- 1Department of Food Science, Cornell University, Ithaca, NY, United States
- 2Department of Food Science, Penn State University, University Park, PA, United States
- 3Department of Food Science, Wageningen University, Wageningen, Netherlands
The Bacillus cereus group comprises 18 different species, including human pathogens as well as psychrotolerant strains that are an important cause of fluid milk spoilage. To enhance our understanding of the genetic markers associated with psychrotolerance (defined here as > 1 log10 increase in cfu/mL after 21 days incubation at 6∘C) among dairy-associated B. cereus group isolates, we used genetic (whole genome sequencing) and phenotypic methods [growth in Skim Milk Broth (SMB) and Brain Heart Infusion (BHI) broth] to characterize 23 genetically-distinct representative isolates from a collection of 503 dairy-associated isolates. Quality threshold clustering identified three categories of psychrotolerance: (i) 14 isolates that were not psychrotolerant in BHI or SMB, (ii) 6 isolates that were psychrotolerant in BHI but not in SMB, and (iii) 2 isolates that were psychrotolerant in BHI and SMB. One isolate, which was psychrotolerant in BHI broth but was just below the cut-off of >1 log10 cfu/mL increase in SMB was not assigned to a cluster. A maximum likelihood phylogeny constructed with core genome single nucleotide polymorphisms classified all psychrotolerant isolates (i.e., psychrotolerant in BHI) into clade VI (representing B. mycoides/weihenstephanensis). Analysis of correlations between gene ortholog presence or absence patterns and psychrotolerance identified 206 orthologous gene clusters that were significantly overrepresented among psychrotolerant strains, including two clusters of cold shock proteins, which were identified in 8/9 and 7/9 psychrotolerant isolates. Gene ontology analyses revealed 36 gene ontology terms that were overrepresented in psychrotolerant isolates, including putrescine catabolic processes and putrescine transmembrane transporter activity. Lastly, Hidden Markov Model searches identified three protein family motifs, including cold shock domain proteins and fatty acid hydroxylases that were significantly associated with psychrotolerance in BHI broth. Analyses of CspA sequences revealed a positive association between psychrotolerant strains and a previously identified “psychrotolerant” CspA sequence. Overall, our data highlight genetic and phenotypic differences in psychrotolerance among B. cereus group dairy-associated isolates and show that psychrotolerance is dependent on the growth medium. We also identified a number of gene targets that could be used for specific detection or control of psychrotolerant B. cereus group isolates.
Introduction
Species within the Bacillus cereus group are associated with a variety of ecological niches, and are highly diverse in terms of their genetic and phenotypic traits (Guinebretière et al., 2008; Ceuppens et al., 2013). While some species are recognized as human pathogens (e.g., B. anthracis, B. cereus) (Didelot et al., 2009; Bottone, 2010), others (e.g., B. thuringiensis) are used as insecticides (Höfte and Whiteley, 1989) or as probiotics (e.g., B. toyonensis) (Jiménez et al., 2013; Kantas et al., 2015). Some species, in particular B. weihenstephanensis, are associated with fluid milk spoilage (Bartoszewicz et al., 2008; Ivy et al., 2012; Hwang and Park, 2015; Saleh-Lakha et al., 2017).
Currently, there are 18 validated species in the B. cereus group, with nine being commonly isolated from food (Liu et al., 2017a). Furthermore, WGS data have revealed that multiple named species do not show sufficient genomic differences to actually be considered individual species. For example, species cereus and thuringiensis should be considered the same genomospecies (i.e., B. cereus/thuringiensis), as should be B. mycoides and B. weihenstephanensis (i.e., B. mycoides/weihenstephanensis) (Kovac et al., 2016, 2017; Liu et al., 2017b). Species classification of B. cereus group isolates often involves characterization using diagnostic species-specific phenotypic characteristics. For example, isolates with rhizoid morphology are classified as either B. mycoides or B. pseudomycoides (Nakamura and Jackson, 1995; Nakamura, 1998; Kovac et al., 2016; Miller et al., 2018). While B. cereus group species and strains are generally mesophilic (Koehler, 2009; Ceuppens et al., 2013; Jiménez et al., 2013), growth temperature ranges differ between species, which has also been used for species classification, with B. weihenstephanensis originally defined by its ability to grow at 7°C but not 43°C (Lechner et al., 1998). While B. weihenstephanensis was the first species in the B. cereus group that was observed to grow at refrigeration temperatures (Lechner et al., 1998), other B. cereus group species have since been reported to grow at or below 7°C (Guinebretière et al., 2008; Soufiane and Côté, 2013; Miller et al., 2016; Liu et al., 2017a). For example, B. wiedmannii isolates have been shown to grow at 5–6°C on BHI agar (Miller et al., 2016, 2018). In contrast to B. weihenstephanensis, B. wiedmannii isolates are also cytotoxic when cultured at 37°C (Miller et al., 2016, 2018), suggesting this species is a novel pathogen in the B. cereus group. Among the B. cereus group species, B. weihenstephanensis represents the species that is most frequently associated with fluid milk spoilage (Páčová et al., 2003; Bartoszewicz et al., 2008). B. weihenstephanensis, as well as potentially other psychrotolerant Bacillus spp., thus represent a particular concern for refrigerated dairy products, such as fluid milk (Huck et al., 2008; Ivy et al., 2012; Gopal et al., 2015).
Attempts to identify signature DNA sequences associated with psychrotolerance have previously identified several housekeeping genes (e.g., gmk, glpF, and tpi) (Francis et al., 1998), as well as genes encoding the conserved cold shock protein CspA that have distinct sequence types for psychrotolerant and mesophilic strains (Lechner et al., 1998). More specifically, comparisons of amino acid sequences of CspA suggested that psychrotolerant strains encode a CspA sequence beginning with “MTV” while mesophilic strains encode for an alanine at the 2nd amino acid (i.e., “MAV”) (Lechner et al., 1998). Other genetic signatures of psychrotolerance include DEAD box RNA helicases, chaperone DnaJ, and the low temperature requirement protein A (LtrA), which were found to be associated with psychrotolerance of Paenibacillus spp. isolates in SMB (growth at 6°C for 21 days) (Switt et al., 2014). While associations between psychrotolerant signatures or motifs have been established, it is important to note that phenotypic analyses characterizing the role that these genetic differences may play (such as a conformational change resulting in enhanced function at low temperatures) have not been performed. Fiedoruk et al. (2017) proposed that these motifs could be a reflection of phylogeny, as isolates from phylogenetically related clades have been shown to encode ‘psychrotolerant signatures’ (Fiedoruk et al., 2017). Furthermore, a number of specific adaptations have also been associated with psychrotolerance in bacteria. For example, alterations in RNA metabolism and activity have been linked with psychrotolerance of Bacillus spp., presumably as secondary structures that form at low temperatures, which interfere with transcription and translation, ultimately reducing or inhibiting the production of cellular proteins at low temperatures (Barria et al., 2013). More specifically, expression of nucleic acid chaperones, such as cold shock proteins (Csp) has been suggested to aid in enhancing transcription at low temperatures (Gualerzi et al., 2003). The fatty acid composition of the cell membrane has also been shown to be associated with thermotolerance among different species within the B. cereus group, with the psychrotolerant groups II (B. wiedmannii) and VI (B. mycoides/weihenstephanensis) expressing higher ratios of C16:1 fatty acids, compared to the mesophilic clades, when cultured at 30°C (Diomandé et al., 2015).
Based on the importance of psychrotolerant B. cereus group species as fluid milk spoilage organisms, and possible concerns about psychrotolerant B. cereus group species that have the potential to cause foodborne illness, we performed genetic and phenotypic characterizations on a diverse set of B. cereus group isolates grown under conditions that simulate refrigerated fluid milk stored over a 21-day shelf-life. The objective of this study was to assess the psychrotolerance of B. cereus group isolates utilizing (i) phenotypic analyses to cluster isolates based on their ability to grow at 6°C in BHI and SMB, and (ii) genetic analyses to associate genes and genetic markers with psychrotolerance. Importantly, we show that psychrotolerance is dependent on the growth medium (i.e., BHI or SMB), and that this growth phenotype is restricted to isolates in clade VI (B. mycoides/weihenstephanensis). We also identified a number of genetic markers that are significantly associated with psychrotolerance.
Materials and Methods
Isolate Selection
Isolate selection was based on an initial “psychrotolerance” screen of isolates representing all 42 rpoB allelic types (AT) (Ivy et al., 2012) found among 503 dairy-associated B. cereus group isolates in the Food Safety Lab collection1. Clade classification was based on rpoB sequence data, which were shown (Kovac et al., 2016) previously to provide for classification consistent with phylogenetic clades proposed by Guinebretière et al. (2008). The initial screen was performed by spread plating 100 μL of overnight cultures grown for 16–20 h at 32°C on BHI agar in two biological replicates. The presence of colonies after a 21-day incubation at 6°C in both biological replicates was considered indicative of psychrotolerance. The screen identified 14 isolates (each representing a different rpoB AT) that were psychrotolerant when grown on BHI agar at 6°C for 21 days. All of these isolates were classified into clades II (B. wiedmannii) and VI (B. mycoides/weihenstephanensis). To allow for comparative studies, we also included nine isolates representing B. cereus group subtypes and clades that are not typically considered psychrotolerant. These additional isolates included (i) one isolate that grouped into clade II (B. wiedmannii), but was not psychrotolerant when grown at 6°C on BHI agar, and (ii) representative isolates of B. cereus group clades I (one isolate; B. pseudomycoides), III (three isolates; B. cereus/anthracis), and IV [four isolates, including the B. cereus s.s. type strain, ATCC 14579T (FSL M8-0473); B. cereus/thuringiensis]. No isolates from clades V or VII (representing B. toyonensis and B. cytotoxicus, respectively) were included in the study, as clade V isolates were rare in our collection of 503 B. cereus group dairy isolates, and clade VII isolates were not represented at all in this collection; both of these species have not been previously characterized as being psychrotolerant (Guinebretière et al., 2013; Jiménez et al., 2013).
Quantification of Growth at 6°C in SMB and BHI Broth
The 23 selected B. cereus group isolates were tested for their ability to grow at 6°C in the nutrient-rich BHI broth, and SMB, which we deemed represents conditions similar to fluid milk. Isolates were streaked in triplicate onto BHI agar from frozen glycerol stocks, followed by incubation for 18–24 h at 32°C. One colony of each replicate was inoculated into 5 mL of BHI broth and incubated for 24 h at 32°C (without shaking). These cultures were used to inoculate tubes with 5 mL of pre-chilled BHI or SMB, to achieve an initial concentration of approximately 2.0–3.0 log10 cfu/mL (median: 2.7 log10 cfu/mL). Tubes were incubated for 21 days (without shaking) at 6°C. On days 0, 14, and 21, a 50 μL aliquot of a serially diluted culture was spiral plated (Autoplate 5000; Advanced Instruments, Inc., Norwood, MA, United States) onto Standard Plate Count (SPC) agar. After spiral plating, SPC agar plates were incubated at 32°C for 24 ± 2 h, colonies were enumerated using a Q-Count Colony Counter (Advanced Instruments, Inc.). Bacterial enumeration data were used to calculate bacterial growth at 6°C after 14 and 21 days, defined as cfu/mL at days 14 and 21, which were then normalized to cfu/mL at day 0. All growth data represent the average of three biological replicates. For data that were below the detection limit of 1 log10 cfu/mL, calculations were performed using the detection limit (i.e., 1 log10), and the final value was reported as “<” (for absolute numbers) or “>” (for expressing the log reduction over the 2-week incubation) the log10 cfu/mL value calculated. Growth patterns were categorized as “psychrotolerant” if an isolate had at least a 1 log10 increase in cfu/mL, or “not psychrotolerant” if an isolate either had a decrease in log10 cfu/mL, or an increase of < 1 log10 cfu/mL, after a 21-day incubation at 6°C.
Quality Threshold Clustering (QTC)
For quality threshold clustering (QTC) analyses, bacterial counts for days 14 and 21 were normalized to day 0 (as described above) and then averaged for each isolate. The qtclust function of the flexclust package (Leisch, 2006) in R statistical software (R Core Team, 2016) was used to cluster data using a radius of 2.2 log10 cfu/mL for 14 and 21 day counts in both BHI and SMB; the radius was chosen to minimize the number of isolates that were not assigned to a cluster and to maximize the number of clusters.
DNA Extraction and Preparation for Whole Genome Sequencing
Among the 23 isolates included in this study (Table 1), whole genome sequence (WGS) data were already available for eight isolates (Kovac et al., 2016; Miller et al., 2016). For the remaining 15 isolates, WGS was performed as part of the study reported here. For these isolates, DNA was extracted from cultures grown in BHI broth using the QIAamp DNA Mini kit (Qiagen, Valencia, CA, United States) according to a modified protocol that included a 45 min lysis step with 180 μL of 20 mg/mL lysozyme in a 37°C water bath. DNA was eluted in 50 μL of 10 mM Tris-HCl (pH 8.0). The concentration of double-stranded DNA was adjusted to 1 ng/μL, and DNA was then submitted to the Cornell University Institute of Biotechnology Genomics Facility (Ithaca, NY, United States) for Nextera XT DNA library preparation and sequencing. Samples were sequenced in two different Illumina HiSeq runs with 2 bp × 100 bp paired-end reads, having 83 and 113x coverage, respectively.
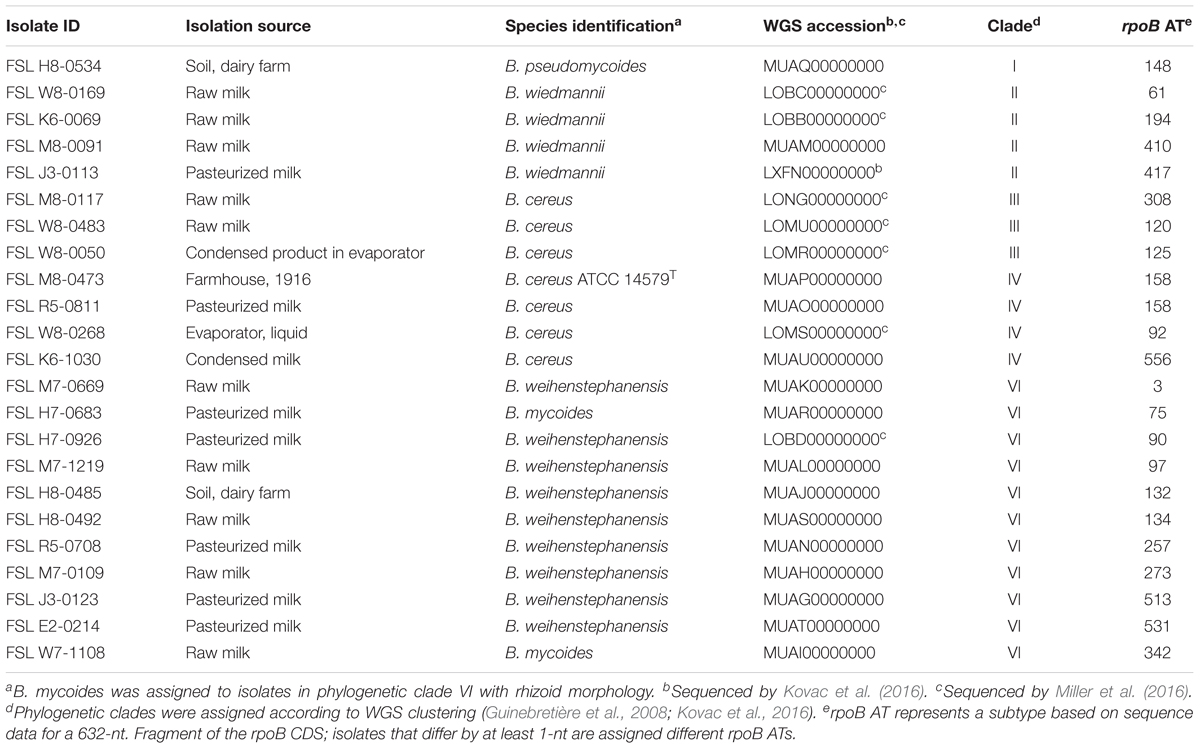
Table 1. Twenty-three B. cereus group isolates selected for cold growth quantification and WGS analyses.
Read Processing, Quality Control, Genome Assembly, and Annotation
Low quality bases, reads, and Nextera XT adapters were trimmed using the default settings of Trimmomatic v0.33 (Bolger et al., 2014). We assessed short read quality using FastQC (v0.11.2) (Babraham Bioinformatics). Genomes were assembled de novo using SPAdes v3.6.2 and a variety of k-mer sizes (21, 33, 55, 77, 99) (Bankevich et al., 2012). QUAST was used to assess the quality of the assembled draft genomes (Gurevich et al., 2013). Average coverage was determined by mapping the reads against draft genomes using BBMap v35.49 and computing the average depth using SAMtools (Li et al., 2009). Sequence reads and assembled draft genomes were submitted to NCBI SRA and WGS databases, respectively (Table 1). Submitted genomes were annotated using the NCBI prokaryotic genome annotation pipeline (Tatusova et al., 2016).
Single Nucleotide Polymorphism (SNP) Detection and Phylogeny Construction
Single nucleotide polymorphisms (SNPs) were called using kSNP3 (Gardner et al., 2015). The k-mer size of 31 was selected using Kchooser. A maximum likelihood (ML) tree was constructed with RAxML v.8.0 (Stamatakis, 2014) using the core SNPs detected by kSNP3. The ML tree was constructed using a general time-reversible (GTR) model with gamma-distributed sites (GAMMA) and 1000 bootstrap repetitions. Phylogenetic clades were assigned according to WGS clustering (Guinebretière et al., 2008; Kovac et al., 2016). The phylogenetic tree was edited using FigTree v.1.4.2.
OrthoMCL and Gene Ontology (GO) Term Annotation
All 23 B. cereus group isolate genomes were analyzed using OrthoMCL (Li et al., 2003) with an inflation value of 2.5 to find groups of orthologous genes found across multiple isolates (i.e., ortholog clusters). A representative protein sequence for each of the 9,885 identified clusters was used for GO annotation using Blast2GO (Conesa and Götz, 2008) searches against SWISS-PROT and RefSeq databases. All protein sequences were searched against the SWISS-PROT database (Bairoch and Apweiler, 2000) and only protein sequences with no GO terms mapped to the SWISS-PROT database were searched against the RefSeq database (O’Leary et al., 2015). The outputs from SWISS-PROT and RefSeq searches were combined and the assigned GO terms were linked to their respective ortholog clusters and to each member of the ortholog cluster. OrthoMCL clusters that were overrepresented among psychrotolerant isolates were assigned gene names by running BLAST using the protein sequences against RefSeq and SWISS-PROT.
Gene Presence/Absence Analysis and Gene Enrichment
Using the ortholog clusters from the OrthoMCL output, the counts of genomes where each gene was present or absent were computed for isolates classified as “psychrotolerant” or “not psychrotolerant” in BHI broth. Presence/absence data for each OrthoMCL cluster (gene) were used to generate 2x2 tables, which were analyzed using two-sided Fisher’s exact tests. Odds ratios were computed, and p-values were adjusted using the false discovery rate (FDR) method.
To identify GO terms that were over- or under-represented among genomes of isolates that were classified as “psychrotolerant” or “not psychrotolerant” in BHI, the number of genes classified as a given GO term were summed for each genome, and then for all genomes classified as showing “psychrotolerant” or “not psychrotolerant.” This approach was used to generate 2x2 tables for each GO term, which were used to run two-sided Fisher’s exact tests and compute odds ratios as described above. FDR-adjusted p-values < 0.05 were considered statistically significant.
Cold Shock Protein Sequence Analysis and Classification
Amino acid sequences of putative cold shock proteins (Csp) identified by SWISS-PROT and Refseq were compared against previously published sequences for five unique Csp proteins from B. cereus and B. subtilis, namely CspA, CspB, CspC, CspD, CspE (Mayr et al., 1996; Francis et al., 1998; Schindler et al., 1999) and an uncharacterized Csp protein described in Mayr et al. (1996). In addition, the distribution of mesophilic and psychrotolerant variants of CspA were characterized by classifying strains based on their having the mesophilic CspA sequence (beginning with “MAV”) or the psychrotolerant sequence (beginning with “MTV”) (Mayr et al., 1996; Francis et al., 1998). In addition, BLAST searches were performed for all Csp proteins using reference sequences for named Csp genes and variants described previously (Mayr et al., 1996; Francis et al., 1998; Fiedoruk et al., 2017) as well as two additional reference sequences representing (i) a novel Csp divergently related to CspC and (ii) a novel Csp annotated by the NCBI pipeline in one of the isolates sequenced here (FSL W8-0050). An e-value of 10-30, with ≤5 amino acid differences between the reference and query sequence match over the full length of the subject sequence, were used as criteria for determining the presence of Csp genes and alleles.
Fisher’s exact tests were used to test for an association between psychrotolerance and the presence of the psychrotolerant CspA sequence. p-Values < 0.05 were considered statistically significant.
Identification of Proteins Related to Psychrotolerance
Twelve HMM protein domains previously shown to be associated with psychrotolerance were obtained from Pfam 26.0 protein families’ database (see Supplementary Table S1; Finn et al., 2015). HMMER v.3.2 (Eddy, 2011), with default parameters was used to search these HMM models against the genomes for the 23 isolates characterized here. Linear regression was performed using R statistical software (2016) to determine whether protein family matches per genome are associated with psychrotolerance after a 21-day incubation at 6°C (calculated as log10 cfu/mL at day 21 – log10 cfu/mL at day 0).
Results
QTC Analyses Identify Three General Psychrotolerance Patterns for B. cereus Group Isolates From Clades Commonly Isolated From Dairy Foods and Environments
To objectively categorize B. cereus group isolates based on their ability to grow under conditions that simulate refrigerated fluid milk (SMB) and a nutrient rich media (BHI broth) we used QTC analyses to group isolates with similar growth patterns, independent of their phylogenetic relationship. Isolates generally showed consistent growth or reduction at both days 14 and 21 (Figure 1 and Table 2). QTC analyses identified three clusters (Figures 1A, 2A–D) of growth patterns after 21 days incubation at 6°C: cluster 1, containing isolates that were not psychrotolerant in BHI or SMB (i.e., <1 log10 increase in cfu/mL in both media); cluster 2, containing isolates that were psychrotolerant in BHI but not SMB, and cluster 3 containing isolates that were psychrotolerant in both SMB and BHI broth. One isolate classified into clade VI (B. mycoides/weihenstephanensis; FSL M7-0109) was not assigned to a cluster; this isolate was psychrotolerant in BHI but was just below the >1 log10 cfu/mL cut-off for psychrotolerance in SMB (Figure 2D).
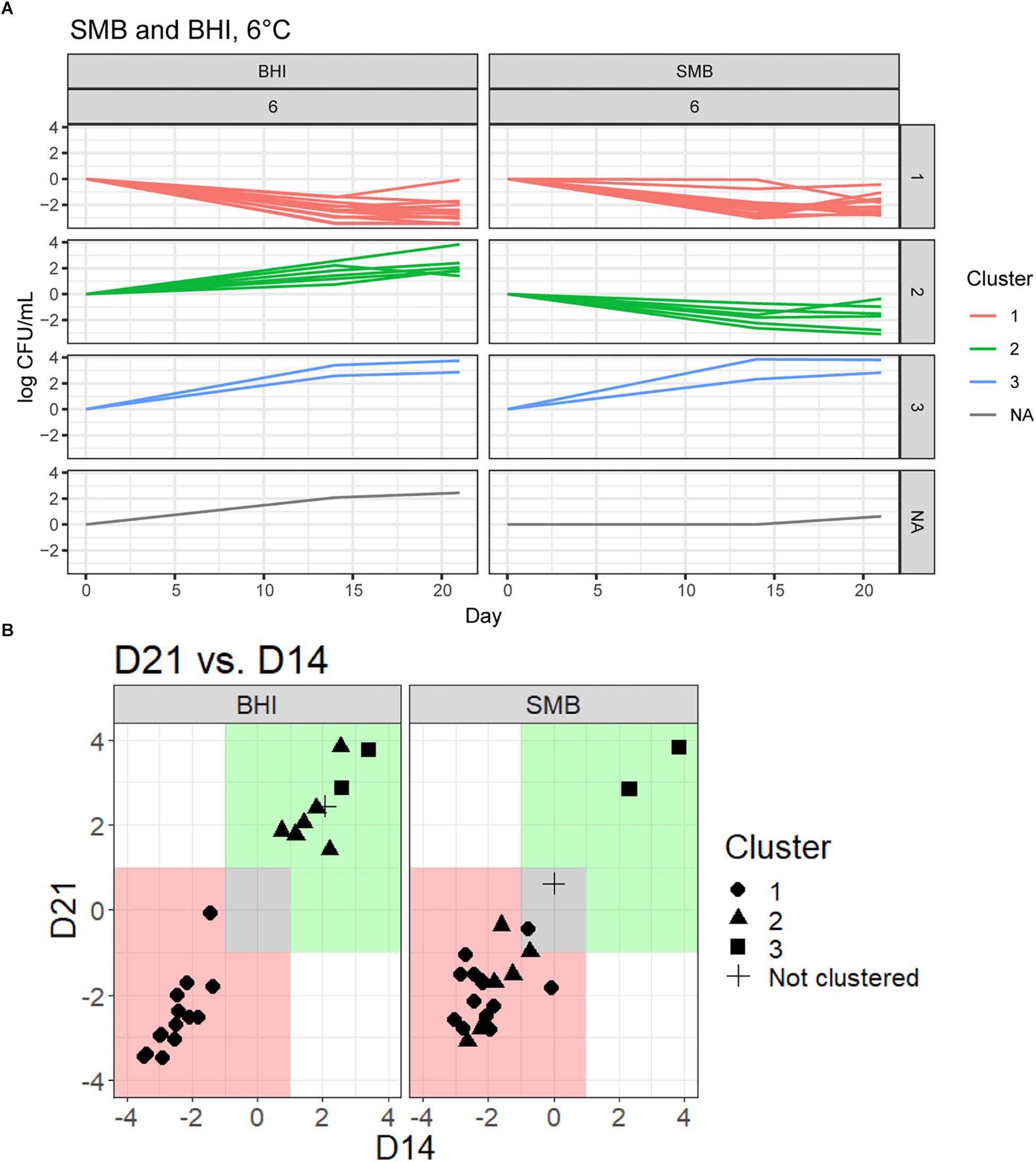
Figure 1. Growth patterns in SMB and BHI broth at 6°C for 23 Bacillus cereus group isolates. (A) Isolates were grouped according to growth in both SMB and BHI broth using quality threshold clustering (QTC). Growth data of three replicates were averaged for each isolate and growth patterns for each isolate were clustered according to their growth under these conditions, clusters are color coded according to their growth patterns. Cluster 1, marked in red, is comprised of 14 isolates that generally showed vegetative cell reduction in both BHI and SMB at 6°C. Isolates (n = 6) in cluster 2, marked in green, generally showed >1 log10 increase in cfu/mL at either day 14 or day 21 in BHI broth, and <1 log10 cfu/mL increase in SMB. Cluster 3, marked in blue, is comprised of two isolates that showed >1 log10 increase in cfu/mL in both BHI and SMB. There was also one isolate, in gray, that did not group using QTC. (B) Growth data (relative to day 0) at each time point were averaged for each of the 23 isolates (three replicates) and plotted for day 14 on the x-axis and for day 21 on the y-axis. Growth patterns for both BHI and SMB are displayed. Different shapes (e.g., circle, triangle) on the graph indicate each QTC group. The area in green represents >1 log10 growth, while the area in red represents >1 log10 reduction. The area shaded in gray represents <1 log10 difference (increase or decrease) from starting inoculum concentration.
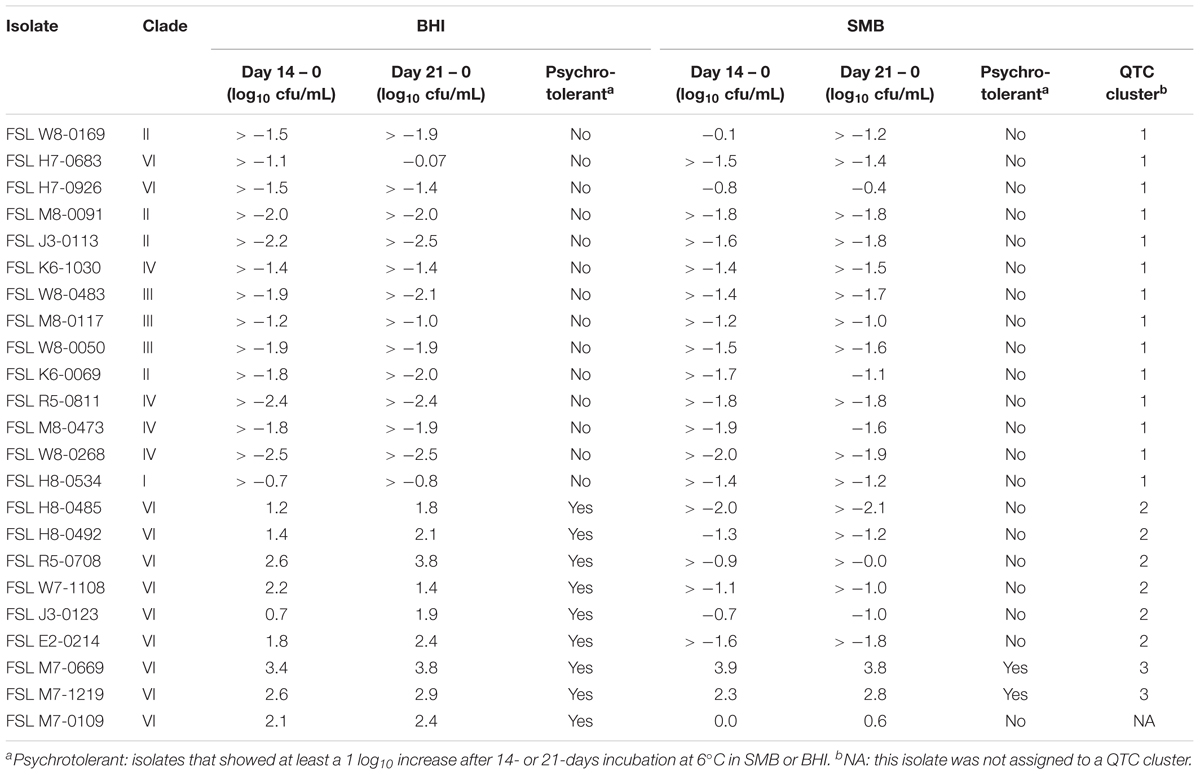
Table 2. Log difference in bacterial count for 14 and 21 days after incubation at 6°C in BHI and SMB.
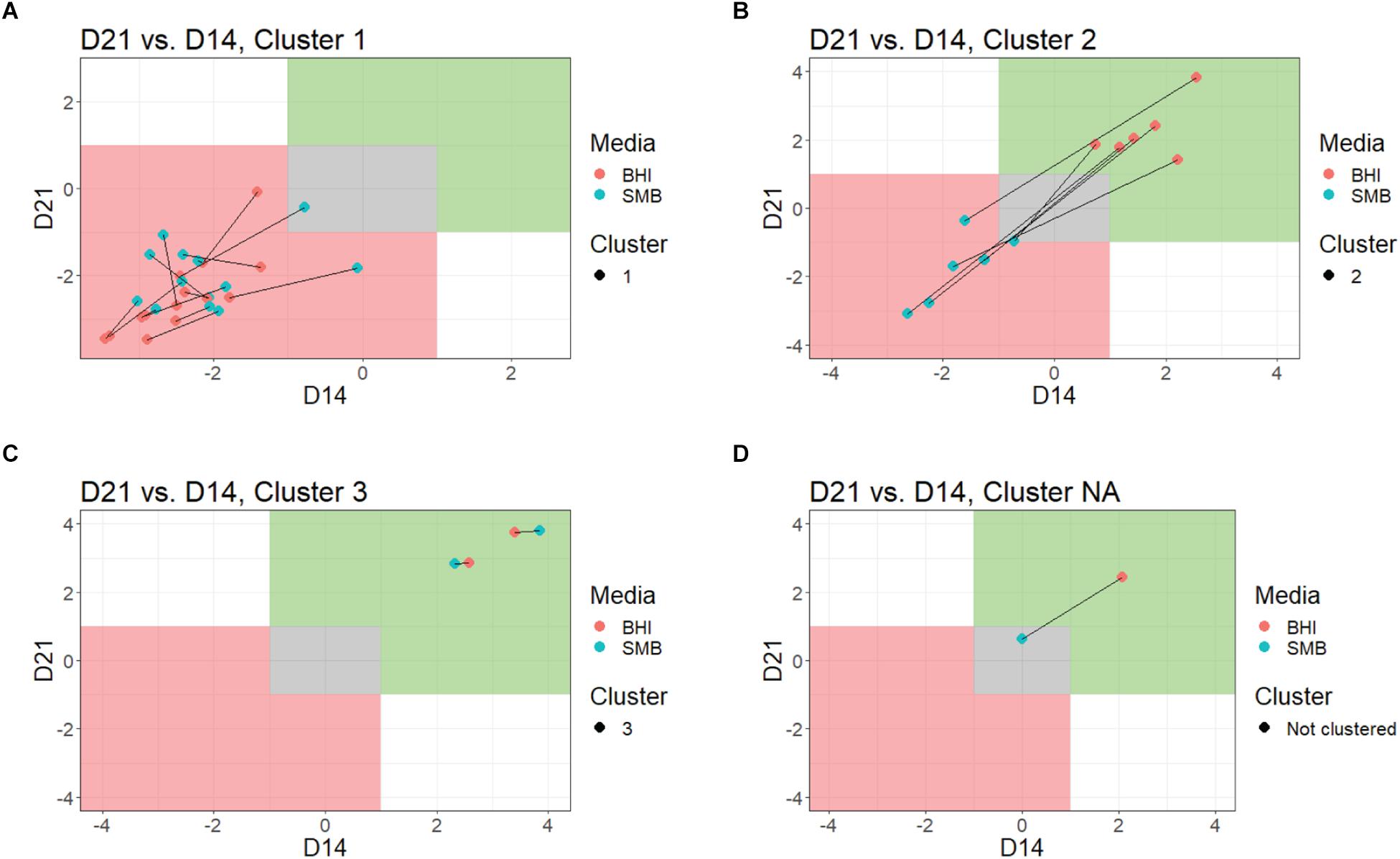
Figure 2. Comparison of growth in BHI broth and SMB, by QTC cluster. Growth data at each time point were plotted for each QTC, individually. Lines connect data from the same isolates, red dots indicate the day 14 vs. day 21 bacterial counts in BHI, blue dots indicating the day 14 vs. day 21 bacterial counts in SMB. The green shaded area is indicative of growth (>1 log10 increase), while gray represents no growth and red indicates reduction (>1 log10 decrease). Data for QTC clusters 1 (A), 2 (B), 3 (C), and isolate FSL M7-0109, which was not assigned to a QTC cluster (D) are shown.
Quality threshold clustering cluster 1 isolates (n = 14) had an average of >1.7 log10 reduction in BHI, and >1.4 log10 reduction in SMB, after a 21-day incubation at 6°C (Figures 1, 2A and Table 2). All isolates from clades I (B. pseudomycoides), II (B. wiedmannii), III (B. anthracis/cereus), IV (B. cereus/thuringiensis), and 2/11 isolates from clade VI (B. mycoides/weihenstephanensis) were assigned to QTC cluster 1.
Six isolates [all from clade VI (B. mycoides/ weihenstephanensis)] were categorized into QTC cluster 2 (Figures 1, 2B and Table 2), which represented isolates that were psychrotolerant in BHI but not in SMB after 21 days at 6°C. All isolates in this cluster showed growth of at least 1 log10 cfu/mL in BHI at both 14 and 21 days (average growth of 1.7 and 2.2 log10 cfu/mL after 14 and 21 days, respectively). In SMB, these isolates had an average of >1.3 and >1.2 log10 cfu/mL reductions after 14 and 21 days, respectively.
Quality threshold clustering cluster 3 (Figures 1, 2C and Table 2) contains two isolates that were psychrotolerant in both BHI and SMB. Both isolates belonged to clade VI (B. mycoides/weihenstephanensis), and grew in both BHI (average growth of 3.0 and 3.4 log10 after 14 and 21 days, respectively) and SMB (3.1 and 3.3 log10 increase after 14 and 21 days) incubated at 6°C.
Psychrotolerance in BHI Broth Is Unique to Isolates in Clade VI
To assess whether there was a relationship between phylogenetic clustering and psychrotolerance, we first mapped the QTC clusters onto a phylogenetic tree based on core genome SNPs of the 23 isolates included in this study. All psychrotolerant isolates clustered in phylogenetic clade VI (B. mycoides/weihenstephanensis); this clade also contained two isolates that were not psychrotolerant in either SMB or BHI (Figure 3). All tested isolates from the other clades (i.e., clades I, II, III, and IV) consistently showed reduction in cell counts in both SMB (mean decrease of >1.5 log10 after 14 and 21 days) and BHI at 6°C (mean decrease of >1.8 and >1.9 log10 after 14 and 21 days, respectively).
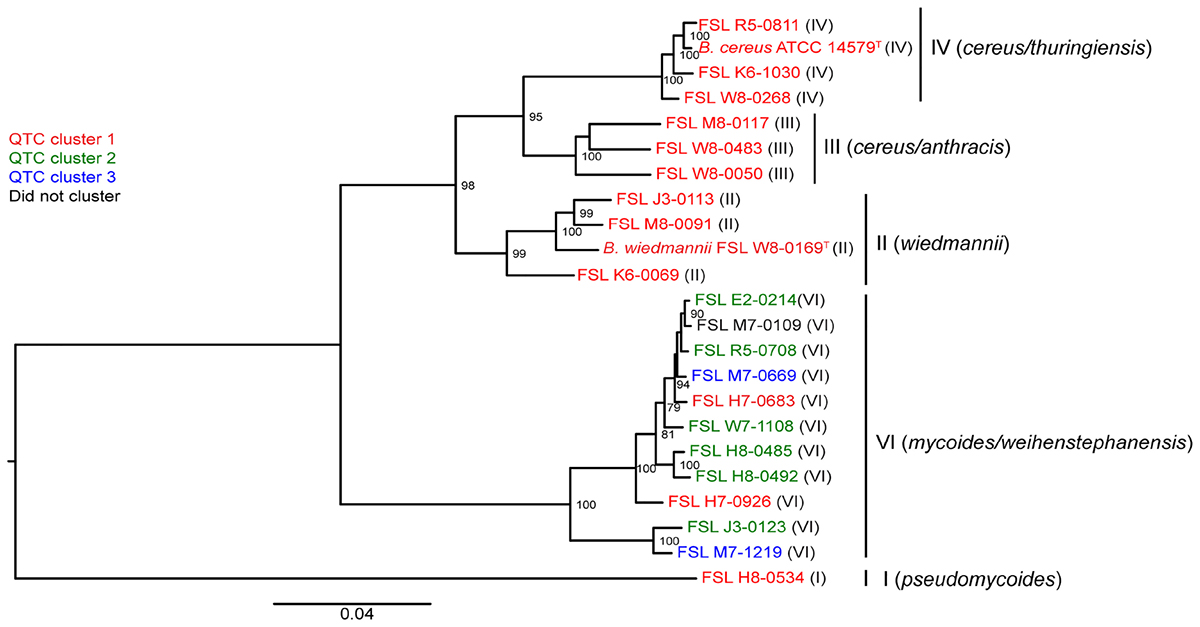
Figure 3. Phylogenetic tree constructed from the core SNPs identified in the genomes of 23 B. cereus isolates. The maximum likelihood tree was constructed using a general time-reversible (GTR) model with gamma-distributed sites and 1,000 bootstrap repetitions. Roman numerals in parentheses represent the phylogenetic clade of the isolate, as defined previously (Kovac et al., 2016). QTC clusters representing isolates with similar growth patterns in BHI and SMB at 6°C (see Figure 1A) are mapped onto the phylogenetic tree. Isolates shown in red represent QTC cluster 1 isolates (non-psychrotolerant in BHI broth or SMB). Isolates shown in green represent QTC cluster 2 isolates (psychrotolerant in BHI broth but not SMB), and isolates shown in blue represent clade 3 isolates (psychrotolerant in both BHI broth and SMB). One isolate (FSL M7-0109) did not cluster with other isolates, and is therefore shown in black font. Numbers at branch points represent bootstrap values; only bootstrap values >70 are shown.
Targeted Searches for Protein Families and Gene Variants Previously Associated With Psychrotolerance Identified Genetic Markers for Growth at 6°C Among B. cereus Group Isolates
To test for specific genetic markers associated with psychrotolerance, we first performed HMM searches for 12 protein families that had previously been identified as being associated with psychrotolerance and cold adaptation. Members of 11 of these 12 protein families were identified in at least one of the B. cereus group genomes tested (see Supplementary Table S2 for details). For eight protein families, genes encoding for these proteins were identified in all 23 genomes, although the number of distinct hits in a given genome sometimes differed considerably (Supplementary Table S2). For example, either six or seven distinct hits for the cold shock domain (listed as “CSD” in Supplementary Table S2) were identified among each of the 23 different genomes. Isolates in QTC cluster 2, which were psychrotolerant in BHI but not in SMB, averaged 6.8 distinct hits whereas all other isolates averaged 6.3 distinct hits. The three HMM families for which we found distinct hits in less than 23 genomes included LtrA, Caps synth-CapC, and FA desaturase 2. LtrA was identified in only two genomes [FSL M8-0473 (ATCC 14579T) and FSL R5-0811], which are both clade IV (B. cereus/thuringiensis) isolates. Caps synth-CapC and FA desaturase 2 were found at least once in 22/23 and 20/23 genomes, respectively (see Supplementary Table S2 for details).
For three HMM, we found a significant positive association between the number of hits and psychrotolerance in BHI; these families include cold shock domain proteins (p = 0.013), FA hydroxylase (p = 0.011), and YdjO (p = 0.002). Psychrotolerant isolates averaged 6.8, 2.1, and 2.7 hits for cold shock domain, FA hydroxylase, and YdjO members, while non-psychrotolerant isolates averaged 6.2, 1.5, and 1.9 distinct hits, respectively. Furthermore, isolates from QTC cluster 2 (psychrotolerant in BHI but not in SMB) averaged more hits of these protein families than isolates in QTC clusters 1 or 3. Surprisingly, the HMM representing the DEAD domain showed a significant negative association (p = 0.025) with psychrotolerance. Isolates categorized as “not psychrotolerant” averaged more hits for the DEAD domain (37.5 hits), than psychrotolerant isolates (35.9 hits).
We also performed BLAST searches for individual named cold shock proteins (CspA, CspB, CspC, CspD, and CspE) and one protein identified through OrthoMCL analyses (defined as “CspM” here) and one protein that was identified as a Csp when the isolate (FSL W8-0050) was annotated using the NCBI annotation pipeline (Supplementary Table S7). Analyses revealed a significant association (p = 0.003; Fisher’s exact test) between psychrotolerant isolates and the presence of the gene encoding the previously defined psychrotolerant CspA signature (Francis et al., 1998). The psychrotolerant CspA sequence was found in 12 isolates, including all 11 clade VI isolates (with 9/11 isolates classified as psychrotolerant) and the 1 clade I isolate (B. pseudomycoides) tested (which was classified as not psychrotolerant). The remaining 11 B. cereus genomes encoded the mesophilic CspA sequence. CspC and CspE (as described in Fiedoruk et al., 2017) were detected in all isolates, regardless of phylogenetic clade. CspB was detected in all clade I and VI isolates, and half of the clade II isolates. CspD1 and CspD2 (Fiedoruk et al., 2017) were detected in all isolates, with clade II and VI isolates encoding CspD1_IIa-c and D2_IV-M, and isolates in clades III and IV encoding CpsD1_IV-I/M and CpsD2_IV-M; the clade I (B. pseudomycoides) isolate encoded the CspD1_IV-I/M and the CspD2_IV-M variants. CpsM was detected in eight isolates (all clade VI) and one isolate encoded CspN (Supplementary Table S7).
Genes Encoding for Invasion Proteins, Cold Shock Proteins, and Protein Families Involved in Putrescine Metabolism Are Overrepresented Among B. cereus Group Isolates That Are Psychrotolerant in BHI Broth
To search for novel genes associated with the observed psychrotolerance we also used OrthoMCL to compare gene presence/absence between psychrotolerant isolates (able to grow at 6°C in BHI broth) and non-psychrotolerant isolates. Overall, OrthoMCL identified 9,885 different orthologous clusters (i.e., clusters of genes assumed to encode for proteins that share the same characteristics) among the 23 genomes analyzed. FDR-corrected Fisher’s exact tests were used to identify genes that were overrepresented among the nine isolates that were psychrotolerant in BHI broth; we did not test for genes overrepresented among isolates that were psychrotolerant in SMB, as only two isolates fit this category. These analyses revealed 206 orthologous gene clusters significantly overrepresented in isolates that were psychrotolerant in BHI broth, and 182 orthologous gene clusters significantly overrepresented in isolates that were not psychrotolerant in BHI broth (Supplementary Tables S3, S4). Importantly, we did not identify any orthologous gene clusters that were unique to all 9 isolates that were psychrotolerant in BHI broth, but were absent from all 14 isolates that were not psychrotolerant in BHI broth. However, we identified 87 clusters (including some discussed in more detail below) that were found among all nine isolates that were psychrotolerant in BHI broth, and only found in 2/14 isolates that were not; all of these gene clusters were specific for clade VI (B. mycoides/weihenstephanensis) and were only found among the 11 clade VI strains characterized here.
Two orthologous clusters representing cold shock proteins (cluster 4979 and cluster 5279) were overrepresented in psychrotolerant isolates (Supplementary Table S3). These clusters were identified as genes encoding YdjO (98% coverage to B. subtilis YdjO) and CspC (100% coverage to B. subtilis CspC), respectively. Further comparison of the amino acid sequence from B. cereus CspC against the newly identified sequence showed that the new sequence does not correspond to B. cereus CspC and was therefore re-named “CspM.” ydjO (cluster 4979) was identified in 8/9 isolates that were psychrotolerant in BHI broth (all except for FSL W7-1108, B. mycoides/weihenstephanensis), while cspM (cluster 5279) was identified in 7/9 psychrotolerant isolates (all except for FSL H8-0492 and FSL M7-0669, both B. mycoides/weihenstephanensis). Interestingly, ydjO was found in two isolates that were not psychrotolerant in BHI broth (FSL H7-0926 and FSL H7-0683) and cspM was identified in one isolate (FSL H7-0926) that was not psychrotolerant in BHI broth. Both of these genes were only identified in isolates from phylogenetic clade VI (B. mycoides/weihenstephanensis), regardless of whether or not the isolate was psychrotolerant; specifically, ydjO and cspM were found in 10/11 and 8/11 clade VI isolates, respectively.
Other orthologous clusters overrepresented in the nine psychrotolerant isolates include: (i) two genes encoding invasion proteins (clusters 4691 and 4799), (ii) two genes annotated as encoding homologs of the damage inducible protein DinB (clusters 4457 and 4953), (iii) two genes encoding glyoxalases (clusters 4700 and 4937), and (iv) two genes encoding GNAT family acetyltransferases (clusters 4699 and 4966) (Supplementary Table S3). Both orthologous clusters encoding for invasion proteins were found in all nine psychrotolerant isolates and in the two non-psychrotolerant isolates (FSL H7-0926 and FSL H7-0683) that are also in clade VI (B. mycoides/weihenstephanensis); no specific pre-existing gene names could be assigned to these two clusters. Orthologous clusters 4457 and 4953, which were annotated as the damage-inducible protein DinB, showed 98 and 99% coverage to dinB in several species in the B. cereus group, respectively. Cluster 4457 was identified in all psychrotolerant strains, as well as in three isolates that were not-psychrotolerant in BHI broth; these three isolates (all clade II, B. wiedmannii) however showed growth at 6°C on BHI agar (Miller et al., 2018). Cluster 4457 was identified in all 11 clade VI isolates, and in isolate FSL J3-0113 (clade II, B. wiedmannii). Cluster 4953 was identified in 8/9 isolates that were psychrotolerant in BHI broth (all except for FSL H8-0482) and in 2/14 isolates that were not psychrotolerant in BHI broth (FSL H7-0926 and FSL H7-0683). Overall, cluster 4953 was identified in 10/11 isolates classified into phylogenetic clade VI (B. mycoides/weihenstephanensis). Among the two orthologous clusters annotated as glyoxalases, no specific gene name could be assigned to cluster 4937, but cluster 4700 was identified as mhqA (98% coverage in B. subtilis with equal coverage matches to paralogs mhqE and mqoO), which encodes a putative ring-cleaving dioxygenase. Cluster 4937 was found in 8/9 isolates that were psychrotolerant in BHI broth (all expect for FSL W7-1108), while cluster 4700 was found in all isolates that were psychrotolerant in BHI broth. Cluster 4937 and cluster 4700 were found in 10/11 and 11/11 clade VI (B. mycoides/weihenstephanensis) isolates, respectively. Among the two clusters identified as GNAT family acetyltransferases, cluster 4699 showed 97% coverage to YoaA of B. subtilis, while cluster 4966 could not be assigned a specific gene name (no coverage match > 95%). yoaA was present in all nine psychrotolerant isolates and in two non-psychrotolerant isolates, while cluster 4966 was identified in 8/9 psychrotolerant isolates and in two non-psychrotolerant isolates. yoaA was found in all 11 clade VI isolates while cluster 4966 was identified in 10/11 clade VI (B. mycoides/weihenstephanensis) isolates (excluding FSL M7-1219); neither of these clusters were identified in isolates outside of clade VI.
GO term analyses revealed 36 terms that were significantly overrepresented in isolates psychrotolerant in BHI broth, and 12 terms that were significantly overrepresented in isolates that were not psychrotolerant in BHI broth (see Supplementary Tables S5, S6 for details). GO terms found to be overrepresented in the genomes of B. cereus group isolates classified as psychrotolerant in BHI broth included the terms “putrescine catabolic process” (GO:0009447) and “putrescine transmembrane transporter activity” (GO:0015489) (see Supplementary Table S5 for a complete list). GO terms found to be overrepresented in the genomes of B. cereus group isolates that were not psychrotolerant in BHI broth include “alkanesulfonate transporter activity” (GO:0042959), “alkanesulfonate monooxygenase activity” (GO:0008726), and “alkanesulfonate catabolic process” (GO:0046306) (see Supplementary Table S6 for a complete list).
Discussion
The B. cereus group includes multiple species that are frequently isolated from dairy foods, including foodborne pathogens (e.g., B. cereus strains expressing emetic or diarrheal toxins) and spoilage organisms, which are of particular importance for refrigerated dairy foods such as fluid milk (e.g., B. weihenstephanensis). A number of recent studies (Kovac et al., 2016; Warda et al., 2016; Zhang et al., 2017; Miller et al., 2018) have combined phenotypic and genomic-based approaches to allow for improved characterization and identification of B. cereus group clonal groups, strains, and isolates that have the ability to cause human disease. However, these studies did not examine the genetic basis for psychrotolerance among B. cereus group isolates. An improved understanding of psychrotolerance and the underlying genetic markers that are associated would allow for improved (i) risk assessments, (ii) detection and characterization methods that can rapidly detect and identify B. cereus group strains that can grow at refrigeration temperatures, and (iii) development of control strategies, specifically for refrigerated dairy products (e.g., fluid milk, certain types of high pH cheeses) that may permit B. cereus group isolates’ growth. Our results provide important data characterizing psychrotolerance among B. cereus group isolates obtained from dairy-associated sources. Our data specifically indicate that among B. cereus species isolates commonly isolated from dairy foods and dairy environments, (i) psychrotolerance in liquid media seems to be limited to strains in B. cereus group clade VI (B. mycoides/weihenstephanensis) and that (ii) a number of genes are linked to psychrotolerance of B. cereus group isolates.
Psychrotolerance in SMB or Rich Media Is Limited to Some, but Not All, Isolates in Phylogenetic Clade VI (B. mycoides/weihenstephanensis)
While our initial screen identified 14 B. cereus group isolates that showed growth on BHI agar at 6°C [11 and 3 isolates from clades VI (B. mycoides/weihenstephanensis) and II (B. wiedmannii), respectively], only nine of these isolates showed growth in BHI broth at 6°C, and just two isolates grew in SMB incubated at 6°C. While 3/4 isolates in phylogenetic clade II (B. wiedmannii), were able to grow on BHI agar incubated at 5–6°C (Miller et al., 2016, 2018), none of these isolates were classified as psychrotolerant in BHI broth or SMB in the results presented here. Interestingly, 4/4 B. wiedmannii isolates included in this study encoded the mesophilic variant of the CspA sequence, whereas the psychrotolerant CspA sequence was only found in the B. mycoides/weihenstephanensis isolates, and the one B. pseudomycoides isolate. B. wiedmannii was first described in 2016 as a psychrotolerant member of the B. cereus group (Miller et al., 2016) where 5/11 isolates showed growth after 21 days of incubation at 5°C on BHI agar and all strains showed growth at 10°C. This suggests that the ability to grow at 6°C may differ between planktonic versus surface growth for this species and suggests a need for further characterization of B. wiedmannii for its ability to grow in different dairy products (e.g., fluid milk, fresh cheeses) under refrigeration conditions, as recent studies (Kovac et al., 2016; Miller et al., 2016, 2018) have shown that B. wiedmannii isolates carry virulence toxin genes associated with diarrheal disease (hblACD and nheABC) and show cytotoxicity in a HeLa cell culture model (Miller et al., 2016, 2018). Additionally, B. wiedmannii isolates have been shown to produce both Hbl and Nhe toxins (Kovac et al., 2016; Miller et al., 2016, 2018). Future studies are needed to combine growth and virulence data to better assess the foodborne illness risk associated with B. wiedmannii contamination of different dairy products.
While nine isolates, which were all classified into B. cereus group clade VI (B. mycoides/weihenstephanensis), showed growth at 6°C in BHI broth, only two of these isolates were also psychrotolerant in SMB. Future experiments may be needed to characterize and compare the ability of psychrotolerant Bacillales to grow in SMB and fluid milk in order to better assess food safety and spoilage risks associated with the presence of different Bacillales (e.g., Paenibacillus, B. wiedmannii, B. weihenstephanensis) in fluid milk. This is important as autoclaving of SMB, as performed here, may result in the production of Maillard reaction products, which have been linked to antimicrobial activity (Ledl and Schleicher, 1990; Hauser et al., 2014). Therefore, SMB may prevent growth of B. cereus group isolates under certain conditions, even though growth may occur in fluid milk. Clade VI (B. mycoides/weihenstephanensis), included all nine isolates that were psychrotolerant in BHI broth. While a number of B. cereus group species have been reported to include isolates that grow at low temperatures (Stenfors and Granum, 2001; Soufiane and Côté, 2013; Miller et al., 2016), our data are consistent with previous reports that B. weihenstephanensis isolates typically are able to grow at refrigeration temperatures (Lechner et al., 1998) and observations that isolates classified into this species have repeatedly been linked to spoilage issues in fluid milk (Páčová et al., 2003; Ivy et al., 2012; Trmčić et al., 2015). Likewise, our data demonstrate the ability of B. mycoides isolates to grow at 6°C. B. mycoides was previously identified as capable of growing at 7°C (Guinebretière et al., 2008), but is not classically defined as being psychrotolerant. Meanwhile, B. weihenstephanensis, which was discovered more than a century after B. mycoides was described in 1886 (Lewis, 1932), has been described as psychrotolerant (Lechner et al., 1998; Stenfors and Granum, 2001). Importantly, WGS shows that B. weihenstephanensis and B. mycoides should be considered the same genomospecies [despite B. mycoides having a rhizhoid morphology (Kovac et al., 2016; Miller et al., 2018)], which would consolidate these Bacillales that are typically psychrotolerant into a single species. It’s noteworthy though, that among the 11 clade VI (B. mycoides/weihenstephanensis) isolates tested here, only two did not grow in either BHI or SMB. Previous studies have also found that not all B. weihenstephanensis show growth at low temperatures. For example, a 2008 study by Guinebretière et al. (2008) reported 7/143 B. weihenstephanensis isolates were unable to grow at 7°C on J-agar.
All isolates outside of clade VI (B. mycoides/weihenstephanensis) showed reductions in vegetative cell numbers in both BHI and SMB after incubation for 14 to 21 days at 6°C, as did some clade VI isolates, particularly when incubated in SMB. While these findings may suggest die-off, reduction of vegetative cell numbers, as determined by plating on SPC agar, could also be caused by B. cereus group cells entering spore form during exposure to cold stress. Although sporulation due to cold stress has not been studied in Bacillus spp., a characterization study of thermophilic Clostridium thermocellum JW20 found that vegetative cells entered spore form when the temperature fell below 45°C for several hours (Freier et al., 1988). In contrast, a more recent study of C. thermocellum found that another strain (ATTC 27405) does not enter spore form at temperatures below 48°C (Mearls et al., 2012), suggesting that sporulation from sub-optimum temperature stress may be a strain-dependent phenotype. Future experiments will thus be needed to assess sporulation of different Bacillales isolates under cold stress exposure, as well as the environmental signals required for these cells to re-enter the vegetative state.
Overrepresentation of Selected Genes Linked to Psychrotolerance in Phylogenetic Clade VI (B. mycoides/weihenstephanensis) Isolates Identifies Potential Targets for Detection of Isolates More Likely to Grow at Low Temperatures
Overall, 206 genes identified using OrthoMCL and three domains identified by HMM had a significant positive association with B. cereus group isolates that were psychrotolerant in BHI broth. Cold shock proteins have been studied in the B. cereus group (Brillard et al., 2010), and a signature sequence of one particular cold shock gene, cspA (encoding CspA) has been used previously to identify B. weihenstephanensis (Francis et al., 1998; Lechner et al., 1998; Stenfors and Granum, 2001). In our study we found a significant association between psychrotolerant isolates and the psychrotolerant CspA signature sequence consistent with a previous study (Lechner et al., 1998). It is important however to note that all isolates that were psychrotolerant in BHI broth in this study were from phylogenetic clade VI (B. mycoides/weihenstephanensis). The CspA signature was also found in a clade I B. pseudomycoides isolate (FSL H8-0534), despite this strain not being psychrotolerant in BHI broth or SMB. It is interesting to note that B. pseudomycoides isolates have not been previously observed to grow at 6°C. Of similar interest, none of the clade II isolates (B. wiedmannii) harbored the psychrotolerant CspA sequence, despite their being psychrotolerant when grown at 6°C on BHI agar. A recent study by Fiedoruk et al. (2017) suggested that the psychrotolerant signature sequences could be a reflection of the phylogenetic ancestry of the strain, rather than an adaptation enabling psychrotolerance. However, further phylogenetic analyses would be required in order to determine the evolutionary history of the B. cereus group species and confirm this hypothesis. Interestingly, analysis of additional cold shock proteins (as detailed by Fiedoruk et al., 2017) among the 23 isolates listed here revealed that all isolates encode CspC and CspE, and therefore detection of these genes does not represent a useful screening method for detection of psychrotolerant strains of B. cereus. Furthermore, we identified two novel cold shock proteins, CspM and CspN (see Supplementary Table S7). CspM was detected in 8/11 clade VI isolates, and CspN was detected in just one clade III strain. Further mutagenesis studies of the psychrotolerant and mesophilic motifs in CspA as well as different cold shock protein genes is needed to determine whether certain motifs or genes enable psychrotolerance or whether their presence solely represent a marker of phylogenetic relatedness and facilitates phenotypes other than the ability to grow at low temperatures.
Aside from the psychrotolerant CspA variant, we also identified a number of (i) protein family motifs and (ii) genes associated with psychrotolerant B. cereus group isolates. HMM analyses revealed three protein families for which the number of hits was positively correlated with isolates that were psychrotolerant in BHI broth, including cold shock domain proteins, fatty acid hydroxylases, and the YdjO protein domain. Cold shock domain proteins have previously been associated with isolates that can grow at low temperatures, especially B. weihenstephanensis (Francis et al., 1998; Lechner et al., 1998). Fatty acid hydroxylases, which have altered expression at low temperatures to allow for adjustments in membrane fluidity, have also been previously associated with psychrotolerance (Mansilla et al., 2004; Chan and Wiedmann, 2008; Diomandé et al., 2015). Finally, the YdjO domain is not only found in the cold shock-induced YdjO protein, which is functionally uncharacterized (Kaan et al., 2002), but is also found in other proteins and may be linked to the ability to grow at lower temperatures. More “hits” of these protein families (cold shock domain, fatty acid hydroxylases, and YdjO) were found in isolates that were psychrotolerant in BHI broth. Surprisingly, DEAD box RNA helicases, which have been previously reported to be associated with psychrotolerance among Bacillales genera (Chan and Wiedmann, 2008; Barria et al., 2013; Switt et al., 2014), had more “hits” among isolates included in this study that were not psychrotolerant in BHI. Genes identified as overrepresented in psychrotolerant B. cereus group isolates included ydjO and cspM, both encoding cold shock proteins. Further functional studies confirming the roles of ydjO and cspM in psychrotolerance, will be beneficial to assess whether these genes represent targets appropriate for rapid identification of psychrotolerant B. cereus strains.
Overall, we identified a number of genetic targets associated with B. cereus group isolates that are psychrotolerant in BHI broth. Future work characterizing the expression and role of these genes in psychrotolerance will be beneficial for predictive assays aimed at detecting psychrotolerant B. cereus group isolates. As psychrotolerant B. cereus group isolates represent important spoilage bacteria in dairy foods, the genetic targets identified here could be used to develop improved DNA-based assays for rapid and specific identification of psychrotolerant B. cereus group isolates throughout the dairy supply chain. Furthermore, our data also demonstrate the importance of assessing psychrotolerance using multiple methods, which, in the context of food isolates, may include media representative of different foods as well as different growth conditions (e.g., liquid culture vs. agar plates).
Conclusion
Isolates in the B. cereus group demonstrate differing capacities for psychrotolerance, which is dependent on phylogeny. While previous studies have suggested that psychrotolerant isolates exist across a variety of B. cereus group species (Stenfors and Granum, 2001; Soufiane and Côté, 2013; Miller et al., 2016), phenotypic characterization of B. cereus group isolates representing the predominant sequence types in dairy foods and environments showed that only isolates from phylogenetic clade VI (B. mycoides/weihenstephanensis) showed growth at 6°C in either SMB or BHI broth. Genetic analyses revealed a number of genes and GO terms significantly associated with isolates that were psychrotolerant in BHI broth, and confirmed that the psychrotolerant CspA amino acid motif is associated with psychrotolerant B. cereus group isolates. While the presence of the psychrotolerant CspA amino acid sequence was positively associated with B. cereus group isolates psychrotolerant in BHI broth, it is important to consider that in this study, only clade VI isolates showed growth at 6°C in either BHI broth or SMB. All clade VI isolates had the psychrotolerant CspA amino acid sequence, regardless of the isolate’s ability to grow at 6°C. The clade I B. pseudomycoides isolate, FSL H8-0534, which did not grow at 6°C in either BHI or SMB, also had the psychrotolerant CspA amino acid sequence. Meanwhile, B. wiedmannii, a previously described psychrotolerant species with pathogenic potential did not have the psychrotolerant CspA sequence. In addition to future research on the functional importance of different genetic markers associated with psychrotolerance in B. cereus groups species, further work on growth at refrigeration temperatures in different matrices and under different environmental conditions will be important, particularly since our data suggest that B. wiedmannii can grow at 6°C on solid media, but not liquid substrates tested.
Data Availability
The datasets generated for this study can be found in NCBI, https://www.ncbi.nlm.nih.gov/bioproject?LinkName=nuccore_bioproject&from_uid=1151525537.
Author Contributions
SB, NM, JK, and MW designed the study. SB and DD conducted growth experiments. SB, RO, DK, JK, and RC performed bioinformatical and statistical analyses. SB, RO, RC, and MW wrote the manuscript. All authors assisted with the revision of this manuscript.
Funding
The New York State Dairy Promotion Board, through the New York State Department of Agriculture and Markets (Albany, NY, United States), provided funding for this project. SB was supported by a USDA National Needs Grant (2013-38420-20519). JK was supported by the USDA NIFA Hatch Appropriations Project PEN04646 (Accession No. 1015787).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge Maureen Gunderson, Cornell University Milk Quality Improvement Program, for preparing all necessary media for these experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00662/full#supplementary-material
Footnotes
References
Bairoch, A., and Apweiler, R. (2000). The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acid. Res. 28, 45–48. doi: 10.1093/nar/28.1.45
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., and Kulikov, A. S. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barria, C., Malecki, M., and Arraiano, C. (2013). Bacterial adaptation to cold. Microbiology 159, 2437–2443. doi: 10.1099/mic.0.052209-0
Bartoszewicz, M., Hansen, B. M., and Swiecicka, I. (2008). The members of the Bacillus cereus group are commonly present contaminants of fresh and heat-treated milk. Food Microbiol. 25, 588–596. doi: 10.1016/j.fm.2008.02.001
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bottone, E. J. (2010). Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 23, 382–398. doi: 10.1128/CMR.00073-09
Brillard, J., Jéhanno, I., Dargaignaratz, C., Barbosa, I., Ginies, C., Carlin, F., et al. (2010). Identification of Bacillus cereus genes specifically expressed during growth at low temperatures. Appl. Environ. Microbiol. 76, 2562–2573. doi: 10.1128/AEM.02348-09
Ceuppens, S., Boon, N., and Uyttendaele, M. (2013). Diversity of Bacillus cereus group strains is reflected in their broad range of pathogenicity and diverse ecological lifestyles. FEMS Microb. Ecol. 84, 433–450. doi: 10.1111/1574-6941.12110
Chan, Y. C., and Wiedmann, M. (2008). Physiology and genetics of Listeria monocytogenes survival and growth at cold temperatures. Crit. Rev. Food Sci. Nutr. 49, 237–253. doi: 10.1080/10408390701856272
Conesa, A., and Götz, S. (2008). Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2018:619832. doi: 10.1155/2008/619832
Didelot, X., Barker, M., Falush, D., and Priest, F. G. (2009). Evolution of pathogenicity in the Bacillus cereus group. Syst. Appl. Microbiol. 32, 81–90. doi: 10.1016/j.syapm.2009.01.001
Diomandé, S. E., Guinebretière, M.-H., De Sarrau, B., Broussolle, V., and Brillard, J. (2015). Fatty acid profiles and desaturase-encoding genes are different in thermo-and psychrotolerant strains of the Bacillus cereus Group. BMC Res. Notes 8:329. doi: 10.1186/s13104-015-1288-4
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Fiedoruk, K., Drewnowska, J. M., Daniluk, T., Leszczynska, K., Iwaniuk, P., and Swiecicka, I. (2017). Ribosomal background of the Bacillus cereus group thermotypes. Sci. Rep. 7:46430. doi: 10.1038/srep46430
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2015). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Francis, K. P., Mayr, R., Von Stetten, F., Stewart, G. S., and Scherer, S. (1998). Discrimination of psychrotrophic and mesophilic strains of the Bacillus cereus group by PCR targeting of major cold shock protein genes. Appl. Environ. Microbiol. 64, 3525–3529.
Freier, D., Mothershed, C. P., and Wiegel, J. (1988). Characterization of Clostridium thermocellum JW20. Appl. Environ. Microbiol. 54, 204–211.
Gardner, S. N., Slezak, T., and Hall, B. G. (2015). kSNP3. 0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Gopal, N., Hill, C., Ross, P. R., Beresford, T. P., Fenelon, M. A., and Cotter, P. D. (2015). The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front. Microbiol. 6:1418. doi: 10.3389/fmicb.2015.01418
Gualerzi, C. O., Giuliodori, A. M., and Pon, C. L. (2003). Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 331, 527–539. doi: 10.1016/S0022-2836(03)00732-0
Guinebretière, M. H., Auger, S., Galleron, N., Contzen, M., De Sarrau, B., De Buyser, M. L., et al. (2013). Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus Group occasionally associated with food poisoning. Int. J. Syst. Evol. Microbiol. 63, 31–40. doi: 10.1099/ijs.0.030627-0
Guinebretière, M. H., Thompson, F. L., Sorokin, A., Normand, P., Dawyndt, P., and Ehling-Schulz, M. (2008). Ecological diversification in the Bacillus cereus Group. Environ. Microbiol. 10, 861–865. doi: 10.1111/j.1462-2920.2007.01495.x
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hauser, C., Müller, U., Sauer, T., Augner, K., and Pischetsrieder, M. (2014). Maillard reaction products as antimicrobial components for packaging films. Food Chem. 145, 608–613. doi: 10.1016/j.foodchem.2013.08.083
Höfte, H., and Whiteley, H. (1989). Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 53, 242–255.
Huck, J. R., Sonnen, M., and Boor, K. J. (2008). Tracking heat-resistant, cold-thriving fluid milk spoilage bacteria from farm to packaged product. J. Dairy Sci. 91, 1218–1228. doi: 10.3168/jds.2007-0697
Hwang, J.-Y., and Park, J.-H. (2015). Characteristics of enterotoxin distribution, hemolysis, lecithinase, and starch hydrolysis of Bacillus cereus isolated from infant formulas and ready-to-eat foods. J. Dairy Sci. 98, 1652–1660. doi: 10.3168/jds.2014-9042
Ivy, R. A., Ranieri, M. L., Martin, N. H., Bakker, H. C., Xavier, B. M., Wiedmann, M., et al. (2012). Identification and characterization of psychrotolerant sporeformers associated with fluid milk production and processing. Appl. Environ. Microbiol. 78, 1853–1864. doi: 10.1128/AEM.06536-11
Jiménez, G., Urdiain, M., Cifuentes, A., López-López, A., Blanch, A. R., and Tamames, J. (2013). Description of Bacillus toyonensis sp. nov., a novel species of the Bacillus cereus group, and pairwise genome comparisons of the species of the group by means of ANI calculations. Syst. Appl. Microbiol. 36, 383–391. doi: 10.1016/j.syapm.2013.04.008
Kaan, T., Homuth, G., Mäder, U., Bandow, J., and Schweder, T. (2002). Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response. Microbiol. 148, 3441–3455. doi: 10.1099/00221287-148-11-3441
Kantas, D., Papatsiros, V., Tassis, P., Giavasis, I., Bouki, P., and Tzika, E. (2015). A feed additive containing Bacillus toyonensis (Toyocerin®) protects against enteric pathogens in postweaning piglets. J. Appl. Microbiol. 118, 727–738. doi: 10.1111/jam.12729
Koehler, T. M. (2009). Bacillus anthracis physiology and genetics. Mol. Aspects Med. 30, 386–396. doi: 10.1016/j.mam.2009.07.004
Kovac, J., Den Bakker, H., Carroll, L. M., and Wiedmann, M. (2017). Precision food safety: a systems approach to food safety facilitated by genomics tools. Trends Anal. Chem. 96, 52–61. doi: 10.1016/j.trac.2017.06.001
Kovac, J., Miller, R. A., Carroll, L. M., Kent, D. J., Jian, J., Beno, S. M., et al. (2016). Production of hemolysin BL by Bacillus cereus group isolates of dairy origin is associated with whole-genome phylogenetic clade. BMC Genomics 17:581. doi: 10.1186/s12864-016-2883-z
Lechner, S., Mayr, R., Francis, K. P., Pruss, B. M., Kaplan, T., Wiessner-Gunkel, E., et al. (1998). Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int. J. Syst. Bacteriol. 48(Pt 4), 1373–1382.
Ledl, F., and Schleicher, E. (1990). New aspects of the Maillard reaction in foods and in the human body. Angew. Chem. Int. Ed. Engl. 29, 565–594. doi: 10.1002/anie.199005653
Leisch, F. (2006). A toolbox for k-centroids cluster analysis. Comput. Stat. Data Anal. 51, 526–544. doi: 10.1016/j.csda.2005.10.006
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., and Homer, N. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, L., Stoeckert, C. J., and Roos, D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
Liu, Y., Du, J., Lai, Q., Zeng, R., Ye, D., Xu, J., et al. (2017a). Proposal of nine novel species of the Bacillus cereus group. Int. J. Syst. Evol. Microbiol. 67, 2499–2508. doi: 10.1099/ijsem.0.001821
Liu, Y., Lai, Q., and Shao, Z. (2017b). Genome analysis-based reclassification of Bacillus weihenstephanensis as a later heterotypic synonym of Bacillus mycoides. Int. J. Syst. Evol. Microbiol. 68, 106–112. doi: 10.1099/ijsem.0.002466
Mansilla, M. C., Cybulski, L. E., Albanesi, D., and De Mendoza, D. (2004). Control of membrane lipid fluidity by molecular thermosensors. J. Bacteriol. 186, 6681–6688. doi: 10.1128/JB.186.20.6681-6688.2004
Mayr, B., Kaplan, T., Lechner, S., and Scherer, S. (1996). Identification and purification of a family of dimeric major cold shock protein homologs from the psychrotrophic Bacillus cereus WSBC 10201. J. Bacteriol. 178, 2916–2925. doi: 10.1128/jb.178.10.2916-2925.1996
Mearls, E. B., Izquierdo, J. A., and Lynd, L. R. (2012). Formation and characterization of non-growth states in Clostridium thermocellum: spores and L-forms. BMC Microbiol. 12:180. doi: 10.1186/1471-2180-12-180
Miller, R. A., Beno, S. M., Kent, D. J., Carroll, L. M., Martin, N. H., Boor, K. J., et al. (2016). Bacillus wiedmannii sp. nov. is a new psychrotolerant and cytotoxic Bacillus cereus group species isolated from dairy foods and environments in the USA. Int. J. Syst. Evol. Microbiol. 66, 4744–4753. doi: 10.1099/ijsem.0.001421
Miller, R. A., Jian, J., Beno, S. M., Wiedmann, M., and Kovac, J. (2018). Genomic and phenotypic characterization of type strains and dairy-associated isolates in the Bacillus cereus group indicates considerable intra-clade variability in toxin production and cytotoxicity. Appl. Environ. Microbiol. 84:e2479-17. doi: 10.1128/AEM.02479-17
Nakamura, L., and Jackson, M. (1995). Clarification of the taxonomy of Bacillus mycoides. Int. J. Syst. Evol. Microbiol. 45, 46–49. doi: 10.1099/00207713-45-1-46
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., Mcveigh, R., et al. (2015). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44,D733–D745. doi: 10.1093/nar/gkv1189
Páčová, Z., Švec, P., Stenfors, L., Vyletělová, M., and Sedláček, I. (2003). Isolation of the psychrotolerant species Bacillus weihenstephanensis from raw cow’s milk. Czech J. Anim. Sci 48, 93–96.
R Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Saleh-Lakha, S., Leon-Velarde, C. G., Chen, S., Lee, S., Shannon, K., Fabri, M., et al. (2017). A study to assess the numbers and prevalence of Bacillus cereus and its toxins in pasteurized fluid milk. J. Food Protect. 80, 1085–1089. doi: 10.4315/0362-028X.JFP-16-521
Schindler, T., Graumann, P. L., Perl, D., Ma, S., Schmid, F. X., and Marahiel, M. A. (1999). The family of cold shock proteins of Bacillus subtilis stability and dynamics in vitro and in vivo. J. Biol. Chem. 274, 3407–3413. doi: 10.1074/jbc.274.6.3407
Soufiane, B., and Côté, J.-C. (2013). Bacillus weihenstephanensis characteristics are present in Bacillus cereus and Bacillus mycoides strains. FEMS Microbiol. Lett. 341, 127–137. doi: 10.1111/1574-6968.12106
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stenfors, L. P., and Granum, P. E. (2001). Psychrotolerant species from the Bacillus cereus group are not necessarily Bacillus weihenstephanensis. FEMS Microbiol. Lett 197, 223–228. doi: 10.1111/j.1574-6968.2001.tb10607.x
Switt, A. I. M., Andrus, A. D., Ranieri, M. L., Orsi, R. H., Ivy, R., Den Bakker, H. C., et al. (2014). Genomic comparison of sporeforming bacilli isolated from milk. BMC Genomics. 15:26. doi: 10.1186/1471-2164-15-26
Tatusova, T., Dicuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Trmčić, A., Martin, N., Boor, K., and Wiedmann, M. (2015). A standard bacterial isolate set for research on contemporary dairy spoilage. J. Dairy Sci. 98, 5806–5817. doi: 10.3168/jds.2015-9490
Warda, A. K., Siezen, R. J., Boekhorst, J., Wells-Bennik, M. H., De Jong, A., Kuipers, O. P., et al. (2016). Linking Bacillus cereus genotypes and carbohydrate utilization capacity. PLoS One 11:e0156796. doi: 10.1371/journal.pone.0156796
Keywords: Bacillus cereus, whole genome sequencing, psychrotolerant, spoilage, skim milk broth
Citation: Beno SM, Orsi RH, Cheng RA, Kent DJ, Kovac J, Duncan DR, Martin NH and Wiedmann M (2019) Genes Associated With Psychrotolerant Bacillus cereus Group Isolates. Front. Microbiol. 10:662. doi: 10.3389/fmicb.2019.00662
Received: 11 January 2019; Accepted: 15 March 2019;
Published: 29 March 2019.
Edited by:
Paula Teixeira, Universidade Católica Portuguesa, PortugalReviewed by:
Izabela Swiecicka, University of Białystok, PolandRamón Alberto Batista-García, Universidad Autónoma del Estado de Morelos, Mexico
Copyright © 2019 Beno, Orsi, Cheng, Kent, Kovac, Duncan, Martin and Wiedmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Wiedmann, bXcxNkBjb3JuZWxsLmVkdQ==
†Present address: Sarah M. Beno, Department of Cell Differential and Integrative Biology, The University of Alabama, Birmingham, AB, United States
 Sarah M. Beno
Sarah M. Beno Renato H. Orsi
Renato H. Orsi Rachel A. Cheng
Rachel A. Cheng David J. Kent
David J. Kent Jasna Kovac
Jasna Kovac Diana R. Duncan
Diana R. Duncan Nicole H. Martin
Nicole H. Martin Martin Wiedmann
Martin Wiedmann