
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 April 2019
Sec. Infectious Agents and Disease
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00901
This article is part of the Research Topic Molecular Adaptations of Vibrionaceae to Changing Environments View all 14 articles
A correction has been applied to this article in:
Corrigendum: Surveillance and Genomics of Toxigenic Vibrio cholerae O1 From Fish, Phytoplankton and Water in Lake Victoria, Tanzania
 Yaovi M. Gildas Hounmanou1*
Yaovi M. Gildas Hounmanou1* Pimlapas Leekitcharoenphon2
Pimlapas Leekitcharoenphon2 Rene S. Hendriksen2
Rene S. Hendriksen2 Tamegnon V. Dougnon3
Tamegnon V. Dougnon3 Robinson H. Mdegela4
Robinson H. Mdegela4 John E. Olsen1
John E. Olsen1 Anders Dalsgaard1
Anders Dalsgaard1The occurrence of toxigenic Vibrio cholerae O1 during a non- outbreak period in Lake Victoria was studied and genetic characteristics for environmental persistence and relatedness to pandemic strains were assessed. We analyzed 360 samples of carps, phytoplankton and water collected in 2017 during dry and rainy seasons in the Tanzanian basin of Lake Victoria. Samples were tested using PCR (ompW and ctxA) with DNA extracted from bacterial isolates and samples enriched in alkaline peptone water. Isolates were screened with polyvalent antiserum O1 followed by antimicrobial susceptibility testing. Whole genome sequencing and bioinformatics tools were employed to investigate the genomic characteristics of the isolates. More V. cholerae positive samples were recovered by PCR when DNA was obtained from enriched samples than from isolates (69.0% vs. 21.3%, p < 0.05), irrespectively of season. We identified ten V. cholerae O1 among 22 ctxA-positive isolates. Further studies are needed to serotype the remaining ctxA-positive non-O1 strains. Sequenced strains belonged to El Tor atypical biotype of V. cholerae O1 of MLST ST69 harboring the seventh pandemic gene. Major virulence genes, ctxA, ctxB, zot, ace, tcpA, hlyA, rtxA, ompU, toxR, T6SS, alsD, makA and pathogenicity islands VPI-1, VPI-2, VSP-1, and VSP-2 were found in all strains. The strains contained Vibrio polysaccharide biosynthesis enzymes, the mshA gene and two-component response regulator proteins involved in stress response and autoinducers for quorum sensing and biofilm formation. They carried the SXT integrative conjugative element with phenotypic and genotypic resistance to aminoglycoside, sulfamethoxazole, trimethoprim, phenicol, and quinolones. Strains contained a multidrug efflux pump component and were resistant to toxic compounds with copper homeostasis and cobalt-zinc-cadmium resistance proteins. The environmental strains belonged to the third wave of the seventh pandemic and most are genetically closely related to recent outbreak strains from Tanzania, Kenya, and Uganda with as low as three SNPs difference. Some strains have persisted longer in the environment and were more related to older outbreak strains in the region. V. cholerae O1 of outbreak potential seem to persist in Lake Victoria through interactions with fish and phytoplankton supported by the optimum water parameters and intrinsic genetic features enhancing survival in the aquatic environment.
Vibrio cholerae, the causative agent of cholera, is one of the oldest pathogens known to cause disease in humans, yet there is still much to be learned regarding its transmission and evolution. Although the majority of the worldś cholera cases occur in Africa, there has been limited research on the occurrence of toxigenic V. cholerae O1 in aquatic environments in countries like Tanzania located around the African Great Lakes (Dalusi et al., 2015a; Hounmanou et al., 2016; Bwire et al., 2018a). Knowledge on occurrence and transmission in and around the lakes are important when establishing preventive and control measures of epidemic cholera, which often affects countries situated around the lakes. Moreover, V. cholerae is ubiquitous in aquatic environments and phytoplankton and fish have been reported as potential reservoirs, e.g., in Tanzania (Haque et al., 2012; Rabia et al., 2017; Hossain et al., 2018a; Nyambuli et al., 2018). The tropical waters of Lake Victoria and Lake Tanganyika are likely natural habitats for the El Tor variant of V. cholerae O1 (Echenberg, 2011).
The incidence of cholera in countries around the Great Lakes varies by season, rainfall, plankton blooms, and level of fishing activities (Nkoko et al., 2011). V. cholerae O1 resistant to common antimicrobials have been isolated in fish and water from the Kenyan side of Lake Victoria (Onyuka et al., 2011). A study in Lake Tanganyika indicated a strong correlation between planktonic blooms, fish abundance and cholera (Plisnier et al., 2015). A recent study in Uganda has also identified V. cholerae O1 in Lake Victoria although the strains were non-toxigenic (Bwire et al., 2018a).
Tanzania has consistently been affected by cholera since 1974 and outbreaks continue to re-occur in various towns, especially in the coastal and Great Lakes regions (Urassa et al., 2009; Reyburn et al., 2011). Since the 7th pandemic reached the country in 1974, cholera has affected more than 230,596 people causing 17,714 deaths, most of which lived in lake zones (Mengel et al., 2014). Despite suggestions of genetic similarity and clonality of environmental and clinical V. cholerae strains (Dalusi et al., 2015b), recent studies have revealed different clonal complexes based on MLVA typing among strains implicated in cholera outbreaks in Tanzania (Kachwamba et al., 2017). Analyses of cholera outbreak strains from Uganda and Mozambique however showed a high level of similarity among strains suggesting a clonal transmission in East African countries (Garrine et al., 2017; Bwire et al., 2018b) probably facilitated by the lakes they share. Thus, there is a need to determine the relatedness of environmental and clinical isolates of V. cholerae and the role of the aquatic lake environments as reservoirs for the pathogen.
Furthermore, V. cholerae has evolved through the emergence of multidrug resistant strains with the acquisition of the SXT integrative conjugative elements (Hendriksen et al., 2011; Spagnoletti et al., 2014; Kaas et al., 2016) and transfer of the cholera toxin gene through phages (Waldor and Mekalanos, 1996). The emergence of the El Tor variant biotype carrying the ctxB1 gene of the Classical biotype and V. cholerae O1 carrying the ctxB7 gene of the Haitian strain is another example of evolution of V. cholerae and stress the importance of a continuous monitoring of the genetic characteristics of environmental V. cholerae (Ghosh-Banerjee et al., 2010; Kim et al., 2014).
The aim of this study was to investigate the occurrence of toxigenic V. cholerae O1 during a non-cholera outbreak period in Lake Victoria and determine the genetic characteristics that support environmental persistence and genetic relatedness to pandemic strains. Whole genome sequencing (WGS) and bioinformatics analysis coupled with environmental surveillance data were applied to determine the occurrence and outbreak potential of V. cholerae O1 in Lake Victoria. The data generated add to our understanding on aquatic reservoirs of toxigenic V. cholerae O1 in Lake Victoria.
During the dry season of June to September 2017 and the rainy season of October to December 2017, 360 samples of lake water, carps (Rastrineobola argentea) and phytoplankton were collected from seven landing sites in the Mwanza Gulf of the Tanzanian basin of Lake Victoria (Figure 1). These included 120 water samples (60 offshore and 60 collected near the lakeshore), 120 carp samples and 120 phytoplankton samples (vertical and horizontal sampling as described below). Samples were collected during both seasons (180 samples in the dry season and 180 samples in the rainy season). Sampling sites were Shadi, Kijiweni and Mkuyuni in Mwanza South and Igombe, Mihama, Bwiru and Kayenze in Mwanza North. Mwanza is located in the Lake Victoria basin between the latitude -2° 31′ 0.01″ S and the longitude 32° 53′ 60.00″ E (Figure 1). Samples of fresh carp (about 200 g per sample, consisting of about 200 individual fish) were purchased from local fishermen immediately after landing at the beach and placed in sterile labeled plastic bags which were transported to the National Fish Quality Control Laboratory (NFQCL) in Mwanza in an insulated box with cooling elements and processed within 4 h of collection.
Surface water samples were collected both onshore and offshore (at least 200 m away from the nearest shore) and were collected in 250 ml sterile glass bottles and transported as shown above for carp samples.
Approximately 100 ml of phytoplankton samples were collected by boat with a phytoplankton towing net of 13-μm mesh size. To maximize chances of recovering V. cholerae from the Lake, phytoplankton samples were obtained from the water surface (horizontal sampling) and from beneath to top (vertical sampling) in order not to miss species/taxa that do not float on the surface water which could harbor V. cholerae of interest. For horizontal samples, the net was set right below the surface of the water after which the boat moved about 50 m. The net was then raised and the phytoplankton sample collected. Vertical samples were collected at a minimum depth of 5 m (up to 10 m depending on the depth offshore) where the net was submerged with a lead to collect the phytoplankton sample by a movement from bottom to top.
In the field, water temperature, pH, electrical conductivity, total dissolved solids and dissolved oxygen were measured using a portable multi-parameter meter (Hengkaituo, Guangdong, China).
Fresh carp samples (25 g consisting of about 20–30 individual carps) were homogenized in a stomacher bag containing 225 ml of alkaline peptone water, pH: 8.5 (APW) (Oxoid Ltd., Hampshire, United Kingdom). The homogenized samples contained carps with all body parts (skin, gills, intestines, flesh etc.). Hundred mL of water samples were filtered through a 0.45 μm pore diameter membrane (Millipore, Bedford, MA, United States) which was directly transferred into 225 mL APW; then the bottle was shaken vigorously before incubation. Similarly, phytoplankton was concentrated on a 0.45 μm pore diameter membrane filter paper (Millipore, Bedford, MA, United States) which was transferred into 225 mL APW. Samples in APW were enriched at 37°C for 6 h. Each sample was then analyzed as described below by a culture-based procedure and by PCR of DNA extracted from enriched APW samples.
After enrichment in APW, samples were streaked onto thiosulfate-citrate-bile salts-sucrose agar (TCBS) plates (Oxoid Ltd) and incubated at 37°C for 18–24 h. Characteristic dark yellow colonies with about 2 mm diameter were selected, purified and subsequently characterized by biochemical tests for identification of V. cholerae (Hounmanou et al., 2016). Pure cultures of presumptive V. cholerae colonies were then subjected to DNA extraction. DNA was extracted from boiled lysates of isolates and subjected to PCR for detection of the outer membrane protein gene (ompW) generating a 588 bp amplicon confirming them as V. cholerae (Dalusi et al., 2015a). Isolates confirmed as V. cholerae were subjected to agglutination with polyvalent V. cholerae O1 antiserum (Bio-Rad, France) and were tested for toxigenicity by PCR targeting the cholera enterotoxin subunit A gene (ctxA) along with the ompW primers in a multiplex reaction.
Strains confirmed as toxigenic V. cholerae O1 were subjected to antimicrobial susceptibility testing by the Kirby-Bauer disk diffusion method with Escherichia coli ATCC 22925 included for quality control. Twelve different antimicrobial disks (Oxoid Ltd) were used and results interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2017). Antimicrobials tested included: streptomycin (10 μg), gentamicin (30 μg), ampicillin (10 μg), nalidixic acid (30 μg), ciprofloxacin (5 μg), trimethoprim (5 μg), ceftazidime (30 μg), tetracycline (30 μg), chloramphenicol (30 μg), amoxicillin-clavulanicc acid (30 μg), cefotaxime (30 μg), and sulfamethoxazole (25 μg).
About 10 ml aliquots of APW enriched samples were stored at -20°C until analysis. Total DNA was extracted using a Zymo DNA extraction Kit (The Epigenetics Company, CA, United States) following the manufacturer’s protocol. DNA was extracted from 200 randomly selected samples of the 360 samples enriched in APW. A multiplex PCR was performed to target the 588-bp region of the ompW gene (V. cholerae species specific) and a 301-bp region of the subunit A of the cholera enterotoxin gene (ctxA). Samples that generated only one band (588 bp) were V. cholerae whereas samples showing both bands (588 and 301 bp) were concluded toxigenic V. cholerae ctxA-positive.
Primers used and PCR conditions were as previously described (Nandi et al., 2000; Dalusi et al., 2015a) with a modification of the annealing temperature which was set at 55.6°C during optimization.
Immediately after sampling in dark bottles, 250 mL phytoplankton samples were fixed with five mL Lugol’s solution and 10% formalin and then transported to the Water Quality Laboratory of the Lake Zone in Mwanza. Using an inverted microscope at resolutions of 40 × 10 and 10 × 10 and following standards keys (Verlecar and Desai, 2004), two ml of fixed sample was used by a trained taxonomist to identify taxons and species groups of phytoplankton present in the water samples.
Proportions of positive V. cholerae samples from the culture-based method and PCR of DNA extracted from APW enriched samples were compared using chi-square in Epi-Info software version 7.21 at a probability of 0.05. Seasonal differences were also tested. Using the statistical software R v3.5.12 logistic regression was performed to estimate the effect of water temperature, pH, conductivity, dissolved oxygen, and total dissolved solids on the occurrence of ctxA-positive V. cholerae between sampling points.
Eight of the ten ctxA-positive V. cholerae O1 recovered from fish, phytoplankton and water and two ctxA-positive non-O1 V. cholerae from phytoplankton and fish (Plankton1, Water1) were selected for WGS analysis. DNA was extracted using the automated Maxwell DNA extraction system (Promega Maxwell RSC, Madison, WI, United States) and the Maxwell DNA extraction and purification kit. Prior to extraction, samples were treated with Proteinase K (Sigma-Aldrich, St. Louis, MO, United States) at 56°C for 1 h followed by 10 min RNase (Sigma-Aldrich) treatment at room temperature. Concentrations of the extracted DNA were determined using a Qubit dsDNA HS assay kit (Invitrogen, United States). The DNA was run in 1% agarose gel to check for quality followed by the preparation for Illumina paired-end WGS using Illumina Miseq (Illumina, Inc., San Diego, CA, United States) according to the procedures previously described (Kaas et al., 2016). Raw sequences are submitted to the European Nucleotide Archive under the project number PRJEB30604 with the accession numbers of each sample indicated in Supplementary Table S1.
The raw paired-end reads were assembled using SPAdes assembler (Bankevich et al., 2012) available online3. Assembled sequences were analyzed in the batch upload pipeline of the CGE platform4as previously described (Thomsen et al., 2016), where KmerFinder 2.1 identified the species and possible contaminations. The species V. cholerae was further confirmed based on the species-specific ompW gene using MyDbFinder 1.2 tool (Siriphap et al., 2017) with a threshold set at 98% identity. This revealed the closest genome to the analyzed sequences as V. cholerae 2010EL-1786 (Reimer et al., 2011). Moreover, the sequences were further analyzed using MyDbFinder 1.2 tool with default options to identify known virulence genes and pathogenicity islands (Supplementary Table S1). This included the identification of V. cholerae serogroup-specific genes (rfbV-O1, wbfZ-O139), biotypes-specific genes (ctxB, rstR, tcpA), putative virulence genes (including ctxA), and VC2346 specific for the 7th pandemic V. cholerae using a threshold of 98% identity (Kaas et al., 2016; Siriphap et al., 2017). MyDbFinder coupled with nucleotides BLAST were used to genotype the strains based on the ctxB of the CTX prophage that they carried. This helps to identify the wave of the seventh pandemic that the strains belonged to using ctxB1, ctxB3, and ctxB7 sequences (Naha et al., 2012; Kaas et al., 2016; Rashid et al., 2016). Detection of genomic islands of V. cholerae mainly VPI-1, VPI-2, VSP-1, VSP-2, was also carried out in silico. Moreover, makA, alsA and Type VI secretion system (T6SS) genes were searched. The CTX prophage genomic region containing the core region and RS1 was also assessed in the samples. Detection of the PICI like elements, i.e., PLE1 and PLE2, responsible for phage susceptibility in V. cholerae was performed using MyDbFinder 1.2. The MLST 2.0 tool reached through the batch upload pipeline analyzed the sequence types (ST) of the V. cholerae strains in order to establish genetic relatedness with known STs that have been implicated in cholera outbreak. The analysis was based on the seven housekeeping genes: adk, gyrB, metE, mdh, pntA, purM, and pyrC (Kaas et al., 2016). Moreover, a ribosomal multi-locus sequence typing which is a more discriminative MLST typing method based on variations of the 53 genes encoding the bacterial ribosome protein subunits (rps genes) was used to investigate differences among the environmental strains as previously described (Jolley et al., 2012).
In the batch upload pipeline (Thomsen et al., 2016), ResFinder 3.0 assessed acquired antimicrobial resistance (AMR) genes in the assembled sequences. In ResFinder with default options, we also searched for beta-lactam resistance genes including blaVCC-1, a carbapenamase gene that is emerging in environmental V. cholerae as recently reported in strains from Canada and Germany (Mangat et al., 2016; Hammerl et al., 2017).
Detection of AMR genotype was further strengthened with the search for mobile genetic elements using MyDbFinder 1.2 (Larsen et al., 2012) for the detection of the SXT integrative conjugative element, the different classes of integrons, and the presence of mutations in the DNA gyrase (gyrA gene) and in the DNA topoisomerase IV (parC gene) (Siriphap et al., 2017). MyDbFinder 1.2 was used to compare sample sequences with the integrating conjugative elements (ICE) of V. cholerae O1 ICEVchHai1(JN648379) and of SXTMO10 (AY034138) at a threshold of 98% identity. Due to high levels of mutations in the SXT element (Wang et al., 2016) the threshold for detection of intSXT was set at 95% for % ID of 40% minimum length.
The search for plasmids was done in three steps. The PlasmidFinder 1.3 tool of the batch upload pipeline in CGE (Thomsen et al., 2016) was used to search for plasmid replicons. Due to the limitation of PlasmidFinder, which detects only replicons that are available in the CGE database, we conducted another specific search. This second step used MyDbFinder 1.2, where a local search was performed between our samples’ genomes and sequences of representative IncA/C plasmids (known for carrying multidrug resistance genes in V. cholerae) available from the Genbank (Accession numbers: KY399978, KM083064, KF551948, CP007636, CP033514, and KJ817377) (Carraro et al., 2014; Folster et al., 2014; Wang et al., 2018). This step also involved two cryptic plasmids recently isolated in V. cholerae strains (Acc. KY486774 and KY486775) (Ceccarelli et al., 2017). In the third step, we used Blast atlas in GView5 where genomes were analyzed against reference plasmids.
Moreover, the genome sequences were annotated in RAST v.2.0 (Brettin et al., 2015) and each annotated file was analyzed through the SEED viewer (Overbeek et al., 2014) for subsystem categorization of the genetic elements involved in survival and persistence in the aquatic environment.
To further identify evolution and genetic similarities within and between the environmental V. cholerae isolates as compared to cholera outbreak strains, assembled genomes were analyzed using CSIPhylogeny version 1.4 with default options (Kaas et al., 2014), where high quality SNPs of the environmental genomes were identified. Sequences of V. cholerae from Uganda and Kenya that have borders to Lake Victoria as well as previous outbreak strains from Tanzania were obtained from the GenBank and compared with our environmental strains to determine genome wide SNPs. Additionally, V. cholerae O1 outbreak strains from other countries of the African Great Lakes region including the Democratic Republic of Congo (DRC), Burundi and Rwanda but also from Zambia were included in the tree to access the spatial-temporal phylogenetic evolution of the environmental strains. Accession numbers of strains used in the SNP tree are reported in Supplementary Table S2. This generated a regional and time-scale phylogenetic tree showing the genetic relatedness and evolution between the environmental non-outbreak V. cholerae O1 strains and pandemic strains from the African Great Lakes region. The reference genome of V. cholerae O1 strain N16961 (Biosample SAMN02603969) was used to root the tree (Kaas et al., 2016). The Newick files obtained in CSIPhilogeny 1.4 were downloaded and the final tree was amended in iTOL6.
Using standard bacteriological procedures followed by PCR for the species-specific gene ompW, 108 isolates were identified as V. cholerae. These strains were isolated from 77 out of 360 samples (21.4%) analyzed with up to three characteristic colonies being selected from each TCBS agar plate for confirmation. However, APW-enriched samples subjected directly to multiplex PCR yielded 138 DNA samples that were positive for V. cholerae (ompW) out of the 200 samples selected for analysis (69.0%). A Chi-square comparison revealed that PCR of DNA extracted directly after enrichment in APW recovered significantly higher proportions of V. cholerae-positive samples than identification of V. cholerae from isolates obtained on TCBS agar plates (69% vs. 21.4%, p < 0.05). A total of 22 V. cholerae isolates out of the 108 isolates originally recovered were positive for the subunit A of the cholera enterotoxin gene (ctxA). Ten of these strains belonged to serogroup O1 and originated from phytoplankton (five isolates), carps (three isolates), and water (two isolates) samples. Phytoplankton collected offshore by the vertical sampling method yielded all five V. cholerae O1 strains from this source. Two water samples collected offshore contained V. cholerae O1 (Table 1).
In DNA extracted from APW enriched samples, 23 samples contained the ctxA gene in phytoplankton (14), water (6) and carps (3). Fifteen of the PCR-positive samples did not yield any colonies by the culture-based technique. The vertical phytoplankton sampling method yielded slightly more toxigenic V. cholerae ctxA-positive samples than the horizontal sampling method (p > 0.05), which was also the case for samples collected onshore compared with offshore water samples (Table 1). The total DNA extracted from samples enriched in APW were however, not used for further characterization because the quality of the DNA does now allow for sequencing analysis.
There was no statistical seasonal difference (p > 0.05) in recovery of V. cholerae when DNA was obtained from pure cultures (32.8 vs. 27.2%, dry and rainy seasons, respectively) compared to when DNA was obtained from APW-enriched samples (71.0 vs. 67.0%). Nevertheless, a statistically higher proportion of ctxA-positive V. cholerae was obtained in the dry season as compared to the rainy season for samples analyzed by culture procedures (p = 0.04).
The water temperature at the different sampling sites ranged from 24.9 to 25.9°C and water pH ranged between 7.5 and 8.5. The electrical conductivity fluctuated between 90 and 150 μS/cm (Supplementary Table S3). None of these parameters varied significantly between sampling points when they were regressed on the total toxigenic V. cholerae ctxA-positive samples (p > 0.05). The analyzed phytoplankton samples contained 45 different taxa belonging to three major phytoplankton groups including green algae (62.2%), cyanobacteria (31.1%), and diatoms (6.7%).
We sequenced ten strains including eight of the isolated ctxA-positive V. cholerae O1 and two ctxA-positive V. cholerae non-O1, which were characterized by analysis of WGS data. Based on WGS analysis, all the ten sequenced strains including the two phenotypically non-O1 contained the rfbV-O1 gene and should therefore be regarded as of serogroup O1. Table 2 shows genomic characteristics of the strains when variations was seen (see further details in the Supplementary Table S1), whereas similar characteristics are described in the text below. Most strains belong to the third wave of the seventh pandemic as they are all atypical El Tor biotype variants of V. cholerae O1, carrying the ctxB7 genotype of the ctxB gene while possessing the rstR and tcpA genes of El Tor biotype. However, strains Water2, Fish1, Fish3 possess ctxB1 of the early third wave and Plankton1 contained ctxB3 of the first wave clustering with older outbreak strains. The in silico MLST revealed that all strains belonged to the same sequence type ST69 and harbored the seventh pandemic-specific gene (VC2346) suggesting that they belong to the same clonal linage.
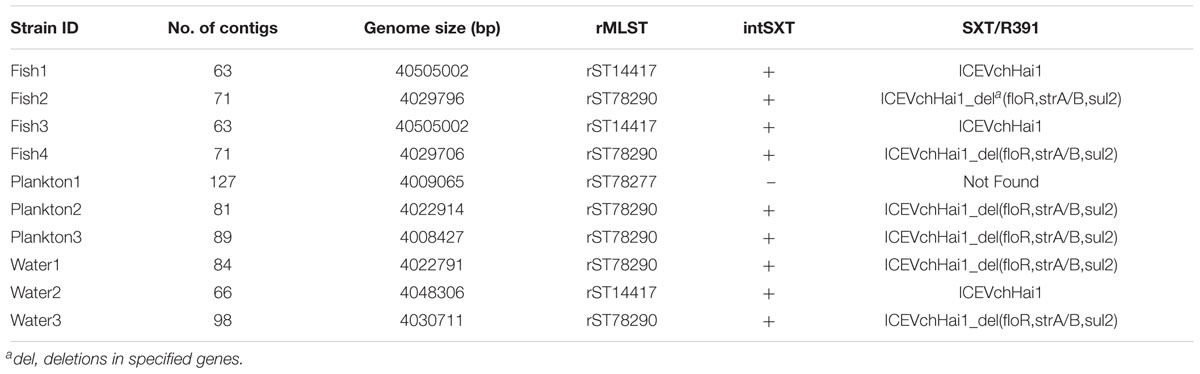
Table 2. Genomic sequence data, virulence profile and occurrence of antimicrobial resistance genes in the V. cholerae strains.
The occurrence of virulence-associated genes and pathogenicity islands among the environmental V. cholerae was similar in all sequenced strains. This included the major virulence-associated genes such as ctxA, ctxB, zot, ace, tcpA, hlyA, mshA, rtxA, ompU, and toxR, whereas stn and chxA genes were absent. Moreover, the strains all contained genes of the Type VI secretion system, the gene involved in glucose metabolism, als and the flagella-mediated cytotoxin gene makA. All strains contained the pathogenicity islands VPI-1, VPI-2, VSP-1, and VSP-2.
All ten sequenced V. cholerae strains showed phenotypic resistance to streptomycin, while eight of them showed resistance to amoxicillin and clavulanic acid. Phenotypic resistance to ampicillin and sulfamethoxazole was observed in 6/10 strains whereas resistance to nalidixic acid and trimethoprim was observed in three strains. All isolates were susceptible to gentamicin, ciprofloxacin, ceftazidime, tetracycline, chloramphenicol and cefotaxime (Figure 2). Sequence analysis revealed, however, that some genes encoding resistance to certain antimicrobial classes were present in strains that are phenotypically susceptible. For instance, all strains harbored the catB9 gene conferring resistance to chloramphenicol, but were phenotypically susceptible. Similarly, all strains except the strain Plankton1 harbored the dfrA1 gene conferring resistance to trimethoprim, while only three of them showed phenotypic resistance.
Moreover, all strains except Plankton1 contained the SXT integrative conjugative element (intSXT gene), but they all lacked specific integrase genes of the class 1 integron (intI gene). Nevertheless, not all the antimicrobial resistance genes normally found on the SXT element such as sul2, dfrA1, dfrA18, floR, strA, and strB which are associated with sulfamethoxazole and trimethoprim, chloramphenicol and streptomycin resistance, respectively were present in the strains according to MyDbFinder 1.2. Therefore, a Blast Atlas analysis was performed where the genomes where compared to the reference V. cholerae strain 2010EL-1786. This revealed that except for strains Fish1, Fish3, and Water2 that were positive for aminoglycoside, phenicol and sulfonamide resistance genes, the remaining strains showed nucleotide deletions in their genomes (Figure 3). A more detailed analysis of the strains that have deletions showed that they contained fragments of the concerned genes (strA, strB, Sul2, floR) on the ICE fragment, with about 1100 bp gaps between bp position 98500–102450 (Figure 3).
Strains Fish1, Fish3, and Water2 showed resistance to nalidixic acid and contained expected amino acid substitutions in gyrA (Ser83-Ile) and parC (Ser85Leu), but none of the ten strains contained the fluoroquinolone resistance gene qnrVC1. Likewise, none of the ten strains contained beta-lactam resistance genes including blaVCC-1, a carbapenamase gene. Nine environmental strains harboring the SXT element had a genomic organization of the integrating conjugative element (ICE) similar to that of the V. cholerae ICEVchHai1 (Reimer et al., 2011) reference strain with some common deletions seen in loci VC1786ICE78 for all strains and in VC1786ICE14 for strains Fish1, Fish3 and Water2. The other six strains had deletions in loci VC1786ICE6-13, VC1786ICE81, and VC1786ICE83-84 (Supplementary Table S4).
Our strains have no plasmid replicons according to PlasmidFinder. Moreover, in MyDbFinder and in Blast atlas, the strains had no copies of the IncA/C plasmid, as well as the cryptic plasmids pSDH1-2. The 10 strains irrespective of their biotype lacked the phage susceptibility region of the PICI like elements (PLE1, PLE2).
All ten analyzed genomes present molecular machinery for attachment, survival and defense for environmental persistence. The magnesium and cobalt efflux protein (CorC), the cobalt-zinc-cadmium resistance protein (CzcD) and the multidrug efflux pump component (MtrF) were conversed in the defense system of all the sequenced strains (Figure 4). Moreover, the strains all possess the mshA gene involved in attachment to chitin, as well as autoinducers (AI-2 LuxP and LuxQ) involved in quorum sensing and biofilm formation for environmental survival. A number of environmental stress response regulator proteins were conserved in the strains mainly the response regulators of the VieSAB transduction system of Vibrio, the two-component response regulator proteins, histidine kinase, and Vibrio polysaccharides (VPS) biosynthesis proteins (Figure 4).
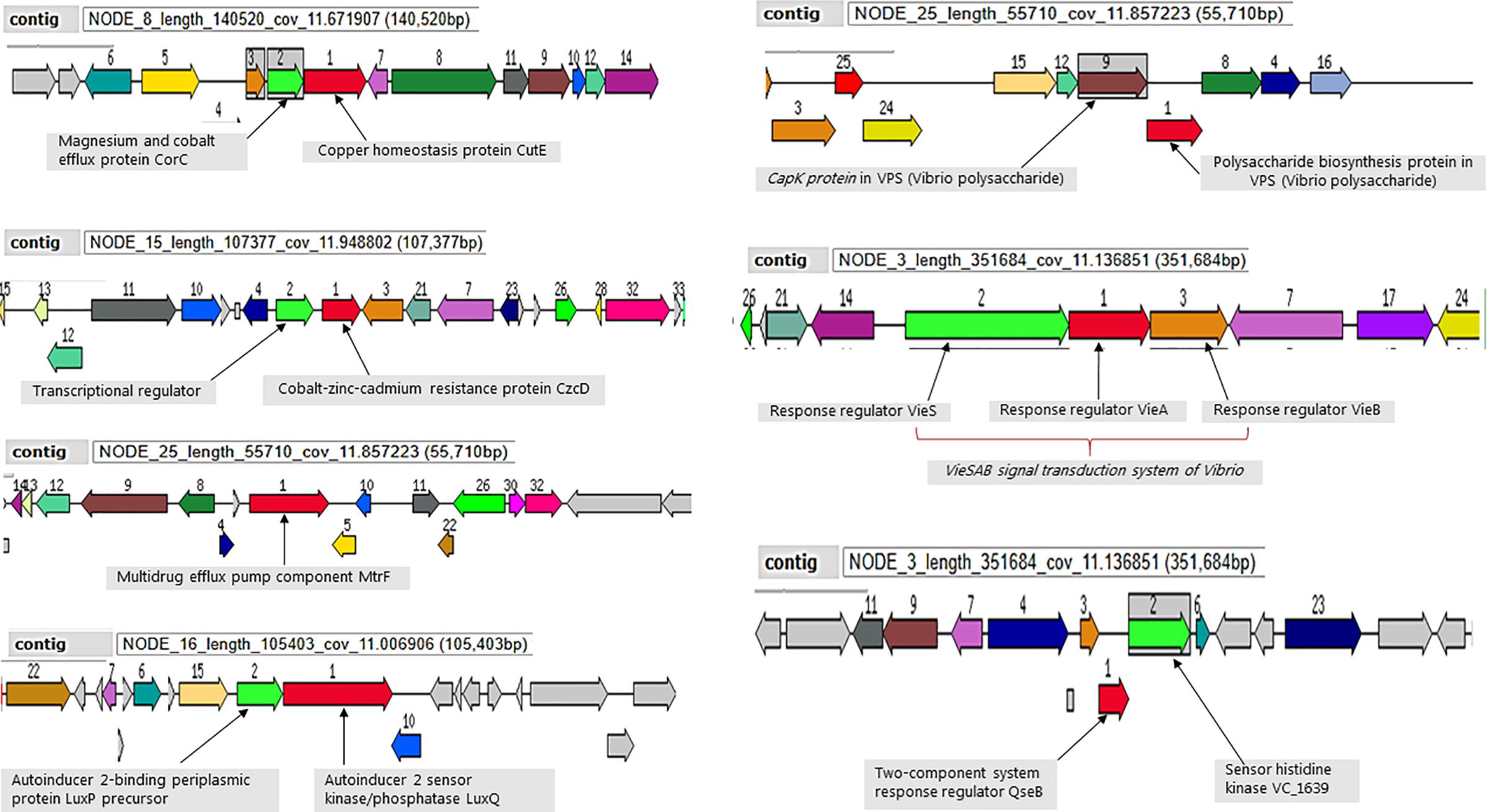
Figure 4. Selected genetic elements encoding for environmental fitness. Strain Plankton2 was used for this illustration. The contig in which the genetic complex are found is indicated.
Overall, the SNP tree revealed a wide diversity among the environmental strains with up to 174 SNP differences (found between strains Plankton1 and Plankton3). Strains Fish1 and Fish3, as well as Fish2 and Fish4 were strictly clonal, respectively. Very close relatedness was also observed between strains Water2, Fish1, and Fish3 (only four SNPs difference), between Water2, Fish2, and Fish4 (only five SNPs difference) and between strains Plankton2, Fish2, and Fish4 (only nine SNPs difference). We further performed a ribosomal MLST typing (Jolley et al., 2012) to assess the diversity of the environmental strains and found little diversity as six of the ten strains belong to the same rMLST type ST78290. The strain Plankton1, which shows above 100 SNP differences compared to its pairs, belongs to a separate rMLST type 78277 whereas strains Fish1, Fish3, and Water2 belong to rMLST type 14417.
When compared to published genomes of the Genbank, the environmental V. cholerae O1 strains were found in three different clusters among the pandemic strains. The strain Plankton1 was distant from all genomes as a singleton but closer to strains associated with cholera outbreaks in Tanzania from 1993 and 1997 with 88 to 95 SNPs (Figure 5A and Supplementary Table S5). Strains Fish1, Fish3, and Water2, however, formed a separate cluster (29–33 SNPs, Supplementary Table S6) with a 2013 pandemic strain from DRC (Figure 5B). This clade also contains older pandemic strains from Burundi (2001) and Tanzania (2009). The other environmental strains were highly related to recent epidemic strains with less than 22 SNPs difference between our environmental O1 strains Fish2, Fish4, Plankton2, Plankton3, Water1, Water 3 and the V. cholerae O1 strains associated with cholera outbreaks in Tanzania and Kenya in 2015 and in Uganda in 2016 (Figure 5C). The strains Fish2 and Fish4 for instance, were highly clonal when compared to the 2015 Tanzanian and Kenyan outbreak strains (only three SNPs apart; Figure 5C) and the Uganda outbreak strain (nine SNP differences) (Biosample Accession SAMN08744331). The largest difference in SNPs between the environmental and pandemic V. cholerae O1 strains was 183, found between strain Plankton1 and one strain associated with the Ugandan cholera outbreak (Biosample Accession: SAMN08744333). The pairwise SNP differences among all strains are reported in Supplementary Table S5.
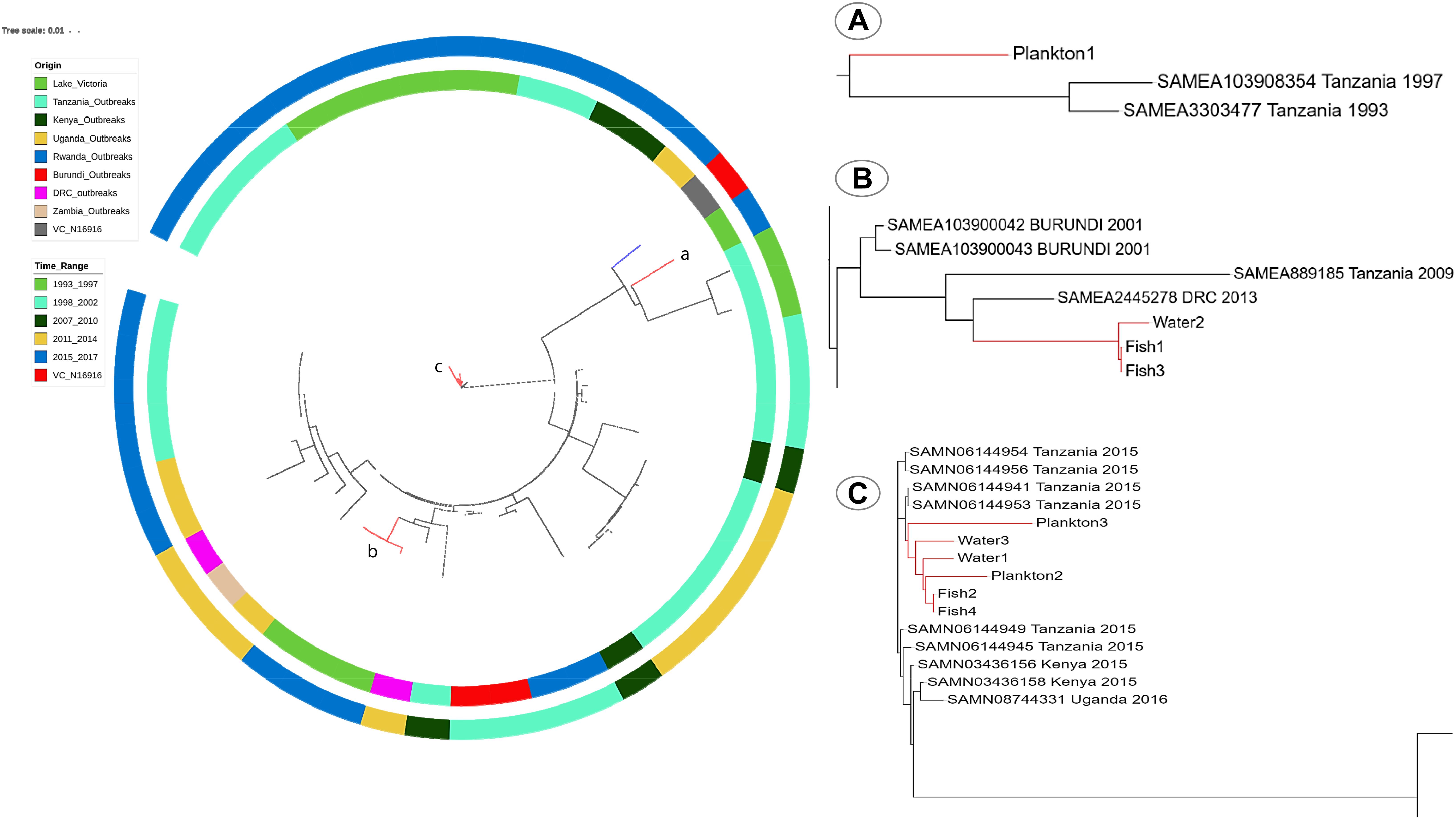
Figure 5. SNP-Tree showing spatial-temporal phylogeny of environmental strains and pandemic V. cholerae from seven countries in the African Great Lakes Region. Colors on the inner lane indicate the origin of strains and the outer lane colors indicate the timeline evolution of the strains as shown in the legend. The tree was rooted with the reference V. cholerae N16916 (blue branch). Red branches identify the position of environmental genomes on the tree. (A) Localization of the strain Plankton1 in a clade of older strains involved in the initial waves of the seventh pandemic; (B) position of Fish1, 3 and Water2, and (C) a clade of recent outbreak strains in Tanzania, Kenya, and Uganda with most environmental strains.
We report the occurrence of toxigenic V. cholerae O1 and non-O1 in the aquatic environment of the Tanzanian basin of Lake Victoria during a non-cholera outbreak period. The presence of cholera toxin-positive V. cholerae non-O1 in Lake Victoria suggests that the ctxA gene is not limited to only the O1/O139 serogroups. It should be noted that we did not agglutinate the non-O1 strains in O139 antiserum. However, V. cholerae O139 do not seem to occur in Africa and a recent report on non-toxigenic O139 needs to be confirmed (Bwire et al., 2018a). Studies have identified V. cholerae O141 and O75 harboring the CTX prophage including the ctxA gene which have been responsible for cholera-like outbreaks, e.g., in the United States (Dalsgaard et al., 2001; Crump et al., 2003; Haley et al., 2014). Moreover, the occurrence of non-O1 toxigenic strains in Lake Victoria underlines that serotyping beyond O1 and O139 antisera should be implemented as the CTX prophage seem to be present in more serogroups than initially documented (Waldor and Mekalanos, 1996).
The two toxigenic non-O1 V. cholerae sequenced in this study showed that they were in fact O1 serogroup strains based on the presence of the rfbV-O1 gene. This discrepancy between the phenotype and the genotype could be due to changes in the O antigen expression leading to delayed and non-conclusive agglutination in these strains during testing. It is unlikely that our phenotypically non-O1 ctxA-positive strains might be of another serogroup than O1 since they possess the rfbV-O1 gene but also belong to the atypical El Tor biotype of serogroup O1.
The most recent cholera outbreak in the study area occurred in Mwanza in March 2016 (Unpublished report from the Tanzanian Ministry of Health and Social Welfare). Thus, the current study was conducted more than a year after the latest reported cholera outbreak; yet toxigenic V. cholerae O1 strains were recovered from phytoplankton, fish (carps) and water samples, suggesting that Lake Victoria is a reservoir of V. cholerae. Isolation of V. cholerae O1 strains between inter-epidemic periods are rare (Alam et al., 2006; Faruque and Mekalanos, 2012) partly because V. cholerae O1 may become dormant, i.e., viable-but-non-culturable (Kamruzzaman et al., 2010; Bari et al., 2013). Nevertheless, our results are consistent with other findings, which also detected toxigenic strains of V. cholerae O1 in aquatic environments during non-cholera outbreak periods (Onyuka et al., 2011; Dalusi et al., 2015a; Hounmanou et al., 2016). Furthermore, the sequenced environmental V. cholerae isolates belong to the seventh cholera pandemic lineage similar to strains that caused recent epidemics (2015–2016) in countries surrounding Lake Victoria (Tanzania, Kenya, and Uganda) with SNPs ranging between 3 and 22 for samples Fish2, Fish4, Plankton2, Plankton3, Water1, and Water 3. This very close genetic relatedness to outbreak strains further substantiates that such environmental isolates are of outbreak potential given that their closest progenitors are epidemic strains. The environmental V. cholerae O1 isolates are likely of outbreak origin and potential progenitors of cholera outbreak strains. On the other hand, the spatial-temporal analysis of our environmental strains revealed a regional spread and long-term environmental persistence of pandemic V. cholerae within the African Great Lakes region. The close genetic relatedness of strain Plankton1 to the 1993 and 1997 pandemic strains from Tanzania suggests that this strain did not emerge from the recent outbreaks like the others but has probably persisted in the lake for at least 20 years. Although Plankton1 is an atypical El Tor strain whereas the 1993 and 1997 pandemic strains were prototype El Tor, their closer phylogenetic relatedness compared to the other environmental and pandemic strains show that this strain most probably underwent ctxB-mediated mutations throughout the years to become atypical El Tor (Rashid et al., 2016). Furthermore, Fish1, Fish3, and Water2 were distant from their environmental counterparts but closely related to older pandemic strains from DRC, Tanzania and Burundi. The finding of strains Fish1, Fish3, and Water2 suggests not only a long-term persistence of V. cholerae in the lake but also an environmental adaptation aided by a human-mediated spread between the countries in the African Great Lakes region enhanced by connecting water bodies. This phylogenetic heterogeneity of V. cholerae isolated in Lake Victoria substantiate that the lake serves as reservoir for long-term persistence of pandemic clones of V. cholerae with a much-dispersed geographical distribution. Sequence analysis revealed that the V. cholerae O1 strains possessed all major virulence-associated genes found in clinical O1 strains, e.g., the cholera enterotoxin genes, toxin co-regulated pilus, hemolysis genes, as well as the flagella-mediated toxin gene makA (Aliabad et al., 2012; Castillo et al., 2018; Dongre et al., 2018). Genes involved in the T6SS that enables V. cholerae to overcome commensals and immune cells in the human gut during infection were also present in our environmental isolates (Unterweger et al., 2014; Logan et al., 2018). Moreover, the environmental strains were V. cholerae O1 El Tor variants of the Haitian ctxB7 genotype and therefore similar to strains implicated in the ongoing third wave of the seventh pandemic known to cause more severe disease as compared to the typical El Tor biotype (Ghosh-Banerjee et al., 2010; Mutreja et al., 2011; Kim et al., 2014). The El Tor biotype variants of V. cholerae O1 are proposed to be common in the African Great Lakes region with potential to cause cholera outbreaks (Echenberg, 2011). Our study provides evidence that cholera outbreak isolates may persist in the aquatic environment during non-outbreak periods. This may explain why clonal isolates are found to cause different outbreaks many years apart, e.g., as seen in Mozambique (Garrine et al., 2017).
Vibrio cholerae of sequence type ST69 belonging to the third wave of the seventh cholera pandemic are the ones responsible for most cholera outbreaks worldwide (Mutreja et al., 2011; Kachwamba et al., 2017; Weill et al., 2017). The presences of the pathogenicity islands VPI-1, VPI-2, VSP-1, and VSP-2 in the strains further confirm their outbreak potential. Our strains clustered in two major ribosomal MLST types mainly rMLST 78290 and 14417, which are identical to rMLST types of cholera outbreaks strains in the region (Mutreja et al., 2011; Kachwamba et al., 2017). We observed that strains of rMLST type 78920 clustered together on the SNP tree with outbreak strains of the same rMLST with strains of rMLST type 14417 clustering in a similar manner (Figure 5). Most cholera outbreak strains isolated from 2015 and the majority of our environmental strains are of an identical rMLST type 78920 suggesting not only a transmission of strains between the aquatic environment and humans, but also a time-scale evolution of V. cholerae from one rMLST type to another. Our results suggests that Lake Victoria is a reservoir for V. cholerae O1 with outbreak potential which are consistent with findings in previous studies (Faruque et al., 1998, 2007; Dalusi et al., 2015b), but are not in agreement with a recent report which questioned whether environmental strains may be progenitors of outbreak strains (Weill et al., 2017). In Haiti, a close phylogenetic relationship has also been reported between clinical and environmental toxigenic V. cholerae O1 strains (Azarian et al., 2014). Moreover, some ctx-negative environmental V. cholerae O1 clustered with strains responsible for clinical cholera and possessed genomic characteristics of the seventh pandemic lineages (Azarian et al., 2016). This provides further arguments that the aquatic environment is a reservoir in the survival and generation of progenitors of outbreak strains of V. cholerae.
The persistence of epidemic V. cholerae O1 strains in the lake can be attributed to a number of environmental factors. One is the presence of suitable hosts. The symbiotic relationship between phytoplankton and V. cholerae is well documented and known to enhance the survival and maintenance of the pathogenicity in these strains even after cholera outbreaks have ceased (Haque et al., 2012; Mitchell et al., 2017). Attachment to aquatic organisms to take advantage of chitin and other nutrients is dependent on the MSHA (mannose-sensitive hemagglutinin) pili (Meibom et al., 2004; Sinha-Ray and Ali, 2017), which were present in our V. cholerae strains and whose expression by the mshA gene increases in warm tropical water with temperatures above 15°C (Lutz et al., 2013). The conductivity measurements and the surface water temperature in the lake averaging 25°C with an alkaline pH averaging 8.5, provides optimum conditions for survival and growth of V. cholerae (Lugomela et al., 2014; Dalusi et al., 2015a; Plisnier et al., 2015). The WGS data analysis further indicates that the V. cholerae strains have survived in and adapted to the lake environment for about two decades. Among phytoplankton groups that may harbor V. cholerae O1 in Lake Victoria, green algae and cyanobacteria were the major species detected in our study which are both documented aquatic reservoirs for V. cholerae O1 and O139 in Asia and Africa (Eiler et al., 2007; Dalusi et al., 2015b; Islam et al., 2015). We noticed that most V. cholerae O1 strains were recovered in phytoplankton samples obtained in deep water which suggests that the vertical sampling technique for phytoplankton should be used in future environmental surveillance studies. Food safety aspects and transmission of V. cholerae O1 in carps to humans are questionable as carps are sun-dried before consumption, but needs to be further explored. V. cholerae O1 was isolated in Nile perch from the Kenyan side of the Lake Victoria (Onyuka et al., 2011), but the role of the small carps as reservoir hosts of V. cholerae O1 remains uncertain. Experimental studies suggest that V. cholerae strains can use accessory toxins and structural components to survive in the gut of zebrafish over long periods (Runft et al., 2014; Mitchell et al., 2017). Moreover, Hilsa fish from Bangladesh and many other species were also reported aquatic hosts of V. cholerae (Halpern and Izhaki, 2017; Hossain et al., 2018a).
Although seasonal variation was not a significant predictor of recovery of toxigenic strains in this study, V. cholerae O1 are generally more frequently isolated during rainy seasons (Reyburn et al., 2011). Our strains were negative for the PICI-like elements responsible for phage susceptibility. These elements are genetic islands of V. cholerae that inhibit lytic phages and therefore serve as a resilience factor toward vibrio phages (Seed et al., 2013). It has been shown that seasonal fluctuations of cholera cases can be associated with predation of V. cholerae O1 by phages in the environment leading to a decrease in the epidemic curve (Faruque and Mekalanos, 2012). A long-term persistence of epidemic V. cholerae O1 clones in the lake can also be attributed to their adaptive genetic machinery made of defense genes against toxic compounds and heavy metals. The strains contained magnesium and cobalt efflux protein (CorC) and the cobalt-zinc- cadmium resistance protein (CzcD). Studies have found different levels of heavy metals and pesticide residues in Lake Victoria (Kishe and Machiwa, 2003; Ogwok et al., 2009), which could be detrimental to bacteria like V. cholerae and limit their growth. The capability of V. cholerae O1 strains to survive in such an environment is justifiable by the genetic elements enhancing their ability to resist these toxic compounds. Moreover, in a complex environment like Lake Victoria, V. cholerae may be subjected to a number of other environmental stresses. The presence of autoinducers, AI-2 LuxP and LuxQ (Joelsson et al., 2007) in the genetic makeup of our strains favors their persistence since they are capable of quorum sensing and forming biofilm to survive and thrive (Kamruzzaman et al., 2010; Bari et al., 2013; Sinha-Ray and Ali, 2017). The strains also contained various stress response regulator proteins like the VieSAB transduction system, the two-component response regulator proteins, and Vibrio polysaccharides (VPS) biosynthesis proteins, which are essential for environmental fitness and persistence in V. cholerae (Fong et al., 2010; Vesth et al., 2010; Lutz et al., 2013). Thus, the molecular characteristics of our V. cholerae strains substantiate their spatial-temporal persistence in the lake from where they may emerge and cause outbreaks.
As the concentration of V. cholerae O1 is low in the aquatic environment between cholera outbreaks, it is important to apply methods with high sensitivity for environmental surveillance (Bwire et al., 2018a; Nyambuli et al., 2018). We observed that PCR of DNA extracted from APW-enriched samples detected more V. cholerae-positive samples than using subculture onto TCBS agar with subsequent confirmation of selected isolates by PCR. Similar observations were reported analyzing sediments and plankton from Tanzanian estuaries using culture-independent PCR techniques (Dalusi et al., 2015a). Viable but non-culturable forms of toxigenic and non-toxigenic V. cholerae are important in the ecological dynamics and epidemiology of cholera (Kamruzzaman et al., 2010; Faruque and Mekalanos, 2012). However, these forms of V. cholerae cells cannot be detected by culture directly on selective agar media. We therefore suggest the detection of toxigenic V. cholerae by PCR of APW-enriched samples as a sensitive and efficient method in routine monitoring of V. cholerae in the environment.
Increasingly, antimicrobials are used in cholera treatment to reduce the volume of diarrhea, and V. cholerae O1 concentrations in stools. Our study revealed that isolates were resistant to commonly used antimicrobials such as streptomycin, amoxicillin + clavulanic acid, ampicillin, trimethoprim and sulfamethoxazole; findings which are similar to earlier studies of V. cholerae from Tanzania (Hounmanou et al., 2016; Rabia et al., 2017). Most of our strains carried the SXT integrative conjugative element containing resistance genes to aminoglycosides, sulfamethoxazole, trimethoprim, phenicols and quinolones with genomic similarity to V. cholerae ICEVchHai1 (Hendriksen et al., 2011; Kaas et al., 2016). Some of our strains carrying genes encoding phenicol resistance were susceptible to chloramphenicol (Siriphap et al., 2017; Hossain et al., 2018b) which shows that the expression of a phenotype is not solely related to the possession of encoding gene. Moreover, the nucleotide deletions that occurred within the SXT element are not rare, as studies have underlined frequent mutation rates in the ICE cassette (Spagnoletti et al., 2014; Wang et al., 2016; Hossain et al., 2018b). This could also explain why the strain Plankton1, which was negative for the SXT element, was genetically distinct from the rest (Figure 5). Plasmids were not a significant factor encoding antimicrobial resistance because antimicrobial resistance genes in our V. cholerae strains was rather related to the SXT element (Wang et al., 2016). The lack of plasmids in our strains may also explain the absence of beta-lactam genes as beta-lactamases are commonly carried by conjugative plasmids (Kudirkiene et al., 2018). Nevertheless, some cryptic plasmids have been found in non-O1 strains (Ceccarelli et al., 2017), while the IncA/C conjugative plasmids responsible for multidrug resistance were detected in some V. cholerae in Haiti (Wang et al., 2018). Acquisition of these resistance profiles not only is of clinical therapeutic relevance but may also be part of the defense mechanisms of V. cholerae in aquatic environments as protection against antimicrobial compounds and antimicrobial residues that may be present in the lake. Besides the ICE element, all sequenced strains contained a multidrug efflux pump component (MtrF) which could support their ability to persist in the lake in case of exposure to antimicrobial residues favoring their environmental persistence as it may also enhance their interaction with aquatic organisms (Alvarez-Ortega et al., 2013), as well as virulence (Alcalde-Rico et al., 2016).
This study reports the occurrence of multidrug resistant V. cholerae O1 in Lake Victoria that are genetically closely related to recent pandemic strains in Tanzania, Kenya and Uganda. The strains identified are also closely related to older pandemic strains recovered in the Democratic Republic of Congo, Burundi and Tanzania up until 1993, suggesting a long-term persistence and wide spatial distribution of pandemic strains within the region with the lake serving as a reservoir. These environmental isolates likely emerged from previous cholera outbreaks and survived in the lake environment for decades through various relations with reservoirs such as phytoplankton and fish. The V. cholerae O1 strains in the lake are potential progenitors of future cholera outbreak strains. Our findings are important for surveillance of V. cholerae O1 and understanding the epidemiology of cholera in countries around the lake.
The present study required no ethical approval since the analyzed samples were collected from the environment, namely water, phytoplankton and carp from Lake Victoria. We, however, obtained a research permit from local authorities in Mwanza, the study area for collection of these samples in the Mwanza basin of Lake Victoria. The permit issued in Kiswahili Language is submitted with the Manuscript only for editorial use.
YH collected samples and carried out the study in the laboratory, analyzed the results, and drafted the manuscript. PL participated in genomic data analysis, critical reviewing and editing of original draft manuscript. RH provided guidance and participated in genomic data analysis, critical reviewing, and editing of the manuscript. TD participated in critical reviewing and editing of the manuscript. RM, JO, and AD conceived, designed, and contributed to the revision of the draft and final approval of the version to be published. AD was the principal supervisor of the project. All authors read and approved the final manuscript.
This study was supported by the Danish International Development Assistance (Danida)-funded project “Innovations and Markets for Lake Victoria Fisheries” (IMLAF), DFC File No. 14-P01-TAN as well as the International Foundation for Sciences (IFS), Stockholm, Sweden through grant 1-2-A-6100-1.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors extend their gratitude to the funders and colleagues at the National Fish Quality Control Laboratory in Mwanza as well as colleagues from the Water Quality Laboratory of the Lake Zone in Mwanza, Tanzania.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00901/full#supplementary-material
Alam, M., Sultana, M., Nair, G. B., Sack, R. B., Sack, D. A., Siddique, A. K., et al. (2006). Toxigenic Vibrio cholerae in the Aquatic Environment of Mathbaria, Bangladesh. Appl. Environ. Microbiol. 72, 2849–2855. doi: 10.1128/AEM.72.4.2849-2855.2006
Alcalde-Rico, M., Hernando-Amado, S., Blanco, P., and Martínez, J. L. (2016). Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front. Microbiol. 7:1483. doi: 10.3389/fmicb.2016.01483
Aliabad, N. H., Bakhshi, B., Pourshafie, M. R., Sharifnia, A., and Ghorbani, M. (2012). Molecular diversity of CTX prophage in Vibrio cholerae: CTX prophage in Vibrio cholerae. Lett. Appl. Microbiol. 55, 27–32. doi: 10.1111/j.1472-765X.2012.03253.x
Alvarez-Ortega, C., Olivares, J., and Martinez, J. L. (2013). RND multidrug efflux pumps: what are they good for? Front. Microbiol. 4:7. doi: 10.3389/fmicb.2013.00007
Azarian, T., Ali, A., Johnson, J. A., Jubair, M., Cella, E., Ciccozzi, M., et al. (2016). Non-toxigenic environmental Vibrio cholerae O1 strain from Haiti provides evidence of pre-pandemic cholera in Hispaniola. Sci. Rep. 6:36115. doi: 10.1038/srep36115
Azarian, T., Ali, A., Johnson, J. A., Mohr, D., Prosperi, M., Veras, N. M., et al. (2014). Phylodynamic analysis of clinical and environmental Vibrio cholerae isolates from haiti reveals diversification driven by positive selection. mBio 5:e1824-14. doi: 10.1128/mBio.01824-14
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bari, S. M. N., Roky, M. K., Mohiuddin, M., Kamruzzaman, M., Mekalanos, J. J., and Faruque, S. M. (2013). Quorum-sensing autoinducers resuscitate dormant Vibrio cholerae in environmental water samples. Proc. Natl. Acad. Sci. U.S.A. 110, 9926–9931. doi: 10.1073/pnas.1307697110
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Bwire, G., Debes, A. K., Orach, C. G., Kagirita, A., Ram, M., Komakech, H., et al. (2018a). Environmental surveillance of Vibrio cholerae O1/O139 in the five african great lakes and other major surface water sources in Uganda. Front. Microbiol. 9:1560. doi: 10.3389/fmicb.2018.01560
Bwire, G., Sack, D. A., Almeida, M., Li, S., Voeglein, J. B., Debes, A. K., et al. (2018b). Molecular characterization of Vibrio cholerae responsible for cholera epidemics in Uganda by PCR, MLVA and WGS. PLoS Negl. Trop. Dis. 12:e0006492. doi: 10.1371/journal.pntd.0006492
Carraro, N., Sauvé, M., Matteau, D., Lauzon, G., Rodrigue, S., and Burrus, V. (2014). Development of pVCR94ΔX from Vibrio cholerae, a prototype for studying multidrug resistant IncA/C conjugative plasmids. Front. Microbiol. 5:44. doi: 10.3389/fmicb.2014.00044
Castillo, D., Kauffman, K., Hussain, F., Kalatzis, P., Rørbo, N., Polz, M. F., et al. (2018). Widespread distribution of prophage-encoded virulence factors in marine Vibrio communities. Sci. Rep. 8:9973. doi: 10.1038/s41598-018-28326-9
Ceccarelli, D., Garriss, G., Choi, S. Y., Hasan, N. A., Stepanauskas, R., Pop, M., et al. (2017). Characterization of two cryptic plasmids isolated in haiti from clinical Vibrio cholerae Non-O1/Non-O139. Front. Microbiol. 8:2283. doi: 10.3389/fmicb.2017.02283
CLSI (2017). Performance Standards for Antimicrobial Susceptibility Testing, 27th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Crump, J. A., Bopp, C. A., Greene, K. D., Kubota, K. A., Middendorf, R. L., Wells, J. G., et al. (2003). Toxigenic Vibrio cholerae Serogroup O141–associated cholera-like diarrhea and bloodstream infection in the United States. J. Infect. Dis. 187, 866–868. doi: 10.1086/368330
Dalsgaard, A., Serichantalergs, O., Forslund, A., Lin, W., Mekalanos, J., Mintz, E., et al. (2001). Clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the CTX phage and the genes encoding the toxin-coregulated pili. J. Clin. Microbiol. 39, 4086–4092. doi: 10.1128/JCM.39.11.4086-4092.2001
Dalusi, L., Lyimo, T. J., Lugomela, C., Hosea, K. M. M., and Sjöling, S. (2015a). Toxigenic Vibrio cholerae identified in estuaries of Tanzania using PCR techniques. FEMS Microbiol. Lett. 362:fnv009. doi: 10.1093/femsle/fnv009
Dalusi, L., Saarenheimo, J., Lyimo, T. J., and Lugomela, C. (2015b). Genetic relationship between clinical and environmental Vibrio cholerae isolates in Tanzania: a comparison using repetitive extragenic palindromic (REP) and enterobacterial repetitive intergenic consensus (ERIC) fingerprinting approach. Afr. J. Microbiol. Res. 9, 455–462. doi: 10.5897/AJMR2014.7307
Dongre, M., Singh, B., Aung, K. M., Larsson, P., Miftakhova, R., Persson, K., et al. (2018). Flagella-mediated secretion of a novel Vibrio cholerae cytotoxin affecting both vertebrate and invertebrate hosts. Commun. Biol. 1:59. doi: 10.1038/s42003-018-0065-z
Echenberg, M. J. (2011). Africa in the Time of Cholera: A History of Pandemics from 1817 to the Present. New York, NY: Cambridge University Press.
Eiler, A., Gonzalez-Rey, C., Allen, S., and Bertilsson, S. (2007). Growth response of Vibrio cholerae and other Vibrio spp. to cyanobacterial dissolved organic matter and temperature in brackish water: Cyanobacterial DOM, temperature and Vibrio growth. FEMS Microbiol. Ecol. 60, 411–418. doi: 10.1111/j.1574-6941.2007.00303.x
Faruque, S. M., Albert, M. J., and Mekalanos, J. J. (1998). Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62, 1301–1314.
Faruque, S. M., and Mekalanos, J. J. (2012). Phage-bacterial interactions in the evolution of toxigenic Vibrio cholerae. Virulence 3, 556–565. doi: 10.4161/viru.22351
Faruque, S. M., Tam, V. C., Chowdhury, N., Diraphat, P., Dziejman, M., Heidelberg, J. F., et al. (2007). Genomic analysis of the Mozambique strain of Vibrio cholerae O1 reveals the origin of El Tor strains carrying classical CTX prophage. Proc. Natl. Acad. Sci. U.S.A. 104, 5151–5156. doi: 10.1073/pnas.0700365104
Folster, J. P., Katz, L., McCullough, A., Parsons, M. B., Knipe, K., Sammons, S. A., et al. (2014). Multidrug-resistant IncA/C plasmid in Vibrio cholerae from Haiti. Infectious Dis. J. 20, 1951–1953. doi: 10.3201/eid2011.140889
Fong, J. C. N., Syed, K. A., Klose, K. E., and Yildiz, F. H. (2010). Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology 156, 2757–2769. doi: 10.1099/mic.0.040196-0
Garrine, M., Mandomando, I., Vubil, D., Nhampossa, T., Acacio, S., Li, S., et al. (2017). Minimal genetic change in Vibrio cholerae in Mozambique over time: multilocus variable number tandem repeat analysis and whole genome sequencing. PLoS Negl. Trop. Dis. 11:e0005671. doi: 10.1371/journal.pntd.0005671
Ghosh-Banerjee, J., Senoh, M., Takahashi, T., Hamabata, T., Barman, S., Koley, H., et al. (2010). Cholera toxin production by the El Tor variant of Vibrio cholerae O1 compared to prototype El Tor and classical biotypes. J. Clin. Microbiol. 48, 4283–4286. doi: 10.1128/JCM.00799-10
Haley, B. J., Choi, S. Y., Grim, C. J., Onifade, T. J., Cinar, H. N., Tall, B. D., et al. (2014). Genomic and phenotypic characterization of Vibrio cholerae Non-O1 isolates from a US gulf coast cholera outbreak. PLoS One 9:e86264. doi: 10.1371/journal.pone.0086264
Halpern, M., and Izhaki, I. (2017). Fish as hosts of Vibrio cholerae. Front. Microbiol. 8:282. doi: 10.3389/fmicb.2017.00282
Hammerl, J. A., Jäckel, C., Bortolaia, V., Schwartz, K., Bier, N., Hendriksen, R. S., et al. (2017). Carbapenemase VCC-1–producing Vibrio cholerae in coastal waters of Germany. Emerg. Infect. Dis. 23, 1735–1737. doi: 10.3201/eid2310.161625
Haque, M. M., Alam, M., and Salam, A. (2012). Frequency of Vibrio cholerae in the water and plankton samples of south- western coastal aquatic habitats of Bangladesh. J. Bangladesh Acad. Sci. 36, 71–78. doi: 10.3329/jbas.v36i1.10922
Hendriksen, R. S., Price, L. B., Schupp, J. M., Gillece, J. D., Kaas, R. S., Engelthaler, D. M., et al. (2011). Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the haitian outbreak. mBio 2:e00157-11. doi: 10.1128/mBio.00157-11
Hossain, Z. Z., Farhana, I., Tulsiani, S. M., Begum, A., and Jensen, P. K. M. (2018a). Transmission and toxigenic potential of Vibrio cholerae in hilsha fish (Tenualosa ilisha) for human consumption in Bangladesh. Front. Microbiol. 9:222. doi: 10.3389/fmicb.2018.00222
Hossain, Z. Z., Leekitcharoenphon, P., Dalsgaard, A., Sultana, R., Begum, A., Jensen, P. K. M., et al. (2018b). Comparative genomics of Vibrio cholerae O1 isolated from cholera patients in Bangladesh. Lett. Appl. Microbiol. 67, 329–336. doi: 10.1111/lam.13046
Hounmanou, Y. M. G., Mdegela, R. H., Dougnon, T. V., Mhongole, O. J., Mayila, E. S., Malakalinga, J., et al. (2016). Toxigenic Vibrio cholerae O1 in vegetables and fish raised in wastewater irrigated fields and stabilization ponds during a non-cholera outbreak period in Morogoro, Tanzania: an environmental health study. BMC Res. Notes 9:466. doi: 10.1186/s13104-016-2283-0
Islam, M. S., Islam, M. S., Mahmud, Z. H., Cairncross, S., Clemens, J. D., and Collins, A. E. (2015). Role of phytoplankton in maintaining endemicity and seasonality of cholera in Bangladesh. Trans. R. Soc. Trop. Med. Hyg. 109, 572–578. doi: 10.1093/trstmh/trv057
Joelsson, A., Kan, B., and Zhu, J. (2007). Quorum sensing enhances the stress response in Vibrio cholerae. Appl. Environ. Microbiol. 73, 3742–3746. doi: 10.1128/AEM.02804-06
Jolley, K. A., Bliss, C. M., Bennett, J. S., Bratcher, H. B., Brehony, C., Colles, F. M., et al. (2012). Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology 158, 1005–1015. doi: 10.1099/mic.0.055459-0
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984
Kaas, R. S., Ngandjio, A., Nzouankeu, A., Siriphap, A., Fonkoua, M.-C., Aarestrup, F. M., et al. (2016). The lake chad basin, an isolated and persistent reservoir of Vibrio cholerae O1: a genomic insight into the outbreak in cameroon, 2010. PLoS One 11:e0155691. doi: 10.1371/journal.pone.0155691
Kachwamba, Y., Mohammed, A. A., Lukupulo, H., Urio, L., Majigo, M., Mosha, F., et al. (2017). Genetic Characterization of Vibrio cholerae O1 isolates from outbreaks between 2011 and 2015 in Tanzania. BMC Infect. Dis. 17:157. doi: 10.1186/s12879-017-2252-9
Kamruzzaman, M., Udden, S. M. N., Cameron, D. E., Calderwood, S. B., Nair, G. B., Mekalanos, J. J., et al. (2010). Quorum-regulated biofilms enhance the development of conditionally viable, environmental Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 107, 1588–1593. doi: 10.1073/pnas.0913404107
Kim, E. J., Lee, D., Moon, S. H., Lee, C. H., Kim, S. J., Lee, J. H., et al. (2014). Molecular insights into the evolutionary pathway of Vibrio cholerae O1 atypical El Tor variants. PLoS Pathog. 10:e1004384. doi: 10.1371/journal.ppat.1004384
Kishe, M. A., and Machiwa, J. F. (2003). Distribution of heavy metals in sediments of Mwanza Gulf of Lake Victoria, Tanzania. Environ. Int. 28, 619–625. doi: 10.1016/s0160-4120(02)00099-5
Kudirkiene, E., Andoh, L. A., Ahmed, S., Herrero-Fresno, A., Dalsgaard, A., Obiri-Danso, K., et al. (2018). The use of a combined bioinformatics approach to locate antibiotic resistance genes on plasmids from whole genome sequences of Salmonella enterica serovars from humans in Ghana. Front. Microbiol. 9:1010. doi: 10.3389/fmicb.2018.01010
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Logan, S. L., Thomas, J., Yan, J., Baker, R. P., Shields, D. S., Xavier, J. B., et al. (2018). The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts. Proc. Natl. Acad. Sci. U.S.A. 115, E3779–E3787. doi: 10.1073/pnas.1720133115
Lugomela, C., Moyo, S., Lyimo, T. J., Namkinga, L. A., Goericke, R., and Sjöling, S. (2014). Co-variations of cholera with climatic and environmental parameters in coastal regions of Tanzania. West. Indian Ocean J. Mar. Sci. 13, 93–105.
Lutz, C., Erken, M., Noorian, P., Sun, S., and McDougald, D. (2013). Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front. Microbiol. 4:375. doi: 10.3389/fmicb.2013.00375
Mangat, C. S., Boyd, D., Janecko, N., Martz, S.-L., Desruisseau, A., Carpenter, M., et al. (2016). Characterization of VCC-1, a novel ambler class A carbapenemase from Vibrio cholerae isolated from imported retail shrimp sold in Canada. Antimicrob. Agents Chemother. 60, 1819–1825. doi: 10.1128/aac.02812-15
Meibom, K. L., Li, X. B., Nielsen, A. T., Wu, C.-Y., Roseman, S., and Schoolnik, G. K. (2004). The Vibrio cholerae chitin utilization program. PNAS 101, 2524–2529. doi: 10.1073/pnas.0308707101
Mengel, M. A., Delrieu, I., Heyerdahl, L., and Gessner, B. D. (2014). “Cholera outbreaks in Africa,” in Cholera Outbreaks, eds G. B. Nair and Y. Takeda (Berlin: Springer), 117–144. doi: 10.1007/82_2014_369
Mitchell, K. C., Breen, P., Britton, S., Neely, M. N., and Withey, J. H. (2017). Quantifying Vibrio cholerae enterotoxicity in a zebrafish infection model. Appl. Environ. Microbiol. 83:e783-17. doi: 10.1128/AEM.00783-17
Mutreja, A., Kim, D. W., Thomson, N., Connor, T. R., Lee, J. H., Kariuki, S., et al. (2011). Evidence for multiple waves of global transmission within the seventh cholera pandemic. Nature 477, 462–465. doi: 10.1038/nature10392
Naha, A., Pazhani, G. P., Ganguly, M., Ghosh, S., Ramamurthy, T., Nandy, R. K., et al. (2012). Development and evaluation of a PCR assay for tracking the emergence and dissemination of haitian variant ctxB in Vibrio cholerae O1 strains isolated from Kolkata. India J. Clin. Microbiol. 50, 1733–1736. doi: 10.1128/JCM.00387-12
Nandi, B., Nandy, R. K., Mukhopadhyay, S., Nair, G. B., Shimada, T., and Ghose, A. C. (2000). Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J. Clin. Microbiol. 38, 4145–4151.
Nkoko, D., Giraudoux, P., Plisnier, P.-D., Tinda, A., Piarroux, M., Sudre, B., et al. (2011). Dynamics of cholera outbreaks in great lakes region of Africa, 1978–2008. Emerg. Infect. Dis. 17, 2026–2034. doi: 10.3201/eid1711.110170
Nyambuli, S., Mhongole, O. J., Katakweba, A. A., Dalsgaard, A., and Mdegela, R. H. (2018). Prevalence, pathogenic markers and antibiotic susceptibility of Vibrio cholerae in sardines, water and phytoplankton in lake Tanganyika, Tanzania. Int. J. Agric. For. Fish. 6:29.
Ogwok, P., Muyonga, J. H., and Sserunjogi, M. L. (2009). Pesticide residues and heavy metals in lake victoria nile perch, lates niloticus, belly flap oil. Bull. Environ. Contam. Toxicol. 82, 529–533. doi: 10.1007/s00128-009-9668-x
Onyuka, J., Kakai, R., Onyango, D., Arama, P. F., Gichuki, J., and Ofulla, A. V. O. (2011). Prevalence and antimicrobial susceptibility patterns of enteric bacteria isolated from water and fish in lake victoria basin of Western Kenya. Int. J. Biol. Med. Sci. 1, 6–13.
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Plisnier, P.-D., Poncelet, N., Cocquyt, C., De Boeck, H., Bompangue, D., Naithani, J., et al. (2015). Cholera Outbreaks at Lake Tanganyika Induced by Climate Change?. Brussels: Belspo.
Rabia, A., Wambura, P., Misinzo, G., Kimera, S., Mdegela, R., Mzula, A., et al. (2017). Molecular epidemiology of Vibrio cholerae recovered from sewage drains, captured fish and humans in 2015/16 cholera outbreak in zanzibar, Tanzania. J. Adv. Microbiol. 5, 1–11. doi: 10.9734/JAMB/2017/36036
Rashid, M., Rashed, S. M., Islam, T., Johura, F.-T., Watanabe, H., Ohnishi, M., et al. (2016). CtxB1 outcompetes CtxB7 in Vibrio cholerae O1, Bangladesh. J. Med. Microbiol. 65, 101–103. doi: 10.1099/jmm.0.000190
Reimer, A., Domselaar, G., Stroika, S., Walker, M., Kent, H., Tarr, C., et al. (2011). Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa. Emerg. Infect. Dis. 17, 2113–2121. doi: 10.3201/eid1711.110794
Reyburn, R., Kim, D. R., Emch, M., Khatib, A., von Seidlein, L., and Ali, M. (2011). Climate variability and the outbreaks of cholera in Zanzibar, East Africa: a time series analysis. Am. J. Trop. Med. Hyg. 84, 862–869. doi: 10.4269/ajtmh.2011.10-0277
Runft, D. L., Mitchell, K. C., Abuaita, B. H., Allen, J. P., Bajer, S., Ginsburg, K., et al. (2014). Zebrafish as a natural host model for Vibrio cholerae colonization and transmission. Appl. Environ. Microbiol. 80, 1710–1717. doi: 10.1128/AEM.03580-13
Seed, K. D., Lazinski, D. W., Calderwood, S. B., and Camilli, A. (2013). A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494, 489–491. doi: 10.1038/nature11927
Sinha-Ray, S., and Ali, A. (2017). Mutation in flrA and mshA Genes of Vibrio cholerae inversely involved in vps-independent biofilm driving bacterium toward nutrients in lake water. Front. Microbiol. 8:1770. doi: 10.3389/fmicb.2017.01770
Siriphap, A., Leekitcharoenphon, P., Kaas, R. S., Theethakaew, C., Aarestrup, F. M., Sutheinkul, O., et al. (2017). Characterization and genetic variation of Vibrio cholerae isolated from clinical and environmental sources in Thailand. PLoS One 12:e0169324. doi: 10.1371/journal.pone.0169324
Spagnoletti, M., Ceccarelli, D., Rieux, A., Fondi, M., Taviani, E., Fani, R., et al. (2014). Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-pandemic Vibrio cholerae lineage. mBio 5, e1356–e1314. doi: 10.1128/mBio.01356-14
Thomsen, M. C. F., Ahrenfeldt, J., Cisneros, J. L. B., Jurtz, V., Larsen, M. V., Hasman, H., et al. (2016). A bacterial analysis platform: an integrated system for analysing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PLoS One 11:e0157718. doi: 10.1371/journal.pone.0157718
Unterweger, D., Miyata, S. T., Bachmann, V., Brooks, T. M., Mullins, T., Kostiuk, B., et al. (2014). The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat. Commun. 5:3549. doi: 10.1038/ncomms4549
Urassa, W., Mhando, Y., Mhalu, F., and Mgonja, S. (2009). Antimicrobial susceptibility pattern of Vibrio cholerae 01 strains during two cholera outbreaks in Dar Es Salaam, Tanzania. East Afr. Med. J. 77, 350–353. doi: 10.4314/eamj.v77i7.46661
Verlecar, X. N., and Desai, S. R. (2004). Phytoplankton Identification Manual. Dona Paula: National Institute of Oceanography.
Vesth, T., Wassenaar, T. M., Hallin, P. F., Snipen, L., Lagesen, K., and Ussery, D. W. (2010). On the origins of a vibrio species. Microb. Ecol. 59, 1–13. doi: 10.1007/s00248-009-9596-7
Waldor, M. K., and Mekalanos, J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914. doi: 10.1126/science.272.5270.1910
Wang, R., Liu, H., Zhao, X., Li, J., and Wan, K. (2018). IncA/C plasmids conferring high azithromycin resistance in vibrio cholerae. Int. J. Antimicrob. Agents 51, 140–144. doi: 10.1016/j.ijantimicag.2017.09.009
Wang, R., Yu, D., Yue, J., and Kan, B. (2016). Variations in SXT elements in epidemic Vibrio cholerae O1 El Tor strains in China. Sci. Rep. 6:22733. doi: 10.1038/srep22733
Keywords: Vibrio cholerae, genomics, aquatic reservoirs, African Great Lakes, microbial ecology
Citation: Hounmanou YMG, Leekitcharoenphon P, Hendriksen RS, Dougnon TV, Mdegela RH, Olsen JE and Dalsgaard A (2019) Surveillance and Genomics of Toxigenic Vibrio cholerae O1 From Fish, Phytoplankton and Water in Lake Victoria, Tanzania. Front. Microbiol. 10:901. doi: 10.3389/fmicb.2019.00901
Received: 07 January 2019; Accepted: 09 April 2019;
Published: 30 April 2019.
Edited by:
Swapan Banerjee, Health Canada, CanadaReviewed by:
Yan Boucher, University of Alberta, CanadaCopyright © 2019 Hounmanou, Leekitcharoenphon, Hendriksen, Dougnon, Mdegela, Olsen and Dalsgaard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaovi M. Gildas Hounmanou, Z2lsQHN1bmQua3UuZGs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.