
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 November 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02546
This article is part of the Research Topic Antimicrobial Resistance As a Global Public Health Problem: How Can We Address It? View all 50 articles
 Bing Bai1,2†
Bing Bai1,2† Zhiwei Lin1,2†
Zhiwei Lin1,2† Zhangya Pu3†
Zhangya Pu3† Guangjian Xu1,2†
Guangjian Xu1,2† Fan Zhang1,2
Fan Zhang1,2 Zhong Chen1,2
Zhong Chen1,2 Xiang Sun1
Xiang Sun1 Jinxin Zheng1
Jinxin Zheng1 Peiyu Li1,2
Peiyu Li1,2 Qiwen Deng1,2*
Qiwen Deng1,2* Zhijian Yu1,2*
Zhijian Yu1,2*Omadacycline (Omad), a new tetracycline (Tet)-class broad-spectrum aminomethylcycline, has been reported to exhibit excellent potency against Gram-positive bacteria, including Staphylococcus aureus and Enterococci. The aim of this study was to evaluate the in vitro activity and heteroresistance characteristics of Omad in clinical S. aureus isolates from China and investigate Omad resistance mechanisms. A sample of 263 non-duplicate clinical S. aureus isolates [127 methicillin-resistant (MRSA) and 136 methicillin-sensitive (MSSA)] were collected retrospectively. Our data indicated that Omad exhibited excellent in vitro activity against both MRSA and MSSA. Omad heteroresistance frequencies were 3.17% (4/126) in MRSA and 12.78% (17/133) in MSSA. No mutations in Tet target sites, (five 16SrRNA copies and 30S ribosomal protein S10) were present in heteroresistance-derived clones, whereas Tet target site mutations contribute to induced Omad resistance in S. aureus in vitro. RNA sequencing (RNA-Seq) revealed that overexpression of branched-chain amino acid transport system II carrier protein and Na/Pi cotransporter family protein contributes to Omad heteroresistance emergence. Whole-genome sequencing demonstrated that the genetic mutation of fibronectin-binding protein (FnBP) could increase the Omad MIC. In conclusion, Omad heteroresistance risk should be considered in clinical isolates with MICs ≥ 0.5 mg/L and Omad susceptibility in S. aureus may be affected by efflux pump proteins (i.e., a branched-chain amino acid transport system II carrier protein and an Na/Pi cotransporter family protein), and FnBP.
Staphylococcus aureus is a pervasive human pathogen that causes infectious diseases ranging in severity from superficial skin abscesses to bacteremia and septic shock (Calfee, 2017). Although the incidence of methicillin-resistant S. aureus (MRSA) infection appears to be declining worldwide, the incidence of bacteremia and severe community-acquired infection caused by methicillin-susceptible S. aureus (MSSA) continues to be an important human health threat (Bal et al., 2017). Both MRSA and MSSA infections remain a major clinical problem exacerbated by the ongoing evolution and transmission of traits engendering resistance or reduced susceptibility to current last-line antimicrobial agents, including linezolid, daptomycin, tigecycline (Tig), and vancomycin. Thus, there remains an urgent need for the development of new antimicrobial agents (Bal et al., 2017; Calfee, 2017).
Omadacycline [7-dimethylamino, 9-(2,2-dimethyl-propyl)-aminomethylcycline; Omad] is a recently developed semisynthetic aminomethylcycline belonging to the tetracycline (Tet) family (Pfaller et al., 2017a). It has extraordinarily broad-spectrum antimicrobial activity against Gram-positive and -negative bacteria, including difficult-to-treat multidrug resistant bacteria, such as MRSA and vancomycin-resistant enterococci (Pfaller et al., 2017a). Like other Tet drugs, Omad is a potent inhibitor of the bacterial ribosome that inhibits bacterial protein synthesis by binding 30S ribosomal subunits during translation (Draper et al., 2014; Honeyman et al., 2015; Heidrich et al., 2016; Villano et al., 2016). Omad has the advantage of being minimally affected by classical Tet resistance mechanisms, including efflux pumps and ribosomal protection mechanisms. Omad also exhibits lower minimum inhibitory concentration (MIC) values against multidrug resistant bacteria than minocycline and Tig (Draper et al., 2014; Honeyman et al., 2015; Heidrich et al., 2016; Villano et al., 2016), making it a novel potential last-resort antibiotic for difficult-to-treat bacteria infections (Noel et al., 2012; Macone et al., 2014; Pfaller et al., 2017b; Zhang et al., 2018).
Heteroresistance means that there are population-wide variable responses to antibiotics. Several reports, including the earliest studies describing the phenomenon, applied this definition without specifying a particular antibiotic concentration range (El-Halfawy and Valvano, 2013, 2015). Previously, we obtained MICs and heteroresistance occurrence data for the new generation Tet-class drug erevacycline in clinical S. aureus isolates from China, underscoring the importance and necessity of investigating the characteristics of new-generation Tet derivatives (Zhang et al., 2018; Zheng et al., 2018). There are limited data regarding Omad activity against clinical S. aureus isolates from China. Heteroresistance development in last-resort antibiotics can hinder efficacy and, ultimately, lead to treatment failure (Claeys et al., 2016; Zhang et al., 2018; Zheng et al., 2018). Thus, it is important to establish potential factors associated with Omad heteroresistance development.
Reduced susceptibility to Tig, an archetype new-generation Tet-class drug, in several species of bacteria has been associated with genetic mutations affecting 30S ribosomal subunits, including altered copy numbers of genes encoding 16S rRNA and 30S ribosomal proteins S3 and S10 (Nguyen et al., 2014; Lupien et al., 2015; Grossman, 2016; Argudin et al., 2018). Tig resistance has been related to regulators of cell envelop proteins, including efflux pumps (e.g., SoxS, MarA, RamA, and Rob) in Gram-negative enterobacteria and MepR/MepA in S. aureus (Nguyen et al., 2014; Grossman, 2016; Linkevicius et al., 2016; Dabul et al., 2018). The possible influences of 30S ribosomal subunit mutations and the overexpression of efflux proteins on Omad heteroresistance and resistance in S. aureus has not been resolved and needs to be further studied.
The main purpose of this study was to investigate the in vitro antimicrobial activity of Omad and to use population analysis profile (PAP) analysis to evaluate the occurrence of Omad heteroresistance in S. aureus isolates from China. We examined Omad heteroresistance mechanisms in S. aureus by conducting polymerase chain reaction (PCR) experiments to detect genetic mutations in 30S ribosome units, administering efflux protein inhibitors (Zhang et al., 2018; Zheng et al., 2018), and conducting RNA sequencing (RNA-Seq) studies. Furthermore, we used in vitro induction of resistance under Omad pressure and next generation sequencing (NGS) to compare Omad-sensitive and -resistant isolates and uncover molecular factors involved in Omad resistance.
A total of 263 non-duplicate clinical S. aureus isolates (127 MRSA and 136 MSSA) were collected from Shenzhen Nanshan People’s Hospital, a tertiary hospital with 1,200 beds in China, between 2008 and 2016. The specimen sources are summarized in Supplementary Figure S1. S. aureus ATCC29213 was used as a quality control organism. All procedures involving human participants were performed in accordance with the ethical standards of Shenzhen University and the 1964 Helsinki declaration and its later amendments, or comparable ethical standards. For this type of study, formal consent is not required.
Staphylococcus aureus antimicrobial susceptibilities to a panel of antibiotics (i.e., amikacin, erythromycin, ciprofloxacin, rifampicin, Tet, tobramycin, vancomycin, linezolid, nitrofurantoin, amoxicillin/clavulanate, and quinupristin) with the VITEK 2 system (BioMérieux, Marcy l’Etoile, France) and susceptibility breakpoints based on CLSI guidelines (2016). Omad was obtained from The Medicines Company (Med Chem Express, Monmouth Junction, NJ, United States). Omad MICs were determined with the agar dilution method according to CLSI guidelines (Klionsky et al., 2016). We employed three Omad MIC levels (≤0.25 mg/L, 0.5 mg/L, and ≥1 mg/L) in our antimicrobial susceptibility analysis. The following Acute Bacterial Skin and Skin Structure Infections Omad susceptibility breakpoints recommended by FDA criteria were adopted: ≤0.5 mg/L for susceptibility, 1 mg/L for intermediate status, and ≥2 mg/L for resistance.
Omad heteroresistance in S. aureus was determined by PAP development as described previously (Zhang et al., 2018; Zheng et al., 2018) with a MIC cut-off criterion of ≤0.5 mg/L. Briefly, 50-μL aliquots (108 bacterial colony forming units) were spread onto Müller-Hinton broth plates containing serial dilutions of Omad (in mg/L): 0.5, 1, 2, and 3. Colonies were counted on Omad-containing plates after 24 h of incubation at 37°C. According to the criteria described above, we defined 2 mg/L as the susceptibility breakpoint for PAP determination of S aureus. For Omad-resistant subpopulations detected among Omad-susceptible S. aureus isolate colonies grown on agar plates with 2 mg/L Omad with a detection limit of ≥5 colony forming units/ml, the parental isolates were considered to have Omad heteroresistance. Two heteroresistance-derived colonies were selected randomly from plates and their Omad and Tig MICs were measured by agar dilution according to CLSI guidelines and then subjected to PCRs, efflux inhibition, and transcriptional analysis (Zhang et al., 2018).
Genomic DNA was extracted from isolates with Lysis Buffer for Microorganisms to Direct PCR (Takara Bio Inc., Japan). Tet resistance genes encoding Tet(K), Tet(L), Tet(M), and Tet(O) were detected by PCR analysis as described previously (Bai et al., 2018). The presence of 30S ribosomal subunit mutations, including five separate copies of the 16S rRNA gene, the genes encoding the 30S ribosomal proteins S3 and S10, and the genes encoding recombinase (RecB) and fibronectin-binding protein (FnBP) were analyzed by PCR and sequence alignment (primer sequences listed in Supplementary Table S1). Multi-locus sequence typing (MLST) was conducted to identify the distributions of sequence types (STs) among MRSA and MSSA isolates. PCR conditions recommended for locus amplification1 were employed.
The role of efflux pumps in Omad heteroresistance was evaluated with the efflux pump inhibitors phenylalanine-arginine-β-naphthylamide (PaβN) and carbonyl cyanidem-chlorophenylhydrazine (CCCP; both from Sigma). Omad MICs were determined by the agar dilution method in the presence and absence of 50 mg/L PAβN or 16 mg/L CCCP. Inhibition was confirmed based on a ≥4-fold MIC reduction (Osei Sekyere and Amoako, 2017; Zhang et al., 2018).
Seven parental S. aureus isolates, including six clinical isolates (MSSAs: CHS221, CHS165, and 149. MRSAs: CHS759, CHS810, and CHS820) and a well-characterized antibiotic-susceptible MS4 strain, were used to select Omad-resistant isolates. These isolates were subcultured serially in Mueller-Hinton broth containing gradual increasing Omad concentrations with the initial concentration being MIC equivalents followed by successive increases to 2×, 4×, 8×, and 16× MICs (Yao et al., 2018), with four passages at each concentration. Isolates from the passages of each concentration were stored at −80°C in Mueller-Hinton broth containing 40% glycerol for subsequent Tet-target site genetic mutation detection, subsequent MIC assays, next generation sequencing, and PCR analysis. Killing curves were performed on the CHS221 (wild-type MSSA strain), CHS221-O (Omad heteroresistance MSSA strain), CHS221-1Δ: (Omad resistance MSSA strain), CHS759 (wild-type MRSA strain), CHS759-O (Omad heteroresistance MRSA strain), and CHS759-1Δ: (Omad resistance MRSA strain). Tubes containing Omad at concentrations corresponding to 0 and 4 mg/L were inoculated with a suspension of each test strain, yielding to a final bacterial density of 8 × 106 cfu/ml. The killing curves shown Omad-resistant strain could grow well at concentrations corresponding to 4 mg/L (Supplementary Figure S2).
Wild-type strain CHS221 (S221) and its heteroresistance-derived S. aureus isolate CHS221-O (S221-O1) were grown and prepared for total RNA extraction with TRIzol reagent (Invitrogen, Carlsbad, CA, United States) as described previously (Zheng et al., 2018). RNA-Seq of the aforementioned parental and heteroresistance-derived isolates was performed as reported previously (Lin et al., 2017). Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. All the downstream analyses were based on the clean data with high quality. The raw data from the samples were analyzed in Subread software; raw counts for each group were normalized and processed in the EdgeR Bioconductor software package. 1.3-fold differences in expression level by RNA-Sequencing were considered to be differentially expressed genes (DEGs). The RNA sequencing outcomes for two strains were deposited in the NCBI database (BioProject accession number PRJNA505108).
We selected 30 DEGs based on our RNA-Seq results and employed qRT-PCR to compare transcriptional expression levels between the parental and heteroresistance-derived strains as described in detail previously (Zheng et al., 2018). The transcriptional expression levels of the eight candidates genes (USA300HOU_RS00705, USA300 HOU_RS03535, USA300 HOU_RS01625, USA300 HOU_RS00550, USA300HOU_ RS13205, USA300HOU_RS13945, USA300HOU_RS10505, and USA300HOU_RS00660) were further analyzed and compared among the CHS165 (MSSA), 149 (MSSA), CHS759 (MRSA), CHS810 (MRSA), and CHS820 (MRSA) parental strains and their derivative heteroresistant and resistant strains. Total RNA of bacteria was extracted using the RNeasyH Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. The extracted RNA was reverse transcribed into cDNA using iScript reverse transcriptase (Bio-Rad, Hercules, CA, United States) with incubation for 5 min at 25°C, followed by 30 min at 42°C and 5 min at 85°C. Subsequently, qRT-PCRs were performed using SYBR green PCR reagents (Premix EX TaqTM, Takara Biotechnology, Dalian, China) in the Mastercycler realplex system (Eppendorf AG, Hamburg, Germany) with amplification conditions of 95°C for 30 s, 40 cycles of 95°C for 5 s and 60°C for 34 s, followed by melting curve analysis. The control gene gyrB was used to normalize gene expression. Threshold cycle (Ct) numbers were determined by detection system software and analyzed with the 2–44Ct method and three replicates have been made. The qRT-PCR primers used are listed in Supplementary Table S2.
An Omad resistant S. aureus strain, MS4O8, was derived from an Omad-susceptible isolate, MS4. Chromosomal DNA was extracted from MS4O8 cells for NGS. Nextera shotgun libraries and whole genome sequencing were performed by Novogene Company (Beijing, China). Illumina PE150 sequencing data were mapped against the CP009828 S. aureus MS4 strain reference genome in bwa mem software (v0.7.5a)2 with standard parameters. Small nucleotide polymorphisms and small insertions/deletions were detected in MS408 cells, relative to MS4, in MUMmer (version 3.23). A custom script was used to detect substitutions, insertions, and deletions that might be impacting protein coding regions. Binary alignment/map files of the sequenced strains were deposited in the NCBI database (BioProject accession number PRJNA511962).
Full-length candidate genes, including USA300HOU_RS00550 (encodes a Na/Pi cotransporter family protein), USA300HOU_ RS01625 (encodes a branched-chain amino acid transport system II carrier protein), USA300HOU_RS03535, USA300HOU_ Tet(K), NI36_12460 (FnBP), and NI36_00170 (RecB), were amplified with extra double enzyme sites from total DNA extracted from USA300HOU and MS4 isolates. RecB-M is RecB with a mutation (R10R, I23V, I23N, H24N, H24Q, V29L, and V35M) and FnBP-M is FnBP with a mutation (T672S and I665V). RecB-M and FnBP-M DNA fragments were amplified from MS4O8 by PCR. The candidate gene fragments were each integrated into separate pIB166 vectors, and their encoded target protein were induced with 2 mM chromium chloride (Wu et al., 2012). The primers used for vector constructs in this study are listed in Supplementary Table S3. Positively vector transformed S. aureus clones were selected with chloramphenicol and verified by PCR and Sanger sequencing. The overexpression plasmids were transformed separately into three to five Omad-sensitive isolates and their integrations was confirmed by PCR and Sanger sequencing. Candidate gene transcriptional levels were measured by qRT-PCR, as described above. Omad and Tig MICs for these derivatives were determined and heteroresistance occurrence in these derivatives was tested by PAP analysis under Omad pressure as described above.
Continuous data were analyzed with Student’s t-tests and one-way factorial analyses of variance (ANOVAs) in SPSS software package (version 17.0, Chicago, IL, United States). P-values < 0.05 were regarded as statistically significant.
Of the 127 MRSA isolates examined, 46 (36.22%), 80 (62.99%), and 1 (0.78%) were found to have Omad MIC levels of ≤0.25 mg/L (sensitive), 0.5 mg/L (sensitive), and 1 mg/L (intermediate), respectively. Of the 136 MSSA isolates examined, 23 (16.91%), 110 (80.88%), and 3 (2.20%) were categorized into these levels, respectively. Thus, there were higher frequencies of MSSA isolates than MRSA isolates with Omad MICs in the 0.5 and ≥1 mg/L levels. We analyzed the distribution of the above three Omad MIC levels among strains with sensitive and intermediate status relative to other common antibiotics (amikacin, erythromycin, ciprofloxacin, rifampicin, Tet, tobramycin, nitrofurantoin, quinupristin, and amoxicillin/clavulanate, vancomycin, and linezolid). The frequencies of isolates at each Omad MIC level found to be resistant to these antibiotics are reported in Table 1, together with the resistance breakpoints used. All S. aureus isolates in this study were susceptible to vancomycin and linezolid, and all of the MSSA isolates were susceptible to amoxicillin/clavulanate. Interestingly, as reported in Table 1, Tet-resistant MRSA were more frequent than Tet-resistant MSSA, and Omad MICs ≥ 0.5 mg/L were more frequent among MSSA isolates than among MRSA isolates, suggesting a non-conformity in the antimicrobial susceptibility dynamics of Tet and Omad. The characteristics of S. aureus with Omad MICs of 1 mg/L are summarized in Supplementary Table S4. Briefly, no genetic mutations in 30S ribosome units were detected and efflux pump inhibition reversed Omad resistance, as evidenced by significant reductions in MICs to ≤0.03 mg/L with CCCP and to 0.25–1 mg/L with PAβN.
The frequencies of genes encoding the Tet-resistance factors Tet(M), Tet(L), Tet(K), and Tet(O), alone and in combination, in MRSA and MSSA isolates are reported in Supplementary Table S5. There were 109 S. aureus isolates harboring at least one Tet-resistance factor gene; their MIC90 values were consistently 0.5 mg/L. Omad exhibited excellent in vitro activity against both Tet-resistance gene carrying and non-carrying S. aureus isolates. Omad MICs for both MRSA and MSSA harboring Tet-resistance factors were ≤0.5 mg/L for all isolates, with the exception of three Tet(K) gene-carrying isolates (1 MRSA and 2 MSSA), indicating that overexpression of Tet(K) might impact Omad susceptibility. It is noteworthy that the Omad MIC values obtained for all 46 MSSA isolates carrying the Tet(K) gene, Tet(L) gene, or both were ≥0.5 mg/L. Meanwhile, of the 63 MRSA isolates with the Tet(M) gene, Tet(K) gene, Tet(L) gene, or some combination of these genes, just 47 (74.60%) had Omad MIC values ≥0.5 mg/L.
MLST results for the 263 isolates are summarized in Supplementary Figure S3. To evaluate the relationship of ST clonality with Omad MIC distribution, we examined ST distributions relative to Tet and Omad MICs (Supplementary Table S6). Omad MICs ≥ 0.5 mg/L were found for 72.58% (45/62) of ST239-MRSA, 57.5% (23/40) of ST59-MRSA, and 57.14% (4/7) of ST1-MRSA isolates. Meanwhile, Omad MICs ≥ 0.5 mg/L were found for 89.66% (26/29) of ST7-MSSA, 89.47% (17/19) ST59-MSSA, 76.92% (10/13) of ST398-MSSA, 85.71% (6/7) of ST88-MSSA, and 71.43% (5/7) of ST120-MSSA isolates. Hence, Omad sensitivity differed between MRSA and MSSA of the same ST (e.g., ST59-MRSA vs. ST59-MSSA). MLST indicated that 72/81 (88.89%) MRSA isolates with Omad MICs ≥ 0.5 mg/L belonged to the top three MRSA STs (ST239, ST59, and ST1), whereas only 52/113 (46.12%) of MSSA isolates with Omad MICs ≥ 0.5 mg/L belonged to the top three MSSA STs (Supplementary Table S6 and Table 1), revealing a more pronounced clustering of Omad MIC creep in the top three MRSA STs than in the top three MSSA STs (nearly 90% vs. less than half).
Omad heteroresistance was identified in 0.0% (0/46) of MRSA with an Omad MIC ≤ 0.25 mg/L and 3.75% (3/80) of MRSA with an Omad MIC of 0.5 mg/L. Omad heteroresistance was identified in 0.0% (0/30) of MSSA with an Omad MIC ≤ 0.25 mg/L and 17.48% (18/103) of MSSA with an Omad MIC of 0.5 mg/L. We determined the Omad and Tig MICs of two clones from each heteroresistant subpopulation and found that their Omad MICs were in the range of 1–8 mg/L and their Tig MICs were in the range of 2–8 mg/L (shown in Supplementary Table S7 and data for six isolates subjected to in vitro resistance induction are shown in Table 2). Moreover, no genetic mutations were found in 30S ribosomal subunit genes (five 16SrRNA gene copies, 30S ribosomal protein S3 and S10 genes) in heteroresistance-derived clones (Table 2 and Supplementary Table S7). In efflux pump inhibition experiments, Omad MICs in heteroresistance-derived S. aureus clones were reduced to ≤0.03 mg/L by CCCP and reduced to 0.25–1 mg/L by PAβN (Supplementary Table S7).
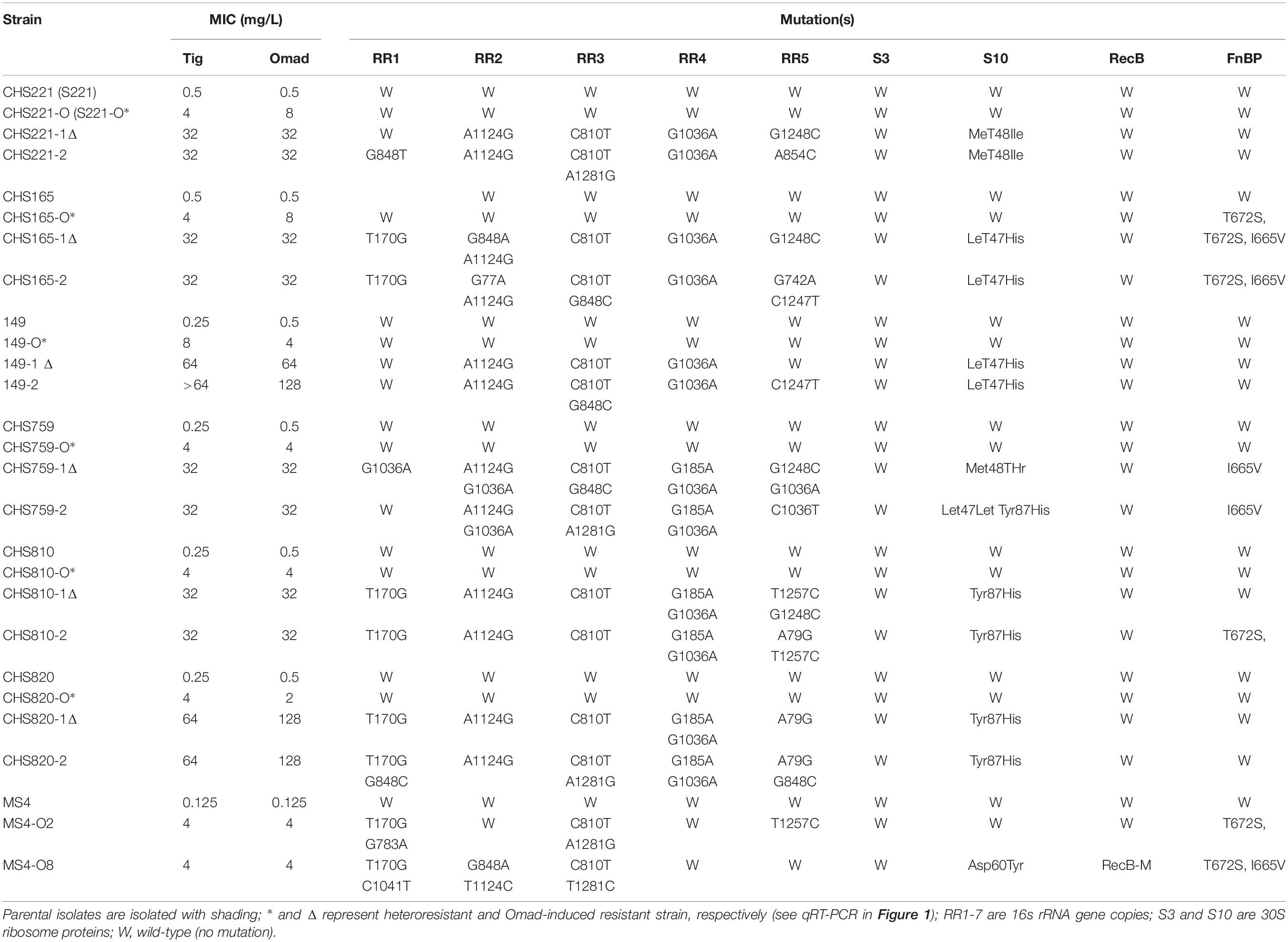
Table 2. Antimicrobial susceptibility and resistance mechanism of seven groups of parental, heteroresistant, and Omad-induced resistant strains.
Efflux pump inhibition experiments indicated that efflux pumps or membrane proteins might participate in the development of heteroresistance. Therefore, RNA-Seq was performed and unigene transcription levels were compared between the parental strain CHS221 (S221) and its heteroresistant derivative CHS221-O1 (S221-O). Ninety six DEGs were found by this approach between S221 and S221-O, including 58 upregulated and 38 down-regulated genes in the derivative strain (Supplementary Figure S4). KEGG pathway analysis showed that the pathways most frequently linked to DEGs were related to phosphate ion transport (3 DEGs), inorganic anion transport (3 DEGs), dihydrofolate reductase activity (2 DEGs), and glycine biosynthesis process (2 DEGs).
Subsequent qRT-PCRs for 30 candidate genes were carried out to test the accuracy of our transcriptomic analyses implicated eight efflux-pump encoding DEGs in heteroresistance. The expression levels of these eight candidate genes determined by RNA-Seq and qRT-PCR are shown in Table 3. The results of qRT-PCRs performed to quantitate transcription in six S. aureus strain groups—inclusive of parental strains, their heteroresistance derivatives, and resistant isolates (Table 2)—enabled us to probe the relationship of their expression levels with Omad susceptibility (Figure 1). The data suggest that transcription levels of the three candidate genes USA300HOU_RS03535, USA300HOU_ RS01625 (encodes a Na/Pi cotransporter family protein), and USA300HOU_RS00550 (encodes a branched-chain amino acid transport system II carrier protein) may impact heteroresistance occurrence.
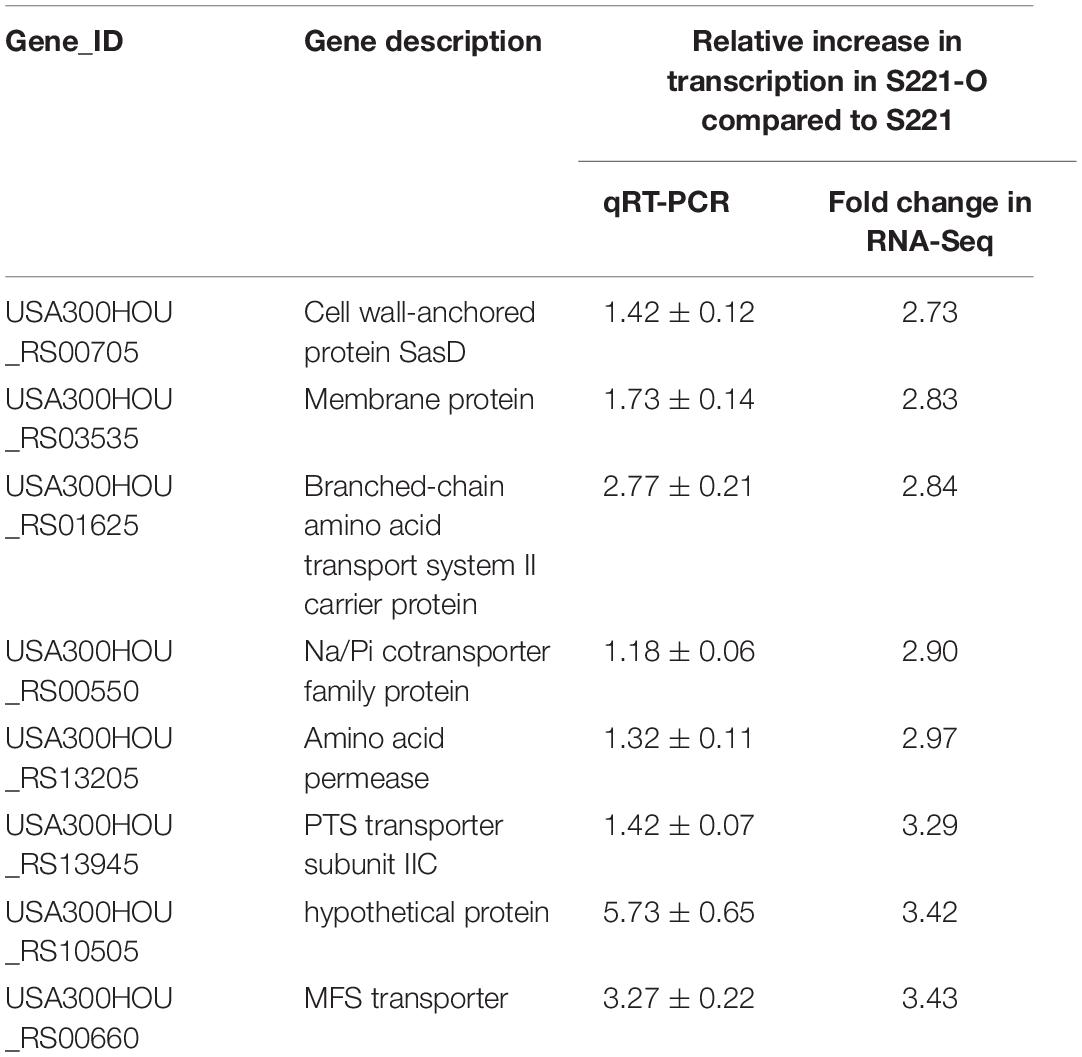
Table 3. Transcriptional expression levels of eight DEGs between S221-O and S221 analyzed by RNA-Seq and qRT-PCR.
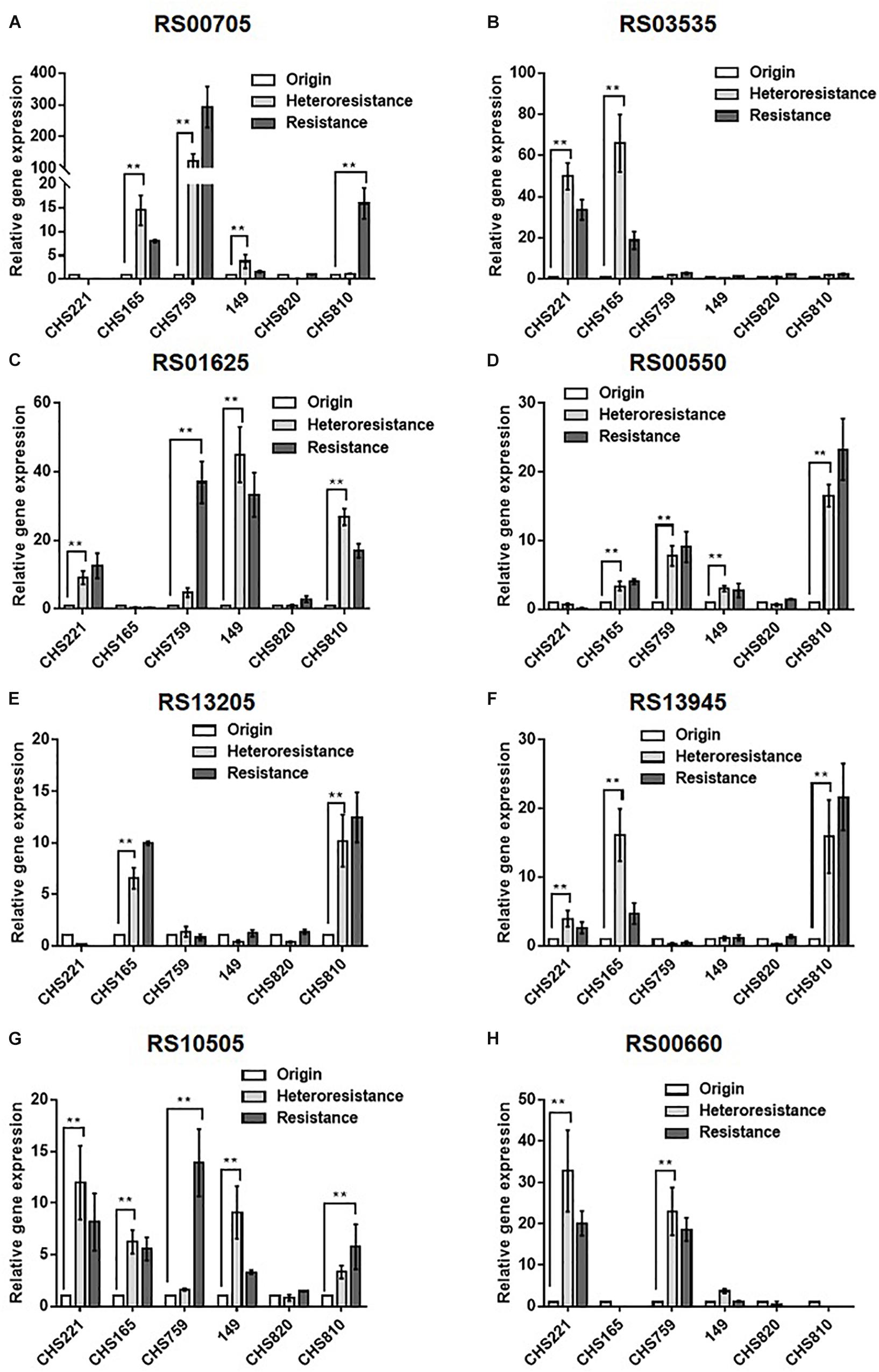
Figure 1. Comparison of the relative transcription of eight candidate DEGs among parental, heteroresistant derivative, and resistant isolates. Relative expression of USA300HOU_RS00705 (A), USA300HOU_RS03535 (B), USA300HOU_RS01625 (C), USA300HOU_RS00550 (D), USA300HOU_RS13205 (E), USA300HOU_RS13945 (F), USA300HOU_RS10505 (G), and USA300HOU_RS00660 (H) were demonstrated by qRT-PCR analysis. The housekeeping gene gyrB was used as the endogenous reference gene. The original strain was used as the reference strain (expression = 1.0). All qRT-PCRs were carried out in triplicate. ∗∗p < 0.01, ∗p < 0.0.5. Parental strains are identified below the X axis and the relative folds increased are shown on the Y axis. The parental, heteroresistant, and resistant isolates are described in Table 2.
To evaluate Omad resistance mechanisms and Omad-Tig cross-resistance, in vitro induction experiments were carried out under Omad pressure with the following Omad-resistant S. aureus: CHS221 (MSSA), CHS165 (MSSA), 149 (MSSA), CHS759 (MRSA), CHS810 (MRSA), CHS820 (MRSA), and MS4. The Omad-resistant isolates were characterized with respect to MICs and resistance mechanisms (Table 2). Importantly, increasing Omad MICs were accompanied by increasing Tig MICs in Omad-resistant S. aureus isolates, indicating that Omad-Tig cross-resistance can be induced under Omad pressure. Moreover, upregulation of Omad MICs was related to increasing numbers of 16SrRNA copies with a genetic mutation. The mutation sites varied among the five 16SrRNA copies, with high frequencies of the T170G polymorphism in RR1, A1124G in RR2, C810T in RR3, and G1036A in RR4. Leu47His and Tyr87His amino acid substitutions in the 30S ribosomal protein S10 were relatively frequent in Omad-resistant bacteria.
To identify the genetic mutations that correlate with Omad resistance, whole genome sequencing of MS4O8 was performed and variants relative to MS4 were detected by NGS in MUMmer, version 3.23 (Supplementary Table S8 and Table 4). Non-synonymous mutations were found in NI36_11090 (encodes 30S ribosomal protein S10), NI36_12460 (fnbp encoding FnBP protein), and NI36_ 00170 (recB encoding recombinase). Mutations affecting these three genes also emerged in our induced Omad-resistance experiment above (Table 2). Notably, 30S ribosomal protein S10 has been widely reported to be associated with Tet-class resistance and the impact of FnBP and RecB protein on Omad susceptibility need to be further verified.

Table 4. Non-synonymous mutations of candidate proteins correlated with Omad resistance found between MS4 and MS4O8.
The impacts of following candidate genes on Omad susceptibility in Omad-sensitive S. aureus isolates was conducted: USA300HOU_RS03535, USA300HOU_RS01625 (encodes a branched-chain amino acid transport system II carrier protein), USA300HOU_RS00550 (encodes a Na/Pi cotransporter family protein), tet(K), NI36_12460/NI36_12465 (fnbp), and NI36_00170 (recB). The former three were candidately implicated in Omad heteroresistance in our qRT-PCR experiments. Meanwhile, tet(K) was found frequently among S. aureus isolates with an Omad MIC of 1 mg/L, and the proteins encoded by fnbp and recB have been hypothesized to participate in antimicrobial resistance evolution.
The overexpression plasmids pRS00550, pRS01625, pRS03535, pTet(K), pRecB, pRecB-M, pFnBP, and pFnBP-M, where -M suffix indicates a mutated variant, were transformed into clinical isolates with low expression of the target gene (Supplementary Tables S9, S10). Stable overexpression of the candidate genes was confirmed by qRT-PCR (Supplementary Figure S5). The influence of the overexpression of these six genes on Omad susceptibility was reported in Table 5. Briefly, although RS00550, RS01625, and RS03535 did not elevate Omad or Tig MICs in the absence of antibiotic pressure, PAP experiments showed that RS00550 or RS01625 overexpression could lead to Omad heteroresistance compared with negative findings in controls (Table 5). RS00550 and RS01625 homology analysis results are reported in Supplementary Tables S11, S12.
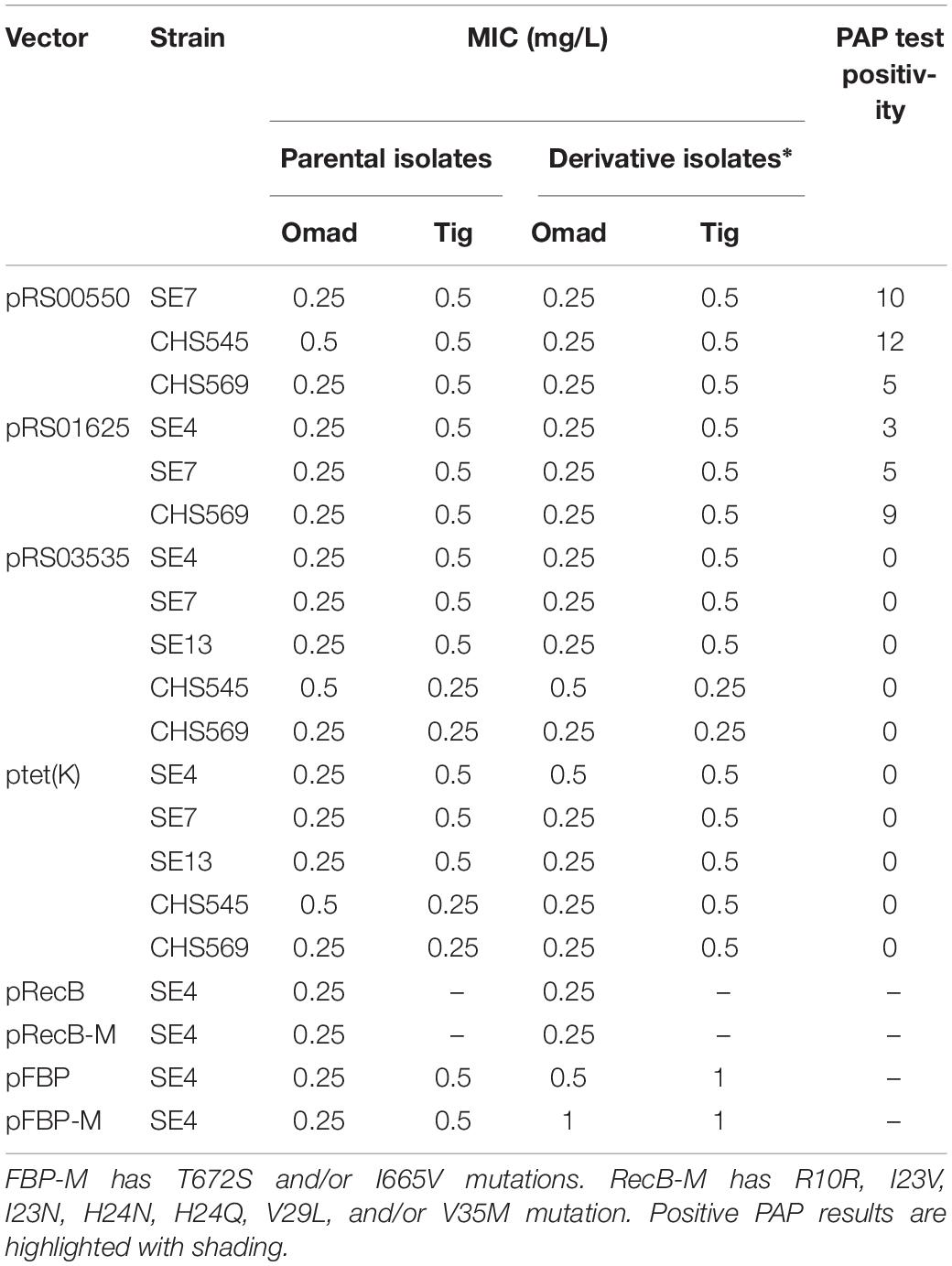
Table 5. Omad and Tig MICs in S. aureus derivatives with overexpression of candidate genes and their impact on PAPs.
Overexpression of tet(K) did not elevate Omad or Tig MICs and had no apparent contribution to heteroresistance development in S. aureus. The overexpression of fnbp (NI36_12 460) and its mutated type led to MIC creep (Table 5), supporting the possibility that FnBPs may participate in Omad resistance development. Homology analysis results for fnbp are shown in Supplementary Table S13. Overexpression of recB and its mutants did not impact Omad susceptibility in S. aureus.
The presently observed low Omad MICs in this study support the supposition that Omad should be considered a prospective preferential choice for S. aureus infection treatment. Omad MICs ≥ 0.5 mg/L were more frequent among MSSA than MRSA in this study, and moreover, Tet-specific resistance genes, particularly tet(K) and tet(L), were found to be more common among MSSA than MRSA isolates, indicating that Omad may have greater efficacy against MRSA than MSSA. Our Omad MICs were higher than previously reported, perhaps due to regional variation and environmental factors (Villano et al., 2016; Pfaller et al., 2017a). We also obtained higher MICs for the new-generation Tet-class drug eravacycline in isolates from China than had been reported for isolates from the United States and Europe, suggesting that Tet-class drug MIC dynamics should monitored across global regions with particular attention to MIC creep in China (Zhang et al., 2018; Zheng et al., 2018).
Major mechanisms of Tet resistance in both Gram-positive and -negative microorganisms have been linked to ribosomal protection proteins and efflux pumps, most of which can be overcome with new generation tetracyclines, including Tig and Omad (Jones et al., 2006; Noel et al., 2012; Draper et al., 2014; Macone et al., 2014; Honeyman et al., 2015; Grossman, 2016; Heidrich et al., 2016; Villano et al., 2016; Pfaller et al., 2017b; Argudin et al., 2018; Zhang et al., 2018). In this study, we obtained low Omad MICs for S. aureus, even among isolates harboring a ribosomal protection protein, namely Tet(M), or an efflux pump factor, namely Tet(K) and Tet(L), uncovering an apparent advantage of using Omad to overcome Tet-specific resistance mechanisms, particularly those mediated by Tet(M), Tet(K), and Tet(L). Notwithstanding, Tig MICs were shown recently to be increased by a high transcriptional level of tet(M) and tet(L) in Enterococcus faecium (Fiedler et al., 2016). Because we observed a higher frequency of Tet-specific genes in MSSA than in MRSA from China and three isolates with MICs ≥ 1 mg/L harbored tet(K) in this study, we hypothesize that tet(K) overexpression may elevate Omad MICs, as was found in E. coli (Linkevicius et al., 2016). Our data demonstrate that tet(K) overexpression does not impact S. aureus susceptibility to Omad in vitro, and thus indicate that Omad can overcome the Tet(K)-mediated resistance in S. aureus.
Prior epidemiological data have revealed ST239 and ST59 to be predominant MRSA STs internationally, with MSSA ST predominance being more variable across regions (Atshan et al., 2013; Leon-Sampedro et al., 2016; Recker et al., 2017). S. aureus clonality of drug-resistance and virulence factors has been reported (Atshan et al., 2013; Leon-Sampedro et al., 2016; Recker et al., 2017). The present examination of the relationship of ST clonality with Omad susceptibility revealed a far higher frequency of the top-three MRSA STs with MICs ≥ 0.5 mg/L than of the top-three such MSSA STs. Although a definite relationship of ST clustering with Omad MICs has not been established, it is noteworthy that ST59-MSSA were much more likely to have Omad MICs ≥ 0.5 mg/L than were ST59-MRSA.
Heteroresistance frequency is an important harbinger of last-resort antibiotic resistance risk (Zhang et al., 2018; Zheng et al., 2018). The present findings of heteroresistance in 16.98% of MSSA and 3.75% of MRSA with Omad MICs ≥ 0.5 mg/L suggest a need to be alert to selection resistance under Omad pressure for S. aureus, especially in strains from China. Moreover, we observed relatively high Tig MICs for Omad heteroresistance-derived clones (2–8 mg/L), indicating a potential risk of Omad-Tig cross-resistance. Mutations affecting 30S ribosomal subunits, which have been reported to participate in Tet or Tig resistance, were not found in our heteroresistance-derived clones or clinical isolates with Omad MICs ≥ 1 mg/L, indicating that 30S ribosomal subunit mutations cannot explain Omad MIC creep and heteroresistance occurrence (Nguyen et al., 2014; Grossman, 2016; Argudin et al., 2018).
The progression of reduced Tig susceptibility in S. aureus has been linked with mepR/mepA encoded efflux pumps (Lupien et al., 2015). However, the possible role of efflux pumps and cell envelopes in Omad heteroresistance in S. aureus is unclear. Multiple reports have shown that protonophore efflux pump inhibitors (e.g., CCCP and PaβN) can be used to evaluate interactions between antibiotics and cell envelope components in bacteria (Klionsky et al., 2016; Bai et al., 2018; Yao et al., 2018). Here, we found that the efflux pump inhibitor PAβN and the cell envelope component inhibitor CCCP reduced Omad MICs of heteroresistance-derived S. aureus clones to as low as ≤0.03 mg/L and 0.12–1 mg/L, respectively, pointing to involvement of efflux pumps or cell envelopes in the progression of Omad heteroresistance in S. aureus (Klionsky et al., 2016; Bai et al., 2018). Omad MICs of isolates with Omad MICs ≥ 1 mg/L could also be reduced by CCCP and PAβN, supporting the notion that efflux pumps or cell envelopes may play an important role in reducing susceptibility to Omad. Our findings showing that overexpression of RS00550 or RS01625 can lead to Omad heteroresistance occurrence, despite having no impact on Omad MICs in the absence of Omad pressure, indicate that expression of genes can facilitate the formation of Omad resistance under antibiotic exposure. Our phylogenic analysis showed that both RS00550 and RS01625 encode efflux pump family proteins, supporting our hypothesis that efflux pump or membrane proteins contribute to Omad heteroresistance.
Crystallographic studies of the Thermus thermophilus 30S ribosomal subunit revealed at least one high-occupancy Tet-binding site and five other minor binding sites in 16S rRNA (Draper et al., 2014; Honeyman et al., 2015; Heidrich et al., 2016). Crystallographic studies with Tet, Tig, and Omad showed that, although they produced slightly different patterns of RNA cleavage and dimethylsulfate modification, all three antibiotics associated with the same binding site, albeit in somewhat different orientations. In several bacterial species, Tig and Omad have been shown to exhibit higher binding affinities and greater antitranslational potencies than Tet or minocycline, and 16SrRNA mutations have been shown to affect Tet binding sites in the 30S ribosomal subunit, which may confer Tet/Tig resistance (Nguyen et al., 2014; Grossman, 2016; Zheng et al., 2018). Consistent with previous reports, we found that greater numbers of 16S rRNA copies with genetic mutations were associated with higher levels Omad/Tig resistance in S. aureus isolates with Omad-resistance induced under Omad pressure (Nguyen et al., 2014; Grossman, 2016). This finding implicates the participation of 16SrRNA mutations in the development of the Omad resistance. Additionally, our finding of frequent 30S ribosomal protein S10 mutations in Omad-derived resistant isolates indicates that such mutations may be an important factor in Omad resistance evolution. It will be important to examine the unknown mechanism(s) underlying MIC elevation during Omad resistance evolution in S. aureus. In this study, NGS was performed to identify candidate genes that may be involved in Omad resistance development and FnBP was identified as a novel membrane molecule that may contribute to Omad MIC elevation. Mechanistically, we hypothesized that the overexpression of FnBP could alter the penetration potential of cell membranes.
Omad exhibited excellent in vitro activity against clinical S. aureus isolates from China and might represent a preferential choice for the treatment of S. aureus infections. However, we must be alert to the potential risk of Omad heteroresistance in S. aureus, especially in strains with MICs ≥ 0.5 mg/L. Compared with MRSA, MSSA had relatively low MICs with a more facile tendency for the occurrence of Omad heteroresistance. Omad heteroresistance in both MSSA and MRSA could be reversed by CCCP and PaβN, indicating involvement of efflux pumps in Omad heteroresistance development in S. aureus. Furthermore, both RS01625 and RS00550, which encode efflux pump family proteins (a branched-chain amino acid transport system II carrier protein and an Na/Pi cotransporter family protein, respectively), were found to contribute to Omad heteroresistance. FnBP emerged as a novel molecule to be associated with Omad resistance and Omad MIC elevation in S. aureus. The present data contribute to understanding potential resistance mechanisms that may impact clinical applications of Omad.
All datasets generated for this study are included in the article/Supplementary Material.
QD and ZY initiated and designed the project. BB, ZP, and ZL performed the molecular biological experiments with bacteria. GX, XS, JZ, and ZC performed the PCRs. FZ, PL, and GX performed the bacterial culturing and MIC testing. All authors participated in the data analysis. QD, ZY, and BB wrote the manuscript incorporating comments from all authors.
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81170370 and 81601797); Science, Technology, and Innovation Commission of Shenzhen Municipality of basic research funds (No. JCYJ20170412143551332); Shenzhen Health and Family Planning Commission (SZXJ2018027 and SZXJ2017032); Sanming Project of Medicine in Shenzhen; the Shenzhen Nanshan District Scientific Research Program of the People’s Republic of China (No. 2018010, 2018011, and 2018021); and provincial medical funds of Guangdong (No. A2018163).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02546/full#supplementary-material
Argudin, M. A., Roisin, S., Dodemont, M., Nonhoff, C., Deplano, A., and Denis, O. (2018). Mutations at the ribosomal S10 gene in clinical strains of Staphylococcus aureus with reduced susceptibility to tigecycline. Antimicrob. Agents Chemother. 62:e01852-17. doi: 10.1128/AAC.01852-17
Atshan, S. S., Nor Shamsudin, M., Lung, L. T., Sekawi, Z., Pei Pei, C., Karunanidhi, A., et al. (2013). Genotypically different clones of Staphylococcus aureus are diverse in the antimicrobial susceptibility patterns and biofilm formations. Biomed. Res. Int. 2013:515712. doi: 10.1155/2013/515712
Bai, B., Hu, K., Li, H., Yao, W., Li, D., Chen, Z., et al. (2018). Effect of tedizolid on clinical Enterococcus isolates: in vitro activity, distribution of virulence factor, resistance genes and multilocus sequence typing. FEMS Microbiol. Lett. 365:fnx284. doi: 10.1093/femsle/fnx284
Bal, A. M., David, M. Z., Garau, J., Gottlieb, T., Mazzei, T., Scaglione, F., et al. (2017). Future trends in the treatment of methicillin-resistant Staphylococcus aureus (MRSA) infection: an in-depth review of newer antibiotics active against an enduring pathogen. J. Glob. Antimicrob. Resist. 10, 295–303. doi: 10.1016/j.jgar.2017.05.019
Calfee, D. P. (2017). Trends in community versus health care-acquired methicillin-resistant Staphylococcus aureus infections. Curr. Infect. Dis. Rep. 19:48. doi: 10.1007/s11908-017-0605-6
Claeys, K. C., Lagnf, A. M., Hallesy, J. A., Compton, M. T., Gravelin, A. L., Davis, S. L., et al. (2016). Pneumonia caused by methicillin-resistant Staphylococcus aureus: does vancomycin heteroresistance matter? Antimicrob. Agents Chemother. 60, 1708–1716. doi: 10.1128/AAC.02388-15
Dabul, A. N. G., Avaca-Crusca, J. S., Van Tyne, D., Gilmore, M. S., and Camargo, I. (2018). Resistance in in vitro selected tigecycline-resistant methicillin-resistant Staphylococcus aureus sequence type 5 is driven by mutations in mepR and mepA genes. Microb. Drug Resist. 24, 519–526. doi: 10.1089/mdr.2017.0279
Draper, M. P., Weir, S., Macone, A., Donatelli, J., Trieber, C. A., Tanaka, S. K., et al. (2014). Mechanism of action of the novel aminomethylcycline antibiotic omadacycline. Antimicrob. Agents Chemother. 58, 1279–1283. doi: 10.1128/AAC.01066-13
El-Halfawy, O. M., and Valvano, M. A. (2013). Chemical communication of antibiotic resistance by a highly resistant subpopulation of bacterial cells. PLoS One 8:e68874. doi: 10.1371/journal.pone.0068874
El-Halfawy, O. M., and Valvano, M. A. (2015). Antimicrobial heteroresistance: an emerging field in need of clarity. Clin. Microbiol. Rev. 28, 191–207. doi: 10.1128/CMR.00058-14
Fiedler, S., Bender, J. K., Klare, I., Halbedel, S., Grohmann, E., Szewzyk, U., et al. (2016). Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J. Antimicrob. Chemother. 71, 871–881. doi: 10.1093/jac/dkv420
Grossman, T. H. (2016). Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 6:a025387. doi: 10.1101/cshperspect.a025387
Heidrich, C. G., Mitova, S., Schedlbauer, A., Connell, S. R., Fucini, P., Steenbergen, J. N., et al. (2016). The novel aminomethylcycline omadacycline has high specificity for the primary tetracycline-binding site on the bacterial ribosome. Antibiotics 5:32. doi: 10.3390/antibiotics5040032
Honeyman, L., Ismail, M., Nelson, M. L., Bhatia, B., Bowser, T. E., Chen, J., et al. (2015). Structure-activity relationship of the aminomethylcyclines and the discovery of omadacycline. Antimicrob. Agents Chemother. 59, 7044–7053. doi: 10.1128/AAC.01536-15
Jones, C. H., Tuckman, M., Howe, A. Y., Orlowski, M., Mullen, S., Chan, K., et al. (2006). Diagnostic PCR analysis of the occurrence of methicillin and tetracycline resistance genes among Staphylococcus aureus isolates from phase 3 clinical trials of tigecycline for complicated skin and skin structure infections. Antimicrob. Agents Chemother. 50, 505–510. doi: 10.1128/AAC.50.2.505-510.2006
Klionsky, D. J., Abdelmohsen, K., Abe, A., Abedin, M. J., Abeliovich, H., Acevedo Arozena, A., et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. doi: 10.1080/15548627.2015.1100356
Leon-Sampedro, R., Novais, C., Peixe, L., Baquero, F., and Coque, T. M. (2016). Diversity and evolution of the Tn5801-tet(M)-like integrative and conjugative elements among Enterococcus, Streptococcus, and Staphylococcus. Antimicrob. Agents Chemother. 60, 1736–1746. doi: 10.1128/AAC.01864-15
Lin, Z., Cai, X., Chen, M., Ye, L., Wu, Y., Wang, X., et al. (2017). Virulence and stress responses of Shigella flexneri regulated by PhoP/PhoQ. Front. Microbiol. 8:2689. doi: 10.3389/fmicb.2017.02689
Linkevicius, M., Sandegren, L., and Andersson, D. I. (2016). Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob. Agents Chemother. 60, 789–796. doi: 10.1128/AAC.02465-15
Lupien, A., Gingras, H., Leprohon, P., and Ouellette, M. (2015). Induced tigecycline resistance in Streptococcus pneumoniae mutants reveals mutations in ribosomal proteins and rRNA. J. Antimicrob. Chemother. 70, 2973–2980. doi: 10.1093/jac/dkv211
Macone, A. B., Caruso, B. K., Leahy, R. G., Donatelli, J., Weir, S., Draper, M. P., et al. (2014). In vitro and in vivo antibacterial activities of omadacycline, a novel aminomethylcycline. Antimicrob. Agents Chemother. 58, 1127–1135. doi: 10.1128/AAC.01242-13
Nguyen, F., Starosta, A. L., Arenz, S., Sohmen, D., Donhofer, A., and Wilson, D. N. (2014). Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 395, 559–575. doi: 10.1515/hsz-2013
Noel, G. J., Draper, M. P., Hait, H., Tanaka, S. K., and Arbeit, R. D. (2012). A randomized, evaluator-blind, phase 2 study comparing the safety and efficacy of omadacycline to those of linezolid for treatment of complicated skin and skin structure infections. Antimicrob. Agents Chemother. 56, 5650–5654. doi: 10.1128/AAC.00948-12
Osei Sekyere, J., and Amoako, D. G. (2017). Carbonyl cyanide m-chlorophenylhydrazine (CCCP) reverses resistance to colistin, but Not to Carbapenems and tigecycline in multidrug-resistant Enterobacteriaceae. Front. Microbiol. 8:228. doi: 10.3389/fmicb.2017.00228
Pfaller, M. A., Huband, M. D., Rhomberg, P. R., and Flamm, R. K. (2017a). Surveillance of omadacycline activity against clinical isolates from a global collection (North America, Europe, Latin America, Asia-Western Pacific), 2010-2011. Antimicrob. Agents Chemother. 61:e00018-e17. doi: 10.1128/AAC.00018-17
Pfaller, M. A., Rhomberg, P. R., Huband, M. D., and Flamm, R. K. (2017b). Activities of omadacycline and comparator agents against Staphylococcus aureus isolates from a surveillance program conducted in North America and Europe. Antimicrob. Agents Chemother. 61:e02411-16. doi: 10.1128/AAC.02411-16
Recker, M., Laabei, M., Toleman, M. S., Reuter, S., Saunderson, R. B., Blane, B., et al. (2017). Clonal differences in Staphylococcus aureus bacteraemia-associated mortality. Nat. Microbiol. 2, 1381–1388. doi: 10.1038/s41564-017-0001-x
Villano, S., Steenbergen, J., and Loh, E. (2016). Omadacycline: development of a novel aminomethylcycline antibiotic for treating drug-resistant bacterial infections. Future Microbiol. 11, 1421–1434. doi: 10.2217/fmb-2016-0100
Wu, Y., Wang, J., Xu, T., Liu, J., Yu, W., Lou, Q., et al. (2012). The two-component signal transduction system ArlRS regulates Staphylococcus epidermidis biofilm formation in an ica-dependent manner. PLoS One 7:e40041. doi: 10.1371/journal.pone.0040041
Yao, W., Xu, G., Bai, B., Wang, H., Deng, M., Zheng, J., et al. (2018). In vitro-induced erythromycin resistance facilitates cross-resistance to the novel fluoroketolide, solithromycin, in Staphylococcus aureus. FEMS Microbiol. Lett. 365:fny116. doi: 10.1093/femsle/fny116
Zhang, F., Bai, B., Xu, G. J., Lin, Z. W., Li, G. Q., Chen, Z., et al. (2018). Eravacycline activity against clinical S. aureus isolates from China: in vitro activity, MLST profiles and heteroresistance. BMC Microbiol. 18:211. doi: 10.1186/s12866-018-1349-7
Keywords: omadacycline, Staphylococcus aureus, antimicrobial activity, multilocus sequence typing, tetracycline specific resistance genes
Citation: Bai B, Lin Z, Pu Z, Xu G, Zhang F, Chen Z, Sun X, Zheng J, Li P, Deng Q and Yu Z (2019) In vitro Activity and Heteroresistance of Omadacycline Against Clinical Staphylococcus aureus Isolates From China Reveal the Impact of Omadacycline Susceptibility by Branched-Chain Amino Acid Transport System II Carrier Protein, Na/Pi Cotransporter Family Protein, and Fibronectin-Binding Protein. Front. Microbiol. 10:2546. doi: 10.3389/fmicb.2019.02546
Received: 24 May 2019; Accepted: 22 October 2019;
Published: 08 November 2019.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Agnese Lupo, ANSES Site de Lyon, FranceCopyright © 2019 Bai, Lin, Pu, Xu, Zhang, Chen, Sun, Zheng, Li, Deng and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiwen Deng, cWl3ZW5kZW5nQGhvdG1haWwuY29t; Zhijian Yu, eXV6aGlqaWFuc211QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.