- 1Applied Microbial Processes and Environmental Health Research Group, Department of Microbiology, Faculty of Life Sciences, University of Benin, Benin City, Nigeria
- 2Department of Environmental Management and Toxicology, Faculty of Life Sciences, University of Benin, Benin City, Nigeria
- 3Sustainable Development Office, University of Benin, Benin City, Nigeria
Gastrointestinal illnesses continue to be a global public health risk. Exposure to foodborne Salmonella directly or indirectly through consumption of ready-to-eat seafood can be an important route of infection to humans. This study was designed to estimate the population cell density, prevalence, virulence gene signatures, and antibiotic resistance of Salmonella serovars from ready-to-eat shrimps. Ready-to-eat (RTE) shrimp samples were obtained from different open markets in Delta and Edo States, Nigeria from November 2016 to October 2017. We employed classical and polymerase chain reaction (PCR) approaches. The mean Salmonella species enumerated from the RTE shrimps ranged from −0.301 to 5.434 log10 cfu/g with 210/1440 (14.58%) of the RTE shrimp samples harbored Salmonella species. After biochemical and PCR approach, the identified isolates were Salmonella Enteritidis 11(24.4%), Salmonella Typhimurium 14 (31.1%) and other Salmonella spp. 20 (44.4%). All Salmonella species recovered were resistant to penicillin and erythromycin with 100% sensitivity to cefotaxime, cephalothin, colistin, and polymyxin B. Findings on the multidrug-resistant (MDR) profile showed that a total of 9/14 (64.3%) of Salmonella Enteritidis were resistant to 5 antibiotics which belongs to 3 different groups of antimicrobials with a multiple antibiotic-resistant (MAR) index of 0.21; while 3/11 (27.3%) of Salmonella Typhimurium were resistant to 11 antibiotics which belongs to 7 different groups of antimicrobials with a MAR index of 0.46. Virulence genes (spiA, sipB, invA, sif A, fljB, and sefA) and resistance genes (class 1 and II integrase, sul2, catB3, flor, tmp, blaTEM, strB, dfr1, and tetC) were also detected in some of the Salmonella species with variable percentage. This study indicates that ready-to-eat shrimps are probable reservoirs harboring Salmonella strains. The identified Salmonella isolates which exhibited virulence determinants and antibiotic-resistant coupled with high MAR index constitute a consumer health risk to the communities.
Introduction
Shrimps constitute a large proportion of crustaceans which varies in sizes (Orji et al., 2016) and have been described as the most significant seafood traded on a global scale (Oosterveer, 2006). The world shrimp production for both farmed and captured shrimp is ~6 million tons with 60% entering the world market. Shrimp has been reported to be the most essentially traded fishery product internationally as it translates to value. Yearly shrimp exports presently value above US$10 billion, or 16% of total fish product exports (Food Agriculture Organization of the United Nations, 2008). Shrimp makes up 20% value of exported fishery products for more than 20 years (CAC, 2002). Imports of shrimps into developed nations are responsible for about 40% trade of intra-developed countries, while approximately 60% comes from developing nations. From developing nation exports, 80% goes to developed nation with only 20% left behind (Josupeit, 2005). Shrimps are one of the important exported aquaculture products from the tropics. The interaction of microbial diversity that comes in connection with shrimps during harvesting and processing is a prospective public health threats as a consequence of disease and spoilage transmission.
The main disease causing serovars of Salmonella enterica subspecies enterica which are pathogenic to humans as a result of diverse seafood and non-seafood products include Salmonella Typhimurium and Salmonella Enteritidis (Ed-dra et al., 2017). Salmonellosis which is an infection of the intestinal epithelium is instigated by the Salmonella genus (Igbinosa and Beshiru, 2017; Beshiru et al., 2018). Within the United States more than 40,000 cases of salmonellosis are recounted yearly with seafood considered as one of the most significant source of Salmonella (Brands et al., 2005; Duran and Marshall, 2005). Contamination results when the salmonellae enter RTE food and replicate within the food, as a result of inadequate food preparation, poor storage temperatures, or cross-contamination (Skyberg et al., 2006). Hence, the occurrence of Salmonella in RTE seafood from open market is an important food safety risk.
Antimicrobial-resistant Salmonella serovars may result from the continuous usage of antimicrobials in food animal production, where these antimicrobial resistant Salmonella are therefore disseminated to humans, usually through contaminated food. The application of antimicrobials in aquaculture systems has led to the accumulation of antibiotic-resistance genes and antibiotic-resistant bacteria (Yano et al., 2011; Igbinosa, 2016). Antibiotics commonly used in agricultural/aquaculture systems in Nigeria are gentamycin, ivermectin, oxytetracycline, tylosin, septinomycin, and cephalosporin. Food and Drug Administration (FDA) has permitted the use of five different drugs (sulfamerazine, chorionic gonadotropin, florfenicol, oxytetracycline hydrochloride, oxytetracycline dihydrate, as well as combination of sulfadimethoxine and ormetoprim) in aquaculture so long as the seafood harbors less than the required maximum residue limit (Serrano, 2005). FDA has also approved two drugs: hydrogen peroxide and formalinwith no tolerance level set a (Serrano, 2005). Multidrug resistance (MDR) in Salmonella is of significant concern as treatment regimens may be challenging, thus making management of these disease difficult. Salmonella Typhimurium is one of the most widespread MDR Salmonella serovars recovered from humans and animals in the United States (Brunelle et al., 2013). The continuous rise and dissemination of antibiotic resistance phenotypes and determinants among Salmonella serovars has metamorphosed into a public health space. Notably, strains of Salmonella which have clinically phenotypic and genotypic resistance to antibiotic agents such as extended spectrum cephalosporins and fluoroquinolones, have been recovered from food animals (Li et al., 2013; Igbinosa, 2015). Within developing countries, overuse and misuse of antibiotics has led to the upsurge of MDR in Salmonella strains (Ed-dra et al., 2017).
Antibiotic-resistant Salmonella connected with cultivated Litopenaeus vannamei have been reported in Malaysia where Salmonella enterica serovar Corvallis recovered from shrimp revealed multiple and individual antibiotic resistance profiles (Banerjee et al., 2012). In Nigeria there are some reports that have revealed the occurrence of Salmonella species from numerous food types, with no study on the surveillance of Salmonella Typhimurium, Salmonella Enteritidis and other Salmonella spp. from RTE shrimps. Hence, the objective of this research was to determine the prevalence, multiple antibiotic resistance, virulence and antibiotic resistance genes of Salmonella serovars recovered from retail RTE-shrimps in Nigeria.
Materials and Methods
Study Area
The RTE shrimp were obtained from major open markets in Delta and Edo States, Nigeria. There are 3 Senatorial Districts in each of Edo and Delta State. Six different markets were assessed from each state which makes it a total of 12 markets with 2 markets from each Senatorial District. In Delta State, markets include Ughelli main market, Sapele market (Delta Central Senatorial District), Ogbegonogo market, Ashafor market Aniocha Asaba market (Delta North Senatorial District), main market Oleh Isoko, and Igbudu market Warri (Delta South Senatorial District). For Edo State, markets include New Benin market, Oba market (Edo South Senatorial District), Igarra market, Jattu market (Edo North Senatorial District), Uromi main market and Ekpoma market (Edo Central Senatorial District). The respective markets were chosen based on the strategic locations in their respective communities and are highly dense due to the population of individuals that patronizes these markets. The RTE shrimp that were collected from these markets were mainly tiger shrimps (Penaeus monodon) and pink shrimp (Penaeus notialis) and included smoked shrimps, dried shrimps, fried shrimps, sauced shrimps, and boiled shrimps.
Sample Collection
One thousand four hundred and forty RTE shrimp samples were obtained between November 2016 and October, 2017. Ten samples each were obtained from each of the respective 12 selected open markets (6 each from Delta and Edo States) culminating in the 1440 RTE samples. Samples were obtained based on the type of RTE shrimps available with respect to the sampling location. The RTE shrimp samples were obtained from the selected open markets with the aid of a sterile polythene bag. The polythene bags were immediately placed on ice pack and conveyed to the laboratory where microbiological analyses were carried out within 24 h after collection.
Enrichment, Enumeration and Isolation of Salmonella Species
This was carried out in line with the International Organization for Standardization (2017). Twenty-five grams of individual RTE shrimp samples was weighed and placed in a sterile homogenizer bag containing 225 mL of tryptone soy broth (TSB) (Merck, Darmstadt, Germany), as pre-enrichment and incubated at 37°C for 18–24 h. Before incubation, the stock suspension was serially diluted using sterile distilled water from 10−1 to 10−9. Dilution with 100 μL of each diluent aseptically platted in triplicates into xylose lysine deoxycholate (XLD) agar (Lab M, Lancashire, United Kingdom) and hektoen enteric agar (HEA) (Lab M, Lancashire, United Kingdom). This was followed with incubated at 37°C for 24–48 h where presumptive Salmonella species which appear as distinct green colonies with or without black centers were enumerated and expressed in log10 colony forming units per gram (log10 cfu/g). After incubation with the pre-enrichment broth with TSB, 100 μL were inoculated into 9.0 mL of selenite cysteine F Broth (Lab M, Lancashire, United Kingdom) and incubated at 37°C for 18–24 h. After incubation, 100 μL of the turbid suspension was inoculated into XLD and HEA and incubated at 37°C for 18–24 h where a maximum of 2 presumptive Salmonella colonies were selected and sub-cultured on a fresh XLD and HEA and incubated at 37°C for 18–24 h. Distinct colonies were further purified on tryptone soy agar (TSA) (Merck, Darmstadt, Germany) incubated at 37°C for 18–24 h. Isolates were transferred into a 1 mL TSA in an Eppendorf tube and incubated at 37°C for 24 h and stored in the refrigerator at 4°C until ready for further use.
Presumptive Identification of Salmonella Species
All Salmonella species were screened via biochemical (oxidase, catalase, indole, and sugar fermentation test, citrate), morphological (Gram reaction with 3% KOH test), and cultural (colony) characterization. Analytical Profile Index 20E (API 20E) was also used for the Salmonella species respectively according to the manufacturer's instructions (BioMerieux, Marcy-l'Étoile, France) using API lab plus software (bioMerieux, Marcy l'Etoile, France).
Genomic Deoxyribonucleic Acid (gDNA) Extraction Procedure
Genomic DNA from Salmonella species were extracted via boiling method described by Igbinosa et al. (2017). The Salmonella isolates were inoculated in 5.0 mL TSB incubated at 37°C for 18–24 h (Beshiru et al., 2018). A 100 μL of the turbid suspension was combined with 100 μL of sterilized distilled water in a 2.0 mL Eppendorf tube and subjected to a dry bath (MK200-2, Shanghai, China) for 15 min at 100°C for cell lyses. The lysed cell mixture was then centrifuged with a mini centrifuge (Mini 14k, Zhuhai, Guangdong, China) at 14 500 r/min for 15 min. The cell fragments were carefully separated from the supernatant. The supernatant was stored at −20°C as the template gDNA.
Polymerase Chain Reaction Amplification Procedure
All reactions were carried out in 25.0 μL volume of reaction (10 × Buffer 2.5 μL; MgCl2 1.0 μL; dNTP-Mix 3.0 μL; Taq polymerase 0.2 μL; Reverse primer 1.25 μL; Forward primer 1.25 μL; sterile double distilled H20 10.8 μL and gDNA 5.0 μL). Primers used for the detection of Salmonella species are shown in Table S1. The reaction was performed via a Peltier-based Thermal Cycler (BioSeparation System, Shanxi, China) with an initial denaturation at 95°C for 10 min; 35 cycles of denaturation at 94°C for 60 s, primer annealing as indicated in Table S1 and extension at 72°C for 90 s; final extension at 72°C for 10 min. Salmonella enterica serovar Typhymurium ATCC 14028, Salmonella Enteritidis ATCC 13076, were used as positive controls while deionized water was used as a negative control for each test procedure. Thermal cyclic conditions for the detection of antibiotic-resistance genes for Salmonella species were as follows; initial denaturation at 94°C for 2 min followed by 35 cycles of 94°C for 1 min, annealing condition as in Table S2 and extension at 72°C for 1 min with a final extension at 72°C for 10 min and cooling to 4°C. The PCR conditions for amplification of the virulence genes were as follows: 5 min of initial denaturation at 95°C, followed by 30 cycles of denaturation at 94°C for 30 s, annealing as described in Table S3, and extension at 72°C for 60 s, ending with a final extension period of 72°C for 2 min. Electrophoresis of the amplified PCR products were loaded on 1.2% agarose gel (CLS-AG100, Warwickshire, United Kingdom) in 0.5 × TAE buffer (pH 8.5, 20 mM Na acetate, 40 mM Tris-HCl, 1 mM EDTA) and allowed to run for 1 h at 100 V. The gels were viewed via a UV transilluminator (EBOX VX5, Vilber Lourmat, France).
Antimicrobial Susceptibility Profile of the Salmonella Isolates
Antimicrobial susceptibility profile of the Salmonella species was carried out using Kirby-Bauer disc diffusion method. Briefly, the purified isolates were inoculated in 5.0 mL Mueller-Hinton Broth (MHB) (Lab M, Lancashire, United Kingdom) and incubated overnight. The optical density (OD) of the turbidity of the broth was adjusted to McFarland standard 0.5 equivalent to 108 cfu/mL. Using a sterile swab sticks, respective broth cultures were aseptically swabbed on Mueller Hinton Agar (Lab M, Lancashire, United Kingdom). A total of 24 antibiotic discs (Mast Diagnostics, Merseyside, United Kingdom) which included kanamycin (30 μg), gentamycin (10 μg), streptomycin (25 μg), erythromycin (15 μg), tobramycin (10 μg), ampicillin (10 μg), amoxicillin (25 μg), imipenem (10 μg), ampicillin/sulbactam (30 μg), meropenem (10 μg), cefotaxime (30 μg), sulfamethoxazole (30 μg), cephalothin (30 μg), trimethoprim (25 μg), erythromycin (15 μg), amoxicillin/clavulanate (30 μg), colistin (20 μg), chloramphenicol (30 μg), penicillin G (10 μnits), polymyxin B (300 units), oxytetracycline (30 μg), doxycycline (30 μg), tetracycline (30 μg), ofloxacin (5 μg), and ciprofloxacin (10 μg) were used for the susceptibility testing. The respective discs were also aseptically impregnated on the agar plates using a sterile forceps equidistant apart. Plates were allowed to stand at room temperature for 5 min and incubated at 37°C for 18–24 h. Resistance, intermediate or susceptibility profile of the isolates were elucidated by determining zone of inhibition and matched with the interpretative chart of Clinical Laboratory Standards Institute (2017) to determine the sensitivity, intermediate and resistance profiles of the isolates to the antibiotics used.
Statistical Analysis
All data were analyzed using the statistical package SPSS (Version 21.0) and Microsoft Excel 2013. Descriptive statistics were carried out to determine the mean population density and expressed in Log10 CFU/g. One Way Analysis of Variance was applied to the densities from open markets while Duncan Multiple Range test was used to show significant difference between mean variables. The p < 0.05 were considered statistically significant.
Results
Population Cell Density of Salmonella Species From the RTE Shrimps
The mean Salmonella species counts from RTE shrimps obtained from open markets are presented in Table S4. The mean Salmonella species counts from the RTE shrimps are all expressed in log10 cfu/g. The values ranged from 0.079 to 3.516 (November), 0.613–3.817 (December), 0.255–4.492 (January), 0.602–4.841 (February), 0.959–4.822 (March), 1.562–5.118 (April), 1.573–5.434 (May), 2.003–5.274 (June), 2.001–5.356 (July), 1.782–4.555 (August), 0.944–4.754 (September), and −0.301 −3.748 (October) during the 12 month sampling regimen. Significant differences were observed across the respective markets as p < 0.01. For the respective markets, values ranged from 0.977 to 2.391 (Oba Market), 0.944–3.283 (New Benin Market), 0.613–3.231 (Jattu Market), 0.079–2.075 (Igarra Market), −0.301 −3.318 (Ekpoma Market), 1.272–3.484 (Uromi Market), 2.572–4.428 (Sapele Market), 3.053–4.481 (Ughele Market), 3.083–5.434 (Ogbegonogo Market), 3.185–5.205 (Ashafor Market), 3.161–4.435 (Igbudu Market), and 3.236–5.356 (Main Market, Oleh). Significant differences were observed across the respective months as p < 0.01.
Prevalence of Positive Salmonella Samples
The distribution of positive Salmonella samples from respective markets include, 14/120 (11.7%) for Oba market, 10/120 (8.33%) for New Benin market, 8/120 (8.33%) for Jattu market, 9/120 (7.5%) for Igarra market, 7/120 (5.83%) for Ekpoma market, 10/120 (8.33%) for Uromi market, 23/120 (19.17%) for Sapele market, 27/120 (22.5%) for Ughele market, 26/120 (21.67%) for Ogbegonogo market, 25/120 (20.83%) for Ashafor market, 27/120 (22.5%) for Igbudu market, 24/120 (20%) for main Market Oleh. Overall, 210/1440 (14.58%) were positive for Salmonella species.
Salmonella Detection From RTE Shrimps
This study revealed, 210/1440 (14.58 %) of the RTE shrimp samples were positive for Salmonella species. All the tentatively 210 Salmonella isolates were characterized via culture-based and biochemical procedures using Gram-reaction with 3% KOH test, oxidase, urease reactions, indole and motility tests. The Salmonella isolates that appear negative for urease, oxidase, indole and Gram-negative rods were selected as presumptive Salmonella. Only 67 Salmonella isolates were positive using this culture-based approach. Analytical profile index (API 20E) were further employed to confirmed the identity of 49 Salmonella isolates. From the 49 Salmonella isolates positive from the API test, Salmonella genus-specific primer was only positive for 45 isolates. This was further identified using the species-specific primer that target Salmonella Enteritidis 11 (24.4%), Salmonella Typhimurium 14 (31.1%) and other Salmonella spp. 20 (44.4%). In Oba Market, 1/4 (25%) were confirmed to be Salmonella Enteritidis, 1/4 (25%) were confirmed to be Salmonella Typhimurium, 2/4 (50%) were confirmed to be other Salmonella species (Table S5).
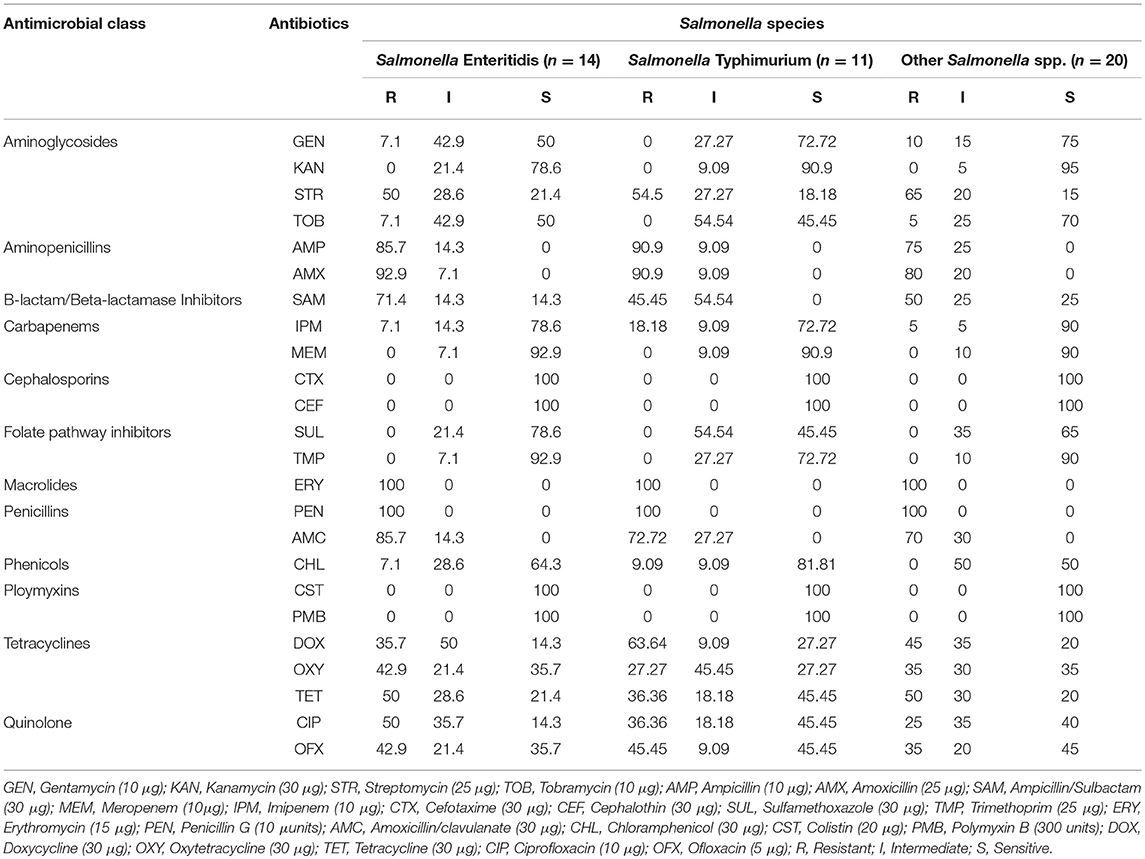
Antimicrobial Susceptibility Profiles of the Salmonella Species From RTE Shrimps
The distribution of antimicrobial susceptibility profile of Salmonella species is presented in Table 1. For Salmonella Enteritidis, 100% (14/14) were resistant to erythromycin and penicillin, 85.7% (12/14) were resistant to amoxicillin/clavulanate and ampicillin, 92.9% (13/14) were resistant to amoxicillin, 71.4% (10/14) were resistant to ampicillin/sulbactam. For Salmonella Typhimurium, 100% (11/11) were resistant to erythromycin and penicillin, 90.9% (10/11) were resistant to ampicillin and amoxicillin, 72.7% (8/11) were resistant to amoxicillin/clavulanate, 63.6% (7/11) were resistant to doxycycline. For other Salmonella species, 100% (20/20) were resistant to erythromycin and penicillin, 80% (16/20) were resistant to amoxicillin, 75% (15/20) were resistant to ampicillin, 70% (14/20) were resistant to amoxicillin/clavulanate, 65% (13/20) were resistant to streptomycin. Number of resistant + intermediate Salmonella species as shown in Table 1 include 0/45 (cefotaxime, cephalothin, polymycin B and colistin), 45/45 (ampicillin, amoxicillin, erythromycin, penicillin, and amoxicillin/clavulanate), 38/45 (ampicillin/sulbactam), 37/45 (streptomycin), 36/45 (doxycline), 33/45 (tetracycline), 30/45 (oxytetracycline and ciprofloxacin), 26/45 (ofloxacin).
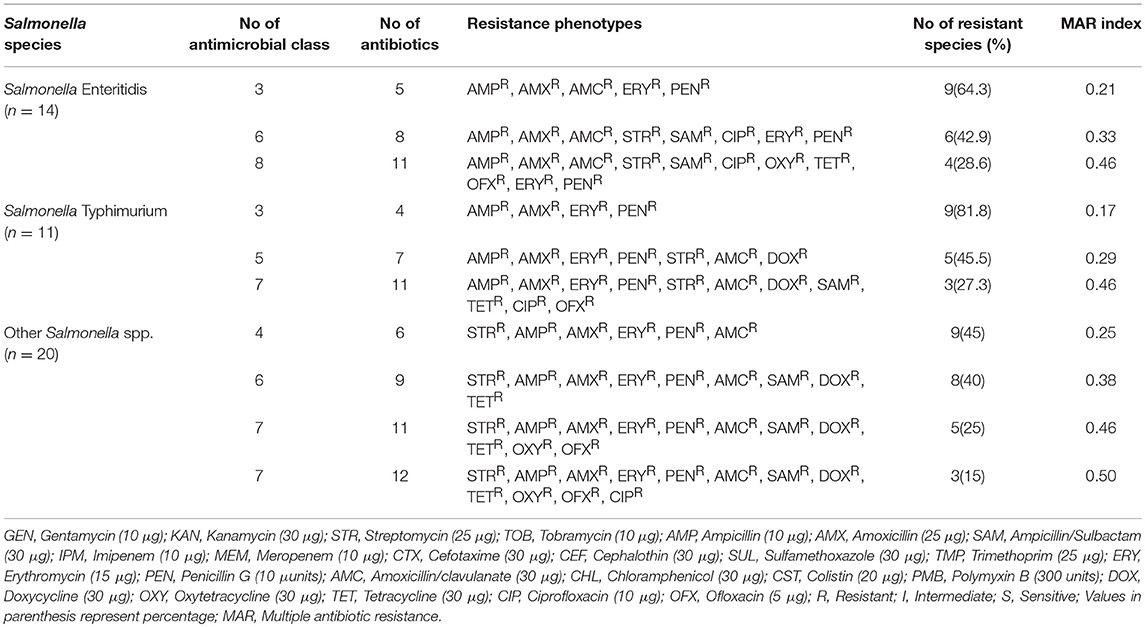
Distribution of Multiple Antibiotic-Resistance Characteristics of the Salmonella Species
The MDR and MAR index distribution of Salmonella species is presented in Table 2. A total of 9/14 (64.3%) of Salmonella Enteritidis were resistant to 5 antibiotics (AMPR, AMXR, AMCR, ERYR, PENR) which belonged to 3 different groups of antimicrobials with a MAR index of 0.21. Furthermore, 4/14 (28.6%) of Salmonella Enteritidis were resistant to 11 antibiotics (AMPR, AMXR, AMCR, STRR, SAMR, CIPR, OXYR, TETR, OFXR, ERYR, PENR) which belonged to 8 different groups of antimicrobials with a MAR index of 0.46. A total of 9/11 (81.8%) of Salmonella Typhimurium were resistant to 4 antibiotics (AMPR, AMXR, ERYR, PENR) which belonged to 3 different groups of antimicrobials with a MAR index of 0.17. Furthermore, 3/11 (27.3%) of Salmonella Typhimurium were resistant to 11 antibiotics (AMPR, AMXR, ERYR, PENR, STRR, AMCR, DOXR, SAMR, TETR, CIPR, OFXR) which belonged to 7 different groups of antimicrobials with a MAR index of 0.46. A total of 9/20 (45%) of other Salmonella spp. were resistant to 6 antibiotics (STRR, AMPR, AMXR, ERYR, PENR, AMCR) which belonged to 4 different groups of antimicrobials with a MAR index of 0.25. Furthermore, 3/20 (15%) of other Salmonella spp. were resistant to 12 antibiotics (STRR, AMPR, AMXR, ERYR, PENR, AMCR, SAMR, DOXR, TETR, OXYR, OFXR, CIPR) which belonged to 7 different groups of antimicrobials with a MAR index of 0.50.
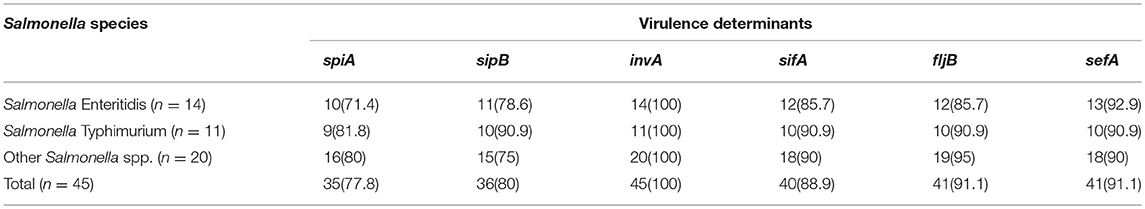
Distribution and Proportion of Virulence Gene Elements Among the Salmonella Species
The distribution of virulence genes among Salmonella species is presented in Table 3. For Salmonella Enteritidis 10/14 (71.4%) harbored spiA (involved in both biofilm formation and virulence), 11/14 (78.6%) revealed sipB (allows easy entering of non-phagocytic cells and lysing of macrophages), 14/14 (100%) harbored invA (Salmonella invasion gene), 12/14 (85.7%) revealed sifA (for the development of filamentous assemblies) and fljB (flagellin gene), 13/14 (92.9%) harbored sefA (fimbrial subunit of Salmonella antigen) (Table 3).
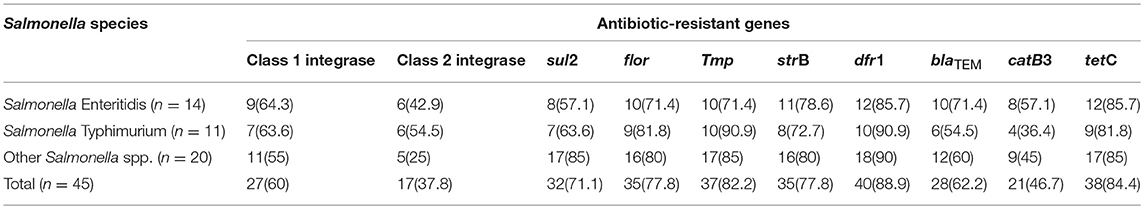
Distribution of Antibiotic-Resistant Elements Among the Salmonella Species
The distribution of antibiotic-resistant elements amongst Salmonella species is presented in Table 4. For Salmonella Enteritidis 9/14 (64.3%) harbored Class 1 integrase, 6/14 (42.9%) demonstrated Class 2 integrase, 8/14 (57.1%) revealed sul2 (sulphonamide resistance gene) and catB3 (group B chloramphenicol acetyltransferase gene), 10/14 (71.4%) revealed flor (florfenicol/chloramphenicol resistance gene), tmp (dihydrofolate reductase gene), blaTEM (beta-lactamase resistant gene), 11/14 (78.6%) demonstrated strB (streptomycin inactivating enzyme), 12/14 (85.7%) harbored dfr1 (specific trimethoprim resistance), and tetC (tetracycline resistance protein) (Table 4).
Discussion
Gastrointestinal illnesses continue to be a global and public health menace. Exposure to food borne Salmonella directly or indirectly via consumption of RTE seafood can be an important route of infection to humans. Findings from this study provide an estimation of the prevalence of Salmonella from RTE shrimps in open markets from south-south region in Nigeria. The prevalence of Salmonella positive samples was higher than a previous study from Turkey (Ikiz et al., 2016) (2%), Iran (Rahimi et al., 2013) (1.8%) and China (Yang et al., 2015) (13%). The prevalence of Salmonella spp. from the RTE shrimp samples assessed in this study was also lower compared to those detected from India (Kumar et al., 2008) (29.0%), Saudi Arabia (Elhadi, 2014) (39.9%), Vietnam (Nguyen et al., 2016) (49.1%), Thailand (Woodring et al., 2012) (21%), Brazil (Carvalho et al., 2013) (16.12%), China (Zhang et al., 2015) (29.7%) and India (Kumar et al., 2009) (26.7%); but higher than, findings by Koonse et al. (2005) from six different countries with participating countries not mentioned at their request (two countries are located in southeast Asia, one is in central Asia, one is in Central America, one is in North America, and one is an island in the Pacific Ocean) re-counted a prevalence rate of 1.6% in shrimp samples. It was also reported in Nigeria (Raufu et al., 2014) that a total of 23/200 (11.5%) samples were positive for Salmonella, with three serovars comprising Salmonella serovars Eko, 47: mt:-, and Hadar, recovered. In Brazil (Carvalho et al., 2013) reported that from a total of 186 confirmed Salmonella spp., five serovars were identified and they include: Salmonella Saintpaul, Salmonella Infantis, Salmonella Panama, Salmonella Madelia, and Salmonella Braenderup. Five different Salmonella serotypes including Salmonella Typhi, Salmonella Newport, Salmonella Paratyphi B, Salmonella Enteritidis, and Salmonella Typhimurium were recovered from seafood samples in Iran (Rahimi et al., 2013). The most prevailing Salmonella serovars from China (Zhang et al., 2015) among the 730 seafood samples examined were Salmonella Typhimurium (4.1%), Salmonella Hvittingfoss (4.1%), Salmonella Schwarzengrund (4.6%), Salmonella Stanley (4.6%), Salmonella Singapore (5.5%), Salmonella Thompson (9.2%), Salmonella Wandsworth (12.0%), and Salmonella Aberdeen (18.4%).
The findings from Yang et al. (2015) reported a most probable number (MPN)/g of 0.3–10, with one sample exceeding 110 MPN/g which was somewhat similar to the Salmonella density in this study. The mean Salmonella density in this study varied across the sampling months as higher densities were observed in the wet season (March to October) compared to dry season (November to February) and from one open market to another particularly from open markets in Delta State. Siala et al. (2017) reported that the presence of Salmonella spp. in shrimps is an indicator of contamination in the shrimp industry which happens to be one of the most significant seafood commodities worldwide. The high rate of positive Salmonella species in RTE shrimps in Southern Nigeria is worrisome and a substantial risk to public health. Thus, it is imperative to manage Salmonella infection in the food production process by further strengthening the surveillance of aquatic food products to circumvent the contamination of RTE seafood products. The high prevalence of Salmonella in open markets in the present study indicates poor sanitary condition during processing as well as the environment and poor hygiene of the RTE shrimp handlers during preparation of the products. The difference in the densities and prevalence of Salmonella from RTE seafood could also be ascribed to geographical variation, contaminated raw materials and poor /inadequate detection methods.
Determination of Salmonella resistance to antibiotics is crucial for therapeutic regimen during outbreaks. Salmonella resistance to erythromycin, amoxicillin and penicillin in this study are of public health threat and thus be as a consequence of extensive usage of these antibiotics in the study area. Interestingly, no Salmonella serovars was resistant to cefotaxime, cephalothin, colistin, and polymyxin B. This is very important to public health as these antibiotics could be crucial in threating drug resistant Salmonella pathogens. Public education to enlighten individual not to misuse these antibiotics is essential to circumvent the occurrence and development of resistance to these antibiotics.
Akiyama et al. (2011) reported from the United States that none of the Salmonella isolates showed resistance to ampicillin, gentamicin, chloramphenicol, kanamycin, sulfisoxazole, tetracycline, and streptomycin. The highest antibiotic resistance Salmonella species form seafood observed by Elhadi (2014) from Saudi Arabia were amoxicillin-clavulanic acid (45%), ampicillin (70%) and tetracycline (90.71%). Percentage resistance to nalidixic acid (47.4%) was the predominant report from Iran by Rahimi et al. (2013), prior to others such as ciprofloxacin (5.3%), trimethoprim (15.8%), streptomycin (15.8%), and tetracycline (36.8%). From China, Yang et al. (2015) reported resistance for ampicillin (28.2%), tetracycline (35.9%), trimethoprim-sulfamethoxazole (25.2%), streptomycin (18.4%) and chloramphenicol (20.4%), with 34.0% being resistant to more than three antibiotics. These were somewhat in accordance to the findings in this study. Zhang et al. (2015) also reported resistance of Salmonella from China from retail aquaculture products to tetracycline (34.1%), sulfonamides (56.5%), streptomycin (28.6%) and ampicillin (23.5%) with lower levels of resistance for ciprofloxacin (2.3%), gentamicin (3.2%), ceftazidime (0.5%) cefepime (0.5%), and cefotaxime (0.9%) which was rather similar to the findings in this study. In addition, 43.3% of the Salmonella serovars from a finding of Zhang et al. (2015) were multidrug resistant which is reduced when compared to the results in this study. Salmonella serotypes such as Typhimurium and Enteritidis have historically been reported as the significant causes of non-typhoidal salmonellosis. Though, other serotypes have been revealed to be included to be prevalent with respect to difference in geographical regions (Brands et al., 2005).
The occurrence of resistance to ciprofloxacin in Salmonella serovars is of public health importance as it translates possible misuse in animals and over-prescription in humans. Salmonella isolates in this study that were resistant to ciprofloxacin were also observed to be multidrug resistant strains to other antibiotics which were in accordance to the finding of Vo et al. (2006) from the Netherlands. MDR Salmonella isolates in this study are prevalent in open markets, which necessitates that more attention be ascribed toward the control and supervision of antibiotic usage, particularly in human health care and agriculture divisions in Nigeria. Bacterial virulence is predisposed by both the occurrence of antibiotic resistance and virulence determinants. The advancement of Salmonella strains that are is based particularly on elements of biochemical and genetic mechanisms so as to heighten their survival via preservation of their antibiotic resistance genes. As regards the virulence determinants that were analyzed, Salmonella Enteritidis, Salmonella Typhimurium, and other Salmonella isolates represent a broader range of pathogenicity.
High MAR index was observed in this study which indicates high use/misuse of antibiotics in the study areas. MAR index of Salmonella isolates ranged from 0.14 to 0.45 for different seafood in a study by Budiati et al. (2013) in Malaysia. From Brazil, Carvalho et al. (2013) reported that 23% of Salmonella serovars were resistant to ≤ 1 antibiotic, 20% were resistant to ≤ 2 antibiotics while 3 strains showed multi-resistance characteristics. These were lower compared to the findings of this study. The rapid development of bacterial resistance is ascribed to the selective pressure of antibiotics via evolutionary responses as a consequence of natural selection.
The dissemination of resistant elements in natural ecosystem can alter as well as change the physiology and population dynamics of resident microbial populaces (Igbinosa and Odjadjare, 2015). The emergence of antibiotic resistant determinants in pathogenic Salmonella species has made it more problematic due to the pervasiveness of horizontal gene transfer which is the procedure where bacteria obtain elements/determinants from the environment (Thomas and Nielsen, 2005). Most antibiotic resistance genes are found on intregons, plasmids or transposons, which can be transferred and mobilized to other bacteria of different or the same species. Integrons have been reported to be involved in the acquisition of antibiotic resistance elements. Class 1 integrons which contains numerous resistance elements could play vital roles in the maintenance and spread of antibiotic resistance in Salmonella species both in the absence and presence of selective pressure as reported in India (Deekshit et al., 2012). Meng et al. (2011) from China documented that class 1 integron showed empty regions from strains in serotypes Choleraesuis isolated from seafood. Findings by Meng et al. (2011) also suggest the possible dissemination of class 1 integrons from foodborne pathogens to human inhabited bacteria through horizontal gene transfer.
The occurrence and dissemination of resistant elements to pathogenic and commensal bacteria of human origin as well as gene transfer in human intestinal microbiome have been reported (Slayers et al., 2004). Antibiotic resistance genes such as tetA and catA1 were present in 60 and 57.52%, of Salmonella isolates, respectively in a study by Deekshit et al. (2012) from India. Adesiji et al. (2014) reported that of the 20 tetracycline resistant isolates from India, 20(100%) tetA, 6(30%) tetB, 7(35%) tetC, and 10(50%) tetG encoded resistant elements, respectively. Of 18 cotrimoxazole-resistant strains, 4(22.2%), 14(77.7%), and 18(100%) had sul3, sul2, and sul1genes, respectively (Adesiji et al., 2014). Deekshit et al. (2013) reported the occurrence of three antibiotic resistance determinants sul1, tetG, and floR from seafood some of which were also detected in this study.
Virulence determinants are involved in bacterial pathogenicity, and their occurrence in Salmonella can result in salmonellosis (). Findings from this study revealed that isolates of Salmonella Enteritidis and Salmonella Typhimurium demonstrated a diverse range of pathogenicity elements, which makes these serovars more virulent toward consumers of the RTE shrimp products especially immunocompromised individuals. Antibiotic resistance phenotypes and determinants have also been reported to be positively correlated with Salmonella virulence (Turki et al., 2014). Infections as a consequence of antibiotic-resistant Salmonella with virulence potential have been reported to take longer to recover from by been frequently fatal, when compared with ailments caused by antibiotic-susceptible strains of Salmonella with virulent capabilities.
The spiA gene of Salmonella is essential for its virulence and biofilm formation in host cells (Romling et al., 2003; Socher et al., 2005; Dong et al., 2011; Col et al., 2013; Beshiru et al., 2018). The sipB gene is required by Salmonella to form functional pores during Salmonella infection of erythrocytes for entry into the host cell through the host cell plasma membrane (Miki et al., 2004). The sipB gene is referred to as trans-locators as they translocate Salmonella effector proteins into host cells (consumers of RTE shrimps) which can cause typhoid fever and gastroenteritis (Galan and Wolf-Watz, 2006). The sipB gene in Salmonella serovars induces apoptotic macrophage either by activating or inducing autophagy and disruption of mitochondria, or by binding the proapoptotic enzyme caspase-1 which results in the discharge of interleukin-1 beta active form (Myeni et al., 2013).
A significant step in the cycle of facultative pathogenic intracellular Salmonella serovars on RTE shrimps and by extension the consumers is the incursion of the cells via the intestinal mucosa. Amplification of nucleotide sequences within the invA gene of Salmonella has been evaluated as a means of detecting invasive Salmonella serovars (). The invA gene of the Salmonella species allows the bacteria to invade the host and initiate infection, thereby increasing the degree of pathogenicity of the isolates. PCR analysis of 15 virulence genes by Yang et al. (2015) from retail seafood in China showed that all 103 Salmonella isolates had at least 4 virulence genes (mgtC, ssaQ, siiD, bcf C, and sopB), where the loci that remains were unevenly distributed. In addition, isolates of Salmonella Typhimurium, Salmonella Enteritidis, and Salmonella Weltevreden displayed a broader range of pathogenicity elements when compared with other Salmonella serovars by Yang et al. (2015) which was evident in this study.
A significant number of Salmonella serovars from RTE shrimps in this study harbored the sif A gene. The sif A gene plays a crucial role in Salmonella virulence. The degree of pathogenicity by Salmonella lies predominantly on the phenotypic manifestation of effector proteins released into the bacterial cell. Salmonella gains enterance into eukaryotic cells and exist in a vascular section with which some effector proteins (e.g., sif A) are located (Zhao et al., 2015).
Flagellin occurrence on RTE shrimps is a significant external antigen for numerous species which aids Salmonella virulence. Considerable heterogeneity of sequence exist within alleles which codes for different flagellar antigen from a previous study by McQuiston et al. (2004) while alleles which encodes similar antigenic flagella were homologous, signifying that flagellin determinants may be beneficial to targets for the genotypic resolve of flagellar antigenic type. Fimbriae are an important factor in Salmonella survival and persistence in the host (Kaur and Jain, 2012). The sef A gene encodes the Salmonella Enteritidis fimbrial protein (Mirmomeni et al., 2008). Studies have also revealed that the sef A gene plays a significant part in the adhesion of Salmonella Enteritidis to biotic surfaces (Lopes et al., 2006). Akiyama et al. (2011) reported that all Salmonella strains were positive for 14 virulence genes (sif A, spiA, invA, sopE, spaN, sipB, msgA, iroN, pagC, prgH, orgA, lpf C, tolC, and sitC) and negative for three genes (cdtB, spvB, and pef A). Some of these genes detected by Akiyama et al. (2011) were also detected in this study. Antibiotic resistance is a major public health menace globally, and particularly persistent in developing countries, including Nigeria, where the problem of infectious disease is on the increase with decreased healthcare budget. Though the emergence and dissemination of antibiotic-resistant Salmonella is a significant concern to food processors, cinnamaldehyde and carvacrol which are effective plant-derived antimicrobials have been reported to inactivate antibiotic-resistant Salmonella enterica in oysters, buffer and celery (Ravishankar et al., 2010). Bacteriophages propose effective and highly specific bio-control of antibiotic-resistant Salmonella pathogens from RTE foods (Guenther et al., 2012). Although phage particles keep their infectious capabilities, they are immobilized freely by the RTE food, which result in loss of their capacity to infect and diffuse target cells. Short-chain fatty acids have found application in animal diets to manage pathogens with Salmonella serovars inclusive (Van-Immerseel et al., 2002). Another alternative to eliminating pathogens is the precise suppression of functions vital to cause infection in the host (Clatworthy et al., 2007). Gene regulation mechanism via quorum sensing, where bacteria regulate the manifestation of numerous genes in reaction to the occurrence of small signal molecules is also very crucial (Defoirdt et al., 2011).
Other management strategies for antibiotic resistance includes the following: limiting the non-therapeutic usage of antibiotics for agriculture; improved information to strengthen resolutions on standard therapeutic regimen, education, other actions, coupled with continuous monitoring and validating effectiveness of management strategies; strengthening infection control boards in hospitals; nutrient management and runoff control; and improved diagnostic procedures, which requires developmental variations and infrastructure upgrades, enhancements in microbiological laboratory equipment and personnel (Global Antibiotic Resistance Partnership - India Working Group, 2011; Pruden et al., 2013). These recommendations could assist in the reduced of antibiotic resistance, directly advance public health, advantageous to the populace and decrease pressure on healthcare system. Finally, enhancing the coverage and types of juvenile vaccines administered by government agencies would enormously decrease the disease burden and circumvent the misuse of antibiotics (Global Antibiotic Resistance Partnership - India Working Group, 2011).
Conclusion
Findings indicate that RTE shrimps act as reservoirs in harboring multiple Salmonella strains. The recovered Salmonella serovars which exhibits multiple virulence and antibiotic resistance genes coupled with high MAR index constitute a risk to consumers. Hence, it is crucial to monitor the usage of antibiotics and hygiene status in processing and post-processing handling to circumvent the acquisition and dissemination of virulent Salmonella serovars. Furthermore, maintenance, and implementation of control measures such as good manufacturing practices (GMP), and hazard analysis and critical control point (HACCP) coupled with education of the RTE shrimp processors is necessary, for reducing and/or spreading Salmonella contamination.
Author Contributions
AB and II carried out the sampling, laboratory procedures, data interpretation, and writing of the manuscript. EI conceptualized, designed, and supervised the research, contributed in the laboratory methodologies and data interpretation, as well as the writing of the manuscript. All authors have read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to The World Academy of Sciences (Grant No. 14-091 RG/BIO/AF/AC) and International Foundation for Science (F5081) for the laboratory consumables to support in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01613/full#supplementary-material
References
Adesiji, Y. O., Deekshit, V. K., and Karunasagar, I. (2014). Antimicrobial-resistant genes associated with Salmonella spp. isolated from human, poultry, and seafood sources. Food Sci. Nut. 2, 436–442. doi: 10.1002/fsn3.119
Akiyama, T., Khan, A. A., Cheng, C., and Stefanova, R. (2011). Molecular characterization of Salmonella enterica serovar saintpaul isolated from imported seafood, pepper, environmental and clinical samples. Food Microbiol. 28, 1124–1128. doi: 10.1016/j.fm.2011.03.003
Banerjee, S., Ooi, M. C., Shariff, M., and Khatoon, H. (2012). Antibiotic resistant Salmonella and Vibrio associated with farmed Litopenaeus vannamei. Sci. World J. 2012:130136. doi: 10.1100/2012/130136
Beshiru, A., Igbinosa, I. H., and Igbinosa, E. O. (2018). Biofilm formation and potential virulence factors of Salmonella strains isolated from ready-to-eat shrimps. PLoS ONE 13:e0204345. doi: 10.1371/journal.pone.0204345
Brands, D. A., Inman, A. E., Gerba, C. P., Mare, C. J., Billington, S. J., and Saif, L. A. (2005). Prevalence of Salmonella spp. in oysters in the United States. Appl. Environ. Microbiol. 71, 893–897. doi: 10.1128/AEM.71.2.893-897.2005
Brunelle, B. W., Bearson, S. M. D., and Bearson, B. L. (2013). Tetracycline accelerates the temporally-regulated invasion response in specific isolates of multidrug-resistant Salmonella enterica serovar Typhimurium. BMC Microbiol. 13:202. doi: 10.1186/1471-2180-13-202
Budiati, T., Rusul, G., Wan-Abdullah, W. N., Arip, Y. M., Ahmad, R., and Thong, K. L. (2013). Prevalence, antibiotic resistance and plasmid profiling of Salmonella in catfish (Clarias gariepinus) and tilapia (Tilapia mossambica) obtained from wet markets and ponds in Malaysia. Aquaculture 375, 127–132. doi: 10.1016/j.aquaculture.2012.11.003
CAC (2002). Discussion Paper on Risk Management Strategies for Vibrio spp. Italy: Seafood. Food and Agriculture Organization / World Health Organization, Rome.
Carvalho, F. C. T., Sousa, O. V., Carvalho, E. M. R., Hofer, E., and Vieira, R. H. S. F. (2013). Antibiotic resistance of Salmonella spp. isolated from shrimp farming freshwater environment in northeast region of Brazil. J. Pathog. 685193:5. doi: 10.1155/2013/685193
Clatworthy, A. E., Pierson, E., and Hung, D. T. (2007). Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3, 541–548. doi: 10.1038/nchembio.2007.24
Clinical and Laboratory Standards Institute (2017). Performance Standards for Antimicrobial Susceptibility Testing M02-A12, M07-A10, and M11-A8. 27th Edn, 282.
Col, R. K., Maj, H. S., Maj, M. V., and Maj, R. H. (2013). Outbreak investigation: Salmonella food poisoning. Med. J. Armed Forces India 69, 388–391. doi: 10.1016/j.mjafi.2013.01.005
Deekshit, V. K., Kumar, B. K., Rai, P., Srikumar, S., Karunasagar, I., and Karunasagar, I. (2012). Detection of class 1 integrons in Salmonella Weltevreden and silent antibiotic resistance genes in some seafood associated nontyphoidal isolates of Salmonella in south-west coast of India. J. Appl. Microbiol. 112, 1113–1122. doi: 10.1111/j.1365-2672.2012.05290.x
Deekshit, V. K., Kumar, K. B., Rai, P., Rohit, A., and Karunasagar, I. (2013). Simultaneous detection of Salmonella pathogenicity island 2 and its antibiotic resistance genes from seafood. J. Microbiol. Methods 93, 233–238. doi: 10.1016/j.mimet.2013.03.015
Defoirdt, T., Sorgeloos, P., and Bossier, P. (2011). Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr. Opinion Microbiol. 14, 251–258. doi: 10.1016/j.mib.2011.03.004
Dong, H., Peng, D., Jiao, X., Zhang, X., Geng, S., and Liu, X. (2011). Roles of the spiA gene from Salmonella Enteritidis in biofilm formation and virulence. Microbiol. 157, 1798–1805. doi: 10.1099/mic.0.046185-0
Duran, G. M., and Marshall, D. L. (2005). Ready to eat shrimp as an international vehicle of antibiotic resistant bacteria. J. Food Prot. 68, 2395–2401 doi: 10.4315/0362-028X-68.11.2395
Ed-dra, A., Filali, F. R., Karraouan, B., El-Allaoui, A., Aboulkacem, A., and Bouchrif, B. (2017). Prevalence, molecular and antimicrobial resistance of Salmonella isolated from sausages in Meknes, Morocco. Microb. Pathog. 105, 340–345. doi: 10.1016/j.micpath.2017.02.042
Elhadi, N. (2014). Prevalence and antimicrobial resistance of Salmonella spp. in raw retail frozen imported freshwater fish to Eastern Province of Saudi Arabia. Asian Pacific. J. Trop. Biomed. 4, 234–238. doi: 10.1016/S2221-1691(14)60237-9
Food and Agriculture Organization of the United Nations (2008). Global Study of Shrimp Fisheries. FAO Fisheries Technical Paper. No. 475. Rome: FAO, 331.
Galan, J. E., and Wolf-Watz, H. (2006). Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573. doi: 10.1038/nature05272
Global Antibiotic Resistance Partnership - India Working Group (2011). Rationalizing antibiotic use to limit antibiotic resistance in India. Indian J. Med. Res. 134, 281–294.
Guenther, S., Herzig, O., Fieseler, L., Klumpp, J., and Loessner, M. J. (2012). Biocontrol of Salmonella Typhimurium in RTE foods with the virulent bacteriophage FO1-E2. Int. J. Food Microbiol. 154, 66–72. doi: 10.1016/j.ijfoodmicro.2011.12.023
Igbinosa, E. O. (2016). Detection and antimicrobial resistance of Vibrio isolates in aquaculture environments: Implications for public health. Microb. Drug Res. 22, 238–245. doi: 10.1089/mdr.2015.0169
Igbinosa, E. O., and Beshiru, A. (2017). Isolation and characterization of antibiotic susceptibility profile of Salmonella species isolated from abattoir environment. IFE J. Sci. 19, 389–397. doi: 10.4314/ijs.v19i2.19
Igbinosa, E. O., and Odjadjare, E. E. (2015). “Antibiotics and antibiotic resistance determinants: an undesired element in the environment” in Battle Against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs, eds A. Méndez-Vilas (Extremadura: Formatex Research Center, 858–866.
Igbinosa, I. H. (2015). Prevalence and detection of antibiotic-resistant determinant in Salmonella isolated from food-producing animals. Trop. Animal Health Product. 47, 37–43. doi: 10.1007/s11250-014-0680-8
Igbinosa, I. H., Beshiru, A., and Igbinosa, E. O. (2017). Antibiotic resistance profile of Pseudomonas aeruginosa isolated from aquaculture and abattoir environments in urban communities. Asian Pac. J. Trop. Dis. 7, 930–935. doi: 10.12980/apjtd.7.2017D6-363
Ikiz, S., Dümen, E., Kahraman, B. B., Bayrakal, G. M., Kahraman, T., and Ergin, S. (2016). Investigation of Salmonella spp. and Listeria monocytogenes in seafood by cultural methods and PCR. Kafkas Univ. Vet. Fak. Derg. 22, 397–401. doi: 10.9775/kvfd.2015.14808
International Organization for Standardization (2017). Microbiology of the food chain – Preparation of test samples, initial suspension and decimal dilutions for microbiological examination – Part 1: General rules for the preparation of the initial suspension and decimal dilutions, 2nd Edn. ISO, 6887-1 26. pages
Josupeit, H. (2005). Trade Flows Between Developed and Developing Countries. Rome: FAO. Available online at: http://www.globefish.org/dynamisk.php4?id=2504 (accessed February 17, 2006).
Kaur, J., and Jain, S. K. (2012). Role of antigens and virulence factors of Salmonella enterica serovar Typhi in its pathogenesis. Microbiol. Res. 167, 199–210. doi: 10.1016/j.micres.2011.08.001
Koonse, B., Burkhardt, W., Chirtel, S., and Hoskin, G. P. (2005). Salmonella and the sanitary quality of aquacultured shrimp. J. Food Protect. 68, 2527–2532. doi: 10.4315/0362-028X-68.12.2527
Kumar, R., Surendran, P. K., and Thampuran, N. (2008). Evaluation of culture, ELISA and PCR assays for the detection of Salmonella in seafood. Lett. Appl. Microbiol. 46, 221–226. doi: 10.1111/j.1472-765X.2007.02286.x
Kumar, R., Surendran, P. K., and Thampuran, N. (2009). Distribution and genotypic characterization of Salmonella serovars isolated from tropical seafood of Cochin, India. J. Appl. Microbiol. 106, 515–524. doi: 10.1111/j.1365-2672.2008.04020.x
Li, R., Lai, J., Wang, Y., Liu, S., Li, Y., and Liu, K. (2013). Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province. China. Int. J. Food Microbiol. 163, 14–18. doi: 10.1016/j.ijfoodmicro.2013.01.020
Lopes, C. V., Velayudhan, T. B., Halvorson, D. A., and Nagaraja, V. K. (2006). Preliminary evaluation of the use of the sef A fimbrial gene to elicit immune response against Salmonella enterica serotype Enteritidis in chickens. Avian Dis. 50, 185–190. doi: 10.1637/7438-090905R.1
McQuiston, J. R., Parrenas, R., Ortiz-Rivera, M., Gheesling, L., Brenner, F., and Fields, P. I. (2004). Sequencing and comparative analysis of flagellin genes fliC, fljB, and flpA from Salmonella. J. Clin. Microbiol. 42, 1923–1932. doi: 10.1128/JCM.42.5.1923-1932.2004
Meng, H., Zhang, Z., Chen, M., Su, Y., Li, L., Miyoshi, S., et al. (2011). Characterization and horizontal transfer of class 1 integrons in Salmonella strains isolated from food products of animal origin. I. J. Food Microbiol. 149, 274–277. doi: 10.1016/j.ijfoodmicro.2011.07.006
Miki, T., Okada, N., Shimada, Y., and Danbara, H. (2004). Characterization of Salmonella pathogenicity island 1 type III secretion-dependent haemolytic activity in Salmonella enterica serovar Typhimurium. Microb. Pathog. 37, 65–72. doi: 10.1016/j.micpath.2004.04.006
Mirmomeni, M. H., Sisakhtnezhad, S., and Sharifi, A. (2008). Rapid detection of Salmonella Enteritidis by PCR amplification of the Sef A gene and its cloning. Pak. J. Biol. Sci. 11, 428–432. doi: 10.3923/pjbs.2008.428.432
Myeni, S. K., Wang, L., and Zhou, D. (2013). SipB-SipC complex is essential for translocon formation. PLoS ONE 8:e60499. doi: 10.1371/journal.pone.0060499
Nguyen, D. T. A., Kanki, M., Nguyen, P. D., Le, H. T., Ngo, P. T., Tran, D. N. M., et al. (2016). Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 236, 115–122. doi: 10.1016/j.ijfoodmicro.2016.07.017
Oosterveer, P. (2006). Globalization and sustainable consumption of shrimp: Consumers and governance in the global space of flows. Int. J. Consumer Studies. 30, 465–476. doi: 10.1111/j.1470-6431.2006.00535.x
Orji, J. O., Nnachi, A. U., Egwuatu, C. C., Akujobi, C. N., Iwuafor, A. A., Efunshile, A. M., et al. (2016). Bacteriological quality of vended fresh shrimps harvested from Ndibe Beach, Afikpo North Local Government Area, Ebonyi State, Nigeria. Sch. J. App. Med. Sci. 4, 4058–4063. doi: 10.21276/sjams.2016.4.11.40
Pruden, A., Larsson, D. G. J., Amézquita, A., Collignon, P., Brandt, K. K., Graham, D. W., et al. (2013). Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ. Health Perspect. 121, 878–885. doi: 10.1289/ehp.1206446
Rahimi, E., Shakerian, A., and Falavarjani, A. G. (2013). Prevalence and antimicrobial resistance of Salmonella isolated from fish, shrimp, lobster, and crab in Iran. Comp. Clin. Pathol. 22, 59–62. doi: 10.1007/s00580-011-1368-3
Raufu, I. A., Lawan, F. A., Bello, H. S., Musa, A. S., Ameh, J. A., and Ambali, A. G. (2014). Occurrence and antimicrobial susceptibility profiles of Salmonella serovars from fish in Maiduguri, sub-Saharah, Nigeria. Egypt. J. Aquatic Res. 40, 59–63. doi: 10.1016/j.ejar.2014.01.003
Ravishankar, S., Zhu, L., Reyna-Granados, J., Law, B., Joens, L., and Friedman, M. (2010). Carvacrol and cinnamaldehyde inactivate antibiotic-resistant Salmonella enterica in buffer and on celery and oysters. J. Food Prot. 73, 234–240. doi: 10.4315/0362-028X-73.2.234
Romling, U., Bokranz, W., Rabsch, W., Zogaj, X., Nimtz, M., and Tschape, H. (2003). Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int. J. Med. Microbiol. 293, 273–285. doi: 10.1078/1438-4221-00268
Serrano, P. H. (2005). Responsible Use of Antibiotics in Aquaculture. Food and Agriculture Organization of the United Nations Rome, FAO Fisheries Technical Paper, 469.
Siala, M., Barbana, A., Smaoui, S., Hachicha, S., Marouane, C., Kammoun, S., et al. (2017). Screening and detecting Salmonella in different food matrices in Southern Tunisia using a combined enrichment/real-time PCR method: Correlation with conventional culture method. Front. Microbiol. 8:2416. doi: 10.3389/fmicb.2017.02416
Skyberg, J. A., Logue, C. M., and Nolan, L. K. (2006). Virulence genotyping of Salmonella spp. with multiplex PCR. Avian Dis. 50, 77–81. doi: 10.1637/7417.1
Slayers, A. A., Gupta, A., and Wang, Y. (2004). Human intestinal bacteria as reservoirs of antibiotic resistant genes. Trends Microbiol. 12, 412–416. doi: 10.1016/j.tim.2004.07.004
Socher, K., Romling, U., and Yaron, S. (2005). Effect of heat, acidification, and chlorination on Salmonella enterica serovar Typhimurium cells in a biofilm formed at the air-liquid interface. Appl. Environ. Microbiol. 71, 1163–1168. doi: 10.1128/AEM.71.3.1163-1168.2005
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of and barriers to horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. doi: 10.1038/nrmicro1234
Turki, Y., Mehr, I., Ouzari, H., Khessairi, A., and Hassen, A. (2014). Molecular typing, antibiotic resistance, virulence gene and biofilm formation of different Salmonella enterica serotypes. J. Gen. Appl. Microbiol. 60, 123–130. doi: 10.2323/jgam.60.123
Van-Immerseel, F., Cauwerts, K., Devriese, L. A., Haesebrouck, F., and Ducatelle, R. (2002). Feed additives to control Salmonella in poultry. World Poult. Sci. J. 58, 501–513. doi: 10.1079/WPS20020036
Vo, A. T., van Duijkeren, E., Fluit, A. C., Wannet, W. J., Verbruggen, A. J., Maas, H. M., et al. (2006). Antibiotic resistance, integrons and Salmonella genomic island 1 among non-typhoidal Salmonella serovars in The Netherlands. Int. J. Antimicrob. Agents 28, 172–179. doi: 10.1016/j.ijantimicag.2006.05.027
Woodring, J., Srijan, A., Puripunyakom, P., Oransathid, W., Wongstitwilairoong, B., and Mason, C. (2012). Prevalence and antimicrobial susceptibilities of Vibrio, Salmonella, and Aeromonas isolates from various uncooked seafoods in Thailand. J. Food Prot. 75, 41–47. doi: 10.4315/0362-028X.JFP-11-211
Yang, X., Wu, Q., Zhang, J., Huang, J., Chen, L., Liu, S., et al. (2015). Prevalence, enumeration, and characterization of Salmonella isolated from aquatic food products from retail markets in China. Food Cont. 57, 308–313. doi: 10.1016/j.foodcont.2015.03.046
Yano, Y., Hamano, K., Satomi, K., Tsutsui, I., and Aue-umneoy, D. (2011). Diversity and characterization of oxytetracycline-resistant bacteria associated with nonnative species, white-leg shrimp (Litopenaeus vannamei), and native species, giant tiger shrimp (Penaeus monodon), intensively cultured in Thailand. J. Appl. Microbiol. 110, 713–722. doi: 10.1111/j.1365-2672.2010.04926.x
Zhang, J., Yang, X., Kuang, D., Shi, X., Xiao, W., Zhang, J., et al. (2015). Prevalence of antimicrobial resistance of non-typhoidal Salmonella serovars in retail aquaculture products. Int. J. Food Microbiol. 210, 47–52. doi: 10.1016/j.ijfoodmicro.2015.04.019
Keywords: multidrug resistant, salmonellosis, virulence determinants, seafood, health risk
Citation: Beshiru A, Igbinosa IH and Igbinosa EO (2019) Prevalence of Antimicrobial Resistance and Virulence Gene Elements of Salmonella Serovars From Ready-to-Eat (RTE) Shrimps. Front. Microbiol. 10:1613. doi: 10.3389/fmicb.2019.01613
Received: 04 July 2018; Accepted: 28 June 2019;
Published: 11 July 2019.
Edited by:
Patrícia Poeta, University of Trás-os-Montes and Alto Douro, PortugalReviewed by:
Ashima Kushwaha Bhardwaj, Independent Researcher, Gurgaon, IndiaSekelwa Cosa, University of Pretoria, South Africa
Kamelia Mahmoud Osman, Cairo University, Egypt
Copyright © 2019 Beshiru, Igbinosa and Igbinosa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Etinosa O. Igbinosa, ZWlnYmlub3NhJiN4MDAwNDA7Z21haWwuY29t
 Abeni Beshiru
Abeni Beshiru Isoken H. Igbinosa2
Isoken H. Igbinosa2 Etinosa O. Igbinosa
Etinosa O. Igbinosa