- State Key Laboratory of Microbial Technology, Shandong University, Qingdao, China
Acidithiobacillaceae, an important family of acidophilic and chemoautotrophic sulfur or iron oxidizers, participate in geobiochemical circulation of the elements and drive the release of heavy metals in mining associated habitats. Because of their environmental adaptability and energy metabolic systems, Acidithiobacillus spp. have become the dominant bacteria used in bioleaching for heavy metal recovery. Flagella-driven motility is associated with bacterial chemotaxis and bacterial responses to environmental stimuli. However, little is known about how the flagellum of Acidithiobacillus spp. is regulated and how the flagellum affects the growth of these chemoautotrophic bacteria. In this study, we analyzed the flagellar gene clusters in Acidithiobacillus strains and uncovered the close relationship between flagella and the sulfur-oxidizing systems (Sox system). The σ28 gene (rpoF) knockout and overexpression strains of Acidithiobacillus caldus were constructed. Scanning electron microscopy shows that A. caldus ΔrpoF cells lacked flagella, indicating the essential role of RpoF in regulating flagella synthesis in these chemoautotrophic bacteria. Motility analysis suggests that the deletion of rpoF resulted in the reduction of swarming capability, while this capability was enhanced in the rpoF overexpression strain. Both static cultivation and low concentration of energy substrates (elemental sulfur or tetrathionate) led to weak growth of A. caldus ΔrpoF cells. The deletion of rpoF promoted bacterial attachment to the surface of elemental sulfur in static cultivation. The absence of RpoF caused an obvious change in transcription profile, including genes in flagellar cluster and those involved in biofilm formation. These results provide an understanding on the regulation of flagellar hierarchy and the flagellar function in these sulfur or iron oxidizers.
Introduction
Acidithiobacillus spp. are acidophilic bacteria and widely used in the bioleaching industry. Acidithiobacillus species formerly belonged to the genus “Thiobacillus” and have been reclassified into the new proteobacterial class Acidithiobacillia (Kelly and Wood, 2000; Hudson et al., 2014). All members in Acidithiobacillus have remarkable capability of oxidizing various reduced inorganic sulfur compounds (RISCs) to obtain electrons for autotrophic growth, and some of them also have ferrous iron oxidation ability (Harrison, 1984). The isolated bacteria in Acidithiobacillus have been classified into seven species: Acidithiobacillus thiooxidans (A. thiooxidans), Acidithiobacillus ferrooxidans (A. ferrooxidans), Acidithiobacillus caldus (A. caldus), Acidithiobacillus albertensis (A. albertensis), Acidithiobacillus ferridurans (A. ferridurans), Acidithiobacillus ferriphilus (A. ferriphilus), and Acidithiobacillus ferrivorans (A. ferrivorans) (Waksman and Joffe, 1922; Temple and Colmer, 1951; Bryant et al., 1983; Hallberg and Lindstrom, 1994; Falagan and Johnson, 2016; Nunez et al., 2017). Acidithiobacillus strains widely exist in iron/sulfur-containing habitats on land or in the sea (Sharmin et al., 2016; Nunez et al., 2017; Kamimura et al., 2018), such as soil (Waksman and Joffe, 1922; Deb et al., 2004), sediments (Nealson, 1997; Otte et al., 2018), hot springs (Arce et al., 2019), iron-sulfur mineral deposits (Johnson et al., 2001; Rowe et al., 2007; Song et al., 2011) and acid mine drainage (AMD) (Colmer and Hinkle, 1947). Due to the unique characteristics in energy metabolism and environmental adaptation, Acidithiobacillus strains have been widely used in biohydrometallurgy for non-ferrous metals production (copper, gold, zinc, etc.) (Rawlings, 2005; Valdés et al., 2008a; Donati et al., 2016; Pollmann et al., 2018).
Flagella are important cell appendages for bacterial motility and biofilm formation (Pratt and Kolter, 1998; Lemon et al., 2007; Friedlander et al., 2013). In biomining, the attachment of bacterial cells on mineral surface is a prerequisite for the acquisition of energy-substrates, which also contributes significantly to mineral dissolution process (Africa et al., 2013). The biofilm formation of Acidithiobacillus spp. promotes cell attachment and facilitates the oxidization of sulfur or iron deposited in ores (Harneit et al., 2006; Lara et al., 2013; Bellenberg et al., 2014), resulting in the extremely low pH, high concentration of heavy metals and temperature change in the habitat. The flagella-mediated motility and chemotaxis are vital for cells to respond to environmental stimuli (pH, temperature, osmolarity, etc.) and to find nutrients for growth (e.g., energy substrates and inorganic salts) (Kiiyukia et al., 1993; Li et al., 1993; Kunin et al., 1995; Walker et al., 1999). Flagella were reported in several Acidithiobacillus species, such as A. thiooxidans (Doetsch et al., 1967), A. caldus (Hallberg and Lindstrom, 1994), and A. albertensis (Bryant et al., 1983; Xia et al., 2007). Different numbers and forms of flagella were observed in different species of Acidithiobacillus: A. thiooxidans and A. caldus cells habor monotrichous flagella while A. albertensis possesses lophotrichous flagella (Bryant et al., 1983; Hallberg and Lindstrom, 1994; Xia et al., 2007). However, there is limited data on flagellar molecular biology of Acidithiobacillus, which limits our understanding on the biofilm formation and flagella-mediated motility in these chemoautotrophic sulfur or iron oxidizers.
Flagella have been extensively studied at gene and protein levels in bacteria other than Acidithiobacillus spp., such as Escherichia coli (Kadoya et al., 2015), Salmonella typhimurium (Kutsukake et al., 1990; Koirala et al., 2014), Caulobacter crescentus (Bryan et al., 1990), Pseudomonas aeruginosa (Totten and Lory, 1990; Jyot et al., 2007), and Vibrio cholerae (Klose and Mekalanos, 1998; McCarter, 2001). The assembly and function of bacterial flagella generally require dozens of genes that encode structural subunits, regulatory proteins, motor force generators and the chemosensory system (Chen et al., 2011; Fitzgerald et al., 2014; Zhao et al., 2014; Osterman et al., 2015). These genes are organized into a large complex cluster in some bacteria, and are regulated hierarchically at transcriptional level via diverse regulatory systems (Dasgupta et al., 2000; Jacobi et al., 2004; Schulz et al., 2012; Galeva et al., 2014; Song et al., 2016). RpoF (σ28 or FliA) is one of the σ factors that regulates the synthesis of filaments and attracts wide attentions in various bacterial species, including Legionella pneumophila (Schulz et al., 2012), E. coli (Kundu et al., 1997), P. aeruginosa (Starnbach and Lory, 1992). However, the function of RpoF in Acidithiobacillus spp. is still not well understand.
This research reported the distribution of flagellar gene clusters in different Acidithiobacillus species, studied the function of the flagellum in A. caldus responding to unfavorable environmental conditions, and investigated the influence of RpoF on A. caldus transcription profile.
Materials and Methods
Bacteria, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study were listed in Table 1. E. coli was cultivated in Luria Bertani (LB) medium with a rotation speed of 200 rpm at 37°C or plated on 1.2% agar plates (Sambrook et al., 2001). A. caldus MTH-04, a strain isolated from hot spring in Tengchong, was grown in modified liquid Starkey-S0/K2S4O6 media or on solid Starkey-Na2S2O3 plates with 1% agar at 40°C (Wang et al., 2016). Antibiotics added to LB medium include chloramphenicol (34 mg/mL), streptomycin (100 mg/mL) and kanamycin (100 mg/mL); for A. caldus, the concentrations of antibiotics were doubled in Starkey media. The cultivation standards of A. caldus MTH-04 were described earlier (Wang et al., 2016). Simply, the single colony on the solid plate was inoculated into 30 mL Starkey-S0 liquid medium and cultivated to stationary phase. Then, the culture was transferred into 150 mL Starkey-S0 liquid medium. When the culture reaches to stationary phase, cells were collected by centrifugation, and diluted with Starkey liquid medium to a final concentration of OD600 = 1.0. An aliquot (1 mL) of the cell solution was inoculated into 150 mL Starkey liquid medium for further experiments. Cell density was measured at OD600 after removal of elemental sulfur by centrifugation at 400 × g for 5 min (Yu et al., 2014; Wang et al., 2016).
Molecular Biological Techniques
Taq DNA polymerase, T4 DNA ligase and restriction enzymes (Xba I, Kpn I, Nhe I and Hind III) were purchased from Takara (Kyoto, Japan). Kits for genomic DNA exaction (E.Z.N.A.® Bacterial DNA Kit), plasmid extraction (E.Z.N.A.® Plasmid Mini Kit I) and DNA purification (E.Z.N.A.® Cycle Pure Kit) were purchased from Omega Bio-tek (Georgia, United States). Chemicals and reagents [Na2S2O3, K2S4O6, agar, (NH4)2SO4, KH2PO4, CaCl2, MgSO4, FeSO4, etc.] were purchased from Sangon (Shanghai, China) and Sinopharm (Shanghai, China). Primer synthesis and DNA sequencing were accomplished by GENEWIZ (Suzhou, China). All primers used in this study were listed in Supplementary Table S1. The genomic sequences and all sequences of flagella and chemotaxis related genes were downloaded from the public National Center for Biotechnology Information (NCBI) website https://www.ncbi.nlm.nih.gov/ (Supplementary Table S2). BlastP and PsiBlast (Altschul et al., 1997) were used to further characterize candidate genes and their predicted protein products. Syntenic regions were displayed and analyzed with modification using Genomeviz (Ghai and Chakraborty, 2007).
Construction of A. caldus rpoF Mutants
The A. caldus MTH-04 ΔrpoF strain was constructed using the gene markerless knockout technique (Wang et al., 2016). The upstream flanking arm (U, 1058 bp) of the rpoF gene was amplified by PCR with primer set rpoF-U-F-Xba I and rpoF-U-R-Kpn I. The primer set rpoF-D-F-Kpn I and rpoF-D-R-Nhe I was used to amplify the downstream flanking arm (D, 1369 bp). Fragments U and D were digested with restriction enzymes Xba I/Kpn I and Kpn I/Nhe I, respectively. Then, these two treated fragments were ligated with the Xba I/Nhe I digested plasmid pSDUDI to obtain the suicide plasmid pSDUDI-rpoF-U-D. The resulting plasmid was verified by restriction enzyme digestion (Xba I and Nhe I) and DNA sequencing analysis before transferring into E. coli SM10 competent cells by heat shock transformation. Then, the suicide plasmids were conjugally transferred by filter mating from E. coli SM10 to A. caldus MTH-04 (Liu et al., 2007). The plasmid pSDUDI-rpoF-U-D was integrated into A. caldus MTH-04 chromosome via the homologous recombination, the single crossover mutants were selected on Starkey-Na2S2O3 plate containing 200 mg/mL kanamycin and determined by PCR with primer set rpoF-out-F/R. After that, the plasmid pSDU1-I-Sce I was conjugated into the single crossovers of A. caldus, inducing the second recombination and generating either the rpoF knockout mutants or wildtype cells. Primers P1 F/R, P2 F/R, and P3 F/R were designed to identify A. caldus ΔrpoF strains. The elimination of plasmid pSDU1-I-Sce I in the rpoF knockout strains was achieved by continuous passages (Wang et al., 2016).
The construction of A. caldus rpoF overexpression and complementation strains were done as follows. The fragment (rpoF) was amplified from A. caldus MTH-04 genome with primer set rpoF-F (containing 59-bp homologous sequence of the promoter of tetH gene) and rpoF-Hind III-R. The tetH promoter fragment (PtetH) was amplified from A. caldus MTH-04 chromosome with primer set PtetH-Kpn I -F and PtetH-R. The two fragments PtetH and rpoF were ligated by fusion PCR. The fused fragment PtetH-rpoF was digested with Hind III and Kpn I, and inserted into plasmid pJRD215 linearizing with the same restriction enzymes, forming the plasmid pJRD215-PtetH-rpoF. The generated plasmid was verified by sequencing and transformed into A. caldus wildtype and ΔrpoF, generating the rpoF overexpression strain WT(rpoF) and the rpoF complementation strain ΔrpoF(rpoF), respectively. The control plasmid pJRD215 was also conjugated into A. caldus wildtype and ΔrpoF to construct the control strains WT(215) and ΔrpoF(215), respectively.
Extraction of Total RNA
For RNA extraction, experimental strains were cultivated to mid-log phase (OD600 0.09–0.13) without agitation in 150 mL liquid Starkey-S0 media with addition of 0.4 g elemental sulfur. Cells were harvested by centrifugation (3000 × g, 4°C, 5 min). RNA-protect Bacteria Reagent (Qiagen) was immediately added, and pellets were stored at -80°C for backup. Total RNA was extracted with Trizol® Reagent (Invitrogen) according to improved protocol’s specifications with a little modification (reaction time for each step was halved and the melting RNA was not performed). The integrity of each RNA sample was assessed by RNA formaldehyde degeneration electrophoresis in a 1.5% agarose gel with 90 mM Tris-boric acid containing 2 mM EDTA (TBE). RNA concentration and purity were determined using the NanoPhotometer® spectrophotometer by measuring A260 and A260/A280 ratio. Three independent experiments were set for the purification of total RNA.
RNA-Seq and Data Analysis
The low cell yield of A. caldus resulted in a small amount of total RNA in one sample, thus an aliquot of total RNA (1.0 μg) was equally taken from three biological samples of each independent experiment, and the three aliquots were mixed as one sample to perform RNA-Seq by Novogene corporation (Tianjin, China). Residual genomic DNA in the sample was digested with RNase-free DNase I (New England Biolabs). RNA concentrations were measured using Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, United States) and the integrity of these RNA samples was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, United States). Then, the rRNA was removed from the mRNA using Ribo-ZeroTM Magnetic Kit (Bacteria). Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNaseH-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. In the reaction buffer, dNTPs with dTTP were replaced by dUTP. Remaining overhangs were converted into blunt ends via exonuclease / polymerase activities. After adenylation of 3′ ends of DNA fragments, it was ligated to NEBNext Adaptor with hairpin loop structure for hybridization. In order to select cDNA fragments of preferentially 150∼200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, NJ, United States). Then 3 μl USER Enzyme (NEB, United States) was applied on size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then PCR was performed with Phusion High Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions and sequenced using an Illumina Hiseq platform.
Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts. Clean data (clean reads) were obtained by removing reads containing adapter, the pair-end reads satisfied N > 10%, and low Q-value reads [(quality value < 20) > 50%] from raw reads. Clean reads were mapped to the A. caldus MTH-04 genome (Accession number: LXQG00000000). HTSeq v0.6.1 was used to count the read numbers mapped to each gene. And then the FPKM (expected number of Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced) of each gene was calculated based on the length of the gene and the number of the reads mapped to this gene (Trapnell et al., 2009).
The read counts were adjusted by edgeR program package through one scaling normalized factor before differential gene expression analysis (Wang et al., 2010). Differential expression analysis was performed using the DEGSeq R package (1.20.0). The P-values were adjusted using the Benjamini and Hochberg method (Benjamini and Hochberg, 1995; Klipper et al., 1995). Corrected P-value of 0.005 and log2FC (Fold change) of one were set as the threshold for significantly differential expression.
Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs) was implemented by the GOseq R package (Young et al., 2010), in which gene length bias was corrected. GO terms with corrected P-value less than 0.05 were considered significantly enriched by differential expressed genes. KOBAS software was used to test the statistical enrichment of DEGs in KEGG pathways.
The raw data of RNA-seq is deposited in NCBI with accession number PRJNA528607. And original analysis of DEGs is listed in Supplementary Table S3.
Real-Time Quantitative PCR (RT-qPCR)
The removal of gDNA and reverse transcription of the RNA samples were performed using PrimeScriptTM RT Reagent Kit (Takara) according to instructions. Quantitative amplification was performed with SYRB® Premix Ex TaqTM (Takara) on the Roche LightCycler 480 system (Roche). The gapA gene, encoding glyceraldehyde-3-phosphate dehydrogenase, was used as reference gene for normalization of RT-qPCR data (Chen et al., 2013). The 2-ΔΔCt method was used to analyze relative changes in gene expression, where [ΔCt = (Ct target – Ct gapA) mutantorwildtype, ΔΔCt = ΔCt mutant – ΔCt wildtype] (Livak and Schmittgen, 2001; Li et al., 2017). Each sample was repeated with three independent extractions of RNA and the standard deviations were calculated and showed as error bars. All primers designed are listed in Supplementary Table S1.
Transmission Electron Microscope (TEM)
Bacteria were inoculated on Starkey-Na2S2O3 plate and grown for 8 days. The colony was gently picked out with blunt tip and suspended in water. This suspension was added dropwise on grids for several minutes, then stained with 2% tungstophosphoric acid for a few seconds before being observed under a Quanta FEG 250 (FEI) electron microscope.
Scanning Electron Microscope (SEM)
Sulfur (S0) coupons were produced by heating elemental sulfur powder until melting in a fume hood. Subsequently, the liquid sulfur was poured on a cover glass and cooled. Coupons were sterilized in an autoclave at 103°C for 3 h (Gonzalez et al., 2013). The initial cells of A. caldus prepared according to standard cultivation process (Wang et al., 2016), were equally inoculated into 150 mL Starkey media stuffing 5 g S0 coupons, and then cultivated for another 7 days with or without agitation and other conditions as usual. Sulfur coupons from the culture were dehydrated through a graded ethyl alcohol series and dried with the critical point drier. Specimens were mounted on stubs, coated with gold, and examined under a FEI Quanta250 FEG at 10 kV. Three coupons and five fields were imaged for each sample.
Swarming Assay
Acidithiobacillus caldus cells were grown for 7 days, and cells were collected by centrifugation (10000 × g, 20°C, 5 min). The bacterial suspension was diluted to an OD600 of 0.5, then punctured into semi-solid medium (Starkey-Na2S2O3 medium containing 0.3% agar) and incubated under aerobic conditions at 40°C. Motility was estimated as the diameter of colonies. The areas of at least 12 colonies grown in three independent plates were measured by Image J software (Collins, 2007).
Results
Distribution of Flagellar Encoding Genes in Different Species of Acidithiobacillus
On basis of their energy substrates, Acidithiobacillus species can be grouped into the sulfur oxidizers (A. caldus, A. thiooxidans, and A. albertensis) and the iron/sulfur oxidizers (A. ferrooxidans, A. ferridurans, A. ferriphilus, and A. ferrivorans). Results showed that all the sulfur oxidizers in Acidithiobacillus harbor the flagellar biosynthesis related genes (Figure 1 and Supplementary Tables S2, S4). However, that is not always the case for the sulfur / iron oxidizers of Acidithiobacillus (Supplementary Table S4). Flagellar gene cluster is discovered in A. ferrivorans, one of the sulfur / iron oxidizer species, but not found in either A. ferrooxidans or A. ferridurans. Three A. ferrivorans strains (CF27, PRJEB5721, and YL15) have flagellar gene clusters that are different from those in the sulfur oxidizers of Acidithiobacillus (Figure 1 and Supplementary Table S4). In contrast, the flagellar gene cluster is not found in the published genomes of other three strains of A. ferrivorans (SS3, 21-59-9 and PQ33) (Supplementary Table S4). The absence of the flagellar gene cluster in A. ferrivorans SS3 complete genome suggests that the flagella might not be present in all strains of A. ferrivorans, but not in Acidithiocillus spp. lacking the Sox system. To be specific, the flagellar gene clusters are found in the species that possess the Sox system, such as A. caldus, A. thiooxdans, A. ferrivorans, A. albertensis, and Thermithiobacillus tepidarius (Figure 1), but not in strains lacking the Sox system in A. ferrooxidans or A. ferridurans (Supplementary Tables S2, S4). Flagellar gene clusters are also found in iron oxidizers used in bioleaching, such as Leptospirillum ferrooxidans C2-3 and Leptospirillum. ferriphilum ML-04 (Supplementary Figure S6).
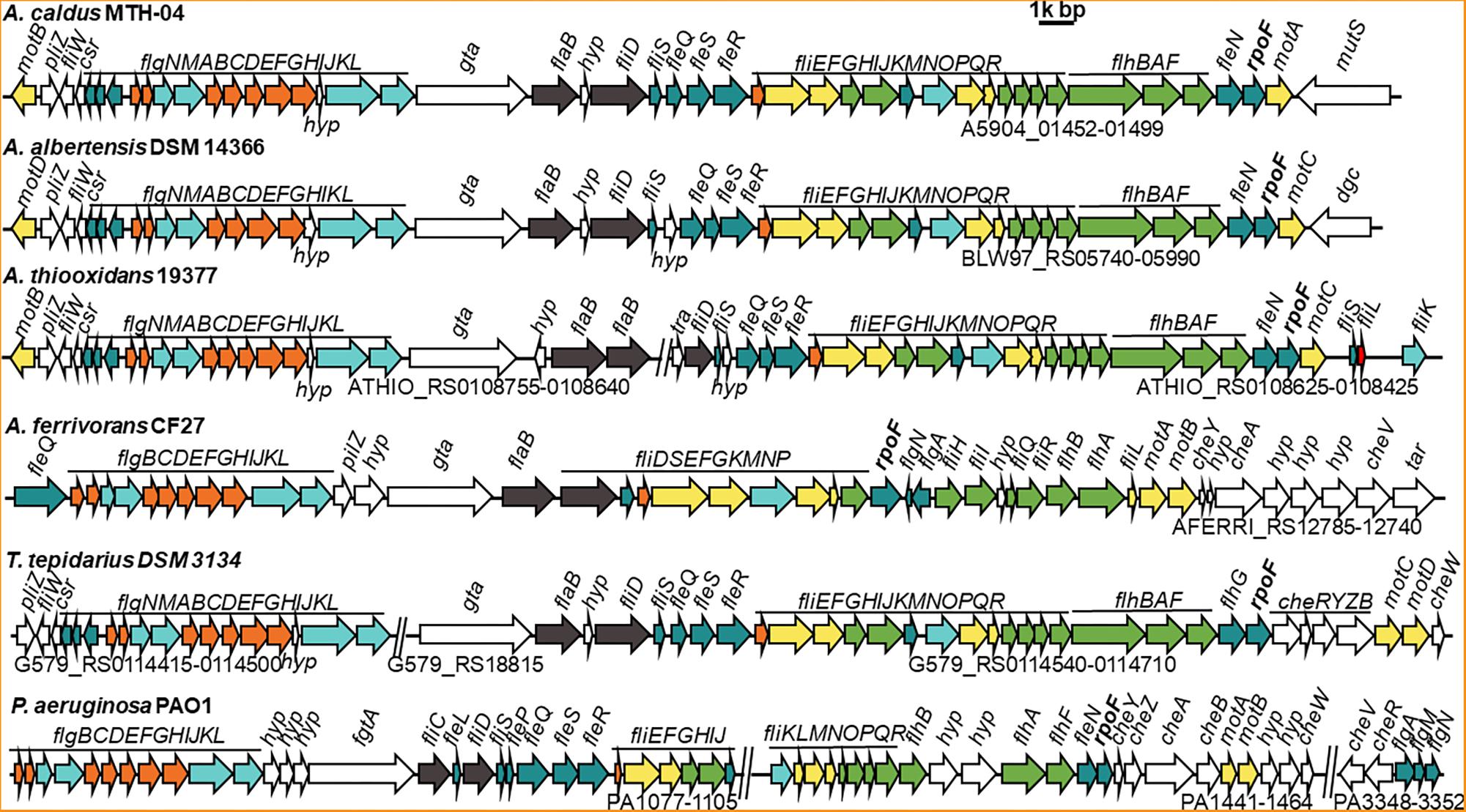
Figure 1. Comparison of flagellar gene clusters among different Acidithiobacillus species and Pseudomonas aeruginosa. GenBank accession numbers for these genomes and the gene IDs for corresponding flagellar cluster are, A. caldus MTH-04 (NZ_CP026328, A5904_01452-01499); A. albertensis DSM 14366 (NZ_MOAD01000001, BLW97_RS06020-RS05990); A. thiooxidans 19377 (NZ_AFOH01000122, ATHIO_RS0108425-RS0108625, ATHIO_RS010860-RS0108755); A. ferrivorans CF27 (NZ_CCCS020000023, AFERRI_RS12740-RS12785); Thermithiobacillus tepidarius DSM 3134 (NZ_KE384096, G579_RS0114415-0114500, G579_RS18815, G579_RS0114540-0114710); Pseudomonas aeruginosa PAO1 (NC_002516, PA1077-1105, PA1441-1464, PA3348-3352). The annotation of each gene is listed in Supplementary Table S2. Different colors mean different structural component of flagellum and other related function: filament is in dark gray, hook is in aquamarine, external basal body (L- and P-ring) is in orange, inner basal body (MS- and C-ring) is in yellow, green represents export apparatus, teal represents regulators and chaperones and white stands for chemotaxis and other unknown function proteins.
All flagellar biosynthesis related genes in A. caldus and A. albertensis, including flagellin genes, regulatory genes and chemotactic genes, are arranged together in one large gene cluster, but these genes are separated into two gene clusters in A. thiooxdans and T. tepidarius DSM3134. Although a flagellar gene cluster is found in A. ferrivorans, the genes and their arrangement orders in A. ferrivorans genomes are distinct from that of A. caldus. No other genes were found in this cluster in A. caldus except for one gene gta, encoding a glycosyl transferase that locates between the basal body/hook gene cluster and the filament gene cluster and is considered to be associated with the glycosylation of flagellins (Figure 1; Arora et al., 2005). In contrast, the flagellar genes of P. aeruginosa, V. cholerae, and E. coli are separated into at least three loci in the genome by other unrelated genes (Figure 1 and Supplementary Figure S6).
FliC is shown to have pleiotropic effect on bacterial motility, growth, biofilm formation and protein secretion (He et al., 2012). The absence of FliC led to the absence of flagella and the deficiency of motility for the cells (Tremblay et al., 2012). There are five filament related genes (flaAC and flaEDB) found in the genome of V. cholerae (Prouty et al., 2001), however, only flaB (a fliC homologous gene) was found in A. caldus. Sequence analysis indicates that FlaB has 31.5% identity to FliC of E. coli, but they differ in immunostimulatory properties (Lee et al., 2003). Besides, phylogenetic analysis indicates that the FlaB protein from Acidithiobacillus was closely related to that of Thiomonas family (Supplementary Figure S1). Interestingly, FlhDC, essential regulators for flagella biosynthesis in E. coli (Wang et al., 2006; Lee et al., 2011), are found in none of A. caldus, A. albertensis, and A. thiooxdans. In contrast, a σ28 factor (RpoF or FliA) and four σ54-dependent regulatory proteins (FleQ, FleR, FleS, and FleN) are encoded in the genome of these chemoautotrophic sulfur or iron oxidizers. The flagellar alternative sigma factor RpoF is located at the end of the flagellar gene clusters in Acidithiobacillus strains and shows 46.9% identity to the RpoF in E. coli and 48.43% to that of P. aeruginosa.
Construction of Different A. caldus rpoF Mutants
Different rpoF mutants were constructed to reveal the role of RpoF in regulating flagellar biosynthesis in A. caldus MTH-04. An “in-out” gene markerless deletion strategy (Figure 2A) was used to generate A. caldus rpoF deletion strain as described in the “Materials and Methods” section. The 2976-bp and 1111-bp fragments were amplified from A. caldus ΔrpoF strain using primer sets P1 F/R specific to the lateral regions of homologous arm and P2 F/R specific to the homologous arm regions, respectively (Figure 2B, lanes 1 and 3). Larger fragments were obtained from A. caldus wildtype with the two primer sets (3,663-bp for P1 F/R and 1,824-bp for P2F/R) (Figure 2B, lanes 2 and 4). No band was detected in A. caldus ΔrpoF strain using primers (P3F/R) specific to rpoF gene (Figure 2B, lane 5), while a 543-bp fragment was amplified from wildtype strain (Figure 2B, lane 6). PCR fragments amplified from genome of A. caldus ΔrpoF strain using primers P1F/R were sequenced to make sure that rpoF gene was markerlessly removed from A. caldus MTH-04 chromosome. A rpoF-expression plasmid was constructed successfully using the mobilizable plasmid pJRD215 as backbone (Figure 2C). The promoter (PtetH) of tetrathionate hydrolase gene (tetH) of A. caldus was selected to induce the transcription of rpoF (Figure 2C). The A. caldus rpoF complemented strain [ΔrpoF(rpoF)], the overexpression strain [WT(rpoF)] and control strains [ΔrpoF(215) and WT(215)] were obtained as described in “Materials and Methods” section (Figure 2C).
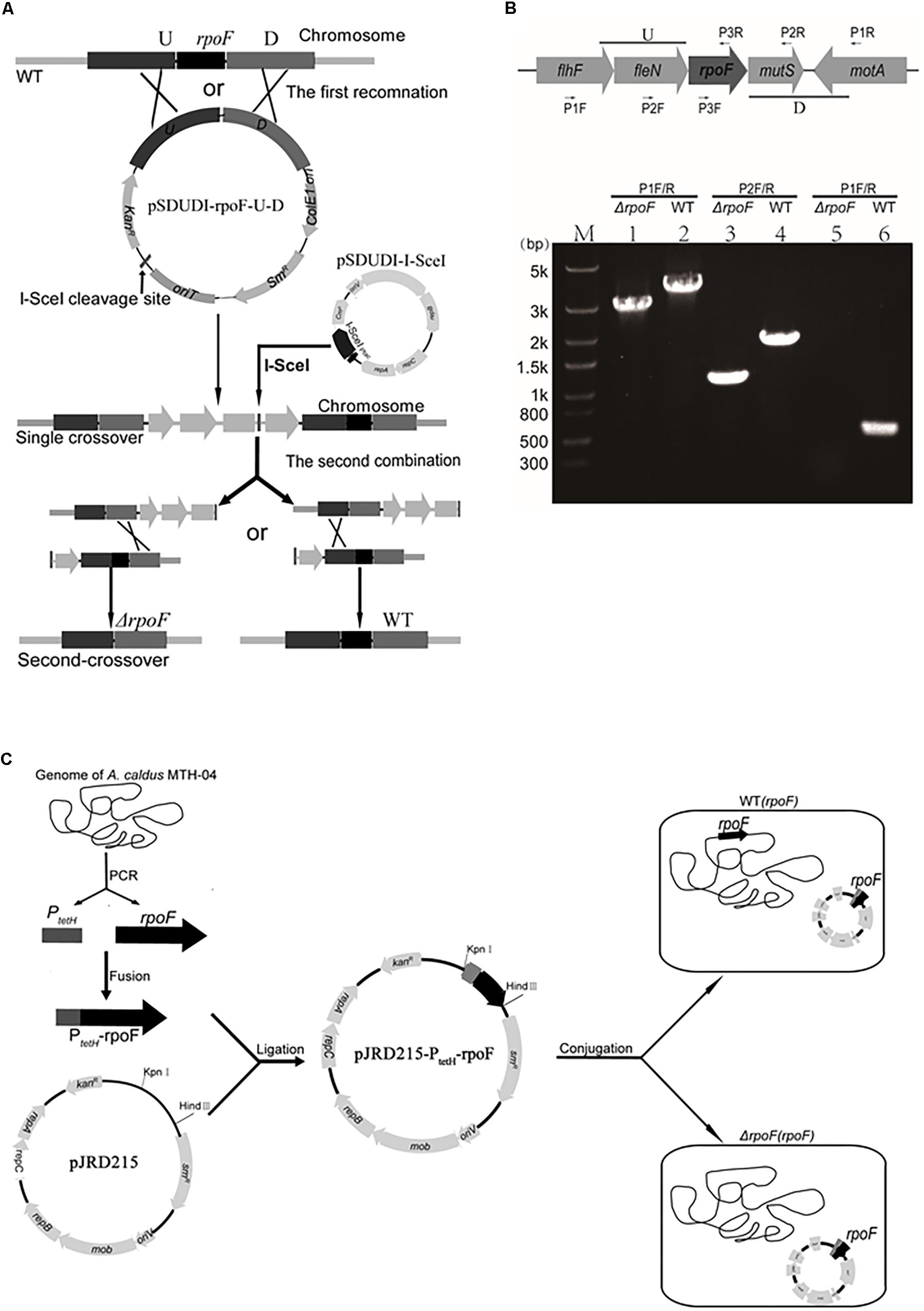
Figure 2. Construction of rpoF deletion, overexpression and complementation strains. (A) The schematic of rpoF double-crossover markerless gene deletion system. (B) Confirmation of rpoF mutant by PCR. Positions of the three sets of primers used for PCR amplifications to confirm rpoF mutagenesis. U, upstream homologous arm of rpoF; D, downstream homologous arm of rpoF. Confirmation of rpoF knockout strain by PCR analyses. flhF (flagellar biosynthesis regulator FlhF), fleN (Cobyrinic acid a,c-diamide synthase), rpoF (flagellar biosynthesis sigma factor), motA (flagellar motor protein MotA) and mutS (DNA mismatch repair protein). Lanes 1, 3, and 5, PCR amplifications from A. caldus ΔrpoF genome with primers P1F/R, P2F/R and P3F/R, respectively. Lanes 2, 4, and 6, PCR fragments amplified from the genome of wildtype strain with primers P1F/R, P2F/R and P3F/R, respectively. (C) Construction of rpoF overexpression and complementation strains. WT(rpoF), wildtype strain harboring plasmid pJRD215-PtetH-rpoF; ΔrpoF(rpoF), rpoF knockout strain containing plasmid pJRD215-PtetH-rpoF (Liu et al., 2007).
RpoF Regulates Flagellar Biosynthesis and Cell Motility of A. caldus
Transmission electron microscope was used to observe the flagella of different A. caldus mutants. No flagellum was found in A. caldus ΔrpoF strain, whereas a single polar flagellum was observed in the wildtype strain, the rpoF complementation strain and the overexpression strain of A. caldus (Figure 3). The result indicates that rpoF is essential for flagellar biosynthesis in A. caldus. Further, the motility of these A. caldus strains was investigated via the swarming assay. The swarming diameters of A. caldus wildtype were approximately twice as large as those of ΔrpoF-formed colonies on semi-solid media (Figures 4A,B). This result suggests that the motility of A. caldus ΔrpoF strain is inhibited significantly. No significant differences were detected between the rpoF complementation strain ΔrpoF(rpoF) and the control strain WT(215) in terms of the swarming diameters (Figures 4C,D). A. caldus rpoF overexpression strain WT(rpoF) showed an increase in colony diameters compared with that of the control strain WT(215) (Figures 4C,D). Thus, the deletion or overexpression of A. caldus rpoF leads to visible effects on the size of colonies, indicating the importance of flagella to A. caldus motility.
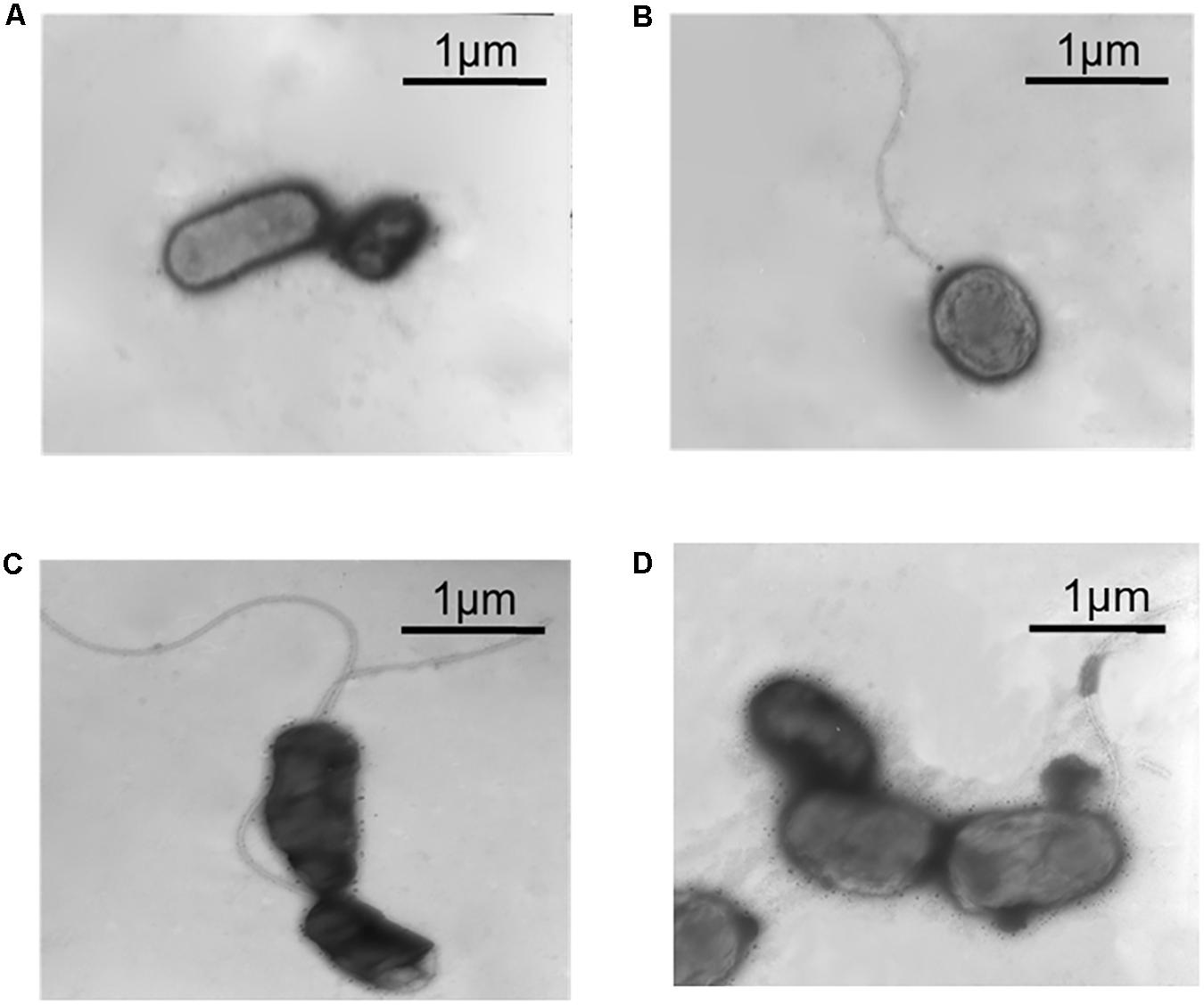
Figure 3. Observation of different A. caldus strains using transmission electron microscope (TEM). (A) A. caldus ΔrpoF(215) strain, the rpoF deletion strain containing blank plasmid pJRD215. (B) A. caldus ΔrpoF(rpoF) strain, the ΔrpoF complemented with the rpoF gene using the plasmid pJRD215-PtetH-rpoF. (C) A. caldus WT(215) strain, the wildtype strain harboring pJRD215. (D) A. caldus WT(rpoF) strain, the overexpression of rpoF gene in A. caldus wildtype using pJRD215-PtetH-rpoF.
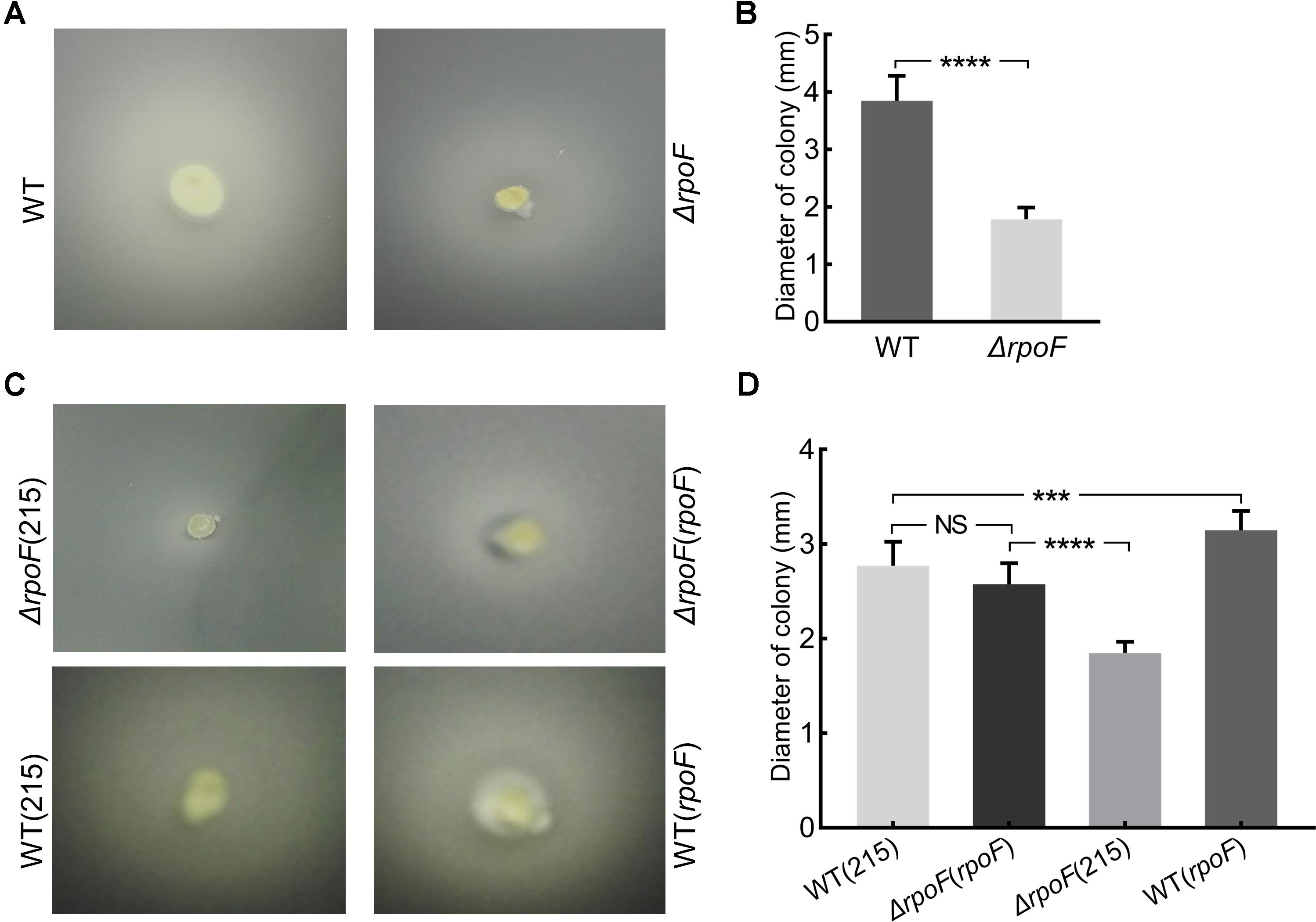
Figure 4. Motility analysis of A. caldus rpoF mutants. (A,C) Colonies of different strains on the semi-solid agar. (B,D) statistical analysis on the swarming diameters. Graphpad Prism7.0 was used to measure the swarming diameters of at least 20 colonies for each strain. The error bars showed Standard Error of the Mean (SEM). The significance was determined using Student’s t-test. NS, no significance; ∗∗∗P < 0.001, significant; ∗∗∗∗P < 0.0001, extremely significant. WT, A. caldus wildtype; ΔrpoF, the rpoF deletion strain; ΔrpoF(215), the rpoF deletion strain carrying blank plasmid pJRD215; ΔrpoF(rpoF), the rpoF deletion strain complemented with plasmid pJRD215-PtetH-rpoF; WT(215), the wildtype strain harboring pJRD215; WT(rpoF), the overexpression of rpoF gene in A. caldus wildtype strain using pJRD215-PtetH-rpoF.
RpoF Influences the Growth of A. caldus in Unfavorable Conditions
Flagella play essential roles in bacterial chemotaxis and motility thus this sophisticated apparatus facilitates the survival of bacteria in various complex environments (Arora et al., 1997; McGowan et al., 2000; Shah et al., 2000; Young et al., 2000; Jahn et al., 2008; Koster et al., 2012; Stevenson et al., 2015). To investigate the influence of RpoF absence on A. caldus growth, we created several unfavorable conditions, including static cultivation, low concentration of S0 and low-energy-density substrate (S4O62-). When A. caldus rpoF deletion and wildtype strains were cultivated in the optimal growth conditions (150 rpm, 1.2 g S0 added in 150 mL media), no growth differences were observed in either their growth curves or growth rates (Figure 5A and Supplementary Figure S2A). When the cells were cultivated without agitation, A. caldus ΔrpoF showed an obvious longer lag phase and lower OD600 value compared to those of wildtype strain (Figure 5B). The OD600 of wildtype and mutant strains reached 0.135 (±0.009) and 0.104 (±0.004) at the stationary phase, respectively (Figure 5B), and A. caldus wildtype strain showed an advantage on the growth rate over that of the mutant before entering stationary phase (Supplementary Figure S2B). These results indicate that agitation is an important factor for the growth of A. caldus ΔrpoF. When the amount of elemental sulfur was decreased to 0.27% (weight/volume) (0.4 g S0 per 150 mL), the difference in growth was detected not only at static condition but also at the shaking condition (Figures 5C,D and Supplementary Figures S2C,D). With agitation and 0.4 g S0, the growth disadvantage and lower growth rate of A. caldus ΔrpoF were observed in the stationary phase (Figure 5C and Supplementary Figure S2C), indicating the limited energy-substrate had a negative effect on the growth of rpoF mutant under shaking condition. When tetrathionate was used as the energy-substrate, A. caldus ΔrpoF exhibited much lower cell density and growth rate compared with that of wildtype strain regardless of agitation or stilling cultivation (Figures 5E,F and Supplementary Figures S2E,F). As a result, the absence of RpoF has no obvious influence on A. caldus growth under optimal conditions (1.2 g S0 and 150 rpm), but brings negative effects on its growth in unfavorable conditions (no shaking, low substrate concentration or non-optimal substrate).
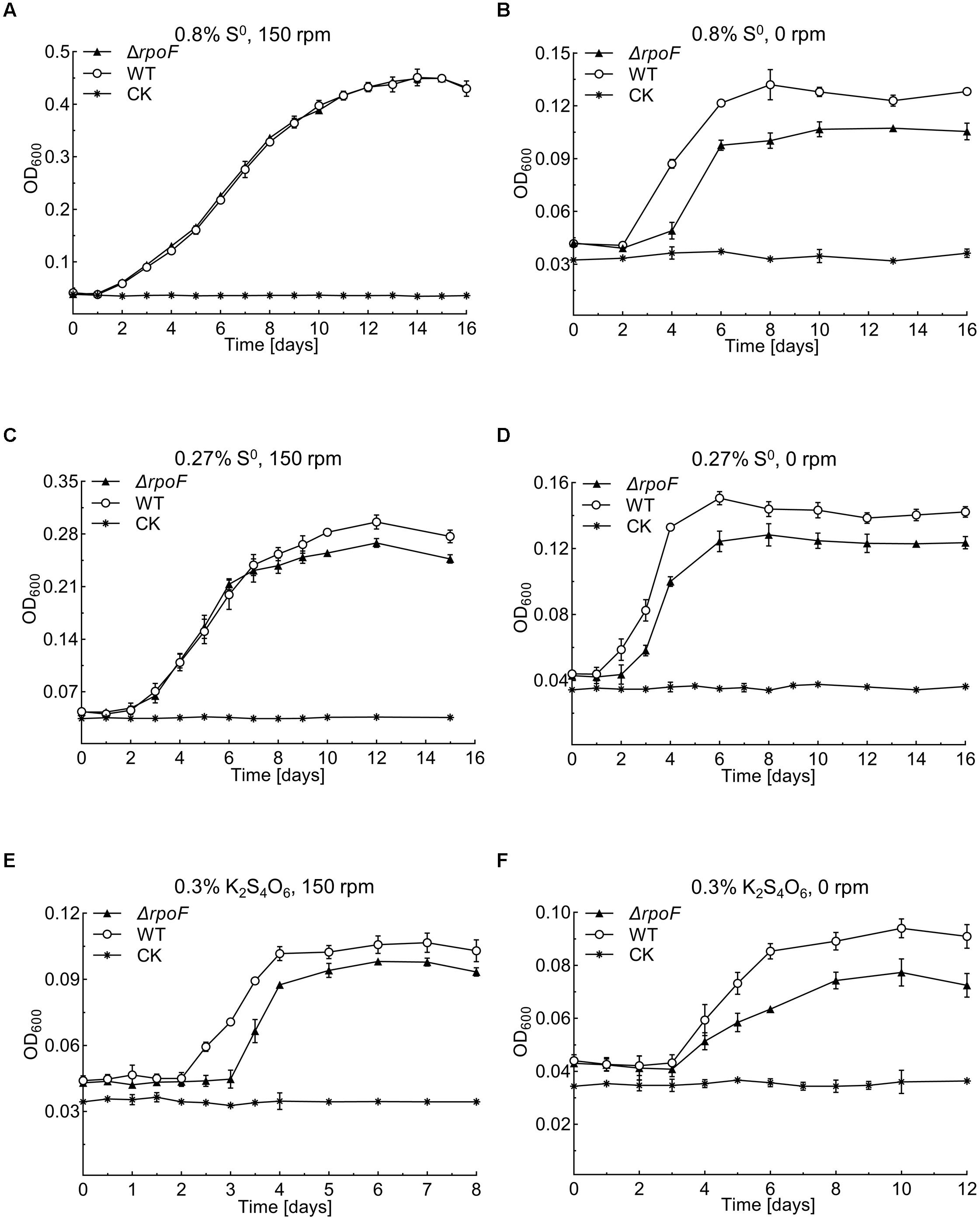
Figure 5. Growth curves of the A. caldus MTH-04 wildtype and ΔrpoF under different growth conditions. All measurements were performed in triplicates. GraphPad Prism 7.0 was used for statistical analysis. (A,B) The growth of strains in Starkey-S0 medium with the addition of 1.2 g S0 at 150 rpm and 0 rpm, respectively. (C,D) The growth of strains in Starkey-S0 medium with the addition of 0.4 g S0 at 150 rpm and 0 rpm, respectively. (E,F) The growth of strains in Starkey-K2S4O6 medium containing 0.1 mM K2S4O6 at 150 rpm and 0 rpm, respectively. CK, control check, medium without cells.
The pH value is an important environmental factor that affects the growth of A. caldus on elemental sulfur. Under the optimal condition of 1.2 g S0 and 150 rpm, the pH of both A. caldus ΔrpoF and wildtype cultures declined from 2.5 (in the 1st day) to approximately 0.7 (in the 9th day) (Supplementary Figure S3A). When the amount of S0 was decreased to 0.4 g in shaking condition, the pH of wildtype culture dropped to 0.97, which is obviously lower than the pH of A. caldus ΔrpoF culture (1.12) (Supplementary Figure S3B). Under the static cultivation, A. caldus ΔrpoF culture containing 0.4 g or 1.2 g S0 showed higher pH values than that of wildtype strain, but the final pH of all these cultures was above 1.0 (Supplementary Figures S3C,D). Therefore, the change patterns of pH values of A. caldus ΔrpoF and wildtype strain cultures were similar to that of cell growth, indicating the correlation between cell growth and the environmental pH changes.
The Absence of RpoF Affects A. caldus Attachment
To investigate the effects of flagella on cell attachment, A. caldus MTH-04 rpoF deletion and overexpression strains were cultivated with S0 coupons (see section “Material and Methods”). Under the static condition, flagella-absent cells tended to attach on the S0 coupon surface (Figures 6A–C), while cells with flagella were inclined to be in the free-living state and the cell density of the rpoF overexpression strain was three times higher than that of the rpoF deletion strain in the liquid culture (Figure 6D). Results indicate that the absence of RpoF results in the tendency of cells to attach to S0-coupons. When the cultivation condition was changed to agitation, no significant differences in cell numbers were detected between the two strains on S0 coupons (Supplementary Figure S4).
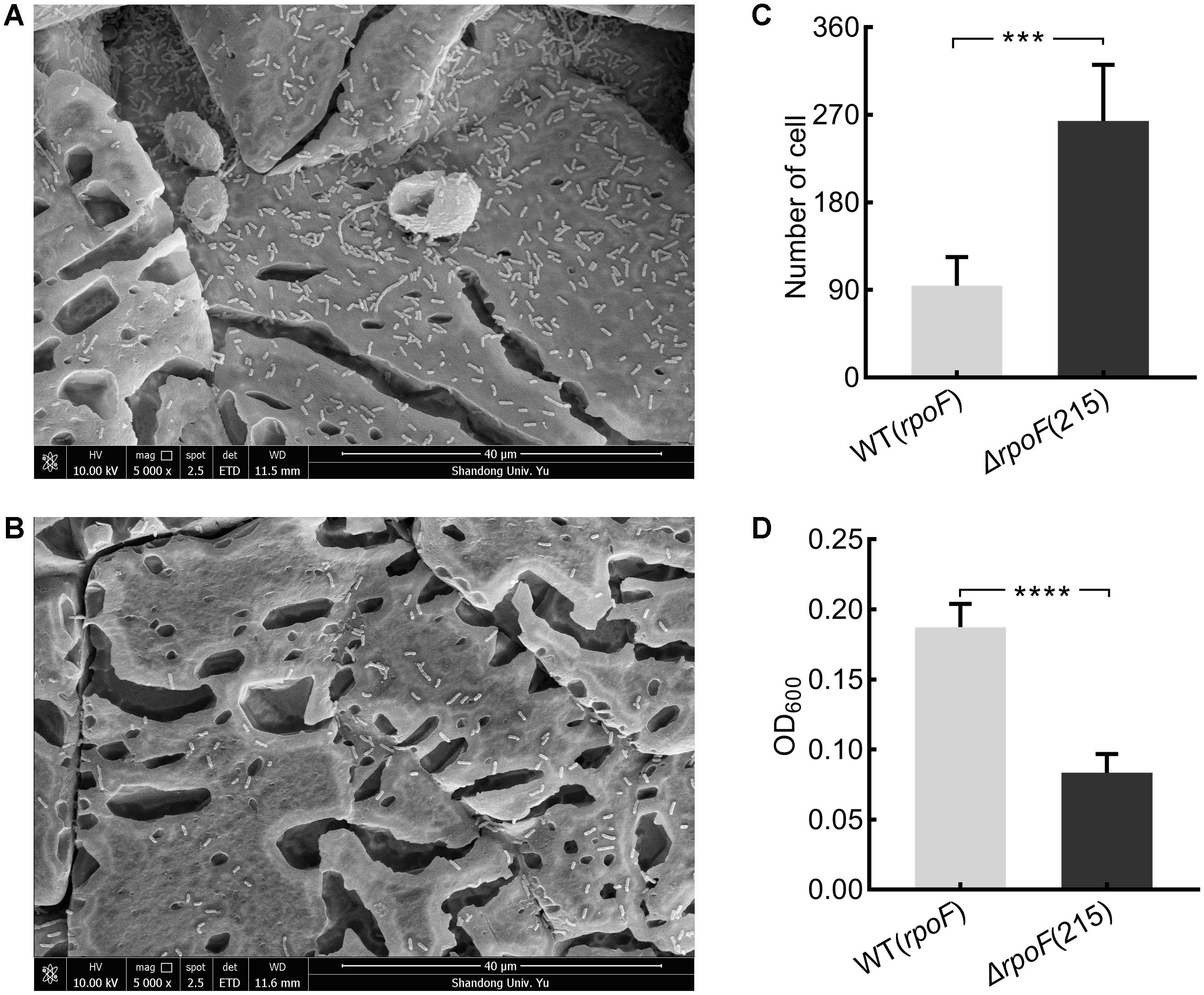
Figure 6. The distribution of A. caldus rpoF deletion and overexpression strains on the S0 coupons. Scanning electron micrographs for strains ΔrpoF(215) (A) and the rpoF overexpression [WT(rpoF)] (B) on sulfur-coupons in static cultivation. Statistical analysis on the number of cells from five different microscopic fields on S0 coupon for ΔrpoF(215) and the rpoF overexpression [WT(rpoF)] strain (C). The OD600 of free cells in liquid Starkey-S0 coupon containing media for ΔrpoF(215) and WT(rpoF) in five duplicates (D). ∗∗∗P < 0.001, significant, ∗∗∗∗P <0.0001, extremely significant.
The Influence of RpoF Mutagenesis on the Transcription of Flagellar Biosynthesis Gene Cluster and the Transcriptome Profiling of A. caldus
To explore the regulatory role of RpoF, A. caldus ΔrpoF and wildtype cells in static cultivation were collected at mid-log growth phase to perform RNA-seq. There were 215 DEGs in A. caldus rpoF knockout strain compared with wildtype strain, including 129 upregulated genes and 86 downregulated genes (Figure 7A and Supplementary Table S3). Forty-one DEGs were selected to perform RT-qPCR assays and the fold changes of these genes were calculated. Statistical analyses of the fold change values from RT-qPCR and RNA-seq indicate that 39 of the 41 genes located in 95% confidence limit (Supplementary Figure S5). The high consistency of the data from RNA-seq and RT-qPCR suggests that the RNA-seq data are reliable. The KEGG pathway enrichment of the DEGs indicates that 20 pathways were influenced by the deletion of rpoF in A. caldus, referring to flagellar assembly, bacterial chemotaxis, section system, two-component system, nitrogen metabolism, oxidative phosphorylation, DNA modification, etc. (Figure 7B and Supplementary Table S5). These results suggest that RpoF plays an important role in regulating the transcription profile of A. caldus.
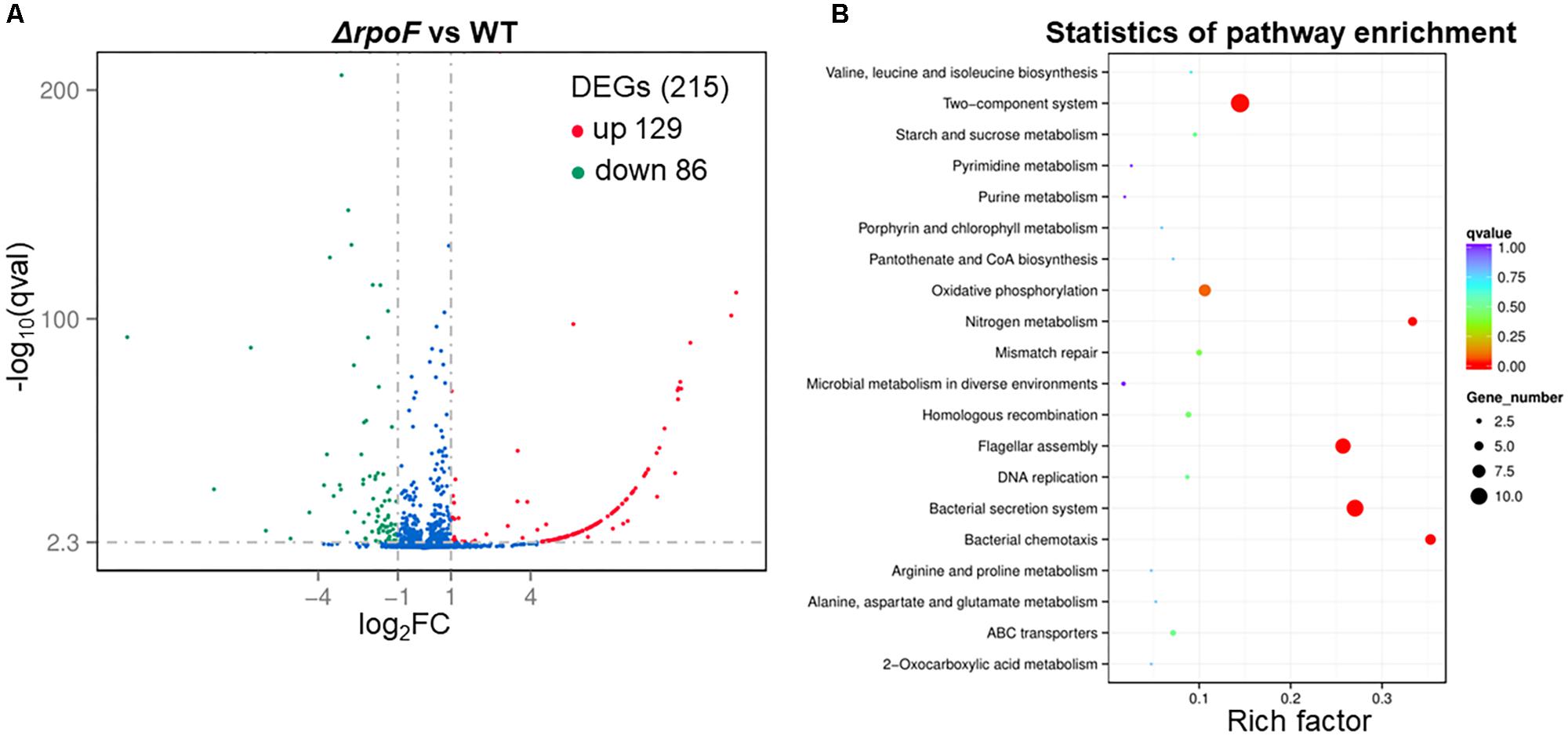
Figure 7. Overall transcriptomic changes of ΔrpoF. (A) Volcano plot showing fold changes and levels of significance for differential expression genes. (B) KEGG pathway enrichment.
The deletion of rpoF in A. caldus MTH-04 resulted in significant downregulation of some genes involved in flagellar synthesis, including fliW (A5904_1454), csrA (A5904_1455), flavoprotein (A5904_1456), hyp (A5904_1457), hyp (A5904_1469), flgK (A5904_1470), flgL (A5904_1471), fliC (A5904_1473), hyp (A5904_1474), fliD(A5904_1475), fliS (A5904_1476), fliA (A5904_1498), fliL (A5904_1536), motA (A5904_1499), and motD (A5904_1452) (Supplementary Table S5). Thus, the regulation of RpoF on the transcription of flagellar gene cluster in A. caldus was verified here. The genes are responsible for the chemotaxis such as cheW (A5904_1445), cheA (A5904_1448), cheZ (A5904_1449), and cheY (A5904_1450), showed different degrees of downregulation in A. caldus rpoF deletion strain (Supplementary Table S5). The low expression levels of these genes in A. caldus rpoF knockout strain implies the close connections between RpoF and chemotaxis in this bacterium. In summary, RpoF controls the expression of flagellar genes, which explains why ΔrpoF does not assemble the flagellin at the transcriptional level. The extremely low transcriptional levels of some genes [A5904_0005 (extracellular solute-binding protein), A5904_0969 (diguanylate cyclase/phosphodiesterase with PAS/PAC and GAF sensor), and A5904_1537 (conserved hypothetical protein)] indicate that the transcription of these genes may be RpoF-dependent.
The deletion of rpoF gave rise to transcriptional changes of genes participating in energy metabolism and respiratory chain in A. caldus. Different types of terminal oxidase encoding genes showed distinct transcriptional changes: the cytochrome c oxidase genes cydB (A5904_1252), cydA (A5904_1253), ccdA (A5904_1799), NADH dehydrogenase genes nuoB (A5904_0714) and ndhF (A5904_2209) were downregulated, while the NADH-ubiquinone oxidoreductase genes nuoG (A5904_0764), nuoH (A5904_0765), nuoI (A5904_0766), and nuoN (A5904_0771) were upregulated (Supplementary Table S5). The transcriptional differences of these terminal oxidase encoding genes indicate that the absence of RpoF could influence the electron transfer in A. caldus. However, almost none of the canonical sulfur-oxidizing genes exhibited transcriptional changes after rpoF mutagenesis in A. caldus, indicating deleting rpoF has no effect on the typical sulfur metabolism (Sox system, S4I pathway, and Dsr system). The influence of RpoF mutagenesis on the other cellular metabolisms was discovered, including the downregulation of genes in the nitrogen metabolism and upregulation of genes in the carbon metabolism (Supplementary Table S5). Unusually, A5904_0981 (glucan 1,4-alpha-glucosidase) and A5904_0114 (bactoprenol glucosyl transferase) were upregulated more than 400 times in A. caldus ΔrpoF strain, suggesting RpoF plays an important role in these corresponding biological processes (energy, nitrogen, and carbon metabolisms).
The absence of RpoF in A. caldus MTH-04 also led to different influences on transcription of genes related to cell permeability and secretion systems. Transporter genes, including A5904_0100 (major facilitator superfamily MFS1), A5904_0983 (O-antigen ABC transporter), A5904_2283 (ExbB proton channel), A5904_2356 (potassium channel protein), A5904_2775 (major facilitator superfamily MFS1), and A5904_2774 (nitrate transporter), exhibited lower transcription levels in the rpoF knockout strain, while 21 conjugation related genes were upregulated in this mutant and some of them even increased more than 100 times at transcription level (Supplementary Table S5). The transcription of genes in type IV secretion system, such as A5904_1857-1865 (in the order of pilN, pilO, pilP, pulE, pulF, pilS, pilU, pilM, and pilV), was upregulated in A. caldus ΔrpoF strain. However, as for pilus synthetic genes, A5904_0039 (pilZ), A5904_0041 (pleD), A5904_0042 (conserved hypothetical protein), and A5904_0043 (prepilin-type N-terminal cleavage/methylation domain-containing protein) were downregulated in A. caldus rpoF deletion strain (Supplementary Table S5).
Some of the transcription regulator genes including a σ factor genes rpoE (A5904_0065), Fis family transcriptional regulator (A5904_1002), LysR family transcriptional regulator (A5904_2210), and two component systems [cheA/Y, cydB/C (A5904_1252/1253)] were downregulated, while some genes like A5904_1597 (helix-turn-helix XRE-family like protein), A5904_2806 (helix-turn-helix XRE-family like protein), and A5904_0628 (looped-hinge helix DNA binding domain, AbrB family) were upregulated in A. caldus ΔrpoF strain (Supplementary Table S5). The similar transcriptional changes were also present among transposase genes. Besides, the DNA replication and modification were affected by the deletion of A. caldus rpoF as well (Supplementary Table S5).
Discussion
In this study, we discovered that the flagellar gene clusters were distributed in almost all Acidithiobacillus species (Supplementary Table S4) with two exceptions, A. ferrooxidan (Valdés et al., 2008b) and A. ferridurans. The sulfur oxidizers that harbor the Sox system tend to form the flagella. Sox system, consists of SoxXA, SoxYZ, and SoxB in Acidithiobacillus spp., is an important and highly efficient pathway for these sulfur oxidizers to obtain electrons for ATP production (Mangold et al., 2011; Chen et al., 2012; Yin et al., 2014; Christel et al., 2016; Wang et al., 2019). The existence of Sox system in these flagella-contained strains in Acidithiobacillus implies the importance of energy supply in the occurrence of flagella in Acidithiobacillus strains (Figure 1 and Supplementary Table S4). Similar rules also applied to Thiobacillus spp. (Supplementary Table S4) which are phylogenetically close to Acidithiobacillus spp.
The flagellum of A. caldus is highly similar to that of P. aeruginosa in morphological structure (McCarter, 2006; Potvin et al., 2008) in addition to the similar flagellar gene arrangement and flagellar regulatory proteins (Figure 1 and Supplementary Table S2). The regulatory role of RpoF for flagellar assembly in A. caldus MTH-04 was determined by TEM observation (Figure 3) and RNA-seq analysis (Figure 8A). These findings indicate that A. caldus probably develops the similar flagellar transcription hierarchy as the reported pattern in P. aeruginosa (Ritchings et al., 1995; Dasgupta et al., 2002, 2003; Soutourina and Bertin, 2003; Luo et al., 2016). As shown in Figure 8B, the flagellar biosynthesis in A. caldus is probably controlled by a four-class hierarchy involving the regulatory proteins RpoN, FleQ, FleS, FleR, and RpoF (FliA). The only Class I gene is fleQ which encodes a σ54-dependent activator that is transcribed by a σ70 promoter. FleQ, along with σ54-holoenzyme, could modulate the transcription of Class II flagellar genes including fleSR, fliDS, fliEFGHIJKMNOPQR, flhBAF, fleN, and flgA. The Class II genes encode flagellar structural components of basal body (MS ring and C ring), export apparatus, switch, as well as the regulatory factors (FleS/FleR, FleN, RpoF, and FlgA). FleS/FleR, a σ54-dependent two component regulatory system (TCS) in which FleS is a sensor kinase for the response regulator, FleR, could regulate the transcription of the Class III genes (flgBCDEFGHIJKL) which encode flagellar basal body rod, P-ring, L-ring and hook. The Class IV genes are σ28-dependent, and are responsible for encoding the alternative flagellins, the putative anti-sigma factor FlgM, the motor components MotAB and chemotactic protein (CheY, CheZ, and CheA).
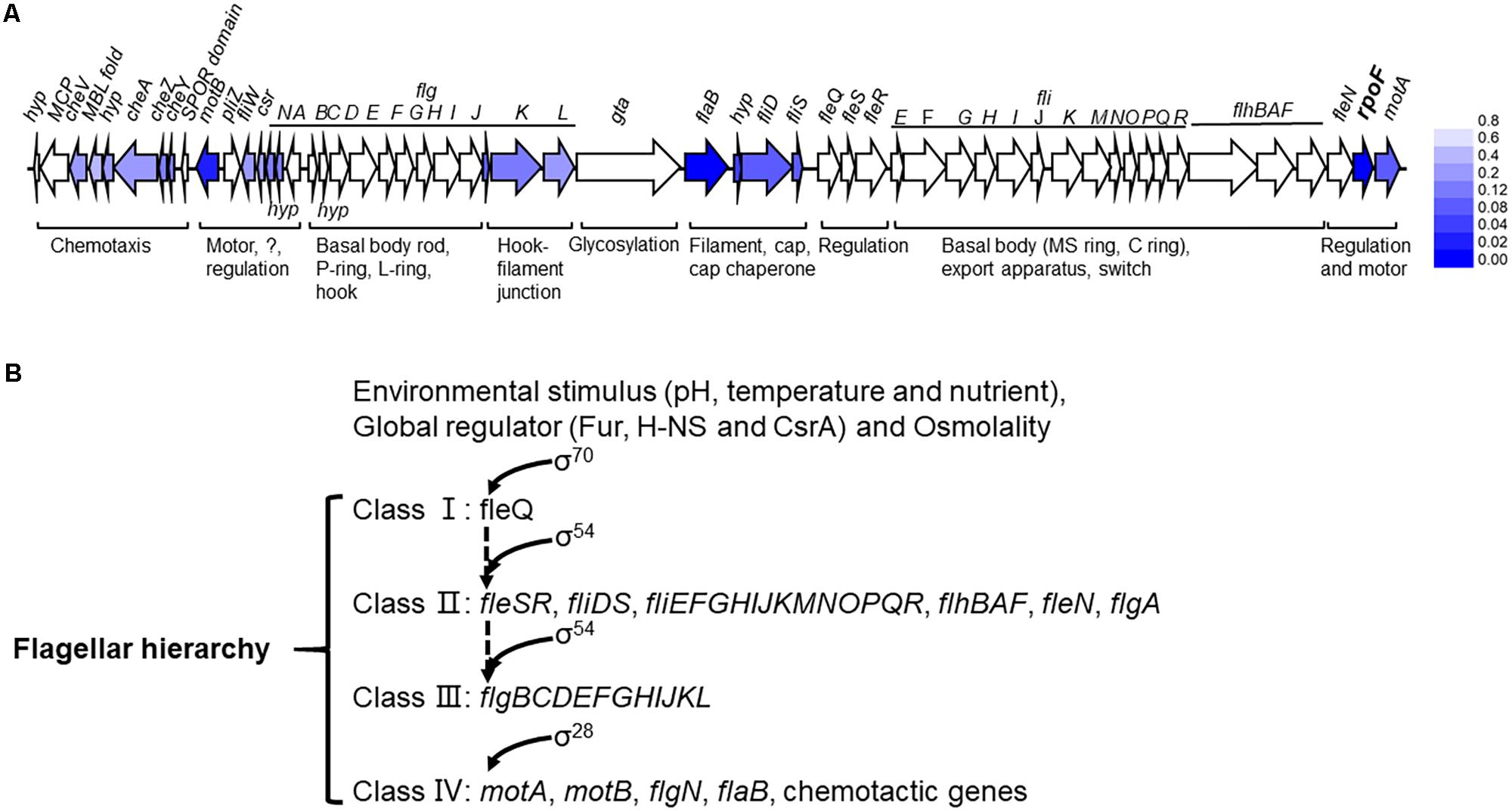
Figure 8. The flagellar transcription hierarchy model of A. caldus. (A) The influence of RpoF on the transcription of flagellar genes. The blue color stands for the downregulation of gene in A. caldus ΔrpoF and the darker color means the greater difference; (B) the proposed model of flagellar regulation hierarchy in A. caldus.
Flagella-driven bacterial motility and chemotaxis are important for microorganisms to find energy substrates and respond to unfavorable environments (Prouty et al., 2001; Soutourina and Bertin, 2003; Liu et al., 2005; Seshasayee et al., 2006; Jonas et al., 2010). A conclusion is drawn from the growth analysis of A. caldus MTH-04 (Figure 5), that flagella endows A. caldus with a strong ability to adapt to the unfavorable environment. Previous works reported that cells still managed to assign the energy for flagella synthesis and assembly even in the unfavorable environments (Liu et al., 2005; Zhao et al., 2007). Wildtype cells of A. caldus grew better than the ΔrpoF in unfavorable conditions (static cultivation or low concentration of energy-substrates) (Figure 5), indicating cells could benefit from flagellar biosynthesis even though it is an energy-consuming process. Recently, an evolutionary tuning of the trade-off between flagella-driven motility and growth proposed that bacterial cells tend to synthesize flagella to gain a better chance for survival when the substrates are insufficient (Ni et al., 2017). The weaker growth of ΔrpoF when little energy substrates were provided (0.4 g, Figures 5C,D) suggests that this trade-off model may apply to chemoautotrophic bacteria as well.
Flagella-mediated cell motility affects the lifestyles of A. caldus in the S0 coupon culture. Under static cultivation conditions, cells with flagella are motile (Figure 4) so that cells tended to be planktonic for easily gaining the energy substrates from solid surfaces and obtaining the oxygen and carbon dioxide from the liquid phase for growth (Figures 6B,D). In contrast, the absence of flagella made the cells immotile (Figure 4), resulting in the tendency of cells attaching on the surface of S0 coupon (Figures 6A,C). When A. caldus strains were cultivated with agitation, it was easy for cells to contact the energy-substrate and air, thus the loss of motility caused by absence of RpoF could not lead to the cell-attachment difference in the S0 coupon cultivation (Supplementary Figure S4). Therefore, the flagella could allow A. caldus to better respond to the environmental stress and obtain growth advantage in the unfavorable condition.
The absence of RpoF influenced the transcription of genes involved in A. caldus biofilm formation. Type IV pili is reported to be responsible for biofilm formation (O’Toole and Kolter, 1998; Pratt and Kolter, 1998; Li et al., 2010; Giltner et al., 2012). Genes of type IV pili (A5904_1858 – 1865) were significantly upregulated in A. caldus rpoF knockout strain, implying the flagella-absent cells are likely to form biofilm. Recent works have shown that the second messenger cyclic diguanylate (c-di-GMP) is another central regulator for biofilm formation in bacteria (Boyd and O’Toole, 2012; Purcell et al., 2012; Romling et al., 2013; Purcell and Tamayo, 2016), and c-di-GMP also plays an important role in the adhesion of Acidithiobacillus spp. to solid surfaces (Ruiz et al., 2012; Castro et al., 2015; Diaz et al., 2018). Several genes related to the c-di-GMP metabolism (A5904_2318, A5904_0969, A5904_2211, and A5904_0041) were differentially expressed in the rpoF deletion strain (Supplementary Table S4). It was shown that A5904_0041 is a pelD homologous gene that is associated with biofilm structure and formation processes in A. thiooxidans (Diaz et al., 2018). Therefore, the differential expression of genes involved in biofilm formation and biofilm regulation in A. caldus ΔrpoF strain, suggests that the absence of RpoF not only influences the flagellum synthesis, but also affects the biofilm formation.
In summary, our experiments focused on the distribution of flagellar gene clusters in Acidithiobacillus spp., the regulatory role of A. caldus RpoF on flagellar biosynthesis and cell adhesion to surfaces, and the effects of flagella-mediated motility on cell responses to unfavorable environments. Therefore, this study provides an overall knowledge and new insights about the flagella in Acidithiobacillus spp., which will promote the study on bacterial environmental adaptation and sulfur metabolism in these acidophilic bacteria.
Author Contributions
C-LY, RW, J-QL (8th Author), and L-XC designed, conducted, and composed the manuscript. C-LY conducted the experiments and performed the bioinformatics analysis. X-KC, J-QL (4th Author), X-ML, XP, and C-JZ analyzed the data and revised the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31570036, 31370138, 31400093, 31570041, 31872621, and 30800011), the Project of Taishan Industry Leading Talent in Shandong Province (LJNY201603), and the State Key Laboratory of Microbial Technology Foundation (M2017-01), People’s Republic of China.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to associate Pro. Qi-Long Qin from Shandong University for suggestions regarding to bioinformatics analysis. We thank Mesfin Angaw for proofreading this manuscript, Chun-Mao Lin and Xiu-Feng Gu for providing some of the medium and agar plates. We also thank the supports from Core Facilities Sharing Platform for Life Sciences of Shandong University, including Hai-Yan Yu for SEM and TEM technical assistance, Zhi-Feng Li and Jing Zhu for RT-qPCR instruction, and Nan-nan Dong for providing bacteriological incubators.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01130/full#supplementary-material
FIGURE S1 | The phylogenetic tree of representative FlaB proteins which are encoded by fliC homologous genes. 35 representative FlaB proteins were used for phylogenetic tree construction with reference sequences. These proteins were labeled with their GenBank accession numbers and bacterial species. ClustalW was used to align these sequences and MEGA 7 was used to build the tree.
FIGURE S2 | Growth rates of the ΔrpoF and wildtype strain of A. caldus MTH-04 under different growth conditions. Each point in the curve was the first derivative of the corresponding points from the growth curve in Figure 5. GraphPad Prism 7.0 was used for statistical analysis. Growth rate curve for cells in the following conditions: Starkey-K2S4O6 medium containing 1 mM K2S4O6 at 150 rpm (A) and 0 rpm (B), Starkey-S0 medium with the addition of 0.4 g S0 at 150 rpm (C) and 0 rpm (D), 1.2 g S0 at 150 rpm (E), and 0 rpm (F).
FIGURE S3 | The pH curves of ΔrpoF and wildtype strain of A. caldus under different growth conditions. The pH changes of Starkey-S0 medium containing 1.2 g S0 at 150 rpm (A) and 0 rpm (B), 0.4 g S0 at 150 rpm (C), and 0 rpm (D), respectively.
FIGURE S4 | The attachment of A. caldus rpoF deletion and wildtype strain on the S0 coupons. Scanning electron micrographs for WT(rpoF) (A) and ΔrpoF(215) (B) on S0 coupons with 150 rpm.
FIGURE S5 | Consistency check of DEGs between RNA-seq and RT-qPCR. (A) RT-qPCR validation for 41 genes. RT-qPCR data are the mean of the results from the three biological replicates. (B) agreement comparison between the RNA-seq and RT-qPCR results indicated the data for 39 of the 41 genes lie within the 95% confidence limit.
FIGURE S6 | Flagellar gene clusters in different bacterial species. GenBank accession number for these genomes and the gene IDs for corresponding flagellar cluster are: Thiomonas intermedia ATCC 15466 (NZ_CP020046, BVH73_RS00605-RS00370, BVH73_ RS00740-RS00670); Thiomonas delicata DSM 16361 (NZ_LT592170, THIARS_RS14765-RS15050, THIARS_RS14615-RS14695); Leptospirillum ferrooxidans C2-3 (NC_017094, LFE_RS10820-RS10725, LFE_RS01330-RS01470); Leptospirillum. ferriphilum ML-04 (NC_018649, LFML04_RS11235-RS12960, LFML04_RS01075-RS01220); Vibrio cholerae O1 (NC_002505, VC0892-0893, VC1008, VC2058-2069, VC2120-2140, VC2187-2205, VC2601); Escherichia coli str. K-12 substr. MG1655 (NC_000913, b1066-1083, b1892-1877, b1920-1950). The annotation of each gene is listed in Supplementary Table S2. Different colors mean different structural component of flagellum and other related function, filament is in dark gray, hook is in aquamarine, external basal body (L- and P-ring) is in orange, inner basal body (MS- and C-ring) is in yellow, green represents export apparatus, teal represents regulators and chaperones and white stands for chemotaxis and other unknown function proteins.
TABLE S1 | All primers used in the experiment.
TABLE S2 | Annotations of flagellar genes for all related strains.
TABLE S3 | The raw data of DEGs.
TABLE S4 | Taxonomic traits of Acidithiobacillus spp., Thiobacillus spp., and Thiomonas spp.
TABLE S5 | Gene classification of RNA-seq.
References
Africa, C. J., Hille, R. P. V., and Harrison, S. T. L. (2013). Attachment of Acidithiobacillus ferrooxidans and Leptospirillum ferriphilum cultured under varying conditions to pyrite, chalcopyrite, low-grade ore and quartz in a packed column reactor. Appl. Microbiol. Biotechnol. 97, 1317–1324. doi: 10.1007/s00253-012-3939-x
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z. H., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Arce, R. A., Puente, S. F., Avendano, R., Martinez, C. M., de Moor, J. M., Pieper, D. H., et al. (2019). Thermoplasmatales and sulfur-oxidizing bacteria dominate the microbial community at the surface water of a CO2-rich hydrothermal spring located in Tenorio Volcano National Park, Costa Rica. Extremophiles 23, 177–187. doi: 10.1007/s00792-018-01072-6
Arora, S. K., Neely, A. N., Blair, B., Lory, S., and Ramphal, R. (2005). Role of motility and flagellin glycosylation in the pathogenesis of Pseudomonas aeruginosa burn wound infections. Infect. Immun. 73, 4395–4398. doi: 10.1128/IAI.73.7.4395-4398.2005
Arora, S. K., Ritchings, B. W., Almira, E. C., Lory, S., and Ramphal, R. (1997). A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179, 5574–5590. doi: 10.1128/jb.179.17.5574-5581.1997
Bellenberg, S., Diaz, M., Noel, N., Sand, W., Poetsch, A., Guiliani, N., et al. (2014). Biofilm formation, communication and interactions of leaching bacteria during colonization of pyrite and sulfur surfaces. Res. Microbiol. 165, 773–781. doi: 10.1016/j.resmic.2014.08.006
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple. J. R. Stat. Soc. Series B Methodol. 57, 389–399. doi: 10.2307/2346101
Boyd, C. D., and O’Toole, G. A. (2012). Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu. Rev. Cell Dev. Biol. 28, 439–462. doi: 10.1146/annurev-cellbio-101011-155705
Bryan, R., Glaser, D., and Shapiro, L. (1990). Genetic regulatory hierarchy in Caulobacter development. Adv. Genet. 27, 1–31.
Bryant, R. D., Mcgroarty, K. M., Costerton, J. W., and Laishle, E. J. (1983). Isolation and characterization of a new acidophilic Thiobacillus species (T. albertis). Can. J. Microbiol. 29, 1159–1170. doi: 10.1139/m83-178
Castro, M., Deane, S. M., Ruiz, L., Rawlings, D. E., and Guiliani, N. (2015). Diguanylate cyclase null mutant reveals that c-di-GMP pathway regulates the motility and adherence of the extremophile bacterium Acidithiobacillus caldus. PLoS One 10:e0116399. doi: 10.1371/journal.pone.0116399
Chen, L., Lina, J., Liu, X., Pang, X., Lin, H., and Lin, J. (2013). Transposition of IS elements induced by electroporation of suicide plasmid in Acidithiobacillus caldus. Enzyme Microb. Technol. 53, 165–169. doi: 10.1016/j.enzmictec.2013.03.002
Chen, L., Ren, Y., Lin, J., Liu, X., Pang, X., and Lin, J. (2012). Acidithiobacillus caldus sulfur oxidation model based on transcriptome analysis between the wild type and sulfur oxygenase reductase defective mutant. PLoS One 7:e39470. doi: 10.1371/journal.pone.0039470
Chen, S., Beeby, M., Murphy, G. E., Leadbetter, J. R., Hendrixson, D. R., Briegel, A., et al. (2011). Structural diversity of bacterial flagellar motors. EMBO J. 30, 2972–2981. doi: 10.1038/emboj.2011.186
Christel, S., Fridlund, J., Buetti-Dinh, A., Buck, M., Watkin, E. L., and Dopson, M. (2016). RNA transcript sequencing reveals inorganic sulfur compound oxidation pathways in the acidophile Acidithiobacillus ferrivorans. FEMS Microbiol. Lett. 363:fnw057. doi: 10.1093/femsle/fnw057
Colmer, A. R., and Hinkle, M. E. (1947). The Role of microorganisms in acid mine drainage: a preliminary report. Science 106, 253–256. doi: 10.1126/science.106.2751.253
Dasgupta, N., Arora, S. K., and Ramphal, R. (2000). fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182, 357–364. doi: 10.1128/JB.182.2.357-364.2000
Dasgupta, N., Ferrell, E. P., Kanack, K. J., West, S. E. H., and Ramphal, R. (2002). fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is σ70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J. Bacteriol. 184, 5240–5250. doi: 10.1128/jb.184.19.5240-5250.2002
Dasgupta, N., Wolfgang, M. C., Goodman, A. L., Arora, S. K., Jyot, J., Lory, S., et al. (2003). A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50, 809–824. doi: 10.1046/j.1365-2958.2003.03740.x
Davison, J., Heusterspreute, M., Chevalier, N., Ha-Thi, V., and Brunei, F. (1987). Vectors with restriction site banks V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene 51, 275–280. doi: 10.1016/0378-1119(87)90316-7
Deb, C., Stackebrandt, E., Pradella, S., Saha, A., and Roy, P. (2004). Phylogenetically diverse new sulfur chemolithotrophs of alpha-proteobacteria isolated from Indian soils. Curr. Microbiol. 48, 452–458. doi: 10.1007/s00284-003-4250-y
Diaz, M., Castro, M., Copaja, S., and Guiliani, N. (2018). Biofilm formation by the acidophile bacterium Acidithiobacillus thiooxidans involves c-di-GMP pathway and Pel exopolysaccharide. Genes 9:E113. doi: 10.3390/genes9020113
Doetsch, R. N., Cook, T. M., and Vaituzis, Z. (1967). On the uniqueness of the flagellum of Thiobacillus thiooxidans. Antonie van Leeuwenhoek 33, 196–202. doi: 10.1007/BF02045551
Donati, E. R., Castro, C., and Urbieta, M. S. (2016). Thermophilic microorganisms in biomining. World J. Microbiol. Biotechnol. 32:179. doi: 10.1007/s11274-016-2140-2
Falagan, C., and Johnson, D. B. (2016). Acidithiobacillus ferriphilus sp. nov., a facultatively anaerobic iron- and sulfur-metabolizing extreme acidophile. Int. J. Syst. Evol. Microbiol. 66, 206–211. doi: 10.1099/ijsem.0.000698
Fitzgerald, D. M., Bonocora, R. P., and Wade, J. T. (2014). Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet. 10:e1004649. doi: 10.1371/journal.pgen.1004649
Friedlander, R. S., Vlamakis, H., Kim, P., Khan, M., Kolter, R., and Aizenberg, J. (2013). Bacterial flagella explore microscale hummocks and hollows to increase adhesion. Proc Natl Acad Sci U.S.A. 110, 5624–5629. doi: 10.1073/pnas.1219662110
Galeva, A., Moroz, N., Yoon, Y. H., Hughes, K. T., Samatey, F. A., and Kostyukova, A. S. (2014). Bacterial flagellin-specific chaperone FliS interacts with anti-sigma factor FlgM. J. Bacteriol. 196, 1215–1221. doi: 10.1128/jb.01278-13
Ghai, R., and Chakraborty, T. (2007). Comparative microbial genome visualization using GenomeViz. Methods Mol. Biol. 395, 97–108. doi: 10.1007/978-1-59745-514-5_6
Giltner, C. L., Nguyen, Y., and Burrows, L. L. (2012). Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. Rev. 76, 740–772. doi: 10.1128/MMBR.00035-12
Gonzalez, A., Bellenberg, S., Mamani, S., Ruiz, L., Echeverria, A., Soulere, L., et al. (2013). AHL signaling molecules with a large acyl chain enhance biofilm formation on sulfur and metal sulfides by the bioleaching bacterium Acidithiobacillus ferrooxidans. Appl. Microbiol. Biotechnol. 97, 3729–3737. doi: 10.1007/s00253-012-4229-3
Hallberg, K. B., and Lindstrom, E. B. (1994). Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology 140(Pt 12), 3451–3456. doi: 10.1099/13500872-140-12-3451
Harneit, K., Göksel, A., Kock, D., Klock, J. H., Gehrke, T., and Sand, W. (2006). Adhesion to metal sulfide surfaces by cells of Acidithiobacillus ferrooxidans, Acidithiobacillus thiooxidans and Leptospirillum ferrooxidans. Hydrometallurgy 83, 245–254. doi: 10.1016/j.hydromet.2006.03.044
Harrison, A. P. (1984). The acidophilic Thiobacilli and other acidophilic bacteria that share their habitat. Annu. Rev. Microbiol. 38, 265–292. doi: 10.1146/annurev.mi.38.100184.001405
He, Y., Xu, T., Fossheim, L. E., and Zhang, X. H. (2012). FliC, a flagellin protein, is essential for the growth and virulence of fish pathogen Edwardsiella tarda. PLoS One 7:e45070. doi: 10.1371/journal.pone.0045070
Hudson, C. M., Williams, K. P., and Kelly, D. P. (2014). Definitive assignment by multigenome analysis of the gammaproteobacterial genus Thermithiobacillus to the class Acidithiobacillia. Polish J. Microbiol. 63, 245–247.
Jacobi, S., Schade, R., and Heuner, K. (2004). Characterization of the alternative sigma factor sigma54 and the transcriptional regulator FleQ of Legionella pneumophila, which are both involved in the regulation cascade of flagellar gene expression. J. Bacteriol. 186, 2540–2547.
Jahn, C. E., Willis, D. K., and Charkowski, A. O. (2008). The flagellar sigma factor fliA is required for Dickeya dadantii virulence. Mol. PlantMicrobe Interact. 21, 1431–1442. doi: 10.1094/MPMI
Johnson, D. B., Rolfe, S., Hallberg, K. B., and Iversen, E. (2001). Isolation and phylogenetic characterization of acidophilic microorganisms indigenous to acidic drainage waters at an abandoned Norwegian copper mine. Environ. Microbiol. 3, 630–637. doi: 10.1046/j.1462-2920.2001.00234.x
Jonas, K., Edwards, A. N., Ahmad, I., Romeo, T., Romling, U., and Melefors, O. (2010). Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ. Microbiol. 12, 524–540. doi: 10.1111/j.1462-2920.2009.02097.x
Jyot, J., Sonawane, A., Wu, W., and Ramphal, R. (2007). Genetic mechanisms involved in the repression of flagellar assembly by Pseudomonas aeruginosa in human mucus. Mol. Microbiol. 63, 1026–1038. doi: 10.1111/j.1365-2958.2006.05573.x
Kadoya, R., Kodama, Y., Matsumoto, K., and Taguchi, S. (2015). Indirect positive effects of a sigma factor RpoN deletion on the lactate-based polymer production in Escherichia coli. Bioengineered 6, 307–311. doi: 10.1080/21655979.2015.1069449
Kamimura, K., Sharmin, S., Yoshino, E., Tokuhisa, M., and Kanao, T. (2018). Draft genome sequence of Acidithiobacillus sp. strain SH, a marine acidophilic sulfur-oxidizing bacterium. Genome Announc. 6:e01603-17. doi: 10.1128/genomeA.01603-17
Kelly, D. P., and Wood, A. P. (2000). Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 50, 511–516. doi: 10.1099/00207713-50-2-511
Kiiyukia, C., Kawakami, H., and Hashimoto, H. (1993). Effect of sodium chloride, pH and organic nutrients on the motility of Vibrio cholerae non-01. Microbios 73, 249–256. doi: 10.1006/mpat.1993.1008
Klipper, A. Y., Wasserman, M., Braunspiegel, W. N., Borstein, D., Peleg, S., Assa, S., et al. (1995). Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med. Hypotheses 45, 486–490. doi: 10.1016/0306-9877(95)90228-7
Klose, K. E., and Mekalanos, J. J. (1998). Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28, 501–510. doi: 10.1046/j.1365-2958.1998.00809.x
Koirala, S., Mears, P., Sim, M., Golding, I., Chemla, Y. R., Aldridge, P. D., et al. (2014). A nutrient-tunable bistable switch controls motility in Salmonella enterica serovar Typhimurium. mBio 5:e01611-14. doi: 10.1128/mBio.01611-14
Koster, D. A., Mayo, A., Bren, A., and Alon, U. (2012). Surface growth of a motile bacterial population resembles growth in a chemostat. J. Mol. Biol. 424, 180–191. doi: 10.1016/j.jmb.2012.09.005
Kundu, T. K., Kusano, S., and Ishihama, A. (1997). Promoter selectivity of Escherichia coli RNA polymerase sigma F holoenzyme involved in transcription of flagellar and chemotaxis genes. J. Bacteriol. 179, 4264–4269. doi: 10.1128/jb.179.13.4264-4269.1997
Kunin, C. M., Hua, T., and Bakaletz, L. O. (1995). Effect of salicylate on expression of flagella by Escherichia coli and Proteus, Providencia and Pseudomonas spp. Infect. Immun. 63, 1796–1799. doi: 10.1007/BF01793865
Kutsukake, K., Ohya, Y., and Iino, T. (1990). Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172, 741–747. doi: 10.1128/jb.172.2.741-747.1990
Lara, R. H., Garcia, M. J. V., Gonzalez, I., and Cruz, R. (2013). Influence of the surface speciation on biofilm attachment to chalcopyrite by Acidithiobacillus thiooxidans. Appl. Microbiol. Biotechnol. 97, 2711–2724. doi: 10.1007/s00253-012-4099-8
Lee, S. K., Stack, A., Katzowitsch, E., Aizawa, S. I., Suerbaum, S., and Josenhans, C. (2003). Helicobacter pylori flagellins have very low intrinsic activity to stimulate human gastric epithelial cells via TLR5. Microbes Infect. 5, 1345–1356. doi: 10.1016/j.micinf.2003.09.018
Lee, Y. Y., Barker, C. S., Matsumura, P., and Belas, R. (2011). Refining the binding of the Escherichia coli flagellar master regulator, FlhD4C2, on a base-specific level. J. Bacteriol. 193, 4057–4068. doi: 10.1128/jb.00442-11
Lemon, K. P., Higgins, D. E., and Kolter, R. (2007). Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 189, 4418–4424. doi: 10.1128/JB.01967-06
Li, C., Louise, C. J., Shi, W., and Adler, J. (1993). Adverse conditions which cause lack of flagella in Escherichia coli. J. Bacteriol. 175, 2229–2235.
Li, L. F., Fu, L. J., Lin, J. Q., Pang, X., Liu, X. M., Wang, R., et al. (2017). The sigma (54)-dependent two-component system regulating sulfur oxidization (Sox) system in Acidithiobacillus caldus and some chemolithotrophic bacteria. Appl. Microbiol. Biotechnol. 101, 2079–2092. doi: 10.1007/s00253-016-8026-2
Li, Y. Q., Wan, D. S., Huang, S. S., Leng, F. F., Yan, L., Ni, Y. Q., et al. (2010). Type IV pili of Acidithiobacillus ferrooxidans are necessary for sliding, twitching motility and adherence. Curr. Microbiol. 60, 17–24. doi: 10.1007/s00284-009-9494-8
Liu, M., Durfee, T., Cabrera, J. E., Zhao, K., Jin, D. J., and Blattner, F. R. (2005). Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J. Biol. Chem. 280, 15921–15927. doi: 10.1074/jbc.M414050200
Liu, X., Lin, J., Zhang, Z. H., Bian, J., Zhao, Q., Liu, Y., et al. (2007). Construction of conjugative gene transfer system between E. coli and moderately thermophilic, extremely acidophilic Acidithiobacillus caldus MTH-04. J. Microbiol. Biotechnol. 17, 162–167. doi: 10.1007/s10295-006-0171-7
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, G., Huang, L., Su, Y., Qin, Y., Xu, X., Zhao, L., et al. (2016). flrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerg. Microbes Infect. 5:e85. doi: 10.1038/emi.2016.82
Mangold, S., Valdes, J., Holmes, D. S., and Dopson, M. (2011). Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front. Microbiol. 2:17. doi: 10.3389/fmicb.2011.00017
McCarter, L. L. (2001). Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65, 445–462. doi: 10.1128/MMBR.65.3.445-462.2001
McCarter, L. L. (2006). Regulation of flagella. Curr. Opin. Microbiol. 9, 180–186. doi: 10.1016/j.mib.2006.02.001
McGowan, C. C., Necheva, A. S., Cover, T. L., and Blaser, M. J. (2000). The role of fliA in motility and acid survival in H. pylori. Gut 47, A43–A44.
Nealson, K. H. (1997). Sediment bacteria: Who’s there, what are they doing and what’s new? Annu. Rev. Earth Planet. Sci. 25, 403–434.
Ni, B., Ghosh, B., Paldy, F. S., Colin, R., Heimerl, T., and Sourjik, V. (2017). Evolutionary remodeling of bacterial motility checkpoint control. Cell Rep. 18, 866–877. doi: 10.1016/j.celrep.2016.12.088
Nunez, H., Moya, B. A., Covarrubias, P. C., Issotta, F., Cardenas, J. P., Gonzalez, M., et al. (2017). Molecular systematics of the genus Acidithiobacillus: insights into the phylogenetic structure and diversification of the taxon. Front. Microbiol. 8:30. doi: 10.3389/fmicb.2017.00030
Osterman, I. A., Dikhtyar, Y. Y., Bogdanov, A. A., Dontsova, O. A., and Sergiev, P. V. (2015). Regulation of flagellar gene expression in bacteria. Biochemistry 80, 1447–1456. doi: 10.1134/S000629791511005X
O’Toole, G. A., and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Otte, J. M., Harter, J., Laufer, K., Blackwell, N., Straub, D., Kappler, A., et al. (2018). The distribution of active iron-cycling bacteria in marine and freshwater sediments is decoupled from geochemical gradients. Environ. Microbiol. 20, 2483–2499. doi: 10.1111/1462-2920.14260
Pollmann, K., Kutschke, S., Matys, S., Raff, J., Hlawacek, G., and Lederer, F. L. (2018). Bio-recycling of metals: recycling of technical products using biological applications. Biotechnol. Adv. 36, 1048–1062. doi: 10.1016/j.biotechadv.2018.03.006
Potvin, E., Sanschagrin, F., and Levesque, R. C. (2008). Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 32, 38–55. doi: 10.1111/j.1574-6976.2007.00092.x
Pratt, L. A., and Kolter, R. (1998). Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30, 285–293. doi: 10.1046/j.1365-2958.1998.01061.x
Prouty, M. G., Correa, N. E., and Klose, K. E. (2001). The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39, 1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x
Purcell, E. B., McKee, R. W., McBride, S. M., Waters, C. M., and Tamayo, R. (2012). Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J. Bacteriol. 194, 3307–3316. doi: 10.1128/JB.00100-12
Purcell, E. B., and Tamayo, R. (2016). Cyclic diguanylate signaling in Gram-positive bacteria. FEMS Microbiol. Rev. 40, 753–773. doi: 10.1093/femsre/fuw013
Rawlings, D. E. (2005). Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microb. Cell Fact. 4:13. doi: 10.1186/1475-2859-4-13
Ritchings, B. W., Almira, E. C., Lory, S., and Ramphal, R. (1995). Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 63, 4868–4876. doi: 10.1016/0928-8244(95)00069-1
Romling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/MMBR.00043-12
Rowe, O. F., Sanchez, E. J., Hallberg, K. B., and Johnson, D. B. (2007). Microbial communities and geochemical dynamics in an extremely acidic, metal-rich stream at an abandoned sulfide mine (Huelva, Spain) underpinned by two functional primary production systems. Environ. Microbiol. 9, 1761–1771. doi: 10.1111/j.1462-2920.2007.01294.x
Ruiz, L. M., Castro, M., Barriga, A., Jerez, C. A., and Guiliani, N. (2012). The extremophile Acidithiobacillus ferrooxidans possesses a c-di-GMP signalling pathway that could play a significant role during bioleaching of minerals. Lett. Appl. Microbiol. 54, 133–139. doi: 10.1111/j.1472-765X.2011.03180.x
Sambrook, J., Fritsch, E. F., and Maniatis, T. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Schulz, T., Rydzewski, K., Schunder, E., Holland, G., Bannert, N., and Heuner, K. (2012). FliA expression analysis and influence of the regulatory proteins RpoN, FleQ and FliA on virulence and in vivo fitness in Legionella pneumophila. Arch. Microbiol. 194, 977–989. doi: 10.1007/s00203-012-0833-y
Seshasayee, A. S. N., Bertone, P., Fraser, G. M., and Luscombe, N. M. (2006). Transcriptional regulatory networks in bacteria: from input signals to output responses. Curr. Opin. Microbiol. 9, 511–519. doi: 10.1016/j.mib.2006.08.007
Shah, D. S., Perehinec, T., Stevens, S. M., Aizawa, S. I., and Sockett, R. E. (2000). The flagellar filament of Rhodobacter sphaeroides: pH-induced polymorphic transitions and analysis of the fliC gene. J. Bacteriol. 182, 5218–5224. doi: 10.1128/JB.182.18.5218-5224.2000
Sharmin, S., Yoshino, E., Kanao, T., and Kamimura, K. (2016). Characterization of a novel thiosulfate dehydrogenase from a marine acidophilic sulfur-oxidizing bacterium, Acidithiobacillus thiooxidans strain SH. Biosci. Biotechnol. Biochem. 80, 273–278. doi: 10.1080/09168451.2015.1088377
Simon, R., Priefer, U., and Pühler, A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechology 1, 784–791. doi: 10.1038/nbt1183-784
Song, J., Lin, J., Ren, Y., and Lin, J. (2011). Competitive adsorption of binary mixture of Leptospirillum ferriphilum and Acidithiobacillus caldus onto pyrite. Biotechnol. Bioprocess Eng. 15, 923–930. doi: 10.1007/s12257-010-0008-0
Song, S., Xue, Y., Liu, E., Wang, K., Zhang, Y., Wu, H., et al. (2016). Comparative analysis of sigma factors RpoS, FliA and RpoN in Edwardsiella tarda. Can. J. Microbiol. 62, 861–869. doi: 10.1139/cjm-2016-0158
Soutourina, O. A., and Bertin, P. N. (2003). Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol. Rev. 27, 505–523. doi: 10.1016/S0168-6445(03)00064-0
Starnbach, M. N., and Lory, S. (1992). The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol. Microbiol. 6, 459–469. doi: 10.1111/j.1365-2958.1992.tb01490.x
Stevenson, E., Minton, N. P., and Kuehne, S. A. (2015). The role of flagella in Clostridium difficile pathogenicity. Trends Microbiol. 23, 275–282. doi: 10.1016/j.tim.2015.01.004
Temple, K. L., and Colmer, A. R. (1951). The autotrophic oxidation of iron by a new bacterium, Thiobacillus ferrooxidans. J. Bacteriol. 62, 605–611.
Totten, P. A., and Lory, S. (1990). Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J. Bacteriol. 172, 7188–7199.
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Tremblay, P. L., Aklujkar, M., Leang, C., Nevin, K. P., and Lovley, D. (2012). A genetic system for Geobacter metallireducens: role of the flagellin and pilin in the reduction of Fe (III) oxide. Environ. Microbiol. Rep. 4, 82–88. doi: 10.1111/j.1758-2229.2011.00305.x
Valdés, J., Pedroso, I., Quatrini, R., Dodson, R. J., Tettelin, H., Blake, R., et al. (2008a). Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC genomics 9:597. doi: 10.1186/1471-2164-9-597
Valdés, J., Pedroso, I., Quatrini, R., and Holmes, D. S. (2008b). Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: insights into their metabolism and ecophysiology. Hydrometallurgy 94, 180–184. doi: 10.1016/j.hydromet.2008.05.039
Waksman, S. A., and Joffe, J. S. (1922). Microörganisms concerned in the oxidation of sulfur in the soil: II. Thiobacillus thiooxidans, a new sulfur-oxidizing organism isolated from the soil. J. Bacteriol. 7, 239–256. doi: 10.1002/path.1700300417
Walker, S., Sojka, M., Dibb, F. M., and Woodward, M. J. (1999). Effect of pH, temperature and surface contact on the elaboration of fimbriae and flagella by Salmonella serotype enteritidis. J. Med. Microbiol. 48, 253–261. doi: 10.1099/00222615-48-3-253
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, R., Lin, J. Q., Liu, X. M., Pang, X., Zhang, C. J., Yang, C. L., et al. (2019). Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Front. Microbiol. 9:3290. doi: 10.3389/fmicb.2018.03290
Wang, S., Fleming, R. T., Westbrook, E. M., Matsumura, P., and McKay, D. B. (2006). Structure of the Escherichia coli FlhDC Complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 355, 798–808. doi: 10.1016/j.jmb.2005.11.020
Wang, Z. B., Li, Y. Q., Lin, J. Q., Pang, X., Liu, X. M., Liu, B. Q., et al. (2016). The two-component system RsrS-RsrR regulates the tetrathionate intermediate pathway for thiosulfate oxidation in Acidithiobacillus caldus. Front. Microbiol. 7:1755. doi: 10.3389/fmicb.2016.01755
Xia, J. L., Peng, A. A., He, H., Yang, Y., Liu, X. D., and Qiu, G. (2007). A new strain Acidithiobacillus albertensis BY-05 for bioleaching of metal sulfides ores. Trans. Nonferrous Metals Soc. China 17, 168–175. doi: 10.1016/s1003-6326(07)60067-3
Yin, H., Zhang, X., Li, X., He, Z., Liang, Y., Guo, X., et al. (2014). Whole-genome sequencing reveals novel insights into sulfur oxidation in the extremophile Acidithiobacillus thiooxidans. BMC Microbiol. 14:179. doi: 10.1186/1471-2180-14-179
Young, G. M., Badger, J. L., and Miller, V. L. (2000). Motility is required to initiate host cell invasion by Yersinia enterocolitica. Infect. Immun. 68, 4323–4326. doi: 10.1128/IAI.68.7.4323-4326.2000
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Yu, Y., Liu, X., Wang, H., Li, X., and Lin, J. (2014). Construction and characterization of tetH overexpression and knockout strains of Acidithiobacillus ferrooxidans. J. Bacteriol. 196, 2255–2264. doi: 10.1128/JB.01472-13
Zhao, K., Liu, M., and Burgess, R. R. (2007). Adaptation in bacterial flagellar and motility systems: from regulon members to ‘foraging’-like behavior in E. coli. Nucleic Acids Res. 35, 4441–4452. doi: 10.1093/nar/gkm417
Keywords: RpoF, Acidithiobacillus caldus, flagellar gene cluster, flagellar synthesis regulation, swarming, Sox system
Citation: Yang C-L, Chen X-K, Wang R, Lin J-Q, Liu X-M, Pang X, Zhang C-J, Lin J-Q and Chen L-X (2019) Essential Role of σ Factor RpoF in Flagellar Biosynthesis and Flagella-Mediated Motility of Acidithiobacillus caldus. Front. Microbiol. 10:1130. doi: 10.3389/fmicb.2019.01130
Received: 25 January 2019; Accepted: 03 May 2019;
Published: 24 May 2019.
Edited by:
Axel Schippers, Federal Institute for Geosciences and Natural Resources, GermanyReviewed by:
Soeren Bellenberg, Linnaeus University, SwedenSophie R. Ullrich, Freiberg University of Mining and Technology, Germany
Jiri Kucera, Masaryk University, Czechia
Copyright © 2019 Yang, Chen, Wang, Lin, Liu, Pang, Zhang, Lin and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Qun Lin, amlhbnF1bmxpbkBzZHUuZWR1LmNu; Lin-Xu Chen, bGlueHVjaGVuQHNkdS5lZHUuY24=
 Chun-Long Yang
Chun-Long Yang Xian-Ke Chen
Xian-Ke Chen Xiang-Mei Liu
Xiang-Mei Liu Lin-Xu Chen
Lin-Xu Chen