- 1State Key Laboratory of Chemical Biology and Drug Discovery, The Hong Kong Polytechnic University, Kowloon, Hong Kong
- 2Department of Infectious Diseases and Public Health, Jockey Club College of Veterinary Medicine and Life Sciences, City University of Hong Kong, Kowloon, Hong Kong
Acinetobacter baumannii is an important clinical pathogen which often causes fatal infections among seriously ill patients. Treatment options for managing infections caused by this organism have become limited as a result of emergence of carbapenem resistant strains. In the current study, whole genome sequencing, gene expression studies and enzyme kinetics analyses were performed to investigate the underlying carbapenem resistance mechanisms in fourteen clinical A. baumannii strains isolated from two hospitals, one each in Hong Kong and Henan Province, People’s Republic of China. A large majority of the A. baumannii strains (11/14) were found to belong to the International Clone II (IC-II), among which six were ST208. Twelve of these strains were carbapenem resistant and found to either harbor blaOXA–23/blaOXA–72, or exhibit over-expression of the blaOXA–51 gene upon ISAba1 insertion. Enzymatic assay confirmed that the OXA variants, including those of blaOXA–51, exhibited strong carbapenem-degrading activities. In terms of other intrinsic mechanisms, a weak correlation was observed between reduced production of outer membrane porin CarO/expression resistance-nodulation-division (RND) efflux AdeB and phenotypic resistance. This finding implied that over-production of carbapenem-hydrolyzing-class D-ß-lactamases (CHDLs), including the intrinsic blaOXA–51 gene and the acquired blaOXA–23 and blaOXA–24 elements, is the key mechanism of carbapenem resistance in A. baumannii. This view is confirmed by testing the effect of NaCl, a known blaOXA inhibitor, which was found to cause reduction in carbapenem MIC by twofolds to eightfolds, suggesting that inhibiting OXA type carbapenemases represents the most effective strategy to control phenotypic carbapenem resistance in A. baumannii.
Introduction
Acinetobacter baumannii is an important Gram-negative pathogen that often causes serious hospital infections, especially among immunocompromised patients in intensive care units (ICUs) (Bergogne-Berezin and Towner, 1996). The increasing mortality due to A. baumannii infections is of major concern as this pathogen exhibits the potential to evolve into carbapenem resistant variants through acquiring antibiotic resistance-encoding mobile genetic elements, which is often exacerbated by the intrinsic low membrane permeability of this organism. These features render A. baumannii one of the bacterial pathogens that exhibits the highest resistance rate in clinical settings (Peleg et al., 2008). In 2013, the United States Center for Disease Control and Prevention estimated that as many as 11,500 A. baumannii infections occurred annually, among which 63% were multidrug resistant, resulting in 500 deaths (Queenan et al., 2012). Likewise, A. baumannii is responsible for more than 1/5 of all clinical Gram-negative bacterial infections in Hong Kong and other Asia-Pacific regions, with a high portion being multidrug resistant (Liu et al., 2012). Recently, the World Health Organization has listed carbapenem-resistant A. baumannii to be “Priority 1: Critical” in its “Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery and Development of New Antibiotics,” further highlighting the worsen situation caused by this pathogen (World Health Organisation, 2017).
Carbapenem resistance in A. baumannii has been attributed to intrinsic cellular mechanisms, including loss of outer membrane porins (OMP) and over-expression of efflux pumps, which could result in alteration of cytoplasmic antimicrobial drug concentration and hence its bactericidal effect (Magnet et al., 2001; Siroy et al., 2005). Several OMPs, including CarO, HMP-AB and OmpW, were found to be involved in transportation of β-lactams across cytoplasmic membrane of this bacterial pathogen (Gribun et al., 2003; Siroy et al., 2006). While OMPs are responsible for the uptake of antibiotics, the multi-drug efflux systems are believed to be involved in removal of drugs by pumping them out of the cell. In particular, the resistance-nodulation-division (RND) type efflux pumps, have long been hypothesized to play a role in rendering resistance toward various antibiotics. In A. baumannii, the most extensively studied RND efflux system is the adeABC gene product, which exhibits substrate specificity toward various β-lactams, including fluoroquinolones, aminoglycosides, tetracyclines and chloramphenicol (Higgins et al., 2004). Nevertheless, evidence confirming a direct linkage between carbapenem susceptibility and the presence/absence of these porin proteins and efflux systems in A. baumannii is currently not available.
Enzymatic mechanisms have been regarded as the key factors that mediate development of carbapenem resistance in Gram negative bacteria, including A. baumannii. Instead of blaIMP, blaVIM and blaNDM which are commonly identified in other bacterial pathogens, the carbapenem-hydrolyzing-class-D β-lactamases (CHDLs) are regarded as key determinants underlying the emergence of carbapenem-resistant A. baumannii (Poirel and Nordmann, 2006). CHDLs denote the OXA-type β-lactamases which exhibit carbapenem hydrolyzing activity. There are various types of blaOXA genes which are known to be harbored by A. baumannii, including blaOXA–51, blaOXA–23, blaOXA–24/40, blaOXA–58, blaOXA–143, and blaOXA–235 (Higgins et al., 2013; Evans and Amyes, 2014). A considerable number of studies have been conducted on enzymes encoded by these resistance genes due to their uniqueness in A. baumannii, particularly the blaOXA–51-like β-lactamases, the genetic determinant of which is inherent in A. baumannii chromosome and can be readily overexpressed as a result of promoter activation by insertion sequences such as ISAba1 (Turton et al., 2006). Apart from this chromosomal resistance gene, plasmid-borne blaOXA–23-like, blaOXA–24/40-like, and blaOXA–58-like elements are also frequently identified in resistant isolates. Among them, blaOXA–23 is the most prevalent CHDL-encoding element in carbapenem-resistant A. baumannii worldwide (Mugnier et al., 2010). A previous study in China reported that 96.5% of carbapenem-resistant A. baumannii isolates carried blaOXA–23-like elements, and that over 96% of those isolates belonged to the Clonal Complex CC92 (Ji et al., 2014). In Hong Kong, the majority of carbapenem-resistant A. baumannii were blaOXA–23-like carrier that belonged to sequence type ST26 (Ho et al., 2010). Similarly, dissemination of blaOXA–23-bearing A. baumannii was also observed in other Asian countries, including Taiwan, Japan, and Korea (Peleg et al., 2008).
A comprehensive study was performed in 2013 to investigate the interplay between intrinsic and extrinsic mechanisms in mediating development of antimicrobial resistance in A. baumannii, including efflux systems, membrane porins, and production of CHDL β-lactamases (Rumbo et al., 2013). The study concluded that the blaOXA–24 and blaOXA–58 are the major determinants of carbapenem resistance in this organism, and that blaOXA–51 and porin proteins were not involved in antimicrobial resistance of blaOXA–24 and blaOXA–58-carrying isolates (Rumbo et al., 2013). Nevertheless, the exact role of intrinsic mechanisms in mediating carbapenem resistant phenotypes in A. baumannii strains carrying blaOXA–23 and blaOXA–51 remained unclear. In this work, we showed that intrinsic resistance mechanisms including RND-mediated efflux and reduced expression of porin proteins did not play a major role in mediating onset of carbapenem resistance in A. baumannii. Instead, the phenotype was mainly conferred by CHDL encoded by the blaOXA–23, blaOXA–72, and blaOXA–51-like genes. In addition, we identified several variants of blaOXA–51, which exhibited carbapenem-hydrolyzing activities that resemble those encoded by blaOXA–23 in terms of substrate profile, as over-expression of these blaOXA–51 variants in A. baumannii upon insertional activation by ISAba1 conferred the host strain a carbapenem resistant phenotype identical to blaOXA–23-carrying strains.
Materials and Methods
Bacterial Isolates
A total of 14 representative Acinetobacter baumannii clinical strains were first included in the genome sequencing, gene expression study, and western blot analysis as described below. The strains were isolated from patients of two hospitals, one each in Hong Kong and Henan Province, People’s Republic of China, during the period between 2000 and 2013. These strains exhibited various carbapenem resistance phenotypes and genotypes. The genetic identity of these isolates was confirmed by the Vitek II bacterial identification system prior to further analysis. The ethic approval for this study was covered by human subject ethic approval, 2018-039, approved by the Second Affiliated Hospital of Zhejiang University, Zhejiang, China. An addition 453 clinical carbapenem-resistant A. baumannii strains isolated from four different regions of China, were included in the screening of blaOXA genes in the latter part of the study. The experimental flow is illustrated by Supplementary Figure S1.
Antimicrobial Susceptibility Test
Minimal inhibitory concentration (MIC) of carbapenems was tested for the 14 Acinetobacter baumannii strains by using the microdilution method and interpreted according to CLSI guidelines (Clinical and Laboratory Standards Institute, 2016). Briefly, bacterial strains were grown on MH agar. Bacterial cell suspensions at a concentration equivalent to a 0.5 McFarland Standard were prepared and inoculated into microplate wells containing different concentrations of carbapenem. The final volume in each well was 150 μL. The effect of the efflux pump inhibitor Phenylalanine-arginine β-naphthylamide (PAβN) and CHDL inhibitor Sodium chloride on the susceptibility of the test strains to carbapenem was determined by adding the compounds to specific wells of the microtiter plate to produce a concentration of 30 μg/ml and 200 mM, respectively. The MIC test was repeated twice to ensure the accuracy of the result.
Whole Genome Sequencing and ST Typing of Acinetobacter baumannii
Whole genomic DNA was extracted from the strains using the InvitrogenTM PureLinkTM Genomic DNA Mini kit, followed by sequencing with an Illumina® NextSeq 550 system. Raw reads generated in this study and the Illumina reads of 11 strains obtained from the NCBI database were trimmed or filtered to remove low-quality sequences and adaptors. Genome data were annotated with the RAST tool (Overbeek et al., 2014) and Prokka (Seemann, 2014). Scaffolds obtained were analyzed by Geneious 9.7. A. baumannii type strain ATCC19606 was used as reference throughout the analysis. ST profiles were determined according to the A. baumannii MLST (Oxford) database using sequences extracted from whole genome sequencing. Discrimination of International Clone II was based on analysis of the blaOXA–51 gene as described previously (Matsui et al., 2013).
qRT-PCR Analysis on Gene Expression
Overnight culture of A. baumannii isolate was inoculated into fresh LB broth until OD 0.6 was reached. Total RNA was extracted using the QIAGEN RNeasy® Mini Kit. The extracted RNA was further treated with the Invitrogen Turbo DNA-freeTM Kit to remove any DNA contaminants. 1 μg of purified mRNA samples were reverse-transcribed to cDNA by using the InvitrogenTM SuperScriptTM III First-Strand Synthesis SuperMix kit for qRT-PCR. Real-time PCR was performed on a Roche LightCycler® 480 System and PowerSYBRTM Green PCR Master Mix was used as the reaction medium. The parameters of PCR were as followed: the reaction mixture was first incubated at 95°C for 10 min for complete denaturation of template and activation of DNA polymerase; followed by 40 cycles of denaturation at 95°C for 10 s, annealing and polymerization at 60°C for 1 min. The melting temperature of PCR product was measured after completion of PCR to ensure the absent of non-specific PCR product. Expression level of endogenous genes was normalized with gyrB and A. baumannii ATCC19606 was used as control for comparison. The gene expression study was performed in triplicate. Primers used in this study are listed in Supplementary Table S1.
Western Blot of blaOXA–51-Like Proteins
Cell lysates were solubilized by boiling with SDS running buffer for 10 min and subsequently separated by SDS-PAGE with 12% separating gel. Proteins were transferred to a PVDF membrane followed by blocking by skimmed milk for 1 h and incubated with mouse anti-OXA-51 antibody at 4°C overnight. The goat anti-mouse antibody was used as the secondary antibody. The signal was generated by HRP substrate and detected by chemiluminator. The broad range anti-GADPH (ABCAM) was used as loading control.
Further Screening of Carbapenem-Resistant Acinetobacter baumannii Clinical Isolates
An addition 453 clinical carbapenem-resistant A. baumannii strains, isolated from four different regions of China, were further screened for the presence of blaOXA–23, presence of ISAba1 in the promoter region of blaOXA–23, blaOXA–51 and presence of ISAba1 in the promoter region of blaOXA–51. The primers sequences are shown in Supplementary Table S1. To screen for the presence of ISAba1 in the promoter region of blaOXA–23 or blaOXA–51, PCR assay was performed using primer ISAba1-F was used together with reverse primer of blaOXA–23 or blaOXA–51. Strains that did not contain blaOXA–23 with insertion of ISAba1 in the promoter region were selected for Western-blot analysis as mentioned above to check for the over-expression of OXA-51 in these strains.
Results and Discussion
Antimicrobial Susceptibility and Sequence Analysis of Acinetobacter baumannii Strains
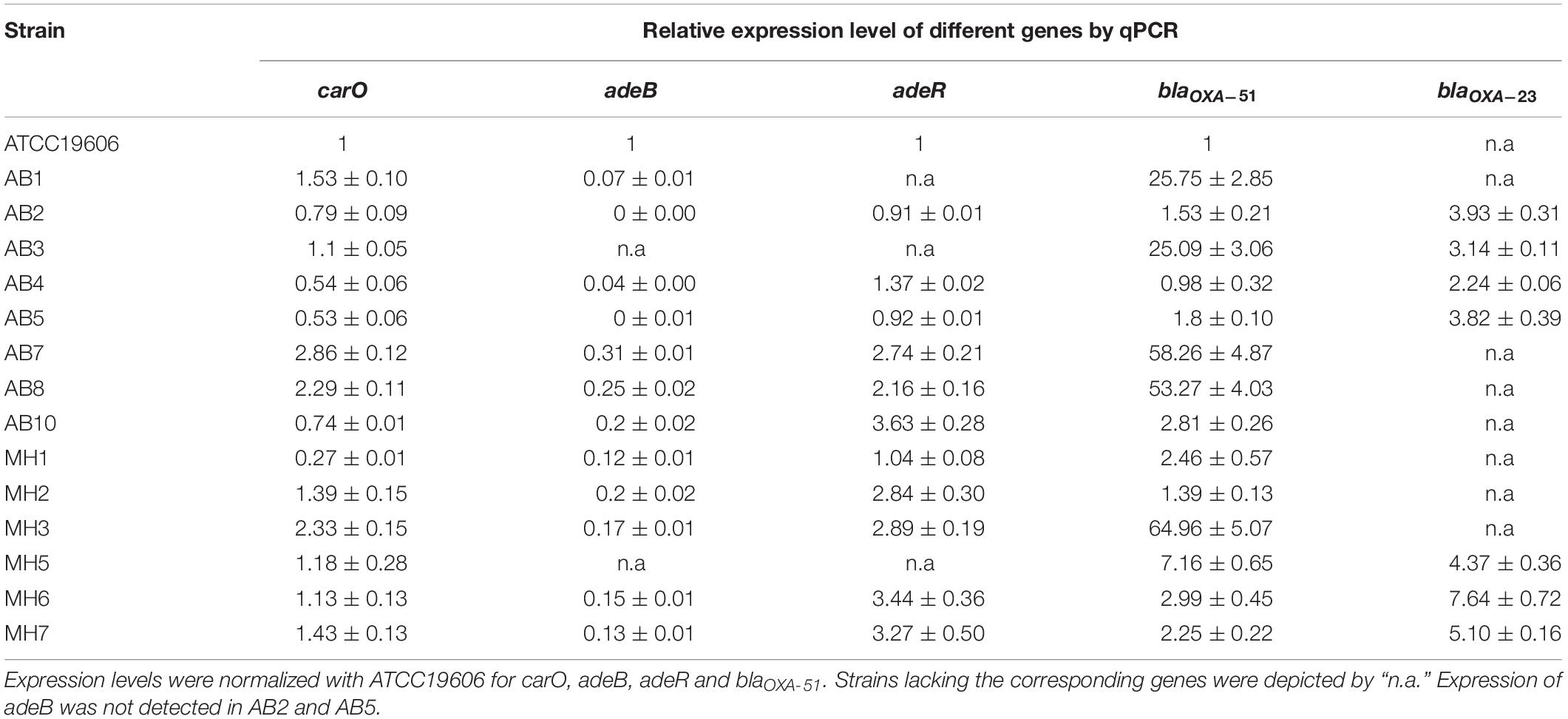
Fourteen clinical A. baumannii strains were first tested for their susceptibility toward carbapenems. Resistance toward imipenem and meropenem was observable in 12 of the 14 strains, with MIC ranging from 8 to ≥64 μg/ml (Table 1). The fourteen strains were then subjected to whole-genome sequencing. Based on the sequences obtained, about half of the strains were found to belong to ST208 (6 out of 14), whereas three strains (AB2, AB5, and MH5) belonged to ST940. For the remaining five strains they belonged to different ST types. Only the three ST940 strains were found to belong to International Clone II (IC II) based on analysis on the intrinsic blaOXA–51 gene harbored by these strains (Table 1 and Supplementary Figure S6). The findings corroborated with those in previous studies which reported the dissemination of A. baumannii Clonal Complex 92 strains within the Asia Pacific region, including Taiwan, South Korea and China (Kim et al., 2013). It has previously been demonstrated that the high genome plasticity of A. baumannii may result in sporadic loss of endogenous genes, altering the physiological status and hence antibiotic susceptibilities of the organism (Roca et al., 2012). In view of the possibility that resistance formation is due to loss of specific physiological functions, the relationship between presence of various intrinsic determinants of carbapenem sensitivity was determined; the determinants tested included the outer membrane porin-encoding genes carO and ompA, as well as the efflux gene adeABC and its regulator adeRS. It was found that all the test strains carried the intact carO and ompA genes without insertion of IS elements; this finding was consistent with that of a previous report (Rumbo et al., 2013). However, three strains (AB1, AB3, and MH5) were found to lack adeRS, with the latter two also lacking adeB. The intrinsic blaOXA–51 gene was detectable in all test strains, among them, the three ST940 strains were found to carry the variant gene blaOXA–99, whereas the blaOXA–66, blaOXA–79, blaOXA–82 and blaOXA–83 variants were identified in the other strains. Interestingly, the ISAba1 element was found to be inserted into the promoter region of blaOXA–51 in five strains, all of which belonged to IC-II. Apart from carriage of various intrinsic CHDL genes, half of the strains were found to have acquired the blaOXA–23 gene, two of which were located in the chromosome. One strain was also found to harbor the blaOXA–72 element (a variant of blaOXA–24) (Table 1). It has been described that alteration of amino acids of the adeRS gene product could result in change of expression level of adeB (Wen et al., 2017). Sequences of carO, adeB and adeRS were then analyzed and aligned in attempt to establish their relationship with carbapenem susceptibility. However, it was shown that the genetic differences detectable in these genes were linked closely to their genetic types (IC-II and non IC-II) rather than carbapenem sensitivity (Supplementary Figures S2–S5). Other known carbapenemase genes such as blaNDM–1 were not detectable in the test strains. The combinational analysis of carbapenem resistance genotypes and their genetic characteristics showed that strains exhibiting carbapenem resistance were either carrying an additional CHDL gene (blaOXA–23 and blaOXA–24) or having an ISAba1 linked upstream region in their blaOXA–51 element. Consistently, two carbapenem-susceptible isolates carried neither blaOXA–23 or blaOXA–24 nor insertion of ISAba1 in the intrinsic blaOXA–51 (Tables 1, 2).
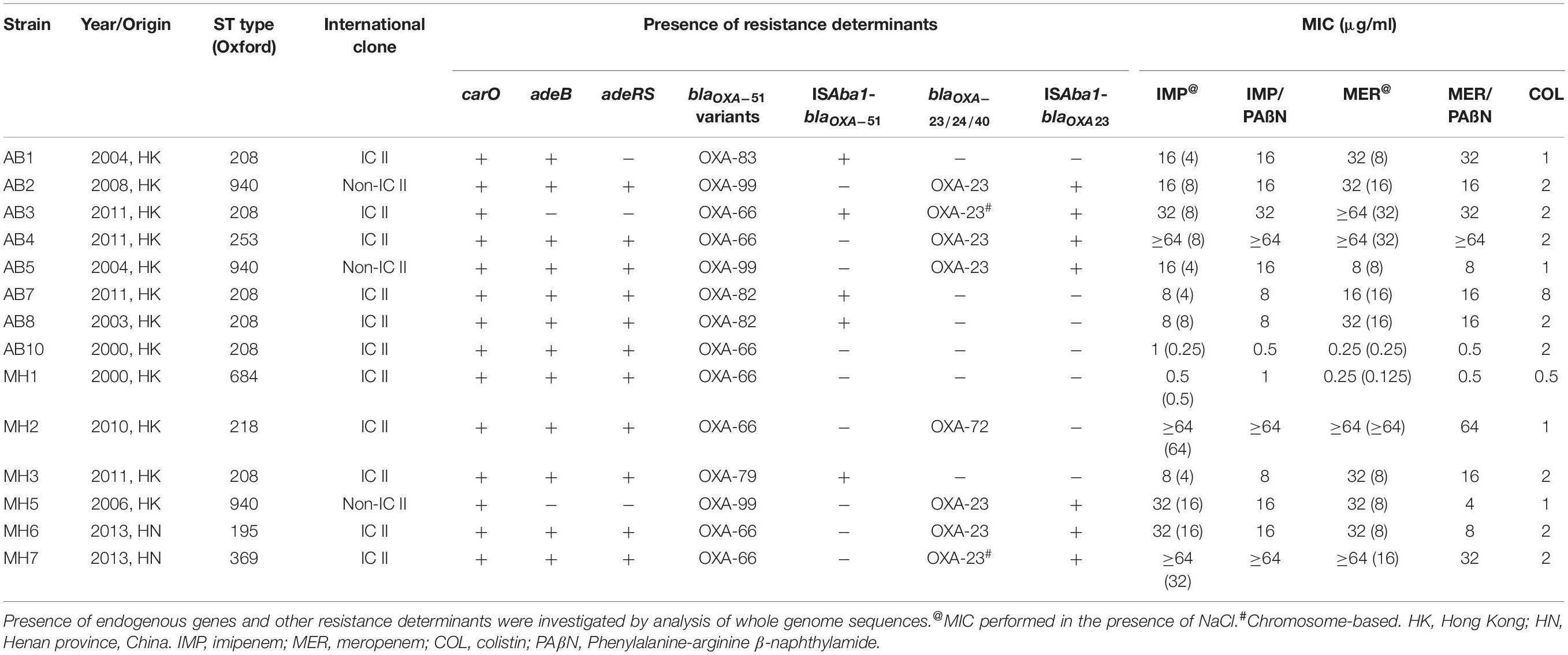
Table 1. Genetic and phenotypic characteristics of 14 Acinetobacter baumannii clinical strains tested in this study.
Efflux Pumps Did Not Mediate Carbapenem Resistance in Acinetobacter baumannii
The effect of RND-efflux systems on carbapenem susceptibility in A. baumannii clinical strains was evaluated by supplementing Phenylalanine-arginine β-naphthylamide (PAβN) at a concentration of 30 μg/ml in determination of MIC toward carbapenem. As shown in Table 1, we found that addition of PAβN exhibited minimal effect on altering carbapenem MIC except for two strains (Table 1). This finding demonstrated that RND efflux pumps only play a minimal role in mediating carbapenem resistance in A. baumannii strains of hospital origins and that the resistance phenotype is mainly conferred by the blaOXA enzymes.
To further investigate the role of various host determinants in mediating changes in carbapenem susceptibility, the expression level of various putative resistance genes, including the RND efflux pump gene adeB, its regulator adeR, the outer membrane porin gene carO, as well as the blaOXA–23 and blaOXA–51-like β-lactamase-encoding elements, were further analyzed by RT-qPCR. The expression level of each gene was normalized by the gene encoding the gyrase protein subunit B (gyrB) of the A. baumannii type strain ATCC19606. The results are shown in Table 2 and Supplementary Figures S7–S11. For carO, varied expression level was observed among the test strains, with most isolates exhibiting a level similar to or lower than that of ATCC19606. For the two susceptible strains, AB10 and MH1, carO expression was lower than that of ATCC19606 and most of the other carbapenem-resistant counterparts. The result is in line with a previous study, in which a decreased expression of carO has been described in carbapenem susceptible isolates recovered from Brazil (Fonseca et al., 2013). It should be noted that although strains AB7, AB8, and MH3 exhibited elevated carO expression by an amplitude >twofolds, their contribution to carbapenem resistance was hard to be addressed due to the presence of other major resistance mechanism, presence of OXA types of carbapenemases (Tables 1, 2).
An unexpected phenomenon regarding the adeB expression level of the test strains was observed. It was proposed that elevated expression of adeABC might contribute to reduced carbapenem susceptibility, possibly due to enhanced extrusion of intracellular antibiotics through efflux activity (Kim et al., 2013). Nevertheless, except for the two test strains AB3 and MH5, which lacked adeB, all other test strains exhibited a very low transcript level of this efflux gene, with a range of 0.07 to 0.31 compared to ATCC19606. Transcript of adeB was not detected in strains AB2 and AB5. Of note, adeB expression in the two carbapenem sensitive strains AB10 and MH1 was similar to most of their resistant counterparts. In order to evaluate whether decreased adeABC expression was due to effect of transcription regulation, expression level of adeR was also examined in an attempt to determine the degree of correlation between the two. Contrary to the data on adeB, similar or higher transcription level of adeR was observed among the majority of the test isolates, ranging from 0.91 to 3.63 folds when compared to ATCC19606 (Table 2). Correlation between expression of adeB and adeR, as well as adeR and carbapenem susceptibility, therefore could not be established. Taken together, the results suggest that the adeABC gene plays a negligible role in mediating changes in carbapenem susceptibility, particularly in strains which harbor additional CHDL genes such as blaOXA–23 and blaOXA–24. Indeed, the results of genotypic analysis corroborated with those of the expression study in that a lack of adeABC efflux system was observed in several resistant strains.
Overexpression of OXA-23 and Intrinsic OXA-51 Through ISAba1 Insertion Was the Major Mechanisms of Carbapenem Resistance
Antimicrobial susceptibility test revealed that all blaOXA–23 carriers were resistant to imipenem and meropenem but exhibited varied MIC values, presumably due to the fact that these strains carried various variants of blaOXA–23 and blaOXA–51. Expression level of blaOXA–51 and blaOXA–23 was examined in an attempt to determine the degree of contribution of carbapenemases encoded by these genes to reduction in carbapenem susceptibility of the A. baumannii strains tested in this study. All blaOXA–23 genes were found to carry insertion in the promoter region by ISAba1. Consistently, expression level of the blaOXA–23 gene was found to be up-regulated 2.24 to 7.64 folds among the strains. Discrepancies between the effects of plasmid-borne and chromosome-based elements were not observed (Tables 1, 2). Among the 6 blaOXA–23 carriers which lacked an over-expressed blaOXA–51, MIC values of imipenem and meropenem ranged from 16 to ≥64 μg/ml and 8 to ≥64 μg/ml, respectively. In particular, the higher transcript level of blaOXA–23 did not necessarily confer higher resistance level toward carbapenems. Since identical blaOXA–23 was harbored by all the strains, the possibility of varied activity toward carbapenem in individual variants can be ruled out. The lack of association between blaOXA–23 expression and susceptibility might then be attributed to physiological status of individual A. baumannii strains. Result of blaOXA–51 expression revealed consistency between ISAba1 insertion and transcript level of this endogenous carbapenemase. For the five test strains which carried the IS element, blaOXA–51 expression ranged from 25.09 to 64.96 folds compared to ATCC19606. To ensure that the overexpression of blaOXA–51 was consistent with their protein production, we took advantage of the monoclonal antibody specific to OXA-51 variants that produced by our lab to examine the production level of these OXA-51 variants by western blotting (Figure 1). It was shown that protein production correlated well with the qRT-PCR data, and that the five strains apparently over-produced the OXA-51 enzymes. The result confirmed that the ISAba1 insertion sequence is a key factor that promotes blaOXA–51 expression. To summarize, although the physiological status of individual strain may affect carbapenem susceptibility to some extent, resistance was observed only among A. baumannii strains which carried either ISAba1-blaOXA–51 or additional CHDL-encoding genes such as blaOXA23 and blaOXA–72.
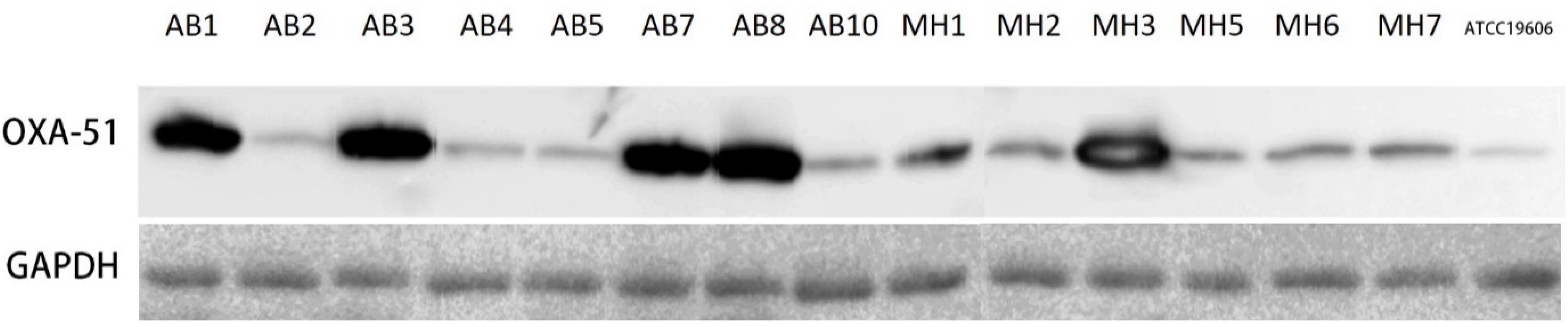
Figure 1. Western blot analysis of Acinetobacter baumannii strains using OXA-51 specific antibody. The broad-range anti-GAPDH was used as loading control.
To confirm that overexpression of OXA-23 and intrinsic OXA-51 is the major mechanisms of carbapenem resistance in clinical A. baumannii, a total of 453 carbapenem-resistant clinical A. baumannii isolated from four different regions of China were screened for the presence of these two mechanisms (Table 3). Our results showed that 404/453 strains carried over-expressed OXA-23 with ISAba1 in the promoter region of blaOXA–23, while the rest of 49 strains that did not carry blaOXA–23 exhibited an insertion of ISAba1 in the promoter region of blaOXA–51 and over-expression of OXA-51, which was confirmed by Western-blot using specific antibody to OXA-51 (Table 3). These data suggested that mechanisms of carbapenem resistance in clinical A. baumannii were mediated by either over-production of OXA-23 or OXA-51 through insertion of ISAba1 in their promoter region.
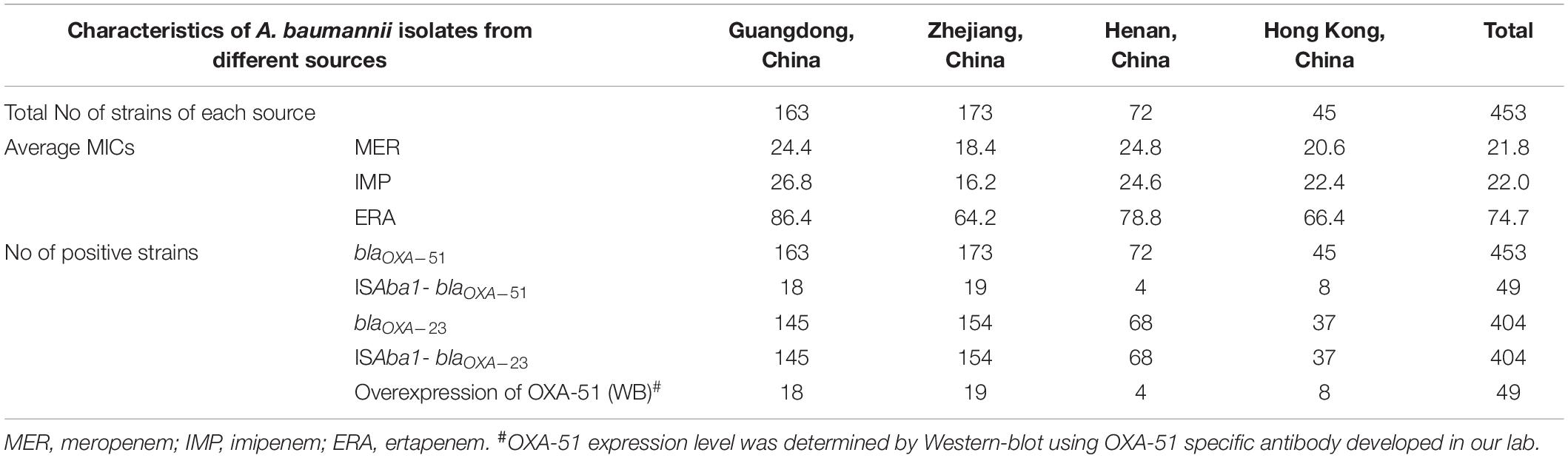
Table 3. Surveillance of mechanisms of carbapenem resistance in clinical carbapenem-resistant A. baumannii isolates from different regions of China.
Correlation analysis of the enzymatic activity of OXA types carbapnemase and meropenem MIC of strains producing these enzymes indicated that: (1) A. baumannii strains that over-expressed OXA-23/OXA-24 regardless of the status of over-expression of OXA-51 variants generally displayed higher MIC of meropenem (≥16 μg/ml); (2) A. baumannii strains that over-expressed OXA-51 variants only displayed relatively low MIC of meropenem, with OXA-83 (32 μg/ml) being higher than OXA-79/OXA-83 (8 μg/ml); and (3) Western-blot experiments indicated that OXA-51 variants exhibited base line expression without the insertion of ISAba1, which is probably true for extrinsic OXAs. OXA variants with lower activity like OXA51 variants are needed to be overexpressed to counter carbapeneme resistance phenotype, while variants with high catalytic activity such as OXA-72 may not need to be overexpressed, its basic line expression might be enough to mediate high carbapenem resistance in A. baumannii in the case of MH2 in this study.
Conclusion
Acinetobacter baumannii is an important pathogen that may cause fatal infections in nosocomial settings. It was thought that the interplay of multiple resistance mechanisms, including over-expression of endogenous efflux systems, suppression of expression of porin proteins, reduced membrane permeability and carriage of CHDL-encoding elements could lead to carbapenem resistance in this bacterial pathogen. In the current study, through systematic characterization of a set of clinical A. baumannii strains with different OXA-23 and MIC profiles, we demonstrated that the RND efflux system and membrane porin proteins were not the key factors that conferred carbapenem resistance in the A. baumannii strains tested. Instead the OXA-23 and OXA-24 enzymes (OXA-72 variant) were found to be the key elements underlying carbapenem resistance in A. baumannii strains as a result of insertion of the ISAba1 element, which provides a promoter for over-expression of the blaOXA–23 and blaOXA–23 genes (Nigro and Hall, 2016). In A. baumannii strains lacking these genes, carbapenem resistance was mainly due to the over-expression of blaOXA–51 variants, again through insertion of the promoter-bearing ISAba1 elements. Findings in this study emphasized the role of blaOXA type enzymes in mediating carbapenem resistance in this major clinical pathogen. These data provided essential information for the design of new strategies to treat clinical infections caused by carbapenem-resistant A. baumannii strains.
Data Availability Statement
The whole genome shotgun sequences of isolates included in this study have been deposited in GenBank with the following accession numbers: AB1, SAMN09667770; AB2, SAMN09667771; AB3, SAMN09667772; AB4, SAMN09667773; AB5, SAMN09667774; AB7, SAMN09667775; AB8, SAMN- 09667776; AB10, SAMN09667777; MH1, SAMN09667778; MH2, SAMN09667779; MH3, SAMN09667780; MH5, SAMN09667781; MH6, SAMN09667782; and MH7, SAMN09667783.
Author Contributions
MW and BC designed and performed the research, and drafted the manuscript. EC edited the manuscript. SC supervised the study and edited the manuscript.
Funding
This work was supported by the Health and Medical Research Fund of the Food and Health Bureau, Hong Kong (15141322) to SC.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02809/full#supplementary-material
References
Bergogne-Berezin, E., and Towner, K. J. (1996). Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9, 148–165. doi: 10.1128/cmr.9.2.148
Clinical and Laboratory Standards Institute (2016). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-sixth Informational Supplement M100-S26. Wayne, PA: CLSI.
Evans, B. A., and Amyes, S. G. (2014). OXA β-lactamases. Clin. Microbiol. Rev. 27, 241–263. doi: 10.1128/CMR.00117-113
Fonseca, E. L., Scheidegger, E., Freitas, F. S., Cipriano, R., and Vicente, A. C. (2013). Carbapenem-resistant Acinetobacter baumannii from Brazil: role of carO alleles expression and blaOXA-23 gene. BMC Microbiol. 13:245. doi: 10.1186/1471-2180-13-245
Gribun, A., Nitzan, Y., Pechatnikov, I., Hershkovits, G., and Katcoff, D. J. (2003). Molecular and structural characterization of the HMP-AB gene encoding a pore-forming protein from a clinical isolate of Acinetobacter baumannii. Curr. Microbiol. 47, 434–443.
Higgins, P. G., Pérez-Llarena, F. J., Zander, E., Fernández, A., Bou, G., and Seifert, H. (2013). OXA-235, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 57, 2121–2126. doi: 10.1128/AAC.02413-2412
Higgins, P. G., Wisplinghoff, H., Stefanik, D., and Seifert, H. (2004). Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J. Antimicrob. Chemother. 54, 821–823. doi: 10.1093/jac/dkh427
Ho, P. L., Ho, A. Y., Chow, K. H., Lai, E. L., Ching, P., and Seto, W. H. (2010). Epidemiology and clonality of multidrug-resistant Acinetobacter baumannii from a healthcare region in Hong Kong. J. Hosp. Infect. 74, 358–364. doi: 10.1016/j.jhin.2009.10.015
Ji, S., Chen, Y., Ruan, Z., Fu, Y., Ji, J., Fu, Y., et al. (2014). Prevalence of carbapenem-hydrolyzing class D β-lactamase genes in Acinetobacter spp. isolates in China. Eur. J. Microbiol. Infect. Dis. 33, 989–997. doi: 10.1007/s10096-013-2037-z
Kim, D. H., Choi, J.-Y. Y., Kim, H. W., Kim, S. H., Chung, D. R., Peck, K. R., et al. (2013). Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob. Agents Chemother. 57, 5239–5246. doi: 10.1128/AAC.00633-613
Liu, Y.-M. M., Chen, Y.-S. S., Toh, H.-S. S., Huang, C.-C. C., Lee, Y.-L. L., Ho, C.-M. M., et al. (2012). In vitro susceptibilities of non-Enterobacteriaceae isolates from patients with intra-abdominal infections in the Asia-Pacific region from 2003 to 2010: results from the study for monitoring antimicrobial resistance trends (SMART). Int. J. Antimicrob. Agents 40(Suppl.), S11–S17. doi: 10.1016/S0924-8579(12)70004-3
Magnet, S., Courvalin, P., and Lambert, T. (2001). Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45, 3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001
Matsui, M., Suzuki, S., Suzuki, M., Arakawa, Y., and Shibayama, K. (2013). Rapid discrimination of Acinetobacter baumannii international clone II lineage by pyrosequencing SNP analyses of bla(OXA-51-like) genes. J. Microbiol. Methods 94, 121–124. doi: 10.1016/j.mimet.2013.05.014
Mugnier, P. D., Poirel, L., Naas, T., and Nordmann, P. (2010). Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 16, 35–40. doi: 10.3201/eid1601.090852
Nigro, S. J., and Hall, R. M. (2016). Structure and context of acinetobacter transposons carrying the oxa23 carbapenemase gene. J. Antimicrob. Chemother. 71, 1135–1147. doi: 10.1093/jac/dkv440
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbio. Rev. 21, 538–582. doi: 10.1128/CMR.00058-57
Poirel, L., and Nordmann, P. (2006). Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12, 826–836. doi: 10.1111/j.1469-0691.2006.01456.x
Queenan, A. M., Pillar, C. M., Deane, J., Sahm, D. F., Lynch, A. S., Flamm, R. K., et al. (2012). Multidrug resistance among acinetobacter spp. in the USA and activity profile of key agents: results from CAPITAL surveillance 2010. Diagn. Microbiol. Infect. Dis. 73, 267–270. doi: 10.1016/j.diagmicrobio.2012.04.002
Roca, I., Espinal, P., Vila-Farres, X., and Vila, J. (2012). The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3:148. doi: 10.3389/fmicb.2012.00148
Rumbo, C., Gato, E., López, M., Ruiz de Alegría, C., Fernández-Cuenca, F., Martínez-Martínez, L., et al. (2013). Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 57, 5247–5257. doi: 10.1128/AAC.00730-713
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Siroy, A., Cosette, P., Seyer, D., Lemaitre-Guillier, C., Vallenet, D., Van Dorsselaer, A., et al. (2006). Global comparison of the membrane subproteomes between a multidrug-resistant Acinetobacter baumannii strain and a reference strain. J. Proteome Res. 5, 3385–3398. doi: 10.1021/pr060372s
Siroy, A., Molle, V., Lemaître-Guillier, C., Vallenet, D., Pestel-Caron, M., Cozzone, A. J., et al. (2005). Channel formation by CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49, 4876–4883. doi: 10.1128/AAC.49.12.4876-4883.2005
Turton, J. F., Ward, M. E., Woodford, N., Kaufmann, M. E., Pike, R., Livermore, D. M., et al. (2006). The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258, 72–77. doi: 10.1111/j.1574-6968.2006.00195.x
Wen, Y., Ouyang, Z., Yu, Y., Zhou, X., Pei, Y., Devreese, B., et al. (2017). Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region. Nucleic Acids Res. 45, 9773–9787. doi: 10.1093/nar/gkx624
Keywords: Acinetobacter baumannii, carbapenem resistance, OXA-23, OXA-51, mechanisms
Citation: Wong MH-y, Chan BK-w, Chan EW-c and Chen S (2019) Over-Expression of ISAba1-Linked Intrinsic and Exogenously Acquired OXA Type Carbapenem-Hydrolyzing-Class D-ß-Lactamase-Encoding Genes Is Key Mechanism Underlying Carbapenem Resistance in Acinetobacter baumannii. Front. Microbiol. 10:2809. doi: 10.3389/fmicb.2019.02809
Received: 24 July 2019; Accepted: 19 November 2019;
Published: 04 December 2019.
Edited by:
Leonardo Neves de Andrade, University of São Paulo, BrazilReviewed by:
Andres Felipe Opazo-Capurro, Universidad de Concepción, ChileJitendraa Vashistt, Jaypee University of Information Technology, India
Copyright © 2019 Wong, Chan, Chan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng Chen, c2hlY2hlbkBjaXR5dS5lZHUuaGs=
†These authors have contributed equally to this work
 Marcus Ho-yin Wong
Marcus Ho-yin Wong Bill Kwan-wai Chan
Bill Kwan-wai Chan Edward Wai-chi Chan
Edward Wai-chi Chan Sheng Chen
Sheng Chen