
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 April 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00789
This article is part of the Research Topic Carbapenem-Resistant Enterobacteriaceae in the Asia Pacific and Beyond View all 13 articles
 Chi-Chung Chen1,2
Chi-Chung Chen1,2 Chih-Cheng Lai3
Chih-Cheng Lai3 Hui-Ling Huang4
Hui-Ling Huang4 Wen-Yu Huang4
Wen-Yu Huang4 Han-Siong Toh5
Han-Siong Toh5 Tzu-Chieh Weng5
Tzu-Chieh Weng5 Yin-Ching Chuang1,6
Yin-Ching Chuang1,6 Ying-Chen Lu2*
Ying-Chen Lu2* Hung-Jen Tang4,6*
Hung-Jen Tang4,6*Objective: This study aims to identify suitable lactobacilli that have anti-carbapenem-resistant Enterobacteriaceae (CRE) activity with in vitro tolerance to pepsin and bile salts.
Methods: Fifty-seven Lactobacillus spp. strains encompassing nine species were collected for investigation. Their viabilities in the presence of pepsin and bile salts were tested using tolerance tests. Their anti-CRE effects were assessed by agar well diffusion and broth microdilution assay, as well as time-kill test.
Results: Of the 57 Lactobacillus isolates collected, 31 had a less than 2-log reduction in their viability in both pepsin and bile salt tolerance tests. Of these 31 isolates, 5 (LUC0180, LUC0219, LYC0289, LYC0413, and LYC1031) displayed the greatest anti-CRE activity with a CRE zone of inhibition greater than 15 mm in agar well diffusion assays. The minimal inhibitory percentages of supernatants from these five strains against CREs ranged from 10 to 30%. With the exception of LUC0180, which had a minimal bactericidal percentage ≥ 40%, the bactericidal percentage of all the strains ranged from 20 to 40%. The inhibitory effect of the cell-free culture supernatants from these Lactobacillus strains did not change after heating but was abolished as the pH changed to 7.0. After a 24-h incubation, five of the Lactobacillus strains at a concentration of 108 CFU/ml totally inhibited the growth of carbapenem-resistant Escherichia coli (CRE316) and Klebsiella pneumoniae (CRE632). After a 48-h incubation, the growth of CRE316 was completely inhibited under each concentration of lactobacilli based on time-kill test. Furthermore, when the concentration of lactobacilli was at 108 CFU/ml, the decline in pH was faster than at other concentrations.
Conclusion: Some Lactobacillus strains exhibit anti-CRE activity, which suggests potential applications for controlling or preventing CRE colonization or infection.
Although Enterobacteriaceae are normal flora of the human intestinal system, they are also common pathogens causing human infections in the setting of both community-acquired and healthcare-associated infections (Hsueh et al., 2010; Toh et al., 2012; Lai et al., 2014; Jean et al., 2016). In this era of widespread antibiotic resistance, Enterobacteriaceae are no exception. Recently, the emergence of carbapenem-resistant Enterobacteriaceae (CRE) has become a more critical issue due to the limited therapeutic options available for these pathogens and the significant morbidity and mortality associated with CRE infections (Tang et al., 2016a; Rodriguez-Bano et al., 2018). Therefore, there is an urgent need for new treatments for these critical CRE-associated conditions.
Lactobacillus is one of a number of probiotics considered to be biological therapeutics and host immune-modulating biologicals that are generally recognized as safe (GRAS). Recent studies demonstrated several antimicrobial mechanisms of Lactobacillus such as nutrient competition, production of inhibitory compounds, immune-stimulation and competition for binding sites. In addition, Lactobacillus can produce lactic acid, acetic acid, formic acid and other acids to reduce intestinal pH, which may be the most important mechanism. These bacteria can also secrete certain antimicrobial molecules, such as ethanol, fatty acid, hydrogen peroxide and bacteriocins to exert the antimicrobial activity (Georgieva et al., 2015; Inglin et al., 2015). Through these mechanisms, Lactobacillus has demonstrated its ability to inhibit several bacterial pathogens, including Clostridium difficile (McFarland, 2015), Escherichia coli (Kumar et al., 2016), Shigella spp. (Mirnejad et al., 2013), Streptococcus mutans (Ahn et al., 2018), Pseudomonas aeruginosa (Jamalifar et al., 2011), and Staphylococcus aureus (Kang et al., 2017). However, no previous studies have assessed the antimicrobial activity of Lactobacillus against CRE. Thus, we conducted this study to identify suitable lactobacilli that have anti-CRE activity with in vitro tolerance to pepsin and bile salts.
Fifty-seven Lactobacillus spp. strains encompassing nine species were obtained from Department of Food Science at the National Chiayi University in Chiayi, Taiwan. Species confirmation was performed by 16S rDNA sequencing. A fragment of the 16S rDNA was amplified by PCR. After amplification, the amplicons were separated by gel electrophoresis and sequenced. Sequences were compared with the NCBI GenBank database using the BLAST search tool to find the closest matches. The basic growth media for LAB were Man-Rogosa-Sharpe (MRS; Oxoid Inc., Ogdensburg, NY, United States).
Twenty clinical strains, including 10 different pulse field gel electrophoresis (PFGE) genotyped carbapenem-resistant Escherichia coli (CREC) and Klebsiella pneumoniae (CRKP) strains were isolated from Chi Mei Medical Center (Lai et al., 2016; Tang et al., 2016b). Species confirmations were performed using the VITEK 2 automated system (bioMérieux, Marcy l’Etoile, France) with standard biochemical methods. Mueller Hinton (MH) broth (Difco Laboratories, Detroit, MI, United States) were used for bacterial pathogens. The isolates were stored at −80°C in Protect Bacterial Preservers (Technical Service Consultants Limited, Heywood, United Kingdom) before use.
Carbapenem susceptibility testing was performed using the disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute [CLSI], 2015). In brief, a 0.5 McFarland turbidity standard inoculum from overnight cultures was followed by incubation of Mueller-Hinton agar plates at 35°C. The antibiotic disks were placed on the agar surface. After 16–18 h of incubation at 35°C, results were interpreted as either sensitive, intermediate, or resistant according to the inhibitory zone diameters around the disks using CLSI breakpoints. Carbapenem resistance was defined as resistance to imipenem, meropenem, doripenem, or ertapenem.
Pulsed-field gel electrophoresis (PFGE) for the Escherichia coli and K. pneumoniae isolates were performed as described previously (Lai et al., 2016) with a CHEF DR II apparatus (Bio-Rad Laboratories, Hercules, CA, United States). Briefly, bacterial chromosomal DNAs were digested using XbaI (New England Biolabs, Beverly, MA, United States). Electrophoresis was carried out for 22 h at 14 μC, with pulse times ranging from 2 to 40 s at 6 V/cm, using a Bio-Rad CHEF MAPPER apparatus (Bio-Rad Laboratories, Richmond, CA, United States). A dendrogram based on the unweighted pair group was generated using methods previously described. The PFGE patterns were visually examined and interpreted according to the criteria of Tenover et al. (1995). The similarities of the PFGE profiles of each strain were compared using a Dice coefficient at 1.0% of tolerance and 1.0% of optimization. Isolates that had < 80% similarity on the PFGE profiles were considered different types (Figure 1).
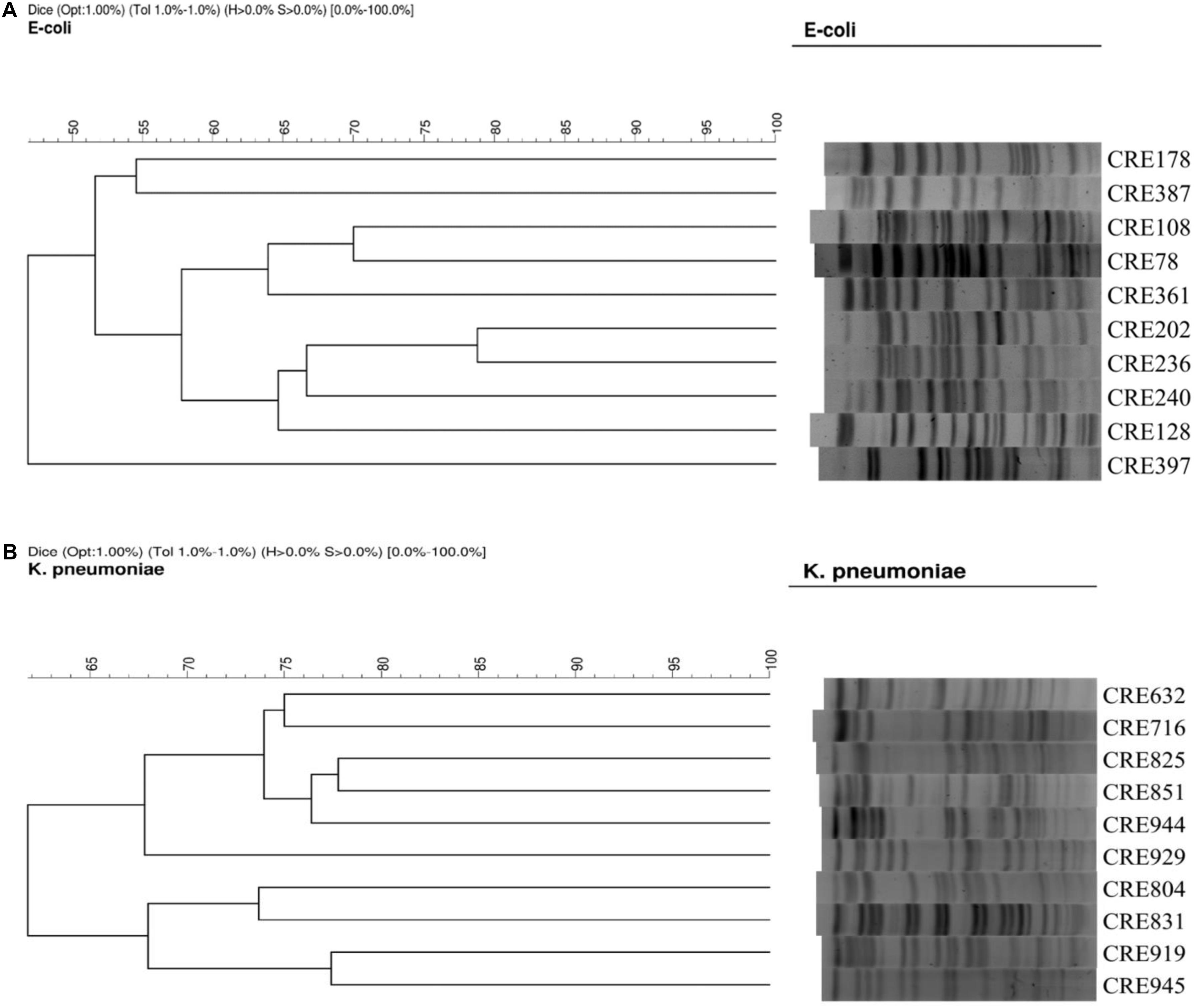
Figure 1. (A) PFGE of 10 isolates of carbapenem-resistant E. coli. (B) PFGE of 10 isolates of carbapenem-resistant K. pneumoniae.
The procedure for broth microdilution was adapted from the CLSI protocol for antimicrobial susceptibility testing (Clinical and Laboratory Standards Institute [CLSI], 2009). Antibiotic solutions were prepared in LAB susceptibility media (90% LSM and 10% MRS broth, adjusted to pH 6.7) (Klare et al., 2005). The following antibiotics were tested: ampicillin, chloramphenicol, clindamycin, erythromycin, fusidic acid, gentamycin, kanamycin, linezolid, streptomycin, teicoplanin, trimethoprim/sulfamethoxazole (1/19), and vancomycin (Sigma-Aldrich, St. Louis, MO, United States). The antibiotic susceptibility tests and their interpretations were carried out according to the CLSI guidelines (British Society for Antimicrobial Chemotherapy [BSAC], 2014; Clinical and Laboratory Standards Institute [CLSI], 2016).
A pepsin solution was prepared by suspending 3 mg/ml pepsin (Sigma-Aldrich, St. Louis, MO, United States) in a 0.85% sterile saline solution adjusted to pH 2.5. To test bacterial viability in the presence of pepsin, the 3 mg/ml pepsin solution, or saline at a pH of 7.0 as control, was inoculated with 106 CFU/ml of Lactobacillus spp. and then incubated at 37°C for 4 h. The viable cell population was determined using the spread plate method. Each experiment was conducted in triplicate (Tokatli et al., 2015).
The bile salt tolerance assay was performed as previously described (Jacobsen et al., 1999). In brief, each strain was adjusted to 1 × 106 CFU/ml in 0.3% Oxgall (Sigma-Aldrich, St. Louis, MO, United States) in PBS pH 7.3, or in PBS pH 7.3 alone as a control. Survival was tested at 37°C after 24 h by the spread plate method. Each experiment was conducted in triplicate.
Lactobacilli cell-free supernatants were cultured in MRS broth at 37°C for 24 h. The cultures were then centrifuged at 4000 rpm at 4°C for 30 min. The supernatants were sterilized by filtration through a 0.22 μm filter (Millipore, Billerica, MA, United States).
The agar well diffusion method (Tagg and McGiven, 1971) was modified to detect antimicrobial activities of supernatants isolated from Lactobacillus strains. First, MH agar plates were swabbed on the surface with CRE bacterial cultures. Then, 6 mm diameter wells were prepared and cell-free supernatants from isolated lactobacilli were loaded in the wells (100 μl/well). Following a 24-h incubation at 37°C, inhibition zones were recorded.
Broth microdilution assay was carried out as previously described (Arena et al., 2016) with some modifications. Overnight cultures of pathogenic bacteria were inoculated into fresh MHB media and seeded into 96-well plates (BD Discovery Labware, Bedford, MA, United States). The CFS of each Lactobacillus culture was separated into three aliquots: The first aliquot received no treatment, the second aliquot was heated at 80°C for 10 min, and the third aliquot was neutralized to pH 7.0 with 1N NaOH. A 200 μl volume of test solution, consisting of 100 μl of the pathogenic bacterial culture (final inoculum was approximately 106 CFU/ml) and 100 μl of one of the CFS aliquots, was mixed into the wells. The CFSs were diluted with MRS broth and used at different percentages (i.e., 10, 20, 30, 40, 50%) in the final 200 μl volume. The minimum inhibitory percentage (MIP), defined as the lowest percentage of supernatant that can inhibit the growth of pathogen, was monitored by measuring optical density (OD600 nm). The minimum bactericidal percentage (MBP) was defined as the lowest percentage of CFS that can kill all the pathogenic bacteria, as detected by subculturing treated samples onto MH agar. All tests were done in triplication.
Carbapenem-resistant Enterobacteriaceae and Lactobacillus strains were individually cultured in their respective broth medium at 37°C for 24 h. The cultures were centrifuged at 6000 rpm, 22°C for 10 min to collect the cell pellet. Then, pathogenic bacteria were inoculated at 1 × 106 CFU/ml and lactobacilli at 1 × 105, 1 × 106, 1 × 107, or 1 × 108 CFU/ml into mono-cultures or 1 × 106 CFU/ml pathogenic bacteria co-culture with 1 × 105, 1 × 106, 1 × 107, or 1 × 108 CFU/ml lactobacilli in tubes containing 10 ml of MRS-MH broth (1:1) (Drago et al., 1997). Mono-cultures and co-cultures were incubated at 37°C for 48 h. Samples were collected at 0, 2, 4, 8, 24, and 48 h for the determination of viable cell count and pH measurements. A 1 ml aliquot of each sample was used to prepare serial dilutions that were poured onto the appropriate agar plates; MRS agar was used for Lactobacillus spp., while MacConkey Agar was used for Enterobacteriaceae. Plates were incubated at 37°C for 24 h and colonies were counted. The assay detection limit was 100 CFU/ml (Shah et al., 2016). All tests were done in triplication.
The paired t-test was used for statistical analysis. The level of significance for all analysis was p < 0.05.
A total of 57 Lactobacillus isolates, including L. plantarum (n = 17), L. paracasei (n = 16), L. rhamnosus (n = 8), L. fermentum (n = 6), L. casei (n = 4), L. brevis (n = 2), L. futsaii (n = 2), L. furfuricola (n = 1), and L. sakei (n = 1) were collected for this study (Table 1). Based on the findings from MIC testing of these Lactobacillus isolates, we determined that these isolates were highly susceptible to chloramphenicol, clindamycin, erythromycin, linezolid and streptomycin with susceptibility rates ranging from 91.2 to 100%. The resistance rates of these isolates against fusidic acid, kanamycin, trimethoprim/sulfamethoxazole, teicoplanin, and vancomycin ranged from 89.5 to 100% (Table 2).
Table 3 displays the results of the pepsin tolerance tests. The viability of 32 of the isolates was reduced by <2 log. Another 19 isolates were totally inhibited in these simulated conditions. Table 4 shows the results of the bile salt tolerance tests. With the exception of LYC1164, which had a 2.1-log reduction in growth, all of the isolates had a less than a 2-log reduction in cell viability after incubation in this simulated intestinal condition for 24 h. Based on the findings of these in vitro viability tests, we chose 31 Lactobacillus isolates with less than 2-log reductions in viability in both pepsin and bile salt tolerance tests. Because of their ability to survive in these simulated gastric and intestinal environments, we selected these strains for the assessment their antibacterial activity.
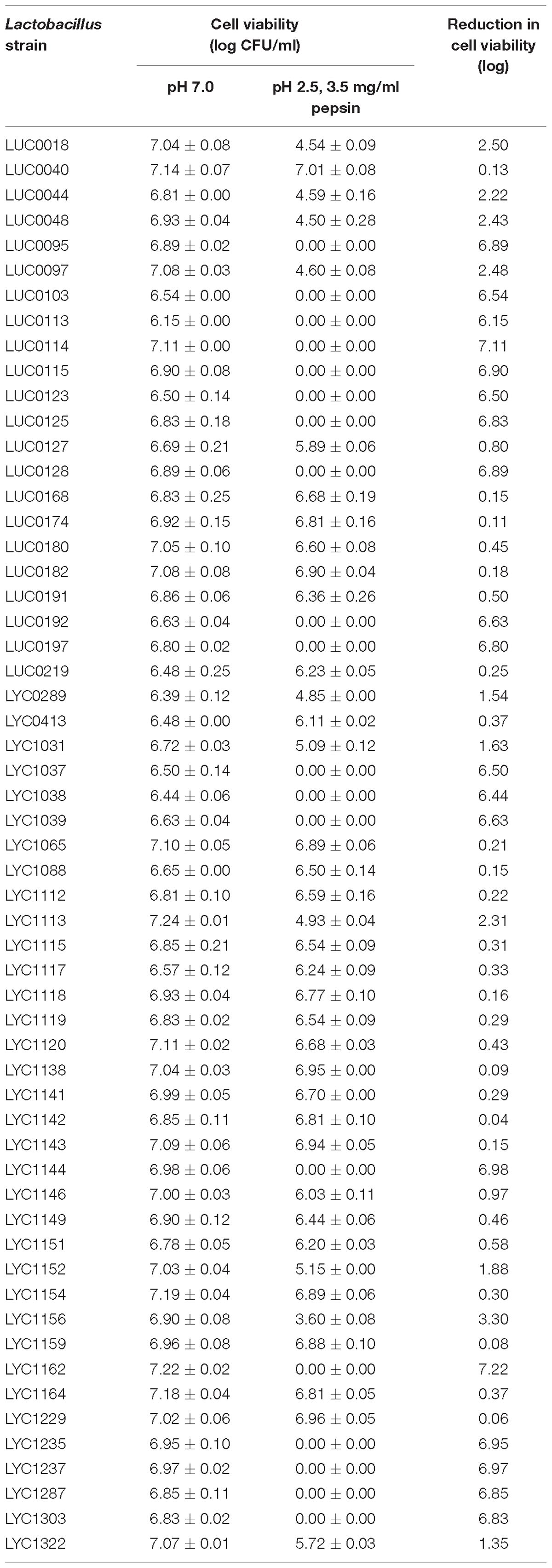
Table 3. Pepsin tolerance test results for 57 Lactobacillus isolates at pH 2.5 with 3.5 mg/ml pepsin.
The 21 isolates displayed the greatest activity against carbapenem-resistant E. coli with zones of inhibition greater than 15 mm (Figure 2A). The greatest activity against carbapenem-resistant K. pneumoniae isolates, with zones of inhibition greater than or equal to 15 mm, were observed in the nine isolates (Figure 2B). Overall, we chose five Lactobacillus strains (LUC0180, LUC0219, LYC0289, LYC0413, and LYC1031), which had the significantly better antibacterial activities than the most of the other strains (p < 0.05) as determined by the agar well diffusion and time-kill assay for further tests of antibacterial activity.
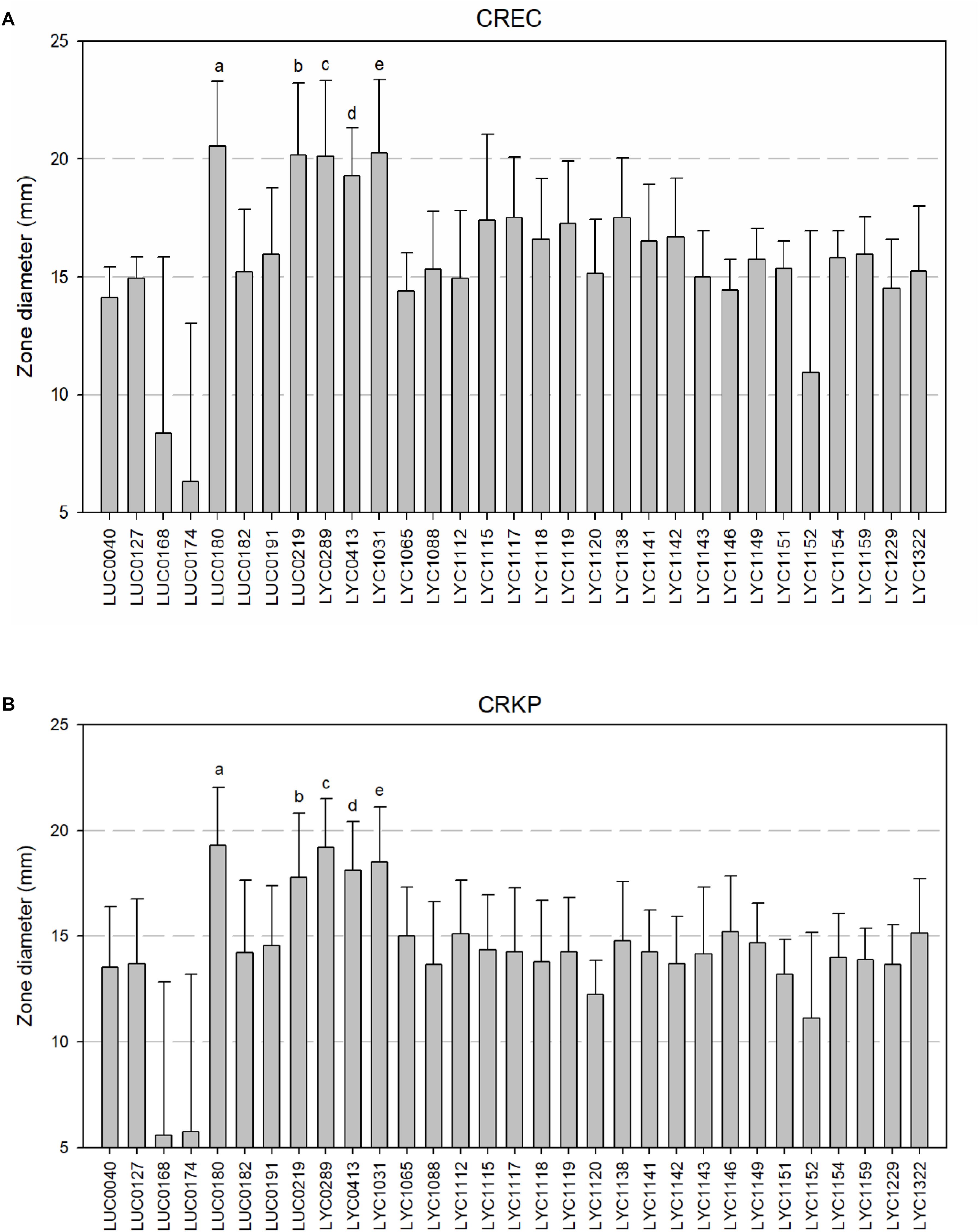
Figure 2. (A) Zone of inhibition of 31 Lactobacillus isolates against carbapenem-resistant E. coli by well diffusion assays. (a, b, c, d, e five isolates showed the significantly better activity than all of the other strains (all p < 0.05). However, there was no significant differences among them). (B) Zone of inhibition of 31 Lactobacillus isolates against carbapenem-resistant K. pneumoniae by well diffusion assays. [a, b, c, d, e five isolates showed the significantly better activity than all of the other strains (all p < 0.05). However, there was no significant differences among them].
Table 5 shows the MIP and MBP for the five most potent Lactobacillus supernatants against CRE. The MIP of these five strains ranged from 10 to 30%. Except for LUC0180, which had an MBP of ≥40%, all of the strains had MBPs ranging from 20 – 40%. The inhibitory effects of these Lactobacillus strains did not change after heating up to 80°C for 10 min. However, the inhibitory effect disappeared once the pH increased to 7.0 (data not show). Additionally, the pH in the presence of LUC0180 seemed to be higher than the other four isolates.
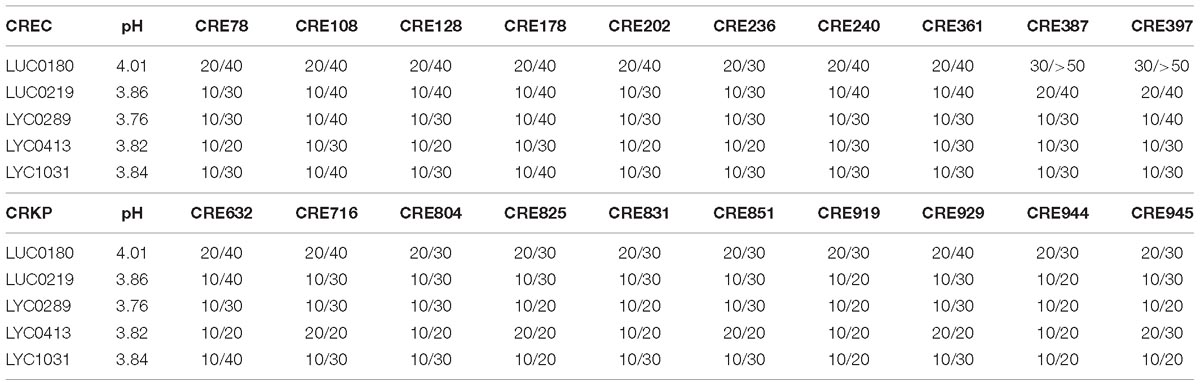
Table 5. The resulting pH and MIPs/MBPs of five Lactobacillus strain supernatants (%) against carbapenem-resistant E. coli (CREC) and K. pneumoniae (CRKP).
Figure 3 shows the results from time-killing test and assessment of the association between pH and antibacterial effects. After a 24-h incubation, there was no significant change between mono-cultures of lactobacilli and co-culture of lactobacilli and carbapenem-resistant E. coli (CRE316) in term of the concentration of lactobacilli (all p > 0.05) (Figure 3A). In contrast, the growth of CRE 316 was significantly inhibited after co-culture with different concentration lactobacilli after 24 h (all p < 0.05) and even 48 h (p < 0.0001) when comparing with mono-culture of CRE (Figure 3B). Furthermore, the decreases in pH were observed in the mono-cultures of lactobacilli,especially at the concentration of 108 and 107 CFU/ml (both p < 0.05) (Figure 3C). The decrease in pH remains significant while in the co-cultures with CRE316 compared with CRE316 monoculture (all p < 0.05, Figure 3D). The similar findings were also noted for the co-culture with carbapenem-resistant K. pneumonia (CR632) (Figures 3E–H).
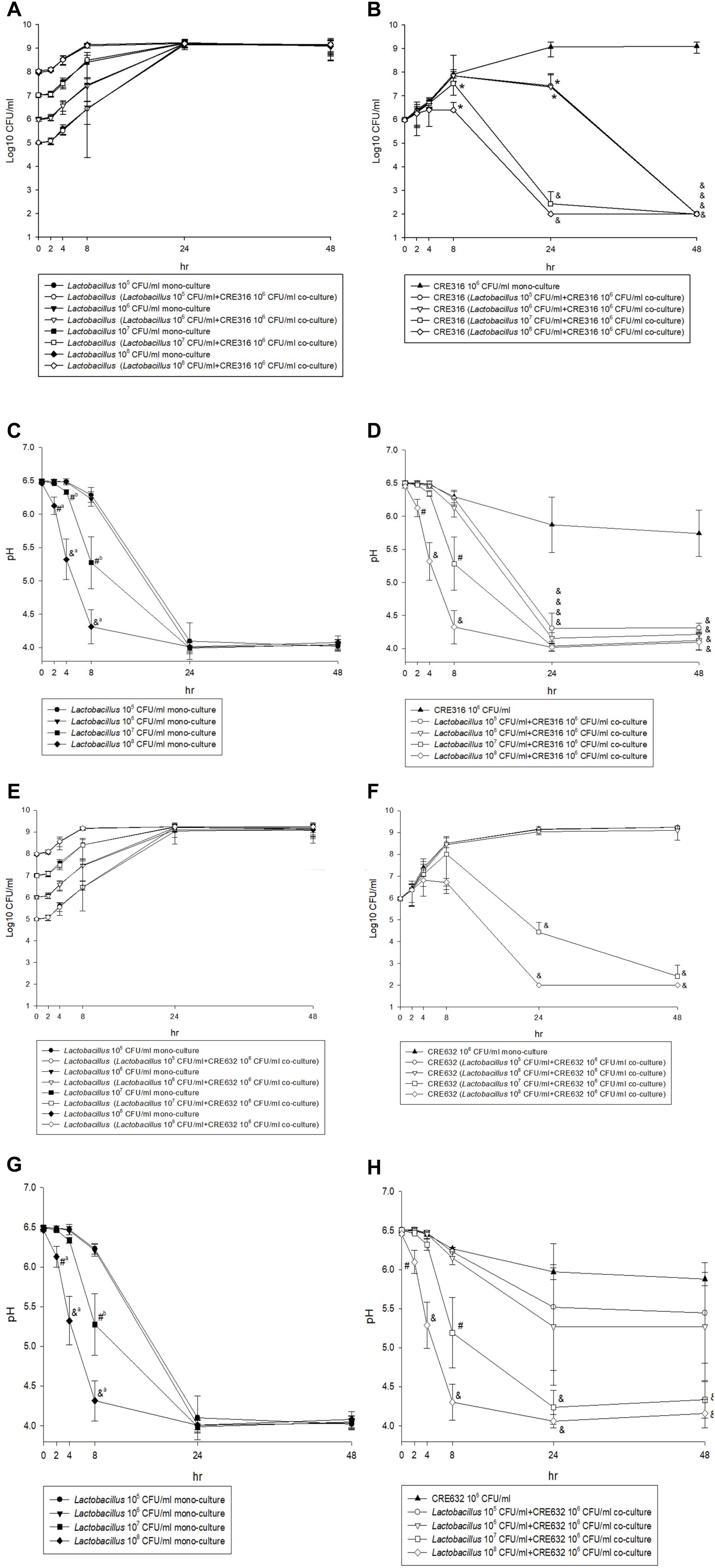
Figure 3. (A,E) The CFU change of five different concentration (105 to 108 CFU/ml) Lactobacillus isolates after mono-culture or co-culture with CRE316/ 632 for 48 h. (B,F) The CFU change of CRE316/ 632 after mono-culture or co-culture with five different concentration (105 to 108 CFU/ml) Lactobacillus isolates for 48 h and comparison between mono-culture and co-culture. (C,G) The pH changes of five different concentration (105 to 108 CFU/ml) Lactobacillus isolates mono-culture for 48 h. (D,H) The pH changes of CRE316/ 632 after mono-culture or co-culture with five different concentration (105 to 108 CFU/ml) Lactobacillus isolates and comparison between mono-culture and co-culture. (a Means the comparisons between 108 and 105,6,7 CFU/ml lactobacillus isolates. b Means the comparisons between 107 and 105,6 CFU/ml Lactobacillus isolates. ∗P-value < 0.05, #P-value < 0.001, &P-value < 0.0001).
In this study, five Lactobacillus strains, including three L. plantarum strains (LUC0219, LYC0289, LYC1031) and one each of L. paracaseri (LUC0180) and L. rhamnosus (LYC0413), exhibited good antibacterial activity against CREs. These inhibitory effects were demonstrated in through various tests including the agar well diffusion method, broth microdilution assay, and time-kill test. Lactobacillus spp. are non-pathogenic Gram-positive rods that are also recognized as normal human flora. Recently, several studies have reported that Lactobacillus strains can exhibit antibacterial activity through several mechanisms such as the production of antimicrobial substances or metabolites (hydrogen peroxide, lactic acid, bacteriocin), competition for nutrients, inhibition of bacterial adhesion to the mucosa, and enhancement of the immune response (Gill et al., 2001; Reid and Burton, 2002; Servin, 2004; Saulnier et al., 2009). Some in vitro studies have also shown that Lactobacillus strains can exhibit antimicrobial activity against C. difficile, E. coli, Shigella spp., S. mutans, P. aeruginosa, and S. aureus (Jamalifar et al., 2011; Mirnejad et al., 2013; Kumar et al., 2016; Kang et al., 2017; Ahn et al., 2018). To the best of our knowledge, this is the first study to document evidence of Lactobacillus antimicrobial activity against CRE. Although in vitro activity cannot be directly translated to in vivo effect, our findings suggest a promising role for Lactobacillus strains in the prevention and treatment of CRE colonization or infection. However, further animal studies are warranted to clarify this issue.
In the time-kill studies, the co-cultures with CREs did not influence the growth of lactobacilli. Additionally, effective inhibitory activity was generally observed at a pH of less than 4.2. These results were comparable with what we observed in our MIP/MBP experiments. By contrast, the antibacterial activity of Lactobacillus strains was dependent on an acidic environment and the inhibitory effect disappeared once the pH became greater than 6.5 during the MIP/MBP tests. Importantly, antimicrobial activity was not influenced by heating. This indicates that the inhibitory effect is mostly due to the acidic conditions, and not the production of bacteriocin-like substances. Similar findings were noted in the time-kill test. Previous reports have shown that the production of organic acids by probiotic organisms and the resulting decrease in culture pH is considered to be the principal antimicrobial effect (Zhang et al., 2011; Tejero-Sarinena et al., 2012). In addition to lowering the culture pH, Alakomi et al. (2000) reported that organic acids could also function as outer membrane permeabilizers of some gram-negative bacteria and enhance the activity of other antimicrobial metabolites. These findings suggest that acidic pH and/or the presence of organic acids may be essential for the antibacterial activity observed in the present study.
In this study, only 31 of the 57 Lactobacillus strains that were initially collected for assessment were found to be able to survive in the simulated gastric and intestinal environment. For clinical application, these characteristics are extremely important. To be functional in the lower enteric tract, the tolerance of lactobacilli to pepsin, low pH and bile salts is essential.
This study had several limitations. First, in addition to the effect of acid environment, we did not investigate the detail mechanism or molecular effectors of these function traits. Second, we used separated tests instead of a unique trait by using in vitro GIT system to evaluate the GIT resistance in this study. Further study is warranted to clarify these issues.
Several Lactobacillus strains exhibit antibacterial activity against CRE. We suggest that this effect may have potential applications through the use of Lactobacillus strains as starter cultures in fermented foods or as food preservatives for controlling or preventing CRE infections. Additional studies are required to determine the effects of complex nutrients on the synthesis of the antibacterial substance, as well as to elucidate the mechanisms and genetic basis of the bactericidal activity. Further animal models are also necessary to document the in vivo survival of these acid, pepsin and bile salt tolerant lactobacilli in the lower enteric tract. If the same inhibitory effect can be documented in vivo, further studies including vancomycin-resistant Enterococcus should be performed in the future.
H-JT and Y-CL are the guarantors of this manuscript. C-CC, C-CL, H-LH, W-YH, H-ST, T-CW, and Y-CC contributed to the conception and design of the study. C-CC and H-JT analyzed and interpreted the data. C-CC, C-CL, and H-JT drafted the manuscript. All authors reviewed the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ahn, K. B., Baik, J. E., Park, O. J., Yun, C. H., and Han, S. H. (2018). Lactobacillus plantarum lipoteichoic acid inhibits biofilm formation of Streptococcus mutans. PLoS One 13:e0192694. doi: 10.1371/journal.pone.0192694
Alakomi, H. L., Skytta, E., Saarela, M., Mattila-Sandholm, T., Latva-Kala, K., and Helander, I. M. (2000). Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 66,2001–2005.
Arena, M. P., Silvain, A., Normanno, G., Grieco, F., Drider, D., Spano, G., et al. (2016). Use of Lactobacillus plantarum strains as a bio-control strategy against food-borne pathogenic microorganisms. Front. Microbiol. 7:464. doi: 10.3389/fmicb.2016.00464
British Society for Antimicrobial Chemotherapy [BSAC] (2014). British Society for Antimicrobial Chemotherapy. Standing Committee on Susceptibility Testing. Version 13.0.
Clinical and Laboratory Standards Institute [CLSI] (2009). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically-. Approved Standard M07-A9, 9th Edn. Wayne, PA: CLSI.
Clinical and Laboratory Standards Institute [CLSI] (2015). Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard—CLSI Document M02-A12, 12th Edn. Wayne, PA: CLSI.
Clinical and Laboratory Standards Institute [CLSI] (2016). Performance Standards for Antimicrobial Susceptibility Testing; Sixteenth Informational Supplement M100-S26. Wayne, PA: Clinical & Laboratory Standards Institute.
Drago, L., Gismondo, M. R., Lombardi, A., de Haën, C., and Gozzini, L. (1997). Inhibition of in vitro growth of enteropathogens by new Lactobacillus isolates of human intestinal origin. FEMS Microbiol. Lett. 153, 455–463.
Georgieva, R., Yocheva, L., Tserovska, L., Zhelezova, G., Stefanova, N., Atanasova, A., et al. (2015). Antimicrobial activity and antibiotic susceptibility of Lactobacillus and Bifidobacterium spp. intended for use as starter and probiotic cultures. Biotechnol. Biotechnol. Equip. 29, 84–91.
Gill, H. S., Rutherfurd, K. J., Cross, M. L., and Gopal, P. K. (2001). Enhancement of immunity in the elderly by dietary supplementation with the probiotic Bifidobacterium lactis HN019. Am. J. Clin. Nutr. 74, 833–839.
Hsueh, P. R., Badal, R. E., Hawser, S. P., Hoban, D. J., Bouchillon, S. K., Ni, Y., et al. (2010). Epidemiology and antimicrobial susceptibility profiles of aerobic and facultative gram-negative bacilli isolated from patients with intra-abdominal infections in the Asia-Pacific region: 2008 results from SMART (Study for Monitoring Antimicrobial Resistance Trends). Int. J. Antimicrob. Agents 36, 408–414. doi: 10.1016/j.ijantimicag.2010.07.002
Inglin, R. C., Stevens, M. J., Meile, L., Lacroix, C., and Meile, L. (2015). High-throughput screening assays for antibacterial and antifungal activities of Lactobacillus species. J. Microbiol. Methods 114, 26–29. doi: 10.1016/j.mimet.2015.04.011
Jacobsen, C. N., Rosenfeldt Nielsen, V., Hayford, A. E., Møller, P. L., Michaelsen, K. F., Paerregaard, A., et al. (1999). Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 65, 4949–4956.
Jamalifar, H., Rahimi, H., Samadi, N., Shahverdi, A., Sharifian, Z., Hosseini, F., et al. (2011). Antimicrobial activity of different Lactobacillus species against multi-drug resistant clinical isolates of Pseudomonas aeruginosa. Iran J. Microbiol. 3, 21–25.
Jean, S. S., Coombs, G., Ling, T., Balaji, V., Rodrigues, C., and Mikamo, H. (2016). Epidemiology and antimicrobial susceptibility profiles of pathogens causing urinary tract infections in the Asia-Pacific region: results from the study for monitoring antimicrobial resistance trends (SMART), 2010-2013. Int. J. Antimicrob. Agents 47, 328–334. doi: 10.1016/j.ijantimicag.2016.01.008
Kang, M. S., Lim, H. S., Oh, J. S., Lim, Y. J., Wuertz-Kozak, K., Harro, J. M., et al. (2017). Antimicrobial activity of Lactobacillus salivarius and Lactobacillus fermentum against Staphylococcus aureus. Pathog. Dis. 75:ftx009.
Klare, I., Konstabel, C., Muller-Bertling, S., Reissbrodt, R., Huys, G., Vancanneyt, M., et al. (2005). Evaluation of new broth media for microdilution antibiotic susceptibility testing of Lactobacilli, Pediococci, Lactococci, and Bifidobacteria. Appl. Environ. Microbiol. 71, 8982–8986.
Kumar, M., Dhaka, P., Vijay, D., Vergis, J., Mohan, V., Kumar, A., et al. (2016). Antimicrobial effects of Lactobacillus plantarum and Lactobacillus acidophilus against multidrug-resistant enteroaggregative Escherichia coli. Int. J. Antimicrob. Agents 48, 265–270. doi: 10.1016/j.ijantimicag.2016.05.014
Lai, C. C., Chen, C. C., Huang, H. L., Chuang, Y. C., and Tang, H. J. (2016). The role of doxycycline in the therapy of multidrug-resistant E. coli - an in vitro study. Sci. Rep. 6:31964. doi: 10.1038/srep31964
Lai, C. C., Chen, Y. H., Lin, S. H., Chung, K. P., Sheng, W. H., Ko, W. C., et al. (2014). Changing aetiology of healthcare-associated bloodstream infections at three medical centres in Taiwan, 2000-2011. Epidemiol. Infect. 142, 2180–2185. doi: 10.1017/S0950268813003166
McFarland, L. V. (2015). Probiotics for the primary and secondary prevention of C. difficile infections: a meta-analysis and systematic review. Antibiotics 4, 160–178.
Mirnejad, R., Vahdati, A. R., Rashidiani, J., Erfani, M., and Piranfar, V. (2013). The antimicrobial effect of Lactobacillus casei culture supernatant against multiple drug resistant clinical isolates of Shigella sonnei and Shigella flexneri in vitro. Iran Red Crescent Med. J. 15, 122–126. doi: 10.5812/ircmj.7454
Reid, G., and Burton, J. (2002). Use of Lactobacillus to prevent infection by pathogenic bacteria. Microbes Infect. 4, 319–324.
Rodriguez-Bano, J., Gutierrez-Gutierrez, B., Machuca, I., and Pascual, A. (2018). Treatment of infections caused by extended-spectrum-beta-lactamase-, ampC-, and carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Rev. 31:e00079-17. doi: 10.1128/CMR.00079-17
Saulnier, D. M., Spinler, J. K., Gibson, G. R., and Versalovic, J. (2009). Mechanisms of probiosis and prebiosis: considerations for enhanced functional foods. Curr. Opin. Biotechnol. 20, 135–141. doi: 10.1016/j.copbio.2009.01.002
Servin, A. L. (2004). Antagonistic activities of Lactobacilli and Bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28, 405–440.
Shah, N., Patel, A., Ambalam, P., Holst, O., Ljungh, A., and Prajapati, J. (2016). Determination of an antimicrobial activity of Weissella confusa, Lactobacillus fermentum, and Lactobacillus plantarum against clinical pathogenic strains of Escherichia coli and Staphylococcus aureus in co-culture. Ann. Microbiol. 66, 1137–1143.
Tang, H. J., Hsieh, C. F., Chang, P. C., Chen, J. J., Lin, Y. H., Lai, C. C., et al. (2016a). Clinical significance of community- and healthcare-acquired carbapenem-resistant Enterobacteriaceae isolates. PLoS One 11:e0151897. doi: 10.1371/journal.pone.0151897
Tang, H. J., Lai, C. C., Chen, C. C., Zhang, C. C., Weng, T. C., Chiu, Y. H., et al. (2016b). Colistin-sparing regimens against Klebsiella pneumoniae carbapenemase-producing K. pneumoniae isolates: combination of tigecycline or doxycycline and gentamicin or amikacin. J. Microbiol. Immunol. Infect. doi: 10.1016/j.jmii.2016.03.003 [Epub ahead of print].
Tejero-Sarinena, S., Barlow, J., Costabile, A., Gibson, G. R., and Rowland, I. (2012). In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: evidence for the effects of organic acids. Anaerobe 18, 530–538. doi: 10.1016/j.anaerobe.2012.08.004
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Toh, H. S., Chuang, Y. C., Huang, C. C., Lee, Y. L., Liu, Y. M., Ho, C. M., et al. (2012). Antimicrobial susceptibility profiles of gram-negative bacilli isolated from patients with hepatobiliary infections in Taiwan: results from the study for monitoring antimicrobial resistance trends (SMART), 2006-2010. Int. J. Antimicrob. Agents 40, S18–S23. doi: 10.1016/S0924-8579(12)70005-5
Tokatli, M., Gulgor, G., Bagder Elmaci, S., Arslankoz İsleyen, N., and Özçelik, F. (2015). In vitro properties of potential probiotic indigenous lactic acid bacteria originating from traditional pickles. Biomed. Res. Int. 2015:315819. doi: 10.1155/2015/315819
Keywords: Lactobacillus, carbapenem-resistant, Enterobacteriaceae, in vitro – antibacterial, activity
Citation: Chen C-C, Lai C-C, Huang H-L, Huang W-Y, Toh H-S, Weng T-C, Chuang Y-C, Lu Y-C and Tang H-J (2019) Antimicrobial Activity of Lactobacillus Species Against Carbapenem-Resistant Enterobacteriaceae. Front. Microbiol. 10:789. doi: 10.3389/fmicb.2019.00789
Received: 28 November 2018; Accepted: 27 March 2019;
Published: 18 April 2019.
Edited by:
Benjamin Andrew Evans, University of East Anglia, United KingdomReviewed by:
Alberto Finamore, Council for Agricultural Research and Economics, ItalyCopyright © 2019 Chen, Lai, Huang, Huang, Toh, Weng, Chuang, Lu and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying-Chen Lu, biolyc2016@gmail.com Hung-Jen Tang, 8409d1@gmail.com
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.