- 1Department of Laboratory Medicine, Research Institute of Bacterial Resistance, Yonsei University College of Medicine, Seoul, South Korea
- 2Department of Laboratory Medicine, Chonnam National University Medical School, Gwangju, South Korea
- 3Department of Laboratory Medicine, Paik Institute for Clinical Research, Inje University College of Medicine, Busan, South Korea
The aim of this work was to assess a novel pseudo-staphylococcal cassette chromosome mec (ΨSCCmec) element in methicillin-resistant Staphylococcus aureus (MRSA) blood isolates. Community-associated MRSA E16SA093 and healthcare-associated MRSA F17SA003 isolates were recovered from the blood specimens of patients with S. aureus bacteremia in 2016 and in 2017, respectively. Antimicrobial susceptibility was determined via the disk diffusion method, and SCCmec typing was conducted by multiplex polymerase chain reaction. Whole genome sequencing was carried out by single molecule real-time long-read sequencing. Both isolates belonged to sequence type 72 and agr-type I, and they were negative for Panton-Valentine leukocidin and toxic shock syndrome toxin. The spa-types of E16SA093 and F17SA003 were t324 and t2460, respectively. They had a SCCmec IV-like element devoid of the cassette chromosome recombinase (ccr) gene complex, designated as ΨSCCmecE16SA093. The element was manufactured from SCCmec type IV and the deletion of the ccr gene complex and a 7.0- and 31.9-kb portion of each chromosome. The deficiency of the ccr gene complex in the SCCmec unit is likely resulting in mobility loss, which would be an adaptive evolutionary mechanism. The dissemination of this clone should be monitored closely.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) clinical isolates were first identified in the early 1960s, immediately after the introduction of penicillinase-stable penicillins in the clinical setting (Jevons et al., 1963). Now, the spread of MRSA strains represents a global concern with a recognizable healthcare burden. The mecA and mecC genes encoding penicillin-binding protein (PBP) 2a of low beta-lactam binding affinity confer beta-lactam resistance to the bacterial host by composing a mobile genetic element, namely, the staphylococcal cassette chromosome mec (SCCmec) (Ito et al., 1999).
The SCCmec element harbors two fundamental components: a mec gene complex and a cassette chromosome recombinase (ccr) gene complex. A unique combination of the mec gene complex class and the ccr gene complex allotype determines the type of the SCCmec element, and its variation within the joining- (J-) regions determine the subtypes of each SCCmec type. To date, 13 SCCmec types have been deposited together with numerous subtypes (International Working Group on the Classification of Staphylococcal Cassette Chromosome, 2009; Baig et al., 2018). The mec gene complex includes the mecA or mecC gene, along with the regulatory mecR1 and mecI genes. The ccr gene complex comprises one or more ccr genes (Deurenberg and Stobberingh, 2008) accounting for the integration/excision of the SCCmec element into and out of the orfX gene in the staphylococcal chromosome (Katayama et al., 2000). The SCCmec-like elements devoid of the mecA gene are denominated as an SCC as long as they share the following characteristics with SCCmec: carriage of the ccr gene(s), integration at integrated site sequences in the chromosome, and the presence of flanking direct repeat sequences. And those without the ccr genes are termed as the pseudo-(Ψ) SCCmec element.
Through a nationwide antimicrobial resistance surveillance in South Korea (Lee et al., 2018), two mecA-positive MRSA blood isolates were identified as those carrying a non-typeable SCCmec element. To assess the non-typeable SCCmec, both genomes were entirely sequenced, and a novel ΨSCCmec E16SA093 element was identified.
Materials and Methods
Bacterial Isolates
A total of 586 S. aureus blood isolates collected between May 2016 and April 2017 from six general hospitals in South Korea (Lee et al., 2018) were screened. Among the 319 cefoxitin-resistant isolates, E16SA093 and F17SA003, whose SCCmec elements could not be typified, were selected for further study.
Antimicrobial Susceptibility Testing and the Determination of SCCmec Types
Antimicrobial susceptibility to antimicrobials used for staphylococci infection was determined by disk diffusion tests on Mueller-Hinton agar (Difco Laboratories, Detroit, MI, USA), following the CLSI guidelines (CLSI, 2018). S. aureus ATCC 25923 was simultaneously tested in each batch for quality control. MRSA isolates were subjected to polymerase chain reaction (PCR) for mecA gene and SCCmec typing, as previously described (Oliveira and de Lencastre, 2002).
Multilocus Sequence Typing, agr Typing, and spa Typing
Multilocus sequence typing (MLST) was carried out by PCR and sequencing of the seven housekeeping genes, arcC, aroE, glpF, gmk, pta, tpi, and yqiL. Allelic numbers and sequence types (STs) were determined by comparing the obtained sequences to the database for S. aureus1. The agr type was determined by multiplex PCR (Gilot et al., 2002), and spa typing was conducted by comparing the PCR-amplified nucleotide sequence of the variable repeat region of the spa gene against the Ridom SpaServer2.
Whole Genome Sequencing
Bacterial whole genomes were sequenced with single-molecule real-time (SMRT) sequencing on a PacBio RSII instrument (Pacific Biosciences, Menlo Park, CA, USA) using genomic DNA from the S. aureus isolates extracted by a Wizard Genomic DNA Purification kit (Promega, Madison, WI, USA). SMRTbell template libraries were prepared, and adapter ligation was performed. Acquired sequencing data were de novo assembled by PacBio SMRT, read with the PacBio SMRT analysis software suite (version 2.3.0). Coding sequences (CDS), including tRNA and rRNA, were annotated using the NCBI Prokaryotic Genome Annotation Pipeline3. Nucleic acid sequences were compared using Basic Local Alignment Search Tool,4 and resistance and virulence determinants were searched for using ResFinder5 and VirulenceFinder6, respectively. Prophages were searched for using the PHAge Search Tool Enhanced Release7. For any putative ccr gene, the site-specific serine recombinase motif (Wang and Archer, 2010) and a modified motif by the consensus pattern (Perreten et al., 2013) were searched for against the coding sequences of both genomes.
Nucleotide Accession Numbers
The nucleotide sequences of the entire genomes of S. aureus E16SA093 and F17SA003 were deposited in GenBank under accession numbers CP031130 and CP031131 for F17SA003 and E16SA093, respectively.
Results and Discussion
Epidemiological Features of MRSA ST72
Following the one-year collection of the 586 S. aureus blood isolates, a total of 319 isolates (54.4%) were MRSA conferred by the mecA gene. A total of 176 (30.0%) isolates belonged to ST72; 112 of those isolates (63.6%) were MRSA, 65 were healthcare-associated (HA) MRSA, and 47 were community-associated MRSA (CA-MRSA). All but three MRSA ST72 isolates (97.3%, 109/112) carried SCCmec type IV, while one possessed SCCmec type II and the remaining two isolates (E16SA093 and F17SA003) had non-typeable elements.
The MRSA ST72 harboring SCCmec IV (ST72-MRSA-IV) was one of the top three CA-MRSA clones in the USA until 2002 as a pulse-field type USA700; however, the clone was suddenly eliminated from the country in 2004 (Tenover et al., 2008). In South Korea, the ST72-MRSA-IV acceded a major CA-MRSA clone by 2005 (Kim et al., 2007), and the ST72-MRSA-IV subsequently grew to be a major HA-MRSA clone. This finding supports the idea that the ST72-MRSA-IV clone was disseminating from communities to hospitals (Song et al., 2011).
Two mecA-Positive MRSA ST72 Blood Isolates Carrying a Non-typeable SCCmec Element
The CA-MRSA E16SA093 was recovered in September 2016 from an 86-year-old female patient with acute infective endocarditis and infective spondylopathy. The patient was transferred from an acute care hospital to a general hospital located in Gwangju city, and blood cultures were carried straightaway. The bacteremia originated from a bone infection, and empirical treatment was started with cefazoline. Definitive treatment was followed with teicoplanin within 72 h after the initial blood culture, and the patient was cured. The HA-MRSA 17SA003 was recovered in January 2017 from a 63-year-old male patient with diabetes mellitus and stage 4 chronic kidney disease hospitalized in a general hospital in Busan city. An initial blood culture was performed on the 13th day of hospitalization, and the origin of S. aureus bacteremia was unidentified. Empirical treatment with cefazoline was replaced to vancomycin within 72 h, and the patient was cured.
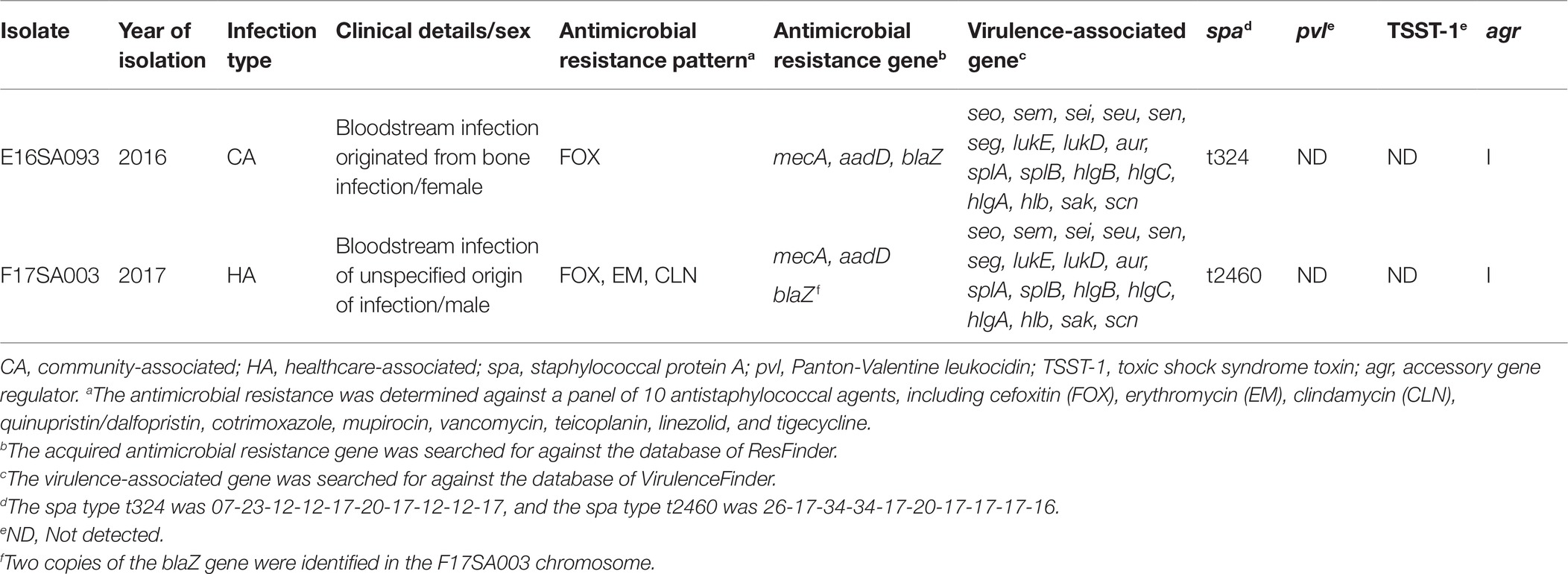
Both MRSA isolates belonged to ST72 and agr-type I. They were negative for both Panton-Valentine leukocidin and toxic shock syndrome toxin (Table 1). The spa-types of E16SA093 and F17SA003 were t324 and t2460, respectively. Among the 10 antimicrobials tested, the E16SA093 isolate was resistant only to cefoxitin, while F17SA003 was resistant not only to cefoxitin but also to erythromycin and clindamycin.
Genome Sequencing and Identification of the Novel ΨSCCmec E16SA093
The de novo assembly of the genome resulted in a 2,767,631,390-bp circularized chromosome, including 2,564 assigned CDSs, 60 tRNAs, and 19 rRNAs for E16SA093, and a 2,849,947,596-bp circularized chromosome, including 2,546 CDSs, 60 tRNAs, and 19 rRNAs for F17SA003. The overall GC contents were 32.9% for both. No plasmid was identified. Acquired genetic elements by both chromosomes were alike, including two intact Staphylococcus prophages (44.1-kb ϕNM3 and 41.2-kb Sap26), 17 virulence factors, and three antimicrobial resistance genes, with an extra copy of blaZ for F17SA003. No known heavy metal resistance genes were identified for either.
For the SCCmec element, a class B mec gene complex lacking the ΨIS1272 upstream from the mecA gene was identified, and neither the ccr gene nor any putative site-specific serine recombinase gene was identified elsewhere in the chromosome (Figure 1). The ΨSCCmec, designated as ΨSCCmecE16SA093, resembled a SCCmec type IV, which is common in MRSA ST72. When compared with the genome of HL1 that is a CA ST72-MRSA-IV recovered in South Korea before 2010 (Chen et al., 2013), a 12.6-kb region was deleted from the SCCmec type IV element, and 7.0- and 31.9-kb chromosomal DNA region was deleted in the E16SA093 and F17SA003, respectively. The Ccr recombinase involves the site-specific integration/excision of SCCmec elements (International Working Group on the Classification of Staphylococcal Cassette Chromosome, 2009), and the CcrA2/CcrB2 in the SCCmec IV is targeting the attB site at the orfX gene (Wang and Archer, 2010). The ΨSCCmecE16SA093 was indeed integrated exactly at attB, suggesting the subsequent elimination of the ccrA2/ccrB2 genes from the SCCmec IV element after the integration event. As the ΨIS1272 was absent, IS-associated recombination was suspected. However, no palindromic sequences were observed in either end of the deleted 19.7- and 44.5-kb DNA fragments targetable by IS1272, with an insertion sequence involved in the structure-dependent transposition or stem-loop replacement (Wan et al., 2017).
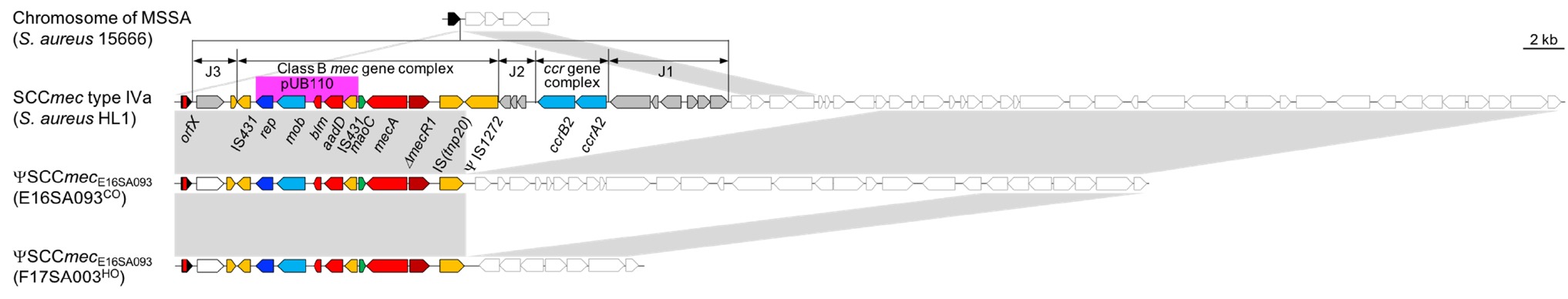
Figure 1. Genomic structures and environments of the ΨSCCmec E16SA093 element in comparison with the SCCmec type IVa in MRSA HL1 (Chen et al., 2013) (GenBank accession, CP003979) and orfX in MSSA 15666 (GenBank accession, EU272085). The conserved region in the gray box showed a resemblance of >99% nt identity. Filled arrows represented the components consisting of the SCCmec element. The open arrows indicated the presence of chromosomal genes in S. aureus. Arrows in black, orfX; red, resistance genes; yellow, transposase; blue, replication origin; sky blue, recombinase; gray, hypothetical coding sequence.
Epidemiology of ΨSCCmec
The fitness benefit of the ccr-gene-loss from SCCmec, resulting in an inherent mecA in the chromosome, has never been assessed, while spontaneous mecA-gene-loss in the absence of selective pressure, driven by the huge biological cost of gene expression, has been demonstrated (Noto et al., 2008). The furnished mecA gene could provide advantages to MRSA in the beta-lactam-abundant habitat, such as the clinical settings, suggesting a course of adaptive evolution for MRSA. While ΨSCCmec is occasionally identified in MRSA (Chen et al., 2010), methicillin-resistant coagulase-negative staphylococci (MRCNS) carrying the element is much more common (Perreten et al., 2013; Shore and Coleman, 2013). The speculation of MRCNS to be a reservoir of SCCmec (Archer et al., 1994), in the MRSA is inspiring.
In this study, we evaluated MRSA ST72 isolates carrying ΨSCCmecE16SA093, which was likely being fabricated from the SCCmec type IV. Excised portions of the chromosomes differed in size, and the event likely occurred independently, indicating that the clonal dissemination of ST72-MRSA-ΨSCCmecE16SA093 has not yet been occurred. The immobile mecA gene could make the MRSA fit the antimicrobial-abundant habitat, even though the mecA gene expression is known to be costly. Further study of the molecular mechanisms driving ccr gene loss is needed, and dissemination of the clone should be surveilled.
Ethics Statement
The research, which has no involvement of human subject but the clinical isolates, does meet the exempt category without approval from Ethics Committee on Human Research of the Health Ministry in South Korea and the study design has not been reviewed by the committee.
Author Contributions
HL and SJ conceived the study. E-JY, HL, and SJ analyzed the data. E-JY and SJ wrote the manuscript. DK, JoS, and JeS collected the strains.
Funding
This research was supported by the Research Program funded by the Korea Centers for Disease Control and Prevention (2017E4400100#).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mina Lee for her technical support with the microbiological experiments.
Footnotes
2. http://www.spaserver.ridom.de
3. http://www.ncbi.nlm.nih.gov/books/NBK174280
4. http://blast.ncbi.nlm.nih.gov
5. https://cge.cbs.dtu.dk/services/ResFinder
References
Archer, G. L., Niemeyer, D. M., Thanassi, J. A., and Pucci, M. J. (1994). Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob. Agents Chemother. 38, 447–454. doi: 10.1128/AAC.38.3.447
Baig, S., Johannesen, T. B., Overballe-Petersen, S., Larsen, J., Larsen, A. R., and Stegger, M. (2018). Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 61, 74–76. doi: 10.1016/j.meegid.2018.03.013
Chen, L., Mediavilla, J. R., Smyth, D. S., Chavda, K. D., Ionescu, R., Roberts, R. B., et al. (2010). Identification of a novel transposon (Tn6072) and a truncated staphylococcal cassette chromosome mec element in methicillin-resistant Staphylococcus aureus ST239. Antimicrob. Agents Chemother. 54, 3347–3354. doi: 10.1128/AAC.00001-10
Chen, Y., Chatterjee, S. S., Porcella, S. F., Yu, Y. S., and Otto, M. (2013). Complete genome sequence of a Panton-Valentine leukocidin-negative community-associated methicillin-resistant Staphylococcus aureus strain of sequence type 72 from Korea. PLoS One 8:e72803. doi: 10.1371/journal.pone.0084522
CLSI (2018). Performance standards for antimicrobial susceptibility testing; twenty-eighth informational supplement. (Wayne, PA, USA: Clinical and Laboratory Standards Institute).
Deurenberg, R. H., and Stobberingh, E. E. (2008). The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8, 747–763. doi: 10.1016/j.meegid.2008.07.007
Gilot, P., Lina, G., Cochard, T., and Poutrel, B. (2002). Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 40, 4060–4067. doi: 10.1128/JCM.40.11.4060-4067.2002
International Working Group on the Classification of Staphylococcal Cassette Chromosome, E (2009). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53, 4961–4967. doi: 10.1128/AAC.00579-09
Ito, T., Katayama, Y., and Hiramatsu, K. (1999). Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43, 1449–1458. doi: 10.1128/AAC.43.6.1449
Jevons, M. P., Coe, A. W., and Parker, M. T. (1963). Methicillin resistance in staphylococci. Lancet 1, 904–907. doi: 10.1016/S0140-6736(63)91687-8
Katayama, Y., Ito, T., and Hiramatsu, K. (2000). A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44, 1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000
Kim, E. S., Song, J. S., Lee, H. J., Choe, P. G., Park, K. H., Cho, J. H., et al. (2007). A survey of community-associated methicillin-resistant Staphylococcus aureus in Korea. J. Antimicrob. Chemother. 60, 1108–1114. doi: 10.1093/jac/dkm309
Lee, H., Yoon, E. J., Kim, D., Jeong, S. H., Won, E. J., Shin, J. H., et al. (2018). Antimicrobial resistance of major clinical pathogens in South Korea, May 2016 to April 2017: first one-year report from Kor-GLASS. Euro Surveill. doi: 10.2807/1560-7917.ES.2018.23.42.1700734
Noto, M. J., Fox, P. M., and Archer, G. L. (2008). Spontaneous deletion of the methicillin resistance determinant, mecA, partially compensates for the fitness cost associated with high-level vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 52, 1221–1229. doi: 10.1128/AAC.01164-07
Oliveira, D. C., and de Lencastre, H. (2002). Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46, 2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002
Perreten, V., Chanchaithong, P., Prapasarakul, N., Rossano, A., Blum, S. E., Elad, D., et al. (2013). Novel pseudo-staphylococcal cassette chromosome mec element (psiSCCmec57395) in methicillin-resistant Staphylococcus pseudintermedius CC45. Antimicrob. Agents Chemother. 57, 5509–5515. doi: 10.1128/AAC.00738-13
Shore, A. C., and Coleman, D. C. (2013). Staphylococcal cassette chromosome mec: recent advances and new insights. Int. J. Med. Microbiol. 303, 350–359. doi: 10.1016/j.ijmm.2013.02.002
Song, J. H., Hsueh, P. R., Chung, D. R., Ko, K. S., Kang, C. I., Peck, K. R., et al. (2011). Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J. Antimicrob. Chemother. 66, 1061–1069. doi: 10.1093/jac/dkr024
Tenover, F. C., Mcallister, S., Fosheim, G., Mcdougal, L. K., Carey, R. B., Limbago, B., et al. (2008). Characterization of Staphylococcus aureus isolates from nasal cultures collected from individuals in the United States in 2001 to 2004. J. Clin. Microbiol. 46, 2837–2841. doi: 10.1128/JCM.00480-08
Wan, T. W., Higuchi, W., Khokhlova, O. E., Hung, W. C., Iwao, Y., and Wakayama, M., et al. (2017). Genomic comparison between Staphylococcus aureus GN strains clinically isolated from a familial infection case: IS1272 transposition through a novel inverted repeat-replacing mechanism. PLoS One 12:e0187288. doi: 10.1371/journal.pone.0187288
Keywords: methicillin-resistant Staphylococcus aureus, sequence type 72, pseudo-SCCmec, ccr gene, blood isolates
Citation: Yoon E-J, Lee H, Kim D, Shin JH, Shin JH and Jeong SH (2019) Methicillin-Resistant Staphylococcus aureus Blood Isolates Harboring a Novel Pseudo-staphylococcal Cassette Chromosome mec Element. Front. Microbiol. 10:540. doi: 10.3389/fmicb.2019.00540
Edited by:
Patrícia Poeta, University of Trás-os-Montes and Alto Douro, PortugalReviewed by:
Kiiyukia Matthews Ciira, Mount Kenya University, KenyaNobumichi Kobayashi, Sapporo Medical University, Japan
Noriko Urushibara, Sapporo Medical University, Japan
Copyright © 2019 Yoon, Lee, Kim, Shin, Shin and Jeong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seok Hoon Jeong, a3NjcGpzaEB5dWhzLmFj
†These authors have contributed equally to this work
 Eun-Jeong Yoon1†
Eun-Jeong Yoon1† Dokyun Kim
Dokyun Kim Jong Hee Shin
Jong Hee Shin Jeong Hwan Shin
Jeong Hwan Shin Seok Hoon Jeong
Seok Hoon Jeong