- 1Department of Microbiology and Cell Biology, Indian Institute of Science, Bangalore, India
- 2International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b), Dhaka, Bangladesh
- 3Pathogen Biology Laboratory, Department of Biotechnology and Bioinformatics, University of Hyderabad, Hyderabad, India
- 4Methodology and Research Infrastructure, Robert Koch Institute, Berlin, Germany
- 5International University of Health and Welfare, Tokyo, Japan
Extended-spectrum β-lactamases (ESBLs) form the most important resistance determinants prevalent worldwide. Data on ESBL-producing Escherichia coli from poultry and livestock are scarce in India. We present data on the functional and genomic characterization of ESBL-producing E. coli obtained from poultry in India. The whole genome sequences of 28 ESBL-producing E. coli were analyzed comprising of 12 broiler chicken E. coli isolates, 11 free-range chicken E. coli isolates, and 5 human extraintestinal pathogenic E. coli. All of the 28 ESBL-producing E. coli isolates were tested for antibiotic susceptibilities, in vitro conjugation, and virulence-associated phenotypic characteristics. A total of 13 sequence types were identified from the poultry E. coli, which included globally successful sequence types such as ST117 (9%), ST131 (4.3%), and ST10 (4.3%). The most common ESBL gene detected in poultry E. coli genomes was blaCTX-M-15 (17%). Also, FIB (73%) and FII (73%) were the most common plasmid replicons identified. Conjugation experiments demonstrated 54 (7/13), 30 (3/10), and 40% (2/5) of broiler, free-range, and human ExPEC E. coli to be able to transfer their ESBL genes, respectively. The in vitro virulence-associated phenotypic tests revealed the broiler, free-range, and human ExPEC isolates to be comparable in biofilm formation, resistance to serum bactericidal activity, adherence, and invasion capabilities. Our overall results showed prevalence of virulence phenotypes among the diverse ESBL-producing E. coli from poultry; while certain E. coli clones from broiler-poultry may indeed have the potential to cause infection in humans.
Introduction
Extended-spectrum-β-lactamases (ESBLs)-producing Enterobacteriaceae have become a serious problem worldwide adversely impacting the treatment of infectious diseases (Miyoshi-Akiyama et al., 2016). Infections with ESBL-producing Escherichia coli are associated with a range of conditions resulting in increased morbidity, mortality, and healthcare costs (Tumbarello et al., 2010). The prevalence of E. coli resistant to extended-spectrum-β-lactam antibiotics is highest among developing countries (Byarugaba, 2004). In India, high levels of cephalosporin resistance have been reported (16–95%) in E. coli following extensive use of ceftriaxone (Miyoshi-Akiyama et al., 2016). ESBLs are beta-lactamases that confer resistance to penicillins, third-generation cephalosporins, and monobactams (Rawat and Nair, 2010). Restricted treatment options due to the emergence of multi-drug resistant phenotypes in ESBL-producing bacteria have led to increased dependence on and subsequent increase in the prevalence of E. coli resistant to last-line antibiotics such as carbapenems (Ranjan et al., 2016).
Although ESBL production was initially reported in hospital-acquired infections caused mainly by Klebsiella pneumoniae, currently, its associations with community-acquired infections, particularly with urinary tract infections caused by E. coli have been frequently reported (Paterson and Bonomo, 2005). The commensal E. coli in the fecal carriage are thought to be one of the main reservoirs of ESBL-E. coli in the community settings (Kluytmans et al., 2013). Scenarios involving transmission of ESBL-producing E. coli in food animals and humans have been reported in several studies (Kluytmans et al., 2013; Mitchell et al., 2015; Robinson et al., 2016). We recently reported ESBL-producing E. coli in retail chicken meat in India, wherein close resemblance of genotypes was also identified between E. coli from chicken meat and human ExPEC pathotypes (Hussain et al., 2017). Several studies have suggested that E. coli from poultry represents significant zoonotic potential (Kluytmans et al., 2013; Mitchell et al., 2015; Robinson et al., 2016; Hussain et al., 2017). Molecular determinants of ESBLs are frequently found on plasmids that could also harbor virulence genes and these can be horizontally transferred to other bacteria, even to bacteria of other genera (Skyberg et al., 2006; Hussain et al., 2012). Presence of virulence factors in ESBL-producing E. coli is an important factor in the epidemiology of drug-resistant infections (Johnson et al., 2006; Jadhav et al., 2011), as exposure to ESBL-producing pathogenic E. coli strains may cause hard-to-treat infections, also in healthy individuals.
Worldwide, significantly large geographic differences are observed in the prevalence of ESBL genes. However, blaCTX-M types and in particular, the blaCTX-M-15 are now the predominant and the most extensively disseminated ESBL genotypes found among E. coli isolates (Hussain et al., 2014; Ranjan et al., 2015, 2017). The ESBL-producing E. coli are also predominantly linked with globally disseminating ST131 clones and are frequently associated with fluoroquinolone resistance (Hussain et al., 2014; Nandanwar et al., 2016; Shaik et al., 2017). Whole genome sequencing (WGS) is an important tool in analyzing the emergence and spread of antimicrobial resistant isolates, and therefore, may help in formulating strategies in controlling antimicrobial resistance.
Although studies have investigated the epidemiology of ESBL-producing E. coli of poultry origins, which may be directly linked to public health (Mellata, 2013; Mitchell et al., 2015; Hussain et al., 2017), the aim of this study is to further obtain a comprehensive understanding of the differences and or similarities in genomic and functional attributes of ESBL-producing E. coli obtained from free-range (country chicken) and farm-raised (broiler) poultry in India by integrating new data regarding phenotypic virulence traits, extensive antimicrobial resistance screening, whole genome sequencing, and comparative genomics. In this study, we identified prevalence of virulent phenotypes among diverse ESBL-E. coli lineages from poultry. Particularly, the whole genome sequencing revealed that the broiler E. coli isolates shared affinities with human ExPEC isolates with regard to virulence, resistance, and phylogenetic backgrounds.
Materials and Methods
Strain Collection
A total of 28 ESBL-producing E. coli isolates were analyzed in this study that included; 23 ESBL E. coli isolates that were randomly selected comprising of 11 free-range E. coli and 12 broiler E. coli, which were a part of an earlier described collection of 168 poultry E. coli from retail chicken meat and carcass isolated from three cities representing three states in India during February 2015 – September 2015 (Hussain et al., 2017). The remaining five ESBL-producing-E. coli used in this study were human ExPEC isolates, which were part of the previously described reports from India (Avasthi et al., 2011; Ranjan et al., 2016; Shaik et al., 2017). Details of all the strains employed in this study are tabulated in Supplementary Table S1. Broiler chicken represents farm-raised broiler chicken category that was fed on commercial feed and raised for meat production; the free-range chicken represents the country (native) birds that were reared in backyard farms and households (not raised in farms).
Whole Genome Sequencing, Phylogenomic Analysis, and Genetic Characterization
Genomic DNA from 13 strains (randomly selected ESBL-E. coli respresenting three geographical locations: Karnataka, Andhra Pradesh and Telangana) was isolated using the Qiagen DNeasy blood and tissue kit (Qiagen, Germany) (Qumar et al., 2017). Illumina MiSeq reads of 13 strains (Supplementary Table S1) were filtered for high quality reads using Trimmomatic (v0.36) (Bolger et al., 2014) and NGS QC Toolkit (v2.3.3) (Patel et al., 2012) and the reads of three Free-range E. coli strains (NAEC1123, NAEC1145, and NAEC1148) were excluded due to poor quality. The high quality reads of 10 strains were then assembled de novo using SPAdes Genome Assembler (v3.6.1) (Bankevich et al., 2012) and further ordered and scaffolded using Contig-Layout-Authenticator (Shaik et al., 2016). The ordered scaffolds were then annotated using RAST server (Overbeek et al., 2014) and the statistics were gleaned using ARTEMIS (Rutherford et al., 2000). The genomic features of 10 poultry E. coli genomes are represented in Supplementary Table S3.
In addition to these newly sequenced 10 genomes (8 Free-range and 2 Broiler), previously studied 10 ESBL E. coli genomes from broiler and 5 genomes from human hosts (with an inclusion criteria of >4,500 CDS and above 80% coding percentage) were also included for the entire analysis (Supplementary Table S1). The sequence type (ST) and plasmid profiles of all the 25 strains were determined in silico.1 Phylogroups of E. coli genomes were determined by an in silico Clermont typing method (Beghain et al., 2018). A Harvest (Treangen et al., 2014) based core-genome phylogenetic tree of 25 E. coli strains was generated by filtering the possible recombining regions, poor alignments, repetitive regions, and the resulting tree was visualized using iTOL (Letunic and Bork, 2016). Virulence and resistance gene screening of these 25 strains was carried out by BLASTp (70% identity and 90% query coverage) analysis against homologous genes (not entire ORFs) present in the Virulence Factors Database (VFDB) and Comprehensive Antibiotic Resistance Database (CARD), respectively. Heatplots depicting the hierarchical clustering were executed in the gplot package of R as described by our group, previously (Ranjan et al., 2016).
Biofilm Formation
Biofilm production was analyzed in flat-bottom 96-well microtiter plates. The overnight LB culture was diluted to optical density 0.05 (OD 600 nm) in fresh M63 minimal medium, and 200 μl of this dilution was added into the wells in triplicates and covered by a breathable sealing and incubated for 48 h at 28°C in a stationary condition. Biofilms were quantified after 48 h of incubation using crystal violet staining, the optical density of the well containing only bacterium-free medium was used as a control. The absorbance was measured at 570 nm. All tests were carried out at least three times, and the results were averaged. Specific biofilm formation was computed using formula specific biofilm formation (SBF) = (AB − CW)/G, where AB = OD 570 nm of the attached and stained bacteria, CW = OD 570 nm of the stained control wells containing only bacteria-free medium, and G = OD 600 nm of bacterial growth in broth immediately after 48 h incubation (Hussain et al., 2014).
Serum Resistance, Adhesion, and Invasion Assays
For serum assay, overnight grown bacterial cultures (5 μl) were added to LB broth (495 μl) and incubated for 1 h in a shaker incubator. These bacterial cultures were then centrifuged, and the pellets were suspended in 1 ml of 1X PBS. About 30 μl of this bacterial suspension was added to 270 μl of 50% (diluted in PBS) human serum (Pan Biotech, Germany) in triplicates in a 96-well microtiter plate. Initially, samples were collected at 0 h, serially diluted in PBS, and plated on LB agar plates for enumerating the colony forming units. The microtiter plate was then incubated for 3 h at 37°C at 100 rpm. After 3 h, samples from each well were aliquoted for serial dilution and plated on LB agar plates. Strains which had equaled or higher colony forming units (CFU) at 3 h compared to 0 h were considered resistant to human serum. The experiment was repeated twice in three technical replicates (Ranjan et al., 2016; Suresh et al., 2018).
Adhesion and invasion assays were performed on T24, human bladder epithelial cells as previously reported (Ranjan et al., 2016). In brief, human bladder epithelial cells were grown in RPMI-1640 in 24 well-plates 48 h prior to cell infection. Following 3 h of infection at 37°C with MOI of 10, the monolayer was washed thrice (using 1X PBS) and lysed (using 0.1% Triton X-100 in 1X PBS). The lysates were serially diluted and plated on LB agar. For the invasion assay, the cells were incubated additionally for 1.5 h with replaced media containing 100 μg/ml gentamicin. The cells were then processed as described for the adhesion assay.
Antimicrobial Susceptibility and Conjugation Assay
Antimicrobial susceptibility of 28 strains (23 poultry and 5 ExPEC strains) against 11 different antibiotics was carried out on Mueller-Hinton agar (MHA) medium using Kirby-Bauer disc diffusion employing following antibiotic discs: colistin (10 μg), tetracycline (30 μg), chloramphenicol (30 μg), oxytetracycline (30 μg), nalidixic acid (30 μg), streptomycin (10 μg), fosfomycin (50 μg), chlortetracycline (30 μg), cotrimoxazole (25 μg), gentamicin (120 μg), and ciprofloxacin (5 μg). All isolates were tested for MBL production employing imipenem with and without EDTA Ezy MIC Strips (HiMedia, India). Results were interpreted as per the criteria of the Clinical and Laboratory Standards Institute (CLSI) (Ranjan et al., 2016). Conjugative transfer of ESBL genes among ESBL producing poultry E. coli was analyzed by in vitro conjugation as previously described by our study group, transconjugants were selected on EMB agar containing 100 μg/ml sodium azide and 4 μg/ml cefotaxime (Hussain et al., 2014).
Statistical Analysis
Statistical analysis for aggregate resistance and virulence gene scores (sum of all resistance/virulence genes for which the isolates were tested positive, respectively) biofilm formation, serum resistance assay, adhesion, and invasion assays were done using the non-parametric Mann-Whitney U-test executed in GraphPad Prism.
Accession Number(s)
The GenBank accession numbers of the 10 genomes sequenced in this study are RDBX00000000 (NAEC1028), RDBY00000000 (NAEC1042), RDBZ00000000 (NAEC1115), RDCA00000000 (NAEC1132), RDCB00000000 (NAEC1134), RDCC00000000 (NAEC1135), RDCD00000000 (NAEC1136), RDCE00000000 (NAEC1146), RDCF00000000 (NAEC1147), and RDCG00000000 (NAEC1154).
Results
Bacterial Isolates
In total, 28 E. coli isolates were analyzed in this study that comprised of 5 human ExPEC, 12 broiler E. coli, and 11 free-range E. coli isolates. All five human ExPEC isolates were originated from urine samples of symptomatic UTI patients, they were collected from Pune city, Maharashtra. Broiler E. coli isolates includes 10 E. coli obtained from cecum (representing 6 isolates from Telangana, 2 from Karnataka, and 2 from Andhra Pradesh) and 2 from chicken meat (from Telangana). Free-range E. coli isolates include six isolates from cecum (representing 4 from Karnataka and 2 from Andhra Pradesh) and five from chicken meat (representing 3 from Telangana, 1 from Karnataka, and 1 from Andhra Pradesh). All the 28 E. coli isolates were tested for the in vitro associated virulence tests. Out of these, only 25 E. coli isolates were used for genomic analysis as three free-range genomes were excluded due to poor quality reads (Supplementary Table S1).
Antimicrobial Susceptibility
A total of 28 ESBL-producing E. coli isolates were screened for their antimicrobial susceptibilities to 11 antibiotics. Disc diffusion-based antimicrobial susceptibility of 11 antibiotics showed high levels of resistance among all tested E. coli isolates irrespective of their host origins (Figure 1). The isolates representing three groups showed comparable resistance rate (100%) towards tetracycline and quinolones class of antibiotics. Free-range E. coli and broiler E. coli isolates, including the human ExPEC, did not differ significantly in resistance with respect to any of the particular antibiotics tested. However, overall resistance scores (resistance to all the 11 antibiotics) only differed marginally among the three groups of E. coli. However, 78.5% of the E. coli isolates analyzed in this study exhibited a multidrug-resistant (MDR) phenotype. Among the 28 ESBL-positive poultry E. coli isolates which were tested for in vitro conjugation, only 12 E. coli isolates (comprising 7 broiler chicken E. coli, 3 free-range chicken E. coli, and 2 ExPEC) successfully transferred cefotaxime resistance to the recipient strain E. coli-J53. The transconjugants showed resistance to one or two antibiotics (mostly to ciprofloxacin and cotrimoxazole) in addition to beta-lactams, but none of the transconjugants was found to be resistant to all the antibiotics to which their parental E. coli isolates were tested resistant (Supplementary Table S2).
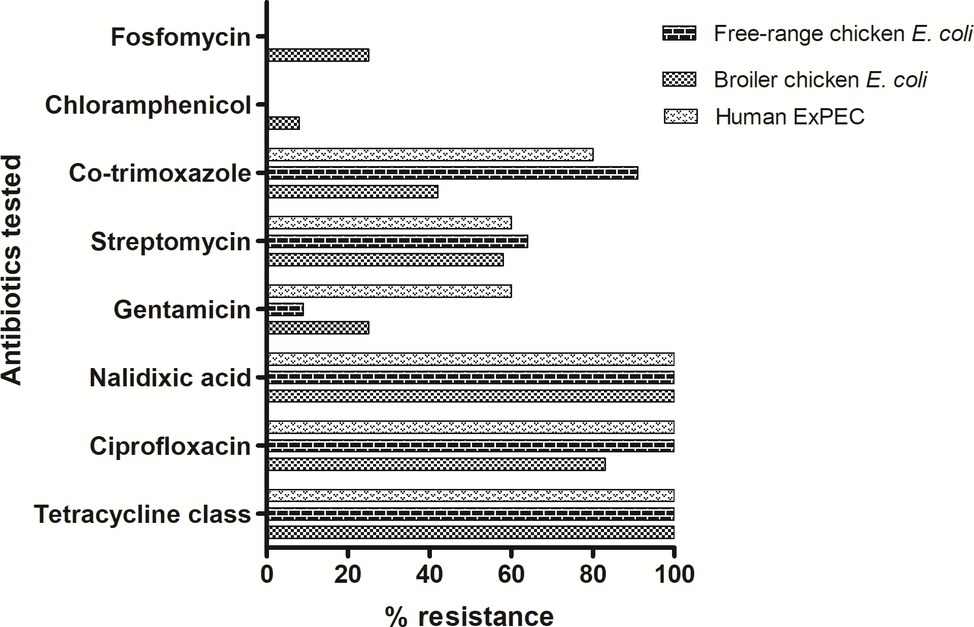
Figure 1. Antibiotic resistance pattern of 28 ESBL-producing E. coli isolates: tetracycline class includes tetracycline, oxytetracycline and chlortetracycline antibiotics. In addition, colistin antibiotic was also tested for which all isolates were found to be susceptible. Out of the 28 ESBL-producing E. coli, 22 (78.5%) were detected to be of MDR phenotype.
Whole Genome-Based Phylogeny, MLST, and Plasmid Profiles
We determined the phylogenetic association among 25 E. coli genomes obtained from broiler (12 genomes), and free-range (8 genomes) chicken samples together with 5 human ExPEC genomes. The phylogenetic tree was broadly classified into four clades: I to IV. Clades I, II, and III represented strains from both broiler and free-range chicken categories (Figure 2). However, none of the human ExPEC strains were included in these three clades. Clade I represented strains that were not clonally clustered together. Nonetheless, the presence of strains belonging to broiler and free-range chicken within clades I, II, and III indicated that the two categories of E. coli strains were related and perhaps shared common ancestors. The clade IV clustered all the five human ExPEC, including the reference strain SE15; this clade also clustered five other broiler chicken E. coli strains that are related to some of well-established ESBL associated sequence types (ST131, ST117, and ST115) and virulent phylotypes (B2, D, and F). Isolates from free-range chicken were not related to this clade. The human ExPEC strains were only related to clade IV possibly because of the small sample size and a limited number of sequence types they belonged to and that they originated from a single geographic location. None of the free-range E. coli belonged to B2, D, and F phylogroups that were represented by the human ExPEC genomes whereas two broiler E. coli isolates were belonging to phylogroup B2 and D each and one to phylogroup F. Using in silico MLST (Larsen et al., 2012), we identified 13 STs, in poultry E. coli which included globally successful sequence types such as ST117 (9%), ST131 (4.3%), and ST10 (4.3%). Using the PlasmidFinder database (Carattoli et al., 2014), we identified a total of 14 different plasmid replicons (including FIA, FIB, FIC, FII, B/O/K/Z, Col, I1, I2, I7, N, po111, Q1, R, and Y) to be present in our collection of E. coli isolates (Supplementary Table S1). FIB (73%) and FII (73%), were the most common plasmid replicons identified among the poultry and human ExPEC genomes analyzed (Supplementary Table S1).
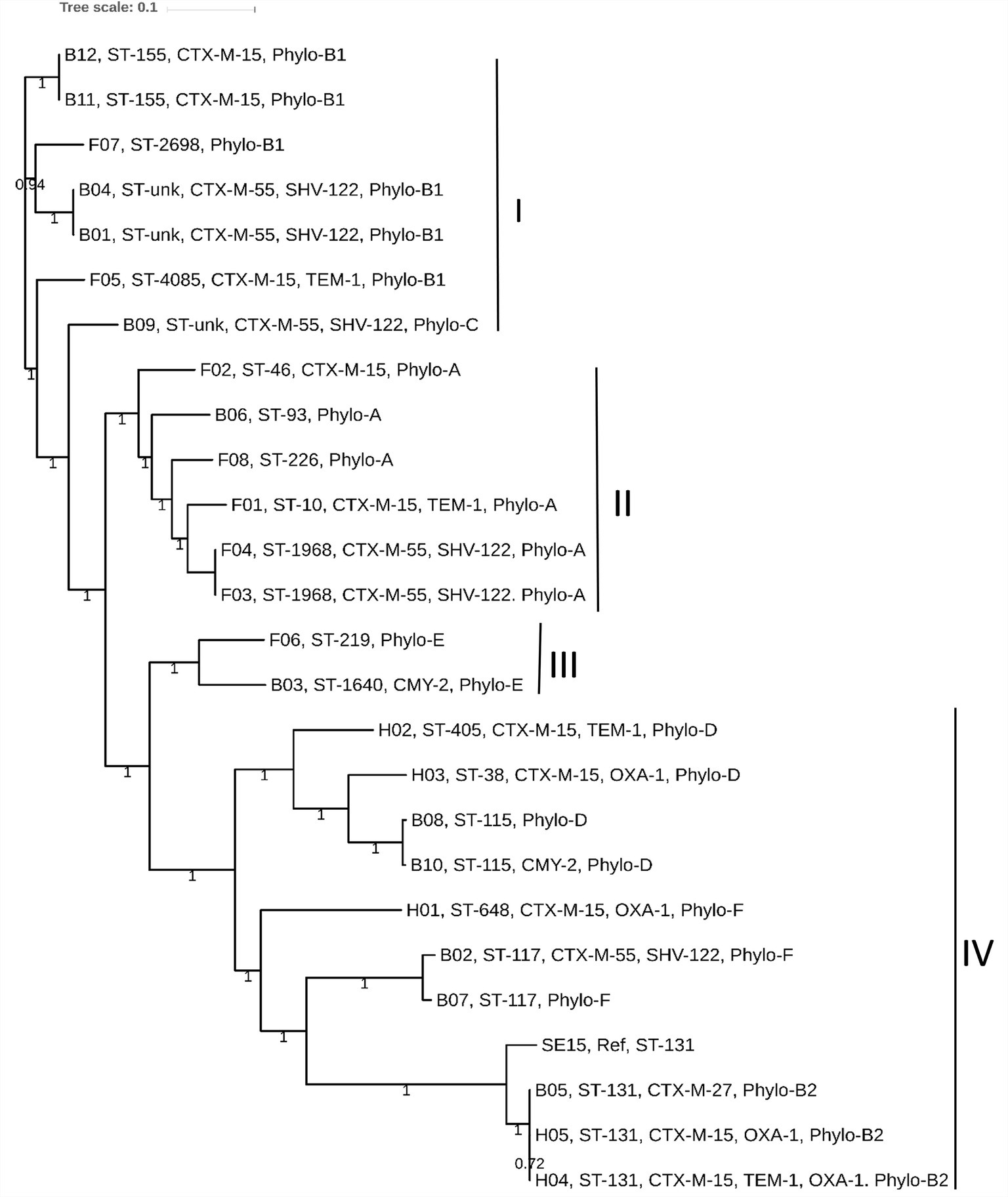
Figure 2. The core genome based consensus Maximum Likelihood phylogenetic tree of 25 E. coli genomes (12 broiler, 8 free-range and 5 human ExPEC genomes) with reference E. coli SE15 genome generated using Harvest. The output of Harvest was visualized using iTOL. The broiler and free-range E. coli isolates were present within same clusters indicating that they share similar genetic backgrounds. However, the human ExPEC isolates largely formed a distinct clade together with the genomes of broiler E. coli and no genome from free-range E. coli was represented within this clade (IV). Labeling at the tips include genome unique IDs (B01 to B12; Broiler E. coli, F01 to F08; Free-range E. coli, H01 to H05; Human ExPEC) followed by sequence types, acquired ESBL genes and the phylogroups.
Whole Genome Sequence-Based Identification of Antibiotic Resistance and Virulence Genes
The presence or absence of different categories of virulence and antimicrobial resistance genes in each of the isolates are displayed in Figures 3, 4. The heat maps indicate the status of antimicrobial resistance and virulence genes and the dendrogram discerns the relationships among the isolates based on the presence/absence profiles of the genes analyzed.
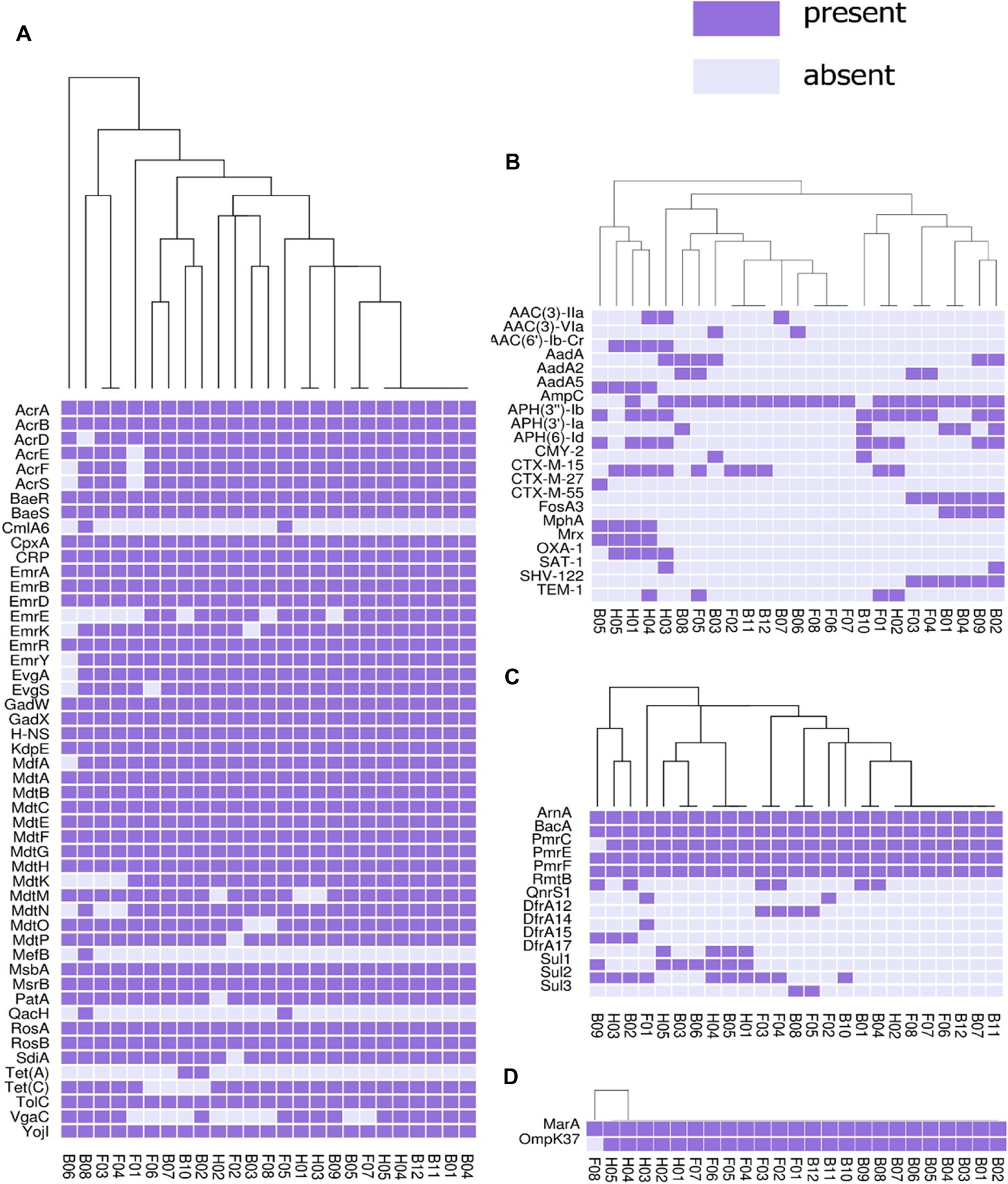
Figure 3. Cluster heat map of 25 E. coli genomes (12 broiler, 8 free-range and 5 human ExPEC genomes) showing the presence and absence of 92 antibiotic resistance genes belonging to different categories (A) antibiotic efflux, (B) antibiotic inactivation, (C) target alteration, protection and replacement, (D) reduced permeability to antibiotics. Results of resistance clustering indicated that the three groups of E. coli isolates including broiler, free-range and human isolates represented clusters with mixed strains which shared resistance gene absence/ presence profile with each other. Gene names are represented on Y axis and the E. coli genomes are listed on the X axis (B01 to B12; Broiler E. coli, F01 to F08; Free-range E. coli, H01 to H05; Human ExPEC). Detail description of genomes can be found in Supplementary Table S1.
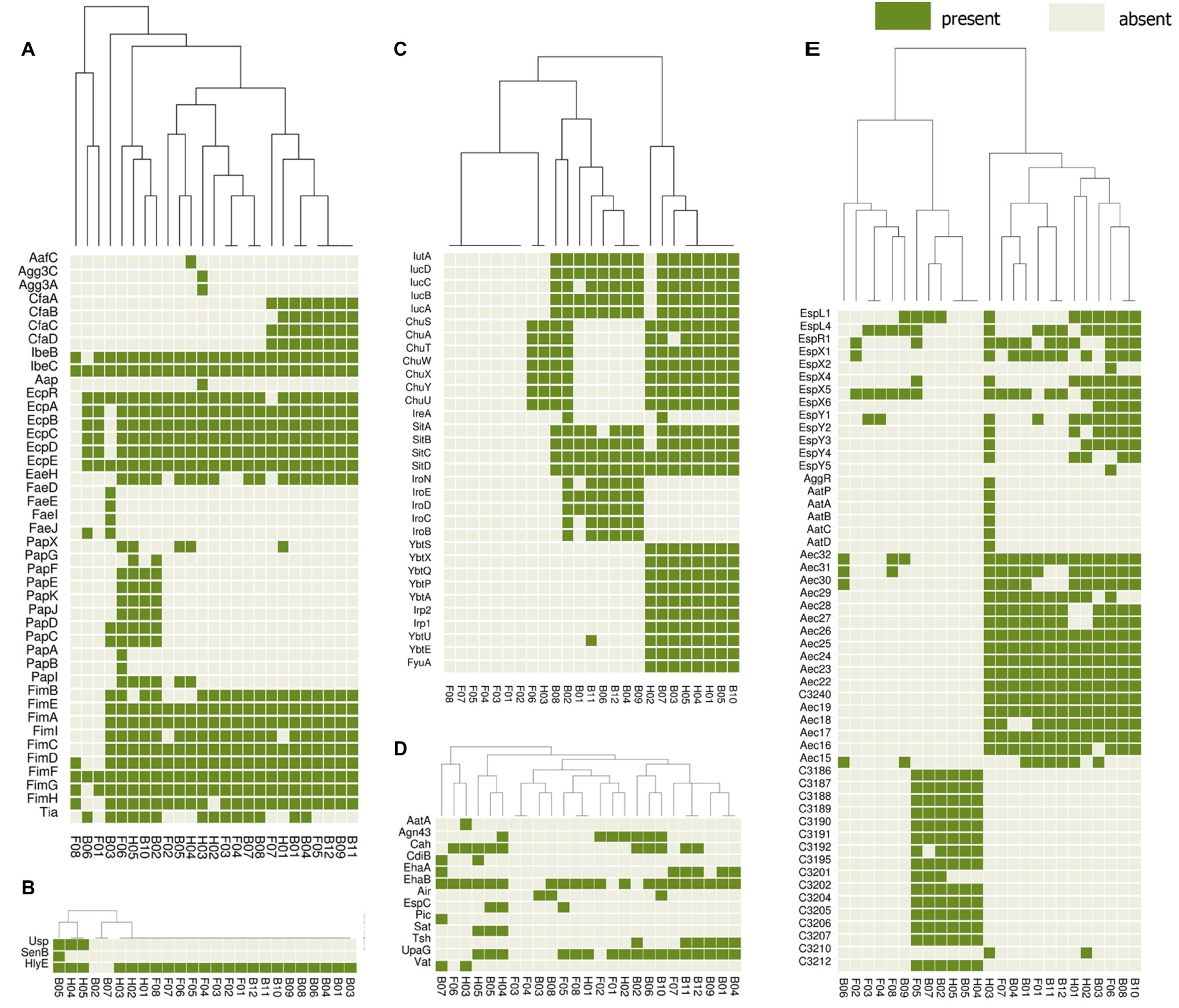
Figure 4. Cluster heat map of 25 E. coli genomes (12 broiler, 8 free-range and 5 human ExPEC genomes) showing the presence and absence of 143 virulence genes belonging to different categories (A) adherence, (B) toxins, (C) iron uptake, (D) auto-transporters, (E) secretion systems. The virulence based clustering indicated that the three groups of E. coli including broiler, free-range and human ExPEC shared virulence gene profiles with each other. Gene names are represented on Y axis and the E. coli genomes are listed on the X axis (B01 to B12; Broiler E. coli, F01 to F08; Free-range E. coli, H01 to H05; Human ExPEC). Detail description of genomes can be found in Supplementary Table S1.
In this analysis (Figure 3), 92 out of 2,678 different resistance genes were detected among 25 E. coli isolates comprising 12 broiler chicken, 8 free-range chicken, and 5 human ExPEC strains. The three groups of E. coli isolates did not demonstrate distinct resistance gene profiles (absence/presence) compared to each other. On the other hand, many strains belonging to the three groups shared resistance gene profiles as evident from the dendrogram. However, when the aggregate resistance gene scores were analyzed, free-range chicken E. coli isolates exhibited significantly lower (p = 0.033) resistance score (median = 54.5) compared to human ExPEC isolates (median = 63) while the resistance gene scores were statistically comparable (p = 0.059) between broiler chicken (median = 56) and human ExPEC isolates (median = 63). Poultry-derived ESBL-producing E. coli isolates in this study harbored different variants of ESBL genes including the blaCTX-M-15 (40%), blaCTX-M-27 (4%), blaCTX-M-55 (24%), blaTEM-1 (16%), and blaSHV-122 (24%) (Figure 3). However, common among them was blaCTX-M-15, which was present in four clonally diverse STs, including ST10, ST46, ST155, and ST4985. The correlation between resistance genotypes (presence of resistance genes) and phenotypes (resistant phenotype) was high for tetracycline (74% agreement), followed by aminoglycosides (65% agreement), sulfonamides (53% agreement), and phenicols (50% agreement) whereas only 6 out of 21 isolates resistant to fluoroquinolone harbored aac (6)-lb-cr (4 isolates) and QnrS1 (2 isolates) genes.
In addition, a second heat map was constructed to screen major groups of known virulence genes across the whole genome sequenced E. coli strains (Figure 4). This analysis revealed presence of 143 out of 2,681 different virulence genes analyzed among the set of 25 E. coli isolates. The heat map demonstrated no unique virulence gene profile for a particular E. coli group (broiler or free-range chicken isolates and human ExPEC), as there was no cluster within the heat map that included only individual strains from the same group. This indicates that strains from all three groups shared virulence gene profiles except for iron acquisition systems as free-range E. coli harbored very few iron acquisition genes. Similar to the resistance scores, the E. coli isolates from free-range chicken showed significantly lower (p = 0.008) aggregate virulence gene scores (median = 27) compared to human ExPEC isolates (median = 68) in contrast to E. coli from broiler chicken (median = 62), which showed statistically comparable values (p = 0.075) with human ExPEC isolates. Similarly, when presence of 15 ExPEC associated genes (pap A/C/E/F/G, fimH, pic, sat, tsh, vat, iutA, ireA, iroN, fyuA, and usp) were analyzed (Pitout, 2012), least number of ExPEC-associated genes (median and range) were detected in free-range E. coli isolates (1, 0–5) whereas, the broiler and human ExPEC isolates were comparable with median and range of 4, 0–9 and 3, 1–9, respectively.
Virulence-Associated Phenotypes of Extended-Spectrum β-Lactamase-Producing E. coli Isolates
Biofilm Formation
Biofilms were produced by all the identified 28 E. coli isolates corresponding to 11 free-range, 12 broiler and 5 human ExPEC isolates (Figure 5A), as compared to the negative control DH5α. Moreover, biofilm formation capability was found to be similarly prevalent among free-range E. coli and broiler isolates (p = >0.05) which indicates their similar propensities to form biofilms. However, free-range E. coli isolates produced optically denser biofilms in comparison to broiler E. coli isolates. Overall, the biofilm formation capabilities of both categories of poultry E. coli (free-range and broiler) isolates were comparable to the biofilm formation capabilities of human extraintestinal pathogenic E. coli (ExPEC) isolates.
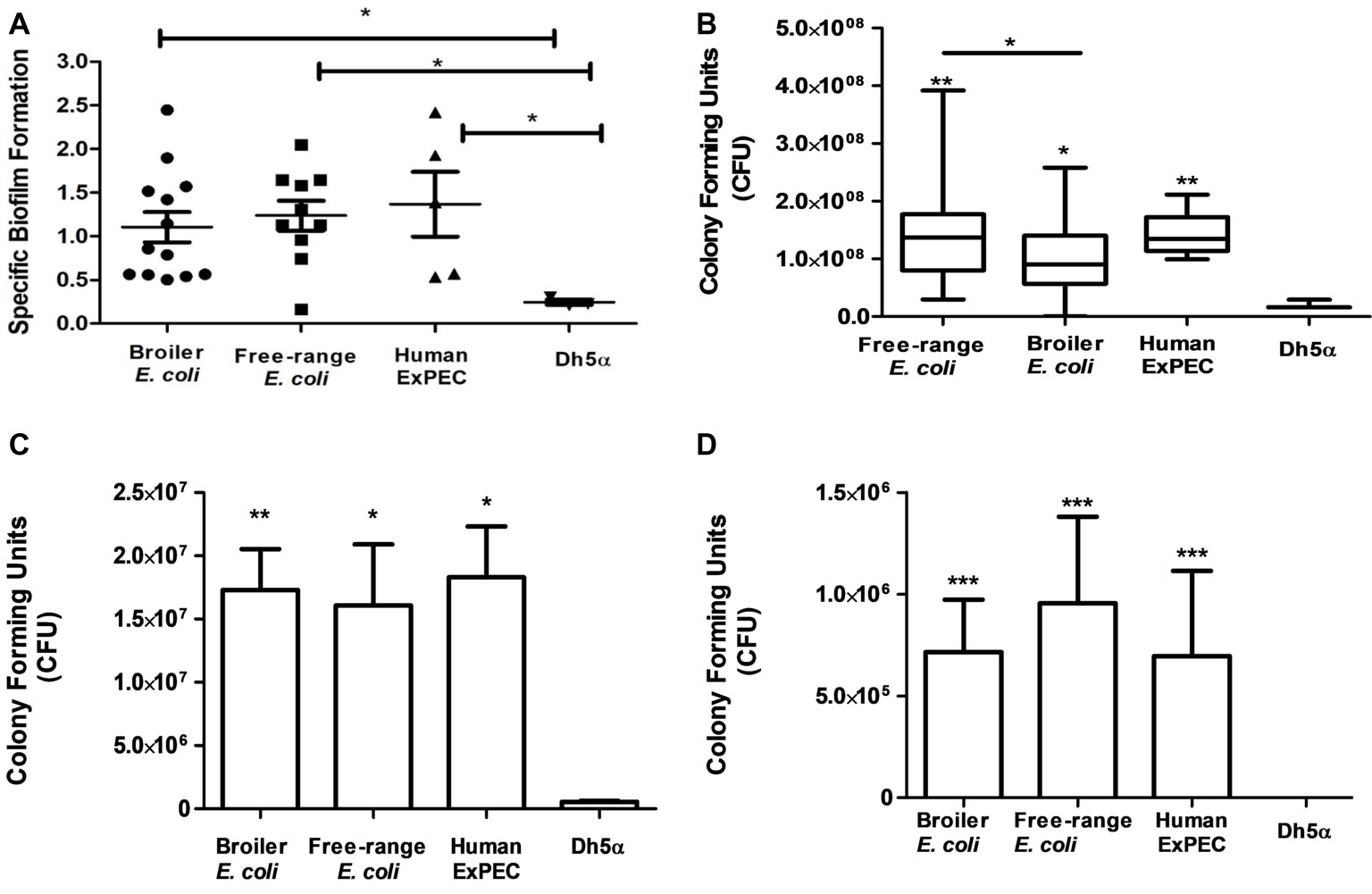
Figure 5. In vitro functional characterization of 11 free-range chicken E. coli isolates, 12 broiler chicken E. coli isolates and 5 human ExPEC including the control E. coli DH5α. (A) Biofilm formation capabilities determined in M-63 minimal media using 96-well microtiter plate method. (B) Resistance to serum bactericidal activity against 50% human serum. (C) Adhesion and (D) invasion capabilities on T24 bladder epithelial cells. All 28 E. coli strains demonstrated significant correlation with the above tested virulence phenotypes compared to the negative control strain, E. coli DH5α. Experiments were performed twice (serum assay)/thrice (adhesion, invasion, and biofilm formation assays) in triplicates and statistical values were obtained using the nonparametric Mann-Whitney U test. 5.01. *p < 0.05; **p < 0.01; ***p < 0.001.
Serum Resistance Assay
All identified E. coli (n = 28) strains were tested for their capacity to resist the serum bactericidal activity when incubated with 50% human serum. Strains that survived from the bactericidal activity of the serum even after 3 h of incubation were defined as serum resistant and those that did not survive or increase their CFU counts were considered serum sensitive. Interestingly, all the 28 E. coli strains, irrespective of free-range, broiler, or human clinical origins, were resistant to human serum as compared to the negative control strain, E. coli DH5α (Figure 5B). Although the broiler group of E. coli demonstrated serum resistance, the mean CFU count of this group was significantly less than free-range and human E. coli groups (p = 0.049 and p = 0.008, respectively). However, no significant difference (p = 0.738) was observed between the mean serum resistance of free-range chicken and human E. coli groups.
Adhesion Property
The E. coli isolates (n = 28) were also tested for their adherence to the human T24 bladder epithelial cells. Free-range and broiler E. coli strains demonstrated significant adherence to T24 cell line in comparison to DH5α strain (Figure 5C). The adhesion capabilities of the poultry E. coli strains (free-range and broiler) were comparable to the adhesion capabilities of human ExPEC strains and lacked any significant differences (p > 0.05). However, the broiler chicken E. coli strains were slightly more (mean 1.73 × 107 CFU) adherent to T24 cells rather than the free-range chicken E. coli isolates (mean 1.6 × 107) but statistically these deviations were non-significant. The mean CFU counts entailing the adherence of human strains to T24 cells were 1.83 × 107.
Invasion Capability
All those strains included in the adhesion assay were also tested for their invasion property in T24 cells by gentamicin (100 μg/ml) protection assay (Figure 5D). Both groups of poultry strains (broiler and free-range chicken) demonstrated prominent invasion into T24 cell line compared to the invasion capabilities of the human ExPEC strains. In contrast to the adhesion capability, the group of free-range chicken E. coli strains showed slightly more invasion rates than the group of broiler chicken E. coli strains (mean 7.1 × 106 CFU) but the two groups did not differ significantly. The mean invasion capability of free-range chicken E. coli in T24 cells was comparable to the mean invasion capability of the human ExPEC strains with 9.5 × 106 CFU and 6.9 × 106 CFU, respectively as the difference was found to be statistically non-significant (p = 0.382).
Discussion
The prevalence of virulence potential in ESBL-producing E. coli originating from healthy production chickens could be a significant factor for both human and animal health. In this baseline study, we tested ESBL-producing E. coli isolates from healthy chickens (broiler and free-range) with regard to their phenotypic virulence traits and extensive antimicrobial resistance. This in vitro virulence and resistance testing revealed that the ESBL-producing E. coli in poultry demonstrated strong virulence and resistance attributes, irrespective of their host origins such as broiler or free-range chicken. E. coli is known to be a highly diverse species that is frequently associated with antimicrobial resistance. Here, we carried out whole-genome sequencing of E. coli isolates obtained from different regions of India, about which such data are largely lacking. We then determined the genomic diversity and/or similarity with respect to core genome phylogeny, STs, virulence and resistance gene profiles. Our observations revealed that E. coli in sampled settings possess high diversity at the level of phylogeny and sequence types.
In our study, about 78.5% of the ESBL-producing E. coli were detected to be of multi-drug resistant phenotype. They were frequently resistant to most of the antibiotics tested such as tetracyclines, fluoroquinolones, aminoglycosides, and sulfonamides. This coincides with the use of tetracyclines (oxytetracycline and chlortetracycline), fluoroquinolones (ciprofloxacin and enrofloxacin), aminoglycosides (gentamicin and streptomycin), and sulfonamides (co-trimoxazole) in meat production industry in India (Laxminarayan and Chaudhury, 2016; Brower et al., 2017; Kakkar et al., 2017). The common ESBL genotype identified was the blaCTX-M-15, which was present in four clonally diverse STs namely, ST10, ST46, ST155, and ST4985. Presence of blaCTX-M-15 in diverse STs has also been recently reported (Mshana et al., 2016; Musicha et al., 2017) and globally, the blaCTX-M-15 gene was reported to be strongly linked with ST131 E. coli lineage (Price et al., 2013). It has been already evident that blaCTX-M-15 can spread by genetic mobile elements such as conjugative plasmids (Inwezerua et al., 2014; Irrgang et al., 2017). Further studies are required in India to accurately estimate the prevalence of blaCTX-M-15 and other variants and correlate its association with different successful lineages causing multi-drug resistant infections. Conjugation experiments suggest that the ESBL production in poultry E. coli isolates may be associated with conjugative plasmids as also reported by others, that E. coli isolates of poultry origin harbor conjugative plasmids (Johnson et al., 2012). Sequence types 155, 115, and 117 constitute the most clonal clades in the population structure of our collection. Interestingly, corroborating with the virulence and resistance gene profiling results, the phylogeny results also revealed that the broiler or free-range chicken E. coli did not form two homogenous groups, instead, they demonstrated mixed clustering. These findings suggest that irrespective of the host origin the E. coli from broiler or free-range chicken share genetic backgrounds with each other and may belong to the same ancestral lineage(s) that could be the natural inhabitants of the poultry. The presence of MDR-bacteria in food sources such as poultry is a matter of public health concern as these MDR bacteria could potentially transfer their resistant traits to food-borne pathogens. Moreover, the presence of MDR-E. coli in the intestines of slaughtered birds can contaminate the carcasses during slaughter and processing, and thereby spread to humans directly or via the food chain.
We then analyzed the ESBL-producing poultry E. coli isolates to characterize the virulence potential of these isolates. Certain in vitro phenotypic traits such as biofilm formation, serum resistance, adhesion and invasion properties may correspond with or give rise to, the pathogenic potential of the E. coli isolates (Nandanwar et al., 2014; Mitchell et al., 2015).
Bacteria produce biofilm in order to protect themselves from host defense elements, antibiotics, and detergents; biofilms allow them to be in close proximity to other pathogens, so as to horizontally acquire genetic material and become either more antibiotic resistant or virulent (Naves et al., 2008). In this study, 100% of tested E. coli strains demonstrated biofilm formation ability and the range of biofilm formation did not vary between the two groups of chicken E. coli isolates (broiler and free-range chicken). Thus, we confirm that E. coli isolates of food origin are being able to survive on polystyrene 96-well microtiter plates. This is an important observation as these E. coli can survive on the food-processing surfaces and potentially persist there withstanding the unfavorable conditions and re-contaminate food products. This is supported by a previous report on E. coli isolates obtained from a poultry slaughterhouse (Rodrigues et al., 2010).
To gain further understanding of the ability of poultry E. coli isolates to cause infections in humans, the 23 poultry E. coli isolates along with 5 human ExPEC isolates were assessed for their ability to attach and invade mammalian T24 bladder epithelial cells in vitro. All 23 poultry E. coli showed stronger adhesion and invasion capabilities comparable to the human ExPEC isolates. Interestingly, the free-range E. coli isolates demonstrated higher invasion strength than the broiler E. coli isolates. In this study, virulence factors, including fim, pap, tsh, upa, and ibeB, were frequently observed (Figure 4) that are reported to be associated with virulence properties (Rodriguez-Siek et al., 2005; Antão et al., 2009; Wang et al., 2012). Effective cellular adhesion followed by invasion are the key events in pathogenicity; the above observation reaffirms the potential of healthy poultry derived E. coli to be of concern. Moreover, the poultry E. coli could use similar mechanisms to strongly adhere to chicken meat (LeStrange et al., 2017), and therefore this could be a potential method of foodborne transmission of pathogenic and or MDR E. coli to humans.
Resistance to the serum bactericidal activity is an important virulence determinant in E. coli strains where it contributes to pathogenesis particularly in extraintestinal infections (Mellata et al., 2003). A strong correlation between serum resistance and the ability of poultry E. coli to infect the internal organs of chicken was reported previously (Mellata et al., 2003). Here, we detected serum resistance for all poultry E. coli isolates including that of human ExPEC isolates. The free-range E. coli isolates exhibited significantly higher serum resistance than the broiler E. coli. In order to survive in extraintestinal sites, microbes, on the one hand, have to overcome harsh environments and on the other hand, have to acquire micronutrients such as iron. Consequently, the poultry E. coli isolates possessed several siderophore genes such as iroN, chuA, and sitD. However, the free-range E. coli isolates harbored very few siderophore genes compared to the broiler E. coli isolates, the reason for such an observation could be found in another study which reported that environmental E. coli isolates are less likely to contain genes associated with aerobactin and yersiniabactin (Searle et al., 2015). The country chicken could have mostly acquired these bacteria from the environment as they feed by free-ranging. All these isolates analyzed in this study were ESBL producers and most of them harbored multiple plasmid types. It is reported that the presence of certain ESBL-plasmids could contribute to competitive fitness and serum survival of the E. coli strains (Ranjan et al., 2018). Plasmid replicon results as reported in this study suggest that this may be the case as we observed the presence of multiple plasmids that have been linked with both resistance and virulence, the most predominant being FIB (73%) and FII (73%) (Johnson et al., 2007).
In conclusion, this baseline study has provided insights into resistance and virulence–related genotypes and phenotypes of ESBL-producing E. coli from broiler and free-range chicken in India. Also, it has revealed a high diversity of lineages of ESBL-producing E. coli circulating in India. Although the human ExPEC isolates could be largely separated from the poultry E. coli isolates when virulence-associated phenotypes were taken into account, the substantial overlap was still evident. Particularly, the broiler chicken E. coli and human ExPEC isolates harbored large number of virulence and resistance genes compared to free-range chicken E. coli. This suggests that the transfer of ESBL-producing isolates from contaminated meat may likely contribute to the colonization of drug-resistant bacteria in humans through consumption of broiler chicken products that are not properly cooked and that certain E. coli clones from broiler-poultry may indeed have the potential to cause infection in humans. Further work is required to ascertain the genomic similarity among large number of geographically matched contemporaneous ESBL-producing E. coli isolates from humans and retail chicken meat. This work significantly contributes to the understanding of the epidemiology of ESBL-producing E. coli particularly from animal foods such as poultry.
Data Availability Statement
The datasets generated for this study can be found in the GenBank accession numbers of the 10 genomes sequenced in this study are RDBX00000000 (NAEC1028), RDBY00000000 (NAEC1042), RDBZ00000000 (NAEC1115), RDCA00000000 (NAEC1132), RDCB00000000 (NAEC1134), RDCC00000000 (NAEC1135), RDCD00000000 (NAEC1136), RDCE00000000 (NAEC1146), RDCF00000000 (NAEC1147), and RDCG00000000 (NAEC1154).
Author Contributions
DC and NA provided overarching supervision to the study. AH designed and conducted the study. AR helped in performing virulence assays. SS, AS, and TS helped in analyzing genome sequence data. MA, HW, LW, and NS helped in preparation of the manuscript.
Funding
AH thanks UGC for Dr. D.S. Kothari post-doctoral fellowship and SERB-DST, for NPDF through its projects F.4-2/2006 (BSR)/ BL/14-15/0273 and PDF/2017/001223, respectively for the financial support. icddr,b would like to thank its core donors: Sweden (SIDA), Govt. of Bangladesh, Canada (CIDA and GAC), and United Kingdom (DFID).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to the members of the Molecular Pathogenesis Lab at the Indian Institute of Science for their help and support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02766/full#supplementary-material
Footnotes
References
Antão, E.-M., Wieler, L. H., and Ewers, C. (2009). Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathog. 1:22. doi: 10.1186/1757-4749-1-22
Avasthi, T. S., Kumar, N., Baddam, R., Hussain, A., Nandanwar, N., Jadhav, S., et al. (2011). Genome of multidrug-resistant uropathogenic Escherichia coli strain NA114 from India. J. Bacteriol. 193, 4272–4273. doi: 10.1128/JB.05413-11
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beghain, J., Bridier-Nahmias, A., Le Nagard, H., Denamur, E., and Clermont, O. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 4. doi: 10.1099/mgen.0.000192
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brower, C. H., Mandal, S., Hayer, S., Sran, M., Zehra, A., Patel, S. J., et al. (2017). The prevalence of extended-spectrum-β-ase-producing multidrug-resistant Escherichia coli in poultry chickens and variation according to farming practices in Punjab, India. Environ. Health Perspect. 125, 1–10. doi: 10.1289/EHP292
Byarugaba, D. (2004). Antimicrobial resistance in developing countries and responsible risk factors. Int. J. Antimicrob. Agents 24, 105–110. doi: 10.1016/j.ijantimicag.2004.02.015
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Hussain, A., Ewers, C., Nandanwar, N., Guenther, S., Jadhav, S., Wieler, L. H., et al. (2012). Multiresistant uropathogenic Escherichia coli from a region in India where urinary tract infections are endemic: genotypic and phenotypic characteristics of sequence type 131 isolates of the CTX-M-15 extended-spectrum-beta-lactamase-producing lineage. Antimicrob. Agents Chemother. 56, 6358–6365. doi: 10.1128/AAC.01099-12
Hussain, A., Ranjan, A., Nandanwar, N., Babbar, A., Jadhav, S., and Ahmed, N. (2014). Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrob. Agents Chemother. 58, 7240–7249. doi: 10.1128/AAC.03320-14
Hussain, A., Shaik, S., Ranjan, A., Nandanwar, N., Tiwari, S. K., Majid, M., et al. (2017). Risk of transmission of antimicrobial resistant Escherichia coli from commercial broiler and free-range retail chicken in India. Front. Microbiol. 8:2120. doi: 10.3389/fmicb.2017.02120
Inwezerua, C., Mendonça, N., Calhau, V., Domingues, S., Adeleke, O. E., and Da Silva, G. J. (2014). Occurrence of extended-spectrum beta-lactamases in human and bovine isolates of Escherichia coli from Oyo state, Nigeria. J. Infect. Dev. Ctries. 8, 774–779. doi: 10.3855/jidc.3430
Irrgang, A., Falgenhauer, L., Fischer, J., Ghosh, H., Guiral, E., Guerra, B., et al. (2017). CTX-M-15-producing Escherichia coli isolates from food products in Germany are mainly associated with an IncF-type plasmid and belong to two predominant clonal E. coli lineages. Front. Microbiol. 8:2318. doi: 10.3389/fmicb.2017.02318
Jadhav, S., Hussain, A., Devi, S., Kumar, A., Parveen, S., Gandham, N., et al. (2011). Virulence characteristics and genetic affinities of multiple drug resistant uropathogenic Escherichia coli from a semi urban locality in India. PLoS One 6:e18063. doi: 10.1371/journal.pone.0018063
Johnson, T. J., Logue, C. M., Johnson, J. R., Kuskowski, M. A., Sherwood, J. S., Barnes, H. J., et al. (2012). Associations between multidrug resistance, plasmid content, and virulence potential among extraintestinal pathogenic and commensal Escherichia coli from humans and poultry. Foodborne Pathog. Dis. 9, 37–46. doi: 10.1089/fpd.2011.0961
Johnson, T. J., Siek, K. E., Johnson, S. J., and Nolan, L. K. (2006). DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188, 745–758. doi: 10.1128/JB.188.2.745-758.2006
Johnson, T. J., Wannemuehler, Y. M., Johnson, S. J., Logue, C. M., White, D. G., Doetkott, C., et al. (2007). Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl. Environ. Microbiol. 73, 1976–1983. doi: 10.1128/AEM.02171-06
Kakkar, M., Walia, K., Vong, S., Chatterjee, P., and Sharma, A. (2017). Antibiotic resistance and its containment in India. BMJ 358:j2687. doi: 10.1136/bmj.j2687
Kluytmans, J. A. J. W., Overdevest, I. T. M. A., Willemsen, I., Kluytmans-Van Den Bergh, M. F. Q., Van Der Zwaluw, K., Heck, M., et al. (2013). Extended-spectrum β-lactamase-producing Escherichia coli from retail chicken meat and humans: comparison of strains, plasmids, resistance genes, and virulence factors. Clin. Infect. Dis. 56, 478–487. doi: 10.1093/cid/cis929
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Laxminarayan, R., and Chaudhury, R. R. (2016). Antibiotic resistance in India: drivers and opportunities for action. PLoS Med. 13:e1001974. doi: 10.1371/journal.pmed.1001974
LeStrange, K., Markland, S. M., Hoover, D. G., Sharma, M., and Kniel, K. E. (2017). An evaluation of the virulence and adherence properties of avian pathogenic Escherichia coli. One Health 4, 22–26. doi: 10.1016/j.onehlt.2017.08.001
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Mellata, M. (2013). Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 10, 916–932. doi: 10.1089/fpd.2013.1533
Mellata, M., Dho-Moulin, M., Dozois, C. M., Curtiss, R., Lehoux, B., and Fairbrother, J. M. (2003). Role of avian pathogenic Escherichia coli virulence factors in bacterial interaction with chicken heterophils and macrophages. Infect. Immun. 71, 494–503. doi: 10.1128/IAI.71.1.494-503.2003
Mitchell, N. M., Johnson, J. R., Johnston, B., Curtiss, R., and Mellata, M. (2015). Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Appl. Environ. Microbiol. 81, 1177–1187. doi: 10.1128/AEM.03524-14
Miyoshi-Akiyama, T., Sherchan, J. B., Doi, Y., Nagamatsu, M., Sherchand, J. B., Tandukar, S., et al. (2016). Comparative genome analysis of extended-spectrum-β-lactamase-producing Escherichia coli sequence type 131 strains from Nepal and Japan. mSphere 1. doi: 10.1128/mSphere.00289-16
Mshana, S. E., Falgenhauer, L., Mirambo, M. M., Mushi, M. F., Moremi, N., Julius, R., et al. (2016). Predictors of blaCTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infect. Dis. 16:187. doi: 10.1186/s12879-016-1527-x
Musicha, P., Feasey, N. A., Cain, A. K., Kallonen, T., Chaguza, C., Peno, C., et al. (2017). Genomic landscape of extended-spectrum β-lactamase resistance in Escherichia coli from an urban African setting. J. Antimicrob. Chemother. 72, 1602–1609. doi: 10.1093/jac/dkx058
Nandanwar, N., Hussain, A., Ranjan, A., Jadhav, S., and Ahmed, N. (2016). Population structure and molecular epidemiology of human clinical multi-drug resistant (MDR) Escherichia coli strains from Pune, India. Int. J. Infect. Dis. 45, 343–344. doi: 10.1016/j.ijid.2016.02.741
Nandanwar, N., Janssen, T., Kühl, M., Ahmed, N., Ewers, C., and Wieler, L. H. (2014). Extraintestinal pathogenic Escherichia coli (ExPEC) of human and avian origin belonging to sequence type complex 95 (STC95) portray indistinguishable virulence features. Int. J. Med. Microbiol. 95, 6–13. doi: 10.1016/j.ijmm.2014.06.009
Naves, P., del Prado, G., Huelves, L., Gracia, M., Ruiz, V., Blanco, J., et al. (2008). Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microb. Pathog. 45, 86–91. doi: 10.1016/j.micpath.2008.03.003
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Patel, R. K., Jain, M., Mardis, E., Wang, Z., Gerstein, M., Snyder, M., et al. (2012). NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619
Paterson, D. L., and Bonomo, R. A. (2005). Extended-spectrum-β-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686. doi: 10.1128/CMR.18.4.657-686.2005
Pitout, J. D. D. (2012). Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front. Microbiol. 3:9. doi: 10.3389/fmicb.2012.00009
Price, L. B., Johnson, J. R., and Aziz, M. (2013). The epidemic of extended-spectrum- β -lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H 30-Rx. MBio 4, e00377–e00313. doi: 10.1128/mBio.00377-13
Qumar, S., Majid, M., Kumar, N., Tiwari, S. K., Semmler, T., Devi, S., et al. (2017). Genome dynamics and molecular infection epidemiology of multidrug-resistant Helicobacter pullorum isolates obtained from broiler and free-range chickens in India. Appl. Environ. Microbiol. 83, e02305–e02316. doi: 10.1128/AEM.02305-16
Ranjan, A., Scholz, J., Semmler, T., Wieler, L. H., Ewers, C., Müller, S., et al. (2018). ESBL-plasmid carriage in E. coli enhances in vitro bacterial competition fitness and serum resistance in some strains of pandemic sequence types without overall fitness cost. Gut Pathog. 10:24. doi: 10.1186/s13099-018-0243-z
Ranjan, A., Shaik, S., Hussain, A., Nandanwar, N., Semmler, T., Jadhav, S., et al. (2015). Genomic and functional portrait of a highly virulent, CTX-M-15-producing H 30-Rx subclone of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 59, 6087–6095. doi: 10.1128/AAC.01447-15
Ranjan, A., Shaik, S., Mondal, A., Nandanwar, N., Hussain, A., Semmler, T., et al. (2016). Molecular epidemiology and genome dynamics of New Delhi metallo-beta-lactamase (NDM) producing extraintestinal pathogenic E. coli (ExPEC) strains from India. Antimicrob. Agents Chemother. 60, 6795–6805. doi: 10.1128/AAC.01345-16
Ranjan, A., Shaik, S., Nandanwar, N., Hussain, A., Tiwari, S. K., Semmler, T., et al. (2017). Comparative genomics of Escherichia coli isolated from skin and soft tissue and other extraintestinal infections. MBio 8, e01070–e01017. doi: 10.1128/mBio.01070-17
Rawat, D., and Nair, D. (2010). Extended-spectrum β-lactamases in gram negative bacteria. J. Global Infect. Dis. 2, 263–274. doi: 10.4103/0974-777X.68531
Robinson, T. P., Bu, D. P., Carrique-Mas, J., Fèvre, E. M., Gilbert, M., Grace, D., et al. (2016). Antibiotic resistance is the quintessential one health issue. Trans. R. Soc. Trop. Med. Hyg. 110, 377–380. doi: 10.1093/trstmh/trw048
Rodrigues, L. B., Dos Santos, L. R., Tagliari, V. Z., Rizzo, N. N., Trenhago, G., de Oliveira, A. P., et al. (2010). Quantification of biofilm production on polystyrene by listeria, Escherichia coli and Staphylococcus aureus isolated from a poultry slaughterhouse. Braz. J. Microbiol. 41, 1082–1085. doi: 10.1590/S1517-83822010000400029
Rodriguez-Siek, K. E., Giddings, C. W., Doetkott, C., Johnson, T. J., and Nolan, L. K. (2005). Characterizing the APEC pathotype. Vet. Res. 36, 241–256. doi: 10.1051/vetres:2004057
Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11120685 (Accessed November 21, 2016).
Searle, L. J., Méric, G., Porcelli, I., Sheppard, S. K., and Lucchini, S. (2015). Variation in siderophore biosynthetic gene distribution and production across environmental and faecal populations of escherichia coli. PLoS One 10:e0117906. doi: 10.1371/journal.pone.0117906
Shaik, S., Kumar, N., Lankapalli, A. K., Tiwari, S. K., Baddam, R., and Ahmed, N. (2016). Contig-layout-authenticator (CLA): a combinatorial approach to ordering and scaffolding of bacterial contigs for comparative genomics and molecular epidemiology. PLoS One 11:e0155459. doi: 10.1371/journal.pone.0155459
Shaik, S., Ranjan, A., Tiwari, S. K., Hussain, A., Nandanwar, N., Kumar, N., et al. (2017). Comparative genomic analysis of globally dominant ST131 clone with other epidemiologically successful extraintestinal pathogenic Escherichia coli (ExPEC) lineages. MBio 8, e01596–e01517. doi: 10.1128/mBio.01596-17
Skyberg, J. A., Johnson, T. J., Johnson, J. R., Clabots, C., Logue, C. M., and Nolan, L. K. (2006). Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect. Immun. 74, 6287–6292. doi: 10.1128/IAI.00363-06
Suresh, A., Ranjan, A., Patil, S., Hussain, A., Shaik, S., Alam, M., et al. (2018). Molecular genetic and functional analysis of pks-harboring, extra-intestinal pathogenic Escherichia coli from India. Front. Microbiol. 9:2631. doi: 10.3389/fmicb.2018.02631
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/PREACCEPT-2573980311437212
Tumbarello, M., Spanu, T., Di Bidino, R., Marchetti, M., Ruggeri, M., Trecarichi, E. M., et al. (2010). Costs of bloodstream infections caused by Escherichia coli and influence of extended-spectrum- β-lactamase production and inadequate initial antibiotic therapy. Antimicrob. Agents Chemother. 54, 4085–4091. doi: 10.1128/AAC.00143-10
Keywords: poultry Escherichia coli, genomics, ESBL, ExPEC, India
Citation: Hussain A, Shaik S, Ranjan A, Suresh A, Sarker N, Semmler T, Wieler LH, Alam M, Watanabe H, Chakravortty D and Ahmed N (2019) Genomic and Functional Characterization of Poultry Escherichia coli From India Revealed Diverse Extended-Spectrum β-Lactamase-Producing Lineages With Shared Virulence Profiles. Front. Microbiol. 10:2766. doi: 10.3389/fmicb.2019.02766
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyMasaki Yamamoto, Kyoto University Hospital, Japan
Copyright © 2019 Hussain, Shaik, Ranjan, Suresh, Sarker, Semmler, Wieler, Alam, Watanabe, Chakravortty and Ahmed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dipshikha Chakravortty, ZGlwYUBpaXNjLmFjLmlu; Niyaz Ahmed, bml5YXouYWhtZWRAaWNkZHJiLm9yZw==
 Arif Hussain
Arif Hussain Sabiha Shaik
Sabiha Shaik Amit Ranjan
Amit Ranjan Arya Suresh
Arya Suresh Nishat Sarker
Nishat Sarker Torsten Semmler
Torsten Semmler Lothar H. Wieler
Lothar H. Wieler Munirul Alam
Munirul Alam Haruo Watanabe5
Haruo Watanabe5 Dipshikha Chakravortty
Dipshikha Chakravortty Niyaz Ahmed
Niyaz Ahmed