- Université Grenoble Alpes, Le Centre National de la Recherche Scientifique, Commissariatá l'Energie Atomique et aux Energies Alternatives, Institut de Biologie Structurale, Grenoble, France
Measles virus is a negative strand virus and the genomic and antigenomic RNA binds to the nucleoprotein (N), assembling into a helical nucleocapsid. The polymerase complex comprises two proteins, the Large protein (L), that both polymerizes RNA and caps the mRNA, and the phosphoprotein (P) that co-localizes with L on the nucleocapsid. This review presents recent results about N and P, in particular concerning their intrinsically disordered domains. N is a protein of 525 residues with a 120 amino acid disordered C-terminal domain, Ntail. The first 50 residues of Ntail extricate the disordered chain from the nucleocapsid, thereby loosening the otherwise rigid structure, and the C-terminus contains a linear motif that binds P. Recent results show how the 5′ end of the viral RNA binds to N within the nucleocapsid and also show that the bases at the 3′ end of the RNA are rather accessible to the viral polymerase. P is a tetramer and most of the protein is disordered; comprising 507 residues of which around 380 are disordered. The first 37 residues of P bind N, chaperoning against non-specific interaction with cellular RNA, while a second interaction site, around residue 200 also binds N. In addition, there is another interaction between C-terminal domain of P (XD) and Ntail. These results allow us to propose a new model of how the polymerase binds to the nucleocapsid and suggests a mechanism for initiation of transcription.
Introduction
Measles virus is a human virus that, although the vaccine is very efficient, still kills about 100,000 people per year (Laksono et al., 2018; WHO, 2019). Measles virus is a single-stranded negative-sense RNA virus belonging to the Paramyxoviridae family. The viral RNA, 15,894 nucleotides long, is bound by thousands of copies of the viral nucleoprotein, forming the helical nucleocapsid (NC). The viral polymerase complex, comprising the viral L protein (L, for Large protein) and phosphoprotein, uses the NC as the template for transcription and replication. The only known structure of L of the viruses of Mononegavirales order, to which also paramyxoviruses belong, is the one from vesicular stomatitis virus (VSV) (Rahmeh et al., 2010; Liang et al., 2015; Qiu et al., 2016), and several reviews have used this structure as a model for the function and structure of measles virus L (Sourimant and Plemper, 2016; Fearns and Plemper, 2017). Figure 1 shows the genes of the measles virus, coding for eight proteins within six genes: nucleoprotein (N, blue), phosphoprotein (P, red), matrix (M), the fusion protein (F), and haemagglutinin (H) (the spike proteins), and the polymerase (L, yellow). The gene of P also encodes the mRNAs for the C and V proteins. These proteins have been found in many other Paramyxoviruses and speculated to be involved in regulation of host cell immune response through different pathways. In addition, it should be mentioned that V of measles virus share the first 231 amino acids with P and thus can also maintain N chaperoning.
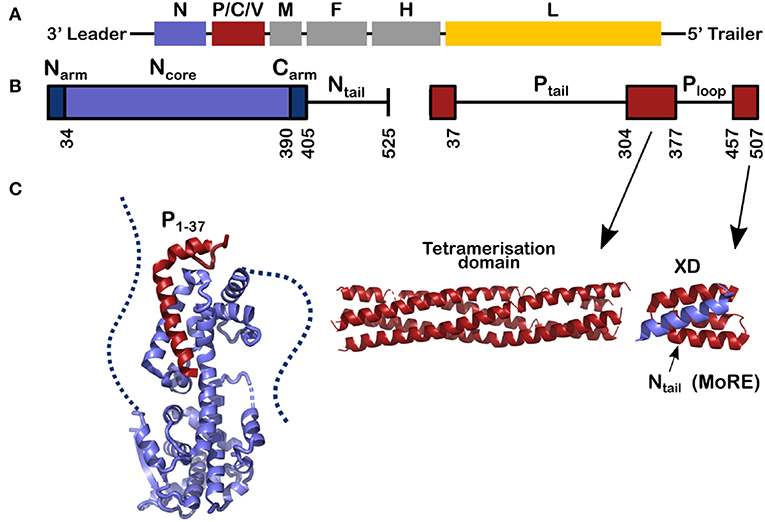
Figure 1. Schemes for the genes and for the nucleoprotein (N) and phosphoprotein (P) of measles virus. (A) The viral RNA of measles virus with its genes; The second gene codes for 3 proteins, P, C, and V and the rest of the genes code for one protein. Throughout this review the structures of N are shown in blue, for P in red and in Figure 8 the polymerase of VSV is shown in yellow. (B) The schemes of N and P proteins show the structured parts with colored bars and the disordered parts with a line. (C) The structured parts of N and P. Left, for the structure of N0P we used the PDB (Protein Data Bank) number 4CO6 and we show the Narm (left) and the Carm (right) with dash; middle, for the tetramerization domain of P we used PDB 3ZDO and right, for the complex between the linear helical motif of Ntail and the X domain of P we used PDB 1T6O.
This review describes recent results on the phosphoprotein (P) and nucleoprotein (N) of measles virus. The classical paradigm that a protein must have a defined and stable structure and that the functional mechanisms of the protein in the cell can be derived from this structure, is not applicable to P and N. This is because most of P is disordered, as is the C-terminal tail of N (Tompa, 2002; Uversky, 2002; Karlin et al., 2003; Longhi et al., 2003; Bourhis et al., 2004; Gerard et al., 2009; Habchi et al., 2014). As shown in Figure 1, 120 residues of N and 265 + 79 residues of P are intrinsically disordered in their functional state. Together this makes up to 1392 RNA bases, almost 9% of the full genome.
Structures and Complexes Between N and P
P and N have multi-domain structures with alternating ordered and disordered regions (Figure 1). P starts with a long disordered tail (304 amino acids) followed by a tetrameric domain of four long, parallel helices between residues 304 and 377 (Figure 1). Tetramer domains of similar length are found in Sendai, Nipah (Tarbouriech et al., 2000; Communie et al., 2013a; Bruhn et al., 2014). P of mumps also forms a tetramer of the same length but with two anti-parallel dimers (Cox et al., 2013; Pickar et al., 2015). For Sendai, Nipah and measles viruses, the polymerase binds to the tetramerization domain (Bowman et al., 1999; Bloyet et al., 2019; Bruhn et al., 2019). At the C-terminus of P there is another folded domain (XD) which is linked to the tetramerization domain via the unfolded Ploop (residues 377–457).
N comprises two distinct parts: Folded Ncore and Ntail, an intrinsically disordered domain, containing a linear motif with α-helical propensity (residues 484–502; Figures 1, 2A). In solution this sequence folds into several helices of different length that exchange with each other and with a purely unfolded form on timescales faster than microseconds (Jensen et al., 2008, 2011; Communie et al., 2013b). This motif binds to the C-terminal domain of P, the X-domain (XD) (Figure 1; Johansson et al., 2003; Longhi et al., 2003; Kingston et al., 2004a,b; Bourhis et al., 2005; Houben et al., 2007; Schneider et al., 2015). Orthomyxoviruses like influenza virus have several separate RNA molecules, each bound to the viral polymerase. Because the nucleocapsids form circular complexes, with the 5′ and 3′ ends bound in close proximity on the polymerase (Ruigrok et al., 2011; Reich et al., 2014), the polymerase stays attached to the same gene while making new mRNAs. In paramyxoviruses such as measles, the polymerase first transcribes the leader RNA and then the mRNAs of N, P, M, F, H, and L. After each mRNA is terminated, L has to move from the end of the gene to the start of the next gene. The binding of Ntail and XD of P may assist in this process such that L remains in contact with the template (Brunel et al., 2014; Bloyet et al., 2016; Cox et al., 2017; Thakkar et al., 2018). The Kd of binding between Ntail and XD in paramyxoviruses is rather weak, between 3 and 8 μM (Schneider et al., 2015; Bloyet et al., 2016). If the affinity is artificially decreased further through mutations in the interacting protein domains, L has been shown to leave the template before transcribing all the genes (Bloyet et al., 2016).
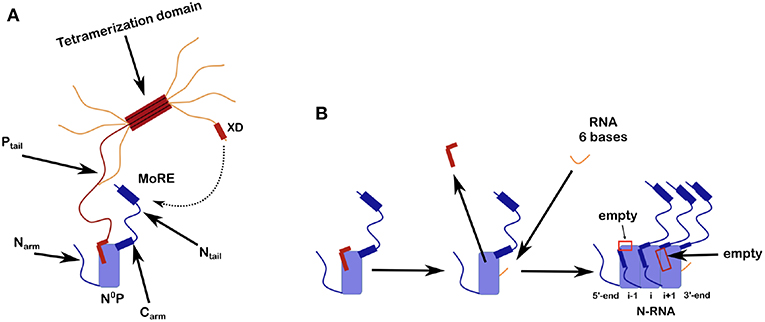
Figure 2. Scheme for N0P and the assembly of nucleocapsid-like particles (NCLP). (A) The N0P complex is made from one full N protomer with the Narm and the Carm that continues in the Ntail plus a tetramer of P. Close to the end of the Ntail there is the helical motif that could bind to the X domain of P. For this picture we show only one full Ptail. The two N-terminal helices of Ptail bind N0 and for the crystal structures of N0P all the Narm, Carm plus Ntail and almost all of P (only the first 2 helices of P are binding to N0) were cut away. (B) Scheme of the assembly of the NCLP with N0P with α1/2 of P and synthesized RNA (Milles et al., 2016). When the RNA binds N0, the two helices of P dissociates from the N0 and the monomer will assemble to the NCLP. In the NCLP, the N-terminal end of the Narm forms a helix that binds to the protomer i-1 and the Carm binds to protomer i+1. In the NCLP the 5′ and 3′ protomers both have one empty site.
Ncore is flanked by N and C-arms which are disordered in the RNA-free form of the protein, and become more structured upon binding to the neighboring Ncore subunits in nucleocapsids. During NC formation the Narm and Carm bind to the neighboring N protomers (Figure 2), with part of the Narm forming a helix (residues 2–14) bound to the surface of the neighbor i-1 and the Carm folding onto neighbor i+1. N can bind RNA, either viral RNA (vRNA) during infection, or cellular RNA during expression in cellular expression systems, making recombinant nucleocapsid-like structures. In order to avoid non-specific binding to cellular RNA, N is chaperoned by the N-terminal residues of P forming an N0P complex (Figures 1, 2A; Masters and Banerjee, 1988; Peluso and Moyer, 1988; Curran et al., 1993, 1995; Mavrakis et al., 2003). When the Narm and Carm from the N0P complex are removed, the complex can be crystallized and the structure can be determined through X-ray diffraction. In this way, the N0P structures of VSV, Nipah, measles, PIV5, HMPV, Marburg and Ebola viruses were determined (Leyrat et al., 2011; Yabukarski et al., 2014; Guryanov et al., 2015; Kirchdoerfer et al., 2015; Leung et al., 2015; Renner et al., 2016; Zhu et al., 2017; Aggarwal et al., 2018).
Up to now, there are no atomic resolution structures of the viral NCs, but NC-like particles (NCLPs) made from measles, Marburg, rabies and VSV viruses, for example, have similar structure as the viral nucleocapsids (Spehner et al., 1991; Fooks et al., 1993; Iseni et al., 1998; Mavrakis et al., 2002; Bhella et al., 2004; Schoehn et al., 2004). Some of the NCLPs assemble into rings rather than helical particles and the first two atomic resolution structures were determined from rings of VSV (10 protomers) and rabies (11 protomers per ring) with the RNA inside the ring (Albertini et al., 2006; Green et al., 2006). The N of RSV (respiratory syncytial virus) makes a ring with 10 to 11 protomers with the RNA on the outside of the ring (Tawar et al., 2009), the ring of PIV5 with 13 protomers with the RNA on the outside the ring (Alayyoubi et al., 2015) and HMPV with 10 protomers as with RSV (Renner et al., 2016). Most of the atomic-resolution structures were determined by crystallization and X-ray diffraction. Other NCLPs form helices of which the structures can be determined by Cryo-EM and single particle-based helical analysis. The first such structure was of measles virus NCLPs comprising 12.3 protomers (Gutsche et al., 2015) and Ebola virus with 24.4 protomers per turn of the helix and the RNA on the outside of the helix (Sugita et al., 2018), as with the rings of RSV and PIV5. Other structures of Ebola virus NCLPs with lower resolution show the same fold of the protein although the structure of the RNA was not visible (Wan et al., 2017; Su et al., 2018). A structure of the NCLP of mumps virus was also determined at a lower resolution (Cox et al., 2014; Severin et al., 2016). The comparison of N0P and the NCLPs structures reveals that, upon RNA binding the relative orientation of the lower and the upped lobes change by ~20o (Green et al., 2006; Alayyoubi et al., 2015; Gutsche et al., 2015; Sugita et al., 2018) and became more closed than the N in the N0P from VSV, measles, PIV5 and Ebola viruses (Leyrat et al., 2011; Guryanov et al., 2015; Kirchdoerfer et al., 2015; Leung et al., 2015; Renner et al., 2016; Aggarwal et al., 2018) suggesting that in the N0P structures the RNA binding groove is primed for binding the RNA.
The structures of nucleoproteins of measles, PIV5, HMPV and Ebola viruses were determined in the absence of Ntail. The NCs of measles virus are very flexible when Ntail is present, but form tighter helices when Ntail is removed, and these NCs can be used for higher resolution image analysis (Figure 3; Heggeness et al., 1980; Bhella et al., 2002, 2004; Schoehn et al., 2004; Renner et al., 2016). The structure of the NCLP of measles virus shows that the Carm of N is close to the center of the helix (Desfosses et al., 2011; Gutsche et al., 2015) indicating that Ntail starts inside the helix. Solution state NMR showed that the first 50 residues of the tail are conformationally restricted before the chain escapes to the outside of the helix (Figure 3; Jensen et al., 2008, 2011). The disordered chain could interfere with stacking of turns, resulting in loosening of the helix, facilitating the functions of the transcription and replication from L. The helical linear motif is part of the C-terminal end of Ntail although N has been shown to function also when this helical motif was moved to different sites within N (Cox et al., 2017).
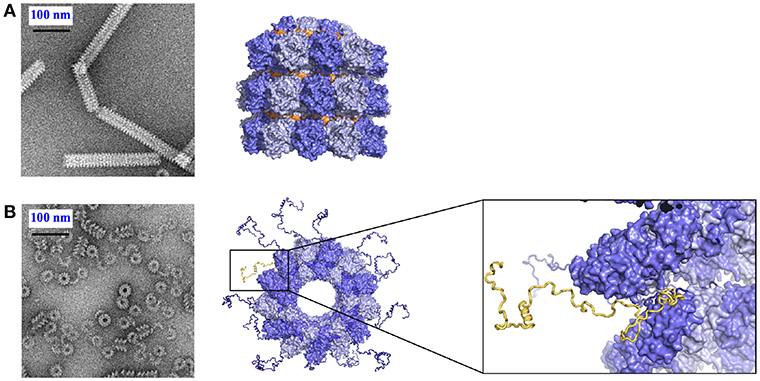
Figure 3. Role of Ntail in the nucleocapsid helix. (A) Tight helical NCLP with cleaved N with negative staining EM on the left and with the near-atomic structure on the right. The figure was made using PDB 4UFT. (B) Loose helical NCLP with full N with negative staining EM at left and with near-atomic structure with several tails. One Ntail with its helical motif is highlighted in yellow.
Binding of the RNA by N
All existing paramyxoviral NCLPs structures were determined from recombinant protein bound to RNA from the cells in which N was expressed. This means that the RNA is probably of random sequence, and the structures have only been able to reveal binding of the phosphates and 2' oxygen of the ribose but not interactions involving sequence-specific bases. Milles and coworkers constructed a fusion N0P complex where the P peptide was fused on the Narm with a TEV cleavage site (Milles et al., 2016). The purified complex was then cleaved to make the P peptide (residues 1–50) and either the full N (residues 1–525) or N up to the Narm (residues 1–405), with no RNA bound (Figures 2B, 4). During incubation of this complex with synthesized RNA, the RNA binds, the P peptide comes off and N associates into NCLPs (Figure 2B). The kinetics of the assembly process can be studied by nuclear magnetic resonance (NMR) spectroscopy because the signal of P becomes narrow (peaks of P appear in the spectrum) when the assembly process is initiated and P peptide dissociates from N. The signal for the residues of Narm and Carm become broader (the peaks disappear) as they bind to the neighboring N particles in the NC. In addition, using RNA labeled with a fluorescent probe, the kinetics of the assembly can be followed by measuring the changes in fluorescence anisotropy. Free RNA tumbles rapidly, however once it is bound to N and then assembles into NCs, the anisotropy increases. The resulting assembly can then be visualized with negative staining EM and NC growth can be followed during several hours (Milles et al., 2016; Figure 4). The nucleoprotein from Sendai and measles viruses bind 6 RNA bases per protomer (Calain and Roux, 1993; Pelet et al., 1996) and micrometer long NCLPs could be made with RNA molecules of only 6 bases in length; either with the 5′ viral genomic RNA (HO-ACCAGA-OH) or a polyA (HO-AAAAAA-OH) sequence and the resulting NCLPs structures were determined with Cryo-EM and image analysis (Desfosses et al., 2019). The structures show the RNA with a gap between the hexamers because the RNA molecules do not contain a 5′ phosphate (Figure 5A). Whereas, the previous structure (Gutsche et al., 2015) mainly showed only N residues binding the sugar and the phosphate of the RNA (residues K180, T183, R194, R195, and R354) the new structures also show residues binding the RNA bases; R195, Q202, Y260, E263, N351 (Figure 5B; Desfosses et al., 2019).
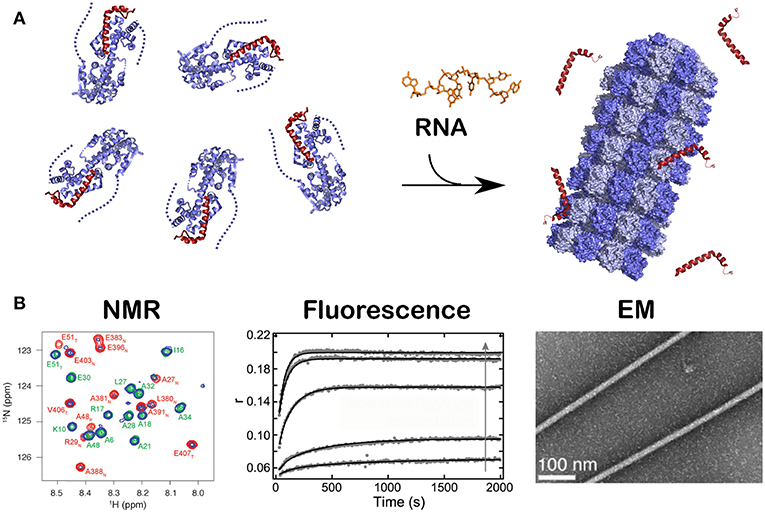
Figure 4. Kinetics of the assembly of NCLP by NMR, fluorescence and EM. Just like in Figure 2B, when RNA was given to the N0P complex, NCLP assembly could followed by NMR in real time because the NMR signals for the P peptide appear over time, by fluorescence anisotropy using fluorescein amidite-labeled RNAs, and by negative staining EM showing the nucleocapsids. For (A) we used PDB number 4CO6 for the N0P complex and PDB 4UFT for the NCLP, (B) is reprinted from Milles et al. (2016); Copyright 2016 Wiley-VCH Verlag GmbH Co. KGaA, used with permission.
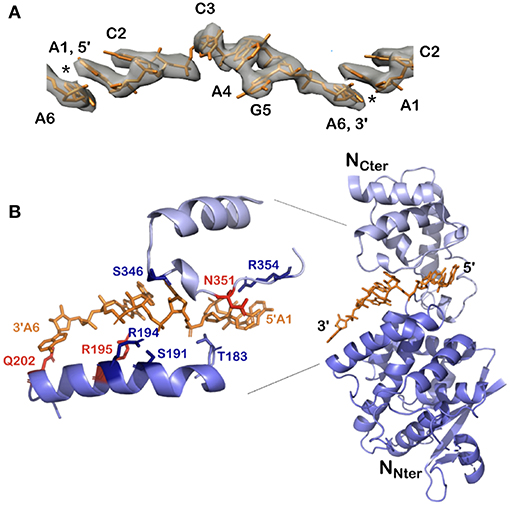
Figure 5. Structure of the 5′ viral genomic RNA (HO-ACCAGA-OH) inside the NCLP. (A) The Cryo-EM electrostatic potential of the 6 bases at the 5′ end of the viral RNA inside the NCLP (EM data base EMD-0142). The structures show gaps (asterisks) between bases 6 and 1, because the RNA (PDB 6H5S) molecules do not have a 5′ phosphate. (B) The protomer at the 3′ end inside the NCLP, showing that the last nucleotide is only bound to the base and not to the phosphate on the 2′ site on the ribose. The figure shows some of the residues binding the RNA, in red those that bind the bases. Reprinted with permission from Desfosses et al. (2019); Copyright 2019 National Academy of Sciences, used with permission. Frontier.
The structures of VSV and rabies virus NCs show that each N protomer can bind 9 bases and RSV can bind 7 bases. The N protomer of measles, PIV5 and Ebola viruses bind 6 bases. In these nucleocapsids the RNA form 2 short A-helices connected by a conformational flip of the backbone RNA; all structures show that 3 bases turn to the nucleoprotein (inward) and the others turn outward (Figure 6). If the RNA followed a long A-helix rather than 2 short helices, then there would be no room for the nucleoprotein and, probably, the new RNA strand made by L during transcription could easily form a low energy double helix RNA structure. These viruses do not code for a helicase; so the nucleoprotein functions as a helicase (Iseni et al., 2000). For VSV/rabies and RSV, NCLP structures show that for each protomer, the last 3′ base stacks with the first 5′ base stabilizing the structure of the nucleocapsids between the two protomers (Albertini et al., 2006; Green et al., 2006; Tawar et al., 2009; Ruigrok et al., 2011). The structure of the NCLP of PIV5 suggests that nucleotides 4–6 turn inward and nucleotides 1–3 outward (3′-in-in-in-out-out-out-5′) and would mean that bases 6 and 1 do not stack (Figure 6A). However, the chemical modification of nucleotide bases in the Sendai viral nucleocapsids (Iseni et al., 2002) showed that bases 1 and 6 are both more open for modification than the other bases. The structure of the NCLP of measles virus (Gutsche et al., 2015) suggested that nucleotides turn 3′-out-in-in-in-out-out-5′. Nucleotides 6 and 1 stack just like the NCLP of rabies virus, VSV, and RSV (Ruigrok et al., 2011; Figure 6B). The structure of the synthesized NCLPs of measles virus with 6 nucleotides (Figure 6C) shows how the 6 nucleotides bind on the protomer and these structures show the same orientation of nucleotides as suggested for the previous structure, with stacking for nucleotides 6 and 1.
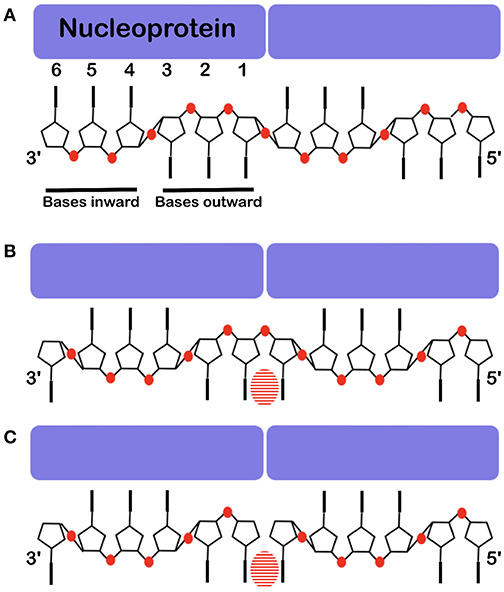
Figure 6. Base stacking over the interface between two protomers. The nucleoprotein binds 6 nucleotides, 3 point inward to the protein and 3 point outward. Often one cannot see clearly which base binds at the interface. (A) The structure of the RNA inside the NCLP of PIV5 (Alayyoubi et al., 2015). (B) The structure of the RNA inside the measles virus NCLP (Gutsche et al., 2015). (C) The structure of 6 nucleotides at the 5′ end of the viral RNA inside of the NCLP of measles virus (Desfosses et al., 2019). Because there is not a phosphate at the 5′, this shows the register of the RNA very clearly. The bases are shown, the phosphates are illustrated in red and the striped ovals show the stacking between two bases over the interface between two protomers.
With the help of NCs formed in vitro, we can identify both the binding of the 5′ end of the RNA to the first N protomer of the NCs, and the binding of the 3′ end on the final protomer of the NCs (Figure 5B): The lower part of N is still bound to the RNA but the upper part of N does not cover the terminal bases and only residue Q202 in the lower edge of the groove binds the last base at the 3′ end. Q202 is important for replication initiation in PIV2 because the mutant Q202A shows increased replication (Matsumoto et al., 2017, 2018). The fact that the 3′ end of the RNA is rather “open” may help the binding of L at the 3′ end of the vRNA.
Structure of the Disordered Phosphoprotein Tail, Residues 1–304
NMR was used to investigate all the 304 disordered residues of the N-terminal domain of P, Ptail, identify the structural behavior of each residue, and characterize the relative flexibility of Ptail (Jensen et al., 2011, 2014; Ozenne et al., 2012; Abyzov et al., 2016; Milles et al., 2016, 2018). Figure 7A shows the rigidity for the residues, with a thick red ribbon for the flexible regions and in thin blue ribbon for the more rigid regions, and 4 regions with a propensity for making helices. The first two helices represent the transient helices α1/2 that bind as helices in the N0P structure, residues 1–37, a short region around residue 110, α3, the STAT binding site (Devaux et al., 2007), and helix α4, around residues 189-198 (Milles et al., 2018). N0P with N0 and the 304 residues of P shows that α-helices 1 and 2 bind on N0 but also helix 4 and a short hydrophobic region just before helix 4, together named δα4 (delta-alpha-4). This region δα4 binds residues 96–127 on N0 (Figure 7B). This binding site and the helix α4 seem to be conserved in many paramyxoviral P proteins. If the “HELL” residues are changed to AAAA in a measles virus transcription/replication assay, the activity is suppressed (Milles et al., 2018). This means that the functional core of N0P is not only N0 plus helices 1 and 2 (Yabukarski et al., 2014; Guryanov et al., 2015) but also up to the δα4 region (Figure 7B). Between δα4 and α3 there is an acidic sequence with many Asp/Glu residues. As N0 cannot be purified in the absence of α1/2, it is complicated to measure the Kd for their binding to N0, but it is estimated to be in the nanomolar range as the complex can be purified on a size-exclusion column. The binding of δα4 to N0 is very weak, the Kd is around 0.6 mM, and we could show by NMR that δα4 binds and dissociates very rapidly while the N-terminal helices α1/2 remain bound, possibly so that the acidic loop blocks the cellular RNA binding to N0P.
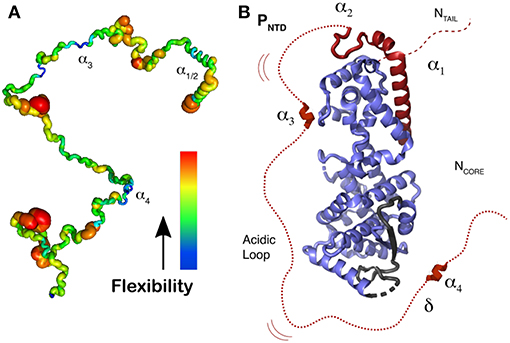
Figure 7. Representation of N0P with N0 and the 304 residues of Ptail. (A) Localization of secondary structure and representation of flexibility/rigidity of Ptail with colors from red (flexible) to blue (rigid) and the transient α-helices from the N-terminal helix (α1 to α4). (B) Representation of N0P with the full Ptail showing α1 and α2 binding on N0, transient α3 and α4 region that weakly binds residues 96–127 of N0 as well the acidic loop between α3 and α4. For the N0 structure we used PDB 4CO6. Adapted from Milles et al. (2018); Copyright 2018 American Association for the Advancement of Science, used with permission. Frontiers.
The measles virus polymerase stutters on the template and adds extra G bases for making several mRNAs and therefore different proteins (Thomas et al., 1988; Cattaneo et al., 1989; Vidal et al., 1990). The V-protein uses the same Ptail as in the P-protein and changes sequence at residues 231 (the editing site, just after δα4) up to residue 299, to form a zinc-binding domain of the V protein (Cattaneo et al., 1989; Liston and Briedis, 1994). For P we know what the domains do: Residues 1-200 bind N0 and the rest of Ptail in another register forms part of V, 304-377 for the tetramerization domain and 457-507 for XD. We do not know the function of the region 378–456 (Ploop) in the transcription and replication processes of measles virus. Additionally it should be mentioned that N and P are both known to be phosphorylated, which can be important for the viral cycle regulation, in particular phosphorylation is involved in the regulation of viral gene expression. However the mechanism of such regulation was not studied yet (Lamb et al., 1976; Sugita et al., 2018).
Model for the Replication Complex of Measles Virus
Cox and Plemper (2015) have suggested a model for the NCs in the presence of N, P and L for the paramyxoviruses. Similarly we have used recent results concerning the structures of P, N, and the nucleocapsids of measles virus (Figure 8) to develop a picture of the functioning polymerase complex. For this we used the structure of the NC with the 6 bases and for the structure of L we used the polymerase of VSV, to indicate the size of this component when bound to the tetramerization domain. In Figure 8 we show only one Ntail and only one Ptail. P could be bound to N with the XD-Ntail complex and with α1 in the free α1-site on N (Figure 2) and L could act on the first bases of the 3′ end of the vRNA (Figure 8A). For the last base at the 3′ end, the 2' of the ribose and the phosphate would be free for binding L. The very long Ptail and Ntail could keep L away from the RNA. However, if δα4 region also binds to its binding site on N, then L could come closer to the vRNA (Figure 8B). To conclude, the current model illustrates the initial genomic RNA register and binding, and suggests a role for N-P interactions in positioning L during RNA processing.
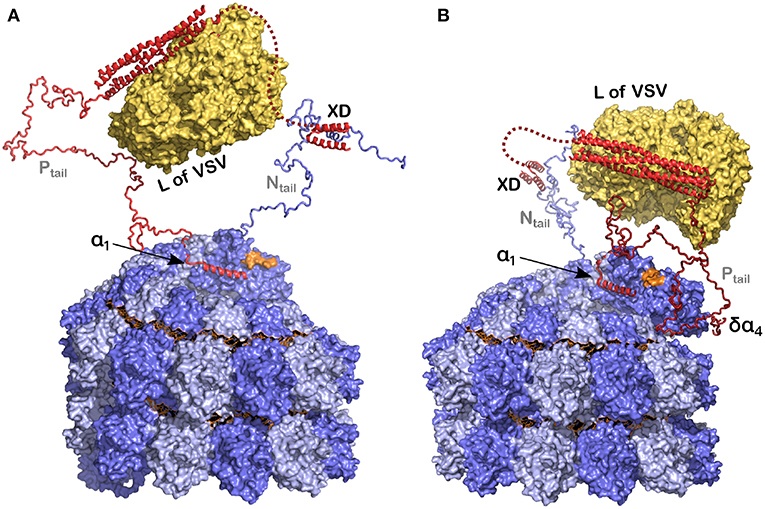
Figure 8. Model of the polymerase binding to the 3′ end of the RNA inside the NC. (A) The polymerase (L) binds to the tetramerization domain of P and P binds to the nucleocapsid, at the linear helical motif of Ntail with the XD, and potentially also with α1 of P on the empty site on the last protomer of the nucleocapsid. In orange is the final 3′ end of the viral RNA. (B) The same as in (A) but now with the δα4 region on residues 96–127 of N0 that may take the polymerase closer to the 3′ end of the RNA. For this figure we used PDB 3ZDO for the tetramerization domain of P, PDB 1T6O for the structure of the complex between the linear helical motif of N and the XD of P, the structure of the polymerase of VSV with PDB 5A22 and the NCLP of measles virus with PDB 6H5S. Modified from Desfosses et al. (2019); Copyright 2019 National Academy of Sciences, used with permission.
Author Contributions
RR and MB wrote the manuscript. SM and SG contributed to reviewing relevant literature, interpretation of studies, drafting the manuscript, and preparation of figures. All authors approved the article for publication.
Funding
This work was supported by GRAL (ANR-10-LABX-49-01), the Finovi (Fondation innovations en infectiologie), and the Fondation de la Recherche Médicale (FRM; Equipe DEQ20170336754). SM acknowledges the European Molecular Biology Organization for a long-term fellowship (ALTF 468-2014) and the European Commission (EMBOCOFUND2012, GA-2012-600394) via Marie Curie Action.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Guy Schoehn, Malene Ringkjøbing Jensen, Irina Gutsche, and Ambroise Desfosses for extensive discussions.
References
Abyzov, A., Salvi, N., Schneider, R., Maurin, D., Ruigrok, R. W. H., Jensen, M. R., et al. (2016). Identification of dynamic modes in an intrinsically disordered protein using temperature-dependent NMR relaxation. J. Am. Chem. Soc. 138, 6240–6251. doi: 10.1021/jacs.6b02424
Aggarwal, M., Leser, G. P., Kors, C. A., and Lamb, R. A. (2018). Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein in complex with an amino-terminal peptide of the phosphoprotein. J. Virol. 92:e01304–17. doi: 10.1128/JVI.01304-17
Alayyoubi, M., Leser, G. P., Kors, C. A., and Lamb, R. A. (2015). Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein-RNA complex. Proc. Natl. Acad. Sci. U.S.A. 112, E1792–E1799. doi: 10.1073/pnas.1503941112
Albertini, A. A., Wernimont, A. K., Muziol, T., Ravelli, R. B. G., Clapier, C. R., Schoehn, G., et al. (2006). Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313, 360–3. doi: 10.2210/pdb2gtt/pdb
Bhella, D., Ralph, A., Murphy, L. B., and Yeo, R. P. (2002). Significant differences in nucleocapsid morphology within the Paramyxoviridae. J. Gen. Virol. 83(Pt 8), 1831–1839. doi: 10.1099/0022-1317-83-8-1831
Bhella, D., Ralph, A., and Yeo, R. P. (2004). Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J. Mol. Biol. 340, 319–331. doi: 10.1016/j.jmb.2004.05.015
Bloyet, L. M., Brunel, J., Dosnon, M., Hamon, V., Erales, J., Gruet, A., et al. (2016). Modulation of re-initiation of measles virus transcription at intergenic regions by PXD to Ntail binding strength. PLoS Pathog. 12:e1006058. doi: 10.1371/journal.ppat.1006058
Bloyet, L. M., Schramm, A., Lazert, C., Raynal, B., Hologne, M., Walker, O., et al. (2019). Regulation of measles virus gene expression by P protein coiled-coil properties. Sci. Adv. 5:eaaw3702. doi: 10.1126/sciadv.aaw3702
Bourhis, J. M., Johansson, K., Receveur-Bréchot, V., Oldfield, C. J., Dunker, K. A., Canard, B., et al. (2004). The C-terminal domain of measles virus nucleoprotein belongs to the class of intrinsically disordered proteins that fold upon binding to their physiological partner. Virus Res. 99, 157–167. doi: 10.1016/j.virusres.2003.11.007
Bourhis, J. M., Receveur-Bréchot, V., Oglesbee, M., Zhang, X., Buccellato, M., Darbon, H., et al. (2005). The intrinsically disordered C-terminal domain of the measles virus nucleoprotein interacts with the C-terminal domain of the phosphoprotein via two distinct sites and remains predominantly unfolded. Protein Sci. 14, 1975–1992. doi: 10.1110/ps.051411805
Bowman, M. C., Smallwood, S., and Moyer, S. A. (1999). Dissection of individual functions of the Sendai virus phosphoprotein in transcription. J. Virol. 73, 6474–6483.
Bruhn, J. F., Barnett, K. C., Bibby, J., Thomas, J. M. H., Keegan, R. M., Rigden, D. J., et al. (2014). Crystal structure of the Nipah virus phosphoprotein tetramerization domain. J. Virol. 88, 758–762. doi: 10.1128/JVI.02294-13
Bruhn, J. F., Hotard, A. L., Spiropoulou, C. F., Lo, M. K., and Saphire, E. O. (2019). A conserved basic patch and central kink in the Nipah virus phosphoprotein multimerization domain are essential for polymerase function. Structure 27, 1–9. doi: 10.1016/j.str.2019.01.012
Brunel, J., Chopy, D., Dosnon, M., Bloyet, L.-M., Devaux, P., Urzua, E., et al. (2014). Sequence of events in measles virus replication: role of phosphoprotein-nucleocapsid interactions. J. Virol. 88, 10851–10863. doi: 10.1128/JVI.00664-14
Calain, P., and Roux, L. (1993). The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J. Virol. 67, 4822–4830.
Cattaneo, R., Kaelin, K., Baczko, K., and Billeter, M. A. (1989). Measles virus editing provides an additional cysteine-rich protein. Cell 56, 759–764.
Communie, G., Crépin, T., Maurin, D., Jensen, M. R., Blackledge, M., and Ruigrok, R. W. H. (2013a). Structure of the tetramerization domain of measles virus phosphoprotein. J. Virol. 87, 7166–7169.
Communie, G., Habchi, J., Yabukarski, F., Blocquel, D., Schneider, R., Tarbouriech, N., et al. (2013b). Atomic resolution description of the interaction between the nucleoprotein and phosphoprotein of Hendra virus. PLoS Pathog. 9:e1003631. doi: 10.1371/journal.ppat.1003631
Cox, R., Green, T. J., Purushotham, S., Deivanayagam, C., Bedwell, G. J., Prevelige, P. E., et al. (2013). Structural and functional characterization of the mumps virus phosphoprotein. J. Virol. 87, 7558–7568. doi: 10.1128/JVI.00653-13
Cox, R., Pickar, A., Qiu, S., Tsao, J., Rodenburg, C., Dokland, T., et al. (2014). Structural studies on the authentic mumps virus nucleocapsid showing uncoiling by the phosphoprotein. Proc. Natl. Acad. Sci. U.S.A. 111, 15208–15213. doi: 10.1073/pnas.1413268111
Cox, R., and Plemper, R. K. (2015). The paramyxovirus polymerase complex as a target for next-generation anti-paramyxovirus therapeutics. Front. Microbiol. 6:459. doi: 10.3389/fmicb.2015.00459
Cox, R. M., Krumm, S. A., Thakkar, V. D., Sohn, M., and Plemper, R. K. (2017). The structurally disordered paramyxovirus nucleocapsid protein tail domain is a regulator of the mRNA transcription gradient. Sci. Adv. 3:e1602350. doi: 10.1126/sciadv.1602350
Curran, J., Homann, H., Buchholz, C., Rochat, S., Neubert, W., and Kolakofsky, D. (1993). The hypervariable C-terminal tail of the Sendai paramyxovirus nucleocapsid protein is required for template function but not for RNA encapsidation. J. Virol. 67, 4358–4364.
Curran, J., Marq, J. B., and Kolakofsky, D. (1995). An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 69, 849–855.
Desfosses, A., Goret, G., Farias Estrozi, L., Ruigrok, R. W., and Gutsche, I. (2011). Nucleoprotein-RNA orientation in the measles virus nucleocapsid by three-dimensional electron microscopy. J. Virol. 85, 1391–1395. doi: 10.1128/JVI.01459-10
Desfosses, A., Milles, S., Jensen, M. R., Guseva, S., Colletier, J.-P., Maurin, D., et al. (2019). Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication. Proc. Natl. Acad. Sci. U.S.A. 116, 4256–4264. doi: 10.1073/pnas.1816417116
Devaux, P., von Messling, V., Songsungthong, W., Springfeld, C., and Cattaneo, R. (2007). Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virology 360, 72–83. doi: 10.1016/j.virol.2006.09.049
Fearns, R., and Plemper, R. K. (2017). Polymerases of paramyxoviruses and pneumoviruses. Virus Res. 234, 87–102. doi: 10.1016/j.virusres.2017.01.008
Fooks, A. R., Stephenson, J. R., Warnes, A., Dowsett, A. B., Rima, B. K., and Wilkinson, G. W. (1993). Measles virus nucleocapsid protein expressed in insect cells assembles into nucleocapsid-like structures. J. Gen. Virol. 74, 1439–1444.
Gerard, F. C., Ribeiro, E. D. A. Jr., Leyrat, C., Ivanov, I., Blondel, D., Longhi, S., et al. (2009). Modular organization of rabies virus phosphoprotein. J. Mol. Biol. 388, 978–96. doi: 10.1016/j.jmb.2009.03.061
Green, T. J., Zhang, X., Wertz, G. W., and Luo, M. (2006). Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313, 357–60. doi: 10.1126/science.1126953
Guryanov, S. G., Liljeroos, L., Kasaragod, P., Kajander, T., and Butcher, S. J. (2015). Crystal structure of the measles virus nucleoprotein core in complex with an N-terminal region of phosphoprotein. J. Virol. 90, 2849–2857. doi: 10.1128/JVI.02865-15
Gutsche, I., Desfosses, A., Effantin, G., Ling, W. L., Haupt, M., Ruigrok, R. W. H., et al. (2015). Structural virology. Near-atomic Cryo-EM structure of the helical measles virus nucleocapsid. Science 348, 704–707. doi: 10.1126/science.aaa5137
Habchi, J., Tompa, P., Longhi, S., and Uversky, V. N. (2014). Introducing protein intrinsic disorder. Chem. Rev. 114, 6561–6588. doi: 10.1021/cr400514h
Heggeness, M. H., Scheid, A., and Choppin, P. W. (1980). Conformation of the helical nucleocapsids of paramyxoviruses and vesicular stomatitis virus: reversible coiling and uncoiling induced by changes in salt concentration. Proc. Natl. Acad. Sci. U.S.A. 77, 2631–2635.
Houben, K., Marion, D., Tarbouriech, N., Ruigrok, R. W. H., and Blanchard, L. (2007). Interaction of the C-terminal domains of sendai virus N and P proteins: comparison of polymerase-nucleocapsid interactions within the paramyxovirus family. J. Virol. 81, 6807–6816. doi: 10.1128/JVI.00338-07
Iseni, F., Barge, A., Baudin, F., Blondel, D., and Ruigrok, R. W. (1998). Characterization of rabies virus nucleocapsids and recombinant nucleocapsid-like structures. J. Gen. Virol. 79(Pt 12), 2909–2919. doi: 10.1099/0022-1317-79-12-2909
Iseni, F., Baudin, F., Blondel, D., and Ruigrok, R. W. (2000). Structure of the RNA inside the vesicular stomatitis virus nucleocapsid. RNA 6, 270–281. doi: 10.1017/S135583820099109X
Iseni, F., Baudin, F., Garcin, D., Marq, J.-B., Ruigrok, R. W. H., and Kolakofsky, D. (2002). Chemical modification of nucleotide bases and mRNA editing depend on hexamer or nucleoprotein phase in Sendai virus nucleocapsids. RNA 8, 1056–10567. doi: 10.1017/S1355838202029977
Jensen, M. R., Communie, G., Ribeiro, E. A. Jr., Martinez, N., Desfosses, A., Salmon, L., et al. (2011). Intrinsic disorder in measles virus nucleocapsids. Proc. Natl. Acad. Sci. U.S.A. 108, 9839–9844. doi: 10.1073/pnas.1103270108
Jensen, M. R., Houben, K., Lescop, E., Blanchard, L., Ruigrok, R. W. H., and Blackledge, M. (2008). Quantitative conformational analysis of partially folded proteins from residual dipolar couplings: application to the molecular recognition element of Sendai virus nucleoprotein. J. Am. Chem. Soc. 130, 8055–8061. doi: 10.1021/ja801332d
Jensen, M. R., Zweckstetter, M., Huang, J. R., and Blackledge, M. (2014). Exploring free-energy landscapes of intrinsically disordered proteins at atomic resolution using NMR spectroscopy. Chem. Rev. 114, 6632–6660. doi: 10.1021/cr400688u
Johansson, K., Bourhis, J. M., Campanacci, V., Cambillau, C., Canard, B., and Longhi, S. (2003). Crystal structure of the measles virus phosphoprotein domain responsible for the induced folding of the C-terminal domain of the nucleoprotein. J. Biol. Chem. 278, 44567–44573. doi: 10.1074/jbc.M308745200
Karlin, D., Ferron, F., Canard, B., and Longhi, S. (2003). Structural disorder and modular organization in Paramyxovirinae N and P. J. Gen. Virol. 84(Pt 12), 3239–3252. doi: 10.1099/vir.0.19451-0
Kingston, R. L., Baase, W. A., and Gay, L. S. (2004a). Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J. Virol. 78, 8630–8640. doi: 10.1128/JVI.78.16.8630-8640.2004
Kingston, R. L., Hamel, D. J., Gay, L. S., Dahlquist, F. W., and Matthews, B. W. (2004b). Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc. Natl. Acad. Sci. U.S.A. 101, 8301–8306. doi: 10.1073/pnas.0402690101
Kirchdoerfer, R. N., Abelson, D. M., Li, S., Wood, M. R., and Saphire, E. O. (2015). Assembly of the Ebola virus nucleoprotein from a chaperoned VP35 complex. Cell Rep. 12, 140–149. doi: 10.1016/j.celrep.2015.06.003
Laksono, B. M., de Vries, R. D., Verburgh, R. J., Visser, E. G., de Jong, A., Fraaij, P. L. A., et al. (2018). Studies into the mechanism of measles-associated immune suppression during a measles outbreak in the Netherlands. Nat. Commun. 9:4944. doi: 10.1038/s41467-018-07515-0
Lamb, R. A., Mahy, B. W., and Choppin, P. W. (1976). The synthesis of Sendai virus polypeptides in infected cells. Virology 69, 116–131.
Leung, D. W., Borek, D., Luthra, P., Binning, J. M., Anantpadma, M., Liu, G., et al. (2015). An intrinsically disordered peptide from Ebola virus VP35 controls viral RNA synthesis by modulating nucleoprotein-RNA interactions. Cell Rep. 11, 376–389. doi: 10.1016/j.celrep.2015.03.034
Leyrat, C., Yabukarski, F., Tarbouriech, N., Ribeiro, E. A. Jr., Jensen, M. R., Blackledge, M., et al. (2011). Structure of the vesicular stomatitis virus N0P complex. PLoS Pathog. 7:e1002248. doi: 10.1371/journal.ppat.1002248
Liang, B., Li, Z., Jenni, S., Rahmeh, A. A., Morin, B. M., Grant, T., et al. (2015). Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162, 314–327. doi: 10.1016/j.cell.2015.06.018
Longhi, S., Receveur-Bréchot, V., Karlin, D., Johansson, K., Darbon, H., Bhella, D., et al. (2003). The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 278, 18638–18648. doi: 10.1074/jbc.M300518200
Masters, P. S., and Banerjee, A. K. (1988). Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J. Virol. 62, 2658–2664.
Matsumoto, Y., Ohta, K., Kolakofsky, D., and Nishio, M. (2017). A point mutation in the RNA-binding domain of human parainfluenza virus type 2 nucleoprotein elicits abnormally enhanced polymerase activity. J. Virol. 91:e02203–16. doi: 10.1128/JVI.02203-16
Matsumoto, Y., Ohta, K., Kolakofsky, D., and Nishio, M. (2018). The control of paramyxovirus genome hexamer length and mRNA editing. RNA 24, 461–467. doi: 10.1261/rna.065243.117
Mavrakis, M., Iseni, F., Mazza, C., Schoehn, G., Ebel, C., Gentzel, M., et al. (2003). Isolation and characterisation of the rabies virus N0-P complex produced in insect cells. Virology 305, 406–414. doi: 10.1006/viro.2002.1748
Mavrakis, M., Kolesnikova, L., Schoehn, G., Becker, S., and Ruigrok, R. W. H. (2002). Morphology of Marburg virus NP-RNA. Virology 296, 300–307. doi: 10.1006/viro.2002.1433
Milles, S., Jensen, M. R., Communie, G., Maurin, D., Schoehn, G., Ruigrok, R. W. H., et al. (2016). Self-assembly of measles virus nucleocapsid-like particles: Kinetics and RNA sequence dependence. Angew. Chem. Int. Ed. Engl. 55, 9356–9360. doi: 10.1002/anie.201602619
Milles, S., Jensen, M. R., Lazert, C., Guseva, S., Ivashchenko, S., Communie, G., et al. (2018). An ultraweak interaction in the intrinsically disordered replication machinery is essential for measles virus function. Sci. Adv. 4:eaat7778. doi: 10.1126/sciadv.aat7778
Ozenne, V., Schneider, R., Yao, M., Huang, J. R., Salmon, L., Zweckstetter, M., et al. (2012). Mapping the potential energy landscape of intrinsically disordered proteins at amino acid resolution. J. Am. Chem. Soc. 134, 15138–15148. doi: 10.1021/ja306905s
Pelet, T., Delenda, C., Gubbay, O., Garcin, D., and Kolakofsky, D. (1996). Partial characterization of a Sendai virus replication promoter and the rule of six. Virology 224, 405–414.
Peluso, R. W., and Moyer, S. A. (1988). Viral proteins required for the in vitro replication of vesicular stomatitis virus defective interfering particle genome RNA. Virology 162, 369–376.
Pickar, A., Elson, A., Yang, Y., Xu, P., Luo, M., and He, B. (2015). Oligomerization of mumps virus phosphoprotein. J. Virol. 89, 11002–11110. doi: 10.1128/JVI.01719-15
Qiu, S., Ogino, M., Luo, M., Ogino, T., and Green, T. J. (2016). Structure and function of the N-terminal domain of the vesicular stomatitis virus RNA polymerase. J. Virol. 90, 715–724. doi: 10.1128/JVI.02317-15
Rahmeh, A. A., Schenk, A. D., Danek, E. I., Kranzusch, P. J., Liang, B., Walz, T., et al. (2010). Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 20075–20080. doi: 10.1073/pnas.1013559107
Reich, S., Guilligay, D., Pflug, A., Malet, H., Berger, I., Crépin, T., et al. (2014). Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516, 361–366. doi: 10.1038/nature14009
Renner, M., Bertinelli, M., Leyrat, C., Paesen, G. C., Saraiva de Oliveira, L. F., Huiskonen, J. T., et al. (2016). Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein. Elife 5:e12627. doi: 10.7554/eLife.12627
Ruigrok, R. W. H., Crépin, T., and Kolakofsky, D. (2011). Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr. Opin. Microbiol. 14, 504–510. doi: 10.1016/j.mib.2011.07.011
Schneider, R., Maurin, D., Communie, G., Kragelj, J., Hansen, D. F., Ruigrok, R. W. H., et al. (2015). Visualizing the molecular recognition trajectory of an intrinsically disordered protein using multinuclear relaxation dispersion NMR. J. Am. Chem. Soc. 137, 1220–1229. doi: 10.1021/ja511066q
Schoehn, G., Mavrakis, M., Albertini, A., Wade, R., Hoenger, A., and Ruigrok, R. W. H. (2004). The 12 å structure of trypsin-treated measles virus N-RNA. J. Mol. Biol. 339, 301–312. doi: 10.1016/j.jmb.2004.03.073
Severin, C., Terrell, J. R., Zengel, J. R., Cox, R., Plemper, R. K., He, B., et al. (2016). Releasing the genomic RNA sequestered in the mumps virus nucleocapsid. J. Virol. 90, 10113–10119. doi: 10.1128/JVI.01422-16
Sourimant, J., and Plemper, R. K. (2016). Organization, function, and therapeutic targeting of the morbillivirus RNA-dependent RNA polymerase complex. Viruses 8, 251–221. doi: 10.3390/v8090251
Spehner, D., Kirn, A., and Drillien, R. (1991). Assembly of nucleocapsid-like structures in animal cells infected with a vaccinia virus recombinant encoding the measles virus nucleoprotein. J. Virol. 65, 6296–6300.
Su, Z., Wu, C., Shi, L., Luthra, P., Pintilie, G. D., Johnson, B., et al. (2018). Electron cryo-microscopy structure of Ebola virus nucleoprotein reveals a mechanism for nucleocapsid-like assembly. Cell 172, 966–978. doi: 10.1016/j.cell.2018.02.009
Sugita, Y., Matsunami, H., Kawaoka, Y., Noda, T., and Wolf, M. (2018). Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 å resolution. Nature 563, 137–140. doi: 10.1038/s41586-018-0630-0
Tarbouriech, N., Curran, J., Ruigrok, R. W., and Burmeister, W. P. (2000). Tetrameric coiled-coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7, 777–781. doi: 10.1038/79013
Tawar, R. G., Duquerroy, S., Vonrhein, C., Varela, P. F., Damier-Piolle, L., Castagné, N., et al. (2009). Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326, 1279–1283. doi: 10.1126/science.1177634
Thakkar, V. D., Cox, R. M., Sawatsky, B., da Fontoura Budaszewski, R., Sourimant, J., Wabbel, K., et al. (2018). The unstructured paramyxovirus nucleocapsid protein tail domain modulates viral pathogenesis through regulation of transcriptase activity. J. Virol. 92:e02064–17. doi: 10.1128/JVI.02064-17
Thomas, S. M., Lamb, R. A., and Paterson, R. G. (1988). Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell 54, 891–902.
Tompa, P. (2002). Intrinsically unstructured proteins. Trends Biochem. Sci. 27, 527–533. doi: 10.1016/S0968-0004(02)02169-2
Uversky, V. N. (2002). Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11, 739–756. doi: 10.1110/ps.4210102
Vidal, S., Curran, J., and Kolakofsky, D. (1990). A stuttering model for paramyxovirus P mRNA editing. EMBO J. 9, 2017–2022.
Wan, W., Kolesnikova, L., Clarke, M., Koehler, A., Noda, T., Becker, S., et al. (2017). Structure and assembly of the Ebola virus nucleocapsid. Nature 551, 394–397. doi: 10.1038/nature24490
WHO (2019). Global Measles and Rubella Update. World Health Organisation, www.who.int/immunization/diseases/measles/ (accessed August 12, 2019).
Yabukarski, F., Lawrence, P., Tarbouriech, N., Bourhis, J. M., Delaforge, E., Jensen, M. R., et al. (2014). Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat. Struct. Mol. Biol. 21, 754–759. doi: 10.1038/nsmb.2868
Keywords: measles virus, nucleoprotein, phosphoprotein, Cryo-EM, NMR, X-ray crystallography, RNA binding
Citation: Guseva S, Milles S, Blackledge M and Ruigrok RWH (2019) The Nucleoprotein and Phosphoprotein of Measles Virus. Front. Microbiol. 10:1832. doi: 10.3389/fmicb.2019.01832
Received: 14 March 2019; Accepted: 25 July 2019;
Published: 21 August 2019.
Edited by:
Mathilde Richard, Erasmus Medical Center, NetherlandsReviewed by:
Richard K. Plemper, Georgia State University, United StatesSara Louise Cosby, Agri-Food and Biosciences Institute (AFBI), United Kingdom
Copyright © 2019 Guseva, Milles, Blackledge and Ruigrok. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Blackledge, bWFydGluLmJsYWNrbGVkZ2VAaWJzLmZy; Rob W. H. Ruigrok, cm9iLnJ1aWdyb2tAaWJzLmZy
 Serafima Guseva
Serafima Guseva Sigrid Milles
Sigrid Milles Martin Blackledge
Martin Blackledge Rob W. H. Ruigrok
Rob W. H. Ruigrok