- 1Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China
- 2School of Life Sciences, Henan University, Kaifeng, China
- 3Jiangsu National Synergetic Innovation Center for Advanced Materials, SICAM, Nanjing, China
- 4College of Life Sciences, Shanghai Normal University, Shanghai, China
Actinobacteria represent one of the most fertile sources for the discovery and development of natural products (NPs) with medicinal and industrial importance. However, production titers of actinobacterial NPs are usually low and require optimization for compound characterization and/or industrial production. In recent years, a wide variety of novel enabling technologies for engineering actinobacteria have been developed, which have greatly facilitated the optimization of NPs biosynthesis. In this review, we summarize the recent advances of synthetic biology approaches for overproducing desired drugs, as well as for the discovery of novel NPs in actinobacteria, including dynamic metabolic regulation based on metabolite-responsive promoters or biosensors, multi-copy chromosomal integration of target biosynthetic gene clusters (BGCs), promoter engineering-mediated rational BGC refactoring, and construction of genome-minimized Streptomyces hosts. Integrated with metabolic engineering strategies developed previously, these novel enabling technologies promise to facilitate industrial strain improvement process and genome mining studies for years to come.
Introduction
As a kind of Gram-positive bacteria with high GC content, actinobacteria undergo complex morphological differentiation and secondary metabolism processes (Barka et al., 2016). Actinobacteria, particularly the Streptomyces genus, have been recognized as the main sources for microbial bioactive natural products (NPs), such as antibiotics, chemotherapeutics, immunosuppressants and anthelmintics, which make important contributions to health care and crop protection (Jensen et al., 2015; Genilloud, 2017; Nepal and Wang, 2019). With the rapid advances in genome sequencing and genome mining methods (Ziemert et al., 2016; Blin et al., 2019), a wealth of hidden NP biosynthetic gene clusters (BGCs) have been revealed by specialized software (i.e., antiSMASH) and are regarded as an untapped treasure trove for the discovery of novel bioactive compounds (Rutledge and Challis, 2015; Niu, 2018). For instance, a single Streptomyces genome usually harbors around 30 NP BGCs, approximately 10-fold more than previously identified by bioactivity screening (Rutledge and Challis, 2015). However, the majority of NP BGCs in actinobacteria are silent or cryptic under laboratory culture conditions, and must be activated for the isolation and characterization of unknown compounds (Rutledge and Challis, 2015; Zarins-Tutt et al., 2016). Furthermore, production titers of many available actinomycete-derived drugs are still low for the economically viable industrial bioprocess. Construction of highly efficient microbial cell factories becomes increasingly critical for commercial application of desired NPs (Kim et al., 2016; Zhang et al., 2016).
With the advents of metabolic engineering and synthetic biology, genetic engineering of actinobacteria could address several major challenges associated with NP discovery, development and large-scale manufacturing. In the last three decades, a variety of genetic engineering strategies have been developed for strain development, including precursor engineering, BGC amplification, deletion of competing pathways, engineering of translational/transcriptional machineries as well as manipulation of pleiotropic/pathway-specific regulators (Baltz, 2016; Kim et al., 2016; Palazzotto et al., 2019). Interested readers are referred to the detailed reviews of systems biotechnology of actinobacteria (Weber et al., 2015; Baltz, 2016; Kim et al., 2016; Zhang et al., 2016; Palazzotto et al., 2019). It is worth noting that with breakthroughs in CRISPR-based genome editing methods, a series of novel enabling technologies have been developed, greatly facilitating the engineering of actinomycetal genomes (i.e., deletion, insertion, replacement and point mutation) as well as NP BGCs (i.e., cloning, editing, deletion and amplification) (Li et al., 2017a; Tong et al., 2018; Alberti and Corre, 2019). In this review, we briefly summarize the most recent synthetic biology approaches and discuss how these technologies enable the generation of microbial cell factories and the discovery of novel therapeutic drug leads. These include dynamic regulation based on metabolite-responsive elements, multiplex site-specific recombination (SSR) system-mediated BGC amplification (MSGE), systematical and rational BGC refactoring, as well as construction of genome-minimized Streptomyces hosts for NP overproduction and discovery. Together with traditional metabolic engineering strategies (Weber et al., 2015; Baltz, 2016), we believe that these newly developed tools will be widely applicable for actinobacteria, providing general strategies for (meta)genome mining-based novel NP discovery as well as for the overproduction of commercially important NPs.
Dynamic Pathway Regulation Based on Metabolite-Responsive Promoters or Regulators
Dynamic metabolic regulation has proved to be an effective strategy to improve production titers of target compounds by balancing bacterial growth and biosynthesis of specific metabolites (Zhang et al., 2012; Dahl et al., 2013; Gupta et al., 2017). Generally, three different approaches – quorum sensing systems, metabolite-responsive promoters and protein/RNA-based biosensors – are used for autonomous control of metabolic pathway flux (Zhang et al., 2015; Polkade et al., 2016; Tan and Prather, 2017). These new concepts were initially developed in Escherichia coli or Saccharomyces cerevisiae, and the latter two have been extended to actinobacteria for the optimization of antibiotic biosynthetic pathways.
Metabolite-Responsive Promoters
In the last three decades, a large variety of metabolic engineering strategies have been developed to optimize production of secondary metabolites in microbes. However, few approaches could enable the coordination between bacterial growth and biosynthesis of target compounds. Recently, Li et al. (2018) employed time-course transcriptome analysis to identify a series of antibiotic-responsible promoters with a transcription profile similar to the inducible promoters, when under the optimal conditions (Figure 1A). These dynamic responsive promoters could be used to efficiently optimize the expression of native actinorhodin and heterogeneous oxytetracycline (OTC) BGCs in Streptomyces coelicolor, subsequently improving the production titers of ACT and OTC by 1.3- and 9.1-fold, respectively, compared with constitutive promoters (Li et al., 2018). Xu et al. (2012) reported a metabolite-responsive promoter, which controlled the transcription of actAB encoding an antibiotic exporter in S. coelicolor. They found that the antibiotic ACT and its biosynthetic intermediates [e.g., (S)-DNPA] could relieve the repression of actAB by binding the transcriptional regulator ActR. That means that the actAB promoter could indirectly respond to intermediates or end-products, thus synergistically regulating ACT biosynthesis and export. The metabolite-responsible promoter based strategy will achieve an autonomous induction of pathway regulation and provide a universal route for titer improvements of desired NPs in actinobacteria.
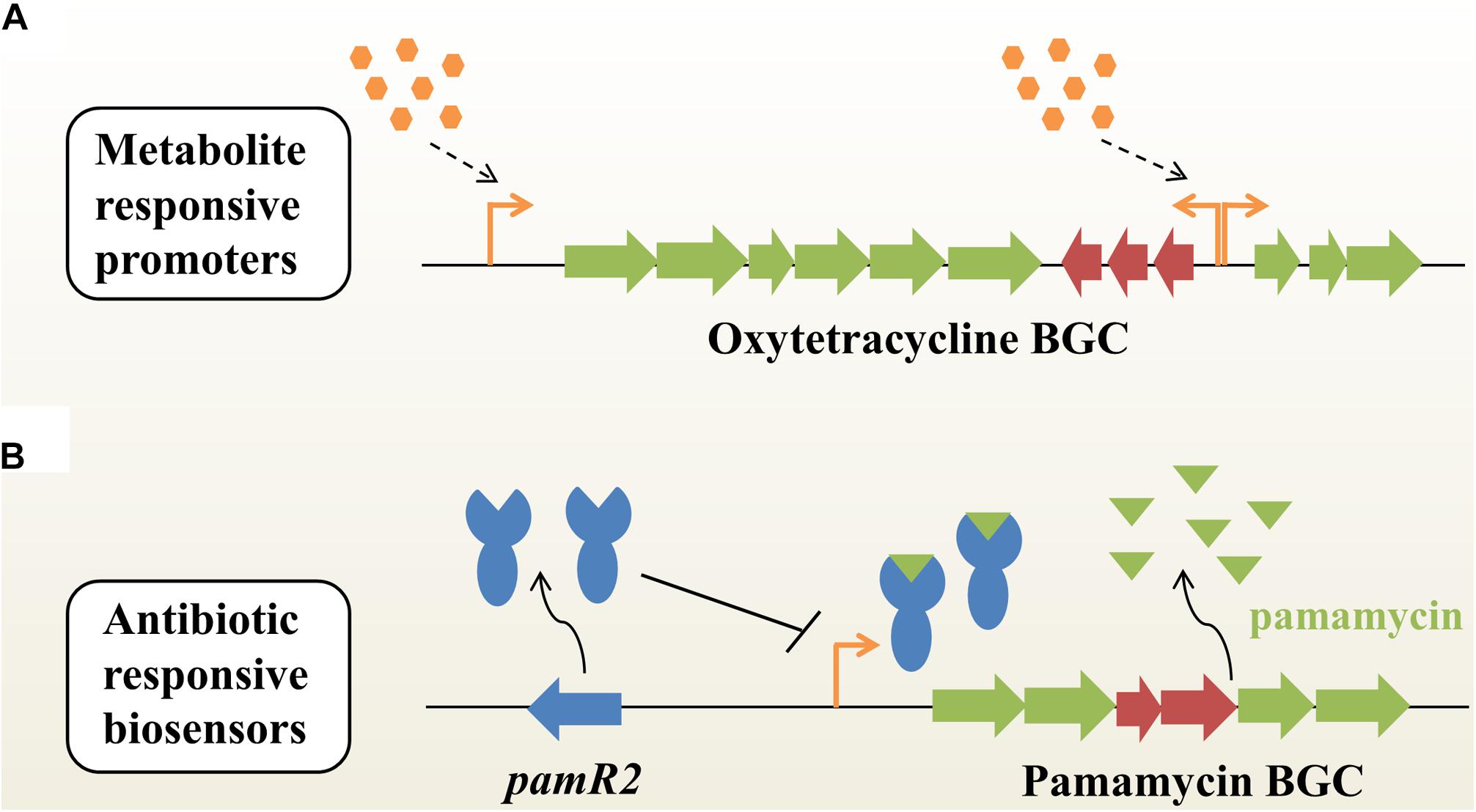
Figure 1. Dynamic pathway regulation balances flux between bacterial growth and antibiotic biosynthesis in actinobacteria. (A) Autoregulated fine-tuning of the expression of an antibiotic (e.g., oxytetracycline) BGC based on metabolite-responsive native promoters without specific transcription factors or additional inducers. BGC, biosynthetic gene cluster. (B) Antibiotic-responsive biosensors. This strategy is engineered from cluster-situated regulators (e.g., PamR2) that bind to their associated promoters upon interaction with the corresponding secondary metabolites (e.g., pamamycins). Placing antibiotic biosynthetic genes under such promoters enables antibiotic dependent gene expression.
Natural Product-Specific Biosensors
Genetically encoded biosensors are a kind of important synthetic biology tool for real-time detection of intracellular metabolites for specific readouts (Zhang et al., 2015). A typical biosensor is composed of three modules: (i) a signal input module, such as transcription factors (TFs) or riboswitches; (ii) a regulatory module, such as TF-dependent promoters; (iii) a signal output module, such as reporter genes (Mahr and Frunzke, 2016; Lim et al., 2018). Until now, three major categories of biosensors - fluorescence resonance energy transfer (FRET)-based sensors, TF-based sensors and riboswitches – have been widely used in metabolic engineering of microbial hosts (Zhang et al., 2015; Lim et al., 2018). As a kind of natural sensory protein, a classical TF has evolved to contain a ligand-binding domain for responding to environmental changes and a DNA-binding domain for regulating gene expression. Interestingly, many NP BGCs encode cluster-situated regulators (CSRs) in actinobacteria, such as TetR-like regulators and Streptomyces antibiotic regulatory proteins (SARPs; Liu et al., 2013; Romero-Rodriguez et al., 2015). These CSRs can be used to dynamically detect developmental state, population density or other environmental changes, and thus determine the onset and production levels of secondary metabolites in actinobacteria. Recently, Rebets et al. (2018) developed a new antibiotic-specific whole-cell biosensor based on a TetR-like repressor for the development of antibiotic-overproducing strains (Figure 1B). Briefly, the highly active macrodiolide antibiotic pamamycins BGC encodes a transporter PamW and a corresponding transcriptional repressor PamR2, which can be deactivated by binding to pamamycins (Rebets et al., 2015). Generally, PamW expression is controlled by PamR2 at low level of pamamycins. However, at very high levels of pamamycins, the native PamR2-based biosensors will reach the detection limit and cannot effectively regulate PamW expression (Rebets et al., 2018). The pamW promoter was directly used to control expression of the kanamycin resistance gene, thus generating the resistance-based G0 whole-cell biosensor system. After UV-induced mutagenesis, strains resistant to a high-concentration of kanamycin showed a significant increase in pamamycins production (up to 15–16 mg/L). Of note is the fact that the native pamamycins-responsive biosensor showed limited operating and dynamic ranges for further applications. To overcome this obstacle, the G1 pamamycins biosensor was further developed by combining different promoters, varying the number and position of operators, as well as using diverse reporter genes. Using the new biosensor with higher sensitivity, three mutated strains were obtained, which could produce up to 30 mg/L of pamamycins. Furthermore, to overcome the low detection limit of the G1 biosensor, the binding affinity of PamR2-pamamycins was also efficiently decreased by designing a panel of PamR2 mutations. As expected, the G2 biosensor showed a better operating dynamic range than that of the G1 biosensor (Rebets et al., 2018). In fact, at least 17% of NP BGCs encode TetR-like regulators and putative transporters simultaneously, which provides possibilities for the development of diverse antibiotic-responsive biosensors (Rebets et al., 2018). With the aid of a metabolite-responsive biosensor system, it will become very convenient to assess the functional expression of NP BGCs after random mutagenesis, genetic manipulation or exposure to various cultivation conditions. In the future, TF-based biosensors will hold great promise for accelerating cell factory development for pharmaceutical production and the activation of silent BGCs for the discovery of novel compounds in actinobacteria (Kim et al., 2016; Zarins-Tutt et al., 2016; Sun et al., 2017).
Multiplex Site-Specific Genome Engineering for NPs Overproduction
In the last three decades, due to their broad host specificities, SSR systems have been widely applied to strain improvements, combinatorial biosynthesis, and heterologous expression of the entire BGCs in Streptomyces, Actinoplanes and other industrial actinobacteria (Baltz, 2012; Stark, 2017; Merrick et al., 2018). To date, at least ten SSR systems have been identified in actinobacteria, which are derived from bacteriophage CBG73463, R4, SV1, TG1, VWB, ΦBT1, ΦC31, ΦJoe, ΦK38-1 or Φ1/6 (Baltz, 2012; Fogg et al., 2017; Li et al., 2017a; Kormanec et al., 2019). In particular, ΦBT1, ΦC31 and TG1 systems have been widely used for cell factory development via multi-copy amplification of target genes or BGCs (Baltz, 2012). For instance, production titers of the antibiotic goadsporin were increased by 2.3-fold by a step-by-step introduction of two extra copies of the goadsporin BGC based on ΦC31 and TG1 integration systems (Haginaka et al., 2014). However, this strategy requires repeated rounds of conjugal transfer and is limited by the number of selection markers. In 2011, Murakami and coworkers demonstrated that the ZouA-RsA/B-mediated recombination system could be used to achieve tandem amplification of the ACT gene cluster with 4–12 copies, resulting in a 20-fold increase in ACT production (Murakami et al., 2011). However, the engineered strains may be genetically unstable in the absence of antibiotic selection due to the presence of tandem amplification. In addition, the method requires the introduction of the ZouA-RsA/B system into both flanks of target BGCs in advance via two-round conjugal transfer before BGC amplification, which is time-consuming and labor-intensive. To address these limitations, our group has recently developed two novel enabling technologies for multi-locus chromosomal integration of target genes or BGCs, including MSGE and aMSGE (Li et al., 2017b, 2019b).
MSGE: Multiplex Site-Specific Genome Engineering Based on the “One Integrase-Multiple attB Sites” Concept
In the actinomycetal genome, there is typically no, or only one, native attB site for each SSR system (Baltz, 2012). In theory, one-step, multi-copy integration of target genes or BGCs could be achieved by introducing multiple artificial attB sites into the host chromosome in advance. Based on this “one integrase-multiple attB sites” concept, an innovative approach MSGE was developed for discrete BGC amplification (Figure 2A; Li et al., 2017b). Using the high-efficiency CRISPR/Cas9 genome editing method (Huang et al., 2015), we sequentially introduced three ΦC31 and two ΦBT1 attB sites into the genomic loci of deleted, non-target secondary metabolite BGCs in the industrial strain Streptomyces pristinaespiralis. Then, five extra copies of the pristinamycin II BGC were integrated into the modified chromosome in two steps using the ΦC31 and ΦBT1 compatible integration systems, which led to significantly improved PII titers (Li et al., 2015, 2017b). Importantly, the novel strategy was also extended to develop a series of powerful Streptomyces coelicolor heterologous expression hosts. Up to four copies of the chloramphenicol or YM-216391 BGCs were simultaneously integrated into these new chassis strains, thus resulting in increased production titers (Li et al., 2017b). Using the highly effective heterologous expression system, YM-216391 BGC was engineered to generate aurantizolicin and a hybrid compound 3, which exhibits significantly increased antitumor activity (Pei et al., 2018). Similarly, Myronovskyi et al. (2018) also constructed a panel of cluster-free, powerful Streptomyces albus chassis strains based on the “one integrase-multiple attB sites” concept. The production titers of a variety of bioactive compounds, including aloesaponarin II, cinnamycin, didesmethylmensacarcin and griseorhodin A, were significantly enhanced in these heterologous expression superhosts (Myronovskyi et al., 2018). It is worth noting that compared with the ZouA-RsA/B recombination system, MSGE-based engineered strains would be genetically stable in the absence of antibiotic selection due to discrete, site-specific integration of target BGCs (Li et al., 2017b). However, when the MSGE method is used to amplify target BGCs with multiple copies in a single step, there is an upper limit to the number of integrated BGCs. For example, we found that the chloramphenicol or YM-216391 BGCs could be simultaneously inserted into S. coelicolor with up to 4 copies. The possible reason is that the high-order amplification of BGCs or the accumulation of target products places an excess burden on bacterial growth (Li et al., 2017b). In the near future, we believe that these versatile Streptomyces hosts will greatly accelerate (meta)genomic mining and combinatorial biosynthesis studies for novel bioactive NP discovery (Li et al., 2017a; Liu et al., 2018; Zhang et al., 2019).
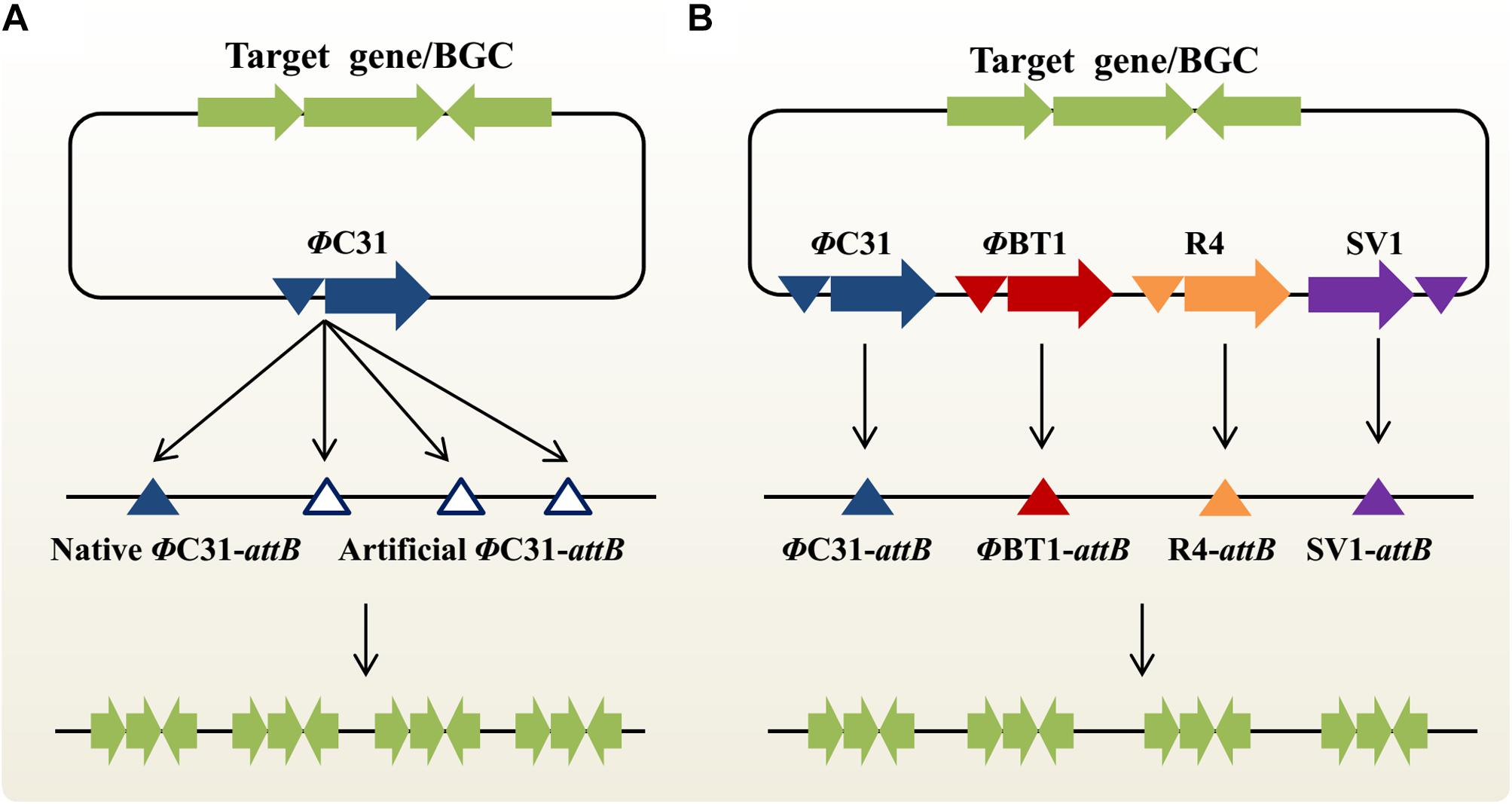
Figure 2. Two emerging synthetic biology approaches for multi-copy integration of target genes or natural products BGCs in actinobacteria. (A) Multiplex site-specific genome engineering (MSGE) for discrete amplification of target genes or BGCs. This method is based on the “one integrase-multiple attB sites” concept. The blue and blank triangles represent the native and artificial ΦC31 attB sites, respectively. (B) Advanced multiplex site-specific genome engineering (aMSGE) for multi-locus chromosomal integration of target genes or BGCs. This method is based on the “multiple integrases-multiple attB sites” concept. Particularly, these discrete attB sites are naturally occurring in the actinomycetal genomes. BGC, biosynthetic gene cluster.
aMSGE: Advanced Multiplex Site-Specific Genome Engineering With Orthogonal Modular Recombinases
Although the MSGE method could be widely used to construct a variety of suitable chassis organisms for NP discovery and overproduction, two major limitations remains to be addressed. On the one hand, it will be difficult or even impossible to initially insert foreign attB sites into the chromosomes of genetically intractable industrial actinobacteria due to the lack of available replicative plasmids and also their low homologous recombination capability, such as spinosad-producing Saccharopolyspora spinosa and epoxomicin-producing Goodfellowiella coeruleoviolacea. On the other hand, repeated introduction of multiple artificial attB sites is still time-consuming and labor-intensive, especially for the slow-growing actinobacteria. To overcome these bottlenecks, our group recently developed an advanced MSGE method (aMSGE) based on the “multiple integrases-multiple attB sites” concept (Figure 2B; Li et al., 2019b). In this improved method, native attB sites of different orthogonal SSR systems in the actinomycetal genome are simultaneously applied to multi-copy integration of target genes or BGCs, rather than introducing foreign attB sites into the host chromosome. Accordingly, a plug-and-play amplification toolkit was designed and constructed, which contains 27 modular recombination plasmids with single or multiple SSR systems. Using this innovative technique, we achieved a high-efficiency introduction of the 5-oxomilbemycin A3/A4 BGC into the parental strain Streptomyces hygroscopicus with up to four extra copies, thus resulting in a significant increase in the production titers of 5-oxomilbemycin A3/A4 (Li et al., 2019b). Compared with previously developed metabolic engineering tools, the aMSGE method doesn’t require the introduction of any genetic modifications before target gene or BGC amplification, which will considerably simplify and accelerate efforts to facilitate NP discovery and overproduction. The whole process for BGC amplification takes only ∼18 days for the construction of high-yield engineered strains (e.g., growth period is ∼6 days). More importantly, the aMSGE method could be applicable to genetically intractable actinobacteria for strain improvements. Given that SSR systems are widely distributed in a variety of microorganisms (Fogg et al., 2014; Stark, 2017), our newly developed methodology should be widely extended to establish more efficient industrial platforms for overproducing valuable chemicals and drugs. However, the aMSGE method could not be used for BGC amplification in actinobacteria without native attB sites.
Rational Pathway Refactoring of NP Biosynthesis
Generally, NP biosynthesis is under the control of highly complicated transcriptional, translational and metabolic regulation, which hampers the ability of systematic BGC engineering to maximize biosynthetic efficiency (Liu et al., 2013). Pathway refactoring provides an effective synthetic biological approach to decouple gene expression from complex native regulation and to achieve precise control of metabolite production by redesigning target BGCs in versatile surrogate hosts (Tan and Liu, 2017; Niu and Li, 2019). The key steps of BGC refactoring are to develop a set of well-characterized genetic control elements and high-efficiency DNA assembly methods (Figure 3).
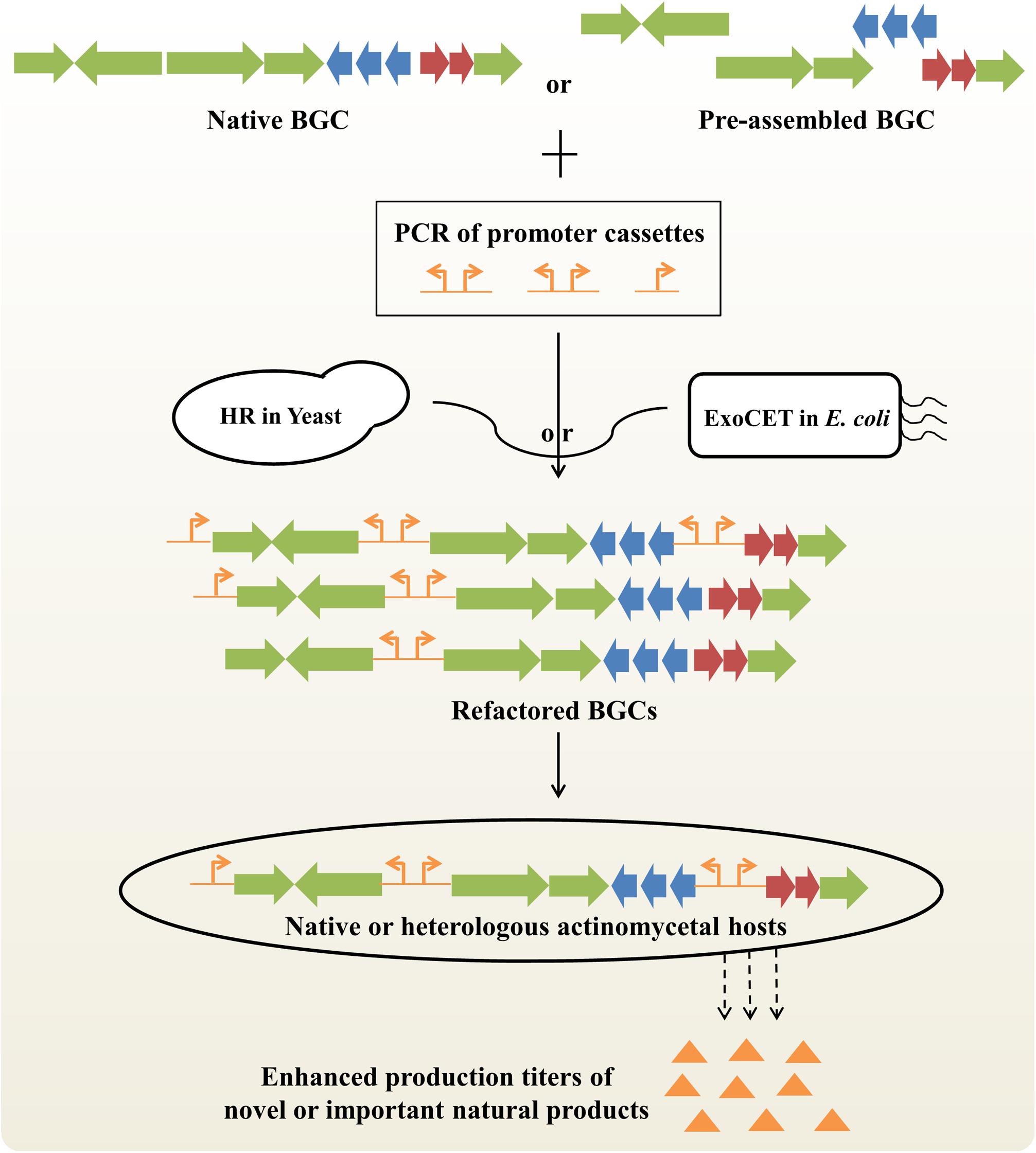
Figure 3. Improved production titers of novel or important natural products by BGC refactoring strategy. Native BGCs can be obtained by in vivo or in vitro BGC cloning/assembly strategies. Preassembled BGC can be obtained by PCR amplification or CRISPR-mediated in vitro digestion of native BGCs. Refactored BGCs can be obtained by partial or complete replacement of native promoters with artificial promoters based on homologous recombination (HR) in S. cerevisiae or exonucleases combined with RecET recombination (ExoCET) in E. coli. Finally, different refactored BGCs will be integrated into native or heterologous actinomycetal hosts for activating silent BGCs or enhancing production titers of clinically important drugs. BGC, biosynthetic gene cluster.
In the last ten years, a series of synthetic regulatory elements have been identified to precisely regulate gene expression with wide dynamic ranges in actinobacteria, including constitutive or inducible promoters, ribosomal binding sites (RBSs), terminators and protein degradation tags (Myronovskyi and Luzhetskyy, 2016; Horball et al., 2018; Ji et al., 2019). For instance, the expression levels of 200 native or synthetic promoters and 200 artificial RBSs were systematically quantitated, which provides a universal toolbox of synthetic modular regulatory elements for the scalable and cost-effective optimization of NP biosynthetic pathway in different Streptomyces (Bai et al., 2015).
Next, a variety of in vitro or in vivo DNA assembly methods have been established to reconstruct target BGCs for diverse applications (Li et al., 2017a; Zhang et al., 2019). On the one hand, a panel of in vivo DNA assembly methods for single-marker or marker-free multiplexed promoter engineering of large BGCs have been developed on the basis of powerful homologous recombination capacity in S. cerevisiae, including DNA assembler (Shao et al., 2011), mCRISTAR (multiplexed Cas9-transformation-associated recombination) (Kang et al., 2016) and miCRISTAR (multiplexed in vitro Cas9-transformation-associated recombination) (Kim et al., 2019). For example, using the miCRISTAR strategy, the activation of a silent BGC led to the characterization of two bacterial cyclic sesterterpenes atolypene A and B (Kim et al., 2019). In another study, yeast-mediated construction of a riboswitch-controlled pathway achieved a 120-fold increase in bottromycin productivity (Eyles et al., 2018). Intriguingly, multiple new bottromycin-related metabolites were also generated by using high-efficiency, flexible BGC modifications. On the other hand, a range of in vitro DNA assembly methods suitable for pathway refactoring have been developed, including modified Gibson assembly (Li et al., 2015), MASTER (methylation-assisted tailorable ends rational) ligation (Chen et al., 2013) and SLIC (Sequence- and Ligation-Independent Cloning) (D’Agostino and Gulder, 2018). Recently, an innovative DNA assembly method, ExoCET (Exonuclease Combined with RecET recombination), was also developed for large-size, multi-operon BGC refactoring (Song et al., 2018; Wang et al., 2018). The artificial 79-kb spinosad BGC with 7 artificial operons under the control of strong constitutive promoters achieved a 328-fold enhanced spinosad production compared to the native BGC (Song et al., 2018). As a simple and robust genetic platform, BGC refactoring will have potentially broad applications in combinatorial biosynthesis and antibiotic overproduction, as well as high-throughput activation of silent BGCs from either metagenomes or uncultured actinobacteria.
Construction of Genome-Reduced Streptomyces Hosts for NPs Discovery and Overproduction
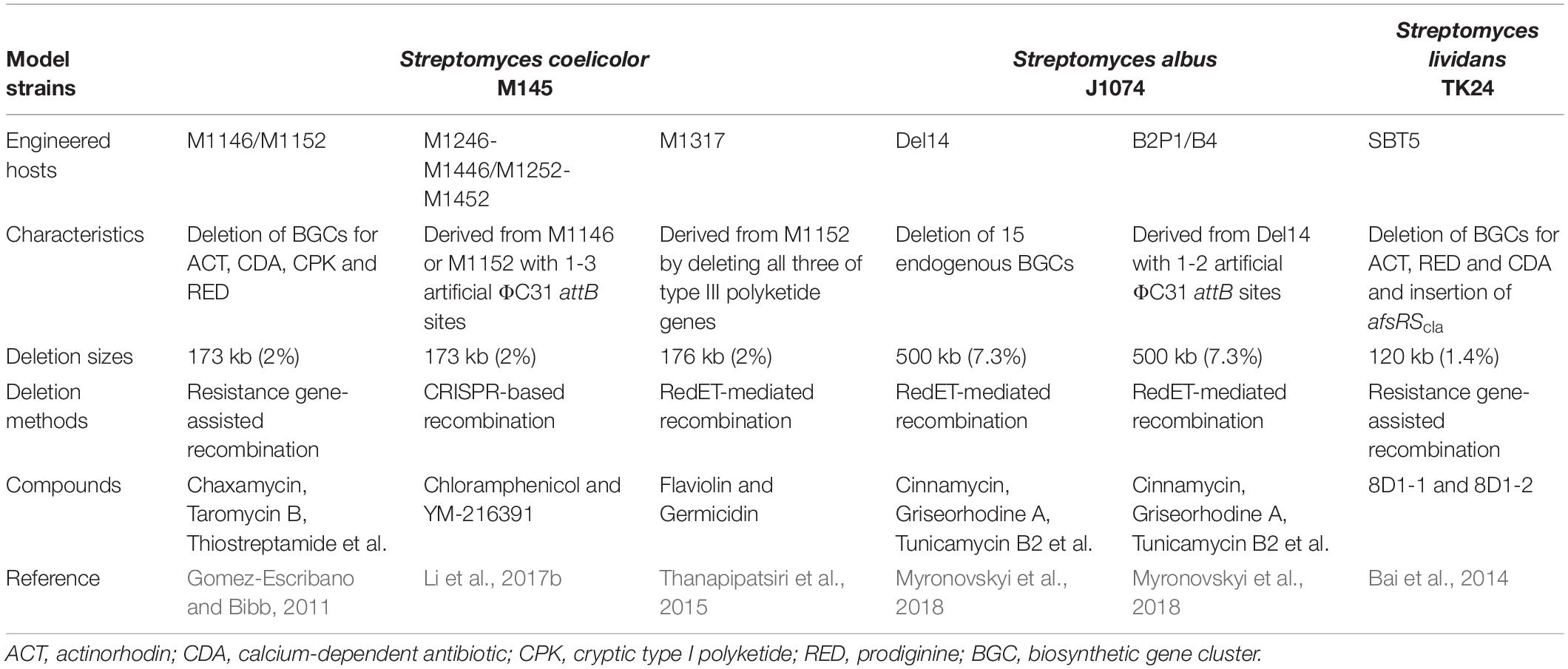
Streptomyces species are capable of producing a wide range of secondary metabolites, such as polyketides, non-ribosomal peptides and terpenes, and possess the unique metabolic background needed for heterologous expression of NP BGCs from actinobacteria or other bacteria with high-GC content (Liu et al., 2018; Nepal and Wang, 2019). Within the streptomycetes, S. coelicolor, S. albus J1074 and Streptomyces lividans have been widely used as surrogate hosts for NPs discovery and overproduction (Beites and Mendes, 2015; Liu et al., 2018). With the continuous advances of novel bioinformatics tools and genetic manipulation techniques (Zhang et al., 2016, 2019; Ziemert et al., 2016), a series of versatile Streptomyces chassis have been developed by deleting non-essential genomic regions, such as redundant BGCs, genome islands and insertion sequences (Liu et al., 2018; Huo et al., 2019; Table 1). Particularly, deletion of redundant BGCs are predicted to increase the supply of primary metabolite precursors, remove competing carbon and nitrogen sinks, and also facilitate the biosynthesis of heterologous BGCs. In addition, engineered strains without redundant BGCs will possess simple extracellular metabolite profiles for convenient structural characterization of novel bioactive compounds.
Along with the emergence of genome mining as a robust approach to discover novel drug leads, the development of reliable Streptomyces chassis for heterologous expression of cloned BGCs is becoming increasingly important (Huo et al., 2019; Nepal and Wang, 2019). As a model strain, S. coelicolor has been widely used to study the molecular regulation of antibiotic biosynthesis and morphological differentiation (Liu et al., 2013; Barka et al., 2016). In the last 10 years, a series of advanced genome editing tools and diverse synthetic regulatory elements have been developed for the optimization of heterologous pathways in S. coeliclor (Myronovskyi and Luzhetskyy, 2016; Alberti and Corre, 2019). In 2011, on the basis of the wild-type strain S. coelicolor M145, two versatile engineered hosts M1146 and M1152 were developed, which contained deletions in four BGCs, responsible for the biosynthesis of ACT, calcium-dependent antibiotic (CDA), cryptic type I polyketides (CPK) and prodiginine (RED), or in combination with a point mutation in rpoB (Gomez-Escribano and Bibb, 2011). The productivity of the chloramphenicol or congocidine BGCs was significantly increased compared with the parental strain (Gomez-Escribano and Bibb, 2011). Nowadays, M1146 and M1152 have been widely used to heterologously express different types of compounds from fastidious original producers or metagenomic DNA (Huo et al., 2019; Nepal and Wang, 2019). Similarly, a genome-reduced host S. lividans SBT5 was also developed by deleting BGCs responsible for ACT, CDA and RED as well as inserting the regulatory gene afsRScla (Bai et al., 2014). Furthermore, to specifically facilitate the study of Type III polyketides, an effective surrogate S. coelicolor host M1317 was constructed by removing all three endogenous Type III polyketide synthase (PKS) genes (gcs, srsA, rppA) on the basis of M1152, which could potentially increase precursor supply and prevent undesirable interference with expression of heterologous Type III PKS genes (Thanapipatsiri et al., 2015). As another important Streptomyces chassis, S. albus J1074 provides high success rates of heterologous BGC expression with rapid growth and high productivity (Nepal and Wang, 2019). Recently, on the basis of S. ablus J1074, 15 endogenous secondary metabolite BGCs were deleted to generate a powerful engineered host Del14 (Myronovskyi et al., 2018). The production yields of heterologously expressed metabolites in S. albus Del14 were higher than those in commonly used S. albus J1074 and S. coelicolor M1152. Furthermore, the authors also introduced 1-2 artificial ΦC31 attB sites into the genomes of S. albus Del14, thus resulting in two another versatile engineered hosts, B2P1 and B4. The two powerful chassis could be easily used for BGC amplification of up to four copies, thus efficiently improving fermentation levels of the encoded compounds (Myronovskyi et al., 2018).
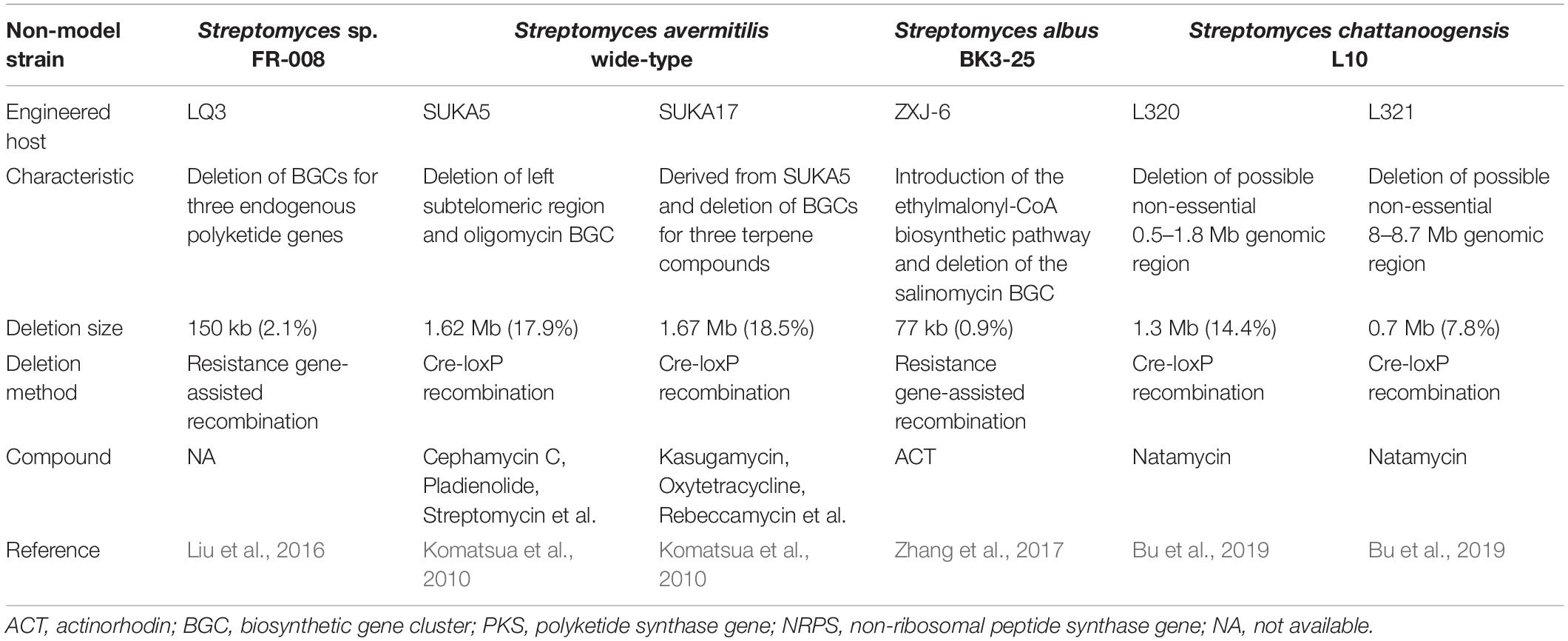
In addition to the construction of a model heterologous expression chassis, the genome-reduced strategy has been frequently used to develop non-model Streptomyces strains for the discovery and overproduction of important NPs (Table 2). Streptomyces sp. FR-008 is a fast-growing, potentially industrial production chassis, which produces the macrolide candicidin and other bioactive compounds. By deleting three endogenous polyketide genes, a mutant strain LQ3 was constructed with a stable and streamlined genome structure, which possibly allows for simple separation and purification of heterologously expressed compounds (Liu et al., 2016). As a producer of the anti-infective avermectin, Streptomyces avermitilis is already optimized to efficiently supply primary precursors. To construct a versatile industrial chassis for heterologous expression of secondary metabolite BGCs, a series of genome-reduced S. avermitilis mutants were obtained by deleting the left subtelomeric region (∼2 Mb) that corresponds to the more variable genome regions (Komatsua et al., 2010). Herein, the SUKA5 and SUKA17 strains were highlighted, which had genome reductions of 17.9 and 18.5%, respectively. S. avermitilis SUKA5 strain has the deletions of both oligomycin BGC and the left subtelomeric region, and the SUKA17 strain is a derivative of SUKA5 with the additional deletions of three terpene BGCs (Komatsua et al., 2010). The feasibility and superiority of these two engineered hosts has been widely confirmed by the efficient production of more than 20 exogenous secondary metabolite BGCs (Komatsu et al., 2013; Ikeda et al., 2014). Similarly, 1.3-Mb and 0.7-Mb possible non-essential genomic regions were deleted in the natamycin-producing industrial strian Streptomyces chattanoogensis L10 by Cre-loxP recombination system, respectively, thus generating two efficient chassis L320 and L321 for the production of valuable polyketides (Bu et al., 2019). In another study, a newly engineered host ZXJ-6 was developed based on a salinomycin-producing industrial strain, Streptomyces albus BK3-25, by deleting the salinomycin BGC and chromosomal integration of a three-gene cassette for the biosynthesis of ethylmalonyl-CoA. It was successfully used for the heterologous expression of polyketide BGCs, such as the ACT BGC from S. coelicolor (Zhang et al., 2017). In the future, the genome-reduced strategy could be widely implemented in a variety of industrial actinobacteria for the improved production of various bioactive compounds.
Concluding Remarks
Actinobacteria, especially bacteria from the genus Streptomyces, have long been employed as an important source of a wide range of novel bioactive small molecules (Barka et al., 2016; Newman and Cragg, 2016; Genilloud, 2017). To harness the production potential of actinobacteria, a variety of innovative metabolic engineering and synthetic biology strategies have been developed in the last 10 years, including dynamic metabolic regulation, BGC amplification, pathway refactoring and genome-minimized Streptomyces chassis (Beites and Mendes, 2015; Tan and Liu, 2017; Tan and Prather, 2017; Li et al., 2019a). We envision that NP discovery and development will be rapidly accelerated by the refactoring and amplification of whole biosynthetic pathways in combination with powerful heterologous expression platforms. Considering that a large number of BGCs are generally silent or expressed at very low levels, it is still important to develop diverse heterologous hosts and universal refactoring approaches to activate silent BGCs or boost production of secondary metabolites. Next, quorum sensing systems are widely distributed in actinobacteria (Polkade et al., 2016), which provide opportunities for bacterial growth-mediated dynamic regulation of metabolic pathways for enhanced production of target compounds. Furthermore, due to the complex regulation between primary and secondary metabolism, iterative application of the design-build-test-learn cycle will be necessary to overproduce different secondary metabolites (Liu et al., 2013; Kim et al., 2016; van der Heul et al., 2018). Finally, the advancements in the fields of genome sequencing, multi-omics and genome editing techniques are paving the way for systems metabolic engineering of industrial actinobacteria, including pathway engineering, regulatory circuit rewiring, host modification and enzyme engineering (Kim et al., 2016; Zhang et al., 2019). Given the requirement for high titers in commercial production, an integrated approach involving traditional mutagenesis screening and rational host/pathway engineering is required to systematically optimize the biosynthesis of target compounds. It is expected that these synthetic biology tools and metabolic engineering strategies presented in this review and future developments will play increasingly important roles in the discovery of novel drug leads, as well as yield improvement for large-scale manufacturing in actinobacteria.
Author Contributions
LL and XL wrote the manuscript. LL, WJ, and YL reviewed, edited, and approved its final version.
Funding
We gratefully acknowledge the financial support from the National Mega-project for Innovative Drugs (2018ZX09711001-006-012), the National Natural Science Foundation of China (31970083, 31770088, and 31630003), and the Science and Technology Commission of Shanghai Municipality (18ZR1446700).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alberti, F., and Corre, C. (2019). Editing streptomycete genomes in the CRISPR/Cas9 age. Nat. Prod. Rep. 36, 1237–1248. doi: 10.1039/c8np00081f
Bai, C., Zhang, Y., Zhao, X., Hu, Y., Xiang, S., Miao, J., et al. (2015). Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces. Proc. Natl. Acad. Sci. U.S.A. 112, 12181–12186. doi: 10.1073/pnas.1511027112
Bai, T.-l., Yu, Y.-f., Xu, Z., and Tao, M.-f. (2014). Construction of Streptomyces lividans SBT5 as an efficient heterologous expression host. J. Huazhong Agric. Univ. 33, 1–6.
Baltz, R. H. (2012). Streptomyces temperate bacteriophage integration systems for stable genetic engineering of actinomycetes (and other organisms). J. Ind. Microbiol. Biotechnol. 39, 661–672. doi: 10.1007/s10295-011-1069-6
Baltz, R. H. (2016). Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J. Ind. Microbiol. Biotechnol. 43, 343–370. doi: 10.1007/s10295-015-1682-x
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Klenk, H. P., et al. (2016). Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/MMBR.00019-15
Beites, T., and Mendes, M. V. (2015). Chassis optimization as a cornerstone for the application of synthetic biology based strategies in microbial secondary metabolism. Front. Microbiol. 6:906. doi: 10.3389/fmicb.2015.00906
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Bu, Q. T., Yu, P., Wang, J., Li, Z. Y., Chen, X. A., Mao, X. M., et al. (2019). Rational construction of genome-reduced and high-efficient industrial Streptomyces chassis based on multiple comparative genomic approaches. Microb. Cell Fact. 18:16. doi: 10.1186/s12934-019-1055-7
Chen, W. H., Qin, Z. J., Wang, J., and Zhao, G. P. (2013). The Master (methylation-assisted tailorable ends rational) ligation method for seamless DNA assembly. Nucleic Acids Res. 41:e93. doi: 10.1093/nar/gkt122
D’Agostino, P. M., and Gulder, T. M. (2018). Direct pathway cloning combined with sequence- and ligation-independent cloning for fast biosynthetic gene cluster refactoring and heterologous expression. ACS Synth. Biol. 7, 1702–1708. doi: 10.1021/acssynbio.8b00151
Dahl, R. H., Zhang, F., Alonso-Gutierrez, J., Baidoo, E., Batth, T. S., Redding-Johanson, A. M., et al. (2013). Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 31, 1039–1046. doi: 10.1038/nbt.2689
Eyles, T. H., Vior, N. M., and Truman, A. W. (2018). Rapid and robust yeast-mediated pathway refactoring generates multiple new bottromycin-related metabolites. ACS Synth. Biol. 7, 1211–1218. doi: 10.1021/acssynbio.8b00038
Fogg, P. C. M., Colloms, S., Rosser, S., Stark, M., and Smith, M. C. M. (2014). New applications for phage integrases. J. Mol. Biol. 426, 2703–2716. doi: 10.1016/j.jmb.2014.05.014
Fogg, P. C. M., Haley, J. A., Stark, W. M., and Smith, M. C. M. (2017). Genome integration and excision by a new Streptomyces bacteriophage, ϕJoe. Appl. Environ. Microbiol. 83:e2767-e16. doi: 10.1128/AEM.02767-16
Genilloud, O. (2017). Actinomycetes: still a source of novel antibiotics. Nat. Prod. Rep. 34, 1203–1232. doi: 10.1039/c7np00026j
Gomez-Escribano, J. P., and Bibb, M. J. (2011). Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 4, 207–215. doi: 10.1111/j.1751-7915.2010.00219.x
Gupta, A., Reizman, I. M. B., Reisch, C. R., and Prather, K. L. J. (2017). Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 35, 273–279. doi: 10.1038/nbt.3796
Haginaka, K., Asamizu, S., Ozaki, T., Igarashi, Y., Furumai, T., and Onaka, H. (2014). Genetic approaches to generate hyper-producing strains of goadsporin: the relationships between productivity and gene duplication in secondary metabolite biosynthesis. Biosci. Biotechnol. Biochem. 78, 394–399. doi: 10.1080/09168451.2014.885824
Horball, L., Siegl, T., and Luzhetskyy, A. (2018). A set of synthetic versatile genetic control elements for the efficient expression of genes in actinobacteria. Sci. Rep. 8:491. doi: 10.1038/s41598-017-18846-1
Huang, H., Zheng, G. S., Jiang, W. H., Hu, H. F., and Lu, Y. H. (2015). One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta. Biochim. Biophys. Sin 47, 231–243. doi: 10.1093/abbs/gmv007
Huo, L., Hug, J. J., Fu, C., Bian, X., Zhang, Y., and Muller, R. (2019). Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. doi: 10.1039/C8NP00091C [Epub ahead of print].
Ikeda, H., Shin-Ya, K., and Omura, S. (2014). Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J. Ind. Microbiol. Biotechnol. 41, 233–250. doi: 10.1007/s10295-013-1327-x
Jensen, P. R., Moore, B. S., and Fenical, W. (2015). The marine actinomycete genus Salinispora: a model organism for secondary metabolite discovery. Nat. Prod. Rep. 32, 738–751. doi: 10.1039/c4np00167b
Ji, C. H., Kim, H., and Kang, H. S. (2019). Synthetic inducible regulatory systemsoptimized for the modulation of secondary metabolite production in Streptomyces. ACS Synth. Biol. 8, 577–586. doi: 10.1021/acssynbio.9b00001
Kang, H. S., Charlop-Powers, Z., and Brady, S. F. (2016). Multiplexed CRISPR/Cas9-and TAR-mediated promoter engineering of natural product biosynthetic gene clusters in yeast. ACS Synth. Biol. 5, 1002–1010. doi: 10.1021/acssynbio.6b00080
Kim, H. U., Charusanti, P., Lee, S. Y., and Weber, T. (2016). Metabolic engineering with systems biology tools to optimize production of prokaryotic secondary metabolites. Nat. Prod. Rep. 33, 933–941. doi: 10.1039/c6np00019c
Kim, S. H., Lu, W. L., Ahmadi, M. K., Montiel, D., Ternei, M. A., and Brady, S. F. (2019). Atolypenes, tricyclic bacterial sesterterpenes discovered using a multiplexed in vitro Cas9-TAR gene cluster refactoring approach. ACS Synth. Biol. 8, 109–118. doi: 10.1021/acssynbio.8b00361
Komatsu, M., Komatsu, K., Koiwai, H., Yamada, Y., Kozone, I., Izumikawa, M., et al. (2013). Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth. Biol. 2, 384–396. doi: 10.1021/sb3001003
Komatsua, M., Uchiyama, T., Omura, S., Cane, D. E., and Ikeda, H. (2010). Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 107, 2646–2651. doi: 10.1073/pnas.0914833107
Kormanec, J., Rezuchova, B., Homerova, D., Csolleiova, D., Sevcikova, B., Novakova, R., et al. (2019). Recent achievements in the generation of stable genome alterations/mutations in species of the genus Streptomyces. Appl. Microbiol. Biotechnol. 103, 5463–5482. doi: 10.1007/s00253-019-09901-0
Li, L., Jiang, W., and Lu, Y. (2017a). New strategies and approaches for engineering biosynthetic gene clusters of microbial natural products. Biotechnol. Adv. 35, 936–949. doi: 10.1016/j.biotechadv.2017.03.007
Li, L., Zheng, G., Chen, J., Ge, M., Jiang, W., and Lu, Y. (2017b). Multiplexed site-specific genome engineering for overproducing bioactive secondary metabolites in actinomycetes. Metab. Eng. 40, 80–92. doi: 10.1016/j.ymben.2017.01.004
Li, L., Liu, X., Wei, K., Lu, Y., and Jiang, W. (2019a). Synthetic biology approaches for chromosomal integration of genes and pathways in industrial microbial systems. Biotechnol. Adv. 37, 730–745. doi: 10.1016/j.biotechadv.2019.04.002
Li, L., Wei, K., Liu, X., Wu, Y., Zheng, G., Chen, S., et al. (2019b). aMSGE: advanced multiplex site-specific genome engineering with orthogonal modular recombinases in actinomycetes. Metab. Eng. 52, 153–167. doi: 10.1016/j.ymben.2018.12.001
Li, L., Zhao, Y., Ruan, L., Yang, S., Ge, M., Jiang, W., et al. (2015). A stepwise increase in pristinamycin II biosynthesis by Streptomyces pristinaespiralis through combinatorial metabolic engineering. Metab. Eng. 29, 12–25. doi: 10.1016/j.ymben.2015.02.001
Li, S., Wang, J., Xiang, W., Yang, K., Li, Z., and Wang, W. (2018). An autoregulated fine-tuning strategy for titer improvement of secondary metabolites using native promoters in Streptomyces. ACS Synth. Biol. 7, 522–530. doi: 10.1021/acssynbio.7b00318
Lim, H. G., Jang, S., Seo, S. W., and Jung, G. Y. (2018). Design and optimization of genetically encoded biosensors for high-throughput screening of chemicals. Curr. Opin. Biotechnol. 54, 18–25. doi: 10.1016/j.copbio.2018.01.011
Liu, G., Chater, K. F., Chandra, G., Niu, G., and Tan, H. (2013). Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143. doi: 10.1128/MMBR.00054-12
Liu, Q., Xiao, L., Zhou, Y., Deng, K., Tan, G., Han, Y., et al. (2016). Development of Streptomyces sp. FR-008 as an emerging chassis. Synth. Syst. Biotechnol. 1, 207–214. doi: 10.1016/j.synbio.2016.07.002
Liu, R., Deng, Z. X., and Liu, T. G. (2018). Streptomyces species: ideal chassis for natural product discovery and overproduction. Metab. Eng. 50, 74–84. doi: 10.1016/j.ymben.2018.05.015
Mahr, R., and Frunzke, J. (2016). Transcription factor-based biosensors in biotechnology: current state and future prospects. Appl. Microbiol. Biotechnol. 100, 79–90. doi: 10.1007/s00253-015-7090-3
Merrick, C. A., Zhao, J., and Rosser, S. J. (2018). Serine integrases: advancing synthetic biology. ACS Synth. Biol. 7, 299–310. doi: 10.1021/acssynbio.7b00308
Murakami, T., Burian, J., Yanai, K., Bibb, M. J., and Thompson, C. J. (2011). A system for the targeted amplification of bacterial gene clusters multiplies antibiotic yield in Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 108, 16020–16025. doi: 10.1073/pnas.1108124108
Myronovskyi, M., and Luzhetskyy, A. (2016). Native and engineered promoters in natural product discovery. Nat. Prod. Rep. 33, 1006–1019. doi: 10.1039/c6np00002a
Myronovskyi, M., Rosenkranzer, B., Nadmid, S., Pujic, P., Normand, P., and Luzhetskyy, A. (2018). Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab. Eng. 49, 316–324. doi: 10.1016/j.ymben.2018.09.004
Nepal, K. K., and Wang, G. (2019). Streptomycetes: surrogate hosts for the genetic manipulation of biosynthetic gene clusters and production of natural products. Biotechnol. Adv. 37, 1–20. doi: 10.1016/j.biotechadv.2018.10.003
Newman, D. J., and Cragg, G. M. (2016). Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661. doi: 10.1021/acs.jnatprod.5b01055
Niu, G. (2018). Genomics-driven natural product discovery in actinomycetes. Trends Biotechnol. 36, 238–241. doi: 10.1016/j.tibtech.2017.10.009
Niu, G., and Li, W. (2019). Next-generation drug discovery to combat antimicrobial resistance. Trends Biochem. Sci. doi: 10.1016/j.tibs.2019.05.005 [Epub ahead of print].
Palazzotto, E., Tong, Y., Lee, S. Y., and Weber, T. (2019). Synthetic biology and metabolic engineering of actinomycetes for natural product discovery. Biotechnol. Adv. 37:107366. doi: 10.1016/j.biotechadv.2019.03.005
Pei, Z. F., Yang, M. J., Li, L., Jian, X. H., Yin, Y., Li, D. H., et al. (2018). Directed production of aurantizolicin and new members based on a YM-216391 biosynthetic system. Org. Biomol. Chem. 16, 9373–9376. doi: 10.1039/c8ob02665c
Polkade, A. V., Mantri, S. S., Patwekar, U. J., and Jangid, K. (2016). Quorum sensing: an under-explored phenomenon in the phylum actinobacteria. Front. Microbiol. 7:131. doi: 10.3389/fmicb.2016.00131
Rebets, Y., Brotz, E., Manderscheid, N., Tokovenko, B., Myronovskyi, M., Metz, P., et al. (2015). Insights into the pamamycin biosynthesis. Angew. Chem. Int. Ed. Engl. 54, 2280–2284. doi: 10.1002/anie.201408901
Rebets, Y., Schmelz, S., Gromyko, O., Tistechok, S., Petzke, L., Scrima, A., et al. (2018). Design, development and application of whole-cell based antibiotic-specific biosensor. Metab. Eng. 47, 263–270. doi: 10.1016/j.ymben.2018.03.019
Romero-Rodriguez, A., Robledo-Casados, I., and Sanchez, S. (2015). An overview on transcriptional regulators in Streptomyces. Biochim. Biophys. Acta. 1849, 1017–1039. doi: 10.1016/j.bbagrm.2015.06.007
Rutledge, P. J., and Challis, G. L. (2015). Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13, 509–523. doi: 10.1038/nrmicro3496
Shao, Z. Y., Luo, Y. Z., and Zhao, H. M. (2011). Rapid characterization and engineering of natural product biosynthetic pathways via DNA assembler. Mol. Biosyst. 7, 1056–1059. doi: 10.1039/c0mb00338g
Song, C., Luan, J., Cui, Q., Duan, Q., Li, Z., Gao, Y., et al. (2018). Enhanced heterologous spinosad production from a 79-kb synthetic multi-operon assembly. ACS Synth. Biol. 8, 137–147. doi: 10.1021/acssynbio.8b00402
Stark, W. M. (2017). Making serine integrases work for us. Curr. Opin. Microbiol. 38, 130–136. doi: 10.1016/j.mib.2017.04.006
Sun, Y. Q., Busche, T., Ruckert, C., Paulus, C., Rebets, Y., Novakova, R., et al. (2017). Development of a biosensor concept to detect the production of cluster-specific secondary metabolites. ACS Synth. Biol. 6, 1026–1033. doi: 10.1021/acssynbio.6b00353
Tan, G. Y., and Liu, T. (2017). Rational synthetic pathway refactoring of natural products biosynthesis in actinobacteria. Metab. Eng. 39, 228–236. doi: 10.1016/j.ymben.2016.12.006
Tan, S. Z., and Prather, K. L. J. (2017). Dynamic pathway regulation: recent advances and methods of construction. Curr. Opin. Chem. Biol. 41, 28–35. doi: 10.1016/j.cbpa.2017.10.004
Thanapipatsiri, A., Claesen, J., Gomez-Escribano, J. P., Bibb, M., and Thamchaipenet, A. (2015). A Streptomyces coelicolor host for the heterologous expression of Type III polyketide synthase genes. Microb. Cell Fact. 14:145. doi: 10.1186/s12934-015-0335-0
Tong, Y., Weber, T., and Lee, S. Y. (2018). CRISPR/Cas-based genome engineering in natural product discovery. Nat. Prod. Rep. 36, 1262–1280. doi: 10.1039/c8np00089a
van der Heul, H. U., Bilyk, B. L., Mcdowall, K. J., Seipke, R. F., and Van Wezel, G. P. (2018). Regulation of antibiotic production in actinobacteria: new perspectives from the post-genomic era. Nat. Prod. Rep. 35, 575–604. doi: 10.1039/c8np00012c
Wang, H., Li, Z., Jia, R., Yin, J., Li, A., Xia, L., et al. (2018). ExoCET: exonuclease in vitro assembly combined with RecET recombination for highly efficient direct DNA cloning from complex genomes. Nucleic Acids Res. 46:e28. doi: 10.1093/nar/gkx1249
Weber, T., Charusanti, P., Musiol-Kroll, E. M., Jiang, X. L., Tong, Y. J., Kim, H. U., et al. (2015). Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes. Trends Biotechnol. 33, 15–26. doi: 10.1016/j.tibtech.2014.10.009
Xu, Y., Willems, A., Au-Yeung, C., Tahlan, K., and Nodwell, J. R. (2012). A two-step mechanism for the activation of actinorhodin export and resistance in Streptomyces coelicolor. mBio 3, e191–e112. doi: 10.1128/mBio.00191-12
Zarins-Tutt, J. S., Barberi, T. T., Gao, H., Mearns-Spragg, A., Zhang, L., Newman, D. J., et al. (2016). Prospecting for new bacterial metabolites: a glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Nat. Prod. Rep. 33, 54–72. doi: 10.1039/c5np00111k
Zhang, F. Z., Carothers, J. M., and Keasling, J. D. (2012). Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol. 30, 354–359. doi: 10.1038/nbt.2149
Zhang, J., Jensen, M. K., and Keasling, J. D. (2015). Development of biosensors and their application in metabolic engineering. Curr. Opin. Chem. Biol. 28, 1–8. doi: 10.1016/j.cbpa.2015.05.013
Zhang, J. J., Tang, X., and Moore, B. S. (2019). Genetic platforms for heterologous expression of microbial natural products. Nat. Prod. Rep. 36, 1313–1332. doi: 10.1039/c9np00025a
Zhang, M. M., Wang, Y. J., Ang, E. L., and Zhao, H. M. (2016). Engineering microbial hosts for production of bacterial natural products. Nat. Prod. Rep. 33, 963–987. doi: 10.1039/c6np00017g
Zhang, X. J., Lu, C. Y., and Bai, L. Q. (2017). Conversion of the high-yield salinomycin producer Streptomyces albus BK3-25 into a surrogate host for polyketide production. Sci. China Life Sci. 60, 1000–1009. doi: 10.1007/s11427-017-9122-8
Keywords: actinobacteria, natural product, synthetic biology, dynamic regulation, BGC amplification, pathway refactoring, genome-minimized host
Citation: Li L, Liu X, Jiang W and Lu Y (2019) Recent Advances in Synthetic Biology Approaches to Optimize Production of Bioactive Natural Products in Actinobacteria. Front. Microbiol. 10:2467. doi: 10.3389/fmicb.2019.02467
Received: 01 September 2019; Accepted: 15 October 2019;
Published: 05 November 2019.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Linquan Bai, Shanghai Jiao Tong University, ChinaBertolt Gust, University of Tübingen, Germany
Copyright © 2019 Li, Liu, Jiang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weihong Jiang, d2hqaWFuZ0BzaWJzLmFjLmNu; Yinhua Lu, eWhsdUBzaG51LmVkdS5jbg==; eWhsdUBzaG51LmVkdQ==
†These authors have contributed equally to this work
 Lei Li1†
Lei Li1† Weihong Jiang
Weihong Jiang