- 1Dipartimento di Scienze per gli Alimenti, la Nutrizione e l′Ambiente (DeFENS), Università degli Studi di Milano, Milan, Italy
- 2Dipartimento di Scienze Agrarie e Ambientali – Produzione, Territorio, Agroenergia (DiSAA), Università degli Studi di Milano, Milan, Italy
- 3Department of Biotechnology, Delft University of Technology, Delft, Netherlands
- 4Department of Earth and Environmental Sciences, University of Milano-Bicocca, Milan, Italy
- 5Microbial Systems Ecology, Department of Freshwater and Marine Ecology, Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam, Netherlands
Arsenic contamination of groundwater aquifers is an issue of global concern. Among the affected sites, in several Italian groundwater aquifers arsenic levels above the WHO limits for drinking water are present, with consequent issues of public concern. In this study, for the first time, the role of microbial communities in metalloid cycling in groundwater samples from Northern Italy lying on Pleistocene sediments deriving from Alps mountains has been investigated combining environmental genomics and cultivation approaches. 16S rRNA gene libraries revealed a high number of yet uncultured species, which in some of the study sites accounted for more of the 50% of the total community. Sequences related to arsenic-resistant bacteria (arsenate-reducing and arsenite-oxidizing) were abundant in most of the sites, while arsenate-respiring bacteria were negligible. In some of the sites, sulfur-oxidizing bacteria of the genus Sulfuricurvum accounted for more than 50% of the microbial community, whereas iron-cycling bacteria were less represented. In some aquifers, arsenotrophy, growth coupled to autotrophic arsenite oxidation, was suggested by detection of arsenite monooxygenase (aioA) and 1,5-ribulose bisphosphate carboxylase (RuBisCO) cbbL genes of microorganisms belonging to Rhizobiales and Burkholderiales. Enrichment cultures established from sampled groundwaters in laboratory conditions with 1.5 mmol L-1 of arsenite as sole electron donor were able to oxidize up to 100% of arsenite, suggesting that this metabolism is active in groundwaters. The presence of heterotrophic arsenic resistant bacteria was confirmed by enrichment cultures in most of the sites. The overall results provided a first overview of the microorganisms inhabiting arsenic-contaminated aquifers in Northern Italy and suggested the importance of sulfur-cycling bacteria in the biogeochemistry of arsenic in these ecosystems. The presence of active arsenite-oxidizing bacteria indicates that biological oxidation of arsenite, in combination with arsenate-adsorbing materials, could be employed for metalloid removal.
Introduction
Natural contamination of groundwater with arsenic (As) represents a serious issue in both developing as well as many developed countries (Smedley and Kinniburgh, 2002). Moreover, in countries where groundwater represents the major source of drinking water, As poses problems to public health, such as in Bangladesh (Cavalca et al., 2013; Yunus et al., 2016). In Europe, As in drinking waters represents a risk for humans in different countries like Greece, Hungary, Romania, Croatia, Serbia, Spain (Katsoyiannis et al., 2015), and Ireland (McGrory et al., 2017). Particularly, in Italy As is present in groundwater at concentrations that are above the WHO limit of 10 μg L-1, due to geological history and substratum rock types, i.e., volcanic rocks in Central Italy and Pleistocene sedimentary rocks in Northern Italy (Mantelli et al., 1999; World Health Organization [WHO], 2001; Ungaro et al., 2008; Carraro et al., 2015; Peña Reyes et al., 2015). Specifically, in South-Eastern part of Lombardy (Po Plain) high levels of As can be measured in groundwaters characterized by low redox and oxygen content, high concentration of iron (Fe), manganese (Mn) and ammonium (NH4), and by the presence of peat (Rotiroti et al., 2014, 2015). In this area, drinking water is supplied through groundwater wells, thus posing a pollution problem of drinking water wells and subsequent health problems to the inhabitants of this area.
Redox processes govern As biogeochemistry of groundwater systems (“aquifers”). Anaerobic conditions leading to As solubilization are driven by the degradation of peat deposits and fuel reductive dissolution of As-bearing amorphous Fe (hydr)oxides by metal-reducing bacteria (Lear et al., 2007; Héry et al., 2008; Sutton et al., 2009; Cai et al., 2013; Herath et al., 2016). Microbial reductive processes are implicated in the dissolution of As from Fe, Mn, and aluminum minerals, either carried out by Fe- and to a lesser extent Mn-reducing bacteria (Islam et al., 2004; Luna et al., 2009), or by dissimilatory arsenate [As(V)]-reducing bacteria (DARB), fueled by the oxidation of organic substrates (Islam et al., 2004; Malasarn et al., 2004; Mladenov et al., 2010). Metal-reducing bacteria, like Geobacter present in Fe- and Mn-rich groundwater, play a role in releasing As to groundwater either by reducing As-bearing Fe-oxides (Giloteaux et al., 2013) or by reducing sediment-bound As(V) to As(III) (Héry et al., 2010). Similarly to Geobacter, also Shewanella (Dhar et al., 2011), Sulfurospirillum (Héry et al., 2008) and Desulfuromonas (Osborne et al., 2015) were found to promote Fe(III) reduction and As release from the sediment to the aquifer via dissimilative reduction processes. In anaerobic and microaerophilic environments, sulfur (S)-oxidizing bacteria can contribute to pyritic-S dissolution by the oxidation of solid S as sulfide, elemental S and thiosulfate (Kodama and Watanabe, 2003; Campbell et al., 2006; Handley et al., 2014; Thiel et al., 2019). On the other hand, sulfate reduction decreases the mobility of As by co-precipitation of sulfide with Fe and As (Kirk et al., 2004).
Once As is solubilized from sediments, it can undergo reductive detoxification (i.e., reduction of As(V) to the more mobile As(III)) conducted by microorganisms bearing the ARS operon (Andres and Bertin, 2016). Conversely, oxidation processes lead to immobilization of As on solid Fe-oxide by chemolithoautotrophic bacteria present in groundwater systems (Alfreider et al., 2009) and able to oxidize As(III) to As(V) (Garcia-Dominguez et al., 2008; Andreoni et al., 2012). In subsurface groundwater at circumneutral pH, Fe-oxidizing bacteria (FeOB) like Gallionella ferruginea (Katsoyiannis and Zouboulis, 2004; Gorra et al., 2012) and anaerobic nitrate-reducing/Fe-oxidizing bacteria contribute to the immobilization of As via their ability to form solid Fe (hydro)oxides minerals (Hohmann et al., 2010).
In subsurface groundwaters, complex bacterial communities were deciphered at the genome level, evidencing inter-organism interactions involved in ecosystem plasticity (Anantharaman et al., 2016). Bacterial communities in As-rich groundwater have been reported to be dominated by Firmicutes, Alpha-, Beta-, Gamma- and Epsilonproteobacteria (Bertin et al., 2011; Liao et al., 2011; Lin et al., 2012; Escudero et al., 2013; Héry et al., 2015; Wang et al., 2016; Crognale et al., 2017). In order to evidence the distribution, phylogeny and activity of As bacteria in groundwater, functional molecular markers have been applied in several groundwater systems (Escudero et al., 2013; Li et al., 2017; Wang et al., 2018). Arsenite-oxidizing bacteria (AOB) have been studied by analyzing the diversity of aioA genes, encoding the large subunit of As(III) oxidase (AioA) with different molecular approaches (Hamamura et al., 2009; Heinrich-Salmeron et al., 2011; Xiao et al., 2016; Crognale et al., 2017). This gene has been retrieved in natural surface waters characterized by As concentration ranging from 0.01 to 4 mg L-1 As (Escudero et al., 2013) and in groundwater with >1 mg L-1 As (Quéméneur et al., 2010). Genes coding for dissimilatory As(V) reductase (ArrA) were used for demonstrating the presence of As-releasing microbial processes in anoxic groundwater in the Coastal Plain in New Jersey (United States) with As ranging from 20 to 80 μg L-1 (Barringer et al., 2010) and in Chinese and Cambodian groundwater sediments (Héry et al., 2015; Li et al., 2017). As resistance by means of As(V) reduction has been investigated widely both in environmental samples as well as in bacterial strains isolated from groundwaters (Guo et al., 2015; Sarkar et al., 2016).
Previous studies focused on the microbiological characterization of As-rich groundwaters in South and South East Asia, including Bangladesh (Hassan et al., 2016; Sultana et al., 2017), China (Guo et al., 2015; Li P. et al., 2015; Wang et al., 2016), West Bengal (Osborne et al., 2015) and Taiwan (Das et al., 2016). Here, As pollution is more severe in shallow aquifers (generally < 50 m below surface) formed by Holocene sediments (Ravenscroft et al., 2009; Fendorf et al., 2010; Zheng et al., 2019). A similar situation has been observed in shallow aquifers in the Mississippi Delta in Southern Louisiana, where microbial respiration has been supposed to be responsible for As dissolution (Yang et al., 2014).
It has been postulated that groundwater As content declines with sediment age and it increases in concomitance with microbial degradation of fresher, and thus more reactive, organic matter (Postma et al., 2012; Stuckey et al., 2016). Conversely, in the Po Plain in Northern Italy, As pollution is more severe in intermediate aquifers (between 50 and 150 m below surface) lying on Pleistocene sediments (Rotiroti et al., 2014, 2015). Here, sediments and the buried organic matter (peat) driving As release to groundwater are older. Therefore, the above-mentioned relationship between groundwater As concentrations and sediment age/freshness of organic matter cannot be applied. A similar phenomenon has been observed in the Mahomet aquifer in Illinois, a Pleistocene glacial aquifer system (Kirk et al., 2004; Kelly et al., 2005).
Hydrogeological and microbial processes related to As dissolution in aquifers lying on recent Holocene sediments, like those in South and South East Asia, have been comprehensively characterized. At the best of authors’ knowledge, microbial characterization of aquifers in ancient floodplain areas, like those lying in the Po Plain at the foot of the Alps, is a novel aspect that cannot be found in previous works.
In this context, the present study aimed to decipher the microbial composition of groundwater residing on Pleistocene sediments in Northern Italy thus expanding knowledge on As dynamics in worldwide contaminated aquifers. Furthermore, the detection of microbial functionality toward As was investigated in order to evidence the potential to exploit resident bacterial populations for metalloid removal.
Materials and Methods
Hydrogeological Features and Physicochemical Characteristics of Groundwaters
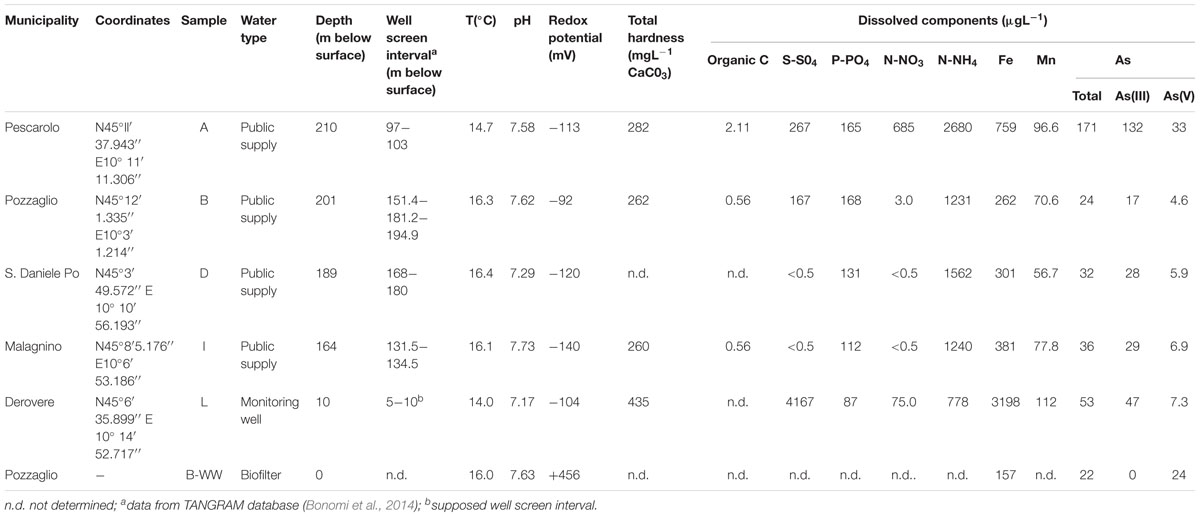
Water samples were collected from six sites located in the province of Cremona (Lombardia, Northern Italy, see Supplementary Figure 1). Samples came from four public-supply wells of drinking water (samples A, B, D, and I), one monitoring well (sample L) and one biofilter unit (sample B-WW) (Table 1). The studied area hosts a multilayer system characterized by vertical alternations of sands (aquifer units) and silty clays (aquitard units), which features the whole lower Po Valley (Ori, 1993). Well A is located in an area closer to the Oglio River where aquifer units are considerably separated by silty clay layers. It taps two overlapping aquifer units separated by a ∼50 m thick silty clay layer. Well D is located in an area closer to the Po River, where sands are abundant and overlapping aquifer units are less separated. Wells B, I and L are located in areas equidistant from both the Po and Oglio Rivers and their hydrogeological features are intermediate with respect to those described for wells A and D. Well B taps a unique deep aquifer unit. Although well I has two screen intervals, it taps a unique aquifer unit; this aquifer is overlaid by a 13 m thick peaty clay layer. Sampling point L is a shallow piezometer tapping the unconfined aquifer (Rotiroti et al., 2014).
Sampling and Physicochemical Analysis of Environmental Parameters
Groundwater samples were collected by purging wells until groundwater temperature, pH, dissolved oxygen and redox potential (Eh) were stabilized. Samples were transported on ice to the laboratory and frozen at -20°C until analyses. All sample bottles and caps were acid-washed and autoclaved before use. pH, dissolved oxygen (DO) and Eh were measured within few hours after sampling, using a pH-meter PCE-228 (PCE Deutschland GmbH, Meschede, Germany), a portable dissolved oxygen-meter-HI 9146 (Hanna Instrument US Inc., Woonsocket, United States) and a mV-meter PCE-228 (PCE Deutschland GmbH, Germany), respectively. Total dissolved carbon was measured by applying the potassium dichromate standard method (ASTM D1252-06, 2012). Ammonium (NH4) and nitrate (NO3) were measured by flow injection analysis and spectrometric detection (FIAstar 5000 Analyzer, Foss Tecator, Denmark). Sulfate was analyzed combining the gravimetric and colorimetric methods according to Murphy and Riley (1962). Samples for As, Fe and Mn analysis were preserved by filtering (0.22 μm) and adding HNO3 to a final concentration of 2% (v/v) and the elements were measured by inductively coupled plasma mass spectrometry (ICP-MS, Agilent Technologies, United States), using a multistandard solution ranging from 0 to 1 mg L-1 (Agilent Technologies, United States). To determine inorganic As speciation, As(V) and As(III) were separated by filtration according to Kim et al. (2007), using WATERS Sep-Pak® Plus Acell Plus QMA cartridge (Waters, MA, United States), and followed by ICP-MS analysis as described above.
Nucleic Acid Extraction
The microbial biomass was collected from 25 L of water that was filtered over 0.22 μm hollow fiber filters (Mediakap-5, SpectrumLabs, United States) and stored at –20°C until nucleic extraction. Biomass was detached from the filter by thoroughly rinsing with 10 mL of 0.2% (w/v) sodium pyrophosphate, and collected from the cartridge with a syringe. The suspension was centrifuged (10,000 rpm for 25 min at 10°C) and total DNA was extracted from the pellets using the PowerSoil® DNA Isolation Kit (MO-BIO Laboratories, Inc., Carlsbad, CA, United States), with two additional lysing steps at 65°C for 30 min, and 90°C for 5 min. The DNA from the enrichment cultures was isolated with UltraCleanTM Microbial DNA Isolation Kit (MO-BIO Laboratories). The extracted DNA was visualized on agarose gel, and the quality and quantity were determined with a ND-1000 spectrophotometer (Nanodrop Inc., Wilmington, DE). PowerWater® RNA Isolation Kit (MO-BIO Laboratories), was used for RNA extraction from groundwater samples, according to manufacturer’s instructions. Despite different attempts, RNA extractions were unsuccessful.
16S rRNA Gene Amplicon Sequencing
Barcoded fragments (∼ 570 bp), spanning the V4 and V6 hypervariable regions (Escherichia coli position 530–1100) of the 16S rRNA gene were amplified from the groundwater DNA extractions using universal bacterial primers 530F (5′-GTGCCAGCMGCNGCGG-3′) and 1100R (5′-GGGTTNCGNTCGTTR-3′), fused to 454 A and B adaptors, respectively (De León et al., 2012). PCR reactions for each sample were carried out in triplicate and then pooled together for pyrosequencing with Life Sciences 454 Genome Sequencer FLX platform (Roche, Switzerland) according to standard protocols. A negative control (blank) was included for each PCR reactions. A 100 ng (1 μL) aliquot of each DNA sample was used for a 50 μL PCR reaction. Taq Master Mix Kit (Qiagen) was used for PCR under the following conditions: 94°C for 3 min followed by 32 cycles of 94°C for 30 s; 60°C for 40 s; and 72°C for 1 min; and a final elongation step at 72°C for 5 min.
Quality treatment was performed by using Pyrotagger (Kunin and Hugenholtz, 2010), removing low quality data. QIIME was applied as sequence quality filter to the original 16S rRNA gene sequence dataset based on the sequence quality log file (Caporaso et al., 2010). Sequences shorter than 200 nucleotides, those with one or more ambiguous bases and those that received a quality score lower than 25 were eliminated. After this, an average of 3000 reads per sample were considered. QIIME was used for phylogenetic analysis of 16S rRNA sequences. Bacterial operational taxonomic units (OTUs) were defined at a value of 97% similarity of the 16S rRNA gene sequences. The number of sequences present in each sample after applying filtering is shown in Supplementary Figure 4A.
Unique pyro-sequences of 16S rRNA genes were aligned using the “align.seqs” command and the Bacterial SILVA SSU Ref database (Release 119). Sequences were taxonomically classified by the RDP-II Naïve Bayesian Classifier using a 60% confidence threshold against the SILVA Database. A phylogenetic tree was constructed using the FastTree with Kimura’s two-parameter model (Price et al., 2009). To investigate patterns in alpha diversity, rarefaction analysis was performed and collector’s curves were calculated (Supplementary Figure 4B), including the Chao1 richness estimator (Chao, 1984) and the Shannon Diversity Index. To investigate patterns in beta-diversity, pairwise distances between bacterial communities were calculated with the UniFrac distance matrix (Lozupone and Knight, 2005). Predictive microbial As, Fe, and S processing profiling within the 16S Amplicon sequencing library was performed according to Zecchin et al. (2017). The reference database used for this analysis included recent updates on literature published within the end of 2018.
The microbial communities of groundwater samples were also analyzed by denaturing gradient gel electrophoresis (DGGE) analysis of 16S rRNA genes using primers Bac341F-GC and Bac907rM according to Schäfer and Muyzer (2001). Details of the PCR reactions, concentration of the reagents, thermal protocol, electrophoresis, and data analysis are described in Supplementary Material Section “Experimental Conditions.”
qPCR of Functional Genes
The presence of As and RuBisCO genes was tested and quantified by real time qPCR reactions. Particularly, genes for arsenite oxidase (aioA), arsenate reductase (arsC), arsenate respiratory reductase (arrA), and for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) Type-I (cbbL) were investigated.
Amplification of arsenite oxidase gene aioA was performed with primers aoxBM1-2F and aoxBM3-2R according to the protocol of Quéméneur et al. (2008). Amplification of arsenate reductase gene arsC was conducted with primers P52F and P323R according to Bachate et al. (2009). Amplification of arsenate respiratory reductase arrA was performed with primers ArrAfwd and ArrArev according to the protocol of Malasarn et al. (2004). Amplification of the gene coding for the large subunit of RuBisCO Type-I (cbbL) was performed with the primers RBCO-1Cf and RBCO-1Cr according to the procedures of Alfreider et al. (2009). PCR reactions were performed in a final volume of 25 μL containing: 2 μL of template, 0.2 mmol L-1 of dNTPs, 1.75 mmoL l-1 of MgCl2, 0.5 μmol L-1 of each primer, 1.5 U of Taq polymerase and 1X PCR buffer. Amplification protocols used for aioA, arsC, arrA and cbbL gene amplifications were the same as reported by the authors. All the reagents were from Invitrogen. PCR reactions were carried out using the T-Gradient apparatus (Biometra, Germany). PCR products were checked on a 1.5% (w/v) agarose gel containing GelRedTM stain (Biotium) at 0.01% (v/v) and visualized using a Bio-Rad Gel Documentation System (Bio-Rad).
The same targets were quantified in the samples by Real Time quantitative PCR (RT-qPCR). To quantify aioA genes, primers aoxBM1-2F and aoxBM2-1R were used, whereas for arsC and arrA quantification the primers were the same used for PCR amplification. Each reaction mixture included 10 μL of 2x SsoFast Evagreen Supermix (Bio-Rad, Hercules, CA), 0.2 ng μL-1 of each primer for aioA and arsC and 0.5 ng μL, and 2 μL of template DNA in a total volume of 20 μL. The thermal protocols used for the amplifications were set up according to Quéméneur et al. (2010) for aioA and to Bachate et al. (2009) for arsC. Amplification of arrA was carried out with the following protocol: 95°C for 10 min, 40 cycles including 95°C for 30 s, 55°C for 1 min and 72°C, and final step for melting curve generation with 55°C for 30 s followed by temperature increasing steps of 0.5°C s-1 each until 95°C. Genomic DNA isolated from Achromobacter sp. strain 1L was used to generate the standard curve for aioA quantification. To obtain the standard curve for arsC quantification, DNA isolated from Bacillus licheniformis strain SeaH-As1w was used. The standard curve for arrA quantification was generated with DNA isolated from Desulfitobacterium hafniense strain DCB-2.
Clone Library Preparation
The purified PCR products were ligated into the pCR2.1-TOPO vector and transformed into TOP10 chemically competent E. coli cells according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA). The transformed cells were plated on Luria-Bertani (LB) agar plates containing 100 μg mL-1 of ampicillin, 80 μg mL-1 of 5-bromo-4-cholo-3-ondolyl-b-D-galactopyranoside (X-Gal). After an overnight incubation at 37°C, white transformants were transferred to LB ampicillin (100 μg mL-1) agar plates and cultured overnight. To ensure the presence of the insert, PCR amplification was performed directly on the colonies by using specific primer pair for each target. The plasmids from a random selection of positive clones were purified and sequenced using M13 universal primers (Invitrogen) and the Taq Dye-Deoxy Terminator Cycle Sequencing kit (Life Technologies Co., Carlsbad, California, United States). The forward and reverse sequences for each target were obtained loading the respective samples on an ABI 310 Genetic Analyzer (Life Technologies Co., United States).
Comparative Sequence Analysis
Nucleotide sequences of aioA, arrA, arsC, and cbbL genes obtained from clone libraries were manually edited, translated into amino acid sequences and searched for closely related amino acid sequences in GenBank database (BlastX). Obtained amino acid sequences and reference sequences for each target were aligned with ClustalX, and trees were built with MEGA 6 using the neighbor-joining distance method based on p-distance (Tamura et al., 2007). A total of 1000 bootstrap replications were calculated.
Selection of Arsenic-Transforming Bacterial Cultures
Microorganisms involved in As cycle (i.e., autotrophic and heterotrophic As(III)-oxidizing bacteria (AAOB and HAOB), detoxifying As(V)-reducing bacteria (ARB), and dissimilatory As(V)-reducing bacteria) were enriched from groundwater samples. To set up the enrichments, BBWM medium (Corsini et al., 2014) added with 1.5 mmol L-1 sodium As(III) (NaAsO2) or with 15 mmol L-1 sodium As(V) (Na2HAsO4⋅7H2O) was prepared. Sodium lactate (NaC3H5O3) was added (0.4 mol L-1) to DARB, HAOB and to ARB enrichments, and omitted to AAOB enrichments. Water samples and culture medium were mixed on a 1:1 proportion and incubated at 28°C under shaking at 150 rpm (AAOB, HAOB and ARB). Set up of DARB enrichments were performed under anaerobic conditions in an anaerobic hood. Arsenic transformation within the enrichments were quantified by permanganate colorimetric analysis performed according to Salmassi et al. (2002) and Dhar et al. (2004).
Results
Physicochemical Characteristics of Groundwater Samples
The physicochemical characterization of water samples is described in Table 1. All groundwater samples were anoxic to mildly oxic, with Eh values ranging from -92 mV in samples B to -140 mV in samples I, whereas Eh in the biofilter unit B-WW was +456 mV. All samples were oligotrophic, with a maximum value of organic carbon of 2.11 μg L-1 measured in sample A. The pH was circumneutral in all samples. Arsenic concentrations ranged from 22 μg L-1 in biofilter sample B-WW to 171 μg L-1 in public supply well A (Table 1). In all samples, As(III) was the dominant As species. In sample B-WW, As content was similar to that of the original groundwater (site B), with As(III) being totally oxidized to As(V). Organic contaminants (i.e., aliphatic or aromatic hydrocarbons) and heavy metals (i.e., cadmium, mercury, selenium, vanadium, and antimony) were absent in all water samples.
Bacterial Community Structure in Groundwater Samples
Barcoded pyrosequencing of 16S rRNA genes (16S Amplicon sequencing) was applied to obtain a detailed description of microbial community composition of the 6 samples.
The rarefaction curve approximated toward a plateau after ∼3000 OTUs were sequenced, indicating that enough sample coverage was obtained in this study (Supplementary Figures 4A,B). The species richness estimator Chao1 and pairwise distances values showed that the samples had different levels of diversity (Supplementary Table 3). Samples formed three separate clusters: public supply wells, biofilter water and monitoring well, with the last one showing a high differentiation, as evidenced by the topology of the weighted UniFrac UPGMA tree (Supplementary Figure 5). Taxonomic classification indicated that 21 phyla of Bacteria and Achaea were present in the 6 samples. In samples A, B, L, and B-WW, 80, 70, 76, and 55%, respectively, of the reads were classified as uncultured taxa, at all taxonomic levels. In samples D and I, the same accounted for 17 and 23%, respectively (Figure 1B). Proteobacteria was the dominant phylum, followed by Bacteroidetes and Chloroflexi in most of the samples. Firmicutes, Nitrospira, Clamydiae, and Acidobacteria were less abundant, although in some wells represented more than 4% of the community (Figure 1A and Supplementary Figure 5). Within the Proteobacteria, Betaproteobacteria dominated in samples A and B (70.1 and 23.8%, respectively), Epsilonproteobacteria predominated in samples I and D (64.8 and 50.2%, respectively), and Gammaproteobacteria predominated in the biofilter sample W (40.0%) (Figure 1A). The most abundant Alphaproteobacteria were members of the genera Sphingopyxis in sample D (8.8%) and Sphingomonas in sample B (7.7%). The main populations of Betaproteobacteria were Thiobacillus (5.4%, in sample W) and Methylophilus (4.5 and 3.0% in samples A and W, respectively). Within Epsilonproteobacteria the only species retrieved was Sulfuricurvum kujiense, which dominated in four out of six groundwater samples and represented 64.8, 50.2, 14.7, and 2.6% of the bacterial community in samples I, D, B, and L, respectively. Within Gammaproteobacteria, Thiotrix (16.3%, in sample B-WW), Pseudomonas (15.1%, in sample D), Azotobacter (5.6%, in sample A) and Methylomonas (3.5%, in sample I) were the most representative genera.
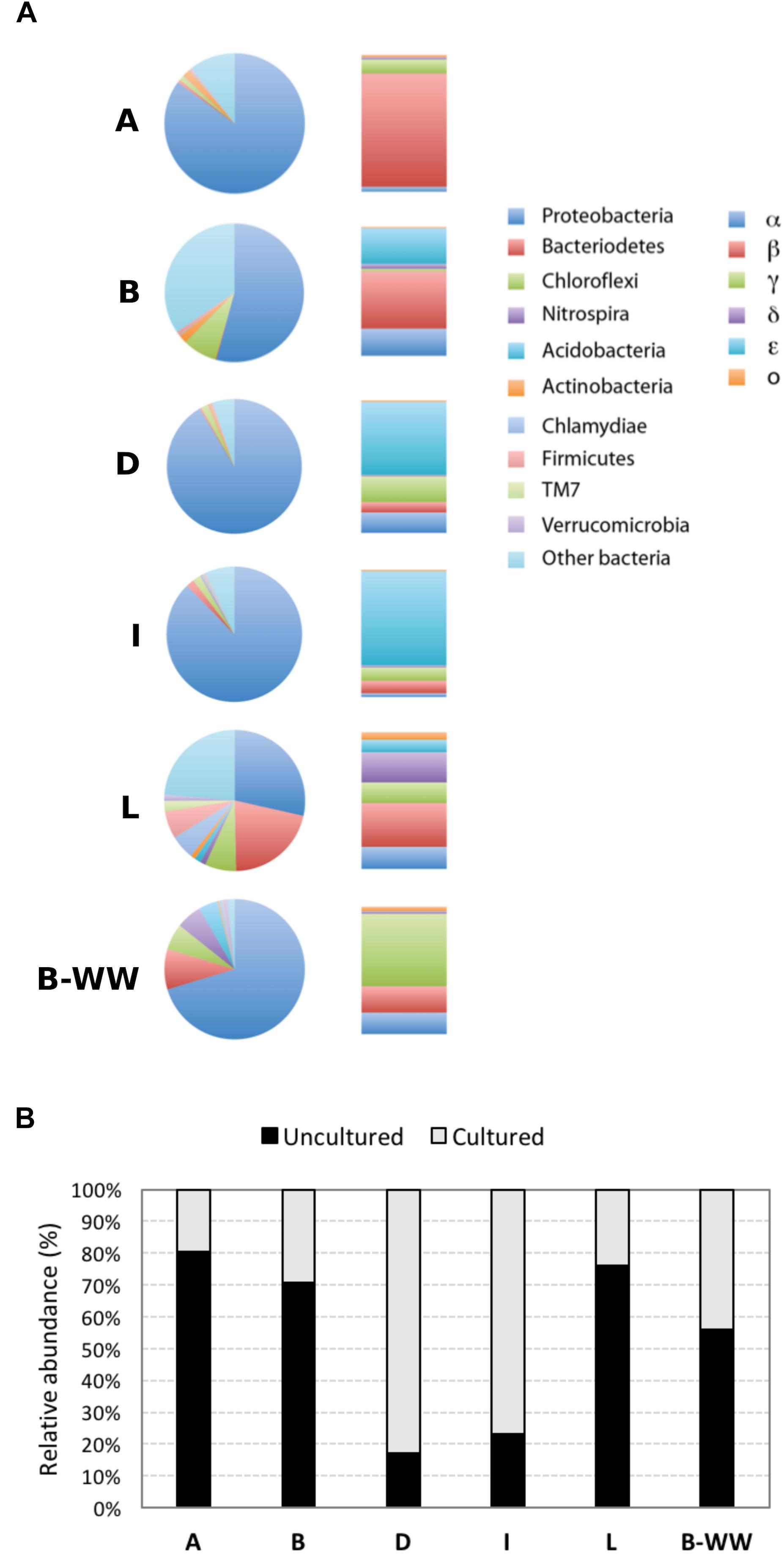
Figure 1. Microbial communities in groundwaters sampled from four drinking water public supplies (A, B, D, and I), one monitoring well (L) and one biofilter unit (B-WW), analyzed by 16S rRNA gene amplicon sequencing. Colored pie charts and bar graphs (A) show the percentage at the phylum level and the percentage of the different classes within the Proteobacteria, respectively. Gray-scale bars (B) represent the percentage of sequences of uncultured vs. cultured species retrieved in the water samples.
Among the retrieved genera, those documented to be involved in As resistance and oxidation, together with S oxidation, were dominant. In these planktonic communities, Fe-cycling bacteria were present although less abundant with respect to S bacteria (Figure 2, Supplementary Table 4, and Supplementary Figure 6). Inferred functionalities of these microbial genera are presented in the Discussion section.
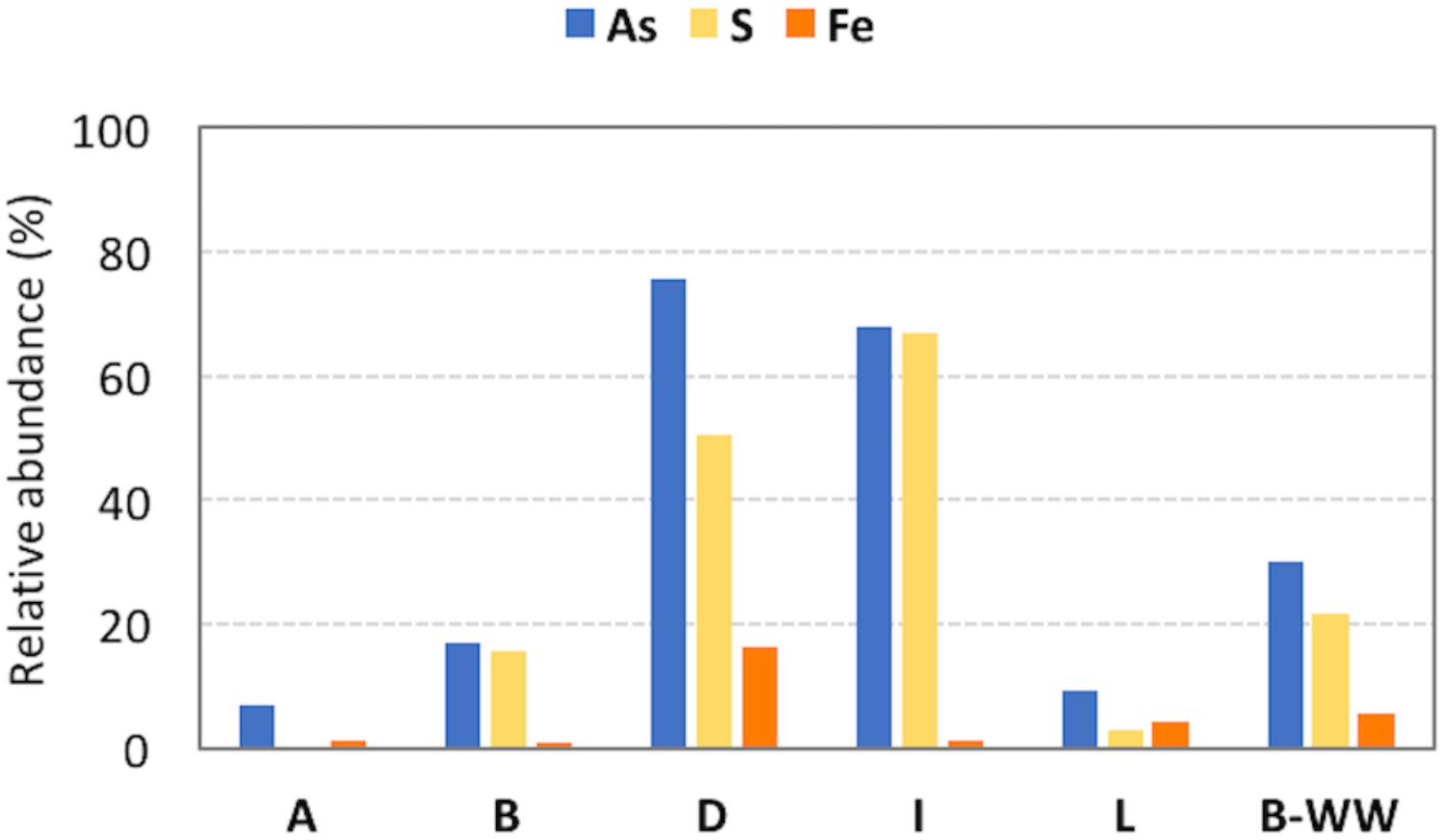
Figure 2. Relative abundances of microorganisms in the 16S rRNA amplicon sequencing library potentially involved in arsenic (As), sulfur (S) and iron (Fe) cycles in the different samples. The vast majority of S cycle-related species belong to Sulfuricurvum spp. in groundwaters B, D and I and to Thiothrix spp. in biofilter unit B-WW.
Data from DGGE analysis (Supplementary Figure 2) performed on the same samples confirmed the presence of a high percentage of uncultured species (Supplementary Table 1), most of which related to S-oxidizing microorganisms (Supplementary Figure 3 and Supplementary Table 2).
Diversity of Arsenic Genes in Groundwaters
In groundwater DNA As-cycling bacteria were quantified and depicted by genetic biomarkers (Table 2). The most abundant were aioA and arsC, with the highest values measured of 4.2 × 102 and 6.8 × 102 gene copies L-1, respectively. ArrA were found only in sample A in low amount (1.8 101 copies L-1). In sample D, the amplification of any functional gene from the DNA was not possible, despite different attempts (Table 2).
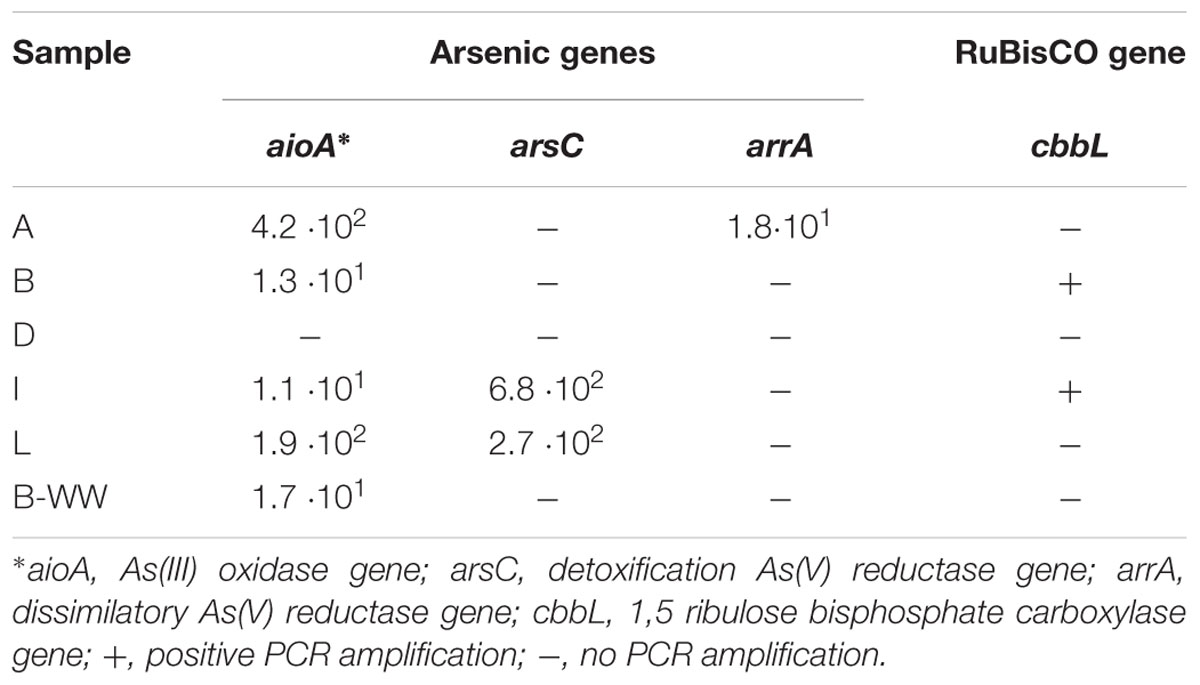
Table 2. Biomarkers quantification (gene copies per L) of arsenic cycle genes and for chemoautotrophy in groundwater DNA.
AioA clone libraries constructed from five water samples contained 140 clones with expected insert size of 1111 bp. Translated amino acid sequences were grouped into two major clusters belonging to Alphaproteobacteria and Betaproteobacteria, regardless the origin of the water (Figure 3A). The cluster associated to Alphaproteobacteria exclusively included members of the order Rhizobiales (95–99% identity). Clones from site A, B, B-WW, and I were similar to the AioA sequence of uncultured bacteria from an As-contaminated soil (97–98% identity with ADF47224, ADF47258, ADF47248, Sultana et al., 2012). Sequences obtained from site I and from site L were related to uncultivated bacteria from different As-contaminated mines or sediments (95–99%, Quéméneur et al., 2008; Heinrich-Salmeron et al., 2011; Yamamura and Amachi, 2014). Finally, clone 55 from site L formed a cluster with the aioA of an uncultivated bacterium (97% to BAP99953) from enrichment cultures with As(III)-spiked paddy soil (Dong et al., 2016) and with the sequence of Nitrobacter hamburgensis X14, a facultative chemolithoautotroph bacterium (Muller et al., 2007). The cluster associated with Betaproteobacteria was related (81–96% identity) to members of the order Burkholderiales. Clone 14 from site A and clones 69, 74, and 96 from site B had 96% similarity to As(III) oxidases of Variovorax sp. 4-2 (ABY19319) which was isolated from a laboratory reactor for biological As(III) oxidation (Battaglia-Brunet et al., 2002), and of an uncultivated bacterium (AGO59191) from As-contaminated soil (Sultana et al., 2012). Finally, clone 24 from site A, closely related to the As(III) oxidase of an uncultured bacterium from As-rich geothermal water (Jiang et al., 2014), and clone 55 from site L, closely related to the aioA of an uncultivated bacterium from sediments (Yamamura and Amachi, 2014), formed a cluster with Thiomonas delicata (ABY19316).
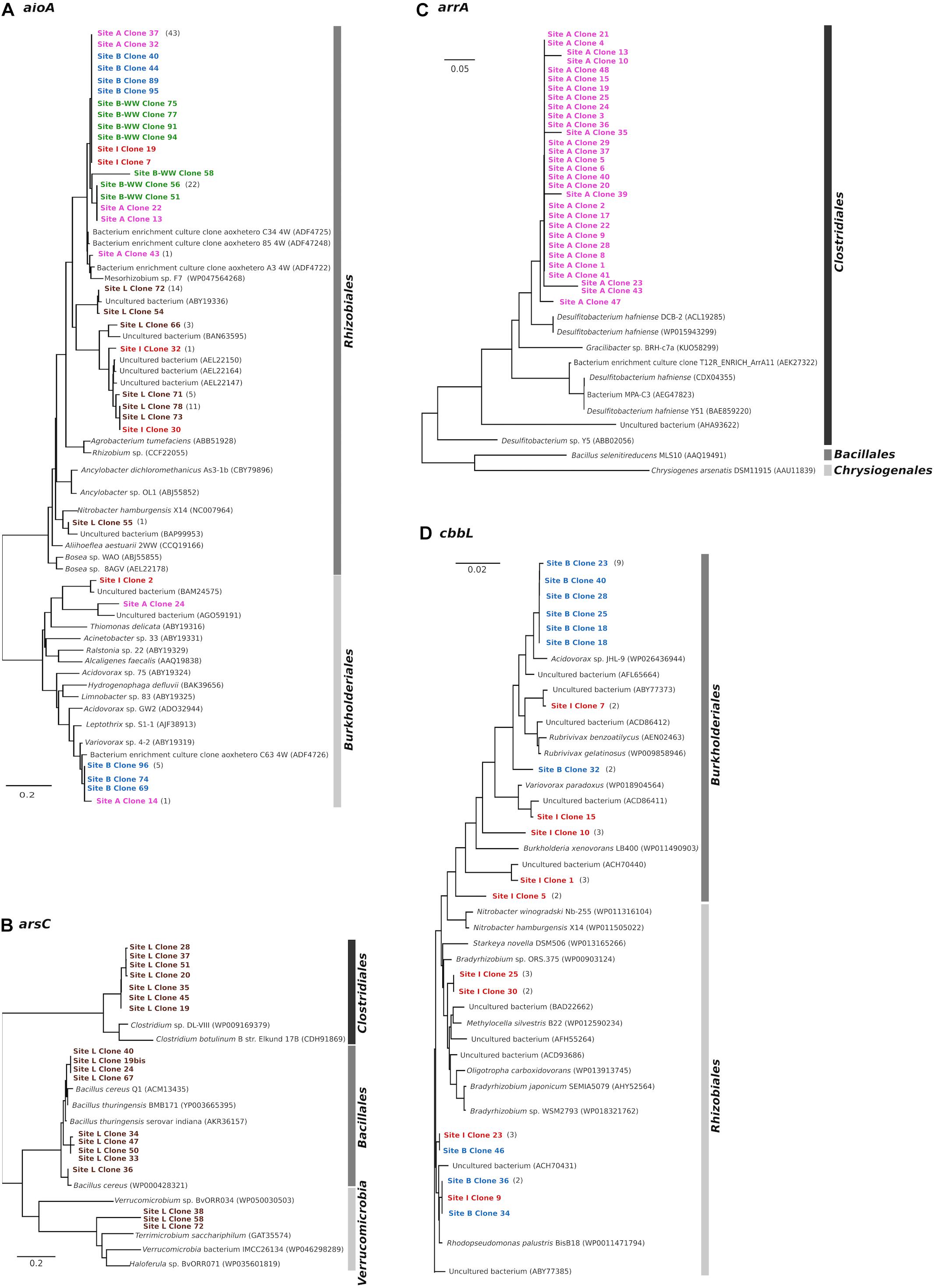
Figure 3. Phylogenetic analysis (neighbor-joining tree) of deduced amino acid sequences of aioA (A), arsC (B), arrA (C) and type I RuBisCO cbbL (D) genes cloned from arsenic-contaminated aquifers and sequences from the NCBI database. As outgroup for the calculations, the AioA sequence of Hoeflea phototrophica DFL-43, the ArsC sequence of Achromobacter xylosoxidans A8, the ArrA sequence of Shewanella sp. ANA-3 and the CbbL sequence of Ancylobacter dichloromethanicus As3-1b were used. The scale bar represents a difference of 0.2 nucleotides per position for AioA and ArsC, of 0.05 for ArrA, and of 0.02 nucleotides per position for CbbL.
Clone libraries constructed with arsC amplicons from sample L contained 20 clones. The translated amino acid sequences of arsC gene were grouped into two major clusters: Firmicutes and Verrucomicrobia (Figure 3B). The cluster associated with Firmicutes was related to members of the orders Bacillales and Clostridiales (78–99% identity). The Bacillales-affiliated group was most closely related to the ArsC of several Bacillus species which were isolated from a deep-subsurface oil reservoir in the Daqing oil field in north-eastern China and from a soil in the United States (Xiong et al., 2009; Li Q. et al., 2015), whereas the Clostridiales-affiliated group was closely related to the ArsC of Clostridium species isolated from Pacific sediments and from an anaerobic sludge (Stringer et al., 2013; Taghavi et al., 2013). A second cluster was associated to ArsC of different members of Verrucomicrobia which were isolated from different environments, such as lake, roots and rice paddy soil (Qiu et al., 2014).
Clone libraries constructed with arrA amplicons from sample A resulted in 30 clones. They formed one cluster with the highest similarity to the ArrA sequences of D. hafniense strains (91% sequence similarity, Figure 3C), known anaerobic bacteria capable of dehalogenation and metal reduction (Pérez-Jiménez et al., 2005; Mumford et al., 2013).
Diversity of RuBisCO Genes in Groundwater Samples
The presence of chemolithoautotrophic strains in the planktonic bacterial communities of two aquifer samples B and I was evidenced by the amplification of the cbbL gene, coding for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) Type-I (Table 2). The cbbL clone libraries from these two samples contained 40 clones. Phylogenetic analysis clearly separated the cbbL sequences into Alpha- and Betaproteobacteria (Figure 3D). The cluster associated with Alphaproteobacteria was related to members of the order Rhizobiales (96–98% identity). Two clones from site B and one clone from site I formed a group together with RuBisCO sequences of an uncultivated bacterium from tar-oil-contaminated porous aquifer (96% to ACH70431, Kellermann et al., 2012) and of a strain of Rhodopseudomonas palustris BisB18 isolated from freshwater sediment (96% to WP0011471794, Oda et al., 2008). Two clones from site I were clustered with members of Beijerinckiaceae and Bradyrhizobiaceae families. Finally, clones 46 from site B and 23 from site I were not clearly affiliated to other RuBisCO sequences and showed the highest homologies (96%) with cbbL genes of uncultured bacteria reported in Alfreider et al. (2009). The cluster associated with Betaproteobacteria was related to members of the order Burkholderiales (95–100% identity). Six out of 10 clones from site B formed one group with 100% similarity to the CbbL sequence of Acidovorax sp. JHL-9 (WP02643944), an autotrophic H2-oxidizing, nitrate-respiring bacterium isolated from a subsurface oxic-anoxic transition zone (Lee et al., 2015). Clones 1, 7, and 15 from site I had high similarity to CbbL of different uncultured Betaproteobacteria from tar-oil-contaminated aquifer (99% to ACH70442), groundwater (98% to ABY77373) and from grassland soil (95% to ACD86411), respectively (Alfreider et al., 2009; Videmsek et al., 2009; Kellermann et al., 2012).
Arsenic-Transforming Bacterial Cultures
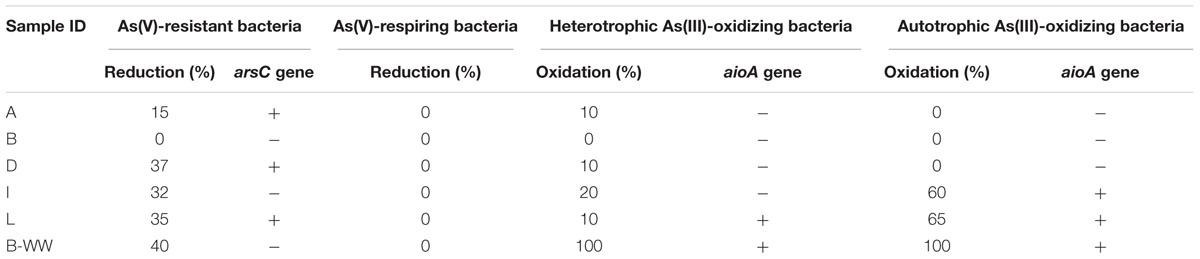
The presence of As-transforming bacteria in groundwaters was confirmed by enrichment cultures after multiple transplants on As(III) or As(V) (Table 3).
Arsenic resistance by means of heterotrophic As(V) reduction and As(III) oxidation was retrieved in most of the samples. On the contrary, As(V) dissimilative reduction was not displayed in any groundwater sample. Under chemolithoautotrophic conditions, As(III) was oxidized in samples I, L, and B-WW. The culture from aerobic biofilter B-WW was the most active one with respect to cultures from groundwaters for all the As transformations. PCR amplifications of arsC and aioA genes confirmed the presence of As-transforming bacteria (Table 3).
Enrichment cultures where As metabolisms were evident, were subjected to DGGE analysis which resulted in the isolation of 29 successfully sequenced bands (see Supplementary Tables 5–7). Autotrophic As(III)-oxidizing enrichments (Supplementary Table 5), were composed of Hydrogenophaga sp. strains, previously reported to be autotrophic As(III) oxidizers (Cavalca et al., 2013) and Acinetobacter sp., Acidovorax sp. and Exiguobacterium auranticum strains previously reported to be heterotrophic As(III) oxidizers (Cavalca et al., 2013; Pandey and Bhatt, 2016). These species are common inhabitants in Chinese high As aquifers (Li et al., 2013; Li P. et al., 2015; Wang et al., 2016). For other species retrieved in autotrophic consortia (i.e., Sphingopyxis chilensis, Luteimonas aestuarii, and Lysobacter capsici), the oxidative activity has never been demonstrated, although in GenBank several As gene sequences are deposited from the genome of related species, including genes for As(III) oxidation. In the heterotrophic As(III)-oxidizing cultures (Supplementary Table 6), several As(III) oxidizers were found, including a close relative of Delftia sp., which was shown to be a facultative chemolithoautotrophic As(III) oxidizer (Sultana et al., 2017). In As(V) reducing enrichments most of the strains already retrieved in As(III) oxidizing cultures were detected, with the exception of Microbacterium hydrocarbonoxydans and Rhodococcus ruber strains (Supplementary Table 7). All were documented to be resistant to As(V) or to carry As genes in their genome.
Discussion
Within the studied area, the content of sand and silt clay varies from the Po River in the Southwest, where sands are prevalent and thus overlapping aquifer units are less separated, to the Oglio River in the Northeast, where silts and clays are abundant generating a high degree of separation between overlapping aquifer units (Beretta et al., 1992; Vassena et al., 2012; Rotiroti et al., 2015). Deeper aquifer units (generally 30 m below the surface) are confined and characterized by extended groundwater circulation, and thus, by longer groundwater residence time. This aspect also influences groundwater chemistry: indeed, longer residence time and no or limited surface infiltration favor reducing environments, which promote the mobilization of As, Fe, Mn, and NH4 driven by the degradation of peat incorporated into semi-permeable silt and clay aquitards (Francani et al., 1994; Zavatti et al., 1995; Carraro et al., 2013; Rotiroti et al., 2014). The peak of As release is expected in intermediate aquifers (generally between 50 and 150 m below surface) since here: (i) reduction of Fe-oxides likely favors the release of As, and (ii) co-precipitation of dissolved As with sulfides is not yet prevailing (Carraro et al., 2015; Rotiroti et al., 2015).
As described by high throughput sequencing, a high number of uncultured species was present in most study sites, evidencing that these environments are still largely unexplored. Species composition in As-affected water samples of Lombardia, resembled those previously described in other planktonic communities of uncontaminated waters in the Netherlands (Roeselers et al., 2015), of metal-contaminated groundwater in the United States (Hemme et al., 2010) and of high As aquifer in Bangladesh (Legg et al., 2012).
A complete C cycle is supported in these oligotrophic environments by the presence of a large number of autotrophic and heterotrophic bacteria. In most samples, autotrophic metabolism could be inferred by different strategies like autotrophic As(III) oxidizing enrichment cultures, PCR amplification of the cbbL gene coding for RuBisCO as well as by detection of autotrophic genera in the 16S Amplicon data. Dissimilative nitrate respiration was largely represented by the presence of Sulfuricurvum, Denitratisoma, Geobacter, Acidovorax, and Dechloromonas genera, which couple S, Fe(II) and As(III) oxidation to NO3 reduction (Finneran et al., 2002; Sun et al., 2009; Carlson et al., 2013; Chakraborty and Picardal, 2013). Nitrogen fixation was represented by a large proportion of Alphaproteobacteria of the order Rhizobiales, which was also evidenced by cbbL gene detection of the Azotobacter genus.
Peculiar of four (I, D, B, and L) out of five studied water samples was the high abundance of Epsilonproteobacteria represented by the chemolithoautotrophic S-oxidizing genus Sulfuricurvum sp., S. kujiense has been detected in terrestrial sulfidic caves (Porter and Engels, 2008) and its predominance was observed in formation waters of oil sands reservoir (Hubert et al., 2012) and in aquifer sediments minimally impacted by residual contamination of uranium and vanadium (Han et al., 2012). High abundance of members of the genus Sulfuricurvum indicates that, in addition to Fe-cycling, chemolithoautotrophic S oxidation at the expense of nitrate or oxygen could be an important process in aquifers of Northern Italy, as in aquifers and peatland freshwaters previously analyzed (Haaijer et al., 2008; Handley et al., 2014). Recently, a large proportion of a single uncultured Sulfuricurvum species was inferred to perform C and N fixation on the basis of groundwater metagenomes (Handley et al., 2014; Anantharaman et al., 2016). Considering the large abundance measured in Northern Italian groundwaters, this species might play a pivotal role in these ecosystems.
Arsenic Cycle
Dissimilative As(V) reduction seemed to be less represented in the aquifer communities as typical As-respiring bacteria were poorly detected, differently from South and South East Asia (Osborne et al., 2015; Das et al., 2016; Gnanaprakasam et al., 2017). Nevertheless, arrA gene belonging to D. hafniense highlighted the presence of this function in sample A. In this sample, DOC (2.11 μg/L) and As content (171 μg L-1) were the highest with respect to other samples. In this confined aquifer, the mobilization of As(V) from sediments could be linked to the degradation of peat incorporated into semi-permeable silty and clayey aquitards. DARB were not enriched from samples, indicating either their absence/low abundance or possible failures in the cultivation strategy.
According to both molecular biomarkers and enrichment cultures, the ARS detoxification system was more represented than the dissimilative As(V) reduction, in accordance with investigations in West Bengal (Paul et al., 2015) and China (Li et al., 2017) groundwaters. ArsC genes detected in the monitoring well sample L belonged to Bacillus, Clostridium and Verrucomicrobium strains in accordance with their presence in bar-coding libraries (0.4, 0.6 and 0.2%, respectively), as previously reported for different environments (Cavalca et al., 2013). These bacteria are responsible of the cycling of soluble As fractions, as As(V) reductases encoded by arsC genes are unable to reduce adsorbed As (Macur et al., 2004) and might be relevant for As(V) reduction to As(III) in the planktonic bacterial communities described in the present study. The Sulfuricurvum genus, dominant in 4 samples, is characterized by the presence of arsC in the genome (Han et al., 2012).
AioA genes coding for arsenite oxidase are present in genomes of the chemolithoautotrophic bacteria retrieved in the studied sites, such as Nitrospira, Alphaproteobacteria (Rhizobiales, Rhodobacterales), Betaproteobacteria (Acidovorax, Hydrogenophaga, Nitrosospira), already detected in As(III)-rich groundwaters of Bangladesh (Hassan et al., 2016) and China (Li et al., 2013; Li P. et al., 2015). The presence of this metabolism was consistent with the phylogeny of aioA genes retrieved in the environmental DNA, as well as with the enrichment of As(III) oxidizing bacteria.
In some of the retrieved bacterial species, a chemolithoautotrophic metabolisms with As(III) as electron donor can be postulated. In fact, cbbL gene sequences belonging to Acidovorax sp., Variovorax sp. and Burkholderia sp. were detected. These microorganisms have been demonstrated to perform As(III) oxidation (Hamamura et al., 2009; Cavalca et al., 2013), while for other species retrieved in the clone library, such as Oligotropha carboxidovorans, Bradyrhizobium japonicum and Rhodopseudomonas palustris, only protein sequences for As(III) oxidases are deposited in the NCBI database.
The presence of As(III)-oxidizing bacteria in the studied sites was confirmed in vivo by enrichment cultivation. These microorganisms could be successfully employed for bioremediation purposes, as previously shown for aquifers and soils (Corsini et al., 2015; Katsoyiannis et al., 2015; Karn and Pan, 2017).
Our analysis indicated that bacteria present in groundwaters were possibly involved in methylation of inorganic As, such as Nitrosomonadales, Desulfovibrio, Methanobacterium and Clostridium, although this metabolism was not investigated in the present study. Under subsurface conditions, the biotic conversion of inorganic to organic forms of As by arsenite methyltransferase ArsM has been recently demonstrated (Maguffin et al., 2015; Wang et al., 2018).
Fe and S Cycles
Directly linked to As cycle, Fe and S bacteria play a significant role in As mobilization in subsurface environments (Héry et al., 2008, 2015). S-oxidizing bacteria represented by Sulfuricurvum genus were abundant in the analyzed aquifers, whereas Thiobacillus and Thiothrix were retrieved in biofilter water. Sediment-associated elemental S and sulfides in the local aquifer may represent a source of electrons for these bacteria and might be responsible of dissolution of metals and metalloids associated to sulfide minerals. Recently, Sulfuricurvum and Thiobacillus were found to be predominant in sediment-associated bacterial communities in high As aquifers (Li P. et al., 2015). The contribution of S-oxidizing bacteria in S cycling has been recognized in subsurface environments, due to their metabolic versatility (Anantharaman et al., 2016). With this respect, S. kujiense (Kodama and Watanabe, 2004) as well as Sulfuricurvum strain RIFRC-1 (Handley et al., 2014) perform anaerobic and microaerobic oxidation of solid S as sulfide, elemental S and thiosulfate, also contributing to pyritic-S oxidation (Campbell et al., 2006). Different sulfate-reducing bacteria (SRB) were detected in all the samples, but with relative abundance always below 1%, probably because sulfate reduction is energetically less favorable and needs a lower redox potential than those measured in the studied sites (∼ -100 mV) (Emmerich et al., 2012). Similar results were obtained in Asian aquifers (Lee et al., 2011; Li et al., 2014). A significant implication of these results from the studied aquifer is that S bacteria could be responsible for the oxidative dissolution of Fe-S-As bearing sediments. Moreover, this trend did not seem to be mitigated by the presence of SRB, that are recognized to limit natural As contamination in groundwater (Kirk et al., 2004; Omoregie et al., 2013).
Fe-oxidizing bacteria Rhodobacter and Leptothrix were retrieved at a lesser extent, whereas Gallionella, a typical neutrophilic freshwater Fe-oxidizing bacterium was not retrieved. Fe-reducing bacteria like Geobacter, Geothrix, Ferribacterium, and Desulfuromonas were detected in samples A, I, D, and L. Among these, only Geobacter was abundant in site L (2.9%), possibly related to the presence of high Fe concentration in water. The other Fe-reducing bacteria accounted for less than 1% of the total community, thus suggesting that the contribution of this population to As release from sediments was limited, in accordance with Anantharaman et al. (2016). Members of the genus Thiobacillus have been shown to reduce Fe (Lovley, 2006). These organisms accounted for 5.4% in B-WW; however, the highly positive redox potential measured makes this metabolism unreliable at the site.
Conclusion
The microbial communities of As-rich groundwaters of Northern Italy were characterized, revealing a wide diversity of uncultured bacterial species, thus deserving these environments of further attention. A complete microbial redox As cycle was present in most aquifers, according to the different As metabolic pathways retrieved by both molecular and cultivation approaches. In Northern Italy Pleistocene aquifers, differently from South and South East Asia aquifers, chemolithotrophy, mainly based on S cycle, might be involved in As mobilization processes. The retrievement of As(III)-oxidizing capacity in the microbiota represents an important source for future studies on the development of decontamination actions.
Data Availability
The datasets generated for this study can be found in GenBank, sequences produced by pyrosequencing were deposited in the Genbank database within the BioProject PRJNA507727. Sequences for aioA genes were deposited under accession number MK497003-30, cbbL genes under accession number MK496989-7002, arrA genes under accession number MK415637-55, DGGE bands under accession number MK312575-88.
Author Contributions
LC and GM contributed to conception and design of the study, to molecular and statistical analyses, and manuscript preparation. SZ and BA contributed to molecular analysis. PZ performed the chemical characterization of groundwater samples. MR and TB contributed to the hydrogeological setting of samples. All authors approved the submitted version.
Funding
The research was supported by the CARIPLO Foundation with projects 2010-2221 and 2014-1301.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the Agenzia Regionale per la Protezione dell’Ambiente (ARPA) Lombardia for providing access to institutional chemical database of the groundwaters and logistic support during sampling. The Department of Food, Environmental and Nutritional Sciences, Universitá degli Studi di Milano, partially covered the open access APC.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01480/full#supplementary-material
References
Alfreider, A., Vogt, C., Geiger-Keiser, M., and Psenner, R. (2009). Distribution and diversity of autotrophic bacteria in groundwater systems based on the analysis of RuBisCO genotypes. Syst. Appl. Microbiol. 32, 140–150. doi: 10.1016/j.syapm.2008.11.005
Anantharaman, K., Brown, C. T., Hug, L. A., Sharon, I., Castelle, C. J. J., Probst, A. J., et al. (2016). Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7:13219. doi: 10.1038/ncomms13219
Andreoni, V., Zanchi, R., Cavalca, L., Corsini, A., Romagnoli, C., and Canzi, E. (2012). Arsenite oxidation in Ancylobacter dichloromethanicus As3-1b strain: detection of genes involved in arsenite oxidation and CO2 fixation. Curr. Microbiol. 65, 212–218. doi: 10.1007/s00284-012-0149-9
Andres, J., and Bertin, P. N. (2016). The microbial genomics of arsenic. FEMS Microbiol. Rev. 40, 299–322. doi: 10.1093/femsre/fuv050
ASTM D1252-06 (2012). Standard Test Methods for Chemical Oxygen Demand (Dichromate Oxygen Demand) of Water. Conshohocken, PA: ASTM International, West.
Bachate, S. P., Cavalca, L., and Andreoni, V. (2009). Arsenic-resistant bacteria isolated from agricultural soils of Bangladesh and characterization of arsenate-reducing strains. J. Appl. Microbiol. 107, 145–156. doi: 10.1111/j.1365-2672.2009.04188.x
Barringer, J. L., Mumford, A., Young, L. Y., Reilly, P. A., Bonin, J. L., and Rosman, R. (2010). Pathways for arsenic from sediments to groundwater to streams: biogeochemical processes in the Inner Coastal Plain, New Jersey, USA. Water Res. 44, 5532–5544. doi: 10.1016/j.watres.2010.05.047
Battaglia-Brunet, F., Dictor, M. C., Garrido, F., Crouzet, C., Morin, D., Dekeyser, K., et al. (2002). An arsenic(III)-oxidizing bacterial population: selection, characterization, and performance in reactors. J. Appl. Microbiol. 93, 656–667. doi: 10.1046/j.1365-2672.2002.01726.x
Beretta, G. P., Francani, V., and Fumagalli, L. (1992). Studio Idrogeologico della Provincia di Cremona [Hydrogeological study of the Province of Cremona]. Bologna. Pitagora Ed.
Bertin, P. N., Heinrich-Salmeron, A., Pelletier, E., Goulhen-Chollet, F., Arsène-Ploetze, F., Gallien, S., et al. (2011). Metabolic diversity among main microorganisms inside an arsenic-rich ecosystem revealed by meta- and proteo- genomics. ISME J. 5, 1735–1747. doi: 10.1038/ismej.2011.51
Bonomi, T., Fumagalli, M., Rotiroti, M., Bellani, A., and Cavallin, A. (2014). The hydrogeological well database TANGRAM: a tool for data processing to support groundwater assessment. Acq. Sott. Ital. J. Groundw. 3, 35–45. doi: 10.7343/as-072-14-0098
Cai, L., Yu, K., Yang, Y., Chen, B., Li, X., and Zhang, T. (2013). Metagenomic explo- ration reveals high levels of microbial arsenic metabolism genes in activated sludge and coastal sediments. Appl. Microbiol. Biotechnol. 97, 9579–9588. doi: 10.1007/s00253-012-4678-8
Campbell, B. J., Engel, A. S., Porter, M. L., and Takai, K. (2006). The versatile 𝜀-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4, 458–468. doi: 10.1038/nrmicro1414
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carlson, H. K., Clark, I. C., Blazewicz, S. J., Iavarone, A. T., and Coates, J. D. (2013). Fe(II) oxidation is an innate capability of nitrate-reducing bacteria that involves abiotic and biotic reactions. J. Bacteriol. 195, 3260–3268. doi: 10.1128/JB.00058-13
Carraro, A., Fabbri, P., Giaretta, A., Peruzzo, L., Tateo, F., and Tellini, F. (2013). Arsenic anomalies in shallow Venetian Plain (Northeast Italy) groundwater. Environ. Earth Sci. 70, 3067–3084. doi: 10.1007/s12665-013-2367-2
Carraro, A., Fabbri, P., Giaretta, A., Peruzzo, L., Tateo, F., and Tellini, F. (2015). Effects of redox conditions on the control of arsenic mobility in shallow alluvial aquifers on the Venetian Plain (Italy). Sci. Total Environ. 532, 581–594. doi: 10.1016/j.scitotenv.2015.06.003
Cavalca, L., Corsini, A., Zaccheo, P., Andreoni, V., and Muyzer, G. (2013). Microbial transformations of arsenic: perspectives for biological removal of arsenic from water. Fut. Microbiol. 8, 753–768. doi: 10.2217/fmb.13.38
Chakraborty, A., and Picardal, F. (2013). Induction of nitrate- dependent Fe(II) oxidation by Fe(II) in Dechloromonas sp. strain UWNR4 and Acidovorax sp. strain 2AN. Appl. Environ. Microbiol. 79, 748–752. doi: 10.1128/AEM.02709-12
Chao, A. (1984). Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270.
Corsini, A., Colombo, M., Muyzer, G., and Cavalca, L. (2015). Characterization of the arsenite oxidizer Aliihoeflea sp. strain 2WW and its potential application in the removal of arsenic from groundwater in combination with Pf-ferritin. Anton. Leeuw. Int. J. G. 108, 673–684. doi: 10.1007/s10482-015-0523-2
Corsini, A., Zaccheo, P., Muyzer, G., Andreoni, V., and Cavalca, L. (2014). Arsenic transforming abilities of groundwater bacteria and the combined use of Aliihoeflea sp. strain 2WW and goethite in metalloid removal. J. Hazard Mater. 269, 89–97. doi: 10.1016/j.jhazmat.2013.12.037
Crognale, S., Zecchin, S., Amalfitano, S., Fazi, S., Casentini, B., Corsini, A., et al. (2017). Phylogenetic structure and metabolic properties of microbial communities in arsenic-rich waters of geothermal origin. Front. Microbiol. 8:2468. doi: 10.3389/fmicb.2017.02468
Das, S., Liu, C. C., Jean, J. S., Lee, C. C., and Yang, H. J. (2016). Effects of microbially induced transformations and shift in bacterial community on arsenic mobility in arsenic-rich deep aquifer sediments. J. Hazard. Mater. 310, 11–19. doi: 10.1016/j.jhazmat.2016.02.019
De León, K. B., Ramsay, B. D., and Fields, M. W. (2012). Quality-score refinement of SSU rRNA gene pyrosequencing differs across gene region for environmental samples. Microb. Ecol. 64, 499–508. doi: 10.1007/s00248-012-0043-9
Dhar, R. K., Zheng, Y., Rubenstone, J., and van Geen, A. (2004). A rapid colorimetric method for measuring arsenic concentrations in groundwater. Anal. Chim. Acta 526, 203–209. doi: 10.1016/j.aca.2004.09.045
Dhar, R. K., Zheng, Y., Saltikov, C. W., Radloff, K. A., Mailloux, B. J., Ahmed, K. M., et al. (2011). Microbes enhance mobility of arsenic in Pleistocene aquifer sand from Bangladesh. Environ. Sci. Technol. 45, 2648–2654. doi: 10.1021/es1022015
Dong, D. T., Yamamura, S., and Amachi, S. (2016). Impact of arsenite on the bacterial community structure and diversity in soil. Microbes Environ. 31, 41–48. doi: 10.1264/jsme2.ME15093
Emmerich, M., Bhansali, A., Lösekann-Behrens, T., Schröder, C., Kappler, A., and Behrens, S. (2012). Abundance, distribution, and activity of Fe(II)-oxidizing and Fe(III)-reducing microorganisms in hypersaline sediments of Lake Kasin, southern Russia. Appl. Environ. Microbiol. 78, 4386–4399. doi: 10.1128/AEM.07637-11
Escudero, L. V., Casamayor, E. O., Chong, G., Pedrós-Alió, C., and Demergasso, C. (2013). Distribution of microbial arsenic reduction, oxidation and extrusion genes along a wide range of environmental arsenic concentrations. PLoS One 8:e78890. doi: 10.1371/journal.pone.0078890
Fendorf, S., Michael, H. A., and van Geen, A. (2010). Spatial and temporal variations of groundwater arsenic in South and Southeast Asia. Science 328, 1123–1127. doi: 10.1126/science.1172974
Finneran, K. T., Housewright, M. E., and Lovley, D. R. (2002). Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediments. Environ. Microbiol. 4, 510–516. doi: 10.1046/j.1462-2920.2002.00317.x
Francani, V., Beretta, G. P., Bareggi, A., Nobile, A., Cremonini Bianchi, M., and Cattaneo, F. (1994). Aspetti idrogeologici del problema della presenza di azoto ammoniacale nelle acque sotterranee della provincia di Cremona [Hydrogeological Aspects of the Occurrence of Ammonium in Groundwater in the Province of Cremona]. Bologna: Pitagora Ed.
Garcia-Dominguez, E., Mumford, A., Rhine, E. D., Paschal, A., and Young, L. Y. (2008). Novel autotrophic arsenite-oxidizing bacteria isolated from soil and sediments. FEMS Microbiol. Ecol. 66, 401–410. doi: 10.1111/j.1574-6941.2008.00569.x
Giloteaux, L., Holmes, D. E., Williams, K. H., Wrighton, C. K., Wilkins, M. J., Montgomery, A. P., et al. (2013). Characterization and transcription of arsenic respiration and resistance genes during in situ uranium bioremediation. ISME J. 7, 370–383. doi: 10.1038/ismej.2012.109
Gnanaprakasam, E. T., Lloyd, J. R., Boothman, C., Matin Ahmed, K., Choudhury, I., Bostick, B. C., et al. (2017). Microbial community structure and arsenic biogeochemistry in two arsenic impacted aquifers in Bangladesh. mBio 8, e1326–e1317. doi: 10.1128/mBio.01326-17
Gorra, R., Webster, G., Martin, M., Celi, L., Mapelli, F., and Weightman, A. J. (2012). Dynamic microbial community associated with iron–arsenic co-precipitation products from a groundwater storage system in Bangladesh. Microb. Ecol. 64, 171–186. doi: 10.1007/s00248-012-0014-1
Guo, H., Liu, Z., Ding, S., Hao, C., Xiu, W., and Hou, W. (2015). Arsenate reduction and mobilization in the presence of indigenous aerobic bacteria obtained from high arsenic aquifers of the Hetao Basin, Inner Mongolia. Environ. Pollut. 203, 50–59. doi: 10.1016/j.envpol.2015.03.034
Haaijer, S. C. M., Harhangi, H. R., Meijerink, B. B., Strous, M., Pol, A., Smolders, A. J. P., et al. (2008). Bacteria associated with iron seeps in a sulfur-rich, neutral pH, freshwater ecosystem. ISME J. 2, 1231–1242. doi: 10.1038/ismej.2008.75
Hamamura, N., Macur, R. E., Korf, S., Ackerman, G., Taylor, W. P., Kozubal, M., et al. (2009). Linking microbial oxidation of arsenic with detection and phylogenetic analysis of arsenite oxidase genes in diverse geothermal environments. Environ. Microbiol. 11, 421–431. doi: 10.1111/j.1462-2910.2008.01781.x
Han, C., Kotsyurbenko, O., Chertkov, O., Held, B., Lapidus, A., Nolan, M., et al. (2012). Complete genome sequence of the sulfur compounds oxidizing chemolithoautotroph Sulfuricurvum kujiense type strain (YK-1T). Stand. Genomic Sci. 6, 94–103. doi: 10.4056/sigs.2456004
Handley, K. M., Bartels, D., O’Loughlin, E. J., Williams, K. H., Trimble, W. L., Skinner, K., et al. (2014). The complete genome sequence for putative H2- and S-oxidizer Candidatus Sulfuricurvum sp., assembled de novo from an aquifer-derived metagenome. Environ. Microbiol. 16, 3445–3462. doi: 10.1111/1462-2920.12453
Hassan, Z., Sultana, M., Westerhoff, H. V., Khan, S. I., and Röling, W. F. M. (2016). Iron cycling potentials of arsenic-contaminated groundwater in Bangladesh as revealed by enrichment cultivation. Geomicrobiol. J. 33, 779–792. doi: 10.1080/01490451.2015.1111471
Heinrich-Salmeron, A., Cordi, A., Brochier-Armanet, C., Halter, D., Pagnout, C., Abbaszadeh-fard, E., et al. (2011). Unsuspected diversity of arsenite-oxidizing bacteria as revealed by widespread distribution of the aoxB gene in Prokaryotes. Appl. Environ. Microbiol. 77, 4685–4692. doi: 10.1128/AEM.02884-10
Hemme, C. L., Deng, Y., Gentry, T. J., Fields, M. W., Wu, L., Barua, S., et al. (2010). Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J. 4, 660–672. doi: 10.1038/ismej.2009.154
Herath, I., Vithanage, M., Bundschuh, J., Maity, J. P., and Bhattacharya, P. (2016). Natural arsenic in global groundwaters: distribution and geochemical triggers for mobilization. Curr. Pollut. Rep. 2, 68–89. doi: 10.1007/s40726-016-0028-2
Héry, M., Gault, A. G., Rowland, H. A. L., Lear, G., Polya, D. A., and Lloyd, J. R. (2008). Molecular and cultivation-dependent analysis of metal-reducing bacteria implicated in arsenic mobilisation in south-east Asian aquifers. App. Geochem. 23, 3215–3223. doi: 10.1016/j.apgeochem.2008.07.003
Héry, M., Rizoulis, A., Sanguin, H., Cooke, D. A., Pancost, R. D., Polya, D. A., et al. (2015). Microbial ecology of arsenic-mobilizing Cambodian sediments: lithological controls uncovered by stable-isotope probing. Environ. Microbiol. 17, 1857–1869. doi: 10.1111/1462-2920.12412
Héry, M., Van Dongen, B. E., Gill, F., Mondal, D., Vaughan, D. J., Pancost, R. D., et al. (2010). Arsenic release and attenuation in low organic carbon aquifer sediments from West Bengal. Geobiology 8, 155–168. doi: 10.1111/j.1472-4669.2010.00233.x
Hohmann, C., Winkler, E., Morin, G., and Kappler, A. (2010). Anaerobic Fe(II)-oxidizing bacteria show As resistance and immobilize As during Fe(III) mineral precipitation. Environ. Sci. Technol. 44, 94–101. doi: 10.1021/es900708s
Hubert, C. R. J., Oldenburg, T. B. P., Fustic, M., Gray, N. D., Larter, S. R., Penn, K., et al. (2012). Massive dominance of Epsilonproteobacteria in formation waters from a Canadian oil sands reservoir containing severely biodegraded oil. Environ. Microbiol. 14, 387–404. doi: 10.1111/j.1462-2920.2011.02521.x
Islam, F. S., Gault, A. G., Boothman, C., Polya, D. A., Charnock, J. M., Chatterjee, D., et al. (2004). Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature 430, 68–71. doi: 10.1038/nature02638
Jiang, Z., Li, P., Jiang, D., Wu, G., Dong, H., Wang, Y., et al. (2014). Diversity and abundance of the arsenite oxidase gene aioA in geothermal areas of Tengchong. Yunnan China. Extremophiles 18, 161–170. doi: 10.1007/s00792-013-06080-7
Karn, S. K., and Pan, X. (2017). Bacterial oxidation and stabilization of As(III) in soil. Environ. Engin. Sci. 34, 158–164. doi: 10.1089/ees.2015.0390
Katsoyiannis, I. A., Mitrakas, M., and Zouboulis, A. I. (2015). Arsenic occurrence in Europe: emphasis in Greece and description of the applied full-scale treatment plants. Desalin. Water Treat. 54, 2100–2107. doi: 10.1080/19443994.2014.933630
Katsoyiannis, I. A., and Zouboulis, A. I. (2004). Application of biological processes for the removal of arsenic from groundwaters. Water Res. 38, 17–26. doi: 10.1016/j.watres.2003.09.011
Kellermann, C., Selesi, D., Lee, N., Hügler, M., Esperschütz, J., Hartman, A., et al. (2012). Microbial CO2 fixation potential in a tar-oil-contaminated porous aquifer. FEMS Microbiol. Ecol. 81, 172–187. doi: 10.1111/j.1574-6941.2012.01359.x
Kelly, W. R., Holm, T. R., Wilson, S. D., and Roadcap, G. S. (2005). Arsenic in glacial aquifers: sources and geochemical controls. Groundwaters 43, 500–510. doi: 10.1111/j.1745-6584.2005.0058.x
Kim, Y. T., Yoon, H., Yoon, C., and Woo, N. C. (2007). An assessment of sampling, preservation, and analytical procedures for arsenic speciation in potentially contaminated waters. Environ. Geochem. Health 29, 337–346. doi: 10.1007/s10653-007-9091-3
Kirk, M. F., Holm, T. R., Park, J., Jin, Q., Sanford, R. A., Fouke, B. W., et al. (2004). Bacterial sulfate reduction limits natural arsenic contamination in groundwater. Geology 32, 953–956. doi: 10.1130/G20842.1
Kodama, Y., and Watanabe, K. (2003). Isolation and characterization of a sulfur-oxidizing chemolithotroph growing on crude oil under anaerobic conditions. Appl. Environ. Microbiol. 69, 107–112. doi: 10.1128/AEM.69.1.107-112.2003
Kodama, Y., and Watanabe, K. (2004). Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int. J. Syst. Evol. Microbiol. 54, 2297–2300. doi: 10.1099/ijs.0.63243-0
Kunin, V., and Hugenholtz, P. (2010). PyroTagger: A Fast, Accurate Pipeline for Analysis of rRNA Amplicon Pyrosequence Data. Boston: ScienceOpen, Inc.
Lear, G., Song, B., Gault, A. G., Polya, D. A., and Lloyd, J. R. (2007). Molecular analysis of arsenate-reducing bacteria within Cambodian sediments following amendment with acetate. Appl. Environ. Microbiol. 73, 1041–1048. doi: 10.1128/AEM.01654-06
Lee, J., Fredrickson, J. K., Plymale, A. E., Dohnalkova, A. C., Resch, C. T., McKinley, J. P., et al. (2015). An autotrophic H2-oxidizing, nitrate-respiring. Tc(VII)-reducing Acidovorax sp. isolated from a subsurface oxic-anoxic transition zone. Microbiol. Rep. 7, 395–403. doi: 10.1111/1758-2229.12263
Lee, S. D., Lee, D. W., and Ko, Y. (2011). Marmoricola korecus sp. nov. Int. J. Syst. Evol. Microbiol. 61, 1628–1631. doi: 10.1099/ijs.0.025460-0
Legg, T. M., Zheng, Y., Simone, B., Radloff, K. A., Mladenov, N., González, A., et al. (2012). Carbon, metals, and grain size correlate with bacterial community structure in sediments of a high arsenic aquifer. Front. Microbiol. 3:1–15. doi: 10.3389/fmicb.2012.00082
Li, P., Jiang, Z., Wang, Y., Deng, Y., Van Nostrand, J. D., Yuan, T., et al. (2017). Analysis of the functional gene structure and metabolic potential of microbial community in high arsenic groundwater. Water Res. 123, 268–276. doi: 10.1016/j.watres.2017.06.053
Li, P., Li, B., Webster, G., Wang, Y., Jiang, D., Dai, X., et al. (2014). Abundance and diversity of sulfate-reducing bacteria in high arsenic shallow aquifers. Geomicrobiol. J. 31, 802–812. doi: 10.1080/01490451.2014.893181
Li, P., Wang, Y., Dai, X., Zhang, R., Jiang, Z., Jiang, D., et al. (2015). Microbial community in high arsenic shallow groundwater aquifers in Hetao Basin of Inner Mongolia. PLoS One 10:e0125844. doi: 10.1371/journal.pone.0125844
Li, Q., Xu, L. Z., Zou, T., Ai, P., Huang, G. H., Li, P., et al. (2015). Complete genome sequence of Bacillus thuringensis strain HD521. Stand. Genomic Sci. 10:62. doi: 10.1186/s40793-015-0058-1
Li, P., Wang, Y., Jiang, Z., Jiang, H., Li, B., Dong, H., et al. (2013). Microbial diversity in high arsenic groundwater in Hetao Basin of Inner Mongolia. Geomicrobiol. J. 30, 897–909. doi: 10.1080/01490451.2013.791354
Liao, V. H., Chu, Y. J., Su, Y. C., Hsiao, S. Y., Wei, C. C., Liu, C. W., et al. (2011). Arsenite-oxidizing and arsenate-reducing bacteria associated with arsenic-rich groundwater in Taiwan. J. Contam. Hydrol. 123, 2020–2029. doi: 10.1016/j.jconhyd.2010.12.003
Lin, X., McKinley, J., Resch, C. T., Kaluzny, R., Lauber, C. L., Fredrickson, J., et al. (2012). Spatial and temporal dynamics of the microbial community in the Hanford unconfined aquifer. ISME J. 6, 1665–1676. doi: 10.1038/ismej.2012.26
Lovley, D. (2006). Dissimilatory Fe(III)- and Mn(IV)-reducing Prokaryotes. Prokaryotes 2, 635–658. doi: 10.1007/0-387-30742-7_21
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. App. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Luna, G. M., Dell’Anno, A., Corinaldesi, C., Armeni, M., and Danovaro, R. (2009). Diversity and spatial distribution of metal-reducing bacterial assemblages in groundwaters of different redox conditions. Int. Microbiol. 12, 153–159. doi: 10.2436/20.1501.01.93
Macur, R. E., Jackson, C. R., Botero, L. M., Mcdermott, T. R., and Inskeep, W. P. (2004). Bacterial populations associated with the oxidation and reduction of arsenic in an unsaturated soil. Environ. Sci. Technol. 38, 104–111. doi: 10.1021/es034455a
Maguffin, S. C., Kirk, M. F., Daigle, A. R., Hinkle, S. R., and Jin, Q. (2015). Substantial contribution of biomethylation to aquifer arsenic cycling. Nat. Geosci. 8, 290–293. doi: 10.1038/ngeo2383
Malasarn, D., Saltikov, C. W., Campbell, K. M., Santini, J. M., Hering, J. G., and Newman, D. K. (2004). arrA is a reliable marker for As(V) respiration. Science 306:455. doi: 10.1126/science.1102374
Mantelli, F. A., Salutini, A., Grilli Ciclioni, P., Bucci, S., Carrozzino, M., and Iazzelli, M. (1999). Quaderni di Geologia Applicata. Bologna: Pitagora Ed.
McGrory, E. R., Brown, C., Bargary, N., Williams, N. H., Mannix, A., Zhang, C., et al. (2017). Arsenic contamination of drinking water in Ireland: a spatial analysis of occurrence and potential risk. Sci. Total Environ. 579, 1863–1875. doi: 10.1016/j.scitotenv.2016.11.171
Mladenov, N., Zheng, Y., Miller, M. P., Nemergut, D. R., Legg, T., Simone, B., et al. (2010). Dissolved organic matter sources and consequences for iron and arsenic mobilization in Bangladesh aquifers. Environ. Sci. Technol. 44, 123–128. doi: 10.1021/es901472g
Muller, D., Médigue, C., Koechler, S., Barbe, V., Barakat, M., Talla, E., et al. (2007). A tale of two oxidation states: bacterial colonization of arsenic-rich environments. PLoS Genetics 3:e53. doi: 10.1371/journal.pgen.0030053
Mumford, C. A., Yee, N., and Young, L. Y. (2013). Precipitation of alacranite (As8S9) by a novel As(V)-respiring anaerobe strain MPA-C3. Environ. Microbiol. 15, 2748–2760. doi: 10.1111/1462-2920.12136
Murphy, J., and Riley, J. P. (1962). A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 27, 31–36. doi: 10.1016/S0003-2670(00)88444-5
Oda, Y., Larimer, F. W., Chain, P. S. G., Malfatti, S., Shin, M. V., Vergez, L. M., et al. (2008). Multiple genome sequences reveal adaptations of a phototrophic bacterium to sediment microenvironments. PNAS 105, 18543–18548. doi: 10.1073/pnas.0809160105
Omoregie, E. O., Couture, R. M., Van Cappellen, P., Corkhill, C. L., Charnock, J. M., Polya, D. A., et al. (2013). Arsenic bioremediation by biogenic iron oxides and sulfides. Appl. Environ. Microbiol. 79, 4325–4335. doi: 10.1128/AEM.00683-13
Ori, G. G. (1993). Continental depositional systems of the Quaternary of the Po Plain (Northern Italy). Sediment Geol. 83, 1–14. doi: 10.1016/S0037-0738(10)80001-6
Osborne, T. H., McArthur, J. M., Sikdar, P. K., and Santini, J. M. (2015). Isolation of an arsenate-respiring bacterium from a redox front in an arsenic-polluted aquifer in West Bengal, Bengal Basin. Environ. Sci. Technol. 49, 4193–4199. doi: 10.1021/es504707x
Pandey, N., and Bhatt, R. (2016). Arsenic removal and biotransformation potential of Exiguobacterium isolated from an arsenic-rich soil of Chhattisgarh, India. Clean Soil Air Water 44, 211–218. doi: 10.1002/clen.201500095
Paul, D., Kazy, S. K., Das Banerjee, T., Gupta, A. K., Pal, T., and Sar, P. (2015). Arsenic biotransformation and release by bacteria indigenous to arsenic contaminated groundwater. Bioresour. Technol. 188, 14–23. doi: 10.1016/j.biortech.2015.02.039
Peña Reyes, F. A., Crosta, G. B., Frattini, P., Basiricò, S., and Della Pergola, R. (2015). Hydrogeochemical overview and natural arsenic occurrence in groundwater from alpine springs (upper Valtellina, Northern Italy). J. Hydrol. 529, 1530–1549. doi: 10.1016/j.jhydrol.2015.08.029
Pérez-Jiménez, J. R., DeFraia, C., and Young, L. Y. (2005). Arsenate respiratory reductase gene (arrA) for Desulfosporosinus sp. strain Y5. Biochem. Biophys. Res. Commun. 338, 825–829. doi: 10.1016/j.bbrc.2005.10.011
Porter, M. L., and Engels, A. S. (2008). Diversity of uncultured Epsilonproteobacteria from terrestrial sulfidic caves and springs. Appl. Environ. Microbiol. 74, 4973–4977. doi: 10.1128/AEM.02915-07
Postma, D., Larsen, F., Thai, N. T., Trang, P. T. K., Jakobsen, R., Nhan, P. Q., et al. (2012). Groundwater arsenic concentrations in Vietnam controlled by sediment age. Nat. Geosci. 5, 656–661. doi: 10.1038/ngeo1540
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large mini- mum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Qiu, Y. L., Kuang, X. Z., Shi, X. S., Yuan, X. Z., and Guo, R. B. (2014). Terrimicrobium sacchariphilum gen. nov., sp. nov., an anaerobic bacterium of the class ‘Spartobacteria’ in the phylum Verrucomicrobia, isolated from a rice paddy field. Int. J. Syst. Evol. Microbiol 64, 1718–1723. doi: 10.1099/ijs.0.060244-0
Quéméneur, M., Cébron, A., Billard, P., Battaglia-Brunet, F., Garrido, F., Leyval, C., et al. (2010). Population structure and abundance of arsenite-oxidizing bacteria along an arsenic pollution gradient in waters of the Upper Isle River Basin. France. Appl. Environ. Microbiol. 76, 4566–4570. doi: 10.1128/AEM.03104-09
Quéméneur, M., Heinrich-Salmeron, A., Muller, D., Lièvremont, D., Jauzein, M., Bertin, P. N., et al. (2008). Diversity surveys and evolutionary relationships of aoxB genes in aerobic arsenite-oxidizing bacteria. Appl. Environ. Microbiol. 74, 4567–4573. doi: 10.1128/AEM.02851-07
Ravenscroft, P., Brammer, H., and Richards, K. (2009). Arsenic Pollution: A Global Synthesis. Chichester: Wiley-Blackwell.
Roeselers, G., Coolen, J., van der Wielen, P. W. J. J., Jaspers, M. C., Atsma, A., de Graaf, B., et al. (2015). Microbial biogeography of drinking water: patterns in phylogenetic diversity across space and time. Environ. Microbiol. 17, 2505–2514. doi: 10.1111/1462-2920.12739
Rotiroti, M., Jakobsen, R., Fumagalli, L., and Bonomi, T. (2015). Arsenic release and attenuation in a multilayer aquifer in the Po Plain (Northern Italy): reactive transport modeling. Appl. Geochem. 63, 599–609. doi: 10.1016/j.apgeochem.2015.07.001
Rotiroti, M., Sacchi, E., Fumagalli, L., and Bonomi, T. (2014). Origin of arsenic in groundwater from the multi-layer aquifer in Cremona (Northern Italy). Environ. Sci. Technol. 48, 5395–5403. doi: 10.1021/es405805v
Salmassi, T. M., Venkateswaren, K., Satomi, M., Nealson, K. H., Newman, D. K., and Hering, J. G. (2002). Oxidation of arsenite by Agrobacterium albertimagni, AOL15, sp. nov., isolated from hot creek, California. Geomicrobiol. J. 19, 53–66. doi: 10.1080/014904502317246165
Sarkar, A., Paul, D., Kazy, S. K., and Sar, P. (2016). Molecular analysis of microbial community in arsenic-rich groundwater of Kolsor, West Bengal. J. Environ. Sci. Health A 51, 229–239. doi: 10.1080/10934529.2015.1094339
Schäfer, H., and Muyzer, G. (2001). Denaturing gradient gel electrophoresis in marine microbial ecology. Methods Microbiol. 30, 425–468. doi: 10.1016/S0580-9517(01)30057-0
Smedley, P. L., and Kinniburgh, D. G. (2002). A review of the source, behaviour and distribution of arsenic in natural waters. Appl. Geochem. 17, 517–568. doi: 10.1016/S0883-2927(02)00018-5
Stringer, S. C., Carter, A. T., Webb, M. D., Wachnicka, E., Crossman, L. C., Sebaihia, M., et al. (2013). Genomic and physiological variability within Group II (non-proteolytic) Clostridium botulinum. BMC Genomics 14:333. doi: 10.1128/genomeA.006605-13
Stuckey, J. W., Schaefer, M. V., Kocar, B. D., Benner, S. G., and Fendorf, S. (2016). Arsenic release metabolically limited to permanently water-saturated soil in Mekong Delta. Nat. Geosci. 9, 70–76. doi: 10.1038/ngeo2589
Sultana, M., Mou, T. J., Sanyal, S. K., Diba, F., Mahmud, Z. H., Parvez, A. K., et al. (2017). Investigation of arsenotrophic microbiome in arsenic-affected Bangladesh groundwater. Groundwater 55, 736–746. doi: 10.1111/gwat.12520
Sultana, M., Vogler, S., Zargar, K., Schmidt, A. C., Saltikov, C., Seifert, J., et al. (2012). New clusters of arsenite oxidase and unusual bacterial groups in enrichments from arsenic-contaminated soil. Arch. Microbiol. 194, 623–635. doi: 10.1007/s00203-011-0777-7
Sun, W., Sierra-Alvarez, R., Milner, L., Oremland, R., and Field, J. A. (2009). Arsenite and ferrous iron oxidation linked to chemolithotrophic denitrification for the immobilization of arsenic in anoxic environments. Environ. Sci. Technol. 43, 6585–6591. doi: 10.1021/es900978h
Sutton, N. B., van der Kraan, G. M., van Loosdrecht, M. C., Muyzer, G., Bruining, J., and Schotting, R. J. (2009). Characterization of geochemical constituents and bacterial populations associated with As mobilization in deep and shallow tube wells in Bangladesh. Water Res. 43, 1720–1730. doi: 10.1016/j.watres.2009.01.006
Taghavi, S., Izquierdo, J. A., and van der Lelie, D. (2013). Complete genome sequence of Clostridium sp. strain DL-VIII, a novel solventogenic Clostridium species isolated from anaerobic sludge. Genome Announc. 1, e605–e613. doi: 10.1128/genomeA.00605-13
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Thiel, J., Byrne, J. M., Kappler, A., Schink, B., and Pester, M. (2019). Pyrite formation from FeS and H2S is mediated through microbial redox activity. Proc. Natl. Acad. Sci. U.S.A. 116, 6897–6902. doi: 10.1073/pnas.1814412116
Ungaro, F., Ragazzi, F., Cappellin, R., and Giandon, P. (2008). Arsenic concentration in the soils of the Brenta Plain (Northern Italy): mapping the probability of exceeding contamination thresholds. J. Geochem. Explor. 96, 117–131. doi: 10.1016/j.gexplo.2007.03.006
Vassena, C., Rienzner, M., Ponzini, G., Giudici, M., Gandolfi, C., Durante, C., et al. (2012). Modeling water resources of a highly irrigated alluvial plain (Italy): calibrating soil and groundwater models. Hydrogeol. J. 20, 449–467. doi: 10.1007/s10040-011-0822-2
Videmsek, U., Hagn, A., Suhadolc, M., Radl, V., Knicker, H., Schloter, M., et al. (2009). Abundance and diversity of CO2-fixing bacteria in grassland soils close to natural carbon dioxide springs. Microbial Ecol. 58, 1–9. doi: 10.1007/s00248-008-9442-3
Wang, Y., Li, P., Jiang, Z., Liu, H., Wei, D., Wang, H., et al. (2018). Diversity nd abundance of arsenic methylating microorganisms in high arsenic groundwater from Hetao Plain of Inner Mongolia, China. Ecotoxicology 27, 1047–1057. doi: 10.1007/s10646-018-1958-9
Wang, Y., Li, P., Jiang, Z., Sinkkonen, A., Wang, S., Tu, J., et al. (2016). Microbial community of high arsenic groundwater in agricultural irrigation area of Hetao Plain, Inner Mongolia. Front. Microbiol. 7:1917. doi: 10.3389/fmicb.2016.01917
World Health Organization [WHO] (2001). Arsenic in Drinking Water. Washington, DC, National Academy Press
Xiao, K., Li, L., Ma, L., Zhang, S., Bao, P., Zhang, T., et al. (2016). Metagenomic analysis revealed highly diverse microbial arsenic metabolism genes in paddy soils with low-arsenic contents. Environ. Poll. 211, 1–8. doi: 10.1016/j.envpol.2015.12.023
Xiong, Z., Jiang, Y., Qi, D., Lu, H., Yang, F., Chen, L., et al. (2009). Complete genome sequence of the extremophilic Bacillus cereus Strain Q1 with industrial applications. J. Bacteriol. 191, 1120–1121. doi: 10.1128/JB.01629-08
Yamamura, S., and Amachi, S. (2014). Microbiology of inorganic arsenic: from metabolism to bioremediation. J. Biosci. Bioeng. 1, 1–9. doi: 10.1016/j.jbiosc.2013.12.011
Yang, N., Winkel, L. H. E., and Johannesson, K. H. (2014). Predicting geogenic arsenic contamination in shallow groundwater of South Louisiana. United States. Environ. Sci. Technol. 48, 5660–5666. doi: 10.1021/es405670g
Yunus, F. M., Khan, S., Chowdhury, P., Milton, A. H., Hussain, S., and Rahman, M. (2016). A review of groundwater arsenic contamination in Bangladesh: the millennium development goal era and beyond. Int. J. Environ. Res. Public Health 13, 215. doi: 10.3390/ijerph13020215
Zavatti, A., Attramini, D., Bonazzi, A., Boraldi, V., Malagò, R., Martinelli, G., et al. (1995). La presenza di Arsenico nelle acque sotterranee della Pianura Padana: evidenze ambientali e ipotesi geochimiche ”occurrence of groundwater arsenic in the Po plain: environmental evidences and geochemical hypothesis”. Quaderni di Geologia Applicata 2, 2301–2324.
Zecchin, S., Corsini, A., Martin, M., and Cavalca, L. (2017). Influence of water management on the active root-associated microbiota involved in arsenic, iron, and sulfur cycles in rice paddies. Appl. Microbiol. Biotechnol. 101, 6725–6738. doi: 10.1007/s00253-017-8382-6
Keywords: arsenic, groundwater, arsenate-reducing bacteria, arsenite-oxidizing bacteria, sulfur bacteria, iron bacteria
Citation: Cavalca L, Zecchin S, Zaccheo P, Abbas B, Rotiroti M, Bonomi T and Muyzer G (2019) Exploring Biodiversity and Arsenic Metabolism of Microbiota Inhabiting Arsenic-Rich Groundwaters in Northern Italy. Front. Microbiol. 10:1480. doi: 10.3389/fmicb.2019.01480
Received: 21 February 2019; Accepted: 13 June 2019;
Published: 02 July 2019.
Edited by:
Qiaoyun Huang, Huazhong Agricultural University, ChinaReviewed by:
Ping Li, China University of Geosciences, ChinaSantosh Kr. Karn, Sardar Bhagwan Singh Post Graduate Institute of Biomedical Science & Research, India
Copyright © 2019 Cavalca, Zecchin, Zaccheo, Abbas, Rotiroti, Bonomi and Muyzer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lucia Cavalca, bHVjaWEuY2F2YWxjYUB1bmltaS5pdA==
 Lucia Cavalca
Lucia Cavalca Sarah Zecchin
Sarah Zecchin Patrizia Zaccheo
Patrizia Zaccheo Ben Abbas
Ben Abbas Marco Rotiroti
Marco Rotiroti Tullia Bonomi
Tullia Bonomi Gerard Muyzer
Gerard Muyzer