- 1School of Life Sciences, Anhui Agricultural University, Hefei, China
- 2State Key Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei, China
- 3Biotechnology Center, Anhui Agricultural University, Hefei, China
Three chemical epigenetic modifiers [5-azacytidine, nicotinamide, and suberoylanilide hydroxamic acid (SAHA)] were applied to induce the metabolites of Penicillium mallochii CCH01, a fungus isolated from the gut of Ectropis oblique. Metabolite profiles of P. mallochii CCH01 were obviously changed by SAHA treatment. Four metabolites (1–4), including two new natural sclerotioramine derivatives, isochromophilone XIV (1) and isochromophilone XV (2), and two known compounds, sclerotioramine (3) and (+)-sclerotiorin (4), were isolated and purified from P. mallochii CCH01 treated with SAHA. Their structures were determined by spectroscopic analyzes. Anti-phytopathogenic activities of the isolated compounds 1–4 were investigated under laboratory conditions, and compound 4 showed broad and important inhibition activities against Curvularia lunata (IC50 = 2.1 μg/mL), Curvularia clavata (IC50 = 21.0 μg/mL), Fusarium oxysporum f. sp. Mornordica (IC50 = 40.4 μg/mL), and Botryosphaeria dothidea (IC50 = 27.8 μg/mL), which were comparable to those of referenced cycloheximide, with IC50 values of 0.3, 5.0, 12.4, and 15.3 μg/mL, respectively. Ingredients 2 and 3 showed selective and potent activities against Colletotrichum graminicola with IC50 values of 29.9 and 9.7 μg/mL, respectively. Furthermore, the antibacterial bioassays showed that compounds 3 and 4 exhibited strong inhibition activities against Bacillus subtilis, with disc diameters of zone of inhibition (ZOI) of 9.1 mm for both compounds, which were a bit weaker than that of referenced gentamycin with a ZOI of 10.8 mm. Additionally, the new metabolite 1 showed a promising activity against Candida albicans (ZOI = 10.5 mm), comparable to that of positive amphotericin B with a ZOI of 23.2 mm. The present results suggest that chemical epigenetic modifier induction was a promising approach to obtaining antimicrobial metabolites encoded by silent biosynthetic genes of P. mallochii.
Introduction
Fungi produce a wealth of valuable natural products like alkaloids, phenolic acids, flavonoids, quinones, terpenoids, steroids, benzopyranones, tetralones, xanthones, and peptides with a remarkable range of biological activities in medicine and agriculture. However, sequenced fungi genomes reveal that there are far more secondary metabolite biosynthesis gene clusters than were clear in chemical studies (Cichewicz, 2010; Sanchez et al., 2012; Ochi and Hosaka, 2013). This suggests that the potential of generating untapped chemical of fungi metabolite under standard laboratory conditions is limited, as a result of transcriptional suppression of numerous biosynthetic gene clusters (BGCs). To improve the rate of discovering novel bioactive compounds, epigenetic perturbation approaches are used to activate or modify these silent BGCs. There have been many successful examples of the use of DNA methyltransferase (DMT) inhibitors to recall cryptic biosynthetic clusters in fungi (Asai et al., 2012; He et al., 2015; Chen et al., 2016; Yu et al., 2019). Like DMT inhibitors, histone deacetylase (HDAC) inhibitors were often employed to manipulate the fungal epigenome, too. Exposing cultures of Graphiopsis chlorocephala to the NAD+-dependent HDAC inhibitor, nicotinamide triggered dramatic changes in the fungal metabolite profile, enabling the isolation of new polyketides (Asai et al., 2012). Cultivating Eupenicillium sp. LG41 in the presence of the same HDAC inhibitor led to enhanced production of two new decalin-containing compounds, eupenicinicols C and D (Li et al., 2017). Similarly, treating Daldinia sp. (Du et al., 2014), Aspergillus wentii (Miao et al., 2014), Chalara sp. 6661 (Adpressa et al., 2017), and Cladosporium sphaerospermum with the Zn (II) NAD+-independent HDAC inhibitor [suberoylanilide hydroxamic acid (SAHA)] contributed to the discovery of new polyketides, diterpenes, xanthones, and tetramic acids (Zhang et al., 2018).
We have been devoting our efforts toward discovering bioactive compounds produced by insect-associated fungi (Zhang et al., 2008, 2011; Li et al., 2014). In order to maximize the opportunity for detecting novel secondary metabolites, three chemical epigenetic modifiers were employed to stimulate the metabolite production of Penicillium mallochii CCH01 from the gut of Ectropis oblique, a pest living on tea plants. We found that metabolite profiles of the fungal strain showed obvious changes, when treated with SAHA. In this paper, we report the details of the isolation, structure elucidation, and biological activities of the metabolites produced by P. mallochii CCH01 cultivated in the presence of SAHA.
Materials and Methods
Isolation and Identification of Fungus From E. oblique
The E. oblique was collected from the tea plantation of Anhui Agricultural University, Hefei, China (latitude: 31.86 N, longitude: 117.27 E, altitude: 20 m above mean sea level). Insects (eight individuals) were kept without food for 1 day and then dissected under sterile conditions. Individual guts were suspended in a vial of 1 ml of 1 × phosphate buffer and further diluted at proportions of 10–1, 10–2, 10–3 in 1 × phosphate buffer. One hundred microliters of the diluted gut solutions was plated on PDA (20 g L–1 potato, 20 g L–1 glucose, and 20 g L–1 agar) plates supplemented with potassium dichromate (50 mg L–1) and nalidixic acid (25 mg L–1). The plates were incubated at 28°C for 4 days for fungus growth and any fungal colonies that formed were sub-cultured on new PDA medium to obtain pure cultures. The isolated strain CCH01 was deposited at the China Center for Type Culture Collection (CCTCC) as CCTCC M2018235, and also preserved on PDA slants at 4°C until use. Morphological character and molecular identification (based on the DNA sequence of ITS region using ITS1 and ITS4 primers after PCR amplification) were implemented to identify the strain.
Effect of Epigenetic Modifying Compounds on the Metabolite Production of P. mallochii CCH01
From a 7-day-old MEA (consisting of 20 g of malt extract, 20 g of sucrose, 1 g of peptone, and 20 g of agar in 1 L of distilled water) culture plate, a conidial spore suspension was prepared by adding 15 mL of distilled water containing 0.2% Tween 80 (w/v) under agitation by glass rod. To study the effect of epigenetic modifiers, aliquots (2 mL) of spore suspension were added into 100 mL of ME production media (consisting of 20 g of malt extract, 20 g of sucrose, and 1 g of peptone in 1 L of distilled water) separately supplemented with DMSO-dissolved SAHA, 5-azacytidine, nicotinamide, resulting in the same final concentrations of 1 mM. Equal amounts of DMSO were added to the control group. The cultures were fermented at 28°C in a shaker rotating at 180 rpm for 10 days. The filter of each fermentation broth was extracted three times with equal amounts of EtOAc. The EtOAc extracts were fast analyzed by TLC.
Fermentation, Extraction, and Isolation
The fungus CCH01 was grown on a ME media (twenty 1000-mL Erlenmeyer flasks, each containing 400 mL of ME media) supplemented with SAHA (in a final concentration of 1 mM) at 28°C in a shaker rotating at 180 rpm for 10 days. Total filter of fermentation broth was extracted with EtOAc (3 × 8 L) at room temperature. The EtOAc phase was evaporated in vacuo to afford a crude extract (14.36 g) and then chromatographed on a silica gel column eluting with a step gradient of CH2Cl2/MeOH (100:0–100:3, v/v) to give seven fractions (Fr1–Fr3). Fr1 (CH2Cl2/MeOH, 100:0, v/v) was further fractionated on a silica gel column, eluting with petroleum ether (PE)–ethyl acetate (EA) (50:1, v/v) to yield compound 4 (35 mg). Fr2 (CH2Cl2/MeOH, 100:1, v/v) was repeatedly subjected to a silica gel column (CH2Cl2/MeOH, 100:0, 100:1, v/v) to give three subfractions (R1–R3); compound 3 (48 mg) crystallized from the EtOAc solution of subfraction R1. Subfractions R2 and R3 were loaded onto a Sephadex LH-20 column (MeOH) to give compounds give compound 1 (15 mg) and 2 (20 mg), separately.
Structure elucidations of the secondary metabolites were made according to the spectroscopic analysis. The NMR spectra were recorded at 25°C with Agilent 600 MHz DD2 spectrometer NMR.
Analysis of Compounds in Extracts by UHPLC-Q-TOF-MS
To determine that the isolated compounds were generated by SAHA, the EtOAc extracts were accurately analyzed by ultra-high-performance liquid chromatography coupled with Q-TOF mass spectrometer (Agilent Technologies, Santa Clara, CA, United States). Chromatographic analysis was performed by using a C18 reverse-phase analytical column: Phenomenex Kinetex 2.6 μ XB-C18 100A (Torrance, CA, United States). UHPLC-Q-TOF parameters were as follows: the column oven temperature was set at 40°C and the injection volume was 5 μL, with a flow rate of 0.4 mL/min. The mobile phase consisted of 0.4% acetic acid in water and 100% acetonitrile; the gradient of the latter increased linearly from 10 to 60% (v/v) at 10 min, to 100% at 15 min, to 60% at 18 min, and to 10% at 20 min. Samples were analyzed in the fast polarity switching mode at a fragmentation voltage of 175 V, over the range of m/z 100–1700. A drying gas flow rate of 11 L/min under a temperature of 350°C and a capillary voltage of 3.5 kV were used.
Antimicrobial Activity in vitro
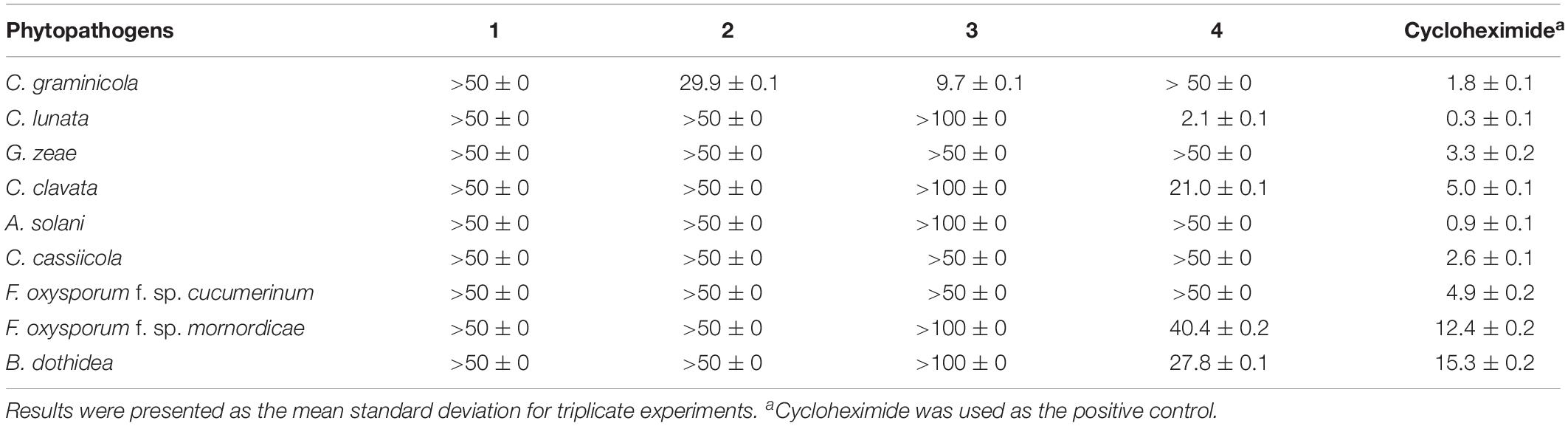
Purified metabolites were screened for their antimicrobial activity. We used nine different phytopathogenic fungi, i.e., Colletotrichum graminicola, Curvularia lunata, Gibberella zeae, Curvularia clavata, Alternaria solani, Corynespora cassiicola, Fusarium oxysporum f. sp. cucumerinum, F. oxysporum f. sp. mornordicae, and Botryosphaeria dothidea, and four human pathogens (Escherichia coli, Staphylococcus aureus, Bacillus subtilis, and Candida albicans) as the tested strains.
The antifungal activity in vitro against phytopathogenic fungi was assayed by the growth rate method (Bibi et al., 2012; Li et al., 2014), with slight modifications. Purified metabolites were dissolved in aqueous solution described previously (Yin et al., 2018) and then mixed with PDA in a Petri dish (9 cm in diameter). Cycloheximide was used as the positive control. The tested phytopathogenic fungi were inoculated onto the center of the medium and then incubated in the dark at 28°C. When the fungal mycelium reached the edges of the control dishes, the antifungal activities were calculated. The percentage of growth inhibition was calculated using the following formula:
where Da meant the diameter of growth zone in the experimental dish (mm) and Db meant the diameter in the control dish (mm). IC50 values were calculated by probit analysis based on percentage of radical growth.
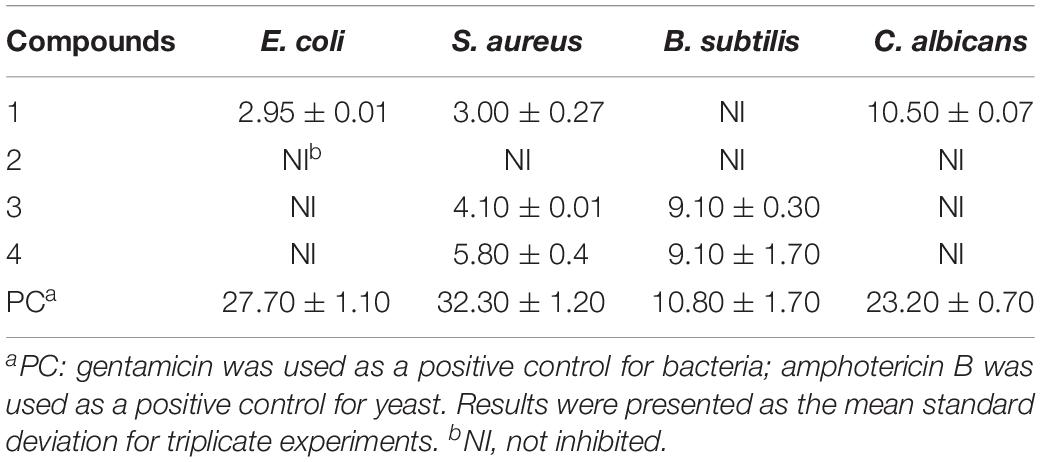
The disc diffusion method was applied to evaluate the antibacterial activities and anti-yeast activities of isolated metabolites. Filter paper disks with metabolite dissolved in DMSO in a concentration (30 μg/filter paper) were added to the culture medium, and the plates were incubated at 37°C (E. coli, S. aureus, and B. subtilis) or 28°C (C. albicans) for 24 h. Filter papers with DMSO, streptomycin sulfate, gentamycin sulfate, and amphotericin B were set as negative and positive controls, respectively. The zone of inhibition (ZOI) was determined by measuring the distance from the center of the disk to the end of the clear zone.
All experiments were performed in triplicate, and data were shown as mean values and standard deviation.
Results
Identification of the Fungus CCH01
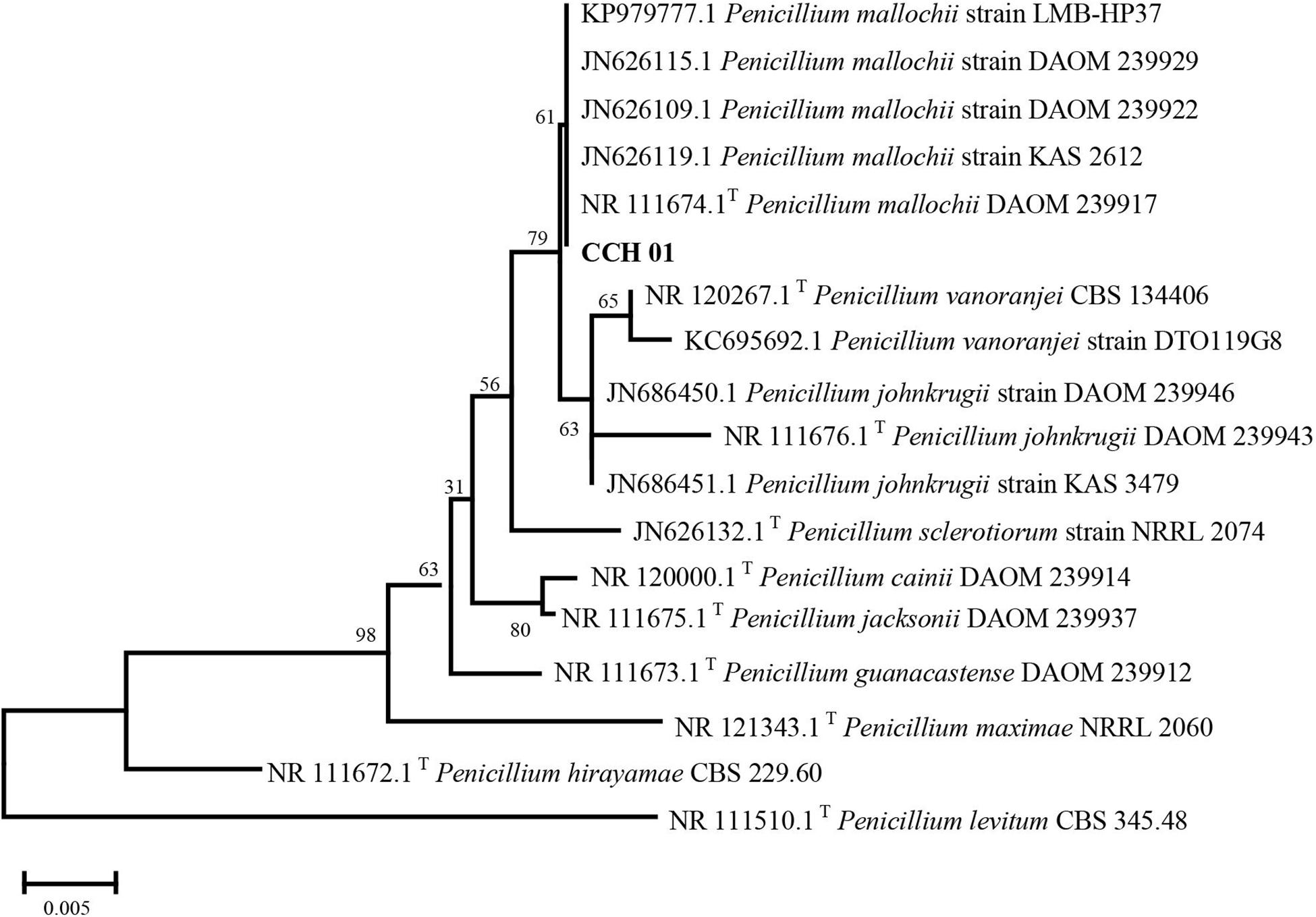
Colonies of CCH01 on the PDA plate grew quickly at 28°C, covering the whole plate (9 cm in diameter) in 7 days. Strain CCH01 showed morphological characteristics similar to members of section Sclerotiora of the Penicillium genus, i.e., monoverticillate conidiophores, greenish gray conidial masses and orange pigment in PDA plates (Visagie et al., 2013; Supplementary Figure S1). Phylogenetic tree (Figure 1) constructed based on ITS rDNA sequences of the fungus using MEGA5 according to the neighbor-joining method indicated that CCH01 was closely related to P. mallochii (NR_11674.1T), with the ITS sequence similarity of 99.8%. Combined with morphological characteristics, the fungus was identified as P. mallochii CCH01.
Effect of SAHA on the Metabolic Profile of P. mallochii CCH01
Crude EtOAc extracts obtained from both the epigenetic treated and the control (DMSO) fermentation broths of the P. mallochii CCH01 were compared by TLC in mobile phase (CH2Cl2/MeOH, 40:1, v/v). A UV visible spot at an Rf value of 0.3 was observed in SAHA-treated extract, which was not visible in the extracts of the culture grown without SAHA (Supplementary Figure S2).
Isolation of Secondary Metabolites From P. mallochii CCH01
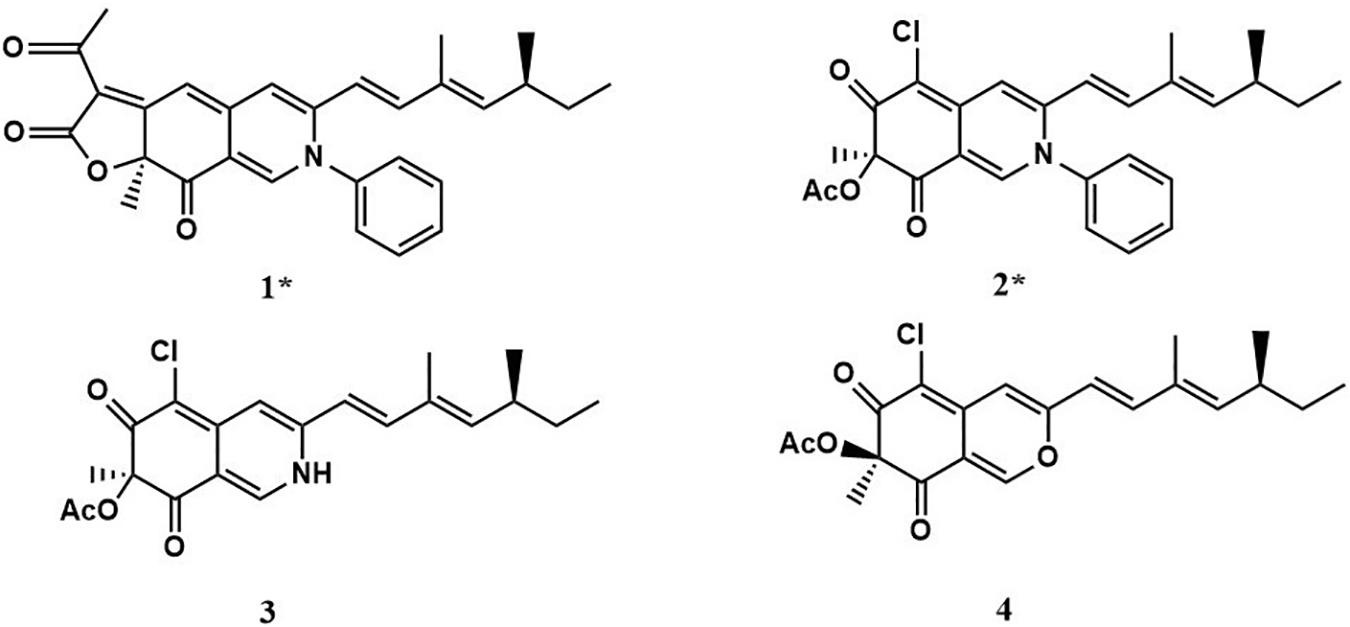
Two new compounds (1 and 2), along with two known metabolites (3 and 4) were isolated from the culture of P. mallochii CCH01 treated with SAHA. Chemical structures of compounds 1–4 are shown in Figure 2.
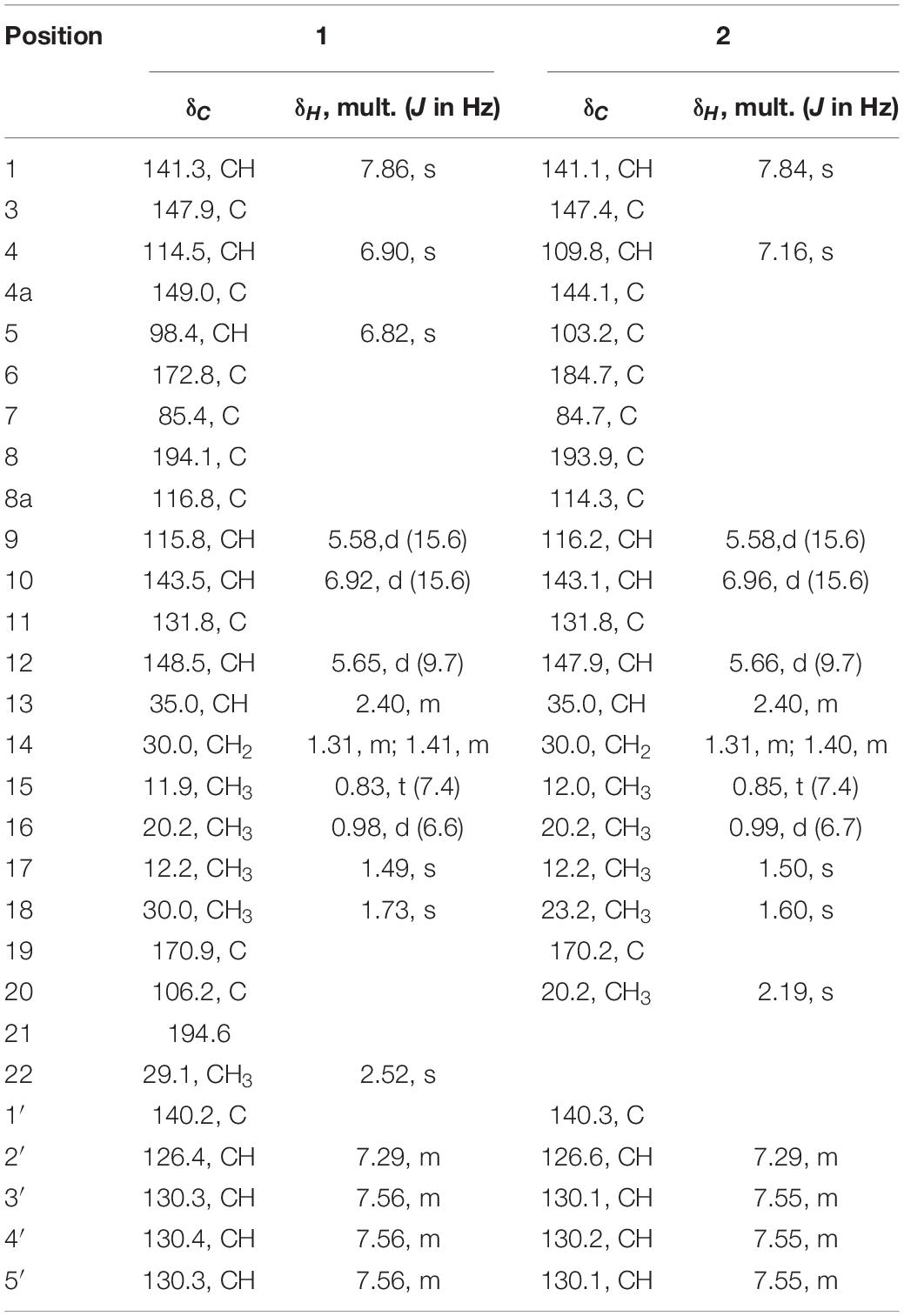
Compound 1 (isochromophilone XIV) was obtained as a purple powder, and its molecular formula C29H29NO4 was deduced from HRESIMES ion peak at m/z 456.2196 [M + H]+, which was consistent with 1H and 13C NMR data (Table 1 and Supplementary Figures S3–S9). 13C NMR data were similar to those of isochromophilone IV (Kanokmedhakul et al., 2006), except ethyl signal (δC 55.4, 36.6). This speculation was confirmed by HMBC correlations from H-1 to C-1 (δC 141.3), C-3 (δC 147.9), C-4a (δC 149.0), and C-8a (δC 116.8); H-4 to C-3 (δC 147.9), C-5 (δC 98.4), C-8a (δC 116.8), and C-9 (δC 115.8); Me-18 to C-6 (δC 172.8); H-9 to C-4 δC 114.5) and C-11 (δC 131.8); H-10 to C-3 (δC 147.9) and C-17 (δC 12.2); H-12 to C-10 (δC 143.5), C-13 (δC 35.0), and C-17 (δC 12.2). 1H–1H COSY indicated that H-9 had a relationship with H-10, H-12, H-13, and H-14 (me-16); me-15 had a relationship with H-2′and H-3′. Thus, the structure of compound 1 was determined as a new isochromophilone derivative (isochromophilone XIV).
Compound 2 (isochromophilone XV) was obtained as a yellow powder, and its molecular formula C27H28ClNO4 was deduced from HRESIMES ion peak at m/z 466.1816 [M + H]+, which was consistent with 1H and 13C NMR data (Table 1 and Supplementary Figures S10–S15). The 1H NMR and 13C NMR data were similar to those of isochromophilone IV (Lucas et al., 2007; Wang et al., 2010), except that the imino at position 2 in isochromophilone IV appeared to be aniline, which corresponded to the increase in molecular weight of 2 by 92 amu compared to 4 (δC 55.4, 36.6). This was further confirmed by the HMBC correlation of H-1 to C-1 (δC 141.1), C-3 (δC 147.4), C-4a (δC 144.1), C-8a (δC 114.3), and C-8 (δC 193.9); H-4 to C-3 (δC 147.4), C-5 (δC 103.2), C-8a (δC 114.3), and C-9 (δC 116.2); Me-18 to C-6 (δC 184.7) and C-8 (δC 193.9); H-9 to C-4 (δC 109.8) and C-1 (δC 141.1); H-10 to C-3 (δC 147.4) and C-17 (δC 12.2); H-12 to C-10 (δC 143.1), C-13 (δC 35.0), and C-17 (δC 12.2); and 1H–1H COSY correlation of H-9 to H-10, H-12, H-13, H-14, Me-16, and Me-15; H-2′ to H-3′. To our knowledge, this compound was first discovered as a natural product, but reported as semisynthetic derivatives (Wei et al., 2017).
Compound 3 was obtained as a red crystal, and its molecular formula C21H24ClNO4 was deduced from HRESIMES m/z 390.1518 [M + H]+. 1H NMR and 13C NMR data were as follows: 1H NMR (600 MHz, CDCl3): δ:7.68 (1H, s, H-1), 6.70 (1H, s, H-4), 6.00 (1H, d, J = 16.0 Hz, H-9), 6.82 (1H, d, J = 16.0 Hz, H-10), 5.65 (1H, d, J = 9.84 Hz, H-12), 2.47 (1H, m, H-13), 1.32 (2H, m, H-14), 0.85 (3H, t, m, H-15), 1.04 (3H, d, J = 6.7 Hz, H-16), 1.83 (3H, s, H-17), 1.66 (3H, s, H-18), 2.25 (3H, s, H-20); 13C NMR (151 MHz, CDCl3) δ: 138.8 (CH, C-1), 147.1 (C, C-3), 110.73 (CH, C-4), 148.25 (C, C-4a), 100.67 (C, C-5), 183.76 (C, C-6), 84.88 (C, C-7), 193.29 (C, C-8), 116.65 (CH, C-9), 143.30 (CH, C-10), 132.04 (C, C-11), 114.67 (CH, C-12), 35.06 (CH, C-13), 30.03 (CH2, C-14), 11.95 (CH3, C-15), 20.16 (CH3, C-16), 12.36 (CH3, C-17), 23.7 (CH3, C-18), 170.23 (C, C-19), 20.37 (CH3, C-20). These showed almost no difference with sclerotioramine (C21H24ClNO4) described in literature (Wang et al., 2010).
Compound 4 was obtained as a yellowish powder, and its molecular formula C21H23O5Cl was deduced from HRESIMES at ion peak at m/z 389.1325 [M-H]–. 1H NMR and 13C NMR data were as follows: 1H NMR (600 MHz, CDCl3) δ: 7.92 (1H, s, H-1), 6.63 (1H, s, H-4), 6.08 (1H, d, J = 15.72 Hz, H-9), 7.06 (1H, d, J = 15.66 Hz, H-10), 5.69 (1H, d, J = 9.72 Hz, H-12), 2.47 (1H, m, H-13), 1.42 (2H, m, H-14), 0.85 (3H, t, J = 7.26 Hz, H-15), 1.83 (1H, s, H-16), 1.00 (3H, d, J = 6.6 Hz, H-17), 1.56 (1H, s, H-18), 2.16 (3H, s, 7-OCOCH3); 13C NMR (151 MHz, CDCl3) δ: 152.67 (CH, C-1), 158.03 (C, C-3), 106.35 (CH, C-4), 138.60 (C, C-4a), 114.51 (C, C-5), 191.74 (C, C-6), 84.53 (C, C-7), 185.90 (C, C-8), 110.78 (C, C-8a), 115.63 (CH, C-9), 142.79 (C, C-10), 131.93 (C, C-11), 148.79 (CH, C-12), 35.10 (CH, C-13), 30.02 (CH2, C-14), 11.92 (CH3, C-15), 12.33 (CH3, C-16), 20.17 (CH3, C-17), 22.49 (CH3, C-18), 170.04 (COCH3), 20.04 (COCH3). It indicated that compound 4 was (+) sclerotiorin (C21H23O5Cl) (Chidananda and Sattur, 2007).
UHPLC-Q-TOF-MS Analysis of Purified Metabolites From Crude Extracts
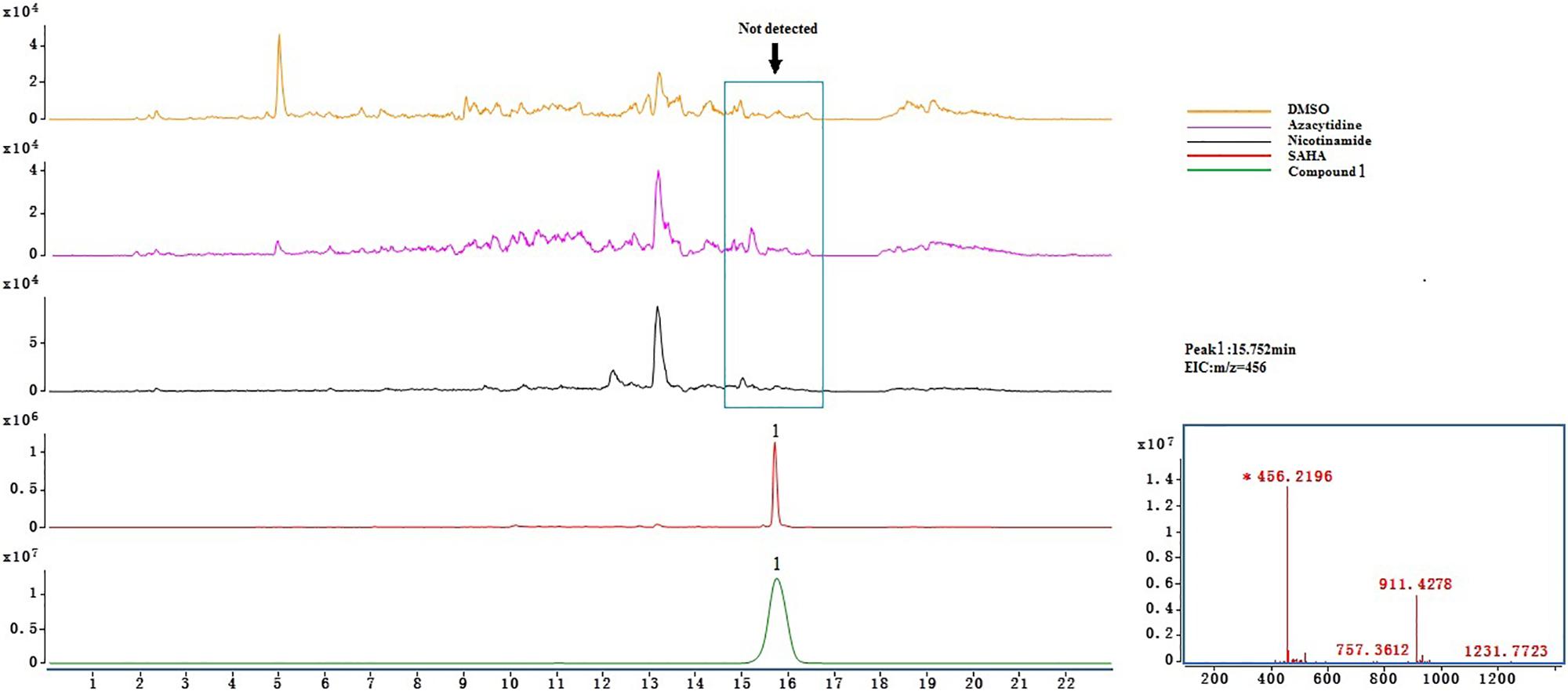
Crude extract samples were detected by UHPLC-Q-TOF-MS. As a result, compounds 2, 3, and 4 existed in all of the samples (Supplementary Figure S16); this suggests that the total of these four ingredients was not the product of SAHA inducing. However, only compound 1 was detected (Figure 3) in the extract of SAHA-treated fermentation broth, but not in the control that showed an intense peak at a retention time of 15.752 min with m/z of 456.2196[M + H]+, which was consistent with purified compound 1 as a standard. This indicated SAHA may activate the dormant gene clusters in the biosynthetic pathways of compound 1.
In vitro Anti-phytopathogenic Activity
The inhibition activities of compounds 1–4 against mycelial growth of nine phytopathogenic fungi were tested in vitro (Table 2). The results showed (+) that sclerotiorin (4) displayed broad and important inhibition activities against C. lunata (IC50 = 2.1 μg/mL), C. clavata (IC50 = 21.0 μg/mL), F. oxysporum f. sp. mornordicae (IC50 = 40.4 μg/mL), and B. dothidea (IC50 = 27.8 μg/mL), which were comparable to those of referenced cycloheximide with IC50 values of 0.3, 12.4, and 15.3 μg/mL, respectively. Ingredient 3 exhibited a selective inhibition activity against C. graminicola with an IC50 value of 9.7 μg/mL, which was comparable to that of cycloheximide (IC50 = 1.8 μg/mL). Meanwhile, metabolite 2 showed a relatively weak activity against C. graminicola with an IC50 value of 29.9 μg/mL.
In vitro Anti-bacterial and Anti-yeast Activities
Compounds 1–4 were tested for their antibacterial and anti-yeast activity against a panel of representative pathogens. As shown in Table 3, both compounds 3 and 4 exhibited potent activities against B. subtilis, with the same ZOI values of 9.10 mm compared with that of positive gentamycin (ZOI = 10.80 mm). Notably, metabolite 1 displayed a moderate activity against C. albicans with a ZOI value of 10.50 mm, compared with that of referenced amphotericin B (ZOI = 23.20 mm).
Discussion
Sclerotiorin, first isolated from Penicillium sclerotiorum, belongs to the azaphilone-type family of natural products, with a γ-lactone, a conjugated ketone, a chlorine atom at C-5, and a branched C-7 side chain (Maccurin and Reilly, 1940). Sclerotiorin and its derivatives were mostly isolated from Penicillium species from soil (Arai et al., 1995; Nam et al., 2000; Celestino et al., 2014), sea (Cheung et al., 2014; Chen et al., 2017), and plant endophytes (Giridharan et al., 2012). However, to the best of our knowledge, it was the first reported that the two new natural products 1 and 2 and the following metabolites 3 and 4 were obtained from the title strain P. mallochii CCH01, a fungus residing in the gut of E. oblique.
Azacytidine, a DNA methylation-modifying agent, was verified as an effective epigenetic modifier that altered secondary metabolites of an Atlantic-forest-soil-derived P. citreonigrum and led to the production of six azaphilones (Wang et al., 2010). As far as we know, there are no references about new azaphilones obtained from Penicillium treated with HDAC inhibitors. In the present study, the effect of three different epigenetic modifiers, i.e., 5-azacytidine, SAHA, and nicotinamide on the metabolic profile of P. mallochii CCH01 was studied. Interestingly, a significant variation in the metabolome of the fungus was observed when treated with SAHA, an HDAC inhibitor. On the basis of further LC-MS analysis experiments, metabolites 2–4 were detected in the crude extracts of both control and treated cultures; only compound 1 was found exclusively in the extract of SAHA-treated fungus (Figure 3). This indicated that the epigenetic modifier SAHA can be used to generate new azaphilones from the gut fungus P. mallochii CCH01.
Conclusion
Here, one strain identified as P. mallochii CCH01 was isolated from the gut of E. oblique, a pest living on tea plants. For developing the potential of P. mallochii CCH01 for the production of new compounds, three chemical epigenetic reagents were applied to stimulate the strain. As a result, SAHA brought significant changes in the secondary metabolites of the fungus. Subsequently, four compounds, including 2 new isochromophilone derivatives, were isolated from the culture treated with SAHA. This result suggests that SAHA can be used to identify diverse natural products hidden in silent biosynthetic pathways from gut fungus P. mallochii CCH01. Sclerotiorin (4) possessed a good prospect in crop and plant protection, with important activities against phytopathogenic fungi. Interestingly, as two simple amine derivatives of sclerotiorin (4), both metabolites 2 and 3 exhibited modest inhibition activities against C. graminicola, which were different with sclerotiorin. Additionally, the new compound 1 showed a promising activity against C. albicans, which was comparable to that of referenced amphotericin B. Ingredients 3 and 4 possessed strong antibacterial activities against B. subtilis in vitro. Thus, our results highlight the potential of epigenetic modification as powerful strategies for generating the production of cryptic fungal metabolites.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
YZ and CW designed the research. YZ supervised the study. SZ, HF, and JH performed the experiments and analyzed the data. SZ wrote the manuscript together with CY. All authors revised the manuscript and approved the final version for submission.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) (31770007) and the Open Fund of State Key Laboratory of Tea Plant Biology and Utilization (SKLTOF20170111).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02186/full#supplementary-material
References
Adpressa, D. A., Stalheim, K. J., Proteau, P. J., and Loesgen, S. (2017). Unexpected biotransformation of the HADC inhibitor vorinostat yields aniline-containing fungal metabolites. ACS Chem. Biol. 12, 1842–1847. doi: 10.1021/acschembio.7b00268
Arai, N., Shiomi, K., Tomoda, H., Tabata, N., Yang, D. J., Masuma, R., et al. (1995). Isochromophilones III-VI, inhibitors of acyl-CoA:cholesterol acyltransferase produced by Penicillium multicolor FO-3216. J. Antibiot. 48, 696–702. doi: 10.7164/antibiotics.48.696
Asai, T., Morita, S., Shirata, N., Taniguchi, T., and Oshima, Y. (2012). Structural diversity of new C13-polyketides produced by Chaetomium mollipilium cultivated in the presence of a NAD+-dependent histone deacetylase inhibitor. Org. Lett. 14, 5456–5459. doi: 10.1021/ol302539s
Bibi, F., Yasir, M., Song, G. C., Lee, S. Y., and Chung, Y. R. (2012). Diversity and characterization of endophytic bacteria associated with tidal flat plants and their antagonistic effects on oomycetous plant pathogens. Plant Pathol. J. 28, 20–31. doi: 10.5423/ppj.oa.06.2011.0123
Celestino, J. d. R., Carvalho, L. E. d., Lima, M. d. P., Lima, A. M., Ogussku, M. M., and Souza, J. V. B. d. (2014). Bioprospecting of amazon soil fungi with the potential for pigment production. Process Biochem. 49, 569–575. doi: 10.1016/j.procbio.2014.01.018
Chen, D., Ma, S., He, L., Yuan, P., She, Z., and Lu, Y. (2017). Sclerotiorin inhibits protein kinase G from Mycobacterium tuberculosis and impairs mycobacterial growth in macrophages. Tuberculosis 103, 37–43. doi: 10.1016/j.tube.2017.01.001
Chen, M., Zhang, W., Shao, C. L., Chi, Z. M., and Wang, C. Y. (2016). DNA methyltransferase inhibitor induced fungal biosynthetic products: diethylene glycol phthalate ester oligomers from the marine-derived fungus Cochliobolus lunatus. Mar. Biotechnol. 18, 409–417. doi: 10.1007/s10126-016-9703-y
Cheung, R. C. F., Wong, J. H., Pan, W. L., Chan, Y. S., Yin, C. M., Dan, X. L., et al. (2014). Antifungal and antiviral products of marine organisms. Appl. Microbiol. Biot. 98, 3475–3494. doi: 10.1007/s00253-014-5575-0
Chidananda, C., and Sattur, A. P. (2007). Sclerotiorin, a novel inhibitor of lipoxygenase from Penicillium frequentans. J. Agric. Food Chem. 55, 2879–2883. doi: 10.1021/jf062032x
Cichewicz, R. H. (2010). Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat. Prod. Rep. 27, 11–22. doi: 10.1039/B920860G
Du, L., King, J. B., and Cichewicz, R. H. (2014). Chlorinated polyketide obtained from Daldinia sp. treated with the epigenetic modifier suberoylanilide hydroxamic acid. J. Nat. Prod. 77, 2454–2458. doi: 10.1021/np500522z
Giridharan, P., Verekar, S. A., Khanna, A., Mishra, P. D., and Deshmukh, S. K. (2012). Anticancer activity of sclerotiorin, isolated from an endophytic fungus Cephalotheca faveolata Yaguchi, Nishim. & Udagawa. Indian J. Exp. Biol. 50, 464–468.
He, X., Zhang, Z., Chen, Y., Che, Q., Zhu, T., Gu, Q., et al. (2015). Varitatin A, a highly modified fatty acid amide from Penicillium variabile cultured with a DNA methyltransferase inhibitor. J. Nat. Prod. 78, 2841–2845. doi: 10.1021/acs.jnatprod.5b00742
Kanokmedhakul, S., Kanokmedhakul, K., Nasomjai, P., Louangsysouphanh, S., Soytong, K., Isobe, M., et al. (2006). Antifungal azaphilones from the fungus Chaetomium cupreum CC3003. J. Nat. Prod. 69, 891–895. doi: 10.1021/np060051v
Li, G., Kusari, S., Golz, C., Laatsch, H., Strohmann, C., and Spiteller, M. (2017). Epigenetic modulation of endophytic Eupenicillium sp. LG41 by a histone deacetylase inhibitor for production of decalin-containing compounds. J. Nat. Prod. 80, 983–988. doi: 10.1021/acs.jnatprod.6b00997
Li, S., Shao, M. W., Lu, Y. H., Kong, L. C., Jiang, D. H., and Zhang, Y. L. (2014). Phytotoxic and antibacterial metabolites from Fusarium proliferatum ZS07 isolated from the gut of long-horned grasshoppers. J. Agric. Food Chem. 62, 8997–9001. doi: 10.1021/jf502484n
Lucas, E. M. F., Castro, M. C. M. d., and Takahashi, J. A. (2007). Antimicrobial properties of sclerotiorin, isochromophilone VI and pencolide, metabolites from a Brazilian cerrado isolate of Penicillium sclerotiorum van Beyma. Braz. J. Microbiol. 38, 785–789. doi: 10.1590/S1517-83822007000400036
Maccurin, T., and Reilly, J. (1940). Sclerotiorine, a chlorinated metabolic product of Penicillium sclerotiorum, van Beyma. Nature 146:335. doi: 10.1038/146335b0
Miao, F. P., Liang, X. R., Liu, X. H., and Ji, N. Y. (2014). Aspewentins A-C, norditerpenes from a cryptic pathway in an algicolous strain of Aspergillus wentii. J. Nat. Prod. 77, 429–432. doi: 10.1021/np401047w
Nam, J.-Y., Son, K.-H., Kim, H.-K., Han, M.-Y., Kim, S.-U., Choi, J.-D., et al. (2000). Sclerotiorin and isochromophilone IV: inhibitors of Grb2-Shc interaction, isolated from Penicillium multicolor F1753. J. Microbiol. Biotechn. 10, 544–546.
Ochi, K., and Hosaka, T. (2013). New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl. Microbiol. Biot. 97, 87–98. doi: 10.1007/s00253-012-4551-9
Sanchez, J. F., Somoza, A. D., Keller, N. P., and Wang, C. C. (2012). Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat. Prod. Rep. 29, 351–371. doi: 10.1039/c2np00084a
Visagie, C. M., Houbraken, J., Rodriques, C., Silva Pereira, C., Dijksterhuis, J., Seifert, K. A., et al. (2013). Five new Penicillium species in section Sclerotiora: a tribute to the Dutch Royal family. Persoonia 31, 42–62. doi: 10.3767/003158513X667410
Wang, X. R., Sena Filho, J. G., Hoover, A. R., King, J. B., Ellis, T. K., Powell, D. R., et al. (2010). Chemical epigenetics alters the secondary metabolite composition of guttate excreted by an Atlantic-forest-soil-derived Penicillium citreonigrum. J. Nat. Prod. 73, 942–948. doi: 10.1021/np100142h
Wei, M. Y., Wang, C. F., Wang, K. L., Qian, P. Y., Wang, C. Y., and Shao, C. L. (2017). Preparation, structure, and potent antifouling activity of sclerotioramine derivatives. Mar. Biotechnol. 19, 372–378. doi: 10.1007/s10126-017-9760-x
Yin, C., Jin, L., Sun, F., Xu, X., Shao, M., and Zhang, Y. L. (2018). Phytotoxic and antifungal metabolites from Curvularia crepinii QTYC-1 isolated from the gut of Pantala flavescens. Molecules 23:E951. doi: 10.3390/molecules23040951
Yu, G., Wang, Q., Liu, S., Zhang, X., Che, Q., Zhang, G., et al. (2019). Methylsulfonylated polyketides produced by Neosartorya udagawae HDN13-313 via exogenous addition of small molecules. J. Nat. Prod. 82, 998–1001. doi: 10.1021/acs.jnatprod.9b00035
Zhang, Y. L., Ge, H. M., Zhao, W., Dong, H., Xu, Q., Li, S. H., et al. (2008). Unprecedented immunosuppressive polyketides from Daldinia eschscholzii, a mantis-associated fungus. Angew. Chem. Int. Ed. Engl. 120, 5907–5910. doi: 10.1002/ange.200801284
Zhang, Y. L., Zhang, J., Jiang, N., Lu, Y. H., Wang, L., Wang, L., et al. (2011). Immunosuppressive polyketides from mantis-associated Daldinia eschscholzii. J. Am. Chem. Soc. 133, 5931–5940. doi: 10.1021/ja110932p
Keywords: gut fungus, Ectropis oblique, Penicillium mallochii CCH01, suberoylanilide hydroxamic acid, antimicrobial activities
Citation: Zhang S, Fang H, Yin C, Wei C, Hu J and Zhang Y (2019) Antimicrobial Metabolites Produced by Penicillium mallochii CCH01 Isolated From the Gut of Ectropis oblique, Cultivated in the Presence of a Histone Deacetylase Inhibitor. Front. Microbiol. 10:2186. doi: 10.3389/fmicb.2019.02186
Received: 21 May 2019; Accepted: 05 September 2019;
Published: 02 October 2019.
Edited by:
Marie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Zaixiang Lou, Jiangnan University, ChinaStefan Taubert, University of British Columbia, Canada
Bury-Moné Germaine Stéphanie, Université Paris-Sud, France
Copyright © 2019 Zhang, Fang, Yin, Wei, Hu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaoling Wei, d2VpY2hsQGFoYXUuZWR1LmNu; Yinglao Zhang, emhhbmd5bEBhaGF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Shuxiang Zhang
Shuxiang Zhang Han Fang1†
Han Fang1† Chaoling Wei
Chaoling Wei Yinglao Zhang
Yinglao Zhang