- ScienceTheEarth, Mesa, AZ, United States
Approximately four billion years ago, the first microorganisms to thrive on earth were anaerobic chemoautotrophic thermophiles, a specific group of extremophiles that survive and operate at temperatures ∼50 – 125∘C and do not use molecular oxygen (O2) for respiration. Instead, these microorganisms performed respiration via dissimilatory metal reduction by transferring their electrons extracellularly to insoluble electron acceptors. Genetic evidence suggests that Gram-positive thermophilic bacteria capable of extracellular electron transfer (EET) are positioned close to the root of the Bacteria kingdom on the tree of life. On the contrary, EET in Gram-negative mesophilic bacteria is a relatively new phenomenon that is evolutionarily distinct from Gram-positive bacteria. This suggests that EET evolved separately in Gram-positive thermophiles and Gram-negative mesophiles, and that EET in these bacterial types is a result of a convergent evolutionary process leading to homoplasy. Thus, the study of dissimilatory metal reducing thermophiles provides a glimpse into some of Earth’s earliest forms of respiration. This will provide new insights for understanding biogeochemistry and the development of early Earth in addition to providing unique avenues for exploration and discovery in astrobiology. Lastly, the physiological composition of Gram-positive thermophiles, coupled with the kinetic and thermodynamic consequences of surviving at elevated temperatures, makes them ideal candidates for developing new mathematical models and designing innovative next-generation biotechnologies.
KEY CONCEPTS
Anaerobe: organism that does not require oxygen for growth.
Chemoautotroph: organism that obtains energy by oxidizing inorganic electron donors.
Convergent Evolution: process in which organisms which are not closely related independently evolve similar traits due to adapting to similar ecological niches and/or environments.
Dissimilatory Metal Reduction: reduction of a metal or metalloid that uses electrons from oxidized organic or inorganic electron donors.
Exoelectrogen: microorganism that performs dissimilatory metal reduction via extracellular electron transfer.
Extremophiles: organisms that thrive in physical or geochemical conditions that are considered detrimental to most life on Earth.
Homoplasy: a character shared by a set of species that is not shared by a common ancestor
Non-synonymous Substitutions (Ka): a substitution of a nucleotide that changes a codon sequence resulting in a change in the amino acid sequence of a protein.
Synonymous Substitutions (Ks): a substitution of a nucleotide that may change a codon sequence, but results in no change in the amino acid sequence of a protein.
Thermophiles: a specific group of extremophiles that survive and operate at temperatures ∼50–125∘C.
Perhaps You’d Better Start from the Beginning
Approximately four billion years ago (Ga), the first microorganisms to thrive on earth were likely anaerobic chemoautotrophic thermophiles, a specific group of extremophiles that survive and operate at temperatures ∼50–125°C and do not use molecular oxygen (O2) for respiration (Pace, 1991; Kashefi and Lovley, 2003; Seckbach, 2004; Sleep, 2018); although this is under investigation (Islas et al., 2003; Boussau and Gouy, 2006; Glansdorff et al., 2008; Weiss et al., 2018). These early microorganisms lived underwater near hydrothermal vents- where modern thermophiles persist (Seckbach, 2004, 2006; Slobodkin et al., 2006; Onyenwoke et al., 2007; Zavarzina et al., 2007; Niu et al., 2009; Slepova et al., 2009). These locations were ideal since they protected microorganisms from ultraviolet radiation and contained plentiful amounts of oxidized compounds including SO4-, Fe(III) oxides, NO3-, and Mn(IV) that were used as terminal electron acceptors (Lovley, 1993a; Nealson and Conrad, 1999; Amend and Shock, 2001; Knoll, 2003; Slobodkin, 2005; Seckbach, 2006). Since many of these minerals are insoluble, microorganisms, or exoelectrogens, adapted mechanisms to perform EET for their survival. Positioned close to the root of the Bacteria kingdom on the tree of life, thermophiles provide a glimpse into some of Earth’s earliest forms of respiration via dissimilatory metal reduction (Ciccarelli et al., 2006; Seckbach, 2006; Logan, 2009).
Within the kingdom Bacteria, there are two distinct classifications – Gram-positive and Gram-negative – that span across many bacterial phyla (Ventura et al., 2007; Vesth et al., 2013). This classification has profound implications on the physiology and structure of the bacterial cell, specifically the structure of the cell wall and membrane. This is significant because exoelectrogens are required to transfer their electrons externally through their cell membranes (Lovley, 1993b; Bond and Lovley, 2003; Leang et al., 2003; Lies et al., 2005; Richter et al., 2008; Coursolle et al., 2010; Holmes et al., 2016).
In Gram-negative bacteria, the cell contains two membranes, an inner membrane and an outer membrane, that are separated by a periplasm. Within the periplasmic space is a thin layer (∼5–10 nm) of peptidoglycan/murein, which is a polymer of sugars, NAG, NAM, and amino acids that accounts for approximately 10% of the dry weight of the cell (Koch, 2006). For EET to occur, electrons must traverse a series of peripheral and integral proteins and cytochromes that are imbedded in the inner membrane, span across the periplasm, and are docked to the outer membrane (Bird et al., 2011).
In contrast, Gram-positive bacteria have a cell wall composed of a single membrane and a relatively thick (20–80 nm) peptidoglycan layer which are separated by a periplasmic space. The peptidoglycan consists of many layers and can weigh as much as 60% of the cell’s total dry weight (Koch, 2006; Ehrlich, 2008; Pham et al., 2008,). In addition, Gram-positive bacteria have teichoic acids that have been implicated as the metal binding sites for the cells, are embedded in the cell wall, and extend from the cell membrane to the outer surface of the peptidoglycan layer (Beveridge and Murray, 1980; Beveridge et al., 1982; Ehrlich, 2008). The presence of a thick peptidoglycan layer, sometimes surrounded by an S-layer, makes it necessary for many metal reducing Gram-positive bacteria to conduct EET via proteins and cytochromes that are packed into fissures within the cell wall, anchored to the peptidoglycan, or positioned along teichoic acids (Ehrlich, 2008; Carlson et al., 2012).
There are three known methods through which exoelectrogens perform EET; with the method employed varying by bacterial species (Torres et al., 2010; Mohan et al., 2014). One method for mediated, or indirect, electron transfer (MET) and two methods for direct electron transfer (DET) (Torres et al., 2010; Mohan et al., 2014). The MET method involves redox mediators, or extracellular shuttles, that are produced by the bacteria and transfer electrons between the bacterial cell and an external metal oxide (Schröder, 2007; Mohan et al., 2014). The two methods for DET to an extracellular electron acceptor include direct contact of a redox protein, often a cytochrome, imbedded on a cell’s outer membrane or peptidoglycan layer to a metal oxide, and long-range (several μm) direct contact of an electrically conductive, or semiconductive, extracellular matrix. The extracellular matrix is hypothesized to confer DET via “nanowires” that are either pili, membranous extensions embedded with redox proteins (often Cyts), or potentially filaments composed entirely of cytochrome monomers (Lovley, 2008; Torres et al., 2010; Carlson et al., 2012; Parameswaran et al., 2013; Pirbadian et al., 2014; Wang et al., 2019). See Figure 1 for an overview of EET in Gram-negative and Gram-positive bacteria.
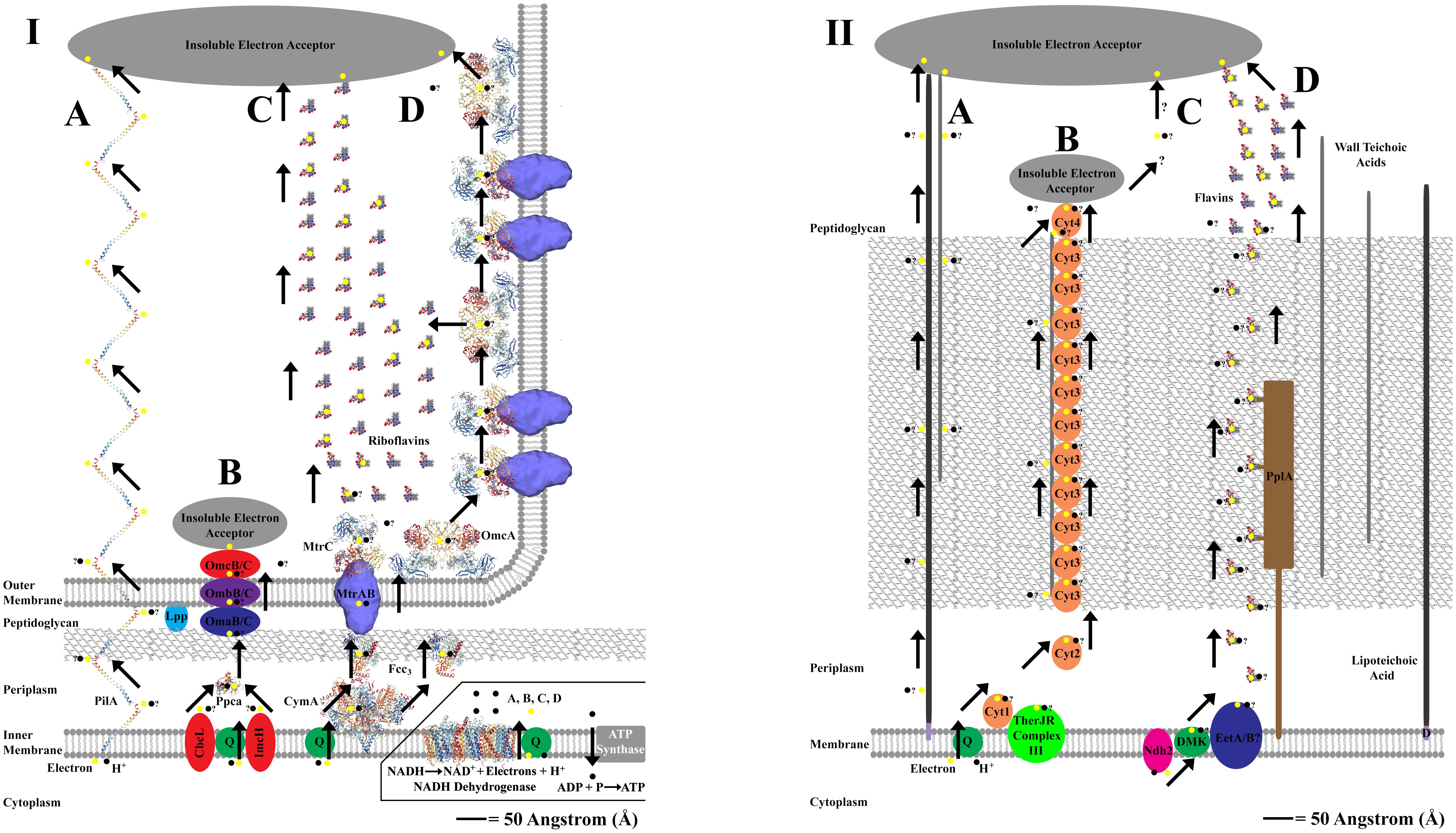
Figure 1. Panel (I,II) shows proposed extracellular electron transfer (EET) mechanisms for Gram-negative and Gram-positive bacteria. An angstrom scale is used to show relative distances for EET from the cytoplasm to external cell environment for each cellular physiology (Hobot et al., 1984; Zuber et al., 2006). Where molecular structure of a protein is used, the protein image was acquired from the Research Collaboratory for Structural Bioinformatics Protein Database (RCSB PDB) and is represented accurately to scale. Cartoon representatives are used to indicate the presence of an enzyme with an unresolved molecular structure, and thus the sizes cannot be drawn to scale. “?” is used to represent uncertainty in the proposed pathway. The inlay in panel (I) is used to show the source of electrons and protons coming from NADH oxidation via NADH dehydrogenase located on the cellular membrane of Gram-negative and Gram-positive bacteria (NADH dehydrogenase shown is from Escherichia coli, Efremov and Sazanov, 2011). This inlay applies for panel (I,II), and is only shown once to conserve space. Panel (IA) represents the PilA pathway in Geobacter sulfurreducens that is proposed to use conductive nanowires to perform direct long range EET at distances up to several microns (Reardon and Mueller, 2013; Mulvankar et al., 2015). Panel (IB) represents an alternative “OMC” pathway for EET in G. sulfurreducens that used a series on redox active proteins which bind directly to an extracellular electron acceptor (Morgado et al., 2011; Zacharoff et al., 2016). Panel (IC) represents the Mtr and OmcA pathway(s) for EET in Shewanella oneidensis that use a series of redox active proteins which can either bind directly to an extracellular electron acceptor or reduce extracellular flavins which shuttle electrons to an extracellular electron acceptor, resulting in mediated EET (Taylor et al., 1999; Marsili et al., 2008; Firer-Sherwood et al., 2011; Edwards et al., 2014, 2015). The MtrAB complex structure is adjusted from Edwards et al. (2018). The CymA structure is represented by the analogous NrfH from Desulfovibrio vulgaris (Rodrigues et al., 2006). Panel (ID) represents the production of membrane based nanowires in S. oneidensis that use the Mtr and OmcA pathway(s) for direct long range EET at distances up to several microns (Pirbadian et al., 2014; Subramanian et al., 2018). Panel (IIA) represents a putative EET pathway that uses a potential combination of lipoteichoic acids [which are bound to the cellular membrane via diacylglycerol (D)] and wall teichoic acids to transfer electron across the peptidoglycan layer (Beveridge and Murray, 1980; Beveridge et al., 1982; Ehrlich, 2008). Panel (IIB) represents the MHC pathway from Thermincola potens which proposes that EET via direct contact to an extracellular electron acceptor occurs via a series of multiheme cytochromes (Cyts) that are embedded in the peptidoglycan layer, perhaps bound to teichoic acids (Carlson et al., 2012). Panel (IIC) shows that the MHC pathway currently does not account for direct long range EET that can transfer electrons at distances up to several microns in T. ferriacetica (Parameswaran et al., 2013; Lusk et al., 2016). Panel (IID) shows a flavin based model for mediated EET that transfers electrons through the peptidoglycan layer using a series of redox shuttles that may or may not bind to proteins embedded in the peptidoglycan layer (Light et al., 2018; Pankratova et al., 2018). In Listeria monocytogenes, this pathway uses a novel NADH dehydrogenase called Ndh2 (Light et al., 2018).
Through the Looking Glass
The research concerning the study of EET has focused primarily on Gram-negative mesophilic microorganisms including Geobacter sulfurreducens (Bond and Lovley, 2003; Zacharoff et al., 2016), Geobacter metallireducens (Lovley, 1993b), and Shewanella oneidensis (Lies et al., 2005), with little emphasis on Gram-positive thermophiles. This is a significant distinction since the EET pathways of Gram-negative mesophilic bacteria including Geobacter are relatively new adaptations, suggesting that they have a separate evolutionary lineage from those of thermophiles (Holmes et al., 2016). Microbial life on Earth has evolved to produce a bimodal distribution of growth, with the highest growth rates congregating at 42°C and 67°C, implying a potential evolutionary transition around 50°C (Amend and Shock, 2001; Corkrey et al., 2016a,b). In addition, although the genome sizes of thermophiles and mesophiles are roughly equivalent, thermophilic genomes have less intergenic regions and reduced variability in their size (Sabath et al., 2013; Van Noort et al., 2013; Wang et al., 2015). For example, all known microorganisms that thrive at temperatures in excess of 60°C have genomes smaller than 4 megabases (Mb) while all microorganisms with genomes larger than 6 Mb thrive at temperature less than 45°C (Sabath et al., 2013; Van Noort et al., 2013). As a consequence, thermophilic cells tend to have less volume, giving them a higher surface area to volume ratio with their surroundings (Dufresne et al., 2005; Giovannoni et al., 2005; Moya et al., 2009; Dill et al., 2011). Finally, an increase in optimal growth temperature has a large impact on the GC content of the bacterial RNA, affects the amino acid sequence of almost every protein within the thermophilic proteome including the frequency and distribution of amino acid residues in the transmembrane domain, and reduces protein length and family size (Hurst and Merchant, 2001; Saunders et al., 2003; Das et al., 2006; Puigbò et al., 2009; Burra et al., 2010; Dutta and Chaudhuri, 2010; Meruelo et al., 2012; Sabath et al., 2013).
It is well documented that thermophilic enzymes (thermozymes) have higher thermostability than mesophilic enzymes (Vieille and Zeikus, 1996, 2001; Friedman et al., 2004; Li et al., 2005). In addition, thermophiles also have a much lower rate of non-synonymous substitutions in protein coding regions of their DNA than do mesophiles (Friedman et al., 2004; Van Noort et al., 2013; Wang et al., 2015). This is observed when measuring the ratio of non-synonymous substitutions (Ka) –changes in codon sequence that affect amino acid sequence- to synonymous substitutions (Ks)–changes in codon sequence that do not affect amino acid sequence (Yang and Bielawski, 2000; Hurst, 2002). The smaller Ka/Ks ratio (ω) (Supplementary Equation S1) observed in thermophiles favors stabilizing selection, implying a strong selective pressure for conservation of thermophilic protein structure and function (Sabath et al., 2013; Wang et al., 2015). The functional stability of thermophilic microorganisms resulting from low non-synonymous mutation rates makes them ideal candidates for conducting studies with high reproducibility and constructing bioreactors for long-term bioremediation (Al-Maghrabi et al., 1999; Chaalal and Islam, 2001; Margesin and Schinner, 2001) or bioenergy production (Bergquist et al., 1987; Patel et al., 2019).
The Gram-positive nature of many thermophiles, coupled with the genomic and proteomic impacts of surviving at high temperature, makes it probable that they use alternative mechanism(s) for EET than those present in Gram-negative mesophilic bacteria. The field of study for exoelectrogens has only a cursory understanding of the mechanisms for EET in Gram-positive thermophiles; however, research suggests that the mechanisms substantially differentiate from mesophilic bacteria. For example, the truncated pilA gene which codes for the formation of electrically conductive pili (Richter et al., 2008; Holmes et al., 2016) that transfer electrons in Geobacter is not found in Gram-positive thermophiles including Thermincola potens (Wrighton et al., 2011; Carlson et al., 2012), Thermincola ferriacetica (Lusk et al., 2015a), Thermoanaerobacter ethanolicus (Holmes et al., 2016), or Thermoanaerobacter pseudethanolicus (Lusk et al., 2015b). In addition, proteomic and genomic data indicate that these microorganisms also lack the Mtr (Coursolle et al., 2010) and Omc (Leang et al., 2003) gene clusters that code for the production of pathways containing multiheme c-type cytochromes (Cyts) that transfer electrons extracellularly in Gram-negative mesophiles (Bird et al., 2011; Carlson et al., 2012; Vecchia et al., 2014; Lusk et al., 2015a, 2018b; Wang et al., 2019).
Currently, the proposed mechanism for EET in many Gram-positive thermophiles, derived from studies with T. potens, is a “MHC” pathway consisting of observed and putative Cyts which transcend the entire cell envelope and bind directly to extracellular oxides (Wrighton et al., 2011; Carlson et al., 2012; Shi et al., 2016). Nevertheless, there is not consensus on the precise mechanism(s) for EET in Gram-positive thermophiles since the MHC hypothesis does not explain the observation of direct long range EET present in some, including T. ferriacetica, because it does not consider the transfer of electrons through a conductive or semi-conductive extracellular matrix (Parameswaran et al., 2013; Lusk et al., 2016, 2018b). Finally, the crux of the proposed model for an MHC pathway containing Cyt relies on the presence of Cyts. However, some Gram-positive thermophilic exoelectrogens, including T. pseudethanolicus, have been discovered that contain no Cyts (Lusk et al., 2015b).
This genetic data, in concert with the physical limitations and ohmic resistance associated with transferring electrons across a thick peptidoglycan layer and eventually through an extracellular matrix, suggests that the EET present in distinct bacterial phyla are the result of a convergent evolutionary process leading to homoplasy. Considering that prokaryotes have been around for 3.5–4 billion years and survived for ∼1.5–2.0 billion years (Sleep, 2018) before atmospheric oxygen (Partin et al., 2013), convergent EET phenomena in prokaryotes is likely. Furthermore, EET likely evolved with similar selective pressures (i.e., the availability of insoluble metal oxides as electron acceptors) across many bacterial phyla. Thus, despite the fundamentally different genetic and proteomic infrastructures of the extracellular matrices contained by Gram-negative mesophiles and Gram-positive thermophiles, similar EET and proton transport limitations are observed in either bacterial physiology (Torres et al., 2008; Wrighton et al., 2011; Carlson et al., 2012; Lusk et al., 2016, 2018b; Holmes et al., 2016).
Some Like It Hot: Modeling with Thermophilic Microorganisms
In lieu of the genetic and proteomic differences between Gram-negative mesophiles and Gram-positive thermophiles, the development and verification of mathematical models can be used to enhance our understanding of EET and its limitations, and potentially to devise productive next generation biotechnologies. Obtaining data in order to construct models for EET often proceeds by cultivating dissimilatory metal reducing microbes on electrodes attached to voltammeters or potentiostats in bioelectrochemical systems (BES) or microbial electrochemical cells (MxCs) (Marcus et al., 2007, 2011; Hamelers et al., 2011; Peng et al., 2013; Renslow et al., 2013). Once grown on electrodes, electrochemical techniques including CV, CA, and EIS can be used to monitor and characterize the influence of a Gram-positive thermophilic cellular physiology on the conductivity of the extracellular matrix (Kbio) and changes in the redox potential (EKA) of electron channeling proteins during EET (Srikanth et al., 2008; Marsili et al., 2010; Strycharz et al., 2011; Yang et al., 2012; Badalamenti et al., 2013; Parameswaran et al., 2013; Yoho et al., 2014, 2015; Tan et al., 2017; Lusk et al., 2018b). For example, research using these electrochemical techniques has already shown the presence of multiple redox pathways and proton coupled EET in exoelectrogens, including Gram-positive thermophiles (Badalamenti et al., 2013; Fu et al., 2013; Yoho et al., 2014, 2015; Zacharoff et al., 2016; Lusk et al., 2018b).
The reasons for these phenomena are worth investigating given that metal-like conductance along teichoic acids or electron hopping along cytochromes embedded in peptidoglycan, particularly at high temperatures, ought to have influences on the limitations of EET in Gram-positive thermophiles compared to Gram-negative mesophiles due to changes in Kbio and EKA (Marcus et al., 2007; Torres et al., 2010; Yates et al., 2016; Lusk et al., 2018b). In addition, with increasing temperature, the Michaelis–Menten (Supplementary Equation S2) saturation curve predicts a higher substrate concentration at which the reaction rate is half of the maximum (KM). Nevertheless, proteins produced by thermophiles maintain similar reaction kinetics and binding affinities to those observed in mesophilic organisms (Marcus et al., 2011; Dong et al., 2018). This is because thermophiles maintain a robust repertoire of genes and proteins that provide them with marginal stability via the regulation of ions, organic molecules, and other osmolytes (Marcus et al., 2007; Okur et al., 2017; Lusk et al., 2018b). Thus, despite the case that the redox potential of cytochromes is highly influenced by pH, ohmic resistance, and temperature, thermophiles have similar redox potentials when compared to their mesophilic counterparts (Chang and Kim, 2005; Liu et al., 2011; Wrighton et al., 2011; Carlson et al., 2012; Deng et al., 2013; Pirbadian et al., 2014; Vecchia et al., 2014; Lusk et al., 2018b). As a consequence, currently no mathematical models of EET draw definitive distinctions concerning limitations in EET resulting from either Gram-positive or Gram-negative bacteria (Marcus et al., 2007, 2011; Hamelers et al., 2011; Peng et al., 2013; Renslow et al., 2013).
A common hypothesis assumes that the evolutionary selection for small genomes and smaller, thermally stable proteins in thermophiles minimizes energy expenditures for cell maintenance and protein synthesis (Harold, 1986; Wagner, 2005; Lane and Martin, 2010; Sabath et al., 2013). Although genome replication only accounts for 2% of a microbe’s energy budget, translation of genetic information into proteins is a high energy cost to the cell, accounting for ∼75% of a cell’s energy expenditures (Harold, 1986; Wagner, 2005; Lane and Martin, 2010). Measurements garnered from MxC experiments involving Gram-positive thermophiles have corroborated this hypothesis, showing that thermophilic exoelectrogens have increased growth (μ) (Supplementary Equation S3) and metabolic rates (rut) (Supplementary Equation S4) while reducing the fraction of electrons received from electron donors that are used for cell maintenance and synthesis of cellular material (fs°), resulting in lower biomass production per electron consumed [measured as yield (Y)] (Supplementary Equation S5) and a greater number of electrons from the electron donor that can be captured from the biomass (measured as CE) (Supplementary Equation S6) (Makarieva et al., 2005; Lee et al., 2009; Parameswaran et al., 2013; Lusk et al., 2018a).
A consequence of cultivating thermophilic microbes is that increasing temperature has a significant impact on the thermodynamics of electron donor – electron acceptor interactions (Supplementary Table S1). For example, when growing microorganisms in bioreactors, including MxCs, it is vital to add buffer in order to mitigate pH inhibition and proton transport limitations (Torres et al., 2008; Lusk et al., 2016); however, operating at 60°C compared to 30°C impacts the pKa of the buffer added to the system (Supplementary Table S2). For example, in a sodium bicarbonate buffered system, the pKa1 drops from 6.33 to 6.30 and the pKa2 drops from 10.29 to 10.14 (Supplementary Equation S7) (Mook and Koene, 1975; Lusk et al., 2015c). Furthermore, the potential of the external electron acceptor for exoelectrogens is dependent upon the temperature and the pH of their environment (Supplementary Table S3) (Logan et al., 2006). At 30°C, the change in potential is ∼60 mV per pH unit; however, this change is 66 mV per pH unit at 60°C resulting in about a 1 mV per 5°C change in the energetics of EET (Supplementary Equation S8) (Katuri et al., 2010; Brownson and Banks, 2014; Lusk et al., 2015c, 2018b). In addition, using a simplified Einstein-Stokes equation, the rate for diffusion at 60°C is ∼2 × faster than 30°C (Supplementary Equation S9) (Lusk et al., 2015c). Also, Henry’s Law constants and the Van’t Hoff equation reveal that solubility of O2 in water is 35% less at 60°C compared to 30°C, reducing O2 contamination issues in anaerobic bioreactors (Supplementary Equation S10) (Lusk et al., 2015c, 2018a). As a result, research with thermophilic bacteria grown as biofilms in MxCs indicates that they may be good candidates for developing next generation biotechnologies since they are able to send more electrons from substrates to extracellular electron acceptors (measured as current density, or j) and at lower buffer concentration than are mesophiles (Torres et al., 2008; Parameswaran et al., 2013; Lusk et al., 2016, 2018a).
Nevertheless, there are practical limitations to consider when operating biotechnologies at thermophilic conditions. For example, it is important to mitigate evaporation given that the Antoine equation shows that the vapor pressure for water is ∼4.5 × greater at 60°C compared to 30°C (Supplementary Equation S11) (Rodgers and Hill, 1978). In addition, certain essential gasses are less soluble at 60°C compared to 30°C which may impact microbial growth (Varshney et al., 2016). Carbon dioxide (CO2), for example, is essential for the growth of many photosynthetic microbes (Singh and Singh, 2014) and is ∼55% less soluble at 60°C compared to 30°C (Carroll et al., 1991). The high energy input required to maintain thermophilic conditions makes the implementation of thermophiles in biotechnologies practical for applications involving thermophilic liquids, including the removal of organic material (measured as COD or BOD) from compost, brewery, sugarcane, beet sugar, or molasses wastes (Skjelhaugen, 1999; Zupančič et al., 2007; Bhatnagar et al., 2016).
All in the Family?
More data is needed on the genealogy of EET pathways and the dynamic genomics, transcriptomics, proteomics, and metabolomics of exoelectrogens performing dissimilatory metal reduction (Zhang et al., 2014; Leary et al., 2015; Wang et al., 2015; Eddie et al., 2017). Furthermore, investigating the role of horizontal gene transfer (HGT) from thermophilic archaea to bacteria, which has been shown to make up as much as 24% of the thermophilic bacterial genome, may reveal EET mechanisms that persist across multiple domains of life and thus provide additional insight into the evolutionary history of EET (Aravind et al., 1998; Nelson et al., 1999; Gu and Hilser, 2009; Yilmazel et al., 2018). More information is needed to draw distinctive conclusions regarding the precise mechanisms for EET in exoelectrogens. Nevertheless, understanding the mechanisms and physiology for EET across diverse microbial species will elucidate the path(s) through which selective pressure to reduce extracellular metal oxides may have led to independent evolutionary adaptations.
To acquire a more fundamental understanding of the processes occurring during dissimilatory metal reduction, it is important to recover RNA from the biomass of respiring exoelectrogens for a transcriptomic analysis of the genes that are being transcribed during EET (Qiao et al., 2009). In addition, it is also essential to probe into the proteomic data of these bacteria to gather a more complete understanding of which proteins are being translated and expressed during dissimilatory metal reduction (Vecchia et al., 2014; Costa et al., 2015, 2018). For example, observing upregulation and downregulation of transcribed genes and translated proteins, and production or cessation of metabolites in the presence of varying electron acceptors and donors will elucidate which respiratory pathways are essential for EET in Gram-positive thermophilic and Gram-negative mesophilic exoelectrogens (Zhang et al., 2014; Leary et al., 2015; Wang et al., 2015; Eddie et al., 2017).
Furthermore, given the pivotal role thermophilic exoelectrogens perform in the Earth’s biogeochemical cycles (Wiegel and Michael, 1998; Fenchel et al., 2012; Partin et al., 2013), increasing our understanding of these microorganisms allows us to gain insight into the processes which formed the composition of early Earth and continue to form its composition in the present day, and perhaps present new opportunities for investigating the presence of exobiological phenomenon on distant planets (Bulat et al., 2011; Girguis and Fundis, 2018). Finally, the discovery of novel thermophilic microorganisms containing thermozymes has large economic potential for the development of novel and existing biotechnologies (Brock and Freeze, 1969; Turner et al., 2007; Mathis et al., 2008; Dopson et al., 2016; Lusk et al., 2018a). These factors make the study of Gram-positive thermophilic exoelectrogens, in addition to well documented studies in Gram-negative mesophilic and thermophilic (Fu et al., 2013) exoelectrogens, essential for increasing our understanding of the function and origins for respiration on Earth, optimizing emerging biotechnologies, and creating new solutions to mitigate contamination remediation and energy development.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
The funding for the publication of this manuscript was provided by ScienceTheEarth, a 501(c)(3) Non-profit Organization which advocates International Science Literacy.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author would like to acknowledge Bruce E. Rittmann and Cesar I. Torres for their support in assisting him through his extreme transition to thermophilic microorganisms. The author would like to thank Shawn P. Mulvaney for his support in easing the transition from the thermophilic Arizona summers to the psychrophilic Washington DC winters. The author would also like to thank various cinematic and literary works whose influences can be found in the section titles. The author would also like to acknowledge Mary Shelley, whose book Frankenstein; or, the Modern Prometheus is currently in its bicentennial and was the inspiration for the title of this piece.
Some sections and ideas in this manuscript were first discussed in the author’s dissertation titled Thermophilic Microbial Electrochemical Cells. The complete reference can be found in the references section.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00818/full#supplementary-material
Abbreviations
BES, bioelectrochemical systems; BOD, biochemical oxygen demand; CA, chronoamperometry; CE, coulombic efficiency; COD, chemical oxygen demand; CV, cyclic voltammetry; Cyt, multiheme c-type cytochrome; D, diacylglycerol; DET, non-mediated direct electron transfer; DMK, demethylmenaquinone; DNA, deoxyribonucleic acid; EET, extracellular electron transfer; EIS, electrochemical impedance spectroscopy; EKA, redox potential of respiratory pathway; Fcc3, flavocytochrome C3; fs°, fraction of electrons received from electron donors that are used for cell maintenance and synthesis of cellular material; Ga, gigaannum or one billion years; GC content, guanine and/or cytosine content; HGT, horizontal gene transfer; Ka, non-synonymous substitutions; Kbio, conductivity of biomass; KM, substrate concentration at which the reaction rate is half of the maximum; Ks, synonymous substitutions; Lpp, lipoprotein; Mb, megabases or million base pairs; MET, mediated or indirect electron transfer; MHC, multiheme cytochrome; mV, millivolts; MxC, microbial electrochemical cell; NAG, N-Acetylglucosamine; NAM, N-Acetylmuramic acid; nm, nanometer; ω, Ka/Ks ratio; pKa, logarithmic acid dissociation constant; Q, quinone; RNA, ribonucleic acid; rut, metabolic rate; μ, growth rate; μm, micrometer; Y, biomass yield.
References
Al-Maghrabi, I. M. A., Bin Aqil, A. O., Chaalal, O., and Islam, M. R. (1999). Use of thermophilic bacteria for bioremediation of petroleum contaminants. Energy Sources 21, 17–19. doi: 10.1080/00908319950014920
Amend, J. P., and Shock, E. L. (2001). Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 25, 175–243. doi: 10.1016/S0168-6445(00)00062-0
Aravind, L., Tatusov, R. L., Wolf, Y. I., Walker, D. R., and Koonin, E. V. (1998). Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 14, 442–444. doi: 10.1016/s0168-9525(98)01553-4
Badalamenti, J. P., Krajmalnik-Brown, R., and Torres, C. I. (2013). Generation of high current densities by pure cultures of anode-respiring Geoalkalibacter spp. Under alkaline and saline conditions in microbial electrochemical cells. mBio 4:e00144-13. doi: 10.1128/mBio.00144-13
Bergquist, P. L., Lov, D. R., Croft, J. E., Streiff, M. B., Daniel, R. M., and Morgan, W. H. (1987). Genetics and potential biotechnological applications of thermophilic and extremely thermophilic microorganisms. Biotechnol. Genet. Eng. Rev. 5, 199–244. doi: 10.1080/02648725.1987.10647838
Beveridge, T. J., Forsberg, C. W., and Doyle, R. J. (1982). Major sites of metal binding in Bacillus licheniformis walls. J. Bacteriol. 150, 1438–1448.
Beveridge, T. J., and Murray, R. G. (1980). Sites of metal deposition in the cell wall of Bacillus subtilis. J. Bacteriol. 141, 876–887.
Bhatnagar, A., Kesari, K., and Shurpali, K. (2016). Multidisciplinary approaches to handling wastes in sugar industries. Water Air Soil Pollut. 227, 1–30.
Bird, L. J., Bonnefoy, V., and Newman, D. K. (2011). Bioenergetic challenges of microbial iron metabolisms. Trends Microbiol. 19, 330–340. doi: 10.1016/j.tim.2011.05.001
Bond, D., and Lovley, D. (2003). Electricity production by Geobacter sulfurreducens attached to electrodes. Appl. Environ. Microbiol. 69, 1548–1555. doi: 10.1128/AEM.69.3.1548-1555.2003
Boussau, B., and Gouy, M. (2006). Efficient likelihood computations with nonreversible models of evolution. Syst. Biol. 55, 756–768. doi: 10.1080/10635150600975218
Brock, T. D., and Freeze, H. (1969). Thermus aquaticus gen. n. and sp. n., a Nonsporulating Extreme Thermophile. J. Bacteriol. 98, 289–297.
Brownson, D. A. C., and Banks, C. E. (2014). The Handbook of Graphene Electrochemistry. London: Springer.
Bulat, S. A., Alekhina, I. A., Marie, D., Martins, J., and Petit, J. R. (2011). Searching for life in extreme environments relevant to Jovian’s Europa: lessons from subglacial ice studies at Lake Vostok (East Antarctica). Adv. Space Res. 48, 697–701. doi: 10.1016/j.asr.2010.11.024
Burra, P., Kalmar, L., Tompa, P., and Haslam, N. (2010). Reduction in structural disorder and functional complexity in the thermal adaptation of prokaryotes (Protein Disorder in Adaptation). PLoS One 5:e12069. doi: 10.1371/journal.pone.0012069
Carlson, H. K., Iavarone, A. T., Gorur, A., Yeo, B. S., Tran, R., Melnyk, R. A., et al. (2012). Surface multiheme c-type cytochromes from Thermincola potens and implications for respiratory metal reduction by Gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 109, 1702–1707. doi: 10.1073/pnas.1112905109
Carroll, J., Slupsky, J., and Mather, A. (1991). The solubility of carbon dioxide in water at low pressure. J. Phys. Chem. Ref. Data 20,1201–1209.
Chaalal, O., and Islam, M. R. (2001). Integrated management of radioactive strontium contamination in aqueous stream systems. J. Environ. Manage. 61, 51–59. doi: 10.1006/jema.2000.0399
Chang, M., and Kim, J. (2005). Improvement of a microbial fuel cell performance as a BOD sensor using respiratory inhibitors. Biosens. Bioelectron. 20, 1856–1859. doi: 10.1016/j.bios.2004.06.003
Ciccarelli, F. D., Doerks, T., Von Mering, C., Creevey, C. J., Snel, B., and Bork, P. (2006). Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–1287. doi: 10.1126/science.1123061
Corkrey, R., Mcmeekin, T., Bowman, J., Ratkowsky, D., Olley, J., and Ross, T. (2016a). Correction: the biokinetic spectrum for temperature. PLoS One 11:e0157804. doi: 10.1371/journal.pone.0157804
Corkrey, R., Mcmeekin, T., Bowman, J., Ratkowsky, D., Olley, J., and Ross, T. (2016b). The biokinetic spectrum for temperature. PLoS One 11:e0153343. doi: 10.1371/journal.pone.0153343
Costa, N. L., Carlson, H. K., Coates, J. D., Louro, R. O., and Paquete, C. M. (2015). Heterologous expression and purification of a multiheme cytochrome from a Gram-positive bacterium capable of performing extracellular respiration. Protein Expr. Purif. 111, 48–52. doi: 10.1016/j.pep.2015.03.007
Costa, N. L., Clarke, T. A., Philipp, A. L., Gescher, J., Louro, R. O., and Paquete, C. M. (2018). Electron transfer process in microbial electrochemical technologies: the role of cell-surface exposed conductive proteins. Bioresour. Technol. 255, 308–317. doi: 10.1016/j.biortech.2018.01.133
Coursolle, D., Baron, D. B., Bond, D. R., and Gralnick, J. A. (2010). The Mtr respiratory pathway is essential for reducing flavins and electrodes in Shewanella oneidensis. J. Bacteriol. 192, 467–474. doi: 10.1128/JB.00925-09
Das, S., Paul, S., Bag, S., and Dutta, C. (2006). Analysis of Nanoarchaeum equitans genome and proteome composition: Indications for hyperthermophilic and parasitic adaptation. BMC Genomics 7:186. doi: 10.1186/1471-2164-7-186
Deng, Y., Josberger, E., Jin, J., Roudsari, A. F., Helms, B. A., Zhong, C., et al. (2013). Corrigendum: H+-type and OH–type biological protonic semiconductors and complementary devices. Sci. Rep. 3:2481.
Dill, K. A., Ghosh, K., and Schmit, J. D. (2011). Physical limits of cells and proteomes. Proc. Natl. Acad. Sci. U.S.A. 108, 17876–17882. doi: 10.1073/pnas.1114477108
Dong, Y., Liao, M., Meng, X., and Somero, G. (2018). Structural flexibility and protein adaptation to temperature: molecular dynamics analysis of malate dehydrogenases of marine molluscs. Proc. Natl. Acad. Sci. U.S.A. 115, 1274–1279. doi: 10.1073/pnas.1718910115
Dopson, M., Ni, G., and Sleutels, T. H. (2016). Possibilities for extremophilic microorganisms in microbial electrochemical systems. FEMS Microbiol. Rev. 40, 164–181. doi: 10.1093/femsre/fuv044
Dufresne, A., Garczarek, L., and Partensky, F. (2005). Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 6:R14.
Dutta, A., and Chaudhuri, K. (2010). Analysis of tRNA composition and folding in psychrophilic, mesophilic and thermophilic genomes: indications for thermal adaptation. FEMS Microbiol. Lett. 305, 100–108. doi: 10.1111/j.1574-6968.2010.01922.x
Eddie, B. J., Wheng, Z., Hervey, W. J., Leary, D. H., Malanoski, A. P., Tender, L. M., et al. (2017). Metatranscriptomics supports the mechanism for biocathode electroautotrophy by “Candidatus tenderia” electrophaga. mSystems 2:e00002-17. doi: 10.1128/mSystems.00002-17
Edwards, M. J., Baiden, N. A., Johs, A., Tomanicek, S. J., Liang, L., Shi, L., et al. (2014). The X-ray crystal structure of Shewanella oneidensis OmcA reveals new insight at the microbe–mineral interface. FEBS Lett. 588, 1886–1890. doi: 10.1016/j.febslet.2014.04.013
Edwards, M. J., White, G. F., Lockwood, C. W., Lawes, M. C., Martel, A., Harris, G., et al. (2018). Structural modeling of an outer membrane electron conduit from a metal-reducing bacterium suggests electron transfer via periplasmic redox partners. J. Biol. Chem. 293, 8103–8112. doi: 10.1074/jbc.RA118.001850
Edwards, M. J., White, G. F., Norman, M., Tome-Fernandez, A., Ainsworth, E., Shi, L., et al. (2015). Redox linked flavin sites in extracellular decaheme proteins involved in microbe-mineral electron transfer. Sci. Rep. 5:11677. doi: 10.1038/srep11677
Efremov, R. G., and Sazanov, L. A. (2011). Structure of the membrane domain of respiratory complex I. Nature 476, 414–420. doi: 10.1038/nature10330
Ehrlich, H. L. (2008). Are gram-positive bacteria capable of electron transfer across their cell wall without an externally available electron shuttle? Geobiology 6, 220–224. doi: 10.1111/j.1472-4669.2007.00135.x
Fenchel, T., King, G., and Blackburn, T. H. (2012). Bacterial Biogeochemistry: The Ecophysiology of Mineral Cycling, 3rd Edn. Boston, MA: Academic Press. doi: 10.1111/j.1472-4669.2007.00135.x
Firer-Sherwood, M., Ando, N., Drennan, C., and Elliott, S. (2011). Solution-based structural analysis of the decaheme cytochrome, MtrA, by small-angle X-ray scattering and analytical ultracentrifugation. J. Phys. Chem. B 115, 11208–11214. doi: 10.1021/jp203603r
Friedman, R. L., Hughes, A. W., and Drake, J. (2004). Genome-wide patterns of nucleoticle substitution reveal stringent functional constraints on the protein sequences of thermophiles. Genetics 167, 1507–1512. doi: 10.1534/genetics.104.026344
Fu, Q., Kobayashi, H., Kawaguchi, H., Wakayama, T., Maeda, H., and Sato, K. (2013). A thermophilic gram-negative nitrate-reducing bacterium, Calditerrivibrio nitroreducens, exhibiting electricity generation capability. Environ. Sci. Technol. 47, 12583–12590. doi: 10.1021/es402749f
Giovannoni, S. J., Tripp, H. J., Givan, S., Podar, M., Vergin, K. L., Baptista, D., et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309, 1242–1245. doi: 10.1126/science.1114057
Girguis, P. R., and Fundis, A. (2018). Peering into the abyss: studying our own ocean to advance astrobiology. Oceanography 31, 32–34.
Glansdorff, N., Xu, Y., and Labedan, B. (2008). The last universal common ancestor: emergence, constitution and genetic legacy of an elusive forerunner. Biol. Direct 3:29. doi: 10.1186/1745-6150-3-29
Gu, J., and Hilser, V. (2009). Sequence-based analysis of protein energy landscapes reveals nonuniform thermal adaptation within the proteome. Mol. Biol. Evol. 26, 2217–2227. doi: 10.1093/molbev/msp140
Hamelers, H. V. M., ter Heijne, A., Stein, N., Rozendal, R. A., and Buisman, C. J. N. (2011). Butler–Volmer–Monod model for describing bio-anode polarization curves. Bioresour. Technol 102, 381–387. doi: 10.1016/j.biortech.2010.06.156
Harold, F. M. (1986). The Vital Force: A Study of Bioenergetics. New York, NY: Freeman. doi: 10.1016/j.biortech.2010.06.156
Hobot, J. A., Carlemalm, E., Villiger, W., and Kellenberger, E. (1984). Periplasmic gel: new concept resulting from the reinvestigation of bacterial cell envelope ultrastructure by new methods. J. Bacteriol. 160, 143–152.
Holmes, D. E., Walker, D. J. F., Dang, Y., and Lovley, D. R. (2016). The electrically conductive pili of Geobacter species are a recently evolved feature for extracellular electron transfer. Microb. Genom. 2:e000072. doi: 10.1099/mgen.0.000072
Hurst, L. (2002). The Ka/ Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18, 486–487. doi: 10.1016/s0168-9525(02)02722-1
Hurst, L. D., and Merchant, A. R. (2001). High guanine-cytosine content is not an adaptation to high temperature: a comparative analysis amongst prokaryotes. Proc. R. Soc. B Biol. Sci. 268, 493–497. doi: 10.1098/rspb.2000.1397
Islas, S., Velasco, A., Becerra, A., Delaye, L., and Lazcano, A. (2003). Hyperthermophily and the origin and earliest evolution of life. Int. Microbiol. 6, 87–94. doi: 10.1007/s10123-003-0113-4
Kashefi, K., and Lovley, D. R. (2003). Extending the upper temperature limit for life. Science 301:934. doi: 10.1126/science.1086823
Katuri, K. P., Kavanagh, P., Rengaraj, S., and Leech, D. (2010). Geobacter sulfurreducens biofilms developed under different growth conditions on glassy carbon electrodes: insights using cyclic voltammetry. Chem. Commun. 46, 4758–4760. doi: 10.1039/c003342a
Knoll, A. H. (2003). Life on a Young Planet: the First Three Billion Years of Evolution on Earth. Princeton, NJ: Princeton University Press.
Koch, A. L. L. (2006). The Bacteria: Their Origin, Structure, Function and Antibiosis. Berlin: Springer.
Leang, C., Coppi, M., and Lovley, D. (2003). OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 185, 2096–2103. doi: 10.1128/JB.185.7.2096-2103.2003
Leary, D. H., Hervey, W. J., Malanoski, A. P., Wang, Z., Eddie, B. J., Tender, G. S., et al. (2015). Metaproteomic evidence of changes in protein expression following a change in electrode potential in a robust biocathode microbiome. Proteomics 15, 3486–3496. doi: 10.1002/pmic.201400585
Lee, H., Torres, C., and Rittmann, B. (2009). Effects of substrate diffusion and anode potential on kinetic parameters for anode-respiring bacteria. Environ. Sci. Technol. 43, 7571–7577. doi: 10.1021/es9015519
Li, W. F., Zhou, X. X., and Lu, P. (2005). Structural features of thermozymes. Biotechnol. Adv. 23, 271–281. doi: 10.1016/j.biotechadv.2005.01.002
Lies, D., Hernandez, M., Kappler, A., Mielke, R., Gralnick, J., and Newman, D. (2005). Shewanella oneidensis MR-1 uses overlapping pathways for iron reduction at a distance and by direct contact under conditions relevant for biofilms. Appl. Environ. Microbiol. 71, 4414–4426. doi: 10.1128/AEM.71.8.4414-4426.2005
Light, S., Su, L., Rivera-Lugo, R., Cornejo, J., Louie, A., Iavarone, A., et al. (2018). A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature 562, 140–144. doi: 10.1038/s41586-018-0498-z
Liu, Y., Climent, V., Berná, A., and Feliu, J. M. (2011). Effect of temperature on the catalytic ability of electrochemically active biofilm as anode catalyst in microbial fuel cells. Electroanalysis 23, 387–394. doi: 10.1002/elan.201000499
Logan, B. E. (2009). Exoelectrogenic bacteria that power microbial fuel cells. Nat. Rev. Microbiol. 7, 375–381. doi: 10.1038/nrmicro2113
Logan, B. E., Hamelers, B., Rozendal, R., and Schroder, U. (2006). Microbial fuel cells: methodology and technology. Environ. Sci. Technol. 40, 5181–5192.
Lovley, D. R. (1993a). Dissimilatory metal reduction. Annu. Rev. Microbiol. 47, 263–290. doi: 10.1146/annurev.mi.47.100193.001403
Lovley, D. R. (1993b). Geobacter metallireducens gen. nov. sp. Nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159, 336–344. doi: 10.1007/bf00290916
Lovley, D. R. (2008). The microbe electric: conversion of organic matter to electricity. Curr. Opin. Biotechnol. 19, 564–574. doi: 10.1016/j.copbio.2008.10.005
Lusk, B. G., Badalamenti, J. P., Parameswaran, P., Bond, D. R., and Torres, C. I. (2015a). Draft genome sequence of the gram-positive Thermophilic iron reducer Thermincola ferriacetica strain Z-0001T. Genome Announc. 3:e01072-15. doi: 10.1128/genomeA.01072-15
Lusk, B. G., Colin, A., Parameswaran, P., Rittmann, B. E., and Torres, C. I. (2018a). Simultaneous fermentation of cellulose and current production with an enriched mixed culture of thermophilic bacteria in a microbial electrolysis cell. Microb. Biotechnol. 1751–7915. doi: 10.1111/1751-7915.12733
Lusk, B. G., Khan, Q. F., Parameswaran, P., Hameed, A., Ali, N., Rittmann, B. E., et al. (2015b). Characterization of electrical current-generation capabilities from Thermophilic bacterium Thermoanaerobacter pseudethanolicus using xylose, glucose, cellobiose, or acetate with fixed anode potentials. Environ. Sci. Technol. 49, 14725–14731. doi: 10.1021/acs.est.5b04036
Lusk, B. G., Parameswaran, P., Popat, S. C., Rittmann, B. E., and Torres, C. I. (2016). The effect of pH and buffer concentration on anode biofilms of Thermincola ferriacetica. Bioelectrochemistry. 112, 47–52. doi: 10.1016/j.bioelechem.2016.07.007
Lusk, B. G., Peraza, I., Albal, G., Marcus, A. K., Popat, S. C., and Torres, C. I. (2018b). pH dependency in anode Biofilms of Thermincola ferriacetica suggests a proton-dependent electrochemical response. J. Am. Chem. Soc. 140, 5527–5534. doi: 10.1021/jacs.8b01734
Lusk, B. G., Torres, C. I., Krajmalnik-Brown, R., and Rittmann, B. E. (2015c). Thermophilic Microbial Electrochemical Cells. Ph.D. theses, Arizona State University, Tempe, AZ. doi: 10.1021/jacs.8b01734
Makarieva, A., Gorshkov, V., and Li, B. (2005). Energetics of the smallest: Do bacteria breathe at the same rate as whales? Proc. Biol. Sci. 272, 2219–2224. doi: 10.1098/rspb.2005.3225
Marcus, A. K., Torres, C. I., and Rittmann, B. E. (2007). Conduction-based modeling of the biofilm anode of a microbial fuel cell. Biotechnol. Bioeng. 98, 1171–1182. doi: 10.1002/bit.21533
Marcus, A. K., Torres, C. I., and Rittmann, B. E. (2011). Analysis of a microbial electrochemical cell using the proton condition in biofilm (PCBIOFILM) model. Bioresour. Technol. 102, 253–262. doi: 10.1016/j.biortech.2010.03.100
Margesin, R., and Schinner, F. (2001). Biodegradation and bioremediation of hydrocarbons in extreme environments. Appl. Microbiol. Biotechnol. 56, 650–663. doi: 10.1007/s002530100701
Marsili, E., Baron, D. B., Shikhare, I. D., Coursolle, D., Gralnick, J. A., and Bond, D. R. (2008). Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U.S.A. 105, 3968–3973. doi: 10.1073/pnas.0710525105
Marsili, E., Sun, J., and Bond, D. R. (2010). Voltammetry and growth physiology of Geobacter sulfurreducens biofilms as a function of growth stage and imposed electrode potential. Electroanalysis 22, 865–874. doi: 10.1002/elan.200800007
Mathis, B., Marshall, C., Milliken, C., Makkar, R., Creager, S., and May, H. (2008). Electricity generation by thermophilic microorganisms from marine sediment. Appl. Microbiol. Biotechnol. 78, 147–155. doi: 10.1007/s00253-007-1266-4
Meruelo, A., Han, S., Kim, S., and Bowie, J. (2012). Structural differences between thermophilic and mesophilic membrane proteins. Protein Sci. 21, 1746–1753. doi: 10.1002/pro.2157
Mohan, V. S., Velvizhi, G., Krishna, V. K., and Babu, L. M. (2014). Microbial catalyzed electrochemical systems: a bio-factory with multi-facet applications. Bioresour. Technol. 165, 355–364. doi: 10.1016/j.biortech.2014.03.048
Mook, W. G., and Koene, B. K. S. (1975). Chemistry of dissolved inorganic carbon in estuarine and coastal brackish waters. Estuar. Coast. Mar. Sci. 3, 325–336. doi: 10.1016/0302-3524(75)90032-8
Morgado, L., Paixao, V. B., Schiffer, M., Pokkuluri, P. R., Bruix, M., and Salgueiro, C. A. (2011). Revealing the structural origin of the redox-Bohr effect: the first solution structure of a cytochrome from Geobacter sulfurreducens. Biochem. J. 441, 179–187. doi: 10.1042/BJ20111103
Moya, A., Gil, R., Latorre, A., Peretó, J., Pilar Garcillán-Barcia, M., and De La Cruz, F. (2009). Toward minimal bacterial cells: evolution vs. design. FEMS Microbiol. Rev. 33, 225–235. doi: 10.1111/j.1574-6976.2008.00151.x
Mulvankar, N. S., Vargas, M., Nevin, K., Tremblay, P., Evans-Lutterodt, K., Nykypanchuk, D., et al. (2015). Structural basis for metallic-like conductivity in microbial nanowires. mBio 6:e00084. doi: 10.1128/mBio.00084-15
Nealson, K. H., and Conrad, P. G. (1999). Life: past, present and future. Philos. Trans. R. Soc. B Biol. Sci. 354, 1923–1939. doi: 10.1098/rstb.1999.0532
Nelson, K. E., Clayton, R. A., Gill, S. R., Gwinn, M. L., Dodson, R. J., Haft, D. H., et al. (1999). Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399, 323–329. doi: 10.1038/20601
Niu, L., Song, L., Liu, X., and Dong, X. (2009). Tepidimicrobium xylanilyticum sp. nov., an anaerobic xylanolytic bacterium, and emended description of the genus Tepidimicrobium. Int. J. Syst. Evol. Microbiol. 59, 2698–2701. doi: 10.1099/ijs.0.005124-0
Okur, H., Hladílková, J., Rembert, K., Cho, Y., Heyda, J., Dzubiella, J., et al. (2017). Beyond the Hofmeister series: ion-specific effects on proteins and their biological functions. J. Phys. Chem. B 121, 1997–2014. doi: 10.1021/acs.jpcb.6b10797
Onyenwoke, R., Kevbrin, V., Lysenko, A., and Wiegel, J. (2007). Thermoanaerobacter pseudethanolicus sp. nov., a thermophilic heterotrophic anaerobe from Yellowstone National Park. Int. J. Syst. Evol. Microbiol. 57, 2191–2193. doi: 10.1099/ijs.0.65051-0
Pace, N. (1991). Origin of life-facing up to the physical setting. Cell 65, 531–533. doi: 10.1016/0092-8674(91)90082-a
Pankratova, G., Leech, D., Gorton, L., and Hederstedt, L. (2018). Extracellular electron transfer by the gram-positive bacterium Enterococcus faecalis. Biochemistry 57, 4597–4603. doi: 10.1021/acs.biochem.8b00600
Parameswaran, P., Bry, T., Popat, S. C., Lusk, B. G., Rittmann, B. E., and Torres, C. I. (2013). Kinetic, electrochemical, and microscopic characterization of the thermophilic, anode-respiring bacterium Thermincola ferriacetica. Environ. Sci. Technol. 47, 4934–4940. doi: 10.1021/es400321c
Partin, C. A., Bekker, A., Planavsky, N. J., Scott, C. T., Gill, B. C., Li, C., et al. (2013). Large-scale fluctuations in precambrian atmospheric and oceanic oxygen levels from the record of U in Shales. Earth Planet. Sci. Lett. 284–293. doi: 10.1016/j.epsl.2013.03.031
Patel, A., Matsakas, L., Rova, U., and Christakopoulos, P. (2019). A perspective on biotechnological applications of thermophilic microalgae and cyanobacteria. Bioresour. Technol. 278, 424–434. doi: 10.1016/j.biortech.2019.01.063
Peng, S., Liang, D., Diao, P., Liu, Y., Lan, F., Yang, Y., et al. (2013). Nernst-ping-pong model for evaluating the effects of the substrate concentration and anode potential on the kinetic characteristics of bioanode. Bioresour. Technol. 136, 610–616. doi: 10.1016/j.biortech.2013.03.073
Pham, T. H., Boon, N., Aelterman, P., Clauwaert, P., De Schamphelaire, L., Vanhaecke, L., et al. (2008). Metabolites produced by Pseudomonas sp. enable a Gram-positive bacterium to achieve extracellular electron transfer. Appl. Microbiol. Biotechnol. 77, 1119–1129. doi: 10.1007/s00253-007-1248-6
Pirbadian, S., Barchinger, S. E., Kar, M. L., Hye, S. B., Jangir, Y., Bouhenni, R. A., et al. (2014). Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc. Natl. Acad. Sci. U.S.A. 111, 12883–12888. doi: 10.1073/pnas.1410551111
Puigbò, P., Wolf, Y. I., and Koonin, E. V. (2009). Search for a ’Tree of Life’ in the thicket of the phylogenetic forest. J. Biol. 8:59. doi: 10.1186/jbiol159
Qiao, Y., Li, C. M., Lu, Z., Ling, H., Kang, A., and Chang, M. W. (2009). A time-course transcriptome analysis of Escherichia coli with direct electrochemistry behavior in microbial fuel cells. Chem. Commun. 41, 6183–6185. doi: 10.1039/b912003c
Reardon, P., and Mueller, K. (2013). Structure of the type IVa major pilin from the electrically conductive bacterial nanowires of Geobacter sulfurreducens. J. Biol. Chem. 288, 29260–29266. doi: 10.1074/jbc.M113.498527
Renslow, R., Babauta, J., Kuprat, A., Schenk, J., Ivory, C., Fredrickson, J., et al. (2013). Modeling biofilms with dual extracellular electron transfer mechanisms. Phys. Chem. Chem. Phys. 15, 19262–19283. doi: 10.1039/c3cp53759e
Richter, H., Mccarthy, K., Nevin, K. P., Johnson, J. P., Rotello, V. M., and Lovley, D. R. (2008). Electricity generation by Geobacter sulfurreducens attached to gold electrodes. Langmuir 24, 4376–4379. doi: 10.1021/la703469y
Rodgers, R., and Hill, G. (1978). Equations for vapour pressure versus temperature: derivation and use of the Antoine equation on a hand-held programmable calculator. Br. J. Anaesth. 50, 415–424. doi: 10.1093/bja/50.5.415
Rodrigues, M., Oliveira, T., Pereira, I., and Archer, M. (2006). X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination. EMBO J. 25, 5951–5960. doi: 10.1038/sj.emboj.7601439
Sabath, N., Ferrada, E., Barve, A., and Wagner, A. (2013). Growth temperature and genome size in bacteria are negatively correlated, suggesting genomic streamlining during thermal adaptation. Genome Biol. Evol. 5, 966–977. doi: 10.1093/gbe/evt050
Saunders, N. F., Thomas, T., Curmi, P. M., Mattick, J. S., Kuczek, E., Slade, R., et al. (2003). Mechanisms of thermal adaptation revealed from the genomes of the Antarctic Archaea Methanogenium frigidum and Methanococcoides burtonii. Genome Res. 13, 1580–1588. doi: 10.1101/gr.1180903
Schröder, U. (2007). Anodic electron transfer mechanisms in microbial fuel cells and their energy efficiency. Phys. Chem. Chem. Phys. 9, 2619–2629. doi: 10.1039/b703627m
Seckbach, J. (2004). Origins Genesis, Evolution and Diversity of Life. Dordrecht: Kluwer. doi: 10.1039/b703627m
Shi, L., Dong, H., Reguera, G., Beyenal, H., Lu, A., Liu, J., et al. (2016). Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 14, 651–662. doi: 10.1038/nrmicro.2016.93
Singh, S. P., and Singh, P. (2014). Effect of CO2 concentration on algal growth: a review. Renew. Sustain. Energy Rev. 38, 172–179. doi: 10.1016/j.rser.2014.05.043
Skjelhaugen, O. (1999). Thermophilic aerobic reactor for processing organic liquid wastes. Water Res. 33, 1593–1602. doi: 10.1016/s0043-1354(98)00422-9
Sleep, N. H. (2018). Geological and geochemical constraints on the origin and evolution of life. Astrobiology 18, 1199–1219. doi: 10.1089/ast.2017.1778
Slepova, T. V., Sokolova, T. G., Kolganova, T. V., Tourova, T. P., and Bonch-Osmolovskaya, E. (2009). Carboxydothermus siderophilus sp. nov., a thermophilic, hydrogenogenic, carboxydotrophic, dissimilatory Fe(III)-reducing bacterium from a Kamchatka hot spring. Int. J. Syst. Evol. Microbiol. 59, 213–217. doi: 10.1099/ijs.0.000620-0
Slobodkin, A. (2005). Thermophilic microbial metal reduction. Microbiology 74, 501–514. doi: 10.1007/s11021-005-0096-6
Slobodkin, A. I., Tourova, T., Kostrikina, N., Lysenko, A., German, K., Bonch-Osmolovskaya, E., et al. (2006). Tepidimicrobium ferriphilum gen. nov., sp. nov., a novel moderately thermophilic, Fe(III)-reducing bacterium of the order Clostridiales. Int. J. Syst. Evol. Microbiol. 56, 369–372. doi: 10.1099/ijs.0.63694-0
Srikanth, S., Marsili, E., Flickinger, M. C., and Bond, D. R. (2008). Electrochemical characterization of Geobacter sulfurreducens cells immobilized on graphite paper electrodes. Biotechnol. Bioeng. 99, 1065–1073. doi: 10.1002/bit.21671
Strycharz, S., Malanoski, A., Snider, R., Yi, H., Lovley, D., and Tender, L. (2011). Application of cyclic voltammetry to investigate enhanced catalytic current generation by biofilm-modified anodes of Geobacter sulfurreducens strain DL1 vs. variant strain KN400. Energy Environ. Sci. 4, 896–913. doi: 10.1039/c0ee00260g
Subramanian, P., Pirbadian, S., El-Naggar, M., and Jensen, G. (2018). Ultrastructure of Shewanella oneidensis MR-1 nanowires revealed by electron cryotomography. Proc. Natl. Acad. Sci. U.S.A. 115, E3246–E3255. doi: 10.1073/pnas.1718810115
Tan, Y., Adhikari, R. Y., Malvankar, N. S., Ward, J. E., Woodard, T. L., Nevin, K. P., et al. (2017). Expressing the Geobacter metallireducens PilA in Geobacter sulfurreducens Yields Pili with Exceptional Conductivity. mBio 8:e02203-16 doi: 10.1128/mBio.02203-16
Taylor, P., Pealing, S. L., Reid, G. A., Chapman, S. K., and Walkinshaw, M. D. (1999). Structural and mechanistic mapping of a unique fumarate reductase. Nat. Struct. Biol. 6, 1108–1112.
Torres, C. I., Kato Marcus, A., and Rittmann, B. E. (2008). Proton transport inside the biofilm limits electrical current generation by anode-respiring bacteria. Biotechnol. Bioeng. 100, 872–881. doi: 10.1002/bit.21821
Torres, C. I., Marcus, A. K., Lee, H., Parameswaran, P., Krajmalnik-Brown, R., and Rittmann, B. E. (2010). A kinetic perspective on extracellular electron transfer by anode-respiring bacteria. FEMS Microbiol. Rev. 34, 3–17. doi: 10.1111/j.1574-6976.2009.00191.x
Turner, P., Mamo, G., and Karlsson, E. (2007). Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb. Cell Fact. 6, 9–21. doi: 10.1186/1475-2859-6-9
Van Noort, V., Bradatsch, B., Arumugam, M., Amlacher, S., Bange, G., Creevey, C., et al. (2013). Consistent mutational paths predict eukaryotic thermostability. BMC Evol. Biol. 13:7. doi: 10.1186/1471-2148-13-7
Varshney, P., Sohoni, S., Wangikar, P., and Beardall, P. (2016). Effect of high CO2 concentrations on the growth and macromolecular composition of a heat- and high-light-tolerant microalga. J. Appl. Phycol. 28, 2631–2640. doi: 10.1007/s10811-016-0797-4
Vecchia, D. E., Shao, P. P., Suvorova, E., Chiappe, D., Hamelin, R., and Bernier-Latmani, R. (2014). Characterization of the surfaceome of the metal-reducing bacterium Desulfotomaculum reducens. Front. Microbiol. 5:432. doi: 10.3389/fmicb.2014.00432
Ventura, M., Canchaya, C., Tauch, A., Chandra, G., Fitzgerald, G. F., Chater, K. F., et al. (2007). Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548. doi: 10.1128/MMBR.00005-07
Vesth, T., Ozen, A., Andersen, S., Kaas, R., Lukjancenko, O., Bohlin, J., et al. (2013). Veillonella, Firmicutes: microbes disguised as Gram negatives. Stand. Genomic Sci. 9, 431–448. doi: 10.4056/sigs.2981345
Vieille, C., and Zeikus, G. J. (2001). Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65, 1–43. doi: 10.1128/mmbr.65.1.1-43.2001
Vieille, C., and Zeikus, J. (1996). Thermozymes: identifying molecular determinants of protein structural and functional stability. Trends Biotechnol. 14, 183–190. doi: 10.1016/0167-7799(96)10026-3
Wagner, A. (2005). Energy constraints on the evolution of gene expression. Mol. Biol. Evol. 22, 1365–1374. doi: 10.1093/molbev/msi126
Wang, F., Gu, Y., O’Brien, J. P., Yi, S. M., Yalcin, S. E., Srikanth, V., et al. (2019). Structure of microbial nanowires reveals stacked hemes that transport electrons over micrometers. Cell 177, 361–369. doi: 10.1016/j.cell.2019.03.029
Wang, Q., Cen, Z., and Zhao, J. (2015). The survival mechanisms of thermophiles at high temperatures: an angle of Omics. Physiology 30, 97–106. doi: 10.1152/physiol.00066.2013
Weiss, M., Preiner, M., Xavier, J., Zimorski, V., and Martin, W. (2018). The last universal common ancestor between ancient Earth chemistry and the onset of genetics. PLoS Genet. 4:e1007518. doi: 10.1371/journal.pgen.1007518
Wiegel, J., and Michael, A. W. W. (eds) (1998). Thermophiles: the Keys to the Molecular Evolution and the Origin of Life? London: Taylor and Francis. doi: 10.1371/journal.pgen.1007518
Wrighton, K. C., Thrash, J. C., Melnyk, R. A., Bigi, J. P., Byrne-Bailey, K., Remis, J. P., et al. (2011). Evidence for direct electron transfer by a gram-positive bacterium isolated from a microbial fuel cell. Appl. Environ. Microbiol. 77, 7633–7639. doi: 10.1128/AEM.05365-11
Yang, Y., Xu, M., Guo, J., and Sun, G. (2012). Bacterial extracellular electron transfer in bioelectrochemical systems. Process Biochem. 47, 1707–1714. doi: 10.1016/j.procbio.2012.07.032
Yang, Z., and Bielawski, J. P. (2000). Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15, 496–503. doi: 10.1016/s0169-5347(00)01994-7
Yates, M. D., Strycharz-Glaven, S. M., Golden, J. P., Roy, J., Tsoi, S., Erickson, J. S., et al. (2016). Measuring conductivity of living Geobacter sulfurreducens biofilms. Nat. Nanotechnol. 11, 910–913. doi: 10.1038/nnano.2016.186
Yilmazel, Y. D., Zhu, X., Kim, Y., Holmes, D. E., and Logan, B. E. (2018). Electrical current generation in microbial electrolysis cells by hyperthermophilic archaea Ferroglobus placidus and Geoglobus ahangari. Bioelectrochemistry 119, 142–149. doi: 10.1016/j.bioelechem.2017.09.012
Yoho, R., Popat, S., and Torres, C. (2014). Dynamic potential-dependent electron transport pathway shifts in anode biofilms of Geobacter sulfurreducens. Chemsuschem 7, 3413–3419. doi: 10.1002/cssc.201402589
Yoho, R. A., Popat, S. C., Rago, L., Guisasola, A., and Torres, C. I. (2015). Anode Biofilms of Geoalkalibacter ferrihydriticus exhibit electrochemical signatures of multiple electron transport pathways. Langmuir 31, 12552–12559. doi: 10.1021/acs.langmuir.5b02953
Zacharoff, L., Chan, C. H., and Bond, D. R. (2016). Reduction of low potential electron acceptors requires the CbcL inner membrane cytochrome of Geobacter sulfurreducens. Bioelectrochemistry 107, 7–13. doi: 10.1016/j.bioelechem.2015.08.003
Zavarzina, D., Sokolova, T., Tourova, T., Chernyh, N., Kostrikina, N., and Bonch-Osmolovskaya, E. (2007). Thermincola ferriacetica sp. nov., a new anaerobic, thermophilic, facultatively chemolithoautotrophic bacterium capable of dissimilatory Fe(III) reduction. Extremophiles 11, 1–7. doi: 10.1007/s00792-006-0004-7
Zhang, H., Chen, X., Braithwaite, D., He, Z., and Mormile, M. (2014). Phylogenetic and metagenomic analyses of substrate-dependent bacterial temporal dynamics in microbial fuel cells. PLoS One 9:e107460. doi: 10.1371/journal.pone.0107460
Zuber, B., Haenni, M., Ribeiro, T., Minnig, K., Lopes, F., Moreillon, P., et al. (2006). Granular layer in the periplasmic space of gram-positive bacteria and fine structures of Enterococcus gallinarum and Streptococcus gordonii septa revealed by cryo-electron microscopy of vitreous sections. J. Bacteriol. 188, 6652–6660. doi: 10.1128/jb.00391-06
Keywords: extremophile, thermophile, extracellular electron transfer, bioelectrochemical system, Gram-positive, biotechnology, biogeochemistry, astrobiology
Citation: Lusk BG (2019) Thermophiles; or, the Modern Prometheus: The Importance of Extreme Microorganisms for Understanding and Applying Extracellular Electron Transfer. Front. Microbiol. 10:818. doi: 10.3389/fmicb.2019.00818
Received: 26 November 2018; Accepted: 01 April 2019;
Published: 26 April 2019.
Edited by:
Pascal E. Saikaly, King Abdullah University of Science and Technology, Saudi ArabiaReviewed by:
Tim Magnuson, Idaho State University, United StatesSuman Bajracharya, King Abdullah University of Science and Technology, Saudi Arabia
Yasemin Dilsad Yilmazel, Rochester Institute of Technology, United States
Copyright © 2019 Lusk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bradley G. Lusk, YnJhZGxleS5sdXNrQGFzdS5lZHU=
 Bradley G. Lusk
Bradley G. Lusk