- 1Department of Translation Research and of New Technologies in Medicine and Surgery, University of Pisa, Pisa, Italy
- 2Unidad de Investigación en Enfermedades Infecciosas, Unidades Médicas de Alta Especialidad Pediatría, Instituto Mexicano del Seguro Social, Mexico City, Mexico
- 3Instituto de Investigaciones en Ciencias de la Salud, National University of Asunción, Asunción, Paraguay
- 4Grupo de Investigación en Biología del Cáncer, Instituto Nacional de Cancerología, Bogotá, Colombia
- 5Genomic Epidemiology Group, German Cancer Research Center (DKFZ), Heidelberg, Germany
- 6Department of Oncology and Pathology, Wayne State University School of Medicine, Detroit, MI, United States
Bacteria are highly social organisms that communicate via signaling molecules and can assume a multicellular lifestyle to build biofilm communities. Until recently, complications from biofilm-associated infection have been primarily ascribed to increased bacterial resistance to antibiotics and host immune evasion, leading to persistent infection. In this theory and hypothesis article we present a relatively new argument that biofilm formation has potential etiological role in the development of digestive tract cancer. First, we summarize recent new findings suggesting the potential link between bacterial biofilm and various types of cancer to build the foundation of our hypothesis. To date, evidence has been particularly convincing for colorectal cancer and its precursor, i.e., polyps, pointing to several key individual bacterial species, such as Bacteroides fragilis, Fusobacterium nucleatum, and Streptococcus gallolyticus subsp. Gallolyticus. Then, we further extend this hypothesis to one of the most common bacterial infection in humans, Helicobacter pylori (Hp), which is considered a major cause of gastric cancer. Thus far, there has been no direct evidence linking in vivo Hp gastric biofilm formation to gastric carcinogenesis. Yet, we synthesize the information to support an argument that biofilm associated-Hp is potentially more carcinogenic, summarizing biological characteristics of biofilm-associated bacteria. We also discuss mechanistic pathways as to how Hp or other biofilm-associated bacteria control biofilm formation and highlight recent findings on Hp genes that influence biofilm formation, which may lead to strain variability in biofilm formation. This knowledge may open a possibility of developing targeted intervention. We conclude, however, that this field is still in its infancy. To test the hypothesis rigorously and to link it ultimately to gastric pathologies (e.g., premalignant lesions and cancer), studies are needed to learn more about Hp biofilms, such as compositions and biological properties of extracellular polymeric substance (EPS), presence of non-Hp microbiome and geographical distribution of biofilms in relation to gastric gland types and structures. Identification of specific Hp strains with enhanced biofilm formation would be helpful not only for screening patients at high risk for sequelae from Hp infection, but also for development of new antibiotics to avoid resistance, regardless of its association with gastric cancer.
Introduction
Over the past two decades, it has been increasingly appreciated that bacteria present in most biological systems exist in biofilms, which are defined as matrix-enclosed microbial accretions adhering to biological or non-biological surfaces and to each other (Hall-Stoodley et al., 2004; Gabrilska and Rumbaugh, 2015; Flemming et al., 2016). Biofilm formation is a key factor for survival in diverse environments and is viewed as an ancient and integral component of the prokaryotic life cycle, as researchers found biofilm formation early in fossil records (∼3.25 billion years ago) (Hall-Stoodley et al., 2004). In biofilms, unicellular bacteria assume a temporary multicellular lifestyle through prolific intercellular interactions, both social and physical, immersing in a complex and specialized matrix formed by both the bacteria and the host (Kostakioti et al., 2013; Watters et al., 2016). These group behaviors represent a microbial social support system to foster survival in hostile environments, share limited resources, favor long-term persistent colonization and maintain the ability to colonize to new niches (Hall-Stoodley et al., 2004; Majumdar and Pal, 2017). Accordingly, this specific mode of living provides strong fitness advantage to biofilm-associated bacteria compared to their planktonic counterparts (Gabrilska and Rumbaugh, 2015; Liu et al., 2016). In particular, biofilm-associated bacteria exhibit increased resistance to chemical disinfectants, antibiotic therapies and human immune responses and thus are associated with long-term persistence (Elias and Banin, 2012; Kostakioti et al., 2013). The self-produced extracellular polymeric substance (EPS) matrix, which is typically composed of polysaccharides, carbohydrate-binding proteins, lipids, extracellular DNA (eDNA), pili, flagella, and other adhesive fibers (Kostakioti et al., 2013; Flemming et al., 2016), plays an important role in antibiotic resistance, enhanced horizontal gene transfer as well as altered gene expressions within the biofilm community, which results in enhanced bacterial virulence (Elias and Banin, 2012; Madsen et al., 2012; Flemming et al., 2016). Furthermore, it has been described that some extracellular pathogens adopt intracellular lifestyle through the formation of bacterial communities with biofilm-like properties, enabling them to persist inside the host cells (Kostakioti et al., 2013). Thus, these biofilm-associated infections in humans on both abiotic (medical devices) and biotic surfaces (e.g., gum, heart valves, lungs, etc.) pose significant challenges to the medical community (Costerton et al., 1999; Hall-Stoodley et al., 2004; Kostakioti et al., 2013).
Chronic inflammation, often caused by chronic microbial infection, has been decisively linked to several stages of carcinogenesis (Grivennikov et al., 2010; Armstrong et al., 2018). A recent estimate also suggests that about 15% of worldwide incident cases of cancer are attributable to chronic infection (Plummer et al., 2016). As discussed above, one way for bacteria to achieve persistent colonization in the hosts is to form biofilms. Despite well-established causal links between certain infectious agents, such as Helicobacter pylori (Hp), and cancer (Plummer et al., 2016), knowledge concerning the association between bacterial biofilm formation and cancer development has been sparse.
Despite the well-founded association between persistent Hp infection and gastric carcinogenesis and growing knowledge concerning unique properties of biofilm-associated bacteria, little is known about biological consequence of Hp in vivo biofilm formation beyond antibiotic resistance. In the subsequent sections, we present our hypothesis that biofilm formation has potential etiological role in the development of digestive tract cancer, which may be particularly relevant to infection-associated cancer, such as Hp-induced gastric cancer. We synthesize recent new findings suggesting the potential link between bacterial biofilm and various types of cancer, as well as the information to support an argument that biofilm associated-Hp is potentially more carcinogenic. We also discuss technical details and challenges in Hp biofilm studies to aid interpretation of the results from various different experimental platforms and designing future studies and highlight recent findings on Hp genes and virulence factors that influence biofilm formation.
Evidence to Support Possible Links Between Biofilm Formation and Human Cancer
Recently, a group from Johns Hopkins University in the US published a series of studies addressing the potential link between biofilm-associated bacteria and colorectal cancer and its precursor, i.e., polyps. The investigators found more frequent polymicrobial biofilm formation in colorectal mucosa of the patients with colorectal cancer or adenoma, compared with that of control subjects who had negative findings at screening colonoscopy. This phenomenon was striking for the right sided tumors compared to left sided lesions (Dejea et al., 2014; Drewes et al., 2017). Importantly, biofilms were present not only on tumors but also at normal surgical margins. Furthermore, biofilm formation was associated with diminished epithelial cell E-cadherin, enhanced IL-6 and Stat3 activation, and increased crypt cell proliferation in normal mucosa (Dejea et al., 2014). The subsequent analysis revealed that these tumor-associated biofilms were enriched with Bacteroides fragilis (Bf) and several periodontal pathogens, including Fusobacterium nucleatum (Fn) and Peptostreptococcus stomatits, and accompanied with altered functions, compared to those in planktonic bacteria, e.g., increases in cytoskeletal proteins, peptidoglycan (PG) biosynthesis and sporulation, and a decrease in flagellar assembly (Drewes et al., 2017). Biofilms were detected also in colorectal mucosa of genetically predisposed individuals, i.e., familial polyposis coli patients, regardless of the locations at the colorectum, but their appearance and composition were different (Dejea et al., 2018). They were rather patchy (as opposed to continuous as observed for sporadic cases) and primarily composed of Bf and polyketide-peptide genotoxin producing pks island positive Escherichia coli, with little periodontal pathogens (Dejea et al., 2018). It was also reported previously that Bf was the main component of inflammatory bowel disease-associated biofilms (Swidsinski et al., 2005).
Another piece of corroborative information arises from growing recognition of the association between oral microbiome, specifically Fn, and colorectal cancer (Sun and Kato, 2016), which was predominantly observed for proximal colon tumors (Hussan et al., 2017). Fn is a normal residential member of dental plaques (biofilms) and well-known periodontal pathogen (Larsen and Fiehn, 2017). Fn is considered to be a co-aggregation expert, with an ability of co-aggregating with a broad range of bacteria, nearly all bacterial species involved in oral plaque formation (Kolenbrander et al., 1989; Allen-Vercoe et al., 2011), a very important property in biofilm formation. Moreover, Fn can bind to and transport otherwise non-invasive bacterial species into host cells, acting as a shuttle in this respect (Edwards et al., 2006). Fn produces outer membrane vesicles (OMV) to facilitate coaggregation and isolated OMVs alone have been shown to exert an equivalent ability to coaggregate other bacteria compared to whole bacterial culture (Kinder and Holt, 1993). OMV production appears to depend on external stimuli and resulting changes in biofilm formation are strain-specific (Musrati et al., 2016). In this context, the putative association with colorectal cancer may be a function of the microbes that Fn gathers in its biofilms, rather than a direct effect of its own virulence. In fact, Flemer et al. (2018) more recently reported that several microbes commonly found in oral biofilms were enriched in colonic mucosa from colorectal cancer patients. Finally, Fn has recently found to be one of the gut microbes linked to pancreatic cancer (del Castillo et al., 2019), although little has been known about biofilm formation in the pancreatic ducts.
Moreover, growing evidence suggest a link between colorectal cancer and Streptococcus gallolyticus subsp. Gallolyticus (SGG), formerly known as S. bovis, an opportunistic pathogen causing biofilm-associated infections, e.g. infective endocarditis on cardiac valves (Boleij et al., 2010, 2012; Sun and Kato, 2016; Butt et al., 2018; Jans and Boleij, 2018). Martins el al. have revealed that this bacterium exploits Pil3 pilus for adhesion to colonic mucus and for colonization of mouse distal colon (Danne et al., 2011; Martins et al., 2016), while Pil1 pilus allows S. gallolyticus to bind to collagen type I and plays a role in biofilm formation (Danne et al., 2011; Martins et al., 2016). Pil3 pilus has been shown to bind not only to human colonic mucins and to human stomach mucins, but also to human fibrinogen (Martins et al., 2016). Accordingly, both pilus proteins play an important role in biofilm formation. Binding to fibrinogen is also known to contribute to increased biofilm formation in other Streptococcus species (Bedran et al., 2013). Comparative genomics of 8 SGG strains form human blood and feces recently revealed that complete pil1-3 loci are only present in virulent strains, causing bacteremia and/or endocarditis translocated through impaired mucosal barrier, while other fecal SGGs do not carry these full pil1-3 loci (Jans and Boleij, 2018). Interestingly, several epidemiological studies, including our own, have found that individuals who express high antibody titers to several S. gallolyticus pilus proteins have increased risk of colorectal cancer (Boleij et al., 2012; Butt et al., 2016; Butt et al., 2018).
Gallbladder is another potential niche where bacteria can form biofilm. In fact, certain strains of Salmonella typhi are known to produce biofilms on the surfaces of cholesterol gallstone (Crawford et al., 2010; Gonzalez-Escobedo and Gunn, 2013), which is in support of the known etiological link between the carriages of S. typhi and gallstones and the development of gallbladder cancer (Di Domenico et al., 2017).
Gastric lumen is one of the most hostile environments in human body, which kills many bacteria within a few minutes. Hp survives in this environment with its urease activity that neutralizes gastric acid (Abadi, 2017), whereas biofilm formation may be more vital for its persistent colonization. Surprisingly, to date, little has been known about in vivo biofilm formation on human gastric mucosa. Three pioneer studies employed high powered electron microscope and demonstrated dense clusters of Hp (Carron et al., 2006; Coticchia et al., 2006; Cellini et al., 2008), primarily in coccoid forms, which are known to be viable but non-cultivable (Cellini, 2014; Percival and Suleman, 2014). It is interesting to note that Hp isolated from gastric cancer patients is often non-cultivable, despite the fact that the bacteria are detectable by other methods (i.e., PCR or histology), and that coccoid forms have been indeed more frequently found in gastric mucosa of gastric cancer patients than in that of peptic ulcer patients (Chan et al., 1994).
Evidence to Support Hp Ability in In Vitro and In Vivo Biofilm Formation
In vitro Abiotic Models
In 1999, Stark et al. (1999) reported the formation of biofilm by Hp in a continuous culture of Hp NCTC 11637 in Brucella broth supplemented with B-cyclodextrin and glucose in a glass fermentor. Hp formed a biofilm containing polysaccharides at the air-liquid interface. Later, Cole et al. (2004) and Bellack et al. (2006) described the progression of biofilm formation with clinical isolates of Hp showing that all strains tested were able to form biofilm at the air-liquid interface on a glass surface, although Hp biofilms have also been detected by molecular methods on the surface of environmental water supply system. Further studies have clarified that biofilms are not simply passive aggregates of cells attached to a surface, but they do complex communitarian functions and should be considered as biological systems.
The ability of in vitro biofilm formation by Hp has been described in several systems. These studies have demonstrated important differences in the in vitro growth conditions of the bacteria and in the abiotic surfaces used in the assay. Microtiter polystyrene plates are the most commonly-used substrate (Azeredo et al., 2017). The biomass attached to the surface of the wells and the formed biofilm is quantitated by staining with violet crystal (Wilson et al., 2017). Yonezawa et al. (2009) observed biofilm formation at the air-liquid interface of microtiter plates. More specific pieces of information regarding the structure of Hp biofilms in vitro on abiotic surface have also been made available by use of scanning electron microscopy (SEM) (Queralt and Araujo, 2007; Yonezawa et al., 2009, 2013). These include Hp morphological changes associated with biofilm formation in an aquatic culture model as well as how culture conditions and nutrient supply affect biofilm density, structures and abundance of OMVs. It is worth noting that OMVs have been found to enhance oxidative stress and genomic damages to host cells and thus possess carcinogenic potential in in vitro culture systems (Chitcholtan et al., 2008). Using transmission electron microscope analysis (TEM), Grande et al. demonstrated the presence of eDNA, which was associated with OMVs and plays a role in Hp aggregation (Grande et al., 2015).
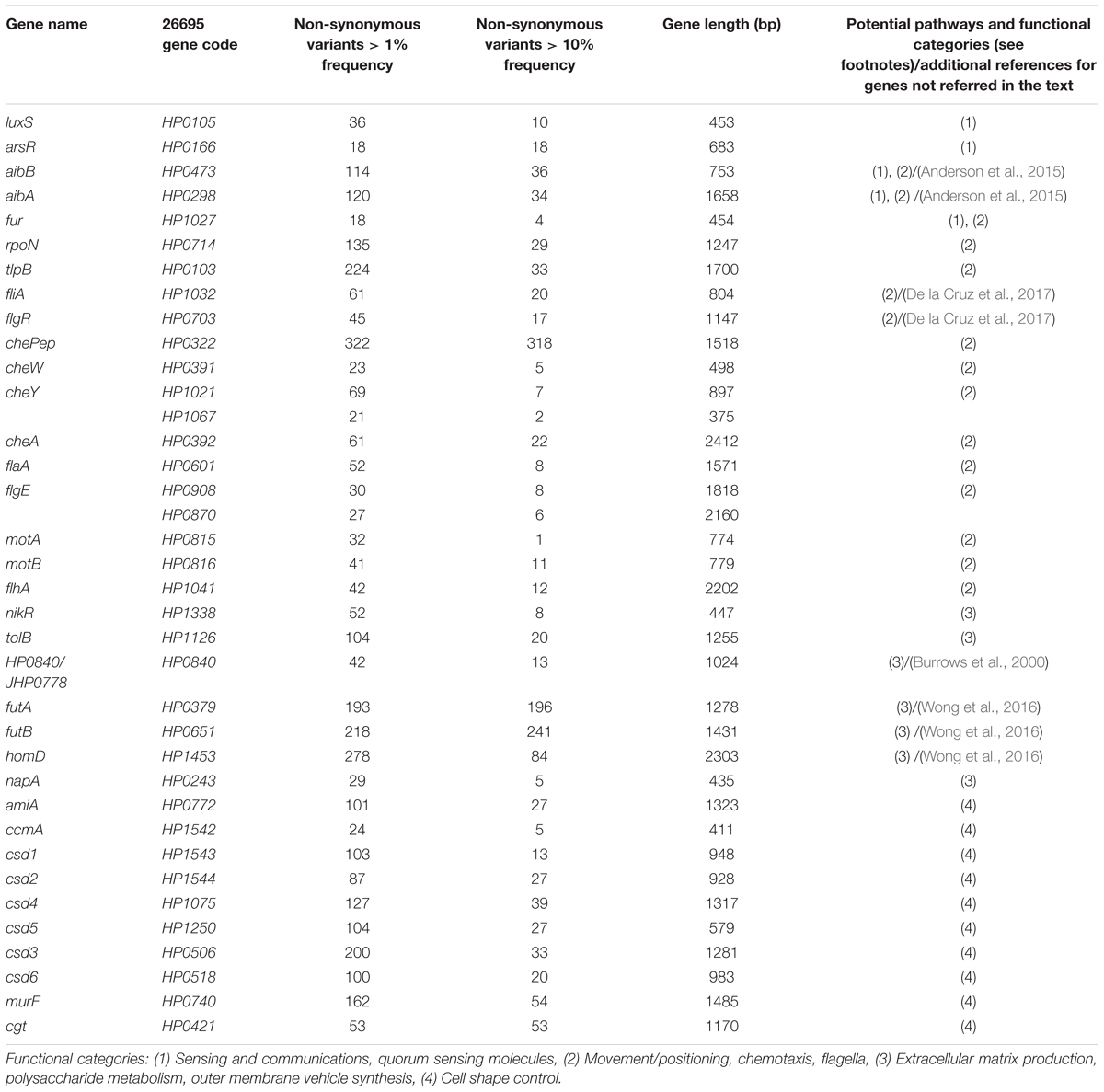
To demonstrate the interaction of bacteria and the role of cellular appendices in the formation of biofilm using SEM, we cultured Hp on glass cover slips for 48 h in RPMI medium (JT, MC, and J Girón, unpublished results). We observed a dense growth of bacteria attached to the substrate (Figure 1A), with an extensive expression of flagella. The images suggest that flagella might have a role in both, adhesion to the substrate and in the interaction between bacteria (Figures 1A,B). Images also show a high formation of smaller pili by bacteria forming the biofilm (Figure 1C). The role of flagella and other pili in the formation of biofilm has been well documented in other bacteria (Belas, 2014; Maldarelli et al., 2016; Zeng et al., 2017).
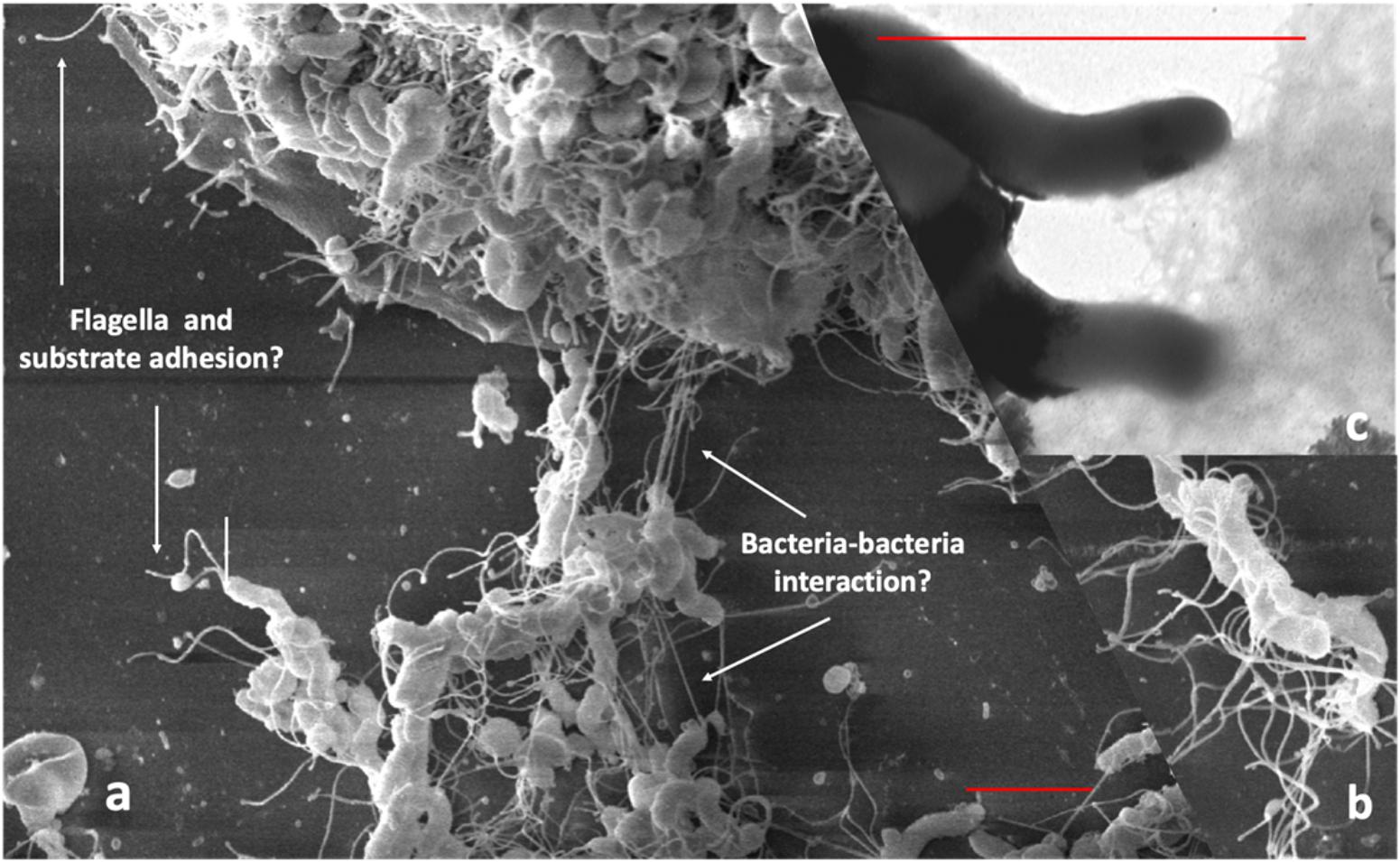
Figure 1. Scanning electron microscopy of Hp strain ATCC 43504 biofilm on a glass substrate. A bacterial suspension of 3 × 108 CFU/ml in Brucella broth was applied to a glass slide in a 24-well plate, incubated, dried, fixed with methanol and processed for SEM, (a) Images showing flagella probably adhering to the substrate; (b) possible bacteria-bacteria interaction with the flagella; (c) pilli produced by H. pylori during biofilm formation. Red bars in (a,c) indicate approximately three-micrometer scale.
In vitro Biotic Models
Hp forms biofilms not only on abiotic surfaces, but also on biotic surfaces. Human cell lines may mimic the in vivo cell behavior and can be used for ex vivo biofilm assays. Ex vivo models also include biofilm growth on natural tissues in a minimally altered environment, offering more strictly controlled experimental conditions than those of in vivo models, which allows more detailed studies. The ex vivo models also facilitate studies of biofilm association with virulence, or therapeutic assays for experimental antibiofilm treatments and biofilm inhibition (Salas-Jara et al., 2016; Magana et al., 2018).
One environmental condition to which Hp is reactive is the endogenously produced quorum-sensing molecule autoinducer-2 (AI-2) that Hp senses as a chemorepellent. Anderson et al. (2015) studied the role of AI-2 chemotactic responses during Hp biofilm formation on biotic and abiotic surfaces. In the biotic assay, they adapted an in vivo model, with conditions similar to those previously described (Tan et al., 2011) using polarized Madin Darby canine kidney (MDCK) epithelial cells seeded onto a transwell filter. Experimental inoculation of Hp strains resulted in the formation of microcolonies on MDCK monolayers, in an AI-2 chemotaxis- dependent manner; similar results were observed in an abiotic model. Salas-Jara et al. (2016) evaluated biofilm formation of two probiotic strains, L. fermentum UCO-979C and L. casei Shirota, on the surfaces of two gastrointestinal cell lines, AGS and Caco cells. Sterile glass coverslips were placed in a 24-well polystyrene plate. The coverslips were treated with poly-L-lysine at 1 mg ml-1 to improve cell adhesion. Subsequently Lactobacillus strains were inoculated and the highest biofilm density was observed 12 h later. The biofilm formed by either L. fermentum UCO-979C or L. casei Shirota strains inhibited the adherence of Hp ATCC 43504 to both cells lines. SEM images of the formed biofilms are shown in their paper (Salas-Jara et al., 2016).
In vivo Biofilm Formation in Animals
A recent study evaluated the potential of isolates of Hp to form biofilm in the stomach of C57BL/6J mice model. Mice were infected through gastric gavage with 108 UFC of Hp strain. Infected mice were examined after 1 and 2 weeks. One week after the last challenge, the mice were sacrificed. For examination, the stomachs were removed and fixed in 4% paraformaldehyde (PFA) and Hp biofilm was demonstrated by immunofluorescence and SEM (Attaran et al., 2016).
Detection of Biofilm-Associated Hp in Human Gastric Mucosa
The first evidence of biofilm formation by Hp during colonization in human gastric mucosa was photographic documentation by Carron et al. (2006). Using endoscopically obtained biopsy specimens and SEM analyses, they demonstrated the presence of dense mature biofilm-associated bacteria, attached to the cell surface of Hp-positive specimens. Hp-negative specimens, in contrast, had smooth mucosa with little evidence of a bacterial community (Carron et al., 2006). SEM also was used to quantify bacterial biofilm density on human gastric mucosa. Among patients with peptic ulcer disease, surface area covered by biofilms was 97.3% in Hp-positive patients, as compared to only 1.64% in Hp-negative patients (Coticchia et al., 2006).
To assess the biofilm-associated Hp in gastric biopsy specimens it is necessary to use methods other than routine ancillary stains, such as immunohistochemistry and fluorescence in situ hybridization (FISH). Preservation of three-dimensional structure is critical in order to analyze the spatial organization of the gut microbiota relative to mucin, host tissue and luminal contents (Hasegawa et al., 2017). Therefore, one of the crucial steps for gastrointestinal pathological laboratory is the fixation and embedment. The most common fixative is formaldehyde, an aqueous fixative, but several authors report that this fixative results in collapse or loss of the mucus layer and a widely used protocol is to process samples with non-aqueous Carnoy fixation to preserve the mucus layer for detection of bacteria adherent to the mucosal surface (Swidsinski et al., 2005; Johansson and Hansson, 2012). Johansson and Hansson (2012) observed a thick inner mucus layer firmly attached to the intestinal epithelia in non-aqueous Carnoy fixative, in contrast with a thin streak of collapsed mucus of less than 1 μm on the same tissue processed with formaldehyde fixative. Using confocal microscopy, Hasegawa et al. (2017) compared preservation of three-dimensional structure of four different embedding media: paraffin wax; polyester wax; optimal cutting temperature (OCT) compound; and glycol methacrylate resin. They conclude that hydrophobic embedding materials as paraffin require organic solvents to remove embedding resin and redistribution, collapse or loss of luminal contents could have occurred. In summary, although all of the examined embedments were capable of producing two-dimensional images, only glycol methacrylate resin enabled both retention and visualization of the three-dimensional luminal distribution of bacteria and food particles (Hasegawa et al., 2017).
The gastric surface mucous cells and gland mucous cells express secretory mucins, MUC5AC and MUC6, respectively. On the other hand, aberrant expression of the secretory mucin MUC2 in gastric mucosa is closely related to intestinal metaplasia and intestinal goblet cells (Ho et al., 1995; Reis et al., 1999; Babu et al., 2006; Matsuda et al., 2008). Hp may inhibit MUC5AC expression by the human gastric epithelium, and thus facilitate colonization. In contrast, increased MUC6 expression may help inhibiting colonization due to its antibiotic properties (Niv, 2015). FISH combines the molecular identification of bacteria with the direct visualization of the bacteria and the mucosa, which provides a significant advantage over culture, PCR, and histological methods alone (Swidsinski et al., 2005). Hasegawa et al. (2017) evaluated microbial FISH and two mucus labeling methods simultaneously to visualize microbial cells and host-derived mucus in intestinal sections following both Carnoy and PFA fixation and embedded in methacrylate or paraffin. Mucus was visualized with a fluorescent wheat germ agglutinin (WGA) and by indirect immunofluorescence using a primary antibody raised against mouse colonic mucin and both fixation methods allowed the visualization of intestinal mucus layers (Hasegawa et al., 2017).
We attempted to obtain additional evidence of Hp biofilm formation in human gastric mucosa fixed in Carnoy, using FISH and confocal laser scanning microscopy (CLSM). We showed that Hp colonization is not limited to the surface of the human gastric glands, but can colonize deep in the glands, particularly in the region of the neck where cells are in constant proliferation and interestingly, also at the bottom of the glands, in intimate contact with the stem cells (Sigal et al., 2015). The identification of colonization of niches deep in the glands was possible because we studied large pieces of stomach coming from patients subjected to bypass surgery for control of weight. Using the same approach, we have confirmed the ability of Hp to form large aggregates suggestive of biofilm formation in the surface of the glands (Figure 2A), in the neck in close contact with proliferative cells and deep in the glands (Figure 2B), in the vicinity of stem cells (Figure 2C). In these studies, Hp was stained with antibodies specific to the bacteria, and confirmed the observed in vivo microbial aggregates were formed exclusively by Hp. However, it was not determined whether these aggregates were embedded in host mucins, bacterial EPSs or their combination. The challenge still remains to study the nature of the biofilm formation by Hp in vivo, in the gastric mucosa of humans and its role in the decades-lasting persistent colonization. The mechanisms of interaction of this biofilm-communities with the gastric mucosa at different niches in the glands may help us understand why in a few cases (<2%) the outcome of this interaction may be a severe disease such as gastric cancer. In particular, our observation of large aggregates of Hp growing in intimate contact with stem cells raises the possibility of damage to these cells as a result of direct interaction with the bacteria.
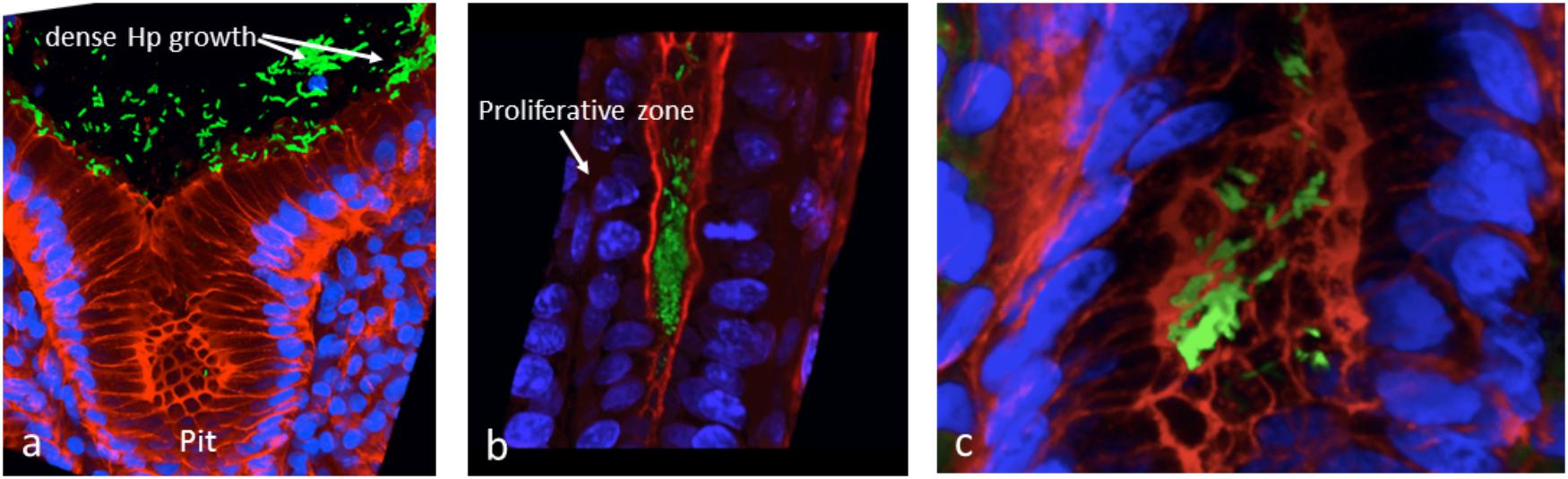
Figure 2. In vivo evidences suggestive of biofilm formation by H. pylori in the gastric glands of humans. (a) Large aggregates of H. pylori colonizing the surface of gastric glands; (b) H. pylori aggregates colonizing the neck of gastric glands, with proliferative cells; (c) colonies of H. pylori deep in the gland, in the vicinity of stem cells. H. pylori in green, actin in red and DNA nucleus in blue.
Quantification of Mucosal Biofilms in Clinical Specimens
Quantification of mucosal biofilm bacteria is rather complex, and it has been performed almost exclusively for gut mucosal biofilms (Swidsinski et al., 2005; Dejea et al., 2014), Swidsinski et al. defined three distinct microscopic fields of the biofilm: (1) adherent bacteria (±1 μm of the epithelial border); (2) mucus-scattered bacteria present within mucus next to the epithelial surface; and (3) mucus ceiling bacteria present in the outer portions of the mucus, at least 10 μm away from the epithelial surface, and found marked increases in fields (1) and (3) for patients with inflammatory bowel disease, using several arithmetic assumptions (Swidsinski et al., 2005). Their data suggest that the average biofilm density in healthy gut mucosa is in a range of 1–5 × 108 per ml (Swidsinski et al., 2005; Dejea et al., 2014), which reflects the fact that the inner mucus layer of normal human colon is impenetrable to bacteria (Matsuo et al., 1997; Johansson et al., 2008; Johansson et al., 2014). This number in healthy gastric mucosa may be even lower due to the high acidity, but to date, exact quantitation of in vivo biofilm in human stomach is lacking. We would need adjustment to the mathematical formula and bacterial probes used for gut biofilms in order to quantify Hp gastric biofilm.
Genetic Regulatory Pathways of Hp Biofilm Formation
Candidate Genes Involved in Biofilm Formation
Yonezawa et al. (2010) and Wong et al. (2016) report substantial Hp strain variability identifying strong biofilm producers and poor biofilm producers in both reference strains as well as clinical isolates. These strain differences are likely to reflect variability in bacterial genes involved in biofilm formation and may be important for targeted intervention to reduce consequences from such infection. Existing literature suggests that three essential traits are required for bacteria to form and grow in biofilm in vivo: (1) inter-bacterial communication, (2) coordinated movements, and (3) aggregation. Furthermore, genes that control bacterial shape to fit biofilm environment may have an additional role in biofilm formation and maintenance.
Inter-bacterial communication is mediated through quorum sensing (QS) system, which is mediated by production and release of chemical signaling molecules in response to cell density and other physiological conditions (Elias and Banin, 2012; Gölz et al., 2012; Majumdar and Pal, 2017). These signaling molecules alter the expression of bacterial QS dependent genes to induce bacterial phenotype changes in virulence, motility, chemotaxis and biofilm formation, thus contributing to adaptation and colonization to the host (Elias and Banin, 2012; Gölz et al., 2012; Majumdar and Pal, 2017). To date, multiple systems based on different groups of molecules have been identified, some very bacteria-specific and others more general. Among those, signaling molecules called autoinducers (AI), AI-2 system, are most universally used by many bacteria, including Hp (Elias and Banin, 2012; Gölz et al., 2012; Majumdar and Pal, 2017). Another molecule produced by multiple bacterial families is N-acylhomoserine lactone (AHL) (He and Zhang, 2008; Elias and Banin, 2012; Papenfort and Bassler, 2016), although there have been no data to support that Hp utilizes the AHL-based QS. The other group of molecules more recently discovered is termed diffusible signal factor (DSF) that consists of fatty acids of various chain lengths and branching and is known to be used by several groups of bacteria (He and Zhang, 2008; Elias and Banin, 2012; Papenfort and Bassler, 2016). One well-characterized DSF is 2-(Z)-tetradecenoic acid (TDA) produced by the xylem-limited plant pathogen Xylella fastidiosa (Ryan and Dow, 2011; Ryan et al., 2015). Recently, Yamashita et al. reported two self-growth-inhibiting compounds in Hp and one of them was identified as 7-(Z)-TDA (Yamashita et al., 2015). Although it has not been reported as a DSF, 2-(Z)-TDA is a positional isomer of 7-(Z)-TDA. Thus, 7-(Z)-TDA probably also acts as a signaling molecule to control the cell density of Hp. Currently, it is not known which Hp genes are responsible for synthesizing this compound. There are no other Hp-specific QS molecules characterized thus far. Several studies have also revealed that signaling pathways induced by QS molecules are followed by the phospho-relay cascade transmitted from the membrane-bound receptors to the cytosolic second-messenger system (Bharati and Chatterji, 2013; Wolska et al., 2016; Jakobsen et al., 2017). Second messengers based on mono (cAMP and cGMP) and di-cyclic or modified nucleotide (ppGpp, c-di-GMP and c-di-AMP) play a crucial role in transmitting the signals received from the surface receptor to the target molecule in the cell. Cyclic dimeric guanosine monophosphate (c-di-GMP) has evolved as a key activator of biofilm formation in almost all bacteria (Römling and Balsalobre, 2012; Bharati and Chatterji, 2013; Wolska et al., 2016; Jakobsen et al., 2017). c-di-GMP is synthesized by the GGDEF domain proteins and degraded by the unrelated EAL and HD-GYP domain proteins. The c-di-GMP signaling network is the most complex secondary signaling system found in bacteria with more than 100 c-di-GMP-metabolizing proteins in some species. This signaling network is especially prominent in γ-Proteobacteria, including the major human pathogens Pseudomonas aeruginosa, Salmonella typhimurium, Escherichia coli, and Vibrio cholerae, but little is known about this network in Hp (Römling and Balsalobre, 2012).
After bacteria reach a suitable niche to live, guided by chemotaxis, bacterial congregation requires adhesive materials, known as EPS. EPS are mostly produced by bacteria and consist of polysaccharides, proteins, enzymes, nucleic acids, lipids, other biopolymers, extracellular bacterial structures such as flagella, pili, fimbriae, and OMV (Flemming and Wingender, 2010). Interestingly, in contrast to many other bacteria, proteins appear to be the central player in Hp biofilm matrix (Windham et al., 2018). Recent studies also indicate that these three processes are not only controlled by Hp protein-coding genes, but also by a number of non-coding small regulatory RNAs (Mika and Hengge, 2013; Fazli et al., 2014; Bak et al., 2015; Wolska et al., 2016). Finally, it is also noteworthy that host molecules may contribute to EPS and host-bacterial co-aggregation. Specifically, secretory immunoglobulin A (IgA) has been shown to facilitate biofilm formation by normal gut flora in human tissue culture and by Escherichia coli in an intestinal cell line (Randal et al., 2003), as well as in a mouse model mono-infected with commensal Bacteroides fragilis (Donaldson et al., 2018). Interestingly, a higher systemic IgA response has been reported in mice infected with a high biofilm producing Hp than in those infected with low biofilm producing Hp strain, although mucosal secretory IgA levels were not quantified (Attaran et al., 2016).
Biofilm-Associated Hp Genes
Inter-bacterial Communication (QS Molecules)
AI-2 is produced as a metabolic byproduct of the reaction carried out by LuxS, which cleaves S-ribosylhomocysteine, producing homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD) and AI-2 as a metabolic byproduct of this reaction. DPD undergoes rapid dehydration and cyclization, existing in equilibrium as several molecules collectively termed as AI-2 (Schauder et al., 2001). LuxS uses methionine as a reduced sulfur source in the processes of de novo cysteine biosynthesis pathway. The luxS gene maps within an operon encoding cysK, metB, and luxS, which are necessary for the de novo cysteine biosynthesis pathway (Doherty et al., 2010). Some luxS mutants show reduced motility, and present a reduced number and length of each flagella. These differences result from a reduced transcription of genes of the flagella biosynthesis pathway (such as flaA, flgE, motA, motB, flhA, and fliI but not flaB genes) (Osaki et al., 2006; Rader et al., 2007; Shen et al., 2010). Gölz and colleagues demonstrated that a normal motility could be restored by adding AI-2. All described defects affecting motility of Hp luxS mutants were restored by the addition of AI-2 or DPD (Gölz et al., 2012). In Hp, AI-2 functions as a signaling molecule up-stream of the flagellar regulator flhA. High amounts of AI-2 increases motility, but it also reduces biofilm formation by Hp, thereby leading to colonization of niches with a small number of bacteria, but with a better provision of nutrients. AI-2 influences flagellar gene expression up-stream of flhA (Tsang and Hoover, 2014) and functions as chemorepellant via TlpB (Gölz et al., 2012). Both mechanisms obviously contribute to regulation of biofilm formation vs. planktonic growth, which in turn promotes bacterial colonization and persistence in the stomach (Rader et al., 2011).
The Hp genome carries few regulatory elements. Three of those are Fur, regulating the ferric uptake; NikR, regulating the response to the presence of nickel response regulator; and ArsRS, a two component system, which responses to an acid environment (Danielli and Scarlato, 2010). In particular, ArsR is essential for in vitro survival (Loh et al., 2010). Mutation studies of ArsS have been performed by mutating arsR into a non-phosphorylatable form (ArsR-D52N mutant). Involvement of ArsS in biofilm formation has been suggested by proteomic analysis and because ArsS mutants have a significant increase cellular aggregation and adherence to the flask at the air-liquid interface. Further studies showed that strains carrying ArsRD52N mutation or a combination of nikR and arsS deletions had a quicker transition to the coccoid form. Similar to biofilm formation, the transition to the coccoid form is known to happen in case of environmental stressors, such as nutrient deprivation. In summary, the data of these studies suggest that the lack of ArsRS system increases stress response, leading to an increased and faster biofilm formation. The simultaneous deletion of arsS and nikR genes decreases the normal Hp functionality and results in increasing of the coccoid forms and increased biofilm formation (Servetas et al., 2016). This hyper-biofilm forming phenotype of arsS mutants is found to be mediated by an outer membrane protein, homB (Servetas et al., 2018).
Chemotaxis That Guides Movement to Reach the Right Position
In the colonization of gastric mucosa by Hp, coordinated movements are required to reach and position in the right topology of the stomach horizontally and vertically to form stable biofilms, guided by chemotaxis and powered by flagella and pili (Harshey, 2003; Howitt et al., 2011; Persat et al., 2015). In fact, aflagellated Hp mutants exhibit impaired biofilm formation (Hathroubi et al., 2018b). Hp species regulate their motility by chemotactic signaling systems, which allow the bacteria to follow favorable chemical gradients in their host environment. A number of transcriptional factors are known to be involved in these processes (De la Cruz et al., 2017). For chemotactic signal transduction, four different groups of proteins are necessary: (1) chemoreceptors, (2) core signal proteins, (3) accessory proteins, and (4) flagellar switch proteins (Lertsethtakarn et al., 2011). In Hp, the chemotactic behavior to low pH is dependent on the chemoreceptor TlpB (Croxen et al., 2006). A luxS mutant of Hp strain G27 showed a reduced stopping frequency in liquid media, which was restored by the addition of AI-2 or DPD (Rader et al., 2011). Analyzing the chemotactic behavior of double and single mutants, these authors confirmed that AI-2 is perceived as a chemorepellant signal via TlpB. In fact, an Hp strain deficient for the chemoreceptor TlpB failed to move away from a source of synthetic DPD and did not display increased stopping behavior upon addition of synthetic DPD. These behaviors were restored upon genetic complementation of the tlpB gene. Hp chemoreceptor TlpB is required for recognition of AI-2, which confirms its role in negative pH taxis (Rader et al., 2011). ChePep mediates another signaling system that controls the flagellar switching in a chemotaxis signaling protein-dependent manner. ChePep localizes to the flagellar pole of Hp, and chePep mutants show aberrant flagellar rotation, although flagella are correctly assembled and motile. In fact, ChePep does not control directly flagellar switching, but functions as a chemotaxis regulator, as it has been shown by studying Hp chemotactic behavior in a pH gradient. Hp cells increase their turning frequency in presence of a chemorepellent (i.e., acid) because of a predicted increase in phosphate signaling in the chemotaxis system. ChePep mutants constantly switch flagellar rotation also in normal culture condition and respond to acid condition by increasing their turning frequency. This behavior can be explained by high levels of phosphorylated CheY protein as obtained in some enteric bacteria by the mutation of CheZ and CheB that normally function as chemotaxis regulators by reducing the phosphorylation state of CheY (Howitt et al., 2011).
Extracellular Matrix, Outer Membrane Vehicle, and Adhesins Involved in Biofilm Formation
The Tol Pal gene cluster has been extensively studied in other organism such as Escherichia (E) coli and its role in the formation of OMV and in bacterial cell integrity is well known. Hp genomes encode homologous genes to E. coli tolB and pal (HP1126 and HP1125, respectively), but no Hp homolog for tolA has been described (Turner et al., 2015). Moreover, Hp possesses two coding regions (HP1127 and HP1128), with no known homologs in the databases, and other two coding sequences, HP1130 and HP1129, putative homologs of tolQ and tolR, respectively. Hp tolB mutants displayed extensive “blebbing” and had very few flagella, which were markedly shorter in length and appeared to form clumps, suggesting a division defect. The morphology of the double mutant tolB-pal was not very different to that of wild type (WT) bacteria, although mutants lacked flagella completely. This phenotype could be due to the fact that the peptidoglycan-associated outer membrane proteins, including those of the Pal family, share sequence homology in the C-terminal region with the E. coli motility protein, MotB (Mot and Vanderleyden, 1994). This region of MotB is particularly important in anchoring the flagella structure to the peptidoglycan. It is therefore possible that any disruption of either the TolB or Pal protein may affect the functioning of the Tol-Pal complex as a whole, thereby altering flagella synthesis or hindering the anchoring of the flagella to the peptidoglycan layer. Furthermore, Hp ΔtolB and Δpal mutants produce >600- and 22-fold more OMVs than WT bacteria, suggesting a strong co-regulation of flagella and OMV (Turner et al., 2015).
Efflux pump functions in the formation of biofilms and multidrug resistance (Soto, 2013). gluP has been shown to be involved in the biofilm formation and multidrug resistance of Hp and the expression of gluP is upregulated by SpoT, which is a known global regulator (Ge et al., 2018). In addition to SpoT, there are other genes involved in both biofilm formation and multidrug resistance in Hp, which is mostly mediated through an increased expression of RND efflux pump genes (Yonezawa et al., 2013; Attaran et al., 2017). Furthermore, The gluP encodes a glucose/galactose transporter that belongs to the major facilitator superfamily of the efflux pump machinery (Tomb et al., 1997), which is mainly responsible for the physiological uptake of sugars, such as D-glucose (Psakis et al., 2009). Its inactivation has also the effect to alter the biofilm formation because glucose is a component of polysaccharides (Limoli et al., 2015) that are part of biofilm matrix (Ge et al., 2018).
Using nuclear magnetic resonance, Yang et al. (2011) identified mannose-related proteoglycans (proteomannans) as a major contributor to Hp EPS in in vitro culture. They detected the presence of NapA protein upregulation in the biofilm, suggesting a mechanism to increase adhesiveness of Hp biofilm. An isogenic mutant of napA revealed an altered biofilm structure with reduced aggregates, when compared to the WT. Adhesins are another class of genes involved in biofilm and the Hp genome contains two adjacent homolog genes, alpA-alpB, annotated as omp20-omp21 and hopC-hopB in the 26695 and G27 genomes, respectively, that may be involved in biofilm formation. A recent study (Yonezawa et al., 2017) compared OMV protein profiles between a Hp strain and its spontaneous weak biofilm-forming mutants and demonstrated that AlpB was an important protein for biofilm formation. alpB mutants derived from various strains have also been shown to be incapable of inducing cell aggregation (Senkovich et al., 2011; Yonezawa et al., 2017). Furthermore, these studies suggest that particularly the variable region of alpB is involved in the attachment to various substrates, including gastric cells.
Our preliminary studies on several known Hp virulence genotypes and biofilm formation in vitro support potential involvement of some adhesins. We tested a total of 45 clinical isolates from Mexican patients, including 15 each for non-atrophic gastritis, intestinal metaplasia and gastric cancer, using 96-well polystyrene microtiter plates. Each well was filled with 135 μl of Brucella broth. The formation of biofilm was started by inoculating 15 μl of pre-cultured bacterial suspension with 3 × 108 CFU/ml into each well. The cultures were incubated under microaerobic conditions during 48 h, at 37°C and 10% of CO2. Uninoculated Brucella broth served as a negative control. After incubation, the supernatant was removed, the plates were washed 3 times with PBS and dried for 30 min. One hundred and fifty microliter of 0.1% crystal violet was added to each well and the plate was incubated at room temperature for 20 min. The excess crystal violet stain was removed, and the plate washed 3 times with PBS. The crystal violet staining biofilm was extracted with 150 μl of ethanol (70%) and measured at OD 595 nm. The assay was done 3 times and the final results were the mean of absorbance values. We tested strains positive or negative for adhesins reported important for Hp colonization. Strains lacking either oipA or sabA showed a significantly reduced ability to form biofilm. In contrast, strains lacking hopZ were in fact more efficient producing biofilm (Figure 3), and we found no differences between strains with or without the babA2 gene. These results need to be confirmed with the study of additional strains, including isogenic strains mutated in the gene under study. On the other hand, our initial results indicate that cagA and vacA genotype may have no influence in the intensity of biofilm formation (Figure 4); formation of biofilm was similar in both, cagA-postive strains (J99 and 26695) and cagA-negative strains (TX30). Formation was also similar between vacA s1 and s2, or m1 and m2 strains; although we observed a tendency for reduced biofilm formation by m2 Hp strains (Figure 4). These observations ensure that the observed association with some adhesines was not mediated through the correlation with the presence of cagA and vacAs1m1 genotypes.
Figure 3. The presence of adhesins in H. pylori strains may influence the formation of biofilm. See text for details on the assay.
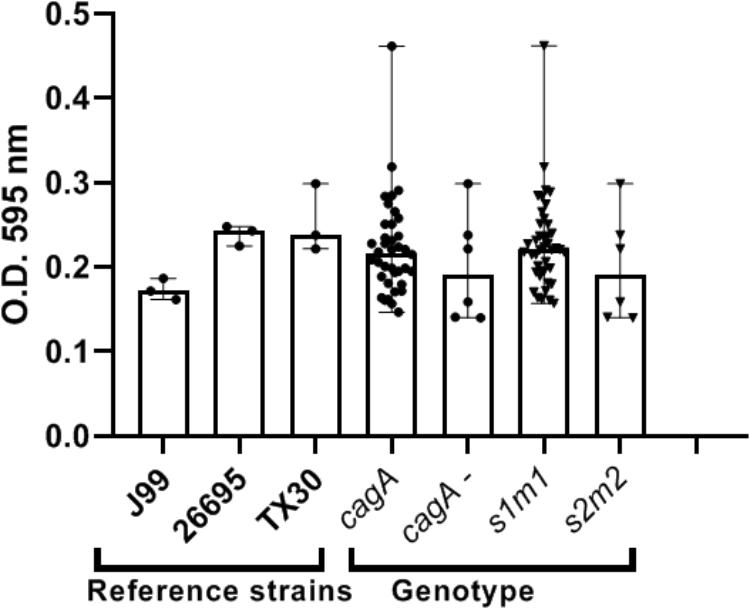
Figure 4. cagA or vacA genotypes did not influence the capacity to form biofilm by H. pylori strains. See text for details on the assay.
Genes Regulating the Shape of Hp
The morphological transition of Hp leads from a spiral rod–shaped organism to a coccoid organism by changes in the peptidoglycan of the bacterial cell wall, These modifications are mediated by AmiA (Chaput et al., 2006, 2016). Moreover these changes influence the ability of the coccoid form to escape the immune system. Studies have shown that peptidases Csd1-4 and Csd6, as well as potential regulators Csd5 and CcmA are required to tailor the PG layer to generate the helical cell shape characteristic of Hp (Sycuro et al., 2010, 2012, 2013). Csd4 cleaves the gamma-D-Glu2-mDAP3 bond of the muramyl tripeptide to produce the muramyl dipeptide (member of M14 metallopeptidase family, Zn2+-dependent). Csd3 (also known as HdpA, member of the M23 metallopeptidase family) has D,Dendopeptidase and D,D-carboxypeptidase (D,D-CPase) activities. Csd6 (HP0518) belongs to the peptidoglycan trimming pathway and also influences the cell shape of Hp. Disruption of the csd6 gene by transposon-induced mutation or its deletion resulted in a straight rod shape and in an increase in tetrapeptide-containing muropeptides (Kim et al., 2015).
More recently Blair et al. (2018) studied the mechanisms by which Csd5 promotes helical cell shape. It has been shown that the N-terminal cytoplasmic (NT) and transmembrane (TM) domains, together with a C-terminal SH3 domain in Csd5 are each required to promote helical shape. Csd5 interacts directly with peptidoglycan via its C-terminal SH3 domain, whereas the N-terminal transmembrane domain promotes interactions with CcmA, MurF that catalyzes the synthesis of a PG precursor, and F1F0 ATP synthase. The recognition of the interaction of Csd5 protein with CcmA, a known cell-shape protein and putative cytoskeletal bactofilin, and with MurF, a known cell elongation factor, was unexpected but not surprising given the connections between PG synthesis, cell shape and intermediate filament proteins in helical organisms. Another study investigated the cholesteryl glucosides (CGs), one of the major components of the cell wall, by the deletion of the hp0421 gene, which encodes cholesteryl α-glucoside transferase that integrates (CGs) into the cell wall of Hp. This determines a deficiency of cholesterol, an alteration in the morphology and shape (“c”-shaped cells were prevalent), and of cell walls components like LPS, which leads to less virulent strain and an increasing of susceptibility to antibiotics (Qaria et al., 2018).
Hp Genetic Variabilities in Biofilm-Associated Genes
To elucidate the diversity of biofilm-associated genes among Hp strain, we used whole genome data (Muñoz-Ramírez et al., 2017) of 74 Latin American Hp strains to analyze 33 genes reported to be involved in biofilm formation (Table 1). This analysis revealed a mean variation of 6.78% in the amino-acidic sequence, ranging from a minimum of 0.39% in motA gene (HP0815, 1 non-synonymous variant in 258 amino acids) to a maximum of 23.12% in chePep gene (HP0322; 318 non-synonymous variants in 506 aa). For luxS (HP0105) that encodes an enzyme to produce AI-2 (He and Zhang, 2008; Gölz et al., 2012; Wolska et al., 2016), we found 10 non-synonymous variants with a variant frequency >10% on a total of 453 bp. Further phylogenetic analysis of the 33 genes in the 74 Hp strains did not show evidence of geographical clustering by country for most of these genes with the only exception of the murF gene (HP0740), which clearly formed separated clusters for Mexico and Colombia (Figure 5). This result would suggests that murF is under constant positive selection to adapt to the human host population they are colonizing. Whether this diversity in murF contributes to differential gastric cancer risk in these two populations is something that deserves further studies.
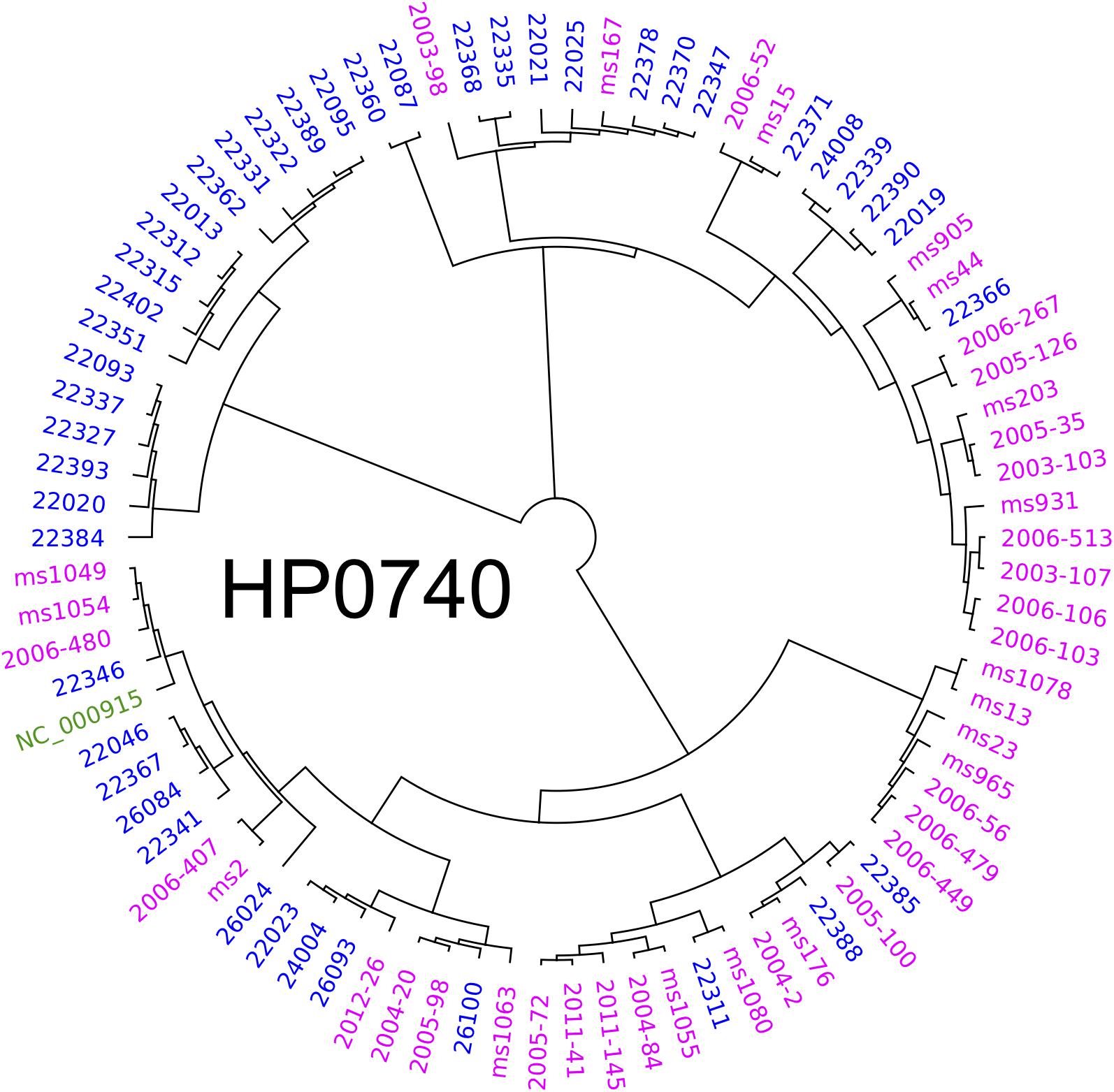
Figure 5. Phylogenetic analyses of HP0740 for 74 Mexican and Colombian strains. The alignment and graphics was produced with Geneious and graphically modified with Inkscape software.
Functional and Morphological Changes Associated With Biofilm Formation
Growth Rate
Slow growth rate has been recognized as one of the major contributing factors to antibiotic resistance of biofilm-associated bacteria (Stewart, 2015). Matured biofilms are known to contain variable fractions of slow growing or non-dividing metabolically inactive cells, which gives rise to persistent colonization (Lewis, 2005; Kostakioti et al., 2013). In vitro mono-microbial biofilm studies demonstrated that Streptococcus mutans biofilms grew about 35% slower than planktonic culture before the stationary phase (Welch et al., 2012). Using transcriptional profiles, Folsom et al. estimated that Pseudomonas aeruginosa grew at just 10% of planktonic growth rate in the stationary biofilms (Folsom et al., 2010). Applying an agarose gel as artificial EPS, Pabst et al. confirmed that the growth rate of Staphylococcus aureus in the stationary biofilms was also about 10% of that of planktonic culture (Pabst et al., 2016). Altered gene expression leading to reduced transcription and to translation and DNA replication arrest, has been identified as molecular characteristics of the stationary biofilm-associated cells (Lewis, 2005; Stewart, 2015). Others suggest that bacterial synthesis of antibacterial compounds against own or neighboring organisms (Elias and Banin, 2012; Yamashita et al., 2015) may also contribute to the slow growth of biofilm-associated bacteria. To date, however, there have been no direct in vitro or in vivo data to support that Hp forming stationary biofilms exhibits a reduced growth rate.
Metabolism
Biofilms are formed by groups of cells in different states, growing or non-growing and metabolically active or inactive in variable fractions, depending on maturity and on chemical gradients (O2 and nutrients) of the biofilms (Stewart, 2015; Flemming et al., 2016). It has been postulated that dormant non-growing metabolically inactive cells reach these metabolic states in order to reduce cell permeability and protect themselves against oxidative stress (Stewart, 2015). Bacterial cells in stationary biofilms have also been reported to increase synthesis of pili, fimbriae and exopolysaccharides as well as lipopolysaccharide modification and production of stress-response enzymes (Prigent-Combaret et al., 1999; Folsom et al., 2010). Wong et al. recently profiled metabolites of high and low biofilm forming Hp strains from Malaysia using liquid chromatography/quadrupole time-of-flight mass spectrometry (Wong et al., 2018). The study compared four high and four low biofilm forming strains cultured up to 3 days in duplicate. Interestingly, low-biofilm-formers produced more metabolites than high-biofilm-formers, consistent with lower overall metabolic activities in biofilm-associated bacteria. Further analysis indicated that the metabolites significantly lower in high biofilm producers belonged to major categories of lipids, with an important role in bacterial-bacterial communications, and metabolites involved in prostaglandin synthesis (Wong et al., 2018).
Morphology
A morphological change described as small colony variants (SCVs) has been linked to various biofilm associated infections, such as cystic fibrosis, osteomyelitis and device-related infections (Proctor et al., 2006). Since its first description for Salmonella enterica serovar typhi (S. typhi) almost 100 years ago, SCVs have now been reported for a wide range of bacterial genera and species, including Staphylococci, Escherichia coli, Pseudomonas aeruginosa, Vibrio cholerae, Shigella spp., Lactobacillus acidophilus, and Neisseria gonorrhoeae (Proctor et al., 2006). Currently, the connection between the SCV phenotype and persistent, recurrent infections has become a hot topic in clinical microbiology. Phenotypically, the size of SCVs are about one tenth of that in regular colonies, with a slow growth rate, atypical colony morphology and unusual biochemical characteristics and are less susceptible to antibiotics than their wild-type counterparts. An altered cellular morphology in the cell wall structure and the emergence of intercellular EPS have also been demonstrated for SCVs from Staphylococcus aureus. These characteristics have been associated with mutations in the nupC gene that encodes a protein involved in thymidine uptake and in the thyA gene that encodes thymidylate synthase (Proctor et al., 2006). A group of Norwegian researchers reported spontaneous appearance of Hp SCVs, although they did not clarify its size relative to normal colonies (Bukholm et al., 1997). This variant was induced by acid exposure and showed enhanced adherence and invasion to epithelial cells (Bukholm et al., 1997; Tannæs et al., 2001). It was found that these morphological changes are a consequence of a shift in cell wall lipid composition, specifically, an increase in lysophospholipids, due to phase variation in the pldA gene, resulting in production of an active form of the outer membrane phospholipase A (OMPLA) (Bukholm et al., 1997; Tannæs et al., 2001). Another colony variant associated with biofilm is the rough colony variant arising from smooth colonies of P. putida (Hansen et al., 2007; Elias and Banin, 2012). This was caused by two independent mutations in wapH (PP4943), a gene involved in lipopolysaccharide (LPS) biosynthesis (Hansen et al., 2007). Rough/smooth phenotype variants due to altered LPS composition were also observed for Hp, yet its association with biofilm formation and virulence is unclear (Bertram-Drogatz et al., 1999).
At the cellular level, the best-described morphological change in Hp associated with biofilm in vivo is coccoid transformation (Carron et al., 2006; Coticchia et al., 2006; Cellini et al., 2008). Coccoid transformation is considered a common feature of Gram-negative rod bacteria under conditions of stress (Andersen and Rasmussen, 2009; van Teeseling et al., 2017; Krzyzek and Gosciniak, 2018). In vitro, coccoid forms of Hp can be induced by prolonged culture or use of suboptimal levels of antibiotics (Krzyzek and Gosciniak, 2018; Poursina et al., 2018). The Hp coccoid forms have been defined as viable but non-culturable (VBNC), thus undetectable by conventional cultures, and are known to have an increased capacity to aggregate into monomicrobial clusters embedded in thick EPS (Cellini et al., 2008; Cellini, 2014; Krzyzek and Gosciniak, 2018). In many ways, VBNC coccoid Hp resembles the characteristics of persister cells documented in biofilms of other bacteria (Lewis, 2005; Pinto et al., 2015) and they can survive up to 1 year just in fresh water (Cellini, 2014; Percival and Suleman, 2014). An accumulation of N-acetylglucosaminyl-N-acetylmuramyl–l-Ala–d-Glu in PG of the cell wall (Costa et al., 1999), catalyzed by AmiA (PG hydrolase), has been described to precede this morphological transformation and also to lead to immune evasion by escaping from NOD1 detection (Chaput et al., 2006). Yet, since PG dictates bacterial cell shape (Rubin and Trent, 2013; van Teeseling et al., 2017) and since many other PG remodeling enzymes have been described (Caccamo and Brun, 2018), genes other than amiA are likely to be involved in this process. In fact, genes encoding the enzymes, pgp1 and pgp2 for Campylobacter spp, and csd1, csd2, csd3, csd4, csd6, murF and related non-enzymatic proteins, csd5 and ccmA for Hp have been identified to be critical in the maintenance of helical shape (Sycuro et al., 2010, 2012, 2013; Kim et al., 2015; Esson et al., 2016; Blair et al., 2018) as detailed in the previous section. In addition, coccoid Hp harvested in vitro from clinical isolates overexpressed spoT, a global transcriptional regulator for stress responses (Poursina et al., 2018). A homologous gene in Mycobacterium smegmatis has been linked to its cell shape control (Gupta et al., 2016) and a Hp spoT-mutant exhibits a reduced ability to form biofilm (Ge et al., 2018). To date, despite these growing observations, it remains to be elucidated whether coccoid transformation indeed prompts biofilm formation, or whether biofilm formation promotes Hp coccoid transformation.
Virulence Factors
It is yet unknown whether biofilm-associated bacteria exert virulence more potently than those living planktonically. There are several factors that affect virulence of biofilm-associated bacteria. As discussed above, the stage (maturity) of biofilms, chemical composition (O2, acidity and nutrients gradients) and microbial (mono- vs. poly-biofilms) environment within biofilms are important determinants of bacterial gene expression (Lewis, 2005; Folsom et al., 2010; Stewart, 2015). In addition, how to quantify or compare virulence is not so straightforward, because life expectancy of bacteria living in biofilms and that of planktonic bacteria are different, as the former is expected to live longer causing persistent infection (Lewis, 2005; Pinto et al., 2015). Furthermore, even if biofilm-associated bacteria express lower virulence per time compared with planktonic bacteria, cumulative effects on host cells may be higher. In addition, aggregative nature of biofilm-forming bacteria may enhance virulence through increased adherence to host cells, even if the same amount and potency of virulence molecules are produced or released. Part of biofilm-associated virulence is also derived from surrounding EPS, not bacterial cells themselves. EPS contains secreted enzymes, toxins and outer membrane vehicles (Flemming and Wingender, 2010; Hathroubi et al., 2018a) and can activate neutrophils without opsonization to induce inflammatory reactions and exert cytotoxicity (Meyle et al., 2012; Fulsundar et al., 2015; Susanne et al., 2015; Kim et al., 2016; Nascimento et al., 2016).
Pinto et al. reviewed virulence of VBNC bacteria predominantly found in biofilms (Pinto et al., 2015). They concluded that most bacteria continue to express their virulence and toxin genes, but expression levels are downregulated or protein secretion may not be detectable. However, altered gene/protein expression seems to vary with types of bacteria, types of virulence genes, or co-colonization with other bacteria. For example, when Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus (MRSA) were grown together as a mixed culture, biofilm development was accelerated and production of P. aeruginosa exotoxin A was increased by 1839-fold in comparison to their respective monocultures (Goldsworthy, 2008). Moreover, P. aeruginosa exoproduct 4-hydroxy-2-heptylquinoline-N-oxide (HQNO) was found to stimulate S. aureus biofilm and SCV formation, leading to an increase in the expression of the fibronectin-binding protein A and a decrease in the expression of the α-hemolysin gene (Mitchell et al., 2010). It was also demonstrated that a spontaneous SCV of Hp released VacA and urease from the cells, while its parent large colony cells retain these toxins within the cells (Bukholm et al., 1997).
Other studies specific to Hp have attempted to identify differential virulence profiles between coccoid and helical/spiral forms. A proteomic study by Bumann et al. (2004) reported differential virulence profiles, i.e., cagA/vacA dominance in helical and ureA/B and groEL dominance in coccoid forms. The lack of CagA protein expression in coccoid Hp was consistent with an earlier report by Roe et al. (1999). However, a more recent high resolution proteomic study reached a different conclusion (Loke et al., 2016). When compared with helical forms, coccoid forms of Hp were found to express higher levels of proteins that are involved in virulence and carcinogenesis, such as secretion system machinery proteins, CagE, CagV, and YidC and proinflammatory proteins, such as OipA and Hps. Poursina et al. tested the mRNA expression of two Hp virulence genes, babA and cagE in coccoid forms harvested in vitro and concluded that both genes were expressed but in lower rates than those of helical forms. Also, coccoid forms of clinical isolates from peptic ulcer patients showed higher cagE expression than the reference Hp strain 26695, indicating the presence of strain variability (Poursina et al., 2013). More recently, Hirukawa et al. (2018) found that all forms of Hp (helical, coccoid and fragmented) expressed certain pathogenic proteins, including CagA, other components of the cag-Type IV secretion system (VirB7 and VirB9), the blood group antigen-binding adhesin BabA, and UreA, to similar levels. However, phosphorylation of CagA in gastric cancer cells was only seen by the helical form (Hirukawa et al., 2018). In contrast, Segal et al. (1996) reported, using the same cells but a different strain, that coccoid Hp was capable of binding and inducing cellular changes of the same sort as spiral Hp, including tyrosine phosphorylation of host proteins and that coccoid Hp induced a stronger cytoskeletal rearrangement than spiral Hp. Co-culture of a gastric non-cancer epithelial cell line with coccoid or helical forms of Hp revealed that helical Hp led to more severe inflammatory and apoptotic responses, while coccoid Hp maintained host cell in a high proliferation rate (Liu et al., 2006). Interestingly, in animal models, colonization efficiency of Hp mutants in coccoid associated genes, aimA and spoT, was markedly reduced (Sun et al., 2012; Chaput et al., 2016). Despite substantial contrasting findings in the observations, overall, accumulated data suggest that coccoid Hp retains virulence, can survive better in vivo and may contribute to carcinogenesis, while helical Hp can be more easily eliminated by acute host immunoinflammatory responses.
The toxin–antitoxin (TA) systems have emerged as important virulence factors in many pathogenic bacteria, which have been described to be beneficial in bacterial fitness, persistence, and virulence (Kędzierska and Hayes, 2016). Importantly, we recently documented the presence of a novel TA system in Hp encoded by HP0968-HP0967 (Cárdenas-Mondragón et al., 2016), which was strongly expressed in bacteria forming mature biofilm on abiotic surface, but not in planktonic growth. In addition, the expression of this TA was significantly increased after contact with gastric epithelial cell line AGS. These results suggest that HP0968-HP0967 may be expressed in the gastric mucosa and have a role in the gastric carcinogenic pathways.
Potential Role of Polymicrobial Biofilms in Carcinogenesis
Gastric Microbiome of Hp-Infected Humans
Bacterial-bacterial interactions are a major driver of pathophysiological consequences of poly-microbial biofilms, as observed in niches like oral cavity and intestine (Larsen and Fiehn, 2017; Li et al., 2017). It is not known whether this is also the case for the stomach, although there is evidence that supports the presence of such interactions in animal models. Interestingly, in Hp-infected INS-GAS mouse models, gastric intraepithelial neoplasia (GIN) developed much earlier in the presence of commensals, than in mice lacking commensals (Lofgren et al., 2011). A subsequent study confirmed that the presence of only a few other bacterial species had a similar impact to that of full commensals on GIN development (Lertpiriyapong et al., 2014) and that co-infection specifically promoted progression of metaplasia and foveolar hyperplasia to dysplasia (Pinzon-Guzman et al., 2018). However, to date clinical observations or in vitro and in vivo experiments for Hp biofilm studies have been primarily limited to be mono-microbial, exclusively to Hp. There are only a couple of studies that demonstrated the presence of non-Hp bacterial aggregates in the gastric mucus layer and crypts of the patients during acid suppression treatment or with gastric primary lymphoma, using chemical and immunochemical staining (Jonkers et al., 1997; Sanduleanu et al., 2001). These studies reported that almost two thirds of the patients harbored non-Hp bacteria and that co-existence of Hp and non-Hp bacteria was common (Jonkers et al., 1997; Sanduleanu et al., 2001), although the presence of EPS between bacteria was not examined.
Recent studies based on 16S rRNA gene sequencing or other high dimensional approaches to characterize the gastric microbiome have revealed that the gastric lumen is inhabited by a wide range of commensal bacteria, contrary to the previous belief that high gastric acidity kills most microorganisms (Alarcón et al., 2017; Yu et al., 2017). Gastric microbiome comprises commensals from the ororespiratory tract through ingestion, as well as in a smaller fraction from the intestinal tract by biliary reflux (Sanduleanu et al., 2001; Yu et al., 2017). Some of these bacteria are just transient, and data pointing to which are truly resident of the gastric mucosa, other than Hp, are still limited. Multiple studies have found that the dominant genera in the gastric mucosa comprise Streptococcus, Lactobacillus, Rothia, Prevotella, Veillonella, Neisseria, and Haemophilus, including over 100 species (Sheh and Fox, 2013; Alarcón et al., 2017; Yu et al., 2017). There is also a consensus that Hp infection, especially before the development of gastric atrophy, substantially reduces diversity and richness of gastric microbiome by dominating others (Alarcón et al., 2017; Parsons et al., 2017). Yet, these studies raise the possibility that some non-Hp gastric bacteria may cohabit with Hp in biofilms and that the interactions between them may play a role in gastric carcinogenesis. Differential compositions of non-Hp gastric microbiome have been reported between normal individuals, pre-malignant lesions and gastric cancer, although the data have been inconsistent concerning specific bacteria associated with the stage of gastric lesions and it is still unclear whether the observed differences in microbiota are a cause or consequence of carcinogenesis (Sheh and Fox, 2013; Aviles-Jimenez et al., 2014; Dias-Jacome et al., 2016; Alarcón et al., 2017). This could suggest a progressive shift in gastric microbiota structure in carcinogenesis, possibly resulting from a complex cross-talk between gastric microbiota and Hp, which are likely to occur within biofilms where they live in proximity. In fact, abundance of non-Hp bacteria may not be important if they act like a keystone pathogen, as it is well known for Porphyromonas gingivalis in dental biofilms (Hajishengallis et al., 2011; Lamont and Hajishengallis, 2015). Keystone pathogens are typically present at very low abundance in a particular microbial niche; still, they can alter commensal structure to become a dysbiotic community and by attracting more pathogenic microbes (Hajishengallis et al., 2011; Lamont and Hajishengallis, 2015). Such biofilms are thought to promote host inflammatory responses and immune suppression, leading to tissue damage and development of precancerous lesions.
Commensal-Hp Interactions
Potential interactions between Hp and other commensals or pathogens have been investigated mainly in co-culture studies and also by virtual bioinformatic models. Das et al. analyzed 16S rRNA gene pyrosequencing data from 39 Indian patients with suspected Hp infection using a network analyses (Das et al., 2017). Their results suggest that Hp has negative interactions with most members of the gastric microbiota, while other microbes interacted positively with each other, showing frequent intra-cluster co-occurrence/co-operation and increased network density. Major microbes that showed negative interactions with Hp include Ralstonia, Bradyrhizobium, Cloacibacterium, Acidovorax, Aeromonas, Halomonas, Bacillus, Methylobacterium, and Meiothermus. On the other hand, Krausse et al. (2005) examined the effects of other bacteria found in the gastric lumen on the growth of 30 Hp clinical isolates and one reference strain, using cross-streak in vitro culture. Among 29 bacteria tested, Staphylococcus epidermidis, Staphylococcus aureus, Pseudomonas aeruginosa, Stenotrophomonas maltophilia, Morganella morganii, Serratia marcescens, Bf, Fn, and Clostridium difficile showed the strongest growth inhibition, with varied degree on the different Hp strains (Krausse et al., 2005). Other researchers have focused on bacteria more common in oral cavity, Streptococcus and Lactobacillus. When a cagA positive Hp strain was co-cultured with human monocyte-derived dendritic cells (DC) in the presence of Lactobacillus, maturation of DC was stimulated, producing more inflammatory cytokines, compared with Hp alone (Wiese et al., 2015). This suggests that lactic acid producing bacteria may enhance gastric inflammatory reactions caused by Hp and may also promote Hp-induced carcinogenesis. These results were consistent with a gastric microbiome study in humans showing increased abundance of Lactobacillus in Hp-associated intestinal metaplasia and intestinal type of gastric cancer, compared with non-atrophic gastritis (Aviles-Jimenez et al., 2014) as well as the increased gastric Lactobacillus population in INS-GAS mouse model co-infected with Hp and limited commensals (Bacteroides, Clostridium, and Lactobacillus) that developed GIN (Lertpiriyapong et al., 2014). However, others have reported a probiotic Lactobacillus strain that inhibited the colonization of Hp in a Mongolian gerbil model (Merino et al., 2018). More relevant to biofilm-associated Hp, Khosravi et al. (2014) reported that Streptococcus mitis induced Hp conversion to coccoid cells in co-culture studies and their proteomic analysis revealed a metabolic crosstalk between these two bacteria, suggesting a probable impact on Hp-associated carcinogenesis (Khosravi et al., 2016; Krzyżek, 2017). Furthermore, a recent animal experiment demonstrated that INS-GAS mice co-infected with Hp and Streptococcus salivarius developed more severe inflammation, hyperplasia, and dysplasia in the stomach when compared with Hp only at 5 months post-infection. These studies address only a small fraction of possible interactions between Hp and other bacteria, but provide evidence suggesting that bacterial-bacterial interactions may modify Hp-associated carcinogenesis (Shen et al., 2018).
Importantly, biofilms are considered to foster horizontal gene transfer between and within species through transduction and transformation and increase fitness of organisms (Madsen et al., 2012). Mixed infection with multiple Hp strains has been found to produce biofilms with higher adherent capacity compared with each strain alone (Grande et al., 2012). Indeed, Hp exhibits high homologous recombination rates (Suerbaum et al., 1998; Falush et al., 2001) and is competent for DNA uptake (Chattopadhyay et al., 2018) as the genome often contains pathogenicity islands (cagPAI) to encode a DNA transfer apparatus, such as tfs4, to facilitate conjugation (Fischer et al., 2010; Fernandez-Gonzalez and Backert, 2014). Conjugative DNA transfer occurred between Campylobacter jejuni and Hp in an experimental system (Oyarzabal et al., 2007) and inter-species horizontal gene transfer and DNA recombinatorial events have been demonstrated within Hp species or with closely related genera (Eppinger et al., 2004; Saunders et al., 2005). Thus, such heightened genetic exchanges are likely to promote bacterial persistence and virulence, as evidenced by acquisition of cagPAI more than 60,000 years ago (Grande et al., 2012; Fernandez-Gonzalez and Backert, 2014).
Concluding Remarks
As discussed above, emerging evidence suggests that certain low abundance gut bacteria, such as Fn and SGG, which are capable of causing biofilm-associated infection, may promote the development of colorectal cancer in humans. This notion is supported by not only epidemiological studies, but also biological data to corroborate molecular pathways to mediate biofilm formation and bacterial aggregation. On the other hand, despite a substantial number of studies to support the ability of Hp to form biofilms in in vitro environments (Garcia et al., 2014; Hathroubi et al., 2018a), in vivo observations of gastric biofilms from human subjects are still very sparse. Studies based on other anatomic niches, such as gut and oral cavity indicate that behavior of biofilm-associated bacteria is different from that of planktonic counterparts (Gabrilska and Rumbaugh, 2015; Liu et al., 2016) and that bacterial-bacterial and host-bacterial interactions taking place in the biofilm community enhance virulence and adverse host responses, probably contributing to cancer initiation and progression (Li et al., 2017). Accordingly, it is highly plausible that the presence of Hp biofilms predisposes individuals to progression to gastric cancer, compared to infection with planktonic Hp, directly through altered virulence or/and indirectly through prolonged exposure due to persistent infection. However, information available to date is insufficient to test these hypotheses and thus more clinical observations are urgently needed. New histological techniques to preserve mucus layers of gastro-intestinal mucosa may be explored to gain better resolution and quantitation of Hp-biofilms in such studies. Furthermore, we need to accumulate more information regarding characteristics of Hp biofilms including compositions and biological properties of EPS and non-Hp microbiome, as well as geographical distribution of biofilms in relation to gastric gland types and structures and to gastric pathologies (e.g., premalignant lesions). We need also to address the possible effects of Hp morphological changes on host oncogenic signaling pathways. Identification of specific Hp gene variants resulting into enhanced biofilm formation would be helpful not only for screening patients at high risk for sequelae from Hp infection, but also for development of new antibiotic regimens to avoid resistance, regardless of its association with gastric cancer. Given putative auxiliary functions of type IV secretion system pili in the contact and adhesion to host and other bacterial cells (Backert et al., 2008; Maldarelli et al., 2016) and because of well established oncogenic properties of CagA, studies focused on cagA-positive Hp in high risk populations would be particularly advantageous in addressing potential effects of Hp biofilms on gastric carcinogenesis.
Ethics Statement
The original clinical studies where human samples were collected were approved by the respective institutional ethical committee in Mexico and Colombia.
Author Contributions
IK conceptualized the manuscript. JT and MC-P carried out the experiments presented in Figures 1–4. FC, JT, MB, and CR contributed to Hp sequence analyses. IK, EK, JT, MC-P, and CR conducted literature review and prepared the first draft. FC and MB participated in editing the draft. All authors approved the final manuscript for submission.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abadi, A. T. B. (2017). Strategies used by Helicobacter pylori to establish persistent infection. World J. Gastroenterol. 23, 2870–2882. doi: 10.3748/wjg.v23.i16.2870
Alarcón, T., Llorca, L., and Perez-Perez, G. (2017). “Impact of the microbiota and gastric disease development by Helicobacter pylori,” in Molecular Pathogenesis and Signal Transduction by Helicobacter pylori, eds N. Tegtmeyer and S. Backert (Cham: Springer), 253–275. doi: 10.1007/978-3-319-50520-6_11
Allen-Vercoe, E., Strauss, J., and Chadee, K. (2011). Fusobacterium nucleatum. Gut Microbes 2, 294–298. doi: 10.4161/gmic.2.5.18603
Andersen, L. P., and Rasmussen, L. (2009). Helicobacter pylori– coccoid forms and biofilm formation. FEMS Immunol. Med. Microbiol. 56, 112–115. doi: 10.1111/j.1574-695X.2009.00556.x
Anderson, J. K., Huang, J. Y., Wreden, C., Sweeney, E. G., Goers, J., Remington, S. J., et al. (2015). Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal. mBio 6:e00379. doi: 10.1128/mBio.00379-15
Armstrong, H., Bording-Jorgensen, M., Dijk, S., and Wine, E. (2018). The complex interplay between chronic inflammation, the microbiome, and cancer: understanding disease progression and what we can do to prevent it. Cancers 10:E83. doi: 10.3390/cancers10030083
Attaran, B., Falsafi, T., and Ghorbanmehr, N. (2017). Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics. World J. Gastroenterol. 23, 1163–1170. doi: 10.3748/wjg.v23.i7.1163
Attaran, B., Falsafi, T., and Moghaddam, A. (2016). Study of biofilm formation in C57Bl/6J mice by clinical isolates of Helicobacter pylori. Saudi J. Gastroenterol. 22, 161–168. doi: 10.4103/1319-3767.178529
Aviles-Jimenez, F., Vazquez-Jimenez, F., Medrano-Guzman, R., Mantilla, A., and Torres, J. (2014). Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 4:4202. doi: 10.1038/srep04202
Azeredo, J., Azevedo, N. F., Briandet, R., Cerca, N., Coenye, T., Costa, A. R., et al. (2017). Critical review on biofilm methods. Crit. Rev. Microbiol. 43, 313–351. doi: 10.1080/1040841X.2016.1208146
Babu, S. D., Jayanthi, V., Devaraj, N., Reis, C. A., and Devaraj, H. (2006). Expression profile of mucins (MUC2, MUC5AC and MUC6) in Helicobacter pylori infected pre-neoplastic and neoplastic human gastric epithelium. Mol. Cancer 5:10. doi: 10.1186/1476-4598-5-10
Backert, S., Fronzes, R., and Waksman, G. (2008). VirB2 and VirB5 proteins: specialized adhesins in bacterial type-IV secretion systems? Trends Microbiol. 16, 409–413. doi: 10.1016/j.tim.2008.07.001
Bak, G., Lee, J., Suk, S., Kim, D., Young Lee, J., Kim, K.-S., et al. (2015). Identification of novel sRNAs involved in biofilm formation, motility, and fimbriae formation in Escherichia coli. Sci. Rep. 5:15287. doi: 10.1038/srep15287
Bedran, T. B. L., Azelmat, J., Spolidorio, D. P., and Grenier, D. (2013). Fibrinogen-induced Streptococcus mutans biofilm formation and adherence to endothelial cells. BioMed Res. Int. 2013:431465. doi: 10.1155/2013/431465
Belas, R. (2014). Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 22, 517–527. doi: 10.1016/j.tim.2014.05.002
Bellack, N. R., Koehoorn, M. W., MacNab, Y. C., and Morshed, M. G. (2006). A conceptual model of water’s role as a reservoir in Helicobacter pylori transmission: a review of the evidence. Epidemiol. Infect. 134, 439–449. doi: 10.1017/S0950268806006005
Bertram-Drogatz, P. A., Sobek-Klocke, I., Moller, C., Wingbermuhle, D., Beil, W., Sewing, K. F., et al. (1999). Growth characteristics and influence of antibiotics on rough/smooth phenotypic variants of Helicobacter pylori. Eur. J. Clin. Microbiol. Infect. Dis. 18, 490–495. doi: 10.1007/s100960050329
Bharati, B. K., and Chatterji, D. (2013). Quorum sensing and pathogenesis: role of small signalling molecules in bacterial persistence. Curr. Sci. 105, 643–656.
Blair, K. M., Mears, K. S., Taylor, J. A., Fero, J., Jones, L. A., Gafken, P. R., et al. (2018). The Helicobacter pylori cell shape promoting protein Csd5 interacts with the cell wall, MurF, and the bacterial cytoskeleton. Mol. Microbiol. 110, 114–127. doi: 10.1111/mmi.14087
Boleij, A., Roelofs, R., Danne, C., Bellais, S., Dramsi, S., Kato, I., et al. (2012). Selective antibody response to Streptococcus gallolyticus pilus proteins in colorectal cancer patients. Cancer Prev. Res. 5, 260–265. doi: 10.1158/1940-6207.CAPR-11-0321
Boleij, A., Roelofs, R., Schaeps, R. M., Schulin, T., Glaser, P., Swinkels, D. W., et al. (2010). Increased exposure to bacterial antigen RpL7/L12 in early stage colorectal cancer patients. Cancer 116, 4014–4022. doi: 10.1002/cncr.25212
Bukholm, G., Tannæs, T., Nedenskov, P., Esbensen, Y., Grav, H. J., Hovig, T., et al. (1997). Colony variation of Helicobacter pylori: pathogenic potential is correlated to cell wall lipid composition. Scand. J. Gastroenterol. 32, 445–454. doi: 10.3109/00365529709025079
Bumann, D., Habibi, H., Kan, B., Schmid, M., Goosmann, C., Brinkmann, V., et al. (2004). Lack of stage-specific proteins in coccoid Helicobacter pylori cells. Infect. Immun. 72, 6738–6742. doi: 10.1128/IAI.72.11.6738-6742.2004
Burrows, L. L., Urbanic, R. V., and Lam, J. S. (2000). Functional conservation of the polysaccharide biosynthetic protein WbpM and its homologues in Pseudomonas aeruginosa and other medically significant bacteria. Infect. Immun. 68, 931–936. doi: 10.1128/iai.68.2.931-936.2000
Butt, J., Romero-Hernández, B., Pérez-Butt, J., Blot, W. J., Teras, L. R., Visvanathan, K., et al. (2018). Antibody responses to Streptococcus Gallolyticus subspecies Gallolyticus proteins in a large prospective colorectal cancer cohort consortium. Cancer Epidemiol. Biomark. Prev. 27, 1186–1194. doi: 10.1158/1055-9965.EPI-18-0249
Butt, J., Romero-Hernández, B., Pérez-Gómez, B., Willhauck-Fleckenstein, M., Holzinger, D., Martin, V., et al. (2016). Association of Streptococcus gallolyticus subspecies gallolyticus with colorectal cancer: serological evidence. Int. J. Cancer 138, 1670–1679. doi: 10.1002/ijc.29914
Caccamo, P. D., and Brun, Y. V. (2018). The molecular basis of noncanonical bacterial morphology. Trends Microbiol. 26, 191–208. doi: 10.1016/j.tim.2017.09.012
Cárdenas-Mondragón, M. G., Ares, M. A., Panunzi, L. G., Pacheco, S., Camorlinga-Ponce, M., Girón, J. A., et al. (2016). Transcriptional profiling of type II toxin–antitoxin genes of Helicobacter pylori under different environmental conditions: identification of HP0967–HP0968 system. Front. Microbiol. 7:1872. doi: 10.3389/fmicb.2016.01872
Carron, M. A., Tran, V. R., Sugawa, C., and Coticchia, J. M. (2006). Identification of Helicobacter pylori biofilms in human gastric mucosa. J. Gastrointest. Surg. 10, 712–717. doi: 10.1016/j.gassur.2005.10.019
Cellini, L. (2014). Helicobacter pylori: a chameleon-like approach to life. World J. Gastroenterol. 20, 5575–5582. doi: 10.3748/wjg.v20.i19.5575
Cellini, L., Grande, R., Di Campli, E., Traini, T., Di Giulio, M., Lannutti, S. N., et al. (2008). Dynamic colonization of Helicobacter pylori in human gastric mucosa. Scand. J. Gastroenterol. 43, 178–185. doi: 10.1080/00365520701675965
Chan, W. Y., Hui, P. K., Leung, K. M., Chow, J., Kwok, F., and Ng, C. S. (1994). Coccoid forms of Helicobacter pylori in the human stomach. Am. J. Clin. Pathol. 102, 503–507.
Chaput, C., Ecobichon, C., Cayet, N., Girardin, S. E., Werts, C., Guadagnini, S., et al. (2006). Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog. 2:e97. doi: 10.1371/journal.ppat.0020097
Chaput, C., Ecobichon, C., Pouradier, N., Rousselle, J.-C., Namane, A., and Boneca, I. G. (2016). Role of the N-Acetylmuramoyl-l-Alanyl Amidase, AmiA, of Helicobacter pylori in peptidoglycan metabolism, daughter cell separation, and virulence. Microb. Drug Resist. 22, 477–486. doi: 10.1089/mdr.2016.0070
Chattopadhyay, S., Chi, P. B., Minin, V. N., Berg, D. E., and Sokurenko, E. V. (2018). Recombination-independent rapid convergent evolution of the gastric pathogen Helicobacter pylori. BMC Genomics 19:835. doi: 10.1186/s12864-018-5231-7
Chitcholtan, K., Hampton, M. B., and Keenan, J. I. (2008). Outer membrane vesicles enhance the carcinogenic potential of Helicobacter pylori. Carcinogenesis 29, 2400–2405. doi: 10.1093/carcin/bgn218
Cole, S. P., Harwood, J., Lee, R., She, R., and Guiney, D. G. (2004). Characterization of monospecies biofilm formation by Helicobacter pylori. J. Bacteriol. 186, 3124–3132. doi: 10.1128/jb.186.10.3124-3132.2004
Costa, K., Bacher, G., Allmaier, G., Dominguez-Bello, M. G., Engstrand, L., Falk, P., et al. (1999). The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J. Bacteriol. 181, 3710–3715.
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Coticchia, J. M., Sugawa, C., Tran, V. R., Gurrola, J., Kowalski, E., and Carron, M. A. (2006). Presence and density of Helicobacter pylori biofilms in human gastric mucosa in patients with peptic ulcer disease. J. Gastrointest. Surg. 10, 883–889. doi: 10.1016/j.gassur.2005.12.009
Crawford, R. W., Reeve, K. E., and Gunn, J. S. (2010). Flagellated but not hyperfimbriated Salmonella enterica serovar typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J. Bacteriol. 192, 2981–2990. doi: 10.1128/jb.01620-09
Croxen, M. A., Sisson, G., Melano, R., and Hoffman, P. S. (2006). The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J. Bacteriol. 188, 2656–2665. doi: 10.1128/JB.188.7.2656-2665.2006
Danielli, A., and Scarlato, V. (2010). Regulatory circuits in Helicobacter pylori: network motifs and regulators involved in metal-dependent responses. FEMS Microbiol. Rev. 34, 738–752. doi: 10.1111/j.1574-6976.2010.00233.x
Danne, C., Entenza, J. M., Mallet, A., Briandet, R., Debarbouille, M., Nato, F., et al. (2011). Molecular characterization of a Streptococcus gallolyticus genomic island encoding a pilus involved in endocarditis. J. Infect. Dis. 204, 1960–1970. doi: 10.1093/infdis/jir666
Das, A., Pereira, V., Saxena, S., Ghosh, T. S., Anbumani, D., Bag, S., et al. (2017). Gastric microbiome of Indian patients with Helicobacter pylori infection, and their interaction networks. Sci. Rep. 7:15438. doi: 10.1038/s41598-017-15510-6
De la Cruz, M. A., Ares, M. A., von Bargen, K., Panunzi, L. G., Martinez-Cruz, J., Valdez-Salazar, H. A., et al. (2017). Gene expression profiling of transcription factors of Helicobacter pylori under different environmental conditions. Front. Microbiol. 8:615. doi: 10.3389/fmicb.2017.00615
Dejea, C. M., Fathi, P., Craig, J. M., Boleij, A., Taddese, R., Geis, A. L., et al. (2018). Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597. doi: 10.1126/science.aah3648
Dejea, C. M., Wick, E. C., Hechenbleikner, E. M., White, J. R., Mark Welch, J. L., Rossetti, B. J., et al. (2014). Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. U.S.A. 111, 18321–18326. doi: 10.1073/pnas.1406199111
del Castillo, E., Meier, R., Chung, M., Koestler, D. C., Chen, T., Paster, B. J., et al. (2019). The microbiomes of pancreatic and duodenum tissue overlap and are highly subject specific but differ between pancreatic cancer and noncancer subjects. Cancer Epidemiol. Biomark. Prev. 28, 370–383. doi: 10.1158/1055-9965.epi-18-0542
Di Domenico, E. G., Cavallo, I., Pontone, M., Toma, L., and Ensoli, F. (2017). Biofilm producing Salmonella Typhi: chronic colonization and development of gallbladder cancer. Int. J. Mol. Sci. 18:E1887. doi: 10.3390/ijms18091887
Dias-Jacome, E., Libanio, D., Borges-Canha, M., Galaghar, A., and Pimentel-Nunes, P. (2016). Gastric microbiota and carcinogenesis: the role of non-Helicobacter pylori bacteria - A systematic review. Rev. Esp. Enferm. Dig. 108, 530–540. doi: 10.17235/reed.2016.4261/2016
Doherty, N. C., Shen, F., Halliday, N. M., Barrett, D. A., Hardie, K. R., Winzer, K., et al. (2010). In Helicobacter pylori, LuxS is a key enzyme in cysteine provision through a reverse transsulfuration pathway. J. Bacteriol. 192, 1184–1192. doi: 10.1128/JB.01372-09
Donaldson, G. P., Ladinsky, M. S., Yu, K. B., Sanders, J. G., Yoo, B. B., Chou, W.-C., et al. (2018). Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800. doi: 10.1126/science.aaq0926
Drewes, J. L., White, J. R., Dejea, C. M., Fathi, P., Iyadorai, T., Vadivelu, J., et al. (2017). High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 3:34. doi: 10.1038/s41522-017-0040-3
Edwards, A. M., Grossman, T. J., and Rudney, J. D. (2006). Fusobacterium nucleatum transports noninvasive Streptococcus cristatus into human epithelial cells. Infect. Immun. 74, 654–662. doi: 10.1128/IAI.74.1.654-662.2006
Elias, S., and Banin, E. (2012). Multi-species biofilms: living with friendly neighbors. FEMS Microbiol. Rev. 36, 990–1004. doi: 10.1111/j.1574-6976.2012.00325.x
Eppinger, M., Baar, C., Raddatz, G., Huson, D. H., and Schuster, S. C. (2004). Comparative analysis of four Campylobacterales. Nat. Rev. Microbiol. 2, 872–885. doi: 10.1038/nrmicro1024
Esson, D., Mather, A. E., Scanlan, E., Gupta, S., de Vries, S. P. W., Bailey, D., et al. (2016). Genomic variations leading to alterations in cell morphology of Campylobacter spp. Sci. Rep. 6:38303. doi: 10.1038/srep38303
Falush, D., Kraft, C., Taylor, N. S., Correa, P., Fox, J. G., Achtman, M., et al. (2001). Recombination and mutation during long-term gastric colonization by Helicobacter pylor: estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. U.S.A. 98, 15056–15061. doi: 10.1073/pnas.251396098
Fazli, M., Almblad, H., Rybtke, M. L., Givskov, M., Eberl, L., and Tolker-Nielsen, T. (2014). Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ. Microbiol. 16, 1961–1981. doi: 10.1111/1462-2920.12448
Fernandez-Gonzalez, E., and Backert, S. (2014). DNA transfer in the gastric pathogen Helicobacter pylori. J. Gastroenterol. 49, 594–604. doi: 10.1007/s00535-014-0938-y
Fischer, W., Windhager, L., Rohrer, S., Zeiller, M., Karnholz, A., Hoffmann, R., et al. (2010). Strain-specific genes of Helicobacter pylori: genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. 38, 6089–6101. doi: 10.1093/nar/gkq378
Flemer, B., Warren, R. D., Barrett, M. P., Cisek, K., Das, A., Jeffery, I. B., et al. (2018). The oral microbiota in colorectal cancer is distinctive and predictive. Gut 67, 1454–1463. doi: 10.1136/gutjnl-2017-314814
Flemming, H.-C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Folsom, J. P., Richards, L., Pitts, B., Roe, F., Ehrlich, G. D., Parker, A., et al. (2010). Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol. 10:294. doi: 10.1186/1471-2180-10-294
Fulsundar, S., Kulkarni, H. M., Jagannadham, M. V., Nair, R., Keerthi, S., Sant, P., et al. (2015). Molecular characterization of outer membrane vesicles released from Acinetobacter radioresistens and their potential roles in pathogenesis. Microb. Pathog. 8, 12–22. doi: 10.1016/j.micpath.2015.04.005
Gabrilska, R. A., and Rumbaugh, K. P. (2015). Biofilm models of polymicrobial infection. Future Microbiol. 10, 1997–2015. doi: 10.2217/fmb.15.109
Garcia, A., Salas-Jara, M. J., Herrera, C., and Gonzalez, C. (2014). Biofilm and Helicobacter pylori: from environment to human host. World J. Gastroenterol. 20, 5632–5638. doi: 10.3748/wjg.v20.i19.5632
Ge, X., Cai, Y., Chen, Z., Gao, S., Geng, X., Li, Y., et al. (2018). Bifunctional enzyme SpoT is involved in biofilm formation of Helicobacter pylori with multidrug resistance by upregulating efflux pump Hp1174 (gluP). Antimicrob. Agents Chemother. 62:e00957-18. doi: 10.1128/AAC.00957-18
Goldsworthy, M. J. H. (2008). Gene expression of Pseudomonas aeruginosa and MRSA within a catheter-associated urinary tract infection biofilm model. Biosci. Horiz. 1, 28–37. doi: 10.1093/biohorizons/hzn008
Gölz, G., Sharbati, S., Backert, S., and Alter, T. (2012). Quorum sensing dependent phenotypes and their molecular mechanisms in Campylobacterales. Eur. J. Microbiol. Immunol. 2, 50–60. doi: 10.1556/EuJMI.2.2012.1.8
Gonzalez-Escobedo, G., and Gunn, J. S. (2013). Identification of Salmonella enterica serovar typhimurium genes regulated during biofilm formation on cholesterol gallstone surfaces. Infect. Immun. 81, 3770–3780. doi: 10.1128/iai.00647-13
Grande, R., Di Campli, E., Di Bartolomeo, S., Verginelli, F., Di Giulio, M., Baffoni, M., et al. (2012). Helicobacter pylori biofilm: a protective environment for bacterial recombination. J. Appl. Microbiol. 113, 669–676. doi: 10.1111/j.1365-2672.2012.05351.x
Grande, R., Di Marcantonio, M. C., Robuffo, I., Pompilio, A., Celia, C., Di Marzio, L., et al. (2015). Helicobacter pylori ATCC 43629/NCTC 11639 outer membrane vesicles (OMVs) from biofilm and planktonic phase associated with extracellular DNA (eDNA). Front. Microbiol. 6:1369. doi: 10.3389/fmicb.2015.01369
Grivennikov, S. I., Greten, F. R., and Karin, M. (2010). Immunity, inflammation, and cancer. Cell 140, 883–899. doi: 10.1016/j.cell.2010.01.025
Gupta, K. R., Baloni, P., Indi, S. S., and Chatterji, D. (2016). Regulation of Growth, Cell Shape, Cell Division, and Gene Expression by Second Messengers (p)ppGpp and Cyclic Di-GMP in Mycobacterium smegmatis. J. Bacteriol. 198, 1414–1422. doi: 10.1128/jb.00126-16
Hajishengallis, G., Liang, S., Payne, M. A., Hashim, A., Jotwani, R., Eskan, M. A., et al. (2011). A low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and the complement pathway. Cell Host Microbe 10, 497–506. doi: 10.1016/j.chom.2011.10.006
Hall-Stoodley, L., Costerton, J. W., and Stoodley, P. (2004). Bacterial biofilms: from the Natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108. doi: 10.1038/nrmicro821
Hansen, S. K., Rainey, P. B., Haagensen, J. A. J., and Molin, S. (2007). Evolution of species interactions in a biofilm community. Nature 445, 533–536. doi: 10.1038/nature05514
Harshey, R. M. (2003). Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57, 249–273. doi: 10.1146/annurev.micro.57.030502.091014
Hasegawa, Y., Mark Welch, J. L., Rossetti, B. J., and Borisy, G. G. (2017). Preservation of three-dimensional spatial structure in the gut microbiome. PLoS One 12:e0188257. doi: 10.1371/journal.pone.0188257
Hathroubi, S., Servetas, S. L., Windham, I., Merrell, D. S., and Ottemann, K. M. (2018a). Helicobacter pylori biofilm formation and its potential role in pathogenesis. Microbiol. Mol. Biol. Rev. 82:e00001-18. doi: 10.1128/mmbr.00001-18
Hathroubi, S., Zerebinski, J., and Ottemann, K. M. (2018b). Helicobacter pylori biofilm involves a multigene stress-biased response, including a structural role for flagella. mBio 9:e1973-18. doi: 10.1128/mBio.01973-18
He, Y.-W., and Zhang, L.-H. (2008). Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS Microbiol. Rev. 32, 842–857. doi: 10.1111/j.1574-6976.2008.00120.x
Hirukawa, S., Sagara, H., Kaneto, S., Kondo, T., Kiga, K., Sanada, T., et al. (2018). Characterization of morphological conversion of Helicobacter pylori under anaerobic conditions. Microbiol. Immunol. 62, 221–228. doi: 10.1111/1348-0421.12582
Ho, S. B., Shekels, L. L., Toribara, N. W., Kim, Y. S., Lyftogt, C., Cherwitz, D. L., et al. (1995). Mucin gene expression in normal, preneoplastic, and neoplastic human gastric epithelium. Cancer Res. 55, 2681–2690.
Howitt, M. R., Lee, J. Y., Lertsethtakarn, P., Vogelmann, R., Joubert, L.-M., Ottemann, K. M., et al. (2011). ChePep controls Helicobacter pylori infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. mBio 2:e00098-11. doi: 10.1128/mBio.00098-11
Hussan, H., Clinton, S. K., Roberts, K., and Bailey, M. T. (2017). Fusobacterium’s link to colorectal neoplasia sequenced: a systematic review and future insights. World J. Gastroenterol. 23, 8626–8650. doi: 10.3748/wjg.v23.i48.8626
Jakobsen, T., Tolker-Nielsen, T., and Givskov, M. (2017). Bacterial biofilm control by perturbation of bacterial signaling processes. Int. J. Mol. Sci. 18:E1970. doi: 10.3390/ijms18091970
Jans, C., and Boleij, A. (2018). The road to infection: host-microbe interactions defining the pathogenicity of Streptococcus bovis/Streptococcus equinus complex members. Front. Microbiol. 9:603. doi: 10.3389/fmicb.2018.00603
Johansson, M. E. V., Gustafsson, J. K., Holmén-Larsson, J., Jabbar, K. S., Xia, L., Xu, H., et al. (2014). Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 63, 281–291. doi: 10.1136/gutjnl-2012-303207
Johansson, M. E. V., and Hansson, G. C. (2012). “Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH,” in Mucins: Methods and Protocols, eds M. A. McGuckin and D. J. Thornton (Totowa, NJ: Humana Press), 229–235. doi: 10.1007/978-1-61779-513-8_13
Johansson, M. E. V., Phillipson, M., Petersson, J., Velcich, A., Holm, L., and Hansson, G. C. (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 15064–15069. doi: 10.1073/pnas.0803124105
Jonkers, D., Gisbertz, I., De Bruine, A., Bot, F., Arends, J. W., Stobberingh, E., et al. (1997). Helicobacter pylori and non-Helicobacter pylori bacterial flora in gastric mucosal and tumour specimens of patients with primary gastric lymphoma. Eur. J. Clin. Invest. 27, 885–892. doi: 10.1046/j.1365-2362.1997.1940756.x
Kędzierska, B., and Hayes, F. (2016). Emerging roles of toxin-antitoxin modules in bacterial pathogenesis. Molecules 21:E790. doi: 10.3390/molecules21060790
Khosravi, Y., Dieye, Y., Loke, M. F., Goh, K. L., and Vadivelu, J. (2014). Streptococcus mitis induces conversion of Helicobacter pylori to coccoid cells during co-culture in vitro. PLoS One 9:e112214. doi: 10.1371/journal.pone.0112214
Khosravi, Y., Loke, M. F., Goh, K. L., and Vadivelu, J. (2016). Proteomics analysis revealed that crosstalk between Helicobacter pylori and Streptococcus mitis may enhance bacterial survival and reduces carcinogenesis. Front. Microbiol. 7:1462. doi: 10.3389/fmicb.2016.01462
Kim, H. S., Im, H. N., An, D. R., Yoon, J. Y., Jang, J. Y., Mobashery, S., et al. (2015). The cell shape-determining Csd6 protein from Helicobacter pylori constitutes a new family of L,D-Carboxypeptidase. J. Biol. Chem. 290, 25103–25117. doi: 10.1074/jbc.M115.658781
Kim, S. W., Oh, M. H., Jun, S. H., Jeon, H., Kim, S. I., Kim, K., et al. (2016). Outer membrane Protein A plays a role in pathogenesis of Acinetobacter nosocomialis. Virulence 7, 413–426. doi: 10.1080/21505594.2016.1140298
Kinder, S. A., and Holt, S. C. (1993). Localization of the Fusobacterium nucleatum T18 adhesin activity mediating coaggregation with Porphyromonas gingivalis T22. J. Bacteriol. 175, 840–850. doi: 10.1128/jb.175.3.840-850.1993
Kolenbrander, P. E., Andersen, R. N., and Moore, L. V. (1989). Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect. Immun. 57, 3194–3203.
Kostakioti, M., Hadjifrangiskou, M., and Hultgren, S. J. (2013). Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the Postantibiotic Era. Cold Spring Harb. Perspect. Med. 3: a010306. doi: 10.1101/cshperspect.a010306
Krausse, R., Piening, K., and Ullmann, U. (2005). Inhibitory effects of various micro-organisms on the growth of Helicobacter pylori. Lett. Appl. Microbiol. 40, 81–86. doi: 10.1111/j.1472-765X.2004.01632.x
Krzyżek, P. (2017). Commentary: proteomics analysis revealed that crosstalk between Helicobacter pylori and Streptococcus mitis may enhance bacterial survival and reduces carcinogenesis. Front. Microbiol. 8:2381. doi: 10.3389/fmicb.2017.02381
Krzyzek, P., and Gosciniak, G. (2018). A proposed role for diffusible signal factors in the biofilm formation and morphological transformation of Helicobacter pylori. Turk. J. Gastroenterol. 29, 7–13. doi: 10.5152/tjg.2017.17349
Lamont, R. J., and Hajishengallis, G. (2015). Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 21, 172–183. doi: 10.1016/j.molmed.2014.11.004
Larsen, T., and Fiehn, N. E. (2017). Dental biofilm infections - an update. APMIS 125, 376–384. doi: 10.1111/apm.12688
Lertpiriyapong, K., Whary, M. T., Muthupalani, S., Lofgren, J. L., Gamazon, E. R., Feng, Y., et al. (2014). Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut 63, 54–63. doi: 10.1136/gutjnl-2013-305178
Lertsethtakarn, P., Ottemann, K. M., and Hendrixson, D. R. (2011). Motility and chemotaxis in Campylobacter and Helicobacter. Annu. Rev. Microbiol. 65, 389–410. doi: 10.1146/annurev-micro-090110-102908
Lewis, K. (2005). Persister cells and the riddle of biofilm survival. Biochemistry 70, 267–274. doi: 10.1007/s10541-005-0111-6
Li, S., Konstantinov, S. R., Smits, R., and Peppelenbosch, M. P. (2017). Bacterial biofilms in colorectal cancer initiation and progression. Trends Mol. Med. 23, 18–30. doi: 10.1016/j.molmed.2016.11.004
Limoli, D. H., Jones, C. J., and Wozniak, D. J. (2015). Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 3. doi: 10.1128/microbiolspec.MB-0011-2014
Liu, W., Røder, H. L., Madsen, J. S., Bjarnsholt, T., Sørensen, S. J., and Burmølle, M. (2016). Interspecific bacterial interactions are reflected in multispecies biofilm spatial organization. Front. Microbiol. 7:1366. doi: 10.3389/fmicb.2016.01366
Liu, Z.-F., Chen, C.-Y., Tang, W., Zhang, J.-Y., Gong, Y.-Q., and Jia, J.-H. (2006). Gene-expression profiles in gastric epithelial cells stimulated with spiral and coccoid Helicobacter pylori. J. Med. Microbiol. 55, 1009–1015. doi: 10.1099/jmm.0.46456-0
Lofgren, J. L., Whary, M. T., Ge, Z., Muthupalani, S., Taylor, N. S., Mobley, M., et al. (2011). Lack of commensal flora in H. pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology 140, 210.e–220.e. doi: 10.1053/j.gastro.2010.09.048
Loh, J. T., Gupta, S. S., Friedman, D. B., Krezel, A. M., and Cover, T. L. (2010). Analysis of protein expression regulated by the Helicobacter pylori ArsRS two-component signal transduction system. J. Bacteriol. 192, 2034–2043. doi: 10.1128/JB.01703-08
Loke, M. F., Ng, C. G., Vilashni, Y., Lim, J., and Ho, B. (2016). Understanding the dimorphic lifestyles of human gastric pathogen Helicobacter pylori using the SWATH-based proteomics approach. Sci. Rep. 6:26784. doi: 10.1038/srep26784
Madsen, J. S., Burmølle, M., Hansen, L. H., and Sørensen, S. J. (2012). The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 65, 183–195. doi: 10.1111/j.1574-695X.2012.00960.x
Magana, M., Sereti, C., Ioannidis, A., Mitchell, C. A., Ball, A. R., Magiorkinis, E., et al. (2018). Options and limitations in clinical investigation of bacterial biofilms. Clin. Microbiol. Rev. 31:e00084-16. doi: 10.1128/cmr.00084-16
Majumdar, S., and Pal, S. (2017). Bacterial intelligence: imitation games, time-sharing, and long-range quantum coherence. J. Cell Commun. Signal. 11, 281–284. doi: 10.1007/s12079-017-0394-6
Maldarelli, G. A., Piepenbrink, K. H., Scott, A. J., Freiberg, J. A., Song, Y., Achermann, Y., et al. (2016). Type IV pili promote early biofilm formation by Clostridium difficile. Pathog. Dis. 74:ftw061. doi: 10.1093/femspd/ftw061
Martins, M., Porrini, C., du Merle, L., Danne, C., Robbe-Masselot, C., Trieu-Cuot, P., et al. (2016). The Pil3 pilus of Streptococcus gallolyticus binds to intestinal mucins and to fibrinogen AU - Martins, Mariana. Gut Microbes 7, 526–532. doi: 10.1080/19490976.2016.1239677
Matsuda, K., Yamauchi, K., Matsumoto, T., Sano, K., Yamaoka, Y., and Ota, H. (2008). Quantitative analysis of the effect of Helicobacter pylori on the expressions of SOX2, CDX2, MUC2, MUC5AC, MUC6, TFF1, TFF2, and TFF3 mRNAs in human gastric carcinoma cells. Scand. J. Gastroenterol. 43, 25–33. doi: 10.1080/00365520701579795
Matsuo, K., Ota, H., Akamatsu, T., Sugiyama, A., and Katsuyama, T. (1997). Histochemistry of the surface mucous gel layer of the human colon. Gut 40, 782–789. doi: 10.1136/gut.40.6.782
Merino, J. S., Garcia, A., Pastene, E., Salas, A., Saez, K., and Gonzalez, C. L. (2018). Lactobacillus fermentum UCO-979C strongly inhibited Helicobacter pylori SS1 in Meriones unguiculatus. Benef. Microbes 9, 625–627. doi: 10.3920/bm2017.0160
Meyle, E., Brenner-Weiss, G., Obst, U., Prior, B., and Hänsch, G. M. (2012). Immune defense against S. epidermidis biofilms: components of the extracellular polymeric substance activate distinct bactericidal mechanisms of phagocytic cells. Int. J. Artif. Organs 35, 700–712. doi: 10.5301/ijao.5000151
Mika, F., and Hengge, R. (2013). Small regulatory RNAs in the control of motility and biofilm formation in E. coli and Salmonella. Int. J. Mol. Sci. 14:4560. doi: 10.3390/ijms14034560
Mitchell, G., Séguin, D. L., Asselin, A.-E., Déziel, E., Cantin, A. M., Frost, E. H., et al. (2010). Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N- oxide. BMC Microbiol. 10:33. doi: 10.1186/1471-2180-10-33
Mot, R., and Vanderleyden, J. (1994). The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12, 333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x
Muñoz-Ramírez, Z. Y., Mendez-Tenorio, A., Kato, I., Bravo, M. M., Rizzato, C., Thorell, K., et al. (2017). Whole genome sequence and phylogenetic analysis show Helicobacter pylori strains from latin america have followed a unique evolution pathway. Front. Cell. Infect. Microbiol. 7:50. doi: 10.3389/fcimb.2017.00050
Musrati, A. A., Fteita, D., Paranko, J., Könönen, E., and Gürsoy, U. K. (2016). Morphological and functional adaptations of Fusobacterium nucleatum exposed to human neutrophil Peptide-1. Anaerobe 39, 31–38. doi: 10.1016/j.anaerobe.2016.02.008
Nascimento, R., Gouran, H., Chakraborty, S., Gillespie, H. W., Almeida-Souza, H. O., Tu, A., et al. (2016). The type II secreted lipase/esterase LesA is a key virulence factor required for Xylella fastidiosa Pathogenesis in Grapevines. Sci. Rep. 6:18598. doi: 10.1038/srep18598
Niv, Y. (2015). Helicobacter pylori and gastric mucin expression: a systematic review and meta-analysis. World J. Gastroenterol. 21, 9430–9436. doi: 10.3748/wjg.v21.i31.9430
Osaki, T., Hanawa, T., Manzoku, T., Fukuda, M., Kawakami, H., Suzuki, H., et al. (2006). Mutation of luxS affects motility and infectivity of Helicobacter pylori in gastric mucosa of a Mongolian gerbil model. J. Med. Microbiol. 55(Pt 11), 1477–1485. doi: 10.1099/jmm.0.46660-0
Oyarzabal, O. A., Rad, R., and Backert, S. (2007). Conjugative transfer of chromosomally encoded antibiotic resistance from Helicobacter pylori to Campylobacter jejuni. J. Clin. Microbiol. 45, 402–408. doi: 10.1128/jcm.01456-06
Pabst, B., Pitts, B., Lauchnor, E., and Stewart, P. S. (2016). Gel-Entrapped Staphylococcus aureus bacteria as models of biofilm infection exhibit growth in dense aggregates, oxygen limitation, antibiotic tolerance, and heterogeneous gene expression. Antimicrob. Agents Chemother. 60, 6294–6301. doi: 10.1128/aac.01336-16
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Parsons, B. N., Ijaz, U. Z., D’Amore, R., Burkitt, M. D., Eccles, R., Lenzi, L., et al. (2017). Comparison of the human gastric microbiota in hypochlorhydric states arising as a result of Helicobacter pylori-induced atrophic gastritis, autoimmune atrophic gastritis and proton pump inhibitor use. PLoS Pathog. 13:e1006653. doi: 10.1371/journal.ppat.1006653
Percival, S. L., and Suleman, L. (2014). Biofilms and Helicobacter pylori: dissemination and persistence within the environment and host. World J. Gastrointest. Pathophysiol. 5, 122–132. doi: 10.4291/wjgp.v5.i3.122
Persat, A., Nadell, C. D., Kim, M. K., Ingremeau, F., Siryaporn, A., Drescher, K., et al. (2015). The mechanical world of bacteria. Cell 161, 988–997. doi: 10.1016/j.cell.2015.05.005
Pinto, D., Santos, M. A., and Chambel, L. (2015). Thirty years of viable but nonculturable state research: unsolved molecular mechanisms. Crit. Rev. Microbiol. 41, 61–76. doi: 10.3109/1040841X.2013.794127
Pinzon-Guzman, C., Meyer, A. R., Wise, R., Choi, E., Muthupalani, S., Wang, T. C., et al. (2018). Evaluation of lineage changes in the gastric mucosa following infection with Helicobacter pylori and specified intestinal flora in INS-GAS mice. J. Histochem. Cytochem. 67:002215541878562. doi: 10.1369/0022155418785621
Plummer, M., de Martel, C., Vignat, J., Ferlay, J., Bray, F., and Franceschi, S. (2016). Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob. Health 4, e609–e616. doi: 10.1016/S2214-109X(16)30143-7
Poursina, F., Faghri, J., Moghim, S., Zarkesh-Esfahani, H., Nasr-Esfahani, B., Fazeli, H., et al. (2013). Assessment of cagE and babA mRNA expression during morphological conversion of Helicobacter pylori From spiral to coccoid. Curr. Microbiol. 66, 406–413. doi: 10.1007/s00284-012-0280-7
Poursina, F., Fagri, J., Mirzaei, N., and Safaei, H. G. (2018). Overexpression of spoT gene in coccoid forms of clinical Helicobacter pylori isolates. Folia Microbiol. 63, 459–465. doi: 10.1007/s12223-017-0557-0
Prigent-Combaret, C., Vidal, O., Dorel, C., and Lejeune, P. (1999). Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181, 5993–6002.
Proctor, R. A., von Eiff, C., Kahl, B. C., Becker, K., McNamara, P., Herrmann, M., et al. (2006). Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4, 295–305. doi: 10.1038/nrmicro1384
Psakis, G., Saidijam, M., Shibayama, K., Polaczek, J., Bettaney, K. E., Baldwin, J. M., et al. (2009). The sodium-dependent d-glucose transport protein of Helicobacter pylori. Mol. Microbiol. 71, 391–403. doi: 10.1111/j.1365-2958.2008.06535.x
Qaria, M. A., Kumar, N., Hussain, A., Qumar, S., Doddam, S. N., Sepe, L. P., et al. (2018). Roles of cholesteryl-α-glucoside transferase and cholesteryl glucosides in maintenance of Helicobacter pylori morphology, cell wall integrity, and resistance to antibiotics. mBio 9:e01523-18. doi: 10.1128/mBio.01523-18
Queralt, N., and Araujo, R. (2007). Analysis of the survival of H. pylori within a laboratory-based aquatic model system using molecular and classical techniques. Microb. Ecol. 54, 771–777. doi: 10.1007/s00248-007-9242-1
Rader, B. A., Campagna, S. R., Semmelhack, M. F., Bassler, B. L., and Guillemin, K. (2007). The quorum-sensing molecule autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J. Bacteriol. 189, 6109–6117. doi: 10.1128/JB.00246-07
Rader, B. A., Wreden, C., Hicks, K. G., Sweeney, E. G., Ottemann, K. M., and Guillemin, K. (2011). Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology 157, 2445–2455. doi: 10.1099/mic.0.049353-0
Randal, B. R., Everett, M. L., Palestrant, D., Love, S. D., Lin, S. S., and Parker, W. (2003). Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology 109, 580–587. doi: 10.1046/j.1365-2567.2003.01700.x
Reis, C. A., David, L., Correa, P., Carneiro, F., Bolós, C. D., Garcia, E., et al. (1999). Intestinal metaplasia of human stomach displays distinct patterns of mucin (MUC1, MUC2, MUC5AC, and MUC6) expression. Cancer Res. 59, 1003–1007.
Roe, I. H., Son, S. H., Oh, H. T., Choi, J., Shin, J. H., Lee, J. H., et al. (1999). Changes in the evolution of the antigenic profiles and morphology during coccoid conversion of Helicobacter pylori. Korean J. Intern. Med. 14, 9–14. doi: 10.3904/kjim.1999.14.1.9
Römling, U., and Balsalobre, C. (2012). Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 272, 541–561. doi: 10.1111/joim.12004
Rubin, E. J., and Trent, M. S. (2013). Colonize, evade, flourish. Gut Microbes 4, 439–453. doi: 10.4161/gmic.25721
Ryan, R. P., An, S.-Q., Allan, J. H., McCarthy, Y., and Dow, J. M. (2015). The DSF family of cell–cell signals: an expanding class of bacterial virulence regulators. PLoS Pathog. 11:e1004986. doi: 10.1371/journal.ppat.1004986
Ryan, R. P., and Dow, J. M. (2011). Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol. 19, 145–152. doi: 10.1016/j.tim.2010.12.003
Salas-Jara, M. J., Sanhueza, E. A., Retamal-Diaz, A., Gonzalez, C., Urrutia, H., and Garcia, A. (2016). Probiotic Lactobacillus fermentum UCO-979C biofilm formation on AGS and Caco-2 cells and Helicobacter pylori inhibition. Biofouling 32, 1245–1257. doi: 10.1080/08927014.2016.1249367
Sanduleanu, S., Jonkers, D., De Bruine, A., Hameeteman, W., and Stockbrügger, R. W. (2001). Non-Helicobacter pylori bacterial flora during acid-suppressive therapy: differential findings in gastric juice and gastric mucosa. Aliment. Pharmacol. Ther. 15, 379–388. doi: 10.1046/j.1365-2036.2001.00888.x
Saunders, N. J., Boonmee, P., Peden, J. F., and Jarvis, S. A. (2005). Inter-species horizontal transfer resulting in core-genome and niche-adaptive variation within Helicobacter pylori. BMC Genomics 6:9. doi: 10.1186/1471-2164-6-9
Schauder, S., Shokat, K., Surette, M. G., and Bassler, B. L. (2001). The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41, 463–476. doi: 10.1046/j.1365-2958.2001.02532.x
Segal, E. D., Falkow, S., and Tompkins, L. S. (1996). Helicobacter pylori attachment to gastric cells induces cytoskeletal rearrangements and tyrosine phosphorylation of host cell proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 1259–1264. doi: 10.1073/pnas.93.3.1259
Senkovich, O. A., Yin, J., Ekshyyan, V., Conant, C., Traylor, J., Adegboyega, P., et al. (2011). Helicobacter pylori AlpA and AlpB bind host laminin and influence gastric inflammation in gerbils. J. Infect. Immun. 79, 3106–3116. doi: 10.1128/IAI.01275-10
Servetas, S. L., Carpenter, B. M., Haley, K. P., Gilbreath, J. J., Gaddy, J. A., and Merrell, D. S. (2016). Characterization of key Helicobacter pylori regulators identifies a role for ArsRS in biofilm formation. J. Bacteriol. 198, 2536–2548. doi: 10.1128/jb.00324-16
Servetas, S. L., Doster, R. S., Kim, A., Windham, I. H., Cha, J.-H., Gaddy, J. A., et al. (2018). ArsRS-dependent regulation of homB contributes to Helicobacter pylori biofilm formation. Front. Microbiol. 9:1497. doi: 10.3389/fmicb.2018.01497
Sheh, A., and Fox, J. G. (2013). The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes 4, 505–531. doi: 10.4161/gmic.26205
Shen, F., Hobley, L., Doherty, N., Loh, J. T., Cover, T. L., Sockett, R. E., et al. (2010). In Helicobacter pylori auto-inducer-2, but not LuxS/MccAB catalysed reverse transsulphuration, regulates motility through modulation of flagellar gene transcription. BMC Microbiol. 10:210. doi: 10.1186/1471-2180-10-210
Shen, Z., Dzink-Fox, J., Wilson, K. T., Whary, M. T., Muthupalani, S., Piazuelo, M. B., et al. (2018). Tu1288 - co-colonization of Helicobacter Pylori with Staphylococcus Epidermidis or Streptococcus Salivarius differ in the progression of gastritis in ins-gas mice. Gastroenterology 154(6, Suppl. 1), S–924–S–925. doi: 10.1016/S0016-5085(18)33114-7
Sigal, M., Rothenberg, M. E., Logan, C. Y., Lee, J. Y., Honaker, R. W., Cooper, R. L., et al. (2015). Helicobacter pylori activates and expands Lgr5+ stem cells through direct colonization of the gastric glands. Gastroenterology 148, 1392.e–1404.e. doi: 10.1053/j.gastro.2015.02.049
Soto, S. M. (2013). Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 4, 223–229. doi: 10.4161/viru.23724
Stark, R. M., Gerwig, G. J., Pitman, R. S., Potts, L. F., Williams, N. A., Greenman, J., et al. (1999). Biofilm formation by Helicobacter pylori. Lett. Appl. Microbiol. 28, 121–126. doi: 10.1046/j.1365-2672.1999.00481.x
Stewart, P. S. (2015). Antimicrobial tolerance in biofilms. Microbiol. Spectr. 3. doi: 10.1128/microbiolspec.MB-0010-2014
Suerbaum, S., Smith, J. M., Bapumia, K., Morelli, G., Smith, N. H., Kunstmann, E., et al. (1998). Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 95, 12619–12624. doi: 10.1073/pnas.95.21.12619
Sun, J., and Kato, I. (2016). Gut microbiota, inflammation and colorectal cancer. Genes Dis. 3, 130–143. doi: 10.1016/j.gendis.2016.03.004
Sun, Y., Li, X., Li, W., Zhao, M., Wang, L., Liu, S., et al. (2012). ). Proteomic analysis of the function of spot in Helicobacter pylori anti-oxidative stress in vitro and colonization in vivo. J. Cell. Biochem. 113, 3393–3402. doi: 10.1002/jcb.24215
Susanne, M., Philippe, F., Eva, M., Birgit, P., Gertrud, M. H., and Ulrike, D. (2015). Activation of neutrophils by the extracellular polymeric substance of S.epidermidis biofilms is mediated by the bacterial heat shock protein Groel. J. Biotechnol. Biomater. 5:176. doi: 10.4172/2155-952x.1000176
Swidsinski, A., Weber, J., Loening-Baucke, V., Hale, L. P., and Lochs, H. (2005). Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43, 3380–3389. doi: 10.1128/jcm.43.7.3380-3389.2005
Sycuro, L. K., Pincus, Z., Gutierrez, K. D., Biboy, J., Stern, C. A., Vollmer, W., et al. (2010). Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell 141, 822–833. doi: 10.1016/j.cell.2010.03.046
Sycuro, L. K., Rule, C. S., Petersen, T. W., Wyckoff, T. J., Sessler, T., Nagarkar, D. B., et al. (2013). Flow cytometry-based enrichment for cell shape mutants identifies multiple genes that influence Helicobacter pylori morphology. Mol. Microbiol. 90, 869–883. doi: 10.1111/mmi.12405
Sycuro, L. K., Wyckoff, T. J., Biboy, J., Born, P., Pincus, Z., Vollmer, W., et al. (2012). Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog. 8:e1002603. doi: 10.1371/journal.ppat.1002603
Tan, S., Noto, J. M., Romero-Gallo, J., Peek, R. M. Jr., and Amieva, M. R. (2011). Helicobacter pylori perturbs iron trafficking in the epithelium to grow on the cell surface. PLoS Pathog. 7:e1002050. doi: 10.1371/journal.ppat.1002050
Tannæs, T., Dekker, N., Bukholm, G., Bijlsma, J. J. E., and Appelmelk, B. J. (2001). Phase variation in the Helicobacter pylori phospholipase a gene and its role in acid adaptation. Infect. Immun. 69, 7334–7340. doi: 10.1128/iai.69.12.7334-7340.2001
Tomb, J.-F., White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., et al. (1997). The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388, 539–547.
Tsang, J., and Hoover, T. R. (2014). Themes and variations: regulation of RpoN-dependent flagellar genes across diverse bacterial species. Scientifica 2014:681754. doi: 10.1155/2014/681754
Turner, L., Praszkier, J., Hutton, M. L., Steer, D., Ramm, G., Kaparakis-Liaskos, M., et al. (2015). Increased outer membrane vesicle formation in a Helicobacter pylori tolB mutant. Helicobacter 20, 269–283. doi: 10.1111/hel.12196
van Teeseling, M. C. F., de Pedro, M. A., and Cava, F. (2017). Determinants of bacterial morphology: from fundamentals to possibilities for antimicrobial targeting. Front. Microbiol. 8:1264. doi: 10.3389/fmicb.2017.01264
Watters, C., Fleming, D., Bishop, D., and Rumbaugh, K. P. (2016). Host responses to biofilm. Prog. Mol. Biol. Transl. Sci. 142, 193–239. doi: 10.1016/bs.pmbts.2016.05.007
Welch, K., Cai, Y., and Strømme, M. (2012). A method for quantitative determination of biofilm viability. J. Funct. Biomater. 3, 418–431. doi: 10.3390/jfb3020418
Wiese, M., Eljaszewicz, A., Helmin-Basa, A., Andryszczyk, M., Motyl, I., Wieczyska, J., et al. (2015). Lactic acid bacteria strains exert immunostimulatory effect on H. pylori-induced dendritic cells. J. Immunol. Res. 2015:106743. doi: 10.1155/2015/106743
Wilson, C., Lukowicz, R., Merchant, S., Valquier-Flynn, H., Caballero, J., Sandoval, J., et al. (2017). Quantitative and qualitative assessment methods for biofilm growth: a mini-review. Res. Rev. J. Eng. Technol. 6.
Windham, I. H., Servetas, S. L., Whitmire, J. M., Pletzer, D., Hancock, R. E. W., and Merrell, D. S. (2018). Helicobacter pylori biofilm formation is differentially affected by common culture conditions, and proteins play a central role in the biofilm matrix. Appl. Environ. Microbiol. 84:e00391-18. doi: 10.1128/aem.00391-18
Wolska, K. I., Grudniak, A. M., Rudnicka, Z., and Markowska, K. (2016). Genetic control of bacterial biofilms. J. Appl. Genet. 57, 225–238. doi: 10.1007/s13353-015-0309-2
Wong, E. H. J., Ng, C. G., Chua, E. G., Tay, A. C. Y., Peters, F., Marshall, B. J., et al. (2016). Comparative genomics revealed multiple helicobacter pylori genes associated with biofilm formation in vitro. PLoS One 11:e0166835. doi: 10.1371/journal.pone.0166835
Wong, E. H. J., Ng, C. G., Goh, K. L., Vadivelu, J., Ho, B., and Loke, M. F. (2018). Metabolomic analysis of low and high biofilm-forming Helicobacter pylori strains. Sci. Rep. 8:1409. doi: 10.1038/s41598-018-19697-0
Yamashita, S., Igarashi, M., Hayashi, C., Shitara, T., Nomoto, A., Mizote, T., et al. (2015). Identification of self-growth-inhibiting compounds lauric acid and 7-(Z)-tetradecenoic acid from Helicobacter pylori. Microbiology 161, 1231–1239. doi: 10.1099/mic.0.000077
Yang, F.-L., Hassanbhai, A. M., Chen, H.-Y., Huang, Z.-Y., Lin, T.-L., Wu, S.-H., et al. (2011). Proteomannans in biofilm of Helicobacter pylori ATCC 43504. Helicobacter 16, 89–98. doi: 10.1111/j.1523-5378.2010.00815.x
Yonezawa, H., Osaki, T., Fukutomi, T., Hanawa, T., Kurata, S., Zaman, C., et al. (2017). Diversification of the AlpB outer membrane protein of Helicobacter pylori affects biofilm formation and cellular adhesion. J. Bacteriol. 199:e00729-16. doi: 10.1128/JB.00729-16
Yonezawa, H., Osaki, T., Hanawa, T., Kurata, S., Ochiai, K., and Kamiya, S. (2013). Impact of Helicobacter pylori biofilm formation on clarithromycin susceptibility and generation of resistance mutations. PLoS One 8:e73301. doi: 10.1371/journal.pone.0073301
Yonezawa, H., Osaki, T., Kurata, S., Fukuda, M., Kawakami, H., Ochiai, K., et al. (2009). Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol. 9:197. doi: 10.1186/1471-2180-9-197
Yonezawa, H., Osaki, T., Kurata, S., Zaman, C., Hanawa, T., and Kamiya, S. (2010). Assessment of in vitro biofilm formation by Helicobacter pylori. J. Gastroenterol. Hepatol. 25, S90–S94. doi: 10.1111/j.1440-1746.2009.06213.x
Yu, G., Torres, J., Hu, N., Medrano-Guzman, R., Herrera-Goepfert, R., Humphrys, M. S., et al. (2017). Molecular characterization of the human stomach microbiota in gastric cancer patients. Front. Cell. Infect. Microbiol. 7:302. doi: 10.3389/fcimb.2017.00302
Keywords: Helicobacter pylori, biofilm, persistent infection, cancer, virulence
Citation: Rizzato C, Torres J, Kasamatsu E, Camorlinga-Ponce M, Bravo MM, Canzian F and Kato I (2019) Potential Role of Biofilm Formation in the Development of Digestive Tract Cancer With Special Reference to Helicobacter pylori Infection. Front. Microbiol. 10:846. doi: 10.3389/fmicb.2019.00846
Received: 04 January 2019; Accepted: 02 April 2019;
Published: 29 April 2019.
Edited by:
Nezar Al-hebshi, Temple University, United StatesReviewed by:
Rossella Grande, Università degli Studi G. d’Annunzio Chieti e Pescara, ItalyMun Fai Loke, University of Malaya, Malaysia
Copyright © 2019 Rizzato, Torres, Kasamatsu, Camorlinga-Ponce, Bravo, Canzian and Kato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ikuko Kato, a2F0b2lAa2FybWFub3Mub3Jn
 Cosmeri Rizzato
Cosmeri Rizzato Javier Torres
Javier Torres Elena Kasamatsu
Elena Kasamatsu Margarita Camorlinga-Ponce
Margarita Camorlinga-Ponce Maria Mercedes Bravo
Maria Mercedes Bravo Federico Canzian
Federico Canzian Ikuko Kato
Ikuko Kato