- 1Department of Biology, Botanical Institute, University of Cologne, Cologne, Germany
- 2Department of Biology, University of Rostock, Rostock, Germany
Biological soil crusts (BSCs) are complex communities of autotrophic, heterotrophic, and saprotrophic (micro)organisms. In the polar regions, these biocrust communities have essential ecological functions such as primary production, nitrogen fixation, and ecosystem engineering while coping with extreme environmental conditions (temperature, desiccation, and irradiation). The microalga Klebsormidium is commonly found in BSCs all across the globe. The ecophysiological resilience of various Klebsormidium species to desiccation and other stresses has been studied intensively. Here we present the results of transcriptomic analyses of two different Klebsormidium species, K. dissectum and K. flaccidum, isolated from Antarctic and Arctic BSCs. We performed desiccation stress experiments at two different temperatures mimicking fluctuations associated with global change. Cultures grown on agar plates were desiccated on membrane filters at 10% relative air humidity until the photosynthetic activity as reflected in the effective quantum yield of photosystem II [Y(II)] ceased. For both species, the response to dehydration was much faster at the higher temperature. At the transcriptome level both species responded more strongly to the desiccation stress at the higher temperature suggesting that adaptation to cold conditions enhanced the resilience of both algae to desiccation stress. Interestingly, the two different species responded differently to the applied desiccation stress with respect to the number as well as function of genes showing differential gene expression. The portion of differentially expressed genes shared between both taxa was surprisingly low indicating that both Klebsormidium species adapted independently to the harsh conditions of Antarctica and the Arctic, respectively. Overall, our results indicate that environmental acclimation has a great impact on gene expression and the response to desiccation stress in Klebsormidium.
Introduction
Biological soil crusts (BSCs) are the dominant vegetation cover on the temporarily snow- and ice-free soil surfaces of the Arctic and Antarctica (Yoshitake et al., 2010; Williams et al., 2017). These communities are complex agglomerations formed by diverse autotrophic and heterotrophic organisms such as microalgae, lichens, bryophytes, fungi, bacteria, and micro fauna (Belnap, 2006; Darby and Neher, 2016). The streptophyte green microalga Klebsormidium (Klebsormidiophyceae) is commonly associated with BSCs in both the Arctic and Antarctica (Hayashi and Shinozaki, 2012; Pushkareva et al., 2016; Borchhardt et al., 2017a,b; Rippin et al., 2018). In general, Klebsormidium is one of the most widespread green algal genera and can be found in numerous habitats around the globe (Elster et al., 2008; Škaloud and Rindi, 2013). Members of this genus are poikilohydric and, hence, unable to actively regulate their cellular water content which in turn may lead to nearly complete dehydration upon desiccation (Holzinger et al., 2014; Karsten and Holzinger, 2014).
The polar regions are characterized by a pronounced seasonality, low water availability for most of the year, and generally low temperatures resulting in harsh environmental conditions forcing organisms to develop adaptive mechanisms. The desiccation tolerance of Klebsormidium had already been described 100 years ago by Piercy (1917) and its ecophysiology has been widely studied on many biogeographically diverse isolates (e.g., Holzinger et al., 2014; Herburger and Holzinger, 2015; Blaas and Holzinger, 2017; Donner et al., 2017). Elster et al. (2008) showed that Arctic and Antarctic Klebsormidium strains are resistant to desiccation-induced injuries. Holzinger et al. (2014) studied the underlying molecular mechanisms in an alpine isolate of Klebsormidium crenulatum and found that dehydration caused an inhibition of photosynthesis while photosynthetic gene transcripts were up-regulated to prepare for rehydration. Furthermore, pathways leading to the synthesis of sucrose and raffinose, both important osmolytes, were up-regulated (Holzinger et al., 2014). Increasing the amount of osmolytes in the cell leads to a more negative osmotic potential which helps to retain water within the cell and protects against membrane damage and protein aggregation (Bisson and Kirst, 1995).
In addition, Klebsormidium enhances the expression of heat-shock proteins (HSPs) and chaperones [e.g., late embryogenesis abundant (LEA) proteins] to prevent protein aggregation and misfolding (Holzinger et al., 2014). As dehydration also enhances the formation of harmful reactive oxygen species (ROS), organisms have to activate appropriate scavenging mechanisms such as the synthesis of antioxidants (Cruz de Carvalho, 2008). Indeed, the desiccation transcriptome of K. crenulatum suggests an accumulation of the antioxidant glutathione and carotenoids in response to water withdrawal (Holzinger et al., 2014).
Besides low water availability, polar strains of Klebsormidium also have to endure low temperatures and long periods of freezing. Manabu et al. (2008) analyzed cold-acclimated filaments of K. flaccidum and found that this microalga had accumulated soluble sugars, such as glucose and sucrose, free amino acids, such as γ-aminobutyric acid, as well as starch grains and oil droplets. Changes in carbohydrate homeostasis may also enable the alga to cope with elevated ROS levels induced by low temperatures (Pommerrenig et al., 2018). Moreover, this species modified its cellular structure, reduced the size of its vacuole, and enlarged both its chloroplasts and cytoplasm (Manabu et al., 2008). Molecular data on the cold-responsive pathways and coping mechanisms are still lacking for Klebsormidium. However, studies on other algae exposed to low temperatures are available. Valledor et al. (2013) investigated the lipid profiles in the chlorophyte Chlamydomonas reinhardtii upon cold stress and found an increase in polyunsaturated fatty acids that is likely occurring to maintain membrane fluidity. The streptophyte Spirogyra varians (Han et al., 2012) accumulated antioxidants at 4°C suggesting an accelerated ROS generation (Han et al., 2012).
Klebsormidium plays an important role in the formation of BSCs, owing to its filamentous morphology and exopolysaccharide production, and also contributes to the primary production of the crust (Barberousse et al., 2006; Büdel et al., 2016). In general, biological crusts are essential components of polar ecosystems as they fix nitrogen and carbon dioxide, protect the soil against erosion, and enhance its fertilization (Evans and Johansen, 1999). However, climate change poses a major threat to these communities as it may affect their composition and abundance substantially (Johnson et al., 2012; Zelikova et al., 2012). For example in Antarctica, permanently ice- and snow-covered ground sets natural species boundaries (Lee et al., 2017). With increasing temperatures these zones will expand and eventually amalgamate which could offset those limits (Lee et al., 2017). Besides warming, higher levels of radiation and altered precipitation patterns are predicted for the Arctic and Antarctica, in particular the Antarctic Peninsula, which may cause an invasion of foreign species (Chown et al., 2012; Pushkareva et al., 2016). Populations of natives, that are unable to compete with those alien species, are likely to become diminished or even die (Bellard et al., 2016).
Here we present a transcriptomic study on two species of Klebsormidium, isolated from Arctic Svalbard and Ardley Island, Antarctica. Both taxa were tested for their desiccation tolerance. We cultivated both species at 5 and 20°C prior to treatment to elucidate how temperature influences the organism’s ability to cope with dehydration stress. Afterward, the filaments were rewetted with water and allowed to recover. Transcriptomes were analyzed at two time points during recovery to assess the underlying molecular mechanisms.
Materials and Methods
Algal Isolates and Cultivation
Two culture isolates of Klebsormidium were used in this study: K. dissectum (EiE-15a; Borchhardt et al., 2017a) was isolated from a BSC collected at Svalbard, Norway, and K. flaccidum (A1-1a; Borchhardt et al., 2017b) was isolated from an Antarctic BSC sampled at Ardley Island, South Shetland Islands. K. dissectum belongs to the Klebsormidium clade E in the clade system, described by Mikhailyuk et al. (2015), while K. flaccidum is affiliated with clade B/C. Both strains are part of the Culture Collection at the University of Rostock.
Both microalgae were cultured on solid 1.5% DifcoTM Agar (Becton Dickinson GmbH, Heidelberg, Germany) enriched with Bold’s basal medium supplemented with vitamins and a tripled nitrate concentration (3N-BBM+V, Starr and Zeikus, 1993) at both 5 and 20°C, and 35 μmol photons m–2 s–1 (Daylight Lumilux Cool White Lamps L36W/840, OSRAM Licht AG, Munich, Germany) under a 16:8-h light–dark cycle.
Growth in Response to Temperature
In vivo chlorophyll a fluorescence was used as a proxy for biomass accumulation according to Gustavs et al. (2009). Growth experiments were carried out at 12 different temperatures (6, 8, 10, 12, 13, 14, 17, 18, 20, 23, 26, and 28°C) at 35 μmol photons m–2 s–1 and a 16:8-h light–dark cycle using a self-designed algal incubator (Kunststoff-Technik GmbH, Rostock, Germany) as described in Woelfel et al. (2014). For acclimation purposes all measurements were always done with log-phase cultures that were pre-incubated at experimental conditions for 10 days. Chlorophyll a fluorescence (excitation 440 nm, emission 680 nm) was measured each day with a SpectraMax M2e multiplate reader (MPR; Molecular Devices, Biberach, Germany) using the software SoftMax Pro version 5.4 (Molecular Devices, LCC, San Jose, CA, United States). Increasing fluorescence was detected as relative fluorescence units (RFUs) and the fluorescence measured directly after the inoculation serving as a starting value. Before each measurement, a dark-incubation of 10 min was performed in order to open all reaction centers of photosystem (PS) II. Calculation of growth rate for each individual replicate was performed as described in Gustavs et al. (2009).
Desiccation Treatment and Rehydration
Both species were cultivated in triplicates in liquid cultures for 10 days under a light regime of 35 μmol photons m–2 s–1 and a 16:8-h light–dark cycle (log-phase) at 5 or 20°C. Following cultivation, 15 ml of each culture was blotted onto WhatmanTM GF six glass fiber filters (GE Healthcare, Little Chalfont, United Kingdom). Three filters of each species and temperature were directly harvested (C = control). The remaining filters were placed into desiccation chambers, developed by Karsten et al. (2014), and dried over 100 g of activated silica gel (Carl Roth, Karlsruhe, Germany), which led to a constant, relative air humidity of about 10%. Desiccation experiments were carried out at 5 or 20°C. During treatment, the effective quantum yield of PS II [Y(II)] was monitored using the pulse amplitude modulated fluorometer PAM 2500 (Heinz Walz, Effeltrich, Germany). Once Y(II) reached zero, another triplicate of filters was sampled (D = desiccated). Immediately afterward, the silica gel was replaced by 100 ml of tap water and the dried filaments were wetted with 200 μl of medium. The relative air humidity in the chamber was now between 95 and 98%. Filter triplicates were harvested 2 h after the wetting (RI = rehydration I) and 24 h after the start of the experiment (RII = rehydration II).
RNA Isolation and Sequencing
All harvested filters were instantly quenched in liquid nitrogen and homogenized in a chilled mortar. Subsequently, the powder was processed with the Spectrum Kit (Sigma–Aldrich, St. Louis, MO, United States) according to Rippin et al. (2016). The quality of the resulting RNA samples was assessed on a BioAnalyzer instrument (Agilent Technologies, Santa Clara, CA, United States) and subsequently used to prepare libraries. Eukaryotic mRNA was enriched by means of oligo-(dT) beads, and fragmented and reverse-transcribed into cDNA using random hexamers. Prior to sequencing on an Illumina HiSeq (2 × 75 bp) at the Cologne Center for Genomics, appropriate adapters were ligated. All 48 libraries were multiplexed to eliminate batch effects during sequencing. All raw reads were submitted to the SRA database under the BioProject ID PRJNA500592.
Bioinformatic Workflow
All raw reads were quality-trimmed and filtered using Trimmomatic 0.35 (Bolger et al., 2014) and PrinSeq Lite 0.20.4 (Schmieder and Edwards, 2011). rRNA sequence reads were separated by SortMeRNA 2.1 (Kopylova et al., 2012) using SILVA SSU NR Ref 119 and LSU Ref 119. Before assembling the reads, COPE 1.2.5 (Liu et al., 2012) was utilized to stitch overlapping reads together. The assembly of the K. dissectum and K. flaccidum transcriptomes was done with Trinity 2.0.6 (Grabherr et al., 2011) and the quality was assessed with scripts from the Trinity package and BUSCO 3.0.2 combined with the embryophyta database (Simão et al., 2015). The assemblies were annotated using the Trinotate pipeline 3.0.01, including TransDecoder 2.12, NCBI BLAST+ 2.3.0 (Altschul et al., 1990), HMMER 3.1 b (Finn et al., 2011), SignalP 4.1 (Petersen et al., 2011), TMHMM 2.0 c (Krogh et al., 2001), RNAmmer 1.2 (Lagesen et al., 2007), and the databases Swiss-Prot (Bairoch and Apweiler, 1999), PFAM 3.1b2 (Sonnhammer et al., 1997), RefSeq Plant 149, the transcriptomes of K. crenulatum (Holzinger et al., 2014) and K. subtile from the 1KP data repository (Matasci et al., 2014) as well as the genome of K. nitens (Hori et al., 2014). The expect value cutoff was set to E −10. The gene map plot was created using the R package circlize (Gu et al., 2014).
The raw reads, obtained from the different conditions tested, were mapped onto the assembled contigs of both organisms using Bowtie 1.1.2 (Langmead et al., 2009) and abundance was estimated with RSEM 1.2.30 (Li and Dewey, 2014). Subsequently, the R package edgeR (Robinson et al., 2010) was employed to identify differentially expressed genes. Genes possessing an FDR (Hochberg, 1995) of >0.001 and a fold change of <4 were excluded from further analysis. Overlapping gene sets between the same comparisons at different temperatures were tested for significance using a hypergeometric distribution in R. Two types of gene set enrichment analyses were carried out in R: GO term enrichment with GoSeq 1.26.0 (Young et al., 2010) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment with clusterProfiler 3.2.11 (Yu et al., 2012), setting the FDR in both cases to 0.05. The online tool REVIGO (Supek et al., 2011) was used to create the basis data for the GO network plots.
Results
Ecophysiological Analysis
Klebsormidium dissectum and K. flaccidum exhibited biomass accumulation between 6 and 28°C (Figure 1). The optimal growth temperature was 18 and 20°C for K. dissectum and K. flaccidum, respectively, which classifies both taxa as psychrotolerant. At the optimal temperatures both strains divided every 2.5 days. However, K. dissectum growed very slowly at low temperatures, with a generation time of approximately 20 days at 6 and 10°C. In contrast, K. flaccidum cells divided on average every 5–10 days between 6 and 15°C.
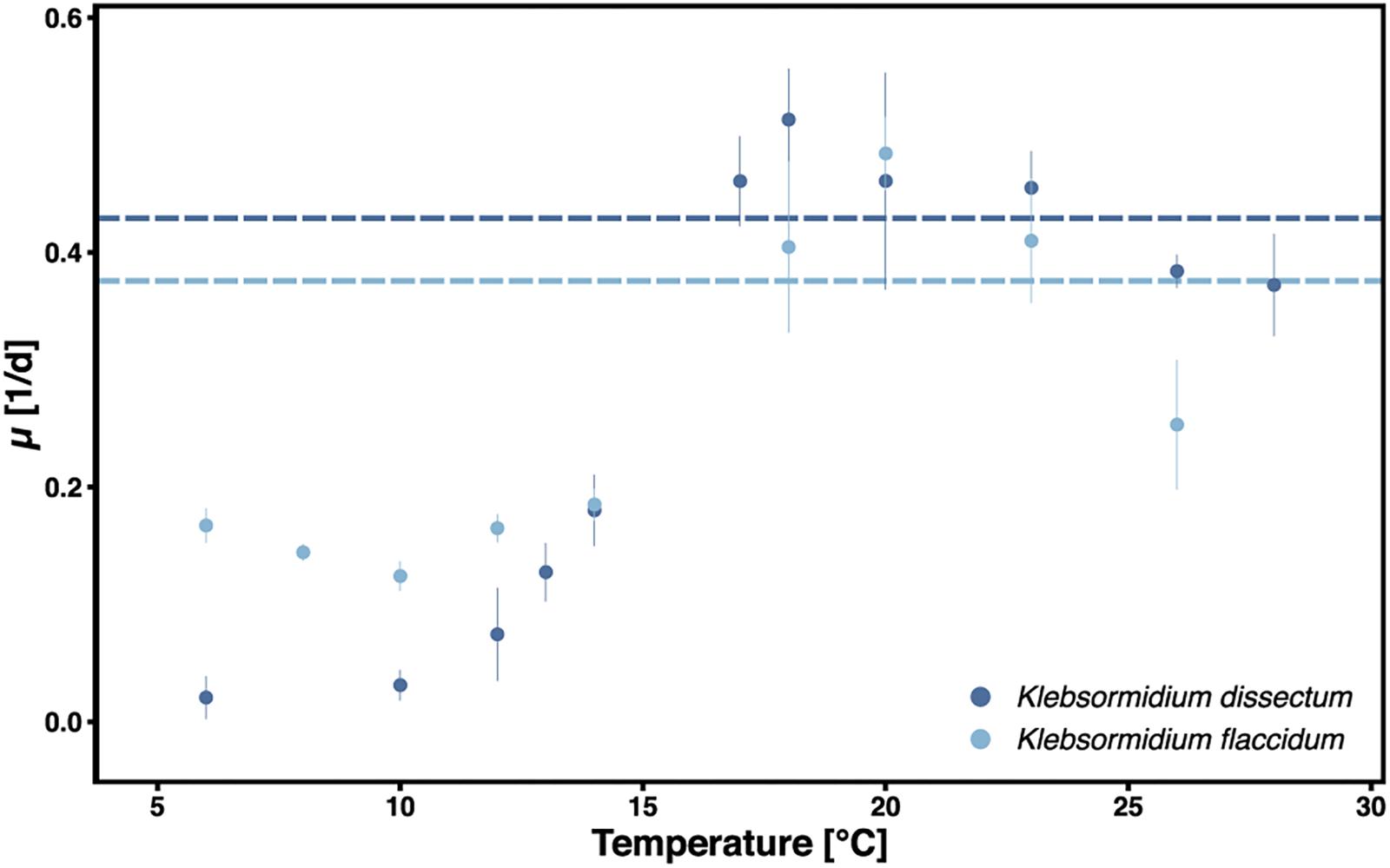
Figure 1. Growth of K. dissectum and K. flaccidum at different temperatures. The growth rate μ was determined for exponential cultures. Bars indicate standard deviation. Horizontal lines indicate the mean maximal growth rate between 12 and 28°C.
Figure 2 shows the experimental setup of the desiccation experiment. As in earlier studies (Holzinger et al., 2014; Rippin et al., 2017), we monitored the desiccation process indirectly by measuring the photosynthetic quantum yield of PS II [Y(II)] with a pulse amplitude modulated fluorometer (PAM). For both taxa, K. flaccidum and K. dissectum, the desiccation time was similar at the same temperature (Figure 3). At 5°C, it took about 270 min to record a minimum Y(II) = 0, while at 20°C the time required to reach a Y(II) = 0 decreased to approximately 150 min. A partial recovery of Y(II) (50–80%) was observed 24 h after the start of the experiment for both Klebsormidium species. While the recovery of K. flaccidum was independent from temperature (approximately 50%), K. dissectum showed a better recovery at 5°C (approximately 75%) compared to 20°C (approximately 50%).
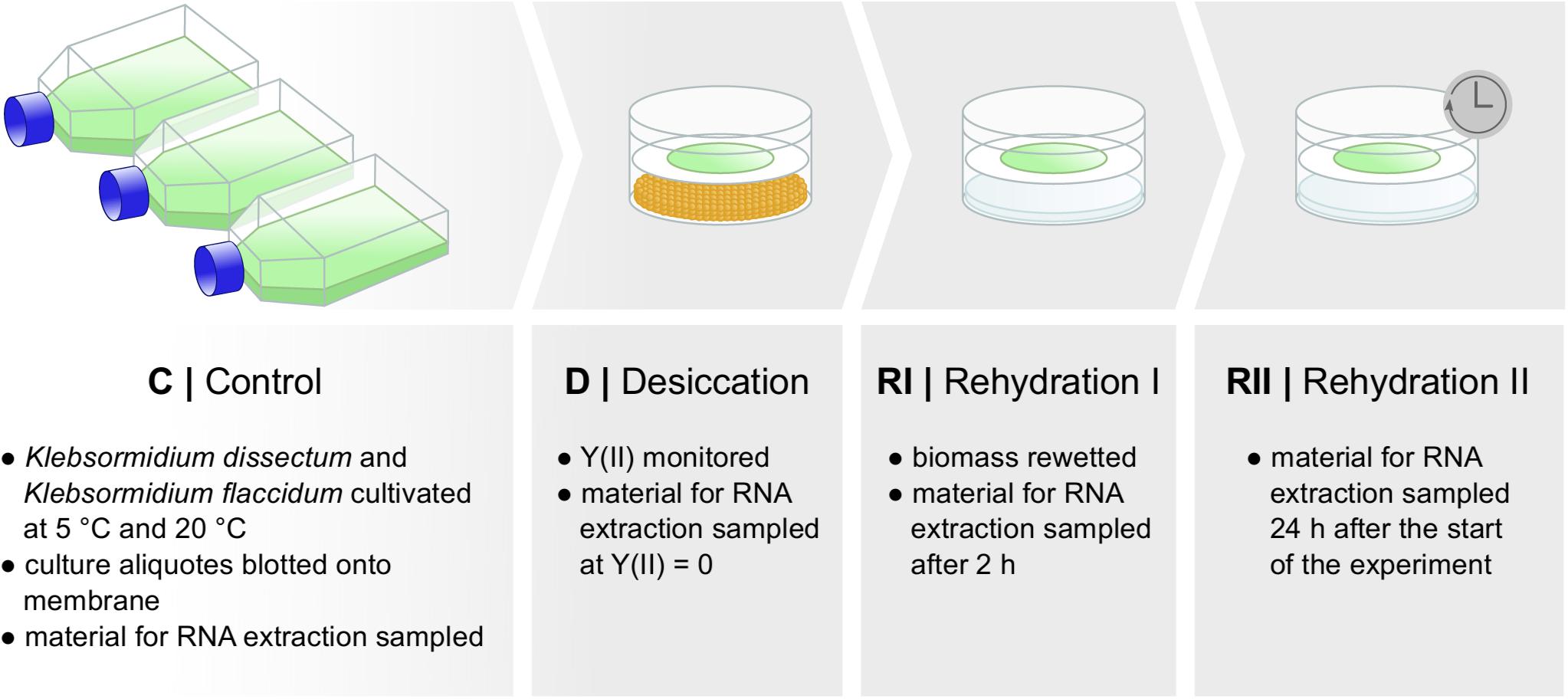
Figure 2. Experimental set up. Two different species of Klebsormidium were desiccated at two different temperatures [5 and 20°C until Y(II) reached zero] for the time indicated in Figure 3 and allowed to recover for 2 h and approximately 24 h. Samples were taken at several time points for control, desiccation treatment, and during the recovery phase. RNA was subsequently extracted.
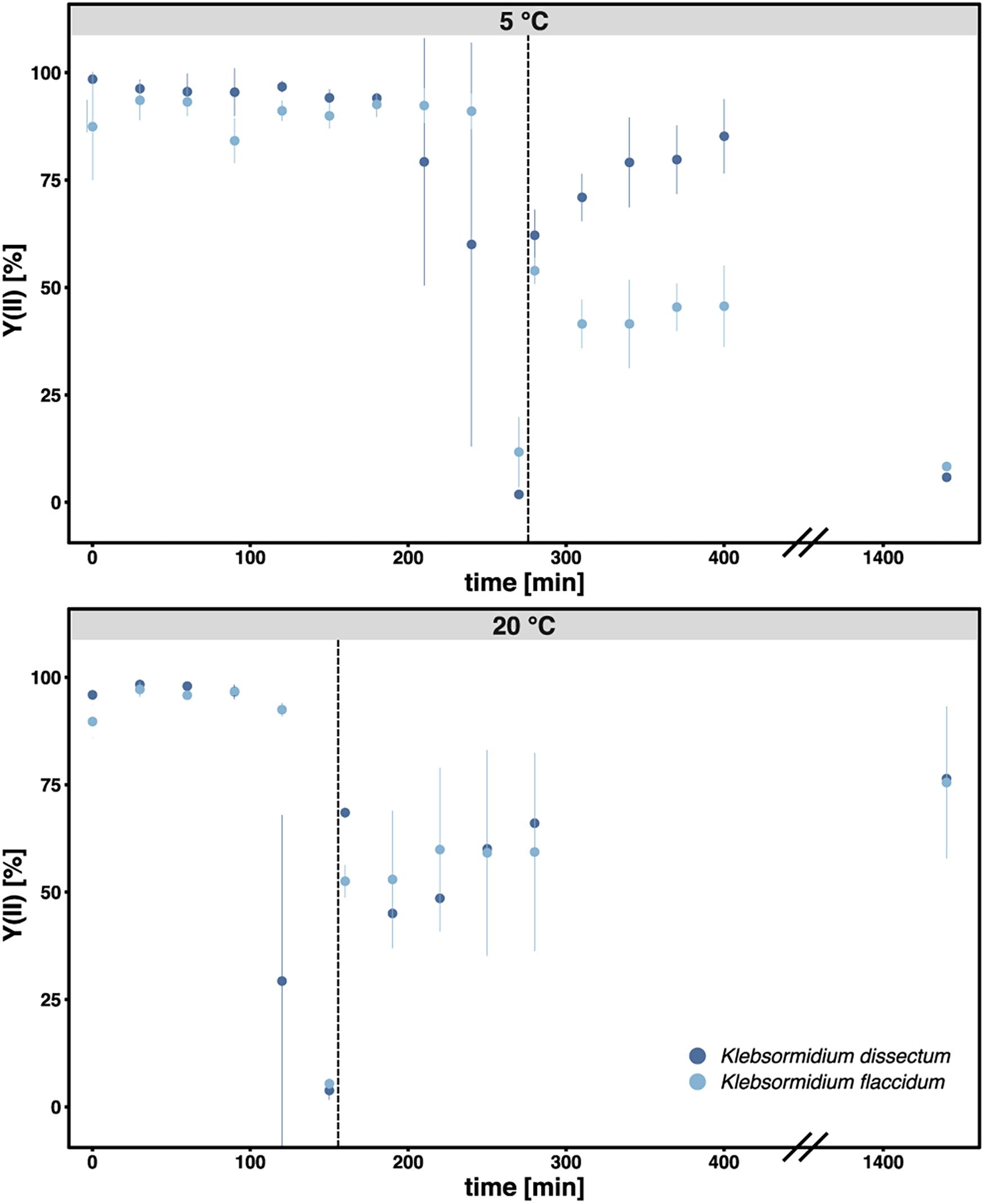
Figure 3. The photosynthetic quantum yield of PS II [Y(II)] was measured over time during the course of desiccation for all samples. Each triplicate was measured three times at different positions of the filter. The bar indicates standard deviation while the dots indicate the mean value for each treatment. Dark blue: K. dissectum; light blue: K. flaccidum. The vertical lines indicate the start of the recovery phase.
Sequencing Outcome and Assembly
About 5.7 ± 0.57 Gbp, which corresponds to approximately 38 ± 4 Mio read pairs (Supplementary Table S1), per library were successfully sequenced. After quality trimming and filtering, 14 ± 6.4 Mio read pairs per library remained and were assembled to 91,678 and 99,957 contigs for K. dissectum and K. flaccidum, respectively. The N50 value for K. dissectum was 2,805 bp and for K. flaccidum 3,021 bp.
A BUSCO analysis was performed for both assemblies as well as for the genome of K. nitens, the transcriptome of K. crenulatum and the transcriptome of K. subtile for comparison (Figure 4A). In the case of K. dissectum, 48.5% of the universal single-copy orthologs was found to be complete (either single or duplicated), 3.5% was fragmented, and 48% was missing. The situation was similar for K. flaccidum with 48.8% of the BUSCO orthologs to be complete, 3.8% fragmented, and 47.4% missing. Running the same analysis on the genome of K. nitens revealed that both assembled transcriptomes were in the same range as the genome. However, the large number of duplicated orthologs in the transcriptomes suggest that the transcriptomes probably still contain artificial isoforms, produced by the Trinity assembler that the BUSCO analysis classifies as duplications. The BUSCO ortholog coverage for the transcriptomes from K. crenulatum and K. subtile was lower when compared to K. dissectum and K. flaccidum.
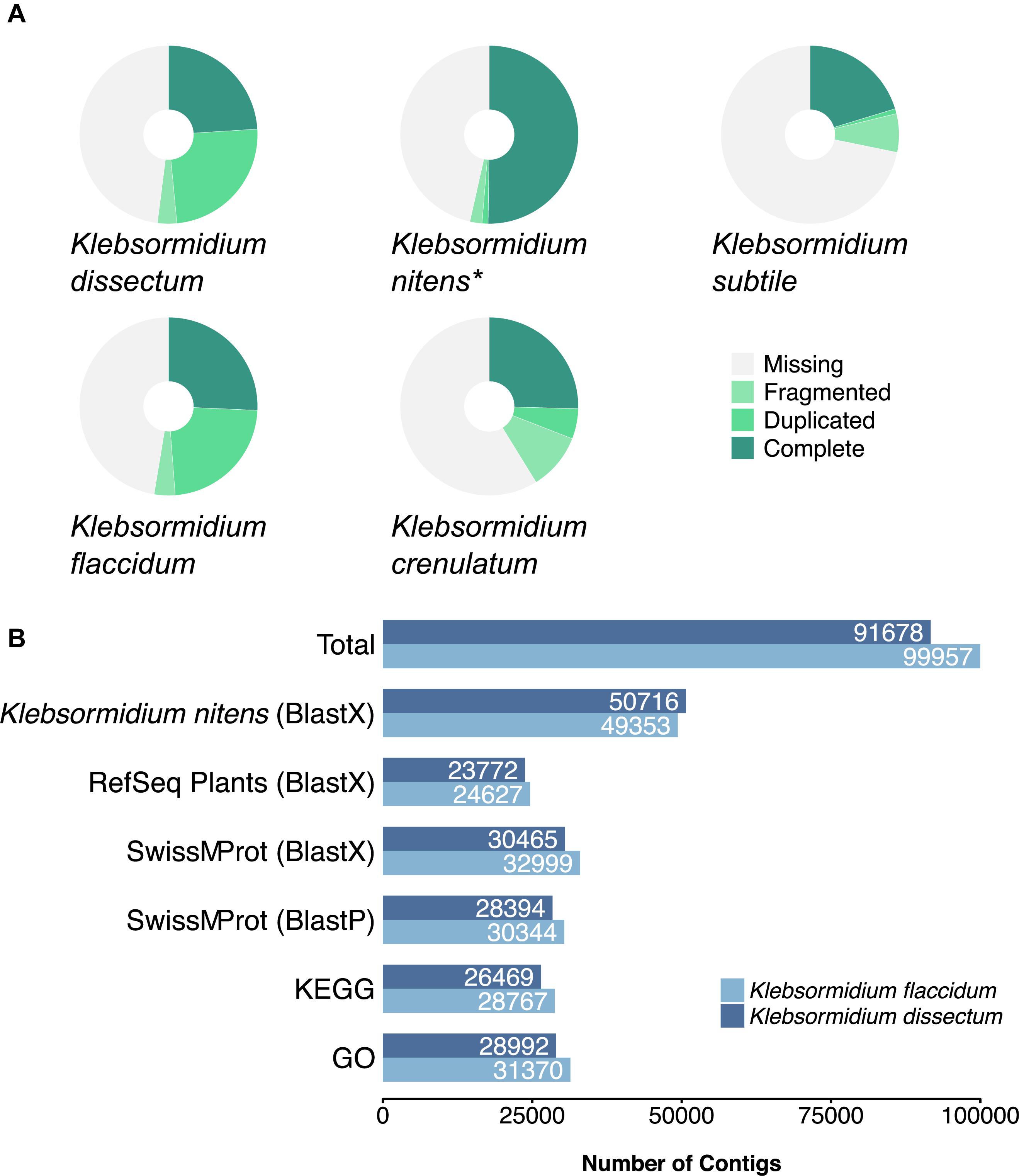
Figure 4. (A) BUSCO analysis of the assembled transcriptomes from K. dissectum and K. flaccidum. The pie chart displays the results of the BUSCO analysis with the categories complete, duplicated, fragmented, and missing. For comparison the transcriptomes of K. crenulatum and K. subtile, as well as the genome (indicated with an asterisk) of K. nitens were included in the BUSCO analysis. (B) Summary of the annotation results for K. dissectum and K. flaccidum. The assembled contigs were blasted against the genome of K. nitens and the indicated databases using the indicated BLAST program and an e-value cut-off of E –10.
Both of the assembled transcriptomes were annotated against Swiss-Prot and RefSeq Plant databases with annotation rates from 24.6 to 33.2% (Figure 4B). The annotation against the genome of K. nitens resulted in 55 and 49.4% identified transcripts. The gene map in Figure 5 shows the nucleotide sequence similarities of both species to each protein of K. nitens. The expect values and coverage for K. dissectum were generally higher than for K. flaccidum. The Venn diagram in the center shows the BlastN results of the transcriptomes of K. dissectum, K. flaccidum, K. crenulatum, and K. subtile against K. nitens. All transcriptomes shared a total of 2,665 genes with the K. nitens genome at the mRNA level with an expect value threshold of at least E −10. The transcriptomes of K. dissectum and K. crenulatum shared the largest and smallest fraction with K. nitens, respectively.
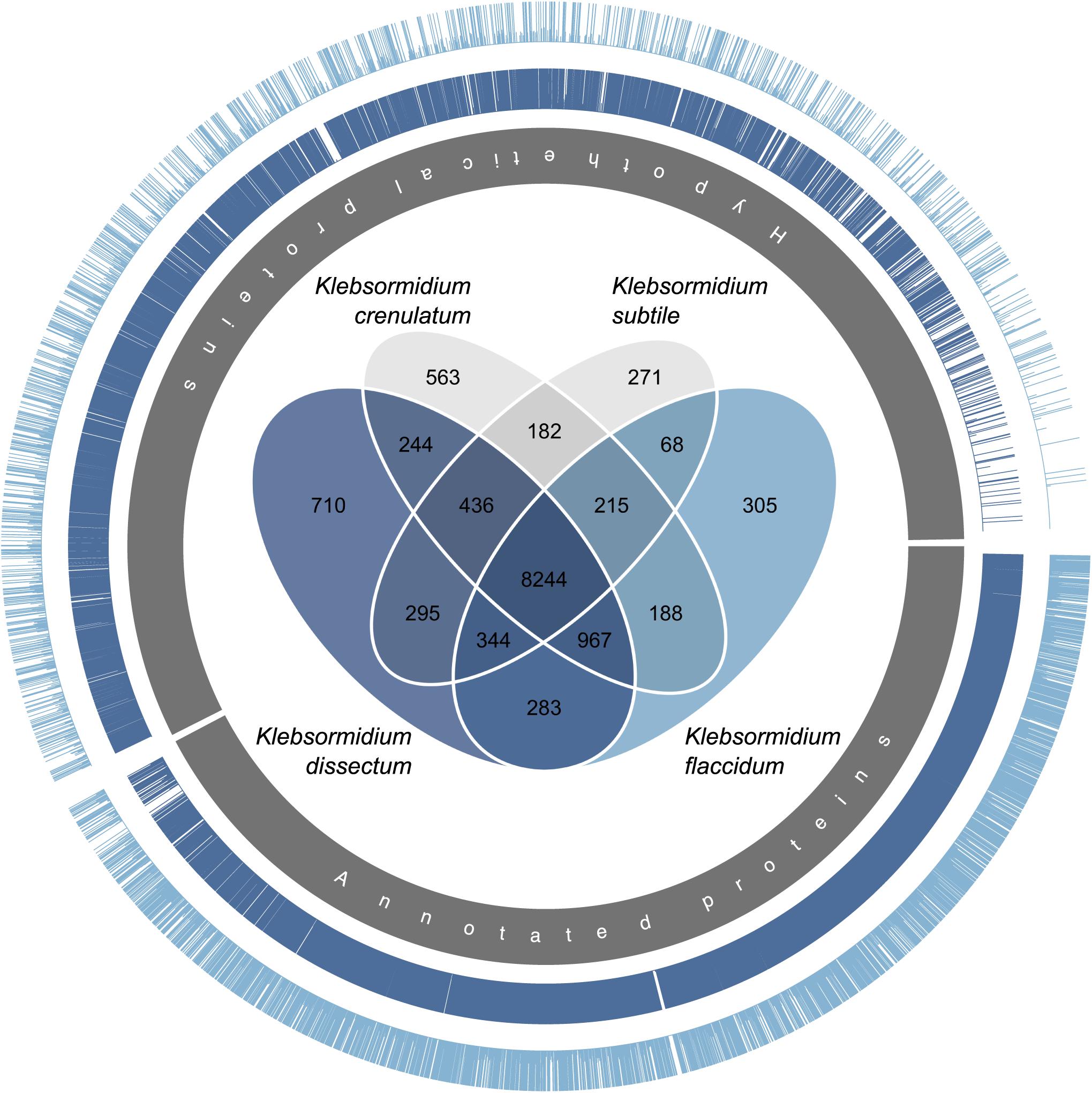
Figure 5. Gene map comparing the genome of K. nitens with the transcriptomes from K. dissectum and K. flaccidum. The inner gray circle indicates the annotated and hypothetical proteins of K. nitens. Each bar in the two outer circles represents a protein of K. nitens being a BLAST top hit when the two transcriptomes were blasted against the K. nitens genome. The bar length indicates the conservation of the protein. The proteins encoded by the K. dissectum transcriptome are generally much more similar to K. nitens proteins than proteins encoded the K. flaccidum transcriptome. The Venn diagram in the center illustrates the similarity of the available Klebsormidium transcriptomes when the transcriptomes were blasted against the K. nitens genome using the BLASTN algorithm and an e-value cut-off of E –10.
Moreover, the assembled transcriptomes were annotated for KEGG IDs 17.4 and 12.1% of the K. dissectum contigs were successfully assigned KO (KEGG orthology) terms and Arabidopsis identifiers, respectively. For K. flaccidum, 17.6% of the assembled transcripts could be assigned KO terms and 13.3% Arabidopsis annotations. The KO terms from both assemblies were mapped onto the KEGG metabolic pathway map (ko01100). The assembled transcriptomes of K. dissectum and K. flaccidum showed a good coverage of the most important pathways (e.g., carbohydrate metabolism, amino acid metabolism, fatty acid metabolism, nucleotide metabolism, respiration) as illustrated in Supplementary Figures S1, S2. For functional characterization, GO (gene ontology) terms were assigned which yielded annotation rates of 31.6 and 31.4% for K. dissectum and K. flaccidum, respectively (Figure 4B).
Differential Gene Expression
To assess which genes were differentially expressed, the raw reads of all libraries were mapped onto the assembled transcripts with a mapping efficiency of 98.99 ± 0.25% (Supplementary Table S1). Figure 6 shows that the overall strongest response for both species was triggered by the desiccation treatment at 20°C while the weakest response was observed for the second rehydration period at 5°C. Furthermore, K. dissectum responded much stronger at 20°C to all treatments compared to K. flaccidum.
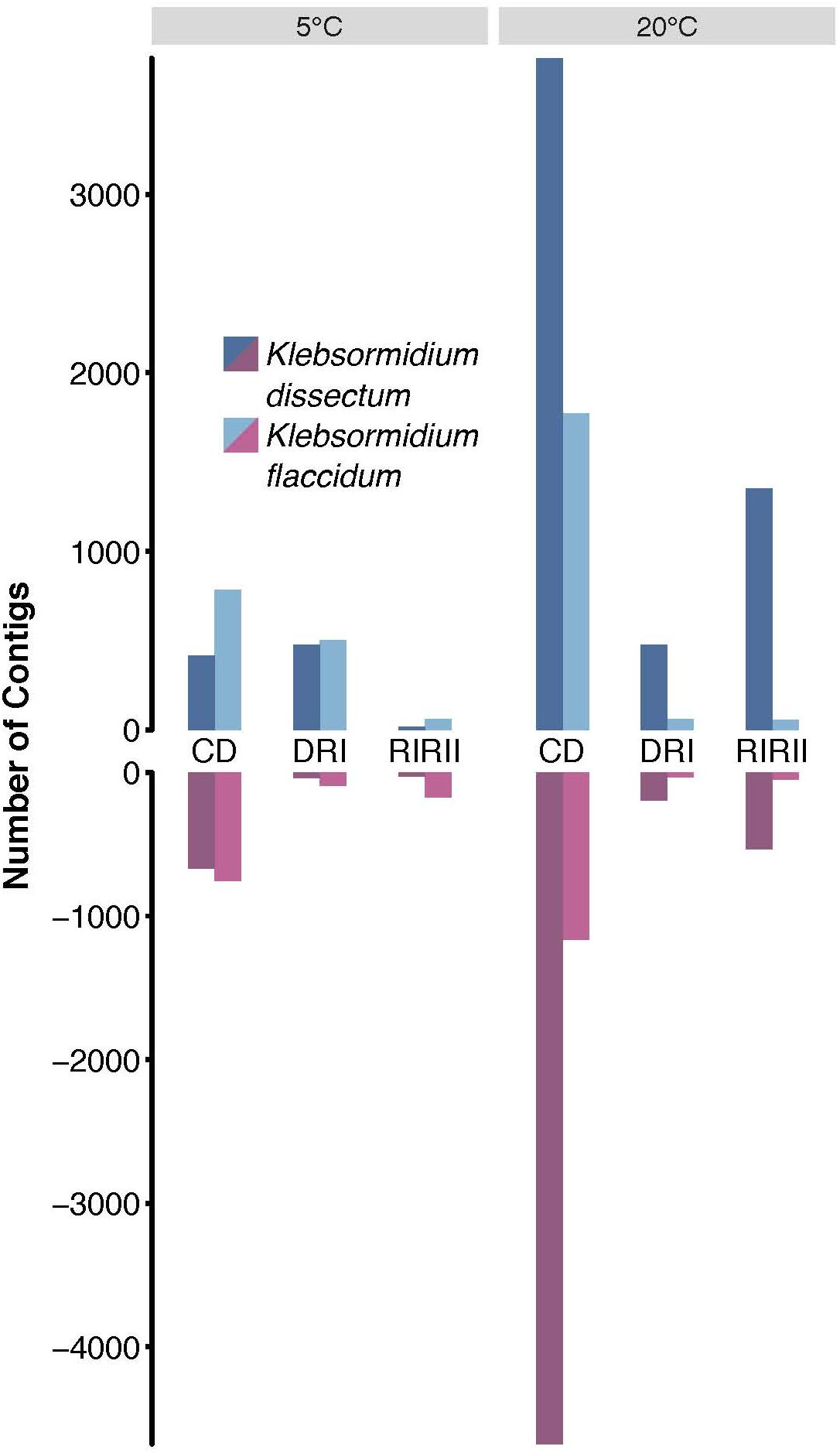
Figure 6. Differential gene expression analysis. The total of upregulated (dark and light blue) and downregulated (dark and light red) contigs of K. dissectum and K. flaccidum upon desiccation and during recovery at 5 and 20°C. Only contigs with a FDR of less or equal to ≤0.001 and a fold change of at least >4 were considered.
The largest overlap of differentially expressed transcripts at 5 and 20°C was observed for the comparison between controls to desiccated samples (Table 1). For K. dissectum, 204 and 329 genes were up- and down-regulated at both temperatures, respectively. K. flaccidum, on the other hand, increased the transcript pool of 311 genes and repressed 164 genes at both temperatures. Comparing desiccation and the first time point of rehydration, K. dissectum shared 66 up-regulated genes between the two temperatures while K. flaccidum exhibited an overlap of 16 up-regulated transcripts. Otherwise, no significant overlap was found.
Gene Set Enrichment Analysis
For KEGG enrichment analysis, Arabidopsis IDs were used and the results are displayed in Table 2. K. dissectum showed almost exclusively all enrichments at 20°C while K. flaccidum solely exhibited enriched pathways at 5°C. Upon desiccation, the down-regulated transcripts of K. dissectum showed an enrichment for the aminoacyl-tRNA biosynthesis, DNA replication, and photosynthesis while the ether lipid metabolism, the fatty acid degradation, the glycerophospholipid metabolism, and the butanoate metabolism were enriched in the up-regulated fraction of the transcriptome. At 5°C, K. flaccidum exhibited enrichment of the glycine, serine, and threonine metabolism in the down-regulated contigs during dehydration.
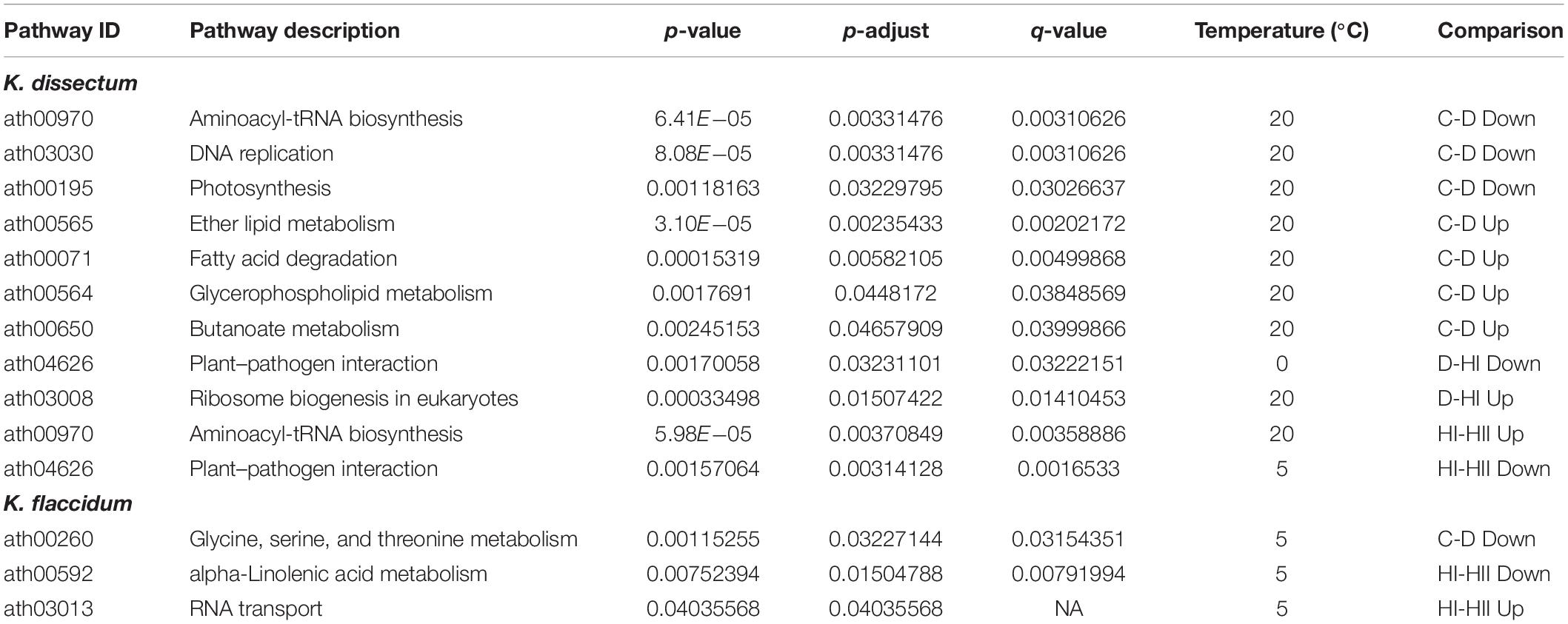
Table 2. A KEGG pathway enrichment analysis based on KO annotations was performed for the up- and downregulated transcripts in all analyzed groups using the ath_pathway map at KEGG.
The GO term enrichment analysis (Supplementary Table S2) revealed a complex regulation during treatment in both K. dissectum and K. flaccidum. Figure 7 displays the number of enriched GO terms in the root categories (BP, biological process; CC, cellular component; MF, molecular function) within the up- and down-regulated fraction of the transcriptomes in response to desiccation and rehydration. Dehydration treatment caused most enrichment in all three categories while the different time points after rehydration only showed a smaller number of enriched terms or none at all. In Figure 8, the relationship between the enriched GO terms is depicted as a network. Both species exhibited an enrichment of photosynthetic and related terms in the repressed transcripts at 20°C when desiccated (Figure 8A). K. dissectum also showed a high number of terms belonging to membrane modification and the carbohydrate metabolism in the up-regulated fraction of the transcriptome at 20°C upon dehydration. The desiccation treatment at 5°C caused an enrichment of cytoskeleton associated terms in the repressed transcript pool of both species. Rehydration resulted in an enrichment of transcription and translation-related GO terms in K. dissectum at 20°C (Figure 8B). At 5°C, K. flaccidum exhibited enriched terms associated with signaling and replication in the up- and down-regulated differentially expressed genes, respectively. Figure 8C indicates accumulation of photosynthetic terms at 20°C in K. flaccidum during the second recovery period.
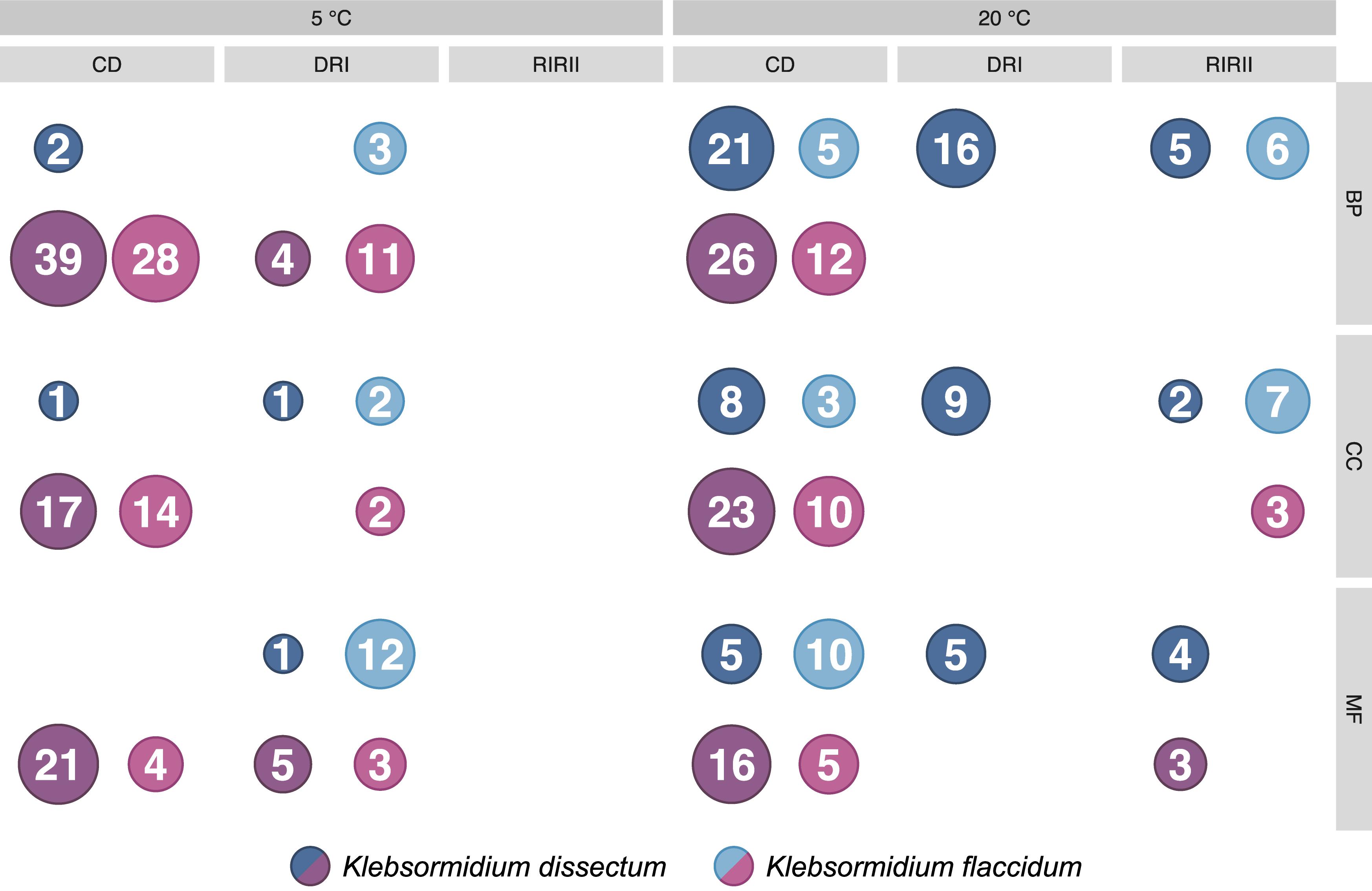
Figure 7. Summary of the GO enrichment analysis. The number of enriched categories, classified according to the three root categories, in the up- and down-regulated part of the transcriptomes are indicated. The detailed GO enrichment analysis is presented in Supplementary Table S2.
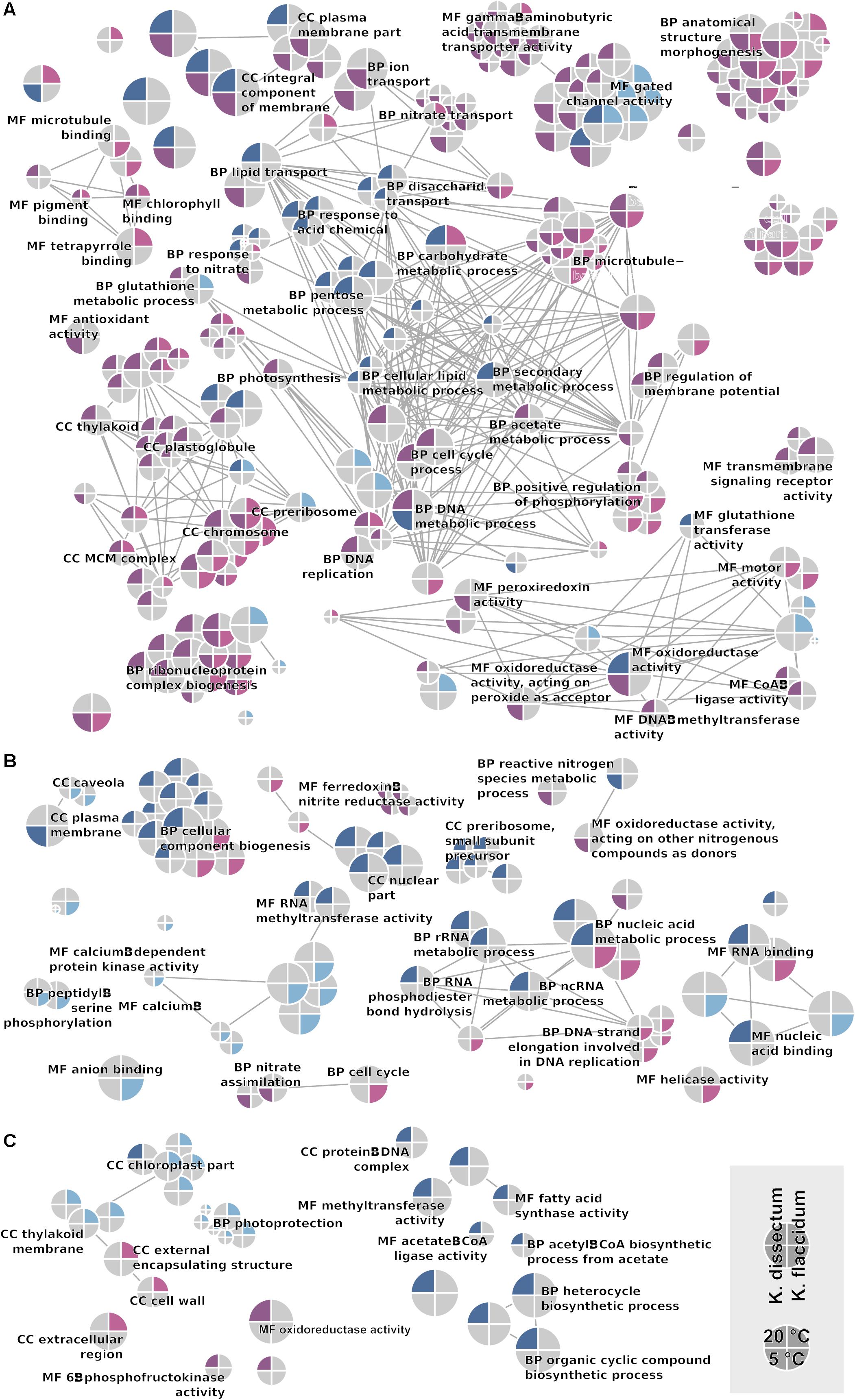
Figure 8. GO network displaying all enriched categories in both Klebsormidium species. Terms regulated at 5°C are indicated as the lower half of the circle, while terms regulated at 20°C are depicted in the upper half. Upregulation and downregulation are indicated with blue and red colors. K. dissectum is on the right while K. flaccidum is on the left side. The root categories are “biological process” (BP), “molecular function” (MF), and “cellular component” (CC). Edges depict shared higher categories. (A) Comparison of control cells with desiccated cells. (B) Comparison of desiccated cells with cell rehydrated for 2 h. (C) Comparison of cells with short (2 h) and long rehydration (24 h after start of the experiment) periods.
Functional Context of Individual Genes
Klebsormidium dissectum showed a strong repression of photosynthetic transcripts at 20°C upon desiccation (Supplementary Table S3). Different parts of PS I and II, subunits of the light harvesting complex, and the magnesium chelatase subunit were down-regulated and partly up-regulated during the second recovery phase. In contrast at 5°C, K. dissectum exhibited almost no response in photosynthetic gene expression. The PS II oxygen-evolving enhancer protein 2 is 2.4-fold down-regulated upon water withdrawal. Moreover, the expression of early light-induced proteins (ELIPs) was elevated both during desiccation treatments as well as recovery at 20°C. At 5°C the same genes were down-regulated upon dehydration. K. flaccidum induced the transcription of ELIPs during the second recovery phase at 20°C and the desiccation treatment at 5°C. ELIP1a.2 was up-regulated fivefold at 20°C when the stress treatment was applied. In contrast to K. dissectum, the expression of photosynthetic genes appeared to be almost unaffected in K. flaccidum.
Both Klebsormidium species indicated a strong decline in cell cycle-associated transcripts when desiccated at 20°C (Supplementary Table S3). Affected genes included condensin, the spindle assembly checkpoint (SAC) protein, the leishmanolysin-like peptidase, the replication machinery in case of K. dissectum, the centromere-associated protein HEC1, the TPR domain containing protein, the pre-initiation complex, and the DNA helicase for K. flaccidum as well as several mini-chromosome maintenance proteins in both cases. At 5°C, the effect for both organisms was minor during desiccation treatment. In the first recovery phase, K. flaccidum represses the pre-initiation complex and some of the mini-chromosome maintenance proteins.
Transcription- and translation-related transcripts in K. dissectum showed a decrease at 20°C upon water stress while rewetting had an increasing effect on these processes (Supplementary Table S3). The U3 small nucleolar RNA-associated protein 10, a transcription elongation and a splicing factor, a RNA recognition motif containing protein, the ribosomal biogenesis regulatory protein, as well as multiple tRNA synthetases responded to the treatment and recovery with changes in their expression patterns. In contrast, K. flaccidum experienced an enhanced expression of an S1 RNA-binding domain containing protein, the U3 small nucleolar RNA-associated protein 24, a WD domain containing protein, and a N-terminal acetyltransferase at 20°C when exposed to dehydration.
At 20°C, a high number of genes belonging to the carbohydrate metabolism of K. dissectum were differentially expressed upon desiccation (Supplementary Table S3). This trend was not observed at 5°C neither for K. flaccidum at any temperature. Both α- and β-amylase, 4-α-glucanotransferase, hexokinase, phosphoglucomutase, sucrose phosphorylase, sucrose synthase, sucrose-phosphate synthase, and sucrose-phosphatase were up-regulated.
Changes in gene expression were also observed in the glycerophospholipid and glycerolipid pathway as well as the fatty acid metabolism for K. dissectum (Supplementary Table S3). This alga increased the transcript pool of phospholipase D, phosphoethanolamine N-methyltransferase, digalactosyldiacylglycerol synthase, diacylglycerol O-acyltransferase, and membrane-bound O-acyl transferase family protein at 20°C upon dehydration. The former two genes were also up-regulated at 5°C during desiccation treatment. Moreover, the expression of acyl-CoA oxidase and long-chain acyl-CoA synthase was enhanced at 20°C in response to dehydration while the acetyl-CoA carboxylase 1 was repressed. During the second recovery phase an approximately ninefold increase of acyl-CoA dehydrogenase and acetyl-CoA carboxylase 1 transcripts was detected.
Treatment affected cytoskeleton-related genes in both K. dissectum and K. flaccidum (Supplementary Table S3). Desiccation caused decrease in expression of actin-related protein Arp2/3 complex subunit C3 (p21), a calcium-binding actin-bundling protein, tubulin, γ-, δ-, and ε-tubulin in K. dissectum at 20°C. Calcium-binding actin-bundling protein, tubulin, and γ-tubulin were up-regulated during the second recovery phase. At 5°C, the transcript pool of ε-tubulin and β-tubulin folding cofactor C was reduced during water stress. Desiccation at 20°C also caused katanin p60 ATPase-containing subunit to become up-regulated. K. flaccidum showed a down-regulation of actin and related proteins, kinesin-like protein and dynamin family protein at 5°C during water withdrawal while the katanin p60 ATPase-containing subunit was up-regulated.
Several stress-related genes were regulated during the desiccation treatment and recovery (Supplementary Table S3). K. dissectum enhanced the expression of the following proteins: HSP70, chaperone DnaJ-domain superfamily protein, ATP-dependent chaperone ClpB, DNA repair protein, catalase, NADPH-dependent thioredoxin reductase, thioredoxin superfamily protein, dehydroascorbate reductase, glutathione-S-transferase (GST), glutathione transferase, early-responsive to dehydration stress (ERD) family protein, ERD4, LEA proteins, ζ-carotene desaturase, phytoene dehydrogenase, and carotenoid isomerase at 20°C upon desiccation. During the recovery and at 5°C, some of these genes showed complex regulation patterns. In K. flaccidum, HSP70, chaperone DnaJ-domain superfamily protein, ATP-dependent chaperone, catalase, peroxiredoxin, dehydroascorbate reductase, GST family protein, cupin superfamily protein, and LEA proteins also have complex expression patterns at both temperatures and during the different treatments.
Discussion
Genetic Similarity of the Klebsormidium Species
The two Klebsormidium species examined in this study were not closely related. The phylogeny/systematics of the genus Klebsormidium has been investigated in recent years and all known taxa can be grouped into 14 different clades (Škaloud and Rindi, 2013; Mikhailyuk et al., 2015). K. dissectum belongs to clade E, the largest clade of morphologically similar strains. This clade also includes the previously studied K. nitens, for which the genome is available (Hori et al., 2014), while the earlier studied K. crenulatum (Holzinger et al., 2014) belongs to clade F. The other investigated strain K. flaccidum belongs to clade B/C. Both transcriptomes showed considerable differences to each other when compared with the published genome. The transcriptome sequences obtained for K. dissectum were more similar to the published genome in terms of genome coverage and sequence similarity (Figure 5) than the sequences obtained for K. flaccidum. Other Klebsormidium transcriptomes available were from one other species within clade F (K. crenulatum, Holzinger et al., 2014) and within clade E (K. subtile, Matasci et al., 2014). Therefore, the two new transcriptomes reveal for the first time the intra-genus genomic variability within Klebsormidium.
In the following sections we will discuss various cellular processes and their responses to the applied desiccation stress and recovery phases. However, before looking at the different processes it might be import to point out that changes in the transcriptome do not cover the complete response of a cell to a changing environment. Pathways might be switched on or off completely posttranslationally and changes in the proteome might be due to changes in the mRNA concentration, translation efficiencies, and/or protein degradation rates (Payne, 2015). Thus, for a complete picture, we would additionally need the proteomes, metabolomes, etc. under the different conditions.
Photosynthesis
Water is crucial for photosynthetic activity as it is necessary to maintain the structural integrity as well as the functionality of algal cells. Furthermore, water molecules play an essential role as electron donors in the electron transport chain of the photosynthesis. In the case of desiccation, photosynthetic organisms may exhibit one of three different coping strategies: escape, avoidance, or tolerance (Holzinger and Karsten, 2013; Fernández-Marín et al., 2016). Klebsormidium is a desiccation-tolerant organism which stalls photosynthetic activity during water stress and resumes the process as soon as water becomes available again (Elster et al., 2008; Karsten et al., 2013; Fernández-Marín et al., 2016). Our results from monitoring the effective quantum yield of PS II Y(II) for K. dissectum and K. flaccidum were in agreement with these studies as Y(II) dropped to zero during desiccation and rose again after water was supplied to the organisms. Moreover, we observed a temperature effect as it took longer to reach a Y(II) of zero at 5°C than at 20°C. The delayed photosynthetic inhibition at 5°C might have occurred due to the reduced water loss rates at lower temperatures (Yoder et al., 2010). However, the delay might also be explained by an increased dehydration tolerance which is caused by cold acclimation as both Klebsormidium species were cultivated at 5°C prior to the experiment. An increased desiccation tolerance resulting from an acclimation to lower temperatures has been described previously for higher plants (Cloutier and Siminovitch, 1982; Siminovitch and Cloutier, 1982; Koster and Bryant, 2006).
In response to desiccation at 20°C, K. dissectum repressed the expression of photosynthetic transcripts strongly while K. flaccidum did not respond as pronounced. In contrast, an alpine strain of K. crenulatum exhibited an up-regulation of photosynthetic genes upon desiccation at 20°C (Holzinger et al., 2014). Holzinger et al. (2014) argued that the increase in transcripts related to photosynthesis is a preparation for the recovery process after rehydration. However, the latter response is rather unusual. Carniel et al. (2016) compared a plethora of studies and found that most desiccation tolerant photosynthetic organisms, for example, the moss Syntrichia ruralis and the resurrection plant Craterostigma plantagineum, respond to water withdrawal by repression of genes involved in photosynthesis. The same holds true for the resurrection plant Myrothamnus flabellifolia which exhibits a down-regulation of transcripts encoding PSI and II probably to decrease the excitation energy and the associated ROS formation (Ma et al., 2015). This might also be the case for K. dissectum at 20°C. During the second phase of rehydration, K. dissectum reactivated the expression of those transcripts. The cyanobacterium Anabaena showed a similar recovery upon rewetting (Higo et al., 2007). Rather surprising is the observation that desiccation induced only minor changes in expression patterns regarding photosynthetic genes in K. dissectum at 5°C and in K. flaccidum at both temperatures. Hence, we assume that both Klebsormidium species cope with the water stress on different molecular levels, e.g., post-translational, which is invisible to expression studies.
Early light-induced proteins belong to the chlorophyll a/b-binding superfamily and act as photoprotectants upon abiotic stress, such as high light and ultraviolet (UV) radiation, but also desiccation (Zeng et al., 2002; Hutin et al., 2003; Hayami et al., 2015; Ma et al., 2015). These proteins are incorporated in the thylakoid membrane which they protect against photooxidative damage by binding free chlorophyll molecules and acting as sinks for excitation energy (Zeng et al., 2002; Hutin et al., 2003; Heddad et al., 2012; Hayami et al., 2015; Ma et al., 2015). K. dissectum at 20°C and K. flaccidum at 5°C exhibited an up-regulation of some ELIPs when dehydrated, most likely to protect the thylakoid membranes. Similar responses are known from other algae, such as the chlorophytes C. reinhardtii and Dunaliella bardawil, when exposed to high light stress (Lers et al., 1991; Teramoto et al., 2004). Other studies found that low temperatures induce ELIP expression too, for example, in the chlorophyte Dunaliella salina (Krol et al., 1997) and the streptophyte S. varians (Han and Kim, 2013). On the other hand, K. dissectum decreased the transcript pool of some ELIPs at 5°C upon desiccation treatment and K. flaccidum enhanced the expression of some ELIPs during the second phase of recovery at 20°C. The streptophytes Zygnema circumcarinatum (Rippin et al., 2017) and K. crenulatum (Holzinger et al., 2014) also showed a complex regulation of ELIPs when exposed to water stress which is typical for a multigene family (Zeng et al., 2002; Hutin et al., 2003).
Cell Cycle
Several studies have described the inhibiting effect of water stress on cell division and cell cycle processes (e.g., Mansour and Hallet, 1981; Bagniewska-Zadworna, 2008; Kakumanu et al., 2012). Desiccation causes the moss Polytrichum formosum to arrest the mitotic phase upon completion and, consequently, all cells enter the interphase (Mansour and Hallet, 1981). An arrest of the cell cycle is also visible in the desiccation transcriptome of K. crenulatum. This alga repressed both DNA replication and the cell cycle upon water withdrawal (Holzinger et al., 2014). The same holds true for several transcripts of K. dissectum and K. flaccidum when these algae were desiccated at 20°C. Condensin, a chromosome condensation complex which is required to maintain the structural integrity of the chromosomes (Smith et al., 2014), is down-regulated in K. dissectum. Moreover, the transcript pool of the SAC protein is decreased. SAC proteins assure equal segregation of chromosomes during cell division (Komaki and Schnittger, 2017). Dehydration also caused a repression of transcripts that are part of the replisome and several DNA replication licensing factors (RLFs) in both K. dissectum and K. flaccidum at 20°C. These RLFs are essential to the cell as they limit the duplication of DNA to exactly once per cell cycle (Julian Blow and Chong, 1996). In addition, K. flaccidum exhibited a down-regulation of RLF transcripts at 5°C during the first recovery phase.
Transcription and Translation
Severe desiccation can slow down and inhibit transcription and translation processes until water becomes available again (Farrant et al., 2007). At 20°C, K. dissectum showed signs of decelerated transcription and translation as a splicing factor and a number of aminoacyl-tRNA synthetases were down-regulated during dehydration treatment. Splicing is essential to convert pre-mRNA into mature mRNA (Chen and Cheng, 2012) while aminoacyl-tRNA synthetases are necessary to couple the correct amino acids to the corresponding tRNA (McClain, 1993). Holzinger et al. (2014) also observed a repressed gene expression of the tRNA-aminoacyl biosynthesis in K. crenulatum during desiccation. During recovery, K. dissectum induces the expression of both transcription- and translation-related transcripts, such as the transcription elongation factor and the ribosomal biogenesis regulatory protein (RRS1), again. Transcription elongation factors stimulate the elongation of the RNA strand by means of different mechanisms (Kim et al., 2007) while RRS1 affects the processing of pre-rRNA and ribosome assembly (Tsuno et al., 2000). Surprisingly, transcripts related to transcription and translation were up-regulated at 20°C in the desiccated filaments of K. flaccidum. The S1 RNA-binding domain protein (Young and Karbstein, 2011) and the U3 small nucleolar RNA-associated protein 24 (Zhang et al., 2013), for example, are involved in the ribosome assembly and WD domain containing proteins play a role in RNA processing (Stirnimann et al., 2010).
Carbohydrate Metabolism
Many plants and algae accumulate certain sugars, such as sucrose and trehalose, in response to desiccation as these carbohydrates serve as low-molecular-weight osmolytes which counteract water potential stress (Ma et al., 2015; Fernández-Marín et al., 2016) and also have a role in ROS protection (Pommerrenig et al., 2018). Concentrating osmoprotectants in the cell establishes a negative osmotic potential, retains water within the cell, and protects both membranes and proteins (Karsten, 2012). An Antarctic strain of the chlorophyte Trebouxia (Sadowsky et al., 2016), for example, increased, similar to desiccation-tolerant plants (Ramanjulu and Bartels, 2002; Dinakar and Bartels, 2013), the sucrose concentration in the cell when desiccated. The same holds true for the streptophyte Z. circumcarinatum which enhanced starch degradation and sucrose formation upon dehydration treatment (Rippin et al., 2017). Our results for K. dissectum at 20°C are in agreement with these studies as the complete pathway from starch to sucrose appeared to be up-regulated when the alga is exposed to water withdrawal. Holzinger et al. (2014) reported similar findings for K. crenulatum. This streptophyte increased the transcript pool of sucrose synthase and sucrose phosphate synthase upon desiccation (Holzinger et al., 2014). However, no effect was visible at 5°C in K. dissectum which could point to a previous sucrose accumulation during the cultivation at lower temperature (e.g., frost hardening). Manabu et al. (2008) reported that cold-acclimated filaments of K. flaccidum contained an increased amount of sucrose, which enhanced the freezing tolerance of the alga. A similar effect is known from Chlorella vulgaris which accumulated raffinose in response to cold shock treatment (Salerno and Pontis, 1989).
Membranes and Lipid Metabolism
As biomembranes are the primary target of stressors, such as low temperatures and dehydration, they are often modified in order to maintain membrane integrity and fluidity (Dinakar and Bartels, 2013; Valledor et al., 2013; Perlikowski et al., 2016). In C. reinhardtii, low temperature leads to two major changes in membrane composition (decrease of the lipophilic fraction and an increase in polyunsatturated fatty acids, Valledor et al., 2013). Similarly, an Antarctic ice microalga Chlamydomonas sp. increased the percentage of unsaturated fatty acids in the chloroplast membrane (Yi-Bin et al., 2017). A similar trend is visible in the desiccation transcriptome of Z. circumcarinatum as the glycero- and glycerophospholipid metabolism is activated (Rippin et al., 2017). Our data indicate that K. dissectum also responded with membrane modifications during water withdrawal as certain enzymes, for example the digalactosyldiacylglycerol synthase and diacylglycerol O-acyltransferase which are both part of the glycerolipid pathway, were up-regulated. Holzinger et al. (2014) detected an induction of the same genes in K. crenulatum upon dehydration. Another important protein involved in stress protection is phospholipase D which has been reported to be up-regulated in the chlorophyte Asterochloris erici (Gasulla et al., 2013) as well as in the streptophyte Z. circumcarinatum during desiccation (Rippin et al., 2017). K. dissectum also showed an induction of phospholipase D both at 5 and 20°C when dehydrated. Moreover, the desiccation transcriptome of K. dissectum suggested changes in fatty acid metabolism at 20°C. The acyl-CoA oxidase, which is part of both the β-oxidation and biosynthesis of unsaturated fatty acids, was up-regulated (Ci et al., 2015). The same holds true for two long chain acyl-CoA synthetases. These enzymes activate fatty acids and may prepare them for elongation, desaturation, lipid synthesis, and β-oxidation (Jia et al., 2016). In contrast, the acetyl-CoA carboxylase 1 is repressed upon water stress but up-regulated again during recovery. Acetyl-CoA carboxylase plays a major role in the biosynthesis of lipids as the enzyme carboxylates acetyl-CoA to form malonyl-CoA (Huerlimann and Heimann, 2013). Recovery also leads to an increase in expression of the acyl-CoA dehydrogenase, another enzyme involved in β-oxidation of fatty acids (Tan and Lee, 2016). K. flaccidum did not show significant regulations of these metabolic pathways.
Cytoskeleton
In response to desiccation, tolerant plants and algae dismantle their cytoskeleton to survive the shrinkage of the cell (Proctor et al., 2007; Pressel and Duckett, 2010; Blaas and Holzinger, 2017). For example, the streptophyte alga K. crenulatum started to disintegrate the F-actin network after 20 min of dehydration (Blaas and Holzinger, 2017). A similar effect is probably also occurring in K. dissectum at both temperatures and in K. flaccidum at 5°C during water stress as transcripts encoding components of the cytoskeleton, such as actin and tubulin, are down-regulated. Moreover, in K. dissectum other proteins regulating the actin filament network are also downregulated. The expression of components of the Arp2/3 complex, which is responsible for the initiation of filament polymerization (Goley and Welch, 2006), was repressed. In addition the calcium-binding actin-bundling protein, an enzyme organizing actin filaments into dense bundles (Pikzack et al., 2005), was down-regulated. K. flaccidum, on the other hand, decreased the expression level of the motor protein kinesin which is an essential part of intracellular transport. The expression of the microtubule-severing enzyme katanin (Roll-Mecak and McNally, 2010) was enhanced in K. dissectum and K. flaccidum at both investigated temperatures providing a clear indication for the dismantling of the cytoskeleton during desiccation. During recovery only minor effects were visible in the transcriptomes except for K. dissectum at 20°C which increased the transcript pool of the above calcium-binding actin-bundling protein and two tubulin proteins.
Protection Mechanisms
Chaperones and HSPs are an important part of the stress response as they assist with the refolding of misfolded proteins and protect them from aggregation which is generally associated with abiotic stress (Wang et al., 2004; Al-Whaibi, 2011). Heat shock, for instance, causes elevated transcript levels of HSPs in the green alga C. reinhardtii (Schulz-Raffelt et al., 2007; Kobayashi et al., 2014) and the red alga Cyanidioschyzon merolae (Kobayashi et al., 2014). HSPs are also induced by other stressors such as light stress in Chlamydomonas (von Gromoff et al., 1989), and desiccation stress in the green algae A. erici (Gasulla et al., 2013) and Z. circumcarinatum (Rippin et al., 2017). Our data indicate that chaperones and HSPs were also involved in the desiccation stress response of K. dissectum and K. flaccidum. HSP70, for example, was up-regulated in K. dissectum upon dehydration at 20°C and repressed during the second recovery phase. Dehydration had a similar effect in the mosses Physcomitrella patens (Tang et al., 2016) and Sanionia uncinata (Park et al., 2018). However, HSPs are not always accumulated during water stress, e.g., in the lichen photobiont Trebouxia gelatinosa (Banchi et al., 2018). The same holds true for K. flaccidum during desiccation treatment which increased the HSP70 transcript pool during the first recovery phase at 20°C. This trend was also observed in the moss Bryum argenteum 2 h after rehydration (Gao et al., 2015).
Many other chaperones also respond to desiccation stress. For example, the chaperone DnaJ was up-regulated during water stress in K. dissectum at 20°C and in K. flaccidum at 5°C. DnaJ proteins are important co-chaperones that are induced upon stress exposure (Wang et al., 2014). Similarly, Z. circumcarinatum increased DnaJ expression upon water withdrawal (Rippin et al., 2017).
Another important group of chaperones are ClpB proteins which are able to reverse protein misfolding caused by abiotic stressors (Lee et al., 2003). Drought stress, for example, induces the expression of ClpB proteins in Oryza sativa (Hu et al., 2009) and the desiccation transcriptome of Z. circumcarinatum showed an up-regulation of the chaperone ClpB1 (Rippin et al., 2017). Upon dehydration and rehydration at 20°C, K. dissectum revealed an increased transcript level of the chaperone ClpB probably to counteract the effect of protein misfolding and aggregation. At 5°C, the alga induced the expression during the first recovery phase.
Abiotic stressors, such as high light, UV radiation, cold stress, and desiccation, may cause the formation of harmful ROS making the synthesis of appropriate scavengers crucial for the survival of the cell (Cruz de Carvalho, 2008). Superoxide dismutase and catalase, for example, convert to H2O2 and, subsequently, to H2O which is no longer harmful to the cell. K. dissectum induced the expression of catalase at 20°C during stress treatment while K. flaccidum increased the transcript level at 5°C upon desiccation and decreased it immediately during the first phase of recovery. These data suggest an accumulation of ROS during stress treatment.
Plant thioredoxins are major players in the oxidative stress response (Vieira Dos Santos and Rey, 2006). We observed that thioredoxins as well as the thioredoxin reductases are up-regulated by K. dissectum exposed to water stress at 20°C. An elevated expression of peroxiredoxin was detected for K. flaccidum during the second recovery phase at 20°C. Another important cellular ROS scavenger is ascorbate which is regenerated by the dehydroascorbate reductase (Cruz de Carvalho, 2008). Upon dehydration, the transcript pool of this enzyme was increased by K. dissectum at 20°C as well as by K. flaccidum at both temperatures supporting an enrichment of ROS in the stressed filaments. Moreover, Le Martret et al. (2011) found that overexpression of the dehydroascorbate reductase and the GST in tobacco leads to an enhanced tolerance against salt and cold stress. GST is involved in the detoxification of the cell by conjugating glutathione to hydrophobic substances (Strange et al., 2001). Z. circumcarinatum also enhances GST expression upon desiccation stress (Rippin et al., 2017). At 20°C, the dehydration treatment resulted in an up-regulation of GST in K. dissectum while K. flaccidum induced the expression at both temperatures. ROS formation is also linked to the denaturation of biomolecules such as lipids, proteins, and nucleic acids (Cruz de Carvalho, 2008). Thus, the transcription of DNA repair proteins, such as the Nijmegen breakage syndrome 1 protein by Z. circumcarinatum upon desiccation, becomes necessary to minimize the destructive effect of ROS (Rippin et al., 2017). During water stress at both temperatures, K. dissectum induced the DNA repair protein confirming this effect. K. dissectum also showed an up-regulation of the phytoene desaturase, the ζ-carotene desaturase, and the carotenoid isomerase during dehydration at 20°C while the latter two are repressed at 5°C. These three enzymes are involved in the biosynthesis of carotenoids (Wurtzel et al., 2003), which protect the cell against oxidative damage (Ramel et al., 2012).
Cupins are a highly diverse protein family that may be involved in floral development, embryogenesis, as well as biotic and abiotic stress response (Dunwell et al., 2001). A cupin superfamily protein was up-regulated in K. flaccidum during the first recovery phase at 20°C and upon desiccation and the first rewetting at 5°C. We propose that cupins originally played a role in desiccation stress response. Possible also in plant seeds, cupins may not only play a role as storage protein, but also to achieve desiccation tolerance as in the algal ancestors of embryophytes. However, more studies are required to fully elucidate their function in Klebsormidium.
Early-responsive to dehydration stress genes (Kiyosue et al., 1994; Alves et al., 2011; Rai et al., 2012) have been studied and characterized in detail. These genes respond rapidly to abiotic stress, especially dehydration, and are functionally and spatially diverse (Alves et al., 2011; Rai et al., 2012). ERD2, for example, is a cytosolic HSP (Kiyosue et al., 1994) while ERD4 is a transmembrane protein, located in the chloroplast membrane, containing two tandem RNA-recognition motifs (Rai et al., 2012). During desiccation treatment at 20°C, K. dissectum enhanced the expression of ERD4 and another ERD protein to counteract the applied water stress. Holzinger et al. (2014) found, that in response to water withdrawal, K. crenulatum also induces the expression of ERD4 as well as a number of LEA proteins. LEA proteins play an essential role in the desiccation stress response as they may prevent protein aggregation and facilitate refolding (Hand et al., 2011). In response to dehydration, K. dissectum increased the transcription level of LEA proteins at both temperatures while K. flaccidum only responded at 5°C. Rippin et al. (2017) also observed an enhanced expression of LEA proteins in Z. circumcarinatum when subjected to desiccation. During the first recovery phase of both Klebsormidium species at 5°C, the LEA protein expression is further induced suggesting that these algae are still in a stressed state. The moss Tortula ruralis showed a similar response during desiccation and rehydration with LEA proteins being up-regulated during both treatments (Oliver et al., 2004).
Conclusion
Overall, the applied desiccation stress has a strong effect on gene expression in both K. dissectum and K. flaccidum compared to the recovery phases. Our findings suggest that Klebsormidium prepares for rehydration during desiccation stress and is able to protect most of its cellular structures from damage and denaturation. Several protection mechanisms, such as osmolyte accumulation, lipid modification, ROS scavenging, and chaperones, are activated to counteract the effects of water withdrawal. However, the degree of activation differed depending on the species and temperature. Higher temperature generates a more pronounced change of expression level while cold acclimated filaments are more persistent and show a weaker reaction. We hypothesize that Klebsormidium obtains an increased desiccation resilience and tolerance when cultivated at low temperatures. Furthermore, K. flaccidum appeared to respond less to desiccation at 20°C compared to K. dissectum. This observation might be linked to the environmental conditions at both sampling sites. The Arctic Svalbard has a more fluctuating climate (higher summer temperature, lower winter temperature) compared to the South Shetland Islands. In addition, there exist strong environmental differences between the Arctic and Antarctica as reflected in their cold water/low temperature history (Zacher et al., 2009). Antarctica is considered to have a much longer cold water history of about 23 Mio years (Sabbe et al., 2003), compared to the “young” geological cold water history of the Arctic (ca. 2 Mio years). These striking differences in temperature history have supported the development of many endemic, cold-adapted organisms in Antarctica (e.g., Gómez et al., 2009). Our data indicate that the Antarctic Klebsormidium is better adapted (as measured by the lower number of genes regulated upon application of desiccation stress) in coping with desiccation than its Arctic counterpart which is likely due to a number of protection mechanisms that are permanently in place. In conclusion, we could show that environmental acclimation as well as species have a major influence on gene expression and desiccation stress response in Klebsormidium.
Data Availability
All raw reads and the assemblies were submitted to the SRA database under the BioProject ID PRJNA500592.
Author Contributions
MR and NB performed the experiments, analyzed the data, and wrote the manuscript. UK and BB conceived the study, analyzed the data, and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the Deutsche Forschungsgemeinschaft (DFG) within the project “Polarcrust” (BE1779/18-1, KA899/23-1) as a part of the Priority Program 1158 “Antarctic Research.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are very grateful to Inga Hennl (Rostock) for her technical support during the RNA isolation. Furthermore, we would like to express our gratitude to Maroua Bouzid Elkhessairi (Cologne) for her assistance with the bioanalyzer and Janine Altmüller (Cologne) for her advice on the sequencing design.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01730/full#supplementary-material
FIGURE S1 | KEGG map overview showing all the mapped annotated KO IDs for K. dissectum.
FIGURE S2 | KEGG map overview showing all the mapped annotated KO IDs for K. flaccidum.
TABLE S1 | Statistics of the sequencing outcome. The number of read pairs, quality pairs, quality merged reads, quality single reads, and the mapping efficiency on the assembled transcripts are given for each replicate. Kd and Kf indicate K. dissectum and K. flaccidum. Five and 20 indicate the temperatures at which the experiment was performed. C, control cells; D, desiccated cells; HI, rewetted sample I (1 h); HII, rewetted sample II (24 h after the start of experiment).
TABLE S2 | GO enrichment analysis. All GO terms enriched are shown for all comparisons.
TABLE S3 | Selected genes regulated in K. dissectum and K. flaccidum upon desiccation and/or rehydration.
Footnotes
References
Altschul, S., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1006/jmbi.1990.9999
Alves, M. S., Fontes, E. P. B., and Fietto, L. G. (2011). EARLY RESPONSIVE to DEHYDRATION 15, a new transcription factor that integrates stress signaling pathways. Plant Signal. Behav. 6, 1993–1996. doi: 10.4161/psb.6.12.18268
Al-Whaibi, M. H. (2011). Plant heat-shock proteins: a mini review. J. King Saud Univ. Sci. 23, 139–150. doi: 10.1016/j.jksus.2010.06.022
Bagniewska-Zadworna, A. (2008). The root microtubule cytoskeleton and cell cycle analysis through desiccation of Brassica napus seedlings. Protoplasma 233, 177–185. doi: 10.1007/s00709-008-0001-z
Bairoch, A., and Apweiler, R. (1999). The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucleic Acids Res. 27, 49–54. doi: 10.1007/BF02248854
Banchi, E., Candotto Carniel, F., Montagner, A., Petruzzellis, F., Pichler, G., Giarola, V., et al. (2018). Relation between water status and desiccation-affected genes in the lichen photobiont Trebouxia gelatinosa. Plant Physiol. Biochem. 129, 189–197. doi: 10.1016/j.plaphy.2018.06.004
Barberousse, H., Ruiz, G., Gloaguen, V., Lombardo, R. J., Djediat, C., Mascarell, G., et al. (2006). Capsular polysaccharides secreted by building façade colonisers: characterisation and adsorption to surfaces. Biofouling 22, 361–370. doi: 10.1080/08927010601035803
Bellard, C., Cassey, P., and Blackburn, T. M. (2016). Alien species as a driver of recent extinctions. Biol. Lett. 12:20150623. doi: 10.1098/rsbl.2015.0623
Belnap, J. (2006). The potential roles of biological soil crusts in dryland hydrologic cycles. Hydrol. Process. 20, 3159–3178. doi: 10.1002/hyp.6325
Bisson, M. A., and Kirst, G. O. (1995). Osmotic acclimation and turgor pressure regulation in algae. Naturwissenschaften 82, 461–471. doi: 10.1007/BF01131597
Blaas, K., and Holzinger, A. (2017). F-actin reorganization upon de- and rehydration in the aeroterrestrial green alga Klebsormidium crenulatum. Micron 98, 34–38. doi: 10.1016/j.micron.2017.03.012
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible read trimming tool for Illumina NGS data. Bioinformatics 30, 2114–2120. doi: 10.1080/10473289.1999.10463977
Borchhardt, N., Baum, C., Mikhailyuk, T., and Karsten, U. (2017a). Biological soil crusts of Arctic Svalbard - water availability as potential controlling factor for microalgal biodiversity. Front. Microbiol. 8:1485. doi: 10.3389/fmicb.2017.01485
Borchhardt, N., Schiefelbein, U., Abarca, N., Boy, J., Mikhailyuk, T., Sipman, H. J. M., et al. (2017b). Diversity of algae and lichens in biological soil crusts of Ardley and King George islands, Antarctica. Antarct. Sci. 29, 229–237. doi: 10.1017/S0954102016000638
Büdel, B., Dulić, T., Darienko, T., Rybalka, N., and Friedl, T. (2016). “Cyanobacteria and algae of biological soil crusts,” in Biological Soil Crusts: An Organizing Principle in Drylands, eds B. Weber, B. Büdel, and J. Belnap (Cham: Springer International Publishing), 55–80. doi: 10.1007/978-3-319-30214-0_4
Carniel, F. C., Gerdol, M., Montagner, A., Banchi, E., De Moro, G., Manfrin, C., et al. (2016). New features of desiccation tolerance in the lichen photobiont Trebouxia gelatinosa are revealed by a transcriptomic approach. Plant Mol. Biol. 91, 319–339. doi: 10.1007/s11103-016-0468-5
Chen, H., and Cheng, S. (2012). Functional roles of protein splicing factors. Biosci. Rep. 32, 345–359. doi: 10.1042/BSR20120007
Chown, S. L., Huiskes, A. H. L., Gremmen, N. J. M., Lee, J. E., Terauds, A., Crosbie, K., et al. (2012). Continent-wide risk assessment for the establishment of nonindigenous species in Antarctica. Proc. Natl. Acad. Sci. U.S.A. 109, 4938–4943. doi: 10.1073/pnas.1119787109
Ci, D., Song, Y., Tian, M., and Zhang, D. (2015). Methylation of miRNA genes in the response to temperature stress in Populus simonii. Front. Plant Sci. 6:921. doi: 10.3389/fpls.2015.00921
Cloutier, Y., and Siminovitch, D. (1982). Correlation between cold- and drought-induced frost hardiness in winter wheat and rye varieties. Plant Physiol. 69, 256–258. doi: 10.1104/pp.69.1.256
Cruz de Carvalho, M. H. (2008). Drought stress and reactive oxygen species. Plant Signal. Behav. 3, 156–165. doi: 10.4161/psb.3.3.5536
Darby, B. J., and Neher, D. A. (2016). Microfauna within biological soil crusts. Ecol. Stud. 226, 139–157. doi: 10.1007/978-3-319-30214-0_8
Dinakar, C., and Bartels, D. (2013). Desiccation tolerance in resurrection plants: new insights from transcriptome, proteome and metabolome analysis. Front. Plant Sci. 4:482. doi: 10.3389/fpls.2013.00482
Donner, A., Glaser, K., Borchhardt, N., and Karsten, U. (2017). Ecophysiological response on dehydration and temperature in terrestrial Klebsormidium (Streptophyta) isolated from biological soil crusts in central European grasslands and forests. Microb. Ecol. 73, 850–864. doi: 10.1007/s00248-016-0917-3
Dunwell, J. M., Culham, A., Carter, C. E., Sosa-Aguirre, C. R., and Goodenough, P. W. (2001). Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26, 740–746. doi: 10.1016/s0968-0004(01)01981-8
Elster, J., Degma, P., Kováčik, L., Valentová, L., Šramková, K., and Batista Pereira, A. (2008). Freezing and desiccation injury resistance in the filamentous green alga Klebsormidium from the Antarctic, Arctic and Slovakia. Biologia 63, 843–851.
Evans, R. D., and Johansen, J. R. (1999). Microbiotic crusts and ecosystem processes. Crit. Rev. Plant Sci. 18, 183–225. doi: 10.1080/07352689991309199
Farrant, J. M., Brandt, W. F., Lindsey, G. G., Wolf Brandt, B., and George Lindsey, B. G. (2007). An overview of mechanisms of desiccation tolerance in selected angiosperm resurrection plants. Plant Stress 1, 72–84. doi: 10.1002/9780470376881.ch3
Fernández-Marín, B., Holzinger, A., and García-Plazaola, J. (2016). “Photosynthetic strategies of desiccation-tolerant organisms,” in Handbook of Photosynthesis, ed. M. Pessarakli (Boca Raton, FL: CRC Press), 663–681. doi: 10.1201/b19498-49
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Gao, B., Zhang, D., Li, X., Yang, H., Zhang, Y., and Wood, A. J. (2015). De novo transcriptome characterization and gene expression profiling of the desiccation tolerant moss Bryum argenteum following rehydration. BMC Genomics 16:416. doi: 10.1186/s12864-015-1633-y
Gasulla, F., Jain, R., Barreno, E., Guéra, A., Balbuena, T. S., Thelen, J. J., et al. (2013). The response of Asterochloris erici (Ahmadjian) Skaloud et Peksa to desiccation: a proteomic approach. Plant Cell Environ. 36, 1363–1378. doi: 10.1111/pce.12065
Goley, E. D., and Welch, M. D. (2006). The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7, 713–726. doi: 10.1038/nrm2026
Gómez, I., Wulff, A., Roleda, M. Y., Huovinen, P., Karsten, U., Quartino, M. L., et al. (2009). Light and temperature demands of marine benthic microalgae and seaweeds in polar regions. Bot. Mar. 52, 593–608. doi: 10.1515/BOT.2009.073
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Gu, Z., Gu, L., Eils, R., Schlesner, M., and Brors, B. (2014). Circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. doi: 10.1093/bioinformatics/btu393
Gustavs, L., Schumann, R., Eggert, A., and Karsten, U. (2009). In vivo growth fluorometry: accuracy and limits of microalgal growth rate measurements in ecophysiological investigations. Aquat. Microb. Ecol. 55, 95–104. doi: 10.3354/ame01291
Han, J. W., and Kim, G. H. (2013). An ELIP-like gene in the freshwater green alga, Spirogyra varians (Zygnematales), is regulated by cold stress and CO2 influx. J. Appl. Phycol. 25, 1297–1307. doi: 10.1007/s10811-013-9975-9
Han, J. W., Yoon, M., Kupper, F. D., Klochkova, T. A., Oh, J. S., Rho, J. R., et al. (2012). Accumulation of galloyl derivatives in a green alga, Spirogyra varians, in response to cold stress. J. Appl. Phycol. 25, 1279–1286. doi: 10.1007/s10811-011-9776-y
Hand, S. C., Menze, M. A., Toner, M., Boswell, L., and Moore, D. (2011). LEA proteins during water stress: not just for plants anymore. Annu. Rev. Physiol. 73, 115–134. doi: 10.1146/annurev-physiol-012110-142203
Hayami, N., Sakai, Y., Kimura, M., Saito, T., Tokizawa, M., Iuchi, S., et al. (2015). The responses of Arabidopsis Early Light-Induced Protein2 to ultraviolet B, high light, and cold stress are regulated by a transcriptional regulatory unit composed of two elements. Plant Physiol. 169, 840–855. doi: 10.1104/pp.15.00398
Hayashi, Y., and Shinozaki, A. (2012). Visualization of microbodies in Chlamydomonas reinhardtii. J. Plant Res. 125, 579–586. doi: 10.1007/s10265-011-0469-z
Heddad, M., Engelken, J., and Adamska, I. (2012). Light stress proteins in viruses, cyanobacteria and photosynthetic eukaryota. Adv. Photosynth. Respir. 34, 299–317. doi: 10.1007/978-94-007-1579-0_14
Herburger, K., and Holzinger, A. (2015). Localization and quantification of callose in the streptophyte green algae Zygnema and Klebsormidium: correlation with desiccation tolerance. Plant Cell Physiol. 56, 2259–2270. doi: 10.1093/pcp/pcv139
Higo, A., Suzuki, T., Ikeuchi, M., and Ohmori, M. (2007). Dynamic transcriptional changes in response to rehydration in Anabaena sp. PCC 7120. Microbiology 153(Pt 11), 3685–3694. doi: 10.1099/mic.0.2007/009233-0
Hochberg, B. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple zesting. J. R. Stat. Soc. 57, 289–300. doi: 10.2307/2346101
Holzinger, A., Kaplan, F., Blaas, K., Zechmann, B., Komsic-Buchmann, K., and Becker, B. (2014). Transcriptomics of desiccation tolerance in the streptophyte green alga Klebsormidium reveal a land plant-like defense reaction. PLoS One 9:e110630. doi: 10.1371/journal.pone.0110630
Holzinger, A., and Karsten, U. (2013). Desiccation stress and tolerance in green algae: consequences for ultrastructure, physiological and molecular mechanisms. Front. Plant Sci. 4:327. doi: 10.3389/fpls.2013.00327
Hori, K., Maruyama, F., Fujisawa, T., Togashi, T., Yamamoto, N., Seo, M., et al. (2014). Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5:3978. doi: 10.1038/ncomms4978
Hu, S.-P., Zhou, Y., Zhang, L., Zhu, X.-D., Li, L., Luo, L.-J., et al. (2009). Correlation and quantitative trait loci analyses of total chlorophyll content and photosynthetic rate of rice (Oryza sativa) under water stress and well-watered conditions. J. Integr. Plant Biol. 51, 879–888. doi: 10.1111/j.1744-7909.2009.00846.x
Huerlimann, R., and Heimann, K. (2013). Comprehensive guide to acetyl-carboxylases in algae. Crit. Rev. Biotechnol. 33, 49–65. doi: 10.3109/07388551.2012.668671
Hutin, C., Nussaume, L., Moise, N., Moya, I., Kloppstech, K., and Havaux, M. (2003). Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc. Natl. Acad. Sci. U.S.A. 100, 4921–4926. doi: 10.1073/pnas.0736939100
Jia, B., Song, Y., Wu, M., Lin, B., Xiao, K., Hu, Z., et al. (2016). Characterization of long-chain acyl-CoA synthetases which stimulate secretion of fatty acids in green algae Chlamydomonas reinhardtii. Biotechnol. Biofuels 9:184. doi: 10.1186/s13068-016-0598-7
Johnson, S. L., Kuske, C. R., Carney, T. D., Housman, D. C., Gallegos-Graves, L. V., and Belnap, J. (2012). Increased temperature and altered summer precipitation have differential effects on biological soil crusts in a dryland ecosystem. Glob. Change Biol. 18, 2583–2593. doi: 10.1111/j.1365-2486.2012.02709.x
Julian Blow, J., and Chong, J. P. J. (1996). Chapter 2 DNA replication and its control. Princ. Med. Biol. 5, 11–31. doi: 10.1016/S1569-2582(96)80106-1
Kakumanu, A., Ambavaram, M. M. R., Klumas, C., Krishnan, A., Batlang, U., Myers, E., et al. (2012). Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol. 160, 846–867. doi: 10.1104/pp.112.200444
Karsten, U. (2012). “Seaweed acclimation to salinity and desiccation stress,” in Seaweed Ecophysiology and Ecology, eds C. Wiencke and K. Bischof (Berlin: Springer), 87–107. doi: 10.1007/978-3-642-28451-9_5
Karsten, U., Herburger, K., and Holzinger, A. (2014). Dehydration, temperature, and light tolerance in members of the aeroterrestrial green algal genus Interfilum (Streptophyta) from biogeographically different temperate soils. J. Phycol. 50, 804–816. doi: 10.1111/jpy.12210
Karsten, U., and Holzinger, A. (2014). Green algae in alpine biological soil crust communities: acclimation strategies against ultraviolet radiation and dehydration. Biodivers. Conserv. 23, 1845–1858. doi: 10.1007/s10531-014-0653-2
Karsten, U., Pröschold, T., Mikhailyuk, T., and Holzinger, A. (2013). Photosynthetic performance of different genotypes of the green alga Klebsormidium sp. (Streptophyta) isolated from biological soil crusts of the Alps. Algol. Stud. 142, 45–62. doi: 10.1127/1864-1318/2013/0102
Kim, B., Nesvizhskii, A. I., Rani, P. G., Hahn, S., Aebersold, R., and Ranish, J. A. (2007). The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc. Natl. Acad. Sci. U.S.A. 104, 16068–16073. doi: 10.1073/pnas.0704573104
Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). Cloning of cDNAs for genes that are early-responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: identification of three ERDs as HSP cognate genes. Plant Mol. Biol. 25, 791–798. doi: 10.1007/BF00028874
Kobayashi, Y., Harada, N., Nishimura, Y., Saito, T., Nakamura, M., Fujiwara, T., et al. (2014). Algae sense exact temperatures: small heat shock proteins are expressed at the survival threshold temperature in Cyanidioschyzon merolae and Chlamydomonas reinhardtii. Genome Biol. Evol. 6, 2731–2740. doi: 10.1093/gbe/evu216
Komaki, S., and Schnittger, A. (2017). The spindle assembly checkpoint in Arabidopsis is rapidly shut off during severe stress. Dev. Cell 43, 172–185.e5. doi: 10.1016/j.devcel.2017.09.017
Kopylova, E., Noé, L., and Touzet, H. (2012). SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217. doi: 10.1093/bioinformatics/bts611
Koster, K. L., and Bryant, G. (2006). “Dehydration in model membranes and protoplasts: contrasting effects at low, intermediate and high hydrations,” in Cold Hardiness in Plants: Molecular Genetics, Cell Biology and Physiology, eds T. H. H. Chen, M. Uemura, and S. Fujikawa (Wallingford: CABI Publishing), 219–234. doi: 10.1079/9780851990590.0219
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Krol, M., Maxwell, D. P., and Huner, N. P. A. (1997). Exposure of Dunaliella salina to low temperature mimics the high light-induced accumulation of carotenoids and the carotenoid binding protein (Cbr). Plant Cell Physiol. 38, 213–216. doi: 10.1080/00220670109598764
Lagesen, K., Hallin, P., Rødland, E. A., Stærfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Bowtie: an ultrafast memory-efficient short read aligner. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Le Martret, B., Poage, M., Shiel, K., Nugent, G. D., and Dix, P. J. (2011). Tobacco chloroplast transformants expressing genes encoding dehydroascorbate reductase, glutathione reductase, and glutathione-S-transferase, exhibit altered anti-oxidant metabolism and improved abiotic stress tolerance. Plant Biotechnol. J. 9, 661–673. doi: 10.1111/j.1467-7652.2011.00611.x
Lee, J. R., Raymond, B., Bracegirdle, T. J., Chadès, I., Fuller, R. A., Shaw, J. D., et al. (2017). Climate change drives expansion of Antarctic ice-free habitat. Nature 547, 49–54. doi: 10.1038/nature22996
Lee, S., Sowa, M. E., Watanabe, Y. H., Sigler, P. B., Chiu, W., Yoshida, M., et al. (2003). The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell 115, 229–240.
Lers, A., Levy, H., and Zamir, A. (1991). Co-regulation of a gene homologous to early light-induced genes in higher plants and β-carotene biosynthesis in the alga Dunaliella bardawil. J. Biol. Chem. 266, 13698–13705. doi: 10.1039/b311328k
Li, B., and Dewey, C. N. (2014). “RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome,” in Bioinformatics: The Impact of Accurate Quantification on Proteomic and Genetic Analysis and Research, ed. R. Liu (Palm Bay, FL: Apple Academic Press), 295–336. doi: 10.1201/b16589
Liu, B., Yuan, J., Yiu, S. M., Li, Z., Xie, Y., Chen, Y., et al. (2012). COPE: an accurate k-mer-based pair-end reads connection tool to facilitate genome assembly. Bioinformatics 28, 2870–2874. doi: 10.1093/bioinformatics/bts563
Ma, C., Wang, H., Macnish, A. J., Estrada-Melo, A. C., Lin, J., Chang, Y., et al. (2015). Transcriptomic analysis reveals numerous diverse protein kinases and transcription factors involved in desiccation tolerance in the resurrection plant Myrothamnus flabellifolia. Hortic. Res. 2:15034. doi: 10.1038/hortres.2015.34
Manabu, N., Kenji, M., Matsuo, U., Nagao, M., Matsui, K., and Uemura, M. (2008). Klebsormidium flaccidum, a charophycean green alga exhibits cold acclimation that is closely associated with compatible solute accumulation and ultrastructural changes. Plant Cell Environ. 31, 872–885. doi: 10.1111/j.1365-3040.2008.01804.x
Mansour, K. S., and Hallet, J. N. (1981). Effect of desiccation on DNA synthesis and the cell cycle of the moss Polytrichum formosum. New Phytol. 87, 315–324. doi: 10.1111/j.1469-8137.1981.tb03202.x
Matasci, N., Hung, L.-H., Yan, Z., Carpenter, E. J., Wickett, N. J., Mirarab, S., et al. (2014). Data access for the 1,000 Plants (1KP) project. Gigascience 3:17. doi: 10.1186/2047-217X-3-17
McClain, W. H. (1993). Rules that govern tRNA identity in protein synthesis. J. Mol. Biol. 234, 257–280. doi: 10.1006/jmbi.1993.1582
Mikhailyuk, T., Glaser, K., Holzinger, A., and Karsten, U. (2015). Biodiversity of Klebsormidium (Streptophyta) from alpine biological soil crusts (Alps, Tyrol, Austria, and Italy). J. Phycol. 51, 750–767. doi: 10.1111/jpy.12316
Oliver, M. J., Dowd, S. E., Zaragoza, J., Mauget, S. A., and Payton, P. R. (2004). The rehydration transcriptome of the desiccation-tolerant bryophyte Tortula ruralis: transcript classification and analysis. BMC Genomics 5:89. doi: 10.1186/1471-2164-5-89
Park, M., Hong, S. G., Park, H., Lee, B., and Lee, H. (2018). Identification of reference genes for RT-qPCR in the Antarctic moss Sanionia uncinata under abiotic stress conditions. PLoS One 13:e0199356. doi: 10.1371/journal.pone.0199356
Payne, S. H. (2015). The utility of protein and mRNA correlation. Trends Biochem. Sci. 40, 1–3. doi: 10.1016/j.tibs.2014.10.010
Perlikowski, D., Kierszniowska, S., Sawikowska, A., Krajewski, P., Rapacz, M., Eckhardt, Ä., et al. (2016). Remodeling of leaf cellular glycerolipid composition under drought and re-hydration conditions in grasses from the Lolium-Festuca complex. Front. Plant Sci. 7:1027. doi: 10.3389/fpls.2016.01027
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Piercy, A. (1917). The structure and mode of life of a form of Hormidium flaccidum, A. Braun. Ann. Bot. 31, 513–537. doi: 10.1093/oxfordjournals.aob.a089661
Pikzack, C., Prassler, J., Furukawa, R., Fechheimer, M., and Rivero, F. (2005). Role of calcium-dependent actin-bundling proteins: characterization of Dictyostelium mutants lacking fimbrin and the 34-kilodalton protein. Cell Motil. Cytoskeleton 62, 210–231. doi: 10.1002/cm.20098
Pommerrenig, B., Ludewig, F., Cvetkovic, J., Trentmann, O., Klemens, P. A. W., and Neuhaus, H. E. (2018). In concert: orchestrated changes in carbohydrate homeostasis are critical for plant abiotic stress tolerance. Plant Cell Physiol. 59, 1290–1299. doi: 10.1093/pcp/pcy037
Pressel, S., and Duckett, J. G. (2010). Cytological insights into the desiccation biology of a model system: moss protonemata. New Phytol. 185, 944–963. doi: 10.1111/j.1469-8137.2009.03148.x
Proctor, M. C. F., Oliver, M. J., Wood, A. J., Alpert, P., Stark, L. R., Cleavitt, N. L., et al. (2007). Desiccation-tolerance in bryophytes: a review. Bryologist 110, 595–621.
Pushkareva, E., Johansen, J. R., and Elster, J. (2016). A review of the ecology, ecophysiology and biodiversity of microalgae in Arctic soil crusts. Polar Biol. 39, 2227–2240. doi: 10.1007/s00300-016-1902-5
Rai, A., Suprasanna, P., D’Souza, S. F., and Kumar, V. (2012). Membrane topology and predicted RNA-binding function of the ‘Early Responsive to Dehydration (ERD4)’ plant protein. PLoS One 7:e32658. doi: 10.1371/journal.pone.0032658
Ramanjulu, S., and Bartels, D. (2002). Drought- and desiccation-induced modulation of gene expression in plants. Plant Cell Environ. 25, 141–151. doi: 10.1046/j.0016-8025.2001.00764.x
Ramel, F., Birtic, S., Cuiné, S., Triantaphylidès, C., Ravanat, J.-L., and Havaux, M. (2012). Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiol. 158, 1267–1278. doi: 10.1104/pp.111.182394
Rippin, M., Becker, B., and Holzinger, A. (2017). Enhanced desiccation tolerance in mature cultures of the streptophytic green alga Zygnema circumcarinatum revealed by transcriptomics. Plant Cell Physiol. 58, 2067–2084. doi: 10.1093/pcp/pcx136
Rippin, M., Komsic-Buchmann, K., and Becker, B. (2016). RNA isolation from biological soil crusts: methodological aspects. Algol. Stud. 151/152, 21–37. doi: 10.1127/algol_stud/2016/0256
Rippin, M., Lange, S., Sausen, N., and Becker, B. (2018). Biodiversity of biological soil crusts from the polar regions revealed by metabarcoding. FEMS Microbiol. Ecol. 94:fiy036. doi: 10.1093/femsec/fiy036
Robinson, M., Mccarthy, D., and Smyth, G. K. (2010). edgeR: a differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Roll-Mecak, A., and McNally, F. J. (2010). Microtubule-severing enzymes. Curr. Opin. Cell Biol. 22, 96–103. doi: 10.1016/j.ceb.2009.11.001
Sabbe, K., Verleyen, E., Hodgson, D. A., Vanhoutte, K., and Vyverman, W. (2003). Benthic diatom flora of freshwater and saline lakes in the Larsemann Hills and Rauer Islands, East Antarctica. Antarct. Sci. 15, 227–248. doi: 10.1017/S095410200300124X
Sadowsky, A., Mettler-Altmann, T., and Ott, S. (2016). Metabolic response to desiccation stress in strains of green algal photobionts (Trebouxia) from two Antarctic lichens of southern habitats. Phycologia 55, 703–714. doi: 10.2216/15-127.1
Salerno, G. L., and Pontis, H. G. (1989). Raffinose synthesis in Chlorella vulgaris cultures after a cold shock. Plant Physiol. 89, 648–651. doi: 10.1104/pp.89.2.648
Schmieder, R., and Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026
Schulz-Raffelt, M., Lodha, M., and Schroda, M. (2007). Heat shock factor 1 is a key regulator of the stress response in Chlamydomonas. Plant J. 52, 286–295. doi: 10.1111/j.1365-313X.2007.03228.x
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Siminovitch, D., and Cloutier, Y. (1982). Twenty-four-hour induction of freezing and drought tolerance in plumules of winter rye seedlings by desiccation stress at room temperature in the dark. Plant Physiol. 69, 250–255. doi: 10.1104/pp.69.1.250
Škaloud, P., and Rindi, F. (2013). Ecological differentiation of cryptic species within an asexual protist morphospecies: a case study of filamentous green Alga Klebsormidium (Streptophyta). J. Eukaryot. Microbiol. 60, 350–362. doi: 10.1111/jeu.12040
Smith, S. J., Osman, K., and Franklin, F. C. H. (2014). The condensin complexes play distinct roles to ensure normal chromosome morphogenesis during meiotic division in Arabidopsis. Plant J. 80, 255–268. doi: 10.1111/tpj.12628
Sonnhammer, E. L. L., Eddy, S. R., and Durbin, R. (1997). Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28, 405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l
Starr, R. C., and Zeikus, J. A. (1993). UTEX - the culture collection of algae at the university of Texas at Austin 1993 list of cultures. J. Phycol. 29, 1–106. doi: 10.1111/j.0022-3646.1993.00001.x
Stirnimann, C. U., Petsalaki, E., Russell, R. B., and Müller, C. W. (2010). WD40 proteins propel cellular networks. Trends Biochem. Sci. 35, 565–574. doi: 10.1016/j.tibs.2010.04.003
Strange, R. C., Spiteri, M. A., Ramachandran, S., and Fryer, A. A. (2001). Glutathione-S-transferase family of enzymes. Mutat. Res. 482, 21–26.
Supek, F., Bošnjak, M., Škunca, N., and Šmuc, T. (2011). Revigo summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. doi: 10.1371/journal.pone.0021800
Tan, K. W. M., and Lee, Y. K. (2016). The dilemma for lipid productivity in green microalgae: importance of substrate provision in improving oil yield without sacrificing growth. Biotechnol. Biofuels 9:255.
Tang, T., Yu, A., Li, P., Yang, H., Liu, G., and Liu, L. (2016). Sequence analysis of the Hsp70 family in moss and evaluation of their functions in abiotic stress responses. Sci. Rep. 6:33650. doi: 10.1038/srep33650
Teramoto, H., Itoh, T., and Ono, T. A. (2004). High-intensity-light-dependent and transient expression of new genes encoding distant relatives of light-harvesting chlorophyll-a/b proteins in Chlamydomonas reinhardtii. Plant Cell Physiol. 45, 1221–1232. doi: 10.1093/pcp/pch157
Tsuno, A., Miyoshi, K., Tsujii, R., Miyakawa, T., and Mizuta, K. (2000). RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 2066–2074. doi: 10.1128/MCB.20.6.2066-2074.2000
Valledor, L., Furuhashi, T., Hanak, A.-M., and Weckwerth, W. (2013). Systemic cold stress adaptation of Chlamydomonas reinhardtii. Mol. Cell. Proteomics 12, 2032–2047. doi: 10.1074/mcp.M112.026765
Vieira Dos Santos, C., and Rey, P. (2006). Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci. 11, 329–334. doi: 10.1016/j.tplants.2006.05.005
von Gromoff, E. D., Treier, U., and Beck, C. F. (1989). Three light-inducible heat shock genes of Chlamydomonas reinhardtii. Mol. Cell. Biol. 9, 3911–3918. doi: 10.1128/MCB.9.9.3911
Wang, G., Cai, G., Kong, F., Deng, Y., Ma, N., and Meng, Q. (2014). Overexpression of tomato chloroplast-targeted DnaJ protein enhances tolerance to drought stress and resistance to Pseudomonas solanacearum in transgenic tobacco. Plant Physiol. Biochem. 82, 95–104. doi: 10.1016/j.plaphy.2014.05.011
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Williams, L., Borchhardt, N., Colesie, C., Baum, C., Komsic-Buchmann, K., Rippin, M., et al. (2017). Biological soil crusts of Arctic Svalbard and of Livingston Island. Polar Biol. 40, 399–411. doi: 10.1007/s00300-016-1967-1
Woelfel, J., Schoknecht, A., Schaub, I., Enke, N., Schumann, R., and Karsten, U. (2014). Growth and photosynthesis characteristics of three benthic diatoms from the brackish southern Baltic Sea in relation to varying environmental conditions. Phycologia 53, 639–651. doi: 10.2216/14-019.1
Wurtzel, E. T., Matthews, P. D., and Luo, R. (2003). Maize phytoene desaturase and ζ-carotene desaturase catalyse a poly-Z desaturation pathway: implications for genetic engineering of carotenoid content among cereal crops. J. Exp. Bot. 54, 2215–2230. doi: 10.1093/jxb/erg235
Yi-Bin, W., Fang-Ming, L., Xiu-Fang, Z., Ai-Jun, Z., Bin, W., Zhou, Z., et al. (2017). Composition and regulation of thylakoid membrane of Antarctic ice microalgae Chlamydomonas sp. ICE-L in response to low-temperature environment stress. J. Mar. Biol. Assoc. UK 97, 1241–1249. doi: 10.1017/S0025315416000588
Yoder, J. A., Chambers, M. J., Tank, J. L., and Keeney, G. D. (2010). High temperature effects on Water loss and survival examining the hardiness of female adults of the spider beetles, Mezium affine and Gibbium aequinoctiale. J. Insect Sci. 9, 1–8. doi: 10.1673/031.009.6801
Yoshitake, S., Uchida, M., Koizumi, H., Kanda, H., and Nakatsubo, T. (2010). Production of biological soil crusts in the early stage of primary succession on a High Arctic glacier foreland. New Phytol. 186, 451–460. doi: 10.1111/j.1469-8137.2010.03180.x
Young, C. L., and Karbstein, K. (2011). The roles of S1 RNA-binding domains in Rrp5’s interactions with pre-rRNA. RNA 17, 512–521. doi: 10.1261/rna.2458811
Young, M. D., Davidson, N., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). goseq: gene Ontology testing for RNA-seq datasets Reading data. Genome Biol. 11:R14.
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi: 10.1089/omi.2011.0118
Zacher, K., Rautenberger, R., Hanelt, D., Wulff, A., and Wiencke, C. (2009). The abiotic environment of polar marine benthic algae. Bot. Mar. 52, 483–490. doi: 10.1515/BOT.2009.082
Zelikova, T. J., Housman, D. C., Grote, E. E., Neher, D. A., and Belnap, J. (2012). Warming and increased precipitation frequency on the Colorado Plateau: implications for biological soil crusts and soil processes. Plant Soil 355, 265–282. doi: 10.1007/s11104-011-1097-z
Zeng, Q., Chen, X., and Wood, A. J. (2002). Two early light-inducible protein (ELIP) cDNAs from the resurrection plant Tortula ruralis are differentially expressed in response to desiccation, rehydration, salinity, and high light. J. Exp. Bot. 53, 1197–1205. doi: 10.1093/jexbot/53.371.1197
Keywords: cold acclimation, desiccation stress, Klebsormidium, polar regions, transcriptomics
Citation: Rippin M, Borchhardt N, Karsten U and Becker B (2019) Cold Acclimation Improves the Desiccation Stress Resilience of Polar Strains of Klebsormidium (Streptophyta). Front. Microbiol. 10:1730. doi: 10.3389/fmicb.2019.01730
Received: 03 April 2019; Accepted: 12 July 2019;
Published: 06 August 2019.
Edited by:
Jesse G. Dillon, California State University, Long Beach, United StatesReviewed by:
Haitham Sghaier, National Center for Nuclear Science and Technology, TunisiaMarc Warwick Van Goethem, Lawrence Berkeley National Laboratory, United States
Copyright © 2019 Rippin, Borchhardt, Karsten and Becker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ulf Karsten, dWxmLmthcnN0ZW5AdW5pLXJvc3RvY2suZGU=; Burkhard Becker, Yi5iZWNrZXJAdW5pLWtvZWxuLmRl
 Martin Rippin
Martin Rippin Nadine Borchhardt
Nadine Borchhardt Ulf Karsten
Ulf Karsten Burkhard Becker
Burkhard Becker