- Office of Research and Development, Center for Environmental Measurement and Modeling, US Environmental Protection Agency, Cincinnati, OH, United States
Antimicrobial resistance (AMR) is a global concern, pertaining not only to human health but also to the health of industry and the environment. AMR research has traditionally focused on genetic exchange mechanisms and abiotic environmental constraints, leaving important aspects of microbial ecology unresolved. The genetic and ecological aspects of AMR, however, not only contribute separately to the problem but also are interrelated. For example, mutualistic associations among microbes such as biofilms can both serve as a barrier to antibiotic penetration and a breeding ground for horizontal exchange of antimicrobial resistance genes (ARGs). In this review, we elucidate how species interactions promote and impede the establishment, maintenance, and spread of ARGs and indicate how management initiatives might benefit from leveraging the principles and tools of community ecology to better understand and manipulate the processes underlying AMR.
Introduction
As pathogens and other microbes become increasingly and more frequently resistant to antibiotics, concern is growing world-wide that the use of antibiotics for treating and preventing diseases in humans, animals, and plants is rapidly becoming less effective and unsustainable (Nerlich and James, 2009; Defoirdt et al., 2011; Stockwell and Duffy, 2012; Wellington et al., 2013; Aćimović et al., 2015). Antimicrobial resistance (AMR) may, in addition, threaten the health of ecosystems (Grenni et al., 2018) and the performance of businesses that rely on large-scale maintenance of microbial monocultures or AMR-related reporter genes for culinary and industrial fermentation processes (Teuber et al., 1999; Bourdichon et al., 2012; Shaw et al., 2016), food and nutritional supplement cultivation (Richmond and Preiss, 1980; Hallmann and Rappel, 1999; Mišurcová et al., 2012; Wells et al., 2017), bioremediation (Jebelli et al., 2018), energy harvesting and biofuel production (Arora et al., 2015; Wang et al., 2015; Oliver et al., 2016), and the derivation of bioprospects (Strobel and Daisy, 2003; Ferrer et al., 2016) such as dyes (Tuli et al., 2015; Sen et al., 2019) and self-healing concrete (Seifan et al., 2016). Although many strategies to mitigate the threat and current impacts of AMR are presently being explored and enacted, such as altering how antibiotics are administered and regulated to sustain or improve their effectiveness (Fridkin and Gaynes, 1999; Drusano, 2003; Pruden et al., 2013; Chakradhar, 2016), discovering or developing new classes of natural and synthetic antibiotics to supplement or replace the old (Livermore, 2011; Moloney, 2016; Wiese and Imhoff, 2019), and employing bacteriocins and other scalable alternatives to the use of antibiotics (Joerger, 2003; Reardon, 2015; Czaplewski et al., 2016; Willing et al., 2018), the problem remains. At the root of this problem is the One Health nature of AMR; i.e., the interconnectedness among human, animal, and environmental systems (Figure 1A; Collignon and McEwen, 2019).
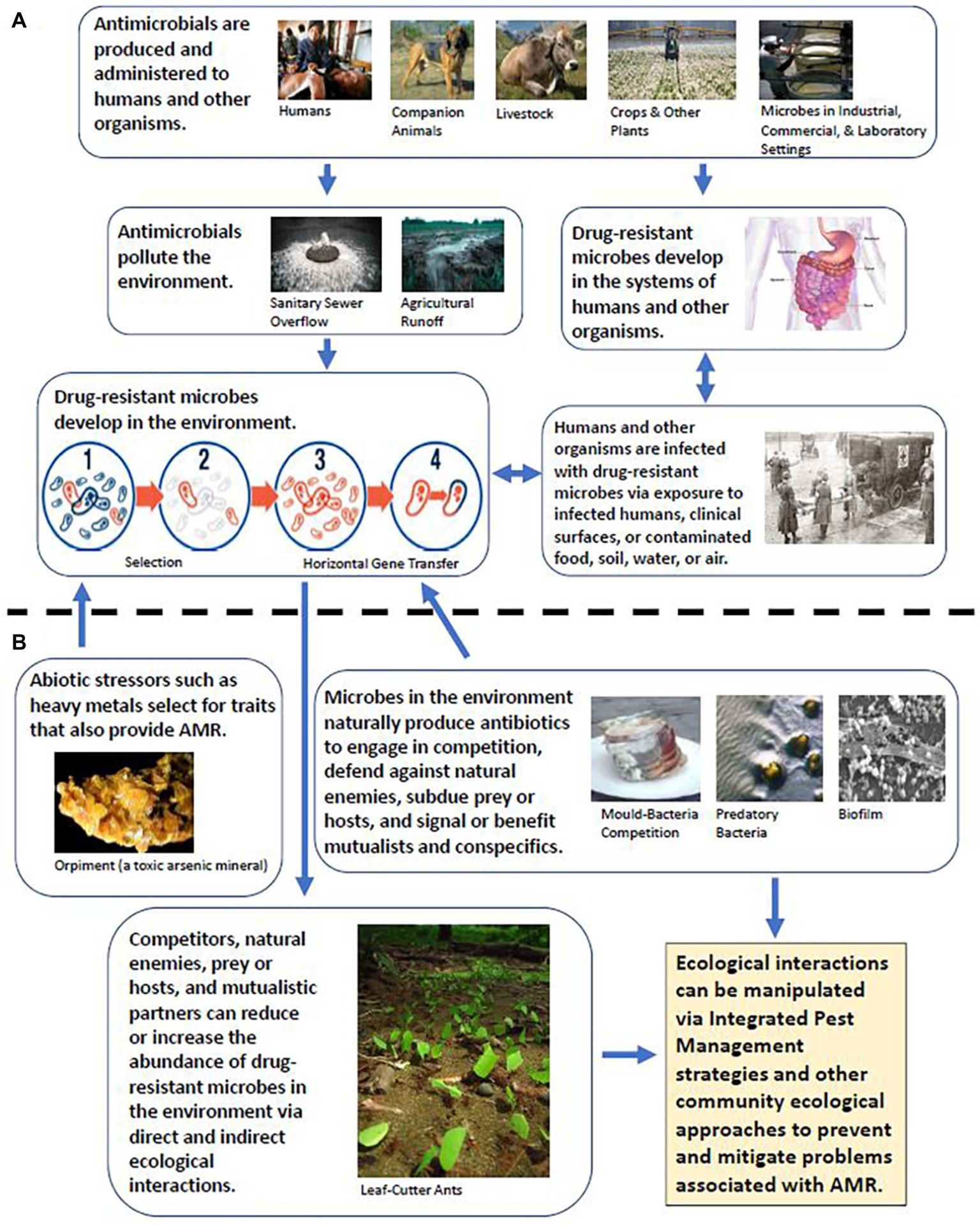
Figure 1. (A) Traditional view of AMR, wherein the environment and wildlife mostly represent opportunities for exposure and/or reservoirs of drug-resistant microorganisms. (B) Expansion of the traditional view of AMR to include the effects and potential management implications of species interactions.
To begin to address this interconnectedness, we must acknowledge that resistance to antibiotics may not be the primary or original (evolutionary) purpose of antimicrobial resistance genes (ARGs). ARGs occur naturally in the environment, where they have been shown or posited to confer protection against toxins such as heavy metals and host-produced biocides and to perform important roles in cellular processes such as quorum sensing (the detection of conspecific or cooperative cells in the environment, associated with cell-to-cell communication/signaling and group activity) and biosynthesis (the enzymatic conversion of simple compounds into more complex products by living things; Figure 1B; Allen et al., 2010; Martinez, 2018). Indeed, ARGs are detected even in habitats that have historically been sheltered from antibiotic pollution and other forms of anthropogenic disturbance (McArthur et al., 2016; Van Goethem et al., 2018). Many antibiotic-producing microbes possess ARGs, ensuring that they are resistant or immune to their own antibiotics (Benveniste and Davies, 1973). Antibiotic-producing microbes may thus be a source of ARGs in sympatric species via horizontal gene transfer (Jiang et al., 2017; Ringel et al., 2017) or select for AMR in the targets of their antibiotics (Wellington et al., 2013), resulting in an “arms race” of cyclical coevolutionary dynamics known as “Red Queen” dynamics (Baron et al., 2018; Decaestecker and King, 2019). From the standpoint of managing AMR, this poses two separate challenges. First, for any new class of antibiotic discovered in nature, there may already be a corresponding suite of ARGs among or transferrable to pathogens (Bengtsson-Palme and Larsson, 2015). Second, efforts to monitor changes in ARG prevalence and identify areas of concern based on comparison to a “least disturbed” reference site may need to account for natural environmental variations in the reference site that favor the natural producers or production of antibiotics. Reference sites may themselves become areas of concern under certain conditions and should, in any case, be critically reviewed to ensure their appropriateness as a baseline of comparison (White and Walker, 1997; Whittier et al., 2007; Berendonk et al., 2015; Rothrock et al., 2016; Vikesland et al., 2017).
High-throughput “-omics” techniques, including genomics, transcriptomics, metabolomics, and proteomics, have greatly enhanced our ability to detect and quantify presence of antibiotic-resistant strains, identify modes of transmission of ARGs, and illuminate complex expression pathways and epigenetic mechanisms (Cockerill, 1999; Cohen et al., 2015; Huijbers et al., 2015; Motta et al., 2015; dos Santos et al., 2016; Anjum et al., 2017). These techniques can be useful for establishing best management practices to reduce antimicrobial resistance (Durante-Mangoni and Zarrilli, 2011; Cohen et al., 2015), estimating and monitoring risk of exposure to antibiotic-resistant pathogens (Brul et al., 2012; Tyson et al., 2015; Haddad et al., 2018; Brockhurst et al., 2019), and assessing environmental impacts of antibiotic pollution (Cairns et al., 2018b; Danner et al., 2019). They have helped to uncover critical roles that the environment plays in determining the establishment, maintenance, and spread of ARGs via mechanisms such as co-selection, co-resistance, cross-resistance, hypermutation, and exchanges of plasmids, transposons, and integrons (Seiler and Berendonk, 2012; Singer et al., 2016; Pal et al., 2017). However, while the value of these insights and of the as-yet untapped potential of -omics techniques in general cannot be overstated, elucidation of the molecular biology of AMR should complement, not overshadow, our understanding of underlying ecological processes, such as the relationships that microbes have with other species in the environment (US Centers for Disease Control and Prevention and UK Science and Innovation Network, 2018; Brockhurst et al., 2019). Here, we review some of these ecological relationships, discuss their implications for AMR management, and highlight existing frameworks in community ecology that could be used to address the problem of AMR holistically and robustly.
Species Interactions Involving Antimicrobial Resistance
Competition
Although the intracellular and ecological functions of antibiotics in nature are varied and, in many cases, uncertain, at least some of the organisms that produce antibiotics appear to use them for allelopathy, “chemical warfare” with competing species (Sturz et al., 1998; Baquero et al., 2009; Raaijmakers and Mazzola, 2012; Chevrette and Currie, 2019). Penicillium notatum, for example, the mold famously discovered by Alexander Fleming to produce penicillin, was found, in that instance, to compete for resources and space with the bacterium Staphylococcus aureus (Demain and Elander, 1999; Bennett and Chung, 2001). This situation could increase the prevalence of ARGs in the environment, as ARG-possessing antibiotic producers outcompete their targets or as targets evolve ARGs in response to their competitors’ antibiotics. Even if microbes do not engage in allelopathy, possessing ARGs may increase their biological fitness due to co-benefits of the resistance mechanisms (e.g., ARGs coding for or regulating efflux pumps simultaneously provide resistance to heavy metals and other toxic compounds; Allen et al., 2010). On the other hand, if resource limitation is the predominant driver of microbial population dynamics and phenotypic plasticity in the expression of ARGs is insufficient to reduce costs (Auld et al., 2009), then microbes that do not house ARGs may have a competitive advantage over microbes that do, due to the energetic expenses or other physiological tradeoffs associated with ARGs (Kang and Park, 2010; Basra et al., 2018).
Moreover, under certain conditions, ARG-possessing microbes might facilitate rather than competitively exclude other microbes (Klümper et al., 2019). This has been shown to occur in one of two ways: through mutualism, which we describe in the next section, or through exploitation of “leaky” or “public” resistance mechanisms. For example, susceptible microbes can benefit from neighboring ARG-possessing competitors’ production and release of signaling compounds such as indole, a compound which activates drug efflux pumps and oxidative-stress protective mechanisms (Lee et al., 2010). As previously mentioned, antibiotics themselves may serve as signaling compounds, though they are more likely to be utilized by conspecifics and mutualistic symbionts than by competitors (Allen et al., 2010). Relative costliness of ARGs may create a selection pressure favoring exploiters over resistance builders, so that only the microbes that require ARGs for essential life processes retain their ARGs over time. Such “race to the bottom” coevolutionary dynamics are known as “Black Queen” dynamics (Morris et al., 2012; Cairns et al., 2018a). Microbes exhibiting Black Queen dynamics would represent a best-case scenario for human priorities regarding the evolution of AMR in pathogens, since it would entail selection against ARGs even under conditions favoring AMR.
Mutualism
The term “mutualism” refers to a relationship of mutually beneficial exchanges between different species (Hoeksema and Bruna, 2000). Certain mutualisms can enable microbes to acquire AMR without necessarily possessing their own independent ARGs. Two such mutualisms that have garnered attention in clinical settings and nutritional science are microbial biofilms and syntrophic consortia. Biofilms are structured associations of microbes that form on surfaces, including within food processing and water treatment facilities, various medical devices, and the human body (Donlan, 2002; Chmielewski and Frank, 2006; Ling et al., 2015; Kovach et al., 2017; Hu et al., 2018). They provide microbes with defenses against adverse conditions, increased efficiency in sequestering and assimilating nutrients, and other cooperative benefits (Jefferson, 2004; Nadell et al., 2016; Zhang et al., 2018). Syntrophic consortia are symbiotic associations of two or more microbial groups that allow for synthesis or degredation of substances that few or none of the constituent microbes would be able to synthesize or degrade on their own (Madigan et al., 2009; Morris et al., 2013; Bradáčová et al., 2019). Note that these characterizations are not mutually exclusive of one another: a biofilm can be a consortium and vice versa, depending on its location, form, and benefits.
The formation of biofilms can not only enhance the horizontal exchange of ARGs among constituent microbes (Molin and Tolker-Nielsen, 2003) but also confer AMR based on the physical and chemical structure of exopolysaccharides and other features of biofilm architecture shielding the constituent microbes’ cell envelopes (Mah and O’Tool, 2001; Jałowiecki et al., 2018). In addition, biofilms can provide resistance based on less intuitive mechanisms, such as slowed growth and inhibition of targeted metabolic processes (Olsen, 2015), achieved through the release of toxins by core constituent microbes (Lewis, 2005). Similarly, syntrophic consortia can allow microbes to gain AMR by creating enzymes in assembly-line fashion that degrade or inhibit antibiotics (e.g., β-lactamases; Olsen, 2015). This can be based on a small number of constituent microbes synthesizing the enzymes and the rest either supplying essential resources (Fan and He, 2011; Liu et al., 2017) or ameliorating factors such oxidative stress (Shatalin et al., 2011), or it can be based on multiple constituent microbes each synthesizing complementary parts of the enzymes (Burmølle et al., 2006; Islas-Espinoza et al., 2012). Mutualisms giving rise to exogenous or emergent AMR may call for the development of new methods of assessing antibiotic susceptibility that go beyond screening for conventional ARGs, at least in the case of the microbes known to engage in such mutualisms.
Predation and Parasitism
Microbes are often the prey or hosts of other species, including other microbes. Use of these natural enemies as biological control agents to combat clinical pathogens is a hot-topic area of research (which we delve deeper into in a later section) and has been shown to be effective under certain circumstances (Kutateladze and Adamia, 2010), including where pathogens already exhibit AMR (Willis et al., 2016). In natural environmental contexts, the influence of predation and parasitism on ARGs and AMR is complicated by the fact that certain predators and parasites produce antibiotics to subdue their prey or hosts and certain prey and hosts produce antibiotics to defend themselves from predators and parasites. For instance, predatory myxobacteria utilize antibiotics (myxovirescin and corallopyronin) to subdue prey such as Escherichia coli (Xiao et al., 2011), as does the non-obligate predator Aristabacter necator Strain 679-2 (pyrrolnitrin, maculosin, and banegasine; Cain et al., 2003). Antarctic sponges of the genus Crella produce antibiotic steroids (norselic acids A–E) that deter predators such as the amphipod Gondogeneia antarctica, as well as protozoan parasites of the genus Leishmania (Ma et al., 2009). Antibiotic-producing consumers and resources, like other antibiotic-producing organisms, may be sources of ARGs in sympatric species and select for AMR in the targets of their antibiotics.
Complex Associations and Indirect Effects
Although the broad categories of competition, mutualism, and predation/parasitism accommodate the entire spectrum of fundamental pairwise relationships within ecology (from mutually detrimental to mutually beneficial), these pairwise relationships do not exist in a vacuum. In the context of real-world ecological communities, mutualisms can enable predators and parasites to subdue their prey or hosts (Hwang et al., 1989; Mlot, 1997; Jones and Nishiguchi, 2004; Shiga, 2005), enable prey or hosts to fend off their predators and parasites (Soler et al., 2010; Pauli et al., 2014; Flórez et al., 2015; Van Arnam et al., 2018; Chevrette and Currie, 2019), and enable competitors to exclude their rivals (Preer et al., 1953; Brown et al., 2008; Mangla et al., 2008). Some of these complex associations and indirect interactions are mediated by antibiotics and ARGs. For example, the medicinal plant Leptospermum scoparium relies on endophytic bacteria to produce antibiotics such as phenazine and 2,4-diacetylphloroglucinol, which inhibit infection of the plant by pathogens such as Pseudomonas syringae pv. actinidiae (Wicaksono et al., 2018). Wicaksono et al. (2018) found that these bacteria were transmissible to other plants and could therefore be used for biological control of the plant diseases. Similarly, the pest beetle Lagria villosa relies on Burkholderia gladioli to produce icosalide, a lipocyclopeptide antibiotic that protects the beetle’s offspring from entomopathogenic bacteria (Dose et al., 2018), bacteria which predatory nematodes often rely on to envenomate prey (Mlot, 1997). Fungus-growing attine ants weed out microfungal parasites (competitors for the ants’ food) of the genus Escovopsis from their fungal gardens using antibiotics produced by a streptomycete bacterium that resides within their cuticles (Currie et al., 2003; Little and Currie, 2007). There is evidence to suggest that the ants might deliberately (arguably “artificially”) select their antibiotic-producing bacteria (Barke et al., 2011) and that the nature of the relationship has permitted the antibiotics to remain effective in controlling the parasitic fungi despite millions of years of coevolution (Pathak et al., 2019). In a study that had implications for agriculture, Li and Alexander (1988) found that use of antibiotic-producing soil inoculants can enhance crop yield and resilience by increasing colonization and nodulation by rhizobia.
Antibiotics and ARGs associated with symbionts of macroscopic hosts seem, at first glance, unlikely to be an important source of antibiotics and ARGs in clinical, industrial, or water treatment settings. If they have shared a long co-evolutionary history with their macroscopic hosts, the symbionts will likely have lost certain functional traits that are important to the fitness of their free-living counterparts (Bennett and Moran, 2015) and have as restricted a geographical distribution as their hosts (Martiny et al., 2006; Joseph et al., 2016). Three caveats to consider, however, are that: (1) even if the symbionts die when their hosts die, the symbionts’ antibiotics and genes are released into the environment; (2) animal migration, human globalization, and other modes of dispersal all permit spread to management-relevant environments (Brown and Barker, 1999; Molmeret et al., 2005; Allen et al., 2010; Forsberg et al., 2012; Zurek and Ghosh, 2014); and (3) many hosts of microbial symbionts are known vectors of disease. Investigating what Galimand et al. (2006) have referred to as a “clinically ominous event,” Hinnebusch et al. (2002) demonstrated that horizontal gene transfer among the microbes inside the Oriental rat flea Xenopsylla cheopis may have been the cause of AMR in Yersinia pestis strains isolated from bubonic plague patients in Madagascar.
Community Ecological Approaches to Addressing Antimicrobial Resistance
While using methods such as -omics to comprehensively profile microbial communities both in and out of clinical settings is still a major priority, the biological data produced through these methods must ultimately be interpreted and synthesized for there to be progress in addressing the problem. The frameworks for this synthesis currently exist within community ecology. Two that are well-established and appropriate for the task are “integrated pest management” and “ecological succession.”
Integrated Pest Management
Integrated pest management (IPM) is a strategy designed to safeguard human health and the environment while avoiding, attenuating, or delaying pest outbreaks or associated damages (Kogan, 1998; Liang et al., 2015). Historically, the major focus of IPM has been minimization of pesticide use and enhancement of its efficacy in targeting herbivorous insects, weeds, and vectors of disease. Part of the motivation for this focus is that pesticides, globally, have promoted pesticide resistance among target species, adversely impacted non-target and beneficial organisms, and, in some cases, proven to be persistent in the environment (Barzman et al., 2015). The issue is analogous to that of antibiotics promoting AMR and other adverse outcomes. Although Magarey et al. (2019) and others (Fidler, 1998; Orzech and Nichter, 2008) have noted that complex cultural, legal, and socioeconomic factors underlie both issues and must be addressed for management practices to be successful, we focus here on providing basic understanding of relevant IPM approaches and elucidating how they might be applied in the context of AMR. Specifically, we consider the approaches of biological control and habitat manipulation.
Biological control is the use of living organisms to control pests. As previously stated, microbes, including those possessing ARGs, have natural enemies such as predators and parasites. A few studies highlight the potential of using these natural enemies to control pathogens responsible for post-harvest diseases of fruits and vegetables (Wilson et al., 1993), tree and woody plant diseases (Cazorla and Mercado-Blanco, 2016), and even diseases within humans and domesticated animals (Negus et al., 2017). Among the presently favored candidate biological control agents are protozoan bacterivores and plasmid-dependent bacteriophages. By decreasing microbial population abundance (the prerequisite for heritable variation and population viability) and exacting opposing selection pressures (compared to those exacted by antibiotics, other natural enemies, or competing microbes), these consumers can theoretically replace antibiotics or enhance the effectiveness of antibiotics and reduce the likelihood of AMR (Hiltunen et al., 2017; Cairns et al., 2018a,b).
Unfortunately, evidence is conflicting as to whether there are tradeoffs between AMR and defense against natural enemies. For example, whereas Chen et al. (2017b) found that AMR coincides with increased susceptibility to bacteriophages, Allen et al. (2017) found that AMR is associated with increased resistance to bacteriophages. If indeed AMR provides the co-benefit of defense against natural enemies, introduction of natural enemies may amplify the effectiveness and prevalence of AMR rather than decrease it. Moreover, in the environment of a gut or epiphytic microbiome, bacteriophages or other natural enemies of microbes may affect microbes that are necessary or beneficial for overall health more than they affect ARG-possessing pathogens. Equivalently, the natural enemies may prevent competitively inferior ARG-possessing microbes from being excluded by other microbes (Bohannan et al., 2002), in a manner akin to “keystone predation” (where apex predators preferentially feed on superior resource competitors, promoting biodiversity among the prey; Amarasekare, 2008).
Due to these concerns, biological control of pathogens, especially within humans and domesticated animals, requires forethought and the means to ensure that “ecological release” of the biological control agents and other potential adverse outcomes do not occur within or on the patients (Miller and Aplet, 1993; Louda et al., 2003; Shanmuganathan et al., 2009). Barratt et al. (2010) noted that numerous recent advances in risk assessment methodologies have helped to minimize the possibility of adverse outcomes of biological control. These include multi-factorial assessments of control agent viability, quarantine laboratory host range testing, pre- and post-release studies incorporating mechanistic population dynamics models, and meta-analysis of cumulative data from past introductions. Each of these advances (with the possible exception of meta-analysis of past introductions) can be tailored to evaluate the efficacy of biological control of pathogens and AMR. For example, comprehensive profiles of microbial communities both in and out of clinical settings can be assembled via sampling and manipulative experiments and be used to establish the host/prey specificity of putative biological control agents or select the most appropriate stage of infection or population growth at which to introduce biological control agents.
Habitat manipulation entails altering the physical, chemical, or biological structure of the landscape to inhibit pests or to benefit or manipulate the natural enemies and competitors of the pests. This includes creating or manipulating breeding grounds and refuges against adverse abiotic conditions, such as overwintering sites in terrestrial systems (Griffiths et al., 2008) or zones of oxygenated hypolimnion in aquatic systems (Klumb et al., 2004). It also includes providing incentives for pests to leave or reduce their impacts (e.g., trap crops; Badenes-Perez et al., 2004), targeting the pests’ symbionts and vectors (Beard et al., 2002; Pocquet et al., 2014), and introducing alternative prey or hosts to amplify the effects of natural enemies via apparent competition (Holt and Bonsall, 2017). In the context of managing AMR, the “landscape” may be outdoor environments and open public spaces serving as source pools of ARGs or hubs of transmission; indoor facilities and equipment; or individual vector, host, or patient microbiomes. At the larger scale, habitat manipulation might thus entail familiar tactics such as regulating and remediating pollutants, employing safe and sustainable hygiene practices, and removing conditions that favor human, plant, or animal exposure to vectors of disease. In the case of microbiomes, analogues of altering abiotic conditions, targeting of pests’ symbionts, and introducing alternative prey or hosts to amplify natural enemies’ effects might include manipulating dietary nutrients, utilizing prophylactics and probiotics (see, however, Gueimonde et al., 2013 for a cautionary perspective on this approach), and targeting non-pathogenic (or less pathogenic) mutualistic partners of pathogens rather than the pathogens themselves. Recent preliminary studies have even suggested that microbes not ordinarily present in the hosts’ organs could be employed for this purpose; e.g., photosynthetic cyanobacteria could be injected into the heart to convert blood carbon dioxide to oxygen and thereby inhibit the causes and symptoms of heart disease and necrosis (Cohen et al., 2017). Treatment strategies that counteract the benefit of exogenous or emergent AMR, such as the use of agents that disrupt quorum sensing or target specific components of the biofilm matrix and the use of biofilm-penetrating antibiotics, would also be a worthwhile extension of the principle of habitat manipulation (Donlan, 2000; Sully et al., 2014).
Ecological Succession
Ecological succession is the phenomenon of biological communities forming or re-assembling and changing over time following natural or anthropogenic disturbances. These disturbances can include influx or loss of resources or habitat structure (e.g., due to urban development), invasion of the ecological community by exotic species, and changes in abiotic conditions due to seasonality or pollution. They can also often be autochthonous, as in the case of pioneer lichen creating soil from bare rock (Shure and Ragsdale, 1977). Patterns of succession affect functional traits within species and interaction strengths among species and depend on factors such as disturbance severity and species’ dispersal limitations (Chang and Turner, 2019). Ecological succession theory has been developed within classical microbiology, even including versions of the concepts of r- and K-strategists (Dorodnikov et al., 2009). However, two popular microbiological paradigms have created some divergence between microbial ecological succession theory and classical ecological succession theory: that of “everything is everywhere, and the environment selects” (O’Malley, 2007) and of microbial utilization of resources being determined thermodynamically by reduction-oxidation reaction-related processes (a.k.a. the “Redox Tower”; Delattre et al., 2019). Through the lens of these paradigms, antibiotics constitute two separate forms of disturbance: a selection pressure against non-resistant microbes (which would release some of their metabolic resources and space, making them available to other microbes) and a metabolic resource for AMR-exhibiting microbes. Combining this concept with an understanding of the previously mentioned syntrophic consortia, it may be possible to predict the rate at which and extent to which antibiotics would be enzymatically degraded in the presence of particular microbial assemblages and the likelihood of ARG spread within those assemblages. This information could be used to assess or predict environmental impacts and legacy effects of antibiotics or to develop novel mitigation strategies. To some extent, it has already been used to develop or enhance certain bioremediation techniques (US Environmental Protection Agency, 2013).
However, real-world microbial succession is typically more complicated (Chen et al., 2017a). Within microbiomes of macroscopic hosts, various potential metabolic resources can become available simultaneously (e.g., via mixed diets), and host immune responses, natural enemies, and competitors of various kinds may also be present. Furthermore, as stated previously, microbes can obtain exogenous or emergent AMR via exploitation of ARG-possessing microbes, potentially promoting Black Queen coevolutionary dynamics (see Competition subsection of Species Interactions Involving AMR). Depending on the level of impact it has on the ARG-possessing microbes, this could allow for coexistence or be analogous to successional replacement of “pioneer” and “nurse” plants (Whittaker, 1993; Tewksbury and Lloyd, 2001). Conceptual models within classical ecological succession theory that could be used to predict such outcomes include that of “alternative stable states” (Fukami and Nakajima, 2011) and that of the “Stress-Gradient Hypothesis” (Bertness and Callaway, 1994). The latter, originally developed for terrestrial plant communities (and later refined by Malkinson and Tielbörger, 2010), posits that, among potential competitors, intense grazing pressure or abiotic stress can increase the prevalence and importance of facilitative interactions such as “associational defense” and “neighborhood habitat amelioration,” provided that the competitors possess adequate complementarity in their physiological traits. Studies of AMR across gradients of antibiotic pollution, other abiotic stress, and bacterivore biodiversity (e.g., Baquero and Negri, 1997; Dunivin and Shade, 2018; Harmand et al., 2018) could be used to parameterize numerical simulations based on the hypothesis and test whether facilitative interactions in the form of AMR-conferring multi-species syntrophic consortia and biofilms occur as predicted along these gradients.
Discussion
AMR is a global human health concern, exacerbated by human population growth and socioeconomic inequity (Allcock et al., 2017; Jensen et al., 2019), climate change (MacFadden et al., 2018), and biological warfare/terrorism (Inglesby et al., 2000). It is also a pressing concern within the realms of industry, food production, and environmental health (Figure 1). The approach of “kill everything susceptible for as long as it is susceptible” has proven to be short-sighted and costly, with far-reaching implications. The relationships that microbes have with other species are an integral component of AMR ecology and should therefore be accounted for in both management initiatives and basic research. As we have described above, these relationships can have significant and sometimes multifaceted roles in determining the prevalence and distribution of AMR and ARGs. They and their effects call for subtle changes in our thinking regarding the boundaries of the problem that allow for innovations in the management of AMR, innovations that could easily and appropriately draw from existing frameworks within the field of community ecology. Priorities that would enable this progress include further characterizing AMR ecology in complex microbial communities and formally articulating and evaluating the links between this ecology and the risks to human and environmental health via mechanistic models and experimental tests of ecological theory.
Among the many questions still needing to be addressed regarding AMR ecology is what impact ARGs have on the longevity and infectivity of pathogens in the environment, outside of their hosts. Although many studies have traced ARGs introduced through human activities such as the generation of wastewaters, few have examined how the microbes associated with these ARGs persist and interact with other species in their new surroundings (Rodriguez-Mozaz et al., 2015; Bengtsson-Palme et al., 2017; Smalla et al., 2018). Knowing how frequently horizontal gene transfer of ARGs occurs among mutualistic symbionts, across trophic levels of microbial food webs, and in general (and, more importantly, knowing why) would enable researchers to better estimate the probability of non-pathogenic organisms with ARGs spreading resistance to the pathogens of humans, plants, and domesticated animals (Cairns et al., 2018a,b). Ideally, future research and mitigation efforts will leverage the fundamentals of modern microbial ecology, bringing together the principles and tools of molecular genetics, classical microbiology, and community ecology to allow us to better understand and manage the processes driving this complex challenge.
Author Contributions
AB provided the premise and framework of the manuscript and contributed the bulk of the writing and literature review pertaining to ecological theory and applications. MJ, MH, NB, and SK each contributed to the background regarding molecular/genetic aspects of AMR and AMR-related exposure risks, assisted in proof-reading, and helped to finalize the overall scope of the manuscript.
Funding
The U.S. Environmental Protection Agency, through its Office of Research and Development, partially funded and participated in the research described herein. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Jay Garland, Eric Villegas, Mark Bagley, Jorge Santo Domingo, and our reviewers for their insights. All photographic images were obtained from the public domain (Wikimedia Commons, the free media repository).
References
Aćimović, S. G., Zeng, Q., McGhee, G. C., Sundin, G. W., and Wise, J. C. (2015). Control of fire blight (Erwinia amylovora) on apple trees with trunk-injected plant resistance inducers and antibiotics and assessment of induction of pathogenesis-related protein genes. Front. Plant Sci. 6:16. doi: 10.3389/fpls.2015.00016
Allcock, S., Young, E. H., Holmes, M., Gurdasani, D., Dougan, G., Sandhu, M. S., et al. (2017). Antimicrobial resistance in human populations: challenges and opportunities. Glob. Health Epidemiol. Genom. 2:e4. doi: 10.1017/gheg.2017.12
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Allen, R. C., Pfrunder-Cardozo, K. R., Meinel, D., Egli, A., and Hall, A. R. (2017). Associations among antibiotic and phage resistance phenotypes in natural and clinical Escherichia coli isolates. MBio 8, e01341–e01317. doi: 10.1128/mBio.01341-17
Amarasekare, P. (2008). Spatial dynamics of keystone predation. J. Anim. Ecol. 77, 1306–1315. doi: 10.1111/j.1365-2656.2008.01439.x
Anjum, M. F., Zankari, E., and Hasman, H. (2017). Molecular methods for detection of antimicrobial resistance. Microbiol. Spectr. 5. doi: 10.1128/microbiolspec.ARBA-0011-2017
Arora, R., Behera, S., and Kumar, S. (2015). Bioprospecting thermophilic/thermotolerant microbes for production of lignocellulosic ethanol: a future perspective. Renew. Sust. Energ. Rev. 51, 699–717. doi: 10.1016/j.rser.2015.06.050
Auld, J. R., Agrawal, A. A., and Relyea, R. A. (2009). Re-evaluating the costs and limits of adaptive phenotypic plasticity. Proc. R. Soc. B 277, 503–511. doi: 10.1098/rspb.2009.1355
Badenes-Perez, F. R., Shelton, A. M., and Nault, B. A. (2004). Evaluating trap crops for diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). J. Econ. Entomol. 97, 1365–1372. doi: 10.1093/jee/97.4.1365
Baquero, F., Alvarez-Ortega, C., and Martinez, J. L. (2009). Ecology and evolution of antibiotic resistance. Environ. Microbiol. Rep. 1, 469–476. doi: 10.1111/j.1758-2229.2009.00053.x
Baquero, F., and Negri, M. C. (1997). Selective compartments for resistant microorganisms in antibiotic gradients. BioEssays 19, 731–736. doi: 10.1002/bies.950190814
Barke, J., Seipke, R. F., Yu, D. W., and Hutchings, M. I. (2011). A mutualistic microbiome: how do fungus-growing ants select their antibiotic-producing bacteria? Commun. Integr. Biol. 4, 41–43. doi: 10.4161/cib.4.1.13552
Baron, S. A., Diene, S. M., and Rolain, J.-M. (2018). Human microbiomes and antibiotic resistance. Hum. Microbiome J. 10, 43–52. doi: 10.1016/j.humic.2018.08.005
Barratt, B. I. P., Howarth, F. G., Withers, T. M., Kean, J. M., and Ridley, G. S. (2010). Progress in risk assessment for classical biological control. Biol. Control 52, 245–254. doi: 10.1016/j.biocontrol.2009.02.012
Barzman, M., Bàrberi, P., Birch, A. N. E., Boonekamp, P., Dachbrodt-Saaydeh, S., Graf, B., et al. (2015). Eight principles of integrated pest management. Agron. Sustain. Dev. 35, 1199–1215. doi: 10.1007/s13593-015-0327-9
Basra, P., Alsaadi, A., Bernal-Astrain, G., O’Sullivan, M. L., Hazlett, B., Clarke, L. M., et al. (2018). Fitness tradeoffs of antibiotic resistance in extraintestinal pathogenic Escherichia coli. Genome Biol. Evol. 10, 667–679. doi: 10.1093/gbe/evy030
Beard, C. B., Cordon-Rosales, C., and Durvasula, R. V. (2002). Bacterial symbionts of the triatominae and their potential use in control of Chagas disease transmission. Annu. Rev. Entomol. 47, 123–141. doi: 10.1146/annurev.ento.47.091201.145144
Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. J. (2017). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 42:p.fux053. doi: 10.1093/femsre/fux053
Bengtsson-Palme, J., and Larsson, D. G. J. (2015). Antibiotic resistance genes in the environment: prioritizing risks. Nat. Rev. Microbiol. 13:396. doi: 10.1038/nrmicro3399-c1
Bennett, J. W., and Chung, K.-T. (2001). Alexander Fleming and the discovery of penicillin. Adv. Appl. Microbiol. 49, 163–184. doi: 10.1016/S0065-2164(01)49013-7
Bennett, G. M., and Moran, N. A. (2015). Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc. Natl. Acad. Sci. USA 112, 10169–10176. doi: 10.1073/pnas.1421388112
Benveniste, R., and Davies, J. (1973). Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA 70, 2276–2280.
Berendonk, T. U., Manaia, C. M., Merlin, C., Fatta-Kassinos, D., Cytryn, E., Walsh, F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13, 310–317. doi: 10.1038/nrmicro3439
Bertness, M. D., and Callaway, R. (1994). Positive interactions in communities. Trends Ecol. Evol. 9, 191–193.
Bohannan, B. J. M., Kerr, B., Jessup, C. M., Hughes, J. B., and Sandvik, G. (2002). Trade-offs and coexistence in microbial microcosms. Antonie Van Leeuwenhoek 81, 107–115. doi: 10.1023/A:1020585711378
Bourdichon, F., Casaregola, S., Farrokh, C., Frisvad, J. C., Gerds, M. L., Hammes, W. P., et al. (2012). Food fermentations: microorganisms with technological beneficial use. Int. J. Food Microbiol. 154, 87–97. doi: 10.1016/j.ijfoodmicro.2011.12.030
Bradáčová, K., Florea, A. S., Bar-Tal, A., Minz, D., Yermiyahu, U., Shawahna, R., et al. (2019). Microbial consortia versus single-strain inoculants: an advantage in PGPM-assisted tomato production? Agronomy 9:105. doi: 10.3390/agronomy9020105
Brockhurst, M. A., Harrison, F., Veening, J.-W., Harrison, E., Blackwell, G., Iqbal, Z., et al. (2019). Assessing evolutionary risks of resistance for new antimicrobial therapies. Nat. Ecol. Evol. 3, 515–517. doi: 10.1038/s41559-019-0854-x
Brown, M. R. W., and Barker, J. (1999). Unexplored reservoirs of pathogenic bacteria: protozoa and biofilms. Trends Microbiol. 7, 46–50.
Brown, S. P., Le Chat, L., and Taddei, F. (2008). Evolution of virulence: triggering host inflammation allows invading pathogens to exclude competitors. Ecol. Lett. 11, 44–51. doi: 10.1111/j.1461-0248.2007.01125.x
Brul, S., Bassett, J., Cook, P., Kathariou, S., McClure, P., Jasti, P. R., et al. (2012). ‘Omics’ technologies in quantitative microbial risk assessment. Trends Food Sci. Technol. 27, 12–24. doi: 10.1016/j.tifs.2012.04.004
Burmølle, M., Webb, J. S., Rao, D., Hansen, L. H., Sørensen, S. J., and Kjelleberg, S. (2006). Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 72, 3916–3923. doi: 10.1128/AEM.03022-05
Cain, C. C., Lee, D., Waldo, R. H. III, Henry, A. T., Casida, E. J. Jr., Wani, M. C., et al. (2003). Synergistic antimicrobial activity of metabolites produced by a nonobligate bacterial predator. Antimicrob. Agents Chemother. 47, 2113–2117. doi: 10.1128/AAC.47.7.2113-2117.2003
Cairns, J., Koskinen, K., Penttinen, R., Patinen, T., Hartikainen, A., Jokela, R., et al. (2018a). Black queen evolution and trophic interactions determine plasmid survival after the disruption of the conjugation network. mSystems 3, e00104–e00118. doi: 10.1128/mSystems.00104-18
Cairns, J., Ruokolainen, L., Hultman, J., Tamminen, M., Virta, M., and Hiltunen, T. (2018b). Ecology determines how low antibiotic concentration impacts community composition and horizontal transfer of resistance genes. Commun. Biol. 1:35. doi: 10.1038/s42003-018-0041-7
Cazorla, F. M., and Mercado-Blanco, J. (2016). Biological control of tree and woody plant diseases: an impossible task? BioControl 61, 233–242. doi: 10.1007/s10526-016-9737-0
Chakradhar, S. (2016). What’s old is new: reconfiguring known antibiotics to fight drug resistance. Nat. Med. 22, 1197–1199. doi: 10.1038/nm1116-1197
Chang, C. C., and Turner, B. L. (2019). Ecological succession in a changing world. J. Ecol. 107, 503–509. doi: 10.1111/1365-2745.13132
Chen, J., Hanke, A., Tegetmeyer, H. E., Kattelmann, I., Sharma, R., Hamann, E., et al. (2017a). Impacts of chemical gradients on microbial community structure. ISME J. 11, 920–931. doi: 10.1038/ismej.2016.175
Chen, L.-K., Kuo, S.-C., Chang, K.-C., Cheng, C.-C., Yu, P.-Y., Chang, C.-H., et al. (2017b). Clinical antibiotic-resistant Acinetobacter baumannii strains with higher susceptibility to environmental phages than antibiotic-sensitive strains. Sci. Rep. 7:6319. doi: 10.1038/s41598-017-06688-w
Chevrette, M. G., and Currie, C. R. (2019). Emerging evolutionary paradigms in antibiotic discovery. J. Ind. Microbiol. Biotechnol. 46, 257–271. doi: 10.1007/s10295-018-2085-6
Chmielewski, R. A. N., and Frank, J. F. (2006). Biofilm formation and control in food processing facilities. Compr. Rev. Food Sci. Food Saf. 2, 22–32. doi: 10.1111/j.1541-4337.2003.tb00012.x
Cockerill, F. R. III. (1999). Genetic methods for assessing antimicrobial resistance. Antimicrob. Agents Chemother. 43, 199–212.
Cohen, A., Bont, L., Engelhard, D., Moore, E., Fernández, D., Kreisberg-Greenblatt, R., et al. (2015). A multifaceted ‘omics’ approach for addressing the challenge of antimicrobial resistance. Future Microbiol. 10, 365–376. doi: 10.2217/fmb.14.127
Cohen, J. E., Goldstone, A. B., Paulsen, M. J., Shudo, Y., Steele, A. N., Edwards, B. B., et al. (2017). An innovative biologic system for photon-powered myocardium in the ischemic heart. Sci. Adv. 3:e1603078. doi: 10.1126/sciadv.1603078
Collignon, P. J., and McEwen, S. A. (2019). One health—its importance in helping to better control antimicrobial resistance. Trop. Med. Infect. Dis. 4:22. doi: 10.3390/tropicalmed4010022
Currie, C. R., Wong, B., Stuart, A. E., Schultz, T. R., Rehner, S. A., Mueller, U. G., et al. (2003). Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science 299, 386–388. doi: 10.1126/science.1078155
Czaplewski, L., Bax, R., Clokie, M., Dawson, M., Fairhead, H., Fischetti, V. A., et al. (2016). Alternatives to antibiotics—a pipeline portfolio review. Lancet Infect. Dis. 16, 239–251. doi: 10.1016/S1473-3099(15)00466-1
Danner, M. C., Robertson, A., Behrends, V., and Reiss, J. (2019). Antibiotic pollution in surface fresh waters: occurrence and effects. Sci. Total Environ. 664, 793–804. doi: 10.1016/j.scitotenv.2019.01.406
Decaestecker, E., and King, K. (2019). “Red queen dynamics” in Reference module in earth systems and environmental sciences, volume 3 – encyclopedia of ecology. 2nd Edn. ed. B. Fath (Amsterdam, The Netherlands: Elsevier), 185–192.
Defoirdt, T., Sorgeloos, P., and Bossier, P. (2011). Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr. Opin. Microbiol. 14, 251–258. doi: 10.1016/j.mib.2011.03.004
Delattre, H., Quéméner, E. D.-L., Duquennoi, C., Filali, A., and Bouchez, T. (2019). Consistent microbial dynamics and functional community patterns derived from first principles. ISME J. 13, 263–276. doi: 10.1038/s41396-018-0272-0
Demain, A. L., and Elander, R. P. (1999). The β-lactam antibiotics: past, present, and future. Antonie Van Leeuwenhoek 75, 5–19. doi: 10.1023/A:1001738823146
Donlan, R. M. (2000). Role of biofilms in antimicrobial resistance. ASAIO J. 46, S47–S52. doi: 10.1097/00002480-200011000-00037
Donlan, R. M. (2002). Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890. doi: 10.3201/eid0809.020063
Dorodnikov, M., Blagodatskaya, E., Blagodatsky, S., Fangmeier, A., and Kuzyakov, Y. (2009). Stimulation of r- vs. K-selected microorganisms by elevated atmospheric CO2 depends on soil aggregate size. FEMS Microbiol. Ecol. 69, 43–52. doi: 10.1111/j.1574-6941.2009.00697.x
dos Santos, B. S., da Silva, L. C. N., da Silva, T. D., Rodrigues, J. F. S., Grisotto, M. A. G., dos Santos Correia, M. T., et al. (2016). Application of omics technologies for evaluation of antibacterial mechanisms of action of plant-derived products. Front. Microbiol. 7:1466. doi: 10.3389/fmicb.2016.01466
Dose, B., Niehs, S. P., Scherlach, K., Flórez, L. V., Kaltenpoth, M., and Hertweck, C. (2018). Unexpected bacterial origin of the antibiotic icosalide: two-tailed depsipeptide assembly in multifarious burkholderia symbionts. ACS Chem. Biol. 13, 2414–2420. doi: 10.1021/acschembio.8b00600
Drusano, G. L. (2003). Prevention of resistance: a goal for dose selection for antimicrobial agents. Clin. Infect. Dis. 36, S42–S50. doi: 10.1086/344653
Dunivin, T. K., and Shade, A. (2018). Community structure explains antibiotic resistance gene dynamics over a temperature gradient in soil. FEMS Microbiol. Ecol. 94:fiy016. doi: 10.1093/femsec/fiy016
Durante-Mangoni, E., and Zarrilli, R. (2011). Molecular epidemiology and management of antimicrobial resistance. Future Microbiol. 6. doi: 10.2217/fmb.11.23
Fan, C., and He, J. (2011). Proliferation of antibiotic resistance genes in microbial consortia of sequencing batch reactors (SBRs) upon exposure to trace erythromycin or erythromycin-H2O. Water Res. 45, 3098–3106. doi: 10.1016/j.watres.2011.03.025
Ferrer, M., Martínez-Martínez, M., Bargiela, R., Streit, W. R., Golyshina, O. V., and Golyshin, P. N. (2016). Estimating the success of enzyme bioprospecting through metagenomics: current status and future trends. Microb. Biotechnol. 9, 22–34. doi: 10.1111/1751-7915.12309
Fidler, D. P. (1998). Legal issues associated with antimicrobial drug resistance. Emerg. Infect. Dis. 4, 169–177. doi: 10.3201/eid0402.980204
Flórez, L. V., Biedermann, P. H., Engl, T., and Kaltenpoth, M. (2015). Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 32, 904–936. doi: 10.1039/C5NP00010F
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O. A., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. doi: 10.1126/science.1220761
Fridkin, S. K., and Gaynes, R. P. (1999). Antimicrobial resistance in intensive care units. Clin. Chest Med. 20, 303–316. doi: 10.1016/S0272-5231(05)70143-X
Fukami, T., and Nakajima, M. (2011). Community assembly: alternative stable states or alternative transient states? Ecol. Lett. 14, 973–984. doi: 10.1111/j.1461-0248.2011.01663.x
Galimand, M., Carniel, E., and Courvalin, P. (2006). Resistance of Yersinia pestis to antimicrobial agents. Antimicrob. Agents Chemother. 50, 3233–3236. doi: 10.1128/AAC.00306-06
Grenni, P., Ancona, V., and Caracciolo, A. B. (2018). Ecological effects of antibiotics on natural ecosystems: a review. Microchem. J. 136, 25–39. doi: 10.1016/j.microc.2017.02.006
Griffiths, G. J. K., Holland, J. M., Bailey, A., and Thomas, M. B. (2008). Efficacy and economics of shelter habitats for conservation biological control. Biol. Control 45, 200–209. doi: 10.1016/j.biocontrol.2007.09.002
Gueimonde, M., Sánchez, B., de los Reyes-Gavilán, C. G., and Margolles, A. (2013). Antibiotic resistance in probiotic bacteria. Front. Microbiol. 4:202. doi: 10.3389/fmicb.2013.00202
Haddad, N., Johnson, N., Kathariou, S., Métris, A., Phister, T., Pielaat, A., et al. (2018). Next generation microbiological risk assessment—potential of omics data for hazard characterization. Int. J. Food Microbiol. 287, 28–39. doi: 10.1016/j.ijfoodmicro.2018.04.015
Hallmann, A., and Rappel, A. (1999). Genetic engineering of the multicellular green alga Volvox: a modified and multiplied bacterial antibiotic resistance gene as a dominant selectable marker. Plant J. 17, 99–109.
Harmand, N., Gallet, R., Martin, G., and Lenormand, T. (2018). Evolution of bacteria specialization along an antibiotic dose gradient. Evol. Lett. 2, 221–232. doi: 10.1002/evl3.52
Hiltunen, T., Virta, M., and Laine, A.-L. (2017). Antibiotic resistance in the wild: an eco-evolutionary perspective. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 372:20160039. doi: 10.1098/rstb.2016.0039
Hinnebusch, B. J., Rosso, M.-L., Schawn, T. G., and Carniel, E. (2002). High-frequency conjugative transfer of antibiotic resistance genes to Yersinia pestis in the flea midgut. Mol. Microbiol. 46, 349–354. doi: 10.1046/j.1365-2958.2002.03159.x
Hoeksema, J. D., and Bruna, E. M. (2000). Pursuing the big questions about interspecific mutualism: a review of theoretical approaches. Oecologia 125, 321–330. doi: 10.1007/s004420000496
Holt, R. D., and Bonsall, M. B. (2017). Apparent competition. Annu. Rev. Ecol. Evol. Syst. 48, 447–471. doi: 10.1146/annurev-ecolsys-110316-022628
Hu, X., Huang, Y.-Y., Wang, Y., Wang, X., and Hamblin, M. R. (2018). Antimicrobial photodynamic therapy to control clinically relevant biofilm infections. Front. Microbiol. 9:1299. doi: 10.3389/fmicb.2018.01299
Huijbers, P. M. C., Blaak, H., de Jong, M. C. M., Graat, E. A. M., Vandenbroucke-Grauls, C. M. J. E., and de Roda Husman, A. M. (2015). Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ. Sci. Technol. 49, 11993–12004. doi: 10.1021/acs.est.5b02566
Hwang, D. F., Arakawa, O., Saito, T., Noguchi, T., Simidu, U., Tsukamoto, K., et al. (1989). Tetrodotoxin-producing bacteria from the blue-ringed octopus Octopus maculosus. Mar. Biol. 100, 327–332. doi: 10.1007/BF00391147
Inglesby, T. V., Dennis, D. T., Henderson, D. A., Bartlett, J. G., Ascher, M. S., Eitzen, E., et al. (2000). Plague as a biological weapon: medical and public health management. JAMA 283, 2281–2290. doi: 10.1001/jama.283.17.2281
Islas-Espinoza, M., Reid, B. J., Wexler, M., and Bond, P. L. (2012). Soil bacterial consortia and previous exposure enhance the biodegradation of sulfonamides from pig manure. Microb. Ecol. 64, 140–151. doi: 10.1007/s00248-012-0010-5
Jałowiecki, L., Żur, J., Chojniak, J., Ejhed, H., and Płaza, G. (2018). Properties of antibiotic-resistant bacteria isolated from onsite wastewater treatment plant in relation to biofilm formation. Curr. Microbiol. 75, 639–649. doi: 10.1007/s00284-017-1428-2
Jebelli, M. A., Maleki, A., Amoozegar, M. A., Kalantar, E., Gharibi, F., Darvish, N., et al. (2018). Isolation and identification of the native population bacteria for bioremediation of high levels of arsenic from water resources. J. Environ. Manag. 212, 39–45. doi: 10.1016/j.jenvman.2018.01.075
Jefferson, K. K. (2004). What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236, 163–173. doi: 10.1111/j.1574-6968.2004.tb09643.x
Jensen, C. S., Nielsen, S. B., and Fynbo, L. (2019). “Risking antimicrobial resistance: a one health study of antibiotic use and its societal aspects” in Risking antimicrobial resistance. eds. C. Jensen, S. Nielsen, and L. Fynbo (Cham: Palgrave Macmillan), 1–24.
Jiang, X., Ellabaan, M. M. H., Charusanti, P., Munck, C., Blin, K., Tong, Y., et al. (2017). Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 8:15784. doi: 10.1038/ncomms15784
Joerger, R. D. (2003). Alternatives to antibiotics: bacteriocins, antimicrobial peptides and bacteriophages. Poult. Sci. 82, 640–647. doi: 10.1093/ps/82.4.640
Jones, B. W., and Nishiguchi, M. K. (2004). Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar. Biol. 144, 1151–1155. doi: 10.1007/s00227-003-1285-3
Joseph, M. B., Stutz, W. E., and Johnson, P. T. J. (2016). Multilevel models for the distribution of hosts and symbionts. PLoS One 11:e0165768. doi: 10.1371/journal.pone.0165768
Kang, Y.-S., and Park, W. (2010). Trade-off between antibiotic resistance and biological fitness in Acinetobacter sp. strain DR1. Environ. Microbiol. 12, 1304–1318. doi: 10.1111/j.1462-2920.2010.02175.x
Klumb, R. A., Bunch, K. L., Mills, E. L., Rudstam, L. G., Brown, G., Knauf, C., et al. (2004). Establishment of a metalimnetic oxygen refuge for zooplankton in a productive Lake Ontario embayment. Ecol. Appl. 14, 113–131. doi: 10.1890/02-5054
Klümper, U., Recker, M., Zhang, L., Yin, X., Zhang, T., Buckling, A., et al. (2019). Selection for antimicrobial resistance is reduced when embedded in a natural microbial community. ISME J. doi: 10.1038/s41396-019-0483-z
Kogan, M. (1998). Integrated pest management: historical perspectives and contemporary developments. Annu. Rev. Entomol. 43, 243–270. doi: 10.1146/annurev.ento.43.1.243
Kovach, K., Davis-Fields, M., Irie, Y., Jain, K., Doorwar, S., Vuong, K., et al. (2017). Evolutionary adaptations of biofilms infecting cystic fibrosis lungs promote mechanical toughness by adjusting polysaccharide production. npj Biofilms Microbiomes 3:1. doi: 10.1038/s41522-016-0007-9
Kutateladze, M., and Adamia, R. (2010). Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 28, 591–595. doi: 10.1016/j.tibtech.2010.08.001
Lee, H. H., Molla, M. N., Cantor, C. R., and Collins, J. J. (2010). Bacterial charity work leads to population-wide resistance. Nature 467, 82–85. doi: 10.1038/nature09354
Lewis, K. (2005). Persister cells and the riddle of biofilm survival. Biochemistry 70, 267–274. doi: 10.1007/s10541-005-0111-6
Li, D.-M., and Alexander, M. (1988). Co-inoculation with antibiotic-producing bacteria to increase colonization and nodulation by rhizobia. Plant Soil 108, 211–219. doi: 10.1007/BF02375651
Liang, J., Tang, S., Cheke, R. A., and Wu, J. (2015). Models for determining how many natural enemies to release inoculatively in combinations of biological and chemical control with pesticide resistance. J. Math. Anal. Appl. 422, 1479–1503. doi: 10.1016/j.jmaa.2014.09.048
Ling, F., Hwang, C., LeChevallier, M. W., Andersen, G. L., and Liu, W.-T. (2015). Core-satellite populations and seasonality of water meter biofilms in a metropolitan drinking water distribution system. ISME J. 10, 582–595. doi: 10.1038/ismej.2015.136
Little, A. E. F., and Currie, C. R. (2007). Symbiotic complexity: discovery of a fifth symbiont in the attine ant-microbe symbiosis. Biol. Lett. 3, 501–504. doi: 10.1098/rsbl.2007.0253
Liu, Y., Chang, H., Li, Z., Feng, Y., Cheng, D., and Xue, J. (2017). Biodegradation of gentamicin by bacterial consortia AMQD4 in synthetic medium and raw gentamicin sewage. Sci. Rep. 7:11004. doi: 10.1038/s41598-017-18286-x
Livermore, D. M. (2011). Discovery research: the scientific challenge of finding new antibiotics. J. Antimicrob. Chemother. 66, 1941–1944. doi: 10.1093/jac/dkr262
Louda, S. M., Pemberton, R. W., Johnson, M. T., and Follett, P. A. (2003). Nontarget effects — the Achilles’ heel of biological control? Retrospective analyses to reduce risk associated with biocontrol introductions. Annu. Rev. Entomol. 48, 365–396. doi: 10.1146/annurev.ento.48.060402.102800
Ma, W. S., Mutka, T., Vesley, B., Amsler, M. O., McClintock, J. B., Amsler, C. D., et al. (2009). Norselic acids A-E, highly oxidized anti-infective steroids that deter mesograzer predation, from the Antarctic sponge Crella sp. J. Nat. Prod. 72, 1842–1846. doi: 10.1021/np900382x
MacFadden, D. R., McGough, S. F., Fisman, D., Santillana, M., and Brownstein, J. S. (2018). Antibiotic resistance increases with local temperature. Nat. Clim. Chang. 8, 510–514. doi: 10.1038/s41558-018-0161-6
Madigan, M. T., Bender, K. S., Buckley, D. H., Sattley, W. M., and Stahl, D. A. (2009). Brock biology of microorganisms. San Francisco: Pearson.
Magarey, R. D., Chappell, T. M., Trexler, C. M., Pallipparambil, G. R., and Hain, E. F. (2019). Social ecological system tools for improving crop pest management. J. Integr. Pest Manag. 10, 1–6. doi: 10.1093/jipm/pmz004
Mah, T.-F., and O’Tool, G. A. (2001). Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39. doi: 10.1016/S0966-842X(00)01913-2
Malkinson, D., and Tielbörger, K. (2010). What does the stress-gradient hypothesis predict? Resolving the discrepancies. Oikos 119, 1546–1552. doi: 10.1111/j.1600-0706.2010.18375.x
Mangla, S., Inderjit,, and Callaway, R. M. (2008). Exotic invasive plant accumulates native soil pathogens which inhibit native plants. J. Ecol. 96, 58–67. doi: 10.1111/j.1365-2745.2007.01312.x
Martinez, J. L. (2018). Ecology and evolution of chromosomal gene transfer between environmental microorganisms and pathogens. Microbiol. Spectr. 6. doi: 10.1128/microbiolspec.MTBP-0006-2016
Martiny, J. B. H., Bohannan, B. J. M., Brown, J. H., Colwell, R. K., Fuhrman, J. A., Green, J. L., et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112. doi: 10.1038/nrmicro1341
McArthur, J. V., Fletcher, D. E., Tuckfield, R. C., and Baker-Austin, C. (2016). Patterns of multi-antibiotic-resistant Escherichia coli from streams with no history of antimicrobial inputs. Microb. Ecol. 72, 840–850. doi: 10.1007/s00248-015-0678-4
Miller, M., and Aplet, G. (1993). Biological control: a little knowledge is a dangerous thing. Rutgers Law Rev. 45, 285–334.
Mišurcová, L., Škrovánková, S., Samek, D., Ambrožová, J., and Machů, L. (2012). Chapter 3 – health benefits of algal polysaccharides in human nutrition. Adv. Food Nutr. Res. 66, 75–145. doi: 10.1016/B978-0-12-394597-6.00003-3
Molin, S., and Tolker-Nielsen, T. (2003). Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14, 255–261. doi: 10.1016/S0958-1669(03)00036-3
Molmeret, M., Horn, M., Wagner, M., Santic, M., and Kwaik, Y. A. (2005). Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71, 20–28. doi: 10.1128/AEM.71.1.20-28.2005
Moloney, M. G. (2016). Natural products as a source for novel antibiotics. Trends Pharmacol. Sci. 37, 689–701. doi: 10.1016/j.tips.2016.05.001
Morris, B. E. L., Henneberger, R., Huber, H., and Moissl-Eichinger, C. (2013). Microbial syntrophy: interaction for the common good. FEMS Microbiol. Rev. 37, 384–406. doi: 10.1111/1574-6976.12019
Morris, J. J., Lenski, R. E., and Zinser, E. R. (2012). The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio 3, e00036–e00012. doi: 10.1128/mBio.00036-12
Motta, S. S., Cluzel, P., and Aldana, M. (2015). Adaptive resistance in bacteria requires epigenetic inheritance, genetic noise, and cost of efflux pumps. PLoS One 10:e0118464. doi: 10.1371/journal.pone.0118464
Nadell, C. D., Drescher, K., and Foster, K. R. (2016). Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 14, 589–600. doi: 10.1038/nrmicro.2016.84
Negus, D., Moore, C., Baker, M., Raghunathan, D., Tyson, J., and Sockett, R. E. (2017). Predator versus pathogen: how does predatory Bdellovibrio bacteriovorus interface with the challenges of killing Gram-negative pathogens in a host setting? Annu. Rev. Microbiol. 71, 441–457. doi: 10.1146/annurev-micro-090816-093618
Nerlich, B., and James, R. (2009). “The post-antibiotic apocalypse” and the “war on superbugs”: catastrophe discourse in microbiology, its rhetorical form and political function. Public Underst. Sci. 18, 574–590. doi: 10.1177/0963662507087974
O’Malley, M. A. (2007). The nineteenth century roots of ‘everything is everywhere’. Nat. Rev. Microbiol. 5, 647–651. doi: 10.1038/nrmicro1711
Oliver, N. J., Rabinovitch-Deere, C. A., Carroll, A. L., Nozzi, N. E., Case, A. E., and Atsumi, S. (2016). Cyanobacterial metabolic engineering for biofuel and chemical production. Curr. Opin. Chem. Biol. 35, 43–50. doi: 10.1016/j.cbpa.2016.08.023
Olsen, I. (2015). Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 34, 877–886. doi: 10.1007/s10096-015-2323-z
Orzech, K. M., and Nichter, M. (2008). From resilience to resistance: political ecological lessons from antibiotic and pesticide resistance. Annu. Rev. Anthropol. 37, 267–282. doi: 10.1146/annurev.anthro.37.081407.085205
Pal, C., Asiani, K., Arya, S., Rensing, C., Stekel, D. J., Larsson, D. G. J., et al. (2017). Metal resistance and its association with antibiotic resistance. Adv. Microb. Physiol. 70, 261–313. doi: 10.1016/bs.ampbs.2017.02.001
Pathak, A., Kett, S., and Marvasi, M. (2019). Resisting antimicrobial resistance: lessons from fungus farming ants. Trends Ecol. Evol. S0169-5347, 30256–30253. doi: 10.1016/j.tree.2019.08.007
Pauli, J. N., Mendoza, J. E., Steffan, S. A., Carey, C. C., Weimer, P. J., and Peery, M. Z. (2014). A syndrome of mutualism reinforces the lifestyle of a sloth. Proc. R. Soc. B Biol. Sci. 281:20133006. doi: 10.1098/rspb.2013.3006
Pocquet, N., Darriet, F., Zumbo, B., Milesi, P., Thiria, J., Bernard, V., et al. (2014). Insecticide resistance in disease vectors from Mayotte: an opportunity for integrated vector management. Parasit. Vectors 7:299. doi: 10.1186/1756-3305-7-299
Preer, J. R., Siegel, R. W., and Stark, P. S. (1953). The relationship between kappa and paramecin in Paramecium aurelia. Proc. Natl. Acad. Sci. USA 39, 1228–1233. doi: 10.1073/pnas.39.12.1228
Pruden, A., Larsson, D. G. J., Amézquita, A., Collignon, P., Brandt, K. K., Graham, D. W., et al. (2013). Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ. Health Perspect. 121, 878–885. doi: 10.1289/ehp.1206446
Raaijmakers, J. M., and Mazzola, M. (2012). Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 50, 403–424. doi: 10.1146/annurev-phyto-081211-172908
Reardon, S. (2015). Antibiotic alternatives rev up bacterial arms race. Nature 521, 402–403. doi: 10.1038/521402a
Richmond, A., and Preiss, K. (1980). The biotechnology of algaculture. Interdiscip. Sci. Rev. 5, 60–70.
Ringel, P. D., Hu, D., and Basler, M. (2017). The role of type VI secretion system effectors in target cell lysis and subsequent horizontal gene transfer. Cell Rep. 21, 3927–3940. doi: 10.1016/j.celrep.2017.12.020
Rodriguez-Mozaz, S., Chamorro, S., Marti, E., Huerta, B., Gros, M., Sànchez-Melsió, A., et al. (2015). Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 69, 234–242. doi: 10.1016/j.watres.2014.11.021
Rothrock, M. J. Jr., Hiett, K. L., Guard, J. Y., and Jackson, C. R. (2016). Antibiotic resistance patterns of major zoonotic pathogens from all-natural, antibiotic-free, pasture-raised broiler flocks in the southeastern United States. J. Environ. Qual. 45, 593–603. doi: 10.2134/jeq2015.07.0366
Seifan, M., Samani, A. K., and Berenjian, A. (2016). Bioconcrete: next generation of self-healing concrete. Appl. Microbiol. Biotechnol. 100, 2591–2602. doi: 10.1007/s00253-016-7316-z
Seiler, C., and Berendonk, T. U. (2012). Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 3:399. doi: 10.3389/fmicb.2012.00399
Sen, T., Barrow, C. J., and Deshmukh, S. K. (2019). Microbial pigments in the food industry—challenges and the way forward. Front. Nutr. 6, 7–21. doi: 10.3389/fnut.2019.00007
Shanmuganathan, T., Pallister, J., Doody, S., McCallum, H., Robinson, T., Sheppard, A., et al. (2009). Biological control of the cane toad in Australia: a review. Anim. Conserv. 13, 16–23. doi: 10.1111/j.1469-1795.2009.00319.x
Shatalin, K., Shatalina, E., Mironov, A., and Nudler, E. (2011). H2S: a universal defense against antibiotics in bacteria. Science 334, 986–990. doi: 10.1126/science.1209855
Shaw, A. J., Lam, F. H., Hamilton, M., Consiglio, A., MacEwen, K., Brevnova, E. E., et al. (2016). Metabolic engineering of microbial competitive advantage for industrial fermentation processes. Science 353, 583–586. doi: 10.1126/science.aaf6159
Shiga, D. (2005). Poisonous partnership: parasitoid wasps use viruses as a weapon. Sci. News 167, 136–137. doi: 10.2307/4015898
Shure, D. J., and Ragsdale, H. L. (1977). Patterns of primary succession on granite outcrop surfaces. Ecology 58, 993–1006. doi: 10.2307/1936920
Singer, A. C., Shaw, H., Rhodes, V., and Hart, A. (2016). Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 7:1728. doi: 10.3389/fmicb.2016.01728
Smalla, K., Cook, K., Djordjevic, S. P., Klümper, U., and Gillings, M. (2018). Environmental dimensions of antibiotic resistance: assessment of basic science gaps. FEMS Microbiol. Ecol. 94:p.fiy195. doi: 10.1093/femsec/fiy195
Soler, J. J., Martín-Vivaldi, M., Peralta-Sánchez, J. M., and Ruiz-Rodríguez, M. (2010). Antibiotic-producing bacteria as a possible defence of birds against pathogenic microorganisms. Open Ornithol. J. 3, 93–100. doi: 10.2174/1874453201003010093
Stockwell, V. O., and Duffy, B. (2012). Use of antibiotics in plant agriculture. Rev. Sci. Tech. 31, 199–210. doi: 10.20506/rst.31.1.2104
Strobel, G., and Daisy, B. (2003). Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 67, 491–502. doi: 10.1128/mmbr.67.4.491-502.2003
Sturz, A. V., Christie, B. R., and Matheson, B. G. (1998). Associations of bacterial endophyte populations from red clover and potato crops with potential for beneficial allelopathy. Can. J. Microbiol. 44, 162–167. doi: 10.1139/w97-146
Sully, E. K., Malachowa, N., Elmore, B. O., Alexander, S. M., Femling, J. K., Gray, B. M., et al. (2014). Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 10:e1004174. doi: 10.1371/journal.ppat.1004174
Teuber, M., Meile, L., and Schwarz, F. (1999). “Acquired antibiotic resistance in lactic acid bacteria from food” in Lactic acid bacteria: Genetics, metabolism and applications. eds. W. N. Konings, O. P. Kuipers, and J. H. J. H. In’t Veld (Dordrecht: Springer).
Tewksbury, J. J., and Lloyd, J. D. (2001). Positive interactions under nurse-plants: spatial scale, stress gradients and benefactor size. Oecologia 127, 425–434. doi: 10.1007/s004420000614
Tuli, H. S., Chaudhary, P., Beniwal, V., and Sharma, A. K. (2015). Microbial pigments as natural color sources: current trends and future perspectives. J. Food Sci. Technol. 52, 4669–4678. doi: 10.1007/s13197-014-1601-6
Tyson, G. H., McDermott, P. F., Li, C., Chen, Y., Tadesse, D. A., Mukherjee, S., et al. (2015). WGS accurately predicts antimicrobial resistance in Escherichia coli. J. Antimicrob. Chemother. 70, 2763–2769. doi: 10.1093/jac/dkv186
US Centers for Disease Control and Prevention and UK Science and Innovation Network (2018). Initiatives for addressing antimicrobial resistance in the environment: Current situation and challenges. Available at: https://wellcome.ac.uk/sites/default/files/antimicrobial-resistance-environment-report.pdf (Accessed July 31, 2019).
US Environmental Protection Agency (2013). Introduction to in situ bioremediation of groundwater. Available at: https://www.epa.gov/sites/production/files/2015-04/documents/introductiontoinsitubioremediationofgroundwater_dec2013.pdf (Accessed July 31, 2019).
Van Arnam, E. B., Currie, C. R., and Clardy, J. (2018). Defense contracts: molecular protection in insect-microbe symbioses. Chem. Soc. Rev. 47, 1638–1651. doi: 10.1039/C7CS00340D
Van Goethem, M. W., Pierneef, R., Bezuidt, O. K. I., Van De Peer, Y., Cowan, D. A., and Makhalanyane, T. P. (2018). A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 6:40. doi: 10.1186/s40168-018-0424-5
Vikesland, P. J., Pruden, A., Alvarez, P. J. J., Aga, D., Bürgmann, H., Li, X.-D., et al. (2017). Toward a comprehensive strategy to mitigate dissemination of environmental sources of antibiotic resistance. Environ. Sci. Technol. 51, 13061–13069. doi: 10.1021/acs.est.7b03623
Wang, H., Park, J.-D., and Ren, Z. J. (2015). Practical energy harvesting for microbial fuel cells: a review. Environ. Sci. Technol. 49, 3267–3277. doi: 10.1021/es5047765
Wellington, E. M. H., Boxall, A. B. A., Cross, P., Feil, E. J., Gaze, W. H., Hawkey, P. M., et al. (2013). The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 13, 155–165. doi: 10.1016/S1473-3099(12)70317-1
Wells, M. L., Potin, P., Craigie, J. S., Raven, J. A., Merchant, S. S., Helliwell, K. E., et al. (2017). Algae as nutritional and functional food sources: revisiting our understanding. J. Appl. Phycol. 29, 949–982. doi: 10.1007/s10811-016-0974-5
White, P. S., and Walker, J. L. (1997). Approximating nature’s variation: selecting and using reference information in restoration ecology. Restor. Ecol. 5, 338–349. doi: 10.1046/j.1526-100X.1997.00547.x
Whittaker, R. J. (1993). Plant population patterns in a glacier foreland succession: pioneer herbs and later-colonizing shrubs. Ecography 16, 117–136. doi: 10.1111/j.1600-0587.1993.tb00064.x
Whittier, T. R., Stoddard, J. L., Larsen, D. P., and Herlihy, A. T. (2007). Selecting reference sites for stream biological assessments: best professional judgment or objective criteria. J. N. Am. Benthol. Soc. 26, 349–360. doi: 10.1899/0887-3593(2007)26[349:SRSFSB]2.0.CO;2
Wicaksono, W. A., Jones, E. E., Casonato, S., Monk, J., and Ridgway, H. J. (2018). Biological control of Pseudomonas syringae pv. actinidiae (Psa), the causal agent of bacterial canker of kiwifruit, using endophytic bacteria recovered from a medicinal plant. Biol. Control 116, 103–112. doi: 10.1016/j.biocontrol.2017.03.003
Wiese, J., and Imhoff, J. F. (2019). Marine bacteria and fungi as promising source for new antibiotics. Drug Dev. Res. 80, 24–27. doi: 10.1002/ddr.21482
Willing, B. P., Pepin, D. M., Marcolla, C. S., Forgie, A. J., Diether, N. E., and Bourrie, B. C. T. (2018). Bacterial resistance to antibiotic alternatives: a wolf in sheep’s clothing? Anim. Front. 8, 39–47. doi: 10.1093/af/vfy003
Willis, A. R., Moore, C., Mazon-Moya, M., Krokowski, S., Lambert, C., Till, R., et al. (2016). Injections of predatory bacteria work alongside host immune cells to treat Shigella infection in zebrafish larvae. Curr. Biol. 26, 3343–3351. doi: 10.1016/j.cub.2016.09.067
Wilson, C. L., Wisniewski, M. E., Droby, S., and Chalutz, E. (1993). A selection strategy for microbial antagonists to control postharvest diseases of fruits and vegetables. Sci. Hortic. 53, 183–189. doi: 10.1016/0304-4238(93)90066-Y
Xiao, Y., Wei, X., Ebright, R., and Wall, D. (2011). Antibiotic production by myxobacteria plays a role in predation. J. Bacteriol. 193, 4626–4633. doi: 10.1128/JB.05052-11
Zhang, S., Merino, N., Okamoto, A., and Gedalanga, P. (2018). Interkingdom microbial consortia mechanisms to guide biotechnological applications. Microb. Biotechnol. 11, 833–847. doi: 10.1111/1751-7915.13300
Keywords: antibiotic, biofilm, competition, consortia, mutualism, predation, indirect effects
Citation: Banerji A, Jahne M, Herrmann M, Brinkman N and Keely S (2019) Bringing Community Ecology to Bear on the Issue of Antimicrobial Resistance. Front. Microbiol. 10:2626. doi: 10.3389/fmicb.2019.02626
Edited by:
Giuseppantonio Maisetta, University of Pisa, ItalyReviewed by:
Jinxin Liu, University of California, Davis, United StatesHenry P. Godfrey, New York Medical College, United States
Copyright © 2019 Banerji, Jahne, Herrmann, Brinkman and Keely. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aabir Banerji, bHljYW50aHJvcHVzbG9yQGNvbWNhc3QubmV0
 Aabir Banerji
Aabir Banerji Michael Jahne
Michael Jahne Scott Keely
Scott Keely