- Department of Analytical Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Kraków, Poland
The use of antibiotics on a mass scale, particularly in farming, and their release into the environment has led to a rapid emergence of resistant bacteria. Once emerged, resistance determinants are spread by horizontal gene transfer among strains of the same as well as disparate bacterial species. Their accumulation in free-living as well as livestock and community-associated strains results in the widespread multiple-drug resistance among clinically relevant species posing an increasingly pressing problem in healthcare. One of these clinically relevant species is Staphylococcus aureus, a common cause of hospital and community outbreaks. Among the rich diversity of mobile genetic elements regularly occurring in S. aureus such as phages, pathogenicity islands, and staphylococcal cassette chromosomes, plasmids are the major mean for dissemination of resistance determinants and virulence factors. Unfortunately, a vast number of whole-genome sequencing projects does not aim for complete sequence determination, which results in a disproportionately low number of known complete plasmid sequences. To address this problem we determined complete plasmid sequences derived from 18 poultry S. aureus strains and analyzed the prevalence of antibiotic and heavy metal resistance determinants, genes of virulence factors, as well as genetic elements relevant for their maintenance. Some of the plasmids have been reported before and are being found in clinical isolates of strains typical for humans or human ones of livestock origin. This shows that livestock-associated staphylococci are a significant reservoir of resistance determinants and virulence factors. Nevertheless, nearly half of the plasmids were unknown to date. In this group we found a potentially mobilizable plasmid pPA3 being a unique example of accumulation of resistance determinants and virulence factors likely stabilized by a presence of a toxin–antitoxin system.
Introduction
Growing antibiotic resistance among clinically relevant bacteria is currently becoming a grave concern of global health. The shrinking array of effective antibacterial drugs elevates mortality and increases frequency and scale of outbreaks (De Jager et al., 2015; Strauß et al., 2017; Dogramachy, 2018; Gu et al., 2018; Douglas et al., 2019). One of the most commonly addressed bacterial species in this context is Staphylococcus aureus. As an opportunistic pathogen S. aureus can colonize healthy individuals and reside asymptomatically as a commensal. The estimated carriage rate in communities reaches between 25 and 30% (Charlebois et al., 2002; Levy et al., 2005; Kuehnert et al., 2006; Li et al., 2014; Morgenstern et al., 2016). Regardless of its commensal nature, in predisposed individuals, such as newborns, young children, the elderly, immuno-compromised, post-surgical, or hospitalized ones, S. aureus is often a cause of difficult to treat and not rarely fatal as well as chronic infections, especially in clinical setting (Anstead et al., 2014; Savini et al., 2016). Community and livestock-associated S. aureus populations are of utmost importance for genetic elements exchange, in particular for dissemination of antibiotic determinants, and thus reservoirs of potentially life-threatening strains (Vandenesch et al., 2003; Armand-Lefevre et al., 2005; Voss et al., 2005; Nimmo, 2012; Planet et al., 2013; Strauß et al., 2017). For all these reasons S. aureus is listed by the World Health Organization as one among several bacterial species of high clinical relevance. These species form a group called ESKAPE (Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter). Each of them poses a serious threat to people’s health (Pendleton et al., 2013). With regard to S. aureus, the greatest concerns are methicillin-resistant S. aureus (MRSA), resistant to an array of β-lactams, as well as vancomycin intermediate/resistant S. aureus (VISA and VRSA), resistant to one of last resort drugs, vancomycin. However, the spread of other antibiotic resistance determinants is not to be underestimated.
Continuing emergence of resistance determinants against novel drugs seems to be only a smaller part of a bigger picture. In fact, their rapid spread is what truly aggravates the problem in healthcare. When speaking about dissemination of resistance determinants, the role of mobile genetic elements (MGEs) and mechanisms determining their mobility should be considered. In staphylococci these include predominantly: transposons, insertion sequences, and integrons (Malachowa and Deleo, 2010; Domingues et al., 2012; Alibayov et al., 2014; Partridge et al., 2018), which shuffle genetic material within a cell and facilitate spread and acquisition of diverse determinants and virulence factors. For this sort of MGEs, is it necessary to be located in autonomously replicating units such as bacterial chromosome or other MGEs such as plasmids or phages. The latter are vectors for inter-cellular exchange of genetic material. Plasmids are usually incorporated by bacterial cells from the environment in an active manner by transformation (Fagerlund et al., 2014) or exchanged between cells either by conjugation (O’Brien et al., 2014; Ramsay et al., 2016) or phage transduction, which appears to be of particular relevance for plasmid transfer in staphylococci, including clinically relevant strains as USA300 (Varga et al., 2012, 2016). In turn, phages and likely phage-derived pathogenicity islands and staphylococcal cassette chromosomes seem to be mostly spread by transduction (Deghorain and Van Melderen, 2012; Scharn et al., 2013). Such a wide range of possibilities may justify a supposition that any resistance determinant may emerge anywhere and then be quickly spread among different bacterial species of diverse host or environment-specificity. Mass scale use of antibacterial drugs, especially in farming, increases the pressure for resistance determinants spread in natural environment and their exchange among free-living bacteria as well as animal commensals (Martinez, 2009; Fluit, 2012; He et al., 2014; Cuny et al., 2017; Kumar et al., 2017; Lau et al., 2017). Temporary colonization of humans by animal strains leading to the interaction with human microbiota or stable colonization (host jump) are not a rare phenomenon and the last part of the route (Armand-Lefevre et al., 2005; Voss et al., 2005; Bosch et al., 2016). Indeed, commensal strains of the same species or genus constitute a vast reservoir for antibiotic resistance determinants for those displaying pathogenic potential as it is observed for staphylococci (Tulinski et al., 2012; Hung et al., 2015; Morgenstern et al., 2016; Savini et al., 2016).
Apart from antibiotic resistance determinants, MGEs facilitate spread of heavy metal resistance and genes encoding virulence factors. The latter are a diverse group of mostly proteins and peptides that facilitate colonization and infection progress. These include for instance: cell-wall-anchored surface proteins interacting with host proteins, extracellular proteases, lipases, nucleases, super-antigens or cytotoxic proteins, and peptides (Marques et al., 1989; Takeuchi et al., 1999; Argudín et al., 2010; Berends et al., 2010; Bukowski et al., 2013; Thammavongsa et al., 2013; Wladyka et al., 2015; Bonar et al., 2016). Acquisition of genes coding for host-specific virulence factors, for instance in a plasmid, seems to be of importance for successful colonization of a new host by staphylococci and contributes to a phenomenon referred to as a host jump (Lowder et al., 2009; Fluit, 2012; McCarthy et al., 2014). Stable or transient livestock-to-human colonization events are continually being reported, particularly in farming environments where workers are exposed to frequent interaction with animals and the environment of their living. Importantly, such strains are also able to spread via human-to-human interaction (Armand-Lefevre et al., 2005; Voss et al., 2005; Huijsdens et al., 2006; Fluit, 2012; Wendlandt et al., 2013).
Nonetheless, emergence and dissemination of either antibiotic resistance determinants or virulence factors’ genes do not comprise the full picture. The last piece, out of the crucial ones, seems to be their stable maintenance. This is of particular importance to those encoded on low-copy-number plasmids (Sengupta and Austin, 2011; Dmowski and Jagura-Burdzy, 2013). Once acquired, in the absence of selective environmental pressure, plasmids carrying resistance determinants are either gradually lost or stably maintained when internal pressure is exerted. It has been conclusively demonstrated that such self-maintenance may be provided by toxin–antitoxin (TA) systems. In fact, the role of TA systems, in the context of stable genetic material maintenance in bacteria, is well-documented (Tsuchimoto et al., 1988; Sobecky et al., 1996; del Solar et al., 1998; Van Melderen and Saavedra De Bast, 2009; Bukowski et al., 2011, 2013). A number of recent reports point out their possible importance for drug resistance dissemination as well, for instance among enterococci (Moritz and Hergenrother, 2007; Rosvoll et al., 2010; Bukowski et al., 2017). However, it still seems that their role in this phenomenon has yet not received due attention.
In this study, complete sequences of 16 different plasmids, including 7 unknown to date, derived from 18 poultry S. aureus strains are reported along with a detailed analysis of distribution of antibiotic and heavy metal resistance determinants as well as other relevant factors among them. Moreover, the physiological effect of such determinants’ presence is carefully examined. Finally, a unique example is presented of plasmids’ shuffling and fragment exchange leading to accumulation of drug and heavy metal resistance determinants along with a TA operon in a likely mobilizable plasmid pPA3.
Materials and Methods
Bacterial Strains and Cultures
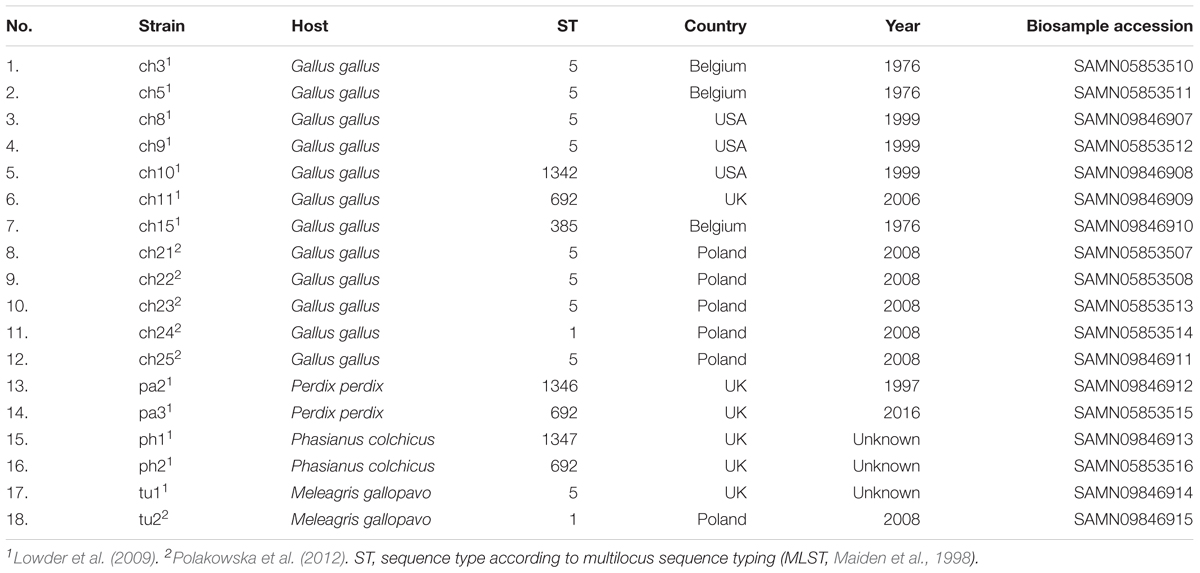
Poultry strains of S. aureus used in this study have been described previously elsewhere (Lowder et al., 2009; Polakowska et al., 2012; Bonar et al., 2016, 2018). The summary is included in Table 1. Bacteria were cultured in tryptic soy broth (TSB) or on tryptic soy agar (TSA) at 37°C unless otherwise described.
Plasmid DNA Isolation
Three 2 ml samples of overnight cultures were collected and centrifuged at 5000 RCF. Supernatants were discarded and each pellet was re-suspended in 200 μl of EC buffer (Smith and Cantor, 1987) supplemented with 1 μl of RNAse A (10 mg/ml, Thermo Scientific) and 20 μl of lysostaphin (1 mg/ml, Madry et al., Unpublished), and then incubated in 37°C for 30 min. Samples were further subjected to plasmid isolation by alkaline lysis and purification on silica spin columns using Plasmid Mini kit (A&A Biotechnology) according to the manufacturer’s protocol. All lysates were pooled on one spin column and plasmid DNA was eluted in 50 μl of sterile, double-distilled, and filtered water. The obtained DNA concentrations ranged from 300 to 900 ng/μl.
Sequencing and de novo Assembly
Plasmid sequencing was performed by Genomed (Warsaw, Poland) using the Illumina MiSeq system. The obtained data were processed in CLC Genomics Workbench, version 8.5.1 (Qiagen/CLC Bio). Reads were analyzed, trimmed, and filtered using Trim Sequences tool from the NGS Core Tools package, at default parameter values, and subsequently assembled into contigs using de novo Assembly tool. The assembly was carried out with scaffolding and automatic pair distance detection and default values for other parameters. Contig sequences were examined for homology to existing staphylococcal plasmids and their fragments using in-house Python 3 scripts, which utilized nucleotide BLAST from the NCBI BLAST+ toolkit, version 2.3.0 (Camacho et al., 2009), and ordered. In case of absence of an adequate reference sequence, the order of contigs was determined by PCR amplification. Each primer was designed to hybridize ca. 200 bp upstream each end of a contig. The primers were used in different combinations to assess the possible arrangement of contigs. The gaps were closed by Sanger sequencing.
Sequence Comparative Analysis
Annotation was performed by using in-house Python 3 scripts and protein BLAST search on staphylococcal protein sequences available in NCBI Protein database, accessed on 14 January 2019, as well as RPS-BLAST search on NCBI Conserved Domain Database, accessed on 14 January 2019 (Marchler-Bauer et al., 2017). The annotation was manually revised and corrected. The complete and annotated plasmid sequences were deposited in GenBank. The accession numbers are included in Table 2. Plasmid pPA3 and homology of its fragments to existing plasmids were determined using nucleotide BLAST and visualized using Circos, version 0.69-3 (Krzywinski et al., 2009). Putative oriT mimics were searched using the core sequence of those from conjugative plasmids (O’Brien et al., 2015). The structure of oriT mimic was generated using mfold web server for single-stranded linear DNA at default parameters (Zuker, 2003) and visualized in CLC Main Workbench, version 12.0.1 (Qiagen/CLC Bio). Other visualizations were prepared using CLC Main Workbench and finished in GIMP image editor.
Phylogenetic Analyses
Multiple sequence alignments were prepared using CLC Main Workbench with the following parameter values for both DNA and protein sequences: gap open cost, 20.0; gap extension cost, 1.0; end gap cost, free; alignment mode, very accurate. Phylogenetic trees were constructed and visualized in CLC Main Workbench for gapless segments of multiple sequence alignments using maximum-likelihood phylogeny tool with the following parameter values: construction method, neighbor joining; nucleotide substitution model, general time reversible, GTR; transition/transversion ratio, 2.0; include rate variation, yes; number of substitution rate categories, 8; gamma distribution parameter, 1.0; estimate substitution rate parameter(s), yes; estimate topology, yes; estimate gamma distribution parameter, yes; perform bootstrap analysis, yes; and replicates, 100.
Detection of Alternative pAvY Plasmids
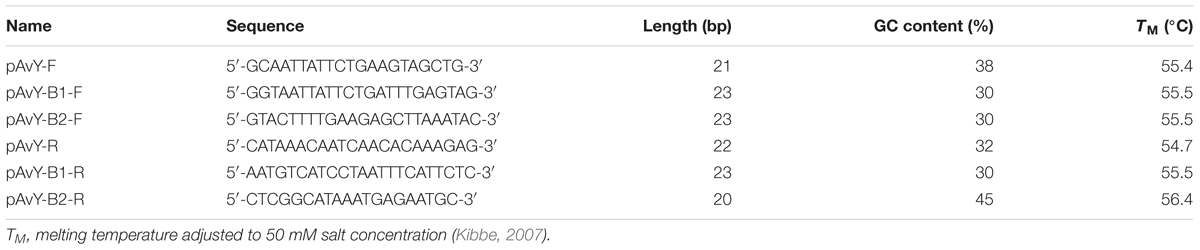
The detection of pAvY, pAvY-B1, and pAvY-B2 plasmids was carried out by PCR reaction using primers listed in Table 3. RUN DNA Polymerase (A&A Biotechnology) was used according to the manufacturer protocol with elongation time 1 min and annealing temperature 60°C.
Antibiotic Resistance Determination
A few bacterial colonies were sampled from an overnight agar plate, suspended in 0.9% NaCl solution and diluted to 0.5 density in McFarland scale using DEN-1B McFarland Densitometer (Biosan). Suspensions were streaked on Mueller Hinton Agar (MHA) plates (Oxoid) using Eddy Jet 2 automatic spiral plater (IUL) in the Lawn 3000 mode. Plates were dried shortly and MIC Test Strips (Liofilchem) were carefully placed on them. The incubation conditions and measurement were carried out according to the manufacturer’s protocol and the interpretation of the results according to EUCAST 2015 (European Committee for Antibiotic Susceptibility Testing) and CLSI 2015 (Clinical and Laboratory Standards Institute) recommendations, which in most cases meant that the results were interpreted after 24 h incubation in 37°C. The analyzed strains always fell into the sensitive or resistant group regardless of whether CLSI or EUCAST interpretation was followed. For the lack of CLSI or EUCAST interpretation for streptomycin, a previous report was used to interpret the results (Archer, 1978).
Heavy Metal Resistance Determination
Fresh liquid cultures were prepared by dilution of overnight ones at 1:100 ratio followed by incubation in the same conditions to reach the optical density at 600 nm close to 0.6. All cultures were adjusted to the optical density of 0.6 and diluted 100-fold. 100 μl samples were transferred to 96-well plates and mixed with 100 μl of heavy metal inorganic compound (CdCl2, Na2HAsO4) solutions in TSB. Twofold serial dilutions in the concentration range from 10 to 160 μM in case of CdCl2 and 10-fold dilutions in range of 20 μM to 20 mM in case of Na2HAsO4 were used. Subsequently plates were incubated at 37°C for 20 h and the minimal inhibitory concentration (MIC) was determined as the lowest heavy metal compound concentration where no bacterial growth was observed. S. aureus RN4220 was used as a reference non-resistant to heavy metals. The results were interpreted according to previous reports (Ji and Silver, 1992; Crupper et al., 1999; Chudobova et al., 2015).
Results
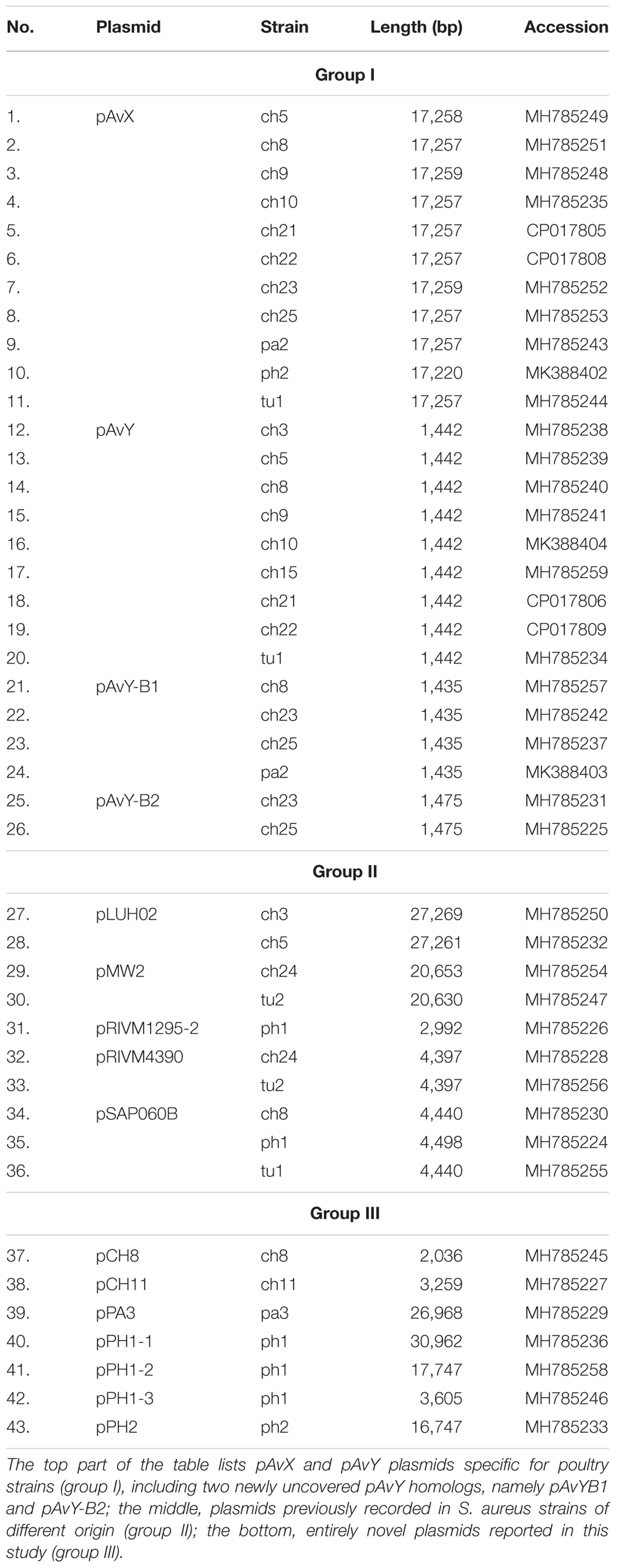
Among 18 strains that were analyzed in the present study 43 occurrences of 16 different plasmids have been reported. Based on their novelty and prevalence among analyzed strains, the plasmids were divided into three distinct groups: (I) poultry-associated plasmids; (II) plasmids of previously known sequences occurring in S. aureus strains of diverse host-specificity; and (III) plasmids entirely unknown to date and characterized in this study (Table 4).
Poultry-Associated pAvX and pAvY (Group I)
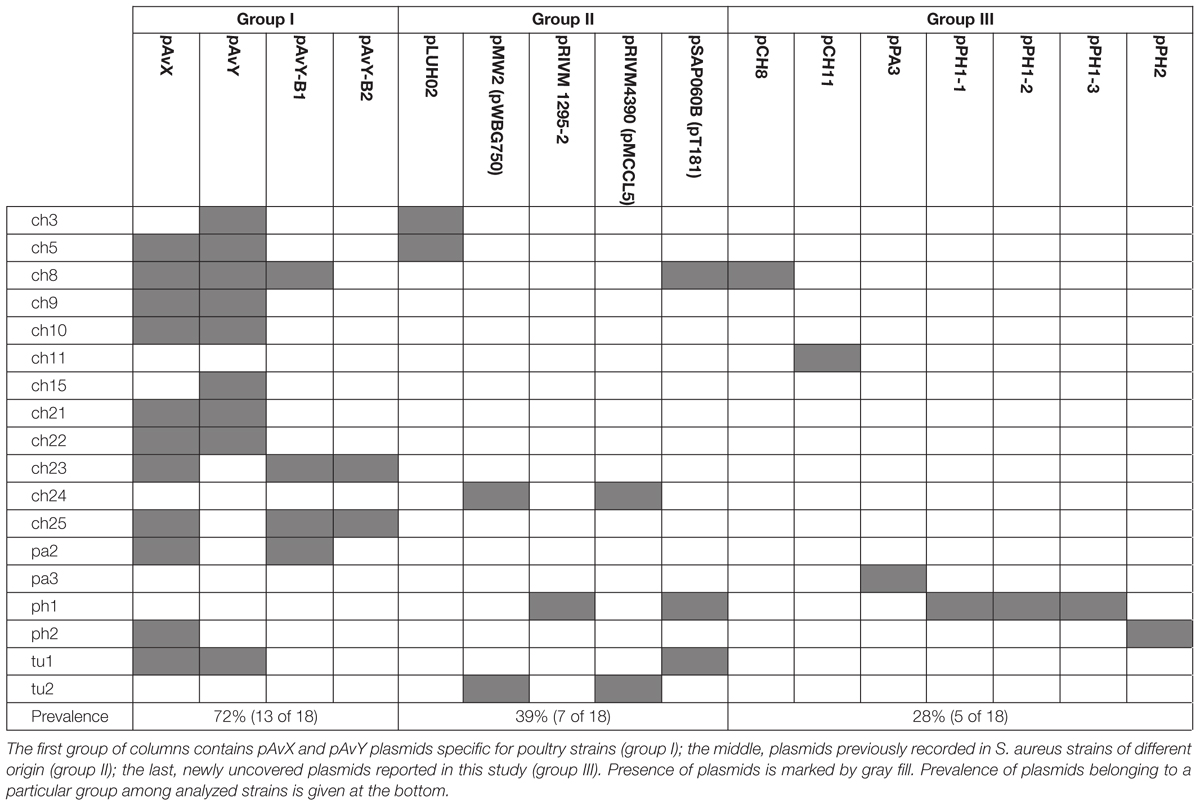
The first group contains known plasmids characteristic for poultry strains. These are pAvX and pAvY (Lowder et al., 2009) as well as two of pAvY’s variants (pAvY-B1 and pAvY-B2), uncovered in this study, which clearly belongs to distinct phylogenetic groups (Figure 1). Remarkably, all three pAvY group variants occur independently and can co-exist next to each other in the same strain, which was independently verified by PCR (Figure 2) to exclude unlikely, however possible, NGS assembly artifacts. In general, the poultry-associated group contains the most frequently occurring plasmids with total prevalence of 72% of the analyzed strains, 61% for pAvX, and 67% for pAvYs. Although frequently occurring, these plasmids do not carry any resistance determinants. However, their presence correlates with virulence (Lowder et al., 2009; Polakowska et al., 2012).
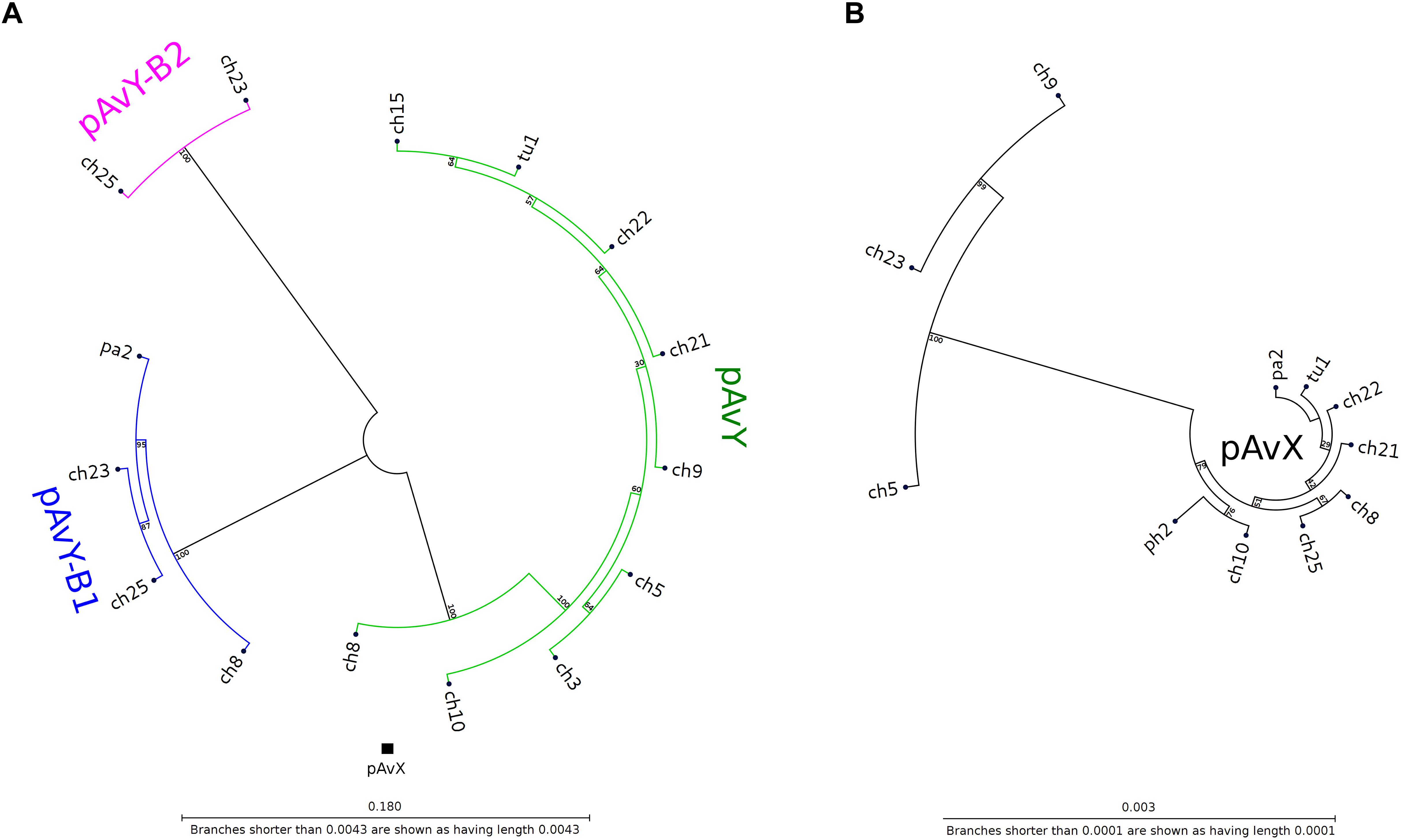
Figure 1. Phylogenetic trees of pAvY group and pAvX plasmids. Panel (A): based on the phylogenetic analysis, pAvY group plasmids might be divided into three distinctive subgroups: common pAvY next to less frequently occurring pAvY-B1 and pAvY-B2. Panel (B): a corresponding tree for pAvX plasmids. These plasmids are highly conserved among all strains when compared to pAvY group. Note that for readability the tree is presented in 60-fold bigger scale. A small black box under the tree in the panel A shows its size in the scale used for pAvY plasmids. On each branch the bootstrap value is given.
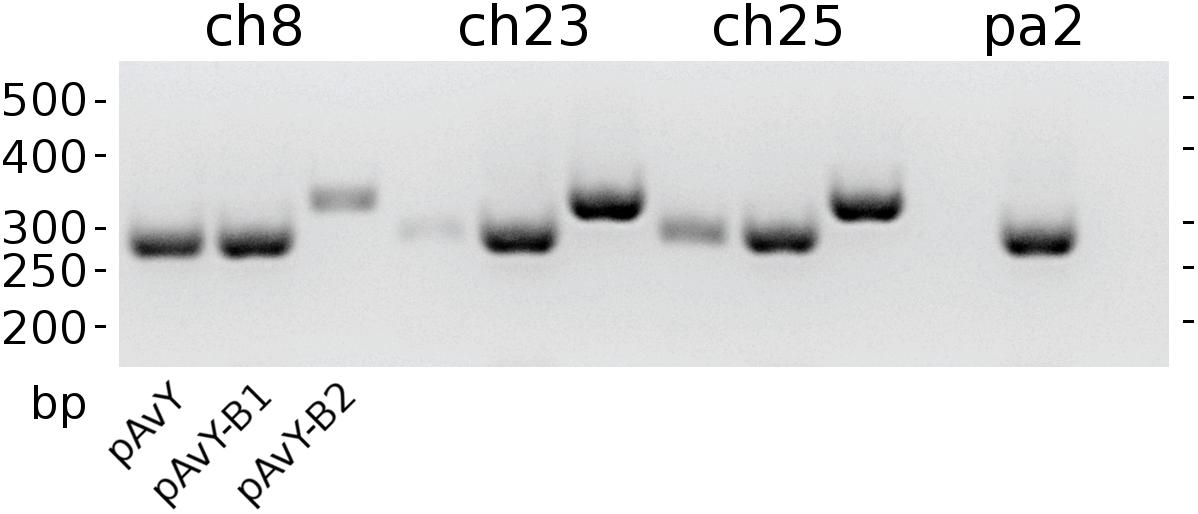
Figure 2. The occurrence of pAvY group plasmids. Expected products’ lengths are: 278 bp for pAvY and pAvY-B1; 321 bp for pAvY-B2. Specific PCR products are visible for pAvY and pAvY-B1 for ch8 strain; pAvY-B1 and pAvY-B2 for ch23 and ch25 strains; and pAvY-B1 for pa2 strain. For ch8, ch23, and ch25, there are clearly weaker signals from PCR products for the remaining variants, which likely are unspecific products.
As regards virulence factors, in pAvX there are genes coding for those of well-documented role (Table 5) such as staphopain A2 (Wladyka et al., 2011a,b; Kalińska et al., 2012) and lysophospholipase (Doery et al., 1965; Marques et al., 1989; Daugherty and Low, 1993). Next to them there is also an operon of pemIK-Sa1, which is the most frequently occurring in staphylococcal plasmids TA system of mazEF/pemIK family (Bukowski et al., 2017) and belongs to class II TA systems with the toxin being a sequence-specific RNase (Bukowski et al., 2013).
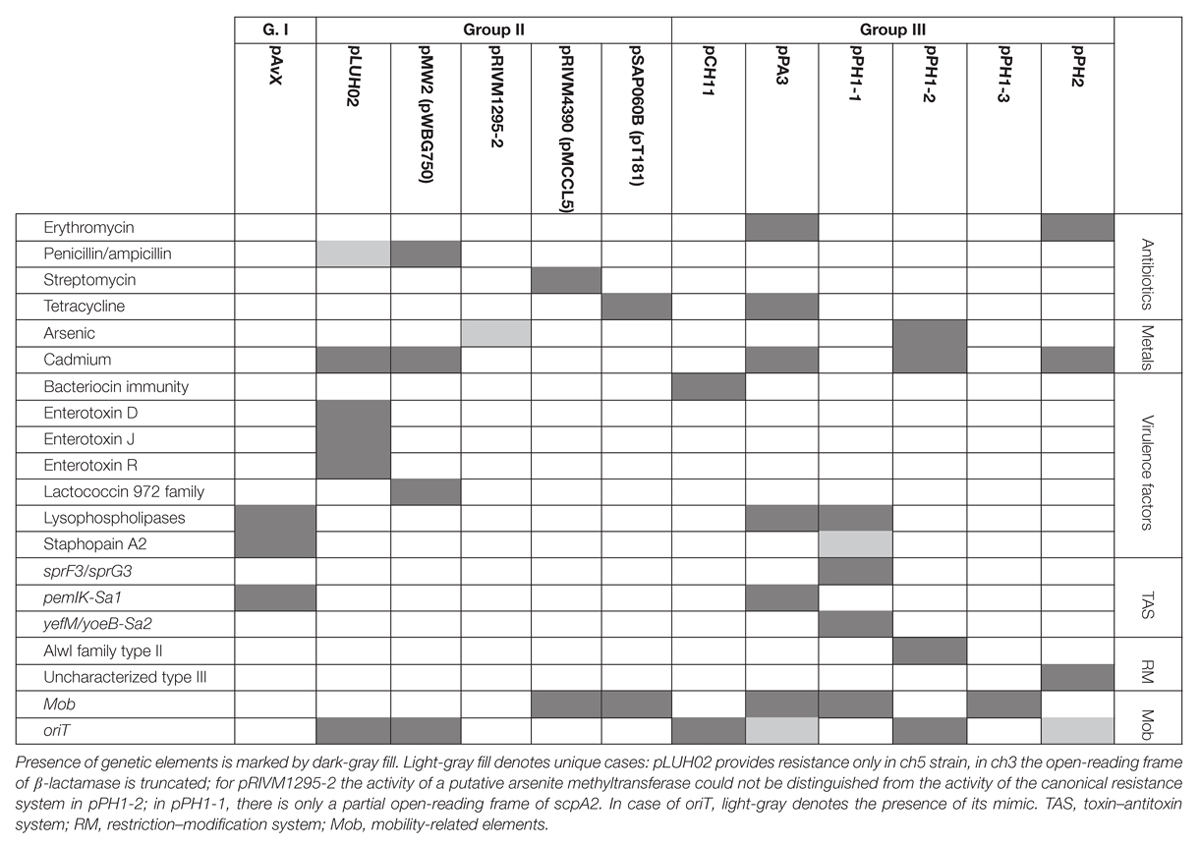
Table 5. Distribution of antibiotic and heavy metal resistance determinants, genes of virulence factors, as well as genetic elements relevant for their maintenance among the three groups of analyzed plasmids.
Plasmids of Known Sequences (Group II)
The second group includes five known plasmids that have been reported so far in S. aureus strains of different origin. The prevalence of these plasmids reaches 39% of analyzed strains.
While the prevalence rate of 39% for this group is by more than one-third lower than that for pAvX and pAvYs (72%), these plasmids are frequent carriers of antibiotic resistance determinants against drugs from groups of aminoglycosides (streptomycin, pRIVM4390), β-lactams (penicillin, pMW2), macrolides (erythromycin, pRIVM1295-2), and tetracyclines (tetracycline, pSAP060B), next to resistance determinants to cadmium (pLUH02 and pMW2) or putatively to arsenic (pRIVM1295-2). Except the strain ch3, for the reasons discussed further, all the remaining carrier strains displayed corresponding resistant phenotypes (Table 5 and Supplementary Tables S1, S2).
Regarding genes of known virulence factors, only pLUH02 carries a cluster of genes of enterotoxins D, J, and R. In pMW2, however, there is a gene of a yet uncharacterized lactococcin 972 family bacteriocin. Bacteriocins facilitate the producer host or environment colonization by eliminating other bacteria. Nevertheless, some bacteriocins exhibit properties of virulence factors (Wladyka et al., 2015). The neighborhood of the aforementioned gene suggests that it is a part of the whole operon, providing immunity proteins and transporters necessary for excretion. Interestingly, plasmids pLUH02 and pMW2 possess oriT, which potentially renders them mobilizable.
Plasmids Unknown to Date (Group III)
The last group consists of plasmids that have not as yet been reported. Prevalence of these plasmids among the analyzed strains reaching 28% is the lowest when compared to the previous groups. However, these are as frequent carriers of genes and determinants, whose prevalence is analyzed in this study, as the second group. Their sequences have been entirely unknown to date with the exception of pPA3. The sequence of pPA3 is, to a considerable extent, a mosaic of sequences derived from other plasmids, each of them provides a unique element and each is flanked by shuffle-inducing sequences such as genes of transposases or invertases (Figure 3).
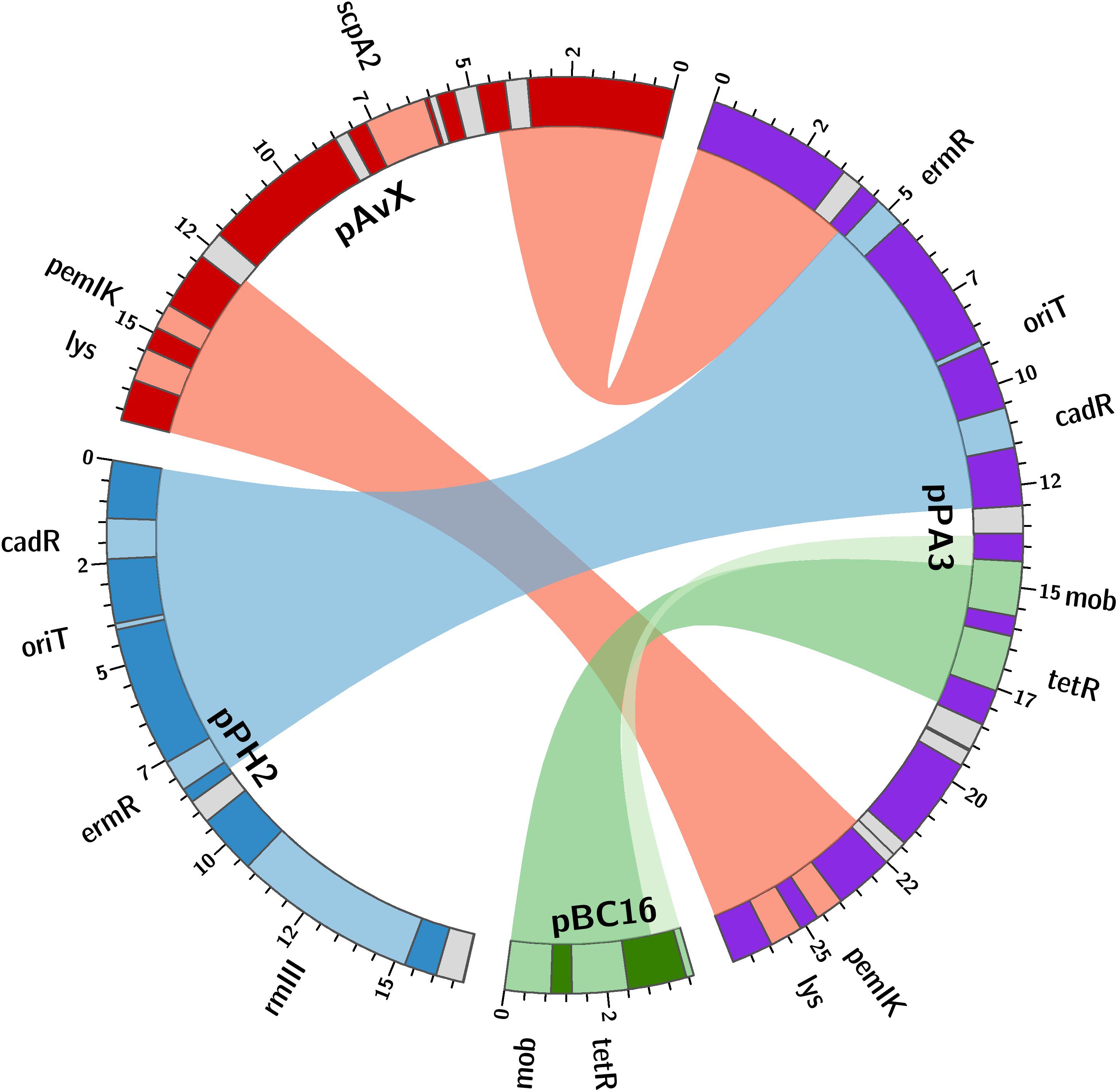
Figure 3. The structure of pPA3 plasmid. The plasmid is composed of elements originating from three different plasmids: common for staphylococcal poultry strains pAvX donates lysophospholipase gene (lys) and pemIK-Sa1 TA system operon (pemIK); pPH2, reported in this study, donates a functional erythromycin and cadmium resistance determinants (ermR and cadR) as well as the uncovered in this study oriT mimic (oriT); pBC16 is entirely incorporated into pPA3 and next to mobilization protein gene (mob) provides a functional tetracycline resistance determinant (tetR). The uncharacterized type III RM system (rmIII) and staphopain A2 (scpA2) operons are also depicted in pPH2 and pAvX, respectively. Light gray boxes denote genes of transposases and recombinases.
As regards resistance determinants, pPA3 carries two of such, against erythromycin and tetracycline. The erythromycin resistance determinant is located in a fragment originating from another unknown to date and reported in this study plasmid, namely pPH2, whereas the tetracycline resistance determinant originates from pBC16, a broad-host plasmid. Both, pPA3 and pPH2, contain determinants of cadmium resistance. Cadmium resistance determinants, together with one of resistance against arsenic, are also located in pPH1-2.
Concerning genes of possible virulence factors, pCH11 carries an immunity determinant against a bacteriocin. However, the authors were unable to find any gene coding for a bacteriocin in this plasmid, which would mean the existing gene provides immunity to one or more bacteriocins produced by other strains. Apart from this gene, pCH11, pPA3, and pPH1-1 possess one encoding lysophospholipase of very high similarity to the one encoded in pAvX.
As for TA systems and functionally related to them restriction–modification (RM) systems (Makarova et al., 2013; Mruk and Kobayashi, 2014), pPA3 contains an operon of pemIK-Sa1 localized in a fragment originating from pAvX. Strikingly, pPH1-1 possesses operons of two unrelated TA system. These are yefM/yoeB-Sa2, which is also class II TA system with the toxin being an RNase (Yoshizumi et al., 2009; Nolle et al., 2013), and a homolog of sprF3/sprG3, which is a class I TA system with the toxin being a bactericidal peptide and the antitoxin an antisense RNA for the toxin transcript (Germain-Amiot et al., 2018; Riffaud et al., 2018). Noteworthy, whereas pemIK-Sa1 prevail mostly in plasmids (Bukowski et al., 2017), yefM/yoeB-Sa2 is mostly chromosome-located (Yoshizumi et al., 2009; Nolle et al., 2013) and sprF/sprG type systems have been reported so far in staphylococcal pathogenicity islands that carry virulence factors and antibiotic resistance (Pichon and Felden, 2005; Riffaud et al., 2018). Plasmids pPH1-2 and pPH2 carry operons of RM systems. The former of type II and AlwI family, whose close homologs are present in a few known staphylococcal genomes, and more distant ones in genomes of other closely-related Gram-positive genera of Firmicutes phylum such as Bacillus, Enterococcus, and Streptococcus. The latter a type III RM system whose closest homologs may be found in Salinicoccus, a genus of free-living, halotolerant bacteria from Staphylococcaceae family, and more distant ones in genera of Bacillus, Enterococcus, Geobacillus, Staphylococcus, and Streptococcus. Neither of these RM systems nor their close homologs have yet been characterized.
Regarding the possibility of horizontal transfer, pCH11, pPA3, pPH1-2, and pPH2 are likely mobilizable as all possess oriT or its mimic. Plasmid pPA3 also contains a gene of mobilization protein. Notably, in case of pPA3, oriT mimic is located within a fragment originating from pPH2 but the gene of mobilization protein comes from pBC16 (Figure 3). Two other plasmids co-existing with pPH1-2, which are pPH1-1 and pPH1-3, encode mobilization protein in trans in respect to pPH1-2.
Discussion
Plasmids seem to be the principle means of dissemination of antibiotic and heavy metal resistance as well as host-specific virulence factors. However, the contemporary abundance of genomic data does not necessarily facilitate tracing their spread in bacterial populations. The main reason is the incompleteness of the genomic sequences. Out of 10,268 assemblies available for S. aureus on 29 January 2019, only 6.3%, are complete sequences (Supplementary Data S1). The remaining genomic sequences are deposited as sets of contigs assembled based on NGS raw data. For contigs it is challenging or even impossible to assess whether a genome contains plasmids. As a result, for more than 10,000 genomic sequences there are only 430 complete plasmid sequences available for S. aureus today (Supplementary Data S1). The magnitude of this estimation closely corresponds with recent reports (O’Brien et al., 2015; Kwong et al., 2017) and has not changed for last 7 years (Lozano et al., 2012).
The frequent presence of group I plasmids, namely pAvX and pAvY, in poultry strains has been reported before (Lowder et al., 2009). The former is a carrier of an operon encoding for cysteine protease staphopain A2 (Takeuchi et al., 1999, 2002; Wladyka et al., 2011a; Kalińska et al., 2012). This protease has been suggested to be a host-specific virulence factor for poultry strains and the transfer of its operon in pAvX to strains of human origin to facilitate a documented interspecies jump from a human to a poultry host (Lowder et al., 2009). The presence of TA system pemIK-Sa1 likely stabilizes its maintenance as the system has been demonstrated to display required properties (Bukowski et al., 2013). Conversely, homologs of group II plasmids, pRIVM4390 (pMCCL5) and pRIVM1295-2 (GenBank accessions CP013623 and CP013618), were reported recently in an opposite context, namely in livestock-associated MRSA (LA-MRSA) strains transmitted from farmed animals to humans in Netherlands, which dated back to 2003 (Bosch et al., 2016). It is noteworthy that pRIVM1295-2 found here in ph1 strain, which belongs to sequence type 1347 (ST1347) according to multilocus sequence typing (MLST, Maiden et al., 1998), was reported before by Lowder et al. (2009) regarding a single human-to-poultry host jump and radiation among broiler chicken of unique to Poland, human ST5 clonal lineage that was roughly estimated to have occurred around 1970s. Two other strains analyzed in our study, ch24 and tu2 (ST1), are MRSA isolated in Poland in 2008 (Polakowska et al., 2012) and carry pRIVM4390. It suggests that these two plasmids are spread among LA-MSSA as well as LA-MRSA and have migrated among different lineages for many decades contributing to resistance determinants dissemination. Plasmid pRIVM4390 was linked by Bosch et al. (2016) to resistance against aminoglycosides: neomycin and kanamycin; and in this study against streptomycin as well (Table 5). The other plasmid, pRIVM1295-2, was not documented to provide any antibiotic resistance. However, we found that one of its genes codes for a protein distantly similar to arsenite methyltransferases, which may comprise an accessory detoxification system to the one present in group III plasmid pPH1-2 of the same strain ph1 (Table 5), which provides resistance against arsenite as well as arsenate. Strain ph1 indeed displays elevated resistance to arsenate (Supplementary Table S2).
The analyzed in this study group II encompasses plasmids of known sequences that were originally reported in human isolates. The plasmid named here pSAP060B carries a tetracycline resistance determinant and occurs in three strains: ch8 (ST5), ph1 (ST1347), and tu1 (ST1). Its exact copy (GenBank accession GQ900417) was reported originally in 2010 in S. aureus SAP060B strain in a research focused on emergence of plasmid-related resistance against non-β-lactams among isolates of S. aureus USA300 strain (McDougal et al., 2010). S. aureus USA300 (ST8) remains of a particular interest as it is a community-associated MRSA (CA-MRSA) responsible for invasive infections across the United States (Moran et al., 2006; Tenover et al., 2006). The sequence of pSAP060B is nearly identical to one of the earliest sequenced staphylococcal plasmid pT181, GenBank accession J01764 (Khan and Novick, 1983). In both cases, it was reported for human clinical subjects and appears to be a frequently occurring plasmid among S. aureus clinical isolates (Águila-Arcos et al., 2017). Nevertheless, pT181 has been reported in human-origin poultry strains before (Lowder et al., 2009), and in this study among three strains of different sequence types, which demonstrates its frequent prevalence among unrelated lineages. A similar example is plasmid pMW2 (pWBG750) carrying a penicillinase gene. It was first reported in a study on the whole genome sequence of highly virulent human CA-MRSA strain MW2 (ST1), which was a cause of fatal septicemia and septic arthritis in a 16-month-old child in the United States in 1998 (Baba et al., 2002). Here we found the plasmid in two aforementioned LA-MRSA strains, namely ch24 and tu2, which markedly are of the same sequence type (ST1). This shows that plasmids that provide resistance determinants and were initially associated with human strains may be stably maintained in the bacterial population after a host jump, spread among livestock, and contribute to this vast reservoir of resistance determinants (Armand-Lefevre et al., 2005; Voss et al., 2005; He et al., 2014; Lau et al., 2017). The next plasmid that supports this conclusion is pLUH02, which we found in ch3 and ch5 strains of the ST5 lineage originating from a human-to-poultry host jump (Lowder et al., 2009). A homolog of pLUH02 (GenBank accession FR714929) has been uncovered among multi-resistant methicillin-susceptible S. aureus (MR-MSSA) isolates from a clinical outbreak in Sweden in 2009 (Lindqvist et al., 2009, 2012). Despite the fact that the isolates were MSSA, the authors argue that they likely originated from MRSA in which the major extent of SCCmec cassette, a MGE carrying determinants of resistance against multiple β-lactams, was lost from the chromosome. For ch5, pLUH02 provides penicillin/ampicillin resistance. In case of ch3 this resistance is not manifested, which is likely a result of a single nucleotide deletion within β-lactamase gene. Nevertheless, in both cases, pLUH02 possesses functional determinants providing resistance against cadmium and an array of genes coding for virulence factors, namely for enterotoxins D, J, and R (Casman et al., 1967; Zhang et al., 1998; Omoe et al., 2003). A series of plasmids carrying the same cluster of enterotoxin genes was reported before Lindqvist et al. (2012) by Omoe et al. (2003) as pIB485-like plasmids. Indeed, pLUH02 belongs to this group (Shearer et al., 2011). Worth mentioning in this context is also a novel plasmid pCH8. It is the most similar to a small plasmid pHSSA0406 (GenBank accession KR870311) described recently in a study analyzing distribution of fosfomycin resistance among MRSA strains isolated in a clinical setting from blood or cerebrospinal fluid (Fu et al., 2016). Nevertheless, the determinant of fosfomycin resistance is lost from pCH8.
Next to known plasmids, previously unknown ones, comprising the group III, are reported in this study as a considerable part of the whole plasmid pool, which may suggests that there is still a significant number of yet uncovered plasmids in S. aureus strains. Among 16 different plasmid sequences reported here, 7 are novel. Among these sequences, there are a few of particular interest. The first one is the newly established group of pAvY plasmids, which replaces what was reported before as one pAvY plasmid (Lowder et al., 2009). The meaning of plasmids from pAvY group for S. aureus strains is elusive. The originally reported pAvY carries only two genes. One coding for a short hypothetical protein, unique to pAvY, and another for a replication protein. Two remaining variants of the plasmid uncovered in this study, namely pAvY-B1 and B2, carry only the gene for the replication protein. Sequence relatedness of these plasmids is strictly reflected in sequences of this protein (Supplementary Figure S1), which may render their replication machineries independent and could explain their stable co-maintenance. The role of pAvY group plasmids remains a mystery. They appear not to carry any relevant genes, yet frequently occur in poultry strains. Presumably they support the maintenance of pAvX plasmid they co-exist with. It has been observed in this study that 10 out of 11 strain carrying pAvX carry one or two plasmids from pAvY group as well (Table 4), which remains consistent with the original study (Lowder et al., 2009). Nevertheless, a possibility of pAvYs being simply selfish genetic elements is also feasible.
Regarding the mechanism of antibiotic resistance and other determinants dissemination, strikingly, a considerable number of plasmids reported in this study, i.e., 6 out of 16, that belong to the group II and III, carry an oriT element or its mimic. Such a sequence may render plasmids mobilizable in the presence of a conjugative plasmid, which facilitate their transfer and accelerate their spread among bacterial cells. In many reported cases a gene of mobilization protein (mob), also known as nicking relaxase, is also present. The gene is provided either in cis, as the oriT mimic in pPA3, or in trans in co-existing plasmids, as in the pair of pMW2/pRIVM4390 or among pSAP060B/pPH1-1/pPH1-3 and pPH1-2 plasmids. However, alongside oriT and a mobilization protein the presence of a conjugative plasmid carrying accessory genes, as one for a coupling protein, and an essential, large gene array encoding a type IV secretion system (T4SS) responsible for creating the mating pore and the physical transfer, is also required (O’Brien et al., 2015; Ramsay and Firth, 2017). Nevertheless, it is clear that a considerable number of plasmids reported in this study are potentially mobilizable. This shows why the spread of antibiotic resistance and other determinants may progress quickly as it is observed (O’Brien et al., 2014; Ramsay et al., 2016; Ramsay and Firth, 2017).
Among group III plasmids, which are the newly uncovered in this study, pPA3 is exceptionally compelling. In-depth analysis of its sequence demonstrates the major mechanisms that underpin resistance determinants and virulence factors dissemination brought up in this work. The fragment of pAvX, the plasmid of group I common among poultry strains, provides the gene of lysophospholipase and the operon of pemIK-Sa1 TA system. Another uncovered in this study group III plasmid pPH2 donates erythromycin and cadmium resistance determinants as well as an oriT mimic, whereas pBC16 is a source of a determinant of tetracycline resistance and a gene of mobilization protein. Strikingly, pBC16, which does not occur autonomously in the analyzed strains, is a broad-host plasmid that has been recorded so far in genera of Bacillus, Enterococcus, Staphylococcus, and Streptococcus. Hence, the observed sequence of pPA3 is a prime example of resistance determinants accumulation by interspecies transfer and DNA shuffling. Markedly, pAvX-derived pemIK-Sa1 TA system likely stabilizes the maintenance of pPA3 together with the resistance determinants it carries and may contribute to their dissemination in bacterial populations, similarly as yefM/yoeB-Sa2 and sprF3/sprG3 in pPH-1. Such a process might be further accelerated by the presence of mob from pBC16 and the oriT mimic from pPH2, which may render pPA3 mobilizable. Mimics of oriT are sequences that apart from the core fragment differ from the original oriT sequences derived from conjugative plasmids. However, in a form of single-stranded DNA they assume a structure mimicking the structure of original oriT sequences. Hence, it is suggested that their presence renders a far greater number of plasmids mobilizable than had initially been expected based on the prevalence of oriT sequences from conjugative plasmids (O’Brien et al., 2015). Here a completely novel sequence of oriT mimic is reported in the shared fragment of unknown to date staphylococcal pPH2 and pPA3 plasmids (Figure 4).
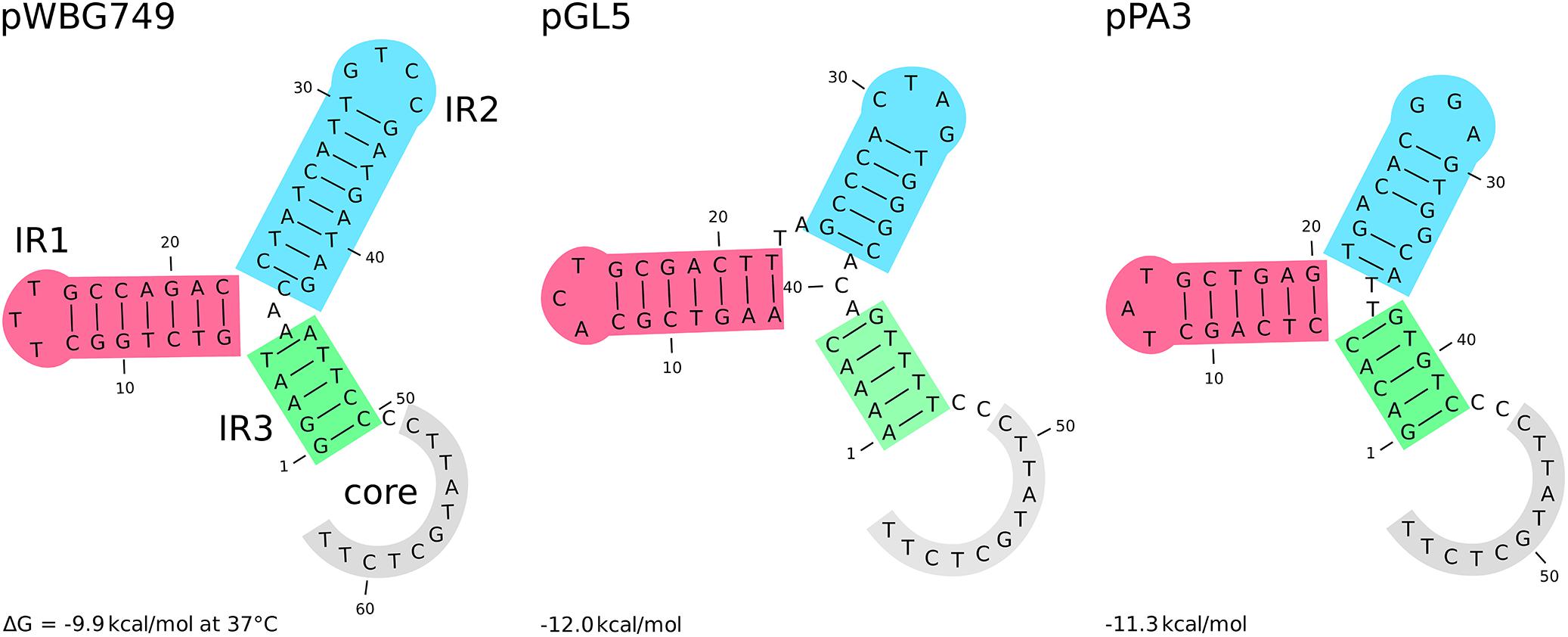
Figure 4. The secondary structure of different oriT elements. From the left: oriT and its mimic derived, respectively, from the conjugative plasmids pWBG747 and mobilizable plasmid pGL5 followed by the novel oriT mimic found in the fragment shared by the uncovered in this study pPH2 and pPA3 plasmids. Three inverted repeats and the core part are highlighted. The secondary structure and their Gibbs free-energy are similar across presented elements and others reported before (O’Brien et al., 2015).
Conclusion
The role of plasmids in dissemination of antibiotic and heavy metal resistance as well as host-specific virulence factors is commonly recognized. Unfortunately, a vast number of sequencing projects do not aim for complete sequence determination, which frequently requires a considerable amount of extra workload. In consequence, the number of known complete plasmid sequences is disproportionately low. To address this problem, we undertook an effort to determine complete plasmids sequences from 18 S. aureus strains of poultry origin. The results show that most of plasmids occurring in the analyzed strains and reported beforehand are being found in clinical isolates of strains typical for humans or human ones of livestock origin. This shows that livestock-associated staphylococci are a significant reservoir of resistance determinants and virulence factors which are spread to human strains. Strikingly, nearly half of the plasmids presented here were unknown to date. In this group we found a plasmid named here as pPA3, which is a unique example of accumulation of resistance determinants and virulence factors likely stabilized by the presence of a TA system. These facts clearly demonstrate that there is a pressing need for studies aimed at complete plasmid sequence determination.
Data Availability
The datasets generated for this study can be found in GenBank.
Auhtor Contributions
MB and BW designed the study. MH grew bacterial cultures and isolated plasmid DNA. RP and MB analyzed the NGS data, performed de novo assembly, and determined the final sequence of pPA3 plasmid. MB and RZ-P performed de novo assembly and determined the final sequences of the remaining plasmids. MB prepared annotations and carried out comparative sequence analyses. AM did antibiotic and heavy metal resistance screening. All authors analyzed and interpreted the results. MB prepared figures and tables. MB and BW wrote the manuscript. All authors revised the manuscript and agreed to be accountable for all aspects of the presented work.
Funding
This research was supported by funds granted by the National Science Centre (NCN, Poland) based on the decision No. DEC-2017/25/B/NZ6/01056 (to BW).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Professor Jacek Miedzobrodzki and Dr. Maja Kosecka-Strojek for providing bacterial strains from the collection of the Department of Microbiology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University. We would also like to thank James Wilson for his professional language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00805/full#supplementary-material
References
Águila-Arcos, S., Álvarez-Rodríguez, I., Garaiyurrebaso, O., Garbisu, C., Grohmann, E., and Alkorta, I. (2017). Biofilm-forming clinical Staphylococcus isolates harbor horizontal transfer and antibiotic resistance genes. Front. Microbiol. 8:2018. doi: 10.3389/fmicb.2017.02018
Alibayov, B., Baba-Moussa, L., Sina, H., Zdeňková, K., and Demnerová, K. (2014). Staphylococcus aureus mobile genetic elements. Mol. Biol. Rep. 41, 5005–5018. doi: 10.1007/s11033-014-3367-3
Anstead, G. M., Cadena, J., and Javeri, H. (2014). Treatment of infections due to resistant Staphylococcus aureus. Methods Mol. Biol. 1085, 259–309. doi: 10.1007/978-1-62703-664-1_16
Archer, G. L. (1978). Antimicrobial susceptibility and selection of resistance among Staphylococcus epidermidis isolates recovered from patients with infections of indwelling foreign devices. Antimicrob. Agents Chemother. 14, 353–359. doi: 10.1128/AAC.14.3.353
Argudín, M. Á., Mendoza, M. C., and Rodicio, M. R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2, 1751–1773. doi: 10.3390/toxins2071751
Armand-Lefevre, L., Ruimy, R., and Andremont, A. (2005). Clonal comparison of Staphylococcus from healthy pig farmers, human controls, and pigs. Emerg. Infect. Dis. 11, 711–714. doi: 10.3201/eid1105.040866
Baba, T., Takeuchi, F., Kuroda, M., Yuzawa, H., Aoki, K., Oguchi, A., et al. (2002). Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359, 1819–1827. doi: 10.1016/S0140-6736(02)08713-5
Berends, E. T. M., Horswill, A. R., Haste, N. M., Monestier, M., Nizet, V., and Von Köckritz-Blickwede, M. (2010). Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J. Innate Immun. 2, 576–586. doi: 10.1159/000319909
Bonar, E., Wojcik, I., Jankowska, U., Kedracka-Krok, S., Bukowski, M., Polakowska, K., et al. (2016). Identification of secreted exoproteome fingerprints of highly-virulent and non-virulent Staphylococcus aureus strains. Front. Cell. Infect. Microbiol. 6:51. doi: 10.3389/fcimb.2016.00051
Bonar, E. A., Bukowski, M., Hydzik, M., Jankowska, U., Kedracka-Krok, S., Groborz, M., et al. (2018). Joint genomic and proteomic analysis identifies meta-trait characteristics of virulent and non-virulent Staphylococcus aureus strains. Front. Cell. Infect. Microbiol. 8:313. doi: 10.3389/fcimb.2018.00313
Bosch, T., Witteveen, S., Haenen, A., Landman, F., and Schouls, L. M. (2016). Next-generation sequencing confirms presumed nosocomial transmission of livestock-associated methicillin-resistant Staphylococcus aureus in the Netherlands. Appl. Environ. Microbiol. 82, 4081–4089. doi: 10.1128/AEM.00773-16
Bukowski, M., Hyz, K., Janczak, M., Hydzik, M., Dubin, G., and Wladyka, B. (2017). Identification of novel mazEF/pemIK family toxin-antitoxin loci and their distribution in the Staphylococcus genus. Sci. Rep. 7:13462. doi: 10.1038/s41598-017-13857-4
Bukowski, M., Lyzen, R., Helbin, W. M., Bonar, E., Szalewska-Palasz, A., Wegrzyn, G., et al. (2013). A regulatory role for Staphylococcus aureus toxin-antitoxin system PemIKSa. Nat. Commun. 4:2012. doi: 10.1038/ncomms3012
Bukowski, M., Rojowska, A., and Wladyka, B. (2011). Prokaryotic toxin-antitoxin systems–the role in bacterial physiology and application in molecular biology. Acta Biochim. Pol. 58, 1–9.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Casman, E. P., Bennett, R. W., Dorsey, A. E., and Issa, J. A. (1967). Identification of a fourth staphylococcal enterotoxin, enterotoxin D. J. Bacteriol. 94, 1875–1882.
Charlebois, E. D., Bangsberg, D. R., Moss, N. J., Moore, M. R., Moss, A. R., Chambers, H. F., et al. (2002). Population-based community prevalence of methicillin-resistant Staphylococcus aureus in the urban poor of San Francisco. Clin. Infect. Dis. 34, 425–433. doi: 10.1086/338069
Chudobova, D., Dostalova, S., Ruttkay-Nedecky, B., Guran, R., Rodrigo, M. A. M., Tmejova, K., et al. (2015). The effect of metal ions on Staphylococcus aureus revealed by biochemical and mass spectrometric analyses. Microbiol. Res. 170, 147–156. doi: 10.1016/j.micres.2014.08.003
Crupper, S. S., Worrell, V., Stewart, G. C., and Iandolo, J. J. (1999). Cloning and expression of cadD, a new cadmium resistance gene of Staphylococcus aureus. J. Bacteriol. 181, 4071–4075.
Cuny, C., Arnold, P., Hermes, J., Eckmanns, T., Mehraj, J., Schoenfelder, S., et al. (2017). Occurrence of cfr-mediated multiresistance in staphylococci from veal calves and pigs, from humans at the corresponding farms, and from veterinarians and their family members. Vet. Microbiol. 200, 88–94. doi: 10.1016/j.vetmic.2016.04.002
Daugherty, S., and Low, M. G. (1993). Cloning, expression, and mutagenesis of phosphatidylinositol-specific phospholipase C from Staphylococcus aureus: a potential staphylococcal virulence factor. Infect. Immun. 61, 5078–5089.
De Jager, P., Chirwa, T., Naidoo, S., Perovic, O., and Thomas, J. (2015). Nosocomial outbreak of New Delhi metalloβ-lactamase-1-producing Gram-negative bacteria in South Africa: a case-control study. PLoS One 10:e0123337. doi: 10.1371/journal.pone.0123337
Deghorain, M., and Van Melderen, L. (2012). The staphylococci phages family: an overview. Viruses 4, 3316–3335. doi: 10.3390/v4123316
del Solar, G., Giraldo, R., Ruiz-Echevarría, M. J., Espinosa, M., and Díaz-Orejas, R. (1998). Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62, 434–464.
Dmowski, M., and Jagura-Burdzy, G. (2013). Active stable maintenance functions in low copy-number plasmids of Gram-positive bacteria I. Partition systems. Pol. J. Microbiol. 62, 3–16.
Doery, H. M., Magnusson, B. J., Gulasekharam, J., and Pearson, J. E. (1965). The properties of phospholipase enzymes in staphylococcal toxins. J. Gen. Microbiol. 40, 283–296. doi: 10.1099/00221287-40-2-283
Dogramachy, N. (2018). Prevalence of nasal carriage rate for methicillin-resistant Staphylococcus aureus and its antibiotic susceptibility profiles in health care workers at Nanakaly Hospital, Erbil, Iraq. Zanco J. Med. Sci. 22, 411–419. doi: 10.15218/zjms.2018.053
Domingues, S., da Silva, G. J., and Nielsen, K. M. (2012). Integrons. Mob. Genet. Elements 2, 211–223. doi: 10.4161/mge.22967
Douglas, A. P., Marshall, C., Baines, S. L., Ritchie, D., Szer, J., Madigan, V., et al. (2019). Utilizing genomic analyses to investigate the first outbreak of van A vancomycin-resistant Enterococcus in Australia with emergence of daptomycin non-susceptibility. J. Med. Microbiol. 68, 303–308. doi: 10.1099/jmm.0.000916
Fagerlund, A., Granum, P. E., and Håvarstein, L. S. (2014). Staphylococcus aureus competence genes: mapping of the SigH, ComK1 and ComK2 regulons by transcriptome sequencing. Mol. Microbiol. 94, 557–579. doi: 10.1111/mmi.12767
Fluit, A. C. (2012). Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 18, 735–744. doi: 10.1111/j.1469-0691.2012.03846.x
Fu, Z., Liu, Y., Chen, C., Guo, Y., Ma, Y., Yang, Y., et al. (2016). Characterization of fosfomycin resistance gene, fosB, in methicillin-resistant Staphylococcus aureus isolates. PLoS One 11:e0154829. doi: 10.1371/journal.pone.0154829
Germain-Amiot, N., Augagneur, Y., Camberlein, E., Nicolas, I., Lecureur, V., Rouillon, A., et al. (2018). A novel Staphylococcus aureus cis-trans type I toxin-antitoxin module with dual effects on bacteria and host cells. Nucleic Acids Res. 47, 1759–1773. doi: 10.1093/nar/gky1257
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
He, T., Wang, Y., Schwarz, S., Zhao, Q., Shen, J., and Wu, C. (2014). Genetic environment of the multi-resistance gene cfr in methicillin-resistant coagulase-negative staphylococci from chickens, ducks, and pigs in China. Int. J. Med. Microbiol. 304, 257–261. doi: 10.1016/j.ijmm.2013.10.005
Huijsdens, X. W., van Dijke, B. J., Spalburg, E., van Santen-Verheuvel, M. G., Heck, M. E., Pluister, G. N., et al. (2006). Community-acquired MRSA and pig-farming. Ann. Clin. Microbiol. Antimicrob. 5:26. doi: 10.1186/1476-0711-5-26
Hung, W. C., Chen, H. J., Lin, Y. T., Tsai, J. C., Chen, C. W., Lu, H. H., et al. (2015). Skin commensal staphylococci may act as reservoir for fusidic acid resistance genes. PLoS One 10:e0143106. doi: 10.1371/journal.pone.0143106
Ji, G., and Silver, S. (1992). Regulation and expression of the arsenic resistance operon from Staphylococcus aureus plasmid pI258. J. Bacteriol. 174, 3684–3694. doi: 10.1128/jb.174.11.3684-3694.1992
Kalińska, M., Kantyka, T., Greenbaum, D. C., Larsen, K. S., Władyka, B., Jabaiah, A., et al. (2012). Substrate specificity of Staphylococcus aureus cysteine proteases - Staphopains A, B and C. Biochimie 94, 318–327. doi: 10.1016/j.biochi.2011.07.020
Khan, S. A., and Novick, R. P. (1983). Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid 10, 251–259. doi: 10.1016/0147-619X(83)90039-2
Kibbe, W. A. (2007). OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 35, W43–W46. doi: 10.1093/nar/gkm234
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kuehnert, M. J., Kruszon-Moran, D., Hill, H. A., McQuillan, G., McAllister, S. K., Fosheim, G., et al. (2006). Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. J. Infect. Dis. 193, 172–179. doi: 10.1086/499632
Kumar, D., Pornsukarom, S., Sivaraman, G. K., and Thakur, S. (2017). Environmental dissemination of multidrug methicillin-resistant Staphylococcus sciuri after application of manure from commercial swine production systems. Foodborne Pathog. Dis. 15, 210–217. doi: 10.1089/fpd.2017.2354
Kwong, S. M., Ramsay, J. P., Jensen, S. O., and Firth, N. (2017). Replication of staphylococcal resistance plasmids. Front. Microbiol. 8:2279. doi: 10.3389/fmicb.2017.02279
Lau, C. H. F., van Engelen, K., Gordon, S., Renaud, J., and Topp, E. (2017). Novel antibiotic resistance determinants from agricultural soil exposed to antibiotics widely used in human medicine and animal farming. Appl. Environ. Microbiol. 83:e00989-17. doi: 10.1128/AEM.00989-17
Levy, R. M., Leyden, J. J., and Margolis, D. J. (2005). Colonisation rates of Streptococcus pyogenes and Staphylococcus aureus in the oropharynx of a young adult population. Clin. Microbiol. Infect. 11, 153–155. doi: 10.1111/j.1469-0691.2004.01042.x
Li, S., Li, J., Qiao, Y., Ning, X., Zeng, T., and Shen, X. (2014). Prevalence and invasiveness of community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis. Indian J. Pathol. Microbiol. 57, 418–422. doi: 10.4103/0377-4929.138737
Lindqvist, M., Isaksson, B., Grub, C., Jonassen, T. O., and Hällgren, A. (2012). Detection and characterisation of SCCmec remnants in multiresistant methicillin-susceptible Staphylococcus aureus causing a clonal outbreak in a Swedish county. Eur. J. Clin. Microbiol. Infect. Dis. 31, 141–147. doi: 10.1007/s10096-011-1286-y
Lindqvist, M., Isaksson, B., Samuelsson, A., Nilsson, L. E., and Hällgren, A. (2009). A clonal outbreak of methicillin-susceptible Staphylococcus aureus with concomitant resistance to erythromycin, clindamycin and tobramycin in a Swedish county. Scand. J. Infect. Dis. 41, 324–333. doi: 10.1080/00365540902801202
Lowder, B. V., Guinane, C. M., Ben Zakour, N. L., Weinert, L. A., Conway-Morris, A., Cartwright, R. A., et al. (2009). Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 19545–19550. doi: 10.1073/pnas.0909285106
Lozano, C., García-Migura, L., Aspiroz, C., Zarazaga, M., Torres, C., and Aarestrup, F. M. (2012). Expansion of a plasmid classification system for gram-positive bacteria and determination of the diversity of plasmids in Staphylococcus aureus strains of human, animal, and food origins. Appl. Environ. Microbiol. 78, 5948–5955. doi: 10.1128/AEM.00870-12
Maiden, M. C. J., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2013). Comparative genomics of defense systems in Archaea and bacteria. Nucleic Acids Res. 41, 4360–4377. doi: 10.1093/nar/gkt157
Malachowa, N., and Deleo, F. R. (2010). Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 67, 3057–3071. doi: 10.1007/s00018-010-0389-4
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Marques, M. B., Weller, P. F., Parsonnet, J., Ransil, B. J., and Nicholson-Weller, A. (1989). Phosphatidylinositol-specific phospholipase C, a possible virulence factor of Staphylococcus aureus. J. Clin. Microbiol. 27, 2451–2454.
Martinez, J. L. (2009). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902. doi: 10.1016/j.envpol.2009.05.051
McCarthy, A. J., Loeffler, A., Witney, A. A., Gould, K. A., Lloyd, D. H., and Lindsay, J. A. (2014). Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol. Evol. 6, 2697–2708. doi: 10.1093/gbe/evu214
McDougal, L. K., Fosheim, G. E., Nicholson, A., Bulens, S. N., Limbago, B. M., Shearer, J. E. S., et al. (2010). Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrob. Agents Chemother. 54, 3804–3811. doi: 10.1128/AAC.00351-10
Moran, G. J., Krishnadasan, A., Gorwitz, R. J., Fosheim, G. E., McDougal, L. K., Carey, R. B., et al. (2006). Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355, 666–674. doi: 10.1056/NEJMoa055356
Morgenstern, M., Erichsen, C., Hackl, S., Mily, J., Militz, M., Friederichs, J., et al. (2016). Antibiotic resistance of commensal Staphylococcus aureus and coagulase-negative staphylococci in an international cohort of surgeons: a prospective point-prevalence study. PLoS One 11:e0148437. doi: 10.1371/journal.pone.0148437
Moritz, E. M., and Hergenrother, P. J. (2007). Toxin-antitoxin systems are ubiquitous and plasmid-encoded in vancomycin-resistant enterococci. Proc. Natl. Acad. Sci. U.S.A. 104, 311–316. doi: 10.1073/pnas.0601168104
Mruk, I., and Kobayashi, I. (2014). To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. 42, 70–86. doi: 10.1093/nar/gkt711
Nimmo, G. R. (2012). USA300 abroad: global spread of a virulent strain of community-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 18, 725–734. doi: 10.1111/j.1469-0691.2012.03822.x
Nolle, N., Schuster, C. F., and Bertram, R. (2013). Two paralogous yefM-yoeB loci from Staphylococcus equorum encode functional toxin-antitoxin systems. Microbiology 159, 1575–1585. doi: 10.1099/mic.0.068049-0
O’Brien, F. G., Eto, K. Y., Murphy, R. J. T., Fairhurst, H. M., Coombs, G. W., Grubb, W. B., et al. (2015). Origin-of-transfer sequences facilitate mobilisation of non-conjugative antimicrobial-resistance plasmids in Staphylococcus aureus. Nucleic Acids Res. 43, 7971–7983. doi: 10.1093/nar/gkv755
O’Brien, F. G., Ramsay, J. P., Monecke, S., Coombs, G. W., Robinson, O. J., Htet, Z., et al. (2014). Staphylococcus aureus plasmids without mobilization genes are mobilized by a novel conjugative plasmid from community isolates. J. Antimicrob. Chemother. 70, 649–652. doi: 10.1093/jac/dku454
Omoe, K., Hu, D. L., Takahashi-Omoe, H., Nakane, A., and Shinagawa, K. (2003). Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect. Immun. 71, 6088–6094. doi: 10.1128/IAI.71.10.6088-6094.2003
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17. doi: 10.1128/CMR.00088-17
Pendleton, J. N., Gorman, S. P., and Gilmore, B. F. (2013). Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 11, 297–308. doi: 10.1586/eri.13.12
Pichon, C., and Felden, B. (2005). Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. U.S.A. 102, 14249–14254. doi: 10.1073/pnas.0503838102
Planet, P. J., Larussa, S. J., Dana, A., Smith, H., Xu, A., Ryan, C., et al. (2013). Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. mBio 4:e00889-13. doi: 10.1128/mBio.00889-13
Polakowska, K., Lis, M. W., Helbin, W. M., Dubin, G., Dubin, A., Niedziolka, J. W., et al. (2012). The virulence of Staphylococcus aureus correlates with strain genotype in a chicken embryo model but not a nematode model. Microbes Infect. 14, 1352–1362. doi: 10.1016/j.micinf.2012.09.006
Ramsay, J. P., and Firth, N. (2017). Diverse mobilization strategies facilitate transfer of non-conjugative mobile genetic elements. Curr. Opin. Microbiol. 38, 1–9. doi: 10.1016/j.mib.2017.03.003
Ramsay, J. P., Kwong, S. M., Murphy, R. J. T., Yui Eto, K., Price, K. J., Nguyen, Q. T., et al. (2016). An updated view of plasmid conjugation and mobilization in Staphylococcus. Mob. Genet. Elements 6:e1208317. doi: 10.1080/2159256X.2016.1208317
Riffaud, C., Pinel-Marie, M.-L., Pascreau, G., and Felden, B. (2018). Functionality and cross-regulation of the four SprG/SprF type I toxin-antitoxin systems in Staphylococcus aureus. Nucleic Acids Res. 47, 1740–1758. doi: 10.1093/nar/gky1256
Rosvoll, T. C. S., Pedersen, T., Sletvold, H., Johnsen, P. J., Sollid, J. E., Simonsen, G. S., et al. (2010). PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTβ-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol. Med. Microbiol. 58, 254–268. doi: 10.1111/j.1574-695X.2009.00633.x
Savini, V., Kosecka, M., Siegwart, E., Marrollo, R., Polilli, E., Palmieri, D., et al. (2016). Daptomycin-resistant Staphylococcus pettenkoferi of human origin. Acta Biochim. Pol. 63, 297–301. doi: 10.18388/abp.2015_1113
Scharn, C. R., Tenover, F. C., and Goering, R. V. (2013). Transduction of staphylococcal cassette chromosome mec elements between strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 5233–5238. doi: 10.1128/AAC.01058-13
Sengupta, M., and Austin, S. (2011). Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect. Immun. 79, 2502–2509. doi: 10.1128/IAI.00127-11
Shearer, J. E. S., Wireman, J., Hostetler, J., Forberger, H., Borman, J., Gill, J., et al. (2011). Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 1, 581–591. doi: 10.1534/g3.111.000760
Smith, C. L., and Cantor, C. R. (1987). Purification, specific fragmentation, and separation of large DNA molecules. Methods Enzymol. 155, 449–467. doi: 10.1016/0076-6879(87)55030-3
Sobecky, P. A., Easter, C. L., Bear, P. D., and Helinski, D. R. (1996). Characterization of the stable maintenance properties of the par region of broad-host-range plasmid RK2. J. Bacteriol. 178, 2086–2093.
Strauß, L., Stegger, M., Akpaka, P. E., Alabi, A., Breurec, S., Coombs, G., et al. (2017). Origin, evolution, and global transmission of community-acquired Staphylococcus aureus ST8. Proc. Natl. Acad. Sci. U.S.A. 114, E10596–E10604. doi: 10.1073/pnas.1702472114
Takeuchi, S., Kinoshita, T., Kaidoh, T., and Hashizume, N. (1999). Purification and characterization of protease produced by Staphylococcus aureus isolated from a diseased chicken. Vet. Microbiol. 67, 195–202. doi: 10.1016/S0378-1135(99)00034-6
Takeuchi, S., Matsunaga, K., Inubushi, S., Higuchi, H., Imaizumi, K., and Kaidoh, T. (2002). Structural gene and strain specificity of a novel cysteine protease produced by Staphylococcus aureus isolated from a diseased chicken. Vet. Microbiol. 89, 201–210. doi: 10.1016/S0378-1135(02)00171-2
Tenover, F. C., McDougal, L. K., Goering, R. V., Killgore, G., Projan, S. J., Patel, J. B., et al. (2006). Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44, 108–118. doi: 10.1128/JCM.44.1.108-118.2006
Thammavongsa, V., Missiakas, D. M., and Schneewind, O. (2013). Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science 342, 863–866. doi: 10.1126/science.1242255
Tsuchimoto, S., Ohtsubo, H., and Ohtsubo, E. (1988). Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J. Bacteriol. 170, 1461–1466.
Tulinski, P., Fluit, A. C., Wagenaar, J. A., Mevius, D., van de Vijver, L., and Duim, B. (2012). Methicillin-resistant coagulase-negative staphylococci on pig farms as a reservoir of heterogeneous staphylococcal cassette chromosome mec elements. Appl. Environ. Microbiol. 78, 299–304. doi: 10.1128/AEM.05594-11
Van Melderen, L., and Saavedra De Bast, M. (2009). Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5:e1000437. doi: 10.1371/journal.pgen.1000437
Vandenesch, F., Naimi, T., Enright, M. C., Lina, G., Nimmo, G. R., Heffernan, H., et al. (2003). Community-acquired methicillin-resistant Staphylococcus aureus carrying panton-valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9, 978–984. doi: 10.3201/eid0908.030089
Varga, M., Kuntová, L., Pantůček, R., Mašlaòová, I., Růžičková, V., and Doškař, J. (2012). Efficient transfer of antibiotic resistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiol. Lett. 332, 146–152. doi: 10.1111/j.1574-6968.2012.02589.x
Varga, M., Pantůček, R., Růžiěková, V., and Doškař, J. (2016). Molecular characterization of a new efficiently transducing bacteriophage identified in meticillin-resistant Staphylococcus aureus. J. Gen. Virol. 97, 258–268. doi: 10.1099/jgv.0.000329
Voss, A., Loeffen, F., Bakker, J., Klaassen, C., and Wulf, M. (2005). Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11, 1965–1966. doi: 10.3201/eid1112.050428
Wendlandt, S., Kadlec, K., Feßler, A. T., Mevius, D., Van Essen-Zandbergen, A., Hengeveld, P. D., et al. (2013). Transmission of methicillin-resistant Staphylococcus aureus isolates on broiler farms. Vet. Microbiol. 167, 632–637. doi: 10.1016/j.vetmic.2013.09.019
Wladyka, B., Dubin, G., and Dubin, A. (2011a). Activation mechanism of thiol protease precursor from broiler chicken specific Staphylococcus aureus strain CH-91. Vet. Microbiol. 147, 195–199. doi: 10.1016/j.vetmic.2010.06.002
Wladyka, B., Kozik, A. J., Bukowski, M., Rojowska, A., Kantyka, T., Dubin, G., et al. (2011b). 1-Antichymotrypsin inactivates staphylococcal cysteine protease in cross-class inhibition. Biochimie 93, 948–953. doi: 10.1016/j.biochi.2011.01.014
Wladyka, B., Piejko, M., Bzowska, M., Pieta, P., Krzysik, M., Mazurek, Ł., et al. (2015). A peptide factor secreted by Staphylococcus pseudintermedius exhibits properties of both bacteriocins and virulence factors. Sci. Rep. 5:14569. doi: 10.1038/srep14569
Yoshizumi, S., Zhang, Y., Yamaguchi, Y., Chen, L., Kreiswirth, B. N., and Inouye, M. (2009). Staphylococcus aureus YoeB homologues inhibit translation initiation. J. Bacteriol. 191, 5868–5872. doi: 10.1128/JB.00623-09
Zhang, S., Iandolo, J. J., and Stewart, G. C. (1998). The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 168, 227–233. doi: 10.1016/S0378-1097(98)00423-6
Keywords: antibiotic resistance (AMR), Staphylococcus aureus, plasmid, heavy metal resistance, virulence factor, toxin–antitoxin (TA)
Citation: Bukowski M, Piwowarczyk R, Madry A, Zagorski-Przybylo R, Hydzik M and Wladyka B (2019) Prevalence of Antibiotic and Heavy Metal Resistance Determinants and Virulence-Related Genetic Elements in Plasmids of Staphylococcus aureus. Front. Microbiol. 10:805. doi: 10.3389/fmicb.2019.00805
Received: 20 February 2019; Accepted: 29 March 2019;
Published: 24 April 2019.
Edited by:
Zhiyong Zong, West China Hospital, ChinaReviewed by:
Jiri Doskar, Masaryk University, CzechiaPeter Kinnevey, Dublin Dental University Hospital, Ireland
Dingqiang Chen, Zhujiang Hospital, China
Copyright © 2019 Bukowski, Piwowarczyk, Madry, Zagorski-Przybylo, Hydzik and Wladyka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michal Bukowski, bS5idWtvd3NraUB1ai5lZHUucGw= Benedykt Wladyka, YmVuZWR5a3Qud2xhZHlrYUB1ai5lZHUucGw=
 Michal Bukowski
Michal Bukowski Rafal Piwowarczyk
Rafal Piwowarczyk Anna Madry
Anna Madry Rafal Zagorski-Przybylo
Rafal Zagorski-Przybylo Marcin Hydzik
Marcin Hydzik Benedykt Wladyka
Benedykt Wladyka