
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 May 2019
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.01118
 Priscilla Branchu1†
Priscilla Branchu1† Oliver J. Charity1†
Oliver J. Charity1† Matt Bawn1‡
Matt Bawn1‡ Gaetan Thilliez1
Gaetan Thilliez1 Timothy J. Dallman2
Timothy J. Dallman2 Liljana Petrovska3
Liljana Petrovska3 Robert A. Kingsley1,4*
Robert A. Kingsley1,4*A multi drug resistant Salmonella enterica 4,[5],12:i- of sequence type 34 (monophasic S. Typhimurium ST34) is a current pandemic clone associated with livestock, particularly pigs, and numerous outbreaks in the human population. A large genomic island, termed SGI-4, is present in the monophasic Typhimurium ST34 clade and absent from other S. Typhimurium strains. SGI-4 consists of 87 open reading frames including sil and pco genes previously implicated in resistance to copper (Cu) and silver, and multiple genes predicted to be involved in mobilization and transfer by conjugation. SGI-4 was excised from the chromosome, circularized, and transferred to recipient strains of S. Typhimurium at a frequency influenced by stress induced by mitomycin C, and oxygen tension. The presence of SGI-4 was associated with increased resistance to Cu, particularly but not exclusively under anaerobic conditions. The presence of silCBA genes, predicted to encode an RND family efflux pump that transports Cu from the periplasm to the external milieu, was sufficient to impart the observed enhanced resistance to Cu, above that commonly associated with S. Typhimurium isolates. The presence of these genes resulted in the absence of Cu-dependent induction of pco genes encoding multiple proteins linked to Cu resistance, also present on SGI-4, suggesting that the system effectively limits the Cu availability in the periplasm, but did not affect SodCI-dependent macrophage survival.
Salmonella enterica serovar Typhimurium (S. Typhimurium), including monophasic variants, accounts for approximately 25% of all human cases of non-typhoidal Salmonella (NTS) infection in Europe, and is widespread in multiple animal reservoirs (Hugas and Beloeil, 2014; Animal and Plant Health Agency [APHA], 2017; Branchu et al., 2018). The epidemiological record of human multidrug-resistant (MDR) S. Typhimurium infections in Europe is characterized by successive waves of dominant MDR variants that persist for 10–15 years (Rabsch et al., 2001; Rabsch, 2007). S. Typhimurium DT104 emerged around 1990, becoming a globally pandemic clone that affected numerous domesticated and wild animal species (Threlfall, 2000). Subsequently, in 2007, a monophasic S. Typhimurium variant (S. 4,[5],12:i-) of sequence type 34 (ST34) emerged in European pig populations and spread globally (hereafter referred to as monophasic S. Typhimurium ST34; Hauser et al., 2010; Antunes et al., 2011; Hopkins et al., 2012; Arguello et al., 2013; Mourao et al., 2015; Andres-Barranco et al., 2016; Bonardi, 2017). The mechanisms that drive succession of S. Typhimurium variants are not known, but selection by commonly used antibiotics is unlikely since successive variants share similar AMR profiles (Rabsch, 2007): ACSSuT (ampicillin, chloramphenicol, streptomycin, sulphonamide, tetracycline) for S. Typhimurium DT104 and ASSuT for monophasic S. Typhimurium ST34.
Cu is both an essential nutrient, due to its role as a cofactor in multiple enzymes in all aerobic organisms, and highly toxic due to its ability to displace iron from iron-sulfur clusters in dehydratases (Macomber and Imlay, 2009). To reduce toxicity, bacteria control the amount of free Cu in the cytoplasm and periplasm using transport systems, and by the oxidation of cuprous (Cu1+) to less toxic cupric (Cu2+) ions (Rensing and Grass, 2003). Escherichia coli, a close relative of Salmonella, encodes multiple transport systems on its chromosome to maintain Cu homeostasis. To overcome the toxicity of Cu, E. coli transports Cu from the cytoplasm into the periplasm via a P1B-type ATPase, CopA, and from the cytoplasm and periplasm to the external milieu via a multicomponent Cu RND family efflux pump, CusCFBA, and oxidizes cuprous ions by the action of the multi-Cu oxidase, CueO (Grass and Rensing, 2001; Franke et al., 2003). CueO and CopA, which are co-regulated by the cytosolic CueR, are the primary Cu homeostasis systems active during aerobic growth (Stoyanov et al., 2001), while CusCFBA is important during anaerobic growth, under the transcriptional control of the periplasmic CusRS two component regulator (Outten et al., 2001). In addition, some E. coli isolated from Cu-rich environments have additional plasmid-encoded genes pcoABCDRSE, that encode several proteins including a multicopper oxidase system which is active in the periplasm, Cu transporters, and a Cu binding protein. The pco locus is regulated by a two-component system (PcoR/PcoS) with a sensor kinase that extends into the periplasm (Brown et al., 1995; Rensing and Grass, 2003).
Cu homeostasis in the genus Salmonella appears to be fundamentally different from that in E. coli due to the deletion of the cusRSCFBA genes from the chromosome noted in a number of commonly used lab strains (McClelland et al., 2001; Espariz et al., 2007; Pontel and Soncini, 2009; Fookes et al., 2011). Therefore, although Salmonella encode CopA and CueO, the lack of the CusCFBA RND family efflux pump means they lack the ability to transport Cu out of the cell entirely, which is likely to have a significant impact on the distribution of Cu within the cell. However, plasmid-borne silRSECBAP genes have been described in a S. Typhimurium strain associated with an outbreak in burn patients that had been treated topically with silver nitrate; these genes encode an RND family efflux pump that is closely related to the CusCFBA system and conferred resistance to silver and Cu (McHugh et al., 1975; Gupta et al., 1999). Although the sil and cus genes are generally absent from the whole genome sequence of reference strains of Salmonella, these genes have been reported in multiple distinct monophasic S. Typhimurium and S. Rissen clones associated primarily with pigs in the past two decades (Mourao et al., 2015; Campos et al., 2016; Mastrorilli et al., 2018).
When the whole genome sequence of monophasic S. Typhimurium ST34 was compared with other S. Typhimurium whole genome sequence, an 80kb genomic island was identified and designated as Salmonella genomic island 4 [SGI-4, note addendum for nomenclature change from SGI-3 (Petrovska et al., 2016)]; this genomic island was present in over 95% of monophasic S. Typhimurium ST34 strains from the United Kingdom and Italy, but absent from a diverse collection of S. Typhimurium including DT104 (Petrovska et al., 2016). Ancestral reconstruction analysis was consistent with acquisition of SGI-4 by horizontal transfer concomitant with clonal expansion of monophasic S. Typhimurium ST34, and rare sporadic loss of the genetic island (Petrovska et al., 2016). SGI-4 contained three clusters of genes predicted to be involved in resistance to Cu and silver or arsenic metal ions (Petrovska et al., 2016).
Here we addressed the hypothesis that SGI-4 is a mobile genetic element (MGE) that encodes multiple metal ion resistance determinants that alter the growth of Salmonella in concentrations of Cu relevant to the host and farm environments. Furthermore, we test the hypothesis that transferable Cu and silver resistance genes alter expression of endogenous Cu homeostasis genes that sense Cu levels in the periplasm and that this affects SocCI-mediated survival in macrophages.
Salmonella Typhimurium strain SL1344 was isolated from a calf in 1973 as described previously (Kroger et al., 2012). Monophasic S. Typhimurium ST34 strains (S04698-09, S04332-09, L00857-09, S05092-07, S06578-07, and L0938-09), S. Typhimurium U288 strains S01960-05, S05968-02 and 11020-1996, S. Typhimurium DT104 strains (NCTC13348, 4582-1995 and S00914-05), and two strains that were closely related to strain SL1344 (9115-1996 and 6164-1997) were isolated from various host species and their whole genome sequences were determined as described previously (Mather et al., 2016; Petrovska et al., 2016). For planktonic culture, bacteria were inoculated from single colonies subcultured on LB agar plates into 10ml of growth medium and incubated at 37oC in aerobic (normal atmospheric) conditions, or microaerobic (10% CO2, 5% H2, 5% O2, and 80% N2) or anaerobic (10% CO2, 10% H2, and 80% N2) conditions using a Whitely A95 Anaerobic Workstation (don whitley scientific). Strains of S. Typhimurium or monophasic S. Typhimurium in which specific genes were replaced with either the cat gene conferring chloramphenicol resistance or the aph gene conferring resistance to kanamycin were constructed using allelic exchange methodology as described previously (Datsenko and Wanner, 2000). Briefly, pairs of oligonucleotide primers specific for amplification of the cat gene or the aph gene from plasmids pKD3 or pKD4 were synthesized with 50 nucleotide sequences on the 3′ end that were identical to sequence immediately proximal to the ATG start and distal to the stop codon of each gene targeted for deletion (Supplementary Table S1). Bacterial strains were routinely cultured in Luria Bertani broth (Oxoid) supplemented with 0.03 mg/l chloramphenicol or 0.05 mg/l kanamycin as appropriate.
A gene model for SGI-4 was constructed using Prokka (Seemann, 2014) with a minimum open reading frame (ORF) size of 100. Annotation of SGI-4 was achieved by aligning ORF sequences to those in the NCBI database using BLASTn to identify genes with the greatest sequence identity.
Phylogenetic trees from whole genome sequence data were constructed and single nucleotide polymorphisms (SNPs) identified in the whole genome sequences by aligning reads using BWA-MEM (Li, 2013), variant calling with Freebayes (Garrison and Marth, 2012) and SNP filtering using vcflib/vcftools (Danecek et al., 2011), combined as a pipeline using Snippy v3.0.1 Maximum-likelihood trees were constructed using a general time-reversible substitution model with gamma correction for amongst-site rate variation with RAxML v8.0.20 with 1000 bootstraps (Stamatakis, 2006). Representative isolates of S. Typhimurium phage types described previously (Petrovska et al., 2016), and whole genome sequences of S. Typhimurium isolates from routine clinical surveillance by Public Health England are available in public databases with accession numbers reported in Supplementary Table S2.
Candidate SGI-4-like elements (SLEs) were identified in genome assemblies in the NCBI non-redundant sequence database (accessed June 2018) by aligning to SGI-4 excluding the ars, sil, and pco loci, using discontiguous megaBLAST. This analysis identified nucleotide sequences from Edwardsiella ictaluri (accession CP001600, 326500..380800) Erwinia tracheiphila (accession CP013970, 1856073..1916737), Enterobacter cloacae (accession CP012162, 4040884..4152906), Enterobacter hormaechei (accession CP010376, 3932000..4028800), Enterobacter hormaechei (accession CP012165, 395200..536000), Pluralibacter gergoviae (accession CP009450, 1604785..1778603) and Salmonella Cubana (accession CP006055, 4214040..4311200). For reconstruction of the phylogeny of putative SLEs, sequence were aligned using clustalW-2.1 (Larkin et al., 2007) to determine sequence identity and conserved regions. A maximum likelihood tree was constructed from aligned nucleotide sequence using RAxML (Stamatakis, 2006). The relationship of SLEs was also investigated by determining the proportion of each SLE aligned by carrying out a pair-wise comparison with discontinuous megaBLAST (Morgulis et al., 2008). Each SLE was used as the query sequence against each SLE as the subject to determine the percent of sequence that aligned and the mean percent nucleotide sequence identity.
For reconstruction of a maximum likelihood tree to investigate the relationship of the SGI-4 encoded and previously described silRSECBAF and cusRSCFBA genes, sequence extracted from 17 whole genome sequences was aligned using ClustalW-2.1 (Larkin et al., 2007) and a tree constructed using 1000 bootstraps and the GTRCAT substitution matrix with RAxML-8.0.22 software (Stamatakis, 2006). The loci were from 15 Cu homeostasis and silver resistance island (CHASRI) sequences (Staehlin et al., 2016), of which nine were chromosomal and six on plasmids, as well as the originally described silRSECFBAP from S. Typhimurium plasmid pMG101, and chromosomal cusSRCFBA from E. coli K-12.
To estimate the distribution of the silA gene in 926 Salmonella whole genome sequences, one from each eBurst group (similar to serovar groups) and therefore representing the diversity of the genus (Alikhan et al., 2018), query sequence was aligned using BLASTn with an 80% coverage threshold and 2,000 maximum hits. BIGSI (Bradley et al., 2019) was used to query the presence of the silA and invA genes in over 450,000 bacterial sequence entries in the European Nucelotide Archive (ENA; accessed on March 29, 2019 with an api url search).
In order to provide a convenient selectable marker for the presence of SGI-4, we constructed a strain of monophasic S. Typhimurium S04698-09 in which the bar gene on SGI-4 was replaced by the cat gene (S04698-09 SGI-4 Δbar::cat), conferring resistance to chloramphenicol (Supplementary Table S1). To provide a selectable marker for the recipient strain, we constructed a strain of S. Typhimurium SL1344 in which the copA gene was deleted and replaced by an aph gene, conferring resistance to kanamycin (Supplementary Table S1). Donors and recipients were cultured in LB broth for 18 h at 37°C with shaking. The OD600nm of each culture was adjusted to 0.1 with fresh LB broth and 2.5 ml of each added to a 50ml tube and incubated for 18 h at 37°C with shaking in aerobic or anaerobic atmosphere, and in the presence or absence of 0.5 mg/l mitomycin C. The number of CFUs per ml of donors and recipients were quantified by culturing serial dilutions on LB agar supplemented with 0.03 mg/l chloramphenicol or 0.05 mg/l kanamycin, respectively. The presence of SGI-4 Δbar::cat in recipient strains was quantified by serial dilution on LB agar supplemented with chloramphenicol and kanamycin. Transfer frequency was defined as the number of recipients containing SGI-4 Δbar::cat as a proportion of donor cells in the culture. To determine whether transconjugant recipient strains contained SGI-4 in the same chromosomal location as the donor, the predicted right junction was amplified using primers that annealed on either side of the right junction of SGI-4 by PCR (Supplementary Table S1). To detect circularization of SGI-4 after excision, outward facing primers that annealed at the left and right junction of SGI-4 were used for PCR amplification (Supplementary Table S1). The sequence of the amplicons was determined by Sanger sequencing using the same primers (Eurofins sequencing service) and aligned to the genome of strain S04698-09.
Fifteen bacterial strains (monophasic S. Typhimurium ST34 strains S04698-09, S04332-09, L00857-09, S05092-07, S06578-07, L0938-09, and S. Typhimurium strains S01960-05, S05968-02, 11020-1996, NCTC13348, 4582-1995, S00914-05, SL1344, 9115-1996, and 6164-1997) were cultured for 18 h in LB broth at 37°C with shaking. A stock solution of 20 mM CuSO4 in LB broth and 25 mM HEPES pH7 using NaOH. Serial dilutions in LB broth (pH7) were performed to generate a range of concentrations in 1mM intervals from 1 to 20 mM CuSO4. 0.2 ml of LB broth buffered with 25mM HEPES pH7 and containing a range of concentrations of Cu sulfate were added to a polystyrene 96-well plate (Nunc) and incubated at 37°C for 24 h in normal atmospheric, microaerobic, or anaerobic conditions to equilibrate. After 24 h, each well was inoculated with 1 × 107 colony-forming units (CFUs) of the test bacterial strain cultured for 18 h in 10 ml LB Broth with shaking. Plates were incubated at 37°C for 24 h in normal atmospheric, microaerobic, or anaerobic conditions. The OD600nm of each well was measured using a BIORAD Benchmark Plus microplate spectrophotometer. The MIC was defined as the mean concentration of Cu sulfate for which the OD600nm of the culture was <0.2 from four biological replicates.
Expression relative to transcript abundance of a constitutively expressed housekeeping gene was determined as previously described (Branchu et al., 2014). Total RNA was extracted from 2 ml samples of overnight cultures of S04698-09 and S04698-09ΔsilCBA strains grown in LB broth supplemented with 20, 100, 200 μM CuSO4, or without CuSO4 supplementation to mid-log phase (OD600nm of 0.2) in anaerobic atmosphere (pcoA) or aerobic atmosphere (copA). Bacteria were harvested by centrifugation and RNA extracted using a FastRNATM spin kit for microbes (MPBio) according to the manufacturer’s instructions. RNA was treated with a TURBO DNA-freeTM kit (ambion, life technologiesTM) according to the manufacturer’s instructions before being reverse-transcribed using the QuantitTect® Reverse Transcription kit (Qiagen®). The resulting cDNA or serial dilutions of a known quantity of genomic DNA for generation of standard curves were amplified using a QuantiFast® SYBR® Green PCR kit (Qiagen®) with specific primers (Supplementary Table S1) for copA or pcoA test genes, and rpoD as a control housekeeping gene using the Applied Biosystems® 7500 real-time PCR system. The expression of copA or pcoA is presented relative to transcript abundance of the rpoD gene.
Murine macrophages (RAW 264.7, ATCC, Rockville, MD) were grown in minimum essential medium (Sigma Aldrich) supplemented with 10% fetal bovine serum, L-glutamine (2 mM), and 1 × nonessential amino acids. For infection studies, 2 × 105 RAW 264.7 cells were seeded per well into 24-well plates and incubated at 37°C for 48 h. Cells were infected with S04698-09 wild-type strain, S04698-09ΔsodCI, S04698-09ΔsilCBApco or S04698-09ΔsodCIΔsilCBApco from an overnight culture in LB broth at a multiplicity of infection of 20. Plates were centrifuged at 1500 rpm for 3 min followed by incubation of the cells for 30 min. The medium was then exchanged for a fresh one of the same compositions with the exception of the addition of gentamicin (100 μg/ml) to kill the extracellular bacteria. After either 2 or 24 h, the cells were washed with PBS and lysed with 1% Triton in PBS. The number of bacterial CFUs was determined by culturing lysate serial dilutions on LB agar. The data were expressed as the proportion of CFUs at 24 h relative to the CFU determined at 2 h.
Where indicated in each figure with lines and asterisks, the Mann-Whitney U test was used to test the null hypothesis that randomly selected values in a sample were equally likely to be greater or smaller than from a second sample, using an alpha level of 0.05 to reject the null hypothesis.
We investigated the presence of candidate genes within the SGI-4 coding for proteins capable of enabling mobilization of SGI-4 to recipient bacteria. The DNA excision, DNA processing, and conjugative transfer mechanisms we considered are common to MGEs such as ICE, also known as conjugative self-transmissible integrative (CONSTIN) elements (Hochhut and Waldor, 1999), integrative mobilizable elements (IMEs), and cis-mobilizable elements (CIMEs; Wozniak and Waldor, 2010). A number of ORFs that exhibited sequence similarity to DNA processing enzymes were identified on SGI-4 by sequence alignment to available databases (Figure 1A and Supplementary Table S3). Sequences of two of these exhibited similarity to site-specific tyrosine recombinases xerC and xerD, which are predicted to mediate integration and excision. However, we were unable to identify a recombination direction factor (RDF), which are known to exhibit little sequence conservation. ORF86 had sequence similarity to traI that encodes a relaxase protein. This relaxase protein is capable of binding to dsDNA at the origin of transfer (oriT) and inducing a single strand nick in conjunction with putative uvrB DNA helicase (ORF85) and topB topoisomerase (ORF 13), this facilitates unwinding of the DNA helix (Salyers et al., 1995). At the left end of SGI-4, parA and parB orthologs (ORFs 1 and 5) encode putative chromosome partitioning proteins. SGI-4 also contained a number of ORFs with similarity to genes encoding components of type IV secretion systems (T4SSs) involved in conjugative transfer of DNA: ORF26 of SGI-4 encoded a putative type IV coupling protein (T4CP), TraD, which initiates conjugal transfer; ORF51 encodes a putative inner membrane protein, TraG, for T4SS stabilization; ORF53 encodes a TraU ortholog involved in pilus assembly; ORF19 encodes a PilL F-type pilus protein; and ORF45 encoding an F-pilus assembly protein. These ORFS were present within several apparent operons (ORFs 14–27, 37–46, 48–53), consisting of multiple genes encoding proteins with no significant similarity to proteins in available databases but often associated with MGEs (Figure 1A and Supplementary Table S3). We were unable to identify a putative oriT sequence in SGI-4 using oriTfinder (Li et al., 2018). However, two pairs of inverted repeats, each with 80% identity to one another, were present in ORF19 and in an intergenic region between ORF64 and ORF65.
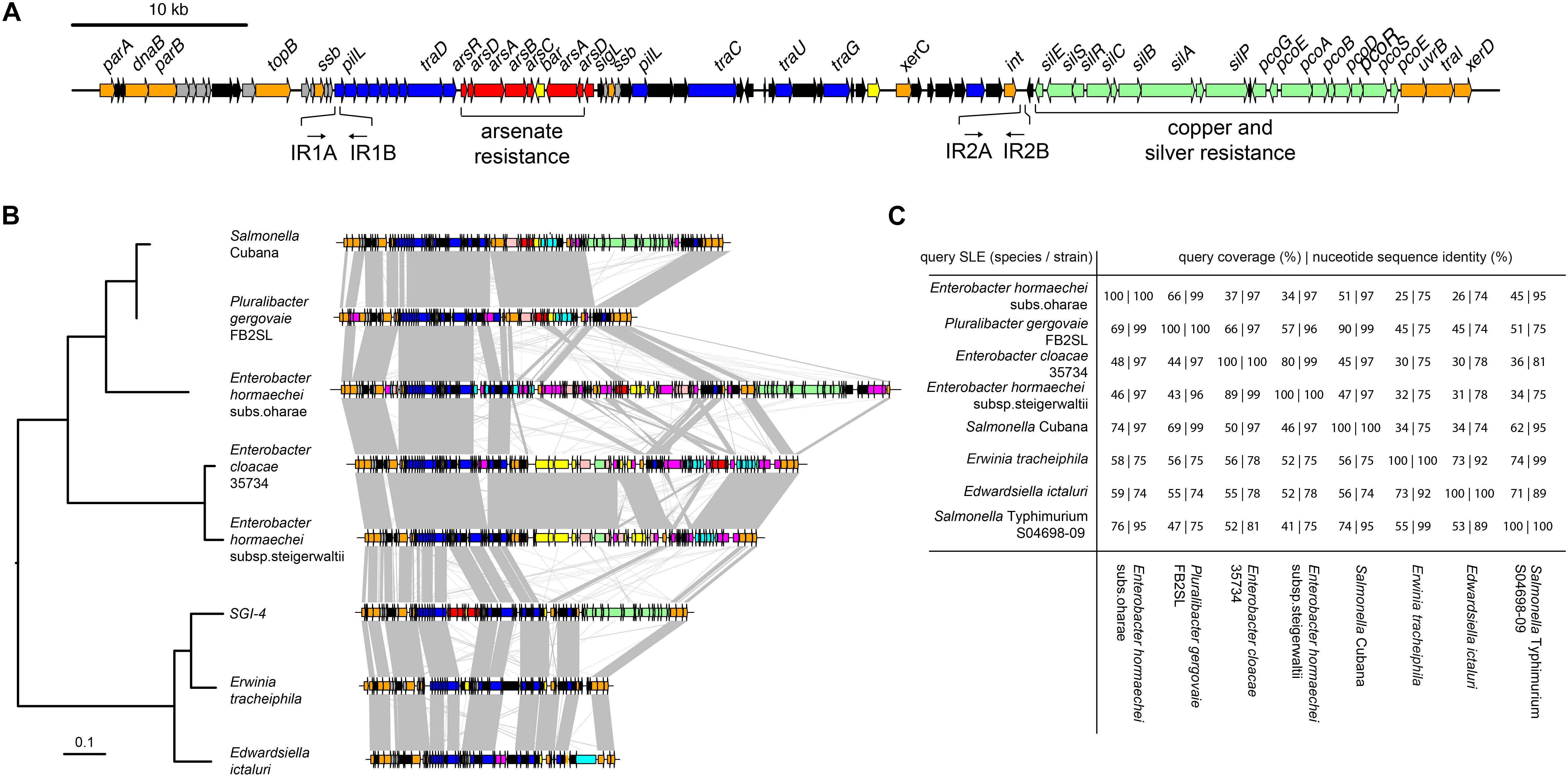
Figure 1. Genetic map of Salmonella Genomic Island 4 (SGI-4) and phylogenetic relationship and gene synteny with SLEs. (A) Genetic diagram of Salmonella Genomic Island 4 (SGI-4); filled arrows indicate open reading frames with predicted functions based on sequence similarity for integration, excision and DNA processing (orange), type 4 secretion systems and conjugal transfer (blue), Cu and silver resistance (green), arsenic resistance (red), genes of unknown function commonly associated with ICE (gray), genes of unknown function not commonly associated with ICE (black). (B) Mid-point rooted maximum likelihood tree of eight SGI-4-like elements (SLEs) constructed using sequence variation in the core nucleotide sequence alignment of SLEs from diverse Enterobacteriaceae and genetic diagram of SLEs with regions of >75% nucleotide sequence identity (gray shading); filled arrows indicate open reading frames with the same designations as (A), with additional functions, cadmium, cobalt, zinc or mercury resistance (pink), transposase and insertion elements (purple), and a repressor protein lexA (brown). (C) Pair-wise sequence comparison of SLEs indicating the percentage of sequence exhibiting >60% identity and the mean nucleotide sequence identity of the shared sequence.
To determine whether SGI-4 was similar to known MGEs we aligned the sequence of SGI-4 with non-redundant nucleotide sequences in the NCBI database using discontinuous megaBLAST. Seven assembled contiguous sequences in the NCBI database contained a core set of ICE genes with a modular arrangement with complete synteny to SGI-4, none of which had been described previously as MGEs (Figure 1B). These exhibited at least 75% nucleotide sequence identity with over 40% of SGI-4, and we therefore refer to these as SGI-4-like elements (SLEs). SLEs were inserted in the genome adjacent to a phenylalanine phe-tRNA in strains from a diverse range of Enterobacteriaceae, indicating this was the common attachment site (attB/attP). SGI-4 was most closely related to SLEs from Erwinia tracheiphila and Edwardsiella ictaluri, and more distantly related to SLEs from two Enterobacter hormachei strains from subspecies oharae and steigerwaltii, Enterobacter cloacae, Pluralibacter gergoviae and an SLE present in a strain of Salmonella enterica serovar Cubana (Figures 1B,C). All shared a number of regions of at least 75% sequence identity and synteny that encoded putative DNA processing enzymes or components of a T4SS, and were interspersed amongst apparent cargo genes involved in diverse functions capable of modifying the phenotype of the host bacterium.
Since SGI-4 encoded many of the genes normally associated with ICE, we determined whether it was capable of mobilization to a recipient S. Typhimurium (strain SL1344) during co-culture. To enable identification of recipients containing SGI-4 we inserted a chloramphenicol resistance gene (cat) in the arsenic resistance locus of SGI-4. Transfer frequency was low under aerobic culture conditions (<1 × 10-8 CFU per donor CFU) but was substantially increased in the presence of mitomycin C, as described previously for the SXT ICE (Beaber et al., 2004), or by culture in anaerobic conditions (Figure 2). Mitomycin C and anaerobiosis had an additive effect on transfer since the transfer rate in anaerobic conditions with mitomycin C was significantly higher (p < 0.005) than any other condition tested (∼1 × 10-4 CFU per donor CFU). PCR amplification and sequence analysis of the junction site of the donor strain indicated that, on transfer, SGI-4 inserts into the same position on the chromosome (phe-tRNA locus) as the donor strain (data not shown). Furthermore, in the monophasic S. Typhimurium strain S04698-09 cultured in the presence of mitomycin C, PCR amplification and sequence analysis of an amplicon generated using specific outward facing primers at each end of SGI-4, was consistent with circularization of excised SGI-4 (data not shown).
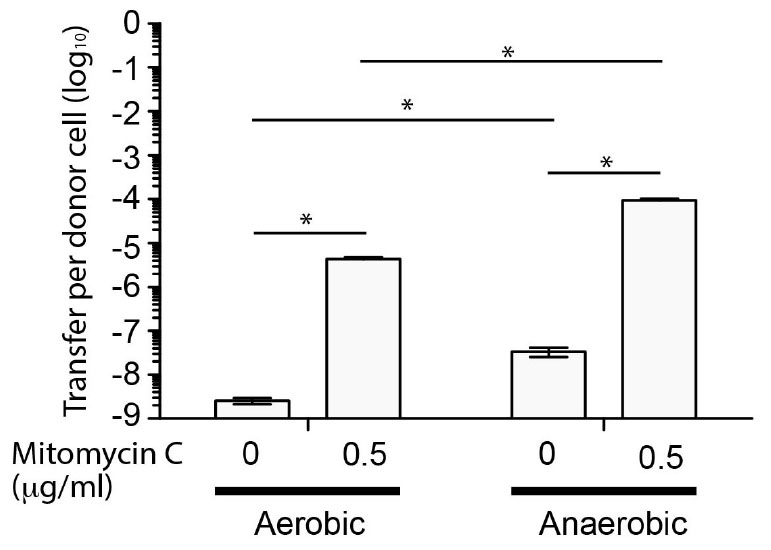
Figure 2. Transfer frequency of SGI-4::cat to S. Typhimurium during co-culture is enhanced by mitomycin C and anaerobic conditions. Bars indicate the mean number of chloramphenicol resistant CFU of S. Typhimurium strain SL1344 per CFU of monophasic S. Typhimurium ST34 strain S04698-09 SGI-4 silCBA::cat, following co-culture. The mean from six biological replicates ± standard deviation are shown. ∗Indictaes that groups indicated by lines were significantly different (p < 0.05).
The SGI-4 sequence was present in 23 of 24 monophasic S. Typhimurium ST34 clade strains, but absent from all other strains, in a collection of representative S. Typhimurium (Figure 3). SGI-4 sequence was also present almost exclusively in isolates of the monophasic S. Typhimurium ST34 clade in the whole genome sequence of 1697 S. Typhimurium and monophasic variant S. Typhimurium isolates from human clinical cases in England and Wales during 2014 and 2015 (Supplementary Figure S1). Within the monophasic S. Typhimurium ST34 clade just 38 isolates (4%) lacked the SGI-4 sequence, and these were distributed sporadically throughout the clade in 28 small clusters or individual leaves of the phylogenetic tree (Supplementary Figure S1). In contrast, of 797 S. Typhimurium isolates that were present outside the monophasic S. Typhimurium ST34 clade, just five contained the SGI-4 sequence, and these all shared a recent common ancestor with the monophasic S. Typhimurium ST34 clade.
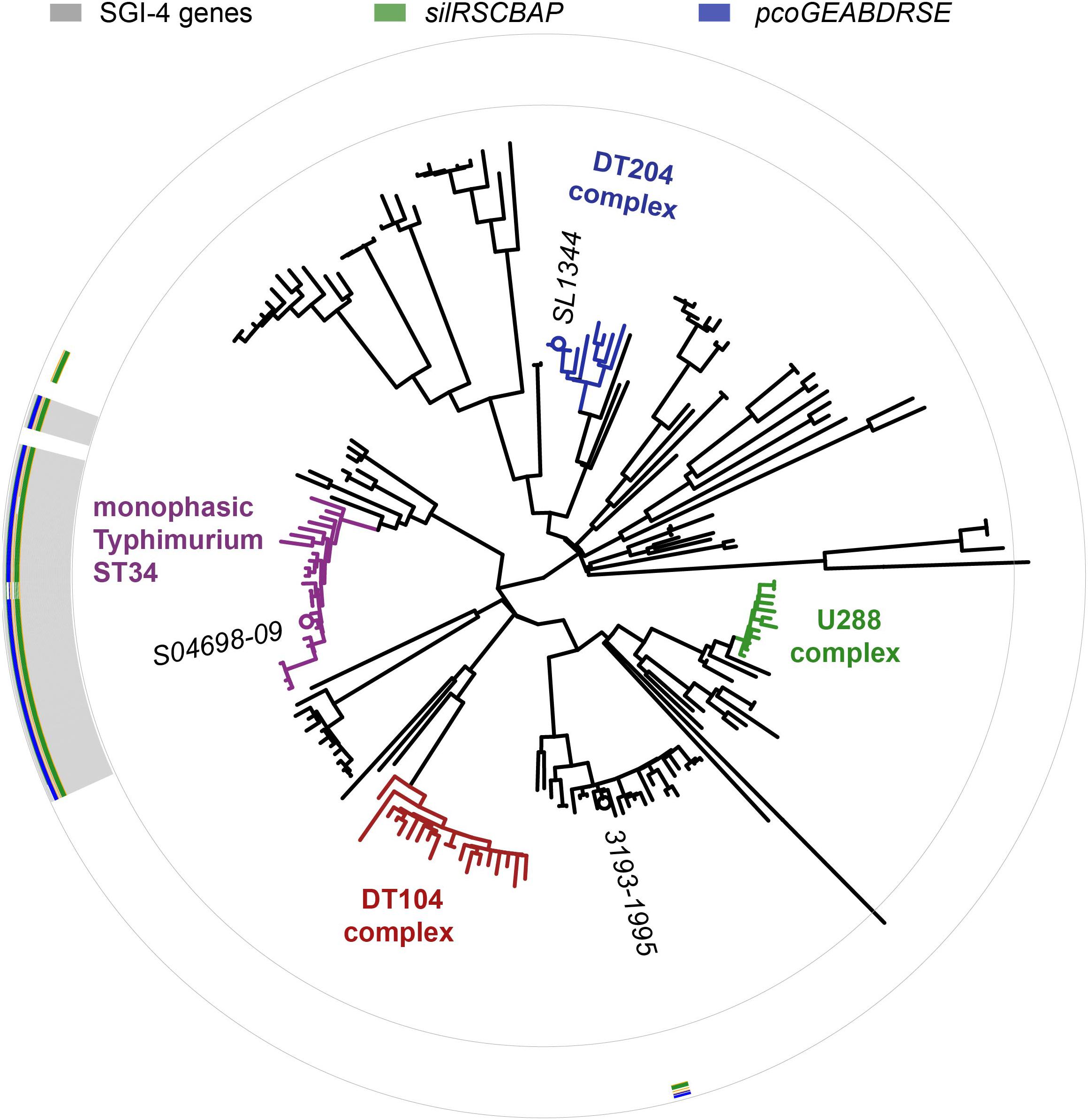
Figure 3. Distribution of SGI-4 in monophasic S. Typhimurium ST34 and representative strains of S. Typhimurium. A maximum likelihood tree constructed using sequence variation in the core genome of 24 monophasic S. Typhimurium ST34 strains and 153 representative strains of S. Typhimurium with reference to the whole genome sequence of strain SL1344 (accession FQ312003). Selected strains are identified (italicized text). The presence of sequences that mapped to 87 SGI-4 ORFs from S04698-09 are represented as filled boxes in 87 concentric circles: sil genes (green), pco genes (blue), or other SGI-4 ORFs (gray).
The nomenclature for copper RND-family efflux pump genes is confusing with sil and cus being used without reference to ancestry. A cluster of 18 ORFS on SGI-4 included 15 genes that exhibited sequence similarity to genes predicted to encode an RND-family efflux pump previously designated as either cusRS cusCFBA (Outten et al., 2001) or silRSE silCBAP (Gupta et al., 1999), and pcoABDRSE pcoEG (pco locus) involved in Cu and silver resistance. Alignment of the RND-family efflux pump genes of SGI-4 with sequences from previously characterized homologs indicated that the cusRSCFBA genes on the E. coli chromosome form a distinct outgroup from a closely related cluster that included the SGI-4 genes, that evolved from a common ancestor closely related to the silRSE silCBAP on pMG101 (Supplementary Figure S2). For this reason, we designated SGI-4 encoded RND-family efflux pump genes as silRSE silCBAP (sil locus).
Investigation of the distribution of the SGI-4 silA genes in two sequence databases indicated that these genes are rare in the genus Salmonella, consistent with deletion early in the evolution of the genus, and infrequent reacquisition. Alignment using BLASTn of the silA gene on SGI-4 with 926 Salmonella genomes, one from each eBurst group (largely corresponding serovar) representing all known genotypic diversity of Salmonella (Alikhan et al., 2018), indicated that just 16 genomes (2%) contained an ortholog. In a second search of the ENA, the silA gene was in 3439 sequence entries, while the invA gene in 51026 data entries, indicating that silA was present in approximately 7% of S. Typhimurium genomes in the database.
In order to investigate the impact of the acquisition of SGI-4 on metal ion resistance, we compared the MICs of Cu sulfate for five strains of monophasic S. Typhimurium ST34 with three strains each of DT204, U288 and DT104 that lacked SGI-4, and a single monophasic S. Typhimurium ST34 strain that also lacked SGI-4 due to deletion (Figure 4). During culture in aerobic or microaerobic conditions the presence of SGI-4 had a small but significant impact on the MIC for Cu. However, under anaerobic conditions, the MICs for strains lacking SGI-4 decreased by around five-fold; in contrast, SGI-4-containing monophasic S. Typhimurium ST34 strains had similar MICs under both aerobic and microaerobic conditions.
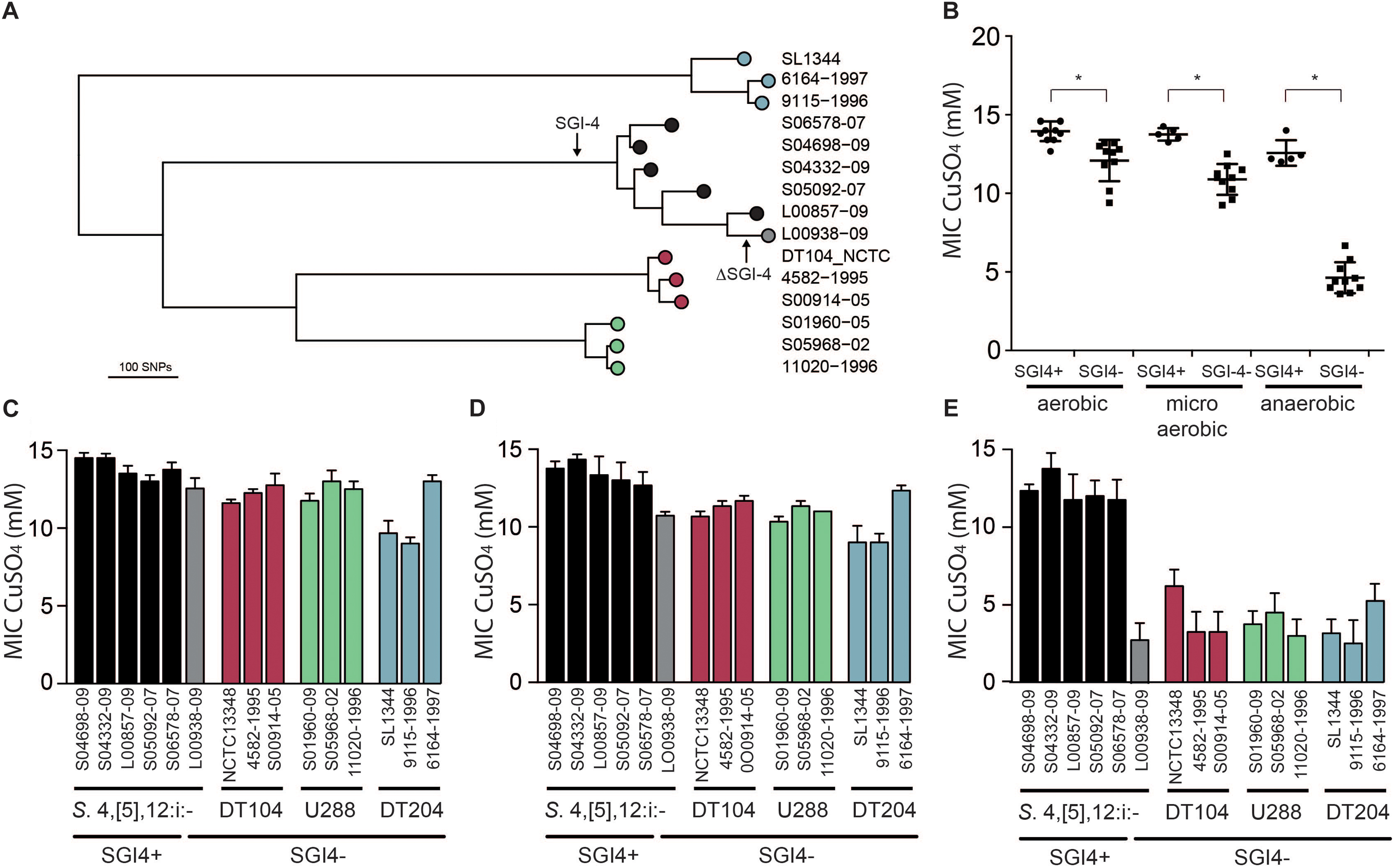
Figure 4. Phylogenetic relationship and Cu sulfate MIC of representative strains of monophasic S. Typhimurium ST34 and S. Typhimurium. (A) A mid-point rooted phylogenetic tree constructed using sequence variation in the core genome with reference to S. Typhimurium strain SL1344 whole genome sequence (accession FQ312003). Leaves of the tree corresponding to representative strains are labeled as filled circles color coded for SGI-4+ monophasic S. Typhimurium ST34 (black) and SGI-4- monophasic S. Typhimurium ST34 (gray), and S. Typhimurium DT104 complex (red), U288 complex (green), and DT204 complex (blue) are shown. (B) Mean MIC for Cu sulfate of SGI-4+ (filled circles) and SGI-4- (filled squares) monophasic S. Typhimurium ST34 and S. Typhimurium strains during aerobic, microaerobic and anaerobic culture. Bars indicate the mean MIC for Cu sulfate for each strain ± standard deviation in aerobic (C), microaerobic (D), and anaerobic (E) atmosphere. Bar colors match the tree leaf circles in (A). ∗Indicates that groups indicated by lines were significantly different (p < 0.05).
SGI-4 has two clusters of genes predicted to encode an RND family efflux pump (silCBA), and a second that includes a multicopper oxidase system (pcoABDE), both previously implicated in resistance to Cu. To determine the relative role of these two loci in Cu resistance we determined the MICs of mutants of monophasic S. Typhimurium ST34 strain S04698-09 that had deletions of either silCBA, pcoABC or both these loci (Figure 5). When functional silCBA genes were present, deletion of pcoABDE genes alone had no effect on resistance to Cu under any of the conditions evaluated. Deletion of silCBA genes resulted in a small but significant (p < 0.05) decrease in the MIC for Cu under aerobic and microaerobic conditions, and a more substantial decrease under anaerobic conditions. However, even under anaerobic conditions, the effect of the silCBA deletion on the MIC for Cu was not comparable to the absence of SGI-4 in U288, DT104 and DT204 isolates which had a MIC that was below 5 mM Cu sulfate. Cu resistance at a similar level to these strains was only observed in monophasic S. Typhimurium ST34 strain S04698-09 when both silCBA and pcoABDE were deleted.
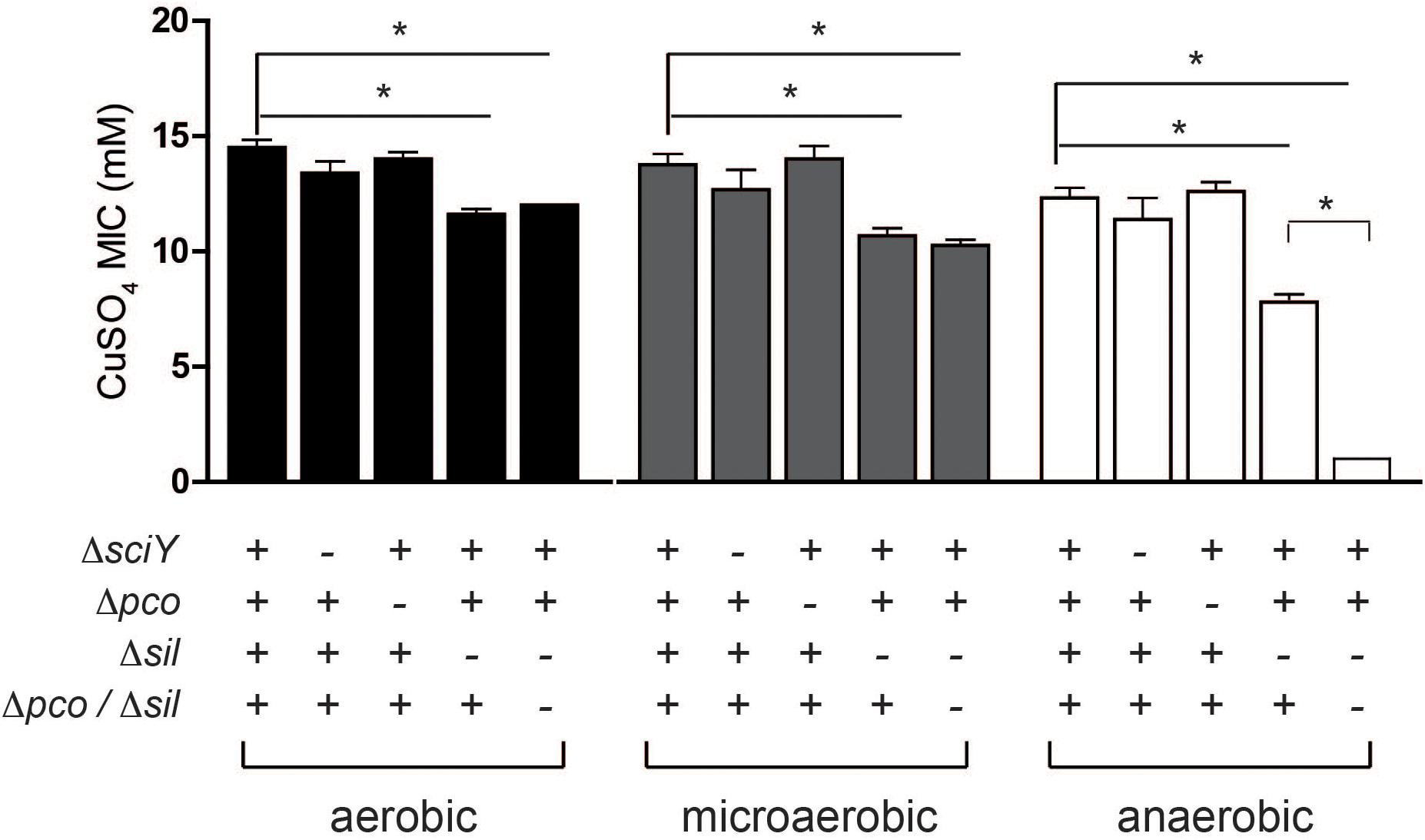
Figure 5. MIC for Cu sulfate of monophasic S. Typhimurium ST34 strain S04698-09 containing defined deletions in sil and pco genes. Bars indicate the mean MIC ± standard deviation of four biological replicates during culture in either aerobic, microaerobic or anaerobic conditions. The genotype with respect to sciY (negative control), pco and sil are indicated as present (+) or deleted and replaced with a cat gene (–). ∗ indicates the MIC was significantly different from wild type strain S04698-09 (p < 0.05).
To investigate any apparent redundancy of pcoABDE in the presence of silCBA, we determined the expression of pcoA in the presence or absence of silCBA. The expression of pcoA was not affected by increasing concentrations of Cu, unless the silCBA genes were deleted from monophasic S. Typhimurium ST34 strain S04698-09 (Figure 6A). In the absence of silCBA induction was around ten-fold that achieved in the absence of Cu in the growth medium.
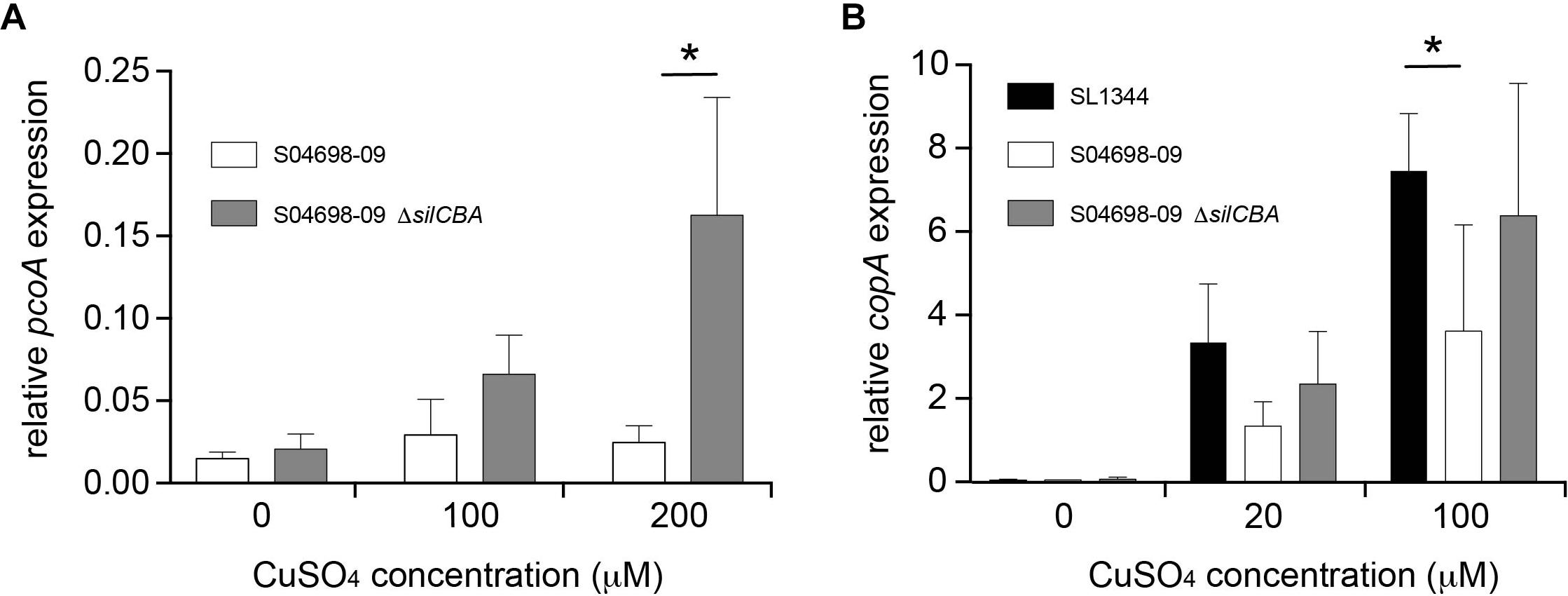
Figure 6. Presence of the silCBA genes decreases relative transcript abundance of pcoA and copA. The abundance of pcoA (A) or copA (B) transcript relative to the housekeeping gene rpoD quantified by quantitative RT-PCR from total RNA prepared from mid-log phase cultures in LB broth supplemented with Cu sulfate. Bars indicate mean relative transcript from five biological replicates ± standard deviation. ∗Indicates that groups indicated by lines were significantly different (p < 0.05).
To determine the impact of SGI-4 acquisition on typical Cu homeostasis in S. Typhimurium, we investigated the expression of the copA gene that encodes the native ATPase protein involved in transport of Cu from the cytoplasm into the periplasm in Salmonella. The expression of copA increased in a dose-dependent manner in the presence of Cu in S. Typhimurium strain SL1344, which lacked SGI-4, and in the monophasic S. Typhimurium ST34 strain S04698-09 that encoded the island. However, expression of copA was significantly higher in S. Typhimurium strain SL1344 than in monophasic S. Typhimurium ST34 strain S04698-09 during culture in media with 100 μM Cu. Deletion of cusACFBA genes in strain S04698-09 resulted in increased expression of copA approaching the expression level observed in SL1344 in 100 μM of CuSO4 (Figure 6B).
We then tested the hypothesis that the presence of silCBA affected the function of SodCI, a periplasmic superoxide dismutase that uses Cu as co-factor that is supplied by the CopA ATPase Cu transporter and the periplasmic Cu binding protein CueP (Osman et al., 2013). SodCI dismutates oxygen free radicals produced by phagosome associated NADPH-dependent oxidase (Phox) of macrophages, contributing to intracellular survival of Salmonella. We expected that the expression of silCBA that encoded a copper RND efflux pump on SGI-4 would result in transport of Cu from the cytoplasm and periplasm to the external milieu (Gupta, 1994), depleting the pool of Cu available in the periplasm. Consistent with this, expression of pcoA, a periplasmic multicopper oxidase gene whose expression is controlled by PcoR that senses copper in the periplasm, was not induced during culture in 100 μM Cu, unless the silCBA was deleted from SGI-4.
To test the hypothesis we infected gamma interferon-activated RAW macrophages with mutants of monophasic S. Typhimurium ST34 strain S04698-09 that encoded either silCBA and sodCI, or lacked one or both of these loci and determined the change in intracellular viable counts of S. Typhimurium in macrophages between 2 and 24 h post-inoculation (Figure 7). Strain S04698-09 exhibited a net replication of nearly ten-fold after 24 h in RAW macrophages, and deletion of the sodCI gene resulted in a small but significant (p < 0.05) reduction. This was consistent with previous reports that SodCI is required by the bacterium to evade the killing mechanisms of macrophages. However, deletion of the silCBA genes did not result in a significant decrease in net replication in RAW macrophages, but additional deletion of sodCI resulted in a similar decrease in net replication to that observed in the presence of the silCBA genes. These data are consistent with a fully functional SodCI gene in the presence of altered Cu homeostasis due to the presence of the silCBA genes.
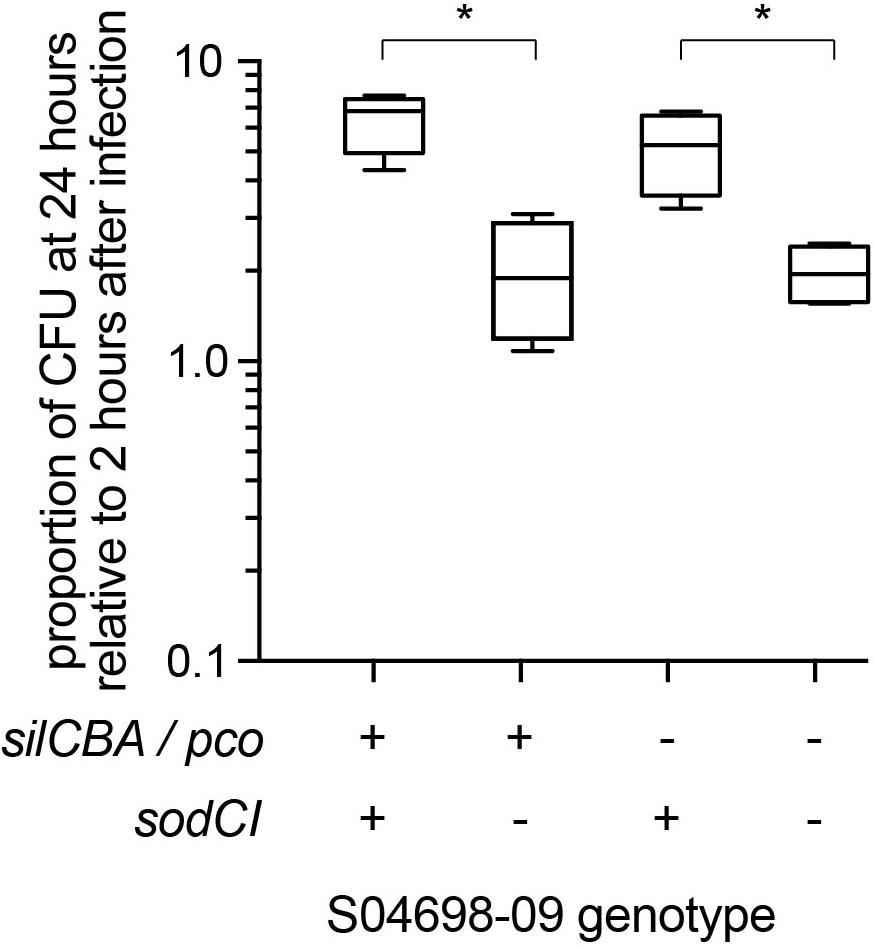
Figure 7. Net replication of monophasic S. Typhimurium ST34 in activated RAW macrophages is sodCI dependent but sil / pco independent. Box and whiskers plot indicating the mean number of bacterial CFUs 24 h post-infection, relative to 2 h post infection of wild type monophasic S. Typhimurium strain S04698-09 and otherwise isogenic strains containing deletions of the sodCI gene, the sil/pco genes together, or all three loci deleted. ∗Indicates that groups indicated by lines were significantly different (p < 0.05).
For vertical transfer ICE undergo replication within the chromosome; for horizontal transfer they undergo excision from the genome (Johnson and Grossman, 2015). Horizontal transfer of an ICE involves common processes including excision from the host chromosome, circularization, conjugative transfer by a type IV secretion system (T4SS), and integration within a recipient genome at an attachment site (attB; Guglielmini et al., 2011). A number of ORFs exhibiting sequence similarity to genes encoding DNA processing enzymes that are predicted to be involved in excision, integration and conjugative transfer of DNA via a type IV secretion system (T4SSs) were also present on SGI-4. Together these data were consistent with the idea that SGI-4 is an ICE.
In an analysis of 1,124 taxonomically diverse complete prokaryotic genomes, 335 putative ICEs were detected based on the presence of T4SS and a relaxase gene in close proximity, indicating the widespread nature of these MGEs (Guglielmini et al., 2011). Despite the presence of multiple genes with sequence similarity to those with functions required for the transfer of ICEs, SGI-4 did not exhibit extensive similarity to previously characterized MGEs, and is currently not present in the ICEberg database (Liu et al., 2018). Seven complete genomes from diverse species of Enterobacteriaceae that were added to available databases since the analysis by Guglielmini et al. (2011), had between 75 and 99% nucleotide sequence identity with SGI-4 and between 41 and 76% shared core sets of genes with SGI-4. As is common for ICEs, core genes were interspersed with cargo genes with functions that were unrelated to mobilization but had the potential to modify the phenotype of the host bacterium (Wozniak and Waldor, 2010). In particular, genes involved in resistance to heavy metals were present on SLEs, with five containing at least one locus associated with resistance to Cu, silver, mercury or arsenic. Two SLEs from Erwinia tracheiphila and Edwardsiella ictaluri were particularly closely related to SGI-4, but contained unrelated cargo genes, highlighting the rapid divergence achieved by horizontal gene transfer in this family of ICE. Our analysis is consistent with SGI-4 evolving from a common ancestor of these ICEs by acquisition of ars, sil and pco cargo genes involved in resistance to metal ions.
With the exception of a small outbreak in a burns unit associated with a S. Typhimurium strain in the 1970s, historically, the sil and pco loci encoding resistance to silver and Cu, have rarely been associated with Salmonella enterica isolates. Indeed, we found that sil genes were present in just 2% of whole genome sequences representing the genotypic diversity of S. enterica and S. bongori, and 7% of more than 50,000 Salmonella whole genome sequences in the ENA. However, sil and pco genes, have been commonly associated with three lineages of monophasic S. Typhimurium in the past 20 years: the “European clone” (monophasic S. Typhimurium ST34); the “Spanish clone”; and the “Southern European clone” (Mourao et al., 2015; Mastrorilli et al., 2018). However, only 74 and 26% of the latter two, respectively, encoded the sil and/or the pco genes and these were plasmid-borne (Mourao et al., 2015), suggesting that they may be lost relatively frequently. In contrast, monophasic S. Typhimurium ST34 encodes sil and pco genes on the chromosome, and although on a MGE, we found these genes in 96% of the 797 clinical isolates from England and Wales that we evaluated. Furthermore, loss of SGI-4 and the silCBA and pco genes was sporadic, and mostly as singleton taxa on the phylogenetic tree, consistent with the absence of selection for their loss (Petrovska et al., 2016).
Given the general paucity of genes encoding Cu/silver RND efflux pumps in Salmonella enterica and the evolutionary history of the genus, the apparent lack of a selective advantage for the loss of silCBA genes may indicate an important role for these genes in the monophasic S. Typhimurium ST34 clone. Deletion of cusCFBA in the Salmonella ancestor is likely to have had a profound effect on Cu distribution in the cell because Salmonella has no alternative mechanism to remove Cu from cell. While E. coli can remove Cu from the cell entirely, Salmonella transports Cu from the cytoplasm, where it is especially toxic, to the periplasm, where it is detoxified by the Cu oxidase CueO or bound to the Cu binding protein, CueP, the major reservoir of Cu in Salmonella (Osman et al., 2010). CueP is thought to play an important role in supplying Cu to superoxide dismutase SodCI, important for survival in the face of the oxidative burst generated by macrophage. The evolution of Salmonella pathogenesis must, therefore, have proceeded in the context of fundamentally altered Cu homeostasis. The reintroduction of the silCBA locus on SGI-4 did indeed appear to alter the Cu distribution and levels in Salmonella as indicated by the lack of pcoA expression in the presence of the RND-family of efflux genes, which are regulated in response to Cu levels in the periplasm. The consequences of altered Cu levels in monophasic S. Typhimurium ST34 is not clear but, at least in RAW macrophages in vitro, the dependency of SodCI Cu as a cofactor did not affect resistance to macrophage killing. This is consistent with the report that limitation of Cu in the periplasm by deletion of the copA and golT genes, and the supply of Cu to SodCI by deletion of the cueP gene in S. Typhimurium had no effect on systemic infection in the murine model of infection (Fenlon and Slauch, 2017).
Strong selection pressure may be important for retention of SGI-4 since our data is consistent with its presence altering Cu distribution and levels. Monophasic S. Typhimurium ST34 is primarily associated with pigs, and it has been suggested that the success of this clone may, in part, have been driven by the extensive use of Cu as a growth promotor in pig rearing (Mourao et al., 2015; Petrovska et al., 2016). Consistent with this idea, the presence of SGI-4 is correlated with enhanced resistance to Cu; this appears to be particularly apparent under anaerobic conditions which are similar to those encountered by Salmonella in the host intestinal tract. Salmonella strains that lacked SGI-4, exhibited MICs for Cu of around 2–3 mM under anaerobic conditions, which is approximately 15% of the values achieved under aerobic conditions. Importantly, this is in the range of the concentrations of Cu found in pig manure effluent and sludge on farms where Cu-supplemention of diet was common (Nicholson et al., 1999; Bolan et al., 2003; Cang et al., 2004). The presence of SGI-4, however, increased the MIC for Cu (under anaerobic conditions) by approximately 500%, elevating it to levels above the concentrations likely to be encountered on pig farms (Nicholson et al., 1999; Bolan et al., 2003; Cang et al., 2004). The sil genes encoded on SGI-4 were entirely responsible for the observed increase in MICs for Cu, since deletion of pco genes alone did not result in a decrease in the MICs. The pco locus encodes a multiCu oxidase which is thought to detoxify the more damaging Cu+ to Cu2+ ions by oxidation (Lee et al., 2002). Monophasic S. Typhimurium ST34 encodes a native Cu oxidase, CueO, and the presence of this protein may, in part, mask the activity of the pco locus. However, the pco genes were capable of enhancing resistance to Cu in the absence of silCFBA. Furthermore, since there was no evidence for loss of pco genes during clonal expansion of the monophasic S. Typhimurium ST34 clade, it remains possible that the Pco system may play a more prominent role in Cu resistance under environmental conditions that we did not evaluate in our in vitro experiments.
RK, PB, and OC conceived the study. PB and OC performed the experiments and generated the data. TD and LP provided materials and expert advice. RK, PB, and OC drafted the manuscript. All authors analyzed the data, provided critical input into the final manuscript, and approved the final version.
RK was supported by research grants BB/N007964/1 and BB/M025489/1, and by the BBSRC Institute Strategic Programme Microbes in the Food Chain BB/R012504/1 and its constituent project(s) BBS/E/F/000PR10348 and BBS/E/F/000PR10349. OC was supported by a BBSRC DTP studentship (BB/M011216/1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This manuscript has been released as a Pre-Print at bioRxiv (Branchu et al., 2019).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01118/full#supplementary-material
FIGURE S1 | Distribution of SGI-4 in monophasic S. Typhimurium ST34 and representative strains of S. Typhimurium. A maximum likelihood tree constructed using sequence variation in the core genome of 1814 monophasic S. Typhimurium ST34 strains or S. Typhimurium isolates with reference to the whole genome sequence of strain SL1344 (accession FQ312003). The presence of sequence in each genome that mapped to 787 SGI-4 ORFs from S04698-09 are represented as filled boxes in 87 concentric circles: sil genes (green), pco genes (blue), or other SGI-4 ORFs (gray).
FIGURE S2 | A maximum likelihood tree showing the relationship of cus and sil loci.
TABLE S1 | Oligonucleotide primers used for the construction of mutant strains of Salmonella, determination of genotype, or qRT-PCR.
TABLE S2 | Isolate name, accession number and the figure(s) in which the data were used to construct maximum likelihood trees.
TABLE S3 | Gene model for SGI-4 including gene designation, annotation, SLE core genome and nucleotide sequence of predicted coding sequences.
Alikhan, N. F., Zhou, Z., Sergeant, M. J., and Achtman, M. (2018). A genomic overview of the population structure of Salmonella. PLoS Genet. 14:e1007261. doi: 10.1371/journal.pgen.1007261
Andres-Barranco, S., Vico, J. P., Marin, C. M., Herrera-Leon, S., and Mainar-Jaime, R. C. (2016). Characterization of Salmonella enterica serovar typhimurium isolates from pigs and pig environment-related sources and evidence of new circulating monophasic strains in Spain. J. Food Prot. 79, 407–412. doi: 10.4315/0362-028X.JFP-15-430
Animal and Plant Health Agency [APHA] (2017). Salmonella in livestock production in Great Britain, 2016. Available at: https://www.gov.uk/government/publications/salmonella-in-livestock-production-in-great-britain-2016 (accessed October 31, 2017).
Antunes, P., Mourao, J., Pestana, N., and Peixe, L. (2011). Leakage of emerging clinically relevant multidrug-resistant Salmonella clones from pig farms. J. Antimicrob. Chemother. 66, 2028–2032. doi: 10.1093/jac/dkr228
Arguello, H., Sorensen, G., Carvajal, A., Baggesen, D. L., Rubio, P., and Pedersen, K. (2013). Prevalence, serotypes and resistance patterns of Salmonella in Danish pig production. Res. Vet. Sci. 95, 334–342. doi: 10.1016/j.rvsc.2013.04.001
Beaber, J. W., Hochhut, B., and Waldor, M. K. (2004). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72.
Bolan, N. S., Khan, M. A., Donaldson, J., Adriano, D. C., and Matthew, C. (2003). Distribution and bioavailability of copper in farm effluent. Sci. Total Environ. 309, 225–236. doi: 10.1016/S0048-9697(03)00052-4
Bonardi, S. (2017). Salmonella in the pork production chain and its impact on human health in the European Union. Epidemiol. Infect. 145, 1513–1526. doi: 10.1017/S095026881700036X
Bradley, P., den Bakker, H. C., Rocha, E. P. C., McVean, G., and Iqbal, Z. (2019). Ultrafast search of all deposited bacterial and viral genomic data. Nat. Biotechnol. 37, 152–159. doi: 10.1038/s41587-018-0010-1
Branchu, P., Bawn, M., and Kingsley, R. A. (2018). Genome variation and molecular epidemiology of Salmonella Typhimurium pathovariants. Infect. Immun. 86:e0079-18. doi: 10.1128/IAI.00079-18
Branchu, P., Charity, O. J., Bawn, M., Thilliez, G., Dallman, T. J., Petrovska, L., et al. (2019). SGI-4 in monophasic Salmonella Typhimurium ST34 is a novel ICE that enhances resistance to copper. bioRxiv doi: 10.1101/518175
Branchu, P., Matrat, S., Vareille, M., Garrivier, A., Durand, A., Crepin, S., et al. (2014). NsrR, GadE, and GadX interplay in repressing expression of the Escherichia coli O157:H7 LEE pathogenicity island in response to nitric oxide. PLoS Pathog 10:e1003874. doi: 10.1371/journal.ppat.1003874
Brown, N. L., Barrett, S. R., Camakaris, J., Lee, B. T., and Rouch, D. A. (1995). Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 17, 1153–1166.
Campos, J., Mourao, J., Marcal, S., Machado, J., Novais, C., Peixe, L., et al. (2016). Clinical Salmonella Typhimurium ST34 with metal tolerance genes and an IncHI2 plasmid carrying oqxAB-aac(6′)-Ib-cr from Europe. J. Antimicrob. Chemother. 71, 843–845. doi: 10.1093/jac/dkv409
Cang, L., Wang, Y., Zhou, D., and Dong, Y. (2004). Heavy metals pollution in poultry and livestock feeds and manures under intensive farming in Jiangsu Province, China. J. Environ. Sci. 16, 371–374.
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645.
Espariz, M., Checa, S. K., Audero, M. E., Pontel, L. B., and Soncini, F. C. (2007). Dissecting the Salmonella response to copper. Microbiology 153(Pt 9),2989–2997. doi: 10.1099/mic.0.2007/006536-0
Fenlon, L. A., and Slauch, J. M. (2017). Cytoplasmic copper detoxification in Salmonella can contribute to sodc metalation but is dispensable during systemic infection. J. Bacteriol. 199:e00437-17. doi: 10.1128/JB.00437-17
Fookes, M., Schroeder, G. N., Langridge, G. C., Blondel, C. J., Mammina, C., Connor, T. R., et al. (2011). Salmonella bongori provides insights into the evolution of the Salmonellae. PLoS Pathog. 7:e1002191. doi: 10.1371/journal.ppat.1002191
Franke, S., Grass, G., Rensing, C., and Nies, D. H. (2003). Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185, 3804–3812.
Garrison, E., and Marth, G. (2012). Haplotype-based variant detection from short-read sequencing. arXiv
Grass, G., and Rensing, C. (2001). CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 286, 902–908. doi: 10.1006/bbrc.2001.5474
Guglielmini, J., Quintais, L., Garcillán-Barcia, M. P., De La Cruz, F., and Rocha, E. P. (2011). The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS genet. 7:e1002222. doi: 10.1371/journal.pgen.1002222
Gupta, A. (1994). Multidrug-resistant typhoid fever in children: epidemiology and therapeutic approach. Pediatr. Infect. Dis. J. 13, 134–140.
Gupta, A., Matsui, K., Lo, J.-F., and Silver, S. (1999). Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 5:183.
Hauser, E., Tietze, E., Helmuth, R., Junker, E., Blank, K., Prager, R., et al. (2010). Pork contaminated with Salmonella enterica serovar 4,[5],12:i:-, an emerging health risk for humans. Appl. Environ. Microbiol. 76, 4601–4610. doi: 10.1128/AEM.02991-09
Hochhut, B., and Waldor, M. K. (1999). Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32, 99–110.
Hopkins, K. L., de Pinna, E., and Wain, J. (2012). Prevalence of Salmonella enterica serovar 4,[5],12:i:- in England and Wales, 2010. Euro surveill. 17, 10–16.
Hugas, M., and Beloeil, P. (2014). Controlling Salmonella along the food chain in the European Union - progress over the last ten years. Euro Surveill. 19:20804.
Johnson, C. M., and Grossman, A. D. (2015). Integrative and conjugative elements (ICEs): what they do and how they work. Ann. Rev. Genet. 49, 577–601. doi: 10.1146/annurev-genet-112414-055018
Kroger, C., Dillon, S. C., Cameron, A. D., Papenfort, K., Sivasankaran, S. K., Hokamp, K., et al. (2012). The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. U.S.A. 109, E1277–E1286. doi: 10.1073/pnas.1201061109
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, S. M., Grass, G., Rensing, C., Barrett, S. R., Yates, C. J., Stoyanov, J. V., et al. (2002). The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem. Biophys. Res. Commun. 295, 616–620.
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv doi: 10.1303/3997v2
Li, X., Xie, Y., Liu, M., Tai, C., Sun, J., Deng, Z., et al. (2018). oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46, W229–W234. doi: 10.1093/nar/gky352
Liu, M., Li, X., Xie, Y., Bi, D., Sun, J., Li, J., et al. (2018). ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47, D660–D665. doi: 10.1093/nar/gky1123
Macomber, L., and Imlay, J. A. (2009). The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349. doi: 10.1073/pnas.0812808106
Mastrorilli, E., Pietrucci, D., Barco, L., Ammendola, S., Petrin, S., Longo, A., et al. (2018). A comparative genomic analysis provides novel insights into the ecological success of the monophasic Salmonella serovar 4,[5],12:i. Front. Microbiol. 9:715. doi: 10.3389/fmicb.2018.00715
Mather, A. E., Lawson, B., de Pinna, E., Wigley, P., Parkhill, J., Thomson, N. R., et al. (2016). Genomic Analysis of Salmonella enterica serovar typhimurium from wild passerines in england and wales. Appl. Environ. Microbiol. 82, 6728–6735. doi: 10.1128/AEM.01660-16
McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., et al. (2001). Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852. doi: 10.1038/35101614
McHugh, G. L., Moellering, R. C., Hopkins, C. C., and Swartz, M. N. (1975). Salmonella typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin. Lancet 1, 235–240.
Morgulis, A., Coulouris, G., Raytselis, Y., Madden, T. L., Agarwala, R., and Schaffer, A. A. (2008). Database indexing for production MegaBLAST searches. Bioinformatics 24, 1757–1764. doi: 10.1093/bioinformatics/btn322
Mourao, J., Novais, C., Machado, J., Peixe, L., and Antunes, P. (2015). Metal tolerance in emerging clinically relevant multidrug-resistant Salmonella enterica serotype 4,[5],12:i:- clones circulating in Europe. Int. J. Antimicrob. Agents 45, 610–616. doi: 10.1016/j.ijantimicag.2015.01.013
Nicholson, F. A., Chambers, B. J., Williams, J. R., and Unwin, R. J. (1999). Heavy metal contents of livestock feeds and animal manures in England and Wales. Biores. Technol. 70, 23–31. doi: 10.1016/S0960-8524(99)00017-6
Osman, D., Patterson, C. J., Bailey, K., Fisher, K., Robinson, N. J., Rigby, S. E., et al. (2013). The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P(1B)-type ATPase copper efflux and periplasmic CueP. Mol. Microbiol. 87, 466–477. doi: 10.1111/mmi.12107
Osman, D., Waldron, K. J., Denton, H., Taylor, C. M., Grant, A. J., Mastroeni, P., et al. (2010). Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J. Biol. Chem. 285, 25259–25268. doi: 10.1074/jbc.M110.145953
Outten, F. W., Huffman, D. L., Hale, J. A., and O’Halloran, T. V. (2001). The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276, 30670–30677. doi: 10.1074/jbc.M104122200
Petrovska, L., Mather, A. E., AbuOun, M., Branchu, P., Harris, S. R., Connor, T., et al. (2016). Microevolution of monophasic Salmonella Typhimurium during epidemic, United Kingdom, 2005–2010. Emerg. Infect. Dis. 22:617. doi: 10.3201/eid2204.150531
Pontel, L. B., and Soncini, F. C. (2009). Alternative periplasmic copper-resistance mechanisms in Gram negative bacteria. Mol. Microbiol. 73, 212–225. doi: 10.1111/j.1365-2958.2009.06763.x
Rabsch, W. (2007). “Salmonella Typhimurium Phage Typing for Pathogens,” in Salmonella, Methods and Protocols, 394 Edn, eds H. Schatten and A. Eisenstark (Totowa, NJ: Humana Press), 177–212.
Rabsch, W., Tschape, H., and Baumler, A. J. (2001). Non-typhoidal salmonellosis: emerging problems. Microb. Infect. 3, 237–247.
Rensing, C., and Grass, G. (2003). Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27, 197–213.
Salyers, A. A., Shoemaker, N. B., Stevens, A. M., and Li, L.-Y. (1995). Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59, 579–590.
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Staehlin, B. M., Gibbons, J. G., Rokas, A., O’Halloran, T. V., and Slot, J. C. (2016). Evolution of a heavy metal homeostasis/resistance island reflects increasing copper stress in enterobacteria. Genome Biol. Evol. 8, 811–826. doi: 10.1093/gbe/evw031
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690.
Stoyanov, J. V., Hobman, J. L., and Brown, N. L. (2001). CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol. Microbiol. 39, 502–511.
Threlfall, E. J. (2000). Epidemic Salmonella typhimurium DT 104 - a truly international multiresistant clone. J. Antimicrob. Chemother. 46, 7–10. doi: 10.1093/Jac/46.1.7
Keywords: Salmonella, monophasic, integrative conjugative element, SGI-4, copper resistance
Citation: Branchu P, Charity OJ, Bawn M, Thilliez G, Dallman TJ, Petrovska L and Kingsley RA (2019) SGI-4 in Monophasic Salmonella Typhimurium ST34 Is a Novel ICE That Enhances Resistance to Copper. Front. Microbiol. 10:1118. doi: 10.3389/fmicb.2019.01118
Received: 06 February 2019; Accepted: 03 May 2019;
Published: 24 May 2019.
Edited by:
Giovanna Suzzi, University of Teramo, ItalyReviewed by:
Qingping Wu, Guangdong Institute of Microbiology (CAS), ChinaCopyright © 2019 Branchu, Charity, Bawn, Thilliez, Dallman, Petrovska and Kingsley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert A. Kingsley, cm9iLmtpbmdzbGV5QHF1YWRyYW0uYWMudWs=
†These authors have contributed equally to this work
‡Present address: Matt Bawn, Earlham Institute, Norwich, United Kingdom
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.