
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 08 March 2019
Sec. Microbiotechnology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00338
This article is part of the Research Topic Microorganisms Degrading Organic Pollutants and Their Potential for the Bioremediation of Contaminated Environments View all 48 articles
Antibiotics play a key role in the management of infectious diseases in humans, animals, livestock, and aquacultures all over the world. The release of increasing amount of antibiotics into waters and soils creates a potential threat to all microorganisms in these environments. This review addresses issues related to the fate and degradation of antibiotics in soils and the impact of antibiotics on the structural, genetic and functional diversity of microbial communities. Due to the emergence of bacterial resistance to antibiotics, which is considered a worldwide public health problem, the abundance and diversity of antibiotic resistance genes (ARGs) in soils are also discussed. When antibiotic residues enter the soil, the main processes determining their persistence are sorption to organic particles and degradation/transformation. The wide range of DT50 values for antibiotic residues in soils shows that the processes governing persistence depend on a number of different factors, e.g., physico-chemical properties of the residue, characteristics of the soil, and climatic factors (temperature, rainfall, and humidity). The results presented in this review show that antibiotics affect soil microorganisms by changing their enzyme activity and ability to metabolize different carbon sources, as well as by altering the overall microbial biomass and the relative abundance of different groups (i.e., Gram-negative bacteria, Gram-positive bacteria, and fungi) in microbial communities. Studies using methods based on analyses of nucleic acids prove that antibiotics alter the biodiversity of microbial communities and the presence of many types of ARGs in soil are affected by agricultural and human activities. It is worth emphasizing that studies on ARGs in soil have resulted in the discovery of new genes and enzymes responsible for bacterial resistance to antibiotics. However, many ambiguous results indicate that precise estimation of the impact of antibiotics on the activity and diversity of soil microbial communities is a great challenge.
Antibiotics are complex molecules with different functional groups in their chemical structures and are divided into several classes (Figure 1, Table 1) depending on the mechanisms of action, i.e., inhibition of cell wall synthesis, alteration of cell membranes, inhibition of protein synthesis, inhibition of nucleic acids synthesis, competitive antagonism, and antimetabolite activity (Kümmerer, 2009). Antibiotics are widely prescribed for treatment of infectious diseases in humans and animals. Moreover, they are used in livestock to increase meat production by preventing infections or outbreaks of diseases and promoting growth at a global scale. The production of antibiotics is still increasing, and the total annual usage has reached from 100,000 to 200,000 tons worldwide (Gelband et al., 2015). Between 2000 and 2015 antibiotic consumption in 76 countries around the world, expressed in defined daily doses (DDDs), increased 65% and, in 2015, reached 42 billion DDDs. Among high-income countries, the leading consumers of antibiotics in 2015 were the United States, France, and Italy. Leading consumers of antibiotics between low and middle-income countries were India, China, and Pakistan (Klein et al., 2018). It has been predicted that in 2030 global antibiotics consumption will be 200% higher than in 2015, with the greatest increase coming from low and middle-income countries. There are significant differences in trends in the antibiotic consumption in European countries. According to the Antimicrobial Consumption—Annual Epidemiological Report for 2016 published by the European Center for Disease Prevention and Control (ECDC), a statistically significant trend of increasing antibiotics usage was observed for Greece and Spain from 2012 to 2016, while over the same time period a statistically significant decreasing antibiotics usage trends were observed for Finland, Luxembourg, Norway and Sweden (ECDC, 2018). The most prescribed antibiotic classes in the US and EU are penicillins, macrolides, cephalosporins, and fluoroquinolones (CDC Centers for Disease, 2015; ECDC, 2018). More detailed information about consumption of different antibiotics in some countries of the EU and US was presented by Singer et al. (2016).
Despite their benefits, a continuous release of antibiotics into the environment and their potential adverse impact on living organisms is of great concern (Fatta-Kassinos et al., 2011; De la Torre et al., 2012; Larsson, 2014; Barra Caracciolo et al., 2015; Brandt et al., 2015). Because the majority of antibiotics are not completely metabolized in the bodies of humans and animals, a high percentage of administered drugs is discharged into water and soil through municipal wastewater, animal manure, sewage sludge, and biosolids (nutrient-rich organic materials resulting from the treatment of sewage) that are frequently used to irrigate and fertilize agricultural lands (Bouki et al., 2013; Daghrir and Drogui, 2013; Wu et al., 2014) (Figure 2). It has been reported that 75–80, 50–90, and 60% of the doses of tetracyclines, erythromycin, and lincomycin, respectively, are excreted in urine and feces (Kumar et al., 2005a; Sarmah et al., 2006). Reported antibiotic concentrations in wastewater vary significantly and range from nanograms to micrograms per mL (Gulkowska et al., 2008; Michael et al., 2013; Kulkarni et al., 2017). Though some wastewater treatment processes can degrade antibiotics, there is notable variability in antibiotic removal rates. This can be attributed to differences in treatment processes, such as nature of influent, treatment plant capacity, and the type of technology used (Forsberg et al., 2012; Wu et al., 2014).
The concentrations of antibiotic residues in manure, sewage sludge, biosolids, and soil show large variations (Table 2) and depend on the type of drug, metabolism of the drug in animals, the duration of treatment and the time of sampling relative to the treatment period. Tetracyclines have the highest concentrations and are most frequently reported antibiotic residues in manure (Pan et al., 2011; Chen et al., 2012; Massé et al., 2014). Other groups of antibiotics with considerable concentrations in manure are fluoroquinolones (Zhao et al., 2010; Van Doorslaer et al., 2014) and sulfonamides (Martínez-Carballo et al., 2007). Among the macrolide antibiotics, the highest concentration in manure was measured for tylosin (Dolliver et al., 2008). Compared to manure, biosolids contain much lower amounts of antibiotics (Jones-Lepp and Stevens, 2007) (Table 2).
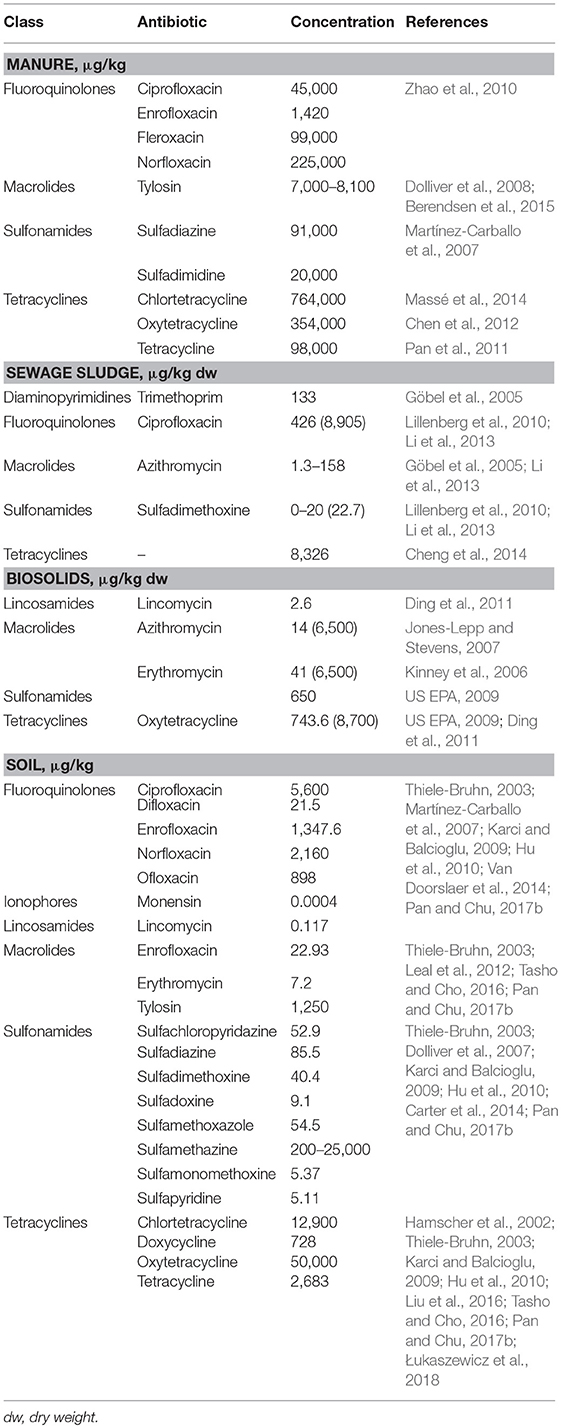
Table 2. Maximum reported concentrations of selected antibiotics detected in manure, sewage sludge, biosolids, and soil.
Scientists have long been aware of potential problems from the presence of antibiotics in soil. Determined antibiotic concentrations in soil matrices have ranged from a few nanograms to milligrams per kg of soil (Table 2). The highest concentrations are usually found in areas treated with manure or used for livestock (Kay et al., 2004; Zhou et al., 2013a; Hou et al., 2015; DeVries and Zhang, 2016). The concentrations of oxytetracycline and chlortetracycline in some agricultural lands may reach extremely high levels, whereas the concentrations of ciprofloxacin, norfloxacin, and tetracycline are typically significantly lower. Accurate quantification of antibiotics and their transformation products in the soil is of the utmost importance and requires advanced analytical methods, such as high-performance liquid chromatography with tandem mass spectrometry (HPLC/MS) (Aga et al., 2016).
Elevated concentrations of antibiotics in the soil selects for preferential outgrowth of antibiotic-resistant bacteria, which results in changes to antibiotics sensitivity of entire microbial populations (Halling-Sørensen et al., 2005; Binh et al., 2007; Ghosh and LaPara, 2007; Kyselková et al., 2013; Ma et al., 2014; Wepking et al., 2017; Atashgahi et al., 2018) (Figure 3). Even very low concentrations of antibiotics in the soil [below the minimum inhibitory concentration (MIC)], creates conditions for genetic changes in bacterial genomes and transfer of antibiotic resistance genes (ARGs) and associated mobile genetic elements (MGEs), such as plasmids, transposons, and genomic islands, between and among microbial populations (Ghosh et al., 2009; Heuer et al., 2011a,b; Du and Liu, 2012; Keen and Patrick, 2013; Tang et al., 2015; Grenni et al., 2018). In addition, co-selection and expression of resistance genes on MGEs may promote the spread of ARGs, even between distantly related bacterial species (Wellington et al., 2013). Autochthonous bacteria in soil may also represent a reservoir of resistance genes in the environment that can be transferred to the bacteria that colonize the human body (Zhang et al., 2015; Zhou et al., 2017). Such genes, e.g., tetracycline-resistance genes, were found in three different soils collected from Yunnan, Sichuan, and Tibet in China (Wang et al., 2017).
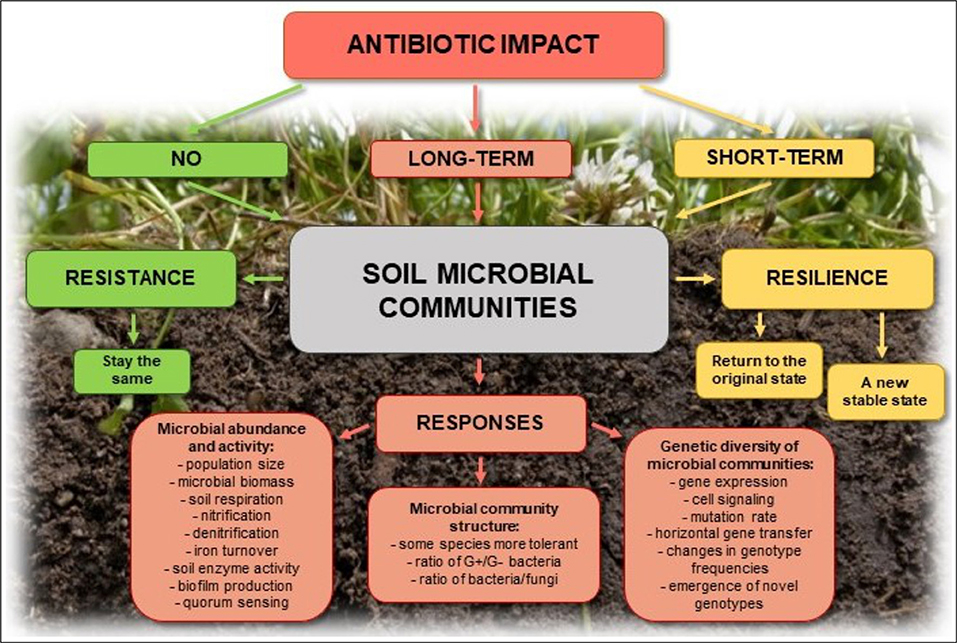
Figure 3. Potential effects of antibiotics on soil microbial communities and their possible responses.
Apart from the selection of antibiotic-resistant microorganisms and the spread of resistance genes in the soil environment, antibiotics may also affect the abundance of soil microorganisms (Pinna et al., 2012; Akimenko et al., 2015; Xu et al., 2016), overall microbial activity (Schmitt et al., 2005; Brandt et al., 2009; Cui et al., 2014; Liu et al., 2015), enzyme activity (Liu et al., 2009; Wei et al., 2009; Chen et al., 2013; Ma et al., 2016), and carbon mineralization and nitrogen cycling (Thiele-Bruhn, 2005; Rosendahl et al., 2012). The impacts of antibiotics on the functional, structural and genetic diversity of soil microorganisms have also been reported (Zielezny et al., 2006; Hammesfahr et al., 2008; Unger et al., 2013; Reichel et al., 2015; Cycoń et al., 2016b; Xu et al., 2016).
The aim of this review is to evaluate recent literature on (1) the degradation of antibiotics in soil and (2) their impact on the microbial community function, (3) the structural and genetic diversity as well as (4) the abundance and diversity of ARGs in soil.
In the soil environment, antibiotics may be subject to different abiotic and/or biotic processes, including transformation/degradation (Reichel et al., 2013; Cui et al., 2014; Manzetti and Ghisi, 2014; Duan et al., 2017), sorption-desorption (Tolls, 2001; Lin and Gan, 2011; Kong et al., 2012; Leal et al., 2013; Vaz et al., 2015; Martínez-Hernández et al., 2016), uptake by plants (Kumar et al., 2005b; Dolliver et al., 2007; Grote et al., 2007; Kuchta et al., 2009; Carter et al., 2014), and runoff and transport into groundwater (Carlson and Mabury, 2006; Davis et al., 2006; Kuchta et al., 2009; Park and Huwe, 2016; Pan and Chu, 2017a) (Figure 2). Hydrolysis is generally considered one of the most important pathways for abiotic degradation of antibiotics. β-lactams are especially susceptible to hydrolytic degradation, whereas macrolides and sulfonamides are known to be less susceptible to hydrolysis (Braschi et al., 2013; Mitchell et al., 2015). Photo-degradation, which contributes to degradation of antibiotics (e.g., quinolones and tetracyclines) spread on the soil surface during application of manure and slurry to agricultural land, in another important abiotic degradation process (Thiele-Bruhn and Peters, 2007). Antibiotics may also be degraded via reductive or oxidative transformation; however, data on these processes are still scarce.
As many authors have suggested that environmental transformation or degradation depends on the molecular structure (Figure 1) and physicochemical properties (Table 1) of antibiotics are the most important properties governing the fate of different antibiotics in soils (Thiele-Bruhn, 2003; Sassman and Lee, 2007; Crane et al., 2010; Leal et al., 2013; Awad et al., 2014; Pan and Chu, 2016; Zhang et al., 2017a). Since many abiotic and biotic factors affect the degradation of antibiotics, different groups of these pharmaceuticals differ in their rates of degradation in soils, as is evidenced by the large range of half-lives in soil, between < 1 and 3,466 days (Figure 4). For example, Braschi et al. (2013) found that amoxicillin was easily degradable, with a half-life of 0.43–0.57 days. Similarly, a study by Liu et al. (2014) revealed that chlortetracycline (10 mg/kg soil) was quickly degraded in soil with a DT50 value < 1 day. In contrast, an outdoor mesocosm study showed long-term persistence of azithromycin, ofloxacin, and tetracycline in soils, with half-lives of 408−3466, 866–1733, and 578 days, respectively (Walters et al., 2010). Detailed information about the degradation rates of antibiotics and obtained DT50 values in different soils is presented in Table 3. Notable, even for antibiotics within the same groups or, in some cases for particular antibiotics, DT50 values differ significantly. Observed differences in persistence or similar compounds are probably due to variations in soil composition, doses of antibiotics and conditions used in these studies. However, based on a review of the literature, it can be concluded that fluoroquinolones, macrolides, and tetracyclines are characterized by high DT50 or half-life values (Figure 4).
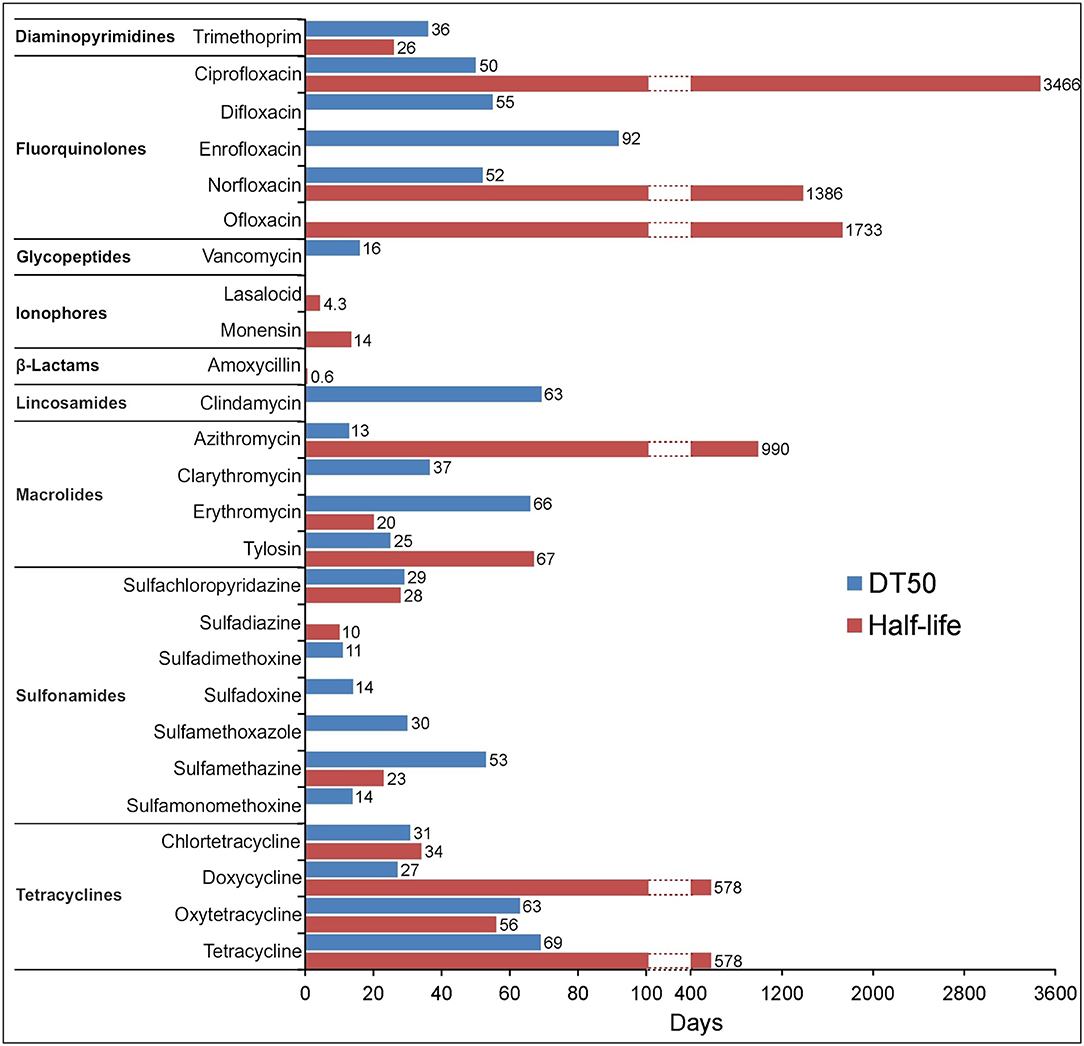
Figure 4. Maximum DT50 and half-life values for the selected antibiotics obtained from available degradation studies (Table 3) irrespective of the type of soil and the concentration of antibiotic used. The absence of bars in some cases means no data available.
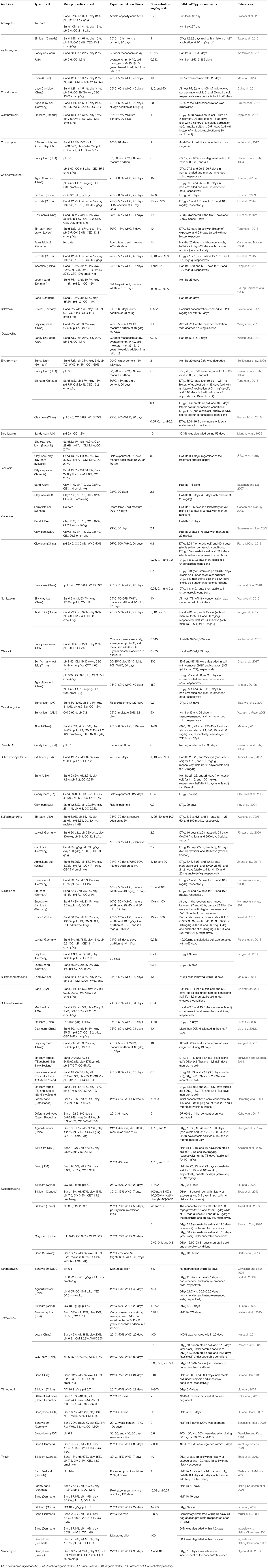
Table 3. Detailed data on the degradation of the selected antibiotics in soils with different characteristics.
The sorption coefficient (Koc) is a very important parameter for estimating environmental distribution and environmental exposure level of antibiotics. As was proposed by Crane et al. (2010), antibiotics with values of Koc > 4,000 l/kg are non-mobile and degradable to a very low degree in soils (very persistent) and the length of time needed for the degradation of 50% of an initial dose is >60 days. In contrast, antibiotics characterized by values of Koc < 15 l/kg are highly mobile and are more easily degraded; these can be classified as compounds with low persistence in soils (DT50 < 5 days). The relatively low persistence of some antibiotics in soils may be related to their low affinity to various organic and non-organic soil components. In contrast, due to their properties tetracyclines, fluoroquinolones, macrolides, and sulfonamides, bind strongly to soil components and form stable residues. For example, Hammesfahr et al. (2008) showed that the recovery rate of sulfadiazine on day 1 of an experiment was 27 and 45% of the initial antibiotic dosages of 10 and 100 mg/kg soil, respectively. Similarly, sulfamethazine applied at concentrations of 20 and 100 mg/kg soil strongly absorbed to soil components and measured concentrations of antibiotic were 62.1 and 31.5 μg/kg soil at the beginning of the experiment and, 255.5 and 129.8 μg/kg soil on day 56, respectively (Awad et al., 2016). Due to the binding of antibiotics to soil particles or the occurrence in a form of complexes, sorbed antibiotics often cannot be detected and may lose their antibacterial activity (Kümmerer, 2009). The adsorption and desorption of antibiotics are also associated with other soil parameters, such as pH and water content. For example, sulfonamides show a decrease in sorption with an increase of soil pH, whereas the binding of macrolides by soil components increases with a decrease of pH. In the first case, the sorption behavior is consistent with changes in the fraction of ionization of the sulfonamide as it converts from its cationic form to the neutral and anionic forms. The behavior of sulfonamides contrasts with tetracyclines and fluoroquinolones, which interact with soil primarily through cation exchange, surface complexation and cation bridging sorption mechanisms. In general, decreases in pH resulted in increased sorption of the cationic forms of antibiotics, suggesting that electrostatic interactions are the favored sorption mechanism for sulfonamides and macrolides (Schauss et al., 2009; Hammesfahr et al., 2011; Kong et al., 2012; Teixidó et al., 2012; Sittig et al., 2014; Wegst-Uhrich et al., 2014; Fernández-Calviño et al., 2015; Wang and Wang, 2015; Liu et al., 2017).
In addition to abiotic processes, microbial degradation may contribute to disappearance of antibiotics in soil. Some bacteria that degrade antibiotics have been isolated from antibiotics-contaminated soils. For example, strains belonging to the genera Microbacterium (Topp et al., 2013), Burkholderia (Zhang and Dick, 2014), Stenotrophomonas (Leng et al., 2016), Labrys (Mulla et al., 2018), Ochrobactrum (Zhang et al., 2017b; Mulla et al., 2018), and Escherichia (Mulla et al., 2018; Wen et al., 2018) were capable of degrading sulfamethazine, penicillin G, tetracycline, erythromycin and doxycycline in liquid cultures, respectively. Other bacteria belonging to the genera Klebsiella (Xin et al., 2012), Acinetobacter, Escherichia (Zhang et al., 2012), Microbacterium (Kim et al., 2011), Labrys (Amorim et al., 2014), and Bacillus (Rafii et al., 2009; Erickson et al., 2014) that were capable of degrading chloramphenicol, sulfapyridine, sulfamethazine, ciprofloxacin, norfloxacin, and ceftiofur have been isolated from patients, sediments, sludge, animal feces, and seawater. In particular, a Microbacterium sp. exhibited degradation of sulfamethazine in soil and, when introduced into agricultural soil, increased the mineralization of that antibiotic by 44–57% (Hirth et al., 2016). The central role of microorganisms in antibiotic degradation or transformation in soil has been confirmed by results of many studies carried out in sterile and non-sterile soils. As depicted in Table 3, the half-life or DT50 values of antibiotics were much lower in soils with autochthonous microorganisms compared to those obtained from sterilized soils. For example, Pan and Chu (2016) showed that when applied to soil at a dosage of 0.1 mg/kg soil, erythromycin disappeared faster in non-sterile soil compared to sterile soil, with DT50 of 6.4 and 40.8 days, respectively. Sulfachloropyridazine applied at 10 mg/kg soil was degraded almost three times faster in soils with autochthonous microorganisms (half-life 20–26 days) compared to sterile soils (half-life 68–71 days) (Accinelli et al., 2007). Zhang et al. (2017a) also showed that microbial activity plays a major role in the biotransformation of sulfadiazine in soil s, with a DT50 of 8.48, 8.97, and 10.22 days (non-sterile soil) and 30.09, 26.55, and 21.21 days (sterile soil) for concentrations of 4, 10, and 20 mg/kg soil, respectively. A similar phenomenon has also been observed for other antibiotics, such as norfloxacin (Pan and Chu, 2016), sulfamethoxazole (Lin and Gan, 2011; Srinivasan and Sarmah, 2014; Zhang et al., 2017a), sulfamethazine (Accinelli et al., 2007; Pan and Chu, 2016), and tetracycline (Lin and Gan, 2011; Pan and Chu, 2016).
Special attention should be paid to the analytical methods for extraction and determination of antibiotics residue when evaluating and comparing degradation experiments. Some techniques may not always be capable of differentiating between degradation and sorption, and insufficient or improper extraction procedures may result in incorrect interpretation of the fate of antibiotics in the soil as antibiotics that are tightly bound to soil particles could be erroneously considered to have been transformed or degraded.
Degradation of antibiotics depends not only on the catabolic activity of soil microorganisms, but also, to a large extent, on the properties of soil, i.e., organic matter content, pH, moisture, temperature, oxygen status, and soil texture (Table 3). For example, Li et al. (2010a) observed differences in the degradation rate of oxytetracycline in two agricultural soils with different characteristics. The calculated DT50 for this antibiotic reached values of 30.2 and 39.4 days for soils with a low or high organic carbon content, respectively. Soil type was also found to significantly affect antibiotics degradation, as demonstrated by Koba et al. (2017) who studied the degradation rate of clindamycin, sulfamethoxazole, and trimethoprim in 12 different soils. The authors characteristics, 44–98, 25–99, and 13–84% of clindamycin, sulfamethoxazole and trimethoprim (2 mg/kg soil), respectively, were degraded within 61 days during the experiment. A similar dependence of the rate of antibiotics degradation on soil type of has also been found for chlortetracycline (Halling-Sørensen et al., 2005; Li et al., 2010a), sulfachloropyridazine (Accinelli et al., 2007), sulfadiazine (Förster et al., 2009; Sittig et al., 2014), sulfamethoxazole (Accinelli et al., 2007; Lin and Gan, 2011; Srinivasan and Sarmah, 2014), and tetracycline (Li et al., 2010a). Another study revealed a significant decrease in the rate of chlortetracycline, erythromycin, and tylosin degradation introduced at a dosage of 5.6 mg/kg soil into a sandy loam soil. These degradation experiments showed that 56, 12, and 0%, 100, 75, and 0%, or 100, 100, and 60% of the initial concentration of chlortetracycline, erythromycin, and tylosin were degraded at temperatures of 30, 20, and 4°C, respectively, within 30 days (Gavalchin and Katz, 1994). In addition, Srinivasan and Sarmah (2014) showed that a lower temperature resulted in a reduced decomposition rate of sulfamethoxazole, irrespective of the soil depth. Some published data have also shown the influence of oxygen in soils on the degradation of antibiotics. For example, Pan and Chu (2016) showed that the half-lives for erythromycin, norfloxacin, sulfamethazine, and tetracycline, all of which had been applied at 0.1 mg/kg soil, increased from 6.4, 2.9, 24.8, and 31.5 days under aerobic conditions to 11.0, 5.6, 34.7, and 43.3 days under anaerobic conditions, respectively. Anaerobic conditions caused an increase in the half-life for sulfamethoxazole by seven days compared to 11.4 days for aerobic degradation of this antibiotic. In turn, no significant effects of incubation conditions on degradation rate were shown for trimethoprim, for which the calculated half-life was about 26 days under both aerobic and anaerobic conditions (Lin and Gan, 2011). The degradation of antibiotics may also be influenced by the pH and moisture content of soils. Weerasinghe and Towner (1997) showed that the half-life of for virginiamycin was negatively correlated with the pH of soils and ranged from 87 to 173 days for different agricultural soils. A study by Wang et al. (2006) revealed that the half-life of sulfadimethoxine in soil decreased from 10.4 days to 6.9, and again to 4.9 days when the soil moisture content was increased from 15, to 20, and 25%, respectively. A similar effect of temperature was shown in the case of the degradation of norfloxacin (Yang et al., 2012).
Degradation of antibiotics strongly depends on their concentration in soil. Increasing dosages of ciprofloxacin (from 1 to 5 and 50 mg/kg soil) led to a reduction of degradation from 75, to 62, and 40% within 40 days (Cui et al., 2014). A similar tendency was also shown by Demoling et al. (2009), who applied sulfamethoxazole at 500, 20, and 1 mg/kg soil and observed a reduction of its removal from 153, to 1.5, and 0.04 mg/kg after 5 weeks. The same trend was reported in degradation of azithromycin, ofloxacin, tetracycline (Walters et al., 2010), sulfadimethoxine (Wang et al., 2006), chlortetracycline (Fang et al., 2016), and SDZ (Zhang et al., 2017a). These results suggest that high concentrations of antibiotics may prolong their persistence in soils, due to the inhibition of the activity of soil microorganisms (Yang et al., 2009; Pan and Chu, 2016). However, application of unrealistically high concentrations tends to overestimate half-lives, which may not reflect realistic situations (Pan and Chu, 2016). The history of antibiotics application also plays a role in the further disappearance of antibiotics in soils. Repeated application of the macrolide antibiotics clarithromycin and erythromycin into soil resulted in a decrease of the DT50 values from 36.48 and 69.93 days to 15.85 and 4.36 days (soil with a history of clarithromycin and erythromycin application at 0.1 mg/kg soil) and to 9.51 and 0.94 days (soil with a history of clarithromycin and erythromycin application at 10 mg/kg soil), respectively (Topp et al., 2016). This phenomenon of pre-adaptation of soil microorganisms was also found for the degradation of chlortetracycline, sulfamethazine and tylosin (Topp et al., 2013).
Most published data have also indicated that the addition of different organic compounds, such as manure, biosolids, slurry, sludge, and compost, into soils may contribute to changes in the rate of antibiotics degradation (Table 3). Sassman and Lee (2007) showed that the addition of manure (20 mg/kg soil) increased the half-life of lasalocid from 3.6 to 4.3 days. In contrast, Yang et al. (2012) showed that norfloxacin (10 mg/kg soil) was degraded faster (half-life 24–39 days) in soil with manure (3–9%) compared to the control soil (half-life 48 days). In turn, Li et al. (2010a) found no significant effects from the addition of manure on degradation of chlortetracycline in soil.
Soil microorganisms perform many vital processes and participate in the maintenance of soil health and quality. They play a crucial role in organic matter turnover, nutrients release, and stabilization of the soil structure and ensure soil fertility. Moreover, many microorganisms act as biological control agents by inhibiting the growth of pathogens (Varma and Buscot, 2005). The homeostasis of soil may be disturbed by biotic and abiotic factors, including bacteriophages, predation, competition, pesticide, heavy metals, toxic hydrocarbons, and antibiotics (Cycoń et al., 2011; Cycoń and Piotrowska-Seget, 2015, 2016; Sułowicz and Piotrowska-Seget, 2016; Xu et al., 2016; Chen et al., 2017; Wepking et al., 2017; Orlewska et al., 2018a). The high antimicrobial activity of antibiotics in soil should differentially inhibit the growth of soil microorganisms and thus influence the soil microbial community composition, which may result in alterations of the ecological functionality of the soil (Kotzerke et al., 2008; Keen and Patrick, 2013; Molaei et al., 2017) (Figure 3).
There is abundant data on the impact of antibiotics on microorganisms and on soil processes mediated by bacteria and fungi. Both the effects of antibiotics on individual microbial populations as well as on the composition of entire microbial communities have been documented. A wide range of methods based on measurements of parameters that reflect the activity and abundance of total microbial communities, such as soil organic matter turnover, respiration, and microbial biomass have been used to characterize such effects. Other methods have focused on selected microbial processes such as nitrification, denitrification, sulfate, and iron reduction, methanogenesis and the activity of enzymes responsible for C, N, and P turnover (Brandt et al., 2009; Chen et al., 2013; Cui et al., 2014; Liu et al., 2015; Ma et al., 2016). A large number of studies have instead focused on assessing changes in microbial diversity, using metagenomics or 16S rRNA gene amplicon sequencing, as well as analysis of phospholipid fatty acids (PLFAs) isolated from soil (Zielezny et al., 2006; Hammesfahr et al., 2008; Reichel et al., 2015; Xu et al., 2016). In the following sections, the methods and parameters utilized to assess effects of antibiotics on the function and structure of soil microbial communities are presented and discussed (Figure 3).
Many studies have found that even low concentrations (below the MIC) of antibiotics influence various soil processes mediated by microorganisms (Table 4). A significant decrease in soil respiration (SR) was reported for soils containing sulfamethoxazole, sulfamethazine, sulfadiazine, and trimethoprim (Kotzerke et al., 2008; Liu et al., 2009), however, this effect was transient and depended on the disappearance rate of these antibiotics (DT50 2–5 days). The authors concluded that the effects of antibiotics on SR diminished due to decreased bioavailability of the antibiotic. In a study by Wepking et al. (2017), the response of microbial respiration to cephapirin, tetracycline, or erythromycin was dependent on both the type of antibiotic and the exposure of soil to dairy cattle manure. In other studies, no obvious effects of tetracycline, chlortetracycline, oxytetracycline, sulfadiazine, sulfapyridine, or tylosin on SR were observed (Thiele-Bruhn and Beck, 2005; Liu et al., 2009; Toth et al., 2011).
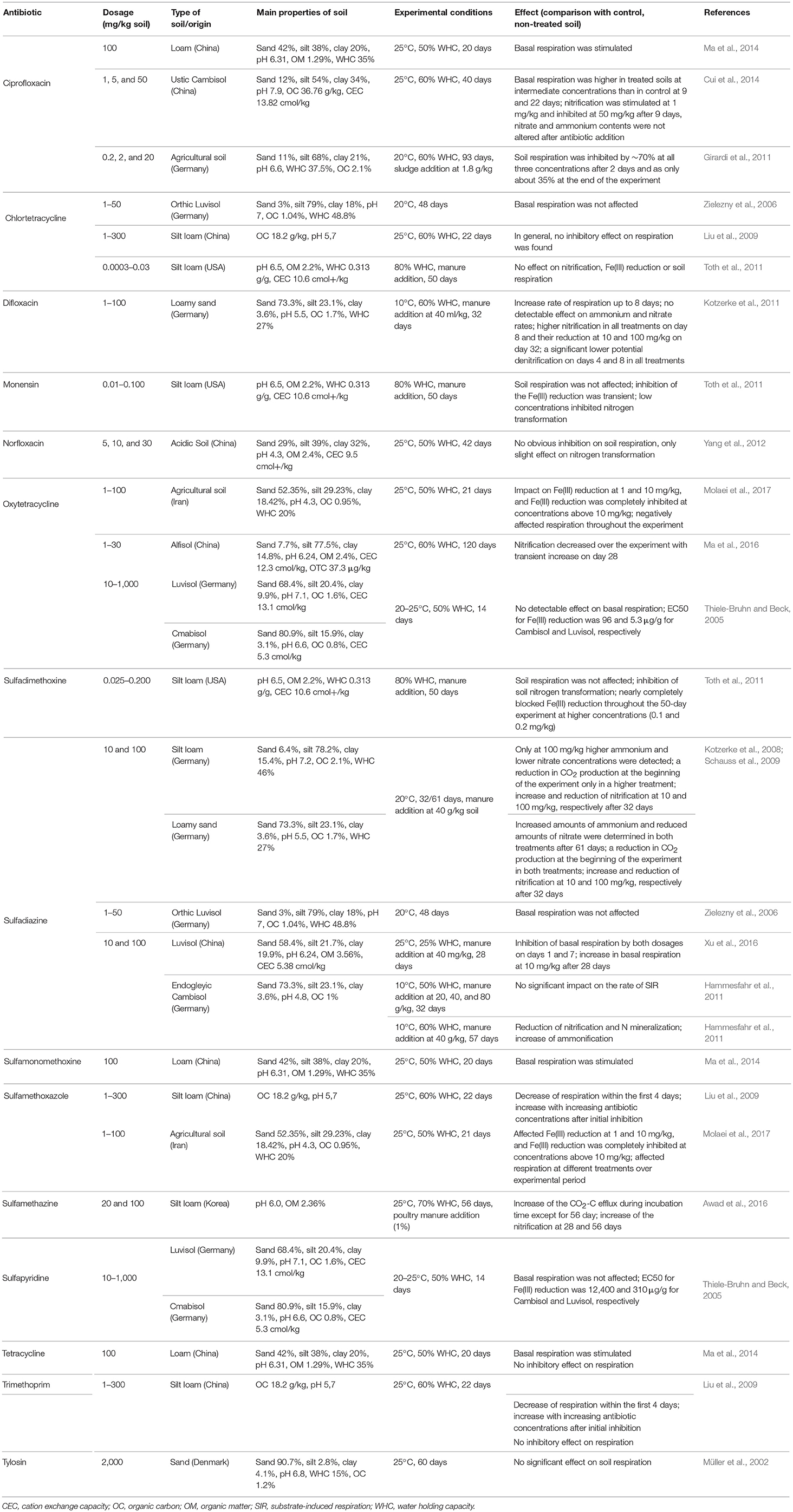
Table 4. Effects of the selected antibiotics on the microbial processes in soils with different characteristics.
Nitrification and/or denitrification rates were also influenced by antibiotic exposure, and the effects were strongly dependent on the type of antibiotic and the length of exposure. For example, sulfadimethoxine inhibited soil nitrification; however, this effect was only observed on some sampling days and only for a high sulfadimethoxine treatment (Toth et al., 2011). A decrease in the nitrification rate caused by a high oxytetracycline concentration (30 mg/kg) and sulfadiazine (100 mg/kg) in a single application was also observed by Ma et al. (2016) and Kotzerke et al. (2008), whereas ciprofloxacin and norfloxacin were reported to stimulate the rate of nitrification in a soil microcosm, but only at the lowest concentration of antibiotic (1 mg/kg soil) (Yang et al., 2012; Cui et al., 2014). A study by DeVries et al. (2015) revealed that low doses of sulfamethoxazole, sulfadiazine, narasin, or gentamicin (500 μg/kg soil) inhibited denitrification, but dosages < 1 μg/kg soil actually stimulated the process transiently. Toth et al. (2011) did not observe any effects of monensin and chlortetracycline, on soil nitrification at concentrations of 0.01–0.1 and 0.0003–0.3 mg/kg soil, respectively.
Antibiotics may also change the turnover rate of iron in soil. Sulfadiazine and monensin blocked Fe(III) reduction in soil over periods ranging from a few days to the end of a 50-day experiment (Toth et al., 2011). Strong inhibition of Fe(III) reduction was also observed in soil contaminated with sulfamethoxazole and oxytetracycline at concentrations >10 mg/kg soil (Molaei et al., 2017). Thiele-Bruhn and Beck (2005) calculated the EC50 value of sulfapyridine for the microbial reduction of Fe(III) in two different soils at 12.4 and 0.310 mg/kg soil. It should be noted that the lack of standardized tests hinders comparisons that would lead to general conclusions about the effects of antibiotics on biogeochemical cycles and the turnover of iron.
Specific enzyme activity is considered to be a useful indicator of the response of microorganisms to stress caused in the soil by antibiotics (Unger et al., 2013; Liu et al., 2014, 2015; Ma et al., 2016) (Table 5). Enzyme activity indicates the potential of microbial communities to carry out biochemical processes that are essential to maintain soil quality. Any application of a toxicant that might affect the growth of soil microorganisms can induce alterations in the general activity of enzymes, such as dehydrogenases (DHAs), phosphatases (PHOSs), and urease (URE) (Gil-Sotres et al., 2005; Hammesfahr et al., 2011; Cycoń et al., 2016a). Inhibition of DHAs and URE was observed in soil amended with tetracycline at a concentration of 1 μg/kg soil, however this dosage did not affect acid phosphatase (PHOS-H) activity. Sulfamethazine applied at 53.6 μg/g soil had a significant short-term negative impact on the activities of DHAs and URE (Pinna et al., 2012). Inhibition of DHAs and arylsulphatase activities with increasing concentration of oxytetracycline at 1 to 200 mg/kg over 7 weeks was also reported by Chen et al. (2013). Benzylpenicillin, tylosin and sulfadiazine inhibited soil DHAs and PHOSs from 35 to 70% compared to the non-antibiotic amended control (Reichel et al., 2014; Akimenko et al., 2015). The results obtained by Unger et al. (2013) showed that the application of oxytetracycline or lincomycin (50 and 200 mg/kg soil) resulted in a temporary decrease in DHA activity in soil. A temporary increase, followed by a decrease, in DHA activity was found in soils containing chlortetracycline (1, 10, and 100 mg/kg soil) by Liu et al. (2015). In another study, DHA activity was unaffected by sulfapyridine or oxytetracycline, even at a dosage of 1 mg/kg soil (Thiele-Bruhn and Beck, 2005).
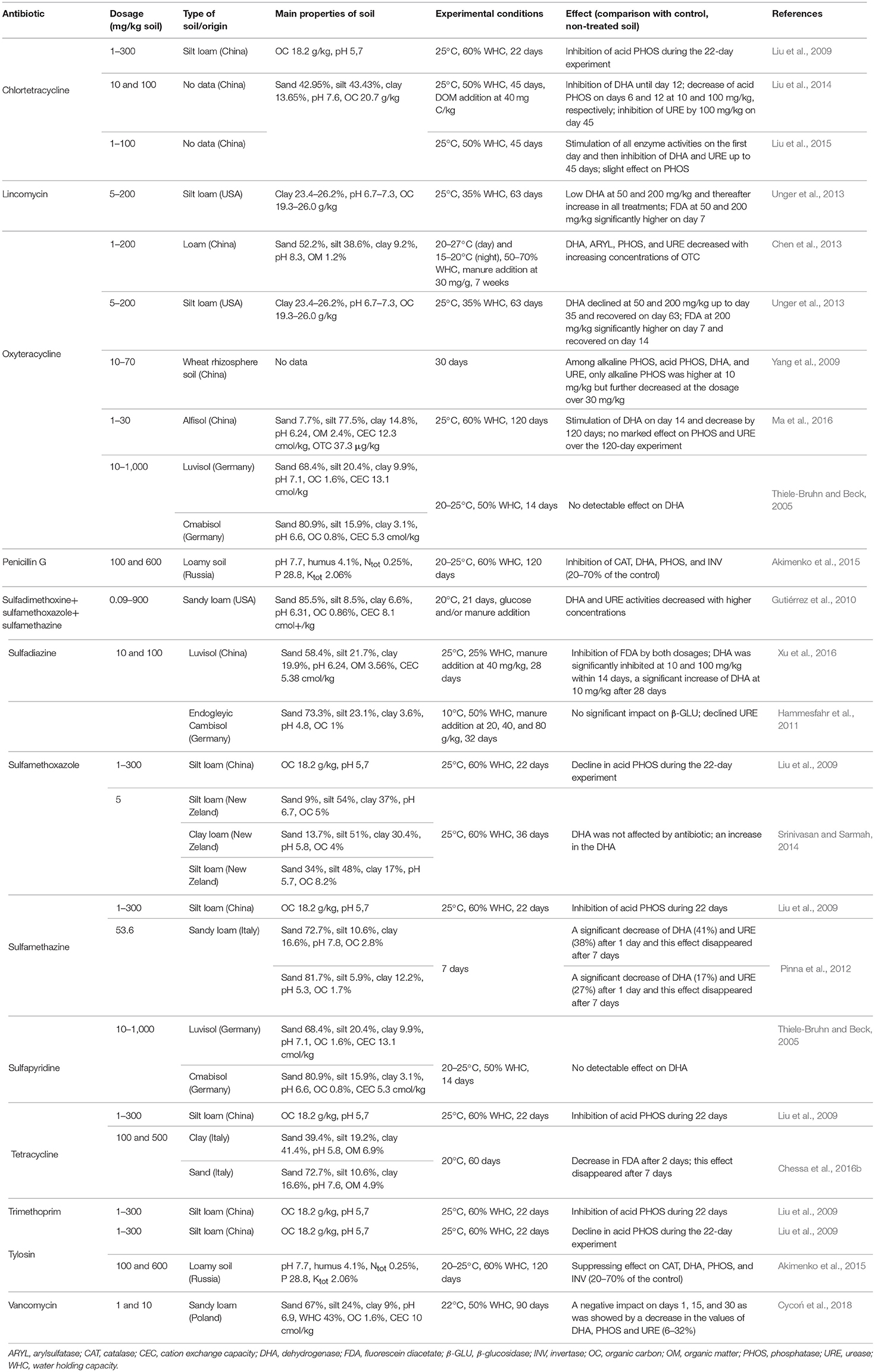
Table 5. Effects of the selected antibiotics on the enzyme activities in soils with different characteristics.
Various effects on the activity of PHOSs from different antibiotics applied to soil have been shown. For example, six antibiotics, i.e., chlortetracycline, tetracycline, tylosin, sulfamethoxazole, sulfamethazine, and trimethoprim at dosages of 1–300 mg/kg soil inhibited the activity of PHOS-H (Liu et al., 2009, 2015). In contrast, results from Yang et al. (2009) indicated that only alkaline phosphatase (PHOS-OH) was sensitive to the application of oxytetracycline, with a 41.3% decline in enzyme activity at a concentration of 10 mg/kg soil and a further decrease of 64.3–80.8% when the dose of the antibiotic exceeded 30 mg/kg. Ma et al. (2016) found that OTC at dosages up to 30 mg/kg soil had no effect on the activity of neutral PHOS over a 120-day experimental period.
The inhibition of enzyme activity in antibiotic-treated soils may be related to inhibition of growth or death of sensitive microorganisms (Boyd and Mortland, 1990; Gianfreda et al., 1994; Alef, 1995; Marx et al., 2005). In turn, the increased activity of enzymes under antibiotics pressure may result from the ability of many bacteria to subsist on antibiotics as a carbon source (Dantas et al., 2008). We can speculate that enzymes produced by such bacteria compensate for the negative effects of antibiotics on enzyme activity by increasing the activity of the antibiotic-resistant microbial community. Similarly, an increase in the activity of some enzymes in soils treated with different antibiotics might be related to the capability of some microorganisms to make use of the antibiotics in their metabolisms, thus resulting in an increase in the abundance of some soil microorganisms and enzyme production. In addition, the presence of some antibiotics in soil may cause an overgrowth of fungi, which are generally less sensitive to antibiotics than bacteria. Fungi are a major producer of enzymes in soils and so may be responsible for observed increases in enzyme activity (Tabatabai and Bremner, 1969; Westergaard et al., 2001; Hammesfahr et al., 2011; Ding et al., 2014).
Functional microbial diversity reflects the ability of an entire microbial community to utilize a suite of substrates (Zak et al., 1994). The response of microorganisms to the presence of antibiotics in soils, expressed as the community level physiological profile (CLPP), can be evaluated using the Biolog (EcoPlates™) or MicroResp™ methods, which are based on the utilization of 31 or 15 carbon sources, respectively, by organisms present in soil extracts (Toth et al., 2011; Ma et al., 2014: Xu et al., 2016) (Table 6). Although the exact numbers and taxonomic identities of the bacterial species responsible for the Biolog reactions remain uncertain, patterns of functional diversity indicate how biodiversity affects specific ecosystem functions (Laureto et al., 2015). For determination of CLPP, the biodiversity (Shannon H') and average well-color development (AWCD) indices have been used as indicators of changes in the catabolic potential of soil microorganisms exposed to antibiotics. For example, chlortetracycline (1 and 10 mg/kg soil) caused a decrease in AWCD values, showing low catabolic potential in the microbial community, however, this effect was seen only at the beginning of the experiment. After 35 days, irrespective of the frequency of chlortetracycline application, the AWCD values gradually recovered to the control level (Fang et al., 2014, 2016). A slight inhibitory effect on microbial activity (expressed as the H′ index) in soil was observed over a range from 1 to 300 mg oxytetracycline /kg soil. Even lower concentrations of this antibiotic (0.5 to 90 mg/kg of soil) significantly decreased the functional activity (expressed as AWCD) of the soil microbial communities (Kong et al., 2006). Sulfonamide antibiotics, such as sulfamethoxazole and sulfamethazine, can also alter the activity of microbial populations, however, they were observed to only have short-term detrimental effects (Demoling et al., 2009; Pinna et al., 2012; Pino-Otín et al., 2017). In turn, the application of chlortetracycline or sulfadimethoxine to soil did not significantly change AWCD, whereas monensin slightly increased the value of the H' index (Toth et al., 2011). In many studies, changes in the preferential degradation of groups of substrates in Eco-plates, e.g., amino acids, carbohydrates, carboxylic acids, aromatic acids, and miscellaneous, by microorganisms were observed over the course of an experimental period. Xu et al. (2016) found that a high sulfadiazine concentration decreased utilization rates of amino acids, carbohydrates, carboxylic acids, and aromatic acids by bacteria present in a soil suspension. A significant decrease in the utilization of carbohydrates and miscellaneous substrates was also observed in soil that had been treated with sulfamethoxazole (Liu et al., 2012a). However, this effect was only observed 7 days after the antibiotic application; by day 21 substrate utilization had increased compared to the first sampling day. The negative impact of antibiotics on CLPP may be mitigated by the enrichment of soil with organic matter from animal wastes, antibiotic-free manure or plant wastes (Demoling et al., 2009; Pinna et al., 2012; Liu et al., 2014). Contrarily, Wang et al. (2016) found that doxycycline application at 10 mg/kg soil increased utilization of the substrates.
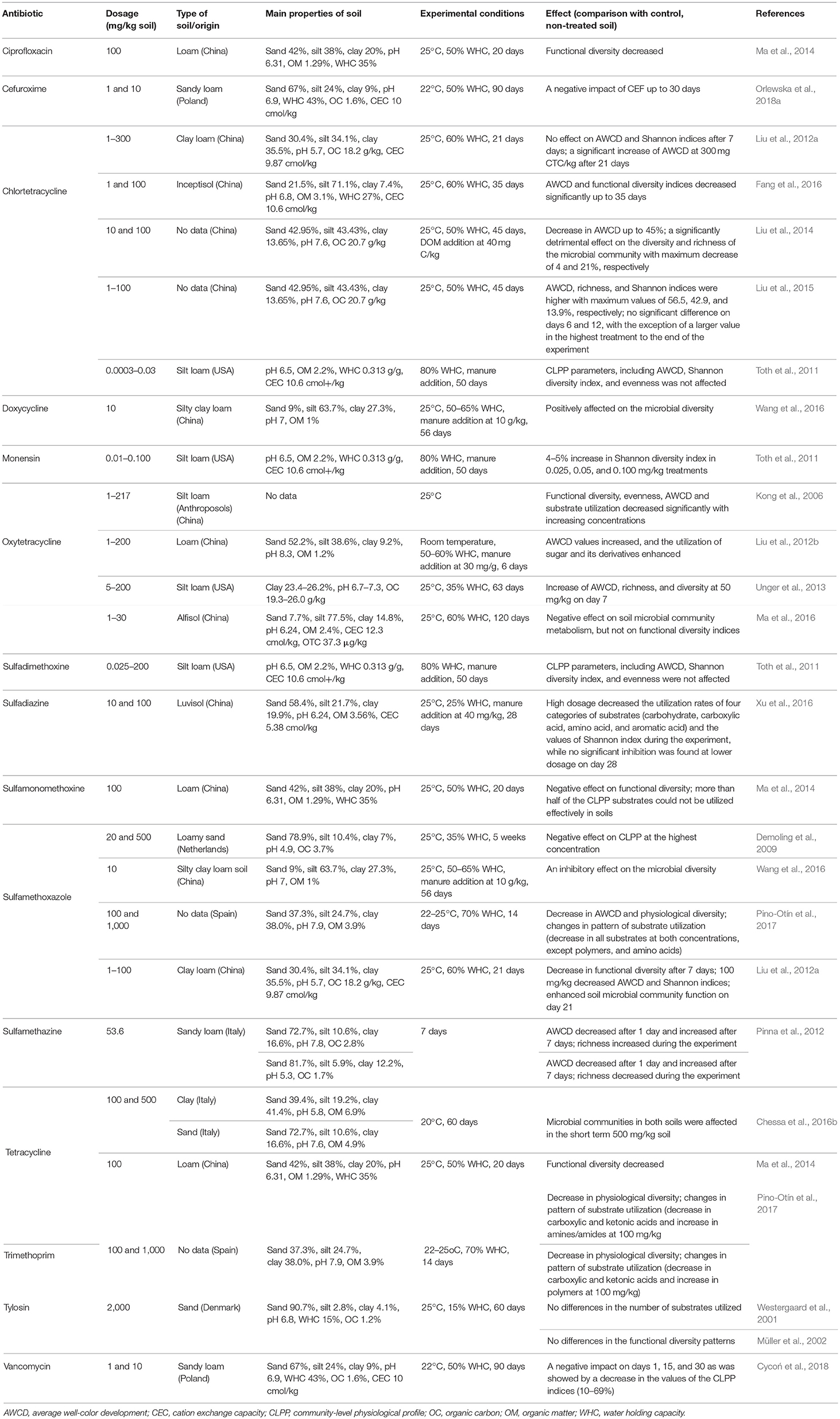
Table 6. Effects of the selected antibiotics on the community-level physiological profile (CLPP) in soils with different characteristics.
CLPP patterns do not always show changes in the functional diversity of microorganisms in response to antibiotic. In such situations, the lack of observable effects in CLPP might be because indices expressing total microbial activity do not detect possible changes caused by low antibiotic concentrations. Moreover, repeated application of antibiotics leads to the adaptation of bacteria to these compounds and proliferation of antibiotic-resistant microorganisms. Another limitation is that the Biolog technique measures only the activity of catabolically active cells and omits non-culturable populations as well as microorganisms in a dormant state. Moreover, fast-growing microorganisms are mainly responsible for effects observed by this test (Floch et al., 2011).
The PLFA method is a rapid tool for assessing the biomass and composition of microbial communities in soil, because various phospholipid-derived fatty acids (PLFAs) are indicative of species or microbial groups in soil. In addition, the ratios of biomass of bacteria/fungi and Gram-positive/Gram-negative bacteria are often used to evaluate the response of microorganisms to organic pollutants (Frostegård et al., 1993, 2011).
In general, application of antibiotics into soil show strong and dose-dependent effects on the microbial structure expressed as the total PLFA biomass (Table 7). For example, Chen et al. (2013) found a significant increase in bacterial and fungal PLFA biomass 7 weeks after oxytetracycline contamination of 1 and 15 mg/kg soil, but a strong decrease in the case of 200 mg antibiotic/kg treatment. A decrease in microbial PLFA biomass content was also found in soils that had been treated with penicillin G (10 and 100 mg/kg soil) and tetracycline (100 mg/kg soil) (Zhang et al., 2016a). Simultaneously, the ratios of Gram-negative/Gram-positive bacteria increased, indicating that the bacteria that were resistant to antibiotics in tested soils were more likely to be Gram-negative. A study by Unger et al. (2013) found only short-term effects (35 days) from lincomycin or oxytetracycline applications (5–200 mg/kg soil), suggesting that the resilience of soil microbial communities toward effects of these antibiotics.
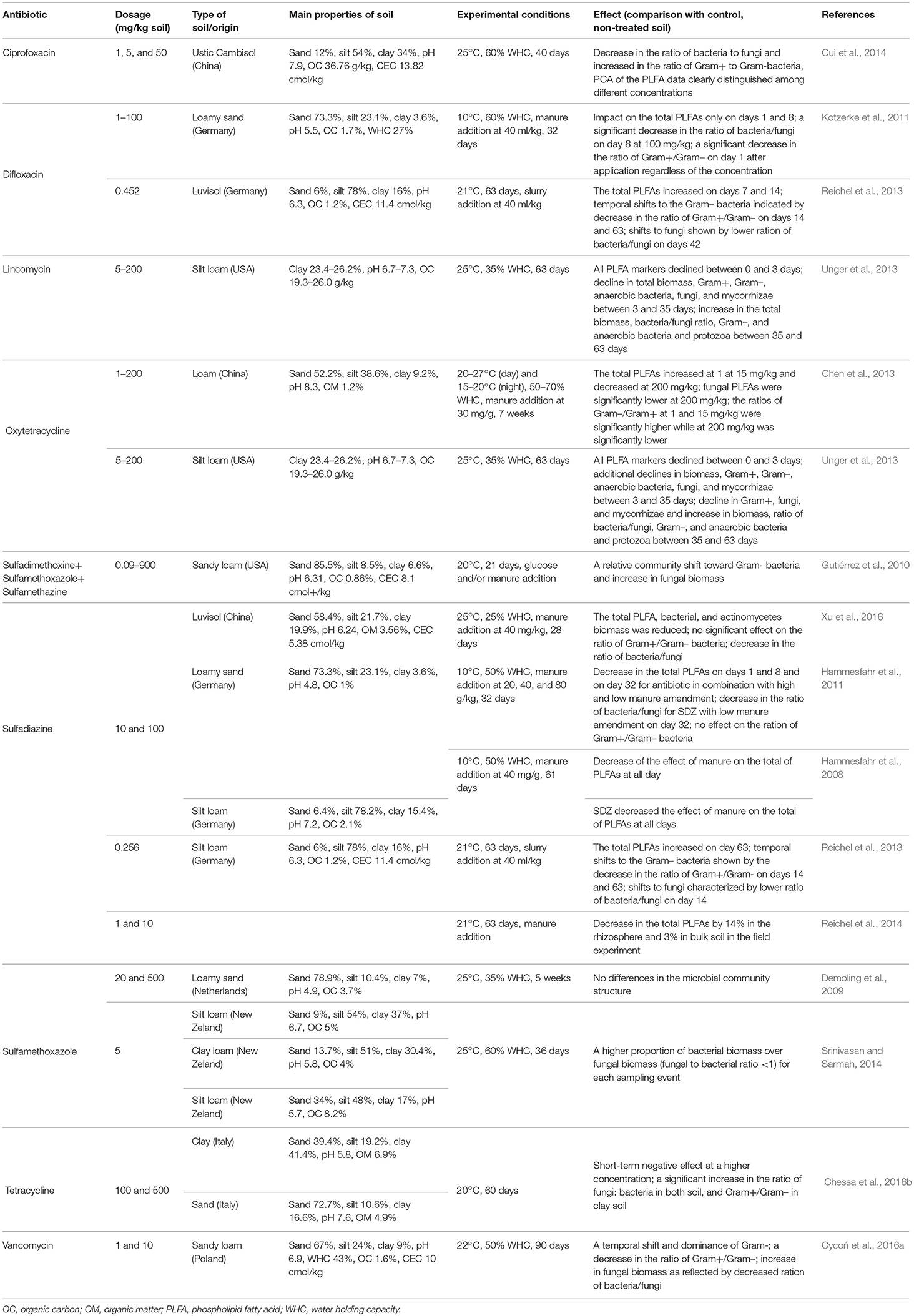
Table 7. Effects of the selected antibiotics on the community structure of microorganisms based on the PLFA analysis in soils with different characteristics.
An increase in total PLFA biomass was also observed in soils treated with sulfadiazine and difloxacin in slurry from pigs medicated with these antibiotics (Reichel et al., 2013). Influences from the difloxacin-slurry on PLFA biomass were observed at the beginning of the experiment (7 and 14 days), but only on the last sampling day (63 day) in the sulfadiazine-slurry. The authors hypothesized that the delayed effect of sulfadiazine was a result of continuous remobilization of antibiotic residues bound to soil particles by microorganisms. Hammesfahr et al. (2008) and Kotzerke et al. (2011), in contrast, found no changes in soil microbial communities upon addition of the above-mentioned antibiotics into soil spiked with slurry.
Several studies showed changes in the soil fungal PLFA signatures upon amendment with antibiotics. For example, the content of fungal marker fatty acids (18:1ω9 and 18:2ω6,9) significantly increased in alfalfa-amended soil and both alfalfa- and manure-amended soils with 20 and 500 mg sulfamethoxazole /kg of soil (Demoling et al., 2009). Gutiérrez et al. (2010) mixed three sulfonamides, sulfadimethoxine, sulfamethoxazole, and sulfamethazine, and observed a general increase in the proportion of fungal PLFAs among the total soil biomass PLFAs. A shift in the soil microbial community toward more fungi because of sulfapyridine application was also noted by Thiele-Bruhn and Beck (2005). The increase in the abundance of fungi may ultimately result in a decline in the productivity and quality of soils and agricultural products (Ding and He, 2010).
The PLFA method, previously used extensively for determination of microbial community composition, has been largely replaced by techniques based on the analysis of nucleic acids extracted from soil. However, PLFA-based analyses remain an adequately sensitive, efficient way to rapidly screen whether a microbial community has been affected by antibiotics.
Many authors have used PCR-dependent DNA fingerprinting techniques such as a terminal restriction fragment length polymorphism (T-RFLP) or denaturing gradient gel electrophoresis (DGGE) to evaluate the impact of antibiotics on diversity of soil microorganisms (Müller et al., 2002; Reichel et al., 2014; Chessa et al., 2016a; Orlewska et al., 2018a,b). Most of these studies target 16S rRNA genes and show altered diversity upon antibiotics applications. These changes may be explained by the disappearance of sensitive bacteria and outgrowth of resistant bacteria present in the antibiotic-contaminated soils (Table 8). For example, the application of tylosin (2 mg/kg soil) strongly modified the DGGE pattern, reducing the number of bands observed on days 15 and 22 of the experiment compared to non-exposed soil. However, on day 33 the difference was smaller, yet detectable (Westergaard et al., 2001). In a study by Müller et al. (2002), the DGGE pattern of 16S rDNA in tylosin-treated soil differed slightly to the diversity to the control soil, although colony morphology typing and potential of microbial communities to utilize selected substrates did not reveal any differences. Cycoń et al. (2016a) found that vancomycin dosed to soil at 10 mg/kg resulted in quantitative changes in microbial community patterns, suggesting selection exerted by the antibiotic. The changes in DGGE patterns suggest a decrease in populations of a species or group of species; revealing that some bacterial species were more sensitive to vancomycin than others were. This was corroborated by a decline in the values of H' and S (richness) indices. Another study showed significant effects of chlortetracycline on the microbial community that lasted for no longer than 1 week after the application of the antibiotic as the communities recovered over time until the end of the experiment (42 day) (Nelson et al., 2011). Contrarily, using PCR-DGGE Zielezny et al. (2006) showed that the application of sulfadiazine and chlortetracycline at concentrations of 1, 10, and 50 mg/kg soil had no significant effect on the structure of the soil microbial community. In another study, changes in DGGE profiles showed that sulfadiazine (10 and 100 μg/g soil) applied to soil in manure altered bacterial diversity (Hammesfahr et al., 2008). Although the DGGE profiles proved the impact of manure and sulfadiazine on the total soil bacterial communities, these effects were less distinct for pseudomonads and β-Proteobacteria. This observation may be explained by the resistance of many strains to sulfonamides, which may be only indirectly affected by sulfadiazine. Genetic changes within the β-Proteobacteria and the pseudomonads, expressed by the appearance or loss of a band in the DGGE profiles from manure, and sulfadiazine-amended soils were also reported by Reichel et al. (2014). However, statistical analysis showed that the β-Proteobacteria significantly responded to the time and moisture regimes, whereas the pseudomonads responded to the factors of time and treatment. The effects of sulfadiazine-containing manure on microbial diversity resulted in a low stability of soil bacterial communities and dominance of taxa that are known to contain human pathogens, such as Gemmatimonas, Leifsonia, Devosia, Clostridium, Shinella, and Peptostreptococcus (Ding et al., 2014). No apparent effects on microbial diversity in response to streptomycin and difloxacin were observed in similar studies (Shade et al., 2013; Jechalke et al., 2014a).
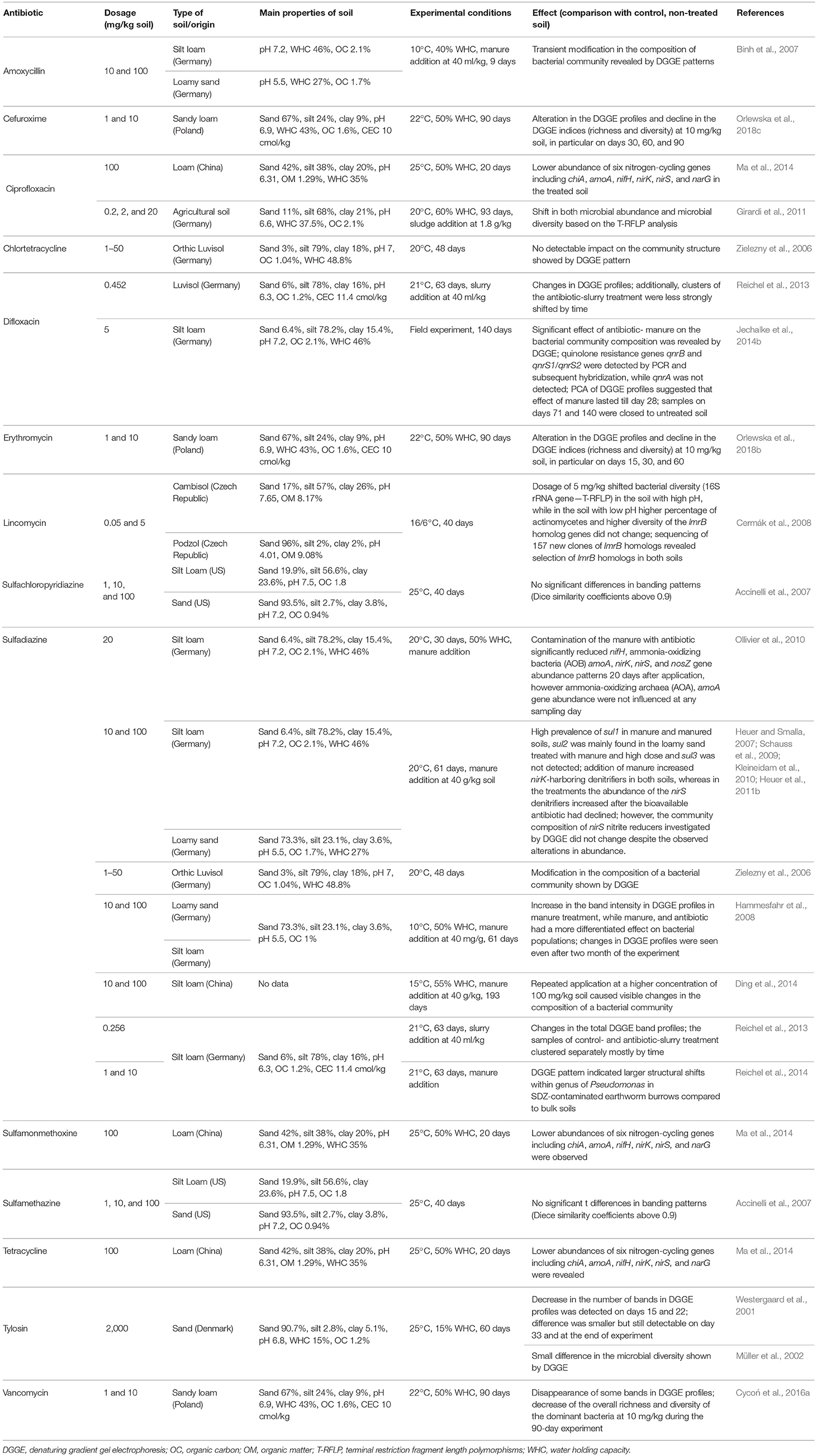
Table 8. Effects of the selected antibiotics on the community structure of microorganisms based on the genetic analyses in soils with different characteristics.
Studies are increasingly focusing on genes for antimicrobial resistance itself, which can be followed by metagenomic sequencing approaches. Resistance determinants present in the soil, defined as the soil resistome, include genes that confer antibiotic resistance to pathogenic and non-pathogenic species present in the environment, as well as proto-resistance genes that serve as a source of resistance elements (D'Costa et al., 2006, 2007; Perry et al., 2014). The potential transfer of antibiotic resistance among microbial populations in soil and the risk of animal and human infection are major concerns (Figure 5).
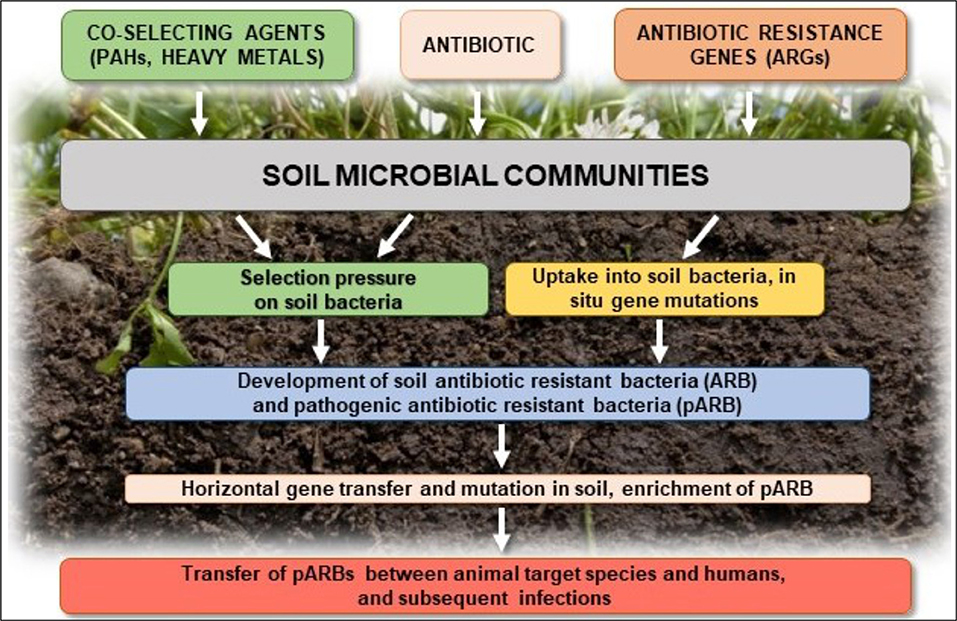
Figure 5. Transfer of antibiotic resistance in soil and risk of animal and human infection (based on Ashbolt et al., 2013).
Special attention is given to the detection and quantitation of ARGs, identifying resistance mechanisms, discovery of novel enzymes with unexpected activities, and the impact of pollutants and agricultural practices on the abundance of ARGs in soils. Resistance to antibiotics has been documented even in bacteria inhabiting pristine soils never exposed to these compounds, for example genes conferring resistance to common antibiotics (β-lactams, fluoroquinolones, and tetracyclines) were found in the Tibet (Chen et al., 2016). Interestingly, a tiny fraction of ARGs located on MGEs has been discovered, indicating a low, but real, potential for these ARGs to be transferred among bacteria (Chen et al., 2016). Additionally, some ARGs that have been discovered encode efflux pumps that are completely different from previously known efflux pumps. A large number (177) of ARGs encoding mostly single or multi-drug efflux pumps conferring resistance to aminoglycosides, chloramphenicol and β-lactams were reported in pristine Antarctic soils (Van Goethem et al., 2018). A low number and diversity of ARGs was detected in glacial soil and permafrost as compared to environments highly impacted by human activities in a study by Zhang et al. (2018). It has been proposed that ARGs in pristine environments most likely represent functional historical genes conferring resistance to natural antibiotics. Antibiotic resistance appeared to be transferred vertically over generations with limited to no horizontal transfer of ARGs between species (Van Goethem et al., 2018).
Diversity and abundance of ARGs is strongly influenced by agricultural practices (Wepking et al., 2017; Xiong et al., 2018). It has been documented that the spreading of manure from animals treated with different antibiotics on fields affects the abundance and diversity of ARGs (Su et al., 2014; Kyselková et al., 2015). However, it has also been documented that organic matter content, pH, and the history of soil management are important factors influencing the fate and abundance of ARGs in the soil (Cermák et al., 2008; Popowska et al., 2012).
The genes conferring resistance to minocycline, tetracycline, streptomycin, gentamycin, kanamycin, amikacin, chloramphenicol, and rifampicin in soils subjected to manure application accounted for ~70% of the total ARGs. More than half of the ARGs shared low similarity, < 60%, with the sequences of their closest proteins in GeneBank (Su et al., 2014). Among the ARGs studied (ampC, tetO, tetW, and ermB), the average abundance of ampC and tetO in the manure-treated soil was 421 and 3.3% greater in comparison with a non-treated control soil, respectively (Wepking et al., 2017). A broad-spectrum profile of ARGs has been found in 5 paddy soils of South China (Xiao et al., 2016). Based on Antibiotic Resistance Genes Database and next generation sequencing, 16 types of ARGs corresponding to 110 ARG subtypes were identified. Multi-drug resistance genes dominated in all soils (up to 47.5%), followed mainly by specific resistance genes for acriflavine, macrolide-lincosamide-streptogramin and bacitracin. Efflux pumps, antibiotic deactivation, and cellular protection were three major resistance mechanisms inferred from the resistome characterized Xiao et al. (2016).
An increase in the number and proportions of ARGs in the bacterial community was also shown by Nõlvak et al. (2016), who studied the effect of animal manure on the rate of the dissemination of ARGs (sul1, tetA, blaCTX-M, blaOXA2, and qnrS) and integron-integrase genes in soil. The abundance of genes for tetracycline resistance (tetM, tetO, and tetW) in soil that had had manure applied was significantly higher than in untreated soil (Xiong et al., 2018). It seems that an increasing abundance of ARGs resulted from inputs of ARGs already present in manure. This is because antibiotics may provide a significant selective advantage for some species or groups of antibiotic-resistant bacteria that then come to dominate the microbial community.
By contrast, many studies have shown that the dissemination of ARGs in soils is more correlated with the application of manure than with the presence of antibiotics. Though no effect on the abundance of tetG, tetO, and tetW was observed in one study, despite the presence of these genes in manure applied to the soil (Fahrenfeld et al., 2013), other studies have found an impact of manure from never antibiotic-treated animals on the number, diversity, and spread potential of ARGs (Udikovic-Kolic et al., 2014; Hu et al., 2016). This phenomenon can be explained by the stimulation of growth autochthonous bacteria and accelerated spread of ARGs by fertilizers free from antibiotics. Due to the ambiguous results of studies concerning the spread of ARGs in manure-treated soil, there is a need for future research to clearly assess the relative contribution of manure and antibiotics on microbial community structure and processes mediated by soil microorganisms.
The studies on the fate of ARGs in soil allowed not only to assess the diversity and abundance of genes of interest, but also to discover new ARGs, and described new resistance mechanisms and novel enzymes responsible for resistance of bacteria to antibiotics (Lau et al., 2017; Wang et al., 2017). Functional metagenomics for the investigation of antibiotic resistance led to the discovery of several tetracycline resistance genes from different classes in soil. Efflux pump genes, including 21 major facilitator superfamily efflux pump genes, were found to dominate the sequence libraries derived from studied soils. In addition, two new genes involved in the enzymatic inactivation of tetracycline were identified (Wang et al., 2017). Similarly, Torres-Cortés et al. (2011) described a new type of dihydrofolate reductase (protein Tm8-3; 26.8 kDa) conferring resistance to trimethoprim in soil microbiome. Using functional metagenomics they also identified three new antibiotic resistance genes conferring resistance to ampicillin, two to gentamicin, two to chloramphenicol, and four to trimethoprim in libraries generated from three different soil samples. Nine carbapenem-hydrolizing metallo-beta-lactamases (MBLs) including seven novel enzymes showing 33–59% similarity to previously described MBLs were discovered. Six originated from Proteobacteria, two from Bacteroidetes, and one from Gemmatimonadetes. MBLs were detected more frequently and exhibited higher diversity in grassland than in agricultural soil (Gudeta et al., 2016). In a study of resistome of bacterial communities in Canadian soils by functional metagenomics, 34 new ARGs with high homology to determinants conferring resistance aminoglycosides, sulfonamides and broad range of β-lactams were identified (Lau et al., 2017). High-resolution proteomics in combination with functional metagenomics resulted in the discovery of a new proline-rich peptide PPPAZI promoting resistance to various macrolides, but not to other ribosome-targeting antibiotics (Lau et al., 2017). Similarly, Donato et al. (2010) identified two novel bifunctional proteins responsible for bacteria resistance to ceftazidime and kanamycin, respectively.
Other synthetic pollutants can also influence the acquisition and maintenance of ARGs that can confer resistance not only to antibiotics but also to a number of structurally unrelated contaminants present in soil. As an example for this, ARGs were ~15 times more abundant in polycyclic aromatic hydrocarbon (PAH) contaminated soils than in similar non-contaminated soils (Chen et al., 2017). It resulted from the selection by PAHs of Proteobacteria containing ARGs encoded multi-drug efflux pumps leading to simultaneously enriching of ARGs carried by them in the soils. Moreover, most of ARGs (70%) found in the PAHs-contaminated soils were not carried by plasmids, indicating a low possibility of horizontal gene transfer (HGT) between bacteria (Chen et al., 2017).
It has recently been reported that heavy metals are also as a factor contributing to maintenance of ARGs in the environment. A study of dairy farms showed significant correlations between the abundance of ARGs, metal resistance genes and the content of Cu and Zn in cattle feces (Zhou et al., 2016). The role of heavy metals can be explained by cross-resistance, tolerance of bacteria to more than one antimicrobial agent (Chapman, 2003; Baker-Austin et al., 2006), and co-resistance, the localization of ARGs and genes encoding resistance to heavy metals and other antibacterial agents on mobile genetic elements (Chapman, 2003), mechanisms. Multi-drug efflux pumps, which mediate rapid extrusion of antibiotics and heavy metals from the cell decrease susceptibility toward these compounds, are an example of cross-resistance (Martinez et al., 2009). Co-resistance can occur due to the close arrangement of genes on a chromosome or extrachromosomal element, which increases the likelihood that genes are subject to combined transmission via HGT. Moreover, heavy metals not only trigger co-selection processes, but also increase the level of tolerance to antibiotics due to co-regulation of resistance genes (Baker-Austin et al., 2006). Such mechanisms may affect the spread of ARGs from environmental populations to bacteria of clinical importance even in the absence of direct antibiotics selection (Herrick et al., 2014).
Antibiotics concentrations in the environment are much lower than that used in therapeutic doses. However, even at sub-inhibitory concentrations (below the MIC), antibiotics may select for resistant phenotypes (Gullberg et al., 2011, 2014). It has been reported that resistant bacteria can be selected at antibiotic concentrations even 100-fold below lethal concentrations. Using data on MICs from the EUCAST database for 111 antibiotics, Bengtsson-Palme and Larsson (2016) estimated that the predicted no-effect concentrations (PNECs) range from 8 ng/L to 64 μg/L. Mutants selected by antibiotics at minimum selective concentration appeared to be more fit than those selected at high concentration and still highly resistant (Gullberg et al., 2011; Sandegren, 2014). Antibiotics at such concentrations play multifaceted roles in the soil, including influencing inter-species competition, signal communication, host–parasite interactions, virulence modulation, and biofilm formation (Sengupta et al., 2013). Moreover, they induce mutations, genetic recombination, and HGT processes (Andersson and Hughes, 2012, 2014). Sub-lethal concentrations of drugs can also increase the mutation rate and potential for enrichment of stable genetic mutants by the induction of SOS responses, increasing translation misreading, and generation of oxygen radicals. It has been reported that sub-lethal concentrations of tetracycline stimulated up to 1,000-fold higher horizontal transfer rates of mobile genetic elements in Listeria monocytogenes (Bahlm et al., 2004) and integron recombination rates in Vibrio cholerae (Guerin et al., 2009). However, the rate of spread of ARGs depends on the antibiotic concentration in the soil. Huang et al. (2016) showed that the dissipation rate of plasmid-located genes [i.e., qnrS, oqxA, aac(6′)-Ib-cr] was significantly lower in soil treated with ciprofloxacin at concentrations of 0.04 and 0.4 mg/kg soil compared to soil treated with antibiotic at 4 mg/kg and controls. Forsberg et al. (2012) also found little evidence for HGT in soil.
Because of the risks posed by the dissemination of ARGs among bacteria and plants, studies investigating new practices designed to decrease the accumulation and transport of ARGs within soil are being conducted. For example, the use of biochar as a soil amendment significantly reduced the abundance of tetracyclines (tetC, tetG, tetW, and tetX) and sulfonamides (sulI and sulII) resistance genes in soil and lettuce tissues (Ye et al., 2016; Duan et al., 2017).
Environmental antibiotic pollution is a problem that is expected to gain more attention in the near future since antibiotic consumption is still increasing around the world. A review of available literature shows that transformation and/or degradation are the most important processes that determine the fate of antibiotics in soils and that soil microorganisms play an important role in these processes. However, the rate of these transformation and degradation depends largely on the antibiotic structure and is affected by many abiotic and biotic factors, as is most evident in the large range of DT50 values of antibiotics in different soils. Studies have shown that within groups of similar antibiotics or even particular antibiotics, DT50 values differ significantly because of various soil properties, antibiotic dosage, and environmental conditions.
Current literature reviewed here indicates that the input of antibiotics into soil alters the structure and activity of microbial communities and the abundance of ARGs. However, results from studies in this field are often ambiguous, making environmental risk assessments related to the presence of antibiotics depend upon too many different factors to be reliable. Interactions between soil, antibiotics, and microorganisms are multifarious and many environmental factors may influence the value of tested parameters. The effect of antibiotics on the activity and diversity of microbial communities depends on the physicochemical parameters of the soil, the antimicrobial activity, and dosage of the antibiotic, as well as the time of exposure. It has become clear that microorganisms that are sensitive to different antibiotics are killed or inhibited in the presence of antibiotics, which may result in outgrowth of resistant bacteria resistant. In turn, this may alter the diversity of the soil microbial communities. Contrarily, there is some evidence that certain microorganisms can adapt to and possibly transform antibiotics. The less toxic transformation products would favor recovery of the original microbial communities from the initial disturbances caused by antibiotics exposure. Several studies found transient negative effects of antibiotics on the functional, structural and genetic diversity of soil microbial communities; a temporary loss of soil functionality with subsequent recovery.
Existing OECD and ISO ecotoxicological tests used to evaluate the toxicity of chemicals on the activity of a single species (e.g., Microtox, MARA, measurements of selected enzyme activities) and some of the processes mediated by soil microorganisms are not sufficient to gauge the effects of antibiotics on soil microbial communities. Therefore, ecotoxicological tests that combine various microbial community parameters should be developed. Moreover, future studies should be based on “omics” approaches such as genomics, transcriptomics, and proteomics, that allow for deeper examination of microbial communities than CLPP, PLFA, and PCR-DGGE methods. “Omics” methods are better tools for tracking the fate and determining dissipation rate of ARGs, as well as for recognizing new mechanisms of antibiotic resistance. Future frameworks for antibiotics should focus on the effect of soil properties on the maintenance and development of antibiotic resistance, the fate of antibiotics and ARGs in manure-fertilized soils, the rate of HGT between antibiotic-resistant and autochthonous bacteria, the link between antibiotic concentrations and genetic changes in soil resistomes, and the role of various contaminants and co-selecting agents on maintenance of ARGs in soil. Moreover, there is a need to complement biological assays with advanced analytical methods (such as LC-MS/MS, isotope dilution mass-spectrometry) and proper sample preparation, allowing assessment of antibiotic concentrations and, especially, of bioavailability in complex environmental matrices. Other problems that should be taken into consideration are variations on the procedures for estimating the limits of antibiotic detection and the lack of standard analytical methods for monitoring antibiotic in the environment. Future studies should also be focused on design and management practices that minimize the spread of ARGs from harvested crops in antibiotic-treated soils into the food chain. Finally, though it is rarely studied, the genetic potential of microorganisms involved in the degradation of antibiotics in soil warrant wider investigation.
MC, AM, and ZP-S conceived and designed content of the paper and wrote the paper. MC prepared the figures and tables.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer UNR and handling Editor declared their shared affiliation.
This work was financed by the National Science Center, Poland (No. DEC-2014/15B/NZ9/04414).
Accinelli, C., Koskinen, W. C., Becker, J. M., and Sadowsky, M. J. (2007). Environmental fate of two sulfonamide antimicrobial agents in soil. J. Agric. Food Chem. 55, 2677–2682. doi: 10.1021/jf063709j
Aga, D. S., Lenczewski, M., Snow, D., Muurinen, J., Sallach, J. B., and Wallace, J. S. (2016). Challenges in the measurement of antibiotics and in evaluating their impacts in agroecosystems: a critical review. J. Environ. Qual. 45, 407–419. doi: 10.2134/jeq2015.07039
Akimenko, Y. V., Kazeev, K. S., and Kolesnikov, S. I. (2015). Impact assessment of soil contamination with antibiotics (For example, an ordinary chernozem). Am. J. Appl. Sci. 12, 80–88. doi: 10.3844/ajassp.2015.80.88
Alef, K. (1995). “Dehydrogenase activity,” in Methods in Applied Soil Microbiology and Biochemistry, eds K. Alef and P. Nannipieri. (London: Academic), 228–231.
Amorim, C. L., Moreira, I. S., Maia, A. S., Tiritan, M. E., and Castro, P. M. L. (2014). Biodegradation of ofloxacin, norfloxacin, and ciprofloxacin as single and mixed substrates by Labrys portucalensis F11. Appl. Microbiol. Biotechnol. 98, 3181–3190. doi: 10.1007/s00253-013-5333-8
Andersson, D. I., and Hughes, D. (2012). Evolution of antibiotic resistance at non-lethal drug concentration. Drug Resist. Updat. 15, 162–172. doi: 10.1016/j.drup.2012.03.005
Andersson, D. I., and Hughes, D. (2014). Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12, 465–478. doi: 10.1038/nrmicro3270
Ashbolt, N. J., Amézquita, A., Backhaus, T., Borriello, P., Brandt, K. K., Collignon, P., et al. (2013). Human health risk assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ. Health Perspect. 121, 993–1001. doi: 10.1289/ehp.1206316
Atashgahi, S., Sánchez-Andrea, I., Heipieper, H. J., van der Meer, J. R., Stams, A. J.M., and Smidt, H. (2018). Prospects for harnessing biocide resistance for bioremediation and detoxification. Science 360, 743–746. doi: 10.1126/science.aar3778
Aust, M.-O., Godlinski, F., Travis, G. R., Hao, X., McAllister, T. A., Leinweber, P., et al. (2008). Distribution of sulfamethazine, chlortetracycline and tylosin in manure and soil of Canadian feedlots after subtherapeutic use in cattle. Environ. Pollut. 156, 1243–1251. doi: 10.1016/j.envpol.2008.03.011
Aust, M.-O., Thiele-Bruhn, S., Seeger, J., Godlinski, F., Meissner, R., and Leinweber, P. (2010). Sulfonamides leach from sandy loam soils under common agricultural practice. Water. Air. Soil Pollut. 211, 143–156. doi: 10.1007/s11270-009-0288-1
Awad, Y. M., Kim, S.-C., Abd El-Azeem, S. A. M., Kim, K.-H., Kim, K.-R., Kim, K., et al. (2014). Veterinary antibiotics contamination in water, sediment, and soil near a swine manure composting facility. Environ. Earth Sci. 71, 1433–1440. doi: 10.1007/s12665-013-2548-z
Awad, Y. M., Ok, Y. S., Igalavithana, A. D., Lee, Y. H., Sonn, Y.-K., Usman, A. R. A., et al. (2016). Sulphamethazine in poultry manure changes carbon and nitrogen mineralisation in soils. Chem. Ecol. 32, 899–918. doi: 10.1080/02757540.2016.1216104
Bahlm, M. I., Sørensen, S. J., Hansen, L. H., and Licht, T. R. (2004). Effect of tetracycline on transfer and establishment of the tetracycline-inducible conjugative transposon Tn916 in the guts of gnotobiotic rats. Appl. Environ. Microbiol. 70, 758–764. doi: 10.1128/AEM.70.2.758-764.2004
Bak, S. A., Hansen, M., Krogh, K. A., Brandt, A., Halling-Sørensen, B., and Björklund, E. (2013). Development and validation of an SPE methodology combined with LC-MS/MS for the determination of four ionophores in aqueous environmental matrices. Int. J. Environ. Anal. Chem. 93, 1500–1512. doi: 10.1080/03067319.2013.763250
Baker-Austin, C., Wright, M. S., Stepanauskas, R., and McArthur, J. V. (2006). Co-selection of antibiotic and metal resistance. Trends Microbiol.14, 176–182. doi: 10.1016/j.tim.2006.02.006
Barra Caracciolo, A., Topp, E., and Grenni, P. (2015). Pharmaceuticals in the environment: biodegradation and effects on natural microbial communities. A review. J. Pharm. Biomed. Anal. 106, 25–36. doi: 10.1016/j.jpba.2014.11.040
Bengtsson-Palme, J., and Larsson, D. G. J. (2016). Concentrations of antibiotics predicted to select for resistant bacteria: proposed limits for environmental regulation. Environ. Int. 86, 140–149. doi: 10.1016/j.envint.2015.10.015
Berendsen, B. J. A., Wegh, R. S., Memelink, J., Zuidema, T., and Stolker, A. A. M. (2015). The analysis of animal feces as a tool to monitor antibiotic usage. Talanta 132, 258–268. doi: 10.1016/j.talanta.2014.09.022
Binh, C. T. T., Heuer, H., Gomes, N. C. M., Kotzerke, A., Fulle, M., Wilke, B. M., et al. (2007). Reichel et aShort-term effects of amoxicillin on bacterial communities in manured soil. FEMS Microbiol. Ecol. 62, 290–302. doi: 10.1111/j.1574-6941.2007.00393.x
Blackwell, P. A., Kay, P., and Boxall, A. B. A. (2007). The dissipation and transport of veterinary antibiotics in a sandy loam soil. Chemosphere 67, 292–299. doi: 10.1016/j.chemosphere.2006.09.095
Bouki, C., Venieri, D., and Diamadopoulos, E. (2013). Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: a review. Ecotoxicol. Environ. Saf. 91, 1–9. doi: 10.1016/j.ecoenv.2013.01.016
Boxall, A. B. A., Blackwell, P., Cavallo, R., Kay, P., and Tolls, J. (2002). The sorption and transport of a sulphonamide antibiotic in soil systems. Toxicol. Lett. 131, 19–28. doi: 10.1016/S0378-4274(02)00063-2
Boxall, A. B. A., Johnson, P., Smith, E. J., Sinclair, C. J., Stutt, E., and Levy, L. S. (2006). Uptake of veterinary medicines from soils into plants. J. Agric. Food Chem. 54, 2288–2297. doi: 10.1021/jf053041t
Boyd, S. A., and Mortland, M. M. (1990). “Enzyme interactions with clays and clay-organic matter complexes,” in Soil Biochemistry Vol. 6, eds J. M. Bollag and G. Stotzky (New York, NY: Marcel Dekker Inc.), 1–28.
Brandt, K. K., Amézquita, A., Backhaus, T., Boxall, A., Coors, A., Heberer, T., et al. (2015). Ecotoxicological assessment of antibiotics: a call for improved consideration of microorganisms. Environ. Int. 85, 189–205. doi: 10.1016/j.envint.2015.09.013
Brandt, K. K., Sjøholm, O. R., Krogh, K. A., Halling-Sørensen, B., and Nybroe, O. (2009). Increased pollution-induced bacterial community tolerance to sulfadiazine in soil hotspots amended with artificial root exudates. Environ. Sci. Technol. 43, 2963–2968. doi: 10.1021/es803546y
Braschi, I., Blasioli, S., Fellet, C., Lorenzini, R., Garelli, A., Pori, M., et al. (2013). Persistence and degradation of new β-lactam antibiotics in the soil and water environment. Chemosphere 93, 152–159. doi: 10.1016/j.chemosphere.2013.05.016
Carlson, J. C., and Mabury, S. A. (2006). Dissipation kinetics and mobility of chlortetracycline, tylosin, and monensin in an agricultural soil in Northumberland County, Ontario, Canada. Environ. Toxicol. Chem. 25, 1–10. doi: 10.1897/04-657R.1
Carter, L. J., Harris, E., Williams, M., Ryan, J. J., Kookana, R. S., and Boxall, A. B. A. (2014). Fate and uptake of pharmaceuticals in soil-plant systems. J. Agric. Food Chem. 62, 5955–5963. doi: 10.1021/jf404282y
CDC Centers for Disease (2015). Control and Prevention. Outpatient Antibiotic Prescription–United States, (Atlanta).
Cermák, L., Kopecký, J., Novotná, J., Omelka, M., Parkhomenko, N., Plháčková, K., et al. (2008). Bacterial communities of two contrasting soils reacted differently to lincomycin treatment. Appl. Soil Ecol. 40, 348–358. doi: 10.1016/j.apsoil.2008.06.001
Chapman, J. S. (2003). Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int. Biodeterior. Biodegr. 51, 271–276. doi: 10.1016/S0964-8305(03)00044-1
Chen, B., He, R., Yuan, K., Chen, E., Lin, L., Chen, X., et al. (2017). Polycyclic aromatic hydrocarbons (PAHs) enriching antibiotic resistance genes (ARGs) in the soils. Environ. Pollut. 220, 1005–1013. doi: 10.1016/j.envpol.2016.11.047
Chen, B., Yuan, K., Chen, X., Yang, Y., Zhang, T., Wang, Y., et al. (2016). Metagenomic analysis revealing antibiotic resistance genes (ARGs) and their genetic compartments in the Tibetan Environment. Environ. Sci. Technol. 50, 6670–6679. doi: 10.1021/acs.est.6b00619
Chen, C., Li, J., Chen, P., Ding, R., Zhang, P., and Li, X. (2014). Occurrence of antibiotics and antibiotic resistances in soils from wastewater irrigation areas in Beijing and Tianjin, China. Environ. Pollut. 193, 94–101. doi: 10.1016/j.envpol.2014.06.005
Chen, W., Liu, W., Pan, N., Jiao, W., and Wang, M. (2013). Oxytetracycline on functions and structure of soil microbial community. J. Soil Sci. Plant Nutr. 13, 967–975. doi: 10.4067/S0718-95162013005000076
Chen, Y. S., Zhang, H. B., Luo, Y. M., and Song, J. (2012). Occurrence and assessment of veterinary antibiotics in swine manures: a case study in East China. Chinese Sci. Bull. 57, 606–614. doi: 10.1007/s11434-011-4830-3
Cheng, M., Wu, L., Huang, Y., Luo, Y., and Christie, P. (2014). Total concentrations of heavy metals and occurrence of antibiotics in sewage sludges from cities throughout China. J. Soils Sediments 14, 1123–1135. doi: 10.1007/s11368-014-0850-3
Chessa, L., Jechalke, S., Ding, G.-C., Pusino, A., Mangia, N. P., and Smalla, K. (2016a). The presence of tetracycline in cow manure changes the impact of repeated manure application on soil bacterial communities. Biol. Fertil. Soils 52, 1121–1134. doi: 10.1007/s00374-016-1150-4
Chessa, L., Pusino, A., Garau, G., Mangia, N. P., and Pinna, M. V. (2016b). Soil microbial response to tetracycline in two different soils amended with cow manure. Environ. Sci. Pollut. Res. 23, 5807–5817. doi: 10.1007/s11356-015-5789-4
Conkle, J. L., Lattao, C., White, J. R., and Cook, R. L. (2010). Competitive sorption and desorption behavior for three fluoroquinolone antibiotics in a wastewater treatment wetland soil. Chemosphere 80, 1353–1359. doi: 10.1016/j.chemosphere.2010.06.012
Crane, M., Boxall, A. B. A., and Barret, K. (2010). Veterinary Medicines in the Environment. Boca Raton, FL: CRC Press.
Cui, H., Wang, S.-P., Fu, J., Zhou, Z.-Q., Zhang, N., and Guo, L. (2014). Influence of ciprofloxacin on microbial community structure and function in soils. Biol. Fertil. Soils 50, 939–947. doi: 10.1007/s00374-014-0914-y
Cycoń, M., Borymski, S., Orlewska, K., Wasik, T. J., and Piotrowska-Seget, Z. (2016b). An analysis of the effects of vancomycin and/or vancomycin-resistant Citrobacter freundii exposure on the microbial community structure in soil. Front. Microbiol. 7:1015. doi: 10.3389/fmicb.2016.01015
Cycoń, M., Borymski, S., Zołnierczyk, B., and Piotrowska-Seget, Z. (2016a). Variable effects of non-steroidal anti-inflammatory drugs (NSAIDs) on selected biochemical processes mediated by soil microorganisms. Front. Microbiol. 7:1969. doi: 10.3389/fmicb.2016.01969
Cycoń, M., Orlewska, K., Markowicz, A., Zmijowska, A., Smolen-Dzirba, J., Bratosiewicz-Wasik, J., et al. (2018). Vancomycin and/or multidrug-resistant Citrobacter freundii altered the metabolic pattern of soil microbial community. Front. Microbiol. 9:1047. doi: 10.3389/fmicb.2018.01047
Cycoń, M., and Piotrowska-Seget, Z. (2015). Biochemical and microbial soil functioning after application of the insecticide imidacloprid. J. Environ. Sci. 27, 147–158. doi: 10.1016/j.jes.2014.05.034
Cycoń, M., and Piotrowska-Seget, Z. (2016). Pyrethroid-degrading microorganisms and their potential for the bioremediation of contaminated soils: a review. Front. Microbiol. 7:1463. doi: 10.3389/fmicb.2016.01463
Cycoń, M., Zmijowska, A., and Piotrowska-Seget, Z. (2011). Biodegradation kinetics of 2,4-D by bacterial strains isolated from soil. Cent. Eur. J. Biol. 6, 188–198. doi: 10.2478/s11535-011-0005-0
Daghrir, R., and Drogui, P. (2013). Tetracycline antibiotics in the environment: a review. Environ. Chem. Lett. 11, 209–227. doi: 10.1007/s10311-013-0404-8
Dantas, G., Sommer, M. O. A., Oluwasegun, R. D., and Church, G. M. (2008). Bacteria subsisting on antibiotics. Science 320, 100–103. doi: 10.1126/science.1155157
Davis, J. G., Truman, C. C., Kim, S. C., Ascoughd, J. C., and Carlsone, K. (2006). Antibiotic transport via runoff and soil loss. J. Environ. Qual. 35, 2250–2260. doi: 10.2134/jeq2005.0348
D'Costa, V. M., Griffith, E., and Wright, G. D. (2007). Expanding the soil antibiotic resistome: exploring environmental diversity. Curr. Opin. Microbiol. 10, 481–489. doi: 10.1016/j.mib.2007.08.009
D'Costa, V. M., McGrann, K. M., Hughes, D. W., and Wright, G. D. (2006). Sampling the antibiotic resistome. Science 311, 374–377. doi: 10.1126/science.1120800
De la Torre, A., Iglesias, I., Carballo, M., Ramírez, P., and Muñoz, M. J. (2012). An approach for mapping the vulnerability of European Union soils to antibiotic contamination. Sci. Total Environ. 414, 672–679. doi: 10.1016/j.scitotenv.2011.10.032
Demoling, L. A., Bååth, E., Greve, G., Wouterse, M., and Schmitt, H. (2009). Effects of sulfamethoxazole on soil microbial communities after adding substrate. Soil Biol. Biochem. 41, 840–848. doi: 10.1016/j.soilbio.2009.02.001
DeVries, S. L., Loving, M., Li, X., and Zhang, P. (2015). The effect of ultralow-dose antibiotics exposure on soil nitrate and N2O flux. Sci. Rep. 5:16818. doi: 10.1038/srep16818
DeVries, S. L., and Zhang, P. (2016). Antibiotics and the terrestrial nitrogen cycle: a review. Curr. Pollut. Rep. 2, 51–67. doi: 10.1007/s40726-016-0027-3
Ding, C., and He, J. (2010). Effect of antibiotics in the environment on microbial populations. Appl. Microbiol. Biotechnol. 87, 925–941. doi: 10.1007/s00253-010-2649-5
Ding, G. C., Radl, V., Schloter-Hai, B., Jechalke, S., Heuer, H., Smalla, K., et al. (2014). Dynamics of soil bacterial communities in response to repeated application of manure containing sulfadiazine. PLoS ONE 9:e92958. doi: 10.1371/journal.pone.0092958
Ding, Y., Zhang, W., Gu, C., Xagoraraki, I., and Li, H. (2011). Determination of pharmaceuticals in biosolids using accelerated solvent extraction and liquid chromatography/tandem mass spectrometry. J. Chromatogr. A 1218, 10–16. doi: 10.1016/j.chroma.2010.10.112
Dolliver, H., Gupta, S., and Noll, S. (2008). Antibiotic degradation during manure composting. J. Environ. Qual. 37:1245. doi: 10.2134/jeq2007.0399
Dolliver, H., Kumar, K., and Gupta, S. (2007). Sulfamethazine uptake by plants from manure-amended soil. J. Environ. Qual. 36:1224. doi: 10.2134/jeq2006.0266
Donato, J. J., Moe, L. A., Converse, B. J., Smart, K. D., Berklein, E. C., McManus, P. S., et al. (2010). Metagenomic analysis of apple orchard soil reveals antibiotic resistance genes encoding predicted bifunctional proteins. Appl. Environ. Microbiol. 76, 4396–4401. doi: 10.1128/AEM.01763-09
Du, L., and Liu, W. (2012). Occurrence, fate, and ecotoxicity of antibiotics in agro-ecosystems. A review. Agron. Sustain. Dev. 32, 309–327. doi: 10.1007/s13593-011-0062-9
Duan, M., Li, H., Gu, J., Tuo, X., Sun, W., Qian, X., et al. (2017). Effects of biochar on reducing the abundance of oxytetracycline, antibiotic resistance genes, and human pathogenic bacteria in soil and lettuce. Environ. Pollut. 224, 787–795. doi: 10.1016/j.envpol.2017.01.021
ECDC (2018). ESAC-Net surveillance data (2017). “Summary of the latest data on antibiotic consumption in the European Union. European Centre for Disease Prevention and Control. Antimicrobial consumption,” in ECDC Annual Epidemiological Report for 2016. (Stockholm: ECDC).
Erickson, B. D., Elkins, C. A., Mullis, L. B., Heinze, T. M., Wagner, R. D., and Cerniglia, C. E. (2014). A metallo-β-lactamase is responsible for the degradation of ceftiofur by the bovine intestinal bacterium Bacillus cereus P41. Vet. Microbiol. 172, 499–504. doi: 10.1016/j.vetmic.2014.05.032
Fahrenfeld, N., Ma, Y., O'Brien, M., and Pruden, A. (2013). Reclaimed water as a reservoir of antibiotic resistance genes: distribution system and irrigation implications. Front. Microbiol. 4:130. doi: 10.3389/fmicb.2013.00130
Fan, Z., Casey, F. X. M., Hakk, H., Larsen, G. L., and Khan, E. (2011). Sorption, fate, and mobility of sulfonamides in soils. Water Air. Soil Pollut. 218, 49–61. doi: 10.1007/s11270-010-0623-6
Fang, H., Han, L., Cui, Y., Xue, Y., Cai, L., and Yu, Y. (2016). Changes in soil microbial community structure and function associated with degradation and resistance of carbendazim and chlortetracycline during repeated treatments. Sci. Total Environ. 572, 1203–1212. doi: 10.1016/j.scitotenv.2016.08.038
Fang, H., Han, Y., Yin, Y., Pan, X., and Yu, Y. (2014). Variations in dissipation rate, microbial function and antibiotic resistance due to repeated introductions of manure containing sulfadiazine and chlortetracycline to soil. Chemosphere 96, 51–56. doi: 10.1016/j.chemosphere.2013.07.016
Fatta-Kassinos, D., Kalavrouziotis, I. K., Koukoulakis, P. H., and Vasquez, M. I. (2011). The risks associated with wastewater reuse and xenobiotics in the agroecological environment. Sci. Total Environ. 409, 3555–3563. doi: 10.1016/j.scitotenv.2010.03.036
Fernández-Calviño, D., Bermúdez-Couso, A., Arias-Estévez, M., Nóvoa-Muñoz, J. C., Fernández-Sanjurjo, M. J., Álvarez-Rodríguez, E., et al. (2015). Competitive adsorption/desorption of tetracycline, oxytetracycline and chlortetracycline on two acid soils: stirred flow chamber experiments. Chemosphere 134, 361–366. doi: 10.1016/j.chemosphere.2015.04.098
Floch, C., Chevremont, A.-C., Joanico, K., Capowiez, Y., and Criquet, S. (2011). Indicators of pesticide contamination: soil enzyme compared to functional diversity of bacterial communities via Biolog® Ecoplates. Eur. J. Soil Biol. 47, 256–263. doi: 10.1016/j.ejsobi.2011.05.007
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O. A., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. doi: 10.1126/science.1220761
Förster, M., Laabs, V., Lamshöft, M., Groeneweg, J., Zühlke, S., Spiteller, M., et al. (2009). Sequestration of manure-applied sulfadiazine residues in soils. Environ. Sci. Technol. 43, 1824–1830. doi: 10.1021/es8026538
Frostegård, Å., Tunlid, A., and Bååth, E. (2011). Use and misuse of PLFA measurements in soils. Soil Biol. Biochem. 43, 1621–1625. doi: 10.1016/j.soilbio.2010.11.021
Frostegård, Å., Tunlid, A., and Baath, E. (1993). Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl. Environ. Microbiol. 59, 3605–3617.
Gao, L., Shi, Y., Li, W., Liu, J., and Cai, Y. (2015). Occurrence and distribution of antibiotics in urban soil in Beijing and Shanghai, China. Environ. Sci. Pollut. Res. 22, 11360–11371. doi: 10.1007/s11356-015-4230-3
Gavalchin, J., and Katz, S. E. (1994). The persistence of fecal-borne antibiotics in soil. J. Assoc. Anal. Chem. Int. 77, 481–485.
Gelband, H., Miller-Petrie, M., Pant, S., Gandra, S., Levinson, J., Barter, D., et al. (2015). “State of the world's antibiotics,” in Center for Disease Dynamics, Economics & Policy, eds CDDEP (Washington, DC: CDDEP), 1–84.
Ghosh, S., and LaPara, T. M. (2007). The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 1, 191–203. doi: 10.1038/ismej.2007.31
Ghosh, S., Sadowsky, M. J., Roberts, M. C., Gralnick, J. A., and LaPara, T. M. (2009). Sphingobacterium sp. strain PM2-P1-29 harbours a functional tet(X) gene encoding for the degradation of tetracycline. J. Appl. Microbiol. 106, 1336–1342. doi: 10.1111/j.1365-2672.2008.04101.x
Gianfreda, L., Sannino, F., Ortega, N., and Nannipieri, P. (1994). Activity of free and immobilized urease in soil: effects of pesticides. Soil Biol. Biochem. 26, 777–784. doi: 10.1016/0038-0717(94)90273-9
Gil-Sotres, F., Trasar-Cepeda, C., Leirós, M. C., and Seoane, S. (2005). Different approaches to evaluating soil quality using biochemical properties. Soil Biol. Biochem. 37, 877–887. doi: 10.1016/j.soilbio.2004.10.003
Girardi, C., Greve, J., Lamshöft, M., Fetzer, I., Miltner, A., Schäffer, A., et al. (2011). Biodegradation of ciprofloxacin in water and soil and its effects on the microbial communities. J. Hazard. Mater. 198, 22–30. doi: 10.1016/j.jhazmat.2011.10.004
Göbel, A, Thomsen, A., McArdell, C. S., Alder, A. C, Giger, W., Theiß, N., Löffler, D., et al. (2005). Extraction and determination of sulfonamides, macrolides, and trimethoprim in sewage sludge. J. Chromatogr. A 1085, 179–189. doi: 10.1016/j.chroma.2005.05.051
Grenni, P., Ancona, V., and Barra Caracciolo, A. (2018). Ecological effects of antibiotics on natural ecosystems: a review. Microchem. J. 136, 25–39. doi: 10.1016/j.microc.2017.02.006
Grote, M., Schwake-Anduschus, C., Michel, R., Stevens, H., Heyser, W., Langenkämper, G., et al. (2007). Incorporation of veterinary antibiotics into crops from manured soil. Landbauforsch. Volkenrode 57, 25–32.
Gudeta, D. D., Bortolaia, V., Pollini, S., Docquier, J.-D., Rossolini, G. M., Amos, A., et al. (2016). Expanding the repertoire of carbapenem-hydrolyzing metallo-β-lactamases by functional metagenomic analysis of soil microbiota. Front. Microbiol. 7:1985. doi: 10.3389/fmicb.2016.01985
Guerin, E., Cambray, G., Sanchez-Alberola, N., Campoy, S., Erill, I., Da Re, S., et al. (2009). The SOS response controls integron recombination. Science 324:1034. doi: 10.1126/science.1172914
Gulkowska, A., Leung, H. W., So, M. K., Taniyasu, S., Yamashita, N., Yeung, L. W. Y., et al. (2008). Removal of antibiotics from wastewater by sewage treatment facilities in Hong Kong and Shenzhen, China. Water Res. 42, 395–403. doi: 10.1016/j.watres.2007.07.031
Gullberg, E., Albrecht, L. M., Karlsson, C., Sandegren, L., and Andersson, D. I. (2014). Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio 5, e01918–e01914. doi: 10.1128/mBio.01918-14
Gullberg, E., Cao, S., Berg, O. G., Ilbäck, C., Sandegren, L., Hughes, D., et al. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158. doi: 10.1371/journal.ppat.1002158
Gutiérrez, I. R., Watanabe, N., Harter, T., Glaser, B., and Radke, M. (2010). Effect of sulfonamide antibiotics on microbial diversity and activity in a Californian Mollic Haploxeralf. J. Soils Sediments 10, 537–544. doi: 10.1007/s11368-009-0168-8
Halling-Sørensen, B., Jacobsen, A.-M., Jensen, J., Sengeløv, G., Vaclavik, E., and Ingerslev, F. (2005). Dissipation and effects of chlortetracycline and tylosin in two agricultural soils: a field-scale study in southern Denmark. Environ. Toxicol. Chem. 24, 802–810. doi: 10.1897/03-576.1
Hammesfahr, U., Bierl, R., and Thiele-Bruhn, S. (2011). Combined effects of the antibiotic sulfadiazine and liquid manure on the soil microbial-community structure and functions. J. Plant Nutr. Soil Sci. 174, 614–623. doi: 10.1002/jpln.201000322
Hammesfahr, U., Heuer, H., Manzke, B., Smalla, K., and Thiele-Bruhn, S. (2008). Impact of the antibiotic sulfadiazine and pig manure on the microbial community structure in agricultural soils. Soil Biol. Biochem. 40, 1583–1591. doi: 10.1016/j.soilbio.2008.01.010
Hamscher, G., Pawelzick, H. T., Höper, H., and Nau, H. (2005). Different behavior of tetracyclines and sulfonamides in sandy soils after repeated fertilization with liquid manure. Environ. Toxicol. Chem. 24, 861–868. doi: 10.1897/04-182R.1
Hamscher, G., Sczesny, S., Höper, H., and Nau, H. (2002). Determination of persistent tetracycline residues in soil fertilized with liquid manure by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. Anal. Chem. 74, 1509–1518. doi: 10.1021/ac015588m
Herrick, J. B., Haynes, R., Heringa, S., Brooks, J. M., and Sobota, L. T. (2014). Coselection for resistance to multiple late-generation human therapeutic antibiotics encoded on tetracycline resistance plasmids captured from uncultivated stream and soil bacteria. J. Appl. Microbiol. 117, 380–389. doi: 10.1111/jam.12538
Heuer, H., Schmitt, H., and Smalla, K. (2011a). Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14, 236–243. doi: 10.1016/j.mib.2011.04.009
Heuer, H., and Smalla, K. (2007). Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9, 657–666. doi: 10.1111/j.1462-2920.2006.01185.x
Heuer, H., Solehati, Q., Zimmerling, U., Kleineidam, K., Schloter, M., Müller, T., et al. (2011b). Accumulation of sulfonamide resistance genes in arable soils due to repeated application of manure containing sulfadiazine. Appl. Environ. Microbiol. 77, 2527–2530. doi: 10.1128/AEM.02577-10
Hirth, N., Topp, E., Dörfler, U., Stupperich, E., Munch, J. C., and Schroll, R. (2016). An effective bioremediation approach for enhanced microbial degradation of the veterinary antibiotic sulfamethazine in an agricultural soil. Chem. Biol. Technol. Agric. 3:29. doi: 10.1186/s40538-016-0080-6
Ho, Y. B., Zakaria, M. P., Latif, P. A., and Saari, N. (2014). Occurrence of veterinary antibiotics and progesterone in broiler manure and agricultural soil in Malaysia. Sci. Total Environ. 488–489, 261–267. doi: 10.1016/j.scitotenv.2014.04.109
Hou, J., Wan, W., Mao, D., Wang, C., Mu, Q., Qin, S., et al. (2015). Occurrence and distribution of sulfonamides, tetracyclines, quinolones, macrolides, and nitrofurans in livestock manure and amended soils of Northern China. Environ. Sci. Pollut. Res. 22, 4545–4554. doi: 10.1007/s11356-014-3632-y
Hu, D., and Coats, J. R. (2007). Aerobic degradation and photolysis of tylosin in water and soil. Environ. Toxicol. Chem. 26, 884–889. doi: 10.1897/06-197R.1
Hu, H.-W., Han, X.-M., Shi, X.-Z., Wang, J.-T., Han, L.-L., Chen, D., et al. (2016). Temporal changes of antibiotic-resistance genes and bacterial communities in two contrasting soils treated with cattle manure. FEMS Microbiol. Ecol. 92, 1–13. doi: 10.1093/femsec/fiv169
Hu, X., Zhou, Q., and Luo, Y. (2010). Occurrence and source analysis of typical veterinary antibiotics in manure, soil, vegetables and groundwater from organic vegetable bases, northern China. Environ. Pollut. 158, 2992–2998. doi: 10.1016/j.envpol.2010.05.023
Huang, T., Xu, Y., Zeng, J., Zhao, D.-H., Li, L., Liao, X.-P., et al. (2016). Low-concentration ciprofloxacin selects plasmid-mediated quinolone resistance encoding genes and affects bacterial taxa in soil containing manure. Front. Microbiol. 7:1730. doi: 10.3389/fmicb.2016.01730
Huang, X., Liu, C., Li, K., Liu, F., Liao, D., Liu, L., et al. (2013). Occurrence and distribution of veterinary antibiotics and tetracycline resistance genes in farmland soils around swine feedlots in Fujian Province, China. Environ. Sci. Pollut. Res. 20, 9066–9074. doi: 10.1007/s11356-013-1905-5
Ingerslev, F., and Halling-Sørensen, B. (2001). Biodegradability of metronidazole, olaquindox, and tylosin and formation of tylosin degradation products in aerobic soil-manure slurries. Ecotoxicol. Environ. Saf. 48, 311–320. doi: 10.1006/eesa.2000.2026
Jacobsen, A. M., Halling-Sørensen, B., Ingerslev, F., and Hansen, S. H. (2004). Simultaneous extraction of tetracycline, macrolide and sulfonamide antibiotics from agricultural soils using pressurised liquid extraction, followed by solid-phase extraction and liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 1038, 157–170. doi: 10.1016/j.chroma.2004.03.034
Jechalke, S., Focks, A., Rosendahl, I., Groeneweg, J., Siemens, J., Heuer, H., et al. (2014a). Structural and functional response of the soil bacterial community to application of manure from difloxacin-treated pigs. FEMS Microbiol. Ecol. 87, 78–88. doi: 10.1111/1574-6941.12191
Jechalke, S., Heuer, H., Siemens, J., Amelung, W., and Smalla, K. (2014b). Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 22, 536–545. doi: 10.1016/j.tim.2014.05.005
Jones-Lepp, T. L., and Stevens, R. (2007). Pharmaceuticals and personal care products in biosolids/sewage sludge: the interface between analytical chemistry and regulation. Anal. Bioanal. Chem. 387, 1173–1183. doi: 10.1007/s00216-006-0942-z
Kang, D. H., Gupta, S., Rosen, C., Fritz, V., Singh, A., Chander, Y., et al. (2013). Antibiotic uptake by vegetable crops from manure-applied soils. J. Agric. Food Chem. 61, 9992–10001. doi: 10.1021/jf404045m
Karci, A., and Balcioglu, I. A. (2009). Investigation of the tetracycline, sulfonamide, and fluoroquinolone antimicrobial compounds in animal manure and agricultural soils in Turkey. Sci. Total Environ. 407, 4652–4664. doi: 10.1016/j.scitotenv.2009.04.047
Kay, P., Blackwell, P. A., and Boxall, A. B. A. (2004). Fate of veterinary antibiotics in a macroporous tile drained clay soil. Environ. Toxicol. Chem. 23, 1136–1143. doi: 10.1897/03-374
Kay, P., Blackwell, P. A., and Boxall, A. B. A. (2005). A lysimeter experiment to investigate the leaching of veterinary antibiotics through a clay soil and comparison with field data. Environ. Pollut. 134, 333–341. doi: 10.1016/j.envpol.2004.07.021
Keen, P. L., and Patrick, D. M. (2013). Tracking change: a look at the ecological footprint of antibiotics and antimicrobial resistance. Antibiotics 2, 191–205. doi: 10.3390/antibiotics2020191
Kim, D.-W., Heinze, T. M., Kim, B.-S., Schnackenberg, L. K., Woodling, K. A., and Sutherland, J. B. (2011). Modification of norfloxacin by a Microbacterium sp. strain isolated from a wastewater treatment plant. Appl. Environ. Microbiol. 77, 6100–6108. doi: 10.1128/AEM.00545-11
Kinney, C. A., Furlong, E. T., Zaugg, S. D., Burkhardt, M. R., Werner, S. L., Cahill, J. D., et al. (2006). Survey of organic wastewater contaminants in biosolids destined for land application. Environ. Sci. Technol. 40, 7207–7215. doi: 10.1021/es0603406
Klein, E,. Y., Van Boeckel, T. P., Martinez, E. M., Pant, S., Gandra, S., Levin, S. A., et al. (2018). Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. U.S.A. 115, E3463–E3479. doi: 10.1073/pnas.1717295115
Kleineidam, K., Sharma, S., Kotzerke, A., Heuer, H., Thiele-Bruhn, S., Smalla, K., et al. (2010). Effect of sulfadiazine on abundance and diversity of denitrifying bacteria by determining nirK and nirS genes in two arable soils. Microb. Ecol. 60, 703–707. doi: 10.1007/s00248-010-9691-9
Koba, O., Golovko, O., Kodešová, R., Fér, M., and Grabic, R. (2017). Antibiotics degradation in soil: a case of clindamycin, trimethoprim, sulfamethoxazole and their transformation products. Environ. Pollut. 220, 1251–1263. doi: 10.1016/j.envpol.2016.11.007
Kong, W., Li, C., Dolhi, J. M., Li, S., He, J., and Qiao, M. (2012). Characteristics of oxytetracycline sorption and potential bioavailability in soils with various physical-chemical properties. Chemosphere 87, 542–548. doi: 10.1016/j.chemosphere.2011.12.062
Kong, W.-D., Zhu, Y.-G., Fu, B.-J., Marschner, P., and He, J.-Z. (2006). The veterinary antibiotic oxytetracycline and Cu influence functional diversity of the soil microbial community. Environ. Pollut. 143, 129–137. doi: 10.1016/j.envpol.2005.11.003
Kotzerke, A., Hammesfahr, U., Kleineidam, K., Lamshöft, M., Thiele-Bruhn, S., Schloter, M., et al. (2011). Influence of difloxacin-contaminated manure on microbial community structure and function in soils. Biol. Fertil. Soils 47, 177–186. doi: 10.1007/s00374-010-0517-1
Kotzerke, A., Sharma, S., Schauss, K., Heuer, H., Thiele-Bruhn, S., Smalla, K., et al. (2008). Alterations in soil microbial activity and N-transformation processes due to sulfadiazine loads in pig-manure. Environ. Pollut. 153, 315–322. doi: 10.1016/j.envpol.2007.08.020
Kuchta, S. L., Cessna, A. J., Elliott, J. A., Peru, K. M., and Headley, J. V. (2009). Transport of lincomycin to surface and ground water from manure-amended cropland. J. Environ. Qual. 38, 1719–1729. doi: 10.2134/jeq2008.0365
Kulkarni, P., Olson, N. D., Raspanti, G. A., Goldstein, R. E. R., Gibbs, S. G., Sapkota, A., et al. (2017). Antibiotic concentrations decrease during wastewater treatment but persist at low levels in reclaimed water. Int. J. Environ. Res. Public Health 14:668. doi: 10.3390/ijerph14060668
Kumar, K., Gupta, C., SChander, Y., and Singh, A. K. (2005a). Antibiotic use in agriculture and its impact on the terrestrial environment. Adv. Agron. 87, 1–54. doi: 10.1016/S0065-2113(05)87001-4
Kumar, K., Gupta, S. C., Baidoo, S. K., Chander, Y., and Rosen, C. J. (2005b). Antibiotic uptake by plants from soil fertilized with animal manure. J. Environ. Qual. 34, 2082–2085. doi: 10.2134/jeq2005.0026
Kumar, R. R., Lee, J. T., and Cho, J. Y. (2012). Fate, occurrence, and toxicity of veterinary antibiotics in environment. J. Korean Soc. Appl. Biol. Chem. 55, 701–709. doi: 10.1007/s13765-012-2220-4
Kümmerer, K. (2001). Drugs in the environment: Emission of drugs, diagnostic aids and disinfectants into wastewater by hospitals in relation to other sources - A review. Chemosphere 45, 957–969. doi: 10.1016/S0045-6535(01)00144-8
Kümmerer, K. (2009). Antibiotics in the aquatic environment–a review–Part, I. Chemosphere 75, 417–434. doi: 10.1016/j.chemosphere.2008.11.086
Kyselková, M., Jirout, J., Chronáková, A., Vrchotová, N., Bradley, R., Schmitt, H., et al. (2013). Cow excrements enhance the occurrence of tetracycline resistance genes in soil regardless of their oxytetracycline content. Chemosphere 93, 2413–2418. doi: 10.1016/j.chemosphere.2013.08.058
Kyselková, M., Kotrbová, L., Bhumibhamon, G., Chronáková, A., Jirout, J., Vrchotová, N., et al. (2015). Tetracycline resistance genes persist in soil amended with cattle feces independently from chlortetracycline selection pressure. Soil Biol. Biochem. 81, 259–265. doi: 10.1016/j.soilbio.2014.11.018
Larsson, D. G.J. (2014). Antibiotics in the environment. Ups. J. Med. Sci. 119, 108–112. doi: 10.3109/03009734.2014.896438
Lau, C. H.-F., van Engelen, K., Gordon, S., Renaud, J., and Topp, E. (2017). Novel antibiotic resistance determinants from agricultural soil exposed to antibiotics widely used in human medicine and animal farming. Appl. Environ. Microbiol. 83, e00989–e00917. doi: 10.1128/AEM.00989-17
Laureto, L. M. O., Cianciaruso, M. V., and Samia, D. S. M. (2015). Functional diversity: an overview of its history and applicability. Braz. J. Natur. Conserv. 13, 112–116. doi: 10.1016/j.ncon.2015.11.001
Leal, R. M. P., Alleoni, L. R. F., Tornisielo, V. L., and Regitano, J. B. (2013). Sorption of fluoroquinolones and sulfonamides in 13 Brazilian soils. Chemosphere 92, 979–985. doi: 10.1016/j.chemosphere.2013.03.018
Leal, R. M. P., Figueira, R. F., Tornisielo, V. L., and Regitano, J. B. (2012). Occurrence and sorption of fluoroquinolones in poultry litters and soils from São Paulo State, Brazil. Sci. Total Environ. 432, 344–349. doi: 10.1016/j.scitotenv.2012.06.002
Leng, Y., Bao, J., Chang, G., Zheng, H., Li, X., Du, J., et al. (2016). Biotransformation of tetracycline by a novel bacterial strain Stenotrophomonas maltophilia DT1. J. Hazard. Mater. 318, 125–133. doi: 10.1016/j.jhazmat.2016.06.053
Li, C., Chen, J., Wang, J., Ma, Z., Han, P., Luan, Y., et al. (2015). Occurrence of antibiotics in soils and manures from greenhouse vegetable production bases of Beijing, China and an associated risk assessment. Sci. Total Environ. 521–522, 101–107. doi: 10.1016/j.scitotenv.2015.03.070
Li, L.-L., Huang, L.-D., Chung, R.-S., Fok, K.-H., and Zhang, Y.-S. (2010a). Sorption and dissipation of tetracyclines in soils and compost. Pedosphere 20, 807–816. doi: 10.1016/S1002-0160(10)60071-9
Li, W., Shi, Y., Gao, L., Liu, J., and Cai, Y. (2013). Occurrence, distribution and potential affecting factors of antibiotics in sewage sludge of wastewater treatment plants in China. Sci. Tot. Environ. 445–446, 306–313. doi: 10.1016/j.scitotenv.2012.12.050
Li, Y.-W., Mo, C.-H., Zhao, N., Tai, Y.-P., Bao, Y.-P., Wang, J.-Y., et al. (2009). Investigation of sulfonamides and tetracyclines antibiotics in soils from various vegetable fields. Huanjing Kexue/Environmental Sci. 30, 1762–1766.
Li, Y.-W., Wu, X.-L., Mo, C.-H., Tai, Y.-P., Huang, X.-P., and Xiang, L. (2011). Investigation of sulfonamide, tetracycline, and quinolone antibiotics in vegetable farmland soil in the pearl river delta area, Southern China. J. Agric. Food Chem. 59, 7268–7276. doi: 10.1021/jf1047578
Li, Z., Chang, P.-H., Jean, J.-S., Jiang, W.-T., and Wang, C.-J. (2010b). Interaction between tetracycline and smectite in aqueous solution. J. Colloid Interface Sci. 341, 311–319. doi: 10.1016/j.jcis.2009.09.054
Lillenberg, M., Yurchenko, S., Kipper, K., Herodes, K., Pihl, V., Lõhmus, R., et al. (2010). Presence of fluoroquinolones and sulfonamides in urban sewage sludge and their degradation as a result of composting. Int. J. Environ. Sci. Technol. 7, 307–312. doi: 10.1007/BF03326140
Lin, K., and Gan, J. (2011). Sorption and degradation of wastewater-associated non-steroidal anti-inflammatory drugs and antibiotics in soils. Chemosphere 83, 240–246. doi: 10.1016/j.chemosphere.2010.12.083
Liu, B., Li, Y., Zhang, X., Wang, J., and Gao, M. (2014). Combined effects of chlortetracycline and dissolved organic matter extracted from pig manure on the functional diversity of soil microbial community. Soil Biol. Biochem. 74, 148–155. doi: 10.1016/j.soilbio.2014.03.005
Liu, B., Li, Y., Zhang, X., Wang, J., and Gao, M. (2015). Effects of chlortetracycline on soil microbial communities: comparisons of enzyme activities to the functional diversity via Biolog EcoPlates™. Eur. J. Soil Biol. 68, 69–76. doi: 10.1016/j.ejsobi.2015.01.002
Liu, F., Wu, J., Ying, G.-G., Luo, Z., and Feng, H. (2012a). Changes in functional diversity of soil microbial community with addition of antibiotics sulfamethoxazole and chlortetracycline. Appl. Microbiol. Biotechnol. 95, 1615–1623. doi: 10.1007/s00253-011-3831-0
Liu, F., Ying, G.-G., Tao, R., Zhao, J.-L., Yang, J.-F., and Zhao, L.-F. (2009). Effects of six selected antibiotics on plant growth and soil microbial and enzymatic activities. Environ. Pollut. 157, 1636–1642. doi: 10.1016/j.envpol.2008.12.021
Liu, L., Liu, Y-H, Liu, C-X., and Huang, X. (2016). Accumulation of antibiotics and tet resistance genes from swine wastewater in wetland soils. Environ. Eng. Manage. J. 15, 2137–2145. doi: 10.30638/eemj.2016.231
Liu, W., Pan, N., Chen, W., Jiao, W., and Wang, M. (2012b). Effect of veterinary oxytetracycline on functional diversity of soil microbial community. Plant Soil Environ. 58, 295–301. doi: 10.1016/j.ibiod.2012.06.010
Liu, Z., Han, Y., Jing, M., and Chen, J. (2017). Sorption and transport of sulfonamides in soils amended with wheat straw-derived biochar: effects of water pH, coexistence copper ion, and dissolved organic matter. J. Soils Sediments 17, 771–779. doi: 10.1007/s11368-015-1319-8
Łukaszewicz, P., Białk-Bielinska, A., Dołzonek, J., Kumirska, J., Caban, M., and Stepnowski, P. (2018). A new approach for the extraction of tetracyclines from soil matrices: application of the microwave-extraction technique. Anal. Bioanal. Chem. 410, 1697–1707. doi: 10.1007/s00216-017-0815-7
Ma, J., Lin, H., Sun, W., Wang, Q., Yu, Q., Zhao, Y., et al. (2014). Soil microbial systems respond differentially to tetracycline, sulfamonomethoxine, and ciprofloxacin entering soil under pot experimental conditions alone and in combination. Environ. Sci. Pollut. Res. 21, 7436–7448. doi: 10.1007/s11356-014-2685-2
Ma, T., Pan, X., Chen, L., Liu, W., Christie, P., Luo, Y., et al. (2016). Effects of different concentrations and application frequencies of oxytetracycline on soil enzyme activities and microbial community diversity. Eur. J. Soil Biol. 76, 53–60. doi: 10.1016/j.ejsobi.2016.07.004
Manzetti, S., and Ghisi, R. (2014). The environmental release and fate of antibiotics. Mar. Pollut. Bull. 79, 7–15. doi: 10.1016/j.marpolbul.2014.01.005
Martens, R., Wetzstein, H.-G., Zadrazil, F., Capelari, M., Hoffmann, P., and Schmeer, N. (1996). Degradation of the fluoroquinolone enrofloxacin by wood-rotting fungi. Appl. Environ. Microbiol. 62, 4206–4209.
Martinez, J. L., Sánchez, M. B., Martínez-Solano, L., Hernandez, A., Garmendia, L., Fajardo, A., et al. (2009). Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33, 430–449. doi: 10.1111/j.1574-976.2008.00157.x
Martínez-Carballo, E., González-Barreiro, C., Scharf, S., and Gans, O. (2007). Environmental monitoring study of selected veterinary antibiotics in animal manure and soils in Austria. Environ. Pollut. 148, 570–579. doi: 10.1016/j.envpol.2006.11.035
Martínez-Hernández, V., Meffe, R., Herrera López, S., and de Bustamante, I. (2016). The role of sorption and biodegradation in the removal of acetaminophen, carbamazepine, caffeine, naproxen and sulfamethoxazole during soil contact: a kinetics study. Sci. Total Environ. 559, 232–241. doi: 10.1016/j.scitotenv.2016.03.131
Marx, M.-C., Kandeler, E., Wood, M., Wermbter, N., and Jarvis, S. C. (2005). Exploring the enzymatic landscape: distribution and kinetics of hydrolytic enzymes in soil particle-size fractions. Soil Biol. Biochem. 37, 35–48. doi: 10.1016/j.soilbio.2004.05.024
Massé, D. I., Saady, N. M. C., and Gilbert, Y. (2014). Potential of biological processes to eliminate antibiotics in livestock manure: an overview. Animals 4, 146–163. doi: 10.3390/ani4020146
McFarland, J. W., Berger, C. M., Froshauer, S. A., Hayashi, S. F., Hecker, S. J., Jaynes, B. H., et al. (1997). Quantitative structure-activity relationships among macrolide antibacterial agents: in vitro and in vivo potency against Pasteurella multocida. J. Med. Chem. 40, 1340–1346. doi: 10.1021/jm960436i
Michael, I., Rizzo, L., McArdell, C. S., Manaia, C. M., Merlin, C., Schwartz, T., et al. (2013). Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: a review. Water Res. 47, 957–995. doi: 10.1016/j.watres.2012.11.027
Mitchell, S. M., Ullman, J. L., Teel, A. L., and Watts, R. J. (2015). Hydrolysis of amphenicol and macrolide antibiotics: chloramphenicol, florfenicol, spiramycin, and tylosin. Chemosphere 134, 504–511. doi: 10.1016/j.chemosphere.2014.08.050
Molaei, A., Lakzian, A., Haghnia, G., Astaraei, A., Rasouli-Sadaghiani, M., Ceccherini, M. T., et al. (2017). Assessment of some cultural experimental methods to study the effects of antibiotics on microbial activities in a soil: an incubation study. PLoS ONE 12:e0180663. doi: 10.1371/journal.pone.0180663
Mulla, S. I., Hu, A., Sun, Q., Li, J., Suanon, F., Ashfaq, M., et al. (2018). Biodegradation of sulfamethoxazole in bacteria from three different origins. J. Environ. Manage. 206, 93–102. doi: 10.1016/1.jenvman.2017.10.029
Müller, A. K., Westergaard, K., Christensen, S., and Sørensen, S. J. (2002). The diversity and function of soil microbial communities exposed to different disturbances. Microb. Ecol. 44, 49–58. doi: 10.1007/s00248-001-0042-8
Muñoz, I., Gómez-Ramos, M. J., Agüera, A., Fernández-Alba, A. R., García-Reyes, J. F., and Molina-Díaz, A. (2009). Chemical evaluation of contaminants in wastewater effluents and the environmental risk of reusing effluents in agriculture. Trends Anal. Chem. 28, 676–694. doi: 10.1016/j.trac.2009.03.007
Nelson, K. L., Brözel, V. S., Gibson, S. A., Thaler, R., and Clay, S. A. (2011). Influence of manure from pigs fed chlortetracycline as growth promotant on soil microbial community structure. World, J. Microbiol. Biotechnol. 27, 659–668. doi: 10.1007/s11274-010-0504-6
Nõlvak, H., Truu, M., Kanger, K., Tampere, M., Espenberg, M., Loit, E., et al. (2016). Inorganic and organic fertilizers impact the abundance and proportion of antibiotic resistance and integron-integrase genes in agricultural grassland soil. Sci. Total Environ. 562, 678–689. doi: 10.1016/j.scitotenv.2016.04.035
Nowara, A., Burhenne, J., and Spiteller, M. (1997). Binding of fluoroquinolone carboxylic acid derivatives to clay minerals. J. Agric. Food Chem. 45, 1459–1463.
Ollivier, J., Kleineidam, K., Reichel, R., Thiele-Bruhn, S., Kotzerke, A., Kindler, R., et al. (2010). Effect of sulfadiazine-contaminated pig manure on the abundances of genes and transcripts involved in nitrogen transformation in the root-rhizosphere complexes of maize and clover. Appl. Environ. Microbiol. 76, 7903–7909. doi: 10.1128/AEM.01252-10
Orlewska, K., Markowicz, A., Piotrowska-Seget, Z., Smolen-Dzirba, J., and Cycoń, M. (2018a). Functional diversity of soil microbial communities in response to the application of cefuroxime and/or antibiotic-resistant Pseudomonas putida strain MC1. Sustain 10:3549. doi: 10.3390/su10103549
Orlewska, K., Piotrowska-Seget, Z., Bratosiewicz-Wasik, J., and Cycoń, M. (2018b). Characterization of bacterial diversity in soil contaminated with the macrolide antibiotic erythromycin and/or inoculated with a multidrug-resistant Raoultella sp. strain using the PCR-DGGE approach. Appl. Soil Ecol. 126, 57–64. doi: 10.1016/j.apsoil.2018.02.019
Orlewska, K., Piotrowska-Seget, Z., and Cycoń, M. (2018c). Use of the PCR-DGGE method for the analysis of the bacterial community structure in soil treated with the cephalosporin antibiotic cefuroxime and/or inoculated with a multidrug-resistant Pseudomonas putida strain MC1. Front. Microbiol. 9:1387. doi: 10.3389/fmicb.2018.01387
Pan, M., and Chu, L. M. (2016). Adsorption and degradation of five selected antibiotics in agricultural soil. Sci. Total Environ. 545–546, 48–56. doi: 10.1016/j.scitotenv.2015.12.040
Pan, M., and Chu, L. M. (2017a). Leaching behavior of veterinary antibiotics in animal manure-applied soils. Sci. Total Environ. 579, 466–473. doi: 10.1016/j.scitotenv.2016.11.072
Pan, M., and Chu, L. M. (2017b). Fate of antibiotics in soil and their uptake by edible crops. Sci. Total Environ. 599–600, 500–512. doi: 10.1016/j.scitotenv.2017.04.214
Pan, M., Wong, C. K. C., and Chu, L. M. (2014). Distribution of antibiotics in wastewater-irrigated soils and their accumulation in vegetable crops in the Pearl River Delta, Southern China. J. Agric. Food Chem. 62, 11062–11069. doi: 10.1021/jf503850v
Pan, X., Qiang, Z., Ben, W., and Chen, M. (2011). Residual veterinary antibiotics in swine manure from concentrated animal feeding operations in Shandong Province, China. Chemosphere 84, 695–700. doi: 10.1016/j.chemosphere.2011.03.022
Park, J. Y., and Huwe, B. (2016). Effect of pH and soil structure on transport of sulfonamide antibiotics in agricultural soils. Environ. Pollut. 213, 561–570. doi: 10.1016/j.envpol.2016.01.089
Park, S., and Choi, K. (2008). Hazard assessment of commonly used agricultural antibiotics on aquatic ecosystems. Ecotoxicology 17, 526–538. doi: 10.1007/s10646-008-0209-x
Perry, J. A., Westman, E. L., and Wright, G. D. (2014).The antibiotic resistome: what's new? Curr. Opin. Microbiol. 21, 45–50. doi: 10.1016/j.mib.2014.09.002
Pinna, M. V., Castaldi, P., Deiana, P., Pusino, A., and Garau, G. (2012). Sorption behavior of sulfamethazine on unamended and manure-amended soils and short-term impact on soil microbial community. Ecotoxicol. Environ. Saf. 84, 234–242. doi: 10.1016/j.ecoenv.2012.07.006
Pino-Otín, M. R., Muñiz, S., Val, J., and Navarro, E. (2017). Effects of 18 pharmaceuticals on the physiological diversity of edaphic microorganisms. Sci. Total Environ. 595, 441–450. doi: 10.1016/j.scitotenv.2017.04.002
Popowska, M., Rzeczycka, M., Miernik, A., Krawczyk-Balska, A., Walsh, F., and Duffy, B. (2012). Influence of soil use on prevalence of tetracyclinestreptomycin, and erythromycin resistance and associated resistance genes. Antimicrob. Agents Chemother. 56, 1434–1443. doi: 10.1128/AAC.05766-11
Rabølle, M., and Spliid, N. H. (2000). Sorption and mobility of metronidazole, olaquindox, oxytetracycline and tylosin in soil. Chemosphere 40, 715–722. doi: 10.1016/S0045-6535(99)00442-7
Rafii, F., Williams, A. J., Park, M., Sims, L. M., Heinze, T. M., Cerniglia, C. E., et al. (2009). Isolation of bacterial strains from bovine fecal microflora capable of degradation of ceftiofur. Vet. Microbiol. 139, 89–96. doi: 10.1016/j.vetmic.2009.04.023
Reichel, R., Michelini, L., Ghisi, R., and Thiele-Bruhn, S. (2015). Soil bacterial community response to sulfadiazine in the soil-root zone. J. Plant Nutr. Soil Sci. 178, 499–506. doi: 10.1002/jpln.201400352
Reichel, R., Radl, V., Rosendahl, I., Albert, A., Amelung, W., Schloter, M., et al. (2014). Soil microbial community responses to antibiotic-contaminated manure under different soil moisture regimes. Appl. Microbiol. Biotechnol. 98, 6487–6495. doi: 10.1007/s00253-014-5717-4
Reichel, R., Rosendahl, I., Peeters, E. T. H. M., Focks, A., Groeneweg, J., Bierl, R., et al. (2013). Effects of slurry from sulfadiazine- (SDZ) and difloxacin- (DIF) medicated pigs on the structural diversity of microorganisms in bulk and rhizosphere soil. Soil Biol. Biochem. 62, 82–91. doi: 10.1016/j.soilbio.2013.03.007
Rosendahl, I., Siemens, J., Groeneweg, J., Linzbach, E., Laabs, V., Herrmann, C., et al. (2011). Dissipation and sequestration of the veterinary antibiotic sulfadiazine and its metabolites under field conditions. Environ. Sci. Technol. 45, 5216–5222. doi: 10.1021/es200326t
Rosendahl, I., Siemens, J., Kindler, R., Groeneweg, J., Zimmermann, J., Czerwinski, S., et al. (2012). Persistence of the fluoroquinolone antibiotic difloxacin in soil and lacking effects on nitrogen turnover. J. Environ. Qual. 41, 1275–1283. doi: 10.2134/jeq2011.0459
Rutgersson, C., Fick, J., Marathe, N., Kristiansson, E., Janzon, A., Angelin, M., et al. (2014). Fluoroquinolones and qnr genes in sediment, water, soil, and human fecal flora in an environment polluted by manufacturing discharges. Environ. Sci. Technol. 48, 7825–7832. doi: 10.1021/es501452a
Sandegren, L. (2014). Selection of antibiotic resistance at very low antibiotic concentrations. Upsala J. Med. Sci. 119, 103–110. doi: 10.3109/03009734.2014.90445
Sarmah, A. K., Meyer, M. T., and Boxall, A. B. A. (2006). A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65, 725–759. doi: 10.1016/j.chemosphere.2006.03.026
Sassman, S. A., and Lee, L. S. (2007). Sorption and degradation in soils of veterinary ionophore antibiotics: Monensin and lasalocid. Environ. Toxicol. Chem. 26, 1614–1621. doi: 10.1897/07-073R.1
Schauss, K., Focks, A., Heuer, H., Kotzerke, A., Schmitt, H., Thiele-Bruhn, S., et al. (2009). Analysis, fate and effects of the antibiotic sulfadiazine in soil ecosystems. TrAC - Trends Anal. Chem. 28, 612–618. doi: 10.1016/j.trac.2009.02.009
Schlüsener, M. P., Von Arb, M. A., and Bester, K. (2006). Elimination of macrolides, tiamulin, and salinomycin during manure storage. Arch. Environ. Contam. Toxicol. 51, 21–28. doi: 10.1007/s00244-004-0240-8
Schmitt, H., Haapakangas, H., and Van Beelen, P. (2005). Effects of antibiotics on soil microorganisms: time and nutrients influence pollution-induced community tolerance. Soil Biol. Biochem. 37, 1882–1892. doi: 10.1016/j.soilbio.2005.02.022
Sengupta, S., Chattopadhyay, M. K., and Grossart, H-P. (2013). The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 4:47. doi: 10.3389/fmicb.2013.00047
Shade, A., Klimowicz, A. K., Spear, R. N., Linske, M., Donato, J. J., Hogan, C. S., et al. (2013). Streptomycin application has no detectable effect on bacterial community structure in apple orchard soil. Appl. Environ. Microbiol. 79, 6617–6625. doi: 10.1128/AEM.02017-13
Shi, Y., Gao, L., Li, W., Liu, J., and Cai, Y. (2012). Investigation of fluoroquinolones, sulfonamides and macrolides in long-term wastewater irrigation soil in Tianjin, China. Bull. Environ. Contam. Toxicol. 89, 857–861. doi: 10.1007/s00128-012-0761-1
Singer, H. P., Wossner, A. E., McArdell, C. S., and Fenner, K. (2016). Rapid screening for exposure to “non-target” pharmaceuticals from wastewater effluents by combining HRMS-based suspect screening and exposure modeling. Environ. Sci. Technol. 50, 6698–6707. doi: 10.1021/acs.est.5b03332
Sittig, S., Kasteel, R., Groeneweg, J., Hofmann, D., Thiele, B., Köppchen, S., et al. (2014). Dynamics of transformation of the veterinary antibiotic sulfadiazine in two soils. Chemosphere 95, 470–477. doi: 10.1016/j.chemosphere.2013.09.100
Srinivasan, P., and Sarmah, A. K. (2014). Dissipation of sulfamethoxazole in pasture soils as affected by soil and environmental factors. Sci. Total Environ. 479–480, 284–291. doi: 10.1016/j.scitotenv.2014.02.014
Stoob, K., Singer, H. P., Mueller, S. R., Schwarzenbach, R. P., and Stamm, C. H. (2007). Dissipation and transport of veterinary sulfonamide antibiotics after manure application to grassland in a small catchment. Environ. Sci. Technol. 41, 7349–7355. doi: 10.1021/es070840e
Su, J. Q., Wei, B., Xu, C. Y., Qiao, M., and Zhu, Y. G. (2014). Functional metagenomic characterization of antibiotic resistance genes in agricultural soils from China. Environ. Int. 65, 9–15. doi: 10.1016/j.envint.2013.12.010
Sukul, P., Lamshöft, M., Zühlke, S., and Spiteller, M. (2008). Sorption and desorption of sulfadiazine in soil and soil-manure systems. Chemosphere 73, 1344–1350. doi: 10.1016/j.chemosphere.2008.06.066
Sułowicz, S., and Piotrowska-Seget, Z. (2016). Response of microbial communities from an apple orchard and grassland soils to the first-time application of the fungicide tetraconazole. Ecotoxicol. Environ. Saf. 124, 193–201. doi: 10.1016/j.ecoenv.2015.10.025
Tabatabai, M. A., and Bremner, J. M. (1969). Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol. Biochem. 1, 301–307.
Tang, X., Lou, C., Wang, S., Lu, Y., Liu, M., Hashmi, M. Z., et al. (2015). Effects of long-term manure applications on the occurrence of antibiotics and antibiotic resistance genes (ARGs) in paddy soils: evidence from four field experiments in south of China. Soil Biol. Biochem. 90, 179–187. doi: 10.1016/j.soilbio.2015.07.027
Tasho, R. P., and Cho, J. Y. (2016). Veterinary antibiotics in animal waste, its distribution in soil and uptake by plants: a review. Sci. Total Environ. 563–564, 366–376. doi: 10.1016/j.scitotenv.2016.04.140
Teixidó, M., Granados, M., Prat, M. D., and Beltrán, J. L. (2012). Sorption of tetracyclines onto natural soils: data analysis and prediction. Environ. Sci. Pollut. Res. 19, 3087–3095. doi: 10.1007/s11356-012-0954-5
Thiele, S. (2000). Adsorption of the antibiotic pharmaceutical compound sulfapyridine by a long-term differently fertilized loess Chernozem. J. Plant Nutr. Soil Sci. 163, 589–594. doi: 10.1002/1522-2624(200012)163:6<589::AID-JPLN589>3.0.CO;2-5
Thiele-Bruhn, S. (2003). Pharmaceutical antibiotic compounds in soils - a review. J. Plant Nutr. Soil Sci. 166, 145–167. doi: 10.1002/jpln.200390023
Thiele-Bruhn, S. (2005). Microbial inhibition by pharmaceutical antibiotics in different soils - Dose-response relations determined with the iron(III) reduction test. Environ. Toxicol. Chem. 24, 869–876. doi: 10.1897/04-166R.1
Thiele-Bruhn, S., and Beck, I.-C. (2005). Effects of sulfonamide and tetracycline antibiotics on soil microbial activity and microbial biomass. Chemosphere 59, 457–465. doi: 10.1016/j.chemosphere.2005.01.023
Thiele-Bruhn, S., and Peters, D. (2007). Photodegradation of pharmaceutical antibiotics on slurry and soil surfaces. Landbauforsch. Volkenrode 57, 13–23.
Thiele-Bruhn, S., Seibicke, T., Schulten, H.-R., and Leinweber, P. (2004). Sorption of sulfonamide pharmaceutical antibiotics on whole soils and particle-size fractions. J. Environ. Qual. 33, 1331–1342.
Tolls, J. (2001). Sorption of veterinary pharmaceuticals in soils: a review. Environ. Sci. Technol. 35, 3397–3406. doi: 10.1021/es0003021
Topp, E., Chapman, R., Devers-Lamrani, M., Hartmann, A., Marti, R., Martin-Laurent, F., et al. (2013). Accelerated biodegradation of veterinary antibiotics in agricultural soil following long-term exposure, and isolation of a sulfamethazinedegrading microbacterium sp. J. Environ. Qual. 42, 173–178. doi: 10.2134/jeq2012.0162
Topp, E., Renaud, J., Sumarah, M., and Sabourin, L. (2016). Reduced persistence of the macrolide antibiotics erythromycin, clarithromycin and azithromycin in agricultural soil following several years of exposure in the field. Sci. Total Environ. 562, 136–144. doi: 10.1016/j.scitotenv.2016.03.210
Torres-Cortés, G., Millán, V., Ramírez-Saad, H. C., Nisa-Martínez, R., Toro, N., and Martínez-Abarca, F. (2011). Characterization of novel antibiotic resistance genes identified by functional metagenomics on soil samples. Environ. Microbiol. 13, 1101–1114. doi: 10.1111/j.1462-2920.2010.02422.x
Toth, J. D., Feng, Y., and Dou, Z. (2011). Veterinary antibiotics at environmentally relevant concentrations inhibit soil iron reduction and nitrification. Soil Biol. Biochem. 43, 2470–2472. doi: 10.1016/j.soilbio.2011.09.004
Udikovic-Kolic, N., Wichmann, F., Broderick, N. A., and Handelsman, J. (2014). Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc. Natl. Acad. Sci. U.S.A. 111, 15202–15207. doi: 10.1073/pnas.1409836111
Unger, I. M., Goyne, K. W., Kennedy, A. C., Kremer, R. J., McLain, J. E. T., and Williams, C. F. (2013). Antibiotic effects on microbial community characteristics in soils under conservation management practices. Soil Sci. Soc. Am. J. 77, 100–112. doi: 10.2136/sssaj2012.0099
US EPA (2009). Targeted National Sewage Sludge Survey. EPA-822-R-08-018. Washington, DC: U.S. Environmental Protection Agency.
Van Doorslaer, X., Dewulf, J., Van Langenhove, H., and Demeestere, K. (2014). Fluoroquinolone antibiotics: an emerging class of environmental micropollutants. Sci. Total Environ. 500–501, 250–269. doi: 10.1016/j.scitotenv.2014.08.075
Van Goethem, M. W., Pierneef, R., Bezuidt, O. K. I., Van De Peer, Y., Cowan, D. A., and Makhalanyane, T. P. (2018). A reservoir of “historical” antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 6:40. doi: 10.1186/s40168-018-0424-5
Varma, A., and Buscot, F. (2005). Microorganisms in Soils: Roles in Genesis and Functions. Berlin Heidelberg: Springer-Verlag.
Vaz, S., Lopes, W. T., and Martin-Neto, L. (2015). Study of molecular interactions between humic acid from Brazilian soil and the antibiotic oxytetracycline. Environ. Technol. Innov. 4, 260–267. doi: 10.1016/j.eti.2015.09.004
Vazquez-Roig, P., Segarra, R., Blasco, C., Andreu, V., and Picó, Y. (2010). Determination of pharmaceuticals in soils and sediments by pressurized liquid extraction and liquid chromatography tandem mass spectrometry. J. Chromatogr. A 1217, 2471–2483. doi: 10.1016/j.chroma.2009.11.033
Walters, E., McClellan, K., and Halden, R. U. (2010). Occurrence and loss over three years of 72 pharmaceuticals and personal care products from biosolids-soil mixtures in outdoor mesocosms. Water Res. 44, 6011–6020. doi: 10.1016/j.watres.2010.07.051
Wang, J., Lin, H., Sun, W., Xia, Y., Ma, J., Fu, J., et al. (2016). Variations in the fate and biological effects of sulfamethoxazole, norfloxacin and doxycycline in different vegetable-soil systems following manure application. J. Hazard. Mater. 304, 49–57. doi: 10.1016/j.jhazmat.2015.10.038
Wang, Q., Guo, M., and Yates, S. R. (2006). Degradation kinetics of manure-derived sulfadimethoxine in amended soil. J. Agric. Food Chem. 54, 157–163. doi: 10.1021/jf052216w
Wang, Q., and Yates, S. R. (2008). Laboratory study of oxytetracycline degradation kinetics in animal manure and soil. J. Agric. Food Chem. 56, 1683–1688. doi: 10.1021/jf072927p
Wang, S., Gao, X., Gao, Y., Li, Y., Cao, M., Xi, Z., et al. (2017). Tetracycline resistance genes identified from distinct soil environments in China by functional metagenomics. Front. Microbiol. 8:1406. doi: 10.3389/fmicb.2017.01406
Wang, S., and Wang, H. (2015). Adsorption behavior of antibiotic in soil environment: a critical review. Front. Environ. Sci. Eng. 9, 565–574. doi: 10.1007/s11783-015-0801-2
Watanabe, N., Bergamaschi, B. A., Loftin, K. A., Meyer, M. T., and Harter, T. (2010). Use and environmental occurrence of antibiotics in freestall dairy farms with manured forage fields. Environ. Sci. Technol. 44, 6591–6600. doi: 10.1021/es100834s
Weerasinghe, C. A., and Towner, D. (1997). Aerobic biodegradation of virginiamycin in soil. Environ. Toxicol. Chem. 16, 1873–1876. doi: 10.1002/etc.5620160916
Wegst-Uhrich, S. R., Navarro, D. A. G., Zimmerman, L., and Aga, D. S. (2014). Assessing antibiotic sorption in soil: a literature review and new case studies on sulfonamides and macrolides. Chem. Cent. J. 8:5. doi: 10.1186/1752-153X-8-5
Wei, X., Wu, S. C., Nie, X. P., Yediler, A., and Wong, M. H. (2009). The effects of residual tetracycline on soil enzymatic activities and plant growth. J. Environ. Sci. Heal. 44, 461–471. doi: 10.1080/03601230902935139
Wellington, E. M. H., Boxall, A. B. A., Cross, P., Feil, E. J., Gaze, W. H., Hawkey, P. M., et al. (2013). The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 13, 155–165. doi: 10.1016/S1473-3099(12)70317-1
Wen, X., Wang, Y., Zou, Y., Ma, B., and Wu, Y. (2018). No evidential correlation between veterinary antibiotic degradation ability and resistance genes in microorganisms during the biodegradation of doxycycline. Ecotoxicol. Environ. Saf. 147, 759–766. doi: 10.1016/j.ecoenv.2017.09.025
Wepking, C., Avera, B., Badgley, B., Barrett, J. E., Franklin, J., Knowlton, K. F., et al. (2017). Exposure to dairy manure leads to greater antibiotic resistance and increased mass-specific respiration in soil microbial communities. Proc. R. Soc. B Biol. Sci. 284, 20162233. doi: 10.1098/rspb.2016.2233
Westergaard, K., Müller, A. K., Christensen, S., Bloem, J., and Sørensen, S. J. (2001). Effects of tylosin as a disturbance on the soil microbial community. Soil Biol. Biochem. 33, 2061–2071. doi: 10.1016/S0038-0717(01)00134-1
Wu, L., Pan, X., Chen, L., Huang, Y., Teng, Y., Luo, Y., et al. (2013). Occurrence and distribution of heavy metals and tetracyclines in agricultural soils after typical land use change in east China. Environ. Sci. Pollut. Res. 20, 8342–8354. doi: 10.1007/s11356-013-1532-1
Wu, X.-L., Xiang, L., Yan, Q.-Y., Jiang, Y.-N., Li, Y.-W., Huang, X.-P., et al. (2014). Distribution and risk assessment of quinolone antibiotics in the soils from organic vegetable farms of a subtropical city, Southern China. Sci. Total Environ. 487, 399–406. doi: 10.1016/j.scitotenv.2014.04.015
Xiao, K.-Q., Li, B., Ma, L., Bao, P., Zhou, X., Zhang, T., et al. (2016). Metagenomic profiles of antibiotic resistance genes in paddy soils from South China. FEMS Microbiol. Ecol. 92:fiw023. doi: 10.1093/femsec/fiw023
Xin, Z., Fengwei, T., Gang, W., Xiaoming, L., Qiuxiang, Z., Hao, Z., et al. (2012). Isolation, identification and characterization of human intestinal bacteria with the ability to utilize chloramphenicol as the sole source of carbon and energy. FEMS Microbiol. Ecol. 82, 703–712. doi: 10.1111/j.1574-6941.2012.01440.x
Xiong, W., Wang, M., Dai, J., Sun, Y., and Zeng, Z. (2018). Application of manure containing tetracyclines slowed down the dissipation of tet resistance genes and caused changes in the composition of soil bacteria. Ecotoxicol. Environ. Saf. 147, 455–460. doi: 10.1016/j.ecoenv.2017.08.061
Xu, Y., Yu, W., Ma, Q., Wang, J., Zhou, H., and Jiang, C. (2016). The combined effect of sulfadiazine and copper on soil microbial activity and community structure. Ecotoxicol. Environ. Saf. 134, 43–52. doi: 10.1016/j.ecoenv.2016.06.041
Yang, J.-F., Ying, G.-G., Liu, S., Zhou, L.-J., Zhao, J.-L., Tao, R., et al. (2012). Biological degradation and microbial function effect of norfloxacin in a soil under different conditions. J. Environ. Sci. Heal. 47, 288–295. doi: 10.1080/03601234.2012.638886
Yang, J.-F., Ying, G.-G., Zhao, J.-L., Tao, R., Su, H.-C., and Chen, F. (2010). Simultaneous determination of four classes of antibiotics in sediments of the Pearl Rivers using RRLC-MS/MS. Sci. Total Environ. 408, 3424–3432. doi: 10.1016/j.scitotenv.2010.03.049
Yang, J.-F., Ying, G.-G., Zhou, L.-J., Liu, S., and Zhao, J.-L. (2009). Dissipation of oxytetracycline in soils under different redox conditions. Environ. Pollut. 157, 2704–2709. doi: 10.1016/j.envpol.2009.04.031
Yang, Y., Owino, A. A., Gao, Y., Yan, X., Xu, C., and Wang, J. (2016). Occurrence, composition and risk assessment of antibiotics in soils from Kenya, Africa. Ecotoxicology 25, 1194–1201. doi: 10.1007/s10646-016-1673-3
Ye, M., Sun, M., Feng, Y., Wan, J., Xie, S., Tian, D., et al. (2016). Effect of biochar amendment on the control of soil sulfonamides, antibiotic-resistant bacteria, and gene enrichment in lettuce tissues. J. Hazard. Mater. 309, 219–227. doi: 10.1016/j.jhazmat.2015.10.074
Zak, J. C., Willing, M. R., Moorhead, D. L., and Wildman, H. G. (1994). Functional diversity of microbial communities: a quantitative approach. Soil Biol. Biochem. 26, 1101–1108. doi: 10.1016/0038-0717(94)90131-7
Zhang, H., Zhou, Y., Huang, Y., Wu, L., Liu, X., and Luo, Y. (2016a). Residues and risks of veterinary antibiotics in protected vegetable soils following application of different manures. Chemosphere 152, 229–237. doi: 10.1016/j.chemosphere.2016.02.111
Zhang, J., Wei, Y., Chen, M., Tong, J., Xiong, J., and Ao, Z. (2015). Occurrence and fate of antibiotic and heavy metal resistance genes in the total process of biological treatment and land application of animal manure: a review. Huanjing Kexue Xuebao/Acta Sci. Circum. 35, 935–946. doi: 10.13671/j.hjkxxb.2014.0843
Zhang, J. Q., and Dong, Y. H. (2008). Effect of low-molecular-weight organic acids on the adsorption of norfloxacin in typical variable charge soils of China. J. Hazard. Mater. 151, 833–839. doi: 10.1016/j.jhazmat.2007.11.046
Zhang, Q., and Dick, W. A. (2014). Growth of soil bacteria, on penicillin and neomycin, not previously exposed to these antibiotics. Sci. Total Environ. 493, 445–453. doi: 10.1016/j.scitotenv.2014.05.114
Zhang, Q., Shamsi, I. H., Zhou, H., Wei, L., Zhu, Z., Adil, M. F., et al. (2016b). Soil microbial communities and mineralization responses to penicillin and tetracycline loads. Res. Rev. J. Microbiol. Biotechnol. 5, 7–15.
Zhang, S., Yang, G., Hou, S., Zhang, T., Li, Z., and Liang, F. (2018). Distribution of ARGs and MGEs among glacial soil, permafrost, and sediment using metagenomic analysis. Environ. Pollut. 234, 339–346. doi: 10.1016/j.envpol.2017.11.031
Zhang, W., Qiu, L., Gong, A., and Yuan, X. (2017b). Isolation and characterization of a high-efficiency erythromycin A-degrading Ochrobactrum sp. strain. Mar. Pollut. Bull. 114, 896–902. doi: 10.1016/j.marpolbul.2016.10.076
Zhang, W.-W., Wen, Y.-Y., Niu, Z.-L., Yin, K., Xu, D.-X., and Chen, L.-X. (2012). Isolation and characterization of sulfonamide-degrading bacteria Escherichia sp. HS21 and Acinetobacter sp. HS51. World J. Microbiol. Biotechnol. 28, 447–452. doi: 10.1007/s11274-011-0834-z
Zhang, Y., Hu, S., Zhang, H., Shen, G., Yuan, Z., and Zhang, W. (2017a). Degradation kinetics and mechanism of sulfadiazine and sulfamethoxazole in an agricultural soil system with manure application. Sci. Total Environ. 607–608, 1348–1356. doi: 10.1016/j.scitotenv.2017.07.083
Zhao, L., Dong, Y. H., and Wang, H. (2010). Residues of veterinary antibiotics in manures from feedlot livestock in eight provinces of China. Sci. Total Environ. 408, 1069–1075. doi: 10.1016/j.scitotenv.2009.11.014
Zhou, B., Wang, C., Zhao, Q., Wang, Y., Huo, M., Wang, J., et al. (2016). Prevalence and dissemination of antibiotic resistance genes and coselection of heavy metals in Chinese dairy farms. J. Hazard. Mater. 320, 10–17. doi: 10.1016/j.jhazmat.2016.08.007
Zhou, L.-J., Ying, G.-G., Liu, S., Zhang, R.-Q., Lai, H.-J., Chen, Z.-F., et al. (2013a). Excretion masses and environmental occurrence of antibiotics in typical swine and dairy cattle farms in China. Sci. Total Environ. 444, 183–195. doi: 10.1016/j.scitotenv.2012.11.087
Zhou, L.-J., Ying, G.-G., Liu, S., Zhao, J.-L., Yang, B., Chen, Z.-F., et al. (2013b). Occurrence and fate of eleven classes of antibiotics in two typical wastewater treatment plants in South China. Sci. Total Environ. 452–453, 365–376. doi: 10.1016/j.scitotenv.2013.03.010
Zhou, L.-J., Ying, G.-G., Zhao, J.-L., Yang, J.-F., Wang, L., Yang, B., et al. (2011). Trends in the occurrence of human and veterinary antibiotics in the sediments of the Yellow River, Hai River and Liao River in northern China. Environ. Pollut. 159, 1877–1885. doi: 10.1016/j.envpol.2011.03.034
Zhou, Y., Niu, L., Zhu, S., Lu, H., and Liu, W. (2017). Occurrence, abundance, and distribution of sulfonamide and tetracycline resistance genes in agricultural soils across China. Sci. Total Environ. 599–600, 1977–1983. doi: 10.1016/j.scitotenv.2017.05.152
Zielezny, Y., Groeneweg, J., Vereecken, H., and Tappe, W. (2006). Impact of sulfadiazine and chlorotetracycline on soil bacterial community structure and respiratory activity. Soil Biol. Biochem. 38, 2372–2380. doi: 10.1016/j.soilbio.2006.01.031
Keywords: antibiotics, degradation, DT50, microbial activities, microbial community structure, antibiotic resistance genes, metagenomics, soil
Citation: Cycoń M, Mrozik A and Piotrowska-Seget Z (2019) Antibiotics in the Soil Environment—Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 10:338. doi: 10.3389/fmicb.2019.00338
Received: 05 July 2018; Accepted: 08 February 2019;
Published: 08 March 2019.
Edited by:
Matthias E. Kaestner, Helmholtz Centre for Environmental Research (UFZ), GermanyReviewed by:
Ulisses Nunes da Rocha, Helmholtz Centre for Environmental Research (UFZ), GermanyCopyright © 2019 Cycoń, Mrozik and Piotrowska-Seget. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mariusz Cycoń, bWN5Y29uQHN1bS5lZHUucGw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.