- 1Spatial Science for Public Health Center, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD, United States
- 2Department of Epidemiology, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD, United States
- 3Office of Environmental Health and Safety, Division of Environmental Public Health, Washington State Department of Health, Olympia, WA, United States
- 4Public Health Laboratories, Division of Disease Control and Health Statistics, Washington State Department of Health, Shoreline, WA, United States
- 5Office of Analytics and Outreach, Center for Food Safety and Applied Nutrition, Food and Drug Administration, College Park, MD, United States
- 6Angelo DePaola Consulting, Coden, AL, United States
Vibrio parahaemolyticus is a naturally occurring bacterium in estuarine waters and is a major cause of seafood-borne illness. The bacterium has been consistently identified in Pacific Northwest waters and elevated illness rates of vibriosis in Washington State have raised concerns among growers, risk managers, and consumers of Pacific oysters (Crassostrea gigas). In order to better understand pre-harvest variation of V. parahaemolyticus in the region, abundance of total and potentially pathogenic strains of the bacterium in a large number of Washington State Pacific oyster samples were compared with environmental conditions at the time of sampling. The Washington Department of Health regularly sampled oysters between June and September at over 21 locations from 2014 to 2018, resulting in over 946 samples. V. parahaemolyticus strains carrying three genetic markers, tlh, trh, and tdh, were enumerated in oyster tissue using a most probable number-PCR analysis. Tobit regressions and seemingly unrelated estimations were used to formally assess relationships between environmental measures and genetic markers. All genetic markers were found to be positively associated with temperature, independent of the abundance of other genetic markers. Surface water temperature displayed a non-linear relationship, with no association observed between any genetic marker in the warmest waters. There were also stark differences between surface and shore water temperature models. Salinity was not found to be substantially associated with any of the genetic variables. The relative abundance of tdh+ strains given total V. parahaemolyticus abundance (pathogenic ratio tdh:tlh) was negatively associated with water temperature in colder waters and decreased exponentially as total V. parahaemolyticus abundance increased. Strains carrying the trh gene had a pronounced positive association with strains carrying the tdh gene but was also negatively associated with the tdh:tlh pathogenic ratio. These results suggest that there are ecological relationships of competition, growth, and survival for V. parahaemolyticus strains in the oyster tissue matrix. This work also improves the overall understanding of environmental associations with V. parahaemolyticus in Washington State Pacific oysters, laying the groundwork for future risk mitigation efforts in the region.
Introduction
Vibrio parahaemolyticus is a Gram-negative, halophilic bacterium naturally present in many coastal waters throughout the world (CDC, 1998; Ansaruzzaman et al., 2005; Su and Liu, 2007; Kirs et al., 2011; Velazquez-Roman et al., 2014; Wu et al., 2014; Martinez-Urtaza et al., 2016). It is the leading cause of seafood-borne bacterial gastroenteritis (Scallan et al., 2011). While symptoms are often mild and self-limiting, V. parahaemolyticus infections (i.e., vibriosis) can at times lead to life-threatening septicemia. The most common cause of vibriosis in humans is the consumption of raw or undercooked seafood (Su and Liu, 2007). The bacterium can accumulate in marine organisms, especially in shellfish due to their filter-feeding behavior (DePaola et al., 2003).
The presence of V. parahaemolyticus in the environment and its capacity for human infection is widespread. The first recorded outbreak occurred in Osaka, Japan, in 1950 (Fujino et al., 1953), and since 1996, there have been more frequent vibriosis outbreaks in North America, South America, Western and Eastern Europe, and Asia (CDC, 1998; Velazquez-Roman et al., 2014; Wu et al., 2014; Gonzalez-Escalona et al., 2016; Haendiges et al., 2016; Martinez-Urtaza et al., 2016). The Pacific Northwest (PNW) region of the United States is an environment in which pathogenic V. parahaemolyticus strains are relatively abundant. A large outbreak of cases first occurred in this region in 1997, when 209 people became ill from eating shellfish harvested in PNW and one person died (CDC, 1998). Since that time, there have been additional outbreaks and increased incidence of sporadic cases in the region, particularly in Washington State where the majority of shellfish are harvested (McLaughlin et al., 2005; National Marine Fisheries Service et al., 2017; Washington Disease Control and Health Statistics, 2018). Cases have continued to rise over time despite multiple efforts by the Washington State Department of Health (WDOH) and shellfish growers to restrict harvesting by time and place and to implement austere post-harvest controls (Paranjpye et al., 2012; Washington State Board of Health, 2015).
While all V. parahaemolyticus bacteria possess a thermolabile hemolysin gene (tlh), not all strains can cause vibriosis. Common indicators of pathogenicity among V. parahaemolyticus bacteria are the thermostable direct hemolysin and the thermostable direct-related hemolysin genes (tdh and trh) (Shirai et al., 1990; Panicker et al., 2004). While neither of these genes are required or consistently predict pathogenicity, they have been shown to be regularly associated with type III secretion systems that lead to human infection, and so are frequently used to measure virulence factors in environmental isolates (Paranjpye et al., 2012; Zhang and Orth, 2013). It is worth noting that type VI secretion systems, while not associated with the hemolysin genes, also appear to play an important role in the pathogenicity of some strains (Salomon et al., 2013; Raghunath, 2014). PNW waters and oysters have relatively high levels of tdh and trh genetic material compared to other North American bodies of water, with the bacterium population being comprised of a wide array of strains that participate in regular genetic recombination (Hazen et al., 2010; Paranjpye et al., 2012; Turner et al., 2013). Many of the bacterium strains common to PNW waters, particularly those that are tdh+ /trh+, have also been frequently associated with vibriosis cases in Washington and throughout the continent (Martinez-Urtaza et al., 2013; Turner et al., 2013; Velazquez-Roman et al., 2014; Xu et al., 2017). It should be noted that observed trh levels in PNW may be due in part to the presence of Vibrio alginolyticus bacteria strains, calling into question the utility of the marker (Gonzalez-Escalona et al., 2008).
There has been extensive research on the range of environmental conditions that are associated with the presence and abundance of V. parahaemolyticus (DePaola et al., 2003; Turner et al., 2014; Johnson, 2015; Paranjpye et al., 2015; Davis et al., 2017). There is a well-documented positive correlation between water temperature and V. parahaemolyticus abundance, and consequently water temperature is used as the primary environmental input in the U.S Food and Drug Administration’s risk assessment for V. parahaemolyticus in oysters (USFDA, 2005). The bacteria cannot survive in freshwater, but concentrations at higher salinity levels vary widely by other water column conditions. Water clarity has been negatively associated with V. parahaemolyticus abundance, potentially due to the surface area suspended particles provide for the bacteria to adhere and grow. High levels of plankton and low levels of dissolved oxygen have also been found to be associated with greater bacterium abundance. Unfortunately, many of these associations vary significantly across geographic regions, time periods, and by the genetic marker of interest, making it difficult to identify broadly applicable relationships between environmental determinants and abundance of the bacterium (DePaola et al., 2003; USFDA, 2005; Turner et al., 2013; Johnson, 2015; Paranjpye et al., 2015; Davis et al., 2017). A notable example of such variation is the reported negative associations between the relative abundance of tdh+ strains in Gulf Coast oysters with both water temperature and the total abundance of V. parahaemolyticus (DePaola et al., 2003). Some of the variation in environmental associations may be contextualized as additional datasets of V. parahaemolyticus abundance are collected in pre-harvest settings that encompass large periods of time and geographic areas.
Given the health concerns related to V. parahaemolyticus in Washington, expected increases in illness rates as higher latitude waters continue to warm over time (Baker-Austin et al., 2017), and the economic impact to one of the largest groups of oyster producers in the country, further investigation into the environmental determinants of the bacterium in the region is warranted. The current study utilizes a large dataset of regularly monitored harvesting sites and public beaches in Washington State. The dataset contains one of the largest collections of oyster samples ever analyzed for V. parahaemolyticus and spans a substantial range of locations and time periods. The size and extent of the current dataset further allows for investigation into environmental associations independent of the concentrations of other genetic markers as well as statistical interactions between determinants. Overall, this analysis evaluated the relationships between absolute and relative abundance of V. parahaemolyticus strains carrying selected genetic markers and water temperature and salinity in Washington State.
Materials and Methods
Since 2006, WDOH has been regularly sampling Pacific oysters (Crassostrea gigas) to measure V. parahaemolyticus abundance. Each year, intertidal shellfish harvesting beds were sampled as frequently as every week from June to September, primarily in the Hood Canal and South Puget Sound but also less frequently in the coastal bays and northern shellfish harvesting waters resulting in over 200 samples per year. For each sample, 13–18 live oysters approximately 10 cm in length were collected, when possible, after being exposed to the air by the receding tide. Oysters were rinsed in seawater before being sealed in a bag and then chilled with ice in an insulated container. Shoreline water and ambient air temperature were recorded with a thermometer. Surface water temperature was also measured where the total water depth was at least 0.6 m. An additional oyster was shucked and its tissue temperature recorded before being discarded. A refractometer was used to measure salinity of the surface water.
Oyster samples were analyzed by the WDOH Public Health Laboratory for V. parahaemolyticus abundance using a previously described most-probable-number (MPN)-based TaqMan real-time PCR assay (Glover, 2015). Three genetic markers were utilized: the species-specific thermolabile hemolysin gene (tlh), the virulent thermostable direct hemolysin gene (tdh), and the thermostable direct-related hemolysin gene (trh). Upon receipt at the laboratory, an oyster from each sample was immediately shucked and the internal temperature was recorded to ensure proper temperature control during transit. Oysters were stored at 2–8°C until sample processing began. All shells were cleaned and shucked following guidelines set by the American Public Health Association (APHA, 1970). At least 12 oysters per sample were washed and all debris removed prior to aseptically shucking from the shell (instead of the hinge) in order to prevent contamination of the oyster tissue. Oyster tissue was then homogenized and inoculated into a MPN dilution series, all within 24 h of initial sample collection. The MPN portion of the assay was conducted using a three-tube six decimal dilution series ranging from 1 to 0.00001 g of oyster tissue enriched in alkaline peptone water (APW) to measure concentrations of the targets (Blodgett, 2017). Samples were incubated for 18–24 h at 35°C. Once removed from the incubator, tubes were read for turbidity and lysed within 1 h.
The real-time PCR assay was used as a qualitative determination of the absence or presence of each targeted gene in the individual tubes of the MPN dilution series. DNA isolation was performed on the Roche MagNA Pure LC instrument using the MagNA Pure LC DNA Isolation Kit III (Roche, Indianapolis, IN, United States). Real-time PCR analysis was performed on the Life Technologies ViiA7 real-time PCR instrument (Thermo Fisher Scientific, Waltham, MA, United States) using a 384-well platform. Commercially prepared mastermix was utilized in the assay (Bioline SensiFASTTM Probe Hi-ROX Kit BIO-82020, Meridian Bioscience, Cincinnati, OH, United States) with the following cycling parameters: an initial denaturation of template at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 s, and combined annealing and extension at 59°C for 60 s. An artificially created internal control (IC) plasmid was added to the PCR mastermix in order to detect any inhibitors within each sample. In addition to the IC plasmid, extraction controls were included on every Magna Pure cartridge and run on the PCR assay. Amplification controls were also included on every real-time PCR run. The positive extraction control was V. parahaemolyticus ATCC 49398TM and the positive amplification control was a combination of previously extracted DNA from V. parahaemolyticus ATCC BAA-240TM and from a trh+ Washington State clinical isolate. The negative extraction and amplification controls were both sterile molecular grade water. If PCR results did not demonstrate expected values for all controls, samples were reprocessed. There have been no reported instances by the WDOH Public Health Laboratory of an incorrect control value not being resolved during reprocessing.
Due to the artificial enrichment of the bacteria in APW, the real-time PCR resulted in positive tubes displaying consistent amplification across all dilutions of the MPN. When late amplification occurred (after 40 cycles), the PCR was repeated in duplicate to ensure appropriate reporting. In instances where trh was detected in the absence of tlh, indicating the presence of only V. alginolyticus (Gonzalez-Escalona et al., 2006), trh values were not reported. The positive and negative results generated on the PCR assay were applied back to the MPN dilution series in order to enumerate the amount of each genetic marker present in the oyster sample. The limit of detection for all genetic markers from the MPN assay was 0.3 MPN/g and the upper limit of quantification was 110,000 MPN/g.
Current statistical analyses are limited to 2014–2018 samples as the trh genetic marker was not targeted before this time period. A total of 1,025 oyster samples were tested for V. parahaemolyticus at 25 sites during these years. In order to limit analysis to well-sampled locations, observations were excluded from the current analysis if taken from sites that were sampled on less than 12 occurrences throughout the 5-year period, or if only one sample was taken at a site for a given year. Furthermore, records that had missing information for any of the environmental or genetic variables were removed. This reduced the current analysis to 879 samples taken at 21 sampling sites (Figure 1). Note that only one sampling site from the northern waters, Samish Bay, was included in the analysis after the reduction in sample size.
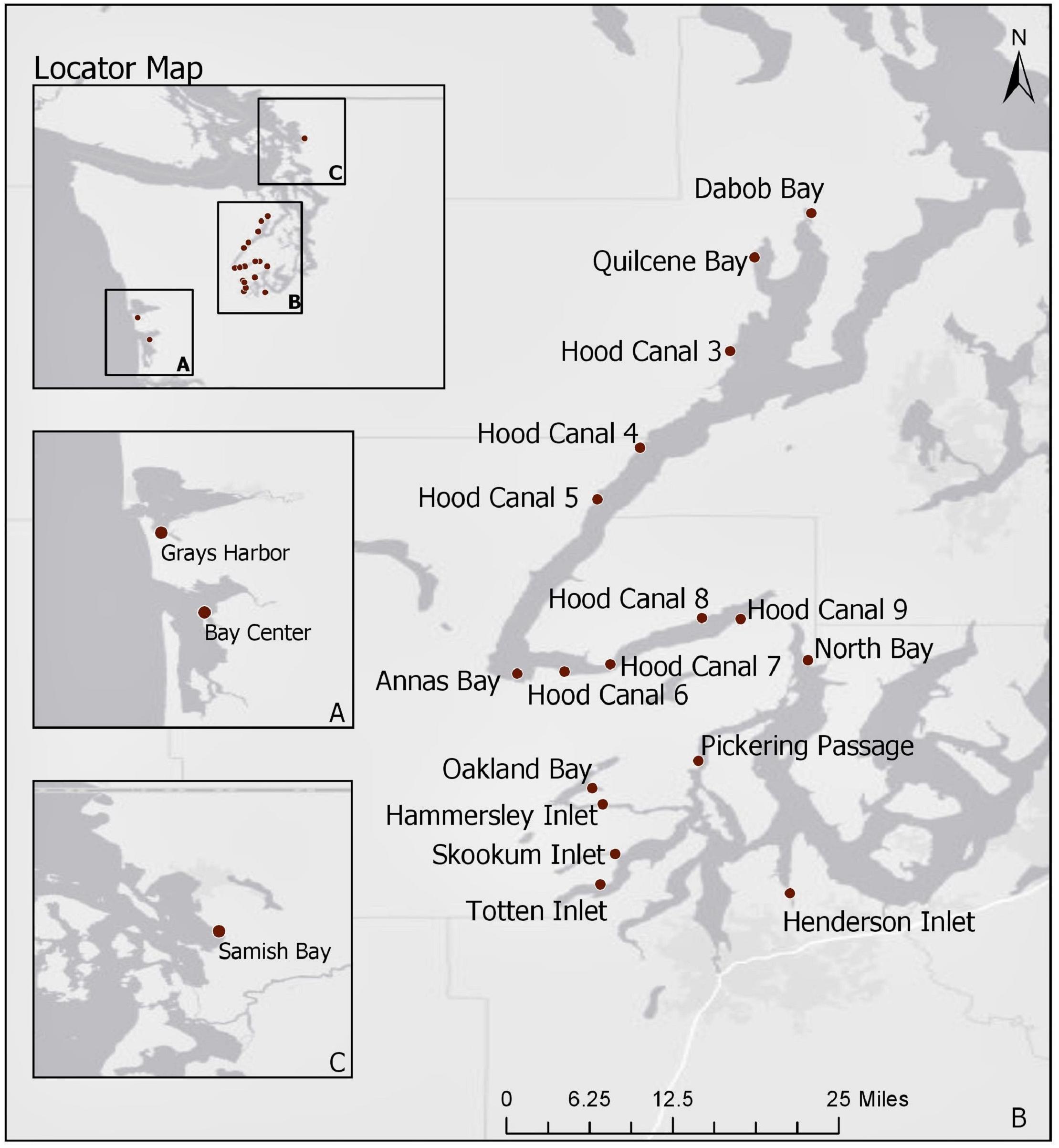
Figure 1. Map of Washington State shellfish sampling sites between 2014 and 2018. Panel A displays the coastal bays, panel B (main map) includes South Puget Sound and Hood Canal, and panel C displays Samish Bay in the northern waters. Basemap attributed to ESRI (2019b).
An additional genetic variable, a tdh:tlh pathogenic ratio, was created to assess the relative abundance of tdh+ strains for each oyster sample collected. All genetic variables were positively skewed and non-negative and so were log-transformed for all statistical analyses. Natural logs were used when variables were the outcome of interest in regression analysis, but in all other instances, log10-transformations were used to facilitate interpretation.
Exploratory analyses included developing boxplots and histograms of each environmental and genetic variable and scatter plots to compare bivariate relationships. Non-linear trends were visually identified using non-parametric local regressions (LOESSs). Pearson correlation were used to quantify the relationships between all genetic and environmental variables. For exploratory analyses, and when used as explanatory variables in regression models, censored genetic values were imputed: those below the limit of detection were halved and those above the upper limit of quantification were doubled.
Tobit regression models were used to characterize the linear associations between environmental measures and V. parahaemolyticus abundance while accounting for left- and right-censored genetic data (limit of detection and limit of quantification, respectively). Separate models were developed for each genetic marker. For the pathogenic ratio, a more sophisticated regression analysis was undertaken in order to preserve the individual censored data for both the numerator (tdh) and denominator (tlh). Zellner’s seemingly unrelated regression (SUR) model was first performed simultaneously on linear tdh and tlh regressions using only non-censored data; residuals were evaluated using a Pearson’s correlation along with the Breusch–Pagan test of independence (Zellner, 1962, 1963; Breusch and Pagan, 1980). If residuals were found to be independent, Tobit regressions for tdh and for tlh could be estimated simultaneously to obtain the joint parameter vector and variance-covariance matrix for coefficients by using a seemingly unrelated estimation technique (Weesie, 1999). Given that all genetic variables were log-transformed, these estimations could be used to calculate the tdh:tlh ratio’s regression parameters and standard errors by subtracting the two sets of estimations via linear combination. The results of all genetic marker models are reported in their exponentiated form, and as such can be interpreted as the relative change in the geometric mean of gene abundance.
For each genetic marker, univariate models were compared to multivariate models that included year, region, and all environmental covariates (Multivariate 1), or that further included all non-redundant genetic covariates (Multivariate 2). Year and region were both treated as categorical variables, and as such indicator variables were included in each multivariate model. Multivariate 1 models were used to determine whether any environmental associations were redundant or unique to certain locations and time periods. Multivariate 2 models were used to determine if environmental associations were mediated by the abundance of total V. parahaemolyticus or potentially pathogenic strains. Note that a Multivariate 2 model was not created for regressions with tlh as the outcome, as potentially pathogenic strains make up a part of the total bacterium abundance. Potential interactions within and between environmental and genetic variables were formally tested by adding them to the regression models. When interactions were deemed to be statistically significant (α = 0.05), linear combinations of the original variable term and the interaction term were used to quantify the impact and statistical significance of the effect modification.
Given the potential redundancy of surface water temperature with shore water temperature, multicollinearity was assessed using the variance inflation factor on sensitivity regression analyses. Model fit of the two covariates was also assessed using the Akaike information criterion (AIC). An additional sensitivity analysis was conducted by removing Henderson Inlet observations, as the sampling site is connected to a freshwater inlet and had substantially lower salinity levels than other locations.
All statistical analyses were performed in Stata 13 (StataCorp, 2011). Maps were created in ArcGIS Pro version 2.3.2 and additional plots were created in R statistical software version 3.5.2 using the ggplot package (Wickham, 2009; R Core Team, 2018; ESRI, 2019a).
Results
The map of Washington State shellfish sampling sites is shown in Figure 1. The mean number of samples taken at each site across the 5-year sampling period was 50.5 with a range of 14–72. The number of samples across years was roughly similar for each site. Total ranges of environmental variables are as follows: ambient air temperature (5.2, 35.0°C); surface water temperature (12.5, 35.4°C); tissue temperature (7.7, 38.4°C); salinity (3.0, 35.0‰). All genetic variables had values below the limit of detection (Ntlh = 9, Ntrh = 101, Ntdh = 469). Only tlh had values above the upper limit of quantification (N = 4) while trh had a maximum observable value of 46,000 MPN/g and tdh a value of 430 MPN/g. The tdh:tlh ratio ranged from 0 to 100%. Using a stratified random sampling approach across all monitoring years (n = 150 for each target), the proportion of oyster samples that experienced late amplification in duplicate PCR runs was estimated to be 2.0% (95% CI: [0.7, 3.3%]) occurring in eight samples for the tlh target and one sample for the trh target. Late amplification increased abundance on average by a factor of 3.4 MPN/g (95% CI: [1.6, 7.0 MPN/g]).
Additional descriptions of the environmental and genetic variables can be found in Table 1 and Figure 2. All temperatures showed similar trends across years, although tissue temperature frequently reached higher values in the Hood Canal and South Puget Sound relative to Samish Bay and the coastal bays. Salinity showed no noticeable variation across region or year. tlh values were slightly lower in 2016 and seemed to be noticeably higher in Hood Canal and lower in Samish and the coastal bays. trh abundance was over an order of magnitude lower than tlh overall and was particularly low in Samish Bay. When tdh was observable, abundance was usually under 1 MPN/g and no samples in Samish Bay had detectable levels of tdh. The pathogenic marker also appeared to be slightly higher in 2016 and 2018. The coastal bays appeared to have overall much higher tdh:tlh values than other sampling regions. Salinity appeared negatively skewed, likely due to the lower salinity measurements in Dabob Bay, Henderson Inlet, and Hood Canal 3 and 8 (results not shown). Log-transformed tdh appeared positively skewed, and this pattern persisted across sampling location and year.
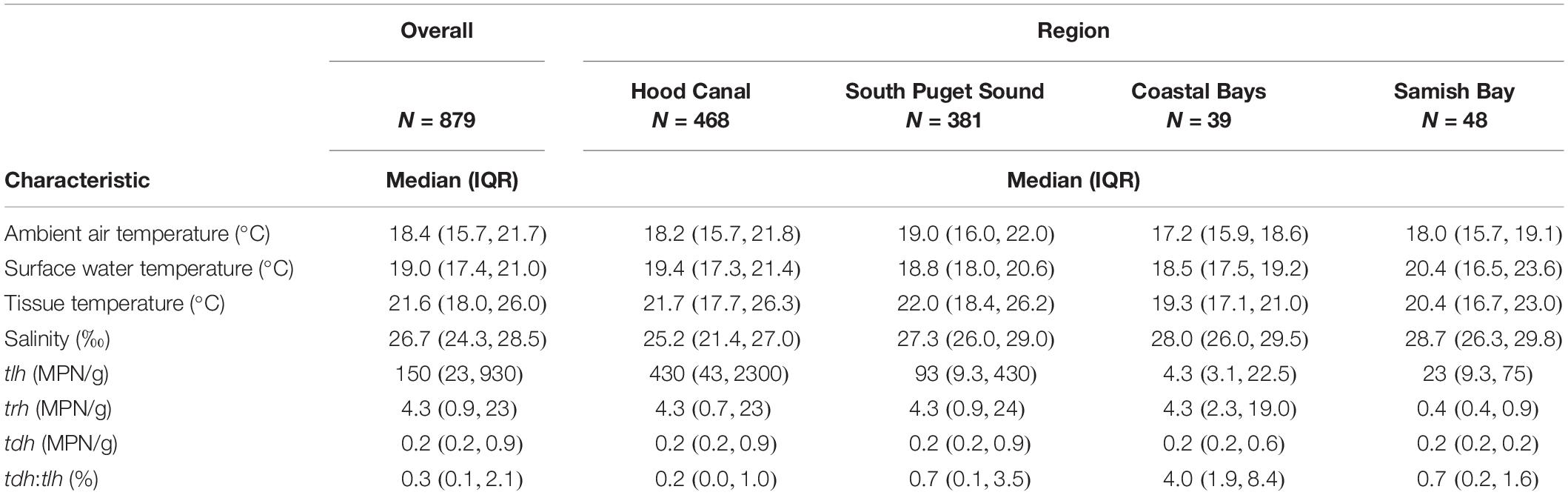
Table 1. Descriptive characteristics of environmental and genetic variables, as well as their variation across sampling regions.
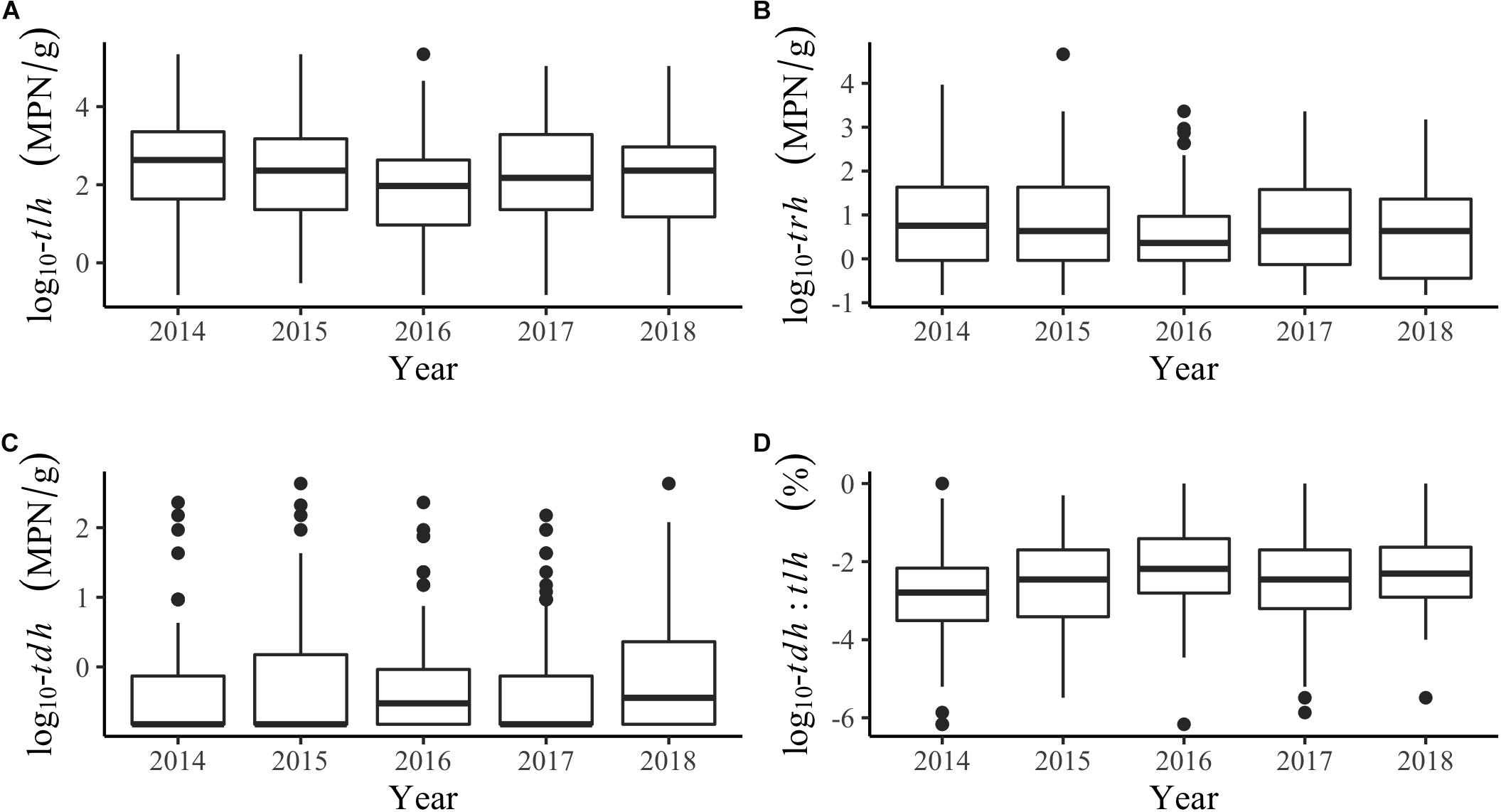
Figure 2. Boxplots of genetic markers by year for (A) tlh, (B) trh, (C) tdh, and (D) the tdh:tlh pathogenic ratio.
Surface water temperature and shore water temperature were highly correlated (ρ = 0.87) with variance inflation factors in sensitivity regression models consistently reaching above 4.0, indicating that coefficient standard errors more than doubled when both variables were included in the same model. In order to avoid this collinearity, shore water temperature was excluded from multivariate regression analyses as surface water temperature provided superior model fit (ΔAIC = 121.6). Furthermore, the surface water temperature variable is more similar to water measurements used in other environmental assessments of V. parahaemolyticus (Duan and Su, 2005; USFDA, 2005; Paranjpye et al., 2015; Davis et al., 2017).
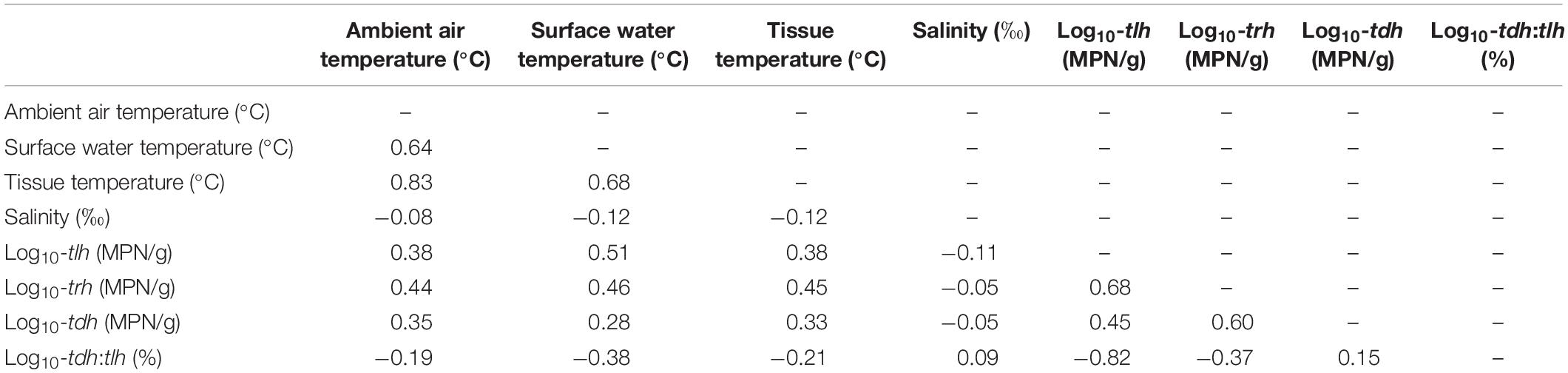
All remaining temperature measures were well-correlated (ρ≥0.64) and were moderately positively correlated with tlh, trh, and tdh (Table 2). Salinity was not correlated with temperature or any genetic variables. tlh, trh, and tdh were positively correlated with one another, with trh displaying the strongest correlations. All temperature measures and trh were slightly negatively correlated with the tdh:tlh ratio, while tlh was strongly negatively correlated with the ratio, indicating that the highest relative abundance of tdh+ strains occurred when total abundance was close to zero. Upon further investigation, this relationship was confirmed to be log-linear; the pathogenic proportion decreased exponentially as total abundance increased (Figure 3). All aforementioned correlations were statistically significant (p < 0.05).
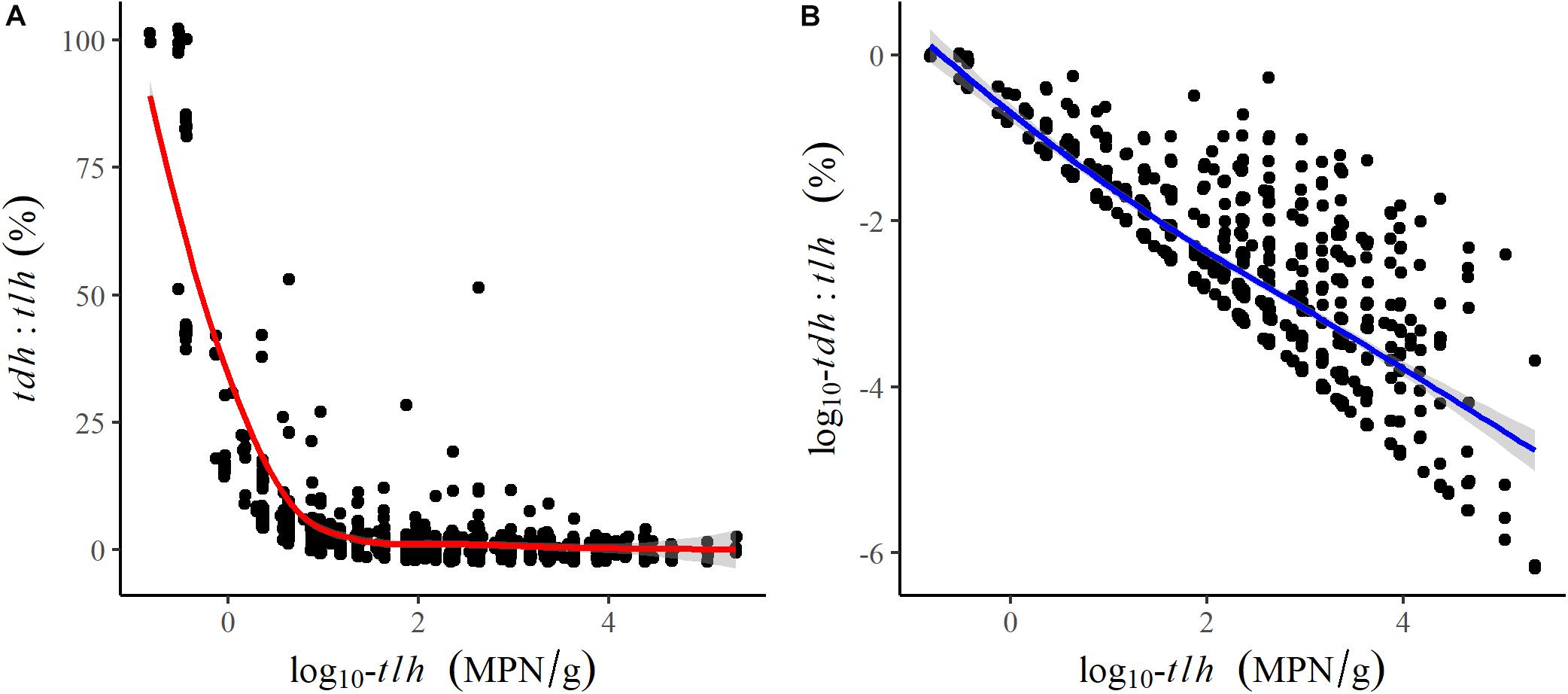
Figure 3. Scatter plots and fitted LOESS lines comparing the association of log-transformed tlh to (A) the tdh:tlh ratio and (B) the log-transformed tdh:tlh ratio.
Tobit regression model results for total V. parahaemolyticus abundance (tlh) and strains carrying the two pathogenic markers (trh, tdh) are reported in Table 3. For all three genetic markers, each temperature variable displayed a statistically significant positive association in the univariate models. LOESS exploratory analysis identified a non-linear relationship with surface water temperature such that a strong positive association was observed up to 22°C and after which no relationship was observed (Figure 4). This non-linear threshold also improved model fit in regression models (ΔAIC = 30.2). The positive association with surface temperature was slightly attenuated for trh when compared to tlh and moderately attenuated for tdh. All temperature associations became attenuated in the multivariate models for tlh, trh, and tdh. In the multivariate tlh model, tissue and air temperature were no longer significantly associated with total abundance while the positive association with water temperature was only slightly attenuated, still indicating almost a doubling in total abundance for every 1°C increase up to 22°C while holding all other environmental covariates constant. For the trh and tdh multivariate models adjusting only for environmental covariates (Multivariate 1), all three temperature measures had attenuated but still statistically significant associations. For the trh model further adjusting for tlh and tdh (Multivariate 2), only tissue temperature maintained a statistically significant association, indicating that on average trh abundance approximately doubled for every 12°C increase in tissue temperature when holding all other covariates constant.
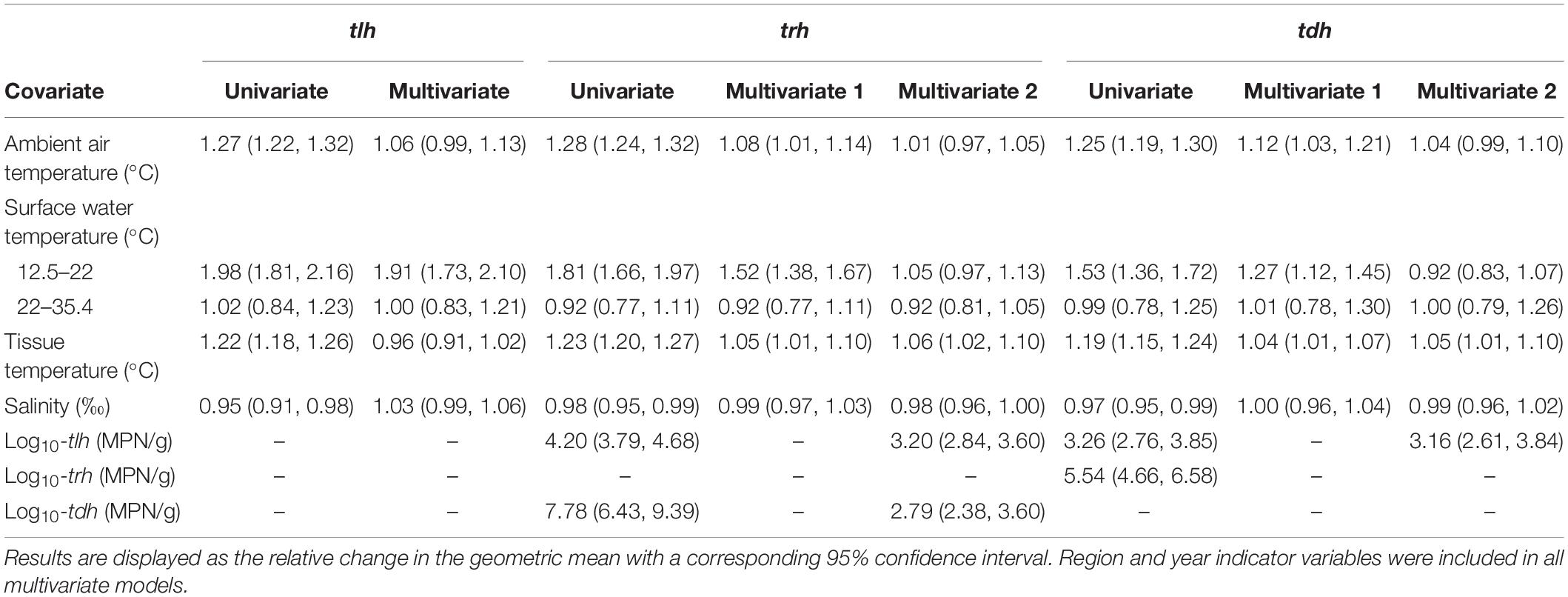
Table 3. Univariate and multivariate Tobit regression models of tlh, trh, and tdh and environmental/genetic covariates.
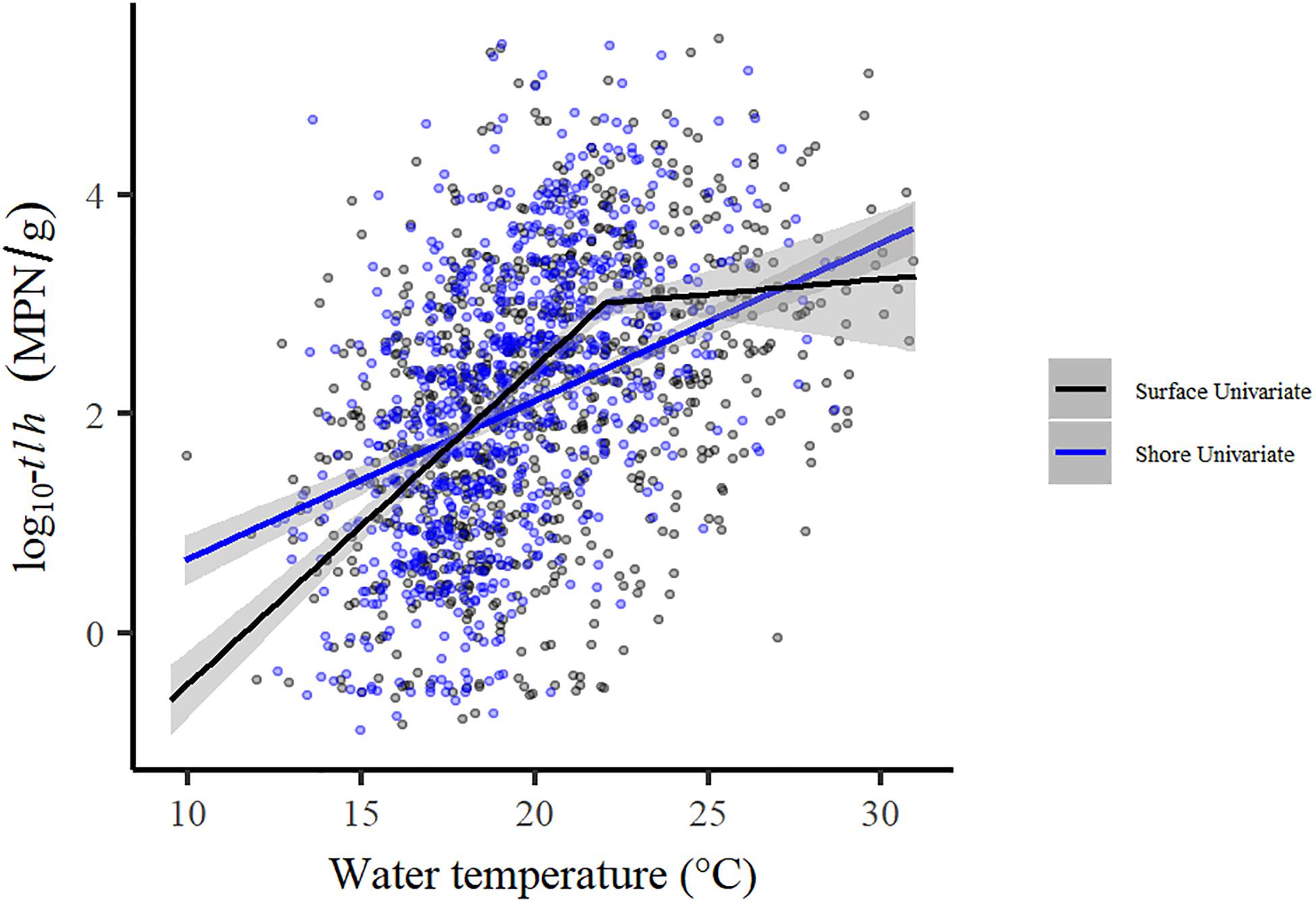
Figure 4. Comparison of univariate regressions for shore versus surface water temperature and total V. parahaemolyticus.
tlh and tdh were both strongly positively associated with trh before and after adjustment, although the association with tdh became noticeably attenuated (Table 3). For the tdh model, further adjusting for tlh (Multivariate 2) left only the tissue temperature association statistically significant, mirroring the relationship of tissue temperature with trh. While tlh maintained a strong positive association with tdh in the univariate and multivariate models, including trh as a covariate resulted in both tissue temperature and tlh no longer displaying statistically significant associations while trh maintained a particularly strong association (eβ = 3.86, 95% CI: [3.33, 4.51]). While salinity displayed a slight but statistically significant negative association in each of the univariate models for tlh, trh, and tdh, salinity no longer displayed a significant association with the three genetic markers in any of the multivariate models.
Each of the three genetic marker models displayed a positive statistical interaction between air and tissue temperature. The interaction suggests that at relatively low temperatures of both media no relationship between either air or tissue temperature and any genetic marker was observed, but at high temperatures of both media at least one of these temperature measures (air or tissue) had a statistically significant positive association with each genetic marker. A representative example of this interaction is shown in Table 4. When adjusting for environmental covariates, the association with surface water temperature was unaffected by this interaction. However, the slopes of air and tissue temperature were negatively transposed due to this adjustment, suggesting a negative slope at overall lower temperatures of both air and tissue (data not shown). These negative relationships should not be interpreted directly, however, as the Tobit model forces this relationship to keep the positive interaction between air and tissue temperature while also accounting for slope attenuation in both temperature measures due to the overwhelming effect of water temperature. An additional positive interaction for the tlh model was independently observed between surface water temperature and salinity such that the positive slope of water temperature in cooler waters (< 22°C) was steeper in more saline waters (Table 5).
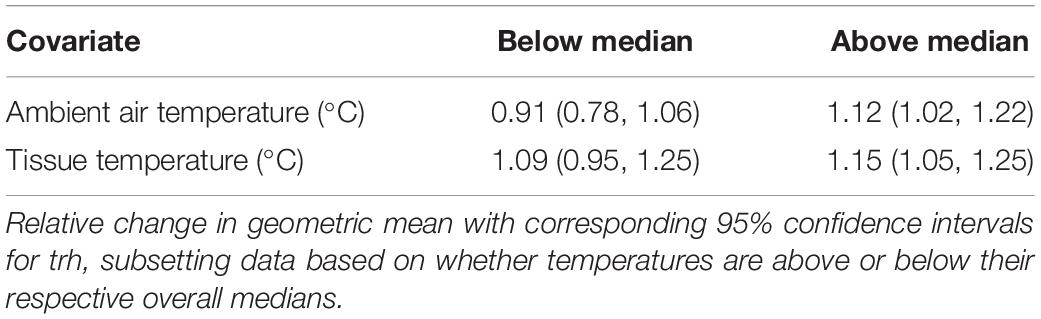
Table 4. Representative example of interaction (effect modification) between tissue temperature and air temperature on genetic marker abundance.

Table 5. Relative change in geometric mean with corresponding 95% confidence intervals of surface water temperature (<22°C) conditioned on water salinity.
Residuals from the SUR model for tlh and tdh did not appear to be correlated (ρ = 0.05; Breusch–Pagan χ2 = 2.02, p = 0.16). Therefore, all tdh:tlh models were estimated using linear combinations of seemingly unrelated estimations of the two Tobit models (Table 6). For the pathogenic ratio, all three temperature measures had statistically significant negative associations, while salinity did not have a statistically significant association. Surface water temperature also displayed a similar non-linear relationship with this ratio, with no significant trend observed at temperatures above 22°C. After adjusting for environmental covariates (Multivariate 1) only water temperature maintained a statistically significant negative association, indicating that on average the tdh:tlh ratio approximately halved for every 2.0°C increase in water temperature up to 22°C. Tissue temperature displayed a statistically significant positive association with the pathogenic ratio, similar to the adjusted relationships observed when modeling trh and tdh concentrations but still noticeably more attenuated than the negative association with water temperature. Further adjusting for trh (Multivariate 2) did not alter the associations for either water or tissue temperature. In contrast to the previous multivariate models, salinity did appear to have a statistically significant negative association with tdh:tlh in the fully adjusted model. The association, however, was very slight: on average approximately halving the ratio given a 23% increase in salinity. trh displayed a negative association with the ratio with and without adjusting for environmental measures, although this association was much more attenuated than the other aforementioned genetic marker associations. An inverted form of the interaction between surface water temperature and salinity observed in the tlh models was seen for the tdh:tlh models, such that the negative slope of water temperature became steeper in more saline waters (Table 5).
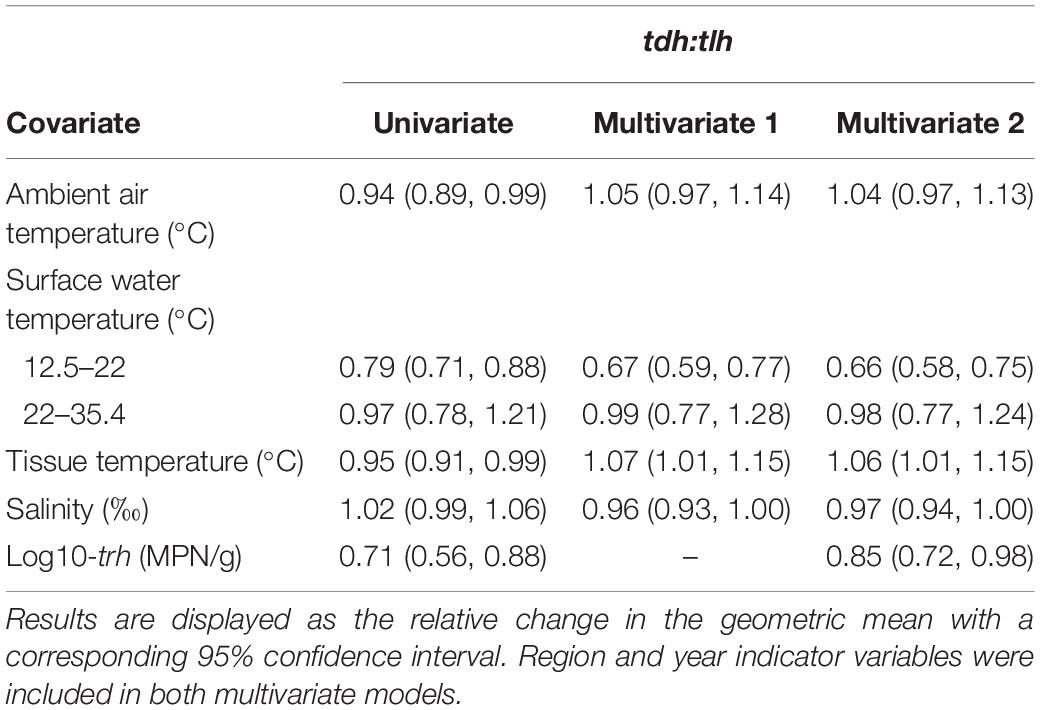
Table 6. Univariate and multivariate associations between the tdh:tlh ratio and environmental/genetic covariates based on linear combinations of seemingly unrelated estimations.
The sensitivity analysis removing samples taken in Henderson Inlet produced no substantial changes in any of the reported results. When directly compared, the univariate shore water model for tlh had a much more gradual slope than the surface model (eβ = 1.40, 95% CI: [1.33, 1.47]), but total abundance became more similar at higher water temperatures, primarily due to the non-linear relationship identified in the surface water model (Figure 4). Additional exploratory analysis revealed that the exponential decrease of tdh:tlh with increasing total V. parahaemolyticus was moderately attenuated in warmer waters (results not shown). Year and region indicator variables remained statistically significant in all multivariate regression models, consistently displaying substantial variation in total or potentially pathogenic strain abundance across space and time (results not shown). These significant trends indicate that the environmental and genetic variables considered in this study did not sufficiently explain spatial and temporal variation of each genetic marker.
Discussion
The current analysis assessed the relationships between environmental measures and genetic markers of V. parahaemolyticus across space and time in a large number of Pacific oyster samples in Washington State. This study confirmed frequently reported positive associations both between temperature and absolute genetic markers as well as among the genetic markers, while also highlighting a previously unobserved upper threshold in the positive relationship with water temperature and genetic markers. This study has also confirmed strong negative associations between the tdh:tlh pathogenic ratio and both total abundance and water temperature, suggesting that an ecological relationship of competition between V. parahaemolyticus strains exists within the oyster tissue matrix. As such, increased attention should be given to the pathogenic ratio in future research.
Temperature, in some medium, was found to be consistently positively associated with tlh, trh, and tdh even after adjustment for similar environmental covariates or other genetic markers. These findings suggest that temperature is at least a moderately strong determinant of total and pathogenic V. parahaemolyticus abundance in oysters, independent of its impact on total abundance or the concentration of another pathogenic marker. Surface water temperature displayed the strongest relationship for all three genetic markers, however, the effect observed for tdh and trh was largely mediated by total V. parahaemolyticus abundance. The non-linear relationship with surface water temperature, such that no association is observed in waters warmer than 22°C, is an important addition to the understanding of ecological relationships for V. parahaemolyticus in PNW. While previous research has frequently shown a consistent positive relationship with water temperature in most bodies of water (Johnson, 2015), prior work in Washington State did not find that highest temperatures correlated with greatest V. parahaemolyticus abundance (Paranjpye et al., 2015). Current findings support this previous work by identifying a threshold relationship, such that increasing temperature beyond sufficiently warm waters (∼22°C) does not result in an increase in average total bacterium abundance. The difference in estimated abundance based on surface versus shore water temperature is also noteworthy. While the two variables are collinear, surface water temperature provides superior model fit and so appears to be the preferred environmental measure for estimating total and pathogenic V. parahaemolyticus abundance. However, shore water temperature was measured more frequently, likely due to the relative ease of sampling compared to surface water temperature, especially at low tide. While shore water can be used as a suitable replacement to surface water temperature when necessary, it is recommended that surface water temperature be collected during V. parahaemolyticus monitoring when possible.
The interaction between air and tissue temperature for all three genetic markers implies that these associations between temperature and V. parahaemolyticus occur primarily in sufficiently warm conditions. In other words, the highest overall abundance of tlh and tdh will likely be found when air and tissue temperatures are both near their maxima, but the lowest overall abundance will not necessarily be found at their minima. The relationship between air, water, and tissue temperature is somewhat complex given the relatively higher heat capacity of water and the strong semidiurnal tides in PNW. When oysters are exposed to ambient air during the intertidal period, there is an opportunity for oyster tissue temperatures to rise considerably and for V. parahaemolyticus to flourish (Jones et al., 2016). While distances between sampling sites and shifts in tidal windows did not always ensure oysters used in this study were collected soon after the tide receded, the results indicate that the deviation between tissue and water temperature across samples was mostly negligible. Bacterium associations with tissue temperature would likely strengthen if samples were consistently taken later in the intertidal period. Regardless, these findings indicate that the highest average levels of abundance for many V. parahaemolyticus strains occur when multiple media have higher than average temperatures.
Despite moderate variation in environmental measures, there was little to no association in the current analysis between water salinity and tlh, trh, or tdh. This finding is consistent with previous research in PNW (Duan and Su, 2005; Paranjpye et al., 2015). The Salish Sea and Washington coastal bays are relatively saline and none of the harvesting waters sampled in this study approached freshwater levels. The unadjusted negative associations, primarily due to lower abundance at sampling sites and time periods with waters above 30‰, became non-significant after accounting for site temperatures. This is likely due to the fact that oceanic water entering into these estuarine systems is often cooler than freshwater and therefore less hospitable to V. parahaemolyticus bacteria (Davis et al., 2017). The statistical interaction identified for both tlh and tdh:tlh indicates that salinity modifies the relationship between total V. parahaemolyticus abundance and surface water temperature, such that more saline waters enable warmer waters to host higher overall bacterium abundance. This finding is consistent with the consensus that V. parahaemolyticus thrives in environments that are both warm and saline (Naughton et al., 2009; Davis et al., 2017).
A limitation of this study is that the implemented MPN-PCR analysis is only able to quantify the overall abundance of each targeted hemolysin gene in oyster tissue samples rather than specific strains of V. parahaemolyticus. The results also include samples that had late amplification (i.e., real-time PCR results above 40 cycles) and therefore may have been non-specific. Although enumeration was performed via the serial dilution test, non-specific amplification results may overestimate abundance of genetic markers. However, the occurrence of late amplification was uncommon, and therefore the impact of such measurement error on the reported findings is expected to be minimal.
An additional limitation of the current analysis is that trh-carrying V. alginolyticus strains may be common in PNW waters and therefore cast doubt upon the gene’s utility as a pathogenic marker of V. parahaemolyticus in the region (Gonzalez-Escalona et al., 2006). While Vibrio spp. are often correlated in environment isolates, the strong association between the two pathogenic markers in this study could also indicate the presence of tdh+ /trh+ strains of V. parahaemolyticus, or alternatively both tdh+ /trh- and tdh-/trh+ strains, rather than substantial abundance of trh+ V. alginolyticus. Nevertheless, it should be noted that there are certain WDOH survey sites where samples frequently result in enumerated trh but not tlh, indicating the presence of only V. alginolyticus (Washington Public Health Laboratory, Personal Communication). While the trh concentration in these instances is often low, the potential impact of V. alginolyticus contributions to reported trh values in other samples could be substantial and therefore must be further investigated. Additional microbial analysis, such as whole-genome sequencing, would be needed to accurately determine the extent of total trh abundance originating from V. alginolyticus in PNW oyster samples. However, regardless of its source, the strong association between tdh and trh, such that its inclusion negated the relationship between tdh and tlh, suggests that the presence of the tdh gene is more dependent on the level of trh than the total abundance of V. parahaemolyticus in the environment.
The exponential relationship between the tdh:tlh pathogenic ratio and total abundance of V. parahaemolyticus, as well as its negative association with water temperature, confirm previous findings observed in Eastern oysters (Crassostrea virginica) in Alabama (DePaola et al., 2003). The results of the current study, which includes a larger sample size, expand upon this original finding by showing that the log-linear relationship is systematic and does not vary by sampling region or year. The consistency of these relationships in a different oyster species, decade, and region of North America suggests that they may be ubiquitous throughout the environment. Given the statistical redundancy of including the tlh gene in a regression model of tdh:tlh, especially for the seemingly unrelated estimation technique used in this analysis, it was impossible to formally evaluate if the negative association between water temperature and the ratio is fully explained by the relationship between temperature and total V. parahaemolyticus abundance. Regardless, these findings suggest that tdh+ strains of V. parahaemolyticus are more likely to be prominent in the total bacterium community in relatively cooler waters. The Food and Drug Administration’s risk model assumed that the ratio between pathogenic and total V. parahaemolyticus in oysters was temperature independent (USFDA, 2005). Therefore, an update to the model to account for temperature-dependent pathogenic ratio variation may improve risk predictions.
Certain strains of V. parahaemolyticus endemic to PNW that contain the tdh marker have been found to be more resilient to competitive environmental conditions (Matz et al., 2011; Nilsson et al., 2019). These findings suggest that cooler waters may provide an opportunity for tdh+ strains of V. parahaemolyticus to better establish itself among the bacterial community. The general resilience of these strains to cooler waters is of great interest, as most V. parahaemolyticus harvesting regulations assume that warmer temperatures result in greater risk of harvested oysters (USFDA, 2017). While previous work has identified only a marginal correlation between illness rates and tdh+ concentrations (Paranjpye et al., 2012), little attention has been given to the relative proportion of these strains in the environment. It may be that lower total V. parahaemolyticus abundance and/or cooler water temperatures provide a unique opportunity for pathogenic strains to cause infections in oyster consumers.
The marginal positive association between tissue temperature and the tdh:tlh pathogenic ratio further suggests that sudden changes in temperature may allow pathogenic strains the opportunity to establish themselves in the V. parahaemolyticus community. Washington State waters are generally cool given their distance from the equator. However, due to recent changes in ocean circulation, warmer waters more frequently enter into the region’s estuarine systems, which can allow Vibrio spp. to flourish (Martinez-Urtaza et al., 2010). A similar pattern may occur on a smaller time-scale with regard to the semidiurnal tides in PNW, where sudden warming of the surfaced oysters allows tdh+ strains to increase proportionately. While comparisons of genetic markers in oysters during high and low tide have been conducted (Jones et al., 2016), the study did not specifically assess the ratio of tdh to total abundance within samples. A similar study or re-analysis of the generated data is therefore suggested.
The moderate negative association between the tdh:tlh pathogenic ratio and total trh indicates that when tdh+ strains make up a high proportion of the local bacterium community, they may be more likely to be tdh+ /trh- strains. These strains are relatively common in the PNW environment, particularly the pandemic O3:K6 strain, although its role in vibriosis cases from consuming PNW oysters is unclear (Paranjpye et al., 2012; Turner et al., 2013; Nilsson et al., 2019). Future research evaluating the association between tdh:tlh and reported vibriosis cases in the region may help determine its utility as an indicator of the risk of foodborne illness. Such findings could also be relevant for other parts of North America given the spread of PNW strains in recent years (Martinez-Urtaza et al., 2013; Xu et al., 2017).
Seemingly unrelated estimations, while commonly applied in other scientific fields (Fiebig, 2001), has, to the authors’ knowledge, not previously been used in microbial research. Its implementation in this work allowed the regression analysis to preserve the censored values of both the numerator and denominator of the tdh:tlh ratio resulting from the limits of detection of the MPN-PCR technique. While most of the censored data from the pathogenic ratio were due to the high proportion of tdh measurements below the limit of detection, exploratory statistical analysis indicated that the corresponding values for tlh varied across the entire range of observable data. A more simplified approach, such as treating all aforementioned ratio measures as left-censored, would therefore have introduced bias. The seemingly unrelated estimation of the separate Tobit models was a preferable alternative, allowing for the linear relationships between the latent Tobit variables and regression covariates to be simultaneously compared for the numerator and denominator of the ratio. The technique applied in this work is unbiased when the residuals of the two regressions are uncorrelated. If this had not been the case, a more complex approach, such as SUR for Tobit regressions (Arouna and Dabbert, 2010), would have been more appropriate. Seemingly unrelated statistical techniques should be considered in future microbial research that examines ratios of different genetic markers, particularly when data are censored.
The simple imputation for censored genetic data implemented in this work is likely why the log-transformed tdh variable appeared positively skewed and in general may have biased reported correlations and regression associations toward the null. This statistical bias is unlikely to impact total V. parahaemolyticus abundance given the low rate of censoring for tlh. However, trh and particularly tdh may be prone to this bias. While a stochastic imputation following the lognormal distributions of the genetic variables could have been implemented as an alternative approach (Wei et al., 2018), this statistical technique seemed unnecessary as the only non-significant relationship between genetic markers was for the tlh covariate in the tdh model after adjusting for trh. Therefore, such an imputation would not be expected to substantially alter the results of the current analysis.
The environmental findings reported in this study arise from one of the largest samples of oysters enumerated for V. parahaemolyticus ever collected. To the authors’ knowledge, it is the largest dataset of its kind available for the PNW and covers a wide array of growing areas in Washington State. The strength of the reported associations and the lack of effect modification by harvesting region increase the likelihood that these findings are generalizable to Washington State and likely other shellfish harvesting waters in the PNW. However, it is also likely that the temperature and salinity associations with the genetic markers reported here are unique to this region of North America, as other research has shown consistently linear positive relationships with water temperature and complex non-linear relationships with salinity in other bodies of water (DePaola et al., 2003; USFDA, 2005; Johnson, 2015; Davis et al., 2017). It is recommended that similar analyses be conducted on different oyster samples in PNW to evaluate the consistency of the current findings across this region.
While the associations reported in this work are noteworthy, additional environmental information, such as water clarity and quality (e.g., turbidity, oxygenation, presence of nutrients, plankton abundance) as well as watershed precipitation and land use, was not readily available for analysis. This missing information may explain some of the associations observed in this work, particularly the residual spatial and temporal variation in the models (Paranjpye et al., 2015; Davis et al., 2017; Nilsson et al., 2019). Therefore, the current results from the multivariate models should be interpreted with caution. Future work can address this limitation by drawing from additional environmental datasets in Washington State and using spatial–temporal statistical techniques to predict these environmental measures at the same place and time as oyster sampling. Similar statistical techniques could also be used to account for potential similarities across oyster samples based upon time of year and specific sampling coordinates. Characterizing these space–time statistical dependencies in a regression framework may further modify the associations observed in this work. These aforementioned models will be utilized in future work.
Conclusion
Abundance of total V. parahaemolyticus and potentially pathogenic strains in Pacific oysters from Washington State were found to have clear positive associations with a variety of measured temperatures and with each other. The relative abundance of tdh+ strains was negatively associated with water temperature, total V. parahaemolyticus, and trh+ strains. This work further characterizes the environmental determinants of the bacterium including potentially pathogenic strains in Pacific oysters. The inclusion of tissue and air temperature, as well as accounting for the non-linear trend in surface water temperature, will also likely improve V. parahaemolyticus estimations used for shellfish safety risk management. These findings lay the groundwork for future characterizations of the bacterium in the region that will attempt to demonstrate how pre-harvest variation relates to the risk of foodborne illness among oyster consumers.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
AF implemented the data analysis and drafted the original manuscript for this work. BD mentored AF, conceived and designed the study, and edited analysis and writing as needed. EA and GO represent the Washington State Department of Health, which collected all oyster samples and environmental measures, conducted microbial analysis, and performed quality assurance and quality control on the dataset. JB assisted in the design and implementation of the regression analysis. AD assisted with study design, writing, and interpretation of the findings. FC advised all authors throughout, specifically supervising the conceptual design, data analysis, and manuscript writing.
Conflict of Interest
AD is a retired USFDA employee, now sole proprietor consultant of Angelo DePaola Consulting.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by the National Institute of Allergy and Infectious Diseases through the grant “Characterizing the Spatial Temporal Dynamics and Human Health Risks of Vibrio parahaemolyticus Bacteria in Estuarine Environments” (PI: Curriero, 1R01AI123931–01A1). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgments
The authors would like to thank Lawrence Sullivan, Laura Johnson, and Clara Hard from the Washington State Department of Health for their collaboration and assistance in coordinating and cleaning the Vibrio oyster monitoring data. Additional thanks are given to the many summer interns who conducted the oyster sampling over the years. The authors are indebted to all the staff and students at the Washington State Public Health Laboratory, particularly William Glover for designing the shellfish microbial methodology. The authors would also like to thank Rohinee Paranjpye and Linda Rhodes at the Northwest Fisheries Science Center for their guidance and support. Finally, the authors would like to thank Anne Corrigan from the Spatial Science for Public Health Center for helping to design the map displayed in this paper.
References
Ansaruzzaman, M., Lucas, M., Deen, J. L., Bhuiyan, N. A., Wang, X. Y., Safa, A., et al. (2005). Pandemic serovars (O3:K6 and O4:K68) of Vibrio parahaemolyticus associated with diarrhea in Mozambique: spread of the pandemic into the African continent. J. Clin. Microbiol. 43, 2559–2562. doi: 10.1128/JCM.43.6.2559-2562.2005
APHA (1970). Recommended Procedures for the Examination of Sea Water and Shellfish. New York, NY: American Public Health Assocation.
Arouna, A., and Dabbert, S. (2010). Determinants of domestic water use by rural households without access to private improved water sources in Benin: a seemingly unrelated Tobit approach. Water Resour. Manage. 24, 1381–1398. doi: 10.1007/s11269-009-9504-4
Baker-Austin, C., Trinanes, J., Gonzalez-Escalona, N., and Martinez-Urtaza, J. (2017). “Non-cholera vibrios: the microbial barometer of climate change,” in Trends Microbiol., 25, 76–84. doi: 10.1016/j.tim.2016.09.008
Blodgett, R. (2017). “Most probable number determination from serial dilutions,” in Bacteriological Analytical Manual (BAM), eds S. Chirtel and Z. Guodong (College Park, MD: USFDA). Available at: www.fda.gov/food/laboratory-methods-food/bam-appendix-2-most-probable-number-serial-dilutions
Breusch, T. S., and Pagan, A. R. (1980). The Lagrange multiplier test and its applications to model-specification in econometrics. Rev. Econ. Stud. 47, 239–253. doi: 10.2307/2297111
CDC (1998). Outbreak of Vibrio parahaemolyticus infections associated with eating raw oysters–Pacific Northwest, 1997. Morb. Mortal. Weekly Rep. 47, 457–462.
Davis, B. J. K., Jacobs, J. M., Davis, M. F., Schwab, K. J., DePaola, A., and Curriero, F. C. (2017). Environmental determinants of Vibrio parahaemolyticus in the Chesapeake Bay. Appl. Environ. Microbiol. 83:e1147-17. doi: 10.1128/AEM.01147-17
DePaola, A., Nordstrom, J. L., Bowers, J. C., Wells, J. G., and Cook, D. W. (2003). Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69, 1521–1526. doi: 10.1128/aem.69.3.1521-1526.2003
Duan, J. Y., and Su, Y. C. (2005). Occurrence of Vibrio parahaemolyticus in two Oregon oyster-growing bays. J. Food Sci. 70, M58–M63. doi: 10.1111/j.1365-2621.2005.tb09047.x
ESRI (2019a). ArcGIS Desktop: ArcGIS Pro Release 2.32. Redlands, CA: Environmental Systems Research Institute.
ESRI (2019b). Garmin, © OpenStreetMap Contributors, and the GIS User Community World Light Gray Base. Redlands, CA: ESRI.
Fiebig, D. G. (2001). “Seemingly unrelated regression,” in A Companion to Theoretical Econometrics, ed. B. H. Baltagi, (Malden, MA: Blackwell Publishing), 101–121. doi: 10.1002/9780470996249.ch6
Fujino, T., Okuno, Y., Nakada, D., Aoyama, A., Fukai, K., Mukai, T., et al. (1953). On the bacteriological examination of shirasu-food poisoning. Med. J. Osaka Univ. 4, 299–304.
Glover, W. A. II (2015). Vibrio parahaemolyticus Enumeration and Detection Through MPN and Real-Time PCR. Columbia, SC: Interstate Shellfish Sanitation Conference.
Gonzalez-Escalona, N., Blackstone, G. M., and DePaola, A. (2006). Characterization of a Vibrio alginolyticus strain, isolated from Alaskan oysters, carrying a hemolysin gene similar to the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 72, 7925–7929. doi: 10.1128/AEM.01548-06
Gonzalez-Escalona, N., Gavilan, R. G., Toro, M., Zamudio, M. L., and Martinez-Urtaza, J. (2016). Outbreak of Vibrio parahaemolyticus sequence type 120, Peru, 2009. Emerg. Infect. Dis. 22, 1235–1237. doi: 10.3201/eid2207.151896
Gonzalez-Escalona, N., Martinez-Urtaza, J., Romero, J., Espejo, R. T., Jaykus, L. A., and DePaola, A. (2008). Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190, 2831–2840. doi: 10.1128/Jb.01808-07
Haendiges, J., Jones, J., Myers, R. A., Mitchell, C. S., Butler, E., Toro, M., et al. (2016). A nonautochthonous U.S. strain of Vibrio parahaemolyticus isolated from Chesapeake Bay oysters caused the outbreak in Maryland in 2010. Appl. Environ. Microbiol. 82, 3208–3216. doi: 10.1128/AEM.00096-16
Hazen, T. H., Pan, L., Gu, J. D., and Sobecky, P. A. (2010). The contribution of mobile genetic elements to the evolution and ecology of Vibrios. FEMS Microbiol. Ecol. 74, 485–499. doi: 10.1111/j.1574-6941.2010.00937.x
Johnson, C. N. (2015). Influence of environmental factors on Vibrio spp. in coastal ecosystems. Microbiol. Spectr. 3, 1–18. doi: 10.1128/microbiolspec.VE-0008-2014
Jones, J. L., Kinsey, T. P., Johnson, L. W., Porso, R., Friedman, B., Curtis, M., et al. (2016). Effects of intertidal harvest practices on levels of Vibrio parahaemolyticus and Vibrio vulnificus bacteria in oysters. Appl. Environ. Microbiol. 82, 4517–4522. doi: 10.1128/AEM.00721-16
Kirs, M., Depaola, A., Fyfe, R., Jones, J. L., Krantz, J., Van Laanen, A., et al. (2011). A survey of oysters (Crassostrea gigas) in New Zealand for Vibrio parahaemolyticus and Vibrio vulnificus. Int. J. Food Microbiol. 147, 149–153. doi: 10.1016/j.ijfoodmicro.2011.03.012
Martinez-Urtaza, J., Baker-Austin, C., Jones, J. L., Newton, A. E., Gonzalez-Aviles, G. D., and DePaola, A. (2013). Spread of Pacific Northwest Vibrio parahaemolyticus strain. N. Engl. J. Med. 369, 1573–1574. doi: 10.1056/NEJMc1305535
Martinez-Urtaza, J., Bowers, J. C., Trinanes, J., and DePaola, A. (2010). Climate anomalies and the increasing risk of Vibrio parahaemolyticus and Vibrio vulnificus illnesses. Food Res. Int. 43, 1780–1790. doi: 10.1016/j.foodres.2010.04.001
Martinez-Urtaza, J., Powell, A., Jansa, J., Rey, J. L., Montero, O. P., Campello, M. G., et al. (2016). Epidemiological investigation of a foodborne outbreak in Spain associated with U.S. West Coast genotypes of Vibrio parahaemolyticus. Springerplus 5:87. doi: 10.1186/s40064-016-1728-1
Matz, C., Nouri, B., McCarter, L., and Martinez-Urtaza, J. (2011). Acquired type III secretion system determines environmental fitness of epidemic Vibrio parahaemolyticus in the interaction with bacterivorous protists. PLoS One 6:e20275. doi: 10.1371/journal.pone.0020275
McLaughlin, J. B., DePaola, A., Bopp, C. A., Martinek, K. A., Napolilli, N. P., Allison, C. G., et al. (2005). Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N. Engl. J. Med. 353, 1463–1470. doi: 10.1056/NEJMoa051594
National Marine Fisheries Service Office of Science and Technology, and Fisheries Statistics Division (2017). Commercial Fisheries Statistics: Annual Commercial Landings Statistics. Silver Spring, MD: NOAA.
Naughton, L. M., Blumerman, S. L., Carlberg, M., and Boyd, E. F. (2009). Osmoadaptation among Vibrio species and unique genomic features and physiological responses of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 75, 2802–2810. doi: 10.1128/AEM.01698-08
Nilsson, W. B., Paranjpye, R. N., Hamel, O. S., Hard, C., and Strom, M. S. (2019). Vibrio parahaemolyticus risk assessment in the Pacific Northwest: it’s not what’s in the water. FEMS Microbiol. Ecol. 95:fiz027. doi: 10.1093/femsec/fiz027
Panicker, G., Call, D. R., Krug, M. J., and Bej, A. K. (2004). Detection of pathogenic Vibrio spp. in shellfish by using multiplex PCR and DNA microarrays. Appl. Environ. Microbiol. 70, 7436–7444. doi: 10.1128/AEM.70.12.7436-7444.2004
Paranjpye, R., Hamel, O. S., Stojanovski, A., and Liermann, M. (2012). Genetic diversity of clinical and environmental Vibrio parahaemolyticus strains from the Pacific Northwest. Appl. Environ. Microbiol. 78, 8631–8638. doi: 10.1128/AEM.01531-12
Paranjpye, R. N., Nilsson, W. B., Liermann, M., Hilborn, E. D., George, B. J., Li, Q., et al. (2015). Environmental influences on the seasonal distribution of Vibrio parahaemolyticus in the Pacific Northwest of the USA. FEMS Microbiol. Ecol. 91:fiv121. doi: 10.1093/femsec/fiv121
R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Raghunath, P. (2014). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. doi: 10.3389/fmicb.2014.00805
Salomon, D., Gonzalez, H., Updegraff, B. L., and Orth, K. (2013). Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One 8:e61086. doi: 10.1371/journal.pone.0061086
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Shirai, H., Ito, H., Hirayama, T., Nakamoto, Y., Nakabayashi, N., Kumagai, K., et al. (1990). Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58, 3568–3573.
Su, Y. C., and Liu, C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558. doi: 10.1016/j.fm.2007.01.005
Turner, J. W., Malayil, L., Guadagnoli, D., Cole, D., and Lipp, E. K. (2014). Detection of Vibrio parahaemolyticus, Vibrio vulnificus and Vibrio cholerae with respect to seasonal fluctuations in temperature and plankton abundance. Environ. Microbiol. 16, 1019–1028. doi: 10.1111/1462-2920.12246
Turner, J. W., Paranjpye, R. N., Landis, E. D., Biryukov, S. V., Gonzalez-Escalona, N., Nilsson, W. B., et al. (2013). Population structure of clinical and environmental Vibrio parahaemolyticus from the Pacific Northwest coast of the United States. PLoS One 8:e55726. doi: 10.1371/journal.pone.0055726
USFDA (2005). Quantitative Risk Assessment on the Public Health Impact of Pathogenic Vibrio parahaemolyticus In Raw Oysters. College Park, MD: USFDA.
USFDA (2017). National Shellfish Sanitation Program (NSSP), Guide for the Control of Molluscan Shellfish. College Park, MD: USFDA.
Velazquez-Roman, J., Leon-Sicairos, N., de Jesus Hernandez-Diaz, L., and Canizalez-Roman, A. (2014). Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front. Cell. Infect. Microbiol. 3:110. doi: 10.3389/fcimb.2013.00110
Washington Disease Control and Health Statistics (2018). Washington State Communicable Disease Report 2017. Shoreline, WA: Washington Department of Health.
Washington State Board of Health (2015). Washington State Vibrio parahaemolyticus Control Plan in WAC 246-282-006. Washington, DC: Washington Administrative Code.
Weesie, J. (1999). Seemlingly unrelated estimation and the cluster-adjusted sandwich estimator. Stata Tech. Bull. 9, 34–47.
Wei, R., Wang, J., Su, M., Jia, E., Chen, S., Chen, T., et al. (2018). Missing value imputation approach for mass spectrometry-based metabolomics data. Sci. Rep. 8:663. doi: 10.1038/s41598-017-19120-0
Wu, Y., Wen, J., Ma, Y., Ma, X., and Chen, Y. (2014). Epidemiology of foodborne disease outbreaks caused by Vibrio parahaemolyticus, China, 2003–2008. Food Control 46, 197–202. doi: 10.1016/j.foodcont.2014.05.023
Xu, F., Gonzalez-Escalona, N., Drees, K. P., Sebra, R. P., Cooper, V. S., Jones, S. H., et al. (2017). Parallel evolution of two clades of a major Atlantic endemic Vibrio parahaemolyticus pathogen lineage by independent acquisition of related pathogenicity islands. Appl. Environ. Microbiol. 83:e1168-17. doi: 10.1128/AEM.01168-17
Zellner, A. (1962). An efficient method of estimating seemingly unrelated regressions and tests for aggregation bias. J. Am. Stat. Assoc. 57, 348–368. doi: 10.1080/01621459.1962.10480664
Zellner, A. (1963). Estimators for seemingly unrelated regression equations: some exact finite sample results. J. Am. Stat. Assoc. 58, 977–992. doi: 10.1080/01621459.1963.10480681
Keywords: Vibrio parahaemolyticus, Crassostrea gigas, Pacific oysters, Washington, temperature, genetic markers, seafood-borne illness
Citation: Flynn A, Davis BJK, Atherly E, Olson G, Bowers JC, DePaola A and Curriero FC (2019) Associations of Environmental Conditions and Vibrio parahaemolyticus Genetic Markers in Washington State Pacific Oysters. Front. Microbiol. 10:2797. doi: 10.3389/fmicb.2019.02797
Received: 21 July 2019; Accepted: 18 November 2019;
Published: 04 December 2019.
Edited by:
Min Suk Rhee, Korea University, South KoreaReviewed by:
Adrian Canizalez-Roman, Autonomous University of Sinaloa, MexicoRachel Todd Noble, The University of North Carolina at Chapel Hill, United States
Copyright © 2019 Flynn, Davis, Atherly, Olson, Bowers, DePaola and Curriero. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benjamin J. K. Davis, QmRhdmlzNjRAamhtaS5lZHU=; Frank C. Curriero, ZmN1cnJpZXJvQGpodS5lZHU=
 Aspen Flynn1
Aspen Flynn1 Benjamin J. K. Davis
Benjamin J. K. Davis Erika Atherly
Erika Atherly John C. Bowers
John C. Bowers