- 1Division of Virology, Defence Research and Development Establishment, Gwalior, India
- 2Division of Biotechnology, Defence Research and Development Establishment, Gwalior, India
- 3Office of Deputy Director of Animal Husbandry, FMD Typing Scheme, Ahmedabad, India
Crimean-Congo hemorrhagic fever (CCHF) is a re-emerging zoonotic viral disease prevalent in many parts of Asia, Europe, and Africa. The causative agent, Crimean-Congo hemorrhagic fever orthonairovirus (CCHFV), is transmitted through hard ticks. Tick vectors especially belonging to the Hyalomma species serve as the reservoir and amplifying host. The vertebrate animals including sheep, goat, and bovine act as a short-lasting bridge linking the virus and ticks. CCHFV causes fatal hemorrhagic fever in humans. Humans are usually infected with CCHFV either through the bite of infected ticks or by close contact with infected animals. Immunological assays, primarily enzyme-linked immunosorbent assay (ELISA) using whole viral antigen, are widely used for serosurveillance in animals. However, the whole virus antigen poses a high biohazard risk and can only be produced in biosafety level 4 laboratories. The present study focuses on the development and evaluation of safe, sensitive, and specific IgG indirect enzyme-linked immunosorbent assay (iELISA) using recombinant nucleoprotein (NP) of CCHF virus as an antigen. The codon-optimized NP gene sequence was synthesized, cloned, and expressed in pET28a+ vector. The recombinant NP was purified to homogeneity by affinity chromatography and characterized through Western blot and MALDI-TOF/MS analysis. The characterized protein was used to develop an indirect IgG microplate ELISA using a panel of animal sera. The in-house ELISA was comparatively evaluated vis-à-vis a commercially available ELISA kit (Vector-Best, Russia) with 76 suspected samples that revealed a concordance of 90% with a sensitivity and specificity of 79.4 and 100%, respectively. The precision analysis revealed that the assay is robust and reproducible in different sets of conditions. Further, the assay was used for serosurveillance in ruminants from different regions of India that revealed 18% seropositivity in ruminants, indicating continued circulation of virus in the region. The findings suggest that the developed IgG iELISA employing recombinant NP is a safe and valuable tool for scalable high-throughput screening of CCHFV-specific antibodies in multiple species.
Introduction
Crimean Congo hemorrhagic fever orthonairovirus (CCHFV), an emerging tick-borne virus, is considered as a biothreat agent because of its potential to cause deadly hemorrhagic fever. A tick-borne virus, CCHFV belongs to the family Nairoviridae and genus Orthonairovirus (Adams et al., 2017). It is endemic in different parts of the world including Asia, Africa, and Europe with a fatality rate up to 80% (Ergönül, 2006). In humans, it causes acute hemorrhagic illness, leading to hypovolemic shock and death in extreme cases. CCHFV is primarily transmitted to humans through the bite of an infected tick. In animals, CCHF is asymptomatic and does not cause any disease but is a major threat to humans especially to those who are in close contact with infected animals particularly in farms, clinic, and abattoir (Mostafavi et al., 2017).
The genome of CCHFV is triple segmented, consisting of small (S), medium (M), and large (L) segments. Electron microscopy depicts viral particles of 90–100 nm in diameter (Zivcec et al., 2016). The open reading frame (ORF) of the S segment encodes a 482-amino-acid-long nucleocapsid protein that is highly conserved in nature and is an attractive target for development of diagnostics and vaccine (Burt et al., 1994; Carter et al., 2012; Vanhomwegen et al., 2012).
Many of the CCHF cases were reported from remote settings, where there is a lack of proper detection facility that leads to underreporting. Limited serosurveillance studies have shown high IgG positivity in livestock in different parts of endemic areas including India (Mourya et al., 2015). Ruminants play a crucial role in providing blood meal to ticks, their transportation across different territorial regions, and transmitting virus to naïve ticks and humans (Mertens et al., 2015; Spengler et al., 2016). Efforts are needed to detect the infection burden in animals using a safe, stable, and sensitive screening test that can be easily accessible in rural health care facilities.
The laboratory detection of CCHFV relies on virus isolation, RT-PCR, and serology. Virus isolation and RT-PCR are limited to biosafety level 4 (BSL-4) laboratory and highly sophisticated laboratory, respectively, having technical expertise (Al-Abri et al., 2017). Further, serological assays, viz., hemagglutination inhibition and complement fixation tests, suffer from various issues including reproducibility and sensitivity (Casals and Tignor, 1974). The currently used serological tests include immunofluorescence (IFA) and enzyme-linked immunosorbent assay (ELISA), though IFA demands specialized equipment, technique, and complexity in data interpretation, whereas ELISA can be performed in laboratory with minimum resources (Vanhomwegen et al., 2012). The widely used serological assays rely on whole virus as the antigen, primarily inactivated viral lysate, or hyper immune mouse ascitic fluid, which is used to capture antibodies (Mourya et al., 2012). Such antigen poses a high biohazard risk and their production is limited to maximum containment facilities (BSL-4), which are limited in the developing world.
The recombinant protein-based antigen is slowly replacing the viral diagnosis over the last few decades (Cuzzubbo et al., 2001; Álvarez-Rodríguez et al., 2012; Souza et al., 2012; Saxena et al., 2013). Bacterial expression systems are the preferred method of choice due to their inexpensive scale up and ease of utility (Babaeipour et al., 2010; Huang et al., 2012). The ease and reproducibility of antigen production make these antigens reliable for commercialization. Attempts were made to develop IgG ELISA for CCHF using recombinant nucleoprotein (NP) in baculovirus (Saijo et al., 2002; Samudzi et al., 2012). However, such expression systems are expensive, need to reinfect with fresh culture for each round of protein synthesis, and thus are inefficient for commercial-scale production. Recombinant NP protein was also attempted to express in prokaryotic system but protein instability was the major concern of the reported system (Liu et al., 2014). Recently, HRP-conjugated recombinant NP-based ELISA was developed for IgG detection, but the HRP-conjugated antigens are liable to lose their epitopes during conjugation and thus lack sensitivity; also, the stability of HRP-conjugated antigen is questionable until experimentally proven (Ge et al., 2017; Emmerich et al., 2018; Sas et al., 2018). Earlier, Schuster et al. (2016) reported the development of a competitive ELISA format using monoclonal antibodies against NP for serosurveillance. However, the involvement of specific monoclonal antibody and the competitive format make the process complex and costlier. The production of recombinant protein in the eukaryotic system is complex and expensive compared to the bacterial system. Keeping in view the above facts, the present study was envisaged to produce a safe and stable recombinant antigen in the prokaryotic system for the development of IgG iELISA assay. Recombinant NP was produced under native conditions in order to express its conformational epitopes to recognize all the possible antibodies. The soluble r-NP was used as a diagnostic intermediate for development of IgG iELISA. The assay was further evaluated with commercial system, and concordance, sensitivity, along with specificity were determined. Both intra- and inter-assay variations were also analyzed to study the precision of assay. Finally, the assay was applied for screening of animal sera from different regions in India for epidemiological serosurveillance.
Materials and Methods
Media, Chemicals, and Solutions
Escherichia coli strain DH5α was procured from Invitrogen, United States. E. coli host strain BL21(DE3) for expression of plasmid was purchased from Thermo Fisher Scientific, United States. The cloning vector pUC57 (ampr) and expression vector pET28a+ (kanr) were purchased from Novagen, United States. Ni-NTA superflow resin was procured from Qiagen, Germany. Protease inhibiting cocktail (Sigma-Aldrich) and TB were from HiMedia. Secondary HRP conjugates (anti-sheep HRP, anti-goat HRP and anti-bovine HRP) and HRPO substrate 3,3′,5,5′-tetramethylbenzidine (TMB) were procured from Sigma-Aldrich, United States. VectoCrimea CHF IgG ELISA kit was purchased from Vector-Best, Russia.
Construction of Recombinant Plasmid
The ORF encoding NP of CCHF virus from Indian isolate ID NIV 112143 (GenBank Accession no. JN572089) was codon optimized for E. coli and custom synthesized from M/s Bio Basic Inc., Canada. The custom synthesized gene (1458 bp) was confirmed by nucleotide sequencing and cloned in pUC 57 cloning vector.
Sub-Cloning of Recombinant NP Gene in pET28a+ Vector
Both recombinant plasmid of NP in pUC57 cloning vector and pET28a+ expression vector were digested with BamHI and HindIII restriction enzymes (RE) at 37°C for 1 h. The insert (NP) and the vector (pET28a+ expression vector) were purified using agarose gel and ligated. The ligated product was transformed into competent E. coli DH5α cells as per standard protocol. Transformed positive clones were selected by RE digestion and colony lysis PCR. The recombinant plasmid was isolated and further transformed into E. coli BL21 (DE3) cells for expression. The recombinant transformed clone was also confirmed by RE digestion, colony lysis PCR, and nucleotide sequencing.
Expression and Localization of Recombinant Protein
The logarithmic phase culture of positive colony was induced with varying concentrations of isopropyl-β-D-thiogalactopyranoside (IPTG) (0.5, 1, 1.5, and 2 mM) in Terrific broth (TB) for different time periods (1–4 h). Further, another lot was induced at 18°C for 18 h. The localization of recombinant protein was identified through sonication of cell suspension followed by centrifugation at 18,600 × g for 30 min at 4°C. The expression profile was studied by 10% SDS-PAGE.
Purification of Recombinant Nucleoprotein Under Native Conditions
Following induction, cells were harvested by centrifugation at 4,000 × g for 20 min. Harvested cells were then resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0) supplemented with phenylmethylsulfonylfluoride (PMSF), lysozyme, and protease inhibiting cocktail (Sigma-Aldrich, United States) and incubated at 4°C for 30 min, followed by sonication at 9 pulse on/off using microprobe at 40% frequency of sonicator (Sonics, United States) for 30 min. The sonicated culture was centrifuged at 10,000 × g for 20 min. The supernatant was filtered with a 0.45-μm filter and loaded on the pre-equilibrated polypropylene column containing Ni-NTA slurry for 2 h at 4°C for efficient binding. After passing the supernatant through a column, it was washed with 10 bed volumes of wash buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8.0) to remove unbound protein. Later on, the bound protein was eluted through elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8.0). Eluted protein was concentrated using an Amicon Ultra centrifugal filter device with a 30-kDa MWCO membrane. Purified protein was dialyzed overnight against dialysis buffer (50 mM NaH2PO4 and 300 mM NaCl). Protein was analyzed on 10% SDS-PAGE followed by staining with Coomassie Brilliant Blue. The concentration of purified protein was estimated by bicinchoninic acid assay (Thermo Fisher Scientific, United States) and stored at −80°C with 5% glycerol to enhance the stability of recombinant NP.
Characterization of r-NP Through Western Blot Analysis
Characterization of r-CCHF-NP was carried out using commercial rabbit polyclonal antibody against NP of CCHFV (Abcam, United States) and anti-His antibody (Sigma-Aldrich, United States) with Western blot. The purified r-NP protein was resolved on 10% SDS and transferred to nitrocellulose membrane. Membranes were blocked with 2% SMP for 2 h, followed by washing with 1 × PBS. Then, the membranes were incubated with 1:500 dilution of commercial polyclonal antibody and 1:1,000 dilution of anti-His antibody diluted in 1% SMP overnight, respectively. After washing thrice with 0.1% PBST, membranes were incubated with their respective HRP conjugate (anti-rabbit and anti-mice HRP conjugate). The conjugates were diluted in 1% Skim milk powder (1:1000) for 2 h followed by washing with 0.1% PBST. The membrane was finally developed using 3,3-diaminobenzidine (DAB) as substrate and H2O2.
In-Gel Protein Digestion
Gel pieces were excised from the SDS-PAGE and destained by three washes of 50% ACN/50 mM NH4HCO3 for 30 min each at room temperature. The protein was subjected to reduction with 10 mM DTT followed by an alkylation with 50 mM iodoacetamide. Gel pieces were dried after treatment with ACN and 100 ng of trypsin in 50 mM NH4HCO3 was added to the gel pieces. Tryptic digestion was carried out overnight at 37°C and the peptides were extracted with 60% ACN and 0.1% TFA. The peptides were vacuum dried and resuspended in 0.5% TFA before the MALDI-MS/MS analysis.
MALDI-TOF-TOF Analysis of Recombinant Nucleoprotein of CCHF
The recombinant NP of CCHF was identified by Applied Biosystems 4800 plus MALDI TOF/TOF Analyzer (AB Sciex, United States) using the tryptic digest of the protein. The digested peptides were spotted onto the target plate following mixing with 1:1 volume of CHCA matrix solution (10 mg/ml). MS mass spectrum was recorded in the reflector positive mode using a laser with accelerated voltage of 2 kV (200 Hz repetition rate with a wavelength of 355 nm). A default calibration was applied for 13 calibration points on 384-well MALDI plate, using a six-component peptide standard in a mass range of 905–3,660 Da. The MS/MS mass spectra were acquired by the data-dependent acquisition method by collision-induced dissociation (CID) at 1 kV voltage. MS/MS fragment ions were generated for 20 strongest precursors selected between the m/Z range of 850–4,000 Da and filtered with a signal-to-noise (S/N) ratio >20 from one MS scan. The precursor ions were selected for fragmentation by a timed ion selector (TIS), using air as collision gas at 1 kV energy and a recharge pressure threshold of 1.5e-006. MS and MS/MS spectra were accumulated for at least 1,200 and 1,600 laser shots, respectively. Peak list was generated using the 4000 Series Explorer Software v. 3.5 (Applied Biosystems) wherein the MS/MS peaks were selected on the basis of a S/N ratio greater than 10. The data thus obtained were explored employing Protein Pilot version 4.0 (Applied Biosystems) and MASCOT 2.0 search engine (Matrix Science, London, United Kingdom). Finally, the peak list was searched against all entries at the non-redundant protein sequence database of NCBI with 16,338,050 sequences using the following search parameters: mass tolerance of 50 ppm for precursor ion and ±0.6 Da for fragment ion with +1 charge state, trypsin digestion with one missed cleavage, and variable modifications of oxidation of methionine and carbamidomethylation of cysteine.
Recombinant Nucleoprotein-Based IgG ELISA
Recombinant CCHF NP protein was tested for its antigenicity in terms of recognition by anti-CCHFV antibody in bovine, sheep, and goat serum samples. For standardization of indirect IgG ELISA, each well of a 96-well maxisorp microtiter plate (Nunc, Denmark) was coated with 300 ng of recombinant CCHF r-NP protein in carbonate–bicarbonate buffer (pH-9.6). Following overnight incubation, the plate was washed thrice with PBST (PBS containing 0.05% Tween 20) and blocked with 5% SMP overnight at 4°C. Test sera were heat inactivated at 56°C for 30 min prior to dilution (1:1,000) in 1% SMP in 0.1% PBS-T. The diluted sera were added to each well and incubated at 37°C for 60 min. The plate was washed with wash buffer (0.1% PBST) five times and then incubated with 1:10,000 dilution of respective HRP-conjugated antibody (Sigma-Aldrich, United States) in 0.05% PBST at 37°C for 60 min. This was followed by washing five times with wash buffer and the reaction was developed by adding 100 μl of TMB (Sigma-Aldrich, United States). Following incubation at room temperature for 10 min, the reaction was stopped using 100 μl of 1N HCl as stop solution and the absorbance was recorded at 450 nm using an ELISA plate reader (BioTek, United States). Thirty negative control sera from healthy animals were included for the standardization of IgG iELISA. All these parameters including coating, dilution of serum and conjugate, washing, and incubation were optimized by taking into account the S/N ratio.
Comparative Evaluation of In-House ELISA With Commercial ELISA Kit
A panel of 76 animal samples from the CCHF-affected region of Gujarat, India, were tested in parallel with in-house iELISA and commercial VectoCrimea CHF IgG ELISA kit (Vector-Best, Russia). The protocol standardized above was used for in-house iELISA, whereas that recommended by M/s Vector-Best was followed during performance of VectoCrimea IgG ELISA kit, with optimization of animal conjugates through checkerboard titration.
Data Analysis
Data were analyzed with SigmaStat 4.0 software by applying confidence of interval at 95%. Vector-Best ELISA was considered as reference test in this study. The cutoff value of in-house iELISA was determined employing receiver operating characteristic (ROC) curve and area under the curve (AUC) analysis. The ROC curve of in-house and VectoCrimea was eventually compared (Hewson et al., 2004) to determine the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the in-house iELISA. The correlation and percent agreement between the two assays were calculated using Spearman correlation coefficient. The inter-rater agreement of two assays was measured using Cohen’s kappa coefficient.
Assay Reproducibility
Robustness of the assay is a crucial parameter for validation of assay, which is defined as the ability of a method to remain unaffected by small variation (Chan et al., 2013). Intra-assay variation (i.e., variation of replicates in one experiment), inter-assay variation (i.e., variation between experiments performed on different days by same operator), and inter-operator variation (i.e., variation between experiments performed by different operators) were performed to study the validation and reproducibility of the assay in different sets of conditions. Intra-assay was performed to monitor the variation in replicates within an assay. Each of six samples (three positive and three negative) were tested in five replicates within an assay, and intra-assay coefficient of variation (standard deviation/average × 100) was calculated. Kruskal–Wallis and repeated measures (RM) one-way ANOVA were calculated for intra-assay. The test was then performed using these calibrators in duplicate on eight different days for inter-assay variation. A total of 16 replicates were tested using the in-house IgG iELISA. The inter-operator assay variation was performed at different laboratories with different operators using the panel of calibrators of low, medium, and high IgG positive samples along with IgG negative sera samples. The values thus obtained were statistically analyzed with Kruskal–Wallis, one-way ANOVA, and percent coefficient of variation for comparing two or more groups.
Serosurveillance for CCHF in Animal Species
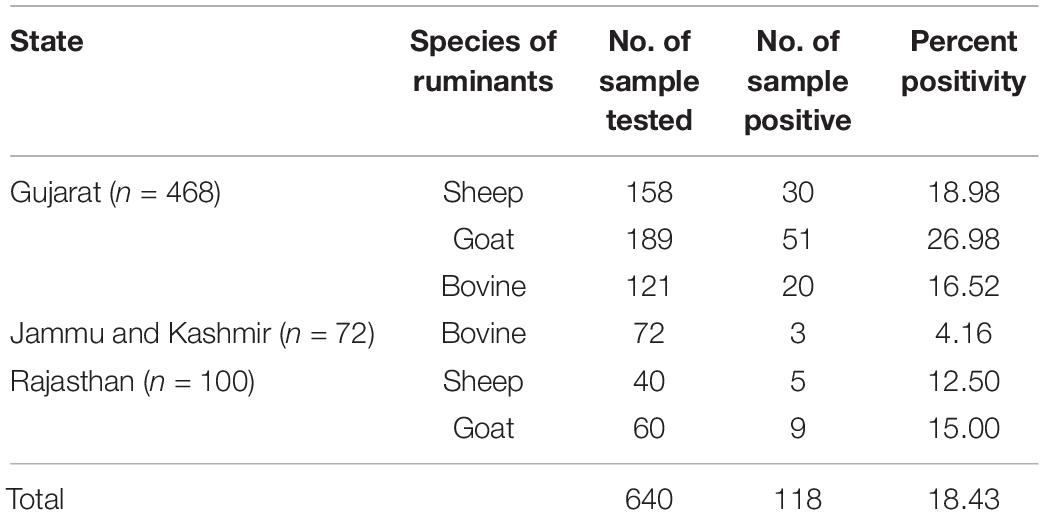
In-house IgG iELISA was then applied to screen a total of 640 animal samples including sheep (n = 198), goat (n = 249), and bovine (n = 193), collected from different Indian states such as Gujarat, Jammu and Kashmir, and Rajasthan.
Results
Cloning, Expression, and Solubility Optimization of CCHF NP Gene
The synthetic gene (1,458 bp) coding for CCHF NP cloned in pUC57 vector was used to release the insert using BamHI and HindIII RE (Figure 1A). The insert was gel purified revealing a size of 1458 bp (Figure 1B). The insert was cloned into pET28a+ expression vector at BamHI and HindIII sites and transformed in E. coli BL21 (DE3) cells. The presence of insert was confirmed through RE analysis, which revealed the release of a 1,458-bp insert (Figure 1C). The expression of NP from positive clone was analyzed in SDS-PAGE. A clear band of 56 kDa was observed in induced culture compared to uninduced cells (Figure 2A). Optimization of induction profile with varying IPTG concentration led to an optimum IPTG concentration of 1 mM (Supplementary Figure S1A). SDS-PAGE analysis revealed that the fraction of r-NP was found in insoluble form as inclusion bodies in post-induced culture up to 4 h at 37°C. To shift the expression to soluble form, conditions were further optimized by lowering the temperature of post-induction (from 36 to 18°C) (Figure 2B and Supplementary Figure S1B). After optimization, transformed BL21 (DE3) cells were grown in TB broth supplemented with 1% glycerol (Vagenende et al., 2009) and 50 μg/ml kanamycin. The expression of mid-log phase culture was induced by IPTG and grown post-induction at 18°C overnight that revealed expression of r-NP of 56 kDa size in soluble form.
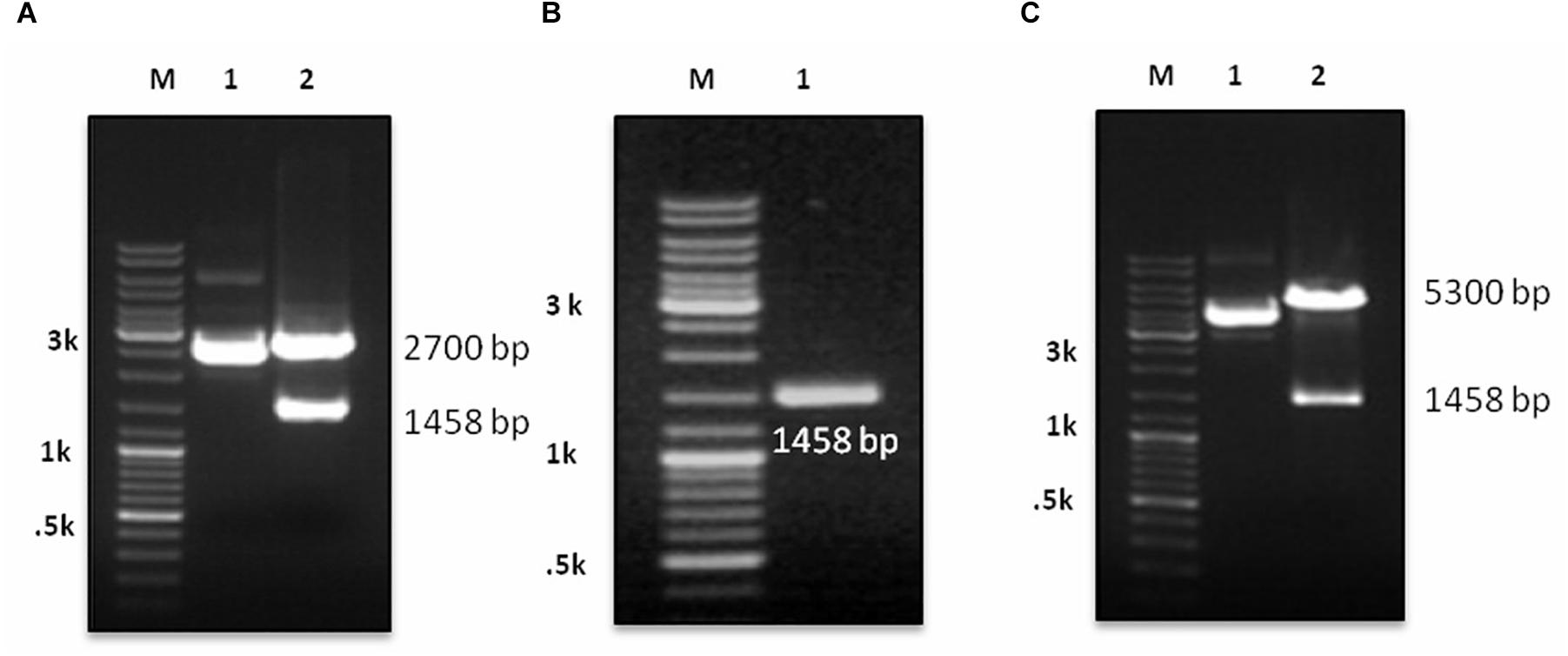
Figure 1. (A) M: DNA Marker (GeneRuler DNA Ladder, Cat. # SM0333, Thermo Fisher Scientific), lane 1: undigested pUC 57 with cloned gene, lane 2: digestion of recombinant plasmid with BamHI and HindIII revealing release of insert. (B) M: DNA Marker; lane 1: LMP purified gene insert (1458 bp). (C) M: DNA Marker; lane 1: ligated gene insert (1458 bp) with pET28a+ expression vector, lane 2: confirmation of ligation through RE digestion of recombinant plasmid with BamHI and HindIII, revealing fragment 5300 bp of vector and 1458 bp of gene insert.
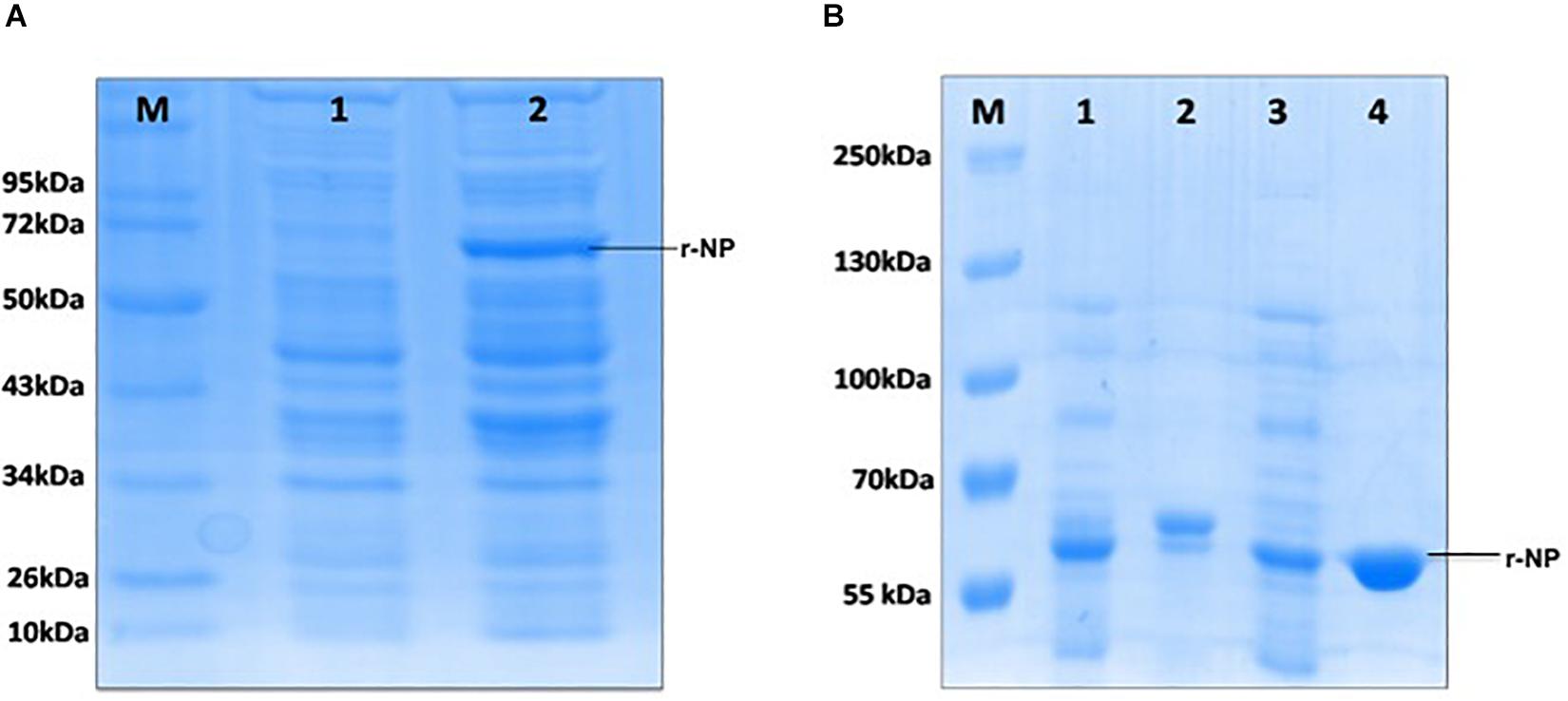
Figure 2. (A) M: Protein molecular weight marker (Cat. # SM0661, Fermentas); lane 1: uninduced BL21 (pET28a+), lane 2: induced BL21 (pET28a+) with 1 mM IPTG. (B) M: Protein molecular weight marker (Cat. #26619, Thermo Fisher Scientific); lane 1: induced bacterial lysate in insoluble form, lane 2: purified protein from insoluble lysate (under denaturing condition), lane 3: induced bacterial lysate in soluble form, lane 4: purified protein from soluble lysate (under native condition).
Purification of Recombinant CCHF Nucleoprotein
To maintain the conformational epitopes of recombinant NP, the purification process was carried out employing native conditions. The protein was mixed with Ni-NTA slurry and finally eluted with 250 mM imidazole. The gel analysis revealed a 56-kDa recombinant NP, as a single intense band (Figure 3). The quantity of purified r-NP was 8 mg/L of shake flask culture. The protein r-NP was stored at -80°C to enhance the stability of r-NP in terms of its aggregation and thus functionality.
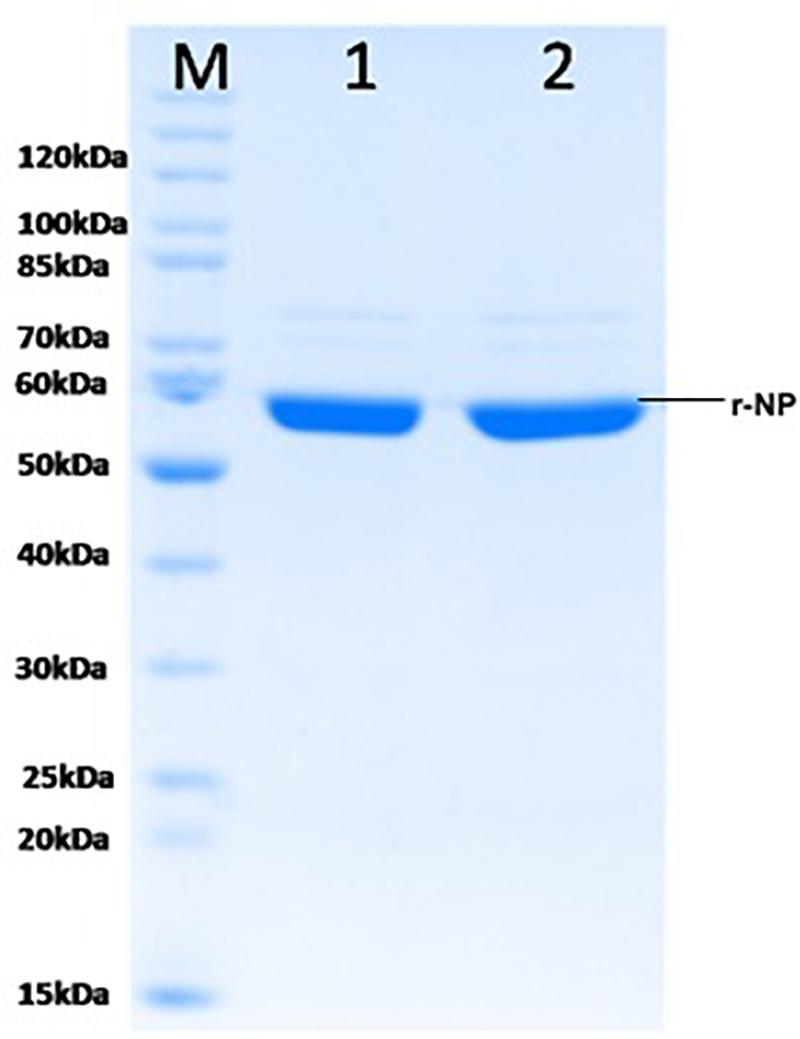
Figure 3. SDS-PAGE analysis of purified r-NP protein. M: Protein molecular weight marker (Cat. # SM0661, Fermentas); lanes 1 and 2, purified recombinant nucleoprotein expressed in BL21 (DE3) cells (56 kDa) using Ni-NTA affinity chromatography.
Characterization of Recombinant CCHF Nucleoprotein
Recombinant purified NP was characterized using Western blot and MALDI-TOF/MS analysis. In Western blot and chemiluminescent analysis, recombinant NP from both soluble and insoluble fractions was probed with commercial rabbit anti CCHF NP polyclonal antibody and mouse anti-His antibody that showed reactivity at 56-kDa protein band, representing the immunological activity of recombinant NP protein (Figures 4A–C). The recombinant NP was identified by MS/MS analysis of tryptic peptides using MALDI-TOF-TOF. The peak list was searched against the complete non-redundant protein sequence database of NCBI with 16,338,050 sequence entries. The protein was identified as a NP from CCHF virus [gi| 15788941] with a MASCOT score of 637 and a sequence coverage of 20% on MS/MS data (Table 1).
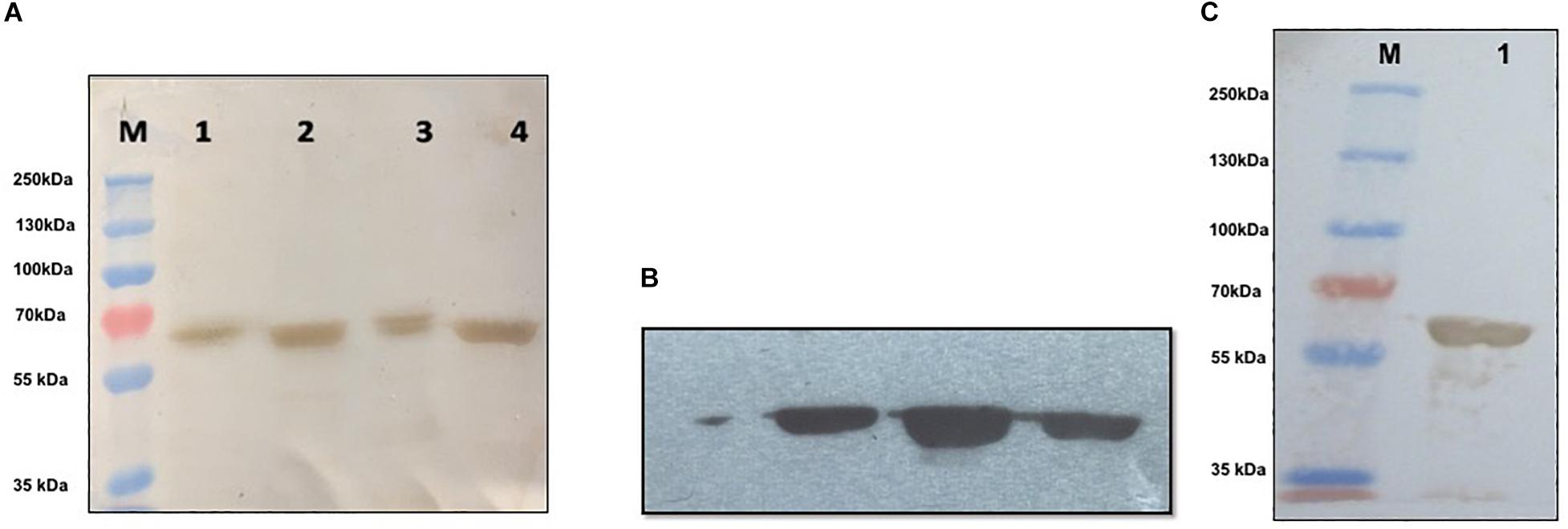
Figure 4. Western blot and chemiluminescent analysis of purified recombinant nucleoprotein (rNP). (A) Characterization of purified nucleoprotein of CCHFV. M: Prestained marker (ColorBurstTM Electrophoresis Marker, Cat # C1992, Sigma-Aldrich), lane 1: induced bacterial lysate generated in denaturing condition, lane 2: Purified rNP in insoluble condition, lane 3: induced bacterial lysate generated in soluble condition, lane 4: purified rNP in soluble condition showing immunological reactivity with commercial polyclonal antibody raised in rabbit against CCHFV (Abcam, United States) at 56 kDa. (B) Chemiluminescent analysis showing reaction of rNP from three different lots with commercial antibody against CCHFV (Abcam, United States). (C) M: Prestained marker (PageRulerTM Plus Prestained Protein Ladder, Cat # 26619, Thermo Fisher Scientific); lane 1: specific binding between rNP and mouse monoclonal anti-His antibody (Sigma-Aldrich, United States).
Table 1. Identification of recombinant nucleoprotein of CCHFV by MS/MS analysis of tryptic peptides using MALDI-TOF-TOF.
Standardization of Indirect IgG ELISA
Crimean-Congo hemorrhagic fever iELISA was standardized with well-characterized antigen, conjugate, and a panel of positive and negative animal sera obtained from Gujarat and Madhya Pradesh. No cross-reactivity was observed with FMD and PPR positive and apparently healthy animal sera. The cutoff of IgG iELISA was set as thrice the average OD value of negative control sera.
Comparative Evaluation and Statistical Analysis
A panel of 76 suspected samples from different animals (sheep, goat, and bovine) were tested in parallel using in-house iELISA assay and commercial VectoCrimea IgG ELISA kit. The comparative evaluation of both the assays is shown in Table 2.
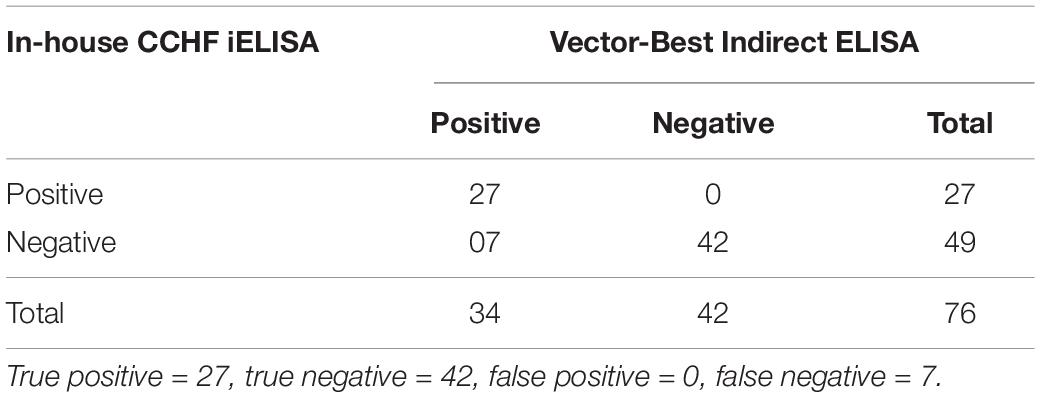
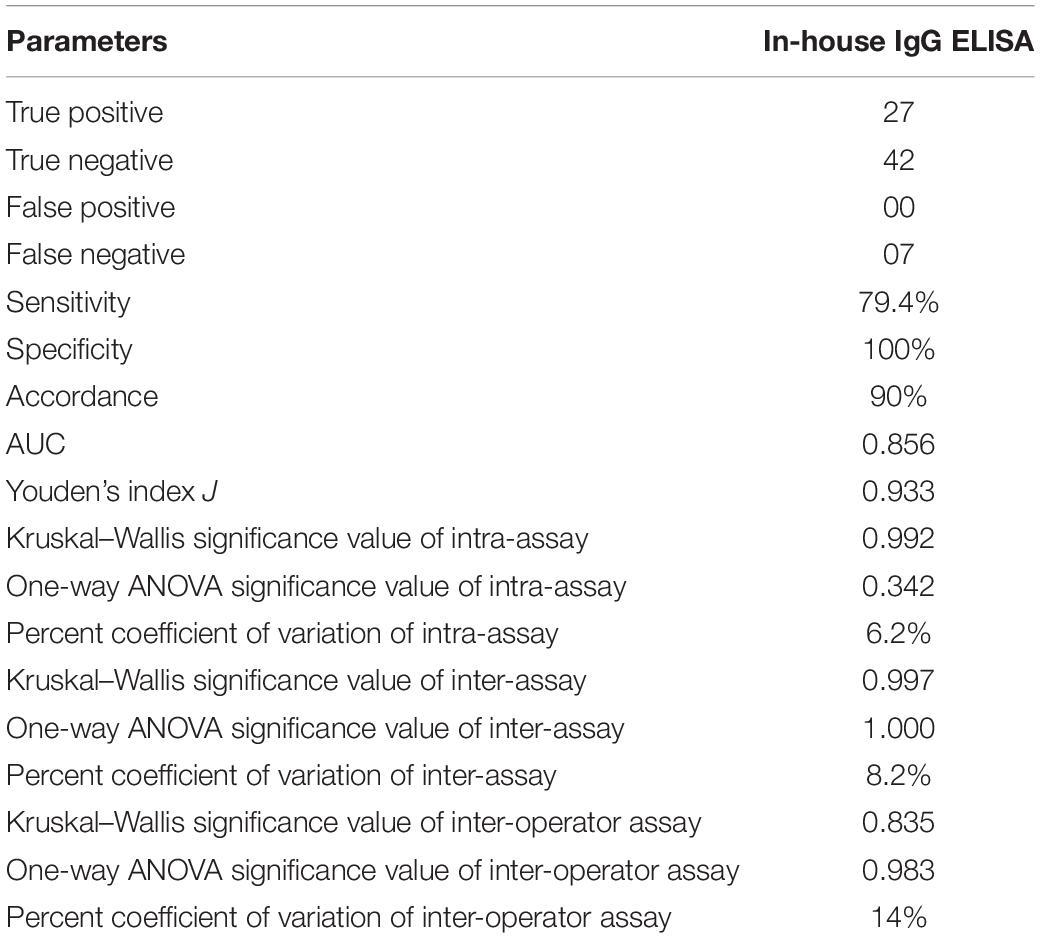
Table 2. Comparative evaluation of the in-house CCHF r-NP IgG ELISA with commercially available kit (Vector-Best, Novosibirsk, Russia) revealing true positive, false positive, true negative, and false negative.
The cutoff was further determined using the ROC curve analysis of in-house iELISA assay and commercial VectoCrimea IgG ELISA assay. The different cutoff was compared in respect to sensitivity and specificity of iELISA. The OD value of 0.414 was recommended as cutoff based on the 95% confidence interval (92.22 to 100%) of in-house assay. The cutoff OD value of 0.4 for the commercial VectoCrimea kit was calculated as per manufacturer instructions. At this threshold value, sensitivity, specificity, PPV, and NPV were calculated. The area under curve (AUC) within ROC showed a value of 0.856 with a Youden’s index (J) of 0.933, reflecting a virtuous accuracy of in-house iELISA (Figure 5). The difference between the ROC curves of in-house and Vector-Best was 0.166, with lowest significance level (p = 0.0008) (Figure 6A). The comparative evaluation of both the assays indicated a concordance of 90.7%. The sensitivity and specificity of the assay were found to be 79.4 and 100%, while the PPV and NPV of the assay were 100 and 85.7%, respectively (Table 3 and Figure 6B). Rho factor generated through Spearman correlation coefficient using SigmaStat 4.0 was 0.78, whereas the Cohen’s kappa coefficient was found to be 0.748, depicting the strong correlation between the in-house and commercial assays (Figure 7).

Figure 5. Receiver operating characteristic (ROC) analysis. ROC curve of in-house iELISA was generated with 52 negative sera and 24 positive sera. The area under ROC (AUC) was 0.856 (95% confidence interval). Youden’s index (J) for the optimal cutoff was 0.933. Orange dotted blue line represents ROC at 95% confidence interval.
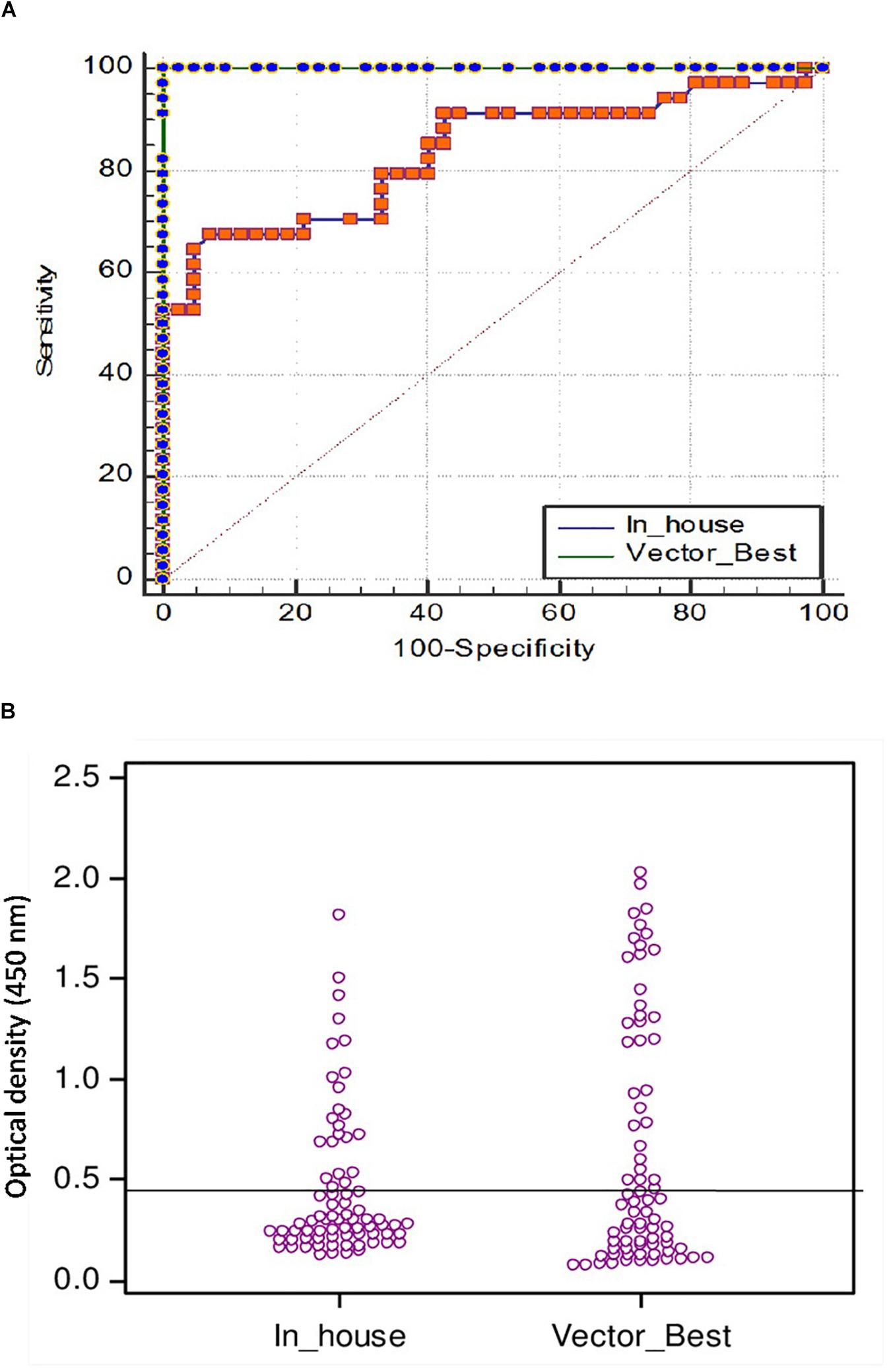
Figure 6. (A) Comparison of ROC curves of VectoCrimea IgG ELISA and in-house IgG ELISA using SigmaStat 4.0 with a panel of 76 positive and negative samples. Vector-Best [blue dotted green line; area under the curve (AUC) = 1.00], In-house (Orange dotted blue line; AUC = 0.856). The red dotted line represents the reference diagonal line of non-discrimination and is plotted from point 0,0 to point 1,1. The difference between two curves was 0.166 with a significance value of p = 0.0008. (B) Threshold of in-house iELISA was determined in comparison to VectoCrimea ELISA assay, estimated as 0.414 at 79.4% sensitivity and 100% specificity.
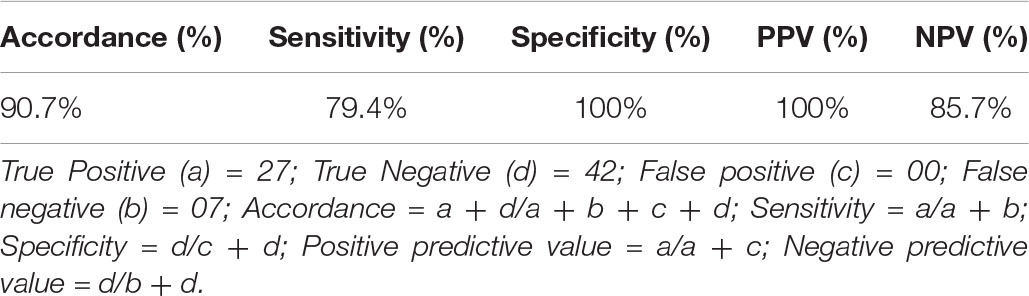
Table 3. Statistical analysis of comparative evaluation of the CCHF r-NP IgG ELISA with commercially available kit (Vector-Best, Novosibirsk, Russia).
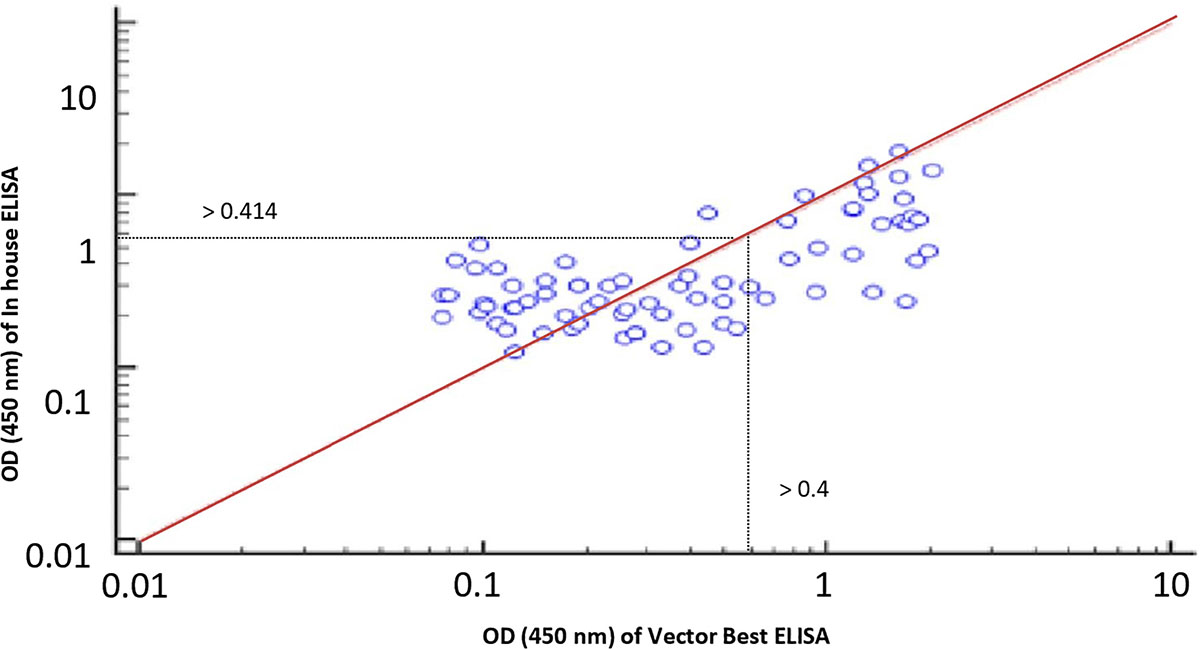
Figure 7. Rho factor was calculated with a Spearman correlation coefficient at 0.78 depicting strong correlation between in-house and commercial assays with 0.414 and 0.4 cutoff of respective assays.
Assay Reproducibility
A panel of calibrators with low, medium, and high IgG positive samples along with negative sera samples were used to study the intra-assay, inter-assay and inter-operator variation analysis. Intra-assay CV was estimated to be 6.4%, depicting the lower level of dispersion around the mean (Figure 8A). Kruskal–Wallis value for intra-assay variation was found to be 0.993, and RM one-way ANOVA p value for intra-assay was found to be 0.342, stating no significant difference within the replicates. Kruskal–Wallis value and one-way ANOVA significance was found to be 0.997 and 1.0 for inter-assay variation, respectively, which shows that there is no statistical difference when test was performed in different days (Figure 8B). Percent coefficient of variation for inter-assay was found to be 8.2%.
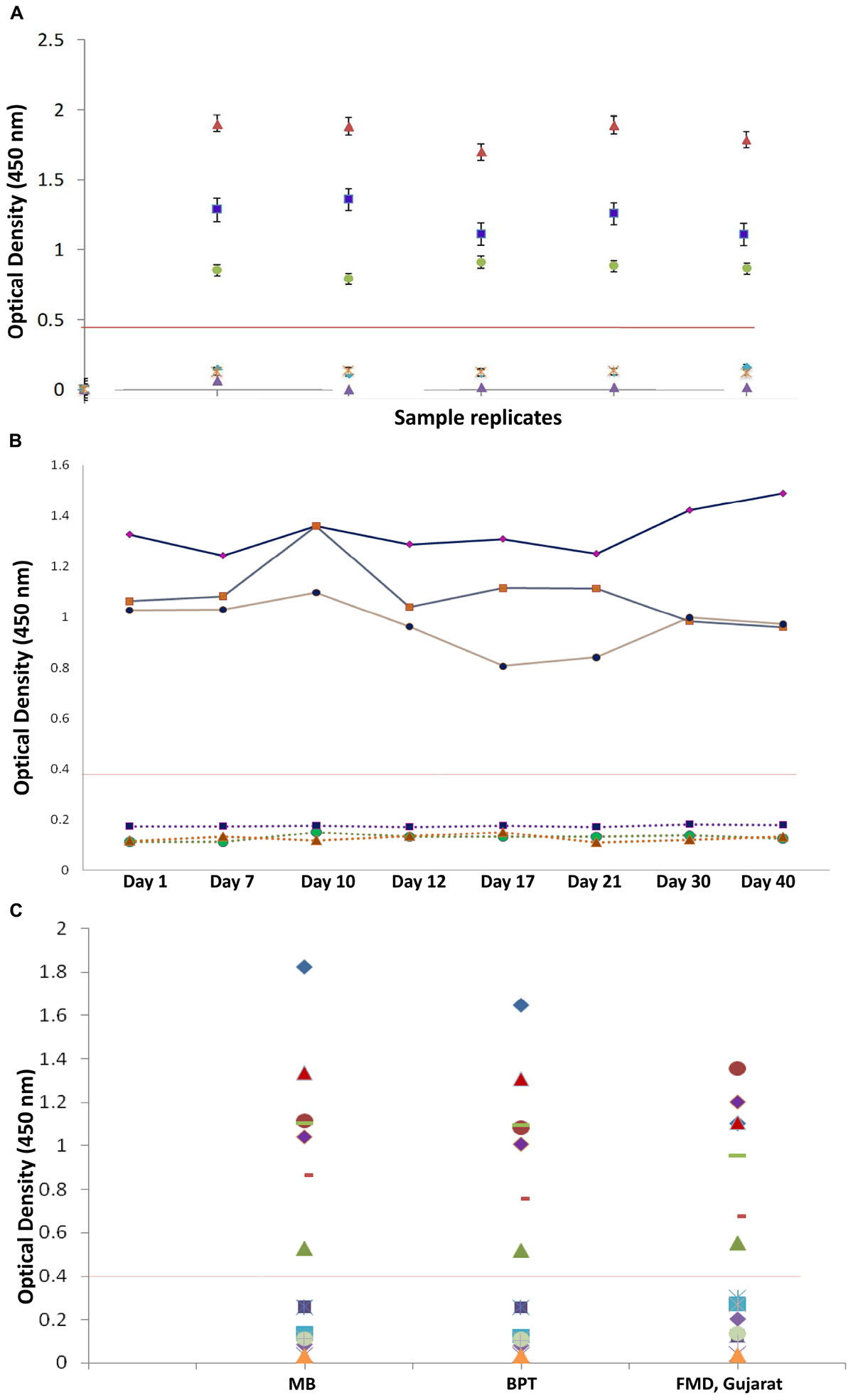
Figure 8. Reproducibility of in-house indirect IgG ELISA. (A) Intra-assay variation (variation of replicates in one experiment) of IgG iELISA was performed with a panel of one from each low, medium, and high positive sample and three negative samples and five replicates within a plate. (B) Inter-assay-variation (variation (between experiments) performed on eight different days (days 1, 7, 10, 12, 17, 21, 30, and 40) in duplicates by the same operator. (C) Inter-operator variation (variation between experiments performed by different operators) and assay precision were conducted at different institutional laboratories and at FMD Centre, Gujarat, with a panel of seven positive and nine negative samples with different operators. Data interpretation was performed with Kruskal–Wallis and one-way ANOVA.)
In the inter-operator assay, the Kruskal–Wallis significance value was 0.835 and one-way ANOVA significance value was 0.983 (Figure 8C). Percent coefficient of variation for inter-operator assay was found to be 14%, which should lie within 15% for inter assay variations. The overall performance of assay is shown in Table 4.
Serosurveillance for CCHF in Animal Species
The CCHF iELISA test was applied to screen a total of 640 samples from different domestic ruminants. The result revealed a seropositivity of 17.67% (n = 35), 24% (n = 60), and 12% (n = 23) in sheep, goat, and bovine, respectively. The region (state) and species-wise seropositivity is provided in Table 5.
Discussion
Crimean-Congo hemorrhagic fever virus is considered as the one of the most emerging tick-borne viral diseases with recurrent sporadic human cases in various countries of the world. CCHF virus circulates in multiple vertebrate hosts in nature but humans are the dead end host. Hard ticks are the principal vector and infected ticks serve as the lifelong reservoir of the virus. The virus is maintained through transovarial and transstadial transmission. Multiple wild and domestic animals including sheep, goat, and bovine are exposed to the virus through the bite of an infected tick. These ruminants remain asymptomatic, but develop a short viremia followed by seroconversion. CCHFV is maintained through a silent tick–vertebrate–tick enzootic cycle (Spengler et al., 2016). Humans in close contact of these animals or their products are at high risk of acquiring infection. Delayed confirmation of CCHF infection is associated with a risk of nosocomial infection leading to outbreak situations (Aradaib et al., 2010).
In spite of widespread circulation, there is a lack of safe and effective surveillance system in various developing countries (Al-Abri et al., 2017). ELISA is a preferred method for serosurveillance of various diseases in animals. In order to minimize the disease burden in the form of mortality and morbidity, rapid and high-throughput screening is paramount. Availability of a field-based detection system is critical for early and efficient management of CCHF, which is often reported from rural settings. Currently available ELISA systems utilize inactivated virus or suckling mouse brain antigen that poses high biohazard risk. With the advent of recombinant DNA technology, various ELISA systems based on recombinant proteins are developed that provides a safe alternative to infectious viral antigen. In the present study, NP was targeted as a diagnostic intermediate, as it is one of the most important and conserved structural protein of CCHFV. The NP of CCHFV reveals more than 95% conservancy across different genotypes (Hewson et al., 2004; Hawman and Feldmann, 2018). This high conservancy enables applicability of NP from a specific genotype for other genotypes in serodiagnosis testing (Saijo et al., 2002; Emmerich et al., 2018). The NP is also the predominantly expressed antigen after infection and is thus a good candidate for detection of viral antibodies (Schmaljohn and Nichol, 2007).
Though NP has been earlier expressed in several expression systems including baculovirus and prokaryote, its expensive production and instability limit its application for serodiagnosis (Tang et al., 2003; Saijo et al., 2005; Dowall et al., 2012). Therefore, in this study, we have developed a safe, stable, and soluble recombinant NP-based indirect IgG ELISA for serosurveillance. As r-NP does not need any posttranslational modification, a bacterial system was preferred. The recombinant NP was expressed with 6 × histidine tag in E. coli BL21 and was purified under native conditions using Ni-NTA affinity chromatography. Since rNP was found in insoluble form when induced at 37°C, the expression was shifted to soluble form by lowering the post-induction temperature to 18°C. This led to the expression of rNP in soluble form, which maintained the conformational integrity of the epitopes. The maintenance of native condition of rNP is crucial as that will enable recognition of antibodies against both linear and conformational epitopes. The high degree of purity (∼95%) and yield (8 mg/L) achieved through shake flask culture in this study shows the potential of the process for commercial exploitation. Further, the immunoreactivity of purified rNP was confirmed using both anti-CCHFV antibody and anti-His antibody in Western blot. The authenticity of expressed rNP was also confirmed through MALDI/TOF-MS. In-house ELISA was optimized through checkerboard titration of antigen, serum, and conjugate with a panel of known positive and negative sera. The cutoff was decided through ROC analysis. Currently, there is no validated commercial serodiagnostic kit available for use in animals. The commercial, Vector-Best kit, which is intended and widely used for human, was adapted in the current study for comparative evaluation of in-house assay as reported earlier (Schuster et al., 2016; Papa et al., 2018; Sas et al., 2018). Further, comparative evaluation of in-house assay and commercial kit was performed, which revealed 90% correlation. The result indicated a diagnostic sensitivity of 79.4% and a diagnostic specificity of 100%, thus minimizing the false-positive result that is crucial in serosurveillance. The stability and performance of in-house assay were also studied with intra-assay, inter-assay, and inter-operator assay variations. In all the validation tests, assay was found to be stable and robust in different sets of conditions such as different days, laboratories, and operators with no statistically significant difference, indicating the robustness and potential utility of in-house iELISA for large-scale applications.
Serosurveillance remains one of the important factors in tracing the circulation of virus in new areas and assists in prevention of infections from animals to human (Bente et al., 2013). This becomes important particularly when there is noprophylaxis and treatment is only supportive for CCHFV. The serosurveillance was carried out using 640 sera from three different ruminant species belonging to three different geographical regions of India. It revealed a seropositivity of 18%, thereby indicating endemicity of CCHFV in India. The relatively higher seropositivity in Gujarat samples is due to the fact that this region is a hotspot for CCHF in India as regular outbreaks have been reported since 2011 (Mourya et al., 2012). The Gujarat samples used in this study were collected from areas reporting human outbreaks with Asia-II lineage (Yadav et al., 2014). The lowest seropositivity in Jammu and Kashmir may be linked to the isolated high-altitude Himalayan region. Interestingly, the successful detection of antibody in Rajasthan samples, where a different lineage (Asia-I) is circulating, clearly demonstrates the utility of in-house iELISA for different lineages (Singh et al., 2016; Yadav et al., 2016). The successful demonstration of the single assay for detection of anti CCHFV antibodies in different animals makes this format amenable for commercial multispecies application. However, this format needs to be further evaluated with a larger sample size from different animals and human.
Conclusion
The present study led to the development of a process for expression and purification of immunogenic recombinant NP of CCHFV in native conformation. Further, the soluble rNP was explored for the development of a safe, stable, and scalable indirect ELISA that can be used in primary health care facilities to trace the circulation of virus in both human and animals. This assay can also be explored and converted into an on-site point of care testing (POCT) device that eases the diagnosis and surveillance program in resource-limited settings. Further directed and regular serosurveillance programs need to be undertaken in combating the emerging threats of CCHF virus in developing world.
Data Availability
All datasets generated for this study are included in the manuscript/Supplementary Files.
Author Contributions
PD conceived the idea. NS performed all the experiments. AS and SN contributed to statistical analysis. JK and SS contributed to sample analysis. SA performed and analyzed MALDI-TOF-TOF. SKS and AK collected and provided suspected animal samples. NS and PD wrote the manuscript. AS and SS critically revised the manuscript. All authors read and approved the final manuscript.
Funding
The authors thank Defence Research and Development Organisation (DRDO), Ministry of Defence, Government of India for providing necessary financial support for this study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Dr. D. K. Dubey, Director, DRDE, Gwalior, for his keen interest, constant support, and provision of necessary facilities for this study. This manuscript is assigned DRDE accession no. DRDE/VIRO/17/2019. We would like to thank Director, DIHAR, Leh for providing animal serum samples. We are also thankful to Dr. Subodh Kumar, Head, Microbiology Division and Dr. A. K. Goel, Head, BioProcess Technology Division, DRDE, for their help in inter-assay analysis. NS would also like to thank Department of Science and Technology, Government of India for providing INSPIRE fellowship.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01822/full#supplementary-material
FIGURE S1 | (A) Induction of E. coli BL21 cells at different IPTG concentrations. M: protein molecular weight marker; lane 1: uninduced BL21 culture, lane 2: induction was given at 0.5 mM IPTG, lane 3: induced at 1.0 mM IPTG, lane 4: induced at 1.5 mM IPTG, lane 5: induced at 2.0 mM IPTG, lane 6: induced at 2.5 mM IPTG. (B) Induction of BL21 cells at different temperature and time. M: protein molecular weight marker; lane 1: induced BL21 cells for 1 h at 37°C (protein in inclusion bodies), lane 2: induced BL21 cells for 2 h at 37°C (protein in inclusion bodies), lane 3: induced BL21 cells for 3 h at 37°C (protein in inclusion bodies), lane 4: induced BL21 cells for 4 h at 37°C (protein in inclusion bodies), lane 5: induced BL21 cells for 18 h at 18°C (protein in soluble form).
Abbreviations
ACN, acetonitrile; AUC, area under curve; BSL 4, biosafety level 4; CCHF, Crimean-Congo hemorrhagic fever; CHCA, α-cyano -4-hydroxycinnamic acid; CID, collision-induced dissociation; DAB, 3,3’-diaminobenzidine; DTT, dithiothreitol; FMD, foot-and-mouth disease; H2O2, Hydrogen peroxide; HRP, horseradish peroxidase; iELISA, indirect enzyme-linked immunosorbent assay; IFA, immunofluorescence; IPTG, isopropyl- β -D-thiogalactopyranoside; MALDI/MS, matrix-assisted laser desorption ionization/mass spectrometry; NP, nucleoprotein; NPV, negative predictive value; PBS, phosphate buffer saline; PMSF, phenyl methyl sulfonyl fluoride; PPR, peste des petits ruminants; PPV, positive predictive value; RE, restriction enzyme; ROC, receiver operating characteristic; SMP, skim milk powder; TFA, trifluoroacetic acid; TIS, timed ion selector.
References
Adams, M. J., Lefkowitz, E. J., King, A. M. Q., Harrach, B., Harrison, R. L., Knowles, N. J., et al. (2017). Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses. Arch. Virol. 162, 2505–2538. doi: 10.1007/s00705-017-3358-5
Al-Abri, S. S., Abaidani, I. A., Fazlalipour, M., Mostafavi, E., Leblebicioglu, H., and Pshenichnaya, N. (2017). Current status of crimean-congo haemorrhagic fever in the world health organization eastern mediterranean region: issues, challenges, and future directions. Int. J. Infect. Dis. 58, 82–89. doi: 10.1016/j.ijid.2017.02.018
Álvarez-Rodríguez, L. M., Ramos-Ligonio, A., Rosales-Encina, J. L., Martínez-Cázares, M. T., Parissi-Crivelli, A., and López-Monteon, A. (2012). Expression, purification, and evaluation of diagnostic potential and immunogenicity of a recombinant NS3 protein from all serotypes of dengue virus. J. Trop. Med. 2012:956875. doi: 10.1155/2012/956875
Aradaib, I. E., Erickson, B. R., Mustafa, M. E., Khristova, M. L., Saeed, N. S., Elageb, R. M., et al. (2010). Nosocomial outbreak of crimean-congo hemorrhagic fever. Sudan. Emerg. Infect. Dis. 16:837. doi: 10.3201/eid1605.091815
Babaeipour, V., Shojaosadati, S. A., Khalilzadeh, R., Maghsoudi, N., and Farnoud, A. M. (2010). Enhancement of human γ-interferon production in recombinant E. coli using batch cultivation. Appl. Biotechnol. Biochem. 160, 2366–2376. doi: 10.1007/s12010-009-8718-5
Bente, D. A., Forrester, N. L., Watts, D. M., McAuley, A. J., Whitehouse, C. A., and Bray, M. (2013). Crimean-congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res. 100, 159–189. doi: 10.1016/j.antiviral.2013.07.006
Burt, F. J., Leman, P. A., Abbott, J. C., and Swanepoel, R. (1994). Serodiagnosis of crimean-congo haemorrhagic fever. Epidemiol. Infect. 113, 551–562. doi: 10.1017/s0950268800068576
Carter, S. D., Surtees, R., Walter, C. T., Ariza, A., Bergeron, É, and Nichol, S. T., et al. (2012). Structure, function, and evolution of the crimean-congo hemorrhagic fever virus nucleocapsid protein. J. Virol. 86, 10914–10923.
Casals, J., and Tignor, G. H. (1974). Neutralization and hemagglutination-inhibition tests with crimean hemorrhagic fever-Congo virus. Proc. Soc. Exp. Biol. Med. 145, 960–966. doi: 10.3181/00379727-145-37933
Chan, W. T., Verma, C. S., Lane, D. P., and Gan, S. K. (2013). A comparison and optimization of methods and factors affecting the transformation of Escherichia coli. Biosci. Rep. 33:e00086. doi: 10.1042/BSR20130098
Cuzzubbo, A. J., Endy, T. P., Nisalak, A., Kalayanarooj, S., Vaughn, D. W., Ogata, S. A., et al. (2001). Use of recombinant envelope proteins for serological diagnosis of dengue virus infection in an immunochromatographic assay. Clin. Vaccine Immunol. 8, 1150–1155. doi: 10.1128/cdli.8.6.1150-1155.2001
Dowall, S. D., Richards, K. S., Graham, V. A., Chamberlain, J., and Hewson, R. (2012). Development of an indirect ELISA method for the parallel measurement of IgG and IgM antibodies against crimean-congo haemorrhagic fever (CCHF) virus using recombinant nucleoprotein as antigen. J. Virol. Methods 179, 335–341. doi: 10.1016/j.jviromet.2011.11.020
Emmerich, P., Mika, A., von Possel, R., Rackow, A., Liu, Y., Schmitz, H., et al. (2018). Sensitive and specific detection of crimean-congo hemorrhagic fever virus (CCHFV)—Specific IgM and IgG antibodies in human sera using recombinant CCHFV nucleoprotein as antigen in μ-capture and IgG immune complex (IC) ELISA tests. PLoS Negl. Trop. Dis. 12:e0006366. doi: 10.1371/journal.pntd.0006366.
Ergönül, Ö (2006). Crimean-congo haemorrhagic fever. Lancet Infect. Dis. 6, 203–214. doi: 10.1016/s1473-3099(06)70435-2
Ge, M., Li, R. C., Qu, T., Gong, W., and Yu, X. L. (2017). Construction of an HRP-streptavidin bound antigen and its application in an ELISA for porcine circovirus 2 antibodies. AMB Express 7:177. doi: 10.1186/s13568-017-0473-3
Hawman, D. W., and Feldmann, H. (2018). Recent advances in understanding crimean–congo hemorrhagic fever virus. F1000Res. 7:1715. doi: 10.12688/f1000research.16189.1
Hewson, R., Chamberlain, J., Mioulet, V., Lloyd, G., Jamil, B., Hasan, R., et al. (2004). Crimean-congo haemorrhagic fever virus: sequence analysis of the small RNA segments from a collection of viruses worldwide. Virus Res. 102, 185–189. doi: 10.1016/j.virusres.2003.12.035
Huang, C. J., Lin, H., and Yang, X. (2012). Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. Ind. Microbiol. Biotechnol. 39, 383–399. doi: 10.1007/s10295-011-1082-9
Liu, D., Li, Y., Zhao, J., Deng, F., Duan, X., Kou, C., et al. (2014). Fine epitope mapping of the central immunodominant region of nucleoprotein from crimean-congo hemorrhagic fever virus (CCHFV). PLoS One 9:e108419. doi: 10.1371/journal.pone.0108419
Mertens, M., Vatansever, Z., Mrenoshki, S., Krstevski, K., Stefanovska, J., Djadjovski, I., et al. (2015). Circulation of crimean-congo hemorrhagic fever virus in the former yugoslav republic of macedonia revealed by screening of cattle sera using a novel enzyme-linked immunosorbent assay. PLoS Negl. Trop. Dis. 9:e0003519. doi: 10.1371/journal.pntd.0003519
Mostafavi, E., Pourhossein, B., Esmaeili, S., Bagheri Amiri, F., Khakifirouz, S., Shah-Hosseini, N., et al. (2017). Seroepidemiology and risk factors of crimean-congo hemorrhagic fever among butchers and slaughterhouse workers in southeastern Iran. Int. J. Infect. Dis 64, 85–89. doi: 10.1016/j.ijid.2017.09.008
Mourya, D. T., Yadav, P. D., Shete, A. M., Gurav, Y. K., Raut, C. G., Jadi, R. S., et al. (2012). Detection, isolation and confirmation of crimean-congo hemorrhagic fever virus in human, ticks and animals in ahmadabad, india, 2010-2011. PLoS Negl. Trop. Dis. 6:e1653. doi: 10.1371/journal.pntd.0001653
Mourya, D. T., Yadav, P. D., Shete, A. M., Sathe, P. S., Sarkale, P. C., Pattnaik, B., et al. (2015). Cross-sectional serosurvey of crimean-congo hemorrhagic fever virus igg in livestock, india, 2013–2014. Emerg. Infect. Dis. 21, 1837–1839. doi: 10.3201/eid2110.141961
Papa, A., Papadopoulou, E., Tsioka, K., Kontana, A., Pappa, S., Melidou, A., et al. (2018). Isolation and whole-genome sequencing of a crimean-congo hemorrhagic fever virus strain. Greece. Ticks Tick Borne Dis. 9, 788–791. doi: 10.1016/j.ttbdis.2018.02.024
Saijo, M., Qing, T., Niikura, M., Maeda, A., Ikegami, T., Prehaud, C., et al. (2002). Recombinant nucleoprotein based enzyme linked immunosorbent assay for detection of immunoglobulin G to crimean–congo hemorrhagic fever virus. J. Clin. Microbiol. 40, 1587–1591. doi: 10.1128/jcm.40.5.1587-1591.2002
Saijo, M., Tang, Q., Shimayi, B., Han, L., Zhang, Y., Asiguma, M., et al. (2005). Antigen-capture enzyme-linked immunosorbent assay for the diagnosis of crimean-congo hemorrhagic fever using a novel monoclonal antibody. J. Med. Virol. 77, 83–88. doi: 10.1002/jmv.20417
Samudzi, R. R., Leman, P. A., Paweska, J. T., Swanepoel, R., and Burt, F. J. (2012). Bacterial expression of crimean-congo hemorrhagic fever virus nucleoprotein and its evaluation as a diagnostic reagent in an indirect ELISA. J. Virol. Methods 179, 70–76. doi: 10.1016/j.jviromet.2011.09.023
Sas, M. A., Comtet, L., Donnet, F., Mertens, M., Vatansever, Z., Tordo, N., et al. (2018). A novel double-antigen sandwich ELISA for the species-independent detection of crimean-congo hemorrhagic fever virus-specific antibodies. Antiviral Res. 151, 24–26. doi: 10.1016/j.antiviral.2018.01.006
Saxena, D., Parida, M., Rao, P. V. L., and Kumar, J. S. (2013). Cloning and expression of an envelope gene of west nile virus and evaluation of the protein for use in an IgM ELISA. Diagn. Microbiol. Infect. Dis. 75, 396–401. doi: 10.1016/j.diagmicrobio.2012.12.007
Schmaljohn, C. S., and Nichol, S. T. (2007). ‘Bunyaviridae”‘, in Fields Virology. Philadelphia: Lippincott Williams & Wilkins, 1741–1789.
Schuster, I., Mertens, M., Köllner, B., Korytáø, T., Keller, M., Hammerschmidt, B., et al. (2016). A competitive ELISA for species-independent detection of crimean-congo hemorrhagic fever virus specific antibodies. Antiviral Res. 134, 161–166. doi: 10.1016/j.antiviral.2016.09.004
Singh, P., Chhabra, M., Sharma, P., Jaiswal, R., Singh, G., Mittal, V., et al. (2016). Molecular epidemiology of crimean-congo haemorrhagic fever virus in India. Epidemiol. Infect. 144, 3422–3425. doi: 10.1017/s0950268816001886
Souza, I. I., Melo, E. S., Ramos, C. A., Farias, T. A., Osório, A. L., Jorge, K. S., et al. (2012). Screening of recombinant proteins as antigens in indirect ELISA for diagnosis of bovine tuberculosis. Springer plus 1:77. doi: 10.1186/2193-1801-1-77
Spengler, J. R., Estrada-Peña, A., Garrison, A. R., Schmaljohn, C., Spiropoulou, C. F., and Bergeron, É (2016). A chronological review of experimental infection studies of the role of wild animals and livestock in the maintenance and transmission of crimean-congo hemorrhagic fever virus. Antiviral Res. 135, 31–47. doi: 10.1016/j.antiviral.2016.09.013
Tang, Q., Saijo, M., Zhang, Y., Asiguma, M., Tianshu, D., Han, L., et al. (2003). A patient with crimean-congo hemorrhagic fever serologically diagnosed by recombinant nucleoprotein-based antibody detection systems. Clin. Vaccine Immunol. 10, 489–491. doi: 10.1128/cdli.10.3.489-491.2003
Vagenende, V., Yap, M. G. S., and Trout, B. L. (2009). Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry 48, 11084–11096. doi: 10.1021/bi900649t
Vanhomwegen, J., Alves, M. J., Županc, T. A., Bino, S., Chinikar, S., Karlberg, H., et al. (2012). Diagnostic assays for crimean-congo hemorrhagic fever. Emerg. Infect. Dis. 18, 1958–1965. doi: 10.3201/eid1812.120710
Yadav, P., Patil, D. Y., Shete, A. M., Kokate, P., Goyal, P., Jadhav, S., et al. (2016). Nosocomial infection of CCHF among health care workers in rajasthan. india. BMC Infect. Dis. 16:624. doi: 10.1186/s12879-016-1971-7
Yadav, P., Raut, C., Patil, D., Majumdar, T. D., and Mourya, D. T. (2014). Crimean-congo hemorrhagic fever: current scenario in india. Proc. Natl. A. Sci. India Sec B Biol. Sci. 84, 9–18. doi: 10.1007/s40011-013-0197-3
Keywords: Crimean-Congo hemorrhagic fever, antigen, protein purification, IgG, seroepidemiology, diagnosis
Citation: Shrivastava N, Shrivastava A, Ninawe SM, Sharma S, Kumar JS, Alam SI, Kanani A, Sharma SK and Dash PK (2019) Development of Multispecies Recombinant Nucleoprotein-Based Indirect ELISA for High-Throughput Screening of Crimean-Congo Hemorrhagic Fever Virus-Specific Antibodies. Front. Microbiol. 10:1822. doi: 10.3389/fmicb.2019.01822
Received: 21 May 2019; Accepted: 24 July 2019;
Published: 23 August 2019.
Edited by:
Mario Poljak, University of Ljubljana, SloveniaReviewed by:
Agustín Estrada-Peña, University of Zaragoza, SpainChristina Deschermeier, Bernhard-Nocht-Institut für Tropenmedizin (BMITM), Germany
Aykut Ozkul, Ankara University, Turkey
Copyright © 2019 Shrivastava, Shrivastava, Ninawe, Sharma, Kumar, Alam, Kanani, Sharma and Dash. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paban Kumar Dash, cGFiYW5kYXNoQGRyZGUuZHJkby5pbg==
 Neha Shrivastava1
Neha Shrivastava1 Sandeep M. Ninawe
Sandeep M. Ninawe Paban Kumar Dash
Paban Kumar Dash