- Beijing Key Laboratory for Forest Pest Control, College of Forestry, Beijing Forestry University, Beijing, China
The Gram-negative bacterium Lonsdalea populi causes a lethal disease known as bark canker on Populus × euramericana in China and Europe. Typical symptoms of bark canker include an abundant white-colored fluid, which oozes from the infected tissues. The availability of the genomic sequence of the bacterium provided the necessary resource to launch genome-scale investigations into the mechanisms fundamental to pathogenesis. Functional analyses of a diverse group of genes encoding virulence factors and components of signaling pathways indicate that successful bark infection depends on specific responses by the pathogen to various stresses, including oxidative stress. Although physiology of resistance is well studied, the molecular processes underlying the defense responses and the genetic basis of resistance to L. populi and in other poplar species remain largely unknown. Control of the disease has relied on chemical measures. Due to the genetic amenability of Lonsdalea and poplar, this pathosystem will become an important model system to unravel molecular mechanisms of bacterial pathogenicity on woody plants. Increased understanding of pathogenesis and signaling in the interaction will facilitate the management of this kind of poplar canker.
Introduction
Poplars are predominantly distributed throughout the northern hemisphere. Because of their rapid growth, wild Populus spp. and their hybrids are currently planted over huge areas worldwide as ornamental plants for landscape greening, production of wood, and multiple industrial uses (Stettler et al., 1996; Jansson and Douglas, 2007). Populus × euramericana, an interspecific hybrid poplar, is one of the most widely grown poplars in China. However, poplars are often attacked by insects and various microbial pathogens. Among them, cankers represent the most destructive poplar diseases, which are localized in the bark of poplar trunk and branches. Cytospora chrysosperma and Botryosphaeria dothidea are the main fungal pathogen of poplar cankers worldwide (Sinclair and Lyon, 2005; Fan et al., 2019). In the 1990s, the bacterial genus Brenneria was also noted to cause canker of trees in Spain, including poplar (Biosca et al., 2006). Currently, a large portion of the P. × euramericana plantation area in China and Hungary is affected by one potentially lethal bacterium, Lonsdalea populi (formerly Lonsdalea quercina subsp. populi; Toth et al., 2013; Li et al., 2014).
This causal agent of poplar canker was first detected by Toth et al. (2013) in Hungary and later described by Li et al. (2014) in China as Lonsdalea populi, formerly known as Lonsdalea quercina subsp. populi (Li et al., 2017). The bacterium belongs to the genus Lonsdalea, which is a novel genus following the removal of the species Brenneria quercina from this classification (Brady et al., 2012). Furthermore, B. quercina was renamed from Erwinia quercina, which caused drippy nut disease of native live oaks in California (Hauben et al., 1998). There are distinctive biochemical and physiological characteristics among the genera of Lonsdalea, Brenneria, and Erwinia, which distinguish each from one another (Toth et al., 2013). Until recently, Lonsdalea quercina contains four subspecies (L. quercina subsp. populi, L. quercina subsp. quercina, L. quercina subsp. iberica, and L. quercina subsp. britannica), which are all pathogens of woody trees (Toth et al., 2013). Due to the genome sequence-derived average nucleotide identity (ANI) values between the four subspecies were 88.71–93.38%, respectively, lower than the proposed species boundary ANI cut-off (95–96%) that is considered the most important criterion to reclassify these subspecies at the species level. It is proposed that three subspecies were elevated to the species level, among them L. quercina subsp. populi was substituted to L. populi sp. nov. (Li et al., 2017). Using 16 s rDNA analysis, we reconstructed the phylogenetic tree (Figure 1) containing those sequences from four Lonsdalea species and other related sequences. This tree also suggests the close relationship of Lonsdalea species to Erwinia amylovora and Pantoea agglomerans, two major phytopathogenic bacteria.

Figure 1. Phylogenetic tree based on 16S ribosomal RNA gene sequences of Lonsdalea species and their closest phylogenetic neighbors. Sequences of 16 s rDNA of Lonsdalea populi N-5-1 (JF424600), Lonsdalea iberica (JF311441), Lonsdalea quercina (AJ223469), Lonsdalea britannica (JF311446), Brenneria alni (AJ233409), Brenneria nigrifluens (U80203), Erwinia amylovora (Z96088), Erwinia pyrifoliae (AJ009930), Dickeya paradisiaca (AF520710), Dickeya chrysanthemi (NR_117738), Pantoea anthophila (EF688010), Pantoea stewartii subsp. stewartii (KM508089), Pectobacterium carotovorum subsp. brasiliense (NR_118228), Pectobacterium atrosepticum (NR_118295), and Xanthomonas citri pv. citri (LC202834). The scale bar indicates a 0.1 nucleotide changes per site. X. citri pv. citri was included as outgroups. The principal host plants and diseases were shown next to the tree branches.
In 2006, severe bark canker symptom on P. × euramericana was observed for the first time in Henan and Shandong provinces of China. In 2009, the same symptom was also detected in P. ×euramericana stands in the central part of Hungary (Toth et al., 2013). Currently, a large part of the P. × euramericana plantation, including in Ningxia and Tianjin of China, is affected by this disease. Although a preliminary report showed that the fungus Fusarium solani was identified as a pathogen of this canker disease in 2009 (He et al., 2009), it is now beyond dispute that L. populi is the causal agent of P. × euramericana canker in China and Europe (Li et al., 2017). The availability of the genome sequence and genetic manipulation platform has accelerated rapid advances into functional genomics of pathogenicity and stress responses. Both the poplar host and the bacterium serve as excellent models for studying the molecular basis of poplar and woody tree-bacterial interactions.
The main aim of this review is to summarize the current state of knowledge concerning the emerging poplar canker. The review provides information on the following topics: (1) disease symptoms and host range; (2) techniques for pathogen detection; (3) functional characterization of virulence factors such as type III secretion and two-component systems; (4) disease management; and (5) perspectives and implications from other pathosystems. This review incorporates what is currently known on the molecular aspects of the poplar-bacterium interaction.
Symptoms of Lonsdalea Canker and Host Range
Poplar canker caused by L. populi is essentially lethal disease, affecting stems or branches of P. × euramericana that are typically more than 3-year olds in the field (Figure 2). Symptom development on P. × euramericana “74/76” starts in early summer with the appearance of abundant, white, sour exudates on the stem (Figure 2A), which is associated with lesions. The lesions expand vertically, causing cracking and also forming a sticky brown-colored fluid oozes with the rotten smell (Figure 2B). Most diseased poplars show a cracked stem or branch, exuding frothy fluid (Figure 2C; Li et al., 2014). The cankers expand rapidly, and in serious cases, poplars with these cankers may die, or entire branches may be broken off by wind (Figure 2D).

Figure 2. Symptoms and damage of canker disease of Populus × euramericana caused by the bacterial pathogen, Lonsdalea populi. (A) Early symptoms with abundant sour and white, sour exudate (black arrow) on poplar trunk; (B) Late brown bark symptoms in the summer month, brownish exudations (red arrow). (C) Overwintering canker with large bark necrosis (white arrow) and fresh necrotic bark (blue arrow). (D) Diseased poplar blown down by the wind in the field.
The incubation period until the appearance of symptoms, following bacterial inoculation of excised stems of poplar, lasts about 3 days, although water-cultured excised stems show initial symptoms after 4 days (Shang et al., 2014). As soon as the whitish-yellow-colored liquid flowed from the inoculation site, the necrotic lesion preferentially expanded longitudinally. The infection spreads within the central tissues including via xylem, tissue which is frequently associated with a brownish discoloration. L. populi was detected by PCR amplification with species-specific primers ahead of the visible necrotic lesion in apparently healthy inner bark tissues and reached peak quantities, however, when associated with the severe necrotic symptom (Shang et al., 2015). Real-time PCR analysis showed that L. populi mainly exists in the phloem tissue, as well as exudates of canker, but not in xylem tissue or the leaves, and the peak bacterial accumulation occurs at 7 days after infection when the phloem tissue was seriously damaged and parenchyma cells were disaggregated (Shang et al., 2014). In field conditions, lesions on the bark associated with this disease mostly become visible during late spring and summer. The beetles, Librodor japonicus and Protaetia brevitarsis, were found with the ooze associated with the cankers. Furthermore, DNA of L. populi can be detected in the beetles from infected trees but not uninfected trees by PCR amplification, thus suggesting that these insects could be helpful in spreading the disease to other trees.
Lonsdalea canker was initially found on four cultivars of P. × euramericana in China, such as “Zhonglin 46,” “74/76,” “Zhonghe 1,” and “Robusta.” Among them, “Zhonglin 46” is the most susceptible, and the incidence on this cultivar can rise up to 70% (Li et al., 2014). Although L. populi is primarily restricted to P. × euramericana, inoculation with the bacterial inocula in the lab leads to bark canker of a wide range of poplar cultivars, including Populus tomentosa. In 2017, Lonsdalea poplar canker was recorded in Portugal (Abelleira et al., 2019). Moreover, Populus × interamericana “Beaupre” and P. × euramericana “I-214” and “MC” were infected by L. populi in nine plantations in Spain (Berruete et al., 2016). Recently, willow trees (Salix matsudana) infected naturally, and stem inoculation experiments using healthy willow trees showed that S. matsudana was also susceptible to L. populi (Li et al., 2019).
Techniques for Pathogen Detection
Isolation of L. populi from necrotic lesions on the bark is straightforward, although F. solani has sometimes been found (He et al., 2009; Li et al., 2014). A tiny tissue sample is aseptically taken from the xylem close to the necrotic bark after the samples were shaken in sterile distilled water for 10 min and incubated on Luria-Bertani medium. Bacterial isolation success largely depends on the season and the age of the canker lesion. Fresh canker usually appears in the late spring and summer, and isolation yields good success. During the winter months, the ratio of bacterial isolation remains lower.
16S rRNA and three housekeeping genes (gyrB, atpD, and infB) can be used as target sequences to differentiate the bacterium from the close species, including L. iberica and L. britannica (Toth et al., 2013; Li et al., 2014). Based on comparative genomic sequence analyses, two species-specific primers (LqfF/LqfR and LqgF/LqgR) are available for the rapid and accurate detection of the bacterium in diseased plant tissues, and a real-time PCR assay for in planta detection at the minimum amount of 0.1 pg. genomic DNA of L. populi has been developed (Shang et al., 2015). These methods were developed to monitor asymptomatic poplar before typical symptoms appeared.
Functional Characterization of Virulence Genes Associated With the Type III Secretion and Two-Component Systems
Research into the molecular biology of virulence has been spurred forward by the availability of genomic resources and the ease of genetic transformation to obtain mutants of L. populi. The genome size of L. populi N-5-1 is 3.9 Mb with G + C content of 55.42% and contains 3,746 predicted protein-encoding genes (Table 1). The bacterium is highly amendable to molecular genetic manipulation, especially insertional inactivation and in-frame deletion methods. Below, we provide and discuss an overview of the current knowledge of L. populi pathogenesis from a genomic perspective.
The type III secretion system (T3SS) comprises major virulence factors in many Gram-negative bacterial pathogens and is usually encoded by a gene cluster associated with pathogenicity islands (Hacker et al., 1997). Bacteria deploy T3SS to deliver various effectors into the host cell, which can manipulate various plant processes to promote infection or activate plant immunity system. Genome analyses of the strain N-5-1 revealed a 23 kb T3SS gene cluster. The genetic organization of this cluster is highly similar to that of the cognate gene clusters in Dickeya dadantii 3,937 and Erwinia amylovora CFBP1430, whereas it is strikingly different from that observed in Ralstonia solanacearum GMI100 (Yang et al., 2014b). This cluster contains 26 Hrp genes related to the hypersensitive response on non-hosts and that contributes to pathogenicity. Nine of these genes encode highly conserved Hrc proteins, while others act as effectors and regulators of T3SS. HrcW mutants exhibited reduced virulence on poplar and heterogeneous expression of N-terminal but not C-terminal HrpW triggered hypersensitive response (HR) on tobacco leaves (Yang et al., 2014a). In addition, HrcV encoding a T3SS structural protein was required for virulence on poplar stem and HR on tobacco leaves, while HrcV played no obvious roles in bacterial growth, motility, and biofilm formation (Yang et al., 2014b). A third Hrp gene, HrcJ, was characterized by insertional mutagenesis and was required for pathogenicity of L. populi on poplar and to elicit an HR response in tobacco. The HrcJ deletion mutant did not affect growth and biofilm formation but significantly decreased the motility (Li et al., 2015). These data document the importance of T3SS in pathogenesis, but it remains to be shown how T3SS is connected to virulence and xylem colonization of poplar plants in L. populi, as shown in Figure 3.
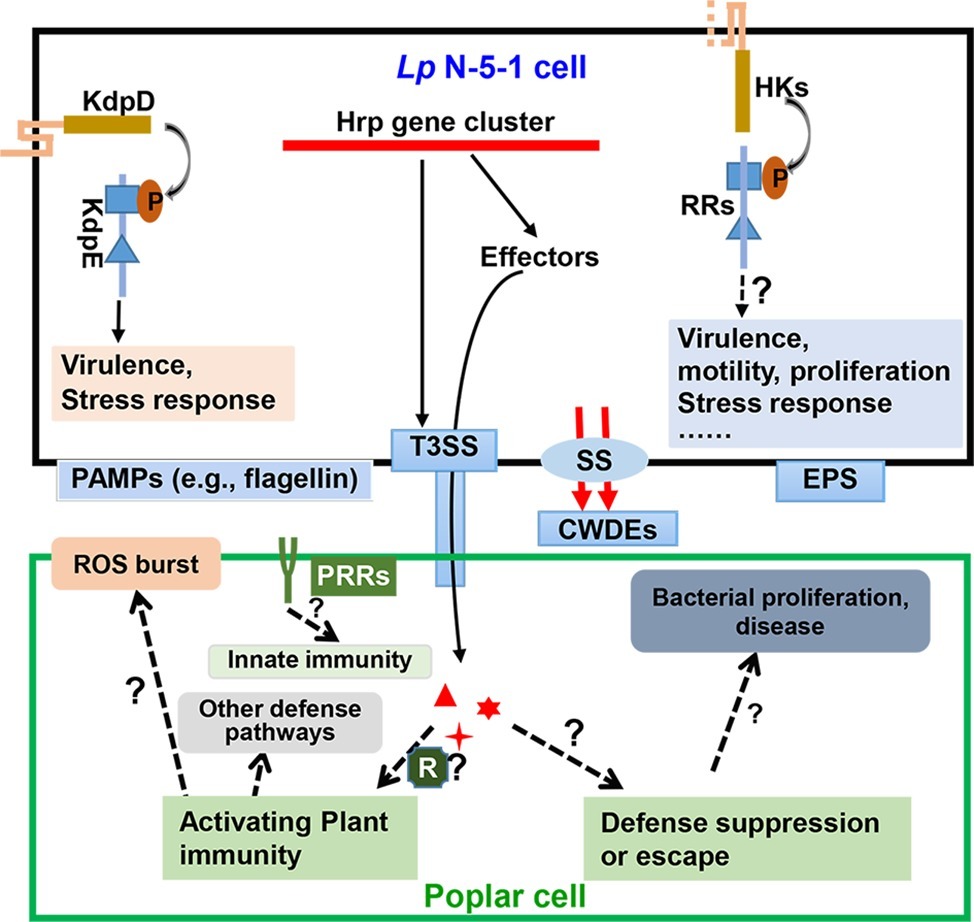
Figure 3. A diagram of the two-component signaling and secretion systems during interaction of Lonsdalea populi and Populus × euramericana. The bacterium L. populi N-5-1 mainly perceives external signals via two-component systems (TCS) comprising histine kinases (HKs) and response regulators (RRs). This leads to activation of TCS signaling that modulates expression of genes associated with virulence, motility, proliferation, stress response, etc. A well-characterized TCS of L. populi N-5-1, KdpD-KdpE, has been demonstrated to control virulence and stress response (Yang et al., 2018). Moreover, L. populi N-5-1 contains a 23 kb gene cluster encoding a type III secretion system (T3SS), which is deployed along with other secretion systems (SS) to deliver effectors and plant cell wall-degrading enzymes (CWDEs), which may activate poplar plant immunity. In response, the host may release a reactive oxygen species (ROS) burst and other defense pathways, or some effectors in susceptible hosts may promote bacterial infection and cause canker disease. Exopolysaccharides (EPS) also facilitate bacterial virulence. On the other hand, plant pattern recognition receptor (PRR) complexes recognize pathogen-associated molecular patterns (PAMPs), which result in activation of plant innate immunity. In this view, an undetermined number of transmembrane domains of HK is shown by dashed lines. Dashed arrows and lines indicate potential regulation. Question marks indicate predicted characteristics that are yet to be substantiated.
In bacteria, two-component signal transduction systems (TCSs) play essential roles in the regulation of physiological and cellular processes, including virulence and adaptation (Stock et al., 2000). A typical TCS comprises a membrane-bound histidine kinase (HK) and a cytosolic response regulator (RR) (Capra and Laub, 2012). BLASTp searches were undertaken to determine how many genes are involved in conserved HK-RR phosphorylation, uncovering 18 HKs and 24 RRs (Yang et al., 2018). Interestingly, TCS genes are thought to account for approximately 2 or 3% of the annotated genes of a bacterial genome (Wuichet et al., 2010). Compared with the number of TCSs with other bacteria of the Pectobacteriaceae family, the genome of L. populi encodes a relatively smaller number of TCSs. Further analyses of TCSs with the set of 18 HKs and 24 RRs revealed 15 pairs of HK and RR genes that were located close to each other in the genome and 12 orphan TCS genes (3 HKs and 9 RRs), which were not clustered with any of the other HK and RR genes in the genome of L. populi. A large-scale insertional mutagenesis study resulted in 32 mutants (16 HK and 16 RR mutants) and further demonstrated that 19 TCS genes regulated bacterial virulence against poplar trees. Among these, a novel TCS, KdpD-KdpE, formed a bi-cistronic operon, which not only controls bacterial virulence, growth, and swimming motility but also regulates bacterial resistance to oxidative and chloramphenicol stresses (Yang et al., 2018). Furthermore, KdpE functions as a transcription factor, which binds to the promoters of 44 genes, including stress response genes. Recently, an orphan RR, LqRR2, has been further characterized, revealing that it is required for virulence and bacterial motility, possibly regulating the expression of flgB, flgC, and flgE, rather than being directly involved in the biofilm formation and extracellular polysaccharide production (Li et al., 2018). Consequently, L. populi TCSs, more HK-RR-like proteins, and KdpD-KdpE or orphan proteins require identification and characterization and also further analyzes the environmental or host stimuli that are recognized and transduced by TCSs. Biochemical and structural investigation could be applied to comprehensively characterize the molecular mechanisms orchestrated by TCSs.
The recent studies into the molecular aspects of virulence in L. populi have addressed a range of questions from T3SS effectors to TCS and stress responses. Future research into the functional genomics of pathogenicity and host adaptation will include a better understanding of molecular bases underlying multiple secretion pathways, secondary metabolite production, biofilm formation, and manipulation of host susceptibility, as summarized in Figure 3.
Poplar-Bacterium Interaction
To better characterize the interaction between L. populi and poplar plants, physiological and molecular responses in that plant have been assessed. Initially, the expression of PR1-1, PR1-2, NPR1-1, NPR1-2, TGA1, TGA2, MYC2-1, and MYC2-2 was recorded as higher in a resistant poplar variety (Populus tomentosa) than the susceptible one (P. deltoids cv. “Zhonghe 1”), while JAZ1, COI1-1, and COI1-2 were downregulated in the resistant variety, suggesting the involvement of SA and JA signal transduction during the infection of the bacterium (Liu et al., 2015). Whether the SA and JA pathways are necessary or sufficient for resistance remains to be determined, the transcriptome analysis is clearly consistent with their involvement. The transcriptomic profiles of poplar in response to infection revealed a potential role of the biosynthesis of secondary metabolites, plant hormone signal transduction, regulation of autophagy, and ABC transporters during L. quercina infection (Hou et al., 2016). In addition, the study also demonstrated that flavonoid biosynthesis pathway and plant hormone signal transduction might play important roles in poplar response to the infection (Hou et al., 2016). Recently, RNA-Seq analysis at the early stage of poplar response upon L. quercina infection showed that Vitamin B6 metabolism, selenocompound metabolism, and benzoxazinoid biosynthesis pathways of poplar were induced and also indicated that flavonoid and phenylpropanoid would be involved in the poplar defense response to the infection at the early stage (Zhou et al., 2019). While the transcriptomic analysis of the poplar response to bacterial infection has been characterized in some detail, significantly less is known regarding the response of the bacterium during infection and colonization of poplar. Dual-RNA sequencing of host and pathogen will provide novel insights into the interaction between poplar and bacterium. Moreover, a striking gap of knowledge on the interaction between poplar R genes and bacterial effectors. Although hundreds of R genes of poplar and a dozen of putative T3SS effectors in the pathogen have been identified, what are the molecular bases of the interaction and how does the interaction determine poplar resistance or susceptibility (Figure 3)?
Disease Management
Integrated disease management strategies for Lonsdalea canker need to minimize poplar losses without raising the specter of environmental concerns. Due to the limited host range of Lonsdalea canker, breeding resistant poplar varieties or deployment of resistant species such as P. tomentosa would be a preferred solution to control the disease. At present, little is known about R-mediated resistance against Lonsdalea canker, although resistant poplar does exist in the field. Until now, P. tomentosa is the most resistant, and 74/76 is medium resistant, while “313,” “Tianyan,” and “599” are the most susceptible. Other control measures have also been explored. For example, Xu reported a potential approach of biological control to use Bacillus subtilis P2 or Streptomyces venezuelae P4, which was isolated from rhizosphere soil of Lonsdalea canker of poplar (Xu et al., 2013). Typically, however, practical implementation of biocontrol organisms under field conditions remains a bottleneck. Some antibiotics, such as polyoxin, streptomycin, ethylicin, and amobam, were screened and exhibited obvious effectiveness to inhibit the pathogen and canker expansion in the lab and under field conditions (Xu et al., 2013). Meanwhile, the application of antibiotics to control this disease on a large scale and the potential for the development of antibiotic resistance in the pathogen should be carefully considered.
Implication From Other Pathosystems and Perspectives
Although the underlying mechanisms of the poplar – L. populi pathosystem appear unexplored, comparisons with other bacterial pathosystems might provide a useful reference of the understanding and control of Lonsdalea canker. Citrus canker disease is caused by Xanthomonas strains (Brunings and Gabriel, 2003), and the disease that these strains cause is one of the most economically damaging diseases affecting citrus products worldwide, and the presence of citrus canker in an area triggers immediate quarantine restrictions (Graham et al., 2004). Integrated approaches for prevention and control of citrus canker have been used, but outbreaks of the disease continue to occur (Graham et al., 2004). Fire blight disease, caused by E. amylovora disease, is incredibly destructive to the fruit industry worldwide (Eastgate, 2000). A lesson from both tree bacterial diseases is the importance of avoiding further introductions of pathogenic strains, the significance of discovering pathogenicity determinants of pathogens, and the major resistance genes of hosts. Therefore, even if L. populi has only been discovered in relatively small regions across the globe, prohibiting the importation of plant material from canker-endemic areas is advised. In addition, it is important to study the genetics, physiology, and molecular mechanisms of the pathogen and poplar, especially in how the molecular interactions between the two can affect disease outcomes.
Although this review has revealed considerable advances in our understanding of the molecular basis of L. populi virulence and stress response, many questions remain unsolved. The molecular events underlying the infection process and overwintering in cankers have caught little consideration. On the one hand, many molecular tools of the bacterium have been established. On the other hand, poplar is an excellent model for trees. Therefore, the development of more effective controls for this devastating disease is a worthy goal for the future, and the poplar – L. populi pathosystem might become a model of forest pathology.
Author Contributions
AL designed the experiments. AL and WH analyzed data and wrote the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31870626).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We deeply appreciated Dr. Steven Klosterman (United States Department of Agriculture, Agricultural Research Service) for carefully reading and editing the manuscript.
References
Abelleira, A., Moura, L., Aguin, O., and Salinero, C. (2019). First report of Lonsdalea populi causing bark canker disease on poplar in Portugal. Plant Dis. 103, 2121–2122. doi: 10.1094/PDIS-12-18-2230-PDN
Berruete, I. M., Cambra, M. A., Collados, R., Monterde, A., Lopez, M. M., Cubero, J., et al. (2016). First report of bark canker disease of poplar caused by Lonsdalea quercina subp. populi in Spain. Plant Dis. 100, 2159–2159. doi: 10.1094/PDIS-03-16-0405-PDN
Biosca, E., Martín, S., Zuriaga, P., Monton, C., Lopez-Ocana, L., and Lopez, M. (2006). “Characterization of Brenneria sp. from poplar cankers in Spain” in Modern multidisciplinary applied microbiology. ed. A. Mendez-Vilas 385–389.
Brady, C. L., Cleenwerck, I., Denman, S., Venter, S. N., Rodriguez-Palenzuela, P., Coutinho, T. A., et al. (2012). Proposal to reclassify Brenneria quercina (Hildebrand and Schroth 1967) Hauben et al. 1999 into a new genus, Lonsdalea gen. nov., as Lonsdalea quercina comb. nov., descriptions of Lonsdalea quercina subsp. quercina comb. nov., Lonsdalea quercina subsp. iberica subsp. nov. and Lonsdalea quercina subsp. britannica subsp. nov., emendation of the description of the genus Brenneria, reclassification of Dickeya dieffenbachiae as Dickeya dadantii subsp. dieffenbachiae comb. nov., and emendation of the description of Dickeya dadantii. Int. J. Syst. Evol. Microbiol. 62, 1592–1602. doi: 10.1099/ijs.0.035055-0
Brunings, A. M., and Gabriel, D. W. (2003). Xanthomonas citri: breaking the surface. Mol. Plant Pathol. 4, 141–157. doi: 10.1046/j.1364-3703.2003.00163.x
Capra, E. J., and Laub, M. T. (2012). Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347. doi: 10.1146/annurev-micro-092611-150039
Eastgate, J. A. (2000). Erwinia amylovora: the molecular basis of fireblight disease. Mol. Plant Pathol. 1, 325–329. doi: 10.1046/j.1364-3703.2000.00044.x
Fan, X. L., Bezerra, J. D. P., Tian, C. M., and Crous, P. W. (2019). Cytospora (Diaporthales) in China. Persoonia. 45, 1–45. doi: 10.3767/persoonia.2020.45.01
Graham, J. H., Gottwald, T. R., Cubero, J., and Achor, D. S. (2004). Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 5, 1–15. doi: 10.1046/j.1364-3703.2004.00197.x
Hacker, J., Blum-Oehler, G., Muhldorfer, I., and Tschape, H. (1997). Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23, 1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x
Hauben, L., Moore, E. R. B., Vauterin, L., Steenackers, M., Mergaert, J., Verdonck, L., et al. (1998). Phylogenetic position of phytopathogens within the Enterobacteriaceae. Syst. Appl. Microbiol. 21, 384–397. doi: 10.1016/S0723-2020(98)80048-9
He, W., Ren, F. J., Guo, L. M., Li, Y., and Chang, J. (2009). Pathogen identification of Populus × euramericana canker disease. Sci. Silvae Sin. 45, 104–108. doi: 10.3321/j.issn:1001-7488.2009.06.018
Hou, J., Wu, Q. M., Zuo, T., Guo, L. M., Chang, J. P., Chen, J. H., et al. (2016). Genome-wide transcriptomic profiles reveal multiple regulatory responses of poplar to Lonsdalea quercina infection. Trees 30, 1389–1402. doi: 10.1007/s00468-016-1376-7
Jansson, S., and Douglas, C. J. (2007). Populus: a model system for plant biology. Annu. Rev. Plant Biol. 58, 435–458. doi: 10.1146/annurev.arplant.58.032806.103956
Li, Y., He, W., Ren, F., Guo, L., Chang, J., Cleenwerck, I., et al. (2014). A canker disease of Populus × euramericana in China caused by Lonsdalea quercina subsp. populi. Plant Dis. 98, 368–378. doi: 10.1094/PDIS-01-13-0115-RE
Li, B., Li, A., Wei, Q., Wang, H., and Wei, H. (2015). Functional analysis of hrc J gene in Lonsdalea quercina subsp. populi. Sci. Silvae Sin. 51, 71–78. doi: 10.11707/j.1001-7488.20151209
Li, Y., Wang, H., Chang, J., Guo, L., Yang, X., and Xu, W. (2019). A new bacterial bark canker disease of Salix matsudana caused by Lonsdalea populi in China. Plant Dis. doi: 10.1094/PDIS-03-19-0583-RE
Li, Y., Xue, H., Guo, L. M., Koltay, A., Palacio-Bielsa, A., Chang, J., et al. (2017). Elevation of three subspecies of Lonsdalea quercina to species level: Lonsdalea britannica sp. nov., Lonsdalea iberica sp. nov. and Lonsdalea populi sp. nov. Int. J. Syst. Evol. Microbiol. 67, 4680–4684. doi: 10.1099/ijsem.0.002353
Li, A., Yang, R., Wei, Q., and He, W. (2018). Functional analysis of orphan LqRR2 gene in Lonsdalea quercina subsp. populi. Acta Phytopathol. Sin., 492–500. doi: 10.13926/j.cnki.apps.000150
Liu, Z., Su, Y., Zuo, T., Chang, J., Guo, L., Wei, H., et al. (2015). The differential expression analysis of the SA and JA signal transduction related genes in poplar varieties susceptible and resistant to canker. Chin. Agric. Sci. Bull. 31, 156–163. doi: 10.11924/j.issn.1000-6850.casb15020033
Shang, J., Li, Y., Li, A., He, W., Guo, L., and Chang, J. (2014). Dynamic changes of Lonsdalea quercina subsp. populi in infection process of Populus × euramericana. J Northeast For. Univ. 42, 120–123, 137. doi: 10.13759/j.cnki.dlxb.20141106.025
Shang, J., Liu, B. L., and He, W. (2015). A new method to detect Lonsdalea quercina in infected plant tissues by real-time PCR. For. Pathol. 45, 28–35. doi: 10.1111/efp.12125
Sinclair, W. A., and Lyon, H. H. (2005). Diseases of trees and shrubs. 2nd Edn. Ithaca, NY: Cornell University Press.
Stettler, R., Bradshaw, J. H., Heilman, P., and Hinckley, T. (1996). Biology of Populus and its implications for management and conservation. Ottawa, ON: NRC Research Press.
Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. doi: 10.1146/annurev.biochem.69.1.183
Toth, T., Lakatos, T., and Koltay, A. (2013). Lonsdalea quercina subsp. populi subsp. nov., isolated from bark canker of poplar trees. Int. J. Syst. Evol. Microbiol. 63, 2309–2313. doi: 10.1099/ijs.0.042911-0
Wuichet, K., Cantwell, B. J., and Zhulin, I. B. (2010). Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 13, 219–225. doi: 10.1016/j.mib.2009.12.011
Xu, F., He, W., Chang, J., Guo, L., Xie, S., and Yang, Y. (2013). Primary study for control of Lonsdalea canker of poplar. Chin. Agric. Sci. Bull. 29, 12–17. doi: 10.11924/j.issn.1000-6850.2013-0174
Yang, R., Deng, C., Wei, J., He, W., Li, A., and Qian, W. (2018). A large-scale mutational analysis of two-component signaling systems of Lonsdalea quercina revealed that KdpD-KdpE regulates bacterial virulence against host poplar trees. Mol. Plant Microbe Interact. 31, 724–736. doi: 10.1094/MPMI-10-17-0248-R
Yang, L., Xu, B., He, W., and Zhang, L. (2014a). The HrpW protein of Lonsdalea quercina N-5-1 has pectate lyase activity and is required for full bacterial virulence. J. Basic Microbiol. 54, 1126–1135. doi: 10.1002/jobm.201300342
Yang, L., Zhang, L., He, W., Chang, J., and Guo, L. (2014b). Type III secretion system is an essential pathogenic factor in poplar canker pathogen Lonsdalea quercina. Acta Phytopathol. Sin. 512–520. doi: 10.13926/j.cnki.apps.2014.05.009
Keywords: poplar canker, Lonsdalea populi, virulence, virulence factors, poplar-bacterium interaction
Citation: Li A and He W (2019) Molecular Aspects of an Emerging Poplar Canker Caused by Lonsdalea populi. Front. Microbiol. 10:2496. doi: 10.3389/fmicb.2019.02496
Edited by:
Brigitte Mauch-Mani, Université de Neuchâtel, SwitzerlandCopyright © 2019 Li and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aining Li, bGlfYWluaW5nQGJqZnUuZWR1LmNu
 Aining Li
Aining Li Wei He
Wei He