
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 May 2019
Sec. Microbial Physiology and Metabolism
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00960
This article is part of the Research Topic Microbial Sulfur Metabolism - From Symbiosis to Global Nutrient Cycling View all 37 articles
 Yi Jie Chelsea Tan
Yi Jie Chelsea Tan Chengzhi Zhao
Chengzhi Zhao Marufa Nasreen
Marufa Nasreen Leo O’Rourke
Leo O’Rourke Rabeb Dhouib
Rabeb Dhouib Leah Roberts
Leah Roberts Ying Wan
Ying Wan Scott A. Beatson
Scott A. Beatson Ulrike Kappler*
Ulrike Kappler*Although sulfite, a by-product of the degradation of many sulfur compounds, is highly reactive and can cause damage to DNA, proteins and lipids, comparatively little is known about the regulation of sulfite-oxidizing enzyme (SOEs) expression. Here we have investigated the regulation of SOE-encoding genes in two species of α-Proteobacteria, Sinorhizobium meliloti and Starkeya novella, that degrade organo- and inorganic sulfur compounds, respectively, and contain unrelated types of SOEs that show different expression patterns. Our work revealed that in both cases, the molecular signal that triggers SOE gene expression is sulfite, and strong up-regulation depends on the presence of a sulfite-responsive, cognate Extracytoplasmic function (ECF) sigma factor, making sulfite oxidation a bacterial stress response. An additional RpoE1-like ECF sigma factor was also involved in the regulation, but was activated by different molecular signals, taurine (Sm) and tetrathionate (Sn), respectively, targeted different gene promoters, and also differed in the magnitude of the response generated. We therefore propose that RpoE1 is a secondary, species-specific regulator of SOE gene expression rather than a general, conserved regulatory circuit. Sulfite produced by major dissimilatory processes appeared to be the trigger for SOE gene expression in both species, as we were unable to find evidence for an increase of SOE activity in stationary growth phase. The basic regulation of bacterial sulfite oxidation by cognate ECF sigma factors is likely to be applicable to three groups of alpha and beta-Proteobacteria in which we identified similar SOE operon structures.
Sulfite is a highly reactive sulfur oxyanion that occurs in both pro- and eukaryotes as a by-product of sulfur compound degradation or external exposure to sulfite (Kappler and Enemark, 2015; Kappler and Schwarz, 2016). Free sulfite can damage proteins, DNA and lipids through formation of adducts, and sulfite-oxidizing enzymes (SOEs) are found in nearly all forms of life (Zhang et al., 2011; Kappler and Enemark, 2015; Kappler and Schwarz, 2016). Interestingly, although SOEs from vertebrates and plants have been shown to be detoxification mechanisms that protect cells from sulfur stress, in bacteria SOEs have mostly been described as elements of energy conserving pathways, e.g., during chemolithotrophy (Hänsch et al., 2007; Simon and Kroneck, 2013; Kappler and Schwarz, 2016).
Bacterial growth on organosulfur compounds and chemolithotrophic growth on inorganic sulfur compounds leads to the formation of significant amounts of sulfite, and the structural and functional diversity of bacterial sulfite dehydrogenases that carry out this process has been well established (Kappler, 2008, 2011; Kappler and Schwarz, 2016). In contrast, comparatively little is known about how expression of these enzymes is regulated, which is key to revealing their cellular and physiological roles.
While in some bacteria such as Deinococcus radiodurans SOE genes appear to be always highly expressed (D’errico et al., 2006), in many bacteria SOEs have complex regulatory patterns where upregulation usually occurs in the presence of metabolizable sulfur substrates, but activity may also undergo growth phase dependent induction and show varying levels of basal expression in different bacterial strains and species (Kappler et al., 2000, 2001; Wilson and Kappler, 2009; Bastiat et al., 2012).
In some bacteria an extracytoplasmic function (ECF) sigma factor/anti-sigma factor (ASF) pair is encoded directly upstream of the genes encoding SOEs and has been proposed to be involved in regulating SOE expression (Kappler et al., 2001; Wilson and Kappler, 2009; Bastiat et al., 2012). ECF sigma factor- based gene regulation is essentialy controlled by the ASF that sequesters the sigma factor in the absence of an activating signal. The involvement of an ECF sigma factor in SOE gene regulation has been confirmed in a recent study of Sinorhizobium meliloti strain GMI11495 (Bastiat et al., 2012). In Sm. meliloti GMI11495 the SOE-associated ECF sigma factor RpoE4 was induced during stationary growth phase and was identified as the key regulator of the sorT gene that encodes the Sm. meliloti SorT SOE (Wilson and Kappler, 2009; Bastiat et al., 2012). The work also uncovered that a second ECF sigma factor, RpoE1, plays a role in inducing sorT expression during growth on taurine, but not in the presence of thiosulfate, the second RpoE4-activating substrate that was identified. Based on these results a model was proposed where during stationary phase and during growth on taurine activation of sorT occurs through the action of both RpoE4 and RpoE1, while during growth on thiosulfate activation of sorT expression required only the action of RpoE4. The molecular signal sensed by RpoE4 was proposed to be sulfite, as sulfite accumulated in the cultures of strains carrying mutations in the sorT gene.
We have previously described the presence of genes encoding ECF sigma factors upstream of SOE encoding operons for two soil bacteria, the Sinorhizobium meliloti 1021 type strain and Starkeya novella (Figure 1; Kappler et al., 2001; Wilson and Kappler, 2009). Both of these bacteria are α-Proteobacteria of the order Rhizobiales, but belong to the families Rhizobiaceae and Xanthobacteraceae, respectively. Sm. meliloti and St. novella have the ability to degrade organosulfur compounds and inorganic sulfur compounds, contain distinct types of SOEs, the homodimeric SorT and the heterodimeric SorAB, (Kappler et al., 2000; Kappler and Bailey, 2005; Mcgrath et al., 2015), and also show different SOE regulatory patterns (Kappler et al., 2001; Wilson and Kappler, 2009; Low et al., 2011). While in St. novella SorAB SOE activity is nearly undetectable in the absence of degradable sulfur compounds (∼0.05 U/mg, Figure 1 and Supplementary Table S1), in Sm. meliloti 1021 SOE activity is always detectable at basal levels of 0.7–1 U/mg (Figure 1 and Supplementary Table S1; Kappler et al., 2001; Wilson and Kappler, 2009). This differs from both the SOE activity pattern in St. novella and the observations made for SOE gene expression in Sm. meliloti strain GMI11495 by Bastiat et al. (2012).
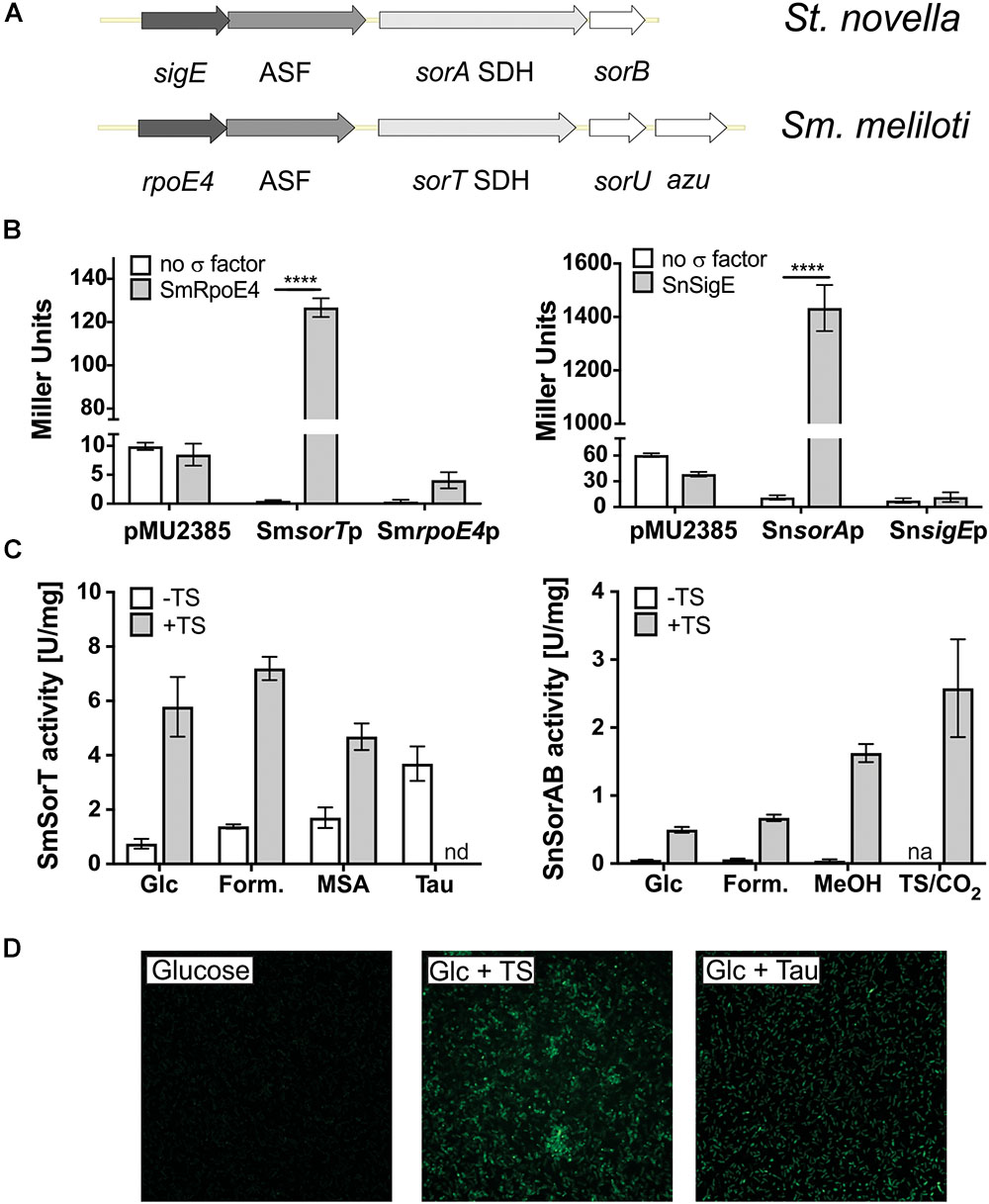
Figure 1. ECF sigma factor-based regulation of SOE gene expression in Sm. meliloti 1021 and St. novella. (A) Schematic representation of the Sm. meliloti rpoE4/sorT and St. novella sigE/sorAB gene regions, highlighting the similarity of the gene arrangement. (B) Reporter gene assays showing the ability of SmRpoE4 and SnSigE to activate the promoters of the cognate SOE encoding genes sorT and sorA. Controls shown test the activity of promoter gene fusions in the absence of a specific transcription factor (white columns) and the ability of the transcription factors to increase expression of the promoterless lacZ gene in pMu2385 (Label: pMu2385), respectively. Positive interactions between test promoters and sigma factors lead to increase beta-Galactosidase activity, 2-way ANOVA: ∗∗∗∗p < 0.0001. (C) Carbon and sulfur substrate dependent changes in SOE activity in Sm. meliloti 1021 and St. novella, highlighting the different modes of basal regulation. Enzyme activities were determined in cell extracts from cultures grown to late exponential growth phase on the indicated substrates using ferricyanide-based (Sm. meliloti SorT) or cytochrome c-based (St. novella SorAB) assays. The growth substrates chosen reflect the different substrate preferences of the bacteria. (D) In vivo induction of sorT promoter-gfp mut2 fusions in Sm. meliloti growing on media with different growth substrates. Fluorescence changes correspond to SOE activity changes shown in (C). Data in (B,C) represent averages of at least three repeat assays (n = 3 biological replicates), error bars represent standard deviation. Abbreviations: ASF, antisigma factor; Azu, azurin; cyt.c, cytochrome c; Form., formate; Glc,-glucose; MeOH, methanol; MSA, methanesulfonic acid; n.d., not determined; n.a., not applicable; Tau, taurine; TS, thiosulfate.
Here we have used Sm. meliloti 1021 and St. novella to investigate the conservation of SOE regulatory patterns across species, the signaling molecule(s) that trigger SOE expression, and to determine whether regulation of SOE expression by two ECF sigma factors is unique to Sm. meliloti strains. Our investigations show that up-regulation of genes encoding SOEs is triggered specifically by the presence of sulfite in both species, making sulfite oxidation a bacterial stress response and negating a major role in energy conservation. The main mode of SOE regulation in both species depended on the cognate ECF sigma factors SmRpoE4 and SnSigE. An additional, overlapping regulatory circuit depending on an RpoE1-like ECF sigma factor was also found in both species, but the respective RpoE1 sigma factors targetted different promoters within the SOE and ECF sigma factor operons (Sm. meliloti rpoE4 promoter, St. novella sorA promoter). Additionally, RpoE1-based activation was based on species-specific stimuli and appeared to account only for a comparatively small fraction of SOE induction under very specific conditions. Phylogenetic analyses revealed at least three clades of ECF sigma factors found in association with SOE genes, one of which is unstudied to date and is comprised of species that appear to lack an RpoE1 homolog, making regulation by the cognate ECF sigma factor the main mode of SOE regulation.
Bacterial strains and plasmids used in this study are listed in Supplementary Table S2. E. coli strains were routinely grown aerobically on Luria Bertani (LB) medium (Ausubel et al., 2005) at 37°C. Starkeya novella DSMZ506T and Sinorhizobium meliloti strain 1021 were cultivated aerobically on either TYS medium (Beringer, 1974) or modified DSMZ medium no 69 at 30°C (Wilson and Kappler, 2009). The DSMZ 69 basal medium was supplemented with either 80 mM methanol, 20 or 40 mM thiosulfate, 20 mM tetrathionate, 10 mM glucose, 20 mM formate, 20 mM methanesulfonic acid, 20 mM taurine, or combinations of these compounds. Where applicable the following antibiotics were added to the growth media (μg/mL): E. coli: ampicillin and kanamycin 100, gentamicin and tetracycline 10, trimethoprim 30; Sm. meliloti: streptomycin 25, tetracycline 5, kanamycin 200.
Standard methods were used throughout (Ausubel et al., 2005). The PureLink Quick plasmid prep and PCR purification kits (Life Technologies) were used for the purification of plasmid DNA, PCR products and preparative restriction enzyme digests, restriction enzymes were from Life Technologies, T4 DNA ligase from Promega. GoTaq green Mastermix (Promega) was used for all standard PCR reactions, Pfu (Stratagene) or Phusion (Finnzymes) polymerases were used for all cloning applications and to generate probes for EMSA assays. Oligonucleotide primers (Supplementary Table S3) were from Life Technologies or IDT DNA technologies.
For gene expression experiments cultures were grown to early/mid–exponential growth phase on glucose or methanol containing medium 69 before sulfur compounds were added (20 mM taurine, 20 mM thiosulfate, or 1 mM sulfite). For SOE gene induction experiments samples for RNA isolation (2 mL) were taken just before addition of the compounds (t = 0) and at 15, 30, 60, 90, 120, and 180 min post-addition. All samples were preserved with RNAProtect Bacteria reagent (Qiagen), RNA was isolated using the Illustra RNAspin mini kit (GE Biosciences) and stored at -80°. gDNA was removed by DNAse treatment (TurboDNA free, Life Technologies), all samples were tested for the absence of gDNA using PCR amplification of the 16S rDNA gene. cDNA was prepared from 500 ng of DNA-free RNA using Superscript III or IV enzymes (Life Technologies) and random hexamer primers.
Quantitative RT-PCR used the SYBR green mastermix (Applied Biosystems) and was essentially carried out and analyzed as in Ang et al. (2017). The final reaction volume was 10 μL and 384 well plates were used throughout. An epMotion workstation (Eppendorf) was used to set up reactions, data was collected using a Quantstudio 6 (Life Technologies) thermal cycler. Gene expression was normalized to 16S gene expression, PCR efficiencies were determined using LinReg (Ramakers et al., 2003).
Transcription start sites were determined using the Life Technologies 5′RACE system according to the manufacturer’s instructions. DNA sequencing at the Australian Equine Genetics Research Centre (University of Queensland) used BigDye v3.1 (Applied Biosystems). EMSA experiments used the Dig-Gelshift Kit v2 (Roche Applied Science). EMSA reactions used a buffer containing 40 mM HEPES pH 7.9, 60 mM KCl, 1 mM MgCl2, 12% glycerol, and 1 mM DTT (Rhodes et al., 1997), RNA polymerase core (Epicenter), 0.25 pmoles DIG-labeled probe, 20 pmoles purified SnSigE, salmon sperm DNA or poly d[IC] (Sigma-Aldrich) were added where applicable. Binding reactions were incubated for 40 min at 28°C before being separated on a 5% 0.5× TBE acrylamide gel at 4°C followed by blotting and detection according to the manufacturer’s instructions.
Gene knockout plasmids pKnock-Km-rpoE4 and pKnock-Km-sorT were created by insertion of 300–400 bp gene fragments into the pKnock-Km vector (Alexeyev, 1999). Electrocompetent Sm. meliloti cells were prepared using 200 mL TYS-based cultures grown at 30°C, 200 rpm to an OD600 of ∼0.4–0.6. Cells were harvested by centrifugation (4000 × g, 4°C, 15 min), washed twice in 50 mL sterile water before resuspending in 1 mL of sterile 10% glycerol. To a 100 μL aliquot of these cells, 0.5–1 μg of plasmid DNA were added, followed by electroporation (2500 V, 25 μF, 400 Ω, 2 mm cuvette) using a Bio-RAD genepulser. Electroporated cells were taken up in 1 mL TYS and incubated at 30°C with shaking for 12 h before plating on selective media. St. novella is not amenable to genetic manipulation (Davidson and Summers, 1983; Davidson et al., 1985; Kappler et al., unpublished), precluding similar experiments with this bacterium.
SDS-PAGE was performed as in Laemmli (1970). Small volume (2–3 mL) cell extracts for enzyme assays were prepared from cultures (10–20 mL) grown to mid/late exponential growth phase using BugBuster Mastermix (Novagen). SOE activity assays were carried out as in Kappler et al. (2000), Wilson and Kappler (2009) and Low et al. (2011) with ferricyanide or cytochrome c as electron acceptors for SOEs from Sm. meliloti and St. novella, respectively. Sulfite concentrations in growth media were determined using fuchsin as in Grant (1947). Pfu (Stratagene) and Phusion (Finnzymes) polymerases were used to amplify gene fragments with high fidelity, e.g., the SnsigE and SmrpoE genes and promoter regions for cloning into pQE30 (Qiagen, SnsigE) or pProex HTB (Life Technologies, SmrpoE4, SmrpoE1, SnrpoE1) or for cloning of promoter regions into pMU2385 or for use in EMSA experiments. All protein expression plasmids were tested for successful protein expression in small scale expression experiments. 6xHis SnSigE was expressed in DH5α at 30°C, using 1 mM IPTG followed by incubation for 4 h before harvesting. The recombinant protein was purified under native conditions using Ni-NTA resin (Qiagen) as per the manufacturer’s instructions or following refolding from inclusion bodies using the protocols of Burgess (1996) and Burgess (2009). Under non-denaturing conditions, 6xHis SnSigE co-purified with E. coli RNA Polymerase.
Beta-galactosidase activity present in E. coli cells carrying fusions of test promoters to the promoterless lacZ gene in pMU2385 (Praszkier et al., 1992) and a second plasmid expressing the relevant sigma factors was determined using the method of Kidd et al. (2005). The sigma factor expression plasmids were the same that were used for protein purification. Plasmids without inserts were used for control reactions that test either the activity of promoters in the absence of a transcription factor or the ability of a sigma factor to alter expression of the pMU2385 promoterless lacZ gene. Promoter fragments used were between 300 and 500 bp and located directly upstream of the coding regions. E. coli cultures for reporter gene assays were inoculated from overnight cultures into 5 mL of LB, grown at 37°C to an OD600 of ∼ 0.4–0.6. Cells were harvested and resuspended in 1xPBS before being used for enzyme assays (Kidd et al., 2005). All assays were repeated at least once and carried out in triplicates for each repeat, enzyme activities are reported in Miller units.
For in vivo monitoring of SOE promoter activities the same promoter fragments that were used in pMU2385 were cloned into pBluescript-gfpmut2 followed by subcloning of the entire expression cassette into pRK415 (Ditta et al., 1985) for transfer into Sm. meliloti as described above. The in vivo activity of promoter gene fusions to gfpmut2 was detected by either epifluorescence or confocal laser microscopy at the SCMB microscopy facility in cultures grown to mid-exponential phase on DSMZ medium 69 with or without the addition of a sulfur compound.
Protein homologs of SnSigE and SmRpoE4 were identified using the BLAST algorithm (Altschul et al., 1997). Analysis of gene environments for ECF26 sigma factors used data available in public databases (e.g., GenBank)1, the Vector Nti Advance 11 program suite (Life Technologies) was used to display and compare the sequence data. The Mist database (v 2.0) (Ulrich and Zhulin, 2007) was used to confirm the detection of ECF sigma factors in the genomes of the model organisms, the ECF finder tool2 was employed to check the classifications of sigma factor sequences. The complete list of sigma factor sequences is available in Supplementary Table S4. Full length protein sequences were aligned ClustalW subsequently analyzed in MEGA 7.0 (Tamura et al., 2011), phylogenetic trees were constructed using Neighbor Joining (NJ), Minimum Evolution (ME), UPGMA, and Maximum Likelihood (ML) algorithms. All analyses assumed uniform rates of evolution of the amino acid sequences. NJ, ME, and UPGMA used a Poisson model of substitution and pairwise deletion of gaps/ missing data while ML analyses used the Jones- Taylor Thornton Model and partial deletion for gaps/ missing data. Robustness testing was carried out using the bootstrap method with 500 resampling cycles. Sequences belonging to ECF groups 12 and 15 as defined in Staron et al. (2009) were used as reference groups (Supplementary Table S4). All of these sequences were clearly located on separate branches outside of the three ECF groups analyzed (data not shown). The consensus ECF26 promotor profile was generated using the MEME suite of motif-based sequence analysis tools3 (Bailey et al., 2009). Specifically, glam2 was used to create the promoter consensus profile and logo, glam2scan was used to search the publicly available genome sequences for the presence of this profile, tomtom was used to compare this profile against the prodoric database of gene regulation (version 8.9) (Muench et al., 2003; Gupta et al., 2007; Frith et al., 2008).
Basic characterization showed that in St. novella and Sm. meliloti 1021 the ECF sigma factor and cognate ASF encoding genes located upstream of the SOE encoding genes are co-transcribed (Supplementary Figure S1), and reporter gene assays (Figure 1) confirmed a specific induction of both the Sm_sorT and Sm_rpoE4 promoters by the SmRpoE4 sigma factor (260 and 16 times induction, respectively), and the ability of the SnSigE sigma factor to bind to and induce expression of the Sn_sorA promoter (129 times induction) but not the Sn_sigE promoter (Figure 1 and Supplementary Figure S2).
Transcription start site mapping by 5′RACE revealed that in keeping with this observation, the Sn_sigE promoter lacks the consensus (-35 GGAAT, -10 CGTC) found upstream of the SmsorT, SnsorA, and SmrpoE4 genes (Supplementary Figure S1). This consensus essentially matches the one determined for the sorT and rpoE4 genes of Sm. meliloti GMI11495 (Bastiat et al., 2012; Schlüter et al., 2013), however, we noticed that the spacing of the -10 and -35 elements differed by 1 bp between some promoters which could alter relative promoter strength (Supplementary Figure S1). Bioinformatic searches of the St. novella and Sm. meliloti 1021 genomes (Galibert et al., 2001; Kappler et al., 2012) did not reveal additional genes with a strong match to this consensus sequence.
As already indicated in the introduction, carbon and sulfur source dependent induction of SOE activity in both bacteria was limited to substrates that can give rise to sulfite either through abiotic processes in the medium (e.g., thiosulfate), or as a result of degradation by the bacteria (SM: taurine, SN: thiosulfate and tetrathionate) (Figure 1 and Supplementary Table S1). A Sm_sorT promoter GFPmut2 fusion introduced into Sm. meliloti 1021 confirmed this sorT gene induction pattern in vivo, with GFP fluorescence being strongest for growth media containing taurine or both glucose and thiosulfate (Figure 1).
The molecular signal sensed by the ECF type sigma factor/ASF pairs that regulate SOE gene expression could either be a metabolizable sulfur source, if induction were induced by degradation of a particular substrate, or sulfite itself if the main purpose of the SOE reaction is to detoxify sulfite. Sm. meliloti and St. novella cultures growing exponentially on glucose-containing media were exposed to taurine, thiosulfate or sulfite, which revealed an immediate, strong increase in SmsorT and SnsorA expression only in response to sulfite exposure (Figure 2 and Supplementary Figure S3). This increase in sorA and sorT gene expression was transient, not lasting beyond 2 h post-exposure, which is in keeping with the relatively low sulfite challenge concentration (1 mM) chosen to avoid toxicity and the rapid chemical and enzymatic turnover of sulfite in solution (Kappler et al., 2000; Wilson and Kappler, 2009).
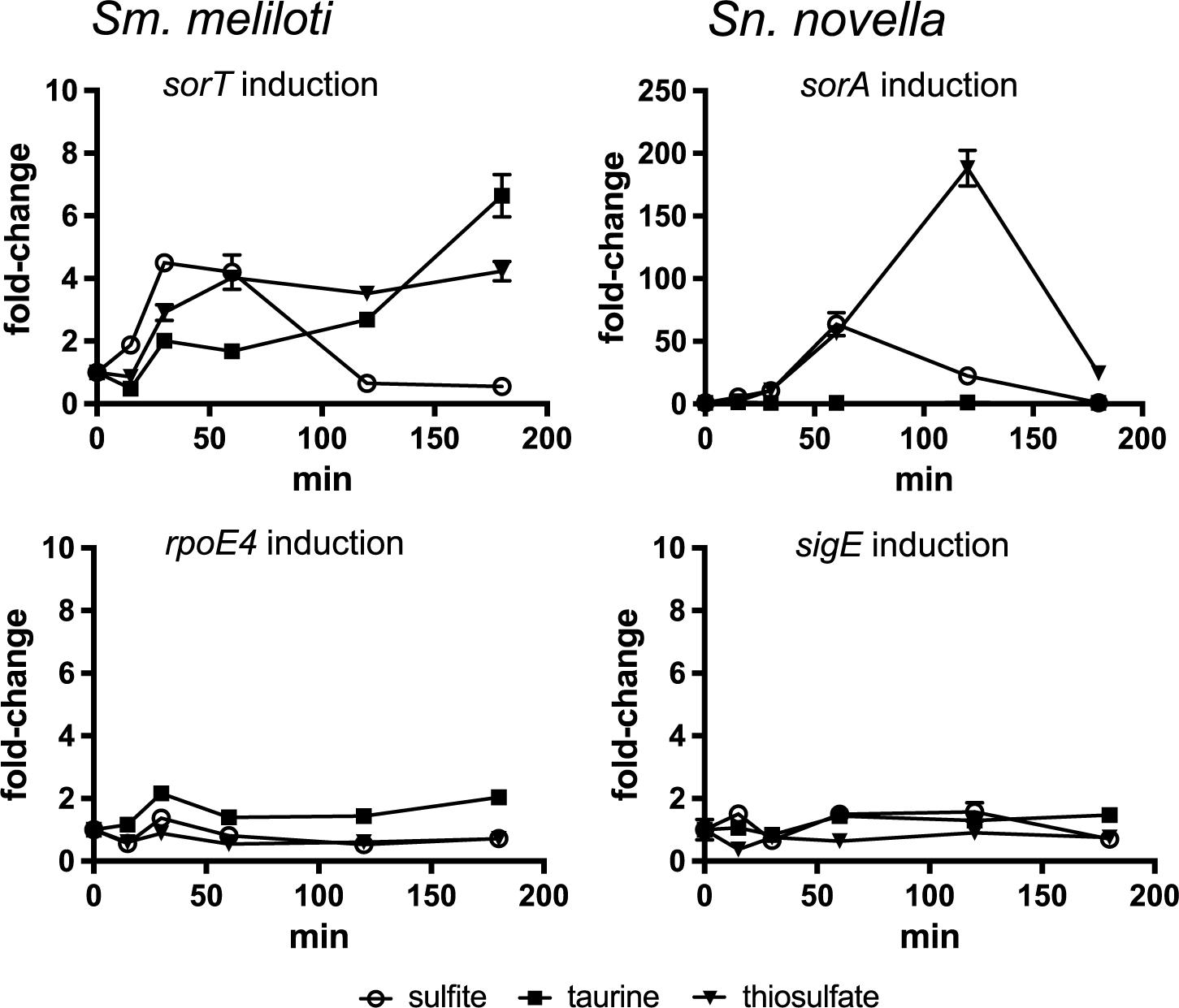
Figure 2. Fold-change in SOE and ECF sigma factor gene expression following exposure to sulfite, taurine and thiosulfate in Sm. meliloti 1021 and St. novella 506T. Left: Sm. meliloti gene expression induction, Top: induction of the sorT SOE gene, Bottom: induction of the rpoE4 ECF sigma factor gene. Right: St. novella gene expression induction, Top: induction of the sorA SOE gene, Bottom: induction of the rpoE4 ECF sigma factor gene. Data are shown as the average and standard deviation of at least three assays. The underlying gene expression data is shown in Supplementary Figure S3. 2-Way ANOVA of changes in rel. normalized gene expression compared to the t = 0 value showed that for sulfite addition changes were statistically significant (alpha = 0.05) from t = 15 min (Sm) and t = 30 min (Sn) with p = 0.0032 (∗∗) (Sn, 30 min) to p < 0.0001 (∗∗∗∗) for all other values, for thiosulfate addition from t = 30 min (Sm) and t = 60 min (Sn) with p < 0.0001 (∗∗∗∗) for all datapoints, for taurine all values were not significant except Sm 120 min, p = 0.0232 (∗), and 180 min (p < 0.0001, ∗∗∗∗). For clarity the p-values are not shown in the figure.
Addition of thiosulfate also gave rise to an induction of Sm_sorT/Sn_sorA gene expression which, however, took about 60–120 min to peak, while taurine exposure caused SmsorT gene expression to increase after >120 min incubation while no effect was observed in St. novella which is unable to metabolize taurine (Kappler et al., 2012; Figure 2). These observations are consistent with sulfite being the signal that induces SOE gene expression in both species. While Sm. meliloti is unable to metabolize thiosulfate, addition of thiosulfate to the DSMZ69 growth medium leads to the abiotic production of sulfite (∼120–160 μM) (Wilson and Kappler, 2009), explaining the thiosulfate-based induction of Sm_sorT gene expression in this species. In contrast, thiosulfate is also a major substrate for St. novella chemolithotrophic growth, and the St. novella thiosulfate degrading enzyme complex that leads to the formation of sulfite is always expressed at high levels (Kappler and Nouwens, 2013), allowing fast degradation of thiosulfate. This explains the strong upregulation of Sn_sorA gene expression observed in St. novella following addition of thiosulfate. In contrast, taurine degradation by S. meliloti that also gives rise to the formation of sulfite (Cook and Denger, 2002) is an inducible process, and would only produce sulfite following production of taurine-degrading enzymes and degradation of significant amounts of taurine, which is consistent with the observed lag of 120 min before induction for Sm_sorT was observed.
Despite sulfite being the common molecular signal inducing SOE gene expression in Sm. meliloti and St. novella, with regards to the basal levels of activity the expression patterns for the SmSorT and SnSorAB SOEs differ clearly (Figure 1 and Supplementary Table S1), and one possible explanation for this could be that unlike what was found in Sm. meliloti (Bastiat et al., 2012), in St. novella only the cognate ECF sigma factor, SnSigE, might be involved in the regulation of SOE expression. To establish if regulation by two ECF sigma factors is present we first tested whether a homolog of the SmRpoE1 sigma factor exists in St. novella. The identified SnRpoE1-ASF pair is encoded by genes Snov_0992/0993 and SnRpoE1 is able to activate the promoter of Snov_0990 (Sn_duf305), a gene encoding a protein containing a domain of unknown function (DUF305), while a 270 bp region directly upstream of the SnrpoE1 gene (Snov_0992) showed no activation (not shown). The Snov_0990 (Sn_duf305) promoter likely also controls expression of the Snov_0991 gene that encodes a YVTN-beta propeller repeat protein from which the Snov_0990 gene is only separated by 12 bp.
In reporter gene assays SnRpoE1 induced strong expression of the Sn_duf305 and Sn_sorA promoters but not Sn_sigEp, while SnSigE strongly induced Sn_sorAp and also showed a weak ability to induce Sn_duf305p activity (Figure 3).
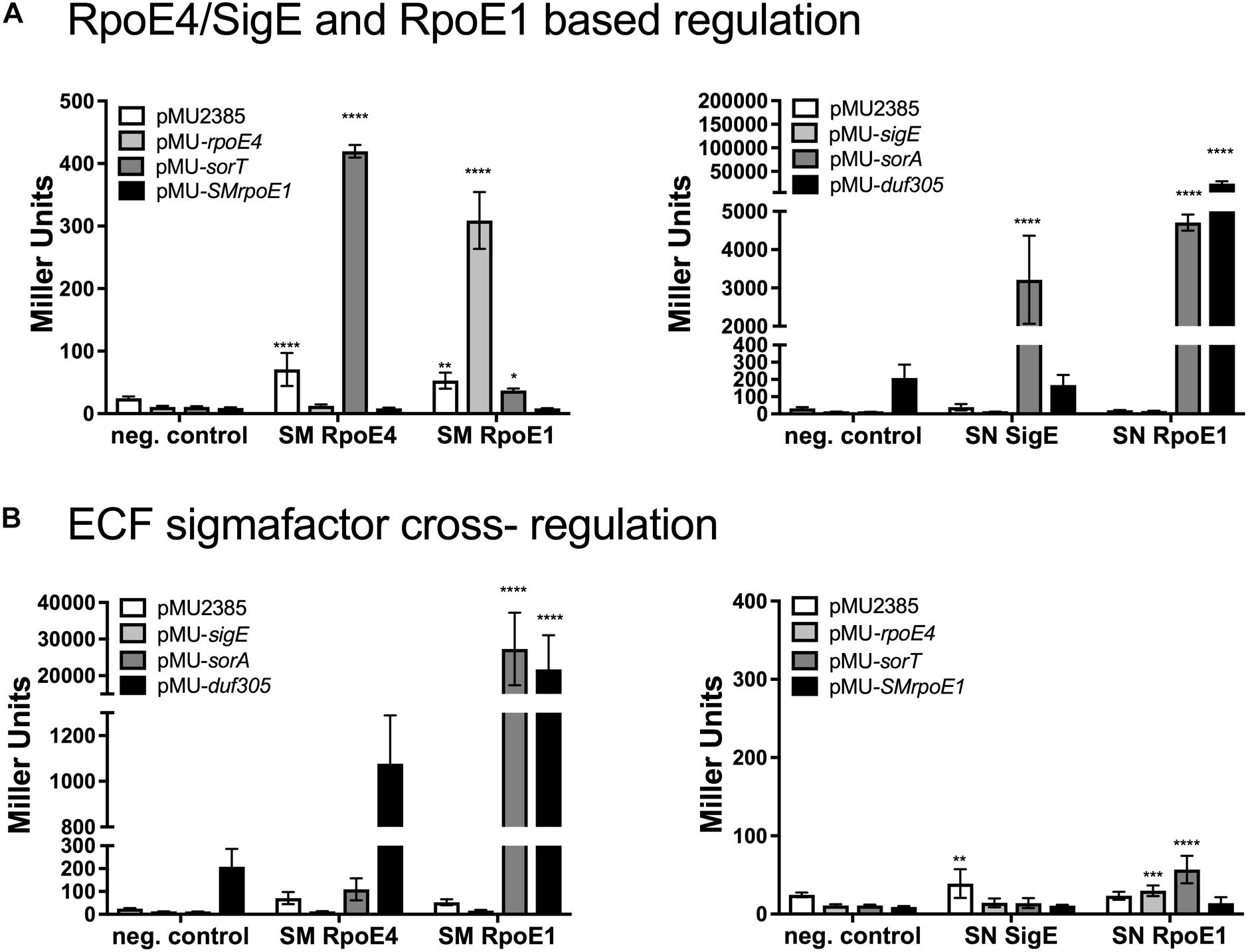
Figure 3. Reporter gene assays investigating promoter activation by Sm. meliloti and St. novella ECF sigma factors SmRpoE4/SnSigE and Sm/SnRpoE1. (A) Sm. meliloti RpoE4 and RpoE1activation of Sm. meliloti rpoE4, sorT, rpoE1 promoters (left) and St. novella SigE and SnRpoE1 activation of St. novella sigE, sorA and duf305 promoters (right). (B) SmRpoE4/SnSigE and Sm/SnRpoE1-mediated regulation of SOE gene and ECF sigma factor promoters species. Activation of St. novella promoters by Sm. meliloti ECF sigma factors (left), and activation of Sm. meliloti promoters by St. novella ECF sigma factors (right). Data are shown as the averages and standard deviation of at least three assays. Statistical analyses used 2-Way ANOVA comparing data for each promoter to the negative control (no sigma factor present). ∗∗∗∗p < 0.0001, ∗∗∗p = 0.0006; ∗∗p = 0.0093; ∗p = 0.0106.
Matching experiments with SmRpoE1 revealed strong induction of the Sm_rpoE4 promoter and very low level induction of the Sm_sorT promoter, while only SmRpoE4 showed strong induction of the Sm_sorT promoter (Figure 3).
Interestingly, the Sm. meliloti RpoE1 and RpoE4 sigma factors were also able to interact with the three St. novella promoters tested here (Figure 3). SmRpoE4 induced low level Sn_sorA promoter activity and medium level expression of the Sn_duf305 promoter, while SmRpoE1, similar to SnRpoE1, gave rise to high levels of Sn_sorAp and Sn_duf305p activation, while no changes in Sn_sigEp activity were observed.
In contrast, SnSigE and SnRpoE1 were unable to recognize the Sm. meliloti sorT, rpoE4, or rpoE1 gene promoters (Figure 3). The ability of SmRpoE1 for interspecies cross-regulation was confirmed using an Sn_sorA promoter-GFPmut2 fusion introduced into Sm. meliloti, where even on media containing only glucose, i.e., non-inducing conditions, strong fluorescence was observed (Supplementary Figure S4). The constant activity of the Sn_sorA promoter in Sm. meliloti 1021 (Figure 3 and Supplementary Figure S4), suggests that in this strain SmRpoE1 was active under all conditions tested.
We then analyzed expression of the SOE, sigma factor and ASF genes in St. novella grown on glucose (heterotrophic growth), methanol (C1-compound growth), thiosulfate and tetrathionate (both chemolithoautotrophic growth). The gene expression data confirmed the strong induction of sorA gene expression in the presence of thiosulfate (∼18 times rel. to glucose levels) also reflected in the enzyme activities (Figures 1, 4). Both SnsigE and SnrpoE1 genes were expressed at similar levels, with sigma factor gene expression levels significantly exceeding (∼10×) those of the respective cognate ASF (SnSigE ASF: Snov_3267, SnRpoE1 ASF: Snov_0993). Expression of SnrpoE1 and associated genes (Snov_0990-0993) was low in glucose, methanol and thiosulfate samples, indicating that RpoE1 was probably not active under these conditions. However, during growth on tetrathionate, activation of SnRpoE1 occurred as indicated by high levels of expression of the Snov_0991 gene that is associated with the gene encoding the Snov_0990 DUF305 protein used in promoter studied (Figure 4). As reporter gene assays with SnRpoE1 showed strong interactions with the Sn_sorA promoter, this suggests that in vivo during growth on tetrathionate SnRpoE1 may have activated Sn_sorA transcription. However, a contribution of the sulfite-responsive SnSigE sigma factor to Sn_sorA expression on tetrathionate cannot be excluded as the degradation pathway for tetrathionate in St. novella is unknown and may lead to the formation of sulfite which would trigger the SnSigE response. This then suggests that the regulatory pattern of the St. novella SorAB SOE depends mostly on the activation of the cognate SnSigE sigma factor.
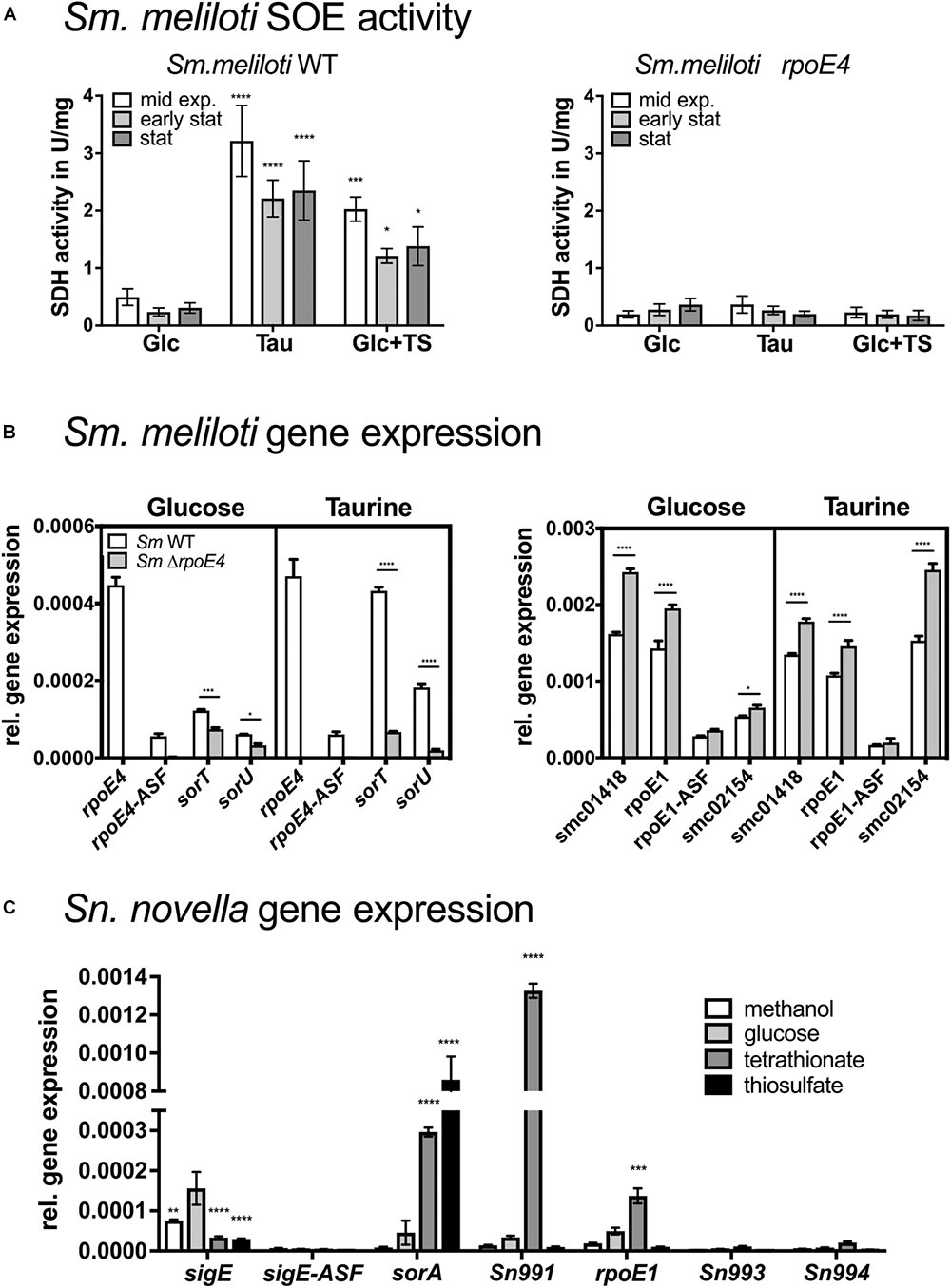
Figure 4. SOE activities and expression of SOE and ECF sigma factor-associated genes in Sm. melilotiWT and Sm. melilotiΔrpoE4 strains (A,B) and in St. novellaWT (C). (A) Growth- phase dependent changes in SOE activity in Sm. melilotiWT and Sm. melilotiΔrpoE4 on glucose, taurine and glucose and thiosulfate. (B) Expression of SmRpoE4 and SmRpoE1 associated genes in Sm. melilotiWT (white) Sm. melilotiΔrpoE4 (gray) during growth on glucose or taurine. (C) Expression of genes associated with the SnSigE and SnRpoE1 encoding genes in St. novellaWT under different growth conditions. Abbreviations: Glc, glucose; TS, thiosulfate; TT, tetrathionate. Growth with TS and TT as substrate was under chemolithoautotrophic conditions. All data are shown as averages and standard deviations of at least three replicate determinations. 2-Way ANOVA was used to analyse data. ∗∗∗∗p < 0.0001, ∗∗∗p = 0.0002–0.0005, ∗∗p = 0.013, ∗p = 0.0101–0.0362.
The constant activation of the RpoE1 ECF sigma factor in Sm. meliloti 1021 indicated by the constant activation of the Sn_sorAp (Supplementary Figure S4) is likely to impact expression patterns of sorT and the rpoE4 associated genes, and we assessed this using Sm. meliloti 1021 sorT or rpoE4 gene knockout strains. All strains showed similar growth rates, with growth on taurine being slower than on glucose- containing media, on which, interestingly, the Sm. melilotiΔrpoE4 strain showed slightly reduced growth (p < 0.01) (Table 1 and Supplementary Figure S2), although this was not the case in the presence of taurine.
SorT enzyme activity levels in Sm. meliloti 1021WT were as expected, with low activities for glucose-containing media and high activity when taurine or glucose and thiosulfate were used as substrates (Figure 4 and Supplementary Figure S5). In contrast, only basal SorT activity levels of ∼0.2–0.3 U/mg were present at all times in the Sm. melilotiΔrpoE4 strain, while in the Sm. melilotiΔsorT strain, as expected, no activity was detected. Under all conditions tested, SmSorT SOE activity was maximal during exponential growth, while in stationary phase SorT activity levels were reduced by 20–40% relative to exponential growth (Figure 4 and Supplementary Figure S5). This was also true for glucose-containing medium, where a stationary-phase dependent induction due to increased internal sulfite formation should have been most apparent as a result of the absence of a growth substrate generating sulfite.
The basal activity observed in the rpoE4 knockout strain likely resulted from the weak interaction of SmRpoE1 with the Sm_sorT promoter observed in reporter gene assays. The comparatively higher basal levels of sorT expression in Sm. meliloti 1021WT (0.7–1.0 U/mg, Supplementary Table S1) is likely due to activation of the Sm_rpoE4 promoter by SmRpoE1, which was seen in the in vitro experiments.
Gene expression analysis in Sm. meliloti 1021WT and Sm. melilotiΔrpoE4 strains matched expectations for the rpoE4 and sorT operons. Maximal SmsorT gene expression occurred in the presence of thiosulfate, while during growth on glucose only low levels of SmsorT expression were observed (Figure 4). SmrpoE4 and the cognate antisigma factor gene SMc_04050 were expressed at low but near constant levels regardless of the growth medium with ASF expression reduced to ∼12–17% of SmrpoE4 expression levels (Figure 4 and Supplementary Figure S6).
The same pattern of expression was also seen for SmrpoE1 and SMc_01420 (RpoE1 ASF) where despite overall very high levels of gene expression under all conditions, expression of the ASF was reduced by ∼80–85%, while expression of the associated SMc_01418 gene was high throughout and showed very little variation with growth conditions (Figure 4).
For the SmrpoE4 knockout strain expression patterns for genes in the SmrpoE1 gene region were unaffected, while expression of SmrpoE4 and SMc_04050 was absent. Expression of SmsorT was reduced to basal levels confirming the results from enzyme assays (Figure 4 and Supplementary Figures S5, S6).
Of the additional genes proposed to be part of the SmRpoE1 regulon by Bastiat et al. (2012) only SMc_02154 showed significant expression, with expression levels and patterns being similar to those seen for SmrpoE1 itself. In contrast, the genes proposed to be controlled by SmRpoE4 (SMc04164, SMc00821, and SMc00108) were expressed at very low levels throughout and showed no variation between the Sm. melilotiWT and Sm. melilotiΔrpoE4 strains, indicating that they are not subject to direct regulation by SmRpoE4 in Sm. meliloti 1021 (Figure 4 and Supplementary Figure S6).
Sulfite oxidation is an essential process in all bacteria that degrade sulfur compounds as either carbon or energy sources, and here we have shown that despite clearly differing types of sulfur metabolism, the presence of structurally distinct SOEs and differing patterns of SOE basal regulation, the basic mechanism of SOE regulation is conserved between two different species of soil bacteria, Sm. meliloti and St. novella (Kappler and Bailey, 2005; Mcgrath et al., 2015).
In both species induction of SOE gene expression is triggered by sulfite, and depends mostly on the respective, cognate ECF sigma factor. With sulfite as the trigger, induction of SOE expression in both species is specific to substrates that lead to sulfite formation either via abiotic processes (e.g., thiosulfate) or are metabolized with sulfite as an intermediate (Sn: thiosulfate and possibly tetrathionate, Sm: taurine). Induction of SOE gene expression was tightly linked to the presence of sulfite produced externally or as a result of energy conserving metabolic processes during exponential growth, and we found no evidence for significant SOE gene expression during stationary growth phase as a result of increased internal digestion processes. We therefore propose a simplified model for SOE activation that is likely to be applicable to many bacterial species where ECF sigma factors are found up- or downstream of SOE encoding genes, and where regulation depends essentially on the presence of sulfite which leads to release of the cognate ECF sigma factors from their respective ASFs by an as yet unknown mechanism (Figure 5 and Supplementary Figures S7, S10).
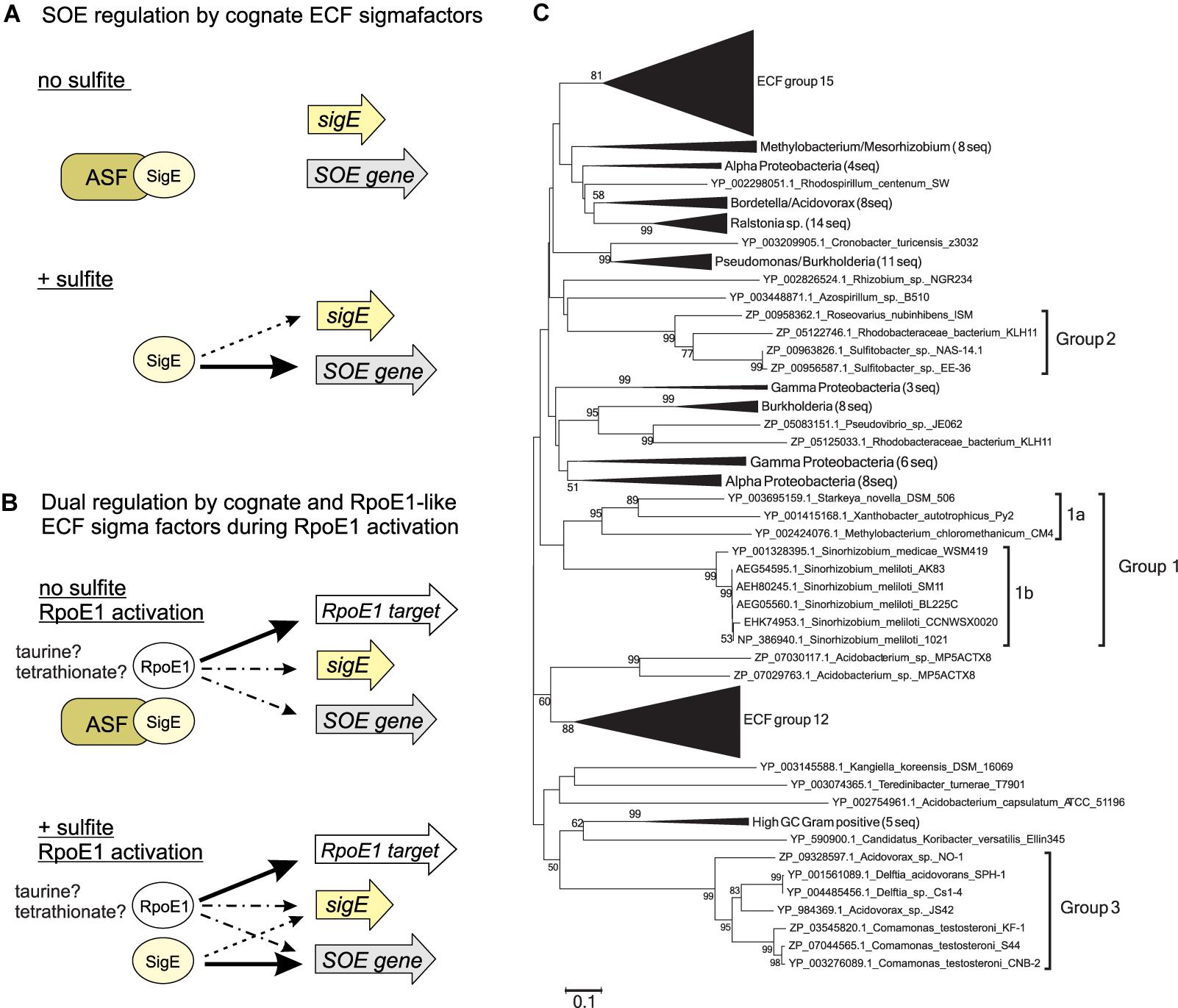
Figure 5. Model of SOE gene regulation (A,B) and phylogenetic tree showing different groups of ECF sigma factors associated with SOE-encoding genes (C). (A) Conserved mode of regulation of SOE gene expression by cognate ECF sigma factors. (B) Regulation of SOE gene expression by two ECF sigma factor regulatory circuits – magnitude of response and induction signal appear to be species-specific. (C) Neighbor -joining tree of ECF sigma factors belonging to ECF group 26 with ECF groups 12 and 15 used reference as groups. The three groups of SOE associated ECF sigma factors are indicated by thicker lines and labels, St. novella SigE and Sm. meliloti RpoE4 are highlighted through use of a larger font and underlining. Bootstrap values are given as percent conservation over 500 replications. Abbreviations: ASF, antisigma factor; SigE, SOE-gene associated ECF sigma factor; SOE gene, gene encoding a sulfite oxidizing enzyme; RpoE1 target, gene controlled by RpoE1 ECF sigma factor; solid arrows, indicate positive regulation with arrow thickness indicating level of activation; dotted arrows/dot and dash arrows, proposed interactions or interactions that were not consistent between the organisms studied here. Dotted lines: Interaction of SOE cognate sigma factors with their own promoters which are not always present; dots and dashes: RpoE1 based activation of SOE associated genes, not all of them are realized in each species studied here (Supplementary Figure S8).
An additional layer of complexity can be added to SOE gene regulation by the presence of a second regulatory circuit that involves RpoE1-type ECF sigma factors. RpoE1 sigma factors from both Sm. meliloti and St. novella were able to interact with either the promoters of the SOE encoding genes and/ or the promoters of the cognate ECF sigma factors (SmRpoE4/SnSigE) in vitro, but the interaction strength and promoter preferences differed clearly between species (Figure 3). While SnRpoE1 strongly activated the Sn_sorA promoter, but did not interact with the Sn_sigE promoter, SmRpoE1 was able to primarily activate Sm_rpoE4p and, to a much lower extent, the Sm_sorT promoter. This indicates that in St. novella, where this sigma factor has never been studied, RpoE1 might exert its effect by directly activating the Sn_sorA promoter, while in Sm. meliloti RpoE1-based regulation would occur mainly via activation of SmrpoE4 expression. SnRpoE1 may thus be responsible for activation of SnsorA gene expression when the SnRpoE1 activating substrate, tetrathionate, is present in the growth medium.
In Sm. meliloti strain 1021 SmRpoE1 appeared to be constantly activated as well as overexpressed (Figures 1, 4 and Supplementary Figure S4), confirming that the C-terminal truncation of the SnRpoE1 ASF encoding gene (SMc_01420) discovered in earlier studies abolishes its ability to effectively sequester RpoE1 (Schlüter et al., 2013). However, the constant activation of SmRpoE1 clearly demonstrated that SMRpoE1 is able to activate SmsorT gene expression in vivo, and explains the unusual, high basal SOE activity present in this strain (Figure 1). In Sm. meliloti GMI11495 where, as in most Sm. meliloti strains, the SmRpoE1 ASF is not truncated, RpoE1-based SmsorT gene expression was only observed in the presence of taurine (Bastiat et al., 2012).
Overall, while RpoE1 mediated regulation of SOE gene expression is present in both species, the specific conditions under which this regulatory circuit is required vary significantly, as does the magnitude and mode of SOE gene regulation in the two species. We therefore propose that RpoE1 based regulation is a secondary mechanism of SOE gene regulation that will have to be studied in the context of sulfur metabolism present in species that contain both RpoE4/SigE-like and RpoE1-like ECF sigma factors (Figure 5 and Supplementary Figure S7).
In summary we have here presented an updated model of activation of SOE gene expression by ECF sigma factors that shows that the presence of sulfite is the main driver of SOE gene expression in two species of α-Proteobacteria that represent different types of bacterial sulfur metabolism, and that co-regulation of SOE gene expression by a second ECF sigma factor occurs in both species but uses distinct mechanisms of regulation. ECF sigma factor genes found in close proximity to SOE encoding genes occur in at least three phylogenetically distinct bacterial groups, two of which contain α-proteobacterial species (group 1 – general and plant-associated soil bacteria such as Rhodobacterales and Rhizobiales, group 2 – mostly marine Rhodobacterales), while the third one exclusively contains β-proteobacterial species (group 3 – mostly Burkholderiales sequences, group 3a Delftia sp., Comamonas sp. and Bordetella sp., group 3b: Acidovorax sp., Thauera sp., and Burkholderia sp.; total of 130 sequences in all three groups) (Figure 5 and Supplementary Figures S8, S9). The promoter consensus sequence (-35 GGAAT and -10 CGTC) (Supplementary Figure S1) identified in Sm. meliloti and St. novella (representatives of groups 1a and 1b, respectively) was present upstream of SOE-encoding genes from phylogenetic groups 1 and 2 (both α-Proteobacterial clusters). There is a strong similarity between the predicted consensus for the SmRpoE1 sigma factor (Schlüter et al., 2013) and the promoter consensus found upstream of the SOE-encoding genes, which may explain the observed co-regulation. This promoter consensus sequence did not appear to be present upstream of any of the group 3 SOE gene sequences and the three groups can also be distinguished based on the positioning of the ECF sigma factor encoding gene relative to the SOE encoding gene (Supplementary Figure S10).
Our phylogenetic analyses further suggest that the recruitment of the ECF sigma factors to SOE gene regions may have occurred at least twice, as the group 3 ECF sigma factors are only distantly related to those found in the α-Proteobacterial groups 1 and 2 investigated here (∼40% amino acid identity), and preliminary investigations also indicate that no close homolog of the RpoE1 ECF sigma factors exists in group 3 species. Given the large number of known ECF sigma factor groups (Staron et al., 2009) it is likely that additional groups of SOE associated ECF sigma factors exist, and future research should focus on these as well as SOE regulation in bacteria known to harbor SOEs not associated with ECF sigma factors. Further work is also needed to understand the molecular details of sulfite sensing by ASFs and release/activation of the sequestered sigma factor, and regulation of ECF sigma factors such as SnSigE that are not subject to autoregulation.
YT, CZ, MN, LO’R, RD, LR, and YW carried out the experimental work. SB contributed to the bioinformatic analyses. YT, RD, and LO contributed to the writing of the manuscript. UK supervised student experiments and was responsible for the preparation of the final manuscript and figures.
This work was supported by a Grant (DP0878525) and fellowship from the Australian Research council to UK and a DoE/Joint Genome Institute Community Sequencing Grant (CSP_787087) to UK and SB. MN was supported by a Faculty for the Future (Schlumberger Foundation) Scholarship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Alexander Bui for assistance with selected aspects of the reporter gene assays.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00960/full#supplementary-material
Alexeyev, M. F. (1999). The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of Gram-negative bacteria. BioTechniques 26, 824–826.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Ang, W. K., Mahbob, M., Dhouib, R., and Kappler, U. (2017). Sulfur compound oxidation and carbon co-assimilation in the haloalkaliphilic sulfur oxidizers Thioalkalivibrio versutus and Thioalkalimicrobium aerophilum. Res. Microbiol. 168, 255–265. doi: 10.1016/j.resmic.2016.12.004
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., et al. (2005). “Current protocols in molecular biology,” in Current Protocols in Molecular Biology, ed. K. Janssen (Hoboken, NJ: John Wiley & Sons Inc.).
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bastiat, B., Sauviac, L., Picheraux, C., Rossignol, M., and Bruand, C. (2012). Sinorhizobium meliloti sigma factors RpoE1 and RpoE4 are activated in stationary phase in response to sulfite. PLoS One 7:e50768. doi: 10.1371/journal.pone.0050768
Beringer, J. E. (1974). R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84, 188–198. doi: 10.1099/00221287-84-1-188
Burgess, R. R. (1996). Purification of overproduced Escherichia coli RNA Polymerase sigma factors by solubilizing inclusion bodies and refolding from sakosyl. Methods Enzymol. 273, 145–149. doi: 10.1016/s0076-6879(96)73014-8
Burgess, R. R. (2009). Refolding solubilized inclusion body proteins. Methods Enzymol. 463, 259–282. doi: 10.1016/S0076-6879(09)63017-2
Cook, A. M., and Denger, K. (2002). Dissimilation of the C-2 sulfonates. Arch. Microbiol. 179, 1–6. doi: 10.1007/s00203-002-0497-0
Davidson, M. S., Roy, P., and Summers, A. O. (1985). Transpositional mutagenesis of Thiobacillus novellus and Thiobacillus versutus. Appl. Environ. Microbiol. 49, 1436–1441.
Davidson, M. S., and Summers, A. O. (1983). Wide-host-range plasmids function in the genus Thiobacillus. Appl. Environ. Microbiol. 46, 565–572.
D’errico, G., Di Salle, A., La Cara, F., Rossi, M., and Cannio, R. (2006). Identification and characterization of a novel bacterial sulfite oxidase with no heme binding domain from Deinococcus radiodurans. J. Bacteriol. 188, 694–701. doi: 10.1128/jb.188.2.694-701.2006
Ditta, G., Schmidhauser, T., Yakobson, E., Lu, P., Liang, X. W., Finlay, D. R., et al. (1985). Plasmids related to the broad host range vector, pRK 290, useful for gene cloning and for monitoring gene expression. Plasmid 13, 149–153. doi: 10.1016/0147-619x(85)90068-x
Frith, M. C., Saunders, N. F. W., Kobe, B., and Bailey, T. L. (2008). Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 4:e1000071. doi: 10.1371/journal.pcbi.1000071
Galibert, F., Finan, T., Long, S., Puhler, A., Abola, P., Ampe, F., et al. (2001). The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293, 668–672. doi: 10.1126/science.1060966
Grant, W. M. (1947). Colorimetric determination of sulfur dioxide. Ind. Engg. Chem. Anal. Edit. 19, 345–346. doi: 10.1021/ac60005a023
Gupta, S., Stamatoyannopoulos, J., Bailey, T., and Noble, W. (2007). Quantifying similarity between motifs. Genome Biol. 8:R24.
Hänsch, R., Lang, C., Rennenberg, H., and Mendel, R. R. (2007). Significance of plant sulfite oxidase. Plant Biol. 9, 589–595. doi: 10.1055/s-2007-965433
Kappler, U. (2008). “Bacterial sulfite dehydrogenases - enzymes for chemolithtrophs only?,” in Microbial Sulfur Metabolism, eds C. Dahl and C. G. Friedrich (Berlin: Springer), 151–169. doi: 10.1007/978-3-540-72682-1_13
Kappler, U. (2011). Bacterial sulfite-oxidizing enzymes. Biochim. Biophys. Acta 1807, 1–10. doi: 10.1016/j.bbabio.2010.09.004
Kappler, U., and Bailey, S. (2005). Molecular basis of intramolecular electron transfer in sulfite-oxidizing enzymes is revealed by high resolution structure of a heterodimeric complex of the catalytic molybdopterin subunit and a c -type cytochrome subunit. J. Biol. Chem. 280, 24999–25007. doi: 10.1074/jbc.m503237200
Kappler, U., Bennett, B., Rethmeier, J., Schwarz, G., Deutzmann, R., Mcewan, A. G., et al. (2000). Sulfite: cytochrome c oxidoreductase from Thiobacillus novellus - purification, characterization and molecular biology of a heterodimeric member of the sulfite oxidase family. J. Biol. Chem. 275, 13202–13212. doi: 10.1074/jbc.275.18.13202
Kappler, U., Davenport, K., Beatson, S., Lucas, S., Lapidus, A., Copeland, A., et al. (2012). Complete genome sequence of the facultatively chemolithoautotrophic and methylotrophic alpha-Proteobacterium Starkeya novella type strain (ATCC 8093T). Stand. Genomic Sci. 7, 44–58. doi: 10.4056/sigs.3006378
Kappler, U., and Enemark, J. H. (2015). Sulfite-oxidizing enzymes. J. Biol. Inorg. Chem. 20, 253–264. doi: 10.1007/s00775-014-1197-3
Kappler, U., Friedrich, C. G., Truper, H. G., and Dahl, C. (2001). Evidence for two pathways of thiosulfate oxidation in Starkeya novella (formerly Thiobacillus novellus). Arch. Microbiol. 175, 102–111. doi: 10.1007/s002030000241
Kappler, U., and Nouwens, A. S. (2013). Metabolic adaptation and trophic strategies of soil bacteria - C1- metabolism and sulfur chemolithotrophy in Starkeya novella. Front. Microbiol. 4:304. doi: 10.3389/fmicb.2013.00304
Kappler, U., and Schwarz, G. (2016). “The sulfite oxidase family of molybdenum enzymes,” in Molybdenum and Tungsten Enzymes, eds R. Hille, C. Schulzke, and M. L. Kirk (Cambridge: Royal Society of Chemistry), 240–273. doi: 10.1039/9781782623915-00240
Kidd, S. P., Potter, A. J., Apicella, M. A., Jennings, M. P., and Mcewan, A. G. (2005). NmlR of Neisseria gonorrhoeae: a novel redox responsive transcription factor from the MerR family. Mol. Microbiol. 57, 1676–1689. doi: 10.1111/j.1365-2958.2005.04773.x
Laemmli, U. K. (1970). Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Low, L., Kilmartin, J. R., Bernhardt, P. V., and Kappler, U. (2011). How are ‘atypical’ sulfite dehydrogenases linked to cell metabolism? - Interactions between the SorT sulfite dehydrogenase and small redox proteins. Front. Microbiol. 2:58. doi: 10.3389/fmicb.2011.00058
Mcgrath, A. P., Laming, E. L., Casas Garcia, G. P., Kvansakul, M., Guss, J. M., Trewhella, J., et al. (2015). Structural basis of interprotein electron transfer in bacterial sulfite oxidation. eLife 4:e09066. doi: 10.7554/eLife.09066
Muench, R., Hiller, K., Barg, H., Heldt, D., Linz, S., Wingender, E., et al. (2003). PRODORIC: prokaryotic database of gene regulation. Nucleic Acids Res. 31, 266–269. doi: 10.1093/nar/gkg037
Praszkier, J., Wilson, I. W., and Pittard, A. J. (1992). Mutations affecting translational coupling between the rep genes of an IncB miniplasmid. J. Bacteriol. 174, 2376–2383. doi: 10.1128/jb.174.7.2376-2383.1992
Ramakers, C., Ruijter, J. M., Deprez, R. H. L., and Moorman, A. F. M. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. doi: 10.1016/s0304-3940(02)01423-4
Rhodes, D., Fairall, L., and Creighton, T. E. (1997). “Analysis of sequence-specific DNA-binding proteins,” in Protein Function, ed. B. D. Hames (Oxford: IRL Press), 215–244.
Schlüter, J.-P., Reinkensmeier, J., Barnett, M. J., Lang, C., Krol, E., Giegerich, R., et al. (2013). Global mapping of transcription start sites and promoter motifs in the symbiotic α-proteobacterium Sinorhizobium meliloti1021. BMC Genomics 14:156. doi: 10.1186/1471-2164-14-156
Simon, J., and Kroneck, P. M. H. (2013). “Chapter two - microbial sulfite respiration,” in Advances in Microbial Physiology, ed. K. P. Robert (Cambridge, MA: Academic Press), 45–117. doi: 10.1016/b978-0-12-410515-7.00002-0
Staron, A., Sofia, H. J., Dietrich, S., Ulrich, L. E., Liesegang, H., and Mascher, T. (2009). The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 74, 557–581. doi: 10.1111/j.1365-2958.2009.06870.x
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Ulrich, L. E., and Zhulin, I. B. (2007). MiST: a microbial signal transduction database. Nucleic Acids Res. 35, D386–D390.
Wilson, J. J., and Kappler, U. (2009). Sulfite oxidation in Sinorhizobium meliloti. Biochim. Biophys. Acta 1787, 1516–1525. doi: 10.1016/j.bbabio.2009.07.005
Keywords: sulfite oxidation, gene expression, gene regulation, extracytoplasmic function sigma factor, microorganisms
Citation: Tan YJC, Zhao C, Nasreen M, O’Rourke L, Dhouib R, Roberts L, Wan Y, Beatson SA and Kappler U (2019) Control of Bacterial Sulfite Detoxification by Conserved and Species-Specific Regulatory Circuits. Front. Microbiol. 10:960. doi: 10.3389/fmicb.2019.00960
Received: 24 December 2018; Accepted: 16 April 2019;
Published: 14 May 2019.
Edited by:
Marc Mussmann, University of Vienna, AustriaReviewed by:
Ralf R. Mendel, Technische Universität Braunschweig, GermanyCopyright © 2019 Tan, Zhao, Nasreen, O’Rourke, Dhouib, Roberts, Wan, Beatson and Kappler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ulrike Kappler, dS5rYXBwbGVyQHVxLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.