- Division of Infectious Diseases, Department of Medicine, University of Pittsburgh School of Medicine, Pittsburgh, PA, United States
HIV-1 viremia persists at low-levels despite clinically effective antiretroviral therapy (ART). Here we review new methods to quantify and characterize persistent viremia at the single genome level, and discuss the mechanisms of persistence including clonal expansion of infected cells and tissue origins of viremia. A deeper understanding of how viremia persists on ART is critically important to the design of therapies to eliminate viremia and achieve a functional cure for HIV-1.
Introduction
Over the last 30 years there have been great strides made in the diagnosis and treatment of HIV-1 infection. The broad implementation of antiretroviral therapy (ART) has saved the lives of many millions of persons with HIV (Palella et al., 1998), but has also uncovered the persistence of HIV-1 on ART, both as a latent reservoir and as an expression of low-level viremia (Chun et al., 1995, 1997a,b; Finzi et al., 1997; Wong et al., 1997; Dornadula et al., 1999). Studies of patients on ART revealed two sources of plasma viremia: (1) short-lived, productively infected CD4+ T-cells that produce bursts of virus and then die; and (2) long-lived cells capable of producing viremia that is below the limit of detection of commercial assays (Ho et al., 1995; Wei et al., 1995; Perelson et al., 1996, 1997; Dornadula et al., 1999). Longitudinal studies of persistent plasma HIV-1 viremia below the limit of detection of commercial assays in persons on long-term ART have provided a biphasic model of viremia decay, including an initial decay phase with a half-life of 39 weeks and a more slowly decaying phase with a half-life of 11 years (Maldarelli et al., 2007; Palmer et al., 2008; Riddler et al., 2016). These studies showed a positive association between persistent plasma viremia and pre-ART plasma HIV-1 RNA. Riddler et al. (2016) reported that persistent plasma viremia was associated with higher CD8 T-cell counts and a lower CD4/CD8 ratio on ART, both markers of incomplete immune recovery. The source(s) and mechanisms of persistent HIV-1 plasma viremia are still largely uncharacterized despite improved detection methods. Because persistent viremia represents a major barrier to HIV-1 cure, its characterization and clearance remain a high priority. Here we review some recent advances in measuring and identifying the origins of persistent viremia.
Persistent Hiv-1 Viremia on Art
Measurement and Recent Improvements
The development of more sensitive, reverse-transcriptase initiated quantitative PCR (RT qPCR) assays revealed that more than half of individuals on ART with plasma HIV-1 RNA suppressed below the limit of detection of commercial assays (20–40 copies/ml) still have detectable HIV-1 RNA in plasma, averaging around 1–3 copies/mL (Dornadula et al., 1999; Palmer et al., 2003, 2008; Maldarelli et al., 2007; Zheng et al., 2013; Riddler et al., 2016). Recent innovations have improved the measurement of this low-level plasma HIV-1 RNA. The first-generation two-step qRT-PCR assay with single copy sensitivity targeted HIV-1 gag (gSCA) and required 6–7 mL of plasma (Palmer et al., 2003). A subsequent single copy qRT-PCR assay targeted a highly conserved region of integrase in HIV-1 pol (iSCA v1.0) and enhanced nucleic acid recovery from a smaller volume of plasma (Cillo et al., 2014). Despite highly successful implementation in many clinical studies, iSCA v1.0 required ultracentrifugation and only assayed about half of the total extracted nucleic acid for HIV-1 RNA. In the most recent iteration of a single-copy assay (iSCA v2.0), ultracentrifugation is replaced with tabletop centrifugation and a greater proportion (∼80%) of the total extracted nucleic acid is tested for HIV-1 RNA. Importantly, when equal volumes of the same donor plasma were tested using versions of iSCA, 55% of the samples that had no HIV-1 RNA detected by iSCA v1.0 had HIV-1 RNA detected by iSCA v2.0 (Tosiano et al., 2019a).
Automated, next-generation commercial platforms can reproducibly measure HIV-1 RNA in plasma above the limit of quantification (20–200 copies/mL depending on the platform) (Wiesmann et al., 2018). Although individual measurements using commercial platforms do not provide the sensitivity of manual single copy assays, automated platforms have potential as a screening tool. For example, results reported by either Roche or Abbott automated platforms as <20 or <40 copies/mL respectively (also known as “detected but not quantifiable”) are almost always detected and quantified by manual single copy assay (Margot et al., 2018; Tosiano et al., 2019b), whereas automated platform results indicating “no target detected” are associated with a significantly lower frequency of HIV-1 RNA detection by manual single copy assays. In addition, Bakkour et al. (2019) have reported that reasonable estimates of HIV-1 RNA copies/mL below the limit of quantification can be obtained using automated platforms to test multiple replicates of plasma to generate a combination of detected, non-detected, and detected but not quantifiable results. Each sample can be assigned a value for HIV-1 RNA by applying a mathematical algorithm based upon the qualitative readouts. Comparisons are in progress of HIV-1 RNA levels obtained by manual single copy assays versus multiple replicates on automated platforms. An automated platform with single copy sensitivity would have distinct advantages over more labor intensive and lower throughput, manual single copy assays.
Association of Persistent Viremia With Cell-Associated HIV-1 DNA
The half-life of persistent plasma viremia on stable ART, calculated using decay dynamics modeling, is more than 11 years (Riddler et al., 2016). Interestingly, decay of HIV-1 proviral DNA-containing cells on ART was recently reported to have a similar half-life of 13 years (Gandhi et al., 2017). Although it is enticing to suggest that the similar half-lives of total cell-associated HIV-1 DNA and plasma HIV-1 RNA on ART represent a direct association between infected cells and persistent plasma viremia, it is important to note that proviral DNA-containing cells rarely contain full-length, intact proviruses. In fact, less than 1–10% of proviruses that persist on ART are capable of producing infectious virus (Fourati et al., 2012; Ho et al., 2013; Bruner et al., 2019). Despite this data, many have reported direct associations of varying degrees between qPCR measures of the proviral reservoir (total cell-associated HIV-1 DNA) and persistent plasma viremia, suggesting that they are related (Chun et al., 2011; Mexas et al., 2012; Hong et al., 2018). The recent development of an assay capable of quantifying intact proviral DNA (Intact Proviral DNA Assay, IPDA) will help address questions regarding the degree of correlation between total and intact cell-associated DNA and plasma viremia (Bruner et al., 2019). Intact proviral DNA correlated modestly with ex vivo measurements of inducible, infectious virus outgrowth. However, such quantitative viral outgrowth assays (qVOAs) have not correlated with levels of persistent plasma viremia in individuals on ART (Siliciano et al., 2003; Eriksson et al., 2013). qVOAs have also been shown to underestimate the size of the reservoir by missing the fraction of intact provirus that is non-inducible ex vivo; this fraction could contribute to plasma viremia in vivo (Ho et al., 2013; Bruner et al., 2019). As such, assays that quantify intact proviruses may show stronger correlations with plasma viremia than total HIV-1 DNA. Studies are currently in progress to assess this possibility.
Association of Persistent Viremia With Cell-Associated HIV-1 RNA
Measurements of various forms of cell-associated bulk HIV-1 RNA have been used to estimate proviral transcriptional activity, both at steady-state and in response to latency reversal agents (Pasternak et al., 2008; Strain and Richman, 2013; Kiselinova et al., 2014; Kearney et al., 2015; Procopio et al., 2015; Hong et al., 2016; Li et al., 2016; Yukl et al., 2018). Whether cell-associated HIV-1 RNA correlates with production of plasma virus is debated. Typically, PCR-based approaches have targeted a single small region of bulk HIV-1 RNA. Although important information about HIV-1 pathogenesis has been garnered from these assays, their utility for assessing latency reversal or changes in viremia have been questioned (Eriksson et al., 2013; Archin et al., 2014; Elliott et al., 2014). In a recent study, cell-associated unspliced HIV-1 RNA strongly correlated with plasma viremia in untreated individuals, but not in individuals on ART (Hong et al., 2018). These conflicting results could be attributed to the accumulation of defective provirus after ART initiation (Bruner et al., 2016) leading to production of defective transcripts that do not result in virion production (Imamichi et al., 2016; Pollack et al., 2017; Wiegand et al., 2017). Interestingly, an analysis of various forms of cell-associated HIV-1 RNA transcripts that are expressed following treatment with latency reversal agents showed very different expression profiles depending upon the conditions and agent used, most of which did not lead to production of full-length polyadenylated transcripts (Yukl et al., 2018; Moron-Lopez et al., 2019). Given the uncertain value of measuring cell-associated HIV-1 RNA in all infected cells, assays that measure HIV-1 RNA transcribed only from intact proviruses (and therefore more likely to produce plasma virus) are likely to be more useful for assessing interventions aimed at perturbing and/or eliminating the HIV-1 reservoir (Wiegand et al., 2017; Yucha et al., 2017).
Sources of Persistent Hiv-1 Viremia on Art
Cell and Tissue Sources
Understanding the source of persistent viremia is critically important for the design of interventions to eliminate it. While the source of persistent viremia is likely multi-faceted and variable across individuals, ongoing studies are seeking common sources of viremia that could serve as more specific therapeutic targets. In virologically suppressed individuals, most HIV-1 proviral DNA is found in resting CD4+ T-cells (Chun et al., 1997a; Finzi et al., 1997; Wong et al., 1997). Though not considered a major contribution to the reservoir, other cells types such as monocytes, macrophages, and hematopoietic stem cells have been reported to harbor proviral DNA (Sonza et al., 2001; Zhu et al., 2002; Zaikos et al., 2018; Mitchell et al., 2019). The propensity for white blood cells to circulate throughout the body and penetrate various tissues and lymphoid organs provides an opportunity for infected cells to access ordinarily anatomically protected and/or immune-privileged sites. As such, HIV-1 RNA and/or DNA have been detected widely across tissues in virologically suppressed individuals, including in lymph nodes (Perreau et al., 2013), cerebrospinal fluid (Dahl et al., 2014; Spudich et al., 2019), adipose tissue (Couturier et al., 2015; Damouche et al., 2015), gut-associated lymphoid tissue (GALT) (Lampinen et al., 2000; Anton et al., 2003; Belmonte et al., 2007); and most recently, in the urethra (Ganor et al., 2019).
By contrast, the cellular and/or tissue reservoir(s) that contribute to persistent plasma viremia have not been clearly identified. It is possible that multiple sources contribute to variable degrees, and that sources vary between individuals. Historically, comparison of sequences of persistent plasma virus and total cell-associated proviral DNA in circulating CD4+ T-cells have rarely identified matches. This finding is likely because limited sequencing methods may not detect rare intact provirus that are producing virus. Indeed, in a study in which intensive sampling and single genome sequencing were performed, proviral sequences were detected that did match persistent plasma virus (Bailey et al., 2006). Another likely explanation for the discrepancy between proviral and plasma viral sequences is that most of the cellular reservoir of HIV-1 is found in anatomically protected and immune-privileged sites and is absent from the peripheral CD4+ T-cells. A combination of assays that can detect and characterize intact proviruses and HIV-1 mRNA in both tissues and blood samples should help identify the likely source(s) of persistent viremia.
Viral Replication or Proviral Expression as the Major Source of Persistent Viremia?
A longstanding debate is whether low-level viremia on ART results from ongoing, complete cycles of viral replication or is from clonally expanded infected T cells that produce virions but that do not infect new cells because they are protected by antiretrovirals (Finzi et al., 1997; Wong et al., 1997; Lorenzo-Redondo et al., 2016). As shown in Figure 1, complete cycles of viral replication give rise to rapid accumulation of mutations attributed to the error-prone nature of reverse transcriptase. An additional consequence of productive cycles of viral replication is multiple integrations of proviral DNA into different locations in chromosomal DNA. Conversely, expansion of infected cells through cellular proliferation produces identical HIV proviral sequences and identical integration sites in cell progeny. Importantly, a subset of these cell progeny can contain transcriptionally active proviruses that produce virions (Figure 1; Bailey et al., 2006; Wagner et al., 2014, Wiegand et al., 2017). For example, Simonetti et al. (2016) reported the detection of a highly expanded CD4+ T-cell clone containing an intact provirus that was a source of persistent viremia on ART.
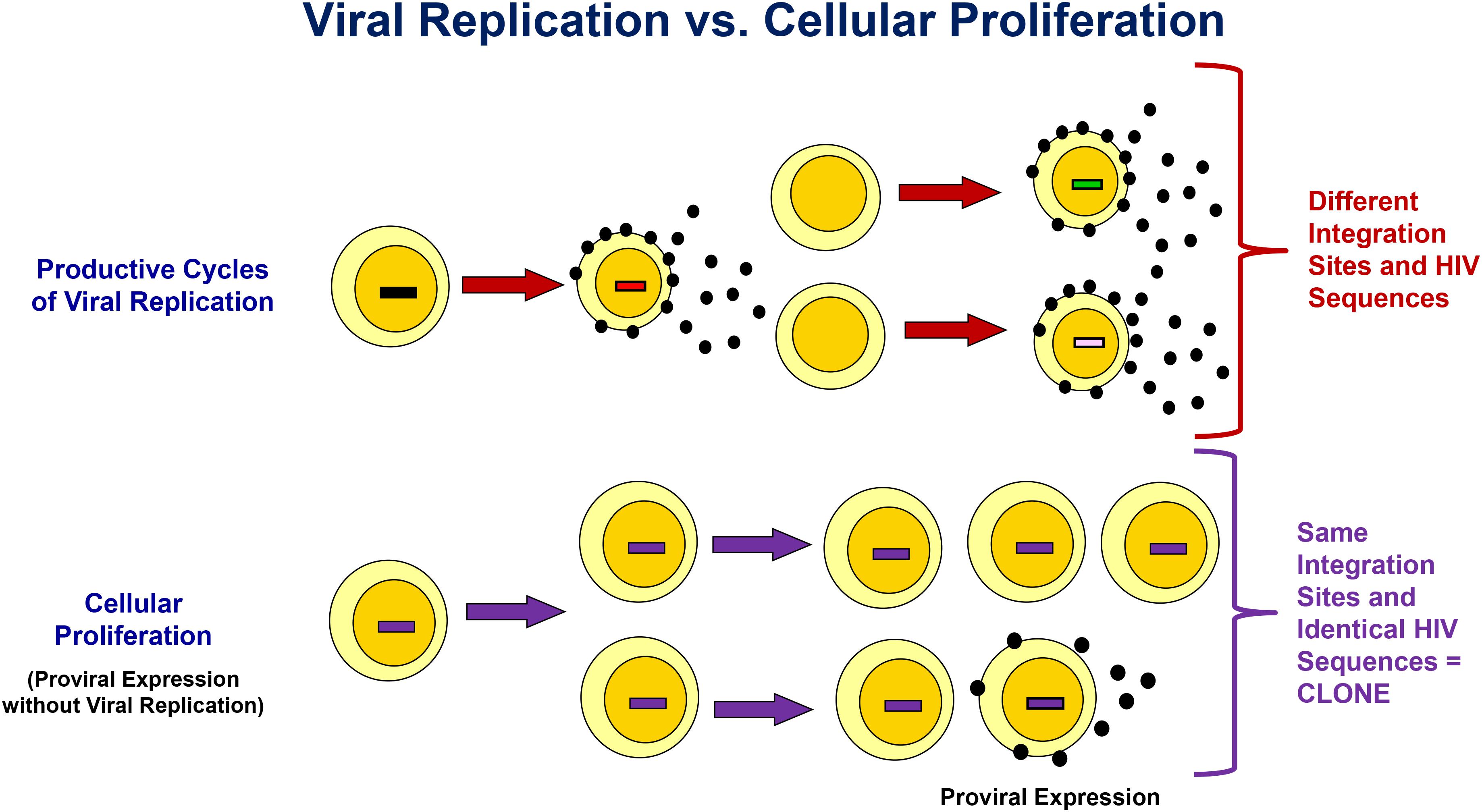
Figure 1. Differentiating Viral Replication From Clonal Expansion with Proviral Expression as the Source of Persistent Viremia on ART. Integration site and HIV sequence analyses can be used to assess the origin of proviral expansion and/or viremia. Productive cycles of viral replication result in both genetic heterogeneity due to errors introduced by reverse transcriptase, and variation in the chromosomal integration site of the HIV provirus. Conversely, clonal expansion results in identical chromosomal HIV integration sites in cell progeny, identical proviral sequences, and identical viral sequences from the subset of cell in the clone that produce virus. See text for more details.
It has been reported that anatomical sanctuary sites such as the lymph nodes can allow residual viral replication on ART, contributing to maintenance of the HIV reservoir (Lorenzo-Redondo et al., 2016). However, a reanalysis of this data revealed (1) a limited data set after adjusting for PCR resampling and hyper-mutated sequences, (2) limited unique HIV RNA and DNA sequences that were available (median of 5 per sample, range 0–37), (3) sampling time points that may not have taken into account the shifting dynamics of the HIV DNA population within the first year of ART, and (4) inconsistent evidence of viral evolution using more complex analyses (Kearney et al., 2017). These results, along with previous studies reporting no evidence of HIV-1 evolution on suppressive ART in chronically infected adults, as well as in children treated shortly after birth when viral diversity is low, argue against active viral replication being the major source of persistent viremia (Joos et al., 2008; Kearney et al., 2014; van Zyl et al., 2017). Additionally, the presence of invariant sequences and the absence of sequence divergence during prolonged ART, or during and after analytical treatment interruption, is indicative of long-lived cells infected and argues against viral replication. As there have been additional reports of viral replication on ART due to low drug penetration and exclusion of immune cells in anatomical sanctuary sites (Buzón et al., 2010, 2011; Sigal et al., 2011; Hatano et al., 2013; Luo et al., 2013; Patterson et al., 2013; Cardozo et al., 2014; Fletcher et al., 2014; Piovoso and Zurakowski, 2014; Puertas et al., 2014), some residual low-level viral replication on ART cannot be definitively ruled out. Nevertheless, the weight of the evidence discussed above argues against viral replication as the major source of persistent viremia.
Finally, the recent discovery by multiple groups that most of the inducible, infectious virus comes from clonally expanded T-cells argues for cellular proliferation and against ongoing viral replication as the major mechanism for persistence of HIV-1 reservoirs (Lorenzi et al., 2016; Bui et al., 2017; Hosmane et al., 2017). Additional studies are in progress to determine whether most persistent viremia is of clonal cell origin, as has already been described in one instance of an individual with advanced malignancy (Simonetti et al., 2016). Identifying the clonal origin of viremia requires in depth analyses, including full-length single genome sequencing of HIV-1 RNA from plasma and viral outgrowth cultures, and HIV DNA from infected cells, to identify possible clones, with confirmation of clonality by integration site analyses (Palmer et al., 2005; Maldarelli et al., 2014; Bui et al., 2017).
Clearing Persistent Viremia
Many interventions are being investigated for their ability to clear the HIV-1 reservoir and achieve a functional or sterilizing cure. Among the strategies being studied are: (1) latency reversal to induce viral protein production and expose infected cells to the immune system; (2) engineering immune cells for artificial priming of an HIV-specific immune response or targeted killing of infected cells; (3) gene therapy for alteration of target cell susceptibility to prevent HIV-1 infection; (4) passive immunotherapy with antibodies identified as broadly HIV neutralizing (bnAbs) to clear viremia and infected cells; and combinations of these approaches. These diverse strategies are reviewed elsewhere (Deeks et al., 2016). A promising monoclonal bnAb targeting the CD4 binding site of the HIV-1 envelope (VRC01) has been extensively evaluated for safety, neutralization capacity and pre-existence or development of resistance. Given its high efficacy in neutralizing free virus and the property of bnAbs to promote antibody-dependent cell-mediated cytotoxicity, VRC01 was evaluated in individuals on long-term ART for its effect on persistent plasma viremia and infected cells. Intravenous infusions of VRC01 in individuals on long-term ART did not lead to any change in markers of the reservoir such as cell-associated proviral DNA and RNA, or in levels of persistent plasma viremia (Lynch et al., 2015; Riddler et al., 2018). These disappointing results suggest that persistent virus is either resistant to VRC01 binding or VRC01 effector functions are impaired, such as Fc-mediated clearance or antibody-dependent cellular cytoxicity (ADCC). Work is ongoing to identify the reason(s) for the lack of effect of VRC01 on reservoir markers and on improving the breadth and effector function of bnAbs.
Unanswered Questions and Concluding Remarks
Whether persistent plasma viremia in individuals on long-term ART consists of infectious virus remains unclear. It has been shown, at least in some instances, that a portion of the persistent viremia produced by infected cell clones is infectious and contributes to rebound virus when ART is stopped (Simonetti et al., 2016; Kearney et al., 2017). Since only an intact gag gene is required for virion production and budding (Delchambre et al., 1989), a scenario in which non-infectious virus is released into the plasma and contributes to persistent plasma viremia is also possible. This possibility may help explain the lack of correlation between levels of persistent plasma viremia and the quantity of inducible, infectious virus ex vivo (Siliciano et al., 2003; Eriksson et al., 2013; van Zyl et al., 2018). Studies evaluating the ability of virions present in persistent plasma viremia to infect target cells would be useful to address this question.
It is also unclear whether viremia that rebounds following ART interruption is coming from the same source as that producing persistent viremia on ART. Interestingly, a recent study linked clonal proviral populations in infected cells to clonal sequences in rebounding viremia after treatment interruption, showing that infected cell clones are an important viral reservoir (De Scheerder et al., 2019). The report also described preliminary evidence of linkage between virus in plasma and rebound virus in a subset of individuals. Additional studies examining sequences from persistent viremia and rebound virus will further inform the question of the relevance of persistent viremia to rebound off ART.
In conclusion, although unanswered questions remain, remarkable progress has been made toward measuring and characterizing persistent plasma viremia in individuals on ART since it was first reported in 1999. Mounting evidence indicates that persistent HIV-1 viremia on ART largely arises from clonally expanded CD4+ T-cells, although some contribution of ongoing viral replication cannot be excluded. Future and ongoing studies to further characterize clonal populations producing low-level viremia and their mechanisms of escape from immune clearance will be important to achieve a functional cure.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by grant funding to JM from the National Cancer Institute (contract number HHSN261200800001E), the National Institute of Allergy and Infectious Diseases (UM1AI126603), the Bill & Melinda Gates Foundation award (OPP1115715). The content of this manuscript is solely the responsibility of authors and does not necessarily represent the official views of funding agencies, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States Government.
Conflict of Interest
JM is a consultant for Gilead Sciences and Xi’an Yufan Biotechnologies, has received research grants to the University of Pittsburgh from Gilead Sciences and Janssen Pharmaceutica, and owns shares in Co-Crystal Pharma, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank the study volunteers and investigators who contributed to the work cited in this review, and to Lorraine Pollini for her excellent editorial assistance.
References
Anton, P. A., Mitsuyasu, R. T., Deeks, S. G., Scadden, D. T., Wagner, B., Huang, C., et al. (2003). Multiple measures of HIV burden in blood and tissue are correlated with each other but not with clinical parameters in aviremic subjects. AIDS 17, 53–63. doi: 10.1097/00002030-200301030-00008
Archin, N. M., Bateson, R., Tripathy, M. K., Crooks, A. M., Yang, K. H., Dahl, N. P., et al. (2014). HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J. Infect. Dis. 210, 728–735. doi: 10.1093/infdis/jiu155
Bailey, J. R., Sedaghat, A. R., Kieffer, T., Brennan, T., Lee, P. K., Wind-Rotolo, M., et al. (2006). Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 80, 6441–6457. doi: 10.1128/jvi.00591-06
Bakkour, S., Deng, X., Bacchetti, P., Stone, M., Worlock, A., Hauenstein, S., et al. (2019). “Replicate Aptima Assay For Quantifying Residual Plasma Viremia In Individuals On ART,” in Proceedings of the Conference on Retroviruses and Opportunistic Infections (CROI), Seattle, WA.
Belmonte, L., Olmos, M., Fanin, A., Parodi, C., Bare, P., Concetti, H., et al. (2007). The intestinal mucosa as a reservoir of HIV-1 infection after successful HAART. AIDS 21, 2106–2108. doi: 10.1097/qad.0b013e3282efb74b
Bruner, K. M., Murray, A. J., Pollack, R. A., Soliman, M. G., Laskey, S. B., Capoferri, A. A., et al. (2016). Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 2, 1043–1049. doi: 10.1038/nm.4156
Bruner, K. M., Wang, Z., Simonetti, F. R., Bender, A. M., Kwon, K. J., Sengupta, S., et al. (2019). A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566, 120–125. doi: 10.1038/s41586-019-0898-8
Bui, J. K., Sobolewski, M. D., Keele, B. F., Spindler, J., Musick, A., Wiegand, A., et al. (2017). Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 13:e1006283. doi: 10.1371/journal.ppat.1006283
Buzón, M. J., Codoñer, F. M., Frost, S. D., Pou, C., Puertas, M. C., Massanella, M., et al. (2011). Deep molecular characterization of HIV-1 dynamics under suppressive HAART. PLoS Pathog. 7:e1002314. doi: 10.1371/journal.ppat.1002314
Buzón, M. J., Massanella, M., Llibre, J. M., Esteve, A., Dahl, V., Puertas, M. C., et al. (2010). HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat. Med. 16, 460–465. doi: 10.1038/nm.2111
Cardozo, E. F., Luo, R., Piovoso, M. J., and Zurakowski, R. (2014). Spatial modeling of HIV cryptic viremia and 2-LTR formation during raltegravir intensification. J. Theor. Biol. 345, 61–69. doi: 10.1016/j.jtbi.2013.12.020
Chun, T. W., Carruth, L., Finzi, D., Shen, X., Digiuseppe, J. A., Taylor, H., et al. (1997a). Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387, 183–188. doi: 10.1038/387183a0
Chun, T. W., Stuyver, L., Mizell, S. B., Ehler, L. A., Mican, J. A., Baseler, M., et al. (1997b). Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A. 94, 13193–13197. doi: 10.1073/pnas.94.24.13193
Chun, T. W., Finzi, D., Margolick, J., Chadwick, K., Schwartz, D., and Siliciano, R. F. (1995). In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 1, 1284–1290. doi: 10.1038/nm1295-1284
Chun, T. W., Murray, D., Justement, J. S., Hallahan, C. W., Moir, S., Kovacs, C., et al. (2011). Relationship between residual plasma viremia and the size of HIV proviral DNA reservoirs in infected individuals receiving effective antiretroviral therapy. J. Infect. Dis. 204, 135–138. doi: 10.1093/infdis/jir208
Cillo, A. R., Vagratian, D., Bedison, M. A., Anderson, E. M., Kearney, M. F., Fyne, E., et al. (2014). Improved single-copy assays for quantification of persistent HIV-1 viremia in patients on suppressive antiretroviral therapy. J. Clin. Microbiol. 52, 3944–3951. doi: 10.1128/JCM.02060-14
Couturier, J., Suliburk, J. W., Brown, J. M., Luke, D. J., Agarwal, N., Yu, X., et al. (2015). Human adipose tissue as a reservoir for memory CD4+ T cells and HIV. AIDS 29, 667–674. doi: 10.1097/QAD.0000000000000599
Dahl, V., Peterson, J., Fuchs, D., Gisslen, M., Palmer, S., and Price, R. W. (2014). Low levels of HIV-1 RNA detected in the cerebrospinal fluid after up to 10 years of suppressive therapy are associated with local immune activation. AIDS 28, 2251–2258. doi: 10.1097/QAD.0000000000000400
Damouche, A., Lazure, T., Avettand-Fenoel, V., Huot, N., Dejucq-Rainsford, N., Satie, A. P., et al. (2015). Adipose tissue is a neglected viral reservoir and an inflammatory site during chronic HIV and SIV infection. PLoS Pathog. 11:e1005153. doi: 10.1371/journal.ppat.1005153
De Scheerder, M. A., Vrancken, B., Dellicour, S., Schlub, T., Lee, E., Shao, W., et al. (2019). HIV rebound is predominantly fueled by genetically identical viral expansions from diverse reservoirs. Cell Host Microbe 26, 347–358.e7. doi: 10.1016/j.chom.2019.08.003
Deeks, S. G., Lewin, S. R., Ross, A. L., Ananworanich, J., Benkirane, M., Cannon, P., et al. (2016). International AIDS society global scientific strategy: towards an HIV cure 2016. Nat. Med. 22, 839–850. doi: 10.1038/nm.4108
Delchambre, M., Gheysen, D., Thines, D., Thiriart, C., Jacobs, E., Verdin, E., et al. (1989). The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 8, 2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x
Dornadula, G., Zhang, H., VanUitert, B., Stern, J., Livornese, L.Jr., Ingerman, M. J., et al. (1999). Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA 282, 1627–1632.
Elliott, J. H., Wightman, F., Solomon, A., Ghneim, K., Ahlers, J., Cameron, M. J., et al. (2014). Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog. 10:e1004473. doi: 10.1371/journal.ppat.1004473
Eriksson, S., Graf, E. H., Dahl, V., Strain, M. C., Yukl, S. A., Lysenko, E. S., et al. (2013). Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 9:e1003174. doi: 10.1371/journal.ppat.1003174
Finzi, D., Hermankova, M., Pierson, T., Carruth, L. M., Buck, C., Chaisson, R. E., et al. (1997). Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300. doi: 10.1126/science.278.5341.1295
Fletcher, C. V., Staskus, K., Wietgrefe, S. W., Rothenberger, M., Reilly, C., Chipman, J. G., et al. (2014). Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc. Natl. Acad. Sci. U.S.A. 111, 2307–2312. doi: 10.1073/pnas.1318249111
Fourati, S., Lambert-Niclot, S., Soulie, C., Malet, I., Valantin, M. A., Descours, B., et al. (2012). HIV-1 genome is often defective in PBMCs and rectal tissues after long-term HAART as a result of APOBEC3 editing and correlates with the size of reservoirs. J. Antimicrob. Chemother. 67, 2323–2326. doi: 10.1093/jac/dks219
Gandhi, R. T., Mcmahon, D. K., Bosch, R. J., Lalama, C. M., Cyktor, J. C., Macatangay, B. J., et al. (2017). Levels of HIV-1 persistence on antiretroviral therapy are not associated with markers of inflammation or activation. PLoS Pathog. 13:e1006285. doi: 10.1371/journal.ppat.1006285
Ganor, Y., Real, F., Sennepin, A., Dutertre, C. A., Prevedel, L., Xu, L., et al. (2019). HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat. Microbiol. 4, 633–644. doi: 10.1038/s41564-018-0335-z
Hatano, H., Strain, M. C., Scherzer, R., Bacchetti, P., Wentworth, D., Hoh, R., et al. (2013). Increase in 2-long terminal repeat circles and decrease in D-dimer after raltegravir intensification in patients with treated HIV infection: a randomized, placebo-controlled trial. J. Infect. Dis. 208, 1436–1442. doi: 10.1093/infdis/jit453
Ho, D. D., Neumann, A. U., Perelson, A. S., Chen, W., Leonard, J. M., and Markowitz, M. (1995). Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373, 123–126. doi: 10.1038/373123a0
Ho, Y. C., Shan, L., Hosmane, N. N., Wang, J., Laskey, S. B., Rosenbloom, D. I., et al. (2013). Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551. doi: 10.1016/j.cell.2013.09.020
Hong, F., Aga, E., Cillo, A., Yates, A. L., Besson, G., Fyne, E., et al. (2016). Novel assays to measure total cell-associated HIV-1 DNA and RNA. J. Clin. Microbiol. 54, 902–911. doi: 10.1128/JVI.01763-08
Hong, F., Jacobs, J. L., Aga, E., Cillo, A. R., Fyne, E., Koontz, D. L., et al. (2018). Associations between HIV-1 DNA copy number, proviral transcriptional activity, and plasma viremia in individuals off or on suppressive antiretroviral therapy. Virology 521, 51–57. doi: 10.1016/j.virol.2018.05.018
Hosmane, N. N., Kwon, K. J., Bruner, K. M., Capoferri, A. A., Beg, S., Rosenbloom, D. I., et al. (2017). Proliferation of latently infected CD4(+) T cells carrying replication-competent HIV-1: potential role in latent reservoir dynamics. J. Exp. Med. 214, 959–972. doi: 10.1084/jem.20170193
Imamichi, H., Dewar, R. L., Adelsberger, J. W., Rehm, C. A., O’doherty, U., Paxinos, E. E., et al. (2016). Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A. 113, 8783–8788. doi: 10.1073/pnas.1609057113
Joos, B., Fischer, M., Kuster, H., Pillai, S. K., Wong, J. K., Boni, J., et al. (2008). HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc. Natl. Acad. Sci. U.S.A. 105, 16725–16730. doi: 10.1073/pnas.0804192105
Kearney, M. F., Spindler, J., Shao, W., Yu, S., Anderson, E. M., O’shea, A., et al. (2014). Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog. 10:e1004010. doi: 10.1371/journal.ppat.1004010
Kearney, M. F., Wiegand, A., Shao, W., Coffin, J. M., Mellors, J. W., Lederman, M., et al. (2015). Origin of rebound plasma HIV includes cells with identical proviruses that are transcriptionally active before stopping antiretroviral therapy. J. Virol. 90, 1369–1376. doi: 10.1128/JVI.02139-15
Kearney, M. F., Wiegand, A., Shao, W., Mcmanus, W. R., Bale, M. J., Luke, B., et al. (2017). Ongoing HIV replication during ART reconsidered. Open Forum Infect. Dis. 4:ofx173. doi: 10.1093/ofid/ofx173
Kiselinova, M., Pasternak, A. O., De Spiegelaere, W., Vogelaers, D., Berkhout, B., and Vandekerckhove, L. (2014). Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS One 9:e85999. doi: 10.1371/journal.pone.0085999
Lampinen, T. M., Critchlow, C. W., Kuypers, J. M., Hurt, C. S., Nelson, P. J., Hawes, S. E., et al. (2000). Association of antiretroviral therapy with detection of HIV-1 RNA and DNA in the anorectal mucosa of homosexual men. AIDS 14, F69–F75.
Li, J. Z., Etemad, B., Ahmed, H., Aga, E., Bosch, R. J., Mellors, J. W., et al. (2016). The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS 30, 343–353. doi: 10.1097/QAD.0000000000000953
Lorenzi, J. C., Cohen, Y. Z., Cohn, L. B., Kreider, E. F., Barton, J. P., Learn, G. H., et al. (2016). Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc. Natl. Acad. Sci. U.S.A. 113, E7908–E7916.
Lorenzo-Redondo, R., Fryer, H. R., Bedford, T., Kim, E. Y., Archer, J., Pond, S. L. K., et al. (2016). Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 530, 51–56. doi: 10.1038/nature16933
Luo, R., Cardozo, E. F., Piovoso, M. J., Wu, H., Buzón, M. J., Martinez-Picado, J., et al. (2013). Modelling HIV-1 2-LTR dynamics following raltegravir intensification. J. R. Soc. Interface 10:20130186. doi: 10.1098/rsif.2013.0186
Lynch, R. M., Boritz, E., Coates, E. E., Dezure, A., Madden, P., Costner, P., et al. (2015). Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 7:319ra206. doi: 10.1126/scitranslmed.aad5752
Maldarelli, F., Palmer, S., King, M. S., Wiegand, A., Polis, M. A., Mican, J., et al. (2007). ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog. 3:e46. doi: 10.1371/journal.ppat.0030046
Maldarelli, F., Wu, X., Su, L., Simonetti, F. R., Shao, W., Hill, S., et al. (2014). HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183. doi: 10.1126/science.1254194
Margot, N., Koontz, D., Mccallister, S., Mellors, J. W., and Callebaut, C. (2018). Measurement of plasma HIV-1 RNA below the limit of quantification (<20 copies/mL) of commercial assays with the integrase HIV RNA single-copy assay. J. Clin. Virol. 108, 50–52. doi: 10.1016/j.jcv.2018.09.003
Mexas, A. M., Graf, E. H., Pace, M. J., Yu, J. J., Papasavvas, E., Azzoni, L., et al. (2012). Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. AIDS 26, 2295–2306. doi: 10.1097/QAD.0b013e32835a5c2f
Mitchell, B. I., Laws, E. I., and Ndhlovu, L. C. (2019). Impact of myeloid reservoirs in HIV cure trials. Curr. HIV/AIDS Rep. 16, 129–140. doi: 10.1007/s11904-019-00438-5
Moron-Lopez, S., Kim, P., Sogaard, O. S., Tolstrup, M., Wong, J. K., and Yukl, S. A. (2019). Characterization of the HIV-1 transcription profile after romidepsin administration in ART-suppressed individuals. AIDS 33, 425–431. doi: 10.1097/QAD.0000000000002083
Palella, F. J.Jr., Delaney, K. M., Moorman, A. C., Loveless, M. O., Fuhrer, J., Satten, G. A., et al. (1998). Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N. Engl. J. Med. 338, 853–860. doi: 10.1056/nejm199803263381301
Palmer, S., Kearney, M., Maldarelli, F., Halvas, E. K., Bixby, C. J., Bazmi, H., et al. (2005). Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43, 406–413. doi: 10.1128/jcm.43.1.406-413.2005
Palmer, S., Maldarelli, F., Wiegand, A., Bernstein, B., Hanna, G. J., Brun, S. C., et al. (2008). Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A. 105, 3879–3884. doi: 10.1073/pnas.0800050105
Palmer, S., Wiegand, A. P., Maldarelli, F., Bazmi, H., Mican, J. M., Polis, M., et al. (2003). New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 41, 4531–4536. doi: 10.1128/jcm.41.10.4531-4536.2003
Pasternak, A. O., Adema, K. W., Bakker, M., Jurriaans, S., Berkhout, B., Cornelissen, M., et al. (2008). Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J. Clin. Microbiol. 46, 2206–2211. doi: 10.1128/JCM.00055-08
Patterson, K. B., Prince, H. A., Stevens, T., Shaheen, N. J., Dellon, E. S., Madanick, R. D., et al. (2013). Differential penetration of raltegravir throughout gastrointestinal tissue: implications for eradication and cure. AIDS 27, 1413–1419. doi: 10.1097/QAD.0b013e32835f2b49
Perelson, A. S., Essunger, P., Cao, Y., Vesanen, M., Hurley, A., Saksela, K., et al. (1997). Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387, 188–191. doi: 10.1038/387188a0
Perelson, A. S., Neumann, A. U., Markowitz, M., Leonard, J. M., and Ho, D. D. (1996). HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271, 1582–1586. doi: 10.1126/science.271.5255.1582
Perreau, M., Savoye, A. L., De Crignis, E., Corpataux, J. M., Cubas, R., Haddad, E. K., et al. (2013). Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J. Exp. Med. 210, 143–156. doi: 10.1084/jem.20121932
Piovoso, M. J., and Zurakowski, R. (2014). “Evaluation of HIV 2-LTR formation models using monotone system theory,” in Proceedings of the American Control Conference (ACC), Portland, OR, 2747–2752.
Pollack, R. A., Jones, R. B., Pertea, M., Bruner, K. M., Martin, A. R., Thomas, A. S., et al. (2017). Defective HIV-1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe 21:e494. doi: 10.1016/j.chom.2017.03.008
Procopio, F. A., Fromentin, R., Kulpa, D. A., Brehm, J. H., Bebin, A. G., Strain, M. C., et al. (2015). A novel assay to measure the magnitude of the inducible viral reservoir in hiv-infected individuals. Ebiomedicine 2, 874–883. doi: 10.1016/j.ebiom.2015.06.019
Puertas, M. C., Massanella, M., Llibre, J. M., Ballestero, M., Buzón, M. J., Ouchi, D., et al. (2014). Intensification of a raltegravir-based regimen with maraviroc in early HIV-1 infection. AIDS 28, 325–334. doi: 10.1097/QAD.0000000000000066
Riddler, S. A., Aga, E., Bosch, R. J., Bastow, B., Bedison, M., Vagratian, D., et al. (2016). Continued slow decay of residual plasma viremia in HIV-1-infected adults receiving long term antiretroviral therapy. J. Infect. Dis. 213, 556–560. doi: 10.1093/infdis/jiv433
Riddler, S. A., Zheng, L., Durand, C. M., Ritz, J., Koup, R. A., Ledgerwood, J., et al. (2018). Randomized clinical trial to assess the impact of the broadly neutralizing HIV-1 monoclonal antibody VRC01 on HIV-1 persistence in individuals on effective ART. Open Forum Infect. Dis. 5:ofy242. doi: 10.1093/ofid/ofy242
Sigal, A., Kim, J. T., Balazs, A. B., Dekel, E., Mayo, A., Milo, R., et al. (2011). Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature 477, 95–98. doi: 10.1038/nature10347
Siliciano, J. D., Kajdas, J., Finzi, D., Quinn, T. C., Chadwick, K., Margolick, J. B., et al. (2003). Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9, 727–728. doi: 10.1038/nm880
Simonetti, F. R., Sobolewski, M. D., Fyne, E., Shao, W., Spindler, J., Hattori, J., et al. (2016). Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc. Natl. Acad. Sci. U.S.A. 113, 1883–1888. doi: 10.1073/pnas.1522675113
Sonza, S., Mutimer, H. P., Oelrichs, R., Jardine, D., Harvey, K., Dunne, A., et al. (2001). Monocytes harbour replication-competent, non-latent HIV-1 in patients on highly active antiretroviral therapy. AIDS 15, 17–22. doi: 10.1097/00002030-200101050-00005
Spudich, S., Robertson, K., Bosch, R. J., Gandhi, R. T., Cyktor, J. C., Mar, H., et al. (2019). Persistent HIV-infected cells in cerebrospinal fluid are associated with poorer neurocognitive performance. J. Clin. Invest. 129, 3339–3346. doi: 10.1172/JCI127413
Strain, M. C., and Richman, D. D. (2013). New assays for monitoring residual HIV burden in effectively treated individuals. Curr. Opin. HIV AIDS 8, 106–110. doi: 10.1097/COH.0b013e32835d811b
Tosiano, M. A., Jacobs, J. L., Shutt, K. A., Cyktor, J. C., and Mellors, J. W. (2019a). A simpler and more sensitive single-copy HIV-1 RNA assay for quantification of persistent HIV-1 viremia in individuals on suppressive antiretroviral therapy. J. Clin. Microbiol. 57, e1714–e1718. doi: 10.1128/JCM.01714-18
Tosiano, M. A., Mar, H., Cyktor, J. C., Koontz, D. L., Eron, J. J., Gandhi, R. T., et al. (2019b). “HIV-1 RNA Detection By Abbott M2000 Correlates With Integrase Single Copy Assay,” in Proceedings of the Conference on Retroviruses and Opportunistic Infections (CROI), Seattle, WA.
van Zyl, G., Bale, M. J., and Kearney, M. F. (2018). HIV evolution and diversity in ART-treated patients. Retrovirology 15:14. doi: 10.1186/s12977-018-0395-4
van Zyl, G. U., Katusiime, M. G., Wiegand, A., Mcmanus, W. R., Bale, M. J., Halvas, E. K., et al. (2017). No evidence of HIV replication in children on antiretroviral therapy. J. Clin. Invest. 127, 3827–3834. doi: 10.1172/JCI94582
Wagner, T. A., Mclaughlin, S., Garg, K., Cheung, C. Y., Larsen, B. B., Styrchak, S., et al. (2014). HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573. doi: 10.1126/science.1256304
Wei, X., Ghosh, S. K., Taylor, M. E., Johnson, V. A., Emini, E. A., Deutsch, P., et al. (1995). Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373, 117–122.
Wiegand, A., Spindler, J., Hong, F. F., Shao, W., Cyktor, J. C., Cillo, A. R., et al. (2017). Single-cell analysis of HIV-1 transcriptional activity reveals expression of proviruses in expanded clones during ART. Proc. Natl. Acad. Sci. U.S.A. 114, E3659–E3668. doi: 10.1073/pnas.1617961114
Wiesmann, F., Ehret, R., Naeth, G., Daumer, M., Fuhrmann, J., Kaiser, R., et al. (2018). Multicenter evaluation of two next-generation HIV-1 quantitation assays, aptima quant Dx and Cobas 6800, in comparison to the realtime HIV-1 reference assay. J. Clin. Microbiol. 56, e292–e218. doi: 10.1128/JCM.00292-18
Wong, J. K., Hezareh, M., Gunthard, H. F., Havlir, D. V., Ignacio, C. C., Spina, C. A., et al. (1997). Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295. doi: 10.1126/science.278.5341.1291
Yucha, R. W., Hobbs, K. S., Hanhauser, E., Hogan, L. E., Nieves, W., Ozen, M. O., et al. (2017). High-throughput characterization of HIV-1 reservoir reactivation using a single-cell-in-droplet PCR assay. Ebiomedicine 20, 217–229. doi: 10.1016/j.ebiom.2017.05.006
Yukl, S. A., Kaiser, P., Kim, P., Telwatte, S., Joshi, S. K., Vu, M., et al. (2018). HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med. 10:eaa9927. doi: 10.1126/scitranslmed.aap9927
Zaikos, T. D., Terry, V. H., Sebastian Kettinger, N. T., Lubow, J., Painter, M. M., Virgilio, M. C., et al. (2018). Hematopoietic stem and progenitor cells are a distinct hiv reservoir that contributes to persistent viremia in suppressed patients. Cell Rep 25, 3759–3773.e9. doi: 10.1016/j.celrep.2018.11.104
Zheng, L., Bosch, R. J., Chan, E. S., Read, S., Kearney, M., Margolis, D. M., et al. (2013). Predictors of residual viraemia in patients on long-term suppressive antiretroviral therapy. Antivir. Ther. 18, 39–43. doi: 10.3851/IMP2323
Zhu, T., Muthui, D., Holte, S., Nickle, D., Feng, F., Brodie, S., et al. (2002). Evidence for human immunodeficiency virus type 1 replication in vivo in CD14(+) monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J. Virol. 76, 707–716. doi: 10.1128/jvi.76.2.707-716.2002
Keywords: HIV-1 persistence, HIV-1 cure, low-level viremia, antiretroviral therapy, plasma viremia, plasma HIV-1 RNA
Citation: Jacobs JL, Halvas EK, Tosiano MA and Mellors JW (2019) Persistent HIV-1 Viremia on Antiretroviral Therapy: Measurement and Mechanisms. Front. Microbiol. 10:2383. doi: 10.3389/fmicb.2019.02383
Received: 13 July 2019; Accepted: 01 October 2019;
Published: 15 October 2019.
Edited by:
Maria J. Buzon, Vall d’Hebron Research Institute (VHIR), SpainReviewed by:
Guinevere Q. Lee, Cornell University, United StatesMa Angeles Muñoz-Fernandez, Gregorio Marañón Hospital, Spain
Copyright © 2019 Jacobs, Halvas, Tosiano and Mellors. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jana L. Jacobs, amxqOTBAcGl0dC5lZHU=
 Jana L. Jacobs
Jana L. Jacobs Elias K. Halvas
Elias K. Halvas Melissa A. Tosiano
Melissa A. Tosiano John W. Mellors
John W. Mellors