- 1Department of Laboratory Medicine, Affiliated Sixth People’s Hospital South Campus, Shanghai University of Medicine & Health Sciences, Shanghai, China
- 2Department of Laboratory Medicine, Southern Medical University Affiliated Fengxian Hospital, Shanghai, China
- 3Department of Laboratory Medicine, Affiliated Fengxian Hospital, Anhui University of Science and Technology, Anhui, China
- 4Central Laboratory, Department of Laboratory Medicine, Shanghai Tenth People’s Hospital, Tongji University, Shanghai, China
- 5Department of Laboratory Medicine, Shanghai Skin Disease Hospital, Tongji University, Shanghai, China
Objectives: To describe the molecular characteristics of beta-lactamases in bloodstream-infection Escherichia coli isolated from elderly patients, and to determine the genotypic patterns of blaCMY–2 and blaADC–162.
Methods: A total of 50 bloodstream-infection E. coli isolates were obtained from patients aged > 50 years at Shanghai Sixth People’s Hospital South Campus during 2015–2018. The isolates were subjected to beta-lactamase detection using phenotypic and molecular methods. Beta-lactamase genes were verified by sequencing and the phylogenetic relationships of the isolates were analyzed by multilocus sequence typing (MLST). The transferability of plasmids carrying blaCMY–2 and blaADC–162 genes was verified by conjugation experiments and plasmid replicon typing.
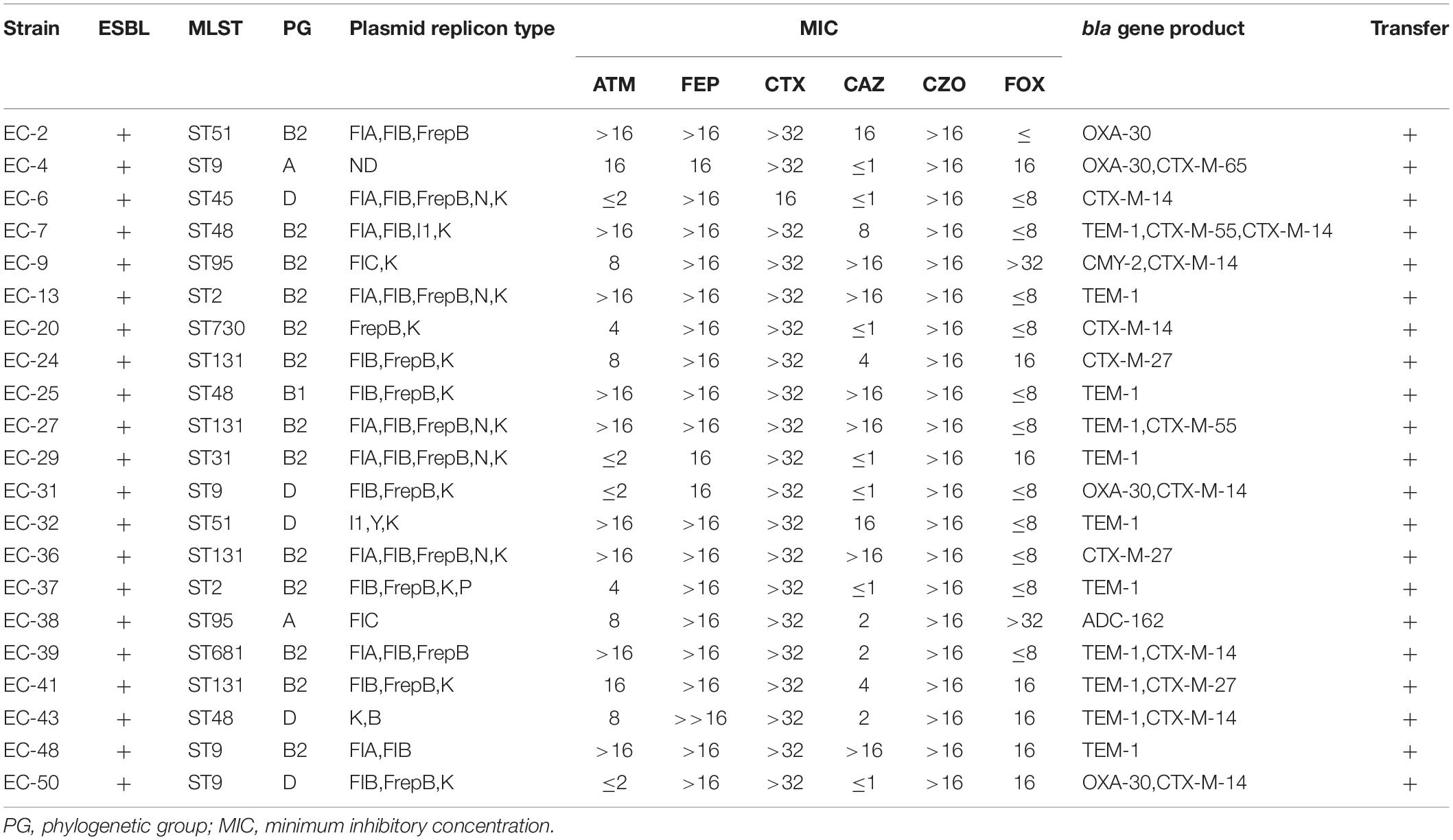
Results: Eight beta-lactamase subtypes were detected in 50 isolates of bloodstream-infection E. coli. blaTEM–1 (21/50) was the most common beta-lactamase gene, followed by blaCTX–M–14 (8/50), blaOXA–27 (5/50), blaCTX–M–27 (3/50), blaCTX–M–65 (1/50), blaADC–162 (1/50), and blaCMY–2 (1/50). Of these, blaADC–162 (ST95-A), and blaCMY–2 (ST95-B2) have not previously been reported in bloodstream-infection E. coli. In 21 isolates, beta-lactamase genes were located on conjugative plasmids belonging to incompatibility groups FrepB (n = 7), FIA (n = 1), FIC (n = 2), K (n = 8), N (n = 1), and I (n = 1), and blaCTX–M was associated with the common elements ISEcp1, IS903, and IS26, but with special sequences (region V, region Y, and region W) for ISEcp1 in 14 isolates.
Conclusion: To the best of our knowledge, this study provides the first molecular characterization of beta-lactamase genes in E. coli isolated from the bloodstream in elderly patients. Beta-lactamase genes were detected at a relatively high frequency in elderly patients with bloodstream E. coli infections. Plasmid replicon analysis showed that horizontal dissemination of beta-lactamase genes was mainly mediated by IncK and IncF plasmids, which could encode multidrug resistance genes. The study also provides the first report of ISAba1-blaADC–162-tnpA and ISEcp1-blaCTX–M–14-IS903-blaCMY–2-blc-sugE in E. coli, and demonstrates IncF plasmid-mediated blaADC–162 and blaCMY–2 gene dissemination among bacteria.
Introduction
Escherichia coli is commonly isolated from clinical bloodstream infections. It is referred to as extraintestinal pathogenic E. coli (Hung et al., 2019) and is usually multidrug-resistant, potentially leading to sepsis and even death of infected patients (van der Mee-Marquet et al., 2015). Antibiotic selection as a result of the extensive clinical application of broad-spectrum antibiotics, especially third-generation cephalosporins, has led to the generation of drug-resistant bacteria (Baron et al., 2014). Bloodstream infection by multidrug-resistant E. coli thus presents difficulties in clinical treatment and has become an important public health problem (Bartoletti et al., 2014). The main resistance mechanism of Gram-negative bacteria such as E. coli involves the production of a variety of hydrolytically active beta-lactamases, from broad- to extended-spectrum enzymes, and the enzymatic hydrolysis profile and host range are constantly changing from chromosome-mediated to plasmid-mediated AmpC beta-lactamases (Du et al., 2002; Razazi et al., 2012).
The major beta-lactamase resistance genes in E. coli are currently members of the blaCTX–M and blaTEM groups, which have been reported in many different countries (Andrea et al., 2013). blaTEM were the first beta-lactamase genes found in Gram-negative bacteria. They are specifically transferred by plasmids, and more than 200 subtypes have been identified, mainly encoding enzymes that hydrolyze penicillin and first generation cephalosporins (Medeiros, 1984; Salverda et al., 2010; Clasen et al., 2019). In contrast, blaCTX–M-encoded enzymes mainly hydrolyze third generation cephalosporins, are mostly located on plasmids of 40–200 kb (Zhao and Hu, 2013), and belong to a wide variety of incompatibility (Inc.) groups. Conjugative plasmids can shuttle between bacteria of the same or different species, thus spreading resistance phenotypes and potentially causing large-scale outbreaks and the prevalence of resistant bacteria. The blaCTX–M group beta lactamases have been widely reported to cause bacterial resistance by conjugative plasmids, though the blaCTX-M group, other ampC beta-lactamase resistance genes, such as blaADC and blaCMY, are also gradually increasing in clinical strains, resulting in greater cephalosporin resistance (Koh et al., 2004). Ceftazidine is the main third generation cephalosporin used in Europe, while cefotaxime and cefoperazone are the most extensively used in China (Lambert et al., 2011). Bacteria with different beta-lactamase genotypes express enzymes with different physical and chemical properties and differences in drug resistance (Medeiros, 1984). Importantly, genotype prevalence also differ among regions. However, the characterization of beta-lactamases in bloodstream-infection E. coli from elderly patients in China has not yet been reported, and whether the upstream and downstream surroundings of these genes in bloodstream-infection E. coli are consistent with other species of bacteria, or if they have special transfer and transmission capabilities, remains unknown. In this study, we carried out phenotypic and genotypic analyses of bloodstream-infection E. coli in a tertiary hospital in China to elucidate the genetic environment in selected isolates in relation to different beta-lactamase types on plasmids in different incompatibility groups. This analysis of the genetic context of the beta-lactamase genes may help to clarify their acquisition, with regard to their origin and further dissemination.
Materials and Methods
Bacterial Strains
A total of 1242 E. coli isolates were recovered from patient samples at Shanghai Sixth People’s Hospital South Campus, China, during 2015–2018. Among these, 50 strains of bloodstream-infection E. coli were isolated from elderly patients and further characterized with regard to extended-spectrum beta-lactamase (ESBL) and ampC genes. E. coli ATCC25922, J53, and E. coli DH5α were maintained in our laboratory.
Antimicrobial Susceptibility Testing
Antibiotic susceptibility was determined by disk diffusion or broth dilution, using E. coli ATCC25922 as a control strain. The tested antibiotics included: amikacin, gentamicin, tobramycin, trimethoprim/sulfamethoxazole, chloramphenicol, meropenem, imipenem, ciprofloxacin, levofloxacin, ampicillin, aztreonam, cefepime, cefotaxime, ceftazidime, cefazolin, ceftriaxone, and cefoxitin. The results were interpreted in accordance with the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2018).
Phenotypic Characterization
ESBL production was determined by double-disc synergy tests and confirmed by E-testing, using cefotaxime-cefotaxime-clavulanic acid and ceftazidime-ceftazidime-clavulanic acid strips, according to the CLSI guidelines. Similarly, phenotypic confirmation of plasmid-mediated AmpC production was performed using the AmpC assay E-test with cefotetan-ceftatan-clozacillin strips, according to the manufacturer’s instructions (BioMerieux, France).
Characterization of Beta-Lactamase Genes
The beta-lactamase genotype of each isolate was determined by polymerase chain reaction (PCR) amplification with specific primers for blaTEM, blaSHV, blaCTX–M–1, blaCTX–M–2, blaCTX–M–8, blaCTX–M–9, blaCTX–M–25, blaOXA–1, blaPER, blaSME, blaKPC, blaVIM, blaNDM, blaIMP, blaGES, blaVEB, blaDHA, blaADC, blaACC, blaCIT, and blaEBC, with bacterially isolated DNA as an amplification template. The total volume of the PCR amplification system was 20 μL, containing 1 μL of genomic DNA template (>50 ng/μL), 10 μL of Premix-rTaq PCR solution (TaKaRa, Japan), 0.4 μL of each primer (10 pmol), and 7 μL of distilled water. PCR was performed using a ProFlex Base Thermal Cycler (Applied Biosystems, Thermo Fisher Scientific, Singapore). The template was denatured at 94°C for 4 min, followed by 35 cycles of 94°C for 40 s, 55°C for 40 s, and 72°C for 40 s, with a final extension stage at 72°C for 5 min. The PCR product was verified by agarose gel electrophoresis and sequencing. All beta-lactamase gene sequencing results were aligned using the BLAST program1.
Conjugation Experiments
Conjugation experiments were performed using sodium azide-resistant E. coli J53 as a receptor. Transconjugants were selected on Luria-Bertani agar plates supplemented with sodium azide (200 μg/mL) (Sigma, Germany) and ampicillin (100 μg/mL). J53 and donor bacteria were resuscitated overnight, and a single colony was selected and enriched for 18 h in LB liquid medium without antibiotics. J53 and donor bacteria were mixed 1:4, and a 0.22 μm pore size filter was applied to the blood plate. Thereafter, 150 μL of the mixed bacteria solution was taken up and added to the filter membrane, and cultured overnight. The filter with the bacteria was then removed and washed in LB liquid medium, diluting 1:100 and 150 μL was then applied to the double-antibody plate, cultured overnight, and a single colony was picked for further experiments. The presence of the beta-lactamase gene in the transconjugant was examined using the primers given in Table 1, and the susceptibility of the strain was tested experimentally, as described above.
Plasmid Replicon Typing
Plasmid DNA was isolated from E. coli using a SanPrep Column Plasmid Mini-Preps Kit plasmid isolation system (Sangon Biotech, Shanghai, China) and stored at −20°C, according to the manufacturer’s instructions. Plasmid replicon typing of E. coli was performed using a multiplex PCR-based method with 18 pairs of primers, as described previously (Carattoli et al., 2005). The typing results were obtained by agarose gel electrophoresis and verified by sequencing.
Analysis of Genetic Environment of Beta-Lactamase Genes
The genetic environment of the beta-lactamase genes was detected by long PCR using LA-Taq PCR solution (TaKaRa) according to the manufacturer’s instructions. Primers to detect the genetic environment surrounding the beta-lactamase genes were designed according to the sequences corresponding to the GenBank Accession numbers listed in Figure 1, using Vector NTI advance 11.0 software. The template was initially denatured at 94°C for 4 min, followed by 35 cycles of 94°C for 40 s, 55°C for 1 min, and 72°C for 5 min, with a final extension at 72°C for 10 min. The PCR products were verified by agarose gel electrophoresis and sequencing, and all sequencing results were aligned using the BLAST program. The primers are listed in Table 1.
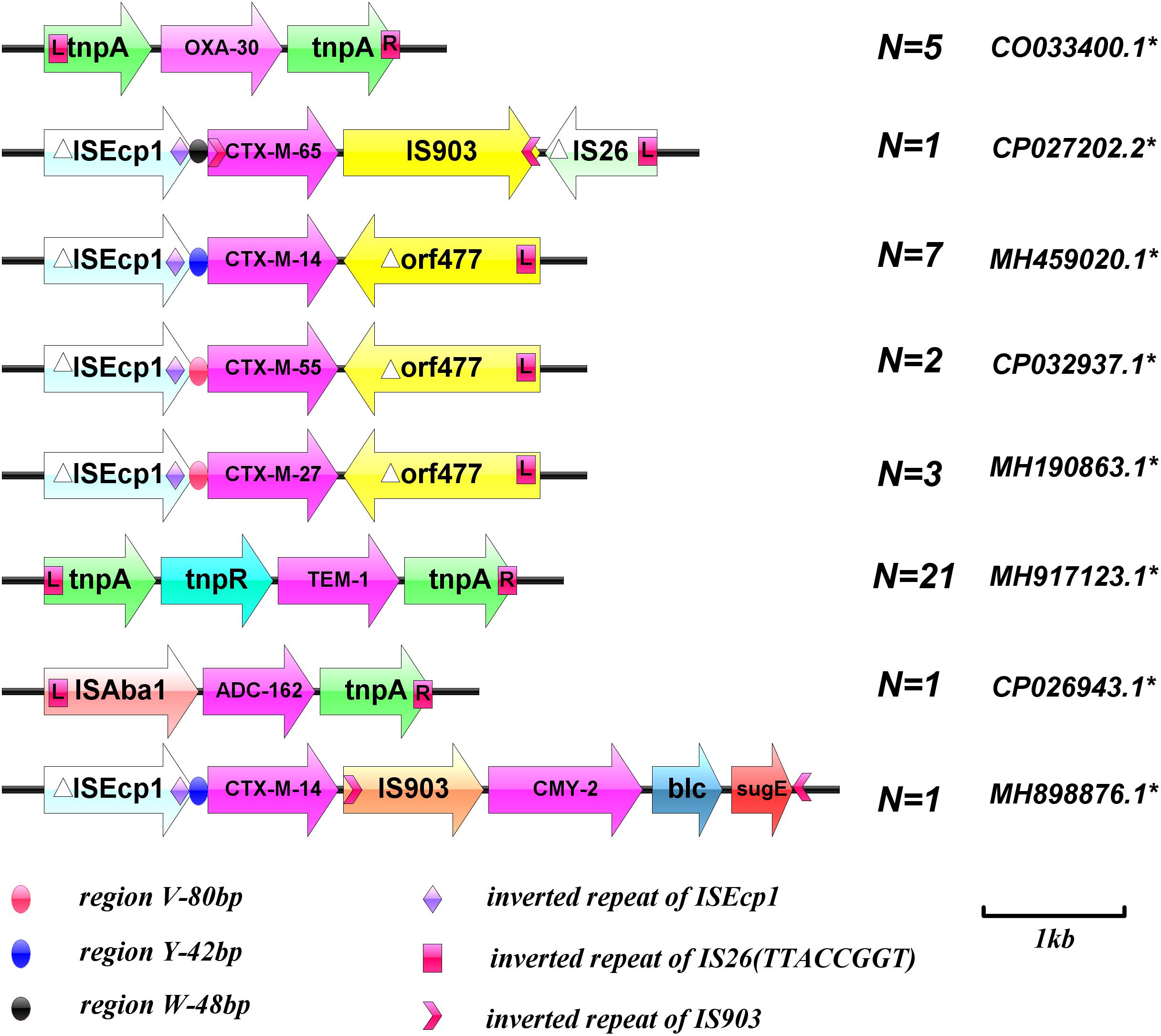
Figure 1. Schematic diagram of the genetic environment surrounding the beta-lactamase genes. ∗GenBank Accession numbers. (Sequences of PCR products were analyzed with BLAST to identify target homologous sequences and their GenBank accession numbers; https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Phylogenetic Grouping and Sequence Type (ST) Determination
The major phylogenetic group of each E. coli strain was determined by multiplex PCR, using the primers listed in Table 1 (Clermont et al., 2000). Multilocus sequence typing (MLST) was performed according to the Pasteur protocol2. Eight conserved housekeeping genes were amplified by PCR using primer sets for dinB, icdA, pabB, polB, putB, trpA, trpB, and uidA, and sequenced (Table 1). Allele profiles and ST assays were performed according to the E. coli MLST website2 protocol.
Results
Antimicrobial Susceptibility
Fifty bloodstream-infection E. coli isolates were obtained from elderly patients (average age, 70.86 years, range 51–92 years; 30% men, 70% women). Most isolates came from the intensive care unit (76%, 38/50), and others from the hematology medical ward (14%, 7/50) and other wards (10%, 5/50). In vitro antimicrobial susceptibility testing showed that most isolates were sensitive to gentamicin (32%), trimethoprim/sulfamethoxazole (38%), chloramphenicol (46%), ciprofloxacin (48%), levofloxacin (50%), ampicillin (82%), aztreonam (36%), cefepime (54%), cefotaxime (54%), ceftazidime (24%), cefazolin (54%), ceftriaxone (21%), and cefoxitin (9%). Moreover, all the isolates were sensitive to imipenem, amikacin, tobramycin, and meropenem.
Genotypes of Beta-Lactamase Genes
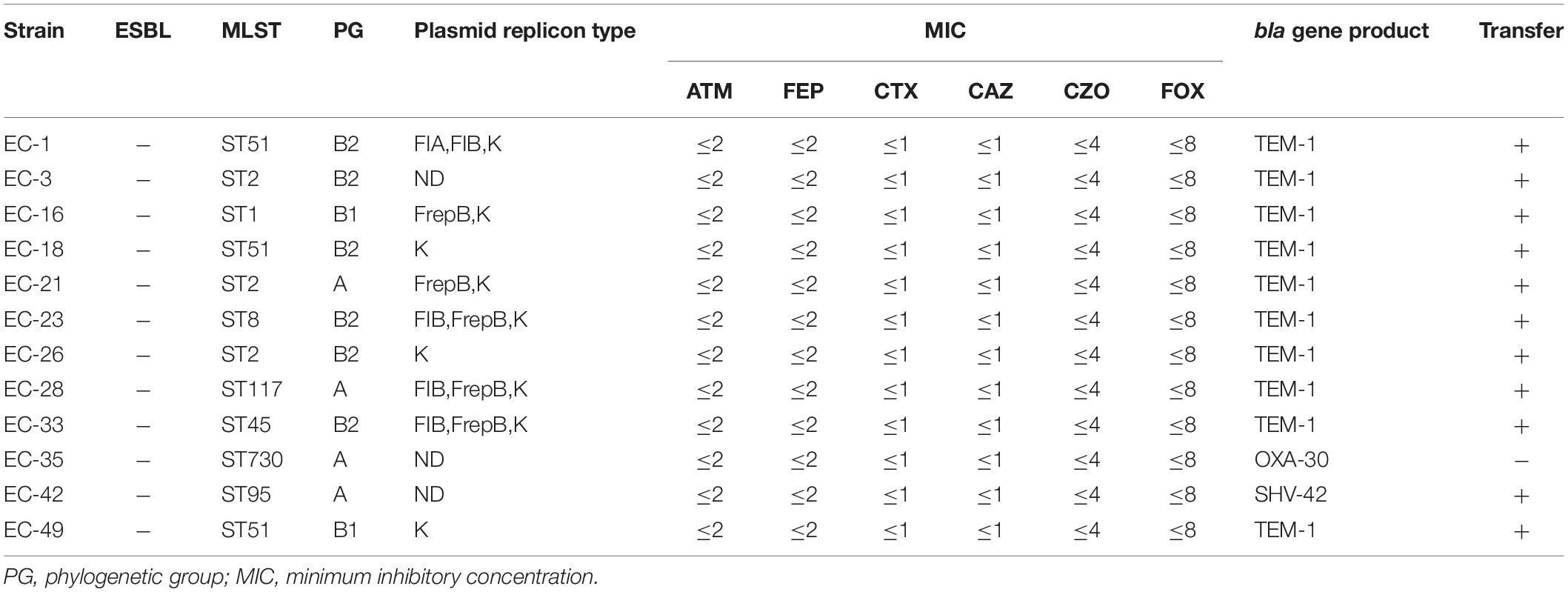
Of the 50 strains, 33 were positive for the beta-lactamase genotype [21 ESBL phenotypes/beta-lactamase genotype positive (Table 2); 12 beta-lactamase genotype positive/phenotype negative (Table 3)], Of these 33 strains, one (3%, 1/33) contained three beta lactamase genes, eight (24%, 8/33) contained two beta lactamase genes, and 24 (73%, 24/33) contained only one beta lactamase gene. Among the beta-lactamase-producing strains, 21 isolates were positive for blaTEM, one for blaSHV, one for blaCIT, two for the blaCTX–M–1 group, 13 for the blaCTX–M–9 group, and five for the blaOXA–1 group. Nucleotide sequence analysis showed that 21 blaTEM-positive isolates carried blaTEM–1 and both blaCTX–M–1 group-positive isolates carried blaCTX–M–55. Of 13 blaCTX–M–9 group-positive isolates, one had blaCTX–M–65, three had blaCTX–M–27, and nine carried blaCTX–M–14. The only blaSHV sequenced was blaSHV–42, the only blaADC sequenced was blaADC–162, the only blaCIT sequenced was blaCMY–2, and all 5 blaOXA–1 group-positive isolates carried blaOXA–30. Meanwhile, all 50 isolates were negative for blaSME, blaPER, blaDHA, blaACC, blaEBC, blaCTX–M–2 group, blaCTX–M–8 group, and blaCTX–M–25 group (Tables 2, 3).
Conjugation Experiments
All 50 E. coli isolates were tested by conjugation and 39 strains were successfully transferred. Thirty-two of the 33 strains carrying beta-lactamase genes were successfully conjugated, but EC-35 was not successfully conjugated. Cefotaxime- and ceftazidime-resistance phenotypes were simultaneously transferred to sodium azide-resistant E. coli J53 recipients by conjugation in beta-lactamase-positive E. coli isolates, respectively.
The conjugate was detected by amplifying the beta lactamase gene primer and the results showed that most of the beta lactamase genes were transferred. However, four OXA-1-group (EC-4,EC-31,EC-35,EC-48), three TEM-group (EC-7,EC-32-EC-49), and four CTX-M-9-group (EC-4,EC-7, EC-27,EC-41) genes did not transfer and were not amplified in the corresponding transconjugants. Resistance to non-beta-lactamase antimicrobials was also co-transferred in some cases, in addition to the transfer of extended-spectrum cephalosporin resistance (Clasen et al., 2019). The characteristics of the E. coli J53 transconjugants carrying beta-lactamase genes are shown in Table 4.
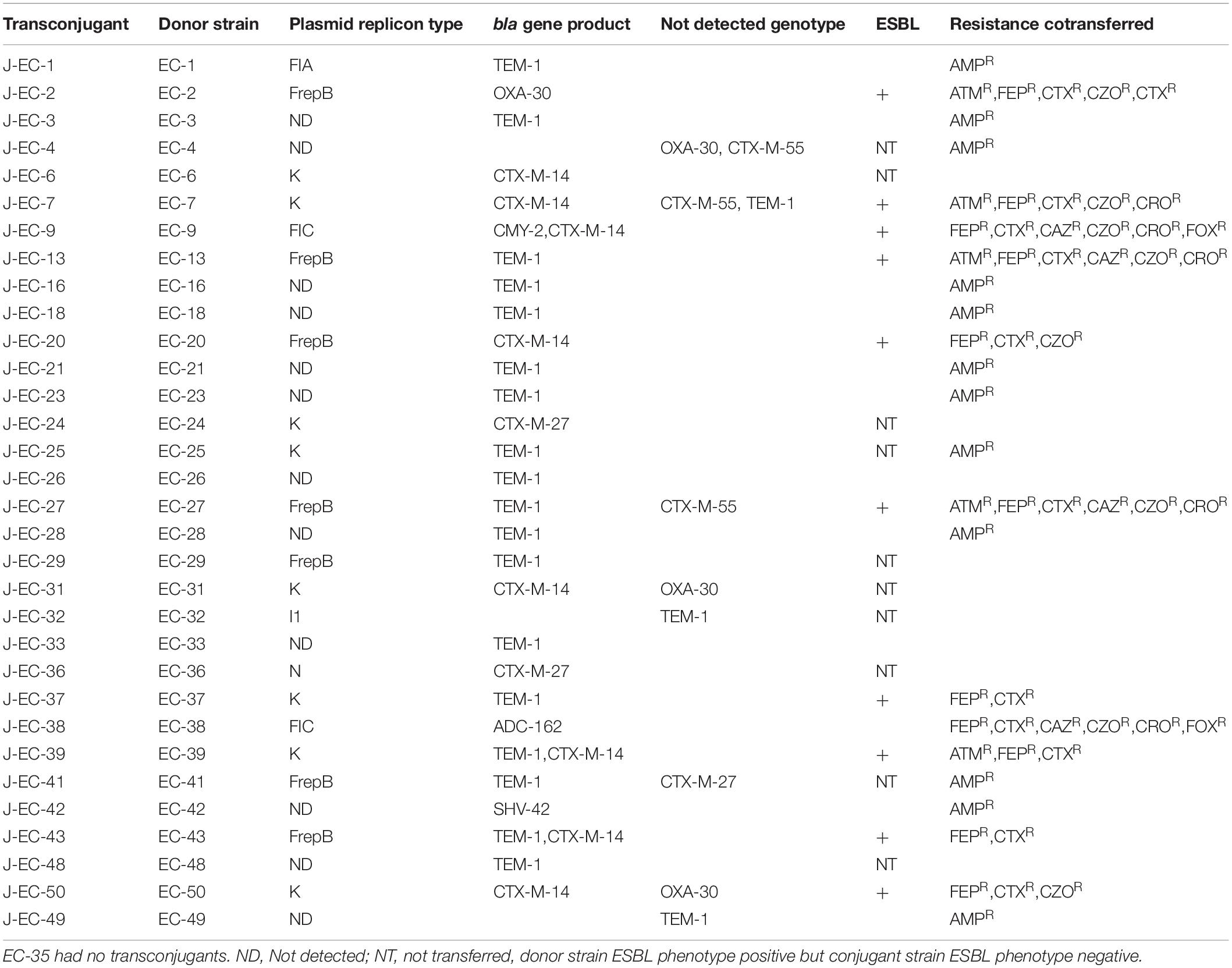
Table 4. Genotypes and drug-resistance phenotypes of bloodstream-infection Escherichia coli conjugates.
Plasmid Analysis
Replicon-typing data for the clinical isolates carrying beta-lactamase genes (Table 2) revealed 10 different replicon types. Among these, two or more plasmid replicon types were detected simultaneously in the 19/21 strain, no plasmid replicon type was detected in the EC-4 strain, and only one plasmid replicon (IncFIC) was carried in the EC-38 strain. The IncF plasmid replicon type was the most common replicon among both types of isolates. Among the ESBL-phenotype-negative strains, plasmid replicon types were not detected in three strains, and the remaining nine strains included only four plasmid replicon types, but all contained IncK plasmid replicons (Table 3).
Plasmid replicon analysis was carried out in the 32 strains with beta-lactamase genes and successful conjugation. Interestingly, no plasmid replicons were detected in 12 transconjugants, and the other 20 transconjugants carried only one plasmid. A total of three plasmid replicon types were detected: IncF (9/19), IncK (9/19), and IncN (1/19) (Table 4). Nine strains were negative for both ESBL genotypes and phenotypes, with no conjugation result and no detection of any plasmid replicon type (Table 5).
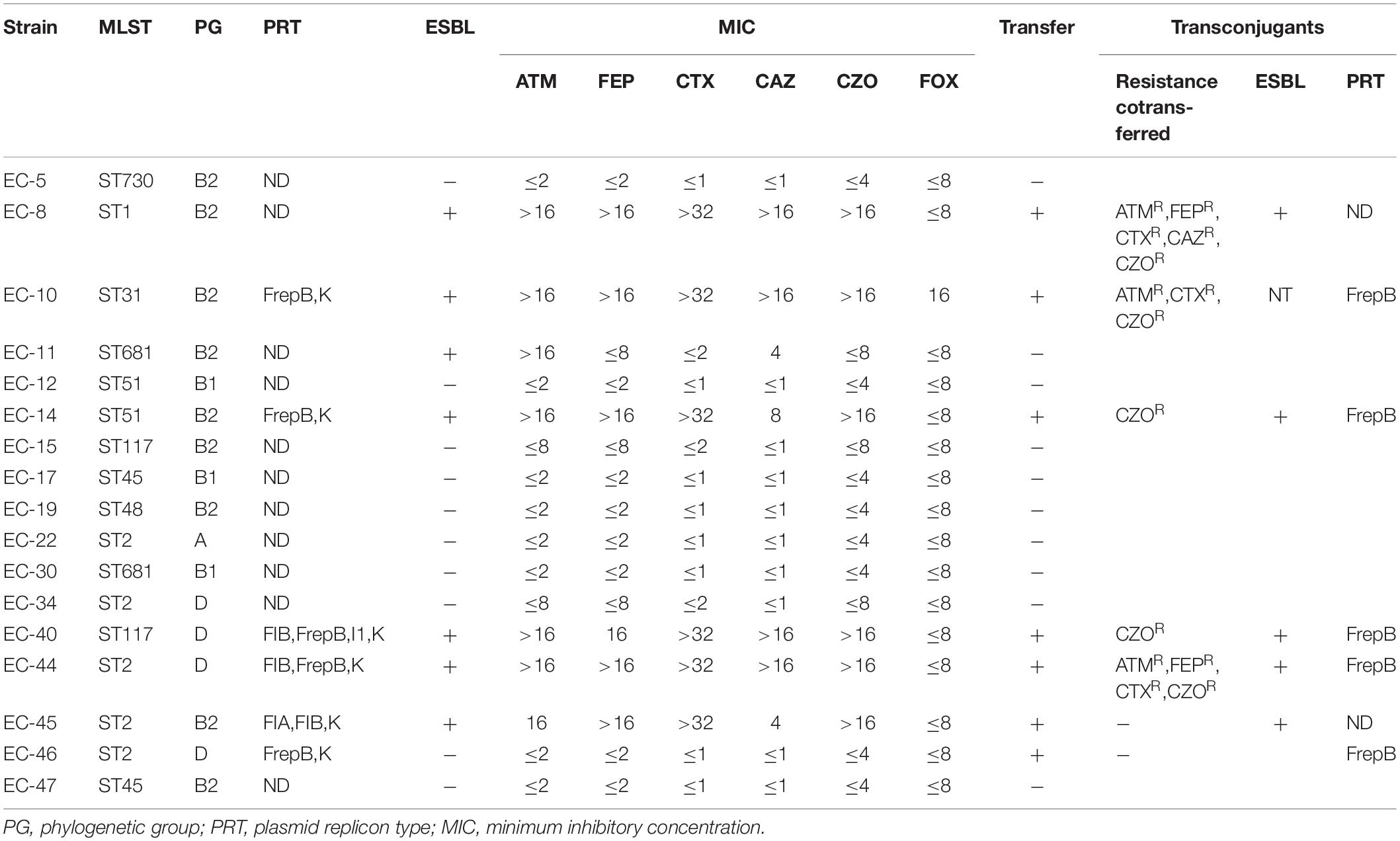
Table 5. ESB-genotype-negative bloodstream-infection Escherichia coli resistance phenotypes and characterization of transconjugants.
Genetic Environment of Beta-Lactamase Genes
The genetic environment surrounding the beta-lactamase genes was verified. Overlapping PCR showed that all blaCTX–M groups were ISEcp1 upstream and orf477 or IS903 downstream. Both blaTEM–1 and blaOXA–30 were located between two transposes and blaADC–162 was located between ISAba1 and tnpA. Unexpectedly, the genetic environment surrounding EC-9 E. coli beta-lactamase genes was relatively unique; blaCTX–M–14 combined with blaCMY–2 through an insertion sequence (IS903), to constitute a composite structure of ISEcp1-blaCTX–M–14-IS903-blaCMY–2-blc-sugE (Figure 1).
Phylogenetic Group and ST Designation
Phylogenetic analysis of E. coli isolates carrying beta-lactamase genes showed that 24 belonged to virulent groups B2 (n = 19) and D (n = 5), and nine to non-toxic groups B1 (n = 3) and A (n = 6). Similarly, analysis of E. coli isolates with non-beta-lactamase genes showed that 13 belonged to virulent groups B2 (n = 9) and D (n = 4), and four belonged to non-toxic groups B1 (n = 3) and A (n = 1).
MLST analysis identified 12 unique STs among the 50 E. coli isolates (Tables 2, 3, 5): ST1 (n = 2), ST2 (n = 10), ST9 (n = 5), ST31 (n = 2), ST45 (n = 4), ST48 (n = 4), ST51 (n = 6), ST95 (n = 3), ST117 (n = 4), ST131 (n = 4), ST681 (n = 3), and ST730 (n = 3) strains.
Discussion
In the present study, we characterized the ESBL and AmpC phenotypes and genotypes of beta-lactamase-producing E. coli blood isolates from patients in China from 2014 to 2018. These results provide the first extensive molecular report of plasmid-mediated ESBL and AmpC beta-lactamase-producing E. coli strains isolated from the bloodstream in elderly patients. Of the 50 E. coli isolates studied, 28 were positive for ESBL phenotypes, 33 were positive for beta-lactamase genes, 21 strains were positive for both, and 10 were negative for both. Thirteen (61.9%) strains had blaCTX–M-type ESBL genes and two (9.5%) produced ampC genes. blaCTX–M-type genes were more common than blaOXA and blaSHV, and ampC genes (blaADC and blaCMY) were observed sporadically. A previous study reported that the blaTEM and blaSHV genes were the most prevalent while the detection frequency of the CTX-M group was low among E. coli isolated from China (Quan et al., 2017), however, the current study found a higher prevalence and variety of blaCTX–M genes than previously reported (Shi et al., 2015; Zhao et al., 2016). The results of our study suggest that previous studies may have underestimated the frequency of blaCTX–M gene transport in Escherichia coli samples isolated from blood, which may be related to increased selective pressure of cephalosporins in China.
The 50 E. coli isolates in the current study carried a variety of blaCTX–M genes (blaCTX–M–14, 8/40; blaCTX–M–27, 3/40; blaCTX–M–55, 2/40; blaCTX–M–65, 1/40), and some also contained blaTEM–1 (21/40). Although these blaCTX–M variants have previously been reported in E. coli strains isolated in many countries (Lambert et al., 2011), few studies have detected the ampC gene in bloodstream-infection E. coli strains in China (Quan et al., 2017), and no previous studies have isolated E. coli from blood samples from elderly patients. In particular, we detected and confirmed a case of the ampC gene blaADC–162. Related studies to date have only detected the ampC gene blaCMY–2 in clinical bloodstream infections of Escherichia coli in Europe, though genome-wide sequencing confirmed that certain strains of chicken carry blaCMY–2 with high homology (Pietsch et al., 2018; Ebmeyer et al., 2019; Seo et al., 2019). To the best of our knowledge, the current study provides the first evidence for blaADC–162 in Escherichia coli strains isolated from clinical samples from patients bloodstream infections. Overall, our results indicated that the diversity of ESBL and/or ampC genes in E. coli strains is increasing, constituting a potential public health problem.
Among the beta-lactamase-genotype-positive strains, most beta-lactamase genes could be transferred to the recipient E. coli J53 strain by conjugation. Interestingly, 32 of the 33 beta-lactamase-genotype-positive strains were also carried conjugant plasmids, while EC-35 failed the conjugation test. EC-35 carried the blaOXA–30 gene, but no plasmid replicon was detected. Additional plasmid extraction experiments indicated that EC-35 carries a plasmid of approximately 15 kb in length, and the plasmid DNA amplified was positive for blaOXA–30 gene. EC-35 was shown to transmit the blaOXA–30 gene through a plasmid, consistent with previous reports (Carattoli, 2009). Eight of the remaining 32 conjugant strains partially or totally lost the beta-lactamase gene. No plasmid replicon was detected in J-EC-4 or J-EC-49, and the beta-lactamase gene was lost completely, and J-EC-7, J-EC-31, and J-EC-50 carried blaCTX–M–14, while the other beta-lactamase genes were lost. All the plasmid replicons were IncK and other plasmid replicons were lost, suggesting that blaCTX–M–14 can be transmitted by the IncK plasmid in bloodstream-infection E. coli. Previous reports indicated that blaCTX–M–14 was mainly related to the IncK plasmid in Spain and Britain (Carattoli, 2009). However, although both J-EC-6 and J-EC-31 had blaCTX–M–14 mediated by the IncK plasmid, the ESBL phenotype of the conjugant strains was lost. The genetic environment around their donor strains was △ISEcp1 (regionY-42 bp) – blaCTX–M–14-orf477. A 42 bp region with an identical sequence (Y sequence) was found upstream of the start codon of the beta-lactamase gene (Eckert et al., 2006). Regarding J-EC-6 and J-EC-31, amplification of the primer pair (ISEcp1-F and CTX-M-9-R) failed when identical primers were used to validate the genetic environment around blaCTX–M–14. Reverse PCR primers (M9R-F and M9R-R) were designed according to CTX-M-9, and sequencing and analysis of the reverse PCR amplification products revealed five and three base mutations in the△ISEcp1 promoter regions of J-EC-6 and J-EC-31, respectively. This may have been due to an increase in the truncated length of ISEcp1, resulting in the loss of promoters in the primer regions and abnormal expression of blaCTX–M–14.
In the present study, only EC-9 and EC-38 strains, with blaCMY–2 and blaADC–162, respectively, were resistant to fosfomycin. EC-9 carried FIC and K plasmid replicates, and EC-38 carried FIC plasmid replicates. Both strains were ST95 but belonged to different developmental groups, B2 and A, respectively. The genetic environments around blaCMY–2 and blaADC–162 were ISEcp1 (regionY-42bp) – blaCTX–M–14-IS903-blaCMY–2-blc-sugE and ISAba1-blaADC–162-tnpA, respectively. IS903 was inserted between two beta-lactamase genes in EC-9 to form a relatively complex tandem structure. Overlapping PCR demonstrated that ISEcp1 (regionY-42bp)-blaCTX–M–14-IS903-blaCMY–2-blc-sugE could be completely transferred by conjugation. ISAba1 in ISAba1-blaADC–162-tnpA is a specific insertion sequence found in Acinetobacter baumannii, which can mediate the transfer of drug-resistance genes. The current study provides the first evidence for the existence of the same ISAba1-blaADC–162-tnpA insertion sequence in E. coli, suggesting that ISAba1 can be transmitted between E. coli and A. baumannii as a mobile genetic structural element.
Interestingly, blaCMY–2 and blaADC–162 were successfully detected in J-EC-9 and J-EC-38, respectively, and both carried only FIC plasmid replicons, indicating that blaCMY–2 and blaADC–162 were both transported by the IncF plasmid. Although only one IncF plasmid-mediated blaCMY–2 and one blaADC–162-positive strain were detected in this study and no evidence of a cloning epidemic was found, the results suggested the need to remain highly vigilant. The IncF plasmid is known to contain three basic replicates: RepFIA, RepFIB, and RepFIC. The IncF plasmid is a narrow host plasmid with a specific region encoding multidrug-resistance genes. It is only prevalent in Enterobacteriaceae bacteria and can be used as a cloning agent (Yang et al., 2015). Multidrug resistance of the IncF plasmid is closely related to its mobile elements, including the insertion sequence, integron, and transposon, allowing it to capture or recombine resistance genes (Saul et al., 1989; Villa et al., 2010; Yang et al., 2015). It is therefore easy to conjugate and transfer, resulting in the dissemination of blaCMY–2 and blaADC–162 clones, with potentially adverse consequences. To the best of our knowledge, this is the first report of IncF-plasmid-mediated blaCMY–2 and blaADC–162 in bloodstream-infection E. coli, indicating the need to be vigilant.
The current results demonstrated that the horizontal transmission of beta-lactamase genes in bloodstream E. coli strains is mainly mediated by IncF and IncK plasmids. Despite the differences in plasmid skeleton and variety, few replicons were detected in bloodstream infection E. coli, and the conjugates carried only one type of replicon at most. In addition, the plasmids and genetic environment of the blaCTX–M group play an important role in regulating the expression, transfer, and transmission of resistance genes. Detection of ISEcp1 upstream of blaCTX–M, blaCMY–2, and blaCMY–M genes with different plasmids showed that ISEcp1 plays an important role in capturing, expressing, and continuously mobilizing blaCTX–M group and blaCMY–2 genes. An ISEcp1 insertion sequence upstream of the gene could result in high levels of expression of blaCTX–M resistance genes and carry these resistance genes between chromosomes and plasmids for transfer, leading to spreading among different strains. Inserted sequences such as IS26, IS903, and orf477 are also frequently associated with blaCTX–M resistance genes. These mobile genetic elements can be distributed randomly upstream and downstream of blaCTX–M resistance genes, forming different genomic components in different blaCTX–M resistance genes, acting on drug-resistance genes either together or separately, regulating their expression, and mediating their transmission.
Conclusion
To the best of our knowledge, this study provides the first molecular characterization of beta-lactamase genes from bloodstream isolates of E. coli from elderly patients. Beta-lactamase genes, especially blaTEM–1, blaCTX–M–14, blaOXA–30, blaCTX–M–27, blaCTX–M–55, and blaCTX–M–65, were widely prevalent in bloodstream-infection E. coli from these patients. Interestingly, blaCMY–2 and blaADC–162 were both transported by IncF plasmids, which are prone to conjugation, indicating the potential for outbreak epidemics related to these genotypes of bloodstream-infection E. coli.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found in GenBank under accession numbers: CO033400.1, CP027202.2, MH459020.1, CP032937.1, MH190863.1, MH917123.1, CP026943.1, and MH898876.1.
Ethics Statement
This study used strains obtained from patient blood. The Ethics Committee of Shanghai University of Medicine & Health Sciences Affiliated Sixth People’s Hospital South Campus waived the need for the study to be reviewed or approved by an ethics committee because none of the strains were cultured in primary culture, and no information could be traced directly to any individual patient.
Author Contributions
LX and QW conceived the study, analyzed the data, and wrote the manuscript. WL coordinated the study. LX, XW, NK, MC, LZ, and MS performed the experiments. LX, QW, and WL revised the manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (Grant Nos. 81572061 and 81572034), and was partly supported by the Outstanding Academic Leaders Plan of Shanghai (Grant No. 2018BR07), the Shanghai University of Medicine and Health Sciences Seed Foundation (Grant No. SFP-18-20-15-003), and the Shanghai Municipal Health and Family Planning Commission Youth Project (Grant No. 20164Y0156), and Fengxian District Science and Technology Commission Youth Project (Grant No. 20181801).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Andrea, D., Mm, Arena, F, Pallecchi, L., and Rossolini, G. M. (2013). CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int. J. Med. Microbiol 303, 305–317. doi: 10.1016/j.ijmm.2013.02.008
Baron, S., Jouy, E., Larvor, E., Eono, F., Bougeard, S., and Kempf, I. (2014). Impact of third-generation-cephalosporin administration in hatcheries on fecal escherichia coli antimicrobial resistance in broilers and layers. Antimicrob, Agents Chemother. 58, 5428–5434. doi: 10.1128/AAC.03106-14
Bartoletti, M., Giannella, M., Caraceni, P., Domenicali, M., Ambretti, S., Tedeschi, S., et al. (2014). Epidemiology and outcomes of bloodstream infection in patients with cirrhosis. J. Hepatol. 61, 51–58. doi: 10.1016/j.jhep.2014.03.021
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/aac.01707-08
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Meth. 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Che, T., Bethel, C. R., Pusztai-Carey, M., Bonomo, R. A., and Carey, P. R. (2014). The different inhibition mechanisms of OXA-1 and OXA-24 beta-lactamases are determined by the stability of active site carboxylated lysine. J. Biol. Chem. 289, 6152–6164. doi: 10.1074/jbc.M113.533562
Clasen, J., Birkegard, A. C., Graesboll, K., and Folkesson, A. (2019). The evolution of TEM-1 extended-spectrum beta-lactamases in E. coli by cephalosporins. J. Glob. Antimicrob. Resist. 19, 32–39. doi: 10.1016/j.jgar.2019.03.010
Clermont, O., Bonacorsi, S., and Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000
CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing. Informational Supplement M100-s28, 28th Edn. Wayne, PA: Clinical Laboratory Standards Institute.
Du, B., Long, Y., Liu, H., Chen, D., Liu, D., Xu, Y., et al. (2002). Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae bloodstream infection: risk factors and clinical outcome. Intens. Care Med. 28, 1718–1723. doi: 10.1007/s00134-002-1521-1
Ebmeyer, S., Kristiansson, E., and Larsson, D. G. J. (2019). CMY-1/MOX-family AmpC β-lactamases MOX-1, MOX-2 and MOX-9 were mobilized independently from three aeromonas species. J. Antimicrob. Chemoth. 74, 1202–1206. doi: 10.1093/jac/dkz025
Eckert, C., Gautier, V., and Arlet, G. (2006). DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemoth. 57, 14–23. doi: 10.1093/jac/dki398
Hung, W. T., Cheng, M. F., Tseng, F. C., Chen, Y. S., Shin-Jung, L. S., Chang, T. H., et al. (2019). Bloodstream infection with extended-spectrum beta-lactamase-producing Escherichia coli: the role of virulence genes. J. Microbiol. Immunol. Infect. doi: 10.1016/j.jmii.2019.03.005 [Epub ahead of print].
Koh, T. H., Wang, G. C., Sng, L. H., and Koh, T. Y. (2004). CTX-M and plasmid-mediated AmpC-producing Enterobacteriaceae Singapore. Emerg. Infect. Dis 10, 1172–1174. doi: 10.3201/eid1006.030726
Lambert, M. L., Suetens, C., Savey, A., Palomar, M., Hiesmayr, M., Morales, I., et al. (2011). Clinical outcomes of health-care-associated infections and antimicrobial resistance in patients admitted to European intensive-care units: a cohort study. Lancet Infect. Dis. 11, 30–38. doi: 10.1016/S1473-3099(10)70258-9
Pietsch, M., Irrgang, A., Roschanski, N., Brenner Michael, G., Hamprecht, A., Rieber, H., et al. (2018). Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genomics 19:601. doi: 10.1186/s12864-018-4976-3
Quan, J., Zhao, D., Liu, L., Chen, Y., Zhou, J., Jiang, Y., et al. (2017). High prevalence of ESBL-producing Escherichia coli and Klebsiella pneumoniae in community-onset bloodstream infections in China. J. Antimicrob. Chemother. 72, 273–280.
Razazi, K., Derde, L. P. G., Verachten, M., Legrand, P., Lesprit, P., and Brun-Buisson, C. (2012). Clinical impact and risk factors for colonization with extended-spectrum β-lactamase-producing bacteria in the intensive care unit. Intens. Care Med. 38, 1769–1778. doi: 10.1007/s00134-012-2675-0
Salverda, M. L., De Visser, J. A., and Barlow, M. (2010). Natural evolution of TEM-1 beta-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol. Rev. 34, 1015–1036. doi: 10.1111/j.1574-6976.2010.00222.x
Saul, D., Spiers, A. J., McAnulty, J., Gibbs, M. G., Bergquist, P. L., and Hill, D. F. (1989). Nucleotide sequence and replication characteristics of RepFIB, a basic replicon of IncF plasmids. J. Bacteriol. 171, 2697–2707. doi: 10.1128/jb.171.5.2697-2707.1989
Seo, K. W., Shim, J. B., and Lee, Y. J. (2019). Emergence of CMY-2-Producing Escherichia coli in Korean layer parent stock. Microb. Drug Resist. 25, 462–468. doi: 10.1089/mdr.2018.0254
Shi, H., Sun, F., Chen, J., Ou, Q., Feng, W., Yong, X., et al. (2015). Epidemiology of CTX-M-type extended-spectrum beta-lactamase (ESBL)-producing nosocomial -Escherichia coli infection in China. Ann. Clin. Microbiol. Antimicrob. 14:4. doi: 10.1186/s12941-015-0063-7
van der Mee-Marquet, N. L., Blanc, D. S., Gbaguidi-Haore, H., Dos Santos Borges, S., Viboud, Q., Bertrand, X., et al. (2015). Marked increase in incidence for bloodstream infections due to Escherichia coli, a side effect of previous antibiotic therapy in the elderly. Front. Microbiol. 6:646. doi: 10.3389/fmicb.2015.00646
Velasova, M., Smith, R. P., Lemma, F., Horton, R. A., Duggett, N., Evans, J., et al. (2019). Detection of extended spectrum beta-lactam (ESBL), AmpC and carbapenem resistance in Enterobacteriaceae in beef cattle in Great Britain in 2015. J. Appl. Microbiol. 126, 1081–1095. doi: 10.1111/jam.14211
Villa, L., Garcia-Fernandez, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Xiao, L., Wang, X., Kong, N., Cao, M., Zhang, L., Wei, Q., et al. (2019). Polymorphisms of gene cassette promoters of the class 1 integron in clinical proteus isolates. Front. Microbiol. 10:790. doi: 10.3389/fmicb.2019.00790
Yang, Q. E., Sun, J., Li, L., Deng, H., Liu, B. T., Fang, L. X., et al. (2015). IncF plasmid diversity in multi-drug resistant Escherichia coli strains from animals in China. Front. Microbiol. 6:964. doi: 10.3389/fmicb.2015.00964
Zhao, S. Y., Zhang, J., Zhang, Y. L., Wang, Y. C., Xiao, S. Z., Gu, F. F., et al. (2016). Epidemiology and risk factors for faecal extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBL-E) carriage derived from residents of seven nursing homes in western Shanghai. China. Epidemiol. Infect. 144, 695–702. doi: 10.1017/S0950268815001879
Keywords: beta lactamase, bloodstream infection, Escherichia coli, extraintestinal pathogenic, AmpC
Citation: Xiao L, Wang X, Kong N, Zhang L, Cao M, Sun M, Wei Q and Liu W (2019) Characterization of Beta-Lactamases in Bloodstream-Infection Escherichia coli: Dissemination of blaADC–162 and blaCMY–2 Among Bacteria via an IncF Plasmid. Front. Microbiol. 10:2175. doi: 10.3389/fmicb.2019.02175
Received: 07 June 2019; Accepted: 05 September 2019;
Published: 01 October 2019.
Edited by:
Vishvanath Tiwari, Central University of Rajasthan, IndiaReviewed by:
Naouel Klibi, Tunis El Manar University, TunisiaMiguel Castañeda, Meritorious Autonomous University of Puebla, Mexico
Copyright © 2019 Xiao, Wang, Kong, Zhang, Cao, Sun, Wei and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quhao Wei, lab_wqh@126.com; Weiwei Liu, huashanvivian@126.com
†These authors have contributed equally to this work
 Linlin Xiao
Linlin Xiao Xiaotong Wang3†
Xiaotong Wang3† Quhao Wei
Quhao Wei