- 1School of Pharmacy, Sungkyunkwan University, Suwon, South Korea
- 2Department of Molecular Science and Technology, Ajou University, Suwon, South Korea
- 3Department of Molecular Cell Biology, School of Medicine, Sungkyunkwan University, Suwon, South Korea
Capsular polysaccharides (CPS), a major virulence factor in Streptococcus pneumoniae, become thicker during blood invasion while not during asymptomatic nasopharyngeal colonization. However, the underlying mechanism controlling this differential pneumococcal CPS regulation remain unclear. Here, we show how VncR, the response regulator of the vancomycin resistance locus (vncRS operon), regulates CPS expression in vncR mutants in three serotype (type 2, 3, and 6B) backgrounds upon exposure to serum lactoferrin (LF). Comparative analysis of CPS levels in the wild type (WT) of three strains and their isogenic vncR mutants after LF exposure revealed a strain-specific alteration in CPS production. Consistently, VncR-mediated strain-specific CPS production is correlated with pneumococcal virulence, in vivo. Electrophoretic mobility-shift assay and co-immunoprecipitation revealed an interaction between VncR and the cps promoter (cpsp) in the presence of serum. In addition, in silico analysis uncovered this protein-DNA interaction, suggesting that VncR binds with the cpsp, and recognizes the strain-specific significance of the tandem repeats in cpsp. Taken together, the interaction of VncR and cpsp after serum exposure plays an essential role in regulating differential strain-specific CPS production, which subsequently determines strain-specific systemic virulence. This study highlights how host protein LF contributes to pneumococcal VncR-mediated CPS production. As CPS plays a significant role in immune evasion, these findings suggest that drugs designed to interrupt the VncR-mediated CPS production could help to combat pneumococcal infections.
Introduction
Streptococcus pneumoniae, usually a commensal in the upper human respiratory tract, is an etiologic agent of pneumonia, sepsis, and meningitis (Bogaert et al., 2004). The successful alteration of a harmless commensal to an invasive pathogen is associated with the trafficking of bacteria across tissue barriers and the subsequent bacterial adaptation to altered host niches. This progression is multifunctional and demands strict regulation (Hammerschmidt et al., 2005).
Among the pneumococcal virulence factors, the capsular polysaccharide (capsule, CPS) is the most significant, and is responsible for disease pathogenesis (Kadioglu et al., 2008). CPS shows a differential expression profile between carriage pneumococci and planktonic pneumococci, capable of causing an invasive disease (Wu et al., 2016). Although several proteins, such as CpsR (Wu et al., 2016), and ComE (Zheng et al., 2017) have been shown to play key roles in the regulation of cps genes, the underlying mechanism is poorly delineated. To date, 94 pneumococcal CPS have been reported (Nurse-Lucas et al., 2016), all of these except two are produced by a Wzy-polymerase-dependent mechanism (Tuomanen et al., 2004; Nurse-Lucas et al., 2016; Zheng et al., 2017). In contrast, the synthesis of the other CPS types (3 and 37) is mediated by a single membrane-bound glycosyltransferase. In these pneumococcal serotypes, the conserved sequences positioned at the 5′ end of all the other loci, which are responsible for the transcription of regulatory proteins, are either absent (type 37) or mutated (type 3) (Moscoso and García, 2009). The cps loci of all Wzy serotypes are positioned at the same chromosomal region (Zheng et al., 2017). cps locus promoter sequences (cpsp) and the first four genes cpsA to cpsD are highly conserved and take part in CPS regulation, whereas, genes downstream of cpsD are serotype-specific (Wu et al., 2016; Ghosh et al., 2018). Moreover, previous studies confirmed that the cps genes are transcribed as an operon from a sole promoter (Guidolin et al., 1994; Aanensen et al., 2007). In contrast, type 3 pneumococcal cpsp is completely different from the other serotypes (Caimano et al., 1998), as a short 87 bp region embracing the cpsp is strictly conserved only among the Wzy serotypes (Moscoso and García, 2009).
The two component signal transduction systems (TCSs) in bacteria are comprised of a membrane-bound histidine kinase protein (HK) and a cytosolic response regulator (RR) (McCluskey et al., 2004). Activation of the TCS by various stimuli causes HK to undergo autophosphorylation, which subsequently transfers a phosphate group to the RR. The phosphorylated RR leads to adaptive responses by altering gene expression (Finlay and Falkow, 1997). TCS10, also known as VncRS, is induced in vancomycin-tolerant clinical pneumococcal samples (Sung et al., 2006), whereas, mutations in vncR did not alter the pneumococcal virulence (Throup et al., 2000), indicating that the role of VncRS in S. pneumoniae virulence is complex and must be explicated.
Previously, we showed that the VncS ligand, serum lactoferrin (LF), induced the type 2 pneumococcal vncRS operon and augmented in vivo mortality rates mediated by pep27, a gene belonging to vncRS operon (Lee et al., 2018). Further, the expression of the cps2A gene, representing the extent of pneumococcal cps transcription, was upregulated in the presence of serum (Ogunniyi et al., 2002). Moreover, BLAST searches revealed that the DNA binding domain (DBD) of VncR is almost homogenous, whereas the cps locus consists of a large number of type-specific genes (McCluskey et al., 2004; Zheng et al., 2017). These considerations have raised our interest in studying the role of VncR in strain-specific CPS-mediated systemic virulence.
Here, we show that VncR regulates CPS synthesis in a strain-specific manner in the presence of LF, which is further associated with pneumococcal virulence. According to our knowledge, we report, for the first time, using both in vitro and in silico analysis, that VncR binds to the cpsp strain-specifically and regulates its synthesis during serum exposure.
Materials and Methods
Bacterial Strains and Growth Conditions
All the reagents used for bacterial culture were purchased from Difco BD (NJ, United States). S. pneumoniae strains D39 (type 2; GenBank: CP000410.2), WU2 (type 3; GenBank: U15171.1), and BG7322 (type 6B; GenBank: JF911505.1) were grown in THY medium (Todd Hewitt medium with 0.5% Yeast extract) at 37°C without aeration. S. pneumoniae strains possessing the erm marker were grown in media supplemented with 2.5 μg/ml erythromycin. In order to see the effect of the human serum or LF on CPS production, the strains were allowed to grow in THY broth until logarithmic phase (OD550 of 0.30) when 10% human serum or 30 mM LF was added and then co-incubated for certain periods. All the strains used in this study are listed in Table 1.
Construction of vncR Deletion Mutants
The S. pneumoniae D39 vncR deletion mutant was constructed in our previous study (Lee et al., 2018). Similarly, in this study, in WU2 and BG7322 pneumococcal strains, a donor DNA fragment in which the deleted vncR genes were substituted with an erythromycin resistance cassette (ermB), was created by overlapping PCR and incorporated into pneumococcal chromosomes by homologous recombination (Kim et al., 2012). The primers used here were listed in Supplementary Table S1.
qRT-PCR
For a comparative study of the transcript levels of all genes under the cps locus of wild type (WT) strains and their respective vncR deletion mutants after serum or LF supplementation, the strains were subjected to RNA isolation using the Trizol method (Invitrogen, United States). cDNA was prepared using 1 μg of total RNA following the manufacturer’s protocol (Takara Clontech, CA, United States). Real time PCR, consisting of 50 ng of cDNA, 10 pmol of each primer (Supplementary Table S2), and SYBR Green master mix (Elpis, Seoul, Korea), was carried out in a 20 μl final volume with the StepOnePlus Real-Time PCR System (Applied Biosystems, CA, United States) under the following cycling conditions: 95°C for 10 min followed by 40 cycles at 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. All qRT-PCR data were normalized to the mRNA level of 16s rRNA genes used as endogenous controls.
Analysis of Pneumococcal CPS by Enzyme-Linked Immunosorbent Assay (ELISA)
Indirect capsule ELISA with slight modifications were performed, as described previously (Xayarath and Yother, 2007), to determine the change in total CPS quantity in pneumococcus strains after serum treatment. S. pneumoniae strains were grown in THY up to A550 = 0.3, and treated with 10% human serum or LF for the desired period. The bacterial cells were pelleted down by centrifugation at 10,000 × g for 10 min, and washed thrice with sterile PBS. All bacterial samples were then normalized to the same optical density at 550 nm. After that, all the samples were heat-killed at 60°C for 20 min. These samples were coated into wells of microtiter plates (Corning Inc.) and incubated for overnight at 4°C. The wells were further blocked for 1 h with 1% (w/v) bovine serum albumin (BSA; Sigma-Aldrich) in PBS (BSA-PBS). After washing thrice with 0.5% Tween-20- containing PBS (PBST), 100 μl serotype-specific CPS rabbit antiserum (Statens Serum Institut, Copenhagen, Denmark) diluted 1:1,000 with BSA-PBS and incubated at room temperature for 1 h. After washing, these samples were incubated with a 1:5,000 dilution of the horseradish peroxidase (HRP) conjugated secondary rabbit antibody at room temperature for 1 h, followed by washing with PBST. Then, the TMB substrate was added to the wells, and the A450 was determined by an ELISA reader (Softmax).
Fluorescence Microscopy
CPS was further observed using fluorescence microscopy as described previously, with slight modifications (Echlin et al., 2016). Different S. pneumoniae strains were grown in THY broth, and after serum treatment, bacteria were collected during the logarithmic phase of growth by centrifugation. Subsequently, the cells were fixed by 1 h incubation with 4% paraformaldehyde, then incubated with 1:100 dilution of the serotype-specific rabbit antiserum (Statens Serum Institut, Copenhagen, Denmark) in 5% FBS and 0.2% saponin for 1 h at room temperature, and then reacted with a 1:200 dilution of Alexa 488 secondary antibody (Thermo Fisher Scientific, MA, United States) for 1 h at room temperature. The sample (15 μl) was mounted on a glass slide and viewed using a confocal microscope (Carl Zeiss LSM 510, Oberkochen, Germany).
Survival Study
Four weeks old male CD1 mice were purchased from Koatech (Pyeongtaek, Korea). Experiments were carried out in a manner which reduces animal suffering and we also used the least numbers associated with effective statistical evaluation. All experimental procedures were sanctioned by the Ethical Committee of Sungkyunkwan University, based on the Helsinki Declaration. To evaluate the role of VncR-regulated CPS thickness on virulence, mice were intranasally infected (n = 10) or intraperitoneally infected (n = 15) with 5 × 107 and 1 × 105 cells, respectively, of WT and their respective vncR deletion mutants after anesthesia with ketamine-xylazine mixture. Mice were observed each day at 4 h intervals, and the survival time for each mouse was recorded.
Protein Purification
VncR comprises of two distinct functional domains, designated N-terminal receiver and C-terminal effector domains. Here, the VncR C-terminal DNA binding domain (VncRc) was purified. The VncRc-coding portion of the gene (position 119–242) was amplified using polymerase chain reaction (PCR) from the D39 WT genomic DNA. The PCR-amplified DNA was restriction enzyme-digested using NcoI/XhoI, cloned into the NcoI/XhoI sites of the pHis Parallel 2 vector and transfected into Escherichia coli XL1-blue and then, into E. coli BL21 (DE3) for overexpression. Recombinant strains were grown at 37°C to an optical density (OD)600 of 0.6–0.8 in Luria Bertani (LB) medium, supplemented with 50 μg/ml ampicillin, with 150 rpm shaking. Protein expression was induced by supplementing the culture medium with 0.5 mM β-D-1-thiogalactopyranoside (IPTG) at 25°C for 24 h. Following this, cells were collected by centrifugation at 3,382 × g for 15 min at 4°C, resuspended in default buffer [50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 5 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride (PMSF)]. The cells were lysed by sonication, and centrifugation at 16,000 × g for 45 min was performed to separate cellular debris and insoluble proteins. Ni-NTA resin (GE Healthcare, United Kingdom) was used to purify supernatant protein fractions as previously described (Sheffield et al., 1999). His-tagged VncRc was then eluted using elution buffer (50 mM Tris-HCl at pH 7.5, 500 mM NaCl, 200 mM Imidazole). The protein was further concentrated by using a centrifugal filter (Centricon, Millipore, Germany) to 5 mg/ml, and then the quality of the purified protein was confirmed by SDS-PAGE.
Electrophoretic Mobility-Shift Assay (EMSA)
DNA fragments of the cpsp of three independent pneumococcal strains were generated by PCR using the respective genomic DNA of those strains as templates (Primers listed in Supplementary Table S3). DNA (200 ng) was incubated with equal amount of purified VncRc protein in buffer containing 10 mM Tris HCl pH 7.9, 10 mM MgCl2, 50 mM NaCl, 100 μg/ml bovine serum albumin (BSA), and 10% serum (Sigma-Aldrich., St. Louis, MO, United States). After a 5 min incubation at 37°C, the samples were loaded on a 6% native polyacrylamide (PAGE gel) pre-run and electrophoresed at 80 and 100 V, respectively, for 30 min at room temperature. The gels were washed once with water and stained using SYBR Gold (Thermo Fisher Scientific, MA, United States).
Western Blotting
The bands from EMSA gel were excised out and subjected to “TCA (trichloroacetic acid) protein precipitation method” (Koontz, 2014). The TCA-precipitated proteins were mixed with 2X SDS-PAGE loading buffer and loaded on a 15% SDS-PAGE gel run at 200 V for 45 min. The gel was subjected to Western Blot transfer in Towbin buffer at 200 V for 1 h. After the transfer was completed, the membrane was blocked with 5% skim milk powder in TBST (Tris-buffered saline with Tween-20) for 1 h followed by overnight incubation with anti-VncR primary antibody “(Kindly provided by Professor Elaine I. Tuomanen)” in 1:2000 dilution. After washing thrice for 10 min with TBST, the membrane was incubated with an anti-rabbit secondary antibody at 1:5000 dilution followed by washing thrice for 10 min with TBST and developed using the picoEPD western blot detection kit (ELPIS-Biotech, Korea).
Co-immunoprecipitation (Co-IP)
For Co-IP experiments, 200 ng of DNA was incubated at 37°C with an equal amount of His-tagged VncRc protein in buffer, containing 10 mM Tris HCl pH 7.9, 10 mM MgCl2, 50 mM NaCl, 100 μg/ml BSA, and 10% serum (Sigma-Aldrich, St. Louis, MO, United States). Anti-His antibody was added after 5 min incubation, and the mixture was incubated overnight at 4°C, with gentle agitation. The next day, protein-A agarose beads were added to the mixture and incubated further for 4 h at 4°C under shaking. The mixture was centrifuged at 370 × g for 2 min and the supernatant was discarded. The pellet was washed with 100 μl H2O and the supernatant was discarded. The pellets were resuspended in the remaining buffer and 10 μl of the Co-IP mix was used for PCR to identify the presence of the cpsp using specific primers. PCR was performed for 35 cycles, with an annealing temperature of 60°C, and the products were run on a 1% agarose gel, stained with ethidium bromide, and visualized using a GelDoc system (Bio-Rad, CA, United States).
Modeling and Protein-DNA Interface Analysis
To predict the 3D structure of the VncR full-length protein, we utilized three different tools; MOE (2018), I-TASSER and SwissModel. These packages predicted almost similarly-folded structures of the VncR with RMSD ranging from 0.5 to 1 Å. These models were further evaluated and optimized to remove all kinds of abnormalities. To predict the DNA binding mechanism of VncR, the fully optimized structure modeled by I-TASSER server was used in protein-DNA docking. The 3D structure of the VncR-binding region of cpsp, containing direct repeats, was modeled through 3D-DART and MOE, and further optimized (using default parameters in MOE). To evaluate the possible binding mechanism of the DBD of VncR and cpsp, the receiver/regulatory domain of the VncR was removed and subjected to protein-DNA docking using ZDOCK online server (Chen et al., 2003) and MOE. Based on the electrostatic complementarity, geometry, and hydrophobicity of the molecular surface, ZDOCK ranks the 100 most probable predictions out of thousands of candidates and has been considered among the top docking servers.
Molecular Dynamics Simulations
To estimate structural changes in the DNA-VncR DBD as a function of time, molecular dynamics simulations (MDS) has been widely utilized. The most accurate and validated complex was simulated in GROMACS v5.0.7 (Abraham et al., 2015). The system was solvated in a dodecahedron box using TIP3P waster model (Jorgensen et al., 1983). The AMBER99SB-ILDN force field has been improved for protein-DNA complexes (Lindorff-Larsen et al., 2010). To mimic the infinite system, periodic boundary conditions were applied to the box in all directions. For long-range electrostatics, the Particle Mesh Ewald approach was employed using 10Å cutoff values (Darden et al., 1993). Bond lengths were constrained using LINC algorithm (Hess et al., 1997), and the system was neutralized using counter ions. Additionally, 100 mM NaCl concentrations were added to mimic physiological conditions. Steepest descents and/or conjugate gradient minimization with a maximum tolerance of 100 kJ/mol/nm were performed to remove steric interactions. To equilibrate the system, it was initially simulated for 100 ps under a constant volume (NVT) ensemble to achieve 300 K by the V-rescale method (Bussi et al., 2007). The equilibrated structures from the NVT ensemble were subjected to constant pressure (NPT) equilibration for 100 ps using the Parrinello-Rahman barostat under an isotropic pressure of 1.0 bar (Parrinello and Rahman, 1981). During data collection, the V-rescale thermostat and Parrinello-Rahman barostat were used to maintain the temperature and pressure at 300 K and 1 bar, respectively, and the atomic coordinates were saved every 2.0 ps for 100 ns total simulation time.
Data Analysis and Graphics
Most of the data were analyzed with built-in modules of GROMACS v5.0.7. Graphical images were produced with PyMol (Rigsby and Parker, 2016) and VMD (Humphrey et al., 1996). Interface analyses were performed in MOE, VMD, and UCSF Chimera, and images were generated in MOE and PyMol. For alanine scanning mutagenesis, a similar protocol was used as described elsewhere (Shah et al., 2016). All computational studies were performed on a Dell PowerEdge server with a CentOS6 GNU/Linux operating system.
Statistical Analysis
Differences in overall survival rates between groups were analyzed by the Log-rank test. Each experiment was carried out at least thrice in duplicates and results were expressed as mean ± standard error of the mean (SEM). Statistical analysis was performed by ANOVA using the Graph Pad Prism software (version 5, Graph Pad Software Inc, CA, United States) and P < 0.05 were considered to be statistically significant.
Results
Strain-Specific Induction of vncR by Serum
In our previous study, mRNA levels of D39 (type 2) vncR were maximally induced by 5 min treatment of 10% human serum (Lee et al., 2018), however, 5 min exposure to 10% serum failed to activate vncR mRNA levels in BG7322 (type 6B) and WU2 (type 3) pneumococci (data not shown). Nonetheless, subsequent prolonged treatment with 10% serum revealed maximal induction of the vncR mRNA in types 3 and 6B in 2 h and 40 min, respectively (Figure 1).
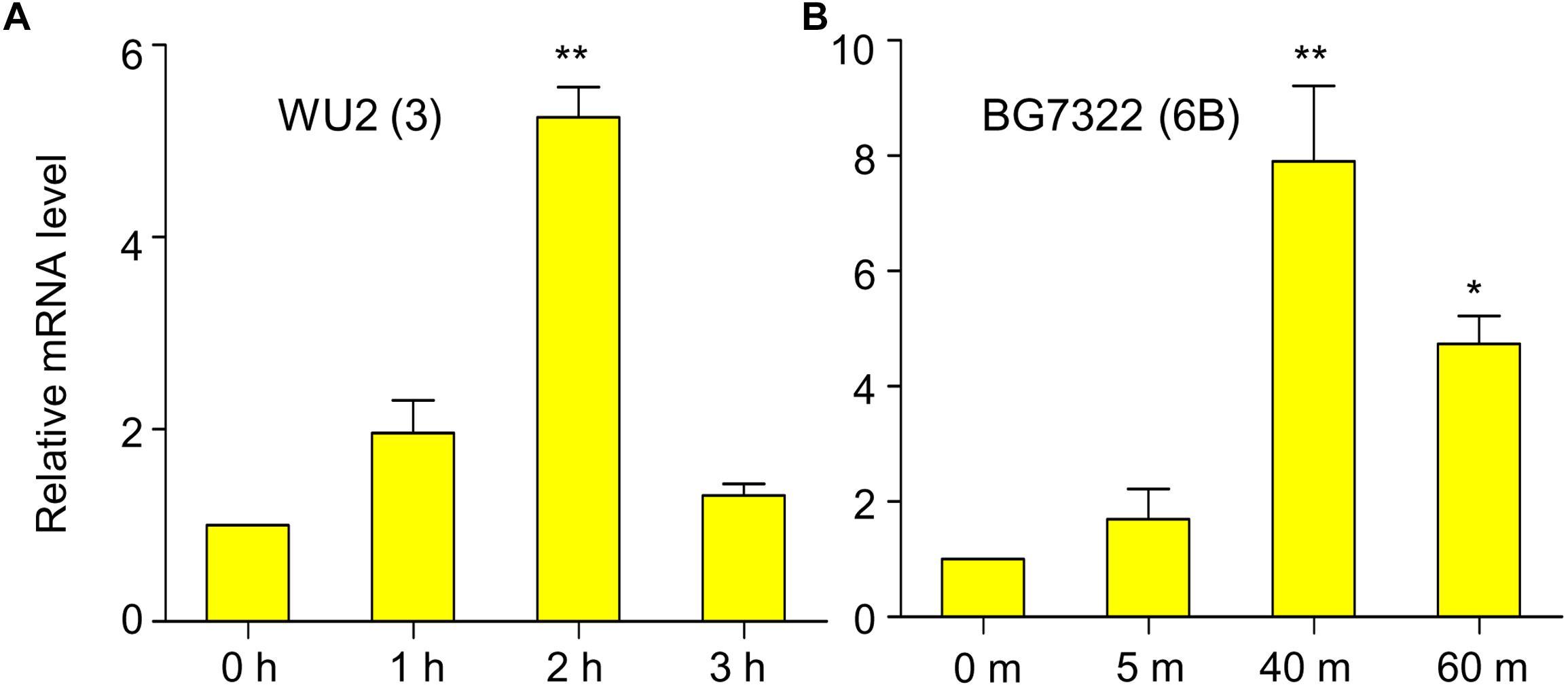
Figure 1. Serum induces vncR expression in a serotype-specific manner. S. pneumoniae strains WU2 (A) and BG7322 (B) were cultured in the THY broth up to mid-log phase, and then incubated with 10% humanprotect serum for 3 h and 60 min respectively. The mRNA expression levels of vncR were analyzed by qRT-PCR. Data are expressed as mean ± standard error of mean (SEM) of 3 experiments in quadruplicates. ∗P < 0.05, ∗∗P < 0.01 (One-way ANOVA) as between compared groups.
VncR Mediates cps Gene Transcription in a Strain-Specific Manner During Serum Exposure
To assess whether the expression of the cps genes is transcriptionally regulated by VncR after serum exposure, comparative qRT-PCR was carried out for all genes under the cps loci, D39 (17 genes), WU2 (2 genes), and BG7322 (14 genes), and their respective isogenic vncR deletion mutants. A schematic representation of these three types of cps loci has been given in Supplementary Figure S1. Results showed that in D39, all the 17 genes were transcriptionally induced ∼4–12-folds within 5 min of serum exposure, which was significantly higher than the induction of its isogenic ΔvncR cps genes (Figure 2A).
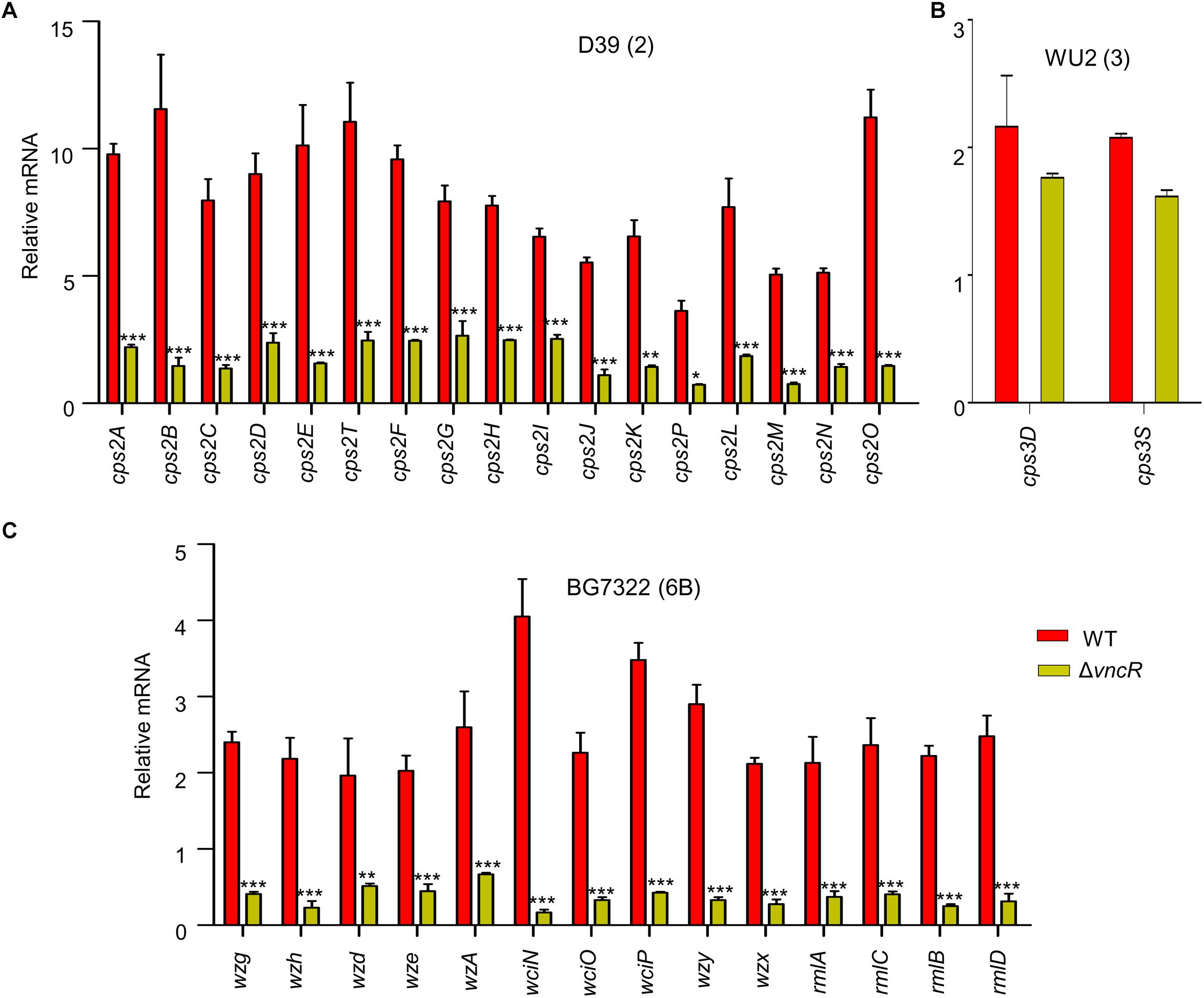
Figure 2. VncR regulates cps gene transcription in a strain-specific manner after serum exposure. S. pneumoniae strains D39 (type 2; A), WU2 (type 3; B), and BG7322 (type 6B; C) and their respective vncR deletion mutants were cultured in THY broth up to mid-log phase, and then incubated with 10% human serum for 5 min, 2 h, and 40 min respectively. The mRNA expression levels of the cps operon were analyzed by qRT-PCR. Data are expressed as mean ± standard error of mean (SEM) of 3 experiments in quadruplicates. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (Two-way ANOVA) as between compared groups.
In type 3, only two genes, cps3D and cps3S, were shown to be responsible for CPS synthesis (Caimano et al., 1998). We found that after 5 min of serum exposure, these cps genes became up-regulated about ∼1.6–2.1-folds from their respective basal levels in both the WT WU2 strain and its ΔvncR mutant (data not shown). Comparative transcription levels of their cps genes were studied for up to 2 h of serum exposure, but they did not show any significant differences between WU2 WT and WU2ΔvncR (Figure 2B).
Among the 14 genes in the cps locus involved in CPS synthesis in type 6B (Elberse et al., 2011), there were no significant differences between BG7322 and its ΔvncR mutant after 5 min of serum exposure (data not shown). After 40 min exposure to the serum, cps genes expression in the WT were upregulated, whereas that of its isogenic ΔvncR showed significant reduction (Figure 2C). These results suggest that pneumococcal cps genes could be regulated strain-specifically by VncR.
VncR Regulates CPS Production in S. pneumoniae Strain-Specifically
To confirm the effect of VncR on type 2, 3, and 6B CPS production, CPS quantities were estimated by indirect ELISA, and visualized by confocal microscopy. ELISA results showed that total CPS amounts were significantly increased after 20 min of serum exposure in D39, whereas, no significant changes were observed in D39ΔvncR (Figure 3A). Unlike in the D39 strain, even after 2 h of serum treatment, CPS amounts in the WU2 WT and its ΔvncR mutant did not show any significant differences, indicating that vncR deletion in type 3 does not affect CPS production (Figure 3B), further supporting our qRT-PCR data.
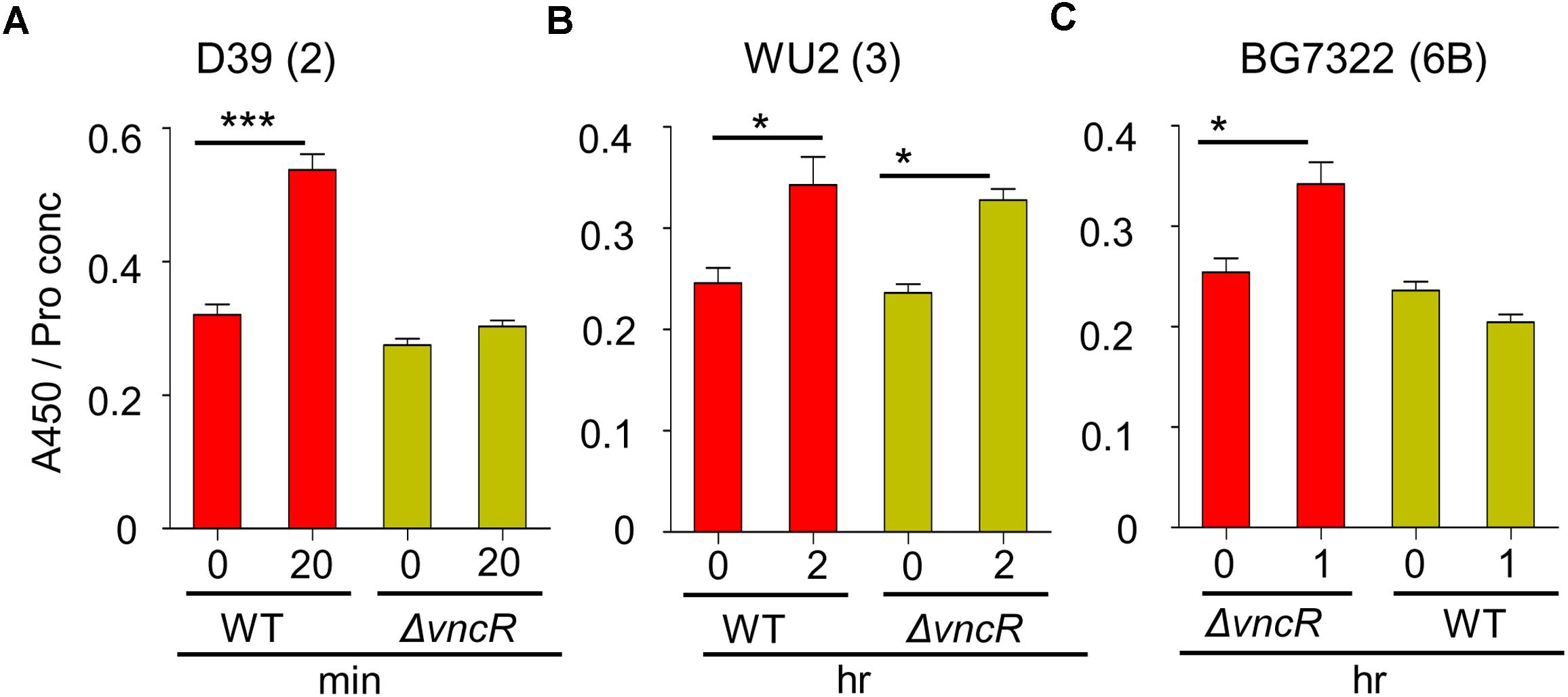
Figure 3. VncR regulates pneumococcal CPS levels in a strain-specific manner after serum exposure. Pneumococcal strains WT D39 (A), WU2 (B), and BG7322 (C) and their isogenic vncR deletion mutants were treated with serum for 20 min, 2 h, and 1 h, respectively. Total CPS quantities in whole cell lysates were determined by ELISA using serotype-specific antiserum. All samples were assayed in quadruplicates and each assay was repeated thrice. Data are expressed as mean ± standard error of mean (SEM) of 3 experiments in quadruplicates. ∗P < 0.05, ∗∗∗P < 0.001 (One-way ANOVA) as between compared groups.
As our qRT-PCR analysis showed maximal differences in the cps mRNA levels of BG7322 WT and ΔvncR mutant after 40 min of serum exposure, the total CPS amounts were compared between these two strains, for up to 1 h of serum exposure, using ELISA. Interestingly, the CPS level in the BG7322 WT was significantly increased, whereas that in the ΔvncR did not change significantly (Figure 3C).
To reconfirm strain-specific CPS regulation by the VncR, the WT D39, WU2, and BG7322, and their respective ΔvncR mutants were grown to log phase and then treated with 10% human serum for a specified time for evaluation using confocal microscopy. Microscopy data revealed that D39 showed stronger fluorescence after 20 min serum exposure, whereas its ΔvncR mutant did not (Supplementary Figure S2). In contrast, no significant differences in fluorescence levels were found between the WU2 and BG7322 WTs, and their respective ΔvncR mutants (data not shown).
LF Is the Differential VncR Regulator for CPS Transcription and Production
Previously, supplementation of 30 mM LF caused the induction of type 2 pneumococcal vncRS, proposing LF as a VncS sensor ligand (Lee et al., 2018). Thus, the effect of LF supplementation was examined in D39, WU2, and BG7322 strains. cps genes in D39 and BG7322 (only representative genes shown here) were induced significantly after 5 and 40 min of 30 mM LF exposure, respectively, whereas, they were not induced by LF depleted serum treatment (Figures 4A,B). ELISA results consistently indicated significant augmentation of total CPS levels in the D39 and BG7322 WT, after 20 min and 1 h of LF serum supplementation, respectively, whereas, no substantial changes were observed after exposure to LF-depleted serum (Figures 4C,D). Moreover, the addition of exogenous LF to the LF-depleted serum resulted in a significant increase in CPS levels (Figures 4C,D), establishing the role of LF in VncR-mediated CPS production during serum exposure. As expected, LF treatment did not show any effect on the CPS production of WU2 strain (Supplementary Figure S3).
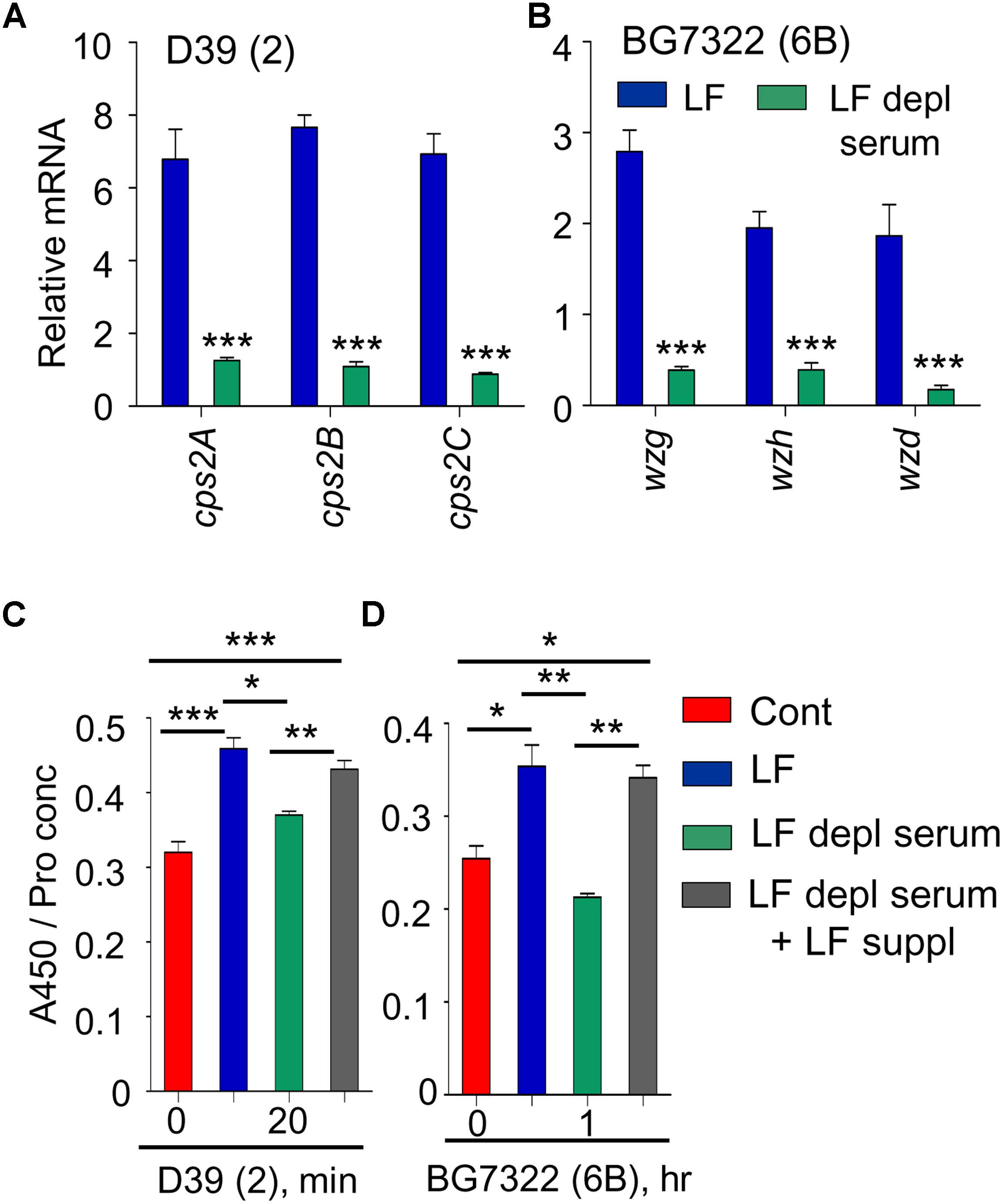
Figure 4. VncR regulates cps gene transcription and CPS levels in a strain-specific manner after serum LF exposure. The D39 (A) and BG7322 (B) WT strains were incubated for 5 and 40 min, respectively, with LF and in LF-depleted serum. The mRNA expression levels of the cps genes were analyzed by qRT-PCR. Additionally, the WT D39 (C) and BG7322 (D) strains were incubated for 20 min and 1 h, respectively, with LF and in the LF-depleted serum and LF was also exogenously added to the LF-depleted serum. Total CPS quantities in whole cell lysates were determined by ELISA using serotype-specific antiserum. All samples were assayed in quadruplicates and each assay was repeated thrice. Data are expressed as mean ± standard error of mean (SEM) of 3 experiments in quadruplicates. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (One-way ANOVA) as between compared groups.
VncR Regulates Pneumococcal Virulence in vivo in a Strain-Specific Manner
To study the effect of ΔvncR on virulence, mice were infected with pneumococci intranasally (i.n.) and survival times were monitored. Survival rate of mice infected with D39ΔvncR was 30%, whereas, all D39WT-infected mice died within 4 days post-infection (Figure 5A). However, the survival rates of WU2ΔvncR infected group and WU2 WT showed no significant differences; all mice in both groups succumbed 7 and 6 days, post-infection, respectively (Figure 5B). Similar to type 2, the survival rate of the BG7322ΔvncR-infected group was 30% compared to 0% for the BG7322 WT-infected group (Figure 5C). For further confirmation, the effect of ΔvncR on virulence in a sepsis model was examined by intraperitoneal (i.p.) infection with D39, WU2, and BG7322 WTs and their ΔvncR mutants. Mice infected with the D39ΔvncR and BG7322ΔvncR mutants consistently showed significantly higher survival rates than mice infected with their respective WTs, 40 h post-infection (Figures 5D,F); whereas, mice infected with the WU2ΔvncR mutant showed no differences in the survival rates compared with the WU2 infected mice (Figure 5E). Overall, D39 and BG7322, comprising the ΔvncR, showed significantly reduced virulence than their respective WTs, while WU2ΔvncR seemed to be almost as equally virulent as its WT.
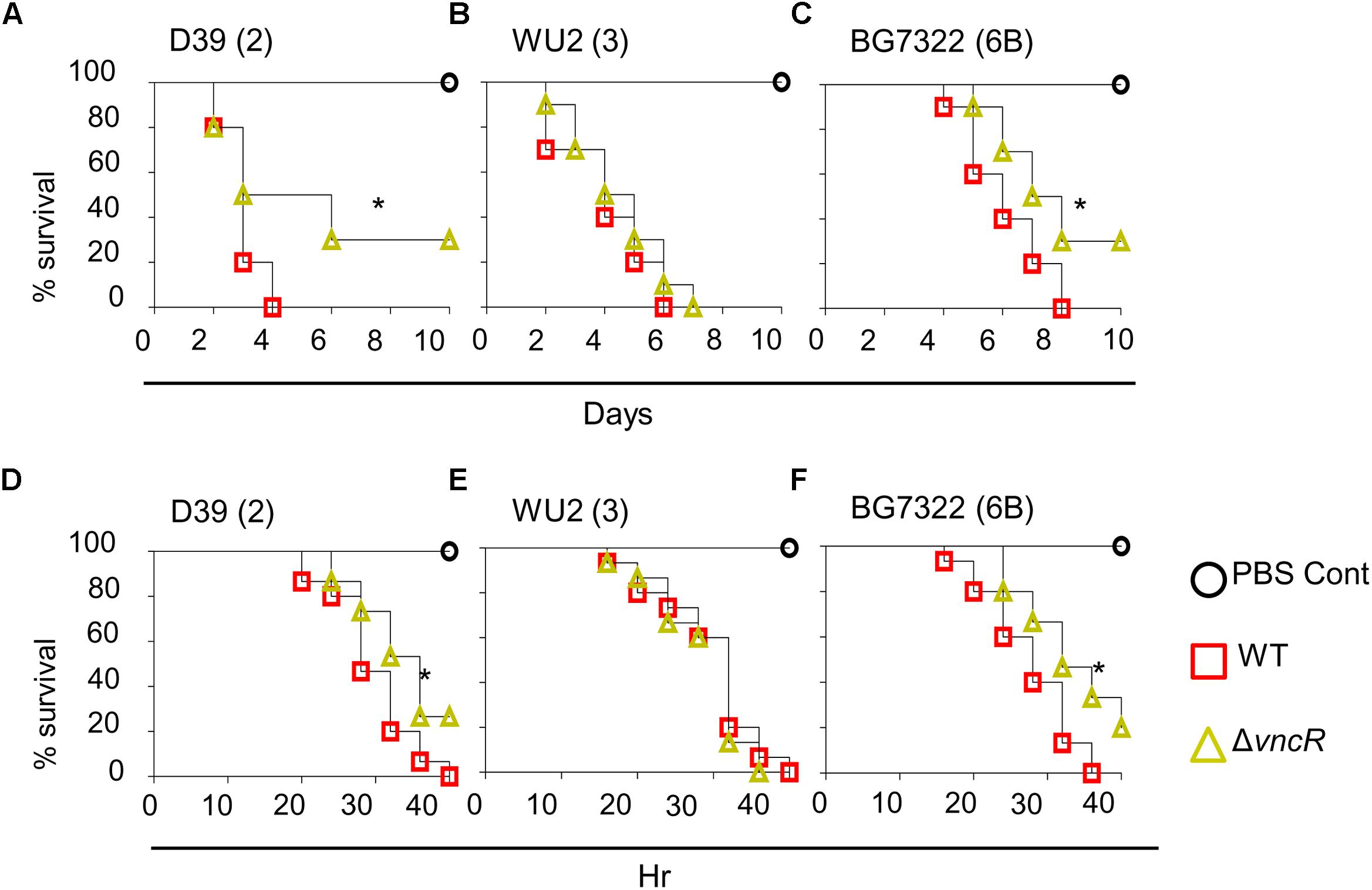
Figure 5. VncR modulates in vivo virulence in a strain-specific manner. Mice (n = 10) were infected intranasally (i.n) with ∼5 × 107 CFU of D39 (A), WU2 (B), BG7322 (C) or isogenic mutants, and survival times were determined. Mice (n = 15) were infected intraperitoneally (i.p) with 1 × 105 CFU of D39 (D), WU2 (E) BG7322 (F) or their isogenic vncR mutants. Mice were observed for 40 h and survival was assayed every 4 h. The results are representative of at least three independent experiments. ∗P < 0.05 (Log-rank test) as between compared groups.
VncR Binds to the cpsp in a Serum-Dependent Manner
To confirm whether the DBD of the VncR protein can bind to the promoter region of cps, we performed EMSA with the purified C-terminal DNA binding domain of VncR (VncRc, Figure 6A) and promoter region of cps genes. Our results revealed that in presence of serum, VncRc could bind to the 218 bp cpsp of types 2 and 6B, as well as, to the 142 bp type 3 cpsp. Interestingly, there were no shift of bands when we incubated cpsp and VncRc without serum (Figure 6C). Control experiments have been done using only cpsp and serum. As among all the strains tested here, the CPS of type 2 39 showed maximal induction after serum treatment within the shortest period, we have included the representative figure of this control experiment using cpsp of type 2 D39 and VncRc protein (Figure 6B). Presence of DNA binding proteins in the human serum is a well-established fact (Sylvia et al., 1975) since a long time, so it is quite possible that when cpsp and serum were incubated, cpsp (DNA) could bind to any of these proteins and showed the shift in the gel. But, when VncRc was incubated along with cpsp and serum, the shift was more uplifted than for the mix of cpsp and serum only. This result confirms that VncRc can bind to cpsp in the presence of serum. Subsequent western blotting using an anti-VncR antibody with these bands (D39 only) of EMSA confirmed the presence of VncRc in the complex (Figure 6D). To corroborate EMSA results, we performed a co-IP experiment using all three cpsp independently and His-tagged VncRc protein. The VncRc-cpsp complexes were pulled down using anti-His antibody and subsequent PCR results indicated the presence of specific cpsp in those complexes (Figure 6E), which was reconfirmed by sequence analysis (Supplementary Figure S4). The supernatants, after pulling down the co-IP complexes, were used as negative controls in PCR reactions.
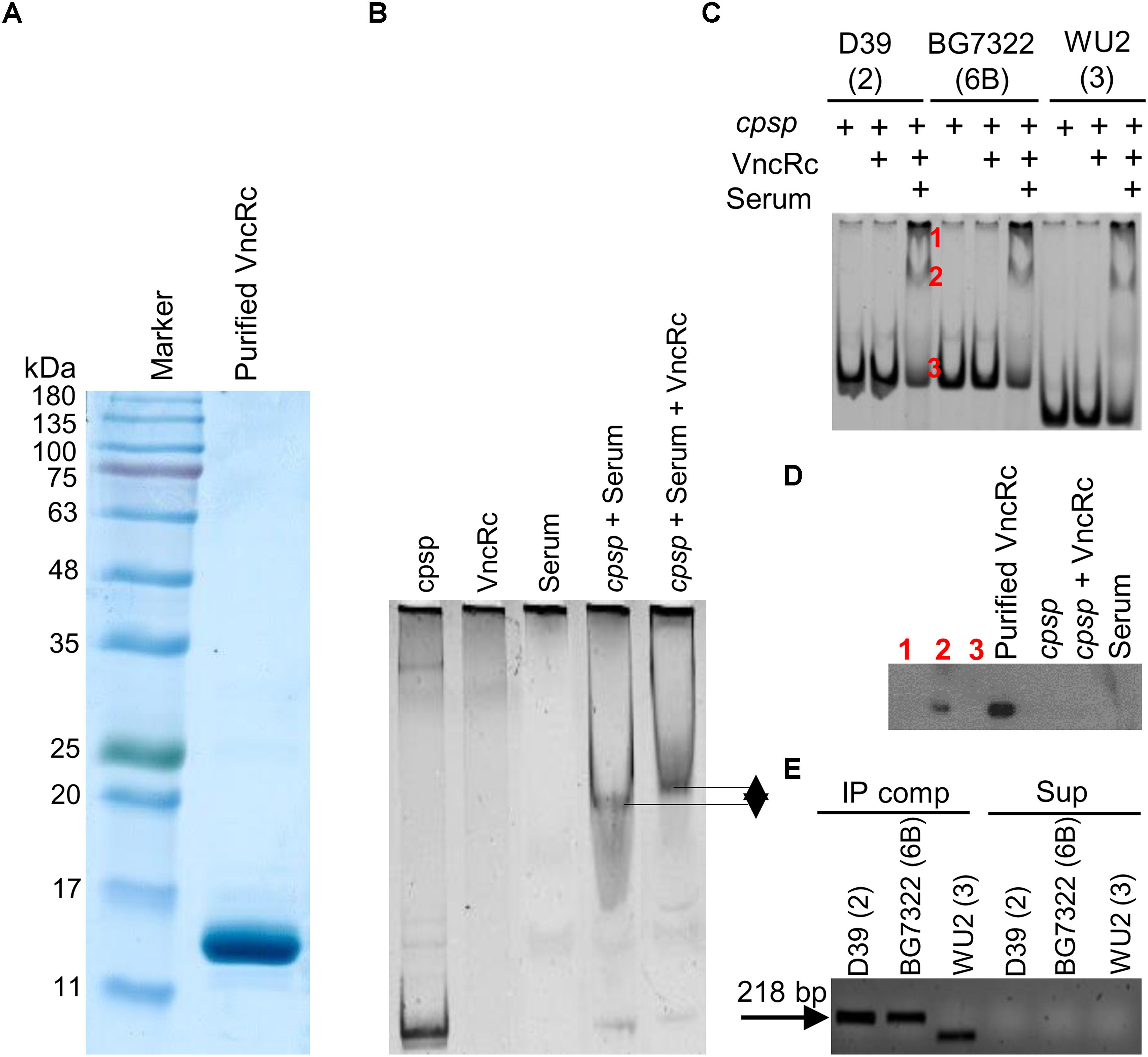
Figure 6. Characterization of the interaction between VncRc and the pneumococcal cpsp. SDS-PAGE analysis to check the purity of the His-tagged VncRc protein (A). EMSA of 6 × His-VncRc protein and D39 cpsp, in the presence of serum. Control experiments have been done using only cpsp and serum. When VncRc was incubated along with cpsp and serum, the shift was more uplifted than the shift of cpsp and serum mix (B). EMSA of 6 × His-VncRc protein and D39, BG7322, and WU2 cpsp, in the presence of serum. The 218 bp D39 and BG7322 cpsp, and 142 bp WU2 cpsp were independently incubated with purified VncRc. Results showed that all three types of cpsp can interact with VncRc in the presence of serum. Bands marked in red color as “1” and “2” are the resultant shifted bands, and band marked in red color as“3” is unshifted band after interaction of D39 cpsp and VncRc in presence of serum (C). Western blot results using anti-VncR antibody of the bands marked in red color as “1,” “2,” and “3” in (B). Purified VncRc, a band of cpsp alone (without VncR), an unshifted band after treatment with VncRc, and serum have been also used as controls. Confirmation of the presence of VncRc in shifted EMSA bands (marked as “2”) – only the shifted bands of D39 were used (D). Detection of cpsp by PCR from the co-immunoprecipitated VncRc-cpsp complex (in the presence of serum) after immunoprecipitation using an anti-His antibody (E). The results in (B–E) are representatives of three independent experiments.
Modeling and Protein-DNA Docking Analyses
Our model suggests that VncR shares sequence and structural similarity with response regulators, including PrrA, VraR, and KdpE, from different bacteria (Leonard et al., 2013; Narayanan et al., 2014). Overall, VncR acquired a stable, folded structure with two distinct domains; the N-terminal domain has a typical (β-α)5 folded structure (Figure 7) that has been reported as analogous to the other receiver domain-containing OmpR/PhoB subfamily proteins (Bourret, 2010). The C-terminal domain acquires a typical helix-turn-helix fold and contains four and two β-sheets at its N- and C-terminuses, respectively, sandwiching the DNA-recognizing helix-turn-helix structure (Figure 7 enhanced part).
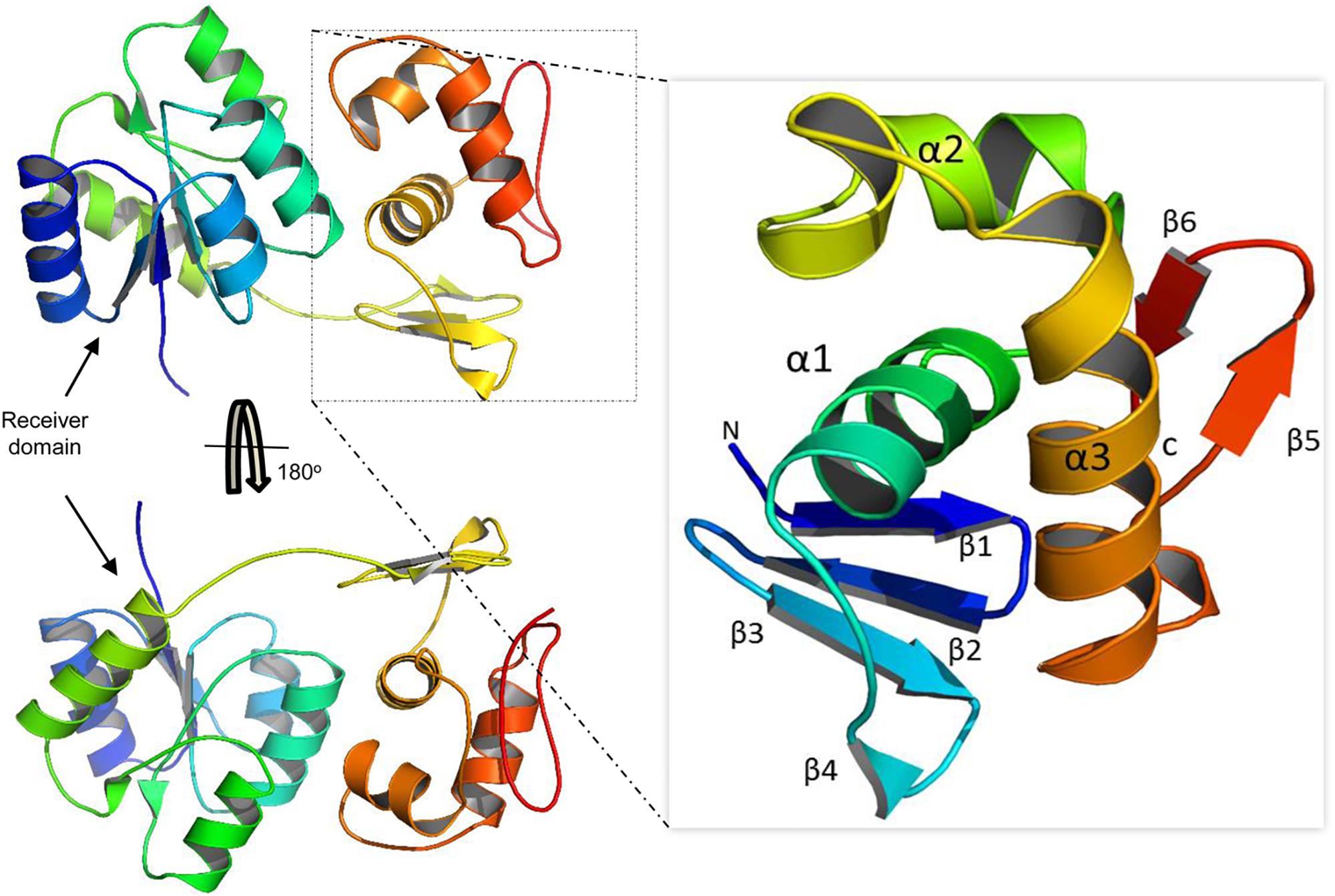
Figure 7. Domain organization of the VncR full-length protein. VncR contains an N-terminal receiver or regulatory domain and a C-terminal DNA-binding domain (DBD). The DBD has a typical helix-turn-helix fold exhibited by OmpR/PhoB subfamily proteins to recognize direct repeats in promoter regions.
Bacterial response regulators recognize the promoter DNA sequence and bind to the direct repeats in tandem (Blanco et al., 2002; Okajima et al., 2008). Similar direct repeats, containing TGTCC/AT, were found in the cpsp of types 2 and 6, but not in type 3 S. pneumoniae. Further, the second repeat in type 6 may contain adenosine (A) or thymine (T) instead of conserved guanosine (G) (Supplementary Figures S5A, S6). Docking procedures suggest that charged residues in the α3 helix established electrostatic interactions with the negatively charged backbone atoms of the major groves (Figure 8). Considering the “G” to “A/T” mutation in the second repeat of type 6, the “GC” base pair of type 2 cpsp was mutated into an “AT” base pair and analyzed for differences. Interestingly, we found that in the type 2 WT, the aromatic base of cytosine establishes a typical cationic-π interaction with a positively-charged amide group of Lys194. However, this interaction was lost in the mutant type 2 cpsp (Supplementary Figure S6). The methyl group moiety at the base of thymine creates a bulky hydrophobic patch (Supplementary Figure S6, electrostatic surface map) in the major grove of the cpsp, suggesting that, a “G” to “A/T” mutation could possibly reduce, or delay, VncR binding and thus CPS production.
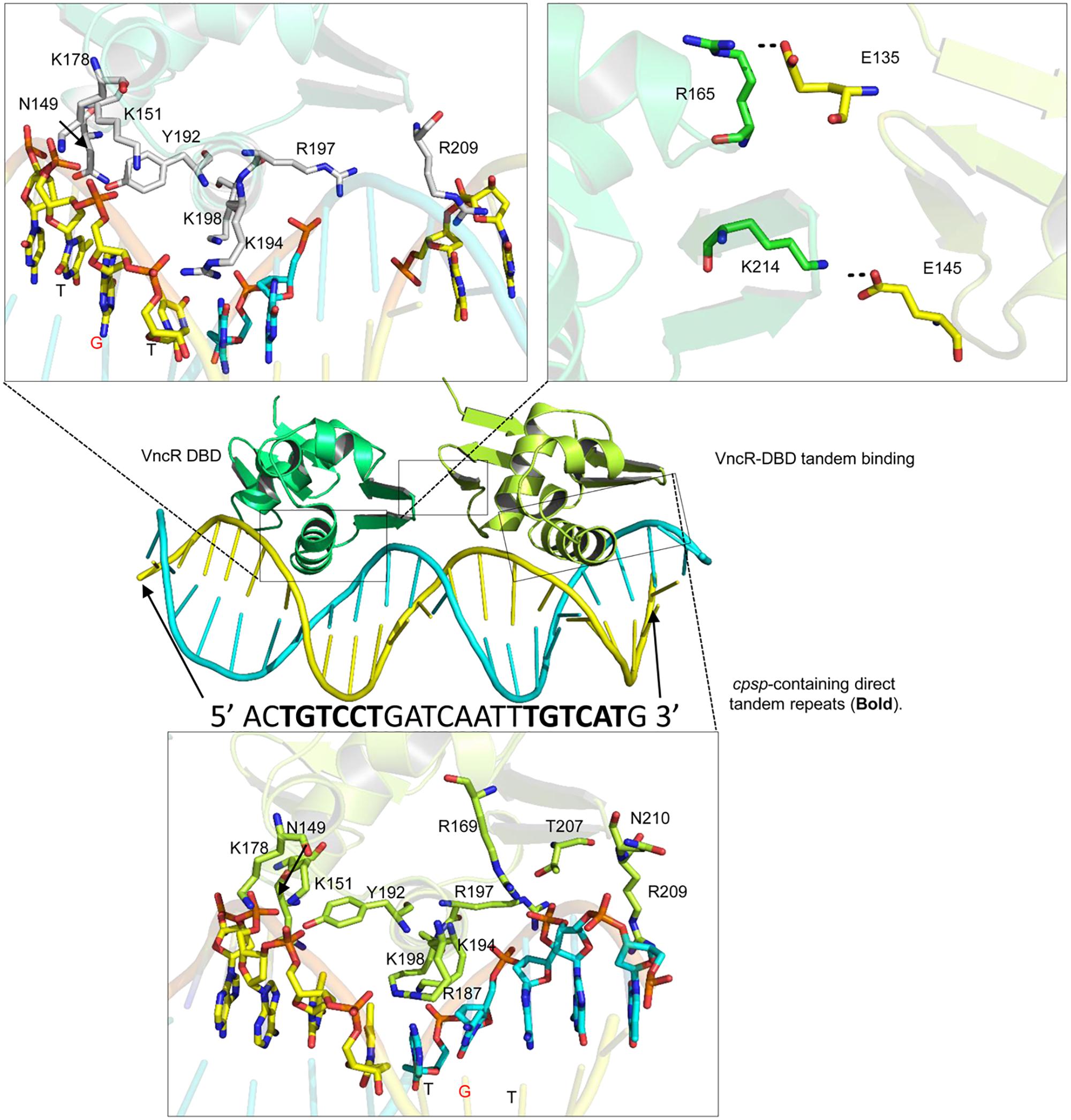
Figure 8. Tandem binding of VncR-DBD to the cps-direct repeat-containing promoter region. The α3 helix of VncR-DBD contains positively charged residues that recognize and interact with the direct repeat-containing cps-promoter in tandem. The salt bridges between the two DBD domains stabilize the tandem binding pattern.
Molecular Dynamics Simulations (MDS) and Interface Analysis
The protein-DNA complex (using the type 2 template) was placed in physiological saline and subjected to 100 ns simulation. The RMSD plot suggests the overall stability of the complex with an average 3Å deviation (Supplementary Figure S5B). To track the structural dynamics and monitor any domain abnormality, the coordinates of the simulating system were extracted from the MD trajectory as a function of time and converted into animation (Supplementary Videos S1, S2). The lowest-energy representative complex was extracted from the MD trajectory and investigated for detailed interface analysis. The α3 helix, containing positively charged arginine (Arg187 and 197) and three lysine residues (Lys194, 198, and 199), established strong electrostatic interaction with the backbone phosphates as well as the basis. Besides, the inter-DBD interface is stabilized by salt bridges established between the Glu145 and Lys214, and Glu135 and Arg165 (Figure 8). A similar interaction pattern has been previously observed in structural studies of the response regulator, RstA (Li et al., 2014).
Alanine Scanning Mutagenesis of the Interfacial Residues
Positively charged and other interfacial residues in the α3 helix were mutated into alanine (using the type 2 template) and their relative effect on the overall complex was measured in a tandem complex. Besides the positively charged residues in the α3 helix, Arg209 has a crucial role in DNA recognition and binding. Overall the DNA-binding affinity of the mutant complexes is drastically reduced if the basic residues are mutated. In addition to the basic residues in the α3 helix, Tyr 192, and Arg169 were found to be crucial for the stability of the VncR-cpsp complex (Supplementary Figure S7).
Discussion
Comparative sequence analysis of VncR exhibited a great similarity to the PhoB of other bacteria (Throup et al., 2000), which has been known to regulate the expression of various bacterial virulence factors (McCluskey et al., 2004). Our results suggested that VncR plays a role in the induction of only type 2 and type 6B pneumococcal cps gene expression in the presence of serum and LF is the main factor behind this induction. We found that the CPS levels were significantly increased both in the type 3 strain and its vncR deletion mutant after a 2 h of serum treatment, although we could not detect the increase in cps genes mRNA levels of type 3 and its vncR deletion mutant after 2 h of serum exposure. The disparity of this VncR-regulated cps transcription may be owed to the fact that serotype 3 strains usually form a simpler polysaccharide structure using a distinct synthesis mechanism (Yother, 2011). Our study proposes that, when pneumococci get exposed to host serum during invasion, VncS senses LF, a serum component, resulting in the induction of VncRS. Induced VncR then binds to the promoters of cps genes to regulate their expression. In line with our in vitro data, we found that mice infected with D39ΔvncR and BG7322ΔvncR showed significantly higher survival rates than mice infected with their respective WTs, but the WU2ΔvncR showed no difference. This may be attributed to the comparatively lesser amounts of CPS of D39ΔvncR and BG7322ΔvncR in serum, which makes them less resistant to opsonophagocytosis, as CPS expression uplifts resistance to the complement C3 deposition and, consequently, to neutrophil-mediated phagocytosis (Kim et al., 1999; Hyams et al., 2010).
In contrast, EMSA results indicated that in presence of serum, the DBD of the VncR protein can bind to the cpsp of all these three serotypes, suggesting that bacterial transcription is dependent upon a coordinate regulation of various factors (Balleza et al., 2009), and that the DNA-binding protein primarily binds somewhere along the DNA non-specifically, followed by diffusion in search of its specific target (Li and Xie, 2011). In agreement with our previous report (Lee et al., 2018), this study proposes that when pneumococci get exposed to host serum during invasion, VncS senses the serum component LF, which results in the phosphorylation of VncS. Phosphorylated VncS then provides a phosphate group to VncR, and phosphorylated VncR binds to the cpsp, which permits the pneumococci to acclimatize to the blood environment. To best mimic in vivo conditions, a purified C-terminal DNA-binding domain of VncR was used in EMSA, so it can bind target DNA without being phosphorylated as the N-terminal domain removal makes it active (Makino et al., 1996). Our findings clearly have some limitations, as it cannot disclose the actual underlying mechanism of this serum-dependent interaction between VncRc with bacterial DNA, and further studies need to be carried out for revealing the underlying mechanism. Under these circumstances, we assume that the presence of serum albumin, which has an amazing ligand-binding capacity and is known to confer stability to many serum components (Sleep, 2015) might somehow facilitate the VncRc-cpsp interaction.
To speculate this differential expression of VncR-induced cps genes in type 2, 3, and 6 strains, we proposed a VncR-DNA model. Interestingly, type 2 and 6 cpsp contain a regular direct repeat, which is absent in type 3. However, all three serotypes share identical VncR protein structures. Thus, we propose that like type 2 and type 6, type 3 may also establish contact with cpsp, supporting our EMSA results, possibly owing to the identical DNA-binding α3, containing positively charged residues and the helix in serotype 3. However, due to the lack of VncR-specific repeats in the type 3 pneumococcal cpsp region, type 3 VncR had the least probability of regulating underlying cps transcription. To further validate our proposed model, the VncR-cpsp complex was simulated under physiologic conditions. The proposed tandem complex retained its stability throughout the simulation. Our in silico molecular simulation data provide evidence in favor of VncR-regulated strain-specific CPS synthesis in the presence of serum, which ultimately influences virulence.
Taken together, our results offer solid indication that pneumococci can regulate cps gene expression strain-specifically in order to familiarize with the varying host environment during invasion through VncR-cpsp interaction. Considering the importance of pneumococcal CPS in invasive diseases, it would be a practical approach to design small molecular inhibitors that target VncR-regulated encapsulation to combat pneumococcal infection.
Data Availability Statement
All datasets generated for this study are included in the manuscript/Supplementary Files.
Ethics Statement
All experimental procedures were reviewed, approved, and sanctioned by the Ethical Committee of Sungkyunkwan University, based on the Helsinki Declaration.
Author Contributions
DR and PG conceived the experiments. PG, MS, SR, S-SP, and HI performed the experiments. PG, MS, SR, SC, KK, and DR analyzed the data. PG, MS, and DR wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Research Foundation grants (NRF-2018R1A2A1A05078102 to DR and NRF-2017R1D1A1B03031019 to PG). The funding body played no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank Professor Elaine I. Tuomanen of St. Jude Children’s Research Hospital for kindly providing us the anti-VncR antibody.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02279/full#supplementary-material
References
Aanensen, D. M., Mavroidi, A., Bentley, S. D., Reeves, P. R., and Spratt, B. G. (2007). Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 189, 7856–7876. doi: 10.1128/jb.00837-07
Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., et al. (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25. doi: 10.1016/j.softx.2015.06.001
Avery, O. T., Macleod, C. M., and Mccarty, M. (1944). Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79, 137–158. doi: 10.1084/jem.79.2.137
Balleza, E., López-Bojorquez, L. N., Martínez-Antonio, A., Resendis-Antonio, O., Lozada-Chávez, I., Balderas-Martínez, Y. I., et al. (2009). Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol. Rev. 33, 133–151. doi: 10.1111/j.1574-6976.2008.00145.x
Blanco, A. G., Sola, M., Gomis-Ruth, F. X., and Coll, M. (2002). Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10, 701–713. doi: 10.1016/s0969-2126(02)00761-x
Bogaert, D., De Groot, R., and Hermans, P. (2004). Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet infect. Dis. 4, 144–154. doi: 10.1016/s1473-3099(04)00938-7
Bourret, R. B. (2010). Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 13, 142–149. doi: 10.1016/j.mib.2010.01.015
Bussi, G., Donadio, D., and Parrinello, M. (2007). Canonical sampling through velocity rescaling. J. Chem. Phys. 126:014101. doi: 10.1063/1.2408420
Caimano, M. J., Hardy, G. G., and Yother, J. (1998). Capsule genetics in Streptococcus pneumoniae and a possible role for transposition in the generation of the type 3 locus. Microb. Drug Resist. 4, 11–23. doi: 10.1089/mdr.1998.4.11
Chen, R., Li, L., and Weng, Z. (2003). ZDOCK: an initial−stage protein−docking algorithm. Proteins 52, 80–87. doi: 10.1002/prot.10389
Darden, T., York, D., and Pedersen, L. (1993). Particle mesh ewald: an N⋅log (N) method for ewald sums in large systems. J. Chem. Phys. 98, 10089–10092. doi: 10.1063/1.464397
Dillard, J. P., Vandersea, M. W., and Yother, J. (1995). Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J. Exp. Med. 181, 973–983. doi: 10.1084/jem.181.3.973
Echlin, H., Frank, M. W., Iverson, A., Chang, T.-C., Johnson, M. D., Rock, C. O., et al. (2016). Pyruvate oxidase as a critical link between metabolism and capsule biosynthesis in Streptococcus pneumoniae. PLos Pathog. 12:e1005951. doi: 10.1371/journal.ppat.1005951
Elberse, K., Witteveen, S., Van Der Heide, H., Van De Pol, I., Schot, C., Van Der Ende, A., et al. (2011). Sequence diversity within the capsular genes of Streptococcus pneumoniae serogroup 6 and 19. PLoS One 6:e25018. doi: 10.1371/journal.pone.0025018
Finlay, B. B., and Falkow, S. (1997). Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61, 136–169.
Ghosh, P., Luong, T. T., Shah, M., Thach, T. T., Choi, S., Lee, S., et al. (2018). Adenylate kinase potentiates the capsular polysaccharide by modulating Cps2D in Streptococcus pneumoniae D39. Exp. Mol. Med. 50:116. doi: 10.1038/s12276-018-0141-y
Guidolin, A., Morona, J. K., Morona, R., Hansman, D., and Paton, J. C. (1994). Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F. Infect. Immun. 62, 5384–5396.
Hammerschmidt, S., Wolff, S., Hocke, A., Rosseau, S., Müller, E., and Rohde, M. (2005). Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect. Immun. 73, 4653–4667. doi: 10.1128/iai.73.8.4653-4667.2005
Hess, B., Bekker, H., Berendsen, H. J. C., and Fraaije, J. G. E. M. (1997). Lincs: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472. doi: 10.1002/(sici)1096-987x(199709)18:12<1463::aid-jcc4>3.3.co;2-l
Humphrey, W., Dalke, A., and Schulten, K. (1996). Vmd: visual molecular dynamics. J. Mol. Graph. 14, 27–28.
Hyams, C., Camberlein, E., Cohen, J. M., Bax, K., and Brown, J. S. (2010). The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect. Immun. 78, 704–715. doi: 10.1128/IAI.00881-09
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., and Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935. doi: 10.1063/1.445869
Kadioglu, A., Weiser, J. N., Paton, J. C., and Andrew, P. W. (2008). The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6:288. doi: 10.1038/nrmicro1871
Kim, E.-H., Choi, S.-Y., Kwon, M.-K., Tran, T. D.-H., Park, S.-S., Lee, K.-J., et al. (2012). Streptococcus pneumoniae pep27 mutant as a live vaccine for serotype-independent protection in mice. Vaccine 30, 2008–2019. doi: 10.1016/j.vaccine.2011.11.073
Kim, J. O., Romero-Steiner, S., Skov Sørensen, U. B., Blom, J., Carvalho, M., Barnard, S., et al. (1999). Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae. Infect. Immun 67, 2327–2333.
Lee, S., Ghosh, P., Kwon, H., Park, S.-S., Kim, G.-L., Choi, S.-Y., et al. (2018). Induction of the pneumococcal VNCRS operon by lactoferrin is essential for pneumonia. Virulence 9, 1562–1575. doi: 10.1080/21505594.2018.1526529
Leonard, P. G., Golemi-Kotra, D., and Stock, A. M. (2013). Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. Proc. Natl. Acad. Sci. U.S.A. 110, 8525–8530. doi: 10.1073/pnas.1302819110
Li, G. W., and Xie, X. S. (2011). Central dogma at the single-molecule level in living cells. Nature 475, 308–315. doi: 10.1038/nature10315
Li, Y. C., Chang, C. K., Chang, C. F., Cheng, Y. H., Fang, P. J., Yu, T., et al. (2014). Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element. Nucleic Acids Res. 42, 8777–8788. doi: 10.1093/nar/gku572
Lindorff-Larsen, K., Piana, S., Palmo, K., Maragakis, P., Klepeis, J. L., Dror, R. O., et al. (2010). Improved side-chain torsion potentials for the amber FF99SB protein force field. Proteins 78, 1950–1958. doi: 10.1002/prot.22711
Makino, K., Amemura, M., Kawamoto, T., Kimura, S., Shinagawa, H., Nakata, A., et al. (1996). DNA binding of PhoB and its interaction with RNA polymerase. J. Mol. Biol. 259, 15–26. doi: 10.1006/jmbi.1996.0298
McCluskey, J., Hinds, J., Husain, S., Witney, A., and Mitchell, T. (2004). A two−component system that controls the expression of pneumococcal surface antigen A (PsaA) and regulates virulence and resistance to oxidative stress in Streptococcus pneumoniae. Mol. Microbiol. 51, 1661–1675. doi: 10.1111/j.1365-2958.2003.03917.x
Moscoso, M., and García, E. (2009). Transcriptional regulation of the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae: a bioinformatic analysis. DNA Res. 16, 177–186. doi: 10.1093/dnares/dsp007
Narayanan, A., Kumar, S., Evrard, A. N., Paul, L. N., and Yernool, D. A. (2014). An asymmetric heterodomain interface stabilizes a response regulator-DNA complex. Nat. Commun. 5:3282. doi: 10.1038/ncomms4282
Nurse-Lucas, M., Mcgee, L., Hawkins, P. A., Swanston, W. H., and Akpaka, P. E. (2016). Serotypes and genotypes of Streptococcus pneumoniae isolates from trinidad and tobago. Int. J. Infect. Dis. 46, 100–106. doi: 10.1016/j.ijid.2016.04.005
Ogunniyi, A. D., Giammarinaro, P., and Paton, J. C. (2002). The genes encoding virulence-associated proteins and the capsule of Streptococcus pneumoniae are upregulated and differentially expressed in vivo. Microbiology 148, 2045–2053. doi: 10.1099/00221287-148-7-2045
Okajima, T., Doi, A., Okada, A., Gotoh, Y., Tanizawa, K., and Utsumi, R. (2008). Response regulator YycF essential for bacterial growth: X-ray crystal structure of the DNA-binding domain and its PhoB-like DNA recognition motif. FEBS Lett. 582, 3434–3438. doi: 10.1016/j.febslet.2008.09.007
Parrinello, M., and Rahman, A. (1981). Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190. doi: 10.1063/1.328693
Rigsby, R. E., and Parker, A. B. (2016). Using the PYMOL application to reinforce visual understanding of protein structure. Biochem. Mol. Biol. Educ. 44, 433–437. doi: 10.1002/bmb.20966
Shah, M., Anwar, M. A., Yesudhas, D., Krishnan, J., and Choi, S. (2016). A structural insight into the negative effects of opioids in analgesia by modulating the TLR4 signaling: an in silico approach. Sci. Rep. 6:39271. doi: 10.1038/srep39271
Sheffield, P., Garrard, S., and Derewenda, Z. (1999). Overcoming expression and purification problems of RHOGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15, 34–39. doi: 10.1006/prep.1998.1003
Sleep, D. (2015). Albumin and its application in drug delivery. Expert Opin. Drug Deliv. 12, 793–812. doi: 10.1517/17425247.2015.993313
Sung, H., Shin, H. B., Kim, M.-N., Lee, K., Kim, E.-C., Song, W., et al. (2006). Vancomycin-tolerant Streptococcus pneumoniae in Korea. J. Clin. Microbiol 44, 3524–3528. doi: 10.1128/jcm.00558-06
Sylvia, P., Brehm, S. O., and Hoch, H. J. A. (1975). DNA-binding proteins in human serum. Biochem. Biophys. Res. Commun. 63, 24–31. doi: 10.1016/s0006-291x(75)80005-2
Throup, J. P., Koretke, K. K., Bryant, A. P., Ingraham, K. A., Chalker, A. F., Ge, Y., et al. (2000). A genomic analysis of two−component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35, 566–576. doi: 10.1046/j.1365-2958.2000.01725.x
Tuomanen, E. I., Mitchell, T. J., Morrison, D. A., and Spratt, B. G. (2004). The Pneumococcus. Washington, D.C: ASM press.
Wu, K., Xu, H., Zheng, Y., Wang, L., Zhang, X., and Yin, Y. (2016). CpsR, a GntR family regulator, transcriptionally regulates capsular polysaccharide biosynthesis and governs bacterial virulence in Streptococcus pneumoniae. Sci. Rep. 6:29255. doi: 10.1038/srep29255
Xayarath, B., and Yother, J. (2007). Mutations blocking side chain assembly, polymerization, or transport of a Wzy-dependent Streptococcus pneumoniae capsule are lethal in the absence of suppressor mutations and can affect polymer transfer to the cell wall. J. Bacteriol. 189, 3369–3381. doi: 10.1128/jb.01938-06
Yother, J. (2011). Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Ann. Rev. Microbiol. 65, 563–581. doi: 10.1146/annurev.micro.62.081307.162944
Zheng, Y., Zhang, X., Wang, X., Wang, L., Zhang, J., and Yin, Y. (2017). ComE, an essential response regulator, negatively regulates the expression of the capsular polysaccharide locus and attenuates the bacterial virulence in Streptococcus pneumoniae. Front. Microbiol. 8:277. doi: 10.3389/fmicb.2017.00277
Keywords: VncR, capsular polysaccharide, strain-specific, lactoferrin, Streptococcus pneumoniae
Citation: Ghosh P, Shah M, Ravichandran S, Park S-S, Iqbal H, Choi S, Kim KK and Rhee DK (2019) Pneumococcal VncR Strain-Specifically Regulates Capsule Polysaccharide Synthesis. Front. Microbiol. 10:2279. doi: 10.3389/fmicb.2019.02279
Received: 27 May 2019; Accepted: 18 September 2019;
Published: 01 October 2019.
Edited by:
Mattias Collin, Lund University, SwedenReviewed by:
Johanna J. Kenyon, Queensland University of Technology, AustraliaGuillaume Goyette-Desjardins, University of Veterinary Medicine Hannover, Germany
Copyright © 2019 Ghosh, Shah, Ravichandran, Park, Iqbal, Choi, Kim and Rhee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong Kwon Rhee, ZGtyaGVlQHNra3UuZWR1
 Prachetash Ghosh
Prachetash Ghosh Masaud Shah
Masaud Shah Subramaniyam Ravichandran
Subramaniyam Ravichandran Sang-Sang Park1
Sang-Sang Park1 Sangdun Choi
Sangdun Choi Kyeong Kyu Kim
Kyeong Kyu Kim Dong Kwon Rhee
Dong Kwon Rhee