- 1Consiglio Nazionale delle Ricerche, Istituto per la Protezione Sostenibile delle Piante, Turin, Italy
- 2Dipartimento di Scienze della Vita e Biologia dei Sistemi, Università di Torino, Turin, Italy
Climate changes include the intensification of drought in many parts of the world, increasing its frequency, severity and duration. However, under natural conditions, environmental stresses do not occur alone, and, in addition, more stressed plants may become more susceptible to attacks by pests and pathogens. Studies on the impact of the arbuscular mycorrhizal (AM) symbiosis on tomato response to water deficit showed that several drought-responsive genes are differentially regulated in AM-colonized tomato plants (roots and leaves) during water deficit. To date, global changes in mycorrhizal tomato root transcripts under water stress conditions have not been yet investigated. Here, changes in root transcriptome in the presence of an AM fungus, with or without water stress (WS) application, have been evaluated in a commercial tomato cultivar already investigated for the water stress response during AM symbiosis. Since root-knot nematodes (RKNs, Meloidogyne incognita) are obligate endoparasites and cause severe yield losses in tomato, the impact of the AM fungal colonization on RKN infection at 7 days post-inoculation was also evaluated. Results offer new information about the response to AM symbiosis, highlighting a functional redundancy for several tomato gene families, as well as on the tomato and fungal genes involved in WS response during symbiosis, underlying the role of the AM fungus. Changes in the expression of tomato genes related to nematode infection during AM symbiosis highlight a role of AM colonization in triggering defense responses against RKN in tomato. Overall, new datasets on the tomato response to an abiotic and biotic stress during AM symbiosis have been obtained, providing useful data for further researches.
Introduction
Drought is a devastating environmental condition that dramatically affects plant growth and crop production. Its adverse impact on global food security is mostly severe in semi-arid and arid regions from many parts of the world (Boyer, 1982). Additionally, climate changes are intensifying the frequency, duration and severity of drought in many agro-environments. In response to drought, domesticated plants rely on a number of physiological and structural adaptations to counteract water deficit or at least to escape most severe effects, as a result of selection for local cropping environments. The understanding of these adaptive mechanisms may be useful to sustain crop production, as well as for developing future breeding strategies. In addition to leaves, also roots, which are the first organ to detect a water deficit, are subjected to several modifications under drought, increasing water uptake and regulating water traffic between plant and soil (Gamboa-Tuz et al., 2018).
Water limiting conditions lead to changes in the expression of drought responsive genes in different plant tissues (Shinozaki and Yamaguchi-Shinozaki, 2007). Advances in technology, i.e., the development of -omics approaches, flanked by a parallel progress for in silico analyses, allowed the elucidation of the molecular mechanisms involved in the response to water deficit, in model and non-model plants. These include several economically important crops such as maize, rice, poplar, tomato, wheat or tropical fruits such as papaya (Kakumanu et al., 2012; Oono et al., 2014; Barghini et al., 2015; Gamboa-Tuz et al., 2018; Iovieno et al., 2018).
Tomato (Solanum lycopersicum L.) is one of the major horticultural crops cultivated worldwide and is a key component of the diet of many billion people. It has been reported that modern cultivars are sensitive to water deficit, which leads to a reduction in seed development and germination, impairing vegetative growth and reproduction (Iovieno et al., 2018). These authors exposed tomato plants to two cycles of prolonged drought stress and a single recovery by re-watering. Transcriptome datasets were generated for multiple time points during the stress and recovery cycles. Results allowed the identification and comprehension of the coordinated responses taking place under drought stress and subsequent recovery in leaves, highlighting the transcriptomic changes that control such physiological modifications (Iovieno et al., 2018).
Apart of abiotic stressors, plants interact in their environment with a complex soil and epiphytic microbiome, including both beneficial and noxious microorganisms. The mutualistic symbiosis established by arbuscular mycorrhizal (AM) fungi with the roots of most crops, have an important role in sustaining yields, as they act as bio-fertilizers and bio-protectors against both abiotic and biotic stresses (Balestrini et al., 2018). The latter include several soil pathogens and pests including plant parasitic nematodes (Schouteden et al., 2015). Root-knot nematodes (RKN; Meloidogyne species) are among the most devastating plant-parasitic nematodes (Jones et al., 2013). Having a wide host range, they cause large economic losses in cultivated plants that are expected to increase as a result of climate change leading water depletion and crop systems intensification. The increasing concern about the environmental impact of traditional nematicides have stimulated research for alternative control practices, including the use of biological control organisms. AM fungi have been reported to be effective against different pathogens and pests (Selosse et al., 2014; Martinez-Medina et al., 2016) and could represent a new environmental-friendly strategy to control nematode infection (Elsen et al., 2008; Vos et al., 2012; Schouteden et al., 2015; Sharma and Sharma, 2017).
Tomato has already been used as a crop model to study AM-colonized plants (Balestrini et al., 2018) as well as nematode–plant interactions (Portillo et al., 2013; Iberkleid et al., 2015; Shukla et al., 2018). Large-scale gene expression analyses have been carried out in mycorrhizal tomato plants using microarrays to identify genes differentially expressed (DE) in roots and shoots of AM-colonized plants (Fiorilli et al., 2009), as well as during the early interaction stages (Dermatsev et al., 2010). A deep sequencing of root transcriptome, using a wild-type tomato and a mutant incapable of supporting a functional AM symbiosis, showed the expression of several genes associated to AM symbiosis (Ruzicka et al., 2013). Comparison between transcriptomic profiles of tomato and Lotus AM-colonized roots has also been performed, suggesting that a certain proportion of AM-responsive genes are conserved across plant species (Sugimura and Saito, 2017). The authors also highlighted the fact that species-dependent AM-responsive genes might be related to specific root features, characterizing each host plant.
Several studies have been carried out on the impact of the AM symbiosis on the tomato response to water deficit (Dell’Amico et al., 2002; Subramanian et al., 2006; Aroca et al., 2008; Wang et al., 2014; Chitarra et al., 2016; Ruiz-Lozano et al., 2016; Rivero et al., 2018; Volpe et al., 2018). Targeted approaches already showed that several drought-responsive genes are differentially regulated in AM-colonized tomato plants (roots and leaves) during water deficit (Chitarra et al., 2016; Ruiz-Lozano et al., 2016). However, global changes in transcripts of mycorrhizal tomato roots, as affected by a water stress condition, have not yet been investigated, as this interaction was thus far studied only in AM-colonized bean roots (Recchia et al., 2018). We hence studied changes in the whole root transcriptome of S. lycopersicum cv San Marzano nano, which was previously tested to evaluate the impact of the AM symbiosis on the tomato water stress responses (Chitarra et al., 2016; Volpe et al., 2018). These previous works showed that the AM symbiosis positively affects the tomato tolerance to water deficit and how the adaptive plant response is dependent on the AM fungal species involved. Additionally, Volpe et al. (2018) have evaluated the impact of the colonization on tomato plants subjected to combined stresses (moderate water stress and aphid infestation) in controlled conditions. A positive effect on the tomato indirect defense toward aphids in terms of enhanced attractiveness toward their natural enemies was observed, as also supported by the characterization of volatile organic compound (VOC) released. In the present study, new information on the role of AM symbiosis to enhance crop tolerance to abiotic and biotic stresses in a global climatic change scenario has been obtained. In detail, changes induced in the transcriptome profile in roots colonized with the AM fungus Rhizophagus intraradices (i) in the presence of a moderate-water stress (abiotic stress), and (ii) following parasitism by the root-knot nematode Meloidogyne incognita (biotic stress) have been evaluated.
Materials and Methods
Plant Material and Growth Conditions
The S. lycopersicum ‘San Marzano nano’ genotype, important for its consumption in Italy, was used. Tomato seeds were surface sterilized in sodium hypochlorite for 20 min, washed five times in sterile water, and germinated on wet paper. Seedlings were then moved to pots containing a mixture of quartz sand (50%), sterile pumice (20%), and an inoculum (30%) of R. intraradices (FR 121), containing AM fungal propagules (spores, mycelium and mycorrhizal root pieces) in a carrier of mixed inert mineral, purchased from MycAgro Lab (Dijon, France). For non-inoculated plants, the sterile inoculum carrier was used alone, instead of the specific inoculum. The plants were maintained in a growth chamber under controlled conditions at 25°C, with a light intensity of 150 μmol/m2/s and a 16 h:8 h light/dark cycle.
Water Stress Treatment
Plants were abundantly irrigated with filtered tap water (twice a week) and Long Ashton solution containing 300 μM of inorganic phosphate (once a week) for 35 days prior the imposition of a moderate water stress treatment. Considered treatments were: (i) AM fungal colonization [non-colonized (C), R. intraradices-colonized (AM)] and (ii) water stress [none (unstressed or NS), moderate (WS) as described in Volpe et al. (2018)]. Nine replicates for each group (C, AM, WS, AM_WS) have been used, arranged in a randomized block design. Before starting the treatments, pots were weighed. Control plants were regularly watered throughout the entire experimental period whereas plants to be stressed were not watered until the pots reached a loss of about 210–220 g of the initial weight, a loss previously described to be needed to reach a moderate WS condition (Volpe et al., 2018). From this moment the plants received the amount of water or nutritive solution required to get them back to their last weight, in order to maintain a moderate stress level. At the end of the experiment (i.e., after 9 weeks), roots were sampled and 60 randomly chosen 1-cm-long root segments per 2 plants were stained with 0.1% cotton blue in lactic acid, to evaluate the presence of the AM fungus, before the RNA extraction. Due to the low quantity of root materials, mainly in the WS treatments, AM fungal colonization has been only qualitatively evaluated.
Nematode Infection Assay
Control and AM-colonized 8-week-old plants were inoculated with 1200 freshly hatched juveniles of Meloidogyne incognita per plant. Juveniles were collected from egg masses of infested tomato roots, which were allowed to hatch in water in a growth chamber at 25°C. At 7 days post-nematode inoculation (dpi) roots were harvested. Root galls from infected colonized (RKN_AM) and uncolonized (RKN) tomato plants were hand-picked and pooled in two biological replicates for each treatment. Samples were immediately frozen in liquid nitrogen and stored at −80°C until RNA-Seq experiments.
Histological Observations
Galls at 7 dpi from AM-colonized and non-colonized tomato roots were hand-dissected under a stereomicroscope. At least five to ten galls were excised from each plant and fixed in a mixture of 1.5% glutaraldehyde and 3% paraformaldehyde (Sigma-Aldrich), dehydrated in ethanol and embedded in acrylic resin LR White (Sigma, St. Louis, MO, United States) according to Melillo et al. (2014). Embedded galls were cut in serial cross-semithin sections (2.5 μm) through all their length, then stained briefly with 1% toluidine blue in 1% borax solution and mounted in Depex. Microscopic observations were performed using bright-field optics on a Leica DM 4500 B light microscope equipped with a Leica DFC 450C camera.
RNA Extraction and Sequencing
Six different treatments (C, WS, AM, RKN, AM_WS, RKN_AM) were set up to study the response in tomato roots. Total RNAs were extracted from root samples (two independent replicates for each treatment) using a RNeasy Plant Mini Kit (Qiagen, La Jolla, CA, United States) with addition of an on-column DNase I digestion, following the manufacturer’s protocol. RNA quantity and quality were determined with a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, United States) and sent to IGA Technology Services (Udine, Italy)1. cDNA libraries were prepared from 4 μg total RNA using TruSeq RNA Sample Preparation Kit v2 (Illumina, Inc., San Diego, CA, United States) and validated according to Illumina’s low-throughput protocol. After normalization, cDNA libraries were pooled for multiplexing before loading onto a flow cell (8–9 samples per lane). The hybridization and cluster generation were performed on a cBot System using TruSeq SR Cluster Kit v3. Sequencing was performed on an Illumina HiScanSQ platform using TruSeq SBS kit v3 (Illumina, Inc.) to obtain Single Reads, 50 nt in length. Due to the scarcity of the collected material in WS treatment, and according to ENCODE standard for RNAseq experiments2, requiring at least two biological replicates, data analysis has then been performed only on C, AM, AM_WS, RKN and RKN_AM. Raw data have been deposited to NCBI and the accession number for this project is PRJNA545411.
RNA-Seq Data Analysis and Differential Gene Expression Quantification
The quality of the raw sequence reads was checked using FastQC3. Raw sequences were processed to eliminate adapters, indexes, as well as genomic sequences added during the sequencing process, using the “RNA-seq analysis” functions included in the CLC Genomics Workbench software v.8.5 (QIAGEN, Aarhus, Denmark)4. Filtered reads from each sample were then separately aligned to the reference genome of S. lycopersicum (SL2.40.26, Sol Genomics Network)5, using CLC (similarity parameter: 0.8; identity parameter: 0.8, mismatch/insertion/deletion penalties: 2/3/3) and employed to quantify the abundance of tomato gene transcripts, measured as the Reads Per Kilobase per Million mapped reads (RPKM) (Mortazavi et al., 2008). A gene was considered to be expressed and included in the downstream analysis if at least five reads were mapped to it and its RPKM value was >0.
A multiple correlation test (Pearson’s correlation) on RPKM values for all pairwise combinations was performed for preliminary batch comparisons of replicates. To identify differentially expressed gene (DEG), statistical analysis was carried out for each treatment group against a reference group (equivalent developmental stages of un-colonized un-stressed roots). RNA-Seq library set differential expression analysis was performed applying “Empirical analysis of Digital Gene Expression (DEG) in R” (EDGE) that implements, in the EdgeR Bioconductor package (Robinson et al., 2010), the ‘Exact Test’ for two-group comparisons (Robinson and Smyth, 2008), able to account for over-dispersion caused by biological variability. Raw counts for each gene were normalized in relation to different sequence depths between duplicate bioassay samples.
Genes in libraries were considered DE when compared with the controls, by applying the Benjamini–Hochberg algorithm for Fold Change (FC) estimation. DEGs displaying at least a P-value (p) ≤ 0.05, in almost one condition, were submitted to further analysis. The terms up-regulation and down-regulation indicate, respectively, transcript levels that were significantly higher or lower than those observed in non-inoculated controls.
Functional Analysis of Tomato DEG
Enrichment analysis of each DEG gene ontology (GO) term was performed by AgriGO device6 (Du et al., 2010) to identify GO category related to single or multiple interactions. GO Enrichment analysis detect functional categories of biological processes, molecular functions and cellular components over-represented, with statistical significance (Fisher’s Exact Test: p-value ≤ 0.05; Hochberg Multi-test adjustment method: FDR ≤ 0.05), in a gene sub set using annotations for that gene set as compared with the remaining genes of the reference organism (S. lycopersicum cDNA library, version 2.47.
Identification of AM Fungal Transcripts in Tomato Roots
To discover AM fungal genes expressed in roots, fungal reads were separated from those of tomato by mapping the complete RNA-Seq data set of AM-colonized roots onto the draft genome sequence of tomato version SL.2.46. Unmapped reads, potentially fungus-derived, were isolated and mapped, with CLC utility and parameter previously described, to the Rhizophagus irregularis DAOM 181602 v1.0 (formerly Glomus intraradices, GLOIN) genome, retrieved from JGI genome portal8 (Tisserant et al., 2013). The RPKM were determined and used to estimate the GLOIN transcripts abundance. To assess differential gene expression between AM_WS and AM samples, proportions-based Z-test statistical analysis (Kal et al., 1999) was applied, assigning the samples different weights depending on library size (total counts). A GO functional enrichment analysis was conducted using Fisher’s exact test with a weight algorithm in AgriGo. The GO annotations of R. irregularis genes were obtained from the Mycocosm JGI genome portal (Tisserant et al., 2013)8.
Results
To gain a comprehensive understanding of the mechanisms activated in mycorrhizal tomato roots under abiotic and biotic stress conditions, plants were subjected to a moderate water stress and to a 7-days long RKN infection. Transcriptome root profiles were obtained from C, AM, AM_WS treatments, while the biotic stress impact was studied using the infection structure (gall) from non-mycorrhizal (RKN) and mycorrhizal (RKN_AM) plants.
Sequencing Data and Transcriptome Mapping
Transcriptome sequencing generated a total of 187,7⋅106 reads of 50 bp in length, for all samples. Good-quality reads (99%) were retained, which were mapped onto the reference genome with CLC. In total, 76–97% of good-quality reads were mapped onto the S. lycopersicum genome (SL2.40.26 assembly) across all samples (Table 1). Expressed tomato genes arose from 16347 (AM_WS2_7) up to 22959 (RKN1_7) out of 34647 predicted genes in the reference genome (Table 1), using a cutoff value of RPKM > 0 to declare a gene as expressed. A high Pearson’s correlation coefficient (r) was observed between RPKM values of each sample replicates sequenced set (average r = 0.92) (Table 2).
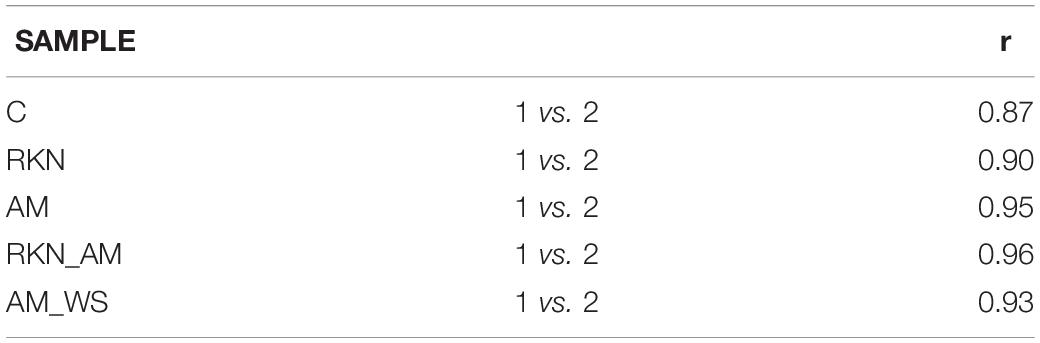
Table 2. Correlation (Pearson’s correlation coefficient) between RPKM values of biological replicate of each sample pairwise combinations of sample set.
Whole Transcriptome Profiles and Differentially Expressed Genes
The genes DE respect to control (p-value < 0.05) arose from 10 (RKN) up to 12 % (AM) of total SL2.40.26 predicted protein coding genes (Table 3). A higher percentage of up-regulated genes was observed only in the AM condition (Figure 1). For further analyses, 7316 differentially expressed genes (DEGs), when compared with un-colonized/un-stressed control, and with a p-value < 0.05 in at least one treatment (AM, AM_WS, RKN, and RKN_AM) were considered (Supplementary Tables S1, S2).
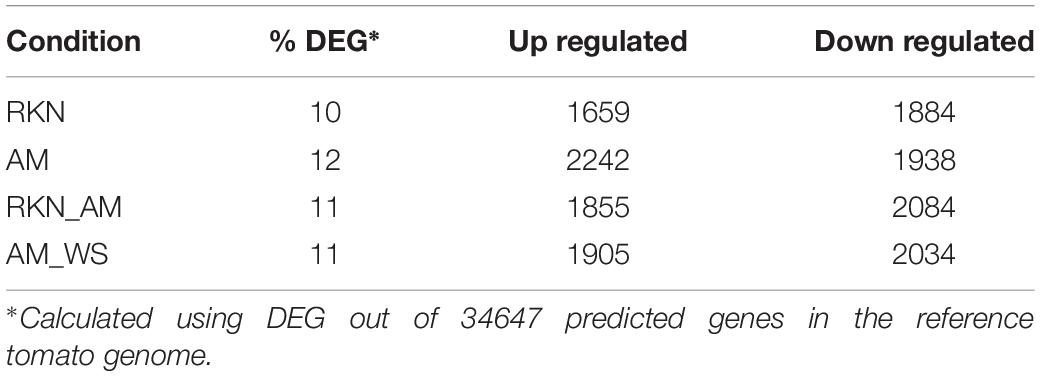
Table 3. Differentially expressed tomato genes comparing to uninoculated unstressed plants (C) (P < 0.05).
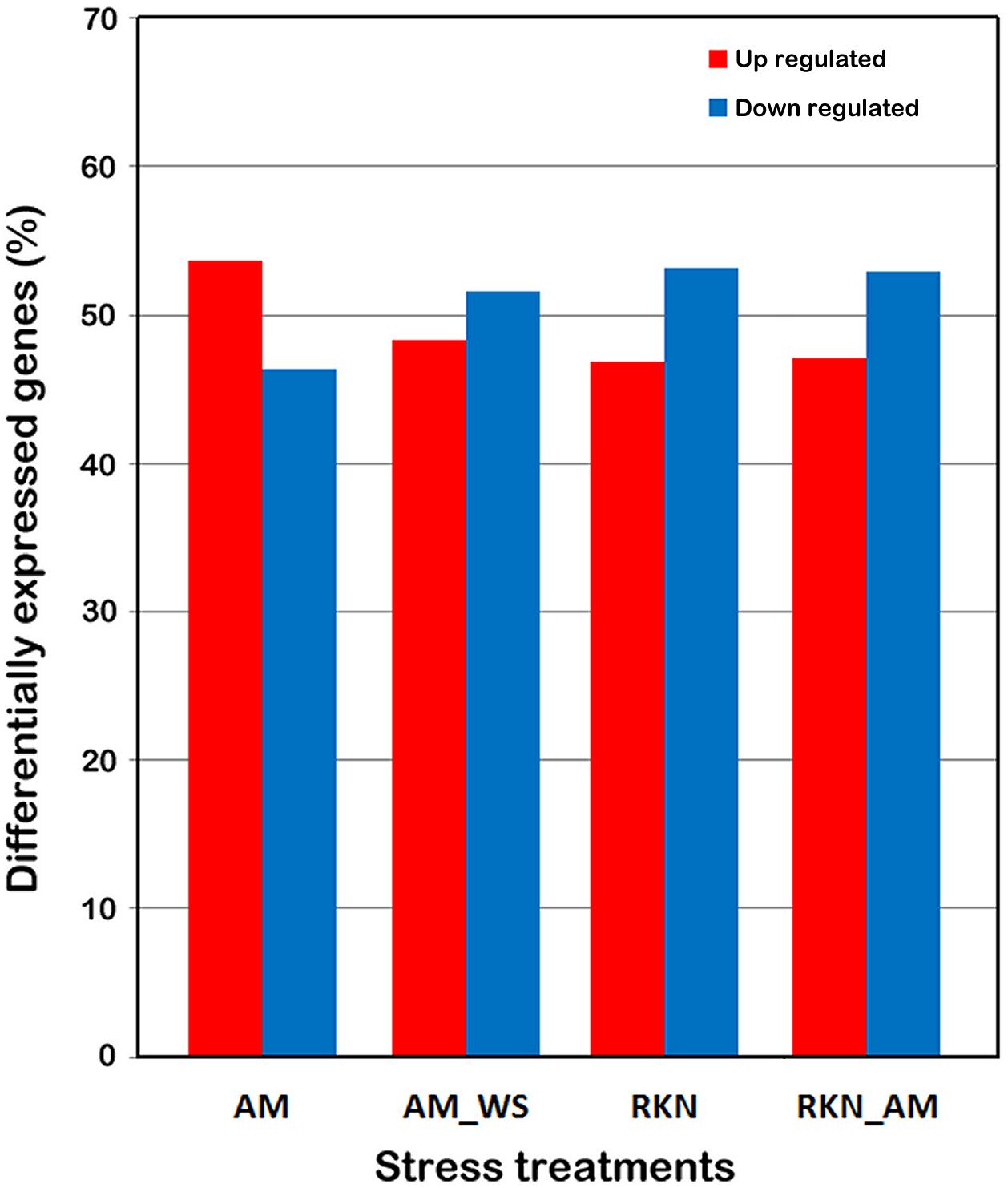
Figure 1. Differentially expressed tomato genes. DEGs were identified by comparing the expression profiles from treated roots (AM, AM_WS, RKN, RKN_AM) with the equivalent developmental stages of control roots (C) (P ≤ 0.05). Each column represents a treatment.
To have an overview of the regulation of the main metabolic processes and signaling pathways involved in the different stress conditions, a GO enrichment analysis was carried out for DEGs in AM_WS and RKN_AM. Data showed that transcripts involved in processes such as “response to oxidative stress,” “functions of peroxidase activity” and “heme binding” were involved in both situations. Specific GO terms enriched during abiotic stress (AM_WS) were related to the molecular function “transcription regulator activity,” while in biotic stress (RKN_AM) to the metabolic process “protein ubiquitination and protein amino acid phosphorylation” and cellular process “microtubule-based movement,” that were particularly over represented (Table 4, Supplementary Tables S3, S4, and Supplementary Figures S1, S2).
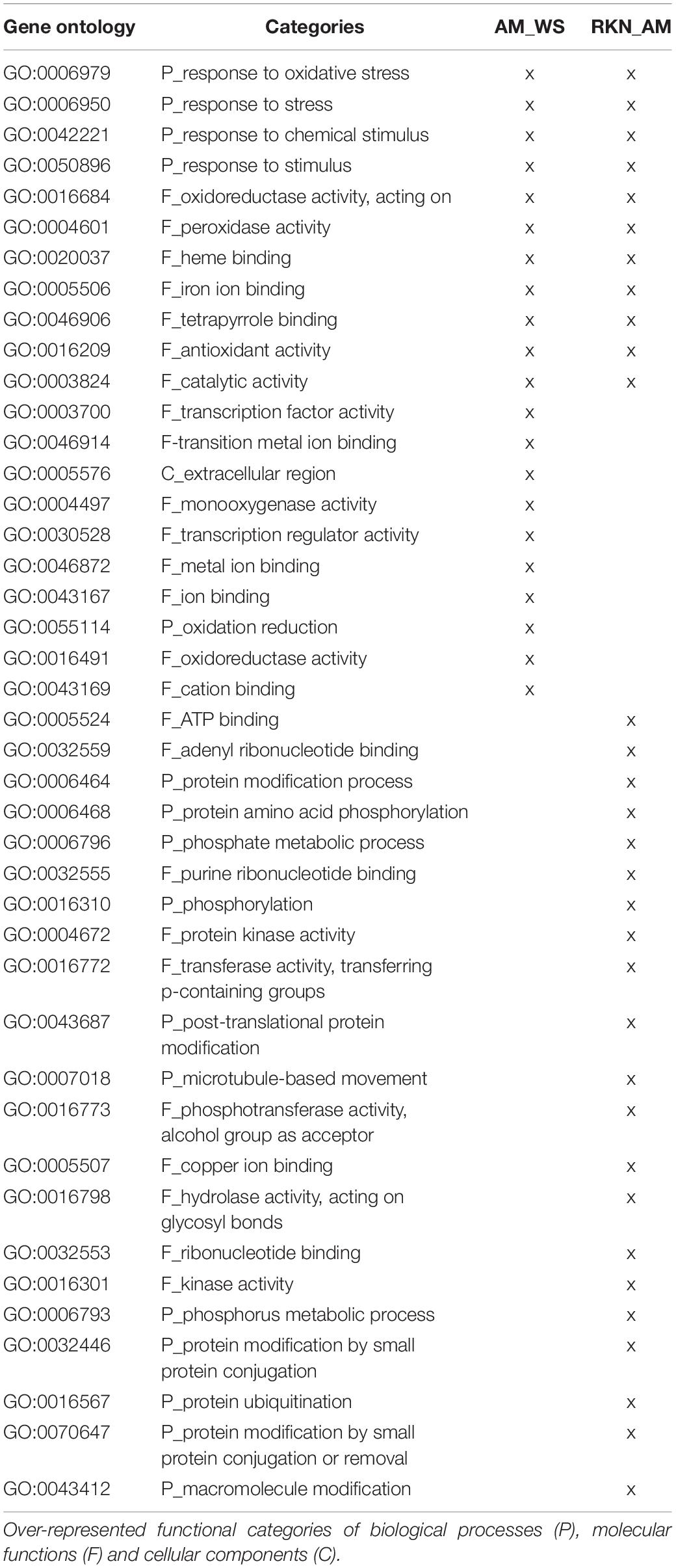
Table 4. GO Enrichment analysis (P-value ≤ 0.05) for differentially expressed genes during abiotic and biotic stress.
In the 7316 DEGs subset, a number of common and exclusive genes were observed among the treatments (Figure 2). Focusing on the 924 transcripts in common among AM, AM_WS, RKN, and RKN_AM treatments, 188 e 689 of them were up- and down-regulated, respectively, whereas the remaining 47 transcripts showed an opposite expression trend, in at least one condition (Supplementary Table S5). GO enrichment analysis conducted for this subset showed up regulated transcripts belonging to processes involved in “biotic stimulus,” “oxidative stress response” and functions like “heme binding,” “transition metal ion binding” and “peroxidase activity.” Processes such as “post-translational protein modification” and “regulation of transcription” were down-regulated, while transcripts ascribed to the “heme binding” function showed different expression trends (Supplementary Table S6). Looking at the AM fungal presence, 196 genes were DE in all the AM-colonized plants, independently from the applied stress condition (Figure 2).
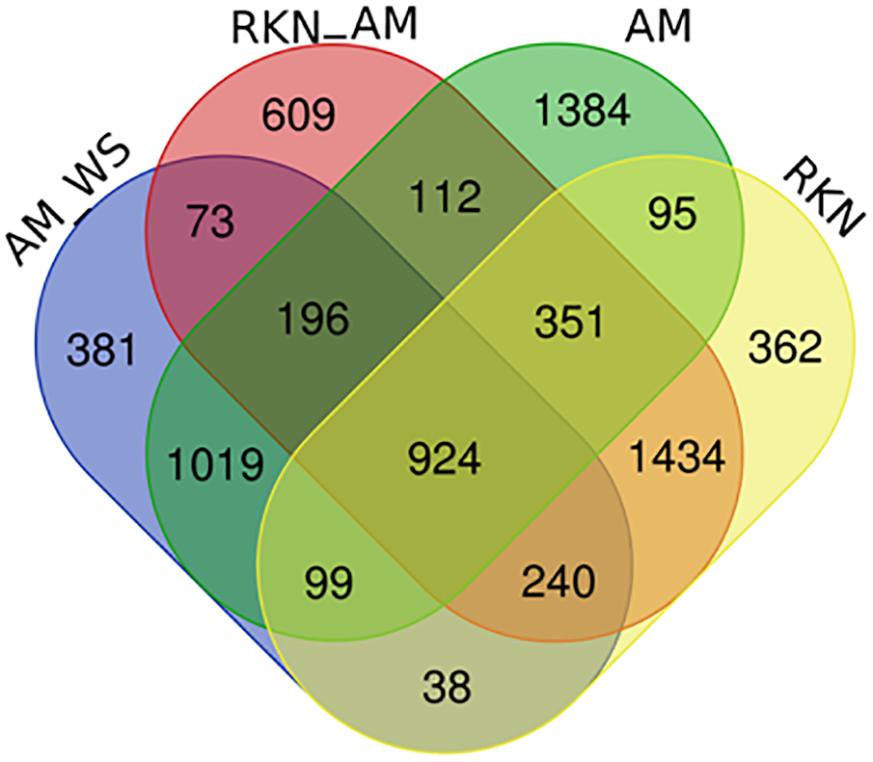
Figure 2. Venn diagram of DEGs (P ≤ 0.05) comparing un-inoculated un-stressed tomato (C) vs. AM, AM_WS, RKN, RKN_AM.
Among the twenty most up-regulated genes found in each condition (Supplementary Table S7), all genes of AM_WS sample were also retrieved in the top 20 DEGs of unstressed AM-colonized plants (AM), although a lower fold change was often recorded in AM_WS plants, probably reflecting a low level of colonization in stressed plants with respect to the AM ones. One transcript (Solyc01g067860.2.1) was up regulated in all stress conditions. Considering the biotic stresses (RKN and RKN_AM), 12 transcripts were in common. The most up-regulated genes in the infection structures (galls) were mainly related to biotic stress response, independently from the AM fungal presence.
Specific Responses to the AM Fungus in Drought Stressed Versus Unstressed Roots
To deeper explore this novel dataset and to further understand the tomato response to WS during AM interaction, as well as the impact of the imposed stress on the symbiosis, the expression profiles of genes described in the literature as specifically involved during AM symbiosis in different plant species have been considered (Fiorilli et al., 2009; Guether et al., 2009; Hogekamp et al., 2011; Hogekamp and Küster, 2013; Fiorilli et al., 2015, 2018; Handa et al., 2015; Balestrini et al., 2017; Li et al., 2018; Recchia et al., 2018; Vangelisti et al., 2018). Particularly, AM symbiosis is known for the improved nutrient exchange established between the two symbionts, involving fine-tuned plant and fungal transporter genes (Casieri et al., 2013; Berruti et al., 2016). A consistent group of plant transporters were identified as DE between unstressed colonized plants (AM) and the control (C) ones (Supplementary Table S8) also confirming that a functional symbiosis was established. Several transporters were significantly up-regulated in AM-colonized plants subjected to a moderate water stress (Supplementary Table S8 and Table 5). Two of them code for two inorganic phosphate transporters (Solyc09g090080.1.1 and Solyc06g051860.1.1), which show homology with the mycorrhizal inducible LePT3 and LePT4 (Balestrini et al., 2007). The other mycorrhiza-inducible LePT5 (Solyc06g051850.1.1) was still up-regulated in AM_WS, although not in a significant way (P > 0.05), in agreement with the fact that this one was the AM-inducible PT gene significantly affected only by the “stress” factor (Volpe et al., 2018). Many other genes coding proteins involved in molecule transport were significantly up-regulated in the presence of the AM fungus upon water stress, such as three ammonium transporters (AMT; i.e., Solyc08g067080.1.1, Solyc03g033300.1.1, Solyc09g065740.1.1) and four potassium (K+) transporters (Solyc03g097860.1.1, Solyc06g051860.1.1 Solyc07g014680.2.1, Solyc05g005740.2.1). Tomato genes coding for three ABC transporter family G (Solyc11g007300.1.1, Solyc01g097430.2.1, Solyc09g098410.1.1), with the last two homologs of the AM symbiosis-induced ABCG transporter LjSTR2 and LjSTR, were among the ten most up-regulated genes in AM_WS, in addition to the three AMT genes, two genes coding for peptide transporters (Solyc01g102360.1.1, Solyc05g009500.2.1) and one gene encoding an high affinity cationic amino acid transporter (CAT; Solyc12g096380.1.1). Sulfate transporters were also regulated and two of them were significantly induced by AM symbiosis (Solyc12g056920.1.1, Solyc10g047170.1.1). In AM_WS they were up- and down-regulated respectively, although not significantly different. Among the down-regulated genes in AM symbiotic roots, both in unstressed and stressed plants, there was a PT gene (Solyc09g066410.1.1) that shows homology with an inorganic phosphate transporter 1-4 (XP_004247235.1) expressed in tomato fruit from mycorrhizal plants (Zouari et al., 2014).
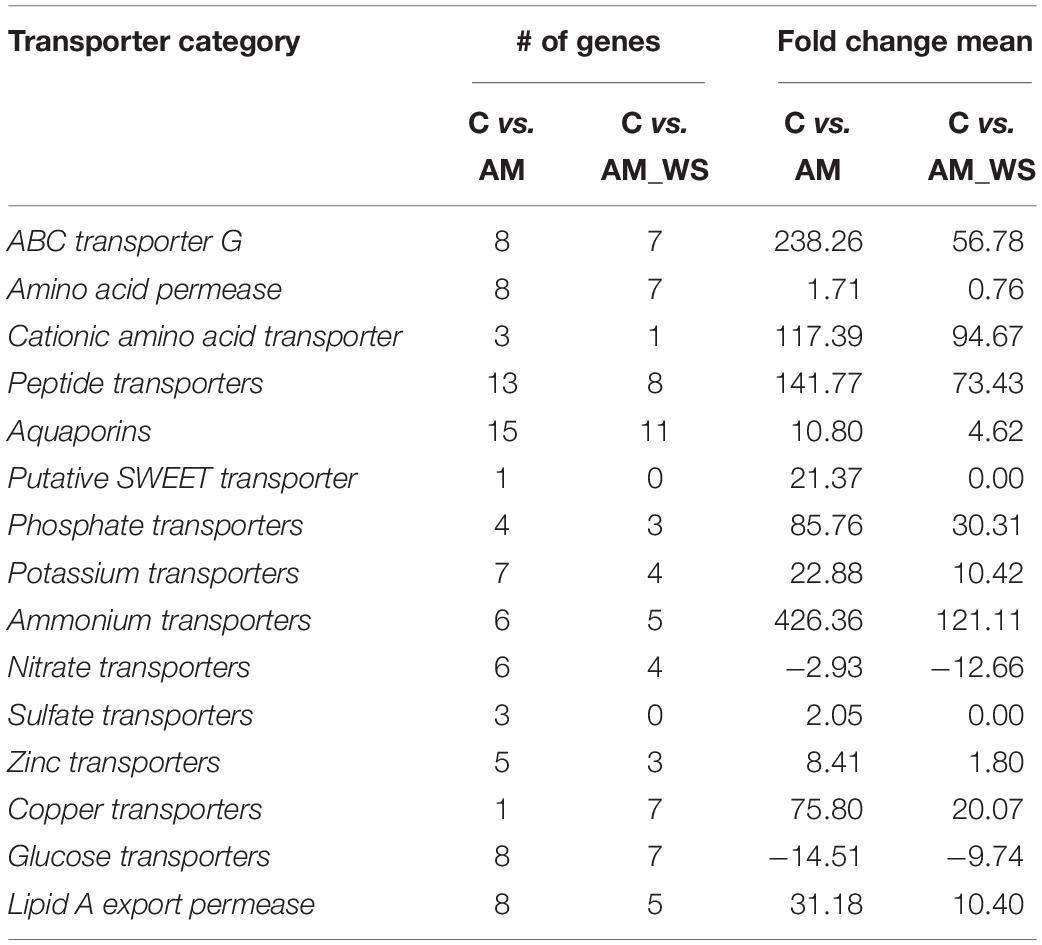
Table 5. Transporter genes significantly regulated by the AM fungus in non-stressed (NS) and stressed (WS) growth conditions.
Almost all the genes reported by Sugimura and Saito (2017) as symbiosis marker genes both in tomato and Lotus were also found strongly up-regulated in AM-colonized roots (Supplementary Table S9), both in unstressed and AM_WS stressed plants. Additionally, other members within gene families already known to be involved in AM symbiosis (e.g., blue copper proteins, germin-like proteins, glutathione-S-transferase, ripening-related proteins, cell wall-related genes etc.) were strongly up-regulated in the presence of the AM fungus (Supplementary Table S10). Generally, expression levels were almost always lower in AM_WS than in AM roots, probably due to an opposite impact of the water stress on their regulation. Among the genes strongly up-regulated in AM_WS (i.e., with fold changes >50), there were those encoding three putative glutathione-S-transferases (Solyc02g081240.1.1, Solyc10g007620.1.1, Solyc10g084960.1.1) in addition to a putative germin-like (Solyc11g068570.1.1), three blue copper (Solyc12g056500.1.1, Solyc10g081520.1.1, Solyc08g079780.1.1), two major allergen (Solyc03g117460.1.1, Solyc03g117450.1.1), four ripening-related (Solyc01g094440.1.1, Solyc05g048780.1.1, Solyc01g094450.1.1, Solyc01g094430.1.1), two putative dirigent-like proteins (Solyc07g052170.1.1, Solyc04g010270.1.1) with three putative cysteine proteinases (Solyc12g056000.1.1, Solyc08g065710.1.1, Solyc08g065690.1.1), and two subtilisin-like proteases (Solyc08g080010.1.1, Solyc02g072370.1.1) (Supplementary Tables S9, S10).
Hormonal related transcript levels have been also already reported to change in the presence of the AM fungus (Gutjahr, 2014; Bedini et al., 2018; McGuiness et al., 2019). The AM marker gene CCD7 (Solyc01g090660.2.1) coding for a carotenoid cleavage dioxygenase 7 with a role in strigolactone (SL) biosynthesis, was found to be still regulated in AM-colonized WS roots (AM_WS) in addition to CCD8 gene (Supplementary Tables S5, S6). Gibberellin-related genes have been also found to be affected by AM symbiosis in Medicago truncatula. In agreement with the data in Ortu et al. (2012), tomato DELLA GAI protein (Solyc11g011260.1.1; Nir et al., 2017) resulted to be up-regulated in the AM treatment, while Gibberellin receptor GID1L2 (Solyc09g075670.1.1) was down-regulated. Gibberellin 20-oxidase-like proteins were up-regulated in AM-colonized roots (Solyc12g013780.1.1 and Solyc01g093980.2.1), while a putative Gibberellin 20-oxidase-1 (Solyc03g006880.2.1) was induced only in the AM treatment. Among the Gibberellin 2-oxidases, one gene (Solyc02g070430.2.1) resulted to be down-regulated in both AM-colonized roots, while Solyc07g061730.2.1 was significantly repressed only in AM_WS and Solyc07g061720.2.1 was up-regulated in AM roots (Supplementary Table S10). Interestingly, genes coding for enzymes with a key regulatory role in the biosynthesis of phenylpropanoid products, which can have diverse functions and are particularly important in plant defense (Mandal et al., 2010), were upregulated in AM plants. In detail, genes coding for two phenylalanine ammonia lyases (PAL; Solyc03g042560.1.1, Solyc09g007900.2.1) and a 4-coumarate-CoA ligase (4CL; Solyc11g007970.1) were significantly up-regulated in AM-colonized plants, suggesting an effect of the AM fungus for enhanced defense ahead of stress occurrence.
In agreement with data already reported (Liu F. et al., 2015), several non-specific lipid transfer proteins were also detected as significantly regulated during AM symbiosis also under WS condition (Supplementary Table S10). A core of differentially regulated genes was related to genes involved in cell wall metabolism and modification, in agreement with previous works (Balestrini and Bonfante, 2014). Among the genes putatively involved in arbuscule development and fungal accommodation, four genes coding for cellulose synthases were significantly up-regulated (Solyc00g030000.1.1, Solyc00g154480.1.1, Solyc07g051820.2.1, Solyc08g076320.2.1), independently from the stress level, in addition to an endo-β-1,4-glucanase (Solyc04g064900.1.1). Several genes encoding expansin proteins were also differentially up- or down regulated in both AM and AM_WS roots, confirming a role for these proteins during the AM colonization.
Transcription factors (TFs) have been reported to be highly involved in the response to drought as well as in AM symbiosis establishment. In our dataset, AM colonization, independently from the growth conditions, elicited the expression of several TF genes belonging to different groups, while other members inside these families were down-regulated (Supplementary Table S9). Looking at the genes expressed in AM and AM_WS, two ethylene responsive transcription factor 2a (Solyc04g071770.2.1, Solyc12g056590.1.1) resulted to be down-regulated, while an opposite trend was observed for the LeERF5 (Solyc03g093560.1.1), up-regulated in AM-colonized roots, independently from the imposed stress. Due to the role in the adventitious roots and in the regulation of auxin responsive genes, the up-regulation in both AM and AM_WS roots of auxin responsive factor (ARF) genes (Solyc07g043610.2.1, Solyc03g118290.2.1) is worth noting since they are considered to be related to the modification of the root apparatus occurring during symbiosis (Vangelisti et al., 2018).
The expression of WS-responsive genes has been considered, to verify the efficacy of the WS stress imposition on tomato transcriptome in the presence of the AM fungus. The relevance of ABA in the response of plants to a water deficit event has already been highlighted by the differential regulation of target genes, directly associated with the ABA biosynthesis and signaling (Kuromori et al., 2018). The co-regulation with negative regulators has already been reported, suggesting the presence of a mechanism to fine-tune plants stress responses (Garcia et al., 2008). The presence of the fungus, independently from the growth condition (NS and WS), induced in roots the expression of a gene (Solyc07g056570.1.1, i.e., LeNCED1) coding a 9-cis-epoxycaratenoid dioxygenase involved in the ABA biosynthetic process, with a fold change value higher in AM than in AM_WS plants. A gene coding for a NAC transcription factor JA2 (Solyc12g013620), reported to be induced by ABA and WS in tomato leaves, and promoting stomatal closure through the induction of the expression of ABA biosynthetic gene NCED1, was down-regulated in AM-colonized roots, both in stressed and unstressed condition, suggesting a specific role in leaves. Solyc03g116390.2.1, coding for a late embryogenesis abundant (LEA) protein already shown to be inducible by drought stress (Gong et al., 2010) was significantly up-regulated in AM_WS, additionally to other two LEA genes (Solyc12g098900.1.1, Solyc10g078770.1.1) and the ABA-responsive gene coding for dehydrin TAS14 (Solyc02g084850.2.1; Sacco et al., 2013), confirming the efficacy of the imposed water stress. Additionally, Solyc06g019170.2.1, the ABA responsive gene Delta l-pyrroline-5-carboxylate synthetase SlP5CS1, involved in proline production, was significantly up-regulated only in AM_WS (Supplementary Tables S1, S2).
As expected, the several tomato aquaporin (AQP) genes showed different regulations among the considered conditions. Several AQP genes (Table 6) were significantly up- or down-regulate in AM roots, and many were significantly regulated also in AM_WS ones. The significant up-regulation observed for the AQP Solyc09g007770.2.1 (SlPIP2;1) in AM_WS roots is worth of noting, suggesting a specific role in WS response for this transcript.
Table 6. List of AQPs regulated in the several conditions (AM, unstressed AM-colonized roots; AM_WS, water stressed AM-colonized roots; RKN, nematode infection; RKN_AM, nematode infection in AM-colonized plants).
AM Fungal Gene Regulation Upon Water Stress
Gene expression analysis of R. irregularis colonizing tomato roots was performed by mapping short reads against the R. irregularis genome. The proportion of R. irregularis-derived reads was less than 12% in AM roots and of 5% in AM_WS (Table 7), consistent with the results of previous studies (Tisserant et al., 2013; Handa et al., 2015; Sugimura and Saito, 2017). We identified 19057 and 16364 R. irregularis expressed genes (RPKM > 0), respectively, in AM and AM_WS roots, with 78.8% of overlapping, corresponding to 52% of the R. irregularis putative protein-coding genes expressed in both conditions. The expression levels of R. irregularis genes were significantly correlated between AM and AM_WS samples (Pearson correlation = 0.85). 77% of the R. irregularis genes found in tomato roots (RPKM > 0) coincided with those expressed by the fungus in Lotus japonicus roots, according to Handa et al. (2015). A total of 1224 R. irregularis genes, 4% of the entire transcriptome, were DE between AM and AM_WS treatments (FC ≥ 2; p-value ≤ 0,01), of which 779 were up-regulated and 445 down- regulated in roots subjected to moderate water stress (Supplementary Table S11). A GO enrichment analysis of the subset of genes highly expressed upon water stress revealed an overrepresentation of GO terms related to molecular function “monooxigenase activity” and “heme binding” (Supplementary Table S12). The most remarkable difference was the higher expression of 18 cytochrome P450 (CYPs), belonging to “heme binding” molecular function, 2 to 12-fold overexpressed in stressed roots (Supplementary Table S11). By contrast, only 5 CYP genes turned out to be down-regulated, with a mean fold change of −2.7 (Supplementary Table S11). Concerning the 20 most up- and down regulated genes, it is worth noting that the majority of them lacks a KOG annotation (16 out of 20 genes both for the up- and the down-regulated). However, the most up-regulated sequence (FC 74.5) contains a CsbD domain, that is present in a bacterial gene induced upon abiotic stresses such as nutrient-limitation (Prágai and Harwood, 2002). Another highly up-regulated gene (FC 65.23) contains a “conidiation protein 6” domain.
Table 7. Summary statistics for Illumina sequencing and mapping against reference R. irregularis DAOM198197 genome assembly (Gloin1).
Several genes encoding proteins containing BTB/POZ and Kelch domains, involved in signaling transduction and regulation at the protein level, were also highly regulated in our dataset (mean up and down FC: 10.7 and −4.3, respectively). Genes encoding for proteins involved in protein turnover (ubiquitin pathway and chaperones) were DE, with 40 transcripts found as up-regulated. Remarkably, three sequences ascribed to glutathione S-transferase were up-regulated in the AM_WS treatment, none of them being down-regulated. As for R. irregularis genes down regulated in stressed tomato roots, they were fewer compared with the up-regulated ones, and no overrepresented GO terms was recorded (Table 8).
Specific Responses to Nematode Infection in Galls From Mycorrhizal and Non-mycorrhizal Roots
During the compatible interaction, RKN trigger complex morphological and physiological changes in parenchymatic cells of the vascular cylinder to establish multinucleate feedings cells, called giant cells (GC), which serve as nutrient sinks for feeding. Nematode feeding sites, surrounded by cortical and epidermal cells, appear in the host root as the typical root-knot or gall.
Microscopic observations were carried out on serial cross sections of AM-colonized and non-colonized galls at 7 dpi. Morphological changes were analyzed to monitor possible alterations in the development of the nematode feeding sites. Non-colonized galls showed large multinucleate GCs occupying the most part of vascular cylinder. The GCs presented small vacuoles and a dense granular cytoplasm containing numerous organelles, acting as a food sink for the growing nematode (Figure 3A). Several sections of AM-colonized galls revealed different features of the feeding sites (Figures 3B–D). Some of them showed GCs comparable with those of non-colonized galls (Figure 3B). Where AM fungal hyphae were observed near to the nematode feeding site (Figures 3D,E), GCs showed scarce cytoplasm with fewer organelles and clear symptoms of early senescence (Figures 3C,D). AM presence supported expression data of AM symbiosis marker genes and reads belonging to the fungus itself in this infection structure. In order to understand the tomato response during AM_RKN interaction, namely the impact of the symbiosis on the parasitism, data analysis was focused on a subset of genes already reported as specifically involved during susceptible RKN-tomato interaction (Iberkleid et al., 2015; Shukla et al., 2018).
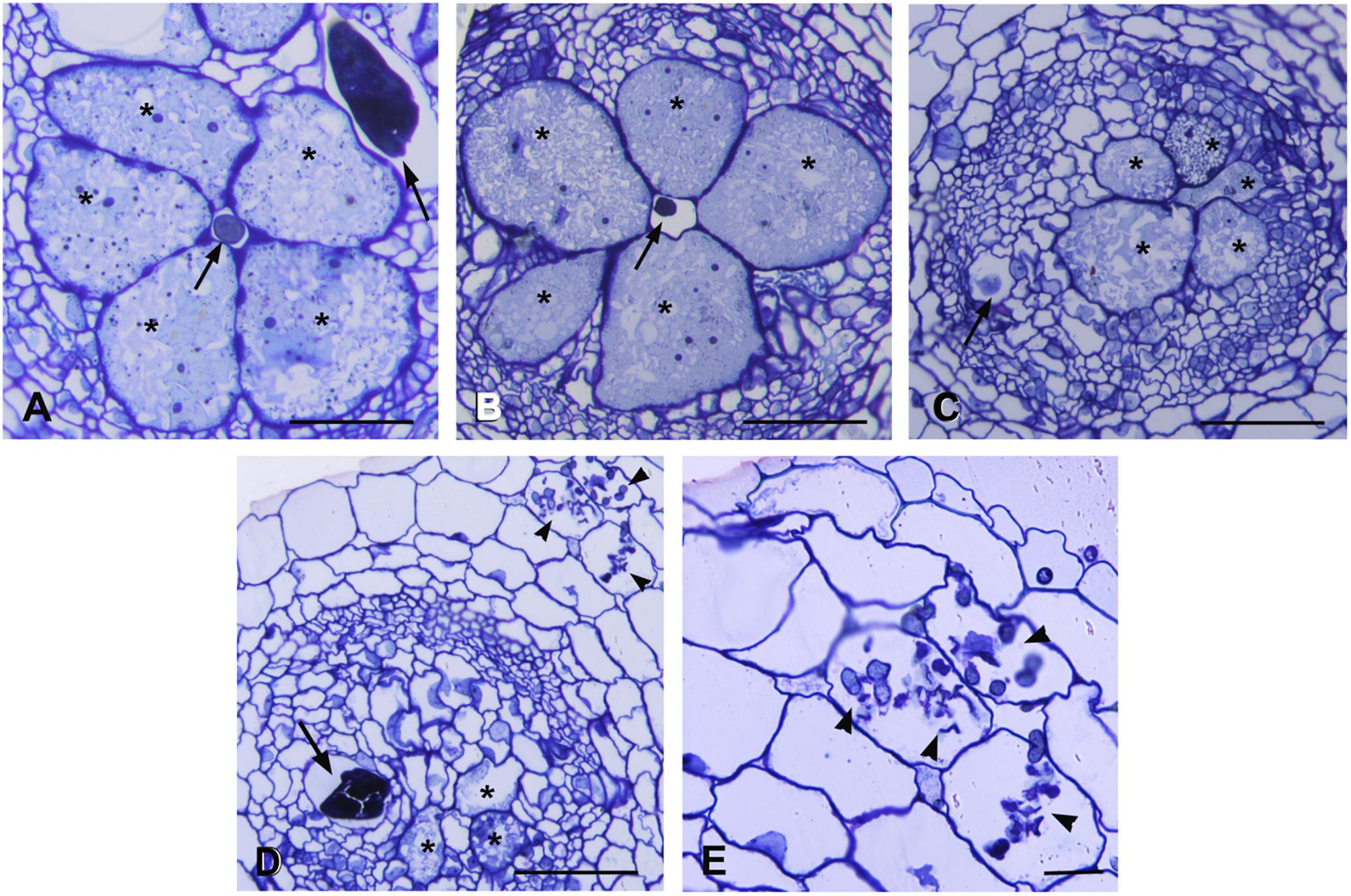
Figure 3. Histological analysis of feeding sites induced by Meloidogyne incognita in non-colonized and in Rhizophagus intraradices-colonized tomato roots at 7 days post inoculation. Cross sections (2.5 μm) were stained with toluidine blue and observed at light microscope. (A) Non-colonized gall contained well developed and metabolically active giant cells surrounding the nematode. (B) Giant cells in a AM-colonized gall with an appearance similar to those in non-colonized gall. (C,D) Feeding sites in AM-colonized roots presented giant cells with degraded cytoplasm and clear symptoms of early senescence. Note the presence of numerous hyphae in cortical cells (D,E). Asterisk, giant cell; arrow, nematode; arrowhead, hyphae. Scale bars: 100 μm in (A–D) and 50 μm in (E).
AM-symbiosis differentially modulated the expression level of a set of genes involved in the parasitism response (Supplementary Table S12). DEGs (P > 0.05) showing an increased transcription level included those related to cell wall (8%), root development (25%), transport (33%), secondary metabolism (13%), hormone (15%), oxidative stress (21%), and stress response (4%). Down-regulated transcripts were mostly related to cell wall (23%), secondary metabolism (13%), hormone (17%), oxidative stress (10%), and stress response (4%) (Supplementary Table S12). The effect on the plant gene expression was evaluated calculating the ratio between AM_RKN/RKN and RKN/AM_RKN fold changes. Genes with ratio > |1.5| (for both up- or down- regulated) were considered in subsequent analyses (Table 9). The expression of all members from categories “root development” (Solyc04g078470.2.1, Solyc01g107710.2.1, Solyc05g009320.2.1) and “transport” (Solyc02g071070.2.1, Solyc04g079510.1.1, and Solyc09g074230.2.1) was induced by the fungus in galls (Supplementary Table S12), and among them the root cap protein 3 (Solyc01g107710.2.1) showed the highest ratio (11.95). Members of other categories (cell wall, hormone, secondary metabolite, oxidative stress response, and stress response-related genes) showed instead an uneven behavior (Table 9). The fungus colonization reduced the expression (negative ratio) of several genes involved in cell-wall metabolism, i.e., those acting on pectin fraction (Solyc03g083730.1.1 and Solyc04g076660.2.1, Solyc09g098270.2.1), two genes coding for an endo-1,4-b-xylanase (Solyc04g077190.2.1) and an endo-1,4-mannosidase (Solyc10g074920.1.1). By contrast, it positively affected the expression of three genes for putative expansins (Solyc07g054170.2.1, Solyc01g090810.2.1, Solyc05g007830.2.1). Among hormone category, genes coding for ethylene-responsive transcription factors (Solyc07g042230.1.1, Solyc08g007820.1.1, Solyc08g007830.1.1, Solyc11g042560.1.1 and Solyc10g050970.1.1) were negatively affected, except Solyc04g012050.2.1. Interestingly, the CCD7 gene (Solyc01g090660.2.1), which is considered also an AM-symbiosis marker, was induced also in colonized galls. Among the genes involved in stress response some members of the cortical cell-delineating protein family (Solyc08g079230.1.1, Solyc08g078900.1.1 and Solyc12g014620.1.1) (Supplementary Table S12) appeared down-regulated in AM-colonized galls whereas others (Solyc03g091020.1.1, Solyc08g079200.1.1) were positively affected by the symbiosis.
Table 9. Effect of AM colonization on gene expression in galls, compared to untreated control (fold changes).
Discussion
Several studies addressed the effect of water stress on growth, yield, secondary metabolite production and gene expression in tomato (Nuruddin et al., 2003; Sánchez-Rodríguez et al., 2011; Giannakoula and Ilias, 2013; Sacco et al., 2013; Iovieno et al., 2018). Previous works also studied some specific aspects of the relationship between tomato and AM fungi under drought stress (Dell’Amico et al., 2002; Subramanian et al., 2006; Aroca et al., 2008; Wang et al., 2014; Chitarra et al., 2016; Ruiz-Lozano et al., 2016; Volpe et al., 2018). An untargeted metabolomic analysis of tomato roots colonized by three AM fungi of different genera, verifying their impact on tolerance to drought or salt stress during symbiosis, showed DEGs from several processes, by looking at genome-wide transcriptional changes (Rivero et al., 2018). Massive transcriptional changes are known to occur during AM symbiosis both in dicots and monocots, and a core of marker genes, considered the functional signature of AM symbiosis, have been identified (Guether et al., 2009; Sugimura and Saito, 2017; Fiorilli et al., 2018).
Previous experiments, using the same biological system and water stress level herein studied, showed that R. intraradices colonization did not lead to a significant difference in plant performance traits. Tomato plants inoculated with R. intraradices showed enhanced internodes/height ratios under water deficit, suggesting a positive AM influence in water stress regimes, likely due to a more compact plant architecture less subject to water dispersion (Volpe et al., 2018). However, in agroecosystems, in addition to water limitation, crops often face other concurrent abiotic and biotic stresses, e.g., insect pests and pathogens (Bai et al., 2018). In the same study, R. intraradices appeared effective in sustaining the plant response to a combined abiotic (moderate WS) and biotic stress (aphid attack), the latter in terms of attractiveness toward aphid natural enemies (Volpe et al., 2018). The impact of AM symbiosis on the transcriptomic profile of nematode infection structure (i.e., the gall), firstly confirms the regulation in AM-colonized plants of diverse metabolic pathways, such as primary and secondary metabolisms, ion transport, transcriptional regulation, including several AM symbiosis markers.
Our dataset emphasizes the putative role of specific gene families during symbiosis establishment and functioning. For example, five out of the six genes coding for putative ripening-related proteins (RRP) were strongly significantly up-regulated both in AM-colonized unstressed and water stressed plants (with FC from 4000 to 395 in AM and from 1000 to 38 in AM_WS). Although their function during symbiosis have not been demonstrated, RRP are known to be regulated in several plant/AM combinations (Fiorilli et al., 2015, 2018; Handa et al., 2015). Genes belonging to this family have been found to be strongly induced in rice large lateral roots colonized by the AM fungus (Fiorilli et al., 2015), including OsAM8, previously identified as a mycorrhiza-responsive gene (Güimil et al., 2005). Additionally, RRP have been reported among the cysteine-rich peptides highly induced genes in AM Lotus roots (Handa et al., 2015). The expression of several Medicago RRP was activated, with respect to wild type plants, in a mutant (pt4) that leads to early arbuscule degeneration (Floss et al., 2017), opening new questions on the role during symbiosis.
Other gene families showed almost all members as significantly regulated in AM-colonized roots (e.g. blue copper-binding proteins, which are considered markers for AM symbiosis and include phytocyanins that play an important role in plant development and stress resistance), suggesting a functional redundancy as already demonstrated for phosphate transport. However, the location of their regulation remains still unknown (i.e., arbuscule-containing cells vs. non-colonized ones). Looking at nutritional aspects, it is worth noting the huge number of up-regulated transporter genes, that indirectly confirm the symbiosis functionality. Interestingly, most of them were significantly up-regulated in AM_WS roots, suggesting that the symbiosis was still functional after imposition of the water stress. The lower gene expression levels in comparison to those in AM roots is in agreement with a decrease in the colonization rate previously reported during water deficit (Chitarra et al., 2016) and also confirmed in this work by a lower percentage of AM fungal reads in AM_WS roots with respect to unstressed plants (AM). However, although symbiosis seems to be affected by water limitation, the impact on root transcriptome is evident also in AM_WS roots. This result reflects the fact that a functional symbiosis seems to be still present during the progression of the water stress, at least for the plant/fungus combination considered in this study.
The 20 most up-regulated genes in AM_WS, compared with those involved in regulation of stress, indicate that AM-colonized roots may differently “sense” the water stress with respect to the uncolonized ones. The most up-regulated genes were in fact still those involved in the symbiosis, at least upon a moderate stress.
Regulation of stress marker genes (i.e., LEA, dehydrin, ABA-responsive genes, P5CS), as observed in AM_WS roots, confirmed the effect of the imposed stress.
The physiological plant response to water deficit is also regulated by the aquaporin proteins (AQPs), controlling water movement through the plant in different physiological conditions. Their regulation is considered as an adaptative mechanism to stress conditions (Kapilan et al., 2018). Molecular analyses already demonstrated a complex transcriptional and post-transcriptional regulation. Data from several studies indicate that AM symbiosis has an impact on host AQPs, altering both plant-water relationships and plant physiology, to better cope with stressful conditions such as drought. However, as reported for other functional aspects related to AM symbiosis, the regulation of AQPs seems to be dependent on the plant and fungal species involved in the symbiosis (Ruiz-Lozano and Aroca, 2017). Data from AM-colonized maize roots, exposed to several growing and water-stressed conditions, showed that the fungus may regulate a large number of AQP genes in the host plants, in several sub-families. Regulation, however, was also dependent on water status and the duration and severity of the imposed stress (Ruiz-Lozano and Aroca, 2017; Balestrini et al., 2018). This can explain the different result with respect to the data already reported by Chitarra et al. (2016) in the same plant/AM fungus combination, but upon a severe water stress level.
If relevant data are nowadays available on the plant protective effect of AM fungi under water stress, the impact of drought on the fungus itself has not been extensively investigated. Our dataset revealed that cytochrome P450 (CYPs) genes were mainly up-regulated in R. intraradices, in presence of water stress. Fungi possess many diverse CYPs, mainly involved in sterol biosynthesis, developmental processes and production of secondary metabolites such as those involved in virulence (Shin et al., 2018). The R. irregularis genome, used for the mapping of fungal genes, contains about 200 CYP-encoding genes, indicating an expansion of such a gene family in AMFs that likely deals with the peculiar AM lifestyle (Tisserant et al., 2013; Handa et al., 2015). As cytochrome P450 is involved in the synthesis of sterols for membrane biogenesis during arbuscule formation (Handa et al., 2015), it could be inferred that their over expression reflects a change in fungal development under water deficit. Other most highly up-regulated fungal genes include a “conidiation protein 6” domain (con-6). In Neurospora crassa con-6 encodes a small protein expressed during the formation of conidia (White and Yanofsky, 1993). Fungal conidiation can be induced by nutrient deprivation or mycelium desiccation, a process worth to verify for R. irregularis propagules upon drought. The sequences most DE in our database included several that were poorly characterized. This situation may reflect the lack of a functional annotation for a consistent part of the AMF genomes so far sequenced, but might also underlie unprecedented, unknown mechanisms exploited to cope with water stress. Several sequences encoding for stress response-related proteins were regulated, suggesting an impact of the water deficit on the AM fungal metabolism, but their mechanism of action could not be hypothesized, based on current annotation data. Fungal genes containing domains involved in signaling transduction, regulation at the protein level and protein turnover were significantly influenced by the WS treatment, being mainly up-regulated. Such gene families underwent a significant expansion in the R. irregularis genome, likely due to the biotrophic lifestyle of the fungus (Zuccaro et al., 2014). Our current data strengthen this idea, suggesting that BTB/POZ and Kelch domain-containing proteins might play a central role in the fungal sensing of environmental stimuli, including the perception of and the response to abiotic stresses.
Three glutathione S-transferases (GSTs) were overexpressed in R. intraradices upon water stress. GSTs are acknowledged players in the cell protection from oxidative damage, and in tomato one GST is involved in osmotic and salt stress tolerance (Xu et al., 2015). Taken together, these data suggest that the up regulation of GST-encoding genes might represent a conserved hallmark of water stress response, from plants to fungi. Additionally, this confirms previously results on the fact that specific categories of genes involved in stress response can be activated in both the symbiotic partners, such as for AQP (Chitarra et al., 2016) and mitogen-activated protein kinase (MAPK) genes (Liu Z. et al., 2015), in tomato and soybean, respectively.
Plant responses to different stresses are highly complex and dynamic and involve changes at the transcriptome, cellular, and physiological level that are related to the specific environmental stress factors encountered. Only a few data are still available on the molecular basis of AM symbiosis-induced resistance against nematodes (Hao et al., 2012; Vos et al., 2012). Data from our study allowed the identification of a core of tomato DEGs related to parasitism response, essential for a successful RKN-tomato association, that was differently modulated in galls from mycorrhizal colonized roots.
RKN induced GCs, that are characterized by cytoskeleton rearrangements, a fragmented vacuolar system, and an increased cytoplasm density with many organelles, may be 100 times as large as normal root parenchyma cells. Thus, an extensive, coordinated remodeling of the cell wall must occur to allow cell expansion (Williamson and Hussey, 1996; Gheysen and Mitchum, 2011). Our data showed different responses in genes associated to cell wall modification in AM-colonized galls.
Several transcripts coding for proteins involved in cell wall biosynthesis and modification were significantly down-regulated in AM-colonized vs. non-colonized galls, suggesting that AM colonization might induce changes in nematode feeding sites by counteracting cell expansion. Conversely, three expansin genes up-regulated during susceptible interaction were highly over expressed in colonized galls. Plant expansins are cell wall loosening proteins involved in the extension of the cell wall (Sampedro and Cosgrove, 2005) and may have a role in the establishment of RKN in tomato (Gal et al., 2006). They have been also reported to be involved in the AM fungal symbiosis development, at different stages of the interaction (Balestrini et al., 2005; Dermatsev et al., 2010). Therefore, we can hypothesize that the highest expression of these genes in R. intraradices colonized galls could be explained by the combined action of RKN infection and AM fungal colonization.
The presence of the AM fungus affected other pathways involved in oxygen species (ROS) homeostasis, and cell cycle/root development and response to stress (Table 7), which can have a role during the establishment of compatible plant-nematode interaction. Nematodes modulate the production of plant ROS that otherwise would be detrimental for their development. Plant peroxidases (POXs) have an important role in ROS homeostasis as they oxidize phenolics, lignin precursors, auxins and secondary metabolites using hydrogen peroxide (Almagro et al., 2009). In addition to the generation/scavenging of ROS, peroxidase activities have been also associated to loosening/stiffening of the cell wall (Shigeto and Tsutsumi, 2016). Due to the large set of peroxidase isoforms, it is difficult to determine their role in the different biological processes and especially in plant defense response. In the compatible tomato-M. incognita interaction genes coding for peroxidases are up- or down-regulated at the same infection time, suggesting that each individual enzyme has its own unique physiological and developmental role (Melillo et al., 2014). Likewise, in galls from AM-colonized roots some genes coding for POXs were differently regulated with respect to galls from non-colonized plants. Peroxidase genes were also regulated in AM-colonized soybean roots, in presence of infection by Fusarium virguliforme (Marquez et al., 2018). The decreased down-regulation of several genes coding for glutathione-S-transferase (GST) in galls from AM-colonized plants compared to non-colonized ones suggests a putative protective role of GST against nematode damage, as previously reported during the R. intraradices-induced biocontrol of the ectoparasitic nematode Xiphinema index in grapevine (Hao et al., 2012).
Members of heat shock protein families (Hsp100, Hsp90, Hsp40, and Hsp20) were down-regulated in RKN and RKN-AM galls, suggesting that plant machinery related to Hsp families is silenced and other pathways may be activated. Interestingly, members of the plant non-specific lipid transfer proteins (LTP), classified as pathogenesis-related proteins, were modulated in the same way in RKN and RKN-AM galls, suggesting different functions in several physiological and stress pathways (Edqvist et al., 2018). Recently, it has been reported that some LTP can induce secondary messengers involved in the induction of different signaling proteins (MAPK family, heat shock factors, etc.) (Li et al., 2014). Another class of LPT is that of the cortical cell delineating proteins, which accumulate in normal root tips and are responsible in the responses of the roots to physical impedance (Huang et al., 1998). Our results showed that they were differentially regulated in RKN and RKN_AM galls and, in particular, the significant increase of the expression of three genes in RKN_AM galls could suggest a protective role of AM in root defense. Other genes associated with stress responses are those encoding major latex protein family (MLP), active in a wide range of developmental processes and in response to biotic and abiotic stresses (Sun et al., 2010; Yang et al., 2015). In our study the Solyc04g007790.2.1 was strongly up-regulated in RKN galls, while in RKN_AM galls the level of this MLP was reduced but still up-regulated, suggesting a role of this protein in the defense response to M. incognita, and that AM could activate other defense pathways.
Galls also showed a different modulation of genes involved in root development and cell cycle, in presence of AM. Expression of cell cycle genes in the plant host appears tightly regulated, implying a strict control of the cell cycle machinery and its molecular components during the plant–nematode interaction (Vieira and de Almeida Engler, 2015, 2017). Our results suggested that in galls from AM-colonized roots a different modulation of gene related to the cell cycle, root tip and lateral root development takes place, indicating a transcriptional reprogramming of the root development, also affected by the fungus. By antagonizing the host plant immune response, RKN manipulate defense pathways in the root galls to promote their own development. Our data agree with previous studies stating that genes involved in JA and ET biosynthesis and signaling are suppressed during fully established RKN compatible interaction (Barcala et al., 2010; Nahar et al., 2011). Several genes involved in JA and ET biosynthesis were in fact less down-regulated by AM, suggesting a complex interaction mediated by the fungus.
Additionally, the carotenoid-derived phytohormones strigolactones (SLs) can enhance symbiosis between plants and AM fungi by inducing hyphal branching (Akiyama et al., 2005) and are also important in tomato defense against RKNs (Xu et al., 2019). It was in fact observed that the silencing of CCD7, coding for an enzyme involved in SLs biosynthesis, increased plant susceptibility to RKNs (Xu et al., 2019). Our data showed that CCD7 was strongly down-regulated in non-colonized galls, while it appeared to be induced in galls from AM-colonized roots, in agreement with their known role in AM-symbiosis, associated to early senescence of AM-colonized galls. In presence of the RKN, a suppression of plant defense responses during parasitism has been also observed, involving an alteration of secondary metabolism. Conversely, in galls from AM-colonized roots, transcription of 4-coumarate CoA ligase and anthocyanidin synthase, i.e., two enzymes involved in the flavonoid pathway leading to anthocyanins and condensed tannins, was activated. Flavonoids act during plant-nematode interactions as defense compounds or signals affecting nematode fitness at different life stages (Chin et al., 2018). They may also be involved in nematode feeding site development by maintaining local auxin accumulation. Therefore, the induction of these two enzymes as well as the reduction of the expression of genes belonging to dihydroflavonol-4-reductase family, up-regulated also in other plant–pathogen interactions (Liu et al., 2010), suggest that AM colonization help plants to respond to nematode challenges.
Conclusion
Our transcriptome dataset offers new information about the symbiotic-responsive genes both from tomato and the AM fungus, representing a solid basis for future investigations. Moreover, data provided new information on the water stress perception by AM-colonized roots as well as on the response to a biotic stress. Several marker genes (for both AM-colonization and stress factors) have been identified, confirming the robustness of the obtained dataset. Lastly, results on the regulation of AM fungal genes upon a moderate water stress condition have been obtained, suggesting a synergy between plant and fungus in this condition. Sequencing data and morphological observations also indicate that the mechanisms involved in the tomato responses to nematode colonization may also be mediated by the AM symbiosis.
Author Contributions
RB, LR, AC, and IP conceived and designed the experiments, and oversaw the manuscript preparation. LR, PV, and MC performed the RNA extraction. PV and MM performed the histopathology and morphological observations. IP performed the bioinformatics analyses. RB, LR, PV, EF, FDL, AdF, and IP performed the data analysis. RB, LR, PV, and AC wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was partially funded by the AQUA project (Progetto Premiale, Consiglio Nazionale delle Ricerche) and by the Eureka!Eurostars Programme, project E-7364 “Poch-art.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01807/full#supplementary-material
FIGURE S1 | Specific GO terms enriched during abiotic stress (AM_WS).
FIGURE S2 | Specific GO terms enriched during biotic stress (RKN_AM).
Footnotes
- ^ www.igatechnology.com
- ^ https://www.encodeproject.org/about/experiment-guidelines/
- ^ https://www.bioinformatics.babraham.ac.uk/projects/fastqc
- ^ http://www.clcbio.com
- ^ ftp://ftp.solgenomics.net
- ^ http://bioinfo.cau.edu.cn/agriGO
- ^ ftp://ftp.solgenomics.net/tomato_genome/annotation/ITAG2.4_release/ITAG2.4_cdna.fasta
- ^ https://genome.jgi.doe.gov
References
Akiyama, K., Matsuzaki, K., and Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. doi: 10.1038/nature03608
Almagro, L., Gómez Ros, L. V., Belchi-Navarro, S., Bru, R., Ros Barceló, A., and Pedreno, M. A. (2009). Class III peroxidases in plant defence reactions. J. Exp. Bot. 60, 377–390. doi: 10.1093/jxb/ern277
Aroca, R., Del Mar Alguacil, M., Vernieri, P., and Ruiz-Lozano, J. M. (2008). Plant responses to drought stress and exogenous ABA application are modulated differently by mycorrhization in tomato and an ABA-deficient mutant (sitiens). Microb. Ecol. 56, 704–719. doi: 10.1007/s00248-008-9390-y
Bai, Y., Sunarti, S., Kissoudis, C., Visser, R. G. F., and van der Linden, C. G. (2018). The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front. Plant Sci. 9:801. doi: 10.3389/fpls.2018.00801
Balestrini, R., and Bonfante, P. (2014). Cell wall remodeling in mycorrhizal symbiosis: a way towards biotrophism. Front. Plant Sci. 5:237. doi: 10.3389/fpls.2014.00237
Balestrini, R., Chitarra, W., Antoniou, C., Ruocco, M., and Fotopoulos, V. (2018). Improvement of plant performance under water deficit with the employment of biological and chemical priming agents. themed issue: assessment and monitoring of crop water use and productivity under present and future climate. J. Agric. Sci. 156, 680–688. doi: 10.1017/s0021859618000126
Balestrini, R., Cosgrove, D. J., and Bonfante, P. (2005). Differential location of α-expansin proteins during the accommodation of root cells to an arbuscular mycorrhizal fungus. Planta 220, 889–899. doi: 10.1007/s00425-004-1431-2
Balestrini, R., Gómez-Ariza, J., Lanfranco, L., and Bonfante, P. (2007). Laser microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells. Mol. Plant Microbe Interact. 20, 1055–1062. doi: 10.1094/mpmi-20-9-1055
Balestrini, R., Salvioli, A., Dal Molin, A., Novero, M., Gabelli, G., Paparelli, E., et al. (2017). Impact of an arbuscular mycorrhizal fungus versus a mixed microbial inoculum on the transcriptome reprogramming of grapevine roots. Mycorrhiza 27, 417–430. doi: 10.1007/s00572-016-0754-8
Barcala, M., Garcia, A., Cabrera, J., Casson, S., Lindsey, K., Favery, B., et al. (2010). Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant J. 61, 698–712. doi: 10.1111/j.1365-313X.2009.04098.x
Barghini, E., Cossu, R. M., Cavallini, A., and Giordani, T. (2015). Transcriptome analysis of response to drought in poplar interspecific hybrids. Genome Data 3, 143–145. doi: 10.1016/j.gdata.2015.01.004
Bedini, A., Mercy, L., Schneider, C., Franken, P., and Lucic-Mercy, E. (2018). Unraveling the initial plant hormone signaling, metabolic mechanisms and plant defense triggering the endomycorrhizal symbiosis behavior. Front. Plant Sci. 9:1800. doi: 10.3389/fpls.2018.01800
Berruti, A., Lumini, E., Balestrini, R., and Bianciotto, V. (2016). Arbuscular mycorrhizal fungi as natural biofertilizers: let’s benefit from Past Successes. Front. Microbiol. 6:1559. doi: 10.3389/fmicb.2015.01559
Boyer, J. S. (1982). Plant productivity and environment. Science 218, 443–448. doi: 10.1126/science.218.4571.443
Casieri, L., Ait Lahmidi, N., Doidy, J., Veneault-Fourrey, C., Migeon, A., Bonneau, L., et al. (2013). Biotrophic transportome in mut, ualistic plant-fungal interactions. Mycorrhiza 23, 597–625. doi: 10.1007/s00572-013-0496-9
Chin, S., Behm, C. A., and Mathesius, U. (2018). Functions of flavonoids in plant-nematode interactions. Plants 7:85. doi: 10.3390/plants7040085
Chitarra, W., Pagliarani, C., Maserti, B., Lumini, E., Siciliano, I., Cascone, P., et al. (2016). Insights on the impact of arbuscular mycorrhizal symbiosis on tomato tolerance to water stress. Plant Physiol. 171, 1009–1023. doi: 10.1104/pp.16.00307
Dell’Amico, J., Torrecillas, A., Rodríguez, P., Morte, A., and Sánchez-Blanco, M. J. (2002). Responses of tomato plants associated with the arbuscular mycorrhizal fungus Glomus clarum during drought and recovery. J. Agric. Sci. 138, 387–393. doi: 10.1017/s0021859602002101
Dermatsev, V., Weingarten Baror, C., Resnick, N., Gadkar, V., Wininger, S., Kolotilin, I., et al. (2010). Microarray analysis and functional tests suggest the involvement of expansins in the early stages of symbiosis of the arbuscular mycorrhizal fungus Glomus intraradices on tomato (Solanum lycopersicum). Mol. Plant Path. 11, 121–135. doi: 10.1111/j.1364-3703.2009.00581.x
Du, Z., Zhou, X., Ling, Y., Zhang, Z., and Su, Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70. doi: 10.1093/nar/gkq310
Edqvist, J., Blomqvist, C., Nieuwland, J., and Salminen, T. A. (2018). Plant lipid transfer proteins: are we finally closing in on the roles of these enigmatic proteins? J.Lipid Res. 59, 1374–1382. doi: 10.1194/jlr.R083139
Elsen, A., Gervacio, D., Swennen, R., and De Waele, D. (2008). AMF-induced biocontrol against plant parasitic nematodes in Musa sp.: a systemic effect. Mycorrhiza 18, 251–256. doi: 10.1007/s00572-008-0173-6
Fiorilli, V., Catoni, M., Miozzi, L., Novero, M., Accotto, G. P., and Lanfranco, L. (2009). Global and cell-type gene expression profiles in tomato plants colonized by an arbuscular mycorrhizal fungus. New Phytol. 184, 975–987. doi: 10.1111/j.1469-8137.2009.03031.x
Fiorilli, V., Vallino, M., Biselli, C., Faccio, A., Bagnaresi, P., and Bonfante, P. (2015). Host and non-host roots in rice: cellular and molecular approaches reveal differential responses to arbuscular mycorrhizal fungi. Front. Plant Sci. 6:636. doi: 10.3389/fpls.2015.00636
Fiorilli, V., Vannini, C., Ortolani, F., Garcia-Seco, D., Chiapello, M., Novero, M., et al. (2018). Omics approaches revealed how arbuscular mycorrhizal symbiosis enhances yield and resistance to leaf pathogen in wheat. Sci. Rep. 8:9625. doi: 10.1038/s41598-018-27622-8
Floss, D. S., Gomez, S. K., Park, H.-J., MacLean, A. M., Müller, L. M., Bhattarai, K.-K., et al. (2017). A Transcriptional program for arbuscule degeneration during AM symbiosis is regulated by MYB1. Curr. Biol. 27, 1206–1212. doi: 10.1016/j.cub.2017.03.003
Gal, T. Z., Aussenberg, E. R., Burdman, S., Kapulnik, Y., and Koltai, H. (2006). Expression of a plant expansin is involved in the establishment of root knot nematode parasitism in tomato. Planta 224, 155–162. doi: 10.1007/s00425-005-0204-x
Gamboa-Tuz, S. D., Pereira-Santana, A., Zamora-Briseño, J. A., Castano, E., Espadas-Gil, F., Ayala-Sumuano, J. T., et al. (2018). Transcriptomics and co-expression networks reveal tissue-specific responses and regulatory hubs under mild and severe drought in papaya (Carica papaya L.). Sci. Rep. 8:14539. doi: 10.1038/s41598-018-32904-2
Garcia, M. E., Lynch, T., Peeters, J., Snowden, C., and Finkelstein, R. (2008). A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 67, 643–658. doi: 10.1007/s11103-008-9344-2
Gheysen, G., and Mitchum, M. G. (2011). How nematodes manipulate plant development pathways for infection. Curr. Opin. Plant Biol. 14, 415–421. doi: 10.1016/j.pbi.2011.03.012
Giannakoula, A. E., and Ilias, I. F. (2013). The effect of water stress and salinity on growth and physiology of tomato. Arch. Biol. Sci. Belgrade 65, 611–620. doi: 10.2298/abs1302611g
Gong, P., Zhang, J., Li, H., Yang, C., Zhang, C., Zhang, X., et al. (2010). Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J. Exp. Bot. 61, 3563–3575. doi: 10.1093/jxb/erq167
Guether, M., Balestrini, R., Hannah, M. A., Udvardi, M. K., and Bonfante, P. (2009). Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol. 182, 200–212. doi: 10.1111/j.1469-8137.2008.02725.x
Güimil, S., Chang, H. S., Zhu, T., Sesma, A., Osbourn, A., Roux, C., et al. (2005). Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc. Nat. Acad. Sci. U. S. A. 102, 8066–8070. doi: 10.1073/pnas.0502999102
Gutjahr, C. (2014). Phytohormone signaling in arbuscular mycorrhiza development. Curr. Op. Plant Biol. 20, 26–34. doi: 10.1016/j.pbi.2014.04.003
Handa, Y., Nishide, H., Takeda, N., Suzuki, Y., Kawaguchi, M., and Saito, K. (2015). RNA-seq transcriptional profiling of an arbuscular mycorrhiza provides insights into regulated and coordinated gene expression in Lotus japonicus and Rhizophagus irregularis. Plant Cell Physiol. 8, 1490–1511. doi: 10.1093/pcp/pcv071
Hao, Z., Fayolle, L., van Tuinen, D., Chatagnier, O., Xiaolin, L., Gianinazzi, S., et al. (2012). Local and systemic mycorrhiza-induced protection against the ectoparasitic nematode Xiphinema index involves priming of defence gene responses in grapevine. J. Exp. Bot. 63, 3657–3672. doi: 10.1093/jxb/ers046
Hogekamp, C., Arndt, D., Pereira, P. A., Becker, J. D., Hohnjec, N., and Küster, H. (2011). Laser microdissection unravels cell-type-specific transcription in arbuscular mycorrhizal roots, including CAAT-box transcription factor gene expression correlating with fungal contact and spread. Plant Physiol. 157, 2023–2043. doi: 10.1104/pp.111.186635
Hogekamp, C., and Küster, H. (2013). A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis. BMC Genomics 14:306. doi: 10.1186/1471-2164-14-306
Huang, Y. F., Jordan, W. R., Wing, R. A., and Morgan, P. W (1998). Gene expression induced by physical impedance in maize roots. Plant Mol. Biol. 37:921.
Iberkleid, I., Sela, N., and Brown Miyara, S. (2015). Meloidogyne javanica fatty acid- and retinol-binding protein (Mj-FAR-1) regulates expression of lipid-, cell wall-, stress- and phenylpropanoid-related genes during nematode infection of tomato. BMC Genomics 16:272. doi: 10.1186/s12864-015-1426-3
Iovieno, P., Punzo, P., Guida, G., Mistretta, C., Van Oosten, M. J., Nurcato, R., et al. (2018). Transcriptomic changes drive physiological responses to progressive drought stress and rehydration in tomato. Front. Plant Sci. 7:371. doi: 10.3389/fpls.2016.00371
Jones, J. T., Haegeman, A., Danchin, E. G., Gaur, H. S., Helder, J., Jones, M. G., et al. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14, 946–961. doi: 10.1111/mpp.12057
Kakumanu, A., Ambavaram, M. M. R., Klumas, C., Krishnan, A., Batlang, U., Myers, E., et al. (2012). Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-seq. Plant Physiol. 160, 846–867. doi: 10.1104/pp.112.200444
Kal, A. J., van Zonneveld, A. J., Benes, V., van den Berg, M., Koerkamp, M. G., Albermann, K., et al. (1999). Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol. Biol. Cell 10, 1859–1872. doi: 10.1091/mbc.10.6.1859
Kapilan, R., Vaziri, M., and Zwiazek, J. J. (2018). Regulation of aquaporins in plants under stress. Biol. Res. 51:4. doi: 10.1186/s40659-018-0152-0
Kuromori, T., Seo, M., and Shinozaki, K. (2018). ABA transport and plant water stress responses. Trends Plant Sci. 23, 513–522. doi: 10.1016/j.tplants.2018.04.001
Li, J., Gao, G. Z., Xu, K., Chen, B. Y., Yan, G. X., Li, F., et al. (2014). Genome-wide survey and expression analysis of the putative non-specific lipid transfer proteins in Brassica rapa L. PLoS One 9:e84556. doi: 10.1371/journal.pone.0084556
Li, M., Wang, R., Tian, H., and Gao, Y. (2018). Transcriptome responses in wheat roots to colonization by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mycorrhiza 28, 747–759. doi: 10.1007/s00572-018-0868-2
Liu, F., Zhang, X., Lu, C., Zeng, X., Li, Y., Fu, D., et al. (2015). Non-specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. J. Exp. Bot. 66, 5663–5681. doi: 10.1093/jxb/erv313
Liu, Z., Li, Y., Ma, L., Wei, H., Zhang, J., He, X., et al. (2015). Coordinated regulation of arbuscular mycorrhizal fungi and soybean MAPK pathway genes improved mycorrhizal soybean drought tolerance. Mol. Plant Microbe Interact. 28, 408–419. doi: 10.1094/MPMI-09-14-0251-R
Liu, H., Du, Y., Chu, H., Shih, C. H., Wong, Y. W., Wang, M., et al. (2010). Molecular dissection of the pathogen-inducible 3-deoxyanthocyanidin biosynthesis pathway in sorghum. Plant Cell Physiol. 51, 1173–1185. doi: 10.1093/pcp/pcq080
Mandal, S. M., Chakraborty, D., and Dey, S. (2010). Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal. Behav. 5, 359–368. doi: 10.4161/psb.5.4.10871
Marquez, N., Giachero, L., Adrien, G., Debat, H., Declerck, S., and Ducasse, D. A. (2018). Transcriptome analysis of mycorrhizal and non-mycorrhizal soybean plantlets upon infection with Fusarium virguliforme, one causal agent of sudden death syndrome. Plant Pathol. 68, 470–480. doi: 10.1111/ppa.12964
Martinez-Medina, A., Flors, V., Heil, M., Mauch-Mani, B., Pieterse, C. M. J., Pozo, M. J., et al. (2016). Recognizing plant defense priming. Trends Plant Sci. 21, 818–822. doi: 10.1016/j.tplants.2016.07.009
McGuiness, P. N., Reid, J. B., and Foo, E. (2019). The role of gibberellins and brassinosteroids in nodulation and arbuscular mycorrhizal associations. Front. Plant Sci. 10:269. doi: 10.3389/fpls.2019.00269
Melillo, M. T., Leonetti, P., and Veronico, P. (2014). Benzothiadiazole effect in the compatible tomato-Meloidogyne incognita interaction: changes in giant cell development and priming of two root anionic peroxidases. Planta 240, 841–854. doi: 10.1007/s00425-014-2138-7
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nahar, K., Kyndt, T., De Vleesschauwer, D., Hofte, M., and Gheysen, G. (2011). The jasmonate pathway is a key player in systemically induced defence against root-knot nematodes in rice. Plant Physiol. 157, 305–316. doi: 10.1104/pp.111.177576
Nir, I., Shohat, H., Panizel, I., Olszewski, N., Aharoni, A., and Weiss, D. (2017). The tomato DELLA protein PROCERA acts in guard cells to promote stomatal closure. Plant Cell 29, 3186–3197. doi: 10.1105/tpc.17.00542
Nuruddin, M. M., Madramootoo, C. A., and Dodds, G. T. (2003). Effects of water stress at different growth stages on greenhouse tomato yield and quality. HortScience 38, 1389–1393. doi: 10.21273/hortsci.38.7.1389
Oono, Y., Yazawa, T., Kawahara, Y., Kanamori, H., Kobayashi, F., Sasaki, H., et al. (2014). Genome-wide transcriptome analysis reveals that cadmium stress signaling controls the expression of genes in drought stress signal pathways in rice. PLoS One 9:e96946. doi: 10.1371/journal.pone.0096946
Ortu, G., Balestrini, R., Pereira, P. A., Becker, J. D., Küster, H., and Bonfante, P. (2012). Plant genes related to gibberellin biosynthesis and signaling are differentially regulated during the early stages of AM fungal interactions. Mol. Plant 5, 951–954. doi: 10.1093/mp/sss027
Portillo, M., Cabrera, J., Lindsey, K., Topping, J., Andrés, M. F., Emiliozzi, M., et al. (2013). Distinct and conserved transcriptomic changes during nematode-induced giant cell development in tomato compared with Arabidopsis: a functional role for gene repression. New Phytol. 197, 1276–1290. doi: 10.1111/nph.12121
Prágai, Z., and Harwood, C. R. (2002). Regulatory interactions between the Pho and σB-dependent general stress regulons of Bacillus subtilis. Microbiology 148, 1593–1602. doi: 10.1099/00221287-148-5-1593
Recchia, G. H., Konzen, E. R., Cassieri, F., Caldas, D. G. G., and Tsai, S. M. (2018). Arbuscular mycorrhizal symbiosis leads to differential regulation of drought-responsive genes in tissue-specific root cells of common bean. Front. Microbiol. 9:1339. doi: 10.3389/fmicb.2018.01339
Rivero, J., Álvarez, D., Flors, V., Azcón-Aguilar, C., and Pozo, M. J. (2018). Root metabolic plasticity underlies functional diversity in mycorrhiza-enhanced stress tolerance in tomato. New Phytol. 220, 1322–1336. doi: 10.1111/nph.15295
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Robinson, M. D., and Smyth, G. K. (2008). Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9, 321–332. doi: 10.1093/biostatistics/kxm030
Ruiz-Lozano, J. M., and Aroca, R. (2017). “Plant aquaporins and mycorrhizae: their regulation and involvement in plant physiology and performance,” in Plant Aquaporins. Signaling and Communication in Plants, eds F. Chaumont and S. Tyerman (Cham: Springer).
Ruiz-Lozano, J. M., Aroca, R., Zamarreño, A. M., Molina, S., Andreo-Jiménez, B., Porcel, R., et al. (2016). Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Environ. 39, 441–452. doi: 10.1111/pce.12631
Ruzicka, D., Chamala, S., Barrios-Masias, F. H., Martin, F., Smith, S., Jackson, L. E., et al. (2013). Inside arbuscular mycorrhizal roots - molecular probes to understand the symbiosis. Plant Genome 6, 1–13.
Sacco, A., Greco, B., Di Matteo, A., De Stefano, R., and Barone, A. (2013). Evaluation of tomato genetic resources for response to water deficit. Am. J. Plant Sci. 4, 131–145. doi: 10.4236/ajps.2013.412a3016
Sánchez-Rodríguez, E., Moreno, D. A., Ferreres, F., del Mar Rubio-Wilhelmi, M., and Ruiz, J. M. (2011). Differential responses of five cherry tomato varieties to water stress: changes on phenolic metabolites and related enzymes. Phytochemistry 72, 723–729. doi: 10.1016/j.phytochem.2011.02.011
Schouteden, N., Waele, D. D., Panis, B., and Vos, C. M. (2015). Arbuscular mycorrhizal fungi for the biocontrol of plant-parasitic nematodes: a review of the mechanisms involved. Front. Microbiol. 6:1280. doi: 10.3389/fmicb.2015.01280
Selosse, M.-A., Bessis, A., and Pozo, M. J. (2014). Microbial priming of plant and animal immunity: symbionts as developmental signals. Trends Microbiol. 22, 607–613. doi: 10.1016/j.tim.2014.07.003
Sharma, I. P., and Sharma, A. K. (2017). Co-inoculation of tomato with an arbuscular mycorrhizal fungus improves plant immunity and reduces root-knot nematode infection. Rhizosphere 4, 25–28. doi: 10.1016/j.rhisph.2017.05.008
Shigeto, J., and Tsutsumi, Y. (2016). Diverse functions and reactions of class III peroxidases. New Phytol. 209, 1395–1402. doi: 10.1111/nph.13738
Shin, J., Kim, J.-E., Lee, Y.-W., and Son, H. (2018). Fungal cytochrome P450s and the P450 complement (CYPome) of Fusarium graminearum. Toxins 10:112. doi: 10.3390/toxins10030112
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Shukla, N., Yadav, R., Kaur, P., Rasmussen, S., Goel, S., Agarwal, M., et al. (2018). Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 19, 615–633. doi: 10.1111/mpp.12547
Subramanian, K. S., Santhanakrishnan, P., and Balasubramanian, P. (2006). Responses of field grown tomato plants to arbuscular mycorrhizal fungal colonization under varying intensities of drought stress. Sci. Hortic. 107, 245–253. doi: 10.1016/j.scienta.2005.07.006
Sugimura, Y., and Saito, K. (2017). Comparative transcriptome analysis between Solanum lycopersicum L. and Lotus japonicus L. during arbuscular mycorrhizal development. Soil Sci. Plant Nutr. 63, 127–136. doi: 10.1080/00380768.2017.1280378
Sun, H., Kim, M., Pulla, R. K., Kim, Y., and Yang, D. (2010). Isolation and expression analysis of a novel major latex-like protein (MLP151) gene from panax ginseng. Mol. Boil Rep. 37, 2215–2222. doi: 10.1007/s11033-009-9707-z
Tisserant, E., Malbreil, M., Kuo, A., Kohler, A., Symeonidi, A., Balestrini, R., et al. (2013). Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Ac. Sci. U. S. A. 110, 20117–20122. doi: 10.1073/pnas.1313452110
Vangelisti, A., Natali, L., Bernardi, R., Sbrana, C., Turrini, A., Hassani-Pak, K., et al. (2018). Transcriptome changes induced by arbuscular mycorrhizal fungi in sunflower (Helianthus annuus L.) roots. Sci. Rep. 8:4. doi: 10.1038/s41598-017-18445-0
Vieira, P., and de Almeida Engler, J. (2015). The plant cell inhibitor KRP6 is involved in multinucleation and cytokinesis disruption in giant-feeding cells induced by root-knot nematodes. Plant Signal. Behav. 10:e1010924. doi: 10.1080/15592324.2015.1010924
Vieira, P., and de Almeida Engler, J. (2017). Plant cyclin-dependent kinase inhibitors of the KRP family: potent inhibitors of root-knot nematode feeding sites in plant roots. Front. Plant Sci. 8:1514. doi: 10.3389/fpls.2017.01514
Volpe, V., Chitarra, W., Cascone, P., Volpe, M. G., Bartolini, P., Moneti, G., et al. (2018). The association with two different arbuscular mycorrhizal fungi differently affects water stress tolerance in tomato. Front. Plant Sci. 9:1480. doi: 10.3389/fpls.2018.01480
Vos, C., Tesfahun, A. N., Panis, B., De Waele, D., and Elsen, A. (2012). Arbuscular mycorrhizal fungi induce systemic resistance in tomato against the sedentary nematode Meloidogyne incognita and the migratory nematode Pratylenchus penetrans. Appl. Soil Ecol. 61, 1–6. doi: 10.1016/j.apsoil.2012.04.007
Wang, B., Yao, Z., Zhao, S., Guo, K., Sun, J., and Zhang, H. (2014). Arbuscular mycorrhizal fungal application to improve growth and tolerance of processing tomato (Lycopersicon esculentum Miller) under drought stress. J. Food Agric. Environ. 12, 452–457.
White, B. T., and Yanofsky, C. (1993). Structural characterization and expression analysis of the Neurospora conidiation egne con-6. Develop. Biol. 160, 254–264. doi: 10.1006/dbio.1993.1303
Williamson, V. M., and Hussey, R. S. (1996). Nematode pathogenesis and resistance in plants. Plant Cell 8, 1735–1745. doi: 10.1105/tpc.8.10.1735
Xu, J., Xing, X.-J., Tian, Y.-S., Peng, R.-H., Xue, Y., Zhao, W., et al. (2015). Transgenic Arabidopsis plants expressing tomato glutathione S-transferase showed enhanced resistance to salt and drought stress. PLoS One 10:e0136960. doi: 10.1371/journal.pone.0136960
Xu, X., Fang, P., Zhang, H., Chi, C., Song, L., Xia, X., et al. (2019). Strigolactones positively regulate defense against root-knot nematodes in tomato. J. Exp. Bot. 70, 1325–1337. doi: 10.1093/jxb/ery439
Yang, C., Liang, S., Wang, H., Han, L., Wang, F., Cheng, H., et al. (2015). Cotton Major Latex Protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dalhliae. Mol. Plant 8, 399–411. doi: 10.1016/j.molp.2014.11.023
Zouari, I., Salvioli, A., Chialva, M., Novero, M., Miozzi, L., Tenore, G. C., et al. (2014). From root to fruit: RNA-Seq analysis shows that arbuscular mycorrhizal symbiosis may affect tomato fruit metabolism. BMC Genomics 15:221. doi: 10.1186/1471-2164-15-221
Keywords: abiotic stress, AM symbiosis, RKN, transcriptomics, stress response
Citation: Balestrini R, Rosso LC, Veronico P, Melillo MT, De Luca F, Fanelli E, Colagiero M, Salvioli di Fossalunga A, Ciancio A and Pentimone I (2019) Transcriptomic Responses to Water Deficit and Nematode Infection in Mycorrhizal Tomato Roots. Front. Microbiol. 10:1807. doi: 10.3389/fmicb.2019.01807
Received: 24 May 2019; Accepted: 22 July 2019;
Published: 13 August 2019.
Edited by:
Carolina Escobar, University of Castilla–La Mancha, SpainReviewed by:
Tina Kyndt, Ghent University, BelgiumJuan Manuel Ruiz-Lozano, Spanish National Research Council (CSIC), Spain
Copyright © 2019 Balestrini, Rosso, Veronico, Melillo, De Luca, Fanelli, Colagiero, di Fossalunga, Ciancio and Pentimone. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raffaella Balestrini, cmFmZmFlbGxhLmJhbGVzdHJpbmlAaXBzcC5jbnIuaXQ=
†These authors have contributed equally to this work
 Raffaella Balestrini
Raffaella Balestrini Laura C. Rosso
Laura C. Rosso Pasqua Veronico
Pasqua Veronico Maria Teresa Melillo
Maria Teresa Melillo Francesca De Luca
Francesca De Luca Elena Fanelli1
Elena Fanelli1 Mariantonietta Colagiero
Mariantonietta Colagiero Alessandra Salvioli di Fossalunga
Alessandra Salvioli di Fossalunga Aurelio Ciancio
Aurelio Ciancio Isabella Pentimone
Isabella Pentimone