- 1Department of Veterinary and Animal Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark
- 2Department of Veterinary Medicine and Public Health, College of Veterinary Medicine and Biomedical Sciences, Sokoine University of Agriculture, Morogoro, Tanzania
- 3Research Unit in Applied Microbiology and Pharmacology of Natural Substances, Research Laboratory in Applied Biology, Polytechnic School of Abomey-Calavi, University of Abomey-Calavi, Abomey-Calavi, Benin
- 4Department of Biochemistry, Microbiology, and Immunology, Wayne State University School of Medicine, Detroit, MI, United States
Studies have reported the occurrence of Vibrio cholerae in fish but little is known about the interaction between fish and toxigenic V. cholerae as opposed to phytoplankton, which are well-established aquatic reservoirs for V. cholerae. The present study determined the role of tilapia (Oreochromis niloticus) as a reservoir host for survival and transmission of V. cholerae in aquatic environments. Three experiments were performed with one repetition each, where O. niloticus (∼2 g) kept in beakers were inoculated with four V. cholerae strains (5 × 107 cfu/mL). Firstly, infected tilapia were kept in stagnant water and fed live brine shrimp (Artemia salina) larvae daily. Secondly, infected tilapia were kept without feeding and water was changed every 24 h. Thirdly, infected tilapia were fed and water was renewed daily. Infected tilapia and non-infected controls were sacrificed on days 1, 2, 3, 7, and 14 post-inoculation and V. cholerae were enumerated in intestinal content and water. Another experiment assessed the transmission of V. cholerae from infected to non-infected tilapia. The study revealed that El Tor biotype V. cholerae O1 and V. cholerae non-O1 colonized tilapia intestines and persisted at stable concentrations during the second week of the experiment whereas the Classical biotype was undetectable after 1 week. In stagnant water with feeding, V. cholerae counts dropped to 105 cfu/ml in water and from 107 to 104 cfu/intestine in fish after 14 days. When water was renewed, counts in water decreased from 107 to 103 cfu/ml and intestinal counts went from 106 to 102 cfu/intestine regardless of feeding. All strains were transmitted from infected to naïve fish after 24 h of cohabitation. Tilapia like other fish may play an essential role in the survival and dissemination of V. cholerae O1 in aquatic environments, e.g., the seventh pandemic strains mostly. In this study, tilapia were exposed to high concentrations of V. cholerae to ensure initial uptake and follow-up studies with lower doses resembling natural concentrations of V. cholerae in the aquatic environment are needed to confirm our findings.
Introduction
Vibrio cholerae is one of the longest recognized human infectious pathogens, yet there is still much to clarify on the emergence and transmission of cholera, the disease for which V. cholerae is the causative agent. V. cholerae O1 and O139 are the only serogroups causing cholera, with the leading strains being the toxigenic V. cholerae O1 El Tor and Classical biotypes (Dalsgaard et al., 2001). The Classical biotype, however, has not been implicated in cholera outbreaks for several decades and has become extremely rare, if not extinct, in the aquatic environment since the beginning of the seventh cholera pandemic (Safa et al., 2006; Nag et al., 2018). The main virulence factor in humans expressed by all biotypes is cholera toxin. The intestinal colonization of V. cholerae in humans requires production of the cholera toxin co-regulated pilus (TCP), whose main transcription activator is ToxT (Faruque et al., 1998; Sanchez and Holmgren, 2011). V. cholerae O1 biotype El Tor that lacks active toxT can therefore be regarded as non-toxigenic. V. cholerae non-O1/O139 strains are ubiquitous in aquatic environments and rarely produce cholera toxin, but can cause sporadic diarrhea (Hounmanou et al., 2016). Most V. cholerae non-O1 do not contain tcpA (regulated by ToxT), but if present, and the role of toxT remains the same which is to regulate the transcription of TCP. In this study, a toxT mutant of V. cholerae O1 El Tor served to assess whether the lack of transcription of tcpA (regulated by ToxT) would affect colonization in tilapia.
Fish are potential carriers for V. cholerae and the occurrence of toxigenic and non-toxigenic strains of V. cholerae in tilapia (Oreochromis niloticus) and African catfish (Clarias gariepinus) has been reported during non-cholera outbreak periods (Hounmanou et al., 2016). V. cholerae non-O1 has also been isolated in carps (Rastrineobola argentea) from Lake Tanganyika (Nyambuli et al., 2018). A study in Bangladesh indicated that fish could serve as a potential vehicle for V. cholerae transmission to humans (Hossain et al., 2018), and genetic analysis of V. cholerae from a cholera outbreak in Zanzibar suggested that marine fish were implicated in pathogen transmission (Rabia et al., 2017).
In sub-Saharan Africa, where the seventh cholera pandemic has a high impact in terms of morbidities and mortalities (Mengel et al., 2014; Weill et al., 2017), frequent epidemics occur around the African Great Lakes Region (AGLR), where fishing, and fish processing represent an essential socio-economic activity (Nkoko et al., 2011; Plisnier et al., 2015; Ajayi and Smith, 2018). Association between cholera and the aquatic environment is well-established in the AGLR (Urassa et al., 2009; Reyburn et al., 2011). Well-recognized aquatic reservoirs of V. cholerae include phytoplanktons, zooplanktons, algae, and cyanobacteria. The role of fish as a reservoir host, however, remain speculative because the mere presence of V. cholerae in fish is not sufficient to confirm that fish is a true reservoir host providing multiplication and persistence of the pathogen (Tamplin et al., 1990; Halpern et al., 2004; Islam et al., 2015; Halpern and Izhaki, 2017).
Studies conducted using zebrafish as models indicated that they can be colonized by V. cholerae and can transmit the bacteria to naïve zebrafish via excretion (Runft et al., 2014; Mitchell et al., 2017). This therefore calls for further studies to explore the fate of V. cholerae in wild caught fish like tilapia.
In the present experimental study, we worked with one of the common edible fish species around the AGLR, namely tilapia (O. niloticus). The aim was to determine the role of tilapia in the survival of V. cholerae in the aquatic environment and in transmitting the pathogen. The results suggest that V. cholerae O1 El Tor (causing the seventh cholera pandemic) do survive in tilapia, which may have important implications in the epidemiology of the ongoing cholera epidemic around the AGLR.
Materials and Methods
Bacterial Strains Used in Tilapia Experiments
Vibrio cholerae O1 El Tor (strain E7946) and Classical (strain O395) biotypes used in this study were clinical strains obtained from a laboratory collection (Table 1). They are all streptomycin resistant, which allowed for the use of selective isolation procedures during the experiment. An environmental V. cholerae non-O1 was isolated from carps (Rastrineobolla agentea) in Lake Victoria, Tanzania, and confirmed streptomycin resistant by disc diffusion following standard methods of isolation and identification of V. cholerae (Hounmanou et al., 2016). The actual serotype of this non-O1 strain was not determined. The strain was included in the study to assess whether colonization in tilapia differs between clinical strains of serogroup O1 and the environmental V. cholerae non-O1 strains. V. cholerae O1 strain JW612, a toxT mutant was included to test whether the lack of transcription of tcpA (regulated by toxT) would affect colonization of the tilapia (Table 1).
Elimination of Natural V. cholerae in Tilapia Prior to Experiment
We used tilapia (O. niloticus) juveniles (approximately 2 g) obtained from a hatchery at the Sokoine University of Agriculture in Morogoro, Tanzania where the experiments were performed. About 20 tilapia juveniles were placed in beakers (2,000 mL) containing autoclaved tap water with constant aeration. To ensure that the tilapia juveniles did not contain environmental V. cholerae, intestinal samples of five tilapia per beaker were plated on thiosulfate-citrate-bile salts-sucrose agar (TCBS) (Oxoid Limited, Hampshire, United Kingdom) after 24, 48, and 72 h. This was followed by a species-specific PCR for the ompW gene (Nandi et al., 2000) using DNA from isolates recovered on TCBS agar and from samples enriched in APW. Water samples from the beakers were also cultured for V. cholerae. No V. cholerae colonies were detected in any water and fish gut samples and none of the samples produced the 588 bp band expected for the ompW gene. Tilapia juveniles confirmed negative for V. cholerae were used in the experiment.
Exposure of Tilapia to V. cholerae Inoculated in Water
The four V. cholerae strains used in the experiments (Table 1) were first grown in Luria-Bertani (LB) broth (Difco, Becton Dickinson and company, Maryland, United States) at 37°C for 24 h with agitation. The growth curves of the strains revealed that the overnight cultures (24 h at 37°C) of all the strains reached on average 1010 cfu/mL (data not shown). Following procedures described for zebrafish models (Runft et al., 2014; Mitchell et al., 2017), the V. cholerae strains were grown overnight in tubes containing 10 ml LB. Tubes with the overnight bacterial cultures were centrifuged at 5,000 g for 2–3 min and the pellets with bacteria were washed twice in normal saline solution (sterile water + 0.9% NaCl). Bacterial cells collected from 10 LB tubes were suspended in one mL normal saline/tube and added to beakers (aquarium) containing 2,000 mL of autoclaved tap water and 20 tilapia juveniles. Thus, the count of each V. cholerae strain was about 5 × 107 cfu/mL in each beaker, similar to concentrations used in the zebrafish models (Runft et al., 2014). This concentration is high compared to the expected concentration of V. cholerae in the natural aquatic environment. However, because gavage is not a natural route of administration in fish, we exposed tilapia to the test strains of V. cholerae by immersion in the beakers; a high dose of inoculum was therefore needed to ensure an uptake of the test strains. The beakers were kept at room temperature (25°C) for 2 weeks with constant aeration by an air pump. The experiment included four beakers with the V. cholerae strains (Table 1) and another beaker containing tilapia juveniles were given 1 mL sterile normal saline solution with no V. cholerae (control). Three tilapia juveniles were sacrificed 1, 2, 3, 7, and 14 days after inoculation of the V. cholerae strains. Fish were infected similarly, in all experiments.
Experimental Design
We conducted three different experiments with two replicates of each. Each experiment was repeated 2 weeks after the end of the first experiment to have a biological replicate that ensures the validity of the study. In each experiment, tilapia were inoculated with 5 × 107 cfu/mL of various V. cholerae strains (Table 1) by immersion in beakers as previously described (Mitchell et al., 2017). Tilapia (∼2 g) were sacrificed on days 1, 2, 3, 7, and 14 post-exposure and the concentration of V. cholerae in water and intestinal content was enumerated as was the water turbidity based on OD600 water measurements. The beakers with tilapia, but no V. cholerae served as controls and one mL of sterile normal saline solution was added at the onset of the experiment.
In Experiment 1, tilapia were starved for 24 h before the experiment. Two hours after the inoculation of the V. cholerae strains, feeding was initiated with fresh hatched brine shrimp (Artemia salina) (JBL GmbH & Co., Neuhofen, Germany) served twice a day until termination of the experiment. The brine shrimp were hatched from dry eggs in sterile water and did not contain V. cholerae as shown by lack of yellow bacterial colonies when grown on TCBS agar plates. Water in the beakers remained unchanged until the experiment was terminated. This experiment aimed to determine the colonization, the survival and shedding of the V. cholerae test strains in tilapia in stagnant water. The control for this experiment was a beaker with tilapia fed with the same brine shrimp and inoculated with sterile normal saline. We chose live feed because of the size of the juveniles being used in the study and also to avoid commercial feeds which may increase water fouling in the beakers.
In Experiment 2, water in the beakers was replaced daily (every 24 h) with fresh sterile water of the same volume (2,000 mL) and fish were not fed. When the water was changed, fish were removed from the initial beaker then washed twice in sterile tap water to remove external V. cholerae by rubbing their surface before placing them in a new beaker with sterile water. Infection procedures were the same as in Experiment 1 but this experiment aimed to assess the impact of feeding and water renewal on the survival and excretion of V. cholerae in tilapia in the absence of feeding.
In Experiment 3, water was changed like in Experiment 2, but fish were fed brine shrimp free of V. cholerae like in the Experiment 1. Tilapia was exposed to V. cholerae by immersion as in the other experiments. This experiment aimed to differentiate between the impact of feeding and water exchange on the survival and excretion of V. cholerae in tilapia.
Enumeration of V. cholerae
Fish were collected every morning and sacrificed. In Experiments 2 and 3 (where water was renewed), fish, and water samples were taken from the beakers before water renewal. After the tilapia juveniles (three fish were collected per time point) were sacrificed, the intestinal content was aseptically removed using sterile scissors and metal blades, and the content placed into 1 mL of sterile normal saline. After homogenization of the intestinal contents by crushing and shaking, serial decimal dilutions of the homogenate were made and 10 μL of dilutions were subsequently spread onto Luria-Bertani Agar (LA) (Difco) plates containing streptomycin (100 μg/mL). One mL water samples from the beakers were collected at each time point and diluted and plated on LA as described for the intestinal content samples. When we obtained 30 or less colonies on LA plates containing streptomycin, all isolates were re-streaked on TCBS agar for confirmation. With higher colony numbers, the identity of at least 30 colonies appearing on the LA plates was confirmed on TCBS agar. Selected El Tor and Classical isolates were collected from TCBS agar plates and confirmed as ctxA-positive by PCR to verify that they did not lose their virulence. In Experiments 2 and 3, colony counts were lower due to the daily change of water and, therefore, 100 μL of each dilution was plated on LA. Colony forming units per fish intestine or mL of water were calculated using counts from all plates based on dilutions with valid counts divided by the sum of offset values. Thus in Experiment 1, the detection limit for one sample (fish intestine or water sample) was 900 cfu (2.95 on a logarithmic scale with base 10) per intestine or per mL of water sample and 150 cfu per intestine for six fish samples combined and 450 cfu pr mL for two water samples. For Experiments 2 and 3 where 100 μL was plated, the detection limit for an individual sample was 9 cfu (1.96 on Log10 scale). At each time point intestines from three tilapia were analyzed individually for V. cholerae.
The use of high concentrations of V. cholerae could increase stress in the fish causing excretion of more waste particles which was evaluated by measuring the optical density values of the water. Like in the zebrafish models (Mitchell et al., 2017), the optical density (OD600) of 1 mL water sample was read at 600 nm in a spectrophotometer using normal saline as the blank. Optical density was measured also in the control groups and at different time points during the 14 days of experiment. OD values of the beaker water were measured at each time point along with V. cholerae counts in intestine and water samples.
Assessing Transmission of V. cholerae Within Tilapia Populations
Ten tilapia juveniles of ∼4 g were exposed to fresh overnight cultures of the V. cholerae strains (Table 1) in beakers containing 1 L autoclaved tap water as described above (approximately 5 × 107 cfu/mL). After 6 h of exposure, a time which previously was found sufficient to allow colonization of V. cholerae in zebrafish (Runft et al., 2014), the tilapia juveniles were washed in autoclaved tap water twice to remove any external V. cholerae present on the fish body. The juveniles were then placed in another beaker with sterile water containing ten naive tilapia juveniles of ∼2 g with the smaller size allowing differentiation from the larger 4 g fish. After 24 h of cohabitation, four of the naïve tilapia juveniles were sacrificed per beaker and intestinal V. cholerae populations were enumerated as described above.
Statistical Analyses
Vibrio cholerae counts (x) were calculated as total count per fish intestine (fish samples) or total count per mL (water samples) for the two repeated trials. Comparisons of bacterial counts in fish samples [log10(x+1)] between strains and over time was done using multiple linear regression where also the interaction between strain and time was assessed. Repetition was not a significant predictor of bacterial counts neither when tested alone nor in the full model and therefore it was left out of the final analysis. V. cholerae counts in water samples were not compared in a similar manner as there was only one sample for each repetition, strain and time point, but those counts were correlated with average counts in the fish using linear regression. Model assumptions were verified using normal probability plot of standardized residuals and histogram of residuals. Homoskedatiscity was checked using rvf-plot plus Breusch-Pagan/Cook-Weisberg test for heteroskedasticity. P-values < 0.05 were taken to indicate significant differences in Stata (Version 12, StataCorp, College Station, TX, United States). Bacterial counts from transmission experiments enumerated 24 h post exposure were only strain dependent. Therefore, one-way ANOVA was performed to compare mean counts for the two trials among strains.
Results
Survival of V. cholerae in Tilapia Kept in Stagnant Water and Given Live Feed
In Experiment 1, tilapia were exposed to 5 × 107 cfu/mL of V. cholerae as described in the Section “Materials Methods.” They fish were kept in stagnant water and fed brine shrimp for the 2 weeks duration of the experiment. Tilapia juveniles exposed to V. cholerae were found to be colonized by V. cholerae within 24 h after exposure, with average intestinal counts for all test strains varying between 107 cfu/intestine on day 1 post infection to 105 cfu/intestine 14 days after infection. V. cholerae counts declined over time for all four strains (Figure 1A), with a significant interaction seen between time and strain (p < 0.001). There was no difference in concentration of different test V. cholerae strains for the first 2 days. However, 3 and 7 days after exposure, counts were lower for the Classical biotype than those of the V. cholerae non-O1 strain (p < 0.05 – p < 0.001 depending on the day). One fish exposed to the V. cholerae O1 Classical biotype did not contain V. cholerae on day 7 (i.e., below the detection limit of 2.95 on a log10-scale). After 14 days, all fish exposed to the V. cholerae O1 Classical biotype no longer contained detectable levels of this strain. Both V. cholerae O1 El Tor and a ΔtoxT V. cholerae O1 El Tor, had lower counts than the V. cholerae non-O1 strain (p < 0.05). Except for the Classical biotype strain, strains survived in tilapia for 2 weeks at constant levels. No V. cholerae was detected in the uninfected control tilapia for the duration of the experiment.
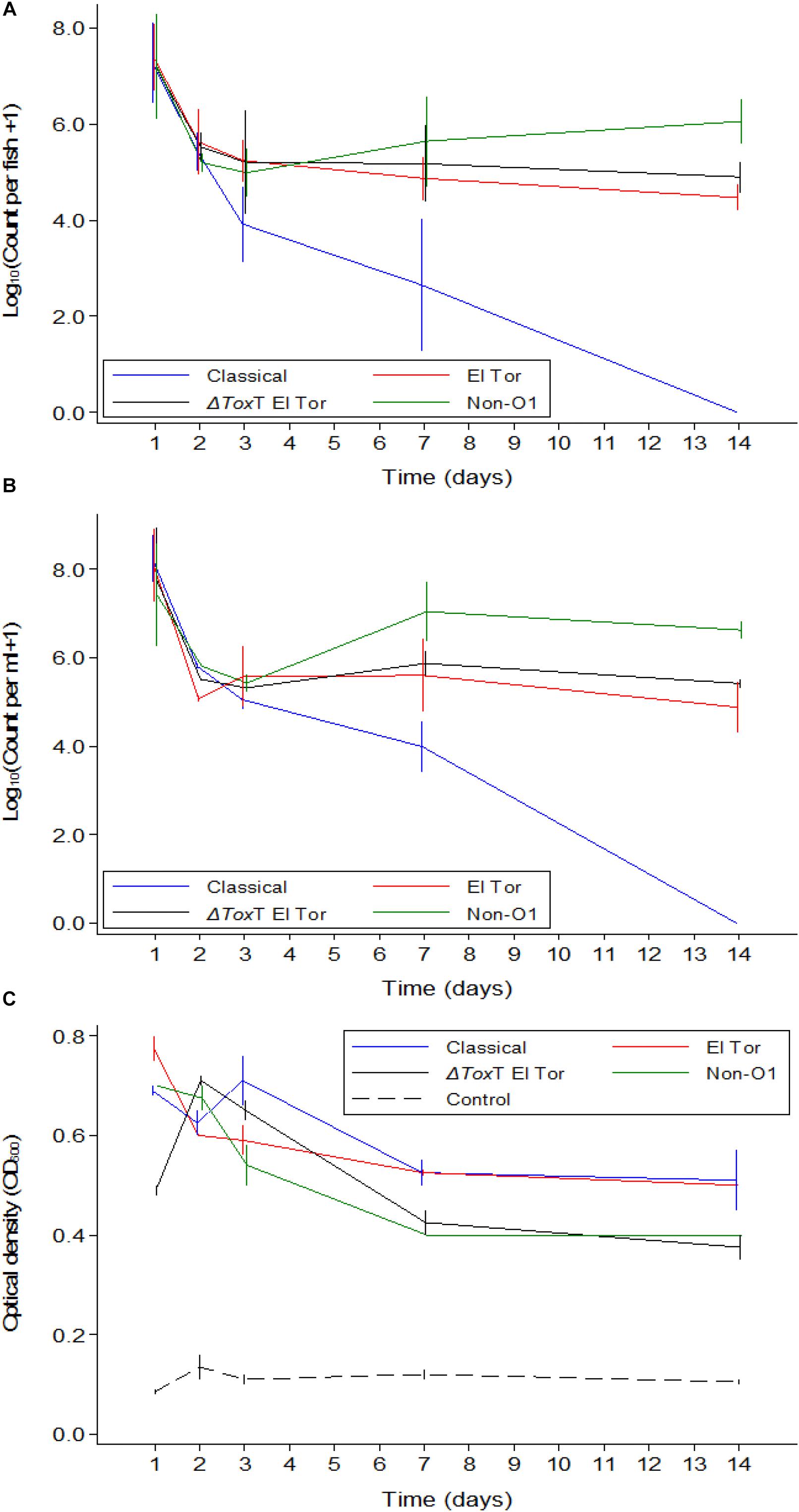
Figure 1. Mean counts of V. cholerae from two repetitions in tilapia intestines (A) count per fish, in water (B) count per mL, and absorbance of water (C) over time when tilapia were kept in the same water and given live feed. Error bars indicate 95% CL. Each strain is slightly off its exact x-value to allow distinction of the error bars. Relevant statistical differences between strains and time points are indicated in the text.
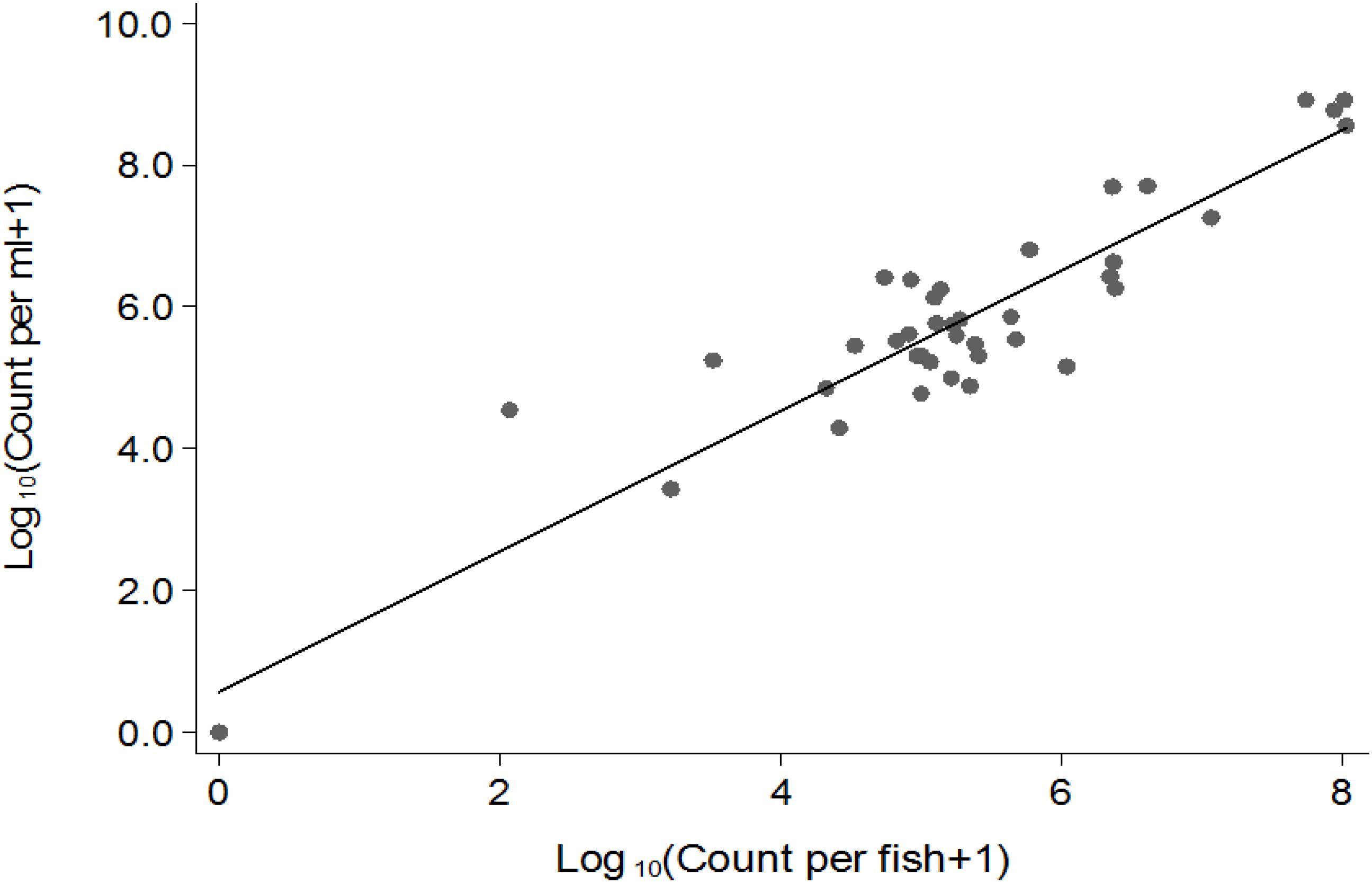
Vibrio cholerae numbers in water were similar to numbers in fish intestinal content. The Classical biotype was not detected in water 1 week after exposure (Figure 1B). Counts in water correlated with average counts in the fish intestine (Figure 2). However, the average V. cholerae counts in water varied between 108 cfu/mL on day 1 to 106 cfu/mL 14 days after inoculation of test strains and was higher than those observed in fish intestines. No V. cholerae was isolated from water in the control beakers.
Optical density of water from beakers with fish exposed to V. cholerae strains was considerably higher than the density in aquaria with unexposed fish, even for the Classical biotype strain after it was no longer detectable. Significant predictors of optical density when analyzing only the four V. cholerae strains were strain (p < 0.05) and time (p < 0.001). The Classical and El Tor biotypes did not differ when adjusting for time, while the ΔtoxT mutant of El Tor, and the non-O1 strains both differed from the Classical strain but did not differ between themselves. Optical water densities were lower at day 7 and 14 than during day 1 (Figure 1C).
Survival of V. cholerae in Tilapia When Water Was Changed Daily and Tilapia Were Not Fed
In Experiment 2, fish were exposed to V. cholerae as described above. However, in contrast to Experiment 1, the tilapia were not fed and the water was changed daily. Thus, after the initial infection only V. cholerae that multiplied in the intestine and excreted by the fish would be detected in the water. With daily water exchange and absence of feeding, tilapia were still colonized by all strains of V. cholerae 24 h post infection with average counts around 106 cfu/intestine (Figure 3A). Up to 2 days post infection, there was no difference in colonization levels between strains (p > 0.05). However, from day 3 post infection, the concentration of the Classical biotype strain decreased significantly in the fish intestines and was undetectable after 1 week (p < 0.001). Despite the absence of feeding and with the constant daily water exchange, tilapia remained colonized with the three other strains of V. cholerae until the end of the 1 weeks, but the counts dropped significantly from day to day, most significantly during the first 7 days (p < 0.05). No V. cholerae growth was detected in the uninfected control group. During the second week, 5–15% mortality was recorded in all beakers probably due to starvation.
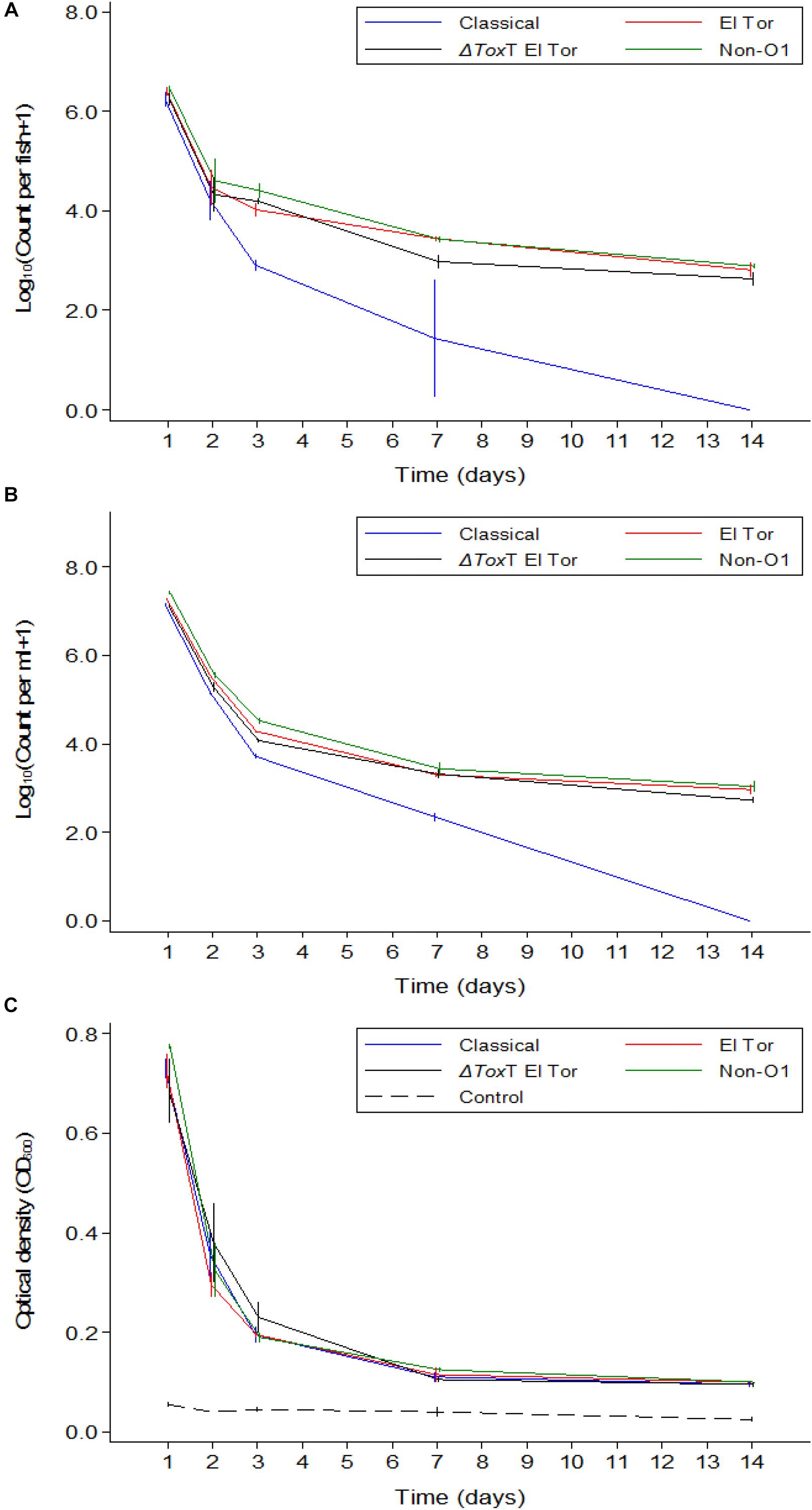
Figure 3. Mean counts of V. cholerae from two repetitions in tilapia intestines (A) count per fish, in water (B) count per mL, and absorbance of water (C) over time when aquarium water was changed daily and tilapia were not fed. Error bars indicate 95% CL. Each strain is slightly off its exact x-value to allow distinction of the error bars. Relevant statistical differences between strains and time points are indicated in the text.
In Experiment 2, V. cholerae concentrations in water were similar to those in the intestine, varying from 107 cfu/mL on day one post infection to 103 cfu/mL 14 days after inoculation, with significant daily decreases (p < 0.05). Like in the fish intestine, the Classical biotype strain could no longer be detected in the water after 1 week (Figure 3B). The other three strains remained present in the water despite the daily water replacement with fresh sterile water, suggesting continuous V. cholerae multiplication, and excretion by the fish. No V. cholerae was detected in the uninfected fish from the control beakers.
Overall, when tilapia were starved and water was exchanged daily, the optical density of water in beakers containing infected tilapia remained significantly higher than in the control beakers where fish were not infected (p < 0.05). The difference was more pronounced in the first week (p < 0.001); however, in the second week of the experiment, excretion levels decreased as the OD values from infected groups became statistically similar to the OD values of the uninfected control even though the numbers were higher than that of the control (p > 0.05). There was no significant difference between the four strains in terms of excretion at any time point (p > 0.05). Despite the absence of the Classical biotype strain in the second week, the OD values in that aquarium remained similar to that of the other strains (Figure 3C).
Survival of V. cholerae in Tilapia When Water Was Changed Daily and Tilapia Were Given Live Feed
In Experiment 3, fish were again infected with V. cholerae and water was renewed daily. In contrast to Experiment 2, these fish were also fed daily. V. cholerae counts in tilapia intestines, counts in water, and the water OD measurements showed similar values and trends as those recorded in Experiment 2 where tilapia were starved (Figures 4A–C). However, there was no fish mortality in Experiment 3 as compared to the Experiment 2 where fish were not fed.
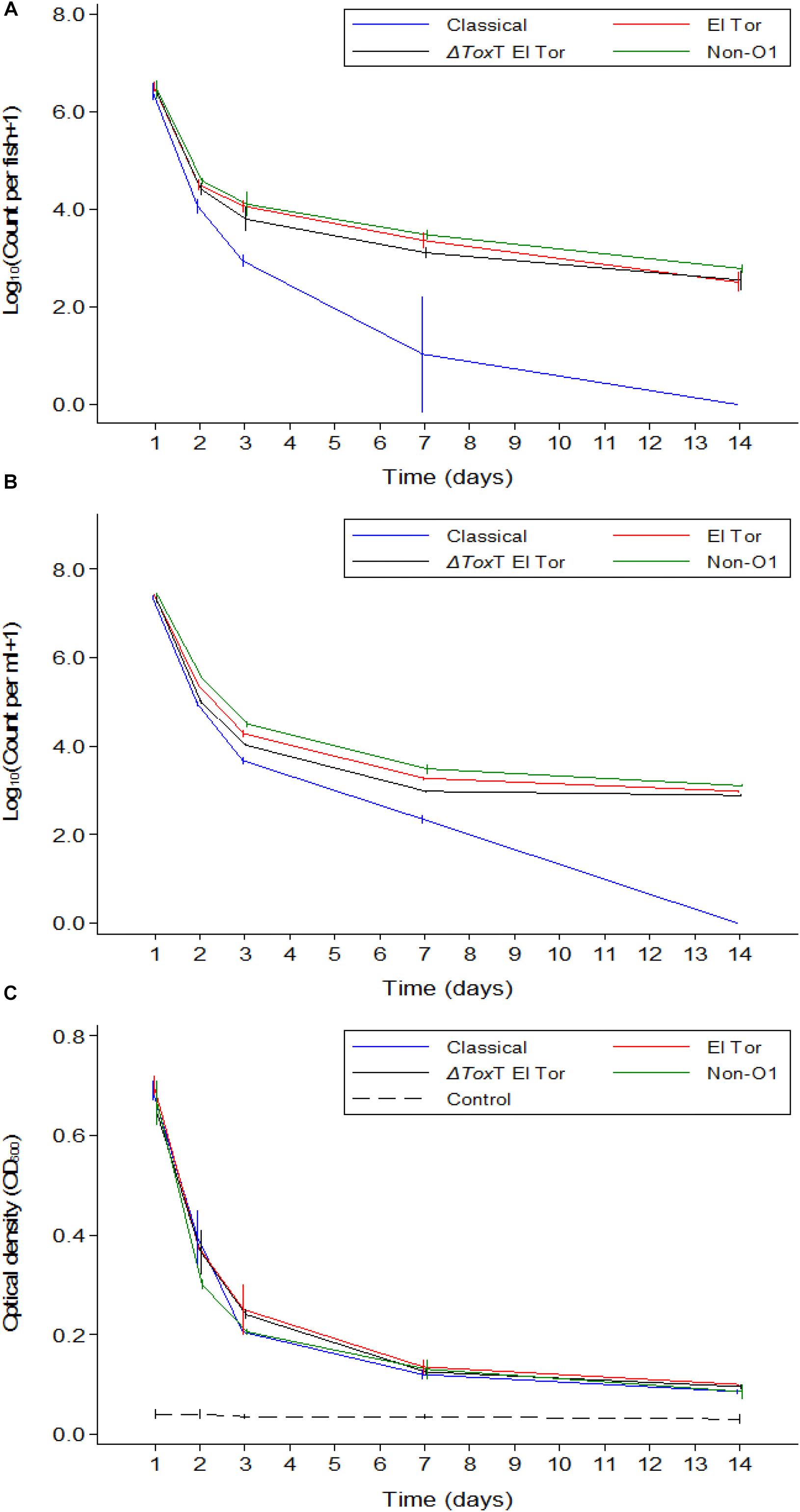
Figure 4. Mean counts of V. cholerae from two repetitions in tilapia intestines (A) count per fish, in water (B) count per mL, and absorbance of water (C) over time when aquarium water was changed daily and tilapia are given live feed. Error bars indicate 95% CL. Each strain is slightly off its exact x-value to allow distinction of the error bars. Relevant statistical differences between strains and time points are indicated in the text.
Comparison of V. cholerae Counts in Fish Guts and in Water Between the Three Experiments
A comparison between the three experiments was made to distinguish if feeding or water renewal influenced the survival of V. cholerae in fish. In stagnant water (Experiment 1), V. cholerae counts dropped from 107 to 105 cfu/mL and from 107 to 104 cfu/intestine in fish. However, in Experiments 2 and 3 where water was changed, V. cholerae in water decreased from 107 to 103 cfu/mL and gut counts ranged between 106 and 102 cfu/intestine, with significant daily decreases (p < 0.05). When water was replaced daily with fresh sterile tap water in Experiments 2 and 3, V. cholerae counts were statistically similar both in water and intestines regardless of presence or absence of feeding (p > 0.05; Figures 5A,B). This indicates that in both water and tilapia intestines, feeding did not have any impact on concentrations of V. cholerae (p > 0.05, Figure 5). In contrast, higher V. cholerae counts were recovered in water and intestine when water was not changed, i.e., comparing Experiment 1 with Experiments 2 and 3 (p < 0.05; Figures 5A,B). This significant variation between the three experiments was due to water renewal.
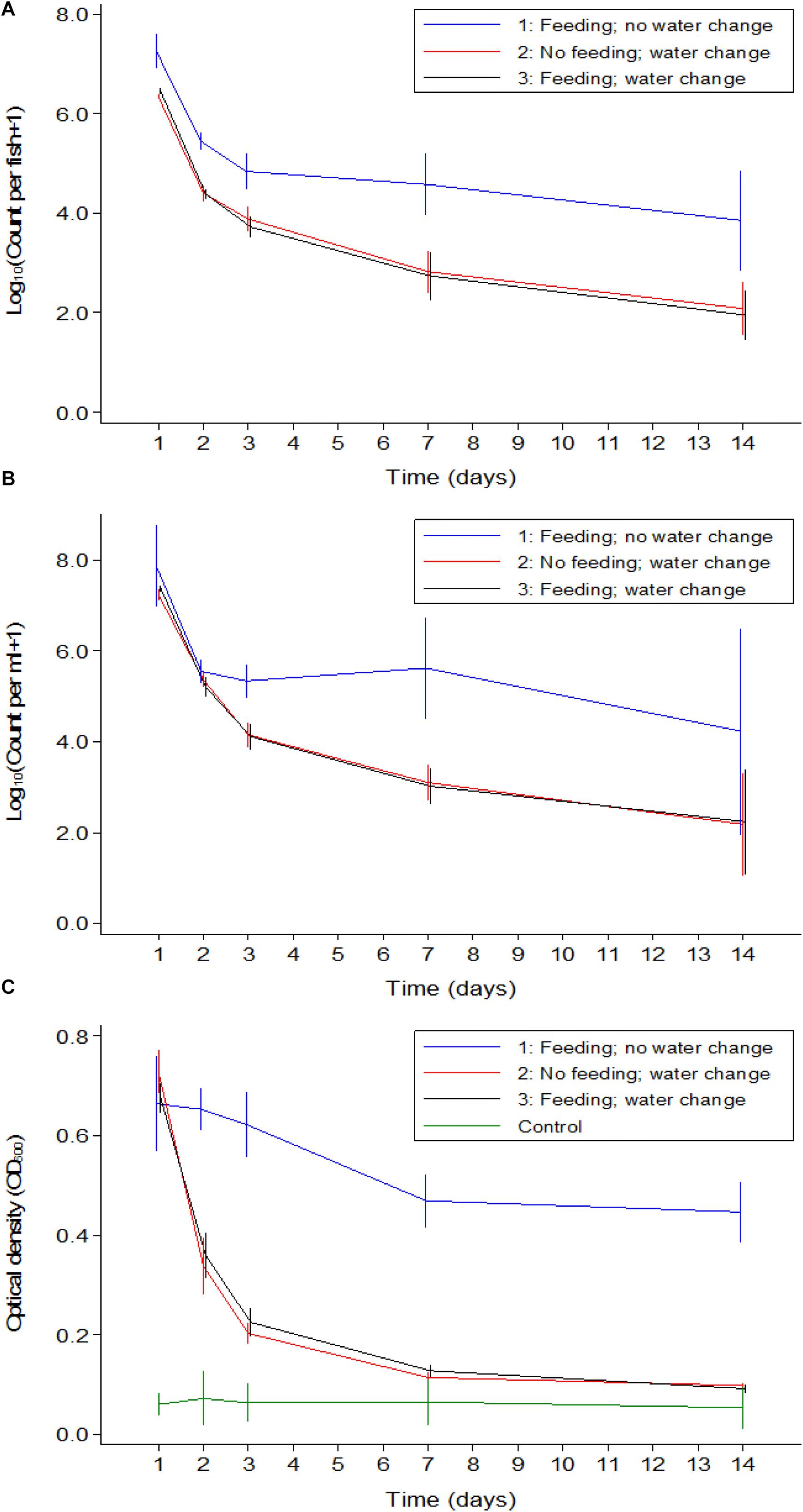
Figure 5. Comparison of V. cholerae counts (all strains combined) in tilapia guts (A), in water (B), and absorbance (C) between the three experiments overtime. Relevant statistical differences between experiments and time points are indicated in the text.
Throughout the experimental period, V. cholerae counts in tilapia intestines were not statistically different between Experiments 2 and 3 (p > 0.05); however, these counts were significantly lower when compared with Experiment 1 at each time point (p < 0.01). Furthermore, in water, the comparison between experiments revealed that there was no significant difference between the three experiments during the two first days, but from day 3 to day 14, the change of water significantly influenced bacterial counts (p < 0.001).
The OD values of water was higher when the experiment was done in the same water as compared to when water was changed daily (p < 0.05, Figure 5C). Apart from day one post infection when OD values from the three experiments were statistically similar, the absorbance of water differed significantly in Experiment 1 when compared to Experiments 2 and 3 (p < 0.001), i.e., OD values in Experiments 2 and 3 were not statistically different (p > 0.05). We conclude that feeding had no significant influence on the OD values but the change of water did reduce the water turbidity overtime as the concentration of V. cholerae decreased.
Transmission of V. cholerae Within Tilapia Populations
As we observed a stable survival of V. cholerae over 2 weeks in infected tilapia compared to the uninfected controls, a question emerged whether the bacteria could be transmitted from infected to naïve tilapia. After 24 h of cohabitation with infected tilapia (without feeding), average V. cholerae counts of 105 cfu/intestine were observed in naïve fish that were initially tested free of V. cholerae. The concentration of V. cholerae found in naïve tilapia were similar for all four test strains (p > 0.05, Figure 6).
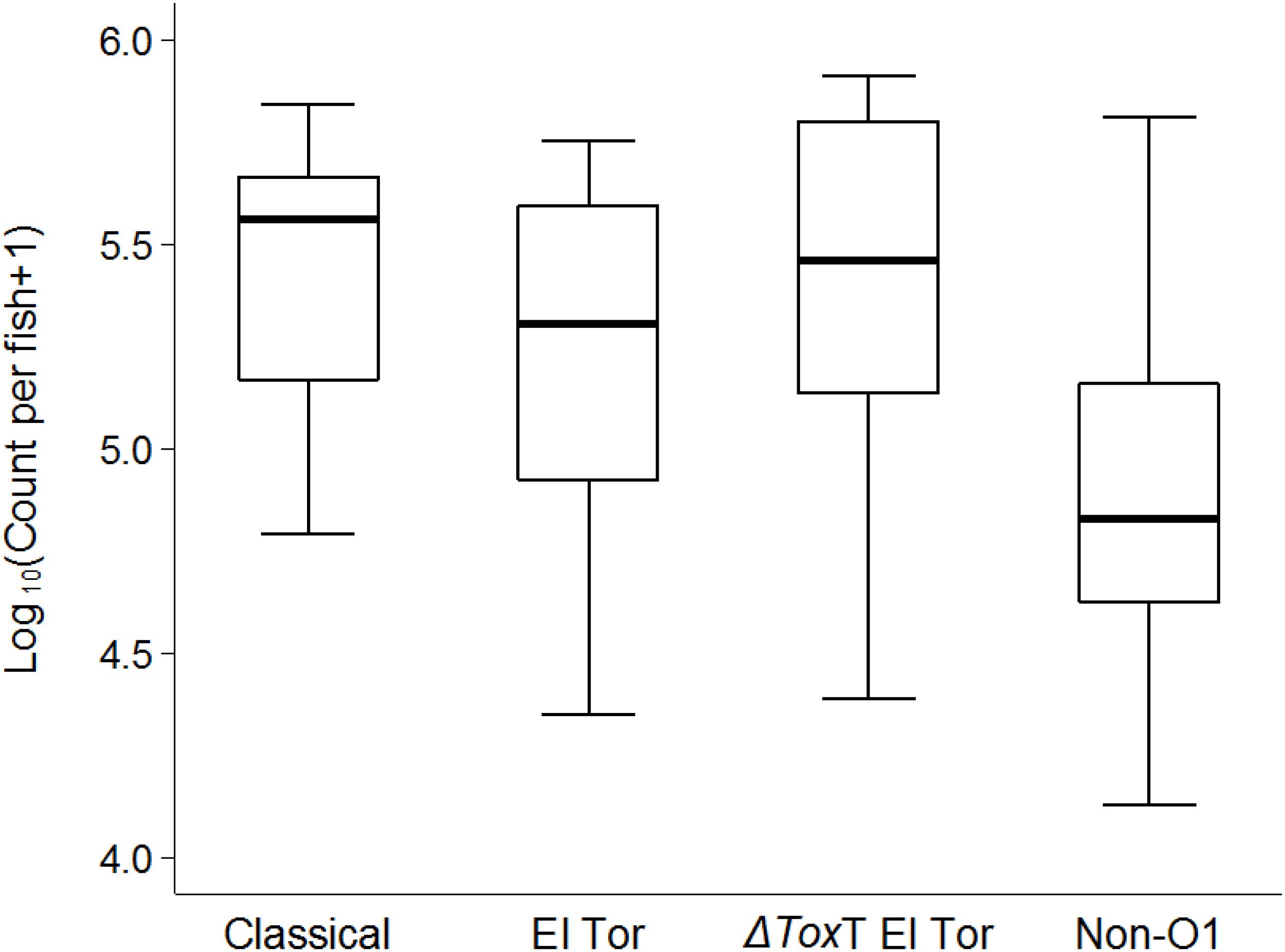
Figure 6. Transmission of V. cholerae from infected to naïve tilapia via excretions 24 h post cohabitation using box and whiskers plot. The thick line is the median and the box is the interquartile range and whiskers the range. Counts are from the two repetitions.
Discussion
Experimental exposure model studies with V. cholerae in zebrafish suggest that V. cholerae can colonize fish guts and be transmitted among zebrafish populations (Runft et al., 2014; Mitchell et al., 2017). Our results in tilapia are consistent with the observations in zebrafish. High counts of the different strains of V. cholerae in tilapia intestines were observed 24 h after exposure as a sign of colonization. The sharp drop in V. cholerae counts in fish and water between 24 and 48 h post inoculum in all three colonization experiments could be due to natural shock of the V. cholerae bacterial cells attributable to changed environments, which may lead to a dormant state also known as viable but not culturable (Kamruzzaman et al., 2010). However, after day 2 to 3 post infection, concentrations of V. cholerae remained more or less stable, demonstrating adaptation and survival in the tilapia. Non-toxigenic strains, notably ΔtoxT of V. cholerae O1 El Tor and the environmental non-O1 strain, were also able to colonize fish and persist over time. Intestinal colonization of V. cholerae in humans requires production of the cholera TCP, whose main virulence transcription activator is ToxT (Faruque et al., 1998; Sanchez and Holmgren, 2011). The fact that toxT mutants and non-O1 strains of V. cholerae were found in tilapia intestine over time is in accordance with observations in zebrafish (Runft et al., 2014) and suggests that TCP is not essential for V. cholerae colonization of fish. Moreover, studies have discovered a novel flagella-mediated cytotoxin MakA which is proposed to be involved in V. cholerae intestinal colonization in zebrafish (Dongre et al., 2018). V. cholerae strains used in our study are wild types and possess a flagellum, so the secretion of MakA protein associated with flagella could be involved in the colonization of tilapia.
In Experiment 1, where fish were kept in stagnant water and fed live brine shrimp (A. salina) free of V. cholerae, there were high V. cholerae counts in intestines and water together with high OD values. Since the infection dose (5 × 107) was the same in all experiments, the higher concentrations of V. cholerae observed in Experiment 1 compared to findings in Experiments 2 and 3, in which water was changed daily, were thought to be associated with the continuous provision of brine shrimp, as their presence could enhance attachment and multiplication of V. cholerae in the fish intestine. The ADP-ribosylating cholix toxin in V. cholerae has been shown to play an important role in the survival of the organism in the aquatic environment and facilitates its attachment to crustaceans, notably the brine shrimp. Moreover, V. cholerae are known for their ability to attach to chitin exoskeletons of shrimp, copepods and other crustaceans that serve as substrate for their survival and multiplication (Huq et al., 1983; Tamplin et al., 1990; Hood and Winter, 1997; Patra and Mohamed, 2003). However, results in Experiment 3 where water was replaced on a daily basis and fish were fed with the same live feed rejected the hypothesis that brine shrimp could enhance colonization, because the change of water was found to be the only significant variable associated with V. cholerae concentrations.
Comparison of the three experiments shows that irrespective of feeding and water exchange, tilapia were colonized by environmental non-O1 V. cholerae as well as V. cholerae seventh pandemic El Tor (7PET) strains and were isolated beyond 2 weeks. This strongly suggests that in natural aquatic environments, where fish can live in stagnant or running water with presence of various feed items, tilapia may constitute a reservoir of toxigenic, and non-toxigenic strains of V. cholerae. It is worth noticing that the concentration of V. cholerae, i.e., 107 cfu per ml water in the beakers, was higher as compared to concentrations that can be expected in natural aquatic environments during non-cholera outbreak periods (Senderovich et al., 2010). In contrast, there are little data available about the actual concentration of V. cholerae O1 in such environments. In a previous study, we did report that tilapia were able to live in raw sewage and were found to carry V. cholerae O1 (Hounmanou et al., 2016). Moreover, tilapia in our experiment were infected by immersion and we therefore used a similar dose of 107 cfu per ml as in the zebrafish experiments (Runft et al., 2014; Mitchell et al., 2017) to ensure an uptake of the test strains. It is not known how many V. cholerae cells fish ingest in natural water systems. The high inoculum of V. cholerae used in this study could enhance colonization and may represent a limitation of our study. Nevertheless, even at low concentrations of V. cholerae similar to natural conditions, like from day 7 in all our experiments (103 CFU/intestine), tilapia remained colonized, with the O1 El Tor biotype maintaining the highest numbers. Furthermore, the transmission experiment with naïve tilapia placed in beakers with tilapia carrying V. cholerae in the intestine showed that when the naïve tilapia were exposed to about 105.5 cfu per mL water they became infected and had similar bacterial concentrations in their intestine after 24 h (Figure 6). It should be noted that we did not determine the V. cholerae concentration in the water in the transmission experiment (Figure 6) and that the stated expected concentration in the water of 105.5 cfu per mL is based on values found at day 2 in Experiment 2 (Figure 3B), where infected tilapia were washed and transferred to beakers with fresh water. Overall, the results indicate that irrespective of the initial concentration of V. cholerae in water, tilapia can become colonized with V. cholerae and act as a reservoir for transmission and long-term survival.
Findings from Experiments 2 and 3 show that the concentration in water after 1 week was around 104 to 103 cfu/mL similar to what has been reported in the natural environmental waters (Senderovich et al., 2010). One week later, the concentration of V. cholerae was around 103 to 102 cfu/mL. Despite a low concentration of V. cholerae in water seen during the last 7 days and the continuous daily water renewal, the OD values of the water in the beakers remained higher than the OD values in the beakers of the control fish. This suggests that the increased OD water values were due to excreted material from the tilapia, i.e., stress-related discharges, probably due to the initial high concentration of V. cholerae in the water. V. cholerae are able to colonize tilapia over an extended time span, multiply in the intestine, and be excreted into the aquatic environment but a high initial concentration could be stressful for the fish. We therefore suggest further studies to explore lower infection doses administered possibly by gavage to ensure sufficient uptake. Moreover, even when the Classical biotype of V. cholerae was no longer detectable in tilapia intestines and in water, the optical density of water in those beakers remained higher than in the control beakers. This is consitent with observations in zebrafish that heat-killed V. cholerae still induced mild diarrhea in zebrafish (Mitchell et al., 2017). This further suggests that the discharges and water turbidity provoked by V. cholerae in tilapia is neither due to cholera toxin genes nor to viability or biotype of V. cholerae but probably caused by the stress generated by the high initial infection dose of V. cholerae. Furthermore, the flagella-mediated secretion of MakA cytotoxin was suggested as a source of toxicity and death in zebrafish infected with wild-type V. cholerae (Dongre et al., 2018).
The absence of Classical biotype V. cholerae after 1 week and the persistence of the seventh cholera pandemic biotype El Tor V. cholerae O1 strains is similar to findings in zebrafish (Runft et al., 2014) and consistent with the rare isolation or extinction of the Classical biotype in the ongoing cholera pandemic (Echenberg, 2011; Weill et al., 2017). The El Tor biotype and the non-O1 serogroup strains seem more fit in the fish gut (aquatic environment) than the Classical O1 biotype strains which may explain the increasing recovery of these strains in most contemporary environmental studies (Hounmanou et al., 2016; Bwire et al., 2018). The persistence of V. cholerae O1 biotype El Tor in tilapia and water is of public health relevance as it provides evidence of environmental survival of the current pandemic El Tor biotype strains where they can emerge from and cause epidemics. Furthermore, in vitro and in vivo experiments have demonstrated that in the presence of glucose, V. cholerae of the Classical biotype generates organic acids that inhibit their growth, while the growth of El Tor biotype is enhanced due to their ability to produce acetoin (2,3-butanediol), a neutral fermentation end product (Yoon and Mekalanos, 2006; Sengupta et al., 2017; Nag et al., 2018). It could therefore, be that carbohydrates present in the water of the beakers, e.g., droppings from the tilapia, did facilitate glucose metabolism of V. cholerae, resulting in loss of viability of the Classical biotype and survival of El Tor biotype. Such unfavorable conditions may also cause the strains, especially the Classical biotypes, to enter a dormant state, known as viable but not culturable (VBNC) (Bari et al., 2013; Xu et al., 2018), which may be one explanation as to why the Classical biotype strains were not detected after 1 week of the experiments.
The concentrations of V. cholerae in tilapia intestines correlated with those in the water. As the number of fish in the beakers decreased over time, V. cholerae counts in water decreased. This strong correlation (p < 0.0001) between counts in water and fish indicates that V. cholerae have reduced ability to multiply in clean water (Colwell et al., 2003) as compared to the intestine and is continuously excreted by the fish host. Thus, the main significant predictor of V. cholerae concentrations in water was the concentration of V. cholerae in the fish intestine. Moreover, the observed similar V. cholerae counts in water and in fish intestines in all experiments is likely attributable to the fact that the beakers used as aquarium provided a restrained space to the fish in a low volume of water (2 L). When V. cholerae-free tilapia cohabitated with infected tilapia in sterile water overnight, their intestine was also colonized, providing evidence of transmission. The transmission was not strain-dependent, as all the four test strains had similar transmission rates, demonstrating that toxigenic and nontoxigenic strains can equally be transmitted between tilapia populations and that V. cholerae can survive and amplify in tilapia, and also be disseminated from tilapia. Our findings are similar to reports in zebrafish models (Runft et al., 2014). Furthermore, the fact that naïve tilapia became infected in the transmission experiments substanciates again that tilapia were effectively and stably colonized by V. cholerae that then was disseminated between fish populations. Although only intestines were studied in our experiments, other organs like gills and skin could also play a significant role in the transmission process as they would provide nutrients favoring colonization of V. cholerae. However, previous studies have demonstrated that intestines are the main factors involved in colonization and transmission of V. cholerae in fish (Runft et al., 2014) and fish were washed twice with sterile saline by rubbing before the transmission experiment to remove external bacteria. The potential epidemiological importance of this study is that during a cholera outbreak, while all efforts are deployed toward containing the epidemics at human level, fish may serve as vehicle of dissemination of the bacteria in other areas, which may subsequently be hit by the same outbreak even when human patients are quarantined in the initial outbreak settings. The possible role of fish in transmitting V. cholerae is further supported by the findings that fish eating birds such as Great cormorants have been found to carry and disperse V. cholerae in space and time as they feed on infected fish and get colonized by V. cholerae (Laviad -Shitrit et al., 2017).
In summary, we have demonstrated that toxigenic V. cholerae O1 biotype El Tor and non-toxigenic strains of V. cholerae colonized the intestines of tilapia and were transmitted to naïve tilapia. This study provides answers to a hypothesis posed in a previous study (Halpern and Izhaki, 2017) that fish can be colonized by Vibrio cholerae and subsequently horizontally transfer V. cholerae to other fish within the same species, and probably to other fish species. This suggests that V. cholerae colonizes and persists in fish, and is transmitted between fish in aquatic environments, which may influence the epidemiology of cholera. Tilapia and other fish are potential reservoir hosts involved in the survival, excretion and transmission of V. cholerae in time and space. Cholera surveillance strategies may need to be updated accordingly including analysis of fish for the presence of V. cholerae O1 in aquatic environments. We furthermore suggest further studies to confirm the role of tilapia as an environmental reservoir host of V. cholerae O1 biotype El Tor using lower infection doses administered possibly by gavage to ensure sufficient uptake and limit stress to the fish.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without under reservation, to any qualified researcher.
Ethics Statement
The present study was approved by the ethical review board through the ethical clearance certificate for conducting animal related research in Tanzania with ethical approval number SUA/CVMBS/018/07 (File submitted to editorial). Euthanasia of fish during the study and disposal of waste materials were performed according to instructions of the above-mentioned ethical approval.
Author Contributions
YH designed the study, carried out the experiments in the laboratory, analyzed the results, and drafted the manuscript. RM supervised the experiments and critically reviewed and edited the original draft of the manuscript. TD participated in critical reviewing and editing of the manuscript. HM contributed to statistical analysis and data interpretation. JW provided the test strains and critical revision of the manuscript. JO and AD conceived the study and reviewed the manuscript. AD validated the data and supervised the study. All authors read and approved the final manuscript.
Funding
The study was supported by the Danish International Development Assistance (Danida) – funded project “Innovations and Markets for Lake Victoria Fisheries” (IMLAF), DFC File No. 14-P01-TAN and the International Foundation for Sciences (IFS), Stockholm, Sweden, through the grant A6100-1 to YH. JW was funded by NIH grant R01AI127390 from the United States Public Health Service.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors extend their gratitude to the funders.
References
Ajayi, A., and Smith, S. I. (2018). Recurrent cholera epidemics in Africa: which way forward? A literature review. Infection doi: 10.1007/s15010-018-1186-5 [Epub ahead of print].
Bari, S. M. N., Roky, M. K., Mohiuddin, M., Kamruzzaman, M., Mekalanos, J. J., and Faruque, S. M. (2013). Quorum-sensing autoinducers resuscitate dormant Vibrio cholerae in environmental water samples. Proc. Natl. Acad. Sci. U.S.A. 110, 9926–9931. doi: 10.1073/pnas.1307697110
Bwire, G., Debes, A. K., Orach, C. G., Kagirita, A., Ram, M., Komakech, H., et al. (2018). Environmental surveillance of Vibrio cholerae O1/O139 in the five african great lakes and other major surface water sources in Uganda. Front. Microbiol. 9:1560. doi: 10.3389/fmicb.2018.1560
Colwell, R. R., Huq, A., Islam, M. S., Aziz, K. M. A., Yunus, M., Khan, N. H., et al. (2003). Reduction of cholera in Bangladeshi villages by simple filtration. Proc. Natl. Acad. Sci. U.S.A. 100, 1051–1055. doi: 10.1073/pnas.0237386100
Dalsgaard, A., Serichantalergs, O., Forslund, A., Lin, W., Mekalanos, J., Mintz, E., et al. (2001). Clinical and environmental isolates of Vibrio cholerae serogroup O141 Carry the CTX phage and the genes encoding the toxin-coregulated pili. J. Clin. Microbiol. 39, 4086–4092. doi: 10.1128/JCM.39.11.4086-4092.2001
Dongre, M., Singh, B., Aung, K. M., Larsson, P., Miftakhova, R., Persson, K., et al. (2018). Flagella-mediated secretion of a novel Vibrio cholerae cytotoxin affecting both vertebrate and invertebrate hosts. Commun. Biol. 1:59. doi: 10.1038/s42003-018-0065-z
Echenberg, M. J. (2011). Africa in the Time of Cholera: a History of Pandemics from 1817 to the Present. New York, NY: Cambridge University Press.
Faruque, S. M., Albert, M. J., and Mekalanos, J. J. (1998). Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62, 1301–1314.
Halpern, M., Broza, Y. B., Mittler, S., Arakawa, E., and Broza, M. (2004). Chironomid egg masses as a natural reservoir of Vibrio cholerae Non-O1 and Non-O139 in freshwater habitats. Microbial. Ecol. 47, 341–349. doi: 10.1007/s00248-003-2007-2006
Halpern, M., and Izhaki, I. (2017). Fish as hosts of Vibrio cholerae. Front. Microbiol. 8:282. doi: 10.3389/fmicb.2017.00282
Hood, M. A., and Winter, P. A. (1997). Attachment of Vibrio cholerae under various environmental conditions and to selected substrates. FEMS Microbiol. Ecol. 22, 215–223. doi: 10.1111/j.1574-6941.1997.tb00373.x
Hossain, Z. Z., Farhana, I., Tulsiani, S. M., Begum, A., and Jensen, P. K. M. (2018). Transmission and toxigenic potential of Vibrio cholerae in Hilsha Fish (Tenualosa ilisha) for human consumption in Bangladesh. Front. Microbiol. 9:222. doi: 10.3389/fmicb.2018.00222
Hounmanou, Y. M. G., Mdegela, R. H., Dougnon, T. V., Mhongole, O. J., Mayila, E. S., Malakalinga, J., et al. (2016). Toxigenic Vibrio cholerae O1 in vegetables and fish raised in wastewater irrigated fields and stabilization ponds during a non-cholera outbreak period in Morogoro, Tanzania: an environmental health study. BMC Res. Notes 9:466. doi: 10.1186/s13104-016-2283-2280
Huq, A., Small, E. B., West, P. A., Huq, M. I., Rahman, R., and Colwell, R. R. (1983). Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45, 275–283.
Islam, M. S., Islam, M. S., Mahmud, Z. H., Cairncross, S., Clemens, J. D., and Collins, A. E. (2015). Role of phytoplankton in maintaining endemicity and seasonality of cholera in Bangladesh. Trans. R. Soc. Trop. Med. Hyg. 109, 572–578. doi: 10.1093/trstmh/trv057
Kamruzzaman, M., Udden, S. M. N., Cameron, D. E., Calderwood, S. B., Nair, G. B., Mekalanos, J. J., et al. (2010). Quorum-regulated biofilms enhance the development of conditionally viable, environmental Vibrio cholerae. Proc. Natl. Acad. Sci. 107, 1588–1593. doi: 10.1073/pnas.0913404107
Laviad -Shitrit, S., Lev-Ari, T., Katzir, G., Sharaby, Y., Izhaki, I., and Halpern, M. (2017). Great cormorants (Phalacrocorax carbo) as potential vectors for the dispersal of Vibrio cholerae. Sci. Rep. 7:7973. doi: 10.1038/s41598-017-08434-8438
Mengel, M. A., Delrieu, I., Heyerdahl, L., and Gessner, B. D. (2014). “Cholera outbreaks in africa,” in Cholera Outbreaks, eds G. B. Nair and Y. Takeda (Berlin: Springer), 117–144. doi: 10.1007/82_2014_369
Mitchell, K. C., Breen, P., Britton, S., Neely, M. N., and Withey, J. H. (2017). Quantifying Vibrio cholerae enterotoxicity in a Zebrafish infection model. Appl. Environ. Microbiol. 83, e783–e717. doi: 10.1128/AEM.00783-717
Nag, D., Breen, P., Raychaudhuri, S., and Withey, J. H. (2018). Glucose metabolism by Escherichia coli inhibits Vibrio cholerae intestinal colonization of Zebrafish. Infect. Immun. 86, e486–e418. doi: 10.1128/IAI.00486-418
Nandi, B., Nandy, R. K., Mukhopadhyay, S., Nair, G. B., Shimada, T., and Ghose, A. C. (2000). Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J. Clin. Microbiol. 38, 4145–4151.
Nkoko, D., Giraudoux, P., Plisnier, P.-D., Tinda, A., Piarroux, M., Sudre, B., et al. (2011). Dynamics of cholera outbreaks in great lakes region of Africa, 1978–2008. Emerg. Infect. Dis. 17, 2026–2034. doi: 10.3201/eid1711.110170
Nyambuli, S., Mhongole, O. J., Katakweba, A. A., Dalsgaard, A., and Mdegela, R. H. (2018). Prevalence, pathogenic markers and antibiotic susceptibility of Vibrio cholerae in Sardines, Water and phytoplankton in lake Tanganyika, Tanzania. Int. J. Agri., Forest. Fish. 6:29.
Patra, S. K., and Mohamed, K. S. (2003). Enrichment of Artemia nauplii with the probiotic yeast Saccharomyces boulardii and its resistance against a pathogenic Vibrio. Aqua. Int. 11, 505–514. doi: 10.1023/B:AQUI.0000004193.40039.54
Plisnier, P.-D., Poncelet, N., Cocquyt, C., De Boeck, H., Bompangue, D., Naithani, J., et al. (2015). Cholera Outbreaks at Lake Tanganyika Induced by Climate Change?. Brussels: Belgian Science Policy.
Rabia, A., Wambura, P., Misinzo, G., Kimera, S., Mdegela, R., Mzula, A., et al. (2017). Molecular Epidemiology of Vibrio cholerae recovered from sewage drains, captured Fish and humans in 2015/16 cholera outbreak in Zanzibar, Tanzania. J. Adv. Microbiol. 5, 1–11. doi: 10.9734/JAMB/2017/36036
Reyburn, R., Kim, D. R., Emch, M., Khatib, A., von Seidlein, L., and Ali, M. (2011). Climate variability and the outbreaks of cholera in Zanzibar, East Africa: a time series analysis. Am. J. Trop. Med. Hyg. 84, 862–869. doi: 10.4269/ajtmh.2011.10-0277
Runft, D. L., Mitchell, K. C., Abuaita, B. H., Allen, J. P., Bajer, S., Ginsburg, K., et al. (2014). Zebrafish as a natural host Model for Vibrio cholerae colonization and transmission. Appl. Environ. Microbiol. 80, 1710–1717. doi: 10.1128/AEM.03580-3513
Safa, A., Bhuyian, N. A., Nusrin, S., Ansaruzzaman, M., Alam, M., Hamabata, T., et al. (2006). Genetic characteristics of Matlab variants of Vibrio cholerae O1 that are hybrids between classical and El Tor biotypes. J. Med. Microbiol. 55, 1563–1569. doi: 10.1099/jmm.0.46689-46680
Sanchez, J., and Holmgren, J. (2011). Cholera toxin - a foe & a friend. Indian J. Med. Res. 133, 153–163.
Senderovich, Y., Izhaki, I., and Halpern, M. (2010). Fish as reservoirs and vectors of Vibrio cholerae. PLoS One 5:e8607. doi: 10.1371/journal.pone.0008607
Sengupta, C., Ekka, M., Arora, S., Dhaware, P. D., Chowdhury, R., and Raychaudhuri, S. (2017). Cross feeding of glucose metabolism byproducts of Escherichia coli human gut isolates and probiotic strains affect survival of Vibrio cholerae. Gut Pathog. 9:3. doi: 10.1186/s13099-016-0153-x
Tamplin, M. L., Gauzens, A. L., Huq, A., Sack, D. A., and Colwell, R. R. (1990). Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 56, 1977–1980.
Urassa, W., Mhando, Y., Mhalu, F., and Mgonja, S. (2009). Antimicrobial susceptibility pattern of Vibrio cholerae 01 strains during two cholera outbreaks in Dar Es Salaam, Tanzania. East Afr. Med. J. 77, 350–353. doi: 10.4314/eamj.v77i7.46661
Weill, F.-X., Domman, D., Njamkepo, E., Tarr, C., Rauzier, J., Fawal, N., et al. (2017). Genomic history of the seventh pandemic of cholera in Africa. Science 358, 785–789. doi: 10.1126/science.aad5901
Xu, T., Cao, H., Zhu, W., Wang, M., Du, Y., Yin, Z., et al. (2018). RNA-seq-based monitoring of gene expression changes of viable but non-culturable state of Vibrio cholerae induced by cold seawater. Environ. Microbiol. Rep. 10, 594–604. doi: 10.1111/1758-2229.12685
Keywords: Vibrio cholerae, tilapia, cholera transmission, microbial ecology, reservoirs
Citation: Hounmanou YMG, Mdegela RH, Dougnon TV, Madsen H, Withey JH, Olsen JE and Dalsgaard A (2019) Tilapia (Oreochromis niloticus) as a Putative Reservoir Host for Survival and Transmission of Vibrio cholerae O1 Biotype El Tor in the Aquatic Environment. Front. Microbiol. 10:1215. doi: 10.3389/fmicb.2019.01215
Received: 20 February 2019; Accepted: 15 May 2019;
Published: 31 May 2019.
Edited by:
Francesca Leoni, Istituto Zooprofilattico Sperimentale dell’Umbria e delle Marche, ItalyReviewed by:
Diane McDougald, University of Technology Sydney, AustraliaThandavarayan Ramamurthy, Translational Health Science and Technology Institute, India
Taviani Elisa, Universidade Eduardo Mondlane, Mozambique
Copyright © 2019 Hounmanou, Mdegela, Dougnon, Madsen, Withey, Olsen and Dalsgaard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaovi Mahuton Gildas Hounmanou, Z2lsQHN1bmQua3UuZGs=; Z2lsbWFodUB5YWhvby5mcg==
 Yaovi Mahuton Gildas Hounmanou
Yaovi Mahuton Gildas Hounmanou Robinson H. Mdegela2
Robinson H. Mdegela2 Tamegnon Victorien Dougnon
Tamegnon Victorien Dougnon Jeffrey H. Withey
Jeffrey H. Withey John E. Olsen
John E. Olsen Anders Dalsgaard
Anders Dalsgaard