- 1Fujian University Key Laboratory for Plant-Microbe Interaction, College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Institute for Plant Genomics and Biotechnology, College of Agriculture and Life Sciences, Texas A&M University, College Station, TX, United States
- 3Department of Plant Pathology and Microbiology, College of Agriculture and Life Sciences, Texas A&M University, College Station, TX, United States
- 4Institute of Oceanography, Minjiang University, Fuzhou, China
The Bcl-2 associated athanogene (BAG) family is an evolutionarily conserved group of co-chaperones that confers stress protection against a variety of cellular insults extending from yeasts, plants to humans. Little is known, however, regarding the biological role of BAG proteins in phytopathogenic fungi. Here, we identified the unique BAG gene (BcBAG1) from the necrotrophic fungal pathogen, Botrytis cinerea. BcBAG1 is the homolog of Arabidopsis thaliana AtBAG4, and ectopic expression of BcBAG1 in atbag4 knock-out mutants restores salt tolerance. BcBAG1 deletion mutants (ΔBcbag1) exhibited decreased conidiation, enhanced melanin accumulation and lost the ability to develop sclerotia. Also, BcBAG1 disruption blocked fungal conidial germination and successful penetration, leading to a reduced virulence in host plants. BcBAG1 contains BAG (BD) domain at C-terminus and ubiquitin-like (UBL) domain at N-terminus. Complementation assays indicated that BD can largely restored pathogenicity of ΔBcbag1. Abiotic stress assays showed ΔBcbag1 was more sensitive than the wild-type strain to NaCl, calcofluor white, SDS, tunicamycin, dithiothreitol (DTT), heat and cold stress, suggesting BcBAG1 plays a cytoprotective role during salt stress, cell wall stress, and ER stress. BcBAG1 negatively regulated the expression of BcBIP1, BcIRE1 and the splicing of BcHAC1 mRNA, which are core regulators of unfolded protein response (UPR) during ER stress. Moreover, BcBAG1 interacted with HSP70-type chaperones, BcBIP1 and BcSKS2. In summary, this work demonstrates that BcBAG1 is pleiotropic and not only essential for fungal development, hyphal melanization, and virulence, but also required for response to multiple abiotic stresses and UPR pathway of B. cinerea.
Introduction
Co-chaperones are proteins that assist chaperones in protein folding, oligomeric assembly, and protein transportation and degradation (Hartl, 1996). The Bcl-2-associated athanogene (BAG) family is a group of broadly conserved co-chaperones of 70-kilodalton heat shock protein (HSP70) (Bracher and Verghese, 2015). In mammals, the BAG family was initially identified by screening mouse cDNA library for Bcl-2 interaction proteins (Takayama et al., 1995). Using the ATPase domain of HSC70/HSP70 as molecular bait in yeast two-hybrid screening, additional BAG family members were identified from human, Caenorhabditis elegans and the fission yeast Schizosaccharomyces pombe (Takayama et al., 1999). The human BAGs contain six members, BAG1 to BAG6, and share a conserved BAG domain (BD) of approximately 45 amino acids, located near the C terminus (Takayama and Reed, 2001). As nucleotide exchange factors (NEFs) of HSP70, BAG proteins play a major role in both positively and negatively modulating HSP70 ATP activity via the BAG domain (Gassler et al., 2001). Moreover, BAGs act as scaffolds between HSP70 and target transcription factors or proteins, thus affecting diverse physiological events (Townsend et al., 2003; Kabbage and Dickman, 2008).
The identification and preliminary characterization of plant BAG proteins is still underway. Using a combination of bioinformatics and structural algorithms, seven BAGs have been identified from Arabidopsis thaliana (AtBAG1–AtBAG7) (Doukhanina et al., 2006), several of which have been selected for functional characterization. The structural and biochemical data of AtBAGs demonstrate that the AtBAGs function as NEFs for HSP70/HSC70 and the regulation mechanism of HSP70/HSC7 is conserved in plants (Fang et al., 2013). Our previous work has uncovered that AtBAG4, 6, and 7 exhibit different cytoprotective specificities. Briefly, AtBAG4 appears to protect plants from various abiotic stress stimuli, e.g., salt and drought (Doukhanina et al., 2006). AtBAG6 is activated via proteolytic cleavage by a specific plant aspartyl protease that is required for autophagic cell death in planta and subsequent resistance to the necrotrophic fungus Botrytis cinerea (Kabbage et al., 2016; Li and Dickman, 2016; Li et al., 2016). Consistently, overexpression of AtBAG6 induced programmed cell death (PCD) in both yeast and plants (Kang et al., 2006). The ER-localized AtBAG7 is an essential component of the unfolded protein response (UPR) and directly interacts with UPR regulator AtBIP2 (Williams et al., 2010). Under heat stress, AtBAG7 is also proteolytically processed in the ER lumen and translocate from the ER to the nucleus, where it interacts with the transcriptional factor WRKY29 for heat tolerance (Li et al., 2017). A number of BAGs have also been reported in other plant species. For example, OsBAG4 from rice interacts with an E3 ubiquitin ligase EBR1 and regulates PCD, which controls plant immunity and broad-spectrum disease resistance (You et al., 2016). Taken together, plant BAGs are multifunctional and modulate numerous physiological and biological processes.
Although the functions of human and plants BAG family members are extensively studied, there is limited knowledge on the roles of the BAGs in fungi. Previous studies showed that SNL1, a mammalian homolog of BAG1 in Saccharomyces cerevisiae, is functionally linked to the nuclear pore complex and plays a role in promoting both protein biogenesis and translation by recruiting ribosomes and HSP70 to the ER membrane (Ho et al., 1998; Verghese and Morano, 2012). Two other BAG1 homologs from Schizosaccharomyces pombe, BAG101 and BAG102, are co-factors of 26S proteasomes, and play as HSP70 chaperones (Kriegenburg et al., 2014). Overexpression of BAG101 and BAG102 inhibit cell growth by triggering HSP70 to release and activate HSF1 (heat shock factor 1) (Poulsen et al., 2017). To date, the only example in filamentous fungi is BAGA from Aspergillus nidulans, which impacts fungal sexual development and modulates secondary metabolism (Jain et al., 2018).
The endoplasmic reticulum (ER) is the central intracellular organelle for protein translocation, protein folding, and protein post-translational modifications, allowing further transport of proteins to the Golgi apparatus and ultimately to vesicles for secretion or display on the plasma surface. Perturbations in ER function, named “ER-stress,” unfolded or misfolded proteins accumulate within the ER and disrupt ER homeostasis to activate an intracellular signaling pathway, known as the UPR eventually culminating in cell death (Malhotra and Kaufman, 2007). As a conserved survival pathway to counteract the lethal effects caused by ER stress, UPR can mitigate accumulation of unfolded proteins and restore ER homeostasis by reducing protein translation and while increasing and misfolded proteins degradation aided by molecular chaperones (e.g., binding immunoglobulin proteins, BiPs) (Sitia and Braakman, 2003; Ron and Walter, 2007). BiPs work as HSP70 chaperones and carry aberrant proteins from the ER to the cytoplasm for degradation by the proteasome (Gething, 1999). The IRE1-HAC1/XBP-1 pathway (HAC1 mRNA in yeast and XBP1 mRNA in metazoans) is a major branch of UPR that is remarkably conserved from yeast to human (Back et al., 2005). UPR is initiated by the activation of the ER stress sensor IRE1, which transmits the signal by removing a non-conventional intron from a transcription factor HAC1/XBP-1 mRNA to produce potent transcriptional activator of UPR targets (Ron and Walter, 2007). Comparing to the extensive studies of UPR in human and plant systems, UPR has only been delineated in small number of fungal pathogens, including Aspergillus fumigatus (Feng et al., 2011), Alternaria brassicicola (Joubert et al., 2011), Ustilago maydis (Heimel et al., 2013; Lo Presti et al., 2016), and all of which demonstrate that UPR regulation is correlated with fungal pathogenicity.
In this study, we identified a unique BAG gene in the necrotrophic fungal pathogen Botrytis cinerea, the causal agent of gray mold diseases to over 1,400 species of cultivated plants worldwide (Elad, 2016). Target gene replacement of BcBAG1 resulted to defect in vegetative growth, conidiation, sclerotial formation, penetration and attenuated virulence in B. cinerea, ΔBcbag1 mutants were more sensitive to various stress conditions indicating that BcBAG1 regulates stress tolerance in B. cinerea. In particular, BcBAG1 deletion mutants significantly increased susceptibility to diverse ER stress-inducer including heat, cold, tunicamycin (Tm), and dithiothreitol (DTT). We demonstrated that BcBAG1 binds to the ER chaperone BcBIP1 and negatively regulate UPR components, including the expression of BcBIP1, BcIRE1 and the splicing of BcHAC1 mRNA, suggesting BcBAG1 is necessary for the maintenance of the UPR in B. cinerea. Collectively, we present the evidence of identification and functional characterization for BcBAG1 a member of BAG family in B. cinerea, which is vital for fungal virulence on host plants and is required for ER stress response with regards to maintenance of UPR.
Materials and Methods
Strains and Culture Conditions
BcBAG1 deletion mutant, ΔBcbag1, was generated from the B. cinerea WT strain B05.10 (Quidde et al., 1999). All strains (Supplementary Table S1) were maintained on potato dextrose agar (PDA) medium at 22°C. Mycelia used for protoplast preparation, genomic DNA and total RNA extraction were grown in YEPD (1% peptone, 0.3% yeast extract, 2% glucose, pH 6.7) at 150 rpm, 22°C for 36 h. B. cinerea protoplast was recovered on SH medium (20% sucrose, 0.5 mM hepes, 1 mM NH4H2PO4, pH 7.0) at 22°C for 12–16 h. The selectable marker, 100 μg/ml hygromycin B (VWR) or 100 μg/ml nourseothricin sulfate (Research Products International) was supplemented to PDA containing 1% agar.
Bioinformatic Analysis
Preliminary BAG protein search and DNA sequence downloading were conducted in B. cinerea B05.10 genome database1. The phylogenetic tree was generated through MEGA v7.0 based on the neighbor-joining method (Kumar et al., 2016). Domain is predicted by Pfam2 and InterPro programs3. The multiple alignment of BAG domain sequence were constructed using Clustal X (Larkin et al., 2007).
BcBAG1 Gene Deletion and Complementation
Primers used in this study are listed in Supplementary Table S2. The replacement constructs for BcBAG1 were generated through the split-marker approach as described before (Goswami, 2012). Briefly, the 800-bp upstream and 855-bp downstream fragments of BcBAG1 were amplified with primer pairs AF/AR and BF/BR (Supplementary Table S2), respectively. The resulting amplicons ligated with the hygromycin phosphotransferase (hph) fragments by using Splicing Overlap Extension (SOE)-PCR. The resulting PCR products (20 μg) were transformed into protoplasts of B. cinerea. Protoplast preparation and PEG-mediated transformation of B. cinerea were performed as the established protocol (Gronover et al., 2001). After transformation, hygromycin-resistant transformants were picked individually and PCR analyses with designated primer pairs OF/OR, UF/UR, and AF/BR were performed to identify transformants that carried the insertion of hph at the BcBAG1 locus. Then all positive transformants were confirmed by subsequent RT-PCR and Southern blotting.
The full length of BcBAG1 was amplified from B05.10 genomic DNA, then ligated into pNAH-OGG with NcoI to create BcBAG1-GFP. GFP-fusion constructs were transformed into B05.10 for subcellular localization analysis. For complementation, the CDS of BcBAG1 with native promoter sequence were amplified with relative primer pairs (CF/CR) and cloned into pNAH-OMG harboring nourseothricin acetyltransferase gene (NAT1) with SpeI/NcoI to make BcBAG1-Com, conferring resistance to antibiotics nourseothricin sulfate. Similar method was used to create the truncated BcBAG1A1-141 and BcBAG1B142-298 constructs. These constructs were transformed into ΔBcbag1 mutants. Transformants with resistance to both nourseothricin and hygromycin were selected and confirmed by PCR and RT-PCR.
Southern Blotting and Real-Time PCR
Fungal genomic DNA was extracted as described (Raeder and Broda, 1985). For Southern blotting, the genomic DNAs were digested with PvuI (NEB) for 24 h at 37°C. Probe labeling, hybridization and detection were performed in accordance with the manufacturer’s instructions for the Digoxigenin High Prime DNA Labeling and Detection Starter Kit I (Roche Applied Science).
Total RNA was isolated using EastepTM Total RNA extraction Kit, following the manufacturer recommendation (Corp). The first-strand cDNA was synthesized with the M-MLV (Moloney Murine Leukemia Virus) reverse transcriptase (Life Technologies). Quantitative RT-PCR (qRT-PCR) was performed with SYBR Green PCR master mix (Applied Biosystems). The fungal actin gene was used as an internal reference. The relative expression levels were calculated by the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Mycelial Growth, Conidiation, and Sclerotial Formation Tests
For vegetative growth assays, 5 mm diameter mycelial plugs were cultured on fresh PDA in the dark at 22°C. Radial growth was measured by colony diameters after 3 days. Determination of the sensitivity of ΔBcbag1 to environmental stresses were performed on modified PDA plates with: 1 M NaCl, 1.2 M D-sorbitol, 2 μg/ml Tm, 2.5 mM DTT, 20 mM H2O2, 0.5 mM tert-butyl hydroperoxide (TBHP), 15 μM bortezomib (Bort), 200 μM MG132, 600 μg/ml calcofluor white (CFW), 0.02% SDS, 1 μg/ml iprodione (Ipro), 0.1 μg/ml fludioxonil (Flud). The percentage of mycelial radial growth inhibition (RGI) was calculated using the formula RGI% = [(C -N)/C ∗ 100], where C is colony diameter of the control, and N is colony diameter of the experimental treatment.
For conidiation assays, conidia of WT and mutants were collected from a 10-day-old PDA plate with 5 ml sterile water and spores were counted microscopically with a hemocytometer. For sclerotial formation, 5 mm diameter mycelial plugs were inoculated to PDA and incubated in the dark at 22°C, the number of mature (melanized) sclerotia were counted after 4 weeks.
Conidial Germination and Fungal Penetration Assays
Conidial germination was conducted as described by Doehlemann et al. (2006). Briefly, conidial suspensions were adjusted to 1.0–1.5 × 105 spores/ml in 10 mM KH2PO4 and 10 mM glucose solution and placed in the center of a glass slide. Incubation was kept in a moist chamber at 22°C for 12 h, 24 h, and 48 h. For the onion infection assay, 20 μl droplets (5.0 × 104 spores/ml) were deposited on the hydrophobic epidermis layers of onion and incubated for 48 h in the dark and moist chamber at 22°C (Viefhues et al., 2014).
Pathogenicity Assay
Three-day-old mycelial plugs with 5 mm diameter or 10-day-old conidial suspensions (1.0–1.5 × 105 spores/ml) were inoculated on 4-week-old detached tomato leaves and grape. Inoculated plant materials were incubated in 16 h daylight humid chamber at 22°C. Results were recorded after 4 days and 7 days. The experiment was repeated at least three times.
Yeast Two-Hybrid (Y2H) Assay
The Y2H assay was conducted according to the manufacturer’s standard instructions (Clontech). The cDNA of BcBAG1 was cloned into pGBKT7 as the bait vector and the cDNAs of HSP70-type chaperones were cloned into pGADT7 as the prey constructs, respectively. The pGBKT7-BcBAG1 and each prey vector were co-transformed into the AH109 yeast strain to evaluate interactions. The positive and negative controls were from the Kit (Clontech).
Arabidopsis Complementation Assays
Arabidopsis thaliana Col-0 and atbag4 T-DNA knock out mutants were obtained from Arabidopsis Stock Center4. Atbag4 homozygous mutants (SALK_027577C) were confirmed by PCR. The BcBAG1 full-length cDNA was cloned into pCB302ES containing the 35S promoter and the HA-epitope tag (Hwang and Sheen, 2002). This construct was transferred into the atbag4 knockout mutants by the floral dipping method (Zhang et al., 2006).
For salt stress assays, seeds were surface sterilized in 70% ethanol for 10 min and in 5% bleach solution for 5 min, and germinated on 1/2 Murashige and Skoog (MS) medium (Invitrogen) at 23°C for 5 days. The seedlings were transferred to fresh 1/2 MS medium containing 100 mM NaCl and grown at 23°C for 2 weeks to 5 weeks.
Results
Identification and Characterization of BcBAG1
To identify BAG proteins in B. cinerea, we searched B. cinerea B05.10 genome database with “BAG” as a query, and obtained one hit (Bcin10g01250.1/BC1G_05107). Additionally, based on Pfam and SMART programs, we search for all gene with BAG domain (BD domain) in genome database. Results indicated that there is only one gene with a single copy (Bcin10g01250.1/BC1G_05107) containing BAG domain, designated BcBAG1 hereafter. Phylogenetic analysis revealed that BcBAG1 shares low similarity with BAGs in yeast, plants and animals while it is closely related to BAG homologs from other filamentous fungi, e.g., Sclerotinia sclerotiorum (86.53%), Magnaporthe oryzae (57.04%), Fusarium oxysporum (52.01%), and Aspergillus nidulans (45.30%) (Supplementary Figure S1A and Supplementary Table S3). BcBAG1 contains a ubiquitin-like domain (UBL) at the N-terminus and a BAG domain at the C-terminus, encoding a 35 kD protein with 298 amino acids (Supplementary Figure S1B). The alignment also showed that most of the key interaction residues for BAG-HSC70/HSP70 binding are conserved in BAG proteins across filamentous fungi, yeast, Arabidopsis, and human (Supplementary Figure S1C).
To address the function of BcBAG1, we generated knockout mutants of BcBAG1 (ΔBcbag1), in the wild-type (WT) strain B05.10 (Supplementary Figure S2A). Two individual ΔBcbag1 lines, K3-7 and K8-6, were validated and selected for later use (Supplementary Figures S2B,C). We obtained three complemented strains by transforming the full-length BcBAG1 under its native promoter into mutants and all strains equally restored the defects of ΔBcbag1. Thus one complemented strain (BcBAG1-Com) was used in the following studies.
BcBAG1 Is Required for Vegetative Growth, Conidiation, and Sclerotial Formation
To evaluate the role of BcBAG1 during vegetative growth of B. cinerea, we examined hyphal growth on PDA. As shown in Figures 1A,C, the diameters of the ΔBcbag1 colonies were similar to WT strain B05.10 and the complementation strain BcBAG1-Com. However, ΔBcbag1 formed a thick hyphal layer on the surface of plates and the amount of aerial hypha drastically increased in comparison with B05.10 and BcBAG1-Com (Figure 1B). These results indicate that BcBAG1 influences on vegetative growth of B. cinerea.
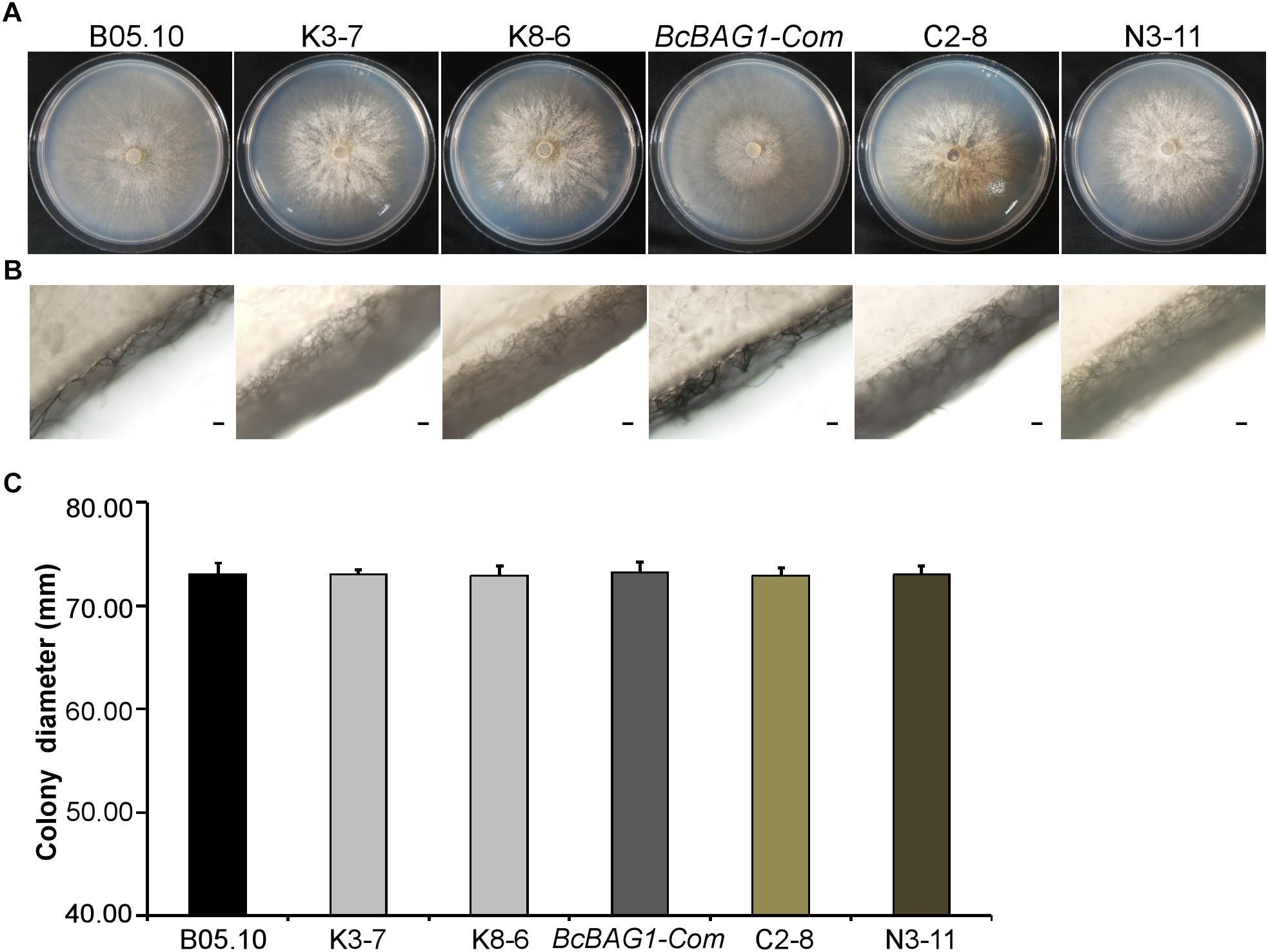
Figure 1. BcBAG1 is required for normal mycelial growth. Colonies (A) and aerial hyphae (B) of all strains were photographed after 3 days growth on PDA medium at 22°C. Scale bar: 100 μm. (C) Statistical analysis of the colony diameters of above strains. B05.10: the wild-type strain; K-3 and K8-6: ΔBcbag1 mutant lines; BcBAG1-Com (full-length BcBAG1), C2-8 (C-terminus of BcBAG1) and N3-11 (N-terminus of BcBAG1): complemented strains. Error bars represent the standard deviations from three independent experiments.
Given that wind-dispersal of conidia determines the severity of gray mold disease in the field (Leroch et al., 2013), we assessed the role of BcBAG1 in conidial production. Conidiation of B05.10, ΔBcbag1 and BcBAG1-Com from 10-day-old PDA culture was measured using microscopic examination. Although ΔBcbag1 still produced aerial mycelia (Figure 1B), conidiogenesis of ΔBcbag1 is significantly reduced (Figure 2A), in detail, the mutants produced approximately 2.1 × 107 conidia/PDA plate, while the WT produced approximately 2.0 × 108 conidia/PDA plate (P < 0.01) (Figure 2D). However, the conidia exhibited normal morphology between ΔBcbag1 and WT (Figure 2B). These results indicate that BcBAG1 involved in conidial production but do not affect conidial morphology in B. cinerea.
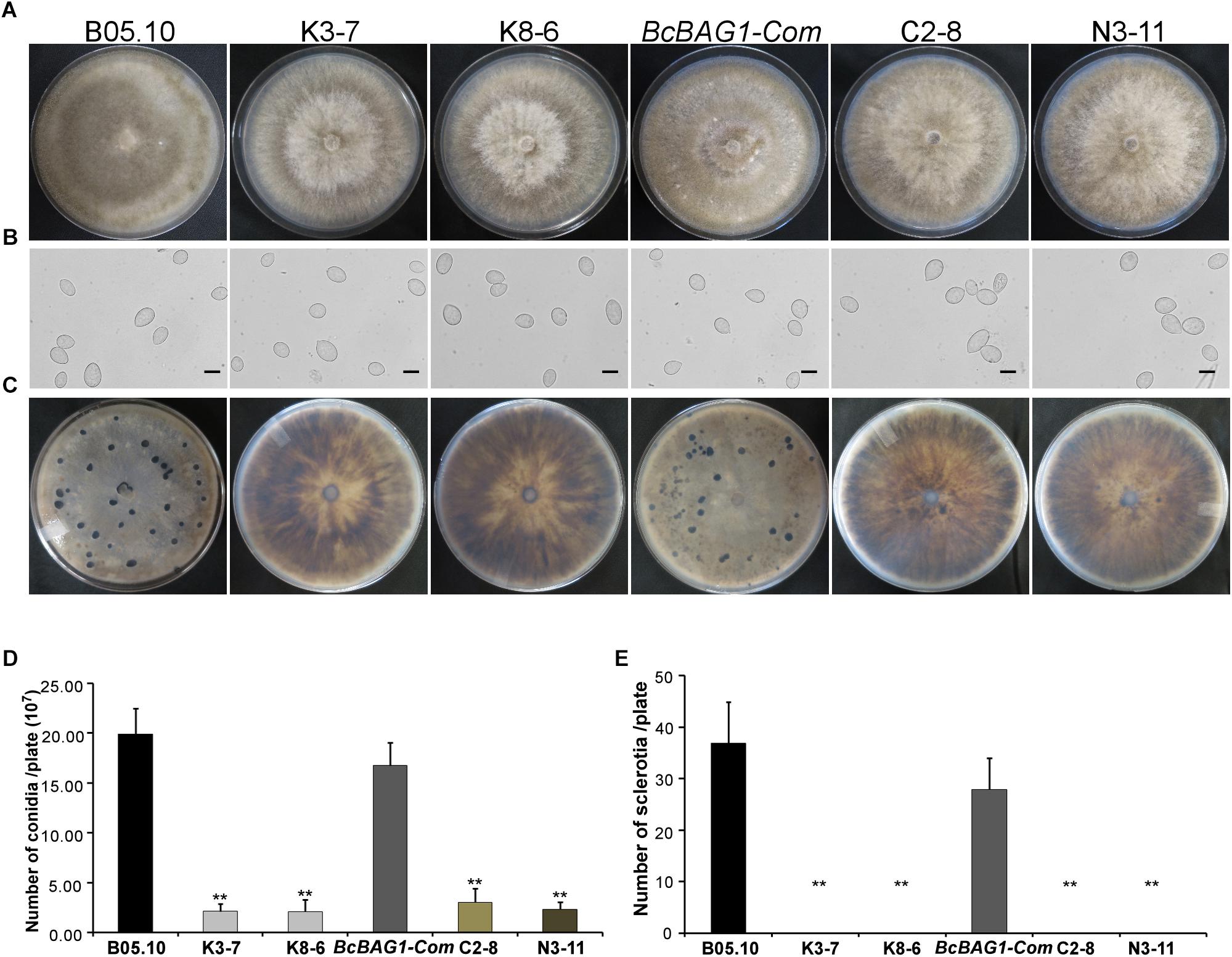
Figure 2. BcBAG1 is essential to conidiation and sclerotial formation. (A) Conidiation is affected by BcBAG1 disruption. Strains grown on PDA for 10 days were examined by light microscopy. (B) Conidia shape comparison. Conidia were collected from 10 days colonies of strains on PDA. Scale bar: 10 μm. (C) Sclerotial formation is abolished in the ΔBcbag1 mutants. Strains were incubated PDA for 10 days in the dark. (D) Quantification of conidia production on PDA plates. (E) Quantification of sclerotia formation on PDA plates. Wild-type strain (B05.10), ΔBcbag1 mutant lines (K3-7 and K8-6), and complemented strains (BcBAG1-Com, C2-8 and N3-11). Error bars represent the standard deviations from three independent experiments and asterisks denote statistical significance (P < 0.01).
Melanization of sclerotia is considered as a survival strategy of various fungi when encountering harsh environments like over-wintering (Williamson et al., 2007). Here, we examined sclerotial formation following BcBAG1 deletion in B. cinerea. After 4-week incubation on PDA in the dark, we observed that ΔBcbag1 mutants were unable to produce sclerotia, while B05.10 and BcBAG1-Com produced abundant sclerotia on PDA (Figures 2C,E), suggesting BcBAG1 is essential for sclerotial formation. Taken together, we reasoned that BcBAG1 plays a crucial role in regulating vegetative growth, conidiation and sclerotial formation in B. cinerea.
BcBAG1 Is Involved in the Regulation of Hyphal Melanization
After incubating on PDA for 7 days, we noticed that ΔBcbag1 displayed increased generation of black pigment when compared to WT (Figure 3A). It has been reported that the dark pigmentation in fungi is due to the accumulation of 1,8 dihydroxynaphthalene (DHN) melanin (Henson et al., 1999). Melanin is a dark durable pigment that protects fungi against diverse environmental stresses, such as UV irradiation and temperature extremes (Bell and Wheeler, 1986; Butler and Day, 1998). We therefore examined whether BcBAG1 participates in melanin biosynthesis. The ΔBcbag1, BcBAG1-Com and the WT strains were cultured on PDA supplemented with 50 μg/ml tricyclazole, a fungicide that specifically inhibits DHN-melanin biosynthesis (Thompson et al., 2000). The result showed tricyclazole was able to repress the massive melanin synthesis in ΔBcbag1 mutants caused by BcBAG1 deletion (Figure 3A). Additionally, we instituted RT-PCR assays to monitor the expression level of THR1 (1,3,8-trihydroxynaphthalene reductase gene), a key component in melanin biosynthesis pathway (Perpetua et al., 1996). Corresponding results obtained from RT-PCR analysis revealed a significantly up-regulation (about 10-folds) in the expression pattern of THR1 in ΔBcbag1 compared to WT (Figure 3B). These data indicate that BcBAG1 negatively regulates melanin biosynthesis pathway to suppress melanin production in B. cinerea.
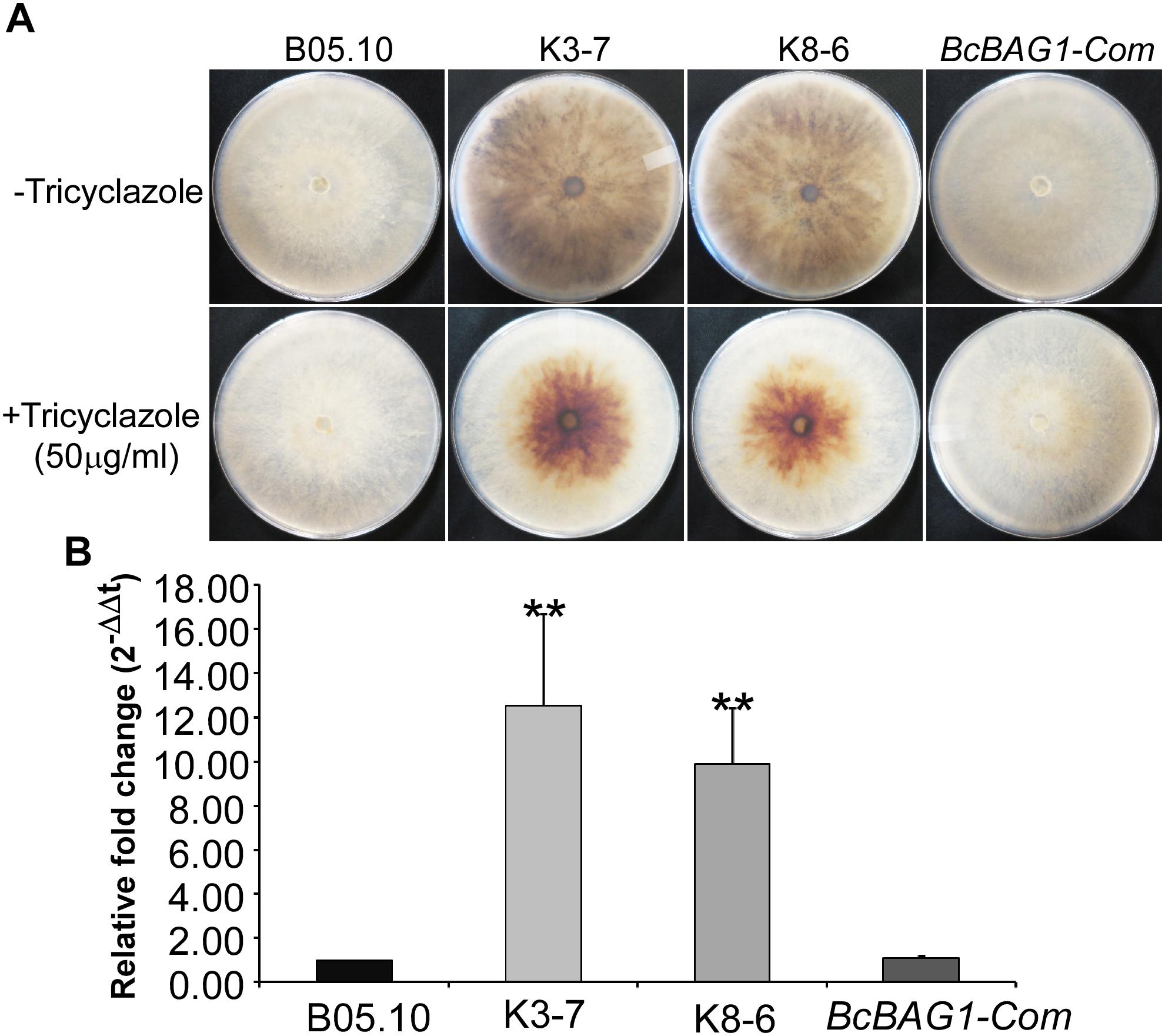
Figure 3. BcBAG1 negatively regulates hypal melanization. (A) Mycelial pigmentation were monitored on PDA for 7 days in darkness without (upper panel) and with (lower panel) tricyclazole (an inhibitor of DHN-melanin biosynthesis) in the wild-type (B05.10), ΔBcbag1 mutants (K-3 and K8-6) and complemented strain (BcBAG1-Com). (B) Transcription analysis of THR1 (1,3,8-trihydroxynaphthalene reductase gene) by RT-PCR. Total RNAs of corresponding strains grown on PDB medium were extracted and conducted for RT-PCR, asterisks denote statistical significance (P < 0.01).
BcBAG1 Is Required for Virulence of B. cinerea
To determine the role of BcBAG1 in pathogenicity and virulence of B. cinerea, we conducted infection assays by inoculating mycelial plugs containing the WT, ΔBcbag1 and BcBAG1-Com, on detached tomato leaves and grapes, respectively. Four days post-inoculation (dpi), ΔBcbag1 only initiated a small localized lesions, whereas the WT and the complemented strains have produced fully expanded lesions that were already at the soft rot stage (Figure 4A). We also performed infection assays on tomato leaves, with conidial suspensions (1.0 × 105 spores/ml). Consistently, ΔBcbag1 showed apparently attenuated virulence compared to the WT and BcBAG1-Com (Figures 4A,B). These results indicate that BcBAG1 functions as a virulence factor by enhancing colonization on the hosts.
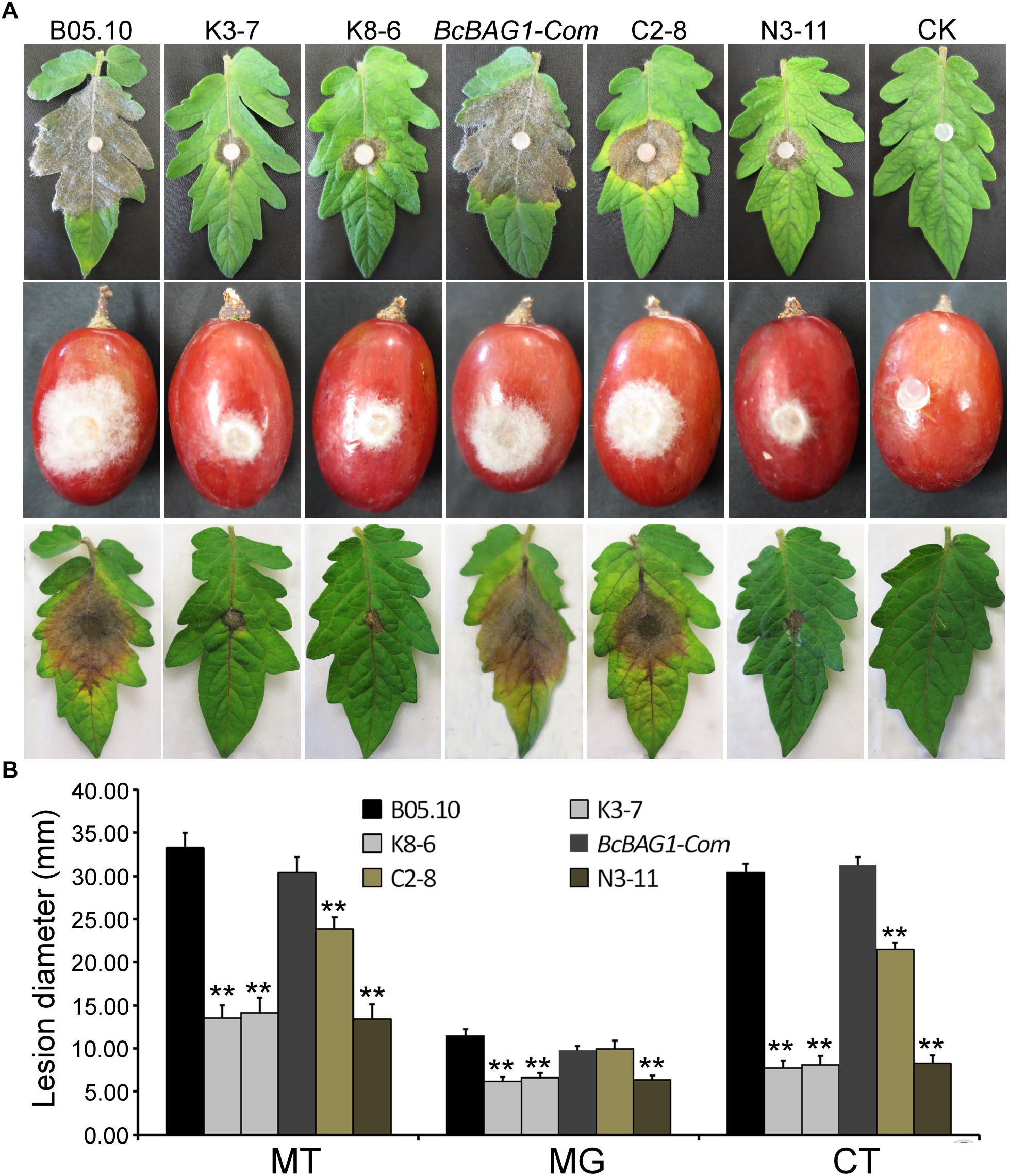
Figure 4. BcBAG1 deletion mutants attenuated pathogenicity. (A) Inoculation assays were implemented on different host plants. Mycelial plugs from PDA after 3 days growth were inoculated on detached tomato leaves (Moneymaker) (upper panel) and wounded grapes (middle panel). The disease phenotype was recorded 4 days post inoculation. Conidial suspensions (1.0–1.5 × 105 spores/ml) were inoculated on detached tomato leaves (Moneymaker) (lower panel) and the disease symptom was recorded 7 days post inoculation. (B) Quantification of lesion diameters of above inoculation. MT, mycelial plugs were inoculated on tomato leaves; MG, mycelial plugs were inoculated on grapes; CT, conidial suspensions were inoculated on tomato leaves. Wild-type strain (B05.10), ΔBcbag1 mutant lines (K3-7 and K8-6), and complemented strains (BcBAG1-Com, C2-8 and N3-11); CK, negative control. Error bars represent the standard deviations from three independent experiments and asterisks indicate statistical significance (P < 0.01).
During infection, B. cinerea produces three types of penetration structures, including germ tube apices (GA), appressoria (HA) and infection cushions (IC) (Vandenheuvel and Waterreus, 1983). To investigate whether the weak virulence of ΔBcbag1 resulted from penetration defects, we evaluated conidial germination using hydrophobic coverslips and the penetration on onion epidermis cells for microscopic observation (Figure 5A). Although all strains initiated germination 12 h post-incubation (hpi), the germination rate of ΔBcbag1 was significantly reduced (by 50%) compared to the germination recorded for the WT and BcBAG1-Com (Figure 5C). Moreover, deletion of BcBAG1 delayed the onset of appressoria formation at 24 hpi and the development of infection cushions (IC) at 48 hpi (Figure 5A). Cotton blue was used to stain the appressorium-like structure, the infection cushion and the hyphae of B. cinerea on onion epidermal cells. As shown in Figure 5B, the conidia of WT and BcBAG1-Com germinated, and the infection cushions surrounded by abundant invasive hyphae were typically developed for penetration on the onion epidermis 48 hpi. In contrast, ΔBcbag1 attenuated the ability to form infection cushions and only few germ tubes and invasive hyphae appeared on onion cells. These results suggest that the reduced virulence of ΔBcbag1 is correlated with the developmental defects of infection and penetration structures.
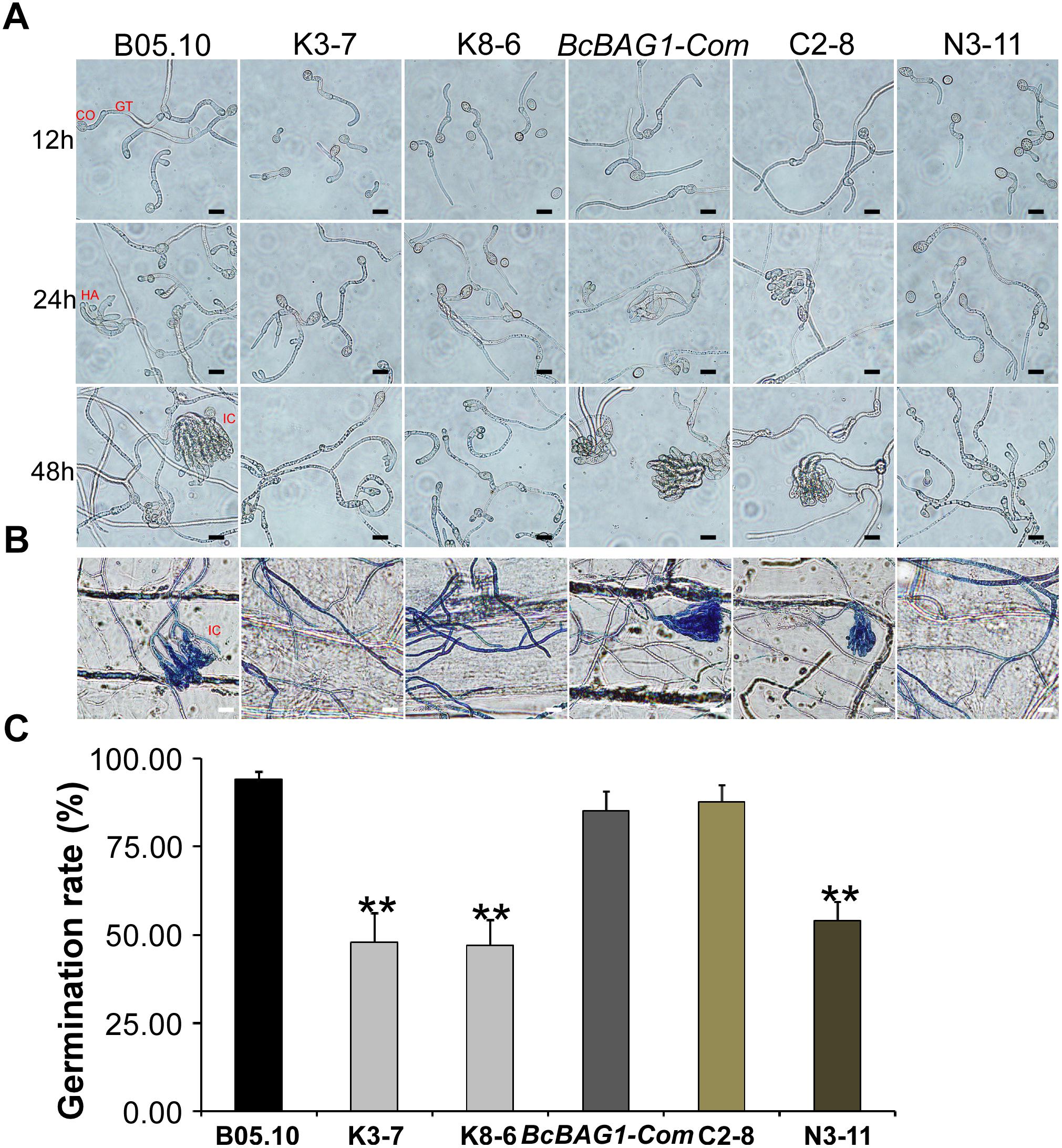
Figure 5. BcBAG1 is required for conidial germination and penetration. (A) Time course of conidial germination on glass slides when conidia were suspended in 10 mM KH2PO4 and glucose solution (pH 6.0). CO, conidia; GT, germ tubes; HA, appressoria; IC, infection cushions. Scale bar: 20 μm. (B) Penetration assay on onion epidermal cells. Onion epidermal cells were inoculated with conidial suspension (5.0 × 104 spores/ml) and the penetration was observed 48 h post incubation (hpi). External fungal structures are stained blue with cotton blue, whereas penetrated hyphae remain unstained. Scale bar: 20 μm. (C) Quantification of conidial germination rates after 12 hpi on hydrophobic glass slide. Error bars represent the standard deviations from three independent experiments (more than 100 conidia per strain). Asterisks denote statistical significance (P < 0.01).
Both UBL and BAG Domains Contribute to BcBAG1 Function
BcBAG1 possesses an N-terminal ubiquitin-like (UBL) domain and a conserved C-terminal BAG domain (BD) (Supplementary Figure S1B). Human BAG-1 interacts with HSP70 via its BAG domain and utilizes the UBL domain in targeting the chaperone cofactor to the 26S proteasome for degradation (Demand et al., 2001). The UBL/BAG domain proteins in S. pombe, SpBAG101 and SpBAG102, display similar interaction pattern to human BAG-1 (Kriegenburg et al., 2014; Poulsen et al., 2017). To validate the functionality of the UBL and the BAG domain of BcBAG1, we generated two truncated forms of BcBAG1; BcBAG1A1-141 and BcBAG1B142-298 (Supplementary Figure S1B), containing UBL and BAG domain, respectively. The BcBAG1A1-141 and BcBAG1B142-298 constructs were subsequently transformed into ΔBcbag1 for complementation, designated as N3-11 and C2-8, respectively. Both N3-11 and C2-8 could not effectively rescue defects in conidia production and sclerotia formation (Figure 2). Interestingly, C2-8 could partially restored conidial germination, infection structure formation and pathogenicity to WT levels (Figures 4, 5). These data suggest that both UBL and BAG domains are necessary for integral BcBAG1 function and we further inferred that the BAG domain plays an indispensable role for pathogenicity of B. cinerea.
BcBAG1 Modulates Multiple Stress Responses in B. cinerea
BAG family members are involved in cell protection during variable biotic and abiotic stress responses (Doukhanina et al., 2006; Behl, 2016). To investigate the function of BcBAG1 in response to environmental stress, we examined the sensitivity of ΔBcbag1 to various abiotic stress stimuli. As shown in Figure 6, ΔBcbag1 was more sensitive to salt stress (1 M NaCl) than WT, but no different with WT to another osmotic stress inducer 1.2 M D-sorbitol. Mycelial growth in response to cell wall stress inducers (0.6 mg/ml CFW; and 0.02% SDS) was measured. ΔBcbag1 showed dramatically increased sensitivity to both CFW and SDS compared to WT (Figure 6). In contrast, when exposing to oxidative stimuli, 20 mM H2O2 and 0.5 mM TBHP, ΔBcbag1 led to an average ∼7% (20 mM H2O2) and ∼8% (0.5 mM TBHP) lower growth inhibition rate than the WT strain (Figure 6), indicating that disruption of BcBAG1 is more resistance to oxidative stress.
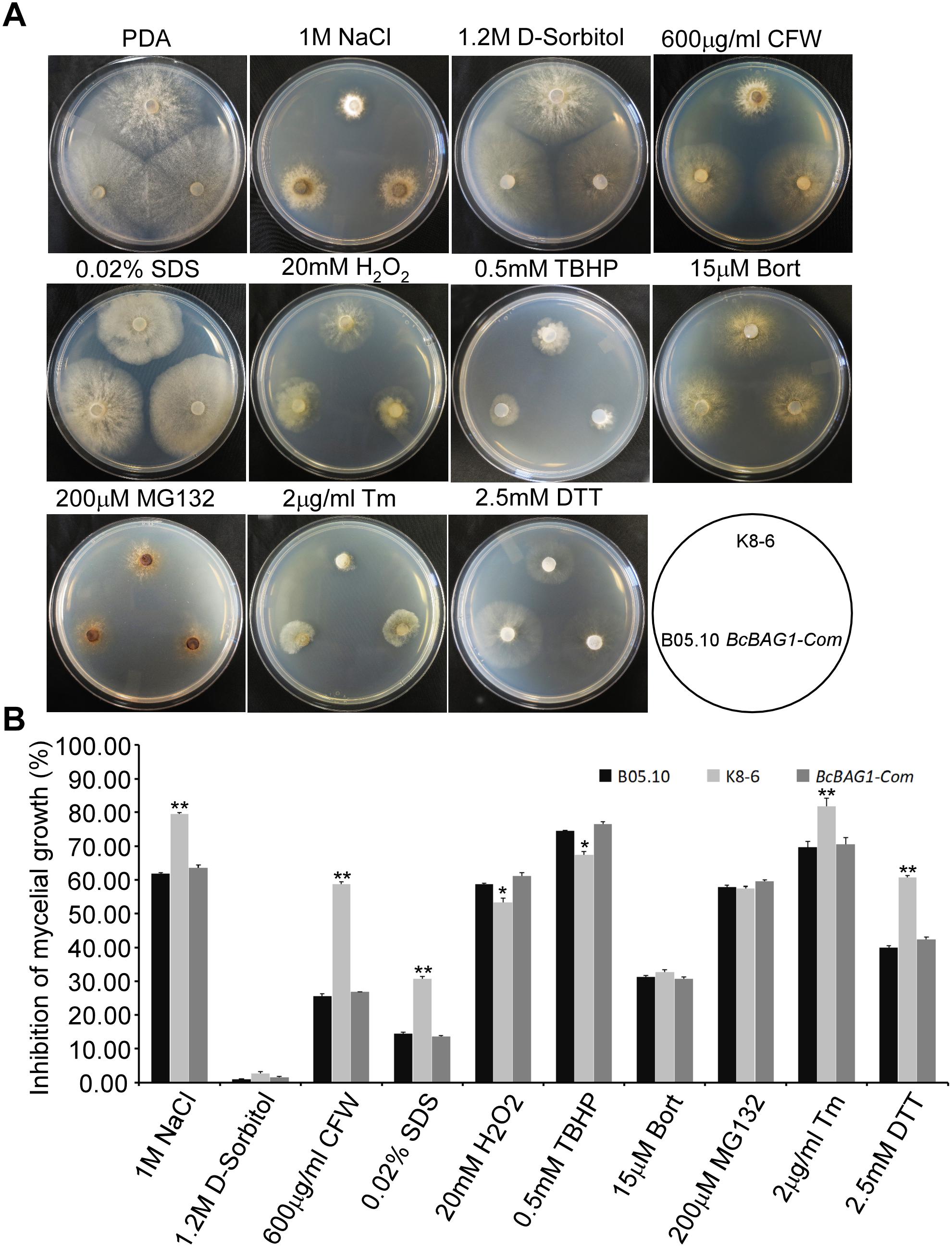
Figure 6. BcBAG1 responses to various stress conditions. (A) Wild-type B05.10, ΔBcbag1 (K8-6) and BcBAG1-Com strains were cultured on PDA medium supplemented with various stresses inducers and photographed after 3 days. (B) Statistical analyses of the inhibition rates of all strains under different stresses. Error bars indicate the standard deviations from three independent experiments and asterisks indicate statistical significance (P < 0.05; P < 0.01).
Human BAG-1 is a coupling factor between HSP70 and 26S proteasome (Luders et al., 2000). BAG3, as a co-chaperone, forms a complex with HSP70 to facilitate the degradation of ubiquitinated proteins via the proteasome or autophagy pathways (Gamerdinger et al., 2011; Minoia et al., 2014). Thus, we examined the role of BcBAG1 in proteasome degradation by testing the sensitivity of ΔBcbag1 to proteasome inhibitors, MG132 and Bort (Huang and Chen, 2009). Unexpectedly, results showed that the growth rate of ΔBcbag1 is not statistically different from WT and BcBAG1-Com (Figure 6), implying that BcBAG1 is not involved in response to proteasome inhibitors, MG132 and Bort. Although human BAG3 mediate the responses to Bort and MG132 (Judge et al., 2017), BcBAG1 does not share the same role in this perspective as the human counterpart.
BcBAG1 Negatively Regulates Unfolded Protein Response (UPR)
Previous work revealed that Arabidopsis BAG7 (AtBAG7) functions as an ER stress co-chaperone to maintain the UPR and protect plants from ER stress-induced cell death (Williams et al., 2010). Except AtBAG4, BcBAG1 shares relative higher identity to AtBAG7 in comparison to other Arabidopsis BAGs (Supplementary Table S3), we therefore examined whether BcBAG1 plays a role in ER stress signaling pathway. ER stress can be induced by chemical compounds, e.g., Tm or DTT (Oslowski and Urano, 2011). Besides, environmental/abiotic stress including excessive heat and cold also trigger ER stress (Williams et al., 2010). Accordingly, we cultured the WT, ΔBcbag1 and BcBAG1-Com on media supplemented with 2 μg/ml Tm and 2.5 mM DTT for 3 days at 22°C. The results showed that the growth of ΔBcbag1 was strongly inhibited by Tm and DTT with a much higher inhibition rate comparing to WT and BcBAG1-Com (Figure 6). In addition, after incubating on PDA under heat (30°C) and cold (4°C) conditions for 7 days, ΔBcbag1 was more sensitive than the WT and BcBAG1-Com to both heat and cold treatments (Figures 7A–C). Quantitative real-time (qRT-PCR) analysis demonstrated that the transcription of BcBAG1 was highly induced upon above ER stress conditions, including heat treatment (50°C for 30 min), Tm (2 μg/ml for 1 h), or DTT (20 mM for 1 h) (Figure 7C), indicating that BcBAG1 is responsible for ER stress tolerance.
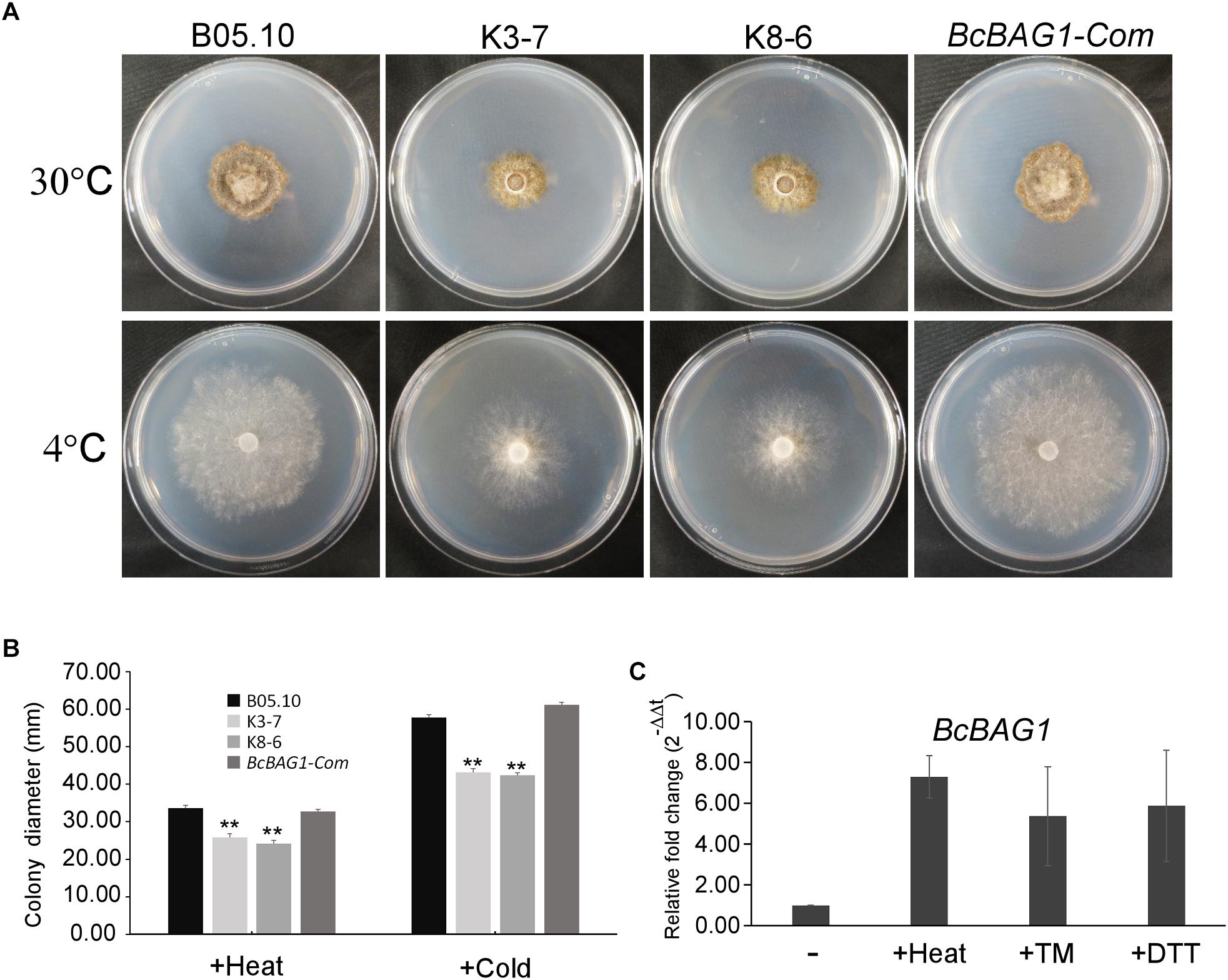
Figure 7. BcBAG1 knock-out mutants are susceptible to ER stress conditions. (A) Wild-type B05.10, ΔBcbag1 (K3-7 and K8-6) and BcBAG1-Com strains were grown on PDA medium at 30°C and 4°C, respectively, for 7 days. (B) Statistical analysis of the colony diameters of above strains. (C) The expression of BcBAG1 is induced by heat, Tm, and DTT treatments. Quantitative RT-PCR was used to evaluate the BcBAG1 transcript levels in the wild-type B05.10 following heat treatment (50°C for 30 min), tunicamycin (Tm, 15 μg/ml for 1 h) and DTT (20 mM for 1 h). Error bars represent the standard deviations from three independent experiments. Asterisks denote statistical significance (P < 0.01).
To cope with ER stress, eukaryotes utilize UPR to alleviate the detrimental effects (Schroder and Kaufman, 2005). In S. cerevisiae, UPR is sensed by transmembrane protein kinase and ribonuclease (RNase) IRE1 and initiated with IRE1-mediated splicing of an unconventional intron (252-nucleotide) from the HAC1 transcript (Cox and Walter, 1996; Ruegsegger et al., 2001). HAC1 encodes a basic leucine zipper-type (bZIP) transcription factor, and the splicing of HAC1 regulates the expression of UPR target genes thus to mitigate ER stress (Mori et al., 1996). To explore the underlying mechanism within the sensitivity of ΔBcbag1 to ER stress, we detected the presence of the spliced and unspliced form of BcHAC1 mRNA by RT-PCR. Spliced form of BcHAC1 (BcHAC1S) in B05.10 was significantly increased upon ER stress, suggesting B. cinerea shares the similar HAC1 splicing process with other organisms during UPR (Figure 8A). Notably, under normal conditions, the amount of BcHAC1S was drastically more abundant in ΔBcbag1 than the WT (Figure 8A, upper panel). More interestingly, BcHAC1 was constitutively spliced in ΔBcbag1 under both conditions that are with/with-out stress (Figure 8A). DNA sequencing confirmed that a 20 nucleotide of the fragment was absent in the spliced form compared to the unspliced form (Figure 8A, lower panel). Meanwhile, we examined the expression of UPR-related genes in B. cinerea, BcBIP1 and BcIRE1, homologs of ER chaperone KAR2/BIP1 and ER stress sensor/transducer IRE1 in S. cerevisiae, respectively (Pincus et al., 2010). Both BcBIP1 and BcIRE1 were induced following ER stress in the WT (Figure 8B). Without stress, the expression levels of BcIRE1 and BcBIP1 were increased by threefold and fivefold, respectively, in ΔBcbag1 (Figure 8B). All these results indicates that, BcBAG1 deletion causes constitutive activation of UPR through negatively regulating the expression of BcBIP1, BcIRE1 and the splicing of BcHAC1 mRNA. We speculate that BcBAG1 effectively repress the harmful and excessive constitutive activation of UPR, thus to maintain the proper UPR level during ER stress signaling pathway.
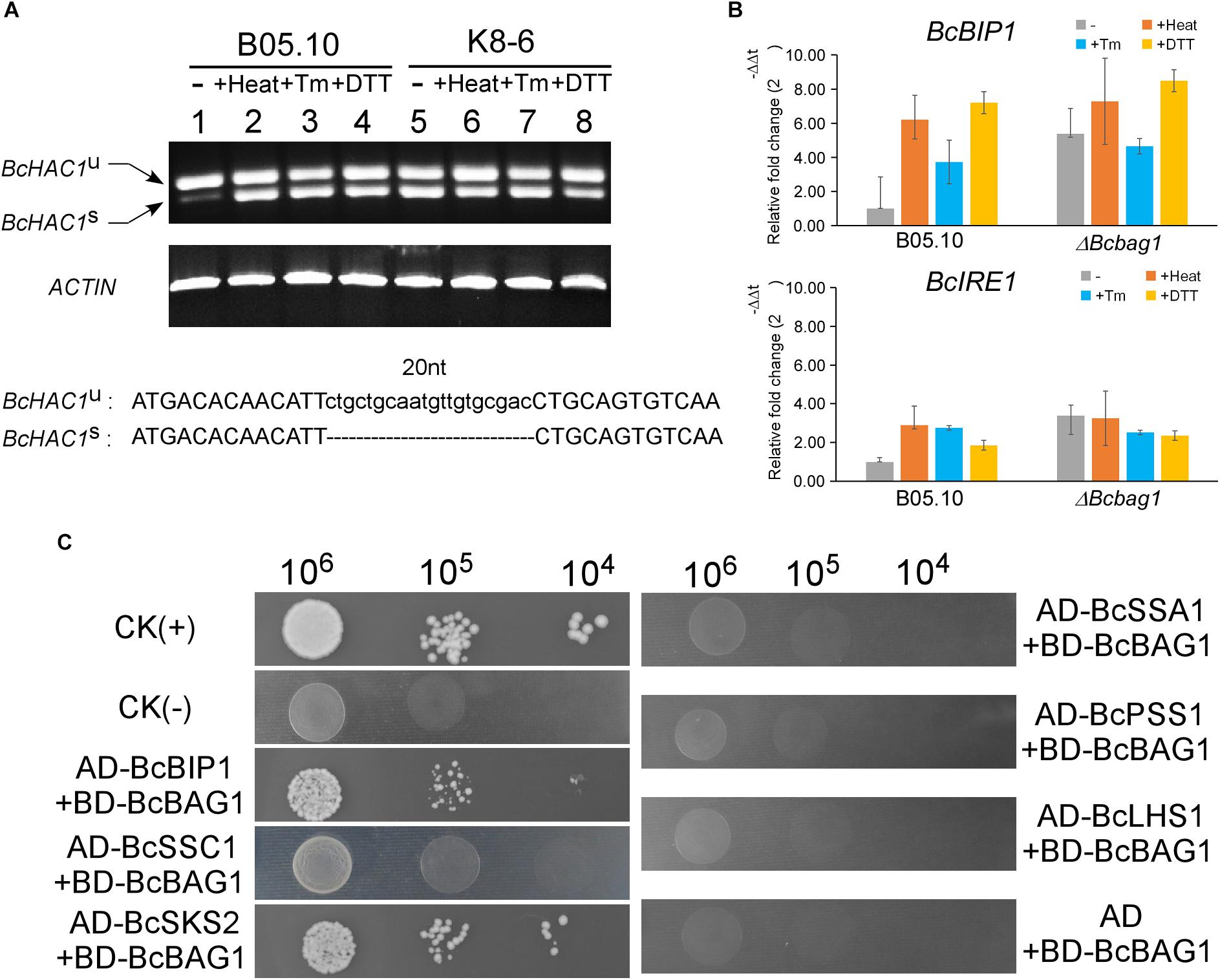
Figure 8. BcBAG1 involves in the activation of unfolded protein response (UPR) and interacts with two HSP70-type chaperones. (A) BcBAG1 negatively regulates the splicing of BcHAC1 mRNA under normal condition. RT-PCR was performed using total RNAs extracted from the wild-type B05.10 and ΔBcbag1 mutant K8-6 grown in liquid PDB for 2 days (lane 1 and lane 5), respectively. The rest of total RNAs were collected from the strains grown in liquid PDB for 2 days then treat with ER stresses. Lane 2 and lane 6: B05.10 and K8-6 subjected to 50°C for 30 min, respectively; lane 3 and lane 7 treated with 15 μg/ml Tm for 1 h, respectively; lane 4 and lane 8: B05.10 and K8-6 treated with 20 mM DTT for 1 h, respectively. The ACTIN gene was used as the internal control. DNA sequence alignment of the BcHAC1U and BcHAC1S. The 20 nt atypical intron is indicated on the bottom panel. (B) The expression profiles of UPR correlated genes, BcBIP1 and BcIRE1 during ER stress by real-time PCR. Total RNAs of corresponding strains grown in PDB medium or after stress treatments were extracted and conducted for real-time PCR. Error bars represent the standard deviations from three independent experiments. (C) Yeast two-hybrid assay between BcBAG1 and HSP70 family members. The yeast transformants diluted to specified concentrations (cell/ml) were plated onto SD/-Leu/-Trp/-His. The interaction of pGBKT7-53 with pGADT7-T was used as the positive control CK (+) and the interaction of pGBKT7-Lam with pGADT7-T as the negative control CK (–), respectively.
BcBAG1 Binds HSP70-Type Chaperones
The heat shock proteins HSP70 family play crucial roles in assisting a variety of protein folding processes (Mayer and Bukau, 2005). Human BAG-1 binds the ATPase domain of Hsc70 to stimulates Hsc70 ATP hydrolysis which results in the release of ADP from Hsc70, thereby regulates specific protein folding and maturation pathways (Hohfeld and Jentsch, 1997). To ascertain whether BcBAG1 bind HSP70-type chaperone in B. cinerea, we performed the yeast two-hybrid assay to establish possible interaction between BcBAG1 and the HSP70 family members, including BcBIP1, BcSSC1, BcSKS2, BcSSA1, BcPSS1, and BcLHS1, which are homologs of HSP70 family in the fission yeast. Results showed that BcBAG1 only interacted with BcBIP1 and BcSKS2 (Figure 8C), demonstrating BcBAG1 does function as a co-chaperone of HSP70 proteins, in accordance with that has been reported in other systems (Gassler et al., 2001; Sondermann et al., 2002; Poulsen et al., 2017).
BcBAG1 Restores Salt Stress Tolerance to atbag4
Using NCBI blastp and blastn programs, we found BcBAG1 is predicted to be the closest homolog of Arabidopsis BAG protein AtBAG4, in light of the amino acid sequences of the BAG domains between BcBAG1 and AtBAG4 share 33% identity and 50% similarity (Supplementary Figures S1A,C and Supplementary Table S3). To determine whether BcBAG1 is able to functionally complement the AtBAG4 T-DNA mutants, full-length BcBAG1 was overexpressed using the cauliflower mosaic virus 35S promoter in the atbag4 T-DNA mutants (atbag4::BcBAG1). Expression levels of BcBAG1 in atbag4 was confirmed by RT-PCR (Figure 9A). Previous studies indicated that atbag4 mutants were more susceptible to salt stress (100 mM NaCl) compared to the WT Col-0 (Doukhanina et al., 2006). Here, we performed the salt tolerance assay for atbag4::BcBAG1, taking Col-0 and atbag4 as the positive and negative controls, respectively. Arabidopsis seedlings were cultured on 1/2 MS medium supplemented with 100 mM NaCl. After 2 weeks treatment, no difference was observed from seedlings. However, 5 weeks later, atbag4 mutants displayed massive chlorosis and bleaching of leaves, while Col-0 and atbag4::BcBAG1 plants grew well and exhibited nearly the same growth tendency under salt conditions (Figure 9B). These observations demonstrate that BcBAG1 can be ectopically expressed in Arabidopsis and fully restore salt tolerance in atbag4.

Figure 9. BcBAG1 restores the salt stress tolerance of AtBAG4 T-DNA knock-out mutant. (A) BcBAG1 ectopically expressed in Arabidopsis plants. BcBAG1 driven by 35S promoter was transformed in to atbag4 mutant (atbag4::BcBAG1) and the expression of BcBAG1 was confirmed by RT-PCR. Taking atbag4 and the wild-type plant Col-0 as controls. CK, negative control. The EF1a gene was used as the internal control. (B) Col-0, atbag4 and atbag4::BcBAG1 were subjected to salt stress. Plants were grown on half-strength Murashige and Skoog (MS) medium with 2% sucrose and subjected to 100 mM NaCl treatment. Plants growth was recorded after 2 weeks and 5 weeks.
Discussion
As an evolutionarily conserved group, BAG proteins from yeast to plants and mammals have been associated with regulation of PCD and cell protection. Recently, they have also been found to play an important role in autophagy, UPR and ubiquitin-proteasome system (Kabbage et al., 2017). However, the identification and characterization of BAG proteins in phytopathogenic fungi is rare. In this study, we explored the B. cinerea genome and identified a unique BAG gene, BcBAG1. Targeted deletion of BcBAG1 exerted strong adverse effects on vegetative growth, conidiation, sclerotia formation, hyphal melanization, stress response, conidial germination, penetration and virulence, suggesting that the pleiotropic function of BAGs delineated in mammals and plants appears to be maintained in B. cinerea. The BAG domain of BcBAG1 shares highest similarity with AtBAG4 in Arabidopsis, also an ectopic expression of BcBAG1 fully restored salt tolerance of atbag4 mutants (Figure 9). In addition, deletion of BcBAG1 resulted in increased sensitivity to salt, cell wall stressors, ER stress inducers, and heat or cold treatments. These results parallel plant AtBAG4 studies in which AtBAG4-overexpressing transgenic tobacco plants confer tolerance to a wide range of stresses such as UV light, cold, salt treatments (Doukhanina et al., 2006). From this perspective, the cytoprotective function of BAGs in response to diverse stresses is relatively conserved between the fungi and plants.
Our previous studies have addressed the importance of AtBAG7 in the maintenance of the UPR and the mechanisms of ER-localized co-chaperone AtBAG7 in stress protection (Williams et al., 2010; Li et al., 2017). Interestingly, BcBAG1 showed a close evolutionary relationship with AtBAG7 among plant BAGs (Supplementary Figure S1A and Supplementary Table S3). Moreover, ΔBcbag1 were more sensitive to ER stress stimuli (Figure 6) and expression of BcBAG1 was induced under ER stress (Figure 7B), suggesting that both BcBAG1 and AtBAG7 are functionally associated with the ER stress. Of note, we found that the deletion of BcBAG1 gives rise to the constitutive activation of UPR with high levels even without stress (Figures 8A,B), indicating that BcBAG1 is necessary for the inhibition of excess UPR under normal condition. Given the defects in fungal development and differentiation caused by deletion of BcBAG1, we speculate that under normal conditions, the abnormal activated UPR is actually harmful to the fungus. It has been reported that moderate activation of UPR is necessary for ER recovery when responding to stress in S. cerevisiae (Chawla et al., 2011). Thus, we suggest that constitutive activation of UPR in ΔBcbag1 results in loss of normal ER protein-folding capacity, which impinges upon the ability to sustain resistance to ER stress. Thus, ΔBcbag1 displayed growth defects during ER stress. In addition, as a multifaceted HSP70 molecular chaperone, BIP ensures an appropriate response to restore protein folding homeostasis to the ER by providing a buffer for inactive IRE1 (Pincus et al., 2010). Our result revealed that disruption of BcBAG1 increases the expression of BcBIP1 and BcIRE1 (Figure 8B) and BcBAG1 interacts with BcBIP1 (Figure 8C). We conclude that the regulation of UPR by BcBAG1 correlates to BcBIP1. The relationship between BcBAG1/BcBIP1/BcIRE1 and how BcBAG1 modulates the ER machinery require further studies.
Previous studies of human BAG-1/HSC70 complex revealed that BAG-1 exploits Glu212, Asp222, Arg237, and Gln245 residues in the BAG domain to bind with Hsc70 ATPase domain (Sondermann et al., 2001). Multiple alignment showed that these residues, with the exception of Glu212, are highly conserved in BcBAG1 BD (Supplementary Figure S1C), implying that BcBAG1 interacts with HSP70 in a similar manner as with human BAG-1. We did examine the interaction between BcBAG1 and HSP70 proteins by Y2H and found that BcBAG1 exclusively interacts with BcBIP1 and BcSKS2 in the HSP70 family, but fails to bind to BcSSC1, BcSSA1, BcPSS1, and BcLHS1 (Figure 8C). Previous studies demonstrated that the S. cerevisiae BAG protein SNL1 interacts with HSP70 family members including SSA1, SSB1, SSB2, SEE1, and SEE2 (Sondermann et al., 2002; Verghese and Morano, 2012; Abrams et al., 2014). While the S. pombe BAG proteins SpBAG101 and SpBAG102 exclusively interact with SSA1, SSA2, and SKS2 (Poulsen et al., 2017). These data suggest BAGs from different origins show different affinity and specificity to HSP70s members. Notably, the interaction between BcBAG1 and BcSKS2 is conserved from that in yeast studies. BcSKS2 is the homolog of fission yeast ribosome-associated chaperone SKS2, and the S. cerevisiae orthologs of fission yeast SKS2, called SSB1 and SSB2. SSB1/2 chaperones play a dual role in de novo protein folding and ribosome biogenesis (Mudholkar et al., 2017). We conjectured that the binding to primary HSP70 chaperone might be responsible for the functional pleiotropy of BcBAG1.
Apart from the BAG domain, BcBAG1 also contains a UBL domain. Human BAG-1 function as a link between HSC/HSP70 and 26S proteasome degradation system via its UBL domain, chaperone-bound substrates are released and degraded (Luders et al., 2000; Alberti et al., 2002). Co-precipitation experiments in fission yeast provide direct evidence that both SpBAG101 and SpBAG102 interact with 26S proteasomes depend on the UBL domain (Poulsen et al., 2017). However, we found that response to proteasome inhibitors, e.g., Bort and MG132, by BcBAG1 deletion was unaffected in fungus (Figure 6). Besides, the result of ubiquitination assay confirmed that deletion of BcBAG1 does not alter the levels of ubiquitination (Supplementary Figure S3). Therefore, we inferred that BcBAG1 is not a key player in the proteasome degradation. However, we cannot exclude the possibility that other proteasome inhibitors may work on BcBAG1 or BcBAG1 can be targeting some proteasome substrate for degradation.
Botrytis cinerea, however, integrates a number of hurdles that must be traversed for successful colonization and defense against the plethora of plant hosts that are encountered (Elad et al., 2007). Therefore, it is relatively difficult to present a precise mechanism for the alteration of pathogenicity and virulence in the host. Based on our data, we supposed that the attenuated pathogenicity of ΔBcbag1 is directly or indirectly related to the following reasons. First, ΔBcbag1 showed a decrease of conidial germination, and delayed the formation of appressoria and infection cushions on an artificial surface and onion epidermis (Figure 6). At the same time, we observed that BcBAG1 resides in the cytoplasm throughout growth stages, but the localization is altered and most likely concentrated at the infection spots during invasion (Supplementary Figure S4), suggesting the expression of BcBAG1 contributes to the formation of penetration structures. Therefore, the impairment of penetration structures in ΔBcbag1 weakens the ability of breaching the host tissues to effectively cause disease. Second, it is reported that cell wall integrity is crucial for B. cinerea virulence and pathogenicity (Soulie et al., 2003, 2006; Cui et al., 2013). ΔBcbag1 increased sensitivity to cell wall stressors, CFW and SDS (Figure 6), indicating BcBAG1 involves in cell wall integrity pathway. Therefore, defect in cell wall integrity is one key to the reduction of virulence. Third, melanin is a factor affecting the virulence of B. cinerea. BcPKS13 and BcBRN1, encoding polyketide synthase and tetrahydroxynaphthalene (THN) reductases, respectively, both are involving in fungal DHN melanin biosynthesis (Zhang et al., 2015). Loss of BcPKS13 and BcBRN1 blocks melanization resulting in enhanced virulence. Conversely, overexpression of BcBRN1 enhances melanization, decreases secretion for virulence factors such as several hydrolytic enzymes and oxalic acid, and attenuated virulence (Zhang et al., 2015). From this perspective, an increment of melanin biosynthesis negatively affects the pathogenesis of B. cinerea. Consistently, our results showed that BcBAG1 negatively regulates melanin biosynthesis (Figure 3), thus the increased mycelial melanin biosynthesis suppress fungal virulence for ΔBcbag1. Fourth, previous studies indicated UPR plays as a central regulator of fungal pathogenesis (Heimel et al., 2013; Guillemette et al., 2014; Krishnan and Askew, 2014a,b). Our study found that BcBAG1 is responsible for proper maintenance of UPR, as a result, abnormal activation of UPR in ΔBcbag1 could cause the attenuated virulence.
In summary, this paper details the biological functions of BcBAG1. We have shown that BcBAG1 exhibits functional versatility and is involved in fungal development, differentiation, stress response, and pathogenicity. BcBAG1 acts as a co-chaperone of HSP70 and is a key regulator required for maintenance of the UPR. In light of these findings, future studies involving translocation of BcBAG1 during infection and identification of other targets are of interest. Taken together, these results demonstrate the importance of the BAG family in filamentous fungus cell death pathways and cytoprotection.
Author Contributions
HZ and MD designed the research. HZ performed the experiments. HZ, YL, and MD analyzed the data. HZ, YL, MD, and ZW wrote the article. MD and ZW revised and approved the manuscript.
Funding
This work was partially supported by NSFC (Grant No. 31471739).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Matthias Hahn (Kaiserslautern University, Germany) for providing vector pNAH-OGG and pNAH-OMG. We also thank the Chinese CSC Scholarship Program for the grant of HZ.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00685/full#supplementary-material
FIGURE S1 | Phylogenetic and sequence analysis of BAG1 in Botrytis cinerea (BcBAG1). (A) The phylogenetic tree of BAG proteins. Evolutionary analyses were conducted in MEGA7.0. The evolutionary history was inferred by using a neighbor-joining method based on the amino acid sequences. The numbers at nodes inferred the percentage of their occurrence in 10,000 bootstrap replicates. Species names and GenBank accession numbers of each sequence are represented as follows: SsBAG1 (Sclerotinia sclerotiorum, XP_001591798.1); MoBAG1 (Magnaporthe oryzae, XP_003710309.1); FoBAG1 (Fusarium oxysporum f. sp. Lycopersici, XP_018239181.1); NcBAG1 (Neurospora crassa, XP_961586.1); AnBAG1 (Aspergillus nidulans, XP_661815.1); UmBAG1 (Ustilago maydis, KIS67500.1); SpBAG101 (Schizosaccharomyces pombe, NP_596760.1); SpBAG102 (Schizosaccharomyces pombe, NP_595316.1); ScSNL1 (Saccharomyces cerevisiae, KZV10602.1); HsBAG-1M (Homo sapiens, NP_001336215.1); HsBAG2 (Homo sapiens, NP_004273.1); HsBAG3 (Homo sapiens, NP_004272.2); HsBAG4 (Homo sapiens, NP_004865.1); HsBAG5 (Homo sapiens, NP_001015049.1); HsBAG6 (Homo sapiens, P46379.2); AtBAG1 (Arabidopsis thaliana, NP_200019.2); AtBAG2 (Arabidopsis thaliana, NP_568950.2); AtBAG3 (Arabidopsis thaliana, NP_196339.1); AtBAG4 (Arabidopsis thaliana, NP_190746.2); AtBAG5 (Arabidopsis thaliana, NP_172670.2); AtBAG6 (Arabidopsis thaliana, AEC10664.1); AtBAG7 (Arabidopsis thaliana, NP_201045.1). (B) Schematic diagram of BcBAG1. Purple red and green boxes indicate the UBL and BAG domain of BcBAG1, respectively. (C) Sequence alignment of the conserved BAG domain from different organisms. Three predicted helixes labeled on the top. Conserved residues which involving in binding of BAG protein to Hsc70 ATPase domain in human HsBAG-1M are indicated by red arrow, and residues critical to packing interactions are highlighted by blue arrow.
FIGURE S2 | Sketch of gene deletion and identification of deletion mutants and complemented transformants. (A) Diagram of targeted gene replacement. (B) RT-PCR confirmation. Lane 1:100 bp DNA Ladder (NEB); lanes 2 and 3: K3-7 and K8-6, respectively; lane 4: B05.10; lanes 5 and 6: complemented transformants; lane 7: PCR negative control. (C) Southern blotting confirmation. Total genomic DNAs were digested by PvuI, and a DNA fragment in the upstream of 5′ terminus of BcBAG1 was selected and labeled as the probe shown in panel (A). M: 1 kb DNA Ladder (NEB).
FIGURE S3 | Disruption of BcBAG1 does not affect protein ubiquitination. Total protein extracts from corresponding strains was western blotted with (upper panel) an anti-Ub antibody (P4D1) and stained with Coomassie brilliant blue (lower panel) as the loading control. B05.10: the wild-type strain; K8-6: ΔBcbag1 mutant lines; C2-8 (C-terminus of BcBAG1) and N3-11 (N-terminus of BcBAG1).
FIGURE S4 | Subcellular localization of BcBAG1. GFP-BcBAG1 was overexpressed in the ΔBcbag1 mutant (OG2-7) and the fluorescence for BcBAG1 localization at different stages was visualized by confocal microscopy. (A) Vegetative hyphae; (B) conidia; (C,D) conidia on hydrophobic glass slides for 12 and 24 h, respectively. (E,F) Both are conidia on onion epidermal cells for 24 h and 48 h, respectively. Scale bars: 20 μm.
TABLE S1 | Wild-type and mutant strains of Botrytis cinerea used in this study.
TABLE S2 | Oligonucleotide primers used in this study.
TABLE S3 | Similarity of full length and BAG domain amino acid sequence between BcBAG1 and other BAGs.
Footnotes
- ^http://fungi.ensembl.org/Botrytis_cinerea/Info/Index
- ^https://pfam.xfam.org/
- ^https://www.ebi.ac.uk/interpro/
- ^https://www.arabidopsis.org/
References
Abrams, J. L., Verghese, J., Gibney, P. A., and Morano, K. A. (2014). Hierarchical functional specificity of cytosolic heat shock protein 70 (Hsp70) nucleotide exchange factors in yeast. J. Biol. Chem. 289, 13155–13167. doi: 10.1074/jbc.M113.530014
Alberti, S., Demand, J., Esser, C., Emmerich, N., Schild, H., and Hohfeld, J. (2002). Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J. Biol. Chem. 277, 45920–45927. doi: 10.1074/jbc.M204196200
Back, S. H., Schroder, M., Lee, K., Zhang, K., and Kaufman, R. J. (2005). ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods 35, 395–416. doi: 10.1016/j.ymeth.2005.03.001
Behl, C. (2016). Breaking BAG: the Co-Chaperone BAG3 in health and disease. Trends Pharmacol. Sci. 37, 672–688. doi: 10.1016/j.tips.2016.04.007
Bell, A. A., and Wheeler, M. H. (1986). Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol. 24, 411–451. doi: 10.1146/annurev.py.24.090186.002211
Bracher, A., and Verghese, J. (2015). GrpE, Hsp110/Grp170, HspBP1/Sil1 and BAG domain proteins: nucleotide exchange factors for Hsp70 molecular chaperones. Subcell Biochem. 78, 1–33. doi: 10.1007/978-3-319-11731-7_1
Butler, M. J., and Day, A. W. (1998). Fungal melanins: a review. Can. J. Microbiol. 44, 1115–1136. doi: 10.1139/cjm-44-12-1115
Chawla, A., Chakrabarti, S., Ghosh, G., and Niwa, M. (2011). Attenuation of yeast UPR is essential for survival and is mediated by IRE1 kinase. J. Cell Biol. 193, 41–50. doi: 10.1083/jcb.201008071
Cox, J. S., and Walter, P. (1996). A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391–404. doi: 10.1016/S0092-8674(00)81360-4
Cui, Z. F., Wang, Y. H., Lei, N., Wang, K., and Zhu, T. H. (2013). Botrytis cinerea chitin synthase BcChsVI is required for normal growth and pathogenicity. Curr. Genet. 59, 119–128. doi: 10.1007/s00294-013-0393-y
Demand, J., Alberti, S., Patterson, C., and Hohfeld, J. (2001). Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 11, 1569–1577. doi: 10.1016/S0960-9822(01)00487-0
Doehlemann, G., Berndt, P., and Hahn, M. (2006). Different signalling pathways involving a G alpha protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol. Microbiol. 59, 821–835. doi: 10.1111/j.1365-2958.2005.04991.x
Doukhanina, E. V., Chen, S., van der Zalm, E., Godzik, A., Reed, J., and Dickman, M. B. (2006). Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J. Biol. Chem. 281, 18793–18801. doi: 10.1074/jbc.M511794200
Elad, S. F. Y. (2016). Botrytis - the Fungus, the Pathogen and its Management in Agricultural Systems. Berlin: springer.
Elad, Y., Williamson, B., Tudzynski, P., and Delen, N. (2007). Botrytis: Biology, Pathology and Control. Berlin: SpringerLink. doi: 10.1007/978-1-4020-2626-3
Fang, S., Li, L., Cui, B., Men, S., Shen, Y., and Yang, X. (2013). Structural insight into plant programmed cell death mediated by BAG proteins in Arabidopsis thaliana. Acta Crystallogr. D Biol. Crystallogr. 69(Pt 6), 934–945. doi: 10.1107/S0907444913003624
Feng, X., Krishnan, K., Richie, D. L., Aimanianda, V., Hartl, L., Grahl, N., et al. (2011). HacA-independent functions of the ER stress sensor IreA synergize with the canonical UPR to influence virulence traits in Aspergillus fumigatus. PLoS Pathog. 7:e1002330. doi: 10.1371/journal.ppat.1002330
Gamerdinger, M., Kaya, A. M., Wolfrum, U., Clement, A. M., and Behl, C. (2011). BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. Embo Rep. 12, 149–156. doi: 10.1038/embor.2010.203
Gassler, C. S., Wiederkehr, T., Brehmer, D., Bukau, B., and Mayer, M. P. (2001). Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J. Biol. Chem. 276, 32538–32544. doi: 10.1074/jbc.M105328200
Gething, M. J. (1999). Role and regulation of the ER chaperone BiP. Semin. Cell Dev. Biol. 10, 465–472. doi: 10.1006/scdb.1999.0318
Goswami, R. S. (2012). Targeted gene replacement in fungi using a split-marker approach. Methods Mol. Biol. 835, 255–269. doi: 10.1007/978-1-61779-501-5_16
Gronover, C. S., Kasulke, D., Tudzynski, P., and Tudzynski, B. (2001). The role of G protein alpha subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol. Plant. Microbe Interact. 14, 1293–1302. doi: 10.1094/MPMI.2001.14.11.1293
Guillemette, T., Calmes, B., and Simoneau, P. (2014). Impact of the UPR on the virulence of the plant fungal pathogen A-brassicicola. Virulence 5, 357–364. doi: 10.4161/viru.26772
Hartl, F. U. (1996). Molecular chaperones in cellular protein folding. Nature 381, 571–579. doi: 10.1038/381571a0
Heimel, K., Freitag, J., Hampel, M., Ast, J., Bolker, M., and Kamper, J. (2013). Crosstalk between the unfolded protein response and pathways that regulate pathogenic development in Ustilago maydis. Plant Cell 25, 4262–4277. doi: 10.1105/tpc.113.115899
Henson, J. M., Butler, M. J., and Day, A. W. (1999). The dark side of the mycelium: melanins of phytopathogenic fungi. Annu. Rev. Phytopathol. 37, 447–471. doi: 10.1146/annurev.phyto.37.1.447
Ho, A. K., Raczniak, G. A., Ives, E. B., and Wente, S. R. (1998). The integral membrane protein snl1p is genetically linked to yeast nuclear pore complex function. Mol. Biol. Cell 9, 355–373. doi: 10.1091/mbc.9.2.355
Hohfeld, J., and Jentsch, S. (1997). GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 16, 6209–6216. doi: 10.1093/emboj/16.20.6209
Huang, L., and Chen, C. H. (2009). Proteasome regulators: activators and inhibitors. Curr Med Chem 16, 931–939. doi: 10.2174/092986709787581860
Hwang, I., and Sheen, J. (2002). Two-component circuitry in Arabidopsis cytokinin signal transduction. Dev. Biol. 247, 484–484.
Jain, S., Wiemann, P., Thill, E., Williams, B., Keller, N. P., and Kabbage, M. (2018). A Bcl-2 Associated Athanogene (bagA) modulates sexual development and secondary metabolism in the filamentous fungus Aspergillus nidulans. Front. Microbiol. 9:1316. doi: 10.3389/fmicb.2018.01316
Joubert, A., Simoneau, P., Campion, C., Bataille-Simoneau, N., Iacomi-Vasilescu, B., Poupard, P., et al. (2011). Impact of the unfolded protein response on the pathogenicity of the necrotrophic fungus Alternaria brassicicola. Mol. Microbiol. 79, 1305–1324. doi: 10.1111/j.1365-2958.2010.07522.x
Judge, L. M., Perez-Bermejo, J. A., Truong, A., Ribeiro, A. J., Yoo, J. C., Jensen, C. L., et al. (2017). A BAG3 chaperone complex maintains cardiomyocyte function during proteotoxic stress. JCI Insight 2:94623. doi: 10.1172/jci.insight.94623
Kabbage, M., and Dickman, M. B. (2008). The BAG proteins: a ubiquitous family of chaperone regulators. Cell Mol. Life Sci. 65, 1390–1402. doi: 10.1007/s00018-008-7535-2
Kabbage, M., Kessens, R., Bartholomay, L. C., and Williams, B. (2017). The life and death of a plant cell. Annu. Rev. Plant Biol. 68, 375–404. doi: 10.1146/annurev-arplant-043015-111655
Kabbage, M., Kessens, R., and Dickman, M. B. (2016). A plant Bcl-2-associated athanogene is proteolytically activated to confer fungal resistance. Microb. Cell 3, 224–226. doi: 10.15698/mic2016.05.501
Kang, C. H., Jung, W. Y., Kang, Y. H., Kim, J. Y., Kim, D. G., Jeong, J. C., et al. (2006). AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ. 13, 84–95. doi: 10.1038/sj.cdd.4401712
Kriegenburg, F., Jakopec, V., Poulsen, E. G., Nielsen, S. V., Roguev, A., Krogan, N., et al. (2014). A chaperone-assisted degradation pathway targets kinetochore proteins to ensure genome stability. PLoS Genet. 10:e1004140. doi: 10.1371/journal.pgen.1004140
Krishnan, K., and Askew, D. S. (2014a). Endoplasmic reticulum stress and fungal pathogenesis. Fungal Biol. Rev. 28, 29–35. doi: 10.1016/j.fbr.2014.07.001
Krishnan, K., and Askew, D. S. (2014b). The fungal UPR A regulatory hub for virulence traits in the mold pathogen Aspergillus fumigatus. Virulence 5, 334–340. doi: 10.4161/viru.26571
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Leroch, M., Kleber, A., Silva, E., Coenen, T., Koppenhofer, D., Shmaryahu, A., et al. (2013). Transcriptome profiling of Botrytis cinerea conidial germination reveals upregulation of infection-related genes during the prepenetration stage. Eukaryot. Cell 12, 614–626. doi: 10.1128/EC.00295-12
Li, Y., and Dickman, M. (2016). Processing of AtBAG6 triggers autophagy and fungal resistance. Plant Signal. Behav. 11:e1175699. doi: 10.1080/15592324.2016.1175699
Li, Y., Kabbage, M., Liu, W., and Dickman, M. B. (2016). Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell 28, 233–247. doi: 10.1105/tpc.15.00626
Li, Y., Williams, B., and Dickman, M. (2017). Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY29. New Phytol. 214, 695–705. doi: 10.1111/nph.14388
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lo Presti, L., Lopez Diaz, C., Turra, D., Di Pietro, A., Hampel, M., Heimel, K., et al. (2016). A conserved co-chaperone is required for virulence in fungal plant pathogens. New Phytol. 209, 1135–1148. doi: 10.1111/nph.13703
Luders, J., Demand, J., and Hohfeld, J. (2000). The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 275, 4613–4617. doi: 10.1074/jbc.275.7.4613
Malhotra, J. D., and Kaufman, R. J. (2007). The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 18, 716–731. doi: 10.1016/j.semcdb.2007.09.003
Mayer, M. P., and Bukau, B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 62, 670–684. doi: 10.1007/s00018-004-4464-6
Minoia, M., Boncoraglio, A., Vinet, J., Morelli, F. F., Brunsting, J. F., Poletti, A., et al. (2014). BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta Implications for a proteasome-to-autophagy switch. Autophagy 10, 1603–1621. doi: 10.4161/auto.29409
Mori, K., Kawahara, T., Yoshida, H., Yanagi, H., and Yura, T. (1996). Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1, 803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x
Mudholkar, K., Fitzke, E., Prinz, C., Mayer, M. P., and Rospert, S. (2017). The Hsp70 homolog Ssb affects ribosome biogenesis via the TORC1-Sch9 signaling pathway. Nat. Commun. 8:937. doi: 10.1038/s41467-017-00635-z
Oslowski, C. M., and Urano, F. (2011). Measuring er stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 490(Pt B), 71–92. doi: 10.1016/B978-0-12-385114-7.00004-0
Perpetua, N. S., Kubo, Y., Yasuda, N., Takano, Y., and Furusawa, I. (1996). Cloning and characterization of a melanin biosynthetic THR1 reductase gene essential for appressorial penetration of Colletotrichum lagenarium. Mol. Plant Microbe Interact. 9, 323–329. doi: 10.1094/MPMI-9-0323
Pincus, D., Chevalier, M. W., Aragon, T., van Anken, E., Vidal, S. E., El-Samad, H., et al. (2010). BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 8:e1000415. doi: 10.1371/journal.pbio.1000415
Poulsen, E. G., Kampmeyer, C., Kriegenburg, F., Johansen, J. V., Hofmann, K., Holmberg, C., et al. (2017). UBL/BAG-domain co-chaperones cause cellular stress upon overexpression through constitutive activation of Hsf1. Cell Stress Chaperones 22, 143–154. doi: 10.1007/s12192-016-0751-z
Quidde, T., Buttner, P., and Tudzynski, P. (1999). Evidence for three different specific saponin-detoxifying activities in Botrytis cinerea and cloning and functional analysis of a gene coding for a putative avenacinase. Eur. J. Plant Pathol. 105, 273–283. doi: 10.1023/A:1008796006051
Raeder, U., and Broda, P. (1985). Rapid preparation Of DNA from filamentous fungi. Lett. Appl. Microbiol. 1, 17–20. doi: 10.1111/j.1472-765X.1985.tb01479.x
Ron, D., and Walter, P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529. doi: 10.1038/nrm2199
Ruegsegger, U., Leber, J. H., and Walter, P. (2001). Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107, 103–114. doi: 10.1016/S0092-8674(01)00505-0
Schroder, M., and Kaufman, R. J. (2005). ER stress and the unfolded protein response. Mutat. Res. 569, 29–63. doi: 10.1016/j.mrfmmm.2004.06.056
Sitia, R., and Braakman, I. (2003). Quality control in the endoplasmic reticulum protein factory. Nature 426, 891–894. doi: 10.1038/nature02262
Sondermann, H., Ho, A. K., Listenberger, L. L., Siegers, K., Moarefi, I., Wente, S. R., et al. (2002). Prediction of novel Bag-1 homologs based on structure/function analysis identifies Snl1p as an Hsp70 co-chaperone in Saccharomyces cerevisiae. J. Biol. Chem. 277, 33220–33227. doi: 10.1074/jbc.M204624200
Sondermann, H., Scheufler, C., Schneider, C., Hohfeld, J., Hartl, F. U., and Moarefi, I. (2001). Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291, 1553–1557. doi: 10.1126/science.291.5508.1553
Soulie, M. C., Perino, C., Piffeteau, A., Choquer, M., Malfatti, P., Cimerman, A., et al. (2006). Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a). Cell. Microbiol. 8, 1310–1321. doi: 10.1111/j.1462-5822.2006.00711.x
Soulie, M. C., Piffeteau, A., Choquer, M., Boccara, M., and Vidal-Cros, A. (2003). Disruption of Botrytis cinerea class I chitin synthase gene Bcchs1 results in cell wall weakening and reduced virulence. Fungal Genet. Biol. 40, 38–46. doi: 10.1016/S1087-1845(03)0005-3
Takayama, S., and Reed, J. C. (2001). Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell Biol. 3, E237–E241. doi: 10.1038/ncb1001-e237
Takayama, S., Sato, T., Krajewski, S., Kochel, K., Irie, S., Millan, J. A., et al. (1995). Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell 80, 279–284. doi: 10.1016/0092-8674(95)90410-7
Takayama, S., Xie, Z. H., and Reed, J. C. (1999). An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J. Biol. Chem. 274, 781–786. doi: 10.1074/jbc.274.2.781
Thompson, J. E., Fahnestock, S., Farrall, L., Liao, D. I., Valent, B., and Jordan, D. B. (2000). The second naphthol reductase of fungal melanin biosynthesis in Magnaporthe grisea: tetrahydroxynaphthalene reductase. J. Biol. Chem. 275, 34867–34872. doi: 10.1074/jbc.M006659200
Townsend, P. A., Cutress, R. I., Sharp, A., Brimmell, M., and Packham, G. (2003). BAG-1: a multifunctional regulator of cell growth and survival. Biochim. Biophys. Acta 1603, 83–98. doi: 10.1016/S0304-419X(03)00002-7
Vandenheuvel, J., and Waterreus, L. P. (1983). Conidial concentration as an important factor determining the type of pre-penetration structures formed by botrytis-cinerea on leaves of french bean (Phaseolus-Vulgaris). Plant Pathol. 32, 263–272. doi: 10.1111/j.1365-3059.1983.tb02833.x
Verghese, J., and Morano, K. A. (2012). A lysine-rich region within fungal BAG domain-containing proteins mediates a novel association with ribosomes. Eukaryot. Cell 11, 1003–1011. doi: 10.1128/EC.00146-12
Viefhues, A., Heller, J., Temme, N., and Tudzynski, P. (2014). Redox systems in botrytis cinerea: impact on development and virulence. Mol. Plant Microbe Interact. 27, 858–874. doi: 10.1094/Mpmi-01-14-0012-R
Williams, B., Kabbage, M., Britt, R., and Dickman, M. B. (2010). AtBAG7, an Arabidopsis Bcl-2-associated athanogene, resides in the endoplasmic reticulum and is involved in the unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 107, 6088–6093. doi: 10.1073/pnas.0912670107
Williamson, B., Tudzynski, B., Tudzynski, P., and van Kan, J. A. (2007). Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8, 561–580. doi: 10.1111/j.1364-3703.2007.00417.x
You, Q., Zhai, K., Yang, D., Yang, W., Wu, J., Liu, J., et al. (2016). An E3 Ubiquitin Ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance. Cell Host Microbe 20, 758–769. doi: 10.1016/j.chom.2016.10.023
Zhang, C. H., He, Y. F., Zhu, P. K., Chen, L., Wang, Y. W., Ni, B., et al. (2015). Loss of bcbrn1 and bcpks13 in Botrytis cinerea not only blocks melanization but also increases vegetative growth and virulence. Mol. Plant Microbe Interact. 28, 1091–1101. doi: 10.1094/Mpmi-04-15-0085-R
Keywords: BAG protein, co-chaperone, Botrytis cinerea, pathogenicity/virulence, unfolded protein response
Citation: Zhang H, Li Y, Dickman MB and Wang Z (2019) Cytoprotective Co-chaperone BcBAG1 Is a Component for Fungal Development, Virulence, and Unfolded Protein Response (UPR) of Botrytis cinerea. Front. Microbiol. 10:685. doi: 10.3389/fmicb.2019.00685
Received: 07 December 2018; Accepted: 19 March 2019;
Published: 09 April 2019.
Edited by:
Marco Catoni, University of Birmingham, United KingdomReviewed by:
Mehdi Kabbage, University of Wisconsin-Madison, United StatesChang Ho Kang, Gyeongsang National University, South Korea
Copyright © 2019 Zhang, Li, Dickman and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zonghua Wang, d2FuZ3poQGZhZnUuZWR1LmNu
†Present address: Yurong Li, Corteva AgriscienceTM, Johnston, IA, United States
 Honghong Zhang
Honghong Zhang Yurong Li
Yurong Li Martin B. Dickman
Martin B. Dickman Zonghua Wang
Zonghua Wang