- 1Institute of Quality Standard and Testing Technology for Agro-Products, Hubei Academy of Agricultural Sciences, Wuhan, China
- 2Key Laboratory of Environment Correlative Dietology, College of Food Science and Technology, Huazhong Agricultural University, Wuhan, China
Ypts (yeast protein transports),also called as ras-associated binding GTPases (Rab), are the largest group of the small GTPases family, which have been extensively studied in model eukaryotic cells and play a pivotal role in membane trafficking, while this study showed potential regulation role of Ypts in fungi. One of Ypts, Ypt7 may be involved in fungal development and secondary metabolism, but the exact mechanism still exists a controversy. In current study, the functions of a Monascus ypt7 homologous gene (mrypt7) from Monascus ruber M7 was investigated by combination of gene-deletion (Δmrypt7), overexpression (M7::PtrpC-mrypt7) and transcriptome analysis. Results showed that the radial growth rate of Δmrypt7 was significantly slower than M. ruber M7, little conidia and ascospores can be observed in Δmrypt7, but the yield of intracellular secondary metabolites was dramatically increased. Simultaneously, the mrypt7 overexpression strain possessed similar capacity for sporulation and secondary metabolism observed in M. ruber M7. Transcriptome results further illustrated that mrypt7 could coordinate with numerous genes involved in the vegetative growth, conidiogenesis, secondary metabolism biosynthesis and transportation of M. ruber M7. Combined with the similar effect of Ypt7 homologs on other fungi, we propose that Ypt7 works more like a global regulatory factor in fungi. To our knowledge, it is the first time to investigate Ypt7 functions in Monascus. It could also improve the understanding of Ypt7 functions in fungi.
Introduction
The largest subfamily of rat sarcoma (Ras) superfamily, ras-associated binding GTPases (Rab), also called as Ypt (yeast protein transport) and Sec (secretion) (Gallwitz et al., 1983; Salminen and Novick, 1987), are involved in the membrane trafficking regulation in all eukaryotes (Maringer et al., 2016; Shinde and Maddika, 2016; Yun et al., 2016; Pfeffer, 2017). As key regulators of membrane transportation, Rab GTPases cycle between GTP-bound (active) and GDP-bound (inactive) conformations which stimulated by guanine nucleotide exchange factors (GEFs). Typical Rab GTPase possess several conserved functional regions, including phosphate/Mg2+ binding domain(PM), GTP/GDP binding domain (G), C-terminal isoprenylation region (C), and so on. For example, the G domain provides phosphate contacts and supplies a Ser/Thr site which is co-ordinated by the Mg2+ ion. The conserved molecular switch mechanism have detailed in some reviews (Lee et al., 2009; Pylypenko et al., 2018). The first Ypt was discovered in Saccharomyces cerevisiae (Schmitt et al., 1986; Pereira-Leal, 2008; Li and Marlin, 2015), and the succedent research results have showed that there are total 11 encoded Ypt proteins in S. cerevisiae, and each of which possesses distinctive function at a particular stage of the membrane transport pathway (Pereira-Leal, 2008; Li and Marlin, 2015). In animal, dozens of Rab/Ypt are proven to regulate vesicle trafficking among organelles (Ohbayashi and Fukuda, 2012; Li and Marlin, 2015; Mignogna and D'Adamo, 2017; Pfeffer, 2017). In plant, Ypts also are required for intracellular trafficking from the trans-Golgi-network to the plasma membrane and/or prevacuolar compartments (Yun et al., 2016; Tripathy et al., 2017). The more detailed Ypts roles for vesicle transports in animal and plant are summarized in the previous reviews (Stenmark, 2009; Ao et al., 2014).
In fungi, the number of Ypt family is stable from 7 to 12 Ypts, each of which may be responsible for a particular stage of the membrane transport pathway (Pereira-Leal, 2008; Li and Marlin, 2015). Among them, Ypt7 is proved as a key regulator of the material movement and transformation among cellular compartments through vacuolar biogenesis and fusion (Ohsumi et al., 2002; Kashiwazaki et al., 2009; Balderhaar et al., 2010; Wickner, 2010), and the Ypt7-mediated vacuolar fission and fusion are proved to be essential for maintaining stabilities of the cytosolic pH and osmolarity, and storing and transferring intermediary metabolites like mammalian lysosomes and plant vacuoles (Richards et al., 2010; de Marcos Lousa and Denecke, 2016; BasuRay et al., 2018), while some investigations have also showed that Ypt7 can influence fungal development and secondary metabolism. For example, the ypt7 gene deletion or overexpression can lead to the variances of conidiogenesis and metabolism in fungi (Chanda et al., 2009a; Xu et al., 2012; Li et al., 2015; Liu et al., 2015; Zheng et al., 2015). However, it is still unclear how Ypt7 regulates fungal development and secondary metabolism, and the relationship among Ypt7-mediated vacuolar changes and fungal development and secondary metabolism.
Monascus spp., as one of the important edible filamentous fungi, can produce many beneficial secondary metabolites (SMs) including Monascus pigments (Mps), monacolin K (MK), γ-aminobutyric acid and so on (Patakova, 2013; Wu et al., 2013). As such, its fermented products, red yeast rice, also named as Hongqu in China have been used as food additives for more than 2,000 years (Chen et al., 2015). What's more, Hongqu has been permitted to use as a food supplement in USA from 1900s due to its cholesterol-lowering effects (Heber et al., 1999). The European Food Safety Authority (EFSA) also published a scientific opinion related to the daily dose of Hongqu containing MK (ESFA, 2011). Although citrinin (CIT), a nephrotoxic toxin produced by some Monascus strains ever hampered Hongqu use, nowadays the control and elimination of CIT in Hongqu have successfully been solved by the strain screenings or molecular biological techniques (Shimizu et al., 2005; He and Cox, 2016).
There were 7 ypt homologous genes (ypt1-ypt7), which functions are predicted (Table S1), have been discovered in the genome of Monascus ruber M7. In current paper, the functions of Monascus ypt7 (mrypt7) gene were investigated by combination of gene disruption, overexpression and transcriptome analysis. The results have revealed that besides the membrane trafficking regulation like other fungi, mrypt7 can also coordinate with numerous genes involved in the development and metabolism of M. ruber M7. Combined with Ypt7 functions in other fungi (Bouchez et al., 2015; Liu et al., 2015; Yang et al., 2018), we discuss and propose that Ypt7 works more like a global regulatory factor in fungi. To our knowledge, it is the first time to investigate Ypt7 functions in Monascus which could help us to improve the understanding of Ypt7 functions in fungi.
Materials and methods
Fungal Strains, Culture Media, and Growth Conditions
M. ruber M7 (CCAM 070120, Culture Collection of State Key Laboratory of Agricultural Microbiology, Wuhan, China), which can produce Mps and CIT, but no MK (Chen and Hu, 2005; Chen et al., 2017), was used to generate the mrypt7 deletion strain (Δmrypt7) and overexpression strain (M7::PtrpC-mrypt7). The potato dextrose agar medium (PDA), Czapek yeast extract agar medium (CYA), glycerol nitrate agar medium (25%) (G25N) and malt extract agar medium (MA) were utilized to observe the strains phenotypic characterization (He et al., 2013). Neomycin-resistant transformants were selected on PDA media containing 15 μg/mL G418 (Sigma-Aldrich, Shanghai, China). All strains were maintained on PDA slant at 28°C.
Cloning and Sequence Analysis of mrypt7 in M. ruber M7
Ypt family genes in M. ruber M7 genome were blast from NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Table S1). Amino acid sequence encoded by mrypt7 was predicted using the SoftBerry's FGENESH program (http://linux1.softberry.com/). mrypt7 homology was compared with 283 fungi Ypt7 downloaded from NCBI to analyze their primary structural features (http://weblogo.threeplusone.com/create.cgi).
Construction and Verification of mrypt7 Gene Deletion and Overexpression Strains
The construction and verification of mrypt7 gene deletion and overexpression strains were implemented according to the literature references (Shao et al., 2009; Liu et al., 2014). Briefly, the mrypt7 gene deletion cassette (5′UTR-G418-3′UTR) and mrypt7 gene overexpression cassette (5′UTR-G418-PtrpC-mrypt7-3′UTR) were constructed by double-joint PCR with the primers listed in Table S2 (Yu et al., 2004), and shown schematically in Figure S1. The mrypt7 gene deletion and overexpression vectors were formed, and transformed to M. ruber M7 using Agrobacterium tumefaciens-mediated transformation system to generate the mrypt7 gene deletion mutants (Δmrypt7) and overexpression transformants (M7::PtrpC-mrypt7), respectively. PCR and southern blot were used to verify the mrypt7 gene deletion and overexpression strains.
Phenotypic Characterization
M. ruber M7, Δmrypt7 and M7::PtrpC-mrypt7 were cultivated on PDA, CYA, MA and G25N for 10 d at 28°C to observe phenotypic characterization. Besides, the three above-mentioned strains were incubated on PDA for 3 d at 28°C for vacuole morphological observation. For a better distinction, the normal vauoles were designated vacuoles (Va), while smaller vauoles were designated fragment vauoles (Fv) (Chanda et al., 2009b).
Detection of Mps and CIT
One milliliter freshly harvested spores (105 cfu/mL) of each strain were inoculated on PDA plate covered with cellophane membranes, and incubated at 28°C for 11 days, the samples were taken every 2 days from the 3rd day to the 11th day of culture to measure the Mps and CIT production. Freeze-dried mycelia and medium powder (0.1 g) was suspended in 1 mL 80 % (v/v) methanol solution, and subjected to 30 min ultrasonication treatment (KQ-250B, Kunshan, China).
The Mps and CIT were separated by an ACQUITY UPLC BEH C18 column (2.1 mm × 100 mm, 1.7 μm), and detected on Waters ACQUITY UPLC I-class system (Waters, Milford, MA, USA). A gradient elution was performed with the mobile phase including solvent A (0.1% formic acid in water) and solvent B (acetonitrile) with a flow rate of 0.3 mL/min and an injection volume of 2 μL. The gradient elution was performed as follows: 60% (v/v) solvent A with 40% (v/v) solvent B maintained for 0.5 min firstly, the content of solvent A was decreased from 60 to 20% for 21 min, and then from 20 to 60% for 0.5 min. Finally, the column was equilibrated with 60% solvent A for 3 min. The temperature of chromatographic column and samples were maintained at 40°C and 4°C, respectively.
RNA Extraction, Library Preparation and Sequencing
Since M. ruber M7 and M7::PtrpC-mrypt7 shared similar phenotype and SMs yield (Figures 2, 3), the mrypt7 functions were further investigated only between M. ruber M7 and Δmrypt7 by transcriptome analysis. Detailly, 1 mL freshly harvested spores (105 cfu/mL) of M. ruber M7 and Δmrypt7 were inoculated on PDA plate covered with cellophane membranes, and incubated at 28°C. Besides, based on our previous results, Monascus ruber M7 starts conidiation at 3rd day on PDA medium, and the secondary metabolited yield reached a relatively high level in 7th day, so the mycelium after cultured 3 days and 7 days were collected and used for the total RNA extraction by TRIzol Reagent (Invitrogen, Life Technologies, USA), two biological replicates were designed for each condition(Muraguchi et al., 2015; Srikumar et al., 2015; Heuston et al., 2018). The RNA purity and integrity were analyzed by Nanodrop NanoPhotometer spectrophotometer (NanoDrop products IMPLEN, CA, USA) and Agilent 2100 BioAnalyzer (Agilent, USA).
For each sample, the cDNA library was constructed using RNA Library Prep Kit for Illumina (NEB, USA). The obtained PCR products were purified by AMPure XP system and library quality was assessed on the Agilent Bioanalyzer 2100 system. The eight samples (M7-3d vs. M7-7d, Δmrypt7-3d vs. Δmrypt7-7d, M7-3d vs. Δmrypt7-3d and M7-7d vs. Δmrypt7-7d, with two repeats in each group) were sequenced using the BGIseq-500RS platform (BGI, Wuhan, China, http://www.mgitech.cn/product/detail/BGISEQ-500.html).
Sequence Quality Evaluation and Validation
The obtained sequence raw reads of above-mentioned 8 samples were saved as FASTQ files, then the clean data were obtained after removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. The expression levels of 10 randomly selected genes in M. ruber M7 were validated by qRT-PCR following the protocol of the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Japan) and the SYBR® Select Master Mix (ABI, USA).
Functional Analysis of Transcriptome Data
The M. ruber M7 genome which contains 8,407 genes was used as a reference genome (Chen, 2015) to calculate the blast rate of genome and clean data by Hierarchical Indexing for Spliced Alignment of Transcripts (HISAT) and Bowtie2 (Langmead and Salzberg, 2012; Kim et al., 2015).
Gene expression levels were estimated by RNA-Seq by Expectation-Maximization (RSEM), the normalized value of f ragments per kilobase of transcript per million mapped reads (FPKM) was used as a parameter to compare the expression levels between M. ruber M7 and Δmrypt7(Li and Dewey, 2011; Van Verk et al., 2013). The orthologs with significantly different expression were identified by NOISeq method with an absolute value of log2−fold change >1 and probability >0.8 (Tarazona et al., 2012).
Gene ontology (GO) (http://www.geneontology.org/) and KEGG pathway (http://www.kegg.jp/) function analysis were implemented to investigate the functions of the differentially expressed genes (DEGs) between M. ruber M7 and Δmrypt7. Moreover, the DEGs involved in fungal growth, sporation and secondary metabolism were further analyzed to illuminate the Mrypt7 role in fungal development and secondary metabolism.
Results
Sequence Analysis and Characterization of mrypt7 in M. ruber M7
The Ypt family genes in M. ruber M7 genome were blasted from NCBI, totally 7 Ypts showed highly homologous with other fungi (Table S1). Among them, Ypt7 homology was further analyzed in this study. Detailly, a 954 bp fragment containing the putative mrypt7 homolog was successfully amplified from the genomic DNA of M. ruber M7. A database searched with softberry (http://linux1.softberry.com/berry.phtml) has been showed that the CDS (coding sequence) length of mrypt7 gene is 591 bp which encodes 196-amino acids and consists of 5 exons (Figure S2). The characteristic motifs or residues of Ypt7 from M7 and other 283 fungi downloaded from NCBI were investigated, and the results illustrated that phosphate/Mg2+ binding domain(PM), GTP/GDP binding domains (G) and C-terminal isoprenylation region (C) are highly conserved in all tested fungi (Figure S2). Besides, a database searched with NCBI-BLAST has been demonstrated that the deduced 196-amino acid sequences encoded by mrypt7 share 91% similarity with the GTP-binding protein Ypt7 of Aspergillus fischeri (Genbank: XP_001259484.1), A. oryzae (Genbank: XP_001824054.1), A. niger (Genban: XP_001398680.2), P. oxalicum (Genbank: EPS32522.1), and P. zonata (Genbank: XP_022585464.1) (Table S1).
Verification of the mrypt7 Deletion and Overexpression Strains
Total 9 putative disruptants (Δmrypt7) were obtained and analyzed, and one of them was displayed here. In PCR analysis as shown in Figure 1A, no DNA band was amplified when the genome of the putative Δmrypt7 strain was used as template with the primer pair Y7-up1/Y7-do1 (Table S1), while a 0.7 kb product appeared using the genome of the wild-type strain M. ruber M7. A 1.2-kb fragment of G418 gene could be amplified from Δmrypt7 using primers G418up/G418do (Table S1), while nothing could be obtained from M. ruber M7. Meanwhile, amplicons of M. ruber M7 (2.3 kb) and Δmrypt7 (2.4 kb) different in size were observed when primers Y7-Zup1/Y7-Ydo1 (Table S1) were used.
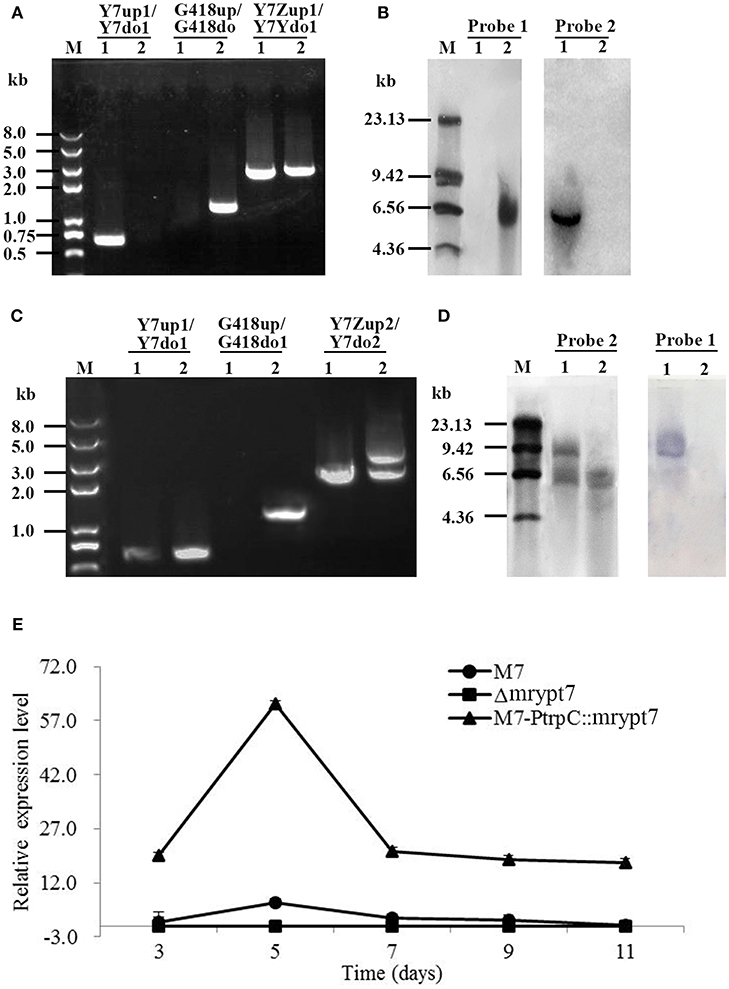
Figure 1. The verification of mrypt7 deletion and overexpression mutants. (A) PCR verification of the mrypt7 deletion mutant; (B) Southern blot verification of the mrypt7 deletion mutant; (C) PCR verification of the mrypt7 overexpression mutant; (D) Southern blot verification of the mrypt7 overexpression mutant; (E) qRT-PCR analysis of mrypt7 gene expression level.
The putative Δmrypt7 was further verified by Southern blot. As showed in Figure 1B, a probe corresponding to the mrypt7 coding region (probe 1,Table S2) yielded a single hybridizing band in a Southern blot of HindIII-digested genomic DNA of the wild-type strain, compared with no band in Δmrypt7, which demonstrated that M. ruber M7 carried a single copy of mrypt7. Meanwhile, no band was detected in the wild-type strain, while a single band occurred in Δmrypt7 using probe 2 (Table S2) which corresponds to the G418 gene. These results proved that Δmrypt7 carried a single integrated copy of the mrypt7 disruption cassette.
Total 16 putative M7::PtrpC-mrypt7 strains with G418 resistance were obtained and analyzed, and one of them was showed as follows. In PCR analysis as shown in Figure 1C, a 1.2-kb product appeared when the genome of the putative M7::PtrpC-mrypt7 strain was used as template with primers G418up/G418do (Table S2), while no DNA band was amplified using the genome of M. ruber M7. Amplicons of M. ruber M7 (3.0-kb) and M7::PtrpC-mrypt7 (4.5 kb and 3.0 kb) were totally different in size when primers Y7-up1/Y7-do1 (Table S2) was used, which proved that there were two copies of the mrypt7 overexpression cassette integrated in M7::PtrpC-mrypt7.
Southern blot analysis (Figure 1D) showed that probe 1 (Table S2) yielded two bands in M7::PtrpC-mrypt7 and a single band in M. ruber M7, while probe 2 (Table S2) generated a single band in M7:: PtrpC-mrypt7 and no band in M. ruber M7, which demonstrated that M7::PtrpC-mrypt7 carried two integrated copies of the mrypt7 and was a successful homologous recombination event.
qRT-PCR was implemented to analyze the transcription levels of the mrypt7 gene in M. ruber M7, Δmrypt7 and M7::PtrpC-mrypt7. As shown in Figure 1E, Δmrypt7 was deficient in the expression of the mrypt7 gene, the average level of mrypt7 expression in M7::PtrpC-mrypt7 was five times higher than that of M. ruber M7. These results further verified the success of gene knockout and overexpression in the putative Δmrypt7 and M7::PtrpC-mrypt7 strains.
Phenotypic Characterization of Δmrypt7, M7::PtrpC-mrypt7 and M. ruber M7
Phenotypes of Monacus ruber were observed on the different media (PDA, CYA, MA, G25N) to investigate the influences of the mrypt7 deletion and overexpression on developmental processes. As showed in Figure 2A, the colony edge of Δmrypt7 was irregular and the growth rates of Δmrypt7 was slower than those of M7::PtrpC-mrypt7 and M. ruber M7. Besides, cleistothecia and conidia formation of Δmrypt7 were obviously inhibited compared with M7::PtrpC-mrypt7 and M. ruber M7. While the colony phenotypes, growth rates and conidia formation of M7::PtrpC-mrypt7 had no significantly difference from those of M. ruber M7 (Figure 2B).
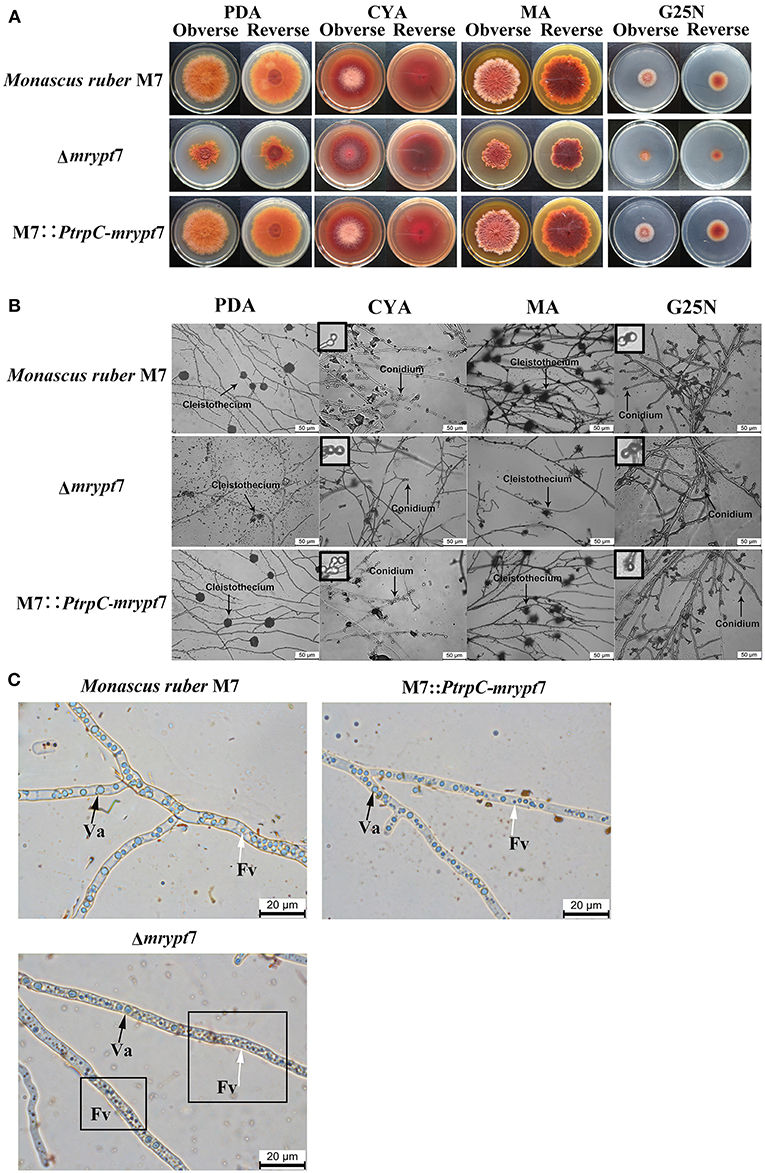
Figure 2. Phenotypic characterization of Δmrypt7, M7::PtrpC-mrypt7 and M. ruber M7. (A) Colony morphologies on PDA, CYA, MA and G25N plates; (B) Microscopic structures on PDA, CYA, MA, and G25N plates, the enlarged area was indicated by arrow, size bar = 50 μm; (C) Vacuole and fragment vacuoles microscopic structures on PDA medium. Va, vacuole (black arrow); Fv, fragment vacuoles (white arrow).
Vacuoles (Va) and fragment vacuoles (Fv) of M. ruber M7, Δmrypt7 and M7::PtrpC-mrypt7 on PDA medium were also observed under microscope. Compared with M7::PtrpC-mrypt7 and M. ruber M7, the number of Fv in Δmrypt7 increased more, while vacuoles reduced relatively and distributed nonuniformly in the mycelia (Figure 2C). The Fv and Va number and distribution between M7::PtrpC-mrypt7 and M. ruber M7 had no big difference, but the more uniform Fv and Va distribution of M7::PtrpC-mrypt7 was apparent (Figure 2C).
Mps and CIT Production Analysis of Δmrypt7, M7::PtrpC-mrypt7 and M. ruber M7
Previous studies (Chen et al., 2017) have demonstrated that M. ruber M7 can produce Mps and CIT, but no MK, so the yields of the 8 main Mps (four yellow pigments, monasfloure A, monascine, monasflore B, ankaflavin; two orange pigments, rubropunctatin, monascuburin; two red pigments, rubropunctamine and monascuburamine) and CIT in M. ruber M7 and its mutants were analyzed in this study to uncover the effect of Mrypt7 on SMs. Generally, all the detected SMs were increased in the mycelia of Δmrypt7 and M7::PtrpC-mrypt7, compared to M. ruber M7 (Figures S3, S4). Take monasfloure A, rubropunctatin, rubropunctamine and CIT production as examples for detail explanation, as showed in Figure 3, the concentration of intracellular yellow, orange and red pigments in Δmrypt7 were 1.8 times, 1.3 times, and 2.8 times of those in M. ruber M7, respectively, while the production of extracellular yellow, orange and red pigments were 63, 45, and 83% of M. ruber M7. In contrast, both intracellular and extracellular Mps in M7::PtrpC-mrypt7 were increased at least 20% compared with M. ruber M7. The intracellular CIT concentration in Δmrypt7 in 11th day was nearly 5 times more than those in M7::PtrpC-mrypt7 and M. ruber M7, while the extracellular CIT was only 20~40% of that in M7::PtrpC-mrypt7 and M. ruber M7. The intracellular and extracellular CIT in M7::PtrpC-mrypt7 and M. ruber M7 possessed the similar concentration.
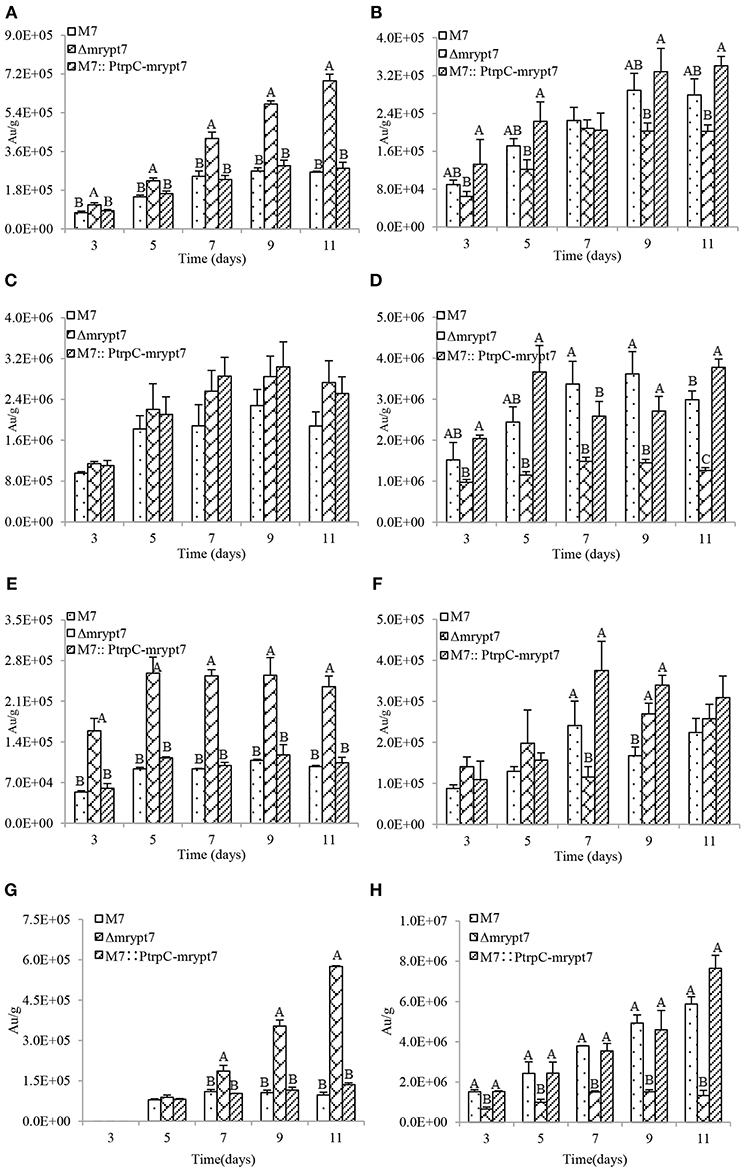
Figure 3. Mps and CIT yield analysis of M.ruber M7, Δmrypt7 and M7::PtrpC-mrypt7. (A) The yield of intracellular Monasfloure A. (B) The yield of extracellular Monasfloure A. (C) The yield of intracellular Rubropunctatin. (D) The yield of extracellular Rubropunctatin. (E) The yield of intracellular Rubropunctamine. (F) The yield of extracellular Rubropunctamine. (G) The yield of intracellular CIT. (H) The yield of extracellular CIT. The error bar represents the standard deviation between the three repeats. Capitals signify p-value < 0.01.
The Mrypt7 Function Elucidation on Development and Secondary Metabolite Production by Transcriptome Analysis
Differentially Expressed Genes Analysis, Annotation and Functional Classification
The transcriptome data obtained by RNA-seq were validated by qRT-PCR, β-actin serving as the reference gene. The expression data of 10 randomly selected genes (GME3693, GME5196, GME5065, GME2292, GME2157, GME5531, GME67, GME3412, GME6749, GME4561, GME2587) which are from the genome of M. ruber M7, fit with the sequencing profiles (Figure S5).
The differentially expressed genes (DEGs) between M7-3d vs. M7-7d, Δmrypt7-3d vs. Δmrypt7-7d, M7-3d vs. Δmrypt7-3d and M7-7d vs. Δmrypt7-7d were analyzed. The DEGs' functions were analyzed through GO function classifications and KEGG pathway. According to GO categories, the DEGs function classifications of the four teams almost belong to biological process, cellular component and molecular function. KEGG pathway analysis manifested that the DEGs were mostly involved in cellular process, environmental information processing, genetic information processing, human diseases and metabolism. For example, the DEGs down-regulated in M7-3d vs. Δmrypt7-3d included ankyrin repeat protein, G protein-coupled receptor and thiazole synthase, meanwhile, the DEGs up-regulated in M7-3d vs. Δmrypt7-3d included syntaxin, Golgi SNAP receptor, Ras GTPase activating like protein and fungal type III polyketide synthase. The DEGs down-regulated in M7-7d vs. Δmrypt7-7d included acyl-CoA synthetase, transposon, ubiquinone biosynthesis protein, exosome complex component and regulator of ribosome biosynthesis; while the DEGs up-regulated in M7-7d vs. Δmrypt7-7d included golgi family apparatus membrane protein, mitochondrial fission protein, vesicular inhibitory amino acid molecule and gama tubulin complex.
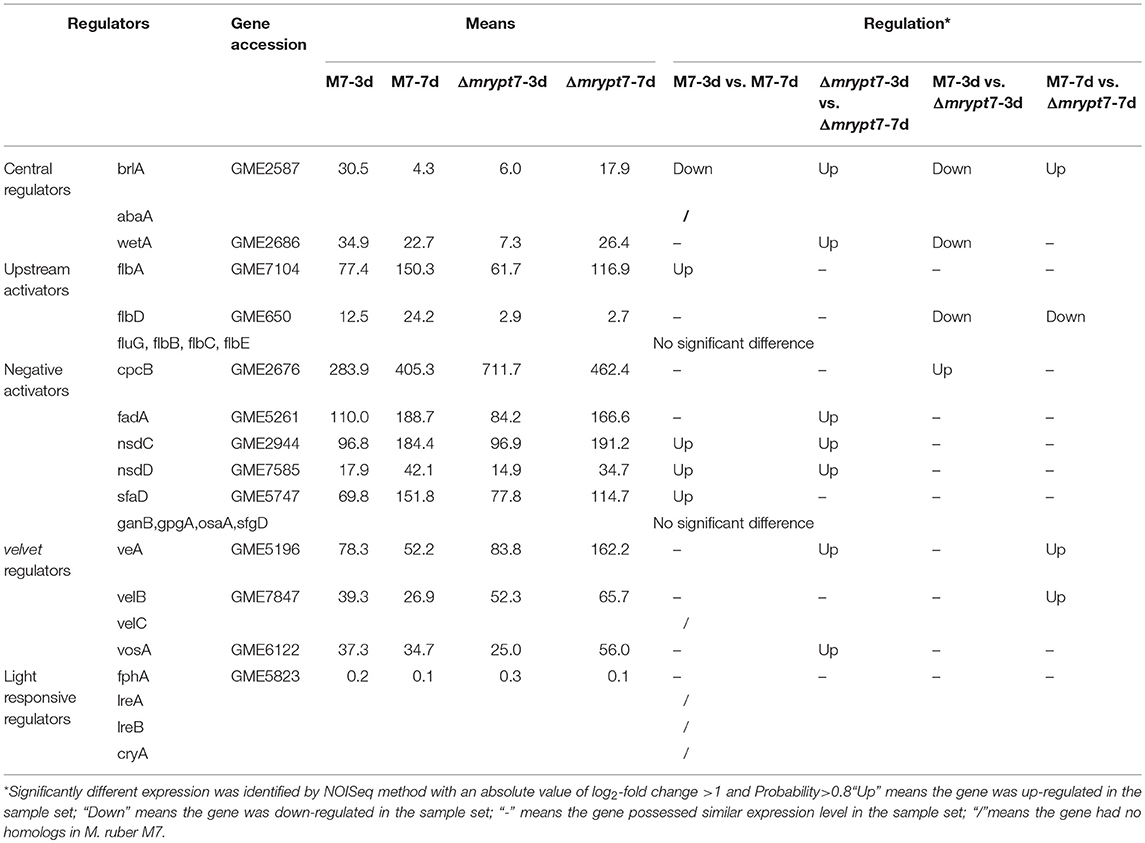
The Fungus-Specific Regulators Coordinating Conidia Were Positively Regulated by Mrypt7
Fungal conidiation regulatory mechanism is very complex, and there are many regulators involved in fungal conidiation which can be divided into central regulators (brlA, abaA, and wetA), upstream activators (fluG, flbA, flbB, flbC, flbD, and flbE), negative regulators (CpcB, NsdC, NsdD, OsaA, SfgA, and VosA, etc.), velvet regulators (VeA, VelB, VelC, and VosA) and light responsive regulators (FphA, CryA, LreA, and LreB) (Park and Yu, 2012, 2016). The putative regulatory DEGs coordinating conidiation in M7 and Δmrypt7 were analyzed in this part to illustrate the regulation of mrypt7 on Monascus conidia (Table 1).
The central regulatory pathway controls conidiation-specific gene expression and asexual developmental processes, very interesting, in M. ruber M7, the central regulatory pathway only includes brlA and wetA without abaA (Chen, 2015). Transcriptome results showed brlA and wetA were significantly down-regulated in Δmrypt7-3d vs. M7-3d, even the brlA gene was up-regulated in Δmrypt7-7d vs. M7-7d, the total expression levels of brlA in Δmrypt7-3d and Δmrypt7-7d were lower than those in M. ruber M7. Besides, the flbD gene belongs to one of the upstream developmental activators which is required for the initiation of conidiation and brlA activation (Kwon et al., 2010) was down-regulated in Δmrypt7-7d. For balancing with upstream activators, on the contrast, the cpcB gene belongs to the negative regulator which inhibits precocious activation of brlA during proliferation (Park and Yu, 2012) was up-regulated in Δmrypt7-3d. What's interesting is that the velvet regulators, veA and velB which suppresses conidiation and activation of sexual development (Bayram and Braus, 2012; Park and Yu, 2016) were up-regulated in Δmrypt7-7d, but little cleistothecia could be found in Δmrypt7 (Figure 2B), which was different from the results found in Aspergillus nidulan (Kim et al., 2002).
Effect of Mrypt7 on the Secondary Metabolites Biosynthesis Process
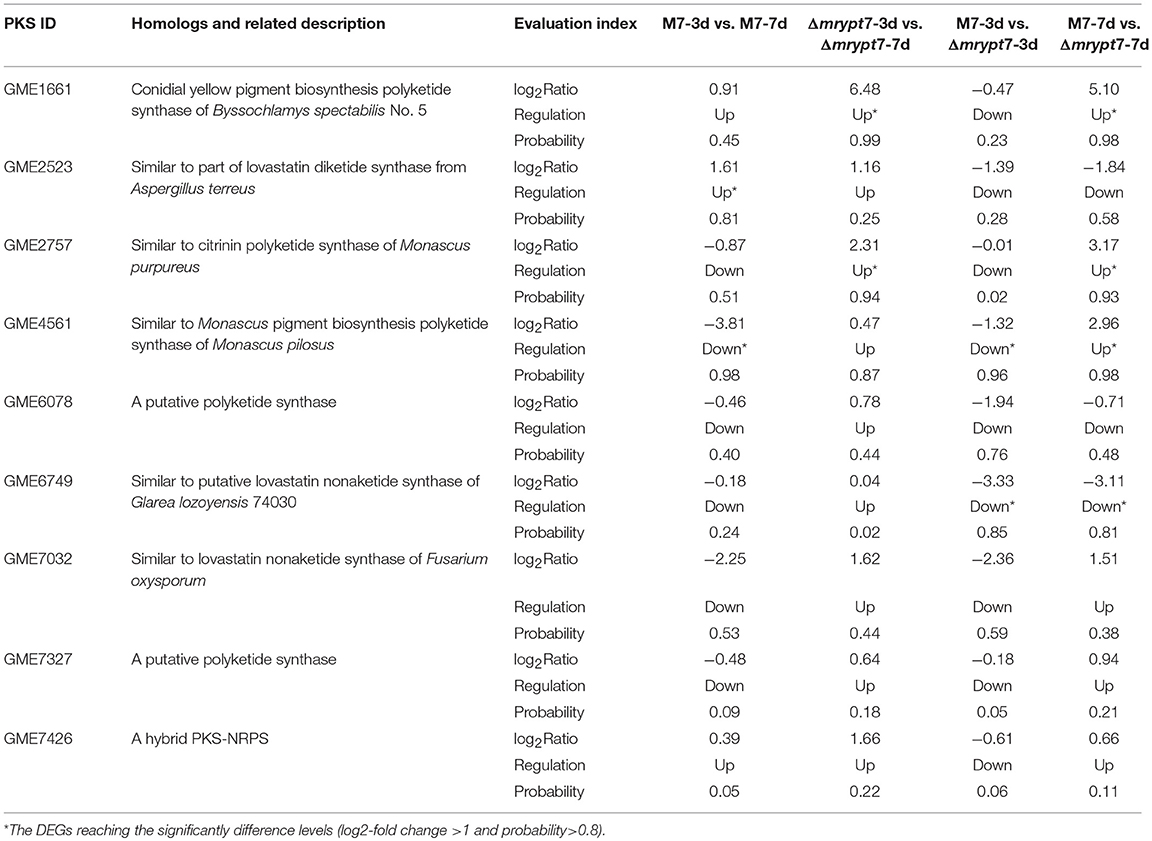
Monascus spp. can produce several secondary metabolites, like Mps, CIT, and so on (Liao et al., 2014; Feng et al., 2016). Previous studies have demonstrated that there are 9 predicted pks (polyketone synthase) genes in M. ruber M7 genome (Chen, 2015), and the different effects of Mrypt7 on these 9 pks genes were listed in Table 2. Among them, only the Mps pks was down-regulated in M7-3d vs. M7-7d, while all pks genes were up-regulated in Δmrypt7-3d vs. Δmrypt7-7d even only conidial yellow pigment pks and Mps pks reaching the significantly difference levels (log2-fold change>1 and probability>0.8). Besides, all pks genes down-regulated in M7-3d vs. Δmrypt7-3d and only Mps pks gene and a putative lovastatin nonaketide pks gene reaching the significantly difference levels; while the putative lovastatin nonaketide synthase down-regulated and conidial yellow pigment, CIT and Mps pks up-regulated in M7-7d vs. Δmrypt7-7d. Combination of these results has revealed that Mrypt7 can remarkably affect the expression of genes involved in SMs biosynthesis, but Mps and CIT pks genes may be more affected by Mrypt7.
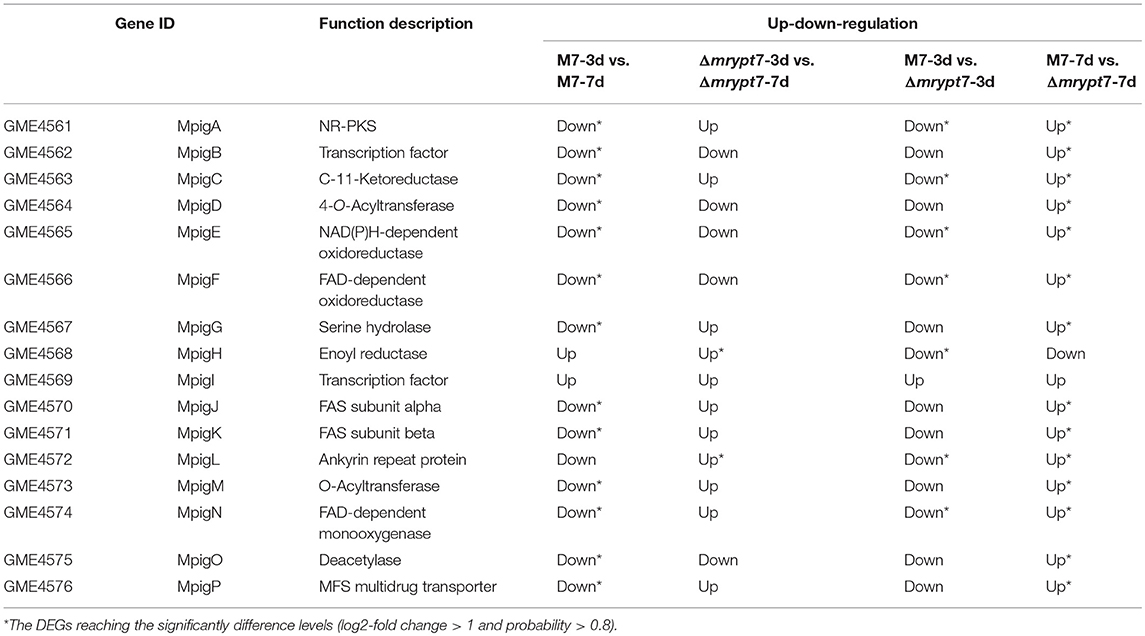
Furture analysis of the expression level of Mps and CIT biosynthesis gene clusters showed that most of these genes down-regulated in M7-3d vs. Δmrypt7-3d and up-regulated in M7-7d vs. Δmrypt7-7d. Generally, for Mps biosynthesis gene cluster (Chen et al., 2017), the expressions of all genes (except MpigH, MpigI, and MpigL) were down-regulated in M7-3d vs. M7-7d, only MpigH and MpigL was up-regulated in Δmrypt7-3d vs. Δmrypt7-7d, the results suggested that the Mps biosynthesis gene cluster in Δmrypt7 maintained a higher expression level compared with M7; nearly all genes down-regulated in Δmrypt7-3d vs. M7-3d but only MpigA, MpigC, MpigE, MpigF, MpigH, MpigL, and MpigN reaching the significantly difference levels, on the contrary, the whole Mps gene cluster (except MpigH and MpigI) were up-regulated in Δmrypt7-7d vs. M7-7d (Table 3).
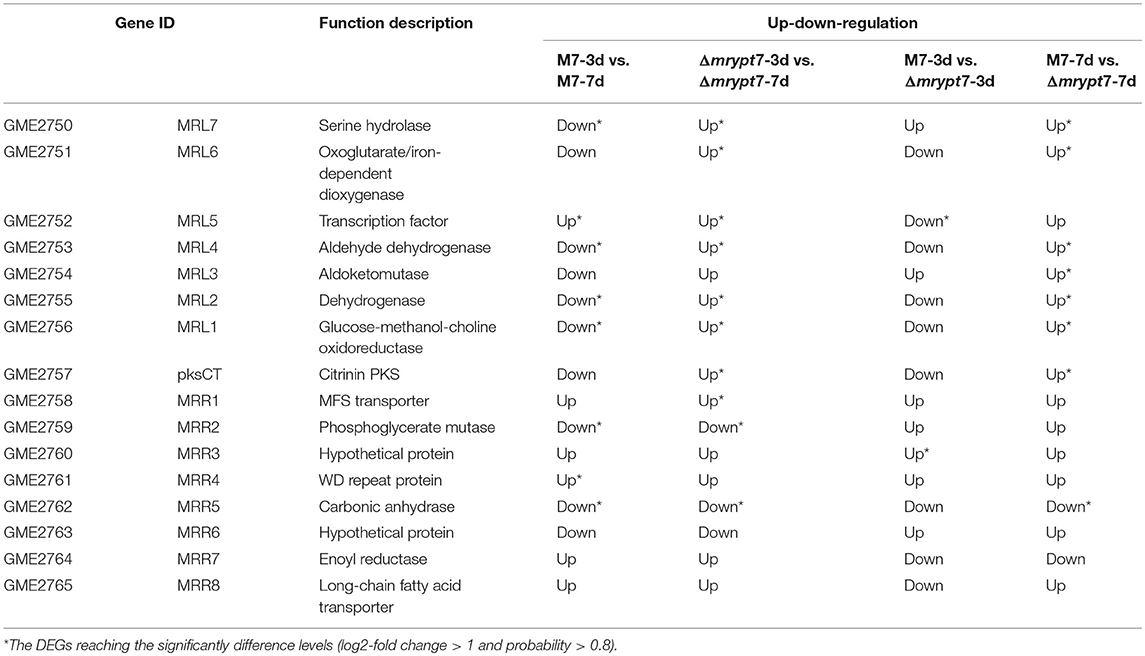
While for the CIT biosynthesis gene cluster (He and Cox, 2016), the expressions of pksCT, MRL7, MRL4, MRL2, MRL1, MRR2, and MRR2 down-regulated and MRL5, MRR4 up-regulated in M7-3d vs. M7-7d; MRL7, MRL6, MRL5, MRL4, MRL2, MRL1, pksCT, and MRR1 up-regulated and MRR2, MRR5 down-regulated in Δmrypt7-3d vs. Δmrypt7-7d; only MRL5 down-regulated and MRR3 up-regulated in Δmrypt7-3d vs. M7-3d, but almost all genes (except MRR5) up-regulated in Δmrypt7-7d vs. M7-7d (Table 4).
Discussion
Ypt/Rab, a single-subunit small GTPase which is related in structure to the Gα subunit of heterotrimeric G proteins (large GTPases) (Santarpia et al., 2012), has been proved to be the key regulators of the membrane trafficking system, endocytosis and exocytosis in all eukaryotes, especially in animals and plants (Fu et al., 2017; Kim et al., 2017; Pfeffer, 2017; Srikanth et al., 2017). In fungi, the functions of Ypt/Rab, especially Ypt7, also only focus on its role of vesicle transport. It's an accepted fact that Ypt7 mainly controls vesicle–vacuolar fusion balance, the disruption and overexpression of Ypt7 caused various vacuole phenotypes (Xu et al., 2012; Li et al., 2015; Liu et al., 2015; Zheng et al., 2015). Besides, the mechanism of Ypt7 mediated fusion interacts with numerous tethering and SNARE (Soluble NSF attachment protein receptor) complexes had been proved (Balderhaar et al., 2010; Ng et al., 2012; Hyttinen et al., 2013). Moreover, conidiogenesis imperfection and SMs production variation can also be found in Ypt7 disruption or overexpression mutants (Chanda et al., 2009a; Li et al., 2015; Liu et al., 2015; Yang et al., 2018), but the mechanism of Ypt7 involved conidial biogenesis and SMs biosynthesis was unclear.
In current study, the functions of mrypt7 (ypt7 homologous) in M. ruber M7 were investigated, we have found that aside from the functions of vesicle fusion, Mrypt7 can synchronously regulate the vegetative growth, conidiogenesis and secondary metabolism in M. ruber M7. Transcriptome results illustrated that the fungus-specific conidiation regulators and SMs biosynthesis genes expression were significantly difference when ypt7 gene was deleted (Tables 1–4). So we propose that Ypt7 works more like a global regulatory factor in fungi, which is first put forward the novel function definition of Rab GTPases.
Fungal conidiation regulatory mechanism is very complex, and there are some differences for the regulatory gene distribution in different fungi. Compared to A. nidulans, the up-to-date regulatory genes were conserved in M. ruber M7, while no homolog hits of abaA (central regulators), VelC (velvet regulators), CryA, LreA, and LreB (light responsive regulators) were searched in M. ruber M7 (Table 1). It seems that a new regulatory network may be owned in Monascus. In current study, the mrypt7 disruption repressed asexual development, meanwhile, the regulators (brlA, wetA and flbD) related to conidia were down-regulated (Figure 2, Table 1), similar results were also found in other fungi like Arthrobotrys oligospora which was testified by qRT-PCR (Yang et al., 2018). These results suggest that Mrypt7 may be a positive regulator for Monascus asexual development and the relative regulation genes. While for Monascus sexual development process, the mrypt7-deletion promoted the expression level of veA and velB, but didn't activate sexual development as expected (Kim et al., 2002). The following tried to interpret a different sexual development regulation of M. ruber M7 may focus on the actual function of veA and velB, and try to find extra regulators coordinate to cleistothecia formation.
In this study, results indicated that the SMs biosynthesis was also regulated by Mrypt7. Transcriptome analysis showed that Mrypt7 had different impact on the expression of the 9 putative pks genes in M. ruber M7 Among them, Mps and CIT biosynthesis gene clusters and biosynthesis pathways have been delineated before (He and Cox, 2016; Chen et al., 2017), even both Mps and CIT in Δmrypt7 were accumulated in the cell, the expression of Mps and CIT biosynthesis gene clusters showed variant expression level when the mrypt7 gene was deleted. Researches presented that vesicle localized enzyme were necessary for SMs biosynthesis until they were eventually turned over in vacuoles (Chanda et al., 2009b; Roze et al., 2011), based on the mycelial morphology and transcriptome results in this study, it's a reasonable statement that the level of the enzyme and the SMs production were affected by these relative genes expression level which regulated by Ypt7.
sBesides, it's proved that Rab/Ypt protein, SNARE, tethering factors and Sec1/Munc18-family protein worked together to mediate the intracellular destination of a transport vesicle (Baker et al., 2015; Milosevic and Sørensen, 2015; Baker and Hughson, 2016). In this study, four SNARE genes which were important for the transportation on Golgi and endosome (bet1, bos1, stx16, and stx7) and three tethering factors genes (golgins, vacuolar protein sorting 22 and transport protein particle complex 10) were differential expressed when mrypt7 gene was deleted (Table S3). Moreover, syntaxin 16 (Stx16), the important members of SNARE complex, which had been proved to mediate early/recycling endosome to trans-Golgi network and late endosome to trans-Golgi network traffic (Chen et al., 2010), was differential expressed when mrypt7 gene was deleted (Figure S6), these results suggest that Mrypt7 is functional in both in early and late endosomes. Mrypt7 and the above mentioned SNARE and tethering factor may work together to finish the fusion process and mediate SMs transportation. The further investigation of the interactions between these proteins could help to develop the detail model of Mrypt7 function in SMs transportation.
It's proved that Ypt family is stable from 7 to 12 Ypts in fungi, except Ypt7, others Ypts (Ypt2, Ypt5, Ypt6, etc.) also affect vegetative growth and conidiogenesis (Wakade et al., 2017; Yang et al., 2017), but had little influence on related genes (Yang et al., 2018). What's more, the Ypt7 disruption had no obvious effect on the expression of the rest of Ypts (Ypt1-Ypt6) in M. ruber M7, only Ypt3 was up-regulated in M7-7d vs. Δmrypt7-7d (Table S3). The results further proved that Ypt7 worked more like a global regulator.
Based on the above results, a model of Ypt7 regulation physiological processes was proposed in this study (Figure 4). Briefly, Ypt7, a single-subunit small GTPase, worked as a global regulatory factor, is required for the development, secondary metabolism and vesicle fusion of Monascus. First, Ypt7 is a positive regulator for fungal development. The conidiogenesis is suppressed combined with the relative genes (brlA, wetA, cpcB, flbD, veA, and velB) differential expression when Ypt7 was deleted, more remarkable, the sexual development is still suppressed even the sexual active regulators (veA and velB) were up-regulated which suggested that the sexual development was more rely on the Ypt7 functional completeness (Yang et al., 2018). Besides, LaeA, the well-known global regulator, impacts asexual and sexual reproduction but has no noticeable effect on these genes (brlA, wetA, cpcB, flbD, veA, and velB) (Liu et al., 2016). Second, Ypt7 is a negative regulator for secondary metabolism, the SMs production remarkable rose when Ypt7 was deleted. Ypt7 disruption caused vesicles quantity significantly increased which may increase the vesicle localized SMs enzymes (Roze et al., 2011), and promoted the expression level of SMs biosynthesis gene (Yang et al., 2018). Third, Ypt7 regulates the early transport and later vesicle fusion simultaneously. the early transport between endoplasmic reticulum (ER) and Golgi apparatus was effected by Ypt7 connecting with some SNARE genes, the up-regulate of Bet1 (blocked early transport) and Bos1 (bet one suppressor) could help to alleviate the lethality associated with disruption of Ypt7 (Newman et al., 1990; Chung et al., 2018). The vesicle fusion and SMs secretion is hampered, but a small quantity of extracellular Mps and CIT can also be detected. Except the known role of Ypt7 in vesicle fusion, two up-regulated syntaxins (Stx7 and Stx16) (Chen et al., 2010) and up-regulated MpigP and MRR1 (multidrug transporters) were supposed to help Mps and CIT secretion.
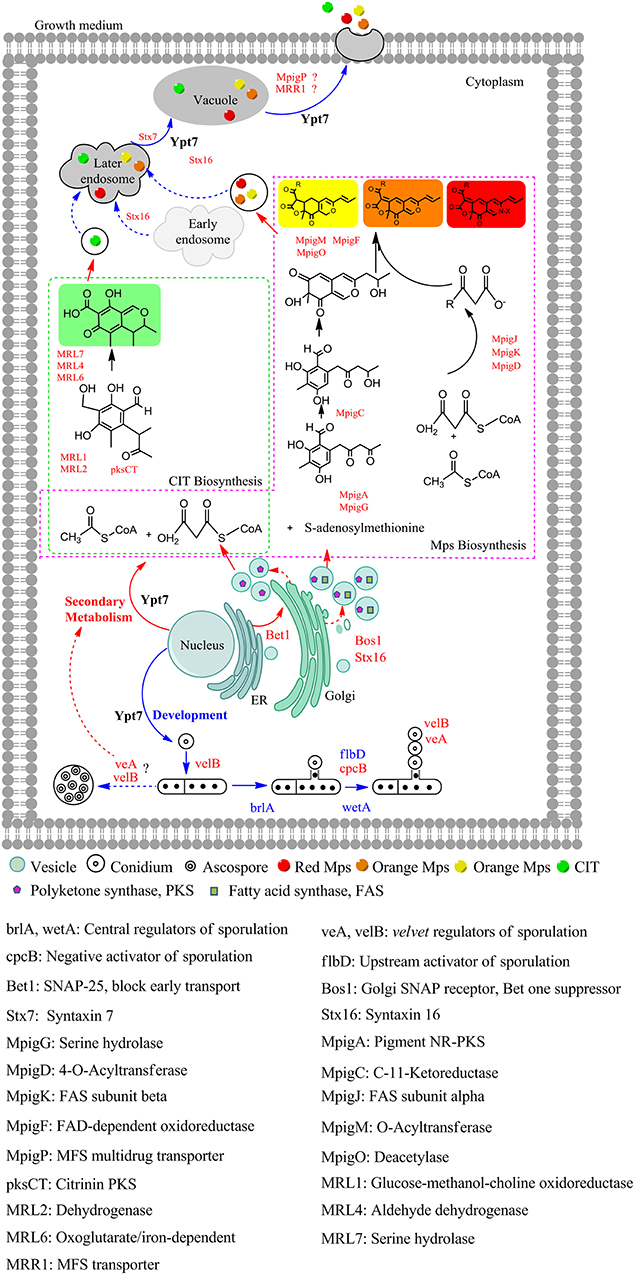
Figure 4. The proposed model of Ypt7 regulation physiological processes in fungi. The proteins and arrows marked in red indicating that they are up-regulated, the proteins and arrows marked in blue indicating that they are down-regulated, the proteins and arrows marked in black indicated the proved pathways. Dotted lines mean the supposed processes, solid lines mean the experimental processes in current study.
In a conclusion, this study has indicated the effect and regulation model of ypt7 gene on vegetative growth, conidiogenesis, vesicle fusion and SMs biosynthesis and transportation in Monascus. This is the first comprehensive analysis of the Rab/Ypt family in Monascus, the results could enrich the understanding of the function of Rab/Ypt family and make some contribution to uncover the SMs biosynthesis and transportation process in filamentous fungi.
Author Contributions
FC and YZ managed the project. JL and ML performed the transformants construction, secondary metabolites analysis and transcriptome results analysis in this work. JL performed the phenotypic characterization, interpreted the analysis results, and wrote the paper. All authors reviewed the manuscript.
Funding
This study was supported by the Major Programs (No. 31730068 and No. 31330059 to FC), General Program (No. 31371824 to YZ) and Young Scientist Program (No. 31701583 to JL) of National Natural Science Foundation of China. I declare all sources of funding received for the research being submitted.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Miss Yangxiaoxiao Shao (The Loomis Chaffee School, 4 Batchelder Rd, Windsor, Connecticut, USA) is thanked for the contribution of PCR reaction, vector construction, and phenotypic characterization in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00452/full#supplementary-material
Figure S1. The schematic diagram of the construction of mrypt7 deletion (A) and overexpression strains (B).
Figure S2. Sequence analysis and characterization of mrypt7 in M. ruber M7. (A) Gene structure analysis by softbarry. CDSf, first (starting with start codon) coding segment; CDSi, internal (internal exon) coding segment; CDSl, last (ending with stop codon) coding segment; CDSo, gene contains the ONE coding exon only; PolA, terminal polyA signal; TSS, transcription start site. (B) Characteristic motifs or residues of Ypt7 in the choosed 285 fungi. phosphate/Mg2+ bingding domain(PM), GTP/GDP bingding domains (G) and C-terminal isoprenylation region (C).
Figure S3. The mainly pigments of M.ruber M7 detected by UPLC. (A) The chromatogram of 4 main yellow pigments at 380 nm which are indicated by 1, 2, 3, and 4; (B) The chromatogram of 2 main orange pigments at 470 nm which are indicated by 5 and 6; (C) The chromatogram of the 2 main red pigments at 520 nm which are indicated by 7 and 8; (D) The chemical structure formula of the 8 pigments.
Figure S4. Monascus pigments yield analysis of M.ruber M7, Δmrypt7 and M7::PtrpC-mrypt7. (A) The yield of intracellular Monasfloure A. (B) The yield of extracellular Monasfloure A. (C) The yield of intracellular Monascine. (D) The yield of extracellular Monascine. (E) The yield of intracellular Monasfluore B. (F) The yield of extracellular Monasfluore B. (G) The yield of intracellular Ankaflavin. (H) The yield of extracellular Ankaflavin. (I) The yield of intracellular Rubropunctatin. (J) The yield of extracellular Rubropunctatin. (K) The yield of intracellular Monascuburin. (L) The yield of extracellular Monascuburin. (M) The yield of intracellular Rubropunctamine. (N) The yield of extracellular Rubropunctamine. (O) The yield of intracellular Monascuburamine. (P) The yield of extracellular Monascuburamine. The error bar represents the standard deviation between the three repeats. Capitals signify p-value < 0.01.
Figure S5. The validation of transcriptome data by qRT-PCR. (A) The expression level of the ten selectived genes in M7-3d VS Δmrypt7-3d. (B) The expression level of the ten selectived genes in M7-7d VS Δmrypt7-7d. The expression level of the genes in M7 was set as 1.
Figure S6. The SNARE interactions in vesicular transport.
Table S1. Ypt homologous genes in the M. ruber M7 genome.
Table S2. Primers used for the deletion and overexpression of mrypt7 gene.
Table S3. The proposed genes involved in vesicle transport.
References
Ao, X., Zou, L., and Wu, Y. (2014). Regulation of autophagy by the rab GTPase network. Cell Death Differ. 21, 348–358. doi: 10.1038/cdd.2013.187
Baker, R. W., and Hughson, F. M. (2016). Chaperoning SNARE assembly and disassembly. Nat. Rev. Mol. Cell Biol. 17, 465–479. doi: 10.1038/nrm.2016.65
Baker, R. W., Jeffrey, P. D., Zick, M., Phillips, B. P., Wickner, W. T., and Hughson, F. M. (2015). A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114. doi: 10.1126/science.aac7906
Balderhaar, H. J. k., Arlt, H., Ostrowicz, C., Bröcker, C., and Sündermann, F. (2010). The rab GTPase Ypt7 is linked to retromer-mediated receptor recycling and fusion at the yeast late endosome. J. Cell Sci. 123:4085–4094. doi: 10.1242/jcs.071977
BasuRay, S., Agola, J. O., Jim, P. A., Seaman, M. N., and Wandinger-Ness, A. (2018). “Rab7a in endocytosis and signaling,” in Encyclopedia of Signaling Molecules, eds. S. Choi. (Cham: Springer International Publishing), 4385–4396.
Bayram, O., and Braus, G. H. (2012). Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol. Rev. 36, 1–24. doi: 10.1111/j.1574-6976.2011.00285.x
Bouchez, I., Pouteaux, M., Canonge, M., Genet, M., Chardot, T., Guillot, A., et al. (2015). Regulation of lipid droplet dynamics in depends on the Rab7-like Ypt7p, HOPS complex and V1-ATPase. Biol. Open 4, 764–775. doi: 10.1242/bio.20148615
Chanda, A., Roze, L. V., Kang, S., Artymovich, K. A., Hicks, G. R., Raikhel, N. V., et al. (2009a). A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 106, 19533–19538. doi: 10.1073/pnas.0907416106
Chanda, A., Roze, L. V., Pastor, A., Frame, M. K., and Linz, J. E. (2009b). Purification of a vesicle-vacuole fraction functionally linked to aflatoxin synthesis in Aspergillus parasiticus. J. Microbiol. Methods 78, 28–33. doi: 10.1016/j.mimet.2009.03.014
Chen, F. S., and Hu, X. Q. (2005). Study on red fermented rice with high concentration of monacolin K and low concentration of citrinin. Int. J. Food Microbiol. 103, 331–337. doi: 10.1016/j.ijfoodmicro.2005.03.002
Chen, W. (2015). Insights Into Biological Characteristics Of Monascus Ruber M7 by Genomics Approaches. (Doctor Thesis) Huazhong Agricultural University, Wuhan, Hubei, China.
Chen, W., Chen, R., Liu, Q., He, Y., He, K., Ding, X., et al. (2017). Orange, red, yellow: biosynthesis of azaphilone pigments in Monascus fungi. Chem. Sci. 8, 4917–4925. doi: 10.1039/C7SC00475C
Chen, W., He, Y., Zhou, Y., Shao, Y., Feng, Y., Li, M., et al. (2015). Edible filamentous fungi from the species Monascus: early traditional fermentations, modern molecular biology, and future genomics. Compr. Rev. Food Sci. F 14, 555–567. doi: 10.1111/1541-4337.12145
Chen, Y., Gan Bin, Q., and Tang Bor, L. (2010). Syntaxin 16: unraveling cellular physiology through a ubiquitous SNARE molecule. J. Cell. Physiol. 225, 326–332. doi: 10.1002/jcp.22286
Chung, K. P., Zeng, Y., Li, Y., Ji, C., Xia, Y., and Jiang, L. (2018). Signal motif-dependent ER export of the Qc-SNARE BET12 interacts with MEMB12 and affects PR1 trafficking in Arabidopsis. J. Cell Sci. 131:jcs202838. doi: 10.1242/jcs.202838
de Marcos Lousa, C., and Denecke, J. (2016). Lysosomal and vacuolar sorting: not so different after all! Biochem. Soc. T 44, 891–897. doi: 10.1042/BST20160050
ESFA (2011). Scientific Opinion on the substantiation of a health claim related to melatonin and reduction of sleep onset latency (ID 1698, 1780, 4080) pursuant to Article 13(1) of Regulation (EC) No 1924/20061. EFSA J. 9:2304. doi: 10.2903/j.efsa.2011.2241
Feng, Y., Shao, Y., Zhou, Y., Chen, W., and Chen, F. (2016). “Monascus Pigments,” in Industrial Biotechnology of Vitamins, Biopigments, and Antioxidants. eds E. J. Vandamme, and J. L. Revuelta (Weinheim: Wiley-VCH Verlag GmbH & Co. KgaA), 497–535.
Fu, Y., Zhu, J. Y., Zhang, F., Richman, A., Zhao, Z., and Han, Z. (2017). Comprehensive functional analysis of rab GTPases in Drosophila nephrocytes. Cell Tissue Res. 368, 615–627. doi: 10.1007/s00441-017-2575-2
Gallwitz, D., Donath, C., and Sander, C. (1983). A yeast gene encoding a protein homologous to the human c-has/bas proto-oncogene product. Nature 306, 704–707. doi: 10.1038/306704a0
He, Y., and Cox, R. J. (2016). The molecular steps of citrinin biosynthesis in fungi. Chem. Sci. 7, 2119–2127. doi: 10.1039/C5SC04027B
He, Y., Liu, Q., Shao, Y., and Chen, F. (2013). Ku70 and ku80 null mutants improve the gene targeting frequency in Monascus ruber M7. Appl. Microbiol. Biotechnol. 97, 4965–4976. doi: 10.1007/s00253-013-4851-8
Heber, D., Yip, I., Ashley, J. M., Elashoff, D. A., Elashoff, R. M., and Go, V. L. W. (1999). Cholesterol-lowering effects of a proprietary Chinese red-yeast-rice dietary supplement. Am. J. Clin. Nutr. 69, 231–236. doi: 10.1093/ajcn/69.2.231
Heuston, E. F., Keller, C. A., Lichtenberg, J., Giardine, B., Anderson, S. M., Hardison, R. C., et al. (2018). Establishment of regulatory elements during erythro-megakaryopoiesis identifies hematopoietic lineage-commitment points. Epigenetics Chromatin 11:22. doi: 10.1186/s13072-018-0195-z
Hyttinen, J. M., Niittykoski, M., Salminen, A., and Kaarniranta, K. (2013). Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim. Biophys. Acta. 1833, 503–510. doi: 10.1016/j.bbamcr.2012.11.018
Kashiwazaki, J., Iwaki, T., Takegawa, K., Shimoda, C., and Nakamura, T. (2009). Two fission yeast Rab7 homologs, Ypt7 and Ypt71, play antagonistic roles in the regulation of vacuolar morphology. Traffic 10, 912–924. doi: 10.1111/j.1600-0854.2009.00907.x
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, H., Han, K., Kim, K., Han, D., Jahng, K., and Chae, K. (2002). The veA gene activates sexual development in Aspergillus nidulans. Fungal. Genet. Biol. 37, 72–80. doi: 10.1016/S1087-1845(02)00029-4
Kim, J. H., Chen, C., Yun, H. R., Lee, Y. S., Yi, Y. B., Kim, T. Y., et al. (2017). Disorder of trafficking system of plasma membrane and vacuole antiporter proteins causes hypersensitive response to salinity stress in Arabidopsis thaliana. J. Plant Biol. 60, 380–386. doi: 10.1007/s12374-017-0042-y
Kwon, N. J., Shin, K. S., and Yu, J. H. (2010). Characterization of the developmental regulator FlbE in Aspergillus fumigatus and Aspergillus nidulans. Fungal Genet. Biol. 47, 981–993. doi: 10.1016/j.fgb.2010.08.009
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lee, M. T. G., Mishra, A., and Lambright, D. G. (2009). Structural mechanisms for regulation of membrane traffic by Rab GTPases. Traffic 10, 1377–1389. doi: 10.1111/j.1600-0854.2009.00942.x
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, G., and Marlin, M. C. (2015). “Rab family of GTPases,” in Rab GTPases: Methods and Protocols, eds G. Li (New York, NY: Springer New York), 1–15.
Li, Y., Li, B., Liu, L., Chen, H., Zhang, H., Zheng, X., et al. (2015). FgMon1, a guanine nucleotide exchange factor of FgRab7, is important for vacuole fusion, autophagy and plant infection in Fusarium graminearum. Sci. Rep. 5:18101. doi: 10.1038/srep18101
Liao, C. D., Chen, Y. C., Lin, H. Y., Chiueh, L. C., and Shih, D. Y. C. (2014). Incidence of citrinin in red yeast rice and various commercial Monascus products in Taiwan from 2009 to 2012. Food Control 38, 178–183. doi: 10.1016/j.foodcont.2013.10.016
Liu, Q., Cai, L., Shao, Y., Zhou, Y., Li, M., Wang, X., et al. (2016). Inactivation of the global regulator LaeA in Monascus ruber results in a species-dependent response in sporulation and secondary metabolism. Fungal Biol. 120, 297–305. doi: 10.1016/j.funbio.2015.10.008
Liu, Q., Xie, N., He, Y., Wang, L., Shao, Y., Zhao, H., et al. (2014). MpigE, a gene involved in pigment biosynthesis in Monascus ruber M7. Appl. Microbiol. Biot. 98, 285–296. doi: 10.1007/s00253-013-5289-8
Liu, X. H., Chen, S. M., Gao, H. M., Ning, G. A., Shi, H. B., Wang, Y., et al. (2015). The small GTPase MoYpt7 is required for membrane fusion in autophagy and pathogenicity of Magnaporthe oryzae. Environ. Microbiol. 17, 4495–4510. doi: 10.1111/1462-2920.12903
Maringer, K., Yarbrough, A., Sims-Lucas, S., Saheb, E., Jawed, S., and Bush, J. (2016). Dictyostelium discoideum RabS and Rab2 colocalize with the Golgi and contractile vacuole system and regulate osmoregulation. J. Biosci. 41, 205–217. doi: 10.1007/s12038-016-9610-4
Mignogna, M. L., and D'Adamo, P. (2017). Critical importance of RAB proteins for synaptic function. Small GTPases 9:145–157. doi: 10.1080/21541248.2016.1277001
Milosevic, I., and Sørensen, J. B. (2015). “Fusion machinery: SNARE protein complex,” in Presynaptic Terminals, ed. S. Mochida. (Tokyo: Springer Japan), 87–127.
Muraguchi, H., Umezawa, K., Niikura, M., Yoshida, M., Kozaki, T., Ishii, K., et al. (2015). Strand-specific RNA-Seq analyses of fruiting body development in Coprinopsis cinerea. PLoS ONE 10:e0141586. doi: 10.1371/journal.pone.0141586
Newman, A. P., Shim, J., and Ferro-Novick, S. (1990). BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol. Cell Biol. 10, 3405–3414. doi: 10.1128/MCB.10.7.3405
Ng, E. L., Gan, B. Q., Ng, F., and Tang, B. L. (2012). Rab GTPases regulating receptor trafficking at the late endosome-lysosome membranes. Cell Biochem. Funct. 30, 515–523. doi: 10.1002/cbf.2827
Ohbayashi, N., and Fukuda, M. (2012). Role of rab family GTPases and their effectors in melanosomal logistics. J. Biochem. 151, 343–351. doi: 10.1093/jb/mvs009
Ohsumi, K., Arioka, M., Nakajima, H., and Kitamoto, K. (2002). Cloning and characterization of a gene (avaA) from Aspergillus nidulans encoding a small GTPase involved in vacuolar biogenesis. Gene 291, 77–84. doi: 10.1016/S0378-1119(02)00626-1
Park, H. S., and Yu, J. H. (2012). Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 15, 669–677. doi: 10.1016/j.mib.2012.09.006
Park, H. S., and Yu, J. H. (2016). Velvet regulators in Aspergillus spp. Korean J. Microbiol. Biotechn. 44:409. doi: 10.4014/mbl.1607.07007
Patakova, P. (2013). Monascus secondary metabolites: production and biological activity. J. Ind. Microbiol. Biotechnol. 40, 169–181. doi: 10.1007/s10295-012-1216-8
Pereira-Leal, J. B. (2008). The Ypt/Rab family and the evolution of trafficking in fungi. Traffic 9, 27–38. doi: 10.1111/j.1600-0854.2007.00667.x
Pfeffer, S. R. (2017). Rab GTPases: master regulators that establish the secretory and endocytic pathways. Mol. Biol. Cell 28, 712–715. doi: 10.1091/mbc.e16-10-0737
Pylypenko, O., Hammich, H., Yu, I. M., and Houdusse, A. (2018). Rab GTPases and their interacting protein partners: structural insights into rab functional diversity. Small GTPases 9, 22–48. doi: 10.1080/21541248.2017.1336191
Richards, A., Veses, V., and Gow, N. A. R. (2010). Vacuole dynamics in fungi. Fungal Biol. Rev. 24, 93–105. doi: 10.1016/j.fbr.2010.04.002
Roze, L. V., Chanda, A., and Linz, J. E. (2011). Compartmentalization and molecular traffic in secondary metabolism: a new understanding of established cellular processes. Fungal Genet. Biol. 48, 35–48. doi: 10.1016/j.fgb.2010.05.006
Salminen, A., and Novick, P. J. (1987). A ras-like protein is required for a post-Golgi event in yeast secretion. Cell 49, 527–538. doi: 10.1016/0092-8674(87)90455-7
Santarpia, L., Lippman, S. M., and El-Naggar, A. K. (2012). Targeting the MAPK–RAS–RAF signaling pathway in cancer therapy. Expert Opin. Ther. Tar. 16, 103–119. doi: 10.1517/14728222.2011.645805
Schmitt, H. D., Wagner, P., Pfaff, E., and Gallwitz, D. (1986). The ras-related YPT1 gene product in yeast: a GTP-binding protein that might be involved in microtubule organization. Cell 47, 401–412. doi: 10.1016/0092-8674(86)90597-0
Shao, Y., Ding, Y., Zhao, Y., Yang, S., Xie, B., and Chen, F. (2009). Characteristic analysis of transformants in T-DNA mutation library of Monascus ruber. World J. Microb. Biot. 25, 989–995. doi: 10.1007/s11274-009-9977-6
Shimizu, T., Kinoshita, H., Ishihara, S., Sakai, K., Nagai, S., and Nihira, T. (2005). Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus. Appl. Environ. Microbiol. 71, 3453–3457. doi: 10.1128/AEM.71.7.3453-3457.2005
Shinde, S. R., and Maddika, S. (2016). A modification switch on a molecular switch: Phosphoregulation of Rab7 during endosome maturation. Small GTPases 7, 164–167. doi: 10.1080/21541248.2016.1175539
Srikanth, S., Woo, J. S., and Gwack, Y. (2017). A large Rab GTPase family in a small GTPase world. Small GTPases 8, 43–48. doi: 10.1080/21541248.2016.1192921
Srikumar, S., Kröger, C., Hébrard, M., Colgan, A., Owen, S. V., Sivasankaran, S. K., et al. (2015). RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella typhimurium. Plos Pathog. 11:e1005262. doi: 10.1371/journal.ppat.1005262
Stenmark, H. (2009). Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Bio. 10, 513–525. doi: 10.1038/nrm2728
Tarazona, S., García, F., Ferrer, A., Dopazo, J., and Conesa, A. (2012). NOIseq: a RNA-seq differential expression method robust for sequencing depth biases. Univer. Southampton 17:18. doi: 10.14806/ej.17.B.265
Tripathy, M. K., Tiwari, B. S., Reddy, M. K., Deswal, R., and Sopory, S. K. (2017). Ectopic expression of PgRab7 in rice plants (Oryza sativa L.) results in differential tolerance at the vegetative and seed setting stage during salinity and drought stress. Protoplasma 254, 109–124. doi: 10.1007/s00709-015-0914-2
Van Verk, M. C., Hickman, R., Pieterse, C. M. J., and Van Wees, S. C. M. (2013). RNA-Seq: revelation of the messengers. Trends Plant Sci. 18, 175–179. doi: 10.1016/j.tplants.2013.02.001
Wakade, R., Labbaoui, H., Stalder, D., Arkowitz, R. A., and Bassilana, M. (2017). Overexpression of YPT6 restores invasive filamentous growth and secretory vesicle clustering in a Candida albicans arl1 mutant. Small GTPases. 6, 1–7. doi: 10.1080/21541248.2017.1378157
Wickner, W. (2010). Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a rab, all dancing in a ring on yeast vacuoles. Annu. Rev. Cell Dev. Biol. 26, 115–136. doi: 10.1146/annurev-cellbio-100109-104131
Wu, M. D., Cheng, M. J., Yech, Y. J., Chen, Y. L., Chen, K. P., Yang, P. H., et al. (2013). Monascus azaphilones A-C, three new azaphilone analogues isolated from the fungus Monascus purpureus BCRC 38108. Nat. Prod. Res. 27, 1145–1152. doi: 10.1080/14786419.2012.715289
Xu, X., Yang, J., An, Y., Pan, Y., and Liu, G. (2012). Over-expression of pcvA involved in vesicle-vacuolar fusion affects the conidiation and penicillin production in Penicillium chrysogenum. Biotechnol. Lett. 34, 519–526. doi: 10.1007/s10529-011-0792-4
Yang, C. D., Dang, X., Zheng, H. W., Chen, X. F., Lin, X. L., Zhang, D. M., et al. (2017). Two Rab5 homologs are essential for the development and pathogenicity of the rice blast fungus Magnaporthe oryzae. Front. Plant Sci. 8:620. doi: 10.3389/fpls.2017.00620
Yang, X., Ma, N., Yang, L., Zheng, Y., Zhen, Z., Li, Q., et al. (2018). Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biot. 102, 4601–4613. doi: 10.1007/s00253-018-8929-1
Yu, J. H., Hamari, Z., Han, K. H., Seo, J. A., Reyes-Domínguez, Y., and Scazzocchio, C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. doi: 10.1016/j.fgb.2004.08.001
Yun, H. R., Rim, Y. G., and Heo, J. B. (2016). Rice small GTPase Rab11 is required for intracellular trafficking from the trans-Golgi-network to the plasma membrane and/or prevacuolar compartments. Appl. Biol. Chem. 59, 163–171. doi: 10.1007/s13765-015-0143-6
Keywords: Ypt7, Monascus, development, secondary metabolism, regulation
Citation: Liu J, Lei M, Zhou Y and Chen F (2019) A Comprehensive Analysis of the Small GTPases Ypt7 Involved in the Regulation of Fungal Development and Secondary Metabolism in Monascus ruber M7. Front. Microbiol. 10:452. doi: 10.3389/fmicb.2019.00452
Received: 15 November 2018; Accepted: 20 February 2019;
Published: 18 March 2019.
Edited by:
Laurent Dufossé, Université de la Réunion, FranceReviewed by:
Wenhui Zheng, Fujian Agriculture and Forestry University, ChinaZonghua Wang, Fujian Agriculture and Forestry University, China
Copyright © 2019 Liu, Lei, Zhou and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youxiang Zhou, emhvdXlvdXhpYW5nQGdtYWlsLmNvbQ==
Fusheng Chen, c3VwZXJ2aXNvci5jaGVuQGFsaXl1bi5jb20=
 Jiao Liu1
Jiao Liu1 Fusheng Chen
Fusheng Chen