
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 September 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02132
This article is part of the Research Topic "One Health" Approach For Revealing Reservoirs And Transmission Of Antimicrobial Resistance View all 27 articles
 Xinjun Hu1,2†
Xinjun Hu1,2† Xiao Yu2†
Xiao Yu2† Yibing Shang1
Yibing Shang1 Hao Xu2
Hao Xu2 Lihua Guo2
Lihua Guo2 Yile Liang1
Yile Liang1 Yixin Kang1
Yixin Kang1 Li Song1
Li Song1 Jifeng Sun1
Jifeng Sun1 Feng Yue1
Feng Yue1 Yimin Mao3*
Yimin Mao3* Beiwen Zheng2*
Beiwen Zheng2*The dissemination of Klebsiella pneumoniae carbapenemases (KPCs) among Gram-negative bacteria is an important threat to global health. However, KPC-producing bacteria from environmental samples are rarely reported. This study aimed to elucidate the underlying resistance mechanisms of three carbapenem-resistant Aeromonas taiwanensis isolates recovered from river sediment samples. Pulsed-field gel electrophoresis (PFGE) and whole genome sequencing (WGS) analysis indicated a close evolutionary relationship among A. taiwanensis isolates. S1-PFGE, Southern blot and conjugation assays confirmed the presence of blaKPC–2 and qnrS2 genes on a non-conjugative plasmid in these isolates. Plasmid analysis further showed that pKPC-1713 is an IncP-6 plasmid with a length of 53,205 bp, which can be transformed into DH5α strain and mediated carbapenems and quinolones resistance. The plasmid backbone of p1713-KPC demonstrated 99% sequence identity to that of IncP-6-type plasmid pKPC-cd17 from Aeromonas spp. and IncP-6-type plasmid: 1 from Citrobacter freundii at 74% coverage. A 14,808 bp insertion sequence was observed between merT gene and hypothetical protein in p1713-KPC, which include the quinolone resistance qnrS2 gene. Emergence of plasmid-borned blaKPC and qnrS2 genes from A. taiwanensis isolates highlights their possible dissemination into the environment. Therefore, potential detection of such plasmids from clinical isolates should be closely monitored.
The global spread of Klebsiella pneumoniae carbapenemases (KPCs) among Gram-negative bacteria, has become a major public health concern in recent decades (Grundmann et al., 2017). Infections caused by KPC-producing bacteria have been associated with a high mortality rate of 53%, which presents tremendous challenges for clinicians and healthcare providers (Munoz-Price et al., 2013). Although most KPCs are commonly found in Enterobacteriaceae, the production of KPCs appeared to be less dominance in Aeromonadaceae spp. (Picao et al., 2013; Montezzi et al., 2015; Lamba and Ahammad, 2017; Paschoal et al., 2017; Tuo et al., 2018; Xu et al., 2018; Zheng et al., 2018b). The emergence of blaKPC gene on broad-host-range plasmids has facilitated its rapid dissemination to Enterobacteriaceae and other Gram-negative families (van Duin and Doi, 2017). More importantly, KPC-type carbapenemases are often associated with quinolone resistance determinants, which can be modulated by qnrS gene (Montezzi et al., 2015). The quinolone resistance gene, qnrS2, shares 92% identity with the qnrS gene, was first detected on a highly mobile plasmid that is isolated from wastewater treatment plant bacterial population (Bonemann et al., 2006). Thus far, the co-existence of blaKPC and qnrS2 in the same plasmid has not been reported.
The objective of the present study was to identify plasmid-borne blaKPC–2 genes in Aeromonas taiwanensis from river sediments in China and to analyze the plasmids carrying them. Three strains were isolated and found to harbor IncP-6 plasmids carrying both blaKPC–2 and qnrS2. We characterized the phenotypic and molecular features of these strains in order to assess their genomic epidemiology profiles. Additionally, the complete nucleotide sequence of p1713-KPC was determined by plasmid sequencing. To the best of our knowledge, this is the first study to indicate the co-occurrence of blaKPC–2 and qnrS2 genes in the same plasmid and this plasmid harboring these two gene were founded in A. taiwanensis isolates for the first time.
The KPC-2-producing A. taiwanensis isolates were previously identified from river sediment samples from China (Xu et al., 2018). A total of three A. taiwanensis isolates were further characterized to uncover the underlying resistance mechanisms. The genomic diversity of A. taiwanensis isolates was assessed by XbaI-pulsed-field gel electrophoresis (PFGE) as described previously (Zheng et al., 2016). A dendrogram of PFGE profiles was constructed with BioNumerics v7.6 by using UPGMA (unweighted pair group method with averages) clustering. Isolates with a similarity cut-off of ≥80% were considered as pulsotypes.
To characterize the genomic features and resistome of A. taiwanensis isolates, WGS was conducted on all the three isolates. Genomic DNA was extracted and sequenced with Hiseq X Ten sequencing platform (Illumina, San Diego, CA, United States). Subsequently, de novo genome assembly and bioinformatics analysis were carried out as previously described (Xu et al., 2018; Zheng et al., 2018a). After obtaining the raw reads, the genome sequence of pKPC-1713 was assembled using plasmidSPAdes (Antipov et al., 2016).
The pan-genome analysis was performed with Roary: the pangenome pipeline (version 3.6.0) using the Prokka annotation (Seemann, 2014; Page et al., 2015). The complete genome sequences of all 20 Aeromonas spp. strains were downloaded from NCBI genome database (current as of March 30, 2019). Roary software was used to cluster the above genomes, The software clustered the genomes based on the genes carried by each strain. According to the distribution of each gene among the strains, the genes were divided into core genes and accessory genes. Core genes were defined as genes carried by 95% or more of the strains. Accessory genes were those carried by less than 95% of the strains. Then the core genome of these strains is obtained. A whole-genome phylogenetic tree was built from the core-genome SNPs of Aeromonas spp. and the three studied isolates. Phylogenetic reconstruction and analysis was performed using the R package phangorn (Schliep, 2011).
The size of plasmid was determined by S1-nuclease PFGE analysis, as previously described (Zheng et al., 2015). Southern blot hybridization of plasmid DNA was performed on Aeromonas hydrophila isolates by using DIG-labeled blaKPC- and qnrS- specific probes, according to manufacturer’s instructions (Roche Diagnostics, Germany) (Zheng et al., 2016). Plasmid conjugation experiments were carried out by filter mating using Escherichia coli J53 and EC600 as recipient strains, at a ratio of 1:1 in broth culture (Zheng et al., 2015). The pKPC-1713 plasmid was transferred into chemically competent E. coli DH5α cells via transformation process. The transformants were selected on Luria-Bertani agar plates supplemented with 2 mg/L meropenem. The presence of blaKPC and qnrS2 genes was then screened by PCR and sequencing. Plasmid DNA was extracted from conjugants with Qiagen Plasmid Midi Kit (Qiagen, Valencia, CA). To obtain the complete sequence of the plasmid co-expressing blaKPC and qnrS2, the combined application of PCR walking method with targeted primers (Supplementary Table S1) and Illumina sequencing technique were performed to specifically investigate the representative plasmid pKPC-1713. The RAST annotation pipeline was chosen to perform rapid annotation of the plasmids (Overbeek et al., 2014). Plasmid replicons and antibiotic resistance genes were identified using CGE server1. The sequence of pKPC-1713 was BLAST for homology against the NCBI plasmids database.
The isolated A. taiwanensis 1713, E. coli DH5α and DH5α: pKPC-1713 were cultured overnight, while E. coli ATCC 25922 was used as a quality control. Minimum inhibitory concentrations (MICs) of amikacin, ampicillin, ampicillin-sulbactam, aztreonam, cefazolin, ceftazidime, cefotetan, ceftriaxone, ciprofloxacin, ertapenem, gentamicin, imipenem, levofloxacin, nitrofurantoin, piperacillin-tazobactam, tobramycin, and trimethoprim antibiotics were determined by VITEK 2 system with AST-GN16 panel. The results of antimicrobial susceptibility testing were interpreted according to 2017’s CLSI guidelines.
Plasmid conjugation was performed as previously described with some modification (Yi et al., 2012). A. taiwanensis isolates were electrotransformed with a plasmid pEC1002-MCR(CP021205) which contains a tra module. Isolates containing the resident and pEC1002-MCR plasmids were used as donor strains. Plasmid conjugative transfer was performed by using donor and recipient sodium azide resistant E. coli J53 cells mixed in a 1:1 ratio as described previously. Transconjugants were selected on MacConkey agar containing 2 mg/L imipenem and 100 mg/L sodium azide. Transconjugants were confirmed by susceptibility testing and PCR.
The whole genome shotgun project of the A. taiwanensis isolates has been deposited into DDBJ/EMBL/GenBank under the Bioproject accession number PRJNA478520. The nucleotide sequence of plasmid p1713-KPC has been deposited into the GenBank database under GenBank accession number MH624132.
Pulsed-field gel electrophoresis analysis revealed that two pulsotypes from A. taiwanensis, namely 198 and 186, exhibited relatively identical PFGE patterns (Supplementary Figure S1). Meanwhile, Roary matrix-based gene sequence analysis generated a large pan-genome matrix of 26,778 gene clusters across 23 genomes (Figure 1). Moreover, the 186, 198, and 1713 isolates were found to be genetically closely related by Roary matrix-based gene sequence analysis (Figure 1). These findings are consistent with the results of PFGE profiles, suggesting a possible clonal spread of KPC-producing A. taiwanensis. So far, only few reports described the detection of plasmid-mediated blaKPC determinants in Aeromonadaceae (Picao et al., 2013; Montezzi et al., 2015). Notably, the presence of blaKPC in Aeromonadaceae considered predominantly environmental is remarkable, and the spread of KPC-producing A. taiwanensis in aquatic environments deserves attention.
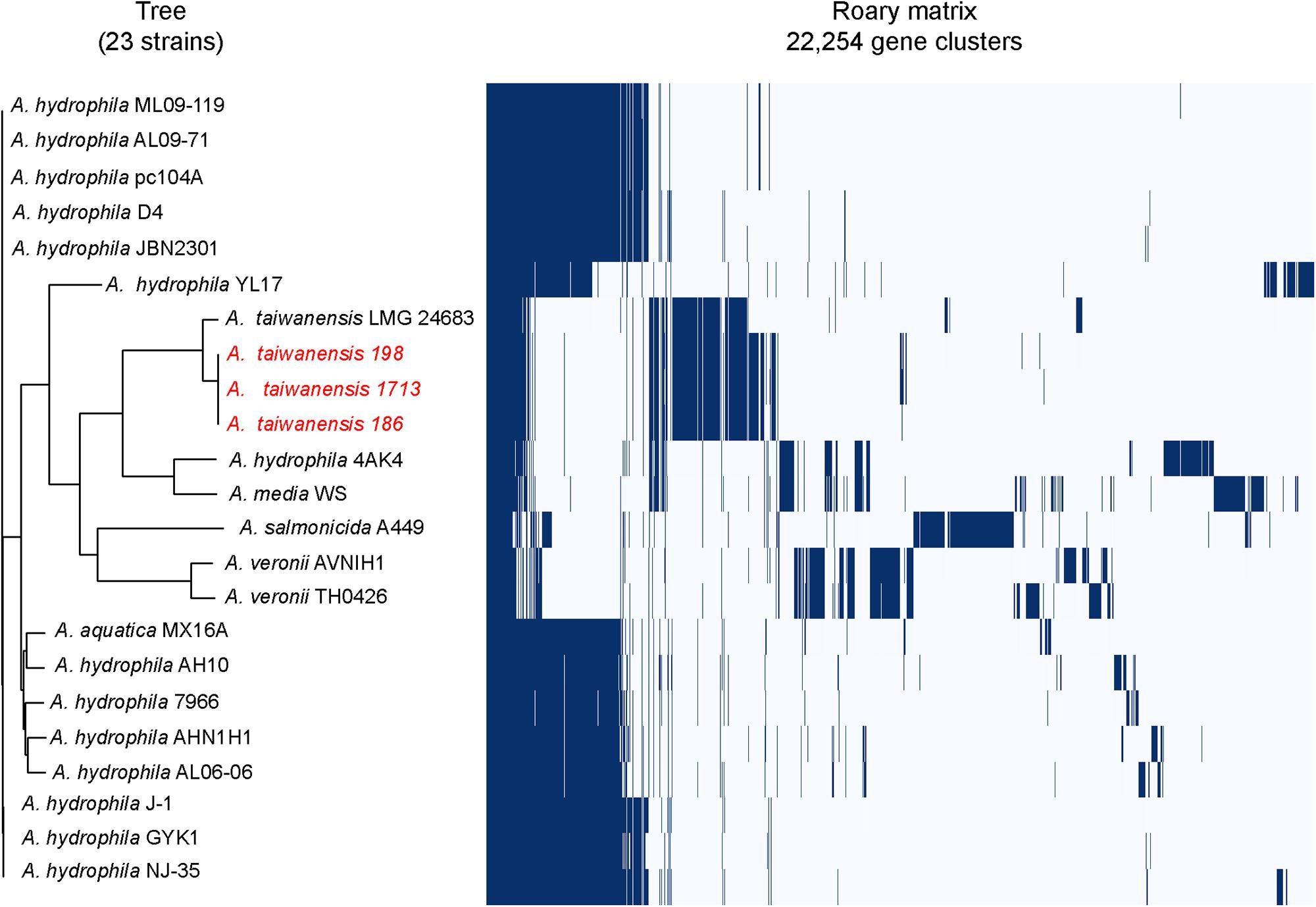
Figure 1. Genetic relatedness analysis of KPC-2-producing Aeromonas spp. isolates. Pan-genome analysis of A. taiwanensis isolates and other Aeromonas isolates by using Roary. The blue bars indicate the pan-genome of Aeromonas isolates, with 26,778 annotated genes detected in all the genomes analyzed. All the complete genome sequences of representative Aeromonas strains were downloaded from NCBI Genome database and used for phylogenetic analysis: Aeromonas aquatica strain MX16A (CP018201), A. hydrophila 4AK4 (CP006579), A. hydrophila strain AH10 (CP011100), A. hydrophila strain AHNIH1 (CP016380), A. hydrophila strain AL06-06 (CP010947), A. hydrophila AL09-71 (CP007566), A. hydrophila subsp. hydrophila ATCC 7966 (NC_008570), A. hydrophila strain D4 (CP013965), A. hydrophila strain GYK1 (CP016392), A. hydrophila J-1 (CP006883), A. hydrophila strain JBN2301 (CP013178), A. hydrophila ML09-119 (NC_021290), A. hydrophila NJ-35 (CP006870), A. hydrophila pc104A (CP007576), A. hydrophila YL17 (CP007518), Aeromonas media WS (CP007567), Aeromonas salmonicida subsp. salmonicida A449 (NC_009348), Aeromonas taiwanensis LMG 24683 (GCA_000820165), Aeromonas veronii strain AVNIH1 (CP014774), and Aeromonas veronii strain TH0426 (CP012504). A. taiwanensis isolates described in this study were colored as red.
S1-PFGE and Southern blot analysis demonstrated that all A. taiwanensis isolates contained a ∼54 kb plasmid, harboring both blaKPC–2 and qnrS2 genes (Figure 2). Moreover, nearly identical plasmid sequences of the three isolates were assembled from WGS data by SPAdes (data not shown), suggesting that these isolates shared the same plasmid profile. Notably, none of the plasmids could be transferred to recipient strains via conjugation, despite repeated attempts, which suggests that blaKPC–2 and qnrS2 are located on a non-conjugative plasmid. Occasionally, qnrS2 has been identified in the plasmids of Aeromonas spp. (Arias et al., 2010; Marti and Balcazar, 2012; Marti et al., 2014; Yang et al., 2017); however, the co-occurrence of blaKPC and qnrS2 genes in the same plasmid has never been described before. To our knowledge, this is the first study to indicate the co-occurrence of blaKPC and qnrS2 genes in the same plasmid. As a consequence, we selected a representative sample of 1713 isolate for further plasmid characterization.
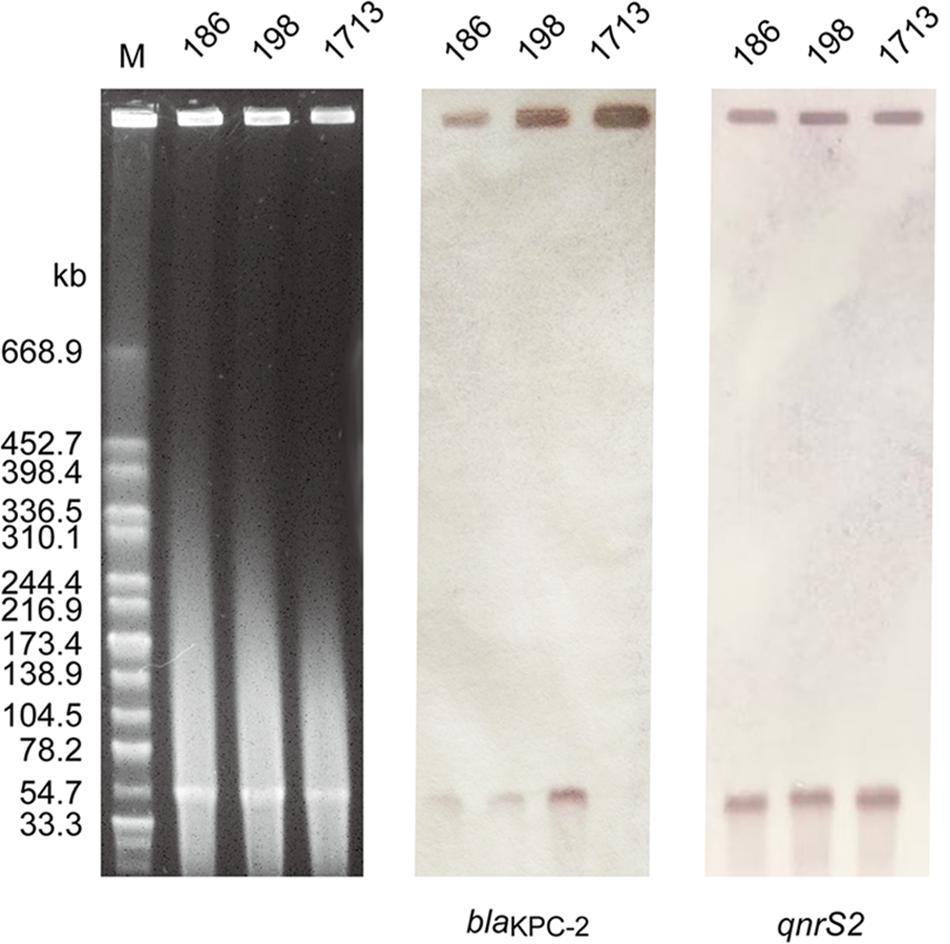
Figure 2. Plasmid profile and Southern blot hybridization of the three A. taiwanensis isolates. The southern blot hybridization of blaKPC and qnrS2 probes using S1 nuclease-digested DNA. Lane M: XbaI-digested genomic DNA fragments of Salmonella enterica serotype Braenderup H9812 as the size marker.
Plasmid transformation and MICs determination revealed that carbapenem and quinolone resistant genes were successfully transferred from a donor strain to E. coli DH5α. Antibiotic susceptibility testing showed that the MIC values of ertapenem, imipenem, ciprofloxacin, and levofloxacin were increased from 0.125 to 8 mg/L, 0.125 to 16 mg/L, 0.25 to 8 mg/L, and 1 to 4 mg/L, respectively (Table 1). These results confirmed that carbapenem and quinolone resistance were successfully transferred to receipt cells.
Sequence analysis of representative pKPC-1713 plasmid revealed the total size of 53,205 bp with an average G + C content of 58% and 66 open reading frames. The plasmid pKPC-1713 harbored five functional regions of genes, including blaKPC–2 encoding region, IS5045 elements, an insertion region, genes involved in plasmid maintenance, and plasmid replication (Figure 3). The search against nr/nt database revealed a 99% identity with Aeromonas sp. ASNIH3 IncP-6-type plasmid pKPC-cd17 at 74% coverage, and a 99% identity with Citrobacter freundii IncP-6-type plasmid: 1 at 74% coverage (Figure 3). In contrast to pKPC-cd17, an insertion of 14,808 bp DNA fragment and IS4321 gene were observed between merT gene and hypothetical protein in p1713-KPC and plasmid: 1, respectively. The insertion sequence of p1713-KPC harbored 18 different genes, which include the quinolone resistance qnrS2 gene. Interestingly, BLAST search of this insertion sequence revealed a 99% identity with A. hydrophila IB101 plasmid AHIB101-pBF7.8 at 100% coverage (Figure 3). We speculate that p1713-KPC has been formed by an IncP-6 plasmid fusing with the qnrS plasmid after one of them has acquired extra DNA with xerD etc. Furthermore, blaKPC–2 gene was found to be located on a 14.5 kb Tn3 transposon element in p1713-KPC with the linear structure: ΔtnpA Tn3-ISApu1-ISApu2-ΔtnpA-Tn3-tnpR Tn3-ISKpn27-ΔblaTEM–1-blaKPC–2-ΔISKpn6 (Figure 3). Previous studies of blaKPC–2 have indicated its association with Tn3-based transposon and responsible for its widespread among Enterobacteriaceae in different geographical locations of China (Shen et al., 2009; Yang et al., 2013).
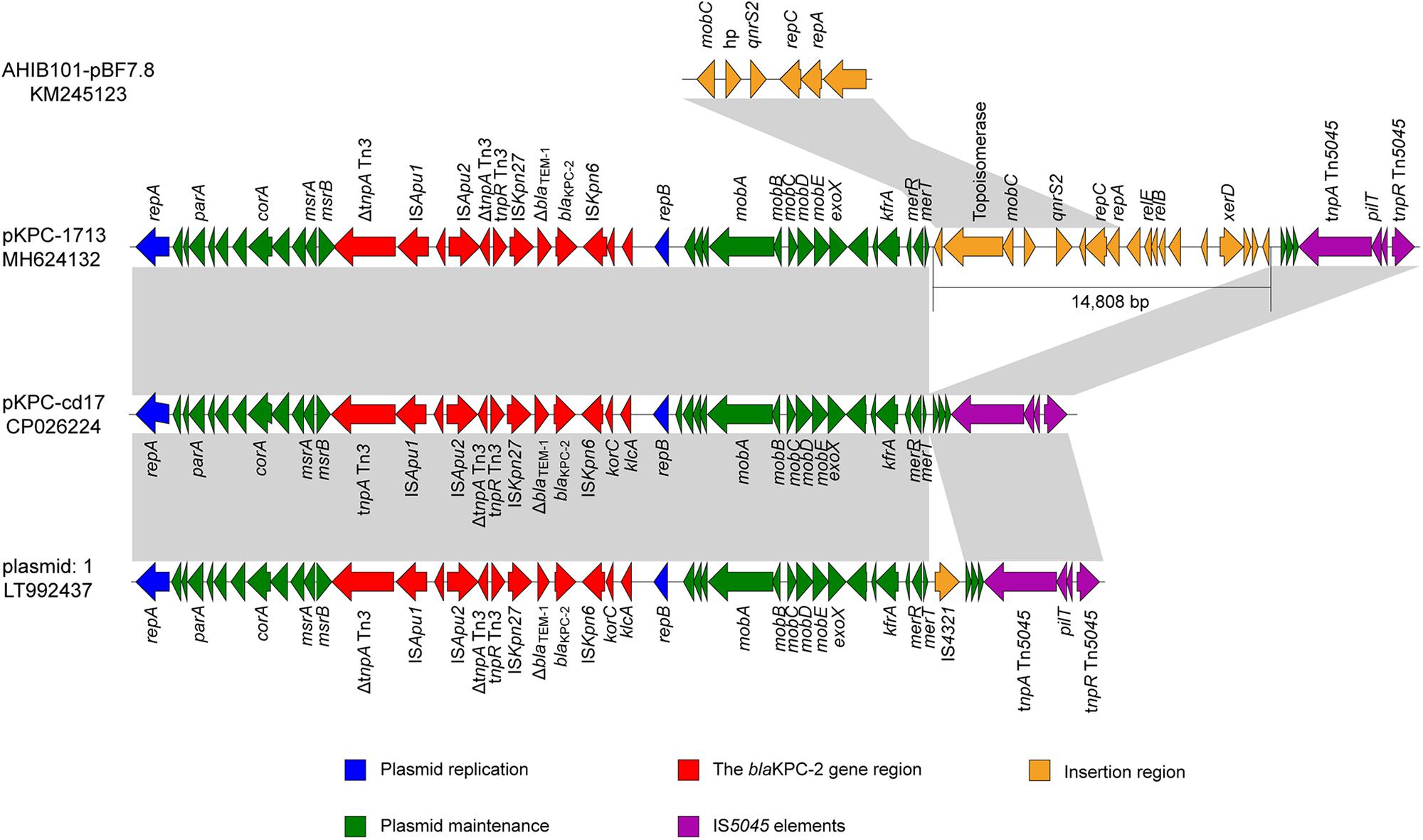
Figure 3. Genomic comparison of p1713-KPC sequence with plasmid pKPC-cd17 (CP026224) from Aeromonas sp. ASNIH3, plasmid 1 (LT992437) from C. Freundii and plasmid AHIB101-pBF7.8 (KM245123) from A. hydrophila. Open reading frames (ORFs) are indicated by arrows, according to their putative functions. The regions sharing a high degree of sequence similarity are shown in gray color.
IncP-6 plasmids are found in various species from both clinical and environmental sources, suggesting that they have a broad host range and potential for long-term persistence in the environment (Naas et al., 2013; Dai et al., 2016; Wang et al., 2017; Yao et al., 2017). Interestingly, most of these blaKPC–2 bearing IncP-6 plasmids were detected in China. Our findings further revealed that blaKPC–2 carrying IncP-6 plasmid has moved into Aeromonas spp. Of note, p1713-KPC is lacking of a tra module encoding primary pilus, which explains the failure of plasmid conjugation. Studies had shown that plasmids could be able to transfer by conjugation if the right self-transmissible plasmids are present (Smillie et al., 2010). However, due to unknown reasons, mobilization transfer from donor A. taiwanensis, which harbored resident plasmid and electrotransformed plasmid containing a tra module, to the recipient cells was not successful. Nevertheless, acquisition of free DNA is a general feature of Aeromonas isolates (Huddleston et al., 2013). And the IncP plasmid, which is a broad-host-range incompatibility plasmid, has also been proved with the potential to mediate the dissemination of antibiotic resistant genes from the Enterobacteriaceae to other Gram-negative bacteria, such as Pseudomonas aeruginosa (Zhao et al., 2017). Since the complex mechanism of plasmid transfer is still not fully understood and these 3 IncP plasmids do encode mobilization functions, potential detection of such incP plasmids from clinical isolates should be closely monitored. Therefore, it is suggested that IncP-6 plasmids may act as an important vector responsible for the genetic transmission of blaKPC and qnrS2 among the aquatic Aeromonas spp.
The analysis of plasmids carrying different resistance genes is pivotal for characterization of bacterial isolates, since they play a significant role in transmission of antibiotic resistance (Galata et al., 2018). In this study, we reported the complete sequence of p1713-KPC, a novel IncP-6 plasmid identified from environmental A. taiwanensis isolates. To our knowledge, this is the first report describing the co-existence of blaKPC–2 and qnrS2 on the same plasmid in A. taiwanensis isolates. The emergence and dissemination of such plasmids in environmental isolates deserve special attention. Given that Aeromonas spp. are ubiquitous organisms isolated from a wide range of environmental niches, they might act as important vectors for the dissemination of plasmid-mediated carbapenem- and quinolone-resistance genes. Therefore, potential detection of such plasmids from clinical isolates should be closely monitored.
The whole genome shotgun project of the A. taiwanensis isolates has been deposited into DDBJ/EMBL/GenBank under the Bioproject accession number PRJNA478520. The complete nucleotide sequence of plasmid p1713-KPC has been deposited into the GenBank database under GenBank accession number MH624132.
XH, YM, and BZ conceived and designed the experiments. XH, XY, YS, HX, YL, YK and LS performed the experiments. LG, JS, and FY analyzed the data. BZ and XH wrote the manuscript.
This study was supported by funding from the National Natural Science Foundation of China (Nos. 81600512 and 81741098); the grant from Henan Science and Technology Department (No. 162102310509); the National Key Research and Development Program of China (No. 2016YFD0501105); and the Zhejiang Provincial Natural Science Foundation of China (No. LY17H190003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02132/full#supplementary-material
FIGURE S1 | PFGE profiles of A. taiwanensis strains. A dendrogram of PFGE profiles was constructed with BioNumerics v7.6 by using UPGMA (unweighted pair group method with averages) clustering. Isolates with a similarity cut-off of ≥80% were considered as pulsotypes. The scale bar indicates the percentage of genetic relatedness.
TABLE S1 | Primers used in the plasmid sequencing of novel IncP-6 plasmid pKPC-1713, harbouring blaKPC–2 and qnrS2 genes.
Antipov, D., Hartwick, N., Shen, M., Raiko, M., Lapidus, A., and Pevzner, P. A. (2016). plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32, 3380–3387.
Arias, A., Seral, C., Navarro, F., Miro, E., Coll, P., and Castillo, F. J. (2010). Plasmid-mediated QnrS2 determinant in an Aeromonas caviae isolate recovered from a patient with diarrhoea. Clin. Microbiol. Infect. 16, 1005–1007. doi: 10.1111/j.1469-0691.2009.02958.x
Bonemann, G., Stiens, M., Puhler, A., and Schluter, A. (2006). Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob. Agents Chemother. 50, 3075–3080. doi: 10.1128/aac.00378-06
Dai, X., Zhou, D., Xiong, W., Feng, J., Luo, W., Luo, G., et al. (2016). The IncP-6 Plasmid p10265-KPC from Pseudomonas aeruginosa carries a novel DeltaISEc33-associated bla KPC-2 gene cluster. Front. Microbiol. 7:310. doi: 10.3389/fmicb.2016.00310
Galata, V., Fehlmann, T., Backes, C., and Keller, A. (2018). PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res. 47, D195–D202. doi: 10.1093/nar/gky1050
Grundmann, H., Glasner, C., Albiger, B., Aanensen, D. M., Tomlinson, C. T., Andrasevic, A. T., et al. (2017). Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect. Dis. 17, 153–163. doi: 10.1016/S1473-3099(16)30257-2
Huddleston, J. R., Brokaw, J. M., Zak, J. C., and Jeter, R. M. (2013). Natural transformation as a mechanism of horizontal gene transfer among environmental Aeromonas species. Syst. Appl. Microbiol. 36, 224–234. doi: 10.1016/j.syapm.2013.01.004
Lamba, M., and Ahammad, S. Z. (2017). Sewage treatment effluents in Delhi: a key contributor of beta-lactam resistant bacteria and genes to the environment. Chemosphere 188, 249–256. doi: 10.1016/j.chemosphere.2017.08.133
Marti, E., and Balcazar, J. L. (2012). Multidrug resistance-encoding plasmid from Aeromonas sp. strain P2G1. Clin. Microbiol. Infect. 18, E366–E368. doi: 10.1111/j.1469-0691.2012.03935.x
Marti, E., Huerta, B., Rodriguez-Mozaz, S., Barcelo, D., Jofre, J., and Balcazar, J. L. (2014). Characterization of ciprofloxacin-resistant isolates from a wastewater treatment plant and its receiving river. Water Res. 61, 67–76. doi: 10.1016/j.watres.2014.05.006
Montezzi, L. F., Campana, E. H., Correa, L. L., Justo, L. H., Paschoal, R. P., da Silva, I. L., et al. (2015). Occurrence of carbapenemase-producing bacteria in coastal recreational waters. Int. J. Antimicrob. Agents 45, 174–177. doi: 10.1016/j.ijantimicag.2014.10.016
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7
Naas, T., Bonnin, R. A., Cuzon, G., Villegas, M. V., and Nordmann, P. (2013). Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J. Antimicrob Chemother. 68, 1757–1762. doi: 10.1093/jac/dkt094
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The seed and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214.
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Paschoal, R. P., Campana, E. H., Correa, L. L., Montezzi, L. F., Barrueto, L. R. L., da Silva, I. R., et al. (2017). Concentration and variety of carbapenemase producers in recreational coastal waters showing distinct levels of pollution. Antimicrob. Agents Chemother. 61:e1963-17. doi: 10.1128/AAC.01963-17
Picao, R. C., Cardoso, J. P., Campana, E. H., Nicoletti, A. G., Petrolini, F. V., Assis, D. M., et al. (2013). The route of antimicrobial resistance from the hospital effluent to the environment: focus on the occurrence of KPC-producing Aeromonas spp. and Enterobacteriaceae in sewage. Diagn. Microbiol. Infect. Dis. 76, 80–85. doi: 10.1016/j.diagmicrobio.2013.02.001
Schliep, K. P. (2011). Phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593. doi: 10.1093/bioinformatics/btq706
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shen, P., Wei, Z., Jiang, Y., Du, X., Ji, S., Yu, Y., et al. (2009). Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53, 4333–4338. doi: 10.1128/AAC.00260-09
Smillie, C., Garcillan-Barcia, M. P., Francia, M. V., Rocha, E. P., and de la Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. doi: 10.1128/MMBR.00020-10
Tuo, H., Yang, Y., Tao, X., Liu, D., Li, Y., Xie, X., et al. (2018). The prevalence of colistin resistant strains and antibiotic resistance gene profiles in Funan River, China. Front. Microbiol. 9:3094. doi: 10.3389/fmicb.2018.03094
van Duin, D., and Doi, Y. (2017). The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8, 460–469. doi: 10.1080/21505594.2016.1222343
Wang, J., Yuan, M., Chen, H., Chen, X., Jia, Y., Zhu, X., et al. (2017). First report of klebsiella oxytoca strain simultaneously producing NDM-1, IMP-4, and KPC-2 carbapenemases. Antimicrob. Agents Chemother. 61:e877-17. doi: 10.1128/AAC.00877-17
Xu, H., Wang, X., Yu, X., Zhang, J., Guo, L., Huang, C., et al. (2018). First detection and genomics analysis of KPC-2-producing citrobacter isolates from river sediments. Environ. Pollut. 235, 931–937. doi: 10.1016/j.envpol.2017.12.084
Yang, J., Ye, L., Guo, L., Zhao, Q., Chen, R., Luo, Y., et al. (2013). A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin. Microbiol. Infect. 19, E509–E515. doi: 10.1111/1469-0691.12275
Yang, Q., Zhao, M., Wang, K. Y., Wang, J., He, Y., Wang, E. L., et al. (2017). Multidrug-resistant Aeromonas veronii recovered from channel catfish (Ictalurus punctatus) in China: prevalence and mechanisms of fluoroquinolone resistance. Microb. Drug Resist. 23, 473–479. doi: 10.1089/mdr.2015.0296
Yao, Y., Lazaro-Perona, F., Falgenhauer, L., Valverde, A., Imirzalioglu, C., Dominguez, L., et al. (2017). Insights into a novel blaKPC-2-encoding IncP-6 plasmid reveal carbapenem-resistance circulation in several Enterobacteriaceae species from wastewater and a hospital source in spain. Front. Microbiol. 8:1143. doi: 10.3389/fmicb.2017.01143
Yi, H., Cho, Y. J., Yong, D., and Chun, J. (2012). Genome sequence of Escherichia coli J53, a reference strain for genetic studies. J. Bacteriol. 194, 3742–3743. doi: 10.1128/JB.00641-12
Zhao, F., Feng, Y., Lu, X., McNally, A., and Zong, Z. (2017). IncP plasmid carrying colistin resistance gene mcr-1 in Klebsiella pneumoniae from hospital sewage. Antimicrob. Agents Chemother. 61:e2229-16. doi: 10.1128/AAC.02229-16
Zheng, B., Dong, H., Xu, H., Lv, J., Zhang, J., Jiang, X., et al. (2016). Coexistence of MCR-1 and NDM-1 in clinical Escherichia coli Isolates. Clin. Infect. Dis. 63, 1393–1395. doi: 10.1093/cid/ciw553
Zheng, B., Feng, C., Xu, H., Yu, X., Guo, L., Jiang, X., et al. (2018a). Detection and characterization of ESBL-producing Escherichia coli expressing mcr-1 from dairy cows in China. J. Antimicrob. Chemother. 74, 321–325. doi: 10.1093/jac/dky446
Zheng, B., Huang, C., Xu, H., Yu, X., Zhang, J., Wang, X., et al. (2018b). Complete nucleotide sequences of two KPC-2-encoding plasmids from the same Citrobacter freundii isolate. J. Antimicrob. Chemother. 73, 531–533. doi: 10.1093/jac/dkx381
Keywords: Aeromonas taiwanensis, blaKPC–2, qnrS2, whole-genome sequencing, plasmid analysis
Citation: Hu X, Yu X, Shang Y, Xu H, Guo L, Liang Y, Kang Y, Song L, Sun J, Yue F, Mao Y and Zheng B (2019) Emergence and Characterization of a Novel IncP-6 Plasmid Harboring blaKPC–2 and qnrS2 Genes in Aeromonas taiwanensis Isolates. Front. Microbiol. 10:2132. doi: 10.3389/fmicb.2019.02132
Received: 05 May 2019; Accepted: 30 August 2019;
Published: 12 September 2019.
Edited by:
Ziad Daoud, University of Balamand, LebanonReviewed by:
Roberto Gustavo Melano, Public Health Ontario, CanadaCopyright © 2019 Hu, Yu, Shang, Xu, Guo, Liang, Kang, Song, Sun, Yue, Mao and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yimin Mao, eWltaW42MTA3QHNpbmEuY29t; Beiwen Zheng, emhlbmdid0B6anUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.