- Guangdong Provincial Key Laboratory of Fermentation and Enzyme Engineering, School of Biology and Biological Engineering, South China University of Technology, Guangzhou, China
Ammonia-oxidizing archaea (AOA) are widely distributed on the earth and play a significant role in the global nitrogen cycle. Although dozens of AOA strains were obtained in the last 13 years, it is still necessary to obtain more AOA strains for the entire exploration of their ecology, physiology, and underlying biochemistry in different environments. In this study, we designed a two-step strategy for the rapid enrichment of Nitrosocosmicus–like AOA from soils. Firstly, combination of kanamycin and ampicillin was chosen as the selective stress for bacteria and quartz sands were used as the attachment of AOA cells during the first step cultivation; only after 40–75 days cultivation, AOA enrichments with abundance >20% were obtained. Secondly, combination of ciprofloxacin and azithromycin was chosen as the selective stress for the following cultivation; it is able to penetrate the biofilms and kill the bacterial cells inside the aggregate, contributing to the AOA enrichments reached high abundances (90%) only after one-time cultivation. Basing on this strategy, three AOA strains were obtained from agricultural soils only after 90–150 days cultivation. Phylogenetic analysis suggested these AOA belong to the soil group I.1b Thaumarchaeota and are closely related to the genus Nitrosocosmicus. In general, AOA enrichment or isolation is very difficult and time-consuming (an average of 2–3 years). Here, we provide a new strategy for the rapid enrichment of high abundance of Nitrosocosmicus-like AOA from soil, which gives a new solution to the AOA enrichment and cultivation in a short period.
Introduction
Autotrophic aerobic ammonia oxidation is the primary step in oxidizing ammonia to nitrate and is therefore central to the global nitrogen cycle (Kowalchuk and Stephen, 2001). This biochemical reaction could be performed by ammonia-oxidizing bacteria (AOB), ammonia-oxidizing archaea (AOA), and comammox bacteria (Beeckman et al., 2018). However, for a long time, AOB were assumed to be the sole drivers of ammonia oxidation in the environment. It was not until 2005, when the first AOA strain, Nitrosopumilus maritimus SCM1, was successfully isolated, that the member of AOA was recognized as one of the contributors to ammonia oxidation (Könneke et al., 2005). From then on, detection, enrichment and cultivation of AOA belonging to the phylum Thaumarchaeota have been widely carried out (Francis et al., 2007; Lehtovirta-Morley, 2018). Autotrophic ammonia-oxidizing microorganisms all possess ammonia monooxygenase (AMO), but the overall stoichiometry of it in AOA is indistinguishable from that of AOB and shows a higher affinity to ammonia (Martens-Habbena et al., 2009; Kits et al., 2017; Kuypers, 2017). AOA appear to be adapted to life under nutrient limitation (Horak et al., 2013; Shiozaki et al., 2016), which suggests that they have a significantly broader habitat range than the characterized AOB. They appear to be the dominant archaeal clade in soil (generally comprising 1–5% of all prokaryotes) (Ochsenreiter et al., 2003; Lehtovirta et al., 2009; Tago et al., 2015), the marine system (comprising 20–40% of all marine bacterioplankton) (Karner et al., 2001; Church et al., 2003), and geothermal habitats (Zhang et al., 2008; Dodsworth et al., 2011).
According to their performances on the ammonia oxidation in most natural systems, AOA have been believed to play a significant role in the global nitrogen cycle (Leininger et al., 2006; Pratscher et al., 2011; He et al., 2012). However, their roles have not been studied as extensively as AOB; it is still necessary to fully explore their ecology, physiology, and underlying biochemistry in environments (Stahl and de la Torre, 2012). Then, it is urgent to obtain more AOA isolates or enrichments. Up to now, 32 different AOA strains distributing in eight archaeal genera (Nitrosopumilus, Nitrosocaldus, Nitrosopelagicus, Nitrososphaera, Nitrosotalea, Nitrosoarchaeum, Nitrosotenuis, and Nitrosocosmicus) have been reported (Supplementary Table S1). Though dozens of AOA isolates and enrichments were obtained in the last 13 years, a time-saving strategy of AOA enrichment and isolation has rarely been reported in this field.
Due to the low maximum specific growth rate (0.011–0.033 h-1) and inhibition by low concentrations of ammonia (2–100 mM) and nitrite (0.028–5.7 mM) (Lehtovirta-Morley et al., 2016), it usually takes a very long time to obtain AOA enrichment from natural samples. Although some organics (such as pyruvate, oxaloacetate, malate, etc.) were reported to be able to promote the growth of AOA (Tourna et al., 2011; Kim et al., 2016; Sauder et al., 2017), they were also consumed by the symbiotic heterotrophic bacteria in the enrichment cultures. More importantly, AOA members related to the Nitrosocosmicus clade can produce extracellular polymeric substances (EPS) to form cell aggregates or biofilms, which provides nutrition and protection for bacterial cells (Flemming et al., 2016; Jung et al., 2016; Kerou et al., 2016). Antibiotics (such as Streptomycin, Kanamycin, and Ampicillin) are often used as the selective stress for the AOA enrichment and purification (Supplementary Table S1). However, the application of antibiotics often stimulates the biofilm formation and the bacterial antibiotic resistance (Hoffman et al., 2005; Kaplan, 2011).
In this study, we designed a two-step strategy for the rapid enrichment of AOA from the environment (Figure 1). During the first step, soil samples were cultivated in the culture media containing no antibiotic; after the identification of nitrite in the culture, 10% of the initial enrichment was transferred into the subculture using kanamycin-Ampicillin as selective stress for the bacterial growth, and quartz sands as attachment for the AOA cells; in the following transfer, the quartz sands in each subculture were obtained and used as inoculums for the next subculture. During the second step, the quartz sands were collected when AOA abundance on the attachment reached 20% and were transferred into a new subculture using Ciprofloxacin-Azithromycin as selective stress; after 2 to 3 subcultures, high abundance of AOA enrichment could be obtained. Using this strategy, three AOA enrichments (abundance >90%) that closely related to the genus Nitrosocosmicus were obtained from agricultural soils, after only 90 to 150 days of cultivation.
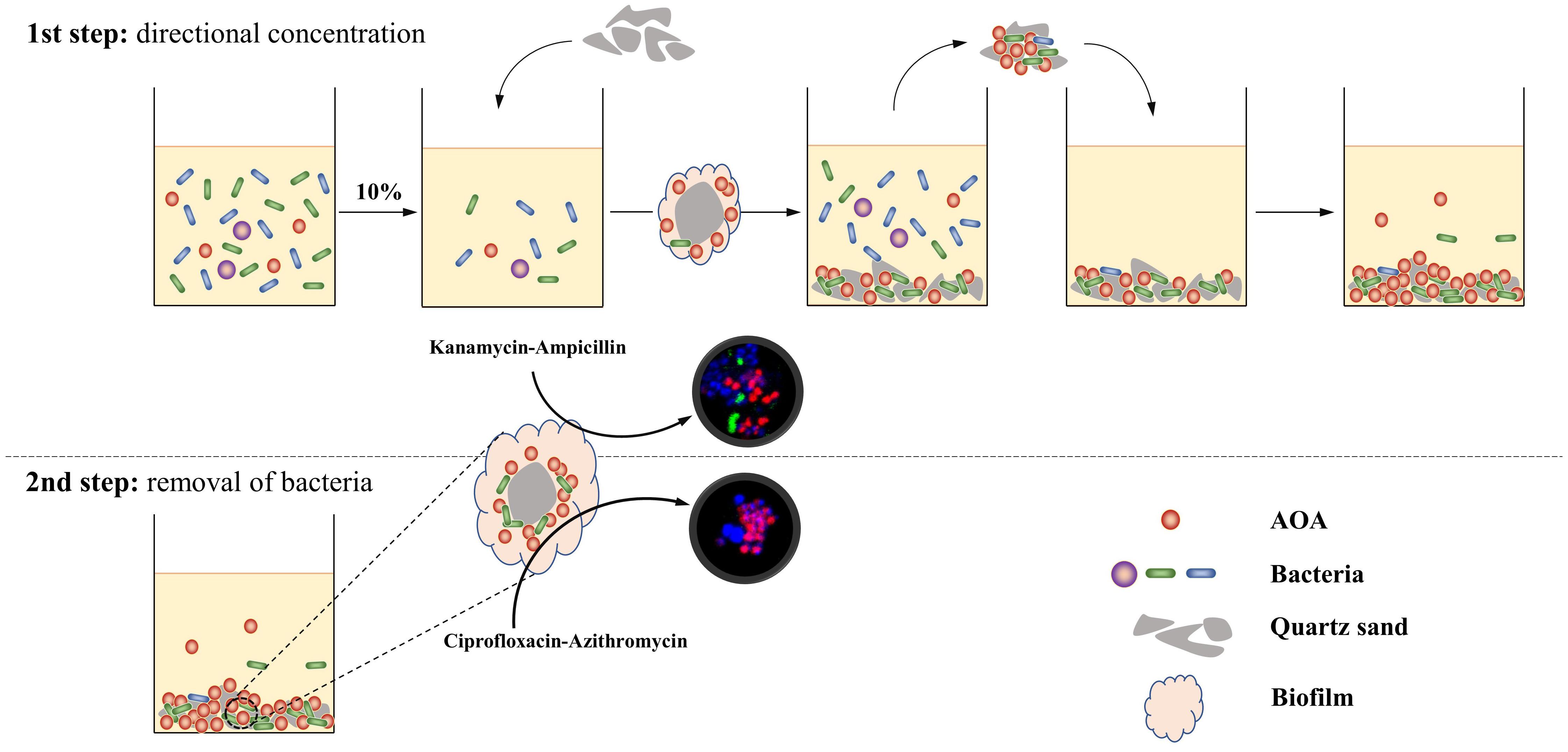
Figure 1. Schematic process of a strategy for rapid enrichment of high abundance AOA. During the first step, the AOA cells were directly absorbed by quartz sands and concentrated on the surface of quartz sands; during the second step, the antibiotic combination ciprofloxacin-azithromycin penetrates biofilms and kills bacteria in the aggregate.
Materials and Methods
Characterization of Soil Sample
Soil samples were collected from paddy fields, garden and vegetable fields (Supplementary Table S2). The diversity of AOA distributed in these soils were studied using high-throughput sequencing archaeal 16S rRNA and amoA genes, which generated an average of 9,109 and 8,838 filtered reads, respectively (Supplementary Table S3). Bioinformatics analysis indicated that the AOA community in these soils mainly consisted of Nitrosocosmicus, Nitrososphaera, Nitrosopumilus, Nitrosotenuis, and Nitrosotalea (Supplementary Figure S1); the Shannon estimator from alpha diversity indices indicated that the SS (Suishi village) soil had the highest AOA diversity and abundance of AOA (Supplementary Table S3). Based on these results, the SS soil was used as an environmental sample for the AOA enrichment in this study.
Cultivation and Enrichment
Five grams of soil collected from the SS site were inoculated into 100 mL of the culture medium and initialized the AOA enrichment in accordance with the two-step strategy. Cultivation of ammonia oxidizer was carried out using an mineral salts medium containing NaCl (1 g L-1), MgCl2⋅6H2O (0.4 g L-1), CaCl2⋅2H2O (0.1 g L-1), KH2PO4 (0.2 g L-1), KCl (0.5 g L-1), and filtration-sterilized solutions including 1 mL L-1 trace element solution (1.5 g L-1 FeCl2⋅4H2O, 190 mg L-1 CoCl2⋅6H2O, 100 mg L-1 MnCl2⋅6H2O, 70 mg L-1 ZnCl2, 62 mg L-1 HBO3, 36 mg L-1 Na2MoO4⋅2H2O, 24 mg L-1 NiCl2⋅6H2O, and 17 mg L-1 CuCl2⋅2H2O), 1 mL L-1 Fe-EDTA solution (details are presented in the Supplemental Materials), 336 mg L-1 NaHCO3, and 107 mg L-1 NH4Cl; the pH value of medium was adjusted to 7.0. Each enrichment culture was incubated at 30°C in the dark without shaking and transferred into fresh media by the time the ammonium concentration was reduced to about 20% of the initial level. 50 mg L-1 of streptomycin, kanamycin, ampicillin, carbenicillin, and tetracycline in different combinations were supplied to the mineral salt medium. Based on the accumulation of nitrite, an effective antibiotic combination was chosen as the selective stress for subcultures. During the cultivation, the concentrations of nitrite and ammonium were determined by Griess-Ilosvay method and indophenol blue method, respectively (ISO/TS 14256–1:2003, 2003). In order to remove fungi contamination, 10 mg L-1 natamycin was added to the initial culture medium. During the first transfer, the cultures were filtered through a 5 μm filter to remove soil debris and the bacterial cells embedded in the massive biofilms.
To assess the effect of quartz sands on the directional concentration of AOA, 10% (v/v) of enrichment cultures were transferred into the fresh liquid media containing 2, 5 or 10% (w/v) of quartz sands (∼ 1 mm diameter). After the ammonium was reduced to 20% of the initial level, the liquid cultures and quartz sands were collected and used for the AOA quantitative analysis, respectively. After the assessment, media containing 10% (w/v) of quartz sands were chosen for the following subcultures. During each transfer, all of the quartz sands in the culture were collected and transferred into fresh media for the next subculture. When the ammonia-oxidizing rate (nitrite-producing rate) and archaea abundance in subcultures reach a stable phase (the relative abundance of AOA in enrichment has no increase), antibiotics including kanamycin, ampicillin, tobramycin, ciprofloxacin, azithromycin, tetracycline, polymyxin, lincomycin, and spiramycin were used to evaluate the effect on the removal of bacterial cells in the biofilm that formed during the AOA enrichment.
Genomic DNA Extraction
To extract the total DNA in liquid culture, 20 mL enrichment culture was collected and filtered through a cellulose filter (0.22 μm, Thermo Fisher Scientific); the filter retaining with cells was collected and cut into pieces and placed in a 2 mL grinding tube containing 1 g quartz sand. To extract the total DNA from the quartz sand, 1 g quartz sand from the enrichment culture was collected and placed in a 2 mL grinding tube. 0.5 mL CTAB extraction buffer (10% CTAB, 0.7 M NaCl, 240 mM potassium phosphate buffer, pH 8.0) and 0.5 mL phenol-chloroform-isoamyl alcohol solution (25:24:1, pH 8.0) was added into the tube and mixed for 5 min using the Vortex Adapter (13000-V1-24, QIAGEN, Germany). After the pretreatment, genomic DNA was extracted according to a protocol that was previously reported by Griffiths (Griffiths et al., 2000). After the DNA extraction, 20 μg glycogen (Thermo Scientific, United States) was used as co-precipitant to deposit DNA from the DNA precipitated solution (30% polyethylene glycol 6000, 1.6 M NaCl). The DNA purity was determined using a UV spectrophotometer (NanoDrop 2000, Thermo Scientific, United States) and by agarose gel electrophoresis. The DNA samples were stored at -20°C for further PCR amplification.
Gene Clone, Identification and Phylogenetic Analysis
Nearly complete 16S rRNA gene and archaeal amoA gene of the AOA enrichments were PCR amplified using primer pairs A21f/1492r and CrenamoA23f/CrenamoA616r (Supplementary Table S4), respectively. PCR products were purified, ligated into pMD®19-T Vector (Takara, Dalian, China), and transformed into Escherichia coli DH5α. Recombinant clones were picked and sequenced with RV-M and M13-47 vector specific primers by IGE Biotechnology (Guangzhou, China).
Evolutionary histories of archaeal 16S rRNA gene nucleotide sequences and amoA gene translated protein sequences were inferred using the maximum likelihood method, based on the Tamura 3-parameter model. All alignments and phylogenetic analyses were conducted by the software MEGA 7 (Kumar et al., 2016).
Quantitative PCR
Archaeal and bacterial 16S rRNA genes were PCR amplified using primer pairs SS16S-1F/SS16S-1R (a primer set specific for Ca. Nitrosocosmicus sp., designed with primer-blast tool in NCBI, according to the 16S rRNA gene sequences from genus Nitrosocosmicus) and 1369F/1492R (Supplementary Table S4), respectively. Agarose gel purified PCR products of archaeal or bacterial 16S rRNA genes were cloned using the TA-cloning kit (Takara, Dalian, China). Plasmids were extracted using the TIANpure Mini Plasmid Kit II (TIANGEN, Beijing, China) and digested using QuickCutTM Hind III (Takara, Dalian, China). The linearized plasmid was used to construct the standard curves. Standard curves were prepared using six serial tenfold dilutions ranging from 102 to 107 gene copies/mL. The DNA was quantified by determining the copy number as well as the concentration and base pair composition of related genes. All quantitative PCR were performed in triplicate on an ABI 7500 Fast real-time PCR system (Applied Biosystems) using TransStart Tip Green qPCR SuperMix (Transgen, Beijing, China). The reaction condition was as follows: 2 min at 94°C; 40 cycles of 10 s at 94°C and 34 s at 60°C. The correlation coefficients (R2) of the standard curves were 0.999. The amplification efficiencies (E) of archaeal and bacterial 16S rRNA gene were 85 to 93%.
Microscopy
To observe the biofilms formed on the surface of quartz sands, glass slides were used as the attachment and laid vertically in the enrichment cultures. After incubation, the slides were directly subjected to the Fluorescence in situ hybridization (FISH) analysis. FISH was performed by a modified method as previously described by Nielsen et al. (2009). In brief, samples were fixed with 50% ethanol (mixed with PBS) at 30°C for 48 h. After resuspending in PBS, samples were immobilized on Poly (L-lysine) slides and dehydrated by graded ethanol (50%, 80% and 98%, each gradient was treated for 3 min); cells were incubated with proteinase K (50 ug/mL) for 30 min at 37°C and washed three times using dH2O. After permeabilized, cells were hybridized at 46°C for 3 h in hybridization buffer with 35% formamide using an Alexa Fluor 488 labeled probe Eub338 and an Alexa Fluor 546 labeled probe Arch915 (Supplementary Table S4). After hybridization, samples were covered with prewarmed washing buffer and incubated at 48 °C for 20 min, followed by covering with ProLongTM Diamond Antifade Mountant with DAPI (Invitrogen, United States). Microscopic observation and documentation were accomplished using a scanning confocal microscope (LSM 710, Carl Zeiss, Germany) and the ZEN 2011 black software.
For scanning electron microscopy (SEM), the fixed cells were mounted on an aluminum stub and sputter-coated with platinum using a sputter coater EMS150T (EMS, United Kingdom), then imaged using a FEI Q25 scanning electron microscope (FEI, United States).
Nucleotide Sequence Accession Number
The nucleotide sequences accession number of the cultured AOA (16S rRNA and amoA gene sequence for phylogenetic analysis) and soil sample (amplicon raw data for community analysis) are summarized in Supplementary Table S5.
Results
Cultivation and Enrichment of AOA
To find a suitable antibiotic for suppressing bacterial growth at the beginning of AOA enrichment, the nitrite that produced in ammonia oxidation was used as the evaluation index for antibiotic selection. As results show in Supplementary Figure S2, addition of kanamycin and ampicillin as the selective stress had 2.44 mg L-1 nitrite accumulation, which was higher than the other antibiotic combinations, indicating that they could be used for the AOA enrichment.
During the first subculture in the first step, the NH4+ consumption contributed to about 19.57 mg L-1 NO2--N production after 24 days cultivation (Figure 2A). The PCR detection performed using the bacterial amoA primers 1F/2R, suggested the absence of AOB in the first step enrichment. Since then, the period time of enrichment cultivation was shortened to 10 days in the third subculture, 8 days in the fifth subculture, and 6 days in the eighth subculture. From the eighth subculture, the ammonia-oxidizing rate and nitrite-producing rate always remained at stable levels, which indicated that the AOA enrichment cultivation reached a stable phase. This result was consistent with the result of a quantitative PCR analysis, that the archaea abundance in the cultures dynamically changed between 34 and 39% after the fourth subculture (Figure 2B). In fact, the archaea abundance had already reached 47% after the third subculture; however, the abundance no longer continuously increased after that time, while only archaeal and bacterial cells increased simultaneously at the same time (Figure 2B). This result suggests that the presences of some bacterial species in the enrichment culture obtained a tolerance or resistance to the activities of kanamycin and ampicillin and then survived under the antibiotic selective stress.
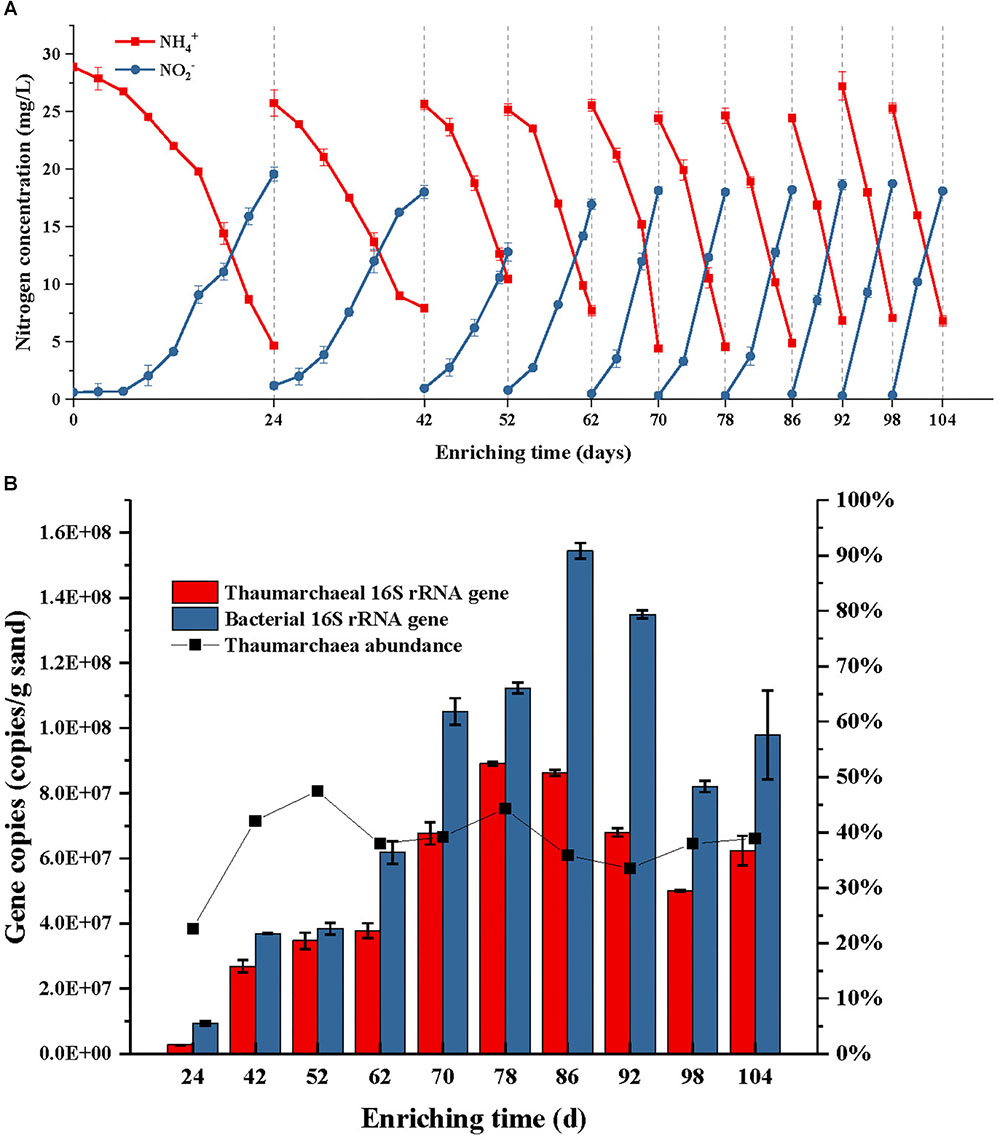
Figure 2. Ammonia consumption and nitrite accumulation (A), as well as thaumarchaeal and bacterial 16S rRNA gene copies (B) of the AOA enrichment during each subculture in use of kanamycin and ampicillin as selective stress; the ammonium with a concentration of 28 mg/L NH4+-N was added into the initial culture medium and each subculture; when the concentration of NH4+-N was reduced to about 20% of the initial level, the sands in the enrichment were obtained and transferred into a fresh medium for subculture. Error bars indicate the standard error of the mean for biological triplicates.
The resistance of bacteria to antibiotics might come from the protection by biofilms that formed by the AOA growing on the surface of quartz sands. Some AOA strains belonging to the Nitrosocosmicus clade have been reported to form aggregates and biofilms (Jung et al., 2016). Similarly, biofilms were also observed to form on the surface of quartz sands (Supplementary Figures S3a,b). Even though the biofilms were not as thick as the biofilms that were formed by many heterotrophic bacteria, they may still have the potential to decrease the activities of antibiotics. To inhibit the biofilm formation or suppress the bacterial growth in the biofilm, some new antibiotics were applied and assessed for their effects. As results shown in Figure 3, addition of tobramycin or polymyxin inhibited the ammonia oxidation of AOA enrichments, as well as the archaeal and bacterial growth in the enrichment cultures; the spiramycin not only inhibited the ammonia oxidation, but also resulted in a large increase of bacteria; tetracycline and lincomycin in some extent had no influence on the ammonia oxidation and showed positive effects on limiting the bacterial growth, which contributed to 61–72% of archaea abundance in the enrichment cultures. In comparison with other antibiotics, ciprofloxacin and azithromycin have no influence on the ammonia oxidation and contributed to the archaea abundances as high as 82% and 87%, respectively (Figure 3B). Moreover, the combination of ciprofloxacin and azithromycin resulted in 91% of the archaea abundance, which was about two times higher than that used in the combination of kanamycin and ampicillin. In the following, subcultures using ciprofloxacin-azithromycin, the archaea abundances remained steady at high levels ranging from 92 to 94% (Figure 3C).
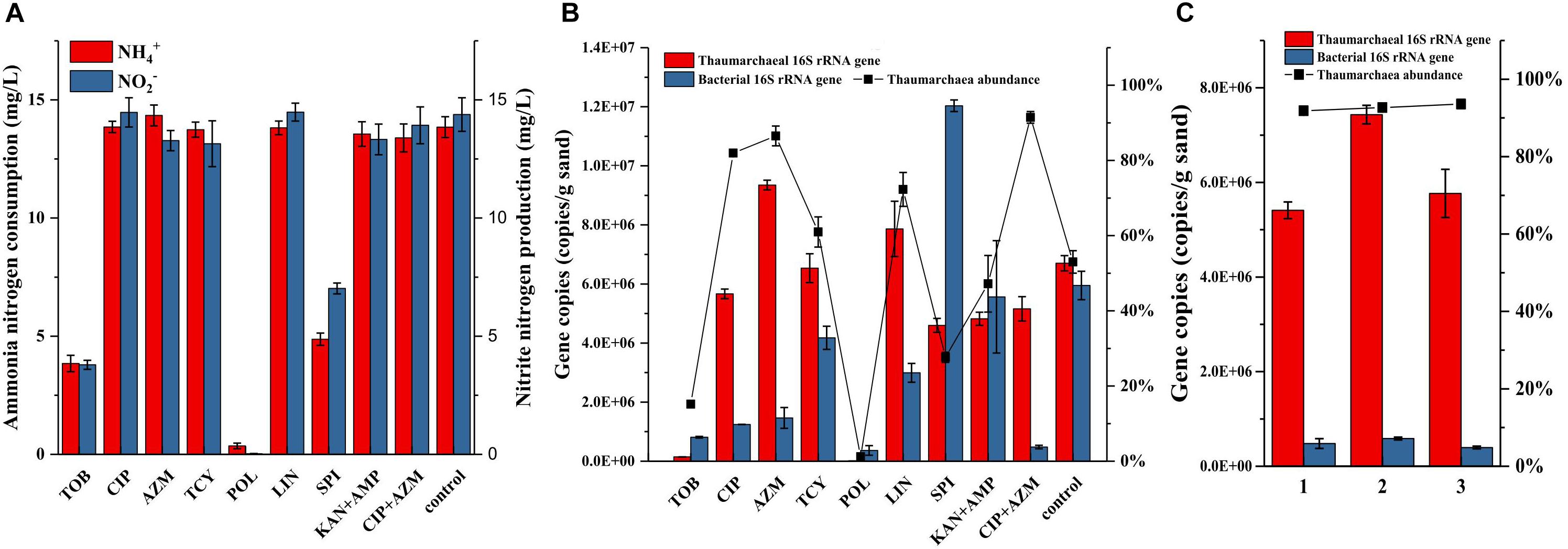
Figure 3. Ammonia consumption and nitrite accumulation (A), as well as thaumarchaeal and bacterial 16S rRNA gene copies (B) in the AOA enrichments using new antibiotics as selective stress; TOB: tobramycin, CIP: ciprofloxacin, AZM: azithromycin, TCY: tetracycline, POL: polymyxin, LIN: lincomycin, SPI: spiramycin, 50 mg/L of each antibiotic was added before inoculation. Thaumarchaeal and bacterial 16S rRNA gene copies, and archaea abundance in the AOA enrichments of three times repetition of the enrichment using ciprofloxacin and azithromycin as selective stress (C). Error bars indicate the standard error of the mean for biological triplicates.
During the AOA cultivation, the bacterial and archaeal cells would aggregate together and form biofilms in the absence of antibiotic (Supplementary Figure S3c). With the presence of kanamycin and ampicillin, thicker biofilms were formed, and more bacteria grew inside the biofilms (Supplementary Figure S3d). Differently, the combination of ciprofloxacin and azithromycin was able to limit the formation of thick biofilms (Supplementary Figure S3e). The comparison of FISH observation indicated that the combination was able to penetrate the biofilms and kill most of the bacterial cells (Supplementary Figures S3c–e). The stable high-abundance of AOA in the enrichment cultures indicated that the addition of ciprofloxacin combined with azithromycin as a selective stress is effective in the removal of bacteria in the aggregate, obtaining high abundance of AOA from the environment in a short-period.
Characterization of High Abundance AOA Enrichment
Based on the two-step strategy, one enrichment with high abundance of AOA was obtained after 150 days of cultivation. This AOA enrichment was able to consume 13.74 mg L-1 NH4+-N and reached a cell density of 6.34 × 106 cells g-1 sand in 14 days (Figure 4a). The FISH profile indicated that only a few bacterial cells was detected in the enrichment culture (Figure 4b), which is consistent with the result of quantitative PCR analysis. The SEM micrograph indicated that these archaeal cells are coccoid and 0.6–1.2 μm in diameter (Figure 4c). They often appeared in groups or aggregation, which may be covered in an extracellular matrix. On the basis of archaeal 16S rRNA and amoA gene sequences, the archaea strain in the enrichment culture belongs to the soil group I.1b Thaumarchaeota, specifically in the Nitrosocosmicus cluster, and is closely related to Ca. Nitrosocosmicus exaquare G61, showing 99.6% and 94.3% similarity, respectively. According to the phylogenetic analysis, the Thaumarchaea is named as Ca. Nitrosocosmicus sp. SS.
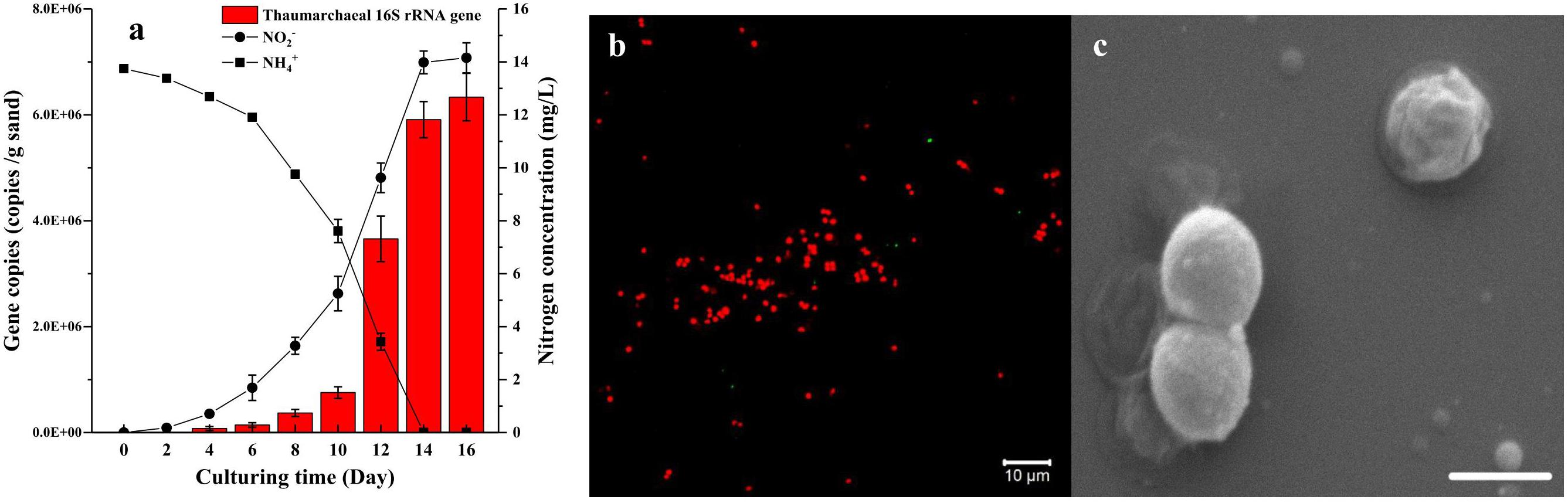
Figure 4. Growth curve (a), FISH profile (b), and SEM micrograph (c) of AOA enrichment culture (AOA abundance >95%) growing in the culture medium containing 14 mg/L NH4+-N and using ciprofloxacin and azithromycin as selective stress. On the FISH profile, archaeal 16S rRNA was labeled with Alexa Fluor 546 Archaea-specific probe (Arc915, red) and bacterial 16S rRNA was labeled with Alexa Fluor 488 labeled Bacteria-specific probe (EUB338, green), objective: Plan-Apochromat 40×/1.3 Oil DIC M27; SEM condition was HV = 5.00 kV, mag = 40000×, scale bar = 1 μm. Error bars indicate the standard error of the mean for biological triplicates.
Application of the Two-Step Strategy to Other Soils
Soil samples that were collected from paddy fields (HN_SD) and banana fields (HN_BJ) were subjected to the AOA enrichment using the methods described above. The newly collected soils were checked for the presence of AOA species using PCR amplification of the amoA gene (Supplementary Figure S4a). During the first-step cultivation, the AOA abundance in the enrichments of HN_SD and HN_BJ soils reached 20% only after 75 days of cultivation (Supplementary Figures S4b–d), similar to the previous enrichment. During the second step, the AOA abundance in the enrichments of HN_SD and HN_BJ soils reached 91% and 89%, respectively, after only 12 days of cultivation (Supplementary Figure S4d). The FISH profiles also indicated that only a small number of bacterial cells were detected in the two enrichment cultures (Supplementary Figures S4e,f). Phylogenetic analysis, based on archaeal 16S rRNA and amoA gene sequences, suggested that the newly obtained archaea in the two enrichments are also affiliated with the genus Nitrosocosmicus, named Ca. Nitrosocosmicus sp. HNSD and Ca. Nitrosocosmicus sp. HNBJ, respectively (Figure 5).
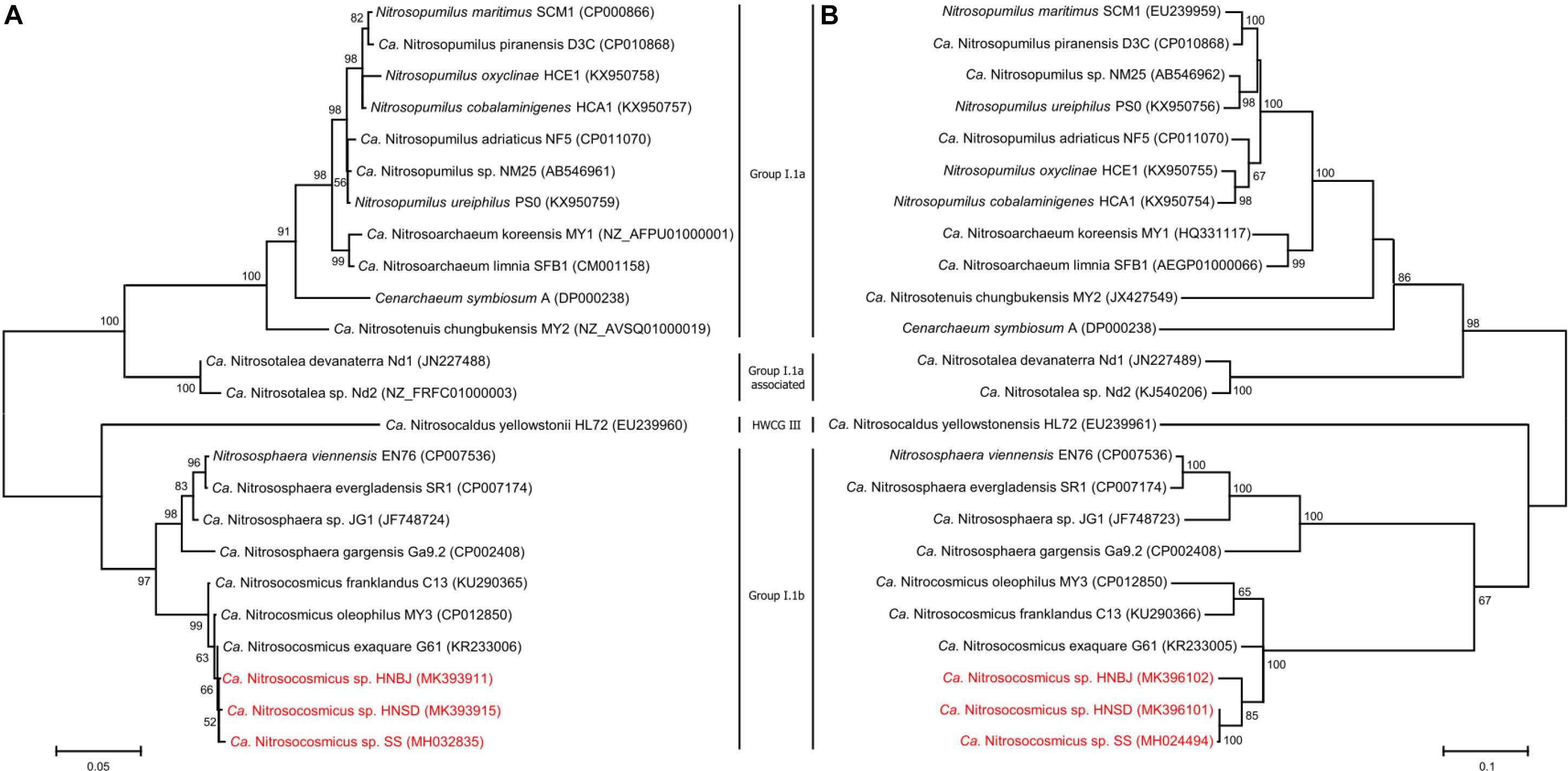
Figure 5. Phylogenetic analyses of AOA 16S rRNA gene (A) and amoA gene (B) by using Maximum Likelihood method. Bootstrap values over 50% based on 1000 replicates are shown.
Discussion
In the last two decades, 32 AOA pure cultures and enrichments were obtained from different environments (Supplementary Table S1). However, the vast majority of AOA in natural ecosystems is uncultured and thus the ecology, physiology, and biochemistry of this vital N-cycling clade is still largely unknown, especially in soils. In this study, a two-step strategy was proposed and used for the rapid enrichment of AOA from soils. During the first step, the AOA cells were directly concentrated on the surface of quartz sands by the formation of biofilms. However, the bacterial cells were also embedded in the biofilms (Supplementary Figure S3a), which contributed to a low abundance of AOA in the enrichments. Then, in the second step, an antibiotic combination ciprofloxacin-azithromycin was applied to penetrate the biofilms and kill the bacterial cells inside the aggregate. Based on the strategy, high abundance AOA enrichments were obtained from agricultural soils after only 90 to 150 days of cultivation. This period is much shorter than other published AOA strains, like the hot spring groups Ca. Nitrosotenuis uzonensis N4 (7 years), Ca. Nitrososphaera gargensis (6 years) and Ca. Nitrosocaldus cavascurensis (4 years), the soil groups Ca. Nitrosotenuis chungbukensis MY2 (3 years), Ca. Nitrosotalea sp. Nd2 (3 years) and Nitrososphaera viennensis EN76 (2 years), and the manmade ecosystem groups Ca. Nitrosocosmicus exaquare G61 (3 years) and Ca. Nitrosotenuis cloacae SAT1 (1 year) (Supplementary Table S1).
In an oligotrophic environment, the CO2 fixing activity of chemolithoautotrophic microorganisms often represents the primary organic source of complex microbial communities (Dattagupta et al., 2009; Denef et al., 2010). Similarly, during cultivation in inorganic media, AOA is the sole primary producers in the system and thus the sole source for organic carbon, which supports growth of heterotrophic bacteria and indirectly initiates the cells aggregate and biofilm formation on the surface of quartz sands. Notably, the member of the Nitrosocosmicus clade has usually been reported to form aggregates and biofilms by the secretion of EPS (Jung et al., 2016; Lehtovirta-Morley et al., 2016). It was reported that 77% and 73% of the cells of Ca. Nitrosocosmicus sp. MY3 were attached to hydrophobic bead and vermiculite, respectively; no significant attachment was observed for Ca. Nitrosotenuis chungbukensis (Jung et al., 2016). In addition, the genomic analysis indicated that members of Nitrososphaerales (such as Nitrososphaera viennensis) encode an extensive repertoire for biofilm formation including EPS production and cell surface modification (Kerou et al., 2016). Although the potential of biofilm formation has not been extensively studied in AOA, these results indicate that the presence of solid attachments (such as quartz sands) might benefit the rapid enrichment of AOA from environmental samples via the directional concentration of biofilms. As results show in Figure 6, the more quartz sands used the more the AOA attached on the quartz sands after 30 days of cultivation; with the addition of 10% (w/v) quartz sands, 46% of the AOA in enrichment culture attached on the surface of quartz sands when the culture inoculated with 10% of liquid seed, while 75% of the AOA attached on the quartz sands when the culture inoculated with quartz sands (attached with AOA cells). Moreover, the addition of kanamycin and ampicillin promoted the formation of biofilms, which might result in the concentration of AOA to the surface of quartz sands in a short time (Figure 4b).
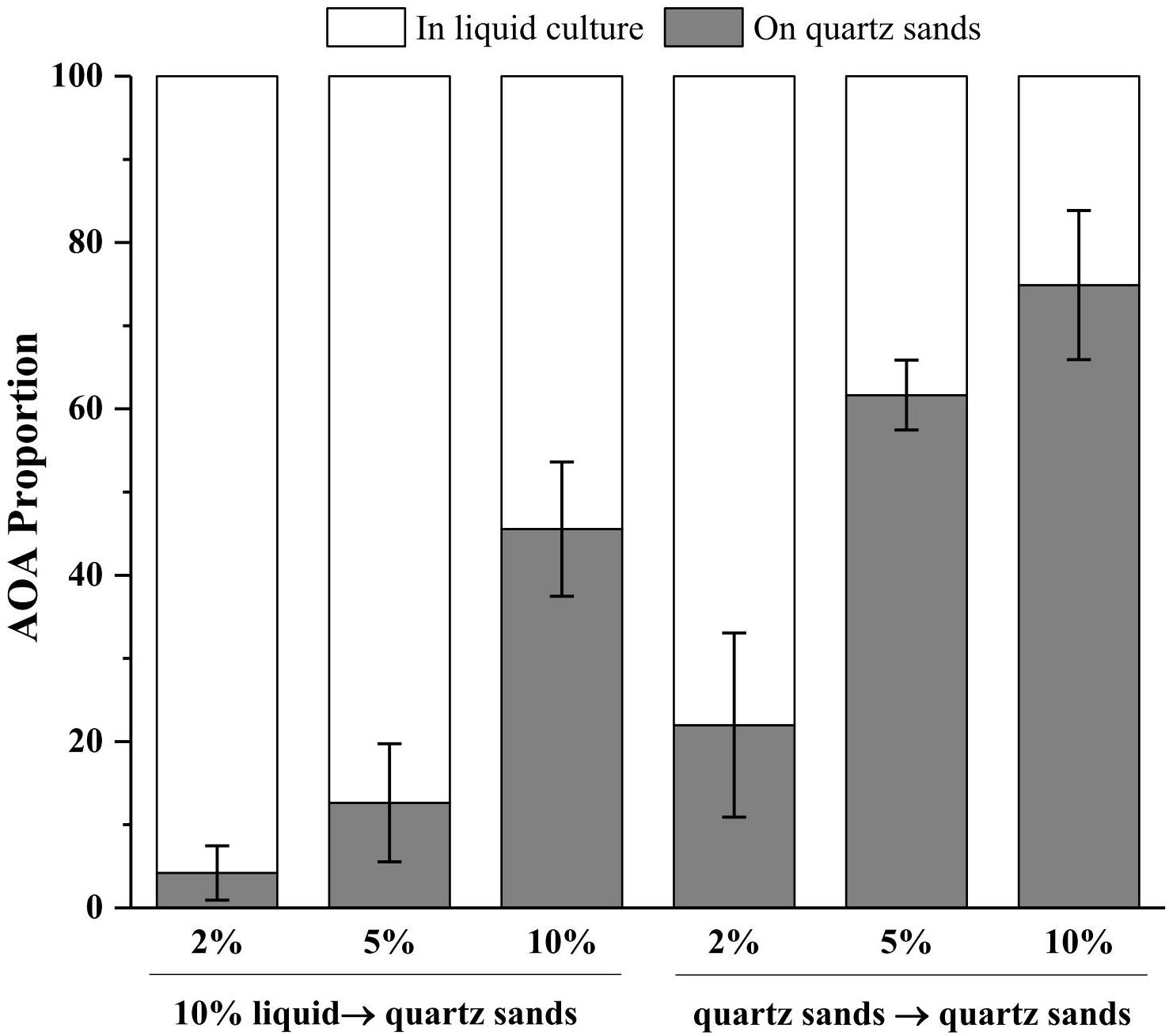
Figure 6. Proportions of AOA in the liquid cultures and on the quartz sands. 10% liquid → quartz sands means 10% enrichment culture was transferred into the fresh media containing 2, 5, or 10% (w/v) quartz sands; quartz sands → quartz sands means all of the quartz sands in the enrichment cultures containing 2, 5, and 10% quartz sands were collected and transferred into the fresh media. Error bars indicate the standard error of the mean for biological triplicates.
During the enrichment cultivation, the AOA and bacterial cells formed a stable aggregate in biofilms, especially in the presence of antibiotics (Supplementary Figures S3a,b,d). Biofilm-induced antibiotic resistance has widely been reported in pathogenic bacteria (Hoffman et al., 2005); however, it is seldomly reported or discussed in AOA cultivation. Though it has become clear that archaeal biofilms are ubiquitous in a natural environment; we are still, however, far from comprehending the molecular mechanisms of interaction among the bacteria and the archaea in biofilm based antibiotic pressure (Orell et al., 2013). It is suggested that the presence of a bacterial partner in the biofilm stimulates AOA growth and enhances aggregate formation (Jung et al., 2016). The EPS synthesized by AOA could be used by heterotrophic bacteria as a substrate and embed the bacterial cells in the biofilm, which contributes to bacterial antibiotic resistance (Hallstoodley et al., 2004; Flemming et al., 2016). Moreover, the bacterial partners can reduce reactive oxygen stress of AOA by scavenging H2O2. This is beneficial for AOA, since they produce H2O2 during their growth, but many species lack a catalase for detoxification of the produced H2O2. Recently, it was shown that the absence of bacterial partners or H2O2 scavenging α-keto acids resulted in the inhibition of AOA growth (Kim et al., 2016). The H2O2 detoxification by bacterial partners harboring catalases is a key mechanism for supporting the AOA growth and would be a reason why it is difficult to remove the bacteria during the AOA enrichment. However, in this study, addition of different concentrations of pyruvate had no significant effect on improving the ammonia oxidation (Supplementary Figure S5). Catalase genes have been reported in the genomes of some soil AOA, such as Nitrososphaera viennensis EN76 (Stieglmeier et al., 2014), Ca. Nitrosocosmicus oleophilus MY3 (Jung et al., 2016) and Ca. Nitrosocosmicus exaquare (Sauder et al., 2017), the produced H2O2 could be scavenged by the catalases that produced by themselves, suggesting their growth independent of bacterial H2O2 scavengers. Overall, the detection of catalase genes in Nitrosocosmicus species and the unaffected ammonia oxidation with added pyruvate as H2O2 scavenger indicates that H2O2 detoxification might play only a minor role in the interaction of bacteria and Nitrosocosmicus-like AOA enrichments.
As discussed above, the interaction between AOA and bacterial partners in the enrichment culture allows them to better adapt to environmental stresses (such as antibiotic). To obtain highly enriched or even pure AOA cultures, this bacterial-archaeal interaction in the biofilm must be halted. In clinical therapy, some antibiotics (such as ciprofloxacin and azithromycin) have been proven to be available when killing pathogenic bacteria by destroying their biofilms (Saini et al., 2015). Azithromycin could slow biofilm formation down, penetrate the biofilms and reduce antibiotic resistance of bacterium as a quorum sensing inhibitor (Gillis and Iglewski, 2004 Persson et al., 2005; Yamamoto et al., 2015); Ciprofloxacin is a broad-spectrum fluoroquinolone antibiotic with good bactericidal activity and can prevent biofilm formation and reduce the preexisting biofilms (Reffuveille et al., 2014). Accordingly, we applied these antibiotics to the AOA enrichment. In comparison with the biofilms that formed in the cultures without addition of antibiotics and with addition of kanamycin and ampicillin, the biofilms in the culture with ciprofloxacin and azithromycin, appeared small and thin (Supplementary Figures S3d,e), which probably proves that the presence of these antibiotics can slow the formation of biofilms down. Higher hybridization rates of FISH also indicated that the presence of ciprofloxacin and azithromycin could increase the permeability of biofilms. Moreover, the dispersed cell that grows without addition of quartz sand but with addition of antibiotics were applied to further explore the potential reason why the combination of ciprofloxacin and azithromycin has a positive effect on improving FISH hybridization. As results show in Supplementary Figure S3, the hybridization rate of Ca. Nitrosocosmicus sp. SS, that cultivated under ciprofloxacin and azithromycin (Supplementary Figure S3f), were higher than that under kanamycin and ampicillin (Supplementary Figure S3g). Ichimiya et al. (1996) had found that the azithromycin was able to inhibit the production of extracellular polysaccharides. It seemed that if fewer EPS coated on the cell surface, more fluorescent probes entered the cells. In summary, the addition of ciprofloxacin combined with azithromycin as selective stress in the cultivation effectively inhibited cell aggregation and biofilms formation, resulting in the removal of most cells and thus to high abundance of AOA in the cultures. Chen et al. (2017) have used ciprofloxacin and/or azithromycin in combination with streptomycin, kanamycin, ampicillin, and tetracycline to cultivate AOA enrichment. Unfortunately, the growth of AOA was significantly reduced or totally inhibited by using the antibiotic combinations. The presence of ciprofloxacin and azithromycin inhibit the cell aggregation and biofilm formation, which are believed to be essential for the AOA survival in the environment, suggesting that these antibiotics cannot be used in initial AOA enrichment. By contrast, in the second step of the AOA enrichment strategy in this study, biofilms with a relatively high abundance of AOA were formed after the first step of concentration by quartz sands; the addition of ciprofloxacin-azithromycin penetrated the biofilms and killed the bacterial cells inside the aggregate, but had no influence on the AOA growth.
It is tempting to speculate that biofilm-forming, terrestrial AOA species, such as members of genera Nitrososphaera, Nitrosotenuis, and Nitrosotalea, can also be enriched using this two-step enrichment strategy. Many AOA species had been detected in the soil samples; however, only the member of Nitrosocosmicus was obtained after the enrichment in this study. It is suggested that this member forms biofilms faster under antibiotic stress and attaches to quartz more easily or grows faster under ciprofloxacin and azithromycin.
Conclusion
In conclusion, the low specific growth rate, bacterial-archaeal interaction, and some other unknown features during the AOA enrichment leads to a lengthy of period time, before obtaining abundance of AOA from the environment. The two-step strategy discussed in this study, that addition of quartz sands to directly concentrate AOA cells and application of ciprofloxacin-azithromycin to effectively remove the bacterial cells inside biofilms, is an effective and timesaving method to obtain high abundance of Nitrosocosmicus–like AOA from the environment. To some extent, with appropriate adjustments, this strategy could be suitable for the enrichment of other AOA members in soils.
Author Contributions
JL, LL, and WL conceived and designed the experiments. LL, JH, and SL performed the experiments and analyzed the data. LL, JL, and WL wrote the manuscript. All authors reviewed, edited, and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 41473072).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00875/full#supplementary-material
References
Beeckman, F., Motte, H., and Beeckman, T. (2018). Nitrification in agricultural soils : impact, actors and mitigation. Curr. Opin. Biotechnol. 50, 166–173. doi: 10.1016/j.copbio.2018.01.014
Chen, H., Yue, Y., Jin, W., Zhou, X., Wang, Q., Gao, S., et al. (2017). Enrichment and characteristics of ammonia-oxidizing archaea in wastewater treatment process. Chem. Eng. J. 323, 465–472. doi: 10.1016/j.cej.2017.04.130
Church, M. J., DeLong, E. F., Ducklow, H. W., Karner, M., Preston, C. M., and Karl, D. M. (2003). Abundance and distribution of planktonic archaea and bacteria in the waters west of the antarctic peninsula. Limnol. Oceanogr. 48, 1893–1902. doi: 10.4319/lo.2003.48.5.1893
Dattagupta, S., Schaperdoth, I., Montanari, A., Mariani, S., Kita, N. T., Valley, J. W., et al. (2009). A novel symbiosis between chemoautotrophic bacteria and a freshwater cave amphipod. ISME J. 3, 935–943. doi: 10.1038/ismej.2009.34
Denef, V. J., Mueller, R. S., and Banfield, J. F. (2010). AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 4, 599–610. doi: 10.1038/ismej.2009.158
Dodsworth, J. A., Hungate, B. A., and Hedlund, B. P. (2011). Ammonia oxidation, denitrification and dissimilatory nitrate reduction to ammonium in two US Great Basin hot springs with abundant ammonia-oxidizing archaea. Environ. Microbiol. 13, 2371–2386. doi: 10.1111/j.1462-2920.2011.02508.x
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Francis, C. A., Beman, J. M., and Kuypers, M. M. (2007). New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1, 19–27. doi: 10.1038/ismej.2007.8
Gillis, R. J., and Iglewski, B. H. (2004). Azithromycin Retards Pseudomonas aeruginosa Biofilm Formation. J. Clin. Microbiol. 42, 5842–5845. doi: 10.1128/jcm.42.12.5842-5845.2004
Griffiths, R. I., Whiteley, A. S., Anthony, G., Donnell, O., Bailey, M. J., and Donnell, A. G. O. (2000). Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66, 5488–5491. doi: 10.1128/aem.66.12.5488-5491.2000
Hallstoodley, L., Costerton, J. W., and Stoodley, P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108. doi: 10.1038/nrmicro821
He, J., Hu, H., and Zhang, L. (2012). Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils. Soil Biol. Biochem. 55, 146–154. doi: 10.1016/j.soilbio.2012.06.006
Hoffman, L. R., D’Argenio, D. A., MacCoss, M. A., Zhang, Z., Jones, R. A., and Miller, S. I. (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436, 1171–1175. doi: 10.1038/nature03912
Horak, R. E. A., Qin, W., Schauer, A. J., Armbrust, E. V., Ingalls, A. E., Moffett, J. W., et al. (2013). Ammonia oxidation kinetics and temperature sensitivity of a natural marine community dominated by Archaea. ISME J. 7, 2023–2033. doi: 10.1038/ismej.2013.75
Ichimiya, T., Takeoka, K., Hiramatsu, K., Hirai, K., Yamasaki, T., and Nasu, M. (1996). The Influence of Azithromycin on the Biofilm Formation of Pseudomonas aeruginosa in vitro. Chemotherapy 42, 186–191. doi: 10.1159/000239440
ISO/TS 14256–1:2003 (2003). Soil Quality-Determination of Nitrate, Nitrite and Ammonium in Field Moist Soils by Extraction with Potassium Chloride Solution. Geneva: International Organisation for Standardisation.
Jung, M. Y., Kim, J. G., Damsté, J. S. S., Rijpstra, W. I. C., Madsen, E. L., Kim, S. J., et al. (2016). A hydrophobic ammonia-oxidizing archaeon of the Nitrosocosmicus clade isolated from coal tar-contaminated sediment. Environ. Microbiol. Rep. 8, 983–992. doi: 10.1111/1758-2229.12477
Kaplan, J. B. (2011). Antibiotic-induced biofilm formation. Int. J. Artif. Organ. 34, 737–751. doi: 10.5301/ijao.5000027
Karner, M., DeLong, E. F., and Karl, D. M. (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510. doi: 10.1038/35054051
Kerou, M., Offre, P., Valledor, L., Abby, S. S., Melcher, M., Nagler, M., et al. (2016). Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc. Natl. Acad. Sci. U.S.A. 113, 7937–7946.
Kim, J. G., Park, S. J., Damsté, J. S. S., Schouten, S., Rijpstra, W. I. C., Jung, M. Y., et al. (2016). Hydrogen peroxide detoxification is a key mechanism for growth of ammonia-oxidizing archaea. Proc. Natl. Acad. Sci. U.S.A. 113, 7888–7893. doi: 10.1073/pnas.1605501113
Kits, K. D., Sedlacek, C. J., Lebedeva, E. V., Han, P., Bulaev, A., Pjevac, P., et al. (2017). Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature 549, 269–272. doi: 10.1038/nature23679
Könneke, M., Bernhard, A. E., de la Torre, J., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Kowalchuk, G. A., and Stephen, J. R. (2001). Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55, 485–529. doi: 10.1146/annurev.micro.55.1.485
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7. 0 for bigger datasets. Mol. Biol. Evol. 33, 1–11. doi: 10.1093/molbev/msw054
Kuypers, M. M. M. (2017). Microbiology: a fight for scraps of ammonia. Nature 549, 162–163. doi: 10.1038/549162a
Lehtovirta, L. E., Prosser, J. I., and Nicol, G. W. (2009). Soil pH regulates the abundance and diversity of Group 1.1c Crenarchaeota. FEMS Microbiol. Ecol. 70, 367–376. doi: 10.1111/j.1574-6941.2009.00748.x
Lehtovirta-Morley, L. E. (2018). Ammonia oxidation: ecology, physiology, biochemistry and why they must all come together. FEMS Microbiol. Lett. 365:fny058. doi: 10.1093/femsle/fny058
Lehtovirta-Morley, L. E., Ross, J., Hink, L., Weber, E. B., Gubry-Rangin, C., Thion, C., et al. (2016). Isolation of “Candidatus Nitrosocosmicus franklandus”, a novel ureolytic soil archaeal ammonia oxidiser with tolerance to high ammonia concentration. FEMS Microbiol. Ecol. 92, 1–10. doi: 10.1093/femsec/fiw057
Leininger, S., Urich, T., Schloter, M., Schwark, L., Qi, J., Nicol, G. W., et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809. doi: 10.1038/nature04983
Martens-Habbena, W., Berube, P. M., Urakawa, H., José, R., and Stahl, D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–979. doi: 10.1038/nature08465
Nielsen, P. H., Daims, H., Lemmer, H., Arslan-Alaton, I., and Olmez-Hanci, T. (2009). FISH Handbook for Biological Wastewater Treatment. London: IWA Publishing.
Ochsenreiter, T., Selezi, D., Quaiser, A., Bonchosmolovskaya, L., and Schleper, C. (2003). Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 5, 787–797. doi: 10.1046/j.1462-2920.2003.00476.x
Orell, A., Fröls, S., and Albers, S. V. (2013). Archaeal biofilms: the great unexplored. Annu. Rev. Microbiol. 67, 337–354. doi: 10.1146/annurev-micro-092412-155616
Persson, T., Givskov, M., and Nielsen, J. (2005). Quorum sensing inhibition: targeting chemical communication in gramnegative bacteria. Curr. Med. Chem. 12, 3103–3115. doi: 10.2174/092986705774933425
Pratscher, J., Dumont, M. G., and Conrad, R. (2011). Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc. Natl. Acad. Sci. U.S.A. 108, 4170–4175. doi: 10.1073/pnas.1010981108
Reffuveille, F., de la Fuente-NúÞnez, C., Mansour, S., and Hancock, R. E. W. (2014). A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob. Agents Chemother. 58, 5363–5371. doi: 10.1128/AAC.03163-14
Saini, H., Chhibber, S., and Harjai, K. (2015). Azithromycin and ciprofloxacin: a possible synergistic combination against Pseudomonas aeruginosa biofilm-associated urinary tract infections. Int. J. Antimicrob. Agents 45, 359–367. doi: 10.1016/j.ijantimicag.2014.11.008
Sauder, L. A., Albertsen, M., Engel, K., Schwarz, J., Nielsen, P. H., Wagner, M., et al. (2017). Cultivation and characterization of Candidatus Nitrosocosmicus exaquare, an ammonia-oxidizing archaeon from a municipal wastewater treatment system. ISME J. 11, 1142–1157. doi: 10.1038/ismej.2016.192
Shiozaki, T., Ijichi, M., Isobe, K., Hashihama, F., Nakamura, K., Ehama, M., et al. (2016). Nitrification and its influence on biogeochemical cycles from the equatorial Pacific to the Arctic Ocean. ISME J. 10, 2184–2197. doi: 10.1038/ismej.2016.18
Stahl, D. A., and de la Torre, J. R. (2012). Physiology and diversity of ammonia-oxidizing archaea. Annu. Rev. Microbiol. 66, 83–101. doi: 10.1146/annurev-micro-092611-150128
Stieglmeier, M., Klingl, A., Alves, R. J. E., Rittmann, S. K. M. R., Melcher, M., Leisch, N., et al. (2014). Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int. J. Syst. Evol. Microbiol. 64, 2738–2752. doi: 10.1099/ijs.0.063172-0
Tago, K., Okubo, T., Shimomura, Y., Kikuchi, Y., Hori, T., Nagayama, A., et al. (2015). Environmental factors shaping the community structure of ammonia-oxidizing bacteria and archaea in sugarcane field soil. Microbes Environ. 30, 21–28. doi: 10.1264/jsme2.ME14137
Tourna, M., Stieglmeier, M., Spang, A., Konneke, M., Schintlmeister, A., Urich, T., et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U.S.A. 108, 8420–8425. doi: 10.1073/pnas.1013488108
Yamamoto, R., Noiri, Y., Yamaguchi, M., Asahi, Y., Maezono, H., Ebisu, S., et al. (2015). Inhibition of polysaccharide synthesis by the sinR orthologue PGN_0088 is indirectly associated with the penetration of Porphyromonas gingivalis biofilms by macrolide antibiotics. Microbiology 161, 422–429. doi: 10.1099/mic.0.000013
Keywords: ammonia-oxidizing archaea, Nitrosocosmicus, biofilms, quartz sand, ciprofloxacin, azithromycin
Citation: Liu L, Li S, Han J, Lin W and Luo J (2019) A Two-Step Strategy for the Rapid Enrichment of Nitrosocosmicus-Like Ammonia-Oxidizing Thaumarchaea. Front. Microbiol. 10:875. doi: 10.3389/fmicb.2019.00875
Received: 26 September 2018; Accepted: 04 April 2019;
Published: 24 April 2019.
Edited by:
Maria J. Delgado, Spanish National Research Council (CSIC), SpainReviewed by:
Lisa Y. Stein, University of Alberta, CanadaZhe-Xue Quan, Fudan University, China
Hanna Koch, Radboud University Nijmegen, Netherlands
Copyright © 2019 Liu, Li, Han, Lin and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weitie Lin, bGZ3dGxpbkBzY3V0LmVkdS5jbg== Jianfei Luo, bGpmamYyMDAyQHNjdXQuZWR1LmNu
 Liangting Liu
Liangting Liu Surong Li
Surong Li Jianfei Luo
Jianfei Luo