- 1Department of Clinical Laboratory, Shanghai General Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China
- 2Institute of Antibiotics, Huashan Hospital, Fudan University, Shanghai, China
- 3Department of Clinical Laboratory, the Third Hospital Affiliated to Nantong University, Wuxi, China
- 4Department of Clinical Laboratory, Xiaoshan Hospital, Hangzhou, China
The toxic shock syndrome toxin-1 (TSST-1), encoded by tst gene, has been proposed to cause staphylococcal toxic shock syndrome (TSS) in a susceptible host, which highlights the need to evaluate the level of tst gene expression and molecular genetic characteristics of the tst-positive isolates. A total of 916 S. aureus isolates collected from seven hospitals in China were screened for the tst gene. The tst positive isolates were characterized by spa, SCCmec, PFGE, and agr typing. Representative strains were also subjected to MLST typing. qRT-PCR was used to quantify tst and major virulence regulator genes expression. We also sequenced the regions of promoter and open reading frame (ORF) of tst to investigate whether they correlate with the variation in tst expression. We found 208 (22.7%) of surveyed isolates including 198 (29.8%) of MRSA and 10 (4.0%) of MSSA isolates harbored the tst gene. The most common clone among tst positive MRSA isolates belonged to ST5 (CC5)-agr2-t002-SCCmecII. The amount of tst mRNA varied 8.4-folds among clinical S. aureus isolates. Sequencing the tst promoter revealed a base T deletion in tst high expressed isolates. As for major virulence regulators, srrA, sarT, RNAIII, and ccpA in four tst differentially expressed strains were detected to be highly expressed, respectively. Our study revealed high prevalence of ST5 (CC5)-agr2-t002-SCCmecII clone among tst positive MRSA in hospitals from China. The levels of tst expression among clinical S. aureus isolates varied, which may be associated with tst promoter and variations in specific virulence regulators.
Introduction
Staphylococcus aureus (S. aureus), as the ubiquitous human pathogen, causes some of the most severe infections in both hospital and community settings. In China, S. aureus has become the main cause of infective osteomyelitis, endocarditis and sepsis (Liu et al., 2014; Yan et al., 2015; Xu et al., 2016). Relying on the selective expression of virulence factors that facilitate tissue invasion, adhesion or immune evasion, S. aureus can colonize almost any human tissue site and survive under highly variable conditions (Iwatsuki et al., 2006; Gorwitz et al., 2008).
S. aureus superantigens (SAgs), remarkably resistant to heat, acids, proteolysis and desiccation, are an extraordinary family of non-glycosylated low-molecular-weight exoproteins (Spaulding et al., 2013). The toxins of this family have the capacity to trigger excessive and non-conventional T-cell activation and cytokines release, and consequently interfere with immune system function systemically (Spaulding et al., 2013; Kulhankova et al., 2014). The biological toxicity of SAgs makes them be critical contributors causing life-threatening infections. The toxic shock syndrome toxin-1 (TSST-1), encoded by tst gene, is a significant member of SAgs and may lead to staphylococcal toxic shock syndrome (TSS) in a susceptible host (Spaulding et al., 2013).
It has been repeatedly documented that 30 to 40% of the population is asymptomatically colonized by S. aureus strains at one or more of their body sites (Blot et al., 1998; Spaulding et al., 2013), and approximate 20% of this organism is TSST-1 producers (Lindsay et al., 1998), indicating a large disease potential. However, the TSS, including both menstrual and non-menstrual categories, is fortunately rarely seen (1–3/100,000) (Brosnahan and Schlievert, 2011). The relatively high rate of tst-positive S. aureus isolates coupled with the low incidence of TSS strongly suggests that sufficient tst expression causes disease only under the appropriate environmental and/or genetic regulation control. Since the virulence of microbe may be dependent on the amount of toxins production (Bronner et al., 2004), quantifying the TSST-1 production by S. aureus strains contributes to offer a basis for developing the reasonable countermeasures to the tst-positive S. aureus infection control. It has been reported that the amounts of TSST-1 produced by clinical MRSA isolates differed up to 170-folds (Nagao et al., 2009). However, the specific regulatory factors or systems accounting for this diversifying gene expression among clinical S. aureus strains remain uncertain.
In this study, we aimed to (i) elucidate genetic background of the tst-positive S. aureus strains collected from seven hospitals in China, (ii) detect the difference at mRNA level of tst in vitro, and (iii) try to find the possible cause of the differential expression of tst.
Materials and Methods
Bacterial Strains
Two hundred and eight tst-positive S. aureus isolates (including 198 MRSA and 10 MSSA) were screened from 7 hospitals collection in Shanghai and Zhejiang province, China. This assemble of bacteria contained 916 non-duplicate S. aureus separated from clinical specimens during December 2008 and January 2013. Shanghai and Zhejiang are in the core position in the Yangtze River Delta, which is an economically advanced and densely populated area in China. Therefore, we think the isolates from these two regions may have a certain representativeness.
DNA Extraction
Genomic DNAs of all the 916 S. aureus isolates were extracted with a TIANamp Bacterial DNA Kit (TIANGEN BIOTECH Co., Ltd., Beijing, China) according to the manufacturer's recommendations, with the modification of adding 10 μl of lysostaphin (1 mg/ml) and incubation at 37°C for 30 min for cell lysis. DNA amount and purity were tested using a NanoDrop spectrometer (Thermo Fisher Scientific, Waltham, MA, USA).
Characterization of tst-Positive Isolates
All the 208 identified tst-positive S. aureus strains were characterized by staphylococcal protein A (spa) typing and pulsed-field gel electrophoresis (PFGE) typing as previously described (Mulvey et al., 2001; Koreen et al., 2004). In addition, the accessory gene regulator (agr) locus typing and staphylococcal cassette chromosome mec (SCCmec) typing had been previously carried out (Zhao et al., 2016) in all the S. aureus isolates with tst gene using methods described by Lina et al. (2003) and Zhang et al. (2005). Based on the diversity of origin and proportion of 30 to 45% of the isolates in each PFGE cluster, 76 (71 MRSA and 5 MSSA) representative isolates were chosen to be characterized by multi-locus sequence typing (MLST). The MLST was implemented by PCR amplification of internal fragments of seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL) (Enright et al., 2000). MLST database (http://saureus.mlst.net/) was used to determine the allelic profile and the consequent sequence type (ST) of tst-positive isolates detected. The clustering of related STs into clonal complexes (CCs) was analyzed using eBURST (http://eburst.mlst.net/) as described previously (Feil et al., 2004).
Total RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
Total RNA extraction and the subsequent qRT-PCR with specific primers were performed to analyze the expression of the tst gene and major virulence regulators in 32 isolates selected based on the proportion of 10 to 30% of the isolates in each PFGE cluster. For RNA extraction, overnight cultures were diluted 1:100 into 2 ml of TSB medium and grown to the post-exponential phase. Cells were deposited by the way of centrifugation. Total RNA samples were extracted using a MiniBEST Universal RNA Extraction Kit (TaKaRa) according to the manufacturer's instructions except a little change in the lysis step as described in the DNA extraction. The extracted RNA was quantified using a NanoDrop spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). cDNA was synthesized with a PrimeScript® RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) according to the manufacturer's protocol. The resulting cDNA products were stored at −20°C until use. The qRT-PCR was performed with 10 μl 2 × SYBR Premix Ex Taq (TaKaRa, Dalian, China), 0.4 μl ROX Reference Dye II (50×), 2 μl cDNA and 0.2 μM each of the forward and reverse primers (Table 1) in a final volume of 20 μl. The thermal cycling programs consisted of 30 s at 95°C, and then 40 cycles of 5 s at 95°C and 34 s at 60°C, followed by a dissociation step of 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s on ABi 7500 Real Time PCR System (Applied Biosystems, Foster, CA, USA). The levels of expression of the target genes (tst and regulatory genes RNAIII, sigB, sarA, ccpA, srrAB, rot, and sarT) in the RNA samples were normalized on the basis of internal standards 16S rRNA. The specificity of the PCR was verified by melting curve analysis.
Sequence Analysis of Variant tst Gene
The tst-gene DNA sequences from the 32 isolates selected with the principle mentioned above were sequenced and analyzed for the identification of mutations. The tst gene, including its promoter region, was amplified by PCR using specific primers (P-tst-F CTCAAAGATAGATTGACCAGCGATG, P-tst-R TTAATTTCTGCTTCTATAGTTT). All the products were sequenced in both directions by Shanghai Sangon Biotech. Sequences were aligned using CLUSTA L X 2.0.
Statistical Analysis
All data were expressed as mean ± standard deviation (SD). Statistically significant differences were determined by ANOVA alone or with a LSD post-hoc test by SAS 9.3 for Windows software (SAS Institute Inc., NC, USA). In each case, statistical significance was indicated by p < 0.05.
Results
Characterization of S. aureus Harboring tst Gene
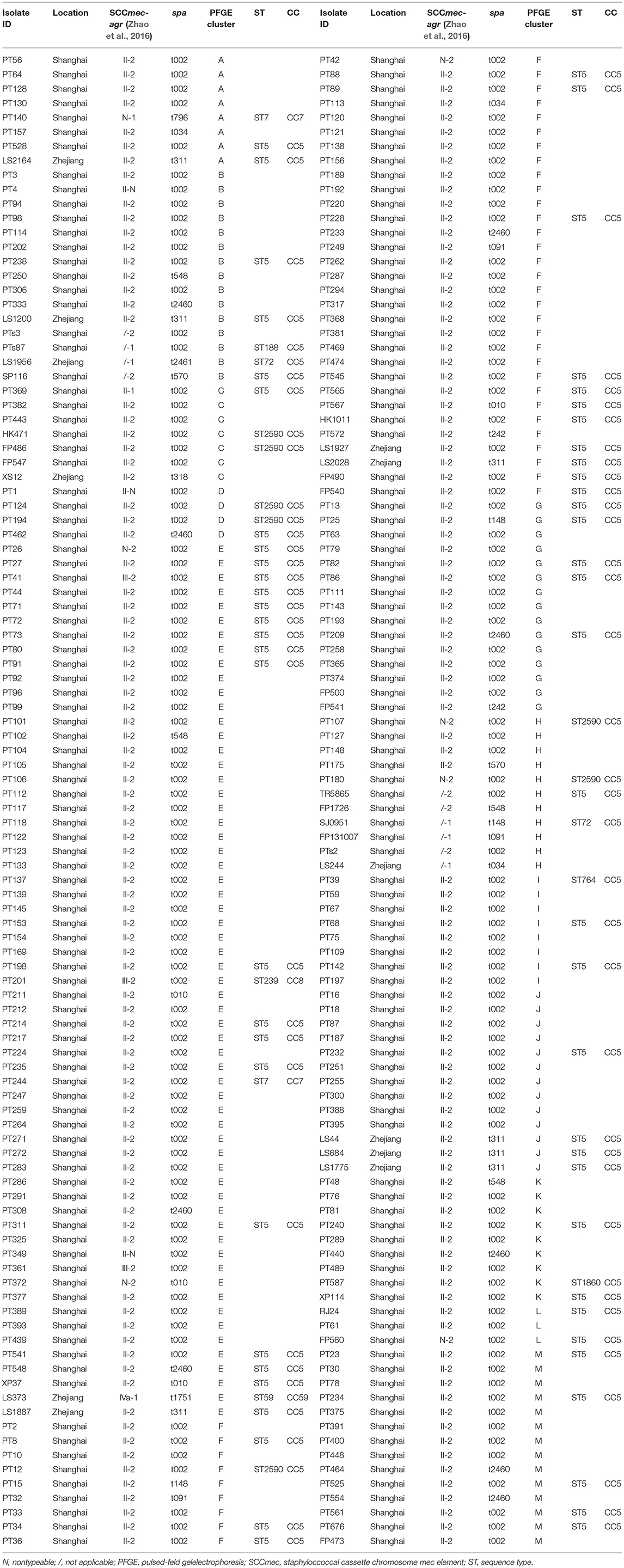
By sequence analysis, a total of 15 spa types were yielded among the 208 tst-positive strains, of which 8 spa types were related exclusively to MRSA, 2 exclusively to MSSA, and 5 to both. The most prevalent spa type in MRSA was t002 (82.3%, 163/198), followed by t2460 (4.5%, 9/198) and t311 (3.5%, 7/198). Other spa types identified in MRSA were detailed as follows: t010 (4/198, 2.0%), t548 (3/198, 1.5%), t037 (2/198, 1.0%), t034 (2/198, 1.0%), t148 (2/198, 1.0%), t242 (2/198, 1.0%), t318 (1/198, 0.5%), t570 (1/198, 0.5%), t796 (1/198, 0.5%), t1751 (1/198, 0.5%). Most of tst-positive MSSA strains were also t002, accounting for 40% (4/10), and the 6 remaining isolates were t034, t091, t148, t2461, t548, t570, respectively.
The 208 tst-positive isolates were classified into 13 PFGE types that were designated by symbols A to M (Figure 1). Most of isolates were clustered into PFGE types E and F, accounting for 29.3% (61/208) and 19.2% (40/208), respectively. Of note, all the MSSA carrying tst gene from different hospitals were grouped into the clusters B and H (Figure 1). Therefore, the high genetic homology of these isolates may demonstrate the existence of a prevalent tst-positive MSSA strain.
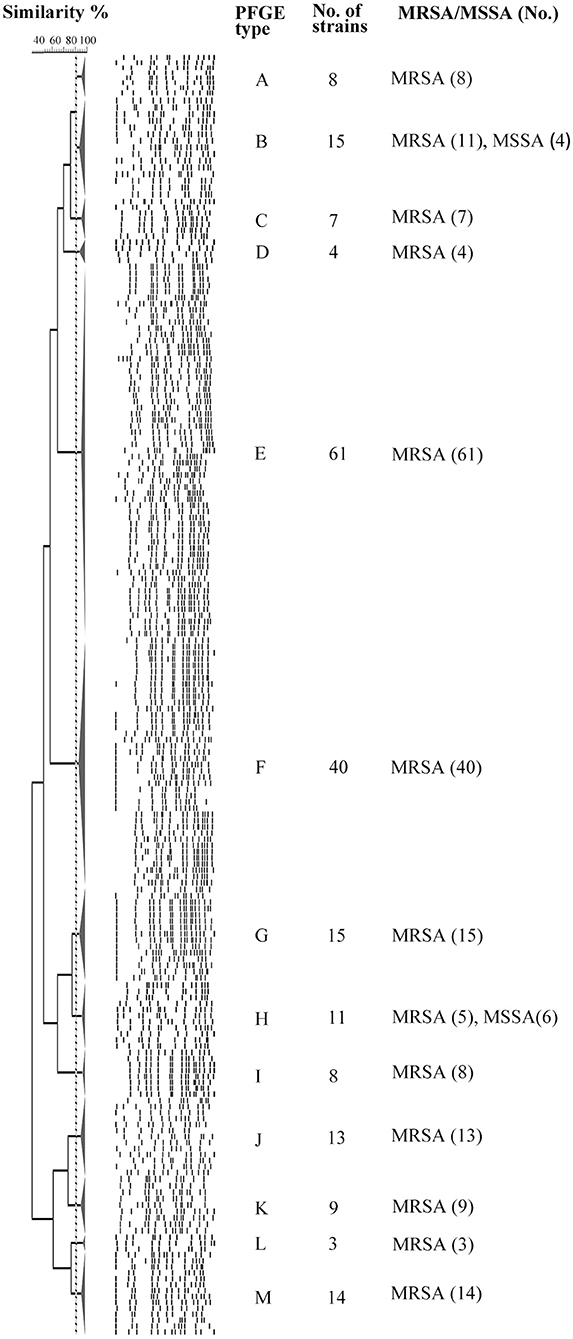
Figure 1. PFGE analysis of 208 tst gene positive S. aureus isolates digested with SmaI. This dendrogram shows the 208 S. aureus strains were clustered into 13 genomic groups (A-M).
The 76 (71 MRSA and 5 MSSA) representative isolates selected on the basis of the aforementioned PFGE dendrogram were detected to contain 9 different STs, of which ST5 (78.9%, 60/76) was most frequently identified, followed by ST2590 (9.2%, 7/76), ST7 (2.6%, 2/76), ST72 (2.6%, 2/76), and 5 additional STs, namely ST59, ST764, ST1860, ST188 and ST239 (1.3%, 1/76 each) (Table 2). Notably, except that 4 MRSA belonging to ST7 (2.6%, 2/76), ST59 (1.3%, 1/76), and ST239 (1.3%, 1/76) clones were clustered to CC7, CC59 and CC8, respectively, all the remaining isolates were clustered to CC5 (94.7%, 72/76). These results indicated a high homogeneity among the tst-positive isolates with regard to their MLST types.
As is apparent from Table 2, all ST2590 strains expressed agr type II and spa type 002. Among the 60 strains of ST5, 59 (98.3%, 59/60) were detected as agr type II, while the one remaining strain belonged to agr type I. Except two untypeable and one SCCmec III isolates, all the ST5 MRSA strains harbored SCCmec II. Both the agr and SCCmec types of all the spa t311 isolates were detected as type II, while their MLST types were identified as ST5. Five SCCmec-untypeable MRSA strains belonged to CC5-spa t002-agr II (4 isolates) and CC7-spa t796-agr I (1 isolate).
We can summarize from Table 2 that the most common clone among MRSA isolates belonged to ST5 (CC5)-agr II-spa t002-SCCmec II (53.9%, 41/76). While the predominant genotype of MSSA was not found because of the less S. aureus isolates.
Direct Transcript Analysis of tst Gene
We performed qRT-PCR with tst-specific primers to assess the tst/l6S ratio on the post-exponential bacterial pellets (32 representative S. aureus isolates). The results showed no obvious variations in the expression levels of tst gene among most of the chosen strains, except several with a significantly different tst mRNA level (Figure 2). The amount of tst mRNA varied 8.4-folds among clinical MRSA isolates in the present study. Two strains with highest and lowest tst mRNA abundances each were chosen to probe into the cause of the differential expression. Interestingly, the two strains (L-RJ24 and L-HK471) lowly expressing tst gene belonged to the same CC, spa, SCCmec and agr types (CC5-agr2-t002-SCCmecII), and the two isolates (H-LS1956 and H-SJ0951) with high amount of tst mRNA were identified as MSSA.
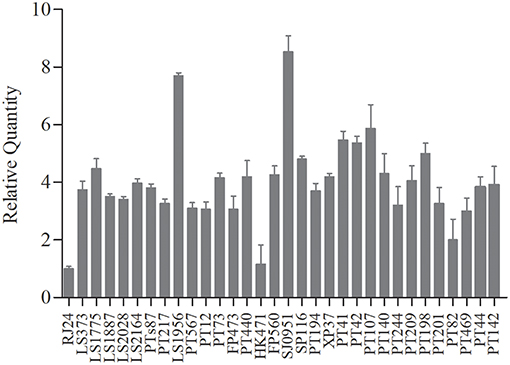
Figure 2. The transcriptional levels of tst gene in 32 chosen S. aureus strains representative. No obvious variations in the expression levels of tst gene among most of the strains, except two MRSA strains (L-RJ24 and L-HK471) with higher and two MSSA strains (H-LS1956 and H-SJ0951) with lower tst mRNA abundances. Data are expressed as the means ± standard deviation (SD). The experiment was independently repeated three times.
Expression of Major Virulence Regulators
Since RNAIII, SigB, SarA, CcpA, SrrAB, Rot, and SarT have been previously shown to have effects on tst transcription (Schmidt et al., 2001; Pragman et al., 2007; Seidl et al., 2008; Andrey et al., 2010, 2015), we measured the transcript levels of these modulator genes in four differentially expressed isolates by qRT-PCR (Figures 3A–H). The results showed comparable expression levels of sigB, sarA, and rot in the four strains (Figures 3B,C,G). Although both strains H-LS1956 and H-SJ0951 produced large amounts of tst mRNA, the expressions of the major virulence regulator genes did not consistently vary in any way from that of strains L-RJ24 and L-HK471, and vice versa. However, RNAIII in strain H-SJ0951 (Figure 3A), ccpA in strain L-HK471 (Figure 3D), srrA in strain H-LS1956 (Figure 3E) and sarT in strain L-RJ24 (Figure 3H) were detected to be highly expressed, and srrB in strain H-LS1956 (Figure 3F) was detected to be lowly expressed in the post-exponential growth phase.
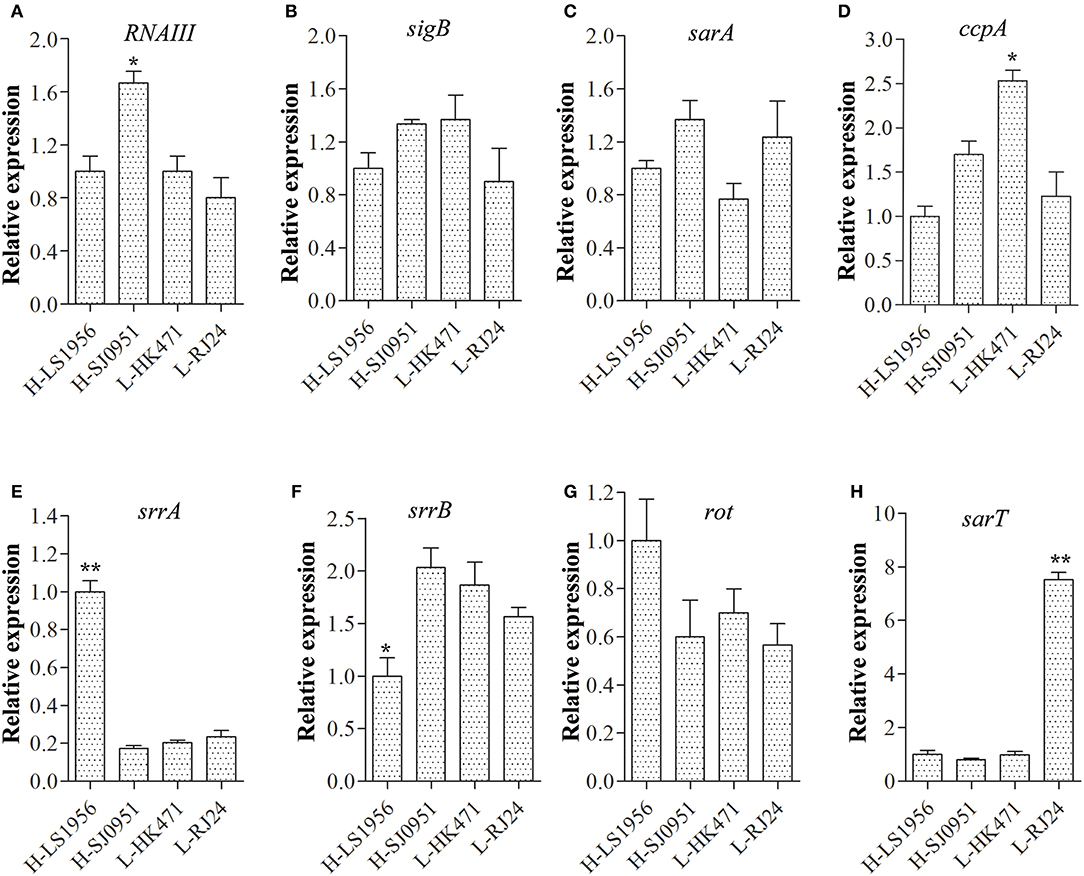
Figure 3. Expression of major virulence regulator genes including RNAIII (A), sigB (B), sarA (C), scpA (D), srrA (E), srrB (F), rot (G), and sarT (H) in four tst differentially expressed S. aureus isolates. RNAIII in strain H-SJ0951, ccpA in strain L-HK471, srrA in strain H-LS1956, and sarT in strain L-RJ24 were detected to be highly expressed and srrB in strain H-LS1956 were detected to be lowly expressed in the post-exponential growth phase. Data are expressed as the means ± standard deviation (SD). Statistically significant differences were determined by ANOVA with LSD post-hoc test. *p < 0.05, ** p < 0.01 compared with the closest tst expression of one of the three strains. The experiment was independently repeated three times.
Detection of Mutations in Open Reading Frame (ORF) and Promoter of tst Gene
To determine how the promoter of tst gene and the structure of TSST-1 itself affect the amount of tst mRNA, the region containing the ORF and promoter of tst gene in the four above-mentioned isolates were sequenced. A comparison of the ORF nucleotide sequences from the four strains with the corresponding sequence of the tst gene from S. aureus strain N315 (GenBank accession no. BA000018.3) revealed no changes. While one mutation for tst promoter was detected between the high and low tst expressed isolates (GenBank accession no. MK537300 and MK537301) (Figure 4). More specifically, a base T deleted in the promoter region (nt-114) was detected in the two isolates with high-expressed tst gene. The mutation occurred in an AT-rich region, which is the homologous sequence of the agr P2-P3 regulatory site SarA binding (Chan and Foster, 1998b) (Figure 4). And it is located in the upstream of SarA binding box 1/2 and a putative catabolite-responsive element (cre) region (Seidl et al., 2008).

Figure 4. Alignment of tst promoters in tst gene high- and low-expression isolates. H, tst high-expression strains; L, tst low-expression strains. Two isolates (H-LS1956 and H-SJ0951) with the tst gene high expression have a base T deletion in the promoter region (nt-114, red rectangular box). This region is AT-rich and the homologous sequence of the agr P2-P3 regulatory site SarA binding (dotted oval) (Chan and Foster, 1998b), and is located in the upstream of SarA binding box 1/2 (black rectangular boxes) and a putative catabolite-responsive element (cre) region (dashed line) (Seidl et al., 2008).
Discussion
TSST-1 is a major virulence factor in S. aureus infections. Here, 208 tst-positive S. aureus clinical isolates collected from multiple hospitals in China were characterized. We found ST5 (CC5)-agr II-t002-SCCmecII was the most common clone among MRSA. Moreover, our results indicated promoter mutation may be one of the factors resulting in the tst gene differential expression. However, it is still need to be verified by site-directed mutagenesis studies. The epidemiology of tst-positive MRSA showed regional variations worldwide. A cross-sectional study in Shiraz, Iran has found 18.1% of surveyed MSSA and 11.6% of MRSA possessed tst gene (Motamedifar et al., 2015). Another study from Japan reported that up to 75% of MRSA harbored tst gene, 96.7% of which belonged to agr 2 (Nagao et al., 2009). In China, the regional differences also exist. A multicenter study showed 31.4% of S. aureus surveyed were tst positive, and CC398, CC15, and CC188 were the most common (He et al., 2013). The CC5 clone is not usually observed in tst gene positive S. aureus isolates in China but reports of this clone are now emerging. A recent study from Suzhou, a city near Shanghai, reported that tst was detected in 18.0% of 150 isolates collected, and is mostly associated with CC5-t002 (Wang et al., 2017). In our strain collection, 29.8% (198/665) of MRSA and 4.0% (10/251) of MSSA isolates were tst gene positive, and ST5 (CC5) isolates (Table 2) with the genotype consisting of agr type 2, spa type t002, SCCmec type II (Zhao et al., 2016), accounted for 53.9 % of 76 representative isolates chosen to be characterized by MLST. It appears therefore that tst prevalence rate is regionally different and CC5 may represent a newly emerging clone in east of China. Additionally, all the chosen tst positive MSSA isolates were CC5 clone, indicating the CC5 tst carrying MRSA likely evolved from the similar CC5 tst carrying MSSA clone.
Due to the high percentage of S. aureus isolates carrying tst gene, understanding of tst expression and the possible expression regulatory mechanism appears to be important. Nagao et al. (2009) identified 170-fold variation in the amount of TSST-1 produced among clinical MRSA isolates. While our results revealed that tst expression varied 8.4-folds on transcriptional level among our clinical S. aureus isolates representative. The magnitude of this difference may derive from detection methods (qRT-PCR vs. western blot). Additionally, we found strains with high tst expression level only accounted for a small part of the isolates detected, generally explaining the phenomenon that relatively high rate of S. aureus harboring tst gene with the low incidence of TSS. This also supported the notion that TSST-1 expression is under the rigorous genetic regulation (Bronner et al., 2004).
MSSA strains have greater potential to secrete toxins, such as Panton-Valentine leukocidin (PVL), than MRSA (Varshney et al., 2010). The cause may be that MSSA isolates have less genetic fitness burden due to the lack of the SCCmec element carriage. However, conflicting conclusions exist in the balance of TSST-1 production and the SCCmec element carriage in S. aureus. Schmitz et al. (1997) have investigated the toxin production of clinical S. aureus isolates and found the TSST-1 expression was independent of the sensitivity of S. aureus to methicillin. While a recent study showed that the tst-positive CC30 MSSA strains produced more TSST-1 toxin when compared with tst-positive CC30 MRSA isolates (Sharma et al., 2018), which is consistent with the findings of this study: the two MSSA isolates (H-LS1956 and H-SJ0951) expressed obviously higher mRNA levels of tst than MRSA with the same genetic background. The contradiction may arise from whether to uniform genetic background of S. aureus when comparing TSST-1 production and carriage of SCCmec element.
Various environmental factors including glucose, oxygen, magnesium ions, α and β chains of hemoglobin, a range of antibiotics (e. x. nafcillin and clindamycin) and TSST-1 itself have been proved to influence the expression of TSST-1 (Kass, 1989; Chan and Foster, 1998a; Yarwood and Schlievert, 2000; Pragman et al., 2007; Schlievert et al., 2007; Stevens et al., 2007; Seidl et al., 2008). These environmental triggers affect TSST-1 expression via a large number of virulence regulators forming a complicated network in S. aureus (Andrey et al., 2015). The accessory gene regulator (agr) system and staphylococcal accessory regulator A (SarA) are thought to be prominent factors to modulate TSST-1 production. The agr system plays a central role in the pathogenesis of S. aureus, and its effector termed RNAIII has been shown to upregulate TSST-1 production (Recsei et al., 1986). SarA affects tst expression through binding to a certain element in tst gene promoter (Andrey et al., 2010). Other regulators controlling tst expression include sigma factor B (SigB) (Andrey et al., 2015), Staphylococcal respiratory response AB (SrrAB) (Pragman et al., 2004), Carbon catabolite protein A (CcpA) (Seidl et al., 2008), SarT (Schmidt et al., 2001), and repressor of toxin (Rot) (Andrey et al., 2015). However, these regulators are only studied in several type strains, whether they take effects in clinical isolates remains unclear. In the present study, we analyzed the expression of RNAIII, sigB, sarA, ccpA, srrAB, rot, and sarT in four tst differentially expressed strains, and found srrA, srrB, RNAIII, sarT, and ccpA were expressed unnormally in specific strains, indicating the regulatory networks in different strains may be not uniformly the same. In this study, we also sequenced the promoter and ORF region of tst. Consequently, a base T deletion located on an AT-rich region binding SarA in tst overexpression isolates was revealed, which was thought to be more likely to lead to tst high expression. Conversely, a previous study reported that no change was found in the sequences of promoter region among differentially expressed strains (Nagao et al., 2009). Overall, the previous and present results indicate that TSST-1 production is controlled in various and complex regulatory systems.
This study had several limitations. Firstly, this study lacks detailed clinical information about the patients, thus we cannot evaluate the association between outcome of patients and tst expression level. Secondly, we only found a mutation in tst promoter in high-expression isolates. Further study, including using dual luciferase report system to determine the function of mutation in tst promoter, will be needed. Thirdly, four differentially expressed virulence regulators in the four chosen isolates were revealed. Whereas, the mechanisms are still unclear and remain to be further demonstrated.
Conclusions
In summary, although we observed a heterogeneity of tst-positive MRSA clones, type ST5 (CC5)-agrII-t002-SCCmecII was proved to be represented one dominated clone in the regions investigated. This highlights the need for prevention and control measures to prevent the spread of this type of strains in China, particularly in Shanghai and Zhejiang regions. Although the tst high-expressing strains rarely occur among the clinical S. aureus isolates, targeting TSST-1, such as using some protein synthesis inhibitory antibiotics or herbal extracts inhibiting the production of this toxin (Qiu et al., 2011, 2012; Hodille et al., 2017; Katahira et al., 2019), for the treatment of S. aureus infection can be considered as an alternative strategy because TSS caused by TSST-1 is fatal. Additionally, the expression of tst has a potential association with the mutation of its promoter and variations in specific virulence regulators expression. Therefore, sequence of the tst promoter and quantification of major virulence regulators expression may provide significant information in survey of MRSA infection.
Author Contributions
QL designed and conceived the investigation. HZ, HY, and SX carried out the experiments. HZ, CH, XX, FH, WS, FG, and CZ analyzed the experiment data. HZ, SX, and QL wrote this manuscript. QL and HZ revised the article and approved the final version to be published.
Key Contribution
These findings implied that ST5 (CC5)-agr2-t002-SCCmecII is the most prevalent tst positive S. aureus clone in the region investigated. In addition, data also showed that tst expression of clinical S. aureus isolates may be associated with tst promoter and variations in specific virulence regulators.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81772247 and No. 81371872), Shanghai Jiaotong University Medical-Engineering Cross Research Youth Fund (YG2016QN31), and Medical Science and Technology Plan (Young Talents) of Zhejiang province (2019RC256).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank all of the researchers for their dedication and contributions in the present study. We thank Dr. Shu Jin (Shanghai People's Hospital of Putuo District) for providing the methods for the lysis of S. aureus (patent number: ZL201310124795.2).
References
Andrey, D. O., Jousselin, A., Villanueva, M., Renzoni, A., Monod, A., Barras, C., et al. (2015). Impact of the regulators SigB, Rot, SarA and sarS on the toxic shock Tst promoter and TSST-1 expression in Staphylococcus aureus. PLoS ONE 10:e0135579. doi: 10.1371/journal.pone.0135579
Andrey, D. O., Renzoni, A., Monod, A., Lew, D. P., Cheung, A. L., and Kelley, W. L. (2010). Control of the Staphylococcus aureus toxic shock tst promoter by the global regulator SarA. J. Bacteriol. 192, 6077–6085. doi: 10.1128/JB.00146-10
Blot, S., Vandewoude, K., and Colardyn, F. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 2025–2026. doi: 10.1056/NEJM199812313392716
Bronner, S., Monteil, H., and Prevost, G. (2004). Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28, 183–200. doi: 10.1016/j.femsre.2003.09.003
Brosnahan, A. J., and Schlievert, P. M. (2011). Gram-positive bacterial superantigen outside-in signaling causes toxic shock syndrome. FEBS J. 278, 4649–4667. doi: 10.1111/j.1742-4658.2011.08151.x
Chan, P. F., and Foster, S. J. (1998a). The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology 144 (Pt 9), 2469–2479. doi: 10.1099/00221287-144-9-2469
Chan, P. F., and Foster, S. J. (1998b). Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180, 6232–6241.
Enright, M. C., Day, N. P., Davies, C. E., Peacock, S. J., and Spratt, B. G. (2000). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38, 1008–1015.
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P., and Spratt, B. G. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186, 1518–1530. doi: 10.1128/jb.186.5.1518-1530.2004
Gorwitz, R. J., Kruszon-Moran, D., McAllister, S. K., McQuillan, G., McDougal, L. K., Fosheim, G. E., et al. (2008). Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J. Infect. Dis. 197, 1226–1234. doi: 10.1086/533494
He, W., Chen, H., Zhao, C., Zhang, F., Li, H., Wang, Q., et al. (2013). Population structure and characterisation of Staphylococcus aureus from bacteraemia at multiple hospitals in China: association between antimicrobial resistance, toxin genes and genotypes. Int. J. Antimicrob. Agents 42, 211–219. doi: 10.1016/j.ijantimicag.2013.04.031
Hodille, E., Rose, W., Diep, B. A., Goutelle, S., Lina, G., and Dumitrescu, O. (2017). The role of antibiotics in modulating virulence in Staphylococcus aureus. Clin. Microbiol. Rev. 30,887–917. doi: 10.1128/CMR.00120-16
Iwatsuki, K., Yamasaki, O., Morizane, S., and Oono, T. (2006). Staphylococcal cutaneous infections: invasion, evasion and aggression. J. Dermatol. Sci. 42, 203–214. doi: 10.1016/j.jdermsci.2006.03.011
Kass, E. H. (1989). Magnesium and the pathogenesis of toxic shock syndrome. Rev. Infect. Dis. 11(Suppl. 1), S167–S173.
Katahira, E. J., Davidson, S. M., Stevens, D. L., and Bolz, D. D. (2019). Subinhibitory concentrations of tedizolid potently inhibit extracellular toxin production by methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 68, 255–262. doi: 10.1099/jmm.0.000905
Koreen, L., Ramaswamy, S. V., Graviss, E. A., Naidich, S., Musser, J. M., and Kreiswirth, B. N. (2004). spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin Microbiol. 42, 792–799. doi: 10.1128/jcm.42.2.792-799.2004
Kulhankova, K., King, J., and Salgado-Pabon, W. (2014). Staphylococcal toxic shock syndrome: superantigen-mediated enhancement of endotoxin shock and adaptive immune suppression. Immunol. Res. 59, 182–187. doi: 10.1007/s12026-014-8538-8
Lina, G., Boutite, F., Tristan, A., Bes, M., Etienne, J., and Vandenesch, F. (2003). Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl. Environ. Microbiol. 69, 18–23. doi: 10.1128/aem.69.1.18-23.2003
Lindsay, J. A., Ruzin, A., Ross, H. F., Kurepina, N., and Novick, R. P. (1998). The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29, 527–543.
Liu, C. L., Chen, Z. J., Wang, F., Hou, H. Y., Wang, Y., Zhu, X. H., et al. (2014). The impact of mgrA on progression of Staphylococcus aureus sepsis. Microb. Pathog. 71–72, 56–61. doi: 10.1016/j.micpath.2014.03.012
Motamedifar, M., Ebrahim-Saraie, H. S., Alfatemi, S. M., Zalipour, M., Kaveh, M., and Khoshkharam-Roodmajani, H. (2015). Frequency of the toxic shock syndrome toxin-1 gene in methicillin-susceptible and -resistant Staphylococcus aureus isolates from teaching hospitals in Shiraz, Iran. Rev. Soc. Bras. Med. Trop. 48, 90–93. doi: 10.1590/0037-8682-0142-2014
Mulvey, M. R., Chui, L., Ismail, J., Louie, L., Murphy, C., Chang, N., et al. (2001). Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J. Clin. Microbiol. 39, 3481–3485. doi: 10.1128/JCM.39.10.3481-3485.2001
Nagao, M., Okamoto, A., Yamada, K., Hasegawa, T., Hasegawa, Y., and Ohta, M. (2009). Variations in amount of TSST-1 produced by clinical methicillin resistant Staphylococcus aureus (MRSA) isolates and allelic variation in accessory gene regulator (agr) locus. BMC Microbiol. 9:52. doi: 10.1186/1471-2180-9-52
Pragman, A. A., Ji, Y., and Schlievert, P. M. (2007). Repression of Staphylococcus aureus SrrAB using inducible antisense srrA alters growth and virulence factor transcript levels. Biochemistry 46, 314–321. doi: 10.1021/bi0603266
Pragman, A. A., Yarwood, J. M., Tripp, T. J., and Schlievert, P. M. (2004). Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186, 2430–2438. doi: 10.1128/jb.186.8.2430-2438.2004
Qiu, J., Li, H., Su, H., Dong, J., Luo, M., Wang, J., et al. (2012). Chemical composition of fennel essential oil and its impact on Staphylococcus aureus exotoxin production. World J. Microbiol. Biotechnol. 28, 1399–1405. doi: 10.1007/s11274-011-0939-4
Qiu, J., Wang, J., Luo, H., Du, X., Li, H., Luo, M., et al. (2011). The effects of subinhibitory concentrations of costus oil on virulence factor production in Staphylococcus aureus. J. Appl. Microbiol. 110, 333–340. doi: 10.1111/j.1365-2672.2010.04888.x
Recsei, P., Kreiswirth, B., O'Reilly, M., Schlievert, P., Gruss, A., and Novick, R. P. (1986). Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol. Gen. Genet. 202, 58–61.
Schlievert, P. M., Case, L. C., Nemeth, K. A., Davis, C. C., Sun, Y., Qin, W., et al. (2007). Alpha and beta chains of hemoglobin inhibit production of Staphylococcus aureus exotoxins. Biochemistry 46, 14349–14358. doi: 10.1021/bi701202w
Schmidt, K. A., Manna, A. C., Gill, S., and Cheung, A. L. (2001). SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect. Immun. 69, 4749–4758. doi: 10.1128/IAI.69.8.4749-4758.2001
Schmitz, F. J., Mackenzie, C. R., Geisel, R., Wagner, S., Idel, H., Verhoef, J., et al. (1997). Enterotoxin and toxic shock syndrome toxin-1 production of methicillin resistant and methicillin sensitive Staphylococcus aureus strains. Eur. J. Epidemiol. 13:699–708.
Seidl, K., Bischoff, M., and Berger-Bachi, B. (2008). CcpA mediates the catabolite repression of tst in Staphylococcus aureus. Infect. Immun. 76, 5093–5099. doi: 10.1128/IAI.00724-08
Sharma, H., Smith, D., Turner, C. E., Game, L., Pichon, B., Hope, R., et al. (2018). Clinical and molecular epidemiology of staphylococcal toxic shock syndrome in the United Kingdom. Emerg. Infect. Dis. 24, 258–266. doi: 10.3201/eid2402.170606
Spaulding, A. R., Salgado-Pabon, W., Kohler, P. L., Horswill, A. R., Leung, D. Y., and Schlievert, P. M. (2013). Staphylococcal and streptococcal superantigen exotoxins. Clin. Microbiol. Rev. 26, 422–447. doi: 10.1128/CMR.00104-12
Stevens, D. L., Ma, Y., Salmi, D. B., McIndoo, E., Wallace, R. J., and Bryant, A. E. (2007). Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 195, 202–211. doi: 10.1086/510396
Varshney, A. K., Martinez, L. R., Hamilton, S. M., Bryant, A. E., Levi, M. H., Gialanella, P., et al. (2010). Augmented production of Panton-Valentine leukocidin toxin in methicillin-resistant and methicillin-susceptible Staphylococcus aureus is associated with worse outcome in a murine skin infection model. J. Infect. Dis. 201, 92–96. doi: 10.1086/648613
Wang, M., Zheng, Y., Mediavilla, J. R., Chen, L., Kreiswirth, B. N., Song, Y., et al. (2017). Hospital dissemination of tst-1-Positive clonal complex 5 (CC5) methicillin-resistant Staphylococcus aureus. Front. Cell Infect. Microbiol. 7:101. doi: 10.3389/fcimb.2017.00101
Xu, H., Cai, S., and Dai, H. (2016). Characteristics of infective endocarditis in a tertiary hospital in east China. PLoS ONE 11:e0166764. doi: 10.1371/journal.pone.0166764
Yan, L., Jiang, D. M., Cao, Z. D., Wu, J., Wang, X., Wang, Z. L., et al. (2015). Treatment of Staphylococcus aureus-induced chronic osteomyelitis with bone-like hydroxyapatite/poly amino acid loaded with rifapentine microspheres. Drug. Des. Devel. Ther. 9, 3665–3676. doi: 10.2147/DDDT.S84486
Yarwood, J. M., and Schlievert, P. M. (2000). Oxygen and carbon dioxide regulation of toxic shock syndrome toxin 1 production by Staphylococcus aureus MN8. J. Clin. Microbiol. 38, 1797–1803.
Zhang, K., McClure, J. A., Elsayed, S., Louie, T., and Conly, J. M. (2005). Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43, 5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005
Keywords: virulence regulators, Staphylococcus aureus, toxic shock syndrome toxin-1, MRSA, gene expression, molecular typing
Citation: Zhao H, Xu S, Yang H, He C, Xu X, Hu F, Shu W, Gong F, Zhang C and Liu Q (2019) Molecular Typing and Variations in Amount of tst Gene Expression of TSST-1-Producing Clinical Staphylococcus aureus Isolates. Front. Microbiol. 10:1388. doi: 10.3389/fmicb.2019.01388
Received: 10 February 2019; Accepted: 03 June 2019;
Published: 19 June 2019.
Edited by:
Mattias Collin, Lund University, SwedenReviewed by:
Iris Spiliopoulou, University of Patras, GreeceXuming Deng, Jilin University, China
Hema Sharma, Imperial College London, United Kingdom
Copyright © 2019 Zhao, Xu, Yang, He, Xu, Hu, Shu, Gong, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingzhong Liu, amlhb2RhbWVkaWNpbmVAZm94bWFpbC5jb20=
†These authors have contributed equally to this work
 Huanqiang Zhao
Huanqiang Zhao Su Xu
Su Xu Han Yang1
Han Yang1 Chunyan He
Chunyan He Xiaogang Xu
Xiaogang Xu Fupin Hu
Fupin Hu Wen Shu
Wen Shu Fang Gong
Fang Gong Chuanling Zhang
Chuanling Zhang Qingzhong Liu
Qingzhong Liu