
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 November 2019
Sec. Microbiotechnology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02586
 Chandan Mukherjee1
Chandan Mukherjee1 Rajojit Chowdhury1
Rajojit Chowdhury1 Mst. Momtaj Begam1
Mst. Momtaj Begam1 Sayak Ganguli2
Sayak Ganguli2 Ritabrata Basak3
Ritabrata Basak3 Basab Chaudhuri4
Basab Chaudhuri4 Krishna Ray1*
Krishna Ray1*Bacterial Pho regulon is a key regulator component in biological phosphorus-uptake. Poly-phosphate accumulating bacteria used in enhanced biological phosphorus removal (EBPR) system encounter negative regulation of the Pho regulon, resulting in reduced phosphorus-uptake from phosphorus-replete waste effluents. This study demonstrates possible trends of overcoming the PhoU negative regulation, resulting in excessive PO43–-P uptake at varying concentrations of NO3–-N through denitrifying phosphorus removal process. We investigated the Pho regulon gene expression pattern and kinetic studies of P-removal by denitrifying phosphate accumulating organisms (DPAOs) which are able to remove both PO43–-P and NO3–-N in single anoxic stage with the utilization of external carbon sources, without the use of stored polyhydroxyalkanoate (PHA) and without any anaerobic-aerobic or anaerobic-anoxic switches. Our study establishes that a minimum addition of 100 ppm NO3–-N leads to the withdrawal of the negative regulation of Pho regulon and results in ∼100% P-removal with concomitant escalated poly-phosphate accumulation by our established DPAO isolates and their artificially made consortium, isolated from sludge sample of PO43– -rich parboiled rice mill effluent, in a settling tank within 12 h of treatment. The same results were obtained when a phosphate rich effluent (stillage from distillery) mixed with a nitrate rich effluent (from explosive industry) was treated together in a single phase anoxic batch reactor, eliminating the need for alternating anaerobic/aerobic or anaerobic/anoxic switches for removing both the pollutants simultaneously. The highest poly-phosphate accumulation was observed to be more than 17% of cell dry weight. Our studies unequivocally establish that nitrate induction of Pho regulon is parallely associated with the repression of PhoU gene transcription, which is the negative regulator of Pho regulon. Based on earlier observations where similar nitrate mediated transcriptional repression was cited, we hypothesize the possible involvement of NarL/NarP transcriptional regulator proteins in PhoU repression. At present, we propose this denitrifying phosphorus removal endeavor as an innovative methodology to overcome the negative regulation of Pho regulon for accelerated unhindered phosphorus remediation from phosphate rich wastewater in India and the developing world where the stringency of EBPR and other reactors prevent their use due to financial reasons.
Phosphorus (P) rich waste effluents are objects of great concern worldwide. Globally, these waste effluents are treated with an enhanced biological phosphorus removal (EBPR) system that is based on an activated sludge process involving a group of microorganisms called poly-phosphate-accumulating organisms (PAOs), which actively take up soluble phosphorus (PO43–-P) from wastewater and accumulate it in the form of poly-phosphate (poly-P) granules. This PO43–-P uptake is enhanced when the organisms are alternated between a carbon-rich anaerobic environment and a carbon-poor aerobic environment. EBPR has the potential to remove 80–90% PO43–-P, with a residual PO43–-P value as low as <1 ppm in the effluent. Minnesota Pollution Control Agency (2006) reported an efficient EBPR system followed by good final clarification which achieved effluent phosphorus concentrations of 0.7 ppm if sufficient VFAs are available in the process. Lower effluent concentrations even down to 0.1 ppm could be achieved through supplemental chemical treatment and advanced effluent filtration techniques applied after EPBR operation. United States Environmental Protection Agency (2007) reported that Biological Nutrient Removal if complemented with chemical addition, tertiary clarification and filtration process, it can achieve average effluent phosphorus concentration as below as 0.06–0.07 ppm. Although a study from China reported 93% PO43–-P removal efficiency by the EBPR process with effluent PO43–-P level as low as 0.253 ppm from municipal wastewater, the influent PO43–-P level was only 3.613 mg L–1 (Li et al., 2011). Similarly, another study on municipal wastewater treatment plants of China reported that 95% P removal was achieved in EBPR, but the influent P conc. was between 3 and 8.7 mg L–1 (Yang et al., 2017).
Phosphate-accumulating organisms usually face a genetic regulatory control that limits their PO43–-P uptake ability. Through a coordinated expression of genes under the Pho regulon, the poly-phosphate-accumulating bacteria sense the external PO43–-P conc., and external low levels allow the cells to uptake PO43–-P, while the reverse takes place under externally PO43–-P replete conc. The Pho regulon is a global regulatory mechanism that is involved in bacterial PO43–-P management and was first characterized in Escherichia coli (E. coli) (Wanner and Chang, 1987). In E. coli, the Pho regulon expresses five major proteins, apart from a two-component system, PhoR-PhoB. Four of the major proteins are phosphate transporters (Pst), namely, PstS, PstC, PstA and PstB, and are the most conserved members of the Pho regulon system in bacteria, while the fifth is a metal-binding protein, PhoU. Under external PO43–-P deplete conditions, PhoB is activated by PhoR acting as a kinase, but under external PO43–-P replete conditions, PhoB activation is interrupted by PhoR acting as a phosphatase, which causes repression of the phosphate regulon (Yamada et al., 1990; Muda et al., 1992; Carmany et al., 2003). PhoU is involved in PhoB dephosphorylation under PO43–-P rich conditions in a yet unknown mechanism. When PhoU is mutated or deleted, PhoR behaves as a constitutive kinase to phosphorylate PhoB, which leads to a high expression of Pho regulon genes (Steed and Wanner, 1993; Rice et al., 2009). This shows that PhoU is involved in the control of the Pst system and prevents uncontrolled PO43–-P uptake, which could be toxic for the cells (Surin et al., 1986; Muda et al., 1992; Carmany et al., 2003; Gardner et al., 2014; Santos-Beneit, 2015). This phenomenon was observed in both E. coli and Synechocystis sp., which suggested that the phoU mutants might contribute to improved biological PO43–-P removal and accumulation in the form of poly-P from wastewaters (Morohoshi et al., 2002). E. coli phoU mutants exhibited 1000-fold higher levels of poly-P accumulation than that of the wild-type under P-rich conditions, and an introduction of the wild-type PhoU gene back into the mutant reinstated poly-P accumulation to the wild-type level, thereby establishing the PhoU gene as the negative regulator for both PO43–-P removal and poly-P accumulation (Kuroda and Ohtake, 2000). An environmental bacterium, Citrobacter freundii, was engineered for overexpression of the polyphosphate kinase gene based on a solo medium-copy plasmid strategy that resulted in 12.7% poly-P accumulation by the cells, thus overcoming the negative regulation of PhoU (Wang et al., 2017). The negative regulation of PhoU is a major limitation to PO43–-P bioremediation when the soluble PO43–-P level is high in waste effluents, e.g., effluents from fertilizer industry (25–308 ppm of PO43–-P) (Abou-Elela et al., 1995), parboiled rice mill industry (34–143 ppm of PO43–-P) (Faria et al., 2006), and rice-based distilleries (223.5 ± 27.5 ppm of PO43–-P) (Bhattacharyya et al., 2014). The screening and development of phoU mutants of PAOs was considered to be a solution to overcome this limitation. However, the growth of phoU mutants was found to be severely defective and they were easily outgrown by revertant(s) that had lost the ability to accumulate poly-P during growth in a nutrient-rich medium (Hirota et al., 2013).
In the EBPR system, the PAOs are able to store phosphorus through sequential anaerobic-aerobic conditions. Carbon sources, particularly volatile fatty acids (VFAs), are taken up anaerobically and stored as poly-β-hydroxyalkanoates (PHA) with the release of P to the outer media and the degradation of glycogen. A higher amount of P is then taken up from the external media when an electron acceptor is supplied (normally oxygen, i.e., aerobic conditions) through PHA oxidation, which is accompanied by biomass growth and the regeneration of glycogen (Carvalho et al., 2007). Soon it was demonstrated that, not only under aerobic conditions but also under anoxic conditions, i.e., with nitrate as the electron acceptor, some PAOs are capable of phosphate uptake and accumulation (Kuba et al., 1993; Egly and Zehnder, 1994; Jørgensen and Pauli, 1995; Barker and Dold, 1997; Mino et al., 1998; Oehmen et al., 2010). It was suggested that nitrate can be used as an electron acceptor (i.e., anoxic conditions) instead of oxygen, which is advantageous because both NO3–-N and PO43–-P are thus removed in the same process (Kerrn-Jespersen and Henze, 1993; Kuba et al., 1993). These organisms came to be known as denitrifying polyphosphate accumulating organisms or DPAOs. Moreover, when compared to conventional EBPR, simultaneous denitrification and P removal can save aeration, minimize sludge disposal and reduce the demand for the often-limiting carbon sources (Kuba et al., 1993, 1996). The activity of DPAOs has often been demonstrated, both in lab-scale and full-scale EBPR systems (Kuba et al., 1993; Ahn et al., 2002; Zilles et al., 2002; Shoji et al., 2003; Kong et al., 2004).
Candidatus accumulibacter has been identified as the most dominant DPAO in the EBPR process in wastewaters (Zeng et al., 2017). The use of 16S rRNA and polyphosphate kinase (ppk1) genes as the genetic markers suggested that Ca. accumulibacter is divided into two main clades: Ca. accumulibacter phosphatis clade I (PAO I) and Ca. accumulibacter phosphatis clade II (PAO II) (Rubio-Rincon et al., 2017). Further studies (Carvalho et al., 2007; Flowers et al., 2009; Oehmen et al., 2010) revealed that the PAO I clade was capable of considerable anoxic phosphate uptake activity using nitrate and/or nitrite as an electron acceptor whrereas PAO II lacked the respiratory nitrate reductase enzyme (nar), but possessed the mechanisms to denitrify nitrite. During the treatment of municipal wastewater by the EBPR system under complete nitrification, clade I which used nitrate as the electron acceptor, was found to be below 5% of the total Ca. accumulibacter population, whereas Clade IID and Clade IIC which used nitrite as the electron acceptor, were always found to be dominant (above 90 and 87.3% of the total population of Ca. accumulibacter, respectively), throughout the operational period (Zeng et al., 2016, 2017).
Tetrasphaera is another one of the most abundant PAOs present in the EBPR systems, but their capacity to achieve denitrifying P removal has not been determined. The capacity of Tetrasphaera to couple denitrification with P uptake has never been established (Marques et al., 2018). Four isolates of Tetrasphaera had been shown to possess the genes for the reduction of nitrate to nitrite and some isolates (T. japonica and T. elongata) had been shown to use nitrate and nitrite as electron acceptors (Lanham et al., 2018). Nevertheless, the amount of anoxic P uptake by a denitrifying Tetrasphaera-PAO has been found to be very low (Marques et al., 2018). Till date, the genera Tetrasphaera and Candidatus accumulibacter appear to be the only known DPAOs to be consistently found in high abundances in full-scale EBPR plants where they could be considered critical to P removal (Nielsen et al., 2019).
In another study, a denitrifying consortium derived from the fluidized bed reactor during the treatment of fish aquaculture effluent, was incubated under laboratory conditions in the presence or absence of nitrate (Barak and van Rijn, 2000a). One of the isolates of denitrifying bacteria (Paracoccus denitrificans) isolated from the fluidized bed reactor was characterized in detail for its denitrifying P-removal ability (Barak and van Rijn, 2000a). It was found that in contrast to PAOs, poly-P synthesis by P. denitrificans took place only in the presence of an external carbon source under either aerobic or anoxic conditions. Furthermore, unlike PAOs, P. denitrificans was unable to utilize PHA as an energy source for poly-P synthesis. Under anaerobic condition, these DPAOs were unable to use external carbon source for PHA synthesis, and degraded glycogen for PHA synthesis; degraded poly-P to release phosphate, which provided energy for growth. Under aerobic/anoxic condition, they produced poly-P and grew utilizing the energy provided by external carbon source and produced glycogen; in the absence of external carbon source, cells with PHA did not grow and did not accumulate poly-P (Kaltwasser et al., 1961; Ferguson and Gadian, 1979; Barak and van Rijn, 2000a). The feasibility of this type of phosphate removal was demonstrated for freshwater as well as marine recirculating systems (Barak and van Rijn, 2000a; Shnel et al., 2002; Barak et al., 2003; Gelfand et al., 2003; Rijn et al., 2006). According to Barak and van Rijn (2000a), the most striking feature of the mode of phosphate uptake and accumulation exhibited by P. denitrificans and other denitrifiers examined in their laboratory, was their ability to synthesize poly-P under either aerobic or anoxic conditions without the need for alternating anaerobic/aerobic (anoxic) switches. It has been repeatedly mentioned by these researchers that unlike conventional PAOs, phosphate uptake by these denitrifiers does not require switches between anaerobic/aerobic or anaerobic/anoxic phases (Barak et al., 2003; Rijn et al., 2006; Shi and Lee, 2006; Zheng et al., 2014). According to Medhi and Thakur (2018), this display of single stage simultaneous N and P removal makes the wastewater treatment by these DPAOs a sustainable process, if suitable technologies can be adopted.
Lee and Park (2008) reported isolation of a denitrifying polyphosphate-accumulating bacterium, Paracoccus sp. YKP-9, from activated sludge of a 5-stage biological nutrient removal process with a step feed system. This strain also accumulated poly-P with the energy provided by an external carbon source under anoxic condition using nitrate, but neither accumulated poly-P nor grew in the absence of an external carbon source under anoxic condition. Moreover, it did not consume intracellular PHA for poly-P accumulation. Similarly, Shi and Lee (2007) reported a denitrifying phosphorus-removing bacterium, Brachymonas sp. strain P12, growing in similar conventional EBPR models, but these models were developed under anoxic or aerobic conditions only, without an anaerobic stage.
In other words, all of the above described DPAOs can combine phosphorus removal and denitrification into a single process using the same amount of organic substrate (Yuan and Oleszkiewicz, 2011). Denitrification is believed to be a strictly anaerobic process (Kaspar, 1982; Tiedje et al., 1982; Payne, 1983) because O2 has been shown to suppress the activity of bacterial dissimilatory nitrate reductase (John, 1977; Tiedje et al., 1982; Payne, 1983) although several studies have established that denitrification could also occur in the presence of O2 (Voets et al., 1975; Meiberg et al., 1980; Bazylinski and Blakemorer, 1983; Robertson and Kuenen, 1983, 1984a,b; Lloyd et al., 1987; Davies et al., 1989). But O2 has no detrimental effect on the denitrifying polyphosphate accumulation activity (Kuba et al., 1996). Yuan and Oleszkiewicz (2011) showed that dissolved O2 conc. of 0.01 ± 0.01 mg L–1 provided favorable condition for DPAOs to grow under anoxic condition. Altogether with its lower COD requirement (Kuba et al., 1996), and reduced aeration costs, denitrifying phosphorus removal (DNPR) can become a cost-effective choice for removing phosphorus and nitrogen simultaneously.
Several researchers (Barak and van Rijn, 2000a, b; Barak et al., 2003; Rijn et al., 2006; Shi and Lee, 2006) have repeatedly emphasized that in the presence of nitrate, the denitrifying consortium is capable of phosphate uptake in excess of their metabolic requirements. It had been shown by them that the denitrifying consortium present in the fluidized bed reactor, assimilated ammonia and phosphate at a molar N:P ratio ranging from 0.5 to 2.4, with an average molar N:P ratio of 1.9. Taking into account that in general the molar N:P ratio of bacterial biomass varies from 5 to 16 (Brock and Madigan, 1991), it was concluded that only under denitrifying conditions i.e., in the presence of nitrate, phosphate is assimilated in excess of the metabolic requirements of the bacteria comprising the consortium. Phosphorus immobilization took place in the anoxic treatment stages of the system where it accumulated up to 19% of the sludge dry weight (Rijn et al., 2006). This fact attracted our attention. This provided us an indication that in the presence of nitrate, DPAOs will prove to be extraordinarily efficient to defy the inhibitory effects of the Pho regulon.
In this connection, we wanted to look at the Pho regulon gene expression profile during phosphorus uptake in some DPAO isolates, which had been isolated from the sludge of parboiled rice mill effluent. Under this background scenario, this study explored the possible trends of overcoming the negative regulation of PhoU resulting in excessive PO43–-P uptake by using DNPR with NO3–-N at a varying range of concentrations. We initiated research to investigate the Pho regulon gene expression pattern and kinetic studies of P-removal by those DPAOs which were able to remove both PO43–-P and NO3–-N in a single anoxic stage with the utilization of external carbon sources, and without any anaerobic-aerobic or anaerobic-anoxic switches.
Sludge sample from a PO43– -rich parboiled rice mill effluent settling tank (containing ∼40 ppm of PO43–-P) was serially diluted (up to 10–9) in sterile water, and 100 μl of each dilution was spread onto a nutrient agar medium and was incubated at 37°C for 2 days. Repeated subculturing of single colonies was performed to obtain a pure culture of the isolates. The morphological and biochemical characteristics (including nitrate reduction ability) of the isolates were determined. Scanning electron microscopy (SEM) of the isolates was also performed (Zeiss SEM, Central Research Facility, Indian Institute of Technology, Kharagpur, India). Molecular identification of the pure bacterial isolates was carried out by a standard procedure of partial 16S rRNA gene amplification by universal primers. The sequences were subjected to blast search for identification and submitted to the NCBI database. The other biochemical tests to characterize the isolates were also carried out following the standard procedures.
(1) Twenty four hour old cultures of the individual isolates and an artificial bacterial consortium (raised from the 24 h old culture of all the isolates by suspending a 100 mg cell pellet of each isolate in 10 ml of 0.84% saline solution) were inoculated into their respective screw capped flask containing 250 ml basal level modified synthetic wastewater SW medium (Supplementary Table S1) (Holler and Trösch, 2001). The final OD at 600 nm after inoculation should be 0.1.
(2) Three sets were prepared for each isolate and the consortium, where in the first set 300 ppm meat extract was used as carbon source and in the second set 1000 ppm acetate (added as sodium acetate) was used as carbon source (where no peptone and meat extract was given) and in the third set no carbon source was added. Fifty ppm of PO43–-P (added as di-potassium hydrogen phosphate, K2HPO4) was added to all the flasks. Nitrate-nitrogen was not added in this anaerobic incubation. All the flasks containing media were flushed with pure nitrogen and all the flasks were put in the HiMedia Anaerobic gas jar with Anaero Gas Pack. All the flasks were incubated with 150 rpm shaking at 30°C for 12 h. After the incubation, culture growth in terms of OD at 600 nm, residual PO43–-P concentrations and residual acetate concentrations (from acetate containing media) and residual dissolved organic carbon (DOC) concentrations (from meat extract containing media) were measured according to the following protocols.
Acetate was measured according to the protocol of Shi and Lee (2006) with some modifications using the GC-FID (Agilent) with Quadrex 007 Series bonded phase fused silica capillary column (15 m, 0.32 mm ID, 0.25 μm film thickness). Samples were acidified with 1 (N) H2SO4 to pH below 3. The flow rate of the carrier gas (N2) was 3 mL min–1. Injector/detector temperature was set to 220/230°C respectively. The column temperature was set in the range of 70°C – 150°C with temperature programing at the rate of 7°C min–1. A five point calibration curve was made between concentration and peak area of 100–2000 ppm acetate (added as sodium acetate). DOC was determined by simple spectrophotometric method (Moore, 1985). Absorbance at 330 nm of the media was measured for DOC calculation. The PO43–-P conc. of the medium was measured by the molybdenum blue method (Krishnaswamy et al., 2009).
(1) The inoculation steps were followed exactly as mentioned earlier (in the above section).
(2) Two main sets were prepared – one was SW with 1000 ppm of NO3–-N (added as potassium nitrate, KNO3) and other one was SW without any NO3–-N. Without NO3–-N sets were incubated aerobically in cotton plugged flasks and with NO3–-N sets were incubated anoxically in screw capped flasks. Both the sets had three sub-sets – without any carbon source, with 1000 ppm acetate (added as sodium acetate) and with 300 ppm meat extract. 50 ppm of PO43–-P (added as di-potassium hydrogen phosphate, K2HPO4) was added to all the flasks. In total six different sets were experimentally set up for each isolate and the consortium.
For creating low DO anoxic environment, medium was dispensed in the reaction container with low surface area to volume ratio which limited the diffusion of O2 in the medium. To maintain efficient DO free environment, 1 cm of liquid paraffin oil was poured on the surface of the medium (according to Sigma-Aldrich nitrate broth product information). Added excess salt in the medium also created low DO level. The DO level was maintained at 0.01 mg L–1 and checked throughout all the experiments by DO meter (Mettler-Tolledo).
All the flasks were incubated with 150 rpm shaking at 30°C for 12 h. After the incubation, culture growth in terms of OD at 600 nm, residual PO43–-P concentrations, residual NO3–-N and NO2–-N concentrations (from NO3–-N containing media), residual acetate concentrations (from acetate containing media) and residual dissolved organic carbon (DOC) concentrations (from meat extract containing media) and poly-P accumulation inside the cell were measured.
The NO3–-N conc. of the medium was determined by measuring the absorbance of nitrosalicylic acid by the spectrophotometric method (Cataldo et al., 1975). NO2–-N conc. was measured by adding 1% sulfanilic acid-0.05% N-naphthylene diamine-HCl dissolved in 1 (M) H3PO4, and measuring the absorbance spectrophotometrically (Baumann et al., 1996). The accumulated poly-P was extracted and quantified according to an established protocol that was standardized in our laboratory (Mukherjee and Ray, 2015), and is very specific for the polyphosphates having chain length not shorter than 10 residues (Lorenz et al., 1999; Kulaev et al., 2004; Omelon and Grynpas, 2008).
Modified SW with complex organic carbon source was used to study maximum PO43–-P and NO3–-N removal efficiency of the individual isolates and the developed consortium. Forty-one combinations of SW were prepared with varying conc. of PO43–-P (0, 10, 50, 100, 250, 500, and 1000 ppm added as K2HPO4) and NO3–-N (0, 50, 100, 500, 1000, and 2000 ppm added as KNO3), except for the combination without any PO43–-P or NO3–-N. The individual isolates and the bacterial consortium (initial OD of 0.04 at 600 nm) was inoculated in 500 ml of SW for each combination and was incubated at 30°C for 12 h. The combinations without NO3–-N were incubated aerobically while combinations with NO3–-N were incubated at low DO anoxic environment. Evaluation of the amount of accumulated poly-P and estimation of the remaining PO43–-P, NO3–-N and NO2–-N concentrations were determined at the end of 12 h. In a similar way, the control non-PAO bacteria like E. coli K 12 ER2925, E. coli K 12 PR1031 (obtained from New England Biolab, NEB) and E. coli DH5α strains were inoculated in all the earlier stated N and P combinations in SW, their P and N removal data were ascertained.
All the above experiments were run for 96 h, and the P and N removal data were collected at the end of 12, 24, 72, and 96 h. The growth of the isolates was also recorded by measuring the absorbance at 600 nm at the intervals of 12, 24, 48, 72, and 96 h. In addition, pH of all the media combinations were recorded at the end of 12, 24, 48, 72, and 96 hrs, for all the isolates including the control non-PAO E. coli strains.
Stillage, a PO43–-P -rich effluent, derived from a rice-based ethanol producing distillery industry was collected from IFB Agro Industries, Noorpur, West Bengal, India. The NO3–-N-rich effluent was collected from an explosive manufacturing industry located in Jharkhand, India. All the effluent samples were transported to the laboratory in ice-cold conditions and were stored at 4°C until further analyses. A maximum of 400 ppm of soluble PO43–-P was found in the stillage and a maximum of 15000 ppm NO3–-N was observed to be present in the explosive industry effluent. Different combinations of the effluent mixture were prepared with varying conc. of stillage (containing 0, 175, 200, 350, and 400 ppm PO43–-P) and explosive industry effluent (containing 0, 750, 1000, 1500, and 3000 ppm NO3–-N), except the combination of 0 + 0. As the consortium was found to grow to a maximum of 3000 ppm of NO3–-N containing explosive effluent, the study was limited within this conc. of NO3–-N. Since the raw explosive industry effluent did not contain any carbon source, 0.05% peptone and 0.03% meat extract were added to the effluent in the combinations prepared without stillage. The batch reactors of dimensions 0.3 m∗0.3 m∗0.1 m were constructed, and each was filled with 5 L of different combinations of the effluent mixture and was inoculated with 2.5 g of 24 h old artificial bacterial consortium. The temperature was maintained at 30°C. Low DO environment for anoxic condition was created as described previously. The residual PO43–-P, NO3–-N and NO2–-N contents were measured at 2, 6, and 12 h intervals.
The rates of PO43– and NO3–-N removal and the order of reaction of the remediation of the SW and effluent mixture by the bacterial consortium were established by a kinetic study by applying the integrated rate laws for the three observed conditions: (i) media combinations containing only PO43–-P in aerobic condition, (ii) media combinations containing only NO3–-N in anoxic condition, and (iii) media combinations containing both PO43–-P and NO3–-N under anoxic condition.
The four PO43– transporter genes, namely, PstS, PstC, PstA, and PstB, and a regulator gene, PhoU, which are the units of the bacterial Pho regulon, were selected for this study. The beta subunit of the DNA directed RNA polymerase gene, RpoB; 16S rRNA gene; alpha subunit of DNA-directed RNA polymerase, RpoA; DNA gyrase A gene, GyrA; Recombinase A gene, RecA were selected to check their stability index (M value) by using the NormFinder software tools (Rocha et al., 2015). For M value detection, the expression of all the referred reference genes were evaluated under identical N and P added combinations of SW that were later utilized to find the expression pattern of our candidate genes. The maximum stable gene was used as the reference gene. Two pure isolates, Escherichia coli isolate SW11 (Accession no. KU740237–KU740238) and Bacillus sp. isolate SW7 (Accession no. KU740235–KU740236), which were found to be the best removers of PO43–-P and NO3–-N (concluded from the experiments performed with individual isolates) and for which the Pho regulon gene sequences were available in public databases, were chosen for this study along with the control non-PAO bacteria, E. coli K 12 ER2925. The primers were designed with the help of the SCITOOLS site of Integrated DNA Technologies1 (Supplementary Table S2). For each of the two isolates, 15 ml of sixteen different combinations of the SW medium were prepared with varying PO43–-P conc. (0, 100, 250, and 500 ppm) and NO3–-N conc. (0, 100, 1000, and 2000 ppm). Five replica sets of each were prepared for five different incubation periods of 1, 2, 4, 6, and 12 h. All of the tubes were inoculated with the overnight grown culture (initial OD of 0.1 at 600 nm) and were incubated at anoxic condition at 30°C for 12 h. Total RNA was extracted from each culture with Purezol RNA isolation reagent (Bio-Rad Laboratories, Hercules, CA, United States) and was quantified by the NanoDrop UV spectrophotometer (Eppendorf Company, Eppendorf, Hamburg, Germany). One μg RNA from each culture was taken for immediate cDNA synthesis by using a cDNA synthesis kit (BioBharati LifeScience Private Limited, Kolkata, West Bengal, India). One μl of each cDNA was used for quantitative real-time PCR (QRT PCR) using iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, United States). The PCR conditions were set as follows: Pre-incubation (1 cycle) – 94°C for 7 min; 3 step amplification (45 cycles) – 94°C for 15 s, 54°C for 40 s, and 72°C for 30 s; melting (1 cycle) – 95°C for 10 s, 65°C for 60 s, and 97°C for 1 s. The final data were analyzed, and accurate normalization was performed with the reference gene expression level.
Studies in triplicate sets were carried out for all of the experiments. All of the represented data were the mean ± standard deviation. The standard deviation was depicted with error bars in each graphical presentation. SigmaPlot 13.0 and Microsoft Excel software were used to prepare the graphical presentations. To observe the significance of changes in different parameters over a number of days, Duncan’s multiple range test was performed for each data with SPSS 13.0 software. Analysis of variance (ANOVA) along with Duncan’s multiple range test was used to determine if there were significant differences in the measured parameters. Values designated with different letters are significantly different at the 5% level.
Scanning electron microscopy images were recorded for all 13 pure bacterial isolates (Supplementary Figure S1). 16S rRNA gene identification of the pure isolates segregated them into three main genera, Bacillus, Escherichia, and Staphylococcus, and the NCBI accession numbers of their sequences are shown in Supplementary Table S3. The SEM images depict long and short rod morphology for the Bacilli, bunch of cocci morphology for the Staphylococci and normal morphology for E. coli. The Gram characters and biochemical characteristics of all the isolates are presented in the Supplementary Figure S2. The most important biochemical character which was found to be common for all the isolates was that all these isolates were positive for nitrate reduction test. This significant finding led us to set up the subsequent experiments to confirm whether this denitrifying ability of the isolates was accompanied by poly-phosphate accumulating ability also. Possession of both the referred properties by the isolates would have confirmed their denitrifying poly-phosphate accumulating (DPAO) nature. The control non-PAO bacteria like E. coli K 12 ER2925, E. coli K 12 PR1031 and E. coli DH5α strains used in this study are established denitrifier strains (Stewart et al., 2002; Clegg et al., 2006).
Under anaerobic condition, no significant difference in growth between the individual isolates and the consortium was observed irrespective of the carbon sources used or without any carbon sources (Figure 1A). No significant difference in growth among the isolates and the consortium was also observed when VFA (acetate) or complex organic carbon source (meat extract) were used as the carbon sources (Figure 1A). When acetate was used as the carbon source, initial acetate concentration added was 1000 ppm in the medium. But no fall from this initial concentration was observed in the media even after 12 h as no isolate including consortium utilized this acetate as carbon source (Figure 2A). But growth rate of all the isolates was appreciable in acetate containing medium (Figure 1A). When meat extract was used as the carbon source, again no utilization of carbon source was observed even after 12 h of growth (Figure 2B). Some of the isolates rather released DOC during their growth. No significant difference in DOC utilization was observed between the individual isolates and the consortium (Figure 2B). Thus, it can be concluded that under anaerobic condition, our experimental bacterial isolates and their artificially made consortium failed to utilize the external carbon sources added. This non-utilization of carbon sources in anaerobic phase is very unusual for conventional DPAOs and has only been reported for P. denitrificans DPAO isolated by Barak and van Rijn (2000a).
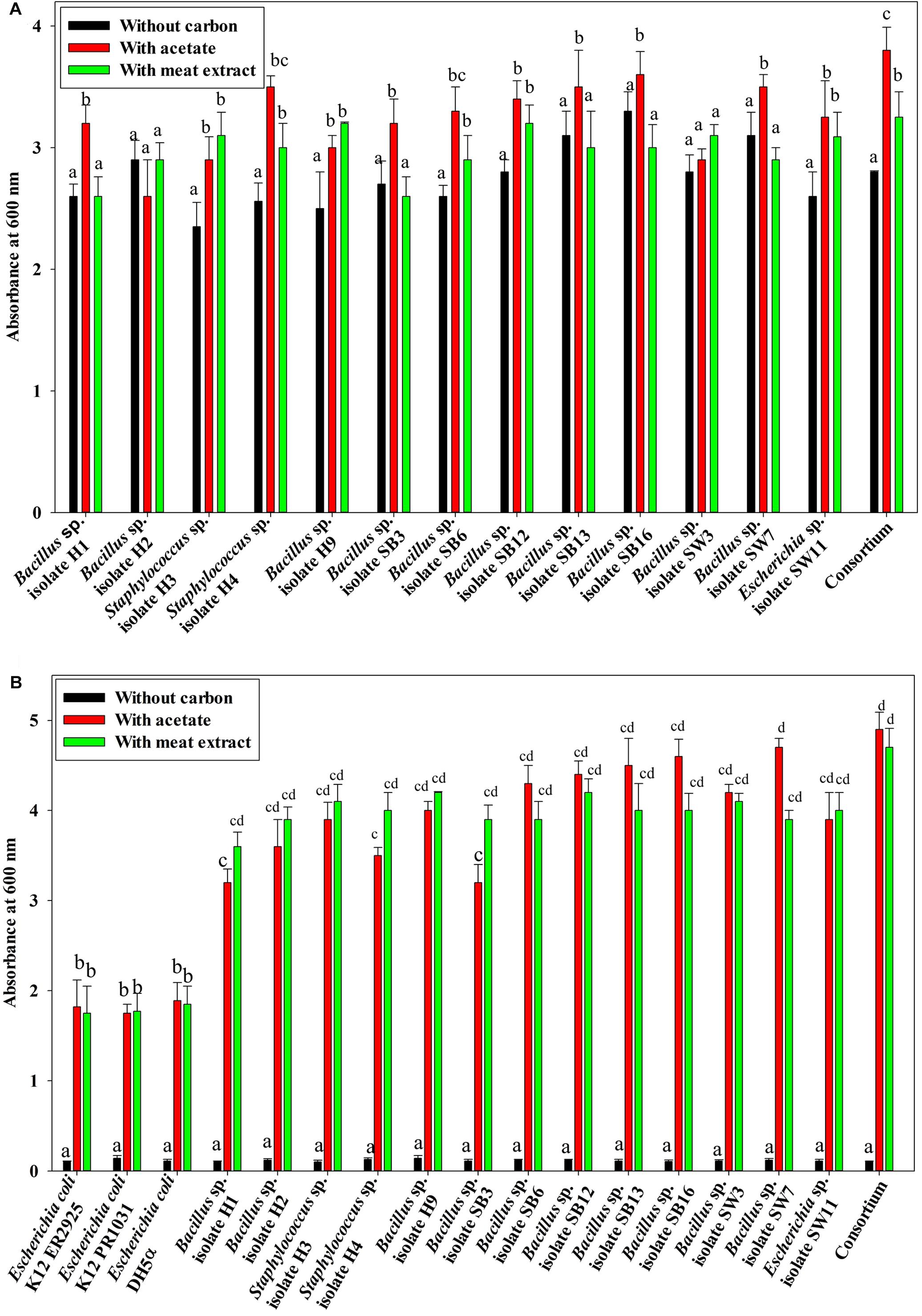
Figure 1. (A) Growth of the 13 DPAO bacterial isolates and their consortium under anaerobic condition with differential carbon sources in 12 h of culture. (B) Average anoxic and aerobic growth of the control non-PAO, E. coli, the 13 DPAO bacterial isolates and their consortium over 12 h in culture conditions with different carbon sources. Values designated with different letters are significantly different at the 5% level.
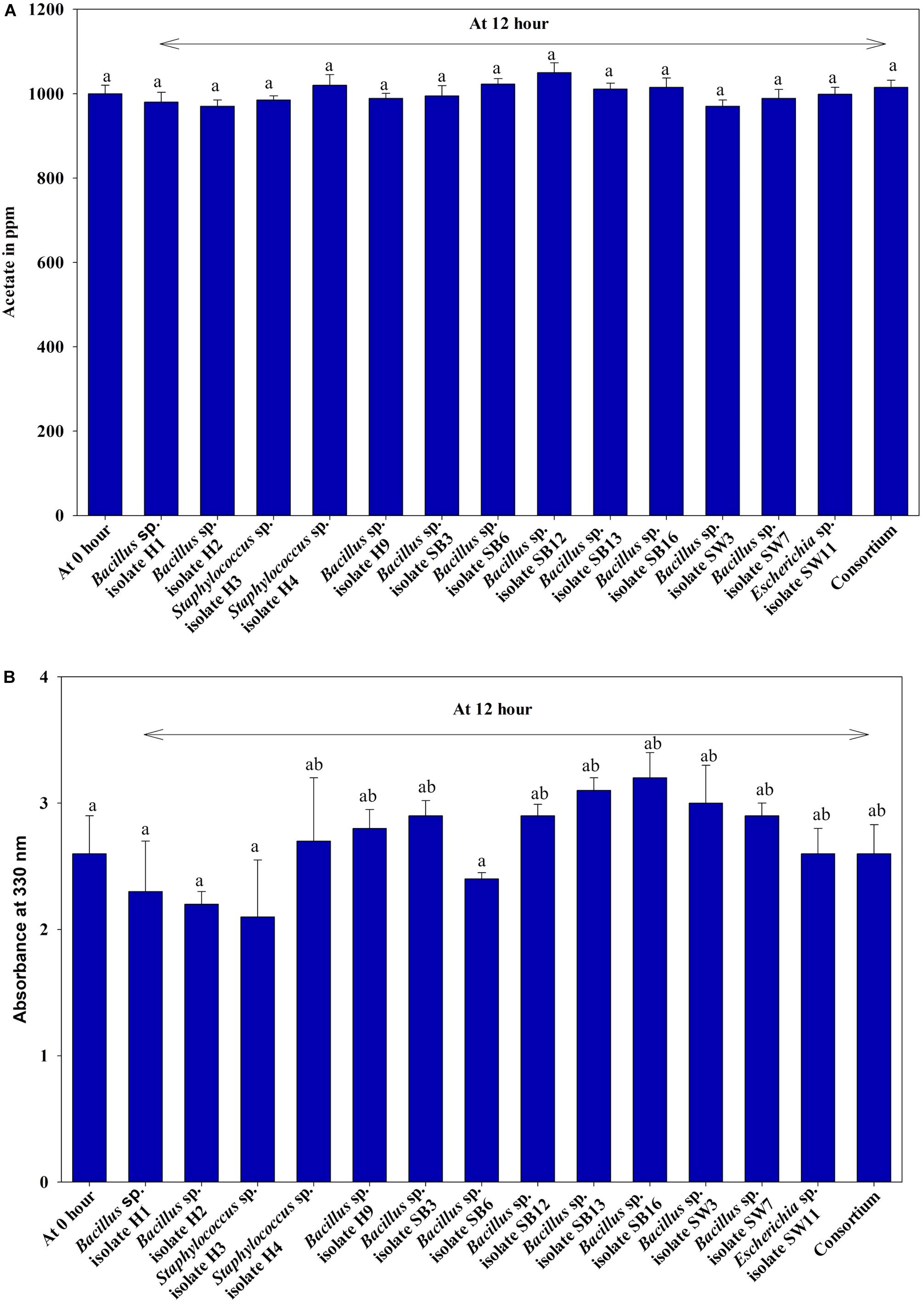
Figure 2. (A) Residual acetate concentration in the medium where only acetate was used as carbon source for 12 h of anaerobic growth of the 13 DPAO isolates and their consortium. (B) Residual dissolved organic carbon (DOC) in the medium supplemented with meat extract as carbon source for 12 h of anaerobic growth of the 13 DPAO isolates and their consortium. Values designated with different letters are significantly different at the 5% level.
Additionally, the PO43–-P removal scenario was also intriguing. Under anaerobic phase, no PO43–-P removal was observed (Figure 3A). No reduction from the initial PO43–-P concentration (50 ppm) was observed even after 12 h of culture (Figure 3A), instead PO43–-P release occurred in all the cases irrespective of the carbon sources present (Figure 3A). Maximum 15 ppm extra PO43–-P was released. No significant difference in PO43–-P release was displayed by the individual isolates or the consortium. It was reported that denitrifying polyphosphate-accumulating bacterium Paracoccus sp. strain YKP-9 also released approximately 15.1 mg L–1 of PO43–-P during the anaerobic phase in 48 h (Lee and Park, 2008).
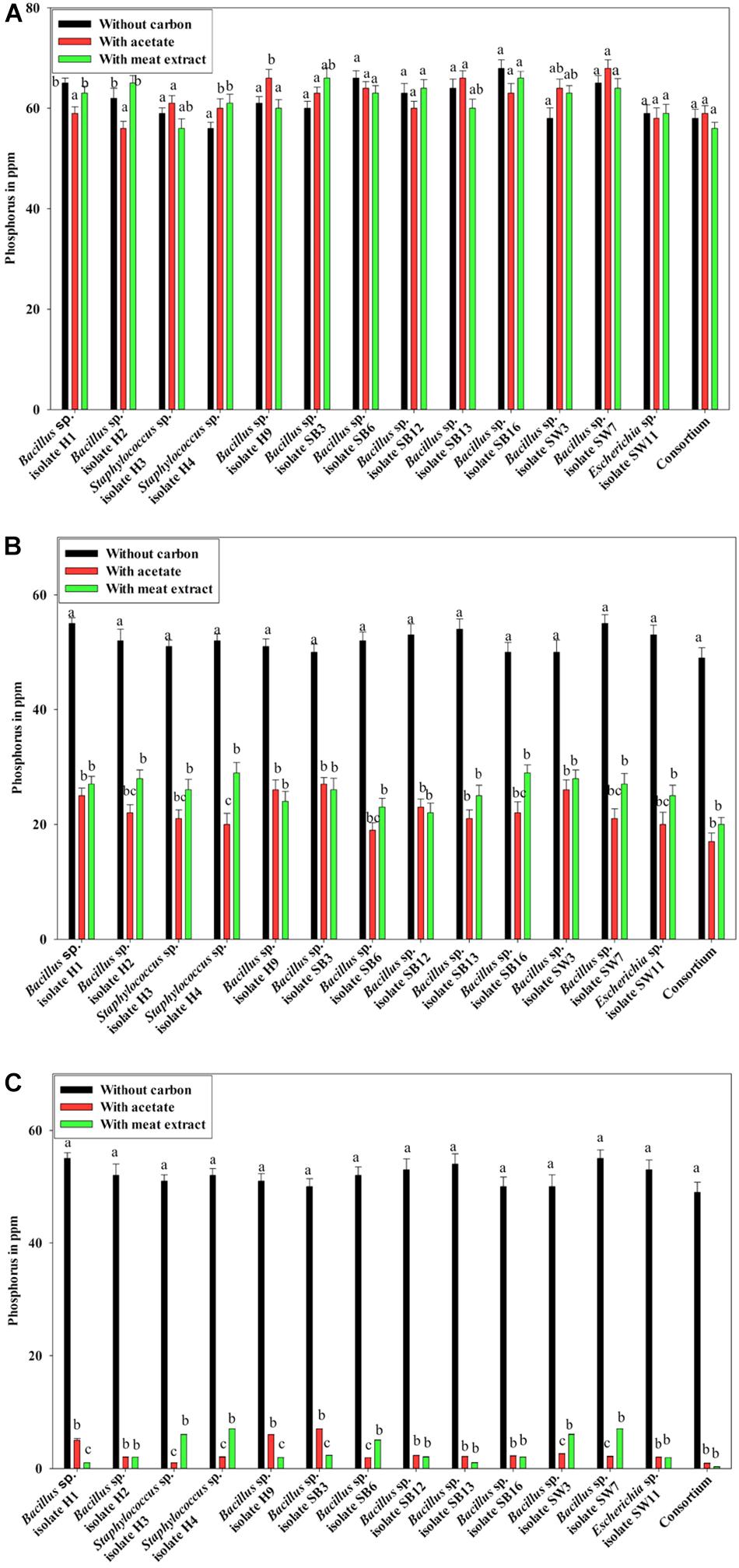
Figure 3. (A) Residual phosphate concentration (starting concentration 50 ppm) in the media after 12 h of anaerobic growth supplemented with different carbon sources for the 13 DPAO isolates and their consortium. (B) Residual phosphate concentration in the media (starting concentration 50 ppm) after 12 h of aerobic growth supplemented with different carbon sources for the 13 DPAO isolates and their consortium. (C) Residual phosphorus (starting concentration 50 ppm) in the medium with different carbon sources/no carbon sources after 12 h of anoxic growth of the 13 DPAO isolates and their consortium in the presence of 1000 ppm NO3–-N. Values designated with different letters are significantly different at the 5% level.
These observations of anaerobic phase strikingly resembled denitrifying PAO P. denitrificans (Barak and van Rijn, 2000a) which was also unable to use external carbon source for PHA synthesis under anaerobic condition and released phosphate by degradation of poly-phosphates that provided energy for growth. The anaerobic phase growth criteria, carbon utilization pattern and PO43–-P release characteristics pointed toward confirmation of the non-conventional DPAO nature of our denitrifying bacterial isolates and their respective consortium.
Under both aerobic and anoxic phases, no growth could be observed in the media with no carbon sources, whereas appreciable growth was observed for media with carbon sources (Figure 1B). Under both aerobic and anoxic phases, media with acetate and media with meat extract as the sole carbon sources, did not exhibit any significant difference in growth parameter (Figure 1B). The consortium showed considerable growth rate in both phases irrespective of the carbon sources used (Figure 1B). All the isolates and the consortium were found to utilize acetate efficiently from the medium (Figure 4A). The H1, H2 isolates efficiently used acetate in aerobic growth phase whereas the SW3, SW7 isolates used it efficiently in anoxic growth phase (Figure 4A). Rest of the isolates including the consortium efficiently used acetate in both the stages. All the isolates and the consortium were also found to efficiently utilize the DOC from the media containing meat extract as the complex organic carbon (Figure 4B). Therefore, it can be inferred that these probable DPAO isolates and their artificially made consortium needed external carbon sources in aerobic as well as in anoxic phase for their growth and to carry out their relevant metabolic activities. This finding is exactly opposite to the carbon utilization mode of the conventional DPAO like Ac. candidatus. The only other DPAOs reported to utilize carbon sources under both aerobic and anoxic conditions are P. denitrificans (Barak and van Rijn, 2000a) and Paracoccus sp. (YKP-9) (Lee and Park, 2008).
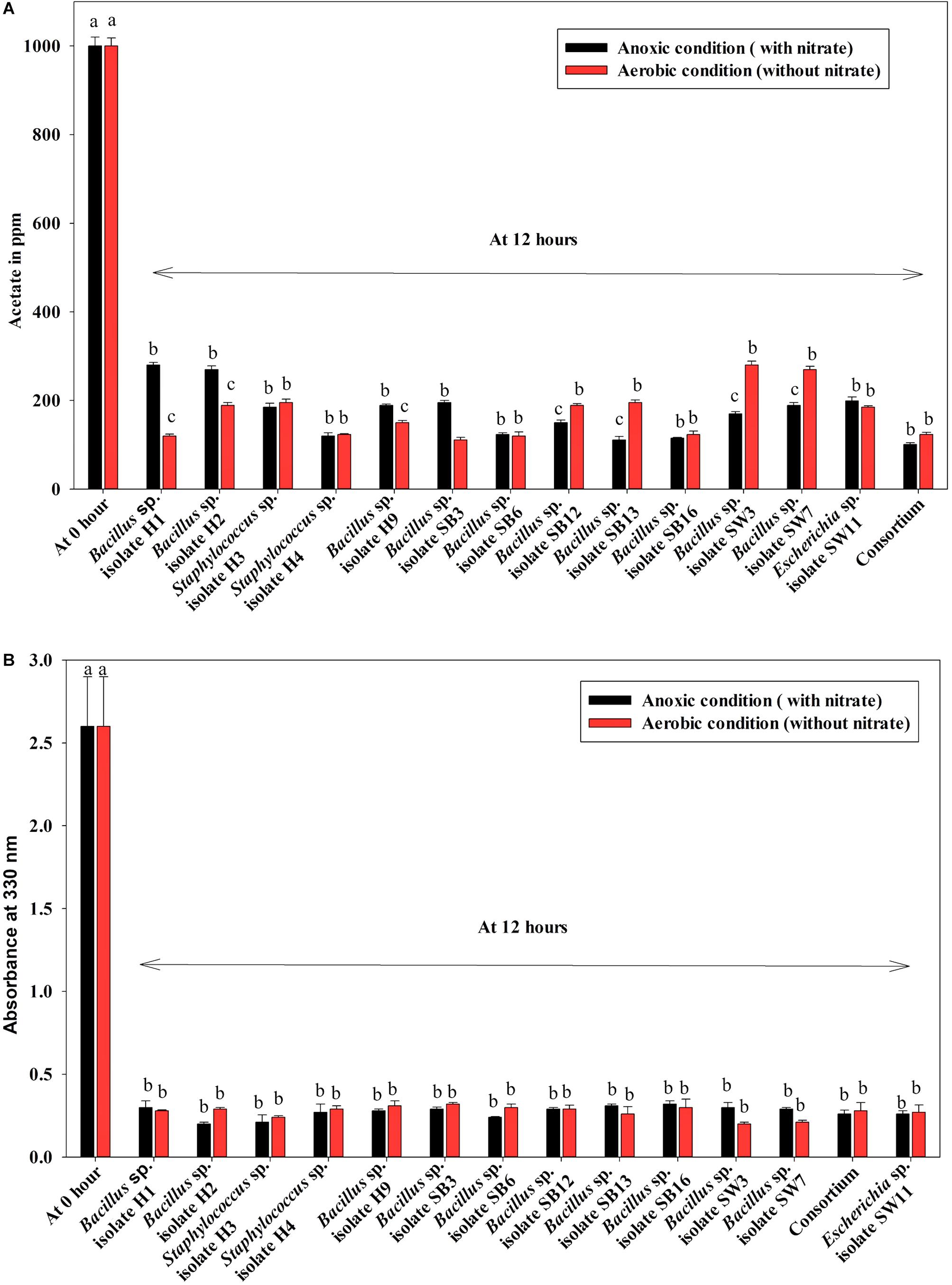
Figure 4. (A) Residual acetate in the medium supplemented with sodium acetate as the carbon source under both anoxic and aerobic growth after 12 h for the 13 DPAO isolates and their consortium. (B) Residual dissolved organic carbon (DOC) in the medium supplemented with meat extract as the carbon source under both anoxic and aerobic growth after 12 h for the 13 DPAO isolates and their consortium. Values designated with different letters are significantly different at the 5% level.
Furthermore, when the PO43–-P removal activity was considered, interesting results were observed. In the media without any carbon source, where no bacterial growth was observed, PO43–-P removal activity was also negative under both the aerobic and anoxic phases. But all the isolates including the consortium removed 50–55% of PO43–-P in 12 h of aerobic growth in the absence of NO3–-N irrespective of the added carbon sources (Figure 3B) while all the isolates including the consortium removed more than 90% of PO43–-P in 12 h of anoxic growth in the presence of 1000 ppm of NO3–-N irrespective of the added carbon sources (Figure 3C). This important finding led us to summarize that these denitrifying isolates and their consortium were capable of growth and PO43–-P removal, both aerobically and anoxically, but they removed PO43–-P at an accelerated rate under anoxic phase in the presence of NO3–-N in the media. Thereafter, all our subsequent experiments on PO43–-P removal by these DPAOs were conducted under single anoxic phase in the presence of varying concentrations of NO3–-N. We assumed that this DNPR which resulted in excessive PO43–-P removal/uptake would have been impossible without being accompanied by the withdrawal of PhoU gene mediated negative regulation.
Denitrification activity under anoxic growth phase was thus observed as follows: (a) no denitrification was observed in media without any carbon sources (Figure 5); (b) almost 100% NO3–-N from the medium was removed irrespective of the carbon sources present in the medium (either acetate or meat extract) in 12 h of anoxic growth (Figure 5). We could not trace the presence of any NO2–-N in the media at the end of 12 h of anoxic growth. It proved that during denitrification NO2–-N was further reduced to its maximum reduced forms.
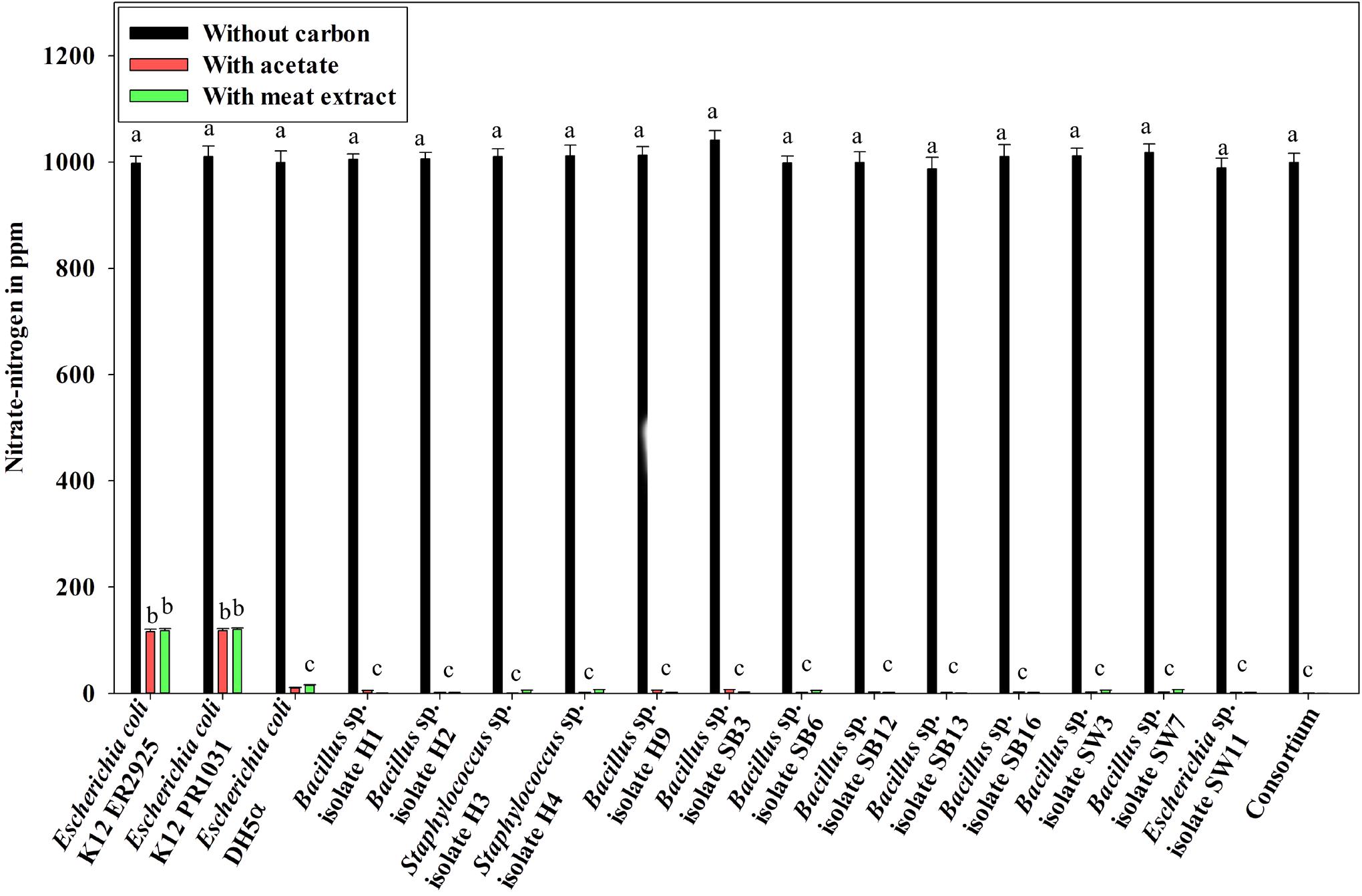
Figure 5. Residual NO3–-N (initial concentration 1000 ppm) in the media with different carbon sources/no carbon sources after 12 h of anoxic growth of the control non-PAO, E. coli, the 13 DPAO isolates and their consortium. Values designated with different letters are significantly different at the 5% level.
For poly-P accumulating activity, about 9–11 fold higher poly-P accumulation than that of the aerobic growth, was observed when the isolates and the consortium were grown anoxically for 12 h in the presence of 1000 ppm of NO3–- N (Figure 6). The consortium showed the highest (2.7% of cell dry weight, CDW) accumulation in 12 h of anoxic growth phase (Figure 6). These results signified that the bacterial isolates and the consortium developed in this study were actually DPAOs, like P. denitrificans (Barak and van Rijn, 2000a), which were capable of simultaneous PO43–-P and NO3–-N removal under single anoxic growth phase without the need for alternating anaerobic/aerobic or anaerobic/anoxic switches. No significant difference in PO43–-P and NO3–-N removal could be detected in the experimental set ups with either acetate or meat extract as the sole carbon sources. For this reason, further studies were carried out with SW media with meat extract as the complex carbon source (Natcheva and Beschkov, 2003). For some of the subsequent remediation experiments, only the artificial DPAO consortium was used because apart from having high PO43–-P and NO3–-N removal capabilities, it also showed the best poly-P accumulation rate over the individual isolates (Figure 6).
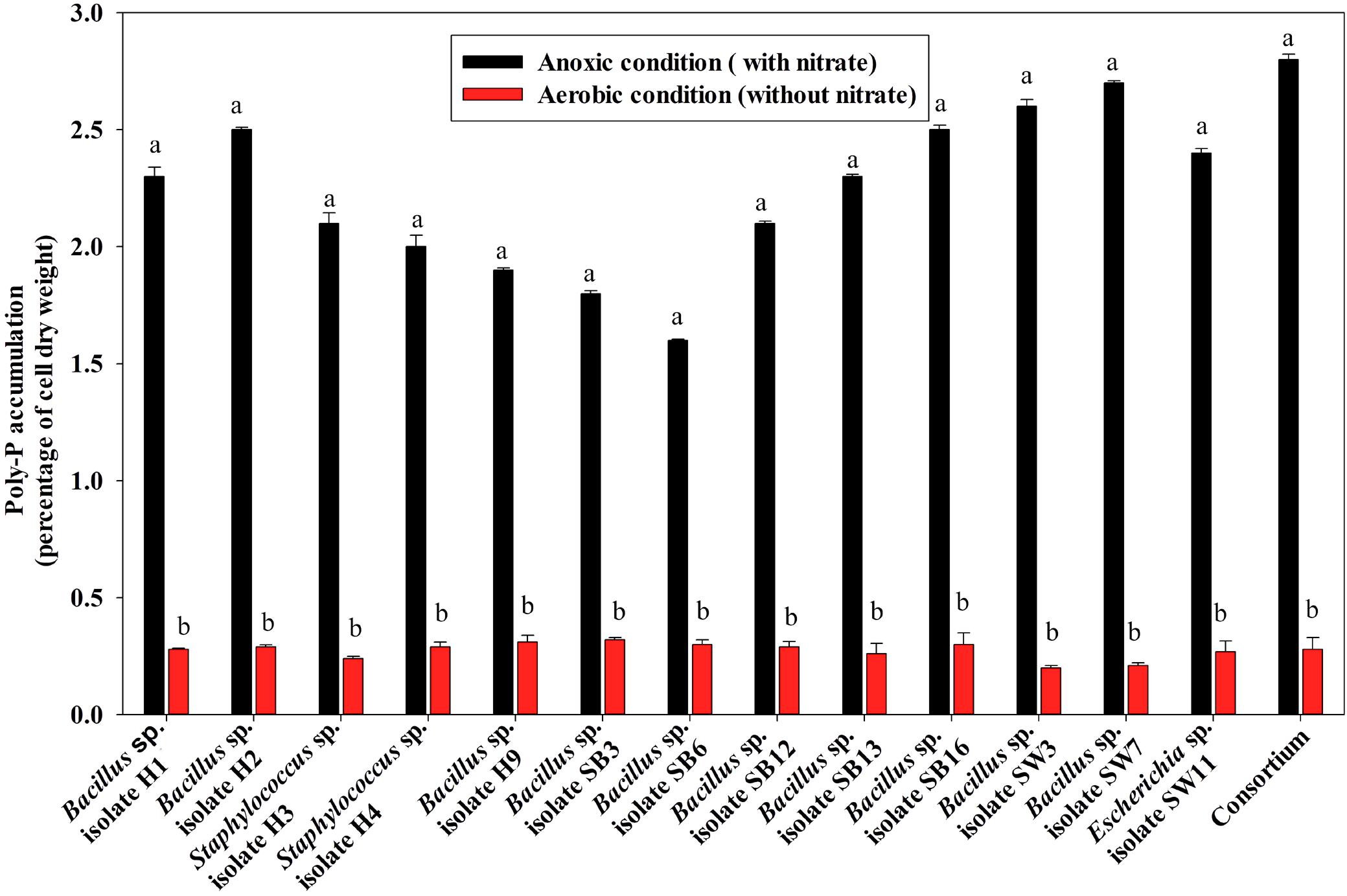
Figure 6. Poly-P accumulation by the 13 DPAO isolates and their consortium after 12 h of anoxic growth (in the presence of 1000 ppm of NO3–-N) and aerobic growth (in absence of NO3–-N) from 50 ppm initial PO43–-P concentration in the medium. Values designated with different letters are significantly different at the 5% level.
Figure 7B reveals an interesting scenario of PO43–-P removal by the DPAOs in the absence of NO3–-N in SW medium, under aerobic condition. The percentage of PO43–-P removal (at the end of 12 h) receded gradually with an increasing conc. of initial PO43–-P, i.e., PO43–-P removal falling from ∼100% when the initial PO43–-P was 10 ppm to 25–30% in case of 1000 ppm of initial PO43–-P. Hence, we considered any external PO43–-P conc. above 10 ppm as P-replete condition in this study. In contrast, the control non-PAO denitrifiers removed only about 15% PO43–-P from 10 ppm initial conc. to only 1–2% P-removal from 1000 ppm of initial PO43–-P conc. in 12 h (Figure 7A). However, when 100, 500, and 1000 ppm of NO3–-N were added to the SW under anoxic environment, a distinct 3–4-fold increase in the PO43–-P removal percentage was observed at the very end of only 2 h, and 85–100% P removal was observed within 12 h, irrespective of the initial PO43–-P conc. (Figures 8B, 9B). By contrast, 0, 50, and 2000 ppm of NO3–-N added combinations failed to elicit any such increase in the PO43–-P removal percentage (Figures 8B, 9B). Similarly, in the three control non-PAO denitrifiers, E. coli K 12 ER2925, E. coli K 12 PR1031 and E. coli DH5α strains, 55–65% PO43–-P removal from 10 ppm initial conc. to 30–35% PO43–-P removal from 1000 ppm initial conc. was observed for 100, 500, and 1000 ppm of NO3–-N concentrations (Figures 8A, 9A). This result provided a clue that the addition of 100–1000 ppm NO3–-N to phosphorus-rich SW under anoxic condition might be critical for achieving the exceptionally high PO43–-P removal (∼85–100%) by the DPAO consortium (Figure 9B). A very congruous poly-P accumulation scenario was also achieved, where the poly-P accrual by the DPAO consortium also increased dramatically to 5–10%, 7.5–10%, 7.5–15%, and 10–22% of CDW from SW containing 100, 250, 500, and 1000 ppm of the initial PO43–-P, respectively, in all of the 100, 500, and 1000 ppm of NO3–-N added combinations (Figure 10). Thus, the fact that 100–1000 ppm of NO3–-N acted as the inducer concentrations, which resulted in a 3–4-fold increase in the PO43–-P uptake and correspondingly accelerated the poly-P accumulation, was constantly proven by these results. The consortium and the control non-PAO E. coli. denitrifiers also demonstrated a very high denitrification rate of approximately 90–100% within 12 h for 50–2000 ppm of initial NO3–-N conc. combined with the 0–1000 ppm of initial PO43–-P combinations (Figures 11A,B), nullifying any effect of the PO43–-P addition on NO3–-N removal from SW. No presence of any NO2–-N in the media at the end of 12 h of anoxic growth was observed repeatedly. We presume that within the 12 h period of denitrification, NO2–-N was reduced to its maximally reduced gaseous forms.
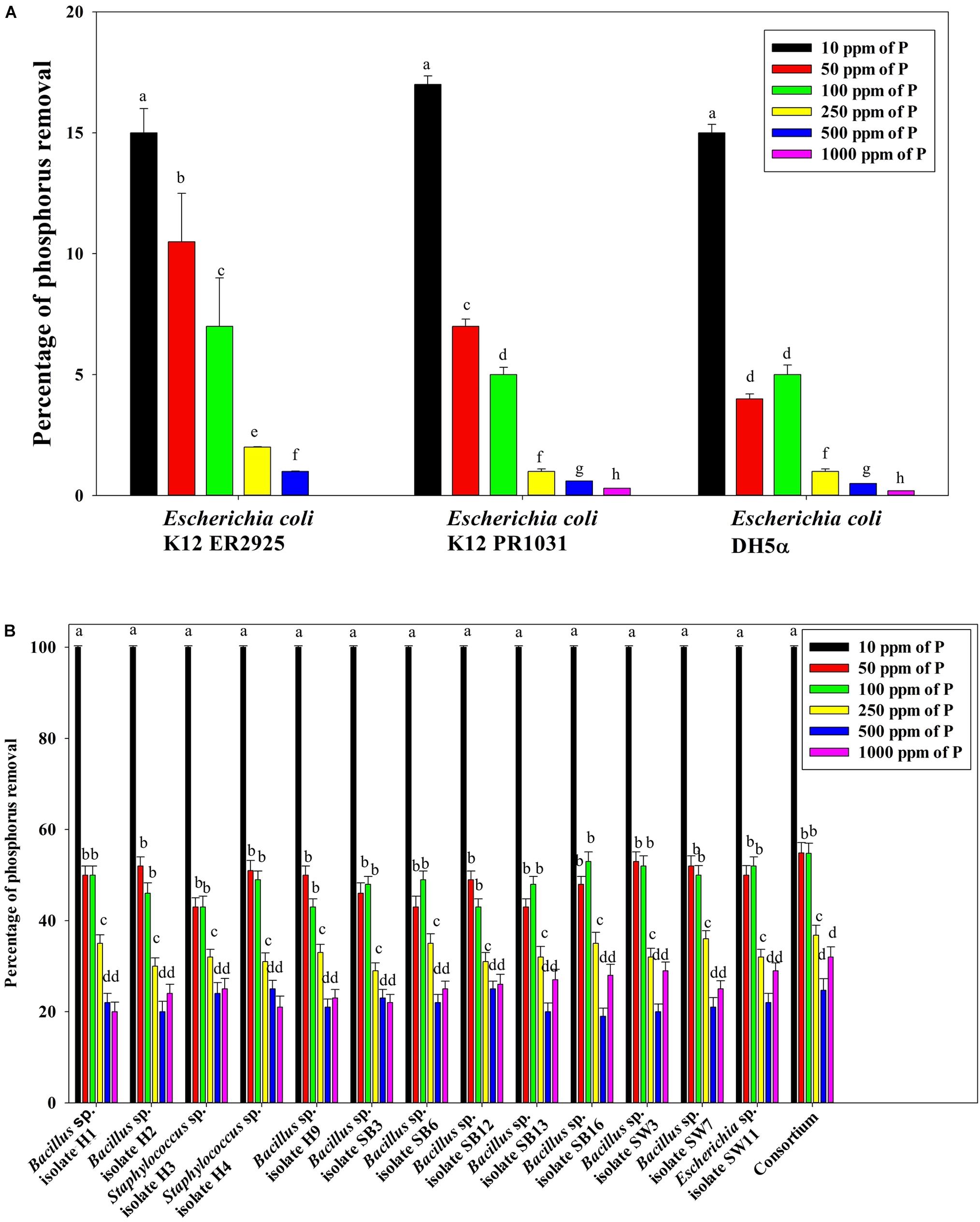
Figure 7. (A) PO43–-P removal by the non-PAO E. coli. strains from different initial PO43–-P concentrations after 12 h of aerobic growth in the absence of NO3–-N. (B) PO43–-P removal by the 13 DPAO isolates and their consortium from different initial PO43–-P concentrations after 12 h of aerobic growth in the absence of NO3–-N. Values designated with different letters are significantly different at the 5% level.
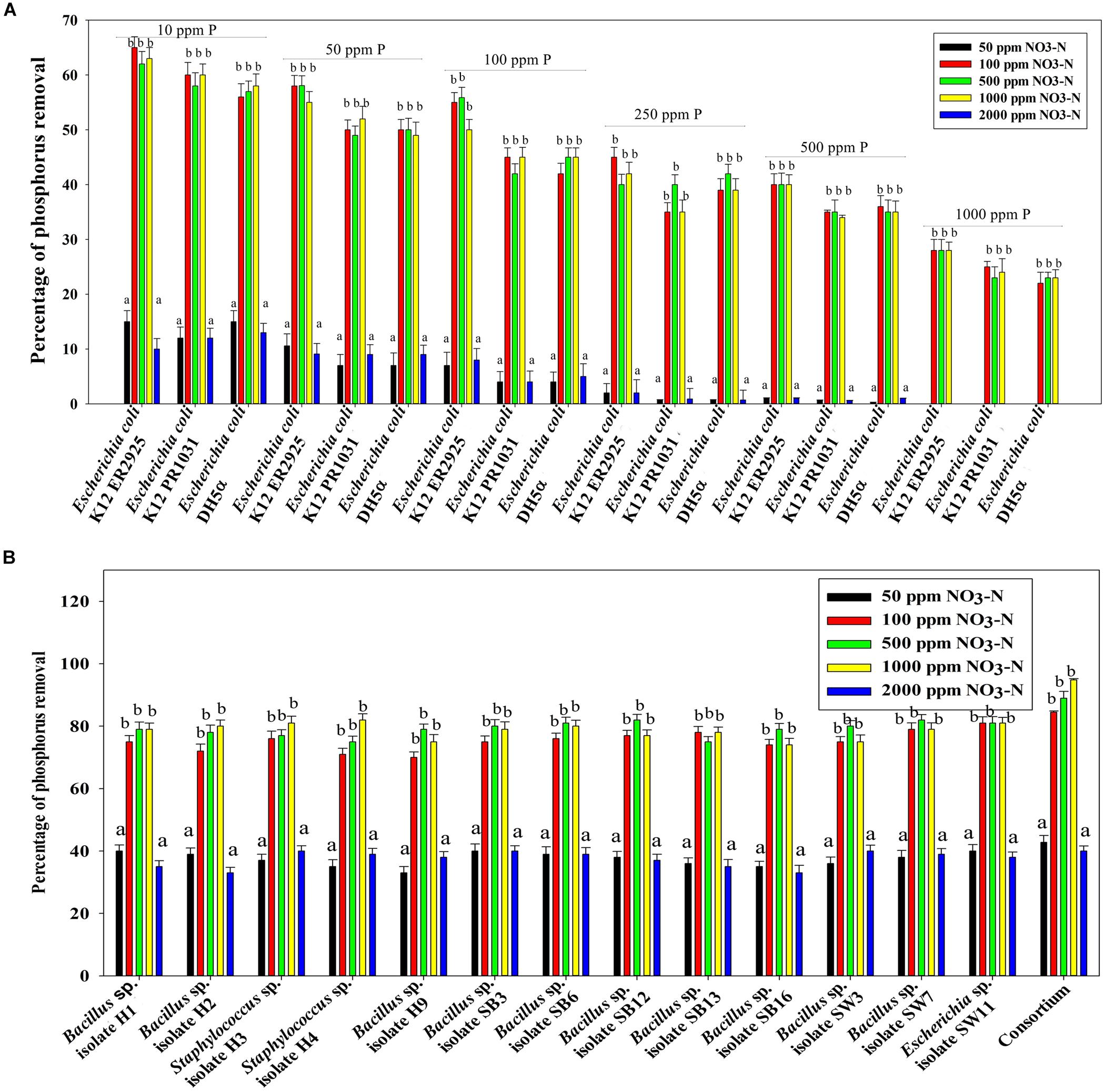
Figure 8. (A) PO43–-P removal by the non-PAO E. coli. strains from different initial PO43–-P concentrations (10–1000 ppm of PO43–-P) after 12 h of anoxic growth in the presence of different initial NO3–-N concentrations. (B) PO43–-P removal by the 13 DPAO isolates and their consortium from 250 ppm initial PO43–-P concentration after 12 h of anoxic growth in the presence of different initial NO3–-N concentrations. Values designated with different letters are significantly different at the 5% level.
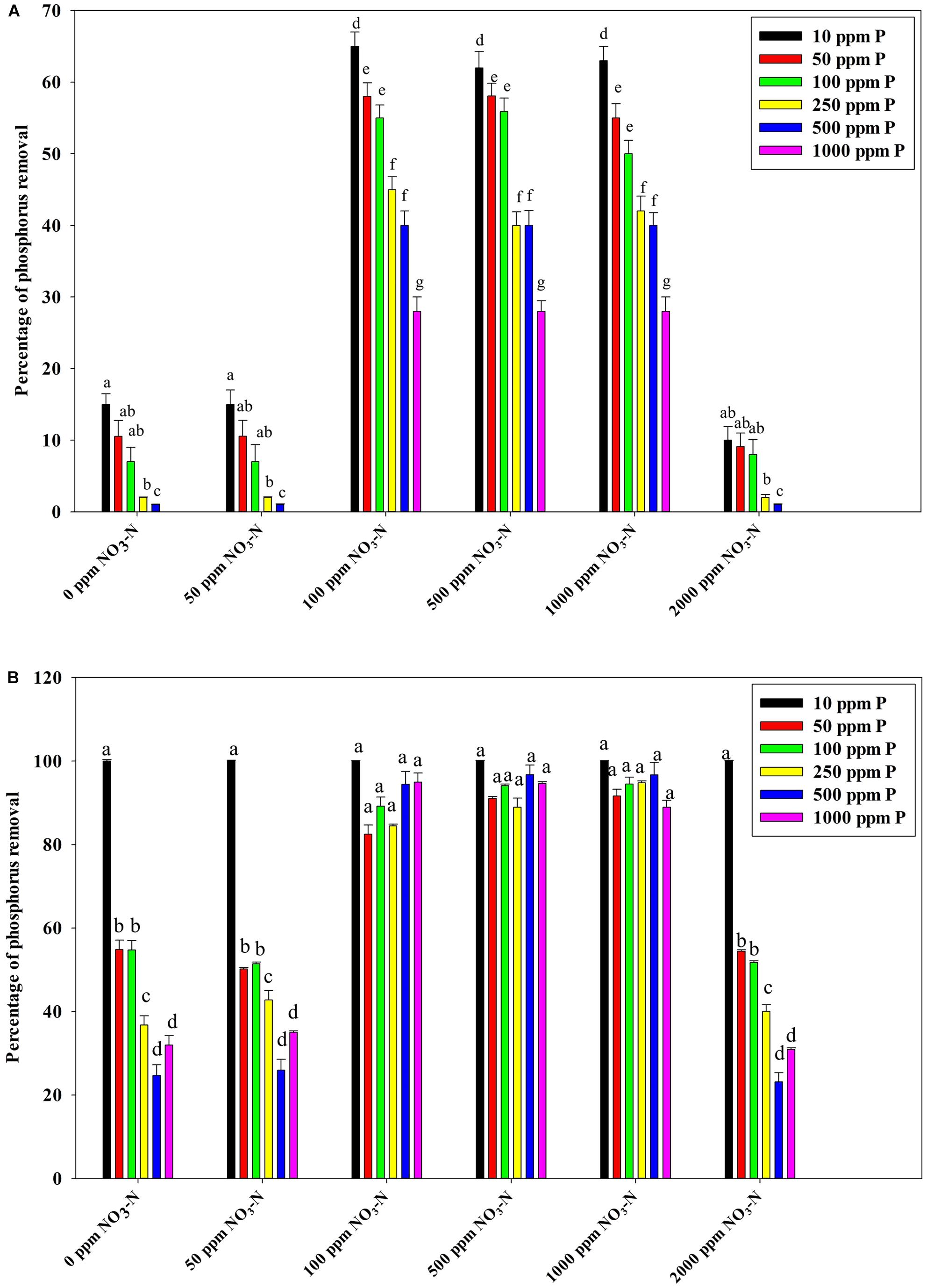
Figure 9. (A) PO43–-P removal by the non-PAO, E. coli. K12 ER 2925 from different initial PO43–-P concentrations after 12 h of anoxic growth in the presence of different initial NO3–-N concentrations. (B) PO43–-P removal by the consortium of the 13 DPAO isolates from different initial PO43–-P concentrations after 12 h of anoxic growth in the presence of different initial NO3–-N concentrations. Values designated with different letters are significantly different at the 5% level.
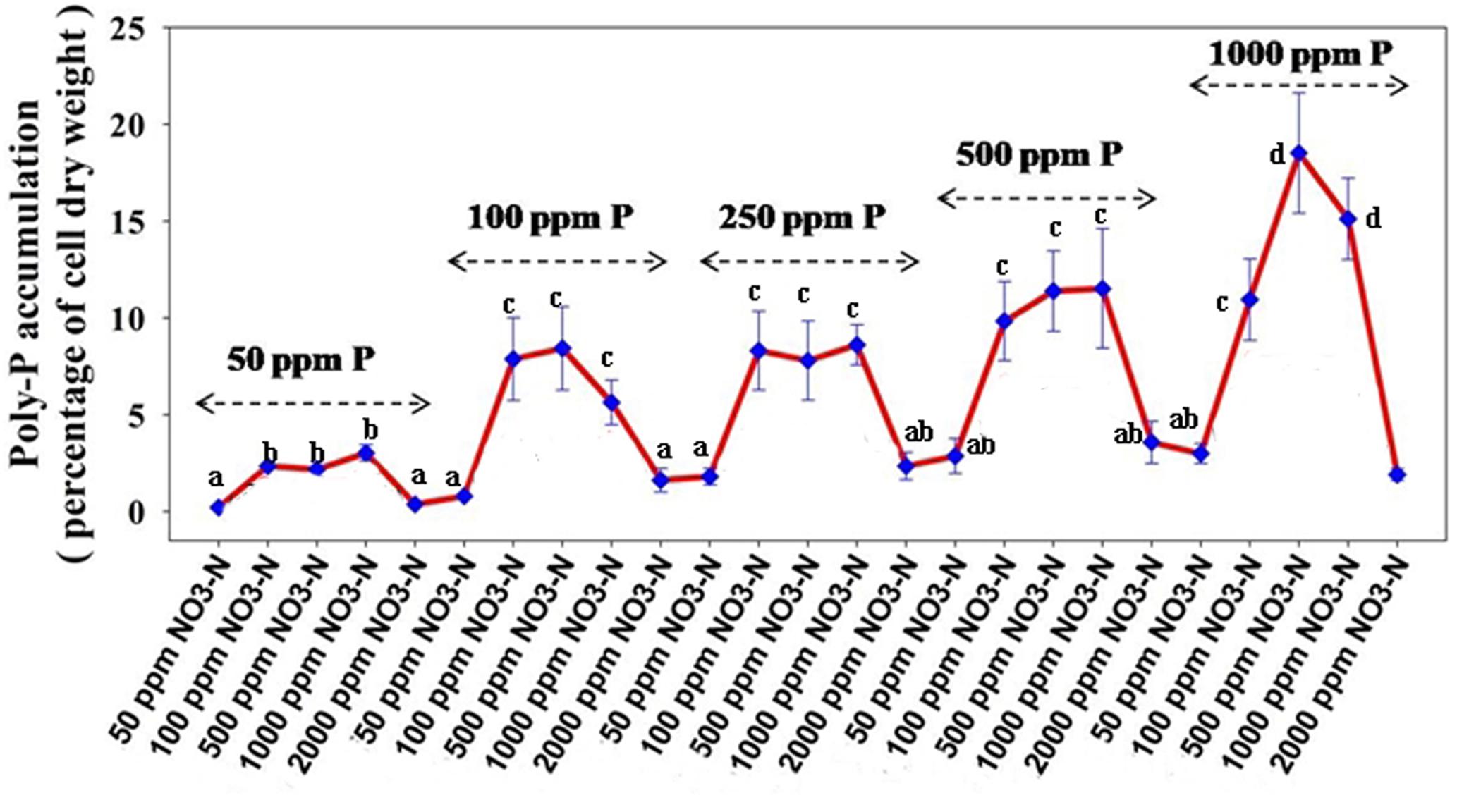
Figure 10. Poly-P accumulation concurrent with PO43–-P removal from synthetic wastewater (SW) in combinations of 50–1000 ppm of PO43–-P and 50–2000 ppm of NO3–-N by the consortium of the 13 DPAO isolates. Values designated with different letters are significantly different at the 5% level.
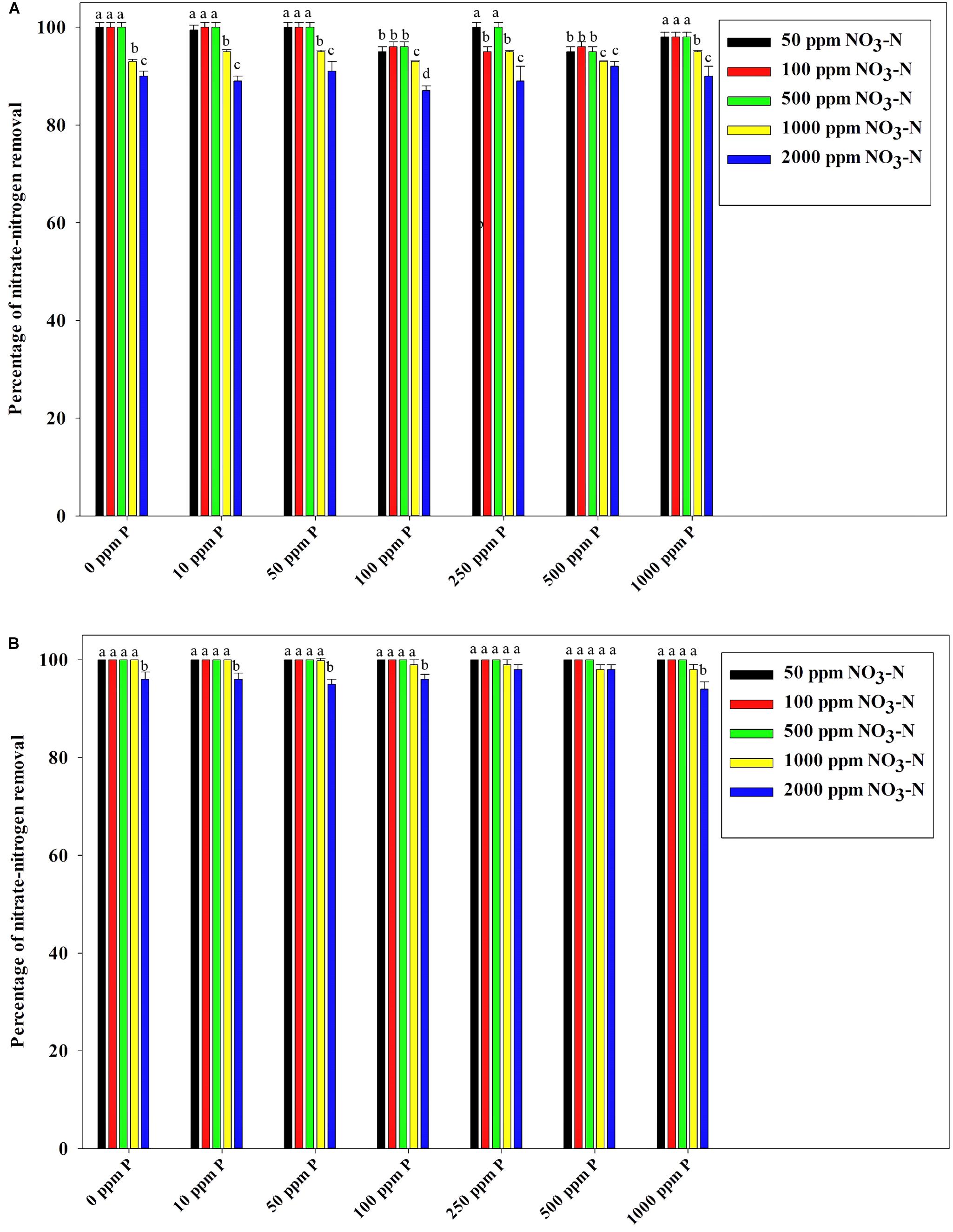
Figure 11. (A) NO3–-N removal by the non-PAO, E. Coli. K12 ER 2925 from different initial NO3–-N concentrations after 12 h of anoxic growth in the presence of different initial PO43–-P concentrations. (B) NO3–-N removal by the consortium of 13 DPAO isolates from different initial NO3–-N concentrations after 12 h of anoxic growth in the presence of different initial PO43–-P concentrations. Values designated with different letters are significantly different at the 5% level.
Although all of the above experiments were continued till 96 h, it was found that insignificant increase in P removal occurred after 12 h (Supplementary Figures S3, S4) with 90–100% nitrate being removed within the 12 h period. Simultaneously, analysis of the growth curves of each of the isolates from each of the P and N combinations over 96 h revealed identical growth increase for each of the isolates till 24 h, after which they reached the stationary phase (Supplementary Figure S5). The growth data demonstrated that the exponential phase of the isolates continued till 24 h.
Increase of pH over 96 h was observed in the media combinations with nitrate because of denitrification, with starting pH at 7 and the maximally increased pH at 7.6 (Supplementary Table S4). Any P-removal caused by chemical precipitation of phosphates at this increased pH can be easily ruled out as this observed pH is not at all favorable for chemical precipitation of insoluble phosphates (Kofina and Koutsoukos, 2005; Daneshgar et al., 2018; Simoes et al., 2018).
A similar situation of the NO3–-N mediated inductive effect on PO43–-P removal was observed when the DPAO consortium was applied to actual wastewaters. Stillage combinations having 175, 200, 350, and 400 ppm of the initial PO43–-P conc. mixed with an explosive industry effluent of 750, 1000, and 1500 ppm of NO3–-N conc., were found to acquire a 6–8-fold increase in PO43–-P removal within the initial 2 h, and finally, within 12 h, 85–100% P removal was achieved in comparison to 20–55% P removal in combinations without NO3–-N (Figure 12). The inductive effect of NO3–-N on PO43–-P removal was again observed along the range of 750–1500 ppm NO3–-N. Similarly, 100% NO3–-N removal was also complete within 12 h from all of the mixed effluent combinations (Supplementary Figure S6), once more justifying the absence of any kind of effect of the initial PO43–-P conc. on NO3–-N removal.
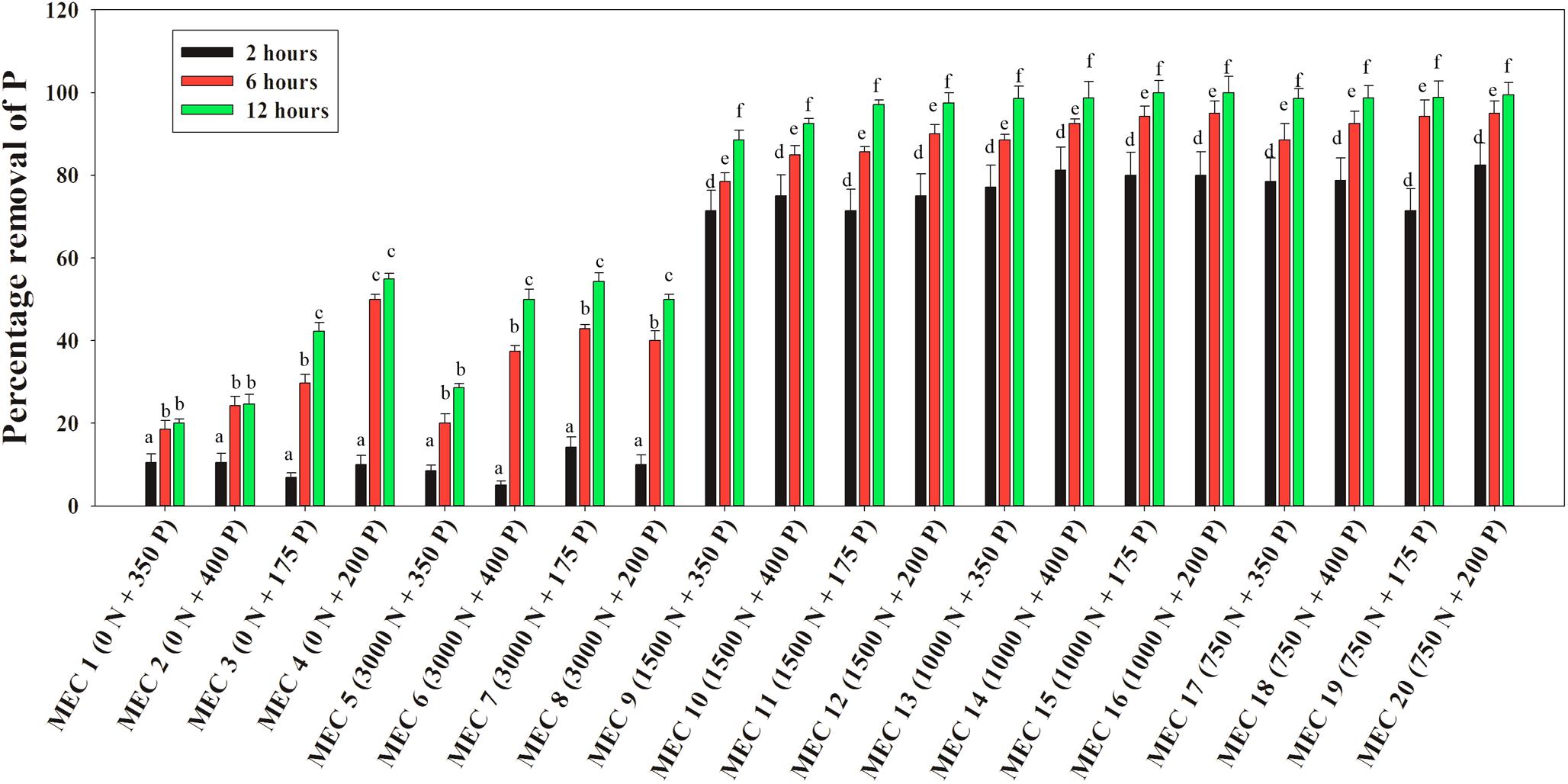
Figure 12. Percentages of PO43–-P removal over 12 h by the consortium of the 13 DPAO isolates from the mixed effluent medium with different combinations of PO43–-P and NO3–-N concentration attained by mixing stillage from the rice-based distillery industry and effluent from the explosives industry where MEC stands for Mixed Effluent Combination. Values designated with different letters are significantly different at the 5% level.
Using classical chemical kinetics, the apparent reaction orders of PO43–-P and NO3–-N removals were established and the dependency of PO43–-P removal on NO3–-N addition was also proven strongly by the kinetic models. The PO43–-P removal pattern from SW and the effluent mixture containing only stillage and no NO3–-N followed the zero-order kinetics for the 12 h period, which was observed from the graph plotted as XA , along the Y-axis and the time (t) along the X-axis, where [P] is the conc. of PO43–-P at a certain time and [P]0 is the initial conc. of PO43–-P at t = 0. The plot with an R2 value that varied from 0.960–0.979 gave straight lines that passed through the origin with slopes (k/[P]0) decreasing from 0.080 for 10 ppm of PO43–-P to 0.007 for 1000 ppm of PO43–-P (Supplementary Figure S7A). In contrast, for the NO3–-N removal pattern from both SW and mixed effluent in combinations with or without PO43–-P, -loge(1-XA) was plotted along the Y-axis and time (t) was plotted along the X-axis. The plots followed first-order kinetics and gave straight lines that passed through the origin with an R2 value that varied from 0.949–0.993 (Supplementary Figure S7B) to 0.913–0.990 (Supplementary Figure S7C). Interestingly, PO43–-P removal in the presence of 50–2000 ppm NO3–-N followed neither zero- nor first- or second-order kinetics. Hence, the method of the initial rates was used to determine the apparent order. The value of R0 (initial rate of the reaction) was calculated from the initial PO43–-P conc. at certain times, t1 and t2 (t2 > t1) following the equation, (Atkins et al., 2018). When the initial PO43–-P removal rate, R0 (for 10–1000 ppm of PO43–-P between times 2 and 1 h) along the Y-axis, was plotted against the initial NO3–-N conc., C0 (50–2000 ppm at time 1 h) along the X-axis, a 4–8-fold increase in the PO43–-P removal rate was noticed (Figure 13). In the graphical representation, a linear plateau achieved for the PO43–-P removal in the NO3–-N addition range of 100–1000 ppm implied that a steady state rate of the PO43–-P uptake was reached, and further NO3–-N addition no longer induced any more appreciable increase. The kinetic analyses again reaffirmed the fact that the addition of a minimum conc. of 100 ppm of initial NO3–-N within the very first hour of remediation was enough to induce the highest PO43–-P removal rate from both the SW and the mixed effluents.
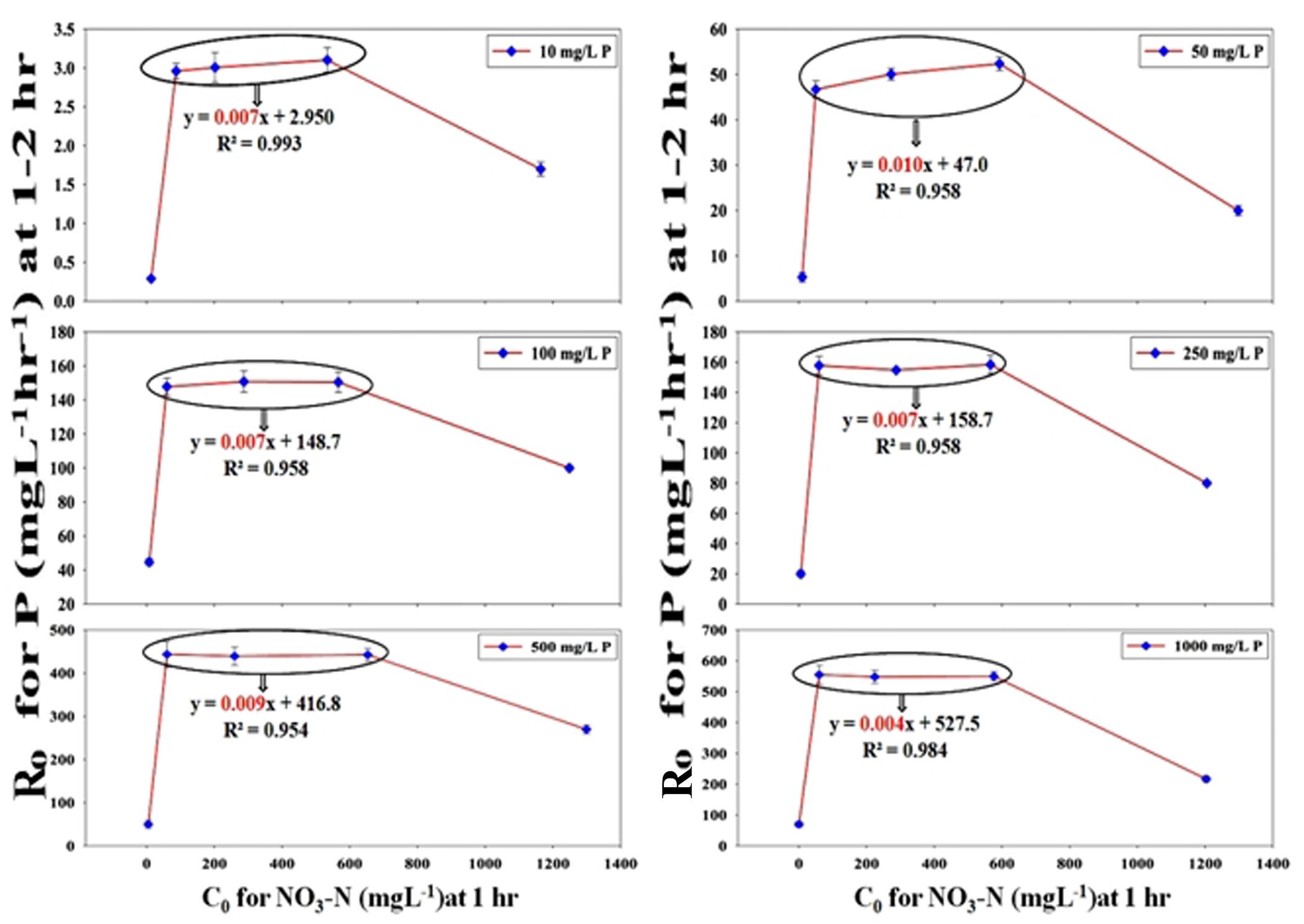
Figure 13. Chemical kinetics of the inductive effect of NO3–-N on PO43−-P removal achieving a stable maximally induced state as observed by plotting the initial rate of PO43−-P removal (R0) at 1–2 h against the initial conc. of NO3–-N (C0) at the end of 1 h for 100, 500 and 1000 ppm of NO3–-N (within the bubble) from SW by the consortium of the 13 DPAO isolates. The straight line equation within the bubble nearing an m value (slope) of almost 0 justified that maximum induction in R0 that already had occurred at C0 attained for 100 ppm of NO3–-N at the end of 1 h and was maintained at the same level for the C0 obtained for 1000 ppm of NO3–-N. An initial PO43−-P conc. of 10–1000 ppm and initial NO3–-N conc. of 50–2000 ppm in different combinations are shown. Uninduced R0 was seen at 1–2 h in combinations with 50 and 2000 ppm of C0 at the end of 1 h.
The average transcription level of the four transporter genes, PstS, PstC, PstA and PstB, and the negative regulator gene, PhoU, relative to P removal with added NO3–-N from the Escherichia coli isolate SW11 and the Bacillus sp. isolate SW7 is depicted in Figure 14. As a comparison the expression level of the same five candidate genes are also displayed in Figure 15 from the identical media combinations for the control non-PAO E. Coli. K 12 ER 2925. RpoB gene was selected as the reference gene for proper normalization as it possessed the lowest M value (highest stability) (Supplementary Figure S8). The expression of the four Pst transporters was always maintained at a high basal level from the initial 1st hour until 12 h in PO43–-P deplete conditions to refute any effect of the added NO3–-N (Figures 14A–D, 15A–D). This supported earlier reports which stated that during P-starved conditions, the Pst genes were highly expressed (Hirota et al., 2013). However, the moment that the PO43–-P replete situation was created by adding 100, 250, or 500 ppm of PO43–-P to the medium, the expression of the Pst genes decreased drastically, even at the very 1st hour, and even when NO3–-N was added to the medium (Figures 14A–D, 15A–D). The repression of the Pho regulon during external PO43–-P rich conditions had also been observed by researchers earlier (Muda et al., 1992). Intriguingly, from the 2nd hour until the 12th hour, PstS, PstC, PstA, and PstB expression levels started to increase and were reinstated to the original level, if not higher, for the combinations of 100, 250, and 500 ppm of PO43–-P with 100–1000 ppm of NO3–-N (Figures 14A–D) in contrast to the restoration of the expression of the same P-transporter genes for E. Coli K 12 2925 to 40–60% of its original basal level (Figures 15A–D). The sustenance of this high expression of Pst transporters under the PO43–-P replete condition was contrary to earlier reported observations. We speculated that the addition of NO3–-N might have played an inductive role in an unknown mechanism to withdraw the repression of the Pho regulon by restoring Pst transporter expression. It was again seen that even a minimum of 100 ppm NO3–-N addition under a highly PO43–-P rich environment (up to 500 ppm PO43–-P in the experiments) could solely induce the Pho regulon by causing a high expression of Pst transporter genes. The 100, 250, and 500 ppm of PO43–-P with 2000 ppm of NO3–-N added failed to elicit any such restoration of PstSCAB expression level (Figures 14A–D, 15A–D). When no PO43–-P was present, the PhoU gene was observed to be downregulated for the studied 12 h period, despite the added NO3–-N (Figures 14E, 15E), with a simultaneous upregulation of the Pst genes. However, the moment that the PO43–-P replete condition was achieved with the addition of 100, 250, or 500 ppm of PO43–-P, the PhoU expression was significantly upregulated to 15–20-fold from the very 1st hour until the 12th hour, even in the absence of NO3–-N in the medium (Figures 14E, 15E), with simultaneous down regulation of the Pst genes. Amazingly, upon the addition of 100 and 1000 ppm of NO3–-N under PO43–-P rich conditions (100, 250, and 500 ppm of P), the expression of PhoU started to reduce significantly from the 2nd hour of incubation, and this 15–20-fold downfall continued steadily until the 12th hour (Figures 14E, 15E). The down regulated PhoU expression level at a high PO43–-P conc. with 100–1000 ppm of added NO3–-N strangely resembled the environment without external PO43–-P. The down regulation of PhoU was accompanied by a concurrent up regulation of Pst transporter genes, which agreed with earlier studies that had established the PhoU protein as a negative regulator of Pho regulon (Gardner et al., 2014). However, the addition of even 100 ppm NO3–-N withdrew the negative regulation by PhoU unexpectedly, even under a PO43–-P replete state, by some unknown mechanism. As long as PhoU was down regulated by adding NO3–-N, the concurrent high expression of PstSCAB transporters could be achieved, which accelerated the PO43–-P uptake (almost 100% uptake even from 1000 ppm of PO43–-P replete environment) from both PO43–-P rich SW and mixed effluents. However, it was again observed that 2000 ppm of NO3–-N addition failed to down regulate the PhoU expression in all the experimental strains (Figures 14E, 15E).
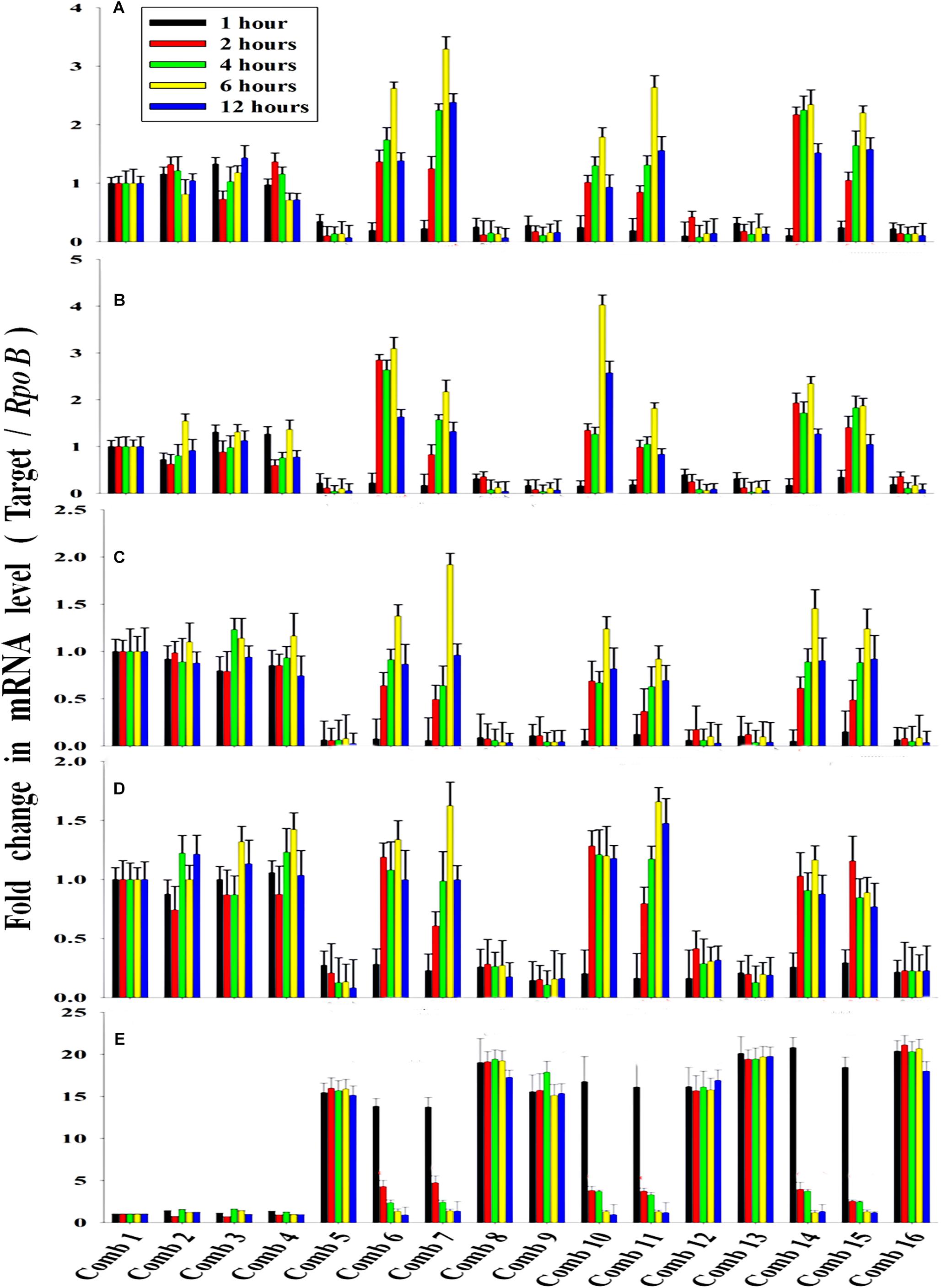
Figure 14. The average transcript level of Pho regulon genes- (A) PstS, (B) PstC, (C) PstA, (D) PstB, (E) PhoU of the Bacillus sp. isolate SW7 (Accession no. KU740235-KU740236) and the Escherichia coli isolate SW11 (Accession no. KU740237-KU740238) over a period of 12 h when grown in SW with the following combinations of PO43–-P and NO3–-N. Comb 1–4: 0 ppm PO43–-P with 0, 100, 1000, 2000 ppm of NO3–-N; Comb 5–8: 100 ppm PO43–-P with 0, 100, 1000, 2000 ppm of NO3–-N; Comb 9–12: 250 ppm PO43–-P with 0, 100, 1000, 2000 ppm of NO3–-N; Comb 13–16: 500 ppm PO43–-P with 0, 100, 1000, 2000 ppm of NO3–-N where Comb stands for Combinations.
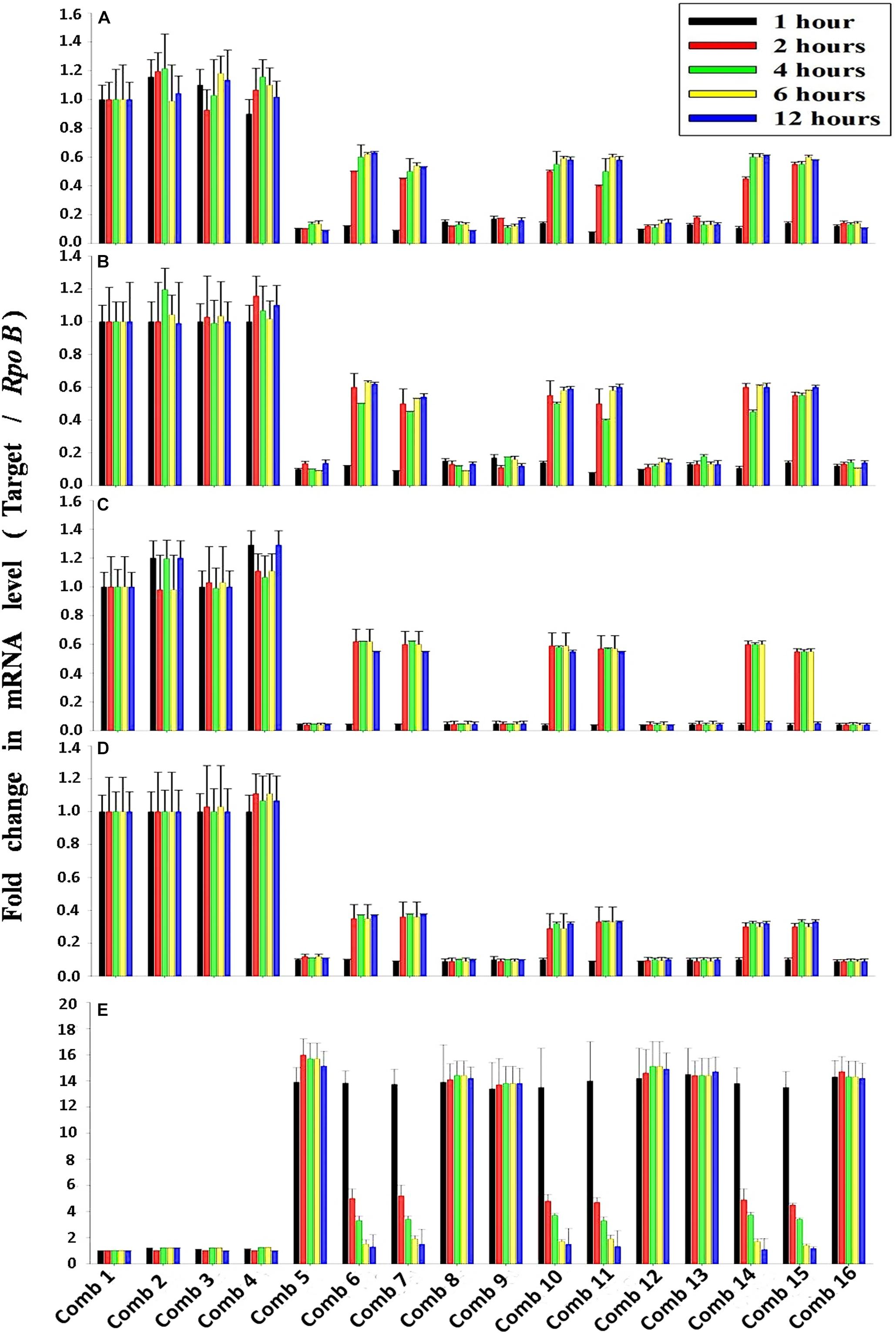
Figure 15. The average transcript level of Pho regulon genes- (A) PstS, (B) PstC, (C) PstA, (D) PstB, (E) PhoU of the non-PAO, E. Coli. K12 ER 2925 over a period of 12 h when grown in SW with the following combinations of PO43–-P and NO3–-N. Comb 1–4: 0 ppm PO43–-P with 0, 100, 1000, 2000 ppm of NO3–-N; Comb 5–8: 100 ppm PO43–-P with 0, 100, 1000, 2000 ppm of NO3–-N; Comb 9–12: 250 ppm PO43–-P with 0, 100, 1000, 2000 ppm of NO3–-N; Comb 13–16: 500 ppm PO43–-P with 0, 100, 1000, 2000 ppm of NO3–-N where Comb stands for Combinations.
In a recent study (Wang et al., 2018) with Dolichospermum flos-aquae, a diazotrophic cyanobacterium, the expression levels of genes involved in PO43–-P uptake, such as the gene for the hydrolysis of phosphomonoesters, PhoD, and the PO43– transporter, PstS, were observed to be upregulated in PO43–-P deficient cultures upon nitrogen (N) addition. N addition also enhanced poly-P formation and alkaline phosphatase activity in PO43–-P deficient cultures. During the PO43–-P deficient treatment, the expression levels of PstS and PhoD were significantly down regulated during the initial 2–4 days, but a 2.8-fold up regulation was observed 6–8 days later in PstS, and a 1.2–8.6-fold up regulation was seen in PhoD, in the absence of N. However, after N addition, the PstS expression was up regulated by 10.7-fold on day 12, and the PhoD expression was up regulated by 17.9-fold after day 8, even in the PO43–-P deficient treatment. Interestingly, in the above study (Wang et al., 2018), the up regulation of PstS and PhoD genes occurred upon the addition of N in PO43–-P deplete conditions, whereas in the case of the DPAO isolates used in this study, NO3–-N addition enhanced the expression of PstSCAB genes even under PO43–-P replete conditions, although the gene was normally reported to be down regulated in bacteria under a PO43–-P replete environment (Muda et al., 1992; Hirota et al., 2013).
The most surprising result presented in this study was the demonstration that NO3–-N at a specific concentration limit, i.e., within 100–1000 ppm, acted as an activator of the Pho regulon even under PO43–-P replete condition. Usually Pho regulon is known to be activated by external PO43–-P starvation signal and by PhoB-PhoR two component system. But in this case, during the DNPR process, in response to a completely opposite signal of excess PO43–-P in external environment (in response to which Pho regulon is normally destined to get shut off), Pho regulon gets activated in presence of 100–1000 ppm of NO3–-N. Under PO43–-P replete external environment, PhoB is dephosphorylated by PhoR-PhoU complex (Wanner, 1992), but this intervention by PhoU seems to be far from possible here as the PhoU expression itself is found to be down regulated since the second hour of NO3–-N addition till the 12 h period (Figures 14E, 15E). Hence, we assume that the sensor PhoB is phosphorylated by normal kinase activity of PhoR at this period without facing any repression from PhoU. We speculate that 100-1000 ppm of NO3–-N by some unknown mechanism is repressing the expression of PhoU for this period, but we also have to remember that during this 12 h period NO3–-N is also being simultaneously removed almost at about 100% removal rate from the wastewater/culture media by the DPAO isolates and their consortium. Hence, 100–1000 ppm of NO3–-N can be considered to be only the initial inducing concentration needed to repress the expression of PhoU gene and the repression is maintained uninterruptedly even after this NO3–-N concentration is gradually decreased to near zero over the 12 h period. At this juncture, we could only assume that NO3–-N must be having an unusual role in the initiation of transcriptional inhibition of PhoU, but how it is being implemented does not fall within the experimental designs of our study. A similar repression was reported in E. coli where PFL gene encoding the enzyme pyruvate formate-lyase, a key enzyme of anaerobic metabolism catalyzing the non-oxidative cleavage of pyruvate to acetyl coenzyme A and formate, was regulated by nitrate (Kaiser and Sawers, 1995). It was demonstrated that the expression of the Pfl operon was negatively regulated by nitrate and when bacterial cells were cultured in minimal medium this regulation was markedly stronger than when they were cultured in rich medium. Nitrate regulation of Pfl operon was proved to be mediated exclusively by the dual sensors, NarX and NarQ and the dual transcription regulators NarL and NarP (Kaiser and Sawers, 1995). Nitrate signaling in bacteria is known to be mediated by the homologous sensor proteins NarX and NarQ (Stewart, 1993). Mutational analyses have defined a heptamer sequence necessary for specific DNA binding by the NarL protein. These heptamers are located at different positions in the regulatory regions of different operons. The response regulator, NarL binds DNA to control nitrate induction and repression of the genes encoding nitrate respiration enzymes and alternate anaerobic respiratory enzymes, respectively (Stewart, 1993). In another instance, repression of gene expression was reported in hyaABCDEF and hybOABCDEFG operons of E. coli encoding for hydrogenase 1 and hydrogenase 2 enzymes respectively, in presence of nitrate as the terminal electron acceptor (Richard et al., 1999). Nitrate repression could however be relieved in a narL narP double mutant, indicating that both NarL and NarP mediated nitrate repression of these two operons. In this study also, nitrate was used as the terminal electron acceptor as an anoxic low DO environment was maintained throughout the experiments. We are still unable to confer at this stage that the nitrate repression of PhoU gene is mediated by NarL/NarP protein binding to the upstream sequences of PhoU, although we could hypothesize that this nitrate mediated repression might involve NarL/NarP protein binding to the upstream elements of PhoU based on earlier observations on similar nitrate mediated transcriptional repression. In Burkholderia pseudomallei, biofilm formation was found to be inhibited in response to exogenous nitrate (Mangalea et al., 2017). Transposon insertional mutants of NarL, the DNA-binding response regulator, NarX, the nitrate sensor, NarG-1 and NarH-1, the alpha and beta subunits of major nitrate reductase and of NarK-1, a nitrate/nitrite transporter, demonstrated insensitivity to biofilm inhibition by nitrate. These results suggested that nitrate sensory cascades and nitrate metabolism pathway were somehow linked to and responsible for biofilm inhibition based on nitrate availability. Apart from these, nitrate was also found to inhibit other alternative respiratory pathways in bacteria. In Dechloromonas aromatica strain RCB, in transition studies from aerobic metabolism through nitrate reduction to perchlorate reduction, increase in the level of transcripts necessary for nitrate and perchlorate reduction was observed concomitantly with decrease in the concentration of exogenous nitrate and perchlorate respectively, suggesting that nitrate negatively regulates transcription of perchlorate reductase thus inhibiting perchlorate reduction leading to preferential utilization of nitrate (Sun and Coates, 2007). All these studies indicated that nitrate mediated transcriptional repression is quite widespread among the microbial community. In eukaryotic system also, supply of exogenous nitrate was found to immediately induce the expression of a transcriptional repressor gene in rice, designated as NIGT1 (Nitrate-Inducible, GARP-type Transcriptional Repressor 1) (Sawaki et al., 2013).
This whole study was based on a single anoxic phase denitrifying phosphorus removal process by the DPAO isolates and their consortium. Our established DPAOs are found to be quite different from the conventional PAOs. PAO needs two stages – anaerobic and aerobic for efficient PO43–-P removal. In anaerobic phase, they use external carbon source like VFA to produce PHA (polyhydroxyalkanoates), and the stored glycogen and polyphosphate is broken down and phosphorus is released to the external environment. In aerobic phase, they degrade the PHA and produce polyphosphate intracellularly through the uptake of PO43–-P from the external environment. Therefore, for PAOs, there is no need of external carbon source in this aerobic phase when actual PO43–-P removal is bound to happen in EBPR system. But our DPAOs produce poly-phosphate only in the presence of external carbon source under anoxic condition where nitrate acts as an electron acceptor. In absence of external carbon sources, cells with PHA do not grow and do not produce poly-phosphate. Not only that, in their anaerobic growth phase, they are also unable to utilize the external carbon sources like PAOs to produce PHA. Rather they produce PHA by degrading internal poly-phosphate or glycogen in anaerobic phase (Barak and van Rijn, 2000a). Our developed DPAOs showed similar characteristics under anaerobic phase growth and only their anoxic growth phase opened up the scope for simultaneous biological PO43–-P and NO3–-N removal carried out in this study with NO3– as the terminal electron acceptor, where external carbon sources present in the effluent/SW could be utilized to its optimum, surpassing the need of anaerobic stage. The EBPR mechanism of DPAO Brachymonas sp. strain P12 was similar to the conventional anaerobic–aerobic (or anaerobic–anoxic) EBPR models, but these models were developed under anoxic or aerobic conditions only, without an anaerobic stage (Shi and Lee, 2007). In their search for DPAO isolates from piggery sludge, enriched sludge, and winery sludge, Shi and Lee (2006) combined anoxic denitrifying ability with aerobic–anoxic PO43–-P removal and compared it with conventional anaerobic–aerobic and anaerobic–anoxic PO43–-P removal. They finally optimized a single-stage anoxic PO43–-P removal process with NO3– as the final electron acceptor for the culture, isolated DPAOs and achieved simultaneous PO43–-P and NO3–-N removal efficiencies of 96 and 86%, respectively from a single stage anoxic reactor. Our results obtained in this study completely matched their findings. This DNPR process, performed in a single stage anoxic phase, also provided evidences of excessive PO43–-P removal, defying the negative regulation of PhoU protein in the presence of a certain conc. range of NO3–-N.
While executing the remediation of mixed effluent, the carbon sources utilized were the complex carbon sources of PO43–-P rich stillage which were easily fermented to produce alcohol along with the unfermented complex carbon sources of VFAs, whereas in experiments with SW, added meat extract were the complex carbon sources utilized. Study of Natcheva and Beschkov (2003) has already established that anoxic denitrification occurs well in the presence of complex carbon sources like meat extract and in our study we did not get any significant differences in terms of PO43–-P and NO3–-N removal where VFA like acetate was used as the sole carbon source instead of meat extract. Our study thus described a single anoxic phase denitrifying PO43–-P removal with NO3– as terminal electron acceptor in the presence of external complex carbon sources like meat extract. The denitrification phenomenon was monitored mainly in terms of decreasing concentration of NO3– from the effluent/SW. We have tested whether this denitrification phenomenon resulted in reduction of nitrate to nitrite. But nitrite could not be traced at the end of 12 h experiments even after repetitive measurements, which made us to conclude that nitrite had been maximally reduced to its gaseous forms through this denitrification process.
Simultaneous removal of PO43–-P and NO3–-N from wastewaters has long been described by several workers (Lee et al., 2001; Hu et al., 2003; Wang et al., 2009; Yang et al., 2010; Yamashita and Yamamoto-Ikemoto, 2014; Delgadillo-Mirquez et al., 2016; Wan et al., 2017; Zhang et al., 2017), but all of those studies relied on a two-reactor system with a continuous switching between the aerobic-anaerobic-anoxic-aerobic cycles. A recent approach that reported PO43–-P and NO3–-N removal efficiencies of 97.8 and 98.2%, respectively, required an obligatory aerobic/anaerobic system for the anammox reaction and a requisite aerobic/anaerobic/anoxic process for denitrifying P removal (Zhang et al., 2017). In another approach, simultaneous PO43–-P and NO3–-N removal efficiencies of 94 and 91%, respectively, were reported from domestic wastewater in a two-sludge sequencing batch reactor following an anaerobic-anoxic/nitrification process with the necessity of maintaining the influent (chemical oxygen demand, COD) COD/P and COD/Total N ratios of 19.9 and 9.9, respectively (Wang et al., 2009). A novel sequencing batch moving bed membrane bioreactor for wastewater treatment was also reported, where the total PO43–-P and NO3–-N removal efficiencies averaged at 82.6 and 84.1%, respectively, with the PO43–-P remediation competence being dependent on the length of the aerobic/anaerobic cycle and varying considerably if the switch did not occur at the appropriate time (Yang et al., 2010). In India, the upflow anaerobic sludge blanket (UASB) reactor was designed especially for PO43–-P removal from industrial effluents based on the anaerobic treatment of the wastewater with methanogenic bacteria, but the maintenance of the anaerobic condition with a removal efficiency of just 10–50 ppm of PO43–-P rendered this system to be comparatively incompetent (Haridas, 2010). These operational complexities and the high maintenance cost of the reactor systems often lead to the closure of wastewater treatment plants in industries, especially in India and in other developing countries. In contrast, in the present study, a simplified single anoxic phase batch reactor system with a compatible denitrifying polyphosphate accumulating bacterial consortium was reported, which is a perfect combination to unequivocally remediate the excess PO43–-P and NO3–-N from wastewater simultaneously or individually. This study explicitly proved that the addition of only 100 ppm of NO3–-N to PO43–-P rich wastewater would result in 100% PO43–-P removal, thereby defying the limitation of the PO43–-P uptake by the repression of Pho regulon in a PO43–-P replete environment. This finding, if exploited in the EBPR system, could overcome the limitation in PO43–-P removal. The corresponding elevated accumulation of poly-P by the bacterial consortium has a high potential to be used as a PO43– biofertilizer. The experiments performed here were restricted to a 5 L system; however, further successful scaling-up to higher volumes that are suitable for industrial purposes is necessary for a worthwhile application of this study.
All datasets generated for this study are included in the article/Supplementary Material.
CM designed the study, performed the experiments, analyzed the results, and contributed toward preparation of the manuscript. RC and MB performed the experiments and helped in manuscript writing. SG helped in designing the primers for real-time PCR. RB and BC performed the kinetic analyses. KR as a supervisor, identified the research problem, conceptualized and designed the work, and wrote the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful for the valuable comments on the earlier version of the manuscript from the reviewers and Associate Editor HS of Frontiers in Microbiology, who helped to improve the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02586/full#supplementary-material
Abou-Elela, S. I., El-Kamah, E. M., Aly, H. I., and Abou-Taleb, E. (1995). Management of waste water from the fertilizer industry. Water Sci. Tech. 32, 45–54.
Ahn, J., Daidou, T., Tsuneda, S., and Hirata, A. (2002). Transformation of phosphorus and relevant intracellular compounds by a phosphorus-accumulating enrichment culture in the presence of both the electron acceptor and electron donor. Biotechnol. Bioeng. 79, 83–93. doi: 10.1002/bit.10292
Atkins, P., De Paula, J., and Keeler, J. (2018). Atkins’ Physical Chemistry, 11th Edn. Oxford: Oxford University Press.
Barak, Y., Cytryn, E., Gelfand, I., Krom, M., and van Rijn, J. (2003). Phosphorus removal in a marine prototype, recirculating aquaculture system. Aquaculture 220, 313–326. doi: 10.1016/S0044-8486(02)00342-3
Barak, Y., and van Rijn, J. (2000a). Atypical polyphosphate accumulation by the denitrifying bacterium Paracoccus denitrificans. Appl. Environ. Microbiol. 66, 1209–1212. doi: 10.1128/aem.66.3.1209-1212.2000
Barak, Y., and van Rijn, J. (2000b). Relationship between nitrite reduction and active phosphate uptake in the phosphate-accumulating denitrifier Pseudomonas sp. Strain JR12. Appl. Environ. Microbiol. 66, 5236–5240. doi: 10.1128/aem.66.12.5236-5240.2000
Barker, P. S., and Dold, P. L. (1997). General model for biological nutrient removal activated-sludge systems: model presentation. Water Environ. Res. 69, 969–984. doi: 10.2175/106143097x125669
Baumann, B., Snozzi, M., Zehnder, A., and Van Der Meer, J. (1996). Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J. Bacteriol. 178, 4367–4374. doi: 10.1128/jb.178.15.4367-4374.1996
Bazylinski, D. A., and Blakemorer, R. P. (1983). Denitrification and assimilatory nitrate reductase in Aquaspirillum magnetotacticum. Appl. Environ. Microbiol. 46, 1118–1124.
Bhattacharyya, A., Saha, J., Haldar, S., Bhowmic, A., Mukhopadhyay, U. K., and Mukherjee, J. (2014). Production of poly-3-(hydroxybutyrate-co-hydroxyvalerate) by haloferax mediterranei using rice-based ethanol stillage with simultaneous recovery and re-use of medium salts. Extremophiles 18, 463–470. doi: 10.1007/s00792-013-0622-9
Brock, T. D., and Madigan, M. T. (1991). The Biology of Microorganisms, 6th Edn. New Jersey, NJ: Prentice-Hall.
Carmany, D. O., Hollingsworth, K., and McCleary, W. R. (2003). Genetic and biochemical studies of phosphatase activity of phoR. J. Bacteriol. 185, 1112–1115. doi: 10.1128/jb.185.3.1112-1115.2003
Carvalho, G., Lemos, P. C., Oehmen, A., and Reis, M. A. M. (2007). Denitrifying phosphorus removal: linking the process performance with the microbial community structure. Water Res. 41, 4383–4396. doi: 10.1016/j.watres.2007.06.065
Cataldo, D. A., Maroon, M., Schrader, L. E., and Youngs, V. L. (1975). Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun. Soil Sci. Plant. 6, 71–80. doi: 10.1080/00103627509366547
Clegg, S. J., Jia, W., and Cole, J. A. (2006). Role of the Escherichia coli nitrate transport protein, NarU, in survival during severe nutrient starvation and slow growth. Microbiology 152, 2091–2100. doi: 10.1099/mic.0.28688-0
Daneshgar, S., Buttafava, A., Capsoni, D., Callegari, A., and Capodaglio, A. G. (2018). Impact of pH and ionic molar ratios on phosphorous forms precipitation and recovery from different wastewater sludges. Resources 7:71. doi: 10.3390/resources7040071
Davies, K. J. P., Lloyd, D., and Boddy, L. (1989). The effect of oxygen on denitrification in Paracoccus denitrificans and Pseudomonas aeruginosa. J. Gen. Microbiol. 135, 2445–2451. doi: 10.1099/00221287-135-9-2445
Delgadillo-Mirquez, L., Lopes, F., Taidi, B., and Pareau, D. (2016). Nitrogen and phosphate removal from wastewater with a mixed microalgae and bacteria culture. Biotechnol. Rep. 11, 18–26. doi: 10.1016/j.btre.2016.04.003
Egly, T., and Zehnder, J. B. (1994). Phosphate and nitrate removal. Curr. Opin. Biotechnol. 5, 275–284. doi: 10.1016/0958-1669(94)90029-9
Faria, O. L. V., Koetz, P. R., dos Santos, M. S., and Nunes, W. A. (2006). Rice parboilization wastewater phosphorus removal by enhanced biological assimilation in sequencing batch reactor (SBR). Cienc. Tecnol. Aliment. 26, 309–317. doi: 10.1590/s0101-20612006000200013
Ferguson, S. T., and Gadian, D. G. (1979). Evidence from 31P nuclear magnetic resonance that polyphosphate synthesis is a slip reaction in Paracoccus denitrificans. Biochem. Soc. Trans. 7, 176–179. doi: 10.1042/bst0070176
Flowers, J. J., He, S., Yilmaz, S., Noguera, D. R., and McMahon, K. D. (2009). Denitrification capabilities of two biological phosphorus removal sludges dominated by different “Candidatus accumulibacter” clades. Environ. Microbiol. Rep. 1, 583–588. doi: 10.1111/j.1758-2229.2009.00090.x
Gardner, S. G., Johns, K. D., Tanner, R., and McCleary, W. R. (2014). The phoU protein from Escherichia coli interacts with phoR, pstB, and metals to form a phosphate-signaling complex at the membrane. J. Bacteriol. 196, 1741–1752. doi: 10.1128/jb.00029-14
Gelfand, I., Barak, Y., Even-Chen, Z., Cytryn, E., van Rijn, J., Krom, M. D., et al. (2003). A novel zero-discharge intensive seawater recirculating system for culture of marine fish. J. World Aquacult. Soc. 34, 344–358. doi: 10.1111/j.1749-7345.2003.tb00072.x
Haridas, A. (2010). Anaerobic Reactor Development for Complex Organic Wastewater. Ph.D Thesis, School of Engineering, Cochin University of Science and Technology, Kochi.
Hirota, R., Motomura, K., Nakai, S., Handa, T., Ikeda, T., and Kuroda, A. (2013). Stable polyphosphate accumulation by a pseudo-revertant of an Escherichia coli phoU mutant. Biotechnol. Lett. 35, 695–701. doi: 10.1007/s10529-012-1133-y
Holler, S., and Trösch, W. (2001). Treatment of urban wastewater in a membrane bioreactor at high organic loading rates. J. Biotechnol. 92, 95–101. doi: 10.1016/S0168-1656(01)00351-0
Hu, J. Y., Ong, S. L., Ng, W. J., Lu, F., and Fan, X. (2003). A new method for characterizing denitrifying phosphorus removal bacteria by using three different types of electron acceptors. Water Res. 37, 3463–3471. doi: 10.1016/s0043-1354(03)00205-7
John, P. (1977). Aerobic and anaerobic bacterial respiration monitored by electrodes. J. Gen. Microbiol. 98, 231–238. doi: 10.1099/00221287-98-1-231
Jørgensen, K. S., and Pauli, A. S. L. (1995). Polyphosphate accumulation among denitrifying bacteria in activated sludge. Anaerobe 1, 161–168. doi: 10.1006/anae.1995.1014
Kaiser, M., and Sawers, G. (1995). Nitrate repression of the Escherichia coli pfl operon is mediated by the dual sensors NarQ and NarX and the dual regulators NarL and NarP. J. Bacteriol. 177, 3647–3655. doi: 10.1128/jb.177.13.3647-3655.1995
Kaltwasser, H., Vogt, G., and Schlegel, H. G. (1961). Polyphosphatsynthese wahrend der nitrat-atmung von Micrococcus denitrificans Stamm 11. Arch. Mikrobiol. 44, 259–265. doi: 10.1007/bf00510947
Kaspar, H. F. (1982). Nitrite reduction of nitrous oxide by propionibacteria : detoxication mechanism. Arch. Microbiol. 133, 126–130. doi: 10.1007/BF00413525
Kerrn-Jespersen, J. P., and Henze, M. (1993). Biological phosphorus uptake under anoxic and anaerobic conditions. Water Res. 27, 617–624. doi: 10.1016/0043-1354(93)90171-d
Kofina, A. N., and Koutsoukos, P. G. (2005). Spontaneous precipitation of struvite from synthetic wastewater solutions. Crystal Growth Design 5, 489–496. doi: 10.1021/cg049803e
Kong, Y., Nielsen, J. L., and Nielsen, P. H. (2004). Microautoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl. Environ. Microbiol. 70, 5383–5390. doi: 10.1128/AEM.70.9.5383-5390.2004
Krishnaswamy, U., Muthusamy, M., and Perumalsamy, L. (2009). Studies on the efficiency of the removal of phosphate using bacterial consortium for the biotreatment of phosphate wastewater. Eur. J. Appl. Sci. 1, 6–15.
Kuba, T., Smolders, G., van Loosdrecht, M. C. M., and Heijnen, J. J. (1993). Biological phosphorus removal from wastewater by anaerobic-anoxic sequencing batch reactor. Water Sci. Tech. 27, 241–252. doi: 10.2166/wst.1993.0504
Kuba, T., van Loosdrechtt, M. C. M., and Heijnen, J. J. (1996). Effect of cyclic oxygen exposure on the activity of denitrifying phosphorus removing bacteria. Water Sci. Tech. 34, 33–40. doi: 10.1016/0273-1223(96)00492-1
Kulaev, I. S., Vagabov, V. M., and Kulakovskaya, T. V. (2004). The Biochemistry of Inorganic Polyphosphates. Chichester: John Wiley & Sons, Ltd.
Kuroda, A., and Ohtake, H. (2000). Molecular analysis of polyphosphate accumulation in bacteria. Biochemistry 65, 304–308.
Lanham, A. B., Oehmen, A., Carvalho, G., Saunders, A. M., Nielsen, P. H., and Reis, M. A. M. (2018). Denitrification activity of polyphosphate accumulating organisms (PAOs) in full-scale wastewater treatment plants. Water Sci. Technol. 78, 2449–2458. doi: 10.2166/wst.2018.517
Lee, D. S., Jeon, C. O., and Park, J. M. (2001). Biological nitrogen removal with enhanced phosphate uptake in a sequencing batch reactor using single sludge system. Water Res. 35, 3968–3976. doi: 10.1016/s0043-1354(01)00132-4
Lee, H. W., and Park, Y. K. (2008). Characterizations of denitrifying polyphosphate-accumulating bacterium Paracoccus sp. Strain YKP-9. J. Microbiol. Biotechnol. 18, 1958–1965.
Li, H., Zhang, Z., Wang, H., Xu, X., Lin, Y., Zhang, J., et al. (2011). Biological phosphorus removal performance and relevant microorganism characteristics of activated sludge in municipal wastewater treatment plants, China. Water Environ. Res. 83, 594–602. doi: 10.2175/106143010X12851009156682
Lloyd, D., Boddy, L., and Davies, K. J. P. (1987). Persistence of bacterial denitrification capacity under aerobic conditions: the rule rather than the exception. FEMS Microbiol. Ecol. 45, 185–190. doi: 10.1016/0378-1097(87)90015-2
Lorenz, B., Schröder, H. C., and Müller, W. E. G. (1999). “Methods of investigation of inorganic polyphosphates in progress in molecular and subcellular biology,” eds Inorganic Polyphosphates Biochemistry Biology Biotechnology, H. C. Schroder, and W. E. G. Muller, (Heidelberg: Springer-Verlag), 23.
Mangalea, M. R., Plumley, B. A., and Borlee, B. R. (2017). Nitrate sensing and metabolism inhibit biofilm formation in the opportunistic pathogen Burkholderia pseudomallei by reducing the intracellular concentration of c-di-GMP. Front. Microbiol. 8:1353. doi: 10.3389/fmicb.2017.01353
Marques, R., Ribera-Guardia, A., Santos, J., Carvalho, G., Reis, M. A. M., Pijuan, M., et al. (2018). Denitrifying capabilities of Tetrasphaera and their contribution towards nitrous oxide production in enhanced biological phosphorus removal processes. Water Res. 137, 262–272. doi: 10.1016/j.watres.2018.03.010
Medhi, K., and Thakur, I. S. (2018). Bioremoval of nutrients from wastewater by a denitrifier Paracoccus denitrificans ISTOD1. Bioresour. Technol. Rep. 1, 56–60. doi: 10.1016/j.biteb.2018.02.006
Meiberg, J. B. M., Bruinenberpg, M., and Harder, W. (1980). Effect of dissolved oxygen tension on the metabolism of methylated amynes in Hyphomicrobium X in the presence and absence of aerobic denitrification. J. Gen. Microbiol. 120, 453–463. doi: 10.1099/00221287-120-2-453
Minnesota Pollution Control Agency (2006). Phosphorus Treatment and Removal Technologies, 2006. Available at: www.pca.state.mn.us (accessed October 2, 2019).
Mino, T., Van Loosdrecht, M. C. M., and Heijnen, J. J. (1998). Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 32, 3193–3207. doi: 10.1016/s0043-1354(98)00129-8
Moore, T. R. (1985). The spectrophotometric determination of dissolved organic carbon in peat waters. Soil Sci. Soc. Am. J. 49, 1590–1592.
Morohoshi, T., Maruo, T., Shirai, Y., Kato, J., Ikeda, T., Takiguchi, N., et al. (2002). Accumulation of inorganic polyphosphate in phoU mutants of Escherichia coli and Synechocystis sp. strain PCC6803. Appl. Environ. Microbiol. 68, 4107–4110. doi: 10.1128/aem.68.8.4107-4110.2002
Muda, M., Rao, N. N., and Torriani, A. (1992). Role of PhoU in phosphate transport and alkaline phosphatase regulation. J. Bacteriol. 174, 8057–8064. doi: 10.1128/jb.174.24.8057-8064.1992
Mukherjee, C., and Ray, K. (2015). An improved method for extraction and quantification of polyphosphate granules from microbial cells. Protoc. Exch. doi: 10.1038/protex.2015.067 [Epub ahead of print].
Natcheva, M., and Beschkov, V. (2003). Denitrification performance of a culture of hermophilic aerobic bacteria NBIMCC 3729. Chem. Biochem. Eng. Q. 17, 123–129.
Nielsen, P. H., McIlroy, S. J., Albertsen, M., and Nierychlo, M. (2019). Re-evaluating the microbiology of the enhanced biological phosphorus removal process. Curr. Opin. Biotechnol. 57, 111–118. doi: 10.1016/j.copbio.2019.03.008
Oehmen, A., Lopez-Vazquez, C. M., Carvalho, G., Reis, M. A., and van Loosdrecht, M. C. (2010). Modelling the population dynamics and metabolic diversity of organisms relevant in anaerobic/anoxic/aerobic enhanced biological phosphorus removal processes. Water Res. 44, 4473–4486. doi: 10.1016/j.watres.2010.06.017
Omelon, S. J., and Grynpas, M. D. (2008). Relationships between polyphosphate chemistry, biochemistry and apatite biomineralization. Chem. Rev. 108, 4694–4715. doi: 10.1021/cr0782527
Payne, W. J. (1983). Bacterial denitrification: asset or defect. Bioscience 33, 319–325. doi: 10.2307/1309319
Rice, C. D., Pollard, J. E., Lewis, Z. T., and McCleary, W. R. (2009). Employment of a promoter-swapping technique shows that PhoU modulates the activity of the PstSCAB2 ABC transporter in Escherichia coli. Appl. Environ. Microbiol. 75, 573–582. doi: 10.1128/aem.01046-08
Richard, D. J., Sawers, G., Sargent, F., McWalter, L., and Boxer, D. H. (1999). Transcriptional regulation in response to oxygen and nitrate of the operons encoding the [NiFe] hydrogenases 1 and 2 of Escherichia coli. Microbiology 145, 2903–2912. doi: 10.1099/00221287-145-10-2903
Rijn, J., Tal, Y., and Schreier, H. (2006). Denitrification in recirculating systems: Theory and applications. Aquacult. Eng. 34, 364–376. doi: 10.1016/j.aquaeng.2005.04.004
Robertson, L. A., and Kuenen, J. G. (1983). Thiosphaera pantotropha gen. nov. sp. nov., a facultatively anaerobic, facultatively autotrophic sulphur bacterium. J. Gen. Microbiol. 129, 2847–2855. doi: 10.1099/00221287-129-9-2847
Robertson, L. A., and Kuenen, J. G. (1984a). Aerobic denitrification - a controversy revived. Arch. Microbiol. 139, 351–354. doi: 10.1007/BF00408378
Robertson, L. A., and Kuenen, J. G. (1984b). Aerobic denitrification : old wine in new bottles? Antonie Van Heuwenhoek 50, 525–544. doi: 10.1007/BF02386224
Rocha, D. J., Santos, C. S., and Pacheco, L. G. (2015). Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek. 108, 685–693. doi: 10.1007/s10482-015-0524-521
Rubio-Rincon, F. J., Lopez-Vazquez, C. M., Welles, L., van Loosdrecht, M. C. M., and Brdjanovic, D. (2017). Cooperation between Candidatus competibacter and Candidatus accumulibacter clade I, in denitrification and phosphate removal processes. Water Res. 120, 156–164. doi: 10.1016/j.watres.2017.05.001
Santos-Beneit, F. (2015). The pho regulon: a huge regulatory network in bacteria. Front. Microbiol. 6:402. doi: 10.3389/fmicb.2015.00402
Sawaki, N., Tsujimoto, R., Shigyo, M., Konishi, M., Toki, S., Fujiwara, T., et al. (2013). A nitrate-inducible GARP family gene encodes an auto-repressible transcriptional repressor in rice. Plant Cell Physiol. 54, 506–517. doi: 10.1093/pcp/pct007
Shi, H., and Lee, C. (2006). Combining anoxic denitrifying ability with aerobic–anoxic phosphorus-removal examinations to screen denitrifying phosphorus-removing bacteria. Int. Biodeterior. Biodegrad. 57, 121–128. doi: 10.1016/j.ibiod.2006.01.001
Shi, H. P., and Lee, C. M. (2007). Phosphate removal under denitrifying conditions by Brachymonas sp. strain P12 and Paracoccus denitrificans PP15. Can. J. Microbiol. 53, 727–737. doi: 10.1139/W07-026
Shnel, N., Barak, Y., Ezer, T., Dafni, Z., and van Rijn, J. (2002). Design and performance of a zero-discharge tilapia recirculating system. Aquacult. Eng. 26, 191–203. doi: 10.1016/s0144-8609(02)00013-4
Shoji, T., Satoh, H., and Mino, T. (2003). Quantitative estimation of the role of denitrifying phosphate accumulating organisms in nutrient removal. Water Sci. Techol. 47, 23–29. doi: 10.2166/wst.2003.0582
Simoes, F., Vale, P., Stephenson, T., and Soares, A. (2018). The role of pH on the biological struvite production in digested sludge dewatering liquors. Sci. Rep. 8:7225. doi: 10.1038/s41598-018-25431-7
Steed, P. M., and Wanner, B. L. (1993). Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU Operon: evidence of a new role for the PhoU protein in the phosphate regulon. J. Bacteriol. 175, 6797–6809. doi: 10.1128/jb.175.21.6797-6809.1993
Stewart, V. (1993). Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol. Microbiol. 9, 425–434. doi: 10.1111/j.1365-2958.1993.tb01704.x
Stewart, V., Lu, Y., and Darwin, A. J. (2002). Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J. Bacteriol. 184, 1314–1323. doi: 10.1128/jb.184.5.1314-1323.2002
Sun, Y., and Coates, J. D. (2007). “Effect of nitrate on microbial perchlorate reduction,” in Proceedings of the American Geophysical Union, Fall Meeting 2007, abstract id. H33E-1680, (San Francisco, CA).
Surin, B. P., Dixon, N. E., and Rosenberg, H. (1986). Purification of the phoU protein, a negative regulator of the pho regulon of Escherichia coli K-12. J. Bacteriol. 168, 631–635. doi: 10.1128/jb.168.2.631-635.1986
Tiedje, J. M., Sexstone, A. J., Myrold, D. D., and Robinson, J. A. (1982). Denitrification; ecological niches, competition and survival. Antonie Van Leeuwenhoek 48, 569–583. doi: 10.1007/bf00399542
United States Environmental Protection Agency (2007). Advanced Wastewater Treatment to Achieve Low Concentration of Phosphorus, EPA 910-R-07-002, April 2007. Washington, DC: United States Environmental Protection Agency.
Voets, J. P., Vanstaen, H., and Verstraete, W. (1975). Removal of nitrogen from highly nitrogenous waste waters. J. Water. Pollut. Control. Fed. 47, 394–398.
Wan, W., He, D., and Xue, Z. (2017). Removal of nitrogen and phosphorus by heterotrophic nitrification-aerobic denitrification of a denitrifying phosphorus-accumulating bacterium Enterobacter cloacae HW-15. Ecol. Eng. 99, 199–208. doi: 10.1016/j.ecoleng.2016.11.030
Wang, S., Xiao, J., Wan, L., Zhou, Z., Wang, Z., Song, C., et al. (2018). Mutual dependence of nitrogen and phosphorus as key nutrient elements: one facilitates Dolichospermum flos-aquae to overcome the limitations of the other. Environ. Sci. Technol. 52, 5653–5661. doi: 10.1021/acs.est.7b04992
Wang, X., Wang, X., Hui, K., Wei, W., Zhang, W., Miao, A., et al. (2017). Highly effective polyphosphate synthesis, phosphate removal, and concentration using engineered environmental bacteria based on a simple solo medium-copy plasmid strategy. Environ. Sci. Technol. 52, 214–222. doi: 10.1021/acs.est.7b04532
Wang, Y., Peng, Y., and Stephenson, T. (2009). Effect of influent nutrient ratios and hydraulic retention time (hrt) on simultaneous phosphorus and nitrogen removal in a two-sludge sequencing batch reactor process. Bioresour. Technol. 100, 3506–3512. doi: 10.1016/j.biortech.2009.02.026
Wanner, B. L. (1992). Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J. Bacteriol. 174, 2053–2058. doi: 10.1128/jb.174.7.2053-2058.1992
Wanner, B. L., and Chang, B. D. (1987). The phoBR operon in Escherichia coli K-12. J. Bacteriol. 169, 5569–5574. doi: 10.1128/jb.169.12.5569-5574.1987
Yamada, M., Makino, K., Shinagawa, H., and Nakata, A. (1990). Regulation of the phosphate regulon of Escherichia coli: properties of phoR deletion mutants and subcellular localization of phoR protein. Mol. Gen. Genet. 220, 366–372. doi: 10.1007/bf00391740
Yamashita, T., and Yamamoto-Ikemoto, R. (2014). Nitrogen and phosphorus removal from wastewater treatment plant effluent via bacterial sulfate reduction in an anoxic bioreactor packed with wood and iron. Int. J. Environ. Res. Public Health 11, 9835–9853. doi: 10.3390/ijerph110909835
Yang, S., Yang, F., Fu, Z., Wang, T., and Lei, R. (2010). Simultaneous nitrogen and phosphorus removal by a novel sequencing batch moving bed membrane bioreactor for wastewater treatment. J. Hazard. Mater. 175, 551–557. doi: 10.1016/j.jhazmat.2009.10.040
Yang, Y., Zhang, L., Shao, H., Zhang, S., Gu, P., and Peng, Y. (2017). Enhanced nutrients removal from municipal wastewater through biological phosphorus removal followed by partial nitritation/anammox. Front. Environ. Sci. Eng. 11:8. doi: 10.1007/s11783-017-0911-0
Yuan, Q., and Oleszkiewicz, J. A. (2011). Effect of dissolved oxygen on biological nutrient removal by denitrifying phosphorus-accumulating organisms in a continuous-flow system. Water Environ. Res. 83, 2107–2114. doi: 10.2175/106143011X12928814444538
Zeng, W., Li, B., Wang, X., Bai, X., and Peng, W. (2016). Influence of nitrite accumulation on “Candidatus accumulibacter” population structure and enhanced biological phosphorus removal from municipal wastewater. Chemophere 144, 1018–1025. doi: 10.1016/j.chemosphere.2015.08.064
Zeng, W., Zhang, L., Fan, P., Guo, J., and Peng, Y. (2017). Community structures and population dynamics of “Candidatus accumulibacter” in activated sludges of wastewater treatment plants using ppk1 as phylogenetic marker. J. Environ. Sci. 67, 237–248. doi: 10.1016/j.jes.2017.09.001
Zhang, M., Qiao, S., Shao, D., Jin, R., and Zhou, J. (2017). Simultaneous nitrogen and phosphorus removal by combined anammox and denitrifying phosphorus removal process. J. Chem. Technol. Biotechnol. 93, 94–104. doi: 10.1002/jctb.5326
Zheng, M., He, D., Ma, T., Chen, Q., Liu, S., Ahmad, M., et al. (2014). Reducing NO and N2O emission during aerobic denitrification by newly isolated Pseudomonas stutzeri PCN-1. Bioresour. Technol. 162, 80–88. doi: 10.1016/j.biortech.2014.03.125
Keywords: denitrifying phosphorus removal, DPAO, single stage anoxic reactor, Pho regulon, negative regulation, transcriptional repression, poly-phosphate accumulation
Citation: Mukherjee C, Chowdhury R, Begam MM, Ganguli S, Basak R, Chaudhuri B and Ray K (2019) Effect of Varying Nitrate Concentrations on Denitrifying Phosphorus Uptake by DPAOs With a Molecular Insight Into Pho Regulon Gene Expression. Front. Microbiol. 10:2586. doi: 10.3389/fmicb.2019.02586
Received: 11 July 2019; Accepted: 25 October 2019;
Published: 08 November 2019.
Edited by:
Hisashi Satoh, Hokkaido University, JapanReviewed by:
Masashi Hatamoto, Nagaoka University of Technology, JapanCopyright © 2019 Mukherjee, Chowdhury, Begam, Ganguli, Basak, Chaudhuri and Ray. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Krishna Ray, a3JheTkxQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.