- 1China Institute of Veterinary Drug Control, Beijing, China
- 2Beijing Advanced Innovation Center for Food Nutrition and Human Health, College of Veterinary Medicine, China Agricultural University, Beijing, China
- 3Key Laboratory of Detection for Veterinary Drug Residue and Illegal Additive, MOA, College of Veterinary Medicine, China Agricultural University, Beijing, China
The objectives of this study were to investigate the prevalence and fluoroquinolone resistant Salmonella isolated from an integrated broiler chicken supply chain and their molecular characterization. In total, 73 Salmonella isolates were recovered from a broiler chicken supply chain in Shanghai. Salmonella isolates were tested for susceptibility to 11 antimicrobial agents using the broth dilution method and were characterized using pulsed-field gel electrophoresis (PFGE). Then, the Salmonella isolates were examined for mutations in quinolone resistance-determining region (QRDR) of gyrA, gyrB, parC, and parE, and were screened for plasmid-mediated quinolone resistance (PMQR) genes. Lastly, we sequenced the plasmids carrying qnrS1 in six Salmonella isolates from three sources (two isolated per source). Among 73 Salmonella isolates, 45 isolates were identified as S. Indiana, 24 were S. Schwarzengrund, 2 were S. Enteritidis, and 2 were S. Stanleyville. In addition, high rates of resistance were detected for nalidixic acid (41.1%) and ciprofloxacin (37.0%), while resistance to other test agents was diverse (2.0–100%). S. Indiana and S. Schwarzengrund isolates from different sources exhibited the same PFGE pattern, suggesting that the Salmonella isolates possessed high potential to spread along the broiler chicken supply chain. gyrA and parC exhibited frequent missense mutations. Moreover, qnrS1 was the most prevalent PMQR gene in the 73 Salmonella isolates, and it was found about a new hybrid plasmid. This study concludes a high prevalence of fluoroquinolone resistant Salmonella in chicken supply chain, threatening the treatment of Salmonella foodborne diseases. In particular, the emergence of a new hybrid plasmid carrying qnrS1 indicates that the recombination of plasmid carrying resistance gene might be a potential risk factor for the prevention and control strategies of drug resistance.
Introduction
Salmonellosis, caused by Salmonella, is one of the most frequently reported foodborne illnesses worldwide. Salmonella is divided into more than 2500 serovars by the White–Kauffman and Le Minor scheme. This classification scheme defines the serogroup according to expression of somatic lipopolysaccharide O antigens, and the serovar by the expression of flagellar H antigens. Salmonella, as an important human pathogen, is a potential public health risk.
It has been estimated that Salmonella causes about 1.2 million illnesses in the United States every year. Food is a major source of these infections, accounting for 1 million illnesses, 19,336 hospitalizations, and 378 deaths (Scallan et al., 2011). The majority of human infections caused by Salmonella is associated with the consumption of food products. Chicken, as one of the most widely consumed meats, is an important reservoir of Salmonella (Adu-Gyamfi et al., 2012; Truong Ha and Yamaguchi, 2012). Importantly, antibiotic-resistant bacteria of animal origin could be transmitted to humans (Piddock, 2002; Khemtong and Chuanchuen, 2008), which adversely affect the treatment of salmonellosis. Therefore, it is necessary to monitor the epidemiology and genetic characteristics of Salmonella in the food chain.
Fluoroquinolones are widely used to treat salmonellosis in human and animal (Folster et al., 2015). Currently, the main mechanism underlying quinolone resistance is the accumulation of mutations in quinolone resistance-determining region (QRDR) of gyrA, gyrB, parC, and parE and plasmid-mediated quinolone resistance (PMQR), which includes five major groups of qnr determinants (qnrA, qnrB, qnrC, qnrD, and qnrS), aac(6′)-Ib-cr and quinolone extrusion such as qepA and oqxAB (Strahilevitz et al., 2009). Some studies focused on fluoroquinolone resistance-related determinants about PMQR and QRDR in Salmonella derived from humans and animals (Wasyl et al., 2014; Wong et al., 2014). However, comprehensive data regarding fluoroquinolone resistance determinants in Salmonella from chicken supply chain are lacking, despite the implication for human health.
Thus, the aims of this study were to investigate the prevalence of Salmonella and their molecular characteristics related to fluoroquinolone resistance determinants, including PMQR and QRDR, in the broiler chicken supply chain in Shanghai. These data provide insight into the quantitative risk of resistant Salmonella from chicken supply chain.
Materials and Methods
Statement of Ethics
This study was carried out in accordance with the ethical guide lines of the College of Veterinary Medicine, China Agricultural University, Beijing. Moreover, before the initiation of this study, formal approval was obtained by the departmental committee of institute. Sampling was carried according to the standard protocols and with prior consent of the farmer/manager of the facilities.
Salmonella Strains and Antimicrobial Susceptibility Testing
Salmonella isolates were recovered from three sources including adult broilers, broiler carcasses and retail chicken, representing vertically integrated commercial broiler chicken supply chain in Shanghai City, China. One sample was collected from each animal or meat product as appropriate. Caecal samples from adult broilers were randomly collected at the abattoir. Whole carcasses or meat samples were aseptically obtained from chicken processing chain. Carcasses from the retail chicken source were sampled from the markets. All samples were immediately transported to the laboratory in an insulated ice boxes containing ice packs. Microbiological procedures were performed immediately upon arrival at the laboratory. All test strains were isolated in CHROMagar Salmonella agar (CHROMagar Company, Paris, France). Suspected Salmonella colonies were confirmed by a PCR assay targeting the invA gene (Rahn et al., 1992). Salmonella serotyping was conducted by performing the slide agglutination test, using Salmonella antisera (S & A Reagents Lab Ltd., Bangkok, Thailand) according to manufacturer’s instructions.
Salmonella isolates were subjected to antimicrobial susceptibility tests using standard broth dilution method of minimum inhibitory concentrations according to the guideline of the Clinical and Laboratory Standards Institute [CLSI] (2015a). Antimicrobial agents included 11 antimicrobials (i.e., amoxicillin/clavulanic acid, nalidixic acid, ampicillin, cefazolin, doxycycline, gentamicin, trimethoprim/sulfamethoxazole, chloramphenicol, ciprofloxacin, meropenem, and ceftriaxone). Escherichia coli ATCC 25922 was used as a quality control strain. The interpretive category for each isolate (susceptible, intermediate, or resistant) was determined according to the CLSI recommendations (Clinical and Laboratory Standards Institute [CLSI], 2015b).
Identification of Fluoroquinolone Resistance-Related Determinant
The DNA templates of isolates were prepared using TIANamp Bacteria DNA Reagent Kit (Tiangen, Beijing, China). The extracted DNAs were amplified by PCR assay. The mutations in gyrA, gyrB, parC, and parE genes were analyzed as described previously (Eaves et al., 2004). Salmonella isolates were screened for oqxA, oqxB, qnrA, qnrB, qnrC, qnrD, qnrS, aac(6′)-Ib-cr, and qepA genes. The primers and amplification conditions were described previously (Chen et al., 2012). PCR products were sequenced and identified.
Pulsed-Field Gel Electrophoresis (PFGE), S1 Nuclease Pulsed-Field Gel Electrophoresis (S1-PFGE), Southern Hybridization, Conjugation and Sequencing by Illumina
All Salmonella isolates were analyzed by PFGE method according to a previous protocol for subtyping Salmonella (Cui et al., 2016). According to the PFGE profile, six Salmonella isolates carrying qnrS1 were randomly selected from three sources (two isolates per source) for analysis by S1-PFGE and Southern hybridization, as previously described (Zhang et al., 2015). For the conjugation assay, six selected Salmonella isolates were used as donor strains, and sodium azide-resistant E. coli J53 was used as recipient strains. Both the donor strain and recipient strain were mixed on Luria-Bertani agar at a ratio of 1:3, and 100 μL mixtures were incubated for 16 h at 37°C. Transconjugants were selected on LB supplemented with sodium azide (100 mg/L) and ciprofloxacin (0.5 mg/L). Plasmids DNA were extracted from six Salmonella isolates transconjugants using by Wizard® Plus SV Minipreps DNA Purification Systems (Promega, Madison, WI, United States), then been sequenced by Illumina HiSeq 2500 system.
Results
Prevalence and Characteristics of Salmonella Isolates in the Chicken Supply Chain
A total of 73 (7.7%) Salmonella isolates were recovered from 715 samples. The highest prevalence of (17.5%, 48 isolates) detected in 200 broiler carcass samples, followed by 127 retail chicken samples (8.7%, 16 isolates), while lowest prevalence (2.3%, 9 isolates) of Salmonella isolates was observed in 388 adult broiler samples. Among these, 45 isolates were identified as S. Indiana, 24 were S. Schwarzengrund, 2 were S. Enteritidis, and 2 were S. Stanleyville. Furthermore, all 73 Salmonella isolates were evaluated for susceptibilities to eleven antibiotics. The rates of resistance were 41.1% for nalidixic acid and 37.0% for ciprofloxacin, while, resistance to other test agents varied substantially (amoxicillin/clavulanic acid: 43.8%, ampicillin: 42.5%, cefazolin: 47.9%, doxycycline: 95.9%, gentamicin: 6.8%, trimethoprim/sulfamethoxazole: 100%, chloramphenicol: 43.8%, meropenem: 2.0%, and ceftriaxone: 12.3%). PFGE profiles are shown in Figure 1. Notably, some S. Indiana isolates from different sources exhibited the same PFGE pattern. Likewise, some S. Schwarzengrund exhibited the same PFGE pattern, suggesting that Salmonella isolates have high potential to spread along the broiler chicken supply chain.
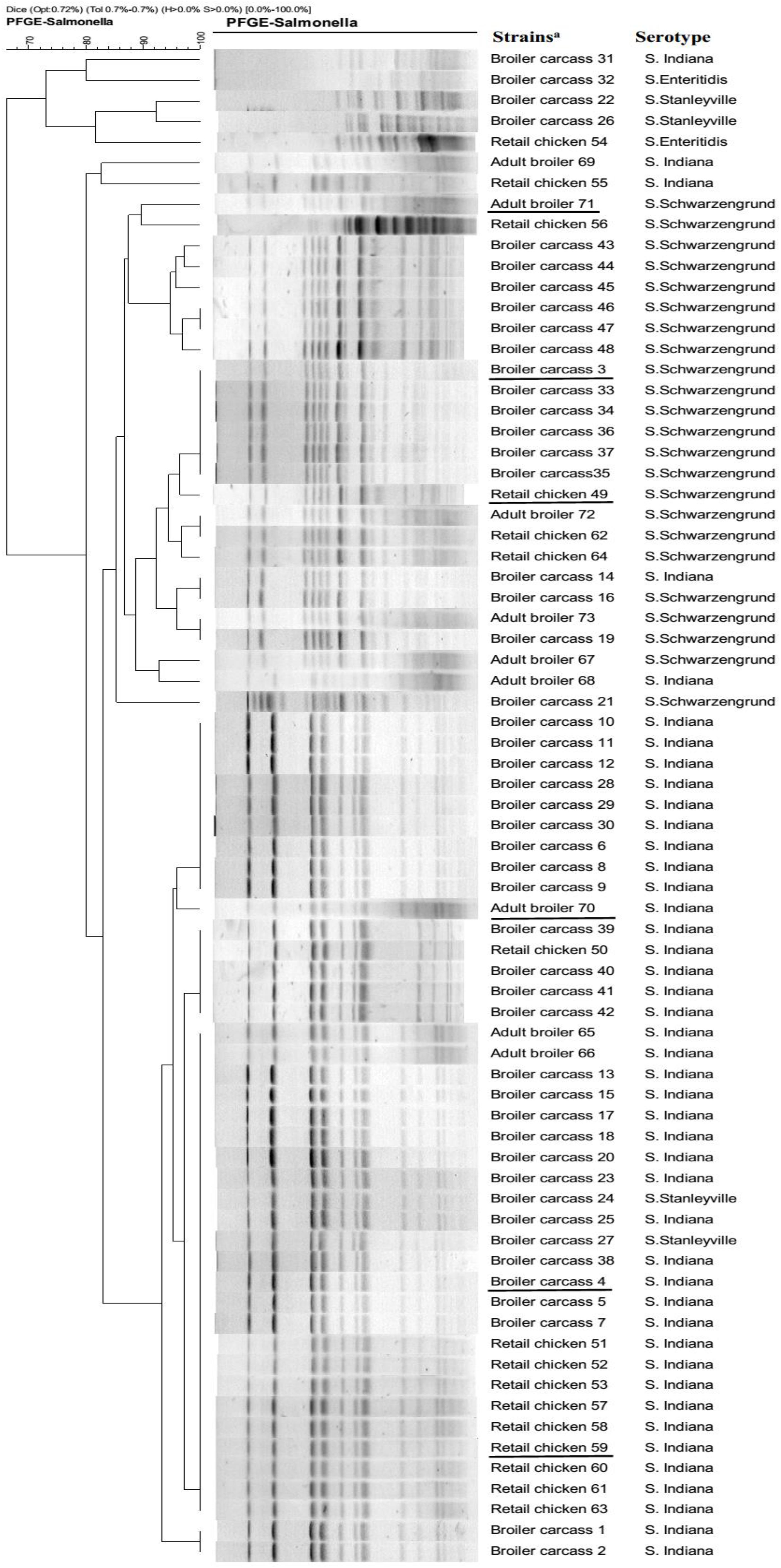
Figure 1. PFGE profiles of Salmonella isolates from the broiler chicken supply chain. Strain codes indicate the source of broiler chicken supply chain and the isolate number. aUnderlined strains were selected to been sequenced by the Illumina HiSeq 2500 system.
Fluoroquinolone Resistance Determinants in the Chicken Supply Chain
Based on the detail information in Supplementary Table S1, mutations within QRDR of gyrA, gyrB, parC, and parE are summarized in terms of serotype in Table 1 The presented data indicated that missense mutations frequently occurred in gyrA and parC, whereas silent mutations were observed in gyrA, gyrB, parC, and parE. Among 73 Salmonella isolates from this broiler chicken supply chain, 47 Salmonella isolates carried the wild-type (no mutation) within gyrA gene, missense mutation (Thr57Ser) within parC gene, 15 of which did not exhibit implicated in fluoroquinolone resistance phenotypes. The remaining 26 isolates contained missense mutations within gyrA (Ser83Phe and Asp87Asn) and parC (Thr57Ser, Ser80Arg). In addition, Table 2 shows the distribution of fluoroquinolone resistance genes (oqxA, oqxB, qnrA, qnrB, qnrC, qnrD, qnrS, aac (6′)-Ib-cr, and qepA). Among them, qnrS1 (30/73) was predominant gene, following by qnrB1 (22/73), oqxA (1/73), oqxB (1/73), and aac (6′)-Ib-cr (1/73). The genes sequences of qnrS1, qnrB1, oqxA, oqxB, and aac (6′)-Ib-cr were deposited in GenBank. Accession numbers were MK990505, MK990506, MK990507, MK990508, and MK990509, respectively. However, the other fluoroquinolone resistance genes were not observed. In total, 53 of 73 Salmonella isolates carried fluoroquinolone resistance genes, and qnrS1 gene was detected in most S. Indiana (23/45), S. Schwarzengrund (5/22), and S. Enteritidis (2/2) isolates, accounting for the majority Salmonella isolates in the broiler supply chain.
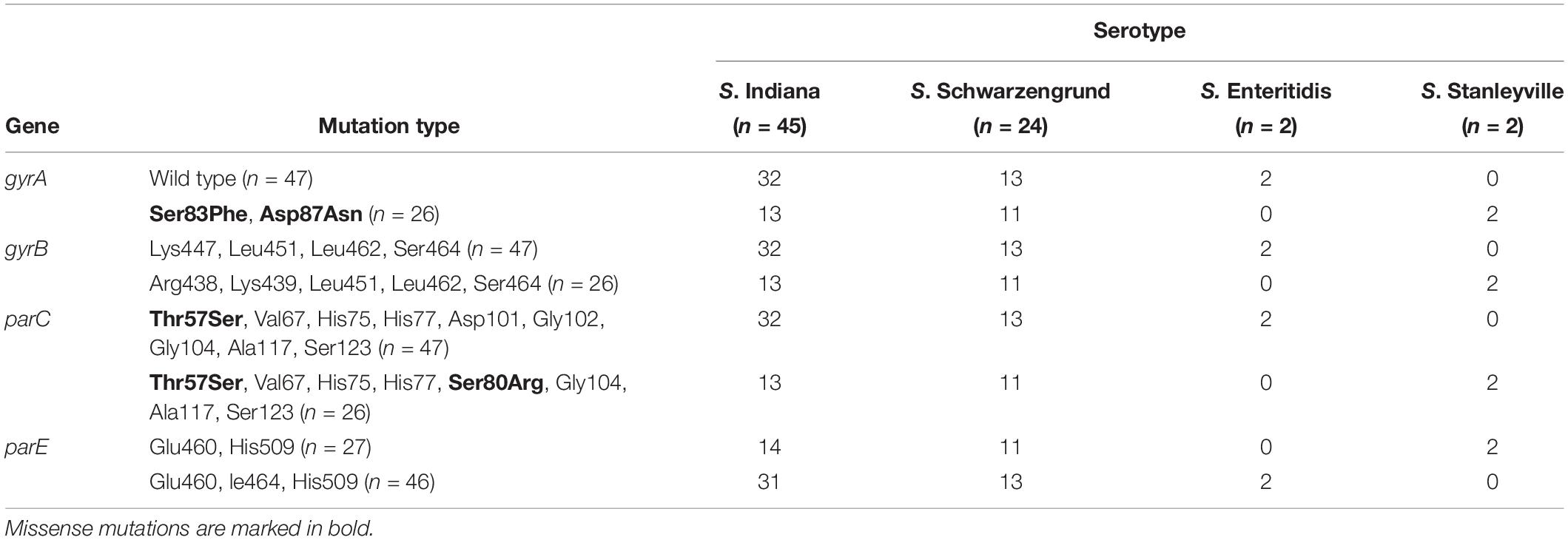
Table 1. Mutations within QRDR of gyrA, gyrB, parC, and parE genes in Salmonella isolates from broiler chicken supply chain.
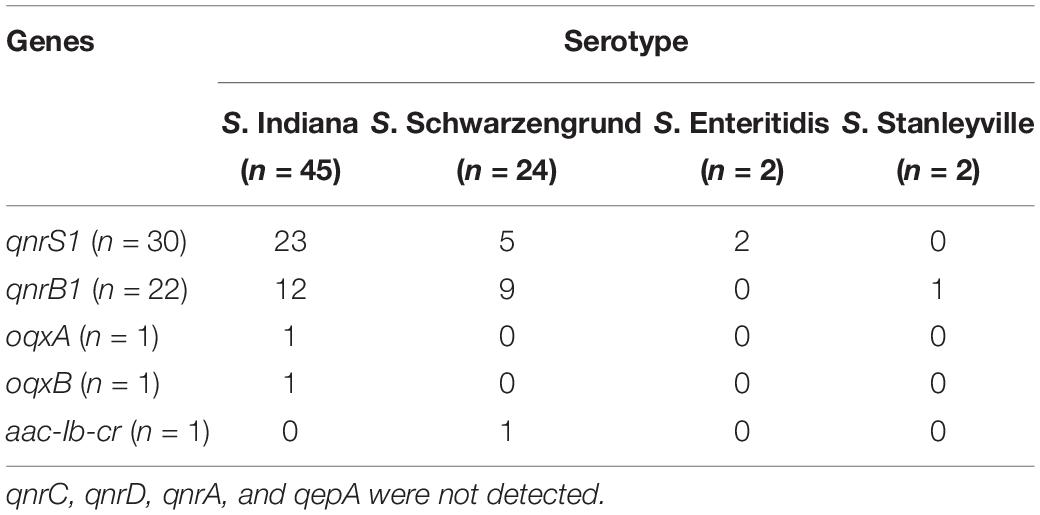
Table 2. The distribution of fluoroquinolone resistance genes about PMQR in Salmonella isolates from broiler chicken supply chain.
Novel Plasmid Associated With qnrS1
The qnrS1 was predominant gene in the chicken supply chain, thus, six Salmonella isolates from three sources (two isolates per source) were randomly selected to analyze the transmission mechanism of qnrS1 according to the PFGE profile. S1-PFGE and Southern blot analyses (Figure 2) indicated that qnrS1 is located on a ∼40 kb plasmid, designated pSH-01. Conjugation experiments by filter mating revealed that qnrS1 could be co-transferred from Salmonella isolates to E. coli J53. S1-PFGE and Southern hybridization confirmed that the DNA probes specific for qnrS1 hybridized to the same plasmids with a size ∼40 kb in both Salmonella isolates and their transconjugants (Figure 2). Then, the plasmids were extracted from transconjugants and sequenced using the Illumina MiSeq system. The analysis of the sequences showed that the plasmids are extremely similar with more than 99% identity, indicating that the plasmids from the different strains are indeed the same plamids. The NCBI BLAST results showed that the hybrid plasmid carrying qnrS1 is a new plasmid type (pSH-01, submission number KY486279.1). It was 43,257 bp in length and harbored 51 predicted open reading frames. Furthermore, a plasmid sequence analysis (Carattoli et al., 2014) of pSH-01 indicated that it is an IncR type hybrid plasmid.
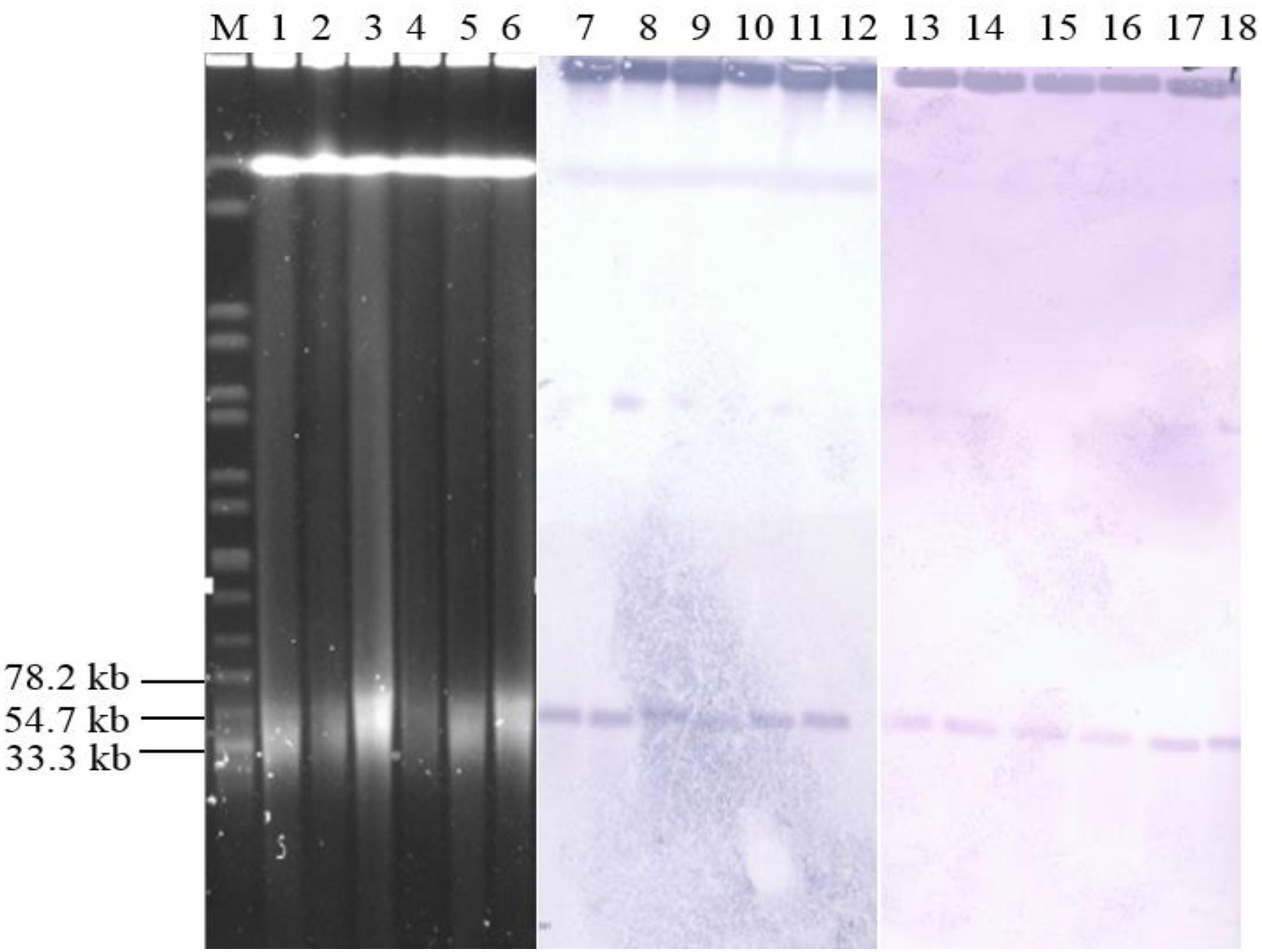
Figure 2. Results of S1-PFGE and Southern blotting. Location of the qnrS1-carrying plasmid pSH-01 in Salmonella by S1-PFGE (lanes M, 1, 2, 3, 4, 5, and 6) and Southern blot hybridization (lanes M, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, and 18). Lane M, serotype Braenderup H9812; lane 1 and 7, Adult broiler 70; lane2 and 8, Adult broiler 71; lane 3 and 9, Broiler carcass 3; lane 4 and 10, Broiler carcass 4; lanes 5 and 11, Retail chicken 49; lanes 6 and 12, Retail chicken 59; lanes 13, Adult broiler 70 transconjugant; lane 14, Adult broiler 71 transconjugant; lane 15, Broiler carcass 3 transconjugant; lane 16, Broiler carcass 4 transconjugant; lanes 17, Retail chicken 49 transconjugant; and lanes 18, Retail chicken 5.
According to the BLAST results of pSH-01 nucleotide sequence against the NCBI database, the two junctions (Figures 3B,C) were often occurred, indicating that the three fragments are usually associated and transferred as a whole. These fragments were almost derived from plasmids. Notably, the genetic features of pSH-01 showed that the fragment carrying qnrS1 (2849–14429) is derived from plasmids of one Shigella flexneri and six E. coli isolates (blast query cover: above 99% and blast identity: 99%). Another fragment (nt14429–31031) was derived from plasmids of Salmonella and other bacterial isolates, and an additional fragment (30758–43257, 1–2851) was mostly derived from plasmids of Klebsiella pneumoniae. The origin of target plasmids covered different bacterial host. According to an alternative sequence analysis, for example, it was postulated that pSH-01 might be hybrids of three plasmids (Figure 3A); the region spanning nt 2849–14429 matched with plasmid pEBG1 (KF738053; nt 37915–26335) of the E. coli strain, nt 14429–31031 shared a nucleotide identity of 99% to the plasmid p33676 (CP012682; nt 25123–41725) from S. Typhimurium, and nt 30758–43257, 1–2851 shared a nucleotide identity of 99% to the plasmid tig00000005_pilon (CP021858; nt 17585–5015, 4958–1790) from K. pneumoniae AR_0125. Similar recombination junctions could also be found in another alternative sequence analysis. Therefore, based on sequence analyses of three recombination junctions (Figure 3B), it was postulated that Tn3, IS6 and homologous recombination played important roles in the formation of pSH-01.
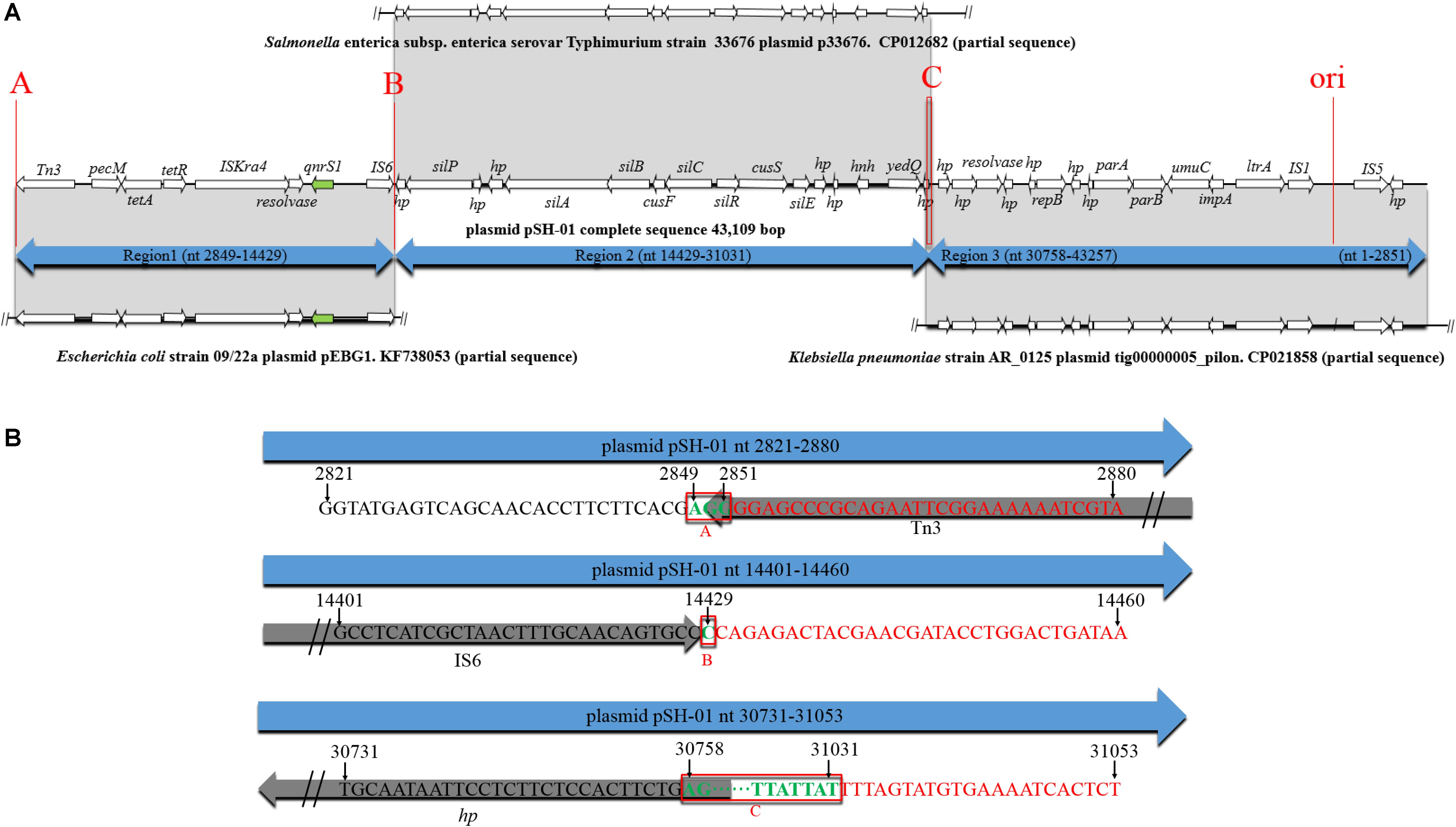
Figure 3. Genetic environment of the qnrS1 gene in plasmid pSH-01. (A–C) The potential recombination junctions in plasmid pSH-01. (A) A structural comparison was made with plasmid pEBG1 from E. coli 09/22a, plasmid p33676 from S. Typhimurium, and plasmid tig00000005_pilon from K. pneumoniae AR_0125. The arrows indicate the positions and directions of transcription of the genes. Regions of 99% nucleotide sequence identity are marked by gray shading. (B) Sequence analysis of three recombination junctions (A–C). The bases in black are upstream sequence of recombination junction in plasmid pSH-01. The bases in red are downstream sequence of recombination junction in plasmid pSH-01. The bases in green and in red boxes represent the common region shared by potential hybridized plasmids.
Discussion
In the broiler supply chain in China, there are geographical differences in the dominance of various Salmonella serotypes and in the prevalence of fluoroquinolones resistant Salmonella. In this study, S. Indiana was the most common serotype isolated from the broiler supply chain, which differs from previous results showing that S. Enteritidis is dominant in Qingdao and S. Weltevreden is dominant in Guangdong, China (Cui et al., 2016; Ren et al., 2016). In addition, several studies have indicated Salmonella could be transmitted along the food chain (Nogrady et al., 2008; Hauser et al., 2012). Our PFGE results showed that there is the potential for the transmission of Salmonella along the broiler chicken chain. With the emergence of antibiotic-resistant bacteria presenting a serious challenge in human and veterinary medicine globally, there is an abundance of evidence showing that the antimicrobial resistance of Salmonella in the chicken supply chain is more possibly attributed to the use of antibiotics in the animal husbandry (Cui et al., 2016). In particular, there are many reports of increasing prevalence of fluoroquinolone-resistant Salmonella (Piddock, 2002; Wasyl et al., 2014), which might be a potential risk for human health. In this study, resistance to ciprofloxacin was detected in 37.0% of the Salmonella isolates, and this resistance rate was relatively high compared to those of previous reports (Cui et al., 2016; Ren et al., 2016; Nhung et al., 2018).
In this study, the same PFGE pattern was shared among the majority of Salmonella isolates (such as S. Indiana and S. Schwarzengrund isolates), which might suggest they are clones of S. Indiana and S. Schwarzengrund, respectively. Compared with the QRDR genotypes in non-clones Salmonella isolates (Eaves et al., 2004), the genetic diversity of Salmonella isolates in this study was lower. In addition, similar to previous investigation (Hopkins et al., 2005), our results indicate that missense mutations occurred frequently in QRDR of gyrA and parC, which are considered major quinolone resistance determinants in Salmonella. In this study, gyrA missense mutations (Ser83Phe and Asp87Asn) were detected in 26/73 Salmonella isolates and these are considered the major target-site mutations in Salmonella (Nüeschinderbinen et al., 2015). Thr57Ser parC substitution was frequently observed in the Salmonella isolates, and a second substitution (Ser80Arg) in parC was also detected in 26 Salmonella isolates. Importantly, Thr57Ser parC substitution was considered not or doubtfully associated to fluoroquinolone resistance phenotypes (Wasyl et al., 2014). The 15 Salmonella isolates with the Thr57Ser parC substitution in this study did not show fluoroquinolone resistance phenotypes, in agreement with previous report (Ceyssens et al., 2015). Although mutation types in gyrA and parC were similar to those in previous studies of Salmonella, it is worth noting the high frequency of silent site mutations in QRDR, which might be developed into potential missense mutations (Heisig, 1993; Hopkins et al., 2005). Furthermore, a recent study has shown that mutations in the target genes gyrA and parC are correlated with an increase of intrinsic fitness in Salmonella (Baker et al., 2013). This indicated that the potential risk that Salmonella isolates with mutations in gyrA and parC may naturally maintain during the broiler chicken supply chain, even if fluoroquinolone use was reduced.
In addition, the predominant PMQR gene varies among bacteria from different sources. The most common PMQR gene was oqxAB in E. coli from chicken (Chen et al., 2012) and in Salmonella from retail meat (Lin et al., 2015), qnrB in Enterobacteriaceae from crows (Halova et al., 2014), and aac(6′)-Ib-cr in bacteria isolated from sewage and surface water (Osinska et al., 2016). However, this study indicated that qnrS was commonly distributed in Salmonella isolates from the broiler chicken supply chain, consistent with the high reported rates in Salmonella isolated from animals, food, and feed (Wasyl et al., 2014).
The recombination of plasmid, to some extent, can provide a mechanism to improve the diversity of plasmids carrying resistance genes. Recombination of hybrid plasmids frequently occurs at insertion sequence (IS) location (Hudson et al., 2014). Recently, NDM-5 and mcr-1 were recombined in the plasmid pCQ02-121 by recombination junctions: IS26 and the nic site of oriT (Sun et al., 2016). Similarly, in this study, the novel plasmid pSH-01 might arise via recombination junction IS6. The other two recombination junctions involve Tn3 and homologous recombination of sequences. Transposons in the Tn3 family can mediate gene reassortment and genomic plasticity owing to their modular organization, and they contribution substantially to antimicrobial drug resistance dissemination or to endowing environmental catabolic capacities (Nicolas et al., 2015).
Genetic features of pSH-01 showed that only region 2 (nt 14429–31031) could matched the plasmid sequences from clinical Salmonella isolates, and the sequences with matches in the other two regions (region 1 and 3) were derived from plasmids from non-Salmonella bacteria. The full-length of pSH-01 did not match an individual plasmid in NCBI. Therefore, this is a new plasmid in Salmonella isolates from the broiler chicken supply chain, suggesting that the diversity of plasmids carrying the resistance gene might be a potential risk factor for the dissemination of qnrS1.
It is worth noting that the new plasmid carrying qnrS1 presented in the six Salmonella isolates (three S. Indiana and three S. Schwarzengrund) from different sources in the broiler chicken supply chain. This suggests that there was a potential epidemic spread of the plasmid in the Salmonella isolates of different serotype from different geographical origin, which is similar to the potential transmission of the plasmids among various serotype of Salmonella and diverse geographical location (Li et al., 2016; Wong et al., 2017). Therefore, we should carefully monitor the new plasmid carrying qnrS1 along the chicken supply chain.
This study provided comprehensive data for the prevalence of Salmonella and their fluoroquinolone resistance determinants associated with QRDR and PMQR in the broiler chicken supply chain. Furthermore, we found that qnrS1, a transmissible PMQR gene, was prevalent in Salmonella isolates from the broiler chicken supply chain. Selective pressure from fluoroquinolones in animals may further promote the recombination and dissemination of the plasmid carrying PMQR genes.
Author Contributions
CW and MC designed the experiments. MC, PZ, JL, and CS carried out the experiments. MC wrote the manuscript. MC, LS, CZ, and QZ reviewed and revised the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2018YFD0500305).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Professor Tariq Ali for help on the manuscript modification. We would like to thank Tami Wang and Hejia Wang for English language suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01865/full#supplementary-material
References
Adu-Gyamfi, A., Torgby-Tetteh, W., and Appiah, V. (2012). Microbiological quality of chicken sold in accra and determination of D10-value of E. coli. Food Nutr Sci. 3, 693–698. doi: 10.4236/fns.2012.35094
Baker, S., Duy, P. T., Nga, T. V. T., Dung, T. T. N., Phat, V. V., Chau, T. T. et al. (2013). Fitness benefits in fluoroquinolone-resistant Salmonella typhi in the absence of antimicrobial pressure. eLife 2:e01229. doi: 10.7554/eLife.01229
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Larsen, M.V., Lund, O., Villa, L. et al. (2014) In silico detection and typing of plasmids using plasmid finder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Ceyssens, P. J., Mattheus, W., Vanhoof, R., and Bertrand, S. (2015). Trends in serotype distribution and antimicrobial susceptibility in Salmonella enterica isolates from humans in Belgium, 2009 to 2013. Antimicrob.AgentsChemother. 59, 544–552. doi: 10.1128/AAC.04203-14
Chen, X., Zhang, W., Pan, W., Yin, J., Pan, Z., Gao, S., et al. (2012). Prevalence of qnr, aac(6 ’)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob. Agents Chemother. 56, 3423–3427. doi: 10.1128/AAC.06191-11
Clinical and Laboratory Standards Institute [CLSI] (2015a). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically CLSI Document M07-A10, 10th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Clinical and Laboratory Standards Institute [CLSI] (2015b). Performance Standard for Antimicrobial Susceptility Testing;Twenty-Fifth Information Suppement. CLSI document M100-S25, Wayne, PA: Clinical and Laboratory Standards Institute.
Cui, M., Xie, M., Qu, Z., Zhao, S., Wang, J., Wang, Y. et al. (2016). Prevalence and antimicrobial resistance of Salmonella isolated from an integrated broiler chicken supply chain in qingdao, China. Food Control. 62, 270–276. doi: 10.1016/j.foodcont.2015.10.036
Eaves, D. J., Randall, L., Gray, D. T., Buckley, A., Woodward, M. J., White, A. P., et al. (2004). Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob. Agents Chemother. 48, 4012–4015. doi: 10.1128/AAC.48.10.4012-4015.2004
Folster, J. P., Campbell, D., Grass, J., Brown, A. C., Bicknese, A., Tolar, B. et al. (2015). Identification and characterization of multidrug-resistant Salmonella enterica serotype albert isolates in the United States. Antimicrob. Agents Chemother. 59, 2774–2779. doi: 10.1128/AAC.05183-14
Halova, D., Papousek, I., Jamborova, I., Masarikova, M., Cizek, A., Janecko, N. et al. (2014). Plasmid-mediated quinolone resistance genes in enterobacteriaceae from american crows: high prevalence of bacteria with variable qnrB genes. Antimicrob. Agents Chemother. 58, 1257–1258. doi: 10.1128/AAC.01849-13
Hauser, E., Tietze, E., Helmuth, R., Junker, E., Prager, R., Schroeter, A. et al. (2012). Clonal dissemination of Salmonella enterica serovar infantis in Germany. Foodborne Pathog. Dis. 9, 352–360. doi: 10.1089/fpd.2011.1038
Heisig, P. (1993). High-level fluoroquinolone resistance in a Salmonella typhimurium isolate due to alterations in both gyrA and gyrB genes. J. Antimicrob. Chemother. 32, 367–377. doi: 10.1093/jac/32.3.367
Hopkins, K. L., Davies, R. H., and Threlfall, E. J. (2005). Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25, 358–373. doi: 10.1016/j.ijantimicag.2005.02.006
Hudson, C. M., Bent, Z. W., Meagher, R. J., and Williams, K. P. (2014). Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS One 9:e99209. doi: 10.1371/journal.pone.0099209
Khemtong, S., and Chuanchuen, R. (2008). Class 1 integrons and Salmonella genomic island 1 among Salmonella enterica isolated from poultry and swine. Microb. Drug Resist. 14, 65–70. doi: 10.1089/mdr.2008.0807
Lin, D., Chen, K., Chan, E. W. -C., and Chen, S. (2015). Increasing prevalence of ciprofloxacin-resistant food-borne Salmonella strains harboring multiple PMQR elements but not target gene mutations. Sci. Rep. 5:14754. doi: 10.1038/srep14754
Li, X. P., Fang, L. X., Song, J. Q., Xia, J., Huo, W., Fang, J. T. et al. (2016). Clonal spread of mcr-1 in PMQR-carrying ST34 Salmonella isolates from animals in China. Sci. Rep. 6:38511. doi: 10.1038/srep38511
Nüeschinderbinen, M., Abgottspon, H., Sägesser, G., Cernela, N., and Stephan, R. (2015). Antimicrobial susceptibility of travel-related Salmonella enterica serovar Typhi isolates detected in Switzerland (2002-2013) and molecular characterization of quinolone resistant isolates. BMC Infect Dis. 15:212. doi: 10.1186/s12879-015-0948-2
Nhung, N. T., Van, N. T. B., Cuong, N. V., Duong, T. T. Q., Nhat, T. T., Hang, T. T. T. et al. (2018). Antimicrobial residues and resistance against critically important antimicrobials in non-typhoidal Salmonella from meat sold at wet markets and supermarkets in Vietnam. Int. J. Food Microb. 266, 301–309. doi: 10.1016/j.ijfoodmicro.2017.12.015
Nicolas, E., Lambin, M., Dandoy, D., Galloy, C., Nguyen, N., Oger, C. A., et al. (2015). The Tn3-family of replicative transposons. Microbiol. Spectr. 3, 1–32.
Nogrady, N., Kardos, G., Bistyak, A., Turcsanyi, I., Meszaros, J., Galantai, Z. et al. (2008). Prevalence and characterization of Salmonella infantis isolates originating from different points of the broiler chicken-human food chain in hungary. Int. J. Food Microbiol. 127, 162–167. doi: 10.1016/j.ijfoodmicro.2008.07.005
Osinska, A., Harnisz, M., and Korzeniewska, E. (2016). Prevalence of plasmid-mediated multidrug resistance determinants in fluoroquinolone-resistant bacteria isolated from sewage and surface water. Environ. Sci. Pollut. Res. Int. 23, 10818–10831. doi: 10.1007/s11356-016-6221-4
Piddock, L. J. (2002). Fluoroquinolone resistance in Salmonella serovars isolated from humans and food animals. FEMS Microbiol. Rev. 26, 3–16. doi: 10.1016/s0168-6445(01)00076-6
Rahn, K., Degrandis, S. A., Clarke, R. C., McEwen, S. A., Galan, J. E., Ginocchio, C. et al. (1992). Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6, 271–279. doi: 10.1016/0890-8508(92)90002-f
Ren, X., Li, M., Xu, C., Cui, K., Feng, Z., and Fu, Y. et al. (2016). Prevalence and molecular characterization of Salmonella enterica isolates throughout an integrated broiler supply chain in China. Epidemiol. Infect. 144, 1–11. doi: 10.1017/S0950268816001515
Scallan, E., Hoekstra, RM., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., and Roy, S. L., et al. (2011) Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101.
Strahilevitz, J., Jacoby, G. A., Hooper, D. C., and Robicsek, A. (2009). Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22, 664–689. doi: 10.1128/CMR.00016-09
Sun, J., Yang, R. S., Zhang, Q., Feng, Y., Fang, L. X., Xia, J. et al. (2016). Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat. Microbiol. 1:16176. doi: 10.1038/nmicrobiol.2016.176
Truong Ha, T., and Yamaguchi, R. (2012). Molecular characterization of antibiotic-resistant Salmonella isolates from retail meat from markets in Northern Vietnam. J. Food Prot. 75, 1709–1714. doi: 10.4315/0362-028X.12-101
Wasyl, D., Hoszowski, A. and Zajä, C. M. (2014). Prevalence and characterisation of quinolone resistance mechanisms in Salmonella spp. Vet. Microbiol. 171, 307–314. doi: 10.1016/j.vetmic.2014.01.040
Wong, M. H., Chan, E. W., and Chen, S. (2017). IS26-mediated formation of a virulence and resistance plasmid in Salmonella enteritidis. J. Antimicrob. Chemother. 32, 367–377. doi: 10.1093/jac/dkx238
Wong, M. H., Chan, E. W., Liu, L. Z., and Chen, S. (2014). PMQR genes oqxAB and aac(6′)Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella typhimurium. Front. Microbiol. 5:521. doi: 10.3389/fmicb.2014.00521
Keywords: Salmonella, fluoroquinolone resistance, qnrS1, hybrid plasmid, broiler chicken supply chain
Citation: Cui M, Zhang P, Li J, Sun C, Song L, Zhang C, Zhao Q and Wu C (2019) Prevalence and Characterization of Fluoroquinolone Resistant Salmonella Isolated From an Integrated Broiler Chicken Supply Chain. Front. Microbiol. 10:1865. doi: 10.3389/fmicb.2019.01865
Received: 07 August 2018; Accepted: 29 July 2019;
Published: 13 August 2019.
Edited by:
Jose L. Martinez, Spanish National Research Council (CSIC), SpainReviewed by:
Lei Dai, Iowa State University, United StatesEliana Guedes Stehling, Universidade de São Paulo Ribeirão Preto, Brazil
Copyright © 2019 Cui, Zhang, Li, Sun, Song, Zhang, Zhao and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Congming Wu, d3VjbUBjYXUuZWR1LmNu
 Mingquan Cui
Mingquan Cui Peng Zhang
Peng Zhang Jiyun Li
Jiyun Li Chengtao Sun
Chengtao Sun Li Song1
Li Song1