- 1Beijing Advanced Innovation Center for Food Nutrition and Human Health, Beijing Engineering and Technology Research Center of Food Additives, Beijing Technology and Business University, Beijing, China
- 2Beijing Academy of Agricultural and Forestry Sciences, Beijing, China
Bacteriocins are ribosomally synthesized antibacterial peptides or proteins from microorganisms. We report a novel bacteriocin producing strain, Enterococcus faecalis Gr17, that was isolated from the Chinese traditional low-salt fermented whole fish product Suan yu. E. faecalis Gr17 displayed potent antibacterial activity against foodborne pathogenic and spoilage bacteria. The complete genome of E. faecalis Gr17 contained one circular chromosome and plasmid. The gene cluster of a novel bacteriocin designated enterocin Gr17 was identified. The enterocin Gr17 structural gene encodes a precursor of the bacteriocin. Two other transporter genes and an immunity gene within two divergent operons were identified as being associated with enterocin Gr17 secretion and protection. The novel enterocin Gr17 was purified by ammonium sulfate precipitation, cation exchange, gel filtration, and reverse-phase high-performance liquid chromatography. The molecular weight of enterocin Gr17 was 4,531.01 Da as determined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and its mature amino acid sequence of enterocin Gr17 was RSYGNGVYCNNSKCWVNWGEAKENIIGIVISGWATGLAGMGR. Sequence alignment revealed that enterocin Gr17 is a class IIa bacteriocin with similarities to enterocin P. The merits of bactericidal activity, sensitivity to enzymes, and pronounced stability to chemicals, temperature (60°C, 30 min and 121°C, 15 min), and pH (2–10) indicated practicality and safety of enterocin Gr17 in the food industry. The complete genome information of E. faecalis Gr17 will improve the understanding of the biosynthetic mechanism of enterocin Gr17, which has potential value as a food biopreservative.
Introduction
Bacteriocins are ribosomally synthesized proteins and protein complexes with a broad spectrum of antibacterial activities against many food-borne pathogens and closely related species (Cleveland et al., 2001). They do not affect cells that produce immune-related proteins (Diep et al., 2007). Bacteriocins from lactic acid bacteria (LAB) are non-toxic, highly potent, and safe, and so have been widely used as preservatives for foods that include vegetables, meats, and other food products (Gálvez et al., 2007; Yang et al., 2014). Bacteriocins may be developed as viable alternatives to antibiotics (Cotter et al., 2013).
Enterococcus is a genus of LAB. The bacteria are Gram-positive, catalase-negative, facultative anaerobic, and non-spore forming (Moraes et al., 2013). Enterococci, the first LAB colonizing the infant gastrointestinal tract (GIT) (Fanaro et al., 2010), are also ubiquitous in fermented foods and the environment (Franz et al., 2003; Foulquie Moreno et al., 2006). Furthermore, some Enterococcus spp. are commercially available and prevent numerous diseases. As examples, Enterococcus faecium SF68® has been used as a food biopreservatives and in the treatment of diarrhea (Kathrani et al., 2016; Holzapfel et al., 2018), Enterococcus faecalis Symbioflor 1 is efficacious for the treatment of sinusitis and bronchitis (Habermann et al., 2002), and E. faecium JWS 833 enhances cytokine production by dendritic cells (Choi et al., 2012). Thus, enterococci are important for the health of humans and animals, as well as in the food industry and the environment.
Lactic acid bacteria produce antimicrobial substances that include organic acids, hydrogen peroxide, and bacteriocins. Bacteriocins have potent inhibitory activity against sensitive strains of bacteria. There are four classes (I–IV) of bacteriocins (Rea et al., 2011; Kumariya et al., 2019). Class I bacteriocins are <5 kDa. They are posttranslationally modified peptides, which contain non-standard amino acids, such as lanthionine and β-methyllanthionine. The class I bacteriocins comprise three subgroups: class Ia (lantibiotics), class Ib (labyrinthopeptins), and class Ic (sanctibiotics) (Parada et al., 2007). Class II bacteriocins (5–10 kDa) are heat-stable unmodified peptides, which comprise four subgroups: class IIa (pediocin-like bacteriocins), class IIb (two-peptide bacteriocins), class IIc (circular bacteriocins), and class IId (linear and non-pediocin-like bacteriocins) (Cui et al., 2012). Class III bacteriocins (>30 kDa) are heat-labile proteins, which include colicins, helveticin M, helveticin J, and enterolysin A (Kumariya et al., 2019). Class IV bacteriocins are large complexes with lipid or carbohydrate moieties. They are now termed bacteriolysins (Liu et al., 2014). Generally, class IIa bacteriocins consist of a conserved YGNGV motif and disulfide bond linkage (Perez et al., 2014).
Enterocin is a bacteriocin obtained from the Enterococcus species. Numerous enterocin-producing enterococci and enterocins have been reported. They include enterocin A from E. faecium CTC492 (Aymerich et al., 1996), enterocin B from E. faecium T136 (Casaus et al., 1997), enterocin P from E. faecium P13 (Cintas et al., 1997), and enterocin Q from E. faecium L50 (Cintas et al., 2000). Class IIa enterocins are synthesized as a precursor with an N-terminal signal peptide, which is cleaved by adenosine triphosphate (ATP)-binding cassette (ABC) transporter (Havarstein et al., 1995) or the Sec secretion system (Cintas et al., 1997). The type of N-terminal signal peptide determines the synthetic mechanism of enterocin. Although bacteriocins and producer cells play an essential role in the food industry, few studies have examined the biosynthetic mechanism and practical application of enterocins.
Virulence factors and antibiotic resistance are important in the pathogenicity of E. faecalis. Many virulence genes have been reported in enterococci, including cytolysins (cylA, cylB, and cylM), gelatinase (gelE), sex pheromones (cpd, cob, and ccf), aggregation substance gene (agg), and extracellular surface protein gene (esp, efaAfs, and efaAfm) (Barbosa et al., 2010; Chajęcka-Wierzchowska et al., 2016). Moreover, several antibiotic resistance genes have been described in enterococci. These include genes conferring resistance to erythromycin (ermB, ermC), tetracycline (tetM, tetS, tetO, tetK, and tetL), ciprofloxacin (gyrA and parC), ampicillin (bla), and vancomycin (vanA, vanB, and vanC) (Ohmomo et al., 2000; Gevers et al., 2003; Comunian et al., 2010; Guo et al., 2017).
In the present study, the complete genome sequence of E. faecalis Gr17, a novel strain isolated from a Chinese traditional low-salt fermented whole-fish product, was determined. The biosynthetic mechanism of enterocin Gr17, a novel bacteriocin from E. faecalis Gr17, was analyzed by bioinformatic analyses. Furthermore, the physicochemical characterization and antibacterial activity of purified enterocin Gr17 were determined. The genome information of E. faecalis Gr17 and the antibacterial properties of enterocin Gr17 provide the theoretical foundation for the potential use of the bacteriocin as a food preservative.
Materials and Methods
Samples and Bacterial Culture Conditions
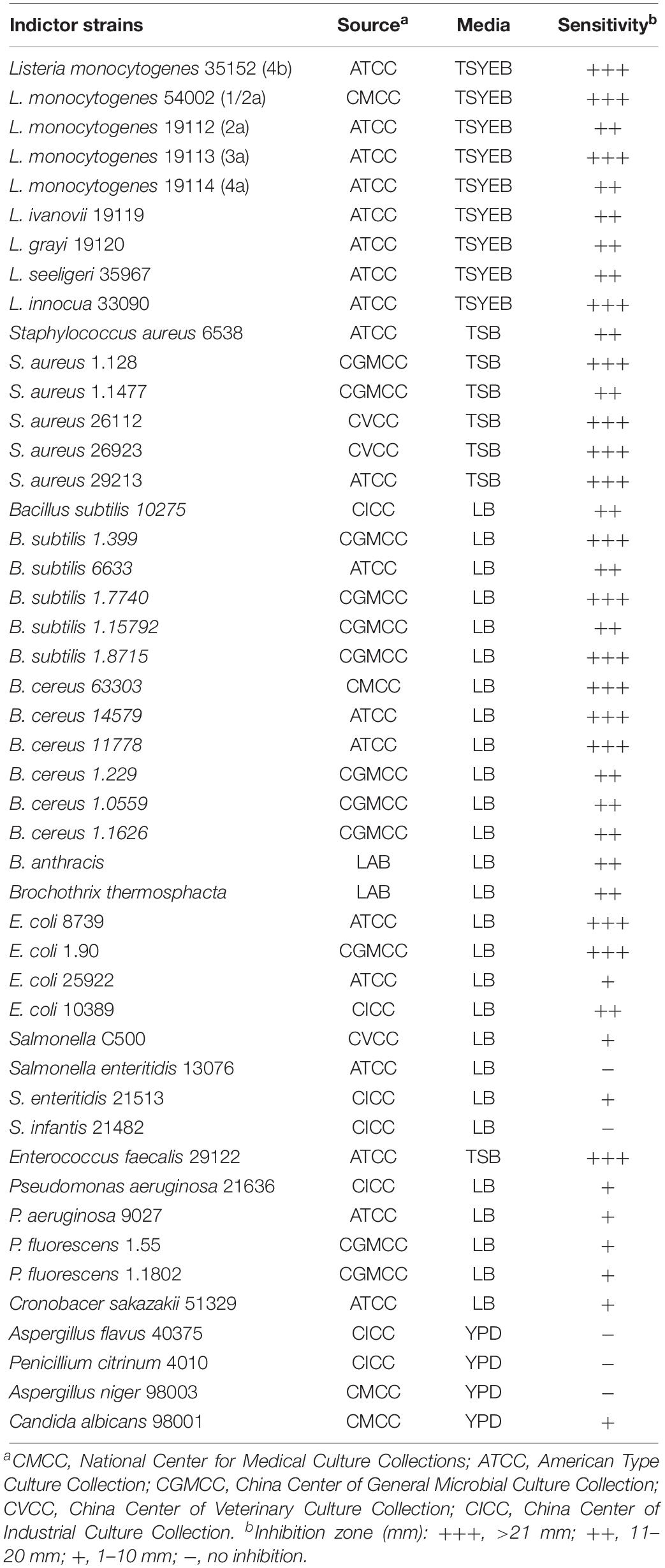
Samples of the Chinese traditional low-salt fermented whole-fish product known as Suan yu were collected from the Dong ethnic minority regions in Liping, Guizhou Province, China. All LAB strains were cultured in MRS medium without agitation at 37°C. The details of each medium used for the indicator strain are provided in Table 1. All bacteria were grown at 37°C and stored at −80°C in culture broth containing 20% glycerol (v/v).
Isolation of Bacteriocin-Producing LAB
Liquid fermented fish samples (three cans) were mixed with 80 mL of sterile 0.9% NaCl. Serial dilutions were made using sterile 0.9% NaCl, and 1 mL of each dilution was spread on MRS agar. The plates were cultured at 37°C for 24 h. The 589 bacterial colonies that developed were each cultured in 2 mL of MRS broth for 24 h at 37°C. Each culture was centrifuged at 8,000 × g for 20 min at 4°C. The supernatant was recovered from each culture, the pH was adjusted to 7.0, and it was filtered through a 0.22-μm filter. The antibacterial activity in the filtered supernatant was determined by the well diffusion method (Voulgari et al., 2010). The 22 selected strains that displayed more pronounced antibacterial activity against the indicator strains (Listeria monocytogenes and Escherichia coli) were further tested with other indicator strains (Staphylococcus aureus, Bacillus subtilis, and Bacillus cereus). E. faecalis Gr17 displayed pronounced antibacterial activity and was selected for the subsequent experiments.
DNA Purification and Identification of Bacteriocin From E. faecalis Gr17
Strain Gr17 was cultured in MRS medium at 37°C without agitation. Genomic DNA was purified using the QIAamp DNA Mini Kit (Qiagen, Germany) according to instructions provided by the manufacturer. The concentration and purity of genomic DNA were assessed using a NanoDrop 2500 spectrophotometer (Thermo Fisher Scientific, MA, United States). Genotypic identification was performed according to the 16S rRNA gene sequence. The extracted genomic DNA was used as the PCR template. The primers were as follows: 16S rRNA-forward, 5′-AGAGTTTGATCCTGGCTCAG-3′; 16S rRNA-reverse: 5′-GGTTACCTTGTTACGACTT-3′. The 16S rRNA amplified as previously described (Rushdy and Gomaa, 2013) was sequenced by Sangon Biotech (Shanghai, China), after which the sequence was used for a BLAST search of the GenBank database.
Genome Sequencing and Assembly
The complete genome of strain Gr17 was prepared using the PacBio platform. According to the protocol, a 20-kb DNA library was constructed and sequenced with Single Molecule, Real-Time (SMRT) technology. The sequences of SMRTCell were assembled by SMRT Pipe version 2.1.1. The reads were de novo assembled and polished using the Hierarchical Genome Assembly Process version 3/Quiver.
Genome Annotation
Gene prediction and annotation were carried out using the Glimmer 3.02 software1 and the Rapid Annotation Search Tool (RAST) (Aziz et al., 2008). Additional gene and function protein identification was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al., 2016) and Clusters of Orthologous Groups (COG) of proteins database (Tatusov et al., 2003).
Purification of Enterocin
Strain Gr17 was grown in 100 mL of MRS broth to an optical density at 600 nm (OD600) of 0.4. A defined portion of the culture (0.5% v/v) was used to inoculate 2 L of MRS broth, which was incubated without agitation for 24 h at 37°C. The bacteria were removed by centrifugation at 8,000 × g for 20 min at 4°C. The supernatant was precipitated using 70% ammonium sulfate at 4°C and desalted by dialysis in sodium phosphate buffer (pH 6.5) with a cellulose semipermeable membrane (molecular weight cutoff, 1,000). The antibacterial activity of crude extracts was assayed, and the antibacterial samples were stored at −80°C.
The active extracts were further purified using the ÄKTATM pure system (GE, MA, United States). A SP-sepharose fast flow cation exchange column (16 × 25 mm) was equilibrated with 20 mM sodium phosphate buffer (pH 5.5), and the samples that had been filtered through a 0.22-μm filter were loaded onto the column and eluted with linear gradient from 0 to 1 M NaCl at a flow rate of 1 mL/min. The fractions were collected according to ultraviolet (UV) absorbance, and the antibacterial activity of collected fractions was assayed.
A Sephadex G25 column (26 × 100 mm) was equilibrated with 20 mM phosphate buffer (pH 5.5) and 2 mL of bacteriocin obtained from the cation exchange column was eluted by elution buffer (20 mM phosphate buffer) at a flow rate of 0.5 mL/min. The antibacterial activity of the collected fractions according to UV absorbance was assayed.
A C18 reverse-phase column (4.6 × 250 mm, 5 μm; Agilent, CA, United States) equipped with a reversed-phase high-performance liquid chromatography (RP-HPLC) system (Agilent) was used for further purification of bacteriocin. A linear gradient elution with 95% water–acetonitrile (5–95%) containing 0.1% trifluoroacetic acid (TFA) was used as the elution phase. The flow rate was 0.5 mL/min, and absorbance was monitored at 280 nm. The purified bacteriocin, which was designated enterocin Gr17, was collected and used to assess its antibacterial activity.
The antibacterial activity was assayed by the agar well diffusion method. The purity was assayed by tricine–sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE). The concentration was determined using a Bicinchoninic Acid Kit (Thermo Fisher Scientific, MA, United States) according to the manufacturer’s instructions.
Molecular Mass of Enterocin Gr17
The molecular mass of purified enterocin Gr17 was determined by AB SCIEX 4700 matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI–TOF–MS) in linear mode (Applied Biosystems, CA, United States). Enterocin Gr17 was spotted on a target plate and left to dry. The dried enterocin was mixed with 0.5 μL of matrix solution containing α-cyano-4-hydroxycinnamic acid dissolved in 0.1% TFA (v/v) and 50% acetonitrile (v/v). Spectrometry was performed in positive ion mode.
Antibacterial Spectrum of Enterocin Gr17
The purified enterocin Gr17 obtained by RP-HPLC was used to determine its antibacterial spectrum against indicator strains containing food spoilage bacteria and food-borne pathogens.
Sensitivity to Heat, pH, Surfactants, and Proteolytic Enzymes
The purified enterocin Gr17 obtained by RP-HPLC was assessed. To determine the effect of temperature on antibacterial activity, enterocin Gr17 was incubated at 60, 80, and 100°C for 30 min, and at 121°C for 15 min. The residual antibacterial activity was tested, and the sample at 37°C was used as the control.
The pH stability of enterocin Gr17 was estimated by adjusting the pH between 2 and 11 with 1 M NaOH or 1 M HCl. After incubation for 3 h at 37°C, the pH was neutralized to pH 6.5 and the residual antibacterial activity was tested.
The effect of ethylenediaminetetraacetic acid (EDTA), SDS, Tween 20, Tween 80, and urea (1%, v/v, final concentration) on enterocin Gr17 was assessed. After incubation for 3 h at 37°C, the residual antibacterial activity was tested.
The sensitivity of enterocin Gr17 to various proteolytic enzymes was determined by mixing 80 μL of enterocin Gr17 with 20 μL of enzymes (1 mg/mL, Sigma–Aldrich, MO, United States) including pepsin (pH 3.0), papain (pH 6.5), proteinase K (pH 7.5), trypsin (pH 7.6), and chymotrypsin (pH 7.8) for 3 h at 37°C. The control lacked enzyme.
Statistical Analyses
All experiments were performed in triplicate. Results were analyzed by analysis of variance (ANOVA) and Duncan’s test with the SPSS 23.0 software (SPSS, IL, United States). All results are presented as mean ± standard deviations (SDs). A P-value < 0.05 was considered statistically significant.
Results
Isolation of Bacteriocin-Producing Strains
A total of 589 single bacterial colonies were isolated from Suan yu. The cell-free supernatant of 22 strains (pH 7.0) showed higher antibacterial activity against indicator strains (L. monocytogenes and E. coli). Strain Gr17 possessed antibacterial activity against S. aureus, B. subtilis, and B. cereus.
Identification and Genome Features of Strain Gr17
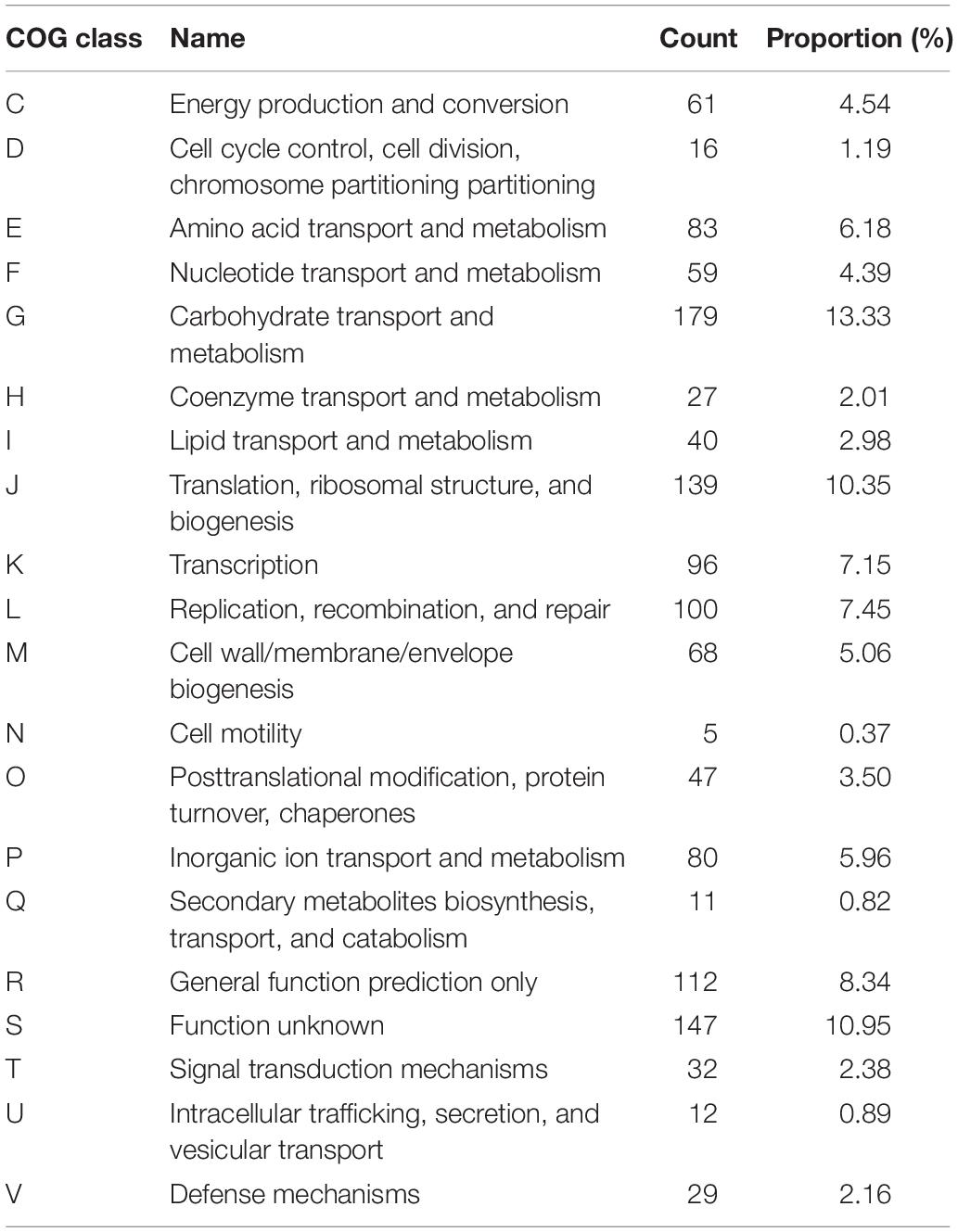
The complete genome and 16S rRNA analysis information identified strain Gr17 as E. faecalis. The strain was designated E. faecalis Gr17. The complete genome of E. faecalis Gr17 consists of a 2,588,149-base pair (bp) circular chromosome and a 49,643-bp circular plasmid designated as pGR-1, with a GC content of 38.47 and 31.51 mol%, respectively. The coding genes, ribosomal RNAs, and transfer RNAs of the chromosome and plasmid pGR17 are listed in Table 2. Additional gene and function protein identification was performed using the KEGG database (Supplementary Table S1) and RAST (Supplementary Table S2). Based on the COG database of proteins (Tatusov et al., 2003), the proteins were divided into functional categories (Table 3). Specially, the immune-related proteins unique for bacteriocin-producing strains belonged to the defense mechanism category. The information of complete genome and COG is listed in Figure 1 and has been deposited at GenBank under the accession numbers CP033376 and CP033377.
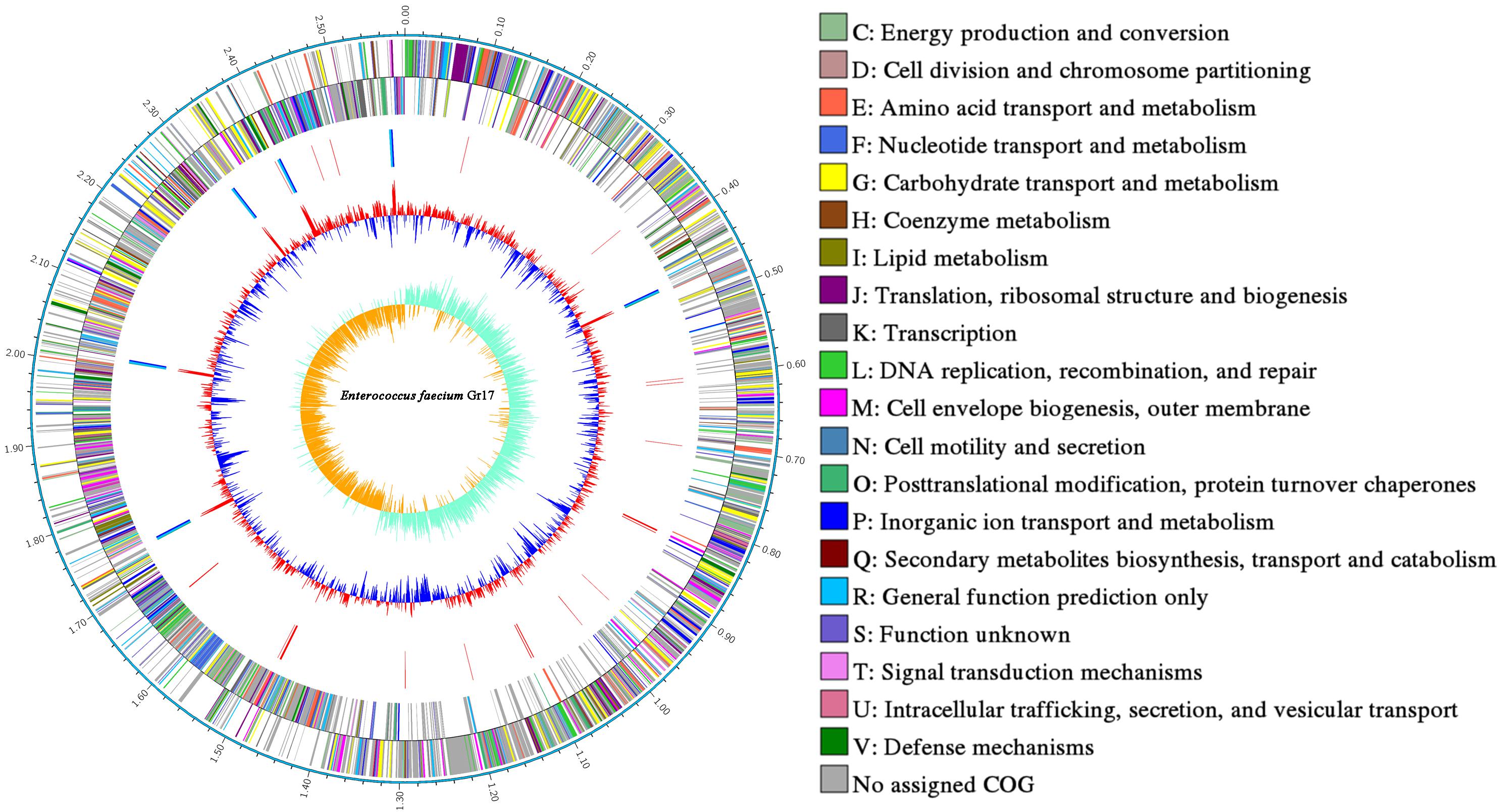
Figure 1. Circular genome map of Enterococcus faecalis Gr17. Ring 1: genome sequences. Rings 2 and 3: COG annotated coding sequences. Ring 4: KEGG enzymes. Ring 5: RNA genes. Ring 6: GC content. Ring 7: GC skew. Very short features were enlarged to enhance visibility. Clustered genes, such as several rRNA genes, may appear as one line due to space limitations. The image was created by using Circos software.
Gene Cluster of Enterocin Gr17
The enterocin Gr17 gene was located on the plasmid. The conserved N-terminal YGNGV motif of mature class IIa bacteriocin was identified in enterocin Gr17 (Figure 2A). Thus, enterocin Gr17 was identified as a class IIa bacteriocin. Also, the type of N-terminal signal peptide of the enterocin Gr17 precursor revealed that the biosynthesis and secretion process of enterocin Gr17 were via the Sec-dependent secretion system. Generally, the biosynthetic gene cluster of enterocin Gr17 contained a structural gene, transporter genes, and immunity gene within three divergent operons (Beckwith, 2013). As shown in Figure 2B, the structural gene was composed of a precursor gene (EA467_13550), whose product could be cleaved to form an N-terminal leader peptide and mature antibacterial bacteriocin (Jack et al., 1995). The transporter genes encoded SecA protein (EA467_12775), an ATPase interacting with SecB protein and SecYEG complex; SecB protein (EA467_00140), which is responsible for recognition of nascent enterocin Gr17; and SecYEG complex (EA467_09045, EA467_06915, and EA467_12635), which facilitates the removal of signal peptide and secretion of mature enterocin Gr17. The immunity gene (EA467_13555) product could protect producer cells from mature enterocin Gr17. The comparison data with other closely related enterocin gene clusters are shown in Figure 2C. The structural gene displayed 97.50, 92.50, and 95.85% homology to enterocin P from E. faecium CWBI_B1430 and E. faecium CRL1385, respectively, and the immunity gene showed 100, 95.52, and 93.29% homology to the immunity genes of enterocin P from E. faecium CWBI_B1430 and E. faecium CRL1385, respectively.
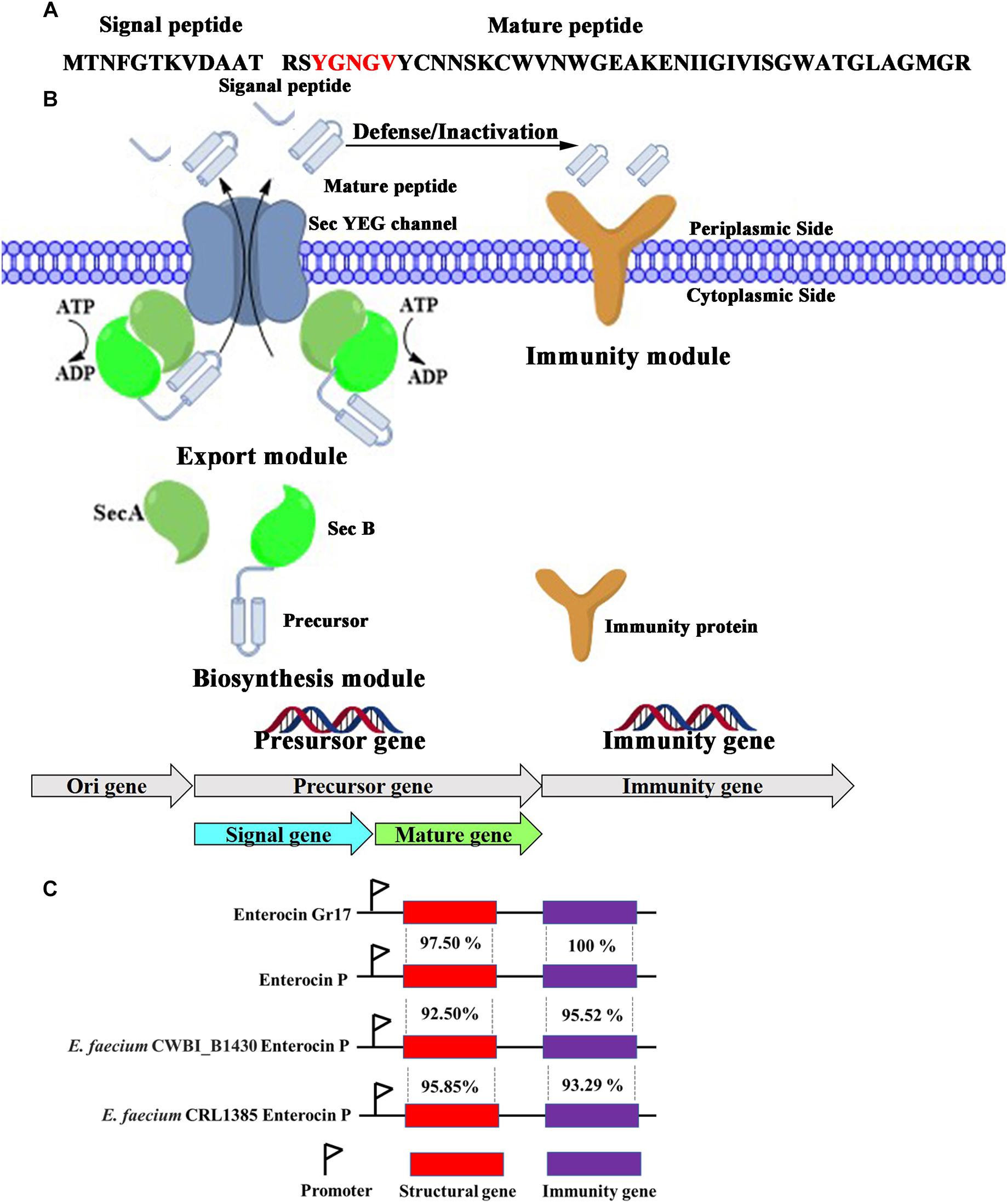
Figure 2. The amino acid sequences of precursor peptides encoded by structural gene (A), biosynthetic mechanism of enterocin Gr17 in E. faecalis Gr17 (B), and the comparison with other closely related enterocin gene clusters (C).
Virulence Factors and Antibiotic Resistance
The genes related to virulence factors, which included those encoding cytolysins (cylA, cylB, and cylM), gelatinase (gelE), sex pheromones (cpd, cob, and ccf), aggregation substance (agg), and extracellular surface proteins (esp, efaAfs, and efaAfm), were not found in the complete genome sequence of E. faecalis Gr17. Also, E. faecalis Gr17 did not contain antibiotic resistance genes encoding resistance to erythromycin (ermB and ermC), tetracycline (tetM, tetS, tetO, tetK, and tetL), ampicillin (bla), and vancomycin (vanA, vanB, and vanC). However, genes encoding resistance to ciprofloxacin (gyrA and parC) were located at EA467_08690 and EA467_03720.
Purification of Enterocin Gr17
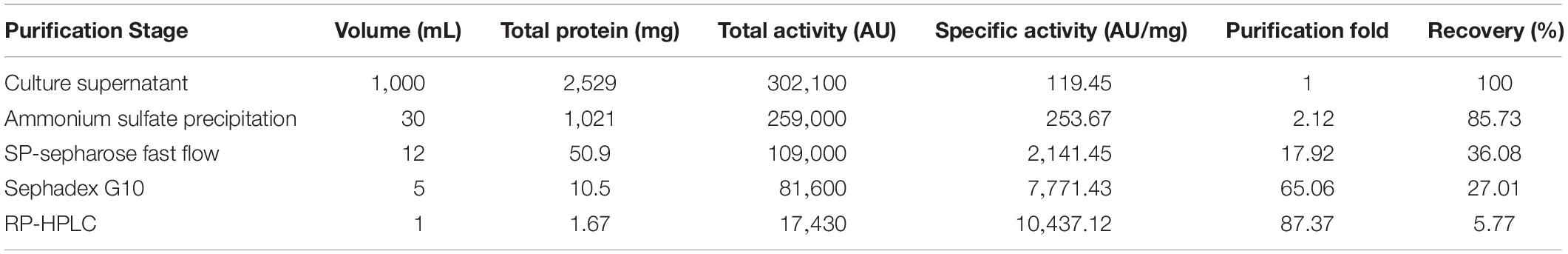
Crude enterocin Gr17 was extracted from fermentation supernatant by ammonium sulfate precipitation. Approximately 2.12-fold purification and 85.73% recovery were achieved (Table 4). SP-sepharose fast flow cation exchange column purification (Figure 3A) detected the active fraction at approximately 40 min elution time. The protein was purified 17.92-fold with 36.08% recovery. Sephadex G10 gel filtration chromatography purification yielded three different peptide fractions, with the active fraction eluted at approximately 30 min (Figure 3B). This process increased the antibacterial activity 65.6-fold, and 27.01% of the initial activity was recovered. RP-HPLC increased antibacterial activity 87.37-fold and the recovery was 5.77% (Figure 3C).
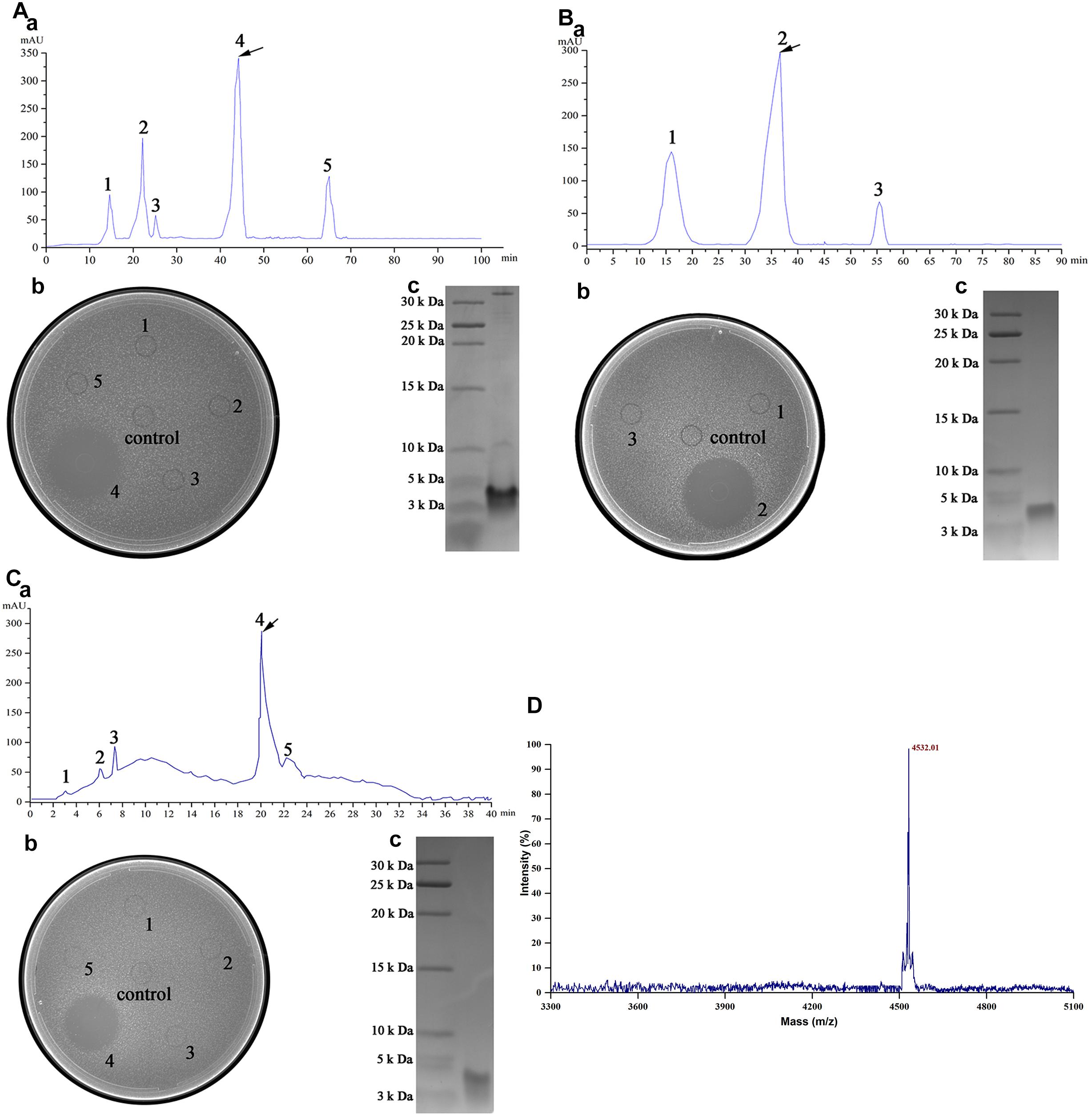
Figure 3. Purification of enterocin Gr17 from E. faecalis Gr17 by column chromatography. (A) Cation exchange column. (B) Gel filtration chromatography. (C) RP-HPLC. (D) Mass spectrum of enterocin Gr17 by MALDI–TOF–MS. a, process of purification; b, antibacterial activity of peaks against indicator strain by agar well diffusion assay; c, tricine–SDS–PAGE of activity fraction.
Molecular Mass and Sequence of Enterocin Gr17
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry revealed that the molecular mass of enterocin Gr17 was 4,531.01 Da (Figure 3D). The complete genome sequence and molecular mass data indicated that the entire amino acid sequence was RSYGNGVYCNNSKCWVNWGEAKENIIGI VISGWATGLAGMGR. Due to the formation of an essential disulfide bond (Drider et al., 2006), the determined molecular mass was similar to the calculated mass. Enterocin Gr17 was different from reported bacteriocins in the protein BLAST search of the GenBank database2. Furthermore, sequence alignment with other mature class IIa bacteriocins showed that enterocin Gr17 has a novel amino acid sequence (Figure 4). The findings indicated that enterocin Gr17 from E. faecalis Gr17 is a novel class IIa bacteriocin with similarities to enterocin P.

Figure 4. Alignment of reported class IIa bacteriocins. Alignments were obtained using DNAMAN 9.0 with default settings.
Antibacterial Spectrum of Enterocin Gr17
Enterocin Gr17 displayed strong antibacterial activity against Gram-positive bacteria (L. monocytogenes, S. aureus, B. subtilis, B. cereus, and E. faecalis) and poor antibacterial activity against Gram-negative bacteria (E. coli, Salmonella enteritidis, Brochothrix thermosphacta, Pseudomonas aeruginosa, Pseudomonas fluorescens, and Cronobacter sakazakii) and against the pathogenic yeast Candida albicans (Table 1).
Stability of Enterocin Gr17
After heating at different temperatures, enterocin Gr17 still possessed antibacterial activity (Figure 5A). Enterocin Gr17 retained inhibitory activity at pH ranging from 2 to 10, but the activity was lost in pH 11 (Figure 5B). Surfactants did not decrease the activity (Figure 5C), and the activity was completely eliminated when incubated with proteolytic enzymes (Figure 5D).
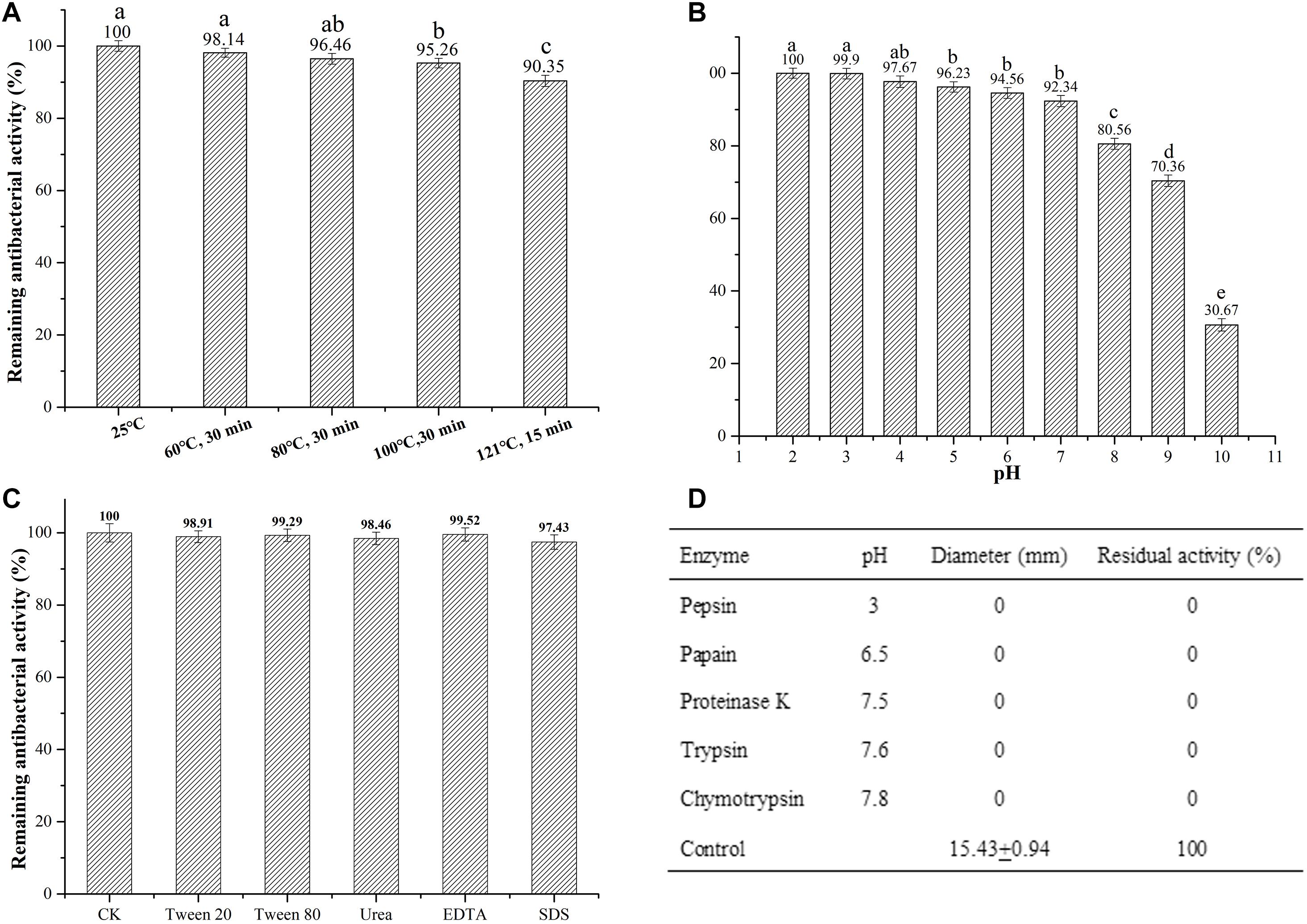
Figure 5. Effects of temperature (A), pH (B), surfactants (C), and protease (D) on the stability of enterocin Gr17.
Discussion
Numerous enterocins from E. faecium have been reported. These include the class IIa enterocin NKR-5-3C from E. faecium NKR-5-3C (Himeno et al., 2012), class IIa enterocin TW21 from E. faecium D081821 (Chang et al., 2013), class IIc enterocin AS-48 (Abengózar et al., 2017), and class IId enterocin K1 (Ovchinnikov et al., 2017). Even though features of the various enterocins have been determined, genome information and details of the biosynthesis mechanisms have been unknown. Also, the genome information of enterocin-producing strains, including E. faecium ICIS 96 (Pashkova et al., 2018), E. faecium CRL1879 (Bonacina et al., 2017), and E. faecium M3K31 (Arbulu et al., 2016), has been identified, but the features and biosynthesis mechanisms have also been unclear. The present findings reveal the biosynthesis mechanism and features of the novel class IIa enterocin Gr17 in the novel strain E. faecalis Gr17.
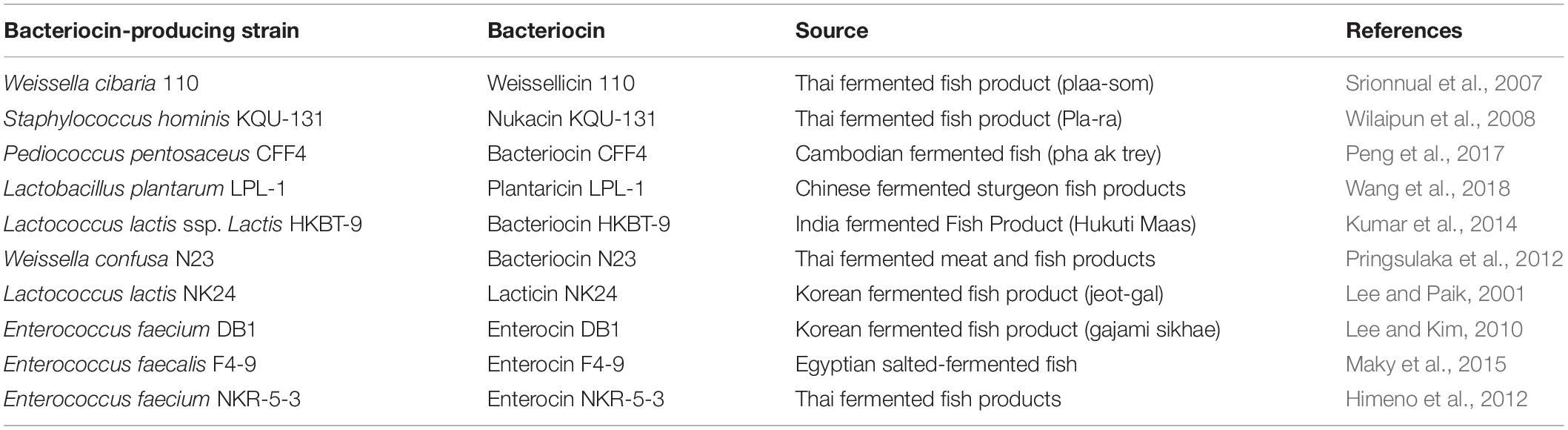
The potential of bacteriocins and bacteriocin-producing LAB as biopreservatives for fermented fish product has inspired searches for strains with potent antagonistic effects against food-poisoning microorganisms from fermented fish. Recent studies have isolated and screened bacteriocin-producing strains from fermented fish in different countries (Table 5). However, bacteriocin-producing strains isolated from Chinese fermented fish have not been adequately studied; as far as we know, the only bacteriocin-producing bacterium is Lactobacillus plantarum LPL-1, which was isolated from Chinese fermented sturgeon fish products (Wang et al., 2018). In this manuscript, we isolated a novel bacteriocin producer, E. faecalis Gr17, from Suan yu, a Chinese traditional low-salt fermented whole-fish product. This is the first report of a novel bacteriocin-producing strain from Suan yu. The antibacterial activity and spectrum of bacteriocins are important factors for the application of bacteriocins in the food industry. The novel amino acid sequence of bacteriocins could provide a foundation for exploring the relationship between bacteriocin structure and antibacterial activity. The characteristics, molecular mass, and amino acid sequence of enterocin Gr17 are obviously different from some well-known enterocins.
Cytolysin encoded by cylA, cylB, and cylM is a bacterial toxin that increases the risks of illness and death. Gelatinase encoded by gelE participates in the initiation and propagation of inflammatory processes. Sex pheromones encoded by cpd, cob, and ccf, and aggregation substance encoded by agg cause cell aggregation and facilitate the transfer of virulence factors and antibiotic resistance genes. Extracellular surface proteins encoded by esp, efaAfs, and efaAfm can promote adhesion of cells and protect the cells from the immune system. These virulence factors were not detected in E. faecalis Gr17, which suggests that the strain is non-virulent and relatively safe for consumers. Also, E. faecalis Gr17 did not contain genes encoding resistance to erythromycin, tetracycline, ampicillin, and vancomycin, but could potentially be resistant to ciprofloxacin. We screened E. faecalis Gr17 from the fermented whole-fish product Suan yu. Ciprofloxacin was often used to protect fish from pathogenic bacteria in aquaculture. It is conceivable that E. faecalis Gr17 may be transmissible from fish by highly efficient gene transfer mechanisms. Still, the available genotypic evidence of potential virulence factors and antibiotic resistance indicates that the strain may be safe to use.
Class IIa enterocins can be exported from cells by the Sec system (e.g., enterocin P in E. faecium P13; Cintas et al., 1997) and the ABC transporter (e.g., enterocin B in E. faecium T136; Casaus et al., 1997). Also, the biosynthetic mechanisms of other enterocins, which are termed leaderless bacteriocins, are unclear; these enterocins include enterocin K1 (Ovchinnikov et al., 2017) and EntEJ97 (Ovchinnikov et al., 2014). The biosynthetic mechanism of enterocin Gr17 belonged to the Sec system according to the complete genome information of E. faecalis Gr17. The genes relevant to the structural gene, transporter genes, and immune-related gene were identified. Moreover, the specific function of relevant genes was predicted by bioinformatic analysis. This genome information of E. faecalis Gr17 provides a better understanding of the biosynthesis mechanism of enterocin Gr17. Further investigations of the relevance between the quorum sensing system and biosynthesis of enterocin Gr17 will be carried out.
The precursor of class IIa enterocins is composed of signal peptide and mature peptide (Jack et al., 1995). Generally, the cleavage site between the signal peptide and mature peptide could be determined by bioinformatic analysis and MALDI–TOF–MS. According to the genome information, the amino acid sequence of the precursor was determined. Enterocin Gr17 was identified as a class IIa bacteriocin because of the N-terminal conserved YGNGV motif of mature class IIa bacteriocin. To determine the molecular mass of the mature peptide, enterocin Gr17 was purified by salt precipitation, cation exchange, gel filtration chromatography, and RP-HPLC. Different from other enterocins, such as enterocin RM6 (7145.0823 Da) (Huang et al., 2013), enterocin TW21 (5302.98 Da) (Chang et al., 2013), and enterocin AS-48 (7149 Da) (Abengózar et al., 2017), the molecular mass of purified enterocin Gr17 was 4,531.01 Da, which corresponded to a calculated molecular mass of 4,533.11 Da due to the formation of a disulfide bond. To some extent, the molecular mass result confirmed the novelty of enterocin Gr17. Especially, the results of genome information and MALDI–TOF–MS revealed that the amino acid sequence of mature enterocin Gr17 was RSY GNGVYCNNSKCWVNWGEAKENIIGIVISGWATGLAGMGR. A BLAST search of the NCBI database3 for mature enterocin Gr17 revealed its difference from reported class IIa enterocins. Therefore, enterocin Gr17 was confirmed as a novel class IIa enterocin. Compared with known class IIa bacteriocins, the enterocin Gr17 possesses a similar N-terminal sequence of xxYGNGVxC.
The antibacterial activity of enterocins can provide a basis for their application as food biopreservatives. In contrast to the narrow spectrum bacteriocins enterocin W (Sawa et al., 2012), enterocin A (Ennahar and Deschamps, 2000), enterocin 416K1 (Sabia et al., 2002), and enterocin CRL35 (Salvucci et al., 2007), enterocin Gr17 possessed antibacterial activity against L. monocytogenes, S. aureus, B. subtilis, B. cereus, B. anthracis, E. coli, S. enteritidis, P. aeruginosa, P. fluorescens, E. faecalis, E. sakazakii, and C. albicans (Table 1). These food-borne pathogenic and spoilage bacteria are also most frequently detected in the food industry and in medical science. Numerous authors also combined bacteriocins with the hurdle technology to inhibit food-borne pathogenic bacteria (Leistner, 2000), such as chemical chelators (sodium tripolyphosphate and EDTA) and physical methods (pH, temperature, high hydrostatic pressure, and pulsed electric field) (Thomas et al., 1998; Ananou et al., 2005; Martínez et al., 2008; Khan et al., 2015; Prudêncio et al., 2015). Thus, enterocin Gr17 has the potential to be applied with the hurdle technology to control food quality. The antibacterial activity of enterocin Gr17 makes it a good candidate for the preservation of various types of foods.
Concerning the application of enterocin Gr17 in the biopreservation of foods, its stability during different chemical treatments is essential. Enterocin Gr17 was very stable to a wide range of pH, high temperatures, and surfactants. These features are typical of the numerous bacteriocins that have been characterized, such as enterocin from E. faecium JCM 5804T (Park et al., 2003), enterocin ON-157 from E. faecium NIAI 157 (Ohmomo et al., 2000), and enterocin RM6 (Huang et al., 2013). The thermal stability of enterocin Gr17 indicates that it is valuable for use in dairy products and heat-processed foods. The pH stability could allow its use with slightly alkaline, neutral, and acidic foods. The surfactant stability could be ideal for use in emulsified foods. The antibacterial activity of enterocin could be destroyed by human digestive enzymes. To a certain extent, enterocin Gr17 could be safely used in the food industry, and safety for the human health remains to be confirmed by toxicity experiments in the future. Therefore, enterocin Gr17 is a good candidate as a safe biopreservative in the food industry.
Generally, the formation of pores in the membrane of cells is lethal. To definitively analyze the antibacterial mechanism of enterocin Gr17 against food-borne pathogenic and spoilage bacteria, further investigations on morphology changes will be done using scanning tunneling microscopy, transmission electron microscopy, and atomic force microscopy, and the exchange of molecules between inner and outer membrane of Gram-negative bacteria will be explored using proton motive force. As well, ATP and inorganic ions will be examined. Metabolomics, transcriptomics, and proteomics data will be combined to clarify the antibacterial mechanism.
Conclusion
Bioinformatic analysis clarified the biosynthetic mechanism of enterocin Gr17 in the novel strain E. faecalis Gr17. Based on bioinformation and MALDI–TOF–MS, enterocin Gr17 from E. faecalis Gr17 was identified. The entire amino acid sequence was determined to be RSYGNGVYCNNSK CWVNWGEAKENIIGIVISGWATGLAGMGR. Enterocin Gr17 exhibited bactericidal activity, sensitivity to enzymes, and stability to chemicals, elevated temperature, and pH. Therefore, enterocin Gr17 is a promising stable and safe biopreservative in various types of foods. Future investigations on the bactericidal mechanism of enterocin Gr17 will be carried out.
Author Contributions
YW, GL, and CW designed the experiments. YW, XL, XH, and DX performed the experiments. YW, GL, YZ, and AM analyzed the results and wrote the manuscript.
Funding
This project was funded by the Natural Science Foundation of China (Nos. 31871772 and 31671832), the Beijing Natural Science Foundation (Grant No. 6192003), high-level teachers in the Beijing Municipal Universities in the period of 13th 5-year plan (No. CIT&TCD201704034), and the Talent Training Quality Construction – First Class Professional Construction (Grant No. PXM2019-014213-000010).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01806/full#supplementary-material
TABLE S1 | Additional gene and function protein identification using the KEGG database.
TABLE S2 | Additional gene and function protein identification by RAST.
Footnotes
- ^ http://www.cbcb.umd.edu/software/glimmer/
- ^ www.ncbi.nlm.nih.gov/BLAST
- ^ www.ncbi.nlm.nih.gov/BLAST
References
Abengózar, M. Á, Cebrián, R., Saugar, J. M., Gárate, T., Valdivia, E., Martínez-Bueno, M., et al. (2017). Enterocin AS-48 as evidence for the use of bacteriocins as new leishmanicidal agents. Antimicrob. Agents Chemother. 61:e02288-16. doi: 10.1128/AAC.02288-16
Ananou, S., Galvez, A., Martinez-Bueno, M., Maqueda, M., and Valdivia, E. (2005). Synergistic effect of enterocin AS-48 in combination with outer membrane permeabilizing treatments against Escherichia coli O157:H7. J. Appl. Microbiol. 99, 1364–1372. doi: 10.1111/j.1365-2672.2005.02733.x
Arbulu, S., Frantzen, C., Lohans, C. T., Cintas, L. M., Herranz, C., Holo, H., et al. (2016). Draft genome sequence of the bacteriocin-producing strain Enterococcus faecium M3K31, isolated from griffon vultures (Gyps fulvus subsp. fulvus). Genome Announc. 4:e00055-16. doi: 10.1128/genomeA.00055-16
Aymerich, T., Holo, H., Havarstein, L. S., Hugas, M., Garriga, M., and Nes, I. F. (1996). Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl. Environ. Microbiol. 62, 1676–1682.
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Barbosa, J., Gibbs, P. A., and Teixeira, P. (2010). Virulence factors among enterococci isolated from traditional fermented meat products produced in the North of Portugal. Food Control 21, 651–656. doi: 10.1016/j.foodcont.2009.10.002
Beckwith, J. (2013). The Sec-dependent pathway. Res. Microbiol. 164, 497–504. doi: 10.1016/j.resmic.2013.03.007
Bonacina, J., Suárez, N., Hormigo, R., Fadda, S., Lechner, M., and Saavedra, L. (2017). A genomic view of food-related and probiotic Enterococcus strains. DNA Res. 24, 11–24. doi: 10.1093/dnares/dsw043
Casaus, P., Nilsen, T., Cintas, L. M., Nes, I. F., Hernandez, P. E., and Holo, H. (1997). Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology 143(Pt 7), 2287–2294. doi: 10.1099/00221287-143-7-2287
Chajęcka-Wierzchowska, W., Zadernowska, A., and Łaniewska-Trokenheim, Ł (2016). Virulence factors, antimicrobial resistance and biofilm formation in Enterococcus spp. isolated from retail shrimps. LWT Food Sci. Technol. 69, 117–122. doi: 10.1016/j.lwt.2016.01.034
Chang, S. Y., Chen, Y. S., Pan, S. F., Lee, Y. S., Chang, C. H., Chang, C. H., et al. (2013). Enterocin TW21, a novel bacteriocin from dochi-isolated Enterococcus faecium D081821. J. Appl. Microbiol. 115, 673–678. doi: 10.1111/jam.12265
Choi, H. J., Shin, M. S., Lee, S. M., and Lee, W. K. (2012). Immunomodulatory properties of Enterococcus faecium JWS 833 isolated from duck intestinal tract and suppression of Listeria monocytogenes infection. Microbiol. Immunol. 56, 613–620. doi: 10.1111/j.1348-0421.2012.00486.x
Cintas, L. M., Casaus, P., Havarstein, L. S., Hernandez, P. E., and Nes, I. F. (1997). Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 63, 4321–4330.
Cintas, L. M., Casaus, P., Herranz, C., Havarstein, L. S., Holo, H., Hernandez, P. E., et al. (2000). Biochemical and genetic evidence that Enterococcus faecium L50 produces enterocins L50A and L50B, the sec-dependent enterocin P, and a novel bacteriocin secreted without an N-terminal extension termed enterocin Q. J. Bacteriol. 182, 6806–6814. doi: 10.1128/JB.182.23.6806-6814.2000
Cleveland, J., Montville, T. J., Nes, I. F., and Chikindas, M. L. (2001). Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71, 1–20. doi: 10.1016/S0168-1605(01)00560-8
Comunian, R., Daga, E., Dupre, I., Paba, A., Devirgiliis, C., Piccioni, V., et al. (2010). Susceptibility to tetracycline and erythromycin of Lactobacillus paracasei strains isolated from traditional Italian fermented foods. Int. J. Food Microbiol. 138, 151–156. doi: 10.1016/j.ijfoodmicro.2009.11.018
Cotter, P. D., Ross, R. P., and Hill, C. (2013). Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. doi: 10.1038/nrmicro2937
Cui, Y., Zhang, C., Wang, Y., Shi, J., Zhang, L., Ding, Z., et al. (2012). Class IIa bacteriocins: diversity and new developments. Int. J. Mol. Sci. 13, 16668–16707. doi: 10.3390/ijms131216668
Diep, D. B., Skaugen, M., Salehian, Z., Holo, H., and Nes, I. F. (2007). Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U.S.A. 104, 2384–2389. doi: 10.1073/pnas.0608775104
Drider, D., Fimland, G., Héchard, Y., Mcmullen, L. M., and Prévost, H. (2006). The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70, 564–582. doi: 10.1128/MMBR.00016-05
Ennahar, S., and Deschamps, N. (2000). Anti-Listeria effect of enterocin A, produced by cheese-isolated Enterococcus faecium EFM01, relative to other bacteriocins from lactic acid bacteria. J. Appl. Microbiol. 88, 449–457. doi: 10.1046/j.1365-2672.2000.00985.x
Fanaro, S., Chierici, R., Guerrini, P., and Vigi, V. (2010). Intestinal microflora in early infancy: composition and development. Acta Paediatr. 92, 48–55. doi: 10.1111/j.1651-2227.2003.tb00646.x
Foulquie Moreno, M. R., Sarantinopoulos, P., Tsakalidou, E., and De Vuyst, L. (2006). The role and application of enterococci in food and health. Int. J. Food Microbiol. 106, 1–24. doi: 10.1016/j.ijfoodmicro.2005.06.026
Franz, C. M., Stiles, M. E., Schleifer, K. H., and Holzapfel, W. H. (2003). Enterococci in foods—a conundrum for food safety. Int. J. Food Microbiol. 88, 105–122. doi: 10.1016/S0168-1605(03)00174-0
Gálvez, A., Abriouel, H., López, R. L., and Ben, O. N. (2007). Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 120, 51–70. doi: 10.1016/j.ijfoodmicro.2007.06.001
Gevers, D., Danielsen, M., Huys, G., and Swings, J. (2003). Molecular characterization of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl. Environ. Microbiol. 69, 1270–1275. doi: 10.1128/AEM.69.2.1270-1275.2003
Guo, H., Pan, L., Li, L., Lu, J., Kwok, L., Menghe, B., et al. (2017). Characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products. J. Food Sci. 82, 724–730. doi: 10.1111/1750-3841.13645
Habermann, W., Zimmermann, K., Skarabis, H., Kunze, R., and Rusch, V. (2002). Reduction of acute recurrence in patients with chronic recurrent hypertrophic sinusitis by treatment with a bacterial immunostimulant Enterococcus faecalis bacteriae of human origin. Arzneimittelforschung 52, 622–627. doi: 10.1055/s-0031-1299941
Havarstein, L. S., Diep, D. B., and Nes, I. F. (1995). A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16, 229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x
Himeno, K., Fujita, K., Zendo, T., Wilaipun, P., Ishibashi, N., Masuda, Y., et al. (2012). Identification of enterocin NKR-5-3C, a novel class IIa bacteriocin produced by a multiple bacteriocin producer, Enterococcus faecium NKR-5-3. Biosci. Biotechnol. Biochem. 76, 1245–1247. doi: 10.1271/bbb.120089
Holzapfel, W., Arini, A., Aeschbacher, M., Coppolecchia, R., and Pot, B. (2018). Enterococcus faecium SF68 as a model for efficacy and safety evaluation of pharmaceutical probiotics. Benef. Microbes 9, 375–388. doi: 10.3920/BM2017.0148
Huang, E., Zhang, L., Chung, Y. K., Zheng, Z., and Yousef, A. E. (2013). Characterization and application of enterocin RM6, a bacteriocin from Enterococcus faecalis. Biomed. Res. Int. 2013:206917. doi: 10.1155/2013/206917
Jack, R. W., Tagg, J. R., and Ray, B. (1995). Bacteriocins of Gram-positive bacteria. Microbiol. Rev. 59, 171–200.
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. doi: 10.1093/nar/gkv1070
Kathrani, A., Larsen, J. A., Kass, P. H., and Fascetti, A. J. (2016). Effect of short-term probiotic Enterococcus faecium SF68 dietary supplementation in over overweight and obese cats without comorbidities. Vet. Rec. Open 3:e000164. doi: 10.1136/vetreco-2015-000164
Khan, A., Vu, K. D., Riedl, B., and Lacroix, M. (2015). Optimization of the antimicrobial activity of nisin, Na-EDTA and pH against Gram-negative and Gram-positive bacteria. LWT Food Sci. Technol. 61, 124–129. doi: 10.1016/j.lwt.2014.11.035
Kumar, M., Jain, A. K., Ghosh, M., and Ganguli, A. (2014). Characterization and optimization of an anti-Aeromonas bacteriocin produced by Lactococcus lactis isolated from hukuti maas, an indigenous fermented fish product. J. Food Process. Pres. 38, 745–947. doi: 10.1111/jfpp.12048
Kumariya, R., Garsa, A. K., Rajput, Y. S., Sood, S. K., Akhtar, N., and Patel, S. (2019). Bacteriocins: classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb. Pathog. 128, 171–177. doi: 10.1016/j.micpath.2019.01.002
Lee, H.-J., and Kim, W. J. (2010). Isolation and characterization of anti-listerial and amylase sensitive enterocin producing Enterococcus faecium DB1 from Gajami-sikhae, a fermented flat fish in Korea. Food Sci. Biotechnol. 19, 373–381. doi: 10.1007/s10068-010-0053-7
Lee, N. K., and Paik, H. D. (2001). Partial characterization of lacticin NK24, a newly identified bacteriocin of Lactococcus lactis NK24 isolated from Jeot-gal. Food Microbiol. 18, 17–24. doi: 10.1006/fmic.2000.0368
Leistner, L. (2000). Basic aspects of food preservation by hurdle technology. Int. J. Food Microbiol. 55, 181–186. doi: 10.1016/S0168-1605(00)00161-6
Liu, W., Pang, H., Zhang, H., and Cai, Y. (2014). “Biodiversity of lactic acid bacteria,” in Lactic Acid Bacteria: Fundamentals and Practice, eds H. Zhang and Y. Cai (Dordrecht: Springer), 103–203. doi: 10.1007/978-94-017-8841-0_2
Maky, M. A., Ishibashi, N., Zendo, T., Perez, R. H., Doud, J. R., Karmi, M., et al. (2015). Enterocin F4-9, a novel O-linked glycosylated bacteriocin. Appl. Environ. Microbiol. 81, 4819–4826. doi: 10.1128/aem.00940-15
Martínez, V. P., Sobrino, L. A., Ben, O. N., Abriouel, H., Lucas, L. R., Valdivia, E., et al. (2008). Enhanced bactericidal effect of enterocin AS-48 in combination with high-intensity pulsed-electric field treatment against Salmonella enterica in apple juice. Int. J. Food Microbiol. 128, 244–249. doi: 10.1016/j.ijfoodmicro.2008.08.014
Moraes, P. M., Perin, L. M., Junior, A. S., and Nero, L. A. (2013). Comparison of phenotypic and molecular tests to identify lactic acid bacteria. Braz. J. Microbiol. 44, 109–112. doi: 10.1590/S1517-83822013000100015
Ohmomo, S., Murata, S., Katayama, N., Nitisinprasart, S., Kobayashi, M., Nakajima, T., et al. (2000). Purification and some characteristics of enterocin ON-157, a bacteriocin produced by Enterococcus faecium NIAI 157. J. Appl. Microbiol. 88, 81–89. doi: 10.1046/j.1365-2672.2000.00866.x
Ovchinnikov, K. V., Kristiansen, P. E., Straume, D., Jensen, M. S., Aleksandrzak-Piekarczyk, T., Nes, I. F., et al. (2017). The leaderless bacteriocin enterocin K1 is highly potent against Enterococcus faecium: a study on structure, target spectrum and receptor. Front. Microbiol. 8:774. doi: 10.3389/fmicb.2017.00774
Ovchinnikov, K. V., Kristiansen, P. E., Uzelac, G., Topisirovic, L., Kojic, M., Nissen-Meyer, J., et al. (2014). Defining the structure and receptor binding domain of the leaderless bacteriocin LsbB. J. Biol. Chem. 289, 23838–23845. doi: 10.1074/jbc.M114.579698
Parada, J. L., Caron, C. R., Medeiros, A. B. P., and Soccol, C. R. (2007). Bacteriocins from lactic acid bacteria: purification, properties and use as biopreservatives. Braz. Arch. Biol. Technol. 50, 512–542. doi: 10.1590/S1516-89132007000300018
Park, S. H., Itoh, K., and Fujisawa, T. (2003). Characteristics and identification of enterocins produced by Enterococcus faecium JCM 5804T. J. Appl. Microbiol. 95, 294–300. doi: 10.1046/j.1365-2672.2003.01975.x
Pashkova, T. M., Vasilchenko, A. S., Khlopko, Y. A., Kochkina, E. E., Kartashova, O. L., and Sycheva, M. V. (2018). Genome sequence of Enterococcus faecium Strain ICIS 96 demonstrating intermicrobial antagonism associated with bacteriocin production. Genome Announc. 6:e00126-18. doi: 10.1128/genomeA.00126-18
Peng, C., Borges, S., Magalhães, R., Carvalheira, A., Ferreira, V., and Teixeira, P. (2017). Characterization of anti-listerial bacteriocin produced by lactic acid bacteria isolated from traditional fermented foods from Cambodia. Int. Food Res. J. 24, 386–393.
Perez, R. H., Zendo, T., and Sonomoto, K. (2014). Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb. Cell Fact. 13:S3. doi: 10.1186/1475-2859-13-S1-S3
Pringsulaka, O., Thongngam, N., Suwannasai, N., Atthakor, W., Pothivejkul, K., and Rangsiruji, A. (2012). Partial characterisation of bacteriocins produced by lactic acid bacteria isolated from Thai fermented meat and fish products. Food Control 23, 547–551. doi: 10.1016/j.foodcont.2011.08.029
Prudêncio, C. V., Mantovani, H. C., Cecon, P. R., and Vanetti, M. C. (2015). Differences in the antibacterial activity of nisin and bovicin HC5 against Salmonella Typhimurium under different temperature and pH conditions. J. Appl. Microbiol. 118, 18–26. doi: 10.1111/jam.12680
Rea, M. C., Ross, R. P., Cotter, P. D., and Hill, C. (2011). “Classification of bacteriocins from Gram-positive bacteria,” in Prokaryotic Antimicrobial Peptides: from Genes to Applications, eds D. Drider and S. Rebuffat (New York, NY: Springer), 29–53. doi: 10.1007/978-1-4419-7692-5_3
Rushdy, A. A., and Gomaa, E. Z. (2013). Antimicrobial compounds produced by probiotic Lactobacillus brevis isolated from dairy products. Ann. Microbiol. 63, 81–90. doi: 10.1007/s13213-012-0447-2
Sabia, C., Manicardi, G., Messi, P., De Niederhausern, S., and Bondi, M. (2002). Enterocin 416K1, an antilisterial bacteriocin produced by Enterococcus casseliflavus IM 416K1 isolated from Italian sausages. Int. J. Food Microbiol. 75, 163–170. doi: 10.1016/S0168-1605(01)00741-3
Salvucci, E., Saavedra, L., and Sesma, F. (2007). Short peptides derived from the NH2-terminus of subclass IIa bacteriocin enterocin CRL35 show antimicrobial activity. J. Antimicrob. Chemother. 59, 1102–1108. doi: 10.1093/jac/dkm096
Sawa, N., Wilaipun, P., Kinoshita, S., Zendo, T., Leelawatcharamas, V., Nakayama, J., et al. (2012). Isolation and characterization of enterocin W, a novel two-peptide lantibiotic produced by Enterococcus faecalis NKR-4-1. Appl. Environ. Microbiol. 78, 900–903. doi: 10.1128/AEM.06497-11
Srionnual, S., Yanagida, F., Lin, L.-H., Hsiao, K.-N., and Chen, Y.-S. (2007). Weissellicin 110, a newly discovered bacteriocin from Weissella cibaria 110, isolated from plaa-som, a fermented fish product from Thailand. Appl. Environ. Microbiol. 73, 2247–2250. doi: 10.1128/AEM.02484-06
Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinform. 4:41. doi: 10.1186/1471-2105-4-41
Thomas, L. V., Davies, E. A., Delves-Broughton, J., and Wimpenny, J. W. (1998). Synergist effect of sucrose fatty acid esters on nisin inhibition of Gram-positive bacteria. J. Appl. Microbiol. 85, 1013–1022. doi: 10.1111/j.1365-2672.1998.tb05266.x
Voulgari, K., Hatzikamari, M., Delepoglou, A., Georgakopoulos, P., Litopoulou-Tzanetaki, E., and Tzanetakis, N. (2010). Antifungal activity of non-starter lactic acid bacteria isolates from dairy products. Food Control 21, 136–142. doi: 10.1016/j.foodcont.2009.04.007
Wang, Y., Qin, Y., Xie, Q., Zhang, Y., Hu, J., and Li, P. (2018). Purification and characterization of plantaricin LPL-1, a novel class IIa bacteriocin produced by Lactobacillus plantarum LPL-1 isolated from fermented fish. Front. Microbiol. 9:2276. doi: 10.3389/fmicb.2018.02276
Wilaipun, P., Zendo, T., Okuda, K., Nakayama, J., and Sonomoto, K. (2008). Identification of the nukacin KQU-131, a new type-A(II) lantibiotic produced by Staphylococcus hominis KQU-131 isolated from Thai fermented fish product (Pla-ra). Biosci. Biotechnol. Biochem. 72, 2232–2235. doi: 10.1271/bbb.80239
Keywords: Enterococcus faecalis, enterocin Gr17, complete genome sequence, gene cluster, antibacterial activity
Citation: Liu G, Wang Y, Li X, Hao X, Xu D, Zhou Y, Mehmood A and Wang C (2019) Genetic and Biochemical Evidence That Enterococcus faecalis Gr17 Produces a Novel and Sec-Dependent Bacteriocin, Enterocin Gr17. Front. Microbiol. 10:1806. doi: 10.3389/fmicb.2019.01806
Received: 08 November 2018; Accepted: 22 July 2019;
Published: 13 August 2019.
Edited by:
Yi-Cheng Sun, Institute of Pathogen Biology (CAMS), ChinaReviewed by:
Venkatesan Arul, Pondicherry University, IndiaCharles M. A. P. Franz, Max Rubner Institut, Germany
Copyright © 2019 Liu, Wang, Li, Hao, Xu, Zhou, Mehmood and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yao Wang, wangyao130897@163.com; Chengtao Wang, wangchengtao@th.btbu.edu.cn
 Guorong Liu
Guorong Liu Yao Wang
Yao Wang Xue Li1
Xue Li1