- 1Limnological Station, Department of Plant and Microbial Biology, University of Zurich, Zurich, Switzerland
- 2Eawag, Swiss Federal Institute of Aquatic Science and Technology, Dübendorf, Switzerland
- 3Institute of Environmental Engineering, ETH Zurich, Institute of Environmental Engineering, Zurich, Switzerland
Microbial biofilms in gravity-driven membrane (GDM) filtration systems can efficiently degrade the cyanotoxin microcystin (MC), but it is unclear if this function depends on the presence of MC-producing cyanobacteria in the source water habitat. We assessed the removal of MC from added Microcystis aeruginosa biomass in GDMs fed with water from a lake with regular blooms of toxic cyanobacteria (ExpL) or from a stream without such background (ExpS). While initial MC removal was exclusively due to abiotic processes, significantly higher biological MC removal was observed in ExpL. By contrast, there was no difference in MC degradation capacity between lake and stream bacteria in separately conducted liquid enrichments on pure MC. Co-occurrence network analysis revealed a pronounced modularity of the biofilm communities, with a clear hierarchic distinction according to feed water origin and treatment type. Genotypes in the network modules associated with ExpS had significantly more links to each other, indicating that these biofilms had assembled from a more coherent source community. In turn, signals for stochastic community assembly were stronger in ExpL biofilms. We propose that the less “tightly knit” ExpL biofilm assemblages allowed for the better establishment of facultatively MC degrading bacteria, and thus for higher overall functional efficiency.
Introduction
Harmful cyanobacterial blooms (cyanoHABs) are increasingly threatening water quality in freshwater and coastal marine ecosystems (Huisman et al., 2018; Paerl, 2018). Some cyanobacterial strains, e.g., from the freshwater genera Microcystis and Planktothrix (Harke et al., 2016; Kurmayer et al., 2016), produce toxic intracellular secondary metabolites, such as microcystins (MCs). These compounds are a particular concern both in drinking and recreational water systems due to their acute and chronic toxicity (Azevedo et al., 2002; Greer et al., 2018). This has led WHO to establish a guideline value of 1 μg L-1 of the ubiquitous MC variant MC-LR in drinking water (WHO, 1998).
A number of studies have reported biological MC degradation in the water column (Zhang et al., 2017; Krishnan et al., 2018; Thees et al., 2018; Yang et al., 2018), in biofilms (Babica et al., 2005; Kohler et al., 2014), and in sediments (Holst et al., 2003; Chen X. et al., 2010) of freshwater habitats, and MC degrading bacterial strains from various phylogenetic lineages have been isolated (Rapala et al., 2005; Amé et al., 2006; Manage et al., 2009; Chen J. et al., 2010). One aerobic pathway of MC-LR degradation mediated by the mlr gene cluster has been identified in Sphingomonas sp. (Bourne et al., 1996). This gene encodes for three enzymes (MlrA, MlrB, and MlrC) that are responsible for the degradation process: first the cyclic peptide is linearized by cleavage of the Adda-Arg bond, and subsequently the MlrB and MlrC enzymes hydrolyze the linear peptide into smaller ones (Bourne et al., 2001; Dziga et al., 2013). Additional degradation pathways have been suggested based on the discovery of novel intermediates and degradation semi-products (Amé et al., 2006; Edwards et al., 2008; Hashimoto et al., 2009; Zhang et al., 2010; Dziga et al., 2017). Moreover, other, hypothetical mechanisms for MC degradation have also been proposed, e.g., alkaline proteases or glutathione S-transferases (Takenaka and Watanabe, 1997; Mou et al., 2013), and the observation of MC degradation under anoxic conditions (Holst et al., 2003; Chen and Chen, 2010; Chen X. et al., 2010) strongly speaks for alternative metabolic strategies. Currently, it is not clear if and to which extent the known MC degrading strains are responsible for this process in complex microbial assemblages.
Physical and/or chemical treatments (e.g., reverse osmosis, chlorine, and ozone) are effective for removal of MCs, and several are regularly used in drinking water treatment facilities (Kumar et al., 2018). In addition, efforts have been made to develop alternative strategies for MC removal, e.g., the heterologous expression of the MlrA enzyme (Wang et al., 2017; Dexter et al., 2018), bioreactors with immobilized Escherichia coli expressing the MlrA enzyme (Dziga et al., 2014), or inoculated with MC degrading bacteria (Tsuji et al., 2006; Phujomjai et al., 2016; Kumar et al., 2019). Moreover, Dziga et al. (2018) have successfully combined a treatment with hydrogen peroxide, MC degrading bacteria, and a recombinant MlrA enzyme to accomplish complete MC removal.
Gravity-driven membrane (GDM) filtration systems are a robust, low-maintenance technology for the small-scale production of drinking water in communities without access to other types of water treatment (Peter-Varbanets et al., 2010; Pronk et al., 2019). After the initial reduction in flux due to biofilm formation on the membranes, a nearly constant and predictable amount of drinking water proportional to total membrane surface area can be produced by GDM systems over extended periods of time (Peter-Varbanets et al., 2017). Moreover, the microbial biofilms have distinct beneficial effects, e.g., they decrease the level of assimilable organic carbon in the filtrates, thereby delaying microbial regrowth (Derlon et al., 2014; Chomiak et al., 2015). Mature GDM biofilms can also be an effective means of removing MCs from toxic cyanobacteria in the feed water (Kohler et al., 2014). This degradation process could be rapidly induced even without extended prior exposure to the toxin, indicating that the original establishment of MC degraders in the biofilm communities was unrelated to this specific metabolic trait (Silva et al., 2018).
The aquatic system that provided the feed water for the above described GDM experiments (Lake Zurich) has a history of mass development of MC-containing cyanobacteria (Posch et al., 2012). Numerous MC degrading bacteria have been isolated from other aquatic habitats with regular cyanoHABs, such as Lake Taihu (Chen J. et al., 2010; Jiang et al., 2011; Yang et al., 2014; Zhang et al., 2017). Thus, it is likely that MC degraders were also present in the microbial assemblages of Lake Zurich that served as the inoculum for the GDM biofilm communities. MC removal possibly also occurs in GDM biofilm communities from source assemblages without prior exposure to toxic cyanobacteria. Numerous examples show the degradation of complex organic compounds by microbes enriched from diverse source communities at appropriate conditions (e.g., Cortes-Tolalpa et al., 2016), adding authority to Baas-Becking’s classical postulate that “everything is everywhere, but, the environment selects” (Baas Becking, 1934). However, while a particular metabolic activity such as biological MC removal might in principle occur in GDM biofilms produced from a wide range of microbial source communities, it is conceivable that this function will be carried out more or less efficiently depending on whether toxic cyanobacteria are present in the system that provides the feed water. Moreover, contrasting enrichment scenarios (i.e., on biofilms or in suspension) might amplify or diminish potential differences in MC degradation between such assemblages. Many planktonic habitats (and enrichment strategies for planktonic microbes) are characterized by turbulent mixing processes (Wüest and Lorke, 2003). By contrast, microbial biofilms are systems with steep vertical gradients and diffusion limitation (Stewart, 2003; Flemming et al., 2016). Microbes may drastically change their overall physiological state when alternating between their free-living and surface-attached life style (Costerton et al., 1995), and the biotic interactions within biofilms are arguably more complex than in the plankton (Moons et al., 2009), likely leading to contrasting patterns of competitive exclusion.
We tested if GDM biofilms fed with source water from a habitat with regular cyanoHABs would result in higher MC removal efficiency than from a habitat without such background. In addition, we hypothesized that potential differences might not merely be related to the presence or absence of MC-degrading bacteria in the source water, but might instead be rooted in community-related aspects of the biofilms. For this purpose, we studied two types of microbial consortia generated with water from Lake Zurich and from a stream without recorded occurrence of toxic cyanobacteria. On the one hand, we established biofilms within GDM filtration systems fed with water from the lake and stream, respectively. Second, we produced planktonic enrichments with pure MC at aerobic and anaerobic conditions.
Experimental Procedures
Gravity-Driven Membrane (GDM) Filtration Experiments
Two GDM experiments with comparable setups were conducted at two sites. The lake experiment (ExpL) was fed with water from Lake Zurich; it was conducted at the Limnological Station of the University of Zurich (location: N 47°19’ 13.24”, E 8°33’ 11.86”). The stream experiment (ExpS) was fed with water from the Chriesbach stream at the Swiss Federal Institute of Aquatic Science and Technology (N 47°24’ 16.30”, E 8°36’ 31.80”). Experiments were conducted in February/March 2017 over a period of 30 days.
Both GDM systems were assembled in a comparable manner (Figure 1), with small differences due to local circumstances. ExpL featured three sedimentation tanks connected to a single 40 L feed tank (Figure 1A), while ExpS had one sedimentation tank that was connected to 11 individual feed tanks (Figure 1B). In ExpL, the lake water was directly pumped into the first sedimentation tank from 5 m depth, while in ExpS, the stream water was pumped from the surface of the Chriesbach stream. In both cases, the feed tanks had an overflow through which excess water could leave the system. For both experiments, the feed tank was connected to the biofouling monitors and placed 65 cm above the monitors to set the transmembrane pressure. Each biofouling monitor was equipped with a 150 kDa polyethersulfone ultrafiltration (UF) membrane (Microdyn Nadir, Wiesbaden, Germany). After passage through the UF membrane, the water was collected in permeate collection bottle. Syringe connections were assembled right above the influx of the biofouling monitor in order to supply the respective biomass without direct contact to the feed water (Figure 1). Before use, the UF membranes were soaked in 40% ethanol for 1.5 h and then rinsed overnight with sterile deionized water (Heffernan et al., 2013). The system between the UF membranes and the permeate bottles was sterilized; however, the bottles were replaced by new ones every 24 h in a non-sterile environment. The temperature in the feed water tank of both experiments was maintained at 20°C. Except for the permeate bottles, the whole system was kept in the dark. In both experiments, the transmembrane flux and MC removal were monitored every 24 h for a period of 30 days. Transmembrane flux was calculated from the water volume in the permeate bottles and the membrane area (19.95 cm2). Portions of 1 mL were taken daily from the permeate bottles and MC was quantified in these samples by HPLC-MS (see below). At the end of the experimental period, the biofilm thickness and structure was evaluated using optical coherence tomography (OCT) (model 930 nm Spectral Domain, Thorlabs GmbH, Germany), and mean thickness was determined by Matlab (MathWorks, United States) according to Derlon et al. (2013). Subsequently, the biofilms were collected and conserved at -20°C for later DNA extraction.
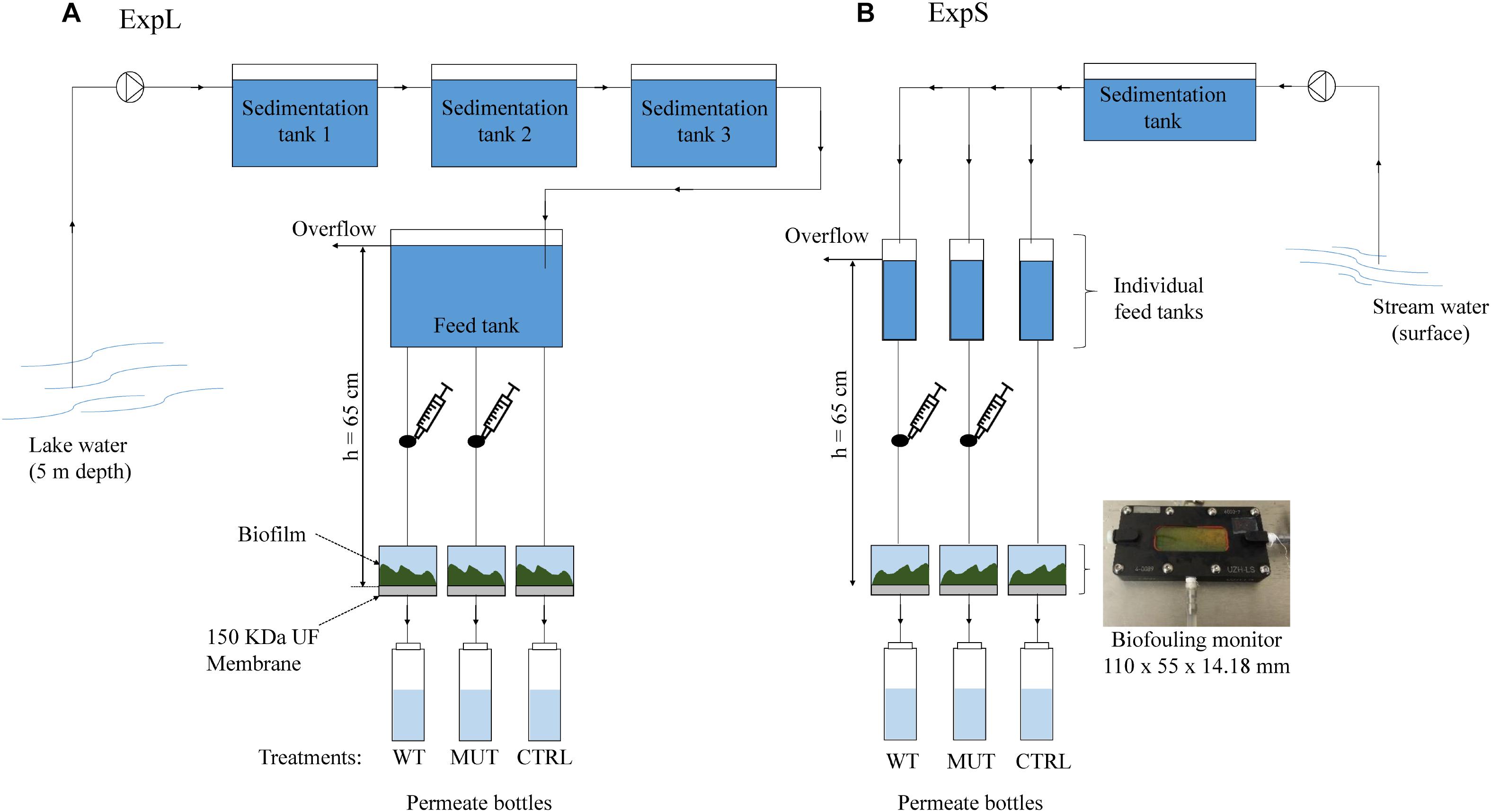
Figure 1. Experimental setup of (A) the lake experiment – ExpL and (B) the stream experiment – ExpS. Each experiment consisted of three treatments: WT – addition of M. aeruginosa biomass containing 1.7 μg MC mg-1 DW day-1; MUT – addition of biomass from the mutant M. aeruginosa strain that does not contain any MC (17 mg DW day-1); and CTRL – only the respective feed water. Each treatment had three biological replicates, except for the WT biomass treatment of ExpS which had five biological replicates. The syringe connection between the feed tank and the biofouling monitor was placed in order to add the correspondent biomass for the WT and MUT biomass treatment.
Each GDM experiment comprised three different treatments: the unamended treatment (CTRL) that only received water from the respective site, the wild-type treatment (WT biomass) that was additionally supplied with MC containing biomass from an axenic strain of Microcystis aeruginosa (see below), and the mutant treatment (MUT biomass) that was supplied with the same amount of MC-free biomass from an axenic mutant strain that does not produce MC (Dittmann et al., 1997). Both, the WT and MUT biomass treatments were supplied with the respective biomass (17 mg day-1 of dry cell weight [DW]) every 24 h. The WT biomass treatment received a MC (MC-LR and [D-Asp3] MC-LR) dose of 1.7 μg MC mg-1 DW day-1. ExpL included three replicates of each treatment (WT biomass, MUT biomass, and CTRL), while ExpS had five replicates for the WT biomass treatment and three replicates for the other two treatments (MUT biomass and CTRL). In ExpL, the conditions of the MUT biomass treatment were changed after day 18 in order to reproduce a previous observation about the MC degradation potential of such biofilms (Silva et al., 2018): the addition of MC-free biomass was replaced by addition of MC-containing biomass. This treatment was subsequently termed MUT+WT biomass.
MC Retention Test
An additional GDM experiment was conducted with the purpose to understand to which extent the apparent initial MC removal in the GDM systems was caused by physicochemical retention on the membrane or adsorption to particles. A GDM system with three treatments (each with six replicates) was set up as described above, albeit using sterile bottles (10 L) in place of a feed tank. These bottles were filled (i) with sterile deionized water, (ii) with pasteurized lake water (80°C, 7 min), and (iii) with raw lake water. Each treatment (sterile, pasteurized, and raw water) was initially supplied with MC-containing biomass from M. aeruginosa (1.7 μg MC mg-1 DW day-1). After 22 h, the amount of MC that had passed through the membrane into the permeate bottle was quantified via HPLC-MS (see below). The proportion of MC retained on the membrane was calculated from the difference between the total MC input and the MC recovered in the permeate bottle.
Production of Cyanobacterial Biomass and MC Quantification
The cultures of wild type M. aeruginosa PCC7806 and a MC-deficient mutant strain (Dittmann et al., 1997) that were used to supplement the GDM biofilms were grown axenically in a medium developed for cyanobacteria cultivation (Jüttner et al., 1983) without addition of chloramphenicol, at constant temperature and light (20°C, 3.5 μmol quanta m-2 s-1). Axenic growth was repeatedly verified by microscopic inspection of the cultures after staining with 4′,6-diamidino-2-phenylindole (DAPI). The wild type strain produces two MC variants, MC-LR and [D-Asp3] MC-LR. The strains were inoculated into replicate 100 mL culture flasks in order to produce sufficient material for the whole experiment, i.e., a total of 4 and 3 L for the wild type and mutant strains, respectively. Biomass was harvested from dense cultures (after ca. 3 weeks of growth) of the two strains, concentrated by centrifugation and then frozen at -20°C in 50 mL aliquots. Three freeze-thaw cycles were performed to destroy cyanobacterial cells and to release the MC from the wild type strain. A subsample was taken to determine dry cell weight and the rest was reserved to supplement the GDM systems. MC concentration in the biomass of the wild type strain was quantified by HPLC-MS, which also served to confirm the absence of MC in biomass from the mutant strain.
Microcystin was quantified as described previously (Silva et al., 2018). Briefly, samples were first filtered (0.2 μm polyethersulfone filter) and then diluted with pure methanol (final concentration 70%). An HPLC 1260 Infinity series system (Agilent Technologies) interfaced with an API 500 triple quadrupole mass spectrometry system (AB Sciex) were used to separate and detect MC. The limit of detection was 0.5 μg L-1 and the limit of quantification 1 μg L-1 for both MC (MC-LR and [D-Asp3] MC-LR). The software Analyst (version 1.6.1, AB Sciex) and MultiQuant (version 2.1, AB Sciex) were used for data acquisition and the quantification of two MC variants: MC-LR (m/z 995.5) and [D-Asp3] MC-LR (m/z 981.5), respectively.
Enrichments Experiment With Pure MC
Enrichments of lake and stream water samples with pure MC-LR were performed to independently investigate the occurrence of (planktonic) MC degraders in both habitats and to obtain additional information about their physiology. Serum bottles (100 mL) were filled either with water from Lake Zurich or from the Chriesbach stream. Three of the six replicate bottles per site were maintained under aerobic and anaerobic conditions, respectively. The aerobic incubations were continuously supplied with sterile (0.2 μm filtered) air. The anaerobic enrichments were maintained inside an anaerobic bench (Anaerobic System Typ 1029, Brouwer AG, Switzerland). The bottles were then spiked with pure MC-LR (purity ≥ 95%, Cayman Chemical, United States) achieving a final concentration of 20 μg L-1 and incubated in the dark at a constant temperature of 20°C. Samples were collected regularly over a period of 15 days and MC-LR was quantified via HPLC-MS as described above. At the end of the experiment, the enriched biomass (altogether ca. 80 mL) was collected and preserved at -20°C for DNA extraction.
DNA Extraction, 16S rRNA Gene Sequencing, and Taxonomic Assignment
DNA was extracted at the end of each experiment (GDM: day 30, enrichments: day 15) from 20 whole membranes of the GDM biofilms and from the remaining volume (∼80 mL) of 12 samples of the enrichment experiment using the DNeasy PowerBiofilm Kit (Qiagen, Germany). The extraction was largely performed according to the manufacturer’s specifications, except for the inhibitors removal step that was extended to 1 hour. The purified DNA was stored at -20°C in 10 mM Tris buffer for further analysis. DNA was amplified with the primer pair 799F-1115R that exclude chloroplasts/cyanobacteria (Chelius and Triplett, 2001; Redford et al., 2010); the reverse primer was modified according to Silva et al. (2018). Partial 16S rRNA gene sequences were obtained using an Illumina MiSeq platform (LGC Genomics, Germany). Raw data processing, the definition of operational taxonomic units (OTUs), and their taxonomic assignment were performed by an in-house pipeline as described previously (Silva et al., 2018). Raw sequencing data have been deposited on the Sequence Read Archive of NCBI under the project number PRJNA529182.
Statistical Analyses
All statistical tests were performed in R (version 3.3.2). Prior to the analyses of the sequencing data, two of the five replicates of the WT treatment from ExpS were randomly excluded to obtain an equal number of replicates for all treatments. The remaining data set was normalized to the sample with the lowest total number of sequence reads as described previously (Silva et al., 2018).
The samples from the GDM experiment were average linkage clustered based on Bray–Curtis dissimilarity scores. The stability of the main branches was tested by cluster-wise stability tests (1000 bootstrap interactions), and significant differences between groups were discovered using similarity profile analyses (α = 0.001). For this, the R packages vegan, clustsig, and fpc were used (Oksanen et al., 2011; Whitaker and Christman, 2014; Hennig, 2015).
For a co-occurrence network of OTUs with the 18 samples of the GDM experiments (ExpL and ExpS), Spearman’s rank correlations were calculated between pairs of OTUs that were present in at least three samples. The p-values were adjusted with the Benjamini–Hochberg method for multiple testing correction (Hochberg and Benjamini, 1995). These calculations were done using the Hmisc R package (Harrell, 2016). Only OTU pairs with statistically significant (p < 0.01) and robust Spearman’s correlation (ρ ≥ 0.6 or ρ ≤-0.6) were considered as a valid positive or negative co-occurrence event. The data were exported in GML format file using the R package igraph (Csardi and Nepusz, 2006). Visualization and analysis (modularity, diameter, average path length, density, and betweenness centrality) were done using the program Gephi (Brandes, 2001; Blondel et al., 2008; Bastian et al., 2009).
Differences between data sets were tested for significance by Student’s t-tests or analyses of variance (ANOVA) followed by Tukey’s honestly significant difference (HSD) test. All proportional data were logit transformed (Warton and Hui, 2011). We tested if there were differences (i) in MC retention/removal between ExpL and ExpS during the first and last week of the experiments, as well as over the complete experimental period (repeated measures ANOVA) (Figures 2B,C); (ii) in MC retention and permeate flux between the treatments raw lake water, pasteurized lake water, and deionized water (Figure 3); (iii) in the degree number (i.e., the number of links between individual OTUs or nodes) between the 5 modules of the co-occurrence network (Figure 5C); (iv) in the proportions of OTUs that occurred at one or both experimental sites and/or treatments; and (v) in the proportions of OTUs that occurred in only one or in all three biological replicates of a particular treatment (Table 2).
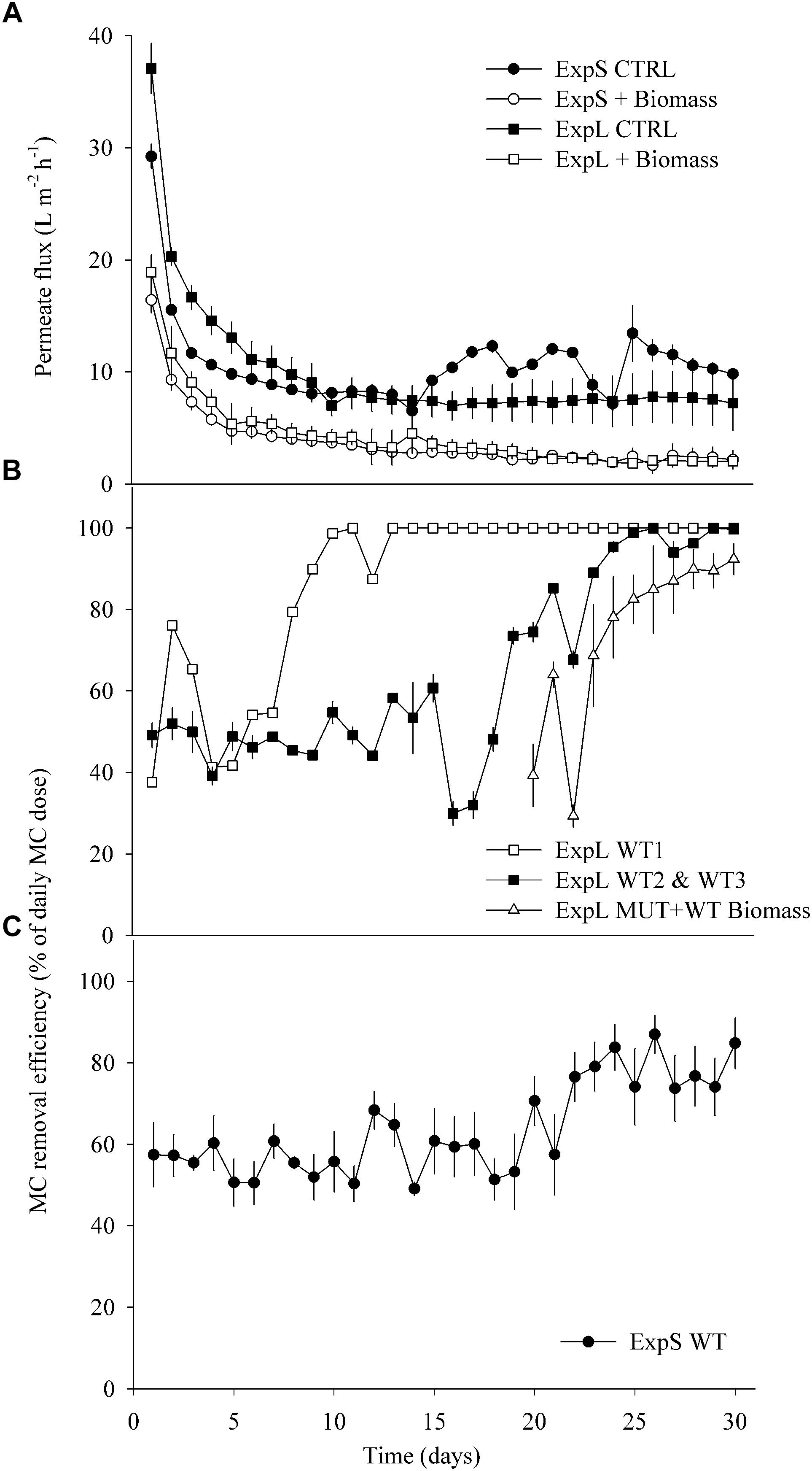
Figure 2. (A) Average permeate flux in ExpL and ExpS. The label “+Biomass” treatment refers to the average of the WT biomass and MUT (+WT biomass) treatments and the label “CTRL” to the treatments that received only lake or stream water. (B) Microcystin removal efficiency in ExpL. Note that the MUT+WT biomass treatment was supplemented with MC-free biomass until day 18 and with MC-containing biomass thereafter. WT 1, 2, and 3 refer to the three biological replicates of the WT treatment. (C) Microcystin removal efficiency in ExpS. Data are depicted as means ± one standard deviation.
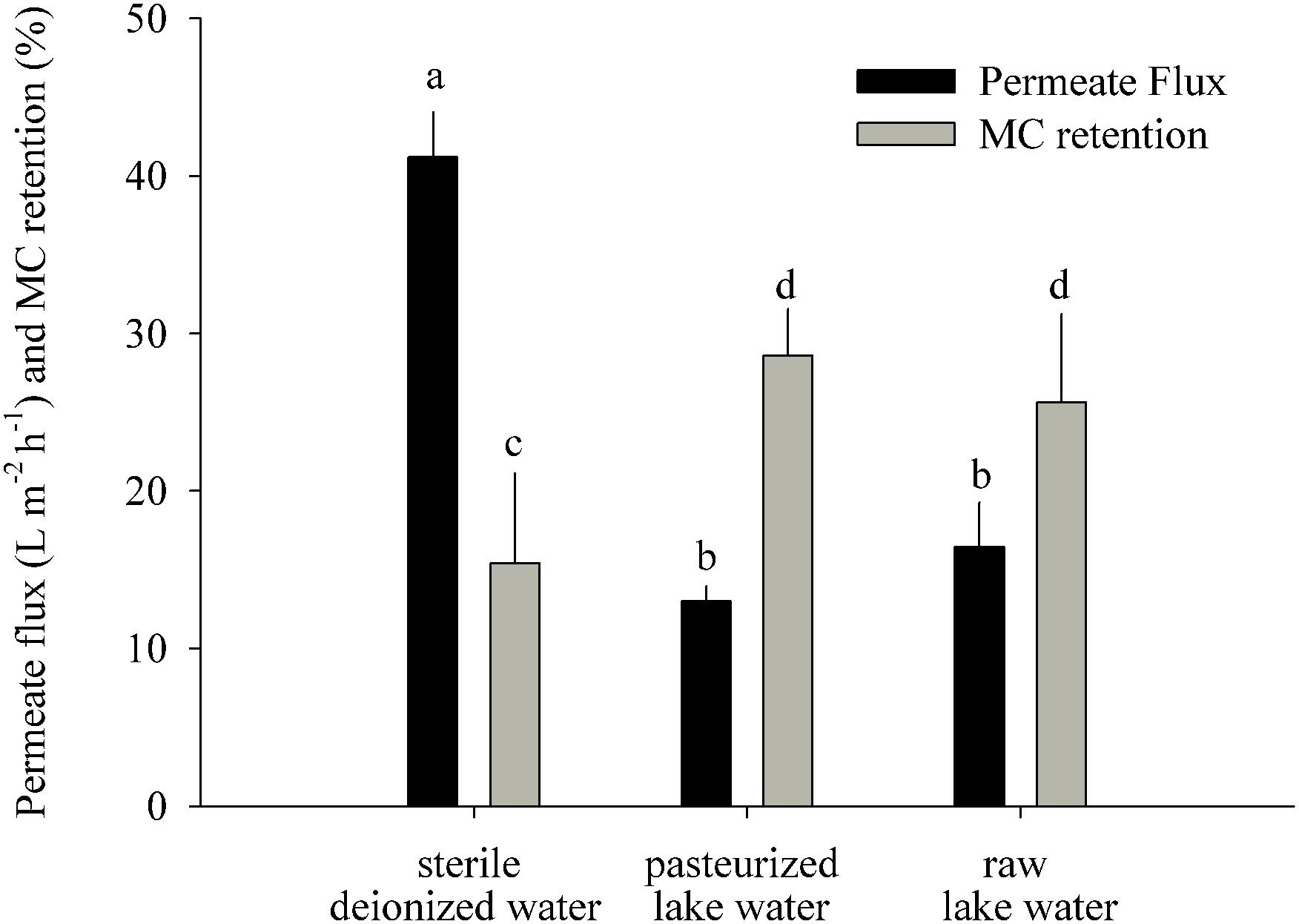
Figure 3. MC retention test: permeate flux and MC retention measured 22 h after addition of MC-containing biomass to the GDM system (means ± one standard deviation). Letters denote treatments that are significantly different for the permeate flux and for the MC retention, respectively (ANOVA followed by Tukey’s HSD tests, p < 0.001–0.05).
Results
GDM Biofilm Properties
The four biomass-amended treatments of both experiments (ExpL and ExpS) behaved similarly in terms of flux decline, by approximately 70% during the first week of operation (Figure 2A). Afterward, all the four biomass-amended treatments stabilized around 2.8 ± 1 L m-2 h-1. In ExpL, the CTRL treatment stabilized at 7.5 ± 0.3 L m-2 h-1, after 80% of flux decline over the first 9 days of operation. In ExpS, this treatment had a small period of stable flux with 8 ± 0.3 L m-2 h-1, but it exhibited irregularities after 13 days of operation, varying between 6 and 13 L m-2 h-1 (Figure 2A).
At the end of the experiment, the biofilm thickness of the CTRL treatments was on average 52 ± 7 and 105 ± 19 μm for ExpL and ExpS, respectively. For the biomass-amended treatments, it was not possible to exactly determine the biofilm thickness because in many replicates the biofouling monitor was completely filled with biofilm/fouling cake (∼1 mm thickness). In terms of horizontal structure, the CTRL biofilms of ExpS exhibited higher heterogeneity compared with ExpL (Supplementary Figure S1).
In ExpL, one of the three replicates of the WT treatment reached 100% of MC removal efficiency after 10 days of operation, while the other two replicates required 25 days until they completely removed the newly added MC (Figure 2B). As observed previously (Silva et al., 2018), the addition of toxic biomass to the MUT treatment led to a rapid upshift of MC removal in these biofilms, to 69 ± 16% after 4 days and to 92 ± 6% after 10 days. The MC removal in each biological replicate of ExpS was generally rather unstable (data not shown). However, the average of all replicates resulted in a reliable MC removal efficiency (Figure 2C): it remained unaltered around 57 ± 6% during the first 20 days of operation, subsequently increased by approximately 20% and fluctuated between 58 ± 10 and 85 ± 6% until the end of the experiment. During the first week of GDM operation, MC removal was significantly lower in ExpL than in ExpS, and significantly higher during the last week, as indicated by both, Repeated Measures ANOVA and Student’s t-tests (n = 7, p < 0.05). MC removal over the entire period was significantly higher in ExpL (Repeated Measures ANOVA, n = 30, p < 0.001).
Microcystin Retention on the GDM Membrane
This experiment was performed to understand how much of the initial MC removal was caused by its abiotic retention on the membrane (Figure 3). The most likely physicochemical processes involved in initial MC retention were the adsorption of MC to the membrane surface and to particles introduced by the influent water and captured on the membrane. The sterile water treatment had a significantly higher permeate flux (41 ± 3 L m-2 h-1) than the pasteurized (13 ± 1 L m-2 h-1) and raw lake water (16 ± 3 L m-2 h-1) treatments (Figure 3) (ANOVA, n = 6, p < 0.001) and significantly lower MC retention (15 ± 6%) than the pasteurized (29 ± 3%) and raw water (26 ± 5%) treatments (ANOVA, n = 6, p < 0.05).
Community Diversity of GDM Lake and Stream Biofilms
After normalization to the smallest sample (4904 reads per sample), the sequence data set comprised 1941 OTUs. Proteobacteria constituted 62% of all reads (52% of OTUs, Supplementary Table S1), followed by Bacteroidetes (11 and 14% of reads and OTUs, respectively), Actinobacteria (7 and 6%), and Firmicutes (2.4 and 4%). Several other phyla also formed > 1% of total reads (Parcubacteria, Acidobacteria, Chloroflexi, Spirochaetae, Deinococcus, Fibrobacteres, and Chlamydiae). There was a significant difference in the relative abundances (read numbers) of the different phyla between sites (paired t-test, n = 13, p < 0.01), but not between treatments (Supplementary Table S1).
Differences Between Treatments
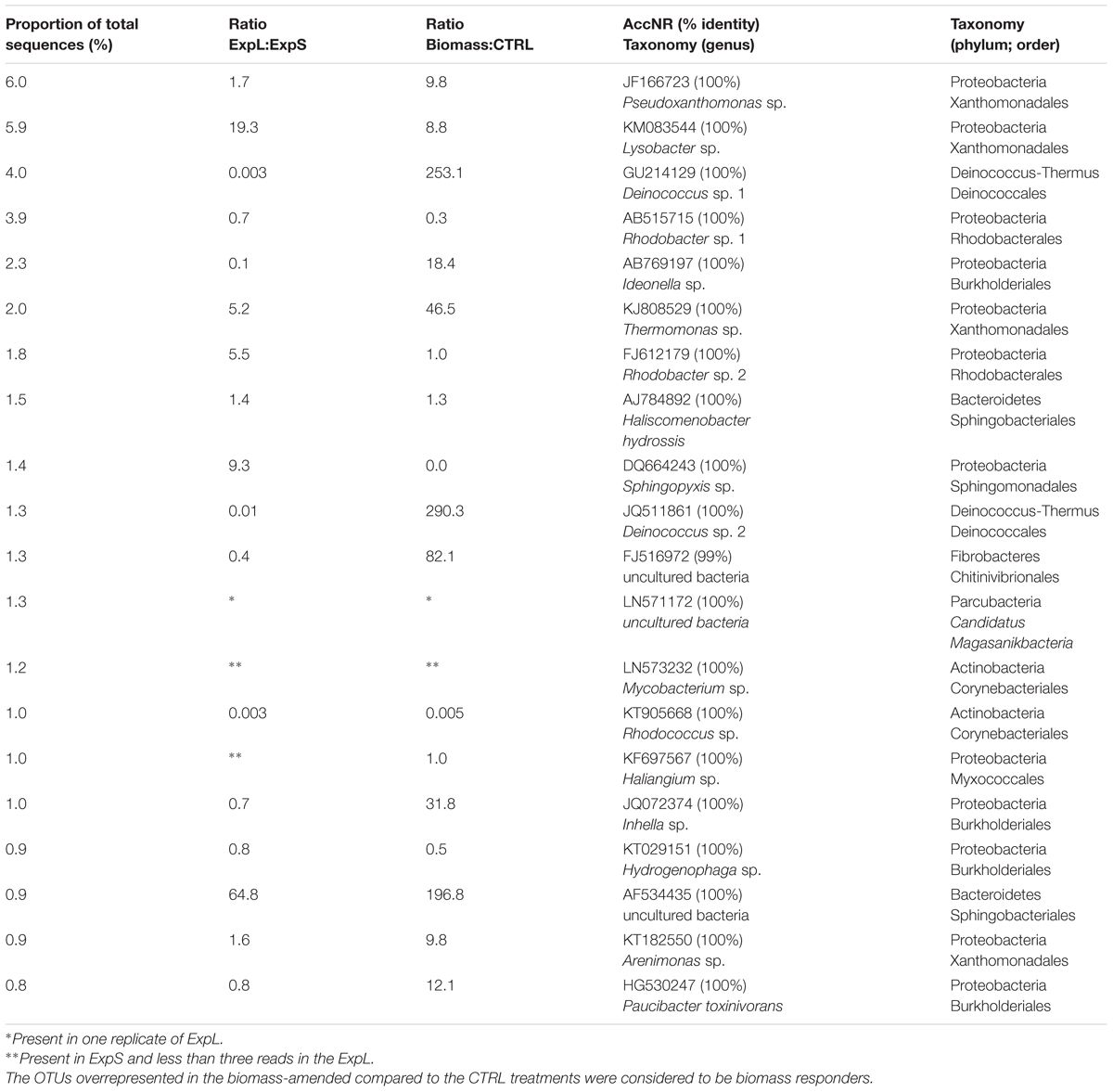
In general, the largest OTUs (most abundant in terms of read numbers) were not exclusive to a particular treatment: <20% of treatment-independent OTUs formed >75% of total reads. However, 11 of these 20 largest OTUs (Table 1) were significantly (>8 times) more abundant in the biomass-amended treatments than in the CTRL. In turn, the CTRL biofilms harbored significantly (>3 times) larger proportions of treatment-exclusive OTUs (35%), but these OTUs together only represented <10% of total reads.
Differences Between Sites
Operational taxonomic unit richness was 1422 and 1069 in ExpS and ExpL, respectively, and significantly higher fraction of OTUs were exclusively found in the stream biofilms (45 vs. 27%). Eight of the top 20 most abundant OTUs (Table 1) were significantly more abundant in ExpL than in ExpS, whereas two OTUs were highly abundant in ExpS but had <3 reads in ExpL. The fractions and read numbers of OTUs that were common to both sites were significantly higher in the WT biomass than in the CTRL biofilms (69 vs. 48% and 90 vs. 62%, respectively, Student’s t-tests, n = 6, p < 0.01). By contrast, the CTRL communities were a significantly better niche for site-specific OTUs (18 vs. 11%, Student’s t-test, n = 12, p < 0.001) that also had significantly more reads (8 vs. 2%, Student’s t-test, n = 12, p < 0.001). Thus, the addition of biomass generally reduced the site-specific differences between the biofilm communities.
Differences Between Biological Replicates
Operational taxonomic unitss were defined as being either “stochastic” or “deterministic” community members if they were present in only one or in all three biological replicates of a particular treatment. On average, only <50% of all OTUs were “deterministic” colonizers of all replicates of a treatment, whereas one-third fell into the “stochastic” category (Table 2). A significantly larger fraction of these “stochastic” OTUs were only found at one of the two experimental sites (17 vs. 12%). By contrast, significantly higher proportions of the “deterministic” OTUs (31 vs. 12%) were present at both sites (Student’s t-tests, n = 12, p < 0.05). Moreover, the proportions of “deterministic” OTUs that occurred at both sites and in every replicate were significantly higher in the WT than in the CTRL treatments (Student’s t-tests, n = 6, p < 0.001), indicating that the addition of biomass favored a more reproducible colonization by cosmopolitan bacterial genotypes.
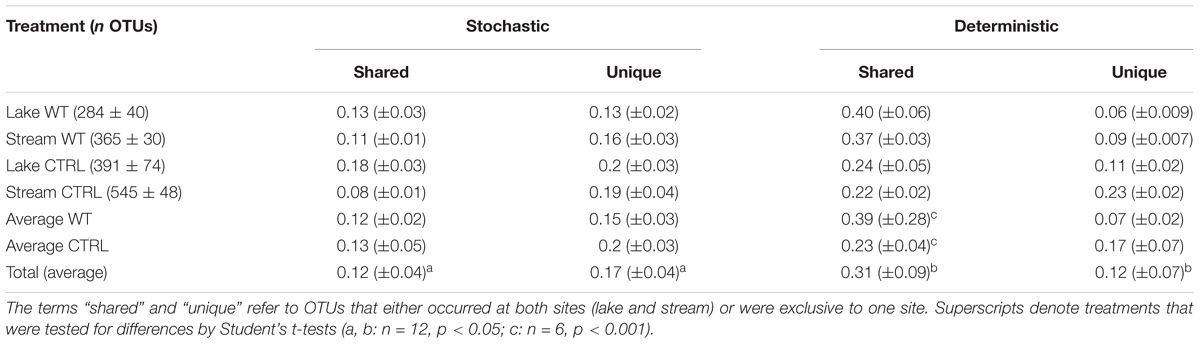
Table 2. Abundances of OTUs in the different treatments (means ± one standard deviation) and proportions of OTUs (means ± one standard error) that were only present in one biological replicate (“stochastic”) or in all the three biological replicates (“deterministic”).
Dissimilarity Analysis
When sequence data were clustered according to Bray–Curtis dissimilarity, the main separation between the communities was according to treatment rather than site (Figure 4). Bootstrap analysis indicated high stability of the four major branchings. Within the two branches of biomass-amended samples, one cluster included all samples of ExpL, and the other two clusters (of approximately 40% similarity) were the WT and MUT treatments of ExpS. The CTRL treatments from both sites fell within the second major branch, with only 20% similarity between each other. A similarity profile analysis revealed five statistically significant groups of samples: (i) the biomass-amended treatments of ExpL, (ii) the WT treatment of ExpS, (iii) the MUT treatment of ExpS, (iv) the CTRL treatment of ExpL, and (v) the CTRL treatment of ExpS. None of the biological replicates were significantly different (α = 0.001) to each other, and neither were the two biomass-amended treatments of ExpL (WT and MUT+WT biomass).
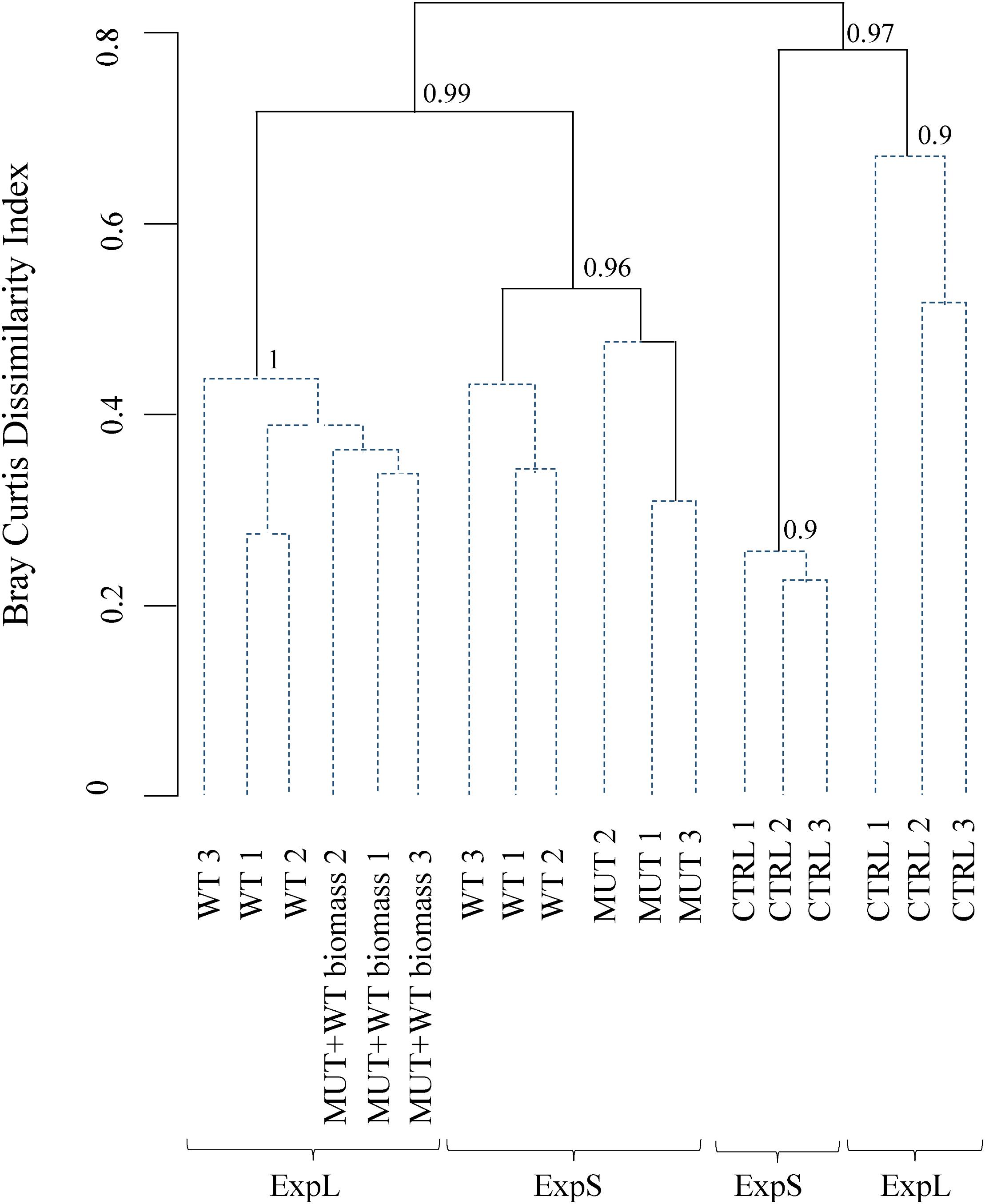
Figure 4. Clustering of microbial communities from both experiments according to Bray–Curtis dissimilarity. Similarity profile analysis identified five significantly different groups (solid lines). Biological replicates were not significant different (α = 0.001, dashed lines) from each other. The cluster stability was tested by bootstrapping (1000 times interaction); bootstrap values are indicated at the branching points.
Microbial Co-occurrence Network
A co-occurrence network was constructed to assess to which extend the individual biofilm communities were composed of the same sets of bacteria at the two sites and within different treatments. The complete network of OTUs from both sites and all treatments comprised 757 nodes (39% of all OTUs and 91% of all reads) linked by 12,198 edges (Figure 5). Ninety-five percent of the network consisted of positive correlations, and negatively correlated OTUs predominantly occurred in different treatments or at different sites. The network had a diameter of 16, an average path length of 5.6, a density of 0.04, and a modularity of 0.5 (Brandes, 2001; Blondel et al., 2008). A more detailed description of network properties and of OTUs specific to the different sites and treatments is given in Supplementary Table S2.
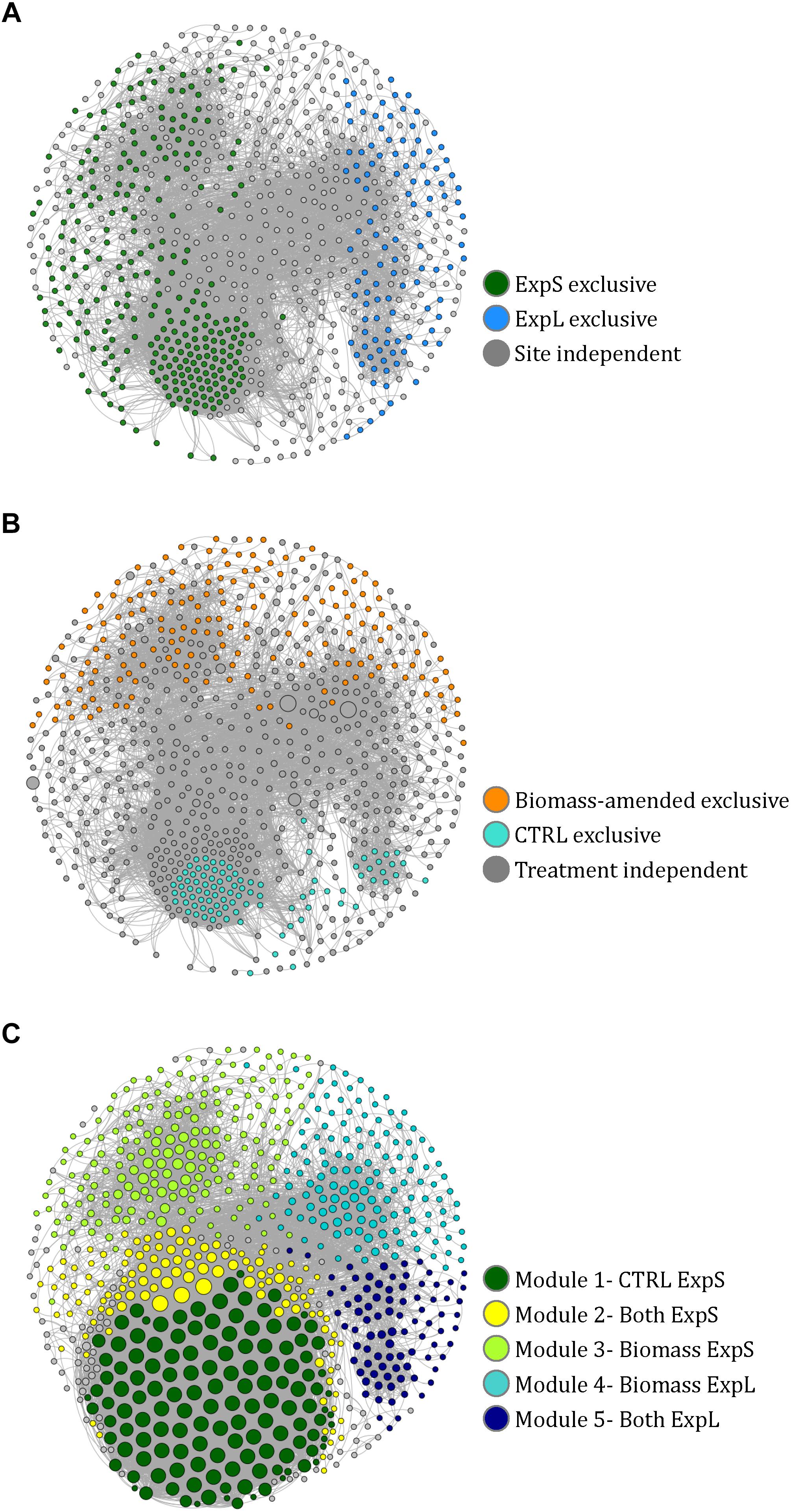
Figure 5. Co-occurrence network of 757 nodes (i.e., significantly correlated OTUs) from the GDM biofilm communities. (A) Nodes colored according to their exclusive occurrence at one site. (B) Nodes colored according to their exclusive occurrence in particular treatments and sized according to read numbers. (C) Nodes colored according to modularity class and sized according to degree (i.e., number of links to other nodes). The naming of the modules in the legend is based on the observations in A and B.
Fifteen distinct modules (coherent sub-networks) were identified based on edge weights and randomization (Blondel et al., 2008). The five major modules represented 91% of all reads in the network (Figure 5C). We could assign these modules to experimental sites and treatments by the location of those OTUs that were only present at one site or in one treatment (Supplementary Table S3): All OTUs that only occurred in ExpS fell into modules 1, 2, and 3 (named ExpS-CTRL, ExpS-Both, and ExpS-Biomass, respectively, Supplementary Table S3), while the modules 4 and 5 (named ExpL-Biomass and ExpL-Both, respectively, Supplementary Table S3) harbored all OTUs that were exclusive to ExpL (Figure 5A). Moreover, modules 3 and 4 included all the exclusive OTUs from the biomass-amended treatments, while the modules 1, 2, and 5 included all the OTUs that were specific to the CTRL treatments (Figure 5B). The ExpS-specific modules 1–3 had significantly higher average degree number (i.e., the number of OTUs that significantly co-occurred with each other) than the ExpL-specific modules 4 and 5 (9660 vs. 3665), indicating a generally more synchronized occurrence of OTUs within ExpS communities (Student’s t-test, n = 226, p < 0.001, Figure 5C).
Communities Enriched With Pure Toxin
Enrichment cultures from both sites amended with pure MC completely consumed the toxin after 11 days of incubation at anaerobic conditions (Figure 6A). By contrast, MC concentrations only decreased by 20 and 36% after 15 days of aerobic incubations of lake and stream water, respectively. Following normalization to the smallest sample (19,029 reads), there were 589 OTUs in the enrichment cultures. About one-quarter of these OTUs (153) were only present in the lake water enrichments, while approximately half (242) were exclusive to the stream water enrichments. Seventy-four percent of all OTUs (13% of reads) were only found in the anaerobic incubations, and 12% (3% of reads) only in the aerobic ones. The 10 most abundant OTUs (>5000 reads, Supplementary Table S4) represented 65% of the total amount of reads.
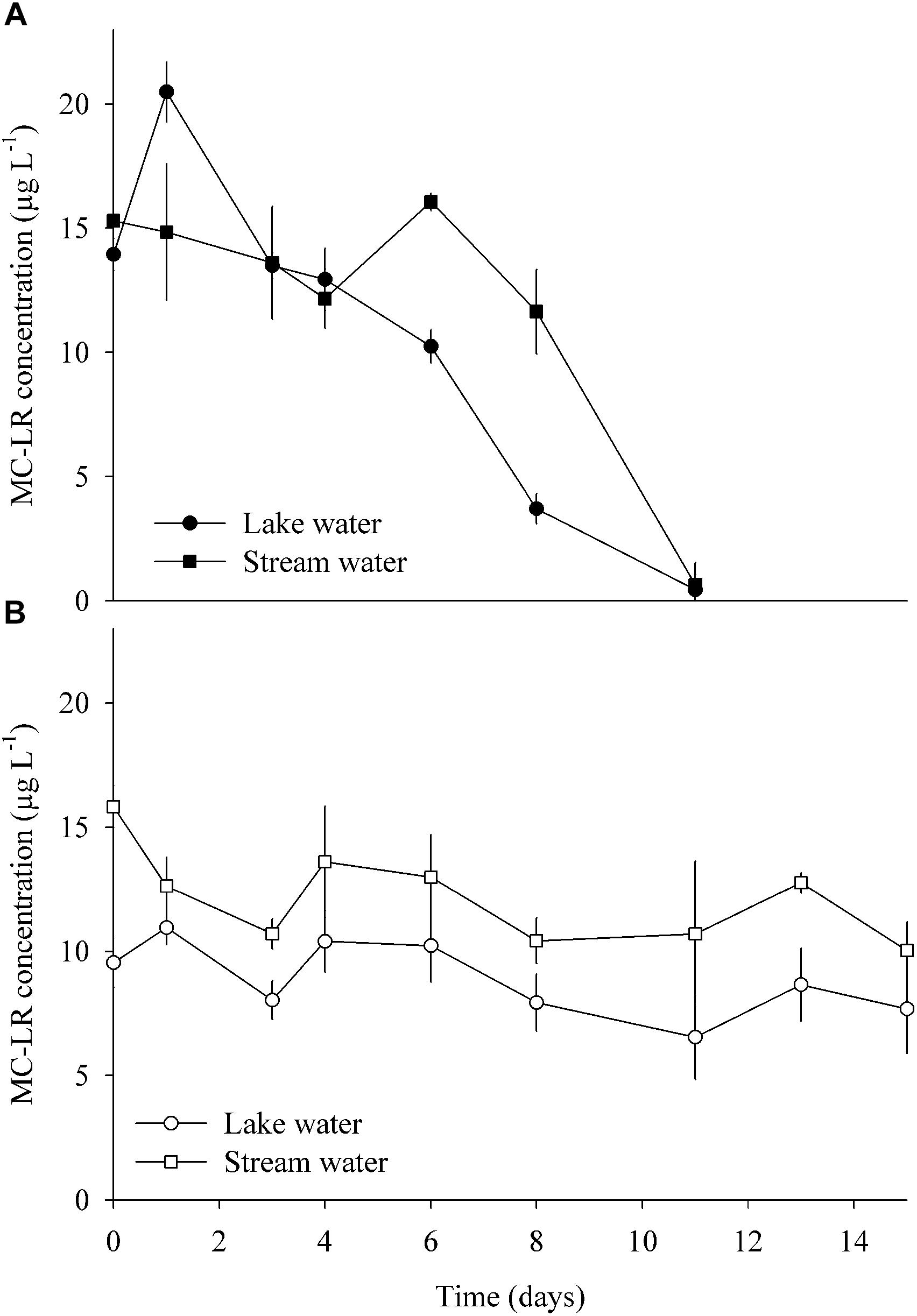
Figure 6. MC degradation in enrichment cultures amended with pure MC, using water from the lake and stream, respectively, and incubated under (A) anaerobic and (B) aerobic conditions (means ± one standard deviation of three replicate incubations).
Thirty-eight percent of all OTUs from the enrichment experiment were also present in the GDM network, including three of the five most abundant enriched OTUs (Limnobacter sp., Acinetobacter sp., and Brevundimonas vesicularis). Contrariwise, the two most abundant OTUs in the GDM network (Pseudoxanthomonas and Lysobacter) were also found in all samples of the enrichment cultures. Two OTUs related to known MC degraders were present in both, the enrichments and the GDM network: Paucibacter toxinivorans (in all enrichment samples) and Stenotrophomonas rhizophila (in 10 of 12 enrichment samples).
Discussion
Physical Properties of Lake and Stream Water Fed GDM Biofilms
As observed previously, permeate flux in GDM systems stabilized after approximately 1 week of operation (Peter-Varbanets et al., 2010). Biomass addition led to twice as much hydraulic resistance and >3 times thicker biofilms (Figure 2A and Supplementary Figure S1), resulting in lower flux (McCarthy et al., 2002; Kohler et al., 2014; Silva et al., 2018). Permeate flux and biofilm thickness of the biomass-amended biofilms in ExpL and ExpS were similar, suggesting that their hydraulic properties were neither affected by the respective characteristics of the lake and stream water influx nor by the slight differences in setup. By contrast, the permeate flux of the two CTRL treatments was markedly different. It is conceivable that the three sedimentation tanks of ExpL allowed for higher particle removal than the single tank of ExpS (Figure 1). Also, the raw water for ExpL was pumped into the system from 5 m depth while it came from the surface of the stream for ExpS, which might have influenced the respective loads of inorganic particles and consequently flux (Chomiak et al., 2014). On the other hand, CTRL biofilms in ExpS were on average twice as thick as in ExpL (Supplementary Figure S1), but had higher and unstable flux rates (Figure 2A). This might be an indication for the activity of larger organisms (Supplementary Figure S1) feeding on the stream water biofilms (Derlon et al., 2012, 2013).
Our short-term experiments suggested that the partial removal of MC from filtrates during the first week of GDM operation was mainly due to retention of the toxin by abiotic processes (Figure 3). About two-thirds of MC in cyanobacterial cells are bound to protein (Zilliges et al., 2011; Wei et al., 2016), and a large proportion of cell fragments is likely physically captured by the membrane. In addition, the passive adsorption to particles or exo-polymeric substances may also play a role, as suggested by significantly higher MC retention in pasteurized lake water samples than in sterile deionized water (Figure 3). A higher loading with inorganic particles in the stream water biofilms might explain the significantly higher levels of initial MC retention in ExpS (Figures 2B,C) at comparable flux rates (Figure 2A) (Morris et al., 2000). Previous GDM experiments with feed water from Lake Zurich showed somewhat lower initial MC retention levels (Kohler et al., 2014; Silva et al., 2018) possibly due to seasonal factors, e.g., the higher abundances of bacteria and other microbes during spring (Salcher et al., 2011; Eckert et al., 2012).
Higher MC Removal in Lake Water Generated Biofilms
Microcystin removal during the last week of GDM operation was a combination of abiotic retention of the toxin on the membrane filters (Figures 2B,C, 3) and its biological degradation. Microbial MC consumption could again be “primed” by addition of non-toxic cyanobacterial biomass in ExpL biofilms (Silva et al., 2018), suggesting that it was a facultative trait of bacteria with a broader metabolic potential, and that their initial establishment in the biofilm communities was unrelated to their ability to consume the toxin. This might be important for understanding the relationship between biofilm function and community structure.
Interestingly, the performance of ExpL biofilms with respect to biological MC removal was strikingly different between biological replicates (Figure 2B) without corresponding differences in permeate flux (Figure 2A). This might be interpreted in the context of stochastic community assembly processes, i.e., communities shaped by immigration and random processes rather than by optimal fitness of community members for a particular environmental niche (Zhou and Ning, 2017): experiments in fermenters (Zhou et al., 2013) and with lake water enrichments (Langenheder et al., 2006) have shown that quasi-identical growth conditions may lead to microbial communities that differ both in composition and functional properties. Notwithstanding these initial differences between replicates, MC removal efficiency in ExpL was significantly higher during the late phase even though permeate flux (Figure 2) and biofilm thickness were indistinguishable: while lake water biofilms increased MC retention by roughly 50% between the first and the last week and eventually completely removed the daily doses of freshly added MC, the biotic degradation in stream water biofilms led to final MC removal levels that were only approximately 20% above the initial, abiotic (Figure 2) “background.” Consequently, the average MC removal rate was significantly lower in ExpS (40 ± 4.3 μg L-1 day-1) than in ExpL (49 ± 2.5 μg L-1 day-1, mean ± standard deviation, n = 3). MC removal rates comparable to those of ExpL were also reported from enrichments of lake water featuring prior blooms of toxic cyanobacteria (Li et al., 2016). The dominant phytoplankton species in Lake Zurich is the MC-containing cyanobacterium Planktothrix rubescens (Posch et al., 2012), whereas there is no recorded history of blooms of toxic cyanobacteria in Chriesbach stream.
The biofilm communities of ExpL and ExpS significantly differed in composition even at the level of phyla (Supplementary Table S1). Yet the lower MC removal in ExpS biofilms cannot be attributed to the absence of equally efficient MC degrading microorganisms in the stream feed water: bacteria from Chriesbach enrichment cultures consumed MC to similar final concentrations as bacteria from Lake Zurich (Figure 6A). Thus, lower MC removal by stream water biofilms must have been due to the less favorable (biotic and abiotic) conditions for the growth of these degraders. The differences in MC removal in ExpL and ExpS might thus reflect that the process is not indigenous in stream water, but that nevertheless, in analogy with Baas Becking’s proposition (Baas Becking, 1934), “every functional potential is everywhere.” A widespread potential for MC degradation has already been noted by Jones and Orr (1994).
Only six of the most abundant genotypes from the biofilms (Table 1) and the enrichment experiment (Supplementary Table S4), have been associated with MC degradation previously: P. toxinivorans, Sphingopyxis sp., Rhodococcus sp., Acinetobacter sp., Methylotenera sp., and Pseudomonas sp. (Li et al., 2017). Involvement of Paucibacter in MC degradation was suggested by its >10-fold higher read numbers in the biomass-amended GDM treatments (Table 1); however, it only occurred in aerobic enrichments, where little MC degradation was observed (Figure 6B). Sphingopyxis and Rhodococcus were mainly enriched in the GDM CTRL treatments (Table 1), excluding them as potential MC degraders, which is in line with previous observations by Li et al. (2016). Our results also support another conclusion of that study, i.e., the putative role of Methylotenera sp. in MC degradation (Supplementary Table S4).
Addition of Biomass Selects for Widely Spread Bacterial Taxa
Bray–Curtis analyses (Figure 4) indicated that the dissimilarity between communities was most strongly driven by the treatments. This was likely related to the occurrence of OTUs at one or at both experimental sites: significantly more bacteria of the WT than of the CTRL biofilms were present in both ExpL and ExpS biofilms. A large fraction of these OTUs, moreover, was found in all three replicates of a particular treatment type (Table 2), suggesting environmental filtering (Cao et al., 2016) as the predominant selection mechanism in biomass-amended biofilms. Indeed, the most abundant of these genotypes were from widely spread bacterial genera: Pseudoxanthomonas spp. have been isolated from sludge, soils (Thierry et al., 2004), hot spring (Chen et al., 2002), and compost (Weon et al., 2006) environments, Lysobacter spp. have been found in soil, the rhizosphere, and freshwater (Reichnbach, 2006), and Arenimonas spp. occur in diverse habitats such as oil-contaminated soil (Young et al., 2007), the seashore (Kwon et al., 2007), an iron mine (Chen et al., 2012), or a eutrophic reservoir (Huy et al., 2013). By contrast, the CTRL treatment harbored significantly more OTUs that only occurred at a single experimental site and/or in only a single biological replicate (Table 2), indicating that these communities were more stochastically assembled from the local species pools. The two most abundant of these “site-specific” genotypes in ExpL biofilms were from groups strictly associated with freshwaters: the novel candidate phylum Parcubacteria (Brown et al., 2015), and Actinobacteria from the ac1 lineage (Newton et al., 2007).
Thus, the richer growth conditions after addition of biomass replaced the “indigenous” (i.e., site-specific) biofilm flora by a more widely spread set of bacteria that, moreover, colonized the replicate biofilms in a more reproducible manner. This change was accompanied by a loss of overall genotypic diversity in the biomass-amended treatments (Table 2), as already observed in our previous experiment (Silva et al., 2018) and other studies (Van Horn et al., 2011). The reduction of diversity and the replacement of indigenous species by rapidly growing “weeds” is a wide-spread consequence of eutrophication in plant and algal assemblages (Hillebrand and Sommer, 2000; Anderson et al., 2002; Hautier et al., 2009). Our data show that a comparable pattern is also found in microbial communities on GDM biofilms.
More Synchronized Biofilm Communities Developing From River Water
Network analysis identified a clear separation between the experimental sites (Figure 5 and Supplementary Table S2). A significantly higher number of co-occurring OTUs (i.e., higher degree number; symbol size in Figure 5C) was found in those modules of the network that could be associated with ExpS from the position of the site-exclusive OTUs (Figures 5A,C and Supplementary Table S3). Thus, bacterial taxa in stream water biofilms in general had more synchronized growth patterns. This might be related to differences in the respective recruitment of biofilm-forming bacteria from lake and stream water. A considerable fraction of the planktonic bacteria in shallow streams such as Chriesbach are released into the water from the biofilms that form in the stream bed and are, in turn, seeds for new biofilm assemblages (Besemer, 2015). By contrast, potentially biofilm-forming bacteria in lake water may originate from a number of different habitats, such as the sediment surface (Dai et al., 2016), the periphyton (Crump and Koch, 2008; Hempel et al., 2008), terrestrial influx (Monard et al., 2016), suspended organic aggregates (Bižić-Ionescu et al., 2015), or aquatic animals (Grossart et al., 2010; Eckert and Pernthaler, 2014).
MC Removing Bacteria in GDM Biofilms: Interplay of Seeding Effects and Biotic Interactions?
Habitat-specific “seeding effects” as outlined above might have contributed to the observed differences in MC degradation between ExpL and ExpS, reflecting the likely greater genotypic and functional diversity of MC degraders in lake water (Krishnan et al., 2018; Thees et al., 2018; Yang et al., 2018). The large Planktothrix population in Lake Zurich (Posch et al., 2012) provides an ample indigenous source of the toxin, and the degradation of toxins from cyanobacterial cells may occur in a variety of niches such as the water column or on sediment surfaces (Chen et al., 2008, Chen X. et al., 2010). One strategy to overcome such differences between feed water communities might thus be to initially seed GDM systems with immobilized bacteria expressing the MlrA enzyme, as has been proposed by Dziga et al. (2014).
Moreover, it is also conceivable that the more variably composed biofilm communities in ExpL allowed for the better establishment of facultative MC degraders than the more tightly knit “deterministic” communities of ExpS (Figure 5): only a single genotype could be unambiguously identified as ExpS-specific MC responder (i.e., >100 times more reads in ExpS than in ExpL, and >100 times more reads in ExpS WT than in ExpS MUT). It was the third most abundant OTU in our experiments (Table 1) and 98% identical to the obligate aerobe Deinococcus cellulosilyticus (Weon et al., 2007). This OTU only had one single partner with the same co-occurrence pattern [identical to a known sulfamethoxazole-degrading Pseudomonas sp. (Herzog et al., 2016)]. Presently, we can only speculate if the “MC responder” OTU was indeed also responsible for MC degradation. However, its conspicuous isolation within the otherwise highly synchronized stream water biofilm community might be an indication that more pronounced competitive interactions in ExpS impeded the establishment of a large variety of facultative MC degrading bacteria.
Author Contributions
MS designed study, performed the experiments, and contributed to data evaluation and co-writing of the manuscript. PD assisted in experiments and discussion of data. ND assisted in experiments and contributed to discussion of data and to manuscript. EM contributed to discussion of data and to manuscript. JP designed study and contributed to data evaluation, discussion of data, and co-writing of the manuscript.
Funding
This work was financially supported by the Swiss National Science Foundation (CR32I2_149648/1; 31003A_182336).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Emil Birnstiel and Cian Cooke for technical assistance and Eugen Loher for assistance with the experimental setup. We thank two reviewers and the editor for their valuable comments on earlier versions of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00843/full#supplementary-material
References
Amé, V. M., Echenique, J. R., Pflugmacher, S., and Wunderlin, D. A. (2006). Degradation of microcystin-RR by Sphingomonas sp. CBA4 isolated from san roque reservoir (Córdoba – Argentina). Biodegradation 17, 447–455. doi: 10.1007/s10532-005-9015-9
Anderson, D. M., Glibert, P. M., and Burkholder, J. M. (2002). Harmful Algal blooms and eutrophication: nutrient sources, composition, and consequences. Estuaries 25, 704–726. doi: 10.1007/BF02804901
Azevedo, S. M. F., Carmichael, W. W., Jochimsen, E. M., Rinehart, K. L., Lau, S., Shaw, G. R., et al. (2002). Human intoxication by microcystins during renal dialysis treatment in Caruaru—Brazil. Toxicology 181–182, 441–446. doi: 10.1016/S0300-483X(02)00491-2
Baas Becking, L. G. M. (1934). Geobiologie Of Inleiding Tot De Milieukunde. Den Haag?: W.P. Van Stockum & Zoon, The Hague.
Babica, P., Bláha, L., and Maršálek, B. (2005). Removal of microcystins by phototrophic biofilms: a microcosm study. Environ. Sci. Pollut. Res. 12, 369–374. doi: 10.1065/espr2005.05.259
Bastian, M., Heymann, S., and Jacomy, M. (2009). “Gephi: an open source software for exploring and manipulating networks,” in Proceedings of the International AAAI Conference on Weblogs and Social Media. Available at: https://gephi.org/users/publications/
Besemer, K. (2015). Biodiversity, community structure and function of biofilms in stream ecosystems. Res. Microbiol. 166, 774–781. doi: 10.1016/j.resmic.2015.05.006
Bižić-Ionescu, M., Zeder, M., Ionescu, D., Orliæ, S., Fuchs, B. M., Grossart, H. P., et al. (2015). Comparison of bacterial communities on limnic versus coastal marine particles reveals profound differences in colonization. Environ. Microbiol. 17, 3500–3514. doi: 10.1111/1462-2920.12466
Blondel, V. D., Guillaume, J.-L., Lambiotte, R., and Lefebvre, E. (2008). Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008:P10008. doi: 10.1088/1742-5468/2008/10/P10008
Bourne, D. G., Jones, G. J., Blakeley, R. L., Jones, A., Negri, A. P., and Riddles, P. (1996). Enzymatic pathway for the bacterial degradation of the cyanobacterial cyclic peptide toxin microcystin LR. Appl. Environ. Microbiol. 62, 4086–4094.
Bourne, D. G., Riddles, P., Jones, G. J., Smith, W., and Blakeley, R. L. (2001). Characterisation of a gene cluster involved in bacterial degradation of the cyanobacterial toxin microcystin LR. Environ. Toxicol. 16, 523–534. doi: 10.1002/tox.10013
Brandes, U. (2001). A faster algorithm for betweeness centrality. J. Math. Sociol. 25, 163–177. doi: 10.1080/0022250X.2001.9990249
Brown, C. T., Hug, L. A., Thomas, B. C., Sharon, I., Castelle, C. J., Singh, A., et al. (2015). Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211. doi: 10.1038/nature14486
Cao, P., Wang, J.-T., Hu, H.-W., Zheng, Y.-M., Ge, Y., Shen, J.-P., et al. (2016). Environmental filtering process has more important roles than dispersal limitation in shaping large-scale prokaryotic beta diversity patterns of grassland soils. Microb. Ecol. 72, 221–230. doi: 10.1007/s00248-016-0762-4
Chelius, M. K., and Triplett, E. W. (2001). The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 41, 252–263. doi: 10.1007/s002480000087
Chen, F., Shi, Z., and Wang, G. (2012). Arenimonas metalli sp. nov., isolated from an iron mine. Int. J. Syst. Evol. Microbiol. 62, 1744–1749. doi: 10.1099/ijs.0.034132-0
Chen, J., and Chen, G. X. (2010). Anaerobic Degradation of Microcystin and Isolation of Anaerobic Degrading Bacterium. Master Thesis, Wuhan University of Technology, Wuhan.
Chen, J., Hu, L., Bin, Z., W., Yan, S. H., Yang, J. D., Xue, Y. F., et al. (2010). Degradation of microcystin-LR and RR by a Stenotrophomonas sp. strain EMS isolated from Lake Taihu, China. Int. J. Mol. Sci. 11, 896–911. doi: 10.3390/ijms11030896
Chen, X., Yang, X., Yang, L., Xiao, B., Wu, X., Wang, J., et al. (2010). An effective pathway for the removal of microcystin LR via anoxic biodegradation in lake sediments. Water Res. 44, 1884–1892. doi: 10.1016/j.watres.2009.11.025
Chen, M.-Y., Tsay, S.-S., Chen, K.-Y., Shi, Y.-C., Lin, Y.-T., and Lin, G.-H. (2002). Pseudoxanthomonas taiwanensis sp. nov., a novel thermophilic, N2O-producing species isolated from hot springs. Int. J. Syst. Evol. Microbiol. 52, 2155–2161. doi: 10.1099/ijs.0.02306-0
Chen, W., Song, L., Peng, L., Wan, N., Zhang, X., and Gan, N. (2008). Reduction in microcystin concentrations in large and shallow lakes: water and sediment-interface contributions. Water Res. 42, 763–773. doi: 10.1016/j.watres.2007.08.007
Chomiak, A., Sinnet, B., Derlon, N., and Morgenroth, E. (2014). Inorganic particles increase biofilm heterogeneity and enhance permeate flux. Water Res. 64, 177–186. doi: 10.1016/j.watres.2014.06.045
Chomiak, A., Traber, J., Morgenroth, E., and Derlon, N. (2015). Biofilm increases permeate quality by organic carbon degradation in low pressure ultrafiltration. Water Res. 85, 512–520. doi: 10.1016/j.watres.2015.08.009
Cortes-Tolalpa, L., Jiménez, D. J., de Lima Brossi, M. J., Salles, J. F., and van Elsas, J. D. (2016). Different inocula produce distinctive microbial consortia with similar lignocellulose degradation capacity. Appl. Microbiol. Biotechnol. 100, 7713–7725. doi: 10.1007/s00253-016-7516-6
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R., and Lappin-Scott, H. M. (1995). Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745. doi: 10.1146/annurev.mi.49.100195.003431
Crump, B. C., and Koch, E. W. (2008). Attached bacterial populations shared by four species of aquatic angiosperms. Appl. Environ. Microbiol. 74, 5948–5957. doi: 10.1128/AEM.00952-08
Csardi, G., and Nepusz, T. (2006). The igraph software package for complex network research. Int. J. Complex Syst. 1695, 1–9.
Dai, Y., Yang, Y., Wu, Z., Feng, Q., Xie, S., and Liu, Y. (2016). Spatiotemporal variation of planktonic and sediment bacterial assemblages in two plateau freshwater lakes at different trophic status. Appl. Microbiol. Biotechnol. 100, 4161–4175. doi: 10.1007/s00253-015-7253-2
Derlon, N., Koch, N., Eugster, B., Posch, T., Pernthaler, J., Pronk, W., et al. (2013). Activity of metazoa governs biofilm structure formation and enhances permeate flux during Gravity-Driven Membrane (GDM) filtration. Water Res. 47, 2085–2095. doi: 10.1016/j.watres.2013.01.033
Derlon, N., Mimoso, J., Klein, T., Koetzsch, S., and Morgenroth, E. (2014). Presence of biofilms on ultrafiltration membrane surfaces increases the quality of permeate produced during ultra-low pressure gravity-driven membrane filtration. Water Res. 60, 164–173. doi: 10.1016/j.watres.2014.04.045
Derlon, N., Peter-varbanets, M., Scheidegger, A., Pronk, W., and Morgenroth, E. (2012). Predation influences the structure of biofilm developed on ultrafiltration membranes. Water Res. 46, 3323–3333. doi: 10.1016/j.watres.2012.03.031
Dexter, J., Dziga, D., Lv, J., Zhu, J., Strzalka, W., Maksylewicz, A., et al. (2018). Heterologous expression of mlrA in a photoautotrophic host – Engineering cyanobacteria to degrade microcystins. Environ. Pollut. 237, 926–935. doi: 10.1016/j.envpol.2018.01.071
Dittmann, E., Neilan, B. A., Erhard, M., von Döhren, H., and Börner, T. (1997). Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol. Microbiol. 26, 779–787. doi: 10.1046/j.1365-2958.1997.6131982.x
Dziga, D., Lisznianska, M., and Wladyka, B. (2014). Bioreactor study employing bacteria with enhanced activity toward cyanobacterial toxins microcystins. Toxins (Basel) 6, 2379–2392. doi: 10.3390/toxins6082379
Dziga, D., Maksylewicz, A., Maroszek, M., Budzyńska, A., Napiorkowska-Krzebietke, A., Toporowska, M., et al. (2017). The biodegradation of microcystins in temperate freshwater bodies with previous cyanobacterial history. Ecotoxicol. Environ. Saf. 145, 420–430. doi: 10.1016/j.ecoenv.2017.07.046
Dziga, D., Maksylewicz, A., Maroszek, M., and Marek, S. (2018). Combined treatment of toxic cyanobacteria Microcystis aeruginosa with hydrogen peroxide and microcystin biodegradation agents results in quick toxin elimination. Acta Biochim. Pol. 65, 133–140. doi: 10.18388/abp.2017_2538
Dziga, D., Wasylewski, M., Wladyka, B., Nybom, S., and Meriluoto, J. (2013). Microbial degradation of microcystins. Chem. Res. Toxicol. 26, 841–852. doi: 10.1021/tx4000045
Eckert, E. M., and Pernthaler, J. (2014). Bacterial epibionts of Daphnia: a potential route for the transfer of dissolved organic carbon in freshwater food webs. ISME J. 8, 1808–1819. doi: 10.1038/ismej.2014.39
Eckert, E. M., Salcher, M. M., Posch, T., Eugster, B., and Pernthaler, J. (2012). Rapid successions affect microbial N-acetyl-glucosamine uptake patterns during a lacustrine spring phytoplankton bloom. Environ. Microbiol. 14, 794–806. doi: 10.1111/j.1462-2920.2011.02639.x
Edwards, C., Graham, D., Fowler, N., and Lawton, L. A. (2008). Biodegradation of microcystins and nodularin in freshwaters. Chemosphere 73, 1315–1321. doi: 10.1016/j.chemosphere.2008.07.015
Flemming, H.-C. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Greer, B., Meneely, J. P., and Elliott, C. T. (2018). Uptake and accumulation of Microcystin-LR based on exposure through drinking water: an animal model assessing the human health risk. Sci. Rep. 8:4913. doi: 10.1038/s41598-018-23312-7
Grossart, H.-P., Dziallas, C., Leunert, F., and Tang, K. W. (2010). Bacteria dispersal by hitchhiking on zooplankton. Proc. Natl. Acad. Sci. U.S.A. 107, 11959–11964. doi: 10.1073/pnas.1000668107
Harke, M. J., Steffen, M. M., Gobler, C. J., Otten, T. G., Wilhelm, S. W., Wood, S. A., et al. (2016). A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 54, 4–20. doi: 10.1016/j.hal.2015.12.007
Harrell, J. F. (2016). Hmisc: HARRELL Miscellaneous. R Packag. Version 3.0- 12, 1–397. Available at: https://cran.r-project.org/web/packages/Hmisc/index.html
Hashimoto, E. H., Kato, H., Kawasaki, Y., Nozawa, Y., Tsuji, K., Hirooka, E. Y., et al. (2009). Further investigation of microbial degradation of microcystin using the advanced marfey method. Chem. Res. Toxicol. 22, 391–398. doi: 10.1021/tx8003517
Hautier, Y., Niklaus, P. A., and Hector, A. (2009). Competition for light causes plant biodiversity loss after eutrophication. Science (80-) 324, 636–638. doi: 10.1126/science.1169512
Heffernan, R., Semião, A. J. C., Desmond, P., Cao, H., Safari, A., Habimana, O., et al. (2013). Disinfection of a polyamide nano filtration membrane using ethanol. J. Memb. Sci. 448, 170–179. doi: 10.1016/j.memsci.2013.07.069
Hempel, M., Blume, M., Blindow, I., and Gross, E. M. (2008). Epiphytic bacterial community composition on two common submerged macrophytes in brackish water and freshwater. BMC Microbiol. 8:58. doi: 10.1186/1471-2180-8-58
Hennig, C. (2015). fpc: Flexible Procedures for Clustering. R Package version 2.1-10. Available at: http://www.homepages.ucl.ac.uk/~{}ucakche/ Herzog, B., Lemmer, H., Horn, H., and Müller, E. (2016). Characterization of pure cultures isolated from sulfamethoxazole-acclimated activated sludge with respect to taxonomic identification and sulfamethoxazole biodegradation potential. Environ. Eng. Act. Sludge Process. 13, 137–142.
Hillebrand, H., and Sommer, U. (2000). Diversity of benthic microalgae in response to colonization time and eutrophication. Aquat. Bot. 67, 221–236. doi: 10.1016/S0304-3770(00)00088-7
Hochberg, Y., and Benjamini, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. R. Stat. Soc. 57, 289–300. doi: 10.2307/2346101
Holst, T., Jørgensen, N. O. G., Jørgensen, C., and Johansen, A. (2003). Degradation of microcystin in sediments at oxic and anoxic, denitrifying conditions. Water Res. 37, 4748–4760. doi: 10.1016/S0043-1354(03)00413-5
Huisman, J., Codd, G. A., Paerl, H. W., Ibelings, B. W., Verspagen, J. M. H. H., and Visser, P. M. (2018). Cyanobacterial blooms. Nat. Rev. Microbiol. 16, 471–483. doi: 10.1038/s41579-018-0040-1
Huy, H., Jin, L., Lee, Y.-K., Lee, K.-C., Lee, J.-S., Yoon, J.-H., et al. (2013). Arenimonas daechungensis sp. nov., isolated from the sediment of a eutrophic reservoir. Int. J. Syst. Evol. Microbiol. 63, 484–489. doi: 10.1099/ijs.0.035410-0
Jiang, Y., Shao, J., Wu, X., Xu, Y., and Li, R. (2011). Active and silent members in the mlr gene cluster of a microcystin-degrading bacterium isolated from Lake Taihu, China. FEMS Microbiol. Lett. 322, 108–114. doi: 10.1111/j.1574-6968.2011.02337.x
Jones, G. J., and Orr, P. T. (1994). Release and degradation of microcystin following algicide treatmetn of a Microcystis aeruginosa bloom in a recreational lake, as determined by HPLC and protein phosphatase inibhition assay. Water Res. 28, 871–876. doi: 10.1016/0043-1354(94)90093-0
Jüttner, F., Leonhardt, J., and Möhren, S. (1983). Environmental factors affecting the formation of mesityloxide, dimethylallylic alcohol and other volatile compounds excreted By Anabaena cylindrica. J. Gen. Microbiol. 129, 407–412. doi: 10.1099/00221287-129-2-407
Kohler, E., Villiger, J., Posch, T., Derlon, N., Shabarova, T., Morgenroth, E., et al. (2014). Biodegradation of microcystins during gravity-driven membrane (GDM) ultrafiltration. PLoS One 9:e111794. doi: 10.1371/journal.pone.0111794
Krishnan, A., Zhang, Y. Q., and Mou, X. (2018). Isolation and characterization of microcystin-degrading bacteria from lake erie. Bull. Environ. Contam. Toxicol. 101, 617–623. doi: 10.1007/s00128-018-2468-4
Kumar, P., Hegde, K., Brar, S. K., Cledon, M., and Kermanshahi Pour, A. (2018). Physico-chemical treatment for the degradation of cyanotoxins with emphasis on drinking water treatment – How far have we come? J. Environ. Chem. Eng. 6, 5369–5388. doi: 10.1016/j.jece.2018.08.032
Kumar, P., Hegde, K., Brar, S. K., Cledon, M., Kermanshahi-pour, A., Roy-Lachapelle, A., et al. (2019). Novel fluidized-bed biofilm reactor for concomitant removal of microcystin-LR and organics. Chem. Eng. J. 359, 99–111. doi: 10.1016/j.cej.2018.11.119
Kurmayer, R., Deng, L., and Entfellner, E. (2016). Role of toxic and bioactive secondary metabolites in colonization and bloom formation by filamentous cyanobacteria Planktothrix. Harmful Algae 54, 69–86. doi: 10.1016/j.hal.2016.01.004
Kwon, S.-W., Kim, B.-Y., Weon, H.-Y., Baek, Y.-K., and Go, S.-J. (2007). Arenimonas donghaensis gen. nov., sp. nov., isolated from seashore sand. Int. J. Syst. Evol. Microbiol. 57, 954–958. doi: 10.1099/ijs.0.64457-0
Langenheder, S., Lindström, E. S., and Tranvik, L. J. (2006). Structure and function of bacterial communities emerging from different sources under identical conditions. Appl. Environ. Microbiol. 72, 212–220. doi: 10.1128/AEM.72.1.212-220.2006
Li, J., Li, J., Shi, G., Mei, Z., Wang, R., and Li, D. (2016). Discerning biodegradation and adsorption of microcystin-LR in a shallow semi-enclosed bay and bacterial community shifts in response to associated process. Ecotoxicol. Environ. Saf. 132, 123–131. doi: 10.1016/j.ecoenv.2016.05.033
Li, J., Li, R., and Li, J. (2017). Current research scenario for microcystins biodegradation – A review on fundamental knowledge, application prospects and challenges. Sci. Total Environ. 595, 615–632. doi: 10.1016/j.scitotenv.2017.03.285
Manage, P. M., Edwards, C., Singh, B. K., and Lawton, L. A. (2009). Isolation and identification of novel microcystin-degrading bacteria. Appl. Environ. Microbiol. 75, 6924–6928. doi: 10.1128/AEM.01928-09
McCarthy, A. A., Walsh, P. K., and Foley, G. (2002). Experimental techniques for quantifying the cake mass, the cake and membrane resistances and the specific cake resistance during crossflow filtration of microbial suspensions. J. Memb. Sci. 201, 31–45. doi: 10.1016/S0376-7388(01)00691-3
Monard, C., Gantner, S., Bertilsson, S., Hallin, S., and Stenlid, J. (2016). Habitat generalists and specialists in microbial communities across a terrestrial-freshwater gradient. Sci. Rep. 6:37719. doi: 10.1038/srep37719
Moons, P., Michiels, C. W., and Aertsen, A. (2009). Bacterial interactions in biofilms. Crit. Rev. Microbiol. 35, 157–168. doi: 10.1080/10408410902809431
Morris, R. J., Williams, D. E., Luu, H. A., Holmes, C. F. B., Andersen, R. J., and Calvert, S. E. (2000). The adsorption of microcystin-LR by natural clay particles. Toxicon 38, 303–308. doi: 10.1016/S0041-0101(99)00149-X
Mou, X., Lu, X., Jacob, J., Sun, S., and Heath, R. (2013). Metagenomic identification of bacterioplankton taxa and pathways involved in microcystin degradation in lake erie. PLoS One 8:e61890. doi: 10.1371/journal.pone.0061890
Newton, R. J., Jones, S. E., Helmus, M. R., and McMahon, K. D. (2007). Phylogenetic ecology of the freshwater actinobacteria acI lineage. Appl. Environ. Microbiol. 73, 7169–7176. doi: 10.1128/AEM.00794-07
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., O’Hara, R. B., Simpson, G. L., et al. (2011). Community Ecology Package. Vegan: CRAN.
Paerl, H. W. (2018). Mitigating toxic planktonic cyanobacterial blooms in aquatic ecosystems facing increasing anthropogenic and climatic pressures. Toxins (Basel) 10:E76. doi: 10.3390/toxins10200076
Peter-Varbanets, M., Dreyer, K., McFadden, N., Ouma, H., Wanyama, K., Etenu, C., et al. (2017). “Evaluating novel gravity-driven membrane (GDM) water kiosks in schools,” in Proceedings of the Conference Paper 40th WEDC, Loughborough.
Peter-Varbanets, M., Hammes, F., Vital, M., and Pronk, W. (2010). Stabilization of flux during dead-end ultra-low pressure ultrafiltration. Water Res. 44, 3607–3616. doi: 10.1016/j.watres.2010.04.020
Phujomjai, Y., Somdee, A., and Somdee, T. (2016). Biodegradation of microcystin [Dha7]MC-LR by a novel microcystin-degrading bacterium in an internal airlift loop bioreactor. Water Sci. Technol. 73, 267–274. doi: 10.2166/wst.2015.482
Posch, T., Köster, O., Salcher, M. M., and Pernthaler, J. (2012). Harmful filamentous cyanobacteria favoured by reduced water turnover with lake warming. Nat. Clim. Change 2, 809–813. doi: 10.1038/nclimate1581
Pronk, W., Ding, A., Morgenroth, E., Derlon, N., Desmond, P., Burkhardt, M., et al. (2019). Gravity-driven membrane filtration for water and wastewater treatment: a review. Water Res. 149, 553–565. doi: 10.1016/j.watres.2018.11.062
Rapala, J., Berg, K. A., Lyra, C., Niemi, R. M., Manz, W., Suomalainen, S., et al. (2005). Paucibacter toxinivorans gen. nov., sp. nov., a bacterium that degrades cyclic cyanobacterial hepatotoxins microcystins and nodularin. Int. J. Syst. Evol. Microbiol. 55, 1563–1568. doi: 10.1099/ijs.0.63599-0
Redford, A. J., Bowers, R. M., Knight, R., Linhart, Y., and Fierer, N. (2010). The ecology of the phyllosphere?: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 12, 2885–2893. doi: 10.1111/j.1462-2920.2010.02258.x
Reichnbach, H. (2006). “The Genus Lysobacter,” in The Prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York, NY: Springer), 939–957.
Salcher, M. M., Pernthaler, J., and Posch, T. (2011). Seasonal bloom dynamics and ecophysiology of the freshwater sister clade of SAR11 bacteria “that rule the waves (LD12). ISME J. 5, 1242–1252. doi: 10.1038/ismej.2011.8
Silva, M. O. D., Blom, J. F., Yankova, Y., Villiger, J., and Pernthaler, J. (2018). Priming of microbial microcystin degradation in biomass-fed gravity driven membrane filtration biofilms. Syst. Appl. Microbiol. 41, 221–231. doi: 10.1016/j.syapm.2017.11.009
Stewart, P. S. (2003). Diffusion in biofilms. J. Bacteriol. 185, 1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003
Takenaka, S., and Watanabe, M. F. (1997). Microcystin LR degradation by Pseudomonas aeruginosa alkaline protease. Chemosphere 34, 749–757. doi: 10.1016/S0045-6535(97)00002-7
Thees, A., Atari, E., Birbeck, J., Westrick, J. A., and Huntley, J. F. (2018). Isolation and characterization of lake erie bacteria that degrade the cyanobacterial microcystin toxin MC-LR. J. Great Lakes Res. 45, 138–149. doi: 10.1016/j.jglr.2018.10.013
Thierry, S., Macarie, H., Iizuka, T., Geißdörfer, W., Assih, E. A., Spanevello, M., et al. (2004). Pseudoxanthomonas mexicana sp. nov. and Pseudoxanthomonas japonensis sp. nov., isolated from diverse environments, and emended descriptions of the genus Pseudoxanthomonas Finkmann et al. 2000 and of its type species. Int. J. Syst. Evol. Microbiol. 54, 2245–2255. doi: 10.1099/ijs.0.02810-0
Tsuji, K., Asakawa, M., Anzai, Y., Sumino, T., and Harada, K. (2006). Degradation of microcystins using immobilized microorganism isolated in an eutrophic lake. Chemosphere 65, 117–124. doi: 10.1016/j.chemosphere.2006.02.018
Van Horn, D. J., Sinsabaugh, R. L., Takacs-Vesbach, C. D., Mitchell, K. R., and Dahm, C. N. (2011). Response of heterotrophic stream biofilm communities to a gradient of resources. Aquat. Microb. Ecol. 64, 149–161. doi: 10.3354/ame01515
Wang, R., Li, J., Jiang, Y., Lu, Z., Li, R., and Li, J. (2017). Heterologous expression of mlrA gene originated from Novosphingobium sp. THN1 to degrade microcystin-RR and identify the first step involved in degradation pathway. Chemosphere 184, 159–167. doi: 10.1016/j.chemosphere.2017.05.086
Warton, D. I., and Hui, F. K. C. (2011). The arcsine is asinine: the analysis of proportions in ecology. Ecology 92, 3–10. doi: 10.1890/10-0340.1
Wei, N., Hu, L., Song, L., and Gan, N. (2016). Microcystin-bound protein patterns in different cultures of Microcystis aeruginosa and field samples. Toxins (Basel) 8:293. doi: 10.3390/toxins8100293
Weon, H.-Y., Kim, B.-Y., Kim, J.-S., Lee, S.-Y., Cho, Y.-H., Go, S.-J., et al. (2006). Pseudoxanthomonas suwonensis sp. nov., isolated from cotton waste composts. Int. J. Syst. Evol. Microbiol. 56, 659–662. doi: 10.1099/ijs.0.63749-0
Weon, H. Y., Kim, B. Y., Schumann, P., Son, J. A., Jang, J., Go, S. J., et al. (2007). Deinococcus cellulosilyticus sp. nov., isolated from air. Int. J. Syst. Evol. Microbiol. 57, 1685–1688. doi: 10.1099/ijs.0.64951-0
Whitaker, D., and Christman, M. (2014). Clustsig: Significant Cluster Analysis. R Packag. version 1.1. Available at: http://www.douglaswhitaker.com WHO (1998). Cyanobacterial Toxins?: Microcystin-LR in Drinking-Water Background Document for Development of WHO Guidelines for Drinking-water Quality. Geneva: WHO
Wüest, A., and Lorke, A. (2003). Small-scale hydrodynamisc in lakes. Annu. Rev. Fluid Mech. 35, 373–412. doi: 10.1146/annurev.fluid.35.101101.161220
Yang, F., Guo, J., Huang, F., Massey, I. Y., Huang, R., Li, Y., et al. (2018). Removal of microcystin-LR by a novel native effective bacterial community designated as YFMCD4 isolated from Lake Taihu. Toxins (Basel) 10:363. doi: 10.3390/toxins10090363
Yang, F., Zhou, Y., Sun, R., Wei, H., Li, Y., Yin, L., et al. (2014). Biodegradation of microcystin-LR and-RR by a novel microcystin-degrading bacterium isolated from Lake Taihu. Biodegradation 25, 447–457. doi: 10.1007/s10532-013-9673-y
Young, C.-C., Kämpfer, P., Ho, M.-J., Busse, H.-J., Huber, B. E., Arun, A. B., et al. (2007). Arenimonas malthae sp. nov., a gammaproteobacterium isolated from an oil-contaminated site. Int. J. Syst. Evol. Microbiol. 57, 2790–2793. doi: 10.1099/ijs.0.64975-0
Zhang, J., Lu, Q., Ding, Q., Yin, L., and Pu, Y. (2017). A novel and native microcystin-degrading bacterium of Sphingopyxis sp. isolated from lake Taihu. Int. J. Environ. Res. Public Health 14:E1187. doi: 10.3390/ijerph14101187
Zhang, M., Pan, G., and Yan, H. (2010). Microbial biodegradation of microcystin-RR by bacterium Sphingopyxis sp. USTB-05. J. Environ. Sci. 22, 168–175. doi: 10.1016/S1001-0742(09)60089-9
Zhou, J., Liu, W., Deng, Y., Jiang, Y. H., Xue, K., He, Z., et al. (2013). Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. MBio 4:e00584-12. doi: 10.1128/mBio.00584-12
Zhou, J., and Ning, D. (2017). Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81:e00002-17. doi: 10.1128/MMBR.00002-17
Keywords: biofilm, microcystin, drinking water, membrane filtration, microbial communities, network analysis, operational taxonomic unit
Citation: Silva MOD, Desmond P, Derlon N, Morgenroth E and Pernthaler J (2019) Source Community and Assembly Processes Affect the Efficiency of Microbial Microcystin Degradation on Drinking Water Filtration Membranes. Front. Microbiol. 10:843. doi: 10.3389/fmicb.2019.00843
Received: 06 December 2018; Accepted: 02 April 2019;
Published: 18 April 2019.
Edited by:
Rainer Kurmayer, University of Innsbruck, AustriaReviewed by:
Dariusz Dziga, Jagiellonian University, PolandLeda Giannuzzi, National University of La Plata, Argentina
Copyright © 2019 Silva, Desmond, Derlon, Morgenroth and Pernthaler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jakob Pernthaler, cGVybnRoYWxlckBsaW1ub2wudXpoLmNo
 Marisa O. D. Silva
Marisa O. D. Silva Peter Desmond
Peter Desmond Nicolas Derlon
Nicolas Derlon Eberhard Morgenroth
Eberhard Morgenroth Jakob Pernthaler
Jakob Pernthaler