Symbiotic microorganisms are omnipresent in nature, ubiquitously associated with animals, plants, fungi, protists, and all other life forms including humans, ranging from having parasitic through commensalistic to mutualistic associations, and affecting every biological aspect of innumerable organisms living on earth (McFall-Ngai et al., 2013; Webster, 2014). Insects represent the majority of macroscopic biodiversity described thus far (Grimaldi and Engel, 2005), and their ubiquitous interactions with microbes underpin their diversity and adaptability in the ecosystem (Bourtzis and Miller, 2003; Zchori-Fein and Bourtzis, 2011).
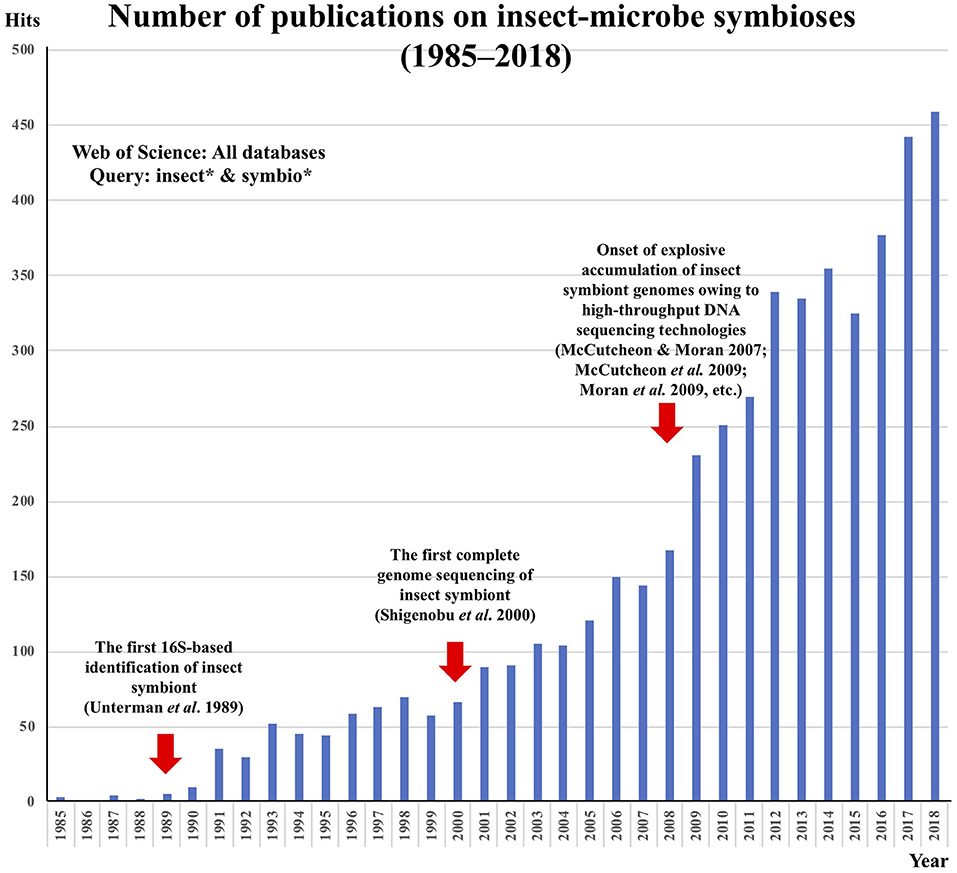
Studies on microbe-insect symbioses have a long history. During the early to mid twentieth century, microbe-insect symbioses were regarded as a focal research area in microbiology. Before the era of molecular biology, high-resolution light microscopy developed by Carl Zeiss and other optic companies was the cutting-edge technology in biology. Using high-quality microscopes, a number of German and other European microbiologists enthusiastically surveyed diverse insects, terrestrial arthropods, and other organisms for their internal microbiota. The enormous number of microscopic observations were compiled by the outstanding German microbiologist, Paul Buchner (Sapp, 2002), in the monumental book “Endosymbiosis of Animals with Plant Microorganisms” (Buchner, 1965). Subsequently, however, advancements in this research area were for decades very slow, mainly because of the general uncultivability of the symbiotic microorganisms—researchers could observe some bacteria residing in and associated with insect cells and tissues cytologically but were unable to characterize or even identify the microbes. In the 1980s, the invention of PCR and the development of DNA sequencing technologies brought about an epoch-making breakthrough in microbiology—environmental microorganisms became identifiable without cultivation on the basis of 16S rRNA gene sequencing. I remember that, in 1989, just before I began my graduate work at the University of Tokyo in the lab of Hajime Ishikawa, who was among the pioneers of the molecular and genomic aspects of aphid endosymbiotic bacteria (Fukatsu, 2006), the first 16S-based molecular phylogenetic identification of an uncultivable aphid endosymbiont [later named Buchnera aphidicola (Munson et al., 1991)] was published by Paul Baumann's group at the University of California, Davis (Unterman et al., 1989). The idea that this extremely specialized insect symbiont is allied to Escherichia coli was a big surprise at that time and served as strong motivation for the field to reach a better understanding of the fastidious microorganisms that are closely allied with insects. In 2000, when I had already started to run my own lab, Ishikawa's group determined the first complete genome of an uncultivable microbial mutualist of the pea aphid, Buchnera aphidicola (Shigenobu et al., 2000), which opened a new era of powerful genomic approaches to microbe-insect symbiosis studies. At that time, Sanger-based DNA sequencing technology was so time-, labor- and cost-intensive that the microbial genomics was not easily accessible for the majority of individual microbiologists. However, from 2007 onward, high-throughput DNA sequencing technologies, initially 454 and Solexa and then Illumina, PacBio, Nanopore, and others, became available, which propelled the explosive accumulation of microbial genome data. I was amazed to see interesting insect genomes coming up one after another from Nancy Moran's group at the University of Arizona (McCutcheon and Moran, 2007; McCutcheon et al., 2009; Moran et al., 2009), and I soon also jumped into the excitement. Sequencing insect symbiont genomes is like opening treasure boxes, uncovering a variety of astonishing evolutionary aspects such as initial massive accumulation of junk DNA elements (Wu et al., 2004; Toh et al., 2006) and subsequent size reduction, often approaching organelle-like sizes (Nakabachi et al., 2006; Pérez-Brocal et al., 2006), catastrophic genome erosion finally leading to symbiont replacements (Campbell et al., 2017; Matsuura et al., 2018), metabolic complementarity between co-evolving reduced symbiont genomes (McCutcheon and Moran, 2007, 2010), extremely tiny symbiont genomes streamlined for specific biological functions (Anbutsu et al., 2017; Salem et al., 2017), dynamic lateral gene transfer and functional fusion across symbiont and host (Dunning-Hotopp et al., 2007; Husnik et al., 2013), and others. The number of publications on microbe-insect symbioses from 1985 to 2018 can be seen in Figure 1, in which the above-mentioned historical trajectory and development are impressively illustrated in relation to the technological innovations.
Inherently, studies on symbiosis are destined to be interdisciplinary, encompassing the fields of ecology, evolution, genomics, and cellular and molecular biology. Reflecting this, despite the large number of scientific journals published in the world, few focus on the publication of microbe-insect symbiosis studies. To my knowledge, Symbiosis (https://link.springer.com/journal/13199) is the only journal whose mission is to publish papers on animal-microbe, plant-microbe, microbe-microbe, and other forms of symbiotic associations. Molecular Plant-Microbe Interactions (https://apsjournals.apsnet.org/loi/mpmi) publishes a considerable number of symbiosis-related papers, though restricted to plant-microbe symbioses. The Invertebrate Microbiology section of Applied and Environmental Microbiology (https://aem.asm.org/) and the Microbe-Microbe and Microbe-Host Interactions field of the ISME Journal (https://www.nature.com/ismej/) are the suitable outlets for papers on microbe-insect symbiotic associations. Journal of Invertebrate Pathology does publish papers on insect symbionts, but the journal mainly focuses on parasites and pathogens (https://www.journals.elsevier.com/journal-of-invertebrate-pathology). Environmental Entomology (https://academic.oup.com/ee) highlights Insect-Symbiont Interactions as a subject area.
Personally, it has been a challenging task for me to find suitable journals in which to publish my own microbe-insect symbiosis studies. For example, dating back to the early 1990s for my master's thesis at the University of Tokyo, I wrote up three papers on a variety of bacterial and fungal symbionts of aphids. I finally published them in entomological journals, Insect Biochemistry and Molecular Biology and Journal of Insect Physiology (Fukatsu and Ishikawa, 1992a,b,c) but could not help feeling that those were not actually a perfect fit. The absence of suitable outlets for insect symbiosis studies has been a long-lasting frustration for me, and this frustration must be shared generally by those who are working on such interdisciplinary research fields related to symbiosis.
In this context, it was a laudable decision of Frontiers in Microbiology to launch the Microbial Symbioses section in 2013. Since then, the Microbial Symbioses section has served as a home to numerous innovative, high-quality research studies, and commentaries on symbiosis. As a newly-appointed Specialty Chief Editor, I am highly motivated to build up an ideal platform for publishing microbe-insect symbiosis studies in the Microbial Symbioses section of Frontiers in Microbiology. To that end, I have contacted the world's most qualified experts who are actively working on microbe-insect symbiotic associations and interactions and have organized a launching Editorial Board for microbe-insect symbioses consisting of 14 Associate Editors as of September 2019. To my knowledge, no other scientific journals specialize in the publication of microbe-insect symbiosis studies. Though not as a journal but as a part of the journal section, I am aiming at manifesting such a publishing body of high quality and visibility, which offers prompt handling, fair and robust evaluation, and qualified publication of submitted manuscripts covering diverse aspects of microbe-insect symbiotic associations and interactions. The open access policy of Frontiers in Microbiology, together with the notable impact factor of 4.259, provides a good reason for consideration. I look forward to the very best works on microbe-insect symbioses being submitted to Frontiers in Microbiology.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JM declared a past collaboration with the author to the handling editor.
Acknowledgments
I thank the Associate Editors, Robert Brucker, Olivier Duron, Nicole Gerardo, Yuval Gottlieb, Allison Hansen, Martin Kaltenpoth, Yoshitomo Kikuchi, Amparo Latorre, Mariana Mateos, Steve Perlman, Michael Poulsen, Akiko Sugio, Christoph Vorburger, and Alex Wilson, constituting the Editorial Board specialized in insect-microbe symbioses in the Microbial Symbioses section of Frontiers in Microbiology, and the Journal Development Manager, Dorina Seitaj, for their valuable support and input.
References
Anbutsu, H., Moriyama, M., Nikoh, N., Hosokawa, T., Futahashi, R., Tanahashi, M., et al. (2017). Small genome symbiont underlies cuticle hardness in beetles. Proc. Natl. Acad. Sci. U.S.A. 114, E8382–E8391. doi: 10.1073/pnas.1712857114
Campbell, M. A., Łukasik, P., Simon, C., and McCutcheon, J. P. (2017). Idiosyncratic genome degradation in a bacterial endosymbiont of periodical cicadas. Curr. Biol. 27, 3568–3575. doi: 10.1016/j.cub.2017.10.008
Dunning-Hotopp, J. C., Clark, M. E., Oliveira, D. C. S. G., Foster, J. M., Fischer, P., Muñoz Torres, M. C., et al. (2007). Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756. doi: 10.1126/science.1142490
Fukatsu, T. (2006). “Obituary: Hajime Ishikawa (1940-2005): the pioneer in the molecular biological studies of insect symbiosis,” in Insect Symbiosis, Vol. 2, eds K. Bourtzis and T. A. Miller (Boca Raton, FL: CRC Press), F1–F6.
Fukatsu, T., and Ishikawa, H. (1992a). Synthesis and localization of symbionin, an aphid endosymbiont protein. Insect. Biochem. Mol. Biol. 22, 167–174. doi: 10.1016/0965-1748(92)90155-8
Fukatsu, T., and Ishikawa, H. (1992b). A novel eukaryotic extracellular symbiont in an aphid, Astegopteryx styraci (Homoptera, Aphididae, Hormaphidinae). J. Insect. Physiol. 38, 765–773. doi: 10.1016/0022-1910(92)90029-D
Fukatsu, T., and Ishikawa, H. (1992c). Soldier and male of an eusocial aphid Colophina arma lack endosymbiont: Implications for physiological and evolutionary interaction between host and symbiont. J. Insect Physiol. 38, 1033–1042. doi: 10.1016/0022-1910(92)90012-3
Grimaldi, D., and Engel, M. S. (2005). Evolution of the Insects. New York, NY: Cambridge University Press.
Husnik, F., Nikoh, N., Koga, R., Ross, L., Duncan, R. P., Fujie, M., et al. (2013). Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153, 1567–1578. doi: 10.1016/j.cell.2013.05.040
Matsuura, Y., Moriyama, M., Łukasik, P., Vanderpool, D., Tanahashi, M., Meng, X. Y., et al. (2018). Recurrent symbiont recruitment from fungal parasites in cicadas. Proc. Natl. Acad. Sci. U.S.A. 115, E5970–E5979. doi: 10.1073/pnas.1803245115
McCutcheon, J. P., McDonald, B. R., and Moran, N. A. (2009). Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. U.S.A. 106, 15394–15399. doi: 10.1073/pnas.0906424106
McCutcheon, J. P., and Moran, N. A. (2007). Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. U.S.A. 104, 19392–19397. doi: 10.1073/pnas.0708855104
McCutcheon, J. P., and Moran, N. A. (2010). Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol. Evol. 2, 708–718.
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Moran, N. A., McLaughlin, H. J., and Sorek, R. (2009). The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science 323, 379–382. doi: 10.1126/science.1167140
Munson, M. A., Baumann, P., and Kinsey, M. G. (1991). Buchnera gen. nov. and Buchnera aphidicola sp. nov., a taxon consisting of the mycetocyte-associated, primary endosymbionts of aphids. Int. J. Syst. Bacteriol. 41, 566–568. doi: 10.1099/00207713-41-4-566
Nakabachi, A., Yamashita, A., Toh, H., Ishikawa, H., Dunbar, H. E., Moran, N. A., et al. (2006). The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314:267. doi: 10.1126/science.1134196
Pérez-Brocal, V., Gil, R., Ramos, S., Lamelas, A., Postigo, M., Michelena, J. M., et al. (2006). A small microbial genome: the end of a long symbiotic relationship? Science 314, 312–323. doi: 10.1126/science.1130441
Salem, H., Bauer, E., Kirsch, R., Berasategui, A., Cripps, M., Weiss, B., et al. (2017). Drastic genome reduction in an herbivore's pectinolytic symbiont. Cell 171, 1520–1531. doi: 10.1016/j.cell.2017.10.029
Sapp, J. (2002). Paul Buchner (1886–1978) and hereditary symbiosis in insects. Int. Microbiol. 5, 145–150. doi: 10.1007/s10123-002-0079-7
Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, Y., and Ishikawa, H. (2000). Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407, 81–86. doi: 10.1038/35024074
Toh, H., Weiss, B. L., Perkin, S. A. H., Yamashita, A., Oshima, K., Hattori, M., et al. (2006). Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16, 149–156. doi: 10.1101/gr.4106106
Unterman, B. M., Baumann, P., and McLean, D. L. (1989). Pea aphid symbiont relationships established by analysis of 16S rRNAs. J. Bacteriol. 171, 2970–2974. doi: 10.1128/jb.171.6.2970-2974.1989
Webster, N. S. (2014). Cooperation, communication, and co-evolution: grand challenges in microbial symbiosis research. Front. Microbiol. 5:164. doi: 10.3389/fmicb.2014.00164
Wu, M., Sun, L. V., Vamathevan, J., Riegler, M., Deboy, R., Brownlie, J. C., et al. (2004). Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:e69. doi: 10.1371/journal.pbio.0020069
Keywords: symbiosis, microbe, insect, publishing, grand challenge
Citation: Fukatsu T (2019) Grand Challenges to Launching an Ideal Platform for Publishing Microbe-Insect Symbiosis Studies. Front. Microbiol. 10:2542. doi: 10.3389/fmicb.2019.02542
Received: 07 September 2019; Accepted: 21 October 2019;
Published: 11 November 2019.
Edited by:
Martin G. Klotz, Washington State University, United StatesReviewed by:
John McCutcheon, University of Montana, United StatesRosario Gil, University of Valencia, Spain
Copyright © 2019 Fukatsu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takema Fukatsu, dC1mdWthdHN1JiN4MDAwNDA7YWlzdC5nby5qcA==
 Takema Fukatsu
Takema Fukatsu