- 1Department of Urology, Taichung Tzu Chi Hospital, The Buddhist Tzu Chi Medical Foundation, Taichung, Taiwan
- 2School of Post-Baccalaureate Chinese Medicine, Tzu Chi University, Hualien, Taiwan
- 3Institute of Biochemistry and Molecular Biology, National Yang-Ming University, Taipei, Taiwan
- 4Department of Biological Science and Technology, China University of Science and Technology, Taipei, Taiwan
- 5Biomedical Industry Ph.D. Program, National Yang-Ming University, Taipei, Taiwan
- 6School of Chinese Medicine, China Medical University, Taichung, Taiwan
Fumarate nitrate reduction regulator (FNR) is a direct oxygen-responsive transcriptional regulator containing an iron-sulfur (Fe–S) cluster. During anaerobic growth, the [4Fe–4S] cluster in FNR (holo-FNR) binds specifically to DNA, whereas exposure to oxygen results in the loss of its DNA-binding activity via oxidation of the [4Fe–4S] cluster. In this study, we aimed to investigate the role of FNR in regulation of capsular polysaccharide (CPS) biosynthesis, serum resistance, and anti-phagocytosis of K. pneumoniae. We found that the CPS amount in K. pneumoniae increased in anaerobic conditions, compared to that in aerobic conditions. An fnr deletion mutant and a site-directed mutant (fnr3CA), with the three cysteines (C20, C23, and C29) replaced with alanines to mimic an FNR lacking the [4Fe-4S] cluster, showed marked increase in CPS amount under anaerobic conditions. A promoter-reporter assay and qRT-PCR confirmed that the transcription of the cps genes was repressed by holo-FNR. In addition, we found that holo-FNR could repress the transcription of rmpA and rmpA2, encoding cps transcriptional activators. Deletion of rmpA or rmpA2 in the Δfnr strain reduced CPS biosynthesis, suggesting that RmpA and RmpA2 participated in the holo-FNR–mediated repression of cps transcription, thereby regulating the CPS amount, serum resistance, and anti-phagocytosis. Taken together, our results provided evidence that RmpA and RmpA2 participated in the holo-FNR–mediated repression of CPS biosynthesis, and resistance to the host defense in response to oxygen availability.
Introduction
Klebsiella pneumoniae is a gram-negative facultative anaerobe that causes both nosocomial and community-acquired infections, including pneumonia, bacteremia, septicemia, and urinary and respiratory tract infections particularly in patients with underlying diseases (Podschun and Ullmann, 1998). In Asian countries, especially Taiwan and Korea, K. pneumoniae is the predominant pathogen responsible for pyogenic liver abscesses in diabetic patients (Han, 1995; Lau et al., 2000; Yang et al., 2009). In recent years, reports of KLA and the spread of hypervirulent strains have increased in western countries (Lederman and Crum, 2005). Furthermore, several K. pneumoniae strains producing ESBL and/or AmpC β-lactamase have been widely identified, thereby increasing the difficulty in clinical treatments (Alicino et al., 2015; Ma et al., 2015; Pitout et al., 2015; Lin et al., 2016). Additionally, hypervirulent K. pneumoniae strains with carbapenem resistance were reported in China (Gu et al., 2017; Zhan et al., 2017). These strains represent a critical threat for human health.
Klebsiella pneumoniae, like many facultative anaerobes in the Enterobacteriaceae family, grow under either aerobic or anaerobic conditions (such as the anaerobic environment of the human colon, micro-aerobic environment of different tissues, and the aerobic external environment). Thus, sensing and responding to oxygen availability is essential for the competitiveness of these bacteria and their survival in vivo. Oxygen plays a critical role in bacteria–host interaction. In the host innate immune system, oxygen is required for the production of reactive oxygen species and NO for defense against bacterial infections (Green et al., 2014). In many facultative anaerobes, including Escherichia coli, Salmonella enterica, Shigella spp., and Pseudomonas aeruginosa., oxygen availability has been reported to modulate the expression of genes involved in metabolic adaption and virulence during infection (Green et al., 2014). However, the effect of oxygen availability on the expression of virulence factors in K. pneumoniae remains largely unknown.
Fumarate Nitrate Reduction regulator (FNR) is a direct oxygen-responsive transcriptional regulator in bacteria. It contains an [4Fe–4S] cluster in the N-terminal sensory domain to modulate the C-terminal DNA binding domain in response to oxygen availability. However, the N-terminal sensory domain of FNR contains four cysteine residues (Cys20, Cys23, Cys29, and Cys122) which are required for coordination with the [4Fe–4S] cluster (Kiley and Beinert, 1998). Under anaerobic conditions, the holo-FNR binds specifically to DNA to form a dimer, whereas exposure to oxygen causes oxidation of the [4Fe–4S] cluster, thereby resulting in the loss of its DNA-binding activity (Melville and Gunsalus, 1996). In E. coli, a metal-free and apo-FNR was formed under aerobic conditions and was degraded by the ATP-dependent protease ClpXP (Mettert and Kiley, 2005). In many bacterial pathogens, FNR not only regulates anaerobic metabolism but also triggers virulence gene expression during infection, such as those involved in iron transport, toxin production, and type III secretion system (Carpenter and Payne, 2014; Green et al., 2014). However, the regulatory role of FNR in K. pneumoniae pathogenesis remains unclear.
Multiple virulence factors, including CPS, lipopolysaccharides, fimbriae, iron-acquisition system, porins, and antibiotic resistance factors, have been identified to be involved in K. pneumoniae infection. These virulence factors are processed or embedded in the cell envelope, thus allowing bacteria to internalize nutrients and adhere to diverse surfaces or niches within the human host for successful infection (Wu and Fives-Taylor, 2001; Kawai et al., 2011). Of these virulence factors, CPS is considered the major determinant of K. pneumoniae pathogenesis (Sahly et al., 2000; Lin et al., 2004). Acapsular K. pneumoniae strains showed less virulence in mouse infection models (Lawlor et al., 2005; Paczosa and Mecsas, 2016). Furthermore, hypervirulent K. pneumoniae isolates often carry heavy CPS loads, which could protect the bacteria from phagocytosis and death due to serum factors (Sahly et al., 2000; Lin et al., 2004). The degree of mucoidy has also been positively correlated with successful establishment of infection (Lin et al., 2004; Regueiro et al., 2006). Therefore, stringent control of CPS biosynthesis to encounter the various environmental stimuli is essential for successful K. pneumonia infection. Previously, we showed that CPS production was affected by iron availability. The coordination of Fur, the CPS regulators RmpA and RcsA, small RNA RyhB, and iron-sulfur cluster regulator (IscR) for regulating CPS biosynthesis was demonstrated to be a crucial mechanism in response to iron availability (Lin et al., 2010; Huang et al., 2012; Wu et al., 2014). In addition, we also found that environmental glucose stimulated CPS production, which was regulated by cAMP signaling pathway (Lin et al., 2013).
A deeper understanding of K. pneumoniae virulence factor expression during infection holds promise for future development of the intervening targets. In this study, we aim to investigate the role of FNR in the regulation of CPS biosynthesis, serum resistance, and anti-phagocytosis of K. pneumoniae under anaerobic conditions.
Materials and Methods
Bacterial Strains, Plasmids, and Media
Bacterial strains and plasmids and the primers used in this study are listed in Tables 1, 2 respectively. Bacteria were routinely cultured at 37°C in LB medium supplemented with the appropriate antibiotics. The antibiotics used include ampicillin (100 μg/mL), kanamycin (25 μg/mL), and streptomycin (500 μg/mL). The aerobic bacteria were cultured in aerated LB broth with agitation (200 rpm) at 37°C for 16 h. The anaerobic bacteria were statically cultured in LB broth at 37°C in an airtight box, filled with 10% CO2 and 90% N2 for 16 h.
Construction of fnr-Deficient Mutants
Specific fnr deletion in K. pneumoniae CG43S3 was performed using the allelic exchange strategy described previously (Lai et al., 2003). In brief, the upstream and downstream flanking regions of fnr were cloned into the suicide vector pKAS46 (Skorupski and Taylor, 1996), a suicide vector containing rpsL, which allows positive selection with streptomycin for vector loss. The resulting plasmid was then mobilized from E. coli S17-1λpir (Miller and Mekalanos, 1988) to K. pneumoniae CG43S3 or CG43S3-derived strains by conjugation. The transconjugants, with the plasmid integrated into the chromosome via homologous recombination, were selected using M9 agar plates containing ampicillin and kanamycin. Several of the colonies were cultured at 37°C in LB broth supplemented with 500 μg/mL streptomycin to the log phase and then spread onto an LB agar plate containing 500 μg/mL streptomycin. The streptomycin-resistant and kanamycin-sensitive colonies were selected, and the deletion was verified by PCR. The resulting K. pneumoniae mutants are listed in Table 1.
Construction of a K. pneumoniae fnr3CA Mutant
A DNA fragment carrying fnr and approximately 1000-bp adjacent regions on either side was amplified by PCR using the primer pairs GT321/GT322 (Table 2) and cloned into the yT&A vector. The resulting plasmid was used as the template for inverse PCR with the primer pair GT315/GT316 (Table 2) to generate a mutant fnr allele encoding the C20A, C23A, and C29A mutations. The recovered PCR product was treated with DpnI for 2 h, subjected to T4 polynucleotide kinase treatment, and self-ligated with T4 DNA ligase. Subsequently, the mutant allele of fnr was subcloned into pKAS46, and the cloning was confirmed by DNA sequencing. Then, the plasmid was mobilized from E. coli S17-1 λpir to the K. pneumoniae Δfnr strain by conjugation, and the subsequent selection was performed as described above.
Construction of the pfnr Complementation Plasmid
To obtain the complementation plasmid (pfnr), a DNA fragment containing the promoter and coding sequence of fnr was amplified by PCR using the primer pair GT311/GT312 (Table 2) and cloned into the pACYC184 shuttle vector. The ligation product was transformed into E. coli DH5α.
Extraction and Quantification of CPS
CPS was extracted and quantified as previously described (Domenico et al., 1989). The glucuronic acid content, representative of the amount of K. pneumoniae K2 CPS, was determined from a standard curve of glucuronic acid (Sigma-Aldrich) and expressed as micrograms per 109 c.f.u. (Blumenkrantz and Asboe-Hansen, 1973).
Construction of the rmpA and rmpA2 Promoter-Reporter Plasmids
To obtain the promoter-reporter plasmids (prmpAZ15 and prmpA2Z15), the DNA fragments containing the promoter sequence of rmpA and rmpA2 were amplified by PCR using the primer pairs GT411/GT412 and GT413/414 respectively (Table 2), and then subcloned into the placZ15 vector.
Measurement of Promoter Activity
The promoter-reporter plasmids, pOrf12, pOrf315, pOrf1617, prmpAZ15, and prmpA2Z15-1 were individually transferred into K. pneumoniae indicated strains by electroporation. The β-galactosidase activity of bacteria when cultured in LB medium under the indicated condition was measured as previously described (Balsalobre et al., 2006).
Quantitative Reverse-Transcription PCR (qRT-PCR)
Total RNA was isolated from bacterial cells cultured overnight under anaerobic condition by using the RNeasy midi-column (QIAGEN) according to the manufacturer’s instructions. RNA was treated with RNase-free DNase I (Roche) to eliminate DNA contamination. Then, 100 ng RNA was reverse-transcribed using the Transcriptor First Strand cDNA Synthesis Kit (Roche) with random primers. qRT-PCR was performed in a Roche LightCycler® 1.5 Instrument using LightCycler TaqMan Master (Roche). Primers and probes were designed for selected target sequences using Universal ProbeLibrary Assay Design Center (Roche Applied Science) and are listed in Table 2. Data were analyzed using the real time PCR software of the Roche LightCycler® 1.5 Instrument. Relative gene expressions were quantified using the comparative threshold cycle 2–ΔΔCT method with 23S rRNA as the endogenous reference.
Bacterial Survival in Serum
Normal human serum, pooled from healthy volunteers, was divided into equal volumes and stored at −70°C before use. Bacterial survival in the serum was determined as previously described (Lai et al., 2003). Briefly, 1 mL of the bacterial cells cultured overnight under anaerobic conditions was washed twice using PBS and resuspended in 1 mL PBS. A mixture containing 250 μL of the cell suspension and 750 μL of pooled human serum was statically incubated at 37°C for 15 min. The number of viable bacteria was then determined by colony counting. The percentage of survival rate was expressed as the number of viable bacteria after incubation with human serum relative to the number of viable bacteria before treatment and then multiplied by 100.
Determination of the Anti-phagocytosis
The phagocytosis of K. pneumoniae strains by RAW264.7 cells was examined as previously described with minor modification (Cheng et al., 2010a). RAW264.7 cells were grown in DMEM (Gibco, Grand Island, NY, United States) containing 10% FBS at 37°C. RAW264.7 cells (4 × 105 cells/well) in 24 well culture plates were co-incubate with anaerobically grown K. pneumoniae strains at a ratio of 25:1 (bacteria to RAW264.7 cells). The cell plates were centrifuged at 500 × g for 5 min to enhance infection. The cells were incubated for 2 h at 37°C to permit phagocytosis. After 2 h incubation, the cells were washed thrice, then 1 mL of DMEM containing 100 μg/mL of gentamycin were added and incubated for another 2 h to kill the extracellular bacteria. After that, the free bacteria outside the cells were washed with PBS (pH 7.4), and then 0.2 mL of sterile 0.025% Triton X-100 was added to lyse the cells of each well. The mixture in each well was then serially diluted and spread onto LB agar plates for 16 h incubation at 37°C. The plates were incubated at 37°C overnight for colony formation. The number of viable bacteria was then determined by colony counting. The percentage of phagocytosis rate was expressed as the number of viable bacteria incubated with the RAW264.7 cells compared with the number of viable bacteria from the pretreatment and multiplied by 100.
Statistical Analysis
The experiments for CPS quantification, promoter activity, qRT-PCR analysis, and bacterial survival in serum and phagocytosis were performed in triplicate. The results are presented as the mean and standard deviation. Differences between groups were evaluated by an unpaired t-test. Differences with P-values < 0.05 and < 0.01 were considered statistically significant.
Results
Holo-FNR Represses CPS Biosynthesis Under Anaerobic Growth Condition
To observe whether FNR affects the CPS biosynthesis in response to oxygen availability, we determined the amount of CPS in K. pneumoniae CG43S3 (WT) and Δfnr strains cultured in aerobic or anaerobic conditions. The CPS amount in anaerobic condition was significantly higher than that in aerobic condition (Figure 1A). Furthermore, fnr deletion in K. pneumoniae increased the CPS amount, compared to that in the WT strain under the anaerobic growth condition; however, the CPS amount in WT and Δfnr strains was remarkably low under the aerobic growth condition. These results indicate that CPS biosynthesis could be inhibited by FNR in K. pneumoniae cultured in anaerobic condition.
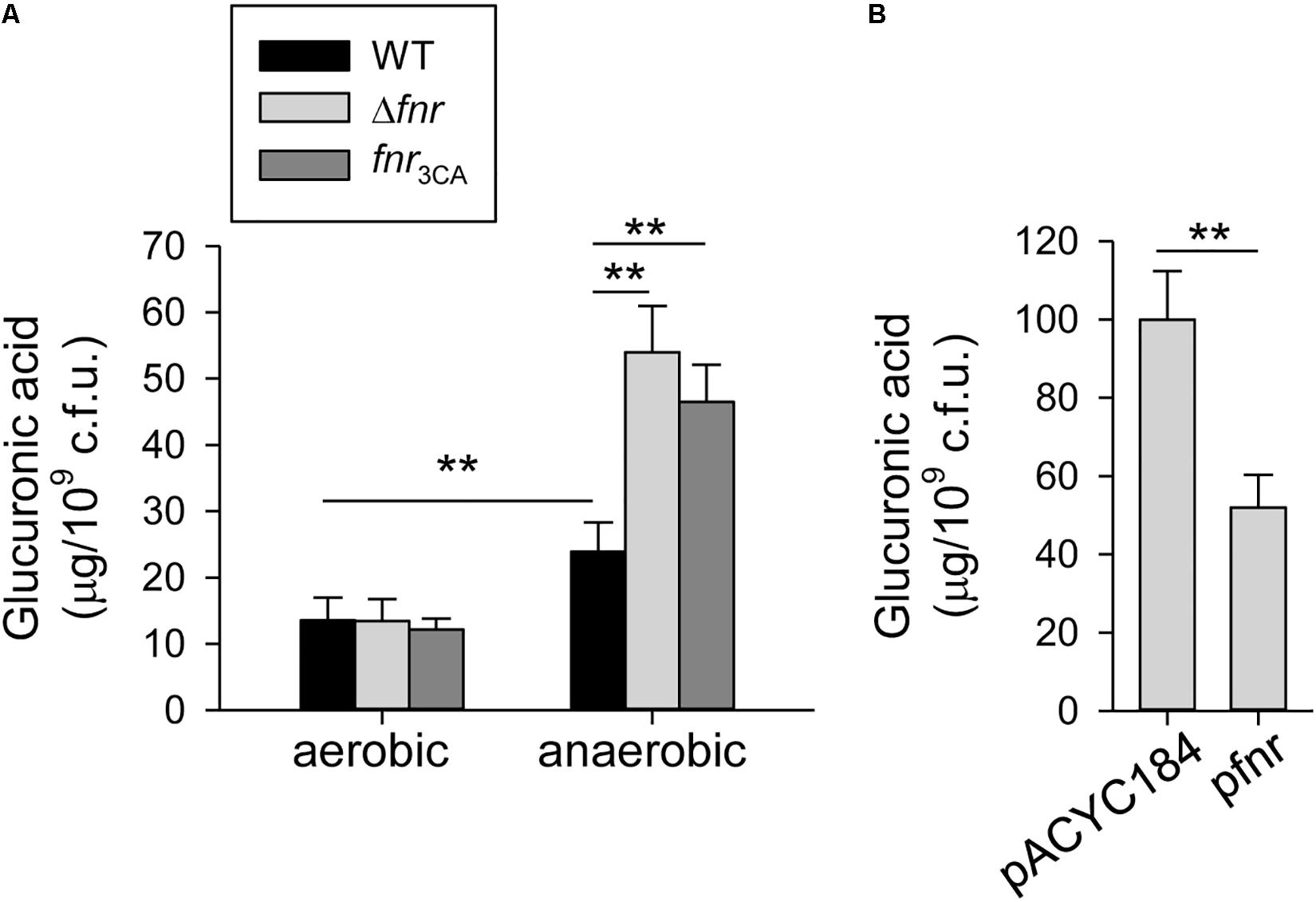
Figure 1. Holo-FNR represses the CPS biosynthesis under anaerobic growth condition. (A) CPS amounts of WT, Δfnr, and fnr3CA strains grown in LB broth under aerobic and anaerobic conditions were measured. (B) CPS amounts of Δfnr carrying pACYC184 or pfnr grown in LB broth under anaerobic condition were determined. After 16 h of growth, the bacterial glucuronic acid content was determined. Error bars indicate standard deviations. ∗∗P < 0.01 compared to the indicated group.
In K. pneumoniae, FNR contains four highly conserved cysteine residues (C20, C23, C29, and C122 in E. coli FNR), which are considered to coordinate the [4Fe–4S] cluster (Kiley and Beinert, 1998). Therefore, to investigate the role of the [4Fe–4S] cluster in FNR regulation of CPS biosynthesis, we created a [4Fe–4S] cluster-deficient fnr mutant, fnr3CA, by replacing the three cysteines (C20, C23, and C29) with alanines and tested whether this mutant affected CPS biosynthesis. No marked effect was observed in the CPS amount in WT and fnr3CA strains under the aerobic growth condition. However, under anaerobic conditions, the CPS amount in the fnr3CA strain was higher than that in the WT, indicating that the [4Fe–4S] cluster is essential for FNR repression during anaerobic growth. For further complementation analysis, the complete fnr were cloned into pACYC184 to yield pfnr. Under anaerobic conditions, the CPS amount in Δfnr [pfnr] was significantly lower than that in Δfnr [pACYC184] (Figure 1B). These results confirmed that FNR in K. pneumoniae has a negative role in the regulation of CPS biosynthesis and that the presence of the [4Fe–4S] cluster in FNR is essential for this regulation under anaerobic growth conditions.
Holo-FNR Acts a Transcriptional Repressor for cps
The K2 cps gene cluster of K. pneumoniae contains 19 ORFs organized into three transcription units orf1–2, orf3–15, and orf16–17 (Arakawa et al., 1995). To investigate the effect of FNR on the expression of the 3 cps transcriptional units, the mRNA level of orf1, orf3, and orf16 were measured by qRT-PCR in WT, Δfnr, and fnr3CA strains grown in LB medium under anaerobic conditions. As shown in Figure 2A, we found that the mRNA levels of orf1, orf3, and orf16 was increased in Δfnr, and fnr3CA strains as compared with the WT. However, the Δfnr strain carrying complement plasmid pfnr could decrease the mRNA levels of orf1, orf3, and orf16, as compared with Δfnr carrying the empty vector (pACYC184) (Figure 2B). In addition, to further observe whether FNR could affect the promoter activity of cps gene cluster, we used the reporter plasmids pOrf12 (Porf1–2:lacZ), pOrf315 (Porf3–15:lacZ), and pOrf1617 (Porf16–17:lacZ), each carrying a promoterless lacZ gene transcriptionally fused to the putative promoter region of the K2 cps gene cluster (Balsalobre et al., 2006), to transform the K. pneumoniae strains ΔlacZ, ΔlacZΔfnr, and ΔlacZ-fnr3CA strains. Under the anaerobic condition, the promoter activity of orf1–2, orf3–15, and orf16–17 in ΔlacZΔfnr and ΔlacZ-fnr3CA strains was higher than that in the ΔlacZ strain (Figure 2C). These results indicate that FNR represses the transcription of cps genes in an [4Fe–4S] cluster-dependent manner.
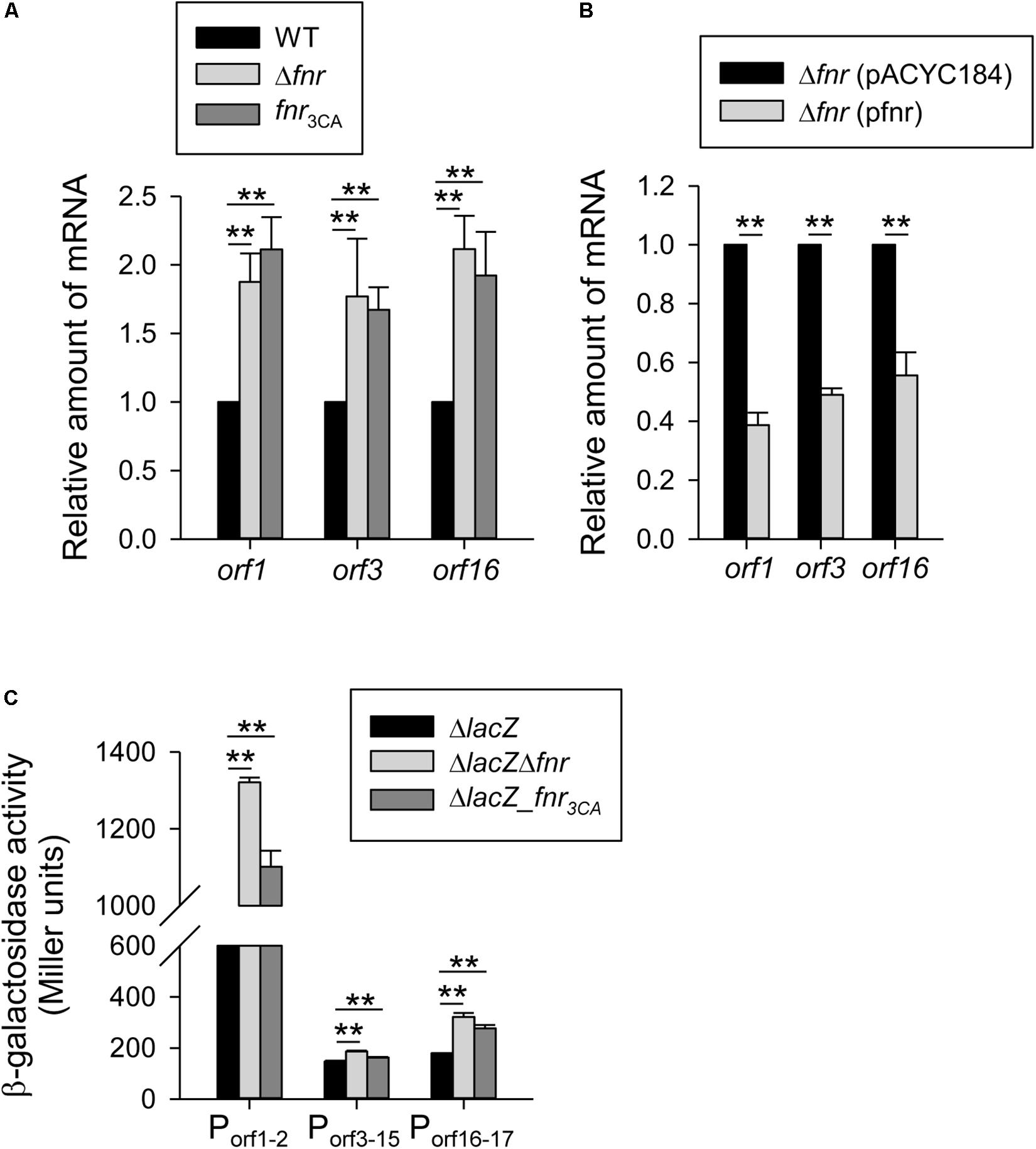
Figure 2. Holo-FNR represses the cps transcription under anaerobic growth condition. The mRNA expression of orf1, orf3, and orf16 was measured in (A) WT, Δfnr, and fnr3CA strains and (B) the complement strain Δfnr carrying pACYC184 or pfnr which grown in LB medium under anaerobic condition by qRT-PCR analysis. (C)β-galactosidase activities of K. pneumoniae CG43S3ΔlacZ and the isogenic strains (ΔlacZΔfnr and ΔlacZ-fnr3CA) carrying the reporter plasmid pOrf12 (Porf1–2:lacZ), pOrf315 (Porf3–15:lacZ), or pOrf1617 (Porf16–17:lacZ) were determined using overnight cultures grown in LB medium under anaerobic condition. Error bars indicate standard deviations. ∗∗P < 0.01 compared to the indicated group.
To further investigate the mechanism of FNR regulation in cps gene transcription, the sequence of the E. coli FNR binding site (TTGAT-N4-ATCAA) (Scott et al., 2003) was used to identify the promoter sequence of the K2 cps gene cluster for K. pneumoniae CG43. Here, the maximum number of possible mismatched nucleotides was set at 2, and only the intergenic regions of the three cps transcriptional units were analyzed. Using these criteria, we found that no typical FNR binding site was located upstream of the three cps transcriptional units, indicating that the FNR represses cps expression indirectly.
Expression of rmpA and rmpA2 Is Repressed by FNR
Multiple transcriptional regulators have been reported to affect CPS biosynthesis in K. pneumoniae CG43, such as CRP, IscR, Fur, RcsA, RcsB, RmpA, RmpA2, KvgA, and KvhR (Balsalobre et al., 2006; Cheng et al., 2010b; Lin et al., 2010, 2013; Wu et al., 2014). Therefore, to further investigate whether these transcription factors are involved in the regulation of cps transcription by FNR, the FNR binding site was searched in the upstream sequence of crp, iscR, fur, rcsA/B, rmpA/A2, kvgA, and kvhR. However, we found that the putative FNR binding site is located at −28 to −15 (5′-TTTAT-ATGT-AACAA-3′) and −333 to −320 (5′-TTGTT-TTTA-ATAAA-3′) relative to the translation start site of RmpA and RmpA2, respectively, but no typical FNR binding site was found upstream of the crp, iscR, fur, rcsA/B, kvgA, and kvhR sequences. Therefore, these results suggest that RmpA and RmpA2 are involved in the FNR-based regulation of cps expression.
To verify this possibility, we first performed qRT-PCR analysis to determine the mRNA levels of rmpA and rmpA2 in the WT, Δfnr, and fnr3CA strains cultured in the anaerobic condition. The mRNA levels of rmpA and rmpA2 in the Δfnr and fnr3CA strains were higher than those in the WT strain (Figure 3A). Furthermore, the Δfnr strain carrying pfnr could decrease the mRNA levels of rmpA and rmpA2, as compared with Δfnr carrying pACYC184 (Figure 3B). Next, to determine whether FNR functions as a transcriptional regulator for rmpA and rmpA2, the reporter plasmids prmpAZ15 (PrmpA:lacZ) and prmpA2Z15 (PrmpA2:lacZ), each carrying a promoterless lacZ gene transcriptionally fused to the putative promoter region of rmpA and rmpA2 respectively, were used to transform the K. pneumoniae strains ΔlacZ, ΔlacZΔfnr, and ΔlacZ-fnr3CA. The promoter activity of rmpA and rmpA2 in the ΔlacZΔfnr and ΔlacZ-fnr3CA strains was higher than that in ΔlacZ strain under the anaerobic growth condition (Figure 3C). These results indicate that holo-FNR acts a transcriptional repressor of rmpA and rmpA2 in K. pneumoniae during anaerobic growth.
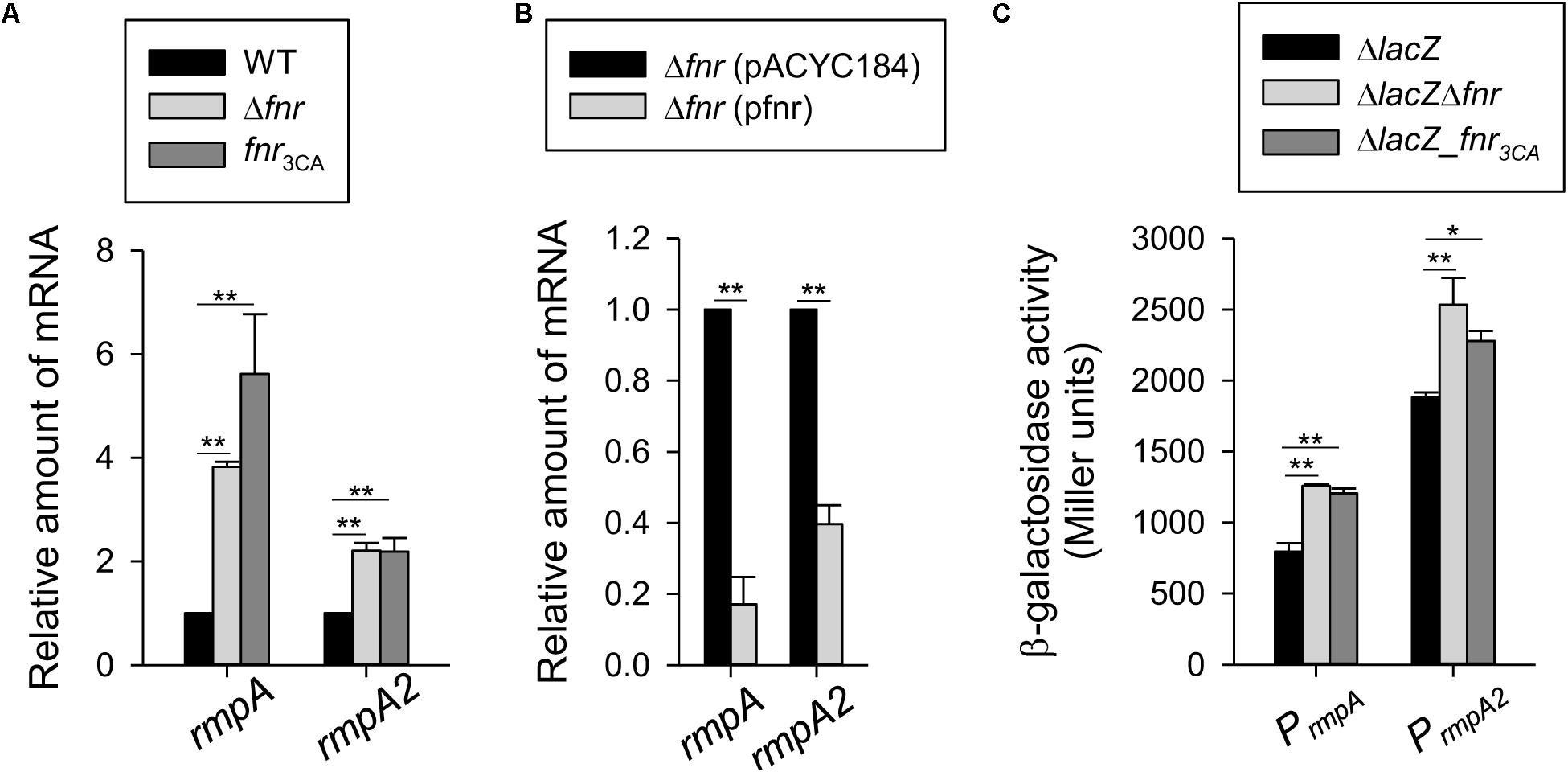
Figure 3. Holo-FNR represses rmpA and rmpA2 transcriptions. (A) The mRNA expression of rmpA and rmpA2 was measured in WT, Δfnr, and fnr3CA strains and (B) the complement strain Δfnr carrying pACYC184 or pfnr which grown in LB medium under anaerobic condition by qRT-PCR analysis. (C) The β-galactosidase activities of K. pneumoniae CG43S3ΔlacZ and the isogenic strains (ΔlacZΔfnr and ΔlacZ-fnr3CA) carrying the reporter plasmids prmpAZ15 (PrmpA:lacZ) and prmpA2Z15 (PrmpA2^*:lacZ) were determined using over-night cultures grown in LB medium under anaerobic condition. Error bars indicate standard deviations. ∗P < 0.05 and ∗∗P < 0.01 compared to the indicated group.
Role of RmpA and RmpA2 in Regulation of FNR on CPS Biosynthesis
To investigate whether RmpA and RmpA2 participate in FNR regulation of CPS biosynthesis, the level of CPS was determined in Δfnr, ΔrmpA, ΔrmpA2, ΔfnrΔrmpA, and ΔfnrΔrmpA2 strains under the anaerobic growth condition. As shown in Figure 4A, the CPS production was reduced in the Δfnr background by the further deletion of rmpA or rmpA2. However, compared with the ΔrmpA or ΔrmpA2 strains, the CPS production was slightly increased in the ΔfnrΔrmpA or ΔfnrΔrmpA2 strains, respectively. Besides, the qRT-PCR analysis indicated that the mRNA levels of orf1 and orf3 were reduced in the Δfnr background by the further deletion of rmpA or rmpA2 (Figure 4B). In addition, compared with the ΔrmpA or ΔrmpA2 strains, the mRNA levels of orf1 and orf3 were increased in the ΔfnrΔrmpA or ΔfnrΔrmpA2 strains, respectively. Deletion of rmpA, but not rmpA2, in the Δfnr strain reduced the mRNA level of orf16. The mRNA level of orf16 was increased in ΔfnrΔrmpA2 compared with that in ΔrmpA2; while no significant difference in the mRNA level of orf16 was found between the ΔrmpA and ΔfnrΔrmpA strains. To further validate the regulation of fnr, rmpA, and rmpA2 on the expression of cps genes, the promoter-reporter assay was performed. As shown in Figure 4C, the promoter activity of orf1-2 and orf3-15 were reduced in the ΔlacZΔfnr background by the further deletion of rmpA or rmpA2. In addition, compared with the ΔlacZΔrmpA or ΔlacZΔrmpA2 strains, the promoter activity of orf1-2 and orf3-15 were increased in the ΔlacZΔfnrΔrmpA or ΔlacZΔfnrΔrmpA2 strains, respectively. Deletion of rmpA, but not rmpA2, in the ΔlacZΔfnr strain reduced the promoter activity of orf16-17. The promoter activity of orf16-17 was increased in ΔlacZΔfnrΔrmpA2 compared with that in ΔlacZΔrmpA2; while no significant difference was found between the ΔlacZΔrmpA and ΔlacZΔfnrΔrmpA strains. These results revealed a complex regulatory circuit in Fnr, RmpA, and RmpA2 that modulate the transcription of cps genes in coordination.
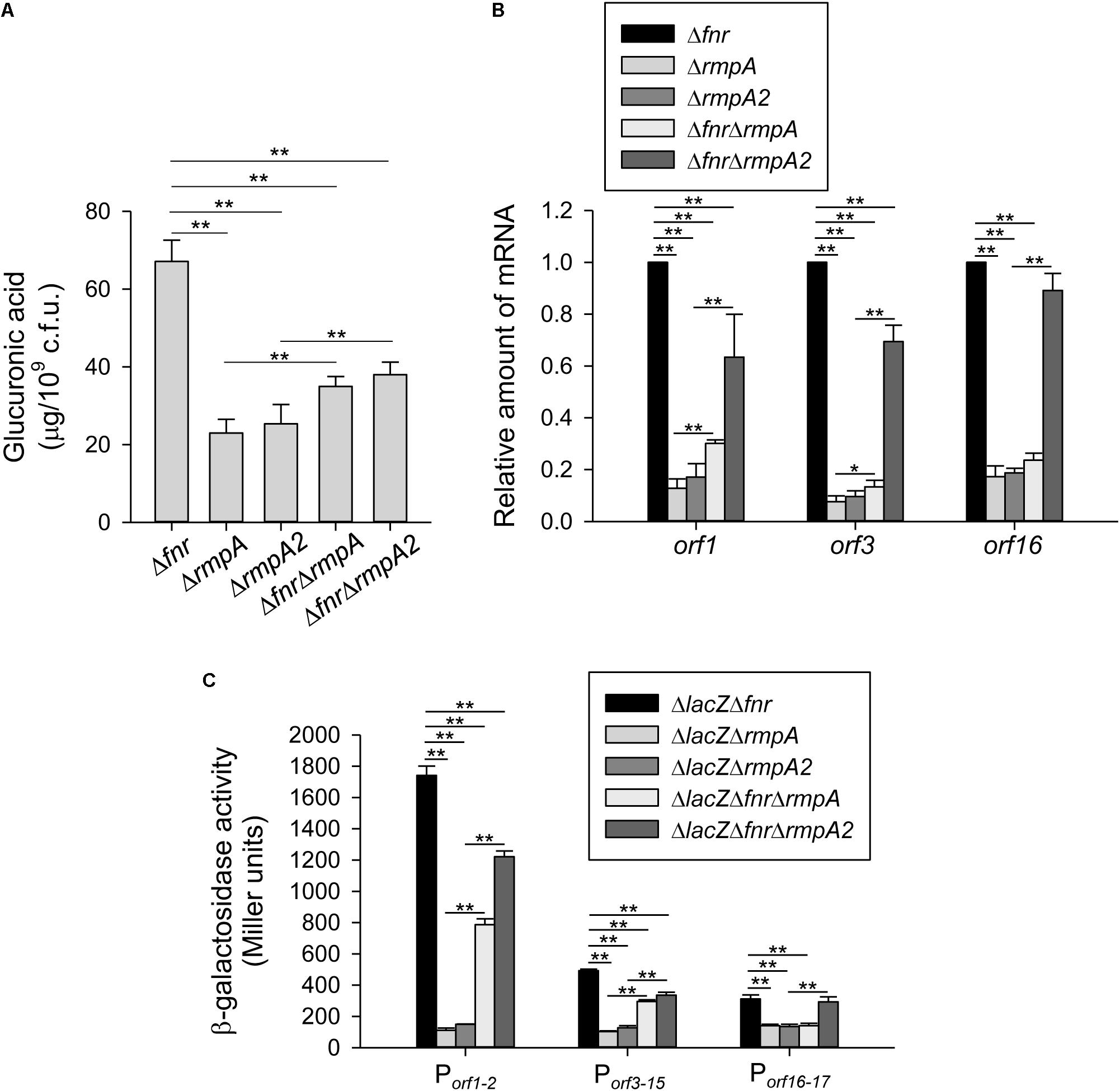
Figure 4. RmpA and RmpA2 are involved in holo-FNR regulation of CPS expression. (A) CPS amounts of Δfnr, ΔrmpA, ΔrmpA2, ΔfnrΔrmpA, and ΔfnrΔrmpA2 strains were determined. Bacterial strains were grown in LB broth under anaerobic condition. After 16 h of growth, the bacterial glucuronic acid content was determined. (B) The mRNA expression of orf1, orf3, and orf16 was measured in Δfnr, ΔrmpA, ΔrmpA2, ΔfnrΔrmpA, and ΔfnrΔrmpA2 strains which grown in LB medium under anaerobic condition by qRT-PCR analysis. (C) The β-galactosidase activities of K. pneumoniae CG43S3ΔlacZΔfnr, ΔlacZΔrmpA, ΔlacZΔrmpA2, and the isogenic strains (ΔlacZΔfnrΔrmpA and ΔlacZΔfnrΔrmpA2) carrying the reporter plasmid pOrf12 (Porf1–2:lacZ), pOrf315 (Porf3–15:lacZ), or pOrf1617 (Porf16–17:lacZ) were determined using overnight cultures grown in LB medium under anaerobic condition. Error bars indicate standard deviations. ∗P < 0.05 and ∗∗P < 0.01 compared to the indicated group.
Effect of FNR on Normal Human Serum Resistance and Anti-phagocytosis
As CPS has been demonstrated to protect K. pneumoniae from serum killing and phagocytosis (Sahly et al., 2000; Lin et al., 2004), FNR may also affect the ability of K. pneumoniae to resist the bactericidal effects of serum and phagocytosis by regulating CPS levels. To test this hypothesis, we determined the survival rate of anaerobically cultured K. pneumoniae strains in 75% normal human serum. Compared with the WT strain, the Δfnr and fnr3CA strains showed a remarkably higher survival rate (Figure 5A), implying the negative role of holo-FNR in the serum resistance of K. pneumoniae. To further investigate whether RmpA and RmpA2 are involved in role of FNR in serum resistance, the survival rates of the ΔfnrΔrmpA and ΔfnrΔrmpA2 strains were observed. The deletion of rmpA or rmpA2 in Δfnr strain reduced the survival rate, compared to that in the Δfnr strain (Figure 5A). This confirms the involvement of RmpA and RmpA2 in FNR-mediated regulation of CPS biosynthesis to influence K. pneumoniae resistance to normal human serum. Next, to investigate the role of FNR in anti-phagocytosis, the survival rates of the WT, Δfnr, and fnr3CA strains were determined. Deletion of fnr and fnr3CA resulted in a marked reduction in the phagocytosis rate, compared to that in the WT strain (Figure 5B). In addition, we also evaluated the phagocytosis rate after the deletion of rmpA and rmpA2 in Δfnr strains. The phagocytosis rate in Δfnr strains lacking rmpA was higher than that in the WT and Δfnr strains (Figure 5B). However, the deletion of rmpA2 in Δfnr strain could restore the effect of phagocytosis to that observed in the WT strain. Collectively, these findings suggest that RmpA and RmpA2 are involved in the negative role of FNR in anti-phagocytosis in K. pneumoniae and that RmpA plays a critical role in anti-phagocytosis of K. pneumoniae.
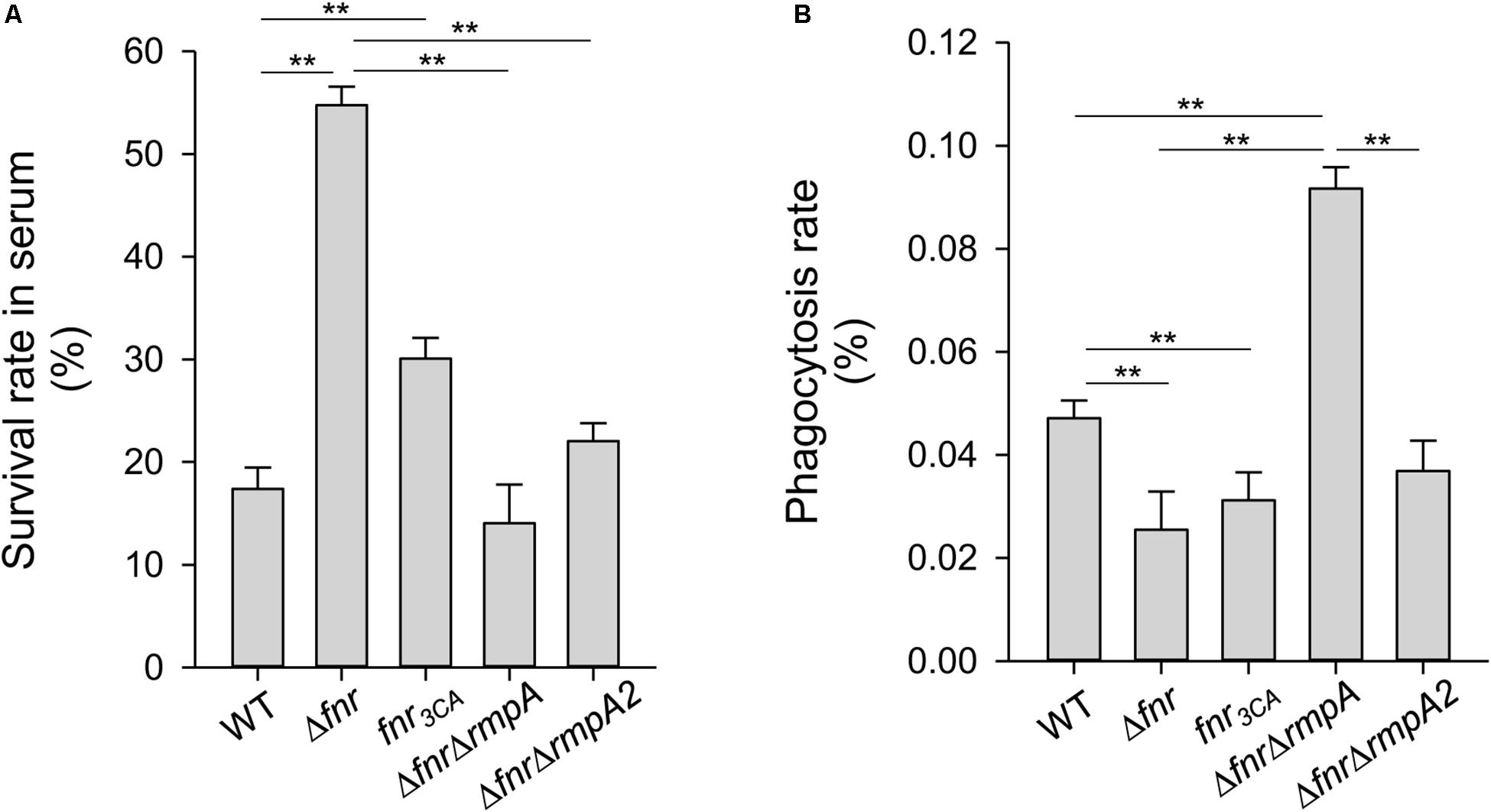
Figure 5. Effects of holo-FNR, RmpA, and RmpA2 on K. pneumoniae susceptibility to normal human serum and phagocytosis. The susceptibility to normal human serum (A) and the phagocytosis from mouse macrophage RAW264.7 (B) of WT, Δfnr, fnr3CA, ΔfnrΔrmpA, and ΔfnrΔrmpA2 strains which overnight grown in LB medium under anaerobic condition were determined. The survival rate in serum and the phagocytosis rate were quantified as described in section “Materials and Methods.” ∗∗P < 0.01 compared to the indicated group.
Discussion
Stringent regulation of CPS amounts in K. pneumoniae is complex and critical for adaptation to the dynamic environmental signals and for successful infection (Lin et al., 2010, 2013; Huang et al., 2012; Ares et al., 2016; Dorman et al., 2018). Here, we demonstrated that holo-FNR acts a repressor of CPS biosynthesis, thereby influencing K. pneumoniae resistance to serum and phagocytosis under anaerobic condition. Furthermore, the involvement of RmpA and RmpA2 in the FNR regulon was also elucidated.
In extraintestinal pathogenic E. coli XM, under low oxygen availability, the biosynthesis of extracytoplasmic polysaccharides increased in 100% serum, as compared to that in LB broth (Ma et al., 2018). Similarly, we found that the CPS amounts of K. pneumoniae were increased in response to anaerobic growth conditions. However, in this context FNR plays an opposite role, by directly activating the biosynthesis of the K-capsule and colanic acid in E. coli (Ma et al., 2018) but repressing the K2 CPS biosynthesis in K. pneumoniae. Since the K2 cps gene clusters (Arakawa et al., 1995) are relatively different from the genes responsible for the biosynthesis of the K-capsule and colanic acid (Ma et al., 2018), it is reasonable that the regulation of CPS biosynthesis differs between the two bacteria. In addition to FNR, ArcA is a well-studied transcriptional regulator in several bacteria and is known to affect the expression of numerous genes in response to oxygen availability (Green and Paget, 2004). During anaerobic growth, ArcB, a membrane-bound sensor, is auto-phosphorylated and the phosphoryl group is transferred to ArcA, a response regulator, to activate or repress the target gene transcription (Green and Paget, 2004). In K. pneumoniae, ArcA promotes persistent colonization in the mouse gastrointestinal tract (Boll et al., 2012); however, the role of ArcBA in K. pneumoniae pathogenesis remains unclear. To investigate whether ArcA is involved in regulation of CPS biosynthesis in K. pneumoniae, the typical ArcA∼P binding site of E. coli (Liu and De Wulf, 2004) was used to analyze the sequences upstream of the three cps transcriptional units in K. pneumoniae. However, no typical ArcA∼P binding site was found in these sequences. Under anaerobic conditions, ArcA-dependent repression of fur transcription has been demonstrated in E. coli and Shigella flexneri (Liu and De Wulf, 2004; Boulette and Payne, 2007). In K. pneumoniae, we found that Fur directly represses the expression of rmpA, rmpA2, and rcsA, subsequently repressing CPS biosynthesis (Lin et al., 2010). Therefore, ArcA may inhibit the Fur-mediated repression of CPS biosynthesis in K. pneumoniae under anaerobic growth conditions. However, further studies are warranted to confirm this.
Several studies have reported RmpA and RmpA2 as important virulence determinants for the mucoid phenotype of K. pneumoniae (Nassif et al., 1989; Arakawa et al., 1991) In K. pneumoniae CG43, deletion of either rmpA or rmpA2 resulted in a marked decreased in cps transcription, thereby repressing the mucoid phenotype (Lai et al., 2003; Cheng et al., 2010b). Consistent with this, we found that deletion of either rmpA or rmpA2 in the Δfnr strain showed a remarkable reduction in the CPS amount as compared to that in Δfnr strain. This indicates that RmpA and RmpA2 act as important activators in the FNR-mediated regulation of CPS biosynthesis during anaerobic growth. Under aerobic conditions, RmpA activates the promoter activity of orf1-2 and orf16-17 in LB medium, whereas RmpA2 only activates the promoter activity of orf1-2 (Cheng et al., 2010b). Nevertheless, in anaerobic conditions, we found that the rmpA-deletion in the Δfnr strain decreased the promoter activity of the three cps transcriptional units; however, the rmpA2-deletion in the Δfnr strain reduced the promoter activity of orf1-2 and orf3-15 but not orf16-17. We considered that the expression of these cps gene clusters were differentially regulated by FNR and many other CPS regulators including RmpA/A2, RcsAB, Fur, IscR, and CRP in response to various environmental stimuli (Lai et al., 2003; Cheng et al., 2010b; Lin et al., 2010, 2013; Wu et al., 2014), which may affect the composition, transportation, and assembly of CPS. The regulatory effect of RmpA and RmpA2 on cps expression in K. pneumoniae cultured in LB medium under anaerobic conditions was similar to that previously observed in K. pneumoniae cultured in M9-glucose minimal medium under aerobic conditions (Cheng et al., 2010b). Oxygen and glucose stimuli are considered to affect global protein acetylation in bacteria (Chohnan et al., 1998; Schilling et al., 2015). In addition, protein acetylation is an abundant post-translational modification in bacteria to control protein structure, stability, and function (Wolfe, 2016; Carabetta and Cristea, 2017). Therefore, whether RmpA and RmpA2 could be acetylated to affect its regulatory activity on cps expression awaits to be investigated.
In E. coli, FNR and CRP bind to a similar DNA sequence, suggesting that the FNR-regulated targets are overlapped to the CRP regulon in anaerobic conditions (Shaw et al., 1983). In K. pneumoniae, CRP also acts a transcriptional repressor for the three cps transcriptional units (Lin et al., 2013). However, CRP could directly bind to the promoter region of orf3-15 and orf16-17 to inhibit the transcription, but CRP-based repression of orf1-2 transcription is required for inhibiting the rcsA expression (Lin et al., 2013). Therefore, although both FNR and CRP repressed the promoter activity of the three cps transcriptional units, their regulatory mechanisms are different.
In heavy encapsulated K. pneumoniae strains, type 3 fimbriae has been demonstrated to play a crucial role in the biofilm formation both on biotic and abiotic surfaces (Di Martino et al., 2003; Jagnow and Clegg, 2003; Wu et al., 2012). However, the thick capsule of K. pneumoniae impedes the assembly and adherence of type 3 fimbriae (Schembri et al., 2005). Thus, tightly controlling the biosynthesis of CPS and fimbriae is critical for successful infection by K. pneumoniae. We consider that FNR cross regulates the biosynthesis of CPS and fimbriae in response to oxygen availability during infection, which awaits to be investigated. In addition, we found that holo-FNR repressed serum resistance in K. pneumoniae under anaerobic conditions. Deletion of rmpA or rmpA2 in the Δfnr strain restored the survival rate in serum to that observed in the WT strain, indicating the complementary functions of RmpA and RmpA2 in serum resistance. Apart from CPS, LPS in K. pneumoniae was also a major determinant in serum resistance in both serotype K1 and K2 strains (Yeh et al., 2016). However, RmpA and RmpA2 have yet been reported to be involved in LPS biosynthesis. Furthermore, we also observed a higher phagocytosis rate in the ΔfnrΔrmpA strain, compared to that in the WT and ΔfnrΔrmpA2 strains. This indicates that compared to RmpA2, RmpA serves a more important role in anti-phagocytosis of K. pneumoniae.
In this study, we found that CPS biosynthesis in K. pneumoniae is affected by oxygen availability. However, holo-FNR acts an important repressor for CPS biosynthesis, subsequently affecting the ability of serum resistance and anti-phagocytosis via inhibition of rmpA and rmpA2 transcription in K. pneumoniae during anaerobic growth. Thus, we demonstrate that FNR plays a critical role in mediating the virulence factor expression in K. pneumoniae pathogenesis in response to oxygen availability.
Data Availability Statement
All datasets generated for this study are included in the manuscript/supplementary files.
Ethics Statement
For isolation of normal human serum from healthy volunteers, the procedure and the respective consent documents were approved by the Ethics Committee of the China Medical University Hospital, Taichung, Taiwan. All healthy volunteers provided written informed consent.
Author Contributions
T-HL, J-TK, C-CW, and C-TL conceived and designed the experiments. J-TK, C-CW, H-FC, and C-TL performed the experiments. T-HL, J-TK, D-YL, and C-TL analyzed the data. T-HL, C-CW, and C-TL contributed to reagents, materials, and analysis tools. C-CW and C-TL wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the grant from Ministry of Science and Technology (MOST 107-2320-B-039-015-), China Medical University (CMU107-S-32), China Medical University Hospital (DMR-107-128), and Taichung Tzuchi Hospital, The Buddhist Tzu Chi Medical Foundation (TTCRD105-09). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Miss Yi-Min Hong for her technical assistance during the study.
Abbreviations
apo-FNR, monomeric FNR; CPS, capsular polysaccharide; DMEM, Dulbecco’s modified Eagle’s medium; ESBL, extended spectrum β-lactamase; FBS, fetal bovine serum; Fur, ferric uptake regulator; holo-FNR, [4Fe-4S] cluster of FNR; KLA, Klebsiella liver abscess; LB, Luria-Bertani; ORF, open reading frame; PBS, phosphate-buffered saline.
References
Alicino, C., Giacobbe, D. R., Orsi, A., Tassinari, F., Trucchi, C., Sarteschi, G., et al. (2015). Trends in the annual incidence of carbapenem-resistant Klebsiella pneumoniae bloodstream infections: a 8-year retrospective study in a large teaching hospital in northern Italy. BMC Infect. Dis. 15:415. doi: 10.1186/s12879-015-1152-0
Arakawa, Y., Ohta, M., Wacharotayankun, R., Mori, M., Kido, N., Ito, H., et al. (1991). Biosynthesis of klebsiella K2 capsular polysaccharide in Escherichia coli HB101 requires the functions of rmpA and the chromosomal cps gene cluster of the virulent strain Klebsiella pneumoniae Chedid (O1:K2). Infect. Immun. 59, 2043–2050.
Arakawa, Y., Wacharotayankun, R., Nagatsuka, T., Ito, H., Kato, N., and Ohta, M. (1995). Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J. Bacteriol. 177, 1788–1796. doi: 10.1128/jb.177.7.1788-1796.1995
Ares, M. A., Fernandez-Vazquez, J. L., Rosales-Reyes, R., Jarillo-Quijada, M. D., Von Bargen, K., Torres, J., et al. (2016). H-NS nucleoid protein controls virulence features of Klebsiella pneumoniae by regulating the expression of type 3 pili and the capsule polysaccharide. Front. Cell Infect. Microbiol. 6:13. doi: 10.3389/fcimb.2016.00013
Balsalobre, C., Johansson, J., and Uhlin, B. E. (2006). Cyclic AMP-dependent osmoregulation of crp gene expression in Escherichia coli. J. Bacteriol. 188, 5935–5944. doi: 10.1128/jb.00235-06
Blumenkrantz, N., and Asboe-Hansen, G. (1973). New method for quantitative determination of uronic acids. Anal. Biochem. 54, 484–489. doi: 10.1016/0003-2697(73)90377-1
Boll, E. J., Nielsen, L. N., Krogfelt, K. A., and Struve, C. (2012). Novel screening assay for in vivo selection of Klebsiella pneumoniae genes promoting gastrointestinal colonisation. BMC Microbiol. 12:201. doi: 10.1186/1471-2180-12-201
Boulette, M. L., and Payne, S. M. (2007). Anaerobic regulation of Shigella flexneri virulence: ArcA regulates Fur and iron acquisition genes. J. Bacteriol. 189, 6957–6967. doi: 10.1128/jb.00621-07
Carabetta, V. J., and Cristea, I. M. (2017). Regulation, function, and detection of protein acetylation in bacteria. J. Bacteriol. 199, e107–e117. doi: 10.1128/JB.00107-17
Carpenter, C., and Payne, S. M. (2014). Regulation of iron transport systems in Enterobacteriaceae in response to oxygen and iron availability. J. Inorg. Biochem. 133, 110–117. doi: 10.1016/j.jinorgbio.2014.01.007
Cheng, H. Y., Chen, Y. F., and Peng, H. L. (2010a). Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J. Biomed. Sci. 17:60. doi: 10.1186/1423-0127-17-60
Cheng, H. Y., Chen, Y. S., Wu, C. Y., Chang, H. Y., Lai, Y. C., and Peng, H. L. (2010b). RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J. Bacteriol. 192, 3144–3158. doi: 10.1128/jb.00031-10
Chohnan, S., Izawa, H., Nishihara, H., and Takamura, Y. (1998). Changes in size of intracellular pools of coenzyme A and its thioesters in Escherichia coli K-12 cells to various carbon sources and stresses. Biosci. Biotechnol. Biochem. 62, 1122–1128. doi: 10.1271/bbb.62.1122
Di Martino, P., Cafferini, N., Joly, B., and Darfeuille-Michaud, A. (2003). Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res. Microbiol. 154, 9–16. doi: 10.1016/s0923-2508(02)00004-9
Domenico, P., Schwartz, S., and Cunha, B. A. (1989). Reduction of capsular polysaccharide production in Klebsiella pneumoniae by sodium salicylate. Infect. Immun. 57, 3778–3782.
Dorman, M. J., Feltwell, T., Goulding, D. A., Parkhill, J., and Short, F. L. (2018). The capsule regulatory network of Klebsiella pneumoniae defined by density-TraDISort. mBio 9:e01863-18. doi: 10.1128/mBio.01863-18
Green, J., and Paget, M. S. (2004). Bacterial redox sensors. Nat. Rev. Microbiol. 2, 954–966. doi: 10.1038/nrmicro1022
Green, J., Rolfe, M. D., and Smith, L. J. (2014). Transcriptional regulation of bacterial virulence gene expression by molecular oxygen and nitric oxide. Virulence 5, 794–809. doi: 10.4161/viru.27794
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2017). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Han, S. H. (1995). Review of hepatic abscess from Klebsiella pneumoniae. An association with diabetes mellitus and septic endophthalmitis. West. J. Med. 162, 220–224.
Huang, S. H., Wang, C. K., Peng, H. L., Wu, C. C., Chen, Y. T., Hong, Y. M., et al. (2012). Role of the small RNA RyhB in the Fur regulon in mediating the capsular polysaccharide biosynthesis and iron acquisition systems in Klebsiella pneumoniae. BMC Microbiol. 12:148. doi: 10.1186/1471-2180-12-148
Jagnow, J., and Clegg, S. (2003). Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology 149, 2397–2405. doi: 10.1099/mic.0.26434-0
Kawai, Y., Marles-Wright, J., Cleverley, R. M., Emmins, R., Ishikawa, S., Kuwano, M., et al. (2011). A widespread family of bacterial cell wall assembly proteins. EMBO J. 30, 4931–4941. doi: 10.1038/emboj.2011.358
Kiley, P. J., and Beinert, H. (1998). Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22, 341–352. doi: 10.1016/s0168-6445(98)00022-9
Lai, Y. C., Peng, H. L., and Chang, H. Y. (2001). Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect. Immun. 69, 7140–7145. doi: 10.1128/iai.69.11.7140-7145.2001
Lai, Y. C., Peng, H. L., and Chang, H. Y. (2003). RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J. Bacteriol. 185, 788–800. doi: 10.1128/jb.185.3.788-800.2003
Lau, Y. J., Hu, B. S., Wu, W. L., Lin, Y. H., Chang, H. Y., and Shi, Z. Y. (2000). Identification of a major cluster of Klebsiella pneumoniae isolates from patients with liver abscess in Taiwan. J. Clin. Microbiol. 38, 412–414.
Lawlor, M. S., Hsu, J., Rick, P. D., and Miller, V. L. (2005). Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol. Microbiol. 58, 1054–1073. doi: 10.1111/j.1365-2958.2005.04918.x
Lederman, E. R., and Crum, N. F. (2005). Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am. J. Gastroenterol. 100, 322–331. doi: 10.1111/j.1572-0241.2005.40310.x
Lin, C. T., Chen, Y. C., Jinn, T. R., Wu, C. C., Hong, Y. M., and Wu, W. H. (2013). Role of the cAMP-dependent carbon catabolite repression in capsular polysaccharide biosynthesis in Klebsiella pneumoniae. PLoS One 8:e54430. doi: 10.1371/journal.pone.0054430
Lin, C. T., Wu, C. C., Chen, Y. S., Lai, Y. C., Chi, C., Lin, J. C., et al. (2010). Fur regulation of the capsular polysaccharide biosynthesis and iron-acquisition systems in Klebsiella pneumoniae CG43. Microbiology 157, 419–429. doi: 10.1099/mic.0.044065-0
Lin, J. C., Chang, F. Y., Fung, C. P., Xu, J. Z., Cheng, H. P., Wang, J. J., et al. (2004). High prevalence of phagocytic-resistant capsular serotypes of Klebsiella pneumoniae in liver abscess. Microbes Infect. 6, 1191–1198. doi: 10.1016/j.micinf.2004.06.003
Lin, W. P., Wang, J. T., Chang, S. C., Chang, F. Y., Fung, C. P., Chuang, Y. C., et al. (2016). The antimicrobial susceptibility of Klebsiella pneumoniae from community settings in taiwan, a trend analysis. Sci. Rep. 6:36280. doi: 10.1038/srep36280
Liu, X., and De Wulf, P. (2004). Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 279, 12588–12597. doi: 10.1074/jbc.m313454200
Ma, J., An, C., Jiang, F., Yao, H., Logue, C., Nolan, L. K., et al. (2018). Extraintestinal pathogenic Escherichia coli increase extracytoplasmic polysaccharide biosynthesis for serum resistance in response to bloodstream signals. Mol. Microbiol. 110, 689–706. doi: 10.1111/mmi.13987
Ma, L., Wang, J. T., Wu, T. L., Siu, L. K., Chuang, Y. C., Lin, J. C., et al. (2015). Emergence of OXA-48-Producing Klebsiella pneumoniae in Taiwan. PLoS One 10:e0139152. doi: 10.1371/journal.pone.0139152
Melville, S. B., and Gunsalus, R. P. (1996). Isolation of an oxygen-sensitive FNR protein of Escherichia coli: interaction at activator and repressor sites of FNR-controlled genes. Proc. Natl. Acad. Sci. U.S.A. 93, 1226–1231. doi: 10.1073/pnas.93.3.1226
Mettert, E. L., and Kiley, P. J. (2005). ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe-4S] cluster. J. Mol. Biol. 354, 220–232. doi: 10.1016/j.jmb.2005.09.066
Miller, V. L., and Mekalanos, J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988
Nassif, X., Honore, N., Vasselon, T., Cole, S. T., and Sansonetti, P. J. (1989). Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol. Microbiol. 3, 1349–1359. doi: 10.1111/j.1365-2958.1989.tb00116.x
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/MMBR.00078-15
Pitout, J. D., Nordmann, P., and Poirel, L. (2015). Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 59, 5873–5884. doi: 10.1128/AAC.01019-15
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/cmr.11.4.589
Regueiro, V., Campos, M. A., Pons, J., Alberti, S., and Bengoechea, J. A. (2006). The uptake of a Klebsiella pneumoniae capsule polysaccharide mutant triggers an inflammatory response by human airway epithelial cells. Microbiology 152, 555–566. doi: 10.1099/mic.0.28285-0
Sahly, H., Podschun, R., Oelschlaeger, T. A., Greiwe, M., Parolis, H., Hasty, D., et al. (2000). Capsule impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect. Immun. 68, 6744–6749. doi: 10.1128/iai.68.12.6744-6749.2000
Schembri, M. A., Blom, J., Krogfelt, K. A., and Klemm, P. (2005). Capsule and fimbria interaction in Klebsiella pneumoniae. Infect. Immun. 73, 4626–4633. doi: 10.1128/iai.73.8.4626-4633.2005
Schilling, B., Christensen, D., Davis, R., Sahu, A. K., Hu, L. I., Walker-Peddakotla, A., et al. (2015). Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol. Microbiol. 98, 847–863. doi: 10.1111/mmi.13161
Scott, C., Partridge, J. D., Stephenson, J. R., and Green, J. (2003). DNA target sequence and FNR-dependent gene expression. FEBS Lett. 541, 97–101. doi: 10.1016/s0014-5793(03)00312-0
Shaw, D. J., Rice, D. W., and Guest, J. R. (1983). Homology between CAP and Fnr, a regulator of anaerobic respiration in Escherichia coli. J. Mol. Biol. 166, 241–247. doi: 10.1016/s0022-2836(83)80011-4
Skorupski, K., and Taylor, R. K. (1996). Positive selection vectors for allelic exchange. Gene 169, 47–52. doi: 10.1016/0378-1119(95)00793-8
Wolfe, A. J. (2016). Bacterial protein acetylation: new discoveries unanswered questions. Curr. Genet. 62, 335–341. doi: 10.1007/s00294-015-0552-4
Wu, C. C., Lin, C. T., Cheng, W. Y., Huang, C. J., Wang, Z. C., and Peng, H. L. (2012). Fur-dependent MrkHI regulation of type 3 fimbriae in Klebsiella pneumoniae CG43. Microbiology 158, 1045–1056. doi: 10.1099/mic.0.053801-0
Wu, C. C., Wang, C. K., Chen, Y. C., Lin, T. H., Jinn, T. R., and Lin, C. T. (2014). IscR regulation of capsular polysaccharide biosynthesis and iron-acquisition systems in Klebsiella pneumoniae CG43. PLoS One 9:e107812. doi: 10.1371/journal.pone.0107812
Wu, H., and Fives-Taylor, P. M. (2001). Molecular strategies for fimbrial expression and assembly. Crit. Rev. Oral. Biol. Med. 12, 101–115. doi: 10.1177/10454411010120020101
Yang, Y. S., Siu, L. K., Yeh, K. M., Fung, C. P., Huang, S. J., Hung, H. C., et al. (2009). Recurrent Klebsiella pneumoniae liver abscess: clinical and microbiological characteristics. J. Clin. Microbiol. 47, 3336–3339. doi: 10.1128/JCM.00918-09
Yeh, K. M., Chiu, S. K., Lin, C. L., Huang, L. Y., Tsai, Y. K., Chang, J. C., et al. (2016). Surface antigens contribute differently to the pathophysiological features in serotype K1 and K2 Klebsiella pneumoniae strains isolated from liver abscesses. Gut Pathog. 8:4. doi: 10.1186/s13099-016-0085-5
Keywords: Klebsiella pneumoniae, oxygen, FNR, capsule polysaccharide, RmpA, RmpA2
Citation: Lin T-H, Wu C-C, Kuo J-T, Chu H-F, Lee D-Y and Lin C-T (2019) FNR-Dependent RmpA and RmpA2 Regulation of Capsule Polysaccharide Biosynthesis in Klebsiella pneumoniae. Front. Microbiol. 10:2436. doi: 10.3389/fmicb.2019.02436
Received: 15 April 2019; Accepted: 10 October 2019;
Published: 29 October 2019.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Miguel A. De la Cruz, Mexican Social Security Institute (IMSS), MexicoYi-Cheng Sun, Institute of Pathogen Biology (CAMS), China
Copyright © 2019 Lin, Wu, Kuo, Chu, Lee and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ching-Ting Lin, Z2luZ3RpbmdAbWFpbC5jbXUuZWR1LnR3
†These authors have contributed equally to this work
 Tien-Huang Lin
Tien-Huang Lin Chien-Chen Wu
Chien-Chen Wu Jong-Tar Kuo
Jong-Tar Kuo Hsu-Feng Chu
Hsu-Feng Chu Ding-Yu Lee
Ding-Yu Lee Ching-Ting Lin
Ching-Ting Lin