
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 27 September 2019
Sec. Microbiological Chemistry and Geomicrobiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02224
This article is part of the Research TopicAstrobiology At The Interface: Interactions Between Biospheres, Geospheres, Hydrospheres And Atmospheres Under Planetary ConditionsView all 10 articles
 Scott D. Hamilton-Brehm1,2*
Scott D. Hamilton-Brehm1,2* Laura E. Stewart3
Laura E. Stewart3 Mavrik Zavarin4
Mavrik Zavarin4 Matt Caldwell5
Matt Caldwell5 Paul A. Lawson5
Paul A. Lawson5 Tullis C. Onstott6
Tullis C. Onstott6 Joseph Grzymski1
Joseph Grzymski1 Iva Neveux1
Iva Neveux1 Barbara Sherwood Lollar7
Barbara Sherwood Lollar7 Charles E. Russell8
Charles E. Russell8 Duane P. Moser1,8*
Duane P. Moser1,8*Deep fractured rock ecosystems across most of North America have not been studied extensively. However, the US Great Basin, in particular the Nevada National Security Site (NNSS, formerly the Nevada Test Site), has hosted a number of influential subsurface investigations over the years. This investigation focuses on resident microbiota recovered from a hydrogeologically confined aquifer in fractured Paleozoic carbonate rocks at 863 – 923 meters below land surface. Analysis of the microorganisms living in this oligotrophic environment provides a perspective into microbial metabolic strategies required to endure prolonged hydrogeological isolation deep underground. Here we present a microbiological and physicochemical characterization of a deep continental carbonate ecosystem and describe a bacterial genus isolated from the ecosystem. Strain DRI-13T is a strictly anaerobic, moderately thermophilic, fumarate-respiring member of the phylum Firmicutes. This bacterium grows optimally at 55°C and pH 8.0, can tolerate a concentration of 100 mM NaCl, and appears to obligately metabolize fumarate to acetate and succinate. Culture-independent 16S rRNA gene sequencing indicates a global subsurface distribution, while the closest cultured relatives of DRI-13T are Pelotomaculum thermopropionicum (90.0% similarity) and Desulfotomaculum gibsoniae (88.0% similarity). The predominant fatty acid profile is iso-C15:0, C15:0, C16:0 and C14:0. The percentage of the straight-chain fatty acid C15:0 is a defining characteristic not present in the other closely related species. The genome is estimated to be 3,649,665 bp, composed of 87.3% coding regions with an overall average of 45.1% G + C content. Strain DRI-13T represents a novel genus of subsurface bacterium isolated from a previously uncharacterized rock-hosted geothermal habitat. The characterization of the bacterium combined with the sequenced genome provides insights into metabolism strategies of the deep subsurface biosphere. Based on our characterization analysis we propose the name Thermoanaerosceptrum fracticalcis (DRI-13T = DSM 100382T = ATCC TSD-12T).
Biomass in the continental crust may be greater than that on the surface of the Earth (Gold, 1992; Whitman et al., 1998; McMahon and Parnell, 2014; Magnabosco et al., 2018). The deep biosphere is an untapped reservoir of microbial biodiversity that may hold novel solutions to industrial, medical, and origin of life questions. While the concept of subsurface life was proposed in the 1920s (Bastin and Greer, 1930), research into deep terrestrial subsurface environments did not begin in earnest until the 1980s (Jorgensen, 2012). Exploration of subsurface microbial biogeochemical processes remain largely unmapped and uncharacterized, due partly to limited access to the terrestrial subsurface and the low biomass densities often found there. Primary sampling opportunities occur through caves, mines, springs, and boreholes. A high level of novel microbial diversity has been discovered in the subsurface through cultivation-independent sequencing of the 16S rRNA gene (Pedersen and Ekendahl, 1990; Kotelnikova and Pedersen, 1997; Takai et al., 2001; Chapelle et al., 2002; Moser et al., 2005; Lin et al., 2006; Osburn et al., 2014). However, only a modest fraction of known lineages are currently represented in culture collections (Daumas et al., 1988; Boone et al., 1995; Ravot et al., 1995; Nilsen et al., 1996; Puspita et al., 2012).
With some notable exceptions (Stevens and McKinley, 1995; Colwell et al., 1997; Fredrickson et al., 1997; Krumholz et al., 1997; Lehman et al., 2001; Osburn et al., 2014), the deep fractured rock ecosystems across most of North America have been relatively little studied for microbiology. However, the US Great Basin, in particular the Nevada National Security Site (NNSS, formerly the Nevada Test Site, Bowen et al., 2001), has hosted a number of influential subsurface investigations over the years (Amy et al., 1992; Haldeman and Amy, 1993; Haldeman et al., 1993; Kieft et al., 1997). Most of the NNSS lies entirely within a geologic extensional zone and is underlain by the “Death Valley Regional Flow System (DVRFS).” This expansive set of groundwater basins is dominated by fractured rock aquifers that comprises ∼100,000 km2 of mountain ranges (up to 3,600 m above mean sea level) and valleys which can reach below sea level (e.g., Death Valley, at −86 m, the lowest point in North America) (D’Agnese et al., 1997; Belcher et al., 2002; Ye et al., 2008; Bushmann et al., 2010). Within this system groundwater flows long distances (i.e., Interbasin Flow), from high-elevation recharge zones in central Nevada to large-discharge springs in and near Death Valley, CA, United States (Winograd and Thordarson, 1975; Winograd and Pearson, 1976; Belcher et al., 2009). Direct access to the DVRFS is achievable through an ongoing Department of Energy (DOE) sponsored environmental management activity tasked with tracking groundwater contamination associated with underground nuclear testing at the NNSS, the Underground Test Area (UGTA) sub-project. The hundreds of monitoring boreholes/wells established and maintained by this program represent a unique regional-scale observatory for deep life study.
This study focuses on resident microbiota in fluids from 863–923 meters below land surface (mbls), accessed via a monitoring borehole, U-3cn#5 (Garber and Johnson, 1967; Bangerter and Giblin, 1998). Intercepting a hydrogeologically confined aquifer within the fractured Paleozoic carbonate rocks of a prominent regional feature termed the “Lower Carbonate Aquifer” (LCA), this well provides a portal into a pristine deep ecosystem that would otherwise be very difficult to access. Analysis of the microorganisms living in this restricted oligotrophic environment provides a perspective into microbial metabolic strategies required to endure prolonged hydrogeological isolation deep underground. The isolation of this unit is demonstrated by the very low radiocarbon content (4.65 pmc 14C) and absence of contamination related to past nuclear testing in the overlying units. Here we present a microbiological and physicochemical characterization of one window into the NNSS portion of the DVRFS and describe the isolation of a representative bacterium from this ecosystem, DRI-13T, a novel, moderately thermophilic, strict fumarate-respiring organism belonging to the phylum Firmicutes from the deep terrestrial biosphere, here designated Thermoanaerosceptrum fracticalcis.
Well U-3cn#5 (Garber and Johnson, 1967; Bangerter and Giblin, 1998) is located in central Yucca Flat, Nevada, United States on the NNSS (latitude 37.06, longitude −116.02, surface elevation 1223 m). Yucca Flat is an arid, intermontane valley partially filled with tuffs and alluvium (260 – 290 m thickness), underlain by partially welded and zeolitized Tertiary-age ash-flow tuffs and bedded tuffs (∼536 m thickness). A highly fractured Paleozoic carbonate layer (colluvium transitioning to dolomite/dolomitic quartzite) of about 41 m thickness completes the sequence and is the source of groundwater for this study. The borehole was drilled in 1965 to a total depth of 923.5 mbls, 120 m southwest of surface ground zero and outside the collapse chimney of the Bilby underground nuclear test conducted in 1963 (249 kt, 63.7 m cavity radius) (Zavarin, 2014) (Figures 1A,B). The hole intersects the local groundwater table at 493.9 mbls and carbonate rocks at 863.2 mbls. Borehole gamma logs conducted within U-3cn#5 indicated the presence of elevated radionuclides within the unsaturated zone tertiary volcanic rock. These radionuclides were thought to have been emplaced by prompt injection during the underground test. Radionuclides have not been detected in the water-saturated fractured carbonates intersected by U-3cn#5, indicating no impact by the nearby underground nuclear test (Thompson, 1999). This volcanic confining unit overlying the carbonate aquifer serves to hydrogeologically isolate a nearly anoxic, slightly geothermal (∼45oC) portion of the LCA. The well is continuously cased through the vadose zone and confining unit (stainless steel below the water table) and was recompleted in 1996/1997, when the permanent 50-horsepower tandem pump used for this study was installed at 691.3 mbls. During pumping, water is obtained from an isolated open hole segment (863.2–923.5 mbls) within the LCA below the casing terminus; most likely derived from distinct fractured zone in core logs at 863.5, 864.4, and 866.2 mbls or a highly fractured zone from 868.7 to 876.3 mbls (Bangerter and Giblin, 1998).
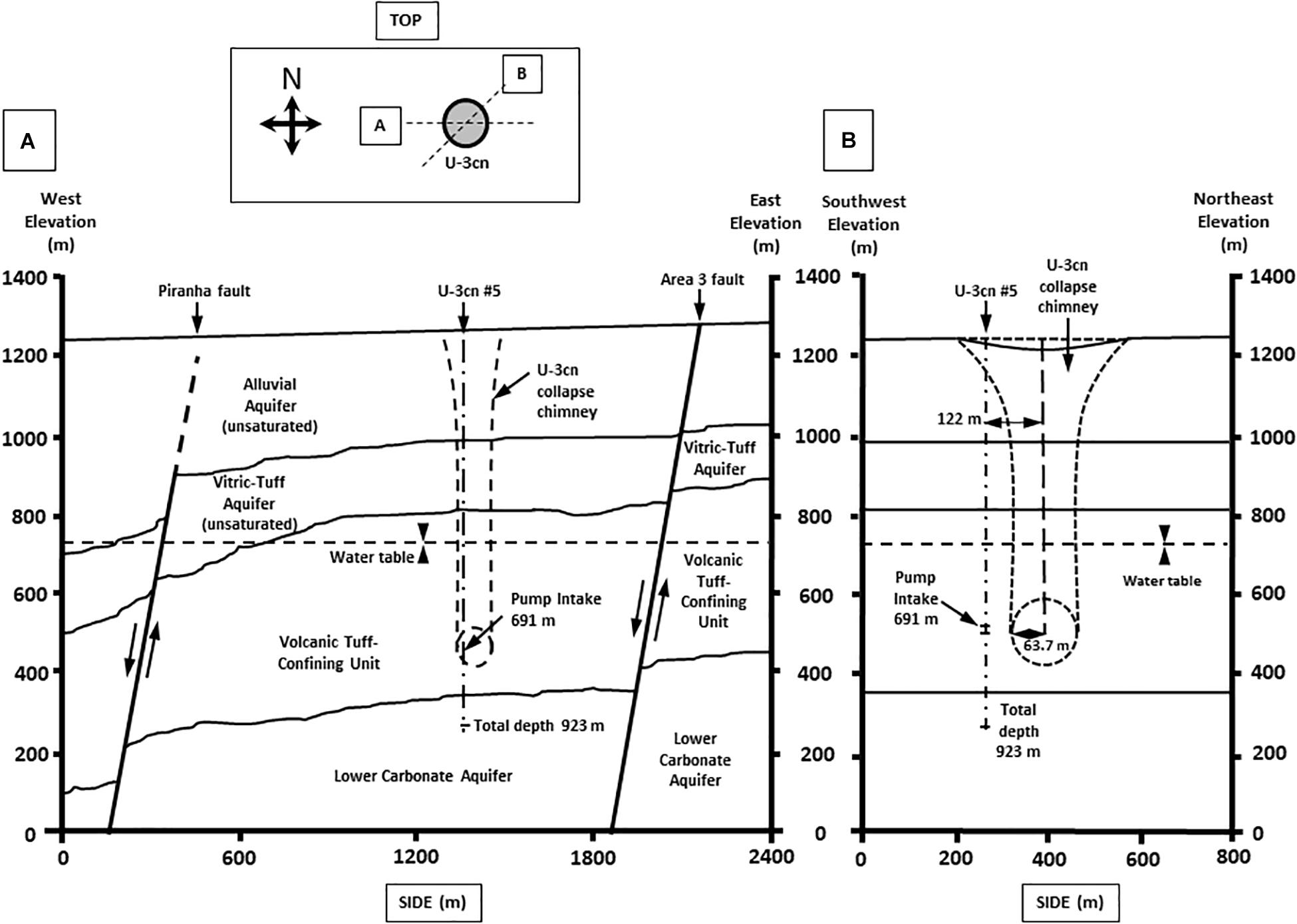
Figure 1. Geological cross section of well U-3cn #5. Cross section from (A) West to East and (B) Southwest to Northeast. Cross sections show borehole U-3cn#5 pump position, Total depth (TD) at 923 m, and relative location below the water table. (Image modified from McCall, RL, Raytheon Services Nevada, 1994, DOE/NV/-500-UC-700) [DOE cleared].
Prior to sampling, U-3cn#5 was pumped and sampled on a semi-regular basis since 1967 (Total of 3 × 106 m3 of water), including March 29, 2011, the subject of this report. Water samples were obtained on the surface from an in-line sampling port affiliated with a hydrologic pump test (104.9 gpm) after removal of 440 m3 of water over a period of days via an autoclaved gas-tight manifold. Water samples for microbial cultivation and dissolved gas sampling were transferred from flowing lines (LS-24 Nuprene Tubing, Masterflex) fitted with 24 Ga needles into sterile crimp-sealed, pre-evacuated, N2 flushed, 120 mL serum bottles (gas samples fixed with 5 μL of saturated HgCl2). Samples for aqueous chemistry were collected from the flowing sample lines and analyzed offsite. Samples for microbial community analysis were collected in two sterile 8 L polypropylene carboys and cells collected offsite on 0.2 μm filters (Sterivex, EMD Millipore, Bedford MA, 2 L per filter).
Field measurements of temperature, dissolved O2 (dO2), conductivity, and pH were made onsite with an MP20 Sonde fitted with a flow cell (Yellow Springs Instruments, Yellow Springs, OH). Most chemical analytes were obtained during routine analysis by the DOE Underground Test Area (UGTA) Program. Additional analyses were performed at the Desert Research Institute (DRI) Water Analysis Laboratory (Reno, NV, United States) or at Princeton University (Princeton, NJ, United States). Nitrite, nitrate, and ammonia were analyzed at DRI using Alpkem RFA 300 and Technicon Automated Colorimetric Analyzers using EPA method SM 4500-NO3 F and SM 4500-NH4 F. Dionex Model ICS 2000 Ion Chromatograph was used for the measurement of Cl–, Br–, and SO42–. A Brinkmann Metrohm Titrando automated titrator, capable of potentiometric titrations to fix inflection end points, was used in the determination of CO32–, HCO3–, pH, EC, and F–. An OI Analytical 1030W Carbon Analyzer was used for determination of dissolved organic carbon (DOC) and total organic carbon (TOC) in water samples. A Thermo Elemental SOLAAR M5 Atomic Absorption Spectrometer with air-acetylene flame and vapor generation capabilities was used for major cation and many metal analyses.
A subset of samples were analyzed in duplicate for anions (F–, Cl–, NO2–, SO42–, Br–, NO3–, PO43–) and for short chain fatty acids (acetate, lactate, formate, and propionate) at Princeton University using a Dionex IC25 ion chromatograph (Thermo Scientific, Waltham, MA, United States) coupled to an MSQ PlusTM ESI-quadrupole mass spectrometer (Thermo Scientific, Waltham, MA, United States) (Lau et al., 2014).
Total and Live/Dead cell counts were determined for groundwater samples using mfgr-provided reagents and protocols with a MicroPro Flow Cytometer (Benton Dickenson).
Planktonic microorganisms, concentrated by filtration, were extracted using the UltraCleanTM Soil DNA Isolation kit (Mo Bio Laboratories Inc., Carlsbad, CA, United States), according to manufacturer’s protocol amended with three freeze/thaw cycles (−80°C/65°C; 20 min each) at the beginning of DNA extraction procedure. Given the proximity of the site to prior nuclear testing, filters were screened for radioactivity by scintillation at the Harry Reid Center for Environmental Studies at the University of Nevada, Las Vegas as a precaution (all were below detection limits). The extracted DNA was used for PCR-amplified clone libraries, universal bacterial primers 27F-YM/149 2R (AGAGTTTGATYMTGGCTCAG/TACCTTGTTACGACTT) and universal archaeal primers were used 21F/149 2R (TTCCGGTTGATCCYGCCGGA/TACCTTGTTACGACTT) to generate near-full-length 16S rRNA gene amplicons (Lane, 1991). PCR was performed using LATaq (Clontech, Mountain View, CA, United States) and thermocycler settings of 95°C, 30 s; 53°C, 30 s; 72°C, 60 s; for 30 cycles. PCR amplicons were purified with an UltraClean GelSpin DNA Purification Kit (Mo Bio Laboratories Inc., Carlsbad, CA, United States), cloned (TOPO-TA, Invitrogen, Carlsbad, CA, United States) and contigs were completely sequenced for bacteria and partially sequenced for archaea by Functional BioSciences (Madison, WI, United States) using vector primers. Contigs were generated using SequencherTM 4.9 (Gene Codes, Ann Arbor, MI, United States), aligned, matched with nearest neighbors and checked for chimeras using Silva (v1.2.11) (Glöckner et al., 2017) and MEGA 5.2 (Tamura et al., 2011).
Microbial cultivation enrichments were performed in 160 mL serum bottles with 25 mL of a custom artificial groundwater medium (AGM), composed of per liter 3.6 g 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES), 1.5 g Na2SO4, 0.174 g K2PO4, 0.138 g Resazurin, 0.4 g MgCl26 H2O, 0.5 g KCl, 0.268 g NH4Cl, 0.25 g NaHCO3, 1 mL ATCC Minimal Vitamins (ATCC, Manassas, VA, United States), and 1 mL ATCC Minimal Minerals. AGM was prepared anaerobically using a modified Hungate technique with 600 mg/L Na2S⋅9H20 as a reducing agent (Miller and Wolin, 1974). The water sample was maintained at 4°C for 2 years prior to attempting enrichments. The primary enrichment consisted of 1 mL inoculum from the environmental sample, incubated in AGM amended with 0.1% (w/v) peptone (BD Bacto, Franklin Lakes, NJ, United States) at 45°C with a headspace of 100% N2. Cell numbers were monitored using a Petroff-Hausser counter (Hausser Scientific Co., Horsham, PA, United States) and an Axioskop2 Plus microscope under phase contrast. Cells reached a density of 106 cells/mL after 1 month and were subsequently transferred three times prior to final isolation by low-intensity heat shock, serial dilution, and streak plating. Heat shocks were performed first on the environmental enrichment by rapidly raising the incubation temperature during logarithmic growth from 45°C to 60°C for 1 h and then rapidly cooling to 45°C. This was repeated for three transfers, the resulting cultures were then serial diluted to sub single cell concentrations. Finally, the putative isolate was streaked on anaerobic 1% w/v agar plates (AGM with agar added) amended with 10 mM fumarate/0.05% (w/v) yeast extract and one of the apparently identical colonies was selected as the type strain.
Growth experiments were conducted in 160-mL serum bottles, containing 25 mL volume of AGM and 10 mM fumarate with a headspace of 100% N2. Cultivation experiments to define temperature optima were incubated at 37, 45, 50, 55, 60, and 65°C in the dark without shaking. Cultivation experiments to determine optimal pH utilized alternative buffers, replacing the HEPES buffer when not appropriate. A final concentration of 10 mM for each buffer was used to achieve the desired pH value. The buffers used were: 1,4-Piperazinediethanesulfonic acid (PIPES) for pH 6.5-7.5, HEPES for pH 8-8.5, and 3-([1,1-Dimethyl-2-hydroxyethyl]amino)-2-hydroxypropanesulfonic acid (AMPSO) for pH 9. All culturing optimization tests were completed in quadruplicate.
Substrate utilization was determined by transferring strain DRI-13T three times at 55°C into AGM media containing 10 mM of the following defined substrates: glucose, formate, fumarate, lactate, acteate, pyruvate, methanol, propionate, ribose, xylose, oxaloacetate, succinate, tartarate, butyrate, malate, citrate, aspartate, methionine, valine, alanine/glycine, leucine/glycine, and glutamate/glycine. Undefined substrates were added to AGM media in concentrations of 0.1% wt/vol for casamino acids, peptone, and yeast extract. Gas phase substrates were added to the serum bottle headspace to a pressure of 30 psi for H2/CO2 (80:20) and CO (100). Electron acceptors 5 mM nitrite, 5 mM nitrate, 10 mM sulfate, 10 mM sulfite, 10 mM thiosulfate, and 0.01% wt/vol elemental sulfur were individually added to AGM medium with each defined and undefined substrate. Success was determined when cultures were successfully transferred three times and reached cell densities of 108 cells/mL by microscopy observations using a Petroff-Hauser cell counting chamber.
Co-culture experiments of strain DRI-13T with wild type Methanothermobacter thermoautotrophicum were conducted in AGM medium containing 10 mM fumarate or 30 psi H2/CO2 (80/20 headspace) at 55°C. Enumeration of both microorganism’s growth was monitored by microscopy based on their distinct morphologies.
Fumarate depletion was monitored by high-performance liquid chromatography (HPLC). Supernatants were filtered and acidified with 200 mM sulfuric acid to a final concentration of 5 mM. A sample of 5 μL was injected into AcclaimTM OA 5 μm column (4 × 250 mm), (Thermo Scientific), housed in Agilent 1100 series HPLC system for each time point. Separation was achieved under an isocratic gradient at a temperature of 38°C and flow of 0.5 mL/min. The DAD detector was adjusted to a wavelength of 210 nm and full scale sensitivity. Retention times of obtained peaks were compared with known standards (Organic Acids Kit, Supelco Analytical) and the quantity calculated using linear standard curves. The results were assessed with Chromeleon 7 software.
Strain DRI-13T isolate was identified by centrifuging 5 mL of planktonic cells during logarithmic growth at 10,000 × g for 15 min at 4°C. DNA was extracted from the resulting cell pellet using an UltraClean Microbial DNA Isolation Kit (Mo Bio, Carlsbad, CA, United States). Universal bacterial primers 27F/1492R (5′-AGAGTTTGATCMTGGCTCAG-3′/5′-ACCTTGTTACGACTT-3′) were used to PCR amplify the small subunit ribosomal RNA (SSU rRNA or 16S rRNA gene) (Weisburg et al., 1991). The PCR product was then sequenced by Functional Biosciences (Madison, WI, United States) to determine identification of the isolate. With other 16S rRNA gene sequences recovered from the National Center for Biotechnology Information (NCBI) website, the sequences were aligned and phylogenetic relationships determined by Silva (v1.2.11) (Glöckner et al., 2017) and MEGA 5.2 (Tamura et al., 2011). Taxonomic classification sequences were obtained from the “List of Prokaryotic names with Standing in nomenclature” (LPSN,1).
High-molecular weight DNA was extracted from DRI-13T by cultivating the organism in five, 1-L batch cultures grown on 10 mM fumarate. Cells were harvested by centrifugation at 15,000 × g for 30 min, and DNA was precipitated and extracted by cetyltrimethyl ammonium bromide (CTAB) buffer/phenol/chloroform (Sambrook and Russell, 2001). DNA was RNase treated, quality-checked on 1% agarose gel and shipped to the DOE Joint Genome Institute (JGI) for draft sequencing using Illumina and PacBio sequencing technology (PacBio RS; Illumina HiSeq 2500). The sequences were assembled according to JGI protocols using Velvet, annotated using a suite of gene characterization tools, and made available as a part of the JGI-IMG data warehouse (Markowitz et al., 2014). The genome of DRI-13T was released to the public as per JGI protocol and is available both through the DOE IMG platform and via NCBI (Accession# PRJNA234897). The full length 16S rRNA gene sequence was submitted to NCBI and assigned accession number KR014122.
For SEM, the samples were moved to a 13 mm Swinney micofilter holder with 0.2 μm Millipore filters and dehydrated. Dehydration of the cells was done with 25% increases of ethanol for 15 min each step from 25 to 95%, then three washes of 100%. Once dehydrated, the filters were critical point dried with a Tousimis Samdri CPD (Tousimis, Rockville MD, United States), mounted on aluminum stubs and coated with approximately 25 nm layer of gold with a SPI sputter coater (SPI supplies, West Chester, PA, United States). Images were taken on a Zeiss 1450EP SEM at 10 kV.
Cells for transmission electron microscopy (TEM) and scanning electron microscopy (SEM) were aseptically removed from anaerobic serum bottles and immediately fixed with 2% v/v glutaraldehyde in 0.1 M sodium cacodylate buffer for 1 h at 4°C. Cells were pelleted by centrifugation (10,000 × g for 10 min) between all steps and then re-suspended each time. The sample was then rinsed with 0.1 M sodium cacodylate buffer for 10 min each for three times and re-suspended in 1% OsO4 in 0.1 M sodium cacodylate buffer for an hour at 4°C, then rinsed in distilled water for 10 min twice. TEM dehydration of the cells was performed with 25% increases of ethanol for 15 min each step from 25 to 95%, then three washes of 100%. Samples were then infiltrated with Spurr’s low viscosity resin (EMS, Hatfield, PA, United States), with increasing concentrations of 100% ethanol and resin. After three changes of 100% resin and 1 h between each change, the samples were then placed in a 70°C oven for polymerization for 12 h. The resin was cut into 50–70 nm thick sections on a MT-X ultramicrotome (RMC, Tuscon, AZ, United States) and images were obtained on a JEOL 1011 TEM (JEOL, United States, Peabody, MA, United States) operating at 80 kV and equipped with an AMT camera system.
For analysis of cellular fatty acids, biomass was harvested from cells grown in fumarate-amended (10 mM) mineral media for 72 h at 55°C. Analysis was performed at the Center for Microbial Identification and Taxonomy (University of Oklahoma). Fatty acid methyl esters were extracted, separated, and analyzed using the Sherlock Microbial Identification System (MIDI) version 6.1 as described previously (Kampfer and Kroppenstedt, 1996; Sasser, 2001). Analysis was performed with an Agilent Technologies 6890N gas chromatograph equipped with a phenyl methyl silicone fused silica capillary column (HP-Ultra 2, 25 m × 0.2 mm × 0.33 μm film thickness) coupled with a flame ionization detector. Hydrogen was used as the carrier gas. The temperature program was preset at 170°C and increased at 5°C min–1 to a final temperature of 270°C. The identification and relative abundance of each fatty acid was expressed in terms of the percentage of total fatty acids using the QTSA1 database.
The physiological and geochemical characteristics of the water samples from the pump test of U-3cn#5 on March 29, 2011 are presented in Table 1. Borehole water was fresh (845 μS/cm or 540 ppm TDS) (Kang and Jackson, 2016), slightly alkaline (pH 7.7), and was 44.7°C, as recorded at the surface. The dissolved oxygen (DO) was measured at 0.19 mg/L (3.2% of saturation), with no detectable sulfide. Reduction/oxidation potential was not measured. Total inorganic carbon (TIC) was 50.3 mg C/L, which at this pH would have been mostly bicarbonate. Major ions were defined by sodium > calcium > sulfate > silica (53.5, 37.2, 36, and 35 mg/L, respectively), followed by lesser amounts of chloride, magnesium, and potassium. Total metals were dominated by iron (1.57 mg/L), followed by lesser amounts of strontium (248 μg/L), manganese (101 μg/L), zinc (10.9 μg/L), and molybdenum (4.8 μg/L). Inorganic nitrogen was dominated by ammonium and nitrate, which were detected at 0.05 mg/L and 0.26 mg/L; while nitrite was below the 0.01 mg/L detection limit. Phosphate was detectable at 0.033 mg/L. Total organic carbon (TOC) was present at 1.5 mg/L, a substantial proportion of which is represented by the short chain fatty acids formate, acetate, and lactate, measured at 0.65, 0.4, and 0.4 mg/L, respectively. Propionate and fumarate were below detection limits. Tritium, a marker for the impact of nuclear device testing, was <7 pCi/L. Stable isotope signatures of d13C for dissolved inorganic carbon (DIC) of −12.8‰, and d 2H of -108‰ were detected as well.
Cell enumeration indicated a total planktonic cell density in borehole water of 1.1 × 104 cells/mL. Of these, a substantial proportion (∼half, 4.2 × 103 cells/mL) were reported as viable based on the output of a flow cytometric Live/Dead assay. To gain a better understanding of microbial community structure in these samples, total DNA from 0.2 micron filters was amplified by PCR and used to construct bacterial and archaeal 16S rRNA gene libraries. A total of 84 complete (∼1400 bp) bacterial sequences were constructed and consolidated into 23 representative taxa that were submitted to NCBI (Accession #:MK682771-MK682793). A total of 35 partial (∼850 bp) archaeal sequences were consolidated into 3 representative taxa that were also submitted to NCBI (Accession #: MK675956-MK675958). Bacterial communities were dominated by Proteobacteria (81% - a mix of Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, and Gammaproteobacteria), Acidobacteria (11%), and Firmicutes (8%) (Supplementary Figure S1). Among the archaea, clones were distributed between Euryarchaeota (71%) predominantly a species related to Methanothermobacter thermautotrophicus and Methanococcus igneus, and Crenarchaeota (29%) (Supplementary Figure S2). Close relatives of strain DRI-13T were not detected in these libraries.
Strain DRI-13T was purified from a 45°C anaerobic enrichment in a synthetic groundwater medium supplemented with 0.1% wt/vol peptone as the sole carbon and energy source. After 3 weeks of isolation techniques (see methods), the DRI-13T colonies were 0.5 mm in diameter, circular (with entire margins), glossy, and convex. The resulting isolate was a Gram-stain-positive bacillus (∼2–6 μm long and ∼0.5 μm wide) which reached a density of ∼106 cells/mL in culture. The strain was motile and was observed by phase contrast microscopy to form a central endospore. Phylogenetic analysis of the full-length 16S rRNA gene (GenBank accession no. KR014122) indicated that DRI-13T possessed 90.0% similarity to Pelotomaculum propionicicum (Imachi et al., 2007) and P. thermopropionicum (Imachi et al., 2002) and 88.0% similarity to Desulfotomaculum gibsoniae (Kuever et al., 1999). The most closely related 16S rRNA gene sequences in available databases affiliated with as-yet uncultured bacteria discovered from subsurface environments in Denmark (99.0% similarity, AY538172.1), Australia (97.0% similarity, EU400652.1), and Japan (97.0% similarity, AB910322.1). By comparing the 16S rRNA gene sequence to selected type strains representing all families under the order Clostridiales, it appears strain DRI-13T is a previously undescribed genus within the family Peptococcacea (Figure 2).
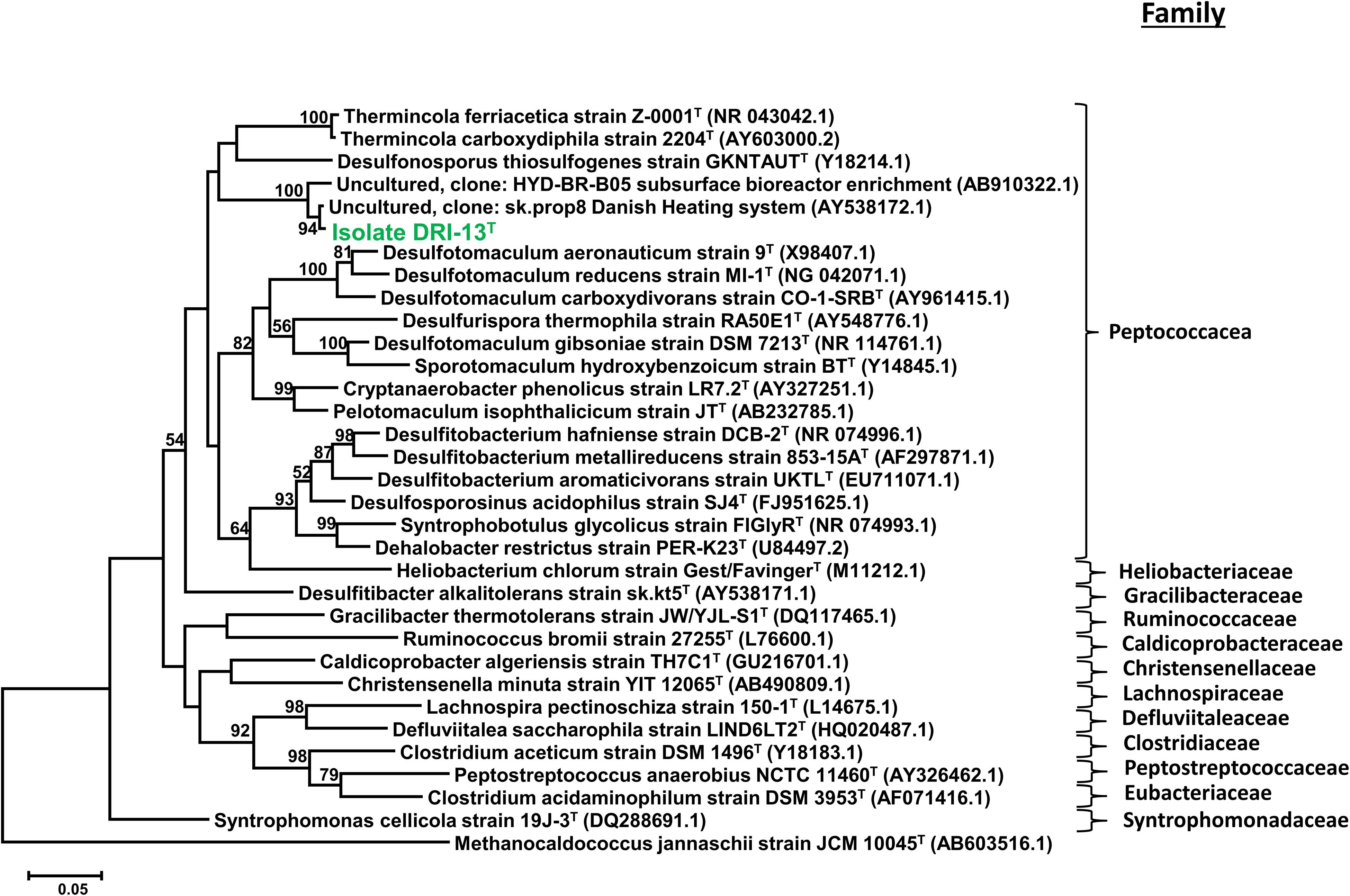
Figure 2. Taxonomic placement of isolate DRI-13T by full length 16S rRNA genes aligned by SILVA (v1.2.11), and maximum-likelihood method based on 1,000 permutations of all bacterial families with nomenclature standing within the class Clostridiales. The scale bar represents 5% identity dissimilarity between sequences.
Analysis by SEM of isolate DRI-13T revealed a distinctive thin rod-shaped morphology characterized by an average length of 4–6 μm and 0.5 μm width (Figure 3A). TEM revealed a distinct inner and outer membrane with an electron-dense matrix and what appears to be a vacuole (Figure 3B).
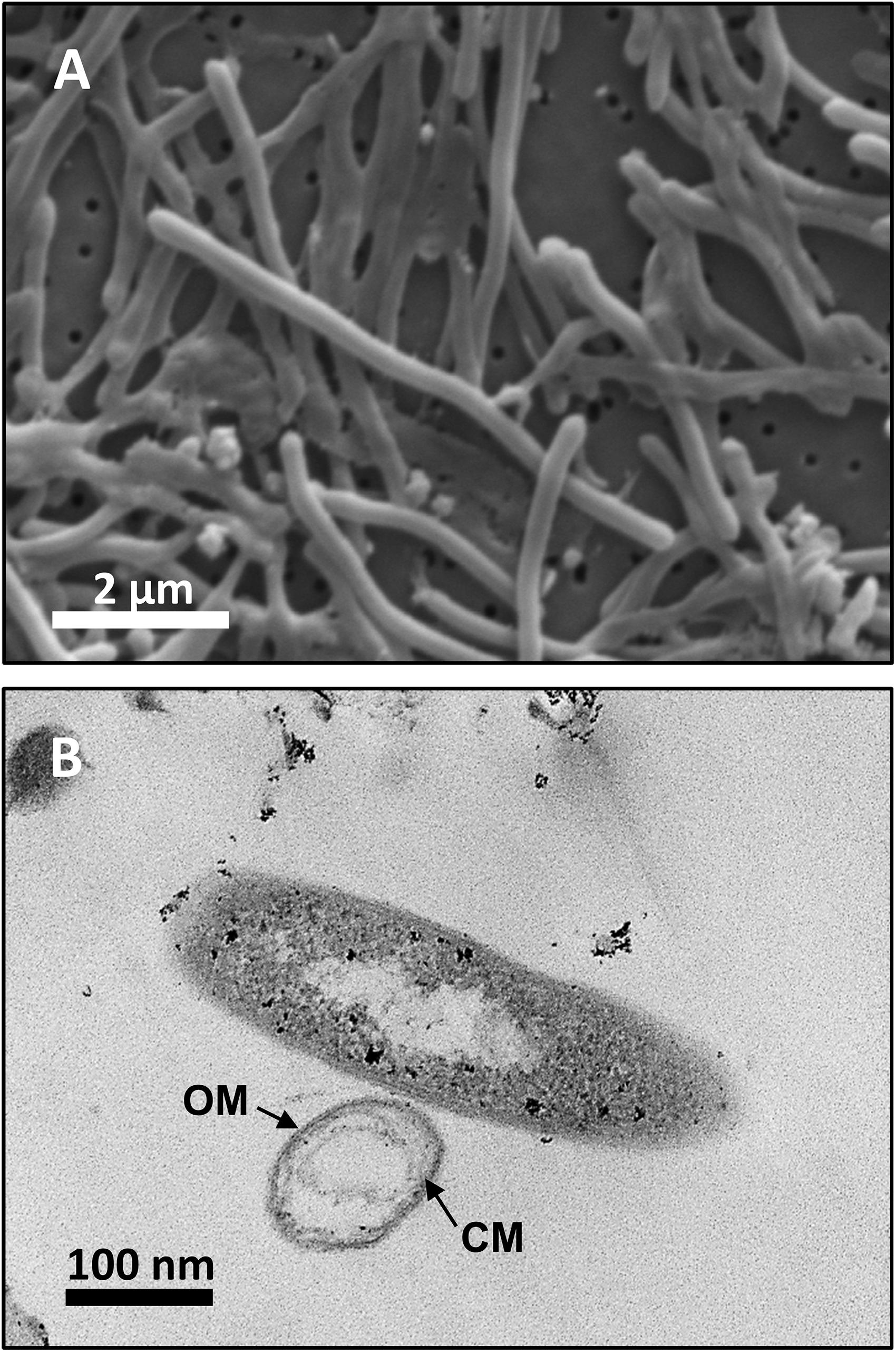
Figure 3. Microscopy images of strain DRI-13T: (A) scanning electron micrograph showing average 4–6 μm length morphology; (B) transmission electron micrograph showing outer membrane (OM) and cytoplasmic membrane (CM).
Strain DRI-13T is a fastidious obligate anaerobe. The isolate grew poorly with 0.1% wt/vol casamino acids, 10 mM glucose, 0.1% wt/vol peptone, or 0.1% wt/vol yeast extract when used as the sole carbon and energy source. No growth was observed when supplemented with H2/CO2 (80:20), CO, formate, lactate, acetate, pyruvate, CH4, methanol, propionate, ribose, xylose, oxaloacetate, succinate, tartarate, butyrate, malate, citrate, aspartate, methionine, and valine. Stickland reactions of alanine/glycine, leucine/glycine, and glutamate/glycine also failed to support growth (Nisman, 1954). Likewise, accessory electron acceptors including nitrite, nitrate, sulfate, sulfite, thiosulfate, and elemental sulfur failed to stimulate growth. The only substrate tested that was successful in supporting growth was fumarate (Table 2).
Specific growth rates of DRI-13T during anaerobic respiration on fumarate display a maximal specific growth rate of 0.4 h–1 under optimal conditions of temperature and pH (55°C and pH 8.0) (Figures 4A,B). Metabolic analysis of DRI-13T shows growth vs. substrate utilization experiment (30 mM fumarate) performed over 70 h (Figure 5). This experiment revealed an initial lag phase of 15–20 h, after which cellular growth commenced along with consumption of fumarate, reaching the lower limit of detection at 65 h. Temporally corresponding to the depletion of fumarate was the coincident production of succinate and acetate, which reached maxima at about 50 h. These experiments revealed a conversion rate of one equivalent of fumarate to approximately 0.83 and 0.33 equivalents of succinate and acetate.
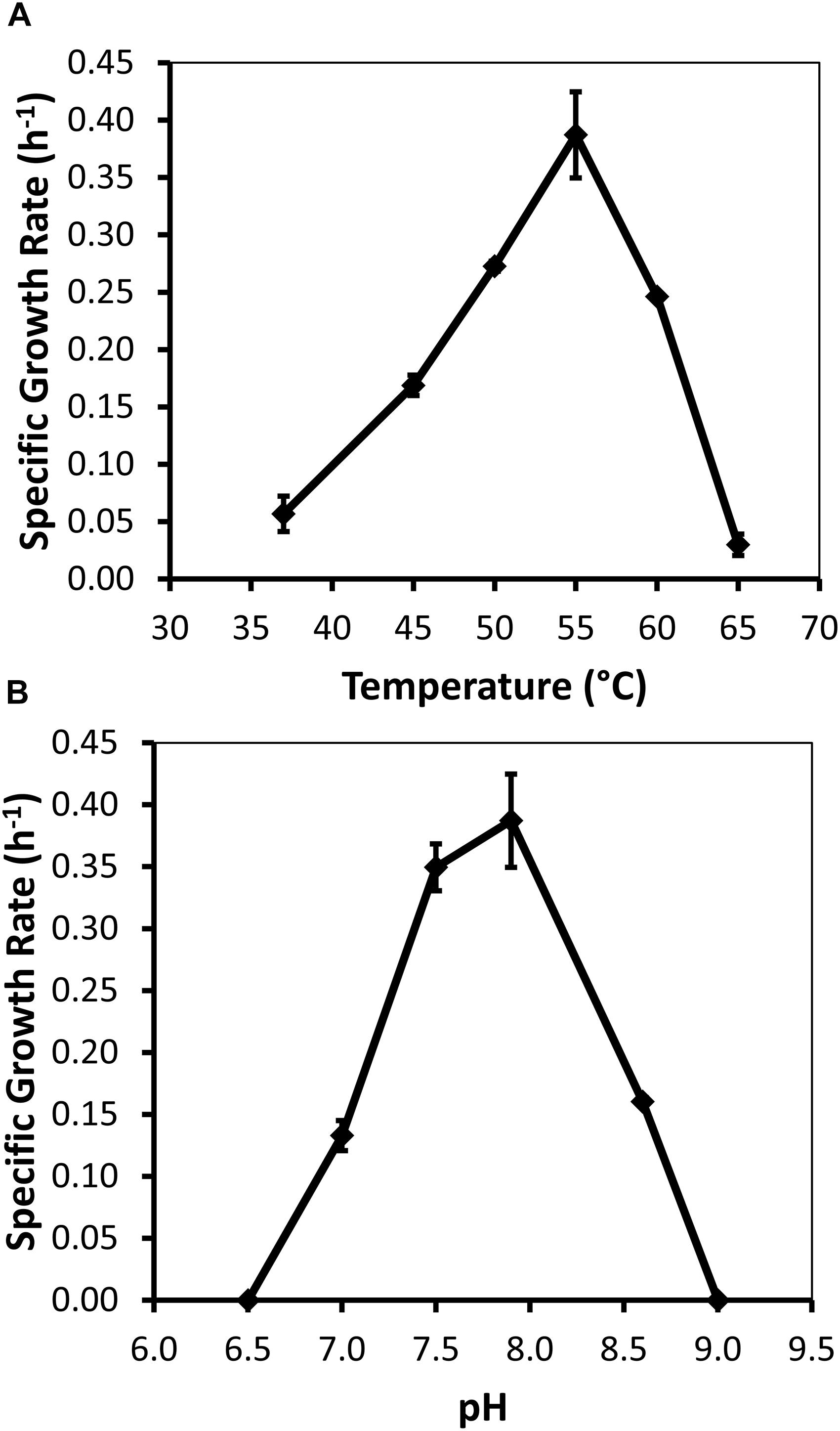
Figure 4. Growth characteristics of strain DRI-13T for: (A) optimal temperature of 55°C; and (B) optimal pH of 8. Data points averaged from quadruplicate cultures using a standardized medium of salts with 10 mM fumarate.
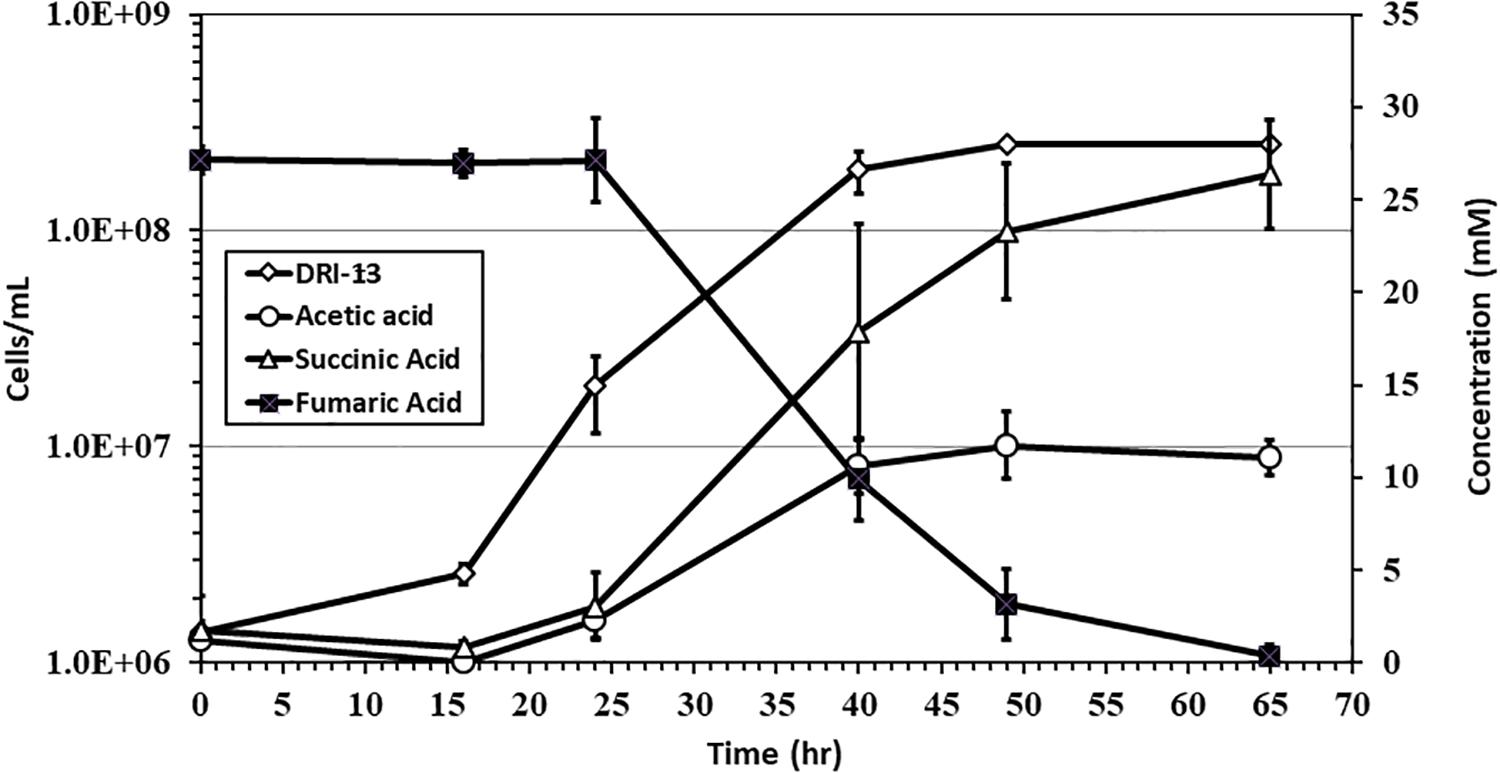
Figure 5. Growth of DRI-13T on 30 mM fumarate and metabolic end products: open diamonds, cell density; filled squares, fumarate depletion; filled triangles, succinate production; filled circles, acetate production. Each data point is an average of triplicate samples with standard deviation error bars.
Lipid analysis indicates that the major fatty acids (>10%) present for strain DRI-13T are the branched-chain fatty acid iso-C15:0 (22.8%), straight-chain C15:0 (15.0%), C16:0 (12.0%), and C14:0 (11.6%), as compared to phylogenetically related bacteria (Table 3).
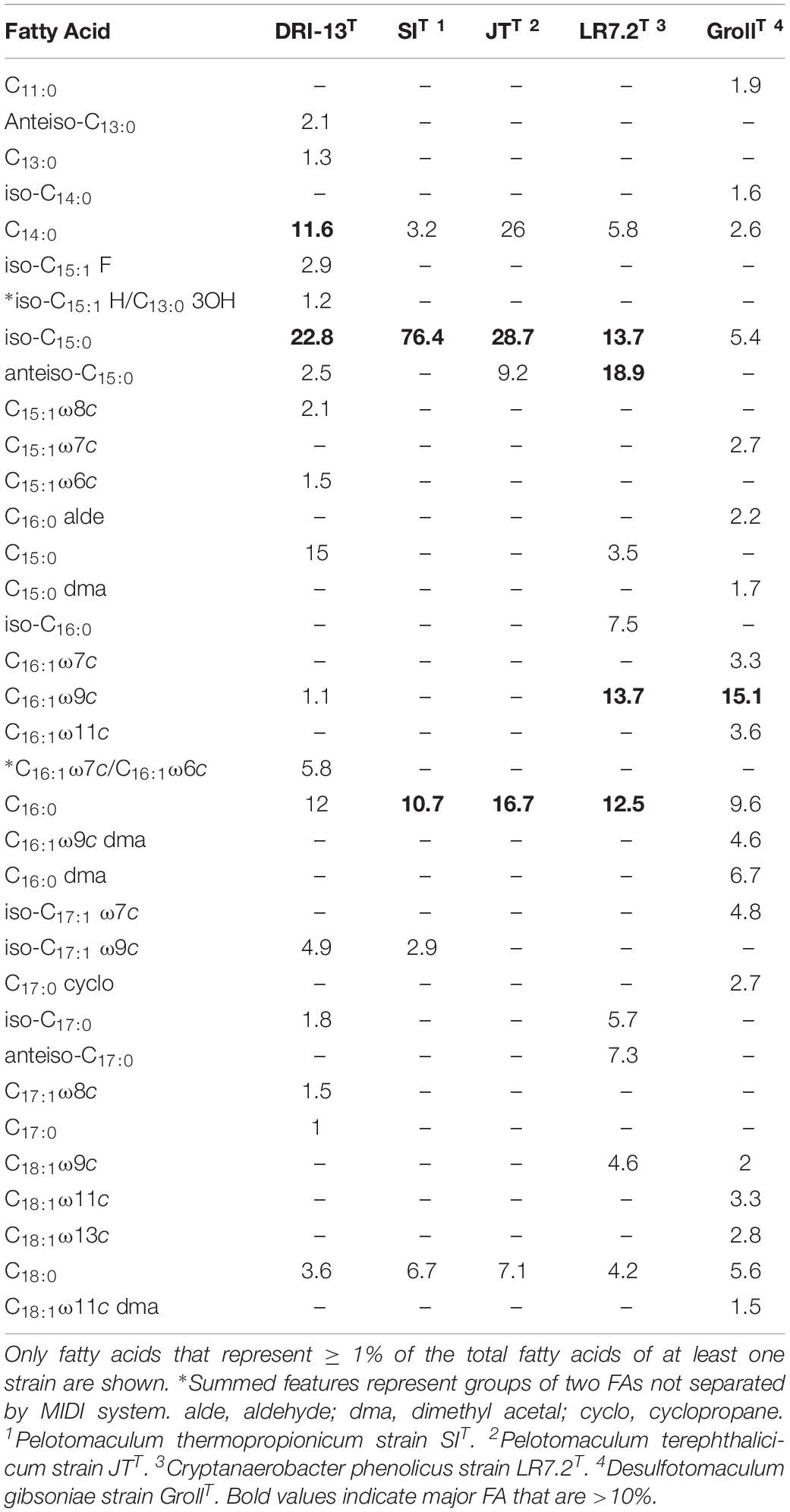
Table 3. Cellular fatty acid composition (%) of strain DRI-13T compared with its closest phylogenetic neighbors.
High-quality genomic DNA was extracted from isolate DRI-13T for genome sequencing by the DOE Joint Genome Institute (JGI, JGI GOLD Gi0054453, NCBI Taxonomic ID 1449126, and Assembly #ASM74602v1). The completed draft genome of DRI-13T is 3,649,665 bp, with an average G + C content of 45% and composed of 105 contigs (Supplementary Table S1). A total of 3,749 genes were identified, of which 3,671 (98%) are predicted to code for proteins (PCGs). Predicted function could be assigned to 2874 (77%) of the total number of PCGs, while 797 (21%) had no known functional prediction2. Genes were identified for assimilatory/dissimilatory sulfate and sulfite reduction, anaerobic fumarate respiration, tripartite ATP-independent secondary passive transport, endospore formation/germination, and flagella biosynthesis (Supplementary Table S2).
Within North America’s Basin and Range (B&R) physiographic province lies the Great Basin (Grayson, 2011), a hydrogeologically defined, internally draining province that comprises portions of the US states of California, Oregon, Idaho, Arizona, and most of Nevada. Fault-controlled flow of deep groundwater within this tectonic extensional zone results in hydrographic basins and sub-basins (Winograd and Thordarson, 1975, Winograd et al., 2005), with the subject of this study, the Death Valley Regional Flow System (DVRFS), being the largest. Borehole U-3cn#5 (Figure 1) intercepts a hydrologically confined lobe of the DVRFS’s and represents a novel window for deep biosphere study of the Basin and Range and extensional zones in general.
The fracture carbonate-hosted aquifer from which DRI-13T was isolated possesses chemistries (e.g., circumneutral pH, relatively high sulfate, low TOC) and overall microbial community structure typical of subsurface fractured carbonate aquifers studied elsewhere (Table 1) (Hohnstock-Ashe et al., 2001; Farnleitner et al., 2005; Gihring et al., 2006; Pfiffner et al., 2006; Park et al., 2009). Nutrients, fixed organic carbon, and energy resources are all relatively limited in this environment. As with other fractured carbonate aquifers, it is unclear how microbial populations are sustained, especially with regard to carbon sources (Hohnstock-Ashe et al., 2001). In U-3cn#5, rRNA gene libraries reveal a microbial community composed of predicted thermophilic anaerobes from the domain Bacteria and Archaea (Supplementary Figures S1, S2). The geothermal/carbonate-controlled nature of this subsurface environment makes it a distinct environment compared to surrounding monitoring wells, which are emplaced in shallower lower temperature systems and other rock types (e.g., volcanics and alluvium), with one exception: borehole BLM-1. Located approximately 75 km South of U-3cn#5 in the discharge zone of the DVRFS, this latter well intersects anoxic geothermal waters (57°C) of the regional fractured carbonate aquifer. In spite of this distance, these two wells share similar bacterial and archaeal profiles, including the presence of important marker subsurface bacteria such as “Candidatus Desulforudis audaxviator,” Desulfotomaculum putei, and the archaeon Methanothermobacter thermautotrophicus (Chivian et al., 2008; Thomas et al., 2013; Sackett, 2018).
Enrichment cultivations from U-3cn#5 water samples in a strictly anaerobic minimal salts medium amended with peptone produced a slowly growing thermophilic bacillus after ∼1 month at 50°C. Sugar and short chain carbohydrate substrates did not promote growth (Table 2). While not directly detected in the subsurface environment of U-3cn#5, the citric acid cycle intermediate fumarate was the only substrate that enabled routine culturing of the microorganism. Isolation techniques yielded a pure culture which was given the strain name, DRI-13T. Analysis of the 1463 nucleotide 16S rRNA gene sequence revealed that DRI-13T affiliates at the species level with uncultured clones from other geothermally influenced subsurface locations including Japan, Australia (unpublished), and Denmark (AB910322.1, EU400652.1, AY538172.1) (Kjeldsen et al., 2007; Baito et al., 2015). Within the family Peptococcaceae, DRI-13T is most closely related (∼90.0% similarity to the 16S rRNA gene) to three type strains Pelotomaculum propionicicum, P. thermopropionicum, and P. isophthalicicum. This establishes DRI-13T as a novel genus using the parameters defined by Yarza et al. (2014) (Figure 2). Of these phylogenetic relatives, only P. thermopropionicum share thermophilic characteristics and utilization of fumarate (Table 2). Unlike DRI-13T, P. thermopropionicum can utilize a wider range of substrates and is described under certain conditions to support a syntrophic interaction with the methanogen Methanothermobacter thermoautotrophicus. The most abundant archaeon in our U-3cn#5 rRNA gene libraries was M. thermoautotrophicus (71% of reads). P. propionicicum and P. isophthalicicum have been reported to have syntrophic interactions with other methanogens as well, though no interactions were established between DRI-13T and an isolated wild type M. thermoautotrophicum from the borehole. Growth combinations only resulted in DRI-13T becoming the majority within the culture bottle and causing no change in growth performance with or without M. thermoautotrophicum (not shown).
Examination of DRI-13T by SEM indicates a bacillus-type morphology, averaging 6 μm in length and 0.5 μm width (Figure 3A). TEM reveals features of the outer membrane and cytoplasmic membrane, including what appears to be a vacuole (Figure 3B), though it is possible that the space within the cell may be an artifact from TEM preparation. If real, the presence of a vacuole may explain an apparent buoyant property of DRI-13T cells requiring centrifugation times greater than 30 min at or above 15,000 × g, which is typical for very small cells or for cells with natural buoyancy. Lab-standard cells from the phylum Firmicutes, such as Desulfotomaculum putei, can be pelleted with less rigorous effort similar to E. coli. While not a feature noted in other subsurface microorganisms, it is conceivable that innate buoyancy control could be advantageous to fracture-dwelling subsurface microorganisms during planktonic growth.
Measureable DRI-13T growth was observed to occur between 37°C (minimum) and 65°C (maximum). While the environmental temperature of U-3cn#5 was recorded at 44.7°C, the optimal temperature for DRI-13T is 55°C (Figure 4A). Although not determined, the in situ temperature from the interval where DRI-13T was obtained was likely higher than that recorded at the wellhead due to the long distance pumped water was lifted through presumably cooler conditions in the aquifer above the confining layer and ∼500 m of vadose zone. As might be expected for an organism adapted to strongly buffered carbonate aquifers, the pH tolerance of DRI-13T is narrow, with observable growth occurring only between 7.0 and 8.5, and optimal growth at pH 8.0 (Figure 4B). As compared to close relatives, the optimal temperature is similar to P. thermopropionicum and P. propionicicum (55°C and 37°C). The growth rate of DRI-13T appears to be substantially faster, doubling every 2.5 h, as opposed to 14.5 and 120 h for P. propionicicum and P. thermopropionicum, respectively (Imachi et al., 2002, 2007). This faster growth rate possibly reflects the independent lifestyle of DRI-13T as opposed to the syntrophic associations of the others. In addition, cells cultured beyond stationary phase (approximately 48 h) exhibit endospore formation, as observed by microscopy and the presence of endospore related genes (Supplementary Table S2). While no discernable cellular movement was consistently observed, DRI-13T genome appears to have flagella related genes as well (Supplementary Table S2).
Lipid analysis indicates that the major fatty acids (>10%) for strain DRI-13T are the branched-chain fatty acid iso-C15:0 (22.8%) along with the straight-chain C15:0 (15.0%), C16:0 (12.0%) and C14:0 (11.6%). The higher proportion of the branched-chain iso-C15:0 in strain DRI-13T is also characteristic of its nearest neighbor, P. thermopropionicum strain SIT in which iso-C15:0 represents the majority of all fatty acids at 76.4% (Imachi et al., 2002), and in P. terephthalicicum strain JTT (28.7%) (Qiu et al., 2006), Cryptanaerobacter phenolicus strain LR7.2T (Juteau et al., 2005), and Desulfotomaculum gibsoniae GrollT (Kuever et al., 1999) (Table 3). While the overall fatty acid methyl ester (FAME) profile for strain DRI-13T clearly differentiates it from its nearest phylogenetic neighbors, the significant percentage of the straight-chain fatty acid C15:0 is a defining characteristic, as it is not present in other closely related species, with the exception of C. phenolicus strain LR7.2T, in which it is only a minor component (Juteau et al., 2005). Interestingly, C15 lipids were not detected in any of 44 fracture and mine water samples from a comprehensive survey of environmental lipids in the ultradeep mines of South Africa (Pfiffner et al., 2006). Another defining characteristic is the detection of C16:1ω 7c/C16:1ω 6c (5.8%), which is not present in other phylogenetic neighbors such as C. phenolicus strain LR7.2T (Juteau et al., 2005), and D. gibsoniae strain GrollT (Kuever et al., 1999). C16:1ω 7c was detected in the South African study, but statistically corresponded with mine service water samples rather than pristine fracture water.
Fumarate catabolism was typical as described previously in other Firmicutes (Dorn et al., 1978; Imachi et al., 2002). High-pressure liquid chromatography (HPLC) detected the removal of fumarate and the concomitant production of succinate and acetate. This supports the idea that fumarate is both being metabolized and used as an electron acceptor in anaerobic respiration (Figure 5). No other observable products such as oxalate, tartrate, lactate, malate, citrate, or propionate were observed by HPLC. Headspace pressure in culture bottles remained stable (∼1 bar) with no obvious over-pressurization. The source of fumarate in the subsurface is unclear. Gelling agents associated with drilling can be a source of fumarate (Borchardt, 1989). However, in this case residue from the drilling of U-3cn#5 may be unlikely given the borehole age (1965) and its flushing with multiple millions of liters of water pumped over the years. Samples of 1 g fractured carbonate from U-3cn#5 (NNSS Core Library), aseptically crushed, extracted with 5 mL boiling water, concentrated, and analyzed on HPLC, did not contain detectable levels of fumarate (data not shown). It is more likely that DRI-13T is cryptically biodegrading the waste products or endogenous decay derived from other members of the microbial community (Béranger et al., 2006). Since DRI-13T does not seem to be a dominant microbial community member (Supplementary Figure S1), it is possible that its relatively low representation is the result of occupying a narrow niche focusing on fumarate respiration or is a relic from a period after drilling when fumarate might have been present.
Assembly of the DRI-13T draft genome from a single library prep resulted in 105 contigs. Complete coverage of the genome failed in regions where repetitive intergenic sequences (mobile elements), hypothetical proteins, or clustered regularly interspaced short palindromic repeats (CRISPR) arrays were present. The estimated genome size is 3,649,665 with a G + C content of 45%. This is comparable to close relatives with sequenced genomes such as D. gibsonia (4.85 Mbp) and P. thermopropionicum (3.02 Mbp). Distribution of nucleotides and genes across the 105 contigs range from 1,045 to 193,268 bases (average 34,759 bases per contig), with 2 to 203 annotated genes (average 36 genes per contig). The JGI/IMG annotation gene calling algorithms predict 3,749 genes in DRI-13T with 77% of them functionally assigned (Supplementary Table S1). Analysis of the metabolic capability of the DRI-13T genome indicates core genes for glycolysis/gluconeogenesis, fatty acid biosynthesis, and amino acid metabolism. Homologs for assimilatory or dissimilatory sulfate and sulfite reduction (dsrA, dsrB, and dsrD) are present. However, sulfide production was never detected when sulfate was present in the media. Amino acid analysis of the dsr genes show an identity of 84% to 99% similarity to Desulfitobacter alkalitolerans, which was isolated from a Danish geothermal heating system, a site where an uncultured 16S rRNA gene with exact homology to DRI-13T had been sequenced (Nielsen et al., 2006; Kjeldsen et al., 2007).
Examination of the anaerobic fumarate respiration pathway revealed that core fumarate catabolic and related genes are present within the genome. Some of the genes for fumarate respiration have undergone duplication generating paralogs (Supplementary Table S2). The metabolic pathway for fumarate respiration converts three fumarate molecules to one acetate and two succinate molecules (Supplementary Figures S3A,B). The gene for the enzyme fumarase is present in the genome and can reversibly convert fumarate by hydration to malate or malate to fumarate by dehydration. The decarboxylation of the malate molecule to pyruvate is completed by NADP dependent malate dehydrogenase (oxaloacetate-decarboxylating), generating carbon dioxide and reductive potential. The further oxidation of pyruvate to acetyl-CoA is accomplished by pyruvate ferredoxin oxidoreductase, generating carbon dioxide and reduced ferredoxin (Thauer et al., 1977). The final reduction of acetyl-CoA with succinate to acetate occurs by succinyl-CoA:acetate CoA-transferase. Concurrently, two molecules of fumarate can be reduced to succinate by succinate dehydrogenase which was also annotated. Generation of ATP by coupling substrate-level phosphorylation is achieved by succinyl-CoA synthetase. DRI-13T generates a proton motive force for ATP synthesis through a NADH ubiquinone-oxidoreductase (Nuo Complex 1) and membrane bound Ni-Fe hydrogenase-4 (Marreiros et al., 2013; Schut et al., 2016; Yu et al., 2018). The FPOF complex allows versatility of energy conservation among anaerobes and is perhaps a recent adaptation to environments rich in sulfide and at an interface with oxygen (e.g., buoyancy), similar to water chemistry conditions found in monitoring well U-3cn#5 (Schut et al., 2016). It is not known what molecule transports electrons to the hydrogenase-4 Ni-Fe complex. Interestingly, seven copies of electron transfer flavoprotein (EtfAB) and quinone oxidoreductase (FixC) were annotated within the genomes, suggesting capacity for high potential electron transfers in bifurcating energy conservation (Costas et al., 2017).
Secondary transporters dependent on substrate binding are ubiquitous in prokaryotes yet poorly characterized. Tripartite ATP-independent periplasmic (TRAP) transporters manage the passive transport of hydrocarbon molecules across the membrane for the cell to use as a source of carbon, electrons, and energy. Isolate DRI-13T has fourteen TRAP 4TM/12TM genes present in its genome, six of which have fused transmembrane subsunits (Supplementary Table S2). TRAP transporters are proposed to be ‘ancient’ secondary transporters that are anchored in the membrane, taking advantage of sodium ion gradients to co-transport 4 carbon molecules from outside the cell wall into the cytoplasm (Rabus et al., 1999; Kelly and Thomas, 2001; Mulligan et al., 2011). The TRAP transporter’s DctP subunit is seemingly specific to particular solutes (Fischer et al., 2015). DRI-13T’s almost exclusive utilization of the dicarboxylate molecule fumarate seems complimentary to the TRAP transport system. It has been demonstrated that the psychrophilic microorganism Psychrobacter arcticus 273-4, exhibited lower growth rates when its TRAP genes were genetically knocked out and afterward fed fumarate (Bakermans et al., 2009). DRI-13T has 41 TRAP annotated transporter genes, compared with only three TRAP genes found in P. thermopropionicum. As for annotated genes, 10.4% of DRI-13T genome fall under the category of transporter (390/3749 genes) as compared to 8.6% (259/3018) for P. thermopropionicum. Three of the fusion TRAP proteins in DRI-13T genome (WP_034425417.1, WP_051965735.1, and WP_51966221.1) have 41-47% amino acid identity to homologs in the deep subsurface bacterium Ca. D. audaxviator (Chivian et al., 2008). The ‘investment’ of DRI-13T genome with ATP independent secondary transporters potentially reveals a methodology of how subsurface microbes manage minimal energy metabolic activities in order to survive in oligotrophic environments. This is seemingly true for Alphaproteobacteria SAR11 that were sequenced from the Sargasso Sea, where TRAP transporters were found to be abundant (Morris et al., 2002; Mulligan et al., 2011).
Spore formation and germination genes were annotated in the genome as well (Brown et al., 1994; Browne et al., 2016). Isolate DRI-13T appears to have 37 spore-related genes (Supplementary Table S2), as compared to microorganisms whose closest amino acid similarities to the GerA gene are Bacillus camelliae, which possesses 77 spore-related genes, and Desulfonispora thiosulfatigenes, which has 29 spore-annotated genes. Interestingly, the number of spore-related genes does not appear to differentiate microorganisms originating from surface and subsurface environments. For example, subsurface microorganisms such as Desulfotomaculum putei (26 spore-related genes), Ca. D. audaxviator (8 spore-related genes), and Bacillus subterraneus (64 spore-related genes) can be compared to surface microorganisms such as Caldicellulosiruptor obsidiansis (11 spore-related genes), and Clostridium difficile BI1 (17 spore related genes).
The thermophilic, anaerobic, fumarate-respiring bacterium DRI-13T was successfully isolated from geothermal water from the Lower Carbonate Aquifer (LCA) of the Death Valley Regional Flow System (DVRFS) collected from fractures at 863 – 923 mbls. Physiological and molecular analysis of strain DRI-13T revealed it to be a new species within a novel genus of the Family Peptococcaceae. Microbial community analysis via 16S rRNA gene libraries of Archaea and Bacteria reveal a community consistent with other anoxic, rock-hosted subsurface aquifers. However, DRI-13T was not present in these samples as a major community member. This study represents one of the first descriptions of indigenous life from the LCA of the DVRFS, and DRI-13T is the first characterized microorganism from this deep subsurface extensional zone habitat. The apparent obligate reliance upon fumarate by this organism is relatively unusual and of uncertain significance in a deep biosphere context. Here we introduce DRI-13T as a representative microorganism from the terrestrial deep biosphere and propose the name Thermoanaerosceptrum fracticalcis DRI-13T gen. nov. sp. nov.
Thermoanaerosceptrum gen. nov. (Ther.mo.an.a.e.ro.scep’trum. Gr. adj. thermos, hot; Gr. prefix an-, not; Gr. n. aer air; Gr. neut. n. skeptron staff; N.L. neut. n. Thermoanaerosceptrum a hot anaerobic staff).
Cells are rod-shaped. Cell wall is Gram-positive type. Central endospores are observed. Thermophilic. Obligate anaerobic chemoorganoheterotroph. Fermentation end products are succinate and acetate. The major fatty acids (>10%) were iso-C15:0, C15:0, C16:0 and C14:0.
The type species is Thermoanaerosceptrum fracticalcis.
(frac.ti.cal’cis. L. part. adj. fractus, broken; L. n. calx, -cis, limestone; N.L. gen. n. fracticalcis, of broken limestone, referring to the origin of the type strain).
Cells are long, straight rods averaging 6 μm long and 0.5 μm wide, occurring singly. The cell wall is Gram-positive type, with central endospores observed. Optimal growth temperature is 55°C, with a maximum of 65°C and minimum of 35°C. Optimal pH is 8.0 with a range of 7.0–8.5. Obligate anaerobic chemoorganoheterotroph that utilizes fumarate as a sole carbon source and electron donor/acceptor for growth. Glucose, casamino acids, peptone, and yeast extract can be weakly utilized as electron/carbon donors. Fermentation end products are succinate and acetate. The estimated genome size of DRI-13T is 3,649,665 bp, of which 87.3% are coding regions. The major fatty acids (>10%) were iso-C15:0, C15:0, C16:0 and C14:0.
The type strain is DRI-13T (DSM 100382T = ATCC TSD-12T), which was isolated from a terrestrial deep biosphere aquifer located in the US Great Basin. The DNA G + C content of the type strain is 45.2 mol%.
The datasets generated for this study can be found in the JGI/IMG and NCBI.
SH-B designed the research project, organized the contributions of all other authors, conducted the experiments, collected the data, and led the writing effort. LS screened the first enrichments and initiated the isolation of microorganism. MZ, TO, and BL provided the geochemical data on U-3cn#5. MC and PL performed and interpreted the lipid analysis. JG and IN performed HPLC and submitted the Illumina genome sequencing analysis. CR contributed to the experimental design and provided the hydrogeological and NNSS logistical expertise. DM secured the site access, collected the samples, secured the major funding, and co-wrote the manuscript.
This work was supported by the Department of Energy grant #DE-PS02-09ER09-07, NASA Astrobiology Institute Cooperative Agreement NNA13AA92A, U.S. National Science Foundation grant DEB-1441717, startup funds for SH-B from Southern Illinois University Carbondale, and JGI Project ‘Genome analyses of microbes integral to the cycling of sulfate and iron’ IDs: Gs0014904 and Gp0046988. Work at Lawrence Livermore National Laboratory (LLNL) was supported by the Subsurface Biogeochemical Research Program of the U.S. Department of Energy’s Office of Biological and Environmental Research (SCW1053).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Russ Shelton, David Finnegan, Jeff Wurtz, and others associated with the DOE UGTA program for logistical support and Bill Wilborn for site access. Special thanks to Brenna R. Hardtner from Henderson Foothill High School for providing the founding scientific name for DRI-13T, and to Craig Rosen from DRI’s Green Power Project for facilitating the interaction with Southern Nevada High Schools. Thanks to Dr. John Shields from the University of Georgia for the SEM and TEM images, Dr. Bernhard Schink from the University of Konstanz for assisting with the proper Greek and Latin name description, Dr. Gerti Schut for the insightful discussions about hydrogenases, and Jenny C. Fisher for assisting with the collection of the U-3cn#5 samples. We also thank Beverly Parker and the reviewers at DOE for the editorial advice and document clearance. Thanks to an undergraduate student David Lynn from the University of Nevada, Las Vegas, NV, United States for the seemingly endless centrifugation for genomic DNA. Work by MZ was prepared by LLNL under contract DE-AC52-07NA27344.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02224/full#supplementary-material
Amy, P., Haldeman, D., Ringelberg, D., Hall, D., and Russell, C. (1992). Comparison of identification systems for classification of bacteria isolated from water and endolithic habitats within the deep subsurface. Appl. Environ. Microbiol. 58, 3367–3373.
Baito, K., Imai, S., Matsushita, M., Otani, M., Sato, Y., and Kimura, H. (2015). Biogas production using anaerobic groundwater containing a subterranean microbial community associated with the accretionary prism. Microb. Biotechnol. 8, 837–845. doi: 10.1111/1751-7915.12179
Bakermans, C., Sloup, R. E., Zarka, D. G., Tiedje, J. M., and Thomashow, M. F. (2009). Development and use of genetic system to identify genes required for efficient low-temperature growth of Psychrobacter arcticus 273-4. Extremophiles 13, 21–30. doi: 10.1007/s00792-008-0193-3
Bangerter, R. M., and Giblin, M. O. (1998). Recompletion Report for BILBY. Nevada Environmental Restoration Project. Springfield, VA: National Technical Information Service.
Bastin, E. S., and Greer, F. E. (1930). Additional data on sulphate-reducing bacteria in soils and waters of Illinois oil fields. AAPG Bull. 14, 153–159.
Belcher, W. R., Bedinger, M. S., Back, J. T., and Sweetkind, D. S. (2009). Interbasin flow in the great basin with special reference to the southern funeral mountains and the source of furnace creek springs, Death Valley, California, U.S. J. Hydrol. 369, 30–43. doi: 10.1016/j.jhydrol.2009.02.048
Belcher, W. R., Sweetkind, D. S., and Elliott, P. E. (2002). Probability distributions of hydraulic conductivity for the hydrogeologic units of the Death Valley regional ground-water flow system, Nevada and California. Water Resour. Investig. Rep. 2:4212.
Béranger, S. C., Sleep, B. E., Sherwood Lollar, B., and Brown, A. J. (2006). Isotopic fractionation of tetrachloroethene undergoing biodegradation supported by endogenous decay. J. Environ. Eng. 132, 725–735. doi: 10.1061/(asce)0733-9372(2006)132:7(725)
Boone, D. R., Liu, Y., Zhao, Z.-J., Balkwill, D. L., Drake, G. R., Stevens, T. O., et al. (1995). Bacillus infernus sp. nov., an Fe (III)-and Mn (IV)-reducing anaerobe from the deep terrestrial subsurface. Int. J. Syst. Bacteriol. 45, 441–448. doi: 10.1099/00207713-45-3-441
Borchardt, J. K. (1989). Chemicals Used in Oil-Field Operations. Houston, TX: Westhollow Research Center, Shell Development Company.
Bowen, S., Finnegan, D., Thompson, J., Miller, C., Baca, P., Olivas, L., et al. (2001). Nevada Test Site Radionuclide Inventory, 1951-1992. Los Alamos, NM: Los Alamos National Laboratory.
Brown, D. P., Ganova-Raeva, L., Green, B. D., Wilkinson, S. R., Young, M., and Youngman, P. (1994). Characterization of spo0A homologues in diverse Bacillus and Clostridium species identifies a probable DNA-binding domain. Mol. Microbiol. 14, 411–426. doi: 10.1111/j.1365-2958.1994.tb02176.x
Browne, H. P., Forster, S. C., Anonye, B. O., Kumar, N., Neville, B. A., Stares, M. D., et al. (2016). Culturing of ‘unculturable’human microbiota reveals novel taxa and extensive sporulation. Nature 533:543. doi: 10.1038/nature17645
Bushmann, M., Nelson, S. T., Tingeya, D., and Eggett, D. (2010). Regional groundwater flow in structurally-complex extended terranes: an evaluation of the sources of discharge at Ash Meadows, Nevada. J. Hydrol. 386, 118–129. doi: 10.1016/j.jhydrol.2010.03.013
Chapelle, F. H., O’Neill, K., Bradley, P. M., Methé, B. A., Ciufo, S. A., Knobel, L. L., et al. (2002). A hydrogen-based subsurface microbial community dominated by methanogens. Nature 415, 312–315. doi: 10.1038/415312a
Chivian, D., Brodie, E. L., Alm, E. J., Culley, D. E., Dehal, P. S., Desantis, T. Z., et al. (2008). Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322, 275–278. doi: 10.1126/science.1155495
Colwell, F. S., Onstott, T. C., Delwiche, M. E., Chandler, D., Fredrickson, J. K., Yao, Q. J., et al. (1997). Microorganisms from deep, high temperature sandstones: constraints on microbial colonization. FEMS Microbiol. Rev. 20, 425–435. doi: 10.1016/s0168-6445(97)00024-7
Costas, A. M. G., Poudel, S., Miller, A.-F., Schut, G. J., Ledbetter, R. N., Fixen, K. R., et al. (2017). Defining electron bifurcation in the electron transferring flavoprotein family. J. Bacteriol. 199:e0440-7. doi: 10.1128/JB.00440-17
D’Agnese, F. A., Faunt, C. C., Turner, A. K., and Hill, M. C. (1997). Hydrogeologic Evaluation and Numerical Simulation of the Death Valley Regional Ground-Water Flow System, Nevada and California. Denver, CO: U.S. Geological Survey.
Daumas, S., Cord-Ruwisch, R., and Garcia, J.-L. (1988). Desulfotomaculum geothermicum sp. nov., a thermophilic, fatty acid-degrading, sulfate-reducing bacterium isolated with H2 from geothermal ground water. Antonie Van Leeuwenhoek 54, 165–178. doi: 10.1007/bf00419203
Dorn, M., Andreesen, J., and Gottschalk, G. (1978). Fermentation of fumarate and L-malate by Clostridium formicoaceticum. J. Bacteriol. 133, 26–32.
Farnleitner, A. H., Wilhartitz, I., Ryzinska, G., Kirschner, A. K., Stadler, H., Burtscher, M. M., et al. (2005). Bacterial dynamics in spring water of alpine karst aquifers indicates the presence of stable autochthonous microbial endokarst communities. Environ. Microbiol. 7, 1248–1259. doi: 10.1111/j.1462-2920.2005.00810.x
Fischer, M., Hopkins, A. P., Severi, E., Hawkhead, J., Bawdon, D., Watts, A. G., et al. (2015). Tripartite ATP-independent periplasmic (TRAP) transporters use an arginine-mediated selectivity filter for high affinity substrate binding. J. Biol. Chem. 290, 27113–27123. doi: 10.1074/jbc.M115.656603
Fredrickson, J. K., McKinley, J. P., Bjornstad, B. N., Long, P. E., Ringelberg, D. B., White, D. C., et al. (1997). Pore-size constraints on the activity and survival of subsurface bacteria in a late Cretaceous shale-sandstone sequence, northwestern New Mexico. Geomicrobiol. J. 14, 183–202. doi: 10.1080/01490459709378043
Garber, M. S., and Johnson, R. H. (1967). A Summary of Lithologic Data, Aquifer Tests, and Contruction of Hydraulic Test Well U-3cn#5, Nevada Test Site. Denver, CO: United States Department of the Interior, Geological Survey.
Gihring, T., Moser, D., Lin, L.-H., Davidson, M., Onstott, T., Morgan, L., et al. (2006). The distribution of microbial taxa in the subsurface water of the Kalahari Shield, South Africa. Geomicrobiol. J. 23, 415–430. doi: 10.1080/01490450600875696
Glöckner, F. O., Yilmaz, P., Quast, C., Gerken, J., Beccati, A., Ciuprina, A., et al. (2017). 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 261, 169–176. doi: 10.1016/j.jbiotec.2017.06.1198
Grayson, D. K. (2011). The Great Basin: A Natural Prehistory. Berkeley, CA: University of California Press.
Haldeman, D., and Amy, P. (1993). Bacterial heterogeneity in deep subsurface tunnels at Rainier Mesa, Nevada Test Site. Microb. Ecol. 25, 183–194. doi: 10.1007/BF00177194
Haldeman, D. L., Amy, P. S., Ringelberg, D., and White, D. C. (1993). Characterization of the microbiology within a 21 m 3 section of rock from the deep subsurface. Microb. Ecol. 26, 145–159. doi: 10.1007/BF00177049
Hohnstock-Ashe, A., Plummer, S., Yager, R., Baveye, P., and Madsen, E. (2001). Further biogeochemical characterization of a trichloroethene-contaminated fractured dolomite aquifer: electron source and microbial communities involved in reductive dechlorination. Environ. Sci. Technol. 35, 4449–4456. doi: 10.1021/es0110067
Imachi, H., Sakai, S., Ohashi, A., Harada, H., Hanada, S., Kamagata, Y., et al. (2007). Pelotomaculum propionicicum sp nov., an anaerobic, mesophilic, obligately syntrophic propionate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 57, 1487–1492. doi: 10.1099/ijs.0.64925-0
Imachi, H., Sekiguchi, Y., Kamagata, Y., Hanada, S., Ohashi, A., and Harada, H. (2002). Pelotomaculum thermopropionicum gen. nov., sp nov., an anaerobic, thermophilic, syntrophic propionate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 52, 1729–1735. doi: 10.1099/ijs.0.02212-0
Jorgensen, B. B. (2012). Shrinking majority of the deep biosphere. Proc. Natl. Acad. Sci. U.S.A. 109, 15976–15977. doi: 10.1073/pnas.1213639109
Juteau, P., Côté, V., Duckett, M.-F., Beaudet, R., Lépine, F., Villemur, R., et al. (2005). Cryptanaerobacter phenolicus gen. nov., sp. nov., an anaerobe that transforms phenol into benzoate via 4-hydroxybenzoate. Int. J. Syst. Evol. Microbiol. 55, 245–250. doi: 10.1099/ijs.0.02914-0
Kampfer, P., and Kroppenstedt, R. M. (1996). Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can. J. Microbiol. 42, 989–1005. doi: 10.1139/m96-128
Kang, M., and Jackson, R. B. (2016). Salinity of deep groundwater in California: water quantity, quality, and protection. Proc. Natl. Acad. Sci. U.S.A. 113, 7768–7773. doi: 10.1073/pnas.1600400113
Kelly, D. J., and Thomas, G. H. (2001). The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol. Rev. 25, 405–424. doi: 10.1016/s0168-6445(01)00061-4
Kieft, T. L., Kovacik, W., Ringelberg, D. B., White, D. C., Haldeman, D. L., Amy, P. S., et al. (1997). Factors limiting microbial growth and activity at a proposed high-level nuclear repository, yucca mountain, nevada. Appl. Environ. Microbiol. 63, 3128–3133.
Kjeldsen, K. U., Kjellerup, B. V., Egli, K., Frolund, B., Nielsen, P. H., and Ingvorsen, K. (2007). Phylogenetic and functional diversity of bacteria in biofilms from metal surfaces of an alkaline district heating system. FEMS Microbiol. Ecol. 61, 384–397. doi: 10.1111/j.1574-6941.2006.00255.x
Kotelnikova, S., and Pedersen, K. (1997). Evidence for methanogenic Archaea and homoacetogenic Bacteria in deep granitic rock aquifers. FEMS Microbiol. Rev. 20, 339–349. doi: 10.1016/s0168-6445(97)00016-8
Krumholz, L. R., McKinley, J. P., Ulrich, G. A., and Suflita, J. M. (1997). Confined subsurface microbial communities in Cretaceous rock. Nature 386, 64–66. doi: 10.1038/386064a0
Kuever, J., Rainey, F. A., and Hippe, H. (1999). Description of Desulfotomaculum sp Groll as Desulfotomaculum gibsoniae sp nov. Int. J. Syst. Bacteriol. 49, 1801–1808. doi: 10.1099/00207713-49-4-1801
Lane, D. J. (1991). “16S/23S rRNA Sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt, and M. Goodfellow, (Hoboken, NJ: John Wiley and Sons).
Lau, M. C., Cameron, C., Magnabosco, C., Brown, C. T., Schilkey, F., Grim, S., et al. (2014). Phylogeny and phylogeography of functional genes shared among seven terrestrial subsurface metagenomes reveal N-cycling and microbial evolutionary relationships. Front. Microbiol. 5:531. doi: 10.3389/fmicb.2014.00531
Lehman, R. M., Roberto, F. F., Earley, D., Bruhn, D. F., Brink, S. E., O’Connell, S. P., et al. (2001). Attached and unattached bacterial communities in a 120-meter corehole in an acidic, crystalline rock aquifer. Appl. Environ. Microbiol. 67, 2095–2106. doi: 10.1128/aem.67.5.2095-2106.2001
Lin, L.-H., Hall, J., Onstott, T., Gihring, T., Lollar, B. S., Boice, E., et al. (2006). Planktonic microbial communities associated with fracture-derived groundwater in a deep gold mine of South Africa. Geomicrobiol. J. 23, 475–497. doi: 10.1080/01490450600875829
Magnabosco, C., Lin, L.-H., Dong, H., Bomberg, M., Ghiorse, W., Stan-Lotter, H., et al. (2018). The biomass and biodiversity of the continental subsurface. Nat. Geosci. 11, 707–717. doi: 10.1038/s41561-018-0221-6
Markowitz, V. M., Chen, I. M., Palaniappan, K., Chu, K., Szeto, E., Pillay, M., et al. (2014). IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 42, D560–D567. doi: 10.1093/nar/gkt963
Marreiros, B. C., Batista, A. P., Duarte, A. M., and Pereira, M. M. (2013). A missing link between complex I and group 4 membrane-bound [NiFe] hydrogenases. Biochim. Biophys. Acta 1827, 198–209. doi: 10.1016/j.bbabio.2012.09.012
McMahon, S., and Parnell, J. (2014). Weighing the deep continental biosphere. FEMS Microbiol. Ecol. 87, 113–120. doi: 10.1111/1574-6941.12196
Miller, T. L., and Wolin, M. J. (1974). A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Microbiol. 27, 985–987.
Morris, R. M., Rappé, M. S., Connon, S. A., Vergin, K. L., Siebold, W. A., Carlson, C. A., et al. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810. doi: 10.1038/nature01240
Moser, D. P., Gihring, T. M., Brockman, F. J., Fredrickson, J. K., Balkwill, D. L., Dollhopf, M. E., et al. (2005). Desulfotomaculum and Methanobacterium spp. dominate a 4- to 5-kilometer-deep fault. Appl. Environ. Microbiol. 71, 8773–8783.
Mulligan, C., Fischer, M., and Thomas, G. H. (2011). Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol. Rev. 35, 68–86. doi: 10.1111/j.1574-6976.2010.00236.x
Nielsen, M. B., Kjeldsen, K. U., and Ingvorsen, K. (2006). Desulfitibacter alkalitolerans gen. nov., sp nov., an anaerobic, alkalitolerant, sulfite-reducing bacterium isolated from a district heating plant. Int. J. Syst. Evol. Microbiol. 56, 2831–2836. doi: 10.1099/ijs.0.64356-0
Nilsen, R. K., Torsvik, T., and Lien, T. (1996). Desulfotomaculum thermocisternum sp. nov., a sulfate reducer isolated from a hot North Sea oil reservoir. Int. J. Syst. Bacteriol. 46, 397–402. doi: 10.1099/00207713-46-2-397
Osburn, M. R., LaRowe, D. E., Momper, L. M., and Amend, J. P. (2014). Chemolithotrophy in the continental deep subsurface: sanford underground research facility (SURF), USA. Front. Microbiol. 5:610. doi: 10.3389/fmicb.2014.00610
Park, J., Sanford, R. A., and Bethke, C. M. (2009). Microbial activity and chemical weathering in the Middendorf aquifer, South Carolina. Chem. Geol. 258, 232–241. doi: 10.1016/j.chemgeo.2008.10.011
Pedersen, K., and Ekendahl, S. (1990). Distribution and activity of bacteria in deep granitic groundwaters of southeastern Sweden. Microb. Ecol. 20, 37–52. doi: 10.1007/BF02543865
Pfiffner, S. M., Cantu, J. M., Smithgall, A., Peacock, A. D., White, D. C., Moser, D. P., et al. (2006). Deep subsurface microbial biomass and community structure in Witwatersrand Basin mines. Geomicrobiol. J. 23, 431–442. doi: 10.1080/01490450600875712
Puspita, I. D., Kamagata, Y., Tanaka, M., Asano, K., and Nakatsu, C. H. (2012). Are uncultivated bacteria really uncultivable? Microbes Environ. 27, 356–366. doi: 10.1264/jsme2.me12092
Qiu, Y.-L., Sekiguchi, Y., Hanada, S., Imachi, H., Tseng, I.-C., Cheng, S.-S., et al. (2006). Pelotomaculum terephthalicum sp. nov. and Pelotomaculum isophthalicum sp. nov.: two anaerobic bacteria that degrade phthalate isomers in syntrophic association with hydrogenotrophic methanogens. Arch. Microbiol. 185, 172–182. doi: 10.1007/s00203-005-0081-5
Rabus, R., Jack, D. L., Kelly, D. J., and Saier, M. H. Jr. (1999). TRAP transporters: an ancient family of extracytoplasmic solute-receptor-dependent secondary active transporters. Microbiology 145, 3431–3445. doi: 10.1099/00221287-145-12-3431
Ravot, G., Magot, M., Fardeau, M.-L., Patel, B., Prensier, G., Egan, A., et al. (1995). Thermotoga elfii sp. nov., a novel thermophilic bacterium from an African oil-producing well. Int. J. Syst. Bacteriol. 45, 308–314. doi: 10.1099/00207713-45-2-308
Sackett, J. D. (2018). Prokaryotic Diversity and Aqueous Geochemistry of Subsurface Environments of the Death Valley Regional Flow System. Ph.D. thesis, University of Nevada Las Vegas, Nevada.
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Sasser, M. (2001). MIDI Technical Note #101: Bacterial Identification by Gas Chromatographic Analysis of Fatty Acids Methyl Esters (GC-FAME). Newark, DE: MIDI, Inc.
Schut, G. J., Zadvornyy, O., Wu, C.-H., Peters, J. W., Boyd, E. S., and Adams, M. W. (2016). The role of geochemistry and energetics in the evolution of modern respiratory complexes from a proton-reducing ancestor. Biochim. Biophys. Acta 1857, 958–970. doi: 10.1016/j.bbabio.2016.01.010
Stevens, T. O., and McKinley, J. P. (1995). Lithoautotrophic microbial ecosystems in deep basalt aquifers. Science 270, 450–455. doi: 10.1038/s41467-017-01288-8
Takai, K., Moser, D. P., DeFlaun, M., Onstott, T. C., and Fredrickson, J. K. (2001). Archaeal diversity in waters from deep South African gold mines. Appl. Environ. Microbiol. 67, 5750–5760. doi: 10.1128/aem.67.21.5750-5760.2001
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thauer, R. K., Jungermann, K., and Decker, K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41, 100–180.
Thomas, J. M., Moser, D. P., Fisher, J. C., Reihle, J., Wheatley, A., Hershey, R. L., et al. (2013). Using water chemistry, isotopes and microbiology to evaluate groundwater sources, flow paths and geochemical reactions in the death valley flow system, USA. Proc. Earth Planet. Sci. 7, 842–845. doi: 10.1016/j.proeps.2013.03.033
Thompson, J. (1999). Laboratory and Field Studies Related to Radionuclide Migration at the Nevada Test Site. Los Alamos, MN: Los Alamos National Laboratory.
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16s ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583. doi: 10.1073/pnas.95.12.6578
Winograd, I. J., Fridrich, C. J., Sweetkind, D., Belcher, W. R., and Thomas, J. M. (2005). Comment on testing the interbasin flow hypothesis at Death Valley, California. EOS Trans. Am. Geophys. Union 86, 295–296.
Winograd, I. J., and Pearson, F. (1976). Major carbon 14 anomaly in a regional carbonate aquifer: possible evidence for megascale channeling, south central Great Basin. Water Resour. Res. 12, 1125–1143. doi: 10.1029/wr012i006p01125
Winograd, I. J., and Thordarson, W. (1975). Hydrogeologic and Hydrochemical Framework, South-Central Great Basin, Nevada-California, With Special Reference to the Nevada Test Site, USGS Professional Paper 712-C.
Yarza, P., Yilmaz, P., Pruesse, E., Glockner, F. O., Ludwig, W., Schleifer, K. H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645. doi: 10.1038/nrmicro3330
Ye, M., Pohlmann, K. F., and Chapman, J. B. (2008). Expert elicitation of recharge model probabilities for the Death Valley regional flow system. J. Hydrol. 354, 102–115. doi: 10.1016/j.jhydrol.2008.03.001
Yu, H., Wu, C.-H., Schut, G. J., Haja, D. K., Zhao, G., Peters, J. W., et al. (2018). Structure of an ancient respiratory system. Cell 173, 1636–1649.e16. doi: 10.1016/j.cell.2018.03.071
Keywords: 16S rRNA, subsurface, deep biosphere, Firmicutes, fractured carbonate, aquifer, borehole, Death Valley Regional Flow System
Citation: Hamilton-Brehm SD, Stewart LE, Zavarin M, Caldwell M, Lawson PA, Onstott TC, Grzymski J, Neveux I, Lollar BS, Russell CE and Moser DP (2019) Thermoanaerosceptrum fracticalcis gen. nov. sp. nov., a Novel Fumarate-Fermenting Microorganism From a Deep Fractured Carbonate Aquifer of the US Great Basin. Front. Microbiol. 10:2224. doi: 10.3389/fmicb.2019.02224
Received: 04 June 2019; Accepted: 11 September 2019;
Published: 27 September 2019.
Edited by:
Akihiko Yamagishi, Tokyo University of Pharmacy and Life Sciences, JapanReviewed by:
Susan Childers, Colby College, United StatesCopyright © 2019 Hamilton-Brehm, Stewart, Zavarin, Caldwell, Lawson, Onstott, Grzymski, Neveux, Lollar, Russell and Moser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Scott D. Hamilton-Brehm, U2NvdHQuSGFtaWx0b24tQnJlaG1Ac2l1LmVkdQ==; Duane P. Moser, RHVhbmUuTW9zZXJAZHJpLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.