- 1State Key Laboratory of Developmental Biology of Freshwater Fish, Hunan Provincial Key Laboratory of Microbial Molecular Biology, College of Life Science, Hunan Normal University, Changsha, China
- 2Shenzhen Key Laboratory of Microbial Genetic Engineering, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
The small heat shock protein plays an important role in response to stresses. We wanted to investigate how Hsp20 affects sporulation and production of insecticidal crystal proteins (ICPs) in Bacillus thuringiensis (Bt) at the stationary growth phase when cells are starved. The hsp20 gene was knocked out in Bt4.0718 (wide type), which is a B. thuringiensis strain screened in our laboratory, using endonuclease I-SceI mediated unmarked gene replacement method. Deletion of Hsp20 resulted in a decrease in both sporulation and ICPs production. Bt4-Δhsp20 cells and its ICP did not have a significant difference in shape and size but entered the decline phase 2 h earlier than the Bt4.0718. In order to find the mechanism that underlies these phenotypes, we completed a proteomic study of differentially expressed proteins (DEPs). In Bt4-Δhsp20 cells, 11 DEPs were upregulated and 184 DEPs downregulated. These affected DEPs are involved in multiple metabolic pathways: (1) six DEPs (two upregulated and four downregulated) are directly related to the sporulation and ICPs synthesis; (2) supply of amino acids including amino acid synthesis and protein recycling; (3) the energy supplementation (the tricarboxylic acid cycle and glycolysis); (4) purine metabolism and mRNA stability. These results suggest that hsp20 may be critical in maintaining the homeostasis of B. thuringiensis during the production of spores and ICPs, and could provide new sight into the sporulation and ICPs formation in B. thuringiensis.
Introduction
Heat shock proteins (HSPs) are a family of proteins that are produced in response to stressful conditions, providing a chaperone function by stabilizing proteins to ensure correct folding, or by helping to refold proteins that are damaged by cell stress (Maio, 1999). HSPs exist in bacteria and in humans and are named according to their molecular weight, such as Hsp60, Hsp70, and Hsp90. These HSPs are the most extensively studied HSPs in humans and plants. The expression of HSPs is transcriptionally regulated and triggered by different environmental stresses, such as infections, inflammation, toxins, starvation, hypoxia, and water deprivation. The principal HSPs include five conserved classes, which are HSP33, HSP60, HSP70, HSP90, and HSP100, and small HSPs (sHSPs) (Schlesinger, 1990).
Small Heat shock proteins are ubiquitous ATP-independent molecular chaperones that prevent protein aggregation upon stress-induced unfolding in nearly all organisms (Bepperling et al., 2012). Hsp20 responds to heat shock or other environmental stresses (Burdon, 1988). Numerous studies have confirmed that Hsp20 plays an important role in the response to chemical or physical conditions. For example, the transcriptional level of hsp20 is strongly induced to resist severe heat and osmotic shocks (Ventura et al., 2007; Khaskheli et al., 2015). In Escherichia coli cells, overexpression of Hsp20 of Deinococcus radiodurans, and GrxC, IscA, and Fdx proteins are involved in the control of cellular redox status, through enhancing tolerance to hydrogen peroxide (H2O2) stress (Singh et al., 2014). Under the stress of water deprivation, Hsp20 is essential for the desiccation resistance of Azotobacter vinelandii (Cocotl-Yañez et al., 2014). Proteomics analysis of the probiotic bacterium, Lactobacillus casei, indicated that Hsp20 may play a crucial role in the adaptation to the physiological processes of lactic acid accumulation (Wu et al., 2009).
Bt, a Gram-positive bacterium and one of the safest microbial pesticides, has been used worldwide in pest control for several decades (Jouzani et al., 2017). It forms characteristic parasporal insecticidal crystal proteins during sporulation (Roh et al., 2007; Van, 2009; Palma et al., 2014; Singh et al., 2018). Moreover, this bacterium can promote plant growth (Armada et al., 2018), bioremediate different heavy metals, biosynthesize metal nanoparticles (Jouzani et al., 2017), produce melanin (Cao et al., 2018), and even kill cancer cells (Moazamian et al., 2018). Due to its important value in agriculture, environment and medicine, mechanisms of ICPs formation and sporulation in B. thuringiensis have been extensively investigated at the transcriptional level (Park et al., 1998; Zhang et al., 1998, 1999; Aronson, 2002; Doruk et al., 2013; Wanapaisan et al., 2013; Hui et al., 2014), as has the study of its post-transcriptional, post-translational, and metabolic pathways (Yousten and Rogoff, 1969; Glatron and Rapoport, 1972; Baum and Malvar, 1995; Mathy et al., 2007; Wang et al., 2013a; Zheng et al., 2015). Heretofore, there are no reports to describe if and how HSPs affect accumulation of ICPs in B. thuringiensis. Since, HSPs are so important for folding as the chaperons, we wanted to explore the effects that the absence of a single protein could have on the metabolism of B. thuringiensis and particularly on its ability to synthesize Cry proteins, one of the most abundant cellular proteins in bacterial cells.
Bt4.0718 is a highly virulent wide-type strain that was screened in our laboratory. It has been subjected to genome sequencing and shot gun proteomics at different stages of growth and sporulation (Huang et al., 2012; Jie et al., 2015). Our data have shown that Hsp20 (WP_001245065.1) protein can be detected in two processes of sporulation. Surprisingly, we found that the FPKM value of hsp20 mRNA increases almost 10-fold from 63.04 to 702.98 from stages of early sporulation to late sporulation, as shown in Supplementary Figure S1. We therefore speculated that it is likely that Hsp20 may play an important role in sporulation and ICPs synthesis in Bt4.0718. In the present study, we deleted the hsp20 gene from the genome and use the iTRAQ proteome (Liu et al., 2015; Wang S. et al., 2016; Zhang et al., 2018) to analyze the Hsp20 null strain. Our results demonstrated that deletion of Hsp20 affected expression of 195 DEPs and reduced synthesis of ICPs.
Materials and Methods
Strains and Culture Conditions
The strains and plasmids used in this study are listed in Table 1. B. thuringiensis strains were routinely cultured at 30°C and 120 rpm in fermentation medium, which was composed of 18 g/L glucose, 14.5 g/L tryptone, 2.5 g/L K2HPO4, 0.02 g/L FeSO4 ⋅ 7H2O, 0.02 g/L MnSO4 ⋅ H2O, and 0.25 g/L MgSO4 ⋅ 7H2O. Necessary antibiotics were added to the culture at the final concentrations of 50 μg/mL for kanamycin, 25 μg/mL for erythromycin, 300 μg/mL for spectinomycin or 60 units for polymyxin. E. coli DH5a and BL21(DE3) were used for cloning and expression of recombinant proteins, respectively. The E. coli strains were grown in Lysogeny broth (LB) medium or LB agar plates at 37°C. The antibiotic concentrations used were 50 μg/mL kanamycin, 100 μg/mL ampicillin and spectinomycin.
DNA Manipulations
The whole genomic DNA of B. thuringiensis 4.0718 was extracted with rapid bacterial genomic DNA isolation kit (B518225, Sangon Biotech), and all steps carried out followed the standard manual. Primers used in the study are listed in Table 2. All polymerase chain reaction (PCR) products were purified using PCR product purification kit (B518141, Sangon Biotech). Recombinant plasmids were constructed in E. coli DH5α, and sequencing services were provided by Invitrogen.
Construction of the Integrating Plasmids for Gene Deletion
To construct the plasmid for hsp20 deletion, we amplified the upstream and downstream homologous arm of hsp20, approximately 700 bp from the B. thuringiensis 4.0718 genomic DNA by PCR using primer pairs of P1/P2, P3/P4. The upstream homologous arm was inserted into plasmid pRP1028 between the SmaI and MluI sites to construct the plasmid pRP1028-hsp20U. The downstream homologous arm was inserted into plasmid pRP1028-hsp20U between the BamHI and SalI sites to construct the integrating plasmid pRP1028-hsp20UD. pRP1028 is a shuttle plasmid with a temperature-sensitive suicide B. thuringiensis replicon. The resulting integrating plasmid was further verified by PCR and sequencing.
Gene Knock-Out Procedure
The gene knock-out system in B. thuringiensis was developed based on homing endonuclease I-SceI mediated unmarked gene replacement method established for B. anthracis (Janes and Stibitz, 2006). To construct the hsp20 deletion mutant Bt4-Δhsp20, we included an I-SceI recognition site for I-SceI restriction endonuclease cleavage and one oriT site for conjugative transfer in pRP1028-hsp20UD. The vector also encodes RFP as a reporter protein and possesses a spectinomycin resistance marker for convenient screening. The subsequent procedures are illustrated in Supplementary Figure S2. The knockout strains and the complemented strains Bt4-Δhsp20::hsp20 were then screened. The related plasmids and strains were kindly provided by the Jin He of Huazhong Agricultural University.
Heterologous Expression and Purification of Hsp20 in E. coli BL21(DE3)
The constructed plasmid pET28a-hsp20 (use the primers P5/P6) was transformed into E. coli BL21(DE3). The overnight culture was diluted 100 times with fresh LB medium supplemented with 100 μgmL–1 kanamycin, then incubated and shaken at 37°C until the OD600 reached 0.6. Hsp20 expression was induced by adding isopropyl-β-d-thiogalactoside to a final concentration of 1 mM and incubated further for 4 h. The induced Hsp20 protein was purified by affinity chromatography in accordance with the protocol of HisTrap FF crude 1-mL column (GE Healthcare, Milwaukee, WI, United States), and was then used for primary anti-serum production in rabbit.
Validation of hsp20 Gene Deletion Strains
Two methods were used to identify whether hsp20 was knocked out; one uses the primers P5/P6 for PCR identification and the other was Western blot (WB) detection. In the latter, 12 μg of proteins at 24 h were separated by NuPAGETM 4% to 12% Bis-Tris Gel (Invitrogen, United States), and was then blotted onto polyvinylidene difluoride membranes (Sigma) using a tank blot apparatus (Toyo, Tokyo, Japan), The transfer, blocking, hybridization, and washing steps were performed according to a previous study (Yang et al., 2015). The incubated primary rabbit antisera, at a dilution of 1:400, was labeled with anti-rabbit secondary antibody conjugated to goat DyLight 680 (Invitrogen, United States) at a dilution of 1:10000. The detection used the Odyssey infrared imaging system (Li-COR Biosciences, Lincoln, NE, United States).
Growth Curve, Morphological Observation
The strains (OD600 = 0.6) were inoculated in approximately 1% in the fermentation medium (30 mL per 250 mL shake flask) and cultured at 30°C at 120 rpm. The growth curve was measured to 50 h and with three biological repetitions. The OD600 was measured to determine the growth rate of the cells by a spectrophotometer, and the samples were taken at each point in time to observe the difference between the strains.
Preparation of Scanning Electron Microscope
Scanning Electron Microscopy of the 10-fold dilution of samples (mixture of spores and ICPs) were washed with sterile ultrapure water 10 times (9,000 × g, 3 min), and were then fixed with 2.5% glutaraldehyde for 12 h at 4°C. After washing three times with sterile ultrapure water, the samples were dehydrated gradually with 30, 50, 70, 80, 90, 95, and 100% ethanol, which was conducted twice for 10 min each. The samples were freeze dried and were then evenly applied on the cover glasses to sputtering gold plating. The samples were subsequently imaged with the scanning electron microscope (Hitachi Su8010, Japan).
Sporulation Assays
Each strain was grown in the fermentation medium at 30°C for 60 h, and diluted to a final OD600 of 1.0. Cells were then heated to 65°C for 30 min, followed by gradient dilution (10×), and a series of 100 μL of diluents were coated onto LB plates. The colony-forming units (cfu) per mL were then counted.
Extraction and Determination of ICPs Concentrations
For the extraction of the ICPs, the strains were grown under the GYS medium for 20 h, when cells started to lyse, 20 mL of each culture was separately collected. The procedures for extracting ICPs were performed according to previous studies (Wang X. et al., 2016; He et al., 2018). Finally, the ICPs were visualized by SDS-PAGE, and the ICPs concentrations were measured by the Bradford method.
Virulence Assays
The larvae of Heliothis armigera were fed at 28°C with a light/dark cycle of 12 h. Artificial diet was comprised of 4 g yeast extraction, 7 g bean flour, 0.5 g vitamin C, 1.5 g agar, 36% acetic acid, and 1 g penicillin per 100 mL of water. The medium was transferred into 24-hole cell culture plates of approximately 1 mL per well (Corning, United States) with gradient concentration of the mixture of spores and crystals. Three strains were fermented for 48 h and suspended in distilled water after centrifugation. Finally, the semi-lethal concentration (LC50) were compared after a 48 h feeding of the SPSS (Inc., version 19.0, United States).
Whole Cell Proteins Extraction
The harvested cells for the 18, 24, and 28 h were washed three times with PBS buffer and added with 300 μL lysis buffer, composed of 10 mL of cell lysate ratio of Urea 4.80 g, 4 mL 50 mM Tris–HCl (pH 8.0), Thiourea 1.52 g, NaCl 0.04 g, and CHAPs 0.40 g dissolved in 50 mM Tris–HCl, and placed with 10 μL protease inhibitor cocktail. The parameters of 3 s, 3 s, and 5 min were used to disrupt the cells by sonicated twice on ice (SONICS VCX150, United States). After cell disruption, the protein solution was separated by centrifugation (12000 × g, 10 min, 4°C). Protein concentration was determined using the Bradford protein assay kit (Thermo Fisher Scientific) and BSA as standard.
iTRAQ Labeling
According to the iTRAQ protocol, 100 μg protein of each sample was digested with Trypsin Gold (Promega, United States) with the following ratio of protein: trypsin = 30:1 at 37°C for 16 h. Peptides were dried by vacuum centrifugation after trypsin digestion, and were then reconstituted in 0.5 M TEAB processed, according to the protocol for 8-plex iTRAQ reagent of the manufacture (AB SCIEX, United States). One unit of iTRAQ reagent was thawed and reconstituted in 24 μL isopropanol. Samples were labeled with the iTRAQ tags as follows: sample Bt4.0718 (119 tag) and hsp20 knockout mutant (121 tag). Samples were tested in duplicates in a single run. The peptides were incubated with the isobaric tags, at room temperature for 2 h. The labeled peptide mixtures were then pooled and dried by vacuum centrifugation.
LC-MS/MS Analysis
First dimensional separation at basic pH reverse phase – tryptic digestion mixture was dissolved in 100 μL mobile phase A (10 mM ammonium hydroxide in water/acetonitrile, 98/2 (v/v), pH = 10.5) and was then injected via 1200 Series HPLC (Agilent) into a Zorbax extend-C18 column (150 × 2.1 mm, Agilent). Mobile phase B was 10 mM ammonium hydroxide in water/acetonitrile, 10/90 (v/v), and the linear gradient elution was performed with 5%–50% B for 50 min at a flow rate of 0.3 ml/min. A total of 10 fractions from the first dimensional separation was collected (one fraction per 4 min) and then lyophilized for further second dimensional online LC-MS analysis.
Reverse-phase nano flow HPLC and tandem mass spectrometry – the second dimensional Chromatography was employed on the Eksigent nanoLC-UltraTM 2D System (AB Sciex, Concord, United States). The lyophilized SCX fractions were re-dissolved in 2% acetonitrile, 0.1% formic acid, and loaded on ChromXP C18 (3 μm, 120 Å) nano LC trap column. The online trapping and desalting procedure were carried out at 2 μL/min for 10 min with 100% solvent A. Solvents were composed of water/acetonitrile/formic acid (A, 98/2/0.1%; B, 2/98/0.1%). An elution gradient of 5%–38% acetonitrile (0.1% formic acid) in 70 min gradient was used on an analytical column (75 μm × 15 cm C18- 3 μm 120 Å, ChromXP Eksigent). LC MS/MS analysis was performed using a triple TOF 5600 System (AB Sciex, Concord, United States) fitted with nano spray III source. Data was acquired using an ion spray voltage of 2.4 kV, curtain gas of 30 PSI, nebulizer gas of 5 PSI, and an interface heater temperature of 150°C. The MS was operated with TOF-MS scans. For IDA, survey scans were acquired in 250 ms and as many as 30 product ion scans (80 ms) were collected if exceeding a threshold of 260 counts per second (counts/s), and with a + 2 to + 5 charge-state. A rolling collision energy setting was applied to all precursor ions for collision-induced dissociation. Dynamic exclusion was set 16 s.
Protein Identification and Quantification – The MS/MS data were analyzed for protein identification and quantification using protein pilot software v.4.5 (Sciex Inc., United States). The local false discovery rate was estimated with the integrated PSPEP tool in the protein pilot software to be approximately 1.0% and the tolerances were specified as ± 0.05 Da for peptides and ± 0.05 Da for MS/MS fragments. Searches were made against Bt DB27 protein database. The Bt DB27 database was read from the UniProt1, approximately 6284 sequences were searched, and parameters included iTRAQ 8-plex quantification, cysteine modified with IAA, trypsin digestion, thorough searching mode, and minimum protein threshold of 95% confidence (unused protein score >1.3). For a protein to be determined as differentially expressed the fold change must be ≥1.5.
Bioinformatics Analysis
For a protein to be determined as differentially expressed the fold change must ≥1.5. The differentially expressed proteins (DEPs) were screened and then analyzed at the online website “OmicsBean,” the GO, and KEGG enrichment analysis were conducted. In the GO and KEGG pathway enrichment analysis of differentially expressed proteins, we compared the DEPs to all of the identified proteins as a background, and the formula used in these analyses was as follows: in which N is the number of GO/KEGG pathway entries in all identified proteins; n is the number of GO/KEGG pathway entries that represent differentially expressed proteins; M is the number of GO/KEGG pathway entries that can be matched to all identified proteins, and m is the number of GO/KEGG pathway entries that can be matched to a differentially expressed protein. If the p-value of the hypergeometric test was less than 0.05, the differentially expressed protein was significantly enriched in the GO/KEGG pathway entry.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from the bacterial cells harvested at the time point of 28 h. Bt4-Δhsp20 and Bt4.0718 were cultivated using Trizol Reagent (15596-026 Invitrogen Biotech). Following digestion of the residual genomic DNA by DNase I (EN0521, Thermo Fisher Scientific), 1 μg of total RNA was reverse transcribed to cDNA utilizing RevertAidTM First Strand cDNA Synthesis Kit (12328040, Thermo Fisher Scientific) according to the instruction of the manufacturer. The two-step real-time RT-PCR was conducted using power SYBR® green PCR master mix (4367659, Thermo Fisher Scientific) in a CFX connect Real-Time PCR detection system (Bio-Rad). The primers used in this study are summarized in Table 2. 16S rRNA was used as a normalization control for gene expression.
Results
Validation of hsp20 Knock-Out and Complemented Strain
To determine the role of Hsp20 in sporulation and ICPs production, we constructed an hsp20 mutant. PCR and WB analyses were performed to confirm the positive clones. First, the upstream and downstream homology arms of hsp20 were confirmed to be ligated to the pRP1028 vector after sequencing. The positive clones of the knockout (Bt4-Δhsp20) and complemented strain (Bt4-Δhsp20::hsp20) were identified by PCR (16S rRNA was used as the blank control). The expression of pET28a-hsp20 was then induced in BL21(DE3) to prepare the polyclonal antibody, and whole proteins of the three strains were extracted for WB analysis verification (30S ribosome protein served as the internal reference). The final results revealed that the knockout and complemented mutations of hsp20 in Bt4.0718 were successful (Supplementary Figure S3).
Growth Curve Comparison of the hsp20 Knockout Strain and Bt4.0718
By comparing the growth curves of Bt4.0718, Bt4-Δhsp20, and Bt4-Δhsp20::hsp20, we found that the stability period of Bt4-Δhsp20 was reduced by nearly 2 h and that the maximum biomass was also slightly reduced. The OD600 of Bt4.0718 peaked at 16.48, and the stationary phase ended after 22 h, which was only slightly different from that of Bt4-Δhsp20::hsp20. The highest OD600 of Bt4-Δhsp20 was 14.43 and entered the decline phase at 20 h (Figure 1).
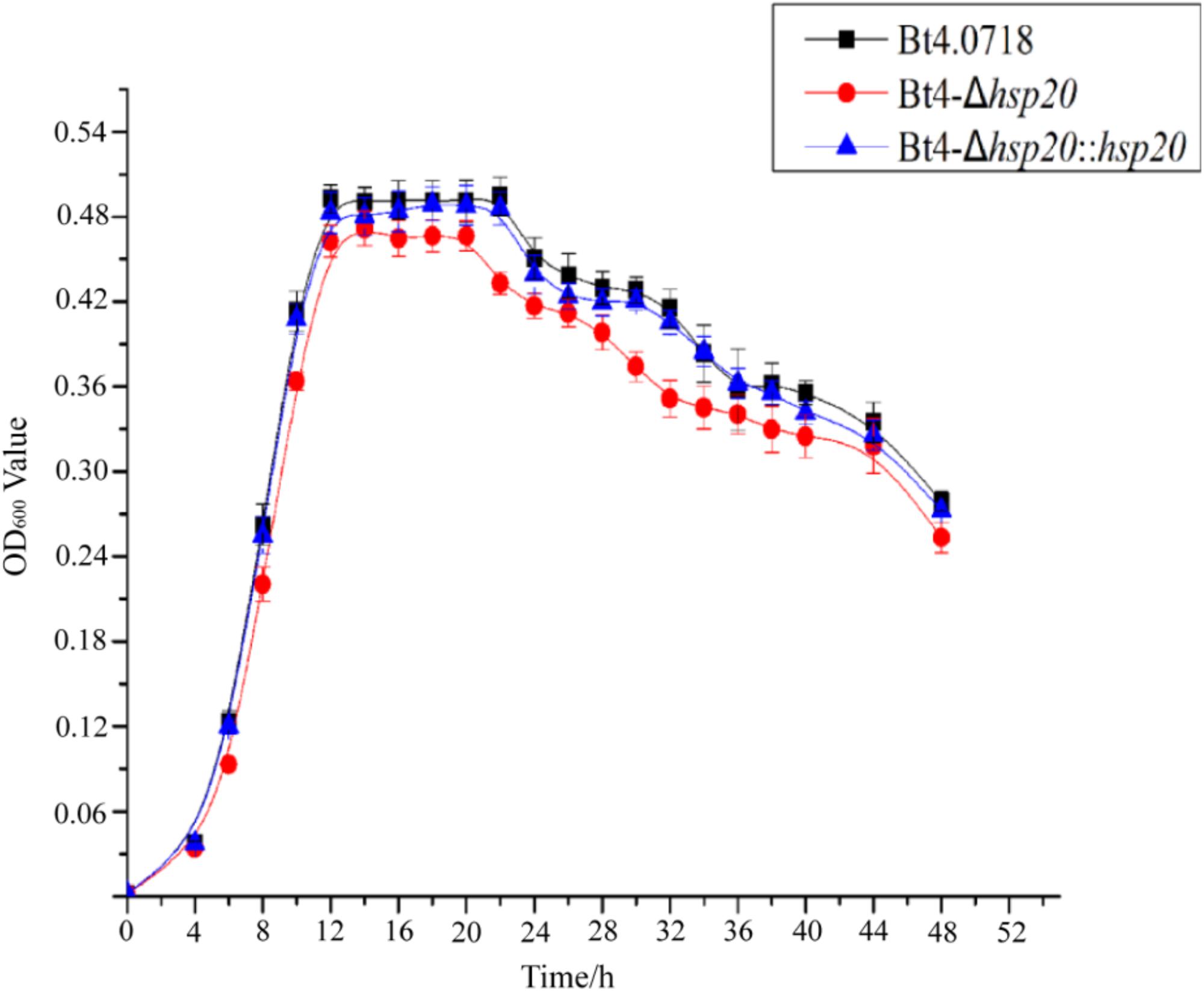
Figure 1. Growth of the hsp20 mutant shows a marked decay at end of stationary phase. Growth curves of the Bt4.0718, Bt4-Δhsp20, and Bt4-Δhsp20::hsp20 strains which a bar for each sampling time point represents the standard error of the mean from three batches.
Effects of hsp20 Deletion on the Sporulation and ICPs
The results of phase contrast microscopy revealed no difference in the phenotypes of the three strains at 12, 24, and 36 h, and only a very small number of spores were produced in Bt4-Δhsp20 compared with that of Bt4.0718 and Bt4-Δhsp20::hsp20. Bt4-Δhsp20 generated few immature spores, whereas Bt4.0718 released a large number of mature spores and obvious rhombic parasporal crystal proteins. A mass of mature spores was also observed in the Bt4-Δhsp20::hsp20 cells at 24 and 36 h. Subsequently, the mixture of spores and crystals of the three strains was observed under a scanning electron microscope (Figure 2A). Aside from the cell and crystals density, no significant difference was observed in the shape and size of the spores and crystal proteins produced by the three strains.
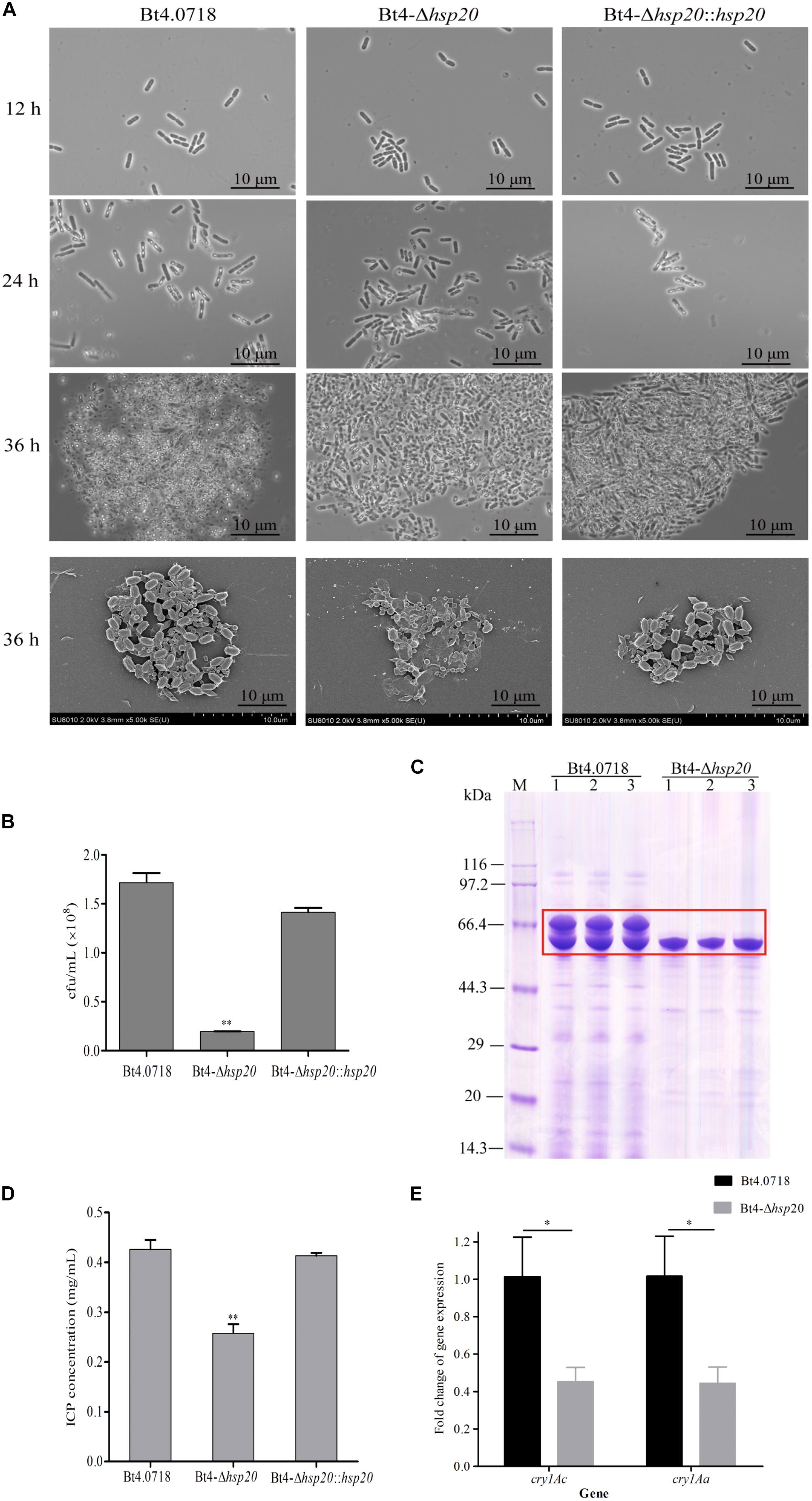
Figure 2. Sporulation and ICPs formation are reduced in Bt4-Δhsp20. (A) Phase contrast microscopy (100 × oil objective) of the strains at 12, 24, and 36 h; scanning electron microscopy of the spores and crystal proteins (10.0 K×) at 36 h. (B) Sporulation of the Bt4-Δhsp20 compared to Bt4.0718, and Bt4-Δhsp20::hsp20 strains in LB at 60 h. Spore numbers per mL were indicated by cfu/mL, and each experiment was carried out in triplicate which the values were means ± standard deviations. Significances of differences by Student’s t-test are indicated (∗∗P ≤ 0.01). (C) ICPs in Bt4.0718 and Bt4-Δhsp20 strains at 20 h analyzed by SDS-PAGE, ICPs were boxed in red. (D) Concentrations of ICPs at 20 h determined by Bradford method in Bt4-Δhsp20 compared to Bt4.0718, and Bt4-Δhsp20::hsp20 strains. The values were means ± standard deviations for triplicate assays, and the significances of differences by Student’s t-test is indicated (∗∗P ≤ 0.01). (E) qRT-PCR analysis the fold-change of cry1Ac and cry1Aa in Bt4-Δhsp20 strain which compared to Bt4.0718 after fermentation for 28 h. The values were means ± standard deviations for triplicate assays, the significances of differences by Student’s t-test are indicated (∗P ≤ 0.05).
Sporulation assays of each strain revealed that Bt4.0718 and the complemented strain produced 1.72 × 108 and 1.41 × 108 cfu/mL, respectively. In contrast, the hsp20 knockout strain produced 1.94 × 107 cfu/mL, which is only 11.8% of that observed in Bt4.0718 (Figure 2B). This result indicated that hsp20 was important in the development and generation of spores. The deletion of hsp20 may lead to the reduction in ICPs yield, as observed in Figure 2C. To be quantitative, we measured the concentration of ICPs, and conducted the qRT-PCR of cry1Ac and cry1Aa, which are the main toxin genes in the Bt4.0718 stain as shown in Figures 2D,E. The concentration of ICPs in Bt4.0718 was 0.426 μg/mL, which was 1.65 times greater than that of knockout strain (0.257 μg/mL), and the complemented strain was 0.413 μg/mL. Whereas, the expression revealed by the qRT-PCR of cry1Ac and cry1Aa in hsp20 knockout strain was nearly half of that exhibited by the Bt4.0718. Due to the quantitative results of iTRAQ on Cry1Ac and Cry1Aa, which are specific proteins of B. thuringiensis with some limitation, the peptides peak area output was calculated as shown in Supplementary Figure S4 (according to the Supplementary Table S1 which from the mass spectrometry proteomics results), and the area level of Cry1Ac and Cry1Aa in Bt4.0718 were approximately 1.623 times that of hsp20 knockout strain (SE = 0.210, ∗P < 0.05).
Virulence assays of the larvae of H. armigera showed that the LC50 of the hsp20 knockout strain was 10.789 μg/mL, whereas the toxicity of Bt4.0718 and the complemented strain were 5.254 and 5.613 μg/mL, respectively (Table 3). This result was consistent with the quantification of ICP.
SDS-PAGE Analysis of Whole Cell Proteins of Bt4.0718 and Bt4-Δhsp20
To detect differences at the proteome level, we carried out the SDS-PAGE analysis of whole proteins of Bt4.0718 and Bt4-Δhsp20 at three time points (18, 24, and 28 h), as shown in Supplementary Figure S5. The protein difference at the T3 period (28 h) increased, and it was selected to run the comparative quantitative proteomic analyses.
Analysis of Differentially Expressed Proteins (DEPs) Between the Bt4.0718 Strain and the hsp20 Knockout Mutant
A total of 1736, 1713, and 1769 proteins were identified in three biological replications (Supplementary Table S2, from the mass spectrometry proteomics results). A fold change ≥1.5 was considered to be differentially expressed. In addition, reproducibility analysis by a Venn diagram showed that the differential proteins shared by three and two groups accounted for 60, 50, and 42% of the total number of differential proteins in groups Q1, Q2, and Q3 with good repeatability, respectively (Figure 3A). A total of 195 DEPs were then identified; approximately 11 were upregulated and 184 were downregulated (Figure 3B), and in the GO and KEGG analysis.
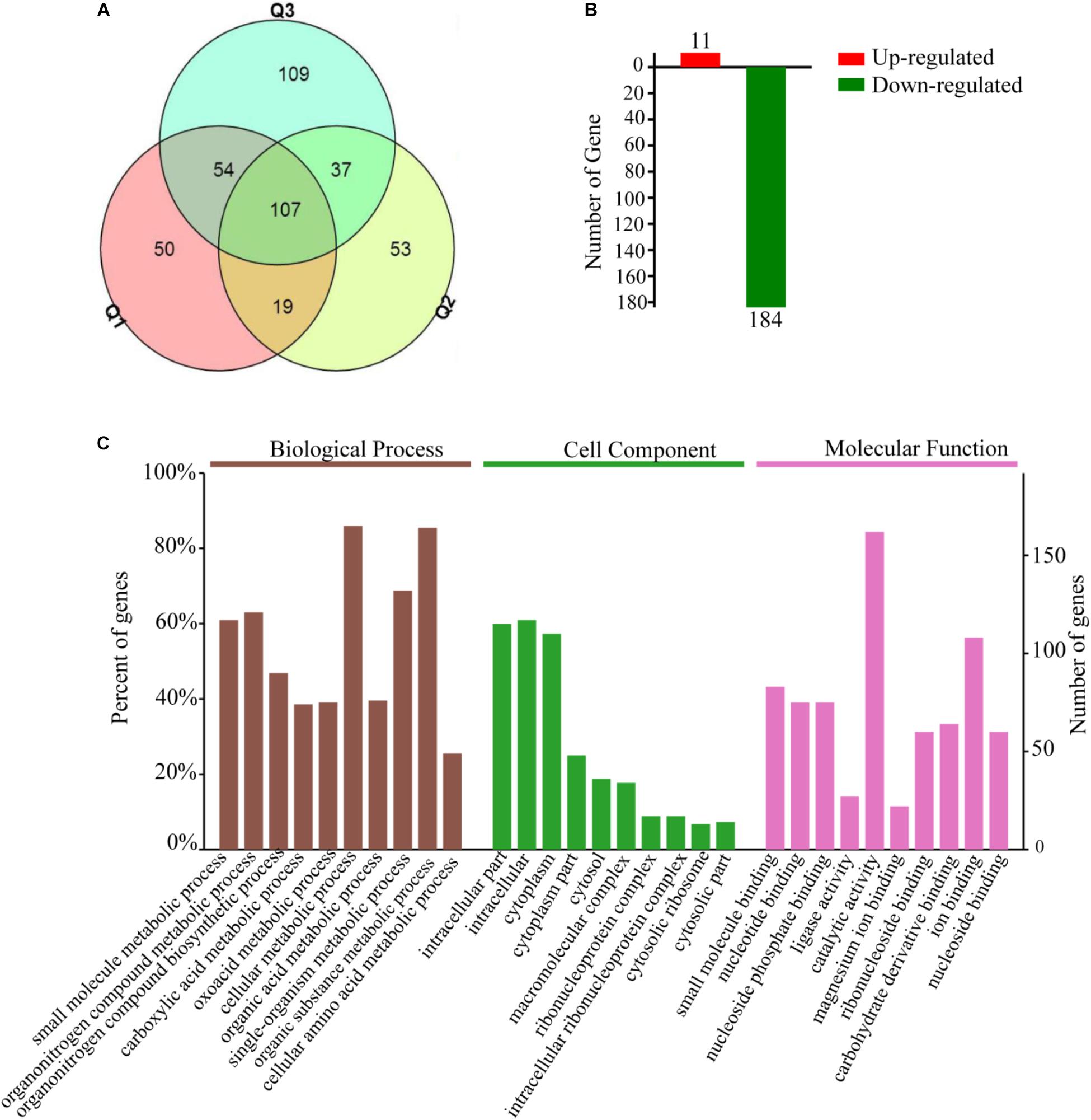
Figure 3. Total of 195 differential expression proteins are screened which Bt4-Δhsp20 compared to Bt4.0718, and most of them are main involved in intracellular substance and cytoplasm metabolism. (A) Venn diagram of differential proteins from every biological repetition (Q1, Q2, and Q3). (B) Number of screened up or downregulated differential expression proteins (11 and 184 were upregulated and downregulated, respectively). (C) GO annotations of the 195 DEPs. Biological processes, cellular component, and molecular function in the three categories significantly enriched and analyzed that ranked the top ten. The entries for each category were arranged according to their p-value size from left to right in order of priority. Based on the vertical axis, each entry can be viewed corresponding to the percentage of total differences in protein expression. A Go term with a p-value of <0.05 was considered significant.
The DEPs were subjected to GO enrichment analysis and were classified as biological process, molecular function or cellular component. The three main biological processes are cellular metabolism, organic substance metabolism, and single-organism metabolism. The molecular function is mainly distributed in catalytic activity and ion binding. The main cellular components are intracellular substance and cytoplasm (Figure 3C).
KEGG Analysis of DEPs
According to the KEGG categories, the proteins were annotated into 18 groups (Figure 4). Carbon metabolism, metabolic pathways, biosynthesis of secondary metabolites, microbial metabolism in diverse environment, and biosynthesis of amino acid were classified as the global and overview maps in metabolism. Whereas, energy, nucleotide, amino acid, cofactors, and vitamins metabolism dominated the cellular or single-organism metabolic processing. Nearly all upregulated genes, such as upp, acoA, dat, hom, leuA, mtnX, valS, and rplM, were classified under energy citric acid cycle (TCA) and amino acid metabolism, including D-alanine, leucine, proline, lysine, cysteine, methionine, and so on, and only sigA and spoIIAB were related to sporulation.
Figure 4. Bar chart of enriched KEGG pathways of the 195 differential expression proteins. The number of involved proteins in a specific pathway and the corresponding p-value are shown on the right side of the column.
Validation of DEPs by qRT-PCR
The proteomic results were verified at the mRNA level. For this purpose, we carried out qRT-PCR quantification of eight genes, including four upregulated genes, namely, dat, odhA, mtnX, and leuA (fold change were 1.66, 2.48, 1.52, and 2.12, respectively), and four downregulated genes, namely, murQ, tig, tpx, and eno (fold change were 0.21, 0.39, 0.34, and 0.38, respectively). The acoA was, additively, 1.54-fold for qRT-PCR quantification. The results showed that the eight proteins exhibited a similar change trend both at the mRNA level and protein level (Figure 5).
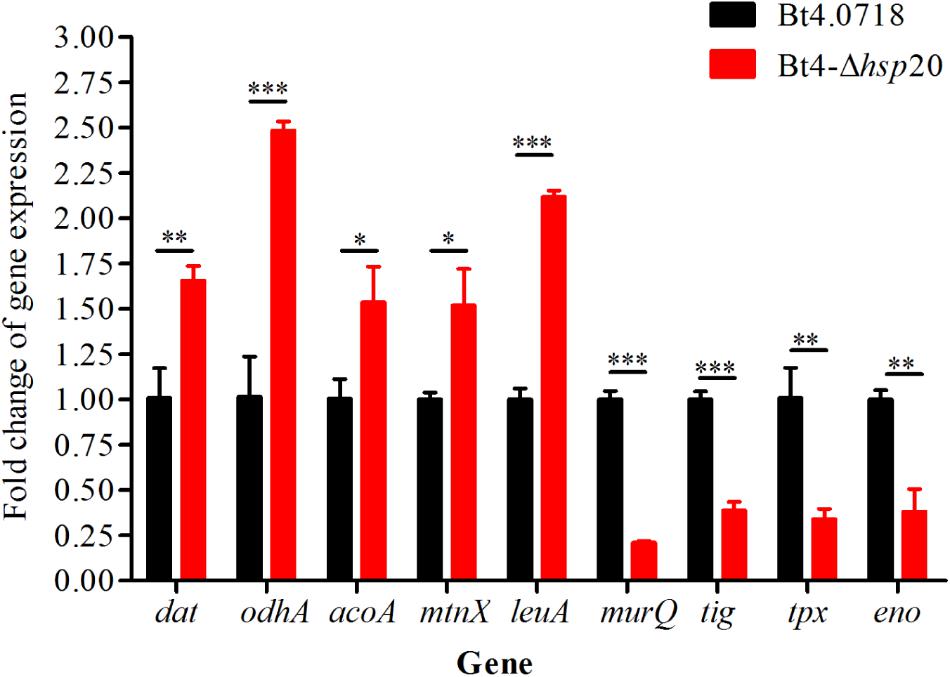
Figure 5. Validation of the differential expression of nine genes. Gene expression level by qRT-PCR of Bt4-Δhsp20 compared with Bt4.0718. The values were means ± standard deviations for triplicate assays, the significances of differences by Student’s t-test are indicated (∗P ≤ 0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001).
Discussion
The Possible Effect of Hsp20 in Early Sporulation
Phase contrast microscopy results suggested that hsp20 influenced early sporulation. Six proteins that are directly related to the sporulation exhibited differential expression in the mutant (two upregulated and four downregulated). SpoIIAB and SigA are upregulated 1.54-fold and 1.56-fold, respectively. SpoIIAB is known to have sigma factor antagonist activity that binds and reduces or prevents the rate sigmaF factor transcription. SigmaF factor subsequently governs the transcription of spoIIIG, which encodes the late prespore-specific regulator sigmaG (Serrano et al., 2004). In Bt4.0718 strain, SigA remains constant during the transition from vegetative growth to sporulation. The sporulation-specific sigma factors appear successively, in the order of SigmaH, SigmaE, and SigmaK (Fujita, 2000), and are overexpressed in sporulation mutants of B. thuringiensis. These mutants are blocked in the phosphorylation of Spo0A, a key regulator of sporulation initiation (Baum and Malvar, 1995). Whereas, the promoters of a number of cry proteins can be driven by SigmaE and SigmaK (Zhang et al., 1998), and cry genes, including cry1, cry4, cry8, cry11, and cry18, which are controlled by both SigE and SigK (Deng et al., 2014). Thus, this may be the reason for the obvious difference in the impact degree of sporulation and ICP formation.
On the other hand, of the downregulated proteins (RacX, SpovG, FtsA, and FtsH), FtsA and FtsH are related to cell division, sporulation and are involved in switching the site of cell division from the mid-cell to polar position (Kemp et al., 2002). FtsA is under the control of SigmaF, whereas FtsH is an ATP-dependent zinc metallopeptidase for both cytoplasmic and membrane proteins. FtsH is initially asymmetrically located in the septa of the sporulation cells and subsequently in the membrane that engulfed the fore-spore during sporulation (Wehrl et al., 2000). Spo0A is important in initiating sporulation; the amount of the master regulator protein Spo0A is reduced in an ftsH knockout and the small amounts of Spo0A protein present are inactive (Ai and Schumann, 2009). Whereas, RacX exhibits an amino acid racemase activity and is probably involved in peptidoglycan modification during cortex synthesis for sporulation from the III to IV stage. spoVG controls the sporulation at the V stage, leading to an asymmetric septation and causing the accumulation of ICPs (Rosenbluh et al., 1981; Crissman et al., 1989; Matsuno and Sonenshein, 1999).
Possible Influences of hsp20 Knockout on Amino Acid Supply at Stationary Phase
During sporulation, some operons and genes involved in amino acid biosynthesis and transport were upregulated in response to amino acid starvation (Wang et al., 2013b). However, among the DEPs, twelve 50S ribosomal protein subunits (rplA/C/D/E, etc.) required for the assembly of 50S ribosomal protein and formation of peptide bond (Nissen et al., 2000) are down-regulated, (0.1 to 0.61-fold) thus the translation level may be decreased. As well as that, 13 kinds of tRNA ligase, which include alaS, argS, asnS, valS, proS, metG, etc., are responsible for the corresponding amino acid transfer in the translation process. Meanwhile, 13 genes involved in the biosynthesis of 9 kinds amino acids, such as proA/B/S, participate in the proline production from glutamate. GlnA catalyzes glutamate into glutamine; hisA/D promote histidine formation; metD/E, argD, glyA, and dapB/H are involved in the syntheses of methionine, arginine, glycine, and lysine, respectively, all of which are also down-regulated. These aspects reflect the shortage of amino acids.
In fact, 80% of amino acids for ICP synthesis mainly came from protein turnover (Monro, 1961), ATP-dependent protease, usually with high expression, could quickly degrade some abnormal polypeptides (Frees et al., 2007; Molière and Turgay, 2009) to provide enough amino acids during sporulation. ATP-dependent protease ClpX, HslU/HslV was observed without significant differences. But, two peptidases observed from the DEPs are down-regulated (0.40-fold, 0.44-fold), which include the cytosol aminopeptidase (PepA), the enzyme CtpA with endopeptidase activity. Therefore, hsp20 could likely contribute to maintain a normal supply of amino acids during sporulation and ICP formation during amino acid starvation.
Energy and Carbon Source Could Be Impacted by Hsp20 During Sporulation
Energy is essential to any microbial life activity, and according to the KEGG analysis of DEPs, some downregulated proteins are involved in TCA cycle and glycolysis, which are required to provide the main ATP for the Bt cells at the stationary phase. The abundant glucose of exponential growth phase mainly flows into glycolysis pathway to produce pyruvate, which can be utilized by other pathways, and the first three enzymes of TCA cycle are critical for sporulation (Jin and Sonenshein, 1994; Ireton et al., 1995). CitZ and CitB participate in the synthesis of citrate. CitZ is involved in the first step of the synthesis of isocitrate from oxaloacetate. CitB catalyzes the reversible isomerization of citrate to isocitrate via cis-aconitate to produce acetyl-coA, which is linked to fatty acid metabolism. If a loss in activity results in sporulation defect, they can be regulated by phosphorylated Spo0A, which is involved in the regulation of citrate concentration and in sporulation (Craig et al., 1997). Furthermore, aconitase (encode by citB) is required for efficient late-sporulation gene expression: its RNA binding activity may stabilize gerE mRNA for proper timing of spore coat assembly (Serio et al., 2006).
In additional, Eno, Pyk, and GpmI are downregulated in the subpathway that catalyzes the reversible conversion of 2-phosphoglycerate into phosphoenolpyruvate. This subpathway is essential for the degradation of carbohydrates via glycolysis. Eno interacts with GroEL, which is involved in sporulation (Endo and Kurusu, 2007). A reduction in the expression of PdhC will decrease the conversion of pyruvate into acetyl-CoA that is involved in glycolysis, ultimately reducing the energy. As a component of the 2-oxoglutarate dehydrogenase complex, down regulated OdhB will reduce the overall conversion of 2-oxoglutarate to succinyl-CoA. The two down regulated proteins, SucD, and SucC are succinate-CoA ligase (ADP-forming) alpha and beta subunits, which are involved in the subpathway that synthesizes succinate from succinyl-CoA and the TCA, which couples the hydrolysis of succinyl-CoA to the synthesis of either ATP or GTP.
For the carbon source, the poly-3-hydroxybutyrate (PHB) and acetoin are commonly produced and excreted during exponential phase as the intercellular and extracellular carbon and energy store in B. thuringiensis. However, PHB metabolism has been indicated to be unrelated to sporulation and parasporal crystal protein formation (Wang X. et al., 2016), and no related DEPs were observed. Moreover, we observe that acoA is upregulated approximately 1.54-fold (only identified with qRT-PCR). This protein mainly catalyzes the 2,6-dichlorophenolindophenol cleavage of acetoin into acetate and acetaldehyde to utilize acetoin as the carbon source for growth (Huang et al., 1999), serving as an alternative to energy supplementation and self-rescue. Hence, acetoin could also be important as the carbon and energy source for sporulation and ICP formation during progressive deficiency of nutrients at stationary phase.
Hsp20 May Cause Some Effects on Purine Metabolism, mRNA Stability During Sporulation and ICP Formation
Down-regulated proteins: PurA/B, adenylate kinase (Adk), and nucleoside diphosphate kinase (Ndk), could diminish purine metabolism. The purA/B/M/N/S code enzymes are involved in both IMP and AMP biosynthesis, which are part of purine biosynthesis (Saxild and Nygaard, 1988; Mäntsälä and Zalkin, 1992). Ndk is involved in the synthesis of nucleoside triphosphates other than ATP. Adk catalyzes the reversible transfer of the terminal phosphate group between ATP and AMP.
Downregulated RpoB and RpoE are DNA-directed RNA polymerase subunits that catalyze the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Moreover, these proteins participate in both the initiation and recycling phases of transcription and are involved in the sigma factor switching that initiates sporulation (Moeller et al., 2012).
For the mRNA stability, two correlated downregulated DEPs were found, namely polyribonucleotide nucleotidyltransferase (Pnp) and ribonuclease J1 (RnjA). Pnp is necessary for competence development in Bacillus subtilis but exerts no effect on sporulation efficiency. By contrast, RnjA exhibits endonuclease and 5′–3′ exonuclease activities, and is involved in both rRNA and mRNA stability and degradation, and its knockout mutant causes major defects in sporulation (Figaro et al., 2013).
Figure 6 shows the pathways by which the DEPs (such as the Table 4) observed in the hsp20 deletion could negatively affect sporulation and ICPs formation. Hsp20 may function in aiding protein folding or aiding translation of these proteins under the starvation conditions leading to sporulation and ICP formation.
Figure 6. Simplified diagram of the effects of hsp20 knockout on the sporulation and ICPs. The genes, proteins, and amino acids are included in the diagram. Black and red indicate downregulation or decrease and upregulation or increase, respectively.
Conclusion
The hsp20 would influence the production of sporulation and ICPs, which are the hallmarks of Bt, and the general metabolism. Stationary growth phase of the Bt4-Δhsp20 was reduced by approximately 2 h, and only a slight decrease was observed in the biomass. By analyzing the DEPs of the iTRAQ proteomics Bt4-Δhsp20 compared to Bt4.0718, it was evident that the deletion of hsp20 caused downregulation of a lot of proteins, which are distributed in multiple metabolic pathways. This most likely cause the decrease in 50S ribosome assembly and its translation level, the short supply of amino acids (the biosynthesis and the protein-turnover), the drop in purine metabolism and some mRNAs stability, the insufficient energy supply (including TCA, glycolysis) during sporulation and ICP formation at the stable phase.
Ethics Statement
This study was approved by the Ethics Committee and Animal Care and Use Committee of Hunan Normal University, and all animals were handled in accordance with the guidelines of Hunan Provincial Council on Animal Care.
Author Contributions
XD and LX conceived and designed all the experiments. JX and JP conducted the proteomic studies (iTRAQ) and drafted the manuscript. TZ, ZY, and XZ performed the qRT-PCR analysis. MQ and SY participated in the Western blot analysis. SL and JL analyzed the proteomic data. PZ contributed to the bacterial culture experiments. XD provided reagents, materials, and analysis tools.
Funding
This work was supported by the National Key Research and Development Program of China (2017YFD0201201), the National Natural Science Foundation of China (31370116), and the Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province (20134486).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02059/full#supplementary-material
FIGURE S1 | Line graph of hsp20 mRNA FPKM value at early sporulation and late sporulation (the bar of two points represents the standard error of mean).
FIGURE S2 | Schematic of the I-SceI mediated marker-less gene knockout method.
FIGURE S3 | Identification of hsp20 knockout. (A) Lanes 1, 2: knockout vector pRP1028-hsp20UD. (B) Colony PCR amplifies the hsp20 gene with P5/P6. Lane 1: wide type strain; Lane 2: complemented strain; Lanes 3, 4: knockout strain; Lanes 5, 6: 16S rRNA sequence of Bt4.0718 and the hsp20 knockout strain. (C) Western blot analysis of hsp20 expression in the three strains. Lane 1: complemented strain; Lane 2: Bt4.0718 strain; Lane 3: hsp20 knockout strain.
FIGURE S4 | Peptides peak area of Cry1Ac and Cry1Aa in Bt4.0718 and hsp20 knockout strain (Std. Error = 0.210, ∗P < 0.05).
FIGURE S5 | SDS-PAGE of the whole cell protein samples of Bt4.0718 and the hsp20 knockout strain at different time (lanes 1–3: 18, 24, and 28 h of Bt4.0718 strain; lanes 4–6: 18, 24, and 28 h of the hsp20 knockout strain).
TABLE S1 | The quantitative ICP peptides results.
TABLE S2 | The quantitative proteome results.
Footnotes
References
Ai, T. T. L., and Schumann, W. (2009). The SpoOE phosphatase of Bacillus subtilis is a substrate of the FtsH metalloprotease. Microbiology 155, 1122–1132. doi: 10.1099/mic.0.024182-0
Armada, E., Leite, M., Medina, A., Azcón, R., and Kuramae, E. (2018). Native bacteria promote plant growth under drought stress condition without impacting the rhizomicrobiome. FEMS Microbiol. Ecol. 94:fiy092. doi: 10.1093/femsec/fiy092
Aronson, A. (2002). Sporulation and delta-endotoxin synthesis by Bacillus thuringiensis. Cell. Mol. Life Sci. 59, 417–425. doi: 10.1007/s00018-002-8434-6
Baum, J. A., and Malvar, T. (1995). Regulation of insecticidal crystal protein production in Bacillus thuringiensis. Mol. Microbiol. 18:1–12. doi: 10.1111/j.1365-2958.1995.mmi_18010001.x
Bepperling, A., Alte, F., Kriehuber, T., Braun, N., Weinkauf, S., Groll, M., et al. (2012). Alternative bacterial two-component small heat shock protein systems. Proc. Natl. Acad. Sci. U.S.A. 109, 20407–20412. doi: 10.1073/pnas.1209565109
Cao, Z., Tan, T., Jiang, K., Mei, S., Hou, X., and Cai, J. (2018). Complete genome sequence of Bacillus thuringiensis L-7601, a wild strain with high production of melanin. J. Biotechnol. 275, 40–43. doi: 10.1016/j.jbiotec.2018.03.020
Cocotl-Yañez, M., Moreno, S., Encarnación, S., López-Pliego, L., Castañeda, M., and Espín, G. (2014). A small heat-shock protein (Hsp20) regulated by RpoS is essential for cyst desiccation resistance in Azotobacter vinelandii. Microbiology 160, 479–487. doi: 10.1099/mic.0.073353-0
Craig, J. E., Ford, M. J., Blaydon, D. C., and Sonenshein, A. L. (1997). A null mutation in the Bacillus subtilis aconitase gene causes a block in Spo0A-phosphate-dependent gene expression. J. Bacteriol. 179, 7351–7359. doi: 10.1128/jb.179.23.7351-7359.1997
Crissman, J. W., Causey, S. C., Thorne, L., and Pollock, T. J. (1989). Accumulation of the insecticidal crystal protein of Bacillus thuringiensis subsp. kurstaki in post-exponential-phase Bacillus subtilis. Appl. Environ. Microbiol. 55, 2302–2307.
Deng, C., Peng, Q., Song, F., and Lereclus, D. (2014). Regulation of cry gene expression in Bacillus thuringiensis. Toxins 6, 2194–2209. doi: 10.3390/toxins6072194
Doruk, T., Avican, U., Camci, I. Y., and Gedik, S. T. (2013). Overexpression of polyphosphate kinase gene (ppk) increases bioinsecticide production by Bacillus thuringiensis. Microbiol. Res. 168, 199–203. doi: 10.1016/j.micres.2012.11.009
Endo, A., and Kurusu, Y. (2007). Identification of in vivo substrates of the chaperonin GroEL from Bacillus subtilis. J. Agric. Chem. Soc. Japan 71, 1073–1077. doi: 10.1271/bbb.60640
Figaro, S., Durand, S., Gilet, L., Cayet, N., Sachse, M., and Condon, C. (2013). Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology. sporulation, and competence. J. Bacteriol. 195, 2340–2348. doi: 10.1128/JB.00164-13
Frees, D., Savijoki, K., Varmanen, P., and Ingmer, H. (2007). Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC. gram-positive bacteria. Mol. Microbiol. 63, 1285–1295. doi: 10.1111/j.1365-2958.2007.05598.x
Fujita, M. (2000). Temporal and selective association of multiple sigma factors with RNA polymerase during sporulation in Bacillus subtilis. Genes Cells 5, 79–88. doi: 10.1046/j.1365-2443.2000.00307.x
Glatron, M. F., and Rapoport, G. (1972). Biosynthesis of the parasporal inclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie 54, 1291–1301. doi: 10.1016/s0300-9084(72)80070-1
He, J., Fu, Y., Yu, Z., Liu, S., Chen, B., Li, Z., et al. (2018). c-di-GMP regulates various phenotypes and insecticidal activity of gram-positive Bacillus thuringiensis. Front. Microbiol. 9:45. doi: 10.3389/fmicb.2018.00045
Huang, M., Oppermann-Sanio, F. B., and Steinbüchel, A. (1999). Biochemical and molecular characterization of the Bacillus subtilis acetoin catabolic pathway. J. Bacteriol. 181, 3837–3841.
Huang, S., Ding, X., Sun, Y., Yang, Q., Xiao, X., Cao, Z., et al. (2012). Proteomic analysis of bacillus thuringiensis at different growth phases by using an automated online two-dimensional liquid chromatography-tandem mass spectrometry strategy. Appl. Environ. Microbiol. 78, 5270–5279. doi: 10.1128/AEM.00424-12
Hui, Y., Wang, P., Qi, P., Rong, R., Liu, C., Lereclus, D., et al. (2014). Weak transcription of the cry1ac gene in nonsporulating bacillus thuringiensis cells. Appl. Environ. Microbiol. 78, 6466–6474. doi: 10.1128/AEM.01229-12
Ireton, K., Jin, S., Grossman, A. D., and Sonenshein, A. L. (1995). Krebs cycle function is required for activation of the Spo0A transcription factor in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 92:2845. doi: 10.1073/pnas.92.7.2845
Janes, B. K., and Stibitz, S. (2006). Routine markerless gene replacement in Bacillus anthracis. Infect. Immun. 74, 1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006
Jie, R., He, H., Wang, T., Ding, X., Zuo, M., Quan, M., et al. (2015). Comparative analysis of genomics and proteomics in Bacillus thuringiensis 4.0718. Plos One 10:e0119065. doi: 10.1371/journal.pone.0119065
Jin, S., and Sonenshein, A. L. (1994). Identification of two distinct Bacillus subtilis citrate synthase genes. J. Bacteriol. 176:4669. doi: 10.1128/jb.176.15.4669-4679.1994
Jouzani, G. S., Valijanian, E., and Sharafi, R. (2017). Bacillus thuringiensis: a successful insecticide with new environmental features and tidings. Appl. Microbiol. Biotechnol. 101, 2691–2711. doi: 10.1007/s00253-017-8175-y
Kemp, J. T., Driks, A., and Losick, R. (2002). FtsA mutants of Bacillus subtilis impaired in sporulation. J. Bacteriol. 184, 3856–3863. doi: 10.1128/JB.184.14.3856-3863.2002
Khaskheli, G. B., Zuo, F., Yu, R., and Chen, S. (2015). Overexpression of small heat shock protein enhances heat- and salt-stress tolerance of bifidobacterium longum NCC2705. Curr. Microbiol. 71, 1–8. doi: 10.1007/s00284-015-0811-0
Liu, X., Zuo, M., Wang, T., Sun, Y., Liu, S., Hu, S., et al. (2015). Proteomic analysis of the influence of Cu(2+) on the crystal protein production of Bacillus thuringiensis X022. Microb. Cell Fact. 14:153. doi: 10.1186/s12934-015-0339-9
Maio, A. D. (1999). De maio a: heat shock proteins: facts, thoughts and dreams. Shock 11, 1–12. doi: 10.1097/00024382-199901000-00001
Mäntsälä, P., and Zalkin, H. (1992). Cloning and sequence of Bacillus subtilis purA and guaA, involved in the conversion of IMP to AMP and GMP. J. Bacteriol. 174, 1883–1890. doi: 10.1128/jb.174.6.1883-1890.1992
Mathy, N., Bénard, L., Pellegrini, O., Daou, R., Wen, T., and Condon, C. (2007). 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129, 681–692. doi: 10.1016/j.cell.2007.02.051
Matsuno, K., and Sonenshein, A. L. (1999). Role of SpoVG in asymmetric septation in Bacillus subtilis. J. Bacteriol. 181:3392.
Moazamian, E., Bahador, N., Azarpira, N., and Rasouli, M. (2018). Anti-cancer parasporin toxins of new Bacillus thuringiensis against human colon (HCT-116) and blood (CCRF-CEM) cancer cell lines. Curr. Microbiol. 75, 1090–1098. doi: 10.1007/s00284-018-1479-z
Moeller, R., Vlašić, I., Reitz, G., and Nicholson, W. L. (2012). Role of altered rpoB alleles in Bacillus subtilis sporulation and spore resistance to heat, hydrogen peroxide, formaldehyde, and glutaraldehyde. Arch. Microbiol. 194, 759–767. doi: 10.1007/s00203-012-0811-4
Molière, N., and Turgay, K. (2009). Chaperone-protease systems in regulation and protein quality control in Bacillus subtilis. Res. Microbiol. 160, 637–644. doi: 10.1016/j.resmic.2009.08.020
Monro, R. E. (1961). Protein turnover and the formation of protein inclusions during sporulation of Bacillus thuringiensis. Biochem. J. 81, 225–232. doi: 10.1042/bj0810225
Nissen, P., Hansen, J., Ban, N., Moore, P. B., and Steitz, T. A. (2000). The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930. doi: 10.1126/science.289.5481.920
Palma, L., Muñoz, D., Berry, C., Murillo, J., and Caballero, P. (2014). Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins 6, 3296–3325. doi: 10.3390/toxins6123296
Park, H. W., Ge, B., Bauer, L. S., and Federici, B. A. (1998). Optimization of Cry3A yields in Bacillus thuringiensis by use of sporulation-dependent promoters in combination with the STAB-SD mRNA sequence. Appl. Environ. Microbiol. 64:3932.
Roh, J. Y., Choi, J. Y., Li, M. S., Jin, B. R., and Je, Y. H. (2007). Bacillus thuringiensis as a specific, safe, and effective tool for insect pest control. J. Microbiol. Biotechnol. 17:547.
Rosenbluh, A., Banner, C. D., Losick, R., and Fitzjames, P. C. (1981). Identification of a new developmental locus in Bacillus subtilis by construction of a deletion mutation in a cloned gene under sporulation control. J. Bacteriol. 148, 341–351.
Saxild, H. H., and Nygaard, P. (1988). Gene-enzyme relationships of the purine biosynthetic pathway in Bacillus subtilis. Mole. Gen. Genet. 211, 160–167. doi: 10.1007/bf00338408
Serio, A. W., Pechter, K. B., and Sonenshein, A. L. (2006). Bacillus subtilis aconitase is required for efficient late-sporulation gene expression. J. Bacteriol. 188, 6396–6405. doi: 10.1128/JB.00249-06
Serrano, M., Neves, A., Soares, C. M., Moran, J. C., and Henriques, A. O. (2004). Role of the anti-sigma factor SpoIIAB in regulation of sigmaG during Bacillus subtilis sporulation. J. Bacteriol. 186, 4000–4013. doi: 10.1128/JB.186.12.4000-4013.2004
Singh, H., Appukuttan, D., and Lim, S. (2014). Hsp20, a small heat shock protein of Deinococcus radiodurans, confers tolerance to hydrogen peroxide in Escherichia coli. J. Microbiol. Biotechnol. 24, 1118–1122. doi: 10.4014/jmb.1403.03006
Singh, S., Kumar, N., Maniraj, R., Lakshmikanth, R., Rao, K., Muralimohan, N., et al. (2018). Expression of Cry2Aa, a Bacillus thuringiensis insecticidal protein in transgenic pigeon pea confers resistance to gram pod borer. Helicoverpa armigera. Sci. Rep. 8:8820. doi: 10.1038/s41598-018-26358-9
Van, K. F. (2009). Insecticidal activity of Bacillus thuringiensis crystal proteins. J. Invertebr. Pathol. 101, 1–16. doi: 10.1016/j.jip.2009.02.009
Ventura, M., Canchaya, C., Zhang, Z., Fitzgerald, G. F., and Sinderen, D. V. (2007). Molecular Characterization of hsp20. Encoding a Small Heat Shock Protein of Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73, 4695–4703. doi: 10.1128/AEM.02496-06
Wanapaisan, P., Chumsakul, O., and Panbangred, W. (2013). Enhanced Cry1Da production in Bacillus thuringiensis by driving expression from the sigma(E) -dependent BtI promoter. J. Appl. Microbiol. 115, 859–871. doi: 10.1111/jam.12276
Wang, J., Mei, H., Qian, H., Tang, Q., Liu, X., Yu, Z., et al. (2013a). Expression profile and regulation of spore and parasporal crystal formation-associated genes in Bacillus thuringiensis. J. Proteome Res. 12, 5487–5501. doi: 10.1021/pr4003728
Wang, J., Mei, H., Zheng, C., Qian, H., Cui, C., Fu, Y., et al. (2013b). The metabolic regulation of sporulation and parasporal crystal formation in Bacillus thuringiensis revealed by transcriptomics and proteomics. Mole. Cell. Proteomics 12:1363. doi: 10.1074/mcp.M112.023986
Wang, S., Yang, Y., Zhao, Y., Zhao, H., Bai, J., Chen, J., et al. (2016). Sub-MIC tylosin inhibits streptococcus suis biofilm formation and results in differential protein expression. Front. Microbiol. 7:384. doi: 10.3389/fmicb.2016.00384
Wang, X., Li, Z., Li, X., Qian, H., Cai, X., Li, X., et al. (2016). Poly-β-hydroxybutyrate metabolism is unrelated to the sporulation and parasporal crystal protein formation in Bacillus thuringiensis. Front. Microbiol. 7:836. doi: 10.3389/fmicb.2016.00836
Wehrl, W., Niederweis, M., and Schumann, W. (2000). The FtsH protein accumulates at the septum of Bacillus subtilis during cell division and sporulation. J. Bacteriol. 182, 3870–3873. doi: 10.1128/jb.182.13.3870-3873.2000
Wu, R., Wang, W., Yu, D., Zhang, W., Li, Y., Sun, Z., et al. (2009). Proteomics analysis of lactobacillus casei zhang, a new probiotic bacterium isolated from traditional home-made koumiss in inner mongolia of china. Mole. Cell. Proteomics 8:2321. doi: 10.1074/mcp.M800483-MCP200
Yang, Q., Li, Y., Yang, H., Jie, R., Tang, S., He, L., et al. (2015). Proteomic insights into metabolic adaptation to deletion of metE in Saccharopolyspora spinosa. Appl. Microbiol. Biotechnol. 99, 8629–8641. doi: 10.1007/s00253-015-6883-8
Yousten, A. A., and Rogoff, M. H. (1969). Metabolism of Bacillus thuringiensis in relation to spore and crystal formation. J. Bacteriol. 100, 1229–1236.
Zhang, B., Struffi, P., and Kroos, L. (1999). sigmaK can negatively regulate sigE expression by two different mechanisms during sporulation of Bacillus subtilis. J. Bacteriol. 181:4081.
Zhang, J., Schairer, H. U., Schnetter, W., Lereclus, D., and Agaisse, H. (1998). Bacillus popilliae cry18Aa operon is transcribed by σE and σK forms of RNA polymerase from a single initiation site. Nucleic Acids Res. 26, 1288–1293. doi: 10.1093/nar/26.5.1288
Zhang, S. F., Zhang, Y., Lin, L., and Wang, D. (2018). iTRAQ-based quantitative proteomic analysis of a toxigenic dinoflagellate alexandrium catenellaand its non-toxigenic mutant exposed to a cell cycle inhibitor colchicine. Front. Microbiol. 9:650. doi: 10.3389/fmicb.2018.00650
Keywords: hsp20, Bacillus thuringiensis, sporulation, insecticidal crystal proteins, iTRAQ
Citation: Xie J, Peng J, Yi Z, Zhao X, Li S, Zhang T, Quan M, Yang S, Lu J, Zhou P, Xia L and Ding X (2019) Role of hsp20 in the Production of Spores and Insecticidal Crystal Proteins in Bacillus thuringiensis. Front. Microbiol. 10:2059. doi: 10.3389/fmicb.2019.02059
Received: 30 November 2018; Accepted: 20 August 2019;
Published: 04 September 2019.
Edited by:
Frank Schreiber, Federal Institute for Materials Research and Testing (BAM), GermanyReviewed by:
Gabriela Olmedo-Alvarez, Unidad Irapuato (CINVESTAV), MexicoJin He, Huazhong Agricultural University, China
Copyright © 2019 Xie, Peng, Yi, Zhao, Li, Zhang, Quan, Yang, Lu, Zhou, Xia and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuezhi Ding, ZGluZ3h1ZXpoaUBodW5udS5lZHUuY24=
†These authors have contributed equally to this work
 Junyan Xie
Junyan Xie Jinli Peng1†
Jinli Peng1† Meifang Quan
Meifang Quan Liqiu Xia
Liqiu Xia Xuezhi Ding
Xuezhi Ding