- 1La Trobe Institute for Molecular Sciences, La Trobe University, Bendigo, VIC, Australia
- 2Department of Physiology, Anatomy and Microbiology, La Trobe University, Bundoora, VIC, Australia
It is now clear that several of the filamentous bacteria in activated sludge wastewater treatment plants globally, are members of the phylum Chloroflexi. They appear to be more commonly found in treatment plants designed to remove nitrogen (N) and phosphorus (P), most of which operate at long sludge ages and expose the biomass to anaerobic conditions. The Chloroflexi seem to play an important beneficial role in providing the filamentous scaffolding around which flocs are formed, to feed on the debris from lysed bacterial cells, to ferment carbohydrates and to degrade other complex polymeric organic compounds to low molecular weight substrates to support their growth and that of other bacterial populations. A few commonly extend beyond the floc surface, while others can align in bundles, which may facilitate interfloc bridging and hence generate a bulking sludge. Although several recent papers have examined the phylogeny and in situ physiology of Chloroflexi in activated sludge plants in Denmark, this review takes a wider look at what we now know about these filaments, especially their global distribution in activated sludge plants, and what their functional roles there might be. It also attempts to outline why such information might provide us with clues as to how their population levels may be manipulated, and the main research questions that need addressing to achieve these outcomes.
Introduction
The Activated Sludge Process
Treatment of domestic and industrial wastewater is essential to reduce potentially harmful levels of organic and inorganic compounds and pathogenic microbes to those allowing the treated water to be run into a receiving water body, such that its self-purification ability is not compromised (Catherine et al., 2013; Gaget et al., 2017). The most popular treatment process globally is activated sludge (Daigger, 2014), and in use now for more than 100 years (Lofrano and Brown, 2010; Jenkins and Wanner, 2014). No longer are these seen as disposal systems, but as valuable sources of recycled water, and the biomass or sludge for recovery of chemicals of value (van Loosdrecht et al., 2014; Puyol et al., 2016). This biomass consists primarily of bacteria and protozoa, which become organized as aggregates called ‘flocs.’ It is generally accepted that the Chloroflexi, the subject of this review, play an important role in providing the filamentous matrix around which desirable strong flocs with rapid settling properties are formed (Kragelund et al., 2007a; Wanner and Jobbagy, 2014; Nierychlo et al., 2019). Through continuous recycling, the populations best suited to treat the raw incoming sewage, are selected (Daigger, 2014; Jenkins and Wanner, 2014).
As discharge license requirements became more stringent, plant configurations evolved to reduce effluent nitrogen (N) and phosphate (P) concentrations to environmentally acceptable levels. In both, further selective pressures are imposed to encourage the growth of bacterial populations whose ecophysiology ensures N and P removal (Barnard and Comeau, 2014; Khunjar et al., 2014; Stensel and Makinia, 2014). These systems were developed largely empirically with no knowledge of the microbes responsible, but subsequently, N removal processes including the SHARON, CANON, N-Damo and ANAMMOX processes have been designed, based on detailed understanding of the bacteria involved (Schmidt et al., 2003; Kartal et al., 2013; van Kessel et al., 2018).
Microbiology of Activated Sludge Communities
Only in the past 30 years have we begun to understand activated sludge microbiology, an outcome coinciding with development of culture independent molecular methods (Nielsen and McMahon, 2014). Development of PCR, cloning and Sanger DNA sequencing allowed compositions of activated sludge communities to be elucidated, with no need to culture individual members, being based instead on using 23S and 16S rRNA gene sequence analyses used as phylogenetic markers (Nielsen and McMahon, 2014). Subsequent design of fluorescently tagged oligonucleotide probes targeting RNA sequences of populations of interest allowed in situ Fluorescence in situ Hybridization (FISH) identification of individual cells (Amann and Fuchs, 2008; Nielsen et al., 2009a; Seviour, 2010b; Noguera et al., 2014). In combination with histochemical staining, stable isotope probing (SIP) and microautoradiography (MAR) (McIlroy et al., 2017a), FISH has elucidated the in situ ecophysiology and possible function/s of individual probed populations at the single cell level. Such data have revealed the true biodiversity of activated sludge communities, and presence of previously unknown populations existing there, including the Chloroflexi discussed in this review, where most have yet to be cultured. The impact of next generation DNA (ngs) amplicon sequencing/metagenomics approaches on our understanding of the Chloroflexi in activated sludge communities will be discussed later.
Nutrient Removal Plants
Microbiology of Plants Designed to Remove Nitrogen
Continuous flow aerobic activated sludge plants removing N have sequential reactors where the influent is passed continuously between aerobic and anoxic zones. In the former, nitrification occurs where ammonia is oxidized first to nitrite and then nitrate by the nitrifying chemolithoautotrophic Bacteria. It was always thought that nitrification involved two different populations, the nitroso bacteria responsible for ammonia oxidation to nitrite, and the nitro bacteria oxidizing nitrite to nitrate (Daims et al., 2015). FISH has revealed that clusters of each co-exist in situ in a mutualistic dependent syntrophic relationship, where the nitroso bacteria supply the nitro bacteria with their energy source nitrite, and in return the nitro bacteria remove this harmful chemical, and supply the nitroso bacteria with ammonia generated from their high urease activity (Daims et al., 2015). We know now that a single population can oxidize ammonia all the way to nitrate. These are the ‘Comammox’ Nitrospira related bacteria, and occur widely, including in N removal wastewater treatment plants (Daims et al., 2015; Roots et al., 2019). Equally unexpectedly, Sorokin et al. (2014) isolated successfully a non-filamentous Chloroflexi nitrifying bacterium, Nitrolancea hollandica, which this review shows occurs in activated sludge systems (see later).
Under ‘anoxic’ conditions, nitrate/nitrite acting as the terminal electron acceptor is then reduced sequentially by denitrifying chemoorganoheterotrophic bacteria using anaerobic respiration to eventually inert dinitrogen gas. In many plants, partially reduced nitrous oxide and nitric oxide are released instead (Foley et al., 2010; Massara et al., 2017). Both are highly persistent greenhouse gases responsible for global warming and climate change.
Molecular techniques have shown that previously unknown Nitrosomonas and diverse Nitrospira and Nitrotoga populations are the most abundant nitrifiers in activated sludge (Daims et al., 2015). Identities of the bacteria important in denitrification, leading to dinitrogen formation and N removal (Lu et al., 2014) show too that most are as yet uncultured bacterial populations (McIlroy et al., 2014).
Microbiology of Plants Designed to Remove Phosphorus
Phosphorus removal is most efficiently achieved biologically by selectively encouraging the proliferation of polyphosphate accumulating organisms (PAO), which accumulate and store intracellular P as polyP granules. The configurations and operating conditions required to achieve this process of Enhanced Biological Phosphate Removal (EBPR) are well proven (Barnard and Comeau, 2014), and will not be detailed here. Essentially the biomass is recycled repeatedly between anaerobic (feed), and aerobic (famine) conditions. Under anaerobic conditions, readily biodegradable substrates in raw sewage are used by PAO to synthesize intracellular storage products using polyphosphate stored under aerobic conditions as main energy source. Under aerobic conditions, with no readily biodegradable substrates available, the PAO use their storage products as energy and carbon sources, and hence are selectively favored. PAO assimilate orthophosphate from the bulk liquid and store it intracellularly as polyphosphate granules (Seviour et al., 2003; Oehmen et al., 2007; Seviour and McIlroy, 2008; McMahon et al., 2010). Their subsequent removal by sludge wasting removes cellular immobilized phosphorus. All plants removing P also remove N.
Equally profound changes in PAO identification have resulted using molecular approaches. Thus, 16S rRNA gene cloning and FISH/MAR/histochemical staining showed that the betaproteobacterial Candidatus ‘Accumulibacter phosphatis’ was a common PAO, the first identified (Oehmen et al., 2007; McMahon et al., 2010; He and McMahon, 2011). Actinobacterial Tetrasphaera species are also PAO, and occupy a different niche in EBPR communities (Marques et al., 2017). Other putative PAO have been proposed, and their claims to be PAO are assessed by Stokholm-Bjerregaard et al. (2017).
The so-called glycogen accumulating bacteria GAO, are thought to share the PAO anaerobic phenotype (McIlroy et al., 2018), but instead of synthesizing polyP aerobically under aerobic famine conditions, they store glycogen (Stokholm-Bjerregaard et al., 2017; Nielsen et al., 2019). They too are phylogenetically and metabolically diverse. While some store PHA anaerobically as many PAO do, the storage products in others can vary (Stokholm-Bjerregaard et al., 2017; McIlroy et al., 2018).
Microbiological Solids Seperation Problems
Most activated sludge plants around the world suffer intermittently from the problems of bulking and foaming (Seviour and Nielsen, 2010; Wanner and Jobbagy, 2014; Rossetti et al., 2017). Both can be caused by excessive proliferation of filamentous bacteria (Jenkins et al., 2003; Martins et al., 2004; Wanner et al., 2014). When filaments extend from the floc surface into the bulk liquid, floc sedimentation velocity is retarded. In extreme cases bulking may result. Alternatively, excessive filament growth can affect floc settlability by rendering them more diffuse (Martins et al., 2004; Burger et al., 2017). In both cases, biomass is then released with the treated liquid effluent into the environment, increasing effluent COD. Membrane reactors are becoming popular because they overcome any floc settling although filamentous bacteria may cause membrane fouling (Crawford et al., 2014).
Activated sludge foam formation is a flotation event, readily visible on the surface of the aerobic reactors. Such foams can lead to serious changes in the operation of the activated sludge process, and in some cases, serve as the source of opportunistic pathogens (de los Reyes, 2010). Stable foams require three components, air bubbles, surfactants and hydrophobic particles (bacterial cells). With only air bubbles and surfactants, a common feature of plant startup, a non-persistent superficial white foam is generated, while in the absence of surfactants, an oily surface scum is generated (Petrovski et al., 2011). Most foams become stabilized by presence of high numbers of filamentous hydrophobic bacteria, but theoretically any sufficiently hydrophobic cells, including unicells will stabilize them (de los Reyes, 2010; Petrovski et al., 2011).
This review deals with a group of filamentous bacteria, the Chloroflexi, who have been associated with bulking and foaming events in activated sludge plants. Although much is known about their identity and ecophysiology in Danish EBPR plants, little information is available regarding their global distribution. This review has as one of its aims to fill that gap in our knowledge.
The Chloroflexi
The phylum ‘Chloroflexi, embraces an ecologically and physiologically diverse group of bacteria, which have been detected in an increasingly wide range of anaerobic habitats including sediments, hot springs, methanogenic anaerobic sludge digesters where they are highly abundant and play an important fermentative role as well as contributing to sludge granulation (Hug et al., 2013; McIlroy et al., 2016; Sun et al., 2016; Xia et al., 2016; Petriglieri et al., 2018; Bovio et al., 2019), the human oral cavity (Campbell et al., 2014), Anammox reactors (Kindaichi et al., 2012; Cao et al., 2016; Wang et al., 2016) and activated sludge communities (Björnsson et al., 2002; Kragelund et al., 2007a, 2011; Speirs et al., 2009, 2011, 2017; Yamada and Sekiguchi, 2009; Hanada, 2014; McIlroy et al., 2018; Andersen et al., 2019; Nierychlo et al., 2019). This is not the place to explore in detail their still evolving systematics, which have been dealt with elsewhere (Hanada, 2014). So only a brief overview is provided here.
It soon became apparent that the original name used to describe these, the Green non-sulfur bacteria’ was inappropriate, as they included a diverse range of mesophilic and thermophilic aerobic and anaerobic chemoorganoheterotrophs as well as photolithoautotrophic bacteria (Hanada, 2014). It was also soon recognized that they were not closely related phylogenetically to the Chlorobia (Green Sulfur Bacteria) or the Purple Non-Sulfur Bacteria. Hence the phylum name Chloroflexi proposed by Garrity et al. (2001) is now accepted.
Based on the 16S rRNA gene sequences of the then available strains, a single class, the ‘Chloroflexi,’ containing two orders, the Chloroflexales and the Herpetosiphonales was proposed by them. All known members were filamentous with an unusual gliding mechanism as a means of motility, and although most stained Gram negatively, none had the characteristic lipopolysaccharide outer membrane of the Gram-negative bacteria. Only four Chloroflexi genera were recognized by Garrity et al. (2001), although it was becoming clear from 16S rRNA gene sequence data retrieved from a wide range of habitats that many uncultured strains existed.
This scheme was modified by Hugenholtz and Stackebrandt (2004) to accommodate the 16S rRNA gene sequence data from newly described strains and those initially classified incorrectly. They proposed four new classes, the Anaerolineae, the Dehalococcoidetes, the Chloroflexia and the Thermomicrobia, corresponding to their earlier groupings labeled [1], [2], [3], and [5], respectively (Hugenholtz et al., 1998). Their phylogenetically distinct group 4 contained only uncultured strains, and so was not named formally. Characterization of three new isolates led Yamada et al. (2006) to propose that the Anaerolineae should be subdivided into two classes, the obligately anaerobic Anaerolineae and the aerobic or facultatively anaerobic Caldilineae, as delineated earlier by Sekiguchi et al. (2003).
These classifications have persisted, and so based on 16S rRNA genesequence data, the phylum Chloroflexi was thought by Hanada (2014) to contain at least six classes. They are the Chloroflexia, which includes all the photolithotrophic members, the Anaerolineae, the Caldilineae, the Ktedonobacteria, all of which appear to be multicellular filamentous bacteria, and the Dehalococcoidetdia. The Thermomicrobia, all members of which were thought to be thermophilic, were once considered to belong to a separate phylum by Garrity et al. (2001). In 2013, Kawaichi et al. (2013) proposed that a new class, the Ardenticatenia, was needed to accommodate a strain they isolated from an iron rich coastal hydrothermal field. This addition was not included in the review of Hanada (2014), and nor was that of Dodsworth et al. (2014), based on their new strain Thermoflexus hugenholtzii. While its 16S rRNA genesequence showed it to be a member of the phylum Chloroflexi, it was not embraced by any of the then six existing classes, and so a new class, the Thermoflexi, was proposed to accommodate it. It seemed then probable that further modifications to the classification of these organisms would be necessary as more strains were characterized using novel characters, and some of these have already suggested the current schemes for Chloroflexi systematics need modifications.
Thus, phylogenetic markers other than 16S rRNA gene sequences have been used. For example, Kunisawa (2011) used genome sequence order comparisons, which reveal shared ancestry between populations. His data gave similar outcomes to those using 16S rRNA gene sequence data, and confirmed that the Thermomicrobia were not a separate phylum, but a class within the Chloroflexi. On the other hand, the application of alternative molecular markers, the signature indels in conserved proteins of the Chloroflexi by Gupta et al. (2013) gave a phylogeny markedly different to those based on 16S rRNA gene sequences. Protein sequences from each of their clades each possessed distinctive indels, but overall their patterns revealed that the phylum Chloroflexi sensu stricto should contain only members of the classes Chloroflexia and the Thermomicrobia, while the other four classes of Hanada (2014) should be considered as taxa related to, but not part of the phylum Chloroflexi. More recently Parks et al. (2018) have looked at possible evolutionary and phylogenetic relationship among the Chloroflexi with their Genome Taxonomic Database (GTDB), based on concatenated alignments of 120 single copy marker genes, and taxa constructed using relative sequence divergence. Among other changes, they suggest that the taxa classes of Caldilineae, Ardenticatenia and Thermoflexia in the SILVA/MiDAS classification should be relegated to the orders Caldilineales, Ardenticatenales and Thermoflexales, respectively. The scheme has already led, for example, to marked changes in the proposed classification of Ca. ‘Amarolinea aalborgensis’ (Andersen et al., 2019), as discussed below.
The Bulking and Foaming Filamentous Bacteria
Originally characterized exclusively on their microscopic features and staining reactions, Eikelboom (1975) separated activated sludge bulking and foaming filaments into several individual ‘morphotypes’ based on their morphology and staining reactions, in the absence then of more discerning methods for their identification. He was unable to culture and appropriately characterize most of these, and the ‘naming’ system he used for them does not follow the strict rules of the International Code for Bacterial Nomenclature. Instead he adopted a morphotype based numerical system (e.g., types 0092, 0851, 0803, etc.), an imprecise system of labeling but one still used widely, inevitably leading to confusion and communication compromises among microbiologists interested in these bacteria.
Furthermore, his widely used morphology based identification manuals (Eikelboom and Van Buijsen, 1983; Eikelboom and Geurkink, 2002; Eikelboom et al., 2006) and those of Jenkins et al. (2003) do not always agree about which feature/s are diagnostic for each filament morphotype. The manuals are also incomplete, and so the more recently characterized filaments including several Chloroflexi (Speirs et al., 2009, 2011, 2017; Andersen et al., 2019; Nierychlo et al., 2019) and “Candidatus Villigracilis,” which data here show (Supplementary Data File S1) is clearly an abundant filament common to activated sludge systems around the world are missing.
Those basing their filament ‘identifications’ exclusively on characters listed in these manuals assume the listings embrace all existing activated sludge filaments. So inevitably they try to match their unknown to one of the filaments described there, even though the match may not be close, and their filament may not be mentioned. This microscopic approach, which is still popular, is quick and simple with practice but uses too few characters, and has one major flaw. It is clear now that a single Eikelboom morphotype may embrace several phylogenetically very different bacteria, which are indistinguishable under the microscope (Seviour, 2010a, b). As the Chloroflexi, like most other filaments, possess no unique morphological feature/s to allow their precise identification in this manner, microscopy alone has failed to reveal whether any of these filament morphotypes are members of this phylum.
During this period, some of the bulking and foaming filament morphotypes of Eikelboom (1975) were cultured successfully, often after micromanipulation, and their 16S rRNA genes sequenced (Nielsen et al., 2009b; Seviour, 2010b; Yoon et al., 2010; Nielsen and McMahon, 2014). Obtaining this information is straightforward for the relatively few filaments isolated into pure culture (see below), but the likelihood of growing more of these filaments, in the absence of any information on their individual growth requirements, seems increasingly small (Seviour, 2010b). The sequence data generated reveal that most bulking filaments represent previously undescribed bacteria. Other filaments have emerged as filamentous forms of previously identified unicellular bacteria, a situation also likely to make their identification by microscopy problematic. Examples are given in Seviour and Nielsen (2010). Consequently, any applied control methods based solely on their microscopic identification are unlikely to work reliably.
The Chloroflexi cultured from activated sludge include a Herpetosiphon sp. (Bradford et al., 1996), which was thought unlikely to cause bulking (Nielsen et al., 2009b), and has not been characterized further. Of the Chloroflexi filaments in activated sludge, “Ca. Kouleothrix aurantiaca.” has been grown and characterized phenotypically (Beer et al., 2002; Kohno et al., 2002; Kragelund et al., 2007a), as has a Chloroflexi filament isolated in Korea (Yoon et al., 2010), named here as a “Ca. Defluviithrix” (see later). No filament in the two identification manuals is a close morphological match to the isolate of Yoon et al. (2010). Earlier claims of success in growing the Chloroflexi morphotype type 0092 (Horan et al., 1988; Buali and Horan, 1989; Bradford et al., 1996) should be viewed cautiously. No convincing evidence has been presented that the filaments they cultured were of this morphotype, and Speirs et al. (2009) showed that the 16S rRNA targeted FISH probe designed by Bradford et al. (1996) to target their type 0092 Bacteroidetes isolate, did not target the common type 0092 morphotype seen in situ in Australia or elsewhere.
Microscopy Based Survey Data
Early plant surveys depended by necessity on using these morphological/staining characters in attempts to see which filaments dominated plants profiles. In most of these (summarized by Seviour and Nielsen, 2010; Rossetti et al., 2017), usually only single samples have been taken, often from a single point in the treatment process, and in many cases from a small selection of treatment plants. The plants sampled are usually located considerable distances from each other, treating a wide diversity of undefined influents, and the filament ‘identifications’ were rarely complemented by comprehensive and detailed operational data (Seviour, 2010b; Rossetti et al., 2017). These compromises limit the usefulness of such survey data, providing as they do a limited snapshot of the filaments present at a single point in time.
Early microscopic surveys by Eikelboom and Van Buijsen (1983), and Jenkins (1992) of European and North American treatment plants respectively, attempted to see if any correlation existed between individual filament morphotypes and plant operational conditions. Their ‘rules of thumb’ are still used widely in attempts to solve bulking and foaming problems in large scale plants around the world, despite many of these relationships being questioned in surveys using molecular methods, as discussed later. More recent survey data (Rossetti et al., 2017) in general support many of their conclusions, but not all similar studies agree (Seviour et al., 1994).
Impact of Molecular Tools on Understanding the Bulking and Foaming Filaments
Which Chloroflexi Filaments Are Present in Activated Sludge Plants?
Most bulking plant communities contain a low biodiversity of filamentous bacteria, among which are the one or sometimes more dominant populations responsible for sporadic incidents of bulking. It is now clear that these communities commonly contain several filamentous members of the phylum Chloroflexi (McIlroy et al., 2015, 2018; Andersen et al., 2019; Nierychlo et al., 2019), and FISH probes, together with helper and competitor probes where employed, have been designed for many of these (Table 1). In writing this review and taking into consideration the recent literature, it was decided that the Eikelboom filaments characterized with molecular methods should no longer be recognized solely by their numerical designations. Thus, together with the phenotypic characters listed in the identification manuals of Jenkins et al. (2003), Eikelboom et al. (2006), and Seviour and Nielsen (2010), it now seems an appropriate time to provide the Chloroflexi filaments with provisional or Candidatus names where appropriate. Switching to a system of filament nomenclature based on using valid names was adopted by McIlroy et al. (2015), and has become the convention used subsequently by McIlroy et al. (2017a), Speirs et al. (2017), Andersen et al. (2019), and Nierychlo et al. (2019).
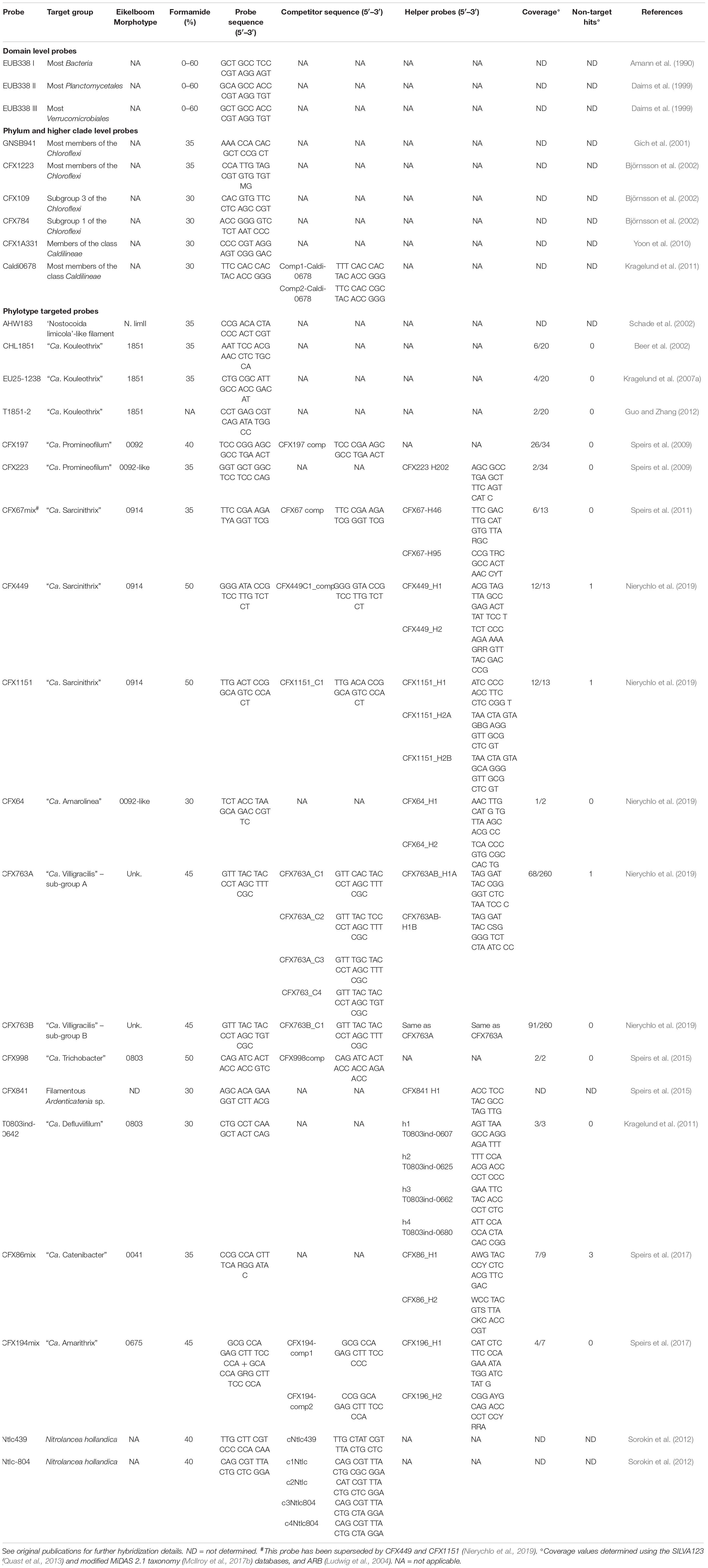
Table 1. FISH probes, oligonucleotide sequences, target coverage detail and hybridization conditions to target members of the filamentous Chloroflexi reported in activated sludge.
In addition to the cultured “Ca. Kouleothrix spp.” mentioned above (Beer et al., 2002; Kohno et al., 2002; Kragelund et al., 2007a), are the uncultured “Ca. Promineofilum breve” (type 0092) (Speirs et al., 2009; McIlroy et al., 2016), “Ca. Defluviifilum” (Danish type 0803) (Kragelund et al., 2011), “Ca. Trichobacter” (Australian type 0803) (Speirs et al., 2015), “Ca. Catenibacter” (type 0041) (Speirs et al., 2017), “Ca. Amarithrix” (type 0675) (Speirs et al., 2017), “Ca. Villigracilis” (not associated with any Eikelboom morphotype) (Nierychlo et al., 2019), and “Ca. Amarolinea aalborgensis,” an atypical type 0092 (Andersen et al., 2019; Nierychlo et al., 2019; Table 1). Attributes of these individual filaments are discussed in detail later in the review.
What Have FISH/PCR Cloning Data Revealed About Chloroflexi in Activated Sludge and Their Impact on Plant Operations?
One of the earliest reports of Chloroflexi in activated sludge was that of Juretschko et al. (2002) in an industrial plant removing N. After PCR/cloning, they recovered 15 Chloroflexi clones, whose sequences placed them all in the Group 1 of Hugenholtz et al. (1998). In the absence then of suitable FISH probes, they could not be identified further. Again in 2002, Björnsson et al. (2002) using FISH analyses, suggested the filamentous Chloroflexi were ubiquitous in non-bulking activated sludge and especially abundant in plants designed to remove N and in some cases P. On the basis of their 16S rRNA gene sequence data, most were members of subgroup 3 (now containing members of the class Chloroflexia). They designed and validated several FISH probes based on their 16S rRNA gene sequences, and some of these are still widely used today. They are the phylum targeted probe CFX1223, the subgroup 3 probe CFX109 and subgroups 1a and 1b targeted probe CFX784 (Table 1). It was recommended that the CFX1223 and the GNSB941 Chloroflexi phylum probes designed earlier by Gich et al. (2001), should be used with them, a practice still followed routinely. Björnsson et al. (2002) also reported for the first time that some of the Chloroflexi in full-scale plants failed to respond to the EUBmix probes designed to target all Bacteria. Unfortunately they were unable to relate their FISH positive Chloroflexi filaments to any of the morphotypes of Eikelboom (1975), because of their location within the flocs. However, a reassessment here of these clone sequences using the MiDAS 2.1 database can now reveal that the Björnsson et al. (2002) and Juretschko et al. (2002) Chloroflexi clone sequences AF234698, AF234710, AF234759, and X84472 were all derived from “Ca. Villigracilis,” while the Björnsson et al. (2002) clone sequence X84565 is here identified as from a “Ca. Kouleothrix.”
Most published FISH surveys have been relatively small scale, and usually carried out on single plant communities, or commonly performed to validate newly designed FISH probes targeting a known individual filament population/s e.g. (Beer et al., 2002; Björnsson et al., 2002; Kragelund et al., 2007b, 2011; Speirs et al., 2009, 2011, 2015, 2017; Nittami et al., 2017; Andersen et al., 2019; Nierychlo et al., 2019). These have often revealed possible relationships between filament abundances and plant configurations, but not always detailed individual plant operational conditions. The general conclusions from these studies are that the Chloroflexi in activated sludge plants are ubiquitous and almost exclusively filamentous. In non-bulking biomass samples they are usually located substantially within the floc, where, as stated earlier, they provide the matrix around which the floc material aggregates (Nielsen et al., 2009b). FISH probing has revealed that some including Ca ‘Amarolinea,” “Ca. Kouleothrix,” “Ca. Amarithrix,” “Ca. Defluviithrix,” “Ca. Sarcinithrix,” and “Ca. Catenibacter” can extend into the mixed liquor and form interfloc bridges (Beer et al., 2002; Speirs et al., 2011, 2015, 2017; Nierychlo et al., 2019).
Yet Wagner et al. (2015) have questioned the role of the Chloroflexi in bulking. Although showing in their qFISH based study that the Chloroflexi were located predominately protruding from the floc surfaces, modeling showed they were less important than the floc bound actinobacterial “Ca. Microthrix parvicella” in determining floc settlabilities. However, only phylum level CFX mix probes were used for Chloroflexi detection and quantification, so no attempt was made to identify which individual Chloroflexi populations might be present in their community. This concern is especially applicable to those mentioned above, whose bundles of filaments act to join flocs together (e.g., Beer et al., 2002), as to whether they were present in the communities studied. Thus, the assumption that all the filamentous Chloroflexi behave in activated sludge as Wagner et al. (2015) describes clearly requires further examination.
According to flotation theory, any hydrophobic cell can stabilize the foams appearing on the surface of activated sludge aerobic reactors (Petrovski et al., 2011), and explains why those of the actinobacterial Mycolata and “Ca. Microthrix parvicella” (de los Reyes, 2010) are commonly seen there. This theory may explain the frequently reported presence of “Ca. Promineofilum” filaments (Rossetti et al., 2017) in foams. However, de los Reyes (2010) has suggested this filament is an ‘accidental’ foam former, carried there by hydrophobic biomass. Furthermore, McIlroy et al. (2016) showed with their genomic data that “Ca. P. breve” cells are not hydrophobic, and nor are those of “Ca. A. aalborgensis” (Andersen et al., 2019) and the filaments described by Nierychlo et al. (2019), a decision based on in situ MAC (microsphere adsorption to cells) assays. However, those of “Ca. Defluviifilum” are, which may explain why these filaments have been observed often in foams, where they probably assist in their stabilization (Kragelund et al., 2011).
As mentioned earlier, many of these Chloroflexi filaments appear to prefer EBPR plants (see earlier), which generally operate at long sludge ages. This characteristic might suggest these are slow growing bacteria, being washed out at shorter sludge ages. Alternatively, because most are associated intimately with flocs, they will be recycled with the settled RAS, and thus stay within the EBPR plants, only leaving in the wasted sludge. Their high abundances may suggest their ecophysiology provides them with some competitive advantage over other bacterial populations in response to the strong selective pressures exerted on the EBPR bacterial community (see earlier). These may include an ability to assimilate substrates anaerobically and use them for synthesis of storage compounds which are then available aerobically for energy production and growth, as seen with the GAO and PAO phenotype (Seviour et al., 2003; Oehmen et al., 2007), or to perform fermentation and/or anaerobic respiration as reported by McIlroy et al. (2016), Andersen et al. (2019), and Nierychlo et al. (2019). Such a facultatively anaerobic chemoheteroorganotrophic lifestyle would provide them with a selective advantage in EBPR plants, where biomass is cycled continuously between anaerobic and aerobic zones (see earlier), and possibly render them less competitive in fully aerobic systems, where they are less common (McIlroy et al., 2016).
Previously undescribed filamentous Chloroflexi members of the Anaerolineae have been detected at high abundances in anaerobic digesters, systems used to stabilize wasted biomass from plants, and to generate methane as an energy source (St-Pierre and Wright, 2014; Kirkgaard et al., 2017; McIlroy et al., 2017a; Petriglieri et al., 2018). It seems reasonable to assume that at least some of these Chloroflexi derive from the aerobic activated sludge community, and consequently might support biomass bulking and foaming there. Kirkgaard et al. (2017) proposed that the Chloroflexi in their digesters consisted mainly of populations migrating from the aerobic bioreactors, although many they identified there were exclusive to the digesters. Petriglieri et al. (2018) also showed that members of the Chloroflexi were highly abundant in digesters, with most of the populations identified there being autochthonous, and thus probably having important roles in sludge digestion.
The question is whether these migrating Chloroflexi can survive and grow in these digesters, being facultative anaerobes (see above), and what their role might be in stabilizing foam formation commonly seen there, and, in having a filamentous morphology, in sludge granule formation. Foam stabilization seems unlikely because most of these flaments are not hydrophobic (Nierychlo et al., 2019). By comparing amplicon 16S rRNA gene sequencing abundances where both cellular and exocellular DNA fragments would be quantified, with those estimated by qFISH, a method requiring intact metabolically active cells, they suggested that the migrating Chloroflexi population steadily decreased in abundance in the digesters. In some cases they were barely detectable by either of these analytical methods. However, their naked DNA appears to survive there, to be detected by amplicon sequencing. These trends would suggest that the migrating Chloroflexi are not metabolically highly active populations in these digesters, and probably play a minor role in sludge stabilization and breakdown. Similar data from other countries are needed to confirm this hypothesis.
How Abundant Are the Chloroflexi in Activated Sludge Plants From FISH Analyses
The most popular FISH quantitative protocol is to express the data as population relative abundances where the biovolume of cells responding to the FISH probe for the filament population of interest is expressed as a percentage of cells responding to the EUBmix FISH probes designed to cover all members of the domain Bacteria (Daims et al., 1999). Unfortunately it is clear that some of the Chloroflexi filaments in activated sludge, and c.a 20% of all Chloroflexi 16S rRNA gene sequences in the SILVA128 NR99 database do not possess a perfect match to any of the domain targeted EUBmix probes (Morgan-Sagastume et al., 2008; Nielsen et al., 2009b; Speirs et al., 2009; Kragelund et al., 2011). However, not all probe target mismatches will be sufficiently destabilizing to prevent probe hybridization (Table 2), as appears to be the case with “Ca. Defluviifilum” (Nierychlo et al., 2019). An overview of the individual EUBmix probe target sites and the corresponding probe sequences in the Chloroflexi filaments described here are given in Table 2. Resolving these problems by designing more EUBmix FISH probes to cover all the known Chloroflexi 16S rRNA target site variants is not straightforward.
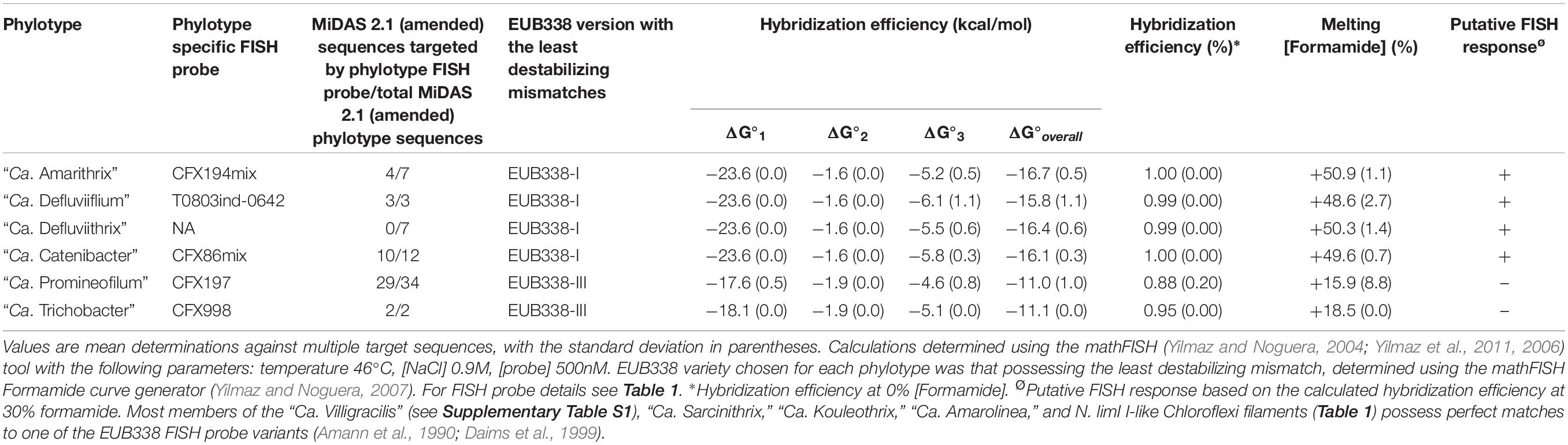
Table 2. Theoretical hybridization efficiency for the EUB338 I, II, III FISH probes, and target sites possessing mismatches within those Chloroflexi filament phylotypes defined by their respective FISH probes (see Table 1).
Most FISH filament analyses show how widely distributed the Chloroflexi are in wastewater treatment plants, although globally the numbers of plants examined are still small. Thus, in a bulking plant in Poland receiving abattoir and dairy milk wastes and designed to remove N and P, Miłobędzka and Muszyński (2015) showed that the Chloroflexi dominated the mixed liquor filament populations (57% of the total bacterial community). Among these, “Ca. Defluviifilum” (Kragelund et al., 2011) were more abundant than “Ca. Kouleothrix” and “Ca. Promineofilum,” but not too surprisingly then, about half of all their Chloroflexi could not be identified below phylum level. In later studies (Miłobędzka et al., 2016) they tried to relate operational parameters like sludge age to abundances of those filaments for which FISH probes were available. Not all these findings agreed with data generated in larger surveys in other countries, probably because their studies were based on sporadic sampling and short survey times, conditions under which such relationships are not always revealed convincingly (Mielczarek et al., 2012).
Filamentous bacteria emerging from aerobic granular surfaces have been identified by FISH over an extended period of operation of reactors supplied with different feeds (Figueroa et al., 2015). These too showed a predominance of Chloroflexi filaments in reactors, especially those fed synthetic wastewater and waste from a fish cannery processing plant, but not in those fed processed marine products. However, as only the phylum level Chloroflexi CFX1223/GNSB941 probes were used, no further taxonomic information was generated. Earlier Morgan-Sagastume et al. (2008) had used only phylum level Chloroflexi probes to detect their presence in a nutrient removal activated sludge plant.
Clearly the most comprehensive and extended filament FISH and q-PCR based survey data published so far are those of Mielczarek et al. (2012), which followed Chloroflexi populations in 56 nutrient (N and P) removal plants in Denmark over 4 years. They set out to reveal if the relative abundances of individual filaments were determined by plant design and operating conditions, and their data show that members of the phylum Chloroflexi were among the most abundant populations, particularly “Ca. Promineofilum” and “Ca. Defluviifilum” (McIlroy et al., 2015), and especially during the summer and autumn. They were then replaced by the actinobacterial “Ca. M. parvicella” during winter and spring, in response to the lower mixed liquor temperatures (Rossetti et al., 2005). “Ca. Kouleothrix” filaments were found in most plants, but always at low relative abundances (Nierychlo and Nielsen, 2014).
Mielczarek et al. (2012) also proposed that each plant had its own unique filament community or fingerprint, which remained generally stable over the sampling period. No evidence suggested that any relationship existed between filament relative abundances and plant operating parameters or wastewater composition over the long term, raising questions about the conclusions reached in the reports discussed above, which were based on small numbers of plants surveyed, limited samples and short operational durations. As reported elsewhere, many of their filaments hybridizing with the CFX1223/GNSB941 probes could not be identified further using the then available targeted Chloroflexi probes, again highlighting the probability that activated sludge communities contain a higher biodiversity, as Speirs et al. (2015, 2017), Andersen et al. (2019), and Nierychlo et al. (2019) have since shown.
A later examination of 128 samples taken from 16 Portuguese plants over a 2 year period (Dos Santos et al., 2015) used FISH only to distinguish between filaments with indistinguishable microscopic morphologies, and so the data they presented were based largely on the Eikelboom/Jenkins methods, with all their limitations (see earlier). Again “Ca. Kouleothrix,” “Ca. Promineofilum,” and now morphotypes 0041/0675 dominated most of their bulking samples. Not unexpectedly, their FISH data exposed phylogenetic variation within a single morphotype. Their attempts to determine which, if any processing parameters were influencing individual filament abundances generated some unexpected outcomes, not the least of which was their conclusion that type 0092 alone among the filament community there was associated with bulking in their plants. This filament rarely extends from the floc surface into the bulk liquid very far, thus ensuring any interfloc bridging is a rare event, although in high abundance, it may affect negatively floc density (Speirs et al., 2009).
Impact of Next Generation Sequencing on Understanding the Biodiversity of the Chloroflexi in Activated Sludge
With the introduction of next generation DNA sequencing (NGS), it became feasible to sequence simultaneously many individually tagged DNA samples quickly and relatively inexpensively (Karst et al., 2016; Hugerth and Andersson, 2017; Nierychlo et al., 2019). It provides a detailed fingerprint of the community population composition and has been used to quantify individual bulking and foaming bacterial populations, as discussed below.
However, no amplicon sequencing protocol provides strictly quantitative estimates of population relative abundances where one population may possess several rrn operons, whose individual 16S rRNAs differ in their sequence (Sun et al., 2016; Espejo and Plaza, 2018). Often this information is not available for the filament under consideration. The limited currently available data suggest that the genomes of “Ca. P. breve” (McIlroy et al., 2016), “Ca. A. aalborgensis.” (Andersen et al., 2019), and “Ca. Kouleothrix” (Ward et al., 2018) Chloroflexi each possess a single rrn operon. Whether this holds true for other activated sludge Chloroflexi filaments is not known.
Furthermore, as with FISH probe design, only those populations whose 16S rRNA gene sequences have been generated and attributed to a specified population in situ, and subsequently included in reference databases will be identifiable. Furthermore, several taxonomic classifiers exist, based on different phylogenetic divisions and groupings, and so sequences will be classified to different phylogenetic groups depending on which classifier is used, as shown in Table 3 for the activated sludge Chloroflexi.
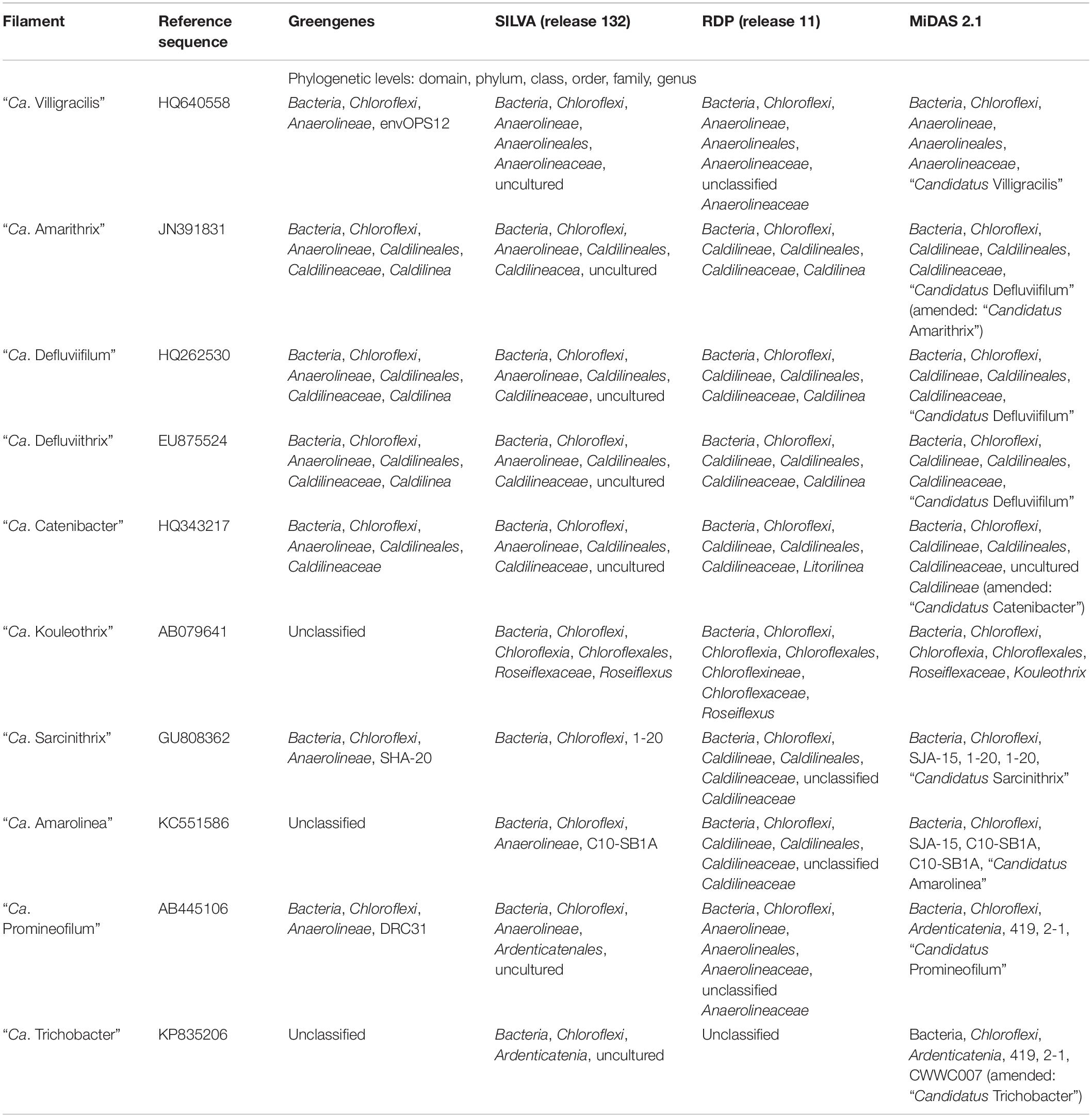
Table 3. Differences in Chloroflexi phylotype classification in reference taxonomies, Greengenes (DeSantis et al., 2006), SILVA (Quast et al., 2013), RDP (Wang et al., 2007; Cole et al., 2014), and MiDAS 2.1 (McIlroy et al., 2017b).
Equally important in amplicon sequencing is to consider critically PCR primer choice. Of the data analyzed here, almost half (5/12) used sequences from the V3–V4 regions of the 16S rRNA gene. Single studies have targeted the V1–V2, the V3, the V4 and V4–V5 regions, while three targeted the V1–V3 region. Optimizing primer choice in such studies is rarely addressed, despite concerns that they are not all equally efficient (Hugerth and Andersson, 2017). Albertsen et al. (2015) showed convincingly that primers targeting the V1–V3 regions better reflected the abundances of Chloroflexi in a community by more than two- fold, compared to some of the primers targeting the V3–4 and V4 regions. Consequently any amplicon based data, including those in Supplementary Data File S1 obtained with suboptimal primers, and not targeting the V1–V3 regions, may under-represent the Chloroflexi present in those samples.
The methodological paper of Karst et al. (2018) describes a protocol for high-throughput 16S rRNA gene sequencing using both DNA and RNA templates. When rRNA is used no PCR amplification step is involved, and so the biases known to be associated with this are removed. This method has the capacity to generate full- length 16S rRNA gene sequences and populations missed by PCR amplification will be detected by it. However, biases in relative abundance value estimates will be introduced by differences in ribosome numbers in members of individual populations. Equally, any variations in sequences of individual 16S rRNA in a single population will affect the community biodiversity data.
NGS Use in Activated Sludge Community Surveys
Amplicon sequencing holds the promise of helping identify many of the as yet unidentified Chloroflexi populations, as demonstrated convincingly for example by Nierychlo et al. (2019). Amplicon sequencing protocols regularly recover data from populations contributing as little as 0.1–0.01% of the total community, compared to the 0.5–2% limitation generally imposed by the PCR cloning approach (a value based on the generation and sequencing output from 50 – 200 cloned 16S rRNA gene sequences). Despite this, most of the currently available amplicon sequence data suggest population distributions observed generally mirror closely those revealed by the earlier molecular methods (Nielsen and McMahon, 2014; Hugerth and Andersson, 2017), and often do not reveal the true depth of community biodiversity (Karst et al., 2018).
To aid in identifying sequences of activated sludge bacteria, the MiDAS database was created (McIlroy et al., 2015, 2017b), whose aim is to consolidate all activated sludge bacteria into a single curated resource1. This resource provides a database for their classification, and a browser containing their curated functional attributes (McIlroy et al., 2015). Its value in identifying the activated sludge Chloroflexi and their ecophysiology will be demonstrated and discussed later.
As with the FISH surveys, many of the early amplicon studies were descriptive inventory fingerprinting or data generating exercises, involving single samples from small numbers of plants (Nielsen and McMahon, 2014). Nevertheless, these revealed predictably high community biodiversity richness, identifying members of the major recovered bacterial phyla, and sometimes extending considerably our understanding of certain functionally important groups, including the nitrifying bacteria, PAO and GAO (Nielsen and McMahon, 2014; Rodríguez et al., 2015; Daims et al., 2016; Ferrera and Sanchez, 2016). They also confirmed the high frequency of occurrences and often abundances of the Chloroflexi in plants of all configurations, but especially in EBPR nutrient removal systems. Therefore, the view was these bacteria should be recognized as core EBPR activated sludge populations, although in an extensive survey of Danish EBPR plants (Saunders et al., 2016), the Chloroflexi were often detected at relatively low abundances, Later examinations of the same plants (Nierychlo et al., 2019) suggested otherwise, which probably reflects the different PCR primers (see above and Supplementary Data File S1) used in the two studies (Albertsen et al., 2015).
As few of the plants examined in these studies were experiencing filamentous bulking or foaming incidents, no special attention was given to the putative bacteria responsible. Several have used amplicon sequencing to examine such communities. Almost all have been carried out in China, and so similar data from plants in other countries are lacking. One exception is the report of Dunkel et al. (2018), who used this approach to look at the 21 filamentous bacteria, including several Chloroflexi morphotypes, in two bulking and foaming German industrial treatment plants over a 3 month period. However, they did not take advantage of the MiDAS database for their filament identifications, instead using the less reliable filament database of Guo and Zhang (2012). They and others e.g. (Yang et al., 2017) have applied their data to suggest the operating factors thought to support the increased abundances of many of these filament clades (Dunkel et al., 2018), but generally these add little to what is known already.
This review has provided the opportunity to re-examine some of these amplicon sequencing data using the MiDAS 2.1 database, to look at the distribution of Chloroflexi filamentous bacteria across multiple treatment plant settings in different countries (McIlroy et al., 2017b). To achieve this, the MiDAS 2.1 taxonomy and sequence database files were amended to include the newly named “Ca. Catenibacter,” “Ca. Amarithrix,” “Ca. Trichobacter,” and “Ca. Defluviithrix” described here (Supplementary Table S1). This exercise has resulted in the group originally representing “Ca. Defluviifilum” (previously MiDAS 1.2 group P2CN44) being divided into three clades each representing “Ca. Defluviifilum,” “Ca. Amarithrix,” and “Ca. Defluviithrix” (Figure 1). This decision was taken because of the propensity of their 16S rRNA sequences to fall into three distinct clades after phylogenetic analysis (Figures 1, 2), where members of each clade generally shared c.a. > 95% 16S rRNA gene sequence similarity (based on a shared 1070 bp 16S rRNA gene alignment), and their reported morphological and physiological differences (Yoon et al., 2010; Kragelund et al., 2011; Speirs et al., 2017; Nierychlo et al., 2019). We believe it is prudent at this time to separate these phylotypes, as there is no convincing evidence to suggest they are representatives of the same genus.
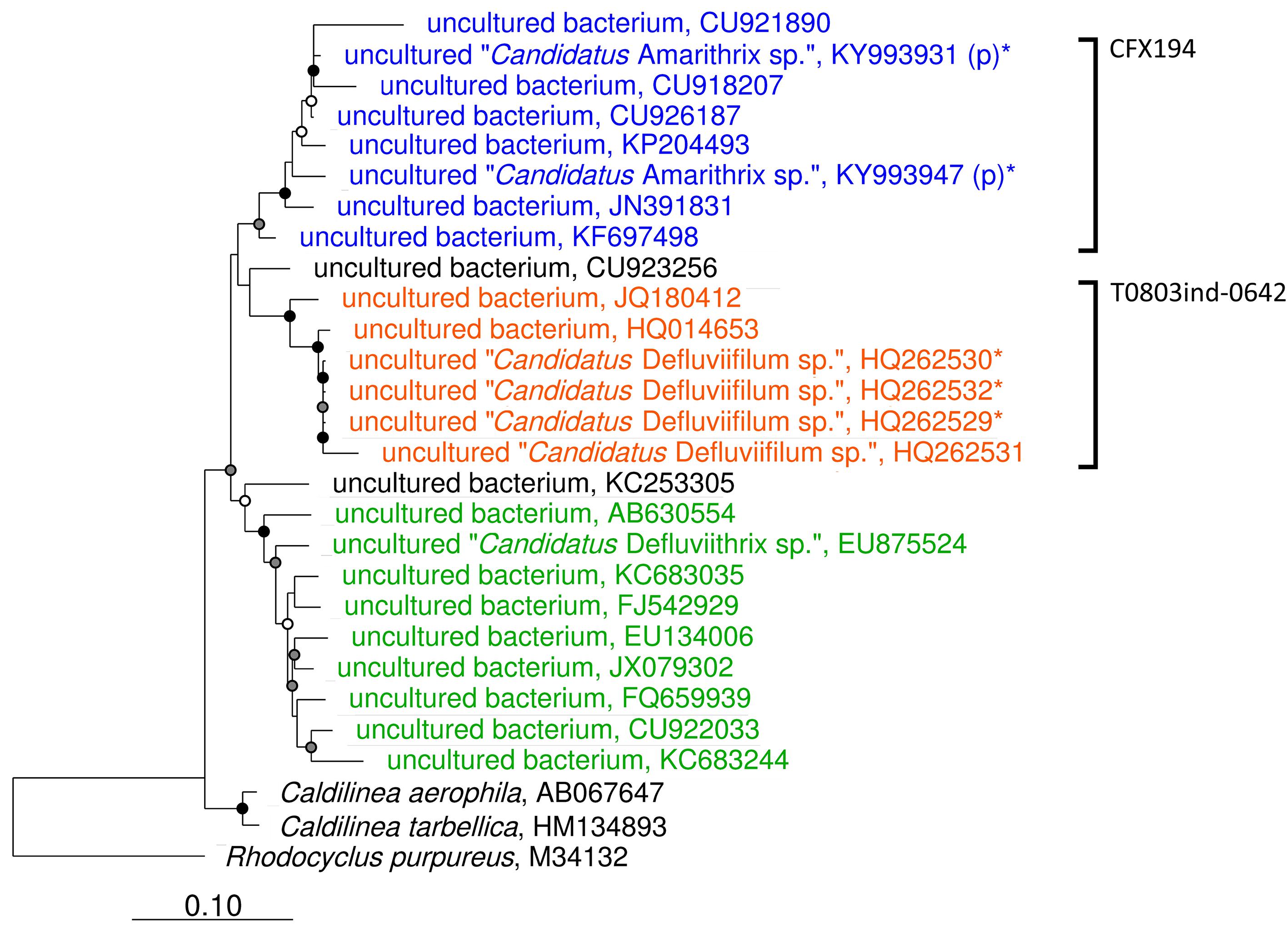
Figure 1. Maximum-likelihood phylogenetic tree showing the Caldilineae clades representing “Ca. Defluviifilum” (orange), “Ca. Amarithrix” (blue), and “Ca. Defluviithrix” (green). The tree was generated by ARB using the PhyMl model, and those sequences present in the MiDAS 2.1 database (McIlroy et al., 2017b). All sequences were >1200 bp in length, except for those partial sequences added using the ‘Quick add’ function in ARB, and identified by ‘(p).’ Sequences marked with ‘∗’ were also included as they derive from their respective filament population (Kragelund et al., 2011; Speirs et al., 2017). The scale bar corresponds to 0.1 substitutions per nucleotide. Parsimony bootstrap values were calculated as percentages of 1000 analyses, and values 50%–75% are indicated with a white circle, 76%–95% with a gray circle, and >96% with a solid black circle. Brackets to the right indicate probe coverage.
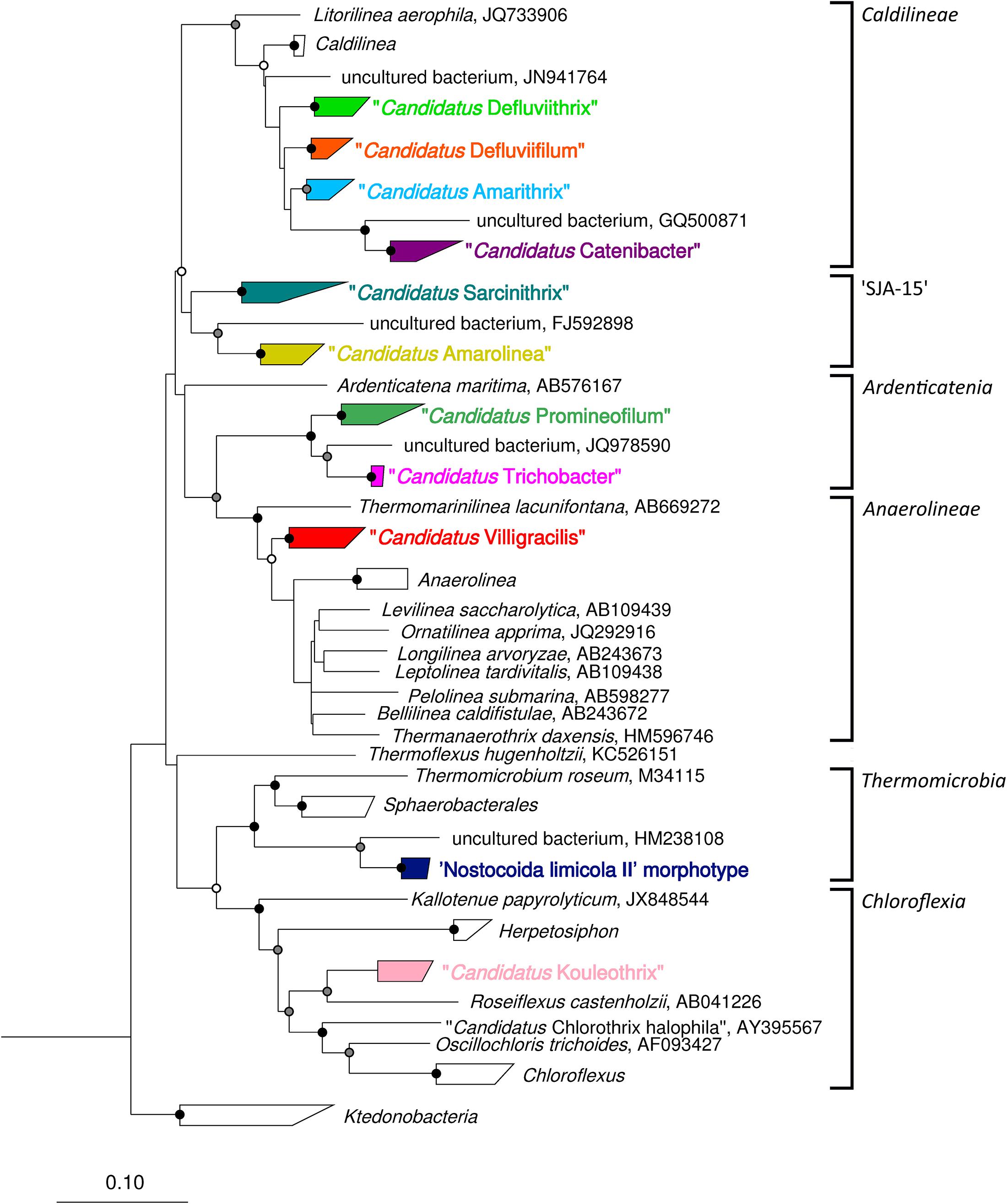
Figure 2. Complete maximum-likelihood phylogenetic tree for the filamentous Chloroflexi phylotypes reported in activated sludge. Sequences representing phylotypes are color coded: “Ca. Catenibacter” (purple), “Ca. Amarithrix” (blue), “Ca. Defluviifilum” (orange), “Ca. Defluviithrix” (light green), “Ca. Sarcinithrix (teal), “Ca. Amarolinea” (yellow), “Ca. Villigracilis” (red), “Ca. Trichobacter” (pink), “Ca. Promineofilum” (dark green), ‘Nostocoida limicola II’ morphotype (dark blue), and “Ca. Kouleothrix” (lilac). For more detail of the “Ca. Amarithrix,” “Ca. Defluviifilum,” and “Ca. Defluviithrix” clades, see Figure 1. The tree was generated by ARB using the PhyMl model, and sequences present in the MiDAS 2.1 database (McIlroy et al., 2017b). All sequences were >1200 bp in length, except for those partial sequences added using the ‘Quick add’ function in ARB. The scale bar corresponds to 0.1 substitutions per nucleotide. Parsimony bootstrap values were calculated as percentages of 1000 analyses, and values 50%–75% are indicated with a white circle, 76%–95% with a gray circle, and >96% with a solid black circle.
Data chosen for these analyses were based on the availability of high quality sequence data and PCR primer details, which had been generated with the Illumina platforms. The survey involved analyses of 288 individual Chloroflexi populations from eight countries in Asia, Europe and North America (Supplementary Data File S1). These data were analyzed using QIIME2 (Bolyen et al., 2018), and protocol details are given as Supplementary Information (Supplementary Data File S2). It was not within the capability of this review to assess which operational, environmental or geographical factors play any part in the distribution of these filaments, as in most cases this information was not provided. However, trends were seen in their distributions in plants both within and between countries, which warrant further investigation. This restricted survey serves as a means to reveal the global distribution and population abundances of these activated sludge Chloroflexi, using the updated version of the MiDAS 2.1 taxonomy database (McIlroy et al., 2017b).
Briefly, the data presented here (Table 4 and Supplementary Data File S1) reveal that “Ca. Villigracilis” and “Ca. Defluviithrix” sequences are widely distributed globally, being detected respectively in 81.3 and 71.2% of all the biomass samples examined. This is despite no mention of either of these in the identification manuals of Eikelboom and Jenkins (see earlier), and neither showed similarities to any of the morphotypes described there (Yoon et al., 2010; Nierychlo et al., 2019). Consequently all surveys based on microscopy and information in these manuals (Rossetti et al., 2017) would miss these. The filaments “Ca. Promineofilum,” “Ca. Sarcinithrix,” “Ca. Catenibacter,” and “Ca. Kouleothrix” were only slightly less frequently seen, occurring in 69%, 69%, 68% and 59% of biomass samples respectively, (Table 4 and Supplementary Data File S1).
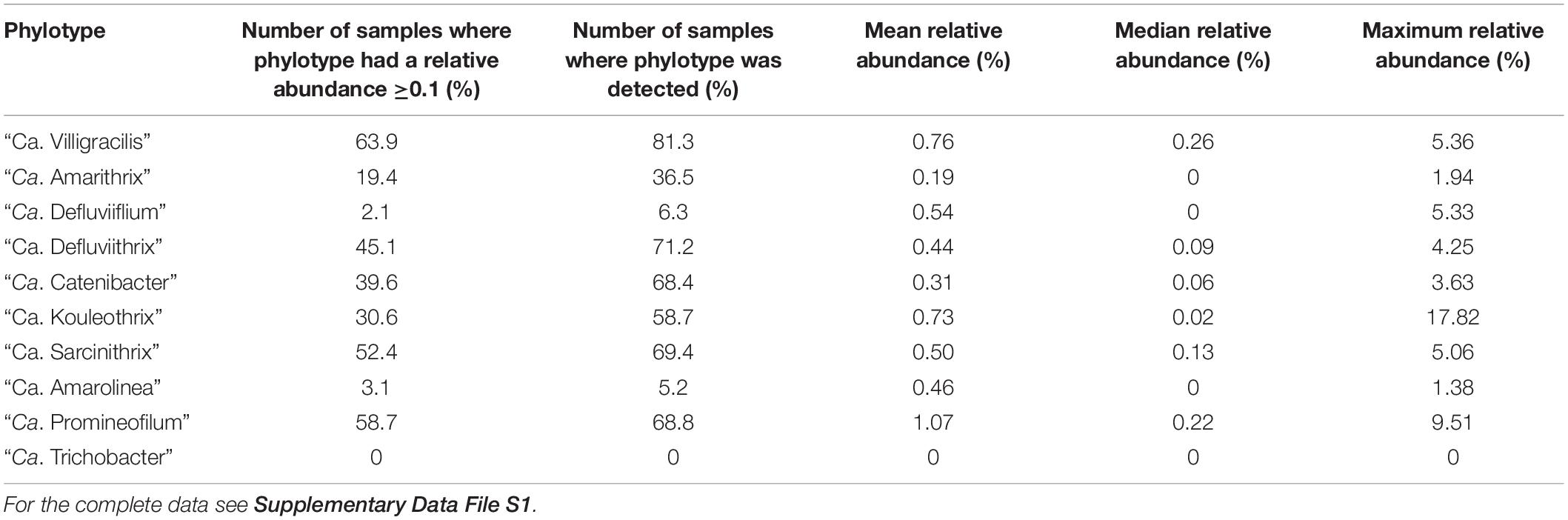
Table 4. Summary of abundances of the activated sludge filamentous Chloroflexi phylotypes distributed across the amplicon data survey.
Conversely, despite it being a highly abundant filament in some samples from Danish plants (Andersen et al., 2019; Nierychlo et al., 2019), “Ca. Amarolinea” was detected sporadically in only 5.2% of the global samples examined here, although only those taken from Danish plants at regular intervals during the yearly sampling period to reveal any marked seasonal changes were analyzed. “Ca Trichobacter” was not detected at all (Table 4 and Supplementary Data File S1). However, these latter two low values may reflect their low representation in the taxonomic database, being limited to two reference sequences each. The average relative abundance of Chloroflexi filaments was usually low at approx. 0.5% of the total population, thus reflecting abundances described in earlier reports. More details on the occurrence of each filament are given below.
Whole Genome Sequencing and Metagenomics of Chloroflexi in Activated Sludge
Activated sludge bacterial whole genome sequencing of pure cultures and metagenomic analyses of mixed communities containing organisms of interest have also been facilitated substantially by DNA ngs protocols, but have not yet been applied widely to the Eikelboom filament morphotypes. With uncultured filaments, analyses of enriched mixed cultures are commonly used to generate metagenomic data. The whole genome sequences of cultured and uncultured bulking filaments have been published (e.g., Lapidus et al., 2011; Kristiansen et al., 2013; McIlroy et al., 2013, 2015; Guo et al., 2015; Sekiguchi et al., 2015). These include the uncultured Chloroflexi filament “Ca. P. breve.” from a plant in Slovenia, and metabolic models have been constructed from it (McIlroy et al., 2016). The whole genome sequence of “Ca. Kouleothrix sp.” is also available (Ward et al., 2018), although incompletely annotated, as is that of “Ca. A. aalborgensis” (Andersen et al., 2019). Their major features will be discussed later. The value of having such information is that they reveal the complete potential metabolic road map of the organism, unlike MAR for example, where only a small snapshot of its physiology/metabolism is exposed. However, as McIlroy et al. (2018) emphasize, while genomes of most activated sludge bacteria of interest contain genes encoding proteins with the same or similar function, MAR is needed to demonstrate in situ whether these genes are expressed under the imposed plant conditions. Such metagenomic studies also provide the basis for transcriptomic studies looking at gene expression in situ in activated sludge populations e.g. (He and McMahon, 2011), but none have been published so far for Chloroflexi filaments.
The In Situ Physiology of the Chloroflexi in Activated Sludge
Attempts have been made to understand the in situ cophysiology of the Chloroflexi in a wide range of habitats, including the different operating conditions encountered in activated sludge treatment processes designed to remove N and P. Early MAR studies with the bulking filaments have been summarized by Nielsen et al. (2009b). Often the only FISH probes used to identify these were the CFX1223/GNSB941 probes, and so little information was generated about the ecophysiology of individual Chloroflexi filament populations. The still limited data available would suggest the Chloroflexi are specialized feeders with very similar metabolic needs, which is surprising in a group so phylogenetically and ecologically diverse (Nierychlo et al., 2019). Data from several FISH/MAR studies generally agree that they have a clear preference especially for simple sugars like sucrose, and for complex polymers and their products and amino acids, not just in activated sludge processes, but also in Anammox and membrane bioreactors (Kragelund et al., 2007a; Miura et al., 2007; Kindaichi et al., 2012). Nierychlo et al. (2019) also suggested that glycerol and long chain fatty acids could be used by some. Genomic information from anaerobic and aerobic Chloroflexi characterized from sediments (Hug et al., 2013) and anaerobic bioreactors (McIlroy et al., 2017a; Petriglieri et al., 2018) suggested similar nutritional preferences for the novel Chloroflexi detected there. Lysed bacterial cell wall debris in the form of the peptidoglycan monomer N-acetyl glucosamine is also commonly utilized (Nielsen et al., 2009b; Nierychlo et al., 2019).
Microautoradiography evidence from activated sludge filaments fails to support any widespread ability to utilize acetate, a major metabolite in nutrient removal plants, although Kragelund et al. (2007a) have evidence that “Ca. Kouleothrix” may. Furthermore, the cultured strain ETI described by Yoon et al. (2010) also grows on acetate as well as lactate and pyruvate as sole carbon sources. Generally these substrates were only assimilated under aerobic conditions, although genomic evidence for “Ca. P. breve” (McIlroy et al., 2016; Nierychlo et al., 2019), and MAR data for “Ca. Defluviifilum” (Kragelund et al., 2011) (see below), would suggest that statement does not apply to all the ‘aerobic’ filaments characterized in activated sludge (see later). Nierychlo et al. (2019) also showed by FISH/MAR that their four previously uncharacterized Chloroflexi in Danish plants assimilated some but not all substrates anaerobically. They shared a similar physiology in being chemoorganoheterotrophic facultatively anaerobic fermenters, using predominantly carbohydrates as substrates, and hence are similar essentially to “Ca. P. breve” (McIlroy et al., 2016). It is still unclear whether these can denitrify. All assimilated sugars under anoxic conditions with NO2/NO3 as terminal electron acceptors, but none could nitrify. Their individual attributes are discussed in more detail in Nierychlo et al. (2019) and below.
Methods used to monitor the synthesis in situ of exocellular depolymerase enzymes reveal how well suited these filaments are for degradation of complex polymers, although the assay systems used did not always reveal individual enzyme substrate specificities. Thus, Kragelund et al. (2007a) showed with “Ca. Kouleothrix” and FISH probes targeting this filament, that it produced a range of polysaccharide degrading enzymes including chitinases, glucuronidases, as well as proteases (Nielsen et al., 2009b). Interesting, their in situ MAR determined metabolic attributes did not always agree with those obtained with the corresponding pure cultures of this filaments (Kragelund et al., 2007a). Later, “Ca. Defluviifilum” (Kragelund et al., 2011) was shown also to synthesizes exocellular enzymes able to metabolize a range of macromolecules. Such attributes suggest an active role for them in the hydrolysis of particulate and colloidal material suspended in the mixed liquor (Nierychlo et al., 2019). They could also assimilate glucose under aerobic and anoxic conditions with nitrite or nitrate acting as electron acceptors, consistent with them behaving as denitrifying bacteria in situ (but see above). With so few Chloroflexi cultured and so few in situ FISH/MAR studies performed outside of Denmark, it seems probable that globally their ecophysiology will eventually reflect more closely their considerable phylogenetic diversity, as already suggested by Ward et al. (2018) for “Ca. Kouleothrix” and by Sorokin et al. (2014) for the chemolithoautotrophic nitrifying non-filamentous Nitrolancea hollandica.
Description of the Chloroflexi Present in Activated Sludge Communities
Sufficient FISH and 16S rRNA amplicon sequence data are now available to make the task of identifying novel members of activated sludge Chloroflexi more straightforward. Thus, retrieved cloned 16S rRNA sequences, in combination with appropriate FISH probe analyses allow probed plant samples to be screened, revealing which of the positive and negative probe target sites the clones of interest may possess (Speirs et al., 2009). Then new FISH probes can be designed against these clone sequences, and applied to samples, where only the filaments of interest will fluoresce. This approach has been used successfully to identify “Ca. Promineofilum,” “Ca. Defuviifilum,” “Ca. Sarcinithrix,” and “Ca. Trichobacter” (Speirs et al., 2009, 2011, 2015; Kragelund et al., 2011; McIlroy et al., 2016; Table 1). “Ca. Amarithrix” and “Ca. Catenibacter” were identified by a similar method, but using amplicon sequencing to retrieve the 16S rRNA gene sequences of interest instead (Speirs et al., 2017). Correspondingly, amplicon sequence data have been used to help identify and map the distribution of Chloroflexi in plants of different configurations in several countries, and in combination with new FISH probes designed against these sequences. Used with MAR, these have contributed toward the elucidation of the ecophysiology of novel Chloroflexi in Danish treatment plants (Andersen et al., 2019; Nierychlo et al., 2019).
The features of interest of individual Chloroflexi filaments detected initially by the phylum level FISH probes CFX1223 and GNSB941 in activated sludge plant samples, and from amplicon sequencing data are described next. Several schemes are available to classify and identify 16S rRNA gene sequences. These are all ‘special purpose’ systems based on different classificatory principles for individual user groups (Seviour, 2010b; Balvočiūtė and Huson, 2017), and often differ in how they nominate and name individual Chloroflexi clades (see McIlroy et al., 2015, 2017a). Hence, the classifications of their 16S rRNA sequences have in some cases changed as later systems have become available (see earlier, Table 3). For example, the “Ca Sarcinithrix” morphotype 0914 Chloroflexi filaments (Speirs et al., 2011) were considered initially to belong to the subgroup 1 of the Chloroflexi, a clade thought to embrace predominantly sequences derived from activated sludge (Björnsson et al., 2002). Later, these filaments were classified as members of the Chloroflexi class Caldilineae, using the quality checked and manually curated SILVA taxonomy (Quast et al., 2013; Speirs et al., 2015), and then lastly attributed to the Chloroflexi group ‘SJA-15’ with the more recent MiDAS taxonomies (McIlroy et al., 2015, 2017b). The MiDAS scheme is based on the SILVA taxonomy, but differs in that it has been curated to include more attributes pertaining to activated sludge organisms, and thus for use specifically by those investigating activated sludge microbial communities (McIlroy et al., 2015). Hence, the MiDAS taxonomy has become clearly the best option for identifying the filamentous Chloroflexi discussed here, and this taxonomy and nomenclature, where appropriate, has been used in the following section.
Anaerolineae
“Ca. Villigracilis” (Nierychlo et al., 2019).
Vi lli gra ci lis; L. m. n. Villo, tuft of hair; L. fem. Adj. gracilis, slender slim.
Villigracilis, a filamentous bacterium often occurring in bundles.
“Ca. Villigracilis” was described by Nierychlo et al. (2019) as one of the more abundant and frequently seen Chloroflexi filamentous bacteria in Danish wastewater treatment plants, and plants elsewhere (Table 4 and Supplementary Data File S1). This filament was formerly referred to as phylotype ‘SBR1029’ in the MiDAS 1.2 release. It possesses thin (0.3–0.4 μm × 15–50 μm) filaments, which were always located within the flocs, where they probably contribute to the matrix supporting floc formation. They frequently grow as bundles. It was not possible to determine by FISH which of the Eikelboom filaments it most closely resembled because of its location (Björnsson et al., 2002; Nierychlo et al., 2019). “Ca. Villigracilis” is a member of the Anaerolineaceae, in the Anaerolineae, and is the only facultatively aerobic member described so far (Nierychlo et al., 2019), with the remainder being strictly anaerobic (Sekiguchi et al., 2001; Yamada et al., 2006, 2007; Sun et al., 2016; McIlroy et al., 2017a). Two subgroups, A and B are recognized, and probes CFX763A and CFX763B designed to target respectively each (Table 1) collectively cover > 60% of the MiDAS 2.1 database sequences for members of this genus (Nierychlo et al., 2019). The amplicon survey performed here support the view that this phylotype is a core activated sludge member, and is present in 81% of the samples analyzed from around the world (Table 4 and Supplementary Data File S1). The mean and maximum relative abundances were found to be 0.76 and 5.36% of the total population respectively in the plants examined here.
Ardenticatenia
“Ca. Promineofilum” (McIlroy et al., 2016).
Pro mi nee oh L. v. combining form of prominere, to project, to jut out; filum L. n. n. filament. Promineofilum, a short filament protruding from flocs.
“Ca. Promineofilum” is the genus name proposed for the Eikelboom filament morphotype type 0092 (McIlroy et al., 2016). This filament was formerly referred to as phylotype ‘B45’ in the MiDAS 1.2 release. The morphotype appears as blunt ended rods protruding from the floc surface, or less commonly, loose in the bulk liquid. It has been associated with foaming, but more likely ends up there accidentally (de los Reyes, 2010), since its cells are not hydrophobic (McIlroy et al., 2016). The filaments stain a distinctive lilac color with the Neisser stain, and so can be recognized readily microscopically. Two Neisser positive morphological variants differing in their trichome diameters were seen by Speirs et al. (2009) in an EBPR plant in Australia, and the thicker variant is the one described here and by McIlroy et al. (2016). These filaments are seen in most activated sludge plants around the world (Table 4 and Supplementary Data File S1), but probably more frequently and at higher abundances in EBPR plants designed to remove N and P microbiologically, where the thicker variant is more abundant than its thinner relative, at least in Australia (Speirs et al., 2009).
McIlroy et al. (2016) have shown from genomic annotation of the Cfx-K genome of “Ca. P. breve” that this filament possesses two chromosomes and a plasmid. Its phylogeny is confused. Based on 16S rRNA sequence data, it is most closely related to the Anaerolineae or Ardenticatenia, and probably represents at least a novel order. The MiDAS 2.1 taxonomy places it currently within the Ardenticatenia. The Candidatus name “Ca. Promineofilum breve” has been proposed for the species from which the Cfx-K genome was derived (McIlroy et al., 2016). Parks et al. (2018) in their GTDB taxonomy have suggested it should be a founder member of a new order, the Promineofilales.
“Ca. P. breve” is a facultatively anaerobic chemoorganoheterotroph, and thus able to obtain energy by both aerobic respiration and fermentation, probably of carbohydrates. MAR experiments showed it could only assimilate glucose, and not amino acids or short chain fatty acids. Glycogen appears to be stored, allowing the filament to survive the inevitable periods of substrate limitation encountered in activated sludge systems. This ability to respire and ferment sugars may explain its increased competitiveness in EBPR removal plants (Jenkins et al., 2003), where it is able to grow under the anaerobic conditions encountered there. In conventional aerobic plants “Ca. Promineofilum” is seen less frequently, possibly because it is unable to compete with the obligatory aerobic members of the community.
Whether it can denitrify is still unclear. No putative nitrate reductase or nitric oxidase encoding genes were present, although a periplasmic nitrous oxide reductase was seen, which suggests nitrous oxide could act potentially as terminal electron acceptor in denitrification. Some genomic data suggest an ability to grow chemolithoautotrophically, possessing as it does the genes for CO2 fixation using the Calvin-Benson-Bassam cycle. However, no genes were seen to suggest any ability to oxidize inorganic energy sources. Like “Ca. Trichobacter,” these filaments fail to hybridize with any of the EUB338mix FISH probes (Table 2). Speirs et al. (2009) have designed two FISH probes, the CFX197 probe which hybridizes with the thicker 0092 morphotype while the CFX223 probe targeted the thinner variant (Table 1 and Figure 2). Whether these represent different ‘species’ or strains of this filament is unknown. The fluorescence image generated from FISH probing suggests the ribosomes are not distributed evenly in the cells, but in patches (Speirs et al., 2009). It is not clear why, but they may be attached to intracellular membranous structures like those reported for the Chloroflexi filaments described by Yoon et al. (2010).
Results from the amplicon data survey (Table 4 and Supplementary Data File S1) agree with and extend earlier observations, and indicate this filament is a common member of activated sludge communities, being present in 68.8% of biomsass samples examined where its mean relative abundance was 1.07% of the total population, and it attained a maximum relative abundance of 9.51%.
“Ca. Trichobacter” (name proposed in this review; Supplementary Table S2).
Trichobacter; Tri ko bac ter. Gr. neut. n. tricho hair; Gr. n. masc. bac ter rod. Trichobacter, a filamentous bacterium consisting of chains of rod shaped cells.
“Ca. Trichobacter” is the name proposed here for the Eikelboom type 0803 morphotype filaments detected in Australian plants, which do not respond to the FISH probes designed by Kragelund et al. (2011) against “Ca. Defluviifilum,” which possesses the same morphotype. It is usually seen at low levels in Australian plants, with abundances well below those of “Ca. Defluviifilum” (Speirs et al., 2015).
The biomass sample from which it was isolated (Speirs et al., 2015) came from a domestic treatment plant in Victoria, Australia, where this filament was at very high abundance, and was held responsible for the bulking event occurring there. It belongs to the Ardenticatenia, and thus is phylogenetically distant to “Ca. Defluviifilum,” being more closely related to “Ca. Promineofilum” (Speirs et al., 2009; McIlroy et al., 2016), with which it shares the same EUB338mix probe variant target site mismatches (see above).
One probe is available for “Ca. Trichobacter” (Table 1 and Figure 2; Speirs et al., 2015). Seven of the 32 Australian activated sludge samples surveyed contained this filament. “Ca. Trichobacter” sequences were included in the amended MiDAS 2.1 database used here for the global amplicon data survey performed for this review (Table 4 and Supplementary Data File S1). However, no examples of this filament were detected in any other plant.
Caldilineae
“Ca. Defluviifilum” (Nierychlo et al., 2019).
De flu v’ i’ i’ fi’um. L. n. defluvium, sewage; L. neut. N. filum, thread. Defluviifilum, a filamentous bacterium in sewage.
“Ca. Defluviifilum” formerly referred to as the phylotype ‘P2CN44’ in the MiDAS 1.2 release (McIlroy et al., 2015), is the name proposed for the uncultured Eikelboom type 0803 filament described by Kragelund et al. (2011) in plants in Denmark, treating domestic and pulp and paper wastes. Phylogenetic analysis places it within the class Caldilineae. This filament possesses the morphological features considered diagnostic for this morphotype (Jenkins et al., 2003; Eikelboom et al., 2006), although being so few in number makes its morphotype microscopic ‘identification’ problematic (Kragelund et al., 2011). The short, straight hydrophobic filaments consisting of square/rectangular cells are often located on the floc surface, but may also form interfloc bridges. No PHA or polyP inclusions were revealed after staining, and no S0 granules detectable by phase contrast. It can be difficult to distinguish this filament from “Ca. Sarcinithrix” (see later), except that its filaments are thinner, and those in bundles tend to align in parallel, while those in “Ca. Sarcinithrix” often traverse each other. The FISH probes now available should resolve this problem.
FISH based surveys of Danish nutrient removal plants (Kragelund et al., 2011) reveal this filament is a common component of the community, and its relative abundances remained quite stable over time. FISH analyses showed it was always filamentous in situ, and “Ca. Promineofilum” filaments were always present with it. “Ca. Defluviifilum” has also been seen at consistently high abundance in an A2O EBPR plant in Japan over a 12 month period.
It was recommended (Kragelund et al., 2011) that their T-0803-0654 FISH probe be used to detect “Ca. Defluviifilum” in plants treating domestic wastes, while the T-0803ind-0642 probe was recommended for those treating industrial wastes. However, Speirs et al. (2015, 2017) have shown that the T0803-0654 probe target site also occurs in several other 16S rRNA sequences with as little as 73% sequence similarity to the “Ca. Defluviifilum” target group. Because of this, the abundance values for this phylotype have probably been artificially inflated in the data derived from FISH surveys (see above). Thus, we recommend that the T0803-0654 probe be no longer used and the T0803ind-0642 probe be used instead, since it was designed to be more specific (Table 1 and Figure 1).
As mentioned already, it was decided for this review to incorporate the “Ca. Amarithrix” and “Ca. Defluviithrix” phylotypes into the MiDAS 2.1 database (Figure 1), which resulted in the number of sequences attributed to the “Ca. Defluviifilum” decreasing substantially. The survey of amplicon data performed here (Table 4 and Supplementary Data File S1) indicate that “Ca. Defluviifilum” was present in 6% of activated sludge samples, with an average relative abundance of 0.57% of the total population, and a maximum relative abundance of 5.33%. These values are generally lower than those given in previous publications (e.g., McIlroy et al., 2015, 2017a; Nierychlo et al., 2019), although not unexpected, because of our reallocation of reference sequences representing this group. This strategy makes any further comparison with previous data problematic, and additional analyses will be required.
“Ca. Defluviithrix” (name proposed in this review; Supplementary Table S2).
De fluv’ i’ i thrix L. neut. n. defluvium sewage; Gr. fem.n. thrix hair; Defluviithrix a filamentous bacterium in sewage.
In 2010, Yoon et al. (2010) cultured seven strains of an unbranched, aerobic, mesophilic filamentous Chloroflexi isolate ET1 in Korea, which they claimed was the first aerobic Caldilineae retrieved from activated sludge (Nierychlo et al., 2019). No apparent sheath was present, and septa were rarely seen. They were unable to relate it definitively to any of the Eikelboom filament morphotypes, but suggested it most closely resembled morphologically Haliscomenobacter hydrossis, a member of the Bacteroidetes (Seviour and Nielsen, 2010). Its 16S rRNA sequence is most closely related (92%) to that of “Ca. Defluviifilum” (described above) and “Ca. Amarithrix” (described below) (Figure 1). Unusual intracellular membrane arrangements and inclusion bodies of an unknown composition were seen. Evidence was presented suggesting that pure cultures of “Ca. Defluviithrix” strain ETI fitted the common Chloroflexi nutritional profiles, in being chemoorganoheterotrophic, and capable of using a limited range of biopolymers, organic acids and simple sugars (not glucose), but only under aerobic conditions (Yoon et al., 2010). No phylotype specific probes were reported for this filament. However, the CFX1A331 probe (Table 1), designed to target members of the Caldilineae including “Ca. Defluviithrix,” showed this filament was most often located within the flocs (Yoon et al., 2010).
“Ca. Defluviithrix” is widely distributed globally, being found in many plants in other parts of the world, as revealed from amplicon sequencing data where it occurred in 71% of the samples surveyed, at a mean relative abundance of 0.44% of the total population and maximum relative abundance of 4.25% (Table 4 and Supplementary Data File S1). As this phylotype is not included in the MiDAS database, comparisons with previous data are not possible, although it seems likely that “Ca. Defluviithrix” has contributed a substantial portion of the “Ca. Defluviifilum” sequences detected in previous amplicon studies. Indeed, here it was the most widely distributed member of the MiDAS 1.2 ‘P2CN44’ group that also includes “Ca. Defluviifilum” and “Ca. Amarithrix.” No success has been reported in culturing any bulking Chloroflexi filaments other than this strain and “Ca. Kouleothrix spp.,” although several anaerobic Anaerolineae filaments from a range of habitats other than activated sludge have been obtained in pure culture (Sekiguchi et al., 2003; Yamada et al., 2006, 2007; Sun et al., 2016; McIlroy et al., 2017a).
“Ca. Amarithrix” (name proposed in this review; Supplementary Table S2).
A mar i thrix; Gr. n. a mar a, sewage duct; N L. masc. n. thrix, thread; Gr. Amarithrix a filamentous bacterium from sewage.
“Ca. Amarithrix” is the name proposed here for a filament sharing features with the Eikelboom morphotype 0675. In many microscopic surveys, this morphotype is often grouped with the Eikelboom type 0041 filament, because of claims (Seviour et al., 1990), that while both have characteristic abundant attached growth on their trichomes, consisting of almost square cells, they could not be separated reliably on the Eikelboom key attribute of differences in their trichome diameters. Type 0675 is described (Jenkins et al., 2003; Eikelboom et al., 2006) as being thinner than type 0041. Whether they represent different morphotypes of the same filament and hence share similar ecologies, or were two phylogenetically distinct filaments has been resolved for Australian strains. It is now clear that although both are members of the Caldilineae in the phylum Chloroflexi they represent two distinct phylotypes belonging to related taxa (Speirs et al., 2017; Figure 2). The CFXmix FISH probes are recommended for initial probing to eliminate filaments belonging to the Betaproteobacteria, followed by the Caldi-0678 probe for the Caldilineae, before the probes CFX194a and CFX194b designed by Speirs et al. (2017) are applied as a mix (Table 1), together with the helper and competitor probes designed for them. FISH based surveys with these two probes (Speirs et al., 2017) would suggest that “Ca. Amarithrix” is more commonly seen in nutrient removal than conventional plants in Australia. However, a few type 0675 morphotypes responded to the CFXmix probes, but not to the CFX194mix probes, suggesting that further phylogenetic diversity within the Chloroflexi exists for this morphotype. At this point no genome data have been generated for this organism. They fall into the MiDAS 2.1 defined “Ca. Defluviifilum” genus, but Speirs et al. (2017) suggest they should be placed currently into a separate genus, based on 16S rRNA sequence similarity values and morphological differences.
For the purposes of this review, we have amended the MiDAS 2.1 database to include “Ca. Amarithrix” (Supplementary Table S1), where it reallocates several “Ca. Defluviifilum” sequences (Figure 1). This filament was detected in 37% of samples with a mean relative abundance of 0.2% of the total populations and maximum relative abundance of 1.94% (Table 4 and Supplementary Data File S1).
“Ca. Catenibacter” (name proposed in this review; Supplementary Table S2).
Ca ten i bac’ ter; L. fem.n. Ca te n i chain; Gr. hyp. masc. ba’cter rod; Catenibacter a bacterium consisting of chain of rod shaped cells.
“Ca. Catenibacter” is the name proposed here for the Chloroflexi Eikelboom morphotype type 0041. As mentioned above, Eikelboom (1975) separated these from type 0675 on differences in their trichome diameters, and type 0041 has been combined with type 0675 in most microscopy based surveys (Seviour et al., 1990; Kragelund et al., 2011). Both can now be separated and hence identified by FISH (Speirs et al., 2017). They share the same general morphological attributes and are distinguished by abundant attached growth on their trichomes. To date, neither has been cultured. FISH survey studies in the past have suggested that the 0041 morphotype is polyphyletic, since some have been claimed to respond to probes targeting members of the phylum “Ca. Saccharibacteria,” previously named TM7 (Hugenholtz et al., 2001; Thomsen et al., 2002; Müller et al., 2007; Mielczarek et al., 2012), and others with Curvibacter in the Betaproteobacteria (Thomsen et al., 2006).
However, the early surveys were performed before the FISH probes for the Chloroflexi (Björnsson et al., 2002) were available, and doubts have since been raised about the specificity of the TM7 FISH probes of Hugenholtz et al. (2001) against filamentous members of this phylum. Thus, Nittami et al. (2014) showed that none of the type 0041/0675 filaments they saw in either Australian or Japanese plants fluoresced with the TM7-305 probe designed to target the “Ca. Saccharibacteria” phylum, including putative type 0041 “Ca. Saccharibacteria” filaments (Hugenholtz et al., 2001). Instead their TM7 FISH probes hybridized consistently with “Ca. Kouleothrix” filaments responding positively to the CHL1851, EU25-1238 and CFXmix probes (see below).
Speirs et al. (2017) designed a FISH probe, which fluoresced only with filaments with the Chloroflexi type 0041 morphology (Table 1 and Figure 2). This CFX86 probe should be used together with the corresponding helper probes, which enhance fluorescence signal strength (Speirs et al., 2017). Surveys using this probe showed these filaments can attain very long trichome lengths (>1000 μm) and extend from flocs leading to interfloc bridging. It is widely distributed globally, being present in 69% of the samples analyzed (Table 4 and Supplementary Data File S1). Its mean relative abundance was similar to the other phylotypes at 0.31% of the total population and a maximum detected relative abundance of 3.63%. Its presence in plants did not always coincide with that of “Ca. Amarithrix.”
Chloroflexi
“Ca. Kouleothrix” (Kohno et al., 2002).
The Eikelboom morphotype 1851 filaments often staining weakly Gram positive, are now considered to be members of the genus “Ca. Kouleothrix,” containing the filamentous isolate “Ca. K. aurantiaca” (Kohno et al., 2002; Kragelund et al., 2007a), and being only approx. 84% similar in their phylogeny to their nearest isolated relative, Roseiflexus castenholtzii. The Australian isolate shares little of this phenotype, being neither a gliding thermotolerant filament, nor one producing carotenoid pigments (Beer et al., 2002), yet it belongs to the phototrophic members in the class Chloroflexia. This observation confirms that close phylogenetic relatedness is not always reflected in a shared physiology/biochemistry.
The Japanese “Ca. Kouleothrix” cultured strains of Kohno et al. (2002) are 99% similar to the Danish cultured strain Ver9Iso2 (Kragelund et al., 2007a), but as both are only 93–95% similar to the Australian type 1851 cultured isolate of Beer et al. (2002), it seems likely that more than one species/genus exists. This filament is seen commonly in plants of all configurations treating both domestic and industrial wastes around the world (Beer et al., 2002; Kragelund et al., 2007a), and especially in Japan Nittami et al. (2017), where its excessive growth commonly leads to bulking. This often involves interfloc bridges of bundles of thin filaments (Beer et al., 2002; Nittami et al., 2017), which are usually, but not always, covered with attached epiphytic bacterial growth, where the attached cells appear distinctively perpendicular to the filament surface. The proclivity of this filament to cause bulking appears high, and analyses of 16S rRNA amplicon data (Table 4 and Supplementary Data File S1) show this filament was present in 59% of activated sludge samples. The average relative abundance was 0.7% of the total population, Real time qPCR methods (Nittami et al. (2017) have been applied to quantify “Ca. Kouleothrix” (type 1851) to determine if a threshold value existed for this filament above which bulking occurred, but comparisons with the FISH based data of Liao et al. (2004) are problematic. As well as using different quantitative methodologies, each used a different value for the contentious sludge volume index (Schuler and Jassby, 2007) to define what represents a bulking sludge.
The whole genome sequence of “Ca. K. aurantiaca” COM-B (JCM 19913) has revealed that it does not behave as a phototroph, despite it possessing the genes required for anoxygenic phototrophy (Ward et al., 2018). Thus, encoded are the complete bacteriochlorophyll biosynthetic pathway, a cytochrome bc complex and a Type 2 reaction center (RC2), as well as genes for RuBisCO and phosphoribulokinase. So it appears to be capable of CO2 fixation using the Calvin cycle pathway. However, missing are genes encoding for the 3-hydroxypropionate cycle, the pathway for CO2 fixation in the genera Chloroflexus and Roseiflexus, its closest relatives (Ward et al., 2018).
The probes CHL1851 and EU25-1238 have been used in surveys of European industrial wastewater treatment plants, where Kragelund et al. (2007a) showed that about 50% of EU-1238 positive “Ca. Kouleothrix” filaments failed to respond to the CHL1851 probe, even though the EU25 isolate possessed its target site. No explanation for this is available. In those plants treating largely domestic wastes, the CHL1851 probe could detect this filament morphotype. Thus, for the in situ detection of “Ca. Kouleothrix,” both these probes should be applied together (Table 1 and Figure 2). Guo and Zhang (2012) raised concerns about the apparent low specificity of the CHL1851 probe of Beer et al. (2002), and proposed a replacement FISH probe, T1851-2 for this filament. However, there are no reports of its use. They claimed then only few members of the Chloroflexia possessed the T-1851-2 probe probe target site, but suggested it was unsuitable for the type 1851 strain described by Kohno et al. (2002), having two mismatches with it.
“SJA-15”.
“Ca. Sarcinithrix” (Nierychlo et al., 2019).
“Ca. Sarcinithrix” is the name given by Nierychlo et al. (2019) to the Eikelboom morphotype 0914. It was first identified in a sample taken from a badly bulking sequencing batch reactor activated sludge plant treating domestic wastes in South Australia, operating with a long sludge age of 20 days (Speirs et al., 2011). It is a member of the MiDAS 2.1 class ‘SJA-15.’ The filaments respond to the EUB338 mix and CFX1223/GNSB941 mix probes. FISH based surveys suggested that “Ca. Sarcinithrix” was seen generally at higher relative abundances in nutrient removal plants operating at long sludge ages (Speirs et al., 2011) in Australia and Denmark (McIlroy et al., 2015). The two FISH probes CFX67a and CFX67b were the first designed against this type 0914 morphotype (Table 1 and Figure 2), and both hybridized to filamentous bacteria having the type 0914 morphology. Helper probes were essential for use with the general CFXmix and EUB338-1 probes, as well as for both the CFX67a and CFX67b probes, as discussed by Speirs et al. (2011). Similar requirements were reported by Nierychlo et al. (2019). As with “Ca. Promineofilum” (Speirs et al., 2009), an uneven FISH signal was seen from individual cells, suggesting uneven distributions of ribosomes in them (Speirs et al., 2011).
Nierychlo et al. (2019) showed subsequently, that the CFX67a probe did not impart fluorescence to the majority of “Ca. Sarcinithrix” filaments seen in Danish plants exposed to it, and failed to target several full-length sequences classified to this genus in the MiDAS databases (McIlroy et al., 2015, 2017b). They designed two new FISH probes CFX449 and CFX1151 (Table 1 and Figure 2), which they suggested should be used together, and which individually cover > 85% of the relevant sequences in the MiDAS 2.1 database. Helper probes were also designed, which failed to enhance fluorescence intensity, but generated a more even fluorescence signal (Nierychlo et al., 2019).
“Ca. Sarcinithrix” appears widespread globally and was detected in 69% of the biomass samples analyzed in the global survey (Table 4 and Supplementary Data File S1). Mean relative abundance in these samples was 0.50% of the total population, and a maximum relative abundance of 5.06%. As stated above, “Ca. Sarcinithrix” can cause serious bulking episodes (Speirs et al., 2011) and together with their widespread distribution, suggests this filament is an organism worthy of more attention.
“Ca. Amarolinea” (Andersen et al., 2019).
A ma ro li’ ne. a Gr. n. amaro conduit, channel, sewage duct; L.fem.n. linea, a thread, a line; Amarolinea a thread from a sewer.
“Ca. Amarolinea,” previously referred to as ‘C10 SB1A’ in the MiDAS 1.2 database, replaces the earlier name “Ca. Amarilinum,” and belongs to the MiDAS 2.1 class ‘SJA-15,’ which also contains “Ca. Sarcinithrix,” and with which it shares several metabolic traits (Andersen et al., 2019). The only FISH probe available, CFX64 (Table 1), was designed against abundant amplicon OTUs 3 and 4592 detected in Danish activated sludge samples (Nierychlo et al., 2019) and the available most closely related full-length sequences. Nierychlo et al. (2019) claim this probe should cover most of the “Ca. Amarolinea” filaments in Danish plants, although how common this filament was elsewhere was not known then. The survey of amplicon data conducted here indicates its distribution is not widespread, being detected in only 5.2% of biomass samples examined here (Table 4 and Supplementary Data File S1). Its relative abundance in these samples was comparable to those of other Chloroflexi phylotypes at 0.5% of total cell community, with a maximum relative abundance of 1.4%. Andersen et al. (2019) have shown that relative abundances as high as 30% have been recorded in some EBPR plants in Denmark for this filament, but even in these plants, abundances were often much lower at other sampling times and in some cases undetectable. At high abundance it probably contributes to sludge bulking episodes (Andersen et al., 2019). Thus, few “Ca. Amarolinea” sequences are present in public 16S rRNA gene sequence databases, and a BLAST search of the Genbank database using the “Ca. Amarolinea” 16S rRNA sequence (MH537630) as reference, and including those generated by amplicon sequencing (approx. 100 - 500bp), revealed only 17 entries with greater than 95% similarity to it. Exceptions to this are those in MBR plants in Korea and especially Taiwan, where “Ca. Amarolinea” was at high abundance in the all the samples examined, which were taken in the winter (Table 4 and Supplementary Data File S1). It should also be mentioned here that Jiang et al. (2018) showed it was a highly abundant filament (18% of total 16S rRNA sequences) in a foaming anoxic reactor in Hong Kong, depite the fact that it does not behave as a hydrophobic filament in situ (Andersen et al., 2019).
It is a non-motile Gram-negative filament (1–2.2 μm × 20–140 μm) with rectangular cells and no visible septa, staining violet/blue with the Neisser stain, and lacking polyphosphate and polyhydroxyalkanoate storage granules. Based on the distinctive Neisser staining reaction, this filament most closely resembles the Eikelboom type 0092 morphotype, as it too is Neisser positive in being distinctively violet/blue. However, the short blunt-ended filaments are thicker (1.5 μm) than those seen with “Ca. Promineofilum,” and are usually located entirely within the flocs, making them difficult to see microscopically.
A genome has now been sequenced for a member of this genus, and has been given the name “Ca. Amarolinea aalborgensis” (Andersen et al., 2019). The phylogeny of this filament appears to differ depending on the assessment criteria. 16S rRNA gene sequence analysis indicates it is a member of the class ‘SJA-15’ (Figure 2), while protein analyses place it within the class Anaerolineae. Based on the GTDB database of single copy marker genes (Parks et al., 2018), Andersen et al. (2019) propose that it should now be placed in a new family within the order Caldilineales, the Amaroliniaceae, with “Ca. Amarolinea, aalborgensis” sp. nov. as its sole member.
It is facultatively anaerobic, being able to ferment carbohydrates as well as being capable of aerobic respiration with a fully functional tricarboxylic acid cycle and cytochrome C oxidase as terminal transport chain donor. All the genes needed for glycolysis are present. “Ca Amarolinea” can carry out dissimilatory NO3 reduction to NH3, possessing genes encoding nitrate and nitrite reduction, but lacking the genes for nitrite acid and nitc acid reductases. It can also store anaerobically glycogen, which may be used as a potential energy source (Andersen et al.,2019), and thus enable it to compete anaerobically with the PAO Tetrasphaera spp. (see earlier) in EBPR plants.
Thermomicrobia
Nostocoida limicola II Chloroflexi
While the Eikelboom Nostocoida limicola II morphotype has been embraced within the Alphaproteobacteria and Actinobacteria (Seviour and Nielsen, 2010), it is also shared by a filamentous member of the Chloroflexi, class Thermomicrobia (Hugenholtz and Stackebrandt, 2004), and has been cultured (Schade et al., 2002).
The probe AHW183 (Schade et al., 2002) designed from the 16S rRNA sequences of four cultured isolates is the only probe available (Table 1 and Figure 2), although it has rarely been used in published surveys, and so little is known of the occurrence of this filament globally. Furthermore, the accession for this probe target (HM316086) is not listed in the MiDAS 2.1 database as representing a filamentous population, and its identification proceeds only to order level (‘JG30-KF-CM45’) within the class Thermomicrobia.
Nitrolancea hollandicus (Sorokin et al., 2014)
The isolation of a non-filamentous nitrite oxidizing nitrifying Chloroflexi, in the class Thermomicrobia from a lab-scale nitrifying bioreactor operating at 35°C by Sorokin et al. (2012, 2014) is a reminder of how poor our present understanding of activated sludge Chloroflexi diversity and ecophysiology currently is. FISH probes have also been described for its in situ identification (Table 1). These have shown that, as with the proteobacterial Nitroso and Nitro bacteria discussed earlier, N. hollandica occurs as large clusters located closely adjacent to the Nitroso bacteria, with whom they presumably form a mutualistic relationship similar to that already discussed above for the other nitrifying bacteria.
The key question is whether N. hollandicus is present and active in large-scale nitrifying activated sludge systems. Sorokin et al. (2012) suggest from its ecophysiology that this organism was unlikely to survive there because, being an ‘r-strategist,’ it requires high concentrations of ammonia and nitrite for energy production and growth. Its ability to grow between 25°C and 63°C, higher temperatures than those reached in European treatment plants at least, may also preclude its presence. Certainly FISH probes designed to target its 16S rRNA genes and qPCR protocols failed to detect it in activated sludge treatment plants in European plants, although a 16S rRNA sequence closely related to N. hollandicus was detected in a SHARON reactor running at higher temperatures in Korea (Sorokin et al., 2012).
So unexpectedly, the survey data generated in this study showed it was present on several occasions in four plants of those examined here, (data not shown) Randers, Lundtofte, Haderslev and Ouyang. The first three are EBPR plants in Denmark and the fourth is a N removal plant in China. This finding raises the prospect that activated sludge plants around the world may contain non-filamentous Chloroflexi with important defined functional roles, and requires further changes to the existing paradigm of the microbiology of nitrification.
It is the only known nitrite oxidizing bacterium, which is not a member of the Proteobacteria, and evidence presented by Sorokin et al. (2012) would suggest it did not evolve from them. N. hollandicus lacks any of the intracellular cytoplasmic membranes seen in the proteobacterial nitrifiers, and their distinctively different lipids. The similarity of its nitrite oxidoreductase encoding nrx complex to those in Nitrobacter and Nitrococcus suggests lateral gene transfer of this complex occurred between them.
Conclusion
Application of the molecular methods discussed here and elsewhere have shown repeatedly that so far unidentified and uncultured Chloroflexi are commonly present in activated sludge plants, especially since the data available presently come from a few countries only. Consequently our understanding of their phylogenetic diversity is far from complete. Andersen et al. (2019) and Nierychlo et al. (2019) have made important contributions toward a better understanding of their presence in Danish activated sludge plants, and we believe this review begins to add a global perspective, and will encourage others to pursue this fascinating group of bacteria.
Problems encountered here were often in comparing phylogenetic data generated in different studies. Much of these arose from subjective choices of DNA extraction methods and of PCR primers, often targeting different variable regions of the 16S rRNA. It is recommended that all future studies should use a standardized protocol for both obtaining and handling DNA samples. The experimentally validated suggestions of Albertsen et al. (2015) provide a sound basis for this.
Recently published surveys of 16S rRNA sequence data generated using PCR based protocols from 269 plants around the world supported by the Global Water Microbiome Consortium (Wu et al., 2019) and the imminent release of survey data based on the full-length 16S rRNA gene sequencing method described by Karst et al. (2018) carried out by the Center for Microbial Communities (Aalborg, Denmark) will surely increase dramatically our understanding of the activated sludge microbiome, and raise important questions for the future. The Chloroflexi will figure strongly there.
So there is an opportunity now to consolidate Chloroflexi amplicon data from plants globally, and extend substantially the data presented in this review from smaller numbers of studies. It is hoped that the user- friendly MiDAS database will be expanded to include these new data sets. However, as such surveys do not embrace every known treatment plant, and examine small sample numbers from each plant, there is still a place for long term studies carried out on individual plants. The important role of MIDAS in the future will be devalued unless journals insist that all published amplicon sequence data be deposited in publically available databases, for future compilation and analyses. This does not always occur presently.
Ultimately, the availability of relative abundance and environmental data, combined with the physiological information demonstrated experimentally with MAR and/or inferred from genome sequences, will surely assist in providing clues needed to understand more fully their roles in activated sludge plants. In the near future transcriptomic studies exploiting the rapidly increased generation of whole genome sequence data will extend the limitations of the FISH/MAR approach, allowing elucidation of the dynamics of in situ gene expression of these important bacteria, and help in solving the operational problems they cause.
Dedication
This manuscript is dedicated to the memory of Professor Valter Tandoi, who made an important contribution to our present understanding of the bulking filamentous bacteria in activated sludge processes.
Author Contributions
RS and LS conceived the idea and its elements for this review. LS designed and undertook the amplicon, 16S rRNA sequence, and FISH probe analyses, and prepared the tables and figures with the assistance from DR and SP. All authors discussed and contributed to the preparation of the final manuscript.
Funding
SP’s laboratory received support from the La Trobe University, Research Focus Area (RFA) securing food, water, and the environment.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank our colleagues who have made their data available for us to review for this publication and Dr. Aharon Oren for taxonomic assistance. We would also like to thank the reviewers for their insightful and constructive comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02015/full#supplementary-material
TABLE S1 | Summary of updates to the MiDAS 2.1 database undertaken for the additional Chloroflexi filaments named and described in this review. The phylogenetic clustering these changes are based upon are those shown in Figure 1.
TABLE S2 | Suggested descriptive nomenclature and characteristics of the newly proposed Candidatus Chloroflexi genera described in the review.
DATA FILE S1 | Data for the amplicon data sets surveyed for this review. The relative abundance for each Chloroflexi filament phylotype is expressed as a percentage of the total number of 16S rRNA gene sequences from each sample remaining after quality control. Analyses were undertaken using QIIME 2 (release 2017.12). Samples retaining fewer than 4000 features after quality control are marked by red text (total features column), since the detection threshold fell below 0.05% relative abundance. Relative abundance values are given and also shaded by their respective heatmap color.
DATA FILE S2 | Step-wise protocol used for retrieving relative abundances of Chloroflexi from published databases of NGS 16S rRNA amplicon sequence data presented Supplementary Data File S1.
Footnotes
References
Albertsen, M., Karst, S. M., Ziegler, A. S., Kirkegaard, R. H., and Nielsen, P. H. (2015). Back to basics–the influence of DNA extraction and primer choice on phylogenetic analysis of activated sludge communities. PLoS One 10:e0132783. doi: 10.1371/journal.pone.0132783
Amann, R. I., Binder, B. J., Olson, R. J., Chisholm, S. W., Devereux, R., and Stahl, D. A. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56, 1919–1925.
Amann, R. I., and Fuchs, B. M. (2008). Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6, 339–348. doi: 10.1038/nrmicro1888
Andersen, M. H., McIlroy, S. J., Nierychlo, M., Nielsen, P. H., and Albertsen, M. (2019). Genomic insights into Candidatus Amarolinea aalborgensis gen.nov., sp. nov., associated with settleability problems in wastewater treatment plants. Syst. Appl. Microbiol. 42, 77–84. doi: 10.1016/j.syapm.2018.08.001
Balvočiūtė, M., and Huson, D. H. (2017). SILVA, RDP, greengenes, NCBI and OTT—how do these taxonomies compare? BMC Genomics 18:114. doi: 10.1186/s12864-017-3501-4
Barnard, J., and Comeau, Y. (2014). “Maco-nutrient removal (phosphorus),” in Activated Sludge - 100 Years and Counting, eds D. Jenkins and J. Wanner (London: IWA Publishing), 93–116.
Beer, M., Seviour, E. M., Kong, Y., Cunningham, M., Blackall, L. L., and Seviour, R. J. (2002). Phylogeny of the filamentous bacterium Eikelboom Type 1851, and design and application of a 16S rRNA targeted oligonucleotide probe for its fluorescence in situ identification in activated sludge. FEMS Microbiol. Lett. 207, 179–183. doi: 10.1016/s0378-1097(01)00561-4
Björnsson, L., Hugenholtz, P., Tyson, G. W., and Blackall, L. L. (2002). Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removal. Microbiology 8, 2309–2318. doi: 10.1099/00221287-148-8-2309
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C., Al-Ghalith, G. A., et al. (2018). QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ 6:e27295v2.
Bovio, P., Cabezas, A., and Etchebehere, C. (2019). Preliminary analysis of Chloroflexi populations in full-scale UASB methanogenic reactors. J. Appl. Microbiol. 126, 667–683. doi: 10.1111/jam.14115
Bradford, D., Hugenholtz, P., Seviour, E. M., Cunningham, M. A., Stratton, H., Seviour, R. J., et al. (1996). 16S rRNA analysis of isolates obtained from gram-negative, filamentous bacteria micromanipulated from activated sludge. Syst. Appl. Microbiol. 3, 334–343. doi: 10.1016/s0723-2020(96)80060-9
Buali, A., and Horan, N. (1989). Variable morphology in certain filamentous bacteria and the implications of this for theories of activated sludge bulking. Environ. Technol. 11, 941–950. doi: 10.1080/09593338909384816
Burger, W., Krysiak-Baltyn, K., Scales, P. J., Martin, G. J. O., Strickland, A. D., and Gras, S. L. (2017). The influence of protruding filamentous bacteria on floc stability and solid-liquid separation in the activated sludge process. Water Res. 123, 578–585. doi: 10.1016/j.watres.2017.06.063
Campbell, A. G., Schwientek, P., Vishnivetskaya, T., Woyke, T., Levy, S., Beall, C. J., et al. (2014). Diversity and genomic insights into the uncultured Chloroflexi from the human microbiota. Environ. Microbiol. 9, 2635–2643. doi: 10.1111/1462-2920.12461
Cao, S., Du, R., Li, B., Ren, N., and Peng, Y. (2016). High-throughput profiling of microbial community structures in an ANAMMOX-USAB reactor treating high-strength wastewater. Appl. Microbiol. Biotechnol. 100, 6457–6467. doi: 10.1007/s00253-016-7427-6
Catherine, Q., Susanna, W., Isidora, E.-S., Mark, H., Aurelie, V., and Jean-Francois, H. (2013). A review of current knowledge on toxic benthic freshwater cyanobacteria–ecology, toxin production and risk management. Water Res. 15, 5464–5479. doi: 10.1016/j.watres.2013.06.042
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, 633–642. doi: 10.1093/nar/gkt1244
Crawford, G. V., Judd, S., and Zsirai, T. (2014). “Membrane systems,” in Activated Sludge - 100 Years and Counting, eds D. Jenkins and J. Wanner (London: IWA Publishing), 319–342.
Daigger, G. T. (2014). “Ardern and Lockett remembrance,” in Activated Sludge - 100 Years and Counting, eds D. Jenkins and J. Wanner (London: IWA Publishing), 1–15.
Daims, H., Brühl, A., Amann, R., Schleifer, K. H., and Wagner, M. (1999). The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 3, 434–444. doi: 10.1016/s0723-2020(99)80053-8
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
Daims, H., Lücker, S., and Wagner, M. (2016). A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 9, 699–712. doi: 10.1016/j.tim.2016.05.004
de los Reyes, F. L. (2010). “Foaming,” in Microbial Ecology of Activated Sludge, eds R. J. Seviour and P. H. Nielsen (London: IWA Publishing), 215–258.
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/aem.03006-05
Dodsworth, J. A., Gevorkian, J., Despujos, F., Cole, J. K., Murugapiran, S. K., Ming, H., et al. (2014). Thermoflexus hugenholtzii gen. nov., sp. nov., a thermophilic, microaerophilic, filamentous bacterium representing a novel class in the Chloroflexi, Thermoflexia classis nov., and description of Thermoflexaceae fam. nov. and Thermoflexales ord. nov. Int. J. Syst. Evol. Microbiol. 6, 2119–2127. doi: 10.1099/ijs.0.055855-0
Dos Santos, L. A., Ferreira, V., Neto, M. M., Pereira, M. A., Mota, M., and Nicolau, A. (2015). Study of 16 Portuguese activated sludge systems based on filamentous bacteria populations and their relationships with environmental parameters. Appl. Microbiol. Biotechnol. 12, 5307–5316. doi: 10.1007/s00253-015-6393-8
Dunkel, T., de León Gallegos, E., Bock, C., Lange, A., Hoffmann, D., Boenigk, J., et al. (2018). Illumina sequencing for the identification of filamentous bulking and foaming bacteria in industrial activated sludge plants. Int. J. Environ. Sci. Technol. 15, 1139–1158. doi: 10.1007/s13762-017-1484-y
Eikelboom, D. H. (1975). Filamentous organisms observed in activated sludge. Water Res. 4, 365–388. doi: 10.1016/0043-1354(75)90182-7
Eikelboom, D. H., and Geurkink, B. (2002). Filamentous micro-organisms observed in industrial activated sludge plants. Water Sci. Technol. 46, 535–542. doi: 10.2166/wst.2002.0531
Eikelboom, D. H., Tandoi, V., Krooneman, J., Borger, A., Thelen, K., Kragelund, C., et al. (2006). Identification and Control of Filamentous Micro-Organisms in Industrial Wastewater Treatment Plants. London: IWA Publishing.
Eikelboom, D. H., and Van Buijsen, H. (1983). Microscopic Sludge Investigation Manual. ıThe Hague: TNO Publishing.
Espejo, R. T., and Plaza, N. M. (2018). Multiple ribosomal RNA operons in bacteria; their concerted evolution and potential consequences on the rate of evolution of their 16S rRNA. Front. Microbiol. 9:1232. doi: 10.3389/fmicb.2018.01232
Ferrera, I., and Sanchez, O. (2016). Insights into microbial diversity in wastewater treatment systems: how far have we come? Biotechnol. Adv. 5, 790–802. doi: 10.1016/j.biotechadv.2016.04.003
Figueroa, M., Del Río, A. V., Campos, J. L., Mendez, R., and Mosquera-Corral, A. (2015). Filamentous bacteria existence in aerobic granular reactors. Bioprocess Biosyst. Eng. 5, 841–851. doi: 10.1007/s00449-014-1327-x
Foley, J., De Haas, D., Yuan, Z., and Lant, P. (2010). Nitrous oxide generation in full-scale biological nutrient removal wastewater treatment plants. Water Res. 3, 831–844. doi: 10.1016/j.watres.2009.10.033
Gaget, V., Lau, M., Sendall, B., Froscio, S., and Humpage, A. R. (2017). Cyanotoxins: which detection technique for an optimum risk assessment? Water Res. 118, 227–238. doi: 10.1016/j.watres.2017.04.025
Garrity, G. M., Holt, J. G., and Castenholz, R. W. (2001). Bergey’s Manual® of Systematic Bacteriology. Berlin: Springer-verlag.
Gich, F., Garcia-Gil, J., and Overmann, J. (2001). Previously unknown and phylogenetically diverse members of the green nonsulfur bacteria are indigenous to freshwater lakes. Arch. Microbiol. 1, 1–10. doi: 10.1007/s00203-001-0354-6
Guo, F., Wang, Z.-P., Yu, K., and Zhang, T. (2015). Detailed investigation of the microbial community in foaming activated sludge reveals novel foam formers. Sci. Rep. 5:7637. doi: 10.1038/srep07637
Guo, F., and Zhang, T. (2012). Profiling bulking and foaming bacteria in activated sludge by high throughput sequencing. Water Res. 8, 2772–2782. doi: 10.1016/j.watres.2012.02.039
Gupta, R. S., Chander, P., and George, S. (2013). Phylogenetic framework and molecular signatures for the class Chloroflexi and its different clades; proposal for division of the class Chloroflexi class. nov. into the suborder Chloroflexineae subord. nov., consisting of the emended family Oscillochloridaceae and the family Chloroflexaceae fam. nov., and the suborder Roseiflexineae subord. nov., containing the family Roseiflexaceae fam. nov. Antonie van Leeuwenhoek 1, 99–119. doi: 10.1007/s10482-012-9790-3
Hanada, S. (2014). “The phylum Chloroflexi, the family Chloroflexaceae, and the related phototrophic families Oscillatoriaceae and Roseiflexaceae,” in The Prokaryotes-Other Major Lineages of Bacteria and the Archaea, ed. E. Rosenburg (Berlin: Springer-Verlag), 515–532. doi: 10.1007/978-3-642-38954-2_165
He, S., and McMahon, K. D. (2011). Microbiology of ‘Candidatus Accumulibacter’ in activated sludge. Microb. Biotechnol. 4, 603–619. doi: 10.1016/j.chemosphere.2017.01.123
Horan, N., Bu’Ali, A., and Eccles, C. (1988). Isolation, identification and characterisation of filamentous and floc-forming bacteria from activated sludge flocs. Environ. Technol. 5, 449–457. doi: 10.1080/09593338809384589
Hug, L. A., Castelle, C. J., Wrighton, K. C., Thomas, B. C., Sharon, I., Frischkorn, K. R., et al. (2013). Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1:22. doi: 10.1186/2049-2618-1-22
Hugenholtz, P., Goebel, B. M., and Pace, N. R. (1998). Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180, 4765–4774.
Hugenholtz, P., and Stackebrandt, E. (2004). Reclassification of Sphaerobacter thermophilus from the subclass Sphaerobacteridae in the phylum Actinobacteria to the class Thermomicrobia (emended description) in the phylum Chloroflexi (emended description). Int. J. Syst. Evol. Microbiol. 54, 2049–2051. doi: 10.1099/ijs.0.03028-0
Hugenholtz, P., Tyson, G. W., Webb, R. I., Wagner, A. M., and Blackall, L. L. (2001). Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 67, 411–419. doi: 10.1128/aem.67.1.411-419.2001
Hugerth, L. W., and Andersson, A. F. (2017). Analysing microbial community composition through amplicon sequencing: from sampling to hypothesis testing. Front. Microbiol. 8:1561. doi: 10.3389/fmicb.2017.01561
Jenkins, D. (1992). Towards a comprehensive model of activated sludge bulking and foaming. Water Sci. Technol. 25, 215–230. doi: 10.2166/wst.1992.0124
Jenkins, D., Richard, M. G., and Daigger, G. T. (2003). Manual on the Causes and Control of Activated Sludge Bulking, Foaming, and other Solids Separation Problems. London: IWA Publishing.
Jenkins, D., and Wanner, J. (2014). Activated Sludge-100 Years and Counting. London: IWA Publishing.
Jiang, X.-T., Ye, L., Li, B., Ma, L. P., and Zhang, T. (2018). Temporal dynamics of activated sludge bacterial communities in two diversity variant full-scale sewage treatment plants. Appl. Microbiol. Biotechnol. 102, 9278–9287. doi: 10.1007/s00253-018-9287-8
Juretschko, S., Loy, A., Lehner, A., and Wagner, M. (2002). The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25, 84–99. doi: 10.1078/0723-2020-00093
Karst, S. M., Albertsen, M., Kirkegaard, R. H., Dueholm, M. S., and Nielsen, P. H. (2016). “Molecular methods,” in Experimental Methods in Wastewater Treatment, eds M. Van Loosdrecht, P. H. Nielsen, C. M. Lopez-Vazquez, and D. Brdjanovic (London: IWA Publishing), 285–323.
Karst, S. M., Dueholm, M. S., McIlroy, S. J., Kirkegaard, R. H., Nielsen, P. H., and Albertsen, M. (2018). Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nat. Biotechnol. 36, 195–195. doi: 10.1038/nbt.4045
Kartal, B., de Almeida, N. M., Maalcke, W. J., Op den Camp, H. J., Jetten, M. S., and Keltjens, J. T. (2013). How to make a living from anaerobic ammonium oxidation. FEMS Microbiol. Rev. 373, 428–461. doi: 10.1111/1574-6976.12014
Kawaichi, S., Ito, N., Kamikawa, R., Sugawara, T., Yoshida, T., and Sako, Y. (2013). Ardenticatena maritima gen. nov., sp. nov., a ferric iron-and nitrate-reducing bacterium of the phylum ‘Chloroflexi’ isolated from an iron-rich coastal hydrothermal field, and description of Ardenticatenia classis nov. Int. J. Syst. Evol. Microbiol. 638, 2992–3002. doi: 10.1099/ijs.0.046532-0
Khunjar, W. O., Pitt, P., Bott, C. B., and Chandran, K. (2014). “Macro-nutrient removal (nitrogen),” in Activated Sludge - 100 Years and Counting, eds D. Jenkins and J. Wanner (London: IWA Publishing), 77–92.
Kindaichi, T., Yuri, S., Ozaki, N., and Ohashi, A. (2012). Ecophysiological role and function of uncultured Chloroflexi in an anammox reactor. Water Sci. Technol. 66, 2556–2561. doi: 10.2166/wst.2012.479
Kirkgaard, R. H., McIlroy, S. J., Kristensen, J. M., Nierychlo, M., Karst, S. M., Dueholm, M. S., et al. (2017). The impact of immigration on microbial community composition in full-scale anaerobic digesters. Sci. Rep. 7:9343. doi: 10.1038/s41598-017-09303-0
Kohno, T., Sei, K., and Mori, K. (2002). Characterization of type 1851 organism isolated from activated sludge samples. Water Sci. Technol. 46, 111–114. doi: 10.2166/wst.2002.0464
Kragelund, C., Levantesi, C., Borger, A., Thelen, K., Eikelboom, D., Tandoi, V., et al. (2007a). Identity, abundance and ecophysiology of filamentous Chloroflexi species present in activated sludge treatment plants. FEMS Microbiol. Ecol. 59, 671–682. doi: 10.1111/j.1574-6941.2006.00251.x
Kragelund, C., Remesova, Z., Nielsen, J. L., Thomsen, T. R., Eales, K., Seviour, R., et al. (2007b). Ecophysiology of mycolic acid-containing Actinobacteria (Mycolata) in activated sludge foams. FEMS Microbiol. Ecol. 61, 174–184. doi: 10.1111/j.1574-6941.2007.00324.x
Kragelund, C., Thomsen, T. R., Mielczarek, A. T., and Nielsen, P. H. (2011). Eikelboom’s morphotype 0803 in activated sludge belongs to the genus Caldilinea in the phylum Chloroflexi. FEMS Microbiol. Ecol. 76, 451–462. doi: 10.1111/j.1574-6941.2011.01065.x
Kristiansen, R., Nguyen, H. T. T., Saunders, A. M., Nielsen, J. L., Wimmer, R., Le, V. Q., et al. (2013). A metabolic model for members of the genus Tetrasphaera involved in enhanced biological phosphorus removal. ISME J. 7, 543–554. doi: 10.1038/ismej.2012.136
Kunisawa, T. (2011). The phylogenetic placement of the non-phototrophic, Gram-positive thermophile ‘Thermobaculum terrenum’ and branching orders within the phylum ‘Chloroflexi’ inferred from gene order comparisons. Int. J. Syst. Evol. Microbiol. 61(Pt 8), 1944–1953. doi: 10.1099/ijs.0.026088-0
Lapidus, A., Nolan, M., Lucas, S., Glavina, D. T. R., Tice, H., Cheng, J. F., et al. (2011). Genome sequence of the filamentous, gliding Thiothrix nivea neotype strain (JP2T). Stand. Genomic Sci. 5, 398–406. doi: 10.4056/sigs.2344929
Liao, J., Lou, I., and de Los Reyes, F. L. (2004). Relationship of species-specific filament levels to filamentous bulking in activated sludge. Appl. Environ. Microbiol. 70, 2420–2428. doi: 10.1128/aem.70.4.2420-2428.2004
Lofrano, G., and Brown, J. (2010). Wastewater management through the ages: a history of mankind. Sci. Total Environ. 408, 5254–5264. doi: 10.1016/j.scitotenv.2010.07.062
Lu, H., Chandran, K., and Stensel, D. (2014). Microbial ecology of denitrification in biological wastewater treatment. Water Res. 64, 237–254. doi: 10.1016/j.watres.2014.06.042
Ludwig, W., Stunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, et al. (2004). ARB: a software environment for sequence data. Nucleic Acid Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
Marques, R., Santos, J., Nguyen, H., Carvalho, G., Noronha, J. P., Nielsen, P. H., et al. (2017). Metabolism and ecological niche of Tetrasphaera and Ca. Accumulibacter in enhanced biological phosphorus removal. Water Res. 122, 159–171. doi: 10.1016/j.watres.2017.04.072
Martins, A. M., Pagilla, K., Heijnen, J. J., and van Loosdrecht, M. C. (2004). Filamentous bulking sludge—a critical review. Water Res. 38, 793–817. doi: 10.1016/j.watres.2003.11.005
Massara, T. M., Malamis, S., Guisasola, A., Baeza, J. A., Noutsopoulos, C., and Katsou, E. (2017). A review on nitrous oxide (N2O) emissions during biological nutrient removal from municipal wastewater and sludge reject water. Sci. Total Environ. 596, 106–123. doi: 10.1016/j.scitotenv.2017.03.191
McIlroy, S. J., Albertsen, M., Andresen, E. K., Saunders, A. M., Kristiansen, R., Stokholm-Bjerrgaard, M., et al. (2014). ‘Candidatus Competibacter’-lineage genomes retrieved from metagenomes reveal functional metabolic diversity. ISME J. 8, 613–624. doi: 10.1038/ismej.2013.162
McIlroy, S. J., Karst, S. M., Nierychlo, M., Dueholm, M. S., Albertsen, M., Kirkegaard, R. H., et al. (2016). Genomic and in situ investigations of the novel uncultured Chloroflexi associated with 0092 morphotype filamentous bulking in activated sludge. ISME J. 10, 2223–2234. doi: 10.1038/ismej.2016.14
McIlroy, S. J., Kirkegaard, R. H., Dueholm, M. S., Fernando, E., Karst, S. M., Albertsen, M., et al. (2017a). Culture-Independent analyses reveal novel Anaerolineaceae as abundant primary fermenters in anaerobic digesters treating waste activated sludge. Front. Microbiol. 8:1134. doi: 10.3389/fmicb.2017.01134
McIlroy, S. J., Kirkegaard, R. H., McIlroy, B., Nierychlo, M., Kristensen, J. M., Karst, S. M., et al. (2017b). MiDAS 2.0: an ecosystem-specific taxonomy and online database for the organisms of wastewater treatment systems expanded for anaerobic digester groups. Database 2017:bax016. doi: 10.1093/database/bax016
McIlroy, S. J., Kristianses, R., Albertsen, M., Karst, M., Rossetti, S., Nielsen, J., et al. (2013). Metabolic model for the filamentous Candidatus Microthrix parvicella based on genomic and metagenomic analyses. ISME J. 7, 1161–1172. doi: 10.1038/ismej.2013.6
McIlroy, S. J., Onetto, C. A., McIlroy, B., Herbst, F. A., Dueholm, M. S., Kirkegaard, R. H., et al. (2018). Genomic and in situ analyses reveal the Micropruina spp. An abundant fermentative glycogen accumulating organisms in enhanced biological phosphorous removal systems. Front. Microbiol. 9:1004. doi: 10.3389/fmicb.2018.01004
McIlroy, S. J., Saunders, A. M., Albertsen, M., Nierychlo, M., McIlroy, B., Hansen, A. A., et al. (2015). MiDAS: the field guide to the microbes of activated sludge. Database 2015:bav062. doi: 10.1093/database/bav062
McMahon, K. D., He, S., and Oehmen, A. (2010). “The microbiology of phosphorous removal,” in Microbial Ecology of Activated Sludge, eds R. J. Seviour and P. H. Nielsen (London: IWA Publishing), 291–319.
Mielczarek, A. T., Kragelund, C., Eriksen, P. S., and Nielsen, P. H. (2012). Population dynamics of filamentous bacteria in Danish wastewater treatment plants with nutrient removal. Water Res. 4612, 3781–3795. doi: 10.1016/j.watres.2012.04.009
Miłobędzka, A., and Muszyński, A. (2015). Population dynamics of filamentous bacteria identified in Polish full-scale wastewater treatment plants with nutrients removal. Water Sci. Technol. 71, 675–684. doi: 10.2166/wst.2014.512
Miłobędzka, A., Witeska, A., and Muszyński, A. (2016). Factors affecting population of filamentous bacteria in wastewater treatment plants with nutrients removal. Water Sci. Technol. 734, 790–797. doi: 10.2166/wst.2015.541
Miura, Y., Watanabe, Y., and Okabe, S. (2007). Significance of Chloroflexi in performance of submerged membrane bioreactors (MBR) treating municipal wastewater. Environ. Sci. Technol. 4122, 7787–7794. doi: 10.1021/es071263x
Morgan-Sagastume, F., Larsen, P., Nielsen, J. L., and Nielsen, P. H. (2008). Characterization of the loosely attached fraction of activated sludge bacteria. Water Res. 424, 843–854. doi: 10.1016/j.watres.2007.08.026
Müller, E., Schade, M., and Lemmer, H. (2007). Filamentous scum bacteria in activated sludge plants: detection and identification quality by conventional activated sludge microscopy versus fluorescence in situ hybridization. Water Environ. Res. 7911, 2274–2286. doi: 10.2175/106143007x183943
Nielsen, P. H., Daims, H., Lemmer, H., Arslan-Alaton, I., and Olmez-Hanci, T. (2009a). FISH Handbook for Biological Wastewater Treatment. London: IWA Publishing.
Nielsen, P. H., Kragelund, C., Seviour, R. J., and Nielsen, J. L. (2009b). Identity and ecophysiology of filamentous bacteria in activated sludge. FEMS Microbiol. Rev. 336, 969–998. doi: 10.1111/j.1574-6976.2009.00186.x
Nielsen, P. H., McIlroy, S. J., Albertsen, M., and Nierychlo, M. (2019). Re-evaluating the microbiology of the enhanced biological phosphorous removal process. Curr. Op. Biotechnol. 57, 111–118. doi: 10.1016/j.copbio.2019.03.008
Nielsen, P. H., and McMahon, K. D. (2014). “Microbiology and microbial ecology of the activated sludge process,” in Activated Sludge – 100 Years and Counting, eds D. Jenkins and J. Wanner (London: IWA Publishing), 53–75.
Nierychlo, M., Miłobędzka, A., Petriglieri, F., McIlroy, B., Nielsen, P. H., and McIlroy, S. J. (2019). The morphology and metabolic potential of the Chloroflexi in full-scale activated sludge wastewater treatment plants. FEMS Microbiol. Ecol. 95:fiy228. doi: 10.1093/femsec/fiy228
Nierychlo, M., and Nielsen, P. H. (2014). “‘Denmark’,” in Activated Sludge - 100 Years and Counting, eds D. Jenkins and J. Wanner (London: IWA Publishing), 197–218.
Nittami, T., Speirs, L. B. M., Fukuda, J., Watanabe, M., and Seviour, R. J. (2014). Fluorescence in situ hybridization probes targeting members of the phylum Candidatus Saccharibacteria falsely target Eikelboom type 1851 filaments and other Chloroflexi members. Environ. Microbiol. Rep. 4, 611–617. doi: 10.1111/1758-2229.12172
Nittami, T., Speirs, L. B. M., Yamada, T., Suzuki, I., Fukuda, J., Kurisu, F., et al. (2017). Quantification of Chloroflexi Eikelboom morphotype 1851 for prediction and control of bulking events in municipal activated sludge plants in Japan. Appl. Microbiol. Biotechnol. 101, 3861–3869. doi: 10.1007/s00253-016-8077-4
Noguera, D. R., Wright, E. S., Camejo, P., and Yilmaz, L. S. (2014). Mathematical tools to optimize the design of oligonucleotide probes and primers. Appl. Microbiol. Biotechnol. 98, 9595–9608. doi: 10.1007/s00253-014-6165-x
Oehmen, A., Lemos, P. C., Carvalho, G., Yuan, Z., Keller, J., Blackall, L. L., et al. (2007). Advances in enhanced biological phosphorus removal: from micro to macro scale. Water Res. 41, 2271–2300. doi: 10.1016/j.watres.2007.02.030
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny revises tree of life. Nature Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Petriglieri, F., Nierychlo, M., Nielsen, P. H., and McIlroy, S. J. (2018). In situ visualisation of the abundant Chloroflexi populations in full-scale anaerobic digesters and the fate of immigrating species. PLoS One 13:e0206255. doi: 10.1371/journal.pone.0206255
Petrovski, S., Dyson, Z. A., Quill, E. S., McIlroy, S. J., Tillett, D., and Seviour, R. J. (2011). An examination of the mechanisms for stable foam formation in activated sludge systems. Water Res. 45, 2146–2154. doi: 10.1016/j.watres.2010.12.026
Puyol, D., Batstone, D. J., Hülsen, T., Astals, S., Peces, M., and Kromer, J. O. (2016). Resource recovery from wastewater by biological technologies: opportunities, challenges, and prospects. Front. Microbiol. 7:2106. doi: 10.3389/fmicb.2016.02106
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Rodríguez, E., García-Encina, P. A., Stams, A. J., Maphosa, F., and Sousa, D. Z. (2015). Meta-omics approaches to understand and improve wastewater treatment systems. Rev. Environ. Sci. Biotechnol. 14, 385–406. doi: 10.1007/s11157-015-9370-x
Roots, P., Wang, Y., Rosenthal, A. F., Griffin, J. S., Sabba, F., Petrovich, M., et al. (2019). Comammox Nitrospira are the dominant ammonia oxidizers in a mainstream low dissolved oxygen nitrification reactor. Water Res. 157, 396–405. doi: 10.1016/j.watres.2019.03.060
Rossetti, S., Levantesi, C., and Tandoi, V. (2017). “The microbiology of the activated sludge process,” in Activated Sludge Separation Problems: Theory, Control Measures, Practical Experiences, eds S. Rossetti, V. Tandoi, and J. Wanner (London: IWA Publishing), 21–52.
Rossetti, S., Tomei, M. C., Nielsen, P. H., and Tandoi, V. (2005). “Microthrix parvicella”, a filamentous bacterium causing bulking and foaming in activated sludge systems: a review of current knowledge. FEMS Microbiol. Rev. 29, 49–64. doi: 10.1016/j.femsre.2004.09.005
Saunders, A. M., Albertsen, M., Vollertsen, J., and Nielsen, P. H. (2016). The activated sludge ecosystem contains a core community of abundant organisms. ISME J. 10, 11–20. doi: 10.1038/ismej.2015.117
Schade, M., Beimfohr, C., and Lemmer, H. (2002). Phylogenetic and physiological characterization of a “Nostocoida limicola”-like organism isolated from activated sludge. Water Sci. Technol. 46, 91–97. doi: 10.2166/wst.2002.0461
Schmidt, I., Sliekers, O., Schmid, M., Bock, E., Fuerst, J., Kuenen, J. G., et al. (2003). New concepts of microbial treatment processes for the nitrogen removal in wastewater. FEMS Microbiol. Rev. 27, 481–492.
Schuler, A. J., and Jassby, D. (2007). Filament content threshold for activated sludge bulking: artifact or reality. Water Res. 41, 4349–4356. doi: 10.1016/j.watres.2007.06.021
Sekiguchi, Y., Ohashi, A., Parks, D. H., Yamauchi, T., Tyson, G. W., and Hugenholtz, P. (2015). First genomic insights into members of a candidate bacterial phylum responsible for wastewater bulking. PeerJ 3:e740. doi: 10.7717/peerj.740
Sekiguchi, Y., Takahashi, H., Kamagata, Y., Ohashi, A., and Harada, H. (2001). In situ, isolation, and physiological properties of a thin filamentous microorganism abundant in methanogenic granular sludges: a novel isolate affiliated with a clone cluster, the green non-sulfur bacteria, subdivision I. Appl. Environ. Microbiol. 67, 5740–5749. doi: 10.1128/aem.67.12.5740-5749.2001
Sekiguchi, Y., Yamada, T., Hanada, S., Hanada, S., Ohashi, A., Harada, H., et al. (2003). Anaerolinea thermophila gen. nov., sp. nov. and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain Bacteria at the subphylum level. Int. J. Syst. Evol. Microbiol. 53, 1843–1851. doi: 10.1099/ijs.0.02699-0
Seviour, R. J., and McIlroy, S. (2008). The microbiology of phosphorus removal in activated sludge processes-the current state of play. J. Microbiol. 46, 115–124. doi: 10.1007/s12275-008-0051-0
Seviour, E. M., Williams, C. J., DeGrey, B., Soddell, J. A., Seviour, R. J., and Lindrea, K. C. (1994). Studies on filamentous bacteria from Australian activated sludge plants. Water Res. 28, 2335–2342. doi: 10.1016/0043-1354(94)90049-3
Seviour, E. M., Williams, C. J., Seviour, R. J., Soddell, J. A., and Lindrea, K. C. (1990). A survey of filamentous bacterial populations from foaming activated sludge plants in eastern states of Australia. Water Res. 24, 493–498. doi: 10.1016/0043-1354(90)90234-w
Seviour, R. J. (2010a). “Factors affecting the bulking and foaming filamentous bacteria in activated sludge,” in Microbial Ecology of Activated Sludge, eds R. J. Seviour and P. H. Nielsen (London: IWA Publishing), 139–168.
Seviour, R. J. (2010b). “The current taxonomic status of the filamentous bacteria found in activated sludge,” in Microbial Ecology of Activated Sludge, eds R. Seviour and P. H. Nielsen (London: IWA Publishing), 139–168.
Seviour, R. J., Mino, T., and Onuki, M. (2003). The microbiology of biological phosphorus removal in activated sludge systems. FEMS Microbiol. Rev. 27, 99–127. doi: 10.1016/s0168-6445(03)00021-4
Seviour, R. J., and Nielsen, P. H. (2010). Microbial Ecology of Activated Sludge. London: IWA Publishing.
Sorokin, D. Y., Lücker, S., Vejmelkova, D., Kostrinkina, N. A., Kleerebezem, R., Rijpstra, W. I., et al. (2012). Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 6, 2245–2256. doi: 10.1038/ismej.2012.70
Sorokin, D. Y., Vejmelkova, D., Lücker, S., Streshinskaya, G. M., Rijpstra, W. I., Damste, S., et al. (2014). Nitrolancea hollandica gen. nov., sp. nov., a chemolithoautotrophic nitrite-oxidizing bacterium isolated from a bioreactor belonging to the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 64, 1859–1865. doi: 10.1099/ijs.0.062232-0
Speirs, L. B. M., Dyson, Z. A., Tucci, J., and Seviour, R. J. (2017). Eikelboom filamentous morphotypes 0675 and 0041 embrace members of the Chloroflexi: resolving their phylogeny, and design of fluorescence in situ hybridisation probes for their identification. FEMS Microbiol. Ecol. 93:fix115. doi: 10.1093/femsec/fix115
Speirs, L. B. M., McIlroy, S. J., Petrovski, S., and Seviour, R. J. (2011). The activated sludge bulking filament Eikelboom morphotype 0914 is a member of the Chloroflexi. Environ. Microbiol. Rep. 3, 159–165. doi: 10.1111/j.1758-2229.2010.00201.x
Speirs, L. B. M., Nittami, T., McIlroy, S., Schroeder, S., and Seviour, R. J. (2009). Filamentous bacterium Eikelboom type 0092 in activated sludge plants in Australia is a member of the phylum Chloroflexi. Appl. Environ. Microbiol. 75, 2446–2452. doi: 10.1128/AEM.02310-08
Speirs, L. B. M., Tucci, J., and Seviour, R. J. (2015). The activated sludge bulking filament Eikelboom morphotype 0803 embraces more than one member of the Chloroflexi. FEMS Microbiol. Ecol. 91:fiv100. doi: 10.1093/femsec/fiv100
Stensel, H. D., and Makinia, J. (2014). “Process development,” in Activated Sludge - 100 Years and Counting, eds D. Jenkins and J. Wanner (London: IWA Publishing), 33–52.
Stokholm-Bjerregaard, M., McIlroy, S. J., Nierychlo, M., Karst, S. M., Albertsen, M., and Nielsen, P. H. (2017). A critical assessment of the microorganisms proposed to be important to enhanced biological phosphorus removal in full-scale wastewater treatment systems. Front. Microbiol. 8:718. doi: 10.3389/fmicb.2017.00718
St-Pierre, B., and Wright, A. D. G. (2014). Comparative metagenomic analysis of bacterial populations in three full-scale mesophilic anaerobic manure digesters. Appl. Microbiol. Biotechnol. 98, 2709–2717. doi: 10.1007/s00253-013-5220-3
Sun, L., Toyonaga, M., Ohashi, A., Matsuura, N., Tourlousse, D. M., Meng, X. Y., et al. (2016). Isolation and characterization of Flexilinea flocculi gen. nov., sp. nov., a filamentous, anaerobic bacterium belonging to the class Anaerolineae in the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 66, 988–996. doi: 10.1099/ijsem.0.000822
Thomsen, T. R., Kjellerup, B. V., Nielsen, J. L., Hugenholtz, P., and Nielsen, P. H. (2002). In situ studies of the phylogeny and physiology of filamentous bacteria with attached growth. Environ. Microbiol. 4, 383–391. doi: 10.1046/j.1462-2920.2002.00316.x
Thomsen, T. R., Kragelund, C., and Nielsen, P. H. (2006). Identity, abundance and physiology of Aquaspirillum-related filamentous bacteria in activated sludge. Water Sci. Tecnol. 54, 237–245. doi: 10.2166/wst.2006.392
van Kessel, M. A., Stultiens, K., Slegers, M. F., Guerrero Cruz, S., Jetten, M. S., Kartal, B., et al. (2018). Current perspectives on the application of N-damo and anammox in wastewater treatment. Curr. Opin. Biotechnol. 50, 222–227. doi: 10.1016/j.copbio.2018.01.031
van Loosdrecht, M., Seah, H., Wah, Y. L., and Cao, Y. (2014). “The next 100 years,” in Activated Sludge - 100 Years and Counting, eds D. Jenkins and J. Wanner (London: IWA Publishing), 407–424.
Wagner, D. S., Ramin, E., Szabo, P., and Dechesne, A. (2015). Microthrix parvicella abundance associated with activated sludge settling velocity and rheology- Quantifying and modelling filamentous bulking. Water Res. 78, 121–132. doi: 10.1016/j.watres.2015.04.003
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, R. J. (2007). Naïve Bayesian Classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/aem.00062-07
Wang, X., Shu, D., and Yue, H. (2016). Taxonomical and functional microbial community dynamics in an Anammox-ASBR system under different Fe(III) supplementation. Appl. Microbiol. Biotechnol. 100, 10147–10163. doi: 10.1007/s00253-016-7865-1
Wanner, J., and Jobbagy, A. (2014). “Solids seperation,” in Activated Sludge - 100 Years and Counting, eds D. Jenkins and J. Wanner (London: IWA Publishing), 171–194.
Wanner, J., Kragelund, C., and Nielsen, P. H. (2014). “Microbiology of bulking,” in Microbial Ecology of Activated Sludge, eds R. J. Seviour and P. H. Nielsen (London: IWA Publishing), 191–214.
Ward, L. M., Hemp, J., Shih, P. M., McGlynn, S. E., and Fischer, W. W. (2018). Evolution of phototrophy in the Chloroflexi phylum driven by horizontal gene transfer. Front. Microbiol. 9:260. doi: 10.3389/fmicb.2018.00260
Wu, L., Ning, D., Zhang, B., Li, Y., Zhang, P., Shan, X., et al. (2019). Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat. Microbiol. 4, 1183–1195. doi: 10.1038/s41564-019-0426-5
Xia, Y., Wang, Y., Wang, Y., Chin, F. Y. L., and Zhang, T. (2016). Cellular adhesiveness and cellulolytic capacity in Anaerolineae revealed by omics-based genome interpretation. Biotechnol. Biofuel 9:111. doi: 10.1186/s13068-016-0524-z
Yamada, T., Imachi, H., Ohashi, A., Harada, H., Hanada, S., Kamagata, Y., et al. (2007). Bellilinea caldifistulae gen. nov. and Longilinea arvoryzae gen. nov., sp. nov., strictly anaerobic, filamentous bacteria of the phylum Chloroflexi isolated from methanogenic propionate-degrading consortia. Int. J. Syst. Evol. Microbiol. 57, 2299–2306. doi: 10.1099/ijs.0.65098-0
Yamada, T., and Sekiguchi, Y. (2009). Cultivation of uncultured Chloroflexi subphyla: significance and ecophysiology of formerly uncultured Chloroflexi ‘subphylum I’ with natural and biotechnological relevance. Microbes Environ. 24, 205–216. doi: 10.1264/jsme2.me09151s
Yamada, T., Sekiguchi, Y., Hanada, S., Imachi, H., Ohashi, A., Harada, H., et al. (2006). Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 56, 1331–1340. doi: 10.1099/ijs.0.64169-0
Yang, Q., Zhao, H., and Du, B. (2017). Bacteria and bacteriophage communities in bulking and on-bulking activated sludge in full-scale municipal wastewater treatment systems. Biochem. Eng. J. 119, 101–111. doi: 10.1016/j.bej.2016.12.017
Yilmaz, L. S., and Noguera, D. R. (2004). Mechanistic approach to the problem of hybridization efficiency in fluorescent in situ hybridization. Appl. Environ. Microbiol. 70, 7126–7139. doi: 10.1128/aem.70.12.7126-7139.2004
Yilmaz, L. S., and Noguera, D. R. (2007). Development of thermodynamic models for simulating probe dissociation profiles in fluorescence in situ hybridization. Biotechnol. Bioeng. 96, 349–363. doi: 10.1002/bit.21114
Yilmaz, L. S., Okten, H. E., and Noguera, D. R. (2006). All regions of the 16S rRNA of Escherichia coli are accessible in situ to DNA oligonucleotides with sufficient thermodynamic affinity. Appl. Environ. Microbol. 72, 733–744. doi: 10.1128/aem.72.1.733-744.2006
Yilmaz, L. S., Parnerkar, S., and Noguera, D. R. (2011). mathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl. Environ. Microbiol. 77, 1118–1122. doi: 10.1128/AEM.01733-10
Keywords: Chloroflexi, activated sludge, bulking, foaming, filamentous bacteria FISH, amplicon sequencing
Citation: Speirs LBM, Rice DTF, Petrovski S and Seviour RJ (2019) The Phylogeny, Biodiversity, and Ecology of the Chloroflexi in Activated Sludge. Front. Microbiol. 10:2015. doi: 10.3389/fmicb.2019.02015
Received: 02 May 2019; Accepted: 16 August 2019;
Published: 13 September 2019.
Edited by:
Simona Rossetti, Water Research Institute (IRSA), ItalyReviewed by:
Simon Jon McIlroy, The University of Queensland, AustraliaSheena Kumari Santhosh Kumar, Durban University of Technology, South Africa
Copyright © 2019 Speirs, Rice, Petrovski and Seviour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steve Petrovski, c3RldmUucGV0cm92c2tpQGxhdHJvYmUuZWR1LmF1; Robert J. Seviour, ci5zZXZpb3VyQGxhdHJvYmUuZWR1LmF1
 Lachlan B. M. Speirs1
Lachlan B. M. Speirs1 Steve Petrovski
Steve Petrovski Robert J. Seviour
Robert J. Seviour