Genome-Based Taxonomic Classification of the Phylum Actinobacteria
A Commentary on
Genome-Based Taxonomic Classification of the Phylum Actinobacteria
by Nouioui, I., Carro, L., García-López, M., Meier-Kolthoff, J. P., Woyke, T., Kyrpides, N. C., et al. (2018). Front. Microbiol. 9:2007. doi: 10.3389/fmicb.2018.02007
Nouioui et al. (2018) have recently reported comprehensive phylogeny and taxonomy of members from the phylum Actinobacteria. Based on genomic-scale analyses, this study makes proposals for many new taxa of Actinobacteria. A significant portion of this study concerns the classification of species from the order Corynebacteriales. This study and another study by Goodfellow and Jones (2012) indicate the order Corynebacteriales as containing several families including Mycobacteriaceae. However, Mycobacteriaceae is a part of the validly published order Mycobacteriales as proposed by Janke (1924) and included in the Approved Lists of bacterial names (Skerman et al., 1980). In contrast, the order Corynebacteriales was proposed in 2012 (Goodfellow and Jones, 2012) and validly published in 2015 (Oren and Garrity, 2015). Thus, according to Rules 23a and 24a of the International Code of Nomenclature of Prokaryotes (Parker et al., 2015), the order name Mycobacteriales Janke 1924 has priority over the name Corynebacteriales Goodfellow and Jones 2015 and all of the families which are part of the order Corynebacteriales should be embedded within the order Mycobacteriales. To correct this anomaly, an emended description of the order Mycobacteriales, based partially on the description of the Corynebacteriales by Goodfellow and Jones (2012) and Nouioui et al. (2018), is provided below.
The genus Mycobacterium previously consisted of 188 named species (Parte, 2014). Our recent comprehensive phylogenomics and comparative analyses of 150 mycobacterial genomes robustly demonstrated the grouping of these species into five distinct clades (Gupta et al., 2018). Besides their distinct separation in genomic-scale phylogenetic trees and in average amino acid identity matrices, the members of these clades were also reliably demarcated based on multiple molecular synapomorphies uniquely shared by members from each clade (Figure 1A). The compelling evidence from these studies formed the basis for our division of the genus Mycobacterium into five genera. In this classification, the genus name Mycobacterium is retained for the clade containing all major human and animal pathogens (e.g., M. tuberculosis, M. leprae, M. bovis, etc.), whereas species from other clades, harboring mainly non-pathogenic species, are transferred into four new genera viz. Mycolicibacterium, Mycolicibacter, Mycolicibacillus, and Mycobacteroides (Figure 1A).
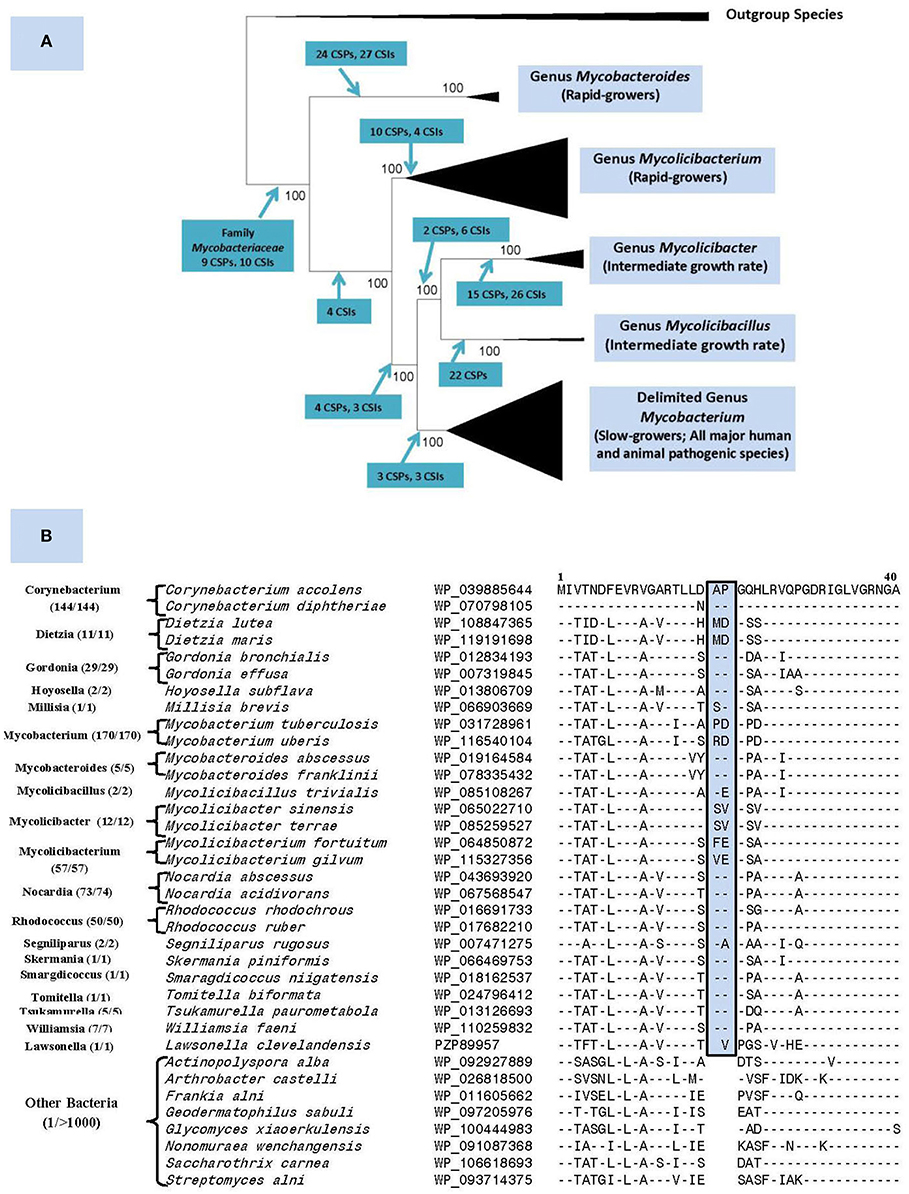
Figure 1. (A) A summary diagram showing the five main clades within the family Mycobacteriaceae, now recognized as distinct genera, seen in different phylogenetic trees. The molecular synapomorphies consisting of conserved signature indels (CSIs) and conserved signature proteins (CSPs), which distinguish different clades are marked on the nodes. The members of these clades/genera also differ in their growth rates. (B) Partial sequence alignment of the ABC-F family ATP-binding cassette domain-containing protein showing a 2 aa insert in a conserved region that is commonly and uniquely present in different members of the order Mycobacteriales. The dashes (–) in the alignment show identity with the amino acids shown on the top line. The numbers with the genus name refer to different unique strains for which sequences were examined. The sequence information is shown for only 1–2 representative species from different genera that are part of this order and very few outgroup species. However, this CSI is not found in other Actinobacteria or other examined bacteria (see Figure S1) with the possible exception of Kroppenstedtia sanguinis (a Firmicute). This CSI as well as a number of other CSIs and CSPs for this order were described in our earlier work (Gao and Gupta, 2012) based on limited sequence information and most of them are still distinctive characteristics of this order.
Nouioui et al. (2018) commend our work for identifying molecular synapomorphies for the five mycobacterial clades and their phylogenetic analysis and those by Tortoli et al. (2017), also support the existence of these clades. However, while not questioning any aspects of our results, Nouioui et al. (2018) question our division of the genus Mycobacterium into five genera for two reasons. First, they indicate that our identification of several synapomorphies for all mycobacteria makes further division of this group into multiple genera arbitrary. However, molecular synapomorphies can exist at different phylogenetic/taxonomic levels ranging from phylum to the genus levels (Gupta, 2016). We have previously identified synapomorphies for the phylum Actinobacteria as well as several classes/orders within this phylum (Gao and Gupta, 2012). For the class Coriobacteriia, synapomorphies were identified for the entire class along with its two orders and three families (Gupta et al., 2013). In fact, we have also described a number of CSIs and CSPs specific for the order Corynebacteriales (now emended order Mycobacteriales) (Gao and Gupta, 2012). Updated sequence information show that all three described CSIs and two CSPs are uniquely shared by different Mycobacteriales species with only isolated exceptions (see Figure 1B). Therefore, the existence of molecular synapomorphies for a higher taxonomic clade should not preclude its further division into lower level taxa, whose monophyly is strongly supported by independent means.
Nouioui et al. (2018) are also critical of our proposal on the grounds that some Mycobacteriales genera viz. Rhodococcus, Gordonia, Nocardia, and Corynebacterium are more divergent than Mycobacterium. However, in comparison to these genera, interspecies relationships within the genus Mycobacterium are now much better understood, because of our comprehensive genomic analyses. Additionally, Mycobacterium species also differ in their growth rates and this distinction is also supported by our classification scheme. Another important rationale for dividing the genus Mycobacterium is that it contains some of the most important human pathogens, including M. tuberculosis, the causative agent of tuberculosis (TB). For developing improved means for detection and treatment of TB, it is crucial to understand how the TB-causing group of bacteria differ from other related bacteria (Gupta, 2018; Gupta et al., 2018). In this context, our classification scheme, which separates all major human and animal pathogenic species (retained within the genus Mycobacterium) from other mycobacteria, constitutes an important step. With this division, attention can be focused on the unique genetic and molecular characteristics of the clinically important group of mycobacteria (Gupta, 2018; Gupta et al., 2018). Thus, we emphasize here that the reservations expressed by Nouioui et al. (2018) of our proposed division of the genus Mycobacterium are not justified.
Emended Description of the Order Mycobacteriales Janke 1924 (Approved Lists 1980)
The order is comprised of aerobic or facultatively anaerobic, chemoorganotrophic species exhibiting Gram-positive or acid-fast staining response. Most members are catalase- positive and form a branched substrate mycelium that fragments into coccoid- to rod-shaped elements or present as branched filaments, cocci, or as pleomorphic forms. Some strains form aerial hyphae. The wall peptidoglycan contains meso-diaminopimelic acid and is of the A1g type. Arabinose and galactose are major wall sugars. Fatty acid profiles are rich in saturated and unsaturated components and usually contain tuberculostearic acid. Mycolic acids are important constituents of the cell envelopes of most members. Members of this order form a monophyletic clade in phylogenetic trees based on 16S rRNA and large datasets of protein sequences. In addition, the following conserved signature indels (CSIs), viz. 2 aa insert in ABC-F family ATP-binding protein (Uup), 1 aa insert in chromosome partitioning protein ParB, and 1 aa deletion in alpha-ketoglutarate decarboxylase (KGD) and the homologs of the two conserved signature proteins (CSPs) with accession numbers NP_301197.1 and NP_301204.1 are also primarily found in the members of this order (Gao and Gupta, 2012). The order contains the families Corynebacteriaceae, Dietziaceae, Gordoniaceae, Lawsonellaceae, Mycobacteriaceae, Nocardiaceae, Segniliparaceae, and Tsukamurellaceae.
Type genus: Mycobacterium Lehmann and Neumann, 1896 (Approved Lists 1980) emend. Gupta et al. 2018
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was supported by the research grant No. 249924 from the Natural Science and Engineering Research Council of Canada awarded to RG.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer VS and handling editor declared their shared affiliation at the time of the review.
Acknowledgments
I thank Dr. Aharon Oren for confirming that the placement of the family Mycobacteriaceae within the order Corynebacteriales contravenes the Code for the Nomenclature of Prokaryotes.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00206/full#supplementary-material
References
Gao, B., and Gupta, R. S. (2012). Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol. Mol. Biol. Rev. 76, 66–112. doi: 10.1128/MMBR.05011-11
Goodfellow, M., and Jones, A. L. (2012). “Order V. Corynebacteriales ord.nov,” in Bergey's Manual of Systematic Bacteriology, Vol. 5, Actinobacteria, eds M. Goodfellow, P. Kampfer, H. J. Busse, M. E. Trujillo, K. Suzuki, W. Ludwig, and W. Whitman (New York, NY: Springer), 235–243.
Gupta, R. S. (2016). Impact of genomics on the understanding of microbial evolution and classification: the importance of Darwin's views on classification. FEMS Microbiol. Rev. 40, 520–553. doi: 10.1093/femsre/fuw011
Gupta, R. S. (2018). Impact of genomics on clarifying the evolutionary relationships amongst Mycobacteria: identification of molecular signatures specific for the tuberculosis-complex of bacteria with potential applications for novel diagnostics and therapeutics. High Throughput 7:E31. doi: 10.3390/ht7040031
Gupta, R. S., Chen, W. J., Adeolu, M., and Chai, Y. (2013). Molecular signatures for the class Coriobacteriia and its different clades; Proposal for division of the class Coriobacteriia into the emended order Coriobacteriales, containing the emended family Coriobacteriaceae and Atopobiaceae fam. nov., and Eggerthellales ord. nov., containing the family Eggerthellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 63, 3379–3397. doi: 10.1099/ijs.0.048371-0
Gupta, R. S., Lo, B., and Son, J. (2018). Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended Genus Mycobacterium and four novel genera. Front. Microbiol. 9:67. doi: 10.3389/fmicb.2018.00067
Janke, A. (1924). Allgemeine Technische Mikrobiologie. I. Teil. Die Mikroorganismen. Dresden; Leipzig: T.Steinkopf.
Lehmann, K. B., and Neumann, R. O. (1896). Atlas und Grundriss der Bakteriologie und Lehrbuch der Speziellen Bakteriologischen Diagnostik.. München: J. F. Lehmann.
Nouioui, I., Carro, L., García-López, M., Meier-Kolthoff, J. P., Woyke, T., Kyrpides, N. C., et al. (2018). Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 9:2007. doi: 10.3389/fmicb.2018.02007
Oren, A., and Garrity, G. M. (2015). Validation list no. 164. List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 65, 2017–2025. doi: 10.1099/ijs.0.000317
Parker, C. T., Tindall, B. J., and Garrity, G. M. (2015). International code of nomenclature of prokaryotes. Int. J. Syst. Evol. Microbiol. 69, S1–S111. doi: 10.1099/ijsem.0.000778
Parte, A. C. (2014). LPSN-list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 42, D613–D616. doi: 10.1093/nar/gkt1111
Skerman, V. B. D., McGowan, V., and Sneath, P. H. A. (1980). Approved lists of bacteria names. Int. J. Syst. Evol. Microbiol. 30, 225–420. doi: 10.1099/00207713-30-1-225
Keywords: family Mycobacteriaceae and the order Mycobacteriales, genus Mycobacterium, order Corynebacteriales, division of the family Mycobacteriaceae into five genera, prokaryotic taxonomy, Priority of taxonomic names
Citation: Gupta RS (2019) Commentary: Genome-Based Taxonomic Classification of the Phylum Actinobacteria. Front. Microbiol. 10:206. doi: 10.3389/fmicb.2019.00206
Received: 23 November 2018; Accepted: 24 January 2019;
Published: 22 February 2019.
Edited by:
Iain Sutcliffe, Northumbria University, United KingdomReviewed by:
Vartul Sangal, Northumbria University, United KingdomPierre-Edouard Fournier, Aix-Marseille Université, France
Copyright © 2019 Gupta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Radhey S. Gupta, Z3VwdGFAbWNtYXN0ZXIuY2E=
 Radhey S. Gupta
Radhey S. Gupta