- 1School of Earth and Atmospheric Sciences, Georgia Institute of Technology, Atlanta, GA, United States
- 2Guangdong Key Laboratory of Agricultural Environment Pollution Integrated Control, Guangdong Institute of Eco-Environmental Science and Technology, Guangzhou, China
- 3School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA, United States
- 4College of Marine Science, University of South Florida, St. Petersburg, St. Petersburg, FL, United States
Dispersant application is a primary emergency oil spill response strategy and yet the efficacy and unintended consequences of this approach in marine ecosystems remain controversial. To address these uncertainties, ex situ incubations were conducted to quantify the impact of dispersant on petroleum hydrocarbon (PHC) biodegradation rates and microbial community structure at as close as realistically possible to approximated in situ conditions [2 ppm v/v oil with or without dispersant, at a dispersant to oil ratio (DOR) of 1:15] in surface seawater. Biodegradation rates were not substantially affected by dispersant application at low mixing conditions, while under completely dispersed conditions, biodegradation was substantially enhanced, decreasing the overall half-life of total PHC compounds from 15.4 to 8.8 days. While microbial respiration and growth were not substantially altered by dispersant treatment, RNA analysis revealed that dispersant application resulted in pronounced changes to the composition of metabolically active microbial communities, and the abundance of nitrogen-fixing prokaryotes, as determined by qPCR of nitrogenase (nifH) genes, showed a large increase. While the Gammaproteobacteria were enriched in all treatments, the Betaproteobacteria and different families of Alphaproteobacteria predominated in the oil and dispersant treatment, respectively. Results show that mixing conditions regulate the efficacy of dispersant application in an oil slick, and the quantitative increase in the nitrogen-fixing microbial community indicates a selection pressure for nitrogen fixation in response to a readily biodegradable, nitrogen-poor substrate.
Introduction
The Deepwater Horizon (DWH) oil spill represents the largest accidental marine oil spill in history (Farrington, 2013; Sammarco et al., 2013). A total of 3.19 million barrels of crude oil along with an unprecedented 1.84 million gallons of dispersant, COREXIT 9500A and COREXIT 9527, were released into the Gulf of Mexico (GOM) (Barbier, 2015). Nearly 60% of the dispersant was applied at the sea surface. Dispersants are complex mixtures of chemical surfactants in a hydrocarbon solvent – the surfactants lower the interfacial tension between oil and water, breaking the oil into small droplets, with an increased surface-to-volume ratio and availability for biodegradation (National Research Council, 2005; Prince, 2015).
Although biodegradation has been studied for decades in seawater, there is as yet no consensus on the rates and controls of the process (Prince et al., 2017). The rate of biodegradation is limited by a range of environmental parameters including oxygen (Leahy and Colwell, 1990), temperature (ZoBell, 1969), nutrient availability (Atlas and Bartha, 1972), pressure (Schwarz et al., 1975; Prince et al., 2016b), and microbial community composition (Joye et al., 2016). Moreover, recent studies reveal that the concentration and form of oil (slick or water accommodated fractions) used in experiments is a major confounding factor in our understanding of biodegradation (Kleindienst et al., 2015b; Prince et al., 2017). The biodegradation rate of dispersed oil slows as the concentration of oil is increased and autochthonous nutrients become limiting at oil concentrations above 250 ppm (Prince et al., 2017). Although dispersant application is widely used as an emergency response strategy, the efficacy of this approach remains under debate (Kleindienst et al., 2015a; Prince et al., 2017). Previous studies indicate that dispersant can enhance (Venosa and Holder, 2007), have little effect (Foght and Westlake, 1982), or diminish (Lindstrom and Braddock, 2002) the rate and extent of petroleum hydrocarbons (PHCs) degradation in seawater. The range in the enhancement of biodegradation by dispersant can be explained in part by differences in the design and approach of previous experiments (Prince and Butler, 2014; Kleindienst et al., 2015a). Experiments must be conducted with concentrations of oil/dispersant resembling those expected during emergency response to an oil spill (Lee et al., 2013). During the DWH discharge, oil concentrations varied from below detection to over 10,000 ppm in surface seawater in the GOM, and the concentration was rapidly diluted with distance from the wellhead (Sammarco et al., 2013). In a survey conducted by NOAA immediately after the DWH oil spill, the majority of observations in the water column were at or below the low ppm level (U. S. National Oceanic and Atmospheric Administration [NOAA], 2010; Wade et al., 2016). At these low ppm concentrations of oil, bioavailable nutrients and oxygen are unlikely to limit microbial activity (Edwards et al., 2011). In contrast, the majority of previous experiments quantified biodegradation in seawater incubations using oil concentrations which are orders of magnitude higher than those expected during active response [i.e., 125 – 2500 ppm (Zahed et al., 2010), 867 ppm (Campo et al., 2013), 83 and 833 ppm (Venosa and Holder, 2007), and 1400 ppm (Lindstrom and Braddock, 2002)]. Using such high oil concentrations in a closed system may cause depletion of nutrients and artifacts which hinder biodegradation (Lee et al., 2013). Further, the dispersion process depends on oceanic mixing conditions, and lab incubations should reflect the mixing conditions in the environment (Venosa et al., 2002; Prince and Butler, 2014). Another uncertainty of dispersant application is the potential impact on indigenous microbial communities (Kleindienst et al., 2015a). Previous studies come to equivocal conclusions by suggesting that dispersants may impede or stimulate the activity of microbial populations (Lindstrom and Braddock, 2002; Chakraborty et al., 2012; Kleindienst et al., 2015b).
The objectives of this study were to investigate the biodegradation of dispersed oil in relation to mixing condition as well as the impacts of dispersant on the dynamics of metabolically active microbial communities in coastal seawater. Surface seawater was amended with artificially weathered Macondo surrogate oil and Corexit 9500A dispersant under as close to in situ conditions as possible. Petroleum hydrocarbon degradation and microbial communities were characterized using a close coupling of analytical chemistry and next generation sequencing techniques. Sequencing analysis of extracted RNA was used as a proxy for the metabolically active microbial communities. The results point to the importance of mixing conditions and alteration of metabolically active microbial communities following dispersant application. Our results under environmentally relevant conditions can be used to improve models that predict the fate of dispersed oil during future emergency response efforts.
Materials and Methods
Sample Collection and Experimental Setup
Surface seawater (top 1 m) was collected from Pensacola beach, FL (30°19′ N, 87°10′ W) with carboys for low mixing and high mixing experiments in June and August 2016, respectively. The sampling site was described in a previous study (Kostka et al., 2011). Seawater was immediately transported to the lab and aerated overnight. Microcosm experiments were conducted as previously described by Prince and Butler with the modifications described below (Prince and Butler, 2014). Two treatments were tested: seawater amended with weathered oil (oil treatment) or seawater amended with weathered oil and dispersant (oil + dispersant treatment) (COREXIT EC9500A; Nalco Environmental Solutions LLC, TX, United States). Surrogate MC252 oil was weathered by evaporation in the lab according to previously published methods (Pelz et al., 2011; Prince and Butler, 2014). Similar to previous work, we gravimetrically measured a 20% weight loss after the weathering process (Prince and Butler, 2014). The chemical composition of the surrogate MC252 crude oil was not confirmed prior to weathering. However, the chemical composition of the surrogate oil has been well characterized in previous work (Pelz et al., 2011) and shown to be highly similar to the MC252 oil, which both contain roughly 20% volatile organic carbon. Analysis of PHCs at the beginning of the incubations confirmed that most volatile PHCs had been removed, and thus, it was concluded that volatile compounds constituted a minor fraction of the weathered oil used as corroborated by previous work (Pelz et al., 2011). Nonetheless, we acknowledge that hydrocarbons in the low range of molecular weight (e.g., C12–C19 alkanes, two and three ring-aromatics), present at the beginning of our incubations, could have been affected by evaporation. Two experiments were conducted under identical conditions, with the exception that the mixing condition was varied. For all enrichments, 1.5 L of seawater was incubated in 2 L glass bottles. In oil treatments under both mixing regimes, 3 μl of oil was added to a floating boom to create a surface oil slick (Supplementary Figure S1; Prince and Butler, 2014). For the low-mixing oil + dispersant treatment, 3 μl of weathered oil was added to a floating boom, followed by 0.2 μl of dispersant. For the high-mixing oil + dispersant treatment, 3.2 μl of premixed oil/dispersant mixture was added to the bottles without booms. The mixture was prepared with a dispersant to oil ratio (DOR) of 1:15, similar to that recommended for emergency oil spill response (Joint Analysis Group [JAG], 2010; Prince and Butler, 2014; Kleindienst et al., 2015a). For each mixing condition, a total of 48 2L bottles were incubated for both chemical (24 bottles) and biological (24 bottles) analysis. Our previous incubations of surface GOM seawater revealed little to no microbial activity detected as respiration to carbon dioxide when no oil was added.
Bottles were incubated unsealed in an incubator at 100 RPM and 25°C in the dark. Autoclaved Nanopure water was routinely added to compensate for evaporation. Based on previous research, the evaporation of weathered oil is expected to be minimal (Prince and Butler, 2014). True duplicate samples for each treatment were sacrificed for both nucleic acid and oil extraction at 0, 7, 15, 22, 30, and 40 days. Additional duplicate bottles were prepared for each treatment and sealed with rubber stoppers for quantification of respiration. Oxygen concentrations were measured by a Presens MicroX 4 optode system with PSt7 non-invasive sensor spots (Presens, Germany) adhered to the inside of the glass bottle. Carbon dioxide concentrations were measured by injecting 100 μl of headspace into a GC-FID equipped with a methanizer (Shimadzu, Japan). Nutrient concentrations were monitored throughout the incubation period. Nitrite/nitrate, ammonia and soluble phosphate measurements were determined using established methods (Murphy and Riley, 1962; Strickland and Parsons, 1972; García-Robledo et al., 2014).
Extraction and Analysis of Hydrocarbons
At each time point, bottles were frozen at −20°C until extraction. For the low-mixing treatments, enrichments were extracted and analyzed according to Prince and Butler (2014). Briefly, the entire volume of each bottle was extracted with chloroform and the solvent was concentrated under a nitrogen purge before analysis. The high-mixing scenario samples were extracted as follows. A 10 μl aliquot of mixed standard was added to each bottle to correct for extraction efficiency. A 200 ml volume of 100% ACS grade dichloromethane (DCM) (BDH Chemicals, United Kingdom) was added to each bottle and gently shaken for 30 s. The DCM layer was then carefully transferred to a separatory funnel and collected in a flask from the bottom. DCM treatment was repeated three times to recover all the residual oil from the bottle. The retrieved DCM layer was then reduced in volume to 5 ml in a Turbovap (Biotage, Uppsala, Sweden) followed by addition of 20 ml of 100% ACS grade hexane (Fisher Chemical, NJ, United States). The volume of the extract was further reduced to 1 ml and stored in brown glass HPLC vials at 4°C until analysis. Extracted oil samples from low-mixing enrichments were analyzed by GC-MS as previously described (Prince and Butler, 2014). The high-mixing samples were analyzed based on previous literature with optimization (Romero et al., 2015; Sørensen et al., 2016). Briefly, samples were analyzed on a GC/MS/MS with a quadrupole mass spectrometer (Agilent, CA, United States), following EPA methods (Method 8270D). DWH crude oil (NIST 2779) was used as a reference standard. The fact that oil measurements were conducted with different instruments at separate institutions was unfortunate and caused by the limited availability of instrument time. Thus, direct comparisons were avoided between the two mixing conditions.
Since PHCs were extracted with different protocols and analyzed on different GC-MS instruments for the two experiments, results are reported as relative values to T0, with respect to 17α(H),21β(H)-hopane as a conserved internal standard (Prince et al., 1994). Half-lives of each class of PHCs were calculated using a first order kinetic equation (Edwards et al., 2011; Zahed et al., 2011; Prince et al., 2013).
Nucleic Acid Extraction and Microbial Community Characterization
Only the high mixing scenario samples were extracted and analyzed for microbial community characterization. All 1.5 L of seawater from each bottle was filtered through 0.2 μm Mobio Powerwater Filter (MoBio Laboratories, CA, United States) and stored at −80°C before extraction. Each half of the filter was extracted for DNA and RNA using the Mobio Powerwater DNA isolation kit (MoBio Laboratories, CA, United States) and Mobio Powerwater RNA isolation kit (MoBio Laboratories, CA, United States), respectively, following the manufacturer’s protocols. RNA was reverse-transcribed to cDNA using the qScript XLT cDNA supermix kit (Quantabio, MA, United States). Extracted DNA or cDNA was quantified with the Qubit HS assay kit (Invitrogen, Carlsbad, CA, United States) and 10 ng per reaction was used to generate SSU rRNA amplicons. Prokaryotic community composition was determined by applying a high-throughput sequencing-based protocol that targets PCR-generated amplicons from the V4 variable regions of the SSU rRNA gene using the primer set CS1_515F (5′-CACTGACGACATGGTTCTACA_GTGCCAGCMGCCGC GGTAA) and CS2_806R (5′-TACGGTAGCAGAGACTT GGTCT_GGACTACHVGGGTWTCTAAT) (Caporaso et al., 2010; Moonsamy et al., 2013). The resulting SSU rRNA gene amplicons were barcoded with unique 10-base barcodes (Fluidigm Corporation, CA, United States), pooled into equal DNA aliquots, and sequenced on an Illumina MiSeq2000 platform at the DNA services facility of the University of Chicago according to established methods (Bybee et al., 2011; Green et al., 2015; Herbold et al., 2015). The generated sequence libraries are available at NCBI under BioProject: PRJNA434233.
Sequence libraries were processed using multiple bioinformatics tools. Paired-end reads were merged with PEAR. The merged sequences were then demultiplexed and trimmed with vsearch and mothur, respectively. Chimeras were detected and removed with vsearch. Dereplicated sequences were clustered into OTUs using SWARM, with d = 1 (Mahé et al., 2014). Representative sequences were then taxonomically assigned via the RDP classifier against the SILVA database (Wang et al., 2007; Quast et al., 2013). Top abundant OTUs were blasted against the NCBI 16S ribosomal RNA database. The OTUs and their closely related neighbors were aligned and screened using the V4 hypervariable region in Mothur. A maximum likelihood tree was generated in MEGA7 with 1000 bootstraps (Kumar et al., 2016).
Quantification of both SSU rRNA genes and dinitrogenase (nifH) genes was performed using quantitative PCR on a StepOnePlus real-time PCR platform (Applied Biosystems, CA, United States) using established methods (Gaby and Buckley, 2017; Kolton et al., 2019; Shin et al., 2019). The qPCR assay employed a PowerUp SYBR Green Master Mix (Applied Biosystems, CA, United States). Quantification of SSU rRNA was performed using the primer set 331F (5′-CCTACGGGAGGCAGCAGT-3′)/518R(5′-ATTACCGCGGCTGCTG-3′) (Muyzer and Waal, 1993). Standard curves were obtained by serial dilution of standard plasmids containing target Escherichia coli k12 SSU rRNA as the insert (2.76 × 103 to 2.76 × 108 copies). The running conditions were: 2 min at 50°C, 2 min at 95°C, followed by 40 cycles of 95°C for 15s, 55°C for 15s and 72°C for 1 min. The primer set used was PolF(5′-TGCGAYCCSAARGCBGACTC-3′)/PolR(5′-ATSGCCATCATYTCRCCGGA-3′) (Poly et al., 2001). Standard curves were obtained by serial dilution of standard plasmids containing target Azotobacter vinelandii nifH gene fragments as the insert (3.2 × 102 to 3.2 × 107 copies). The running conditions were: 2 min at 50°C, 2 min at 95°C, followed by 45 cycles of 95°C for 15s, 63°C for 1 min. In all experiments, negative controls containing no template DNA were subjected to the same qPCR procedure to exclude or detect any possible DNA contamination.
Results
Impact of Dispersant and Mixing Conditions on Hydrocarbon Biodegradation
The current experiments were conducted with freshly collected seawater, and the active microbial community immediately responded to oil addition, with no observed lag phase in activity. The oil used in the experiment was weathered with a 20% weight loss, which is considered representative of surfaced oil which is treated with dispersant in situ during emergency response efforts (National Research Council, 2005).
Microbial respiration was quantified as oxygen consumption and CO2 production (Supplementary Figure S2). In the high mixing experiment, no significant differences in respiration rate were observed between treatments, with measured oxygen consumption rates of 4.39 ± 0.64 and 4.46 ± 0.75 μmol O2/L/day measured in oil only and oil + dispersant treatments, respectively. Similar to the oxygen consumption results, no difference was observed in CO2 production rates between treatments (0.34 ± 0.01 and 0.35 ± 0.02 μmol CO2/L/day for oil only and oil + dispersant treatments, respectively). In the low mixing experiment, a slight increase in respiration rates was found in the dispersant treatment, but the difference was not statistically significant (2.71 ± 0.52 and 3.44 ± 0.52 μmol O2/L/day for oil only and oil + dispersant treatments, respectively). Rate of CO2 production slightly increased from 0.41 ± 0.02 to 0.49 ± 0.03 μmol CO2/L/day (p < 0.05) when dispersant was applied. Further, major nutrients, inorganic nitrogen and phosphorus, remained relatively constant during the incubations, except for nitrate/nitrite slightly increased during the last stages (after 22 days) (Supplementary Figure S3).
Degradation significantly differed between different classes of hydrocarbon compounds (Figure 1). Low molecular weight compounds were degraded first followed by more recalcitrant compounds such as certain PAHs. First order degradation coefficients, obtained by linear regression of log transformed concentration data, suggested a significant difference between the two treatments at high mixing conditions (p < 0.05), while no significant difference between treatments under low mixing conditions. In all incubations, alkanes between C12 and C19 were degraded rapidly. Naphthalene and its homologs were rapidly degraded or volatilized within the first week of incubation in all treatments, whereas phenanthrene and its homologs showed a high recalcitrance to biodegradation in all incubations other than the high mixing dispersant treatment. Duplicates of the same treatment showed minor variations at the majority time points, and thus the results were representative for the scenario.
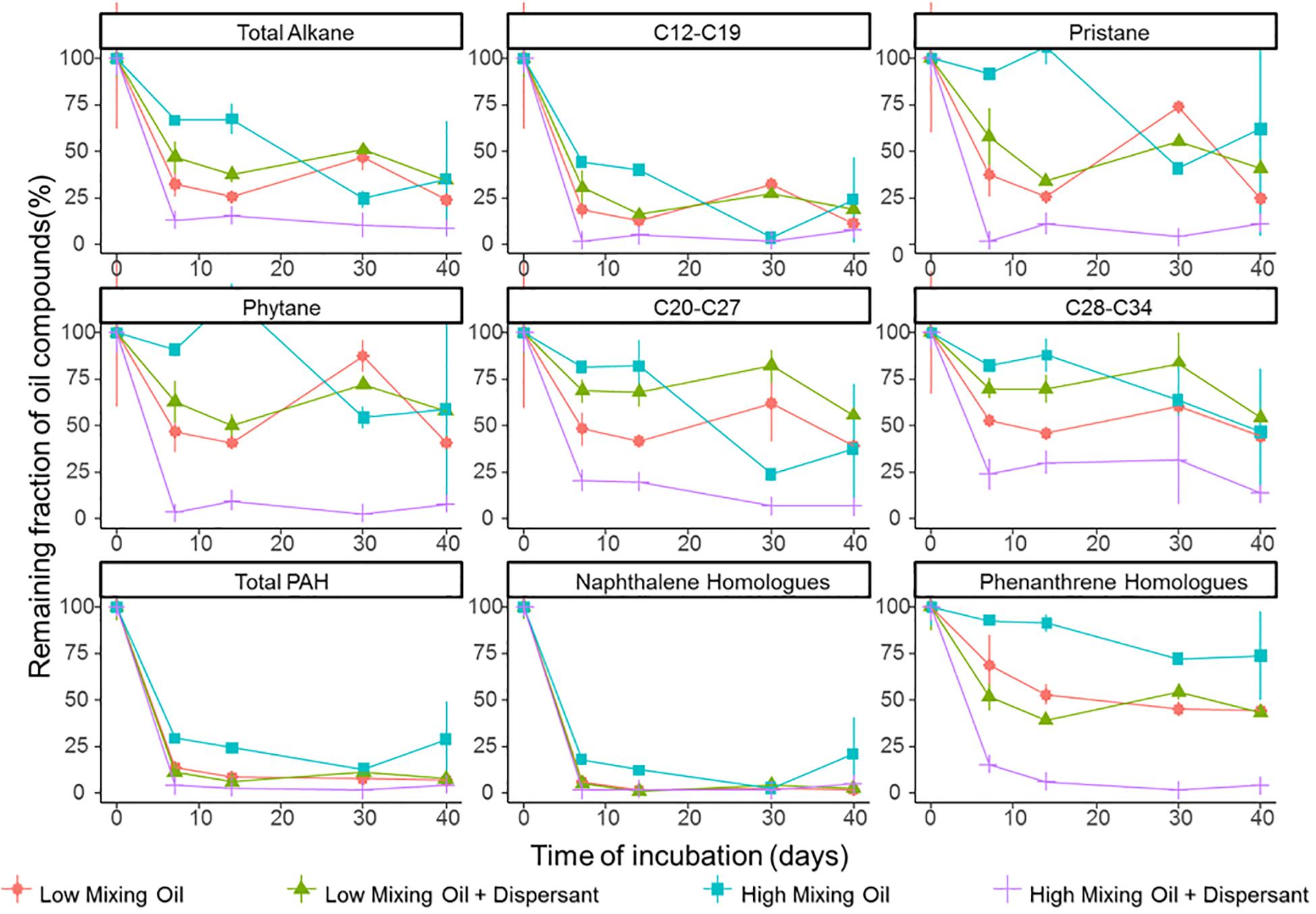
Figure 1. Degradation of petroleum hydrocarbons in microcosms of surface seawater. Hydrocarbon compounds are clustered based on their chemical properties and structure. Values shown are averages of duplicate measurements. Error bars are standard deviations.
The efficacy of dispersant treatment strongly depended on the mixing condition (Figure 1). At low mixing conditions with booms, no statistical difference was observed between treatments with or without dispersant, while the degradation rate was significantly elevated in the dispersant treatment in the high mixing scenario (without booms). Under high mixing conditions, half-lives of total petroleum hydrocarbons (TPH) were 15.4 and 8.8 days for the oil only and oil + dispersant treatments, respectively (Supplementary Figure S4). The effect of dispersant on biodegradation was more pronounced for lower solubility compounds, including phenanthrene and its homologs and long chain alkanes (C ≥ 20).
Diagnostic isomer ratios were used for confirmation of both alkane and PAH degradation. The ratio for n-C17/pristane and n-C18/phytane decreased with time in all treatments (Supplementary Table S1), clearly indicating the loss of alkane compounds was due to degradation (De Jonge et al., 1997). For PAH degradation, preferential degradation of 2-methylphenanthrene over 1-methylphenanthrene was previously established as an indicator of biodegradation (Bastiaens et al., 2000; Leys et al., 2005; Lamberts et al., 2008). The ratio of 2-methylphenanthrene to 1-methylphenanthrene increased with time in the high mixing condition treatments (Supplementary Figure S5) providing further evidence of biodegradation (58).
Response of the Total and Active Microbial Communities
Microbial communities were characterized in high mixing condition treatments to mimic the dynamics of microbial populations that carry out biodegradation of completely dispersed oil in seawater. We used RNA sequences as a proxy to characterize the metabolically active microbial populations in oil-contaminated marine ecosystems. Similar patterns in community compositions were observed over time at the RNA and DNA level (Figures 2, 3, and Supplementary Figure S6), indicating that the majority of groups detected are metabolically active. According to PERMANOVA analysis (Anderson, 2005), both time and treatment were statistically significant factors that shaped microbial communities (p < 0.05).
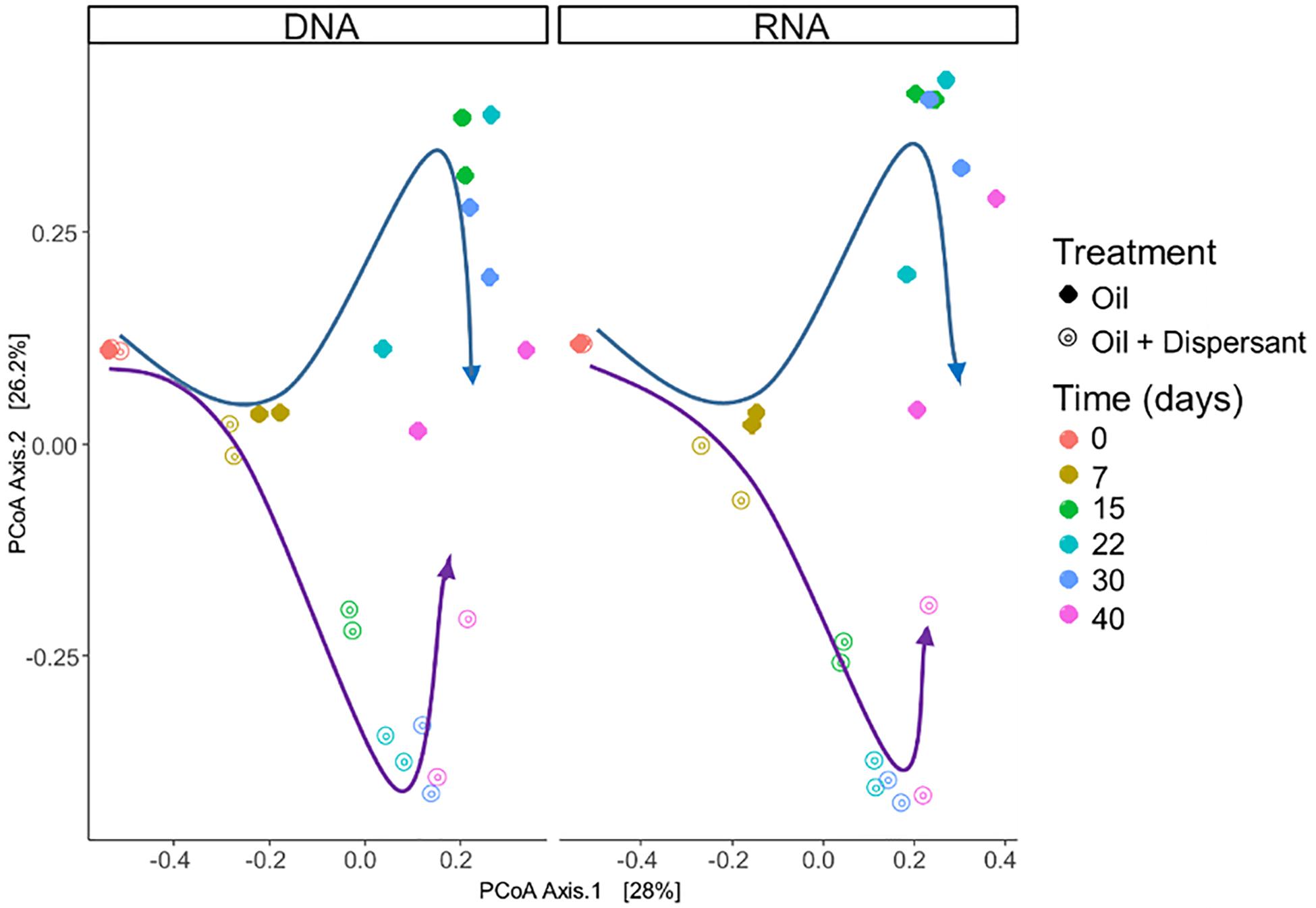
Figure 2. Comparison of microbial community composition with incubation time and treatment in seawater microcosms for both high mixing treatments. Left and right panels show microbial community composition from sequencing of SSU rRNA genes in DNA and RNA extracts, respectively. Arrows highlight the changes in microbial community composition: blue arrows indicate oil treatments, purple arrows indicate the oil + dispersant treatments. Similarities between microbial communities are displayed as the Bray–Curtis distance metric on a PCoA plot.
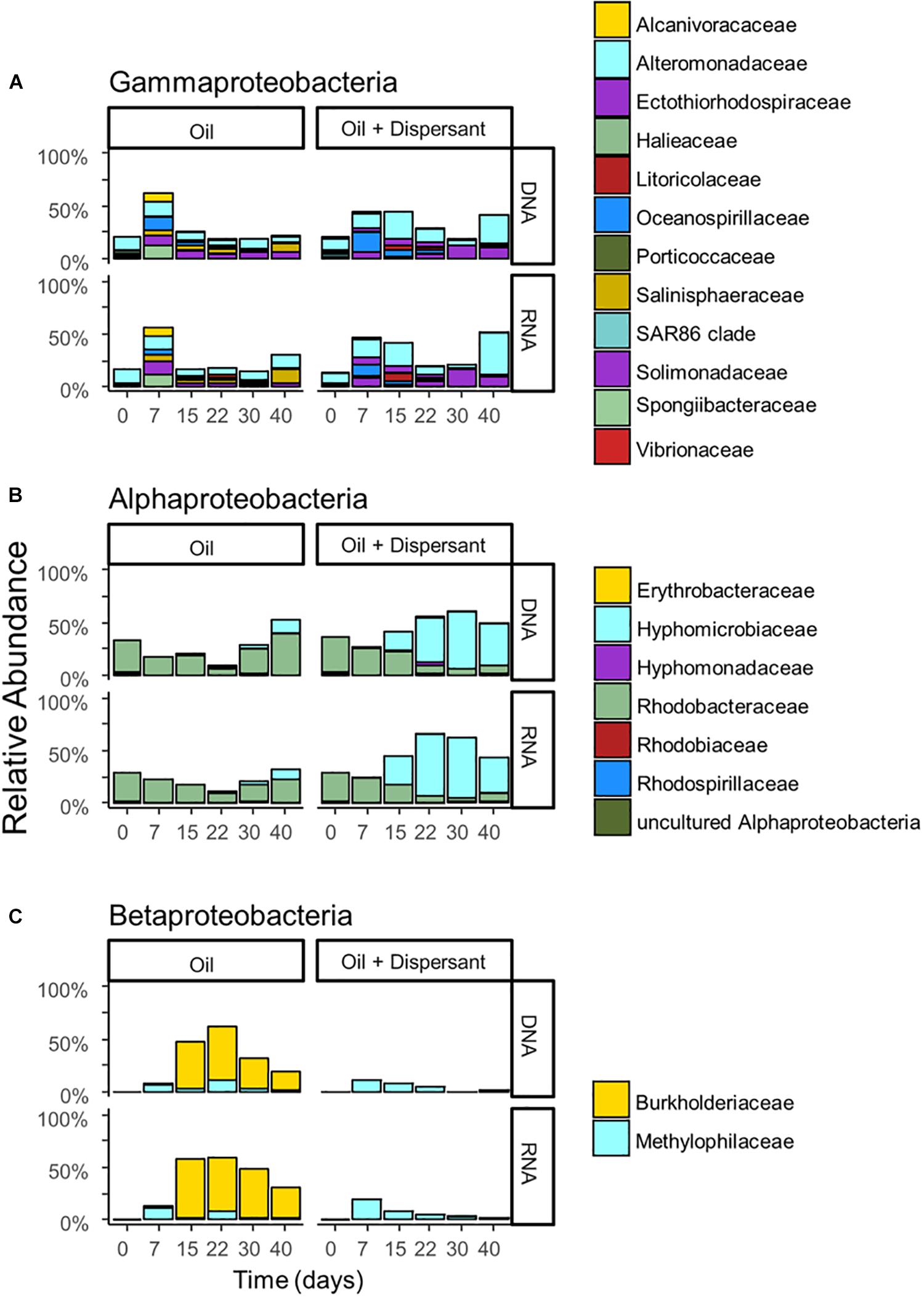
Figure 3. The relative abundance of (A) Gammaproteobacteria, (B) Alphaproteobacteria, and (C) Betaproteobacteria with incubation time and treatment for high mixing treatments. Barplots show mean value of duplicated samples. Taxa are grouped at the family level and relative abundance is calculated relative to total sequences.
A succession of microbial populations, dominated by those affiliated with known hydrocarbon-degrading bacterial groups, was observed in parallel with the chemical evolution of petroleum hydrocarbon compounds. Microbial community composition was consistent between duplicates of the same treatment (Figure 2). Community composition of the oil only and oil + dispersant treatments clearly diverged substantially by day 15. Bray–Curtis distance metrics showed that the two treatments diverged until 2–3 weeks of incubation and then converged by the end of the 40 day incubation, indicating a more similar composition of both total and metabolic active community by the end of the experiment. A large fraction of the retrieved sequences could not be assigned at the genus level, indicating that most microbial populations that develop in dispersed oil remain uncharacterized. Microbial communities during the first 2 weeks in all treatments were dominated by the class Gammaproteobacteria (Figure 3), which contributed up to 63% (DNA) and 58% (RNA) relative sequence abundance in oil only treatments, and 44% (DNA) and 45% (RNA) in dispersant treatments. Succession over time in individual microbial populations followed the changes in hydrocarbon chemistry. Typical oil degraders observed during oil inputs, such as Alcanivorax (denovo30) were observed within in the first week of incubation and then disappeared in the community (Figure 4). A dominant group (denovo4) that was closely related to the Polycyclovorans algicola of the family Solimonadaceae (Figure 3A), showed high relative abundance during the first 2 weeks of the incubation, contributing up to 22.3% in individual samples. A bloom of the family Oceanospirilliceae was also observed on day 7 in both treatments. The dominant OTU (denovo18) showed high sequence identity to Marinobacterium marisflavi. The group Alteromonadaceae was also abundant in both cases, and the closest match of the dominant OTU (denovo9) was closely related to the Marinobacter adhaerens.

Figure 4. The maximum likelihood phylogenetic tree of selected dominant OTUs. Names in red indicated the dominant OTUs from the current study, names in black indicated the closely related taxa that has been previously reported.
An OTU (denovo1) within the Burkholderiaceae of the Betaproteobacteria dominated in oil only treatments during the middle to late stages of incubation, comprising up to 51% of the total DNA library on day 22 and 56% of the total RNA library on day 15 (Figure 3C). Another family of the Betaproteobacteria group responding to the treatment was the Methylophilaceae. The dominant OTU (denovo11) of the family was closely related to the Rivicola pingtungensis and Methylotenera versatilis.
Two major families, Hyphomicrobiaceae and Rhodobacteraceae, of the Alphaproteobacteria were showed high relative abundance within the microbial community (Figure 3B). Multiple OTUs, represented by denovo8, related to the Rhodobacteraceae, were abundant in the early stages of the incubation, contributing up to 22 and 24% at the RNA level in both oil and dispersant treatments at day 7, respectively. Another set of OTUs, including denovo6, within the Rhodobacteraceae dominated the communities during the latter stages of incubation in oil only treatments. An OTU within the family Hyphomicrobiaceae (denovo2) dominated the microbial community from day 15 in the oil + dispersant treatment, constituting up to 55 and 57% at the DNA and RNA level, respectively (Figure 3B). A bloom of Hyphomicrobiaceae was observed at day 15, when both naphthalene and phenanthrene homologs were depleted in the oil + dispersant treatment. Meanwhile, an increase in relative abundance of the family Hyphmicrobiaceae emerged in the oil only treatment on day 40.
Microbial abundance, as determined by qPCR of SSU rRNA genes, was constant throughout all incubations (Figure 5A). In contrast, our qPCR results suggest that certain ecosystem functions are impacted by oil and dispersant treatment. The relative abundance of nitrogenase gene (nifH), which is a widely used molecular marker of nitrogen fixation (Gaby and Buckley, 2014), was up-regulated during the middle stages of incubation (Figure 5B), and the maximum abundance of nifH in the oil + dispersant treatments was over twice that in the oil only treatments. The abundance of nifH in the oil + dispersant treatments peaked on day 15, comprising 18% of the community as estimated by normalizing to the abundance of SSU rRNA genes. In the oil only treatments, a maximum abundance of 8% was observed at day 22.
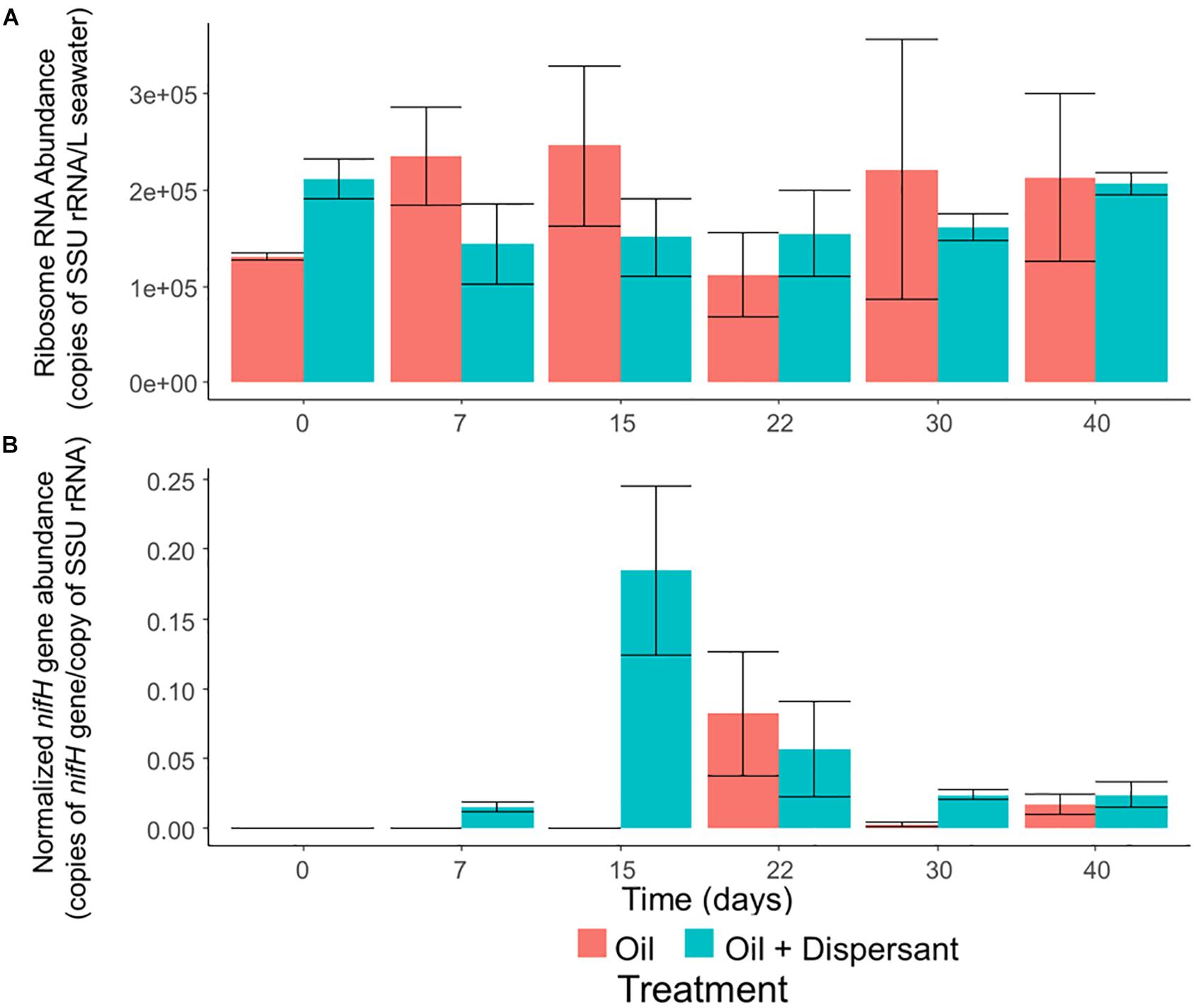
Figure 5. The abundance of overall bacteria and nitrogen-fixing prokaryotes in high mixing seawater microcosms as determined by quantitative PCR of (A) SSU rRNA genes and (B) nifH genes normalized to SSU rRNA genes. Error bars represent the standard deviation of duplicates.
Discussion
The primary objectives of this study were to investigate the influence of mixing condition on the ability of dispersant to enhance PHC biodegradation as well as to identify dispersant impacts on metabolically active microbial communities in surface seawater from the Gulf of Mexico. Boom treatments were used to simulate conditions when an oil slick is not treated with dispersant or under conditions when there is inadequate turbulence to mix dispersant with oil in the slick. The simulated oil slick remained in booms and was visible throughout the 40-day incubation with and without dispersant application. Treatments without booms simulated a scenario, where the dispersant was efficiently mixed with oil by turbulence and thus biodegradation was further enhanced. Mixing energy in the surface ocean is defined by dissipative power, which ranges from 0 W/kg without wind or wave action to 1 W/kg during catastrophic storms such as a hurricane or typhoon (Deane et al., 2016). Our low mixing treatment should resemble the windless/waveless condition, whereas the high mixing treatment mimics the top few meters of surface ocean exposed to substantial wind or wave mixing. Quantification of turbulence energy is beyond the scope of this investigation and warrants further study.
Impact of Dispersant Application on the Rates and Controls of Hydrocarbon Biodegradation
In the current study, addition of low concentrations of oil that mimic response conditions did not stimulate microbial respiration in the mesocosms. This result was corroborated by previous work, which showed no or minor changes in oxygen consumption upon oil + dispersant treatment (McFarlin et al., 2014), and respiration rates from our study are comparable to those measured in the surface oil slick generated by the Deepwater Horizon (DWH) discharge (ranging from 4.8 to 16.3 μmol O2/L/day) (Edwards et al., 2011). Although the O2 consumption to CO2 production ratio was relatively high (∼13), this could be explained by the formation of oxygenated PHCs and other intermediates. Biodegradation was shown to introduce oxygen into PHCs and form polarized intermediates, which are not able to be resolved by GC measurement (Aeppli et al., 2012). Since dispersant plus mixing led to enhanced biodegradation, these results indicate that oil biodegradation represented only a portion of the indigenous respiration occurring in our incubations. Respiration rates observed in the current study are corroborated by previous work, which suggested an average background respiration of 2.2 μmol O2/L/day in surface seawater in the GOM (Edwards et al., 2011). Similarly, the minimal changes in bacterial abundance and major nutrients observed in the current study also agreed with previous experiments conducted at low concentrations of oil expected during emergency response efforts (Edwards et al., 2011; McFarlin et al., 2014; Brakstad et al., 2015). Elevation in nitrate/nitrite concentrations, observed during the later stages of incubation, can be explained by the release of mineralized NH4+ followed by nitrification to NO3– when labile carbon substrate availability decreased. Overall, our observations indicate that overall respiration is neither inhibited nor enhanced by dispersant, and dispersant does not appear to stimulate the total growth of microbial communities at concentrations that resemble those expected during in situ response efforts. These findings agree with field observations during the DWH spill, where bacterial biomass remained constant within and outside the weathered surface oil slick (Edwards et al., 2011). In contrast, at similar oil concentrations, Wang et al. (2016) reported a fivefold increase in microbial abundance during incubation of deeper waters (collected from 1500 m water depth) amended with dispersed oil. Wang et al. (2016) may have observed growth due to the fact that their seawater mesocosms were amended with fresh crude oil along with dispersant, whereas our study employed weathered Macondo surrogate oil, which is more recalcitrant to biodegradation, as it contains a decreased portion of low molecular weight (LMW) compounds. Depletion of the labile portion of oil will reduce substrate quality and therefore diminish bacterial growth efficiency (Eiler et al., 2003). Alternatively, nutrient concentrations were an order of magnitude higher in the deep seawater used by Wang et al. (2016) which could have supported enhanced growth.
In this study, biodegradation followed previously reported patterns, in that labile compounds were degraded first followed by more recalcitrant compounds, especially for multi-methylated PAHs (Hu et al., 2017). The loss of PHCs in low mixing treatments appeared to be faster than the high mixing oil only treatment. Since experimental conditions were consistent between two oil only treatments, this could be explained by the fact that surface water was collected at a different time for each treatment and thus the results could reflect the presence of a different microbial populations, which warrants further study.
The differences in biodegradation rates among the treatments suggested that when directly applied to the oil slick without sufficient mixing, dispersant may not enhance hydrocarbon degradation. Under high mixing conditions, the application of dispersant enhanced the degradation of nearly all hydrocarbon groups. As a result, the decreased half-life of the total PHCs was comparable with previous measurements of 14 days for chemically dispersed oil in surface water (Prince and Butler, 2014) as well as degradation rates measured in simulated oil plumes (Hazen et al., 2010a; Prince et al., 2013; Wang et al., 2016). In addition, our results are in agreement with model predicted half-lives (Prosser et al., 2016). The effectiveness of the dispersant was more pronounced on recalcitrant compounds, possibly due to their low solubility, as these compounds were likely to remain with droplets. Thus, dispersant application could enhance degradation by decreasing droplet size and increasing the surface-to-volume ratio of the oil-water interface (Brakstad et al., 2014, 2017; Prince et al., 2017).
A subset of previous work has observed no increase in hydrocarbon degradation after dispersant application (Lindstrom and Braddock, 2002; Macías-Zamora et al., 2014) or concluded that microorganisms use dispersant as a carbon substrate and therefore attenuate hydrocarbon degradation (Lindstrom and Braddock, 2002; Kleindienst et al., 2015b). Inconsistencies in the effects of dispersant on hydrocarbon degradation should be interpreted with caution and may be attributed to methodological considerations. Studies have often employed oil, nutrient amendments, or dispersant-to-oil ratios that are far from in situ conditions expected during response efforts after a spill (Lee et al., 2013). Experiments that employ a water accommodated fraction (WAF) or a chemically enhanced water accommodated fraction (CEWAF) are difficult to compare to those in which oil and dispersant are directly added to seawater. The WAF method often excludes floating oil, which is likely to have a major impact on the activity of microbial communities in the incubations (Prince et al., 2016a). For example, using WAF and CEWAF treatments in seawater incubations, previous studies suggested that dispersant could suppress hydrocarbon degradation by selecting for dispersant degrading bacteria and against the most effective hydrocarbon degraders (Kleindienst et al., 2015b, 2016). However, it is unclear what portion of the amended oil was contained in the incubations and it is possible that oil droplets that were not entrained during the production of WAF or CEWAF would be excluded and therefore alter the availability of oil for biodegradation (Prince et al., 2016a). A higher oil carbon content in the WAF treatment could explain why microbial activity was higher relative to that of the CEWAF treatment in the study (Kleindienst et al., 2015b).
When dispersant is applied to oil slicks at the ocean surface, undispersed oil will remain in the slick, potentially hindering biodegradation. Therefore, this study focused on the comparison of a simulated oil slick to fully dispersed oil (Bejarano et al., 2013). The results confirm that dispersant application requires sufficient mixing power to disperse oil slicks (National Research Council, 2005; Prince, 2015). With sufficient mixing, dispersant enhances hydrocarbon degradation. At near in situ oil concentrations, the dispersant enhances hydrocarbon degradation under sufficient mixing condition. In contrast, dispersant application did not lead to a significant increase in oil degradation under the low mixing condition. Other environmental parameters, such as oxygen and nutrient availability, are unlikely to limit biodegradation at low oil concentrations (Lee et al., 2013).
Response of the Metabolically Active Microbial Communities to Dispersant Application
Changes in microbial communities in general paralleled PHC degradation patterns. The correspondence between total and active microbial community compositions suggested rapid turnover of microbial biomass. Shifts in community beta diversity with time suggested recovery from oil perturbation at the end of the experiment, agreeing with previous literature (Brakstad et al., 2017). The dominance of Proteobacteria was expected, as this group was often observed in previous PHC contaminated sites (Hazen et al., 2010b; Kostka et al., 2011; Valentine et al., 2014). The Gammaproteobacteria group contains many known aerobic hydrocarbon-degrading bacteria, such as Alcanivorax, that were shown to rapidly respond to oil deposition in marine ecosystems (Harayama et al., 1999; Kasai et al., 2002; Gertler et al., 2009; Hazen et al., 2010a; Kostka et al., 2011; Beazley et al., 2012). For example, similar to our previous field observations in Pensacola beach sands that were impacted by oil from the DWH disaster, a bloom of Alcanivorax dieselolei was observed in the oil treatments of this study (Figure 3A; Kostka et al., 2011). The Alcanivorax observed in the current study was closely related to the Alcanivorax borkumensis, a type strain widely studied for hydrocarbon biodegradation (Yakimov et al., 1998; Naether et al., 2013). However, Alcanivorax was not observed in abundance in the dispersant treatments, which may indicate that Corexit 9500 inhibits the adherence and growth of Alcanivorax, as suggested in a previous study (Bookstaver et al., 2015). Alternatively, dispersant application may have dramatically enhanced alkane availability and resulted in such a rapid response of Alcanivorax that by our first sampling point, day 7, this group was already succeeded by other taxa.
The genus P. algicola was suggested to utilize a wide range of both aliphatic and PAH compounds (Gutierrez et al., 2013, 2015). The substrate range of this group would account for its presence throughout the incubations, including when labile alkanes and low molecular weight PAHs were depleted. The presence of this OTU (denovo4) in both treatments suggested it is insensitive to dispersant. The response of the Oceanospirillaceae and Alteromonadaceae shown here agreed with observations of in situ microbial communities in contaminated seawater during the DWH disaster. Both groups contain known hydrocarbon-degrading bacteria that increased in relative abundance in response to oil spills in various environments (Deppe et al., 2005; Atlas et al., 2015; Yang et al., 2016; Techtmann et al., 2017).
Degradation occurred at a slower rate in the oil only treatment, in concurrence with a more prolonged bloom of microbial populations capable of degrading low molecular weight PAHs, such as members of the Burkholderiaceae. The dominance of this group is noteworthy, as members of the Betaproteobacteria usually comprise a relatively low percentage of marine bacterioplankton. The most abundant OTU, denovo1, was closely related (97% similarity) to Limnobacter litoralis KP1-19 (Lu et al., 2011). Previous work detected the essential genes for aromatic compound degradation in Limnobacter (Vedler et al., 2013), and this microbial group responded to crude oil addition in concurrence with the presence of PAHs (Canul-Chan et al., 2017). The absence of this group in the dispersant treatment may suggest its growth was inhibited by the presence of dispersed oil or an inability to compete with other PAH degraders in dispersed oil. The Methylophilaceae also increased in relative abundance in situ during the DWH discharge (Kessler et al., 2011; Eyice et al., 2015). This group contains well known methylotrophic bacteria, and some members are putative hydrocarbon-degraders (Thompson et al., 2017). Alternatively, Joye et al. (2014) suggested that these C1-oxidizers could be responding to the elevated DOM generated by the hydrocarbon-degraders.
The presence of the Rhodobacteraceae was constant throughout the incubation. The dominant populations, however, shifted with incubation time, and possibly with substrate availability. At early stages of incubation, multiple OTUs were enriched within the community, which could be responsible for the degradation of low molecular weight PAHs (Gutierrez et al., 2011). The closely related strain to dominant OTU denovo8 was Marivivens sp. JLT3646, which was suggested to possess the potential to degrade monoaromatic compounds based on genome annotation (Chen et al., 2018).
After depletion of all low molecular weight PHCs, other members within the Rhodobacteraceae were enriched during the latter stages of incubation, in agreement with previous observations attributing the degradation of more recalcitrant hydrocarbons to members of the Rhodobacteraceae (Kostka et al., 2011). The dominant OTU, denovo6, showed high sequence similarity to Marivita roseacus (99%) of the Roseobacter lineage (Budinoff et al., 2011). This lineage is a ubiquitous marine bacterial group that associates with phytoplankton cells or organic particles in the surface ocean (Slightom and Buchan, 2009). This cell-surface interaction may explain the abundance of this lineage in the oil treatment, possibly attaching to the oil slick, while the absence of slick in the dispersant treatment would be less favorable. Some phytoplankton-associated heterotrophic bacteria utilize hydrocarbons, both alkanes and PAHs, as they grow on natural hydrocarbons produced or accumulated by phototrophs (Gutierrez et al., 2011; Lea-Smith et al., 2015). One recent study suggests that members of the Roseobacter group have the potential to degrade aromatic hydrocarbons (Rodrigo-Torres et al., 2017). In fact, Klotz et al. isolated a new Roseobacter strain, Tritonibacter horizontis, from oil contaminated surface water collected in the Gulf of Mexico during the DWH discharge (Klotz et al., 2018). The strain demonstrated the ability to utilize hydroxylated and substituted aromatic compounds but cannot grow on alkanes (Giebel et al., 2016). Thompson et al. (2017) also found that members of this lineage were enriched by oil, especially in the presence of phytoplankton.
No strains matching sequences in the NCBI library fell within the same genus with denovo2. The closest neighbor was Hyphomicrobium vulgare, with only 93% sequence similarity, which suggested they belong to the same family, Hyphomicrobiaceae. Members of the Hyphomicrobiaceae were previously demonstrated to degrade hydrocarbons in pure culture and were enriched in previous studies of oil contaminated sites (Ozaki et al., 2006; Lafortune et al., 2009; Fahrenfeld et al., 2014). For example, Rodrigues et al. (2017) found members from this group were associated with both fluoranthene and pyrene addition, indicating their ability to degrade more recalcitrant high molecular weight PAHs. In the current study, the dominance of the Hyphomicrobiaceae was observed at different time points between oil only and oil + dispersant treatments. This phenomenon suggested a similar community response to reduced availability of low molecular weight hydrocarbons affected by different treatments.
Results of the current study indicate that dispersant application did not substantially alter bacterial abundance during PHC degradation, which supports previous observations in the surface oil slick after the DWH spill (Edwards et al., 2011). Meanwhile, enrichment in nifH genes suggested nutrient limitation during incubation. Since oil is depleted in major nutrient elements (i.e., nitrogen and phosphorus), biodegradation of this carbon-rich substrate has long been associated with nutrient limitation in marine environments (Atlas, 1981). For example in a metagenomic time series, an increase in the abundance of genes encoding nitrogen fixation and other nutrient acquisition pathways correlated with the abundance of genes for hydrocarbon degradation in oil-contaminated beach sands impacted by the DWH spill (Rodriguez-R et al., 2015). The nitrogen fixation process was followed by the nitrification mentioned earlier. Neither nitrification nor nitrogen fixation processes were shown to be associated with specific OTUs. Our results provide quantitative evidence that links nitrogen fixation to hydrocarbon degradation in microbial communities of surficial seawater, suggesting that these planktonic communities undergo nutrient limitation in response to oil contamination in coastal ecosystems (Rodriguez-R et al., 2015).
Conclusion
The results of this study indicate that when applying dispersant to an oil slick, biodegradation may not be substantially enhanced unless sufficient mixing is provided. When the simulated slick was sufficiently dispersed, a higher rate of removal was observed for more recalcitrant hydrocarbon compounds (such as methylated phenanthrene), suggesting that surface area available for microbial colonization is a primary factor limiting hydrocarbon degradation, and the application of dispersant will likely alleviate this constraint. While microbial growth and respiration were not substantially altered between treatments, RNA analysis revealed that dispersant application resulted in pronounced changes to the composition of metabolically active microbial communities. The quantitative increase in nitrogen-fixing members of the microbial community suggests a selection pressure for nitrogen fixation, likely indicating the robust response of the indigenous microbial communities to a readily biodegradable nitrogen-poor substrate. In order to improve model predictions and the bioremediation of dispersed oil during emergency response efforts, future study is warranted on the coupling of biodegradation to nitrogen fixation.
Data Availability Statement
The datasets generated for this study can be accessed from NCBI, PRJNA434233.
Author Contributions
XS: conceptualization, methodology, formal analysis, investigation, writing – original draft, visualization, and validation. LC and EM: methodology and investigation. IR: methodology, investigation, and resources. DH: resources and funding acquisition. JK: conceptualization, resources, writing – review and editing, supervision, and funding acquisition.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was made possible in part by grants from the Gulf of Mexico Research Initiative to the C-IMAGE II, C-IMAGE III, and Deep-C consortia. The authors thank Roger Prince for help with the petroleum hydrocarbon analysis and the crew of the R/V Weatherbird II for their assistance during the field program. Data are publicly available through the Gulf of Mexico Research Initiative Information and Data Cooperative (GRIIDC) at: https://data.gulfresearchinitiative.org/, doi: 10.7266/N72V2DNW.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02387/full#supplementary-material
References
Aeppli, C., Carmichael, C. A., Nelson, R. K., Lemkau, K. L., Graham, W. M., Redmond, M. C., et al. (2012). Oil weathering after the deepwater horizon disaster led to the formation of oxygenated residues. Environ. Sci. Technol. 46, 8799–8807. doi: 10.1021/es3015138
Anderson, M. J. (2005). PERMANOVA: a FORTRAN Computer Program for Permutational Multivariate Analysis of Variance. Auckland: University of Auckland.
Atlas, R. M. (1981). Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microbiol. Rev. 45, 180–209.
Atlas, R. M., and Bartha, R. (1972). Degradation and mineralization of petroleum in sea water?: limitation by nitrogen and phosphorous. Biotechnol. Bioeng. 14, 309–318. doi: 10.1002/bit.260140304
Atlas, R. M., Stoeckel, D. M., Faith, S. A., Minard-Smith, A., Thorn, J. R., and Benotti, M. J. (2015). Oil biodegradation and oil-degrading microbial populations in marsh sediments impacted by oil from the deepwater horizon well blowout. Environ. Sci. Technol. 49, 8356–8366. doi: 10.1021/acs.est.5b00413
Barbier, C. J. (2015). MDL 2179 Oil Spill by the Oil Rig “Deepwater Horizon. Available at: http://www.laed.uscourts.gov/case-information/mdl-mass-class-action/oilspill
Bastiaens, L., Springael, D., Wattiau, P., Harms, H., DeWachter, R., Verachtert, H., et al. (2000). Isolation of adherent polycyclic aromatic hydrocarbon (PAH)-degrading bacteria using PAH-sorbing carriers. Appl. Environ. Microbiol. 66, 1834–1843. doi: 10.1128/AEM.66.5.1834-1843.2000
Beazley, M. J., Martinez, R. J., Rajan, S., Powell, J., Piceno, Y. M., Tom, L. M., et al. (2012). Microbial community analysis of a coastal salt marsh affected by the deepwater horizon oil spill. PLoS One 7:e41305. doi: 10.1371/journal.pone.0041305
Bejarano, A. C., Levine, E., and Mearns, A. J. (2013). Effectiveness and potential ecological effects of offshore surface dsipersant use during the Deepwater Horizon oil spill: a retrospective analysis of monitoring data. Environ. Monit. Assess. 185, 10281–10295. doi: 10.1007/s10661-013-3332-y
Bookstaver, M., Bose, A., and Tripathi, A. (2015). Interaction of Alcanivorax borkumensis with a surfactant decorated oil - water interface. Langmuir 31, 5875–5881. doi: 10.1021/acs.langmuir.5b00688
Brakstad, O. G., Daling, P. S., Faksness, L. G., Almås, I. K., Vang, S. H., Syslak, L., et al. (2014). Depletion and biodegradation of hydrocarbons in dispersions and emulsions of the Macondo 252 oil generated in an oil-on-seawater mesocosm flume basin. Mar. Pollut. Bull. 84, 125–134. doi: 10.1016/j.marpolbul.2014.05.027
Brakstad, O. G., Nordtug, T., and Throne-Holst, M. (2015). Biodegradation of dispersed Macondo oil in seawater at low temperature and different oil droplet sizes. Mar. Pollut. Bull. 93, 144–152. doi: 10.1016/j.marpolbul.2015.02.006
Brakstad, O. G., Ribicic, D., Winkler, A., and Netzer, R. (2017). Biodegradation of dispersed oil in seawater is not inhibited by a commercial oil spill dispersant. Mar. Pollut. Bull 129, 555–561. doi: 10.1016/j.marpolbul.2017.10.030
Budinoff, C. R., Dunlap, J. R., Hadden, M., and Buchan, A. (2011). Marivita roseacus sp. nov., of the family Rhodobacteraceae, isolated from a temperate estuary and an emended description of the genus Marivita. J. Gen. Appl. Microbiol. 57, 259–267. doi: 10.2323/jgam.57.259
Bybee, S. M., Bracken-Grissom, H., Haynes, B. D., Hermansen, R. A., Byers, R. L., Clement, M. J., et al. (2011). Targeted amplicon sequencing (TAS): A scalable next-gen approach to multilocus, multitaxa phylogenetics. Genome Biol. Evol. 3, 1312–1323. doi: 10.1093/gbe/evr106
Campo, P., Venosa, A. D., and Suidan, M. T. (2013). Biodegradability of corexit 9500 and dispersed south Louisiana crude oil at 5 and 25°C. Environ. Sci. Technol. 47, 1960–1967. doi: 10.1021/es303881h
Canul-Chan, M., Sanchez-Gonzalez, M., Gonzalez-Burgos, A., Zepeda, A., and Rojas-Herrera, R. (2017). Population structures shift during the biodegradation of crude and fuel oil by an indigenous consortium. Int. J. Environ. Sci. Technol. 1, 1–6. doi: 10.1007/s13762-017-1362-1367
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth0510-335
Chakraborty, R., Borglin, S. E., Dubinsky, E. A., Andersen, G. L., and Hazen, T. C. (2012). Microbial response to the MC-252 oil and corexit 9500 in the Gulf of Mexico. Front. Microbiol 3:357. doi: 10.3389/fmicb.2012.00357
Chen, X., Zhan, P., Han, Y., Lin, D., Sun, J., and Tang, K. (2018). Complete genome sequence of Marivivens sp. JLT3646, a potential aromatic compound degrader. Mar. Genomics 38, 9–11. doi: 10.1016/j.margen.2017.06.002
Deane, G. B., Stokes, M. D., and Callaghan, A. H. (2016). The saturation of fluid turbulence in breaking laboratory waves and implications for whitecaps. J. Phys. Oceanogr. 46, 975–992. doi: 10.1175/JPO-D-14-0187.1
De Jonge, H., Freijer, J. I., Verstraten, J. M., Westerveld, J., and Van Der Wielen, F. W. M. (1997). Relation between bioavailability and fuel oil hydrocarbon composition in contaminated soils. Environ. Sci. Technol 31, 771–775. doi: 10.1021/es960456
Deppe, U., Richnow, H.-H., Michaelis, W., and Antranikian, G. (2005). Degradation of crude oil by an arctic microbial consortium. Extremophiles 9, 461–470. doi: 10.1007/s00792-005-0463-462
Edwards, B. R., Reddy, C. M., Camilli, R., Carmichael, C. A., Longnecker, K., and Van Mooy, B. A. S. (2011). Rapid microbial respiration of oil from the Deepwater Horizon spill in offshore surface waters of the Gulf of Mexico. Environ. Res. Lett. 6:035301. doi: 10.1088/1748-9326/6/3/035301
Eiler, A., Langenheder, S., Bertisson, S., and Tranvik, L. J. (2003). Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Appl. Environ. Microbiol. 69, 3701–3709. doi: 10.1128/AEM.69.7.3701
Eyice, Ö, Namura, M., Chen, Y., Mead, A., Samavedam, S., and Schäfer, H. (2015). SIP metagenomics identifies uncultivated Methylophilaceae as dimethylsulphide degrading bacteria in soil and lake sediment. ISME J 9, 2336–2348. doi: 10.1038/ismej.2015.37
Fahrenfeld, N., Cozzarelli, I. M., and Bailey, Z. (2014). Insights into biodegradation through depth-resolved microbial community functional and structural profiling of a crude-oil contaminant plume. Environ. Microbiol. 68, 453–462. doi: 10.1007/s00248-014-0421-426
Farrington, J. W. (2013). Oil pollution in the marine environment I: Inputs, big spills, small spills, and dribbles. Environ. Sci. Policy Sustain. Dev. 55, 3–13. doi: 10.1080/00139157.2013.843980
Foght, J. M., and Westlake, D. W. S. (1982). Effect of the dispersant corexit 9527 on the microbial degradation of prudhoe bay oil. Can. J. Microbiol 28, 117–122. doi: 10.1139/m82-012
Gaby, J. C., and Buckley, D. H. (2014). A comprehensive aligned nifH gene database: a multipurpose tool for studies of nitrogen-fixing bacteria. Database 2014, 1–8. doi: 10.1093/database/bau001
Gaby, J. C., and Buckley, D. H. (2017). The use of degenerate primers in qPCR analysis of functional genes can cause dramatic quantification bias as revealed by investigation of nifH primer performance. Microb. Ecol. 74, 701–708. doi: 10.1007/s00248-017-0968-0
García-Robledo, E., Corzo, A., and Papaspyrou, S. (2014). A fast and direct spectrophotometric method for the sequential determination of nitrate and nitrite at low concentrations in small volumes. Mar. Chem. 162, 30–36. doi: 10.1016/j.marchem.2014.03.002
Gertler, C., Gerdts, G., Timmis, K. N., and Golyshin, P. N. (2009). Microbial consortia in mesocosm bioremediation trial using oil sorbents, slow-release fertilizer and bioaugmentation. FEMS Microbiol. Ecol. 69, 288–300. doi: 10.1111/j.1574-6941.2009.00693.x
Giebel, H.-A., Klotz, F., Voget, S., Poehlein, A., Grosser, K., Teske, A., et al. (2016). Draft genome sequence of the marine Rhodobacteraceae strain O3.65, cultivated from oil-polluted seawater of the Deepwater Horizon oil spill. Stand. Genomic Sci. 11:81. doi: 10.1186/s40793-016-0201-207
Green, S. J., Venkatramanan, R., and Naqib, A. (2015). Deconstructing the polymerase chain reaction: understanding and correcting bias associated with primer degeneracies and primer-template mismatches. PLoS One 10:e0128122. doi: 10.1371/journal.pone.0128122
Gutierrez, T., Green, D. H., Nichols, P. D., Whitman, W. B., Semple, K. T., and Aitken, M. D. (2013). Polycyclovorans algicola gen. nov., sp. nov., an aromatic-hydrocarbon-degrading marine bacterium found associated with laboratory cultures of marine phytoplankton. Appl. Environ. Microbiol. 79, 205–214. doi: 10.1128/AEM.02833-2812
Gutierrez, T., Singleton, D. R., Aitken, M. D., and Semple, K. T. (2011). Stable isotope probing of an algal bloom to identify uncultivated members of the Rhodobacteraceae associated with low-molecular-weight polycyclic aromatic hydrocarbon degradation. Appl. Environ. Microbiol. 77, 7856–7860. doi: 10.1128/AEM.06200-6211
Gutierrez, T., Thompson, H. F., Angelova, A., Whitman, W. B., Huntemann, M., Copeland, A., et al. (2015). Genome sequence of Polycyclovorans algicola strain TG408, an obligate polycyclic aromatic hydrocarbon-degrading bacterium associated with marine eukaryotic phytoplankton. Genome Announc. 3:e207-15. doi: 10.1128/genomeA.00207-15.Copyright
Harayama, S., Kishira, H., Kasai, Y., and Shutsubo, K. (1999). Petroleum biodegradation in marine environments. J. Molec. Microbiol. Biotechnol. 1, 63–70.
Hazen, T. C., Dubinsky, E. A., DeSantis, T. Z., Andersen, G. L., Piceno, Y. M., Singh, N., et al. (2010a). Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330, 204–208. doi: 10.1126/science.1195979
Hazen, T. C., Dubinsky, E. A., DeSantis, T. Z., Andersen, G. L., Piceno, Y. M., Singh, N., et al. (2010b). SOM Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330, 204–208. doi: 10.1126/science.1195979
Herbold, C. W., Pelikan, C., Kuzyk, O., Hausmann, B., Angel, R., Berry, D., et al. (2015). A flexible and economical barcoding approach for highly multiplexed amplicon sequencing of diverse target genes. Front. Microbiol. 6:731. doi: 10.3389/fmicb.2015.00731
Hu, P., Dubinsky, E. A., Probst, A. J., Wang, J., Sieber, C. M. K., Tom, L. M., et al. (2017). Simulation of Deepwater Horizon oil plume reveals substrate specialization within a complex community of hydrocarbon degraders. Proc. Natl. Acad. Sci. U.S.A. 114, 7432–7437. doi: 10.1073/pnas.1703424114
Joint Analysis Group (JAG) (2010). Review of Preliminary Data to Examine Subsurface Oil in the Vicinity of MC252 #1 May 19 to June 19, 2010. Available at: https://repository.library.noaa.gov/view/noaa/130 (accessed September 22, 2017).
Joye, S., Kleindienst, S., Gilbert, J., Handley, K., Weisenhorn, P., Overholt, W., et al. (2016). Responses of microbial communities to hydrocarbon exposures. Oceanography 29, 136–149. doi: 10.5670/oceanog.2016.78
Joye, S. B., Teske, A. P., and Kostka, J. E. (2014). microbial dynamics following the macondo oil well blowout across gulf of mexico environments. Bioscience 64, 766–777. doi: 10.1093/biosci/biu121
Kasai, Y., Kishira, H., Sasaki, T., Syutsubo, K., Watanabe, K., and Harayama, S. (2002). Predominant growth of Alcanivorax strains in oil- contaminated and nutrient-supplemented sea water. Environ. Microbiol. 4, 141–147. doi: 10.1046/j.1462-2920.2002.00275.x
Kessler, J. D., Valentine, D. L., Redmond, M. C., Du, M., Chan, E. W., Mendes, S. D., et al. (2011). A persistent oxygen anomaly reveals the fate of spilled methane in the deep Gulf of Mexico. Science 331, 312–315. doi: 10.1126/science.1199697
Kleindienst, S., Paul, J. H., and Joye, S. B. (2015a). Using dispersants after oil spills: impacts on the composition and activity of microbial communities. Nat. Rev. Microbiol. 13, 388–396. doi: 10.1038/nrmicro3452
Kleindienst, S., Seidel, M., Ziervogel, K., Grim, S., Loftis, K., Harrison, S., et al. (2015b). Chemical dispersants can suppress the activity of natural oil-degrading microorganisms. Proc. Natl. Acad. Sci. U.S.A. 112, 14900–14905. doi: 10.1073/pnas.1507380112
Kleindienst, S., Seidel, M., Ziervogel, K., Grim, S., Loftis, K., Harrison, S., et al. (2016). Reply to Prince et al.: ability of chemical dispersants to reduce oil spill impacts remains unclear. Proc. Natl. Acad. Sci. U.S.A. 113, E1422–E1423. doi: 10.1073/pnas.1600498113
Klotz, F., Brinkhoff, T., Freese, H. M., Wietz, M., Teske, A., Simon, M., et al. (2018). Tritonibacter horizontis gen. nov., sp. nov., a member of the rhodobacteraceae, isolated from the Deepwater Horizon oil spill. Int. J. Syst. Evol. Microbiol. 68, 736–744. doi: 10.1099/ijsem.0.002573
Kolton, M., Marks, A., Wilson, R. M., Chanton, J. P., and Kostka, J. E. (2019). Impact of warming on greenhouse gas production and microbial diversity in anoxic peat from a Sphagnum-dominated bog (Grand Rapids, Minnesota, United States). Front. Microbiol. 10, 1–13. doi: 10.3389/fmicb.2019.00870
Kostka, J. E., Prakash, O., Overholt, W. A., Green, S. J., Freyer, G., Canion, A., et al. (2011). Hydrocarbon-degrading bacteria and the bacterial community response in gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl. Environ. Microbiol 77, 7962–7974. doi: 10.1128/AEM.05402-5411
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lafortune, I., Juteau, P., Déziel, E., Lépine, F., Beaudet, R., and Villemur, R. (2009). Bacterial diversity of a consortium degrading high-molecular-weight polycyclic aromatic hydrocarbons in a two-liquid phase biosystem. Environ. Microbiol. 57, 455–468. doi: 10.1007/S00248-008-9417-9414
Lamberts, R. F., Christensen, J. H., Mayer, P., Andersen, O., and Johnsen, A. R. (2008). Isomer-specific biodegradation of methylphenanthrenes by soil bacteria. Environ. Sci. Technol. 42, 4790–4796. doi: 10.1021/es800063s
Leahy, J. G., and Colwell, R. R. (1990). Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 54, 305–315.
Lea-Smith, D. J., Biller, S. J., Davey, M. P., Cotton, C. A. R., Perez Sepulveda, B. M., Turchyn, A. V., et al. (2015). Contribution of cyanobacterial alkane production to the ocean hydrocarbon cycle. Proc. Natl. Acad. Sci. U.S.A. 112, 13591–13596. doi: 10.1073/pnas.1507274112
Lee, K., Nedwed, T., Prince, R. C., and Palandro, D. (2013). Lab tests on the biodegradation of chemically dispersed oil should consider the rapid dilution that occurs at sea. Mar. Pollut. Bull. 73, 314–318. doi: 10.1016/j.marpolbul.2013.06.005
Leys, N. M., Ryngaert, A., Bastiaens, L., Wattiau, P., Top, E. M., Verstraete, W., et al. (2005). Occurrence and community composition of fast-growing Mycobacterium in soils contaminated with polycyclic aromatic hydrocarbons. FEMS Microbiol. Ecol. 51, 375–388. doi: 10.1007/978-3-319-51055-2_11
Lindstrom, J. E., and Braddock, J. F. (2002). Biodegradation of petroleum hydrocarbons at low temperature in the presence of the dispersant corexit 9500. Mar. Pollut. Bull. 44, 739–747. doi: 10.1016/s0025-326x(02)00050-4
Lu, H., Sato, Y., Fujimura, R., Nishizawa, T., Kamijo, T., and Ohta, H. (2011). Limnobacter litoralis sp. nov., a thiosulfate- oxidizing, heterotrophic bacterium isolated from a volcanic deposit, and emended description of the genus Limnobacter. Int. J. Syst. Evol. Microbiol 61, 404–407. doi: 10.1099/ijs.0.020206-0
Macías-Zamora, J. V., Meléndez-Sánchez, A. L., Ramírez-Álvarez, N., Gutiérrez-Galindo, E. A., and Orozco-Borbón, M. V. (2014). On the effects of the dispersant Corexit 9500© during the degradation process of n-alkanes and PAHs in marine sediments. Environ. Monit. Assess. 186, 1051–1061. doi: 10.1007/s10661-013-3438-3432
Mahé, F., Rognes, T., Quince, C., de Vargas, C., and Dunthorn, M. (2014). Swarm: robust and fast clustering method for amplicon-based studies. PeerJ 2:e593. doi: 10.7717/peerj.593
McFarlin, K. M., Prince, R. C., Perkins, R., and Leigh, M. B. (2014). Biodegradation of dispersed oil in Arctic seawater at −1°C. PLoS One 9:e84297. doi: 10.1371/journal.pone.0084297
Moonsamy, P. V., Williams, T., Bonella, P., Holcomb, C. L., Höglund, B. N., Hillman, G., et al. (2013). High throughput HLA genotyping using 454 sequencing and the fluidigm access ArrayTM system for simplified amplicon library preparation. Tissue Antigens 81, 141–149. doi: 10.1111/tan.12071
Murphy, J., and Riley, J. P. (1962). A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 27, 31–36. doi: 10.1016/s0003-2670(00)88444-5
Muyzer, G., and Waal, E. D. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695.
Naether, D. J., Slawtschew, S., Stasik, S., Engel, M., Olzog, M., Wick, L. Y., et al. (2013). Adaptation of hydrocarbonoclastic Alcanivorax borkumensis SK2 to alkanes and toxic organic compounds - a physiological and transcriptomic approach. Appl. Environ. Microbiol. 79, 4282–4293. doi: 10.1128/AEM.00694-613
National Research Council, (2005). Oil Spill Dispersants: Efficacy and Effects. Washington, DC: National Research Council.
Ozaki, S., Kishimoto, N., and Fujita, T. (2006). Isolation and phylogenetic characterization of microbial consortia able to degrade aromatic hydrocarbons at high rates. Microbes Environ. 21, 44–52. doi: 10.1264/jsme2.21.44
Pelz, O., Brown, J., Huddleston, M., Rand, G., Gardinali, P., Stubblefield, W., et al. (2011). Selection of a Surrogate MC252 Oil as a Reference Material for Future Aquatic Toxicity Tests and Other Studies. Available at: http://gulfresearchinitiative.org/wp-content/uploads/2012/05/Surrogate-Oil-selection-Paper-at-SETAC.pdf (accessed November 11, 2014).
Poly, F., Monrozier, L. J. L., and Bally, R. (2001). Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152, 95–103. doi: 10.1016/S0923-2508(00)01172-1174
Prince, R. C. (2015). Oil spill dispersants: boon or bane? Environ. Sci. Technol. 49, 6376–6384. doi: 10.1021/acs.est.5b00961
Prince, R. C., and Butler, J. D. (2014). A protocol for assessing the effectiveness of oil spill dispersants in stimulating the biodegradation of oil. Environ. Sci. Pollut. Res. 21, 9506–9510. doi: 10.1007/s11356-013-2053-2057
Prince, R. C., Butler, J. D., and Redman, A. D. (2017). The rate of crude oil biodegradation in the sea. Environ. Sci. Technol. 51, 1278–1284. doi: 10.1021/acs.est.6b03207
Prince, R. C., Coolbaugh, T. S., and Parkerton, T. F. (2016a). Oil dispersants do facilitate biodegradation of spilled oil. Proc. Natl. Acad. Sci. U.S.A. 113, E1421–E1421. doi: 10.1073/pnas.1525333113
Prince, R. C., Nash, G. W., and Hill, S. J. (2016b). The biodegradation of crude oil in the deep ocean. Mar. Pollut. Bull. 111, 354–357. doi: 10.1016/j.marpolbul.2016.06.087
Prince, R. C., Elmendorf, D. L., Lute, J. R., Hsu, C. S., Haith, C. E., Senius, J. D., et al. (1994). 17α(H)-21β(H)-Hopane as a conserved internal marker for estimating the biodegradation of crude oil. Environ. Sci. Technol. 28, 142–145. doi: 10.1021/es00050a019
Prince, R. C., McFarlin, K. M., Butler, J. D., Febbo, E. J., Wang, F. C. Y., and Nedwed, T. J. (2013). The primary biodegradation of dispersed crude oil in the sea. Chemosphere 90, 521–526. doi: 10.1016/j.chemosphere.2012.08.020
Prosser, C. M., Redman, A. D., Prince, R. C., Paumen, M. L., Letinski, D. J., and Butler, J. D. (2016). Evaluating persistence of petroleum hydrocarbons in aerobic aqueous media. Chemosphere 155, 542–549. doi: 10.1016/j.chemosphere.2016.04.089
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Rodrigo-Torres, L., Pujalte, M. J., and Arahal, D. R. (2017). Draft genome sequence of Thalassobius gelatinovorus CECT 4357T, a roseobacter with the potential ability to degrade polycyclic aromatic hydrocarbons. Gene Rep. 9, 32–36. doi: 10.1016/j.genrep.2017.08.005
Rodrigues, E. M., Morais, D. K., Pylro, V. S., Redmile-Gordon, M., de Oliveira, J. A., Roesch, L. F. W., et al. (2017). Aliphatic hydrocarbon enhances phenanthrene degradation by autochthonous prokaryotic communities from a pristine seawater. Microb. Ecol. 75, 688–700. doi: 10.1007/s00248-017-1078-1078
Rodriguez-R, L. M., Overholt, W. A., Hagan, C., Huettel, M., Kostka, J. E., and Konstantinidis, K. T. (2015). Microbial community successional patterns in beach sands impacted by the Deepwater Horizon oil spill. ISME J. 9, 1928–1940. doi: 10.1038/ismej.2015.5
Romero, I. C., Schwing, P. T., Brooks, G. R., Larson, R. A., Hastings, D. W., Ellis, G., et al. (2015). Hydrocarbons in deep-sea sediments following the 2010 Deepwater Horizon blowout in the northeast Gulf of Mexico. PLoS One 10:e0128371. doi: 10.1371/journal.pone.0128371
Sammarco, P. W., Kolian, S. R., Warby, R. A. F., Bouldin, J. L., Subra, W. A., and Porter, S. A. (2013). Distribution and concentrations of petroleum hydrocarbons associated with the BP/Deepwater Horizon Oil Spill. Gulf of Mexico. Mar. Pollut. Bull. 73, 129–143. doi: 10.1016/j.marpolbul.2013.05.029
Schwarz, J. R., Walder, J. D., and Colwell, R. R. (1975). Deep-sea bacteria: growth and utilization of hydrocarbons at ambient and In Situ Pressure. Can. J. Microbiol. 28, 982–986. doi: 10.1139/m75-098
Shin, B., Kim, M., Zengler, K., Chin, K. J., Overholt, W. A., Gieg, L. M., et al. (2019). Anaerobic degradation of hexadecane and phenanthrene coupled to sulfate reduction by enriched consortia from northern Gulf of Mexico seafloor sediment. Sci. Rep. 9, 1–13. doi: 10.1038/s41598-018-36567-x
Slightom, R. N., and Buchan, A. (2009). Surface colonization by marine roseobacters: integrating genotype and phenotype. Appl. Environ. Microbiol. 75, 6027–6037. doi: 10.1128/AEM.01508-1509
Sørensen, L., Meier, S., and Mjøs, S. A. (2016). Application of gas chromatography/tandem mass spectrometry to determine a wide range of petrogenic alkylated polycyclic aromatic hydrocarbons in biotic samples. Rapid Commun. Mass Spectrom. 30, 2052–2058. doi: 10.1002/rcm.7688
Strickland, J. D. H., and Parsons, T. R. (1972). A Pratical Handbook of Seawater Analysis. Ottawa: Fisheries Research Board of Canada.
Techtmann, S. M., Zhuang, M., Campo, P., Holder, E., Elk, M., Hazen, T. C., et al. (2017). Corexit 9500 enhances oil biodegradation and changes active bacterial community structure of oil enriched microcosms. Appl. Environ. Microbiol 83:e03462-16.
Thompson, H., Angelova, A., Bowler, B., Jones, M., and Gutierrez, T. (2017). Enhanced crude oil biodegradative potential of natural phytoplankton-associated hydrocarbonoclastic bacteria. Environ. Microbiol. 19, 2843–2861. doi: 10.1111/1462-2920.13811
U. S. National Oceanic, and Atmospheric Administration [NOAA], (2010). Brooks McCall Cruises Log. Silver Spring, MA: NOAA.
Valentine, D. L., Fisher, G. B., Bagby, S. C., Nelson, R. K., Reddy, C. M., Sylva, S. P., et al. (2014). Fallout plume of submerged oil from Deepwater Horizon. Proc. Natl. Acad. Sci. U.S.A. 11, 15906–15911. doi: 10.1073/pnas.1414873111
Vedler, E., Heinaru, E., Jutkina, J., Viggor, S., Koressaar, T., Remm, M., et al. (2013). Limnobacter spp. as newly detected phenol-degraders among Baltic Sea surface water bacteria characterised by comparative analysis of catabolic genes. Syst. Appl. Microbiol. 36, 525–532. doi: 10.1016/j.syapm.2013.07.004
Venosa, A. D., and Holder, E. L. (2007). Biodegradability of dispersed crude oil at two different temperatures. Mar. Pollut. Bull. 54, 545–553. doi: 10.1016/j.marpolbul.2006.12.013
Venosa, A. D., King, D. W., and Sorial, G. A. (2002). The baffled flask test for dispersant effectiveness: A round Robin evaluation of reproducibility and repeatability. Spill Sci. Technol. Bull. 7, 299–308. doi: 10.1016/S1353-2561(02)00072-75
Wade, T. L., Sericano, J. L., Sweet, S. T., Knap, A. H., and Guinasso, N. L. Jr. (2016). Spatial and temporal distribution of water column total polycyclic aromatic hydrocarbons (PAH) and total petroleum hydrocarbons (TPH) from the Deepwater Horizon (Macondo) incident. Mar. Pollut. Bull. 103, 286–293. doi: 10.1016/j.marpolbul.2015.12.002
Wang, J., Sandoval, K., Ding, Y., Stoeckel, D., Minard-Smith, A., Andersen, G., et al. (2016). Biodegradation of dispersed macondo crude oil by indigenous Gulf of Mexico microbial communities. Sci. Total Environ. 55, 453–468. doi: 10.1016/j.scitotenv.2016.03.015
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-67
Yakimov, M. M., Golyshin, P. N., Lang, S., Moore, E. R. B., Abraham, W., Lunsdorf, H., et al. (1998). Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Bacteriol. 48, 339–348. doi: 10.1099/00207713-48-2-339
Yang, T., Nigro, L. M., Gutierrez, T., Ambrosio, L. D., Joye, S. B., Highsmith, R., et al. (2016). Pulsed blooms and persistent oil-degrading bacterial populations in the water column during and after the Deepwater Horizon blowout. Deep. Res. Part II 129, 282–291. doi: 10.1016/j.dsr2.2014.01.014
Zahed, M. A., Aziz, H. A., Isa, M. H., and Mohajeri, L. (2010). Effect of initial oil concentration and dispersant on crude oil biodegradation in contaminated seawater. Bull. Environ. Contam. Toxicol. 84, 438–442. doi: 10.1007/s00128-010-9954-9957
Zahed, M. A., Aziz, H. A., Isa, M. H., Mohajeri, L., Mohajeri, S., and Kutty, S. R. M. (2011). Kinetic modeling and half life study on bioremediation of crude oil dispersed by Corexit 9500. J. Hazard. Mater. 185, 1027–1031. doi: 10.1016/j.jhazmat.2010.10.009
Keywords: hydrocarbon degradation, Deepwater Horizon, dispersant, microbial community, hydrocarbon analysis
Citation: Sun X, Chu L, Mercando E, Romero I, Hollander D and Kostka JE (2019) Dispersant Enhances Hydrocarbon Degradation and Alters the Structure of Metabolically Active Microbial Communities in Shallow Seawater From the Northeastern Gulf of Mexico. Front. Microbiol. 10:2387. doi: 10.3389/fmicb.2019.02387
Received: 24 February 2019; Accepted: 01 October 2019;
Published: 18 October 2019.
Edited by:
Tony Gutierrez, Heriot-Watt University, United KingdomReviewed by:
Andreas Teske, The University of North Carolina at Chapel Hill, United StatesZhanfei Liu, The University of Texas at Austin, United States
Copyright © 2019 Sun, Chu, Mercando, Romero, Hollander and Kostka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joel E. Kostka, joel.kostka@biology.gatech.edu
†ORCID: Joel E. Kostka orcid.org/0000-0003-3051-1330
 Xiaoxu Sun
Xiaoxu Sun Lena Chu
Lena Chu Elisa Mercando
Elisa Mercando Isabel Romero
Isabel Romero David Hollander
David Hollander Joel E. Kostka
Joel E. Kostka