related to the weak alkaline character of seawater (Liu et al., 2015). Intriguingly, Mesonia algae K4-1 has 12 genes encoding Na+/H+ antiporter proteins, including two genes encoding NhaB type Na+/H+ antiporter proteins, a gene encoding an NhaC type Na+/H+ antiporter protein, and three NhaP type Na+/H+ antiporter proteins. These antiporters play a crucial part in maintaining pH homeostasis and empowering salt tolerance (Herz et al., 2003; Klotz et al., 2006). Expect the single subunit Na+/H+ antiporter, Mesonia algae K4-1 contains the multisubunit Mrp-like Na+/H+ antiporter. As a hetero-oligomer, the Mrp system, broadly exists in the phylogenetic process of bacteria and archaea. The physiological effects of the Mrp system have been confirmed in alkaline tolerance, Na+ tolerance and K+ tolerance. Compared with Mesonia algae K4-1, Mesonia sp. HuA40 also encodes the Mrp-like Na+/H+ antiporter. The difference is that in addition to a gene encoding an NhaC type Na+/H+ reverse transport, Mesonia sp. HuA40 possesses a gene encoding an NhaD type Na+/H+ antiporter. This NhaD type Na+/H+ antiporter was nearly identical to that of the moderate halophilic bacterium Psychroflexus salaries and presented 82.4% identity and 95% similarity with the NhaD gene. Almost all halophilic microorganisms have the ability to expel Na+ from the interior of cells and maintain cytoplasmic pH homeostasis under alkaline conditions using Na+/H+ antiporters (Wang Y. et al., 2017). Analysis of these genes helps to understand the regulation of ion concentration and pH homeostasis under environmental change.
Cold Adaptation
Cold-adapted bacteria can be successfully grown under extreme conditions of cold marine environments because of various structural and physiological adjustments in the genome (Russo et al., 2010). Mesonia algae K4-1 was isolated from the Arctic, and our study found that it can thrive at 4°C (data not shown). However, Mesonia sp. HuA40 cannot grow at low temperature. To better understand the cold-adapted mechanism of the strain, we used genomic data to study and compare the traits of two strains, Mesonia algae K4-1 and Mesonia sp. HuA40, for cold adaptation (Table 3). When temperatures drop, bacteria must sense environmental changes and adjust their metabolism. Histidine kinases act as a multifunctional sensory to control numerous cold-responsive genes as well as many different stimuli (Sinetova and Los, 2016; Tang et al., 2019). However, in the Mesonia algae K4-1 genome, 30 genes encoding histidine kinases were found. Only 15 histidine kinases were encoded in the Mesonia sp. HuA40 genome. This result indicated that Mesonia algae K4-1 has a more powerful ability to respond to external stimuli. Glutathione synthase (gshB), a key enzyme for glutathione synthesis that has a key function in maintaining cell redox homeostasis, osmotic stress and protecting membrane lipids from the oxidative stress induced at cold temperatures (Loi et al., 2015; Mocali et al., 2017), was only encoded in the Mesonia algae K4-1 genome. This result suggests that Mesonia algae K4-1 faces high ROS concentrations, and glutathione likely helps Mesonia algae K4-1 to cope with oxidative stress in cold environments. When bacteria are exposed to cold stimulation, their growth will stagnate for a short time. This is because low temperature leads to the formation of more stable secondary structures in the process of translation, which inhibits initiation and extension. However, bacteria overcome growth stagnation by cold-induced RNA helicase, which contributes to the formation of cold-adapted ribosomes and RNA degradosomes, implying a role in unwinding the RNA secondary structure stabilization at low temperature (Owttrim, 2006, 2013). A total of 8 and 3 RNA helicases were annotated in Mesonia algae K4-1 and Mesonia sp. HuA40, respectively, suggesting that RNA helicase may play a role in the normal cellular function of Mesonia algae K4-1 at lower than optimal growth temperature. Furthermore, the organic compatible solutes of Mesonia algae K4-1 also encoded cryoprotection to maintain its cell normal physiological function at low temperatures (Wemekamp-Kamphuis et al., 2004). Some studies have reported that psychrophilic proteins are generally characterized by a higher degree of structural flexibility, lower thermostability, and higher specific activity (De Maayer et al., 2014). Therefore, we analyzed the amino acid composition ratios of different side chain sizes in Mesonia algae K4-1 and Mesonia sp. HuA40 proteins (Table 5). The amino acid composition of Mesonia algae K4-1 protein is characterized by more small residues such as Gly, Ser, Thr, and Asn and less large side chain amino acids, such as Arg, Leu, and Lys. The results are consistent with the amino acid composition of the cold-adapted proteins that are characterized by more small residues such as Gly and non-charged polar amino acids, particularly Gln and Thr, and less hydrophobic amino acids, particularly Leu, compared with their mesophilic counterparts (Yang et al., 2015).
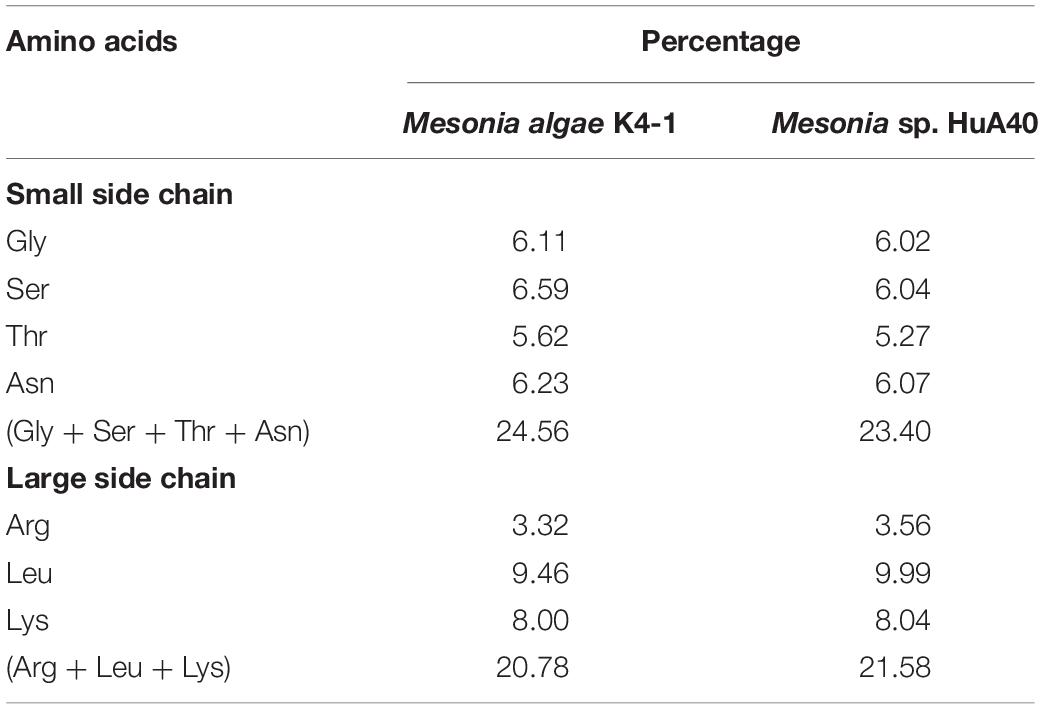
Table 5. Comparison of the side chain size between Mesonia algae K4-1 and Mesonia sp. HuA40 protein.
The growth curve shows that the optimum growth temperature of Mesonia algae K4-1 is close to 20°C, while that of Mesonia sp. HuA40 is close to 40°C (Figure 5). The growth temperature range of Mesonia algae K4-1 of 10–50°C is obviously wider than that of Mesonia sp. HuA40, which is 20–50°C. Moreover, Mesonia algae K4-1 can grow at a low temperature of 4°C, while Mesonia sp. HuA40 growth is stagnant at low temperature. These results suggested that Mesonia algae K4-1 has a stronger cold adaptation ability.
Polysaccharide Biosynthesis
The extracellular polysaccharides produced by many marine bacteria give them the ability to survive in extreme marine environments, helping bacteria concentrate organic matter, absorb metal ions (Zhang et al., 2017), form biofilms, and tolerate high acidity, UV radiation (Nicolaus et al., 2010), low temperatures and high salinities (Nichols et al., 2005; Kazak et al., 2010). To study the ecological role of Mesonia algae K4-1 exopolysaccharide in cold Arctic environments, the saccharide biosynthetic gene clusters were compared and analyzed (Figure 6). Using antiSMASH3.0.5 to predict secondary metabolite biosynthesis gene clusters, we found that the Mesonia algae K4-1 genome has 6 genes clusters related to saccharide biosynthesis, whereas Mesonia sp. HuA40 contains 5. Glycosyltransferase is the most important enzyme in the synthesis of polysaccharides (Cantarel et al., 2009; Liang et al., 2015). The number of glycosyltransferase types determines the number of monosaccharides in polysaccharide repeat units and the types of glycosidic bonds connected to each other. The genome annotation revealed that Mesonia algae K4-1 harbors 54 predicted glycosyl transferases, of which 12 belong to family two and 22 belong to family one. Among the 41 glycosyltransferase genes encoded by Mesonia sp. HuA40, 17 belong to the glycosyltransferase I family, and 10 belong to the glycosyltransferase II family. The discovery of polysaccharide exporter membrane proteins Wza, EpsA, etc., indicated that Mesonia algae K4-1 and Mesonia sp. HuA40 had the ability to secrete extracellular polysaccharides. After grown in fermentation culture for 4 days, the maximum EPS production of Mesonia algae K4-1 reached 9.86 mg/ml, while Mesonia sp. HuA40 was able to produce 7.96 mg/ml (Supplementary Figure S3). Our experimental results show that Mesonia algae K4-1 had higher extracellular polysaccharide production than Mesonia sp. HuA40. The extracellular polysaccharides of the Arctic sea ice bacterium Pseudoalteromonas sp. SM20310 could significantly enhance the high-salinity tolerance of SM20310 and improve the survival of SM20310 after freeze-thaw cycles, which enables the strain to adapt to a low temperature, high salinity sea ice environment (Liu et al., 2013). The large quantities of extracellular polysaccharides are beneficial to Mesonia algae K4-1 for growth in the Arctic ocean.
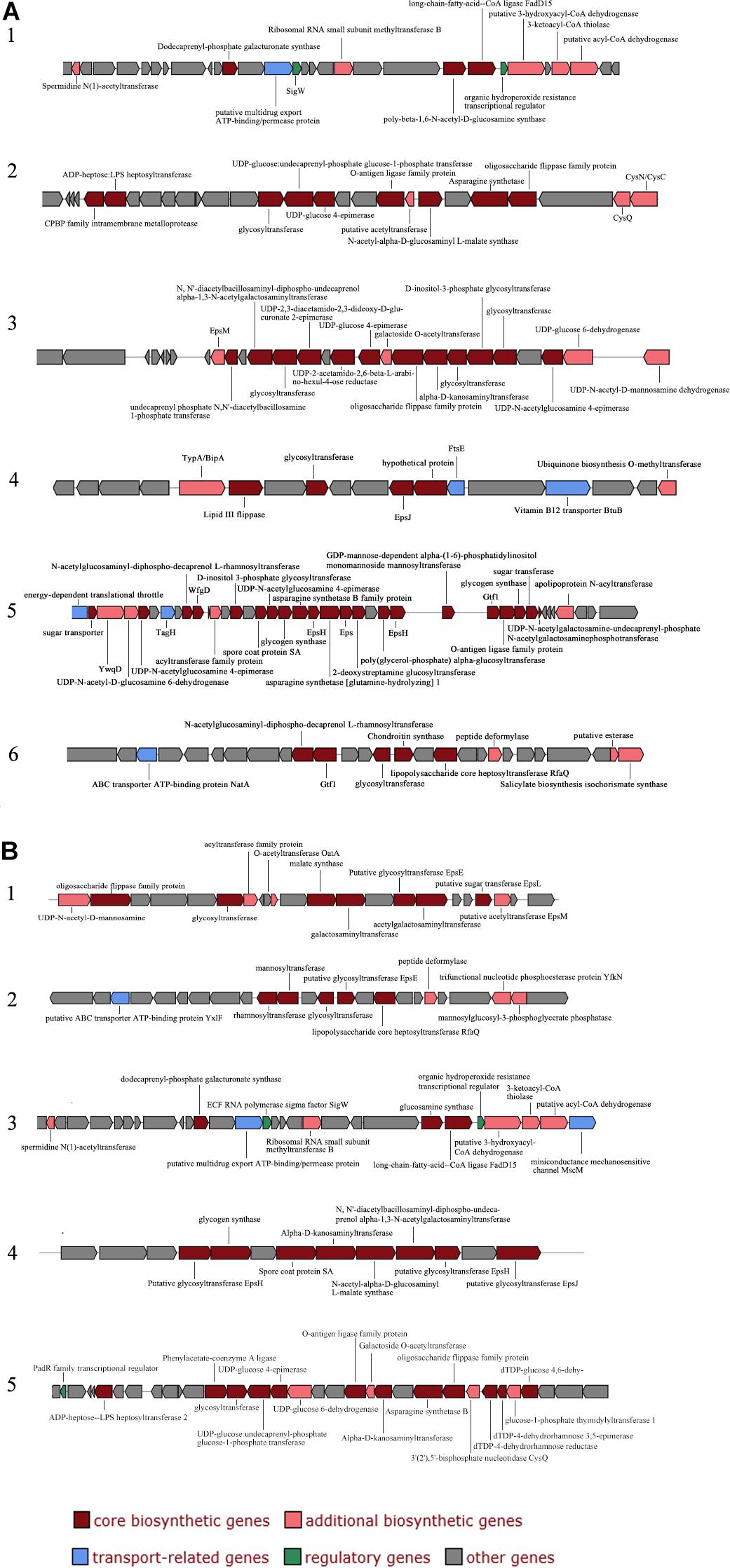
Figure 6. Organization of saccharide cluster structural genes in Mesonia algae K4-1 (A) and Mesonia sp. HuA40 genomes (B).
Proteolytic Capacity
Bacterial proteases are widespread enzymes that play important roles in cell viability, stress response and so on (Culp and Wright, 2017). They can catalyze the conversion of high molecular weight organics into small substrates to be transported into cells for further processing (Ran et al., 2014). To compare the hydrolysis ability of the extracellular proteins of the two strains, the two strains were subjected to fermentation culture, and the enzyme activity of the extracellular protein was measured (Supplementary Figure S4). Both strains reached maximum enzyme activity on the fourth day of fermentation, while the extracellular enzyme activity of Mesonia algae K4-1 was significantly higher than that of Mesonia sp. HuA40. Mesonia algae K4-1 contains 223 peptidases and Mesonia sp. HuA40 has 167. The extracellular peptidases encoded by the two strains were classified using the MEROPS peptidase database; the extracellular peptidases belong mainly to the family of metal peptidases and serine peptidases, as shown in Figure 7. All the extracellular peptidases of Mesonia algae K4-1 were classified into 22 different families, and 41 Mesonia sp. HuA40 peptidases with signal peptides could be assigned to 17 different families. Most of the enzymes of Mesonia algae K4-1 belongs to the M1 and S8 family. The diversity of the extracellular peptidase family and the high activity of extracellular proteases indicated that Mesonia algae K4-1 has a higher ability to degrade proteinaceous organic nitrogen and to acquire nutrients from the complex environment.
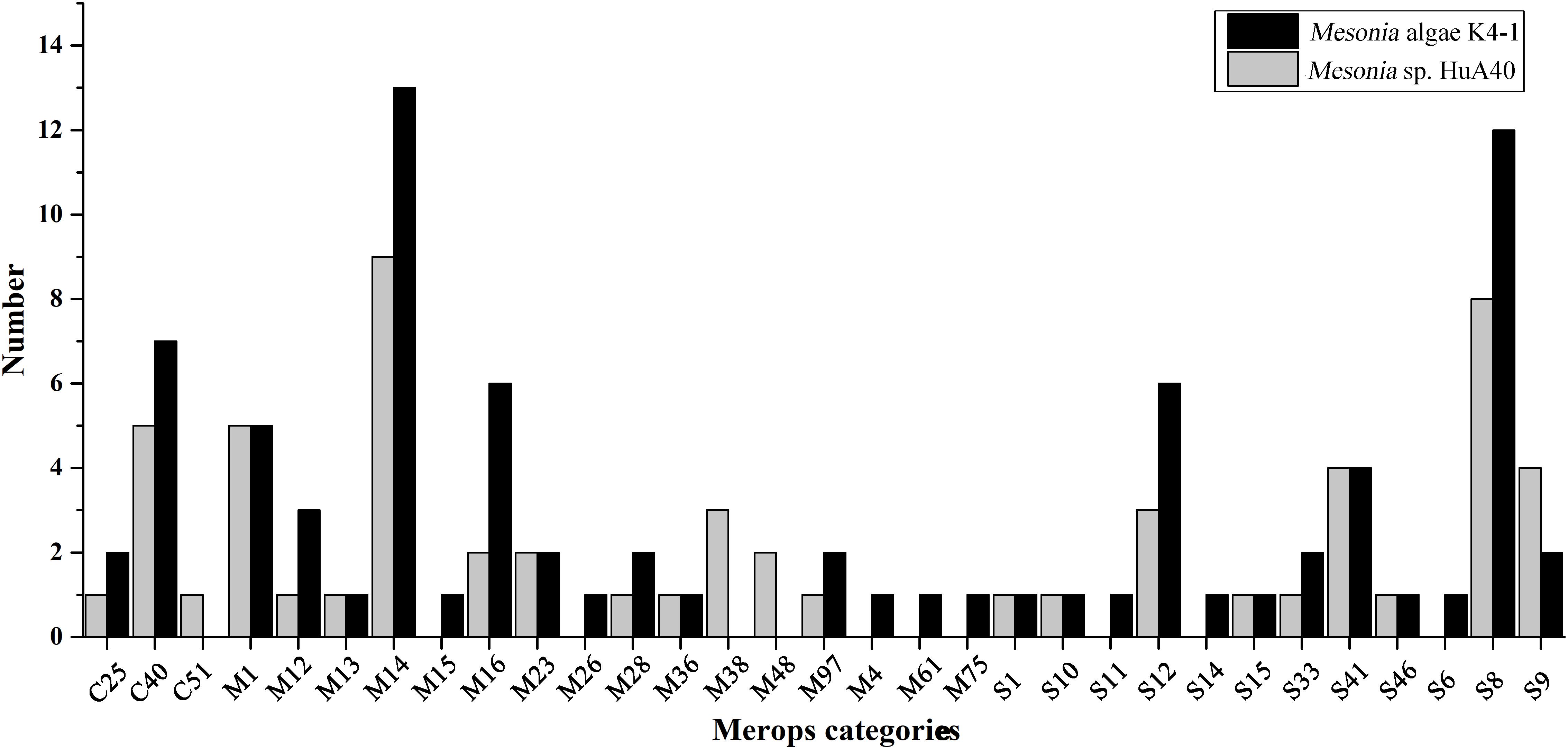
Figure 7. The MEROPS category of the extracellular peptidases from Mesonia algae K4-1 and Mesonia sp. HuA40.
Conclusion
This work presents the first complete genome of the genus Mesonia. The genome of the polar ocean bacterium Mesonia algae K4-1 was described and compared with the tropical sea-water bacterium Mesonia sp. HuA40. Our study found that Mesonia algae K4-1 had a larger genome and a higher proportion of genes involved in transcription and signal transduction, indicating that Mesonia algae K4-1 might have a more powerful ability to respond to various environmental stimuli. Strain Mesonia algae K4-1 can tolerate 3–14% NaCl and high pH conditions, while Mesonia sp. HuA40 can only grow under 3–9% NaCl and pH 6–8. Comparative genomic analysis revealed the salt tolerance mechanism of the moderately halophilic bacterium Mesonia algae K4-1 by synthesizing or accumulating organic solutes to maintain osmotic balance under high osmotic pressure. In addition, Mesonia algae K4-1 thrived at low temperature because Mesonia algae K4-1 harbored a variety of genes related to cold adaptation, such as RNA helicase, glutathione, and organic compatible solutes. In addition, we found that Mesonia algae K4-1 synthesized a large number of extracellular polysaccharides and secreted various families of extracellular proteases to degrade organic nitrogen, which might be related to the adaptation of Mesonia algae K4-1 to extremely oligotrophic environments. Our findings provide new insights into the genomic features of Mesonia and the adaptive characteristics of different microorganisms in extreme environments.
Data Availability Statement
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession VRLS00000000 (Mesonia algae K4-1) and VRLT00000000 (Mesonia sp. HuA40). The version described in this article is XXXX01000000. The genomic data for this study are available at https://www.ncbi.nlm.nih.gov/Traces/wgs/VRLS01?display=contigs, https://www.ncbi.nlm.nih.gov/Traces/wgs/VRLT01?display=contigs.
Author Contributions
RH performed the experiments and prepared the manuscript draft. HH, CY, and DX conceived and designed the experiments. HH and JH helped to revise the manuscript. DL and MW assisted in the formatting of the figures. CL and YZ assisted in data analysis. All authors read and agreed to the final content.
Funding
This work was supported by the National Natural Science Foundation of China (31370104 and 31771899), the Natural Science Foundation of Hunan Province, China (2018JJ2497), and the Fundamental Research Funds for the Central Universities of Central South University (2018zzts392 and 2018gczd041).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank professor Yu-Zhong Zhang (State Key Laboratory of Microbial Technology, Shandong University, China) for support and assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02812/full#supplementary-material
Footnotes
References
Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: signalP 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
Bentkowski, P., Van Oosterhout, C., and Mock, T. (2015). A model of genome size evolution for prokaryotes in stable and fluctuating environments. Genome Biol. Evol. 7, 2344–2351. doi: 10.1093/gbe/evv148
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Caputo, A., Fournier, P. E., and Raoult, D. (2019). Genome and pan-genome analysis to classify emerging bacteria. Biol. Direct 14:5. doi: 10.1186/s13062-019-0234-0
Chen, D. S., Wu, Y. Q., Zhang, W., Jiang, S. J., and Chen, S. Z. (2016). Horizontal gene transfer events reshape the global landscape of arm race between viruses and homo sapiens. Sci. Rep. 6:26934. doi: 10.1038/srep26934
Chen, P. E., Cook, C., Stewart, A. C., Nagarajan, N., Sommer, D. D., Pop, M., et al. (2010). Genomic characterization of the Yersinia genus. Genome Biol. 11:R1. doi: 10.1186/gb-2010-11-1-r1
Choi, A., Baek, K., Lee, H., and Cho, J. C. (2015). Mesonia aquimarina sp. nov., a marine bacterium isolated from coastal seawater. Int. J. Syst. Evol. Microbiol. 65(Pt 1), 135–140. doi: 10.1099/ijs.0.069336-0
Collins, R. E., Rocap, G., and Deming, J. W. (2010). Persistence of bacterial and archaeal communities in sea ice through an Arctic winter. Environ. Microbiol. 12, 1828–1841. doi: 10.1111/j.1462-2920.2010.02179.x
Culp, E., and Wright, G. D. (2017). Bacterial proteases, untapped antimicrobial drug targets. J. Antibiot. 70, 366–377. doi: 10.1038/ja.2016.138
D’Amico, S., Collins, T., Marx, J. C., Feller, G., and Gerday, C. (2006). Psychrophilic microorganisms: challenges for life. EMBO Rep. 7, 385–389. doi: 10.1038/sj.embor.7400662
Daubin, V., Moran, N. A., and Ochman, H. (2003). Phylogenetics and the cohesion of bacterial genomes. Science 301, 829–832. doi: 10.1126/science.1086568
De Maayer, P., Anderson, D., Cary, C., and Cowan, D. A. (2014). Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 15, 508–517. doi: 10.1002/embr.201338170
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Descorps-Declere, S., Lemoine, F., Sculo, Q., Lespinet, O., and Labedan, B. (2008). The multiple facets of homology and their use in comparative genomics to study the evolution of genes, genomes, and species. Biochimie 90, 595–608. doi: 10.1016/j.biochi.2007.09.010
Fischer, S., Brunk, B. P., Chen, F., Gao, X., Harb, O. S., Iodice, J. B., et al. (2011). Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinform. Chapter 6, Unit 6, 11–19. doi: 10.1002/0471250953.bi0612s35
Gerhardt, P. (1994). Methods for General and Molecular Bacteriology. Washington, DC: American Society for Microbiology.
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57(Pt 1), 81–91. doi: 10.1099/ijs.0.64483-0
Gunasekera, T. S., Csonka, L. N., and Paliy, O. (2008). Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J. Bacteriol. 190, 3712–3720. doi: 10.1128/jb.01990-07
Gweon, H. S., Bailey, M. J., and Read, D. S. (2017). Assessment of the bimodality in the distribution of bacterial genome sizes. ISME J. 11, 821–824. doi: 10.1038/ismej.2016.142
Hall, J. P. J., Brockhurst, M. A., and Harrison, E. (2017). Sampling the mobile gene pool: innovation via horizontal gene transfer in bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20160424. doi: 10.1098/rstb.2016.0424
He, H., Chen, X., Li, J., Zhang, Y., and Gao, P. (2004). Taste improvement of refrigerated meat treated with cold-adapted Protease. Food Chem. 84, 307–311. doi: 10.1016/s0308-8146(03)00242-5
Herz, K., Vimont, S., Padan, E., and Berche, P. (2003). Roles of NhaA, NhaB, and NhaD Na+/H+ antiporters in survival of Vibrio cholerae in a saline environment. J. Bacteriol. 185, 1236–1244. doi: 10.1128/jb.185.4.1236-1244.2003
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2016). eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. doi: 10.1093/nar/gkv1248
Husnik, F., and McCutcheon, J. P. (2018). Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 16, 67–79. doi: 10.1038/nrmicro.2017.137
Jeong, S., Vollprecht, R., Cho, K., Leiknes, T., Vigneswaran, S., Bae, H., et al. (2016). Advanced organic and biological analysis of dual media filtration used as a pretreatment in a full-scale seawater desalination plant. Desalination 385, 83–92. doi: 10.1016/j.desal.2016.02.017
Joghee, N. N., and Jayaraman, G. (2016). Biochemical changes induced by salt stress in halotolerant bacterial isolates are media dependent as well as species specific. Prep. Biochem. Biotechnol. 46, 8–14. doi: 10.1080/10826068.2014.970689
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kang, H. S., and Lee, S. D. (2010). Mesonia phycicola sp. nov., isolated from seaweed, and emended description of the genus Mesonia. Int. J. Syst. Evol. Microbiol. 60(Pt 3), 591–594. doi: 10.1099/ijs.0.011338-0
Kazak, H., Öner, E. T., and Dekker, R. F. H. (2010). “Extremophiles as sources of exopolysaccharides,” in Handbook of Carbohydrate Polymers, eds R. Ito and Y. Matsuo (New York, NY: Nova Science Publishers, Inc), 605–619.
Klotz, M. G., Arp, D. J., Chain, P. S., El-Sheikh, A. F., Hauser, L. J., Hommes, N. G., et al. (2006). Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium Nitrosococcus oceani ATCC 19707. Appl. Environ. Microbiol. 72, 6299–6315. doi: 10.1128/AEM.00463-06
Kolberg, J., Busse, H. J., Wilke, T., Schubert, P., Kampfer, P., and Glaeser, S. P. (2015). Mesonia hippocampi sp. nov., isolated from the brood pouch of a diseased Barbour’s Seahorse (Hippocampus barbouri). Int. J. Syst. Evol. Microbiol. 65, 2241–2247. doi: 10.1099/ijs.0.000245
Konstantinidis, K. T., Serres, M. H., Romine, M. F., Rodrigues, J. L., Auchtung, J., McCue, L. A., et al. (2009). Comparative systems biology across an evolutionary gradient within the Shewanella genus. Proc. Natl. Acad. Sci. U.S.A. 106, 15909–15914. doi: 10.1073/pnas.0902000106
Konstantinidis, K. T., and Tiedje, J. M. (2004). Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. U.S.A. 101, 3160–3165. doi: 10.1073/pnas.0308653100
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lagesen, K., Hallin, P., Rodland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lee, S. Y., Lee, M. H., and Yoon, J. H. (2012). Mesonia ostreae sp. nov., isolated from seawater of an oyster farm, and emended description of the genus Mesonia. Int. J. Syst. Evol. Microbiol. 62(Pt 8), 1804–1808. doi: 10.1099/ijs.0.033175-0
Lentzen, G., and Schwarz, T. (2006). Extremolytes: natural compounds from extremophiles for versatile applications. Appl. Microbiol. Biotechnol. 72, 623–634. doi: 10.1007/s00253-006-0553-9
Liang, D. M., Liu, J. H., Wu, H., Wang, B. B., Zhu, H. J., and Qiao, J. J. (2015). Glycosyltransferases: mechanisms and applications in natural product development. Chem. Soc. Rev. 44, 8350–8374. doi: 10.1039/c5cs00600g
Liu, L., Si, L., Meng, X., and Luo, L. (2015). Comparative transcriptomic analysis reveals novel genes and regulatory mechanisms of Tetragenococcus halophilus in response to salt stress. J. Ind. Microbiol. Biotechnol. 42, 601–616. doi: 10.1007/s10295-014-1579-0
Liu, S. B., Chen, X. L., He, H. L., Zhang, X. Y., Xie, B. B., Yu, Y., et al. (2013). Structure and ecological roles of a novel exopolysaccharide from the arctic sea ice bacterium Pseudoalteromonas sp. Strain SM20310. Appl. Environ. Microbiol. 79, 224–230. doi: 10.1128/AEM.01801-12
Loi, V. V., Rossius, M., and Antelmann, H. (2015). Redox regulation by reversible protein S-thiolation in bacteria. Front. Microbiol. 6:187. doi: 10.3389/fmicb.2015.00187
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Magrane, M., and UniProt, C. (2011). UniProt knowledgebase: a hub of integrated protein data. Database 2011:bar009. doi: 10.1093/database/bar009
Masuko, T., Minami, A., Iwasaki, N., Majima, T., Nishimura, S., and Lee, Y. C. (2005). Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 339, 69–72. doi: 10.1016/j.ab.2004.12.001
Miranda, C. D., Rojas, R., Geisse, J., Romero, J., and Gonzalez-Rocha, G. (2015). Scallop larvae hatcheries as source of bacteria carrying genes encoding for non-enzymatic phenicol resistance. Mar. Pollut. Bull. 95, 173–182. doi: 10.1016/j.marpolbul.2015.04.026
Mocali, S., Chiellini, C., Fabiani, A., Decuzzi, S., de Pascale, D., Parrilli, E., et al. (2017). Ecology of cold environments: new insights of bacterial metabolic adaptation through an integrated genomic-phenomic approach. Sci. Rep. 7:839. doi: 10.1038/s41598-017-00876-4
Nedashkovskaya, O. I., Kim, S. B., Han, S. K., Lysenko, A. M., Rohde, M., Zhukova, N. V., et al. (2003). Mesonia algae gen. nov., sp. nov., a novel marine bacterium of the family Flavobacteriaceae isolated from the green alga Acrosiphonia sonderi (Kutz) Kornm. Int. J. Syst. Evol. Microbiol. 53(Pt 6), 1967–1971. doi: 10.1099/ijs.0.02626-0
Nedashkovskaya, O. I., Kim, S. B., Zhukova, N. V., Kwak, J., Mikhailov, V. V., and Bae, K. S. (2006). Mesonia mobilis sp. nov., isolated from seawater, and emended description of the genus Mesonia. Int. J. Syst. Evol. Microbiol. 56(Pt 10), 2433–2436. doi: 10.1099/ijs.0.64376-0
Nichols, C. A., Guezennec, J., and Bowman, J. P. (2005). Bacterial exopolysaccharides from extreme marine environments with special consideration of the southern ocean, sea ice, and deep-sea hydrothermal vents: a review. Mar. Biotechnol. 7, 253–271. doi: 10.1007/s10126-004-5118-2
Nicolaus, B., Kambourova, M., and Oner, E. T. (2010). Exopolysaccharides from extremophiles: from fundamentals to biotechnology. Environ. Technol. 31, 1145–1158. doi: 10.1080/09593330903552094
Nyyssola, A., Kerovuo, J., Kaukinen, P., von Weymarn, N., and Reinikainen, T. (2000). Extreme halophiles synthesize betaine from glycine by methylation. J. Biol. Chem. 275, 22196–22201. doi: 10.1074/jbc.M910111199
Omae, K., Yoneda, Y., Fukuyama, Y., Yoshida, T., and Sako, Y. (2017). Genomic analysis of Calderihabitans maritimus KKC1, a thermophilic, hydrogenogenic, carboxydotrophic bacterium isolated from marine sediment. Appl. Environ. Microbiol. 83:e00832-17. doi: 10.1128/AEM.00832-17
Oren, A. (2008). Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 4:2. doi: 10.1186/1746-1448-4-2
Oren, A. (2013a). Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front. Microbiol. 4:315. doi: 10.3389/fmicb.2013.00315
Oren, A. (2013b). Salinibacter: an extremely halophilic bacterium with archaeal properties. FEMS Microbiol. Lett. 342, 1–9. doi: 10.1111/1574-6968.12094
Owttrim, G. W. (2006). RNA helicases and abiotic stress. Nucleic Acids Res. 34, 3220–3230. doi: 10.1093/nar/gkl408
Owttrim, G. W. (2013). RNA helicases: diverse roles in prokaryotic response to abiotic stress. RNA Biol. 10, 96–110. doi: 10.4161/rna.22638
Padan, E., Bibi, E., Ito, M., and Krulwich, T. A. (2005). Alkaline pH homeostasis in bacteria: new insights. Biochim. Biophys. Acta 1717, 67–88. doi: 10.1016/j.bbamem.2005.09.010
Pittera, J., Jouhet, J., Breton, S., Garczarek, L., Partensky, F., Marechal, E., et al. (2018). Thermoacclimation and genome adaptation of the membrane lipidome in marine Synechococcus. Environ. Microbiol. 20, 612–631. doi: 10.1111/1462-2920.13985
Qin, Q. L., Xie, B. B., Yu, Y., Shu, Y. L., Rong, J. C., Zhang, Y. J., et al. (2014). Comparative genomics of the marine bacterial genus Glaciecola reveals the high degree of genomic diversity and genomic characteristic for cold adaptation. Environ. Microbiol. 16, 1642–1653. doi: 10.1111/1462-2920.12318
Qin, Q. L., Zhang, X. Y., Wang, X. M., Liu, G. M., Chen, X. L., Xie, B. B., et al. (2010). The complete genome of Zunongwangia profunda SM-A87 reveals its adaptation to the deep-sea environment and ecological role in sedimentary organic nitrogen degradation. BMC Genomics 11:247. doi: 10.1186/1471-2164-11-247
Raiger Iustman, L. J., Tribelli, P. M., Ibarra, J. G., Catone, M. V., Solar Venero, E. C., and Lopez, N. I. (2015). Genome sequence analysis of Pseudomonas extremaustralis provides new insights into environmental adaptability and extreme conditions resistance. Extremophiles 19, 207–220. doi: 10.1007/s00792-014-0700-7
Ran, L. Y., Su, H. N., Zhou, M. Y., Wang, L., Chen, X. L., Xie, B. B., et al. (2014). Characterization of a novel subtilisin-like protease myroicolsin from deep sea bacterium Myroides profundi D25 and molecular insight into its collagenolytic mechanism. J. Biol. Chem. 289, 6041–6053. doi: 10.1074/jbc.M113.513861
Rawlings, N. D., Waller, M., Barrett, A. J., and Bateman, A. (2014). MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, D503–D509. doi: 10.1093/nar/gkt953
Ricciardelli, A., Casillo, A., Vergara, A., Balasco, N., Corsaro, M. M., Tutino, M. L., et al. (2019). Environmental conditions shape the biofilm of the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Microbiol. Res. 218, 66–75. doi: 10.1016/j.micres.2018.09.010
Roberts, M. F. (2005). Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 1:5. doi: 10.1186/1746-1448-1-5
Russo, R., Giordano, D., Riccio, A., di Prisco, G., and Verde, C. (2010). Cold-adapted bacteria and the globin case study in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Mar. Genomics 3, 125–131. doi: 10.1016/j.margen.2010.09.001
Sanjukta, R. K., Farooqi, M. S., Rai, N., Rai, A., Sharma, N., Mishra, D. C., et al. (2013). Expression analysis of genes responsible for amino acid biosynthesis in halophilic bacterium Salinibacter ruber. Indian J. Biochem. Biophys. 50, 177–185.
Saum, S. H., Pfeiffer, F., Palm, P., Rampp, M., Schuster, S. C., Muller, V., et al. (2013). Chloride and organic osmolytes: a hybrid strategy to cope with elevated salinities by the moderately halophilic, chloride-dependent bacterium Halobacillus halophilus. Environ. Microbiol. 15, 1619–1633. doi: 10.1111/j.1462-2920.2012.02770.x
Siglioccolo, A., Paiardini, A., Piscitelli, M., and Pascarella, S. (2011). Structural adaptation of extreme halophilic proteins through decrease of conserved hydrophobic contact surface. BMC Struct. Biol. 11:50. doi: 10.1186/1472-6807-11-50
Sinetova, M. A., and Los, D. A. (2016). New insights in cyanobacterial cold stress responses: genes, sensors, and molecular triggers. Biochim. Biophys. Acta 1860(11 Pt A), 2391–2403. doi: 10.1016/j.bbagen.2016.07.006
Sung, H. R., Joh, K., and Shin, K. S. (2017). Mesonia maritima sp. nov., isolated from seawater of the South Sea of Korea. Int. J. Syst. Evol. Microbiol. 67, 2574–2580. doi: 10.1099/ijsem.0.001974
Tang, J., Du, L. M., Liang, Y. M., and Daroch, M. (2019). Complete genome sequence and comparative analysis of Synechococcus sp. CS-601 (SynAce01), a cold-adapted cyanobacterium from an oligotrophic antarctic habitat. Int. J. Mol. Sci. 20:E152. doi: 10.3390/ijms20010152
Thomas, D. N., and Dieckmann, G. S. (2002). Antarctic Sea ice–a habitat for extremophiles. Science 295, 641–644. doi: 10.1126/science.1063391
Urvantseva, A. M., Bakunina, I. Y., Nedashkovskaya, O. I., Kim, S. B., and Zvyagintseva, T. N. (2006). Distribution of intracellular fucoidan hydrolases among marine bacteria of the family Flavobacteriaceae. Appl. Biochem. Microbiol. 42, 484–491. doi: 10.1134/s0003683806050073
Van Trappen, S., Tan, T. L., Yang, J., Mergaert, J., and Swings, J. (2004). Glaciecola polaris sp. nov., a novel budding and prosthecate bacterium from the Arctic Ocean, and emended description of the genus Glaciecola. Int. J. Syst. Evol. Microbiol. 54(Pt 5), 1765–1771. doi: 10.1099/ijs.0.63123-0
Wang, F. Q., Xie, Z. H., Zhao, J. X., Chen, G. J., and Du, Z. J. (2015). Mesonia sediminis sp. nov., isolated from a sea cucumber culture pond. Antonie Van Leeuwenhoek 108, 1205–1212. doi: 10.1007/s10482-015-0575-3
Wang, L., Wang, J., and Jing, C. (2017). Comparative genomic analysis reveals organization, function and evolution of ars genes in Pantoea spp. Front. Microbiol. 8:471. doi: 10.3389/fmicb.2017.00471
Wang, Y., Song, N., Yang, L., Abdel-Motaal, H., Zhang, R., Zhang, Z., et al. (2017). A novel NhaD-type Na(+)/H(+) antiporter from the moderate halophile and alkaliphile Halomonas alkaliphila. Can. J. Microbiol. 63, 596–607. doi: 10.1139/cjm-2017-0104
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H. U., Bruccoleri, R., et al. (2015). antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. doi: 10.1093/nar/gkv437
Welsh, D. T. (2000). Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol. Rev. 24, 263–290. doi: 10.1111/j.1574-6976.2000.tb00542.x
Wemekamp-Kamphuis, H. H., Sleator, R. D., Wouters, J. A., Hill, C., and Abee, T. (2004). Molecular and physiological analysis of the role of osmolyte transporters BetL, Gbu, and OpuC in growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 70, 2912–2918. doi: 10.1128/aem.70.5.2912-2918.2004
Wu, C., Liu, D., Yang, X., Wu, R., Zhang, J., Huang, J., et al. (2016). Improving production of protease from Pseudoalteromonas sp. CSN423 by random mutagenesis. Mar. Biotechnol. 18, 610–618. doi: 10.1007/s10126-016-9721-9
Yang, L. L., Tang, S. K., Huang, Y., and Zhi, X. Y. (2015). Low temperature adaptation is not the opposite process of high temperature adaptation in terms of changes in amino acid composition. Genome Biol. Evol. 7, 3426–3433. doi: 10.1093/gbe/evv232
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1–6. doi: 10.1007/s10482-017-0844-4
Yu, Z. C., Chen, X. L., Shen, Q. T., Zhao, D. L., Tang, B. L., Su, H. N., et al. (2015). Filamentous phages prevalent in Pseudoalteromonas spp. confer properties advantageous to host survival in Arctic sea ice. ISME J. 9, 871–881. doi: 10.1038/ismej.2014.185
Yukimura, K., Nakai, R., Kohshima, S., Uetake, J., Kanda, H., and Naganuma, T. (2010). Spore-forming halophilic bacteria isolated from Arctic terrains: implications for long-range transportation of microorganisms. Polar Sci. 3, 163–169. doi: 10.1016/j.polar.2009.07.002
Zhang, Z., Cai, R., Zhang, W., Fu, Y., and Jiao, N. (2017). A novel exopolysaccharide with metal adsorption capacity produced by a marine bacterium Alteromonas sp. JL2810. Mar. Drugs 15:E175. doi: 10.3390/md15060175
Keywords: Mesonia, complete genome, environmental adaptation, the Arctic Ocean, salt tolerance
Citation: Huan R, Huang J, Liu D, Wang M, Liu C, Zhang Y, Yi C, Xiao D and He H (2019) Genome Sequencing of Mesonia algae K4-1 Reveals Its Adaptation to the Arctic Ocean. Front. Microbiol. 10:2812. doi: 10.3389/fmicb.2019.02812
Received: 07 September 2019; Accepted: 20 November 2019;
Published: 04 December 2019.
Edited by:
Brian D. Lanoil, University of Alberta, CanadaCopyright © 2019 Huan, Huang, Liu, Wang, Liu, Zhang, Yi, Xiao and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: HaiLun He, aGVsZW5oZUBjc3UuZWR1LmNu
 Ran Huan1
Ran Huan1 HaiLun He
HaiLun He