- Department of Plant Sciences, University of California, Davis, Davis, CA, United States
Genus Frankia is comprised primarily of nitrogen-fixing actinobacteria that form root nodule symbioses with a group of hosts known as the actinorhizal plants. These plants are evolutionarily closely related to the legumes that are nodulated by the rhizobia. Both host groups utilize homologs of nodulation genes for root-nodule symbiosis, derived from common plant ancestors. The corresponding endosymbionts, Frankia and the rhizobia, however, are distantly related groups of bacteria, leading to questions about their symbiotic mechanisms and evolutionary history. To date, a stable system of electrotransformation has been lacking in Frankia despite numerous attempts by research groups worldwide. We have identified type IV methyl-directed restriction systems, highly-expressed in a range of actinobacteria, as a likely barrier to Frankia transformation. Here we report the successful electrotransformation of the model strain F. alni ACN14a with an unmethylated, broad host-range replicating plasmid, expressing chloramphenicol-resistance for selection and GFP as a marker of gene expression. This system circumvented the type IV restriction barrier and allowed the stable maintenance of the plasmid. During nitrogen limitation, Frankia differentiates into two cell types: the vegetative hyphae and nitrogen-fixing vesicles. When the expression of egfp under the control of the nif gene cluster promoter was localized using fluorescence imaging, the expression of nitrogen fixation in nitrogen-limited culture was localized in Frankia vesicles but not in hyphae. The ability to separate gene expression patterns between Frankia hyphae and vesicles will enable deeper comparisons of molecular signaling and metabolic exchange between Frankia-actinorhizal and rhizobia-legume symbioses to be made, and may broaden potential applications in agriculture. Further downstream applications are possible, including gene knock-outs and complementation, to open up a range of experiments in Frankia and its symbioses. Additionally, in the transcriptome of F. alni ACN14a, type IV restriction enzymes were highly expressed in nitrogen-replete culture but their expression strongly decreased during symbiosis. The down-regulation of type IV restriction enzymes in symbiosis suggests that horizontal gene transfer may occur more frequently inside the nodule, with possible new implications for the evolution of Frankia.
Introduction
Bacteria in the genus Frankia form nitrogen-fixing root nodule symbioses with a group of host plants, the actinorhizal plants. The actinorhizal plants are evolutionarily closely related to the legumes within the Nitrogen-fixing Clade (NFC; Soltis et al., 1995). The bacterial symbionts are only distantly related to each, however, Frankia belongs to the phylum Actinobacteria, comprised of gram-positive bacteria with high-GC genomes. By contrast the rhizobia, symbionts of the legumes, are gram-negative proteobacteria with genomes generally closer to 50% GC. Additionally, Frankia is a multicellular, hyphal organism with a complex life cycle. In nitrogen limiting conditions, Frankia can produce stalked, ovoid to spherical structures that develop from branch hyphae called vesicles (Benson and Silvester, 1993). Frankia vesicles, known to be the sites of nitrogen fixation, are surrounded by a lamellar envelope comprised of hopanoid lipids (Berry et al., 1993) and have high rates of internal oxygen consumption through respiration. The vesicle envelope increases in number of layers in response to oxygen tension, thereby likely reducing the flow of oxygen into the vesicle interior to protect the nitrogenase complex from deactivation (Benson and Silvester, 1993). Thus, unlike the rhizobia-legume symbioses, where the host nodule tissue regulates the flow of oxygen during nitrogen fixation (Appleby, 1984), most Frankia are capable of fixing nitrogen in the vesicles in atmospheric oxygen conditions under nitrogen limitation (Benson and Silvester, 1993; Gtari et al., 2015). Finally, in symbiosis and in vitro, Frankia is capable of forming sporangia containing thick-walled spores (Benson and Silvester, 1993), which apparently serve as a resting stage and as a source of propagules that can enhance nodule numbers (Burleigh and Torrey, 1990).
Biological nitrogen fixation has potential applications in the development of minimal input strategies for more sustainable agriculture globally, and in nutrient-deficient soils (Gutierrez, 2012; Mus et al., 2016). Genetic improvement has the potential to introduce nitrogen-fixing symbioses into a wider range of crop plants once a broader understanding of the mechanisms involved in these symbioses is achieved (Mus et al., 2016). Actinorhizal and legume hosts share the Common Symbiotic Pathway, a signaling pathway at early stages of interaction that leads to the development of the nodule by the host after recognition of the symbiont (Oldroyd, 2013), in addition to several key gene orthologs involved in the process of nodule development (Battenberg et al., 2018; Griesmann et al., 2018). On the bacterial side the rhizobial signaling molecules that trigger nodulation in many of the legume hosts, called Nod factors, are synthesized by nod genes and have been extensively studied (Oldroyd, 2013). By contrast, the majority of Frankia genomes do not contain clear sets of nod gene homologs, with the exception of certain members of the cluster II Frankia whose genomes contain homologs of the rhizobial nodABC genes. These nod gene homologs are expressed in symbiosis (Persson et al., 2015). Other mechanisms have been found to trigger nodulation in some legumes by rhizobia, including an effector protein injected into the host by a type III secretion system (Okazaki et al., 2013), however genes responsible for these pathways lack homologs in Frankia genomes as well. Signals that elicit responses in host roots, including root-hair curling, an early step in some rhizobial and actinorhizal symbioses (Oldroyd, 2013), have been detected experimentally in some Frankia in clusters I and III. The structures of these molecules, however, have not been determined (Ceremonie et al., 1999; Cissoko et al., 2018).
The major advances in knowledge of rhizobial symbioses have been made possible by the development of genetic tools for dissecting the metabolic and regulatory pathways in the microsymbiont-host interactions. Long et al. (1982) originally demonstrated the role of the nod genes in symbiosis by complementing nodulation-deficient mutants of Sinorhizobium meliloti with nod genes encoded on a replicating, broad host-range plasmid. More recently, expression systems based on marker genes in rhizobia have enabled a wider-range of experiments including the tracking of microsymbionts during the host infection process (Gage, 2002; Fournier et al., 2015) and the identification of regulatory networks involved in symbiotic interactions (Spaepen et al., 2009). However, to date, there has been no stable electroporation-based genetic transformation system for Frankia (Kucho et al., 2009), and little understanding of what the barriers to stable transformation may be. Thus an electrotransformation system in Frankia, as reported here, has the potential to enable functional inquiries into the diverse mechanisms involved in establishing root-nodule symbiosis and maintaining biological nitrogen fixation in actinorhizal symbioses. These inquiries will, in turn, contribute to a wider understanding of the origins of root nodule symbioses and mechanisms of Frankia and rhizobial interaction with hosts in the NFC.
Frankia presents several barriers to transformation. Actinobacteria in general have low rates of homologous recombination due to competition between their homologous recombination pathway and a Non-Homologous End-Joining pathway (Zhang et al., 2012). Additionally, natural vectors for Frankia transformation are limited as no Frankia phages have been discovered (Simonet et al., 1990). Finally, its multicellular hyphae require experiments to be performed on hyphal segments rather than individual cells. Despite these barriers, it has been demonstrated that DNA can be electroporated into Frankia cells (Myers and Tisa, 2003), providing a potential avenue for genetic transformation. Furthermore, Kucho et al. (2009) reported initial success in integrating a non-replicating plasmid into the chromosome of Frankia sp. CcI3. However the recombined plasmid was lost in the following generation, limiting its further use in experiments.
Restriction enzymes pose a barrier to successful transformation in many bacteria due to their role in defense of digesting foreign DNA. In some actinobacteria, the use of unmethylated plasmids has increased transformation efficiency (Ankri et al., 1996; Molle et al., 1999), potentially due to the expression of type IV methyl-directed restriction enzymes. In the majority of bacterial taxa, DNA is methylated during replication by the methyltransferase Dam, to mark parent strands for DNA repair and excision of misincorporated bases from the daughter strand (Sanchez-Romero et al., 2015). The majority of actinobacteria, however, lack dam homologs (Sanchez-Romero et al., 2015) and an investigation of genomes of Streptomyces, Rhodococcus, and Micromonospora spp. found that these genomes were not methylated in the canonical Dam pattern (Novella et al., 1996). While actinobacteria have recently been shown to have a unique mismatch repair pathway that does not involve Dam (Castaneda-Garcia et al., 2017) it is not yet known how this pathway utilizes methylation, if at all.
In this study we report successful electrotransformation of Frankia alni ACN14a by using an unmethylated plasmid to circumvent the methylation-targeting restriction enzyme encoded in the ACN14a genome. This permitted the maintenance of plasmids derived from the very broad host-range replicating plasmid pIP501 (Kurenbach et al., 2003) in F. alni cultures for several months of continuous subculture, and implicates the type IV restriction enzyme as the major barrier to transformation. Use of a replicating plasmid also allowed the maintenance of the plasmid in Frankia without depending on its homologous recombination system. Bioinformatic analysis revealed a differential expression of type IV restriction genes between F. alni transcriptomes in symbiosis vs. culture, which has implications for acquisition of horizontally-transferred genes in endophytic environments by Frankia. Additionally, we show that egfp expressed from a replicating plasmid can be used to label gene expression differences between Frankia cell types (vesicles vs. hyphae) during nitrogen fixation, opening up molecular genetics-based experiments, including marker gene expression, on Frankia symbioses in the future.
Materials and Methods
Restriction Enzyme Analysis
Restriction enzyme genes from all completely sequenced actinobacterial genomes available in the REBASE database (accessed 6/19/17) were downloaded with their annotations (Roberts et al., 2010). Restriction enzymes were categorized by enzyme types: I, II, III, or IV. Bacterial transcriptomes used in this study were downloaded from the NCBI GEO database (listed in Supplementary Table S1). Transcriptomes analyzed were chosen based on the following criteria: (1) transcriptomes were made from actively growing pure cultures, (2) the organism whose transcriptome was sequenced did not have any genetic manipulations, e.g., mutations or exogenous plasmids, and (3) media used for sequencing did not have any additional experimental compounds added including antibiotics or complex carbon sources. For comparisons between transcriptomes, expression levels for each gene were calculated as the percent of genes with lower expression than the gene of interest in each transcriptome (i.e., percentile ranking).
Culture Conditions
E. coli strains DH5α and GM48 (dam- and dcm-) were grown in 50 ml Difco 1.5% (w/v) Luria Broth (LB) (Catalog #241420, Becton Dickinson, Franklin Lakes, NJ), pH 6.8, in 250 ml flasks at 37°C with shaking at 150 rpm overnight. Plates were made with 1.5% (w/v) Bacto Agar (Becton Dickinson, Franklin Lakes, NJ, United States, catalog #214010) in 1.5% LB, and incubated overnight at 37°C. All media were first sterilized by autoclaving for 30 min at 121°C.
Frankia alni ACN14a (Normand and Lalonde, 1982) was cultured in liquid BAPP media modified from Murry et al. (1984) by the addition of 5 mM pyruvate and 5 mM MOPS, adjusted to pH 6.7, in sterile plastic six-well plates (catalog #353046, Corning Inc., Corning, NY, United States) with 4 ml of media per well. We observed that F. alni ACN14a grew in the presence of tetracycline; thus tetracycline was added to a final concentration of 10 μg/ml to prevent contamination. (+)N media included 5 mM ammonium chloride as a nitrogen source whereas (–)N media had no added nitrogen source. For subculturing, hyphae were collected in sterile 2 ml microcentrifuge tubes with a 14G syringe needle, centrifuged at 10,000 rpm for 10 min in a tabletop microcentrifuge (model #5424, Eppendorf, Hamburg, Germany), and re-suspended in 1 ml fresh BAPP media. Cultures were then homogenized by passage through a 21G needle six times. Resulting homogenate equivalent to 50 μl packed-cell volume was added to 4 ml of fresh media in six-well plates and incubated at 28°C with shaking at 50 rpm. Cultures were routinely subcultured once per week. For selection of transformants, chloramphenicol was used at a final concentration of 25 μg/ml.
DNA Extraction
Plasmids were purified from E. coli using a QIAprep® Spin Miniprep Kit (Catalog #27106, Qiagen). Two milliliters of overnight culture were pelleted at 13,000 rpm for 5 min and used for extraction. Plasmids were eluted in EB buffer (Qiagen) and quantified on a NanoDrop Microvolume Spectrophotometer (ThermoFisher). Plasmids were further purified by running on a 0.7% agarose gel, measured against Quick-Load® Purple 1 kb Plus DNA Ladder (catalog #N0550S, New England Biolabs), and extracted from the gel with a Zymoclean Gel DNA Recovery Kit (Catalog #11-300, Genesee Scientific, San Diego, CA, United States). To synthesize unmethylated plasmids, methylated plasmids were first extracted from E. coli DH5α and transformed into E. coli GM48 by heat shock in CaCl2 (Sambrook et al., 1989). Cells were spread on LB plates with chloramphenicol and incubated overnight, then cultured in liquid LB media with chloramphenicol and re-extracted as above.
F. alni DNA extraction was carried out with a CTAB protocol (Feil et al., 2012). Briefly, cells were pelleted at 10,000 rpm for 5 min in a tabletop centrifuge. Cells were then re-suspended in TE buffer and lysed with lysozyme, SDS, Proteinase K, and CTAB. DNA was extracted with 24:1 chloroform:isoamyl alcohol, and 25:24:1 phenol:chloroform:isoamyl alcohol. Extracts were then precipitated in isopropanol followed by washing in ethanol, resuspended in 170 μl DNase-free water, treated with 2 μl 100 mg/ml RNase A for 1 h (catalog #10109169001, Roche Diagnostics, Mannheim, Germany), air-dried overnight at 37°C, resuspended in 50 μl TE buffer, and quantified by NanoDrop.
Plasmid Synthesis
A plasmid designated as pIGSAF (Supplementary Figure S1A) was designed for constitutive expression of egfp and synthesized by ligating a PCR-amplified fragment containing the egfp gene and promoter from plasmid pDiGc (Helaine et al., 2010, Addgene, Cambridge, MA, United States) to plasmid pSA3 (Dao and Ferretti, 1985). Plasmid pSA3 is a broad host-range replicating plasmid originally developed as a shuttle vector by ligating pIP501-derivative pGB305 to the E. coli-specific replicating plasmid pACYC184 (Dao and Ferretti, 1985). The pIP501 origin of replication has been shown to replicate in bacteria of diverse phyla including Firmicutes, from which it was originally isolated (Horodniceanu et al., 1976), as well as Actinobacteria and Proteobacteria (Kurenbach et al., 2003). Plasmid pSA3 contains chloramphenicol and tetracycline resistance genes and an E. coli-specific p15A origin of replication for higher-copy number propagation (Dao and Ferretti, 1985).
To synthesize PCR products for ligation, primers were designed using NCBI Primer-BLAST with default settings (Ye et al., 2012). All primers used in this study are listed in Table 1. For cloning, primers were designed with linkers adding target restriction sites onto their 5′ ends (Table 1) as well as 4–6 additional bases to aid in restriction digestion of the ends. For the synthesis of pIGSAF, primers were designed targeting the egfp coding region as well as the promoter region 200 base pairs upstream, amplifying a fragment 1238 base pairs in length. PCR was performed in a Bio-Rad S1000 Thermal Cycler (Bio-Rad, Hercules, CA, United States) using a Qiagen Taq PCR kit (catalog #201223). Products were synthesized by first performing 10 cycles of amplification using the lower annealing temperature corresponding to the binding site on the target DNA and then an additional 30 cycles using the annealing temperature of the full primer including the linker (Table 1).
The egfp fragment was synthesized with SalI and BamHI restriction sites on the 5′ and 3′ ends, respectively, with primers EGFP_SalI_F and EGFP_BamHI_R. The PCR product and plasmid pSA3 were then digested with both enzymes. Digestion with SalI and BamHI removed a fragment approximately 200 base pairs in size from plasmid pSA3. The remaining 10 kb fragment was purified on a 0.7% agarose gel and extracted. This fragment was then mixed together with the egfp PCR product, denatured by heating for 5 min at 65°C, cooled on ice to allow binding of base-paired overhangs, and then the fragments were ligated together by incubation with T4 ligase (catalog #M0202S, Qiagen) at 16°C overnight. The resulting ligation was transformed into E. coli DH5α by heat shock (Sambrook et al., 1989). Transformants were selected on LB plates with chloramphenicol at a final concentration of 25 μg/ml. Transformed colonies were inoculated into liquid LB media with chloramphenicol and cultured as described above. Plasmid pIGSAF was then re-extracted from transformed E. coli and its composition was confirmed by digestion with SalI and BamHI. The total length of the ligated plasmid was confirmed to be approximately 12 kb by gel electrophoresis.
To study the differential expression of egfp in response to nitrogen limitation, a second plasmid, pIGSAFnif, was synthesized that permitted the egfp coding-region to be expressed under the control of the nif cluster promoter of F. alni ACN14a (Supplementary Figure S1A). The nif cluster promoter is upstream of the nif genes coding for the subunits of nitrogenase. Nitrogenase activity has been demonstrated in vesicle fractions of Frankia cultures but not in the hyphal fraction (Noridge and Benson, 1986; Tisa and Ensign, 1987), suggesting that egfp expression (GFP fluorescence) under control of the nif cluster promoter will be up-regulated in vesicles relative to hyphae. The egfp coding region of pDiGc was amplified without the upstream promoter region using primers GFP_CDS_EcoRI_F and GFP_CDS_SalI_R (Table 1). These primers added an EcoRI restriction site before the start codon and a SalI site 200 bases downstream of the stop codon. Separately, the 344 bases upstream of the nif nitrogenase cluster in the F. alni ACN14a genome were amplified with an XbaI site upstream and an EcoRI site downstream using primers nif_promoter_XbaI_F and nif_promoter_EcoRI_R (Table 1). These two PCR products were digested with EcoRI (catalog #R0101S, New England Biolabs) and ligated together as above to produce a fragment 1134 bp in length. The ligation product was then re-amplified by PCR using the nif promoter forward primer and the egfp reverse primer. The amplified ligation product and plasmid pSA3 were then each digested with SalI and XbaI, ligated together, transformed, and selected on chloramphenicol as above. The final size of plasmid pIGSAFnif was approximately 10.8 kb (Supplementary Figure S1B).
Frankia Transformation
F. alni cells were grown in culture for 1 week prior to transformation. Hyphae equivalent to approximately 50 μl packed-cell volume were collected then pelleted as above and re-suspended in 500 μl of ice-cold sterile deionized (DI) water. This wash step was repeated two more times, but after the third round of centrifugation the pellet was re-suspended in 300 μl ice-cold sterile 10% glycerol instead. Hyphae in the cell suspension were then homogenized by passage through a 21G needle twice.
Three hundred microliter of the F. alni cell suspension was pipetted into an electroporation cuvette with a 2 mm gap (Molecular BioProducts Catalog #5520, San Diego, CA, United States) and mixed with 10 μg of plasmid DNA. Cells were electroporated with unmethylated plasmid prepared as described above, or with methylated plasmid extracted from E. coli DH5α as a control. The cuvette was then incubated on ice for 5 min. Electroporation was carried out in a Bio-Rad Gene PulserTM with Pulse Controller at 2.5 kV, 200 Ω resistance, and 25 μF of capacitance. The cuvette was then immediately filled with 1 ml of ice-cold BAPP media. The cuvette was sealed with Parafilm® (Pechiney Plastic Packaging, Menasha, WI, United States) and incubated overnight without shaking at 28°C.
The following day the F. alni culture was removed from the cuvette and added to 3.5 ml of sterile BAPP media with tetracycline to prevent contamination, but without chloramphenicol, as above, in a glass 25 ml test tube. The culture was incubated at 28°C without shaking until visible hyphae were observed (approximately 10 days after electroporation). At this point F. alni hyphae were homogenized and sub-cultured into fresh media as above. Subculturing was repeated once more 1 week later. The following week (2 weeks after visible hyphae were observed) the hyphae were sub-cultured again into BAPP media, this time with chloramphenicol added for selection to a final concentration of 25 μg/ml. Chloramphenicol was chosen as the selective antibiotic because all Frankia strains tested by Tisa et al. (1998) were susceptible to it. This process was repeated the following week for an additional round of selection. F. alni cells transformed with the methylated plasmid were similarly cultured.
The presence of plasmid pIGSAF electroporated into F. alni cultures was then confirmed by two methods: (1) visualizing total DNA extracts from wild-type and transformed ACN14a on gels and (2) with PCR. For DNA extract visualization, 1 μg each of DNA extract from transformed and untransformed F. alni cultures was loaded into a well on a 0.7% agarose gel. Purified plasmid pIGSAF, extracted with a Qiaprep kit, was loaded into a separate well for comparison. Electrophoresis was run at 80 V and then the gel was visualized with a UVP High Performance Ultraviolet Transilluminator (Analytik Jena, Jena, Germany). Genomic DNA was also amplified with primer pairs pSA3_Cm_F/pSA3_CmR (Table 1) for the chloramphenicol resistance gene of plasmid pSA3 and gfp_qPCR_F/gfp_qPCR_R (Table 1) for the egfp gene of plasmid pIGSAF. Primers nifH_qPCR_F/nifH_qPCR_R (Table 1) for the nifH nitrogenase gene from the F. alni genome were used as a positive control for the quality of DNA extracts. Additionally, purified plasmid pDiGc from E. coli DH5α was used as a DNA template for egfp amplification as a positive control. PCR products were separated on an agarose gel and extracted as in Plasmid Synthesis, above, and sequenced by the UCDNA Sequencing Facility (University of California, Davis, CA, United States). Forward and reverse sequences were combined to make full-length amplicon sequences. These were aligned with MUSCLE (Edgar, 2004) with published reference sequences for the egfp and camR genes. Additionally, the sequences were compared by BLAST against gene sequences obtained from the original plasmids (Supplementary Figure S2).
Confocal Microscopy
Confocal microscopy was used to visualize expression of egfp in F. alni hyphae and vesicles. egfp was expressed both constitutively, from plasmid pIGSAF, and differentially between (+)N (nitrogen replete) and (-)N (nitrogen limited) media under control of the F. alni nif promoter using plasmid pIGSAFnif. Cultures were imaged after on1 week of incubation from the previous transfer, 25 weeks after transformation, as described in Plasmid Maintenance on Selective Media, below. For imaging, F. alni cultures were grown either in (+)N or (–)N BAPP medium as in Culture Conditions, above, and immobilized on glass slides with a drop of 3% molten agarose solution (catalog #A9539, Sigma, St. Louis, MO, United States) maintained in a water bath at 50°C. Slides were pre-heated on a slide warmer (Fisher) at 50°C. Fifteen microliter of Frankia hyphal suspension was then pipetted onto each slide and covered with 35 μl of 3% molten agarose. A #1.5 coverslip was added to the Frankia cells in agarose, and then the slide was allowed to cool to room temperature. The Frankia preparations were visualized on a Leica TCS SP8 STED 3X confocal microscope with either a 20X objective, or a 100X oil-immersion objective, using either brightfield or fluorescence with a HyD detector, in the Advanced Imaging Facility, University of California, Davis. For fluorescence imaging, samples were excited with 488 nm light. The emission wavelengths were collected from 500 to 550 nm. Images were stored as .lif files from Leica LAS X and then viewed in FIJI (Schindelin et al., 2012). For imaging of individual hyphae and vesicles, the 100X objective lens was used and images were taken in Z-stacks with a step size of 0.1 μm. To visualize F. alni colonies at low magnification, Z-stack images were taken with the 20X objective in five steps of 2 μm each and then combined using the highest fluorescent intensity of each pixel (FIJI MAX setting).
qPCR Verification of Differential Gene Expression
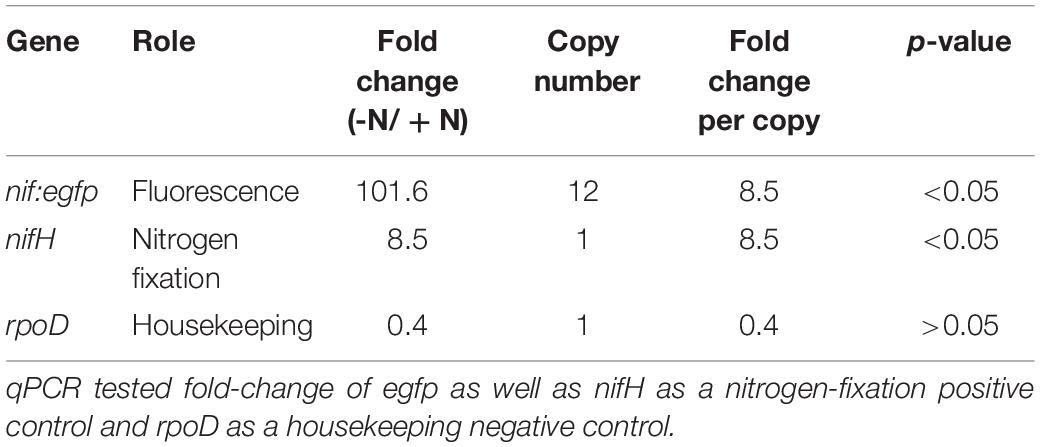
Quantification of gene expression and fold-changes were obtained based on qPCR amplification as an additional line of evidence demonstrating the plasmid transformation and the differential expression of genes (hyphae vs. vesicles) regulated by the nif promoter under nitrogen-fixing conditions. Cultures of transformed cells were grown in 4.0 ml (+)N and (–)N media in sterile six-well plates with shaking at 50 rpm at 28°C. After 5 days, RNA was extracted by bead-beating, following a protocol adapted from Dietrich et al. (2000): F. alni hyphae were pelleted at 9000 rpm for 15 min, resuspended in 1050 μl Buffer RLT (Qiagen), and transferred to 2 ml tubes containing Lysing Matrix B (catalog #6911-100, MP Biomedicals, Burlingame, CA, United States). Samples were processed with a FastPrep FP120 (Thermo Fisher Scientific, Waltham, MA, United States) for 45 s at setting 6.5, then placed on ice for 45 s. The processing step was then repeated twice more with cooling on ice between each step. Supernatants were then transferred to Qiagen RNeasy spin columns and purified with a Qiagen RNeasy Mini Kit (catalog #74104). RNA was eluted in RNase-free water and then contaminating DNA was digested with an Invitrogen TURBO DNA-free Kit (catalog #AM1907, Waltham, MA, United States). Finally, cDNA was synthesized with an Invitrogen Superscript III Kit (catalog #18080051) with random hexamer primers. qPCR was performed on a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) with Fast SYBR Green qPCR Master Mix (catalog #4385612, Applied Biosystems) and primer pairs rpoD_qPCR_F/rpoD_qPCR_R, nifH_qpCR_F/nifH_qpCR_R, infC_qPCR_F/infC_qPCR_R, and gfp_qPCR_F/gfp_qPCR_R (Table 1). One microliter of cDNA was used in each reaction. The egfp gene from plasmid pIGSAF and nifH (FRAAL6813) gene from the F. alni genome were used as experimental targets to confirm differential regulation of egfp under nitrogen limitation. Housekeeping gene infC (FRAAL5216) was used for normalization as in Alloisio et al. (2010) and sigma factor gene rpoD (FRAAL2026) was used as a negative control. To quantify fold-change the ΔΔCt values for each sample were calculated then converted into fold-changes equal to 2–ΔΔCt (Livak and Schmittgen, 2001).
Plasmid Copy Number
The copy number of plasmid pIGSAF in F. alni cultures under antibiotic selection was determined with both the absolute and relative quantification methods of Lee et al. (2006). For absolute quantification standards were created by serial 10-fold dilution, repeated five times, of the plasmid pDiGc extracted from E. coli DH5α and F. alni genomic DNA from an untransformed culture. These standards were quantified with qPCR using primer pair gfp_qPCR_F/gfp_qPCR_R and nifH_qPCR_F/nifH_qPCR_R respectively (Table 1). One microliter of DNA extract was used for each reaction. Standard curves were created by linear regression fit to a plot of log copy number vs. qPCR cycles (Ct) of the standards, resulting in strong correlations (R2 = 0.99) for both curves. The log number of copies of the egfp gene of plasmid pIGSAF per sample was calculated with the Ct value of amplification by primer pair gfp_qPCR_F/gfp_qPCR_R (Table 1) and the regression equation of the standard curve and then back-transformed. The number of copies of the infC and nifH genes of the F. alni genome (FRAAL5216) were calculated the same way with primer pairs infC_qPCR_F/infC_qPCR_R and nifH_qPCR_F/nifH_qPCR_R (Table 1). The resulting copy numbers of the plasmid-bound egfp gene and genome-bound infC (FRAAL5216) and nifH (FRAAL6813) genes per sample were then used to calculate the ratio of plasmid to genome. qPCR reactions were performed in duplicate on DNA extracts from each of three separate cultures and then the copy numbers were averaged with a geometric mean.
For relative quantification, the same DNA extracts as above were used and the F. alni nifH (FRAAL6813) gene was amplified with primer pair nifH_qpCR_F/nifH_qpCR_R (Table 1) as well. The ΔΔCt method of Lee et al. (2006) was used to estimate the difference in fold-change between egfp (on the plasmid) and nifH (on the genome, FRAAL6813) between the samples, using infC (FRAAL5216) quantification as the endogenous control for normalization. The ΔΔCt values for each sample were calculated as in qPCR Verification of Differential Gene Expression, above, and then averaged with a geometric mean.
Plasmid Maintenance on Selective Media
Three separate transformation experiments were carried out, from August 2017 through March 2019, as outlined in Table 2. Transformed cultures were maintained with weekly subculturing into fresh selective media and used in experiments over the course of several months following each transformation. Initial transformations of F. alni with plasmid pIGSAF were performed in August of 2017; the cultures were maintained by subculturing and were visualized by confocal microscopy from December 2017 (17 weeks after transformation) through March 2018 (32 weeks). Transformation of F. alni with plasmid pIGSAFnif was performed in January 2018, selected as above, and imaged by confocal microscopy in April 2018 (15 weeks). These cultures were maintained by subculturing in selective media for RNA extraction for qPCR through July 2018 (27 weeks). A third round of transformants was created for use in the Plasmid Persistence assay detailed below. These cultures were transformed in September 2018 and subcultured weekly in selective media through March 2019. The presence of the plasmid was confirmed by DNA extraction and PCR amplification of the egfp gene in March 2019 (25 weeks) as described in Frankia Transformation, above.
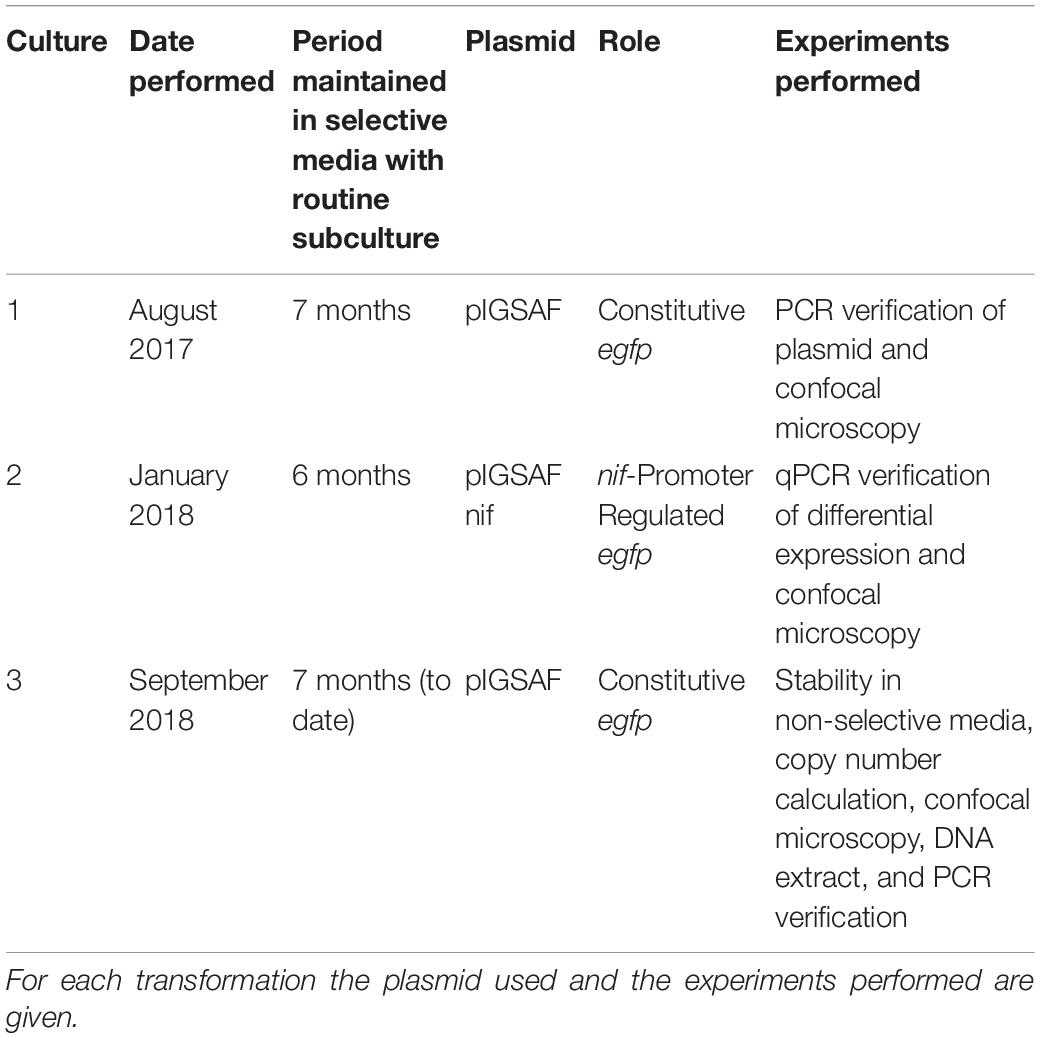
Table 2. Separate transformations performed on Frankia alni ACN14a and cultures maintained on selective media.
Plasmid Persistence in Non-selective Media
To test the persistence of plasmid pIGSAF in transformed F. alni without selection, a time-course of growth in media without antibiotics was performed. Fresh cultures were transformed with plasmid pIGSAF and initially selected as described in Frankia Transformation, above. After this 4-week selection process, cultures were then grown in non-selective media: cells were first pelleted and re-suspended in fresh BAPP media, then subcultured without the addition of chloramphenicol into six-well plates and incubated for 1 week. A separate set of cultures from the same stock was maintained in selective media and subcultured at the same time points as a control. Each week both sets of cultures were pelleted and re-suspended in 500 μl fresh BAPP media. The suspensions were homogenized by passage through a 21G needle twice. 250 μl of each homogenate was transferred to 4 ml fresh BAPP media without chloramphenicol in each well and the remaining 250 μl was used for total genomic DNA extraction as described in DNA Extraction above. This process was repeated once per week for 4 weeks. At each sampling point, the relative amount of plasmid in each sample was quantified by qPCR, performed in duplicate for each of three biological replicates, using egfp primers GFP_qPCR_F and GFP_qPCR_R (Table 1). Fold-change of plasmid between each time point was calculated using the ΔΔCt method (Lee et al., 2006). The infC gene (FRAAL5216), amplified from the same DNA extracts with primers infC_qPCR_F and infC_qPCR_R (Table 1), was used as a control to normalize the amount of DNA in each sample. To determine significant changes in plasmid abundance, two-tailed Welch’s t-tests were performed in R on normalized ΔCt values (p < 0.05). Cultures grown with and without selection for 4 weeks were also imaged with fluorescence according to methods in Confocal Microscopy, above.
Results
Identification of Restriction Enzymes in Frankia and Transcriptome Analysis
Types I, II, and IV restriction enzymes were identified in several Frankia genomes (Figure 1). Examining the transcriptome of Frankia alni ACN14a grown in (+)N culture (Alloisio et al., 2010), we found that three restriction enzyme genes were highly expressed: one type I enzyme (FRAAL4992, 92nd percentile), one type II enzyme (FRAAL0249, 91st percentile) and one type IV enzyme (FRAAL3325, 85th percentile) (Figure 1). The type IV enzyme was annotated in REBASE as a “Type IV Methyl-directed restriction enzyme” of the Mrr methyladenine-targeting family. Other Frankia genomes contained significant homologs of this type IV enzyme as well. The genome of Frankia casuarinae CcI3 contained five type IV enzymes, the most of any Frankia genome examined (Figure 1). In (+)N and (-)N-grown transcriptomes of Frankia sp. CcI3, type IV Mrr enzymes were very highly expressed, up to the 95th percentile (Figure 1). One of the five type IV enzymes in CcI3 (Francci3_2839) was annotated as a Mcr type IV enzyme, which targets 5-methylcytosine instead of methyladenine (REBASE) and its expression was very low, in only the 18th percentile of genes in the transcriptome. Of the five complete genomes examined, only the cluster II Frankia Candidatus Frankia datiscae Dg1 did not contain any putative type IV restriction genes.
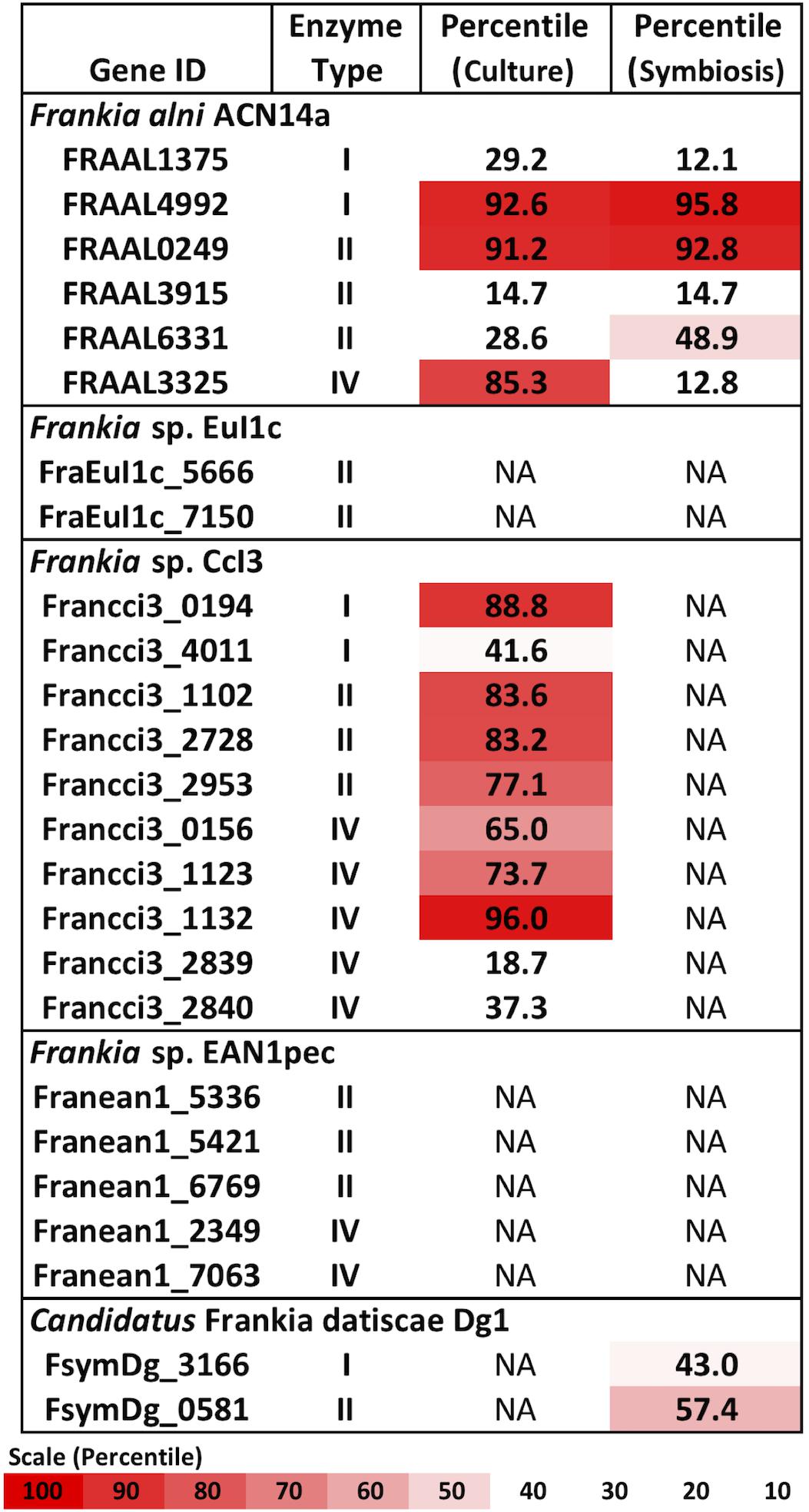
Figure 1. Restriction enzymes annotated in the Frankia genomes and expression levels of each gene in the transcriptome in culture when available (Alloisio et al., 2010; Bickhart and Benson, 2011). NA: Transcriptome not available.
A symbiotic transcriptome of F. alni ACN14a from root nodules is available in addition to transcriptomes of free-living (nitrogen-replete) culture (Alloisio et al., 2010). In symbiosis with Alnus glutinosa the mrr homolog was down-regulated approximately 7.8-fold (p < 0.007), from the 85th percentile in culture to an expression level no higher than 12% of transcriptome genes, while the other restriction enzymes identified in the F. alni genome (type I and type II) were not down-regulated in symbiosis (Figure 1). The other available Frankia symbiotic transcriptome, Frankia sp. Dg1 (Persson et al., 2015), showed very low expression of its two restriction enzymes (type I and type II, Figure 1).
Of the complete actinobacterial genomes outside of genus Frankia with published transcriptomes that were analyzed, Mycobacterium smegmatis, Streptomyces avermitilis, and Rhodococcus jostii also highly expressed type IV restriction enzyme genes in culture, in the upper-70th percentiles of their respective transcriptomes (Figure 2). In comparison, the transcriptomes of proteobacteria and firmicutes examined expressed their corresponding mrr genes around the 50th or 60th percentiles. One notable exception was the actinobacterium Mycobacterium tuberculosis, which expressed its single gene for a methyladenine-directed restriction enzyme (Mrr) at extremely low levels, around the 6th percentile in culture.
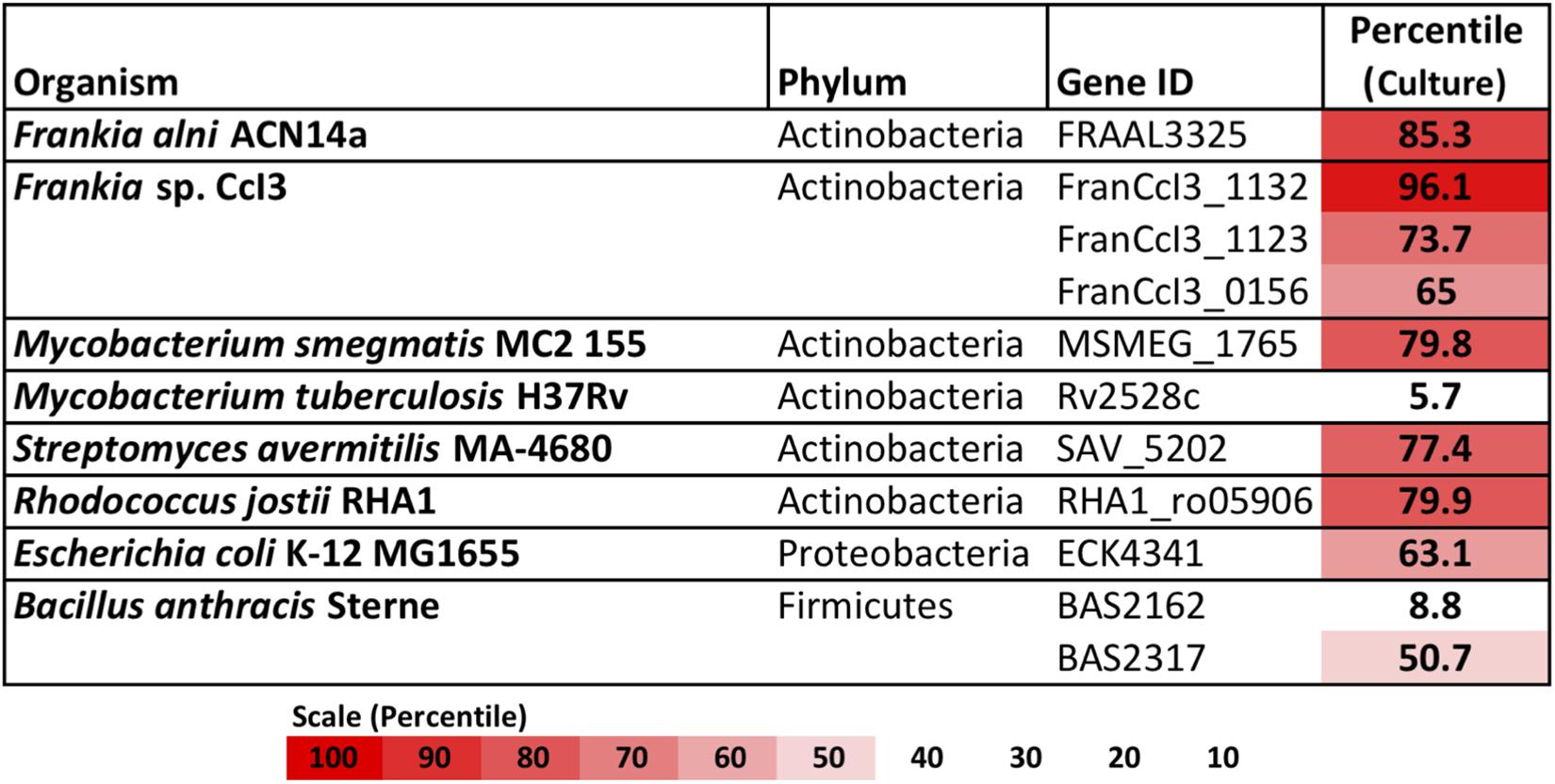
Figure 2. Restriction enzymes annotated in the genomes of Frankia, with other actinobacteria, proteobacteria, and firmicutes for comparison. For each, expression levels for available transcriptomes growing in pure culture are provided.
Genetic Transformation of Frankia alni ACN14a With an Unmethylated Replicating Plasmid
After electroporation F. alni cells formed visible hyphae in culture after about 10 days. When subcultured into chloramphenicol-selective media, F. alni cultures transformed with unmethylated plasmid pIGSAF were able to grow, whereas untransformed cultures and those transformed with methylated plasmid were not. When analyzed on the gel, purified plasmid pIGSAF from E. coli formed bands representing linear (approximately 12 kb), circular (10 kb), and supercoiled (6 kb) plasmid (Figure 3A). Presence of plasmid pIGSAF in transformed cultures was verified by a band present in DNA extracts corresponding to the linear form of plasmid pIGSAF (calculated size 11.8 kb) that was not present in extracts from wild-type F. alni (Figure 3A). PCR amplification and sequencing of the egfp gene from DNA extracts of transformed F. alni cultures confirmed the identity of the plasmid (Figure 3B and Supplementary Figure S2). Absolute copy number quantification with qPCR estimated the plasmid was present at 12.5 copies of plasmid per molecule of genomic DNA; relative quantification gave a similar estimate of 11.8 copies per genome (Supplementary Table S2). Each experiment was performed on cultures maintained with routine weekly subculture for 15–32 weeks post-transformation, as described in the Methods.
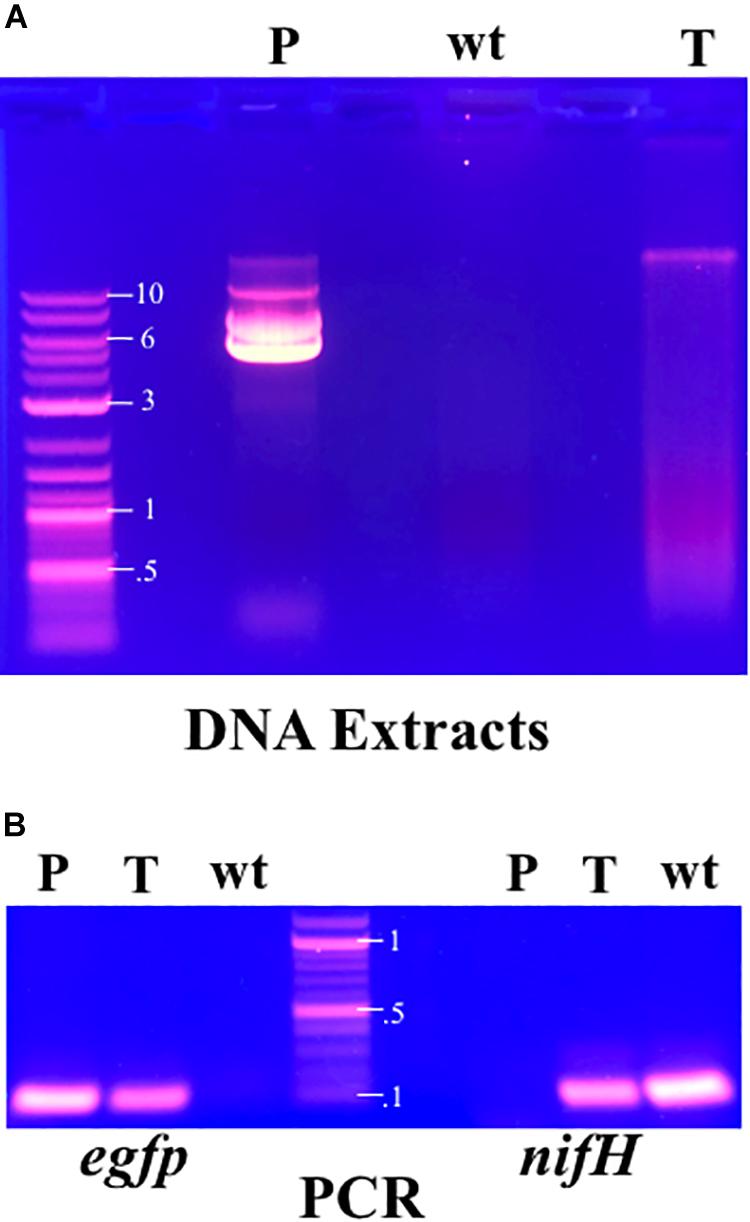
Figure 3. (A) Total DNA extracts from cultures of transformed and untransformed F. alni maintained in culture with routine subculture for 25 weeks from transformation, analyzed by gel electrophoresis. The extract from transformed F. alni (T, right) shows an additional band not present in untransformed, wild-type, cells (wt, middle) equivalent to linearized plasmid pIGSAF from E. coli (P, left). (B) PCR amplification of egfp (left, 120 bp) and nifH (right, 131 bp) genes. Amplification was performed on purified plasmid pDiGc (P, left) and genomic DNA extracts from F. alni transformed with plasmid pIGSAF (T) and wild-type F. alni (wt), respectively. Labels on DNA ladder denote sizes in kilobases (kb). The plasmid carrying egfp is not present in the wt F. alni DNA and the nifH gene is not present in purified plasmid pDiGc.
When imaged under 488 nm wavelength of excitation in the confocal microscope, green fluorescence typical of GFP was observed in hyphae as shown in Figure 4. Wild-type F. alni hyphae displayed autofluorescence around 575 nm as observed by Hahn et al. (1993), but no autofluorescence was observed in the 500–550 nm range (Figure 4).
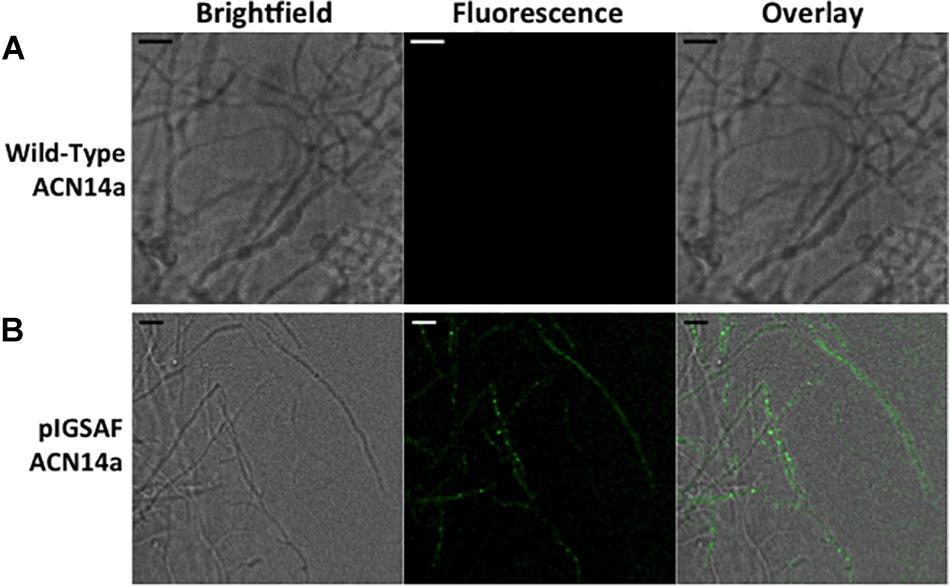
Figure 4. Transformation of Frankia alni ACN14a with a plasmid expressing egfp. (A) Wild-type F. alni ACN14a control. (B) Transformed F. alni ACN14a with plasmid pIGSAF. Images were obtained on a confocal microscope with a 100X objective with both brightfield and fluorescence and then overlaid. Size bars equal 5 μm.
Differential Regulation of egfp Under the Control of the Frankia alni ACN14a nif Cluster Promoter
When grown in (–)N culture, transformants carrying the pIGSAFnif plasmid showed significant up-regulation of the egfp gene conjugated to the nif cluster promoter, approximately 100-fold relative to (+)N culture, or approximately 8.5-fold per copy of plasmid (Table 3). The F. alni nifH gene (FRAAL6813), used as a positive control for nitrogen fixation, was similarly significantly up-regulated approximately 8.5-fold in (–)N media compared with (+)N cultures. Expression of the F. alni rpoD housekeeping gene (FRAAL2026), used as a negative control, was not significantly different between (+)N and (–)N cultures (Table 3).
F. alni containing pIGSAFnif grown in (–)N media fluoresced predominantly in the vesicles, observed at 100X magnification (Figure 5 and Supplementary Figure S3). Little to no fluorescence was observed in hyphae. Fluorescence in the vesicles was present both in the spherical portion as well as in the stalk connecting the vesicle to the hyphae. No fluorescence was observed in hyphae or vesicles of wild-type F. alni grown in (–)N media (Figure 5A). Due to the step size of 0.1 μm bright fluorescence was only observed when vesicles were in the plane of focus (Supplementary Figure S3). Few vesicles were present in images likely since the cells were observed after 1 week of culture in (-)N medium.
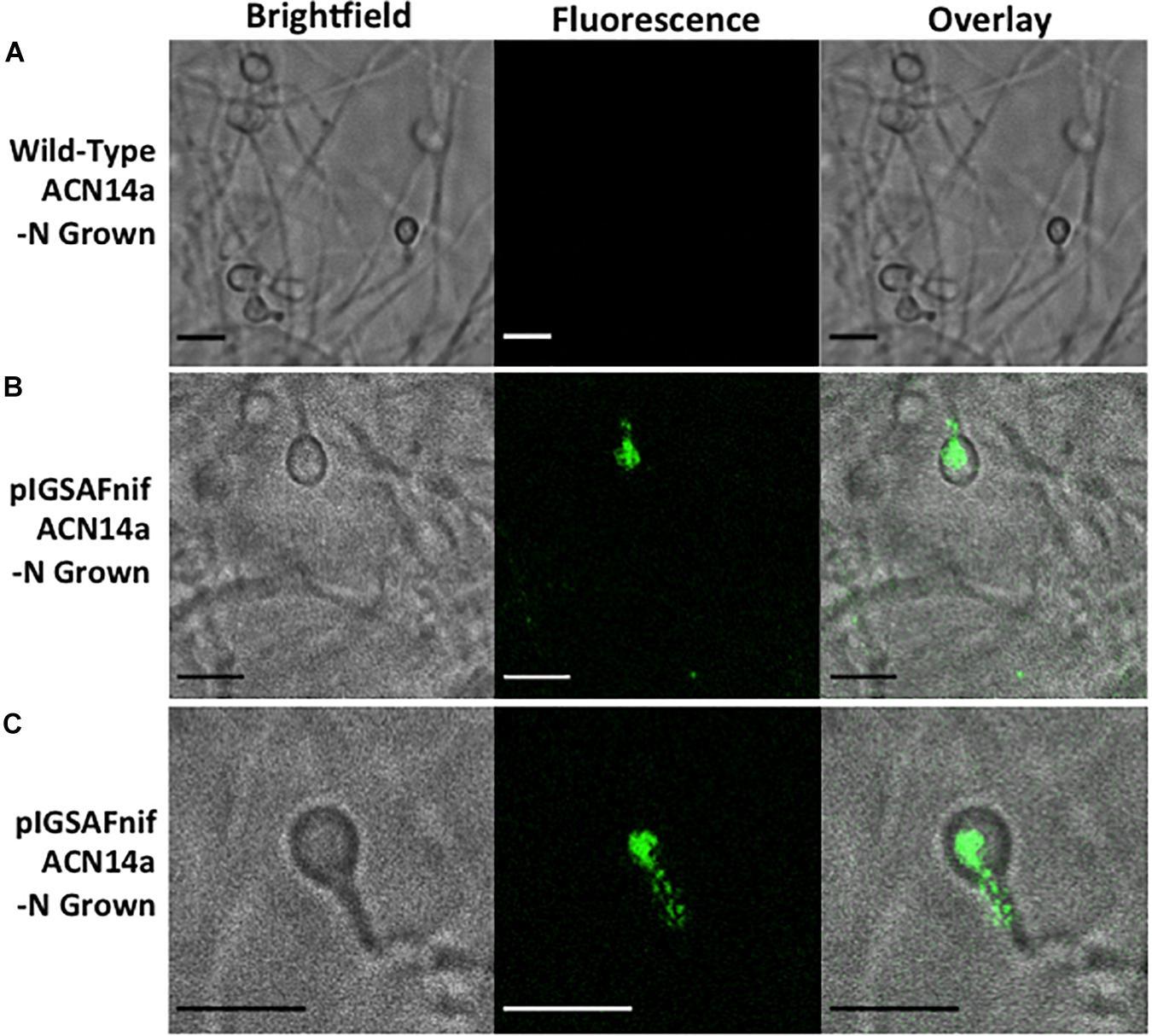
Figure 5. Fluorescence detection of transformation of Frankia alni ACN14a with a plasmid expressing egfp under the control of the nif cluster promoter region from the F. alni genome. Transformed cultures were grown in both (+)N and (–)N media to compare fluorescence with and without nitrogen fixation. (A) F. alni ACN14a transformed with plasmid pIGSAFnif grown in (+)N media. (B) F. alni ACN14a with plasmid pIGSAFnif grown in (–)N media. (C) Image of a vesicle showing fluorescence in the stalk. Images were obtained on a confocal microscope at 100X magnification with brightfield illumination and fluorescence imaging and then overlaid. Size bars equal 5 μm.
Stability of Plasmid pIGSAF in F. alni ACN14a in the Absence of Selection
In cultures grown without chloramphenicol selection, statistical analysis of the qPCR data for Frankia cells transformed with pIGSAF did not show a significant difference in the amount of plasmid relative to genomic DNA after one, two, and three rounds of sub-culturing (Figure 6). Only after the fourth round of sub-culturing was a significant decrease from the initial plasmid concentration detected by qPCR (Figure 6), however, even after 4 weeks without selection, fluorescence throughout the colony was still readily observed (Figure 7B).
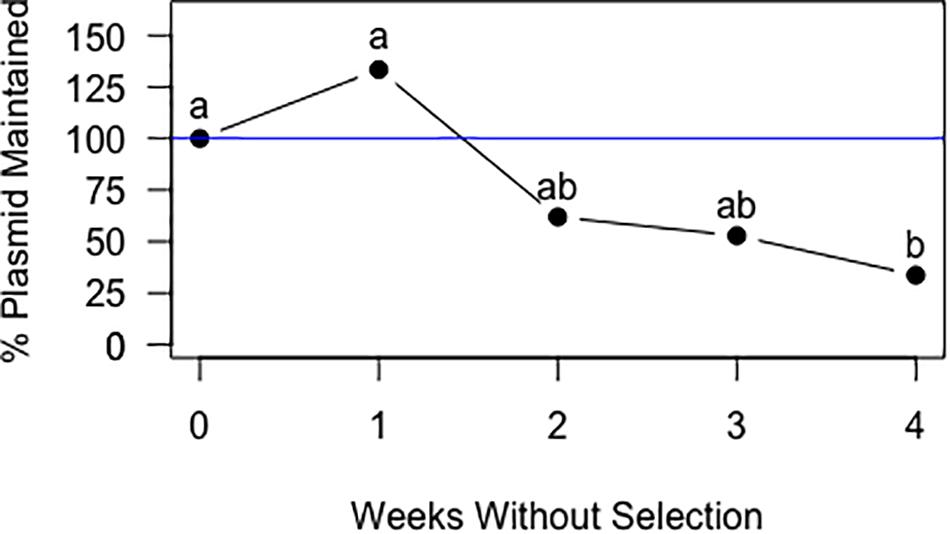
Figure 6. Maintenance of plasmid pIGSAF in Frankia alni in culture without chloramphenicol selection over the course of 4 weeks. Each week the culture was sub-cultured into fresh media and genomic DNA was extracted to measure relative plasmid concentrations via qPCR. Labels “a” and “b” indicate time points that are significantly different from each other (p < 0.05).
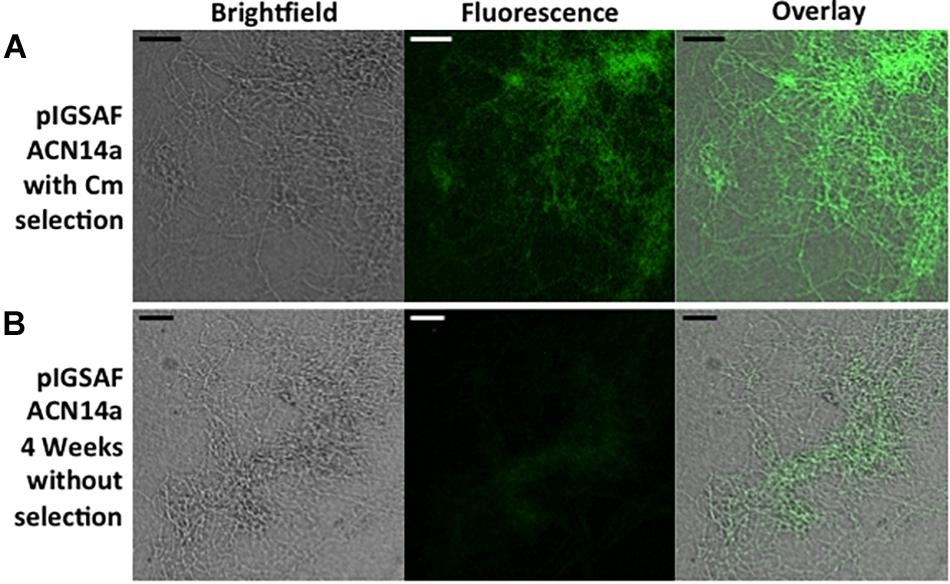
Figure 7. Fluorescence of a Frankia alni hyphal colony transformed with plasmid pIGSAF grown (A) with chloramphicol selection for 10 weeks (B) and without selection for 4 weeks. Colonies were viewed at 20X magnification. Size bars equal 25 μm.
Discussion
Unmethylated DNA Circumvents the Restriction Barrier to Genetic Transformation of Frankia alni ACN14a With a Replicating Plasmid
We have shown that F. alni can be stably transformed with an unmethylated replicating plasmid introduced by electroporation. The methods presented here circumvent two major transformation barriers in Frankia. First, the lack of methylation avoids restriction of the plasmid by type IV methyl-directed restriction enzymes. Second, the use of a plasmid replicated and maintained outside the genome does not rely on the reduced homologous recombination rate in actinobacteria (Zhang et al., 2012). Plasmids pIGSAF and pIGSAFnif (Supplementary Figure S1) were stably maintained in F. alni culture and used to perform routine experiments. Three independent transformations were performed over the course of 2 years and the resulting transformants were maintained on selective media by repeated subculture for at least 7 months (Table 2). The presence and stability of the plasmids were confirmed with gels of whole genome DNA extracts (Figure 3A), PCR and qPCR amplification of plasmid-bound genes (Figure 3B, Table 3, and Supplementary Figure S2), and visualization of GFP expressed from plasmids (Figures 4, 5, 7). The copy number of plasmid pIGSAF per genome in F. alni was determined to be about 12 copies per cell (Table 3), in line with findings for other plasmids derived from plasmid pIP501 that have been estimated to be maintained at approximately 10 copies per cell (Behnke et al., 1981). In addition, plasmid pIGSAF was determined to be stable in non-selective media for a period of at least 3 weeks (Figure 6).
Of the restriction enzymes identified in Frankia genomes (Figure 1) our analysis indicated that the type IV enzymes posed the most likely barrier to transformation. Types I and II enzymes recognize specific sequence motifs of generally six to eight nucleotides (Tock and Dryden, 2005) and hence are statistically highly unlikely to be a broad-range barrier to transformation or horizontal transfer (Thomas and Nielsen, 2005). Additionally, in the genome of F. alni ACN14a the majority of types I and II genes showed very low expression in both (+)N-culture and symbiosis (Figure 1).
A type IV homolog of an mrr type methyladenine-targeting restriction gene was highly expressed in F. alni in culture (Figure 1), suggesting that DNA with methylated adenine bases is degraded in this organism. Actinobacteria, especially Frankia, express type IV methyl-directed restriction enzyme genes more highly in culture than do proteobacteria and firmicutes (Figure 2), a finding that correlates with previous reports of higher transformation efficiencies with unmethylated plasmids than methylated in Corynebacterium (Ankri et al., 1996) and Streptomyces spp. (Molle et al., 1999). Genomes of the majority of actinobacteria are missing homologs of the dam methyltransferase gene (Sanchez-Romero et al., 2015) whose product is used to mark parent DNA strands during replication, and mutS and mutL that form a complex for the removal and repair of mismatched bases on the daughter strand determined by the methylation of adenine residues (Sachadyn, 2010). Together, these factors suggest a preference for unmethylated over methylated DNA among most of the actinobacteria.
Type IV restriction enzymes have been suggested to have evolved as a counter to phage methylation systems that themselves evolved to evade host restriction systems through the methylation of restriction target sites (Westra et al., 2012). Phage genomes adopt the methylation patterns of their previous host (Loenen and Raleigh, 2014) thus increasing the likelihood of digestion by actinobacterial enzymes if replicated in a dam + host. The expression of type IV restriction enzymes in actinobacteria therefore could represent an adaptation to prevent infection by phages based on the methylation state of their genomes. Differences in methylation patterns between actinobacteria and other bacterial phyla (Novella et al., 1996) potentially constitute a barrier to horizontal gene transfer between these groups, including phage-mediated gene transfer.
Of particular interest to the evolution of root nodule symbioses is the possibility of transfer of relevant genes between Frankia and the rhizobia, and vice versa. It has been suggested that the nodA gene involved in Nod factor biosynthesis evolved in the actinobacteria, including some Frankia, and was then horizontally transferred to the rhizobia (Persson et al., 2015). If type IV restriction enzymes create a barrier to horizontal transfer into actinobacteria from dam + bacteria including proteobacteria, it would seem that horizontal transfer from actinobacteria to other phyla would be more likely than the reverse.
However, F. alni was observed to down-regulate its type IV mrr gene substantially in symbiosis (Figure 2). As roots contain much lower concentrations of bacteriophage than the surrounding soil (Ward and Mahler, 1982) this could represent a decreased necessity for restriction enzymes as a defense mechanism during symbiosis. A potential side-effect of this down-regulation, however, is that the barrier to horizontal transfer posed by type IV enzymes is likely lowered during symbiosis. In plants, the endophytic compartment is dominated by actinobacteria, with specific taxa of other phyla including proteobacteria and bacteroidetes (Lundberg et al., 2012). Due to the barrier created by type IV enzymes in the free-living condition (i.e., outside symbiosis), horizontal transfer involving Frankia in the soil is likely limited to other actinobacteria whose genomes similarly lack methylation (Novella et al., 1996), many of whom are endophytes (Trujillo et al., 2015). Inside the host, however, Frankia is likely more receptive to genes from other endophytic taxa as well. Combined, these factors could result in the preferential acquisition of genes involved in growth and host-microbe interactions in planta. In legume symbioses, exudates from hosts into their rhizospheres have been proposed to promote the conjugative transfer of symbiosis genes from a donor group of rhizobia to others, broadening the range of symbionts available to the host (Ling et al., 2016). Down-regulation of type IV restriction genes in actinorhizal nodule symbioses could be another mechanism that enhances horizontal transfer of genes related to interactions with plants by making the recipient Frankia more susceptible in the endophytic environment.
M. tuberculosis showed much lower transcription of its annotated type IV methyladenine targeting restriction enzyme than other actinobacteria. M. tuberculosis expresses an adenine methyltransferase in hypoxic conditions that regulates the expression of genes likely involved with survival during macrophage infection (Shell et al., 2013). For this reason it is likely that M. tuberculosis responds to methylated DNA differently than other actinobacteria; indeed electrotransformation of M. tuberculosis can be readily achieved with methylated plasmids replicated in E. coli DH5α (Pelicic et al., 1997), suggesting that methylated DNA is not digested in M. tuberculosis.
In this study derivatives of broad host-range plasmid pSA3 were capable of replication in F. alni. This shows that the broad host-range origin is capable of replication in Frankia and supports its use as a vector for the manipulation of Frankia spp. The parent plasmid of pSA3, pIP501, replicates in a very broad range of bacteria including Streptomyces lividans and E. coli (Kurenbach et al., 2003) indicating the potential for transformation of additional actinobacteria with these plasmids.
Differential Regulation of egfp Under the Control of the Frankia alni ACN14a nif Cluster Promoter
The expression of the egfp gene of plasmid pIGSAFnif was up-regulated in (–)N media compared with expression in (+)N media, at proportional levels to the expression of the nifH nitrogenase gene (FRAAL6813, Table 3), demonstrating for the first time that expression of reporter genes can be manipulated in Frankia. This transformation system resulted in the ability to visualize the expression of nitrogen fixation genes in vitro by fluorescence microscopy (Figure 5). Interestingly, fluorescence was detected in both the spherical portion of the vesicle as well as in the stalk that connects to the hyphae, suggesting that nitrogen fixation genes are expressed in both parts of the vesicle. Previous studies have shown that the vesicle envelope is deposited around the stalk as well as the spherical part of the vesicle (Lancelle et al., 1985), supporting the observation that nitrogen fixation can occur in the stalk.
Although the fluorescence observed when egfp was expressed under the control of the F. alni nif cluster promoter was predominantly in the vesicles, some fluorescence was occasionally observed in hyphae under nitrogen-fixing conditions whereas in (+)N media there was no observable fluorescence (Figure 5). This suggests that there can be condition-dependent expression of nif genes in the hyphae as well as the vesicles induced by nitrogen limitation. Frankia spp. in symbiosis with members of the Casuarinaceae have been reported to fix nitrogen in hyphae, since no vesicles are differentiated (Murry et al., 1985); this pattern correlated with the formation of a lignified host cell wall in the symbiotic tissue that likely reduces oxygen partial pressure (Berg and McDowell, 1987). In liquid culture, there may be zones of low pO2 that develop in portions of a Frankia hyphal colony where nitrogen fixation could be induced.
Frankia spp. in symbiosis have been suggested to be more autonomous than rhizobial microsymbionts due to their ability to control the flow of oxygen with the formation of vesicles, and due as well to the expression of more metabolic pathways in the microsymbiont in symbiosis. These factors potentially allow Frankia to be more metabolically independent from their hosts (Alloisio et al., 2010; Berry et al., 2011). The development of genetic tools for the manipulation of Frankia will allow further exploration into these and other distinctive molecular aspects of actinorhizal symbioses, which will, in turn, further inform analyses of the evolution and diversity of root nodule symbiosis.
Future Directions
The transformation methods presented here should be applicable for genetic experiments in other Frankia strains. Because plasmid pIGSAF has a broad host-range origin of replication and expresses egfp under the control of a constitutive promoter, the plasmid is likely to be usable in other strains as well. Even in the absence of selection, plasmid pIGSAF was found to be stable in F. alni cultures for at least 3 weeks (Figure 6) and cultures continued to show fluorescence after at least 4 weeks (Figure 7). Three to 4 weeks is a time period that sufficiently spans the stages of nodulation and early nitrogen fixation in a broad-spectrum of hosts. This includes Alnus glutinosa with F. alni ACN14a (Alloisio et al., 2010), and Casuarina cunninghamiana (Torrey, 1976), Discaria trinervis (Valverde and Wall, 1999), Shepherdia argentaea (Racette and Torrey, 1989), and Datisca glomerata (Berry et al., 2004). This suggests that transformants can be used to inoculate plants to study of the role of Frankia and its interactions with hosts during nodule establishment and symbiosis. In future this system could be modified using recombination or site-specific integrases to anchor genes within the genome. Differential regulation of reporter genes such as egfp can be used to localize the expression of genes identified by genomics and transcriptomics in specific Frankia cell types, in different growth conditions, and in symbiosis. Replicating plasmids may also enable the study of gene function by constitutive expression of selected genomic genes, by promoter switching or by knock-down experiments expressing anti-RNAs to genes of interest (Gillaspie et al., 2009). Circumventing the natural restriction systems of Frankia will also increase the transformation rate of non-replicating plasmids and enable higher efficiency recombination, which can be combined with CRISPR systems, for gene knock-out experiments as attempted by Kucho et al. (2009).
While this manuscript was in review a separate method for the transformation of Frankia spp. utilizing conjugation with a methylation-positive E. coli was reported by Pesce et al. (2019). Conjugative transfer has been shown to evade recipient restriction systems (Stein et al., 1988; Baltz, 1994), thus presumably allowing Frankia spp. to circumvent the type IV restriction barrier identified in this study. Nevertheless, the transformation efficiency of Streptomyces spp. by conjugation has been shown to increase over 104-fold with unmethylated DNA (Flett et al., 1997). Thus, in addition to enabling the transformation of Frankia spp. by electroporation, our analysis of restriction systems in Frankia spp. can be further utilized to improve the transformation efficiency of conjugative transfer to Frankia spp. through the use of methylation-deficient E. coli donors. Optimized transformation efficiency will be crucial for future studies involving the generation of recombinants including gene knock-outs.
Author Contributions
IG and AB conceived the project. IG carried out the project and wrote the manuscript. SV and GN assisted with the experiment and designed primer sequences. AB edited the manuscript.
Funding
IG was supported by a UC Davis Department of Plant Sciences graduate student research fellowship. Research funding for this project was provided by UC Davis Henry A. Jastro Research Grants to IG, and by USDA-NIFA-CA-D-PLS-2173-H (AB).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Rebecca Parales for providing plasmid pSA3, Philippe Normand for Frankia alni ACN14a, and Wolf Heyer for E. coli strain GM48. We also thank Ingrid Brust-Mascher for technical assistance with confocal microscopy. A previous version of this manuscript was initially published as a pre-print in bioRxiv (Gifford et al., 2018).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02230/full#supplementary-material
FIGURE S1 | Plasmid maps of pIGSAF (A) and pIGSAFnif (B), derived from plasmid pSA3 (Dao and Ferretti, 1985). Maps depict the egfp gene (green), the promoter of egfp (yellow), the origins of replication (blue), and the chloramphenicol resistance gene (camR, red). ori corresponds to the p15A E. coli-specific high-copy number origin originally derived from pACYC184 (Rose, 1988). Rep and Cop correspond to the broad host-range origin and copy number control regions derived from pGB305 (Dao and Ferretti, 1985), respectively. Also shown are unique restriction sites, and the EcoRI site introduced by ligation of the nif promoter to the egfp coding region in plasmid pIGSAFnif.
FIGURE S2 | Alignments and BLAST e-values of egfp and camR gene PCR products amplified from transformed F. alni DNA extracts. PCR sequences were compared against those of published reference sequences for each gene.
FIGURE S3 | 100X magnification confocal images showing fluorescent vesicles in different planes of focus. Two vesicles shown are in the same confocal view as in Figure 5 (A), but are more visibly fluorescent when focused (B). These vesicles can be seen in Figure 5, but with less fluorescence at that particular plane of focus.
TABLE S1 | Listing of genomes and transcriptomes used in this study. Transcriptomes are listed with their GEO accession number and original reference.
TABLE S2 | Standard curve and copy number calculations of plasmid pIGSAF determined by qPCR with relative and absolute quantification.
References
Alloisio, N., Queiroux, C., Fournier, P., Normand, P., Vallenet, D., Medigue, C., et al. (2010). The Frankia alni symbiotic transcriptome. Mol. Plant Microbe Interact. 23, 593–607. doi: 10.1094/MPMI-23-5-0593
Ankri, S., Reyes, O., and Leblon, G. (1996). Electrotransformation of highly DNA-restrictive Corynebacteria with synthetic DNA. Plasmid 35, 62–66. doi: 10.1006/plas.1996.0007
Appleby, C. A. (1984). Leghemoglobin and Rhizobium respiration. Annu. Rev. Plant Physiol. 35, 443–478. doi: 10.1146/annurev.arplant.35.1.443
Baltz, R. H. (1994). “Gene expression in recombinant Streptomyces,” in Recombinant Microorganisms: Gene Expression, ed. A. Smith (New York, NY: Marcel Dekker), 309–381.
Battenberg, K., Potter, D., Tabuloc, C., Chiu, J. C., and Berry, A. M. (2018). Comparative transcriptomics of two actinorhizal plants and the legume Medicago truncatula support the homology of root nodule symbioses and is congruent with a two-step process of evolution in the nitrogen-fixing clade of angiosperms. Front. Plant Sci. 9:1256. doi: 10.3389/fpls.2018.01256
Behnke, D., Gilmore, M. S., and Ferretti, J. J. (1981). Plasmid pGB301, a new multiple resistance streptococcal cloning vehicle and its use in cloning of a gentamicin/kanamycin resistance determinant. Mol. Gen. Genet. 182, 414–421. doi: 10.1007/bf00293929
Benson, D. R., and Silvester, W. B. (1993). Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol. Rev. 57, 293–319.
Berg, R. H., and McDowell, L. (1987). Endophyte differentiation in Casuarina actinorhizae. Protoplasma 136, 104–117. doi: 10.1007/bf01276359
Berry, A. M., Harriott, O. T., Moreau, R. A., Osman, S. F., Benson, D. R., and Jones, A. D. (1993). Hopanoid lipids compose the Frankia vesicle envelope, presumptive barrier of oxygen diffusion to nitrogenase. Proc. Natl. Acad. Sci. U.S.A. 90, 6091–6094. doi: 10.1073/pnas.90.13.6091
Berry, A. M., Mendoza-Herrera, A., Guo, Y., Hayashi, J., Persson, T., Barabote, R., et al. (2011). New perspectives on nodule nitrogen assimilation in actinorhizal symbioses. Funct. Plant Biol. 38, 645–652.
Berry, A. M., Murphy, T. M., Okubara, P. A., Jacobsen, K. R., Swensen, S. M., and Pawlowski, K. (2004). Novel expression pattern of cytosolic Gln synthetase in nitrogen-fixing root nodules of the actinorhizal host, Datisca glomerata. Plant Physiol. 135, 1849–1862. doi: 10.1104/pp.103.031534
Bickhart, D. M., and Benson, D. R. (2011). Transcriptomes of Frankia sp. strain CcI3 in growth transitions. BMC Microbiol. 11:192. doi: 10.1186/1471-2180-11-192
Burleigh, S., and Torrey, J. G. (1990). Effectiveness of different Frankia cell types as inocula for the actinorhizal plant Casuarina. Appl. Environ. Microbiol. 56, 2565–2567.
Castaneda-Garcia, A., Prieto, A. I., Rodriguez-Beltran, J., Alonso, N., Cantillon, D., Costas, C., et al. (2017). A non-canonical mismatch repair pathway in prokaryotes. Nat. Commun. 8:14246. doi: 10.1038/ncomms14246
Ceremonie, H., Debelle, F., and Fernandez, M. P. (1999). Structural and functional comparison of Frankia root hair deforming factor and rhizobia Nod factor. Can. J. Bot. 77, 1293–1301. doi: 10.3389/fpls.2018.01494
Cissoko, M., Hocher, V., Gherbi, H., Gully, D., Carre-Mlouko, A., Sane, S., et al. (2018). Actinorhizal signaling molecules: Frankia root hair deforming factor shares properties with NIN inducing factor. Front. Plant Sci. 9:1494. doi: 10.3389/fpls.2018.01494
Dao, M. L., and Ferretti, J. J. (1985). Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl. Environ. Microbiol. 49, 115–119.
Dietrich, G., Schaible, U. E., Diehl, K. D., Mollenkopf, H., Wiek, S., Hess, J., et al. (2000). Isolation of RNA from mycobacteria grown under in vitro and in vivo conditions. FEMS Microbiol. Lett. 186, 177–180. doi: 10.1016/s0378-1097(00)00138-5
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Feil, W. S., Feil, H., and Copeland, A. (2012). Bacterial Genomic DNA Isolation Using CTAB. Walnut Creek, CA: Joint Genome Institute.
Flett, F., Mersinias, V., and Smith, C. P. (1997). High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155, 223–229. doi: 10.1016/s0378-1097(97)00392-3
Fournier, J., Teillet, A., Chabaud, M., Ivanov, S., Genre, A., Limpens, E., et al. (2015). Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol. 167, 1233–1242. doi: 10.1104/pp.114.253302
Gage, D. J. (2002). Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J. Bacteriol. 184, 7042–7046. doi: 10.1128/jb.184.24.7042-7046.2002
Gifford, I., Vance, S., Nguyen, G., and Berry, A. M. (2018). Stable genetic transformation and heterologous expression in the nitrogen-fixing plant endosymbiont Frankia alni ACN14a. bioRXiv doi: 10.1101/496703
Gillaspie, D., Perkins, I., Larsen, K., McCord, A., Pangonis, S., Sweger, D., et al. (2009). Plasmid-based system for high-level gene expression and antisense gene knockdown in Bartonella henselae. Appl. Environ. Microbiol. 75, 5434–5436. doi: 10.1128/AEM.00949-09
Griesmann, M., Chang, Y., Liu, X., Song, Y., Haberer, G., Crook, M. G., et al. (2018). Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 24:eaat1743. doi: 10.1126/science.aat1743
Gtari, M., Ghodbane-Gtari, F., Nouioui, I., Ktari, A., Hezbri, K., Mimouni, W., et al. (2015). Cultivating the uncultured: growing the recalcitrant cluster-2 Frankia strains. Sci. Rep. 5:13112. doi: 10.1038/srep13112
Gutierrez, R. A. (2012). Systems biology for enhanced plant nitrogen nutrition. Science 336, 1673–1675. doi: 10.1126/science.1217620
Hahn, D., Amann, R. I., and Zeyer, J. (1993). Whole-cell hybridization of Frankia strains with fluorescence- or digoxigenin-labeled, 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 59, 1709–1716.
Helaine, S., Thompson, J. A., Watson, K. G., Liu, M., Boyle, C., and Holden, D. W. (2010). Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl. Acad. Sci. U.S.A. 107, 3746–3751. doi: 10.1073/pnas.1000041107
Horodniceanu, T., Bouanchaud, D. H., Bieth, G., and Chabbert, Y. A. (1976). R plasmids in Streptococcus agalactiae (Group B). Antimicrob. Agents Chemother. 10, 795–801. doi: 10.1128/aac.10.5.795
Kucho, K. I., Kakoi, K., Yamaura, M., Higashi, S., Uchiumi, T., and Abe, M. (2009). Transient transformation of Frankia by fusion marker genes in liquid culture. Microbes Environ. 24, 231–240. doi: 10.1264/jsme2.me09115
Kurenbach, B., Bohn, C., Prabhu, J., Abudukerim, M., Szewzyk, U., and Grohmann, E. (2003). Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid 50, 86–93. doi: 10.1016/s0147-619x(03)00044-1
Lancelle, S. A., Torrey, J. G., Hepler, P. K., and Callaham, D. A. (1985). Ultrastructure of freeze-substituted Frankia strain HFPCcI3, the actinomycete isolated from root nodules of Casuarina cunninghamiana. Protoplasma 127, 64–72. doi: 10.1007/bf01273702
Lee, C., Kim, J., Shin, S. G., and Hwang, S. (2006). Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J. Biotechnol. 123, 273–280. doi: 10.1016/j.jbiotec.2005.11.014
Ling, J., Wang, H., Wu, P., Li, T., Tang, Y., Naseer, N., et al. (2016). Plant nodulation inducers enhance horizontal gene transfer of Azorhizobium caulinodans symbiosis island. Proc. Natl. Acad. Sci. U.S.A. 113, 13875–13880. doi: 10.1073/pnas.1615121113
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loenen, W. A. M., and Raleigh, E. A. (2014). The other face of restriction: modification-dependent enzymes. Nucleic Acids Res. 42, 56–69. doi: 10.1093/nar/gkt747
Long, S. R., Buikema, W. J., and Ausubel, F. M. (1982). Cloning of Rhizobium meliloti nodulation genes by direct complementation of Nod- mutants. Nature 298, 485–488. doi: 10.1038/298485a0
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Molle, V., Palframan, W. J., Findlay, K. C., and Buttner, M. J. (1999). WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 182, 1286–1295. doi: 10.1128/jb.182.5.1286-1295.2000
Murry, M. A., Fontaine, M. S., and Torrey, J. G. (1984). Growth kinetics and nitrogenase induction in Frankia sp. ArI3 grown in batch culture. Plant Soil 78, 61–78. doi: 10.1007/978-94-009-6158-6_7
Murry, M. A., Zhongze, Z., and Torrey, J. G. (1985). Effect of O2 on vesicle formation, acetylene reduction, and O2-uptake kinetics in Frankia sp. HFPCcI3 isolated from Casuarina cunninghamiana. Can. J. Microbiol. 31, 804–809. doi: 10.1139/m85-151
Mus, F., Crook, M. B., Garcia, K., Costas, A. G., Geddes, B. A., Kouri, E. D., et al. (2016). Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl. Environ. Microbiol. 82, 3698–3710. doi: 10.1128/AEM.01055-16
Myers, A. K., and Tisa, L. S. (2003). Effect of electroporation conditions on cell viability of Frankia EuI1c. Plant Soil 254, 83–88. doi: 10.1007/978-94-017-1601-7_10
Noridge, N. A., and Benson, D. R. (1986). Isolation and nitrogen-fixing activity of Frankia sp. strain CpI1 vesicles. J. Bacteriol. 166, 301–305. doi: 10.1128/jb.166.1.301-305.1986
Normand, P., and Lalonde, M. (1982). Evaluation of Frankia isolated from provenances of two Alnus species. Can. J. Microbiol. 28, 1133–1142. doi: 10.1139/m82-168
Novella, I. S., Marin, I., and Sanchez, J. (1996). Restriction analysis of actinomycetes chromosomal DNA. Can. J. Microbiol. 42, 201–206. doi: 10.1139/m96-031
Okazaki, S., Kaneko, T., Sato, S., and Saeki, K. (2013). Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 110, 17131–17136. doi: 10.1073/pnas.1302360110
Oldroyd, G. E. D. (2013). Speak, friend, and enter: signaling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Pelicic, V., Jackson, M., Reyrat, J., Jacobs, W. R., Gicquel, B., and Guilhot, C. (1997). Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 94, 10955–10960. doi: 10.1073/pnas.94.20.10955
Persson, T., Battenberg, K., Demina, I. V., Vigil-Stenman, T., Vanden Heuvel, B., Pujic, P., et al. (2015). Candidatus Frankia datiscae Dg1, the actinobacterial microsymbiont of Datisca glomerata, expresses the canonical nod genes nodABC in symbiosis with its host plant. PLoS One 10:e0127630. doi: 10.1371/journal.pone.0127630
Pesce, C., Oshone, R., Hurst, S. G., Kleiner, V. A., and Tisa, L. S. (2019). Stable transformation of the actinobacteria Frankia. Appl. Environ. Microbiol. 85, e957–19. doi: 10.1128/AEM.00957-19
Racette, S., and Torrey, J. G. (1989). Root nodule initiation in Gymnostoma (Casuarinaceae) and Shepherdia (Elaeagnaceae) induced by Frankia strain HFPGpI1. Can. J. Bot. 67, 2873–2879. doi: 10.1139/b89-368
Roberts, R. J., Vincze, T., Posfai, J., and Macelis, D. (2010). REBASE-a database for DNA restriction and modification: enzymes, genes, and genomes. Nucleic Acids Res. 38(Suppl._1), D234–D236.
Rose, R. E. (1988). The nucleotide sequence of pACYC184. Nucleic Acids Res. 16:355. doi: 10.1093/nar/16.1.355
Sachadyn, P. (2010). Conservation and diversity of MutS proteins. Mutat. Res. 694, 20–30. doi: 10.1016/j.mrfmmm.2010.08.009
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Second Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Sanchez-Romero, M. A., Cota, I., and Casadesus, J. (2015). DNA methylation in bacteria: from the methyl group to the methylome. Curr. Opin. Microbiol. 25, 9–16. doi: 10.1016/j.mib.2015.03.004
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Shell, S. S., Prestwich, E. G., Baek, S., Shah, R. R., Sassetti, C. M., Dedon, P. C., et al. (2013). DNA methylation impacts gene expression and ensures hypoxic survival of Mycobacterium tuberculosis. PLoS Pathog. 9:e1003419. doi: 10.1371/journal.ppat.1003419
Simonet, P., Normand, P., Hirsch, A. M., and Akkermans, A. D. L. (1990). “The genetics of the Frankia-Actinorhizal symbiosis,” in Molecular Biology of Symbiotic Nitrogen Fixation, ed. P. M. Gresshoff (Boca Raton, FL: CRC-Press), 77–109. doi: 10.1201/9781351074742-4
Soltis, D. E., Soltis, P. S., Morgan, D. R., Swensen, S. M., Mullin, B. C., Dowd, J. M., et al. (1995). Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc. Natl. Acad. Sci. U.S.A. 92, 2647–2651. doi: 10.1073/pnas.92.7.2647
Spaepen, S., Das, F., Luyten, E., Michiels, J., and Vanderleyden, J. (2009). Indole-3-acetic acid-regulated genes in Rhizobium etli CNPAF512. FEMS Microbiol. Lett. 291, 195–200. doi: 10.1111/j.1574-6968.2008.01453.x
Stein, D. C., Gregoire, S., and Piekarowicz, A. (1988). Restriction of plasmid DNA during transformation but not conjugation in Neisseria gonorrhoeae. Infect. Immun. 56, 112–116.
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. doi: 10.1038/nrmicro1234
Tisa, L. S., Chval, M. S., Krumholz, G. D., and Richards, J. (1998). Antibiotic resistance patterns of Frankia strains. Can. J. Bot. 77, 1257–1260. doi: 10.1139/b99-067
Tisa, L. S., and Ensign, J. C. (1987). Isolation and nitrogenase activity of vesicles from Frankia sp. strain EAN1pec. J. Bacteriol. 169, 5054–5059. doi: 10.1128/jb.169.11.5054-5059.1987
Tock, M. R., and Dryden, D. T. F. (2005). The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 8, 466–472. doi: 10.1016/j.mib.2005.06.003
Torrey, J. G. (1976). Initiation and development of root nodules of Casuarina (Casuarinaceae). Am. J. Bot. 63, 335–344. doi: 10.1002/j.1537-2197.1976.tb11820.x
Trujillo, M. E., Riesco, R., Benito, P., and Carro, L. (2015). Endophytic actinobacteria and the interaction of Micromonospora and nitrogen fixing plants. Front. Microbiol. 6:1341. doi: 10.3389/fmicb.2015.01341
Valverde, C., and Wall, L. G. (1999). Time course of nodule development in the Discaria trinervis (Rhamnaceae) – Frankia symbiosis. New Phytol. 141, 345–354. doi: 10.1046/j.1469-8137.1999.00345.x
Ward, R. L., and Mahler, R. J. (1982). Uptake of bacteriophage f2 through plant roots. Appl. Environ. Microbiol. 43, 1098–1103.
Westra, E. R., Swarts, D. C., Staals, R. H. J., Jore, M. M., Brouns, S. J. J., and van der Oost, J. (2012). The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 46, 311–339. doi: 10.1146/annurev-genet-110711-155447
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., and Madden, T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134
Keywords: Frankia, transformation, restriction enzymes, DNA Methylation, nitrogen fixation, root nodule, symbiosis, actinobacteria
Citation: Gifford I, Vance S, Nguyen G and Berry AM (2019) A Stable Genetic Transformation System and Implications of the Type IV Restriction System in the Nitrogen-Fixing Plant Endosymbiont Frankia alni ACN14a. Front. Microbiol. 10:2230. doi: 10.3389/fmicb.2019.02230
Received: 03 January 2019; Accepted: 11 September 2019;
Published: 24 September 2019.
Edited by:
Lorena Carro, Newcastle University, United KingdomReviewed by:
Maher Gtari, Carthage University, TunisiaRichard Jonathan Harrison, NIAB EMR, United Kingdom
Copyright © 2019 Gifford, Vance, Nguyen and Berry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isaac Gifford, isgifford@ucdavis.edu
 Isaac Gifford
Isaac Gifford Summer Vance
Summer Vance Giang Nguyen
Giang Nguyen Alison M. Berry
Alison M. Berry