- MOE Joint International Research Laboratory of Animal Health and Food Safety, College of Veterinary Medicine, Nanjing Agricultural University, Nanjing, China
Linezolid and vancomycin are among the last-resort antimicrobial agents in the treatment of multidrug-resistant Gram-positive bacterial infections. Linezolid- and vancomycin-resistant (LVR) Gram-positive bacteria may pose severe threats to public health. In this study, three optrA- and vanG-positive Streptococcus suis strains were isolated from two farms of different cities. There were only 1 and 343 single-nucleotide polymorphisms in coding region (cSNPs) of HCB4 and YSJ7 to YSJ17, respectively. Mobilome analysis revealed the presence of vanG, erm(B), tet(O/W/32/O), and aadE-apt-sat4-aphA3 cluster on an integrative and conjugative element, ICESsuYSJ17, and erm(B), aphA3, aac(6′)-aph(2″), catpC194, and optrA on a prophage, ΦSsuYSJ17-3. ICESsuYSJ17 exhibited a mosaic structure and belongs to a highly prevalent and transferable ICESa2603 family of Streptococcus species. ΦSsuYSJ17-3 shared conserved backbone to a transferable prophage Φm46.1. A novel composite transposon, IS1216E-araC-optrA-hp-catpC194-IS1216E, which can be circulated as translocatable unit (TU) by IS1216E, was integrated on ΦSsuYSJ17-3. Vancomycin resistance phenotype and vanG transcription assays revealed that the vanG operon was inducible. The LVR strain YSJ17 exhibited moderate virulence in a zebrafish infection model. To our knowledge, this is the first report of LVR isolate, which is mediated by acquired resistance genes optrA and vanG operons in Gram-positive bacteria. Since S. suis has been recognized as an antimicrobial resistance reservoir in the spread of resistance genes to major streptococcal pathogens, the potential risks of disseminating of optrA and vanG from S. suis to other Streptococcus spp. are worrisome and routine surveillance should be strengthened.
Introduction
Streptococcus suis is one of the most important bacterial causes of meningitis and contributes substantially to antimicrobial use in swine industry worldwide (Goyette-Desjardins et al., 2014). As a normal colonizer of the respiratory tract in pigs, S. suis can cause serious invasive infections in both pigs and humans, which posed a major public health challenge in Southeast Asia, including China (Tang et al., 2006; Yu et al., 2006). S. suis is thought to be an important antimicrobial resistance (AMR) reservoir contributing to the spread of AMR genes to major streptococcal pathogens (Palmieri et al., 2011; Huang et al., 2016b).
Avoparcin, a vancomycin analog, was widely used in food animals as a feed additive during 1940–1990s. The misuse of avoparcin had been reported to be associated with the occurrence of vancomycin-resistant enterococcus (VRE) in most parts of Europe (Bager et al., 1997; Hao et al., 2016). For this reason, avoparcin has been banned in food-producing animals in Denmark, 1995, and subsequently in China, 2002. However, vancomycin resistance still persists in pig farms even 20 years after the ban of avoparcin (Bortolaia et al., 2015; Birkegard et al., 2019). Two groups of van resistance operons have been identified according to the key ligase genes that encode either D-Ala-D-Lac ligase (vanA, vanB, vanD, and vanM) or D-Ala-D-Ser ligase (vanC, vanE, vanG, vanL, and vanN) (Lebreton et al., 2011; Binda et al., 2014). The vanG operon confers low-level vancomycin resistance and has been detected in many species, including Enterococcus faecalis, Enterococcus faecium, Clostridium difficile, Clostridium argentinense, Ruminococcus, and Streptococcus agalactiae (Depardieu et al., 2003; Domingo et al., 2007; Ammam et al., 2012; Srinivasan et al., 2014; Berthet et al., 2015; Sassi et al., 2018). Recently, we reported the vanG-type vancomycin resistance in zoonotic pathogen S. suis (Huang et al., 2018).
Oxazolidinones, including linezolid and tedizolid, are recognized as last-resort antimicrobial agents for the control of clinical infections caused by multidrug-resistant Gram-positive pathogens. Linezolid resistance in these bacteria is traditionally associated with mutations in domain V of 23S rRNA gene and ribosomal proteins L3 and L4 (Mendes et al., 2014). However, transferable resistance genes, cfr, cfr(B), and optrA, have been identified in enterococci worldwide, including Jiangsu, China (Long et al., 2006; Deshpande et al., 2015; Wang et al., 2015; Antonelli et al., 2018; Bender et al., 2018; Zhou et al., 2019), which also confer resistance to phenicols and other ribosomal-targeted antibiotics. More recently, a third oxazolidinone resistance gene, poxtA, was identified from MRSA and enterococci (Antonelli et al., 2018; Brenciani et al., 2019; Huang et al., 2019b; Lei et al., 2019). Currently, only cfr and optrA have been reported in streptococci, only in S. suis of animal origin (Wang et al., 2013; Huang et al., 2017), thus suggesting that cfr and optrA have occurred in animal setting under selection, as phenicols and other ribosomal-targeted antibiotics are broadly used in veterinary medicine (Shen et al., 2013; Hao et al., 2016).
Linezolid- and vancomycin-resistant (LVR) enterococci in hospitals have been recently described (O’driscoll et al., 2015; Krull et al., 2016; Bender et al., 2018). However, researches on other Gram-positive bacteria and in other settings have not been documented. In this study, to the best of our knowledge, we present the first LVR S. suis isolate of pig origin, which was mediated by optrA and vanG operons. The genetic basis of optrA and vanG operons was characterized by whole-genome sequencing (WGS) and the virulence was evaluated using a zebrafish infection model.
Materials and Methods
Sample Processing and Bacteria Identification
One hundred and eighty-nine S. suis clinical isolates used in this study were collected from a total of 658 pig samples, including pharyngeal swabs from 600 asymptomatic pigs and tissues (heart, liver, spleen, lung, tonsil, and joint fluid each pig) from 58 diseased pigs, Jiangsu Province, China, 2016–2017. This study aims to survey the current status of oxazolidinone resistance in S. suis from pigs. The swabs and tissues were collected by farm veterinary and delivered to the laboratory within 24 h. The samples were plated on the Todd–Hewitt agar supplemented with 5% fetal calf serum in the presence of nalidixic acid (15 mg/L) and Polymyxin B (10 mg/L). Five colonies each sample were selected and cultured in Todd–Hewitt broth with 5% fetal calf serum followed by PCR identification by using S. suis-specific primers targeting gdh and recN genes (Ishida et al., 2014). Duplicate isolates from samples of the same pig were excluded.
Antimicrobial Susceptibility Assays and LVR Mechanisms
The MICs of vancomycin (VAN), linezolid (LZD), and florfenicol (FFC) as well as penicillin (PEN), enrofloxacin (ENR), gentamicin (GEN), streptomycin (SPT), kanamycin (KAN), tetracycline (TET), erythromycin (ERY), tilmicosin (TIL), and lincomycin (LIN) to S. suis isolates were tested and evaluated by the broth microdilution method according to the Clinical and Laboratory Standards Institute guidelines (VET08-ED4 and M100-ED28). The presence of LVR resistance genes vanG, cfr, cfr(B), cfr(C), optrA, and poxtA was detected by PCR using primers previously described (Supplementary Table S1).
Genome Sequencing and Analysis
Genomic DNA was prepared from overnight cultures of the LVR isolates using the E.Z.N.A.®, Bacteria DNA kit (Omega Bio-Tek, Nanjing, China). Purified genomic DNA was subjected for WGS on the Illumina Hiseq2500 platform (Novogene, Beijing, China). Draft genome was assembled with SOAP de novo version 2.04 by default parameters (Li et al., 2008). For complete genome sequencing, the genomic DNA was further sequenced using the PacBio RSII System (Biozeron, Shanghai, China). The sequences were annotated using the NCBI Prokaryotic Genome Annotation Pipeline.
Average nucleotide identity (ANI) analysis was performed using the pyani software1 using whole genome sequences. The single-nucleotide polymorphisms in coding region (cSNPs) were determined by global alignment and local alignment between sample sequence and the reference genome. The maximum-likelihood (ML) methods were performed for the genome-wide phylogenetic analysis using PhyML 3.0 (Guindon et al., 2010). Nucleotide substitution model selection was estimated with jModelTest 2.1.10 (Darriba et al., 2012) and Smart Model Selection in PhyML 3.0. The model GTR + G was selected for ML analyses with 1,000 bootstrap replicates to calculate the bootstrap values of the topology.
Additional acquired resistance genes were identified in the genomes using ResFinder 3.1 (Zankari et al., 2012). Integrative and conjugative elements (ICEs) and prophages in NCL1 strain YSJ17 were identified by comparison with other NCL1 strains from this study and GenBank, and representative genomes of S. suis serotype 2, 9, and 24. Chromosomal mutations involved in β-lactam resistance and substitutions responsible for fluoroquinolone resistance were identified using BLASTn analysis.
Transferability Assays
Conjugative transfer assays were examined by filter mating experiment as described previously (Huang et al., 2016a). In mating experiments, donor and recipient strains were mixed at a ratio of 1:10 on a nitrocellulose membrane. Selection of transconjugants was performed on Todd–Hewitt agar containing rifampin (25 mg/L), fusidic acid (50 mg/L), and florfenicol (10 mg/L) or vancomycin (1 mg/L). Strain S. suis YSJ17 served as donor and S. suis P1/7RF (also known as BAA-853RF) was used as recipient (Huang et al., 2016b). Control, donor, and recipient strains were plated on selective medium independently. In addition, an overlap and inverse PCR method was introduced to detect the circular intermediate form of the novel optrA- and catpC194-carrying IS1216 composite transposons and the vanG-carrying ARGI2 using primer pairs (Supplementary Table S1).
Inducible Vancomycin Resistance Assays
Inducible vancomycin resistance phenotype assay was performed as previously described (Srinivasan et al., 2014; Huang et al., 2018). In brief, strains were pre-incubated with 1/10 × MIC vancomycin for 1 h in THB supplemented with 0.2% yeast extract (THY). Bacterial cultures were then diluted to 0.05 of OD600 in THY with 1/2 × MIC vancomycin. The growth curve (A600) was measured every hour over a 13-h period. Inducible transcription of the vanG gene was investigated using RT-PCR with RNA templates extracted from bacterial cultures in the absence or incubation with 1/10 × MIC or 1/2 × MIC of vancomycin. RNA isolation and PCR amplification were carried out as described previously (Huang et al., 2018).
Ethics Statement and Zebrafish Infection Model
The zebrafish infection experimental protocols were handled according to the guidelines of Experimental Animal Management Measures of Jiangsu Province and were approved by the Laboratory Animal Monitoring Committee of Jiangsu Province, China [Permit number: SYXK (Su) 2017-0007]. The zebrafish infection experiments were carried out as previously reported (Wu et al., 2010). Five groups of 15 zebrafish each were injected with 20 μL of PBS or bacterial suspensions containing a series of 10-fold serial dilutions (105–108 cfu), respectively, incubated in plastic containers for 72 h at 28°C, and the mortality was monitored from three parallel experiments. The LD50 values at 72 h were calculated by the Reed and Muench (1938) method.
Results
Isolation of vanG- and optrA-Positive S. suis
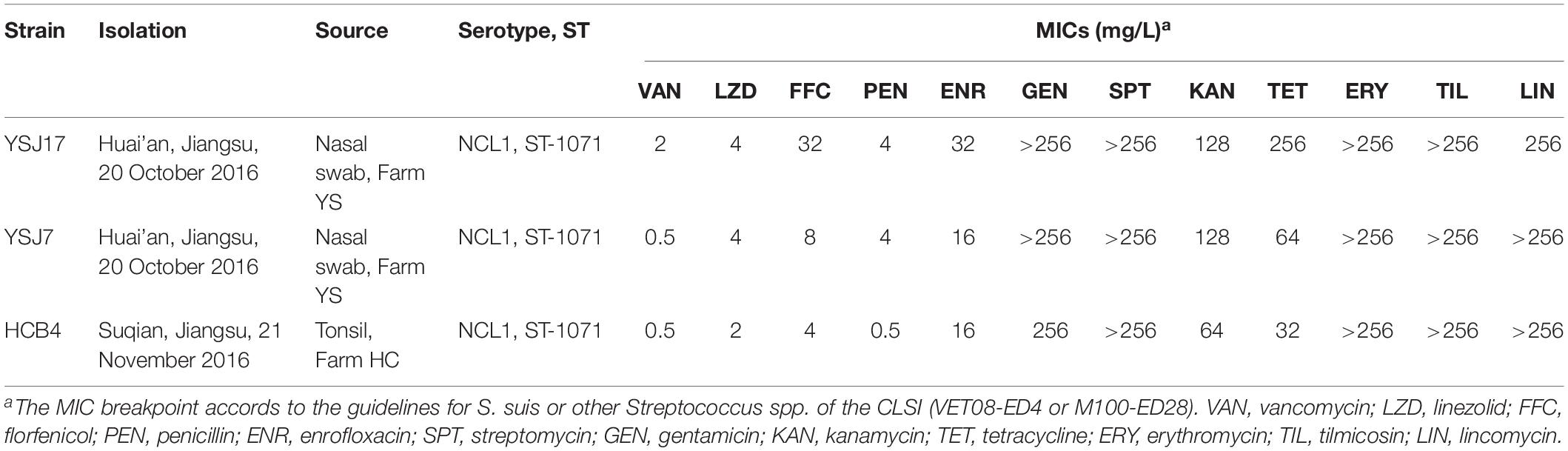
Since the first report of vanG operon in a S. suis serotype 24 isolate (Huang et al., 2018), we began the experiment by looking for the prevalence of vanG operon in S. suis using previously reported primers (Supplementary Table S1). Among 189 S. suis isolates collected from Jiangsu, China, 3 strains of different pig origin were positive for vanG, but exhibited variable phenotype to vancomycin. Strain YSJ17 (Farm YS) was vancomycin non-susceptible (MIC 2 mg/L), while YSJ7 (Farm YS) and HCB4 (Farm HC) were vancomycin susceptible (MIC 0.5 mg/L). On the other hand, 68 (35.98%) of the 189 isolates were optrA-positive (Table 1), with linezolid MIC values ranging from 0.25 to 16 mg/L. Among them, one isolate was also positive for cfr (Huang et al., 2019a) and none of the isolates was positive for cfr(B), cfr(C), or poxtA.
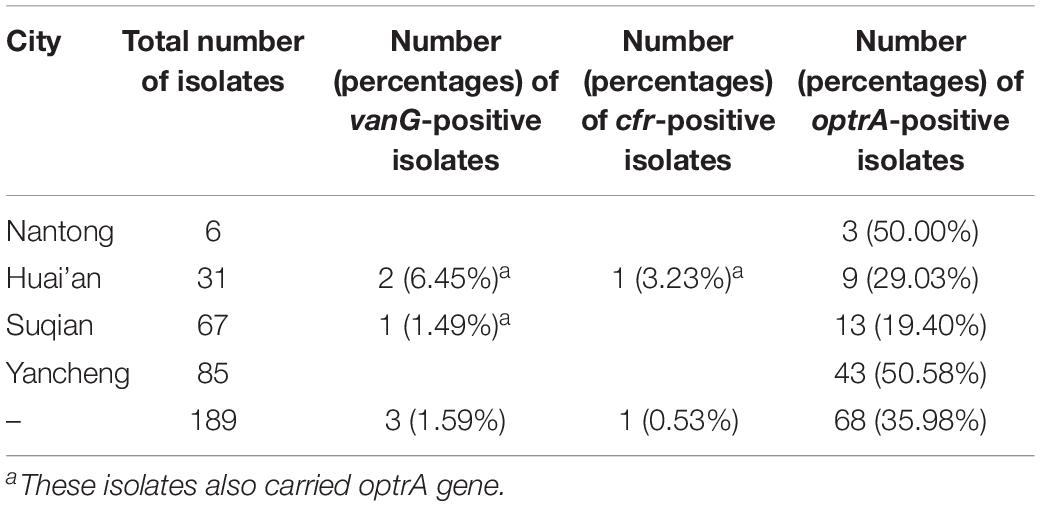
Table 1. Results of the screening of 189 Streptococcus suis of pig origin in Jiangsu, China, for the presence of the vanG, cfr, and optrA genes.
It is noteworthy that all three vanG-carrying isolates were linezolid non-susceptible and carried the optrA gene (Table 2). In addition, they exhibited resistance or elevated MIC values to penicillin, enrofloxacin, gentamicin, streptomycin, kanamycin, tetracycline, erythromycin, tilmicosin, lincomycin, and florfenicol (Table 2). Since, to our knowledge, no vanG- and optrA-positive bacteria have been identified so far, we further analyzed these S. suis strains by WGS.
Genomic and Phylogenetic Analyses
Whole-genome sequencing analysis of the three isolates showed that they were all assigned to multi-locus sequence type ST1071. ANI analysis showed that YSJ17 had 0.9998 and 0.9987 average identity to HCB4 and YSJ7, respectively, but less similar to serotype 24 strain BSB6 (0.9659 of ANI) (Figure 1). Further SNP analysis showed only 1 and 343 cSNPs of HCB4 and YSJ7 to YSJ17, respectively. According to the capsular polysaccharide synthesis locus, the isolates can be classified to a novel capsular polysaccharide loci (NCL) type NCL1 (Okura et al., 2014; Qiu et al., 2016). Remarkable, S. suis NCL1 strains have been frequently isolated from both diseased and healthy pigs (Qiu et al., 2016; Zheng et al., 2017). To analyze the evolution of the three vanG- and optrA-carrying NCL1 isolates, a cSNPs-based phylogenetic tree was generated by comparison with other NCL1 strains from GenBank and representative genomes of S. suis serotype 2 (P1/7 and 05ZYH33), serotype 9 (GZ0565), and serotype 24 (BSB6). The phylogenetic tree demonstrated that YSJ17, HCB4, and YSJ7 clustered together with serotype 24 strain BSB6 and more distinct to other NCL1 strains (Figure 2).
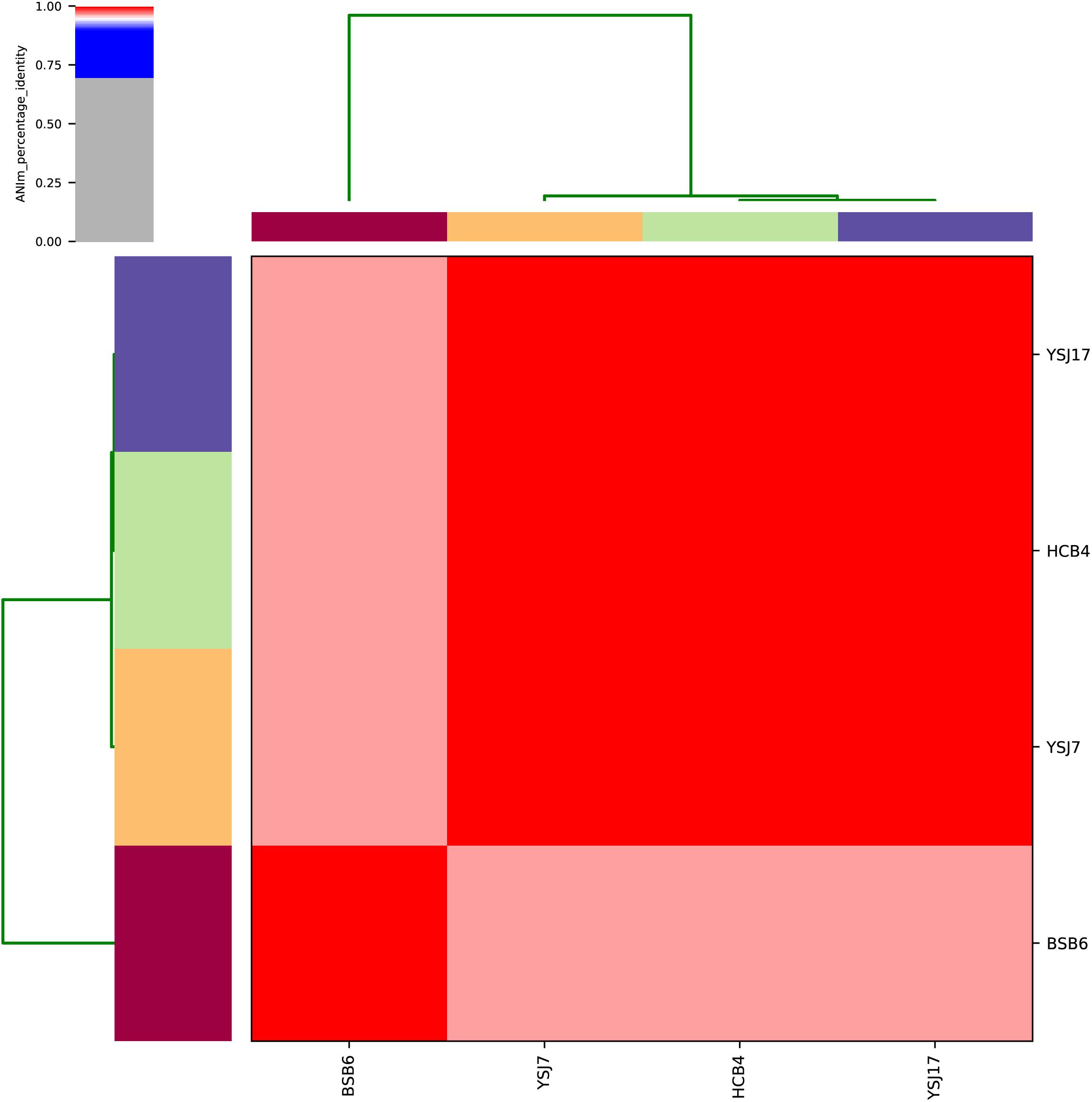
Figure 1. Average nucleotide identity (ANI) analysis of the four vanG-carrying S. suis isolates using whole-genome sequences.
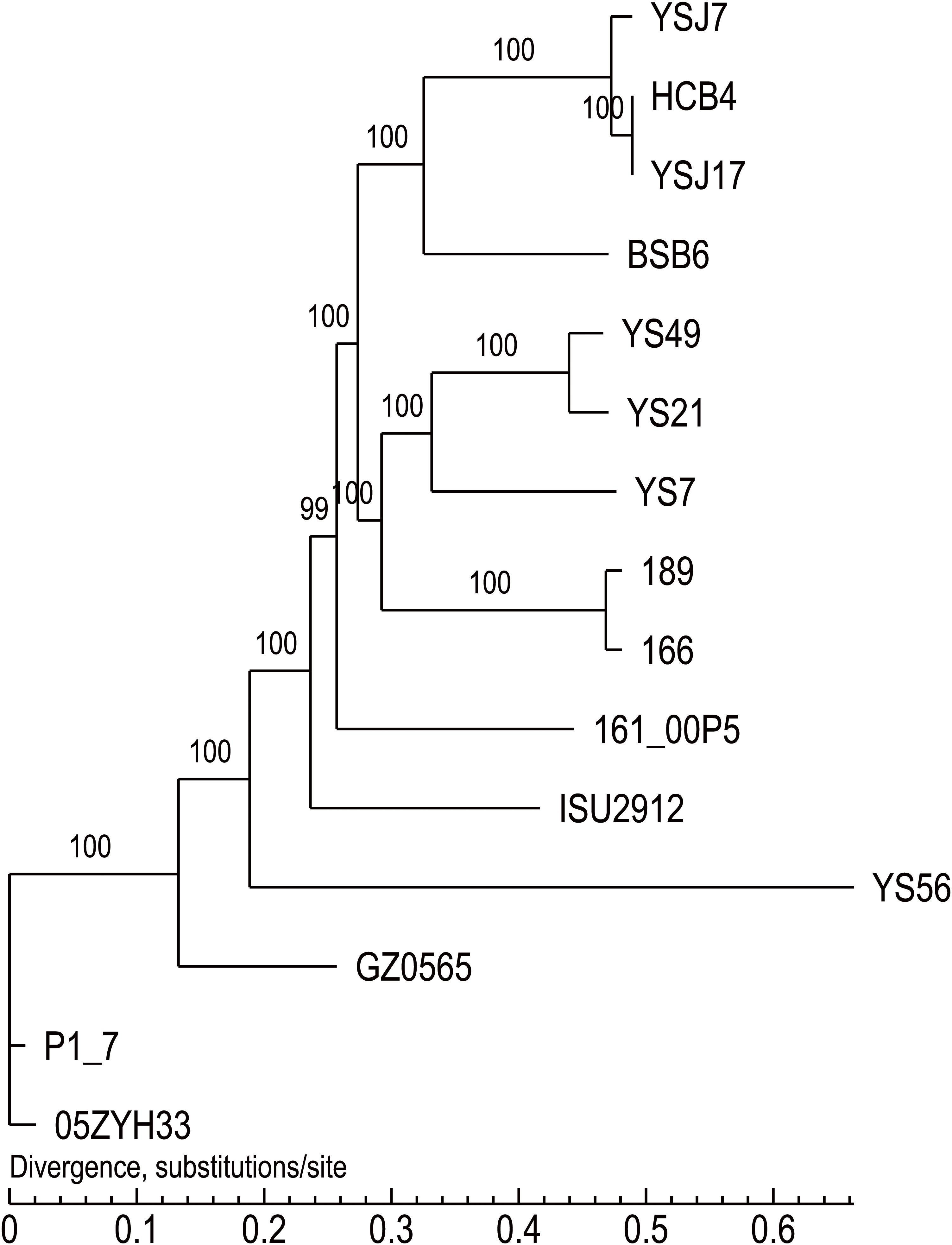
Figure 2. The cSNPs-based phylogenetic trees of the three vanG- and optrA-carrying S. suis isolates generated by comparison with other NCL1 strains from GenBank, and representative genomes of S. suis serotype 2 (P1/7 and 05ZYH33), serotype 9 (GZ0565), and serotype 24 (BSB6).
AMR Molecular Mechanisms and Mobilome Analysis
BLAST search for the acquired AMR genes revealed the presence of optrA, vanG, and also the aminoglycoside-modifying enzyme genes aadE, sat4, apt, aphA3, and aac(6′)–aph(2″); the tetracycline resistance gene tet(O/W/32/O); the macrolide–lincosamide–streptogramin B resistance gene erm(B); and the chloramphenicol resistance gene catpC194 (Supplementary Table S2). Mutations involved in penicillin resistance were found in pbp2x gene encoding penicillin-binding protein PBP2x (Supplementary Figure S1; Ge et al., 2012). Substitutions in GyrA (Ser81–Lys) and ParC (Ser79–Tyr) were observed, which are known to confer fluoroquinolone resistance (Escudero et al., 2007; Yao et al., 2014). No additional characterized genes or mutations for oxazolidinone resistance were observed. These data were in agreement with the AMR phenotype, with the exception of YSJ7 and HCB4, which were susceptible to vancomycin (Table 2).
To characterize further the mobile genetic elements (MGEs) containing these AMR genes, isolate YSJ17 was completely sequenced, with a small plasmid pYSJ17 of 4,065 bp. The YSJ17 chromosome had a size of 2,551,120 bp and encoded 2,489 putative coding sequences. We mapped the ICEs and prophages by comparing 11 publicly available NCL1 strains and serotype 2, 9, and 24 representative strains (Supplementary Figure S2). An ICE (ICESsuYSJ17) and four prophages (ΦSsuYSJ17-1 to -4) were identified. The size, insertion location, att site, and ARGs of the MGEs are summarized in Supplementary Table S2. ICESsuYSJ17 contained aadE, sat4, apt, aphA3, tet(O/W/32/O), erm(B), and vanG operon, while ΦSsuYSJ17-3 harbored erm(B), aphA3, aac(6′)–aph(2″), catpC194, and optrA.
To test the transferability of the vanG-carrying ICESsuYSJ17 and optrA-carrying ΦSsuYSJ17-3, mating experiments between S. suis YSJ17 and recipient strain S. suis P1/7RF were performed. However, we could not obtain transconjugant using florfenicol and vancomycin after more than three independent attempts, with donor and recipient at a ratio of approximately 108 and 109 cfu, respectively.
Genetic Characterization of the vanG-Carrying ICESsuYSJ17
The vanG-carrying ICESsuYSJ17 was 79,886 bp in length and encoded 89 predicted ORFs. Sequence analysis of ICESsuYSJ17 indicated that it is a mosaic ICE similar to ICESsuBSB6 and ICESsuHA681 and belonged to the ICESa2603 family (Figure 3A; Ambroset et al., 2015). The majority (right part, nt 21,302–79,886) of ICESsuYSJ17 was nearly identical to ICESsuBSB6 (Huang et al., 2018), with only the presence of two extra IS elements, IS1533 and IS1216, in the latter ICE (Figure 3A). This includes ARGI1 containing erm(B), tet(O/W/32/O), and aadE-apt-sat4-aphA3 cluster, and ARGI2 carrying vanG operon (Huang et al., 2018). The left part (nt 1–24,281) of ICESsuYSJ17 matched to ICESsuHA681, with a difference of only an inverted fragment (nt 17,784–21,301). This includes two variable regions (HS-1 and HS-2) and an insertion I-2 previously identified (Huang et al., 2018).
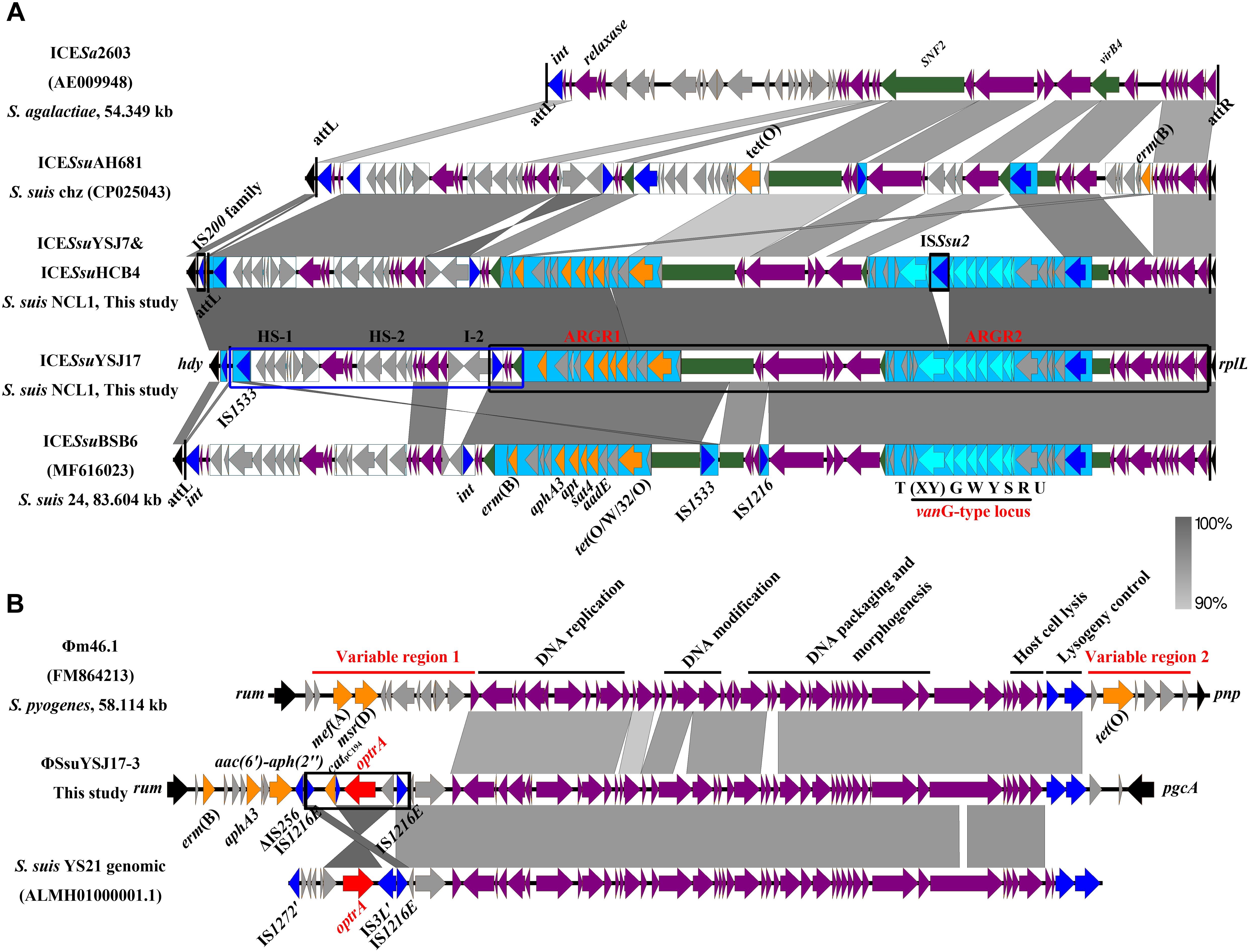
Figure 3. Genetic representation of the vanG-carrying ICESsuYSJ17 and the optrA-harboring ΦSsuYSJ17-3. Regions of >90% identity were marked by gray shading. The MGEs’ flanking chromosomal genes were shown in black arrows, integrases/recombinases/transposases were shown in dark blue, core structure genes were in purple arrows, and accessory genes were in light gray arrows. vanG operon genes, optrA, and other AMR genes were shown in light blue, red, and orange, respectively. (A) Comparison of the S. suis NCL1 ICESsuYSJ17 and ICESsuYSJ7/ICESsuHCB4 with that of S. agalactiae ICESa2603 (AE009948), S. suis Chz ICESsuAH681 (CP025043), and S. suis serotype 24 ICESsuBSB6 (MF616023). SNF2 and vriB4 genes that were inserted with ARGR1 and ARGR2 were highlighted in green. ARGR1, ARGR2, intergenic hotspots HS-1 and HS-2, and insertion site I-2 according to ICESsuBSB6 were indicated. The two-part segments best matched to ICESsuAH681 and ICESsuBSB6 were highlighted in the blue box and black box, respectively. Vertical lines indicate the att sequence sites. (B) Comparison of the S. suisΦSsuYSJ17-3 with S. pyogenes Φm46.1 (FM864213) and S. suis YS21 genomic sequence (ALMH01000001.1). Horizontal lines indicate the module structure genes and variable region genes. The composite transposon IS1216E-araC-optrA-hp-catpC194-IS1216E was highlighted in the black box.
Since S. suis YSJ17 and HCB4 were vancomycin susceptible, we tested if the vanG operon was intact to explore the genetic basis for the phenotype. As shown in Figure 3A, an IS element of 1,503 bp, which showed 97% identity to ISSsu2, was inserted at the base of the 365 base of vanXY gene in strains YSJ17 and HCB4. This, as a result, abolished the expression of vanYWG(XY)T resistance operon, although no effect of the vanURS regulatory operon was observed.
Genetic Characterization of the optrA-Carrying ΦSsuYSJ17-3 and Detection of the IS1216E-araC-optrA-hp-catpC194 Translocatable Unit
The 56,723-bp optrA-carrying ΦSsuYSJ17-3 was integrated at the 1328 base of rum gene, a well-conserved insertion hotspot for other MGEs (Srinivasan et al., 2014; Huang et al., 2016b). Genetic comparison showed that the ΦSsuYSJ17-3 shared conserved prophage backbone to the mef(A)- and tet(O)-carrying Φm46.1 (Brenciani et al., 2010), the cadA/C-tet(W) fragment of S. suis ΦSsUD.1 (Palmieri et al., 2010), and the genomic sequence of the optrA-carrying S. suis NCL1 strains YS21/YS49/YS50 (Figure 3B; Huang et al., 2017). Significantly, ΦSsuYSJ17-3 was present in 10 of 11 current available genomes of NCL1 strains, but absent in other S. suis serotype strains (Supplementary Figure S2).
Different from the genetic context of optrA in plasmids and chromosomes of E. faecalis, which usually occupy the optrA-erm(A)-like or fexA–optrA resistance cluster (He et al., 2016), and in S. suis YS21/YS49/YS50, which occurred in an 8.1-kb optrA and erm(A)-like containing element or a 7.4-kb optrA-carrying fragment (Huang et al., 2017), a 6,568-bp size composite transposon organized in IS1216E-araC-optrA-hp-catpC194-IS1216E structure was present on ΦSsuYSJ17-3. A TU verification PCR using optrA-F2/catpC194-R and optrA-R2/catpC194-F pairs amplified a product of 2,857 and 3,405 bp, respectively (Figure 4). This confirmed the formation of TU of IS1216E-araC-optrA-hp-catpC194 with a size of 5,759 bp in all three isolates.
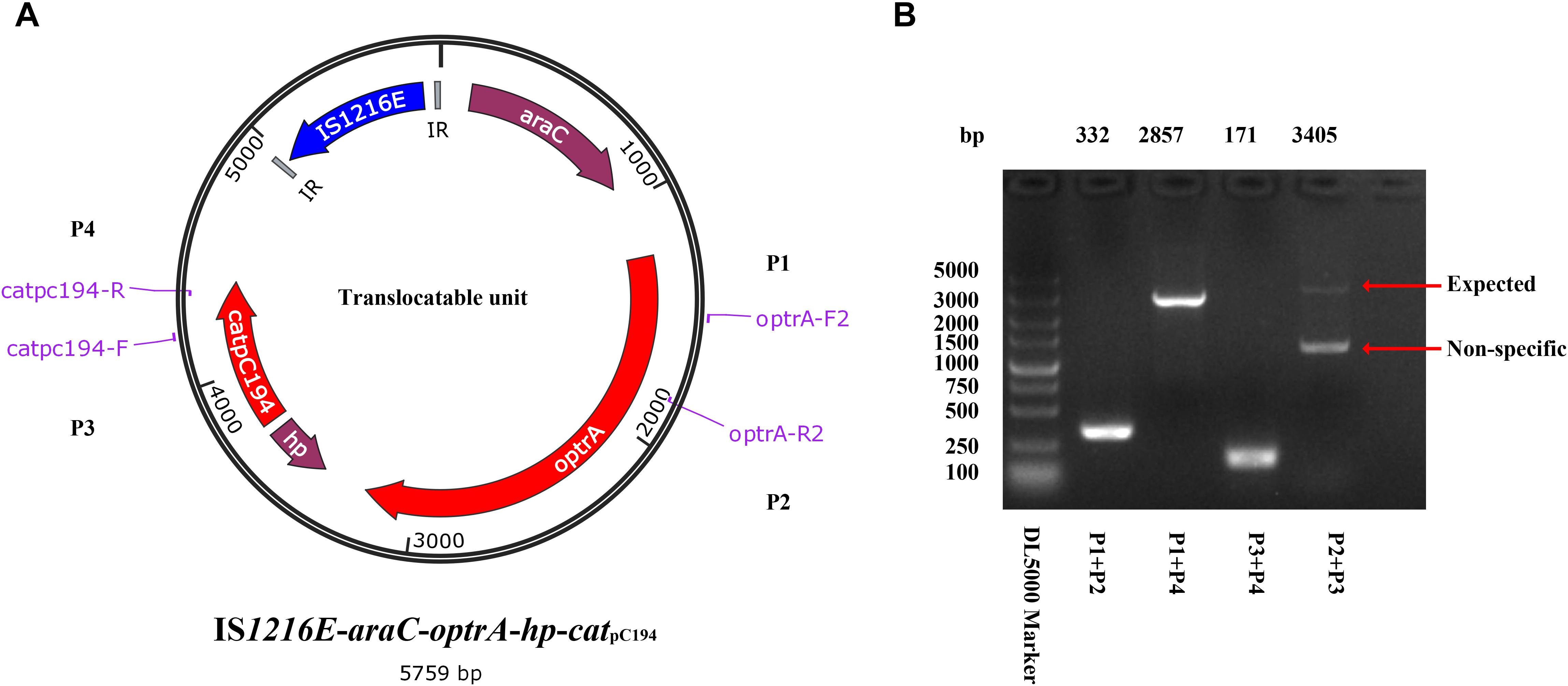
Figure 4. Identification of the translocatable unit of IS1216E-araC-optrA-hp-catpC194. (A) Genetic presentation of the minicircle TU and primers designed. The genes optrA and catpC194 were shown in red arrows, genes araC and hypothetical protein (hp) were shown in purple, and tnpIS1216E was shown in blue. Two IR sequences flanking IS1216E were shown in gray. (B) PCR products using P1–P4 primer pairs by a TU verification PCR method. The products were further sequenced and confirmed the circular form presented in (A).
Inducible Vancomycin Resistance Assays
In order to test if vanG-type resistance to vancomycin was inducible in YSJ7, we performed vancomycin resistance phenotype and vanG transcription assays. As expected, preincubation with 1/10 × MIC vancomycin shortened the growth lag (Figure 5), and the vanG transcription was increased 5.6- and 4.3-fold in the presence of 1/10× or 1/2 × MIC vancomycin, respectively. These suggest that the S. suis YSJ17 vanG-type resistance to vancomycin was inducible and the vanG transcription was similar to that of enterococci (Depardieu et al., 2015; Sassi et al., 2018).
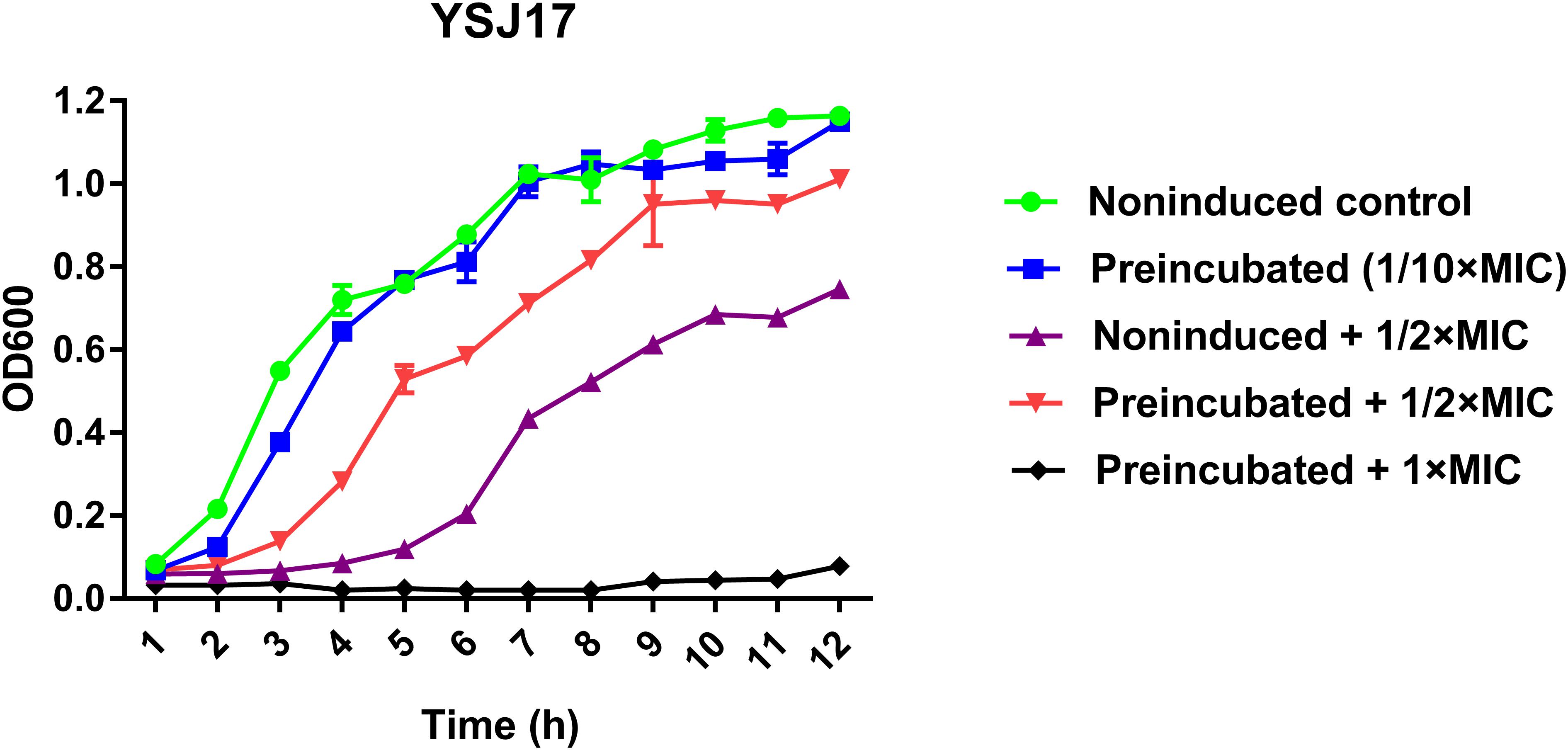
Figure 5. Inducible vancomycin resistance phenotype assay. Strain YSJ17 was tested for inducible vancomycin resistance by preincubation of 1/10 × MIC (0.2 mg/L) vancomycin for 1 h in Todd–Hewitt broth plus 0.2% yeast extract (THY) prior to dilution back to OD600 = 0.05 in the same medium containing 1/2 × MIC (1 mg/L) vancomycin. Growth lag was observed by the non-induced + 1/2 × MIC treatment and partial restoration was observed by the preincubated + 1/2 × MIC treatment.
Virulence of the LVR Strain YSJ17
To evaluate the virulence of the LVR strains, we firstly determined in the genomes for the distribution of 24 virulence-related genes responsible for the virulence of S. suis serotype 2 (Fittipaldi et al., 2012; Dong et al., 2015, 2017). As shown in Supplementary Table S3, 16 of 24 virulence-related genes were detected in all three isolates. A previous study suggested that S. suis serotype 2 strains carrying six genes (epf, sly, rgg, endoD, comR, and scnF) can be predicted as virulent (Dong et al., 2015). In this study, only rgg and endoD were detected in all three NCL1 isolates (Supplementary Table S3).
Although having been frequently isolated from both healthy pigs and lung from diseased pigs, the virulence of NCL1 strains has not been assessed accurately by an animal model. To access this, we measured the virulence of YSJ17 by a zebrafish infection model (Wu et al., 2014). The mortality was 0 and 86.67% after 72-h injection of a dose of 106 and 107 cfu, respectively (Figure 6). While for S. suis serotype 2 virulent strain SC070731, the mortality was 40 and 100% with a dose of 106 and 107 cfu, respectively. The LD50 was 0.9 × 107 cfu/fish for YSJ17 and 1.2 × 106 cfu/fish for SC070731. Zebrafish infection with the avirulent strain SH040917 showed no mortality. These results suggest that the NCL1 strain YSJ17 was less virulent than S. suis serotype 2 virulent strain SC070731. However, the pathogenic mechanism remains to be further explored.
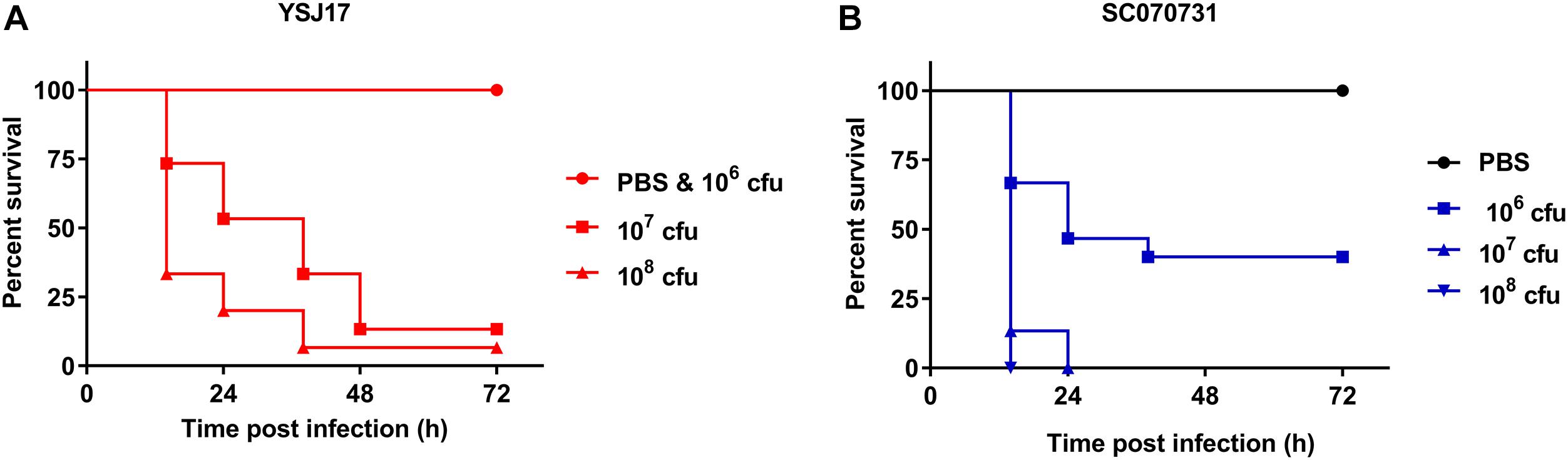
Figure 6. Dose-dependent lethality of zebrafish model infected with (A) LVR S. suis NCL1 strain YSJ17 and (B) the serotype 2 virulent reference strain SC070731. Zebrafish were injected with serial dilutions of 105–109 cfu of S. suis. Survival rate was recorded during a 72 h period after infection. Mortality of YSJ17 and SC070731 with a dose of 106–108 cfu was presented.
Discussion
Antimicrobials have been widely used in animals for prevention, treatment, and also as growth promoters. The indiscriminate use of antimicrobials contributes to the emergence of AMR in commensal bacteria, animal pathogens, and also zoonotic pathogens (Mcewen and Fedorka-Cray, 2002). It is of special concern to animal and human health, as resistant bacteria are likely to be transmitted within farm animals and from farm animals to humans through close contact or food chain, as well as AMR genes may be transferred from commensal bacteria to zoonotic pathogens (Mcewen and Fedorka-Cray, 2002; Marshall and Levy, 2011; Thanner et al., 2016). The extensive use of avoparcin in veterinary medicine has led to an increasing incidence of VRE in animals and healthy people during the 1990s (Bager et al., 1997; Klare et al., 1999; Marshall and Levy, 2011). In addition, exclusive use of florfenicol in veterinary medicine could have co-selected the presence of phenicols-oxazolidinones (PhO)-resistant strains (Long et al., 2006; Wang et al., 2015). However, the knowledge addressing the transfer of linezolid- and/or vancomycin-resistant genes from commensal enterococci to zoonotic pathogens is rare. Recently, we characterized the optrA-mediated linezolid resistance and vanG-type vancomycin resistance in zoonotic pathogen S. suis separately (Huang et al., 2017, 2018). The genetic background of optrA and vanG was highly similar to commensal bacteria enterococci, highlighting the possible transmission from enterococci to S. suis. But the prevalence of these genes in S. suis has not been investigated.
In the present study, we detected the prevalence of the vanG and optrA genes in S. suis during 2016–2017. The vanG gene was detected in S. suis NCL1 strains from different cities aside from serotype 24 isolate (Huang et al., 2018), which suggests that S. suis may be considered as a possible reservoir for vanG, even >20 years after the avoparcin ban in food-producing animals (Bager et al., 1997). In addition, high prevalence of optrA was observed in S. suis, which may have occurred under florfenicol selection in animal settings (Hao et al., 2016). More alarmingly, to our knowledge, we reported the first LVR isolate carrying the transferrable resistance genes optrA and vanG operons, which might initially be acquired from enterococci and may contribute to their transfer from S. suis to other Gram-positive bacteria (Palmieri et al., 2011; Huang et al., 2016b).
The acquisition and dissemination of AMR genes in streptococci is strongly associated with MGEs, mainly the ICEs and prophages (Palmieri et al., 2011; Huang et al., 2016b). The present study showed the co-location of vanG operon with erm(B), tet(O/W/32/O), and aadE-apt-sat4-aphA3 cluster on an ICE (ICESsuYSJ17) of S. suis NCL1 strains of different origin (Figure 3A), which is similar to serotype 24 strain of ICESsuBSB6 (Huang et al., 2018). Genetic characterization showed that ICESsuYSJ17 was a mosaic ICE of the ICESa2603 family. This family of ICEs are highly prevalent and constitute a diverse group of ICEs associated with AMR in major Streptococcus species (Ambroset et al., 2015; Huang et al., 2016b). A variety of AMR determinants for tetracyclines [tet(M), tet(L), tet(O), tet(40), and tet(O/W/32/O)], macrolides [erm(B)], aminoglycosides (aphA3, sat, ant6, and aadE), and phenicols (cat) have been shown on ICEs of the ICESa2603 family (Chen et al., 2007; Palmieri et al., 2012; Huang et al., 2016b). Recently, acquisition of the phenicol–oxazolidinone resistance gene optrA and the vancomycin resistance gene operon vanG within ICESa2603 family ICE has been observed (Huang et al., 2017, 2018). Particularly, horizontal transfer of ICESa2603 family ICEs between Streptococcus species has been documented (Davies et al., 2009; Haenni et al., 2010; Li et al., 2011; Palmieri et al., 2012). Moreover, tandem recombination of ICESsu32457 and S. agalactiae ICESa2603 has been reported (Marini et al., 2015). In this study, the transfer of ICESsuYSJ17 failed. It may be partially due to the inactivation of an essential conjugation protein VirB4 in ICESsuYSJ17 as in S. suis BSB6 (Huang et al., 2018). However, it may also be possible that the experimental conditions were not adapted, with a transfer frequency of <10–9 per recipient, as the potential transfer of these genes with the help of other MGEs cannot be ruled out. Thus, the acquisition of vanG and other co-located AMR genes within the highly transferable ICESa2603 family of ICEs may promote the potential transfer of vanG operon among Gram-positive cocci.
The present study also showed the co-existence of optrA and catpC194 as well as erm(B), aphA3, and aac(6′)–aph(2″) on ΦSsuYSJ17-3. Genetic characterization showed that ΦSsuYSJ17-3 was a Φm46.1-like prophage (Brenciani et al., 2010). Φm46.1-like prophage, which was originally reported in S. pyogenes, has now been reported in and is transferable to other streptococci (Brenciani et al., 2010; Di Luca et al., 2010; Giovanetti et al., 2014; Huang et al., 2016b). Further analysis showed that the acquisition of optrA-catpC194 was mediated by an IS1216 family composite transposon, IS1216E-araC-optrA-hp-catpC194-IS1216E (Figure 3B). IS1216E belongs to the IS6 family and has been increasingly associated with the spread of oxazolidinone resistance genes cfr, optrA, and poxtA; the vancomycin resistance genes vanA and vanM; the macrolide–lincosamide–streptogramin B resistance genes erm(B), erm(T), and lnu(B); the tetracycline resistance genes tet(M), tet(L), tet(S), and tet(S/M); and the aminoglycoside resistance genes spw and aadE in Gram-positive cocci (Xu et al., 2010; Barile et al., 2012; Liu et al., 2012; Ciric et al., 2014; He et al., 2016; Li et al., 2016; Di Sante et al., 2017). As composite transposons have a potential to excise and form as a TU, we developed a TU verification PCR and detected the mini-circle TU of IS1216E-araC-optrA-hp-catpC194 (Figure 4), probably via IS1216E-mediated recombination, highlighting the role of IS1216E in the spread of optrA-catpC194 genes.
Conclusion
In conclusion, to our knowledge, this is the first report of the co-occurrence of optrA and vanG operons in Gram-positive bacteria. The acquisition and persistence of optrA- and vanG-carrying S. suis in pigs may contribute to the potential transfer of these resistance genes to other Gram-positive bacteria.
Data Availability
The S. suis YSJ17 complete genome and its plasmid pYSJ17 have been deposited in GenBank (Accession Nos. CP032064 and CP032065), and the S. suis YSJ7 and HCB4 draft genomes have been deposited in GenBank (Accession Nos. QXEQ00000000 and QXEP00000000).
Author Contributions
JH and LW developed the concept and designed the experiments. JH, FD, XL, and DD performed the experiments and collected the data. JH and LW prepared the manuscript. All authors have contributed to, and seen and approved the manuscript.
Funding
This work was supported by the National College Students Innovation and Entrepreneurship Training Program (201710307054), the National Natural Science Foundation of China (31702292 and 31872517), the Natural Science Foundation of Jiangsu Province (BK20170710), the China Postdoctoral Science Foundation (2017M611841 and 2018T110515), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Zongfu Wu for technical assistance on zebrafish infection experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02026/full#supplementary-material
Footnotes
References
Ambroset, C., Coluzzi, C., Guedon, G., Devignes, M. D., Loux, V., Lacroix, T., et al. (2015). New insights into the classification and integration specificity of Streptococcus integrative conjugative elements through extensive genome exploration. Front. Microbiol. 6:1483. doi: 10.3389/fmicb.2015.01483
Ammam, F., Marvaud, J. C., and Lambert, T. (2012). Distribution of the vanG-like gene cluster in Clostridium difficile clinical isolates. Can. J. Microbiol. 58, 547–551. doi: 10.1139/w2012-002
Antonelli, A., D’andrea, M. M., Brenciani, A., Galeotti, C. L., Morroni, G., Pollini, S., et al. (2018). Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J. Antimicrob. Chemother. 73, 1763–1769. doi: 10.1093/jac/dky088
Bager, F., Madsen, M., Christensen, J., and Aarestrup, F. M. (1997). Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev. Vet. Med. 31, 95–112.
Barile, S., Devirgiliis, C., and Perozzi, G. (2012). Molecular characterization of a novel mosaic tet(S/M) gene encoding tetracycline resistance in foodborne strains of Streptococcus bovis. Microbiology 158, 2353–2362. doi: 10.1099/mic.0.058206-0
Bender, J. K., Cattoir, V., Hegstad, K., Sadowy, E., Coque, T. M., Westh, H., et al. (2018). Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: Towards a common nomenclature. Drug Resist. Update 40, 25–39. doi: 10.1016/j.drup.2018.10.002
Berthet, N., Perichon, B., Mazuet, C., Chapeton-Montes, D., Bouchier, C., Bouvet, P., et al. (2015). A vanG-type locus in Clostridium argentinense. J. Antimicrob. Chemother. 70, 1942–1945. doi: 10.1093/jac/dkv073
Binda, E., Marinelli, F., and Marcone, G. L. (2014). Old and new glycopeptide antibiotics: Action and resistance. Antibiotics 3, 572–594. doi: 10.3390/antibiotics3040572
Birkegard, A. C., Graesboll, K., Clasen, J., Halasa, T., Toft, N., and Folkesson, A. (2019). Continuing occurrence of vancomycin resistance determinants in Danish pig farms 20 years after removing exposure to avoparcin. Vet. Microbiol. 232, 84–88. doi: 10.1016/j.vetmic.2019.04.007
Bortolaia, V., Mander, M., Jensen, L. B., Olsen, J. E., and Guardabassi, L. (2015). Persistence of vancomycin resistance in multiple clones of Enterococcus faecium isolated from Danish broilers 15 years after the ban of avoparcin. Antimicrob. Agents Chemother. 59, 2926–2929. doi: 10.1128/AAC.05072-14
Brenciani, A., Bacciaglia, A., Vignaroli, C., Pugnaloni, A., Varaldo, P. E., and Giovanetti, E. (2010). Phim46.1, the main Streptococcus pyogenes element carrying mef(A) and tet(O) genes. Antimicrob. Agents Chemother. 54, 221–229. doi: 10.1128/AAC.00499-09
Brenciani, A., Fioriti, S., Morroni, G., Cucco, L., Morelli, A., Pezzotti, G., et al. (2019). Detection in Italy of a porcine Enterococcus faecium isolate carrying the novel phenicol-oxazolidinone-tetracycline resistance gene poxtA. J. Antimicrob. Chemother. 74, 817–818.
Chen, C., Tang, J. Q., Dong, W., Wang, C. J., Feng, Y. J., Wang, J., et al. (2007). A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS One 2:e315. doi: 10.1371/journal.pone.0000315
Ciric, L., Brouwer, M. S., Mullany, P., and Roberts, A. P. (2014). Minocycline resistance in an oral Streptococcus infantis isolate is encoded by tet(S) on a novel small, low copy number plasmid. FEMS Microbiol. Lett. 353, 106–115. doi: 10.1111/1574-6968.12410
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9, 772.
Davies, M. R., Shera, J., Van Domselaar, G. H., Sriprakash, K. S., and Mcmillan, D. J. (2009). A novel integrative conjugative element mediates genetic transfer from group G Streptococcus to other {beta}-hemolytic Streptococci. J. Bacteriol. 191, 2257–2265. doi: 10.1128/JB.01624-08
Depardieu, F., Bonora, M. G., Reynolds, P. E., and Courvalin, P. (2003). The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol. Microbiol. 50, 931–948.
Depardieu, F., Mejean, V., and Courvalin, P. (2015). Competition between VanU(G) repressor and VanR(G) activator leads to rheostatic control of vanG vancomycin resistance operon expression. PLoS Genet. 11:e1005170. doi: 10.1371/journal.pgen.1005170
Deshpande, L. M., Ashcraft, D. S., Kahn, H. P., Pankey, G., Jones, R. N., Farrell, D. J., et al. (2015). Detection of a new cfr-like gene, cfr(B), in Enterococcus faecium isolates Recovered from human specimens in the United States as part of the SENTRY antimicrobial surveillance program. Antimicrob. Agents Chemother. 59, 6256–6261. doi: 10.1128/AAC.01473-15
Di Luca, M. C., D’ercole, S., Petrelli, D., Prenna, M., Ripa, S., and Vitali, L. A. (2010). Lysogenic transfer of mef(A) and tet(O) genes carried by Phim46.1 among group A streptococci. Antimicrob. Agents Chemother. 54, 4464–4466. doi: 10.1128/AAC.01318-09
Di Sante, L., Morroni, G., Brenciani, A., Vignaroli, C., Antonelli, A., D’andrea, M. M., et al. (2017). pHTbeta-promoted mobilization of non-conjugative resistance plasmids from Enterococcus faecium to Enterococcus faecalis. J. Antimicrob. Chemother. 72, 2447–2453. doi: 10.1093/jac/dkx197
Domingo, M. C., Huletsky, A., Giroux, R., Picard, F. J., and Bergeron, M. G. (2007). vanD and vanG-like gene clusters in a Ruminococcus species isolated from human bowel flora. Antimicrob. Agents Chemother. 51, 4111–4117.
Dong, W., Ma, J., Zhu, Y., Zhu, J., Yuan, L., Wang, Y., et al. (2015). Virulence genotyping and population analysis of Streptococcus suis serotype 2 isolates from China. Infect. Genet. Evol. 36, 483–489. doi: 10.1016/j.meegid.2015.08.021
Dong, W., Zhu, Y., Ma, Y., Ma, J., Zhang, Y., Yuan, L., et al. (2017). Multilocus sequence typing and virulence genotyping of Streptococcus suis serotype 9 isolates revealed high genetic and virulence diversity. FEMS Microbiol. Lett. 364, fnx192. doi: 10.1093/femsle/fnx1192
Escudero, J. A., San Millan, A., Catalan, A., De La Campa, A. G., Rivero, E., Lopez, G., et al. (2007). First characterization of fluoroquinolone resistance in Streptococcus suis. Antimicrob. Agents Chemother. 51, 777–782.
Fittipaldi, N., Segura, M., Grenier, D., and Gottschalk, M. (2012). Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 7, 259–279. doi: 10.2217/fmb.11.149
Ge, Y., Wu, J., Xia, Y., Yang, M., Xiao, J., and Yu, J. (2012). Molecular dynamics simulation of the complex PBP-2x with drug cefuroxime to explore the drug resistance mechanism of Streptococcus suis R61. PLoS One 7:e35941. doi: 10.1371/journal.pone.0035941
Giovanetti, E., Brenciani, A., Morroni, G., Tiberi, E., Pasquaroli, S., Mingoia, M., et al. (2014). Transduction of the Streptococcus pyogenes bacteriophage Phim46.1, carrying resistance genes mef(A) and tet(O), to other Streptococcus species. Front. Microbiol. 5:746. doi: 10.3389/fmicb.2014.00746
Goyette-Desjardins, G., Auger, J. P., Xu, J., Segura, M., and Gottschalk, M. (2014). Streptococcus suis, an important pig pathogen and emerging zoonotic agent—An update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 3, e45.
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Haenni, M., Saras, E., Bertin, S., Leblond, P., Madec, J. Y., and Payot, S. (2010). Diversity and mobility of integrative and conjugative elements in bovine isolates of Streptococcus agalactiae, S. dysgalactiae subsp dysgalactiae, and S. uberis. Appl. Environ. Microbiol. 76, 7957–7965. doi: 10.1128/AEM.00805-10
Hao, H., Sander, P., Iqbal, Z., Wang, Y., Cheng, G., and Yuan, Z. (2016). The risk of some veterinary antimicrobial agents on public health associated with antimicrobial resistance and their molecular basis. Front. Microbiol. 7:1626.
He, T., Shen, Y., Schwarz, S., Cai, J., Lv, Y., Li, J., et al. (2016). Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J. Antimicrob. Chemother. 71, 1466–1473. doi: 10.1093/jac/dkw016
Huang, J., Chen, L., Li, D., Wang, M., Du, F., Gao, Y., et al. (2018). Emergence of a vanG-carrying and multidrug resistant ICE in zoonotic pathogen Streptococccus suis. Vet. Microbiol. 222, 109–113. doi: 10.1016/j.vetmic.2018.07.008
Huang, J., Chen, L., Wu, Z., and Wang, L. (2017). Retrospective analysis of genome sequences revealed the wide dissemination of optrA in Gram-positive bacteria. J. Antimicrob. Chemother. 72, 614–616.
Huang, J., Liang, Y., Guo, D., Shang, K., Ge, L., Kashif, J., et al. (2016a). Comparative genomic analysis of the ICESa2603 family ICEs and spread of erm(B)- and tet(O)-carrying transferable 89K-subtype ICEs in Swine and Bovine Isolates in China. Front. Microbiol. 7:e00055. doi: 10.3389/fmicb.2016.00055
Huang, J., Ma, J., Shang, K., Hu, X., Liang, Y., Li, D., et al. (2016b). Evolution and diversity of the antimicrobial resistance associated mobilome in Streptococcus suis: A probable mobile genetic elements reservoir for other streptococci. Front. Cell. Infect. Microbiol. 6:118.
Huang, J., Sun, J., Wu, Y., Chen, L., Duan, D., Lv, X., et al. (2019a). Identification and pathogenicity of an XDR Streptococcus suis isolate that harbours the phenicol-oxazolidinone resistance genes optrA and cfr, and the bacitracin resistance locus bcrABDR. Int. J. Antimicrob. Agents 54, 43–48. doi: 10.1016/j.ijantimicag.2019.04.003
Huang, J., Wang, M., Gao, Y., Chen, L., and Wang, L. (2019b). Emergence of plasmid-mediated oxazolidinone resistance gene poxtA from CC17 Enterococcus faecium of pig origin. J. Antimicrob. Chemother. doi: 10.1093/jac/dkz250 [Epub ahead of print].
Ishida, S., Tien Le, H. T., Osawa, R., Tohya, M., Nomoto, R., Kawamura, Y., et al. (2014). Development of an appropriate PCR system for the reclassification of Streptococcus suis. J. Microbiol. Methods 107, 66–70. doi: 10.1016/j.mimet.2014.09.003
Klare, I., Badstubner, D., Konstabel, C., Bohme, G., Claus, H., and Witte, W. (1999). Decreased incidence of VanA-type vancomycin-resistant enterococci isolated from poultry meat and from fecal samples of humans in the community after discontinuation of avoparcin usage in animal husbandry. Microb. Drug Resist. 5, 45–52.
Krull, M., Klare, I., Ross, B., Trenschel, R., Beelen, D. W., Todt, D., et al. (2016). Emergence of linezolid- and vancomycin-resistant Enterococcus faecium in a department for hematologic stem cell transplantation. Antimicrob. Resist. Infect. Control 5, 31. doi: 10.1186/s13756-016-0131-6
Lebreton, F., Depardieu, F., Bourdon, N., Fines-Guyon, M., Berger, P., Camiade, S., et al. (2011). D-Ala-d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 55, 4606–4612. doi: 10.1128/AAC.00714-11
Lei, C. W., Kang, Z. Z., Wu, S. K., Chen, Y. P., Kong, L. H., and Wang, H. N. (2019). Detection of the phenicol-oxazolidinone-tetracycline resistance gene poxtA in Enterococcus faecium and Enterococcus faecalis of food-producing animal origin in China. J. Antimicrob. Chemother. 74, 2459–2461.
Li, M., Cai, C., Chen, J., Cheng, C., Cheng, G., Hu, X., et al. (2016). Inducible Expression of both ermB and ermT conferred high macrolide resistance in Streptococcus gallolyticus subsp. pasteurianus isolates in China. Int. J. Mol. Sci. 17, E1599. doi: 10.3390/ijms17101599
Li, M., Shen, X., Yan, J., Han, H., Zheng, B., Liu, D., et al. (2011). GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol. Microbiol. 79, 1670–1683. doi: 10.1111/j.1365-2958.2011.07553.x
Li, R., Li, Y., Kristiansen, K., and Wang, J. (2008). SOAP: Short oligonucleotide alignment program. Bioinformatics 24, 713–714. doi: 10.1093/bioinformatics/btn025
Liu, Y., Wang, Y., Wu, C., Shen, Z., Schwarz, S., Du, X. D., et al. (2012). First report of the multidrug resistance gene cfr in Enterococcus faecalis of animal origin. Antimicrob. Agents Chemother. 56, 1650–1654. doi: 10.1128/AAC.06091-11
Long, K. S., Poehlsgaard, J., Kehrenberg, C., Schwarz, S., and Vester, B. (2006). The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50, 2500–2505.
Marini, E., Palmieri, C., Magi, G., and Facinelli, B. (2015). Recombination between Streptococcus suis ICESsu32457 and Streptococcus agalactiae ICESa2603 yields a hybrid ICE transferable to Streptococcus pyogenes. Vet. Microbiol. 178, 99–104. doi: 10.1016/j.vetmic.2015.04.013
Marshall, B. M., and Levy, S. B. (2011). Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 24, 718–733. doi: 10.1128/CMR.00002-11
Mcewen, S. A., and Fedorka-Cray, P. J. (2002). Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34(Suppl. 3), S93–S106.
Mendes, R. E., Deshpande, L. M., and Jones, R. N. (2014). Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist. Update 17, 1–12. doi: 10.1016/j.drup.2014.04.002
O’driscoll, C., Murphy, V., Doyle, O., Wrenn, C., Flynn, A., O’flaherty, N., et al. (2015). First outbreak of linezolid-resistant vancomycin-resistant Enterococcus faecium in an Irish hospital, February to September 2014. J. Hosp. Infect. 91, 367–370. doi: 10.1016/j.jhin.2015.09.006
Okura, M., Lachance, C., Osaki, M., Sekizaki, T., Maruyama, F., Nozawa, T., et al. (2014). Development of a two-step multiplex PCR assay for typing of capsular polysaccharide synthesis gene clusters of Streptococcus suis. J. Clin. Microbiol. 52, 1714–1719. doi: 10.1128/JCM.03411-13
Palmieri, C., Magi, G., Mingoia, M., Bagnarelli, P., Ripa, S., Varaldo, P. E., et al. (2012). Characterization of a Streptococcus suis tet(O/W/32/O)-carrying element transferable to major Streptococcal pathogens. Antimicrob. Agents Chemother. 56, 4697–4702. doi: 10.1128/AAC.00629-12
Palmieri, C., Princivalli, M. S., Brenciani, A., Varaldo, P. E., and Facinelli, B. (2010). Different genetic elements carrying the tet(W) gene in two human clinical isolates of Streptococcus suis. Antimicrob. Agents Chemother. 55, 631–636. doi: 10.1128/AAC.00965-10
Palmieri, C., Varaldo, P. E., and Facinelli, B. (2011). Streptococcus suis, an emerging drug-resistant animal and human pathogen. Front. Microbiol. 2:235. doi: 10.3389/fmicb.2011.00235
Qiu, X., Bai, X., Lan, R., Zheng, H., and Xu, J. (2016). Novel capsular polysaccharide loci and new diagnostic tools for high-throughput capsular gene typing in Streptococcus suis. Appl. Environ. Microbiol. 82, 7102–7112.
Reed, L. J., and Muench, H. (1938). A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27, 493–497.
Sassi, M., Guerin, F., Lesec, L., Isnard, C., Fines-Guyon, M., Cattoir, V., et al. (2018). Genetic characterization of a VanG-type vancomycin-resistant Enterococcus faecium clinical isolate. J. Antimicrob. Chemother. 73, 852–855. doi: 10.1093/jac/dkx510
Shen, J., Wang, Y., and Schwarz, S. (2013). Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 68, 1697–1706. doi: 10.1093/jac/dkt092
Srinivasan, V., Metcalf, B. J., Knipe, K. M., Ouattara, M., Mcgee, L., Shewmaker, P. L., et al. (2014). vanG element insertions within a conserved chromosomal site conferring vancomycin resistance to Streptococcus agalactiae and Streptococcus anginosus. MBio 5, e1386–e1314. doi: 10.1128/mBio.01386-14
Tang, J., Wang, C., Feng, Y., Yang, W., Song, H., Chen, Z., et al. (2006). Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 3:e151. doi: 10.1371/journal.pmed.0030151
Thanner, S., Drissner, D., and Walsh, F. (2016). Antimicrobial resistance in agriculture. MBio 7, e2227–e2215. doi: 10.1128/mBio.02227-15
Wang, Y., Li, D., Song, L., Liu, Y., He, T., Liu, H., et al. (2013). First report of the multiresistance gene cfr in Streptococcus suis. Antimicrob. Agents Chemother. 57, 4061–4063. doi: 10.1128/AAC.00713-13
Wang, Y., Lv, Y., Cai, J., Schwarz, S., Cui, L., Hu, Z., et al. (2015). A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 70, 2182–2190. doi: 10.1093/jac/dkv116
Wu, Z., Zhang, W., Lu, Y., and Lu, C. (2010). Transcriptome profiling of zebrafish infected with Streptococcus suis. Microb. Pathog. 48, 178–187. doi: 10.1016/j.micpath.2010.02.007
Wu, Z. F., Wu, C. Y., Shao, J., Zhu, Z. Z., Wang, W. X., Zhang, W. W., et al. (2014). The Streptococcus suis transcriptional landscape reveals adaptation mechanisms in pig blood and cerebrospinal fluid. RNA 20, 882–898. doi: 10.1261/rna.041822.113
Xu, X., Lin, D., Yan, G., Ye, X., Wu, S., Guo, Y., et al. (2010). vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob. Agents Chemother. 54, 4643–4647. doi: 10.1128/AAC.01710-09
Yao, J., Shang, K., Huang, J., Ran, W., Kashif, J., and Wang, L. (2014). Overexpression of an ABC transporter and mutations of GyrA, GyrB, and ParC in contributing to high-level ciprofloxacin resistance in Streptococcus suis type 2. Biosci. Trends 8, 84–92.
Yu, H., Jing, H., Chen, Z., Zheng, H., Zhu, X., Wang, H., et al. (2006). Human Streptococcus suis outbreak, Sichuan, China. Emerg. Infect. Dis. 12, 914–920.
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zheng, H., Qiu, X., Roy, D., Segura, M., Du, P., Xu, J., et al. (2017). Genotyping and investigating capsular polysaccharide synthesis gene loci of non-serotypeable Streptococcus suis isolated from diseased pigs in Canada. Vet. Res. 48, 10. doi: 10.1186/s13567-017-0417-6
Keywords: Streptococcus suis, vancomycin resistance, vanG, linezolid resistance, optrA, mobile genetic elements, pathogenicity
Citation: Du F, Lv X, Duan D, Wang L and Huang J (2019) Characterization of a Linezolid- and Vancomycin-Resistant Streptococcus suis Isolate That Harbors optrA and vanG Operons. Front. Microbiol. 10:2026. doi: 10.3389/fmicb.2019.02026
Received: 21 May 2019; Accepted: 19 August 2019;
Published: 10 September 2019.
Edited by:
Peter Mullany, University College London, United KingdomReviewed by:
Carla Novais, University of Porto, PortugalAna P. Tedim, Institute of Health Sciences Studies of Castilla y León (IECSCYL), Spain
Copyright © 2019 Du, Lv, Duan, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liping Wang, d2xwNzFAMTYzLmNvbQ==; Jinhu Huang, amh1YW5nQG5qYXUuZWR1LmNu
 Fanshu Du
Fanshu Du Xi Lv
Xi Lv Duan Duan
Duan Duan Liping Wang
Liping Wang Jinhu Huang
Jinhu Huang