- 1Department of Physics, School of Science, Tianjin University, Tianjin, China
- 2Key Laboratory of Systems Bioengineering, Ministry of Education, Tianjin University, Tianjin, China
- 3SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering, Tianjin, China
DNA replication initiates from multiple replication origins (ORIs) in eukaryotes. Discovery and characterization of replication origins are essential for a better understanding of the molecular mechanism of DNA replication. In this study, the features of autonomously replicating sequences (ARSs) in Saccharomyces cerevisiae have been comprehensively analyzed as follows. Firstly, we carried out the analysis of the ARSs available in S. cerevisiae S288C. By evaluating the sequence similarity of experimentally established ARSs, we found that 94.32% of ARSs are unique across the whole genome of S. cerevisiae S288C and those with high sequence similarity are prone to locate in subtelomeres. Subsequently, we built a non-redundant dataset with a total of 520 ARSs, which are based on ARSs annotation of S. cerevisiae S288C from SGD and then supplemented with those from OriDB and DeOri databases. We conducted a large-scale comparison of ORIs among the diverse budding yeast strains from a population genomics perspective. We found that 82.7% of ARSs are not only conserved in genomic sequence but also relatively conserved in chromosomal position. The non-conserved ARSs tend to distribute in the subtelomeric regions. We also conducted a pan-genome analysis of ARSs among the S. cerevisiae strains, and a total of 183 core ARSs existing in all yeast strains were determined. We extracted the genes adjacent to replication origins among the 104 yeast strains to examine whether there are differences in their gene functions. The result showed that the genes involved in the initiation of DNA replication, such as orc3, mcm2, mcm4, mcm6, and cdc45, are conservatively located adjacent to the replication origins. Furthermore, we found the genes adjacent to conserved ARSs are significantly enriched in DNA binding, enzyme activity, transportation, and energy, whereas for the genes adjacent to non-conserved ARSs are significantly enriched in response to environmental stress, metabolites biosynthetic process and biosynthesis of antibiotics. In general, we characterized the replication origins from the genome-wide and population genomics perspectives, which would provide new insights into the replication mechanism of S. cerevisiae and facilitate the design of algorithms to identify genome-wide replication origins in yeast.
Introduction
DNA replication is a highly orchestrated process, which is tightly controlled to duplicate the genetic materials into both daughter cells (Bell and Labib, 2016). The specific sites where DNA replication initiates and double-stranded DNA starts unwinding are termed replication origins (ORI) (Jacob et al., 1963; Gilbert, 2001). The identification of ORIs has long been a critical issue, which is helpful to elucidate the molecular mechanism of DNA replication.
The base composition asymmetry widely exists in bacterial genomes (Lobry, 1996; Rocha et al., 1999; Zhang and Gao, 2017; Quan and Gao, 2019). Based on this phenomenon, some strategies to predict replication origin of chromosomes (oriCs) have been developed, for instance, GC skew (Lobry, 1996), cumulative GC skew (Grigoriev, 1998), skewed oligomers (Salzberg et al., 1998) and Z-curve (Zhang and Zhang, 2014). Considering the distributions of DnaA boxes and the conserved oriCs-adjacent genes in different phyla (Mackiewicz et al., 2004; Luo et al., 2018), the web server Ori-Finder (Gao and Zhang, 2008; Luo et al., 2014) has been developed based on the Z-curve method to predict oriCs in bacteria.
For eukaryotes, due to the long linear chromosomes, initiation of DNA replication occurs at multiple discrete sites and activates following the specific timing of DNA replication during the S phase (Taylor, 1960; Hand, 1978; Friedman et al., 1997). The characteristics of eukaryotic replication origins are best understood in the budding yeast Saccharomyces cerevisiae. Sequences conferring the ability of the autonomous replication on circular plasmid molecule are termed autonomously replicating sequences (ARSs) (Stinchcomb et al., 1981) that are regarded as ORIs in yeast chromosomes (Brewer and Fangman, 1987). Taking ARS1 as an example, it consists of the A element (ARS consensus sequence, ACS) (Marahrens and Stillman, 1992) where the ATP-dependent origin recognition complex (ORC) specifically recognizes and binds (Bell and Stillman, 1992; Li et al., 2018), the B1 element partially involved in ORC-DNA interaction (Duderstadt and Berger, 2008; Li et al., 2018), and the B2 element associated with mini-chromosome maintenance (MCM) proteins (Wilmes and Bell, 2002). ARS1 also contains the binding site for site-specific DNA-binding protein ABF1 (ARS binding factor I) (Diffley and Stillman, 1988), whereas ABF1 is not a universal ARS-binding factor. The experimental methods for identifying ARSs in yeast such as the two-dimensional (2D) gel analysis (Brewer and Fangman, 1987; Newlon et al., 1993), microarray-based approaches (Lee et al., 2007), chromatin immunoprecipitation (ChIP) including microarray (ChIP-chip) (Wyrick et al., 2001) and sequencing (ChIP-seq) (Eaton et al., 2010) as well as deep sequencing approaches (Müller et al., 2013) have provided plentiful accurate and reliable results. However, they are costly and time-consuming.
With the accumulation of experimental data and sequencing genomes, the available databases related to replication origins in yeast such as SGD (Cherry et al., 2012), OriDB (Nieduszynski et al., 2006a), DeOri (Gao et al., 2012) and DNA replication (Cotterill and Kearsey, 2008) have been established and updated, which brings new opportunities to study ORIs in yeast genome via more efficient and faster bioinformatic methods. For example, Breier et al. (2004) developed an Oriscan algorithm to predict ORIs in the S. cerevisiae genome utilizing both the ACS motif and its flanking AT-rich region. Consequently, 84% of the top 100 Oriscan predictions matched known ARSs or replication protein binding sites, whereas with the accumulation of predictions, only 56% of the top 350 Oriscan predictions were matched. The result indicated that the algorithm using the similarity to 26 featured origins may limit the discovery of new potential ARSs. The machine learning-based techniques for predicting ORIs in yeast genome have been developed in recent years. Both iRO-3wPseKNC (Liu et al., 2018) and PseKNC2.0 (Dao et al., 2019) web-servers generated the sample formulation based on the mode of PseKNC (pseudo K-tuple nucleotide composition) for describing nucleotide sequences, and utilized the machine learning methods of random forest (RF) and support vector machine (SVM), respectively. Both the web-servers are user-friendly and efficiently performing, whereas direct extraction of the overall ORI sequence information without highlighting the characteristic conservative motifs will undoubtedly dilute the specific features of ORIs, resulting in lowering the prediction accuracy and increasing the false positives. Nieduszynski et al. (2006b) combined the results of ACS motif searches, phylogenetic conservation and microarray data, which enabled the prediction of essential ORIs throughout the S. cerevisiae genome. The result of phylogenetic conservation of replication origin sequences among closely related Saccharomyces species evidently improves the determination of the genome-wide location of replication origins, which suggested that multi-aspect analysis of replication origin sequences will facilitate the performance of prediction models. Although ORIs are essential for the maintenance of S. cerevisiae genome, the yeast chromosome harboring multiple origin deletions has been reported to replicate relatively normally (Dershowitz et al., 2007; Bogenschutz et al., 2014), that is to say, for an individual ORI, it is optional or redundant, which reflects the unexpected flexibility of DNA replication and also implies that there are still a number of potential replication origins to be discovered. Research on replication origins of only one or several strains may provide limited information. In recent years, the accumulation of published S. cerevisiae whole genome sequences (Strope et al., 2015; Zhu et al., 2016; Peter et al., 2018) is unprecedented, hence we can not only comprehensively analyze the ORI features at a genome-wide level, but also have the chance to compare the similarities and differences of replication origin sequences among diverse budding yeast strains from a population genomics respective. However, there are no such reports on the analysis of replication origins based on large-scale genomic data so far.
In this study, we firstly summarized and analyzed the characteristics of replication origin sequences in the reference genome of S. cerevisiae S288C, including classification of ARSs and the specific features in different types. Then we retrieved 104 genome sequences of budding yeasts with high genome integrity, and built a non-redundant dataset based on the published ARSs of S. cerevisiae S288C and supplemented with the confirmed ARSs from OriDB and DeOri databases, which makes it possible for us to conduct a large-scale comparison of replication origin sequences among various yeast strains from a population genomics perspective. By a pan-genome analysis of ARSs among the S. cerevisiae strains, we determined the core ARSs existing in all yeast strains. We also analyzed the distribution bias of various ARSs with different conservation along the chromosomes. To explore whether the ARS-adjacent genes are conserved or not, we extracted genes adjacent to replication origins among the 104 yeast strains. Subsequently, we compared the enriched function of genes adjacent to various ARSs with different conservation and attempted to explain the relationships between replication origins and their adjacent genes.
Materials and Methods
Strains and Datasets
We retrieved the reference genome sequence of S. cerevisiae S288C (version: R64-2-1) as well as 103 well-annotated budding yeast genome sequences with high genome integrity (>95%) from the NCBI FTP site1. The annotation of S. cerevisiae S288C was downloaded from the SGD FTP site2. From literature sources, the information of ecological origins and geographical origins of above S. cerevisiae strains were obtained (Strope et al., 2015; Peter et al., 2018) and the datasets of temporal replication of yeast chromosomes were collected (Raghuraman et al., 2001). We also acquired the available ACS sequences from YeastMine database3 populated by SGD.
Evaluating the Sequence Similarity of ARSs
Sequences similarity analysis of ARSs in S. cerevisiae S288C reference genome was conducted by local BLAST 2.7.1+ (Camacho et al., 2009) (cutoff: E-value ≥ 5e-10, identity ≥90%, coverage ≥90%). Subsequently, the ARS sequences that have multiple alignment results were visualized by ClicO FS (Cheong et al., 2015).
Scan of Homologous ARSs in 104 S. cerevisiae Strains
In this study, a non-redundant dataset consisting of 520 ARSs was constructed based on the available ARSs of S. cerevisiae S288C from SGD database (Cherry et al., 2012) and supplemented with the confirmed ARSs from OriDB database (Nieduszynski et al., 2006a)4 and ARSs of S. cerevisiae from DeOri 6.0 database (Gao et al., 2012)5. For each ARS in this dataset, we performed a BLAST against 104 S. cerevisiae genomes (cutoff: E-value ≥ 5e-10, identity ≥90%, coverage ≥90%). Then, a custom Python script was used to extract the aligned information, which was converted into a GFF3 format file. The conservative profile of ARSs was statistically analyzed according to the frequency of homologous ARSs among 104 S. cerevisiae strains. Finally, we divided the annotation file of sorted homologous ARSs into 104 separate annotation files based on strain names.
Extraction of the Protein-Coding Genes Adjacent to Replication Origins
Firstly, we merged the ARS annotation files with the CDS annotation files of the corresponding 104 S. cerevisiae strains. Then, a custom Python script was used to extract the adjacent genes on both sides of ARSs, and the interval or intersected distance between ARSs and their adjacent genes were calculated. Subsequently, the sequences of these genes adjacent to ARSs among 104 S. cerevisiae strains were collected to BLAST against the dataset obtained from the latest version (February 2019) of UniProtKB6 by the “blastp” program with the e-value cutoff of 1e-5.
Functional Enrichment Analysis
Lists of genes adjacent to conserved ARSs and non-conserved ARSs were prepared through the above steps. Then the gene lists were submitted to DAVID website (Huang et al., 2008) to perform the enrichment analysis of the Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways by Fisher’s exact test. False discovery rate (fdr) was used to filter out the false-positive results with the cutoff of 0.05 for statistical significance.
Results and Discussion
Characteristic Analysis of ARSs in S. cerevisiae Genomes
Overview of ARSs in S. cerevisiae S288C Reference Genome
There are a total of 352 ARSs in S. cerevisiae S288C reference genome available in the SGD database (Supplementary Table 1). The length of these experimentally verified ARSs ranged from 51 to 1324 bp and mainly (63.63%) concentrated in 70–250 bp (Supplementary Figure 1A), and the median length of ARSs in each chromosome is mainly around 240 bp (Supplementary Figure 1B). By linear regression analysis, the fitting line suggested that the count of ARSs was positively correlated to the length of chromosomes with the correlation coefficient of 0.7758 (Supplementary Figure 1C).
The S. cerevisiae S288C genome sequence has an average GC content of 38.38%, while that of ORI sequences is only 29.65%. With the accumulation of experimental data, there are 196 published ACS sequences in the YeastMine database populated by SGD. About 87.43% of ARSs that contain the ACS element are within the length of 300 bp. Here, we took the ACS element as the center to explore the base distribution of ARSs (Supplementary Figure 2) by WebLogo plot (Crooks et al., 2004). The ACS is visible as the high central peak with a high proportion of T residues. With the increasing number of ACS subjects, the degenerate ACS is slightly changed from 5′-WTTTATRTTTW-3′ (Broach et al., 1983) to 5′-WTTTAYRTTTW-3′ (Marahrens and Stillman, 1992). In this study, we collected those published 196 ACS sequences from YeastMine and generated the matrix profile of ACS motif (Supplementary File 1) using “Bio.motifs” package included in Biopython for subsequent prediction of ACS in the candidate sequence. A broad region located directly 3′ to the ACS termed B elements (Lucas and Raghuraman, 2003), which showed low sequence similarity among various ARSs. And only minimal conservations located 3′ to the ACS were detected, for example the conserved 5′-TT-3′ of B1 element and a relatively high frequency of adenine residues start from around 30 to 110 bp, which are consistent with the previous research (Broach et al., 1983; Huang and Kowalski, 1996; Lucas and Raghuraman, 2003; Breier et al., 2004).
In order to investigate the uniqueness of the ARS sequences in S. cerevisiae S288C yeast genome, we conducted a sequence similarity analysis of all these 352 annotated ARSs. The majority of ARS sequences (94.32%) are unique, while only 20 ARSs have multiple alignment results (Supplementary File 2) distributed in intra-chromosomes and inter-chromosomes (Figure 1). Interestingly, all the similar ARS pairs distributed in inter-chromosomes are biased to locate in subtelomeric regions generally within 20 kb of both ends of yeast chromosome (Brown et al., 2010). Yue et al. (2017) found that subtelomeres possess a higher level of copy number variants (CNV) accumulation than those from the internal chromosomal cores and non-reciprocal exchanges and duplications among subtelomeric regions appear to be widespread among eukaryotes (Eichler and Sankoff, 2003), which supported our findings. We subsequently scanned the homologous ARSs of these 20 ARSs among 104 S. cerevisiae strains (Supplementary Table 2), and we found that the most conserved pairs among 104 strains are two pairs internal ARSs compared with those of subtelomeric ARSs. One is the pair of ARS 810 and ARS 811 closed to the tandem array of CUP1 that are associated with resistance to the toxicity of copper (Fogel and Welch, 1982). The other is the pair of ARS1200-1 and ARS 1200-2 known as rARSs (Miller and Kowalski, 1993) that are associated with yeast life span (Kwan et al., 2013). These results suggested that compared to similar ARS pairs distributed in inter-chromosomes, similar ARS pairs located in intra-chromosomes prefer to be shared between strains.
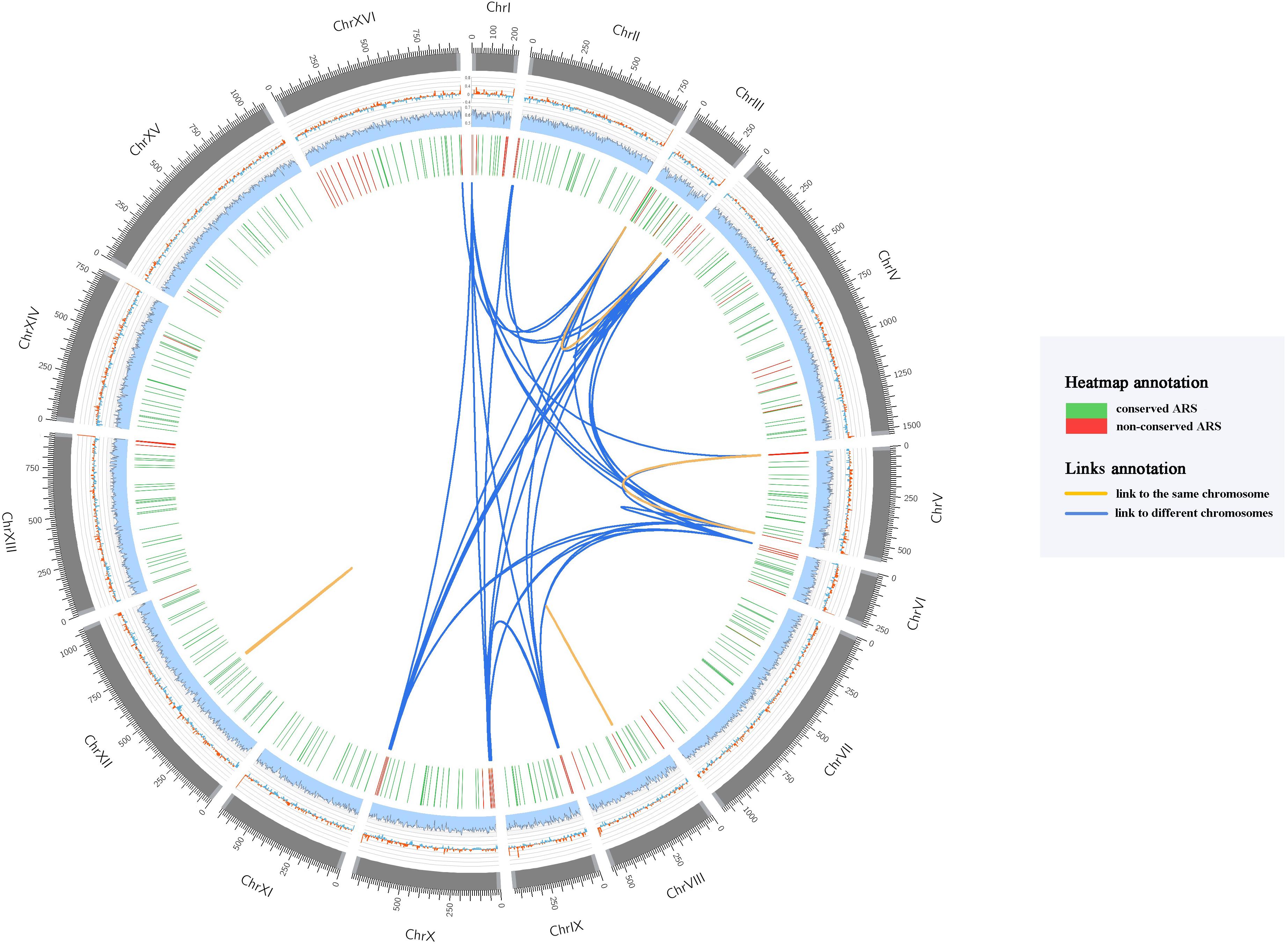
Figure 1. Circos plot showing the ARSs distribution of S. cerevisiae S288C reference genome. Every circle is described in the outermost-innermost direction. (1) The outermost circle represents the S. cerevisiae S288C chromosomes in kb, and the subtelomeric regions are colored in lighter gray; (2) GC-skew (window=3 kb, step=3 kb); (3) AT content (window=3 kb, step=3 kb); (4) Conservation heatmap of 352 ARSs. The location of each bar in the heatmap denotes the position of the ARS in each chromosome of S. cerevisiae S288C, and the number of homologous ARSs in 104 yeast strains is represented by the color of green (conserved ARS) and red (non-conserved ARS); (5) Links showed the result of similar ARS sequences. The orange links represent the similar ARS sequences that are mapped to the identical chromosome, and the blue links represent the similar ARS sequences that are located on different chromosomes.
Population Genomic Analysis of ARSs Among 104 Yeast Genomes
The 104 S. cerevisiae strains that we focused on in this study showed broad genotypic and phenotypic diversity (Strope et al., 2015; Peter et al., 2018). What is the extent of distribution of replication origins in these S. cerevisiae strains with various phylogenetic distance? Therefore, we built a non-redundant dataset with a total of 520 ARSs based on ARSs annotation of S. cerevisiae S288C from SGD and supplemented with the confirmed ARSs from OriDB and DeOri database (Supplementary Table 4).
A marked similarity between two nucleotide sequences may reflect the fact that they come from the same ancestral sequence driven by evolution (Petsko and Ringe, 2004). By mapping ARS sequences from the non-redundant dataset to 104 S. cerevisiae genomes, we determined the homologous ARSs in each chromosome among these yeast strains. Even though the strict alignment conditions had been set, plentiful homologous ARS sequences were found among various yeast genomes and the majority of the homologous ARSs were mapped to the chromosome of the corresponding ARS of the non-redundant dataset. We measured the proportion of homologous ARSs in all strains within the species of S. cerevisiae and displayed it in heat map plot (Supplementary Figure 3), illustrating that the conservation profile of ARSs is not evenly distributed along the chromosomes. It should be noted that only the homologous ARSs located in the corresponding chromosome of the 520 ARS dataset were collected. As for the non-unique ARSs, their chromosomal regions were also taken into consideration. According to the number of homologous ARSs in 104 budding yeast strains, we defined those ARSs existing in more than 90% of the yeast strains as conserved ARSs, and the rest as non-conserved ARSs. We found 430 conserved ARSs accounting for 82.7% of the ARSs from the dataset, and these ARSs are not only conserved in sequence but also relatively conserved in the chromosomal position among various S. cerevisiae genomes (Supplementary Figure 4A), which likely served as the organizational framework for S. cerevisiae genomes. Although large-scale structural variants might exist in the chromosome XII of different yeast strains, the relative position between ARSs in the fragments with structural variants are conserved. Interestingly, about 80% of ARSs located in the subtelomeric regions are non-conserved ARSs. The number of homologous ARSs from subtelomeres is less than those from internal chromosomal regions (one-side Mann–Whitney U test, p-value < 0.01). Subtelomeric regions profoundly contribute to genetic and phenotypic diversity, which recognized as peculiarly dynamic regions of chromosomal evolution (Eichler and Sankoff, 2003; Dujon, 2010; Yue et al., 2017). These areas with rampant genomic rearrangement are the hotspots of reciprocal translocations (Eichler and Sankoff, 2003), which could interpret that ARSs located in subtelomeric regions possess lower conservation than those located in internal chromosomal regions. Subtelomeric regions showed a strong relevance of rapid adaptation to novel niches (Brown et al., 2010; Bergström et al., 2014; Yue et al., 2017), and were known as the hot spots of genetic variation (Peter et al., 2018), which helps accelerate genome evolution and divergence (Cohn et al., 2006). The distribution of non-conserved ARSs in subtelomeres of different S. cerevisiae strains may reflect the strain specificity during genome evolution.
We also performed the pan-genome analysis of ARSs (pan-ARSs) among 104 S. cerevisiae strains by PanGP (Zhao et al., 2014) software. The pan-ARSs size curve illustrated closed pan-ARSs (Supplementary Figure 5). Since we adopted limited ARSs pool to map to yeast strains, it seems that the additional strains could not provide new ARSs to S. cerevisiae pan-ARSs. The result showed that a small number of yeast strains are sufficient to cover the majority of S. cerevisiae pan-ARSs. Core ARSs represent the ARSs exist in all strains of S. cerevisiae. The core ARSs curve showed that the size of core ARS approached to a constant value, suggesting that these 183 core ARSs might serve as the organizational framework for the S. cerevisiae genome.
The total number of homologous ARSs corresponding to the chromosome of ARS from non-redundant dataset for each strain was calculated, and the amount of data we obtained was sufficient for subsequent analysis. Based on the geographic and environmental origins of yeast strains (Supplementary Table 5), we classified the strains into several subsets. By illustrating the distribution of homologous ARSs in each subset (Supplementary Figure 6), we found that the average number of homologous ARSs in each category are relatively similar, however, due to distinct ecological niche and various degree of human association of the isolated strains, the data fluctuation range are various from each class, which may underline a key role of human-driven activities in shaping the distribution of ARSs in S. cerevisiae (Strope et al., 2015; Peter et al., 2018).
In the process of DNA replication, the ORC is recruited to replication origins, followed by the binding of CDC6 (cell division cycle 6) and CDT1 (Cdc10-dependent transcript 1) as well as loading of the MCM helicase complex, which formed the pre-RC (pre-replication complex) proteins (Fragkos et al., 2015). The pre-RCs would bind to all potential origins, however, potential replication origins are in excess and only a small fraction of assembled pre-RCs will be activated at each cell cycle. In addtion, the activation of pre-RCs does not occur simultaneously. Some are fired in the early S phase, and others are activated in the mid or late S phase (Méchali, 2010). The number of corresponding homologous ARSs found in different yeast strains showed the conservation of ARS within the species of S. cerevisiae. To analyze the correlation coefficient between the ARS conservation and the replication fire time, the data of the replication time (Raghuraman et al., 2001) together with the number of homologous ARSs among S. cerevisiae strains were adopted. The result showed that the conservation of ARSs was non-randomly associated with replication time, the Pearson correlation between the conservation of ARSs and replication time shows the value of −0.484 (p-value < 0.01). By comparing the different replication time between the conserved ARSs and non-conserved ARSs (Supplementary Figure 7), we found that the higher conservation of ARSs, the earlier it might initiate (the pairwise Wilcox.test, p-value < 0.001). Combining with the previous findings, we could conclude that in the species of budding yeast, the ARSs biased toward the subtelomeric regions tends to possess weaker conservation and later replicated fire time, which was consistent with the previous conclusion that subtelomeric regions generally possess late DNA replication and low levels of transcription (Barton et al., 2003; Yamazaki et al., 2013). We could also infer that those conservative and earlier replicated replication origins may possess more vital missions than others in chromosomes, for instance their neighboring genes have the priority to early replicate to maintain the growth of yeast strains.
Functional Analysis of Genes Adjacent to Replication Origins in S. cerevisiae Genomes
Genes Adjacent to ARSs in S. cerevisiae S288C Reference Genome
In bacteria, the distribution of oriCs and its corresponding adjacent replication-related genes such as dnaA, dnaN or gidA are highly conserved among different phyla and around 43% of the oriCs are biased close to dnaA among a total of 2740 bacterial chromosomes distributed in various phyla (Luo et al., 2018). The relationship between the oriC and adjacent replication-related genes has been successfully applied to predict the location of oriCs in bacterial chromosomes (Gao and Zhang, 2008). In archaea, replication origins are found to locate next to cdc6/cdc1 (Norais et al., 2007). It is worth surveying the distribution profile of eukaryotic replication origins and their corresponding adjacent genes.
It is generally accepted that the locations of replication origins are exclusively restricted to intergenic regions in eukaryotes (Brewer, 1994; Gilbert, 2001). Here we extracted protein-coding genes adjacent to replication origins of the well-annotated reference genome of S. cerevisiae S288C. In recent years, studies on minimal ARS (miniARS) in yeast have been reported (Liachko et al., 2013; Tsai et al., 2014). However, research on systematically and accurately identifying the precise boundaries of minimal functional replication regions have not been performed due to a large number of replication origins in yeast chromosomes. Please note that the boundaries of the ARSs used in this study are all collected from the original literature, which may be not confined to the minimum essential regions.
According to S. cerevisiae reference genome annotation, we classified the replication origins based on the positional relationships between ARSs and their adjacent genes in the chromosomes. It is defined as the intergenic ORI if there is no intersection between replication origin and its corresponding adjacent protein-coding genes, otherwise as the intersected ORI that the replication origin sequence partially or completely overlaps the adjacent protein-coding genes (Supplementary Figure 8). The result showed that the intergenic ORIs account for 68.18% of known replication origins of S. cerevisiae S288C. Distance distribution among ARSs and their adjacent protein-coding genes showed that their interval distances are mainly less than 1000 bp (Supplementary Figure 9). Although there are 112 intersected ORIs with the average length of 395 bp, about 55.35% of their overlapped segments are less than 30% of their own lengths. Since the ACS element is an essential and conservative element in ARSs, we subsequently scanned the overlapped segments between the intersected ORI and its overlapping protein-coding genes using the matrix profile of ACS motif (Supplementary File 1). The result showed that there are 40 overlapped segments contain ACS motif, suggesting the overlapped segments may be important for these ORIs. We also identified the repeats in ARSs executed by REPuter (Kurtz et al., 2001) program (options:./repfind -c -f -p -r -l 8 -best 50 -h 0 –s) with the e-value cutoff of 5e-2. We found that the majority (92.90%) of ARSs contain repeats (Supplementary Table 3). For intergenic ORIs, repeats (average AT content of 91.10%) are generally characterized by continuous A base, continuous T base, or alternating repeats of A and T base (Supplementary Figure 10). However, for the overlapped segment with ACS motif between the intersected ORI and its overlapping protein-coding gene, repeats in these segments (average AT content of 83.52%) are biased to possess higher GC content (Supplementary Table 3), which suggest that the sequence composition of the overlapped segment between the intersected ORI and its overlapping gene may be constrained by the gene composition.
For S. cerevisiae, the effectiveness of ARSs is various, and restraining the initiation of certain ARSs could affect the expression of neighboring genes. Histone gene pairs (HTA1-HTB1, HHT1-HHF1) are closely positioned to replication origins (ARS428, ARS209) in S. cerevisiae S288C genome. Inactivation of ARSs that are proximal to HTA1–HTB1 gene pairs significantly delayed replication of HTA1 and HTB1, resulting in halving the expression of histone genes (Muller and Nieduszynski, 2017). The delay in replication of centromeric regions (including ARS919 and ARS920) contributes to chromosome instability (Natsume et al., 2013). Nevertheless, S. cerevisiae with multiple origin deletions (ARS600, ARS601/2, ARS603, ARS603.5, ARS604, ARS506, and ARS606) in chromosome VI can replicate relatively normally without detectable growth defects (Dershowitz et al., 2007). Essential genes are those indispensable for the survival of an organism (Giaever et al., 2002), and we found 47 intersected ORIs overlapped with essential genes (data from DEG database7) (Gao et al., 2015; Supplementary Table 1). Any genetic variation occurred in the overlapped region may cause changes in both replication origin and the essential gene, which may disturb the stability and integrity of genomes and even threaten the viability of yeast cells.
Subsequently, in order to assess which factors could determine the difference between the intergenic ORIs and the intersected ORIs, principal component analysis (Supplementary Figure 11) was conducted by integrating the comprehensive features of ARSs in S. cerevisiae S288C including length, GC content, the positional relationship with the adjacent gene, relative chromosomal position, the number of homologous ARSs we obtained among S. cerevisiae species as well as replication time [data from Raghuraman et al. (2001)] and gene expression profiles of S. cerevisiae [data from Arava et al. (2003)]. The intergenic ORIs and intersected ORIs showed relatively visible distinction in the PCA score plot. The average expression of genes adjacent to intersected ORI was significantly lower than that of genes adjacent to intergenic ORI (one-side Mann–Whitney U test, p-value < 0.05). We guessed that replication-related proteins that bind or be recruited at replication origins may interfere with the expression of overlapping genes, and the influence of expression of overlapping genes has to be considered in space as well as time, because only a subset of origins are activated during every cell cycle and the activation of replication origins may vary according to the cell fate or environmental conditions (Méchali, 2010; Fragkos et al., 2015). However, the comparison was only based on the inference of statistical results, and the specific relationships between replication origins and their adjacent genes require more detailed experimental studies. We found that most of the above mentioned factors were linearly uncorrelated to the ORIs that possessed the various positional relationship with the adjacent gene, but the intergenic ORIs and the intersected ORIs could be roughly distinguished through the PCA analysis if these factors were comprehensively considered.
Genes Adjacent to Conserved ORIs Among 104 S. cerevisiae Genomes
In the study of population genomic analysis of ORIs among 104 yeast genomes, we identified 430 conserved ARSs from the ARSs dataset. Based on the homologous ARSs of various yeast strains, we extracted their adjacent genes from the corresponding yeast genomes. According to the number of genes located next to each of the corresponding conserved ORIs in 104 budding yeast strains, we defined the genes existing in more than 90% of the yeast strains as conserved adjacent genes, and the rest as non-conserved adjacent genes. As a result, a total of 662 conserved adjacent genes were collected and 27.64% of them belong to essential genes based on DEG database (Gao et al., 2015; Supplementary Table 4). We also found that the conserved adjacent genes relatively conserved in both chromosomal position and orientation among the various S. cerevisiae genomes (Supplementary Figure 4B), which suggested that the adjacent relationship between ARSs and their corresponding genes are conserved in chromosomes among 104 yeast strains.
In bacteria, the replication-related genes such as dnaA and dnaN, are highly conservatively close to oriCs (Luo et al., 2018). Likewise, genes involved in the initiation of DNA replication in S. cerevisiae are found to conservatively locate next to the replication origins, such as orc3, mcm2, mcm4, mcm6, and cdc45 (Supplementary Table 4). During G1 phase, ORC complex recognizes and bind sequence-specifically to ACS in the presence of ATP (Bell and Stillman, 1992; Watson et al., 2013). And helicase-loading proteins, CDC6 and CDT1, are recruited to load MCM2–7 complexes onto the replication origin (Bell and Labib, 2016). During S phase, loaded helicases are activated by CDK (cyclin dependent kinase) and DDK (Dbf4-dependent kinase). Those two factors, CDC45 (cell division cycle 45) and GINS (Go, Ichi, Ni, and San) complex are tightly associated with MCM2-7 at replication forks to form the activated helicase called the CMG complex (CDC45–MCM–GINS) (Moyer et al., 2006; Pacek et al., 2006). Then the CMG complex would be well assembled and activated to unwind the double-stranded DNA and start to initiate DNA synthesis (Fragkos et al., 2015).
We found that the conserved adjacent genes were significantly enriched in twenty-three GO terms and five KEGG pathways related to DNA binding, enzyme activity, transportation and energy, including sequence-specific DNA binding, nucleotide binding, lyase activity, catalytic activity, ion transport, transmembrane transport and ATP binding (Figures 2A,B and Supplementary Table 6). Obviously, these GO terms and KEGG pathways are strongly correlated with the process of DNA replication. Due to the advantage in chromosomal position, the genes adjacent to replication origins are preferentially replicated after the double-stranded DNA start unwinding. We could infer that replication origin neighboring genes involved in DNA binding, enzyme activity, transportation, and energy might have a higher priority to replicate. It is likely that the aggregation of ORI and its conserved adjacent genes enable the gene-encoded products to be more effectively involved in the DNA replication initiation in the localized cellular space. It is possible that these preferential replicated genes enable normal and efficient DNA replication and possibly make it orderly organized in protein-DNA and protein-protein interactions, which may guarantee stable operations in the yeast cell cycle. With regard to non-conserved ARSs, also considered as strain-specific ARSs, we found that these genes are significantly enriched in response to environmental stress (such as temperature and drug), metabolites biosynthetic process and biosynthesis of antibiotics (Figures 2C,D and Supplementary Table 7). We speculated that the preferential replication related to these adjacent genes is likely to provide raw materials for the active metabolism of yeast strains, and may enhance the adaptability of the strains to survive in the environmental stress.
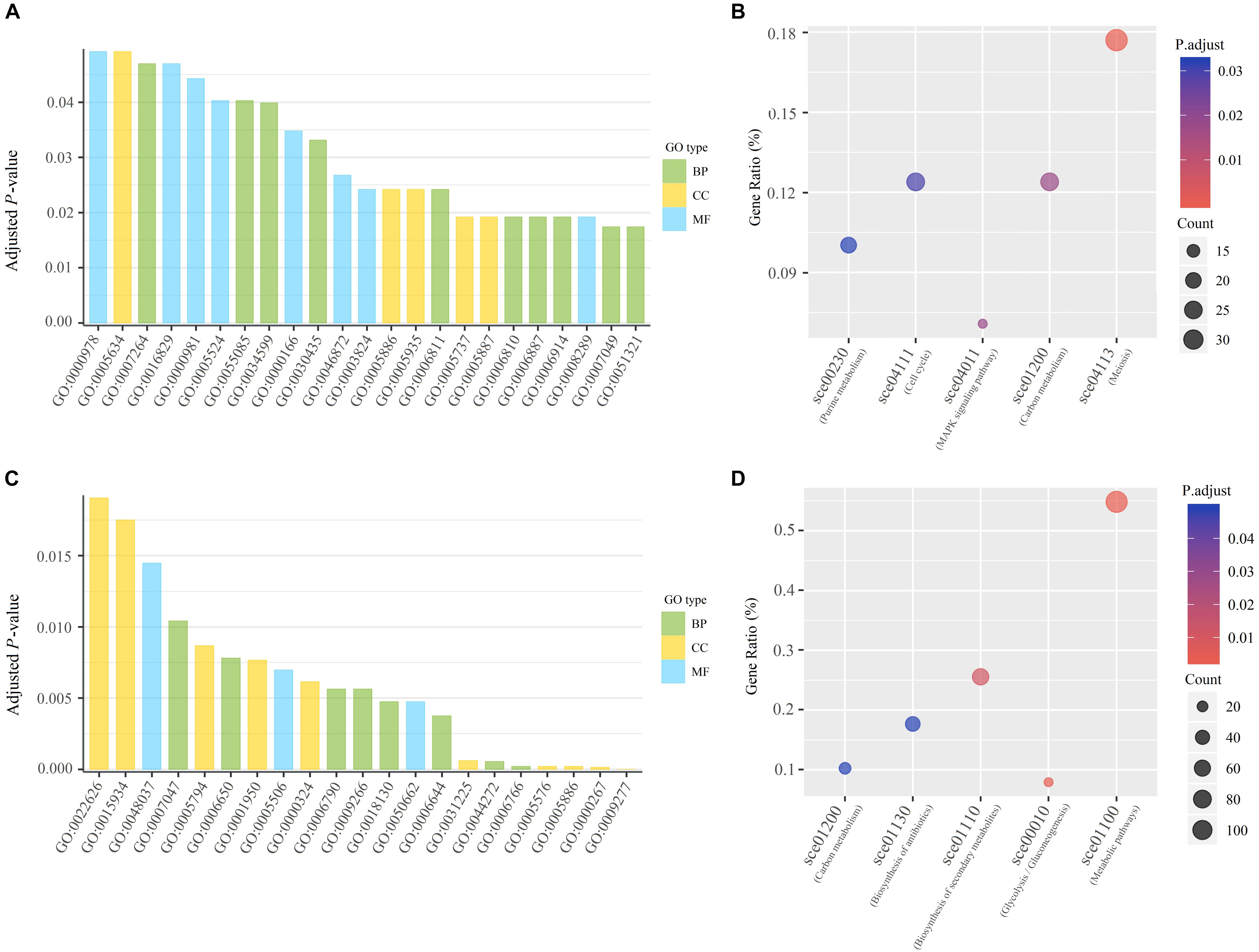
Figure 2. Functional analysis of genes adjacent to replication origins. (A) GO enrichment analysis of 662 conserved genes adjacent to conserved ARSs. GO terms with the adjusted p-value ≤ 0.05 are shown. The statistical significance was assessed by Fisher’s exact test with False discovery rate (fdr) correction. BP for the biological process; MF for molecular function; CC for the cellular component. (B) Scatterplot for significantly enriched KEGG pathways of 662 conserved genes adjacent to conserved ARSs. KEGG pathways with the adjusted p-value ≤ 0.05 are shown. The statistical significance was assessed by Fisher’s exact test with fdr correction. The size and color of dots represent the gene number and the adjusted p-value, respectively. Gene ratio is the proportion of enriched genes among all conserved genes neighboring ORIs. (C) GO enrichment analysis of genes adjacent to non-conserved ARSs. (D) Scatterplot for significantly enriched KEGG pathways of genes adjacent to non-conserved ARSs.
Conclusion
In this study, we comprehensively analyzed features of replication origin sequences of S. cerevisiae from genome-wide and population genomics perspectives. We conducted the data-analytic work for investigating the similarities and genomic positions of the ARS sequences among the diverse budding yeast strains obtained from various ecological and geographical backgrounds. We also performed a characterization of the genes that are adjacent to the conserved and non-conserved ARSs among the 104 yeast strains. These results presented here may provide insights into the replication mechanism of S. cerevisiae and facilitate the development of algorithms for further prediction of replication origins in budding yeast genomes. For examples, the conserved ARS-adjacent genes should be taken into consideration in the design of prediction algorithms, just like Ori-Finder considering the conserved oriC-adjacent genes (such as dnaA, dnaN, and gidA), which would make the prediction more robust and reliable. In addition, as modular parts, the core ARSs and their conserved adjacent genes might provide a useful reference for the rational design of replication origins for the synthetic S. cerevisiae genome. However, the conserved ARSs and their corresponding adjacent genes are obtained based on sequence alignment and statistical results, whereas the biological significance of the positional conservation of the replication origins and their adjacent genes requires more detailed experimental proof. Since DNA replication is one of the highly conserved processes of eukaryotic cell, and almost all the proteins related to the DNA replication in yeast correspond to a single ortholog in humans and other eukaryotic species (Bell and Labib, 2016), the features and rules of DNA replication initiation found in S. cerevisiae genomes may be extended to higher eukaryotes.
Author Contributions
DW conducted the data analysis and drafted the manuscript. FG supervised the study and revised the manuscript. Both authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31571358, 21621004, 31171238, and 91746119).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Prof. Chun-Ting Zhang for the invaluable assistance and inspiring discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02122/full#supplementary-material
Footnotes
- ^ ftp://ftp.ncbi.nih.gov/genomes/all/
- ^ ftp://ftp.yeastgenome.org/sequence/S288C_reference
- ^ http://yeastmine.yeastgenome.org
- ^ http://cerevisiae.oridb.org/
- ^ http://tubic.org/deori/
- ^ ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete
- ^ http://tubic.org/deg/
References
Arava, Y., Wang, Y., Storey, J. D., Liu, C. L., Brown, P. O., and Herschlag, D. (2003). Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 100, 3889–3894. doi: 10.1073/pnas.0635171100
Barton, A. B., Su, Y., Lamb, J., Barber, D., and Kaback, D. B. (2003). A function for subtelomeric DNA in Saccharomyces cerevisiae. Genetics 165, 929–934.
Bell, S. P., and Labib, K. (2016). Chromosome duplication in Saccharomyces cerevisiae. Genetics 203, 1027–1067. doi: 10.1534/genetics.115.186452
Bell, S. P., and Stillman, B. (1992). ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357, 128–134. doi: 10.1038/357128a0
Bergström, A., Simpson, J. T., Salinas, F., Barré, B., Parts, L., Zia, A., et al. (2014). A high-definition view of functional genetic variation from natural yeast genomes. Mol. Biol. Evol. 31, 872–888. doi: 10.1093/molbev/msu037
Bogenschutz, N. L., Rodriguez, J., and Tsukiyama, T. (2014). Initiation of DNA replication from non-canonical sites on an origin-depleted chromosome. PLoS One 9:e114545. doi: 10.1371/journal.pone.0114545
Breier, A. M., Chatterji, S., and Cozzarelli, N. R. (2004). Prediction of Saccharomyces cerevisiae replication origins. Genome Biol. 5:R22.
Brewer, B. J. (1994). Intergenic DNA and the sequence requirements for replication initiation in eukaryotes. Curr. Opin. Genet. Dev. 4, 196–202. doi: 10.1016/s0959-437x(05)80045-0
Brewer, B. J., and Fangman, W. L. (1987). The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51, 463–471. doi: 10.1016/0092-8674(87)90642-8
Broach, J. R., Li, Y. Y., Feldman, J., Jayaram, M., Abraham, J., Nasmyth, K. A., et al. (1983). Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harb. Symp. Quant. Biol. 47(Pt 2), 1165–1173. doi: 10.1101/sqb.1983.047.01.132
Brown, C. A., Murray, A. W., and Verstrepen, K. J. (2010). Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr. Biol. 20, 895–903. doi: 10.1016/j.cub.2010.04.027
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Cheong, W.-H., Tan, Y.-C., Yap, S.-J., and Ng, K.-P. (2015). ClicO FS: an interactive web-based service of Circos. Bioinformatics 31, 3685–3687. doi: 10.1093/bioinformatics/btv433
Cherry, J. M., Hong, E. L., Amundsen, C., Balakrishnan, R., Binkley, G., Chan, E. T., et al. (2012). Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700–D705. doi: 10.1093/nar/gkr1029
Cohn, M., Liti, G., and Barton, D. B. (2006). “Telomeres in fungi,” in Comparative Genomics, ed. Pa. P. J. Sunnerhagen (Heidelberg: Springer), 101–130. doi: 10.1007/4735_108
Cotterill, S., and Kearsey, S. E. (2008). DNAReplication: a database of information and resources for the eukaryotic DNA replication community. Nucleic Acids Res. 37, D837–D839. doi: 10.1093/nar/gkn726
Crooks, G. E., Hon, G., Chandonia, J.-M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Dao, F. Y., Lv, H., Wang, F., Feng, C. Q., Ding, H., Chen, W., et al. (2019). Identify origin of replication in Saccharomyces cerevisiae using two-step feature selection technique. Bioinformatics 35, 2075–2083. doi: 10.1093/bioinformatics/bty943
Dershowitz, A., Snyder, M., Sbia, M., Skurnick, J. H., Ong, L. Y., and Newlon, C. S. (2007). Linear derivatives of Saccharomyces cerevisiae chromosome III can be maintained in the absence of autonomously replicating sequence elements. Mol. Cell. Biol. 27, 4652–4663. doi: 10.1128/mcb.01246-06
Diffley, J. F., and Stillman, B. (1988). Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc. Natl. Acad. Sci. U.S.A. 85, 2120–2124. doi: 10.1073/pnas.85.7.2120
Duderstadt, K. E., and Berger, J. M. (2008). AAA+ ATPases in the initiation of DNA replication. Crit. Rev. Biochem. Mol. Biol. 43, 163–187. doi: 10.1080/10409230802058296
Eaton, M. L., Galani, K., Kang, S., Bell, S. P., and Macalpine, D. M. (2010). Conserved nucleosome positioning defines replication origins. Genes Dev. 24, 748–753. doi: 10.1101/gad.1913210
Eichler, E. E., and Sankoff, D. (2003). Structural dynamics of eukaryotic chromosome evolution. Science 301, 793–797. doi: 10.1126/science.1086132
Fogel, S., and Welch, J. W. (1982). Tandem gene amplification mediates copper resistance in yeast. Proc. Natl. Acad. Sci. U.S.A. 79, 5342–5346. doi: 10.1073/pnas.79.17.5342
Fragkos, M., Ganier, O., Coulombe, P., and Mechali, M. (2015). DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 16, 360–374. doi: 10.1038/nrm4002
Friedman, K. L., Brewer, B. J., and Fangman, W. L. (1997). Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells 2, 667–678.
Gao, F., Luo, H., and Zhang, C. T. (2012). DeOri: a database of eukaryotic DNA replication origins. Bioinformatics 28, 1551–1552. doi: 10.1093/bioinformatics/bts151
Gao, F., Luo, H., Zhang, C.-T., and Zhang, R. (2015). Gene essentiality analysis based on DEG 10, an updated database of essential genes. Methods Mol. Biol. 1279, 219–233. doi: 10.1007/978-1-4939-2398-4_14
Gao, F., and Zhang, C.-T. (2008). Ori-Finder: a web-based system for finding oriC s in unannotated bacterial genomes. BMC Bioinformatics 9:79. doi: 10.1186/1471-2105-9-79
Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391.
Gilbert, D. M. (2001). Making sense of eukaryotic DNA replication origins. Science 294, 96–100. doi: 10.1126/science.1061724
Grigoriev, A. (1998). Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26, 2286–2290. doi: 10.1093/nar/26.10.2286
Hand, R. (1978). Eucaryotic DNA: organization of the genome for replication. Cell 15, 317–325. doi: 10.1016/0092-8674(78)90001-6
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2008). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Huang, R. Y., and Kowalski, D. (1996). Multiple DNA elements in ARS305 determine replication origin activity in a yeast chromosome. Nucleic Acids Res. 24, 816–823. doi: 10.1093/nar/24.5.816
Jacob, F., Brenner, S., and Cuzin, F. (1963). On the regulation of DNA replication in bacteria. Cold Spring Harb. Symp. Quant. Biol. 28, 329–348.
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., and Giegerich, R. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642. doi: 10.1093/nar/29.22.4633
Kwan, E. X., Foss, E. J., Tsuchiyama, S., Alvino, G. M., Kruglyak, L., Kaeberlein, M., et al. (2013). A natural polymorphism in rDNA replication origins links origin activation with calorie restriction and lifespan. PLoS Genet. 9:e1003329. doi: 10.1371/journal.pgen.1003329
Lee, W., Tillo, D., Bray, N., Morse, R. H., Davis, R. W., Hughes, T. R., et al. (2007). A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39, 1235–1244. doi: 10.1038/ng2117
Li, N., Lam, W. H., Zhai, Y., Cheng, J., Cheng, E., Zhao, Y., et al. (2018). Structure of the origin recognition complex bound to DNA replication origin. Nature 559, 217–222. doi: 10.1038/s41586-018-0293-x
Liachko, I., Youngblood, R. A., Keich, U., and Dunham, M. J. (2013). High-resolution mapping, characterization, and optimization of autonomously replicating sequences in yeast. Genome Res. 23, 698–704. doi: 10.1101/gr.144659.112
Liu, B., Weng, F., Huang, D.-S., and Chou, K.-C. (2018). iRO-3wPseKNC: identify DNA replication origins by three-window-based PseKNC. Bioinformatics 34, 3086–3093. doi: 10.1093/bioinformatics/bty312
Lobry, J. (1996). Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 13, 660–665. doi: 10.1093/oxfordjournals.molbev.a025626
Lucas, I. A., and Raghuraman, M. K. (2003). The dynamics of chromosome replication in yeast. Curr. Top. Dev. Biol. 55, 1–73. doi: 10.1016/s0070-2153(03)01001-9
Luo, H., Quan, C. L., Peng, C., and Gao, F. (2018). Recent development of Ori-Finder system and DoriC database for microbial replication origins. Brief. Bioinformatics doi: 10.1093/bib/bbx174 [Epub ahead of print].
Luo, H., Zhang, C. T., and Gao, F. (2014). Ori-Finder 2, an integrated tool to predict replication origins in the archaeal genomes. Front. Microbiol. 5:482. doi: 10.3389/fmicb.2014.00482
Mackiewicz, P., Zakrzewska-Czerwiñska, J., Zawilak, A., Dudek, M. R., and Cebrat, S. (2004). Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 32, 3781–3791. doi: 10.1093/nar/gkh699
Marahrens, Y., and Stillman, B. (1992). A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255, 817–823. doi: 10.1126/science.1536007
Méchali, M. (2010). Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell Biol. 11, 728–738. doi: 10.1038/nrm2976
Miller, C. A., and Kowalski, D. (1993). cis-acting components in the replication origin from ribosomal DNA of Saccharomyces cerevisiae. Mol. Cell Biol. 13, 5360–5369. doi: 10.1128/mcb.13.9.5360
Moyer, S. E., Lewis, P. W., and Botchan, M. R. (2006). Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U.S.A. 103, 10236–10241. doi: 10.1073/pnas.0602400103
Müller, C. A., Hawkins, M., Retkute, R., Malla, S., Wilson, R., Blythe, M. J., et al. (2013). The dynamics of genome replication using deep sequencing. Nucleic Acids Res. 42:e3. doi: 10.1093/nar/gkt878
Muller, C. A., and Nieduszynski, C. A. (2017). DNA replication timing influences gene expression level. J. Cell Biol. 216, 1907–1914. doi: 10.1083/jcb.201701061
Natsume, T., Muller, C. A., Katou, Y., Retkute, R., Gierlinski, M., Araki, H., et al. (2013). Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol. Cell 50, 661–674. doi: 10.1016/j.molcel.2013.05.011
Newlon, C. S., Collins, I., Dershowitz, A., Deshpande, A. M., Greenfeder, S. A., Ong, L. Y., et al. (1993). Analysis of replication origin function on chromosome III of Saccharomyces cerevisiae. Cold Spring Harb. Symp. Quant. Biol. 58, 415–423. doi: 10.1101/sqb.1993.058.01.048
Nieduszynski, C. A., Hiraga, S.-I., Ak, P., Benham, C. J., and Donaldson, A. D. (2006a). OriDB: a DNA replication origin database. Nucleic Acids Res. 35, D40–D46.
Nieduszynski, C. A., Knox, Y., and Donaldson, A. D. (2006b). Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev. 20, 1874–1879. doi: 10.1101/gad.385306
Norais, C., Hawkins, M., Hartman, A. L., Eisen, J. A., Myllykallio, H., and Allers, T. (2007). Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 3:e77. doi: 10.1371/journal.pgen.0030077
Pacek, M., Tutter, A. V., Kubota, Y., Takisawa, H., and Walter, J. C. (2006). Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 21, 581–587. doi: 10.1016/j.molcel.2006.01.030
Peter, J., De Chiara, M., Friedrich, A., Yue, J.-X., Pflieger, D., Bergström, A., et al. (2018). Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344. doi: 10.1038/s41586-018-0030-5
Petsko, G. A., and Ringe, D. (2004). Protein Structure and Function. Chapter 4: From Sequence to Function. London: New Science Press.
Quan, C. L., and Gao, F. (2019). Quantitative analysis and assessment of base composition asymmetry and gene orientation bias in bacterial genomes. FEBS Lett. 593, 918–925. doi: 10.1002/1873-3468.13374
Raghuraman, M., Winzeler, E. A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., et al. (2001). Replication dynamics of the yeast genome. Science 294, 115–121. doi: 10.1126/science.294.5540.115
Rocha, E. P., Danchin, A., and Viari, A. (1999). Universal replication biases in bacteria. Mol. Microbiol. 32, 11–16. doi: 10.1046/j.1365-2958.1999.01334.x
Salzberg, S. L., Salzberg, A. J., Kerlavage, A. R., and Tomb, J.-F. (1998). Skewed oligomers and origins of replication. Gene 217, 57–67. doi: 10.1016/s0378-1119(98)00374-6
Stinchcomb, D. T., Mann, C., Selker, E., and Davis, R. W. (1981). “DNA sequences that allow the replication and segregation of yeast chromosomes,” in The Initiation of DNA Replication, ed. D. S. Ray (New York, NY: Academic Press), 473–488. doi: 10.1016/b978-0-12-583580-0.50036-4
Strope, P. K., Skelly, D. A., Kozmin, S. G., Mahadevan, G., Stone, E. A., Magwene, P. M., et al. (2015). The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 25, 762–774. doi: 10.1101/gr.185538.114
Taylor, J. H. (1960). Asynchronous duplication of chromosomes in cultured cells of Chinese hamster. J. Biophys. Biochem. Cytol. 7, 455–464.
Tsai, H.-J., Baller, J. A., Liachko, I., Koren, A., Burrack, L. S., Hickman, M. A., et al. (2014). Origin replication complex binding, nucleosome depletion patterns, and a primary sequence motif can predict origins of replication in a genome with epigenetic centromeres. mBio 5:e01703-14. doi: 10.1128/mBio.01703-14
Watson, J. D., Baker, T. A., Bell, S. P., Gann, A., Levine, M., and Losick, R. (2013). “The replication of DNA,” in Molecular Biology of the Gene, ed. J. D. Watson (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), 257–312.
Wilmes, G. M., and Bell, S. P. (2002). The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proc. Natl. Acad. Sci. U.S.A. 99, 101–106. doi: 10.1073/pnas.012578499
Wyrick, J. J., Aparicio, J. G., Chen, T., Barnett, J. D., Jennings, E. G., Young, R. A., et al. (2001). Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294, 2357–2360. doi: 10.1126/science.1066101
Yamazaki, S., Hayano, M., and Masai, H. (2013). Replication timing regulation of eukaryotic replicons: Rif1 as a global regulator of replication timing. Trends Genet. 29, 449–460. doi: 10.1016/j.tig.2013.05.001
Yue, J.-X., Li, J., Aigrain, L., Hallin, J., Persson, K., Oliver, K., et al. (2017). Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat. Genet. 49, 913–924. doi: 10.1038/ng.3847
Zhang, G., and Gao, F. (2017). Quantitative analysis of correlation between AT and GC biases among bacterial genomes. PLoS One 12:e0171408. doi: 10.1371/journal.pone.0171408
Zhang, R., and Zhang, C.-T. (2014). A brief review: the z-curve theory and its application in genome analysis. Curr. Genomics 15, 78–94. doi: 10.2174/1389202915999140328162433
Zhao, Y., Jia, X., Yang, J., Ling, Y., Zhang, Z., Yu, J., et al. (2014). PanGP: a tool for quickly analyzing bacterial pan-genome profile. Bioinformatics 30, 1297–1299. doi: 10.1093/bioinformatics/btu017
Keywords: replication origin, DNA replication, Saccharomyces cerevisiae, genome-wide analysis, autonomously replicating sequence
Citation: Wang D and Gao F (2019) Comprehensive Analysis of Replication Origins in Saccharomyces cerevisiae Genomes. Front. Microbiol. 10:2122. doi: 10.3389/fmicb.2019.02122
Received: 16 November 2018; Accepted: 29 August 2019;
Published: 13 September 2019.
Edited by:
John R. Battista, Louisiana State University, United StatesReviewed by:
Gregory Marczynski, McGill University, CanadaKazumasa Yoshida, Kyushu University, Japan
Copyright © 2019 Wang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Gao, Zmdhb0B0anUuZWR1LmNu
 Dan Wang
Dan Wang Feng Gao
Feng Gao